- 1Highly Pathogenic Microorganisms, Centre for Biological Threats and Special Pathogens, Robert Koch Institute, Berlin, Germany
- 2Cellular Interactions of Bacterial Pathogens, Centre for Biological Threats and Special Pathogens, Robert Koch Institute, Berlin, Germany
The Legionella genus comprises more than 60 species. In particular, Legionella pneumophila is known to cause severe illnesses in humans. Legionellaceae are ubiquitous inhabitants of aquatic environments. Some Legionellaceae are motile and their motility is important to move around in habitats. Motility can be considered as a potential virulence factor as already shown for various human pathogens. The genes of the flagellar system, regulator and structural genes, are structured in hierarchical levels described as the flagellar regulon. Their expression is modulated by various environmental factors. For L. pneumophila it was shown that the expression of genes of the flagellar regulon is modulated by the actual growth phase and temperature. Especially, flagellated Legionella are known to express genes during the transmissive phase of growth that are involved in the expression of virulence traits. It has been demonstrated that the alternative sigma-28 factor is part of the link between virulence expression and motility. In the following review, the structure of the flagellar regulon of L. pneumophila is discussed and compared to other flagellar systems of different Legionella species. Recently, it has been described that Legionella micdadei and Legionella fallonii contain a second putative partial flagellar system. Hence, the report will focus on flagellated and non-flagellated Legionella strains, phylogenetic relationships, the role and function of the alternative sigma factor (FliA) and its anti-sigma-28 factor (FlgM).
Introduction and Overview
The Legionellaceae family consists of a single genus: Legionella that comprises more than 60 species so far (Gomez-Valero et al., 2009; Bajrai et al., 2016; Khodr et al., 2016). New species are identified continuously (i.e., Legionella drancourtii, Legionella gresilensis, and Legionella beliardensis), extending the list of known Legionella species (Lo Presti et al., 2001; La Scola et al., 2004; Gomez-Valero et al., 2009; Rizzardi et al., 2015; Bajrai et al., 2016; Khodr et al., 2016). More than 20 pathogenic Legionella species are known today that differ in their ability to infect hosts and to cause severe to mild diseases in humans (Rizzardi et al., 2015). Human pathogens known to cause the Legionnaires' disease—an atypical pneumonia—are for instance Legionella pneumophila, Legionella micdadei, and Legionella longbeachae (Yu et al., 2002; Whiley and Bentham, 2011). Legionella known to cause the Pontiac fever—a mild flu-like disease—are for instance Legionella feelei, L. micdadei and Legionella anisa, but also L. pneumophila. Often Legionella strains of the same species and same serogroup cause one of the mentioned diseases (Swanson and Hammer, 2000; Fields et al., 2002; Wang et al., 2015).
Nevertheless, it was assumed that humans are accidental hosts of Legionella species within which the bacterium replicates (Horwitz and Silverstein, 1980; Cianciotto et al., 1989; Horwitz, 1992; Fields, 1996; Neumeister et al., 1997; Newton et al., 2010). Known natural hosts are protozoa, especially free-living amoebae: Acanthamoeba spp., Naegleria spp., or Hartmanella vermiformis (Barbaree et al., 1986; Rowbotham, 1986; Fields, 1996; Atlas, 1999; Fields et al., 2002; Greub and Raoult, 2004; Abdel-Nour et al., 2013; Richards et al., 2013; Cateau et al., 2014). Accordingly, in general, Legionella species are prevalent inhabitants of soil, mud and above all of aquatic environments (Fliermans et al., 1981; Fields, 1996; Atlas, 1999; Gomez-Valero et al., 2009; Declerck, 2010; Schalk et al., 2014; Currie and Beattie, 2015). The ability of L. pneumophila to grow within biofilms made by Klebsiella pneumoniae or Pseudomonas aeruginosa in aquatic or wet environments raised questions about their host-free persistence (Stewart et al., 2012). In connection with favorable aquatic habitats and potential protozoa hosts, especially flagella-driven motility of some Legionella spp. is an important feature needed to move around, to find new hosts and to form maybe even biofilms (Kirov et al., 2004; Danhorn and Fuqua, 2007; Heuner and Albert-Weissenberger, 2008). For Legionella infecting humans, motility may also be crucial for spreading within lungs of patients, as flagellated forms of L. pneumophila were detected in alveolar spaces (Chandler et al., 1980; Jager et al., 2014). Recently, it was published that L. feelei strains that cause Legionnaires' disease are flagellated while L. feelei strains that cause the Pontiac fever are non-flagellated (Wang et al., 2015). The majority of Legionella species are flagellated (Elliott and Johnson, 1981; Bornstein et al., 1991; Bangsborg et al., 1995; Heuner et al., 1995), but not all pathogenic Legionella have a complete flagellar regulon (i.e., L. longbeachae and Legionella oakridgensis, see below) (Orrison et al., 1983; Heuner et al., 1995; Cazalet et al., 2010; Kozak et al., 2010; Brzuszkiewicz et al., 2013).
The assumption that the expression of flagella and virulence are linked was already made at an early stage (Rowbotham, 1986) and later on confirmed. It was shown that there is a regulatory link between the expression of a virulent phenotype and the flagellum (Pruckler et al., 1995; Byrne and Swanson, 1998; Hammer et al., 2002; Gal-Mor and Segal, 2003; Molofsky et al., 2005; Heuner and Albert-Weissenberger, 2008; Albert-Weissenberger et al., 2010; Schulz et al., 2012). The expression of flagellar genes is regulated on the flagellar regulon, extensively investigated in L. pneumophila due to its biphasic intracellular life cycle during which the bacterium undergoes a shape change. Within the host, after replication inside of Legionella-containing vacuoles (LCVs), when nutrients become limited, L. pneumophila differentiates into a flagellated, non-replicating form. The flagellated, transmissible, mature form is stress-resistant, virulent and metabolically resting as well as infectious (abbr. MIF) (Rowbotham, 1986; Byrne and Swanson, 1998; Heuner et al., 1999; Swanson and Hammer, 2000; Faulkner and Garduno, 2002; Garduno et al., 2002; Hammer et al., 2002; Molofsky and Swanson, 2004; Fonseca and Swanson, 2014; Eisenreich and Heuner, 2016). The actual release process of mature forms is still under discussion: either the bacteria are released from the LCV into the environment by lysis of the host or the bacteria are released first into the cytosol of the host and then after an additional putative round of replication into the environment (Rowbotham, 1986; Molmeret et al., 2004). The latter hypothesis implies that the flagellum is produced inside the cytosol of the host and not in LCVs as proposed earlier. Furthermore, there is also a possibility that the bacteria are released by the host via a non-lytic mechanism (Chen et al., 2004; Bouyer et al., 2007; Berk et al., 2008). However, the released form is well-prepared to reinfect new hosts or to differentiate into a viable-but-nonculturable form (VBNC) meant to enable a long-term survival of the bacteria (Rowbotham, 1986; Steinert et al., 1997; Ohno et al., 2003; Molmeret et al., 2010; Al-Bana et al., 2014). VBNC forms can be resuscitated when they are taken up by amoebae (Steinert et al., 1997; Ohno et al., 2003; Al-Bana et al., 2014). Further, different morphological forms of L. pneumophila have been recently discussed (Robertson et al., 2014). Next to the biphasic intracellular life cycle it was shown that L. pneumophila exhibits also a life stage-specific bipartite metabolism, an area for further investigations (Schunder et al., 2014; Eisenreich and Heuner, 2016; Gillmaier et al., 2016; Hauslein et al., 2016).
Motility of Legionella
Different forms of bacterial motility are known including swarming, twitching and sliding. Notably the flagellum—next to pili—allows bacteria to move. Bacterial motility is often related to chemotactic behavior that enables a bacterium to locate special environmental conditions and to get closer to higher concentrations of attractants (Szurmant and Ordal, 2004; Hazelbauer et al., 2008; Micali and Endres, 2016). Some Legionella have a chemotaxis system (L. longbeachae, Legionella parisiensis, and Legionella bozemanii) but most Legionella do not have the corresponding genes (e.g., L. pneumophila, L. micdadei, and L. oakridgensis). Moreover, the ability of Legionella to swarm and to show a chemotaxis behavior has not yet been reported.
Twitching motility is based on a functional type IV pilus. The ability to move forward by twitching has been reported for L. pneumophila (Coil and Anne, 2009; Hoppe et al., 2017). In addition, sliding motility, a surfactant-mediated motility, has been described for L. pneumophila (Stewart et al., 2009).
The Flagellum and the Flagellar Regulon
The Structure of the Flagellum and Flagellar Systems
Most Legionella species are motile due to a single polar flagellum (Figure 1) (Chandler et al., 1980; Elliott and Johnson, 1982; Heuner et al., 1995). More than 50 genes are involved in the expression of functional flagella, and due to high metabolic costs, a tight regulation is essential (Chilcott and Hughes, 2000; McCarter, 2006; Osterman et al., 2015). The flagellum of Legionella consists of a basal body, a hook structure and a filament (Figure 2; Heuner and Steinert, 2003; Heuner and Albert-Weissenberger, 2008). For the assembly of the flagellum, needed proteins (hook, rod and the filament forming proteins) are exported out of the cell by a flagellum-specific export apparatus, a type III-like secretion system (T3SS) (Heuner and Albert-Weissenberger, 2008; Altegoer and Bange, 2015).
