Loss of Function of Scavenger Receptor SCAV-5 Protects C. elegans Against Pathogenic Bacteria
- Key Laboratory of the Ministry of Education for Medicinal Plant Resources and Natural Pharmaceutical Chemistry, National Engineering Laboratory for Resource Development of Endangered Crude Drugs in the Northwest of China, College of Life Sciences, Shaanxi Normal University, Xi’an, China
Scavenger receptors play a critical role in innate immunity by acting as the pattern-recognition receptors. There are six class B scavenger receptors homologs in C. elegans. However, it remains unclear whether they are required for host defense against bacterial pathogens. Here, we show that, of the six SCAV proteins, only loss of function scav-5 protect C. elegans against pathogenic bacteria S. typhimurium SL1344 and P. aeruginosa PA14 by different mechanism. scav-5 mutants are resistant to S. typhimurium SL1344 due to dietary restriction. While scav-5 acts upstream of or in parallel to tir-1 in conserved PMK-1 p38 MAPK pathway to upregulate the innate immune response to defend worms against P. aeruginosa PA14. This is the first demonstration of a role for SCAV-5 in host defense against pathogenic bacteria. Our results provide an important basis for further elucidating the underlying molecular mechanism by which scav-5 regulates innate immune responses.
Introduction
Scavenger receptors are eight classes transmembrane glycoproteins that first defined by its ability to bind and subsequently internalize modified low-density lipoproteins (mLDL) (PrabhuDas et al., 2017). Later, a large repertoire of ligands such as lipoproteins, cholesterol esters, phospholipids, proteoglycans, and carbohydrates have been identified that can be recognized by Scavenger receptors. Thus, Scavenger receptors are involved in an impressively broad range of functions including lipid metabolism, antigen presentation, phagocytosis, clearance of apoptotic cells, and innate immunity (Silverstein and Febbraio, 2009; Graham et al., 2011; He et al., 2011; Canton et al., 2013). Innate immunity is the first line of defense against pathogens, which is critical to maintain homeostasis, prevent infection, and activate the adaptive immune response. The components of innate immunity include external physical and chemical barriers, mucous membranes, internal humoral and, cellular effector mechanisms (Riera Romo et al., 2016). It is now clear that many scavenger receptors can recognize conserved patterns unique to microbial surfaces that are referred to as pathogen-associated patterns (PAMPs) (Mukhopadhyay and Gordon, 2004; Pluddemann et al., 2006; Pluddemann et al., 2011). On the other hand, scavenger receptors can be used as co-receptors by bacteria and viruses for entry into host cells (Haraga et al., 2008; Hawkes et al., 2010; Que et al., 2013).
The nematode Caenorhabditis elegans has been used as a model for studying bacterial virulence and innate immunity (Irazoqui et al., 2010). Despite lacking both Toll and Imd pathways as well as the adaptive immune system (Anderson et al., 1985; Gottar et al., 2002), C. elegans has developed special mechanisms that include innate immune response, bacteria avoidance behavior, and RNA interference to defend against pathogenic bacteria throughout evolution (Aliyari and Ding, 2009; Taffoni and Pujol, 2015). In fact, the absence of adaptive immune response makes C. elegans very useful for dissecting the innate immune mechanisms in pathogen-host interactions. The primary food source of C. elegans in laboratory is the E. coli strain OP50, but other bacteria and fungi also support its growth and reproduction. Several feed-based infection models have been established, such as Salmonella typhimurium and Pseudomonas aeruginosa (Tan et al., 1999; Aballay et al., 2000), to study the regulation between intestinal infection and innate immunity (Alegado et al., 2011; Kirienko et al., 2013; Curt et al., 2014). Salmonella typhimurium SL1344 proliferates and establishes a persistent infection in the intestine of C. elegans, resulting in the eventual death of the worm by actively inhibiting innate immune pathways (Aballay et al., 2000; Desai et al., 2019). The human pathogen Pseudomonas aeruginosa PA14 kills worms within 4–24 hours by the production several diffusible toxins in ‘fast killing’ model (Cezairliyan et al., 2013), ultimately suppresses the FOXO/DAF-16 innate immunity pathway in the intestine of C. elegans (Zheng et al., 2021). The innate immunity in C. elegans is regulated by several major pathways to defense against pathogens including PMK-1 p38 MAPK pathway. In the PMK-1 pathway, TIR-1 functions upstream of the NSY-1–SEK-1–PMK-1 p38 MAPK cascade (Kim et al., 2002; Couillault et al., 2004; Andrusiak and Jin, 2016). In this cascade, NSY-1 phosphorylates SEK-1, SEK-1 phosphorylates PMK-1, and finally activates PMK-1 to regulate pathogen response genes (Tanaka-Hino et al., 2002).
SCAV-1-6 are the six scavenger receptors homologs in C. elegans, belong to class B scavenger receptors (SRB). There are three members in SRB in mammals including CD36, SRB-I/II (SRB-II is a splicing variant of SRB-I), and lysosomal integral membrane protein 2 (Means et al., 2009). SRB has been reported to be involved in innate immunity (Feng et al., 2011). However, whether SCAV-1-6 have effects on defense against pathogenic bacteria in C. elegans and the underlying mechanism remains unclear.
Here, we investigate the functions of SCAV-1-6 in innate immunity in C. elegans. We find that scav-5 mutants can effectively defend C. elegans against pathogenic bacteria S. typhimurium SL1344 and P. aeruginosa PA14. In addition, we show that the mechanism of protecting C. elegans against the above two pathogenic bacteria is quite different by scav-5 loss of function. The scav-5 mutants displayed reduced pharyngeal pumping after S. typhimurium SL1344 infection, which lead to dietary restriction and extended lifespan. While, after P. aeruginosa PA14 infection, innate immune response was activated through PMK-1 p38 MAPK pathways in scav-5 mutants. Moreover, our genetic epistatic analysis indicates that scav-5 may function upstream of, or in parallel to tir-1 in PMK-1 innate immune response pathway to protect C. elegans against P. aeruginosa PA14. Our results provide an important basis for further elucidating the underlying mechanism of how scav-5 regulates innate immune response.
Materials And Methods
C. elegans and Bacterial Strains
The C. elegans strains used in this study were N2, scav-2(ok877), scav-3(ok1286) and scav-5(ok1606). C. elegans were maintained on NGM plates with E. coli OP50 or HT115 bacteria.
RNAi of scav-1, scav-4, scav-6
Three RNAi constructs were created using 700 bp, 899 bp and 730 bp segments of scav-1, scav-4, scav-6 coding region (bases 1 to 700, 3101 to 4000, and 1to 730, respectively), which were amplified by PCR and cloned into the vector pPD129.36, These plasmids were transformed into the RNAi bacteria strain HT115, and RNAi experiments were carried out with these strains following established protocols (Timmons et al., 2001) by using HT115 bacteria expressing the empty vector pPD129.36 as the control. For all RNAi experiments, the synchronization egg of the old-adult N2 spawn on NGM plates with RNAi bacteria, and L4 stage animals were used for subsequent studies, as described in more detail below.
C. elegans Pathogenic Bacteria Infection Assay
S. typhimurium SL1344 were pipetted on NGM plates and incubated overnight. P. aeruginosa PA14 infect C.elgans with slow killing way, which P. aeruginosa PA14 were pipetted on 0.35% NGM low osmotic pressure medium. Plates were incubated for 24 hours at 37°C, and 24 hours at 25°C (Shivers et al., 2010). C.elegans were cultured on these plates based on eating pathogenic bacteria established infection model.
Measurement of Lifespan and Heat Resistance
The indicated genotypes old-adult animals spawn on NGM plates with E. coli OP50, and L4 stage animals were picked on new NGM plates with E. coli OP50. Each NGM plate culture approximately 25 animals and each group amount have 80 animals at least. Animals of L4 stage as zero-day in lifespan count. Thermotolerance assays were performed as described (McColl et al., 2010). Briefly, WT and scav-5(ok1606) L4 stage hermaphrodites were infected with S. typhimurium SL1344 and P. aeruginosa PA14 for 24 hours respectively, adults were transferred to 35°C and scored as alive or dead based on responding to prodding with a platinum wire after 12 hours.
Bacterial Avoid Behavior
Small, uniform circular bacterial lawns were prepared by pipetting 100 uL of overnight cultures of E. coli OP50 or S. typhimurium SL1344, P. aeruginosa PA14 on the center of NGM dishes and allowing at least one day to dry. To score each animal as inside or outside the lawn (Avoidance=Nout/Ntotal) at larval stages (L1 to L4) and young adults (YA), we transfer 50-80 eggs to the bacterial lawn without disturbing or spreading the lawn and record avoidance of developmental stage.
Pharyngeal Pump
Pharyngeal pumping was assessed by observing the number of pharyngeal contractions during a 10-sec interval for longitudinal studies or a 60-sec interval. The experimental details of observing the number of pharyngeal contractions were determined as described (Kumar et al., 2016).
Bacterial Colonization
We transferred plasmid pET28a into DH5α through heat shock and transferred plasmid ptfLC3 into P. aeruginosa PA14 through electric shock. L4 stage WT and scav-5(ok1606) were cultured on DH5α-GFP or S. typhimurium SL1344-GFP, P. aeruginosa PA14-GFP continuing 48 h and following observation the bacterial colonization in intestine by microscope.
