Tenacibaculosis caused by Tenacibaculum maritimum: Updated knowledge of this marine bacterial fish pathogen
- 1Department of Fish Diseases and Management, Faculty of Veterinary Medicine, Suez Canal University, Ismailia, Egypt
- 2Department of Veterinary Microbiology, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand
- 3Center of Excellence in Fish Infectious Diseases (CE FID), Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand
- 4Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Suez Canal University, Ismailia, Egypt
- 5Department of Medical Microbiology and Immunology, Faculty of Medicine, Assuit University, Assuit, Egypt
- 6Laboratory Medicine Department, Faculty of Applied Medical Sciences, Umm Al-Qura University, Makkah, Saudi Arabia
- 7Directorate of Veterinary Medicine, Ismailia, Egypt
- 8Laboratorio de Patología de Organismos Acuáticos y Biotecnología Acuícola, Facultad de Ciencias Biológicas, Universidad Andrés Bello, Viña del Mar, Chile
- 9Interdisciplinary Center for Aquaculture Research (INCAR), Viña del Mar, Chile
- 10Centro de Investigación Marina Quintay (CIMARQ), Universidad Andrés Bello, Quintay, Chile
Tenacibaculosis occurs due to the marine bacterial pathogen Tenacibaculum maritimum. This ulcerative disease causes high mortalities for various marine fish species worldwide. Several external clinical signs can arise, including mouth erosion, epidermal ulcers, fin necrosis, and tail rot. Research in the last 15 years has advanced knowledge on the traits and pathogenesis mechanisms of T. maritimum. Consequently, significant progress has been made in defining the complex host-pathogen relationship. Nevertheless, tenacibaculosis pathogenesis is not yet fully understood. Continued research is urgently needed, as demonstrated by recent reports on the re-emerging nature of tenacibaculosis in salmon farms globally. Current sanitary conditions compromise the development of effective alternatives to antibiotics, in addition to hindering potential preventive measures against tenacibaculosis. The present review compiles knowledge of T. maritimum reported after the 2006 review by Avendaño-Herrera and colleagues. Essential aspects are emphasized, including antigenic and genomic characterizations and molecular diagnostic procedures. Further summarized are the epidemiological foundations of the T. maritimum population structure and elucidations as to the virulence mechanisms of pathogenic isolates, as found using biological, microbiological, and genomic techniques. This comprehensive source of reference will undoubtable serve in tenacibaculosis prevention and control within the marine fish farming industry. Lastly, knowledge gaps and valuable research areas are indicated as potential guidance for future studies.
1. Introduction
Aquaculture is a prominent and promising food-production industry, globally providing animal-protein sources and helping to cover food shortage gaps arising from population overshoots (Subasinghe et al., 2009). Approximately 17% of animal protein and 7% of all protein for human consumption is derived from fish; indeed, more than 3.3 million people rely on fish for up to 20% of average per capita animal-protein intake (FAO, 2020). Increasing demand requires increased fish production, a particularly acute situation considering the recession of fisheries in recent decades (Molenaar and Caddell, 2019). New technologies are helping to propel the industry, including shrimp and, specifically, fish production (Kumar and Engle, 2016). However, more intensive farming may negatively impact culturing waters, as associated with environmental pollution and the emergence and spread of aquatic pathogens (Leung and Bates, 2013). Water-borne diseases are, a critical issue facing the global aquaculture industry (Melba et al., 2021). Aquatic diseases are not a singular event but, rather, one of a linked chain of events involving interactions among pathogenic microorganisms, aquatic species, and environmental conditions (Lieke et al., 2020).
Relatively few pathogenic bacteria account for most of the financial losses incurred by the industry. However, new “emerging” diseases arise biannually and constantly impose health challenges limiting fish production. Classically, pathogenic bacteria are classified as extracellular, facultative intracellular, and obligate intracellular (Silva, 2012). The survival and dispersion of facultative bacteria are uncertain, particularly as environmental conditions worsen. By contrast, most pathogenic microbes regularly exist as assembled colonies on the host or freely live in the aquatic environment (Hassan et al., 2021). Interestingly, all bacterial types could turn pathogenic if fish immunity is compromised (Kirjusina et al., 2007).
Prominent among the causes for economic losses in global aquaculture are emerging diseases caused by Tenacibaculum species (Family Flavobacteriaceae, Phylum Bacteroidetes). Most available studies involving Tenacibaculum species associate disease in marine fish with infection caused by Tenacibaculum maritimum (see review by Avendaño-Herrera et al., 2006a). This disease was originally described as a Flexibacter infection in association with mortalities among farmed red (Pagrus major) and blackhead (Acanthopagrus schlegelii) seabream in Japan (Masumura and Wakabayashi, 1977). From 1977 to 2006, the disease was variably termed in relation to external lesions, fish species, and fish age. These changes finally included a taxonomic name change for the bacterium (see reviews by Bernardet, 1998; Avendaño-Herrera et al., 2006a).
Tenacibaculum spp. infections are today commonly referred to as marine tenacibaculosis or, simply, tenacibaculosis, a term originally coined to describe the ulcerative disease caused by the filamentous T. maritimum bacterium (Avendaño-Herrera et al., 2006a). Tenacibaculosis is generally associated with gross lesions on the body, including ulcerative and/or necrotic skin lesions, an eroded/hemorrhagic mouth, frayed fins, and tail rot (Avendaño-Herrera et al., 2006a; Elanco, 2018). Despite the association between T. maritimum isolation and the aforementioned clinical signs, consensus as to the primary or opportunistic nature of this pathogen is lacking. Research by numerous groups in recent decades has significantly advanced understandings of the characteristics and pathogenesis of T. maritimum, thereby helping to define complex host-pathogen relationships (Elanco, 2018). However, tenacibaculosis pathogenesis is still not fully elucidated.
Most published reviews on T. maritimum report evidence existing prior to 2006. Therefore, the purpose of the present review is to compile and summarize knowledge obtained on T. maritimum within the last 15 years. Focus is given to phenotypic, antigenic, and genomic characterizations and to recent molecular diagnostic procedures. Additionally summarized are the epidemiological foundations of the T. maritimum population structure (i.e., host and geographical distribution). Finally reported are findings elucidating the virulence mechanisms of pathogenic T. maritimum isolates through biological, microbiological, and genomic techniques. The compilation of this information will prove invaluable for tenacibaculosis prevention and control in marine fish farming. Lastly, emphasis given to still existing knowledge gaps and valuable research areas will serve as guidance for future investigative efforts.
2. Description of Tenacibaculum maritimum
The original taxonomic classification of the previously mentioned Flexibacter bacterium was a topic of controversy and confusion until Suzuki et al. (2001). This research reclassified said bacterium as T. maritimum, in addition to adding Tenacibaculum ovolyticum and two other species to the newly formed genus. Today, T. maritimum is the type species of the Tenacibaculum genus, which includes 33 validly and 3 invalidly published species (https://www.bacterio.net/genus/tenacibaculum). All are exclusively found in marine environments attached to or associated with the surface of marine organisms such as macroalgae, various invertebrates, and fish. In addition to T. maritimum, seven other Tenacibaculum fish pathogens have been identified – T. ovolyticum from Atlantic halibut (Hippoglossus hippoglossus) (Hansen et al., 1992; Avendaño-Herrera et al., 2022a); T. discolor from European and Asian sea bass (Dicentrarchus labrax and Lates calcarifer) (Piñeiro-Vidal et al., 2008a); T. soleae from Senegalese sole (Solea senegalensis) (Piñeiro-Vidal et al., 2008b); T. dicentrarchi from Atlantic salmon (Salmo salar) (Piñeiro-Vidal et al., 2012; Avendaño-Herrera et al., 2016); T. finnmarkense from rainbow trout (Oncorhynchus mykiss) (Olsen et al., 2020); and T. singaporense (Miyake et al., 2020) and T. piscium (Olsen et al., 2020; Avendaño-Herrera et al., 2022b) from Corkwing wrasse (Symphodus melops). Hereinafter, this review will focus only on T. maritimum.
The physical traits of T. maritimum are as follows: filamentous; Gram-negative; usually sized 2 – 30 µm in length with a diameter of 0.5 µm; gliding motility; and positive growth on media containing ≥ 10% seawater (i.e., no growth with only NaCl as the only added salt) (Suzuki et al., 2001). Several traits are common to the Tenacibaculum genus, including the production of a primarily zeaxanthin yellow carotenoid pigment, flexirubin-type pigment are absent; the major respiratory quinone is menaquinone-6 and are catalase- and -oxidase positive (Suzuki, 2015). As a point of difference compared to other Tenacibaculum species, including T. ovolyticum (Hansen et al., 1992) and T. finnmarkense (Olsen et al., 2020), T. maritimum absorb Congo red on solid agar when some colonies on agar are directly flooding with drops of a 0 ± 01% aqueous solution of the dye (Avendaño-Herrera et al., 2004a).
Intraspecific phenotypic similarities among T. maritimum isolates may aid in taxonomic identification when compared to other Tenacibaculum species pathogenic to fish. Indeed, some of the above-mentioned morphotype and physiological properties can serve as initial tube and plate analyses for species differentiation, in addition to the application of traditional, ready-made identification systems (e.g., API ZYM and API 50 CH) (Kolygas et al., 2012; Yardimci and Timur, 2016; Fernández-Álvarez and Santos, 2018). However, API results are not always identical due to factors such as the inoculum, isolate, and incubation temperature, among others. For example, differences are reported for tests on nitrate reduction and hydrogen-sulfide production (Avendaño-Herrera et al., 2004a). The phenotypic and biochemical characteristics used for the precise detection and differentiation of T. maritimum against other fish pathogens are given in Table 1. The complete genome sequence of the type strain NCIMB 2154T consists of a circular chromosome of 3,435,971 base pairs with 2,866 predicted protein-coding genes, excluding any plasmid (Pérez-Pascual et al., 2017). Some of these genes are implicated in T. maritimum virulence and pathogenicity, as discussed in section 8 Virulence Factors.
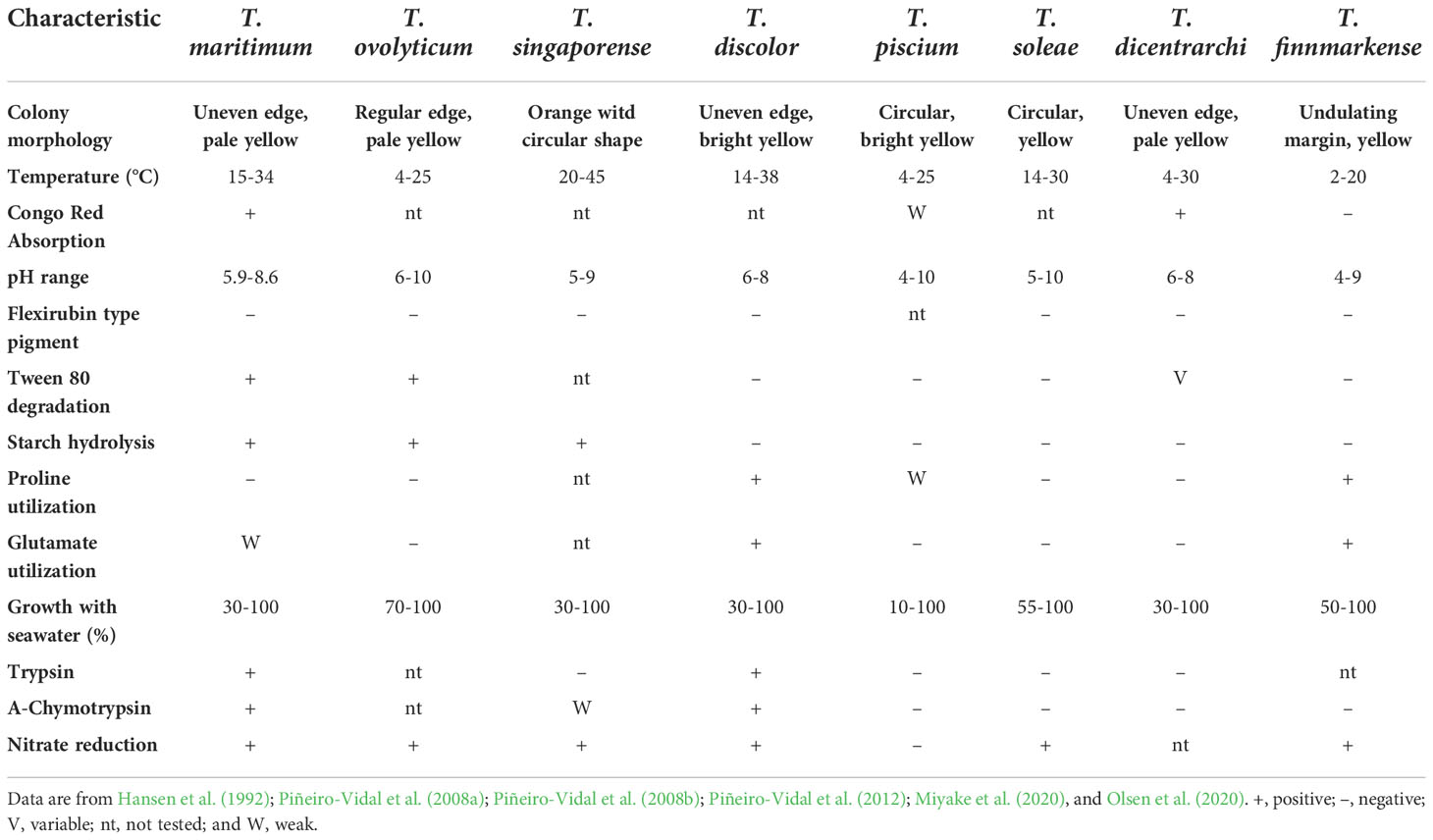
Table 1 Differential phenotypic and biochemical characteristics of the type strains for Tenacibaculum species associated with fish diseases.
3. Antigenic and genetic characterization of Tenacibaculum maritimum
Various typing methods have been applied in epidemiological studies of T. maritimum, including the slide agglutination test; the dot-blot and immunoblotting assays; multiplex PCR (mPCR) serotyping; the randomly amplified polymorphic DNA (RAPD) method; the multi-locus sequence analysis (MLSA); and assembled draft genomes, among others. These methods confirm the existence of intra-specific diversity among T. maritimum isolates. In the early 2000s, three serotypes were identified through immunoblot analyses of lipopolysaccharides with varying degrees of relation to host fish species (Avendaño-Herrera et al., 2004a; Avendaño-Herrera et al., 2005a). Subsequent serotyping research describes up to four serotypes in T. maritimum (Piñeiro-Vidal et al., 2007; Fernández-Álvarez and Santos, 2018). This antigenic diversity makes it difficult to select candidate strain(s) for intra-species vaccine development, as described in section 9 Treatment and Vaccination Programs for T. maritimum.
Despite proposed serotyping schemes for T. maritimum, several disadvantages exist for conventional mammal-antisera-based serotyping, including sera availability and result interpretations (Gorski, 2021). These difficulties have spurred the development of new molecular techniques, such as those proposed by Whitfield et al. (2020), where determination of the O-antigen structure allows for differentiating serotypes. This method is based on genes coding for the biosynthesis of the O-antigen, which are often located in a single genomic cluster (i.e., the O-AGC) and fall into one of the following three classes: (i) nucleotide sugar biosynthesis genes; (ii) sugar transferase genes; or (iii) genes required for O-unit translocations, chain synthesis, and chain-length determination. Application of this knowledge is evidenced by Lopez et al. (2022a), a study that identified the O-AGC gene cluster in T. maritimum using a set of recently published T. maritimum genomes (Pérez-Pascual et al., 2017; Bridel et al., 2020) and primers specifically designed for the O-antigens of representative isolates of the defined serotypes (Avendaño-Herrera et al., 2004a; Avendaño-Herrera et al., 2005a). These breakthroughs allowed Lopez et al. (2022a) to develop an mPCR-based serotyping scheme that combines traditional serotyping with a genome-wide association study.