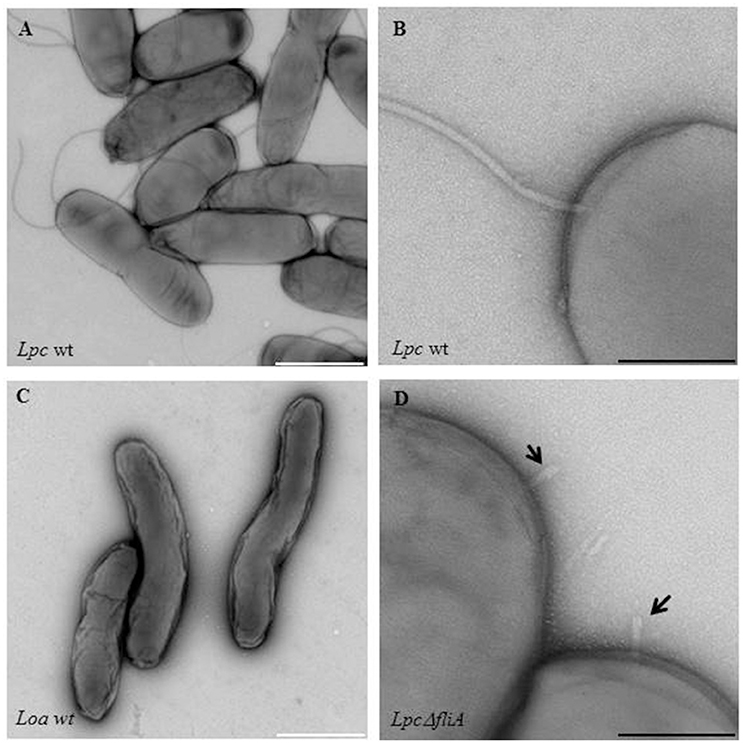
Figure 1. Electron microscopic images of flagellated and non-flagellated Legionella wild-type strains as a flagella mutant strain. Shown are (A,B) Legionella pneumophila Corby (Lpc wt) and (C) Legionella oakridgensis (Loa wt) wild-type strains that are either representative of flagellated or non-flagellated Legionella. An electron microscopic image of a fliA L. pneumophila mutant strain (Lpc ΔfliA), is also shown (D). The straight hook structure of the mutant strain is indicated by black arrows (D). Bacteria were grown in AYE medium at 30°C and the samples were stained with 0.5% uranyl acetate. White scale bars: 1 μm; black scale bars: 200 nm.
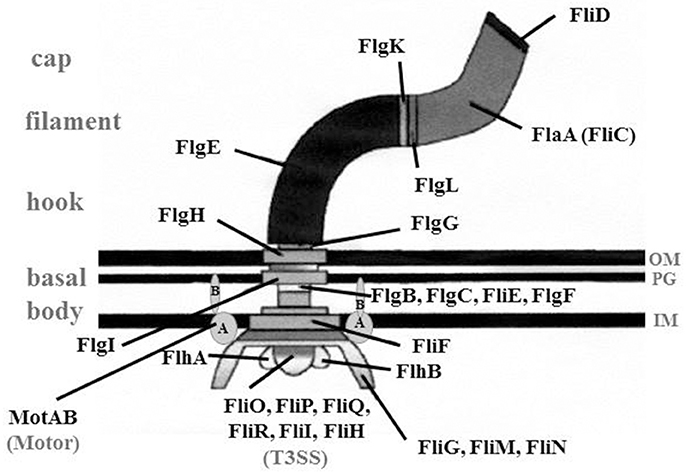
Figure 2. Overview of the structure of a flagellum. Schematic drawing of a flagellum consisting of basal body, hook structure and filament. Cap, FliD protein that is essential for the assembly of the filament, Motor, MotAB proteins that provide energy for the rotation; T3SS, type III-like secretion system; FlgD (not shown), the hook cap protein needed to incorporate FlgE; IM, inner membrane; OM, outer membrane; PG, peptidoglycan layer; (Heuner and Albert-Weissenberger (2008), modified).
The basal body consists of a rod, three rings [“membrane/supramembrane” (MS), “peptidoglycan” (P), and “lipopolysaccharide” (L)] and a motor switch complex (MotAB). The MotAB is providing the energy for the rotation of the flagellum (Minamino and Imada, 2015). For the formation of the slightly curved hook structure, FlgE and FlgD are essential. The hook cap protein FlgD assists when FlgE is incorporated into the hook structure (Altegoer and Bange, 2015).
Interestingly, an uncommonly straight hook has been reported for L. pneumophila mutant strains (ΔfliA, ΔfliD, and ΔflaA, i.e., a flagellin mutant) (Figure 1) (Schulz et al., 2012). The findings give credit to the assumption that L. pneumophila might have a straight hook that is hard to detect in wildtype strains. The filament consists mainly out of a single protein, the flagellin (FlaA or FliC) (Figure 2). The cap protein FliD is essential for the assembly of flagellin subunits into the filament. To assemble the filament, flagellin is exported through the filament structure by a flagellum-specific export apparatus (T3SS) and assembled at the tip of the filament (Heuner and Albert-Weissenberger, 2008; Altegoer and Bange, 2015).
More details about the flagellum structure can be found in dedicated review articles (Aldridge and Hughes, 2002; Heuner and Steinert, 2003; Macnab, 2003; Pallen et al., 2005; Heuner and Albert-Weissenberger, 2008; Altegoer and Bange, 2015).
Next to the regular flagellar system, a second putative flagellar system was suspected for two Legionella species: Legionella fallonii and L. micdadei (Gomez-Valero et al., 2014). Comparative genome analysis led to the suspicion that the strains do have homologs to flagellar genes of L. pneumophila. The identified genetic region is comprised of genes that encode a putative basal body, a secretion system, as well as a putative hook structure. No homologs to flaA or fliD were found in the predicted genomic region. Yet, further investigations are needed to find out if a T3SS or a putative second flagellum is encoded. Additionally, in silico investigation, performed on the draft genome sequence of Legionella israelensis, identified a similar operon. A BLAST search using the operon (10,041 bp, ctg_064, L. israeliensis draft-genome; Burstein et al., 2016) as query identified similar genes in L. drancourtii, L. fallonii, L. worsleiensis, L. quateirensis, L. birminghamensis, and L. drozanskii (Heuner, unpublished results). Initial findings show that the operon is present in two out of three Legionella clades (Figure 3), leaving a margin for additional studies.
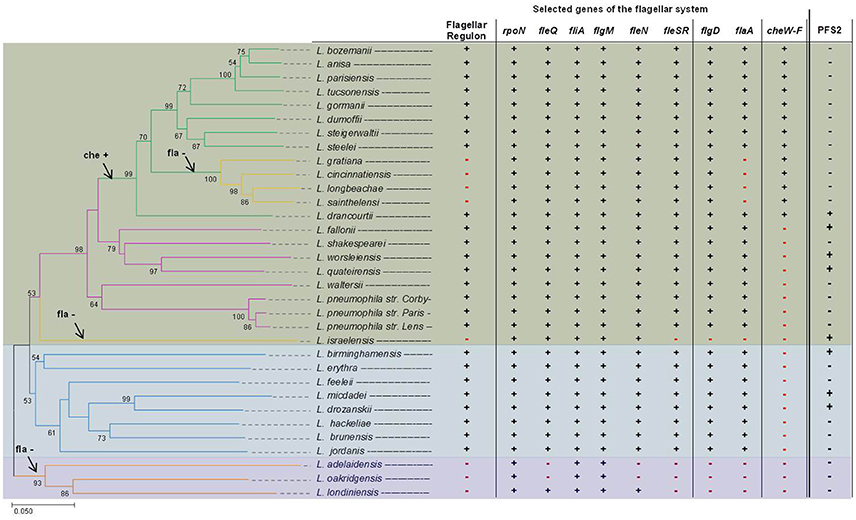
Figure 3. Evolutionary relationships of Legionella species and distribution of flagellar regulon and chemotaxis genes. Shown is a phylogenetic tree of Legionella spp. based on the fliA gene (left side) and also the distribution of flagellar regulon and chemotaxis genes (right side). The phylogenetic tree was reconstructed on the basis of the fliA gene using the Neighbor-Joining method (Saitou and Nei, 1987) and a bootstrap of 1,000 (Felsenstein, 1985). The bootstrap >50 is reported next to the branches. The phylogenetic distances were computed using the Tamura-Kumar method (Tamura and Kumar, 2002) and refers to the units of the number of base substitutions per site. For the evolutional analysis, performed in MEGA7 (Kumar et al., 2016), position containing gaps and/or missing data were eliminated. To determine whether selected genes belonging to the flagellar regulon and chemotaxis genes are present among investigated Legionella, an in silico BLAST search was performed using Legionella draft genomes recently published by Burstein et al. (2016). The complete structure of genes of the flagellar regulon of L. pneumophila strain Corby, L. longbeachae, L. israeliensis, and L. oakridgensis are given in Figure 5. Data for L. pneumophila strains were taken from Cazalet et al. (2010), Glockner et al. (2008), and Albert-Weissenberger et al. (2010). Legionella clade I is highlighted in green, clade II in blue and clade III in purple. On the left, in the phylogenetic tree, possible time points when the flagellin gene (flaA) and the chemotaxis (che) genes were lost (–) or acquired (+) are indicated with arrows. On the right, the presence (+) and/or absence (–) of selected genes belonging to the flagellar regulon, the chemotaxis operon or an operon encoding a putative second flagellum or a putative T3SS (PFS2, based on unpublished data) are indicated, respectively.
