Genetic characteristics, antimicrobial susceptibility, and virulence genes distribution of Campylobacter isolated from local dual-purpose chickens in central China
- 1Key Laboratory of Prevention and Control Agents for Animal Bacteriosis (Ministry of Agriculture and Rural Affairs), Institute of Animal Husbandry and Veterinary, Hubei Academy of Agricultural Sciences, Wuhan, China
- 2Hubei Provincial Key Laboratory of Animal Pathogenic Microbiology, Institute of Animal Husbandry and Veterinary, Hubei Academy of Agricultural Sciences, Wuhan, China
- 3Institute of Animal Husbandry and Veterinary, Hubei Academy of Agricultural Sciences, Hubei Hongshan Laboratory, Wuhan, China
Food-borne antibiotic-resistant Campylobacter poses a serious threat to public health. To understand the prevalence and genetic characteristics of Campylobacter in Chinese local dual-purpose (meat and eggs) chickens, the genomes of 30 Campylobacter isolates, including 13 C. jejuni and 17 C. coli from Jianghan-chickens in central China, were sequenced and tested for antibiotic susceptibility. The results showed that CC-354 and CC-828 were the dominant clonal complexes of C. jejuni and C. coli, respectively, and a phylogenetic analysis showed that three unclassified multilocus sequence types of C. coli were more closely genetically related to C. jejuni than to other C. coli in this study. Of the six antibiotics tested, the highest resistance rates were to ciprofloxacin and tetracycline (100%), followed by lincomycin (63.3%), erythromycin (30.0%), amikacin (26.7%), and cefotaxime (20.0%). The antibiotic resistance rate of C. coli was higher than that of C. jejuni. The GyrA T86I mutation and 15 acquired resistance genes were detected with whole-genome sequencing (WGS). Among those, the GyrA T86I mutation and tet(O) were most prevalent (both 96.7%), followed by the blaOXA-type gene (90.0%), ant(6)-Ia (26.7%), aac(6’)-aph(3’’) (23.3%), erm(B) (13.3%), and other genes (3.3%). The ciprofloxacin and tetracycline resistance phenotypes correlated strongly with the GyrA T86I mutation and tet(O)/tet(L), respectively, but for other antibiotics, the correlation between genes and resistance phenotypes were weak, indicating that there may be resistance mechanisms other than the resistance genes detected in this study. Virulence gene analysis showed that several genes related to adhesion, colonization, and invasion (including cadF, porA, ciaB, and jlpA) and cytolethal distending toxin (cdtABC) were only present in C. jejuni. Overall, this study extends our knowledge of the epidemiology and antibiotic resistance of Campylobacter in local Chinese dual-purpose chickens.
1 Introduction
According to the report of the World Health Organization (WHO), food-borne diseases, ranging from diarrhea to cancer, are a major cause of human morbidity and mortality and affect one in 10 people worldwide every year (WHO, 2022). Campylobacteriosis is one of the most frequently reported food-borne diseases throughout the world (EFSA BIOHAZ Panel [EFSA Panel on Biological Hazards] et al., 2020). The acute infectious diarrhea caused by Campylobacter is mainly treated with antibiotics, such as fluoroquinolones and macrolides (Pham et al., 2016). However, the use of antibiotics in both human treatments and animal breeding hisolated from poultry meat samples.as caused antimicrobial resistance in Campylobacter to become an increasingly serious problem, and has posed a serious threat to public health over the past two decades (Luangtongkum et al., 2009). In 2017, fluoroquinolone-resistant Campylobacter was listed as one of the six high-priority antimicrobial-resistant pathogens by WHO (Romanescu et al., 2023). In China, bacterial antibiotic resistance monitoring data show that Campylobacter has maintained a high level of resistance to ciprofloxacin (> 90%) in various regions, (Li et al., 2016; Wang et al., 2016; Ju et al., 2018).
Poultry is the most important natural host of Campylobacter. In the European Union, the average prevalence of Campylobacter in birds and contaminated broiler carcasses is 71.2% and 75.8%, respectively (Soro et al., 2020), and more than 90% of commercial laying hens are colonized with Campylobacter (Jones et al., 2016). The breed of chicken is directly related to Campylobacter infection. Brena (Brena, 2013) reported that chickens reared indoors under higher welfare standards with decreased stocking density, the prevalence of Campylobacter was lower in a slower-growing breed (Hubbard JA57) than in a standard fast-growing breed (Ross 308). However, Humphrey et al. (Humphrey et al., 2014) demonstrated no intrinsic difference in the susceptibility of broiler breeds to C. jejuni under their experimental conditions.
China has many indigenous poultry resources, and many local chickens are dual-purpose (meat–egg) producers, with a longer growth cycle than broiler chickens. In general, traditional commercial broilers, such as AA broiler, Ross 308, are slaughtered in about 42 days (Fortuoso et al., 2019). However, some of the Chinese local chickens, such as Jianghan-chickens, usually start laying eggs at 140-150 days and then are slaughtered as food around 300 days. The life cycle of this type of production differs from that of commercial chickens, which may make the ecology (including antibiotic resistance) of Campylobacter different in production cycle. Previous studies have reported that under the same breeding conditions, the Huainan partridge chicken had a lower rate of Campylobacter infection than Heihua chickens or Ni-ke hon chickens, but a higher rate than AA+ chickens (Huang et al., 2009). Bai et al. (Bai et al., 2021) found that the isolation rate of Campylobacter was lower in slaughterhouses processing yellow feather broilers (14.2%) than in those processing white feather broilers or turkeys (from 26.3 to 100%). However, there are still few data on the prevalence of Campylobacter in local chickens in China.
The prevalence of antibiotic-resistant Campylobacter in poultry also cannot be ignored. Bacteria usually acquire antimicrobial resistance (AMR) by two main pathways. One involves chromosomal mutations at the target sites of antibiotic action, such as the point mutation in the gyrA gene that causes resistance to fluoroquinolone antibiotics (Iovine, 2013). The second involves the horizontal gene transfer of mobile genetic elements that contain resistance genes (Aksomaitiene et al., 2021). In the past few years, antibiotic-resistant Campylobacter in chicken house environment, eggshell, carcasses, poultry production, and the processing chain have been reported in many countries (Modirrousta et al., 2016; Tang et al., 2020b; Habib et al., 2023). Although several studies have detected antimicrobial-resistant Campylobacter in dual-purpose chickens (Foster-Nyarko et al., 2021; Metreveli et al., 2022; Rangaraju et al., 2022), Jianghan-chicken is a unique resource, which distributed in Central China. At present, the research on Jianghan-chicken is mainly focused on the eradication of Salmonella pullorum and avian leukosis, the overall resistance and virulence of Campylobacter in this chicken are unclear. Notably, the prevalence of Campylobacter, the generation and spread of its antibiotic resistance, and the complexity of its pathogenesis are probably related to the diversity of the Campylobacter genome. Many virulence genes have undergone expansion or contraction in specific lineages, resulting in differences in the content of virulence genes and ultimately leading to the specificity of their pathogenicity (Zhong et al., 2022). Fortunately, DNA sequencing technologies provide efficient methods with which to understand the antibiotic-resistance and pathogenic mechanisms of Campylobacter.
