Roles of Gut Bacteriophages in the Pathogenesis and Treatment of Inflammatory Bowel Disease
- State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, National Medical Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
Inflammatory bowel disease (IBD), including Crohn’s disease and ulcerative colitis, are chronic, relapsing intestinal inflammatory disorders. Although the molecular mechanisms governing the pathogenesis of IBD are not completely clear, the main factors are presumed to be a complex interaction between genetic predisposition, host immune response and environmental exposure, especially the intestinal microbiome. Currently, most studies have focused on the role of gut bacteria in the onset and development of IBD, whereas little attention has been paid to the enteroviruses. Among of them, viruses that infect prokaryotes, called bacteriophages (phages) occupy the majority (90%) in population. Moreover, several recent studies have reported the capability of regulating the bacterial population in the gut, and the direct and indirect influence on host immune response. The present review highlights the roles of gut phages in IBD pathogenesis and explores the potentiality of phages as a therapeutic target for IBD treatment.
Introduction
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are chronic disorders characterized by persistent inflammation in the gastrointestinal tract (Abraham and Cho, 2009). In recent years, the incidence of IBD has increased year by year, with a significant burden on the global economy and public health (Molodecky et al., 2012; Kaplan, 2015). Current drugs can only relieve symptoms, but can’t cure this disease. Therefore, it is urgent to uncover the specific etiology of IBD, and explore effective treatment strategy.
The specific etiology of IBD is complicated and remains unclear so far. However, researches have demonstrated that the genetic factors, the host immune system, and the environmental factors (e.g., intestinal microbiota) are associated with the initiation and development of IBD (Maloy and Powrie, 2011; Geremia et al., 2014; Ananthakrishnan et al., 2018; Turpin et al., 2018). In particular, the roles of gut microbiota in the pathogenesis of IBD has gained a lot of attention. Accumulating evidence has demonstrated that IBD is accompanied with the dysbiosis of intestinal bacteria (Ni et al., 2017; Sartor and Wu, 2017; Vemuri et al., 2017; Guan, 2019). However, except for the bacterial components, abundant viruses, especially prokaryotic viruses (phages) reside in the human gut (Reyes et al., 2010; Minot et al., 2011). More precisely, 90% of all enteroviruses are phages, and the remaining 10% are plant and animal viruses (Breitbart et al., 2003). It is estimated that the human gut contains approximately 1015 phages in total (Dalmasso et al., 2014; Carding et al., 2017), which outnumbering the gut bacteria (1014) by 10 folds (Qin et al., 2010). In addition to occupy the huge population, the gut phages can also shape the bacterial community structure by lysing and killing the host bacteria, modulate the immune response, and mediate the anti-inflammatory response (Ogilvie and Jones, 2015; Łusiak-Szelachowska et al., 2017; Gogokhia and Round, 2021). Hence, further understanding of the alterations of gut phage community, and interactions between gut phages and host immune system are helpful for elucidating the underlying molecular mechanisms of pathogenesis in IBD.
So far, the current traditional medical drugs (e.g., aminosalicylates, corticosteroids, and immunosuppressive agents) and surgical operations for IBD treatment focused mostly on the relief of clinical symptoms and the disease’s secondary effects (Anand B. Pithadia, 2011; Magro et al., 2020). More recently, with the deep understanding of microbiota-related pathogenicity in IBD development, the advent of biological agents opens up a new era for the management of IBD. Moreover, the treatment goal has also evolved from control of intestinal inflammation toward mucosal healing, including a restoration of mucosal barrier function (Mao and Hu, 2016). Apart from probiotics, prebiotics, synbiotics and fecal microbiota transplant, phage therapy has also been considered as a potential tool for microbiota modification (Oka and Sartor, 2020).
In this review, we provide an overview of the biology of phages, interactions between phages and their host bacteria, the gut phages community in healthy individuals, the alterations of gut phages in IBD patients and experimental models, and the underlying molecular mechanisms (e.g., immune and inflammatory regulation) that involved in the initiation and progression of IBD. Besides, phage-based therapeutic approaches for IBD treatment will also be discussed.
Phage Biology and Interactions With Bacterial Host
Phages are viruses that infecting prokaryotic cells (e.g., bacteria and archaea), couldn’t reproduce without the host cellular machinery. Just like eukaryotic viruses, phages are either DNA- or RNA-based viruses, and the vast majority (96.3%) of them belong to the double-stranded DNA (dsDNA) viruses (Ackermann and Prangishvili, 2012).The host range of phage is defined as the strains or species of bacteria that a phage is able to infect (Hyman and Abedon, 2010). Most phages are only capable of infecting a narrow range of bacteria that are closely related. This specific selection depends on various factors, such as the specificity of phage’s host binding proteins, biochemical interactions during infection, presence of related prophages or particular plasmids, and bacterial phage-resistance mechanisms (Weinbauer, 2004; Diaz-Munoz and Koskella, 2014; Letarov and Kulikov, 2017).
The life cycles of phages encompass four patterns: lytic, lysogenic, chronic, and pseudolysogenic cycles (Weinbauer, 2004). In general, obligatory lytic phages (e.g., phage T1, T4 or T7) replicate through the lytic cycle. They bind to specific host-cell receptors, penetrate and infect host cell, hijack host cell’s replication and translation machinery to produce virions, produce lytic enzymes to lyse host cell, and finally release virions into the surrounding environment (Fernandes and Sao-Jose, 2018). Different from lytic phages, temperate phages (e.g., phage λ, P1 or P2) replicate through the lysogenic or lytic cycle. In a lysogenic cycle, they do not kill host cell and produce virions, but instead integrate the phage genome into the host’s chromosome or maintain as a plasmid, replicate with the bacterial host, and transmit to its progeny at each cell division (Carding et al., 2017). However, under some environmental stressors, such as DNA damage response, changes in nutrients, PH or temperature, exposure to antibiotics, hydrogen peroxide or foreign DNA, the temperate phages can be activated and switched to the lytic cycle (Howard-Varona et al., 2017).
Compared with the lytic and lysogenic cycle, the chronic cycle and pseudolysogeny remain poorly described. The chronic life cycle occurs in archaeal viruses or some filamentous phages (e.g., phage M13), which is characterized by continuously releasing the newly formed virions and affecting the growth rate of the infected host cell without causing the cell lysis (Munson-McGee et al., 2018; Stone et al., 2019). However, in a pseudolysogenic cycle, the phage genome neither integrates into the host cell’s chromosome nor propagates to produce newly virions. This phenomenon can be observed in poor nutritional conditions that limited bacterial DNA replication or protein synthesis (Cenens et al., 2013; Mirzaei and Maurice, 2017).
Characteristics of Gut Phages in Healthy Individuals
It has been confirmed that the first colonization and initiation of intestinal microbiota begins during the delivery process (Fanaro et al., 2003; Salazar et al., 2014). Accumulating evidence suggests that the gut bacteria is relatively simple in the beginning of life, then it changes rapidly within the first few days after birth, and then ultimately becomes more diverse and stable over time (Sakata et al., 2005; Klaassens et al., 2007; Yatsunenko et al., 2012; Pilar Manrique et al., 2016). But whether gut phages follow this pattern or not?
The gut phage consists of two elements, the temperate phage located within bacterial genomes and the free virions or virus-like particles (VLPs) (Sutton and Hill, 2019).The first study describing the infant gut phage community was conducted in fresh fecal samples (collected from one week to three months of age) by Breitbart et al. in 2008 (Breitbart et al., 2008). Direct epifluorescence microscope (EFM) counts showed that the meconium (the first fecal excretion of a newborn) couldn’t detect any VLPs. However, about 108 VLPs per gram wet weight of feces were detected by the end of the first week. Moreover, the following metagenomic sequencing analysis demonstrated that the identifiable sequences were dominated by dsDNA phage groups, such as Sipho-, Podo-, and Myoviruses. In addition, they also found that the diversity of the infant fecal viral community was extremely low and dynamic. This finding is consistent with previous studies, that low microbial diversity in infant guts (Magne et al., 2006; Breitbart et al., 2008). Later in 2015, another two studies also showed high dynamics of gut phages community during the first 2 and 2.5 years of life, respectively (Lim et al., 2015; Reyes et al., 2015). In addition to phages from the Caudovirales order (Siphoviridae, Inoviridae, Myoviridae and Podoviridae families), Lim et al. also described the presence of Microviridae family (single-stranded DNA genome) in the dominated phages, and a shift of phage community from Caudovirales-dominated to Microviridae-dominated composition by 24 months of age (Lim et al., 2015). Moreover, they also found that the richness and diversity of gut phages was greatest in the first 4 days of life, and then decreased with age (Lim et al., 2015).