The Flagellar Regulon
The expression of flagellar genes of L. pneumophila is regulated in a hierarchical cascade (Figure 4) (Heuner et al., 1995, 2006; Heuner and Steinert, 2003; Jacobi et al., 2004; Albert-Weissenberger et al., 2010; Schulz et al., 2012). Their expression depends on growth phase, temperature, medium viscosity and nutrient availability (e.g., amino acids and fatty acids) (Ott et al., 1991; Byrne and Swanson, 1998; Heuner et al., 1999, 2006; Heuner and Albert-Weissenberger, 2008).
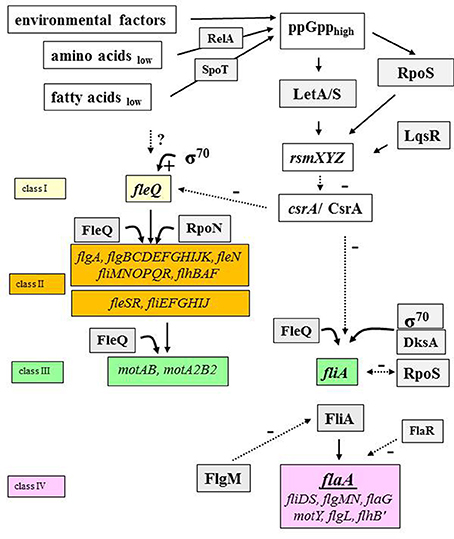
Figure 4. The flagellar regulon cascade. The regulatory cascade which leads to the expression of proteins that are needed to construct a functional flagellum is shown (Heuner and Albert-Weissenberger, 2008; Sahr et al., 2017, modified). Specific environmental factors lead to the activation of the pathway through alarmone (ppGpp) accumulation. Some main players of the flagellar regulon cascade are: the activator protein FleQ, the alternative sigma factor 28 (FliA), the two-component system (LetA/S) and the carbon storage regulator (CsrA) as examples. FleQ is the master regulator of the flagellar genes (Class II and class III). FliA is the regulator of flagellar genes of class IV. Black lined, continuous arrows refer to positive regulation events. The broken lines in black refer to negative regulatory effects of respective factors. Question marks (?) indicate pathway points with putative mode of actions that need to be specified by further investigations. RsmXYZ, regulatory RNAs; FlaA, flagellin; FlaR, transcriptional regulator (LysR family); FleQ and FleR, sigma factor 54 activator protein; FlgM, anti-sigma 28 factor; RpoS, alternative sigma 38 factor.
In short, intracellular alarmone accumulation—ppGpp, a signal molecule, produced when environmental conditions are unfavorable (e.g., limited nutrient supply)—is regulated by RelA and SpoT. RelA senses the amount of available intracellular amino acids and SpoT senses the amount of intracellular fatty acids (Hammer and Swanson, 1999; Dalebroux et al., 2009, 2010).
Alarmone accumulation triggers the activation of an alternative stationary-phase sigma factor (RpoS) and of the two component system LetA/S (Byrne and Swanson, 1998; Hammer et al., 2002; Zusman et al., 2002; Molofsky and Swanson, 2004; Dalebroux et al., 2009, 2010; Edwards et al., 2009; Rasis and Segal, 2009; Sahr et al., 2009) (Figure 4). RpoS and LetA/S promote the transcription of small regulatory RNAs (RsmX, RsmY, RsmZ). RsmX plays a role in the virulence of L. pneumophila (Sahr et al., 2012). The transcription of rsmZ/rmsY is also influenced by a quorum sensing system regulator, LqsR (Tiaden et al., 2007; Schell et al., 2016). The two regulatory RNAs are able to bind a number of carbon storage regulator molecules (CsrA) at once (Sahr et al., 2017). CsrA is a negative regulator and through the binding on RsmY or RsmZ, other targets of the regulatory RNAs can be expressed. The expression of transmissive traits starts and main activator proteins (e.g., FleQ) are produced and flagellar genes are expressed (Zusman et al., 2002; Molofsky and Swanson, 2003; Rasis and Segal, 2009; Sahr et al., 2009, 2017; Albert-Weissenberger et al., 2010). Notably, some parts of the function of the negative regulator CsrA (flaA expression and motility of L. pneumophila) can be “complemented” by ectopically expressed csrT, a CsrA-like regulatory gene associated with integrative conjugative elements (Abbott et al., 2015).
CrsA is also controlling a major regulator involved in the expression of flagella, FleQ (Sahr et al., 2017). FleQ is responsible for the expression of early flagellar genes belonging to class II and III genes in an RpoN-dependent and RpoN-independent pathway (Jacobi et al., 2004; Albert-Weissenberger et al., 2010; Schulz et al., 2012). RpoN is an enhancer-binding protein encoding an alternative sigma factor that initiates transcription when activator proteins like FleQ, FleR, and PilR (Jacobi et al., 2004) are present. When class II and III genes are expressed, the activity of FliA—the alternative sigma factor—leads to the expression of class IV genes and the assembly of the flagellum (Figure 4). RpoN and FleR seem to be responsible for a negative feedback loop on flagellar genes (Albert-Weissenberger et al., 2010). It was found that RpoS and FlaR (transcriptional regulator FlaR, LysR family member) are also involved in the expression of the flagellin gene (Heuner et al., 2000; Bachman and Swanson, 2001, 2004; Rasis and Segal, 2009; Sahr et al., 2009). The production of FlaA is also regulated by cyclic di-GMP, shown by the analysis of a gene (cdgS13) coding for a protein with diguanylate cyclase activity (Levi et al., 2011). The influence of cyclic di-GMP on flagellum-based motility has been shown for other bacteria than Legionella species (Wolfe and Visick, 2008).
The Flagellum and Virulence
Already early on, it has been hypothesized that virulence and flagellum expression are genetically linked with each other. It has been shown that motile, transmissive Legionella were more infectious for amoebae than non-motile replicative phase Legionella (Rowbotham, 1986; Pruckler et al., 1995; Bosshardt et al., 1997; Byrne and Swanson, 1998; Hammer et al., 2002; Heuner et al., 2002; Molofsky et al., 2005; Heuner and Albert-Weissenberger, 2008). Different experiments could show that the motility but not the flagellin promotes the contact with host cells. Motility increases the infectivity and the fitness. Furthermore, it turned out that the flagellum is not necessary for intracellular replication (Pruckler et al., 1995; Dietrich et al., 2001; Polesky et al., 2001; Heuner et al., 2002; Jacobi et al., 2004; Molofsky et al., 2005; Schulz et al., 2012).
The four major regulators of the flagellar regulon (RpoN, FleQ, FleSR, FliA) seems to be involved in the invasion process of L. pneumophila into hosts. These findings point out the proposed link between virulence traits and flagellum expression (Dietrich et al., 2001; Hammer et al., 2002; Molofsky et al., 2005; Heuner and Albert-Weissenberger, 2008; Albert-Weissenberger et al., 2010; Schulz et al., 2012). Especially the FliA regulon plays an important role (please, see the section: FliA and its implication in virulence below). FliA, but not the flagellin (flaA), is involved into the ability of L. pneumophila to form biofilm that allow bacteria to survive whenever environmental conditions are not favorable. (Mampel et al., 2006). Notably unwanted biofilms are a health issue causing a significant amount of nosocomial infections (Bryers, 2008). Legionella are known to survive within biofilm of other bacteria (e.g., Klebsiella pneumophila and Pseudomonas aeruginosa) (Molofsky et al., 2005; Stewart et al., 2012). More recently, findings about L. pneumophila's ability to form biofilms by itself in natural environments and on medical devices have attracted attention (Lau and Ashbolt, 2009; Abu Khweek et al., 2013). Biofilm formation is regulated by temperature, surface material and intracellular growth (Konishi et al., 2006; Piao et al., 2006; Bigot et al., 2013) and biofilm-derived L. pneumophila do not express flagellin (Abu Khweek et al., 2013). More information about biofilms and L. pneumophila can be found in a recent review (Abdel-Nour et al., 2013).
The flagellum also affects the resistance of hosts to Legionnaires' disease and when Legionella do not produce flagellin they can evade the innate immune response in macrophages (Hawn et al., 2003; Molofsky et al., 2006; Ren et al., 2006; Abu Khweek et al., 2013). Resistance is mediated by the Naip5/Ipaf-dependent recognition of flagellin, which induces a protective immunity in non-A/J mouse models (Ricci et al., 2005). Detailed information about addressed points can be found in dedicated reviews (Fontana and Vance, 2011; Schell et al., 2016; Mascarenhas and Zamboni, 2017).
The Alternative Sigma Factor 28
FliA Expression
One of the major regulators involved in the expression of the flagellum is FliA and an increased alarmone level leads to accumulation of functional FliA (Bruggemann et al., 2006; Heuner et al., 2006; Dalebroux et al., 2010). The alternative sigma factor (σ28) is directly involved in the regulation and expression of the flagellin gene (flaA) and others (Figure 4). A fliA mutant of L. pneumophila does not produce flagellin and is consequently non-flagellated. Moreover, a ΔfliA mutant of Escherichia coli can be completed with a fliA gene of L. pneumophila (Heuner et al., 1995, 1997, 2002; Bruggemann et al., 2006; Albert-Weissenberger et al., 2010; Schulz et al., 2012).
