Background
Acinetobacter baumannii (A. baumannii) is a Gram-negative, coccobacilli aerobic pathogen which causes nosocomial infections in hospitals worldwide. A. baumannii has been identified as one of the six important and highly drug resistant hospital pathogens by the “Infectious Disease Society of America (IDSA)” (Talbot et al., 2006; Boucher et al., 2009). A. baumannii rapidly acquired antibiotic resistance, leading to wide-spread multidrug-resistant isolates, which are impervious to nearly all antibiotic treatments (Fournier et al., 2006; Vashist and Rajeswari, 2006; Perez et al., 2007; Roca et al., 2012). Therefore, dissection of the molecular details at gene level of A. baumannii biology is urgently needed which can greatly help in our understanding of the resistance mechanisms that are evolved by the pathogen. Recently, some efforts were made to understand the relation between junk DNA, mutation frequencies with respect to antibiotic resistance in bacteria (Martinez and Baquero, 2000; Gil and Latorre, 2012) and in A. baumannii (Tiwari et al., 2012). However, very little is known about the role of specific virulence factors and cellular processes that are associated with the development of resistance in A. baumannii. A small fraction of genomic DNA is known to adopt non-canonical B-DNA (unusual) structures, and can cause genomic instability. This fact is well known in eukaryotes, however, is also suggested that it can apply in bacteria also (Martín-Lozano et al., 2002). In this connection, the purine repeat sequences attract special attention due to their high ability to convert the double stranded DNA structures to triple stranded DNA structures. The poly(purine) tracts by forming DNA triplex structures are prone to induce mutations as well as interventions in various cellular processes including replication, transcription etc. (Wells, 2008; Singh and Rajeswari, 2015). Therefore, it is interesting to look at the genes containing purine repeats in bacterial infection and in resistance and as an example, we performed a systematic search in susceptible and drug resistant strains of A. baumannii for exclusively poly(purines).
It is important to mention that the DNA triplex was first proposed by Pauling and Corey (1953) which was later proved to exist in solution by Felsenfeld et al. (1957). DNA-triplex structure, as the name suggests, is formed between three strands of DNA, the third strand forms Hoogsteen base-pairs with that strand which is rich in purines; while the Watson-Crick basepairs remains intact. DNA triplexes could be of either intra-molecular where the third strand is part of the DNA or inter-molecular if the third strand is from external DNA. Further, DNA-triplex is known as “Purine motif (R)” if the third strand is rich in purines, alternately it is called “Pyrimidine motif (Y)” where the third strand contains largely pyrimidines (Lyamichev et al., 1986; Frank-Kamenetskii and Mirkin, 1995; Rajeswari, 2012; Dyke, 2013).
It has been reported that purine repeats have potential to form DNA-triplex which can induce mutation and/or genetic instability which can further influence DNA replication, repair and transcription in eukaryotes (Bacolla et al., 2004, 2006; Jain et al., 2008). Keeping in view of the above discussion on the importance of purine repeats in causing genomic instability via DNA-triplex formation, we investigated the whole genome of A. baumannii to see if there are any potential purine repeat sequences which can form intra-molecular DNA triplex in causing drug resistance.
Materials and Methods
Genome Information
Genome sequence in FASTA format and annotation information in GenBank format of all the 14 strains of the Acinetobacter baumannii which include susceptible (SC) and multi-drug resistant (MDR) strains (Accession Number of strains are given in Table 1) were retrieved from the NCBI Nucleotide browser (https://www.ncbi.nlm.nih.gov/nuccore) on February, 2016.