Just like gut bacteria, the gut phage community is relatively stable in adults compared with infants (Lawrence et al., 2019). Studies have indicated that the gut phage community is dominated by temperate phages, and unique to individuals regardless of the genetic relatedness (Reyes et al., 2010). The extreme interpersonal diversity of human enteroviruses derives from two sources, i.e., persistence of a small portion of the global virome within the gut of each individual and rapid evolution of some long-term virome members (e.g., Microviridae) (Samuel Minot et al., 2013). Additionally, another recent research characterized the gut phage community as a mixture of three classes, including a core shared by more than 50% of individuals, another core found in 20-50% of people, and a set of phages that are rarely shared or unique to a person (Pilar Manrique et al., 2016). Furthermore, another study also reported the temporal stability and inter-individual diversity, and the predominance of virulent crAss-like (infecting bacteria of the order Bacteroidales) and Microviridae phages in the gut (Shkoporov et al., 2019). Significantly, the crAss-like phages are the most abundant viruses in the human gut, that accounting for up to 90% of the reads from human fecal viral metagenomes, and about 22% of the reads in the total metagenome (Dutilh et al., 2014; Yutin et al., 2018; Shkoporov et al., 2019). It is estimated that approximately 1015 phages exist in the intestine, which outnumber the commensal bacteria by 10-fold (Lozupone et al., 2012; Carding et al., 2017; Neil and Cadwell, 2018; Lin and Lin, 2019; Matijasic et al., 2020). However, about 108 -109 VLPs were detected in a gram of wet weight feces by direct counting with microscopy (Breitbart et al., 2008; Kim et al., 2011; Hoyles et al., 2014).
In general, current methods for studying the gut virome are dependent on direct observation and counting of VLPs by using EFM and transmission electron microscopy (TEM), isolation of individual phages infecting specific host bacteria strains by culturing, and the newly booming high-throughput metagenomic sequencing and bioinformatics technology (Shkoporov and Hill, 2019). Given that most of the viral metagenomic data can’t be found in the public databases, studies for the gut phage composition and function are just beginning (Reyes et al., 2012; Aggarwala et al., 2017).
Roles of Gut Phages in IBD Pathogenesis
As described above, the gut phage community maintains dynamic equilibrium under normal physiological conditions, whereas this balance can be disturbed by various factors, such as diet, lifestyle modification or pathological status (Minot et al., 2011; Coughlan et al., 2021; Marongiu et al., 2021). Advancing evidence has demonstrated that dysbiosis of gut phages may stimulate the development or aggravate the course of diseases, such as periodontal disease, Parkinson’s disease, type 2 diabetes, cancer or gastrointestinal disease (Azeredo et al., 2016; Manrique et al., 2017; Ma et al., 2018; Tetz et al., 2018; Santiago-Rodriguez and Hollister, 2019; Maronek et al., 2020). Up to now, a number of studies have been conducted in clinical samples and experimental models to demonstrate the relationship between gut phage homeostasis and the development and progression of IBD (Norman et al., 2015; Duerkop et al., 2018; Clooney et al., 2019). According to their experimental results, we concluded that gut phages may contribute to IBD pathogenesis through the following three pathways: alteration of gut phage diversity, regulation of gut bacterial population, and modulation of pro-inflammatory action and local immune response.
Alterations of Gut Phage Community in IBD Patients and Experimental Models
Different from bacteria or fungi, there is no universal marker gene that can be utilized to identify viruses, and the majority of obtained sequences do not exist in publicly available databases (Roux et al., 2015; Krishnamurthy and Wang, 2017). As for the changes of enteric phage community, most current studies are based on metagenomic sequencing of fecal samples and intestinal biopsies. Differences have been found between healthy individuals and IBD patients or experimental models (Table 1).
IBD Patients
Dysbiosis of intestinal bacteria has been implicated in the initiation and deterioration of IBD, but what is the origin of dysbiosis? Phages outnumber bacteria by a factor of 10, and exert a strong influence on bacterial diversity and population structure. In order to ascertain whether bacterial imbalance in IBD is related to gut phages, Lepage et al. conducted the first study in 2008 to measure the total viral community in CD patients (Lepage et al., 2008). Biopsy samples were obtained to detect VLPs in the mucosa by using EFM and TEM. Viral abundance was compared between healthy individuals and patients with CD, and also between the ulcerated and non-ulcerated mucosa of these patients. They found that CD patients harbored significantly more VLPs than healthy individuals (2.9×109 vs. 1.2×108 VLPs/biopsy), whereas CD ulcerated mucosa contained less VLPs than non-ulcerated mucosa (2.1×109 vs. 4.1×109 VLPs/biopsy) (Lepage et al., 2008). More recently, another study described the mucosa virus in patients with UC. Results showed a high abundance of Caudovirales phages, but decreased diversity, richness and eveness of mucosa Caudovirales in UC patients compared with healthy controls (Zuo et al., 2019). Moreover, they also found that abundance of Escherichia phage and Enterobacteria phage was significantly higher in the mucosa of UC patients than heathy controls.
In addition to changes of phage population on the mucosa, alterations in fecal samples and colonic tissues between IBD patients and healthy controls or between different disease types (CD and UC) also have been reported. A metagenomics analysis of gut tissue and wash samples has characterized a large abundance of phages when compared with pediatric CD patients and control individuals (Wagner et al., 2013). Moreover, they also found that the largest proportions of sequences were Bacteroides phage B10-8 and phage B124-14, and the Mycobacterium phage composition was different in ileum tissue samples between CD patients and controls (Wagner et al., 2013). A recent virome sequencing study conducted with the proximal and distal colonic wash samples from pediatric CD patients demonstrated high interpatient diversity and low but significant intra-patient variation between different sites (Yan et al., 2020). In another study, the differences of bacterial and viral communities in different type of samples from adult CD patients at different stages were investigated (Perez-Brocal et al., 2015). They found that phages were more abundant in feces (3 folds) than in biopsies, and the bacterial community reflects the disease status of individuals more accurately than the viral communities. Moreover, numerous viral biomarkers specifically associated with CD disease were identified, that phages infecting bacterial orders Alteromonadales and Clostridiales, including bacterial species Clostridium acetobutylicum and Retroviridae family were increased in subjects with CD (Perez-Brocal et al., 2015). Another metagenomics analysis of colonic biopsies has demonstrated that nearly half of the phages were associated with bacterial strains identified in the colon samples (Wang et al., 2015). Additionally, a recent study has investigated the dynamics of stool virome in very early onset (VEO) IBD, which is defined as onset of IBD before 6 years of age (Liang et al., 2020). Results showed that there is no significant different of total number of VLPs between VEO-IBD and healthy controls, but the VEO-IBD subjects exhibit a higher ratio of Caudovirales versus Microviridae compared to healthy controls (Liang et al., 2020).
As mentioned above, differences of gut phages population between CD and UC patients have also been investigated. A metagenomics analysis of stool samples from CD and UC patients demonstrated that Caudovirales phages significantly increased in IBD patients compared with healthy cohorts. Moreover, the gut phage community of CD and UC patients were disease- and cohort-specific (Norman et al., 2015). Later, Clooney et al. reanalyzed the above published dataset of healthy and IBD gut virome. Evidence has been found to prove that a healthy core of virulent phages is replaced by temperate phages in CD patients (Clooney et al., 2019). In another study, Fernandes et al. investigated the fecal virome in children with CD, UC and age-matched healthy controls. Results indicated that the relative abundance of Caudovirales was greater than that of Microviridae phages in both IBD and healthy controls, and the Caudovirales phages were more abundant in CD than UC (p=0.05) but not controls, and the richness of viral strains in Microvioridae but not Caudovirales was higher in controls than CD (p=0.05) but not UC cases (Fernandes et al., 2019). Another recent study demonstrated the ecological structure of the human gut temperate phage community by using publicly available whole-metagenome shotgun sequencing data (Integrative, 2014; Lloyd-Price et al., 2019; Nishiyama et al., 2020). Consistent with previous reports, they found the compositions of temperate phage community were highly specific to each individual and the abundance was different between IBD patients and non-IBD patients. In addition, temperate phages infecting Bacteroides uniformis and Bacteroides thetaiotaomicron were over-represented in active UC patients, whereas their hosts were under-represented in comparison with non-IBD patients (Nishiyama et al., 2020).
Animal Experiments
Apart from researches conducted in IBD patients, animal experiments have also been an important pathway to explore the roles of enteric phages in IBD pathogenesis. A recent metagenomics study indicated that the intestinal phage population altered and transited from an ordered state to a stochastic dysbiosis in a mice model with colitis (Duerkop et al., 2018). Interestingly, these alterations are similar to those observed in human IBD patients (Norman et al., 2015). Additionally, they observed a decrease in phage community diversity, an expansion of subsets of phages and a decrease of certain phages number (e.g., Clostridiales phages) during colitis (Duerkop et al., 2018).
In conclusion, current researches are focused on describing fecal and mucosal phage communities by using metagenomic sequencing and bioinformatic analysis. Most of the studies demonstrated the increased abundance and decreased diversity of Caudovirales phages both in CD and UC in comparison with health controls. However, another recent study showed no significant difference of gut phage number between IBD and healthy controls (Liang et al., 2020). Some of the experiments also showed alterations of certain phages, such as increased Escherichia phages and Enterobacteria phages in UC mucosa, decreased Clostridials phages during colitis, and increased Alteromonadales and Clostridiales phages in CD subjects (Figure 1).
