Multilocus Genotyping of Giardia duodenalis in Alpine Musk Deer (Moschus chrysogaster) in China
- 1Key Laboratory of Biomarker Based Rapid-Detection Technology for Food Safety of Henan Province, Food and Pharmacy College, Xuchang University, Xuchang, China
- 2College of Animal Science and Technology, Tarim University, Alar, China
- 3College of Veterinary Medicine, Henan Agricultural University, Zhengzhou, China
Giardia duodenalis is the underlying cause of a significant number of outbreaks of gastrointestinal illness in humans and animals worldwide. The purpose of this study was to elucidate the prevalence and genetic diversity of G. duodenalis in captive alpine musk deer (Moschus chrysogaster) in China. A total of 202 fecal samples were collected from three farms in Gansu Province, China. Identification of G. duodenalis was conducted by nested PCR targeting the genes coding for SSU rRNA, β-giardin (bg), glutamate dehydrogenase (gdh) and triosephosphate isomerase (tpi). The overall prevalence of G. duodenalis in captive alpine musk deer in surveyed area was 19.3% (39/202). Two G. duodenalis genetic assemblages were identified, namely assemblage A and E. Mixed genotype infections (A+E) were found in 15.4% (6/39) of positive samples. Multilocus genotyping (MLG) analysis of G. duodenalis isolates revealed six novel assemblage A MLGs formed by two newly-described MLG-subtypes which belonged to sub-assemblage AI. To the best of our knowledge, this is the first report on MLG of G. duodenalis isolates in captive alpine musk deer in China. The presence of zoonotic assemblages and sub-assemblages of G. duodenalis in deer species suggests that these animals may potentially act as a reservoir of this protozoan for humans.
Introduction
Giardia duodenalis (also known as Giardia lamblia and Giardia intestinalis) is the most prevalent protozoan pathogen, commonly found in the intestinal tract of humans and animals worldwide (Adam, 2021). Transmission of G. duodenalis infection occurs by several routes either directly (i.e., person-to-person, animal-to-animal, or zoonotic infection) or indirectly (i.e., water or food) (Dixon, 2021). Approximately 280 million people are considered to be infected with G. duodenalis worldwide, with infection rates at the range of 8.0–30.0% in developing countries and 0.4–7.5% in developed countries (Feng and Xiao, 2011; Ryan and Zahedi, 2019). Giardiasis is generally a self-limiting clinical illness in humans, whereas it can be threatening to infants, young children, the elderly, institutionalized individuals, travelers, and immunocompromised individuals (Cacciò et al., 2018; Cai et al., 2021). Nitroimidazoles (e.g., metronidazole and tinidazole) are the most commonly drugs used to treat giardiasis, although requiring multiple doses and being often associated with adverse effects (Argüello-García et al., 2020).
To date, according to the reservoir and genetic characteristics of the protozoan, eight Giardia species have been recognized (Ryan et al., 2019). However, most studies for both public and veterinary health have focused on the taxonomy, population genetics, and epidemiology of G. duodenalis (Li et al., 2017a; Xiao and Feng, 2017). Based on studies employing iso-enzymatic and nucleic acid polymorphisms, G. duodenalis is known as a multispecies complex, consisting of eight genetic assemblages (A–H) considering different host distribution (Cacciò et al., 2018); assemblages A and B are commonly found in humans and occasionally in other mammals; assemblages C and D are often found in canids; assemblage E mainly infects ungulates; assemblages F, G, and H are specific to cats, rodents and pinnipeds, respectively (Ryan and Cacciò, 2013; Cai et al., 2021). Furthermore, putative sub-assemblages have been identified within assemblage A (AI–III) and assemblage B (BIII and BIV) using a multilocus genotyping (MLG) approach (Capewell et al., 2021).
China has the largest wild and captive populations of alpine musk deer (Moschus chrysogaster) in the world, which are mainly distributed in the Qinghai Tibet Plateau, Sichuan and Gansu Province (Jiang et al., 2021). Musk, produced by adult male alpine musk deer, is an important raw material for preparations of the traditional Chinese medicine and the fragrance industry. However, illegal hunting, habitat fragmentation, and other human activities have decimated wild alpine musk deer populations in China (Cai et al., 2020). For these reasons, the alpine musk deer has been listed as an endangered species by the International Union for Conservation of Nature (IUCN) and as category I-protected wild animal in China. Although the Chinese government has encouraged enterprises to participate in programs of breeding of captive alpine musk deer, gastrointestinal infections are the most significant threats to population growth and breeding scale whose fatality rate is approximately 30% (Li et al., 2017b). Currently, little information is available on the prevalence and genetic characteristics of G. duodenalis in cervids in China.
Thus, the aim of the present study was to investigate the prevalence and genetic diversity of G. duodenalis in captive alpine musk deer. The findings discussed herein provide insights into the development of preventive measures against Giardia infection.
Material and Methods
Ethics Statement
This study was performed with strict adherence to the recommendations of the Guide for the Care and Use of Laboratory Animals of the Ministry of Health, China. The research protocol was reviewed and approved by the Research Ethics Committee of Tarim University (approval no. ECTU 2020-0013). Farm owners’ consent was obtained prior to proceeding to fecal sample collection from selected animals.
Samples
In September 2020, 202 fecal samples were collected from three farms in Gansu Province, China (Figure 1). Alpine musk deer animals were shed-fed and housed in separate breeding houses according to age. Fresh fecal samples were taken from the soil immediately after defecation using a sterile disposal latex glove, ensure absence of exogenous contamination. Subsequently, samples were placed individually into a disposable plastic container, recorded the date, site, age, and number. All animals from which fecal samples were obtained were apparently in good health with no signs of diarrhea at the time of sample collection. Samples were transferred to the laboratory in an insulated container on ice and stored at 4°C prior to DNA extraction.
