Ehrlichia chaffeensis Tandem Repeat Effector Targets Differentially Influence Infection
- 1Department of Pathology, University of Texas Medical Branch, Galveston, TX, USA
- 2Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, TX, USA
- 3Center for Biodefense and Emerging Infectious Diseases, University of Texas Medical Branch, Galveston, TX, USA
- 4Sealy Center for Vaccine Development, University of Texas Medical Branch, Galveston, TX, USA
- 5Institute for Human Infections and Immunity, University of Texas Medical Branch, Galveston, TX, USA
Ehrlichia chaffeensis infects mononuclear phagocytes and survives intracellularly by exploiting host cell processes to evade host defenses. The mechanisms involved are not fully defined, but appear to rely largely on a subset of tandem repeat proteins (TRP) effectors. E. chaffeensis TRPs are type 1 secreted effectors that interact with a functionally diverse group of host cell targets associated with various biological processes. In this study, we investigated the influence of TRP host target proteins on ehrlichial infection by RNA interference. In total, 138 TRP-interacting host proteins identified by yeast two-hybrid were targeted by siRNA and the infection level determined by real-time qPCR. Knockdown of 124 (89%) TRP target proteins had significant influence on infection either by inhibiting (85%) or promoting (15%) ehrlichial infection. Notably, knockdown of 18 host proteins which interacted with TRP120 promoted the infection, suggesting that these targets may be degraded to promote infection. Host proteins that interact with TRPs are involved in cellular processes, including cell signaling, vesicle trafficking and intracellular transport, transcriptional regulation, metabolism, protein posttranslational modification, and apoptosis. Selected host targets were examined by immunofluorescent microscopy during infection and were found to localize with the morulae, or in the host cell cytoplasm adjacent to morulae. This study confirms that the majority of host proteins known to interact with TRP effectors influence infection and further extends the current knowledge that E. chaffeensis TRPs participate in a complex array of host protein interactions in order to reprogram the host cell and promote intracellular survival.
Introduction
Ehrlichia chaffeensis is an obligately intracellular bacterium and the etiologic agent of the emerging life-threatening human zoonosis, human monocytotropic ehrlichiosis (HME) (Paddock and Childs, 2003). E. chaffeensis selectively infects mononuclear phagocytes and resides in endosome-like membrane-bound vacuoles where it replicates and evades innate host defenses (Paddock and Childs, 2003). The mechanisms by which E. chaffeensis enters the host cell, avoids destruction, and establishes persistent infection are not well-understood, but functionally relevant host-pathogen interactions are essential for reprogramming the host cell defense mechanisms. This molecular strategy involves type 1 secreted tandem repeat protein (TRP) effectors (Lina et al., 2016b).
E. chaffeensis TRPs are major immunoreactive proteins that elicit strong host antibody responses during infection. The tandem repeat (TR) domains in TRP120, TRP47, and TRP32 are acidic, serine-rich, and contain protective species-specific epitopes (Doyle et al., 2006; Luo et al., 2008, 2009; Kuriakose et al., 2012). TRP120 and TRP47 are differentially expressed by infectious dense cored cells (DC), while TRP32 is expressed by both DCs and replicating reticulate cells (RC) (Popov et al., 2000; Doyle et al., 2006; Luo et al., 2008). Consistent with type 1 secretion (T1S) signals identified in the C-terminal domains of TRPs, TRPs have been experimentally identified as T1S system substrates through studies using a heterologous T1S apparatus of Escherichia coli (Wakeel et al., 2011).
In order to identify Ehrlichia-host interactions and help define the complex mechanisms by which E. chaffeensis modulates host cells, multiple studies using the yeast two-hybrid (Y2H) approach have been performed to better understand molecular host-pathogen interactions involving TRPs. TRP120, TRP47, and TRP32 have been shown to interact with a diverse network of host proteins involved in many host cellular processes including cell signaling, vesicle trafficking and intracellular transport, transcriptional regulation, metabolism, posttranslational modification and apoptosis, indicating the important roles of TRPs in reprogramming the host cell (Wakeel et al., 2009; Luo et al., 2011; Luo and McBride, 2012).
TRPs are modified by multiple host posttranslational modification pathways, including SUMOylation, ubiquitination and phosphorylation, which appear to mediate functional interactions and extend the number and diversity of interactions with host targets, as well as localization to various subcellular locations, including the nucleus (Wakeel et al., 2010; Dunphy et al., 2014). TRP120 is modified by SUMO at a canonical consensus SUMO conjugation motif located in the C-terminal domain, which has been further confirmed using a high-density microfluidic peptide array (Zhu et al., 2016). TRP120 conjugation with SUMO mediates interactions with host protein targets, and inhibition of the host SUMO pathway significantly decreases interaction between TRP120 and host protein targets, resulting in decreased ehrlichial intracellular survival (Dunphy et al., 2014). TRP120 also interacts with components of the ubiquitin pathways, including the E3 ligases, KLHL12 and FBXW7 as well as ubiquitin (Ub) isoforms UBB and UBC, which suggests TRP120 is a target of Ub conjugation (Luo et al., 2011). TRP47 is phosphorylated and interacts with the Src family tyrosine kinase, Fyn, which may be involved in the tyrosine phosphorylation of TRP47 (Wakeel et al., 2009, 2010). TRPs also contain many additional predicted phosphorylation sites; however, it is not clear which protein kinases are involved and how the phosphorylation affects TRP function or interactions with the host cell.
We have demonstrated the influence of selected TRP120 or TRP32-interacting host proteins on ehrlichial infection by RNA interference (Luo and McBride, 2012; Luo et al., 2016); however, a comprehensive analysis of all TRP-host interactions has not been performed. In this study, we extend the role of TRP-host interactions by investigating the influence of 138 TRP120, TRP47, and TRP32 interacting host target proteins on ehrlichial infection by RNA interference. We directly demonstrate that E. chaffeensis exploits the host cells through complex TRP interactions with a large and diverse array of host targets to promote intracellular survival.
Materials and Methods
Cell Culture and Cultivation of E. chaffeensis
Human monocytic leukemia cells (THP-1, from ATCC) were propagated in RPMI medium 1640 with L-glutamine and 25 mM HEPES buffer (Invitrogen, Carlsbad, Invitrogen), supplemented with 1 mM sodium pyruvate, 2.5 g/L D-(+)-glucose (Sigma, St. Louis, MO), and 10% fetal bovine serum (HyClone, Logan, UT). E. chaffeensis (Arkansas strain) was cultivated in THP-1 cells as previously described (Kuriakose et al., 2011).
siRNAs and Antibodies
All specific siRNAs were MISSION esiRNA from Sigma, which are endoribonuclease-prepared siRNA pools comprised of a heterogeneous mixture of siRNAs that all target the same mRNA sequence. These multiple silencing triggers lead to highly specific and effective gene silencing (guaranteed > 70% knockdown at the mRNA level) with lower off-target effects than single or pooled siRNAs. Targeting sequences of all siRNAs can be found on Sigma website. The control siRNA was ON-TARGETplus non-targeting siRNA from GE Healthcare Dharmacon (Lafayette, CO), which was designed to leverage seed-region optimization and patented modification patterns to have fewer off-targets than traditionally designed, unmodified negative control siRNAs. Thus, changes in mRNA or protein levels in cells treated with the control reflect a baseline cellular response that can be compared to the levels in cells treated with target-specific siRNA. Alexa Fluor 488-labeled negative siRNA was from Qiagen (Germantown, MD). Rabbit and mouse anti-TRP32, TRP47, or TRP120 antibodies have been described previously (Kuriakose et al., 2012). Other antibodies used in this study were mouse anti-human FBXW7 (R&D Systems, Minneapolis, MN), α-tubulin and PTPN2 (Santa Cruz, Dallas, TX), and rabbit anti-human CD14 (Abgent, San Diego, CA), EIF3A, ERA1, KARS, LMAN1, and STAT6 (Proteintech, Rosemont, IL), FYN and SLC43A3 (Santa Cruz), IRF2BP2 and SEPX1 (Pierce, Rockford, IL), KDM6B (Novus, Littleton, CO), and RC3H1 (Sigma).
RNA Interference
THP-1 cells (1 × 105/well on a 96-well plate) were transfected with 5 pmol siRNA using Lipofectamine 3000 (Invitrogen) according to the manufacturer's protocol. An Alexa Fluor 488-labeled negative siRNA was used as a control to monitor transfection efficiency. At 1 day posttransfection, the cells were synchronously infected by cell-free E. chaffeensis at a MOI of ~50, and collected at 1 and 2 days postinfection (p.i.) for Western blot and quantitative PCR (qPCR) to determine knockdown levels and infection status, respectively. The experiment of each siRNA was repeated for at least three times. Infectious dense-cored (DC) ehrlichiae enter the host cell and transform into large replicating reticulate cells (RC). At 1 day p.i. ehrlichiae are represented predominantly by RC and RC ehrlichiae replicate during the next 2 days p.i. before maturing into DC at 3 days. Thus, infected cells were not collected at 3 days p.i. since some cells started to collapse and release Ehrlichia.
Quantification of E. chaffeensis by qPCR
THP-1 cells were pelleted, washed by PBS, lysed in SideStep lysis and stabilization buffer (Agilent, Santa Clara, CA) for 10 min at room temperature with shaking, and analyzed for bacterial load using real-time qPCR. Amplification of the integral ehrlichial disulfide bond formation protein (dsb) gene and human glyceraldehyde-3-phosphate dehydrogenase (gapdh) gene was performed separately using Brilliant II SYBR Green mastermix (Agilent), 200 nM forward primers (dsb: 5′-gctgctccaccaataaatgtatccct-3′; gapdh: 5′-ggagtccactggcgtcttcac-3′) and 200 nM reverse primers (dsb: 5′-gtttcattagccaagaattccgacact-3′; gapdh: 5′-gaggcattgctgatgatcttgag-3′). The qPCR thermal cycling protocol (denaturation at 95°C 10 min, then 40 cycles of 95°C 30 s, 58°C 30 s, 72°C 30 s) was performed on the 7900HT fast real-time PCR system (Applied Biosystems, Foster City, CA). The fold change of dsb copy number relative to the control was normalized to qPCR-detected levels of the host gapdh gene.