The expression of flagellar class III and IV genes is induced in a FleQ-dependent manner. The FliA-regulated class IV genes are involved in the assembly of the filament and flagella motility (flgL, fliD, flaA, motY). Both lead to the complete synthesis of the flagellum (Jacobi et al., 2004; Albert-Weissenberger et al., 2010). The fliA gene itself is expressed in a FleQ-dependent but RpoN-independent manner (Albert-Weissenberger et al., 2010). Nevertheless, FleQ and RpoN are not necessary for a basal expression of fliA. For a basal expression, fliA is transcribed from a putative sigma-70 promoter element and later, during the exponential phase, the expression of fliA is induced in a FleQ-dependent manner (Schulz et al., 2012). Accordingly, it was hypothesized that during the exponential phase the basal fliA promotor activity may be mediated by DksA independent of the ppGpp concentration, whereas during the post-exponential phase DksA cooperates with ppGpp to activate fliA (Dalebroux et al., 2010). The identification of the transcription start point of fliA corroborates the presence of a putative DksA binding site, an A/T rich discriminator site (Schulz et al., 2012).
FliA and Its Implication in Virulence
FliA is a regulator that is also involved in the expression of putative virulence genes (Bruggemann et al., 2006; Albert-Weissenberger et al., 2010; Tlapak et al., 2017).
Several investigations performed on a fliA mutant strain of L. pneumophila pointed out that the mutant (at low MOI) is not replicating in host cells anymore (Dictyostelium discoideum). The fliA mutant seems to be less infectious for macrophages and non-cytotoxic to bone marrow-derived macrophages. Moreover, the mutant has a reduced fitness potential in amoebae (Dietrich et al., 2001; Hammer et al., 2002; Heuner et al., 2002; Jacobi et al., 2004; Molofsky et al., 2005; Heuner and Albert-Weissenberger, 2008; Schulz et al., 2012). Likewise, a fliA mutant strain of L. oakridgensis showed a reduced fitness in its host (Acanthamoeba lenticulata) (Tlapak et al., 2017). In addition, another L. pneumophila fliA mutant strain exhibited a reduced ability to form biofilms (Mampel et al., 2006).
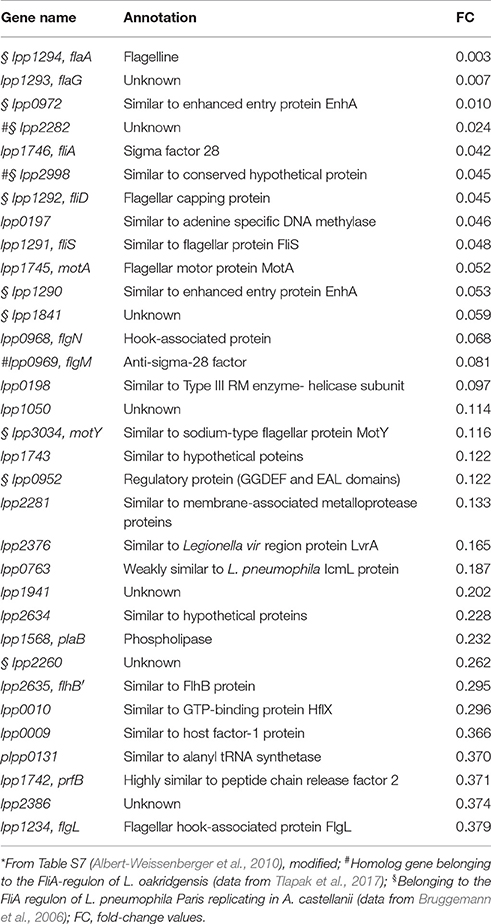
FliA is obviously a virulence factor, and target genes of fliA were investigated to understand its implication for virulence (Bruggemann et al., 2006; Albert-Weissenberger et al., 2010; Tlapak et al., 2017). Target genes of fliA in L. pneumophila strains include genes of the flagellar regulon (e.g., flaA and flgM) and others (e.g., enhA and lvrA), illustrated in Figure 4 and listed in Table 1 (Bruggemann et al., 2006; Albert-Weissenberger et al., 2010; Schulz et al., 2012). Other identified FliA-dependent genes encode for putative virulence factors corroborating the involvement of FliA in the establishment of Legionella infections. Identified putative virulence factors are: lpp0952, lpp1290, and lpp0972. The first one, lpp0952, is coding for a GGDEF/EAL and PAS/PAC domain protein (Bruggemann et al., 2006; Albert-Weissenberger et al., 2010). The two remaining genes are homologs of the enhanced entry proteins EnhA. Respective homologs were also found in L. longbeachae which is non-flagellated, putatively associated with the flagellar system (Kozak et al., 2010).
Recently, especially L. oakridgensis simplified the identification of fliA targets potentially involved in virulence (Tlapak et al., 2017). L. oakridgensis strains are non-flagellated, the entire flagellar regulon is missing and only homologs of FliA and FlgM are present (Figure 5). L. oakridgensis is less infectious then L. pneumophila, but still causes Legionnaires' disease. In addition, L. oakridgensis replicates in guinea pigs, in human cell lines, in Acanthamoeba lenticulata and for growth in media no additional cysteine is needed. (Orrison et al., 1983; Fields et al., 1986; O'Connell et al., 1996; Neumeister et al., 1997; Lo Presti et al., 2001; Brzuszkiewicz et al., 2013). Nevertheless, L. oakridgensis exhibits a functional T4SS, homologs of known virulence factors, as well as newly identified virulence factors (Brzuszkiewicz et al., 2013). L. oakridgensis is used to investigate FliA since a fliA L. oakridgensis knockout will not cause the inactivation of the entire flagellar regulon and target genes of FliA can still be identified as well as genes involved in the expression of virulence traits. However, mutant strain analyses aimed at identifying target genes of FliA in L. oakridgensis yielded no results for putative FliA-dependent virulence genes yet (Table 1; Tlapak et al., 2017). Additional investigations are needed including phenotypic characterizations and deletion analyses of further target genes of FliA.

Figure 5. Structure of the operons of the flagellar regulon of L. pneumophila Corby (Lpc), L. longbeachae (Llo), L. israeliensis (Lis), and L. oakridgensis (Loa). Genes are indicated by arrows and gene names are given above. Flagellar and chemotaxis genes are indicated in red and blue, respectively; other genes in black. Genes not present in a strain are indicated by thin arrows (Brzuszkiewicz et al., 2013, modified).
FliA-FlgM Interaction in Legionella oakridgensis
Flagellated bacteria regulate the FliA activity often post-transcriptionally. For Salmonella, Escherichia, and Vibrio species it is known that an anti-sigma-28 factor (FlgM) binds FliA, preventing the binding of FliA to FliA-dependent promoter sites, and FliA-dependent genes are consequently not translated. After assembling of the hook-basal body structure, FlgM is exported and FliA is not repressed anymore (Gillen and Hughes, 1991; Ohnishi et al., 1992; Chilcott and Hughes, 2000; Aldridge et al., 2006). In Helicobacter pylori, the FlgM protein is inactivated instead of being exported out of the cells (Rust et al., 2009). FliA-FlgM interactions in Legionella are still unknown. Recent findings suggest that FliA-FlgM interaction might be different than in other flagellated bacteria, at least for L. oakridgensis. Respective species do not have a flagellum (Figure 1C), a flagellar regulon (Figure 5) and a basal body although flgM and fliA homologs are present which encode for functional FlgM and FliA proteins (Brzuszkiewicz et al., 2013; Tlapak et al., 2017); consequently, the mechanism controlling FliA-FlgM interactions must be different.
For L. pneumophila as well as for L. oakridgensis it was found that the expression of FlgM or homologs is sigma-28-dependent (Albert-Weissenberger et al., 2010; Tlapak et al., 2017). Although L. oakridgensis has no flagellar system, the expression of fliA-dependent genes is growth phase- and temperature-dependent (Heuner et al., 1999; Tlapak et al., 2017). Moreover, for L. oakridgensis it was demonstrated that FlgM is a negative regulator of FliA-dependent genes and the protein itself seems to be degraded in a growth phase- and temperature-dependent manner (Tlapak et al., 2017). Thus, it seems likely that, as described for H. pylori, FlgM in L. oakridgensis is degraded by protease activity instead of being secreted. However, investigations are needed to show if FlgM in L. pneumophila is effectively secreted in a basal body-dependent manner.
Distribution of the Flagellar System Among Legionella Species
Phylogenetic reconstruction—based on concatenated amino acid alignment of 78 orthologous ORFs—divided the Legionella species into three major clades (clade I and clade II, clade III) (Burstein et al., 2016). Clade I is comprised of most Legionella species including L. pneumophila, L. parisiensis, L. bozemanii, and L. longbeachae (Burstein et al., 2016). Clade II comprises among others L. feelei and L. micdadei and clade III, a deep-branching clade, includes three members: Legionella adelaidensis, L. oakridgensis and Legionella londiniensis (Burstein et al., 2016). Phylogenetic reconstructions performed herein, yielded similae results that are given in Figure 3. As opposed to former investigations, the phylogenetic tree was reconstructed on the basis of the fliA gene.
The flagellar system can be found in Legionella species classified as clade I or II (Cazalet et al., 2004, 2010; Chen et al., 2004; Bruggemann et al., 2006; Glockner et al., 2008; Kozak et al., 2010; Brzuszkiewicz et al., 2013; Gomez-Valero et al., 2014; Burstein et al., 2016), but not in clade III Legionella. Clade III Legionella do not have a functional flagellar system and do not have most of the flagellar regulon genes, except for fliA and its anti-sigma factor flgM (L. oakridgensis, L. adelaidensis, and L. londiniensis) (Cazalet et al., 2010; Brzuszkiewicz et al., 2013; Tlapak et al., 2017) and two additional genes: fleQ and fleN (L. londiniensis) (Figures 3, 5). Also, some clade I Legionella species do not have a functional flagellar system (clade I: L. longbeachae, Figure 5, Legionella gratiana, Legionella cincinnatiensis, Legionella sainthelensi, and L. israelensis). It has been hypothesized that the loss of flagellar genes has not happened recently (Kozak et al., 2010). This is corroborated by the finding that L. longbeachae and all subclade members are negative for the flagellar regulon but positive for genes coding for the sigma factor FliA, the regulator FleN and the two component system comprising of FleR and FleS, as well as FlgD (Figure 3) (Cazalet et al., 2010; Kozak et al., 2010). Also L. israelensis is negative for flaA (Heuner et al., 1995) and most flagellar regulon genes, except: fleQ, fliA, fleN, and flgM (Figure 5). The finding allows to assume that the flagellar system may have been lost at different time points during the evolution of Legionella species (Figure 3). In addition, some genes that have regulatory functions outside of the flagellar system are still present (Albert-Weissenberger et al., 2010; Cazalet et al., 2010; Kozak et al., 2010; Tlapak et al., 2017). The same applies to flgD which is involved in the assembly of the hook structure of the flagellum with unassigned hypothetical alternative functions.
