CtrA activates the expression of glutathione S-transferase conferring oxidative stress resistance to Ehrlichia chaffeensis
- Key Laboratory of Molecular Microbiology and Technology of the Ministry of Education, Department of Microbiology, College of Life Sciences, Nankai University, Tianjin, China
Ehrlichia chaffeensis, the causative agent of human monocytic ehrlichiosis (HME), is a Gram-negative obligatory intracellular bacterium, which infects and multiplies in human monocytes and macrophages. Host immune cells produce reactive oxygen species (ROS) to eliminate E. chaffeensis upon infection. E. chaffeensis global transcriptional regulator CtrA activates the expression of GshA and GshB to synthesize glutathione (GSH), the most potent natural antioxidant, upon oxidative stress to combat ROS damage. However, the mechanisms exploited by E. chaffeensis to utilize GSH are still unknown. Here, we found that in E. chaffeensis CtrA activated the expression of glutathione S-transferase (GST) upon oxidative stress, and E. chaffeensis GST utilizes GSH to eliminate ROS and confers the oxidative stress resistance to E. chaffeensis. We found that CtrA bound to the promoter regions of 211 genes, including gst, in E. chaffeensis using chromatin immunoprecipitation coupled to deep sequencing (ChIP-seq). Recombinant E. chaffeensis CtrA directly bound to the gst promoter region determined with electrophoretic mobility shift assay (EMSA), and activated the gst expression determined with reporter assay. Recombinant GST showed GSH conjugation activity towards its typical substrate 2,4-dinitrochlorobenzene (CDNB) in vitro and peptide nucleic acid (PNA) transfection of E. chaffeensis, which can knock down the gst transcription level, reduced bacterial survival upon oxidative stress. Our results demonstrate that E. chaffeensis CtrA regulates GSH utilization, which plays a critical role in resistance to oxidative stress, and aid in the development of new therapeutics for HME.
Introduction
Ehrlichia chaffeensis is a Gram-negative obligatory intracellular bacterium that preferentially infects human monocytes or macrophages and causes human monocytic ehrlichiosis (HME). HME is one of the most prevalent, life-threatening emerging zoonoses with the clinical signs of headache, fever, myalgia, malaise accompanied by anemia, leukopenia, thrombocytopenia and the elevation of aminotransferases (Paddock and Childs, 2003; Kumagai et al., 2010). Nearly 40-60% of patients need hospitalization and the mortality is approximately 3% (Paddock and Childs, 2003).
CtrA is a response regulator of the two-component regulatory system CckA/CtrA and is conserved among α-proteobacteria including all sequenced members of the family Anaplasmataceae. The consensus CtrA-binding motifs are TTAA-N7-TTAAC (9-mer) and TTAACCAT (8-mer) (Quon et al., 1996; Laub et al., 2002). In Caulobacter crescentus, CtrA directly regulates at least 95 genes (Quon et al., 1996; Domian et al., 1997; Laub et al., 2002). In E. chaffeensis, CtrA binds to the promoter regions of bolA, surE, ompA, gshA and gshB and activates their expression (Cheng et al., 2011; Yan et al., 2022). All of these 5 genes are critical for E. chaffeensis infection and survival (Cheng et al., 2011; Mohan Kumar et al., 2013; Lina et al., 2016; Yan et al., 2022), but are not present in C. crescentus CtrA regulon, indicating that the adaptation of E. chaffeensis to its obligate intracellular growth has led to the emergence of unique CtrA regulon. However, the CtrA regulon in E. chaffeensis has not been fully illustrated.
Upon infection, host immune cells produce reactive oxygen species (ROS) to kill pathogens (Sharma et al., 2017; Sies and Jones, 2020). E. chaffeensis EtpE blocks the generation of ROS and Etf-1 upregulates host manganese superoxide dismutase (MnSOD) to reduce ROS levels in macrophages (Liu et al., 2012; Teymournejad and Rikihisa, 2020), but the production of ROS by neutrophils cannot be suppressed by E. chaffeensis (Lin and Rikihisa, 2007). The mechanisms exploited by E. chaffeensis to cope with the ROS produced by other cells remain largely unknown.
GshA and GshB are two enzymes that catalyze L-glutamate (L-Glu), L-cysteine (L-Cys) and L-glycine (L-Gly) to synthesize glutathione (GSH), the most potent natural antioxidant (Figure S1) (Smirnova and Oktyabrsky, 2005; Morris et al., 2013). In E. chaffeensis we found that the expression of gshA and gshB was activated by CtrA upon oxidative stress (Yan et al., 2022). In bacteria, GSH can be utilized by glutathione peroxidase (GPx) and glutathione S-transferase (GST) to eliminate ROS (Skopelitou et al., 2012a; Morris et al., 2013). However, E. chaffeensis lacks GPx (Rikihisa, 2015) and encodes only one GST (ECH_0847). The function and regulation of E. chaffeensis GST remain to be investigated. In this study, to gain insights into CtrA regulon in E. chaffeensis we performed chromatin immunoprecipitation coupled to deep sequencing (ChIP-seq) and identified that 211 genes, including gst, were regulated by CtrA. Then we characterized GST function by enzymatic assays in vitro and peptide nucleic acid (PNA) knockdown in vivo.
Materials and methods
Bacteria and cell culture
E. chaffeensis Arkansas strain was propagated in THP-1 cells in RPMI 1640 medium supplemented with 2 mM L-glutamine and 10% fetal bovine serum (FBS) (Every Green, Zhejiang, China) at 37°C in 5% CO2 and 95% air, as described previously (Cheng et al., 2006). The THP-1 cell line present in this study was obtained from Fuheng Biotechnology (Cat. number FH0112, Shanghai, China).
Escherichia coli strains (Supplementary Table S1) DH5α and BL21 (DE3) used for general cloning and protein expression, respectively, were cultured in LB broth supplemented with ampicillin (100 μg·mL-1), chloramphenicol (34 μg·mL-1) or kanamycin (50 µg·mL-1), as necessary.
Expression and purification of recombinant proteins
The DNA fragment encoding full-length CtrA was cloned into pET-41a(+) to express recombinant CtrA (GST-rCtrA) as described previously (Cheng et al., 2006). The DNA fragments encoding GshA, GshB and GST were amplified using specific primers (Supplementary Table S1). The gshA fragment was cloned into pET-His-SUMO to express recombinant GshA (SUMO-rGshA) with an N-terminal His-tag. The fragment of gshB or gst was cloned into pET-33b(+) to express recombinant GshB (rGshB) or GST (rGST) with an N-terminal His-tag, respectively. The constructed plasmids were transformed into E. coli DH5α cells, extracted and confirmed by sequencing. E. coli BL21 (DE3) cells were transformed with the resulting plasmids to express SUMO-rGshA, rGshB and rGST respectively, with pET-41a(+) to express GST or pET-His-SUMO to express SUMO. E. coli BL21 (DE3) cells were induced to express recombinant proteins at an OD600 of 0.6 with 0.1 mM isopropyl-thio-β-D-galactoside (IPTG; Solarbio, Beijing, China) at 37°C for 4 h. All proteins were purified from soluble fraction with Ni-affinity chromatography and concentrated using an Amicon Ultra-0.5 Centrifugal Filter Unit (10 kDa MWCO).
Chromatin immunoprecipitation coupled to deep sequencing (ChIP-seq)
Fixed cell sample was prepared from 2.5 × 107 infected THP-1 cells (> 90% infected) following the instructions for suspension cell sample preparation from IGENEBOOK Biotechnology (Wuhan, China). The ChIP-seq experiments and data analyses were performed by IGENEBOOK Biotechnology. Briefly, cell culture was crosslinked with 1% (w/v) formaldehyde and incubated at room temperature for 10 min. Crosslinking was terminated by adding 125 mM L-Gly. Crosslinked cells were washed twice with 1 × PBS at 4°C, pelleted, frozen in liquid nitrogen and subjected to ChIP-seq. Rabbit antiserum against E. chaffeensis CtrA (Yan et al., 2022) was used for immunoprecipitation of E. chaffeensis chromosomal DNA fragments bound by CtrA. Total DNA before immunoprecipitation was used as input DNA. Fold enrichment was expressed as immunoprecipitation signal normalized against input signal.