In this study, we investigated the genetic diversity, antibiotic resistance, and the distributions of the resistance and virulence genes of Campylobacter in local dual-purpose Jianghan-chickens in four regions of central China. We also used whole-genome sequencing (WGS) to evaluate the genetic diversity of Campylobacter and the phenotypic and genetic determinants associated with its intrinsic resistance. This data from this study extends our understanding of the prevalence and genomic characteristics of food-borne Campylobacter in local chickens in China.
2 Materials and methods
2.1 Bacterial isolates and culture conditions
In this study, 30 Campylobacter isolates were isolated from 312 samples collected from eight chicken farms breeding local dual-purpose (meat–egg) chickens in four regions of central China in 2022 (Supplementary Table 1). Freshly collected cloacal swabs were stored in Cary–Blair Modified Transport Medium (Amresco, Englewood, USA) and transported to the laboratory at 4°C for Campylobacter isolation. The samples were pre-enriched in Bolton broth containing Campylobacter growth supplement (Oxoid, Basingstoke, UK) and Campylobacter Bolton broth selective supplement (Oxoid), and cultured at 42°C for 24 h under microaerobic conditions (5% O2, 10% CO2, and 85% N2). Subsequently, 100 µl cultures were inoculated on modified charcoal cefoperazone deoxycholate agar (mCCDA, Oxoid) plates containing Campylobacter CCDA selective supplements at 42°C under microaerobic condition for 48 h. Suspected positive colonies were identified with Gram staining and 16S rDNA PCR (Linton et al., 1997). All isolates were identified with PCR targeting the C. jejuni-specific hipO gene and the C. coli-specific asp gene (Lawson et al., 1998).
2.2 Antimicrobial sensitivity testing
All isolates were tested for antimicrobial susceptibility to ciprofloxacin, tetracycline, cefotaxime, amikacin, erythromycin, and lincomycin with the disk diffusion method on Mueller Hinton Agar (Oxoid), according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (Igwaran and Okoh, 2020). When the isolates were resistant to at least three different types of antibiotics, they were considered multidrug resistant (MDR). Escherichia coli ATCC 25922 was used as a quality control strain.
2.3 Whole-genome sequencing and analysis
The genomic DNA of the Campylobacter species was extracted with the TIANamp Bacteria DNA Kit (Tiangen, Beijing, China). The purity and concentration of the genomic DNA were determined by NanoDrop™ One sectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Genomic DNA (5 μg; OD260/280 = 1.8–2.0) was used for library construction. The Illumina NovaSeq 6000 sequencing platform (MajorBio Co., Shanghai, China) was used to sequence those libraries with a 2 × 150-bp read length. The raw reads obtained after sequencing were filtered with the fastp software (version 0.19.6) (Chen et al., 2018) and clean reads were obtained after the adapter sequences and low-quality sequences (Q < 20) were removed. The clean reads were then assembled with SOAPdenovo version 2.04 (Luo et al., 2012). The assembled contigs were uploaded to PubMLST (https://pubmlst.org/Campylobacter/) to determine their multilocus sequence types (STs) and clonal complexes (CCs). The phylogenetic tree and SNP count matrix heat map based on SNP analysis was obtained by using the online tool “multiple genome analysis” provided by BacWGSTdb 2.0 (http://bacdb.cn/BacWGSTdb/). RM1221_CP000025, which had abundant studies on its genome (Parker et al., 2006; Neal-McKinney et al., 2021; St. Charles et al., 2022), was selected as reference genome in BacWGSTdb tool and the construction of the phylogenetic tree in this tool relies on Neighbor-Joining (NJ) algorithm (Feng et al., 2021). The virulence genes were predicted based on the Virulence Factor Database (VFDB; http://www.mgc.ac.cn/VFs/). The tool ResFinder v.4.1 was used to detect acquired AMR genes and point mutations in specific genes conferring AMR; 90% minimum percentage identity and 60% minimum length coverage were used as the selection criteria. The sequence of the regulatory region of the cmeABC promoter (CmeR-Box) which is a 16-base inverted repeat sequence [TGTAATA (or T) TTTATTACA] (Cheng et al., 2020) and the amino acid sequence of CmeR were obtained by comparing the sequence alignment through BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). RAST Server (Rapid Annotation using Subsystem Technology) was used for Genome annotation of the assembled genome of multi-drug resistant Campylobacter spp, and the annotation scheme was ClassicRAST (http://rast.theseed.org/FIG/rast.cgi). Antibiotic resistance gene were also analyzed by Mobile Element Finder (https://cge.food.dtu.dk/services/MobileElementFinder/), and SnapGene® 2.3.2 was used to visualize gene arrangement.
2.4 Correlation analysis of susceptibility phenotypes and genotypes
The possible link between the Campylobacter resistance phenotype and the genotype predicted with WGS was analyzed by manually comparing the susceptibility test results (resistance or susceptibility) with the presence of known corresponding resistance genes and/or specific mutations. The percentage correlation between the resistance phenotype and genotype was calculated as the sum of true positives and true negatives divided by all the isolates tested. The positive predictive value was calculated by dividing the true positives by the sum of the true positives and false negatives, and the negative predictive value was calculated by dividing the true negatives by the sum of the true negatives and false positives. Sensitivity was calculated by dividing the true positives by the sum of the true positives and false positives, and specificity was calculated by dividing the true negatives by the sum of the true negatives and false negatives (Hodges et al., 2021).