Quantitative RT-PCR
To generate synchronous populations of worms for RNA extraction, we bleached WT and scav-5(ok1606) adults to collect eggs and cultured eggs at 20°C on NGM dishes spread with E. coli OP50. These synchronized worms were washed and collected for RNA isolation. Briefly, RNA was isolated using a Trizol (Invitrogen) and chloroform extraction (reagent Phenol: chloroform: isoamyl alcohol, 25:24:1 was used to chloroform extraction and isopropanol was used to precipitation RNA). RNA was diluted in nuclease-free water and quantified using a NanoDrop. cDNA was synthesized using HiScript II Q RT SuperMix kit (Vazyme). Real-time PCR was performed using an Applied Biosystems Step One Plus Real-Time PCR system and SYBR green master mix. mRNA fold change was calculated using the comparative CT method (Schmittgen and Livak, 2008) by comparing mRNA levels of the internal control gene tbg-1.
Immunoblot Analyses
Harvested worms were put in a glass homogenizer with RIPA buffer for grinding adequately. The lysate was centrifuged and supernatant protein was transferred into the new tube. Protein was quantified using BCA protein assay kit. And the total protein from each sample was electrophoresis on PAGE 10% gels, transferred to nitrocellulose membranes, blocked with 5% powdered milk in TBST, and probed with a 1:1000 dilution of an antibody that recognizes the phosphorylated of PMK-1 (Cell Signaling Technology Corporation). The blot was then stripped and reprobed with a 1:1000 dilution of an anti-actin antibody. Anti-rabbit, and anti-mouse IgG secondary antibodies were used to detect the primary antibodies following the addition of ECL reagents (Thermo Fisher Scientific, Inc.) which were visualized using a luminescence instrument.
Membrane Yeast Two-Hybrid (MYTH) Assay
scav-5 cDNA was inserted between the PstI and XbaI site of vector pMetYCgate and tir-1 cDNA was inserted between the EcroI and XmaI site of vector pNubXgate32-HA. The fusion plasmids were transferred into yeast strain AP4 and picked individual yeast colony into medium and incubated overnight, diluted it into 10-1, 10-2 and 10-3 respectively, dropped onto SD-Trp-Leu-His-Ade medium by 10 uL each drop. Images of yeast were taken after culturing at 30°C for 3 days.
Microscopy
Nematodes were mounted onto 2% agar pads, paralyzed with levamisole and photographed using an AXIO Imager Z1 microscope. Photographs were acquired using the same imaging conditions for a given experiment, and were processed in Photoshop.
Statistical Analyses
All data from lifespan, heat stress resistance, avoidance, pharyngeal pump, immunoblot and qRT-PCR analyses were repeated by three biological duplications, and were analyzed using the unpaired two-tailed Student’s t-test. “*” means P< 0.05; “**” means P<0.01; “***” means P<0.001, “****” means P<0.0001. Error bars in each column diagram represent SEM.
Results
All Members of the Scavenger Receptors Family Except SCAV-3 Are Expressed in the Intestinal Tissue in C. elegans
To investigate the role of scavenger receptors family in innate immunity in C. elegans, we first set out to define the tissues in which they are expressed. We generated plasmids that express green fluorescent protein (GFP) under the control of around 3 kb proximal promoter of scav-1-6, and got exchromosomal transgenic strains by microinjection. We observed strong GFP expression in intestine tissues, which were driven by scav-1, scav-2, scav-4, scav-5, and scav-6 promoter (Figures 1A, B, D–F). While SCAV-3 was expressed in all tissues, which is consistent with a previous study (Figure 1C) (Li et al., 2016). Intestinal epithelial cells provide an essential line for C. elegans against ingested pathogens (Gravato-Nobre and Hodgkin, 2005). Since immune response to pathogenic bacteria was measured from when they reached young adult stage, we further observed in vivo expression of scavenger receptors in L4 worms, and found that SCAV-1, SCAV-2, SCAV-4, SCAV-5 and SCAV-6 were expressed in intestine tissues (Figure S1). Thus, our expression pattern results indicate that scav-1, scav-2, scav-4, scav-5, and scav-6 may be involved in defense against pathogenic bacteria.
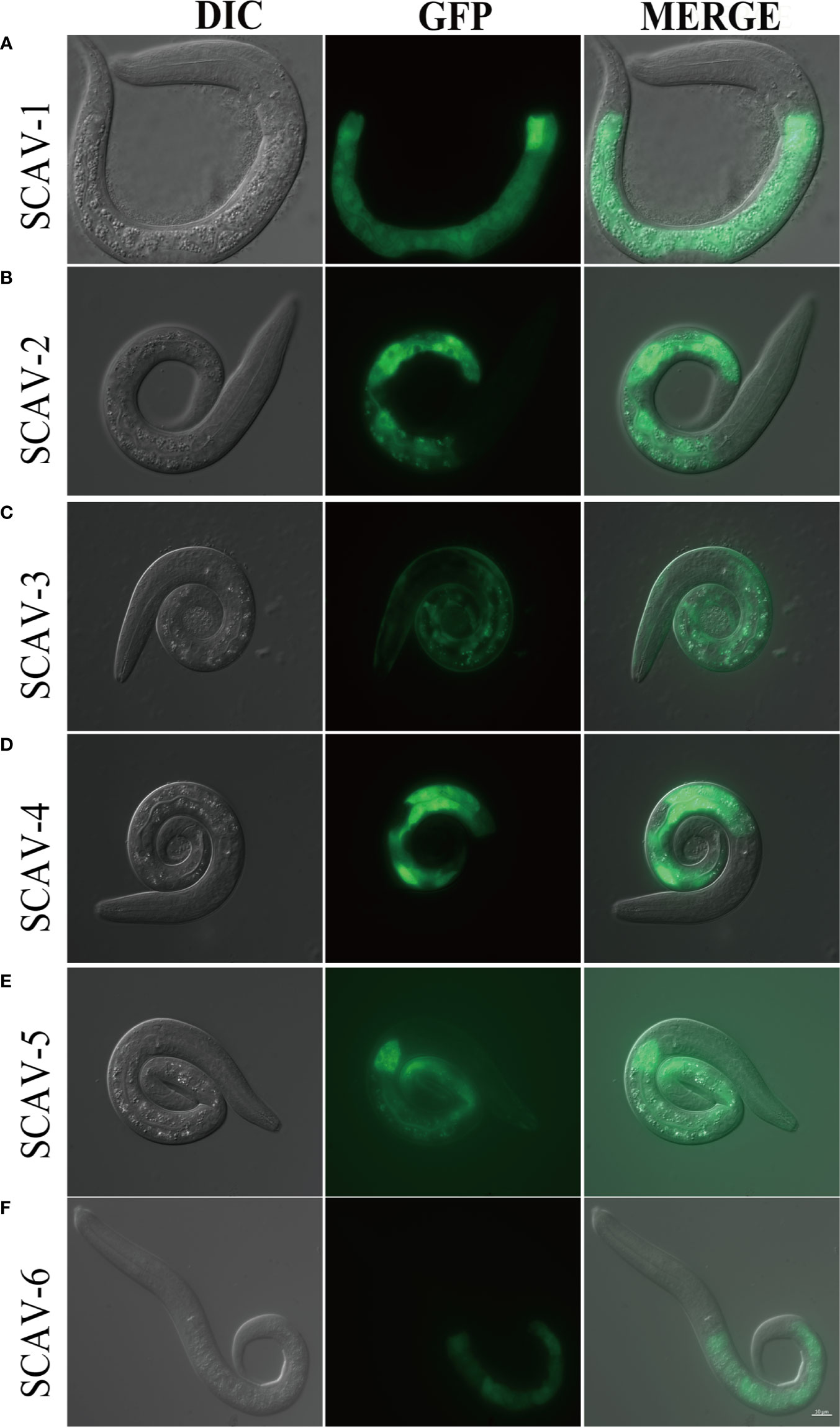
Figure 1 The expression pattern of scavenger receptors in C.elegans. (A–F): Tissue profiling of scavenger receptor SCAV-1-6. DIC images of L1 stage live transgenic animals (left). Green fluorescence images show tissue localization of scavenger receptor SCAV-1-6 (middle). Merge images (right). Scale bar=10 μm.
Loss of Function of scav-5 Protects C. elegans Against Pathogenic Bacteria Infection by P. aeruginosa PA14 and S. typhimurium SL1344
To examine the interaction between scavenger receptors and innate immune response in C. elegans, we used scavenger receptors mutants scav-2(ok877), scav-3(ok1286), scav-5(ok1606) and scav-1, scav-4, scav-6 RNAi-treatment wide type (WT) worms, and analyzed their lifespan when feeding on non-pathogenic bacteria OP50. We observed that only the lifespans of scav-3(ok1286) mutants were significantly reduced compared to the wild-type N2 (Figures 2A, B). Next, we analyzed their lifespan after P. aeruginosa PA14 and S. typhimurium SL1344 infection. We found that only the lifespans of scav-5(ok1606) mutants were significantly extended than control (Figures 2C–F). Many C. elegans mutations that delay aging also increase stress resistance (Wu et al., 2002; McColl et al., 2005; Saier et al., 2018). To test the effect of scav-5 mutation on stress resistance, we monitored heat resistance at 35°C of scav-5 mutants fed on E. coli OP50, S. typhimurium SL1344 and P. aeruginosa PA14 respectively. Our results shown that scav-5 mutant animals have significantly increased thermotolerance compared to WT when feeding on OP50 and S. typhimurium SL1344. We did not observe any survival of wide type and scav-5 mutants by heat stress treatment when feeding on P. aeruginosa PA14, which may due to the strong toxicity of PA14 (Figure S2A). scav-5(ok1606) is likely to be a strong loss of function or null allele based on the lack of detectable mRNA, we used this allele for our further analysis (Figures S2B, S2C). To confirm that loss of function of SCAV-5 was responsible for defense against pathogenic bacteria in scav-5(ok1606) mutants, we generate transgenic strains scav-5(ok1606);Pscav-5SCAV-5 that expresses SCAV-5 by its promoter by injecting plasmids into scav-5(ok1606) mutants, and analyzed their lifespan fed on S. typhimurium SL1344 and P. aeruginosa PA14 respectively. We found that re-expresses SCAV-5 in scav-5(ok1606) mutants abolished its protection effects after S. typhimurium SL1344 and P. aeruginosa PA14 infection (Figure S3). Our results demonstrated that loss of function of SCAV-5 defend C. elegans against pathogenic bacteria infection.