Western Immunoblot
The THP-1 cell lysates were prepared using CytoBuster protein extraction reagent (Novagen/EMD, Gibbstown, NJ), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane. Western immunoblot was performed with horseradish peroxidase-labeled goat anti-rabbit, or mouse IgG (heavy and light chains) conjugate (Kirkegaard & Perry Laboratories, Gaithersburg, MD) and SuperSignal West Dura chemiluminescent substrate (Thermo Scientific).
Immunofluorescence Microscopy
Uninfected or E. chaffeensis-infected THP-1 cells at 2 days p.i. were collected, and the indirect immunofluorescent antibody assay was performed as previously described (Luo et al., 2011). Fluorescence images were obtained using a BX61 epifluorescence microscope (Olympus, Japan) and the Slidebook 5.0 software (Intelligent Imaging Innovations, Denver, CO).
Statistics
The statistical differences between experimental groups were assessed with the two-tailed Student's t-test, and significance was indicated by a P < 0.05.
Results
Demonstration of Knockdown of Protein Expression by Western Blot Analysis
To verify the knockdown of gene expression by siRNA, 11 target proteins of high priority for the Ehrlichia-host interaction study were detected at 2 days posttransfection by Western blot, including six TRP120-interacting proteins eukaryotic translation initiation factor 3 subunit A (EIF3A), Era-like 12S mitochondrial rRNA chaperone 1 (ERA1), F-box and WD repeat domain containing 7 (FBXW7), lysine (K)-specific demethylase 6B (KDM6B), interferon regulatory factor 2 binding protein (IRF2BP2), and selenoprotein X 1 (SEPX1), two TRP47-interacting proteins FYN proto-oncogene Src family tyrosine kinase (FYN) and protein tyrosine phosphatase non-receptor type 2 (PTPN2), and three TRP32-interacting proteins CD14 molecule (CD14), ring finger and CCCH-type domain 1 (RC3H1) and solute carrier family 43 member 3 (SLC43A3). The expression of all these proteins was substantially reduced in specific siRNA-transfected cells, respectively, compared with the unrelated control siRNA-transfected cells, indicating the successful knockdown of gene expression and good efficiency of siRNAs (Figure 1).
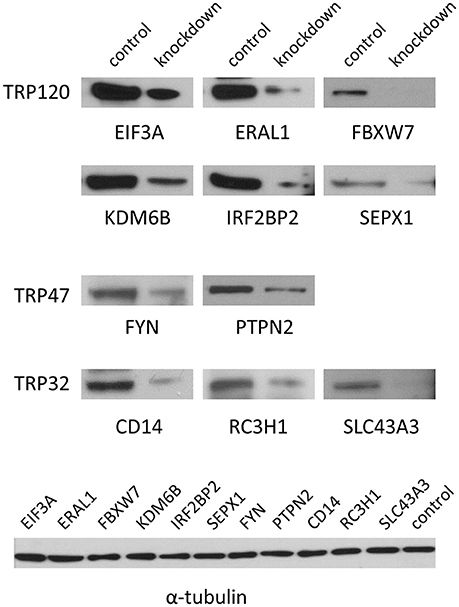
Figure 1. Verification of knockdown of TRP-interacting host proteins by Western blot. THP-1 cells were transfected with each specific or control siRNA and 2 days posttransfection Western blots were performed to determine knockdown efficiency. The α-tubulin was visualized as a control of Western blot.
Impact of Knockdown of TRP120-Interacting Proteins on E. chaffeensis Survival in Host Cells
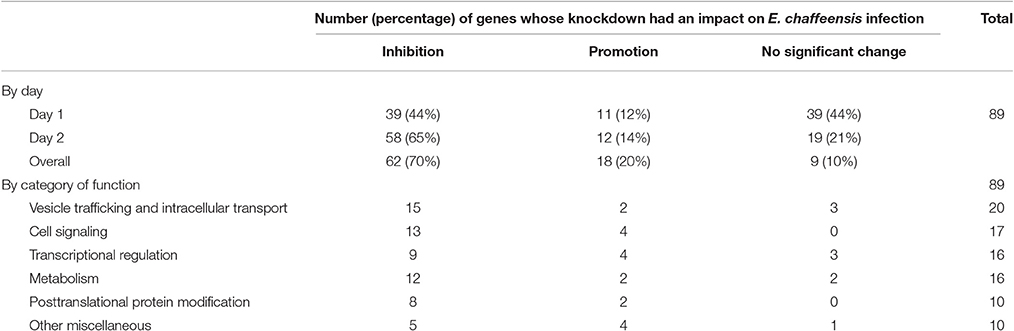
In total, 89 siRNAs available on the market were used to target 89 TRP120-interacting host proteins respectively, and then the impact of each siRNA on ehrlichial infection was assessed by qPCR at 1 and 2 days (Figure 2) p.i. Overall, knockdown of 80 (90% of 89) TRP120 target proteins had significant influence on ehrlichial infection, while only 9 (10%) proteins did not. Knockdown of 62 (70%) proteins inhibited ehrlichial infection, whereas knockdown of 18 (20%) proteins promoted ehrlichial infection. At 1 day p.i., the decrease of 39 (44%) proteins decreased Ehrlichia infection significantly, whereas the decrease of 11 (12%) proteins increased Ehrlichia infection significantly. At 2 days p.i., the decrease of 58 (65%) proteins decreased Ehrlichia infection significantly, whereas the decrease of 12 (14%) proteins increased Ehrlichia infection significantly (Table 1). The results indicated that most TRP120-interacting host proteins influence E. chaffeensis infection, and most TRP120 target proteins promote Ehrlichia infection. Knockdown of some targets largely associated with transcriptional regulation (day 1) increased infection, suggesting that these are downregulated or degraded by Ehrlichia to promote infection.
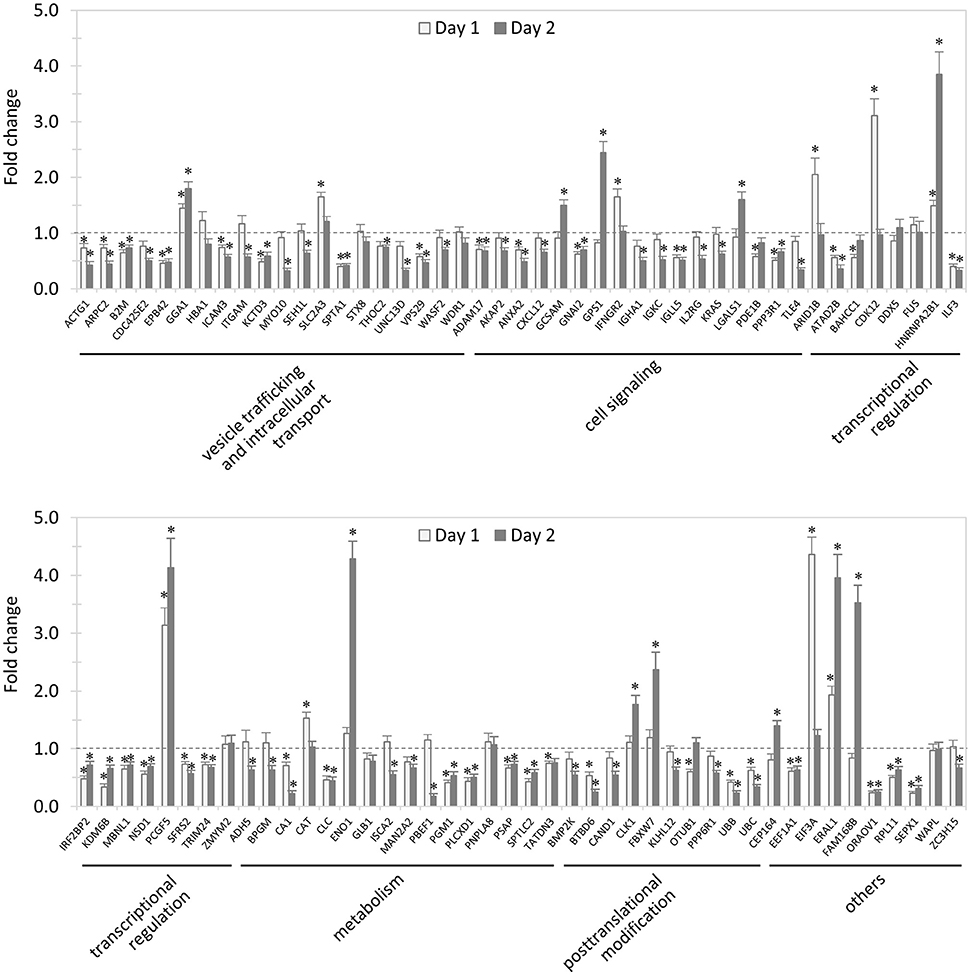
Figure 2. Influence of knockdown of TRP120-interacting proteins on Ehrlichia infection. THP-1 cells were transfected with target or control siRNA, and then infected by E. chaffeensis. Infection status was determined by qPCR at 1 and 2 days postinfection, compared to control scrambled siRNA-transfected cells. E. chaffeensis dsb gene copy numbers were normalized to host gapdh gene. Results were from three independent experiments, and the values were means ± standard deviations (*P < 0.05, significantly different from control infection).