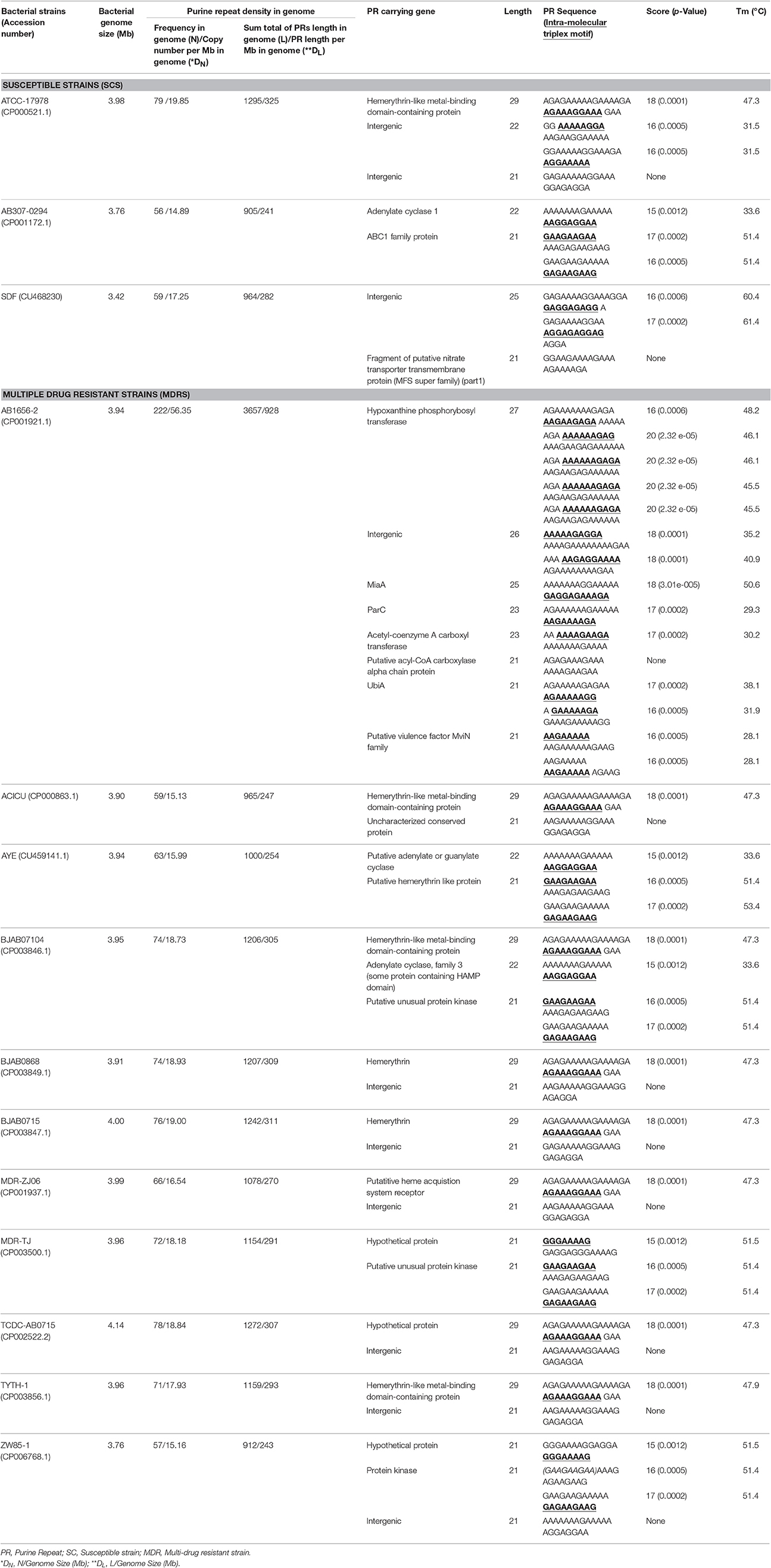
Table 1. Distribution and enrichment of purine repeats and characteristics of DNA-triplex formed by purine (n > 20) repeats; and their associated genes in various strains of Acinetobacter baumannii.
