Persistent Exposure to Fusobacterium nucleatum Triggers Chemokine/Cytokine Release and Inhibits the Proliferation and Osteogenic Differentiation Capabilities of Human Gingiva-Derived Mesenchymal Stem Cells
- 1Department of Human Microbiome, School and Hospital of Stomatology, Shandong University and Shandong Provincial Key Laboratory of Oral Tissue Regeneration and Shandong Engineering Laboratory for Dental Materials and Oral Tissue Regeneration, Jinan, China
- 2Department of Periodontology, School of Stomatology, Shandong University, Jinan, China
- 3Department of Oral Medicine, School of Stomatology, Shandong University, Jinan, China
- 4State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China
Fusobacterium nucleatum is one of the most frequent pathogenic bacteria causing periodontitis. The direct effect of Fusobacterium nucleatum (F. nucleatum) on oral stem cells has rarely been reported. In this study, we aimed to evaluate how gingiva-derived mesenchymal stem cells (GMSCs) respond to a direct challenge with F. nucleatum. GMSCs were isolated by the limiting dilution method and exposed to F. nucleatum at various multiplicities of infection (MOIs; F. nucleatum:cell ratios of 10:1, 50:1, and 100:1) for 24 h to 4 weeks. Our results indicated that F. nucleatum significantly inhibited cell proliferation in a dose-dependent manner and promoted cell migration and the release of chemokines/cytokines, such as CCL2, CXCL1, and IL-6. Additionally, F. nucleatum inhibited GMSC osteogenic differentiation partly by decreasing alkaline phosphatase (ALP) activity, mineralized nodule formation, and osteogenesis-related gene and protein expression. RNA-sequencing analyses indicated that F. nucleatum time-dependently activated cellular signaling pathways during the process of osteogenic differentiation. A total of 64 cell differentiation-related genes were found to be differentially expressed between non-infected and F. nucleatum-infected GMSCs at 3, 7, 14, and 21 d. Intriguingly, we discovered that the 64 cell differentiation-related differentially expressed genes (DEGs) were significantly enriched in cancer-related pathways, such as bone cancer, osteosarcoma and bone marrow cancer, which provides new insight into tumorigenesis during the process of GMSC osteogenic differentiation. In conclusion, this study demonstrates that persistent exposure to F. nucleatum promotes cell migration and chemokine/cytokine release and inhibits the proliferation and osteogenic differentiation of GMSCs. Our study provides a novel and long-time bacteria-cell co-culture in vitro model and makes a foundation for the future mechanistic studies of GMSCs under F. nucleatum infection.
Introduction
Oral microorganisms are abundant and play key roles in maintaining oral health; periodontitis is a typical chronic infectious disease caused by increased oral pathogenic bacterium (Paster and Dewhirst, 2009). Groups of oral pathogens contribute to the occurrence of periodontitis; however, their pathogenesis effects in periodontitis have not been fully illuminated. Fusobacterium nucleatum (F. nucleatum) is a common anaerobe in the oral cavity and has been frequently detected with high abundance in periodontitis patients (Wang et al., 2015). F. nucleatum is an opportunistic pathogen closely associated with the occurrence and development of periodontitis, and its abundance is positively associated with the periodontal pocket depth (Papapanou et al., 1993; Moore and Moore, 1994; Socransky et al., 1998). Studies have shown that F. nucleatum possesses versatile adhesion properties and can co-aggregate with all other strains of oral bacteria (Brook and Walker, 1986; Kinder and Holt, 1993). The dental biofilm, which is composed of a large number of bacteria has been identified as the most difficult therapeutic target for periodontitis (Moore and Moore, 1994; Socransky and Haffajee, 2002). F. nucleatum has its unique pathogenic potential and is dominant in the dental biofilm at a later stage, which play critical roles in the destruction of periodontal supporting tissues (Moore and Moore, 1994; Ebersole et al., 1995). However, if F. nucleatum could directly destruct the periodontal supporting tissues and the potential pathogenic mechanism have not been fully elucidated.
The F. nucleatum in periodontal pockets can easily invade the host tissue and interact with the host cells (Jeng et al., 1999). F. nucleatum has been reported to exert pathogenic effects by producing a group of virulence factors and toxic metabolites, such as lipopolysaccharide, porin, butyrate and propionate ammonia (Takada et al., 1988; Rogers et al., 1991; Bolstad et al., 1996). In peripheral blood mononuclear and polymorphonuclear cells, F. nucleatum induces apoptosis by activating caspases and then plays an immunosuppressive role in host cells (Jewett et al., 2000). In gingival epithelial cells (GECs), F. nucleatum can activate innate immune responses and promote the secretion of proinflammatory cytokines and matrix metalloproteinases (MMPs), which finally play crucial roles in the destruction of the periodontal supporting tissues (Gursoy et al., 2008, 2012). F. nucleatum can promote the secretion of interleukin (IL)-1β in GECs by activating the NLRP3 inflammasome (Bui et al., 2016).
The gingival epithelium is recognized as the first barrier against oral infection (Ji et al., 2009; Signat et al., 2011). However, F. nucleatum can break through the epithelial barrier and penetrate into the gingival connective tissues in patients with severe periodontitis (Signat et al., 2011). It has been reported that F. nucleatum can induce the release of interleukin-6 in gingival fibroblasts and significantly inhibit the cell proliferation (Bartold et al., 1991; Rossano et al., 1993). Recently, researchers have isolated a new type of mesenchymal stem cell from human gingival connective tissue, namely, gingival tissue-derived mesenchymal stem cells (GMSCs) (Fournier et al., 2010; Mitrano et al., 2010). GMSCs possess normal karyotype and telomerase activity and exhibit a stable phenotype in long-term cultures; furthermore, these cells have self-renewal, multipotent differentiation, and immunomodulatory abilities, which have attracted increasing attention from scholars (Fournier et al., 2010; Mitrano et al., 2010; Tomar et al., 2010; Tang et al., 2011). Given their accessibility and relative abundance in gingival tissues, GMSCs have a bright future in cell-based tissue regenerative medicine for the treatment of human dental diseases and the improvement of human health (Zhao et al., 2015). In previous studies, GMSCs have been widely used in nerve regeneration and bone tissue regeneration (Wang et al., 2011; Mao et al., 2019). In addition, Li et al. reported that the inflammatory microenvironment could induce GMSCs differentiation toward a profibrotic phenotype and then lead to inflammatory gingiva hyperplasia, which is detrimental for GMSC-based therapy (Li et al., 2013). However, the detailed biological characteristic changes of GMSCs in the inflammatory environment have not been reported thus far, and the effect of F. nucleatum on GMSCs has not been previously examined. In this study, we aimed to explore how GMSCs respond to a direct challenge with F. nucleatum by evaluating the proliferation, migration, and inflammatory cytokine production and the osteogenic differentiation ability in F. nucleatum infected GMSCs. In addition, we performed a comprehensive gene expression profile analysis of F. nucleatum infected GMSCs during the process of osteogenic differentiation. Our study provides a novel and long-time bacteria-cell co-culture in vitro model and performs comprehensive biological characteristic changes of GMSCs under time-series F. nucleatum infection, which makes a foundation for the future mechanistic studies of GMSCs under F. nucleatum infection.
Materials and Methods
Human Subjects and Ethical Statements
This study was approved by the Medical Ethical Committee of the Stomatology School, Shandong University (Protocol Number: 20170101). Three healthy individuals, aged from 20 to 25 years, who underwent impacted tooth extraction at the Stomatology Hospital of Shandong Province, were recruited. All individuals were informed about the research project and signed the informed consent according to the Helsinki Declaration of 1975.
Bacterial Strains, Cell Cultures, and Identification
Fusobacterium nucleatum subsp. nucleatum ATCC 25586 was provided by Shandong Provincial Key Laboratory of Oral Tissue Regeneration (Jinan, China). Stock cultures were routinely propagated in brain-heart infusion (BHI, Haibo, Qingdao, China) broth supplemented with 5 μg/ml hemin and 1 μg/ml menadione in an anaerobic atmosphere with 85% N2, 5% H2, and 10% CO2 for 24 h in a constant temperature incubator at 37°C. F. nucleatum was tested and verified by polymerase chain reaction (PCR) assay and 16S rRNA gene sequencing. The primers used are shown in Supplementary Table 1. F. nucleatum cultured in the logarithmic growth phase was harvested by centrifugation at 4,500 rpm for 5 min and washed three times with phosphate buffered saline (PBS, HyClone, Logan, UT, USA). The bacterial concentration was spectrophotometrically standardized to optical density (OD) value at 600 nm = 1 F. nucleatum, corresponding to 108 bacteria/ml. Finally, the cells were suspended in Dulbecco's modified Eagle's medium (DMEM, HyClone) for subsequent studies.
Cell Isolation and Culture
The excised gingival tissues were immediately immersed in DMEM with 5% antibiotics (100 mg/ml penicillin, 100 mg/ml streptomycin, Sigma Aldrich, St Louis, MO, USA) and quickly transferred to the laboratory. Then the free gingival tissue samples were washed, and the epithelium was removed and minced into small fragments of ~1–3 mm2 and digested for 2 h at 37°C by 3 mg/ml collagenase I (Sigma Aldrich) and 4 mg/mL dispase II (Invitrogen, Carlsbad, CA, USA). To obtain GMSCs, the single-cell suspension was filtered through a 70 μm cell strainer and seeded into 10 cm Petri dishes at a density of 60 cells per cm2 according to a previous study (Ge et al., 2012). The primary GMSCs were cultured with DMEM containing 20% fetal bovine serum (FBS, BioInd, Kibbutz, Israel) at 37°C in a humidified atmosphere of 5% CO2. Cells were fed fresh medium every 3 days until the cell monolayer reached 80–90% confluence. GMSCs were trypsinized and passaged at a dilution ratio of 1:3 to expand the culture in 10% FBS DMEM medium without antibiotics. The fourth passage cells were used for subsequent experiments.
Colony Formation and Flow Cytometric Analysis
GMSCs were seeded in six-well-plates at a density of 1 × 103 cells/well and cultured with DMEM containing 10% FBS; and the cells were fed fresh medium every 3 days. After 7 d, the cells were fixed in 4% paraformaldehyde (Sigma Aldrich) for 30 min and stained with crystal violet (Solarbio, Beijing, China) for 5 min. The total number of cell colonies was photographed and counted under a high-power microscope (Olympus, Tokyo, Japan). Groups of fifty or more adherent cells derived from the same mother cell were assessed as a colony. For the phenotypic characterization of stem cells, GMSCs infected with or without F. nucleatum at the multiplicity of infection (MOI) of 100 at 3 d were collected and resuspended in blocking buffer with 1% immunoglobulins G and M for 30 min and then incubated with fluorescein-conjugated mouse monoclonal antibodies (10 μg/ml) specific for human CD29, CD44, CD73, and CD45 (Becton Dickinson Biosciences, NY, USA) for 30 min on ice. After being washed, the cells were fixed in PBS and subjected to flow cytometric analyses.
Cell Morphology and Proliferation Assays
GMSCs were seeded in 6-well (1 × 105 cells/well) plates in growth medium with 10% FBS and no antibiotics. Cells were left untreated or incubated with F. nucleatum at multiplicities of infection (MOIs) of 10, 50, and 100 (F. nucleatum:cell ratio of 10:1, 50:1, 100:1) from 1 to 7 d, and cells were fed fresh medium every day. The cell morphology was detected by crystal violet (Solarbio) staining. Cells were fixed in 4% paraformaldehyde for 30 min and stained with crystal violet (Solarbio) for 30 min. The cells were observed immediately under the microscope and photographed (Olympus). Cell proliferation was detected by counting the cell numbers through an automated cell counter (Countstar, Shanghai, China). Briefly, cells infected with or without F. nucleatum were trypsinized, collected in a 1.5 ml microcentrifuge tube and centrifuged for 5 min at 1,000 rpm. After being washed twice with PBS, cells were resuspended in 1 ml of PBS and mixed thoroughly. Then, 20 μl of cell resuspension solution was added to the counting chamber and detected by a cell counter (Countstar). Cell proliferation rate was detected by the 5-ethynyl-20-deoxyuridine (EdU) labeling assay according to the instructions of an EdU Apollo DNA in vitro kit (RIBOBIO, Guangzhou, China).