TRP120-interacting proteins include host targets involved in vesicle trafficking and intracellular transport, cell signaling, transcriptional regulation, posttranslational modification, metabolism, and others. In each category, knockdown of most or all target proteins had significant influence on ehrlichial infection, and the knockdown of most proteins caused inhibition of infection, consistent with the overall impact of TRP120-interacting proteins (Table 1). The results indicated that TRP120 has moonlighting functions through interactions with diverse targets to modulate important host cellular processes to facilitate Ehrlichia survival.
Impact of Knockdown of TRP47-Interacting Proteins on E. chaffeensis Survival in Host Cells
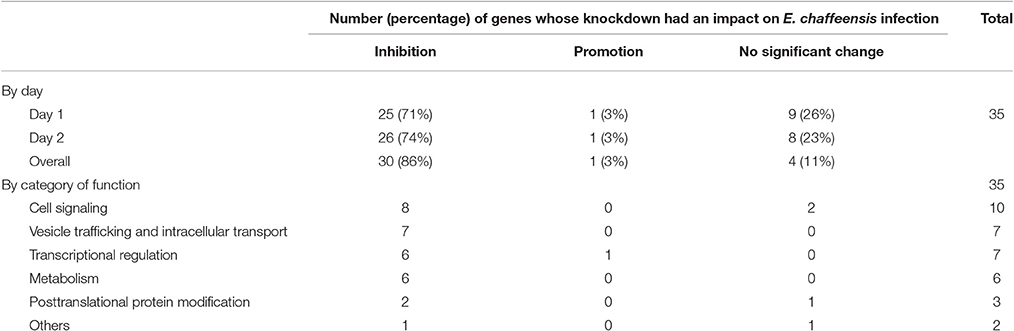
In total, 35 siRNAs available on the market were used to target 35 TRP47-interacting host proteins respectively, and then the impact of each siRNA on ehrlichial infection was assessed by qPCR at 1 and 2 days p.i. (Figure 3). Overall, knockdown of 31 (89% of 35) TRP47 target proteins had significant influence on ehrlichial infection, while only 4 (11%) proteins did not. Knockdown of 30 (86%) proteins inhibited ehrlichial infection, whereas knockdown of only 1 (3%) protein (polycomb group ring finger 5 [PCGF5]) that also interacted with TRP120 promoted ehrlichial infection. At both 1 and 2 days p.i., only PCGF5 knockdown increased E. chaffeensis infection significantly, but the knockdown of 25 (71%) proteins decreased the infection significantly at 1 day p.i., and the knockdown of 26 (74%) proteins negatively affected infection significantly at 2 days p.i. (Table 2). Therefore, consistent with TRP120-interacting proteins, most TRP47-interacting host proteins play a role in E. chaffeensis survival by either inhibiting or promoting the infection; moreover, most TRP47-interacting proteins appear to promote rather than inhibit Ehrlichia infection.
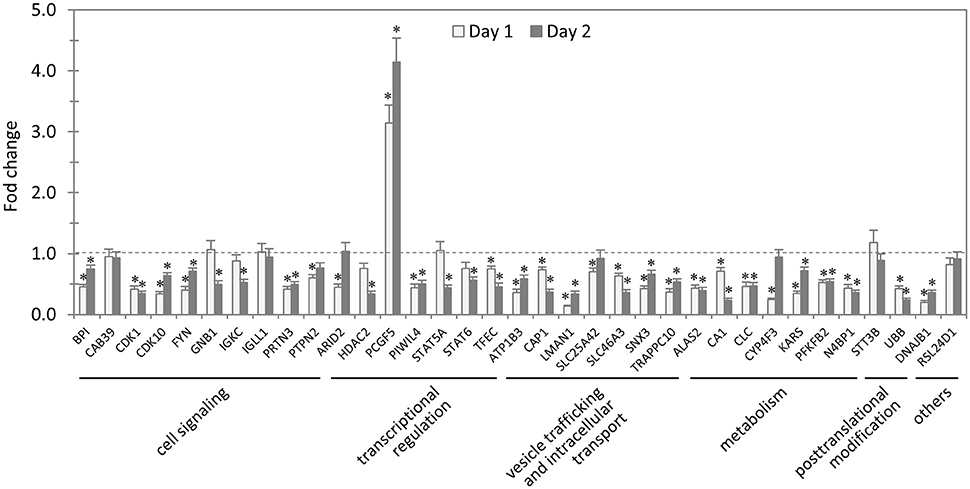
Figure 3. Influence of knockdown of TRP47-interacting proteins on Ehrlichia infection. THP-1 cells were transfected with target or control siRNA, and then infected by E. chaffeensis. Bacterial number changes were determined by qPCR at 1 and 2 days postinfection, compared to control scrambled siRNA-transfected cells. E. chaffeensis dsb gene copy numbers were normalized to host gapdh gene. Results were from three independent experiments, and the values were means ± standard deviations (*P < 0.05, significantly different from control infection).
TRP47-interacting proteins include host targets involved in cell signaling, transcriptional regulation, vesicle trafficking and intracellular transport, metabolism and posttranslational modification, consistent with TRP120-interacting proteins. In each category, knockdown of most or all target proteins had significant influence on ehrlichial infection, and the knockdown of majority of these protein caused inhibition of infection, consistent with the overall influence of TRP47-interacting proteins (Table 2). The results indicated that TRP47 has moonlighting functions through interactions with diverse targets to modulate important host cellular processes to facilitate Ehrlichia survival.
Impact of Knockdown of TRP32-Interacting Proteins on E. chaffeensis Survival in Host Cells
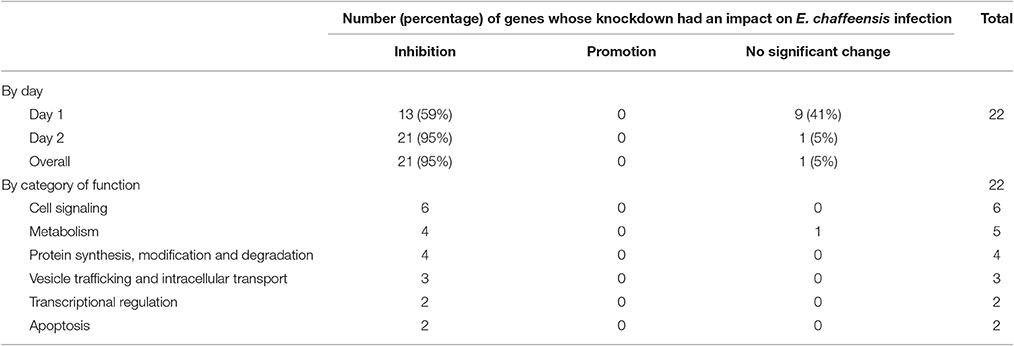
In total, 22 siRNAs were used to target 22 TRP32-interacting host proteins respectively, and then the impact of each siRNA on ehrlichial infection was assessed by qPCR (Figure 4). Overall, knockdown of 21 (95% of 22) TRP32 target proteins significantly reduced ehrlichial infection, while knockdown of only 1 (5%) protein (transketolase [TKT]) did not have significant impact on infection. None of TRP32-interacting protein knockdowns promoted ehrlichial infection. At 1 day p.i., the decrease in 13 (59%) proteins had a significant negative impact on Ehrlichia infection, and at 2 days p.i., the decrease in 21 (95%) proteins had a significant negative impact on Ehrlichia infection (Table 3). The results indicated that almost all TRP32-interacting proteins play a role in E. chaffeensis survival by promoting the infection.
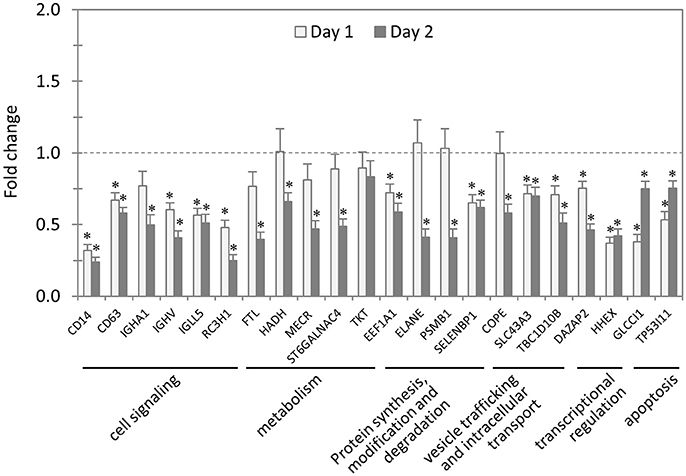
Figure 4. Influence of knockdown of TRP32-interacting proteins on Ehrlichia infection. THP-1 cells were transfected with target or control siRNA, and then infected by E. chaffeensis. Bacterial number changes were determined by qPCR at 1 and 2 days postinfection, compared to control scrambled siRNA-transfected cells. E. chaffeensis dsb gene copy numbers were normalized to host gapdh gene. Results were from three independent experiments, and the values were means ± standard deviations (*P < 0.05, significantly different from control infection).
TRP32-interacting proteins include host targets involved in cell signaling, metabolism, protein synthesis, modification and degradation, vesicle trafficking and intracellular transport, transcriptional regulation, and apoptosis, similar to TRP120/TRP47-interacting proteins. In each category, knockdown of most or all target proteins significantly reduced Ehrlichia infection, consistent with the overall influence of TRP32-interacting proteins (Table 3). The results indicated that TRP32 plays multiple roles in reprogramming important host cellular processes.
Overall Impact of Knockdown of TRP-Interacting Proteins on E. chaffeensis Survival in Host Cells
By Y2H we have previously identified totally 150 host proteins interacting with E. chaffeensis TRPs, including 98 proteins with TRP120, 38 proteins with TRP47, and 22 proteins with TRP32, while TRP120 shares 8 common targets with TRP47 and TRP32 (Wakeel et al., 2009; Luo et al., 2011; Luo and McBride, 2012). In this study, 138 (92% of 150) siRNAs were used to reduce the expression of corresponding genes respectively, including 89 siRNAs for TRP120 targets, 35 siRNAs for TRP47 targets, and 22 siRNAs for TRP32 targets. Overall, knockdown of 124 (89% of 138) TRP target proteins had significant influence on ehrlichial infection. Among 124 TRP target proteins, knockdown of 105 (85%) proteins inhibited ehrlichial infection, whereas knockdown of 19 (15%) proteins promoted ehrlichial infection. The inhibition or promotion of ehrlichial survival occurred at different stages of infection (1 or/and 2 days p.i.). For each TRP, target proteins were consistently involved in similar important host cell processes.