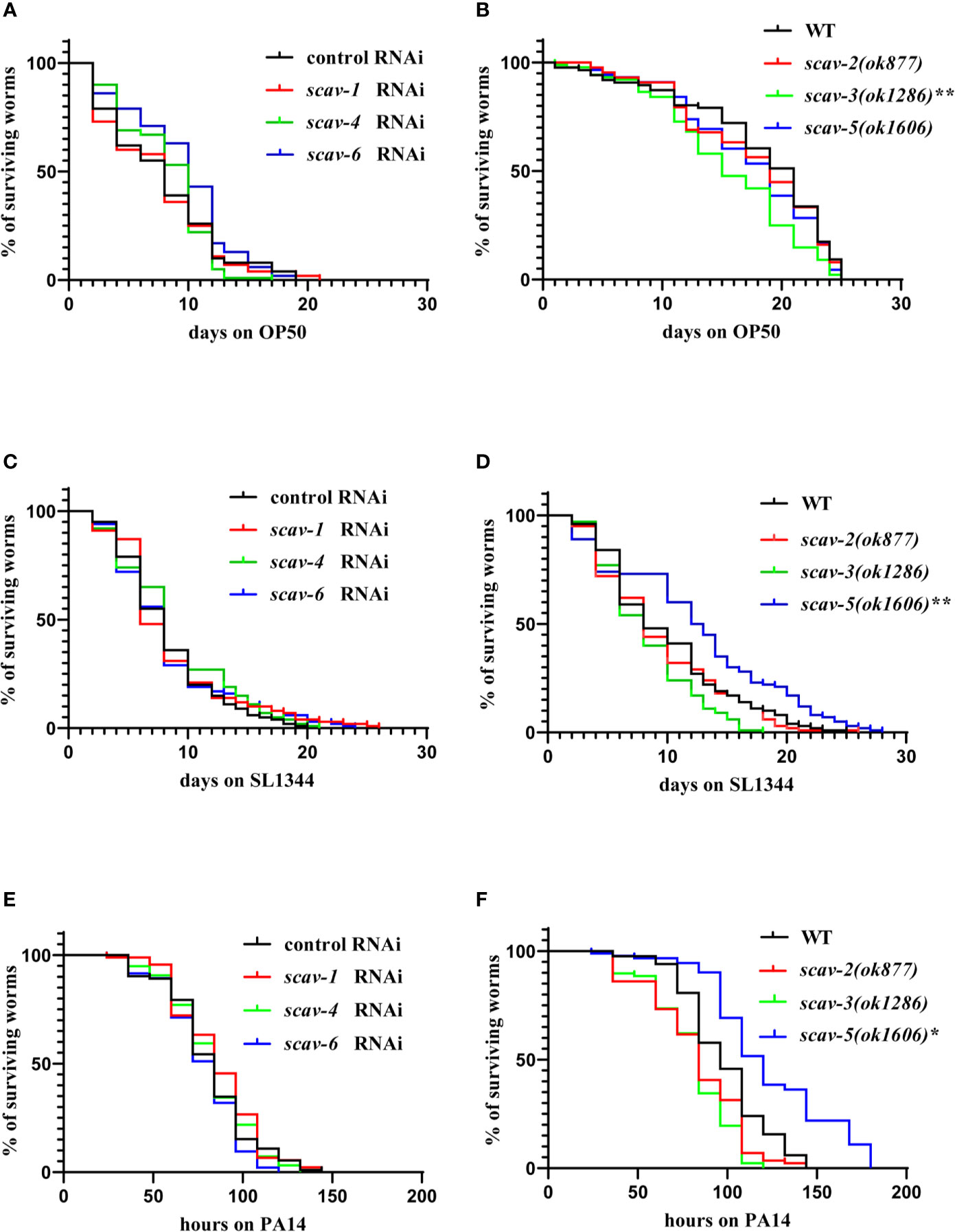
Figure 2 scav-5 mutants are resistant to S. typhimurium SL1344 and P. aeruginosa PA14. (A, B) The lifespan of wild type with scav-1, scav-4, scav-6 RNAi (A) and scav-2, scav-3, scav-5 mutants (B) fed on E. coli OP50. (C, D) The scav-5(ok1606) mutation significantly extended the lifespan of otherwise wild type, scav-1, scav-4, scav-6 RNAi (C) and scav-2(ok877), scav-3(ok1286) mutants (D) fed on S. typhimurium SL1344. (P < 0.01) (E, F) The scav-5(ok1606) mutation significantly extended the lifespan of otherwise wild type, scav-1, scav-4, scav-6 RNAi (E) and scav-2(ok877), scav-3(ok1286) mutants (F) fed on P. aeruginosa PA14. (P < 0.05).
scav-5 Mutants Are Resistant to S. typhimurium SL1344 by Dietary Restriction
To determine the mechanism by which scav-5 mutants are resistant to S. typhimurium SL1344, we first quantified their bacterial lawn avoidance behavior. We found that scav-5 mutants animals did not display bacterial avoidance behavior when cultured on E. coli OP50 or S. typhimurium SL1344 (Figures 3A–D). Next, we analyzed the pharyngeal pumping rate of scav-5 mutants when fed on E. coli OP50 or S. typhimurium SL1344 by using a dissecting microscope. We observed that scav-5 mutants can pump at a rate comparable to wild type when fed on E. coli OP50 (Figure 3E). While scav-5 mutants displayed a substantial age-related reduction of pharyngeal pumping rate when cultured on S. typhimurium SL1344 (Figure 3F). These imply that the reduced pharyngeal pumping rate in scav-5 mutants when feeding on S. typhimurium SL1344 leads to reduced bacterial ingestion, and then results in dietary restriction. To further confirm the dietary restriction in scav-5 mutants when feeding on S. typhimurium SL1344, we detected the E. coli DH5α and S. typhimurium colonization in the intestine of scav-5 mutants by feeding with E. coli DH5α-GFP and S. typhimurium-GFP for 48h after L4 stage respectively. scav-5 mutants intestine displayed significantly decreased fluorescence intensity when cultured on S. typhimurium-GFP (Figures 3G, H), the decreased level of S. typhimurium-GFP in scav-5 mutants was confirmed by western blotting (Figure 3H). Furthermore, we observed the mRNA level of eat-2, which is required for pharyngeal pumping rate and subsequent food intake (McKay et al., 2004), in WT and scav-5 mutants when feeding on S. typhimurium SL1344, and found the transcriptional level of eat-2 was reduced in scav-5 (Figure 3I), these results demonstrate that scav-5 mutation protects worms against S. typhimurium SL1344 by dietary restriction.
