4-Allyl-2-methoxyphenol modulates the expression of genes involved in efflux pump, biofilm formation and sterol biosynthesis in azole resistant Aspergillus fumigatus
- 1Anti-mycotic Drug Susceptibility Laboratory, Amity Institute of Biotechnology, Amity University Uttar Pradesh, Noida, India
- 2Department of Biotechnology and Bioinformatics, Jaypee University of Information Technology, Solan, India
- 3Amity Institute of Biotechnology, Amity University Haryana, Gurugram (Manesar), India
Introduction: Antifungal therapy for aspergillosis is becoming problematic because of the toxicity of currently available drugs, biofilm formation on host surface, and increasing prevalence of azole resistance in Aspergillus fumigatus. Plants are rich source of bioactive molecules and antimicrobial activity of aromatic bioactive compounds draws attention because of its promising biological properties. The present study elucidated the antibiofilm activity of 4-allyl-2-methoxyphenol (eugenol) against azole-resistant environmental A. fumigatus isolates.
Methods: Soil samples were collected from agricultural fields across India; azole-resistant A. fumigatus (ARAF) were isolated followed by their molecular identification. Antibiofilm activity of eugenol was calculated via tetrazolium based-MTT assay. The expression of the multidrug efflux pumps genes MDR1, MDR4, transporters of the MFS gene, erg11A gene encoding 14α demethylase, and transcription regulatory genes, MedA, SomA and SrbA, involved in biofilm formation of A. fumigatus were calculated by quantitative real time PCR.
Results: Out of 89 A. fumigatus isolates, 10 were identified as azole resistant. Eugenol exhibited antibiofilm activity against ARAF isolates, ranging from 312 to 500 µg/mL. Confocal laser scanning microscopy analysis revealed absence of extracellular matrix of ARAF biofilm after eugenol treatment. The gene expression indicated significantly low expression of efflux pumps genes MDR1, MDR4, erg11A and MedA in eugenol treated ARAF isolates when compared with untreated isolates.
Conclusions: Our results demonstrate that eugenol effects the expression of efflux pump and biofilm associated genes as well as inhibits biofilm formation in azole resistant isolates of A. fumigatus.
1 Introduction
Aspergillus infections manifest as chronic pulmonary aspergillosis, invasive aspergillosis, bronchitis, allergic bronchopulmonary aspergillosis and severe asthma with fungal sensitization. Aspergillus infections can cause life-threatening diseases in individuals with weakened immunity, having underlying diseases, or organ transplant (Denning et al., 2016). Invasive manifestation of aspergillosis is fatal, with mortality rate nearly 50% when treated promptly and >80% when the treatment is delayed (Kosmidis and Denning, 2015). Aspergillus fumigatus, a saprophytic pathogenic fungus, causes majority of pulmonary infections in immunocompromised patients (Bongomin et al., 2017). Azoles are currently recommended as first-line drugs to treat A. fumigatus infections. However, a steady global increase in azole resistance in A. fumigatus, resulting in therapeutic failures, has been a matter of serious clinical concern (Ahangarkani et al., 2020a). The resistant strains can be isolated from the patients taking azole drugs for a long time as well as from the environment because of prolonged usage of azole pesticides (Chowdhary et al., 2014; Ahangarkani et al., 2020b). Triazole resistance has been reported in azole naïve patients in India without previous triazole exposure, suggesting a possible role for environmental transmission of resistance (Dabas et al., 2018). Azoles target the erg11A/cyp51A gene; therefore, azole resistance due to mutation in this gene or its overexpression has been previously reported (Price et al., 2015). Several other resistance mechanisms such as overexpression of drug efflux pumps and transporters of the major facilitator superfamily (MFS) have also been reported (Da Silva Ferreira et al., 2004; Fraczek et al., 2013).
A. fumigatus also forms biofilms, thereby increasing the severity of fungal infection (Müller et al., 2011). Biofilm formation protects the fungus from host immune system and reduces the efficacy of antifungal drug therapy leading to development of antimicrobial resistance (Mowat et al., 2007). The minimal inhibitory concentrations (MICs) of antifungals, particularly azoles against Aspergillus biofilms are predominantly high (Beauvais and Latgé, 2015). A. fumigatus biofilms exhibit drug resistance due to biofilm-specific upregulation of efflux proteins and presence of an extracellular matrix (ECM) (Manavathu et al., 2014). Multidrug resistance (MDR) efflux pumps in A. fumigatus have been described and are associated with increased resistance to itraconazole (Nascimento et al., 2003; Da Silva Ferreira et al., 2004). MDR pumps transport a variety of toxic substrates such as azole drugs from the cells. Rajendran et al., 2011 reported the upregulation of the MDR4 gene in A. fumigatus isolate and an in vivo mouse biofilm model after 24-h voriconazole treatment. This study suggested that the overexpression of the MDR4 gene in A. fumigatus biofilm is associated with azole resistance. A. fumigatus regulatory protein MedA and transcription factor SomA are involved in biofilm formation and adherence (Gravelat et al., 2010). The disruption of the MedA gene leads to impairment of conidiation and biofilm formation. The ΔmedA mutant demonstrated reduced adherence to pulmonary epithelial and endothelial cells (Gravelat et al., 2010). Therefore, the treatment of infections caused by Aspergillus biofilms is a serious clinical challenge, emphasizing the need for novel and effective therapeutic agents.
The discovery of a novel antifungal and anti-biofilm agents with ideal features, such as broad-spectrum activity, low toxicity, minimal side effects, and enhanced bioavailability, is a key priority in medical mycology research (Brauer et al., 2019). Plants are extremely rich sources of bioactive molecules with analgesic, anti-inflammatory, anti-carcinogenic, anti-mutagenic, and even repellent properties (Pramod et al., 2010; Chung et al., 2014). 4-allyl-2-methoxyphenol, also known as eugenol, is a volatile naturally occurring phenolic bioactive constituent in clove oil, nutmeg, and basil (Ferreira et al., 2017). It possesses significant antioxidant, antifungal, and anti-inflammatory properties, in addition to analgesic and local anaesthetic activity (Huang et al., 2015; Marchese et al., 2017; Goswami et al., 2022). This compound offers a number of therapeutic benefits via inhibition of generative reactive oxygen and nitrogen species, scavenging of free radicals, and disruption of microbial biofilms (Khalil et al., 2017). Eugenol inhibits Candida spp. via altering fungal cell membrane and cell wall resulting in the release of cellular contents (Bennis et al., 2004; Ahmad et al., 2010). Inhibitory effect of eugenol has been observed in the initial phase of biofilm formation as well as in the mature biofilm of Candida albicans (Khan and Ahmad, 2012). However, inhibitory effect of eugenol on environmental isolates of azole-resistant A. fumigatus has not been reported to the best of our understanding. Therefore, the present study elucidated the antibiofilm activity of 4-allyl-2-methoxyphenol (eugenol) against azole-resistant environmental A. fumigatus isolates.
2 Materials and methods
2.1 Screening for azole-resistant A. fumigatus (ARAF) isolates from soil samples
A total of 110 soil samples were collected from various agricultural fields across India. Briefly, 2 g of each soil sample was suspended in 10 mL of sterile 0.9% normal saline, vortexed, and allowed to settle for 4-5 h. From this stock suspension, 100 μL of the supernatant was inoculated on Potato Dextrose Agar (PDA) plates supplemented with 0.5 mg/mL chloramphenicol and were incubated at 28 ± 2°C for 5 days. All samples were processed as biological triplicates. All A. fumigatus isolates grown on agar plates were identified by macroscopic and microscopic characteristics (de Hoog et al., 1995). A. fumigatus isolates were further screened for azole resistance by plating the spores on PDA containing (a) no drug as control; (b) 1 μg/mL itraconazole (ITC); (c) 1 μg/mL voriconazole (VRC), and (d) 0.5 μg/mL posaconazole (PSZ) (Tangwattanachuleeporn et al., 2017). The plates were incubated at 28 ± 2°C for 5 days. A. fumigatus isolates grown on azole-containing agar plates were further subcultured and maintained on PDA plates.
2.2 Molecular identification of ARAF isolates
Genomic DNA was extracted from the ten ARAF isolates using the cetyl trimethyl ammonium bromide (Lee et al., 1988; Wu et al., 2001). Molecular identification of the isolates as A. fumigatus was confirmed by the amplification and sequencing of full-length 18S internal transcribed spacer (ITS) region using the ITS1 (5’ TCC GTA GGT GAA CCT GCGG 3’) and ITS4 (5’ TCC TCC GCT TAT TGA TATGC 3’) primers (White et al., 1990) and partial sequencing of β-tubulin gene using Tub5 (5’ TGACCCAGCAGATGTT 3’) and Tub6 (5’ GTTGTTGGGAATCCACTC 3’) primers Mellado et al., 2007; Hoda et al., 2020).
The phylogenetic tree was prepared using β-tubulin gene sequence alignments in the MEGA (version11.0.13) program, using maximum likelihood method. The support for each clade was determined by bootstrap method with 1,000 replicates.