At the genetic level, cluster analyses of the RAPD-PCR profiles for 29 isolates and 3 reference strains of T. maritimum showed that, regardless of the oligonucleotide primer employed, the isolates and strains were separated into two main groups that strongly correlated with the host species and/or the described O serotypes (Avendaño-Herrera et al., 2004a; Avendaño-Herrera et al., 2004b). Elucidation of the T. maritimum genome through next generation sequencing techniques has allowed for (i) the design of primers directed at constitutive genes of the pathogen and (ii) the characterization of population diversity. While an advancement, these evolved developments naturally limited the use of random amplification techniques, such as those previously proposed by Avendaño-Herrera et al. (2004a), because they are based on a comparison of amplification patterns. These previous limitations have been overcome by Habib et al. (2014) through the application of MLSA to 114 isolates representatives of the Tenacibaculum genus, including T. maritimum isolates reflecting the global diversity of this species. The proposed method is based on eleven loci located within single-copy protein-coding genes conserved across the Flavobacteriaceae family were used to design generic degenerated PCR primers for the Tenacibaculum genus. Habib et al. (2014) highlighted the existence of a cohesive T. maritimum group and recommended the use thereof to monitor infections caused by Tenacibaculum species. However, not all primer sets given by Habib et al. (2014) had the same efficiency during amplification, leading Småge et al. (2016a) to propose the use of other primer sequences targeting five genes.
Confirmation of the MLSA scheme as a powerful tool for taxonomic affiliation and isolate identification of T. maritimum has been demonstrated with field isolates (Småge et al., 2016a; Valdes et al., 2021; Lopez et al., 2022a). Such power has even been shown for the pathogen T. dicentrarchi (Avendaño-Herrera et al., 2016; Klakegg et al., 2019) and several pathogenic species that have yet to be described (Olsen et al., 2017). In this sense, Frisch et al. (2018a) noted that genotyping in the Canadian region increased their understandings of the genetic profile of T. maritimum. These authors reported two new sequence types among Western Canadian isolates of T. maritimum. Despite the known antigenic and genetic heterogeneity of T. maritimum hindering development of effective biological products (e.g., vaccines), the available investigative techniques for isolates of interest within the T. maritimum population, including mPCR-serotyping and MLSA, represent a significant and robust advancement towards commercial vaccines.
4. Natural reservoirs and prevalence factors
The natural reservoir(s) of T. maritimum are unclear as little ecological data for this microorganism exist. However, the propensity of T. maritimum to survive for a prolonged time in different seawater microcosms (Avendaño-Herrera et al., 2006b), as well as its ability to infect different fish species indicates that water is an important infection route. This may explain why T. maritimum has been isolated from sediments, tank surfaces, and water exposed to infected fish stocks (Santos et al., 1999). Some reports suggest that natural T. maritimum outbreaks occur shortly after transferring fish from hatchery tanks to inshore net cages, could be explained by handling stress and/or skin damage, indicating pathogen-to-host access by horizontal transmission in seawater (see Avendaño-Herrera et al., 2006a).
Certain bacteria may use the ecosystem along with the normal microbiota of marine organisms as reservoirs (Higgins, 2000), as has been described for T. maritimum. To this end, jellyfish (Phialella quadrata) blooms could conceivably provide both a colonization site (via initial nematocyst-related injuries) and a source of Tenacibaculum infection (Ferguson et al., 2010). Barker et al. (2009) also suggested that the sea lice (Lepeophtheirus salmonis) might serve as an organic substrate capable of extending Tenacibaculum persistence in seawater. More recently, Llewellyn et al. (2017) tracked the diversity and composition of S. salar skin surface microbiota throughout a complete L. salmonis infection cycle, finding that the Tenacibaculum genus was perhaps the most abundant taxon across all mucus and water samples. Indeed, the ability of T. maritimum like member of Flavobacteriaceae family to survive in host skin mucus and cope with the bactericidal activity of the skin mucus reflects the potential of this tissue to act as a major reservoir (Magariños et al., 1995; Staroscik and Nelson, 2008; Mabrok et al., 2016; Guardiola et al., 2019).
One factor that plays a key role in the presence of T. maritimum is seawater temperature. Acute and/or chronic exposure to suboptimal temperatures is generally suppressive, especially for the adaptive immunity of fish, thus increasing disease prevalence (Abram et al., 2017). A significant rise in the severity and prevalence of tenacibaculosis has been reported at higher seawater temperatures and salinities (see Avendaño-Herrera et al., 2006a). Other evidence reported by Yamamoto et al. (2010) demonstrates the influence of water temperature on the experimental induction of tenacibaculosis in Japanese olive flounder (Paralichthys olivaceus) using immersion and dilution methods. These authors observed high mortality rates between 17 and 26 °C but not below 17 °C or above 26 °C. In contrast, Brosnahan et al. (2019) declared that warm seawater temperatures have no influence or strong correlation with an increased prevalence of T. maritimum. In turn, the rising seawater temperature around Tasmania has increased tenacibaculosis frequency (van Gelderen et al., 2009a), but, interestingly, massive tenacibaculosis outbreaks have also been recorded during the winter (Soltani et al., 1996; Olsen et al., 2011). Conversely, Abd El-Galil and Hashem (2012a) reported high disease prevalence in black damsel (Neoglyphidodon melas), Picasso triggerfish (Rhinecanthus assasi), and broomtail wrasse (Cheilinus lunulatus) during winter but no T. maritimum infections during summer. Likewise, López et al. (2010) reported three outbreaks of tenacibaculosis in wedge sole fish (Dicologlossa cuneata) cultured at 20.5°C in southwestern Spain. The winter water temperature ranged mainly from 15-20°C, which, as concluded by the authors, is perfect for the growth and propagation of T. maritimum. Taken together, these reports could explain the explosive appearance of tenacibaculosis outbreaks worldwide, since, as is known, climate change has increased the temperature of seawater globally (Marcogliese, 2008). Further studies are required to elucidate this issue further and assess the risks associated with this and other environmental parameters.
In addition to water temperature, tenacibaculosis is often associated with poor management conditions (e.g., high rearing density, reduced or excessive feed administration, and/or mechanical damage of the skin and mucus barrier) (van Gelderen et al., 2011; Escribano et al., 2020). Adverse growing conditions increase the intensity and distribution pattern of the causative agent in various organs of hosts (Cepeda and Santos, 2002). On the other hand, natural outbreaks of tenacibaculosis at farms without a prior history of T. maritimum infection imply that transferred fish may act as asymptomatic vectors. This is consistent with the proposed horizontal transmission of tenacibaculosis, as confirmed by van Gelderen et al. (2010a). Transmission of T. maritimum through seawater and directly from one host to another have been proposed as routes of infection (Fringuelli et al., 2012). Likewise, the successful recovery of bacteria from the intestine of apparently healthy infected fish without any histological and pathological lesions reflects another potential reservoir for T. maritimum (Faílde et al., 2013). Given all these factors, T. maritimum has the potential to harm a wide range of wild, captive, and farmed fish species, making it a pathogen worthy of serious consideration as a threat to the global fish-aquaculture industry (Elanco, 2018).
5. Geographical distribution and host susceptibility
The geographical and host distribution of T. maritimum isolates is presented in depth by Avendaño-Herrera et al. (2006a). Importantly, and as mentioned in section 4 Natural Reservoirs and Prevalence Factors, T. maritimum is a mesophilic microorganism that grows well between 15 and 34°C, a range representative of aquatic environments in which fish are farmed (Suzuki, 2015). Temperature range can likewise vary when considering T. maritimum isolates from warmer-water fish species (Frisch et al., 2018a), even T. maritimum isolates from salmon and other cold-water species appear to have lower optimal temperatures. Although the optimum growth temperature under laboratory culture conditions is 30°C. Incubation temperatures from 15 to 18°C are probably suitable for most salmonid-related isolates. Despite the slow growth of cultures seeded directly from external lesions, this bacterium forms distinctive colonies with mutable rhizoid edges and adherence capacity on low-nutrient media containing sea salts (e.g., Flexibacter maritimus medium [FMM] and Marine Agar 2216) (Pazos et al., 1996). Marine agar supplemented with 50 µg/mL kanamycin has also been devised for the recovery and isolation of T. maritimum directly from skin ulcers (Frisch et al., 2018a). Underscoring the need for suitable culturing media, microbiology companies have developed an FMM broth1 as the medium of choice for laboratories without immediate access to a natural source of seawater.
These advancements in isolation and culturing mean that T. maritimum has now been retrieved and recognized in more geographies and hosts than those described up to 2006. In Europe, T. maritimum has been isolated in Italy from farmed tub gurnard (Chelidonichthys lucerna L.) and wild turbot (Scophthalmus maximus) (Magi et al., 2007); and in Norway from the lumpsucker cleaner fish (Cyclopterus lumpus) (Småge et al., 2016b). In Asia, the pathogen was retrieved from Korean olive flounder (P. olivaceus) (Jang et al., 2009). In Africa, this bacterium has been reported in Egypt in association with gilthead sea bream (Sparus aurata) and European sea bass (D. labrax) (Moustafa et al., 2014; Moustafa et al., 2015). Tenacibaculum maritimum has even extended beyond food-oriented fish to some valuable ornamental fish, including the Picasso triggerfish, black damselfish, and broomtail wrasse in Egypt (Abd El-Galil and Hashiem, 2011; Abd El-Galil and Hashem, 2012a; Haridy et al., 2015). In French Polynesia, this bacterium has been described as the cause of massive mortalities (up to 90%) among the tropical orbicular batfish (Platax orbicularis) (Lopez et al., 2022b). Despite significant efforts to remain free-of T. maritimum infections in Chile, which is the second largest producer of salmon after Norway, outbreaks of T. maritimum have been reported in farmed Atlantic salmon (Apablaza et al., 2017) and rainbow trout (Valdes et al., 2021), and outbreaks have likewise been reported in Canada among Sockeye salmon (Oncorhynchus nerka) (Nekouei et al., 2018). Susceptible hosts for T. maritimum and the respective geographic origins are briefly summarized in Table 2.
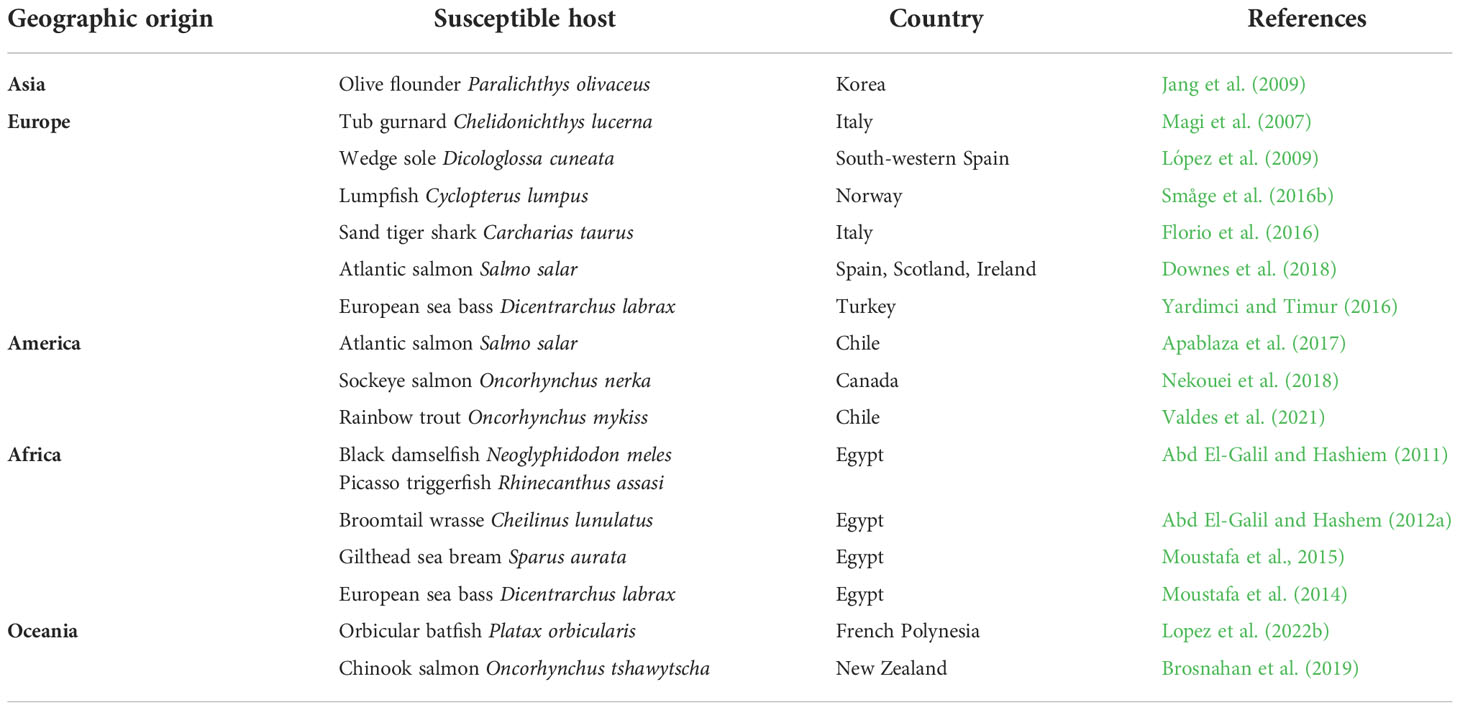
Table 2 Susceptible host species for Tenacibaculum maritimum and the respective geographic origin (updated since 2006).
6. Presumptive and confirmative diagnostic procedures for Tenacibaculum maritimum
The initial diagnosis of tenacibaculosis caused by T. maritimum was, for a long period, based on clinical signs associated with diseased fish. However, field experience indicates that assigning a specific diagnosis based solely on this criterion is quite difficult as the same signs could also be attributable to other pathogens (Fernández-Álvarez and Santos, 2018). Insufficiency as a diagnostic method becomes clearer knowing that different Tenacibaculum species can be found in the same lesion. Indeed, clinical signs from coinfections can be difficult to distinguish since information is lacking as to which pathogen is responsible for which sign of infection. For example, tenacibaculosis is commonly associated with external clinical signs in Atlantic salmon, but the lesions are very different when caused by T. maritimum (i.e., body and caudal fin lesions) or T. dicentrarchi (i.e., head and snout lesions) (Nowlan et al., 2020). An erroneous attribution for the cause of mortality is most worrying when considering that such diagnoses are used to substantiate metaphylactic antibacterial treatments to cages containing healthy, moribund, infected, and/or disease-carrying fish. Another common, yet controversial, practice within Chilean salmon farms is the diagnosis of death caused by Tenacibaculum spp. when signs of mixed infection with the intracellular, facultative fish pathogen Piscirickettsia salmonis are present. Prior to 2018, such cases were classified exclusively as piscirickettsiosis (Irgang and Avendaño-Herrera, 2022). These cases are now classified as infections caused by a “Tenacibaculum-complex.” For instance, the first isolation of T. maritimum together with T. dicentrarchi from Chilean-farmed Atlantic salmon occurred during a harmful algae bloom caused by Pseudochattonella spp. (Apablaza et al., 2017). Similar tenacibaculosis coinfections have been reported in salmon farms in Norway (Olsen et al., 2011; Småge et al., 2016b), Australia (Wilson et al., 2019), and Canada (Nowlan et al., 2021).
Several studies further indicate differences in the susceptibility of certain fish species to T. maritimum infection based on age, and lesions vary greatly between affected fish species (Chen et al., 1995). Crucial to remember is that within a few days, damaged tissues can significantly deteriorate from early- to advanced-stage ulceration (Bernardet et al., 1994). Weight also impacts susceptibility; fish weighing 2-80 g are more susceptible to acute infection, whereas fish over 100 g seem less susceptible (Avendaño-Herrera et al., 2006a). Further influencing infection severity and mortality rates are temperature, water parameters, rearing conditions, stress, and interfering parasitic infestations (Nowlan et al., 2021). In a presumptive diagnosis, the main macroscopic lesions associated with T. maritimum infection are external and include ulcerative skin, tissue necrosis, mouth erosion, tail and fin rots, and necrosis of the gills and eyes [see review by Bernardet (1998); Avendaño-Herrera et al., (2006a)]. All of these tissues are usually infected by T. maritimum, resulting in bacterial networks around dead surfaces, producing potent toxins and attenuating host defense mechanisms (van Gelderen et al., 2009b; Guardiola et al., 2019). The most common pathologic finding internally is paleness of the organs (Toranzo et al., 2005). However, a more serious T. maritimum infection in a gray mullet (Mugil capito) specimen showed moderate erythematous hemorrhaging at the base of the anal and pelvic fins and septicemic lesions manifested as severe congestion in the gills, kidney, and other visceral organs, with bloody hemorrhagic exudates (Abdel-Latif, 2013).