Electrophoresis mobility shift assay
Electrophoresis mobility shift assay (EMSA) was performed as described previously (Cheng et al., 2011). Briefly, the promoter regions of gst (305 bp) and p28 (260 bp) were amplified with PCR using specific primers (Supplementary Table S1). Fifty picomoles of purified GST-rCtrA or GST (negative control) was incubated with 50 ng of DNA probes in a 20-μL reaction mixture containing 10 mM Tris-HCl (pH 7.5), 1 mM DTT and 1% (w/v) L-Gly on ice for 30 min. Samples were loaded onto 8% native polyacrylamide gel in 1 × TBE buffer [89 mM Tris (pH 8.3), 89 mM boric acid and 2 mM EDTA], which had been prerun at 100 V for 1 h, and electrophoresed at 10 mA on ice for 1.5 h. The gel was stained in 1 × TBE containing 0.5 μg·mL-1 ethidium bromide at room temperature for 10 min. Bands were visualized using a ChemiDoc XRS+ molecular imager (Bio-Rad).
Construction of enhanced green fluorescent protein fusions and reporter assay
Enhanced green fluorescent protein (EGFP) fusions were constructed as described previously (Duan et al., 2021). Briefly, the promoter region of gst or p28 was amplified and inserted upstream of the promoter-less egfp gene in a pQE60 vector. E. coli BL21 (DE3) cells containing pACYCDuet-1 harboring ctrA (pACYCDuet-1-rCtrA) or the empty pACYCDuet-1 vector were transformed with the pQE60-promoter-EGFP fusion constructs. After induction of rCtrA expression with 0.05 mM IPTG at 37°C for 3 h, bacterial samples were collected and subjected to Western blotting to measure the amounts of EGFP, RpoA, and rCtrA in each sample.
Western blotting
Western blotting was performed as described previously (Yan et al., 2022). Briefly, to detect the amount of EGFP in the reporter assay, the same numbers of E. coli cells were collected by centrifugation at 12000 × g for 1 min, resuspended in 1 × SDS sample buffer [62.5 mM Tris-HCl (pH 6.8), 2% (w/v) SDS, 0.02% (w/v) bromophenol blue, 1% (v/v) β-mercaptoethanol and 10% (v/v) glycerol] and boiled for 10 min. The samples were then subjected to 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a PVDF membrane (Millipore, Co. Cork, Ireland). The protein levels of EGFP, RpoA, and rCtrA were determined using a mouse monoclonal anti-GFP antibody (GeneTex Inc., North America, GTX628528), mouse monoclonal anti-E. coli RNA polymerase α antibody (BioLegend, San Diego, CA, United States, 663104), and rabbit antiserum against E. chaffeensis CtrA (Yan et al., 2022), respectively. The relative amounts of EGFP or RpoA in E. coli BL21 (DE3) cells expressing rCtrA were normalized against those in E. coli BL21 (DE3) cells containing pACYCDuet-1 vector.
Isolation of host cell-free E. chaffeensis
E. chaffeensis was isolated from infected THP-1 cells as described previously (Yan et al., 2022). Briefly, E. chaffeensis-infected THP-1 cells (2 × 107 cells, > 90% infected) were harvested at 600 × g at room temperature for 5 min. The pellet was suspended in fresh culture medium and passed through a 23-gauge needle with a syringe on ice for 20 times to rupture the host cell membrane. To remove unbroken cells and cell debris, the mixture was centrifuged at 1,000 × g at 4°C for 5 min. Bacteria in supernatant were collected by additional centrifugation at 10,000 × g at 4°C for 10 min. The bacterial pellet was suspended in fresh culture medium for synchronous culture or in 0.3 M sucrose for PNA transfection.
Quantitative RT-PCR
Total RNA was extracted from each sample and reverse transcribed to cDNA as described previously (Duan et al., 2021). The amounts of E. chaffeensis 16S rRNA, ctrA, gst, gshA, gshB, and human GAPDH were determined with qRT-PCR using specific primers (Supplementary Table S1) and the ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) on a StepOnePlus Real-Time PCR System (Applied Biosystems, MA, USA). The expression levels of bacterial genes were normalized against that of E. chaffeensis 16S rRNA. Relative bacterial number was determined as the amount of bacterial 16S rRNA normalized against that of human GAPDH mRNA.
Activity assay of glutathione S-transferase
The activity of rGST was determined as described previously with minor modifications (Habig and Jakoby, 1981). Briefly, the reaction mixture was composed of 100 mM Na3PO4 (pH 6.5), 1 mM 2,4-dinitrochlorobenzene (CDNB; Xiya Reagent, Shandong, China) and varying concentrations of GSH. Reactions were initiated by adding 3 μg of purified rGST or SUMO as a negative control in a final volume of 200 μL, then incubated at 37°C for 30 min. The formation of GSH-CDNB conjugate was measured by OD340.
Activity assay of glutamate-cysteine ligase
The activity of SUMO-rGshA was determined as described previously with minor modifications (Tu and Anders, 1998). Briefly, the reaction mixture was composed of 100 mM Tris-HCl (pH 8.0), 50 mM KCl, 20 mM MgCl2, 2 mM EDTA, 5 mM ATP, 10 mM L-α-aminobutyric acid and varying concentrations of monosodium L-glutamate. Reactions were initiated by adding 1.25 μg of purified SUMO-rGshA or SUMO as a negative control in a final volume of 80 μL, then incubated at 37°C for 30 min. Twenty microliters of 5% (w/v) SDS were added into each reaction mixture to terminate the reaction and the concentration of inorganic phosphate (Pi) generated by the reaction (Figure S1) was determined as described previously with minor modifications (Satoh et al., 2006). Briefly, 100 μL of reagent mixture containing 24 mg·mL-1 ascorbic acid in (NH4)2MoO4-H2SO4 solution [1.145 g (NH4)2MoO4 and 5.95 mL H2SO4 in 250 mL of water] was added to each reaction mixture. Reaction mixtures were incubated at room temperature for 2 h and concentrations of Pi were measured by OD750 using Varioskan Flash (Thermo Fisher Scientific). To calibrate the Pi concentration, 80 μL of increasing concentrations of KH2PO4 from 0 to 1,000 μM was mixed with 20 μL 5% (w/v) SDS and subjected to Pi determination as described above to generate a standard curve for each experiment.
Activity assay of glutathione synthetase
The activity of rGshB was determined as described previously with minor modifications (Oppenheimer et al., 1979). Briefly, the reaction mixture was composed of 100 mM Tris-HCl (pH 8.2), 50 mM KCl, 20 mM MgCl2, 2 mM EDTA, 10 mM Na3ATP, 2.5 mM DTT, 5 mM L-γ-L-glutamyl-L-cysteine (γ-GC) and varying concentrations of L-Gly. Reactions were initiated by adding 0.75 μg of purified rGshB or SUMO as a negative control in a final volume of 80 μL, then incubated at 37°C for 30 min. The concentration of Pi generated by these reactions (Figure S1) was measured as described above.
PNA transfection
PNA transfection to knock down CtrA, GST, GshA or GshB in E. chaffeensis was performed as described previously (Yan et al., 2021). Antisense PNA oligomers targeting 31-46 bp following the start codon of ctrA (CtrA PNA), 10-23 bp following the start codon of gst (GST PNA), 13-28 bp following the start codon of gshA (GshA PNA) and 19-34 bp following the start codon of gshB (GshB PNA) and a control PNA with a scrambled sequence (CTL PNA) were synthesized by KareBay Biochem, Inc. (Ningbo, China; Supplementary Table S1). The knockdown efficiency of CtrA PNA has been verified previously (Yan et al., 2022). To verify the specific binding of PNAs to their targets, ssRNAs derived from gst (GST ssRNA, 3-33 bp following the start codon of gst mRNA), gshA (GshA ssRNA, 6-37 bp following the start codon of gshA mRNA) and gshB (GshB ssRNA, 12-43 bp following the start codon of gshB mRNA) were synthesized by Genewiz (Suzhou, China; Supplementary Table S1). Hybridization of ssRNA with PNA oligomer was performed as described with minor modifications (Yan et al., 2018). Briefly, 80 pmol of ssRNA and 80 pmol of gene-specific PNA or CTL PNA were added into hybridization buffer containing 250 mM Tris-HCl (pH 7.2) in a final volume of 15 μL. Hybridization was performed on a BIO-GENER GE4852T thermocycler (Hangzhou, China) with the following program: 95°C for 3 min, then gradual cooling to 20°C over 75 cycles (20 s per cycle), 20°C for 1 min. Hybridization mixtures were then subjected to 12% native polyacrylamide gel electrophoresis and bands were visualized by GelGreen nucleic acid staining (Biomed, Beijing, China).