2.3 Antifungal susceptibility testing
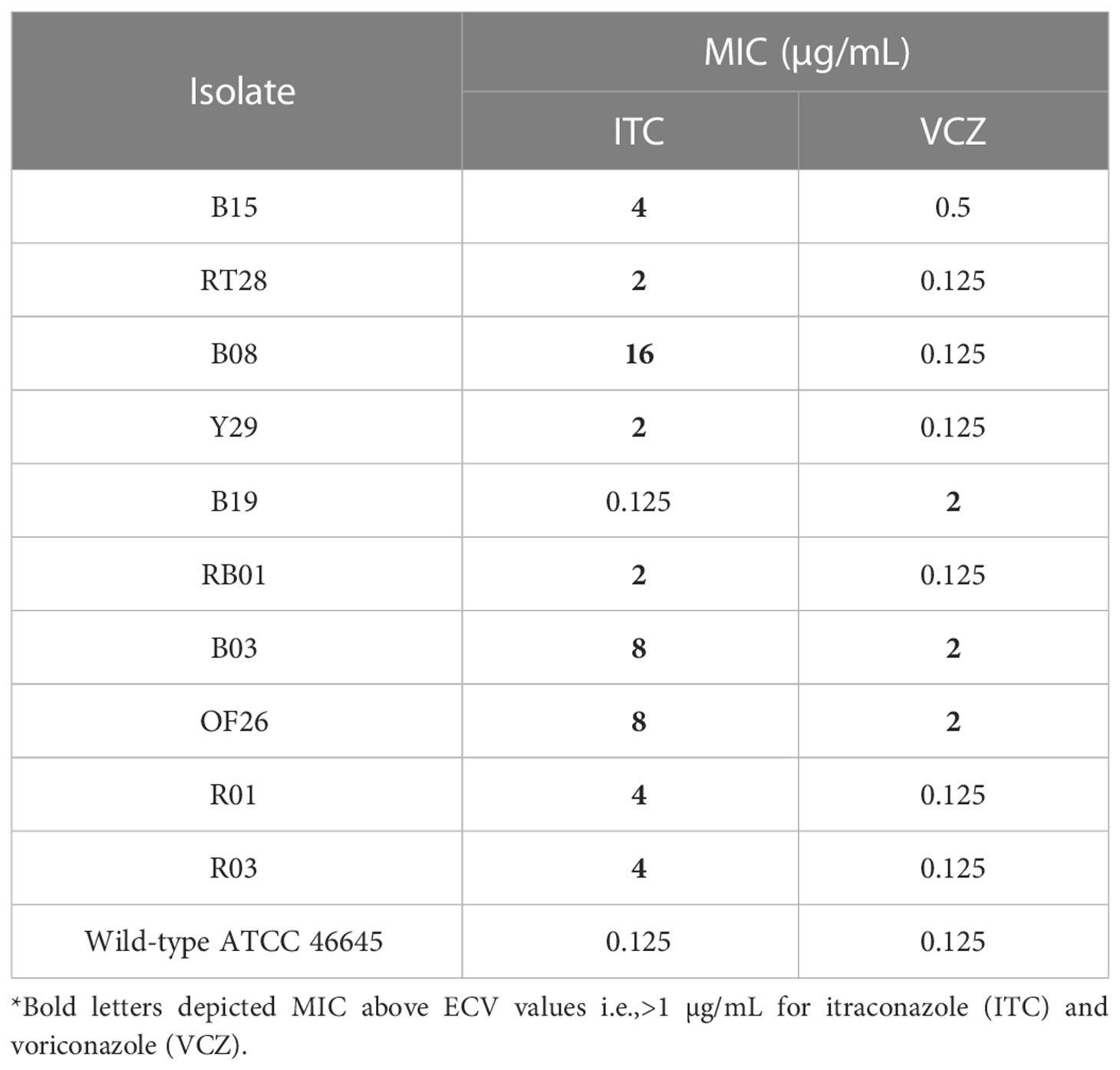
The spores (conidia) of azole-resistant and wild-type A. fumigatus were harvested in sterile phosphate buffered saline (1× PBS) supplemented with 0.05% Tween 20; suspension was adjusted to 1×104 conidia/mL in potato dextrose broth. MIC of ITC and VRC against ARAF with reference to wild-type strain of A. fumigatus (ATCC 46645) was determined using CLSI M38-A2 broth microdilution method (Alexander and Clinical and Laboratory Standards Institute, 2017) in a 96-well polystyrene plate (Tarsons, India). The experiment was carried out in triplicates. The stock of azole drugs was prepared in dimethyl sulfoxide. Two-fold dilutions were prepared in a 96-well microplate to obtain concentrations ranging from 64–0.125 µg/mL for ITC and VRC. Each well was inoculated with 100 µL of the conidial suspension except negative control. Microplate was incubated at 28 ± 2°C for 5 days, and the growth in each well was compared with that of the positive control. The MIC of a drug is determined as the lowest concentration with no visible growth relative to the drug-free control (Alexander and Clinical and Laboratory Standards Institute, 2017). The results were further analyzed with reference to epidemiological cutoff values (ECVs) for ITC: 1 μg/mL; VRC: 1 μg/mL (Pfaller et al., 2011; Lass-Flörl, 2014; CLSI, 2018).
2.4 A. fumigatus biofilm eradication activity of eugenol
The biofilm-eradication concentration (BEC80) of eugenol for a preformed fungal biofilm was calculated using MTT assay in a 96-well flat bottom microtiter plates with minor modifications (Sav et al., 2018). Conidial suspension (100 µL) in RPMI was added to each well of the flat bottom microtiter plate and incubated at 37 °C for 24 h without agitation. After 24 h, non-adherent conidia were removed by washing with 1× PBS. Thereafter, 100 μL of various concentrations of eugenol (2000–39 µg/mL) were added to the respective wells, and further incubated at 37 °C statically for an additional 24 h. Negative control wells contained only RPMI media. Wells with preformed biofilms without any treatment were considered as positive control. The effect of eugenol on the preformed fungal biofilms was estimated using a semi-quantitative viability based MTT reduction assay (Manavathu et al., 2014).
For confocal laser scanning microscopy (CLSM) analysis, the samples were processed as described by González-Ramírez et al. (2016) with minor modifications. The biofilms of ARAF isolates were cultured in a 12-well polystyrene plate at the calculated BEC80 of eugenol (Manavathu et al., 2014). Biofilm samples were stained with calcofluor white M2R (Sigma, St. Louis, MO, USA) and were analyzed using Nikon Instruments A1 Confocal Laser Microscope Series with NIS elements (C software, Japan). BEC80 is the concentration at which nearly 80% of fungal biofilm is eradicated.
2.5 Sequencing of the cyp51A gene
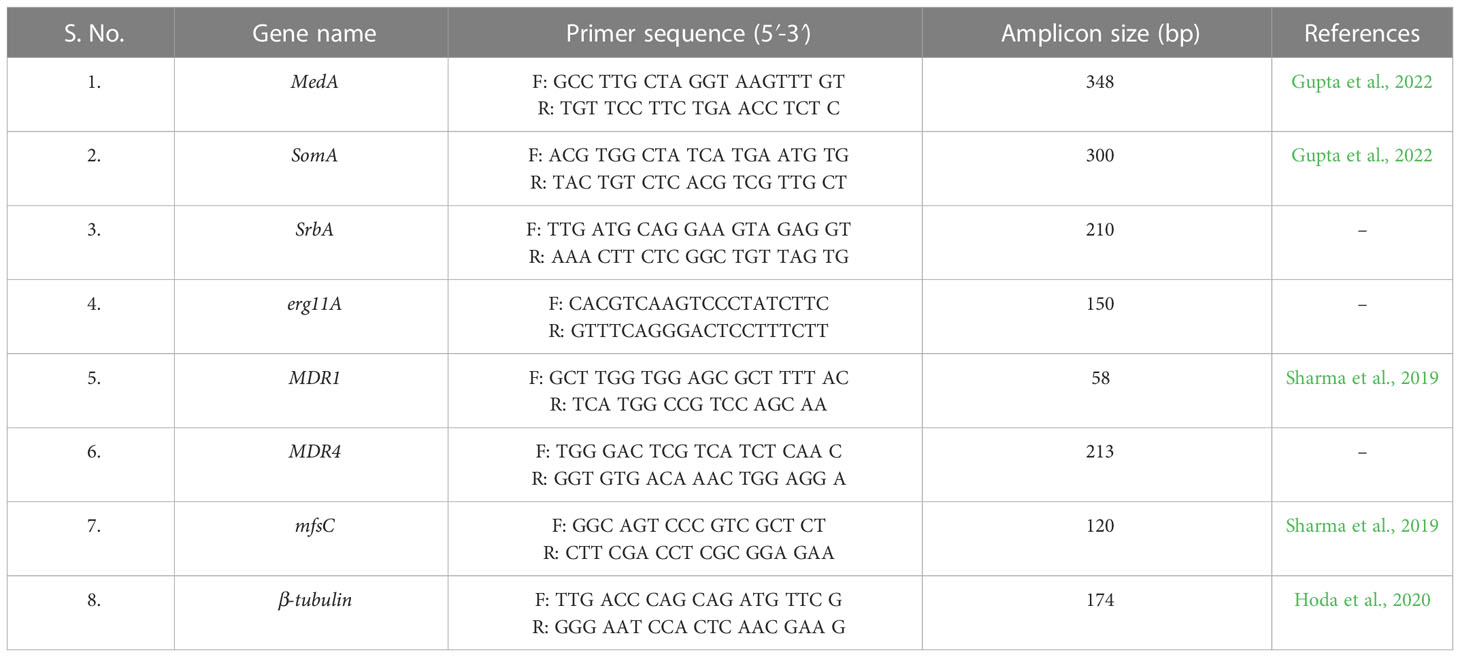
Two different primer sets (Tangwattanachuleeporn et al., 2017) were used for amplification of the cyp51A gene mutations (Table 1). Polymerase chain reaction (PCR) was performed using a final volume of 20 µL PCR buffer, containing 4 µL of fungal genomic DNA, 10 µL of PCR Master Mix (HiMedia, India), 0.50 µL (10 pmol/µL) of each primer and volume makeup by deionized water. Thermal cycling conditions for amplification was as follows: 95°C for 5 min followed by 35 cycles of denaturation at 95˚C for 30 s; annealing at 57.8˚C for 30 s and extension at 72˚C for 30 s; and final extension at 72˚C for 10 min. The amplified products were analyzed using agarose gel electrophoresis and purified using HiMedia quick gel purification kit according to the manufacturer’s instructions. Nucleotide sequencing was performed via Sanger’s sequencing using ABI 370 XL (Applied Biosystems). The sequence of the products was compared to the A. fumigatus wild-type cyp51A sequence accession no. AF338659 (Mellado et al., 2007; Rodriguez-Tudela et al., 2008) using the NCBI alignment service, Align Sequence Nucleotide Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Clustal Omega tool (https://www.ebi.ac.uk/Tools/msa/clustalo/).
