Host Immunity to Malassezia in Health and Disease
- 1Section of Immunology, Vetsuisse Faculty, University of Zurich, Zurich, Switzerland
- 2Institute of Experimental Immunology, University of Zurich, Zurich, Switzerland
The microbiota plays an integral role in shaping physical and functional aspects of the skin. While a healthy microbiota contributes to the maintenance of immune homeostasis, dysbiosis can result in the development of diverse skin pathologies. This dichotomous feature of the skin microbiota holds true not only for bacteria, but also for fungi that colonize the skin. As such, the yeast Malassezia, which is by far the most abundant component of the skin mycobiota, is associated with a variety of skin disorders, of which some can be chronic and severe and have a significant impact on the quality of life of those affected. Understanding the causative relationship between Malassezia and the development of such skin disorders requires in-depth knowledge of the mechanism by which the immune system interacts with and responds to the fungus. In this review, we will discuss recent advances in our understanding of the immune response to Malassezia and how the implicated cells and cytokine pathways prevent uncontrolled fungal growth to maintain commensalism in the mammalian skin. We also review how the antifungal response is currently thought to affect the development and severity of inflammatory disorders of the skin and at distant sites.
Introduction
The skin, one of our body's largest organs, harbors a wide variety of microbial communities, including innocuous symbiotic organisms but also potential pathogens (Findley and Grice, 2014). Advances in our understanding of the molecular mechanisms of microbial virulence and host defense have improved the current view how dysbiosis and dysregulated immune responses against commensal microbes drive pathological conditions (Chen et al., 2018).
Fungi are increasingly being recognized as common members of the microbiota. The most prevalent fungi on the mammalian skin are those of the genus Malassezia, with >90% of all skin fungi belonging to this genus (Findley et al., 2013). Currently, 18 species of Malassezia have been identified, of which 10 were found in humans and the others in a growing number of animal hosts (Guillot and Bond, 2020). Although generally viewed as a commensal, Malassezia has also been associated with various dermatological conditions including mild diseases, such as dandruff and pityriasis versicolor, to more severe inflammatory diseases, such as seborrheic dermatitis and atopic dermatitis (AD) (Saunte et al., 2020). In rare cases, Malassezia has been reported to cause blood stream infections (Iatta et al., 2014). In dogs, overgrowth of Malassezia is associated with otitis and dermatitis and treatment of such disorders with antifungals often improves the conditions (Bond et al., 2020). In contrast to the situation in dogs, the association of Malassezia with skin disorders in humans primarily relies on clinical association studies, while a causative relationship remains a matter of debate and the mechanism of pathogenesis unclear. Dysbiosis with a higher fungal diversity and shifts in the relative abundance of certain Malassezia species have been implicated in AD. A large metagenomic study has reported an increase in the relative abundance of M. dermatis and M. sympodialis and a reduction of M. globosa on the skin of AD patients (Chng et al., 2016). Lack of consensus with other studies on the skin mycobiota in AD (Jo et al., 2017) may at least in part be due to sampling biases and discrepancies between culture- and sequence-based methods. In addition to the reported shifts in the species distribution between normal and diseased skin, intraspecies variations (Wu et al., 2015), which are known to alter phenotype and function in other fungal species (Ropars et al., 2018), may further complicate the situation. The pathogenicity of Malassezia spp. may also be modulated by mycoviruses that were recently identified in some isolates (Clancey et al., 2019; Park et al., 2019). Moreover, the microenvironment of the diseased skin, which is characterized by barrier disruption, lipid deficiency and elevated pH in case of AD (Weidinger and Novak, 2016), can modulate the metabolism and thereby the functional properties of the fungus (Chng et al., 2016). Inter-kingdom communications within the skin microbiota may also influence the capacity of Malassezia in promoting (or possibly preventing) skin disorders (Li et al., 2017). Finally, host factors such as genetics, immune status or comorbidities, can also influence the skin mycobiome composition and pathogenic potential (Jo et al., 2017) (Figure 1).
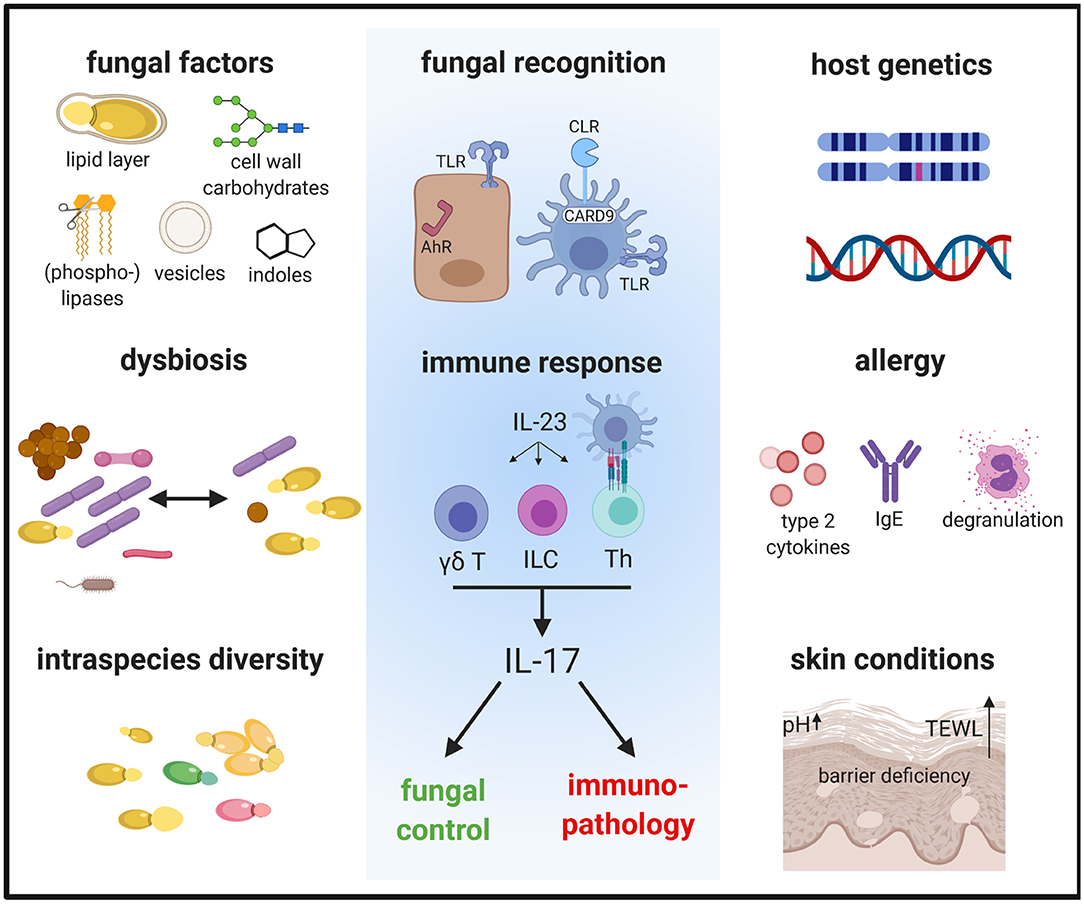
Figure 1. The immune response to Malassezia is influenced by fungal factors including cell wall constituents and secreted components, inter- and intraspecies variations and host factors such as genetics and skin or systemic predisposing conditions. These factors determine how Malassezia is recognized by the host and in turn how the host responds to the fungus. The antifungal response is characterized by the activation of the IL-23/IL-17 axis, which not only controls fungal growth but can also mediate immunopathology. AhR, aryl hydrocarbon receptor; CLR, C-type lectin receptor; γδ T, γδ T cells; ILC, innate lymphoid cells; TLR, Toll-like receptor; TEWL, trans-epidermal water loss; Th, T helper cells.