3 Results
3.1 Genetic diversity analysis
Among the 30 Campylobacter isolates (13 C. jejuni and 17 C. coli) sequenced, nine sequence types (STs) in five clonal complexes (CC) based on multilocus sequence typing (MLST) were identified. Three C. coli and one C. jejuni isolates were not assigned an ST. ST8724 and ST2328 were not defined as a CC (Figure 1; Supplementary Table 2). The dominant ST for C. jejuni was ST354 (53.8%, 7/13), and the other isolates belonged to ST7469, ST8724, ST43, ST3924, or ST2328 (each 7.6%, 1/13). CC-354 was the dominant CC among the C. jejuni isolates. Among the C. coli isolates, ST825 was the most frequent ST (52.9%, 9/17), followed by ST872 (23.5%, 4/17) and ST1586 (5.9%, 1/17), and all of these assigned STs belonged to CC-828.
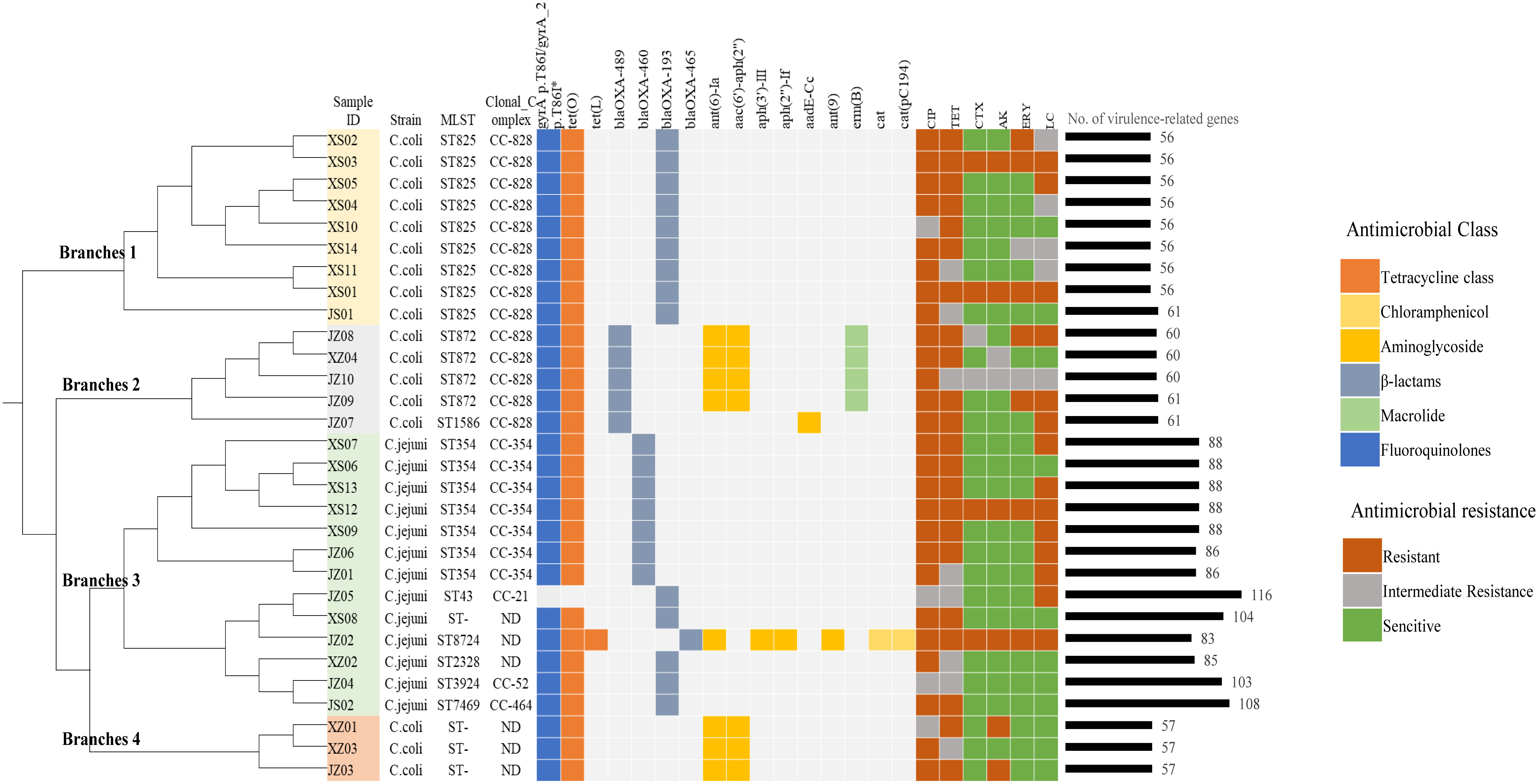
Figure 1 Genetic relationships, antimicrobial-resistance phenotypes, and the distributions of resistance- and virulence-related genes determined in this study. The phylogenetic tree was constructed based on genomic single-nucleotide polymorphisms, and the reference genome was RM1221_CP000025. The genetic determinants of antibiotic resistance are grouped according to their corresponding antibiotic categories and are color coded. The isolates were divided into 4 branches of the tree and distinguished by different colors: yellow (branch 1), gray (branch 2), green (branch 3), red (branch 4). CIP, ciprofloxacin; TET, tetracycline; CTX, cefotaxime; AK, amikacin; ERY, erythromycin; LC, lincomycin. *Point mutation.
SNP analysis was further carried out, and we found that there was a certain genetic diversity among these isolates, and these differences involved SNP differences vary greatly, from a few to thousands (Supplementary Figure 1). All the isolates can be cluster to four main branches of the phylogenetic tree (Figure 1). Branches 1 and 2 contained the major clonal complex CC-828 of C. coli, and branch 3 contained the main clonal complex (CC-354) of C. jejuni. Interestingly, branch 4, which contained the three C. coli isolates with unassigned STs, clustered with the larger branch containing C. jejuni.