Purine Repeat Search
In the present study, purine repeats (PRs) are defined as poly(purine) tracks of size, n ≥ 15, without any single interruption of pyrimidine, which is a strict definition of triplex forming sequences. To identify the PRs, a GUI (graphical user interface) based standalone Windows-based software entitled “nTrackAnnotator” (http://www.aiims.edu/images/depart/biochem/nTrackAnnotator.rar) (Singh and Rajeswari, 2017) was used. The nTrackAnnotator is developed by our group, which can be used for eukaryotic/prokaryotic genome sequences, including PRs. The nTrackAnnotator was developed in C# 4.0 object oriented programming language at. NET Framework 4.0 for Windows-platform specific to NCBI/Genome data format.
Density of Purine Repeats
In order to characterize the repeats, we used two measures of density, the density in number (DN) and the density in length (DL) (Kendall, 1938) and are defined as:
Kendall τ rank test (Kendall, 1938) has been performed to find the association between DN and DL. The Kendall rank correlation coefficient (τ) between the two variables with “n” number of observations is defined as:
Intra-Molecular Triplex Motif Search
The intramolecular triplex motifs were identified using the Biocondcutor package “Triplex,” which is an R interface to the C implementation of a dynamic programming algorithm for triplex search (http://bioconductor.org/packages/release/bioc/html/triplex.html) (Hon et al., 2013). The R package “triplex” detected positions of subsequences in the PRs, where these sub-sequences were likely to fold into an intramolecular triplex. Further, the triplex-score and p-value of the triplex motifs were also calculated to find the probability of occurrence of detected triplexes in a random sequence and to find the quality of intra-molecular triplex. A higher score value indicates a higher quality triplex. Further, the Bioconductor R package was also used for the visualization of exact base-paring in two dimensions.
Prediction of the Stability of Intra-Molecular DNA Triplex
The stability of any DNA-triplex measured in terms of the melting temperature (Tm) depends on various factors, such as the composition of the sequence of DNA-triplex, its concentration and pH. A rough prediction of the melting temperature (Tm) of DNA-triplex in the genome was determined using Roberts and Crothers empirical equations (equations i–iii) (Roberts and Crothers, 1996; Goñi et al., 2004):
where CTP is the concentration of DNA- triplex and R is gas constant.
The enthalpy (ΔH°) is evaluated (in kcal/mol) using equation (ii):
The Gibbs free energy (ΔG°) at 37°C was computed using equation (iii):
where (X) and (Y) represents cytosine and thymine respectively in the triplex forming sequence.
Results and Discussion
The notion that genomic sequences are random in nature is far from truth; interestingly they are made up of systematically ordered and information rich patterns. These repeated sequence patterns such as poly(purine) repeats have been reported to be linked with genome functions and organization (Bucher and Yagil, 1991; Achaz et al., 2002; Gil and Latorre, 2012). Therefore, to understand the significance of intra-molecular triplex and its integration of genetic factors that might be responsible for A. baumannii infection and pathogenecity, we undertook a whole-genome sequence analysis of 14 A. baumannii strains to explore common features of their evolution and the diversity of mechanisms. Our goal was to (i) identify triplex forming purine repeat distribution in the susceptible and clinical isolates, (ii) demonstrate presence of stable intra-molecular triplexes, and (iii) find the impact of triplexes in the infection evolution of the resistance phenotype. A contiguous PR of at least 10 base pairs is required for the duplex acceptor and can form intra-molecular triplexes (Cheng and Van Dyke, 1993; Orson et al., 1999). Since triplex is formed by shorter PRs are not stable under physiological conditions and even single base pyrimidine interruptions are known to greatly destabilized DNA-triplexes (Orson et al., 1999), therefore, we performed the analysis of PR, n ≥ 15 present in SC and MDR strains of A. baumannii.