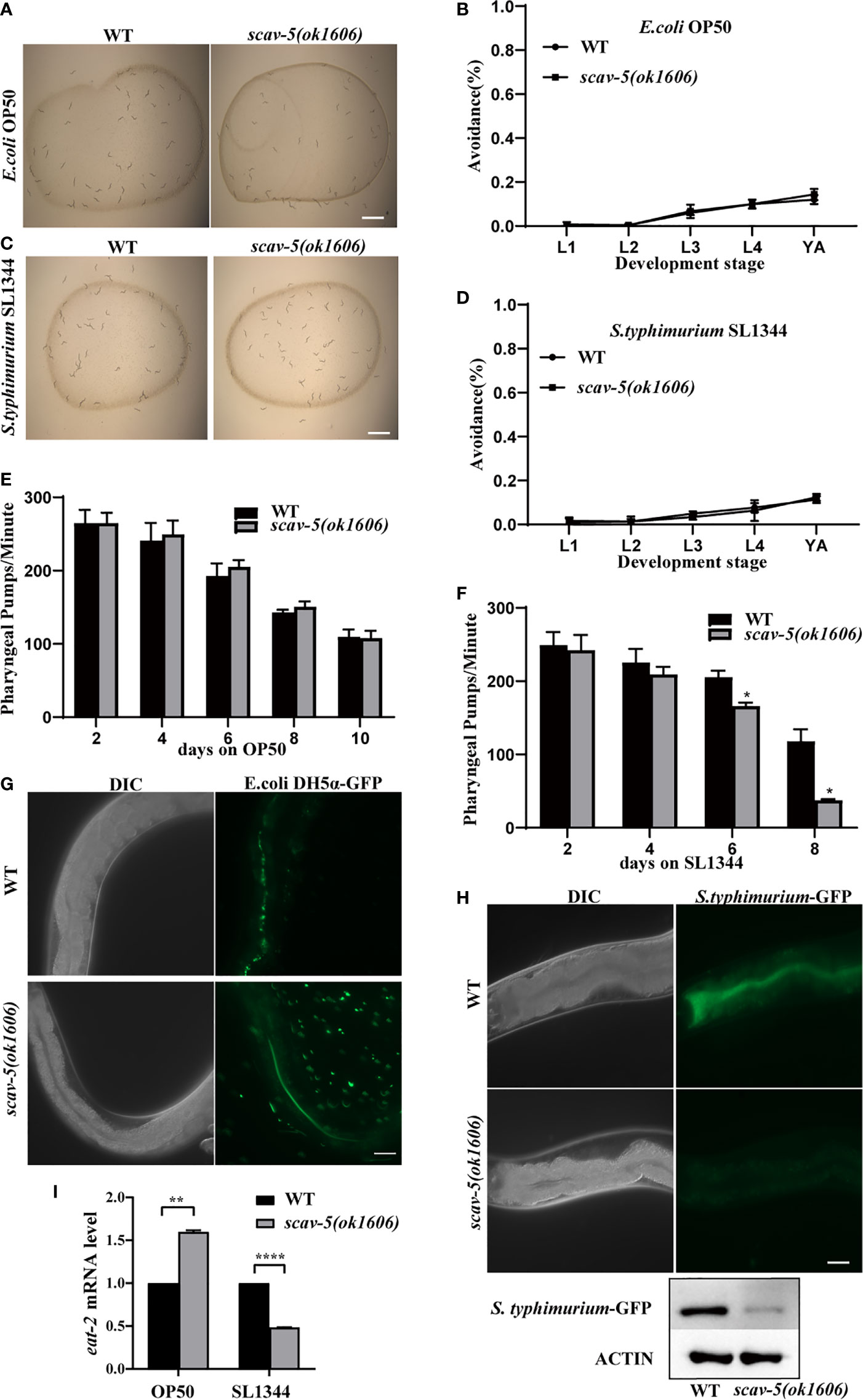
Figure 3 scav-5 mutants display dietary restriction when cultured on S. typhimurium SL1344. (A, C) Bright field photographs of wild type and scav-5(ok1606) mutant cultured on E. coli OP50 (A) and S. typhimurium SL1344 (B) both have no avoidance behavior at L4 stage. Scale bar=100 μm. (B, D) Quantification of avoidance rate of scav-5(ok1606) mutants cultured on E. coli OP50 (B) and S. typhimurium SL1344 (D) from L1 to young adult stage. (E, F) Pharyngeal pumps per min of wild type and scav-5(ok1606) mutants fed with E. coli OP50 (E) and S. typhimurium SL1344 (F). (G, H) The DIC and GFP fluorescence image of partial intestine in wild type and scav-5(ok1606) mutant cultured on E. coli OP50 (G) and S. typhimurium SL1344 (H) 48 h after L4 stage. Scale bar=20 μm. (I) qRT-PCR analysis of transcription level of DR gene eat-2 in wild-type and scav-5(ok1606) animals cultured on E. coli OP50 or S. typhimurium SL1344.
Loss of Function of scav-5 Does Not Upregulate Defense Genes Expression in C. elegans Infected With S. typhimurium SL1344
C. elegans has an innate immune system and responds to pathogenic bacterial by expression of defense genes (Kim et al., 2002; Ermolaeva and Schumacher, 2014). To test whether loss of function of scav-5 activates the innate immune response in C. elegans, we analyzed the mRNA level of pathogen response genes by quantitative RT-PCR (qRT-PCR). In C. elegans, clec-7, clec-60, and clec-82 encode C-type lectin proteins (Schulenburg et al., 2008), lys-5 encodes lysozyme (Boehnisch et al., 2011), and F53A9.8 encode antimicrobial peptides (Zugasti and Ewbank, 2009). Surprisingly, we observed that the expression of pathogen response genes clec-7, clec-60 clec-82, lys-5, and F53A9.8 were lower in scav-5 mutants compared to wild type when cultured on E. coli OP50. And not all but most of the pathogen response genes listed above were downregulated in scav-5 mutants cultured on S. typhimurium SL1344 (Figures 4A–E). These indicate that scav-5 may be a positive regulator for defense genes expression fed on E. coli OP50 or S. typhimurium SL1344. Intriguingly, we also found that p38 PMK-1 MAPK innate immune response pathway was downregulated in scav-5 mutants cultured on E. coli OP50 or S. typhimurium SL1344 by detecting the levels of PMK-1 phosphorylation (Figure 4F), which further confirm that scav-5 is a positive regulator for innate immune response when cultured on E. coli OP50 or S. typhimurium SL1344.
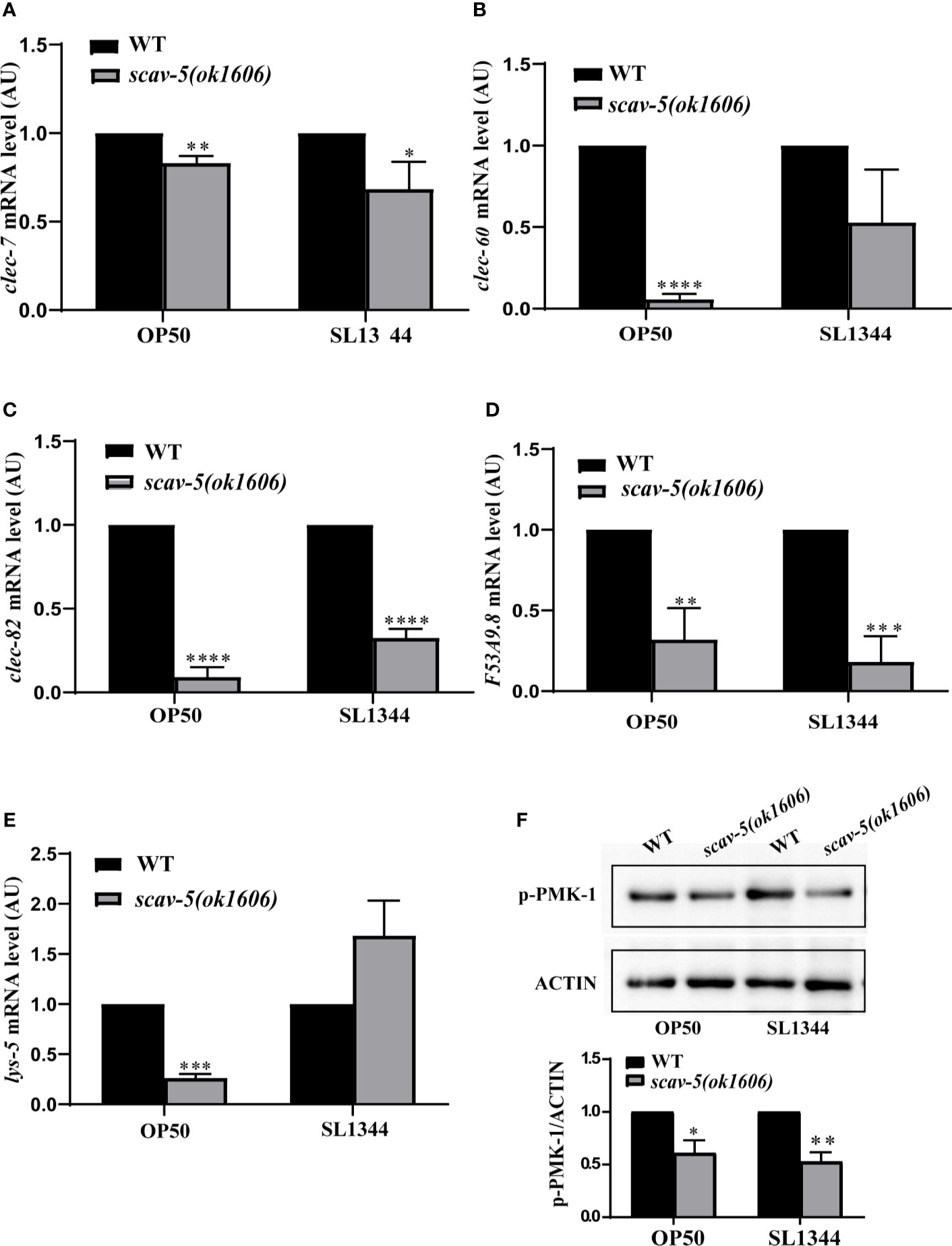
Figure 4 The innate immune response in scav-5 mutants fed on E. coli OP50 or S. typhimurium SL1344 were downregulated. (A) Immunoblot analysis of p-PMK-1 in young adult animals of wild type and scav-5(ok1606) mutants fed on E. coli OP50 or S. typhimurium SL1344 for 48h. (B–F) qRT-PCR analysis of transcription level of pathogenic response genes clec-7, clec-60, and clec-82 (B–D), lys-5 (E), and F53A9.8 (F) in wild-type and scav-5(ok1606) animals cultured on E. coli OP50 or S. typhimurium SL1344.