At the microscopic level, Vilar et al. (2012) described the pathologic changes in S. sole infected with T. maritimum using light and scanning-electron microscopy. The main lesions observed were detachment of the skin dermis and epidermis, hyaline degeneration, and necrosis in the musculature, as well as an inflammatory response around muscle cells. Moreover, scanning electron microscopy revealed the presence of filamentous, Gram-negative bacteria in the dermis layer and over the scales. Other research in a P. orbiculares specimen associated the intensity of ulcerative skin lesions with the number and area of whitish patches caused by T. maritimum (Lopez et al., 2022b). The respective histopathological view thereof for affected skin showed severe degeneration in both the dermis and epidermis layers with an aggregation of filamentous bacteria and leukocyte infiltration. These observations could be associated with the moderate hydrophobicity and rapid ability to form biofilms of T. maritimum isolates, as evidenced under in vitro conditions with live and dead cells in multi-layered bacterial aggregates (Figure 1). These traits may contribute to T. maritimum pathogenicity (Levipan et al., 2019). The aforementioned points complicate diagnosis and mean that macroscopic and microscopic observations are only a hypothetical approach. The use of laboratory protocols is, therefore, essential.
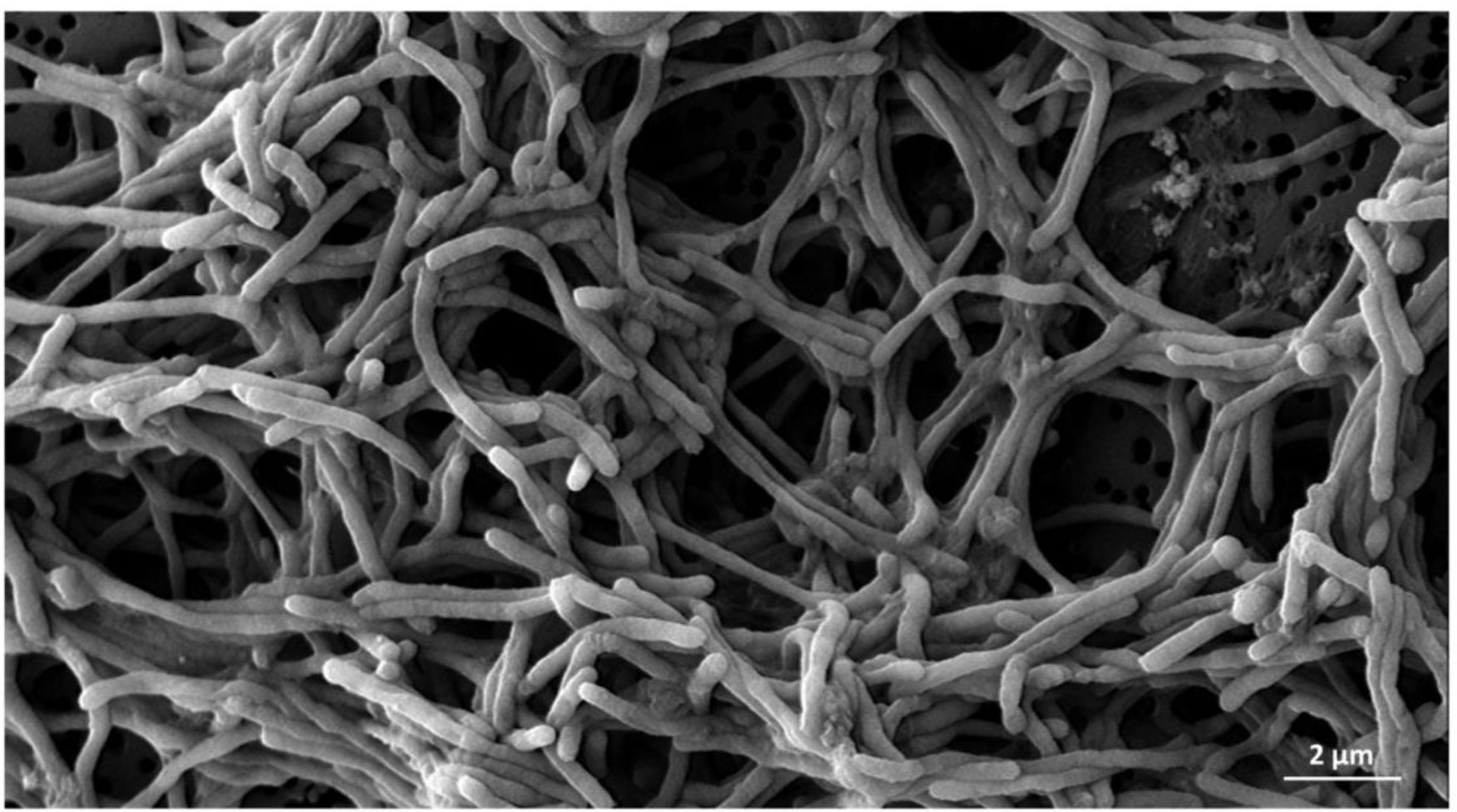
Figure 1 Scanning micrograph of the Tenacibaculum maritimum type strain CECT 4276T at 96 hours of growth. The pathogen clusters are covered by a thick layer of biofilm and exopolysaccharides observed by scanning electron microscopy. The bar corresponds to 2 µm.
Definitive diagnosis requires colony isolation on specific media (i.e., FMM agar or media supplemented with antibiotics), but T. maritimum isolation from diseased fish is not always successful. Thereafter, minimal morphological and biochemical traits must be determined. Tenacibaculum maritimum typically display as flat, pale-yellow colonies with irregular edges on FMM agar, but on marine agar, colonies look round and yellow (Pazos et al., 1996). However, these traditional microbiological isolation and identification methods require a protracted time for precise diagnosis, resulting in high fish mortalities. These phenotyping assessments are becoming less frequently used due to the ever-increasing availability of advanced analytical methods (e.g., PCR, gene sequencing, and proteomic approaches). Moreover, specific molecular detection may be challenging given the diversity of undescribed Tenacibaculum taxa.
Importantly, T. maritimum as a filamentous bacterium presents many obstacles to laboratory work. Cells aggregate and adhere tightly to different substrates, thus impeding the recognition of T. maritimum colonies (Yamamoto et al., 2010). Mabrok et al. (2016) has evaluated several approaches to improve bacterial culture conditions, including treatment with non-ionic surfactants, detergents, cellulase hydrolysis, and shaking. Higher yields without clustering were obtained through continuous shaking during culturing (i.e., 140 rpm). This finding contrasts to other trials that gave obvious bacterial aggregates. However, bacterial growth depends on the employed culture medium. For example, significantly more limited growth is observed for FMM broth as compared to marine broth (Figure 2).
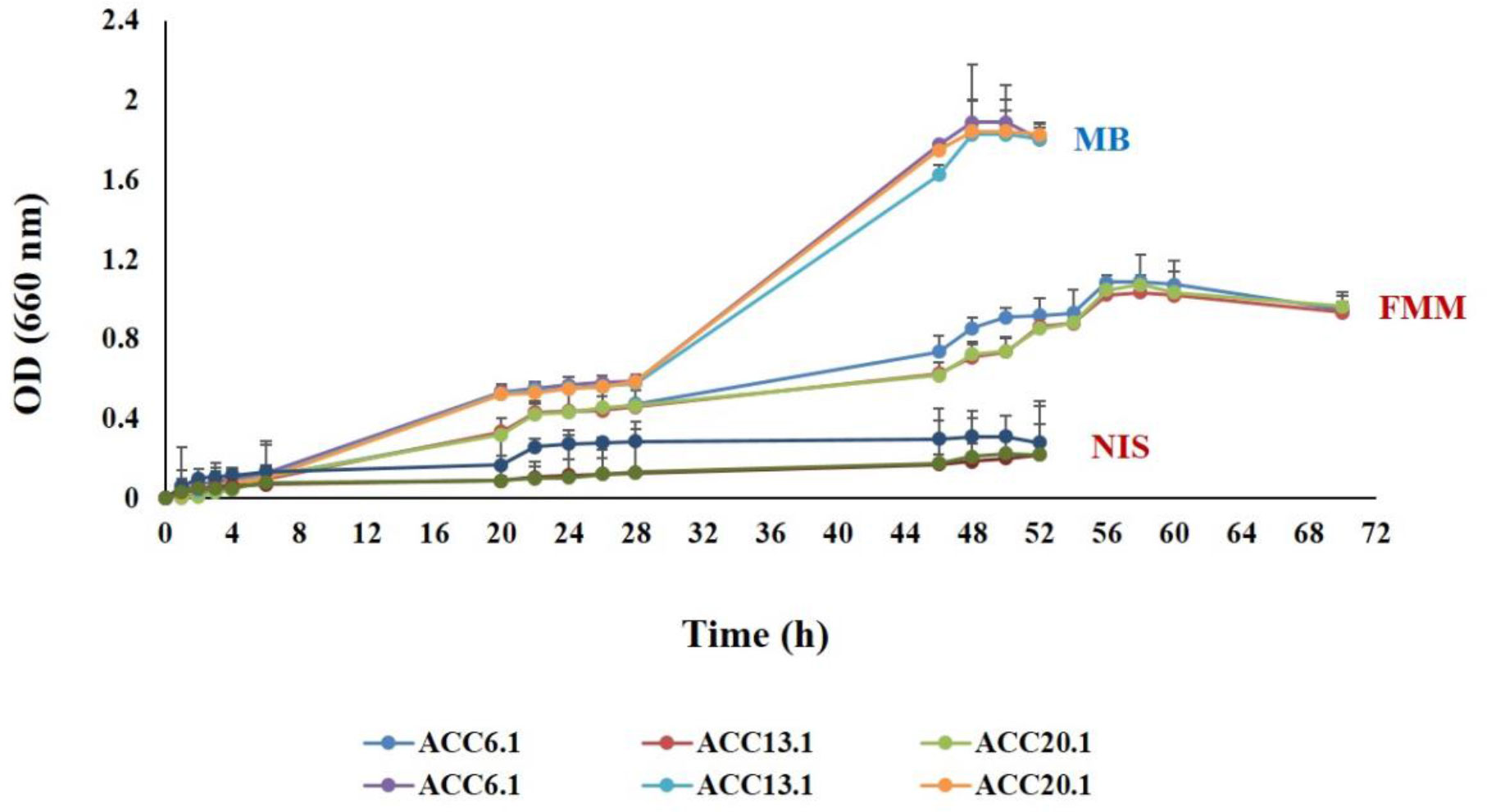
Figure 2 Comparative study to evaluate the bacterial growth curve of T. maritimum strains ACC20.1, ACC13.1, and ACC6.1 recovered from a Portuguese Senegalese sole farm following cultivation on marine broth (MB), marine broth treated with a ten-fold dilution of a detergent Igepal CA-630 (NIS), and Flexibacter maritimus broth (FMM). Data obtained from Abdelaziz (2016). Optical density (OD) readings were taken every 2 hours of incubation at 28°C and were performed in duplicate for each strain at 600 nm.
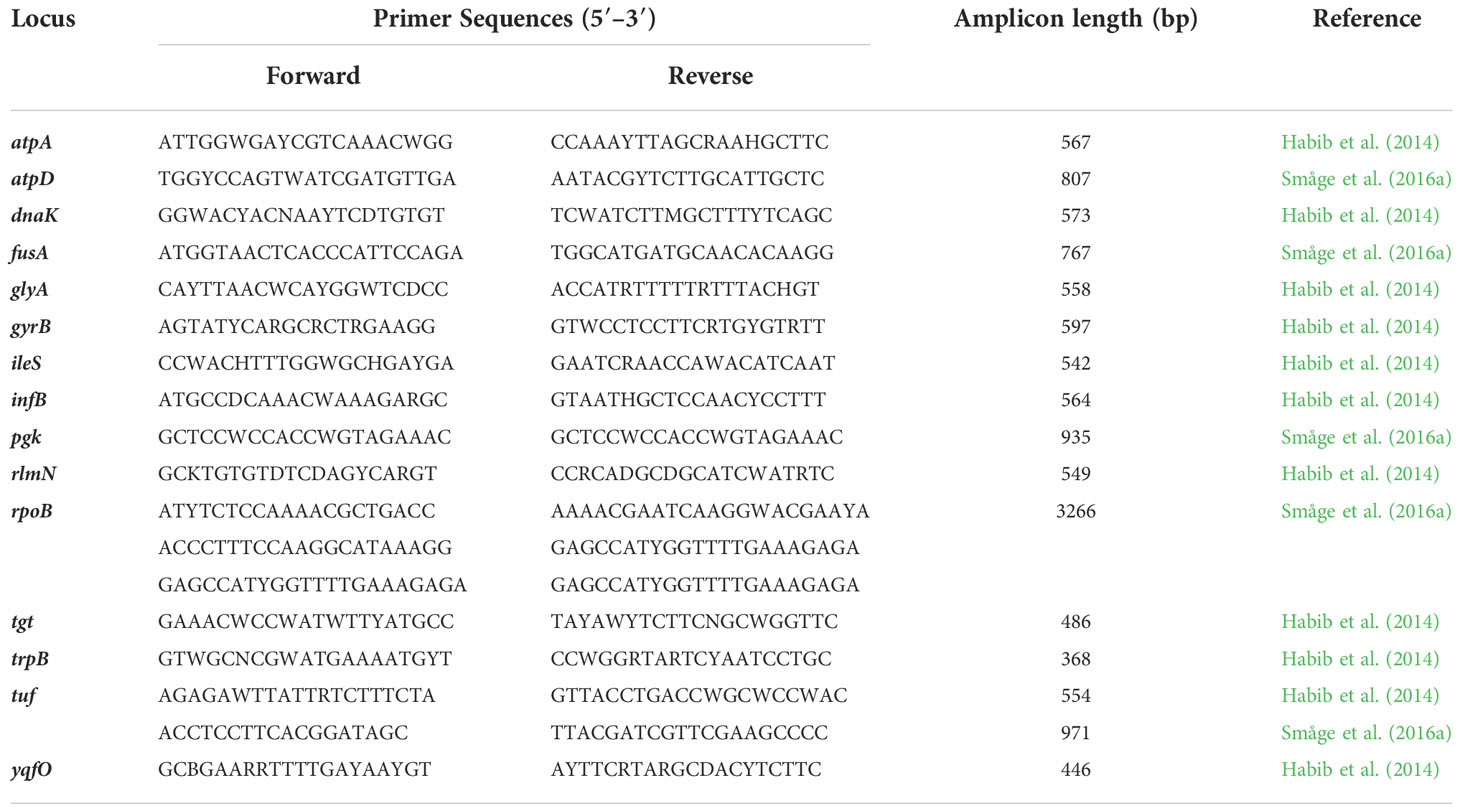
Undoubtedly, PCR protocols can serve as powerful tools to identify fish pathogens from plate cultures and fish tissues (Cunningham, 2002). The high sensitivity of the PCR technology promotes the early detection of pathogens soon after infection and even before disease onset. Since Toyama et al. (1996), various primer sets and PCR protocols (e.g., nested PCR or qPCR) have evolved for the specific detection of T. maritimum from pure or mixed cultures and fish tissues. Many protocols are based on the amplification of ribosomal genes, mainly 16S rRNA. All protocols are summarized in Table 3.