TRP-Expressing Ehrlichiae Colocalize with Human Target Proteins in E. chaffeensis-Infected THP-1 Cells
Previously, we have demonstrated the interactions of many identified target proteins with E. chaffeensis TRP120, TRP47, or TRP32 by multiple approaches (Wakeel et al., 2009; Luo et al., 2011; Luo and McBride, 2012). Here we examined host target proteins with well-defined and interesting functions and confirmed the interactions with E. chaffeensis TRPs by immunofluorescence assay. Eight target proteins were examined, including three TRP120-interacting proteins EIF3A, ERA1, and KDM6B, three TRP47-interacting proteins signal transducer and activator of transcription 6 (STAT6), lectin mannose binding 1 (LMAN1), and lysyl-tRNA synthetase (KARS), and two TRP32-interacting proteins RC3H1 and SLC43A3. Consistent with our previous reports, double-immunofluorescence labeling of E. chaffeensis-infected THP-1 cells revealed seven proteins EIF3A, ERA1, KDM6B, STAT6, KARS, RC3H1, and SLC43A3 exhibited strong colocalization with the morulae (Figures 5A–D,F–H), while LMAN1 localized in the host cell cytoplasm adjacent to the morula membrane (Figure 5E). Redistribution of TRP target proteins in E. chaffeensis-infected cells compared to uninfected cells was observed. For example, EIF3A, ERAL1, KDM6B, STAT6, KARS, and SLC43A3 were associated with morulae in infected THP-1 cells (Figures 5A–D,F–H), while in uninfected THP-1 cells, EIF3A, ERAL1, KDM6B, and KARS had diffused or punctate distribution mainly in the cytoplasm (Figures 5A–C,F, insets), and STAT6 and SLC43A3 were distributed throughout the whole cell (Figures 5D,H, insets).
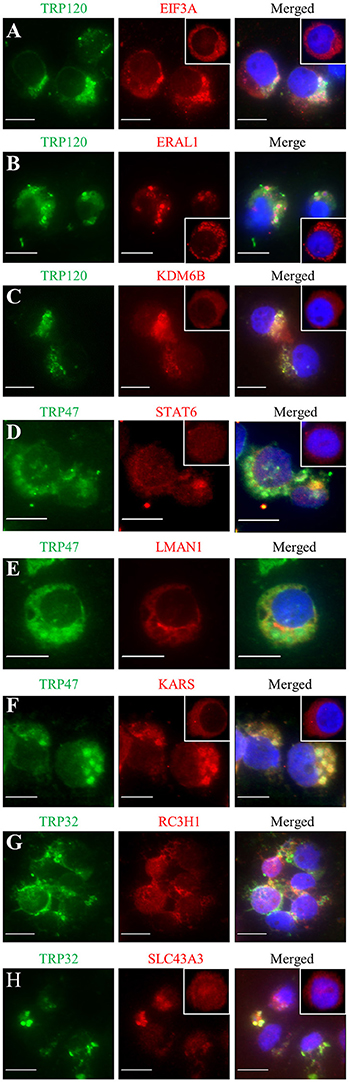
Figure 5. Colocalization of TRP120, TRP47, and TRP32 with their interacting host proteins in E. chaffeensis-infected THP-1 cells. Fluorescence microscopy of infected (2 days postinfection) THP-1 cells stained with 4,6′-diamidino-2-phenylindole (blue, showing the nucleus), TRP antibody (green), and host protein antibody (red; panels A–H, respectively) show co-localization of E. chaffeensis TRP-labeled morulae with host protein. The insets (panels A–D,F,H) show the distribution of EIF3A, ERAL1, KDM6B, STAT6, KARS, and SLC43A3 in uninfected THP-1 cells. Fluorescence images were obtained using an Olympus BX61 epifluorescence microscope. Bar, 10 μm.
Discussion
In recent years multiple studies from our laboratory have identified numerous Ehrlichia-host interactions and determined that Ehrlichia TRPs interact with a diverse network of host proteins involved in many host cellular processes including cell signaling, vesicle trafficking and intracellular transport, transcriptional regulation, metabolism, posttranslational modification, and apoptosis (Wakeel et al., 2009; Luo et al., 2011; Luo and McBride, 2012). Previous studies have helped understand the complex mechanisms by which E. chaffeensis modulates host cell processes. This study found that knockdown of host proteins that interact with Ehrlichia TRPs differentially influence intracellular survival. Figure 6 illustrates important host cellular processes that E. chaffeensis TRPs modulate during infection through interactions with host targets.
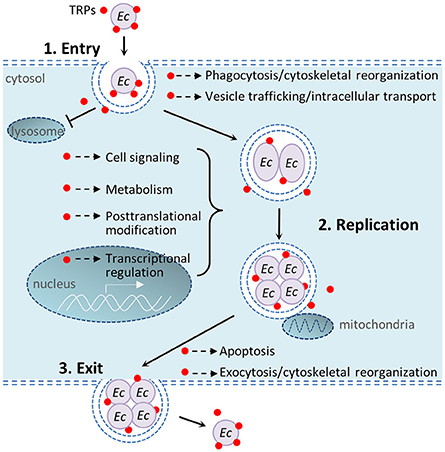
Figure 6. Schematic diagram of important host cellular processes that E. chaffeensis tandem repeat proteins modulate during infection. 1. Bacterial entry: E. chaffeensis binds and enters the host cell by phagocytosis. TRPs interact with host cytoskeletal proteins including actins and actin-related proteins, and also exploit host cell signaling (such as Wnt) to facilitate phagocytosis; TRPs also interact with host vesicular trafficking system to help establish an intracellular niche in a membrane-bound vacuole that does not undergo phagolysosomal fusion. 2. Bacterial replication: TRPs are secreted into the intramorular space and translocate into cytosol and nucleus of the host cell, where these effectors interact with diverse host proteins to modulate cell processes, including vesicle trafficking and intracellular transport, cell signaling, transcriptional regulation, posttranslational modification, metabolism, apoptosis, and others in order to avoid host defense system and permit intracellular replication. 3. Bacterial exit: TRPs interact with many apoptosis-associated proteins (some in mitochondria) to induce host cell apoptosis; TRPs also interact with host cytoskeletal proteins to facilitate exocytosis. Red dots show TRPs, including TRP120, TRP47, and TRP32. Ec, E. chaffeensis.
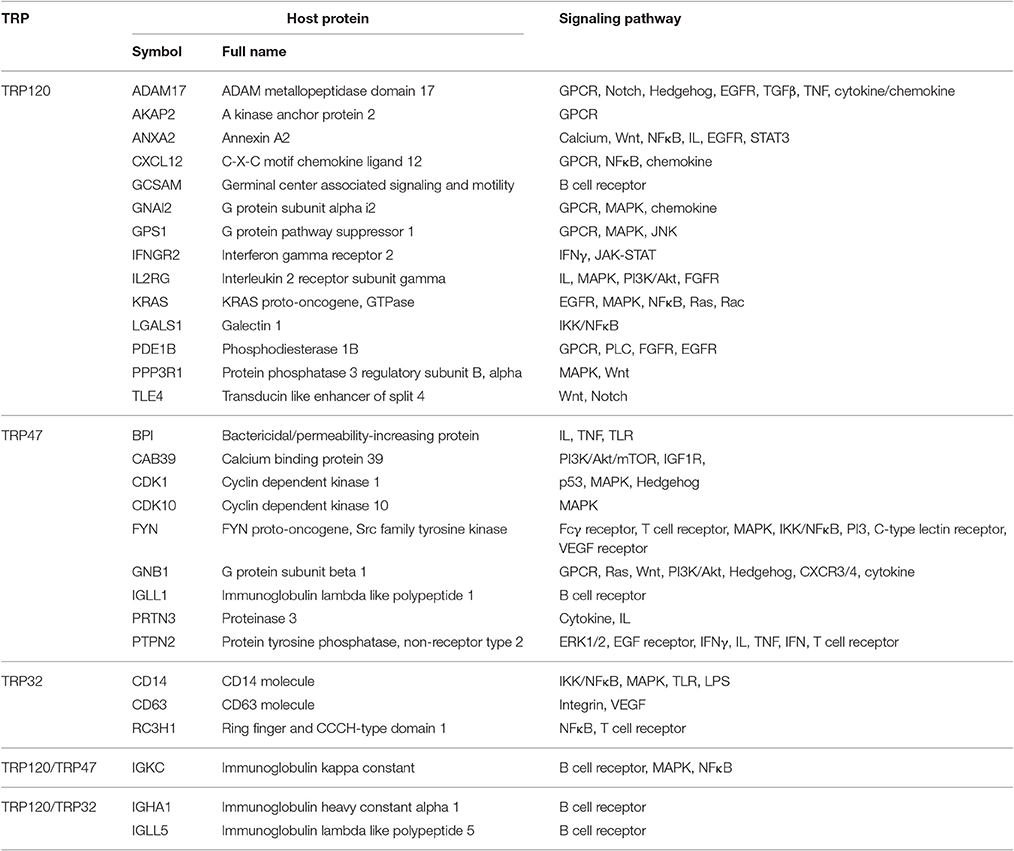