The investigated Legionella genomes were also screened for the presence/absence of genes of major regulators of the flagellar system as well as of the chemotaxis operon (Figure 3). It was found that the genes of the chemotaxis operon are only found in a subclade of the clade I Legionella. L. longbeachae is the first Legionella species described to exhibit chemotaxis genes (Cazalet et al., 2010; Kozak et al., 2010) that do not have flagellar genes. It seemed paradoxical that L. pneumophila is flagella positive but chemotaxis negative and L. longbeachae is flagella negative but chemotaxis positive. The distribution of the chemotaxis operon may indicate that the chemotaxis operon was acquired by a common 'ancestor' of this sub-tree clade (Figure 3).
Conclusion
The review aimed to summarize knowledge gained about flagella and the flagellar regulon of different Legionella species. The majority of Legionella species exhibit genes encoding for a functional flagellum and they are flagellated (Elliott and Johnson, 1981; Bornstein et al., 1991; Bangsborg et al., 1995; Heuner et al., 1995). Motility increases infectivity and fitness, helping the bacteria to reach new hosts after successful replication within protozoan host cells and release into aquatic environments.
Some Legionella—including some pathogenic species (e.g., L. longbeachae, and L. oakridgensis)—are not flagellated and most flagellar regulon genes are absent (Orrison et al., 1983; Heuner et al., 1995; Cazalet et al., 2010; Kozak et al., 2010; Brzuszkiewicz et al., 2013). With the advance in molecular techniques and the ability to produce and to process metagenomics datasets, it was found that some of the non-flagellated Legionella have still parts of the flagellar regulon, mainly genes with regulator functions.
However, in Legionella flagellum synthesis is associated with the expression of a virulence phenotype; and motility can be seen as a virulence and a fitness factor in Legionella and other bacteria. The alternative sigma factor FliA is also involved in the expression of virulence traits. FliA-dependent putative virulence genes were already identified by initial investigations that need to be extended. Also, additional investigations are needed to determine the role of FliA and molecular mechanisms of FliA-FlgM interactions in Legionellae. FlgM and FliA, two main players involved in the expression of the flagellum genes, are still present in non-flagellated Legionella, a promising take-off for future investigations. Nevertheless, the flagellum is not necessarily needed for an intracellular replication within host cells. Moreover, in some hosts the Naip5/Ipaf-dependent recognition of flagellin can cause an innate immune response leading to resistance against Legionella infections (Molofsky et al., 2006; Ren et al., 2006). For example, it was reported that biofilm-derived L. pneumophila without flagellin expression evade the innate immune response in macrophages (Abu Khweek et al., 2013), as it was suggested for the non-flagellated L. longbeachae (Cazalet et al., 2010; Kozak et al., 2010). It seems that under certain conditions, the loss of the flagellum may increase the fitness of bacteria. For instance, L. pneumophila which can be found mainly in aquatic environments, is still flagellated whereas L. longbeachae which can be found predominantly in soil, is non-flagellated (Kozak et al., 2010). Nevertheless, if the loss of the flagellar system from Legionella species depends on the habitat or environmental conditions remains unanswered.
As outlined, flagellated and non-flagellated Legionella are positive for genes belonging to the chemotaxis operon. The ability of Legionella to swarm and to show off a chemotaxis behavior has not yet been reported. Interestingly, some chemotaxis-positive and flagellar operon-negative Legionella (e.g., L. longbeachae) give credit to the assumption that chemotaxis genes may not be involved in flagellum-mediated motility. Recent investigations could even show that chemotaxis sensory systems—different from those found in E. coli—in distinct bacteria (e.g., Myxococcus spp., Geobacter spp.) are not necessarily involved in bacterial flagellum-mediated motility (Kirby, 2009; Kozak et al., 2010). Chemotaxis-like systems seem to be involved among other things in type IV pilus-based motility and cell to cell interaction and/or social motility (Kirby, 2009; Kozak et al., 2010). Accordingly, additional experimentations are needed to investigate the role of the chemotaxis operon in flagellated and non-flagellated, chemotaxis-positive Legionella.
Author Contributions
SA and KH contributed substantial to the conception and design of the work. SA and KH wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Roland Grunow for assistance with different aspects of the presented work and for copy-editing. This work received financial support by grant HE2854/5-1+2/9-1 from the Deutsche Forschungsgemeinschaft DFG (Bonn, Germany) to KH.
References
Abbott, Z. D., Flynn, K. J., Byrne, B. G., Mukherjee, S., Kearns, D. B., and Swanson, M. S. (2015). csrT represents a new class of csrA-like regulatory genes associated with integrative conjugative elements of Legionella pneumophila. J. Bacteriol. 198, 553–564. doi: 10.1128/JB.00732-15
Abdel-Nour, M., Duncan, C., Low, D. E., and Guyard, C. (2013). Biofilms: the stronghold of Legionella pneumophila. Int. J. Mol. Sci. 14, 21660–21675. doi: 10.3390/ijms141121660
Abu Khweek, A., Fernandez Davila, N. S., Caution, K., Akhter, A., Abdulrahman, B. A., Tazi, M., et al. (2013). Biofilm-derived Legionella pneumophila evades the innate immune response in macrophages. Front. Cell. Infect. Microbiol. 3:18. doi: 10.3389/fcimb.2013.00018
Al-Bana, B. H., Haddad, M. T., and Garduno, R. A. (2014). Stationary phase and mature infectious forms of Legionella pneumophila produce distinct viable but non-culturable cells. Environ. Microbiol. 16, 382–395. doi: 10.1111/1462-2920.12219
Albert-Weissenberger, C., Sahr, T., Sismeiro, O., Hacker, J., Heuner, K., and Buchrieser, C. (2010). Control of flagellar gene regulation in Legionella pneumophila and its relation to growth phase. J. Bacteriol. 192, 446–455. doi: 10.1128/JB.00610-09
Aldridge, P. D., Karlinsey, J. E., Aldridge, C., Birchall, C., Thompson, D., Yagasaki, J., et al. (2006). The flagellar-specific transcription factor, sigma28, is the Type III secretion chaperone for the flagellar-specific anti-sigma28 factor FlgM. Genes Dev. 20, 2315–2326. doi: 10.1101/gad.380406
Aldridge, P., and Hughes, K. T. (2002). Regulation of flagellar assembly. Curr. Opin. Microbiol. 5, 160–165. doi: 10.1016/S1369-5274(02)00302-8
Altegoer, F., and Bange, G. (2015). Undiscovered regions on the molecular landscape of flagellar assembly. Curr. Opin. Microbiol. 28, 98–105. doi: 10.1016/j.mib.2015.08.011
Atlas, R. M. (1999). Legionella: from environmental habitats to disease pathology, detection and control. Environ. Microbiol. 1, 283–293. doi: 10.1046/j.1462-2920.1999.00046.x
Bachman, M. A., and Swanson, M. S. (2001). RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40, 1201–1214. doi: 10.1046/j.1365-2958.2001.02465.x
Bachman, M. A., and Swanson, M. S. (2004). Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect. Immun. 72, 2468–2476. doi: 10.1128/IAI.72.5.2468-2476.2004
Bajrai, L. H., Azhar, E. I., Yasir, M., Jardot, P., Barrassi, L., Raoult, D., et al. (2016). Legionella saoudiensis sp. nov., isolated from a sewage water sample. Int. J. Syst. Evol. Microbiol. 66, 4367–4371. doi: 10.1099/ijsem.0.001357
Bangsborg, J. M., Hindersson, P., Shand, G., and Hoiby, N. (1995). The Legionella micdadei flagellin: expression in Escherichia coli K 12 and DNA sequence of the gene. APMIS 103, 869–877. doi: 10.1111/j.1699-0463.1995.tb01446.x
Barbaree, J. M., Fields, B. S., Feeley, J. C., Gorman, G. W., and Martin, W. T. (1986). Isolation of protozoa from water associated with a legionellosis outbreak and demonstration of intracellular multiplication of Legionella pneumophila. Appl. Environ. Microbiol. 51, 422–424.
Berk, S. G., Faulkner, G., Garduno, E., Joy, M. C., Ortiz-Jimenez, M. A., and Garduno, R. A. (2008). Packaging of live Legionella pneumophila into pellets expelled by Tetrahymena spp. does not require bacterial replication and depends on a Dot/Icm-mediated survival mechanism. Appl. Environ. Microbiol. 74, 2187–2199. doi: 10.1128/AEM.01214-07
Bigot, R., Bertaux, J., Frere, J., and Berjeaud, J. M. (2013). Intra-amoeba multiplication induces chemotaxis and biofilm colonization and formation for Legionella. PLoS ONE 8:e77875. doi: 10.1371/journal.pone.0077875
Bornstein, N., Marmet, D., Dumaine, M. H., Surgot, M., and Fleurette, J. (1991). Detection of flagella in 278 Legionella strains by latex reagent sensitized with antiflagellum immunoglobulins. J. Clin. Microbiol. 29, 953–956.