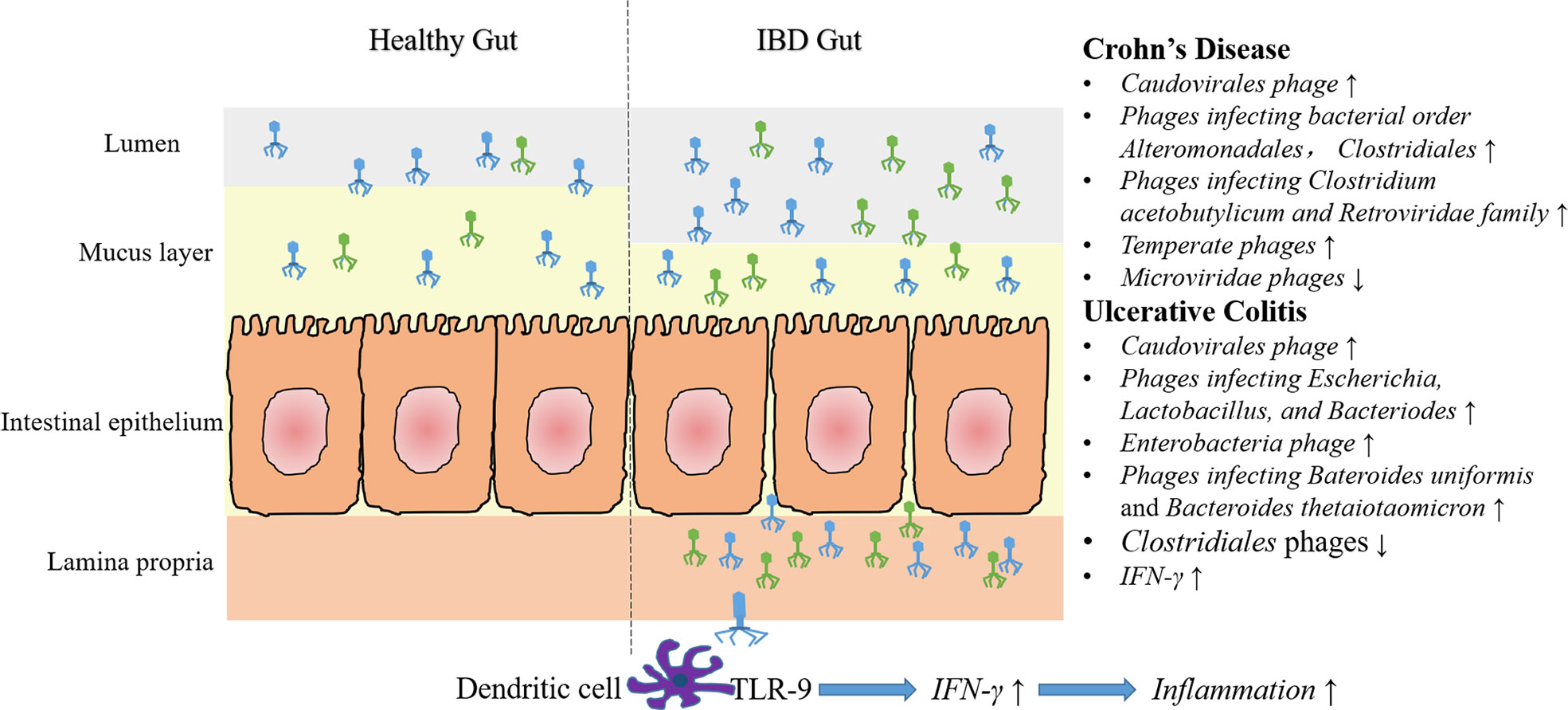
Figure 1 Characteristics of gut phage community and phage associated immune modulation in IBD. Compared with healthy gut, more caudovirales phages are found in the intestinal lumen and mucus layer. Moreover, some specific phage community changes have been demonstrated in CD and UC. In CD, an increased abundance of temperate phages and phages infecting bacterial order Alteromonadales, Clostridiales, Retroviridae family and Clostridium acetobutylicum, and a decreased abundance of Microviridae phages have been reported. In UC, numbers of phages infecting Escherichia, Lactobacillus and Bacteriodes (e.g., Bacteroides uniformis and Bacteroides thetaiotaomicron) and Enterobacteria phages increased, whereas Clostridiales phages decreased. It should be noted, however, some of the above results are just concluded from a single experiment, and that can’t be compared with each other, due to different samples and methods are used. Additionally, impaired epithelial barrier in IBD lead to increased intestinal permeability, that lead to the migration of many phage particles into the lamina propria or even the circulation. In vitro experiments showed that recognition of phage DNA via TLR9 on dendritic cells stimulated the production of IFN-γ, thereby aggravated intestinal inflammation.
Phages Affect IBD Through Bacterial Modulation
Virulent phages that are capable of lysing host bacteria were commonly found in the gut of patients with IBD (Clooney et al., 2019). It has been shown that phage invasion affected host bacteria, resulting in changes in bacterial abundance of specific species (Reyes et al., 2013). Researches have reported the dysbiosis of gut bacteria both in CD and UC patients, with a decreased diversity, expanded potentially pathogenic proteobacteria (e.g., E. coli, Fusobacteria) and reduced potentially protective Firmicutes (e.g., Faecalibacterium prausnitzii, Rumininococci and Clostridium clusters IV and XIVa) (Frank et al., 2007; Hoarau et al., 2016; Lloyd-Price et al., 2019). Nishiyama et al. have demonstrated an increased abundance of phages infecting Bacteroides uniformis and Bacteroides thetaiotaomicron, and a decreased abundance of their host bacteria (Nishiyama et al., 2020). Combining the above two information, we can infer that there is a certain relationship between the phage number and its host bacteria population. Germ-free (GF) animals, without any microbial colonization in their intestines, are perfect model systems for gut microbiota related studies (Uzbay, 2019; Qv et al., 2020). A recent study investigated in GF mice models showed that phage predation directly impacts susceptible bacteria, leading to cascading effects on other bacterial species, with consequences on the gut metabolome (Hsu et al., 2019).
Except for virulent phages, temperate phages can also influence bacterial fitness and diversity (Brussow et al., 2004). Phages substantially contribute to the genetic variability of bacteria by horizon gene transfer and increasing of the mutation rate (Abeles and Pride, 2014; Janka Babickova, 2015). Prophages carrying genes encoding antibiotic resistance may provide evolutionary advantages to pathogenic or probiotic bacteria. Besides, stress-induced activation of a prophage dormant in commensal or pathogenic bacteria might lead to the activation of its lytic cycle, thereby reduce the amount of the host bacteria. A metagenomic study showed a higher prevalence or abundance of phages infecting Faecalibacterium prausnitzii in stools of IBD patients than in those of healthy controls (Cornuault et al., 2018). Since less Faecalibacterium prausnitzii has been reported in IBD patients, they suggested that phages could trigger or aggravate Faecalibacterium prausnitzii depletion. But how or whether these phages being activated into lytic cycle in patients are not being described.
Overall, the current evidence is still limited, how phages directly or indirectly impact bacterial communities are barely unclear and speculatively feasible. A recent review also proposed a theoretical model of temperate phages in mediating the gut bacteria community (Lin et al., 2019). Further experimental studies are needed to reveal the complicated phage-bacteria interactions in IBD.
Phages Affect IBD Through Immune Modulation
It is well acknowledged that IBD is associated with the imbalance of immune response and inflammatory reaction. Studies have showed a relief of enteric viruses (rotavirus) in gut inflammation via toll-like receptor (TLR) 3 and TLR7-mediated interferon-β (IFN-β) production (Yang et al., 2016). While phages do not infect mammalian cells, several studies have demonstrated the direct and indirect influence of these prokaryotic viruses on host immune system (Hodyra-Stefaniak et al., 2015; Bollyky and Secor, 2019; Gogokhia and Round, 2021). Indeed, phages have been suggested to be a key element that helps to shape innate, humoral and cell-mediated immunity (Duerkop and Hooper, 2013).
First, phages act as a prominent defender of the mucosal barrier against bacteria. In vitro studies have showed that phages can adhere to mucus layer, thereby reduced microbial colonization and pathology. The adherence of phages was mediated by interactions between displayed immunoglobulin (Ig)-like domains of phage capsid proteins and glycan residues, such as those in mucin glycoprotein (Barr et al., 2013a).
In addition to provide a non-host-derived immunity, phages can also provide an acquired antimicrobial immunity, and help to control local inflammatory and autoimmune reactions in the gut (Górski and Weber-Dabrowska, 2005; Barr et al., 2013b). A recent study showed that both induction of innate and adaptive immunity was upregulated in phage-treated animals (Gogokhia et al., 2019). Results showed that the percentage of CD4+ and CD8+ T cells in the mesenteric lymph nodes (MLNs) were significantly increased. Besides, the proportions and numbers of CD4+ T cells within the Peyer’s patches (PP) were higher in GF mice that treated with phages compared with GF mice (Gogokhia et al., 2019). And in vitro experiments suggested that dendritic cell recognition of phage DNA can stimulate IFN-γ production through a TLR-9 dependent pathway, which functioned to exacerbate the intestinal inflammation and contribute to disease severity (Gogokhia et al., 2019).