Notably, E. chaffeensis TRPs interact with numerous host proteins associated with cellular signaling pathways, including 17 proteins with TRP120, 10 proteins with TRP47, and six proteins with TRP32, suggesting that infection and replication of E. chaffeensis involves TRP exploitation of conserved cell signaling pathways, such as Wnt, Notch, mitogen-activated protein kinase/extracellular signal-regulated kinases (MAPK/ERK), phosphatidylinositol 3-kinase/Akt/mechanistic target of rapamycin (PI3K/Akt/mTOR), nuclear factor kappa B (NFκB), and cytokine/chemokine-mediated signaling (Table 4). Wnt and Notch are fundamental cell signaling pathways that play major roles in the regulation of gene expression, cell proliferation and differentiation, and embryonic development (Hayward et al., 2008). Five TRP120-interacting proteins including ADAM metallopeptidase domain 17 (ADAM17), annexin A2 (ANXA2), protein phosphatase 3 regulatory subunit B alpha (PPP3R1), transducin like enhancer of split 4 (TLE4), and G protein subunit beta 1 (GNB1) are signaling molecules of Wnt and Notch pathways. This is consistent with recent studies in which we have used different approaches to demonstrate that Wnt and Notch signaling pathways play important roles in ehrlichial infection, and TRPs mediate ehrlichial invasion and survival through activation and modulation of Wnt and Notch pathways (Luo et al., 2016; Lina et al., 2016a). In this study, we further demonstrated that knockdown of nearly all of TRP-interacting proteins involved in host cell signaling, except two TRP47-interacting proteins, calcium binding protein 39 (CAB39) and immunoglobulin lambda like polypeptide 1 (IGLL1), significantly influenced E. chaffeensis survival. These results support the importance of many conserved cellular signaling pathways in ehrlichial survival. For example, G-protein coupled receptor (GPCR) signaling involves a variety of ligands and stimuli and can regulate a highly interconnected network of signal transductions and cellular responses (Roth et al., 2015). We found that six TRP120-interacting proteins including ADAM17, A kinase anchor protein 2 (AKAP2), C-X-C motif chemokine ligand 12 (CXCL12), G protein subunit alpha i2 (GNAI2), G protein pathway suppressor 1 (GPS1), and phosphodiesterase 1B (PDE1B), and one TRP47-interacting protein GNB1 are involved in GPCR signaling. In addition, MAPK signaling is a conserved pathway involved in a variety of fundamental cellular processes (Cargnello and Roux, 2011). It has been reported that p38 MAPK/ERK1/2 are activated at the early stage of E. chaffeensis infection, but the activations are subsequently reduced in infected cells compared with those in uninfected cells (Lin and Rikihisa, 2004). We found that six TRP120-interacting proteins GNAI2, GPS1, immunoglobulin kappa constant (IGKC), interleukin 2 receptor subunit gamma (IL2RG), KRAS proto-oncogene GTPase (KRAS), and PPP3R1, four TRP47-interacting proteins cyclin dependent kinase 1 (CDK1), cyclin dependent kinase 10 (CDK10), FYN, and IGKC, and one TRP32-interacting protein, CD14, are involved in MAPK signaling. Knockdown of all these TRP target proteins significantly influenced infection, indicating that GPCR and MAPK signaling pathways are involved in ehrlichial survival. Based on these studies, Wnt, Notch, and MAPK as well as GPCR signaling pathway appear to be major targets of TRP exploitation. Further investigation is needed to understand the specific role of each TRP target and associated pathway(s) in ehrlichial pathobiology.
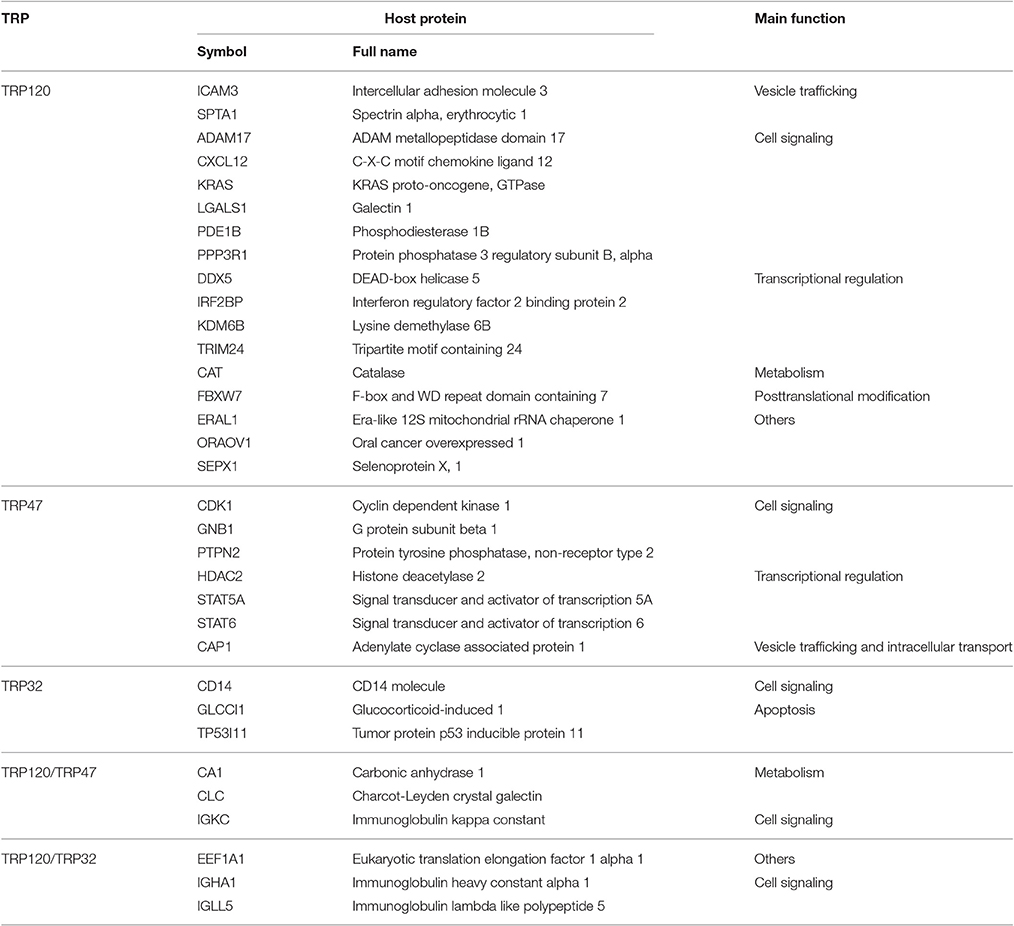
An important host cellular process in which many TRP-interacting host proteins are involved is vesicle trafficking and intracellular transport. This process involves 20 TRP120-interacting proteins, seven TRP47-interacting proteins, and three TRP32-interacting proteins. Remarkably, from this group we found a subset of actin-related proteins interacting with Ehrlichia TRPs (particularly TRP120), including eight TRP120-interacting proteins and one TRP47-interacting protein (Table 5). Ehrlichia TRPs interact directly with host cytosolic actin gamma 1 (ACTG1) and eight other actin-binding or related proteins, including actin related protein 2/3 complex subunit 2 (ARPC2), CDC42 small effector 2 (CDC42SE2), integrin subunit alpha M (ITGAM), myosin X (MYO10), spectrin alpha erythrocytic 1 (SPTA1), WAS protein family member 2 (WASF2), WD repeat domain 1 (WDR1), and adenylate cyclase associated protein 1 (CAP1), suggesting that actin cytoskeleton plays an important role in ehrlichial infection. Most of these TRP-interacting proteins have also been found to be involved in phagocytosis or endocytosis. Multiple studies have shown the importance of TRP120 in ehrlichial binding and internalization, and most recently, we demonstrated that TRPs directly activate Wnt signaling to induce ehrlichial phagocytosis (Popov et al., 2000; Kumagai et al., 2010; Luo et al., 2016). In addition, it has been reported that E. chaffeensis is transported through the filopodia of the host cell and inhibition of actin polymerization in infected cells prevents filopodia formation. Moreover, the spreading process of Ehrlichia canis in macrophages is also dependent on actin cytoskeleton (Thomas et al., 2010; Alves et al., 2014). Other studies reported that E. chaffeensis surface protein EtpE binds GPI-anchored protein DNase X to trigger entry by N-WASP-dependent actin polymerization (Mohan Kumar et al., 2013, 2015). Therefore, interaction of TRPs with actin cytoskeleton might facilitate ehrlichial entry/release and necessary intracellular trafficking during Ehrlichia infection. Our RNAi data showed that knockdown of all these Ehrlichia TRP target proteins except WDR1 significantly reduced bacterial load, demonstrating the importance of actin cytoskeleton organization in ehrlichial entry and/or survival.
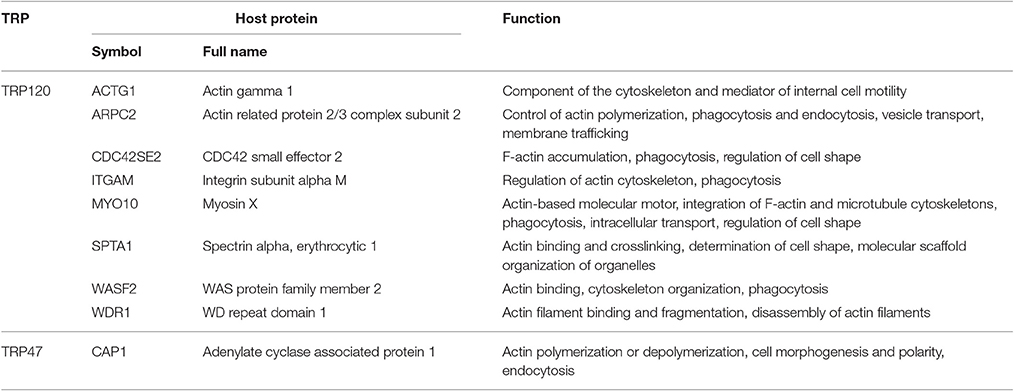
Table 5. E. chaffeensis TRP-interacting host proteins with primary function in actin cytoskeleton organization.