2.6 Quantification of gene expression by qRT-PCR
Ten azole-resistant A.fumigatus isolates (eugenol-treated and untreated) along with a susceptible A.fumigatus (ATCC 46645; control) isolates were investigated for the expression analysis of the genes of interest by quantitative real time PCR (qRT-PCR; Gupta et al., 2022). Two micrograms of total RNA were reverse transcribed into first-stand cDNA using the Hi-cDNA synthesis kit (HiMedia, India) by following the manufacturer’s recommendations. Real-time PCR was performed using an ABI QuantStudio 3 (Applied Biosystems, Streetsville, Canada), and amplification products were detected with SYBR-green master mix (HiMedia, India) for fungal gene expression. The gene expression was estimated using the 2− ΔΔCt method, with β-tubulin as the reference gene. The gene specific primers were designed for MDR4, erg11A and SrbA using Primer 3 software (http://primer3.ut.ee/) (Untergasser et al., 2012). The primer sets used in this study are listed in Table 1.
2.7 Statistical analysis
For biofilm percentage, statistical analysis was performed using non-linear regression in a dose response manner with 95% confidence interval (CI). The range of best fit value at 95% CI for all the ARAF isolates has been listed in Table S1. In gene expression analysis, one-way ANOVA test was used to analyse whether the expression levels were statistically different. The p value of ≤0.05 was considered significant. Statistical analyses were also performed using GraphPad Prism v8.0.2.263.
3 Results
3.1 Screening for ARAF isolates from environmental samples
We have isolated 245 Aspergillus isolates from 110 soil samples and were further identified on the basis of macroscopic and microscopic characteristics. Of these, 89 isolates were identified as A. fumigatus (Table 2). Agar screening assay revealed that 10 isolates were azole resistant.
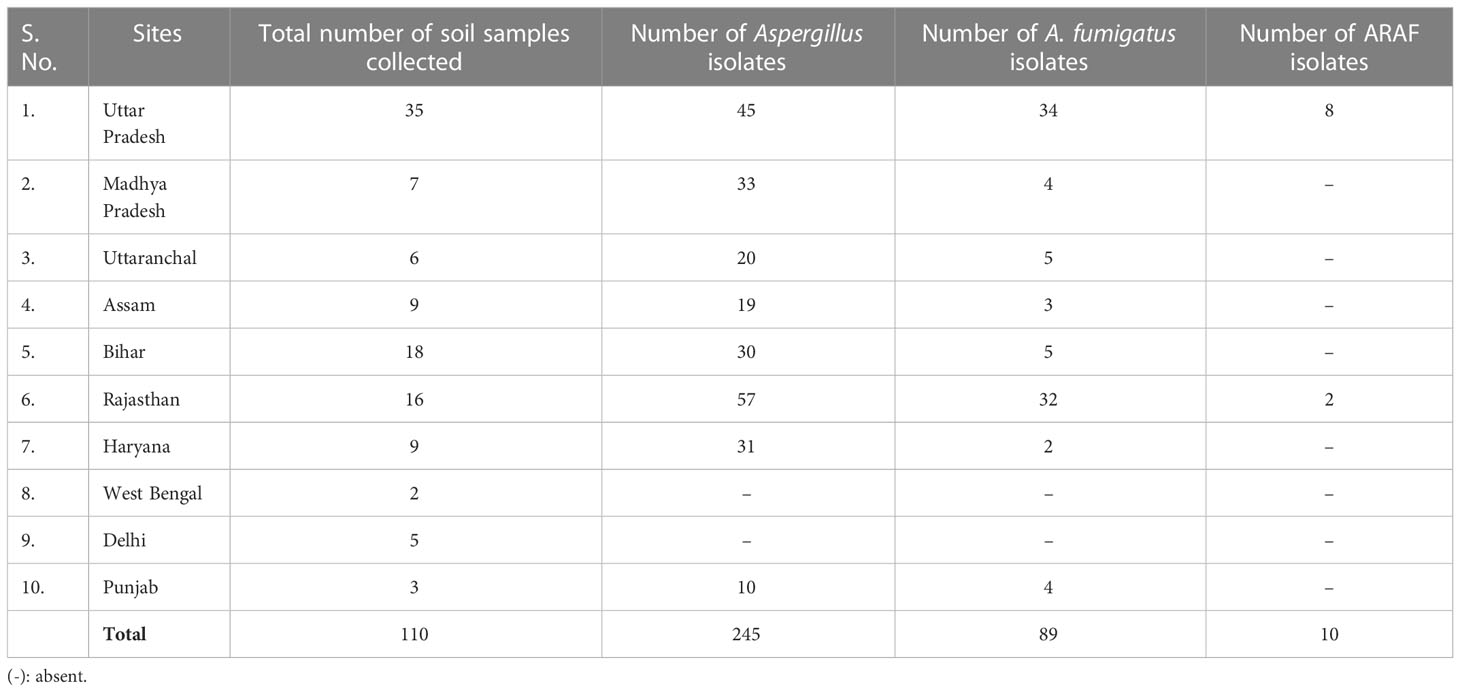
Table 2 Environmental ARAF isolates (identified macroscopically and microscopically) obtained from the soil samples collected from different regions of India.
3.2 Molecular identification of ARAF isolates
The 18S ITS region and β-tubulin gene sequence were amplified using PCR and visualized using ethidium bromide staining on agarose gel. The amplified regions of ITS and β-tubulin gene were further sequenced. The sequences of ten ARAF isolates were 99%–100% identical to the A. fumigatus sequence in the NCBI database. The identified fungal 18S ITS sequences were submitted to GenBank NCBI databases (https://www.ncbi.nlm.nih.gov/). The GenBank accession numbers generated for the submitted ITS sequences are MW485771, MW045410, MT941034, MW045442, MW045443, MW045444, MT941029, MZ284948, MZ379445, MZ379446 and for the β-tubulin gene sequences are ON759330, ON792384, ON792385, ON792386, ON792387, ON792388, ON792389, OP891221, ON792391 and OP891220. Phylogenetic tree of partial β-tubulin gene sequence by maximum likelihood was depicted in Figure S1.
3.3 Antifungal susceptibility testing
The results of antifungal activity of azole drugs against environmental isolates of ARAF are presented in Table 3. MIC of 9/10 isolates against ITC were above its ECV i.e., >1 μg/mL whereas three isolates had MIC >1 μg/mL against VRC (Table 3). The wild-type strain of A. fumigatus ATCC 46645 was sensitive to the azoles.
3.4 A. fumigatus biofilm eradication
The effect of eugenol on the biofilms formed by ARAF isolates as well as wild-type A. fumigatus ATCC 46645 strain was evaluated using semi-quantitative approach via MTT assay. The graphs depicted the percentage of biofilm at different concentrations of eugenol in comparison to wild type ATCC 46645 (Figure 1). The effective concentration of eugenol to eradicate the fungal biofilm was calculated as 312–500 µg/mL. In this range, biofilm eradication was observed up to 80% in a dose-responsive manner with statistical significance (95% CI). The BEC80 of eugenol for the biofilm of azole-resistant isolates was similar to that of wild-type ATCC 46645. In contrast, B19, B15, and RT28 isolates showed BEC80 of >1000 µg/mL.