Last but not least, just like bacteria, phages can pass the intestinal wall and migrate to lymph, peripheral blood and internal organs, and then directly modulate the host immune system (Górski et al., 2006). Besides, results have demonstrated that bacterial translocation from the gut to tissue induces inflammation and development of metabolic disease (Burcelin, 2016). Phages could limit bacterial translocation by directly eliminating sensitive bacteria, thereby indirectly inhibit gut inflammation that caused by bacterial translocation. Moreover, phages may downregulate gut immune cells such as dendritic cells and prevent pro-inflammatory action of these cells (Górski et al., 2012; Górski et al., 2016).
In conclusion, the current available data concerning the influence of gut phages on IBD by immune modulation is limited. The idea we came up above is based on the immune disorders in IBD and the regulation of immune response showed in phages. Further studies focused on the interactions between gut phages and immune response in IBD patients or animal models are urgently needed.
Use of Phages for IBD Treatment
Due to the relapsing inflammatory disorders in IBD patients, controlling of intestinal inflammatory is the primary target of treatment. Currently, traditional therapies includes 5-aminosalicylic acid derivatives (e.g., sulfasalazine and mesalazine), corticosteroides (e.g., prednisone, hydrocortisone, budesonide, prednisolone, dexamethasone) and immunosuppressive agents (e.g., azathioprine, methotrexate, mycophenolate, cyclosporine, tacrolimus, 6-mercaptopurine) (Moja et al., 2015; Wang et al., 2016; Matsumoto et al., 2016; Bots et al., 2018; Sokollik et al., 2018; Damiao et al., 2019; Nielsen et al., 2020; Antunes et al., 2021). Although these therapies have been proved effective in IBD patients, severe adverse events with impaired quality of life can’t be ignored (Quezada et al., 2018). Recent compelling evidence has demonstrated the crucial roles of resident microbiota in driving immune dysfunction and inflammation in IBD, thereby microbial-targeted therapies are being studied for IBD treatment (Oka and Sartor, 2020). Probiotics, prebiotics and fecal microbiota transplant are commonly used for microbiota-targeted therapies, which have been proved to be safe and potentially effective for correcting the dysregulated immune response (Derwa et al., 2017; Laurell and Sjoberg, 2017; Gagliardi et al., 2018; Zuo and Ng, 2018; Cohen et al., 2019). Besides, phage therapy, as another effective method for restoring gut microbiome homeostasis (Dixit et al., 2021), has also been proposed as a therapeutic target for IBD treatment.
Indeed, phages has long been used as a therapy to treat bacterial infectious diseases (e.g., cholera and bacillary dysentery), due to their function of lysing host bacteria (Summers, 1993; Ho, 2001). In particular, with the increasing emergence of multi-drug resistance bacteria, phages are being explored in targeting and killing specific infectious bacteria that associated with intestinal infection, such as Clostridioides difficile in colitis (Park et al., 2019) and Fusobacterium nucleatum in colorectal cancer (Machuca et al., 2010). In addition to their antibacterial activity, current phage therapy also focused on the immunomodulating properties.
Currently, phage therapy used for IBD treatment mainly targeted to the adherent invasive Escherichia coli (AIEC). Compared with healthy individuals, the AIEC strains were much more common in CD patients (Lamps et al., 2003). Moreover, studies have suggested the involvement of AIEC strain in maintaining intestinal inflammation in IBD (Sartor and Wu, 2017; Carolina Palmela et al., 2018; Chervy et al., 2020). Several researches have showed the effectiveness of phages in reducing intestinal E. coli colonization (Vahedi et al., 2018; Yu et al., 2018). Vahedi and his colleagues isolated specific phages against enteropathogenic E. coli from hospital sewage, and proved that single dose of phage cocktail (2×109 PFU/mL) was able to control the bacteria infection (Vahedi et al., 2018). A recent study has tested phage therapy against AIEC strain in a dextran sulfate sodium (DSS) induced colitis mouse model (Galtier et al., 2017). Results showed that a single day of oral treatment with phage cocktail significantly decreased the colonization of AIEC strain LF82, with reduced symptoms over a 2-week period. Additionally, in a double-blinded, placebo-controlled crossover trial, administration of a commercial cocktail of E. coli-targeting phages for 28 days selectively reduced the fecal E. coli loads without globally disrupt the gut microbiota community and increased anti-inflammatory cytokines IL-4 (Febvre et al., 2019).
In addition to the in vivo animal models, in vitro models also play vital roles in testing the therapeutic activity of gut phages. A recent study has developed an intestinal epithelium model able to produce mucus by co-culturing Caco-2 and HT29-MTX (4:1 ratio) cells (Nunez-Sanchez et al., 2020). This mammalian cell model will enable a better understanding of phage-bacteria interactions and the protective effects of phage therapy.
Collectively, researches about the efficacy of phage therapy in IBD treatment is scarce, either in clinical trials or animal experiments. More further studies are needed to clarity the ability of phages in modulate intestinal microbiota balance and immune response. Although the purified formulation of phages has been proved to be non-toxic (Divya Ganeshan and Hosseinidoust, 2019; Dixit et al., 2021), the safety of phage therapy still needed to be taken into consideration. Besides, researchers have also suggested the timing of administration of phage therapy as a challenge in the future studies (Janka Babickova, 2015).
Conclusions and Perspectives
As a widespread and incurable enteric disease, it is urgent and meaningful to reveal the pathogenic mechanisms of IBD. Studies that exploring the roles of gut microbiome in IBD pathogenesis should not focus solely on the bacterial composition, but neglect the enteroviruses, especially gut phages, which are huge number and closely related to the bacteria. The recent fast-growing in high-throughput sequencing and bioinformatics technologies has enabled major advances in human gut phageome researches. Current scattered researches have uncovered an increased abundance and decreased diversity of caudovirales phages in IBD patients or experimental models in comparison with healthy individuals. However, the majority of identified phage sequences are not yet taxonomically classified, that is to say annotated sequences do not exist in the public databases (Dalmasso et al., 2014; Shkoporov and Hill, 2019). Therefore, insights based on viral taxonomy require careful consideration. In the mucosa, CD patients were found to have more phages than healthy individuals, whereas the ulcerated mucosa had fewer phages than unaffected mucosa (Lepage et al., 2008). This paradoxical result may be caused by the incomplete databases. Moreover, methods that used in existing studies vary, that can impede our ability to compare the outputs of different studies. To be noted, whether an altered phageome is the consequence or the cause of IBD is currently not fully understood. Besides, the underlying mechanisms of phages affecting IBD through bacterial modulation and immune regulation still needs more experimental evidence.
Limited understanding of phage biology and disease microbiology suppressed the usage of phage therapy. Current phage therapy for IBD treatment mainly targeted the AIEC strain. The discovery of gut bacteria that directly linked to IBD development will accelerate the application of phage therapy. Besides, the efficacy of phage therapy should also pay attention to the influence of phages on immune response apart from the antibacterial properties.
In conclusion, many key information about the human gut phageome, phage-bacteria interactions, and phage-host interactions remains unclear. In the future, further optimization of techniques for phage isolation and identification may be required. Additionally, more in vivo and in vitro studies are needed to elucidate the roles of gut phages in IBD development and treatment.
Author Contributions
LQ researched the topics and drafted the manuscript. SM, YL, and JZ helped to search the reference during the preparation of the manuscript. LQ and LL revised the manuscript for publication. All authors read and approved the final manuscript.