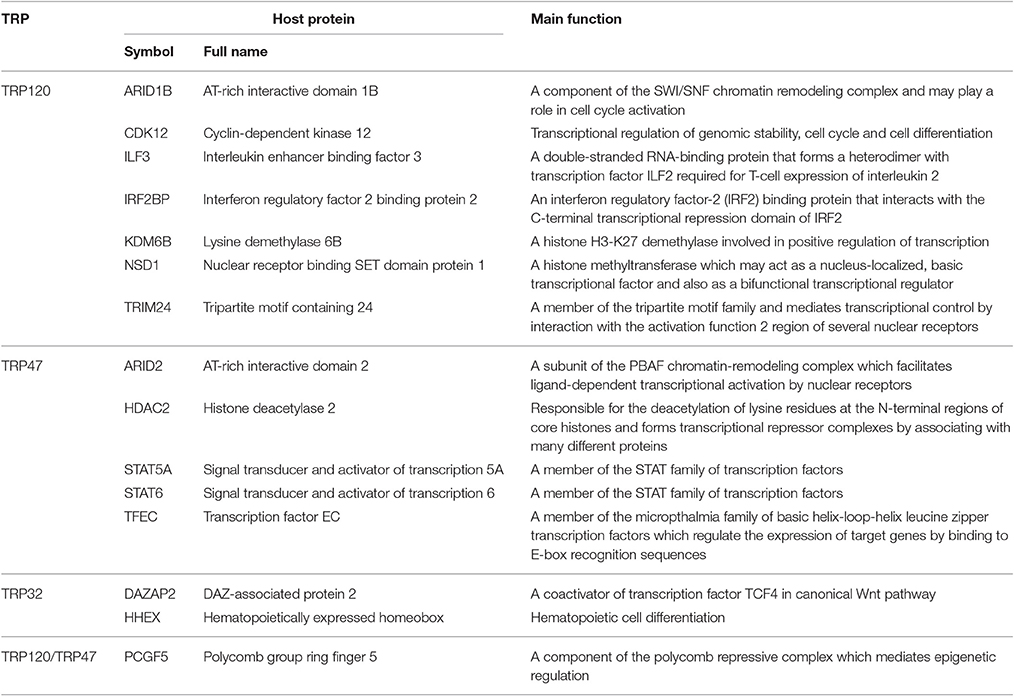
Another group of TRP-interacting proteins with significant interest is host proteins involved in transcriptional regulation, including 16 TRP120-interacting proteins, seven TRP47-interacting proteins, and two TRP32-interacting proteins (Table 6). These proteins are either transcription factors or transcription regulators (coactivator or corepressor). Both TRP120 and TRP47 interact with PCGF5, a component of the polycomb repressive complex which mediates epigenetic regulation (Gao et al., 2014). Other important TRP120-interacting proteins involved in transcriptional regulation include AT-rich interactive domain 1B (ARID1B), cyclin-dependent kinase 12 (CDK12), interleukin enhancer binding factor 3 (ILF3), IRF2BP2, KDM6B, nuclear receptor binding SET domain protein 1 (NSD1), and tripartite motif containing 24 (TRIM24). Important TRP47-interacting proteins involved in transcriptional regulation include AT-rich interactive domain 2 (ARID2), histone deacetylase 2 (HDAC2), signal transducer and activator of transcription 5A and 6 (STAT5A and STAT6), and transcription factor EC (TFEC). Moreover, TRP32 interacts with DAZ-associated protein 2 (DAZAP2) and hematopoietically expressed homeobox (HHEX). We have previously reported that Ehrlichia TRP120 and TRP32 are nucleomodulins that directly bind host DNA and regulate the expression of numerous genes involved with transcriptional regulation, signal transduction, and cell differentiation and proliferation (Zhu et al., 2011; Farris et al., 2016). In this study, our RNAi data showed that knockdown of all Ehrlichia TRP target proteins involved in transcriptional regulation significantly influenced infection level, therefore, TRPs also play roles in regulation of host gene transcription by direct interactions with transcription factors and regulators in multiple cellular processes including survival, growth, and differentiation.
TRP-interacting targets include some host proteins involved in protein posttranslational modification (PTM). In particular, TRP120 interacts with 10 host proteins that have the main function in posttranslational modification machinery, including phosphorylation and ubiquitination. TRP120 interacts with host kinase and phosphatase involved in protein phosphorylation/dephosphorylation, including BMP2 inducible kinase (BMP2K), CDC-like kinase 1 (CLK1), and protein phosphatase 6 regulatory subunit 1 (PPP6R1). TRP120 also interacts with multiple host proteins involved in protein ubiquitination, including BTB domain containing 6 (BTBD6), a putative ubiquitin ligase adaptor protein (Sobieszczuk et al., 2010); cullin-associated and neddylation dissociated 1 (CAND1), an essential regulator of Cullin-RING ubiquitin ligases (Goldenberg et al., 2004); FBXW7, one of the four subunits of ubiquitin protein ligase complex SCFs (SKP1-cullin-F-box) (Hao et al., 2007); kelch-like family member 12 (KLHL12), a substrate adaptor of the Cullin-3 ubiquitin ligase complex (Jin et al., 2012); OTU deubiquitinase, ubiquitin aldehyde binding 1 (OTUB1), a highly specific ubiquitin iso-peptidase (Edelmann et al., 2009); and an ubiquitin precursor protein ubiquitin C (UBC). Ubiquitin B (UBB), another ubiquitin precursor, is a common interacting target of TRP120 and TRP47. TRP47 also interacts with NEDD4 binding protein 1 (N4BP1), an inhibitor of the E3 ubiquitin ligase (Oberst et al., 2007). Moreover, TRP120 has recently been found to be directly modified by SUMO, and more importantly, SUMO conjugation contributes to interactions with defined host proteins, such as PCGF5, actin and myosin cytoskeleton components, and recruitments of host proteins to the ehrlichial vacuole that influence infection (Dunphy et al., 2014). It remains unclear whether the functional consequences of TRP ubiquitination and phosphorylation are different from those associated with TRP SUMOylation, but it is well-established that many bacterial effectors mimic host proteins involved in the host posttranslational machinery to modify host proteins and signaling (Ribet and Cossart, 2010). Thus, PTMs may contribute to the moonlighting function of Ehrlichia TRPs and facilitate many diverse interactions between TRPs and host proteins. Another intracellular bacterium moonlighting effector is Chlamydia trachomatis CPAF, a protease which targets multiple host and bacterial proteins to maintain vacuole integrity, manipulate signaling pathways and promote virulence (Jorgensen et al., 2011; Zhong, 2011). Our RNAi data demonstrate that knockdown of all TRP target proteins involved in PTM significantly influenced infection, indicating that E. chaffeensis exploits major posttranslational modification machineries of host cells to facilitate a survival strategy.
Host proteins involved in metabolism are also frequent interacting targets of E. chaffeensis TRPs, including 16 TRP120-interacting proteins, six TRP47-interacting proteins, and five TRP32-interacting proteins. This category covers a variety of host metabolism processes, including glycometabolism, lipid metabolism, protein metabolism, nucleic acid metabolism, ion, and other inorganic metabolism. Two proteins, carbonic anhydrase I (CA1) and Charcot-Leyden crystal galectin (CLC) are common interacting targets of TRP120 and TRP47, suggesting their importance during infection. Our RNAi data demonstrated that knockdown of most of these Ehrlichia TRP target proteins significantly influenced bacterial load, indicating that TRPs also play a role in regulating host metabolism to favor the survival.
Interestingly, Y2H studies also discovered that a large number of apoptosis-associated proteins of the host cell were TRP targets, although most of these proteins are classified into other categories of function because their primary functions appear not to be related to apoptosis (Table 7). Two TRP32-interacting proteins glucocorticoid-induced 1 (GLCCI1) and tumor protein p53 inducible protein 11 (TP53I11) have main function in cell apoptosis. Expression of GLCCI1 is induced by glucocorticoids and may be an early marker for glucocorticoid-induced apoptosis (Chapman et al., 1996). TP53I11 is a downstream target of p53 and is involved in the regulation of apoptosis (Wu et al., 2009). Three common interacting proteins of TRPs, eukaryotic elongation factor 1 alpha 1 (EEF1A1), CA1, and CLC, play roles in apoptosis (Ejiri, 2002; Kubach et al., 2007; Zheng et al., 2015). In addition, TRPs interact with a wide variety of immunoglobulin molecules, such as IGKC, immunoglobulin heavy constant alpha 1 (IGHA1) and immunoglobulin lambda like polypeptide 5 (IGLL5), which have also been linked to apoptosis (Yang et al., 2009). TRP47 interacts with other seven host proteins associated with apoptosis, for example, STAT5A and STAT6, both of which are members of the STAT family of transcription factors and have been found to induce the expression of anti-apoptotic protein BCL2L1/BCL-XL (Calò et al., 2003); and CAP1, which has been implicated in promoting apoptosis by functioning as an actin shuttle to mitochondria (Wang et al., 2008). TRP120 interacts with relatively more apoptosis-associated proteins of the host cell. For example, CXCL12 is a negative regulator of intrinsic apoptotic signaling pathway in response to DNA damage, and FBXW7, in contrast, is a positive regulator of oxidative stress-induced intrinsic apoptotic signaling pathway (Hattermann et al., 2010). Previous studies have demonstrated that host cell apoptosis is delayed by stabilization of the mitochondrial membrane potential during E. chaffeensis and E. ewingii infection (Xiong et al., 2008; Liu et al., 2011). Our RNAi data showed that knockdown of all apoptosis-associated TRP target proteins (except for DEAD-box helicase 5 [DDX5]) significantly influenced bacterial load, thus, the interactions of TRPs with different apoptosis-associated proteins may serve an opposite function by inhibiting or promoting apoptosis in the different stages of infection, to facilitate ehrlichial survival and release, respectively.