To understand the role of Malassezia in the development and severity of skin diseases it is important to understand the mechanisms of skin-fungus interactions in the (normal) mammalian skin.
Host Response to the Skin Commensal Yeast Malassezia
Constant exposure of the skin to commensal microbes results in a continuous activation of the cutaneous immune system. Active immunosurveillance of the microbiota is critical to prevent dysbiosis, microbial overgrowth and translocation across epithelial barriers as evidenced by the frequent occurrence of opportunistic infections in immunodeficient individuals (Pellicciotta et al., 2019).
Malassezia-host interactions are mediated via direct contacts as well as indirectly via secreted factors (Velegraki et al., 2015) and extracellular vesicles released from the fungus, which may assist the delivery of soluble mediators to host cells (Johansson et al., 2018; Zhang et al., 2019; Vallhov et al., 2020). These interactions have been studied primarily in vitro with cultured cells (Sparber and LeibundGut-Landmann, 2017). Reconstructed human epidermis and ex vivo skin models, which reflect the complexity of the skin more closely, have also been developed to study the cutaneous antifungal response (Corzo-Leon et al., 2019; Pedrosa et al., 2019). More recently, a murine model has become available that provides insights into the host response against Malassezia in vivo. In contrast to in vitro systems, it allows to study the contribution of circulating immune cells, which are not normally resident in the skin, to the cutaneous host response over prolonged periods of time (Sparber et al., 2019).
Toll-like receptors (TLRs) and C-type lectin receptors (CLRs) recognize carbohydrates in the fungal cell wall (Plato et al., 2015). Although the cell wall of Malassezia shows some differences to that of other fungal genera and is covered by a lipid-rich outer layer (Mittag, 1995; Kruppa et al., 2009; Stalhberger et al., 2014), CLRs are thought to play a prominent role in Malassezia sensing. Dectin-2 and Mincle have both been shown to bind Malassezia cell wall constituents resulting in Malassezia phagocytosis and cytokine production (Ishikawa et al., 2013; Haider et al., 2019). However, in vivo the two receptors do not appear to be essential for antifungal defense because knockout mice lacking either of the two receptors do not manifest an impaired immune response in the skin (Sparber et al., 2019). Functional redundancy between the receptors and/or with other CLRs may provide a likely explanation for reconciling the involvement of the downstream signaling adaptor Card9 in the cutaneous response to Malassezia (Sparber et al., 2019). The requirement of MyD88 for the antifungal response (Sparber et al., 2019) may be explained by the involvement of TLR2, which is also activated by Malassezia (Baroni et al., 2006) (Figure 1).
Card9-mediated signaling couples innate fungal recognition to the adaptive immune system, and importantly, it drives polarization of CD4+ T cells into IL-17A-secreting effector cells (LeibundGut-Landmann et al., 2007). While this was first observed with Candida albicans in humans and mice (LeibundGut-Landmann et al., 2007; Glocker et al., 2009), it also applies to Malassezia: Malassezia-specific T helper cells belong preferentially to the Th17 subset in both host species and Malassezia-specific Th17 response is abolished in Card9-deficient mice (Sparber et al., 2019).
αβ T cells are not the only IL-17A-producing cell type. In the skin, γδ T cells and innate lymphoid cells (ILCs) constitute prominent additional sources of IL-17A and respond swiftly to cytokine stimulation in an antigen-independent manner (Cua and Tato, 2010). Indeed, both γδ T cells and ILCs secrete IL-17A in the skin of Malassezia-colonized mice (Sparber et al., 2019) (Figure 1). Whether the activation of γδ T cells and ILCs is a consequence of the experimental infection and/or whether the innate cells contribute to long-term immunosurveillance of Malassezia commensalism, remains to be determined. Intriguingly, the Card9 pathway does not seem to be required for induction of innate IL-17A by Malassezia (Sparber et al., 2019). This is reminiscent of what has been reported for innate IL-17A induction in a murine model of oropharyngeal candidiasis (Bishu et al., 2014), although the underlying cause and the Card9-independent regulatory mechanisms are unclear so far.
Complementary to classical pattern recognition receptors such as CLRs, other classes of receptors are also implicated in the recognition of Malassezia in the skin. They are particularly relevant in keratinocytes, which lack Syk/Card9-coupled CLRs. Examples of keratinocyte-fungal interactions are provided by studies on C. albicans, which implicated receptors like E-cadherin, EphA2, or EGFR in the antifungal response (Phan et al., 2007; Swidergall et al., 2018; Ho et al., 2019). In case of Malassezia, the aryl hydrocarbon receptor (AhR), a ligand-dependent transcription factor, has gained attention because Malassezia-derived metabolites, in particular indoles, can trigger AhR signaling. Indole production was first reported for M. furfur (Gaitanis et al., 2008), but other species of Malassezia may also produce AhR ligands (Magiatis et al., 2013; Buommino et al., 2018) (Figure 1). The activation of AhR signaling can modulate skin homeostasis in manifold ways, including oxidation, epidermal barrier function, melanogenesis, and innate immunity (Furue et al., 2014). Importantly, AhR is implicated in type 17 immunity by promoting Th17 differentiation (Quintana et al., 2008; Veldhoen et al., 2008) and stimulating the production of IL-17A and related cytokines by Th17 cells, innate lymphocytes and ILCs (Veldhoen et al., 2008; Martin et al., 2009; Qiu et al., 2012). Whether and how AhR signaling modulates skin immunity and in particular the type 17 response to Malassezia remains an open question.