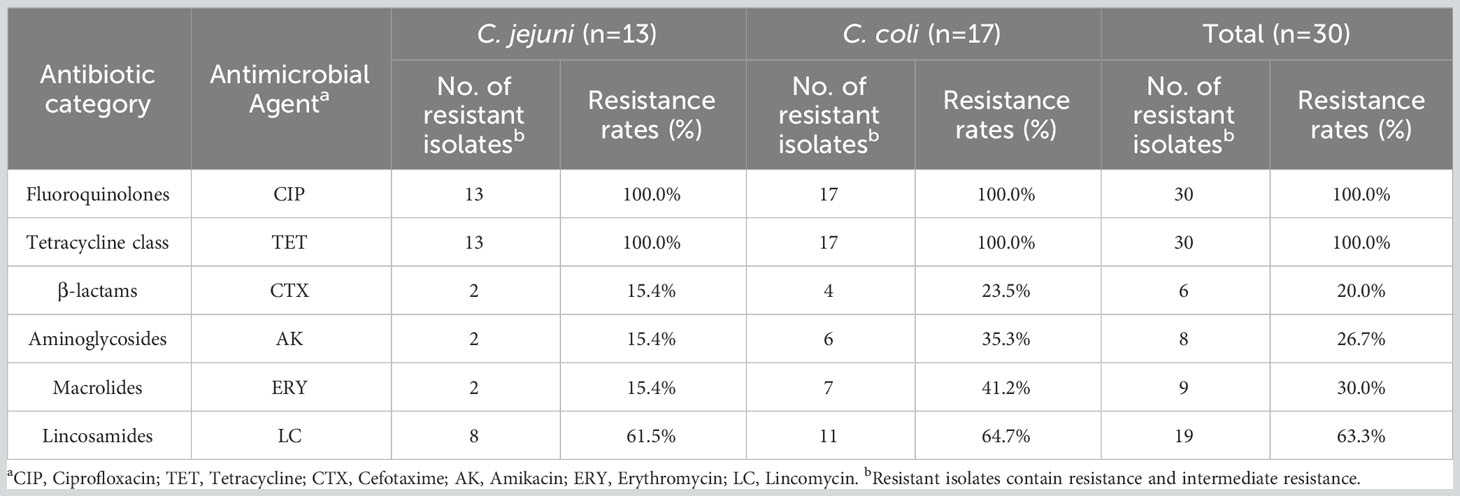
3.2 Antimicrobial susceptibility
All the isolates were tested for susceptibility to six antibiotics. As shown in Table 1, all showed resistance to ciprofloxacin and tetracycline (100% in both C. jejuni and C. coli). More than half the isolates were resistant to lincomycin (61.5% of C. jejuni and 64.7% of C. coli). The resistance rates of Campylobacter to erythromycin, amikacin, and cefotaxime were 30.0%, 26.7%, and 20.0%, respectively. The resistance rate of C. coli to erythromycin was 41.2%, which was more than twice that of C. jejuni (15.4%). The data showed similar trends for amikacin (35.3% in C. coli and 15.4% in C. jejuni). Among the 30 isolates, 22 were resistant to three or more classes of antimicrobial agents, and the most prevalent pattern of MDR was resistance to ciprofloxacin, tetracycline, and lincomycin (45.5%, 10/22) (Figure 1).
3.3 Antibiotic resistance genes and resistance mutations
In this study, a C257T chromosomal point mutation in the gyrA gene, which conferring the Thr-86-Ile substitution, and 15 acquired resistance genes were identified by genome-wide analysis. Figure 1 and Supplementary Table 3 show the distributions of the genetic determinants of resistance detected in each isolate with WGS.
Of all the isolates tested, 96.7% (29/30) carried the gyrA gene point mutation (C257T) along with a ciprofloxacin resistance phenotype. The correlation analysis of resistance phenotype and genotype showed that the gyrA C257T mutation correlated strongly with ciprofloxacin resistance (100% in C. coli and 92.3% in C. jejuni) (Table 2).
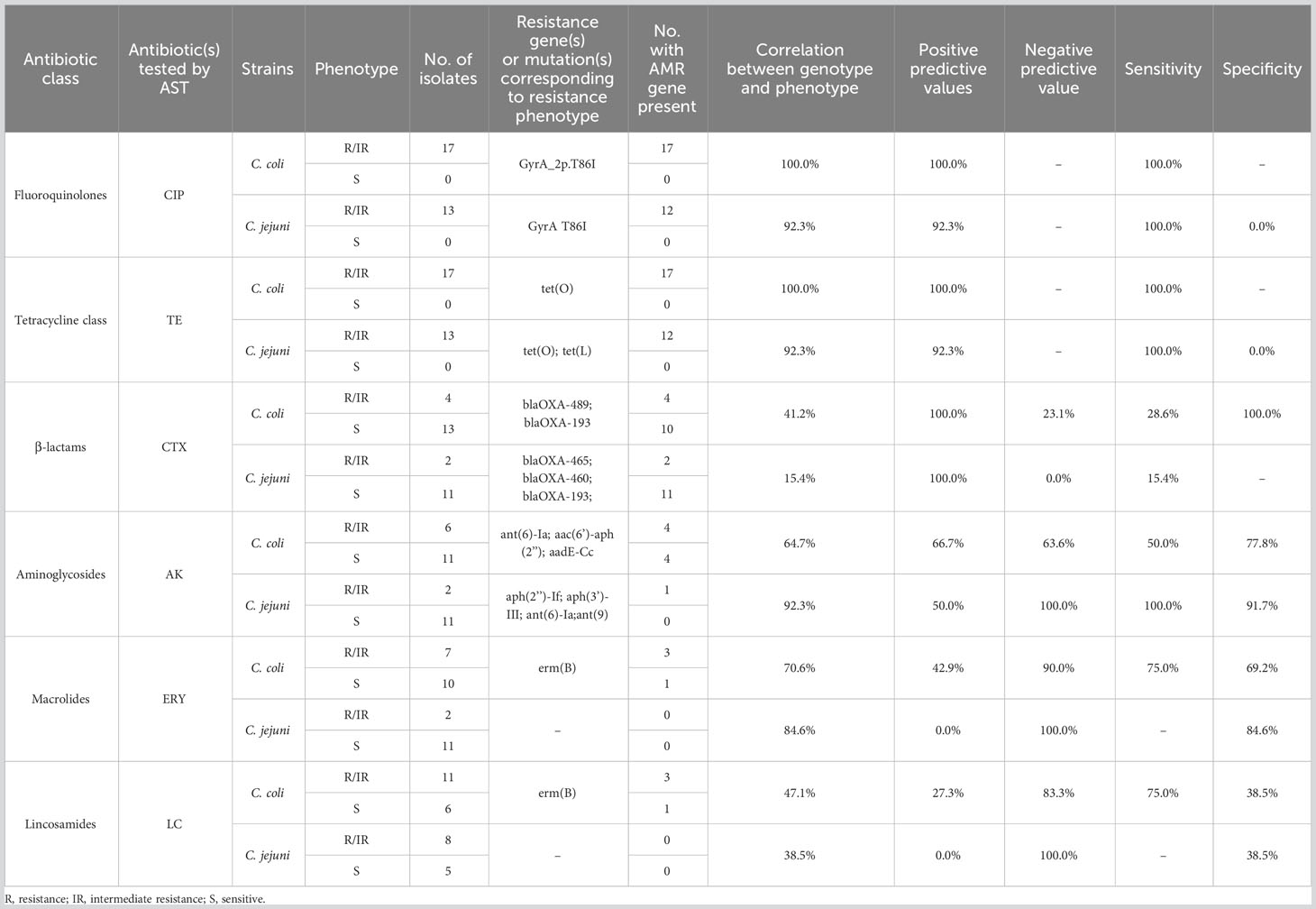
Table 2 Correlation analysis of antibiotic resistance phenotype and antibiotic resistance determinants.