Cell Migration Assay
The effect of F. nucleatum on GMSCs migrations was evaluated by a transwell chamber with an 8.0 μM pore size (Corning, Corning, NY, USA). Approximately 5 × 104 cells cultured in complete medium with no antibiotics were seeded in the upper chamber, and the lower plates were filled with 500 μl of 10% FBS containing medium with or without F. nucleatum at MOIs of 10, 50, and 100. Medium containing 10% FBS was used as a negative control (NC). The chambers were incubated for 20 h at 37°C. The cells that migrated through the membrane were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet. Six high-power microscopic fields (×200) per filter were randomly selected by blind evaluation and the number of cells that had migrated into the undersurface of the membrane was counted.
Enzyme-Linked Immunosorbent Assay (ELISA)
GMSCs (2 × 105 cells/well) were seeded in 6-well-plates, cultured with 10% FBS DMEM without antibiotics and treated with F. nucleatum at MOIs of 10, 50, and 100 for 24, 48, and 72 h. The cell culture supernatants were collected and centrifuged at 12,000 rpm for 5 min at 4°C. The levels of secreted protein IL-6, IL-8, and IL-1β were measured by ELISA (BioLegend, San Diego, CA, USA) according to the manufacturer's instructions. The OD values were measured by a microplate reader (SPECTROstar Nano, BMG Labtech, Offenburg, Germany) at 450 and 570 nm, and the values at 570 nm were subtracted from those at 450 nm in the following data analysis. The experiment was performed in triplicate and repeated three times with cell culture supernatants from 3 different donors.
Alkaline Phosphatase (ALP) Activity Assay
GMSCs (2 × 105 cells/well) were seeded in 6-well-plates and cultured with osteogenic medium without antibiotics [OM, medium supplemented with 10% FBS, 10−8mol/L dexamethasone (Solarbio), 50 mg/L ascorbic acid and 10 mmol/L β-glycerophosphate (Sigma-Aldrich)]. F. nucleatum was used to stimulate cells at MOIs of 10, 50, and 100 for 3, 7, 14, and 21 d. The medium and F. nucleatum were changed every 3 days. Cells were lysed with 1% TritonX-100 (Solarbio) for 30 min. Cell lysates were collected, and the protein concentration was measured according to a bicinchoninic acid (BCA) protein assay kit (CWBIO, Beijing, China). ALP activity was detected according to the instructions of the assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) and the absorbance was measured with a microplate reader (SPECTROstar Nano) at a wavelength of 520 nm.
Alizarin Red Staining
GMSCs (2 × 105 cells/well) were seeded in 6-well-plates and cultured in OM (no antibiotics included) with or without F. nucleatum (MOIs of 10, 50, and 100) for 14, 21, and 28 d. The medium and F. nucleatum were changed every 3 days. The cells were fixed in 4% paraformaldehyde, and extracellular matrix calcification was estimated by 2% alizarin red S (pH 4.3, Sigma-Aldrich). The relative amount of calcium was quantified with 10% (w/v) cetylpyridinium chloride (CPC, Solarbio) and 10 mM sodium phosphate solution. The absorbance was measured at a wavelength of 562 nm.
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
GMSCs (2 × 105 cells/well) were seeded in 6-well-plates and cultured with DMEM without antibiotics and then infected with F. nucleatum (MOIs of 10, 50, and 100) for different lengths of times. Total RNA was extracted with TRIzol (CWBIO) and the mRNA concentration was determined using an ultramicro spectrophotometer (Thermo, Waltham, MA, USA). One microgram of mRNA was reverse-transcribed to cDNA using a cDNA synthesis reagent kit (CWBIO). qRT-PCR was performed with UltraSYBR (CWBIO) on a LightCycler 96 Real-Time PCR System (Roche, Basel, Switzerland) in triplicate. Data were analyzed using the 2−ΔΔCt method. The sequences of the primers used for amplification are shown in Supplementary Table 2.
Western Blot Assay
GMSCs (2 × 105 cells/well) seeded in 6-well-plates-were infected with F. nucleatum (MOIs of 10, 50, and 100) for 3, 7, 14, and 21 d. The medium and F. nucleatum were changed every 3 days. Cells were harvested with RIPA lysis buffer containing 1% protease inhibitors (CWBIO). Protein concentrations were measured by a BCA protein assay and 20 μg/lane of protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The primary antibodies were incubated with the membrane at 4°C overnight at the dilution of 1:1,000–10,000 according to the manufacturer's instructions (Supplementary Table 3) and membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000; Proteintech, Chicago, IN, USA) for 1 h at room temperature. The protein bands were visualized with enhanced chemiluminescence reagents (Millipore) and scanned using an extra sensitive imager (Amersham Imager 600; GE Healthcare Life Sciences, Pittsburgh, PA, USA). Image J 1.44 software (NIH, Bethesda, Maryland, USA) was used to quantify the protein expression levels.
RNA Sequencing (RNA-seq) Analysis
Twenty-four samples from 3 individuals (for each individual, 4 control samples at 3, 7, 14, and 21 d with no F. nucleatum treatment, and 4 experimental samples with F. nucleatum infection at an MOI of 100 at the same time point were included) were sequenced for the analysis of gene expression at the whole-genome level at Novogene (Beijing, China) by RNA-seq. Total RNA was isolated, evaluated for quality, reverse-transcribed to cDNA, and sequenced on the Illumina platform. After quality control (QC), clean reads were mapped to a reference genome (GRCh38) via hierarchical indexing for the spliced alignment of transcripts (HISAT v2.0.5) (Kim et al., 2015) and featureCounts (v1.5.0-p3) was used to count the number of reads mapped to each gene (Liao et al., 2014). Then, the fragments per kilobase of exon per million reads mapped (FPKM) of each gene was calculated based on the length of the gene, and the read count mapped to the gene. Based on the gene expression level, DEseq2 was used to detect the differentially expressed genes (DEGs) between control groups and F. nucleatum-treated groups by an absolute fold-change in DEGs >2 and an adjusted P < 0.01. A Gene Ontology (GO) enrichment analysis of the DEGs was implemented by the clusterProfiler R package, and GO terms with corrected P < 0.05 were considered significantly enriched (Young et al., 2010). Pathway enrichment analysis of DEGs was performed based on the Disease Ontology (DO) (Bello et al., 2018), Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000), Reactome database (Croft et al., 2011), and DisGeNET database (Pinero et al., 2017). The protein-protein interaction network was generated by STRING website with the default parameters (von Mering et al., 2005). The RNA sequencing data have been deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through the GEO series accession number GSE126821.
Statistical Analysis
The experiment was performed in triplicate and repeated three times. All biological data for validating the effect of F. nucleatum on GMSCs were expressed as mean ± standard deviation (SD). First, we checked whether the data confirmed to normal distribution. The normally distributed data were analyzed using GraphPad Prism software (version 6, MacKiev Software, Boston, MA, USA), and differences among more than two groups were analyzed by one-way or two-way ANOVA with Tukey's honestly significant difference (HSD) comparison test. For the data that did not conform to normal distribution, non-parametric analysis with Kruskal-Wallis test was used to analyze the significance of the differences. P < 0.05 was considered as statistically significant.
Results
Isolation and Characteristics of GMSCs
Primary cultures of single-cell suspensions from human gingival tissues exhibited a spindle-shaped fibroblast-like morphology, which was maintained at the fourth generation (Figures 1A,B). All trials of single cell-derived colony formation were successfully performed (Figure 1C). Cell clusters formed from a single GMSC colony (Figure 1D). A crystal violet staining assay was performed to determine GMSC morphology after F. nucleatum infection at MOIs of 10, 50, and 100 for 48 h. The results indicated that F. nucleatum induced no significant change in GMSCs morphology (Figure 1E). The surface markers of GMSCs were detected by flow cytometry. GMSCs were uniformly positive (>99%) for the mesenchymal stem cells markers CD29, CD44, and CD73, and negative for the hematopoietic stem cell marker CD45 (<1%), and F. nucleatum treatment (MOI = 100) did not influence the expression of surface markers on GMSCs (Figure 1F).
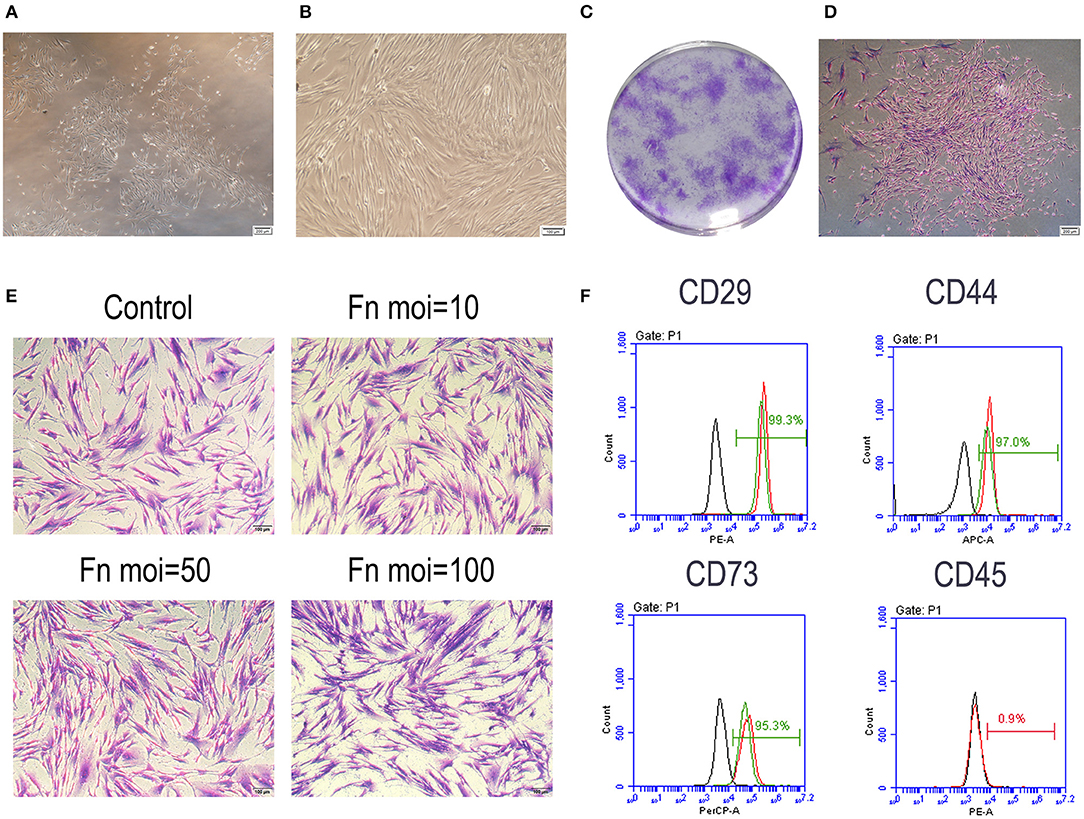
Figure 1. Characteristic of GMSCs. (A) Primary GMSCs under low-power magnification; Scale bar: 200 μm. (B) The fourth passage GMSCs present a spindle-shaped fibroblast-like morphology. Scale bar: 100 μm. (C) Single colonies of GMSCs after 7 d. (D) Cell clusters derived from a single GMSC colony. Scale bar: 200 μm. (E) Crystal violet staining results of GMSCs morphology without or with F. nucleatum infection at MOIs of 10, 50, and 100 at 48 h. Scale bar: 100 μm. (F) Flow cytometric analysis of GMSC surface markers, including CD29, CD44, CD73, and CD45. The black line represents the negative control group, the red line represents GMSCs without F. nucleatum infection (MOI = 100) and the green line represents the F. nucleatum-infected GMSCs.