Three micrograms of CtrA PNA, GST PNA, GshA PNA, GshB PNA or CTL PNA dissolved in nuclease-free water were mixed with 100 μL of host cell-free E. chaffeensis in 0.3 M sucrose, then incubated on ice for 15 min. Electroporation was conducted at 2,000 V, 25 μF, and 400 Ω with a 10-ms pulse using a Gene Pulser Xcell electroporation system (Bio-Rad, Hercules, CA, United States) in a 2-mm electroporation cuvette (Bio-Rad). Then the PNA-transfected E. chaffeensis was transferred to a T25 flask to infect 5 × 105 THP-1 cells and incubated at 37°C for 2 h with gentle shaking every 15 min to facilitate bacterial internalization. To detect the effect of GST PNA, GshA PNA and GshB PNA transfection, infected cells were harvested at 36 hours post infection (h p.i.). The expression levels of gst, gshA and gshB were determined using qRT-PCR.
H2O2 assay
E. chaffeensis was treated with H2O2 as described previously (Yen et al., 2020). THP-1 cells were synchronously infected with CtrA PNA-, GST PNA-, GshA PNA-, GshB PNA- or CTL PNA-transfected E. chaffeensis. At 36 h p.i., 20 μL of 10 mM H2O2 diluted in culture medium was added into 2 mL of the synchronous culture, reaching a final concentration of 100 μM. After 2 h of treatment at 37°C, the bacterial number in each sample was determined with qRT-PCR.
Statistical analysis
CtrA ChIP-seq assay was performed for a single sample and ChIP-seq data were analyzed by IGENEBOOK Biotechnology (Wuhan, China). All other experiments were performed at least three times. Statistical analyses were performed using GraphPad Prism 7.0. Statistical significance of a two-group comparison was assessed using Student’s t test (two-tailed). A value of P < 0.05 was considered significant.
Results
The promoter regions of 211 genes including gst are enriched in CtrA ChIP-seq assay using antiserum against E. chaffeensis CtrA
To identify the genes regulated by CtrA in E. chaffeensis, we first performed ChIP-seq assay. Using rabbit antiserum against E. chaffeensis CtrA, we identified that the promoter regions of 211 genes were enriched by more than 1.5-fold (P-value < 0.01) compared to input DNA (Supplementary Table S2). Among these genes, 69 were annotated and 64 were successfully classified into Cluster of Orthologous Group (COG) functional categories (E-value < 0.01). COG analysis revealed that these genes were involved in coenzyme transport and metabolism, energy production and conversion, replication, recombination, and repair, and various other functions (Figure 1). Consistent with the previous studies (Cheng et al., 2011; Yan et al., 2022), the promoter regions of ctrA, bolA, surE and gshB were enriched by 1.64-, 1.72-, 1.61- and 1.71-fold, respectively. The promoter regions of gshA and ompA were also enriched by 1.45- and 1.30-fold respectively (Supplementary table S2).
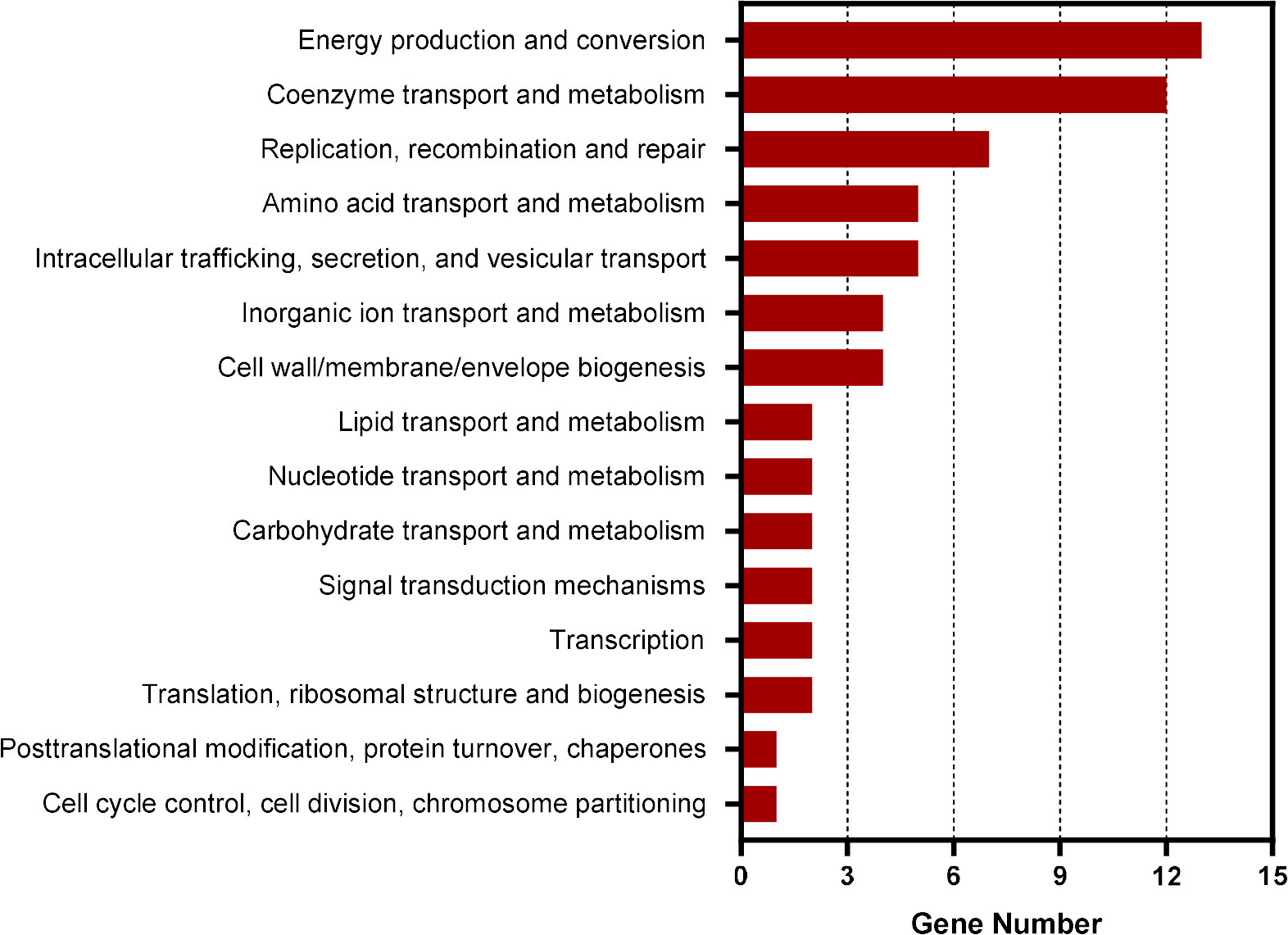
Figure 1 COG analysis of the promoter regions of genes enriched in CtrA ChIP-seq assay. COG analysis was accomplished by COGClassifier (https://github.com/moshi4/COGclassifier). The length of bar indicates the number of genes.
Among these enriched promoter regions, notably, the gst promoter region was enriched 1.81-fold compared to input DNA (Supplementary Table S2). GST is an enzyme that is able to catalyze the reaction of GSH with ROS, then eliminate ROS (Skopelitou et al., 2012a). The amino acid sequence of E. chaffeensis GST showed 34.35% identity with that of Agrobacterium tumefaciens GST (GenBank ID: AAK86108.1) (Figure S2A) (Skopelitou et al., 2012b). We found a 9-mer CtrA binding motif (TTAA-N7-TTAAC) in the gst promoter region (-181 to -166 calculated from the translational start codon) using pDRAW32 DNA analysis software (http://acaclone.com/) (Figure S2B). These results suggest that CtrA regulates the gst expression in E. chaffeensis.
CtrA activates the gst expression in E. chaffeensis upon oxidative stress
We then investigated whether the gst expression was regulated by CtrA. CtrA is expressed at the late stage of E. chaffeensis intracellular growth (Cheng et al., 2011; Yan et al., 2022). Using qRT-PCR, we examined the gst expression pattern in synchronously cultured E. chaffeensis in THP-1 cells. We found that the gst expression was also upregulated at the late stage of E. chaffeensis intracellular growth (Figure 2A), indicating that the gst expression may be regulated by CtrA.