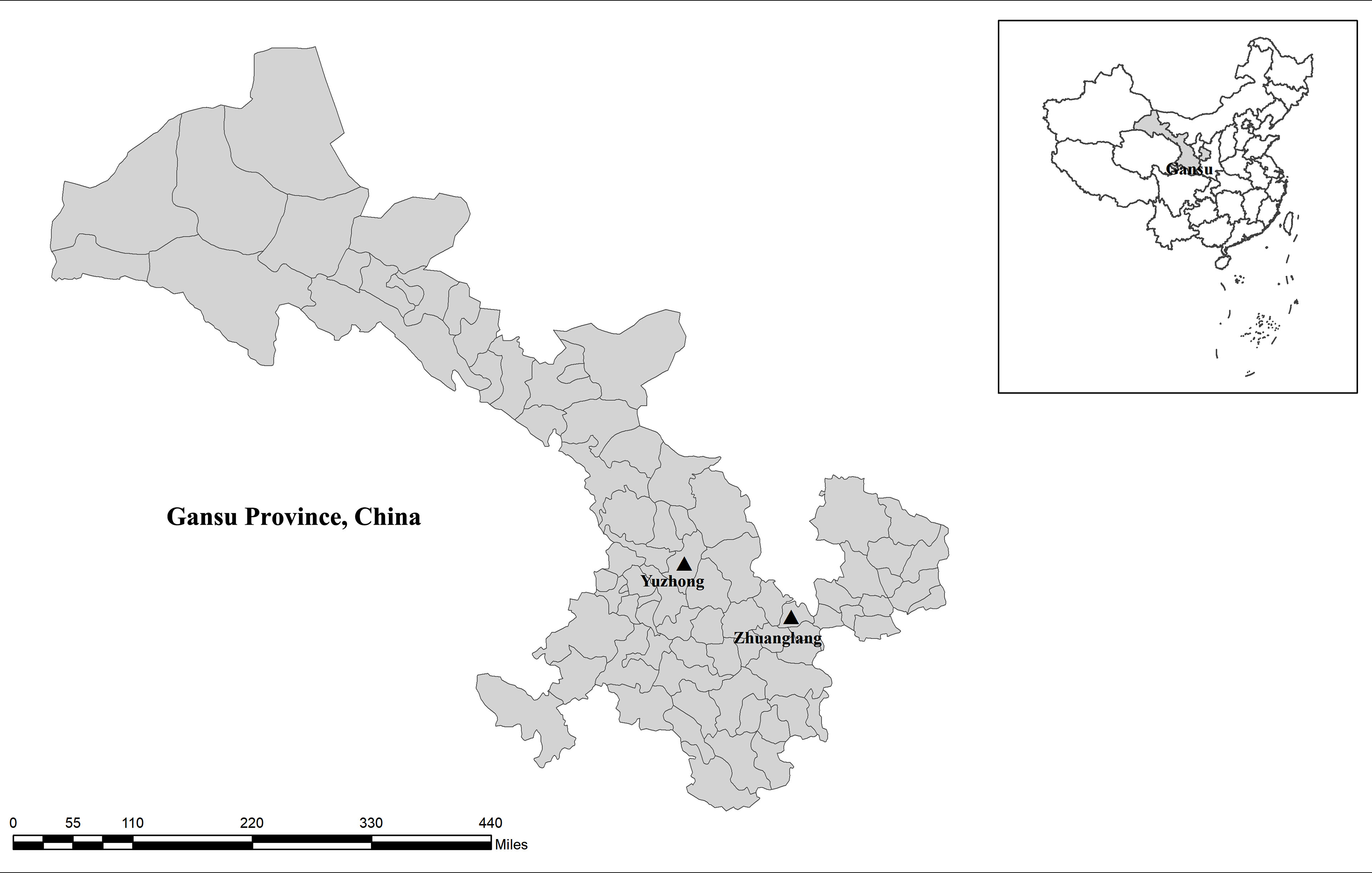
Figure 1 Sampling sites. No copyright permission was required. The figure was designed with the software ArcGIS 10.2. The map has been originally modified and assembled according to permission and attribution guidelines of the National Geomatics Center of China (http://www.ngcc.cn).
DNA Extraction and Genotyping
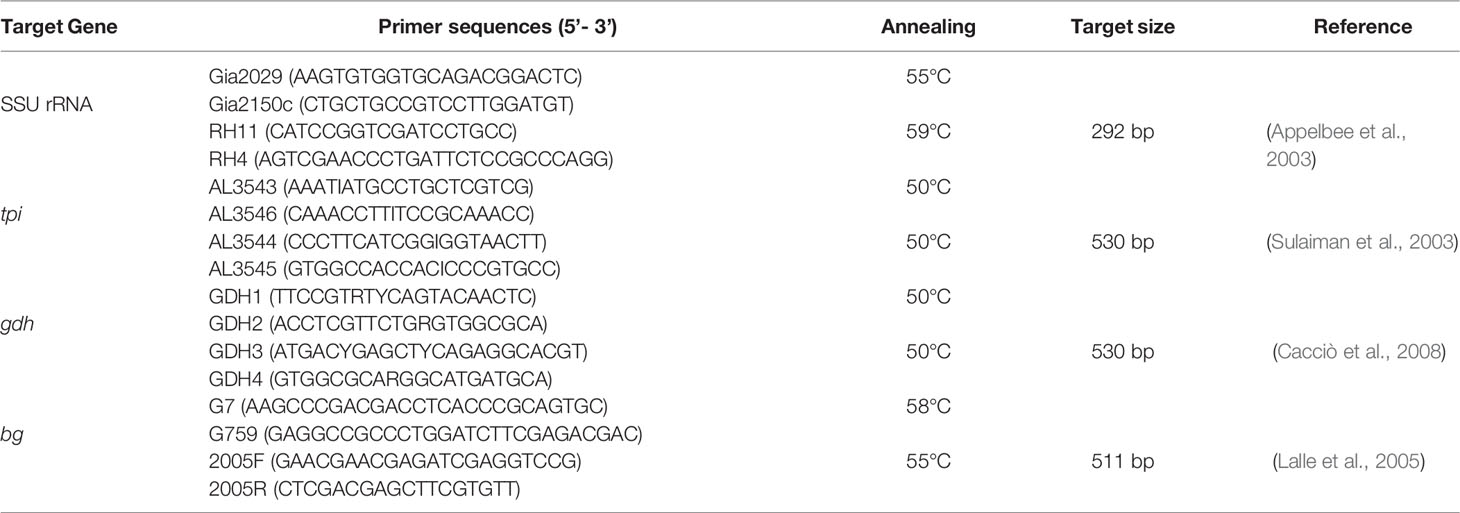
Genomic DNA was extracted from approximately 200 mg of each precipitated sample using the E.Z.N.A.® Stool DNA kit (Omega Bio-tek Inc., Norcross, GA, USA), according to manufacturer’s instructions. The extracted DNA was stored at -20°C until PCR assay. Four genes were used for G. duodenalis genotyping by nested PCR, namely SSU rRNA, β-giardin (bg), glutamate dehydrogenase (gdh), and triosephosphate isomerase (tpi) (Table 1). Positive (DNA from an isolate known to harbor the four surveyed loci) and negative (reagent-grade water) controls were included in each PCR amplification.
