The role of tryptophan in Chlamydia trachomatis persistence
- 1The First School of Clinical Medicine, Chenzhou No.1 People’s Hospital, Southern Medical University, Guangzhou, China
- 2Department of Clinical Microbiology Laboratory, Chenzhou No.1 People’s Hospital, Chenzhou, China
Chlamydia trachomatis (C. trachomatis) is the most common etiological agent of bacterial sexually transmitted infections (STIs) and a worldwide public health issue. The natural course with C. trachomatis infection varies widely between individuals. Some infections clear spontaneously, others can last for several months or some individuals can become reinfected, leading to severe pathological damage. Importantly, the underlying mechanisms of C. trachomatis infection are not fully understood. C. trachomatis has the ability to adapt to immune response and persist within host epithelial cells. Indoleamine-2,3-dioxygenase (IDO) induced by interferon-gamma (IFN-γ) degrades the intracellular tryptophan pool, to which C. trachomatis can respond by converting to a non-replicating but viable state. C. trachomatis expresses and encodes for the tryptophan synthase (TS) genes (trpA and trpB) and tryptophan repressor gene (trpR). Multiple genes interact to regulate tryptophan synthesis from exogenous indole, and persistent C. trachomatis can recover its infectivity by converting indole into tryptophan. In this review, we discuss the characteristics of chlamydial infections, biosynthesis and regulation of tryptophan, the relationship between tryptophan and C. trachomatis, and finally, the links between the tryptophan/IFN-γ axis and C. trachomatis persistence.
Introduction
Chlamydia trachomatis (C. trachomatis) is an aerobic, gram-negative, obligate intracellular bacterial pathogen (Chumduri et al., 2013). The World Health Organization (WHO) estimated that there were 131 million newly infected cases of C. trachomatis infections among adolescents and adults aged 15-49 in 2012, with a global incidence of 38 infections per 1,000 women and 33 infections per 1,000 men (O'Neill et al., 2018). The estimated number of chlamydial infection cases is most likely to be an underestimation of the actual number of cases, because most natural courses are asymptomatic and go undetected. Up to 70–80% of the C. trachomatis infections in women are asymptomatic, and therefore unrecognized and untreated. Studies on the natural course of untreated C. trachomatis lower genital tract infections in women show spontaneous clearance rates of 30% in the first weeks or months, approximately 50% in 1 year, 80% in 2 years, and 94% in 4 years (den Hartog et al., 2006; Ouellette et al., 2021).
C. trachomatis invades the epithelial cells of the endocervix and upper genital tract in women, as well as the conjunctiva, urethra, and rectum in both men and women (Morrison et al., 2020). In females, the cervix is the most commonly infected site and C. trachomatis can enter the uterus and fallopian tubes from the cervix to provoke an inflammatory condition and serious reproductive complications. While the spontaneous clearance of chlamydial lower genital tract infections may lead to an underestimation of the percentage of women at risk of complications, because clearance from the lower genital tract does not mean that the causative agent has not already ascended to the upper genital tract. Most infected women seem to have an adequate immune response, but some infected women will have a long-lasting C. trachomatis infection and, subsequently, an increased risk of sequelae, including the development of pelvic inflammatory disease, tubal scarring, ectopic pregnancy, and infertility (Hua et al., 2015).
C. trachomatis has the ability to adapt to immune response and persists within host epithelial cells, resulting in an irreversible inflammatory damage. A previous study on asymptomatic non-pregnant women showed the resolution rate of C. trachomatis to be 11–45% and could not be treated in a timely manner (Juliana et al., 2021). Untreated women are more likely to get cervicitis, urethritis, pelvic inflammatory diseases, chronic pelvic pain, ectopic pregnancy, miscarriage, and infertility (Yeow et al., 2016; Casillas-Vega et al., 2017). Progression to these sequelae is thought to be the outcome of pathological inflammatory responses such as tissue disruption, fibrosis, and scarring. Moreover, C. trachomatis infection may increase the probability of co-infections with human immunodeficiency virus and Neisseria gonorrhoeae. Further, C. trachomatis impedes human papilloma virus (HPV)-induced mechanisms that maintain cellular and genomic integrity and it may be linked to cervical cancer (Koster et al., 2022). C. trachomatis has also been identified in the tissues of 70% of ovarian tumors and none in benign or normal ovaries, suggesting a potential role in ovarian carcinogenesis (Jonsson et al., 2018). C. trachomatis infection is also associated with adverse outcomes in pregnancy, including chorioamnionitis, preterm delivery, and low birth weight (Tamarelle et al., 2019). Nearly half of babies born vaginally to mothers infected with genital C. trachomatis will get chlamydial conjunctivitis, some babies will also acquire nasopharyngeal infections and even develop chlamydial pneumonia (Redgrove and McLaughlin, 2014). Revealing that the pathogenic mechanisms of persistent C. trachomatis infection is of the utmost importance for developing effective strategies to prevent and control complications.
The genomes of chlamydial strains are remarkably conserved in genetic order and content, except the polymorphic regions of the genome called plasticity zones (PZ). Trp resides in the PZ and encodes tryptophan synthase (TS) (Caldwell et al., 2003; Carlson et al., 2004). TS is a heterotetramer composed of two α subunits (TrpA) and two β subunits (TrpB), which catalyzes the two final biosynthetic reactions of tryptophan (Miles, 2001). A distinct difference between urogenital and ocular isolates of C. trachomatis is that only the former expresses TS and is therefore able to synthesize tryptophan via indole salvage (Carlson et al., 2004; Carlson et al., 2005). Interferon-gamma (IFN-γ) is considered to be a major anti-chlamydial effector cytokine. IFN-γ acts indirectly on cells to limit the tryptophan required for C. trachomatis by activating indoleamine-2,3-dioxygenase (IDO) to deplete the intracellular tryptophan pool, resulting in restricted chlamydial growth and development in human epithelial cells. C. trachomatis can recover from a persistent state into an infectious state by converting indole into tryptophan via TS (Aiyar et al., 2014). We speculate that the synthesis of tryptophan using exogenous indoles in an IFN-γ-rich environment allows the bacteria to escape IFN-γ-mediated immune responses and establish a persistent infection. If true, this would be an important virulence factor and provide a potential mechanism for C. trachomatis to evade host defenses. In this review, we summarize the current understanding of changes in molecular and morphological characteristics of C. trachomatis during tryptophan starvation, the relationship between tryptophan and C. trachomatis, and the influence of the tryptophan/IFN-γ axis on C. trachomatis persistence.
Biphasic life cycle of C. trachomatis
Based on the characterization of the major outer membrane protein (MOMP), C. trachomatis can be divided into several serogroups, including A, B, Ba, C, D, Da, E, F, G, Ga, H, I, Ia, J, K, L1, L2, L2a, and L3. C. trachomatis serovars A–C can cause trachoma that is the dominant cause of blindness, while serovars D–K make Chlamydia the most prevalent bacterial sexually transmitted infection worldwide. Lymphogranuloma venereum (LGV) serovars L1–L3 cause STIs with ulcers and vesicle formation (Bayramova et al., 2018; Murray and McKay, 2021; Sixt, 2021). Genotypes D, E, and F are more prevalent in urogenital infections and are common among heterosexuals. Genotypes D, G, and J are commonly detected in the anorectal tract in women and men who have sex with men (MSM) (Bowden et al., 2021).
C. trachomatis demonstrates a unique biphasic life cycle with two morphologically distinct forms: metabolically inactive elementary bodies (EBs) infect host cells through phagocytosis or endocytosis and differentiate into metabolically active replicating reticulate bodies (RBs). EBs differentiate into RBs and begin to replicate within a membrane-bound compartment of the eukaryotic cell in a process known as inclusion (Howe et al., 2019). RBs divide by binary fission, and consequently grow until the entire cytoplasmic matrix is filled and the nucleus is dislocated. RBs adapt in various ways to promote survival and replication: 1) through selective fusion of vesicles to inhibit lysosomal fusion and promote nutrient-rich extracellular vesicles; 2) through a complex isolation mechanism to acquire nutrients such as phosphatidylcholine and cholesterol; 3) through regulation of the host cells intrinsic immunity and cellular survival, among other mechanisms. After 24–74 hours, RBs re-differentiate into EBs, which are then released to initiate a new infection cycle by the lysis of host cell and/or the extrusion of inclusion (Murray and McKay, 2021).