Distribution and Enrichment of PRs in A. baumannii Strains
The distribution of PRs in both susceptible (SC) and multi-drug resistant (MDR) bacterial genomes is given in Table 1. The distribution of PRs in different strains of A. baumannii was found to be in general comparable to between SC and MDR isolates except MDR strain AB1656-2 (frequency, N, in Table 1) where the frequency is approximately 3-folds higher. The larger PR length of 29 nucleotides was observed in eight bacterial strains (SC strain: ATCC-17978; MDR strains: ACICU, BJAB07104, BJAB0868, BJAB0715, MDR-ZJ06, TCDC-AB0715, ZW85-1). Majority of PRs were found in the range of 15–20 nucleotides length (Table 1). The frequency of PRs was observed to be proportional to the length of PRs in corresponding bacterial strain except in the MDR strains “TYTH-1” and “MDR-TJ.” The TYTH-1 strains carry 71 PRs with 1159 nucleotides purine repeat length, however, “MDR-TJ” carry 72 PRs with 1154 nucleotides length. The observation that the highest PRs (n = 222) found in MDR strain “AB1656-2” indicates the possibility of greater frequency of mutations. The existence of such large density of PRs could be due to massive duplications of short PRs in A. baumannii and also indicates its biological effect on genomic functions.
After identifying the PRs, they were characterized by using two parameters of repeat density; DN and DL. It was not surprising to find the positive correlation between DN and DL (τ = 0.956; p = 2.5034e-06; Kendall τ rank test). As this observation supports the report that the repeat sequences (like transposable elements) have high chance of duplications in the corresponding chromosomes (Coissac et al., 1997; Achaz et al., 2001). However, the biological interpretation of these measures may be quite different. The DN is assimilated to the rate of amplification, which is a fine balance between duplication and deletion processes. While, the DL which is a measure of redundancy tolerated by the chromosome can be extrapolated to the evolutionary changes (Coissac et al., 1997; Achaz et al., 2001, 2002). For example, the DN in ATCC-17978 is only 19.85 per Mb as compared to that of an MDR strain, AB1656-2 is 56.35 per Mb. Similarly, the DL in ATCC-1798 is 325 per Mb vs. AB1656-2 is 928 per Mb. It is clear from the observed high PR density (DN and DL) in the MDR strain; AB1656-2 in A. baumannii hints their critical role in genomic adaptation processes.
Annotation and Functional Importance of Potential PRs (≥ 20) in A. baumannii Strains
A glance at Table 1 reveals that there are a minimum of 56 PRs found in any strain, SC or MDR, of A. baumannii. However, amongst such a large number of PRs (total of 1,106 in 14 strains), only 4% (37 PRs) were found to carry larger than 20 purines. As the stability of DNA-triplex formed by a PR is directly dependent on its length, we chose to characterize the localization of these 37 PRs which are discussed in sequel.
In principle purine repeats can form intra-molecular DNA triple helical structure; however, this may not be always conducive. Lexa et al. (2011) reported that there could be two possible reasons for purine repeats to form not so stable DNA-triplex. There are: (i) analogical to mismatches in DNA duplex, involving nucleotides in triplets that do not readily form Hoogsteen bonds; (ii) geometrically incompatible neighboring bases hindering proper alignment of strands for optimal hydrogen bonding and stacking (Lexa et al., 2011). Therefore, the “Bioconductor R package: triplex,” was used to detect strong intra-molecular triplex forming sequences within the identified potential PRs. Further, stability of DNA-triplex was quantitatively determined by melting temperature (Tm) of the intra-molecular triplexes that were calculated on the basis of Roberts and Crothers empirical equations (Roberts and Crothers, 1996). The rules were derived from van't Hoff analysis using a number of different DNA-triplexes tested at a variety of pH values and to predict the stability of DNA triplex on the basis of the sequence.