Defense Gene Expression Were Upregulated in scav-5 Mutants Infected by P. aeruginosa PA14
To verify the mechanism by which scav-5 mutants are resistant to P. aeruginosa PA14, we first quantified their bacterial lawn avoidance behavior. We found that similar to wild type, scav-5 mutants animals displayed bacterial avoidance behavior when cultured on P. aeruginosa PA14 (Figures 5A, B). Next, we analyzed the pharyngeal pumping rate of scav-5 mutants when fed on P. aeruginosa PA14. We observed that compared to WT, scav-5 mutants displayed significantly higher pharyngeal pumping rate on 48h-72h after L4 stage when cultured on S. P. aeruginosa PA14 (Figure 5C), indicating that scav-5 mutants had no dietary restrictions on P. aeruginosa PA14. To further confirm that the dietary restriction did not exist in scav-5 mutants when feeding on P. aeruginosa PA14, we detected P. aeruginosa PA14 colonization in the intestine of scav-5 mutants by feeding with P. aeruginosa PA14-GFP for 24h after L4 stage. scav-5 mutants intestine displayed significantly increased fluorescence intensity when cultured on P. aeruginosa PA14-GFP (Figure 5D). The increased level of P. aeruginosa PA14-GFP in scav-5 mutants was confirmed by Western blotting (Figure 5D). These results demonstrate that scav-5 mutation defend C. elegans against P. aeruginosa PA14 infection is not by dietary restriction. We then asked whether loss of function of scav-5 activates the innate immune response in C. elegans against P. aeruginosa PA14 infection by analyzing the mRNA level of pathogen response genes. We observed that the expression of pathogen response genes clec-60 clec-82, lys-5, and F53A9.8 were higher in scav-5 mutants when compared to wild type cultured on P. aeruginosa PA14 (Figures 6A–E). These results reveal that defense genes expression were upregulated in scav-5 mutants infected by P. aeruginosa PA14.
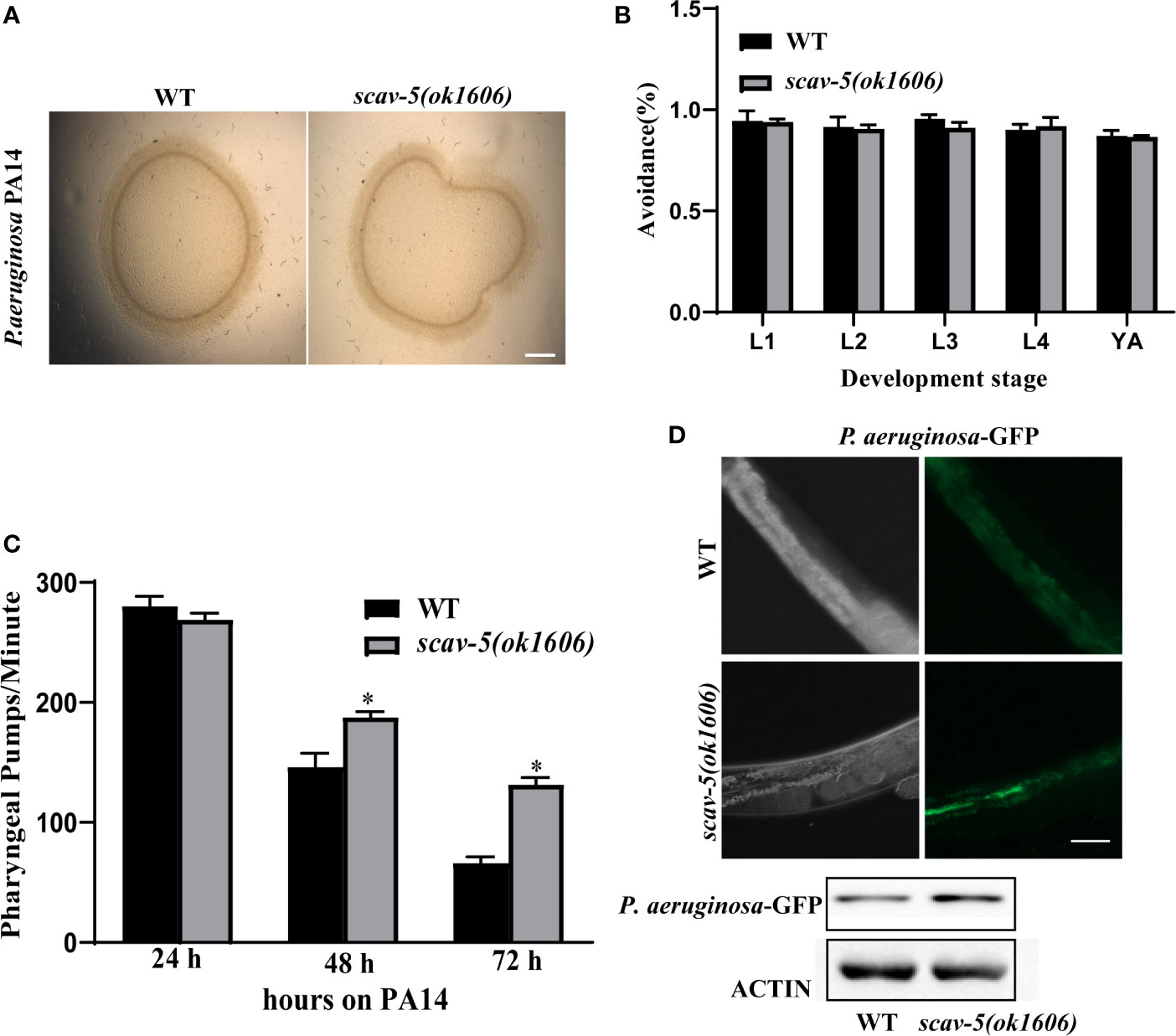
Figure 5 scav-5 mutants do not display dietary restriction when cultured on P. aeruginosa PA14. (A, B) Bright field photographs of wild type and scav-5(ok1606) mutant avoidance from P. aeruginosa PA14 in L4 stage (A) and quantification of avoidance rate from L1 stage to young adult (B), Scale bar=100 μm. (C) Statistics of pharyngeal pumps per min of wild type and scav-5(ok1606) mutants fed with P. aeruginosa PA14. (D) The DIC and GFP fluorescence image of partial intestine in wild type and scav-5(ok1606) mutant cultured on P. aeruginosa PA14-GFP 24 h after L4 stage. And immunoblot analysis of protein from young adult animals fed with P. aeruginosa PA14 24 h using antibodies that recognize the p-PMK-1 and actin. Scale bar=50 μm.
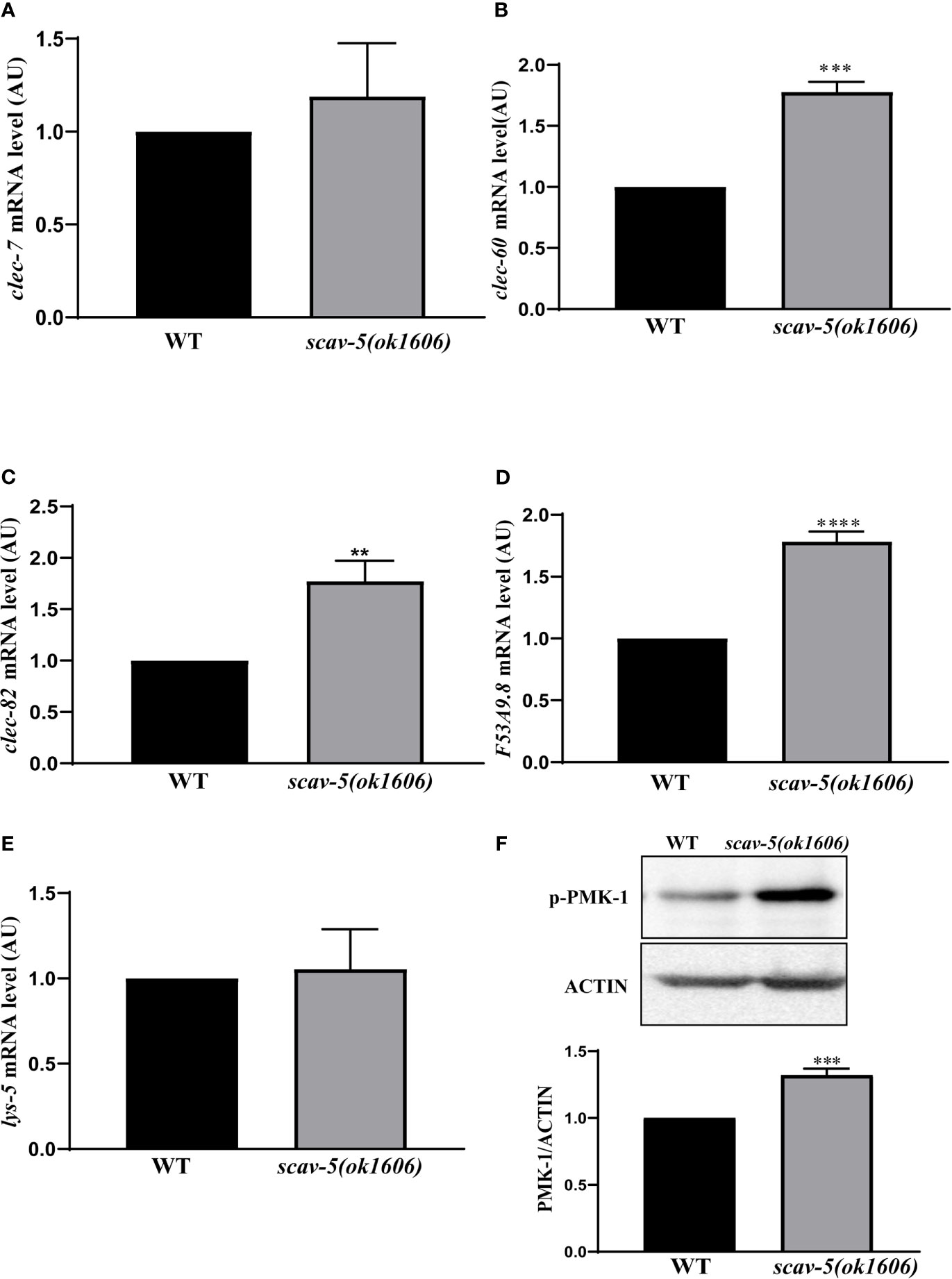
Figure 6 PMK-1 signaling pathways are activated in scav-5 mutants after P. aeruginosa PA14 infection. (A) Immunoblot analysis of protein from young adult animals fed with P. aeruginosa PA14 24 h using antibodies that recognize the p-PMK-1 and actin. (B–F) qRT-PCR analysis of transcription level of pathogenic response genes clec-7, clec-60, and clec-82 (B–D), lys-5 (E), and F53A9.8 (F) in wild-type and scav-5(ok1606) animals cultured on P. aeruginosa PA14.