Another pivotal feature of C. trachomatis is its ability to establish persistence, which is a reversible state that occurs in adverse growth conditions. In this state, RBs stop dividing and they turn into aberrant bodies (ABs), which are obviously larger in size and in this form successfully survive. As a persistent infection, C. trachomatis re-enters the cycle of development when the host’s immune function is restored, drugs are discontinued and the living environment is improved (Chumduri et al., 2013; Bayramova et al., 2018) (Figure 1).
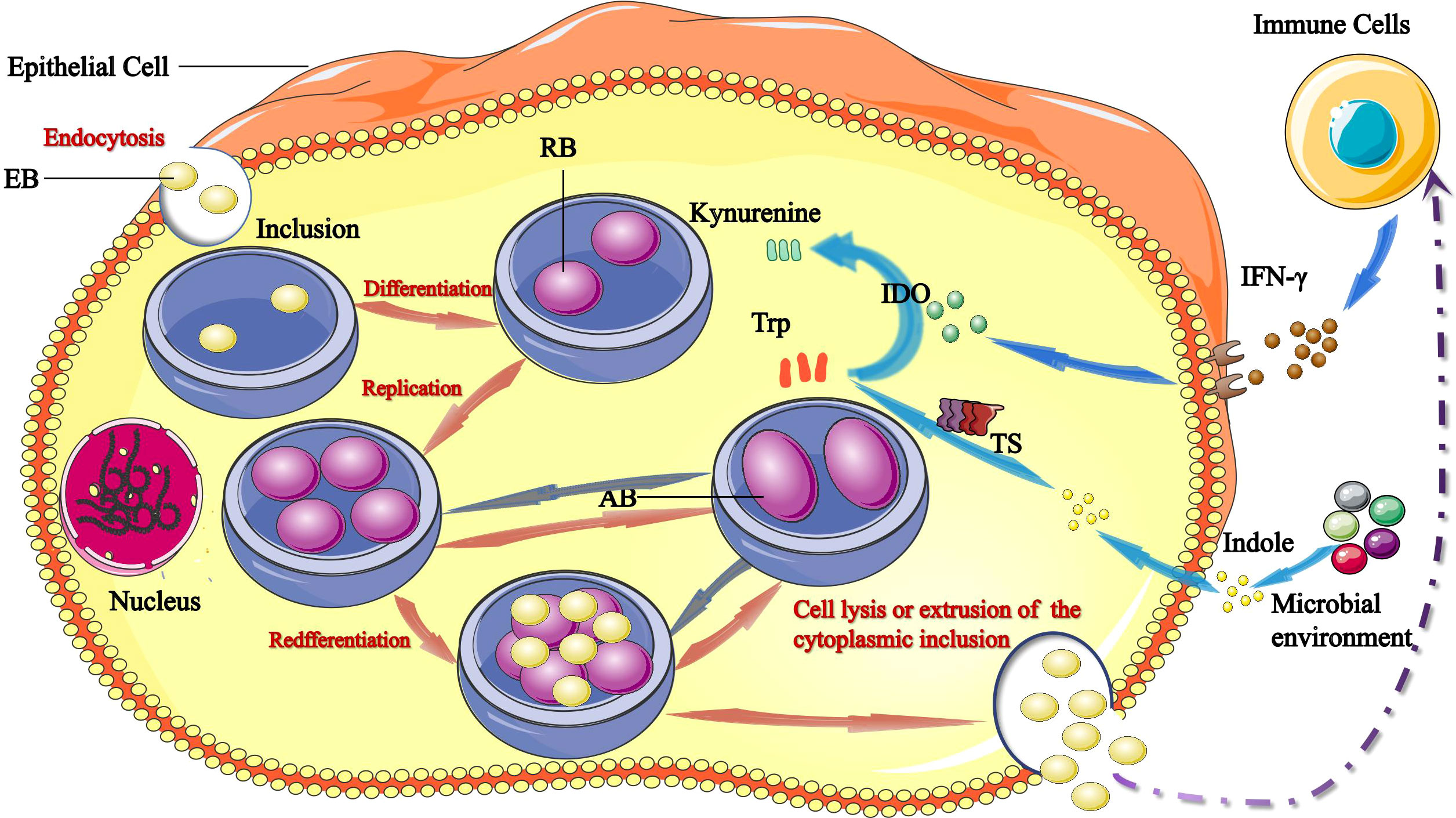
Figure 1 Links between tryptophan/IFN-γ axis and C. trachomatis. The infectious form of C. trachomatis, elementary bodies (EBs) infect epithelial cells via endocytosis. Upon entry, EBs differentiate into reticulate bodies (RBs) and start replicating within the inclusion. RBs divide by binary fission, and consequently grow until the entire cytoplasmic matrix is filled and the nucleus is dislocated. After 24–74 hours, RBs redifferentiate into EBs, EBs are released by host cell lysis and/or the extrusion of inclusion to initiate a new infection cycle. IFN-γ induces production of IDO1, which catabolizes tryptophan into kynurenine. Tryptophan starvation can lead to RBs turn into aberrant bodies (ABs). The chlamydial tryptophan synthase (TS) gene converts indoles produced by other microbiota back into tryptophan. Persistent C. trachomatis can become infectious and re-enter the life cycle by converting indole into tryptophan via TS.
C. trachomatis persistence
Plenty of factors may contribute to persistent infection, including use of antibiotics, nutritional deficiency, cytokine release, viral infection, phages, other diseases, and individual differences (Bayramova et al., 2018). The course of a C. trachomatis infection (i.e., whether the infection will be cleared or persistent) may be determined by pathogenic virulence factors, environmental factors, and/or host immune factors. During treatment, about 10–15% of patients are reinfected leading to development of a persistent infection. C. trachomatis can establish asymptomatic, persistent infections by several mechanisms, including antibiotic resistance, immune evasion, and suppression of apoptosis (Chumduri et al., 2013). Chlamydial persistence represents a means by which the pathogen resists a key immune response, allowing expression of gene products but inhibiting gene products necessary for rapid pathogen proliferation. Persistent C. trachomatis infection is a reversible state under detrimental growth conditions, which is viable but metabolically quiescent, with an enlarged aberrant form. This is a special state that bacteria enter when they are challenged and can regain their pathogenicity when conditions are suitable for survival.
In the host immune system, the most well-recognized biological characteristic of C. trachomatis is its ability to stay in contact with the host for a long period of time, during which the bacteria are apparently quiescent or latent. However, this characteristic is also the least understood and is a matter of controversy in the field. The first known article on human chlamydial infection was written by Ritter in 1879 (Radomski et al., 2016). C. trachomatis can remain quiescent in a host for a long time and subsequently contribute to a chronic clinical disease. This was first recognized in infections of Chlamydia psittaci (C. psittaci). Between 1879 and 1928, outbreaks of C. psittaci that were related to the import of parrots occurred in the United States, Germany, England, France, and Switzerland. People who had close contact with birds would acquire infection from those sick birds. Moulder suggested that this was a “persistent” infection and presumed that it had entered a nonreplicating stage that he termed a “cryptic body”. Unfortunately, he never went on to study these observations further (Rank and Yeruva, 2014).
There has been more evidence of persistent chlamydial infection in the reproductive tract since persistent infection was first defined in 1980, but there is no uniform definition of persistent infection until now. Molecular biology studies suggest that persistent infection is the presence of abnormal body formation in the body, while pathogenesis studies suggest that it is a live but uncultured infection. From a clinical perspective, persistent infection is defined as an asymptomatic infection. The incidence of persistent recurrent chlamydial infection cannot be fully estimated because live chlamydia are difficult to culture (Moulder et al., 1980). C. trachomatis has successfully evolved mechanisms to avoid death by autophagy and the host immune mechanisms to persist within host epithelial cells. In persistent chlamydia infection, biosynthesis of membrane and structural proteins, as well as lipopolysaccharides (LPS) stops, while stress-induced production of chlamydial 60-kDa heat shock protein (Hsp60) increases. In addition, synthesis of MOMP, which is a potentially protective antigen, decreases in persistent infections, while levels of the 57-kDa chlamydial heat shock protein (Hsp57) increase, which may contribute to the pathogenesis of the disease. The expression of chlamydial abnormal bodies includes outer membrane proteins, lower levels of LPS, and Hsp60. Hsp60 stimulates production of inflammatory cytokines and the oxidation of low-density lipoproteins (LDLs). Anti-Hsp60 immune reactivities are associated with the pathological consequence of chlamydial infections (LaVerda et al., 1999; Kinnunen et al., 2001; Witkin et al., 2017). RBs stop dividing, after which they turn into ABs with an obviously larger size and they do not produce a new parent (EB). The AB state is a reversible state that occurs in adverse growth conditions, in which C. trachomatis stops replicating and has a reduced capacity to proliferate and metabolize. It has continued chromosomal replication and active biosynthetic processes. RBs enter a persistent non-replicative but viable state under unfavorable conditions. The infectious form of the organism, the EB, is again generated when immune attack subsides (Witkin et al., 2017).