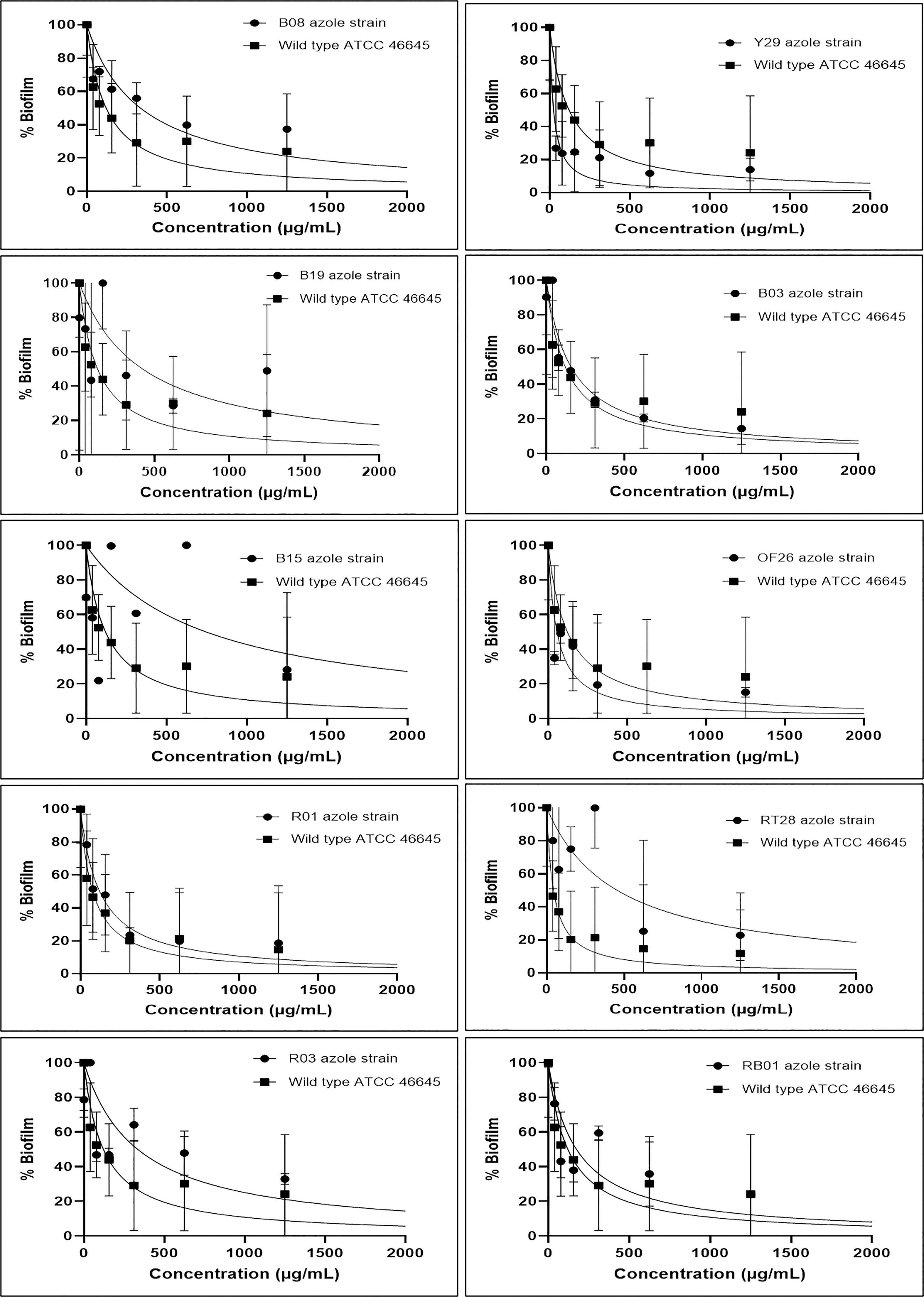
Figure 1 Effect of eugenol on biofilm percentage in azole resistant (B08, Y29, B19, B03, B15, OF26, R01, RT28, R03 and RB01) isolates and susceptible (wild-type ATCC 46645). BEC80 of eugenol ranged from 312–500 µg/mL except B19, B15, and RT28 isolates having BEC80 of >1000 µg/mL. Experiment was carried out in duplicate and analysed for statistical significance using non-linear regression in a dose response manner with 95% confidence interval (CI).
CLSM analysis of the untreated azole-resistant isolates and wild type ATCC 46645 showed aggregated multicellular hyphae enclosed within ECM. All the ARAF isolates indicated similar biofilm morphology as of wild type ATCC 46645. In untreated azole-resistant isolates, highly compact intermingled hyphae were observed, thereby strengthening the structure to reduce the effect of antifungals. When the susceptible and ARAF isolates were treated with eugenol at BEC80, we observed that the eugenol effectively eradicated the ECM and hyphae disintegrated in seven ARAF isolates (B08, Y29, RB01, B03, OF26, R01, and R03) (Figure 2). However, B19, B15, and RT28 isolates showed comparatively less eradication of biofilm when treated with eugenol at BEC80.
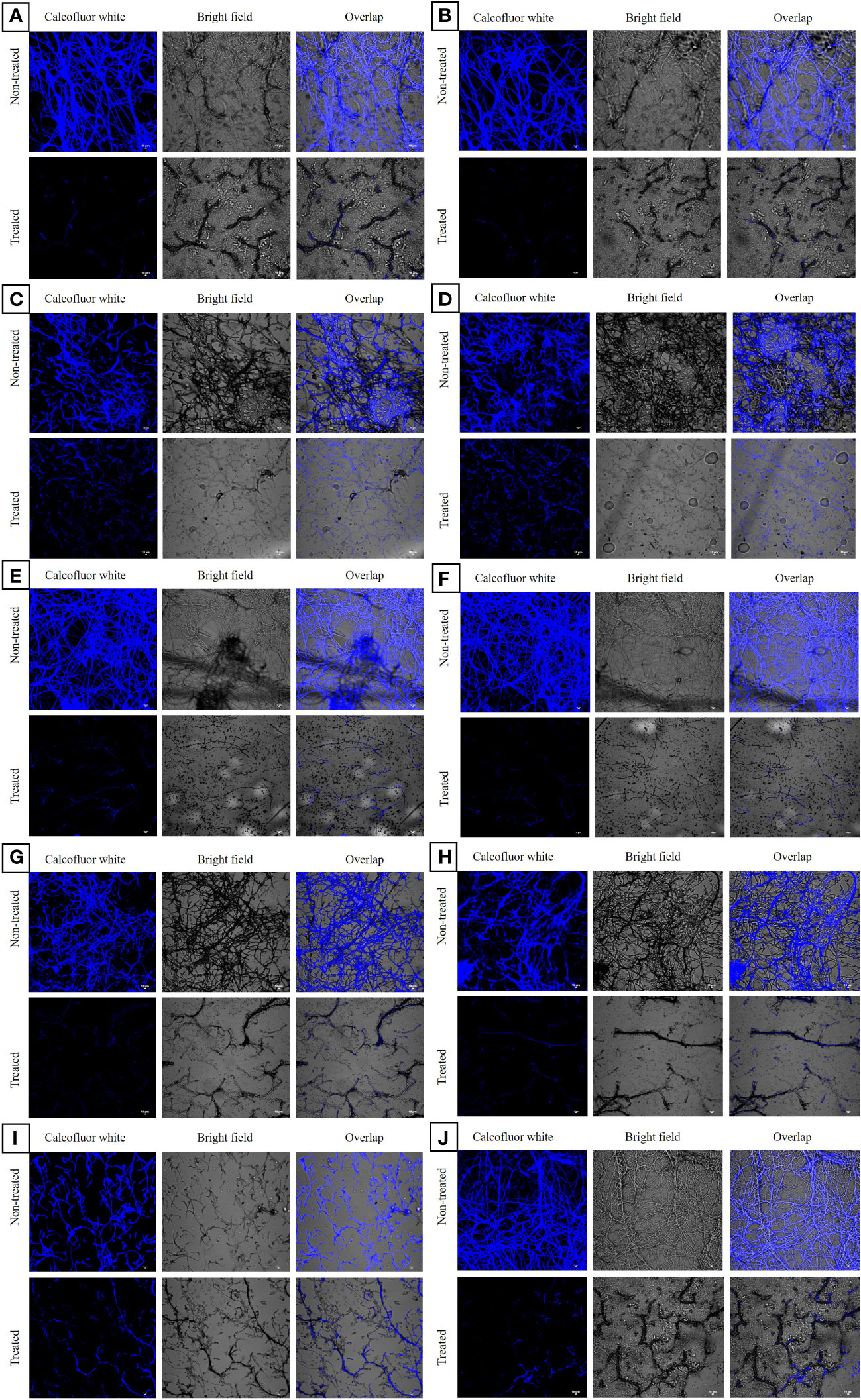
Figure 2 Confocal laser scanning microscopic images of biofilms of azole-resistant A. fumigatus isolates stained with calcofluor white dye. It depicted eugenol untreated and treated azole-resistant (A-J) B08, Y29, RT28, B15, RB01, B03, OF26, R01, B19 and R03 isolates, respectively. In eugenol-treated samples disintegrated hyphae without ECM were observed at 20 × magnification. The chitin component is stained with the calcofluor white dye and shown in three images with and without fluorochrome and overlapping. Scale Bar: 10 μm.
3.5 Mutation analysis of the cyp51A gene
The cyp51A gene of 10 ARAF isolates was sequenced and aligned with that of susceptible A. fumigatus strain accession no. AF338659 to identify non-synonymous mutations (Figure 3). Amino acid alterations were observed as a result of nucleotide substitutions in the cyp51A gene. The amino acid change Q131L was detected in all resistant environmental isolates. Two ITC-resistant isolates (R01 and R03) were found to have M172V non-synonymous nucleotide mutation in the cyp51A region. Some other reported mutations were not found in our tested isolates.