Funding
This research was supported by the Independent Project Fund of the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases (2020) and grants from the National Natural Science Foundation of China (81790631 and 81330011).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abeles, S. R., Pride, D. T. (2014). Molecular Bases and Role of Viruses in the Human Microbiome. J. Mol. Biol. 426 (23), 3892–3906. doi: 10.1016/j.jmb.2014.07.002
Abraham, C., Cho, J. H. (2009). Inflammatory Bowel Disease. N Engl. J. Med. 361 (21), 2066–2078. doi: 10.1056/NEJMra0804647
Ackermann, H. W., Prangishvili, D. (2012). Prokaryote Viruses Studied by Electron Microscopy. Arch. Virol. 157 (10), 1843–1849. doi: 10.1007/s00705-012-1383-y
Aggarwala, V., Liang, G., Bushman, F. D. (2017). Viral Communities of the Human Gut: Metagenomic Analysis of Composition and Dynamics. Mob DNA 8, 12. doi: 10.1186/s13100-017-0095-y
Anand, B., Pithadia, S. J. (2011). Treatment of Inflammatory Bowel Disease (IBD). Pharmacol. Rep. 63, 629–642. doi: 10.1016/S1734-1140(11)70575-8
Ananthakrishnan, A. N., Bernstein, C. N., Iliopoulos, D., Macpherson, A., Neurath, M. F., Ali, R. A. R., et al. (2018). Environmental Triggers in IBD: A Review of Progress and Evidence. Nat. Rev. Gastroenterol. Hepatol. 15 (1), 39–49. doi: 10.1038/nrgastro.2017.136
Antunes, J. C., Seabra, C. L., Domingues, J. M., Teixeira, M. O., Nunes, C., Costa-Lima, S. A., et al. (2021). Drug Targeting of Inflammatory Bowel Diseases by Biomolecules. Nanomaterials (Basel) 11 (8), 2035. doi: 10.3390/nano11082035
Azeredo, J., Pinto, G., Silva, M. D., Peddey, M., Sillankorva, S. (2016). The Role of Bacteriophages in Periodontal Health and Disease. Future Microbiol. 11 (10), 1359–1369. doi: 10.2217/fmb-2016-0081
Barr, J. J., Auro, R., Furlan, M., Whiteson, K. L., Erb, M. L., Pogliano, J., et al. (2013a). Bacteriophage Adhering to Mucus Provide a non-Host-Derived Immunity. Proc. Natl. Acad. Sci. U.S.A. 110 (26), 10771–10776. doi: 10.1073/pnas.1305923110
Barr, J. J., Youle, M., Rohwer, F. (2013b). Innate and Acquired Bacteriophage-Mediated Immunity. Bacteriophage 3 (3), e25857. doi: 10.4161/bact.25857
Bollyky, P. L., Secor, P. R. (2019). The Innate Sense of Bacteriophages. Cell Host Microbe 25 (2), 177–179. doi: 10.1016/j.chom.2019.01.020
Bots, S., Gecse, K., Barclay, M., D’Haens, G. (2018). Combination Immunosuppression in IBD. Inflamm. Bowel Dis. 24 (3), 539–545. doi: 10.1093/ibd/izx065
Breitbart, M., Haynes, M., Kelley, S., Angly, F., Edwards, R. A., Felts, B., et al. (2008). Viral Diversity and Dynamics in an Infant Gut. Res. Microbiol. 159 (5), 367–373. doi: 10.1016/j.resmic.2008.04.006
Breitbart, M., Hewson, I., Felts, B., Mahaffy, J. M., Nulton, J., Salamon, P., et al. (2003). Metagenomic Analyses of an Uncultured Viral Community From Human Feces. J. Bacteriol. 185 (20), 6220–6223. doi: 10.1128/jb.185.20.6220-6223.2003
Brussow, H., Canchaya, C., Hardt, W. D. (2004). Phages and the Evolution of Bacterial Pathogens: From Genomic Rearrangements to Lysogenic Conversion. Microbiol. Mol. Biol. Rev. 68 (3), 560–602, table of contents. doi: 10.1128/MMBR.68.3.560-602.2004
Burcelin, R. (2016). Gut Microbiota and Immune Crosstalk in Metabolic Disease. Mol. Metab. 5 (9), 771–781. doi: 10.1016/j.molmet.2016.05.016
Carding, S. R., Davis, N., Hoyles, L. (2017). Review Article: The Human Intestinal Virome in Health and Disease. Aliment Pharmacol. Ther. 46 (9), 800–815. doi: 10.1111/apt.14280
Carolina Palmela, C. C., Xu, Z., Torres, J., Sevrin, G., Hirten, R., Barnich, N., et al. (2018). Adherent-Invasive Escherichia Coli in Inflammatory Bowel Disease. Gut 67, 574–587. doi: 10.1136/gutjnl-2017-314903
Cenens, W., Makumi, A., Mebrhatu, M. T., Lavigne, R., Aertsen, A. (2013). Phage-Host Interactions During Pseudolysogeny: Lessons From the Pid/Dgo Interaction. Bacteriophage 3 (1), e25029. doi: 10.4161/bact.25029
Chervy, M., Barnich, N., Denizot, J. (2020). Adherent-Invasive E. Coli: Update on the Lifestyle of a Troublemaker in Crohn’s Disease. Int. J. Mol. Sci. 21 (10), 3734. doi: 10.3390/ijms21103734
Clooney, A. G., Sutton, T. D. S., Shkoporov, A. N., Holohan, R. K., Daly, K. M., O’Regan, O., et al. (2019). Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease. Cell Host Microbe 26, 764–778. doi: 10.1016/j.chom.2019.10.009
Cohen, L. J., Cho, J. H., Gevers, D., Chu, H. (2019). Genetic Factors and the Intestinal Microbiome Guide Development of Microbe-Based Therapies for Inflammatory Bowel Diseases. Gastroenterology 156 (8), 2174–2189. doi: 10.1053/j.gastro.2019.03.017
Cornuault, J. K., Petit, M.-A., Mariadassou, M., Benevides, L., Moncaut, E., Langella, P., et al. (2018). Phages Infecting Faecalibacterium Prausnitzii Belong to Novel Viral Genera That Help to Decipher Intestinal Viromes. Microbiome 6 (1), 65–65. doi: 10.1186/s40168-018-0452-1
Coughlan, S., Das, A., O’Herlihy, E., Shanahan, F., O’Toole, P. W., Jeffery, I. B. (2021). The Gut Virome in Irritable Bowel Syndrome Differs From That of Controls. Gut Microbes 13 (1), 1–15. doi: 10.1080/19490976.2021.1887719
Dalmasso, M., Hill, C., Ross, R. P. (2014). Exploiting Gut Bacteriophages for Human Health. Trends Microbiol. 22 (7), 399–405. doi: 10.1016/j.tim.2014.02.010
Damiao, A., de Azevedo, M. F. C., Carlos, A. S., Wada, M. Y., Silva, T. V. M., Feitosa, F. C. (2019). Conventional Therapy for Moderate to Severe Inflammatory Bowel Disease: A Systematic Literature Review. World J. Gastroenterol. 25 (9), 1142–1157. doi: 10.3748/wjg.v25.i9.1142
Derwa, Y., Gracie, D. J., Hamlin, P. J., Ford, A. C. (2017). Systematic Review With Meta-Analysis: The Efficacy of Probiotics in Inflammatory Bowel Disease. Aliment Pharmacol. Ther. 46 (4), 389–400. doi: 10.1111/apt.14203
Diaz-Munoz, S. L., Koskella, B. (2014). Bacteria-Phage Interactions in Natural Environments. Adv. Appl. Microbiol. 89, 135–183. doi: 10.1016/B978-0-12-800259-9.00004-4
Divya Ganeshan, S., Hosseinidoust, Z. (2019). Phage Therapy With a Focus on the Human Microbiota. Antibiotics (Basel) 8 (3), 131. doi: 10.3390/antibiotics8030131
Dixit, K., Chaudhari, D., Dhotre, D., Shouche, Y., Saroj, S. (2021). Restoration of Dysbiotic Human Gut Microbiome for Homeostasis. Life Sci. 278:119622. doi: 10.1016/j.lfs.2021.119622
Duerkop, B. A., Hooper, L. V. (2013). Resident Viruses and Their Interactions With the Immune System. Nat. Immunol. 14 (7), 654–659. doi: 10.1038/ni.2614
Duerkop, B. A., Kleiner, M., Paez-Espino, D., Zhu, W., Bushnell, B., Hassell, B., et al. (2018). Murine Colitis Reveals a Disease-Associated Bacteriophage Community. Nat. Microbiol. 3 (9), 1023–1031. doi: 10.1038/s41564-018-0210-y
Dutilh, B. E., Cassman, N., McNair, K., Sanchez, S. E., Silva, G. G., Boling, L., et al. (2014). A Highly Abundant Bacteriophage Discovered in the Unknown Sequences of Human Faecal Metagenomes. Nat. Commun. 5, 4498. doi: 10.1038/ncomms5498
Fanaro, S., Chierici, R., Guerrini, P., Vigi, V. (2003). Intestinal Microflora in Early Infancy: Composition and Development. Acta Paediatr. Suppl. 91 (441), 48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x
Febvre, H. P., Rao, S., Gindin, M., Goodwin, N. D. M., Finer, E., Vivanco, J. S., et al. (2019). PHAGE Study: Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults. Nutrients 11 (3), 666. doi: 10.3390/nu11030666