scav-5 Mutants Are Resistant to P. aeruginosa PA14 Through Activation of PMK-1 p38 MAPK Pathway
To explore whether SCAV-5 participates in the innate immune response through the PMK-1 p38 MAPK pathway in C. elegans after P. aeruginosa PA14 infection, we first measured activated PMK-1 level by immunoblotting. Our results showed that activated PMK-1 in scav-5 mutants fed on P. aeruginosa PA14 was significantly increased compared to wild type control (Figure 6F). These data proved that defects can activate PMK-1 p38 MAPK pathways in C. elegans infected by P. aeruginosa PA14. Next, we test whether pmk-1 was required for scav-5 mutants lifespan extension after P. aeruginosa PA14 infection. Our results in line with previous studies that loss- and reduction-of-function mutations of p38 MAPK PMK-1 pathway components lead to a reduced lifespan of worms fed on P. aeruginosa PA14 (Figure 7A) (Xu et al., 2013; Head et al., 2017). Besides, we found pmk-1 RNAi treatment suppressed the extended lifespan phenotype of scav-5 mutants when cultured on P. aeruginosa PA14 (Figure 7B), demonstrating the requirement of pmk-1 pathway for scav-5 mutants lifespan extension infected by P. aeruginosa PA14. Furthermore, our genetic epistasis analysis suggested scav-5 function upstream of or in parallel to tir-1 (Figure 7B) (Couillault et al., 2004). Our preliminary analysis of protein structure by the SMART website revealed that SCAV-5 is a membrane protein with two transmembrane domains. We then utilized the split-ubiquitin based membrane yeast two-hybrid (MYTH) system for detecting the interaction of SCAV-5 with TIR-1. However, the result showed that SCAV-5 and TIR-1 did not physically interact (Figure 7C). Taken together, our results reveal that scav-5 mutation protects C. elegans against P. aeruginosa PA14 by upregulating PMK-1 p38 MAPK pathway.
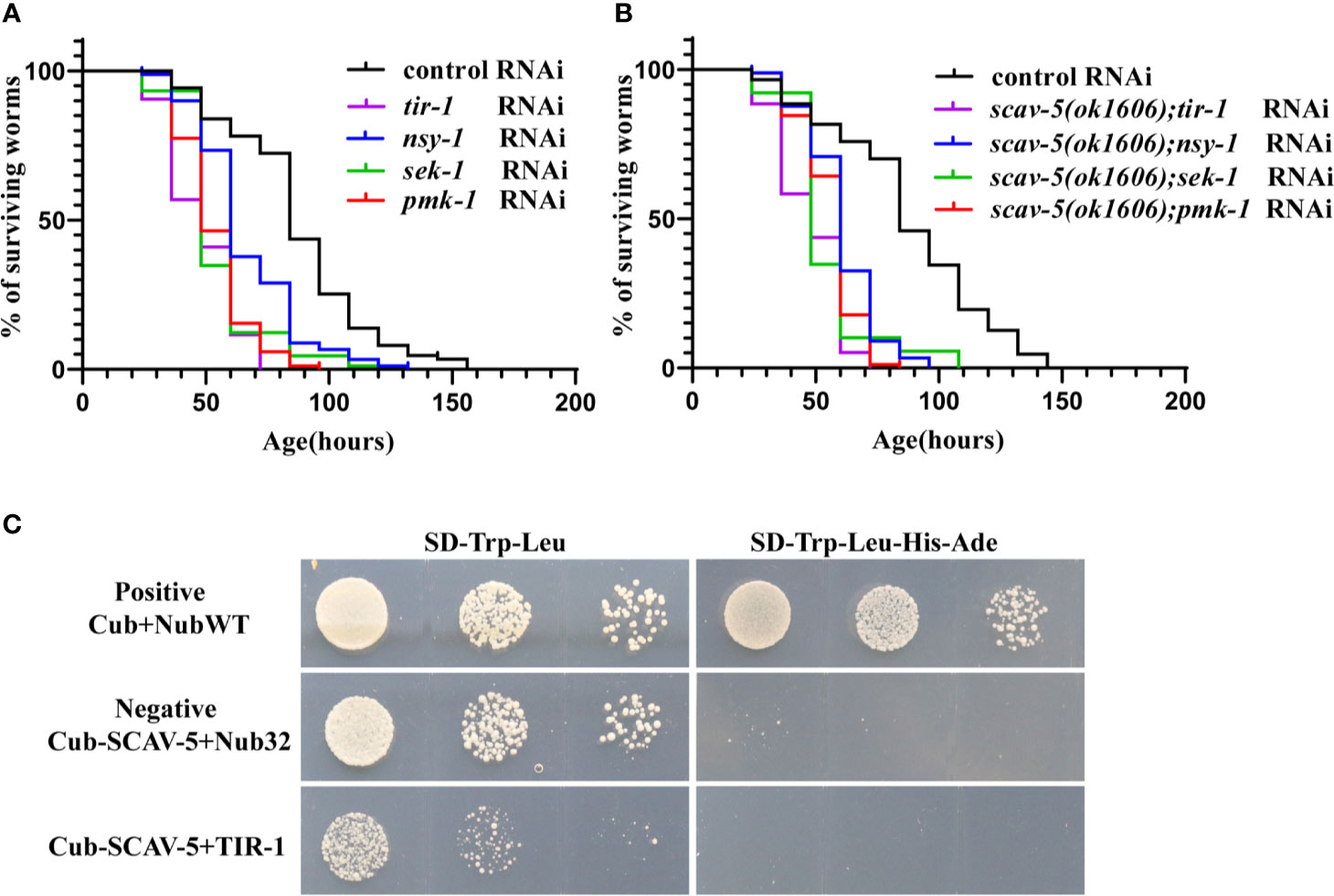
Figure 7 Genetic epistatic analysis of scav-5 in the PMK-1 pathway defense against P. aeruginosa PA14 and test of the interaction SCAC-5 with TIR-1. (A, B) Survival curves of wild type (A) and scav-5(ok1606) mutants (B) with tir-1, nsy-1, sek-1, pmk-1 RNAi cultured on P. aeruginosa PA14. (C) Analysis of interaction between SCAV-5 and TIR-1 by yeast two hybrid. The yeast solution was diluted by three gradients in each group, which were cultured on SD-Trp-Leu medium (left) and SD -Trp-Leu-His-Ade medium (right).
Discussion
Mounting evidence shows that many scavenger receptors, including the prototype class B type scavenger receptor CD36, play an important role in innate immunity by serving as the pattern-recognition receptors, in particular against bacterial pathogens. SCAV-1-6 are the six class B scavenger receptors homologs in C. elegans. However, it is not clear whether they have effect on host defense against bacterial pathogens. Here, we show that defects in scav-5 protect worms against pathogenic bacterial S. typhimurium SL1344 and P. aeruginosa PA14 by different mechanism. scav-5 mutants are resistant to S. typhimurium SL1344 due to dietary restriction. While scav-5 mutation protects worms against P. aeruginosa PA14 by activating the innate immune response through conserved PMK-1 p38 MAPK pathway.
We found that, of the six SCAV proteins homologous, only scav-5 is involved in innate immune response against pathogenic bacterial. scav-1, scav-2, scav-4, scav-5 and scav-6 are expressed in the intestine tissues. It is widely reported that, in response to bacterial infections, C. elegans produces an array in intestinal epithelial cells by expressing related antimicrobial genes (JebaMercy et al., 2011). The damage of intestinal epithelium would cause worms more hypersensitive to pathogenic bacteria, which allows live bacteria to enter the intestinal lumen (Kumar et al., 2019). The expression pattern of scav-5 consistent with its function. Indeed, a previous study demonstrates that scav-1 is necessary for C. elegans survival after fungal pathogens infection (Means et al., 2009). It will be interesting to test whether scav-2, scav-4, scav-5 and scav-6 participate in defense against other pathogens infection. We observed that SCAV-3 was expressed in all tissues and required for lifespan extension of C. elegans when cultured in E. coli OP50. Our results in line with the previous study, which also reveals that SCAV-3 is lysosomal membrane protein and is the key regulator of lysosome integrity, motility, and dynamics.
We found that compared to control, the expression of pathogen response genes were downregulated in scav-5 mutants without infection or cultured on S. typhimurium SL1344, either PMK-1 p38 MAPK pathway was activated. These indicate that scav-5 may be required for the expression of pathogen response genes in worms without infection. This is an important issue for future research. A possible explanation for the downregulation expression of pathogen response genes in scav-5 mutants fed on SL1344 might be dietary restriction. Bacteria as food for C. elegans was ingested by increasing its rate of pharyngeal pumping, which is a process controlled by pharyngeal motor neurons (Kumar et al., 2019). Thus, decreased ingestion of S. typhimurium SL1344 in scav-5 mutants implicate its function may relate to neuron regulation. Further studies will need to determine the underlying mechanism.