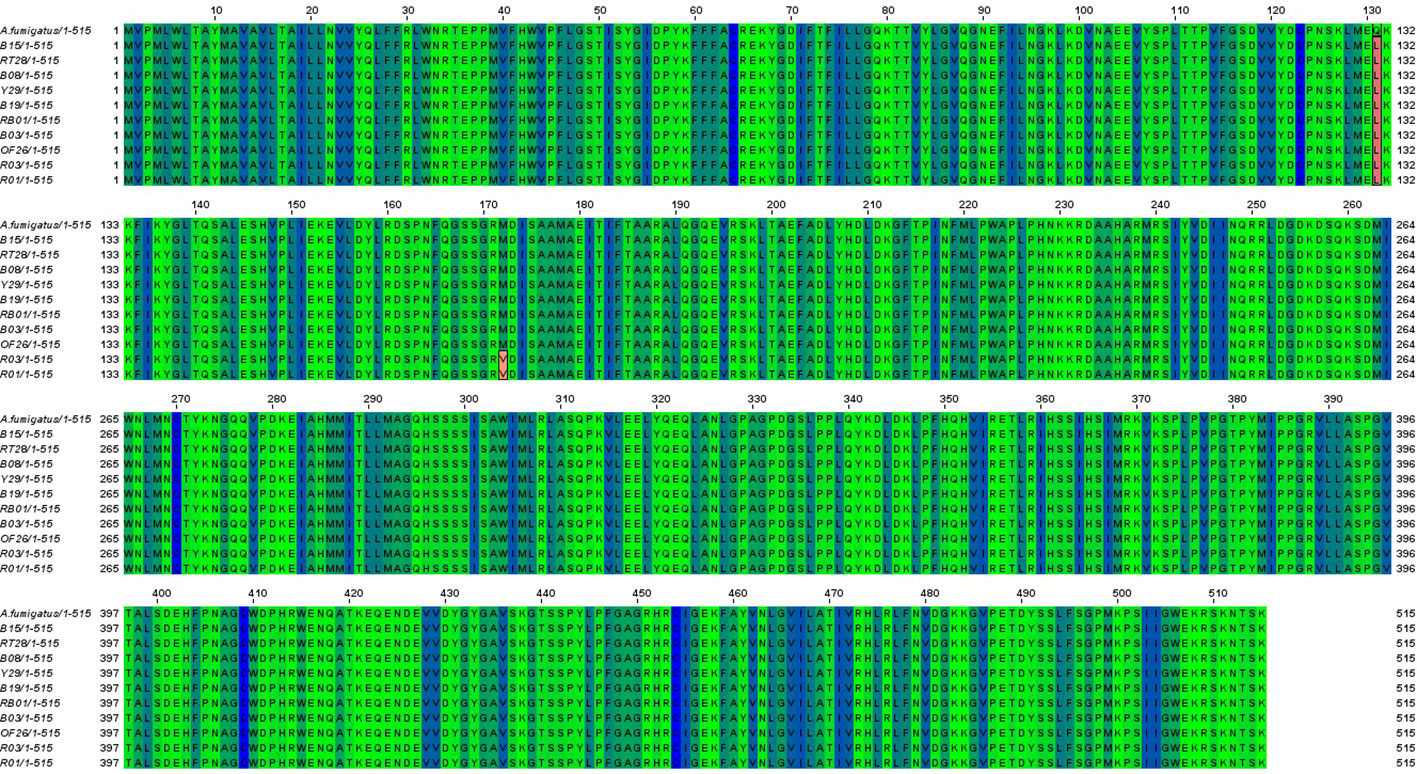
Figure 3 Sequence alignment of Cyp51A proteins from susceptible A. fumigatus (accession no. AF338659) and azole resistant isolates (B08, Y29, B19, B03, B15, OF26, R01, RT28, R03 and RB01). Identical residues were marked in blue and green shade whereas amino acid substitutions found in the Cyp51A sequence of the ARAF isolates were highlighted in red color.
3.6 Gene expression analysis
The impact of eugenol treatment on MDR efflux pump genes MDR1 and MDR4, transporters of the MFS gene mfsC, sterol 14α-demethylase encoding gene erg11A, and transcription regulatory genes, MedA, SomA and SrbA, involved in biofilm formation of A. fumigatus were investigated by reverse transcription followed by qRT-PCR for differential gene expression. Figure 4 demonstrates the two-fold relative expression of gene of interest in eugenol-treated and untreated ARAF isolates compared with the susceptible A. fumigatus (wild type ATCC 46645).
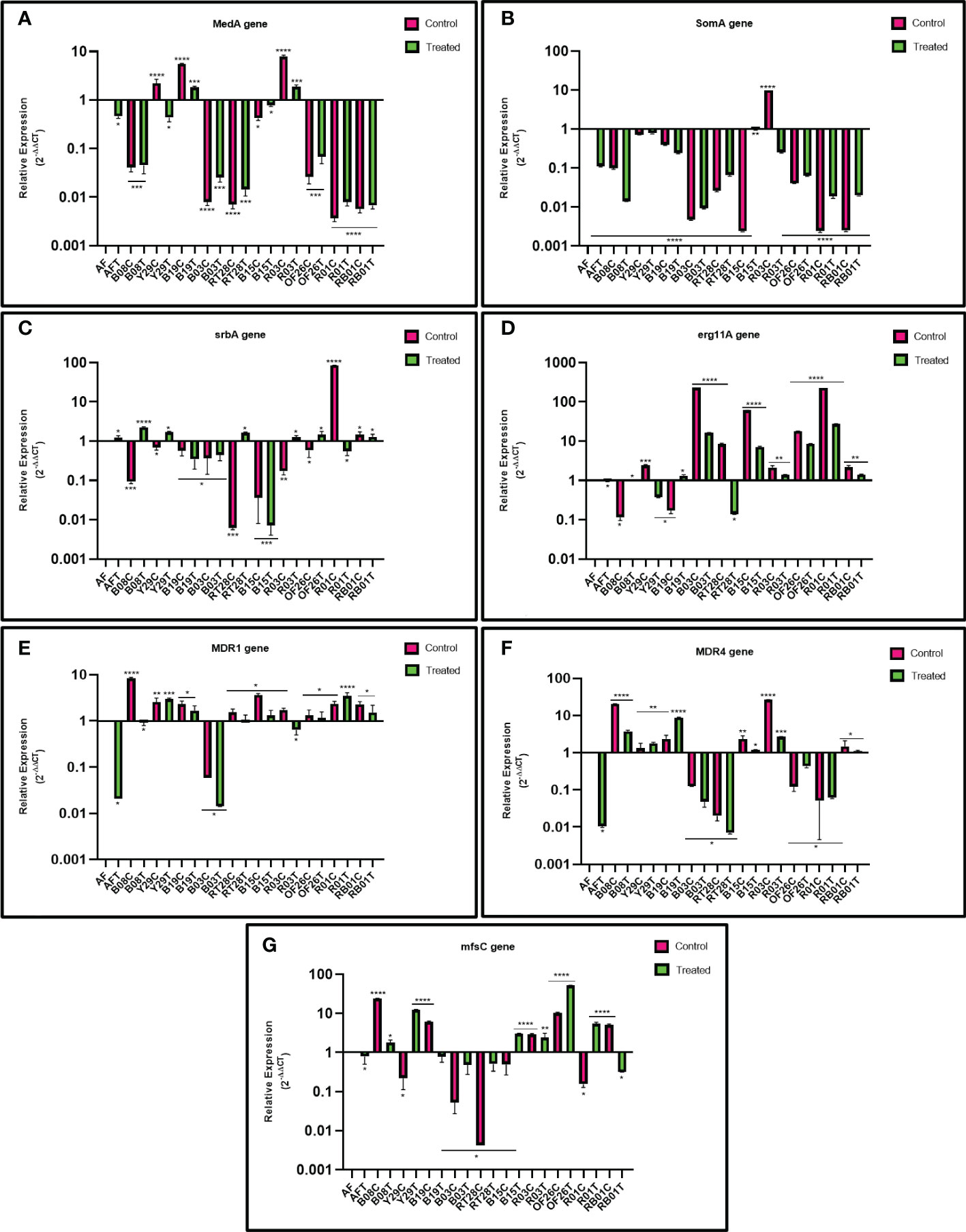
Figure 4 Relative expression of MedA (A) SomA (B) SrbA (C) erg11A (D) MDR1 (E) MDR4 (F) and mfsC (G) genes in azole-resistant environmental isolates relative to that in susceptible A fumigatus (wild type ATCC 46645) in untreated and eugenol-treated conditions. β-tubulin was used as a housekeeping gene to normalize transcription levels.
The MedA gene was upregulated in 3/10 untreated ARAF isolates compared to susceptible A. fumigatus. Except R03 untreated isolate, none of the ARAF isolates showed upregulation of the SomA gene. Expression analysis of the SrbA gene revealed that the gene is downregulated in both conditions, as was observed in three (B19, B03, B15) out of ten isolates. Relative expression of gene MDR4 showed upregulation in six ARAF untreated isolates. Expression was decreased after eugenol treatment in four isolates (B08, B15, R03 and RB01). The MDR1 gene was upregulated in all untreated isolates except B03 and expression was decreased after eugenol treatment in seven isolates. The erg11A gene was highly expressed in all the treated as well as untreated ARAF isolates except Y29, RT28 (treated) and B19, B08 (untreated). mfsC gene expression analysis was downregulated in two treated isolates (B19 and RB01).