By applying the “Bioconductor R package: triplex,” to all the 37 potential PRs, eight sequences found not form intra-molecular DNA-triplex. However, there may be multiple independent intra-molecular DNA triplexes possible within a certain PR, for example, ABC1 family protein (PR of 21 purines) shows two such sequences (Table 1). The results also showed that all PRs were of pyrimidine motif, with an exception of PR present in the protein, hypoxanthine phosphorybosyl transferase, of MDR strain AB1656-2, was found to be forming intra-molecular triplex of both purine and pyrimidine motifs from the same sequence.
The rest of 29 PRs were further evaluated for their relative intra-molecular DNA-triplex forming stability using melting temperature (Tm). The results showed that a large number (19) of PRs were more stable (Tm ≥ 40°C) as compared to those which Tm about 30°C (Table 1). It is tempting to speculate that when the bacteria enters in the host (human), the DNA-triplexes with Tm about 30°C are not be stable and therefore dissociate as the physiological temperature of the body 37°C, is above its Tm. Therefore, intra-molecular DNA-triplexes ≥ 40°C present in 20 genes were considered to be stable (Table 1).
Majority of PRs were limited in the coding regions of the genome and only 10 were located in inter-genic regions (Table 1). It is interesting to note that AB165-2 strain also contains maximum (8) repeats with purine tracks (≥20) as compared to that of rest of 13 strains (Table 1). Further, the purine repeat with maximum number of purines, 29, was found to be conserved (sequence identity: 100%; query coverage: 100%) as single copy in the eight A. baumannii strains. The significant observation is that the PR with 29 purines was restricted to only “hemerythrin” or “hemerythrin like metal binding proteins” (Table 1). In this context, it is important to note that hemerythrin and like proteins are known to be associated with various cellular functions including detoxification, transport, storage of iron and/or oxygen or a role as a sensory protein (Chenier et al., 2008; Eijkelkamp et al., 2011; Fiester and Actis, 2013). Iron is a stress factor that plays a critical role in the virulence of A. baumannii and the pathogenesis of human infections. Under iron stress conditions, hemerythrin and hemerythrin like proteins were reported to be donwregulated in A. baumanni (Eijkelkamp et al., 2011; Fiester and Actis, 2013). Considering the vital role of iron and iron associated proteins like hemerythrin, it appears that the presence of large purines could be one of the major reasons in inducing mutations and DNA triplex mediated downregulation of these proteins. It clearly indicates that there is some relation that exists between hemerythrin gene related iron utilization and DNA triplex formation in the A. baumannii, however, the mechanisms need to be explored. In addition to this, many enzymes were identified carrying potential PR such as hypoxanthine phosphorybosyl transferase, acetyl-coenzyme A carboxyl transferase, adenylate cyclase, protein kinases, and hypothetical proteins (Table 1) could also be affected. The functions of these enzymes are in general are crucial and therefore can directly or in-directly play role in survival of the bacteria.
More importantly, we found several virulence factor genes (parC, MviN family gene, VbiA, MiaA) carrying potential PRs located only in the MDR strains indicating further the possible functions in bacterial pathogenesis (Table 1). Among the several factors that affect the appearance and spread of acquired antibiotic resistance, the mutation frequency and the biological cost of resistance are of special importance (Björkholm et al., 2001; Hammerstrom et al., 2015). It is reported that mutations in parC (topoisomerase IV subunit C) gene contribute in antibiotic susceptibility and for this reason parC gene has been one of the drug targets for A. baumanni infections (Vila et al., 1995; Vakili et al., 2014). The other virulence factor MviN family gene, although its function is not elucidated but bioinformatics analysis indicates that MviN is an integral membrane protein and appeared to be a member of the MviN MATE (multi-antimicrobial extrusion) like super-family shown to function as drug/sodium antiporters (Omote et al., 2006). These proteins have also been shown to mediate resistance to a wide range of antibiotics drugs including fluoroquinolones, aminoglycosides etc. (Omote et al., 2006). Thus, the identification of potential PRs in the above mentioned genes in A. baumannii suggests their crucial role in infections, pathogenesis and drug-resistance either by triplex mediated mutatgenesis or gene deregulation.