Consistently with the host-protective role of the IL-23/IL-17 immune axis in barrier tissues, IL-17 cytokines (including IL-17A and IL-17F, and possibly the epithelial cell-derived family member IL-17C, which is functionally related to IL-17A and IL-17F) prevents Malassezia overgrowth on the murine skin (Sparber et al., 2019). This is reminiscent of the well-known activity of IL-17 against other fungi, in particular C. albicans, on the skin and mucosal surfaces (Sparber and LeibundGut-Landmann, 2019). In contrast to the prominent role of IL-17 in protection from mucocutaneous candidiasis, the mechanisms of immunosurveillance of Malassezia in humans are likely more complex. No case has been reported to date where a genetic defect in the IL-17 pathway or an upstream signaling element manifests in detectable Malassezia overgrowth or development of Malassezia-associated skin disorders. Of note, an increased incidence of seborrheic dermatitis was reported as an adverse event accompanying administration of the anti-IL-17A antibody Ixekuzumab (Saeki et al., 2017), and a genetic variant of IL23R was found associated with a decrease in dandruff (Ehm et al., 2017). These observations support a link between the IL-23/IL-17 immune axis and Malassezia in humans. More data are needed to confirm these findings. Moreover, additional immune mechanisms involved in the control of Malassezia commensalism await to be discovered.
Malassezia-Induced Immunity and Immunopathology in the Skin
While type 17 immunity is primarily host-beneficial and supports a protective state by controlling commensal fungi in barrier tissues, IL-17 can also bear pathological potential if dysregulated. The IL-17 pathway is a strong driver of inflammation in several immune-mediated diseases, the prime example of which is psoriasis (McGeachy et al., 2019). Mechanistically, IL-17 signaling induces not only the production of antimicrobial peptides and tissue repair molecules, but also chemokines that attract inflammatory myeloid cells to the tissue and it promotes hyperproliferation of keratinocytes. The causative role of IL-17 in psoriasis is demonstrated by the therapeutic efficacy of neutralizing antibodies targeting IL-17 family cytokines and receptors (Hawkes et al., 2017). Although the initial trigger of the pathological IL-17 response in psoriasis is unknown, (skin) fungal commensals are likely involved due to their prominent IL-17-inducing capacity.
IL-17 has also been linked to AD (Koga et al., 2008), although the causative relationship between the cytokine and disease pathogenesis is less clear. It appears to be of particular importance in subtypes of the disease (Leonardi et al., 2015; Noda et al., 2015; Esaki et al., 2016). Staphylococcus aureus colonization of the skin is among the most well-known hallmarks of AD (Kong et al., 2012), and the induction of IL-17 in response to epicutaneous S. aureus critically mediates skin inflammation in an experimental setting (Liu et al., 2017; Nakagawa et al., 2017). Likewise, Malassezia promotes skin inflammation under AD-like conditions via the IL-23/IL-17 immune axis (Sparber et al., 2019). Support for the contribution of Malassezia-induced IL-17 to disease pathogenesis is further provided by the observation that Malassezia-specific Th17 cells are enriched in AD patients (Balaji et al., 2011; Sparber et al., 2019). Beyond AD, a link between the IL-23/IL-17 immune axis and Malassezia-associated disorders has also been proposed in seborrheic dermatitis (Wikramanayake et al., 2018) and dandruff (Ehm et al., 2017). Besides the prominent IL-17 profile, Malassezia-responsive T cells in humans also produce IFN-γ (Balaji et al., 2011; Bacher et al., 2019; Sparber et al., 2019), whereby the reason underlying the discrepancy between the two host species remains unclear.
The association of Malassezia with AD was first based on the observation that AD patients are often sensitized to Malassezia with Th2 cells and IgE that are directed against the fungus (Scalabrin et al., 1999; Zargari et al., 2001; Johansson et al., 2002; Balaji et al., 2011). The Malassezia-specific IgE titers were found to correlate with the severity of AD (Zhang et al., 2011; Glatz et al., 2015), albeit their pathogenic role in AD is not well-understood. An increase in Malassezia-specific serum IgE was also documented in canine AD (Khantavee et al., 2019) (Figure 1).
Important questions arise as to how Malassezia-specific Th2 cells are primed and whether they are a cause or a consequence of the allergic response in the atopic skin environment. It also remains unclear how the mixed Th2/Th17 response develops in allergic individuals and how the Th cell subsets are related to each other. Possible non-exclusive scenarios comprise that they may result from differential polarization or selective outgrowth of specific subsets. Clonotypic analysis of C. albicans-specific T cell subsets revealed intraclonal functional heterogeneity in support of a scenario of preferential outgrowth of individual subsets (Becattini et al., 2015). Mixed Th2/Th17 populations may also arise from T cell plasticity with intermediate T cells co-producing IL-4 and IL-17A and co-expressing GATA3 and RORγt as they have been observed in the blood and airways of asthmatic patients (Cosmi et al., 2010; Wang et al., 2010; Irvin et al., 2014) and in the skin of atopic individuals (Roesner et al., 2015).
Regarding the mechanism how Malassezia-specific T cells promote the allergic inflammation, it was speculated that cross-reactivity may be involved and thereby add an autoimmune component to the pathogenicity of AD. Among the Malassezia-derived antigens that have been identified are some that are phylogenetically highly conserved in mammals, such as thioredoxin and manganese-dependent superoxide dismutase (Schmid-Grendelmeier et al., 2005; Glaser et al., 2006; Balaji et al., 2011). Alternatively, Malassezia-specific T cells may also mediate immunopathology via cross-reactivity with other microbes as recently reported for C. albicans-specific T cells (Bacher et al., 2019). The emerging concept of cytokine-mediated and T cell receptor-independent bystander activation of T cells may represent another putative mechanism (Lee et al., 2019).