4 Discussion
Environmental surveillance studies of azole resistance have been reported from many countries including Italy, Austria, India, Thailand, and the USA (Mortensen et al., 2010; Chowdhary et al., 2014; Tangwattanachuleeporn et al., 2017). However, the global prevalence of ARAF is still not clearly determined in routine resistance testing. Additionally, failure of current mainstay antifungals is a serious concern for clinicians as well as researchers. As azoles are the frontline antifungals, resistance could complicate the management of chronic and invasive diseases caused by A. fumigatus leading to treatment failure.
In the present study, the prevalence of ARAF in the Indian environment was 11.23% which is lower than that reported from European countries (Talbot et al., 2018) but higher than that reported from most of the Asian nations (Tangwattanachuleeporn et al., 2017). The management of azole-resistant aspergillosis remains a challenge and there are no guidelines as well as appropriate recommendations for this purpose. Understanding local rates of environmental resistance is critical for efficient patient management and it has been proposed that the level of environmental A. fumigatus azole resistance should be used to guide treatment of Aspergillus infections (Snelders et al., 2011). Triazole antifungals are crucial for the treatment and prophylaxis of infections occurring due to Aspergillus spp. The main route of azole resistance developed in the environment is related to the inappropriate use of azole-based pesticides. Azole pesticides have similar molecular structure as that of triazole drugs and resistant strains that develop in the environment are more efficient for developing cross resistance towards medical triazoles (Snelders et al., 2012). The clinical studies suggest that liposomal amphotericin B or a combination of VRC or PSZ with an echinocandin may be effective treatment (Elefanti et al., 2013). Eugenol, being a phytochemical, has already been reported to have antifungal activity in Candida spp. The MIC of eugenol have been reported in the range of 375–750 µg/mL against Candida dubliniensis and Candida tropicalis and 200–400 µg/mL against Candida krusei (De Paula et al., 2014; Sharifzadeh and Shokri, 2021). Eugenol containing essential oils were found to transform the hyphal ultrasound and virulence factors of A. fumigatus and Trichophyton rubrum (Gayoso et al., 2005). Antifungal activity of eugenol against Fusarium oxysporum, Lycopersici 1322 has also been reported, suggesting that eugenol can be used in preventive and therapeutic applications (Park et al., 2009).
Biofilm-associated infections have very high mortality rate and difficult to cure with existing drug therapies. Mowat et al. (2007) reported the formation of biofilm structures in A. fumigatus cultures which were resistant to antifungal drugs. Eugenol exhibited significantly favourable antifungal activity on various preformed biofilms, adherent cells, subsequent biofilm formation, and cell morphogenesis of C. albicans, without instigating haemolytic activity in human erythrocytes (Palaniappan and Holley, 2010). Similar results have been reported by De Paula et al. (2014) against Candida spp. In the present study, antibiofilm activity of eugenol against environmental isolates of ARAF suggested that effective BEC80 range of eugenol was 312–500 µg/mL against seven ARAF isolates and wild-type azole-susceptible A. fumigatus strain. Eugenol eradicated fungal biofilm along with the elimination of ECM in a dose-dependent manner. Further, the CLSM analysis supported the results of MTT assay with eradication of A. fumigatus biofilm. The biofilms of three ARAF isolates (B19, B15, and RT28) were less affected by eugenol treatment. CLSM analysis of eugenol-treated ARAF isolates revealed eradication of ECM and intermingled hyphae except B19 isolate.
Azoles target 14α-demethylase encoding erg11A gene and block ergosterol biosynthesis (Price et al., 2015). The overexpression of this gene in ARAF isolates could be a possible drug resistance mechanism. The present study calculated the expression of erg11A gene with and without eugenol; the gene was significantly overexpressed in six ARAF isolates under both conditions whereas it was found to be downregulated in two isolates (Y29 and RT28) after eugenol treatment. Ergosterol biosynthesis gene expression increased in biofilms formed by planktonic cultures. In azole resistant strains cyp51A gene plays a crucial role, since the deletion of the cyp51A gene resulted in increased sensitivity to azoles (Garcia-Rubio et al., 2018). Sterol biosynthesis is directly impacted by iron and oxygen levels and reductions in these key molecules stimulates ergosterol biosynthesis gene expression through SrbA dependent mechanisms (Blatzer et al., 2011). In submerged biofilm cultures, oxygen levels are depleted over the course of biofilm development and correspond with an increase in transcription of the SrbA dependent gene erg25A (Kowalski et al., 2020). Thus, as biofilm cell density increases, oxygen levels within the immature biofilm become depleted, leading to both an increase in expression of ergosterol biosynthetic genes and a decrease in metabolism. These two factors may contribute to the localized increase in antifungal resistance within the developing biofilm. Expression analysis of the SrbA gene revealed the downregulation of gene in the presence and absence of eugenol in the following three ARAF isolates B19, B03, B15 and also in untreated ARAF isolates encoded B08, Y29, RT28, and OF26. The role of overexpression of efflux pumps, ATP-binding cassette (ABC) transporters and transporters of the MFS gene, in C. albicans and C. glabrata has been well documented (Cannon et al., 2009; Morschhäuser, 2010). Two major facilitator transporters, AfuMDR1 and AfuMDR2, were identified in A. fumigatus by Tobin et al. (1997). Nascimento et al. (2003) showed upregulation of two transporters, MDR3 and MDR4 in ITC resistant A. fumigatus strains, the latter of which has also been shown to be upregulated in a biofilm phenotype. Overexpression of efflux pumps in azole resistance has also been emphasized in other studies (Da Silva Ferreira et al., 2004; Fraczek et al., 2013). The relative expression of the MDR efflux pumps genes MDR1 and MDR4 was analyzed in this study, and we observed that MDR1 was upregulated in nine isolates except B03. After eugenol treatment, the relative gene expression decreased in two ARAF isolates (B03 and R03). MDR4 gene is reported in C. albicans biofilms and is associated with drug detoxification (Mukherjee et al., 2003). Overexpression of MDR4 gene in azole resistance A. fumigatus strains after 24-h VRC treatment has been reported by Rajendran et al. (2011). Upregulation of MDR4 was analyzed in six untreated isolates and after eugenol decreased in four ARAF (B03, OF26, R01 and RT28) isolates. But overexpression of the gene after eugenol treatment when compared to untreated has not been observed in our study. MDR pumps in A.fumigatus are associated with triazole resistance (Rajendran et al., 2011). In our study, the overexpression of the MDR4 gene suggested that this gene is linked with ITC resistance and after eugenol treatment gene expression was decreased. Further, transcription regulatory genes MedA and SomA were downregulated in all the eugenol-treated ARAF isolates except B19 and R03 for MedA gene expression and similar results were reported by Gupta et al. (2022) for the downregulation of MedA and SomA genes in presence of isoeugenol. Further, mfsC gene expression analysis showed that the gene was downregulated in the four treated ARAF isolates (B19, RT28, B03 and RB01). Our observations suggest that many unidentified molecular mechanisms are associated to biofilm formation in environmental ARAF isolates which still needs to be explored. Further investigation of the relationship between the eugenol and MDR efflux pump genes and their pathways in A.fumigatus virulence and azole susceptibility may be needed to understand the mechanism of the observed interactions.
We observed notable antibiofilm activity of 4-allyl-2-methoxyphenol (eugenol) against azole-resistant A. fumigatus isolates. This activity appears to be mediated in some ARAF isolates via the inhibition of the MedA and efflux pump MDR genes. The use of eugenol alone or in combination with conventional antifungal drugs could be a valuable addition to current therapeutic strategies for treating azole-resistant A fumigatus infections.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
PS and LG performed literature search, conducted all the experiments, and drafted the manuscript. MV also performed experiments and helped in manuscript editing. SH analysed transcriptional data. MS and JS critically reviewed the manuscript and corrected and PV conceptualized the idea as well as critically analysed the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
Authors would like acknowledge FIST facility at AIMT Amity University Uttar Pradesh for confocal microscopy facility and Amity University Uttar Pradesh for providing the infrastructure and facilities for research. PS would like to thank CSIR for providing student scholarship support in the form of Senior Research Fellowship (manpower support) (09/915(0013)/2018-EMR-I).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1103957/full#supplementary-material
References
Ahangarkani, F., Badali, H., Abbasi, K., Nabili, M., Khodavaisy, S., de Groot, T., et al. (2020b). Clonal expansion of environmental triazole resistant Aspergillus fumigatus in Iran. J. Fungi. 6, 199. doi: 10.3390/jof6040199
Ahangarkani, F., Puts, Y., Nabili, M., Khodavaisy, S., Moazeni, M., Salehi, Z., et al. (2020a). First azole-resistant aspergillus fumigatus isolates with the environmental TR46 /Y121F/T289A mutation in Iran. Mycoses. 63, 430–436. doi: 10.1111/myc.13064
Ahmad, A., Khan, A., Manzoor, N., Khan, L. A. (2010). Evolution of ergosterol biosynthesis inhibitors as fungicidal against candida. Microb. Pathog. 48, 35–41. doi: 10.1016/j.micpath.2009.10.001
Alexander, B. D., Clinical and Laboratory Standards Institute (2017). Reference method for broth dilution antifungal susceptibility testing of filamentous fungi. 3rd ed. CLSI standard M38 (ISBN 1-56238-830-4). Pennsylvania USA.