F. nucleatum Inhibited GMSC Proliferation
The effect of F. nucleatum on GMSCs proliferation were detected by counting the total cell numbers or the new-growth cell numbers in GMSCs infected with or without F. nucleatum. A cell counting assay showed that F. nucleatum inhibited GMSC proliferation in a dose-dependent manner. Compared with the control group, F. nucleatum infection at an MOI of 100 significantly decreased the number of GMSCs from 1 to 6 d at each time point, and the cell proliferation rate was noticeably reduced. GMSCs co-cultured with F. nucleatum at an MOI of 10 showed equivalent cell proliferation rates as the control group at each time point. At an MOI of 50, the cell numbers at 1, 3, and 5 d were decreased, whereas they were not significantly different from those in the control group at 2, 4, and 6 d (Figure 2A). The EdU labeling assay demonstrated that F. nucleatum infection at an MOI of 50 and 100 significantly decreased the cell proliferation rate of GMSCs at 24 h (Figures 2B,C).
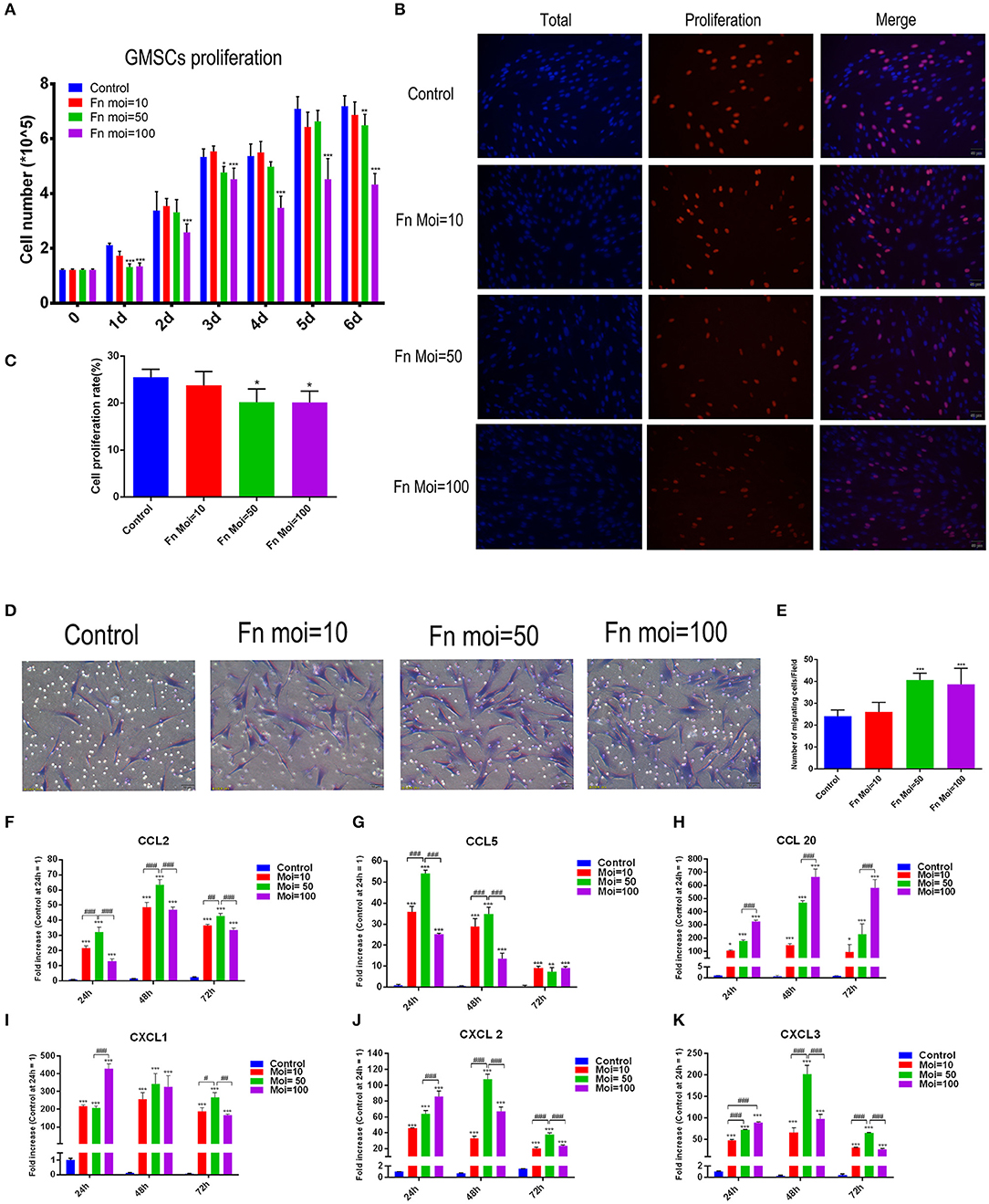
Figure 2. Effects of F. nucleatum on GMSC proliferation, migration and chemokine production. (A) Number of GMSCs detected by electronic cell counter with or without F. nucleatum infection at MOIs of 10, 50, and 100 (n = 6). (B) EdU assay of GMSCs after F. nucleatum infection (MOIs of 0, 10, 50, and 100) at 24 h. Scale bar: 50 μm. (C) Cell proliferation rate of GMSCs detected by EdU assay (n = 4). (D) Migration of human GMSCs infected with F. nucleatum at MOIs of 10, 50, and 100. Scale bar: 50 μm. (E) Statistical analysis of GMSC migration results. CCL2 (F), CCL5 (G), CCL20 (H), CXCL1 (I), CXCL2 (J), CXCL3 (K) chemokine production at the gene expression level in GMSCs co-cultured with F. nucleatum (MOIs of 0, 10, 50, and 100) at 24, 48, and 72 h (n = 3). All data are shown as the mean ± SD. Statistical analyses were performed by one-way ANOVA with Tukey's multiple-comparison test or non-parametric analysis with Kruskal-Wallis test. *P < 0.05; **P < 0.01; and ***P< 0.001 compared with the control at each time point. #P < 0.05, ##P < 0.01, and ###P < 0.001.
F. nucleatum Promoted GMSC Migration and Chemokine Production
To explore the effect of F. nucleatum infection on GMSCs migration, transwell assays were designed and the migration related chemokines were detected. Transwell migration assays indicated that compared with NC groups, GMSC co-cultured groups with F. nucleatum at MOIs of 50 and 100 had a significantly enhanced migration capacity (24 ± 1.138 vs. 40 ± 1.282 and 38 ± 3.007 cells/field, P < 0.001). F. nucleatum at an MOI of 10 also slightly promoted GMSC migration, but the difference was not significant (P > 0.05). These results indicated that F. nucleatum could enhance GMSC migration in a concentration-dependent manner (Figures 2D,E).
To further explore the mechanism by which F. nucleatum promotes cell migration, we detected the gene expression level of chemokines by qRT-PCR after F. nucleatum infection, and the results indicated that F. nucleatum dose-dependently elevated the expression of the chemokines CCL2, CCL5, CCL20, C-X-C motif chemokine ligand (CXCL)-1, CXCL2, and CXCL3 at 24 h, 48, 72 h (Figures 2F–K). In detail, F. nucleatum infection at an MOI of 50 induced the maximum CCL2 gene expression level at 24, 48, and 72 h, whereas the expression of CCL20 reached a peak at 24, 48, and 72 h after F. nucleatum infection at an MOI of 100 (Figures 2F,H). For CCL5, the highest mRNA level was induced by F. nucleatum at an MOI of 50 at 24 and 48 h; however, at 72 h, no significant difference was observed among the three F. nucleatum-infected groups (Figure 2G). The gene expression level of CXCL1 was dramatically elevated by F. nucleatum infection at an MOI of 100 at 24 h, whereas the peak appeared at 72 h in the MOI of 50 group (Figure 2I). When cells were co-cultured with F. nucleatum for 24 h, the maximum gene expression level of CXCL2 and CXCL3 appeared in the MOI of 100 group, while F. nucleatum infection at an MOI of 50 induced the highest mRNA expression of CXCL2 and CXCL3 at 48 and 72 h (Figures 2J,K).
F. nucleatum Promoted Intracellular Enzyme and Inflammatory Cytokine Production in GMSCs
Bacterium infection could directly induce the destruction of periodontal supporting tissues, and to clarify the effect of F. nucleatum on altering the inflammation cytokines production in GMSCs, we performed the in vitro model of F. nucleatum (MOI = 10, 50, and 100) co-cultured with GMSCs at 24, 48, and 72 h. Our results indicated that F. nucleatum dose-dependently elevated the expression of intracellular enzyme genes, including prostaglandin-endoperoxide synthase (PTGS)-2 and superoxide dismutase (SOD)-2. The peak expression of PTGS2 and SOD2 appeared after F. nucleatum infection at an MOI of 50 for 24, 48, and 72 h (Figures 3A,B). In addition, F. nucleatum dose-dependently elevated the production of proinflammatory cytokines, including colony-stimulating factor (CSF)-2, IL-6, IL-8, and IL-1β at the gene level (Figures 3C–F). F. nucleatum infection at an MOI of 100 induced the maximum CSF2 gene expression level at 24 h, whereas at 48 and 72 h, CSF2 expression was significantly elevated by F. nucleatum infection at MOIs of 10, 50, and 100. No significant difference was observed between the MOI of 50 and MOI of 100 groups (Figure 3C). The gene expression level of IL-6 was increased to the greatest extent by F. nucleatum infection at an MOI of 50 at 24 h, whereas no differences existed at 48 h and 72 h with F. nucleatum infection at MOIs of 50 and 100 (Figure 3D). For IL-8, the gene expression was significantly increased by F. nucleatum infection (MOIs of 10, 50, and 100) at 24, 48, and 72 h. F. nucleatum infection at an MOI of 100 induced the highest mRNA expression of IL-8 at 24 h, 48 and 72 h (Figure 3E). The gene expression level of IL-1β was significantly elevated by F. nucleatum infection at MOIs of 10, 50, and 100 at 24, 48, and 72 h, and the maximum gene expression level of IL-1β appeared at 48 h infection with F. nucleatum at an MOI of 100 (Figure 3F). However, at the protein level, compared with the control group, IL-6 and IL-8 levels in the groups co-cultured with F. nucleatum at MOIs of 10, 50, and 100 were significantly elevated; there were no significant differences among the groups infected with different concentrations of F. nucleatum (Figures 3G,H). The protein level of IL-1β in the F. nucleatum-infected groups was not significantly changed compared with that in the control group (Figure 3I).