Given the pathogenic potential of the Malassezia-specific immune response, tight regulation is required to maintain commensalism. Induction of immune tolerance to skin commensal microbes has been investigated in case of Staphylococcus epidermidis. Antigen-specific regulatory T cells (Tregs) that accumulate during a critical developmental window in neonatal life were found important to balance the antimicrobial Th17 effector cells directed against the commensal (Scharschmidt et al., 2015). In a model of fungal commensalism with C. albicans however, Tregs and regulatory cytokines such as IL-10 appeared not essential for immune homeostasis and stable persistence of the fungus in the oral mucosa (Kirchner et al., 2019). Whether and how immune regulation contributes to immunosurveillance and prevents immunopathology in case of Malassezia remains to be determined.
The pathogenic effects of Malassezia in case of AD and other inflammatory skin disorder may also be mediated by T cell-independent mechanisms. Malassezia may promote disease by triggering the production of pro-inflammatory mediators in keratinocytes and tissue-resident immune cells in the atopic skin that are more accessible to the fungus, owing to barrier defects. Malassezia itself produces lipases and phospholipases that generate free fatty acids, which can damage the integrity of the skin and thereby might contribute to irritation and inflammation (Dawson, 2007; Saunders et al., 2012). Malassezia proteases may also interfere with cutaneous wound healing by degrading extracellular matrix components (Poh et al., 2020). While these various Malassezia-derived components have been studied in vitro, their relevance for promoting inflammation in the skin remains unclear. Complementary to the above described scenarios, Malassezia may also influence disease by modulating the virulence of other skin microbes such as S. aureus via inter-kingdom interactions (Li et al., 2017). It was postulated that such interactions could possibly be host beneficial rather than disease promoting by attenuating bacterial biofilm formation (Li et al., 2017), or by mechanisms such as those reported for Staphylococcus interspecies interactions (Nakatsuji et al., 2017).
Malassezia and Disease Beyond the Skin
Aside from its abundance on the skin, Malassezia has also been detected in extracutaneous sites. In particular, Malassezia sequences have been identified in the gastrointestinal tract in association with inflammatory bowel disease (IBD). Fungal dysbiosis in IBD is characterized with an enrichment of fungal taxa belonging to the Basidiomycota phylum, and this effect was balanced by an equivalent decrease in taxa belonging to the Ascomycota phylum (Sokol et al., 2017). More recently, in an ITS1 sequencing analysis of intestinal washings, Malassezia was associated with a subgroup of Crohn's disease patients carrying the CARD9 risk allele (Limon et al., 2019). Among the Malassezia sequences, M. restricta was most abundant. Relevance for the role of M. restricta in gut inflammation was provided by the observation that gastrointestinal delivery of the fungus aggravates the outcome of DSS-induced colitis in mice in a Card9-dependent manner (Limon et al., 2019). This phenotype was further linked to the induction of Th17 cells by M. restricta in the colonic lamina propria (Limon et al., 2019), whereby IL-17 is involved in the pathogenesis of disease in this model (Ito et al., 2008). It remains unclear whether the detection of Malassezia by sequencing approaches reflects transient relocation or true colonization of the gut by the fungus. In addition, it is unknown whether Malassezia-specific Th17 cells are involved in the pathogenesis of Crohn's disease, and if so, whether these cells get primed locally in response to fungal dysbiosis, or whether they translocate from the skin.
That Th17 cells directed against commensal fungi act at distal sites from where they have been induced has also been observed in other contexts with either host-protective or pathogenic consequences. In gut colonization experiments, C. albicans-induced Th17 cells protect from systemic candidiasis in a T cell- and IL-17-dependent manner (Shao et al., 2019). The protective effect extends to other extracellular pathogens, such as S. aureus (Shao et al., 2019). These effects are complemented by harmful consequences such as increased susceptibility to airway inflammation and exacerbation of asthma-like symptoms (Noverr et al., 2004; Shao et al., 2019). Besides these examples from experimental models of C. albicans colonization in mice, C. albicans-specific Th17 cells in human were also shown to cross-react with A. fumigatus and to expand in patients with A. fumigatus-driven lung pathology, suggesting an involvement in allergic asthma (Bacher et al., 2019). Examples of a skin-lung axis have been provided by bacterial skin commensals (Fyhrquist et al., 2014). It is tempting to speculate that Th17 cells primed against Malassezia in the skin may act in a similar way to promote the progression from AD to allergic asthma and rhinitis and thereby contribute to the atopic march.
Beyond its impact on the pathogenesis of inflammatory diseases, Malassezia was recently implicated in carcinogenesis. Among the microbes infiltrating pancreatic ductal adeno-carcinomas (PDA) tumors, Malassezia was markedly enriched (Aykut et al., 2019). In a murine model of PDA, Malassezia exerted a tumor-promoting effect via a mechanism involving mannose-binding lectin, a CLR and activator of complement and implicated in antifungal innate immunity (Aykut et al., 2019). Malassezia is likely a ligand of MBL, although direct binding has not been demonstrated. Whether fungal dysbiosis is cause or consequence of oncogenesis is not fully clear. The link between Malassezia and carcinogenesis may be more general, as fungal dysbiosis was also observed in colitis-associated cancer with an enrichment of Malassezia in the colonic mucosa associated fungal microbiota (Richard et al., 2018).
Concluding Remarks
Recent advances in the field have shed light on the immune response to Malassezia in the skin and on the divergent consequences that the antifungal response can have on the host. IL-17-dependent immunity against Malassezia contributes to the maintenance of fungal commensalism and skin homeostasis. The same pathway however can also have host-adverse effects in predisposed individuals, and this effect is likely not limited to the skin but also affects extracutaneous sites. The factors determining these context-dependent outcomes remain largely unclear. Moreover, it remains unknown to what extent the effects of the antifungal immunity on disease pathogenesis is shared or distinct between different Malassezia-associated disorders. Beyond immunological pathways, non-immunological factors such as neurotransmitters and possibly hormones affect the antifungal response, and this is likely also the case with Malassezia. Consistently, neuroendocrine changes are known to impact the course and severity of Malassezia-associated skin disorders. Future research will inform about the potential of therapeutically targeting fungal communities or the antifungal response for improving patient outcome.
Author Contributions
FS and SL-L wrote the minireview. FR created the figure with BioRender.com. All authors reviewed the manuscript.