We found that the expression of pathogen response genes were upregulated in scav-5 mutants when cultured on P. aeruginosa PA14.This was further confirmed by that scav-5 acts in PMK-1 p38 MAPK pathway after P. aeruginosa PA14 infection, although scav-5 is required for expression of pathogen response genes under normal condition. As PMK-1 controls basal levels of pathogen response genes on E. coli and also induce the upregulation of these genes’ expression upon infection. Our results imply scav-5 may function in another pathway for controlling the basal level of pathogen response genes. We found that scav-5 may function upstream of, or in parallel to tir-1 in PMK-1 innate immune response pathway to protect C. elegans against P. aeruginosa PA14. However, SCAV-5 and TIR-1 did not physically interact. Further research should be undertaken to investigate how scav-5 participate in PMK-1 innate immune response pathway.
In summary, our results provide evidence that loss of function of Scavenger receptor SCAV-5 protects C. elegans against pathogenic bacteria S. typhimurium SL1344 and P. aeruginosa PA14 by different mechanisms, establish the link between the SCAV-5 and the innate immune response. To our knowledge, this is the first demonstration of a role for SCAV-5 in host defense against pathogenic bacteria. SCAV-5 is an ortholog of human SRB-I/II, which is implicated in platelet-type bleeding disorder 10 and progressive myoclonus epilepsy 4 (Dibbens et al., 2009; Silverstein and Febbraio, 2009). Thus, our research provides important clues for further dissecting the mechanism by which SRB-I/II regulates innate immune responses.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
HX and QZ conceived the study. AL did most of the experiments. HJ did the manuscript revision and experimental repeats. LY and YW contributed to materials. HX, QZ, and AL wrote the manuscript with feedback from all authors. All authors contributed to the article and approved the submitted version.
Funding
This work was partially supported by the National Natural Science Foundation of China (Grant No. 31671439 to HX), National Natural Science Foundation of China Youth Program (Grant No. 31801164 to QZ), Natural Science Foundation of Shaanxi Province, China (Grant No. 2020JM-271 to HX), the program of Innovative Research Team for the Central Universities (Grant No. GK202001004 to HX), the Fundamental Research Key Project Funds for the Central Universities (Grant No. GK202007009 to HX).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Drs. Chonglin Yang as well as the C. elegans Genetic Center for C. elegans strains and Dr. Cheng-Gang Zou for P. aeruginosa PA14 and S. typhimurium SL1344 strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.593745/full#supplementary-material
References
Aballay, A., Yorgey, P., Ausubel, F. M. (2000). Salmonella Typhimurium Proliferates and Establishes a Persistent Infection in the Intestine of Caenorhabditis Elegans. Curr. Biol. 10, 1539–1542. doi: 10.1016/S0960-9822(00)00830-7
Alegado, R. A., Chin, C. Y., Monack, D. M., Tan, M. W. (2011). The Two-Component Sensor Kinase KdpD Is Required for Salmonella Typhimurium Colonization of Caenorhabditis Elegans and Survival in Macrophages. Cell Microbiol. 13, 1618–1637. doi: 10.1111/j.1462-5822.2011.01645.x
Aliyari, R., Ding, S. W. (2009). RNA-Based Viral Immunity Initiated by the Dicer Family of Host Immune Receptors. Immunol. Rev. 227, 176–188. doi: 10.1111/j.1600-065X.2008.00722.x
Anderson, K. V., Jurgens, G., Nusslein-Volhard, C. (1985). Establishment of Dorsal-Ventral Polarity in the Drosophila Embryo: Genetic Studies on the Role of the Toll Gene Product. Cell 42, 779–789. doi: 10.1016/0092-8674(85)90274-0
Andrusiak, M. G., Jin, Y. (2016). Context Specificity of Stress-Activated Mitogen-Activated Protein (MAP) Kinase Signaling: The Story as Told by Caenorhabditis Elegans. J. Biol. Chem. 291, 7796–7804. doi: 10.1074/jbc.R115.711101
Boehnisch, C., Wong, D., Habig, M., Isermann, K., Michiels, N. K., Roeder, T., et al. (2011). Protist-Type Lysozymes of the Nematode Caenorhabditis Elegans Contribute to Resistance Against Pathogenic Bacillus Thuringiensis. PloS One 6, e24619. doi: 10.1371/journal.pone.0024619
Canton, J., Neculai, D., Grinstein, S. (2013). Scavenger Receptors in Homeostasis and Immunity. Nat. Rev. Immunol. 13, 621–634. doi: 10.1038/nri3515
Cezairliyan, B., Vinayavekhin, N., Grenfell-Lee, D., Yuen, G. J., Saghatelian, A., Ausubel, F. M. (2013). Identification of Pseudomonas Aeruginosa Phenazines That Kill Caenorhabditis Elegans. PloS Pathog. 9, e1003101. doi: 10.1371/journal.ppat.1003101
Couillault, C., Pujol, N., Reboul, J., Sabatier, L., Guichou, J. F., Kohara, Y., et al. (2004). TLR-Independent Control of Innate Immunity in Caenorhabditis Elegans by the TIR Domain Adaptor Protein TIR-1, an Ortholog of Human SARM. Nat. Immunol. 5, 488–494. doi: 10.1038/ni1060
Curt, A., Zhang, J., Minnerly, J., Jia, K. (2014). Intestinal Autophagy Activity Is Essential for Host Defense Against Salmonella Typhimurium Infection in Caenorhabditis Elegans. Dev. Comp. Immunol. 45, 214–218. doi: 10.1016/j.dci.2014.03.009
Desai, S. K., Padmanabhan, A., Harshe, S., Zaidel-Bar, R., Kenney, L. J. (2019). Salmonella Biofilms Program Innate Immunity for Persistence in Caenorhabditis Elegans. Proc. Natl. Acad. Sci. U.S.A. 116, 12462–12467. doi: 10.1073/pnas.1822018116
Dibbens, L. M., Michelucci, R., Gambardella, A., Andermann, F., Rubboli, G., Bayly, M. A., et al. (2009). SCARB2 Mutations in Progressive Myoclonus Epilepsy (PME) Without Renal Failure. Ann. Neurol. 66, 532–536. doi: 10.1002/ana.21765
Ermolaeva, M. A., Schumacher, B. (2014). Insights From the Worm: The C. Elegans Model for Innate Immunity. Semin. Immunol. 26, 303–309. doi: 10.1016/j.smim.2014.04.005
Feng, H., Guo, L., Wang, D., Gao, H., Hou, G., Zheng, Z., et al. (2011). Deficiency of Scavenger Receptor BI Leads to Impaired Lymphocyte Homeostasis and Autoimmune Disorders in Mice. Arterioscler. Thromb. Vasc. Biol. 31, 2543–2551. doi: 10.1161/ATVBAHA.111.234716
Gottar, M., Gobert, V., Michel, T., Belvin, M., Duyk, G., Hoffmann, J. A., et al. (2002). The Drosophila Immune Response Against Gram-Negative Bacteria is Mediated by a Peptidoglycan Recognition Protein. Nature 416, 640–644. doi: 10.1038/nature734
Graham, S. A., Antonopoulos, A., Hitchen, P. G., Haslam, S. M., Dell, A., Drickamer, K., et al. (2011). Identification of Neutrophil Granule Glycoproteins as Lewis(x)-Containing Ligands Cleared by the Scavenger Receptor C-Type Lectin. J. Biol. Chem. 286, 24336–24349. doi: 10.1074/jbc.M111.244772
Gravato-Nobre, M. J., Hodgkin, J. (2005). Caenorhabditis Elegans as a Model for Innate Immunity to Pathogens. Cell Microbiol. 7, 741–751. doi: 10.1111/j.1462-5822.2005.00523.x
Haraga, A., Ohlson, M. B., Miller, S. I. (2008). Salmonellae Interplay With Host Cells. Nat. Rev. Microbiol. 6, 53–66. doi: 10.1038/nrmicro1788
Hawkes, M., Li, X., Crockett, M., Diassiti, A., Finney, C., Min-Oo, G., et al. (2010). CD36 Deficiency Attenuates Experimental Mycobacterial Infection. BMC Infect. Dis. 10, 299. doi: 10.1186/1471-2334-10-299
Head, B. P., Olaitan, A. O., Aballay, A. (2017). Role of GATA Transcription Factor ELT-2 and P38 MAPK PMK-1 in Recovery From Acute P. Aeruginosa Infection in C. Elegans. Virulence 8, 261–274. doi: 10.1080/21505594.2016.1222334
He, M., Kubo, H., Morimoto, K., Fujino, N., Suzuki, T., Takahasi, T., et al. (2011). Receptor for Advanced Glycation End Products Binds to Phosphatidylserine and Assists in the Clearance of Apoptotic Cells. EMBO Rep. 12, 358–364. doi: 10.1038/embor.2011.28