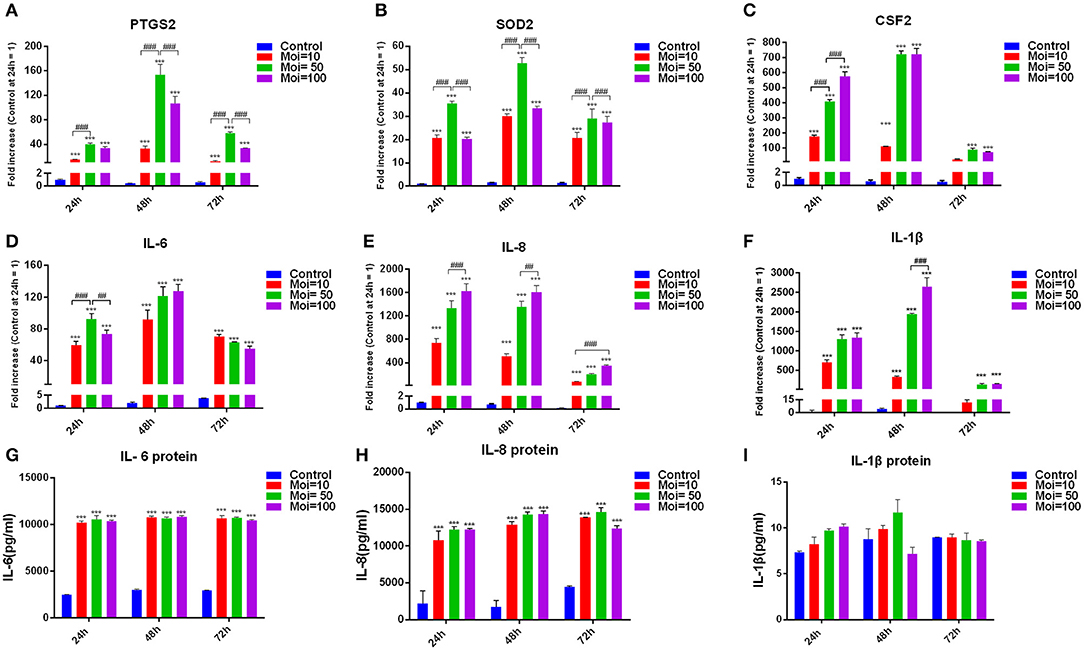
Figure 3. Effects of F. nucleatum on GMSC cytokine production. PTGS2 (A), SOD2 (B), CSF2 (C), IL-6 (D), IL-8 (E), and IL-1β (F) cytokine production at the gene expression level in GMSCs infected with F. nucleatum (MOIs of 0, 10, 50, and 100) at 24, 48, and 72 h (n = 3). Protein levels of IL-6 (G), IL-8 (H), and IL-1β (I) in GMSCs co-cultured with F. nucleatum (MOIs of 0, 10, 50, and 100) at 24, 48, and 72 h (n = 3). The histogram represents the mean ± SD. Statistical analyses were performed by two-way ANOVA with Tukey's multiple-comparison test. ***P < 0.001 compared with the control at each time point. ##P < 0.01 and ###P < 0.001.
F. nucleatum Inhibited the Osteogenic Differentiation of GMSCs by Decreasing the ALP Activity and Mineral Deposition Formation
To explore the effect of F. nucleatum infection on GMSCs osteogenic differentiation, the osteogenic differentiation ability in GMSCs co-cultured with time-series F. nucleatum were detected by ALP activity assay and alizarin red staining assay. The ALP activity of GMSCs was measured at days 3, 7, 14, and 21 after F. nucleatum infection at MOIs of 10, 50, and 100. The results indicated that the ALP activity of uninfected GMSCs peaked on day 7 and F. nucleatum dose-dependently decreased the ALP activity in GMSCs co-cultured with F. nucleatum at 3, 7, 14, and 21 d (Figure 4A). The mineral deposition in GMSCs with or without F. nucleatum infection was measured on days 14, 21, and 28 according to alizarin red staining assay and the relative amount of calcium was quantified by CPC. The results indicated that the mineral deposition of GMSCs increased time-dependently from 14 to 28 d, and large quantities of mineralized nodules were detected at day 28 in the GMSCs without infection by F. nucleatum, whereas in GMSCs co-cultured with F. nucleatum, the osteogenic medium-induced mineral deposition formation on days 21 and 28 were dose-dependently inhibited by F. nucleatum (Figures 4B,C).
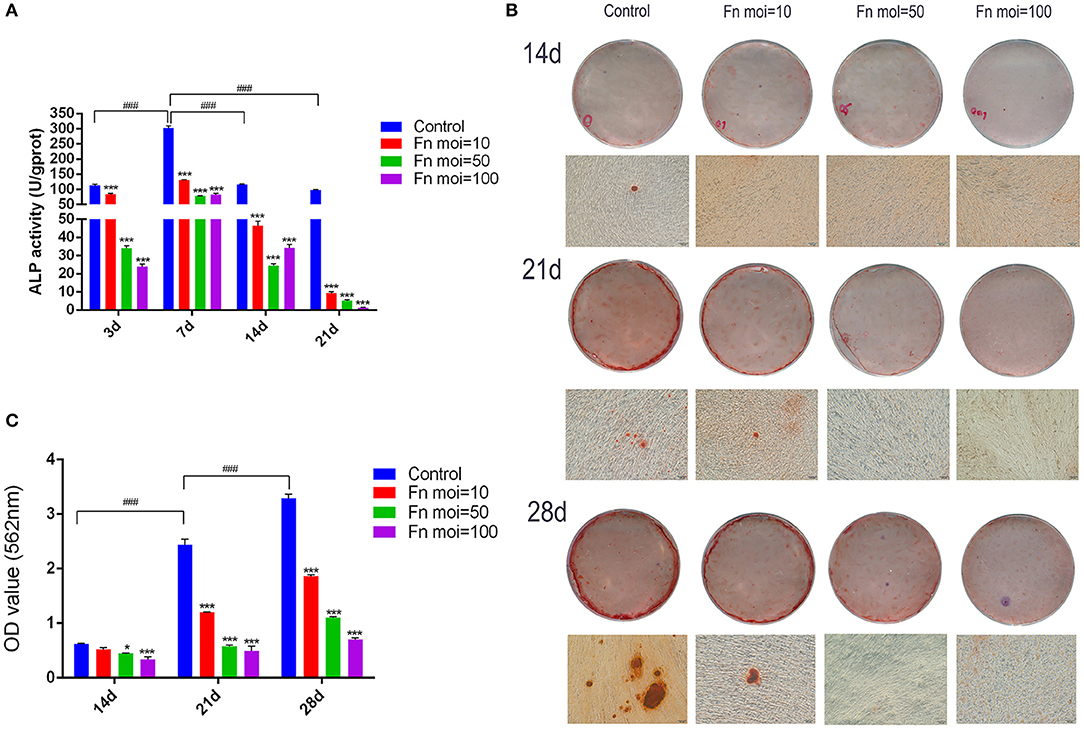
Figure 4. Effects of F. nucleatum on GMSC osteogenic differentiation. (A) ALP activity in GMSCs co-cultured with F. nucleatum (MOIs of 0, 10, 50, and 100) at 3, 7, 14, and 21 d. (B) Representative photographs of alizarin red staining of GMSCs at 14, 21, and 28 d with or without F. nucleatum infection at MOIs of 10, 50, and 100 by digital camera or microscope; scale bar: 100 μm. (C) The concentration of calcium deposited by GMSCs at 14, 21, and 28 d with or without F. nucleatum infection at MOIs of 10, 50, and 100. Histograms represent the mean ± SD (n = 3). Statistical analyses were performed using two-way ANOVA with Tukey's multiple-comparison test. *P < 0.05 and ***P < 0.001 compared with the control at each time point. ###P < 0.001.
F. nucleatum Inhibited the Gene and Protein Expression of Osteogenic Markers in GMSCs
To explore the mechanism of F. nucleatum inhibited GMSCs osteogenic differentiation ability from the molecular levels, osteogenic related genes and proteins in GMSCs free or co-cultured with F. nucleatum were detected. The gene expression levels of ONC, ALP, Runx2, OCN, BSP, and OPN were detected at 3, 7, 14, and 21 d in GMSCs with or without F. nucleatum infection (Figures 5A–F). The results showed that the gene expression levels of ONC were increased at day 3 and day 7, and ALP expression peaked at day 7. F. nucleatum infection at the MOIs of 50 and 100 significantly decreased the gene expression of ONC at 3, 7, and 14 d (Figure 5A), and F. nucleatum dose-dependently decreased the gene expression of ALP at 7, 14, and 21 d (Figure 5B). For Runx2, the gene expression level was increased at day 7; the high expression level was maintained at day 14 and decreased at day 21. F. nucleatum infection (MOIs of 10, 50 and 100) significantly downregulated the expression of this gene level at 7, 14, and 21 d (Figure 5C). The expression of OCN, which is a middle-advanced stage marker, peaked at day 14, and F. nucleatum infection at an MOI of 100 significantly reduced the gene expression level at 7 d, 14 d and 21 d (Figure 5D). BSP is recognized as late osteogenic marker, and the BSP gene expression level was significantly decreased by F. nucleatum (MOIs of 10, 50 and 100) infection at days 7, 14, and 21 (Figure 5E). For OPN, another osteogenic marker, the highest gene expression level appeared on day 21, and F. nucleatum infection (MOIs of 10, 50 and 100) dramatically decreased the gene expression level at 14 d and 21 d (Figure 5F). Based on these results, we confirmed F. nucleatum at the MOI of 100 significantly inhibited the osteogenic gene expression at various stages. In addition, we confirmed that F. nucleatum at an MOI of 100 could dramatically downregulate the expression of the osteogenesis related proteins ALP, COL1, BMP2, Runx2, and Osterix at days 3, 7, 14, and 21. The BSP protein level was decreased by F. nucleatum stimulation at days 14 and 21 (Figures 5G–M).