Funding
Work in the LeibundGut lab was supported by the Swiss National Science Foundation (grant no. 310030_189255 to SL-L).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aykut, B., Pushalkar, S., Chen, R., Li, Q., Abengozar, R., Kim, J. I., et al. (2019). The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574, 264–267. doi: 10.1038/s41586-019-1608-2
Bacher, P., Hohnstein, T., Beerbaum, E., Rocker, M., Blango, M. G., Kaufmann, S., et al. (2019). Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell 176, 1340–1355 e15. doi: 10.1016/j.cell.2019.01.041
Balaji, H., Heratizadeh, A., Wichmann, K., Niebuhr, M., Crameri, R., Scheynius, A., et al. (2011). Malassezia sympodialis thioredoxin-specific T cells are highly cross-reactive to human thioredoxin in atopic dermatitis. J. Allergy Clin. Immunol. 128, 92–99.e4. doi: 10.1016/j.jaci.2011.02.043
Baroni, A., Orlando, M., Donnarumma, G., Farro, P., Iovene, M. R., Tufano, M. A., et al. (2006). Toll-like receptor 2 (TLR2) mediates intracellular signalling in human keratinocytes in response to Malassezia furfur. Arch. Dermatol. Res. 297, 280–288. doi: 10.1007/s00403-005-0594-4
Becattini, S., Latorre, D., Mele, F., Foglierini, M., De Gregorio, C., Cassotta, A., et al. (2015). T cell immunity. Functional heterogeneity of human memory CD4(+) T cell clones primed by pathogens or vaccines. Science 347, 400–406. doi: 10.1126/science.1260668
Bishu, S., Hernandez-Santos, N., Simpson-Abelson, M. R., Huppler, A. R., Conti, H. R., Ghilardi, N., et al. (2014). The adaptor CARD9 is required for adaptive but not innate immunity to oral mucosal Candida albicans infections. Infect. Immun. 82, 1173–1180. doi: 10.1128/IAI.01335-13
Bond, R., Morris, D. O., Guillot, J., Bensignor, E. J., Robson, D., Mason, K. V., et al. (2020). Biology, diagnosis and treatment of Malassezia dermatitis in dogs and cats: clinical consensus guidelines of the world association for veterinary dermatology. Vet. Dermatol. 31:75. doi: 10.1111/vde.12834
Buommino, E., Baroni, A., Papulino, C., Nocera, F. P., Coretti, L., Donnarumma, G., et al. (2018). Malassezia pachydermatis up-regulates AhR related CYP1A1 gene and epidermal barrier markers in human keratinocytes. Med. Mycol. 56, 987–993. doi: 10.1093/mmy/myy004
Chen, Y. E., Fischbach, M. A., and Belkaid, Y. (2018). Skin microbiota-host interactions. Nature 553, 427–436. doi: 10.1038/nature25177
Chng, K. R., Tay, A. S., Li, C., Ng, A. H., Wang, J., Suri, B. K., et al. (2016). Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat. Microbiol. 1:16106. doi: 10.1038/nmicrobiol.2016.106
Clancey, S. A., Ruchti, F., LeibundGut-Landmann, S., Heitman, J., and Ianiri, G. (2019). A novel mycovirus evokes transcriptional rewiring in Malassezia and provokes host inflammation and an immunological response. bioRxiv 12:880518. doi: 10.1101/2019.12.18.880518
Corzo-Leon, D. E., Munro, C. A., and MacCallum, D. M. (2019). An ex vivo human skin model to study superficial fungal infections. Front. Microbiol. 10:1172. doi: 10.3389/fmicb.2019.01172
Cosmi, L., Maggi, L., Santarlasci, V., Capone, M., Cardilicchia, E., Frosali, F., et al. (2010). Identification of a novel subset of human circulating memory CD4(+) T cells that produce both IL-17A and IL-4. J. Allergy Clin. Immunol. 125, 222–230 e1–4. doi: 10.1016/j.jaci.2009.10.012
Cua, D. J., and Tato, C. M. (2010). Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 10, 479–489. doi: 10.1038/nri2800
Dawson, T. L. Jr. (2007). Malassezia globosa and restricta: breakthrough understanding of the etiology and treatment of dandruff and seborrheic dermatitis through whole-genome analysis. J. Investig. Dermatol. Symp. Proc. 12, 15–19. doi: 10.1038/sj.jidsymp.5650049
Ehm, M. G., Aponte, J. L., Chiano, M. N., Yerges-Armstrong, L. M., Johnson, T., Barker, J. N., et al. (2017). Phenome-wide association study using research participants' self-reported data provides insight into the Th17 and IL-17 pathway. PLoS ONE 12:e0186405. doi: 10.1371/journal.pone.0186405
Esaki, H., Brunner, P. M., Renert-Yuval, Y., Czarnowicki, T., Huynh, T., Tran, G., et al. (2016). Early-onset pediatric atopic dermatitis is TH2 but also TH17 polarized in skin. J. Allergy Clin. Immunol. 138, 1639–1651. doi: 10.1016/j.jaci.2016.07.013
Findley, K., and Grice, E. A. (2014). The skin microbiome: a focus on pathogens and their association with skin disease. PLoS Pathog. 10:e1004436. doi: 10.1371/journal.ppat.1004436
Findley, K., Oh, J., Yang, J., Conlan, S., Deming, C., Meyer, J. A., et al. (2013). Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370. doi: 10.1038/nature12171
Furue, M., Takahara, M., Nakahara, T., and Uchi, H. (2014). Role of AhR/ARNT system in skin homeostasis. Arch. Dermatol. Res. 306, 769–779. doi: 10.1007/s00403-014-1481-7
Fyhrquist, N., Ruokolainen, L., Suomalainen, A., Lehtimaki, S., Veckman, V., Vendelin, J., et al. (2014). Acinetobacter species in the skin microbiota protect against allergic sensitization and inflammation. J. Allergy Clin. Immunol. 134, 1301–1309 e11. doi: 10.1016/j.jaci.2014.07.059
Gaitanis, G., Magiatis, P., Stathopoulou, K., Bassukas, I. D., Alexopoulos, E. C., Velegraki, A., et al. (2008). AhR ligands, malassezin, and indolo[3,2-b]carbazole are selectively produced by Malassezia furfur strains isolated from seborrheic dermatitis. J. Invest. Dermatol. 128, 1620–1625. doi: 10.1038/sj.jid.5701252