C. trachomatis is highly dependent on the biochemical processes and nutrient supply from the host cell. Growing evidence suggests that tryptophan plays a significant role in chlamydial growth and pathogenesis, which has generated interest in the regulatory mechanisms of tryptophan expression. Under conditions of tryptophan deficit, C. trachomatis can use its own synthesis to maintain tryptophan metabolism in a stable state, and when host cells are severely deficient in tryptophan, C. trachomatis activates self-protective mechanisms and enters a state of persistent infection to survive (Bommana et al., 2021) (Figure 1).
Abnormal development in chlamydial persistent infection
In chlamydial persistent infection, genes including tryptophan synthesis (trpR, trpB, and trpA), phospholipid biosynthesis (lpd, lpxK, lipA and pgsA.2), DNA repair and recombination (xerC, dnaB, recA and yqgF), translation (trmD, rl21, fusA, rs19, tsf, efp.2, ytgB.2, infC) and many early cycle genes (incD-G and euo) were highly upregulated. FtsW (a chaperone of PBP 3) and AmiA (an amidase) were significantly down-regulated, suggesting that cell division was blocked in persistent infection. In addition, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was involved in many crucial pathways in bacteria. The expression of gapA (GAPDH) changed in the presence of IFN-γ. GAPDH was an essential enzyme in the glycolytic and pentose phosphate pathway that reduced NAD+ to NADH by oxidizing glyceraldehyde-3-phosphate (GAP) to 1,3-diphosphoglycerate. Previous studies have shown that the expression of chlamydial GAPDH was associated with colonization, pathogenesis, virulence and chlamydial growth. Furthermore, GAPDH functioned in both phases of the biphasic life cycle of C. trachomatis (Belland et al., 2003; Barrett et al., 2020; Schormann et al., 2020).
The protein product of euo has been thought to repress late transcription and its expression was significantly up-regulated in persistent C. trachomatis, upon removal of IFN-γ, the product was significantly decreased. Nevertheless, increased expression of euo was currently thought to be correlated with glucose deprivation, but not persistence (Belland et al., 2003).
Furthermore, pH2AX, H3K9me3, gH2AX induced by DNA double-strand breaks (DSBs) and senescence-associated heterochromatin foci (SAHF) showed upregulation during infection. Dysregulation of host cell signal, disrupting host chromatin, repair of DSBs and regulation of cell cycle were likely to form malignant transformation (Chumduri et al., 2013).
Biology of tryptophan
The tryptophan synthase operon
C. trachomatis has developed a mechanism to evade host immune defenses by retaining a subset of trp genes, encoded in the PZ: trpA and trpB, encoding homologs of the α (TrpA) and β (TrpB) subunits of TS; and trpR, encoding a tryptophan repressor (Carlson et al., 2006; Aiyar et al., 2014; Ziklo et al., 2016a). TrpRBA are located in the PZ of the C. trachomatis genome, which is approximately 50 kb in length and accounts for most of the sequence variations in C. trachomatis strains (O'Neill et al., 2018). TrpAB is a bifunctional enzyme catalyzing the final two steps. The enzyme is a heterotetramer consisting of two α subunits and two β subunits (αββα) (McClarty et al., 2007). Upregulation of the trpBA operon converts indole to tryptophan. TrpR binds an operator sequence (trpO) and inhibits trpBA transcription. When sufficient tryptophan is present, trpR inhibits transcription of the tryptophan operon and transcription of other additional genes, including trpR itself, aroH, aroL, and mtr. In the presence of low tryptophan, trpR is released from the trpO to stimulate gene expression (Wood et al., 2003; Akers and Tan, 2006; Banerjee and Nelson, 2019).
In C. trachomatis, catalytically impaired TrpA is an allosteric regulator of TrpB. TrpA can improve TrpB activity by four-fold. TrpAB activity is influenced to promote the open state by monovalent cations, such as NH4+, Cs+, K+, and Na+ (Michalska et al., 2021).
TrpA (subunit α) is a TIM-barrel, whereas TrpB (subunit β) consists of two 3-layer structural domains (αβα). Further, TrpA catalyzes the α reaction, converting indole-3-glycerol phosphate into indole and glyceraldehyde-3-phosphate. Indole is subsequently utilized by TrpB. TrpB catalyzes the β-replacement reaction, in which indole takes the place of the hydroxyl radical of serine into tryptophan. Within each αβ heterodimer, subunit α and subunit β act independently but in a highly coordinated fashion. Both subunits perform two functions: allosteric communication and catalysis (Dunn, 2012; Michalska et al., 2021).
Sherchand et al. have demonstrated that the indole metabolites, indole-3-propionic acid (IPA) and indole-β-acrylic acid (IAA), compete with tryptophan for binding to TrpR to inhibit its binding to TrpO. IPA and IAA are deleterious to urogenital C. trachomatis through de-repressing trpRBA Expression and indole-producing microbiomes prevent chlamydial immune clearance by TS. The presence of indole allows tryptophan biosynthesis to evade starvation and blocks serine deamination (Sherchand and Aiyar, 2019).
Tryptophan synthesis
Tryptophan is an essential aromatic amino acid in the growth and development of C. trachomatis. Among the 20 common essential amino acids, tryptophan is the least abundant by protein content and the largest by molecular weight. Tryptophan is unique in that it is specified by just one codon. The amount of tryptophan is generally low, which is biochemically the most expensive to synthesize in vivo (Lo et al., 2012). Some organisms can synthesize tryptophan de novo via the successive action of a series of enzymes (Wood et al., 2003). The entire tryptophan synthetic pathway requires the sequential action of seven enzymes (TrpA-TrpG) and 78 ATP molecules. Given the high metabolic burden, the pathway is strictly controlled in multiple aspects (Michalska et al., 2021). Comparative genomics of C. pecorum has revealed that C. pecorum possesses a near complete tryptophan operon (trpRDCAB), and C. pecorum isolates can escape IFN-γ effects by depleting tryptophan expression in human epithelial cells (Islam et al., 2018). Most of the sequenced chlamydial genomes do not contain the full complement of genes required to synthesize tryptophan, six (trpA, B, C, D, F, R) in C. psittaci, four (trpA, B, F, R) in C. trachomatis serovar D, one (trpF) in C. muridarum, and none in C. pneumoniae. Different susceptibilities of chlamydial strains to IFN-γ may be due to variations in tryptophan synthesis. As an infant passes through the obstetric canal, genital C. trachomatis can also infect the conjunctivae, which leads to a self-limiting conjunctivitis rather than trachoma (Caldwell et al., 2003). This suggests that genital strains are highly sensitive to the inhibitory effects of IFN-γ in the ocular environment, perhaps because there is no exogenous indole.
Tryptophan metabolism
Tryptophan metabolism follows three major pathways: 1) tryptophan is directly converted into several molecules, including the ligands for the aryl hydrocarbon receptor (AhR); 2) tryptophan is transformed into kynurenine (kynurenine pathway (KP)) via IDO in both immune and epithelial cells; and 3) tryptophan is metabolized to the serotonin (5-hydroxytryptamine (5-HT)) production pathway via tryptophan hydroxylase 1 (TpH1) in enterochromaffin cells (Agus et al., 2018). Metabolic tryptophan pathways, including the 5-HT, KP, and AhR pathways, play an important role in physiology and pathophysiology.
Tryptophan starvation contributes to C. trachomatis persistence
C. trachomatis regulates the tryptophan biosynthetic pathway by encoding trp genes. When the host limits the bioavailability of tryptophan through defense mechanisms, C. trachomatis can use this mechanism to maintain its own tryptophan metabolism at a steady state, and when host cells are severely deficient in tryptophan, C. trachomatis turns on self-protective mechanisms to maintain its integrity and survival by entering a state of persistent infection. The most prominent feature of the AB phenotype is an increase of TrpA and TrpB levels from under 0.05% of total protein in EBs to 9% in ABs. The differentiation process of EBs to RBs, and RBs to ABs is highly dependent on tryptophan, indicating that TrpAB may have an advantage in infection. Furthermore, the total tryptophan content in the AB form is 1.9-fold lower compared to the EB form (Ostergaard et al., 2016).