Other molecular methods that couple PCR amplification with a serological procedure or species-specific oligonucleotide probes are also available to detect T. maritimum, including the PCR-enzyme-linked immunosorbent assay (PCR-ELISA) (Wilson et al., 2002), reverse transcriptase polymerase chain reaction-enzyme hybridization assay (RT-PCR-EHA) (Wilson and Carson, 2003), and DNA microarray probe (Warsen et al., 2004). The incorporation of molecular probes allows pathogen detection in less time and with increased diagnostic specificity and robustness. Following assessments of unpublished T. maritimum genomes, Habib et al. (2014) notably proposed a genotyping scheme based on the multi-locus sequence analysis (MLSA). The resulting phylogenetic tree clearly distinguished T. maritimum from other relevant species, including T. soleae, T. gallaicum, T. discolor, T. dicentrarchi, and T. ovolyticum. Further revealed were the convoluted lineages between pathogenic and environmental strains, supporting the proposal of pathogenicity independently evolving in Tenacibaculum species (Habib et al., 2014; Bridel et al., 2018). Småge et al. (2016a) also propose other specific primers for the atpD, fusA, pgk, rpoB, and tuf genes, which could be used with isolates of other Tenacibaculum species (Table 4).
Multi-locus sequence analysis is now widely used not only for genetic differentiation and the estimation of evolutionary relationships among Tenacibaculum species, but also for the specific identification of T. maritimum from pure cultures (Småge et al., 2016a; Valdes et al., 2021; Lopez et al., 2022a). MLSA provides unambiguous DNA sequence data that can be readily compared through web-based databases globally, thus providing unbiased information similar to or better than that provided by other PCR-based methods (Lin et al., 2014). The recommended primer sets for MLSA are summarized in Table 4.
Most PCR protocols are (a) time-consuming; (b) liable to produce multi-sized and nonspecific amplicons for environmental and field samples; and (c) require thermal cycling devices (Klein, 2002; Yang and Rothman, 2004; Mabrok et al., 2021). A recent breakthrough – Matrix-Assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF) mass spectrometry (MS) – has risen as an alternative diagnostic tool for the in-field and laboratory detection of marine Tenacibaculum species (Fernández-Álvarez et al., 2017). A recent molecular study by Bridel et al. (2020) investigated the genomes of 25 T. maritimum isolates, the average genome sizes of which were ~3.356 Mb. The core genome involved 2,116 protein-coding genes (~75%). An adequate level of nucleotide diversity was observed in the most conserved domains (~0.0071 bp−1). The conducted molecular analyses further evidenced remarkable recombination (r/m ≥ 7) and categorization into various subgroups. Moreover, MALDI-TOF-MS was used to detect biodiversity among the recovered 25 T. maritimum strains. The sequence analyses revealed various mass peaks that resembled ribosomal proteins. Furthermore, variation in one or more amino acids was demonstrated, illustrating the mass shift in the bacterial ribosomal proteins. These detected peak shifts greatly associated with the specific genotype defined by MLSA. Consequently, both MALDI-TOF-MS and MLSA are proposed as techniques highly specific for the diagnosis of T. maritimum when pure bacterial isolates are studied.
7. Pathogenicity of Tenacibaculum maritimum infection
Pathogenicity is defined as the hereditary capacity of a microorganism to actuate infection, as interceded by particular harmful variables. Pathogenicity also alludes to the degree of bacterial destructiveness and harmful impact on host tissues (Casadevall and Pirofski, 2001). This concept is often associated with host susceptibility as well as the virulence of the causative agent (Brock et al., 2003). As presented in Table 2, the distribution of T. maritimum among fish species other than those indicated by Avendaño-Herrera et al. (2006a) has been confirmed. Indeed, T. maritimum shows a lack of strict host specificity, thus expanding previously described geographical and host distributions, as described in depth in section 5 Geographical Distribution and Host Susceptibility. Tenacibaculosis may therefore be a risk for many species of anadromous and marine fish, including in species in which the disease has not yet been described or, unfortunately, simply ignored. Controversy arises in cases where serotypes (Avendaño-Herrera et al., 2004a; Avendaño-Herrera et al., 2005a; Lopez et al., 2022a) and/or genotypes particular of T. maritimum strains (Habib et al., 2014) colonize certain species or are distributed in specific geographical areas.
For example, Avendaño-Herrera et al. (2006c) reported a sole isolate that does not produce mortality in turbot fry specimens challenged by intraperitoneal and immersion routes. Likewise, Frisch et al. (2018c) reported differences in the cumulative mortality of different Canadian isolates of T. maritimum against Atlantic salmon challenged by bathing. The inefficiency in reproducing T. maritimum infection under laboratory conditions using a single infection model (i.e., injection, cohabitation, or immersion) supports scientific research that T. maritimum could be an opportunistic pathogen that primarily causes extensive skin damage and gill abrasion with subsequent systemic infection. Current scientific evidence supports disease transmission through seawater, but limitations presented by the initial variability of inoculum of T. maritimum during growth in microbiological cultures – that is, the formation of bacterial aggregates (Mabrok et al., 2016) – could affect success in the natural simulation of T. maritimum infection.
Initial pathogenicity studies conducted in the 1990s with commercial fish species such as European sea bass (Bernardet et al., 1994), rainbow trout, greenback flounder (Rhombosolea tapiriña) (Soltani et al., 1996), and Atlantic salmon (Powell et al., 2004) found varying degrees of mortality based on infection method and routes of bacterial inoculation, as noted above. By contrast, Avendaño-Herrera et al. (2006c) reported that the tested T. maritimum isolates (regardless of dosage or serotype) did not cause disease when injected intraperitoneally but did cause significant mortality in turbot after an18 h bath challenge. This first approach to establishing a robust challenge model using advances in knowledge of T. maritimum virulence properties has resulted in decreasing bath-challenge exposure times to just a few hours (i.e., 1.5 to 7 h of incubation) (Frisch et al., 2018c; Valdes et al., 2021).
The available scientific evidence shows that seawater is of crucial importance in the horizontal transmission of T. maritimum, but other routes of entry to the host cannot be ruled out, such as direct contact with marine invertebrates colonized by the bacterium (Ferguson et al., 2010). This includes through food, as reported by Chen et al. (1995). Environmental factors (i.e., seawater temperature and salinity) further influence the severity of T. maritimum infections, but more exhaustive studies are required to precisely identify the primary reservoirs of infection and/or the role of culture water in the epidemiology of infections and specificity of risk factors. This compilation of knowledge is needed to control and prevent T. maritimum outbreaks.
8. Virulence factors
Virulence is the relative capacity of a microorganism to cope with the host’s immune response and to replicate and reproduce, thus resulting in disease development (Casadevall and Pirofski, 1999). In the context of this review, virulence is the degree to which T. maritimum can produce tenacibaculosis. A first step for the effective colonization of a pathogenic microorganism within a host is adherence. Interestingly, the name of the bacterial genus [i.e., Te.na.ci.ba’cu.lum. L. adj. n. tenax -acis holding fast; L. neut. n. baculum stick; N.L. neut. n. (Suzuki et al., 2001)] indicates that Tenacibaculum species are rod-shaped bacteria that adhere to the surface of marine organisms. There are two forms of adhesion, specific and non-specific. These adhesion forms allow microbial expansion within host tissues and encourage the dispersion of toxins (Ofek and Doyle, 2012). Specific adhesion is mediated by certain compounds on the bacterial surface that bind to receptors on host tissues, and non-specific adhesion depends on hydrophobic or ionic bonds between the bacterial surface and the supporting substrate (Ofek and Doyle, 1994).
Burchard et al. (1990) reported that the adhesion power of T. maritimum cells is increased constitutivelyon substrates with low-critical energy surfaces (e.g., hydrophobic). This was ascribed to the capacity of the microbes to deliver extracellular polymers or slime, which permit a solid connection to hydrophobic surfaces. Most invading pathogens recognize and bind to fish mucus through sugar-binding proteins such as lectins. This is the first step in initiating defensive responses (Imberty and Varrot, 2008) since the abundance of carbohydrate residues in the skin mucus of hosts has been related to the progression of infection (Estensoro et al., 2013). This hallmark reflects the capacity of pathogens to attach to the external surfaces of many fish species. This hypothesis was affirmed by Magariños et al. (1995), who stated that T. maritimum adheres strongly to the skin mucus of turbot, sea bream, and European sea bass, regardless of the salt solution in which the mucus dissolves.
Moreover, in vitro work established by van Gelderen et al. (2010b) proved the adhesive nature of different T. maritimum strains. Similarly, data regarding mucus and plasma bactericidal activities revealed a considerable lack of immunity in S. sole against T. maritimum and proved the ability of this pathogen to present evading strategies against the infected host (Mabrok et al., 2016). Even in turbot that survived exposure to virulent strains, T. maritimum was only recovered from mucus (Avendaño-Herrera et al., 2006c). The same team of researchers subsequently showed that T. maritimum is part of the autochthonous bacterial population in fish and can survive for long periods in the aquatic environment by using fish mucus as a reservoir (Avendaño-Herrera et al., 2004c; Avendaño-Herrera et al., 2006d). In the case of T. maritimum, this adhesion and colonization capacity cannot be attributed to bacterial structures such as pili, fimbriae, or flagella since T. maritimum cells lack all of these elements (Avendaño-Herrera et al., 2006a). Guardiola et al. (2019) found that T. maritimum can withstand and overcome the skin mucus barrier due to a passive change of the glycosylation pattern.
Besides stickiness, the hemagglutinating activity of T. maritimum may also influence its destructiveness. Further demonstrated by Pazos (1997), was that T. maritimum agglutinates different types of blood cells. Extracellular products, subcellular components, and lipopolysaccharides are additional virulence-related mechanisms of T. maritimum, as detailed in depth in the review by Avendaño-Herrera et al. (2006a). The bacterial toxins present in extracellular products may also play a role during disease either by encouraging bacterial colonization and invasion or by changing and disintegrating host tissues (Baxa et al., 1988). Supporting these points, van Gelderen et al. (2009b) demonstrated that T. maritimum toxins induce gill necrosis, cellular damage in the heart and pyloric caeca, and autolysis and rapid lysis in S. salar cells.
T. maritimum is a proteolytic fish pathogen (Wakabayashi et al., 1986). The release of extracellular proteases is an index of virulence, with effects exerted on the tissues and defense mechanisms of the host. In this sense, the production of toxins and proteolytic enzymes during bacterial invasion is essential for successful colonization and virulence (Rahman et al., 2014). According to Frisch et al. (2018b), T. maritimum proteases act synergistically with other virulence determinants, ultimately resulting in tissue damage or mortality for the host. The involvement of bacterial toxins or extracellular enzymes in the invasion strategy of and successful generalized infection by T. maritimum was further supported through observations of a delayed mucosal immune response and incipient plasma peroxidase and lysozyme activities observed in bath-challenged S. senegalensis (Guardiola et al., 2019).
Despite efforts made in the past few decades, the pathogenesis of T. maritimum is a multifactorial process that is still not fully understood. To further elucidate the cell-level immune responses of and the cytotoxic effects against S. senegalensis head-kidney leukocytes, Abdelaziz (2016) performed in vitro trials through exposure to the extracellular products of different T. maritimum isolates. The leukocytes increased the production of reactive oxygen species but not of nitric oxide. Interestingly, while a low concentration of T. maritimum extracellular products triggered production of reactive oxygen species, a high concentration suppressed production; thus reflecting the ability of T. maritimum to cope with the phagocytic response of S. senegalensis using suppressive toxins or enzymes (Abdelaziz, 2016). Cytotoxic activity against the leukocytes was likewise dose-dependent, providing initial clues into the mechanisms by which Tenacibaculum actuate cell death. To this end, light microscopy of fish leukocytes showed inner vacuolization, cellular detachment and prolongation, cell-membrane weakening, and cell clustering and conglomeration, especially after incubation with high concentrations of extracellular T. maritimum products (Figure 3) (Abdelaziz, 2016).
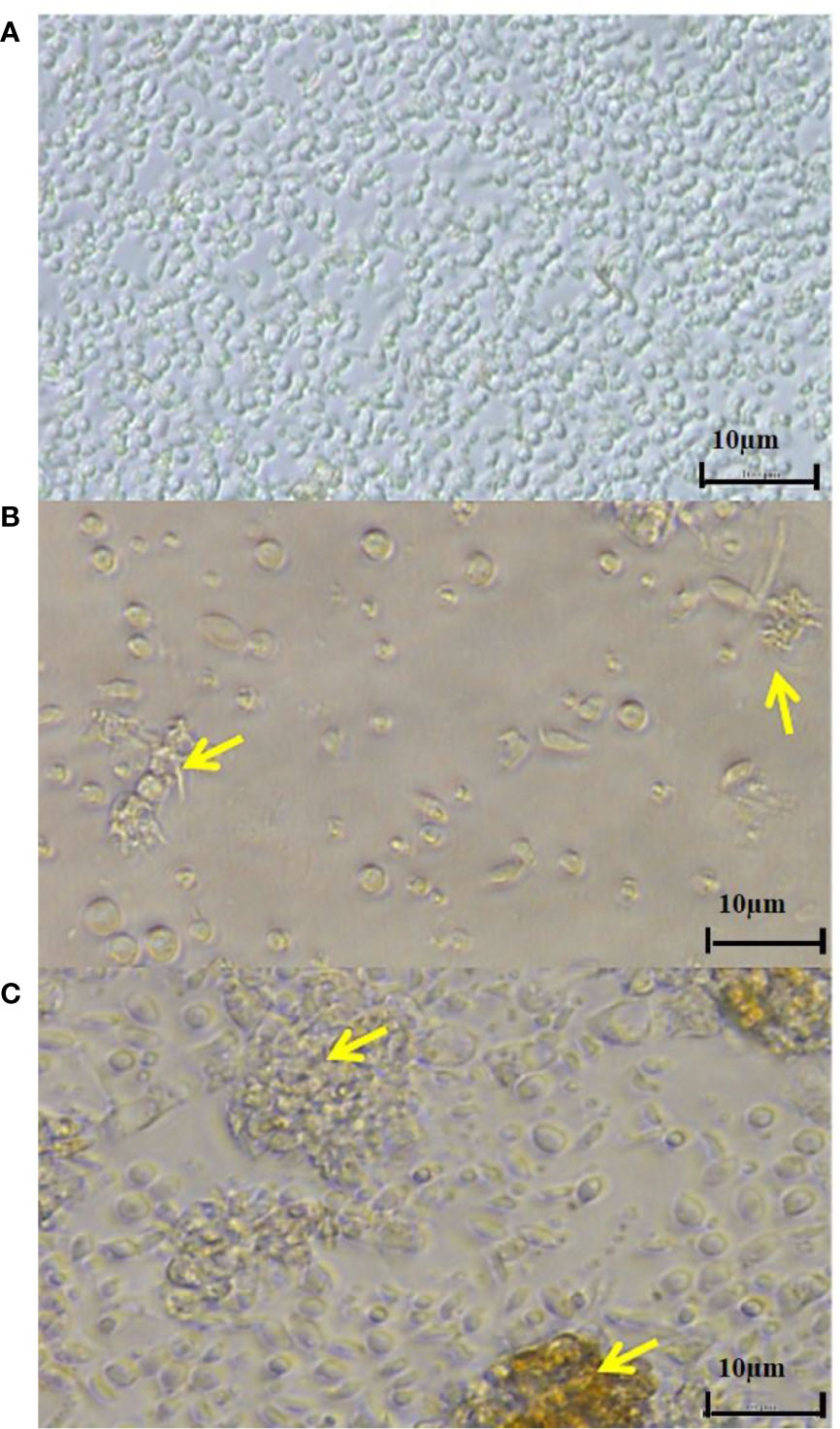
Figure 3 Microscopic pictures of Senegalese sole head-kidney leukocytes after exposure to (A) L15 medium or (B, C) extracellular products of T. maritimum at 100 µg mL-1 for 24 h. Yellow arrows indicate (B) inner vacuolization, cell elongation and/or degradation and (C) cell clustering and conglomeration. These results were obtained from Abdelaziz (2016).