Glaser, A. G., Limacher, A., Fluckiger, S., Scheynius, A., Scapozza, L., and Crameri, R. (2006). Analysis of the cross-reactivity and of the 1.5 A crystal structure of the Malassezia sympodialis Mala s 6 allergen, a member of the cyclophilin pan-allergen family. Biochem. J. 396, 41–49. doi: 10.1042/BJ20051708
Glatz, M., Buchner, M., von Bartenwerffer, W., Schmid-Grendelmeier, P., Worm, M., Hedderich, J., et al. (2015). Malassezia spp.-specific immunoglobulin E level is a marker for severity of atopic dermatitis in adults. Acta Derm. Venereol. 95, 191–196. doi: 10.2340/00015555-1864
Glocker, E. O., Hennigs, A., Nabavi, M., Schaffer, A. A., Woellner, C., Salzer, U., et al. (2009). A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N. Engl. J. Med. 361, 1727–1735. doi: 10.1056/NEJMoa0810719
Guillot, J., and Bond, R. (2020). Malassezia yeasts in veterinary dermatology: an updated overview. Front. Cell. Infect. Microbiol. 10:79. doi: 10.3389/fcimb.2020.00079
Haider, M., Dambuza, I. M., Asamaphan, P., Stappers, M., Reid, D., Yamasaki, S., et al. (2019). The pattern recognition receptors dectin-2, mincle, and FcRgamma impact the dynamics of phagocytosis of Candida, Saccharomyces, Malassezia, and Mucor species. PLoS ONE 14:e0220867. doi: 10.1371/journal.pone.0220867
Hawkes, J. E., Chan, T. C., and Krueger, J. G. (2017). Psoriasis pathogenesis and the development of novel targeted immune therapies. J. Allergy Clin. Immunol. 140, 645–653. doi: 10.1016/j.jaci.2017.07.004
Ho, J., Yang, X., Nikou, S. A., Kichik, N., Donkin, A., Ponde, N. O., et al. (2019). Candidalysin activates innate epithelial immune responses via epidermal growth factor receptor. Nat. Commun. 10:2297. doi: 10.1038/s41467-019-09915-2
Iatta, R., Cafarchia, C., Cuna, T., Montagna, O., Laforgia, N., Gentile, O., et al. (2014). Bloodstream infections by Malassezia and Candida species in critical care patients. Med. Mycol. 52, 264–269. doi: 10.1093/mmy/myt004
Irvin, C., Zafar, I., Good, J., Rollins, D., Christianson, C., Gorska, M. M., et al. (2014). Increased frequency of dual-positive TH2/TH17 cells in bronchoalveolar lavage fluid characterizes a population of patients with severe asthma. J. Allergy Clin. Immunol. 134, 1175–1186 e7. doi: 10.1016/j.jaci.2014.05.038
Ishikawa, T., Itoh, F., Yoshida, S., Saijo, S., Matsuzawa, T., Gonoi, T., et al. (2013). Identification of distinct ligands for the C-type lectin receptors Mincle and Dectin-2 in the pathogenic fungus Malassezia. Cell Host Microbe 13, 477–488. doi: 10.1016/j.chom.2013.03.008
Ito, R., Kita, M., Shin-Ya, M., Kishida, T., Urano, A., Takada, R., et al. (2008). Involvement of IL-17A in the pathogenesis of DSS-induced colitis in mice. Biochem. Biophys. Res. Commun. 377, 12–16. doi: 10.1016/j.bbrc.2008.09.019
Jo, J. H., Kennedy, E. A., and Kong, H. H. (2017). Topographical and physiological differences of the skin mycobiome in health and disease. Virulence 8, 324–333. doi: 10.1080/21505594.2016.1249093
Johansson, C., Eshaghi, H., Linder, M. T., Jakobson, E., and Scheynius, A. (2002). Positive atopy patch test reaction to Malassezia furfur in atopic dermatitis correlates with a T helper 2-like peripheral blood mononuclear cells response. J. Invest. Dermatol. 118, 1044–1051. doi: 10.1046/j.1523-1747.2002.01758.x
Johansson, H. J., Vallhov, H., Holm, T., Gehrmann, U., Andersson, A., Johansson, C., et al. (2018). Extracellular nanovesicles released from the commensal yeast Malassezia sympodialis are enriched in allergens and interact with cells in human skin. Sci. Rep. 8:9182. doi: 10.1038/s41598-018-27451-9
Khantavee, N., Chanthick, C., Sookrung, N., and Prapasarakul, N. (2019). Antibody levels to Malassezia pachydermatis and Staphylococcus pseudintermedius in atopic dogs and their relationship with lesion scores. Vet. Dermatol. 31, 111–118. doi: 10.1111/vde.12802
Kirchner, F. R., Littringer, K., Altmeier, S. V., Tran, D. T., Schonherr, F., Lemberg, C., et al. (2019). Persistence of Candida albicans in the oral mucosa induces a curbed inflammatory host response that is independent of immunosuppression. Front. Immunol. 10:330. doi: 10.3389/fimmu.2019.00330
Koga, C., Kabashima, K., Shiraishi, N., Kobayashi, M., and Tokura, Y. (2008). Possible pathogenic role of Th17 cells for atopic dermatitis. J. Invest. Dermatol. 128, 2625–2630. doi: 10.1038/jid.2008.111
Kong, H. H., Oh, J., Deming, C., Conlan, S., Grice, E. A., Beatson, M. A., et al. (2012). Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859. doi: 10.1101/gr.131029.111
Kruppa, M. D., Lowman, D. W., Chen, Y. H., Selander, C., Scheynius, A., Monteiro, M. A., et al. (2009). Identification of (1–>6)-beta-D-glucan as the major carbohydrate component of the Malassezia sympodialis cell wall. Carbohydr. Res. 344, 2474–2479. doi: 10.1016/j.carres.2009.09.029
Lee, H. G., Lee, J. U., Kim, D. H., Lim, S., Kang, I., and Choi, J. M. (2019). Pathogenic function of bystander-activated memory-like CD4(+) T cells in autoimmune encephalomyelitis. Nat. Commun. 10:709. doi: 10.1038/s41467-019-08482-w
LeibundGut-Landmann, S., Gross, O., Robinson, M. J., Osorio, F., Slack, E. C., Tsoni, S. V., et al. (2007). Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat. Immunol. 8, 630–638. doi: 10.1038/ni1460
Leonardi, S., Cuppari, C., Manti, S., Filippelli, M., Parisi, G. F., Borgia, F., et al. (2015). Serum interleukin 17, interleukin 23, and interleukin 10 values in children with atopic eczema/dermatitis syndrome (AEDS): association with clinical severity and phenotype. Allergy Asthma Proc. 36, 74–81. doi: 10.2500/aap.2015.36.3808