Bosshardt, S. C., Benson, R. F., and Fields, B. S. (1997). Flagella are a positive predictor for virulence in Legionella. Microb. Pathog. 23, 107–112. doi: 10.1006/mpat.1997.0134
Bouyer, S., Imbert, C., Rodier, M. H., and Hechard, Y. (2007). Long-term survival of Legionella pneumophila associated with Acanthamoeba castellanii vesicles. Environ. Microbiol. 9, 1341–1344. doi: 10.1111/j.1462-2920.2006.01229.x
Bruggemann, H., Hagman, A., Jules, M., Sismeiro, O., Dillies, M. A., Gouyette, C., et al. (2006). Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell. Microbiol. 8, 1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x
Brzuszkiewicz, E., Schulz, T., Rydzewski, K., Daniel, R., Gillmaier, N., Dittmann, C., et al. (2013). Legionella oakridgensis ATCC 33761 genome sequence and phenotypic characterization reveals its replication capacity in amoebae. Int. J. Med. Microbiol. 303, 514–528. doi: 10.1016/j.ijmm.2013.07.003
Burstein, D., Amaro, F., Zusman, T., Lifshitz, Z., Cohen, O., Gilbert, J. A., et al. (2016). Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet. 48, 167–175. doi: 10.1038/ng.3481
Byrne, B., and Swanson, M. S. (1998). Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66, 3029–3034.
Cateau, E., Delafont, V., Hechard, Y., and Rodier, M. H. (2014). Free-living amoebae: what part do they play in healthcare-associated infections? J. Hosp. Infect. 87, 131–140. doi: 10.1016/j.jhin.2014.05.001
Cazalet, C., Gomez-Valero, L., Rusniok, C., Lomma, M., Dervins-Ravault, D., Newton, H. J., et al. (2010). Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires' disease. PLoS Genet. 6:e1000851. doi: 10.1371/journal.pgen.1000851
Cazalet, C., Rusniok, C., Bruggemann, H., Zidane, N., Magnier, A., Ma, L., et al. (2004). Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173. doi: 10.1038/ng1447
Chandler, F. W., Thomason, B. M., and Hebert, G. A. (1980). Flagella on Legionnaires' disease bacteria in the human lung. Ann. Intern. Med. 93, 715–716. doi: 10.7326/0003-4819-93-5-715
Chen, J., de Felipe, K. S., Clarke, M., Lu, H., Anderson, O. R., Segal, G., et al. (2004). Legionella effectors that promote nonlytic release from protozoa. Science 303, 1358–1361. doi: 10.1126/science.1094226
Chilcott, G. S., and Hughes, K. T. (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708. doi: 10.1128/MMBR.64.4.694-708.2000
Cianciotto, N., Eisenstein, B. I., Engleberg, N. C., and Shuman, H. (1989). Genetics and molecular pathogenesis of Legionella pneumophila, an intracellular parasite of macrophages. Mol. Biol. Med. 6, 409–424.
Coil, D. A., and Anne, J. (2009). Twitching motility in Legionella pneumophila. FEMS Microbiol. Lett. 293, 271–277. doi: 10.1111/j.1574-6968.2009.01532.x
Currie, S. L., and Beattie, T. K. (2015). Compost and Legionella longbeachae: an emerging infection? Perspect. Public Health 135, 309–315. doi: 10.1177/1757913915611162
Dalebroux, Z. D., Edwards, R. L., and Swanson, M. S. (2009). SpoT governs Legionella pneumophila differentiation in host macrophages. Mol. Microbiol. 71, 640–658. doi: 10.1111/j.1365-2958.2008.06555.x
Dalebroux, Z. D., Yagi, B. F., Sahr, T., Buchrieser, C., and Swanson, M. S. (2010). Distinct roles of ppGpp and DksA in Legionella pneumophila differentiation. Mol. Microbiol. 76, 200–219. doi: 10.1111/j.1365-2958.2010.07094.x
Danhorn, T., and Fuqua, C. (2007). Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61, 401–422. doi: 10.1146/annurev.micro.61.080706.093316
Declerck, P. (2010). Biofilms: the environmental playground of Legionella pneumophila. Environ. Microbiol. 12, 557–566. doi: 10.1111/j.1462-2920.2009.02025.x
Dietrich, C., Heuner, K., Brand, B. C., Hacker, J., and Steinert, M. (2001). Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69, 2116–2122. doi: 10.1128/IAI.69.4.2116-2122.2001
Edwards, R. L., Dalebroux, Z. D., and Swanson, M. S. (2009). Legionella pneumophila couples fatty acid flux to microbial differentiation and virulence. Mol. Microbiol. 71, 1190–1204. doi: 10.1111/j.1365-2958.2009.06593.x
Eisenreich, W., and Heuner, K. (2016). The life stage-specific pathometabolism of Legionella pneumophila. FEBS Lett. 590, 3868–3886. doi: 10.1002/1873-3468.12326
Elliott, J. A., and Johnson, W. (1981). Immunological and biochemical relationships among flagella isolated from Legionella pneumophila serogroups 1, 2, and 3. Infect. Immun. 33, 602–610.
Elliott, J. A., and Johnson, W. (1982). Virulence conversion of Legionella pneumophila serogroup 1 by passage in guinea pigs and embryonated eggs. Infect. Immun. 35, 943–946.
Faulkner, G., and Garduno, R. A. (2002). Ultrastructural analysis of differentiation in Legionella pneumophila. J. Bacteriol. 184, 7025–7041. doi: 10.1128/JB.184.24.7025-7041.2002
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fields, B. S. (1996). The molecular ecology of legionellae. Trends Microbiol. 4, 286–290. doi: 10.1016/0966-842X(96)10041-X
Fields, B. S., Barbaree, J. M., Shotts, E. B. Jr., Feeley, J. C., Morrill, W. E., Sanden, G. N., et al. (1986). Comparison of guinea pig and protozoan models for determining virulence of Legionella species. Infect. Immun. 53, 553–559.
Fields, B. S., Benson, R. F., and Besser, R. E. (2002). Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15, 506–526. doi: 10.1128/CMR.15.3.506-526.2002
Fliermans, C. B., Cherry, W. B., Orrison, L. H., Smith, S. J., Tison, D. L., and Pope, D. H. (1981). Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41, 9–16.
Fonseca, M. V., and Swanson, M. S. (2014). Nutrient salvaging and metabolism by the intracellular pathogen Legionella pneumophila. Front. Cell. Infect. Microbiol. 4:12. doi: 10.3389/fcimb.2014.00012
Fontana, M. F., and Vance, R. E. (2011). Two signal models in innate immunity. Immunol. Rev. 243, 26–39. doi: 10.1111/j.1600-065X.2011.01037.x
Gal-Mor, O., and Segal, G. (2003). The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34, 187–194. doi: 10.1016/S0882-4010(03)00027-5
Garduno, R. A., Garduno, E., Hiltz, M., and Hoffman, P. S. (2002). Intracellular growth of Legionella pneumophila gives rise to a differentiated form dissimilar to stationary-phase forms. Infect. Immun. 70, 6273–6283. doi: 10.1128/IAI.70.11.6273-6283.2002
Gillen, K. L., and Hughes, K. T. (1991). Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J. Bacteriol. 173, 6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991
Gillmaier, N., Schunder, E., Kutzner, E., Tlapak, H., Rydzewski, K., Herrmann, V., et al. (2016). Growth-related metabolism of the carbon storage poly-3-hydroxybutyrate in Legionella pneumophila. J. Biol. Chem. 291, 6471–6482. doi: 10.1074/jbc.M115.693481
Glockner, G., Albert-Weissenberger, C., Weinmann, E., Jacobi, S., Schunder, E., Steinert, M., et al. (2008). Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int. J. Med. Microbiol. 298, 411–428. doi: 10.1016/j.ijmm.2007.07.012
Gomez-Valero, L., Rusniok, C., and Buchrieser, C. (2009). Legionella pneumophila: population genetics, phylogeny and genomics. Infect. Genet. Evol. 9, 727–739. doi: 10.1016/j.meegid.2009.05.004
Gomez-Valero, L., Rusniok, C., Rolando, M., Neou, M., Dervins-Ravault, D., Demirtas, J., et al. (2014). Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires' disease. Genome Biol. 15, 505. doi: 10.1186/PREACCEPT-1086350395137407
Greub, G., and Raoult, D. (2004). Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17, 413–433. doi: 10.1128/CMR.17.2.413-433.2004
Hammer, B. K., and Swanson, M. S. (1999). Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33, 721–731. doi: 10.1046/j.1365-2958.1999.01519.x
Hammer, B. K., Tateda, E. S., and Swanson, M. S. (2002). A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44, 107–118. doi: 10.1046/j.1365-2958.2002.02884.x
Hauslein, I., Manske, C., Goebel, W., Eisenreich, W., and Hilbi, H. (2016). Pathway analysis using (13) C-glycerol and other carbon tracers reveals a bipartite metabolism of Legionella pneumophila. Mol. Microbiol. 100, 229–246. doi: 10.1111/mmi.13313
Hawn, T. R., Verbon, A., Lettinga, K. D., Zhao, L. P., Li, S. S., Laws, R. J., et al. (2003). A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease. J. Exp. Med. 198, 1563–1572. doi: 10.1084/jem.20031220
Hazelbauer, G. L., Falke, J. J., and Parkinson, J. S. (2008). Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33, 9–19. doi: 10.1016/j.tibs.2007.09.014
Heuner, K., and Albert-Weissenberger, C. (2008). “The flagellar regulon of Legionella pneumophila and the expression of virulence traits,” in Legionella: Molecular Microbiology, eds K. Heuner and M. S. Swanson (Norfolk, UK: Horizon Scientific Press), 249.