Sequence and Phylogenetic Analysis
All positive secondary PCR products from SSU rRNA, bg, gdh, and tpi genes were bi-sequenced by GENEWIZ (Suzhou, China). Nucleotide sequences were aligned and edited with DNAstar Lasergene Editseq 7.1.0 (https://www.dnastar.com/software/lasergene/) and Chromas Pro 2.1.10 (http://technelysium.com.au/wp/chromaspro/). Genotypes and subtypes of G. duodenalis were determined by aligning reference sequences available in NCBI GenBank database using ClustalX 2.1 (http://www.clustal.org/). To determine genetic diversity among the isolates, concatenated sequences (bg-tpi-gdh) from each isolate at the three analyzed loci were aligned with reference sequences. Neighbor-joining (NJ) analysis was performed using MEGA 7.0 (http://www.megasoftware.net/) to infer the phylogenetic relationships of concatenated sequences based on the Kimura-2 parameter model.
Nucleotide Sequence Accession Numbers
Representative nucleotide sequences of bg and tpi genes of G. duodenalis are available in the NCBI GenBank database under the accession numbers OM273018-OM273020, respectively.
Results
Prevalence and Assemblages of G. duodenalis
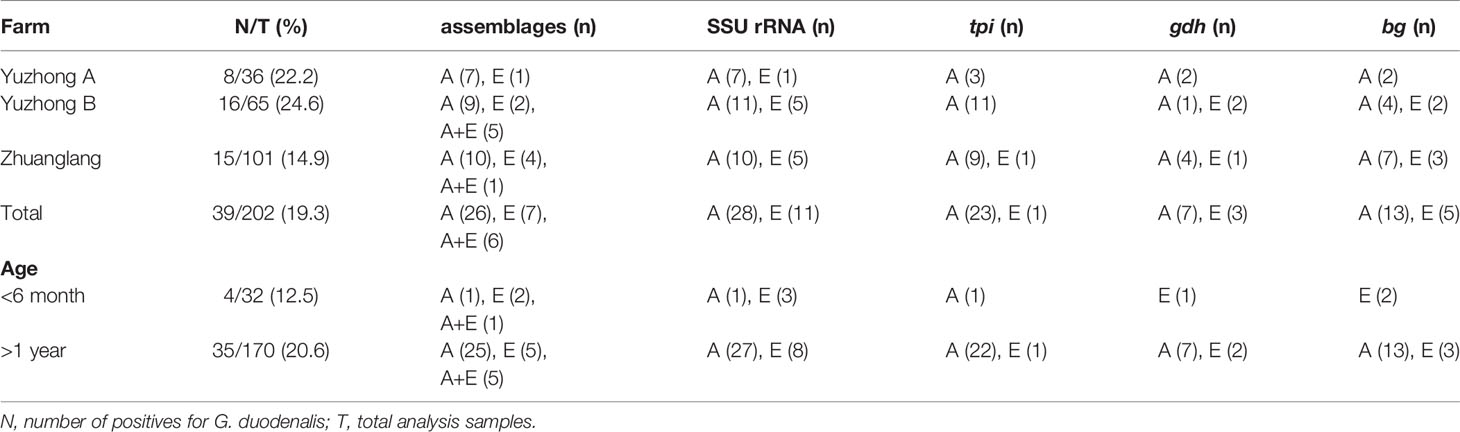
Overall, a total of 39 samples (19.3%, 39/202) were confirmed to be G. duodenalis by PCR at the SSU rRNA locus (Table 2). Prevalence rate by region was as follows: Yuzhong A (22.2%, 8/36), Yuzhong B (24.6%, 16/65), and Zhuanglang (14.9%, 15/101). In addition, the infection was numerically more frequent in adults (>1 year, 20.6%, 35/170) compared to young animals (<6 month, 12.5%, 4/32). Subsequently, all G. duodenalis-positive samples were genotyped by MLG of SSU rRNA, bg, tpi and gdh genes. Two G. duodenalis genetic assemblages were identified among samples: assemblage A (72.2%, 26/39) and assemblage E (17.9%, 7/39). Mixed genotype infections (A+E) were found in 6 of 39 samples.
Polymorphisms at Single Loci
Amplification of the bg gene was obtained from 18 of 39 G. duodenalis-positive isolates; among these, 13/18 (72.2%) of isolates were identified as belonging to genetic assemblage A and 5/18 (27.8%) of assemblage E (Table 3). Within assemblage A isolates, three subtypes were formed and designated as A1 (n=4), A2 (n=1), and A3 (n=8). Compared to the sequence MK610391, A1 sequences exhibited one single-nucleotide polymorphism (SNP) (C327T), whereas A2 sequence contained three SNPs (T302C, G308A, and C327T). A3 sequences were identical to MK610392. Moreover, the five assemblage E sequences were identical to the sequence MK610387.
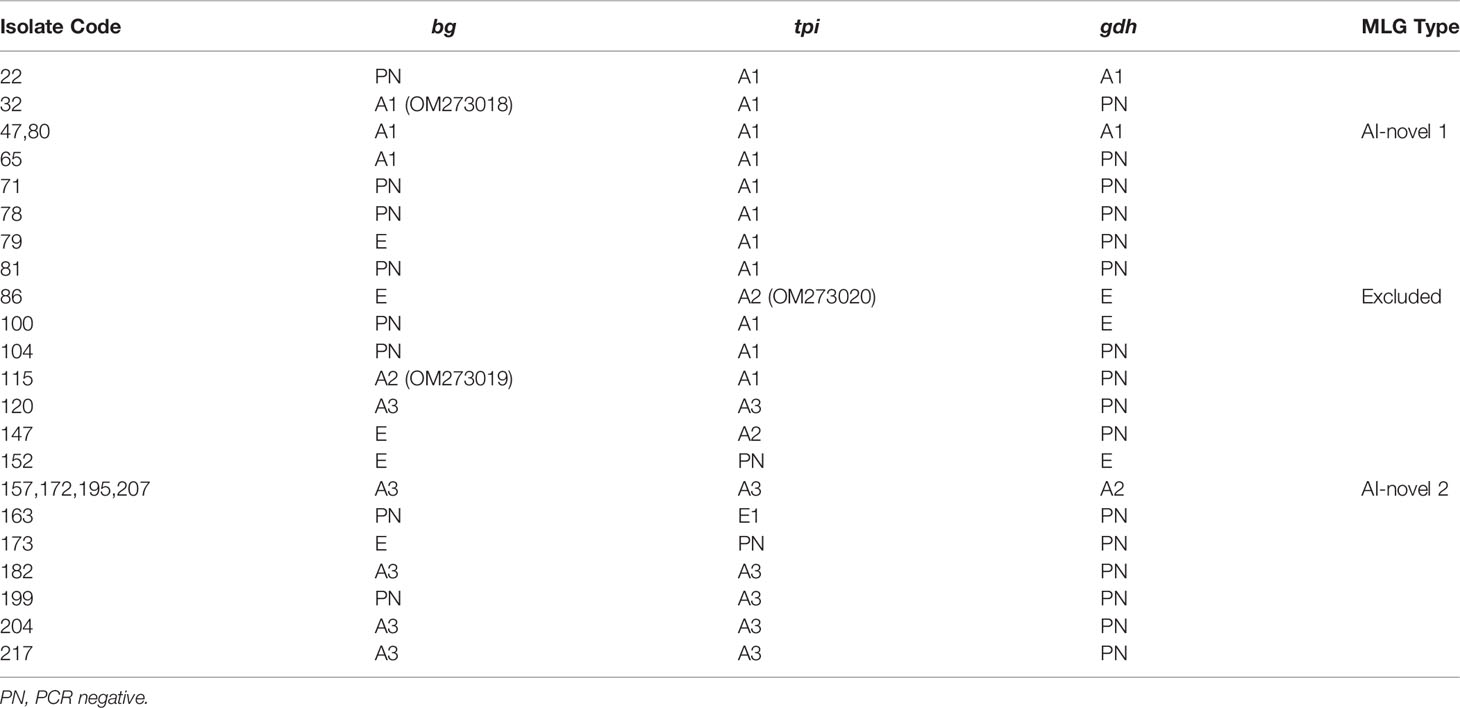
Table 3 Multi-locus characterization of G. duodenalis isolates in alpine musk deer in China based on bg, gdh and tpi genes.