Irazoqui, J. E., Urbach, J. M., Ausubel, F. M. (2010). Evolution of Host Innate Defence: Insights From Caenorhabditis Elegans and Primitive Invertebrates. Nat. Rev. Immunol. 10, 47–58. doi: 10.1038/nri2689
JebaMercy, G., Pandian, S. K., Balamurugan, K. (2011). Changes in Caenorhabditis Elegans Life Span and Selective Innate Immune Genes During Staphylococcus Aureus Infection. Folia Microbiol. (Praha) 56, 373–380. doi: 10.1007/s12223-011-0060-y
Kim, D. H., Feinbaum, R., Alloing, G., Emerson, F. E., Garsin, D. A., Inoue, H., et al. (2002). A Conserved P38 MAP Kinase Pathway in Caenorhabditis Elegans Innate Immunity. Science 297, 623–626. doi: 10.1126/science.1073759
Kirienko, N. V., Kirienko, D. R., Larkins-Ford, J., Wahlby, C., Ruvkun, G., Ausubel, F. M. (2013). Pseudomonas Aeruginosa Disrupts Caenorhabditis Elegans Iron Homeostasis, Causing a Hypoxic Response and Death. Cell Host Microbe 13, 406–416. doi: 10.1016/j.chom.2013.03.003
Kumar, S., Dietrich, N., Kornfeld, K. (2016). Angiotensin Converting Enzyme (ACE) Inhibitor Extends Caenorhabditis Elegans Life Span. PloS Genet. 12, e1005866. doi: 10.1371/journal.pgen.1005866
Kumar, S., Egan, B. M., Kocsisova, Z., Schneider, D. L., Murphy, J. T., Diwan, A., et al. (2019). Lifespan Extension in C. Elegans Caused by Bacterial Colonization of the Intestine and Subsequent Activation of an Innate Immune Response. Dev. Cell 49, 100–117.e6. doi: 10.1016/j.devcel.2019.03.010
Li, Y., Chen, B., Zou, W., Wang, X., Wu, Y., Zhao, D., et al. (2016). The Lysosomal Membrane Protein SCAV-3 Maintains Lysosome Integrity and Adult Longevity. J. Cell Biol. 215, 167–185. doi: 10.1083/jcb.201602090
McColl, G., Rogers, A. N., Alavez, S., Hubbard, A. E., Melov, S., Link, C. D., et al. (2010). Insulin-Like Signaling Determines Survival During Stress via Posttranscriptional Mechanisms in C. Elegans. Cell Metab. 12, 260–272. doi: 10.1016/j.cmet.2010.08.004
McColl, G., Vantipalli, M. C., Lithgow, G. J., The, C. (2005). Elegans Ortholog of Mammalian Ku70, Interacts With Insulin-Like Signaling to Modulate Stress Resistance and Life Span. FASEB J. 19, 1716–1718. doi: 10.1096/fj.04-2447fje
McKay, J. P., Raizen, D. M., Gottschalk, A., Schafer, W. R., Avery, L. (2004). Eat-2 and Eat-18 Are Required for Nicotinic Neurotransmission in the Caenorhabditis Elegans Pharynx. Genetics 166, 161–169. doi: 10.1534/genetics.166.1.161
Means, T. K., Mylonakis, E., Tampakakis, E., Colvin, R. A., Seung, E., Puckett, L., et al. (2009). Evolutionarily Conserved Recognition and Innate Immunity to Fungal Pathogens by the Scavenger Receptors SCARF1 and CD36. J. Exp. Med. 206, 637–653. doi: 10.1084/jem.20082109
Mukhopadhyay, S., Gordon, S. (2004). The Role of Scavenger Receptors in Pathogen Recognition and Innate Immunity. Immunobiology 209, 39–49. doi: 10.1016/j.imbio.2004.02.004
Pluddemann, A., Mukhopadhyay, S., Gordon, S. (2006). The Interaction of Macrophage Receptors With Bacterial Ligands. Expert Rev. Mol. Med. 8, 1–25. doi: 10.1017/S1462399406000159
Pluddemann, A., Mukhopadhyay, S., Gordon, S. (2011). Innate Immunity to Intracellular Pathogens: Macrophage Receptors and Responses to Microbial Entry. Immunol. Rev. 240, 11–24. doi: 10.1111/j.1600-065X.2010.00989.x
PrabhuDas, M. R., Baldwin, C. L., Bollyky, P. L., Bowdish, D. M. E., Drickamer, K., Febbraio, M., et al. (2017). A Consensus Definitive Classification of Scavenger Receptors and Their Roles in Health and Disease. J. Immunol. 198, 3775–3789. doi: 10.4049/jimmunol.1700373
Que, F., Wu, S., Huang, R. (2013). Salmonella Pathogenicity Island 1(SPI-1) at Work. Curr. Microbiol. 66, 582–587. doi: 10.1007/s00284-013-0307-8
Riera Romo, M., Perez-Martinez, D., Castillo Ferrer, C. (2016). Innate Immunity in Vertebrates: An Overview. Immunology 148, 125–139. doi: 10.1111/imm.12597
Saier, C., Gommlich, I., Hiemann, V., Baier, S., Koch, K., Horn, G., et al. (2018). Agrimonia Procera Wallr. Extract Increases Stress Resistance and Prolongs Life Span in Caenorhabditis Elegans via Transcription Factor DAF-16 (FoxO Orthologue). Antioxidants (Basel) 7, 1–11. doi: 10.3390/antiox7120192
Schmittgen, T. D., Livak, K. J. (2008). Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schulenburg, H., Hoeppner, M. P., Weiner, J., 3rd, Bornberg-Bauer, E. (2008). Specificity of the Innate Immune System and Diversity of C-Type Lectin Domain (CTLD) Proteins in the Nematode Caenorhabditis Elegans. Immunobiology 213, 237–250. doi: 10.1016/j.imbio.2007.12.004
Shivers, R. P., Pagano, D. J., Kooistra, T., Richardson, C. E., Reddy, K. C., Whitney, J. K., et al. (2010). Phosphorylation of the Conserved Transcription Factor ATF-7 by PMK-1 P38 MAPK Regulates Innate Immunity in Caenorhabditis Elegans. PloS Genet. 6, e1000892. doi: 10.1371/journal.pgen.1000892
Silverstein, R. L., Febbraio, M. (2009). CD36, a Scavenger Receptor Involved in Immunity, Metabolism, Angiogenesis, and Behavior. Sci. Signal 2, re3. doi: 10.1126/scisignal.272re3
Taffoni, C., Pujol, N. (2015). Mechanisms of Innate Immunity in C. Elegans Epidermis. Tissue Barriers 3, e1078432. doi: 10.1080/21688370.2015.1078432
Tanaka-Hino, M., Sagasti, A., Hisamoto, N., Kawasaki, M., Nakano, S., Ninomiya-Tsuji, J., et al. (2002). SEK-1 MAPKK Mediates Ca2+ Signaling to Determine Neuronal Asymmetric Development in Caenorhabditis Elegans. EMBO Rep. 3, 56–62. doi: 10.1093/embo-reports/kvf001
Tan, M. W., Mahajan-Miklos, S., Ausubel, F. M. (1999). Killing of Caenorhabditis Elegans by Pseudomonas Aeruginosa Used to Model Mammalian Bacterial Pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 96, 715–720. doi: 10.1073/pnas.96.2.715
Timmons, L., Court, D. L., Fire, A. (2001). Ingestion of Bacterially Expressed dsRNAs can Produce Specific and Potent Genetic Interference in Caenorhabditis Elegans. Gene 263, 103–112. doi: 10.1016/S0378-1119(00)00579-5
Wu, Z., Smith, J. V., Paramasivam, V., Butko, P., Khan, I., Cypser, J. R., et al. (2002). Ginkgo Biloba Extract EGb 761 Increases Stress Resistance and Extends Life Span of Caenorhabditis Elegans. Cell Mol. Biol. (Noisy-le-grand) 48, 725–731.
Xu, A., Shi, G., Liu, F., Ge, B. (2013). Caenorhabditis Elegans Mom-4 is Required for the Activation of the P38 MAPK Signaling Pathway in the Response to Pseudomonas Aeruginosa Infection. Protein Cell 4, 53–61. doi: 10.1007/s13238-012-2080-z
Zheng, Z., Zhang, X., Liu, J., He, P., Zhang, S., Zhang, Y., et al. (2021). GABAergic Synapses Suppress Intestinal Innate Immunity via Insulin Signaling in Caenorhabditis Elegans. Proc. Natl. Acad. Sci. U.S.A. 118. doi: 10.1073/pnas.2021063118
Keywords: scavenger receptors, SCAV-5, C. elegans, innate immunity, pathogenic bacteria
Citation: Luo A, Jing H, Yuan L, Wang Y, Xiao H and Zheng Q (2021) Loss of Function of Scavenger Receptor SCAV-5 Protects C. elegans Against Pathogenic Bacteria. Front. Cell. Infect. Microbiol. 11:593745. doi: 10.3389/fcimb.2021.593745
Received: 09 October 2020; Accepted: 08 July 2021;
Published: 03 August 2021.
Edited by:
Rodnei Dennis Rossoni, Sao Paulo State University, BrazilReviewed by:
Gabriela Lima, São Paulo State University, BrazilRajagopal Kammara, Central Food Technological Research Institute (CSIR), India
Copyright © 2021 Luo, Jing, Yuan, Wang, Xiao and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Zheng, zqian@snnu.edu.cn; Hui Xiao, huixiao@snnu.edu.cn
†These authors have contributed equally to this work
 Aixiao Luo
Aixiao Luo Huiru Jing†
Huiru Jing†  Lei Yuan
Lei Yuan Hui Xiao
Hui Xiao Qian Zheng
Qian Zheng