Jordan et al. have demonstrated significantly lower tryptophan levels and a trend towards lower IFN-γ levels in women who cleared their infection compared to others with persistent infection. High levels of cervicovaginal tryptophan were detected in most women who had spontaneously cleared their chlamydial infection (Jordan et al., 2017).
C. trachomatis is auxotrophic for tryptophan and must obtain the essential amino acid from the external environment. C. trachomatis is an obligate intracellular bacterium which scavenges tryptophan from the host. These bacteria obtain this essential amino acid from host cells through a pool of free amino acids in the cytoplasm or by ingesting enzymatic degradation products. Deprivation of tryptophan represses chlamydial replication and prevents the spread of infection by blocking the differentiation of RBs into EBs, making tryptophan synthesis a pathogenic strategy for evading the host defenses within an IFN-γ-rich environment (Ouellette et al., 2021).
Ibana et al. have demonstrated that IDO1 inhibitor 1-methyl-Trp (L-1-MT) limited the number of EBs, reduced the numbers of inclusions, blocked IFN-γ-mediated persistence of C. trachomatis, enhanced efficacy of chlamydial antibiotic clearance and increased the susceptibility of persistent infection in response to doxycycline treatment. Adding L-1-MT to reactivate the persistent C. trachomatis induced by IFN-γ restricted the productive multiplication of the bacterium, which might be due to the inhibition of the enzyme. Furthermore, the available tryptophan was insufficient to support normal late-stage development of bacteria (Ibana et al., 2011).
Recently, there has been increased interest into the correlation between genes encoding tryptophan codons and persistence. Our previous work and that of others have demonstrated that C. muridarum colonizes the genital tract and establishes long-lasting colonization in the intestinal tract (Hongliang et al., 2022). Urogenital C. trachomatis strains can utilize exogenous indole to synthesize tryptophan using trpBA. Several genes interact to regulate the tryptophan synthesis from exogenous indoles, and C. trachomatis can recover its infectivity from persistence by converting indole into tryptophan. If tryptophan is added, ABs can be reactivated and enter a normal growth cycle (Ostergaard et al., 2016; Michalska et al., 2021). Depletion of tryptophan may lead to inhibition of T cell proliferation and increased cell apoptosis (Chen, 2011).
Iron-dependent repressor YtgR regulates the gene expression of trp
The trp operon contains trpR, trpB and trpA and an intergenic region (IGR). TrpR regulates the major promoter (PtrpR), while the IGR has the iron-dependent repressor YtgR and a promoter (PtrpBA) to regulate trpBA expression. YtgR is the only known iron-dependent regulator in C. trachomatis that binds to the trpRBA promoter (PtrpRBA) and inhibits transcriptional initiation of the ptrpBA responding to iron and/or tryptophan starvation. At the same time, binding of YtgR promotes transcriptional termination of the main promoter upstream of trpR. Thus, YtgR represents an alternative strategy to attenuate trpBA expression. When iron levels are high, YtgR locks on to the DNA in the middle of this set of genes, which effectively shuts down the genes on either side of the binding site. When iron levels drop, YtgR is released from the DNA, allowing renewed tryptophan synthesis (Pokorzynski et al., 2019; Pokorzynski et al., 2020).
The decrease in YtgR levels activates trpBA expression from PtrpBA, which is regarded as an alternative attenuation strategy in tryptophan regulated prokaryotes. Tryptophan deprivation limits expression of the YtgR repressor to activate transcription from the alternative YtgR-regulated promoter for trpBA. This regulatory mechanism parallels cis attenuation by trpL in Escherichia coli (E. coli), though notable differences exist: namely, this attenuation mechanism is mediated by a trans-acting transcription factor (Pokorzynski et al., 2020).
Links between tryptophan/IFN-γ axis and C. trachomatis persistence
One of the most important cellular immune responses in the development of protective immunity against chlamydial infection is characterized by antigen-specific IFN-γ- secreting CD4+ and CD8+ T cells. IFN-γ is a critical cytokine in innate and adaptive immune responses against a broad spectrum of bacterial and viral infections, including C. trachomatis (Belland et al., 2003; McClarty et al., 2007; Ziklo et al., 2016a). While high IFN-γ levels were shown to eradicate the infection, lower levels can also drive urogenital C. trachomatis to enter their persistence form, characterized by aberrant, non-infectious bodies in vitro (Ziklo et al., 2018; Ziklo et al., 2019). IFN-γ is a major part of the anti-chlamydial immune response and the most important factor.
One hypothesis suggests that indole-producing bacteria might play an important part in C. trachomatis infections, allowing C. trachomatis to counteract the IFN-γ mediated immune response by providing indole. C. trachomatis bacteria and humans do not produce indole, but microorganisms that colonize the genital and gastrointestinal tract do (Muramatsu et al., 2016). Indole-producing microorganisms such as various anaerobic vaginal floras, including but not limited to, E. coli, Peptostreptococcus spp., Fusobacterium spp., Bacteroides spp., are likely to be widespread in the genital and gastrointestinal tracts, which could be a direct source of indole or tryptophan. Interestingly, indole-producing bacteria are more prevalent in women who experience bacterial vaginosis. The incidence of secondary complications of chlamydial infection increases during vaginosis co-infection with C. trachomatis (McClarty et al., 2007). Previous studies have shown that indole has been detected in genital secretions from women who were infected with C. trachomatis, and women with repeated C. trachomatis infections had significantly increased vaginal kynurenine/tryptophan ratios (Ziklo et al., 2018; Ziklo et al., 2019).
Clinical observations suggest that levels of IFN-γ in the infected intracervical microenvironment were higher than normal levels (Ibana et al., 2018). Indole generation at microbial sites allowed tryptophan production by TS, thus avoiding interferon-induced tryptophan starvation by providing exogenous indole (Sherchand and Aiyar, 2019). Lactobacillus was the dominant and protective vaginal microbiota in females, which could kill numerous vaginal pathogens, including C. trachomatis. Indole was absent or present in very small amounts in the genital secretions when Lactobacillus was dominant. However, when non-lactobacilli were predominant, such as bacterial vaginosis, indoles were easily detectable (Witkin et al., 2017). Indole was detected in the genital secretions of two infected women with C. trachomatis (Ziklo et al., 2018). Under tryptophan-deficient conditions, vaginal specimens with high indole levels had higher recovery of C. trachomatis, which provided preliminary support for the crucial effect of the interferon-γ-tryptophan-indole axis. The introduction of exogenous indoles into cell culture media could rescue urogenital C. trachomatis strains from IFN-γ and allowed them to subsequently produce infective progenies (Ziklo et al., 2016b).
The anti-chlamydial effects of IFN-γ in humans include the induction of nitric oxide from arginine by nitric oxide synthase, thereby triggering iron deprivation as well as conversion of L- tryptophan into L-formyl kynurenine via IDO. L-formyl kynurenine has a wide range of potential roles in the host, such as a precursor to NAD(P) or conversion to acetyl-coenzyme A (acetyl-CoA) in the mitochondria. In addition, IFN-γ strongly induces synthesis of host tryptophanyl-tRNA synthetase, whose elevated level not only favors host access but also tends to sequester the already diminished tryptophan pool away from parasitic metabolism, IFN-γ-induced synthesis of host tryptophan-tRNA synthase not only facilitates host entry but also tends to isolate the already reduced tryptophan pool away from parasitic metabolism (Lo et al., 2012).
IDO is a key protein in human intracellular immune defense against C. trachomatis, which limits the growth of the tryptophan auxotroph. Therefore, the enzyme restricts C. trachomatis growth. The consequences of tryptophan starvation for C. trachomatis are a double-edged sword: the RBs may die leading to clearance of the infection, or the RBs may become persistent (Aiyar et al., 2014; Witkin et al., 2017; Banerjee and Nelson, 2019). C. trachomatis may use TS to synthesize tryptophan inside the inclusion, but IDO cannot enter the inclusion (Banerjee and Nelson, 2019).