Heuner, K., Bender-Beck, L., Brand, B. C., Luck, P. C., Mann, K. H., Marre, R., et al. (1995). Cloning and genetic characterization of the flagellum subunit gene (flaA) of Legionella pneumophila serogroup 1. Infect. Immun. 63, 2499–2507.
Heuner, K., Brand, B. C., and Hacker, J. (1999). The expression of the flagellum of Legionella pneumophila is modulated by different environmental factors. FEMS Microbiol. Lett. 175, 69–77. doi: 10.1111/j.1574-6968.1999.tb13603.x
Heuner, K., Dietrich, C., Skriwan, C., Steinert, M., and Hacker, J. (2002). Influence of the alternative σ28 factor on virulence and flagellum expression of Legionella pneumophila. Infect. Immun. 70, 1604–1608. doi: 10.1128/IAI.70.3.1604-1608.2002
Heuner, K., Dietrich, C., Steinert, M., Gobel, U. B., and Hacker, J. (2000). Cloning and characterization of a Legionella pneumophila-specific gene encoding a member of the LysR family of transcriptional regulators. Mol. Gen. Genet. 264, 204–211. doi: 10.1007/s004380000310
Heuner, K., Hacker, J., and Brand, B. C. (1997). The alternative sigma factor sigma28 of Legionella pneumophila restores flagellation and motility to an Escherichia coli fliA mutant. J. Bacteriol. 179, 17–23. doi: 10.1128/jb.179.1.17-23.1997
Heuner, K., Jacobi, S., Albert, C., Steinert, M., Brüggemann, H., and Buchrieser, C. (2006). “Gene expression and virulence in Legionella: the flagellar regulon,” in Legionella: State of the Art 30 Years After its Recognition, eds N. Cianciotto, Abu, Y. Kwaik, Edelstein, P. Fields, B. Geary, D. Harrison, T. Joseph, C. Ratcliff, R. J. Stout, and Swanson (Washington, DC: ASM), 327–332.
Heuner, K., and Steinert, M. (2003). The flagellum of Legionella pneumophila and its link to the expression of the virulent phenotype. Int. J. Med. Microbiol. 293, 133–143. doi: 10.1078/1438-4221-00259
Hoppe, J., Unal, C. M., Thiem, S., Grimpe, L., Goldmann, T., Gassler, N., et al. (2017). PilY1 Promotes Legionella pneumophila infection of human lung tissue explants and contributes to bacterial adhesion, host cell invasion, and twitching motility. Front. Cell. Infect. Microbiol. 7:63. doi: 10.3389/fcimb.2017.00063
Horwitz, M. A. (1992). Interactions between macrophages and Legionella pneumophila. Curr. Top. Microbiol. Immunol. 181, 265–282. doi: 10.1007/978-3-642-77377-8_10
Horwitz, M. A., and Silverstein, S. C. (1980). Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Invest. 66, 441–450. doi: 10.1172/JCI109874
Jacobi, S., Schade, R., and Heuner, K. (2004). Characterization of the alternative sigma factor sigma54 and the transcriptional regulator FleQ of Legionella pneumophila, which are both involved in the regulation cascade of flagellar gene expression. J. Bacteriol. 186, 2540–2547. doi: 10.1128/JB.186.9.2540-2547.2004
Jager, J., Marwitz, S., Tiefenau, J., Rasch, J., Shevchuk, O., Kugler, C., et al. (2014). Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infect. Immun. 82, 275–285. doi: 10.1128/IAI.00703-13
Khodr, A., Kay, E., Gomez-Valero, L., Ginevra, C., Doublet, P., Buchrieser, C., et al. (2016). Molecular epidemiology, phylogeny and evolution of Legionella. Infect. Genet. Evol. 43, 108–122. doi: 10.1016/j.meegid.2016.04.033
Kirby, J. R. (2009). Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu. Rev. Microbiol. 63, 45–59. doi: 10.1146/annurev.micro.091208.073221
Kirov, S. M., Castrisios, M., and Shaw, J. G. (2004). Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 72, 1939–1945. doi: 10.1128/IAI.72.4.1939-1945.2004
Konishi, T., Yamashiro, T., Koide, M., and Nishizono, A. (2006). Influence of temperature on growth of Legionella pneumophila biofilm determined by precise temperature gradient incubator. J. Biosci. Bioeng. 101, 478–484. doi: 10.1263/jbb.101.478
Kozak, N. A., Buss, M., Lucas, C. E., Frace, M., Govil, D., Travis, T., et al. (2010). Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J. Bacteriol. 192, 1030–1044. doi: 10.1128/JB.01272-09
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
La Scola, B., Birtles, R. J., Greub, G., Harrison, T. J., Ratcliff, R. M., and Raoult, D. (2004). Legionella drancourtii sp. nov., a strictly intracellular amoebal pathogen. Int. J. Syst. Evol. Microbiol. 54(Pt 3), 699–703. doi: 10.1099/ijs.0.02455-0
Lau, H. Y., and Ashbolt, N. J. (2009). The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J. Appl. Microbiol. 107, 368–378. doi: 10.1111/j.1365-2672.2009.04208.x
Levi, A., Folcher, M., Jenal, U., and Shuman, H. A. (2011). Cyclic diguanylate signaling proteins control intracellular growth of Legionella pneumophila. MBio 2:e00316–10. doi: 10.1128/mBio.00316-10
Lo Presti, F., Riffard, S., Meugnier, H., Reyrolle, M., Lasne, Y., Grimont, P. A., et al. (2001). Legionella gresilensis sp. nov. and Legionella beliardensis sp. nov., isolated from water in France. Int. J. Syst. Evol. Microbiol. 51(Pt 6), 1949–1957. doi: 10.1099/00207713-51-6-1949
Macnab, R. M. (2003). How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100. doi: 10.1146/annurev.micro.57.030502.090832
Mampel, J., Spirig, T., Weber, S. S., Haagensen, J. A., Molin, S., and Hilbi, H. (2006). Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl. Environ. Microbiol. 72, 2885–2895. doi: 10.1128/AEM.72.4.2885-2895.2006
Mascarenhas, D. P., and Zamboni, D. S. (2017). Inflammasome biology taught by Legionella pneumophila. J. Leukoc. Biol. 101, 841–849. doi: 10.1189/jlb.3MR0916-380R
McCarter, L. L. (2006). Regulation of flagella. Curr. Opin. Microbiol. 9, 180–186. doi: 10.1016/j.mib.2006.02.001
Micali, G., and Endres, R. G. (2016). Bacterial chemotaxis: information processing, thermodynamics, and behavior. Curr. Opin. Microbiol. 30, 8–15. doi: 10.1016/j.mib.2015.12.001
Minamino, T., and Imada, K. (2015). The bacterial flagellar motor and its structural diversity. Trends Microbiol. 23, 267–274. doi: 10.1016/j.tim.2014.12.011
Molmeret, M., Bitar, D. M., Han, L., and Kwaik, Y. A. (2004). Disruption of the phagosomal membrane and egress of Legionella pneumophila into the cytoplasm during the last stages of intracellular infection of macrophages and Acanthamoeba polyphaga. Infect. Immun. 72, 4040–4051. doi: 10.1128/IAI.72.7.4040-4051.2004
Molmeret, M., Jones, S., Santic, M., Habyarimana, F., Esteban, M. T., and Kwaik, Y. A. (2010). Temporal and spatial trigger of post-exponential virulence-associated regulatory cascades by Legionella pneumophila after bacterial escape into the host cell cytosol. Environ. Microbiol. 12, 704–715. doi: 10.1111/j.1462-2920.2009.02114.x
Molofsky, A. B., Byrne, B. G., Whitfield, N. N., Madigan, C. A., Fuse, E. T., Tateda, K., et al. (2006). Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203, 1093–1104. doi: 10.1084/jem.20051659
Molofsky, A. B., Shetron-Rama, L. M., and Swanson, M. S. (2005). Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect. Immun. 73, 5720–5734. doi: 10.1128/IAI.73.9.5720-5734.2005
Molofsky, A. B., and Swanson, M. S. (2003). Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50, 445–461. doi: 10.1046/j.1365-2958.2003.03706.x
Molofsky, A. B., and Swanson, M. S. (2004). Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53, 29–40. doi: 10.1111/j.1365-2958.2004.04129.x
Neumeister, B., Schoniger, S., Faigle, M., Eichner, M., and Dietz, K. (1997). Multiplication of different Legionella species in Mono Mac 6 cells and in Acanthamoeba castellanii. Appl. Environ. Microbiol. 63, 1219–1224.
Newton, H. J., Ang, D. K., van Driel, I. R., and Hartland, E. L. (2010). Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 23, 274–298. doi: 10.1128/CMR.00052-09
O'Connell, W. A., Dhand, L., and Cianciotto, N. P. (1996). Infection of macrophage-like cells by Legionella species that have not been associated with disease. Infect. Immun. 64, 4381–4384.