Conclusions
Based on the in-silico analysis of A. baumannii genome of clinical strains, the following major conclusions can be drawn. Triplex forming purine repeat sequences (PRs) were found to be randomly distributed (genic and intergenic) across the entire A. baummannii strains. The PRs, n ≥ 20, referred to as “Potential PRs.” Presence of potential PRs in bacterial genome reveal strong possibility to form stable intra-molecular DNA-triplex that can induce mutations and thereby interfere with genome functions in A. baumannii. As, iron plays important role in bacterial survival, the identification of “potential PRs” in genes of hemerythrin and hemerethyrin-like binding proteins, which are associated with iron utilization, is considered to be a significant finding of the present study. Equally important are the potential PR associated genes of enzymes (transferase, cyclase, kinase etc.) and of virulence factors (parC, UbiA, MiaA, MviN); which are known for their functional role in bacterial adaptation and infections.
Author Contributions
MR: designed the experiments; HS: performed the experiments and analyzed the data; MR: analyzed the data, drafted, edited, and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
HS thanks the Indian Council of Medical Research (ICMR), New Delhi, India for providing Senior Research Fellowship (3/1/2/43/Neuro/2013-NCD-I).
References
Achaz, G., Netter, P., and Coissac, E. (2001). Study of intrachromosomal duplications among the eukaryote genomes. Mol. Biol. Evol. 18, 2280–2288. doi: 10.1093/oxfordjournals.molbev.a003774
Achaz, G., Rocha, E. P. C., Netter, P., and Coissac, E. (2002). Origin and fate of repeats in bacteria. Nucleic Acids Res. 30, 2987–2994. doi: 10.1093/nar/gkf391
Bacolla, A., Collins, J. R., Gold, B., Chuzhanova, N., Yi, M., Stephens, R. M., et al. (2006). Long homopurine*homopyrimidine sequences are characteristic of genes expressed in brain and the pseudoautosomal region. Nucleic Acids Res. 34, 2663–2675. doi: 10.1093/nar/gkl354
Bacolla, A., Jaworski, A., Larson, J. E., Jakupciak, J. P., Chuzhanova, N., Abeysinghe, S. S., et al. (2004). Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl. Acad. Sci. U.S.A. 101, 14162–14167. doi: 10.1073/pnas.0405974101
Björkholm, B., Sjölund, M., Falk, P. G., Berg, O. G., Engstrand, L., and Andersson, D. I. (2001). Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 98, 14607–14612. doi: 10.1073/pnas.241517298
Boucher, H. W., Talbot, G. H., Bradley, J. S., Edwards, J. E., Gilbert, D., Rice, L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12. doi: 10.1086/595011
Bucher, P., and Yagil, G. (1991). Occurrence of oligopurine.oligopyrimidine tracts in eukaryotic and prokaryotic genes. DNA Seq. 1, 157–172. doi: 10.3109/10425179109020767
Cheng, A. J., and Van Dyke, M. W. (1993). Monovalent cation effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 21, 5630–5635. doi: 10.1093/nar/21.24.5630
Chenier, D., Beriault, R., Mailloux, R., Baquie, M., Abramia, G., Lemire, J., et al. (2008). Involvement of fumarase C and NADH oxidase in metabolic adaptation of Pseudomonas fluorescens cells evoked by aluminum and gallium toxicity. Appl. Environ. Microbiol. 74, 3977–3984. doi: 10.1128/AEM.02702-07
Coissac, E., Maillier, E., and Netter, P. (1997). A comparative study of duplications in bacteria and eukaryotes: the importance of telomeres. Mol. Biol. Evol. 14, 1062–1074. doi: 10.1093/oxfordjournals.molbev.a025712
Dyke, M. W. V. (2013). Do DNA Triple Helices or Quadruplexes Have a Role in Transcription? Austin, TX: Landes Bioscience.