The aryl hydrocarbon receptor—The downstream product of tryptophan binds to this receptor
The AhR was first characterized as a protein binder and mediator of the toxic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD, dioxin) and dioxin-like compounds (DLCs). AhR is a cytoplasmic receptor and transcription factor, which is activated primarily by binding to a cognate ligand. AhR is vital for a broad array of immune functions. AhR is also an intracellular receptor of the basic helix-loop-helix (bHLH) gene family. Subsequent studies have demonstrated that the AhR plays a pivotal role in maintaining cellular homeostasis and in pathophysiology. The AhR can bind structurally diverse compounds, which may function as endogenous ligands (Beischlag et al., 2008; Avilla et al., 2020). AhR is thought to be an important molecule in various cellular processes, such as tumorigenesis, embryogenesis, and inflammation. However, the molecular mechanisms of AhR in persistent infections is unknown (Murray et al., 2014).
It is an established hypothesis in the field, C. trachomatis infection triggers an IFN-γ response that upregulates IDO1 and IDO1 depletes tryptophan to kynurenine. Kynurenine has been shown to be an important scavenger for free radicals in vitro, facilitating the pathogen’s resistance to host-induced oxidative stress; and kynurenine has been demonstrated to affect the morphology of epithelium, which may provide a more suitable environment for chlamydial to infection (Bellmann-Weiler et al., 2010). The establishment of C. trachomatis model revealed that IDO-mediated effects not only inhibited the growth of C. trachomatis, but might be also involved in the persistence of this pathogen (MacKenzie et al., 2007). Belladonna et al. found that tolerogenic dendritic cells (DCs) could grant suppressive ability in an IDO-dependent fashion to immunogenic DCs, and kynurenine—a downstream metabolic product of tryptophan—can initiate DC-dependent inhibition of T cell growth independent of tryptophan deprivation. (Belladonna et al., 2006).
Kynurenine is an intermediate affinity AhR ligand that has been implicated in regulatory T cell (Treg) functional maturation and suppression of inflammatory cytokine production in dendritic cells. Moreover, several downstream Kyn metabolites such as kynurenic acid, xanthurenic acid, and cinnabarinic acid are potent AhR ligands that have been implicated in modulating cancer cell migration and cytokine production in mouse and human T cells. Tryptophan can also be altered by oxidative reactions to the AhR ligands 2-(1’H-indole-30-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE) and 6- formylindolo[3,2-b] carbazole (FICZ) in mouse and human cells (Shinde and McGaha, 2018; Platten et al., 2019).
Once bound to the ligand, AhR is activated by a conformational change exposing the nuclear localization signal (NLS), which acts as a transcription factor that affects gene expression via recruitment of coactivators and corepressors of targeted DNA regions, promoter binding, and cellular signal transduction mechanisms (Sun et al., 2020). AhR functions by interacting with various ligands, including PAS heterodimerization partners AhR nuclear translocator (ARNT), chaperone proteins, and immunophilin-like proteins, including heat shock protein 90 (Hsp90) and AhR-Interacting Protein p23. The complex releases Hsp90 allowing the receptor to translocate to the nucleus. Subsequently, activated AhR and ARNT form a heterodimer, which binds to the xenobiotic response element (XRE) leading to transcriptional changes in ligand-metabolizing enzymes (CYP1A1, CYP1A2, CYP1B1), immunoregulatory and growth factors (IL-10, ARG1, IL-6, IL-22, VEGF), as well as the negative regulator of the AhR pathway (AHRR) (Avilla et al., 2020; Safe et al., 2020; Sun et al., 2020) (Figure 2).
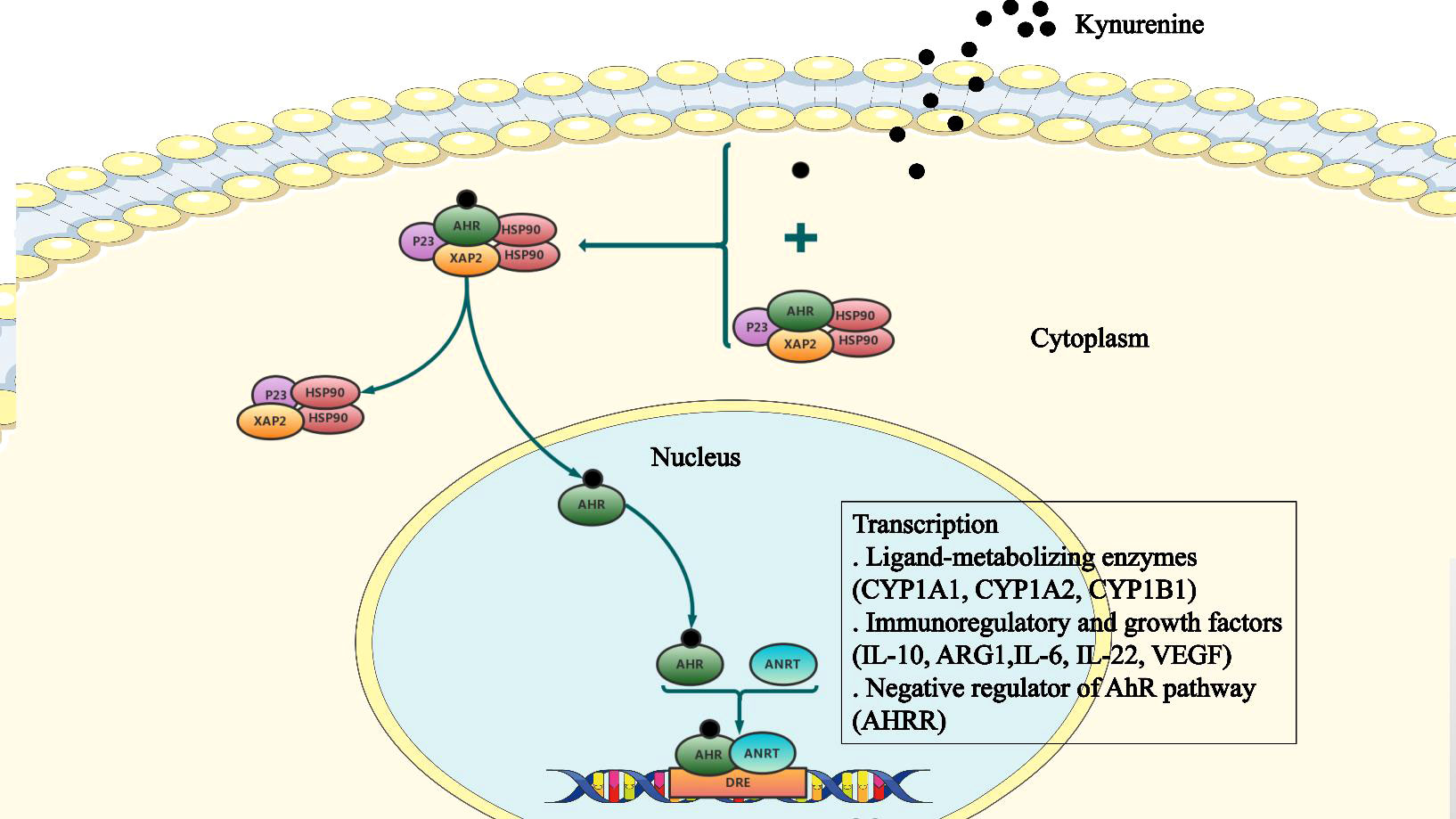
Figure 2 The cellular AhR signaling pathway. The aryl hydrocarbon receptor (AhR) binds to a complex of Hsp90, X-associated protein-2 (XAP2), and P23, which is present in the cytoplasm in a dormant state. Following ligand binding, AhR undergoes conformation changes, the nuclear localization signal (NLS) is exposed, and Hsp90 is released from the complex. Subsequently, AhR forms a heterodimer with the AhR nuclear translocator (ARNT). This heterodimer binds to the xenobiotic response element (XRE) to alter expression of downstream target genes.
AhR is a key factor in limiting macrophage mediated inflammation and may play a dominant role in macrophage polarization and induction of suppressive immune responses. AhR also controls the differentiation of dendritic cells from monocyte precursors, which function as a vital regulator of inflammatory potential for dendritic cells. Moreover, IFN-γ increases the expression and activity of IDO, and in macrophages and dendritic cells may increase kynurenine synthesis, which induces TGF-β1 production and limits the inflammatory response through a feedback mechanism (Shinde and McGaha, 2018).