One of the most studied virulence mechanisms of T. maritimum is its ability to compete effectively with the host for iron (Ratledge and Dover, 2000). Avendaño-Herrera et al. (2005b) suggested at least two systems of iron acquisition for different T. maritimum serotypes: one involving the synthesis of siderophores and another allowing for the utilization of heme groups as iron sources by direct binding. These physiological results are supported by molecular studies that demonstrate a siderophore-biosynthesis gene cluster (tbs gene) in the T. maritimum genome. This gene cluster is responsible for the synthesis of the bisucaberin-class siderophore (Fujita et al., 2012). The tbs genes are encoded to produce Tenacibaculum bisucaberin synthase. Moreover, various TonB-dependent outer membrane receptors were detected in the T. maritimum genome; the MARIT_0185 type is placed in the tbs locus that encodes for the bisucaberin siderophore iron transporter. The genome of T. maritimum has genes encoding proteins that initiate iron uptake and/or storage (Pérez-Pascual et al., 2017). As previously described, T. maritimum is characterized by non-flagellar gliding motility that enables movement over various surfaces. It is no surprise then that the T. maritimum genome contains 14 gliding genes (gldA to gldN) and 10 spr genes that encode for various proteins needed for the gliding motility (Pérez-Pascual et al., 2017). Pérez-Pascual and colleagues (2017) additionally detected numerous genes for adhesins and polysaccharide biosynthesis. The proteins displaying lectin or carbohydrate-binding motifs could be involved in the strong adhesive and hemagglutination properties, as well as biofilm-forming ability, of T. maritimum described by Pazos (1997). Surface-attached biofilms and cell-surface hydrophobicity are fundamental for the environmental persistence and transmission of T. maritimum (Burchard et al., 1990). The kinetics of biofilm formation in T. maritimum, especially on adhesive surfaces may serve as transient reservoirs of virulent strains that can further exacerbate the outbreak of tenacibaculosis after the cell-detachment stage (Levipan et al., 2019).
The T. maritimum genome also has genes encoding the superoxide dismutases manganese-dependent type (sodA), iron-dependent sodB, and zinc-dependent type (sodC), the last of which is only found in virulent strains (Sheng et al., 2014). These enzymes enable the bacteria to transform superoxide anions into molecular oxygen and hydrogen peroxide that can be easily metabolized by peroxidase and catalase enzymes, the end effect of which is tolerance to the oxidative stress exerted by the host immune response (Lee et al., 2012). Similarly, these enzymes act as a group of powerful toxins and virulence determinants mainly involved in damaging host cells, including ceramidase and a sphingomyelinase (Pérez-Pascual et al., 2017). Sphingomyelinase has been described as a highly potent cytotoxin to the host (Oda et al., 2010), and ceramidase acts as a potent exotoxin (Ito et al., 2014). Numerous other genes associated with virulence mechanisms in T. maritimum, especially toxins, were summarized in depth by Pérez-Pascual et al. (2017). These genes include a cholesterol-dependent cytolysin, an alternative hemolysin gene, and the cslA gene (MARIT_2107) encoding a PL8_3 family chondroitin AC lyase. These were very similar to those produced by another fish pathogen, Flavobacterium columnare (Suomalainen et al., 2006), and each plays an invasive role in fish tissues.
Not all genetic information on T. maritimum has reached consensus. For example, controversy surrounds the existence of communication processes mediated by quorum sensing. Romero et al. (2010) identified n-butyryl-L-homoserine lactone (C4-HSL) by liquid chromatography-mass spectrometry in the type strain of T. maritimum and demonstrated degradation activity for long N-acyl-homoserine lactones (C10-HSL). By contrast, Pérez-Pascual et al. (2017) did not detect a homologous gene for homoserine lactone biosynthesis in the genome, nor were any coding genes found for processes involved in the inhibition of bacterial communication (i.e., quorum quenching).
The genome sequences of the T. maritimum type strain provided valuable insights into the existence of many predicted genes potentially implicated in the virulence and pathogenicity of T. maritimum. Through the analysis of 24 T. maritimum-strain genomes recovered from different hosts and geographical areas, Bridel et al. (2020) identified all the predicted toxins in the core-genome and the virulence factors (i.e., cholesterol-dependent cytolysin, collagenase, sphingomyelinase, ceramidase, chondroitin AC lyase, streptopain family protease, sialidase, iron uptake systems, and type IX secretion system [T9SS]) previously identified by Pérez-Pascual et al. (2017). Continued confirmations through physiological studies of the initial work by Pérez-Pascual and colleagues (2017) will help elucidate and describe the genes and pathways likely involved in T. maritimum virulence. Therefore, further physiological research is necessary to understand the pathogenesis of tenacibaculosis.
9. Treatment and vaccination programs for Tenacibaculum maritimum
To date, the only commercially available vaccine for T. maritimum is for turbot, specifically strain LPV1.7 serotype O22. Hence, the control of tenacibaculosis in other cultured species and for other strains is mainly limited to the frequent use of antibiotics and certain disinfectants (Avendaño-Herrera et al., 2008). Antibiotics can be used to treat tenacibaculosis with varying degrees of success. As a preventive or prophylactic measure, surface-acting disinfectants administered by immersion may be effective. Temperature and/or salinity manipulation may also aid in disease mitigation (Santos et al., 2019). Notwithstanding, the susceptibility of tenacibaculosis-causing agents to different antimicrobials commonly used in aquaculture is highly variable and dependent on Tenacibaculum species and the antimicrobial.
An in vitro study using disk diffusion susceptibility testing on FMM plates, showed that 63 T. maritimum isolates from several hosts and geographical regions were resistant to oxolinic acid but susceptible to amoxicillin, nitrofurantoin, florfenicol, oxytetracycline, and trimethoprim-sulfamethoxazole (Avendaño-Herrera et al., 2008). In addition, some isolates presented resistance to enrofloxacin and flumequine. A previous study – the goal of which was to propose a standardized culture medium and procedure for determining the minimum inhibitory concentration – showed that 32 T. maritimum strains were resistant to oxolinic acid but highly susceptible to amoxicillin and trimethoprim-sulfamethoxazole (Avendaño-Herrera et al., 2005c). More recently, Abdelbaky et al. (2021) found that all T. maritimum strains examined from diseased Haffara bream (Rhabdosargus haffara) and marbled spinefoot (Siganus rivulatus) were sensitive to erythromycin, cephalothin, ampicillin, and chloramphenicol but showed high resistance to ofloxacin and tetracycline. More confounding is that field results are not always the same, even if the tested bacteria show a higher sensitivity to the chosen drug in vitro (Cepeda and Santos, 2002).
The widespread use of antimicrobials in aquaculture could cause the appearance of multidrug-resistant T. maritimum strains (Watts et al., 2017). In the related fish pathogen, Flavobacterium psychrophilum, R-plasmids, antimicrobial resistance genes, and mutations in resistance elements are associated with antibiotic resistance (Izumi and Aranishi, 2004). However, T. maritimum does not have plasmids. Therefore, the occurrence of antimicrobial resistance could be associated with some intrinsic resistance mechanism, such as the efflux pump system, permeability of the bacterial outer cell membrane, and/or the synthesis and release of antibiotic-degrading enzymes, especially β -lactamase (Clark et al., 2009; Henríquez-Núñez et al., 2012; Pérez-Pascual et al., 2017).
Enrofloxacin, a second generation fluoroquinolone, usually exhibits promising antimicrobial activity against Flavobacterium spp. (Akinbowale et al., 2006). Two new antimicrobial 2-alkyl-4-quinolones, a common core in synthetic antibactericidals, showed promising antibacterial activity against T. maritimum, thus providing a new opportunity to develop antibacterial drugs for fish farming (Li et al., 2018). However, the frequent use of these compounds for tenacibaculosis treatment has decreased efficacy and resulted in futile usage (Jang et al., 2009). Another widely applied antimicrobial in aquaculture worldwide is florfenicol, a compound specifically developed for veterinary medical use. Florfenicol is widely used for the treatment of tenacibaculosis caused by the different species of the genus (Irgang et al., 2021). Interestingly, the existence of a multidrug efflux pump present in T. maritimum has been reported (Pérez-Pascual et al., 2017), which could cause resistance to florfenicol. Such resistance has been previously reported in Chryseobacterium species, another member of the Flavobacteriaceae family (Michel et al., 2005). These efflux pumps also play a key role in intercellular signaling, bacterial virulence, and detoxification of intracellular bacterial metabolites (Pasqua et al., 2019). Another widely used antimicrobial in fish aquaculture, particularly in developed countries, is oxytetracycline (Avendaño-Herrera et al., 2023). Yet again, extensive usage may soon result in efficacy deterioration (Irgang et al., 2021). Considering the current scenario for global aquaculture, several actions are required to prevent the emergence of multidrug-resistant bacterial pathogens and maintain the efficacy of antibiotics – (i) the continuous monitoring and surveillance of the antimicrobial susceptibility of T. maritimum isolates using laboratory tests, such as those proposed by the Clinical Laboratory Standards Institute3 for bacteria isolated from aquatic animals and (ii) the proper use and rotation of antimicrobial agents.
The emergence of multiple drug resistant pathogens, together with the negative impact of antibiotics on the beneficial gastrointestinal microbiota of fish, has encouraged developed countries to prohibit the use of all subtherapeutic antibiotics (Regulation [EC] No 1831/2003, Europe) (Simon and Klaus, 2008). This has pushed the scientific community to explore non-antibiotic related alternatives, such as probiotics and treatment with herbal dietary medications. For example, Reyad et al. (2013) demonstrated that the marine actinomycetes probiotic (YSCl2334) – isolated from Salwa Beach in the Jazan Region of Saudi Arabia – can generate antimicrobial activity against T. maritimum. Likewise, the probiotic Roseobacter group (Phaeobacter piscinae S26) can be effective in killing pathogenic Tenacibaculum species, including T. maritimum (Tesdorpf et al., 2022). Similarly, twelve indigenous cultured probiotic isolates from the gastrointestinal tract of three temperate flatfish species (Wanka et al., 2018), as well as four spore-former isolates obtained from the heat-treated gut contents of European sea bass (Serra et al., 2019), showed promising inhibition activity towards T. maritimum.
Comparably, plant extracts exhibit potent antimicrobial activity against a variety of Gram-positive and Gram-negative bacteria and are generally regarded as safe by the Food and Drug Administration (Soniya et al., 2013). To this end, in vitro susceptibility studies indicated potential antimicrobial activity of carvacrol, a derivative of the oregano plant (Abd El-Galil and Hashiem, 2012b), and of some native plant extracts from Jeju Island against tested T. maritimum strains (Jang et al., 2009). All these data provide insights into the potential use of non-antibiotic compounds in treating and preventing the pathogenic threat of T. maritimum; however, further research into the applications and appropriate dosages thereof is still being conducted.
Despite recent strategies applied for effective treatment, bacterial fish diseases remain a major economic obstacle to aquaculture globally. The emergence of multidrug-resistant bacterial pathogens increases the need for effective vaccination programs (Sneeringer et al., 2019). Modern vaccine technology targets specific bacterial component, such as novel antigens (Cimica and Galarza, 2017). RNA particle-vaccines provide greater immunity as compared to conventional protocols. Innovations in vaccine manufacturing are indeed promising for the aquaculture industry (Frietze et al., 2016). However, few attempts at commercial-level vaccinations have been described, and, as previously mentioned, only one bacterin is available on the market for turbot (Romalde et al., 2005). The developed vaccine was applied by bathing for fish of 1-2 g, followed by a booster injection dose at a size of 20-30 g. The survival rate of the challenged fish after bath immunization was about 50%, and increased to >85% after booster injection. The systematic application of this vaccine in some turbot farms has decreased the prevalence of tenacibaculosis.
Numerous vaccination trials in salmonids exist, but with contradictory outcomes. The first trials of a T. maritimum vaccine for Atlantic salmon farmed in Australia resulted in little or no protection (Carson et al., 1993; Carson et al., 1994). However, S. salar immunized with an adjuvant-based vaccine performed better and showed a greater survival rate than other tested vaccines at 27 days post-challenge with T. maritimum (van Gelderen et al., 2009a). More recently, an inactivated whole-cell oil-adjuvant vaccine against T. maritimum produced an antibody response in Atlantic salmon, but with no significant protection (Frisch et al., 2018c). The antigenic and genetic variability existing among T. maritimum isolates complicates candidate selection for vaccine development. Supporting this, Salati et al. (2005) emphasized that lipopolysaccharides are the main protective antigens of this pathogen.
To date, no commercial vaccine against tenacibaculosis in salmonids is available, and Toranzo et al. (2004) indicate that the vaccine developed for turbot is not effective in preventing tenacibaculosis in other fish species. Autovaccine or autogenous bacterins are alternatives that several scientists havedeveloped and implemented. However, important information gaps exist regarding the effectiveness of these biological products and the host defense mechanisms. These factors altogether may hinder the development of appropriate vaccination programs for this economically impactful Tenacibaculum species.
10. Conclusion and knowledge gaps
Tenacibaculosis has risen over the past five years to become one of the most devastating bacterial infections impacting various species of farmed fish and geographical regions. The abrupt re-emergence of disease caused by T. maritimum is in addition to the appearance of new Tenacibaculum species, currently placing the global salmonid aquaculture industry at risk. Considering the broader availability of scientific studies on T. maritimum in the last 15 years, we conducted an extensive review on the status of and advancements on this bacterium since 2006. In summary, the taxonomic status of this bacterium is clear, consensus exists regarding intra-specific diversity, and specific, sensitive diagnostic tools are available (i.e., PCR, molecular probes in PCR-ELISA, RT-PCR-EHA, and MALDI-TOF assays, etc.) that are applicable to pure cultures as well as in early detection through infectious signs occurring in farmed fish. Advancements in genome sequencing have paved the way for developing methods invaluable to the genetic (i.e., MLSA) and serotype (i.e., mPCR) tracking of T. maritimum. Both tools could be useful in creating new species-specific vaccines. Other significant advancements include those related to disease replication under laboratory conditions, specifically through the immersion of fish with the pathogen, thus simulating natural conditions. This tool is indispensable in the search for new products for the treatment (e.g., plant-based compounds) and prevention (e.g., vaccines) of tenacibaculosis. Unfortunately, understanding remains lacking regarding the reservoirs for this bacterium and for the risk factors that influence bacterial appearance. Particularly unclear is the influence of environmental factors (e.g., seawater temperature), an important point considering the context of globally increased temperatures due to climate change. The genome has provided information on the existing mechanisms of pathogenicity in T. maritimum, but physiological studies demonstrating the participation of proteins, and not just the presence of the gene, are deficient. Research has also advanced knowledge of the fish response to T. maritimum, but much remains to be explored. This situation is likely the driver behind antibiotics remaining the most used measure to control T. maritimum. As such, there is an urgent need to establish best-practice sanitary measures, to develop new vaccines and autovaccines, and to search for other control measures besides antibiotics, which potentially include herbal-based medications, phages, or probiotics. Lastly, more research is needed to determine why T. maritimum and other Tenacibaculum species are generally considered opportunistic pathogens. This point takes on particular relevance when considering that some Tenacibaculum isolates possess a larger virulence arsenal than other isolates, as specifically due to the diversity within this bacterial genus. Consequently, we must be open to understanding that fish are farmed intensively, and bacterial infections are not restricted and specific to a single microorganism. Rather, coinfections occur, whether with different bacteria or, in many cases, with the same bacterial species but of different genotypes/serotypes. The presence of T. maritimum or other Tenacibaculum species in the aquatic environment can therefore be a factor in the health of farmed fish, regardless of the pathogen being primary, secondary, or, simply, a coexisting microorganism. Future research should focus on resolving the pending questions raised by our review.
Author contributions
MM, AA, HH, ES, AF and RA-H participated in writing and editing of the manuscript. BA, SA and RA-H constructed the contents of the figures and tables. CR and RA-H participated in and supervised the thorough review of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
MM was supported through a Senior Postdoctoral Fellow Grant awarded by the graduate school of Chulalongkorn University, Thailand. This conducted research was further supported by the Thailand Science Research and Innovation (TSRI) Fund year 2021, Chulalongkorn University_FRB640001_01_31_6. RA-H gratefully acknowledges financial support received through the FONDECYT 1190283 and FONDAP INCAR Grant n°15110027 and 1522A0004 from Agencia Nacional de Investigación y Desarrollo (ANID) from the Chilean Government.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ https://www.condalab.com/medios-de-cultivo-deshidratados/1050-5471-caldo-fmm.html#/2-formato-500_g
- ^ https://www.hipra.com/wcm/connect/hipra/7c54199f-880a-43bc-a46e-4e981deaad2f/ICTHIOVAC-TM-ETI-701222-01.4.pdf?MOD=AJPERES&CACHEID=ROOTWORKSPACE.Z18_L26A0J40OGJR60QGTTTS0N3067-7c54199f-880a-43bc-a46e-4e981deaad2f-mSMeSVA
- ^ www.clsi.org
References
Abdelaziz, M. A. M. (2016). The host/pathogen interaction during experimental infection of Senegalese sole (Solea senegalensis) by Tenacibaculum maritimum. PhD thesis (Universidade do Porto (Portugal) ProQuest Dissertations Publishing, Portugal) 10643756.