Sequence analysis of the tpi locus revealed that 23 out of 24 successfully amplified isolates were identified as assemblage A, whereas only one was classified as assemblage E. The single assemblage E sequence was 100% identical to the sequence KT922262. Among assemblage A sequences, A1 (n=12) and A3 (n=9) sequences were identical to the sequences MK639171 and MK639172, respectively. In addition, A2 (n=2) sequences showed a SNP (C21T) compared to the sequence MK639173. At the gdh locus, seven and three isolates were successfully amplified and identified as assemblage A and E, respectively. The subtypes A1 (n=3) and A2 (n=4) were consistent with the sequences MN047217 and the MK645799, respectively. Moreover, the three assemblage E sequences were identical to the sequence MK645786.
MLG and Phylogenetic Analysis
In total, seven isolates were successfully sequenced at bg, tpi and gdh loci, and formed six assemblage A MLGs after removal of sequences of mixed infection samples (Table 3). The six identified assemblage A MLGs were composed of two novel MLG-subtypes (AI-novel 1 and AI-novel 2); AI-novel 1 was found in two isolates, whereas AI-novel 2 was identified in four isolates. Phylogenetic relationships of assemblage A MLGs with reference genotypes are shown in Figure 2. Based on phylogenetic analysis, all assemblage A MLGs identified herein were clustered in the MLG AI branch, with MLG AI-novel 1 clustered closer to MLG AI-1, whereas MLG AI-novel 2 clustered closer to MLG AI-2.
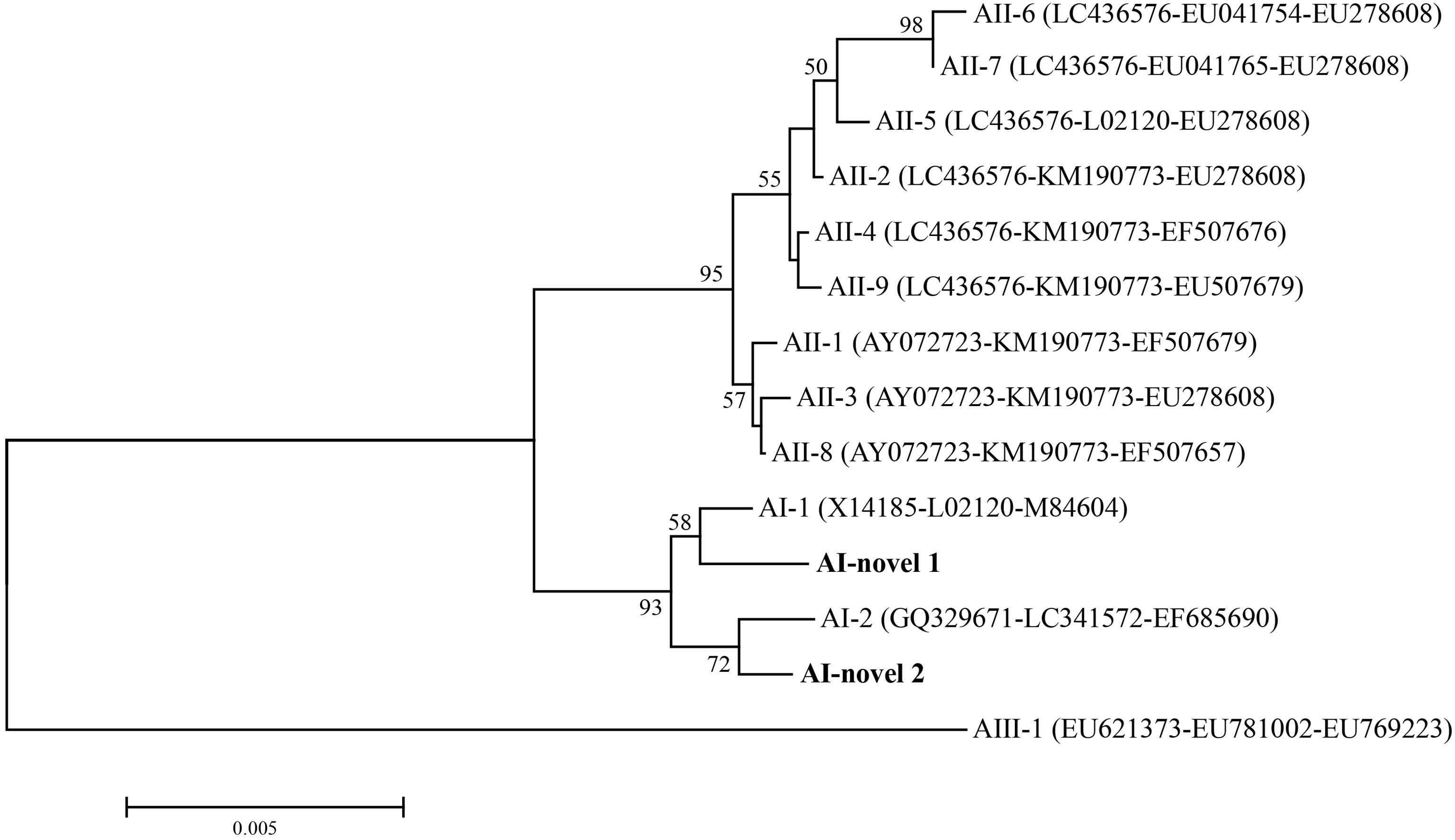
Figure 2 Phylogenetic relationships among G. duodenalis assemblage A isolates inferred by neighbor-joining analysis based on concatenated datasets for bg, tpi and gdh nucleotide sequences. Bootstrap values greater than 50% from 1000 replicates were shown on nodes. The novel MLGs in this study were indicated in bold.