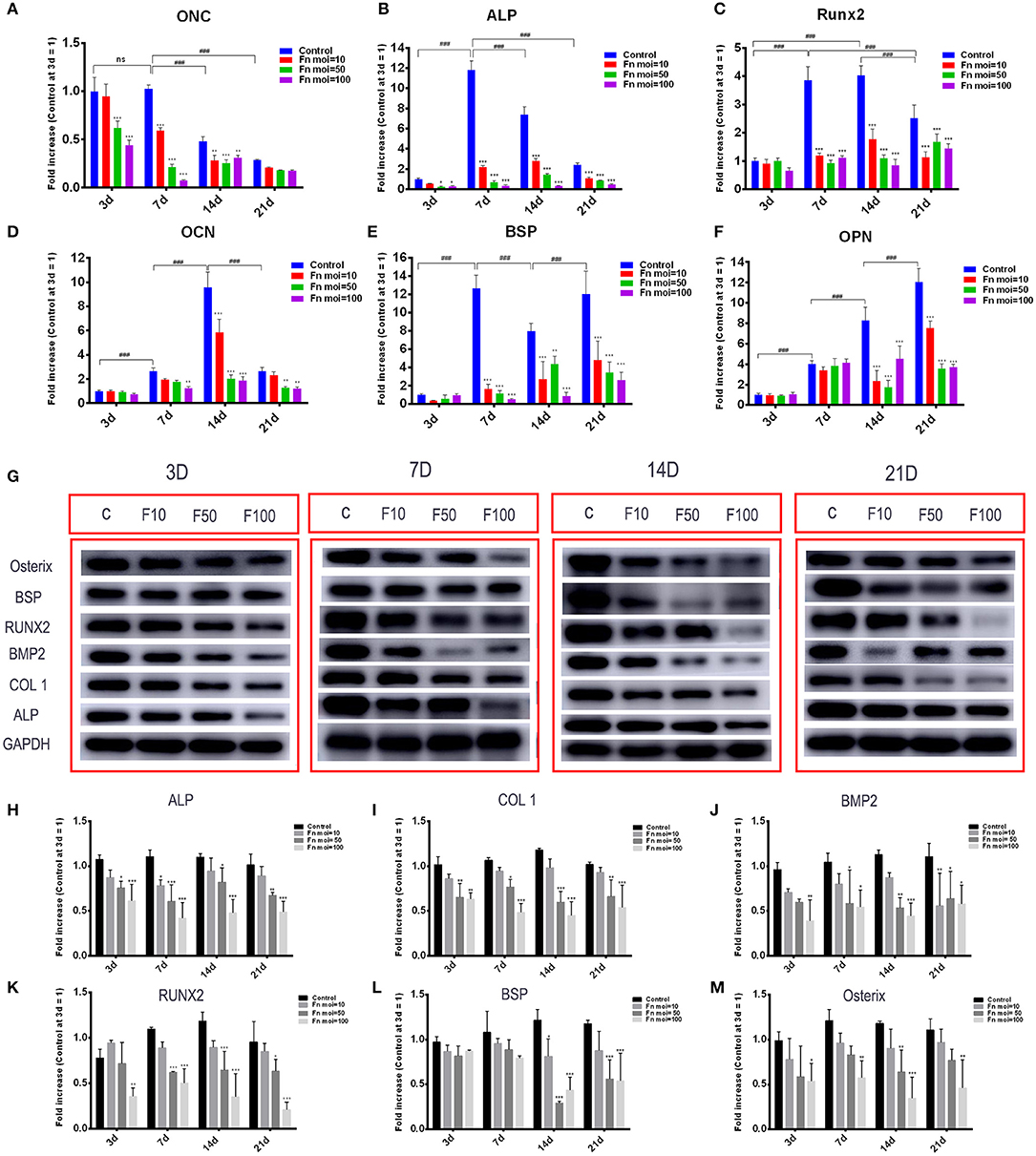
Figure 5. Effects of F. nucleatum on the expression of osteogenesis-related genes and proteins in GMSCs. The gene expression of ONC (A), ALP (B), Runx2 (C), OCN (D), BSP (E), and OPN (F) in GMSCs co-cultured with F. nucleatum (MOIs of 0, 10, 50, and 100) at 3, 7, 14, and 21 d. (G) The protein expression of ALP, COL1, BMP2, RUNX2, BSP, osterix on days 3, 7, 14, 21 detected by western blot. The relative protein levels of ALP/GAPDH (H), COL1/GAPDH (I), BMP2/GAPDH (J), RUNX2/GAPDH (K), BSP/GAPDH (L), osterix/GAPDH (M). The histograms represent the mean ± SD (n = 3). Statistical analyses were performed using two-way ANOVA with Tukey's multiple-comparison test. ns, no statistic significance. *P < 0.05; **P < 0.01; and ***P < 0.001 compared with the control at each time point. ###P < 0.001.
RNA-seq Analysis of GMSCs Under Time Series F. nucleatum Infection
To better understand the global gene expression changes in F. nucleatum-infected GMSCs in OM culture conditions over time, we performed a genome-wide transcriptome analysis by RNA-seq. The pearson correlation coefficient analysis and principal coordinates analysis (PCoA) of the genome-wide gene expression levels showed that F. nucleatum-infected GMSCs significantly separated from normal cells as the infected time increased (Supplementary Figures 1, 2). In addition, the results indicated that one control sample at 14 d (B14C) and two treated samples at the 14 d (B14F) and 21 d (A21F) time points deviated greatly from the other samples in the same group; thus, these three samples were excluded from the subsequent analysis. These results suggest that the effects of F. nucleatum infection on GMSCs may accumulate over time.
To characterize the DEGs affected by F. nucleatum, the gene expression profiles of normal GMSCs vs. infected GMSCs at 3, 7, 14, and 21 d were compared at each time point by DEseq2, and 258, 323, 591, 245 DEGs were identified at each time point (Supplementary Figure 3). The overlapping DEGs among the 4 time points included 8 genes that were differentially expressed between normal GMSCs and F. nucleatum-infected GMSCs throughout the whole process. In addition, 149, 140, 292, and 84 DEGs were specifically identified at 3, 7, 14, and 21 d of F. nucleatum infection, respectively (Figure 6A). The heatmap analysis showed that among the 8 DEGs, the gene expression levels of AK4, PODNL1, ADAMTS7, CFD, and EPHB6 were down-regulated, and those of NRG1, CD163, AL1 were up-regulated (Figure 6B). The GO analysis of the 8 genes indicated that they were involved in kinase regulatory activity, and the extracellular matrix, and enriched in cell migration-related signaling pathways, such as directional guidance of interneurons involved in migration from the subpallium to the cortex (Figure 6C). In addition, the reactome functional enrichment analysis showed that 149 DEGs specifically induced by F. nucleatum infection at 3 d were predominantly enriched in the interferon-related signaling pathway (Figure 6D); 140 DEGs specifically produced after 7 d of F. nucleatum infection were mostly enriched in extracellular matrix organization, collagen formation, collagen biosynthesis, and modifying enzymes (Figure 6E), and 292 unique DEGs generated after 14 d of F. nucleatum infection were mainly enriched in IL-10, IL-4, and IL-13 signaling, chemokine receptors bind chemokines, and O-linked glycosylation signaling pathway (Figure 6F); finally, 84 DEGs specifically produced at 21 d of F. nucleatum infection were largely enriched in class A/1 (rhodopsin-like receptors) and GPCR-related signaling pathway (Figure 6G). Furthermore, we identified a total of 999 DEGs that were generated after F. nucleatum infection at 4 time points, and all these DEGs could be divided into 4 clusters (Supplementary Figure 4 and Supplementary Table 4).
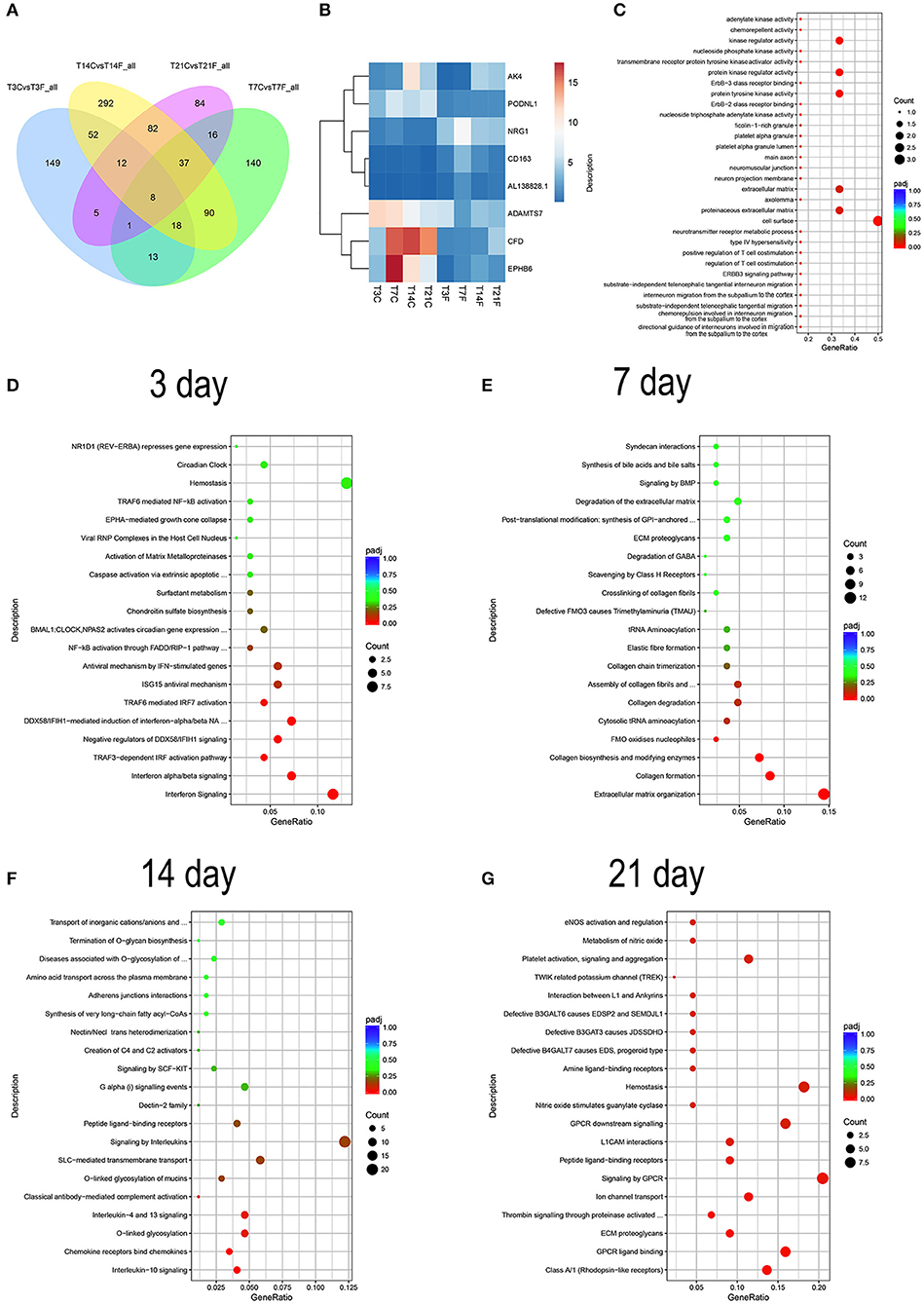
Figure 6. RNA-seq analysis of GMSCs co-cultured with F. nucleatum (MOI of 100). GMSCs from 3 different donors were infected by F. nucleatum at 3, 7, 14, and 21 d, and the whole gene expression were detected by RNA-seq. T3C, T7C, T14C, and T21C represent the GMSCs in control group without F. nucleatum infection at 3, 7, 14, and 21 d; T3F, T7F, T14F, and T21F represent F. nucleatum infected GMSCs group at 3, 7, 14, and 21 d, respectively. (A) Venn diagram summarizing overlapping DEGs in GMSCs among the four F. nucleatum infection time points. T3CvsT3F_all, T7CvsT7F_all, T14CvsT14F_all, and T21CvsT21F_all represent the DEGs generated by F. nucleatum infection at 3, 7, 14, and 21 d, respectively. (B) The average gene expression level of 8 overlapping DEGs at each group was presented by heatmap. The legend show the gene expression level is increased as the color changes from blue to red. (C) The GO enrichment analysis result of the 8 overlapping DEGs. (D) DO analysis of the 149 DEGs specifically generated after 3 d of F. nucleatum infection. (E) DO analysis of the 140 DEGs specifically generated after 7 d of F. nucleatum infection. (F) DO analysis of the 292 DEGs specifically generated after 14 d of F. nucleatum infection. (G) DO analysis of the 84 DEGs specifically generated after 21 d of F. nucleatum infection. In the legend, the size of the dot represents the number of the DEGs enrichment in the relevant signaling pathway and the colors of the dot represent the Padj value decreased from blue to red (C–G).
To further investigate the effect of F. nucleatum on GMSC differentiation, the expression signatures of stem cell differentiation-related DEGs involved in the GO biological processes were selected (Figure 7A). The Venn diagram revealed a total of 64 DEGs; the DEGs varied with increasing F. nucleatum infection time, and no overlapping DEGs were generated (Figure 7B), which suggests that GMSC osteogenic differentiation is regulated by various genes at different stages during F. nucleatum infection. The heatmap comprehensively showed the gene expression levels of 64 genes, including Bcl2, IL-6, and TGFb2, which indicates inflammation and apoptosis related genes were activated by F. nucleatum during the process of GMSCs osteogenic differentiation (Figure 7C). The GO function analysis confirmed the following: (1) at the molecular function level, all of the DEGs were enriched in cytokine activity, cytokines and growth factor binding-related signaling pathway; (2) at the cellular component level, all of the DEGs were enriched in growth factor complex, cell surface, and extracellular matrix signaling pathways; and (3) at the biological process level, all of the DEGs were enriched in stem cell differentiation, and mesenchymal cell differentiation signaling pathways, which indicates that the biological processes of GMSC development and differentiation are closely related to cytokine production and related receptor activation in the F. nucleatum induced microenvironment (Figure 7D). Interestingly, we discovered that 64 cell differentiation-related DEGs were associated with human cancers, such as bone cancer, osteosarcoma, and thyroid cancer according to the DO enrichment analysis (Figure 7E). The KEGG and DisGenet analyses also confirmed that the DEGs were significantly enriched in cancer-related pathways, such as colorectal cancer and fibroid tumor, and so on (Supplementary Figure 5), which suggests that persistent exposure to F. nucleatum greatly increases the risk of GMSC carcinogenesis during the process of osteogenic differentiation. The protein-protein interaction network of 64 DEGs showed a potentially complex interaction relationship among these genes (Supplementary Figure 6).