Fernandes, S., Sao-Jose, C. (2018). Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 10 (8), 396. doi: 10.3390/v10080396
Fernandes, M. A., Verstraete, S. G., Phan, T. G., Deng, X., Stekol, E., LaMere, B., et al. (2019). Enteric Virome and Bacterial Microbiota in Children With Ulcerative Colitis and Crohn Disease. J. Pediatr. Gastroenterol. Nutr. 68 (1), 30–36. doi: 10.1097/MPG.0000000000002140
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., Pace, N. R. (2007). Molecular-Phylogenetic Characterization of Microbial Community Imbalances in Human Inflammatory Bowel Diseases. Proc. Natl. Acad. Sci. U.S.A. 104 (34), 13780–13785. doi: 10.1073/pnas.0706625104
Gagliardi, A., Totino, V., Cacciotti, F., Iebba, V., Neroni, B., Bonfiglio, G., et al. (2018). Rebuilding the Gut Microbiota Ecosystem. Int. J. Environ. Res. Public Health 15 (8), 1679. doi: 10.3390/ijerph15081679
Galtier, M., De Sordi, L., Sivignon, A., de Vallee, A., Maura, D., Neut, C., et al. (2017). Bacteriophages Targeting Adherent Invasive Escherichia Coli Strains as a Promising New Treatment for Crohn’s Disease. J. Crohns Colitis 11 (7), 840–847. doi: 10.1093/ecco-jcc/jjw224
Geremia, A., Biancheri, P., Allan, P., Corazza, G. R., Di Sabatino, A. (2014). Innate and Adaptive Immunity in Inflammatory Bowel Disease. Autoimmun. Rev. 13 (1), 3–10. doi: 10.1016/j.autrev.2013.06.004
Gogokhia, L., Buhrke, K., Bell, R., Hoffman, B., Brown, D. G., Hanke-Gogokhia, C., et al. (2019). Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe 25 (2), 285–299.e288. doi: 10.1016/j.chom.2019.01.008
Gogokhia, L., Round, J. L. (2021). Immune-Bacteriophage Interactions in Inflammatory Bowel Diseases. Curr. Opin. Virol. 49, 30–35. doi: 10.1016/j.coviro.2021.04.010
Górski, A., Międzybrodzki, R., Borysowski, J., Dąbrowska, K., Wierzbicki, P., Ohams, M., et al. (2012). Phage as a Modulator of Immune Responses: Practical Implications for Phage Therapy. Adv. Virus Res. 83, 41–71. doi: 10.1016/b978-0-12-394438-2.00002-5
Górski, A., Międzybrodzki, R., Weber-Dąbrowska, B., Fortuna, W., Letkiewicz, S., Rogóż, P., et al. (2016). Phage Therapy: Combating Infections With Potential for Evolving From Merely a Treatment for Complications to Targeting Diseases. Front. Microbiol. 7:1515. doi: 10.3389/fmicb.2016.01515
Górski, A., Wazna, E., Dabrowska, B. W., Dabrowska, K., Switała-Jeleń, K., Miedzybrodzki, R. (2006). Bacteriophage Translocation. FEMS Immunol. Med. Microbiol. 46 (3), 313–319. doi: 10.1111/j.1574-695X.2006.00044.x
Górski, A., Weber-Dabrowska, B. (2005). The Potential Role of Endogenous Bacteriophages in Controlling Invading Pathogens. Cell Mol. Life Sci. 62 (5), 511–519. doi: 10.1007/s00018-004-4403-6
Guan, Q. (2019). a Comprehensive Review and Update on the Pathogenesis of Inflammatory Bowel Disease. J. Immunol. Res. 2019:7247238. doi: 10.1155/2019/7247238
Ho, K. (2001). Bacteriophage Therapy for Bacterial Infections. Rekindling a Memory From the Pre-Antibiotics Era. Perspect. Biol. Med. 44 (1), 1–16. doi: 10.1353/pbm.2001.0006
Hoarau, G., Mukherjee, P. K., Gower-Rousseau, C., Hager, C., Chandra, J., Retuerto, M. A., et al. (2016). Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. mBio 7 (5), e01250–16. doi: 10.1128/mBio.01250-16
Hodyra-Stefaniak, K., Miernikiewicz, P., Drapala, J., Drab, M., Jonczyk-Matysiak, E., Lecion, D., et al. (2015). Mammalian Host-Versus-Phage Immune Response Determines Phage Fate In Vivo. Sci. Rep. 5:14802. doi: 10.1038/srep14802
Howard-Varona, C., Hargreaves, K. R., Abedon, S. T., Sullivan, M. B. (2017). Lysogeny in Nature: Mechanisms, Impact and Ecology of Temperate Phages. ISME J. 11 (7), 1511–1520. doi: 10.1038/ismej.2017.16
Hoyles, L., McCartney, A. L., Neve, H., Gibson, G. R., Sanderson, J. D., Heller, K. J., et al. (2014). Characterization of Virus-Like Particles Associated With the Human Faecal and Caecal Microbiota. Res. Microbiol. 165 (10), 803–812. doi: 10.1016/j.resmic.2014.10.006
Hsu, B. B., Gibson, T. E., Yeliseyev, V., Liu, Q., Lyon, L., Bry, L., et al. (2019). Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe 25 (6), 803–814.e805. doi: 10.1016/j.chom.2019.05.001
Hyman, P., Abedon, S. T. (2010). Bacteriophage Host Range and Bacterial Resistance. Adv. Appl. Microbiol. 70, 217–248. doi: 10.1016/s0065-2164(10)70007-1
Integrative, H.M.P.R.N.C (2014). The Integrative Human Microbiome Project: Dynamic Analysis of Microbiome-Host Omics Profiles During Periods of Human Health and Disease. Cell Host Microbe 16 (3), 276–289. doi: 10.1016/j.chom.2014.08.014
Janka Babickova, R. G. (2015). Pathological and Therapeutic Interactions Between Bacteriophages, Microbes and the Host in Inflammatory Bowel Disease. World J. Gastroenterol. 21 (40), 11321–31130. doi: 10.3748/wjg.v21
Kaplan, G. G. (2015). The Global Burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 12 (12), 720–727. doi: 10.1038/nrgastro.2015.150
Kim, M. S., Park, E. J., Roh, S. W., Bae, J. W. (2011). Diversity and Abundance of Single-Stranded DNA Viruses in Human Feces. Appl. Environ. Microbiol. 77 (22), 8062–8070. doi: 10.1128/AEM.06331-11
Klaassens, E. S., de Vos, W. M., Vaughan, E. E. (2007). Metaproteomics Approach to Study the Functionality of the Microbiota in the Human Infant Gastrointestinal Tract. Appl. Environ. Microbiol. 73 (4), 1388–1392. doi: 10.1128/AEM.01921-06
Krishnamurthy, S. R., Wang, D. (2017). Origins and Challenges of Viral Dark Matter. Virus Res. 239, 136–142. doi: 10.1016/j.virusres.2017.02.002
Lamps, L. W., Madhusudhan, K. T., Havens, J. M., Greenson, J. K., Bronner, M. P., Chiles, M. C., et al. (2003). Pathogenic Yersinia DNA Is Detected in Bowel and Mesenteric Lymph Nodes From Patients With Crohn’s Disease. Am. J. Surg. Pathol. 27 (2), 220–227. doi: 10.1097/00000478-200302000-00011
Laurell, A., Sjoberg, K. (2017). Prebiotics and Synbiotics in Ulcerative Colitis. Scand. J. Gastroenterol. 52 (4), 477–485. doi: 10.1080/00365521.2016.1263680
Lawrence, D., Baldridge, M. T., Handley, S. A. (2019). Phages and Human Health: More Than Idle Hitchhikers. Viruses 11 (7), 587. doi: 10.3390/v11070587
Lepage, P., Colombet, J., Marteau, P., Sime-Ngando, T., Dore, J., Leclerc, M. (2008). Dysbiosis in Inflammatory Bowel Disease: A Role for Bacteriophages? Gut 57 (3), 424–425. doi: 10.1136/gut.2007.134668
Letarov, A. V., Kulikov, E. E. (2017). Adsorption of Bacteriophages on Bacterial Cells. Biochem. Biokhimiia 82 (13), 1632–1658. doi: 10.1134/S0006297917130053
Liang, G., Conrad, M. A., Kelsen, J. R., Kessler, L. R., Breton, J., Albenberg, L. G., et al. (2020). The Dynamics of the Stool Virome in Very Early Onset Inflammatory Bowel Disease. J. Crohns Colitis. 14 (11), 1600–1610. doi: 10.1093/ecco-jcc/jjaa094
Lim, E. S., Zhou, Y., Zhao, G., Bauer, I. K., Droit, L., Ndao, I. M., et al. (2015). Early Life Dynamics of the Human Gut Virome and Bacterial Microbiome in Infants. Nat. Med. 21 (10), 1228–1234. doi: 10.1038/nm.3950
Lin, D. M. L., Lin, H. C. (2019). A Theoretical Model of Temperate Phages as Mediators of Gut Microbiome Dysbiosis. F1000Res 8, F1000. doi: 10.12688/f1000research.18480.1
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature 569 (7758), 655–662. doi: 10.1038/s41586-019-1237-9