Discussion
Six species of musk deer (Moschus spp.) have historically been bred in China, which include siberian musk deer (M. moschiferus), forest musk deer (M. berezovskii), black musk deer (M. fuscus), alpine musk deer (M. chrysogaster), himalayan musk deer (M. leucogaster), and anhui musk deer (M. anhuiensis) (Fan et al., 2018). In previous studies, deer have been considered as a major reservoir of viruses, bacteria, and parasites for humans and livestock (Böhm et al., 2007; Mehrpad et al., 2018). In particular, a recent study has suggested the potential emergence of a new reservoir of SARS-CoV-2 viruses in free-ranging white-tailed deer, which may open new pathways for evolution, transmission to other wildlife species, and potential spillback of novel variants to humans (Hale et al., 2022).
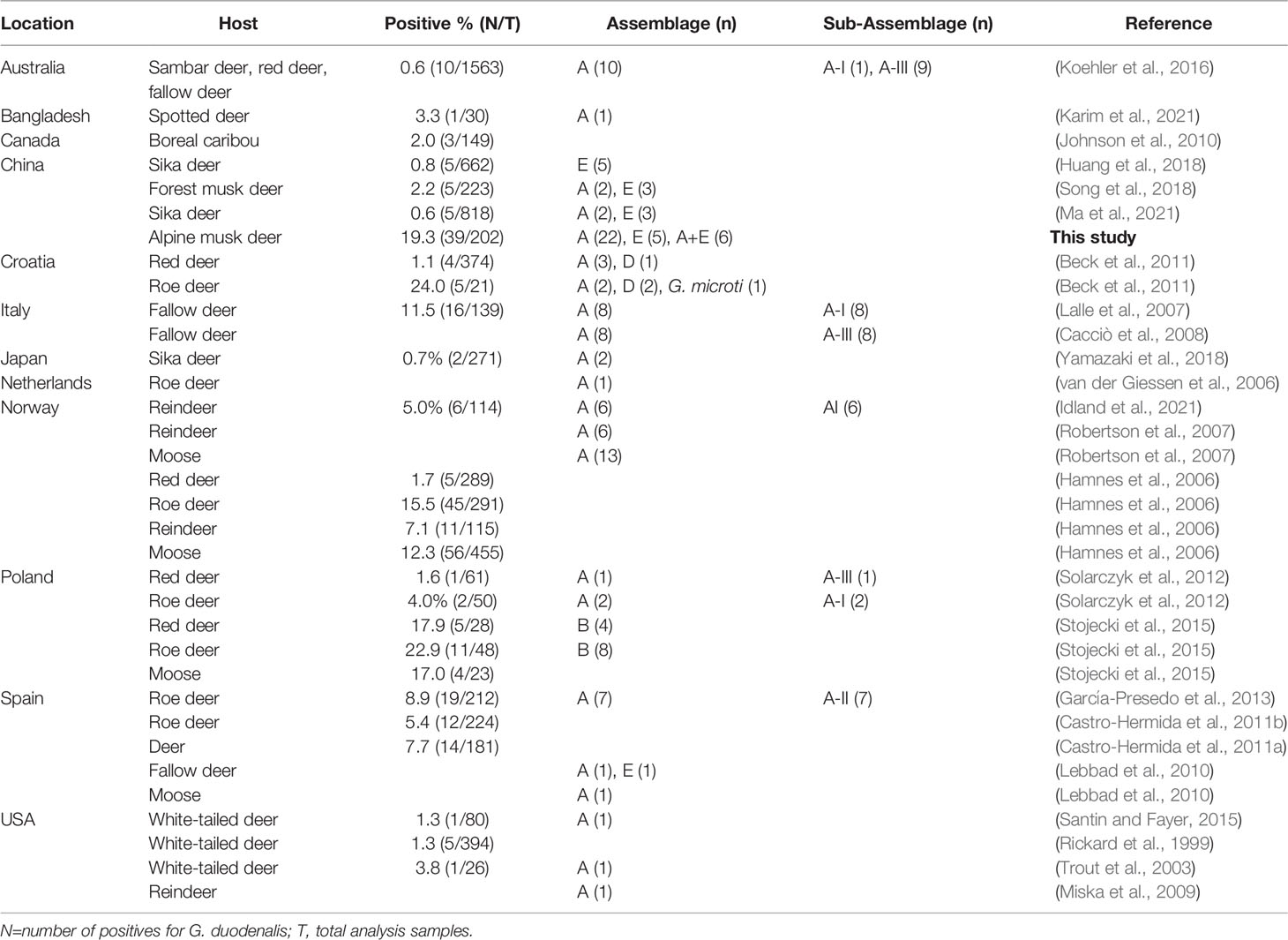
Giardia spp. infects a broad range of hosts including humans, livestock, companion animals, wildlife and birds (Ryan et al., 2021). However, information on the distribution, molecular characteristics and zoonotic potential of Giardia in cervids is scant. To date, G. duodenalis infections in cervids have been reported in several countries, including Australia, Bangladesh, Canada, Croatia, Italy, Japan, Netherlands, Norway, Poland, Spain, Sweden, USA and China, with the infection rates ranging from 0.6% to 24.0% (Table 4). Interestingly, the rodent-specific species Giardia microti has been isolated in roe deer (Capreolus capreolus) in Croatia (Beck et al., 2011). In the present study, the prevalence of G. duodenalis was 19.3% (39/202) in alpine musk deer, which is higher than that reported in sika deer (0.6% and 0.8%) and forest musk deer (2.2%) in China. The discrepancies in infection rates of G. duodenalis may be related to the differences in geographical location, sampling season, animal species, and sample size. To the best of our knowledge, this is the first study to isolate and characterize G. duodenalis from alpine musk deer in China using MLG.
Methods based on sequence analysis of SSU rRNA, gdh, bg and tpi genes have currently been widely used for genotyping G. duodenalis isolates from human and animal samples in order to obtain high-sequencing resolution (Feng and Xiao, 2011). To date, molecular studies have identified G. duodenalis in fallow deer, forest musk deer, moose, red deer, reindeer, roe deer, sambar deer, spotted deer and white-tailed deer with a worldwide distribution (Table 4). In addition to zoonotic assemblages A and B, other G. duodenalis assemblages including E (mainly found in hoofed mammals) and D (mainly found in canines) have also been reported occasionally in these animal hosts (Beck et al., 2011; Huang et al., 2018), which suggests potential transmission routs of G. duodenalis assemblages between humans, livestock, companion animals and cervids. In the present study, both assemblage A and E were identified, and assemblage A was the predominant genotype. Mixed infections were observed in alpine musk deer at both assemblage and sub-assemblage levels, which may be a result of infection with Giardia parasites with different genetic profiles. Assemblages A is responsible for most giardiasis cases in humans, especially in South America and the Middle East (Xiao and Feng, 2017; Ryan et al., 2021). Interestingly, the host-adapted genotype assemblage E which was approximately 87% similar to assemblages A in genome, has been reported in at least 57 human giardiasis cases in Brazil, Egypt, Vietnam, Australia and New Zealand (Abdel-Moein and Saeed, 2016; Fantinatti et al., 2016; Zahedi et al., 2017; Garcia-R et al., 2021; Iwashita et al., 2021). Collectively, the presence of zoonotic assemblages of G. duodenalis in alpine musk deer in China suggests that these animals may potentially act as a reservoir of G. duodenalis for humans.