The tryptophan metabolic pathway promotes C. trachomatis persistence via modulation of host immune responses
Chlamydial infection provokes an IFN-γ response resulting in IDO production. Importantly, IDO inhibits chlamydial growth by converting tryptophan to kynurenine to deplete host tryptophan pools. IFN-γ is also thought to act indirectly on C. trachomatis cells to limit the tryptophan required for C. trachomatis by activating IDO (Lo et al., 2012; Murray and McKay, 2021; Ouellette et al., 2021). Genital C. trachomatis strains can overcome this tryptophan issue by encoding an enzyme for tryptophan synthesis. Leonhardt et al. have found that a proportion of bacteria are unable to enter the persistent state at very low tryptophan levels (Leonhardt et al., 2007).
Tryptophan is essential for T cell growth and proliferation. A metabolic pathway can fine tune host mucosal responses according to tryptophan catabolism to maintain the evolution of immune physiology, through regulation of AhR-mediated transcription of IL-22. AhR facilitates immune homeostasis, which plays an antibacterial role by regulating AhR-mediated transcription of IL-22, and it also confers anti-inflammatory effects by mediating Treg differentiation (Zelante et al., 2013).
Tryptophan depletion can cause T cells to arrest in the G1 phase of the cell cycle leading to reduced proliferation. Moreover, tryptophan catabolites can induce T cell apoptosis. Kynurenine itself and kynurenine derivatives can stimulate primary naive T cells to differentiate to Tregs and promote subsequent development of Tregs via the AhR pathway. IDO can function as a signal transducer to promote the tolerogenic phenotype in plasmacytoid dendritic cells (pDCs) and pDC-induced forkhead box P3 (FoxP3) in the presence of transforming growth factor β (TGF-β) (Chen, 2011; Ziklo et al., 2019).
In addition, products of IDO can induce FoxP3 expression (an essential transcription factor for Treg immunosuppression), promote Treg synthesis, block Th17 cell proliferation and regulation of Th17 cell differentiation. Th17 cells can synthesize IL-17 and they are implicated in host defenses as well as chronic inflammatory diseases. There is also interesting data demonstrating an association between IDO enzyme activity, Th17 differentiation, and Treg from primary naive T cells (Favre et al., 2010).
Conclusions
In this review, we have described the relationship between tryptophan and C. trachomatis, as well as the mechanism by which IFN-γ functions as a protective cytokine against chlamydial infections. We have also discussed the role of IFN-γ-mediated chlamydial virulence responses, and the ways in which different genetic host predispositions vary between host cell types. The protective effect of IFN-γ is due to catabolism of tryptophan by IDO. Tryptophan starvation can stimulate a viable but non-culturable persistent phenotype in C. trachomatis that can be reactivated when the IFN-γ response is diminished and/or tryptophan is available. Therefore, overcoming tryptophan starvation may provide a mechanism by which C. trachomatis can cause persistent and chronic infection, despite the presence of IFN-γ. Genomic analyses suggest that genital C. trachomatis can convert indoles into tryptophan to maintain selective pressure. In contrast, ocular isolates cannot synthesize tryptophan. Given that the trpBA gene is present in the PZ, indole availability may be a selective factor to allow indole salvage.
Faced with similar IFN-γ stress as genital strains, it is important to understand why ocular strains have lost TS activity and what other biological differences may exist that may contribute to tissue tropism and disease pathogenesis. In the meantime, further studies are needed to characterize the microbial environment of the reproductive and gastrointestinal tract during chlamydial infection to assess the full impact of the TS gene on chlamydial diseases pathogenesis.
A more comprehensive understanding of the role of tryptophan in mediating C. trachomatis infection and pathogenesis and the differences among women in factors affecting clinical outcomes is an important goal of future research. Using such knowledge will aid the development of more personalized and targeted treatments to reduce incidence of adverse outcomes.
Author contributions
Conceptualization: LW and HC. Writing—original draft preparation: LW. Writing—review and editing: LW, YH, HY and HC. funding acquisition: HC. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Hunan Provincial Natural Science Foundation of China (2021JJ70002) and Chenzhou Science and technology Bureau (zdyf 2020035 and yfzx201908).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agus, A., Planchais, J., Sokol, H. (2018). Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 23 (6), 716–724. doi: 10.1016/j.chom.2018.05.003
Aiyar, A., Quayle, A. J., Buckner, L. R., Sherchand, S. P., Chang, T. L., Zea, A. H., et al. (2014). Influence of the tryptophan-indole-IFNgamma axis on human genital chlamydia trachomatis infection: role of vaginal co-infections. Front. Cell Infect. Microbiol. 4. doi: 10.3389/fcimb.2014.00072
Akers, J. C., Tan, M. (2006). Molecular mechanism of tryptophan-dependent transcriptional regulation in chlamydia trachomatis. J. Bacteriol. 188 (12), 4236–4243. doi: 10.1128/JB.01660-05
Avilla, M. N., Malecki, K. M. C., Hahn, M. E., Wilson, R. H., Bradfield, C. A. (2020). The ah receptor: Adaptive metabolism, ligand diversity, and the xenokine model. Chem. Res. Toxicol. 33 (4), 860–879. doi: 10.1021/acs.chemrestox.9b00476
Banerjee, A., Nelson, D. E. (2019). How chlamydia trachomatis conquered gut microbiome-derived antimicrobial compounds and found a new home in the eye. Proc. Natl. Acad. Sci. U. S. A. 116 (25), 12136–12138. doi: 10.1073/pnas.1907647116
Barrett, K. F., Dranow, D. M., Phan, I. Q., Michaels, S. A., Shaheen, S., Navaluna, E. D., et al. (2020). Structures of glyceraldehyde 3-phosphate dehydrogenase in neisseria gonorrhoeae and chlamydia trachomatis. Protein Sci. 29 (3), 768–778. doi: 10.1002/pro.3824
Bayramova, F., Jacquier, N., Greub, G. (2018). Insight in the biology of chlamydia-related bacteria. Microbes Infect. 20 (7-8), 432–440. doi: 10.1016/j.micinf.2017.11.008
Beischlag, T. V., Luis Morales, J., Hollingshead, B. D., Perdew, G. H. (2008). The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 18 (3), 207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20
Belladonna, M. L., Grohmann, U., Guidetti, P., Volpi, C., Bianchi, R., Fioretti, M. C., et al. (2006). Kynurenine pathway enzymes in dendritic cells initiate tolerogenesis in the absence of functional IDO. J. Immunol. 177 (1), 130–137. doi: 10.4049/jimmunol.177.1.130
Belland, R. J., Nelson, D. E., Virok, D., Crane, D. D., Hogan, D., Sturdevant, D., et al. (2003). Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. U. S. A. 100 (26), 15971–15976. doi: 10.1073/pnas.2535394100
Bellmann-Weiler, R., Martinz, V., Kurz, K., Engl, S., Feistritzer, C., Fuchs, D., et al. (2010). Divergent modulation of chlamydia pneumoniae infection cycle in human monocytic and endothelial cells by iron, tryptophan availability and interferon gamma. Immunobiology 215 (9-10), 842–848. doi: 10.1016/j.imbio.2010.05.021
Bommana, S., Somboonna, N., Richards, G., Tarazkar, M., Dean, D. (2021). Tryptophan operon diversity reveals evolutionary trends among geographically disparate chlamydia trachomatis ocular and urogenital strains affecting tryptophan repressor and synthase function. mBio 12 (3), e00605-21. doi: 10.1128/mBio.00605-21