We classified TRP-interacting host proteins according to their main functions, but many of them are moonlighting proteins which could be included in multiple categories. For example, EEF1A1, a common target protein of TRP120 and TRP32, is the second most abundant protein in eukaryotes after actin. It is an isoform of the alpha subunit of the elongation factor-1 complex, so its main function is the enzymatic delivery of aminoacyl tRNAs to the ribosome during protein translation; however, EEF1A1 is also one of the most important multifunctional eukaryotic proteins and has been reported to be involved in regulation of transcription, cytoskeletal remodeling, cellular response to epidermal growth factor stimulus, regulation of chaperone-mediated autophagy, and apoptosis (Condeelis, 1995; Ejiri, 2002). IGKC, another common target protein of TRP120 and TRP47, is the immunoglobulin kappa constant domain, so its main function is immune response including antigen binding, immunoglobulin receptor binding and complement activation, but it also appears to be involved in other signaling pathways, vesicle-mediated transport, receptor-mediated endocytosis, and phagocytosis (Bentley and Rabbitts, 1980). It is not clear if all or part of the functions of these proteins are involved in Ehrlichia infection, but some of them are very likely moonlighting when interacting with TRPs, since they belong to multiple important common categories of TRP-interacting proteins, such as cell signaling, transcriptional regulation, vesicle trafficking and apoptosis. In addition, some putative host proteins interact with two TRPs, including EF1A1, IGHA1, IGLL5 (interacting with both TRP32 and TRP120), PCGF5, IGKC, CA1, CLC, and UBB (with TRP47 and TRP120), implicating the importance of not only the moonlighting and overlapping host targets but also the crosstalking and converging cellular networks by Ehrlichia effectors.
Our siRNA experiments revealed that knockdown of TRP-interacting host proteins could either increase or decrease E. chaffeensis load. Most siRNAs decreased the bacterial load, indicating that these proteins are necessary for Ehrlichia infection; however, siRNAs of 18 TRP120-interacting proteins and one TRP47-interacting protein (PCGF5) increased the bacterial load, indicating that these proteins inhibit the Ehrlichia infection. For example, Golgi associated gamma adaptin ear containing ARF binding protein 1 (GGA1) is a ubiquitous coat protein that regulates the protein trafficking between the trans-Golgi network and the endosome/lysosome system (Bonifacino, 2004), so E. chaffeensis may inactivate GGA1 to prevent early endosomal maturation into late endosomes and then fusion with lysosome. PCGF5 is a component of the polycomb repressive complex which controls expression of many developmental regulator genes (Gao et al., 2014), thus, E. chaffeensis may inhibit this transcriptional repressor in order to modulate host cell gene expression to favor ehrlichial survival. Uniquely, knockdown of many TRP120 host targets promoted infection, suggesting that TRP120 plays a role in modulating levels of these targets during infection. This result is likely related to our recent finding that TRP120 can function as ubiquitin ligase and degrade host target proteins such as PCGF5 (Zhu and McBride unpublished). In this study, no siRNA of TRP32-interacting proteins was found to increase the bacterial load significantly, probably because only C-terminus of TRP32 was available as the bait of Y2H screening and resulted in the loss of some host proteins interacting with other domains of TRP32. In addition, siRNAs of 14 TRP-interacting proteins could not change bacterial load significantly. This is probably because these proteins are important at the late stage of Ehrlichia infection, or they play a nonessential role in infection. Some of these proteins may be too abundant to be reduced sufficiently, or they are just nonspecific Y2H targets. We did not perform the late time point of infection in our experiments, since some infected cells started to collapse and release Ehrlichia at 3 day p.i.
Our RNAi data also show that changes in Ehrlichia infection status can occur at different stages of infection for different host proteins, suggesting that Ehrlichia TRPs and host proteins interact at different time points during infection. For example, some siRNAs, such as those targeting CDC42SE2, ITGAM, MYO10, unc-13 homolog D (UNC13D) and TLE4, reduced bacterial load significantly at 2 days p.i. but not significantly at day 1, suggesting that these proteins may be not involved in bacterial internalization or early stage of infection. In contrast, some siRNAs, such as those targeting solute carrier family 2 member 3 (SLC2A3), interferon gamma receptor 2 (IFNGR2), PDE1B, ARID1B, and EIF3A, reduced or increased bacterial load significantly at 1 day p.i. but not significantly at day 2, suggesting that these proteins are more important for the bacterial internalization or early stage of infection. Meanwhile, most siRNAs reduced or increased bacterial load significantly at both 1 and 2 days p.i., suggesting that these proteins are important for both early and intermediate stages of infection. Moreover, knockdown of some TRP-interacting proteins had a dramatic effect on infection, suggesting that they play key roles. For example, knockdown of SPTA1, ILF3, CLC, UBB, SEPX1, and PCGF5 reduced or increase the infection very significantly at both 1 and 2 days p.i., suggesting that they are critical factors exploited by Ehrlichia TRPs for bacterial survival; however, we cannot exclude the possibility that siRNAs may have different knockdown efficiency. We also found that the knockdown of any single target protein by RNA interference could not abolish the ehrlichial growth completely. As observed by Western blot, 100% knockdown of a target protein by siRNA is very difficult, thus, a knockout cell line would be necessary to determine if a host protein is essential for E. chaffeensis infection.
The molecular mechanisms by which Ehrlichia enters and survives in the host cell remain unclear. This study documents the extensive effector-host interactions that contribute to survival of this obligately intracellular pathogen, and will help understand the molecular mechanisms utilized by other intracellular pathogens to infect host cells. Further studies will delineate the specific roles of these complex and diverse TRP-host interactions during infection.
Author Contributions
TL and PD designed, performed the experiments and analyzed the data. TL wrote the manuscript. JM directed and contributed to the writing of the manuscript.
Funding
This work was supported by grants AI106859 and AI126144 from the National Institute of Allergy and Infectious Diseases (NIAID), and by funding from the Clayton Foundation for Research (to JM).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alves, R. N., Levenhagen, M. A., Levenhagen, M. M., Rieck, S. E., Labruna, M. B., and Beletti, M. E. (2014). The spreading process of Ehrlichia canis in macrophages is dependent on actin cytoskeleton, calcium and iron influx and lysosomal evasion. Vet. Microbiol. 168, 442–446. doi: 10.1016/j.vetmic.2013.11.030
Bentley, D. L., and Rabbitts, T. H. (1980). Human immunoglobulin variable region genes–DNA sequences of two V kappa genes and a pseudogene. Nature 288, 730–733. doi: 10.1038/288730a0
Bonifacino, J. S. (2004). The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 5, 23–32. doi: 10.1038/nrm1279
Calò, V., Migliavacca, M., Bazan, V., Macaluso, M., Buscemi, M., Gebbia, N., et al. (2003). STAT proteins: from normal control of cellular events to tumorigenesis. J. Cell. Physiol. 197, 157–168. doi: 10.1002/jcp.10364
Cargnello, M., and Roux, P. P. (2011). Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83. doi: 10.1128/MMBR.00031-10
Chapman, M. S., Askew, D. J., Kuscuoglu, U., and Miesfeld, R. L. (1996). Transcriptional control of steroid-regulated apoptosis in murine thymoma cells. Mol. Endocrinol. 10, 967–978.