Beauvais, A., Latgé, J. P. (2015). Aspergillus biofilm In vitro and In vivo. Microbiol. Spectr 3, 4. doi: 10.1128/microbiolspec
Bennis, S., Chami, F., Chami, N., Bouchikhi, T., Remmal, A. (2004). Surface alteration of Saccharomyces cerevisiae induced by thymol and eugenol. Lett. Appl. Microbiol. 38, 454–458. doi: 10.1111/j.1472-765X.2004.01511.x
Blatzer, M., Barker, B. M., Willger, S. D., Beckmann, N., Blosser, S. J., Cornish, E. J., et al. (2011). SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus. PloS Genet. 7, e1002374. doi: 10.1371/journal.pgen.1002374
Bongomin, F., Gago, S., Oladele, R. O., Denning, D. W. (2017). Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi. (Basel). 18, 57. doi: 10.3390/jof3040057
Brauer, V. S., Rezende, C. P., Pessoni, A. M., De Paula, R. G., Rangappa, K. S., Nayaka, S. C., et al. (2019). Antifungal agents in agriculture: Friends and foes of public health. Biomolecules. 23, 521. doi: 10.3390/biom9100521
Cannon, R. D., Lamping, E., Holmes, A. R., Niimi, K., Baret, P. V., Keniya, M. V., et al. (2009). Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 22, 291–321. doi: 10.1128/CMR.00051-08
Chowdhary, A., Sharma, C., Hagen, F., Meis, J. F. (2014). Exploring azole antifungal drug resistance in Aspergillus fumigatus with special reference to resistance mechanisms. Future Microbiol. 9, 697–711. doi: 10.2217/fmb.14.27
Chung, G., Im, S. T., Kim, Y. H., Jung, S. J., Rhyu, M. R., Oh, S. B. (2014). Activation of transient receptor potential ankyrin 1 by eugenol. Neuroscience. 261, 153–160. doi: 10.1016/j.neuroscience.2013.12.047
Clinical and Laboratory Standards Institute (CLSI) (2018). Epidemiological cut-off values for antifungal susceptibility testing 2nd ed (Wayne P.A. USA: CLSI supplement M59 Clinical and Laboratory Standards Institute).
Dabas, Y., Xess, I., Bakshi, S., Mahapatra, M., Seth, R. (2018). Emergence of azole-resistant Aspergillus fumigatus from immunocompromised hosts in india. antimicrob. Agents. Chemother. 27, e02264–e02217. doi: 10.1128/AAC.02264-17
Da Silva Ferreira, M. E., Capellaro, J. L., Dos Reis Marques, E., Malavazi, I., Perlin, D., Park, S., et al. (2004). In vitro evolution of itraconazole resistance in Aspergillus fumigatus involves multiple mechanisms of resistance. Antimicrob. Agents. Chemother. 48, 4405–4413. doi: 10.1128/AAC.48.11.4405-4413.2004
de Hoog, G., Guarro, J., Tran, C. S., Wintermans, R. G. F., Gene, J. (1995). Hypho- mycetes atlas of clinicalfungi (Chichester England: John Wiley & Sons).
Denning, D. W., Cadranel, J., Beigelman-Aubry, C., Ader, F., Chakrabarti, A., Blot, S., et al. (2016). European Society for clinical microbiology and infectious diseases and European respiratory society. chronic pulmonary aspergillosis: rationale and clinical guidelines for diagnosis and management. Eur. Respir. J. 47, 45–68. doi: 10.1183/13993003
De Paula, S. B., Bartelli, T. F., Di Raimo, V., Santos, J. P., Morey, A. T., Bosini, M. A. (2014). Effect of eugenol on cell surface hydrophobicity adhesion and biofilm of candida tropicalis and Candida dubliniensis isolated from oral cavity of HIV-infected patients. Evid. Based. Complement. Alternat. Med. 2014, 505204. doi: 10.1155/2014/505204
Elefanti, A., Mouton, J. W., Verweij, P. E., Tsakris, A., Zerva, L., Meletiadis, J. (2013). Amphotericin b- and voriconazole-echinocandin combinations against Aspergillus spp.: Effect of serum on inhibitory and fungicidal interactions. Antimicrob. Agents. Chemother. 57 (10), 4656–4663. doi: 10.1128/AAC.00597-13
Ferreira, I. C. F. R., Martins, N., Barros, L. (2017). Phenolic compounds and its bioavailability: In vitro bioactive compounds or health promoters? Adv. Food. Nutr. Res. 82, 1–44. doi: 10.1016/bs.afnr.2016.12.004
Fraczek, M. G., Bromley, M., Buied, A., Moore, C. B., Rajendran, R., Rautemaa, R., et al (2013). The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in aspergillus fumigatus. J. Antimicrob. Chemother. 68, 1486–1496. doi: 10.1093/jac/dkt075
Garcia-Rubio, R., Alcazar-Fuoli, L., Monteiro, M. C., Monzon, S., Cuesta, I., Pelaez, T., et al. (2018). Insight into the significance of Aspergillus fumigatus cyp51A polymorphisms. Antimicrob. Agents. Chemother. 62, e00241–e00218. doi: 10.1128/AAC.00241-18
Gayoso, C. W., Lima, E. O., Oliveira, V. T., Pereira, F. O., Souza, E. L., Lima, I. O., et al. (2005). Sensitivity of fungi isolated from onychomycosis to Eugenia cariophyllata essential oil and eugenol. Fitoterapia 76, 247–249. doi: 10.1016/j.fitote.2004.12.005
González-Ramírez, A. I., Ramírez-Granillo, A., Medina-Canales, M. G., Rodríguez-Tovar, A. V., Martínez-Rivera, M. A. (2016). Analysis and description of the stages of Aspergillus fumigatus biofilm formation using scanning electron microscopy. BMC Microbiol. 16, 243. doi: 10.1186/s12866-016-0859-4
Goswami, L., Gupda, L., Paul, S., Vermani, M., Vijayaraghavan, P., Bhattacharya, A. K.. (2022). Design and synthesis of eugenol/isoeugenol glycoconjugates and other analogues as antifungal agents against Aspergillus fumigatus. RSC Med Chem 13, 955–962. doi: 10.1039/d2md00138a
Gravelat, F. N., Ejzykowicz, D. E., Chiang, L. Y., Chabot, J. C., Urb, M., Macdonald, K. D., et al. (2010). Aspergillus fumigatus MedA governs adherence host cell interactions and virulence. Cell Microbiol. 1, 473–488. doi: 10.1111/j.1462-5822.2009.01408.x
Gupta, L., Sen, P., Bhattacharya, A. K., Vijayaraghavan, P. (2022). Isoeugenol affects expression pattern of conidial hydrophobin gene RodA and transcriptional regulators MedA and SomA responsible for adherence and biofilm formation in Aspergillus fumigatus. Arch. Microbiol. 204, 214. doi: 10.1007/s00203-022-02817-w
Hoda, S., Vermani, M., Joshi, R. K., Shankar, J., Vijayaraghavan, P. (2020). Anti-melanogenic activity of myristica fragrans extract against Aspergillus fumigatus using phenotypic based screening. BMC Complement Med. Ther. 20, 67. doi: 10.1186/s12906-020-2859-z
Huang, X., Liu, Y., Lu, Y., Ma, C. (2015). Anti-inflammatory effects of eugenol on lipopolysaccharide-induced inflammatory reaction in acute lung injury via regulating inflammation and redox status. Int. Immunopharmacol. 26, 265–271. doi: 10.1016/j.intimp.2015.03.026
Khalil, A. A., Rahman, U. U., Khan, M. R., Sahar, A., Mehmood, T., Khan, M. (2017). Essential oil eugenol: sources extraction techniques and nutraceutical perspectives. RSC. Adv. 7, 32669–32681. doi: 10.1039/C7RA04803C
Khan, M. S., Ahmad, I. (2012). Antibiofilm activity of certain phytocompounds and their synergy with fluconazole against Candida albicans biofilms. J. Antimicrob. Chemother. 67, 618–621. doi: 10.1093/jac/dkr512
Kosmidis, C., Denning, D. W. (2015). The clinical spectrum of pulmonary aspergillosis. Thorax. 70, 270–277. doi: 10.1136/thoraxjnl-2014-206291
Kowalski, C. H., Morelli, K. A., Schultz, D., Nadell, C. D., Cramer, R. A. (2020). Fungal biofilm architecture produces hypoxic microenvironments that drive antifungal resistance. Proc. Natl. Acad. Sci. U. S. A. 117, 22473–22483. doi: 10.1073/pnas.2003700117
Lass-Flörl, C. (2014). Susceptibility testing in Aspergillus species complex. Clin. Microbiol. Infect. 20 Suppl 6, 49–53. doi: 10.1111/1469-0691.12514
Lee, S. B., Milgroom, M. G., Taylor, J. W. (1988). A rapid high yield mini-prep method for isolation of total genomic DNA from fungi fungal. Genet. Rep. 35, 23–24. doi: 10.4148/1941-4765.1531