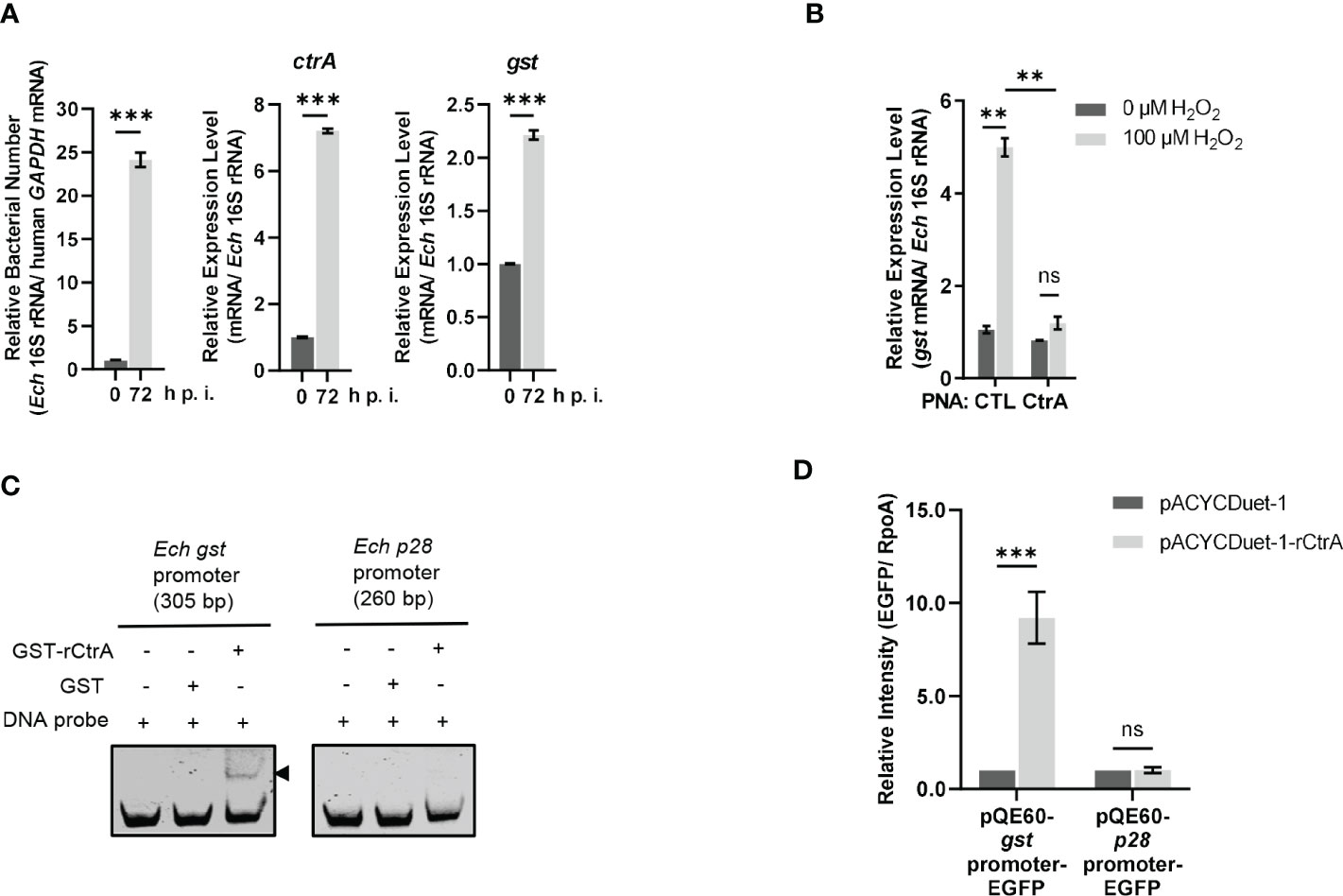
Figure 2 CtrA binds to the gst promoter region and activates the gst expression upon oxidative stress. (A) The expression levels of ctrA and gst and relative bacterial numbers at different stages of E. chaffeensis intracellular growth. RNA samples were prepared from synchronously cultured E. chaffeensis in THP-1 cells at 0 h and 72 h p.i. The expression level of each gene was determined with qRT-PCR and normalized against that of E. chaffeensis 16S rRNA. Relative bacterial number was determined as the amount of bacterial 16S rRNA normalized against that of human GAPDH mRNA. Relative values to the amount at 0 h p.i. are shown. Data indicate means ± standard deviations (n = 3). The significant differences are represented by P-values determined with Student’s t test (*** indicates P < 0.001). (B) CtrA activates the gst expression in E. chaffeensis upon oxidative stress. At 36 h p.i. THP-1 cells synchronously infected with CTL PNA- or CtrA PNA-transfected E. chaffeensis were treated with H2O2 at a final concentration of 100 μM or culture medium (CTL) at 37°C for 2 h. The gst expression level was determined with qRT-PCR and normalized against that of E. chaffeensis 16S rRNA. Relative values to the gst amount of CTL PNA-transfected E. chaffeensis without H2O2 treatment are shown. Data indicate means ± standard deviations (n = 3). The significant differences are represented by P-values determined with Student’s t test (ns indicates P > 0.05, and ** indicates P < 0.01). (C) CtrA binds to the gst promoter. DNA probe (50 ng) was incubated alone, with GST (2.5 µM) or with GST-rCtrA (2.5 µM) on ice for 30 min. Shifted band is indicated by an arrowhead. The p28 promoter region is shown as a negative control. The length (bp) of the probe is shown above each panel. (D) CtrA activates the gst expression. E. coli BL21 (DE3) cells grown to an OD600 of 0.4 were induced to express rCtrA with 0.05 mM IPTG at 37 °C for 3 h. The amount of EGFP or RpoA was determined using Western blotting. The relative intensities of EGFP to those of RpoA were measured by Image J and calculated by setting the value of bacteria containing pACYCDuet-1 and corresponding pQE60-promoter-EGFP fusion construct as 1. Data indicate means ± standard deviations (n = 3). The significant differences are represented by P-values determined with Student’s t test (ns indicates P > 0.05, and *** indicates P < 0.001).
GshA and GshB are glutamate-cysteine ligase and glutathione synthetase respectively that catalyze GSH synthesis from L-Glu, L-Gly, L-Cys and ATP (Figure S1). Previously we found that CtrA activates the expression of gshA and gshB in E. chaffeensis upon oxidative stress (Yan et al., 2022). We then investigated whether the gst expression was activated by CtrA upon oxidative stress. PNA is a DNA mimic that has been shown to bind single- and double-stranded DNA and RNA with high affinity and specificity (Pelc et al., 2015), and can be used to knock down the levels of E. chaffeensis proteins (Sharma et al., 2017; Yan et al., 2018; Yan et al., 2021; Yan et al., 2022). Previously we used PNA to specifically knock down CtrA level in E. chaffeensis (Yan et al., 2022). Host cell-free E. chaffeensis was transfected with CtrA PNA or CTL PNA, then used to infect THP-1 cells. At 36 h p.i. the infected THP-1 cells were treated with 100 μM H2O2 at 37°C for 2 h. We found that the gst expression in E. chaffeensis transfected with CTL PNA was significantly upregulated after H2O2 treatment, suggesting that GST may be required for conferring oxidative stress resistance to E. chaffeensis (Figure 2B). After CtrA level was knocked down by CtrA PNA, the gst expression showed no changes after H2O2 treatment (Figure 2B), indicating that the gst expression upon oxidative stress is activated by CtrA.
We then performed EMSA using GST-rCtrA to investigate whether CtrA directly binds to the gst promoter. Purified GST-rCtrA showed a single band on the SDS-PAGE gel (Figure S3). The DNA probe derived from the gst promoter region shifted upon incubation with GST-rCtrA (Figure 2C). The binding specificity was confirmed by the result that no shifted band was detected when the DNA probe was incubated with purified GST protein or when a DNA probe derived from the p28 promoter region, which is not bound by CtrA, was incubated with GST-rCtrA (Figure 2C). These results indicate that CtrA directly binds to the gst promoter in E. chaffeensis.