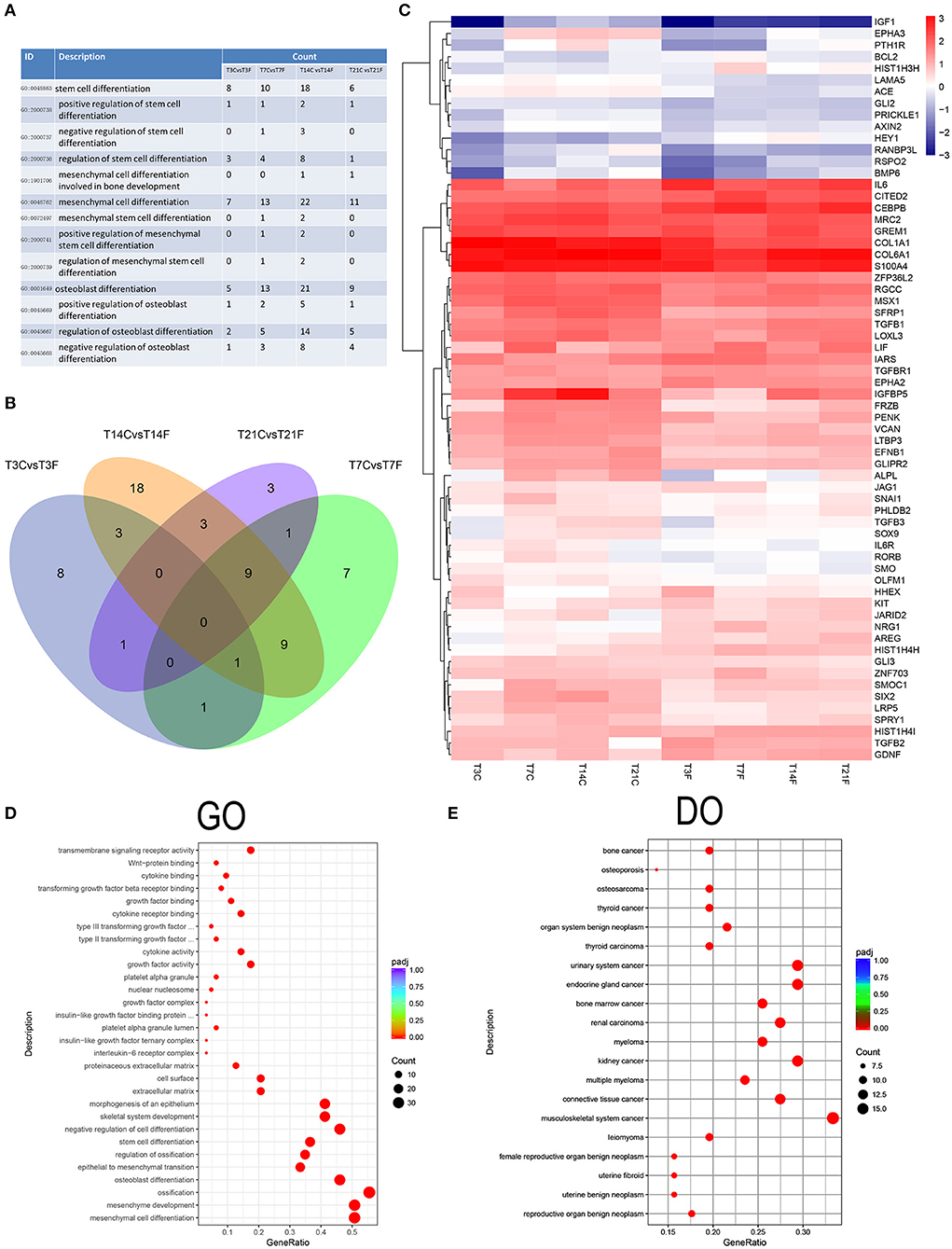
Figure 7. Analysis of the cell differentiation-related DEGs. The GMSCs from 3 different donors were infected by F. nucleatum at 3, 7, 14, and 21 d, and the whole gene expression were detected by RNA-seq. The osteogenic differentiation related DEGs was identified and further analyzed. (A) The cell differentiation-related biological process GO terms were identified and the DEGs number in GMSCs after F. nucleatum infection at 3, 7, 14, and 21 d were calculated compared with control group at each time point. (B) The cell differentiation-related DEGs generated after F. nucleatum infection at 3, 7, 14, and 21 d were overlapped by Venn diagram. T3CvsT3F, T7CvsT7F, T14CvsT14F, and T21CvsT21F represent the osteogenic differentiation related DEGs numbers generated by F. nucleatum infection at 3, 7, 14, and 21 d, respectively. (C) The average genes expression level of the 64 union cell differentiation related DEGs generated by F. nucleatum infection at 4 time points at each group were presented by heatmap. The legend show the gene expression level is increasing as the color changes from blue to red. The GO (D) and DO (E) enrichment analysis of the 64 unique cell differentiation-related DEGs. In the legend, the size of the dot represents the number of the DEGs enrichment in the relevant signaling pathway and the colors of the dot represent the Padj value decreased from blue to red.
Discussion
Polymicrobial infection with Porphyromonas gingivalis and F. nucleatum has been shown to can aggravate periodontal alveolar bone loss and induce an increased inflammatory response in a mouse periodontitis model (Polak et al., 2009). The stimulatory mechanism of inflammatory bone resorption in periodontitis induced by P. gingivalis has been comprehensive reported (Hanazawa, 1998; Jia et al., 2019), whereas the defined effect of F. nucleatum on oral tissue destruction has rarely been explored. Our current study confirms that F. nucleatum significantly alters the biological properties of GMSCs by promoting cell migration and chemokine/cytokine release and decreasing the proliferation and osteogenic differentiation ability. The large number of inflammatory cytokines release and the decreased osteogenic differentiation ability of GMSCs are critical in periodontal supporting tissue destruction.
In this study, we demonstrate that F. nucleatum inhibits the proliferation of GMSCs. It has been reported that F. nucleatum can impair endothelial cell and mononuclear cell proliferation, which is consistent with our finding in GMSCs (Haake and Lindemann, 1997; Mendes et al., 2016). Viable and formaldehyde-treated, but not sonicated or heat-treated F. nucleatum bacteria, can cause severe aggregation and apoptosis of peripheral blood mononuclear cells (PBMCs) (Huynh et al., 2011). Bartold et al. reported that F. nucleatum inhibits human gingival fibroblast proliferation by releasing ammonium and butyrate (Bartold et al., 1991). However, F. nucleatum promotes colorectal cancer cell (CRC) line proliferation by modulating the E-cadherin/β-catenin signaling pathway or activating Toll-like receptor 4 signaling to nuclear factor-kB (Rubinstein et al., 2013; Yang et al., 2017). F. nucleatum could promote CRC line proliferation but inhibit the GMSCs proliferation, which suggested the mechanism of F. nucleatum regulated cell proliferation is different in cancer cell lines and primary somatic cells. In the future, we would explore the mechanism of F. nucleatum on GMSCs proliferation inhibition by separating the detailed bacterial components and evaluating the effect of each component on GMSCs proliferation.
In human leukemic monocyte cells, F. nucleatum can trigger an inflammatory response by upregulating the expression of tumor necrosis factor-α, IL-1β, IL-6, IL-8, and monocyte chemoattractant protein-1 and promote the recruitment and transmigration of monocytes through endothelial cells (Wang et al., 2019). Uitto et al. reported that F. nucleatum increase infected epithelial cell migration and survival by increasing collagenase 3 production (Uitto et al., 2005). F. nucleatum infection leads to the recruitment of macrophages and osteoclasts, finally resulting in gingival inflammation and bone resorption (Johnson et al., 2018). In human GECs, F. nucleatum upregulates the gene expression levels of IL-1, IL-6, IL-8, SOD2, CCL20, CXCL1, CXCL3, and CSF2 partly by activating the NF-κB, mitogen-activated protein kinase (MAPK) p38, and MAPK kinase/extracellular signal-regulated kinase (MEK/ERK) pathways (Huang et al., 2004; Milward et al., 2007; Ramage et al., 2017). F. nucleatum increases the production of PTGS2, PGE2, and the ratio of RANKL/OPG in periodontal ligament (PDL) cells, which play key roles in the inflammation and destruction in periodontitis (Nogueira et al., 2014). Bui et al. reported that F. nucleatum could invasive into GECs and then leads to the activation of NLRP3 inflammasome, which finally resulted in the secretion of IL-1β and the elevated protein level could be detected, which was different with our findings in F. nucleatum infected GMSCs (Bui et al., 2016). In the current study, we preliminarily confirmed that F. nucleatum could promote GMSC migration, which is consist with the results of F. nucleatum on epithelial cell, monocytes, macrophages and osteoclasts. In addition, we confirmed F. nucleatum could increase the chemokines and inflammatory cytokines releases, such as CCL2, CXCL1, PTGS2, IL-6, and IL-8, which is consistent with the previous studies on F. nucleatum infection with GECs and PDL cells. However, the IL-1β expression in F. nucleatum infected GMSCs is inconsistent with GECs, which indicated that the mechanism of F. nucleatum on GMSCs and GECs are different. Our current study is limited for lack of the detailed molecular mechanism by which F. nucleatum promotes GMSC migration and inflammatory cytokine production.
The periodontal supporting tissues regeneration based on stem cells has been regarded as a novel therapeutic strategy to repair the periodontal defects (Iohara et al., 2004; Liu et al., 2008). Tissue regeneration is an organized dynamic process, including hemostasis, an inflammatory phase, proliferation, and maturation/matrix remodeling; external factors and the infection by pathogenic microorganisms could largely impact the whole process of periodontal tissues regeneration (Hammerle and Giannobile, 2014). GMSCs have attracted a great deal of attention in periodontal regenerative therapy research because of their ease of accessibility, well-established self-renewal, multipotent differentiation, and immune modulatory properties (Hammerle and Giannobile, 2014; Diniz et al., 2016; Fawzy and Dorfer, 2016). However, current study on stem cell research is defective for few concerned on the osteogenic differentiation ability of stem cells in the inflammatory microenvironment. In our study, we firstly confirmed that F. nucleatum significantly inhibited GMSC osteogenic differentiation by decreasing the osteogenesis-related gene and protein expression, ALP activity and mineralized nodule formation. Our transcriptome analysis results showed that F. nucleatum disturbed GMSC gene expression during osteogenic differentiation, and some DEGs are significantly enriched in cancer-related signaling pathways, which suggested that persistent F. nucleatum infection had the potential to induce GMSCs progression into cancer cells. Previous studies indicated that F. nucleatum was closely associated with various types of cancer, such as colorectal cancer, gastric cancer, and pancreatic cancer (Amitay et al., 2017; Hsieh et al., 2018), which suggested that it was worth to focus on tumorigenesis during the process of long-term stem cell-based regeneration therapy.