Manavathu, E. K., Vager, D. L., Vazquez, J. A. (2014). Development and antimicrobial susceptibility studies of in vitro monomicrobial and polymicrobial biofilm models with Aspergillus fumigatus and pseudomonas aeruginosa. BMC Microbiol. 3, 14–53. doi: 10.1186/1471-2180-14-53
Marchese, A., Barbieri, R., Coppo, E., Orhan, I. E., Daglia, M., Nabavi, S. F., et al. (2017). Anti-microbial activity of eugenol and essential oils containing eugenol: A mechanistic viewpoint. Crit. Rev. Microbiol. 43, 668–689. doi: 10.1080/1040841X.2017.1295225
Mellado, E., Garcia-Effron, G., Alcázar-Fuoli, L., Melchers, W. J., Verweij, P. E., Cuenca-Estrella, M., et al. (2007). A new aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents. Chemother. 51, 1897–1904. doi: 10.1128/AAC.01092-06
Morschhäuser, J. (2010). Regulation of multidrug resistance in pathogenic fungi. Fungal. Genet. Biol. 47, 94–106. doi: 10.1016/j.fgb.2009.08.002
Mortensen, K. L., Mellado, E., Lass-Flörl, C., Rodriguez-Tudela, J. L., Johansen, H. K., Arendrup, M. C. (2010). Environmental study of azole-resistant Aspergillus fumigatus and other aspergilli in Austria Denmark and Spain. Antimicrob. Agents. Chemother. 54, 4545–4549. doi: 10.1128/AAC.00692-10
Mowat, E., Butcher, J., Lang, S., Williams, C., Ramage, G. (2007). Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J. Med. Microbiol. 56 (Pt 9), 1205–1212. doi: 10.1099/jmm.0.47247-0
Mukherjee, P. K., Chandra, J., Kuhn, D. M., Ghannoum, M. A. (2003). Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect. Immun. 71, 4333–4340. doi: 10.1128/IAI.71.8.4333-4340.2003
Müller, F. M., Seidler, M., Beauvais, A. (2011). Aspergillus fumigatus biofilms in the clinical setting. Med. Mycol. 49 Suppl 1, S96–S100. doi: 10.3109/13693786.2010.502190
Nascimento, A. M., Goldman, G. H., Park, S., Marras, S. A., Delmas, G., Oza, U., et al. (2003). Multiple resistance mechanisms among aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrob. Agents. Chemother. 47, 1719–1726. doi: 10.1128/AAC.47.5.1719-1726.2003
Palaniappan, K., Holley, R. A. (2010). Use of natural antimicrobials to increase antibiotic susceptibility of drug resistant bacteria. Int. J. Food. Microbiol. 140, 164–168. doi: 10.1016/j.ijfoodmicro.2010.04.001
Park, M. J., Gwak, K. S., Yang, I., Kim, K. W., Jeung, E. B., Chang, J. W., et al. (2009). Effect of citral eugenol nerolidol and alpha-terpineol on the ultrastructural changes of. Trichophyton. mentagrophytes. 80, 290–296. doi: 10.1016/j.fitote.2009.03.007
Pfaller, M., Boyken, L., Hollis, R., Kroeger, J., Messer, S., Tendolkar, S., et al. (2011). Use of epidemiological cutoff values to examine 9-year trends in susceptibility of aspergillus species to the triazoles. J. Clin. Microbiol. 49, 586–590. doi: 10.1128/JCM.02136-10
Pramod, K., Ansari, S. H., Ali, J. (2010). Eugenol: a natural compound with versatile pharmacological actions. Nat. Prod. Commun. 5, 1999–2006. doi: 10.1177/1934578X1000501236
Price, C. L., Parker, J. E., Warrilow, A. G., Kelly, D. E., Kelly, S. L. (2015). Azole fungicides-understanding resistance mechanisms in agricultural fungal pathogens. Pest. Manage. Sci. 71, 1054–1058. doi: 10.1002/ps.4029
Rajendran, R., Mowat, E., McCulloch, E., Lappin, D. F., Jones, B., Lang, S., et al. (2011). Azole resistance of aspergillus fumigatus biofilms is partly associated with efflux pump activity. Antimicrob. Agents. Chemother. 55, 2092–2097. doi: 10.1128/AAC.01189-10
Rodriguez-Tudela, J. L., Alcazar-Fuoli, L., Mellado, E., Alastruey-Izquierdo, A., Monzon, A., Cuenca-Estrella, M. (2008). Epidemiological cutoffs and cross-resistance to azole drugs in aspergillus fumigatus. Antimicrob. Agents Chemother. 52, 2468–2472. doi: 10.1128/AAC.00156-08
Sav, H., Rafati, H., Öz, Y., Dalyan-Cilo, B., Ener, B., Mohammadi, F., et al. (2018). Biofilm formation and resistance to fungicides in clinically relevant members of the fungal genus fusarium. J. Fungi. 4, 16. doi: 10.3390/jof4010016
Sharifzadeh, A., Shokri, H. (2021). In vitro synergy of eugenol on the antifungal effects of voriconazole against Candida tropicalis and Candida krusei strains isolated from the genital tract of mares. Equine. Vet. J. 53, 94–101. doi: 10.1111/evj.13268
Sharma, C., Nelson-Sathi, S., Singh, A., Pillai, M. R., Chowdhary, A. (2019). Genomic perspective of triazole resistance in clinical and environmental Aspergillus fumigatus isolates without cyp51A mutations. Fungal. Genet. Biol. 132, 103265. doi: 10.1016/j.fgb.2019.103265
Snelders, E., Camps, S. M., Karawajczyk, A., Schaftenaar, G., Kema, G. H., van der Lee, H. A. (2012). Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PloS One 7, e31801. doi: 10.1371/journal.pone.0031801
Snelders, E., Melchers, W. J., Verweij, P. E. (2011). Azole resistance in Aspergillus fumigatus: A new challenge in the management of invasive aspergillosis? Future Microbiol. 6, 335–347. doi: 10.2217/fmb.11.4
Talbot, J. J., Subedi, S., Halliday, C. L., Hibbs, D. E., Lai, F., Lopez-Ruiz, F. J., et al. (2018). Surveillance for azole resistance in clinical and environmental isolates of aspergillus fumigatus in Australia and cyp51A homology modelling of azole-resistant isolates. J. Antimicrob. Chemother. 73, 2347–2351. doi: 10.1093/jac/dky187
Tangwattanachuleeporn, M., Minarin, N., Saichan, S., Sermsri, P., Mitkornburee, R., Groß, U., et al. (2017). Prevalence of azole-resistant aspergillus fumigatus in the environment of Thailand. Med. Mycol. 55, 429–435. doi: 10.1093/mmy/myw090
Tobin, M. B., Peery, R. B., Skatrud, P. L. (1997). Genes encoding multiple drug resistance like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene. 200, 11–23. doi: 10.1016/s0378-1119(97)00281-3
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115. doi: 10.1093/nar/gks596
White, T. J., Bruns, T. D., Lee, S. B., Taylor, J. W., Innis, M. A., Gelfand, D. H., et al. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols. A guide to methods and applications. (San Diego: Academic Press), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Keywords: Azole-resistant Aspergillus fumigatus, 4-allyl-2-methoxyphenol, biofilms, MDR4, mfsC
Citation: Sen P, Gupta L, Vijay M, Vermani Sarin M, Shankar J, Hameed S and Vijayaraghavan P (2023) 4-Allyl-2-methoxyphenol modulates the expression of genes involved in efflux pump, biofilm formation and sterol biosynthesis in azole resistant Aspergillus fumigatus. Front. Cell. Infect. Microbiol. 13:1103957. doi: 10.3389/fcimb.2023.1103957
Received: 21 November 2022; Accepted: 06 January 2023;
Published: 01 February 2023.
Edited by:
Renátó Kovács, University of Debrecen, HungaryReviewed by:
Kunlong Yang, Jiangsu Normal University, ChinaFatemeh Ahangarkani, Mazandaran University of Medical Sciences, Sari, Iran
Copyright © 2023 Sen, Gupta, Vijay, Vermani Sarin, Shankar, Hameed and Vijayaraghavan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pooja Vijayaraghavan, vrpooja@amity.edu
†These authors have contributed equally to this work and share first authorship
‡ORCID: Pooja Sen, orcid.org.0000-0002-0819-1599
Lovely Gupta, orcid.org.0000-0002-2610-3142
Mukund Vijay, orcid.org.0000-0002-7359-7093
Maansi Vermani Sarin, orcid.org.0000-0003-1635-4463
Jata Shankar, orcid.org.0000-0003-4993-9580
Saif Hameed, orcid.org/0000-0002-6477-1870
Pooja Vijayaraghavan, orcid.org.0000-0001-5943-9462
 Pooja Sen
Pooja Sen Lovely Gupta
Lovely Gupta Mukund Vijay
Mukund Vijay Maansi Vermani Sarin
Maansi Vermani Sarin Jata Shankar
Jata Shankar Saif Hameed
Saif Hameed Pooja Vijayaraghavan
Pooja Vijayaraghavan