Due to the lack of effective genetic manipulation methods in E. chaffeensis, in order to investigate whether CtrA activates the gst expression, we used an E. coli reporter assay system (Cheng et al., 2011; Duan et al., 2021; Yan et al., 2022). The gst promoter region was inserted upstream of the promoter-less egfp gene in the pQE60 plasmid to generate gst promoter-EGFP fusion construct. The p28 promoter region was used to generate p28 promoter-EGFP fusion construct as a negative control. E. coli BL21 (DE3) cells containing pACYCDuet-1 vector harboring E. chaffeensis ctrA gene (pACYCDuet-1-rCtrA) or pACYCDuet-1 vector only (negative control) were transformed with the EGFP fusion constructs, respectively. The rCtrA expression induced by IPTG resulted in a significantly higher expression of EGFP in bacteria harboring gst promoter-EGFP construct compared to the vector control, while it had no effect on bacteria harboring p28 promoter-EGFP construct (Figure 2D; Figure S4). These results indicate that CtrA activates the gst expression in E. chaffeensis.
E. chaffeensis GST, GshA and GshB are functional in vitro
We next determined the GST function in vitro. The purified rGST and the negative control SUMO, which was purified using the same method, showed single bands on the SDS-PAGE gel (Figure S5). The enzymatic activity of rGST was measured by GSH-CDNB conjugate formation (Figure 3A). The Km value of rGST was determined for GSH. The Km and Vmax values of the enzymes are listed in Table 1. The results showed that E. chaffeensis rGST was functional and its Km value for GSH was found to be in a similar range with that of A. tumefaciens GST reported previously (Skopelitou et al., 2012a), indicating that GST is a functional enzyme in E. chaffeensis that catalyzes the conjugation of GSH to its typical substrate CDNB.
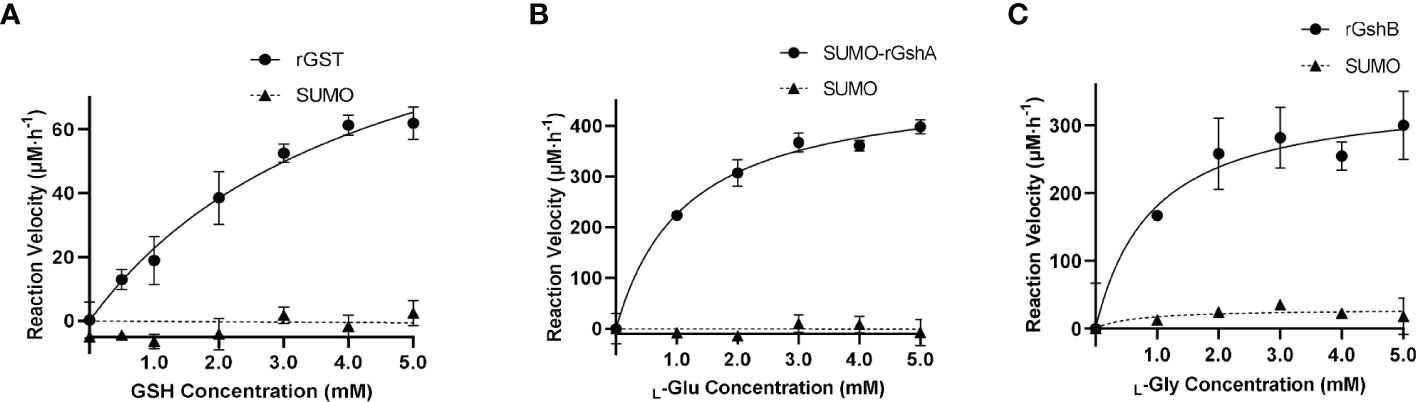
Figure 3 E. chaffeensis GST, GshA and GshB are functional in vitro. (A) Michaelis-Menten plot of rGST reaction velocity versus GSH concentration. (B) Michaelis-Menten plot of SUMO-rGshA reaction velocity versus L-Glu concentration. (C) Michaelis-Menten plot of rGshB reaction velocity versus L-Gly concentration.
In our previous study, we verified the functions of E. chaffeensis GshA and GshB by complementing Pseudomonas aeruginosa respective mutants (Yan et al., 2022). Here, we also determined the functions of E. chaffeensis GshA and GshB in vitro. Since rGshA was unable to be expressed efficiently in E. coli BL21 (DE3) cells, we constructed a plasmid to express SUMO-fused recombinant GshA (SUMO-rGshA). The purified SUMO-rGshA and rGshB showed single bands on the SDS-PAGE gel (Figure S5). The enzymatic activities of SUMO-rGshA and rGshB were measured based on Pi formation (Figures 3B, C). The Km value of SUMO-rGshA or rGshB was determined for L-Glu or L-Gly respectively. The Km and Vmax of the enzymes are listed in Table 1. The results showed that SUMO-rGshA was functional and the Km value for L-Glu was found to be in a similar range with that of A. tumefaciens GshA (Gromes et al., 2008). Also, rGshB was functional and the Km value for L-Gly was found to be in a similar range with that of E. coli GshB (Tanaka et al., 1997). These results indicate that GshA and GshB are functional enzymes in E. chaffeensis, which catalyze the synthesis of GSH.
PNA targeting gst, gshA or gshB reduces the survival ability of E. chaffeensis upon oxidative stress
Since E. chaffeensis GST was functional in vitro, we then investigated its function in vivo. We designed GST PNA that specifically binds to the region following the translation start codon of gst and confirmed the specificity by showing that GST PNA bound to the GST ssRNA (synthesized gst mRNA fragment, 3-33 bp following the start codon) (Figure 4). Transfection of host cell-free E. chaffeensis with GST PNA significantly reduced the gst mRNA level at 36 h p.i. (Figure 4). We then investigated whether the reduction of the gst expression influenced the bacterial survival upon oxidative stress. At 36 h p.i. the infected THP-1 cells were treated with 100 μM H2O2 at 37 °C for 2 h. The survival ability of E. chaffeensis transfected with GST PNA was significantly lower than that of E. chaffeensis transfected CTL PNA with H2O2 treatment (Figure 5A), indicating that GST is critical for E. chaffeensis to cope with oxidative stress.
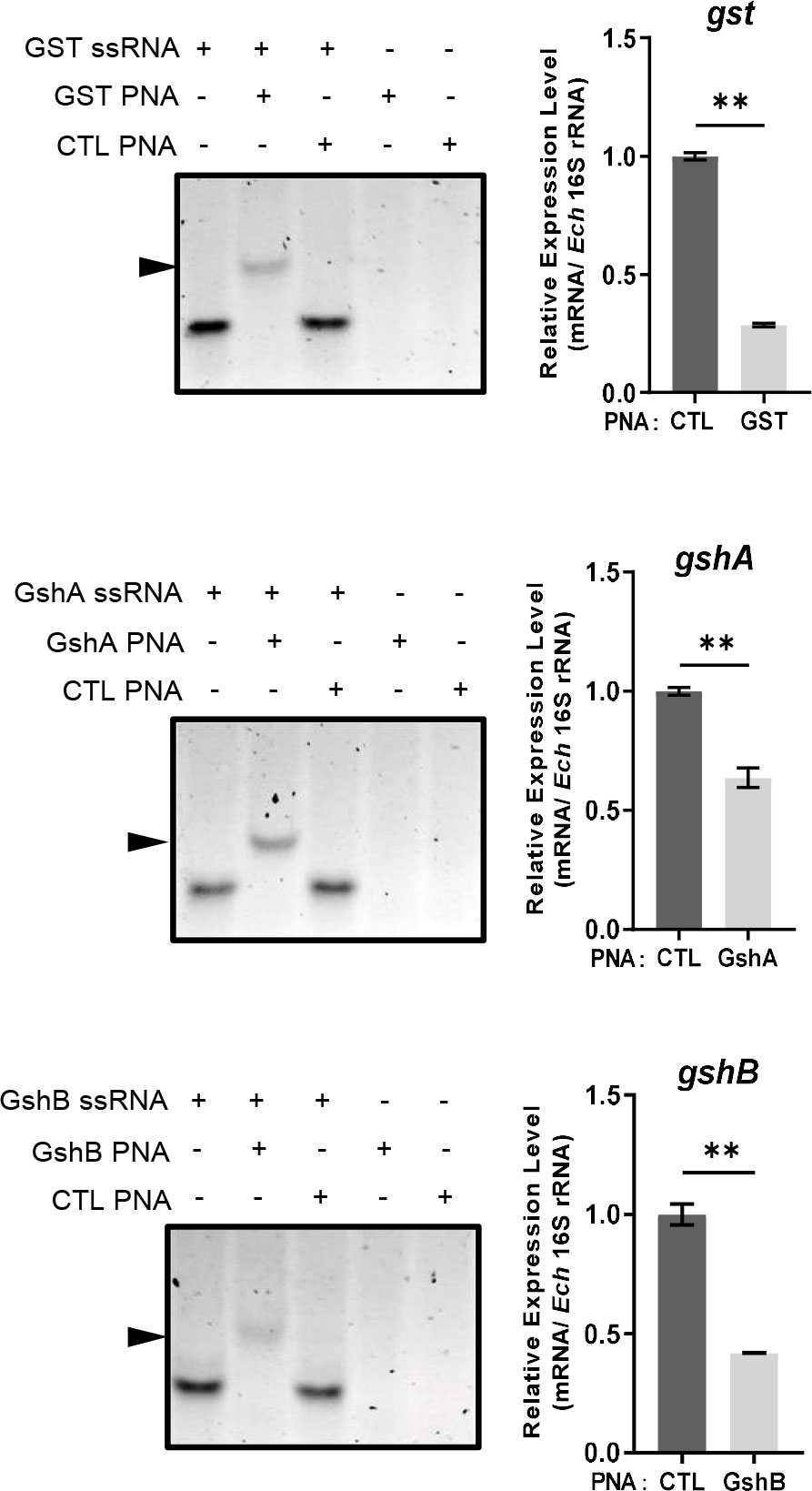
Figure 4 PNA transfection inhibits the expression of gst, gshA, and gshB respectively. GST PNA, GshA PNA or GshB PNA binds to its corresponding ssRNA (left). ssRNA probe (80 pmol) alone, with its corresponding PNA (80 pmol) or with CTL PNA (80 pmol) was annealed from 95 °C to 20 °C. The same amount of corresponding PNA (80 pmol) or CTL PNA (80 pmol) alone was treated at the same condition. Shifted bands are indicated by arrows. The expression levels of gst, gshA and gshB were determined with qRT-PCR and normalized against that of E. chaffeensis 16S rRNA (right). Relative values to the amount of CTL PNA-transfected E. chaffeensis are shown. Data indicate means ± standard deviations (n = 3). The significant differences are represented by P-values determined with Student’s t test (** indicates P < 0.01).