Li, H., Goh, B., Teh, W. K., Jiang, Z.J, Goh, P. Z., Goh, A., et al. (2017). Skin commensal Malassezia globosa secreted protease attenuates Staphylococcus aureus biofilm formation. J. Invest. Dermatol. 138, 1137–1145. doi: 10.1016/j.jid.2017.11.034
Limon, J. J., Tang, J., Li, D., Wolf, A. J., Michelsen, K. S., Funari, V., et al. (2019). Malassezia is associated with crohn's disease and exacerbates colitis in mouse models. Cell Host Microbe 25, 377-388 e6. doi: 10.1016/j.chom.2019.01.007
Liu, H., Archer, N. K., Dillen, C. A., Wang, Y., Ashbaugh, A. G., Ortines, R. V., et al. (2017). Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-mediated T cell responses. Cell Host Microbe 22, 653–666 e5. doi: 10.1016/j.chom.2017.10.006
Magiatis, P., Pappas, P., Gaitanis, G., Mexia, N., Melliou, E., Galanou, M., et al. (2013). Malassezia yeasts produce a collection of exceptionally potent activators of the Ah (dioxin) receptor detected in diseased human skin. J. Invest. Dermatol. 133, 2023–2030. doi: 10.1038/jid.2013.92
Martin, B., Hirota, K., Cua, D. J., Stockinger, B., and Veldhoen, M. (2009). Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity 31, 321–330. doi: 10.1016/j.immuni.2009.06.020
McGeachy, M. J., Cua, D. J., and Gaffen, S. L. (2019). The IL-17 family of cytokines in health and disease. Immunity 50, 892–906. doi: 10.1016/j.immuni.2019.03.021
Mittag, H. (1995). Fine structural investigation of Malassezia furfur. II. The envelope of the yeast cells. Mycoses 38, 13–21. doi: 10.1111/j.1439-0507.1995.tb00003.x
Nakagawa, S., Matsumoto, M., Katayama, Y., Oguma, R., Wakabayashi, S., Nygaard, T., et al. (2017). Staphylococcus aureus virulent PSMalpha peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation. Cell Host Microbe 22, 667–677 e5. doi: 10.1016/j.chom.2017.10.008
Nakatsuji, T., Chen, T. H., Narala, S., Chun, K. A., Two, A. M., Yun, T., et al. (2017). Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9:eaah4680. doi: 10.1126/scitranslmed.aah4680
Noda, S., Suarez-Farinas, M., Ungar, B., Kim, S. J., de Guzman Strong, C., Xu, H., et al. (2015). The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J. Allergy Clin. Immunol. 136, 1254–1264. doi: 10.1016/j.jaci.2015.08.015
Noverr, M. C., Noggle, R. M., Toews, G. B., and Huffnagle, G. B. (2004). Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect. Immun. 72, 4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004
Park, M., Chon, Y. J., Kim, D., Yang, C. S., Lee, S. M., Dawson, T.L. Jr., et al. (2019). A novel totivirus alters gene expression and vacuolar morphology in Malassezia cells and induces a TLR3-mediated inflammatory immune response. bioRxiv 12:880526. doi: 10.1101/2019.12.17.880526
Pedrosa, A. F., Lisboa, C., Branco, J., Pellevoisin, C., Miranda, I. M., and Rodrigues, A. G. (2019). Malassezia interaction with a reconstructed human epidermis: Keratinocyte immune response. Mycoses 62, 932–936. doi: 10.1111/myc.12965
Pellicciotta, M., Rigoni, R., Falcone, E. L., Holland, S. M., Villa, A., and Cassani, B. (2019). The microbiome and immunodeficiencies: lessons from rare diseases. J. Autoimmun. 98, 132–148. doi: 10.1016/j.jaut.2019.01.008
Phan, Q. T., Myers, C. L., Fu, Y., Sheppard, D. C., Yeaman, M. R., Welch, W. H., et al. (2007). Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5:e64. doi: 10.1371/journal.pbio.0050064
Plato, A., Hardison, S. E., and Brown, G. D. (2015). Pattern recognition receptors in antifungal immunity. Semin. Immunopathol. 37, 97–106. doi: 10.1007/s00281-014-0462-4
Poh, S. E., Goh, J., Chen, F., Chua, W., Gan, S. Q., Lim, P. L., et al. (2020). Identification of Malassezia furfur secreted aspartyl protease 1 (MfSAP1) and its role in extracellular matrix degradation. Front. Cell. Infect. Microbiol. 10:148. doi: 10.3389/fcimb.2020.00148
Qiu, J., Heller, J. J., Guo, X., Chen, Z. M., Fish, K., Fu, Y. X., et al. (2012). The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36, 92–104. doi: 10.1016/j.immuni.2011.11.011
Quintana, F. J., Basso, A. S., Iglesias, A. H., Korn, T., Farez, M. F., and Bettelli, E. (2008). Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453, 65–71. doi: 10.1038/nature06880
Richard, M. L., Liguori, G., Lamas, B., Brandi, G., da Costa, G., Hoffmann, T. W., et al. (2018). Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 9, 131–142. doi: 10.1080/19490976.2017.1379637
Roesner, L. M., Heratizadeh, A., Begemann, G., Kienlin, P., Hradetzky, S., and Niebuhr, M. (2015). Der p1 and Der p2-Specific T cells display a Th2, Th17, and Th2/Th17 phenotype in atopic dermatitis. J. Invest. Dermatol. 135, 2324–2327. doi: 10.1038/jid.2015.162
Ropars, J., Maufrais, C., Diogo, D., Marcet-Houben, M., Perin, A., Sertour, N., et al. (2018). Gene flow contributes to diversification of the major fungal pathogen Candida albicans. Nat. Commun. 9:2253. doi: 10.1038/s41467-018-04787-4
Saeki, H., Nakagawa, H., Nakajo, K., Ishii, T., Morisaki, Y., Aoki, T., et al. (2017). Efficacy and safety of ixekizumab treatment for Japanese patients with moderate to severe plaque psoriasis, erythrodermic psoriasis and generalized pustular psoriasis: results from a 52-week, open-label, phase 3 study (UNCOVER-J). J. Dermatol. 44, 355–362. doi: 10.1111/1346-8138.13622
Saunders, C. W., Scheynius, A., and Heitman, J. (2012). Malassezia fungi are specialized to live on skin and associated with dandruff, eczema, and other skin diseases. PLoS Pathog. 8:e1002701. doi: 10.1371/journal.ppat.1002701