Condeelis, J. (1995). Elongation factor 1 alpha, translation and the cytoskeleton. Trends Biochem. Sci. 20, 169–170. doi: 10.1016/S0968-0004(00)88998-7
Doyle, C. K., Nethery, K. A., Popov, V. L., and McBride, J. W. (2006). Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74, 711–720. doi: 10.1128/IAI.74.1.711-720.2006
Dunphy, P. S., Luo, T., and McBride, J. W. (2014). Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect. Immun. 82, 4154–4168. doi: 10.1128/IAI.01984-14
Edelmann, M. J., Iphöfer, A., Akutsu, M., Altun, M., di Gleria, K., Kramer, H. B., et al. (2009). Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 418, 379–390. doi: 10.1042/BJ20081318
Ejiri, S. (2002). Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Biosci. Biotechnol. Biochem. 66, 1–21. doi: 10.1271/bbb.66.1
Farris, T. R., Dunphy, P. S., Zhu, B., Kibler, C. E., and McBride, J. W. (2016). Ehrlichia chaffeensis TRP32 is a nucleomodulin that directly regulates expression of host genes governing differentiation and proliferation. Infect. Immun. 84, 3182–3194. doi: 10.1128/IAI.00657-16
Gao, Z., Lee, P., Stafford, J. M., von Schimmelmann, M., Schaefer, A., and Reinberg, D. (2014). An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 516, 349–354. doi: 10.1038/nature13921
Goldenberg, S. J., Cascio, T. C., Shumway, S. D., Garbutt, K. C., Liu, J., Xiong, Y., et al. (2004). Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119, 517–528. doi: 10.1016/j.cell.2004.10.019
Hao, B., Oehlmann, S., Sowa, M. E., Harper, J. W., and Pavletich, N. P. (2007). Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 26, 131–143. doi: 10.1016/j.molcel.2007.02.022
Hattermann, K., Held-Feindt, J., Lucius, R., Muerkoster, S. S., Penfold, M. E., Schall, T. J., et al. (2010). The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer Res. 70, 3299–3308. doi: 10.1158/0008-5472.CAN-09-3642
Hayward, P., Kalmar, T., and Arias, A. M. (2008). Wnt/Notch signalling and information processing during development. Development 135, 411–424. doi: 10.1242/dev.000505
Jin, L., Pahuja, K. B., Wickliffe, K. E., Gorur, A., Baumgartel, C., Schekman, R., et al. (2012). Ubiquitin-dependent regulation of COPII coat size and function. Nature 482, 495–500. doi: 10.1038/nature10822
Jorgensen, I., Bednar, M. M., Amin, V., Davis, B. K., Ting, J. P., McCafferty, D. G., et al. (2011). The Chlamydia protease CPAF regulates host and bacterial proteins to maintain pathogen vacuole integrity and promote virulence. Cell Host Microbe 10, 21–32. doi: 10.1016/j.chom.2011.06.008
Kubach, J., Lutter, P., Bopp, T., Stoll, S., Becker, C., Huter, E., et al. (2007). Human CD4+CD25+ regulatory T cells: proteome analysis identifies galectin-10 as a novel marker essential for their anergy and suppressive function. Blood 110, 1550–1558. doi: 10.1182/blood-2007-01-069229
Kumagai, Y., Matsuo, J., Hayakawa, Y., and Rikihisa, Y. (2010). Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J. Bacteriol. 192, 4122–4133. doi: 10.1128/JB.00132-10
Kuriakose, J. A., Miyashiro, S., Luo, T., Zhu, B., and McBride, J. W. (2011). Ehrlichia chaffeensis transcriptome in mammalian and arthropod hosts reveals differential gene expression and post transcriptional regulation. PLoS ONE 6:e24136. doi: 10.1371/journal.pone.0024136
Kuriakose, J. A., Zhang, X., Luo, T., and McBride, J. W. (2012). Molecular basis of antibody mediated immunity against Ehrlichia chaffeensis involves species-specific linear epitopes in tandem repeat proteins. Microbes Infect. 14, 1054–1063. doi: 10.1016/j.micinf.2012.05.012
Lin, M., and Rikihisa, Y. (2004). Ehrlichia chaffeensis downregulates surface Toll-like receptors 2/4, CD14 and transcription factors PU.1 and inhibits lipopolysaccharide activation of NF-κB, ERK 1/2 and p38 MAPK in host monocytes. Cell. Microbiol. 6, 175–186. doi: 10.1046/j.1462-5822.2003.00355.x
Lina, T. T., Dunphy, P. S., Luo, T., and McBride, J. W. (2016a). Ehrlichia chaffeensis TRP120 activates canonical Notch signaling to downregulate TLR2/4 expression and promote intracellular survival. MBio 7, e00672–e00616. doi: 10.1128/mBio.00672-16
Lina, T. T., Farris, T., Luo, T., Mitra, S., Zhu, B., and McBride, J. W. (2016b). Hacker within! Ehrlichia chaffeensis effector driven phagocyte reprogramming strategy. Front. Cell. Infect. Microbiol. 6:58. doi: 10.3389/fcimb.2016.00058
Liu, Y., Zhang, Z., Jiang, Y., Zhang, L., Popov, V. L., Zhang, J., et al. (2011). Obligate intracellular bacterium Ehrlichia inhibiting mitochondrial activity. Microbes Infect. 13, 232–238. doi: 10.1016/j.micinf.2010.10.021
Luo, T., Dunphy, P. S., Lina, T. T., and McBride, J. W. (2016). Ehrlichia chaffeensis exploits canonical and noncanonical host Wnt signaling pathways to stimulate phagocytosis and promote intracellular survival. Infect. Immun. 84, 686–700. doi: 10.1128/IAI.01289-15
Luo, T., Kuriakose, J. A., Zhu, B., Wakeel, A., and McBride, J. W. (2011). Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infect. Immun. 79, 4382–4391. doi: 10.1128/IAI.05608-11
Luo, T., and McBride, J. W. (2012). Ehrlichia chaffeensis TRP32 interacts with host cell targets that influence intracellular survival. Infect. Immun. 80, 2297–2306. doi: 10.1128/IAI.00154-12
Luo, T., Zhang, X., and Mcbride, J. W. (2009). Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clin. Vaccine Immunol. 16, 982–990. doi: 10.1128/CVI.00048-09
Luo, T., Zhang, X., Wakeel, A., Popov, V. L., and McBride, J. W. (2008). A variable-length PCR target protein of Ehrlichia chaffeensis contains major species-specific antibody epitopes in acidic serine-rich tandem repeats. Infect. Immun. 76, 1572–1580. doi: 10.1128/IAI.01466-07
Mohan Kumar, D., Lin, M., Xiong, Q., Webber, M. J., Kural, C., and Rikihisa, Y. (2015). EtpE binding to DNase X induces Ehrlichial entry via CD147 and hnRNP-K recruitment, followed by mobilization of N-WASP and actin. MBio 6:e01541. doi: 10.1128/mBio.01541-15
Mohan Kumar, D., Yamaguchi, M., Miura, K., Lin, M., Los, M., Coy, J. F., et al. (2013). Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS Pathog. 9:e1003666. doi: 10.1371/journal.ppat.1003666
Oberst, A., Malatesta, M., Aqeilan, R. I., Rossi, M., Salomoni, P., Murillas, R., et al. (2007). The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase itch. Proc. Natl. Acad. Sci. U.S.A. 104, 11280–11285. doi: 10.1073/pnas.0701773104
Paddock, C. D., and Childs, J. E. (2003). Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16, 37–64. doi: 10.1128/CMR.16.1.37-64.2003
Popov, V. L., Yu, X., and Walker, D. H. (2000). The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb. Pathog. 28, 71–80. doi: 10.1006/mpat.1999.0327
Ribet, D., and Cossart, P. (2010). Post-translational modifications in host cells during bacterial infection. FEBS Lett. 584, 2748–2758. doi: 10.1016/j.febslet.2010.05.012
Roth, S., Kholodenko, B. N., Smit, M. J., and Bruggeman, F. J. (2015). G protein-coupled receptor signaling networks from a systems perspective. Mol. Pharmacol. 88, 604–616. doi: 10.1124/mol.115.100057
Sobieszczuk, D. F., Poliakov, A., Xu, Q., and Wilkinson, D. G. (2010). A feedback loop mediated by degradation of an inhibitor is required to initiate neuronal differentiation. Genes Dev. 24, 206–218. doi: 10.1101/gad.554510
Thomas, S., Popov, V. L., and Walker, D. H. (2010). Exit mechanisms of the intracellular bacterium Ehrlichia. PLoS ONE 5:e15775. doi: 10.1371/journal.pone.0015775
Wakeel, A., Den Dulk-Ras, A., Hooykaas, P. J., and McBride, J. W. (2011). Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Front. Cell. Infect. Microbiol. 1:22. doi: 10.3389/fcimb.2011.00022
Wakeel, A., Kuriakose, J. A., and McBride, J. W. (2009). An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect. Immun. 77, 1734–1745. doi: 10.1128/IAI.00027-09
Wakeel, A., Zhang, X., and McBride, J. W. (2010). Mass spectrometric analysis of Ehrlichia chaffeensis tandem repeat proteins reveals evidence of phosphorylation and absence of glycosylation. PLoS ONE 5:e9552. doi: 10.1371/journal.pone.0009552
Wang, C., Zhou, G. L., Vedantam, S., Li, P., and Field, J. (2008). Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis. J. Cell Sci. 121, 2913–2920. doi: 10.1242/jcs.023911
Wu, Y., Liu, X. M., Wang, X. J., Zhang, Y., Liang, X. Q., and Cao, E. H. (2009). PIG11 is involved in hepatocellular carcinogenesis and its over-expression promotes Hepg2 cell apoptosis. Pathol. Oncol. Res. 15, 411–416. doi: 10.1007/s12253-008-9138-5
Xiong, Q., Bao, W., Ge, Y., and Rikihisa, Y. (2008). Ehrlichia ewingii infection delays spontaneous neutrophil apoptosis through stabilization of mitochondria. J. Infect. Dis. 197, 1110–1118. doi: 10.1086/533457
Yang, S. B., Chen, X., Wu, B. Y., Wang, M. W., Cai, C. H., Cho, D. B., et al. (2009). Immunoglobulin kappa and immunoglobulin lambda are required for expression of the anti-apoptotic molecule Bcl-xL in human colorectal cancer tissue. Scand. J. Gastroenterol. 44, 1443–1451. doi: 10.3109/00365520903369953
Zheng, Y., Xu, B., Zhao, Y., Gu, H., Li, C., Wang, Y., et al. (2015). CA1 contributes to microcalcification and tumourigenesis in breast cancer. BMC Cancer 15:679. doi: 10.1186/s12885-015-1707-x
Zhong, G. (2011). Chlamydia trachomatis secretion of proteases for manipulating host signaling pathways. Front. Microbiol. 2:14. doi: 10.3389/fmicb.2011.00014
Zhu, B., Farris, T., Milligan, S., Chen, H. S., Zhu, R. J., Hong, A., et al. (2016). Rapid identification of ubiquitination and SUMOylation target sites by microfluidic peptide array. Biochem. Biophys. Rep. 5, 430–438. doi: 10.1016/j.bbrep.2016.02.003
Keywords: Ehrlichia chaffeensis, tandem repeat protein, effector-host interaction, infection, cell signaling, vesicle trafficking, transcriptional regulation, posttranslational modification
Citation: Luo T, Dunphy PS and McBride JW (2017) Ehrlichia chaffeensis Tandem Repeat Effector Targets Differentially Influence Infection. Front. Cell. Infect. Microbiol. 7:178. doi: 10.3389/fcimb.2017.00178
Received: 11 February 2017; Accepted: 24 April 2017;
Published: 12 May 2017.
Edited by:
Matthew S. Francis, Umeå University, SwedenReviewed by:
Ulrike G. Munderloh, University of Minnesota, USADamien F. Meyer, Agricultural Research Centre for International Development, France
Cecilia Tamborindeguy, Texas A&M University, USA
Copyright © 2017 Luo, Dunphy and McBride. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jere W. McBride, jemcbrid@utmb.edu
 Tian Luo
Tian Luo Paige S. Dunphy1
Paige S. Dunphy1  Jere W. McBride
Jere W. McBride