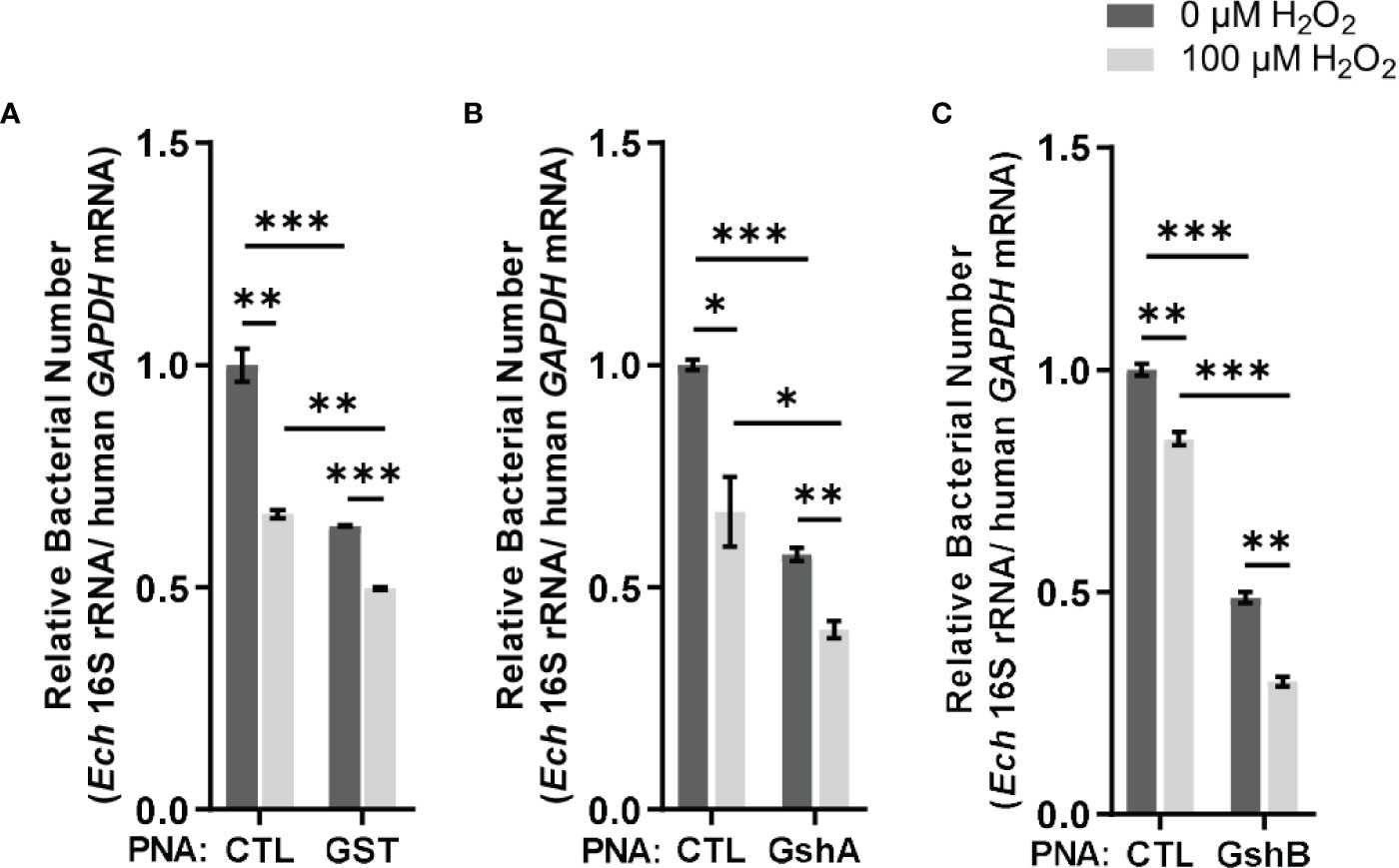
Figure 5 Transfection of GST PNA (A), GshA PNA (B) or GshB PNA (C) reduces the survival ability of E. chaffeensis upon oxidative stress. At 36 h p.i. THP-1 cells synchronously infected with GST PNA-, GshA PNA-, GshB PNA- or CTL PNA-transfected E. chaffeensis were treated with H2O2 at a final concentration of 100 μM or culture medium (CTL) at 37 °C for 2 h. Relative bacterial numbers was determined as the amount of bacterial 16S rRNA normalized against that of human GAPDH mRNA. Relative values to the amount of CTL PNA-transfected E. chaffeensis without H2O2 treatment are shown. Data indicate means ± standard deviations (n = 3). The significant differences are represented by P-values determined with Student’s t test (* indicates P < 0.05, ** indicates P < 0.01, and *** indicates P < 0.001).
We also determined the survival ability of E. chaffeensis transfected with GshA PNA or GshB PNA upon oxidative stress. We designed GshA PNA and GshB PNA and confirmed the specificity by showing that both GshA PNA and GshB PNA bound to their corresponding ssRNA (synthesized gshA or gshB mRNA fragment, 6-37 bp or 12-43 bp following the start codon, respectively) (Figure 4). Transfection of host cell-free E. chaffeensis with GshA PNA or GshB PNA significantly reduced the mRNA level of gshA or gshB at 36 h p.i. (Figure 4). At 36 h p.i. the infected THP-1 cells were treated with H2O2 as described above. The survival ability of E. chaffeensis transfected with GshA PNA or GshB PNA was also significantly lower than that of E. chaffeensis transfected CTL PNA (Figures 5B, C). All these results together indicate that GSH synthesis and utilization are critical for E. chaffeensis to cope with oxidative stress.