Bowden, K. E., Joseph, S. J., Cartee, J. C., Ziklo, N., Danavall, D., Raphael, B. H., et al. (2021). Whole-genome enrichment and sequencing of chlamydia trachomatis directly from patient clinical vaginal and rectal swabs. mSphere 6 (2), e01302-20. doi: 10.1128/mSphere.01302-20
Caldwell, H. D., Wood, H., Crane, D., Bailey, R., Jones, R. B., Mabey, D., et al. (2003). Polymorphisms in chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 111 (11), 1757–1769. doi: 10.1172/jci200317993
Carlson, J. H., Hughes, S., Hogan, D., Cieplak, G., Sturdevant, D. E., McClarty, G., et al. (2004). Polymorphisms in the chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect. Immun. 72 (12), 7063–7072. doi: 10.1128/IAI.72.12.7063-7072.2004
Carlson, J. H., Porcella, S. F., McClarty, G., Caldwell, H. D. (2005). Comparative genomic analysis of chlamydia trachomatis oculotropic and genitotropic strains. Infect. Immun. 73 (10), 6407–6418. doi: 10.1128/IAI.73.10.6407-6418.2005
Carlson, J. H., Wood, H., Roshick, C., Caldwell, H. D., McClarty, G. (2006). In vivo and in vitro studies of chlamydia trachomatis TrpR:DNA interactions. Mol. Microbiol. 59 (6), 1678–1691. doi: 10.1111/j.1365-2958.2006.05045.x
Casillas-Vega, N., Morfin-Otero, R., Garcia, S., Llaca-Diaz, J., Rodriguez-Noriega, E., Camacho-Ortiz, A., et al. (2017). Frequency and genotypes of chlamydia trachomatis in patients attending the obstetrics and gynecology clinics in jalisco, Mexico and correlation with sociodemographic, behavioral, and biological factors. BMC Womens Health 17 (1), 83. doi: 10.1186/s12905-017-0428-5
Chumduri, C., Gurumurthy, R. K., Zadora, P. K., Mi, Y., Meyer, T. F. (2013). Chlamydia infection promotes host DNA damage and proliferation but impairs the DNA damage response. Cell Host Microbe 13 (6), 746–758. doi: 10.1016/j.chom.2013.05.010
den Hartog, J. E., Morre, S. A., Land, J. A. (2006). Chlamydia trachomatis-associated tubal factor subfertility: Immunogenetic aspects and serological screening. Hum. Reprod. Update 12 (6), 719–730. doi: 10.1093/humupd/dml030
Dunn, M. F. (2012). Allosteric regulation of substrate channeling and catalysis in the tryptophan synthase bienzyme complex. Arch. Biochem. Biophys. 519 (2), 154–166. doi: 10.1016/j.abb.2012.01.016
Favre, D., Mold, J., Hunt, P. W., Kanwar, B., Loke, P., Seu, L., et al. (2010). Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci. Transl. Med. 2 (32), 32ra36. doi: 10.1126/scitranslmed.3000632
Hongliang, C., Min, S., Wang, L, Zhao, L., Luo, F., Lei, W., et al. (2022). Lactobacillus modulates chlamydial infectivity and genital tract pathology in vitro and in vivo. Front. Microbiol. 13, 877223. doi: 10.3389/fmicb.2022.877223
Howe, S. E., Shillova, N., Konjufca, V. (2019). Dissemination of chlamydia from the reproductive tract to the gastro-intestinal tract occurs in stages and relies on chlamydia transport by host cells. PloS Pathog. 15 (12), e1008207. doi: 10.1371/journal.ppat.1008207
Hua, Z., Frohlich, K. M., Zhang, Y., Feng, X., Zhang, J., Shen, L. (2015). Andrographolide inhibits intracellular chlamydia trachomatis multiplication and reduces secretion of proinflammatory mediators produced by human epithelial cells. Pathog. Dis. 73 (1), 1–11. doi: 10.1093/femspd/ftu022
Ibana, J. A., Belland, R. J., Zea, A. H., Schust, D. J., Nagamatsu, T., AbdelRahman, Y. M., et al. (2011). Inhibition of indoleamine 2,3-dioxygenase activity by levo-1-methyl tryptophan blocks gamma interferon-induced chlamydia trachomatis persistence in human epithelial cells. Infect. Immun. 79 (11), 4425–4437. doi: 10.1128/IAI.05659-11
Ibana, J. A., Sherchand, S. P., Fontanilla, F. L., Nagamatsu, T., Schust, D. J., Quayle, A. J., et al. (2018). Chlamydia trachomatis-infected cells and uninfected-bystander cells exhibit diametrically opposed responses to interferon gamma. Sci. Rep. 8 (1), 8476. doi: 10.1038/s41598-018-26765-y
Islam, M. M., Jelocnik, M., Huston, W. M., Timms, P., Polkinghorne, A. (2018). Characterization of the in vitro chlamydia pecorum response to gamma interferon. Infect. Immun. 86 (4), e00714-17. doi: 10.1128/IAI.00714-17
Jonsson, S., Oda, H., Lundin, E., Olsson, J., Idahl, A. (2018). Chlamydia trachomatis, chlamydial heat shock protein 60 and anti-chlamydial antibodies in women with epithelial ovarian tumors. Transl. Oncol. 11 (2), 546–551. doi: 10.1016/j.tranon.2018.02.008
Jordan, S. J., Olson, K. M., Barnes, S., Wilson, L. S., Berryhill, T. F., Bakshi, R., et al. (2017). Lower levels of cervicovaginal tryptophan are associated with natural clearance of chlamydia in women. J. Infect. Dis. 215 (12), 1888–1892. doi: 10.1093/infdis/jix240
Juliana, N. C. A., Omar, A. M., Pleijster, J., Aftab, F., Uijldert, N. B., Ali, S. M., et al. (2021). The natural course of chlamydia trachomatis, neisseria gonorrhoeae, trichomonas vaginalis, and mycoplasma genitalium in pregnant and post-delivery women in pemba island, Tanzania. Microorganisms 9 (6), 1180. doi: 10.3390/microorganisms9061180
Kinnunen, A., Paavonen, J., Surcel, H. M. (2001). Heat shock protein 60 specific T-cell response in chlamydial infections. Scand. J. Immunol. 54 (1-2), 76–81. doi: 10.1046/j.1365-3083.2001.00940.x
Koster, S., Gurumurthy, R. K., Kumar, N., Prakash, P. G., Dhanraj, J., Bayer, S., et al. (2022). Modelling chlamydia and HPV co-infection in patient-derived ectocervix organoids reveals distinct cellular reprogramming. Nat. Commun. 13 (1), 1030. doi: 10.1038/s41467-022-28569-1
LaVerda, D., Kalayoglu, M. V., Byrne, G. I. (1999). Chlamydial heat shock proteins and disease pathology: new paradigms for old problems? Infect. Dis. Obstet Gynecol 7 (1-2), 64–71. doi: 10.1155/S1064744999000137
Leonhardt, R. M., Lee, S. J., Kavathas, P. B., Cresswell, P. (2007). Severe tryptophan starvation blocks onset of conventional persistence and reduces reactivation of chlamydia trachomatis. Infect. Immun. 75 (11), 5105–5117. doi: 10.1128/IAI.00668-07
Lo, C. C., Xie, G., Bonner, C. A., Jensen, R. A. (2012). The alternative translational profile that underlies the immune-evasive state of persistence in chlamydiaceae exploits differential tryptophan contents of the protein repertoire. Microbiol. Mol. Biol. Rev. 76 (2), 405–443. doi: 10.1128/MMBR.05013-11
MacKenzie, C. R., Heseler, K., Muller, A., Daubener, W. (2007). Role of indoleamine 2,3-dioxygenase in antimicrobial defence and immuno-regulation: tryptophan depletion versus production of toxic kynurenines. Curr. Drug Metab. 8 (3), 237–244. doi: 10.2174/138920007780362518
McClarty, G., Caldwell, H. D., Nelson, D. E. (2007). Chlamydial interferon gamma immune evasion influences infection tropism. Curr. Opin. Microbiol. 10 (1), 47–51. doi: 10.1016/j.mib.2006.12.003
Michalska, K., Wellington, S., Maltseva, N., Jedrzejczak, R., Selem-Mojica, N., Rosas-Becerra, L. R., et al. (2021). Catalytically impaired TrpA subunit of tryptophan synthase from chlamydia trachomatis is an allosteric regulator of TrpB. Protein Sci. 30 (9), 1904–1918. doi: 10.1002/pro.4143
Miles, E. W. (2001). Tryptophan synthase: a multienzyme complex with an intramolecular tunnel. Chem. Rec 1 (2), 140–151. doi: 10.1002/tcr.4
Morrison, S. G., Giebel, A. M., Toh, E., Banerjee, A., Nelson, D. E., Morrison, R. P. (2020). A genital infection-attenuated chlamydia muridarum mutant infects the gastrointestinal tract and protects against genital tract challenge. mBio 11 (6), e02770-20. doi: 10.1128/mBio.02770-20