Eijkelkamp, B. A., Hassan, K. A., Paulsen, I. T., and Brown, M. H. (2011). Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12:126. doi: 10.1186/1471-2164-12-126
Felsenfeld, G., Davies, D. R., and Rich, A. (1957). Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 79, 2023–2024. doi: 10.1021/ja01565a074
Fiester, S. E., and Actis, L. A. (2013). Stress responses in the opportunistic pathogen Acinetobacter baumannii. Future Microbiol. 8, 353–365. doi: 10.2217/fmb.12.150
Fournier, P. E., Vallenet, D., Barbe, V., Audic, S., Ogata, H., Poirel, L., et al. (2006). Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. doi: 10.1371/journal.pgen.0020007
Frank-Kamenetskii, M. D., and Mirkin, S. M. (1995). Triplex DNA structures. Annu. Rev. Biochem. 64, 65–95. doi: 10.1146/annurev.bi.64.070195.000433
Gil, R., and Latorre, A. (2012). Factors behind junk DNA in bacteria. Genes (Basel). 3, 634–650. doi: 10.3390/genes3040634
Goñi, J. R., de la Cruz, X., and Orozco, M. (2004). Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Res. 32, 354–360. doi: 10.1093/nar/gkh188
Hammerstrom, T. G., Beabout, K., Clements, T. P., Saxer, G., and Shamoo, Y. (2015). Acinetobacter baumannii repeatedly evolves a hypermutator phenotype in response to tigecycline that effectively surveys evolutionary trajectories to resistance. PLoS ONE 10:e0140489. doi: 10.1371/journal.pone.0140489
Hon, J., Martinek, T., Rajdl, K., and Lexa, M. (2013). Triplex: an R/Bioconductor package for identification and visualization of potential intramolecular triplex patterns in DNA sequences. Bioinformatics 29, 1900–1901. doi: 10.1093/bioinformatics/btt299
Jain, A., Wang, G., and Vasquez, K. M. (2008). DNA triple helices: biological consequences and therapeutic potential. Biochimie 90, 1117–1130. doi: 10.1016/j.biochi.2008.02.011
Kendall, M. (1938). A new measure of rank correlation. Biometrika 30, 81–89. doi: 10.1093/biomet/30.1-2.81
Lexa, M., Martínek, T., Burgetová, I., Kopecek, D., and Brázdová, M. (2011). A dynamic programming algorithm for identification of triplex-forming sequences. Bioinformatics 27, 2510–2517. doi: 10.1093/bioinformatics/btr439
Lyamichev, V. I., Mirkin, S. M., and Frank-Kamenetskii, M. D. (1986). Structures of homopurine-homopyrimidine tract in superhelical DNA. J. Biomol. Struct. Dyn. 3, 667–669. doi: 10.1080/07391102.1986.10508454
Martinez, J. L., and Baquero, F. (2000). Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44, 1771–1777. doi: 10.1128/AAC.44.7.1771-1777.2000
Martín-Lozano, D., Cisneros, J. M., Becerril, B., Cuberos, L., Prados, T., Ortíz-Leyba, C., et al. (2002). Comparison of a repetitive extragenic palindromic sequence-based PCR method and clinical and microbiological methods for determining strain sources in cases of nosocomial Acinetobacter baumannii bacteremia. J. Clin. Microbiol. 40, 4571–4575. doi: 10.1128/JCM.40.12.4571-4575.2002
Omote, H., Hiasa, M., Matsumoto, T., Otsuka, M., and Moriyama, Y. (2006). The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 27, 587–593. doi: 10.1016/j.tips.2006.09.001
Orson, F. M., Klysik, J., Bergstrom, D. E., Ward, B., Glass, G. A., Hua, P., et al. (1999). Triple helix formation: binding avidity of acridine-conjugated AG motif third strands containing natural, modified and surrogate bases opposed to pyrimidine interruptions in a polypurine target. Nucleic Acids Res. 27, 810–816. doi: 10.1093/nar/27.3.810