Abdelbaky, A., Soliman, A., Abdelsalam, M., Aboulezz, A. (2021). Genotypic characterization of some dermotropic and systemic bacterial pathogens affecting two commercial red Sea fishes. Egyptian J. Aquat. Biol. Fish. 25, 297–312. doi: 10.21608/ejabf.2021.211886
Abd El-Galil, M. A., Hashem, M. (2012a). Epidemiological and bacteriological studies on tenacibaculosis in some red Sea fishes, Egypt. Int. J. Env. Sci. Eng(IJESE) 3, 25–32.
Abd El-Galil, M. A., Hashiem, M. (2011). Tenacibaculosis in Picasso tigger fish (Rhinecanthus assasi) and black damsel fish (Neoglyphieodon meles) of red sea at hurghada, Egypt. Life Sci. J. Acta Zhengzhou Univ. Overseas Edition 8, 1166–1171.
Abd El-Galil, M. A., Hashiem, M. (2012b). Experimental infection of tenacibaculosis and a trial for treatment by plant extract carvacrol in surge wrasses fish (Thalassoma purpureum). Life Sci. J. Acta Zhengzhou Univ. Overseas Edition 9, 442–447.
Abdel-Latif, H. M. (2013). Assessment of potential pathogenicity of emergent marine bacterium, Tenacibaculum maritimum to thin lipped grey mullet (Mugil capito) farmed in Egypt. Group 1, 10.
Abram, Q. H., Dixon, B., Katzenback, B. A. (2017). Impacts of low temperature on the teleost immune system. Biology 6, 39. doi: 10.3390/biology6040039
Akinbowale, O. L., Peng, H., Barton, M. (2006). Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J. Appl. Microbiol. 100, 1103–1113. doi: 10.1111/j.1365-2672.2006.02812.x
Apablaza, P., Frisch, K., Brevik, Ø. J., Småge, S. B. (2017). Primary isolation and characterization of Tenacibaculum maritimum from Chilean Atlantic salmon mortalities associated with a Pseudochattonella spp. algal bloom. J. Aquat. Anim. Health 29, 143–149. doi: 10.1080/08997659.2017.1339643
Avendaño-Herrera, R., Irgang, R., Magariños, B., Romalde, J. L., Toranzo, A. E. (2006b). Use of microcosms to determine the survival of the fish pathogen Tenacibaculum maritimum in seawater. Environ. Microbiol. 8, 921–928. doi: 10.1111/j.1462-2920.2005.00981.x
Avendaño-Herrera, R., Irgang, R., Núñez, S., Romalde, J. L., Toranzo, A. E. (2005c). Recommendation of an appropriate medium for in vitro drug susceptibility testing of the fish pathogen Tenacibaculum maritimum. Antimicrob. Agents Chemother. 49 (1), 82–87. doi: 10.1128/AAC.49.1.82-87.2005
Avendaño-Herrera, R., Irgang, R., Sandoval, C., Moreno-Lira, P. (2016). Isolation, characterization and virulence potential of Tenacibaculum dicentrarchi in salmonid cultures in Chile. Transbound. Emerg. Dis. 63, 121–126. doi: 10.1111/tbed.12464
Avendaño-Herrera, R., Magariños, B., Irgang, R., Toranzo, A. E. (2006d). Use of hydrogen peroxide against the fish pathogen Tenacibaculum maritimum and its effect on infected turbot (Scophthalmus maximus). Aquaculture 257, 104–110. doi: 10.1016/j.aquaculture.2006.02.043
Avendaño-Herrera, R., Magariños, B., López-Romalde, S., Romalde, J. L., Toranzo, A. E. (2004a). Phenotypic characterization and description of two major O-serotypes in Tenacibaculum maritimum strains from marine fishes. Dis. Aquat. Organisms 58, 1–8. doi: 10.3354/dao058001
Avendaño-Herrera, R., Magariños, B., Moriñigo, M. A., Romalde, J. L., Toranzo, A. E. (2005a). A novel O-serotype in Tenacibaculum maritimum strains isolated from cultured sole (Solea senegalensis). Bull. Eur. Assoc. Fish Pathologists 25, 70–74.
Avendaño-Herrera, R., Magariños, B., Toranzo, A. E., Beaz, R., Romalde, J. L. (2004d). Species-specific polymerase chain reaction primer sets for the diagnosis of Tenacibaculum maritimum infection. Dis. Aquat. Organisms 62, 75–83. doi: 10.3354/dao062075
Avendaño-Herrera, R., Mancilla, M., Miranda, C. D. (2023). Use of antimicrobials in Chilean salmon farming: Facts, myths and perspectives. Rev. Aquacult. 15, 89–111. doi: 10.1111/raq.12702
Avendaño-Herrera, R., Núñez, S., Barja, J. L., Toranzo, A. E. (2008). Evolution of drug resistance and minimum inhibitory concentration to enrofloxacin in Tenacibaculum maritimum strains isolated in fish farms. Aquacult. Int. 16, 1–11. doi: 10.1007/s10499-007-9117-y
Avendaño-Herrera, R., Núñez, S., Magariños, B., Toranzo, A. (2004c). A non-destructive method for rapid detection of Tenacibaculum maritimum in farmed fish using nested PCR amplification. Bull. Eur. Assoc. Fish Pathologists 24, 280–286.
Avendaño-Herrera, R., Olsen, A. B., Saldarriaga-Cordoba, M., Colquhoun, D. J. (2022b). Isolation, identification, virulence potential and genomic features of Tenacibaculum piscium isolates recovered from Chilean salmonids. Transbound. Emerg. Dis. doi: 10.1111/tbed.14606
Avendaño-Herrera, R., Rodríguez, J., Magariños, B., Romalde, J., Toranzo, A. (2004b). Intraspecific diversity of the marine fish pathogen Tenacibaculum maritimum as determined by randomly amplified polymorphic DNA-PCR. J. Appl. Microbiol. 96, 871–877. doi: 10.1111/j.1365-2672.2004.02217.x
Avendaño-Herrera, R., Saldarriaga-Córdoba, M., Irgang, R. (2022a). Draft genome sequence of Tenacibaculum ovolyticum to-7Br, recovered from a farmed Atlantic salmon (Salmo salar). Microbiol. Resource Announcements 11 (7), e0025422. doi: 10.1128/mra.00254-22
Avendaño-Herrera, R., Toranzo, A. E., Magariños, B. (2006a). Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: A review. Dis. Aquat. Organisms 71, 255–266. doi: 10.3354/dao071255
Avendaño-Herrera, R., Toranzo, A., Magariños, B. (2006c). A challenge model for Tenacibaculum maritimum infection in turbot, Scophthalmus maximus (L.). J. Fish Dis. 29, 371–374. doi: 10.1111/j.1365-2761.2006.00712.x
Avendaño-Herrera, R., Toranzo, A. E., Romalde, J. L., Lemos, M. L., Magarinos, B. (2005b). Iron uptake mechanisms in the fish pathogen Tenacibaculum maritimum. Appl. Environ. Microbiol. 71, 6947–6953. doi: 10.1128/AEM.71.11.6947-6953.2005
Bader, J. A., Shotts, Jr. E. B. (1998). Identification of Flavobacterium and flexibacter species by species-specific polymerase chain reaction primers to the 16S ribosomal RNA gene. J. Aquat. Anim. Health 10, 311–319. doi: 10.1577/1548-8667(1998)010<0311:IOFAFS>2.0.CO;2
Barker, D. E., Braden, L. M., Coombs, M. P., Boyce, B. (2009). Preliminary studies on the isolation of bacteria from sea lice, Lepeophtheirus salmonis, infecting farmed salmon in British Columbia, Canada. Parasitol. Res. 105, 1173–1177. doi: 10.1007/s00436-009-1523-9
Baxa, D. V., Kawai, K., Kusuda, R. (1988). In vitro and in vivo activities of Flexibacter maritimus toxins. Bull. Mar. Sci. Fish. Kochi Univ. 10, 1–8.
Bernardet, J.-F. (1998). Cytophaga, Flavobacterium, flexibacter and Chryseobacterium infections in cultured marine fish. Fish Pathol. 33, 229–238. doi: 10.3147/jsfp.33.229
Bernardet, J.-F., Kerouault, B., Michel, C. (1994). Comparative study on Flexibacter maritimus strains isolated from farmed sea bass (Dicentrarchus labrax) in France. Fish Pathol. 29, 105–111. doi: 10.3147/jsfp.29.105
Bridel, S., Bourgeon, F., Marie, A., Saulnier, D. (2020). Genetic diversity and population structure of Tenacibaculum maritimum, a serious bacterial pathogen of marine fish: From genome comparisons to high throughput MALDI-TOF typing. Vet. Res. 51, 1–17. doi: 10.1186/s13567-020-00782-0
Bridel, S., Olsen, A.-B., Nilsen, H., Bernardet, J.-F., Achaz, G., Avendaño-Herrera, R., et al. (2018). Comparative genomics of Tenacibaculum dicentrarchi and “Tenacibaculum finnmarkense” highlights intricate evolution of fish-pathogenic species. Genome Biol. Evol. 10, 452–457. doi: 10.1093/gbe/evy020
Brock, T. D., Madigan, M. T., Martinko, J. M., Parker, J. (2003). Brock Biology of Microorganisms. Upper Saddle River (NJ: Prentice-Hall).
Brosnahan, C. L., Munday, J. S., Ha, H. J., Preece, M., Jones, J. B. (2019). New Zealand rickettsia-like organism (NZ-RLO) and Tenacibaculum maritimum: Distribution and phylogeny in farmed Chinook salmon (Oncorhynchus tshawytscha). J. Fish Dis. 42, 85–95. doi: 10.1111/jfd.12909
Burchard, R. P., Rittschof, D., Bonaventura, J. (1990). Adhesion and motility of gliding bacteria on substrata with different surface free energies. Appl. Environ. Microbiol. 56, 2529–2534. doi: 10.1128/aem.56.8.2529-2534.1990
Carson, J., Schmidtke, L., Lewis, T., Valentine, P. (1994). Development of a vaccine against disease caused by Flexibacter maritimus: Results of efficacy testing of three types of vaccine. In Valentine, P. (Ed.), Barriers and Break-throughs (Hobart, TAS: Research and Development Review Seminar), p. 149–158.
Carson, J., Schmidtke, L., Mccos, P. (1993). A salmonid vaccine against Flexibacter maritimus-dream or reality In Valentine, P. (Ed.), Seeking and solving: Papers from the Saltas 1993 (Hobart, TAS: Research and Development Review Seminar), p. 113–120.
Casadevall, A., Pirofski, L.-a. (1999). Host-pathogen interactions: Redefining the basic concepts of virulence and pathogenicity. Infect. Immun. 67, 3703–3713. doi: 10.1128/IAI.67.8.3703-3713.1999
Casadevall, A., Pirofski, L. A. (2001). Host-pathogen interactions: The attributes of virulence. J. Infect. Dis. 184, 337–344. doi: 10.1086/322044
Cepeda, C., García-Márquez, S., Santos, Y. (2003). Detection of Flexibacter maritimus in fish tissue using nested PCR amplification. J. Fish Dis. 26, 65–70. doi: 10.1046/j.1365-2761.2003.00431.x
Cepeda, C., Santos, Y. (2002). First isolation of Flexibacter maritimus from farmed Senegalese sole (Solea senegalensis, kaup) in Spain. Bull. European Assoc. Fish Pathologists 22, 388–392.
Chen, M., Henry-Ford, D., Groff, J. (1995). Isolation and characterization of Flexibacter maritimus from marine fishes of California. J. Aquat. Anim. Health 7, 318–326. doi: 10.1577/1548-8667(1995)007<0318:IACOMF>2.3.CO;2
Cimica, V., Galarza, J. M. (2017). Adjuvant formulations for virus-like particle (VLP) based vaccines. Clin. Immunol. 183, 99–108. doi: 10.1016/j.clim.2017.08.004
Clark, S. E., Jude, B. A., Danner, G. R., Fekete, F. A. (2009). Identification of a multidrug efflux pump in Flavobacterium johnsoniae. Vet. Res. 40, 1–10. doi: 10.1051/vetres/2009038
Cunningham, C. O. (2002). Molecular diagnosis of fish and shellfish diseases: Present status and potential use in disease control. Aquaculture 206, 19–55. doi: 10.1016/S0044-8486(01)00864-X
Downes, J., Yatabe, T., Marcos-Lopez, M., Rodger, H. (2018). Investigation of co-infections with pathogens associated with gill disease in Atlantic salmon during an amoebic gill disease outbreak. J. Fish Dis. 41, 1217–1227. doi: 10.1111/jfd.12814
Elanco. (2018). Technical Report: An Overview of Emerging Diseases in the Salmonid Farming Industry. Elanco Canada Ltd., Canada. Available online: https://www.vetinst.no/rapporter-og-publikasjoner/rapporter/2019/an-overview-of-emergingdiseases-in-the-salmonid-farming-industry-technical-report/_/attachment/download/879fcbae-cc63-4a88-9391-fff0c18580f9:d79c3cf13885c938419d77c9cca12e5e1ab2b4d6/Emerging%20diseases%20techinical%20report%20-%20january%202019.pdf.