Discussion
Due to the reduction of bacterial genome during evolution, E. chaffeensis only encodes a few transcriptional regulators (Cheng et al., 2015), which leads to merging of genes, especially genes critical for bacterial infection and survival, under the control of these regulators. We found the number of genes regulated by CtrA in E. chaffeensis was higher than that in C. crescentus. Previously it has been reported that CtrA regulates the expression of bolA, ompA, surE, gshA and gshB in E. chaffeensis (Cheng et al., 2011; Yan et al., 2022). Here we found that CtrA regulated the gst expression in E. chaffeensis. All these genes are critical for E. chaffeensis infection and survival (Cheng et al., 2011; Yan et al., 2022), but are not present in CtrA regulons in other bacteria. Thus, identification of CtrA regulon in E. chaffeensis can provide new information on the mechanisms exploited by E. chaffeensis to find its unique niche during infection. Using ChIP-seq, we identified 211 genes bound by CtrA. Among these genes, the annotated genes belong to 15 categories analyzed by COG and may contribute to E. chaffeensis virulence. For example, HupB is a nucleoid-associated protein that promotes the survival of Mycobacterium tuberculosis in iron-limited host cell environment by inducing the biosynthesis of mycobactin, an Fe(III)-specific high affinity siderophore (Pandey et al., 2014; Kriel et al., 2018). TatC is a component of the twin-arginine translocation (Tat) system, which is predicted to export virulence factors from cytoplasm into periplasm in Anaplasma phagocytophilum (Dugat et al., 2014) and is essential for the viability of M. tuberculosis (De Buck et al., 2008). SecF and SecG are components of the Sec translocation system, which may participate in the type IV secretion system assembly in A. phagocytophilum (Dugat et al., 2014). PstC is a component of the phosphate-specific transport (Pst) system involved in phosphate uptake to support bacterial growth in E. coli (Hsieh and Wanner, 2010). The roles of these genes during E. chaffeensis infection remain to be investigated.
GSTs are enzymes that conjugate GSH to various substrates for ROS elimination and detoxification (Skopelitou et al., 2012a; Skopelitou et al., 2012b). E. chaffeensis lacks GPx, and encodes only one GST, unlike that multiple GSTs present in other bacteria. Here, we showed that E. chaffeensis GST played a critical role in GSH-dependent ROS elimination. After interaction with ROS, GSH is converted to oxidized GSH (GSSG). While GSSG is unable to be recycled back to GSH due to the lack of glutathione reductase (GR) in E. chaffeensis (Rikihisa, 2015), other mechanisms may be involved in GSSG processing. In Novosphingobium aromaticivorans, an Atm1/ABCB7/HMT1/ABCB6 family ATP-binding cassette (ABC) exporter, NaAtm1, is able to export various GSH derivatives, including GSSG, out of bacteria (Lee et al., 2014). In E. chaffeensis, ECH_0313, a putative ABC transporter, shows 38% sequence identity with NaAtm1. It is likely that E. chaffeensis exports GSSG via an ABC exporter.
Previously we found that CtrA activates the expression of gshA and gshB, which encode enzymes for GSH synthesis, upon oxidative stress (Yan et al., 2022). Here we found that CtrA activated the gst expression upon oxidative stress. Interestingly, the ChIP-seq results showed that the promoter region of ECH_0313 was bound by CtrA. Together, these results indicate that in E. chaffeensis CtrA regulate the GSH synthesis and utilization pathway to eliminate ROS and maintain redox homeostasis. However, the mechanism by which oxidative stress signal is transmitted to CtrA is still unknown. CtrA activation is regulated by the kinase/phosphatase activity of CckA, the cognate histidine kinase of CtrA. In C. crescentus, the kinase/phosphatase activity of CckA is regulated by the interaction with DivK and DivL or the binding of its PAS domain to cyclic-di-GMP (Iniesta et al., 2010; Tsokos et al., 2011; Mann et al., 2016). In E. chaffeensis, DivK and DivL are missing and CckA lacks PAS domains (Cheng et al., 2006; Cheng et al., 2011). Thus, the kinase/phosphatase activity of CckA may be controlled by other mechanisms. CckA may detect the redox condition or the concentration of GSSG directly, or indirectly through other pathways such as two other two-component regulatory systems, NtrY/NtrX and PleC/PleD (Cheng et al., 2006; Kumagai et al., 2006).
The lack of effective genetic manipulation methods greatly hampered the function research of E. chaffeensis genes (Cheng et al., 2013). In the present study, we characterized E. chaffeensis GST function using purified recombinant protein in vitro and PNA to temporarily knock down the gst expression in vivo. Using the same methods, we also characterized E. chaffeensis GshA and GshB, which suggests that a combination of these two methods can be used to study the function and pathogenesis of E. chaffeensis proteins.
Data availability statement
CtrA ChIP-seq raw data have been deposited in the NCBI Short Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra) under the accession number PRJNA898263. The original data presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
QL, JY and ZC conceived, designed the experiments, and wrote the manuscript. QL, JY, SZ, NY and ML performed the experiments. YJ, FB and WW analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32170199, 31970179, 32170177, 31870130, and 82061148018) and National Key Research and Development Program of China (2021YFE0201300). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgments
We thank Dr. Changhao Bi at the Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences (Tianjin, China), for kindly providing plasmid (pQE60-EGFP).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1081614/full#supplementary-material
Supplementary Table S1 | Bacterial strains, plasmids, primers, PNAs and ssRNAs used in this study.
Supplementary Table S2 | E. chaffeensis CtrA ChIP-seq results.
References
Cheng, Z., Kumagai, Y., Lin, M., Zhang, C., Rikihisa, Y. (2006). Intra-leukocyte expression of two-component systems in Ehrlichia chaffeensis and Anaplasma phagocytophilum and effects of the histidine kinase inhibitor closantel. Cell Microbiol. 8 (8), 1241–1252. doi: 10.1111/j.1462-5822.2006.00704.x
Cheng, Z., Miura, K., Popov, V. L., Kumagai, Y., Rikihisa, Y. (2011). Insights into the CtrA regulon in development of stress resistance in obligatory intracellular pathogen Ehrlichia chaffeensis. Mol. Microbiol. 82 (5), 1217–1234. doi: 10.1111/j.1365-2958.2011.07885.x
Cheng, C., Nair, A. D., Indukuri, V. V., Gong, S., Felsheim, R. F., Jaworski, D., et al. (2013). Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PloS Pathog. 9 (2), e1003171. doi: 10.1371/journal.ppat.1003171
Cheng, C., Nair, A. D., Jaworski, D. C., Ganta, R. R. (2015). Mutations in Ehrlichia chaffeensis causing polar effects in gene expression and differential host specificities. PloS One 10 (7), e0132657. doi: 10.1371/journal.pone.0132657
De Buck, E., Lammertyn, E., Anne, J. (2008). The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol. 16 (9), 442–453. doi: 10.1016/j.tim.2008.06.004
Domian, I. J., Quon, K. C., Shapiro, L. (1997). Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90 (3), 415–424. doi: 10.1016/s0092-8674(00)80502-4
Duan, N., Ma, X., Cui, H., Wang, Z., Chai, Z., Yan, J., et al. (2021). Insights into the mechanism regulating the differential expression of the P28-OMP outer membrane proteins in obligatory intracellular pathogen Ehrlichia chaffeensis. Emerg. Microbes Infect. 10 (1), 461–471. doi: 10.1080/22221751.2021.1899054
Dugat, T., Loux, V., Marthey, S., Moroldo, M., Lagree, A. C., Boulouis, H. J., et al. (2014). Comparative genomics of first available bovine Anaplasma phagocytophilum genome obtained with targeted sequence capture. BMC Genomics 15, 973. doi: 10.1186/1471-2164-15-973