Moreover, in order to elucidate the genetic diversity of G. duodenalis in alpine musk deer, positive samples identified in the present were subjected for sub-genotyping by MLG. Moderate genetic variation was observed within assemblage A sequences, whereas no genetic variation was noticed within assemblage E sequences, which may be due to the low allelic sequence heterozygosity (ASH) in the genomes of assemblages A and E (Kooyman et al., 2019). In addition, the six assemblage A MLGs were composed of two novel MLG-subtypes which belonged to sub-assemblage AI. Among the three sub-assemblages within assemblage A, sub-assemblage AI is most commonly found in animals, whereas sub-assemblage AII is mostly found in humans; sub-assemblage AIII is rare and has been found in wild ruminants and two human giardiasis cases in Romania and New Zealand (Feng and Xiao, 2011; Cai et al., 2021; Ryan et al., 2021). In published studies, both sub-assemblage AI, AII and AIII were identified in various deer (Table 4). Further studies based on MLG analysis are necessary to gain a better understanding on the potential role of deer in the zoonotic transmission of G. duodenalis.
Conclusions
In conclusion, this is the first report of G. duodenalis in alpine musk deer with a high prevalence in China. Two G. duodenalis assemblages were identified, assemblage A and E. Moderate genetic diversity was observed within assemblage A sequences based on MLG analysis. Six assemblage A MLGs were identified which were composed of two novel MLG-subtypes belonging to sub-assemblage AI. Collectively, zoonotic assemblages of G. duodenalis identified in the present study point out that alpine musk deer may potentially act as reservoirs of this protozoan to humans.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by Research Ethics Committee of Tarim University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
ZC, QW, XH, JB, BZ, BW, and XG performed the experiments. ZC drafted the manuscript. MQ and JL critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research was funded by the National Natural Science Foundation of China (32102689), Science and Technology Planning Project of Henan Province (222102110240), the Program for Young and Middle-aged Leading Science, Technology, and Innovation of Xinjiang Production & Construction Group (2018CB034), and Scientific research project of Xuchang University (2022GJPY009 and 2022YB034).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel-Moein, K. A., Saeed, H. (2016). The Zoonotic Potential of Giardia Intestinalis Assemblage E in Rural Settings. Parasitol. Res. 115 (8), 3197–3202. doi: 10.1007/s00436-016-5081-7
Adam, R. D. (2021). Giardia Duodenalis: Biology and Pathogenesis. Clin. Microbiol. Rev. 34 (4), e0002419. doi: 10.1016/s0304-4017(02)00422-3
Appelbee, A. J., Frederick, L. M., Heitman, T. L., Olson, M.E. (2003). Prevalence and Genotyping of Giardia duodenalis From Beef Calves in Alberta, Canada. Vet. Parasitol. 112 (4), 289–294. doi: 10.1016/s0304-4017(02)00422-3
Argüello-García, R., Leitsch, D., Skinner-Adams, T., Ortega-Pierres, M. G. (2020). Drug Resistance in Giardia: Mechanisms and Alternative Treatments for Giardiasis. Adv. Parasitol. 107, 201–282. doi: 10.1016/bs.apar.2019.11.003
Beck, R., Sprong, H., Lucinger, S., Pozio, E., Cacciò, S. M. (2011). A Large Survey of Croatian Wild Mammals for Giardia Duodenalis Reveals a Low Prevalence and Limited Zoonotic Potential. Vector Borne Zoonotic Dis. 11 (8), 1049–1055. doi: 10.1089/vbz.2010.0113
Böhm, M., White, P. C., Chambers, J., Smith, L., Hutchings, M. R. (2007). Wild Deer as a Source of Infection for Livestock and Humans in the UK. Vet. J. 174 (2), 260–276. doi: 10.1016/j.tvjl.2006.11.003
Cacciò, S. M., Beck, R., Lalle, M., Marinculic, A., Pozio, E. (2008). Multilocus Genotyping of Giardia Duodenalis Reveals Striking Differences Between Assemblages A and B. Int. J. Parasitol. 38 (13), 1523–1531. doi: 10.1016/j.ijpara.2008.04.008
Cacciò, S. M., Lalle, M., Svärd, S. G. (2018). Host Specificity in the Giardia Duodenalis Species Complex. Infect. Genet. Evol. 66, 335–345. doi: 10.1016/j.meegid.2017.12.001
Cai, W., Ryan, U., Xiao, L., Feng, Y. (2021). Zoonotic Giardiasis: An Update. Parasitol. Res. 120 (12), 4199–4218. doi: 10.1007/s00436-021-07325-2
Cai, Y., Yang, J., Wang, J., Yang, Y., Fu, W., Zheng, C., et al (2020). Changes in the Population Genetic Structure of Captive Forest Musk Deer (Moschus Berezovskii) With the Increasing Number of Generation Under Closed Breeding Conditions. Animals (Basel) 10 (2), 255. doi: 10.3390/ani10020255
Capewell, P., Krumrie, S., Katzer, F., Alexander, C. L., Weir, W. (2021). Molecular Epidemiology of Giardia Infections in the Genomic Era. Trends Parasitol. 37 (2), 142–153. doi: 10.1016/j.pt.2020.09.013
Castro-Hermida, J. A., García-Presedo, I., Almeida, A., González-Warleta, M., Correia Da Costa, J. M., Mezo, M. (2011a). Cryptosporidium Spp. And Giardia Duodenalis in Two Areas of Galicia (NW Spain). Sci. Total Environ. 409 (13), 2451–2459. doi: 10.1016/j.scitotenv.2011.03.010