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., Knight, R. (2012). Diversity, Stability and Resilience of the Human Gut Microbiota. Nature 489 (7415), 220–230. doi: 10.1038/nature11550
Łusiak-Szelachowska, M., Weber-Dąbrowska, B., Jończyk-Matysiak, E., Wojciechowska, R., Górski, A. (2017). Bacteriophages in the Gastrointestinal Tract and Their Implications. Gut Pathog. 9, 44–44. doi: 10.1186/s13099-017-0196-7
Machuca, P., Daille, L., Vines, E., Berrocal, L., Bittner, M. (2010). Isolation of a Novel Bacteriophage Specific for the Periodontal Pathogen Fusobacterium Nucleatum. Appl. Environ. Microbiol. 76 (21), 7243–7250. doi: 10.1128/AEM.01135-10
Magne, F., Abely, M., Boyer, F., Morville, P., Pochart, P., Suau, A. (2006). Low Species Diversity and High Interindividual Variability in Faeces of Preterm Infants as Revealed by Sequences of 16S Rrna Genes and PCR-Temporal Temperature Gradient Gel Electrophoresis Profiles. FEMS Microbiol. Ecol. 57 (1), 128–138. doi: 10.1111/j.1574-6941.2006.00097.x
Magro, F., Cordeiro, G., Dias, A. M., Estevinho, M. M. (2020). Inflammatory Bowel Disease - non-Biological Treatment. Pharmacol. Res. 160, 105075. doi: 10.1016/j.phrs.2020.105075
Maloy, K. J., Powrie, F. (2011). Intestinal Homeostasis and Its Breakdown in Inflammatory Bowel Disease. Nature 474 (7351), 298–306. doi: 10.1038/nature10208
Manrique, P., Dills, M., Young, M. J. (2017). The Human Gut Phage Community and its Implications for Health and Disease. Viruses 9 (6), 141. doi: 10.3390/v9060141
Mao, R., Hu, P. J. (2016). The Future of IBD Therapy: Where are We and Where Should We Go Next? Dig Dis. 34 (1-2), 175–179. doi: 10.1159/000443135
Maronek, M., Link, R., Ambro, L., Gardlik, R. (2020). Phages and Their Role in Gastrointestinal Disease: Focus on Inflammatory Bowel Disease. Cells 9 (4), 1013. doi: 10.3390/cells9041013
Marongiu, L., Burkard, M., Venturelli, S., Allgayer, H. (2021). Dietary Modulation of Bacteriophages as an Additional Player in Inflammation and Cancer. Cancers (Basel) 13 (9), 2036. doi: 10.3390/cancers13092036
Matijasic, M., Mestrovic, T., Paljetak, H. C., Peric, M., Baresic, A., Verbanac, D. (2020). Gut Microbiota Beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD. Int. J. Mol. Sci. 21 (8), 2668. doi: 10.3390/ijms21082668
Matsumoto, T. M., Watanabe, K., Hisamatsu, T., Nakase, H., Yoshimura, N., Lshida, T., et al. (2016). Adalimumab Monotherapy and a Combination With Azathioprine for Crohn’s Disease: A Prospective, Randomized Trial. J. Crohns Colitis 10 (11), 1259–1266(11), 1259-1266. doi: 10.1093/ecco-jcc/jjw152
Ma, Y., You, X., Mai, G., Tokuyasu, T., Liu, C. (2018). A Human Gut Phage Catalog Correlates the Gut Phageome With Type 2 Diabetes. Microbiome 6 (1), 24. doi: 10.1186/s40168-018-0410-y
Minot, S., Sinha, R., Chen, J., Li, H., Keilbaugh, S. A., Wu, G. D., et al. (2011). The Human Gut Virome: Inter-Individual Variation and Dynamic Response to Diet. Genome Res. 21 (10), 1616–1625. doi: 10.1101/gr.122705.111
Mirzaei, M. K., Maurice, C. F. (2017). Menage a Trois in the Human Gut: Interactions Between Host, Bacteria and Phages. Nat. Rev. Microbiol. 15 (7), 397–408. doi: 10.1038/nrmicro.2017.30
Moja, L., Danese, S., Fiorino, G., Del Giovane, C., Bonovas, S. (2015). Systematic Review With Network Meta-Analysis: Comparative Efficacy and Safety of Budesonide and Mesalazine (Mesalamine) for Crohn’s Disease. Aliment Pharmacol. Ther. 41 (11), 1055–1065. doi: 10.1111/apt.13190
Molodecky, N. A., Soon, I. S., Rabi, D. M., Ghali, W. A., Ferris, M., Chernoff, G., et al. (2012). Increasing Incidence and Prevalence of the Inflammatory Bowel Diseases With Time, Based on Systematic Review. Gastroenterology 142 (1), 46–e30. doi: 10.1053/j.gastro.2011.10.001
Munson-McGee, J. H., Snyder, J. C., Young, M. J. (2018). Archaeal Viruses From High-Temperature Environments. Genes (Basel) 9 (3), 128. doi: 10.3390/genes9030128
Neil, J. A., Cadwell, K. (2018). The Intestinal Virome and Immunity. J. Immunol. 201 (6), 1615–1624. doi: 10.4049/jimmunol.1800631
Nielsen, O. H., Steenholdt, C., Juhl, C. B., Rogler, G. (2020). Efficacy and Safety of Methotrexate in the Management of Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis of Randomized, Controlled Trials. EClinicalMedicine 20, 100271. doi: 10.1016/j.eclinm.2020.100271
Nishiyama, H., Endo, H., Blanc-Mathieu, R., Ogata, H. (2020). Ecological Structuring of Temperate Bacteriophages in the Inflammatory Bowel Disease-Affected Gut. Microorganisms 8 (11), 1663. doi: 10.3390/microorganisms8111663
Ni, J., Wu, G. D., Albenberg, L., Tomov, V. T. (2017). Gut Microbiota and IBD: Causation or Correlation? Nat. Rev. Gastroenterol. Hepatol. 14 (10), 573–584. doi: 10.1038/nrgastro.2017.88
Norman, J. M., Handley, S. A., Baldridge, M. T., Droit, L., Liu, C. Y., Keller, B. C., et al. (2015). Disease-Specific Alterations in the Enteric Virome in Inflammatory Bowel Disease. Cell 160 (3), 447–460. doi: 10.1016/j.cell.2015.01.002
Nunez-Sanchez, M. A., Colom, J., Walsh, L., Buttimer, C., Bolocan, A. S., Pang, R., et al. (2020). Characterizing Phage-Host Interactions in a Simplified Human Intestinal Barrier Model. Microorganisms 8 (9), 1374. doi: 10.3390/microorganisms8091374
Ogilvie, L. A., Jones, B. V. (2015). The Human Gut Virome: A Multifaceted Majority. Front. Microbiol. 6:918. doi: 10.3389/fmicb.2015.00918
Oka, A., Sartor, R. B. (2020). Microbial-Based and Microbial-Targeted Therapies for Inflammatory Bowel Diseases. Digestive Dis. Sci. 65 (3), 757–788. doi: 10.1007/s10620-020-06090-z
Park, H., Laffin, M. R., Jovel, J., Millan, B., Hyun, J. E., Hotte, N., et al. (2019). The Success of Fecal Microbial Transplantation in Clostridium Difficile Infection Correlates With Bacteriophage Relative Abundance in the Donor: A Retrospective Cohort Study. Gut Microbes 10 (6), 676–687. doi: 10.1080/19490976.2019.1586037
Perez-Brocal, V., Garcia-Lopez, R., Nos, P., Beltran, B., Moret, I., Moya, A. (2015). Metagenomic Analysis of Crohn’s Disease Patients Identifies Changes in the Virome and Microbiome Related to Disease Status and Therapy, and Detects Potential Interactions and Biomarkers. Inflamm. Bowel Dis. 21 (11), 2515–2532. doi: 10.1097/mib.0000000000000549
Pilar Manrique, B. B., Walka, S. T., van der Oost, J., de Vos, W. M., Young, M. J. (2016). Healthy Human Gut Phageome. PNAS 113 (37), 10400–10405. doi: 10.1073/pnas.1601060113
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 464 (7285), 59–65. doi: 10.1038/nature08821
Quezada, S. M., McLean, L. P., Cross, R. K. (2018). Adverse Events in IBD Therapy: The 2018 Update. Expert Rev. Gastroenterol. Hepatol. 12 (12), 1183–1191. doi: 10.1080/17474124.2018.1545574
Qv, L., Yang, Z., Yao, M., Mao, S., Li, Y., Zhang, J., et al. (2020). Methods for Establishment and Maintenance of Germ-Free Rat Models. Front. Microbiol. 11:1148. doi: 10.3389/fmicb.2020.01148
Reyes, A., Blanton, L. V., Cao, S., Zhao, G., Manary, M., Trehan, I., et al. (2015). Gut DNA Viromes of Malawian Twins Discordant for Severe Acute Malnutrition. Proc. Natl. Acad. Sci. U.S.A. 112 (38), 11941–11946. doi: 10.1073/pnas.1514285112