Escribano, M., Ramos-Pinto, L., Fernández-Boo, S., Afonso, A., Costas, B., Guardiola, F. (2020). Mucosal immune responses in Senegalese sole (Solea senegalensis) juveniles after Tenacibaculum maritimum challenge: A comparative study between ocular and blind sides. Fish Shellfish Immunol. 104, 92–100. doi: 10.1016/j.fsi.2020.05.080
Estensoro, I., Jung-Schroers, V., Álvarez-Pellitero, P., Steinhagen, D., Sitjà-Bobadilla, A. (2013). Effects of Enteromyxum leei (Myxozoa) infection on gilthead sea bream (Sparus aurata)(Teleostei) intestinal mucus: Glycoprotein profile and bacterial adhesion. Parasitol. Res. 112, 567–576. doi: 10.1007/s00436-012-3168-3
Faílde, L. D., Losada, A. P., Bermúdez, R., Santos, Y., Quiroga, M. I. (2013). Tenacibaculum maritimum infection: Pathology and immunohistochemistry in experimentally challenged turbot (Psetta maxima l.). Microb. Pathogen. 65, 82–88. doi: 10.1016/j.micpath.2013.09.003
FAO (2020). State of world fisheries and aquaculture 2020 (Russian edition): Sustainability in action (Rome, Italy: Food & Agriculture Org). doi: 10.4060/ca9229en
Ferguson, H. W., Christian, M. D., Hay, S., Nicolson, J., Sutherland, D., Crumlish, M. (2010). Jellyfish as vectors of bacterial disease for farmed salmon (Salmo salar). J. Vet. Diagn. Invest. 22, 376–382. doi: 10.1177/104063871002200305
Fernández-Álvarez, C., González, S. F., Santos, Y. (2019). Quantitative PCR coupled with melting curve analysis for rapid detection and quantification of Tenacibaculum maritimum in fish and environmental samples. Aquaculture 498, 289–296. doi: 10.1016/j.aquaculture.2018.08.039
Fernández-Álvarez, C., Santos, Y. (2018). Identification and typing of fish pathogenic species of the genus Tenacibaculum. Appl. Microbiol. Biotechnol. 102, 9973–9989. doi: 10.1007/s00253-018-9370-1
Fernández-Álvarez, C., Torres-Corral, Y., Saltos-Rosero, N., Santos, Y. (2017). MALDI-TOF mass spectrometry for rapid differentiation of Tenacibaculum species pathogenic for fish. Appl. Microbiol. Biotechnol. 101, 5377–5390. doi: 10.1007/s00253-017-8324-3
Florio, D., Gridelli, S., Fioravanti, M. L., Zanoni, R. G. (2016). First isolation of Tenacibaculum maritimum in a captive sand tiger shark (Carcharias taurus). J. Zoo Wildlife Med. 47, 351–353. doi: 10.1638/2015-0064.1
Frietze, K. M., Peabody, D. S., Chackerian, B. (2016). Engineering virus-like particles as vaccine platforms. Curr. Opin. Virol. 18, 44–49. doi: 10.1016/j.coviro.2016.03.001
Fringuelli, E., Savage, P., Gordon, A., Baxter, E., Rodger, H., Graham, D. (2012). Development of a quantitative real-time PCR for the detection of Tenacibaculum maritimum and its application to field samples. J. Fish Dis. 35, 579–590. doi: 10.1111/j.1365-2761.2012.01377.x
Frisch, K., Småge, S. B., Brevik, Ø. J., Duesund, H., Nylund, A. (2018a). Genotyping of Tenacibaculum maritimum isolates from farmed Atlantic salmon in Western Canada. J. Fish Dis. 41, 131–137. doi: 10.1111/jfd.12687
Frisch, K., Småge, S. B., Johansen, R., Duesund, H., Brevik, Ø. J., Nylund, A. (2018b). Pathology of experimentally induced mouthrot caused by Tenacibaculum maritimum in Atlantic salmon smolts. PloS One 13, e0206951. doi: 10.1371/journal.pone.0206951
Frisch, K., Småge, S. B., Vallestad, C., Duesund, H. (2018c). Experimental induction of mouthrot in Atlantic salmon smolts using Tenacibaculum maritimum from Western Canada. J. Fish Dis. 41, 1247–1258. doi: 10.1111/jfd.12818
Fujita, M. J., Kimura, N., Yokose, H., Otsuka, M. (2012). Heterologous production of bisucaberin using a biosynthetic gene cluster cloned from a deep sea metagenome. Mol. Biosyst. 8, 482–485. doi: 10.1039/C1MB05431G
Gorski, L. (2021). Serotype Assignment by Sero-agglutination, ELISA, and PCR. In: Fox, E. M., Bierne, H., Stessl, B. (eds) Listeria Monocytogenes. Methods in Molecular Biology, vol 2220. Humana, New York, NY. doi: 10.1007/978-1-0716-0982-8_5
Gourzioti, E., Kolygas, M., Athanassopoulou, F., Babili, V. (2016). Tenacibaculosis in aquaculture farmed marine fish. J. Hellenic Vet. Med. Soc. 67, 21–32. doi: 10.12681/jhvms.15620
Guardiola, F., Mabrok, M., Machado, M., Azeredo, R., Afonso, A., Esteban, M., et al. (2019). Mucosal and systemic immune responses in Senegalese sole (Solea senegalensis kaup) bath challenged with Tenacibaculum maritimum: A time-course study. Fish Shellfish Immunol. 87, 744–754. doi: 10.1016/j.fsi.2019.02.015
Habib, C., Houel, A., Lunazzi, A., Bernardet, J.-F. (2014). Multilocus sequence analysis of the marine bacterial genus tenacibaculum suggests parallel evolution of fish pathogenicity and endemic colonization of aquaculture systems. Appl. Environ. Microbiol. 80, 5503–5514. doi: 10.1128/AEM.01177-14
Hansen, G. H., Bergh, Ø., Michaelsen, J., Knappskog, D. (1992). Flexibacter ovolyticus sp. nov., a pathogen of eggs and larvae of Atlantic halibut, Hippoglossus hippoglossus l. Int. J. System. Evol. Microbiol. 42, 451–458. doi: 10.1099/00207713-42-3-451
Haridy, M., Hasheim, M., Abd El-Galil, M., Sakai, H., Yanai, T. (2015). Pathological findings of Tenacibaculum maritimus infection in black damselfish, Neoglyphieodon melas and Picasso triggerfish, Rhinecanthus assasi in red Sea, Egypt. Vet. Sci. Technol. 6, 214. doi: 10.4172/2157-7579.1000214
Hassan, M. A., Abd Allah, N. A., Mabrok, M. (2021). Inevitable impact of some environmental stressors on the frequency and pathogenicity of marine vibriosis. Aquaculture 536, 736447. doi: 10.1016/j.aquaculture.2021.736447
Henríquez-Núñez, H., Evrard, O., Kronvall, G., Avendaño-Herrera, R. (2012). Antimicrobial susceptibility and plasmid profiles of Flavobacterium psychrophilum strains isolated in Chile. Aquaculture 354, 38–44. doi: 10.1016/j.aquaculture.2012.04.034
Imberty, A., Varrot, A. (2008). Microbial recognition of human cell surface glycoconjugates. Curr. Opin. Struct. Biol. 18, 567–576. doi: 10.1016/j.sbi.2008.08.001
Irgang, R., Avendaño-Herrera, R. (2022). Evaluation of the in vitro susceptibility of Tenacibaculum dicentrarchi to tiamulin using minimum inhibitory concentration tests. J. Fish Dis. 45 (6), 795–799. doi: 10.1111/jfd.13604
Irgang, R., Mancilla, M., Avendaño-Herrera, R. (2021). Florfenicol and oxytetracycline susceptibility patterns in Chilean isolates of Tenacibaculum dicentrarchi: An emerging pathogen for farmed salmonids. J. Fish Dis. 44, 1043–1046. doi: 10.1111/jfd.13380
Ito, M., Okino, N., Tani, M. (2014). New insight into the structure, reaction mechanism, and biological functions of neutral ceramidase. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 1841, 682–691. doi: 10.1016/j.bbalip.2013.09.008
Izumi, S., Aranishi, F. (2004). Relationship between gyrA mutations and quinolone resistance in Flavobacterium psychrophilum isolates. Appl. Environ. Microbiol. 70, 3968. doi: 10.1128/AEM.70.7.3968-3972.2004
Jang, Y.-H., Jeong, J.-B., Yeo, I.-K., Kim, K.-Y., Harikrishnan, R., Heo, M.-S. (2009). Biological characterization of Tenacibaculum maritimum isolated from cultured olive flounder in Korea and sensitivity against native plant extracts. J. Fish Pathol. 22, 53–65.
Kirjusina, M., Briede, I., Bondad-Reantaso, M. (2007). Extension manual on some important viruses, parasites and bacteria of aquatic animals in Latvia, NDC/LZRA/FAO. Riga. p. 69
Klakegg, Ø., Abayneh, T., Fauske, A. K., Fülberth, M., Sørum, H. (2019). An outbreak of acute disease and mortality in Atlantic salmon (Salmo salar) post−smolts in Norway caused by Tenacibaculum dicentrarchi. J. Fish Dis. 42, 789–807. doi: 10.1111/jfd.12982
Klein, D. (2002). Quantification using real-time PCR technology: Applications and limitations. Trends Mol. Med. 8, 257–260. doi: 10.1016/S1471-4914(02)02355-9
Kolygas, M., Gourzioti, E., Vatsos, I., Athanassopoulou, F. (2012). Identification of Tenacibaculum maritimum strains from marine farmed fish in Greece. Vet. Rec. 170, 623. doi: 10.1136/vr.100778
Kumar, G., Engle, C. R. (2016). Technological advances that led to growth of shrimp, salmon, and tilapia farming. Rev. Fish. Sci. Aquacult. 24, 136–152. doi: 10.1080/23308249.2015.1112357
Lee, M., Jun, S.-Y., Yoon, B.-Y., Song, S., Lee, K., Ha, N.-C. (2012). Membrane fusion proteins of type I secretion system and tripartite efflux pumps share a binding motif for TolC in gram-negative bacteria. PloS One 7, e40460. doi: 10.1371/journal.pone.0040460
Leung, T. L., Bates, A. E. (2013). More rapid and severe disease outbreaks for aquaculture at the tropics: Implications for food security. J. Appl. Ecol. 50, 215–222. doi: 10.1111/1365-2644.12017
Levipan, H. A., Tapia-Cammas, D., Molina, V., Irgang, R., Toranzo, A. E., Magariños, B., et al. (2019). Biofilm development and cell viability: An undervalued mechanism in the persistence of the fish pathogen Tenacibaculum maritimum. Aquaculture 511, 734267. doi: 10.1016/j.aquaculture.2019.734267
Lieke, T., Meinelt, T., Hoseinifar, S. H., Pan, B., Straus, D. L., Steinberg, C. E. (2020). Sustainable aquaculture requires environmental-friendly treatment strategies for fish diseases. Rev. Aquacult. 12, 943–965. doi: 10.1111/raq.12365
Lin, T., Lin, L., Zhang, F. (2014). Review on molecular typing methods of pathogens. Open J. Med. Microbiol. 4, 147. doi: 10.4236/ojmm.2014.43017
Li, D., Oku, N., Hasada, A., Shimizu, M., Igarashi, Y. (2018). Two new 2-alkylquinolones, inhibitory to the fish skin ulcer pathogen Tenacibaculum maritimum, produced by a rhizobacterium of the genus burkholderia sp. beilstein. J. Organic Chem. 14, 1446–1451. doi: 10.3762/bjoc.14.122
Llewellyn, M., Leadbeater, S., Garcia, C., Sylvain, F.-E. (2017). Parasitism perturbs the mucosal microbiome of Atlantic salmon. Sci. Rep. 7, 1–10. doi: 10.1038/srep43465
Lopez, P., Bridel, S., Saulnier, D., David, R., Magariños, B., Torres, B. S., et al. (2022a). Genomic characterization of Tenacibaculum maritimum O-antigen gene cluster and development of a multiplex PCR-based serotyping scheme. Transbound. Emerg. Dis. 69 (5), e2876–e2888. doi: 10.1111/tbed.14637
López, J., Núñez, S., Magariños, B., Castro, N., Navas, J., de la Herran, R., et al. (2009). First isolation of Tenacibaculum maritimum from wedge sole, Dicologoglossa cuneata (Moreau). J. Fish Dis. 32, 603–610. doi: 10.1111/j.1365-2761.2009.01029.x
López, J., Piñeiro-Vidal, M., García-Lamas, N., de la Herran, R., Navas, J., Hachero-Cruzado, I., et al. (2010). First isolation of Tenacibaculum soleae from diseased cultured wedge sole, Dicologoglossa cuneata (Moreau), and brill, Scophthalmus rhombus (L.). J. Fish Dis. 33, 273–278. doi: 10.1111/j.1365-2761.2009.01105.x
Lopez, P., Saulnier, D., Swarup-Gaucher, S., David, R. (2022b). First isolation of virulent Tenacibaculum maritimum isolates from diseased orbicular batfish (Platax orbicularis) farmed in Tahiti island. Pathogens 11, 131. doi: 10.3390/pathogens11020131
Mabrok, M., Elayaraja, S., Chokmangmeepisarn, P., Jaroenram, W. (2021). Rapid visualization in the specific detection of Flavobacterium columnare, a causative agent of freshwater columnaris using a novel recombinase polymerase amplification (RPA) combined with lateral flow dipstick (LFD) assay. Aquaculture 531, 735780. doi: 10.1016/j.aquaculture.2020.735780
Mabrok, M., Machado, M., Serra, C., Afonso, A., Valente, L., Costas, B. (2016). Tenacibaculosis induction in the Senegalese sole (Solea senegalensis) and studies of Tenacibaculum maritimum survival against host mucus and plasma. J. Fish Dis. 39, 1445–1455. doi: 10.1111/jfd.12483
Magariños, B., Pazos, F., Santos, Y., Romalde, J. L., Toranzo, A. E. (1995). Response of Pasteurella piscicida and Flexibacter maritimus to skin mucus of marine fish. Dis. Aquat. Organisms 21, 103–108. doi: 10.3354/dao021103
Magi, G., Avendano-Herrera, R., Magarinos, B., Toranzo, A., Romalde, J. (2007). First reports of flexibacteriosis in farmed tub gurnard (Chelidonichthys lucernus l.) and wild turbot (Scophthalmus maximus) in Italy. Bull. European Assoc. Fish Pathologists 27, 177.
Marcogliese, D. J. (2008). The impact of climate change on the parasites and infectious diseases of aquatic animals. Rev. Scientifique Technique 27, 467–484. doi: 10.20506/rst.27.2.1820
Masumura, K., Wakabayashi, H. (1977). An outbreak of gliding bacterial disease in hatchery-born red seabream (Pagrus major) and gilthead (Acanthopagrus schlegeli) fry in Hiroshima. Fish Pathol. 12, 171–177. doi: 10.3147/jsfp.12.171
Melba, B., Nihad, F., Brett, M., David, H. (2021). A 12-point checklist for surveillance of diseases of aquatic organisms: A novel approach to assist multidisciplinary teams in developing countries. Rev. Aquacult. 13, 1469–1487. doi: 10.1111/raq.12530
Michel, C., Matte-Tailliez, O., Kerouault, B., Bernardet, J. F. (2005). Resistance pattern and assessment of phenicol agents' minimum inhibitory concentration in multiple drug resistant Chryseobacterium isolates from fish and aquatic habitats. J. Appl. Microbiol. 99, 323–332. doi: 10.1111/j.1365-2672.2005.02592.x
Miyake, S., Soh, M., Azman, M. N., Ngoh, S. Y., Orbán, L., Seedorf, H. (2020). Insights into the microbiome of farmed Asian sea bass (Lates calcarifer) with symptoms of tenacibaculosis and description of Tenacibaculum singaporense sp. nov. Antonie van Leeuwenhoek 113, 737–752. doi: 10.1007/s10482-020-01391-9
Molenaar, E. J., Caddell, R. (2019). “International fisheries law: Achievements, limitations and challenges,” in Strengthening international fisheries law in an era of changing oceans hart(Oxford, UK).
Moustafa, M., Eissa, A., Laila, A., Gaafar, A., Abumourad, I., Elgendy, M. (2014). Mass mortalities in mari-cultured European sea bass (Dicentrarchus labrax) at northern Egypt. Res. J. Pharm. Biol. Chem. Sci. 5, 95–109.
Moustafa, M., Eissa, A., Laila, A., Gaafar, A., Abumourad, I., Elgendy, M. (2015). Investigations into the potential causes of mass kills in mari-cultured gilthead sea bream (Sparus aurata) at northern Egypt. Res. J. Pharm. Biol. Chem. Sci. 6, 466–477.
Nekouei, O., Vanderstichel, R., Ming, T., Kaukinen, K. H. (2018). Detection and assessment of the distribution of infectious agents in juvenile Fraser river sockeye salmon, Canada, in 2012 and 2013. Front. Microbiol. 9, 3221. doi: 10.3389/fmicb.2018.03221
Nowlan, J. P., Britney, S. R., Lumsden, J. S., Russell, S. (2021). Application of quantitative-PCR to monitor netpen sites in British Columbia (Canada) for Tenacibaculum species. Pathogens 10, 414. doi: 10.3390/pathogens10040414
Nowlan, J. P., Lumsden, J. S., Russell, S. (2020). Advancements in characterizing Tenacibaculum infections in Canada. Pathogens 9 (12), 1029. doi: 10.3390/pathogens9121029
Oda, M., Takahashi, M., Matsuno, T., Uoo, K., Nagahama, M., Sakurai, J. (2010). Hemolysis induced by Bacillus cereus sphingomyelinase. Biochim. Biophys. Acta (BBA) Biomembr. 1798, 1073–1080. doi: 10.1016/j.bbamem.2010.03.004
Ofek, I., Doyle, R. J. (1994). Principles of bacterial adhesion. In: Bacterial Adhesion to Cells and Tissues. (Boston, MA: Springer). doi: 10.1007/978-1-4684-6435-1_1
Ofek, I., Doyle, R. J. (2012). Bacterial adhesion to cells and tissues. In: Ofek, I., Doyle, R. J., editors. (New York: Chapman & Hall)
Olsen, A. B., Gulla, S., Steinum, T., Colquhoun, D. J., Nilsen, H. K., Duchaud, E. (2017). Multilocus sequence analysis reveals extensive genetic variety within Tenacibaculum spp. associated with ulcers in sea−farmed fish in Norway. Vet. Microbiol. 205, 39–45. doi: 10.1016/j.vetmic.2017.04.028
Olsen, A. B., Nilsen, H., Sandlund, N., Mikkelsen, H., Sørum, H., Colquhoun, D. (2011). Tenacibaculum sp. associated with winter ulcers in sea-reared Atlantic salmon Salmo salar. Dis. Aquat. Organisms 94, 189–199. doi: 10.3354/dao02324
Olsen, A. B., Spilsberg, B., Nilsen, H. K., Lagesen, K. (2020). Tenacibaculum piscium sp. nov., isolated from skin ulcers of sea-farmed fish, and description of Tenacibaculum finnmarkense sp. nov. with subdivision into genomovars finnmarkense and ulcerans. Int. J. System. Evol. Microbiol. 70, 6079–6090. doi: 10.1099/ijsem.0.004501
Pasqua, M., Grossi, M., Zennaro, A., Fanelli, G. (2019). The varied role of efflux pumps of the MFS family in the interplay of bacteria with animal and plant cells. Microorganisms 7, 285. doi: 10.3390/microorganisms7090285
Pazos, F. (1997). Flexibacter maritimus: Estudio fenotípico, inmunológico y molecular (Spain: Universidade de Santiago de Compostela, Spain).