Ohnishi, K., Kutsukake, K., Suzuki, H., and Lino, T. (1992). A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an antisigma factor inhibits the activity of the flagellum-specific sigma factor, sigma F. Mol. Microbiol. 6, 3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x
Ohno, A., Kato, N., Yamada, K., and Yamaguchi, K. (2003). Factors influencing survival of Legionella pneumophila serotype 1 in hot spring water and tap water. Appl. Environ. Microbiol. 69, 2540–2547. doi: 10.1128/AEM.69.5.2540-2547.2003
Orrison, L. H., Cherry, W. B., Tyndall, R. L., Fliermans, C. B., Gough, S. B., Lambert, M. A., et al. (1983). Legionella oakridgensis: unusual new species isolated from cooling tower water. Appl. Environ. Microbiol. 45, 536–545.
Osterman, I. A., Dikhtyar, Y. Y., Bogdanov, A. A., Dontsova, O. A., and Sergiev, P. V. (2015). Regulation of flagellar gene expression in bacteria. Biochem. Mosc. 80, 1447–1456. doi: 10.1134/S000629791511005X
Ott, M., Messner, P., Heesemann, J., Marre, R., and Hacker, J. (1991). Temperature-dependent expression of flagella in Legionella. J. Gen. Microbiol. 137, 1955–1961. doi: 10.1099/00221287-137-8-1955
Pallen, M. J., Penn, C. W., and Chaudhuri, R. R. (2005). Bacterial flagellar diversity in the post-genomic era. Trends Microbiol. 13, 143–149. doi: 10.1016/j.tim.2005.02.008
Piao, Z., Sze, C. C., Barysheva, O., Iida, K., and Yoshida, S. (2006). Temperature-regulated formation of mycelial mat-like biofilms by Legionella pneumophila. Appl. Environ. Microbiol. 72, 1613–1622. doi: 10.1128/AEM.72.2.1613-1622.2006
Polesky, A. H., Ross, J. T., Falkow, S., and Tompkins, L. S. (2001). Identification of Legionella pneumophila genes important for infection of amoebas by signature-tagged mutagenesis. Infect. Immun. 69, 977–987. doi: 10.1128/IAI.69.2.977-987.2001
Pruckler, J. M., Benson, R. F., Moyenuddin, M., Martin, W. T., and Fields, B. S. (1995). Association of flagellum expression and intracellular growth of Legionella pneumophila. Infect. Immun. 63, 4928–4932.
Rasis, M., and Segal, G. (2009). The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol. Microbiol. 72, 995–1010. doi: 10.1111/j.1365-2958.2009.06705.x
Ren, T., Zamboni, D. S., Roy, C. R., Dietrich, W. F., and Vance, R. E. (2006). Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18. doi: 10.1371/journal.ppat.0020018
Ricci, M. L., Torosantucci, A., Scaturro, M., Chiani, P., Baldassarri, L., and Pastoris, M. C. (2005). Induction of protective immunity by Legionella pneumophila flagellum in an A/J mouse model. Vaccine 23, 4811–4820. doi: 10.1016/j.vaccine.2005.05.013
Richards, A. M., Von Dwingelo, J. E., Price, C. T., and Abu Kwaik, Y. (2013). Cellular microbiology and molecular ecology of Legionella-amoeba interaction. Virulence 4, 307–314. doi: 10.4161/viru.24290
Rizzardi, K., Winiecka-Krusnell, J., Ramliden, M., Alm, E., Andersson, S., and Byfors, S. (2015). Legionella norrlandica sp. nov., isolated from the biopurification systems of wood processing plants. Int. J. Syst. Evol. Microbiol. 65(Pt 2), 598–603. doi: 10.1099/ijs.0.068940-0
Robertson, P., Abdelhady, H., and Garduno, R. A. (2014). The many forms of a pleomorphic bacterial pathogen-the developmental network of Legionella pneumophila. Front. Microbiol. 5:670. doi: 10.3389/fmicb.2014.00670
Rowbotham, T. J. (1986). Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22, 678–689.
Rust, M., Borchert, S., Niehus, E., Kuehne, S. A., Gripp, E., Bajceta, A., et al. (2009). The Helicobacter pylori anti-sigma factor FlgM is predominantly cytoplasmic and cooperates with the flagellar basal body protein FlhA. J. Bacteriol. 191, 4824–4834. doi: 10.1128/JB.00018-09
Sahr, T., Bruggemann, H., Jules, M., Lomma, M., Albert-Weissenberger, C., Cazalet, C., et al. (2009). Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 72, 741–762. doi: 10.1111/j.1365-2958.2009.06677.x
Sahr, T., Rusniok, C., Dervins-Ravault, D., Sismeiro, O., Coppee, J. Y., and Buchrieser, C. (2012). Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 9, 503–519. doi: 10.4161/rna.20270
Sahr, T., Rusniok, C., Impens, F., Oliva, G., Sismeiro, O., Coppee, J. Y., et al. (2017). The Legionella pneumophila genome evolved to accommodate multiple regulatory mechanisms controlled by the CsrA-system. PLoS Genet. 13:e1006629. doi: 10.1371/journal.pgen.1006629
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schalk, J. A., Euser, S. M., van Heijnsbergen, E., Bruin, J. P., den Boer, J. W., and de Roda Husman, A. M. (2014). Soil as a source of Legionella pneumophila sequence type 47. Int. J. Infect. Dis. 27, 18–19. doi: 10.1016/j.ijid.2014.05.009
Schell, U., Simon, S., and Hilbi, H. (2016). Inflammasome recognition and regulation of the Legionella flagellum. Curr. Top. Microbiol. Immunol. 397, 161–181. doi: 10.1007/978-3-319-41171-2_8
Schulz, T., Rydzewski, K., Schunder, E., Holland, G., Bannert, N., and Heuner, K. (2012). FliA expression analysis and influence of the regulatory proteins RpoN, FleQ and FliA on virulence and in vivo fitness in Legionella pneumophila. Arch. Microbiol. 194, 977–989. doi: 10.1007/s00203-012-0833-y
Schunder, E., Gillmaier, N., Kutzner, E., Eisenreich, W., Herrmann, V., Lautner, M., et al. (2014). Amino acid uptake and metabolism of Legionella pneumophila hosted by Acanthamoeba castellanii. J. Biol. Chem. 289, 21040–21054. doi: 10.1074/jbc.M114.570085
Steinert, M., Emody, L., Amann, R., and Hacker, J. (1997). Resuscitation of viable but non-culturable Legionella pneumophila philadelphia JR32 by Acanthamoeba castellanii. Appl. Environ. Microbiol. 63, 2047–2053.
Stewart, C. R., Muthye, V., and Cianciotto, N. P. (2012). Legionella pneumophila persists within biofilms formed by Klebsiella pneumoniae, Flavobacterium sp., and Pseudomonas fluorescens under dynamic flow conditions. PLoS ONE 7:e50560. doi: 10.1371/journal.pone.0050560
Stewart, C. R., Rossier, O., and Cianciotto, N. P. (2009). Surface translocation by Legionella pneumophila: a form of sliding motility that is dependent upon type II protein secretion. J. Bacteriol. 191, 1537–1546. doi: 10.1128/JB.01531-08
Swanson, M. S., and Hammer, B. K. (2000). Legionella pneumophila pathogesesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54, 567–613. doi: 10.1146/annurev.micro.54.1.567
Szurmant, H., and Ordal, G. W. (2004). Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68, 301–319. doi: 10.1128/MMBR.68.2.301-319.2004
Tamura, K., and Kumar, S. (2002). Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Mol. Biol. Evol. 19, 1727–1736. doi: 10.1093/oxfordjournals.molbev.a003995
Tiaden, A., Spirig, T., Weber, S. S., Bruggemann, H., Bosshard, R., Buchrieser, C., et al. (2007). The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell. Microbiol. 9, 2903–2920. doi: 10.1111/j.1462-5822.2007.01005.x
Tlapak, H., Rydzewski, K., Schulz, T., Weschka, D., Schunder, E., and Heuner, K. (2017). Functional analysis of the alternative sigma-28 factor FliA and Its Anti-sigma factor FlgM of the non-flagellated Legionella species, L. oakridgensis. J. Bacteriol. 199:e00018-17. doi: 10.1128/JB.00018-17
Wang, C., Saito, M., Tanaka, T., Amako, K., and Yoshida, S. (2015). Comparative analysis of virulence traits between a Legionella feeleii strain implicated in Pontiac fever and a strain that caused Legionnaires' disease. Microb. Pathog. 89, 79–86. doi: 10.1016/j.micpath.2015.09.004
Whiley, H., and Bentham, R. (2011). Legionella longbeachae and legionellosis. Emerg. Infect. Dis. 17, 579–583. doi: 10.3201/eid1704.100446
Wolfe, A. J., and Visick, K. L. (2008). Get the message out: cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol. 190, 463–475. doi: 10.1128/JB.01418-07
Yu, V. L., Plouffe, J. F., Pastoris, M. C., Stout, J. E., Schousboe, M., Widmer, A., et al. (2002). Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186, 127–128. doi: 10.1086/341087
Keywords: Legionella, flagellar regulon, flagellin, virulence, FliA, FleQ, alternative sigma factor
Citation: Appelt S and Heuner K (2017) The Flagellar Regulon of Legionella—A Review. Front. Cell. Infect. Microbiol. 7:454. doi: 10.3389/fcimb.2017.00454
Received: 27 June 2017; Accepted: 06 October 2017;
Published: 20 October 2017.
Edited by:
Matthias P. Machner, National Institutes of Health (NIH), United StatesReviewed by:
Ombeline Rossier, Université Paris-Sud, FranceZachary David Dalebroux, University of Oklahoma Health Sciences Center, United States
Copyright © 2017 Appelt and Heuner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Klaus Heuner, aGV1bmVya0Bya2kuZGU=
 Sandra Appelt1
Sandra Appelt1 Klaus Heuner
Klaus Heuner