Pauling, L., and Corey, R. B. (1953). A proposed structure for the nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 39, 84–97. doi: 10.1073/pnas.39.2.84
Perez, F., Hujer, A. M., Hujer, K. M., Decker, B. K., Rather, P. N., and Bonomo, R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3471–3484. doi: 10.1128/AAC.01464-06
Rajeswari, M. R. (2012). DNA triplex structures in neurodegenerative disorder, Friedreich's ataxia. J. Biosci. 37, 519–532. doi: 10.1007/s12038-012-9219-1
Roberts, R. W., and Crothers, D. M. (1996). Prediction of the stability of DNA triplexes. Proc. Natl. Acad. Sci. U.S.A. 93, 4320–4325. doi: 10.1073/pnas.93.9.4320
Roca, I., Espinal, P., Vila-Farrés, X., and Vila, J. (2012). The Acinetobacter baumannii Oxymoron: commensal Hospital Dweller Turned Pan-Drug-Resistant Menace. Front. Microbiol. 3:148. doi: 10.3389/fmicb.2012.00148
Singh, H. N., and Rajeswari, M. R. (2015). Role of long purine stretches in controlling the expression of genes associated with neurological disorders. Gene 572, 175–183. doi: 10.1016/j.gene.2015.07.007
Singh, H. N., and Rajeswari, M. R. (2017). nTrackAnnotator: software for detection and annotation of sequence tracks of chosen nucleic acid bases with defined length in genome. Gene Rep. 7, 32–34. doi: 10.1016/j.genrep.2017.01.004
Talbot, G. H., Bradley, J., Edwards, J. E. Jr., Gilbert, D., and Bartlett, J. G. (2006). Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42, 657–668. doi: 10.1086/499819
Tiwari, V., Nagpal, I., Subbarao, N., and Moganty, R. R. (2012). In-silico modeling of a novel OXA-51 from ß-lactam-resistant Acinetobacter baumannii and its interaction with various antibiotics. J. Mol. Model. 18, 3351–3361. doi: 10.1007/s00894-011-1346-3
Vakili, B., Khorvash, F., Fazeli, H., and Khaleghi, M. (2014). Detection of quinolone-resistance mutations of parC gene in clinical isolates of Acinetobacter baumannii in Iran. J. Res. Med. Sci. 19, 567–570.
Vashist, J., and Rajeswari, M. R. (2006). Structural investigations on novel porin, OmpAb from Acinetobacter baumannii. J. Biomol. Struct. Dyn. 24, 243–253. doi: 10.1080/07391102.2006.10507116
Vila, J., Ruiz, J., Goñi, P., Marcos, A., and Jimenez de Anta, T. (1995). Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 39, 1201–1203. doi: 10.1128/AAC.39.5.1201
Keywords: Acinetobacter baumannii, DNA-triplex, purine repeat, multi-drug resistant, bioinformatics analysis, genome sequence analysis
Citation: Singh HN and Rajeswari MR (2017) DNA-triplex Forming Purine Repeat Containing Genes in Acinetobacter baumannii and Their Association with Infection and Adaptation. Front. Cell. Infect. Microbiol. 7:250. doi: 10.3389/fcimb.2017.00250
Received: 20 September 2016; Accepted: 26 May 2017;
Published: 16 June 2017.
Edited by:
Yongqun He, University of Michigan Health System, United StatesReviewed by:
Yang Zhang, University of Pennsylvania, United StatesArzucan Ozgur, Boğaziçi University, Turkey
Zhaohui Ni, Jilin University, China
Copyright © 2017 Singh and Rajeswari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Moganty R. Rajeswari, cmFqZXN3YXJpMzAxMUBob3RtYWlsLmNvbQ==
 Himanshu N. Singh
Himanshu N. Singh Moganty R. Rajeswari
Moganty R. Rajeswari