Pazos, F., Santos, Y., Macias, A., Núñez, S., Toranzo, A. (1996). Evaluation of media for the successful culture of Flexibacter maritimus. J. Fish Dis. 19, 193–197. doi: 10.1111/j.1365-2761.1996.tb00701.x
Pérez-Pascual, D., Lunazzi, A., Magdelenat, G., Rouy, Z. (2017). The complete genome sequence of the fish pathogen Tenacibaculum maritimum provides insights into virulence mechanisms. Front. Microbiol. 8, 1542. doi: 10.3389/fmicb.2017.01542
Piñeiro-Vidal, M., Centeno-Sestelo, G., Riaza, A., Santos, Y. (2007). Isolation of pathogenic Tenacibaculum maritimum-related organisms from diseased turbot and sole cultured in the Northwest of Spain. Bulletin of the European Association of Fish Pathologists, 27(1), 29–35.
Piñeiro-Vidal, M., Riaza, A., Santos, Y. (2008a). Tenacibaculum discolor sp. nov. and Tenacibaculum gallaicum sp. nov., isolated from sole (Solea senegalensis) and turbot (Psetta maxima) culture systems. Int. J. System. System. Evol. Microbiol. Microbiol. 58, 21–25. doi: 10.1099/ijs.0.65397-0
Piñeiro-Vidal, M., Carballas, C. G., Gomez-Barreiro, O., Riaza, A., Santos, Y. (2008b). Tenacibaculum soleae sp. nov., isolated from diseased sole (Solea senegalensis kaup). Int. J. System. Evol. Microbiol. 58, 881–885. doi: 10.1099/ijs.0.65539-0
Piñeiro-Vidal, M., Gijón, D., Zarza, C., Santos, Y. (2012). Tenacibaculum dicentrarchi sp. nov., a marine bacterium of the family Flavobacteriaceae isolated from European sea bass. Int. J. System. Evol. Microbiol. 62, 425–429. doi: 10.1099/ijs.0.025122-0
Powell, M., Carson, J., van Gelderen, R. (2004). Experimental induction of gill disease in Atlantic salmon Salmo salar smolts with Tenacibaculum maritimum. Dis. Aquat. Organisms 61, 179–185. doi: 10.3354/dao061179
Rahman, T., Suga, K., Kanai, K., Sugihara, Y. (2014). Biological and serological characterization of a non-gliding strain of Tenacibaculum maritimum isolated from a diseased puffer fish takifugu rubripes. Fish Pathol 49, 121–129. doi: 10.3147/jsfp.49.121
Ratledge, C., Dover, L. G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941. doi: 10.1146/annurev.micro.54.1.881
Reyad, A. M., Atta, H. M., Mahran, H. A. (2013). Antibacterial activity of Nocardiopsis dassonvillei yscl2334 against Tenacibaculum maritimum isolated from diseased fishes in marine aquaculture. Egyptian J. Exp. Biol. (Botany) 9, 183–191.
Romalde, J. L., Ravelo, C., López-Romalde, S., Avendano-Herrera, R., Magariños, B., Toranzo, A. E. (2005). Vaccination strategies to prevent emerging diseases for Spanish aquaculture. Develop. Biologicals 121, 85–95.
Romero, M., Avendaño-Herrera, R., Magariños, B., Cámara, M., Otero, A. (2010). Acylhomoserine lactone production and degradation by the fish pathogen Tenacibaculum maritimum, a member of the Cytophaga–Flavobacterium–Bacteroides (CFB) group. FEMS Microbiol. Lett. 304, 131–139. doi: 10.1111/j.1574-6968.2009.01889.x
Salati, F., Cubadda, C., Viale, I., Kusuda, R. (2005). Immune response of sea bass Dicentrarchus labrax to Tenacibaculum maritimum antigens. Fish. Sci. 71, 563–567. doi: 10.1111/j.1444-2906.2005.01000.x
Santos, Y., Pazos, F., Barja, J. (1999). Flexibacter maritimus, causal agent of flexibacteriosis in marine fish. In: Olivier, G (ed.) ICES Identification Leaflets for Diseases and Parasites of Fish and Shellfish. No. 55, pp. 1–6. (Copenhagen, Denmark: International Council for the Exploration of the Sea).
Santos, Y., Pazos, F., Barja, J. (2019). ICES Tenacibaculum maritimum, causal agent of tenacibaculosis in marine fish.
Serra, C. R., Almeida, E. M., Guerreiro, I., Santos, R. (2019). Selection of carbohydrate-active probiotics from the gut of carnivorous fish fed plant-based diets. Sci. Rep. 9, 1–15. doi: 10.1038/s41598-019-42716-7
Sheng, Y., Abreu, I. A., Cabelli, D. E., Maroney, M. J., Miller, A. F., Teixeira, M., et al. (2014). Superoxide dismutases and superoxide reductases. Chem. Rev. 114, 3854–3918. doi: 10.1021/cr4005296
Silva, M. T. (2012). Classical labeling of bacterial pathogens according to their lifestyle in the host: Inconsistencies and alternatives. Front. Microbiol. 3, 71. doi: 10.3389/fmicb.2012.00071
Simon, O., Klaus, B. (2008). The marketing and the use of feed additives in the European union with particular regard to zootechnical feed additives. Eur. Food Feed Law Rev. 3, 132–154.
Småge, S. B., Brevik, Ø. J, Duesund, H., Ottem, K. F., Watanabe, K., et al. (2016a). Tenacibaculum finnmarkense sp. nov., a fish pathogenic bacterium of the family Flavobacteriaceae isolated from Atlantic salmon. Antonie van Leeuwenhoek 109, 273–285. doi: 10.1007/s10482-015-0630-0
Småge, S. B., Frisch, K., Brevik, Ø. J, Watanabe, K., Nylund, A. (2016b). First isolation, identification and characterisation of Tenacibaculum maritimum in Norway, isolated from diseased farmed sea lice cleaner fish Cyclopterus lumpus l. Aquaculture 464, 178–184. doi: 10.1016/j.aquaculture.2016.06.030
Sneeringer, S., Bowman, M., Clancy, M. (2019). The U.S. and EU Animal Pharmaceutical Industries in the Age of Antibiotic Resistance, ERR-264, U.S. Department of Agriculture, Economic Research Service. Available at: https://www.ers.usda.gov/webdocs/publications/93179/err-264.pdf?v=961.
Soltani, M., Munday, B., Burke, C. (1996). The relative susceptibility of fish to infections by Flexibacter columnaris and Flexibacter maritimus. Aquaculture 140, 259–264. doi: 10.1016/0044-8486(95)01157-9
Soniya, M., Kuberan, T., Anitha, S., Sankareswari, P. (2013). In vitro antibacterial activity of plant extracts against gram positive and gram negative pathogenic bacteria. Int. J. Microbiol. Immunol. Res. 2, 1–5.
Staroscik, A. M., Nelson, D. R. (2008). The influence of salmon surface mucus on the growth of Flavobacterium columnare. J. Fish Dis. 31 (1), 59–69. doi: 10.1111/j.1365-2761.2007.00867.x
Subasinghe, R., Soto, D., Jia, J. (2009). Global aquaculture and its role in sustainable development. Rev. Aquacult. 1, 2–9. doi: 10.1111/j.1753-5131.2008.01002.x
Suomalainen, L. R., Tiirola, M., Valtonen, E. (2006). Chondroitin AC lyase activity is related to virulence of fish pathogenic Flavobacterium columnare. J. Fish Dis. 29, 757–763. doi: 10.1111/j.1365-2761.2006.00771.x
Suzuki, M. (2015). Tenacibaculum. In Bergey’s Manual of Systematics of Archea and Bacteria, John Wiley & Sons. Inc. and Bergey ́s Manual Trust, p. 7. doi: 10.1002/9781118960608.gbm0034
Suzuki, M., Nakagawa, Y., Harayama, S., Yamamoto, S. (2001). Phylogenetic analysis and taxonomic study of marine cytophaga-like bacteria: Proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int. J. System. Evol. Microbiol. 51, 1639–1652. doi: 10.1099/00207713-51-5-1639
Tesdorpf, J. E., Geers, A. U., Strube, M. L., Gram, L., Bentzon-Tilia, M. (2022). Roseobacter group probiotics exhibit differential killing of fish pathogenic Tenacibaculum species. Appl. Environ. Microbiol. 88, e02418–e02421. doi: 10.1128/aem.02418-21
Toranzo, A. E., Magariños, B., Romalde, J. L. (2005). A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246, 37–61. doi: 10.1016/j.aquaculture.2005.01.002
Toranzo, A., Romalde, J., Dopazo, C., Magariños, B., Barja, J. (2004). Disease trends in the primary marine fish species cultured in Spain: A 20-year study. World Aquacul. 35, 35–38.
Toyama, T., Kita-Tsukamoto, K., Wakabayashi, H. (1996). Identification of Flexibacter maritimus, Flavobacterium branchiophilum and Cytophaga columnaris by PCR targeted 16S ribosomal DNA. Fish Pathol. 31, 25–31. doi: 10.3147/jsfp.31.25
Valdes, S., Irgang, R., Barros, M. C., Ilardi, P. (2021). First report and characterization of Tenacibaculum maritimum isolates recovered from rainbow trout (Oncorhynchus mykiss) farmed in Chile. J. Fish Dis. 44, 1481–1490 doi: 10.1111/jfd.13466
van Gelderen, R., Carson, J., Gudkovs, N., Nowak, B. (2010b). Physical characterisation of Tenacibaculum maritimum for vaccine development. J. Appl. Microbiol. 109, 1668–1676. doi: 10.1111/j.1365-2672.2010.04795.x
van Gelderen, R., Carson, J., Nowak, B. (2009a). Experimental vaccination of Atlantic salmon (Salmo salar l.) against marine flexibacteriosis. Aquaculture 288, 7–13. doi: 10.1016/j.aquaculture.2008.11.012
van Gelderen, R., Carson, J., Nowak, B. (2009b). Effect of extracellular products of Tenacibaculum maritimum in Atlantic salmon, Salmo salar l. J. Fish Dis. 32, 727–731. doi: 10.1111/j.1365-2761.2009.01032.x
van Gelderen, R., Carson, J., Nowak, B. (2010a). Experimentally induced marine flexibacteriosis in Atlantic salmon smolts Salmo salar. i. pathogenicity. Dis. Aquat. Organisms 91, 121–128. doi: 10.3354/dao02219
van Gelderen, R., Carson, J., Nowak, B. (2011). Experimentally induced marine flexibacteriosis in Atlantic salmon smolts Salmo salar. II. pathology. Dis. Aquat. Organisms 95, 125–135. doi: 10.3354/dao02329
Vilar, P., Faílde, L., Bermúdez, R., Vigliano, F. (2012). Morphopathological features of a severe ulcerative disease outbreak associated with Tenacibaculum maritimum in cultivated sole, Solea senegalensis (L.). J. Fish Dis. 35, 437–445. doi: 10.1111/j.1365-2761.2012.01360.x
Wakabayashi, H., Hikida, M., Masumura, K. (1986). Flexibacter maritimus sp. nov., a pathogen of marine fishes. Int. J. System. Evol. Microbiol. 36, 396–398. doi: 10.1099/00207713-36-3-396
Wanka, K. M., Damerau, T., Costas, B., Krueger, A., Schulz, C., Wuertz, S. (2018). Isolation and characterization of native probiotics for fish farming. BMC Microbiol. 18, 1–13. doi: 10.1186/s12866-018-1260-2
Warsen, A. E., Krug, M. J., LaFrentz, S., Stanek, D. R., Loge, F. J., Call, D. R. (2004). Simultaneous discrimination between 15 fish pathogens by using 16S ribosomal DNA PCR and DNA microarrays. Appl. Environ. Microbiol. 70, 4216–4221. doi: 10.1128/AEM.70.7.4216-4221.2004
Watts, J. E., Schreier, H. J., Lanska, L., Hale, M. S. (2017). The rising tide of antimicrobial resistance in aquaculture: Sources, sinks and solutions. Mar. Drugs 15, 158. doi: 10.3390/md15060158
Whitfield, C., Williams, D. M., Kelly, S. D. (2020). Lipopolysaccharide o-antigens–bacterial glycans made to measure. J. Biol. Chem. 295 (31), 10593–10609. doi: 10.1074/jbc.REV120.009402
Wilson, T., Carson, J. (2003). Development of sensitive, high-throughput one-tube RT PCR enzyme hybridisation assay to detect selected bacterial fish pathogens. Dis. Aquat. Organisms 54, 127–134. doi: 10.3354/dao054127
Wilson, T., Carson, J., Bowman, J. (2002). Optimisation of one-tube PCR-ELISA to detect femtogram amounts of genomic DNA. J. Microbiol. Methods 51, 163–170. doi: 10.1016/S0167-7012(02)00055-6
Wilson, T. K., Douglas, M., Dunn, V. (2019). First identification in Tasmania of fish pathogens Tenacibaculum dicentrarchi and T. soleae and multiplex PCR for these organisms and T. maritimum. Dis. Aquat. Organisms 136, 219–226. doi: 10.3354/dao03407
Yamamoto, T., Kawai, K., Oshima, S.-i. (2010). Evaluation of an experimental immersion infection method with Tenacibaculum maritimum in Japanese flounder Paralichthys olivaceus. Aquacult. Sci. 58, 481–489.
Yang, S., Rothman, R. E. (2004). PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 4, 337–348. doi: 10.1016/S1473-3099(04)01044-8
Keywords: Tenacibaculum maritimum, tenacibaculosis, pathogenicity, marine fish, aquaculture
Citation: Mabrok M, Algammal AM, Sivaramasamy E, Hetta HF, Atwah B, Alghamdi S, Fawzy A, Avendaño-Herrera R and Rodkhum C (2023) Tenacibaculosis caused by Tenacibaculum maritimum: Updated knowledge of this marine bacterial fish pathogen. Front. Cell. Infect. Microbiol. 12:1068000. doi: 10.3389/fcimb.2022.1068000
Received: 12 October 2022; Accepted: 28 November 2022;
Published: 06 January 2023.
Edited by:
Manuel L. Lemos, University of Santiago de Compostela, SpainReviewed by:
Mark J. McBride, University of Wisconsin–Milwaukee, United StatesDavid Hunnicutt, St. Norbert College, United States
Copyright © 2023 Mabrok, Algammal, Sivaramasamy, Hetta, Atwah, Alghamdi, Fawzy, Avendaño-Herrera and Rodkhum. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Channarong Rodkhum, channarong.r@chula.ac.th; Ruben Avendaño-Herrera, reavendano@yahoo.com; ravendano@unab.cl
 Mahmoud Mabrok
Mahmoud Mabrok Abdelazeem M. Algammal
Abdelazeem M. Algammal Elayaraja Sivaramasamy
Elayaraja Sivaramasamy Helal F. Hetta
Helal F. Hetta Banan Atwah6
Banan Atwah6  Saad Alghamdi
Saad Alghamdi Ruben Avendaño-Herrera
Ruben Avendaño-Herrera Channarong Rodkhum
Channarong Rodkhum