Castro-Hermida, J. A., García-Presedo, I., González-Warleta, M., Mezo, M. (2011b). Prevalence of Cryptosporidium and Giardia in Roe Deer (Capreolus Capreolus) and Wild Boars (Sus Scrofa) in Galicia (NW, Spain). Vet. Parasitol. 179 (1-3), 216–219. doi: 10.1016/j.vetpar.2011.02.023
Dixon, B. R. (2021). Giardia Duodenalis in Humans and Animals - Transmission and Disease. Res. Vet. Sci. 135, 283–289. doi: 10.1016/j.rvsc.2020.09.034
Fan, Z., Li, W., Jin, J., Cui, K., Yan, C., Peng, C., et al (2018). The Draft Genome Sequence of Forest Musk Deer (Moschus Berezovskii). GigaScience 7 (4), giy038. doi: 10.1093/gigascience/giy038
Fantinatti, M., Bello, A. R., Fernandes, O., Da-Cruz, A. M. (2016). Identification of Giardia Lamblia Assemblage E in Humans Points to a New Anthropozoonotic Cycle. J. Infect. Dis. 214 (8), 1256–1259. doi: 10.1093/infdis/jiw361
Feng, Y., Xiao, L. (2011). Zoonotic Potential and Molecular Epidemiology of Giardia Species and Giardiasis. Clin. Microbiol. Rev. 24 (1), 110–140. doi: 10.1128/CMR.00033-10
García-Presedo, I., Pedraza-Díaz, S., González-Warleta, M., Mezo, M., Gómez-Bautista, M., Ortega-Mora, L. M., et al (2013). The First Report of Cryptosporidium Bovis, C. Ryanae and Giardia Duodenalis Sub-Assemblage A-II in Roe Deer (Capreolus Capreolus) in Spain. Vet. Parasitol. 197 (3-4), 658–664. doi: 10.1016/j.vetpar.2013.07.002
Garcia-R, J. C., Ogbuigwe, P., Pita, A. B., Velathanthiri, N., Knox, M. A., Biggs, P. J., et al. (2021). First Report of Novel Assemblages and Mixed Infections of Giardia Duodenalis in Human Isolates From New Zealand. Acta Trop. 220, 105969. doi: 10.1016/j.actatropica.2021.105969
Hale, V. L., Dennis, P. M., McBride, D. S., Nolting, J. M., Madden, C., Huey, D., et al. (2022). SARS-CoV-2 Infection in Free-Ranging White-Tailed Deer. Nature 602 (7897):481–486. doi: 10.1038/s41586-021-04353-x
Hamnes, I. S., Gjerde, B., Robertson, L., Vikøren, T., Handeland, K. (2006). Prevalence of Cryptosporidium and Giardia in Free-Ranging Wild Cervids in Norway. Vet. Parasitol. 141 (1-2), 30–41. doi: 10.1016/j.vetpar.2006.05.004
Huang, J., Zhang, Z., Zhang, Y., Yang, Y., Zhao, J., Wang, R., et al. (2018). Prevalence and Molecular Characterization of Cryptosporidium Spp. And Giardia Duodenalis in Deer in Henan and Jilin, China. Parasit. Vectors 11 (1), 239. doi: 10.1186/s13071-018-2813-9
Idland, L., Juul, A. M., Solevåg, E. K., Tysnes, K. R., Robertson, L. J., Utaaker, K. S. (2021). Occurrence of Faecal Endoparasites in Reindeer (Rangifer Tarandus) in Two Grazing Areas in Northern Norway. Acta Vet. Scand. 63 (1), 13. doi: 10.1186/s13028-021-00578-y
Iwashita, H., Sugamoto, T., Takemura, T., Tokizawa, A., Vu, T. D., Nguyen, T. H., et al. (2021). Molecular Epidemiology of Giardia Spp. In Northern Vietnam: Potential Transmission Between Animals and Humans. Parasite Epidemiol. Control 12, e00193. doi: 10.1016/j.parepi.2020.e00193
Jiang, F., Gao, H., Qin, W., Song, P., Wang, H., Zhang, J., et al. (2021). Marked Seasonal Variation in Structure and Function of Gut Microbiota in Forest and Alpine Musk Deer. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.699797
Johnson, D., Harms, N. J., Larter, N. C., Elkin, B. T., Tabel, H., Wei, G. (2010). Serum Biochemistry, Serology, and Parasitology of Boreal Caribou (Rangifer Tarandus Caribou) in the Northwest Territories, Canada. J. Wildl. Dis. 46 (4), 1096–1107. doi: 10.7589/0090-3558-46.4.1096
Karim, M. R., Li, J., Rume, F. I., Sumon, S. M. R., Selim, A. S. M., Hoda, N., et al (2021). Occurrence and Molecular Characterization of Cryptosporidium Spp. And Giardia Duodenalis Among Captive Mammals in the Bangladesh National Zoo. Parasitol. Int. 84, 102414. doi: 10.1016/j.parint.2021.102414
Koehler, A. V., Haydon, S. R., Jex, A. R., Gasser, R. B. (2016). Cryptosporidium and Giardia Taxa in Faecal Samples From Animals in Catchments Supplying the City of Melbourne With Drinking Water, (2011 to 2015). Parasit. Vectors 9 (1), 315. doi: 10.1186/s13071-016-1607-1
Kooyman, F. N. J., Wagenaar, J. A., Zomer, A. (2019). Whole-Genome Sequencing of Dog-Specific Assemblages C and D of Giardia Duodenalis From Single and Pooled Cysts Indicates Host-Associated Genes. Microb. Genom. 5 (12), e000302. doi: 10.1099/mgen.0.000302
Lalle, M., Frangipane di Regalbono, A., Poppi, L., Nobili, G., Tonanzi, D., Pozio, E., et al. (2007). A Novel Giardia Duodenalis Assemblage A Subtype in Fallow Deer. J. Parasitol. 93 (2), 426–428. doi: 10.1645/GE-983R.1
Lalle, M., Pozio, E., Capelli, G., Bruschi, F., Crotti, D., Cacciò, S. M. (2005). Genetic Heterogeneity at the Beta-Giardin Locus Among Human and Animal Isolates of Giardia Duodenalis and Identification of Potentially Zoonotic Subgenotypes. Int. J. Parasitol. 35 (2), 207–213. doi: 10.1016/j.ijpara.2004.10.022
Lebbad, M., Mattsson, J. G., Christensson, B., Ljungström, B., Backhans, A., Andersson, J. O., et al. (2010). From Mouse to Moose: Multilocus Genotyping of Giardia Isolates From Various Animal Species. Vet. Parasitol. 168 (3-4), 231–239. doi: 10.1016/j.vetpar.2009.11.003
Li, Y., Hu, X., Yang, S., Zhou, J., Zhang, T., Qi, L., et al (2017b). Comparative Analysis of the Gut Microbiota Composition Between Captive and Wild Forest Musk Deer. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01705