Gromes, R., Hothorn, M., Lenherr, E. D., Rybin, V., Scheffzek, K., Rausch, T. (2008). The redox switch of γ-glutamylcysteine ligase via a reversible monomer-dimer transition is a mechanism unique to plants. Plant J. 54 (6), 1063–1075. doi: 10.1111/j.1365-313X.2008.03477.x
Habig, W. H., Jakoby, W. B. (1981). Assays for differentiation of glutathione S-transferases. Methods Enzymol. 77, 398–405. doi: 10.1016/s0076-6879(81)77053-8
Hsieh, Y. J., Wanner, B. L. (2010). Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13 (2), 198–203. doi: 10.1016/j.mib.2010.01.014
Iniesta, A. A., Hillson, N. J., Shapiro, L. (2010). Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc. Natl. Acad. Sci. U.S.A. 107 (15), 7012–7017. doi: 10.1073/pnas.1001767107
Kriel, N. L., Gallant, J., van Wyk, N., van Helden, P., Sampson, S. L., Warren, R. M., et al. (2018). Mycobacterial nucleoid associated proteins: An added dimension in gene regulation. Tuberculosis (Edinb) 108, 169–177. doi: 10.1016/j.tube.2017.12.004
Kumagai, Y., Cheng, Z., Lin, M., Rikihisa, Y. (2006). Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect. Immun. 74 (9), 5014–5022. doi: 10.1128/IAI.00735-06
Kumagai, Y., Matsuo, J., Hayakawa, Y., Rikihisa, Y. (2010). Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J. Bacteriol 192 (16), 4122–4133. doi: 10.1128/JB.00132-10
Laub, M. T., Chen, S. L., Shapiro, L., McAdams, H. H. (2002). Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U.S.A. 99 (7), 4632–4637. doi: 10.1073/pnas.062065699
Lee, J. Y., Yang, J. G., Zhitnitsky, D., Lewinson, O., Rees, D. C. (2014). Structural basis for heavy metal detoxification by an Atm1-type ABC exporter. Science 343 (4), 6175–1136. doi: 10.1126/science.1246489
Lina, T. T., Dunphy, P. S., Luo, T., McBride, J. W. (2016). Ehrlichia chaffeensis TRP120 activates canonical notch signaling to downregulate TLR2/4 expression and promote intracellular survival. mBio 7 (4), e00672–16. doi: 10.1128/mBio.00672-16
Lin, M., Rikihisa, Y. (2007). Degradation of p22phox and inhibition of superoxide generation by Ehrlichia chaffeensis in human monocytes. Cell Microbiol. 9 (4), 861–874. doi: 10.1111/j.1462-5822.2006.00835.x
Liu, H., Bao, W., Lin, M., Niu, H., Rikihisa, Y. (2012). Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell Microbiol. 14 (7), 1037–1050. doi: 10.1111/j.1462-5822.2012.01775.x
Mann, T. H., Seth Childers, W., Blair, J. A., Eckart, M. R., Shapiro, L. (2016). A cell cycle kinase with tandem sensory PAS domains integrates cell fate cues. Nat. Commun. 7, 11454. doi: 10.1038/ncomms11454
Mohan Kumar, D., Yamaguchi, M., Miura, K., Lin, M., Los, M., Coy, J. F., et al. (2013). Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PloS Pathog. 9 (10), e1003666. doi: 10.1371/journal.ppat.1003666
Morris, D., Khurasany, M., Nguyen, T., Kim, J., Guilford, F., Mehta, R., et al. (2013). Glutathione and infection. Biochim. Biophys. Acta 1830 (5), 3329–3349. doi: 10.1016/j.bbagen.2012.10.012
Oppenheimer, L., Wellner, V. P., Griffith, O. W., Meister, A. (1979). Glutathione synthetase. Purification from rat kidney and mapping of the substrate binding sites. J. Biol. Chem. 254 (12), 5184–5190. doi: 10.1016/S0021-9258(18)50577-9
Paddock, C. D., Childs, J. E. (2003). Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16 (1), 37–64. doi: 10.1128/CMR.16.1.37-64.2003
Pandey, S. D., Choudhury, M., Sritharan, M. (2014). Transcriptional regulation of mycobacterium tuberculosis hupB gene expression. Microbiol. (Reading) 160 (Pt 8), 1637–1647. doi: 10.1099/mic.0.079640-0
Pelc, R. S., McClure, J. C., Kaur, S. J., Sears, K. T., Rahman, M. S., Ceraul, S. M. (2015). Disrupting protein expression with peptide nucleic acids reduces infection by obligate intracellular Rickettsia. PloS One 10 (3), e0119283. doi: 10.1371/journal.pone.0119283
Quon, K. C., Marczynski, G. T., Shapiro, L. (1996). Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84 (1), 83–93. doi: 10.1016/s0092-8674(00)80995-2
Rikihisa, Y. (2015). Molecular pathogenesis of Ehrlichia chaffeensis infection. Annu. Rev. Microbiol. 69, 283–304. doi: 10.1146/annurev-micro-091014-104411
Satoh, M., Iwahori, T., Sugawara, N., Yamazaki, M. (2006). Liver argininosuccinate synthase binds to bacterial lipopolysaccharides and lipid a and inactivates their biological activities. J. Endotoxin Res. 12 (1), 21–38. doi: 10.1179/096805106X89062
Sharma, P., Teymournejad, O., Rikihisa, Y. (2017). Peptide nucleic acid knockdown and intra-host cell complementation of Ehrlichia type IV secretion system effector. Front. Cell Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00228
Sies, H., Jones, D. P. (2020). Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 21 (7), 363–383. doi: 10.1038/s41580-020-0230-3
Skopelitou, K., Dhavala, P., Papageorgiou, A. C., Labrou, N. E. (2012a). A glutathione transferase from Agrobacterium tumefaciens reveals a novel class of bacterial GST superfamily. PloS One 7 (4), e34263. doi: 10.1371/journal.pone.0034263
Skopelitou, K., Muleta, A. W., Pavli, O., Skaracis, G. N., Flemetakis, E., Papageorgiou, A. C., et al. (2012b). Overlapping protective roles for glutathione transferase gene family members in chemical and oxidative stress response in Agrobacterium tumefaciens. Funct. Integr. Genomics 12 (1), 157–172. doi: 10.1007/s10142-011-0248-x
Smirnova, G. V., Oktyabrsky, O. N. (2005). Glutathione in bacteria. Biochem. (Mosc) 70 (11), 1199–1211. doi: 10.1007/s10541-005-0248-3
Tanaka, T., Nishioka, T., Oda, J. (1997). Nicked multifunctional loop of glutathione synthetase still protects the catalytic intermediate. Arch. Biochem. Biophys. 339 (1), 151–156. doi: 10.1006/abbi.1996.9821
Teymournejad, O., Rikihisa, Y. (2020). Ehrlichia chaffeensis uses an invasin to suppress reactive oxygen species generation by macrophages via CD147-dependent inhibition of Vav1 to block Rac1 activation. mBio 11 (2), e00267–20. doi: 10.1128/mBio.00267-20
Tsokos, C. G., Perchuk, B. S., Laub, M. T. (2011). A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev. Cell 20 (3), 329–341. doi: 10.1016/j.devcel.2011.01.007
Tu, Z., Anders, M. W. (1998). Identification of an important cysteine residue in human glutamate-cysteine ligase catalytic subunit by site-directed mutagenesis. Biochem. J. 336 (Pt 3), 675–680. doi: 10.1042/bj3360675
Yan, J., Liang, Q., Chai, Z., Duan, N., Li, X., Liu, Y., et al. (2022). Glutathione synthesis regulated by CtrA protects Ehrlichia chaffeensis from host cell oxidative stress. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.846488
Yan, Q., Lin, M., Huang, W., Teymournejad, O., Johnson, J. M., Hays, F. A., et al. (2018). Ehrlichia type IV secretion system effector etf-2 binds to active RAB5 and delays endosome maturation. Proc. Natl. Acad. Sci. U.S.A. 115 (38), E8977–E8986. doi: 10.1073/pnas.1806904115
Yan, Q., Zhang, W., Lin, M., Teymournejad, O., Budachetri, K., Lakritz, J., et al. (2021). Iron robbery by intracellular pathogen via bacterial effector-induced ferritinophagy. Proc. Natl. Acad. Sci. U.S.A. 118 (23), e2026598118. doi: 10.1073/pnas.2026598118
Keywords: Ehrlichia chaffeensis, human monocytic ehrlichiosis, oxidative stress, GST, CtrA
Citation: Liang Q, Yan J, Zhang S, Yang N, Li M, Jin Y, Bai F, Wu W and Cheng Z (2022) CtrA activates the expression of glutathione S-transferase conferring oxidative stress resistance to Ehrlichia chaffeensis. Front. Cell. Infect. Microbiol. 12:1081614. doi: 10.3389/fcimb.2022.1081614
Received: 27 October 2022; Accepted: 24 November 2022;
Published: 12 December 2022.
Edited by:
Hua Niu, Affiliated Hospital of Guilin Medical University, ChinaReviewed by:
Mingqun Lin, The Ohio State University, United StatesYong Qi, Huadong Research Institute for Medicine and Biotechniques, China
Copyright © 2022 Liang, Yan, Zhang, Yang, Li, Jin, Bai, Wu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihui Cheng, zhihuicheng@nankai.edu.cn
†These authors have contributed equally to this work
 Qi’an Liang
Qi’an Liang Jiaqi Yan
Jiaqi Yan Shuwen Zhang
Shuwen Zhang  Meifang Li
Meifang Li Yongxin Jin
Yongxin Jin Fang Bai
Fang Bai Weihui Wu
Weihui Wu Zhihui Cheng
Zhihui Cheng