Saunte, D. M. L., Gaitanis, G., and Hay, R. J. (2020). Malassezia-associated skin diseases, the use of diagnostics and treatment. Front. Cell. Infect. Microbiol. 10:112. doi: 10.3389/fcimb.2020.00112
Scalabrin, D. M., Bavbek, S., Perzanowski, M. S., Wilson, B. B., Platts-Mills, T. A., and Wheatley, L. M. (1999). Use of specific IgE in assessing the relevance of fungal and dust mite allergens to atopic dermatitis: a comparison with asthmatic and nonasthmatic control subjects. J. Allergy Clin. Immunol. 104, 1273–1279. doi: 10.1016/S0091-6749(99)70024-2
Scharschmidt, T. C., Vasquez, K. S., Truong, H. A., Gearty, S. V., Pauli, M. L., Nosbaum, A., et al. (2015). A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43, 1011–1021. doi: 10.1016/j.immuni.2015.10.016
Schmid-Grendelmeier, P., Fluckiger, S., Disch, R., Trautmann, A., Wuthrich, B., Blaser, K., et al. (2005). IgE-mediated and T cell-mediated autoimmunity against manganese superoxide dismutase in atopic dermatitis. J. Allergy Clin. Immunol. 115, 1068–1075. doi: 10.1016/j.jaci.2005.01.065
Shao, T. Y., Wang, X. G., Jiang, T. T., Huang, F. S., Andersen, H., Kinder, J. M., et al. (2019). Commensal Candida albicans positively calibrates systemic Th17 immunological responses. Cell Host Microbe 25, 404–417 e6. doi: 10.1016/j.chom.2019.02.004
Sokol, H., Leducq, V., Aschard, H., Pham, H. P., Jegou, S., Landman, C., et al. (2017). Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. doi: 10.1136/gutjnl-2015-310746
Sparber, F., De Gregorio, C., Steckholzer, S., Ferreira, F. M., Dolowschiak, T., Ruchti, F., et al. (2019). The skin commensal yeast malassezia triggers a Type 17 response that coordinates anti-fungal immunity and exacerbates skin inflammation. Cell Host Microbe 25, 389–403 e6. doi: 10.1016/j.chom.2019.02.002
Sparber, F., and LeibundGut-Landmann, S. (2017). Host responses to Malassezia spp. in the Mammalian Skin. Front. Immunol. 8:1614. doi: 10.3389/fimmu.2017.01614
Sparber, F., and LeibundGut-Landmann, S. (2019). Interleukin-17 in Antifungal Immunity. Pathogens 8:54. doi: 10.3390/pathogens8020054
Stalhberger, T., Simenel, C., Clavaud, C., Eijsink, V. G., Jourdain, R., Delepierre, M., et al. (2014). Chemical organization of the cell wall polysaccharide core of Malassezia restricta. J. Biol. Chem. 289, 12647–12656. doi: 10.1074/jbc.M113.547034
Swidergall, M., Solis, N. V., Lionakis, M. S., and Filler, S. G. (2018). EphA2 is an epithelial cell pattern recognition receptor for fungal beta-glucans. Nat. Microbiol. 3, 53–61. doi: 10.1038/s41564-017-0059-5
Vallhov, H., Johansson, C., Veerman, R. E., and Scheynius, A. (2020). Extracellular vesicles released from the skin commensal yeast Malassezia sympodialis activate human primary keratinocytes. Front. Cell Infect. Microbiol. 10:6. doi: 10.3389/fcimb.2020.00006
Veldhoen, M., Hirota, K., Westendorf, A. M., Buer, J., Dumoutier, L., Renauld, J. C., et al. (2008). The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109. doi: 10.1038/nature06881
Velegraki, A., Cafarchia, C., Gaitanis, G., Iatta, R., and Boekhout, T. (2015). Malassezia infections in humans and animals: pathophysiology, detection, and treatment. PLoS Pathog. 11:e1004523. doi: 10.1371/journal.ppat.1004523
Wang, Y. H., Voo, K. S., Liu, B., Chen, C. Y., Uygungil, B., Spoede, W., et al. (2010). A novel subset of CD4(+) T(H)2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. J. Exp. Med. 207, 2479–2491. doi: 10.1084/jem.20101376
Weidinger, S., and Novak, N. (2016). Atopic dermatitis. Lancet 387, 1109–1122. doi: 10.1016/S0140-6736(15)00149-X
Wikramanayake, T. C., Hirt, P., Almastadi, M., Mitchell, H., Tomic-Canic, M., Romero, L., et al. (2018). Increased IL-17-expressing gammadelta T cells in seborrhoeic dermatitis-like lesions of the Mpzl3 knockout mice. Exp. Dermatol. 27, 1408–1411. doi: 10.1111/exd.13798
Wu, G., Zhao, H., Li, C., Rajapakse, M. P., Wong, W. C., Xu, J., et al. (2015). Genus-wide comparative genomics of malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet. 11:e1005614. doi: 10.1371/journal.pgen.1005614
Zargari, A., Eshaghi, H., Back, O., Johansson, S., and Scheynius, A. (2001). Serum IgE reactivity to Malassezia furfur extract and recombinant M. furfur allergens in patients with atopic dermatitis. Acta Derm. Venereol. 81, 418–422. doi: 10.1080/000155501317208363
Zhang, E., Tanaka, T., Tajima, M., Tsuboi, R., Kato, H., Nishikawa, A., et al. (2011). Anti-malassezia-specific ige antibodies production in japanese patients with head and neck atopic dermatitis: relationship between the level of specific IgE antibody and the colonization frequency of cutaneous malassezia species and clinical severity. J. Allergy 2011:645670. doi: 10.1155/2011/645670
Keywords: mycobiota, commensalism, immunopathology, cutaneous immunity, Malassezia
Citation: Sparber F, Ruchti F and LeibundGut-Landmann S (2020) Host Immunity to Malassezia in Health and Disease. Front. Cell. Infect. Microbiol. 10:198. doi: 10.3389/fcimb.2020.00198
Received: 11 March 2020; Accepted: 15 April 2020;
Published: 05 May 2020.
Edited by:
Amariliz Rivera, New Jersey Medical School, United StatesReviewed by:
Joshua J. Obar, Dartmouth College, United StatesIrina Leonardi, Cornell University, United States
Copyright © 2020 Sparber, Ruchti and LeibundGut-Landmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Florian Sparber, florian.sparber@uzh.ch
 Florian Sparber
Florian Sparber Fiorella Ruchti
Fiorella Ruchti Salomé LeibundGut-Landmann
Salomé LeibundGut-Landmann