Most of the isolates (96.7%, 29/30) contained the tet(O) gene, and one C. jejuni strain carried tet(L) (3.3%, 1/30). All isolates showed tetracycline resistance. The correlation between the tetracycline resistance phenotype and the resistance gene tet(O) or tet(L) was 100% in C. coli and 92.3% in C. jejuni.
The blaOXA-type β-lactamase-encoding gene was identified in 27 strains (90%, 27/30). And 22.2% (6/27) of isolates were resistant to cefotaxime.
Six aminoglycoside antibiotic resistance genes were detected in our isolates: ant(6)-Ia (26.7%, 8/30), aac(6’)-aph(3’’) (23.3%, 7/30), aph(3’)-III (3.3%, 1/30), aph(2’’)-If (3.3%, 1/30), ant(9) (3.3%, 1/30), and aadE-Cc (3.3%, 1/30). In this study, these genes mainly occurred in pairs, such as ant(6)-Ia and aac(6’)-aph(3’’), in C. coli (23.3%, 7/30). These resistance gene combinations did not correlate strongly with the amikacin resistance phenotypic in C. coli (64.7%), but did correlate strongly with it in C. jejuni (92.3%).
The erythromycin and lincomycin resistance gene erm(B) was only identified in four C. coli isolates (13.3%, 4/30). The correlation between erm(B) and the erythromycin or lincomycin resistance phenotype was not strong (70.6% or 47.1%, respectively, in C. coli; and 84.6% or 38.5%, respectively, in C. jejuni). Further analysis of the isolates for point mutations in 23S rRNA revealed eight mutations in total (Supplementary Table 4), although neither the A2075G nor A2074C/G mutation, which reportedly cause erythromycin resistance, was detected.
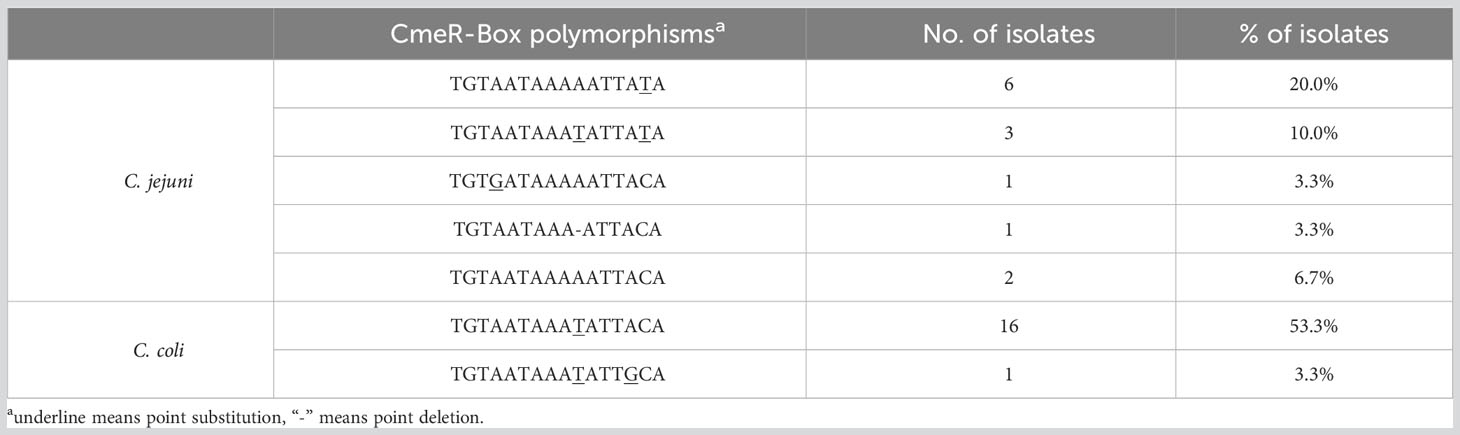
3.4 CmeR-Box polymorphisms
A CmeR-Box polymorphism analysis of all isolates (Table 3) detected six CmeR-Box variants in 28 isolates. Among these, point substitutions were most common (96.4%), involving 17 C. coli and 10 C. jejuni isolates, whereas only one C. jejuni isolate (3.6%) had a point deletion, and no point insertion was detected in the CmeR-Box.
3.5 Genetic environment analysis of antibiotic resistance gene clusters in an MDR C. jejuni isolate
We analyzed the genetic environments of the resistance genes in C. jejuni JZ02, which was resistant to all of six antibiotics tested. Two antibiotic resistance gene clusters were detected (Figure 2). Gene cluster 1 contained the tet(O), tet(L), and cat (pC194) genes (Figure 2A). A transposase was encoded upstream from the tet(L) gene, and a 39-bp repeat and another transposase gene that shared 100% identity with IS1216 family transposase gene, were detected between tet (O) and cat (pC194). Moreover, a transposon encoding the protein TnpV was detected upstream from the tet(O) gene. Gene cluster 2 consisted of the ant (9), aph(3’)-III, aph(2’’)-If, and cat genes (Figure 2B). However, no mobile genetic elements or repetitive sequences were detected in this gene cluster, although a box element and several hypothetical proteins with sequences similar to those of some Gram-positive bacteria were found.