Moulder, J. W., Levy, N. J., Schulman, L. P. (1980). Persistent infection of mouse fibroblasts (L cells) with chlamydia psittaci: evidence for a cryptic chlamydial form. Infect. Immun. 30 (3), 874–883. doi: 10.1128/iai.30.3.874-883.1980
Muramatsu, M. K., Brothwell, J. A., Stein, B. D., Putman, T. E., Rockey, D. D., Nelson, D. E. (2016). Beyond tryptophan synthase: Identification of genes that contribute to chlamydia trachomatis survival during gamma interferon-induced persistence and reactivation. Infect. Immun. 84 (10), 2791–2801. doi: 10.1128/IAI.00356-16
Murray, S. M., McKay, P. F. (2021). Chlamydia trachomatis: Cell biology, immunology and vaccination. Vaccine 39 (22), 2965–2975. doi: 10.1016/j.vaccine.2021.03.043
Murray, I. A., Patterson, A. D., Perdew, G. H. (2014). Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat. Rev. Cancer 14 (12), 801–814. doi: 10.1038/nrc3846
O'Neill, C. E., Skilton, R. J., Pearson, S. A., Filardo, S., Andersson, P., Clarke, I. N. (2018). Genetic transformation of a c. trachomatis ocular isolate with the functional tryptophan synthase operon confers an indole-rescuable phenotype. Front. Cell Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00434
Ostergaard, O., Follmann, F., Olsen, A. W., Heegaard, N. H., Andersen, P., Rosenkrands, I. (2016). Quantitative protein profiling of chlamydia trachomatis growth forms reveals defense strategies against tryptophan starvation. Mol. Cell Proteomics 15 (12), 3540–3550. doi: 10.1074/mcp.M116.061986
Ouellette, S. P., Hatch, N. D., Wood, N. A., Herrera, A. L., Chaussee, M. S. (2021). Codon-dependent transcriptional changes in response to tryptophan limitation in the tryptophan auxotrophic pathogens chlamydia trachomatis and streptococcus pyogenes. mSystems 6 (6), e0126921. doi: 10.1128/mSystems.01269-21
Platten, M., Nollen, E. A. A., Rohrig, U. F., Fallarino, F., Opitz, C. A. (2019). Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 18 (5), 379–401. doi: 10.1038/s41573-019-0016-5
Pokorzynski, N. D., Brinkworth, A. J., Carabeo, R. (2019). A bipartite iron-dependent transcriptional regulation of the tryptophan salvage pathway in chlamydia trachomatis. Elife 8, e42295. doi: 10.7554/eLife.42295
Pokorzynski, N. D., Hatch, N. D., Ouellette, S. P., Carabeo, R. A. (2020). The iron-dependent repressor YtgR is a tryptophan-dependent attenuator of the trpRBA operon in chlamydia trachomatis. Nat. Commun. 11 (1), 6430. doi: 10.1038/s41467-020-20181-5
Radomski, N., Einenkel, R., Muller, A., Knittler, M. R. (2016). Chlamydia-host cell interaction not only from a bird's eye view: some lessons from chlamydia psittaci. FEBS Lett. 590 (21), 3920–3940. doi: 10.1002/1873-3468.12295
Rank, R. G., Yeruva, L. (2014). Hidden in plain sight: chlamydial gastrointestinal infection and its relevance to persistence in human genital infection. Infect. Immun. 82 (4), 1362–1371. doi: 10.1128/IAI.01244-13
Redgrove, K. A., McLaughlin, E. A. (2014). The role of the immune response in chlamydia trachomatis infection of the Male genital tract: A double-edged sword. Front. Immunol. 5. doi: 10.3389/fimmu.2014.00534
Safe, S., Jin, U. H., Park, H., Chapkin, R. S., Jayaraman, A. (2020). Aryl hydrocarbon receptor (AHR) ligands as selective AHR modulators (SAhRMs). Int. J. Mol. Sci. 21 (18), 6654. doi: 10.3390/ijms21186654
Schormann, N., Campos, J., Motamed, R., Hayden, K. L., Gould, J. R., Green, T. J., et al. (2020). Chlamydia trachomatis glyceraldehyde 3-phosphate dehydrogenase: Enzyme kinetics, high-resolution crystal structure, and plasminogen binding. Protein Sci. 29 (12), 2446–2458. doi: 10.1002/pro.3975
Sherchand, S. P., Aiyar, A. (2019). Ammonia generation by tryptophan synthase drives a key genetic difference between genital and ocular chlamydia trachomatis isolates. Proc. Natl. Acad. Sci. U.S.A. 116 (25), 12468–12477. doi: 10.1073/pnas.1821652116
Shinde, R., McGaha, T. L. (2018). The aryl hydrocarbon receptor: Connecting immunity to the microenvironment. Trends Immunol. 39 (12), 1005–1020. doi: 10.1016/j.it.2018.10.010
Sixt, B. S. (2021). Host cell death during infection with chlamydia: a double-edged sword. FEMS Microbiol. Rev. 45 (1), fuaa043. doi: 10.1093/femsre/fuaa043
Sun, M., Ma, N., He, T., Johnston, L. J., Ma, X. (2020). Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit. Rev. Food Sci. Nutr. 60 (10), 1760–1768. doi: 10.1080/10408398.2019.1598334
Tamarelle, J., Thiebaut, A. C. M., de Barbeyrac, B., Bebear, C., Ravel, J., Delarocque-Astagneau, E. (2019). The vaginal microbiota and its association with human papillomavirus, chlamydia trachomatis, neisseria gonorrhoeae and mycoplasma genitalium infections: a systematic review and meta-analysis. Clin. Microbiol. Infect. 25 (1), 35–47. doi: 10.1016/j.cmi.2018.04.019
Witkin, S. S., Minis, E., Athanasiou, A., Leizer, J., Linhares, I. M. (2017). Chlamydia trachomatis: the persistent pathogen. Clin. Vaccine Immunol. 24 (10), e00203-17. doi: 10.1128/CVI.00203-17
Wood, H., Fehlner-Gardner, C., Berry, J., Fischer, E., Graham, B., Hackstadt, T., et al. (2003). Regulation of tryptophan synthase gene expression in chlamydia trachomatis. Mol. Microbiol. 49 (5), 1347–1359. doi: 10.1046/j.1365-2958.2003.03638.x
Yeow, T. C., Wong, W. F., Sabet, N. S., Sulaiman, S., Shahhosseini, F., Tan, G. M., et al. (2016). Prevalence of plasmid-bearing and plasmid-free chlamydia trachomatis infection among women who visited obstetrics and gynecology clinics in Malaysia. BMC Microbiol. 16, 45. doi: 10.1186/s12866-016-0671-1
Zelante, T., Iannitti, R. G., Cunha, C., De Luca, A., Giovannini, G., Pieraccini, G., et al. (2013). Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39 (2), 372–385. doi: 10.1016/j.immuni.2013.08.003
Ziklo, N., Huston, W. M., Hocking, J. S., Timms, P. (2016a). Chlamydia trachomatis genital tract infections: When host immune response and the microbiome collide. Trends Microbiol. 24 (9), 750–765. doi: 10.1016/j.tim.2016.05.007
Ziklo, N., Huston, W. M., Taing, K., Katouli, M., Timms, P. (2016b). In vitro rescue of genital strains of chlamydia trachomatis from interferon-gamma and tryptophan depletion with indole-positive, but not indole-negative prevotella spp. BMC Microbiol. 16 (1), 286. doi: 10.1186/s12866-016-0903-4
Ziklo, N., Huston, W. M., Taing, K., Timms, P. (2019). High expression of IDO1 and TGF-beta1 during recurrence and post infection clearance with chlamydia trachomatis, are independent of host IFN-gamma response. BMC Infect. Dis. 19 (1), 218. doi: 10.1186/s12879-019-3843-4
Keywords: Chlamydia trachomatis, tryptophan, persistent infection, interferon gamma, indoleamine-2,3-dioxygenase
Citation: Wang L, Hou Y, Yuan H and Chen H (2022) The role of tryptophan in Chlamydia trachomatis persistence. Front. Cell. Infect. Microbiol. 12:931653. doi: 10.3389/fcimb.2022.931653
Received: 29 April 2022; Accepted: 14 July 2022;
Published: 02 August 2022.
Edited by:
Subramanian Dhandayuthapani, Texas Tech University Health Sciences Center El Paso, United StatesReviewed by:
Rey Carabeo, University of Nebraska Medical Center, United StatesKevin Hybiske, University of Washington, United States
Copyright © 2022 Wang, Hou, Yuan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongliang Chen, chenhongliang2007@126.com
 Li Wang
Li Wang YingLan Hou1,2
YingLan Hou1,2  Hongliang Chen
Hongliang Chen