Li, J., Wang, H., Wang, R., Zhang, L. (2017a). Giardia Duodenalis Infections in Humans and Other Animals in China. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02004
Ma, N., Wang, H. X., Tao, W. F., Xue, N. Y., Bai, J. Y., Zhao, Q., et al. (2021). Detection of Point Prevalence and Assemblages of Giardia Spp. In Dairy Calves and Sika Deer, Northeast China. Vector Borne Zoonotic Dis. 21 (9), 685–691. doi: 10.1089/vbz.2020.2757
Mehrpad, S., Cleveland, C. A., DeNicola, A., Dubey, J. P., Yabsley, M. J. (2018). Survey for Selected Pathogens in Philippine Deer (Rusa Marianna) From Guam, Marianna Islands, USA. Vet. Parasitol. Reg. Stud. Rep. 11, 36–40. doi: 10.1016/j.vprsr.2017.11.010
Miska, K. B., Jenkins, M. C., Trout, J. M., Santín, M., Fayer, R. (2009). Detection and Comparison of Giardia Virus (GLV) From Different Assemblages of Giardia Duodenalis. J. Parasitol. 95 (5), 1197–1200. doi: 10.1645/GE-1876.1
Rickard, L. G., Siefker, C., Boyle, C. R., Gentz, E. J. (1999). The Prevalence of Cryptosporidium and Giardia Spp. In Fecal Samples From Free-Ranging White-Tailed Deer (Odocoileus Virginianus) in the Southeastern United States. J. Vet. Diagn. Invest. 11 (1), 65–72. doi: 10.1177/104063879901100111
Robertson, L. J., Forberg, T., Hermansen, L., Hamnes, I. S., Gjerde, B. (2007). Giardia Duodenalis Cysts Isolated From Wild Moose and Reindeer in Norway: Genetic Characterization by PCR-Rflp and Sequence Analysis at Two Genes. J. Wildl. Dis. 43 (4), 576–585. doi: 10.7589/0090-3558-43.4.576
Ryan, U., Cacciò, S. M. (2013). Zoonotic Potential of Giardia. Int. J. Parasitol. 43 (12-13), 943–956. doi: 10.1016/j.ijpara.2013.06.001
Ryan, U. M., Feng, Y., Fayer, R., Xiao, L. (2021). Taxonomy and Molecular Epidemiology of Cryptosporidium and Giardia - a 50 Year Perspective, (1971-2021). Int. J. Parasitol. 51 (13-14), 1099–1119. doi: 10.1016/j.ijpara.2021.08.007
Ryan, U., Hijjawi, N., Feng, Y., Xiao, L. (2019). Giardia: An Under-Reported Foodborne Parasite. Int. J. Parasitol. 49 (1), 1–11. doi: 10.1016/j.ijpara.2018.07.003
Ryan, U., Zahedi, A. (2019). Molecular Epidemiology of Giardiasis From a Veterinary Perspective. Adv. Parasitol. 106, 209–254. doi: 10.1016/bs.apar.2019.07.002
Santin, M., Fayer, R. (2015). Enterocytozoon Bieneusi, Giardia, and Cryptosporidium Infecting White-Tailed Deer. J. Eukaryot. Microbiol. 62 (1), 34–43. doi: 10.1111/jeu.12155
Solarczyk, P., Majewska, A. C., Moskwa, B., Cabaj, W., Dabert, M., Nowosad, P. (2012). Multilocus Genotyping of Giardia Duodenalis Isolates From Red Deer (Cervus Elaphus) and Roe Deer (Capreolus Capreolus) From Poland. Folia Parasitol. (Praha) 59 (3), 237–240. doi: 10.14411/fp.2012.032
Song, Y., Li, W., Liu, H., Zhong, Z., Luo, Y., Wei, Y., et al. (2018). First Report of Giardia Duodenalis and Enterocytozoon Bieneusi in Forest Musk Deer (Moschus Berezovskii) in China. Parasit. Vectors 11 (1), 204. doi: 10.1186/s13071-018-2681-3
Stojecki, K., Sroka, J., Caccio, S. M., Cencek, T., Dutkiewicz, J., Kusyk, P. (2015). Prevalence and Molecular Typing of Giardia Duodenalis in Wildlife From Eastern Poland. Folia Parasitol. (Praha) 62, 2015.042. doi: 10.14411/fp.2015.042
Sulaiman, I. M., Fayer, R., Bern, C., Gilman, R. H., Trout, J. M., Schantz, P. M., et al. (2003). Triosephosphate Isomerase Gene Characterization and Potential Zoonotic Transmission of Giardia Duodenalis. Emerg. Infect. Dis. 9 (11), 1444–1452. doi: 10.3201/eid0911.030084
Trout, J. M., Santin, M., Fayer, R. (2003). Identification of Assemblage A Giardia in White-Tailed Deer. J. Parasitol. 89 (6), 1254–1255. doi: 10.1645/GE-3165RN
van der Giessen, J. W., de Vries, A., Roos, M., Wielinga, P., Kortbeek, L. M., Mank, et al. (2006). Genotyping of Giardia in Dutch Patients and Animals: A Phylogenetic Analysis of Human and Animal Isolates. Int. J. Parasitol. 36 (7), 849–858. doi: 10.1016/j.ijpara.2006.03.001
Xiao, L., Feng, Y. (2017). Molecular Epidemiologic Tools for Waterborne Pathogens Cryptosporidium Spp. And Giardia Duodenalis. Food Waterborne Parasitol. 8-9, 14–32. doi: 10.1016/j.fawpar.2017.09.002
Yamazaki, A., Izumiyama, S., Yagita, K., Kishida, N., Kubosaki, A., Hara-Kudo, Y., et al. (2018). The Molecular Detection of Cryptosporidium and Giardia in Sika Deer (Cervus Nippon Centralis) in Japan. Food Saf. (Tokyo) 6 (2), 88–95. doi: 10.14252/foodsafetyfscj.2017029
Keywords: Giardia duodenalis, alpine musk deer, multilocus genotyping, zoonotic potential, China
Citation: Cui Z, Wang Q, Huang X, Bai J, Zhu B, Wang B, Guo X, Qi M and Li J (2022) Multilocus Genotyping of Giardia duodenalis in Alpine Musk Deer (Moschus chrysogaster) in China. Front. Cell. Infect. Microbiol. 12:856429. doi: 10.3389/fcimb.2022.856429
Received: 17 January 2022; Accepted: 28 March 2022;
Published: 20 April 2022.
Edited by:
Razakandrainibe Romy, Université de Rouen, FranceReviewed by:
Mohamed Mammeri, Ecole Nationale Veterinaire D’alfort, FranceAbdelmounaim Mouhajir, Université de Rouen, France
Copyright © 2022 Cui, Wang, Huang, Bai, Zhu, Wang, Guo, Qi and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Qi, qimengdz@163.com; Junqiang Li, lijunqiangcool@126.com
 Zhaohui Cui
Zhaohui Cui Qilin Wang2
Qilin Wang2  Meng Qi
Meng Qi Junqiang Li
Junqiang Li