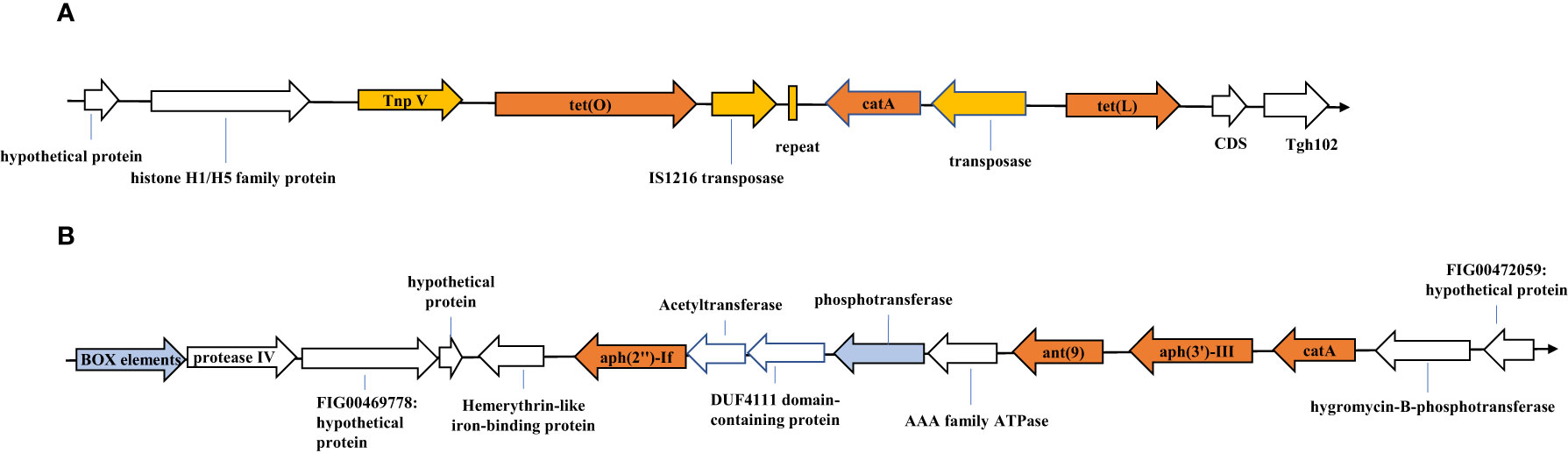
Figure 2 Resistance gene clusters identified in Campylobacter jejuni strain JZ02. (A) Tetracycline resistance gene cluster; (B) Aminoglycosides resistance gene cluster. SnapGene® 2.3.2 made this figure.
3.6 Virulence gene detection
Based on the VFDB, 126 virulence-related genes, involving adhesion, invasion, motility, toxins, and the type IV secretion system, were identified (Supplementary Table 5). We observed more virulence-related genes in C. jejuni (83–116 per isolate) than in C. coli (56–61 per isolate), and among them, isolates C. jejuni JZ05 (CC-21) and JS02 (CC-464) had the most virulence-related genes (Figure 1). Most of the genes only detected in C. jejuni were related to motility and adhesion, including cadF, htrB, pebA, ciaB, jlpA, and cheA, and genes encoding cytolethal distending toxin (cdtABC) were also only detected in C. jejuni. Type IV secretion system genes, including virB11, virB10, virB9, virB8, virB4, and virD4, were detected in one C. jejuni isolate (3.3%), and wlaN was only found in two C. jejuni isolates (6.7%) (Table 4). Campylobacter isolates in most branches (branch 1, 2, 4) of the phylogenetic tree had similar numbers of virulence genes, and the categories of these genes were not quite different. Interestingly, in branch 3, the type and abundance of virulence genes vary greatly from different ST types, and most of the different genes are related to capsular synthesis and immune regulation. Some isolates with more virulence-related genes were distributed in a sub-branch of branch 3.
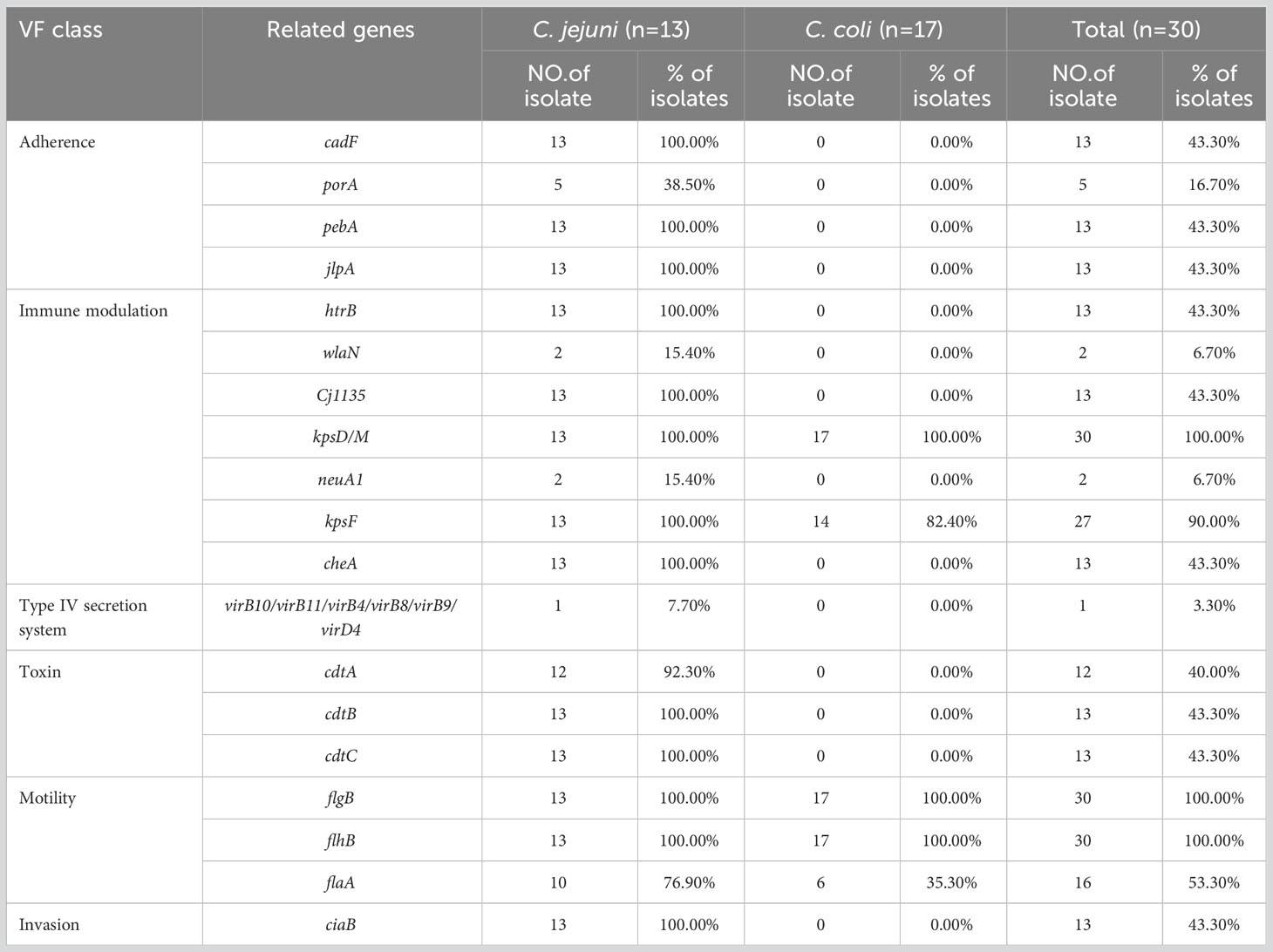
Table 4 Frequencies of parts of predicted virulence-related factors in the genomes of 30 Campylobacter isolates.