In summary, we confirmed that F. nucleatum played multi-dimensional roles in altering the biological properties of GMSCs, including inhibiting GMSC proliferation, promoting cell migration and chemokine/cytokine generation, such as CCL2, CCL20, CXCL1, PTGS2, IL-6, IL-8, and IL-1β. Moreover, F. nucleatum decreases GMSC osteogenic differentiation partly by decreasing ALP activity and mineralized nodule formation, and downregulating osteogenesis-related gene and protein expression. Additionally, a time course RNA-seq analysis indicated that F. nucleatum activated cellular signaling pathways in a time-dependent manner. In addition, we identified a total of 64 cell differentiation-related DEGs in GMSCs with or without F. nucleatum infection at 3, 7, 14, and 21 d. Intriguingly, we discovered that the cell differentiation-related DEGs generated in F. nucleatum-infected GMSCs are significantly enriched in cancer-related pathways, such as bone cancer, osteosarcoma and bone marrow cancer. Our study is the first to illuminate the multidimensional biological effects of F. nucleatum on GMSCs and makes a foundation for future mechanistic studies of GMSCs under F. nucleatum infection. However, our current study is limited due to the lack of detailed molecular experiments to evaluate the mechanism by which F. nucleatum inhibits GMSC proliferation and osteogenic differentiation and promotes migration and cytokine generation. In the future, we will construct a large number of F. nucleatum mutant strains, establish in vivo models, and perform a series of molecular biology experiments, such as skin injury models of migration and cell immunofluorescence assays to explore the comprehensive effects and mechanisms by which F. nucleatum affects GMSC proliferation, migration, and osteogenic differentiation.
Data Availability Statement
The datasets generated for this study can be found in the NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number (GSE126821).
Ethics Statement
This study was approved by the Medical Ethical Committee of Stomatology School, Shandong University (Protocol Number: 20170101). All individuals were informed with this research project and signed the informed consent according to the Helsinki Declaration of 1975.
Author Contributions
QF conceived, designed, and supervised this study. WK and DT collected samples. WK performed experiments and analyzed the data. WK and QF wrote the paper. XJ, XZ, WK, and QF revised the manuscript.
Funding
This study was supported by the Natural Science of Shandong Province (Nos. ZR201702190185 and 2018GSF118231), the National Natural Science Foundation of China (No. 81630072), the Construction Engineering Special Fund of Taishan Scholars of Shandong Province (No. tsqn20161068), and the Program of Qilu young scholars of Shandong University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate the volunteers to provide samples. We thank all colleagues in our laboratory for their kind advice and support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00429/full#supplementary-material
Supplementary Figure 1. RNA-seq analysis of GMSCs co-cultured with F. nucleatum (MOI of 100). GMSCs from 3 different donors were infected by F. nucleatum at 3, 7, 14, and 21 d. The correlation of the 24 samples was showed according to the pearson correlation coefficient analysis. The first letter in the sample name represents the donor ID, the middle number represents the time, and the last number C and F represent control and F. nucleatum, respectively. For example, A3C represent the sample from donor A that growth for 3 days without F. nucleatum infection; A3F represent the sample from donor A that growth for 3 d with F. nucleatum infection.
Supplementary Figure 2. RNA-seq analysis of GMSCs co-cultured with F. nucleatum (MOI of 100). GMSCs from 3 different donors were infected by F. nucleatum at 3, 7, 14, and 21 d. The principal coordinates analysis (PCoA) of the genome-wide gene expression levels showed F. nucleatum-infected GMSCs significantly separated from normal cells as the infected time increased. The first letter in the sample name represents the donor ID, the middle number represents the time, and the last number C and F represent control and F. nucleatum, respectively. In the legend, T3C, T7C, T14C, and T21C represent control group at 3, 7, 14, and 21 d respectively. T3F, T7F, T14F, and T21F represent F. nucleatum-infected group at 3, 7, 14, and 21 d, respectively. For example, T3C represents the union of A3C, B3C, and C3C; T3F represents the union of A3F, B3F, and C3F.
Supplementary Figure 3. RNA-seq analysis of GMSCs co-cultured with F. nucleatum (MOI of 100). GMSCs from 3 different donors were infected by F. nucleatum at 3, 7, 14, and 21 d. The number of DEGs after F. nucleatum infection at 3, 7, 14, and 21 d were calculated. The red dot represents the up-regulated genes; the green dot represents the down-regulated genes; and the blue dot represents the genes not influenced by F. nucleatum infection. The gene numbers regulated by F. nucleatum are showed in the legend.
Supplementary Figure 4. The GMSCs from 3 different donors were infected by F. nucleatum at 3, 7, 14, and 21 d, and the whole gene expression were detected by RNA-seq. Compared with control group at each time point, the DEGs generated in GMSCs after F. nucleatum infection at 3, 7, 14, and 21 d were identified. The average genes expression level of the 999 union DEGs generated by F. nucleatum infection at 4 time points at each group were presented by heatmap (absolute of the fold-change of DEGs > 2 and an adjusted P < 0.01). T3C, T7C, T14C, and T21C represent control group at 3, 7, 14, and 21 d, respectively. T3F, T7F, T14F, and T21F represent F. nucleatum infected group at 3, 7, 14, and 21 d, respectively. For example, T3C represents the union of A3C, B3C, and C3C; T3F represents the union of A3F, B3F, and C3F. The legend show the gene expression level is increasing as the color changes from blue to red.
Supplementary Figure 5. The GMSCs from 3 different donors were infected by F. nucleatum at 3, 7, 14, and 21 d, and the whole gene expression were detected by RNA-seq. The 64 unique osteogenic differentiation-related DEGs were enriched in signaling pathways according to KEGG (A) and DisGeNET (B) database. In the legend, the size of the dot represents the number of the DEGs enrichment in the relevant signaling pathway and the colors of the dot represent the Padj value decreased from blue to red.
Supplementary Figure 6. The GMSCs from 3 different donors were infected by F. nucleatum at 3, 7, 14, and 21 d, and the whole gene expression were detected by RNA-seq. A total of 64 unique osteogenic differentiation-related DEGs were identified after F. nucleatum infection at 3, 7, 14, 21 d, and the complex interactive network of these 64 DEGs were visualized based on the STRING database. All the genes are presented by the color nodes, and the node content represents the 3D structure of the proteins. The known, predicated or other protein-protein interaction associations are presented by different color edges.
Supplementary Table 1. Primers sequences for quantitative PCR of F. nucleatum.
Supplementary Table 2. Primers sequences for quantitative real-time PCR of GMSCs.
Supplementary Table 3. Primary antibodies used in the assay of western blot.
Supplementary Table 4. The union 999 DEGs list of GMSCs infected by F. nucleatum at day 3, 7, 14, and 21 (absolute of the fold-change of DEGs >2 and an adjusted P < 0.01).
References
Amitay, E. L., Werner, S., Vital, M., Pieper, D. H., Hofler, D., Gierse, I. J., et al. (2017). Fusobacterium and colorectal cancer: causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis 38, 781–788. doi: 10.1093/carcin/bgx053
Bartold, P. M., Gully, N. J., Zilm, P. S., and Rogers, A. H. (1991). Identification of components in Fusobacterium nucleatum chemostat-culture supernatants that are potent inhibitors of human gingival fibroblast proliferation. J. Periodont. Res. 26, 314–322. doi: 10.1111/j.1600-0765.1991.tb02069.x
Bello, S. M., Shimoyama, M., Mitraka, E., Laulederkind, S., Smith, C. L., Eppig, J. T., et al. (2018). Disease ontology: improving and unifying disease annotations across species. Dis. Model. Mech. 11:dmm032839. doi: 10.1242/dmm.032839
Bolstad, A. I., Jensen, H. B., and Bakken, V. (1996). Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin. Microbiol. Rev. 9, 55–71. doi: 10.1128/CMR.9.1.55
Brook, I., and Walker, R. I. (1986). The relationship between Fusobacterium species and other flora in mixed infection. J. Med. Microbiol. 21, 93–100. doi: 10.1099/00222615-21-2-93
Bui, F. Q., Johnson, L., Roberts, J., Hung, S. C., Lee, J., Atanasova, K. R., et al. (2016). Fusobacterium nucleatum infection of gingival epithelial cells leads to NLRP3 inflammasome-dependent secretion of IL-1beta and the danger signals ASC and HMGB1. Cell. Microbiol. 18, 970–981. doi: 10.1111/cmi.12560
Croft, D., O'Kelly, G., Wu, G., Haw, R., Gillespie, M., Matthews, L., et al. (2011). Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 39, D691–D697. doi: 10.1093/nar/gkq1018
Diniz, I. M., Chen, C., Ansari, S., Zadeh, H. H., Moshaverinia, M., Chee, D., et al. (2016). Gingival mesenchymal stem cell (GMSC) delivery system based on RGD-coupled alginate hydrogel with antimicrobial properties: a novel treatment modality for peri-implantitis. J. Prosthodont. 25, 105–115. doi: 10.1111/jopr.12316
Ebersole, J. L., Kesavalu, L., Schneider, S. L., Machen, R. L., and Holt, S. C. (1995). Comparative virulence of periodontopathogens in a mouse abscess model. Oral Dis. 1, 115–128. doi: 10.1111/j.1601-0825.1995.tb00174.x
Fawzy, E. K., and Dorfer, C. E. (2016). Gingival mesenchymal stem/progenitor cells: a unique tissue engineering gem. Stem Cells Int. 2016:7154327. doi: 10.1155/2016/7154327
Fournier, B. P., Ferre, F. C., Couty, L., Lataillade, J. J., Gourven, M., Naveau, A., et al. (2010). Multipotent progenitor cells in gingival connective tissue. Tissue Eng. Part A 16, 2891–2899. doi: 10.1089/ten.tea.2009.0796
Ge, S., Mrozik, K. M., Menicanin, D., Gronthos, S., and Bartold, P. M. (2012). Isolation and characterization of mesenchymal stem cell-like cells from healthy and inflamed gingival tissue: potential use for clinical therapy. Regen. Med. 7, 819–832. doi: 10.2217/rme.12.61
Gursoy, U. K., Koenonen, E., and Uitto, V. J. (2008). Stimulation of epithelial cell matrix metalloproteinase (MMP-2,-9,-13) and interleukin-8 secretion by fusobacteria. Oral Microbiol. Immunol. 23, 432–434. doi: 10.1111/j.1399-302X.2008.00453.x
Gursoy, U. K., Pollanen, M., Kononen, E., and Uitto, V. (2012). A novel organotypic dento-epithelial culture model: effect of Fusobacterium nucleatum biofilm on B-defensin-2,-3, and LL-37 expression. J Periodontol. 83, 242–247. doi: 10.1902/jop.2011.110177