Reyes, A., Haynes, M., Hanson, N., Angly, F. E., Heath, A. C., Rohwer, F., et al. (2010). Viruses in the Faecal Microbiota of Monozygotic Twins and Their Mothers. Nature 466 (7304), 334–338. doi: 10.1038/nature09199
Reyes, A., Semenkovich, N. P., Whiteson, K., Rohwer, F., Gordon, J. I. (2012). Going Viral: Next-Generation Sequencing Applied to Phage Populations in the Human Gut. Nat. Rev. Microbiol. 10 (9), 607–617. doi: 10.1038/nrmicro2853
Reyes, A., Wu, M., McNulty, N. P., Rohwer, F. L., Gordon, J. I. (2013). Gnotobiotic Mouse Model of Phage-Bacterial Host Dynamics in the Human Gut. Proc. Natl. Acad. Sci. U.S.A. 110 (50), 20236–20241. doi: 10.1073/pnas.1319470110
Roux, S., Hallam, S. J., Woyke, T., Sullivan, M. B. (2015). Viral Dark Matter and Virus-Host Interactions Resolved From Publicly Available Microbial Genomes. Elife 4, e08490. doi: 10.7554/eLife.08490
Sakata, S., Tonooka, T., Ishizeki, S., Takada, M., Sakamoto, M., Fukuyama, M., et al. (2005). Culture-Independent Analysis of Fecal Microbiota in Infants, With Special Reference to Bifidobacterium Species. FEMS Microbiol. Lett. 243 (2), 417–423. doi: 10.1016/j.femsle.2005.01.002
Salazar, N., Arboleya, S., Valdes, L., Stanton, C., Ross, P., Ruiz, L., et al. (2014). The Human Intestinal Microbiome at Extreme Ages of Life. Dietary Intervention as a Way to Counteract Alterations. Front. Genet. 5:406. doi: 10.3389/fgene.2014.00406
Samuel Minot, A. B., Chehoud, C., Wu, G. D., Lewis, J. D., Bushman, F. D. (2013). Rapid Evolution of the Human Gut Virome. PNAS 110 (30), 12450–12455. doi: 10.1073/pnas.1300833110
Santiago-Rodriguez, T. M., Hollister, E. B. (2019). Human Virome and Disease: High-Throughput Sequencing for Virus Discovery, Identification of Phage-Bacteria Dysbiosis and Development of Therapeutic Approaches With Emphasis on the Human Gut. Viruses 11 (7), 656. doi: 10.3390/v11070656
Sartor, R. B., Wu, G. D. (2017). Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology 152 (2), 327–339 e324. doi: 10.1053/j.gastro.2016.10.012
Shkoporov, A. N., Clooney, A. G., Sutton, T. D. S., Ryan, F. J., Daly, K. M., Nolan, J. A., et al. (2019). The Human Gut Virome is Highly Diverse, Stable, and Individual Specific. Cell Host Microbe 26 (4), 527–541 e525. doi: 10.1016/j.chom.2019.09.009
Shkoporov, A. N., Hill, C. (2019). Bacteriophages of the Human Gut: The “Known Unknown” of the Microbiome. Cell Host Microbe 25 (2), 195–209. doi: 10.1016/j.chom.2019.01.017
Sokollik, C., Fournier, N., Rizzuti, D., Braegger, C. P., Nydegger, A., Schibli, S., et al. (2018). The Use of 5-Aminosalicylic Acid in Children and Adolescents With Inflammatory Bowel Disease. J. Clin. Gastroenterol. 52 (10), e87–e91. doi: 10.1097/MCG.0000000000000864
Stone, E., Campbell, K., Grant, I., McAuliffe, O. (2019). Understanding and Exploiting Phage-Host Interactions. Viruses 11 (6), 567. doi: 10.3390/v11060567
Summers, W. C. (1993). Cholera and Plague in India : The Bacteriophage Inquiry of 1927-193 6. J. Hist. Med. Allied Sci. 48 (3), 275-301. doi: 10.1093/jhmas/48.3.275
Sutton, T. D. S., Hill, C. (2019). Gut Bacteriophage: Current Understanding and Challenges. Front. Endocrinol. (Lausanne) 10:784. doi: 10.3389/fendo.2019.00784
Tetz, G., Brown, S. M., Hao, Y. H., Tetz, V. (2018). Parkinson’s Disease and Bacteriophages as Its Overlooked Contributors. Sci. Rep. 8, 10812. doi: 10.1038/s41598-018-29173-4
Turpin, W., Goethel, A., Bedrani, L., Croitoru Mdcm, K. (2018). Determinants of IBD Heritability: Genes, Bugs, and More. Inflamm. Bowel Dis. 24 (6), 1133–1148. doi: 10.1093/ibd/izy085
Uzbay, T. (2019). Germ-Free Animal Experiments in the Gut Microbiota Studies. Curr. Opin. Pharmacol. 49, 6–10. doi: 10.1016/j.coph.2019.03.016
Vahedi, A., Soltan Dallal, M. M., Douraghi, M., Nikkhahi, F., Rajabi, Z., Yousefi, M., et al. (2018). Isolation and Identification of Specific Bacteriophage Against Enteropathogenic Escherichia Coli (EPEC) and In Vitro and In Vivo Characterization of Bacteriophage. FEMS Microbiol. Lett. 365 (16), fny136. doi: 10.1093/femsle/fny136
Vemuri, R., Gundamaraju, R., Eri, R. (2017). Role of Lactic Acid Probiotic Bacteria in IBD. Curr. Pharm. Des. 23 (16), 2352–2355. doi: 10.2174/1381612823666170207100025
Wagner, J., Maksimovic, J., Farries, G., Sim, W. H., Bishop, R. F., Cameron, D. J., et al. (2013). Bacteriophages in Gut Samples From Pediatric Crohn’s Disease Patients: Metagenomic Analysis Using 454 Pyrosequencing. Inflammation Bowel Dis. 19 (8), 1598–1608. doi: 10.1097/MIB.0b013e318292477c
Wang, W., Jovel, J., Halloran, B., Wine, E., Patterson, J., Ford, G., et al. (2015). Metagenomic Analysis of Microbiome in Colon Tissue From Subjects With Inflammatory Bowel Diseases Reveals Interplay of Viruses and Bacteria. Inflammation Bowel Dis. 21 (6), 1419–1427. doi: 10.1097/MIB.0000000000000344
Wang, Y., Parker, C. E., Bhanji, T., Feagan, B. G., MacDonald, J. K. (2016). Oral 5-Aminosalicylic Acid for Induction of Remission in Ulcerative Colitis. Cochrane Database Syst. Rev. 4:CD000543. doi: 10.1002/14651858.CD000543.pub4
Weinbauer, M. G. (2004). Ecology of Prokaryotic Viruses. FEMS Microbiol. Rev. 28 (2), 127–181. doi: 10.1016/j.femsre.2003.08.001
Yan, A., Butcher, J., Mack, D., Stintzi, A. (2020). Virome Sequencing of the Human Intestinal Mucosal-Luminal Interface. Front. Cell Infect. Microbiol. 10:582187. doi: 10.3389/fcimb.2020.582187
Yang, J. Y., Kim, M. S., Kim, E., Cheon, J. H., Lee, Y. S., Kim, Y., et al. (2016). Enteric Viruses Ameliorate Gut Inflammation via Toll-Like Receptor 3 and Toll-Like Receptor 7-Mediated Interferon-Beta Production. Immunity 44 (4), 889–900. doi: 10.1016/j.immuni.2016.03.009
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human Gut Microbiome Viewed Across Age and Geography. Nature 486 (7402), 222–227. doi: 10.1038/nature11053
Yutin, N., Makarova, K. S., Gussow, A. B., Krupovic, M., Segall, A., Edwards, R. A., et al. (2018). Discovery of an Expansive Bacteriophage Family That Includes the Most Abundant Viruses From the Human Gut. Nat. Microbiol. 3 (1), 38–46. doi: 10.1038/s41564-017-0053-y
Yu, L., Wang, S., Guo, Z., Liu, H., Sun, D., Yan, G., et al. (2018). A Guard-Killer Phage Cocktail Effectively Lyses the Host and Inhibits the Development of Phage-Resistant Strains of Escherichia Coli. Appl. Microbiol. Biotechnol. 102 (2), 971–983. doi: 10.1007/s00253-017-8591-z
Zuo, T., Lu, X. J., Zhang, Y., Cheung, C. P., Lam, S., Zhang, F., et al. (2019). Gut Mucosal Virome Alterations in Ulcerative Colitis. Gut 68 (7), 1169–1179. doi: 10.1136/gutjnl-2018-318131
Keywords: inflammatory bowel disease (IBD), gut phage, immune response, pathogenesis, therapeutic target
Citation: Qv L, Mao S, Li Y, Zhang J and Li L (2021) Roles of Gut Bacteriophages in the Pathogenesis and Treatment of Inflammatory Bowel Disease. Front. Cell. Infect. Microbiol. 11:755650. doi: 10.3389/fcimb.2021.755650
Received: 09 August 2021; Accepted: 09 November 2021;
Published: 25 November 2021.
Edited by:
Paul De Vos, University Medical Center Groningen, NetherlandsReviewed by:
Xingmin Sun, University of South Florida, United StatesClaudio Cermelli, University of Modena and Reggio Emilia, Italy
Copyright © 2021 Qv, Mao, Li, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lanjuan Li, ljli@zju.edu.cn
 Lingling Qv
Lingling Qv Sunbing Mao
Sunbing Mao  Lanjuan Li
Lanjuan Li