4 Discussion
Campylobacter is the main bacterial pathogen causing human diarrhea worldwide, and its increasing prevalence and antibiotic resistance have caused great concern globally in recent years, in both human and veterinary clinics. Poultry is the main host of Campylobacter, but the prevalence of the pathogen varies across different species and different regions. For instance, an investigation in southeastern Italy showed that the prevalence of C. jejuni was higher in broilers than in laying hens (45.7% and 21.1%, respectively) (Parisi et al., 2007). In Europe, the prevalence of broiler flocks colonized with Campylobacter ranged from 18% to > 90% in different countries (Newell and Fearnley, 2003). Meat–egg dual-purpose local chickens may differ from commercial varieties because their breeding modes and breeding cycles differ, and they may pose a potential risk of Campylobacter transmission to both meat and eggs (Ahmed et al., 2021). Therefore, we investigated the phylogenetic relationships, virulence genes, antibiotic resistance, and genetic bases of the resistance phenotypes of Campylobacter isolates collected from local meat–egg dual-purpose chicken in China.
In this study, we identified two main prevalent Campylobacter species, C. jejuni and C. coli, and found strong genetic diversity in the Campylobacter strains transmitted in these chickens. The National Center for Biotechnology Information (NCBI) database indicated that CC-354 strains occur mainly in the United States and the United Kingdom, whereas they are quite dispersed in other countries (Yu et al., 2020). A previous study showed that CC-353 and CC-464 are the dominant CCs of C. jejuni in central China, and CC-354 was the dominant population of C. jejuni detected in the present study. CC-21 is also the most frequently reported C. jejuni genotype in diarrhea patients in China (Zhang et al., 2020b), and in Zhang et al.’s study (Zhang et al., 2020a), CC-21 was also the dominant Campylobacter CC in chickens in southeastern China. However, in the present study, only one strain belonging to CC-21 was isolated, suggesting that the diversity of C. jejuni may vary by region and sample source, and that the epidemic patterns of Campylobacter may differ in local meat–egg dual-purpose chickens. Three ST types, ST1586, ST872, and ST828, were found in the C. coli isolates, which belong to the same clonal complex, CC-828. This was expected because CC-828 is the dominant population of C. coli, and a large number of past studies have reported its prevalence around the world (Zhang et al., 2016; Di Giannatale et al., 2019; Gomes et al., 2019). Based on principal component analysis (PCA) on the evolutionary distances of core gene families, Snipen et al. reported that Campylobacter has a mixed evolutionary pattern characterized by genomes (Snipen et al., 2012). It is noteworthy that the three C. coli strains with undefined STs detected in this study clustered with C. jejuni on the same large branch of a phylogenetic tree based on a genomic SNP analysis, suggesting that their genetic relationship was close. Previous studies have shown that an bidirectional increase in the rate of recombination between C. jejuni and C. coli has led to the gradual convergence of the two species (Sheppard et al., 2008).
Antibiotic resistance has become one of the most important factors threatening human public health globally (Mancuso et al., 2021). It is noteworthy that 73.3% of Campylobacter isolates were multidrug resistant in the present study. The resistance rates of Campylobacter to ciprofloxacin and tetracycline in China are high, and studies have reported rates of 90%–100% in broilers (Ma et al., 2014; Li et al., 2016; Wang et al., 2021). In the present study, there were similar high resistance rates to ciprofloxacin and tetracycline in both C. jejuni and C. coli. In the past, fluoroquinolones have been widely used in the edible animal industry, especially in poultry production, although tetracyclines are also commonly used to treat and prevent bacterial diseases in poultry in China. This may explain the high resistance rates to these two antibiotics in Campylobacter. In the early 1980s, the development and introduction of the third-generation extended-spectrum cephalosporin cefotaxime provided a new treatment for patients infected with Gram-negative bacilli (Hawkey, 2008). Here, we detected a relatively low rate of cefotaxime resistance (20.0%). Although it is not approved for use in food animals in China (Dai et al., 2008), we detected a high rate of amikacin resistance in Campylobacter (26.7%). Nor did the proportion of erythromycin-resistant isolates in our study differ greatly from that reported in previous studies (30.0% and 25.2%, respectively) (Cheng et al., 2020). However, the erythromycin resistance rate of C. jejuni was lower than in previous studies (15.4% and 30.1%, respectively), whereas the rate in C. coli was higher (41.2% and 18.3%, respectively) (Cheng et al., 2020). We detected high rates of resistance to lincomycin in both C. jejuni and C. coli, which may be related to the antibiotics commonly used in the areas from which the isolates were collected. The resistance of C. coli to antibiotics other than tetracycline and ciprofloxacin was greater than that of C. jejuni. These findings are consistent with the results of Tang et al. (Tang et al., 2020a), who reported that the prevalence of antibiotic resistance in chicken-derived C. coli was higher than in chicken-derived C. jejuni. In general, there is a worrying trend that, although the addition of antibiotics to feed supplements was banned in China in 2020, it has not reduced antibiotic resistance. On the contrary, some antibiotic resistance rates are still rising in some regions (Cheng et al., 2020).
Previous studies have shown that there is a strong correlation between the presence of AMR determinants detected with WGS and phenotypic antibiotic resistance (Rokney et al., 2020; Habib et al., 2023). However, it is known that Campylobacter also has many resistance mechanisms other than resistance gene-mediated, such as changes in membrane permeability, modification of the antibiotic efflux pumps, etc. (Iovine, 2013). The determinants of drug resistance do not always confer resistance phenotypes, and single resistance determinant may correlate weakly with certain antibiotics (Šoprek et al., 2022). In this study, we found that the overall correlation between the 16 antibiotic resistance determinants detected with ResFinder v.4.1 and phenotypic resistance was not strong, and that there were huge differences between the different antibiotics. This suggests that current research into the resistance mechanisms of Campylobacter remains to be improved, and that simply analyzing bacterial resistance in terms of the antibiotic resistance determinants predicted with WGS does not provide an accurate assessment.