Haake, S. K., and Lindemann, R. A. (1997). Fusobacterium nucleatum T18 aggregates human mononuclear cells and inhibits their PHA-stimulated proliferation. J Periodontol. 68, 39–44. doi: 10.1902/jop.1997.68.1.39
Hammerle, C. H., and Giannobile, W. V. (2014). Biology of soft tissue wound healing and regeneration–consensus report of Group 1 of the 10th European Workshop on Periodontology. J. Clin. Periodontol. 41(Suppl. 15), S1–S5. doi: 10.1111/jcpe.12221
Hanazawa, S. (1998). [Study on stimulatory mechanism of inflammatory bone resorption induced by Porphyromonas gingivalis]. Nihon saikingaku zasshi. Jpn J Bacteriol. 53, 571–586. doi: 10.3412/jsb.53.571
Hsieh, Y. Y., Tung, S. Y., Pan, H. Y., Yen, C. W., Xu, H. W., Lin, Y. J., et al. (2018). Increased abundance of Clostridium and Fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci. Rep. 8:158. doi: 10.1038/s41598-017-18596-0
Huang, G., Zhang, H. B., Dang, H. N., and Haake, S. K. (2004). Differential regulation of cytokine genes in gingival epithelial cells challenged by Fusobacterium nucleatum and Porphyromonas gingivalis. Microb. Pathog. 37, 303–312. doi: 10.1016/j.micpath.2004.10.003
Huynh, T., Kapur, R. V., Kaplan, C. W., Cacalano, N., Haake, S. K., et al. (2011). The role of aggregation in Fusobacterium nucleatum-induced immune cell death. J. Endodont. 37, 1531–1535. doi: 10.1016/j.joen.2011.06.034
Iohara, K., Nakashima, M., Ito, M., Ishikawa, M., Nakasima, A., and Akamine, A. (2004). Dentin regeneration by dental pulp stem cell therapy with recombinant human bone morphogenetic protein 2. J. Dent. Res. 83, 590–595. doi: 10.1177/154405910408300802
Jeng, J. H., Chan, C. P., Ho, Y. S., Lan, W. H., Hsieh, C. C., and Chang, M. C. (1999). Effects of butyrate and propionate on the adhesion, growth, cell cycle kinetics, and protein synthesis of cultured human gingival fibroblasts. J Periodontol. 70, 1435–1442. doi: 10.1902/jop.1999.70.12.1435
Jewett, A., Hume, W. R., Le, H., Huynh, T. N., Han, Y. W., Cheng, G., et al. (2000). Induction of apoptotic cell death in peripheral blood mononuclear and polymorphonuclear cells by an oral bacterium, Fusobacterium nucleatum. Infect. Immun. 68, 1893–1898. doi: 10.1128/IAI.68.4.1893-1898.2000
Ji, S., Shin, J. E., Kim, Y. S., Oh, J., Min, B., and Choi, Y. (2009). Toll-like receptor 2 and NALP2 mediate induction of human beta-defensins by Fusobacterium nucleatum in gingival epithelial cells. Infect. Immun. 77, 1044–1052. doi: 10.1128/IAI.00449-08
Jia, L., Han, N., Du, J., Guo, L., Luo, Z., and Liu, Y. (2019). Pathogenesis of important virulence factors of Porphyromonas gingivalis via toll-like receptors. Front. Cell. Infect. Microbiol. 9:262. doi: 10.3389/fcimb.2019.00262
Johnson, L., Coutinho Almeida-da-Silva, C. L., Takiya, C. M., Figliuolo, V., Rocha, G. M., Weissmueller, G., et al. (2018). Oral infection of mice with Fusobacterium nucleatum results in macrophage recruitment to the dental pulp and bone resorption. Biomed. J. 41, 184–193. doi: 10.1016/j.bj.2018.05.001
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kinder, S. A., and Holt, S. C. (1993). Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J. Bacteriol. 175, 840–850. doi: 10.1128/jb.175.3.840-850.1993
Li, N., Liu, N., Zhou, J., Tang, L., Ding, B., Duan, Y., et al. (2013). Inflammatory environment induces gingival tissue-specific mesenchymal stem cells to differentiate towards a pro-fibrotic phenotype. Biol. Cell 105, 261–275. doi: 10.1111/boc.201200064
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, Y., Zheng, Y., Ding, G., Fang, D., Zhang, C., Bartold, P. M., et al. (2008). Periodontal ligament stem cell-mediated treatment for periodontitis in miniature swine. Stem Cells 26, 1065–1073. doi: 10.1634/stemcells.2007-0734
Mao, Q., Nguyen, P. D., Shanti, R. M., Shi, S., Shakoori, P., Zhang, Q., et al. (2019). Gingiva-derived mesenchymal stem cell-extracellular vesicles activate schwann cell repair phenotype and promote nerve regeneration. Tissue Eng. Part A 25, 887–900. doi: 10.1089/ten.tea.2018.0176
Mendes, R. T., Nguyen, D., Stephens, D., Pamuk, F., Fernandes, D., Van Dyke, T. E., et al. (2016). Endothelial cell response to Fusobacterium nucleatum. Infect. Immun. 84, 2141–2148. doi: 10.1128/IAI.01305-15
Milward, M. R., Chapple, I. L. C., Wright, H. J., Millard, J. L., Matthews, J. B., and Cooper, P. R. (2007). Differential activation of NF-kappa B and gene expression in oral epithelial cells by periodontal pathogens. Clin. Exp. Immunol. 148, 307–324. doi: 10.1111/j.1365-2249.2007.03342.x
Mitrano, T. I., Grob, M. S., Carrion, F., Nova-Lamperti, E., Luz, P. A., Fierro, F. S., et al. (2010). Culture and characterization of mesenchymal stem cells from human gingival tissue. J. Periodontol. 81, 917–925. doi: 10.1902/jop.2010.090566
Moore, W. E., and Moore, L. V. (1994). The bacteria of periodontal diseases. Periodontol. 2000 5, 66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x
Nogueira, A. V. B., Nokhbehsaim, M., Eick, S., Bourauel, C., Jaeger, A., Jepsen, S., et al. (2014). Biomechanical loading modulates proinflammatory and bone resorptive mediators in bacterial-stimulated PDL cells. Mediat. Inflamm. 2014:425421. doi: 10.1155/2014/425421
Papapanou, P. N., Sellen, A., Wennstrom, J. L., and Dahlen, G. (1993). An analysis of the subgingival microflora in randomly selected subjects. Oral Microbiol. Immunol. 8, 24–29. doi: 10.1111/j.1399-302X.1993.tb00538.x
Paster, B. J., and Dewhirst, F. E. (2009). Molecular microbial diagnosis. Periodontol. 2000 51, 38–44. doi: 10.1111/j.1600-0757.2009.00316.x
Pinero, J., Bravo, A., Queralt-Rosinach, N., Gutierrez-Sacristan, A., Deu-Pons, J., Centeno, E., et al. (2017). DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–D839. doi: 10.1093/nar/gkw943
Polak, D., Wilensky, A., Shapira, L., Halabi, A., Goldstein, D., Weiss, E. I., et al. (2009). Mouse model of experimental periodontitis induced by Porphyromonas gingivalis/Fusobacterium nucleatum infection: bone loss and host response. J. Clin. Periodontol. 36, 406–410. doi: 10.1111/j.1600-051X.2009.01393.x
Ramage, G., Lappin, D. F., Millhouse, E., Malcolm, J., Jose, A., Yang, J., et al. (2017). The epithelial cell response to health and disease associated oral biofilm models. J. Periodont. Res. 52, 325–333. doi: 10.1111/jre.12395
Rogers, A. H., Zilm, P. S., Gully, N. J., Pfennig, A. L., and Marsh, P. D. (1991). Aspects of the growth and metabolism of Fusobacterium nucleatum ATCC 10953 in continuous culture. Oral Microbiol. Immunol. 6, 250–255. doi: 10.1111/j.1399-302X.1991.tb00486.x
Rossano, F., Rizzo, A., Sanges, M. R., Delero, G. C., and Tufano, M. A. (1993). Human monocytes and gingival fibroblasts release tumor-necrosis-factor-alpha, interleukin-1-alpha and interleukin-6 in response to particulate and soluble fractions of prevotelia-melaninogenica and Fusobacterium-nucleatum. Int. J. Clin. Lab. Res. 23, 165–168. doi: 10.1007/BF02592303
Rubinstein, M. R., Wang, X., Liu, W., Hao, Y., Cai, G., and Han, Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206. doi: 10.1016/j.chom.2013.07.012
Signat, B., Roques, C., Poulet, P., and Duffaut, D. (2011). Role of Fusobacterium nucleatum in periodontal health and disease. Curr. Issues Mol. Biol. 13, 25–35.
Socransky, S. S., and Haffajee, A. D. (2002). Dental biofilms: difficult therapeutic targets. Periodontol. 2000 28, 12–55. doi: 10.1034/j.1600-0757.2002.280102.x
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., and Kent, R. J. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x
Takada, H., Ogawa, T., Yoshimura, F., Otsuka, K., Kokeguchi, S., Kato, K., et al. (1988). Immunobiological activities of a porin fraction isolated from Fusobacterium nucleatum ATCC 10953. Infect. Immun. 56, 855–863.
Tang, L., Li, N., Xie, H., and Jin, Y. (2011). Characterization of mesenchymal stem cells from human normal and hyperplastic gingiva. J. Cell. Physiol. 226, 832–842. doi: 10.1002/jcp.22405
Tomar, G. B., Srivastava, R. K., Gupta, N., Barhanpurkar, A. P., Pote, S. T., Jhaveri, H. M., et al. (2010). Human gingiva-derived mesenchymal stem cells are superior to bone marrow-derived mesenchymal stem cells for cell therapy in regenerative medicine. Biochem. Biophys. Res. Commun. 393, 377–383. doi: 10.1016/j.bbrc.2010.01.126
Uitto, V. J., Baillie, D., Wu, Q., Gendron, R., Grenier, D., Putnins, E. E., et al. (2005). Fusobacterium nucleatum increase collagenase 3 production and migration of epithelial cells. Infect. Immun. 73, 1171–1179. doi: 10.1128/IAI.73.2.1171-1179.2005
von Mering, C., Jensen, L. J., Snel, B., Hooper, S. D., Krupp, M., Foglierini, M., et al. (2005). STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33, D433–D437. doi: 10.1093/nar/gki005
Wang, F., Yu, M., Yan, X., Wen, Y., Zeng, Q., Yue, W., et al. (2011). Gingiva-derived mesenchymal stem cell-mediated therapeutic approach for bone tissue regeneration. Stem Cells Dev. 20, 2093–2102. doi: 10.1089/scd.2010.0523
Wang, P., Duan, D., Zhou, X., Li, X., Yang, J., Deng, M., et al. (2015). Relationship between expression of human gingival beta-defensins and levels of periodontopathogens in subgingival plaque. J. Periodontal Res. 50, 113–122. doi: 10.1111/jre.12187
Wang, Q., Zhao, L., Xu, C., Zhou, J., and Wu, Y. (2019). Fusobacterium nucleatum stimulates monocyte adhesion to and transmigration through endothelial cells. Arch. Oral Biol. 100, 86–92. doi: 10.1016/j.archoralbio.2019.02.013
Yang, Y., Weng, W., Peng, J., Hong, L., Yang, L., Toiyama, Y., et al. (2017). Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappa B, and up-regulating expression of MicroRNA-21. Gastroenterology 152, 851–866.e24. doi: 10.1053/j.gastro.2016.11.018
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Keywords: Fusobacterium nucleatum, gingival mesenchymal stem cells, cell proliferation, inflammatory cytokines, osteogenesis differentiation
Citation: Kang W, Ji X, Zhang X, Tang D and Feng Q (2019) Persistent Exposure to Fusobacterium nucleatum Triggers Chemokine/Cytokine Release and Inhibits the Proliferation and Osteogenic Differentiation Capabilities of Human Gingiva-Derived Mesenchymal Stem Cells. Front. Cell. Infect. Microbiol. 9:429. doi: 10.3389/fcimb.2019.00429
Received: 01 June 2019; Accepted: 02 December 2019;
Published: 17 December 2019.
Edited by:
Jan Potempa, University of Louisville, United StatesReviewed by:
Frank C. Gibson III, University of Florida, United StatesDarryl J. Hill, University of Bristol, United Kingdom
Shinsmon Jose, University of Cincinnati, United States
Copyright © 2019 Kang, Ji, Zhang, Tang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Feng, fengqiang@sdu.edu.cn
 Wenyan Kang
Wenyan Kang Xiaoli Ji1,3
Xiaoli Ji1,3  Qiang Feng
Qiang Feng