In the present study, phenotypic resistance to ciprofloxacin and tetracycline correlated well with the presence of the gyrA gene point mutation (C257T) and the tet(O) or tet(L) gene, respectively, confirming that they are the main factors conferring resistance to the corresponding antimicrobial agents. CTX-M type β-lactamases usually are the cause of drug resistance of Gram-negative bacteria to cephalosporin such as cefotaxime, but CTX-M was not found in resistant strains in this study. In our isolates, 90% isolates of our study contained blaOXA-type β-lactamase-encoding gene. Indeed, most Campylobacter strains contain the bla-OXA gene encoding β-lactamase that confers resistance to carbapenems, but not to cephalosporin (Hadiyan et al., 2022). Research has already shown that different β-lactamases have different hydrolysis profiles (Poirel et al., 2011) and that the expression of β-lactamase directly affects the resistance of strains to β-lactam antibiotics (Casagrande Proietti et al., 2020). This may also explain why strains containing the bla-OXA gene but with a β-lactam-sensitive phenotype have been found in several other studies (Griggs et al., 2009; Zeng et al., 2014; Hadiyan et al., 2022). Our study further confirmed that the presence of bla-OXA gene is not related to the resistance of cephalosporin drugs.
The prevalence of aminoglycoside-resistance-related determinants was low in the isolates tested, but these determinants showed relatively high diversity. A previous study demonstrated that the combined action of the aph(3′)-III, aac(6′)-aph(2′′), and ant(6)-Ia genes conferred resistance to aminoglycoside antibiotics on Campylobacter (Zhang et al., 2022), which was confirmed in our study. However, even with the synergistic effect of ant(6)-Ia and aac(6’)-aph(3’’), the correlation between each gene and amikacin resistance was still low. Moreover, a C. coli isolate containing aadE-Cc showed a sensitive phenotype. This finding is consistent with a report by Painset et al. (Painset et al., 2020), who also observed Campylobacter strains carrying the aadE-Cc gene that were not resistant to some aminoglycoside antibiotics. There may be some unknown mechanism that inactivate these genes in Campylobacter.
Erythromycin and lincomycin have similar resistance mechanisms (Zhao et al., 2016; Wang et al., 2022). In the present study, the correlation between erm(B) and resistance to those two antibiotics was not strong. Therefore, we analyzed the sequence of 23S rRNA, and found no A2075G mutation.
Since most of the strains in this study have multi-drug resistance, we analyzed the genetic environment of the resistance gene of JZ02 (resistant to six antibiotics), and try to know how this strain obtained the antibiotic resistance gene. As is known that Campylobacter can acquire exogenous DNA through natural transformation (Wang and Taylor, 1990). The spread of antibiotic resistance genes in Campylobacter isolates from humans, animals, and the environment has previously been reported (Asuming-Bediako et al., 2019). The tetracycline resistance gene tet(O) is believed to have originated in Gram-positive cocci (Zilhao et al., 1988), and the tetracycline resistance mediated by this gene is mainly spread via the horizontal transfer of resistance genes on conjugated plasmids (Wardak et al., 2007). Although we found that resistance gene tet(O) was located on chromosome of isolate JZ02, some tranposase-encoding sequences were detected near tet(O). The presence of these transposases implies that the antibiotic-resistance genes were co-transferred with some mobile genetic elements into the genomes of related strains. Although no relevant mobile elements were found in cluster 2, several genes, such as ant(9) and aph(2’’)-If, which encode aminoglycoside-modifying enzymes, are similar to those of some Gram-positive bacteria, indicating that they may have derived from Gram-positive bacteria in the environment or animal intestines (Fabre et al., 2018).
The ability of Campylobacter to cause human diseases is considered multifactorial, and several genes are closely related to its virulence, including ciaB and cdtABC (Lopes et al., 2021). An analysis of the virulence-related genes of our isolates showed that C. jejuni carried more virulence-related genes than C. coli, which is consistent with the study of Lapierre et al. (Lapierre et al., 2016), and most of these genes were involved in motility (flaA), adhesion (cadF, cheA, jlpA et al), and invasion (ciaB). It is noteworthy that in this study, CC-21 and CC-464 had the most virulence-related genes and these two clonal complexes are also common among the clinical isolates of Campylobacter (Zhang et al., 2020a; Zhang et al., 2020b; Zang et al., 2021). Then we found that the additional genes they carried were mainly involved in immune modulation like bacterial capsule biosynthesis, especially by sugar and aminotransferase enzymes (kfiD, glf, Cj1426c, Cj1432c, Cj1434c, Cj1435c, Cj1436c, Cj1437c) while these genes do not be harbored in other complexes(Supplementary Table 5; Supplementary Figure 2). Although a high prevalence of virulence-associated genes (ciaB and flaA) has been already reported in Campylobacter strains infecting children with moderate to severe diarrhea (Quetz et al., 2012), these genes were only detected in C. jejuni in the present study. This may explain why C. jejuni colonizes its host more readily than C. coli and is responsible for more food-borne bacterial infection events (Moffatt et al., 2019; Callahan Sean et al., 2021; Schirone and Visciano, 2021). Virulence genes related to the type IV secretion system were only found in one strain of C. jejuni, and these genes are less prevalent in Asia and Europe (Panzenhagen et al., 2021). We also detected the wlaN gene, which is involved in Guillain–Barre syndrome in two C. jejuni isolates(Guirado et al., 2020).
In conclusion, in this study, we have demonstrated the genetic diversity and antimicrobial susceptibility of Campylobacter isolated from local dual-purpose chickens in China, and analyzed their resistance- and virulence-related genes. It thus provides important data on the epidemiological characteristics of Campylobacter in this food source.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving animals were reviewed and approved by the Ethics Committee of Institute of Animal Husbandry and Veterinary, Hubei Academy of Agricultural Sciences (Wuhan, China).
Author contributions
JX, YC, QingL, and TZ conceived and designed the experiments. JX, YC, QinL, and YG performed the experiments. GW, WZ, and QH analyzed the data. JX, HS, and TZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key Research and Development Plan of China (2022YFD1800400), the China Agriculture Research System (CARS-41), the Key Projects of Hubei Natural Science Foundation (2021CFA019), and the Key Research and Development Program of Hubei Province (2022BBA0055).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1236777/full#supplementary-material
Supplementary Figure S1 | Matrix of SNP pair counts among 30 isolates. Number of SNP was calculated by comparing the genome sequences to the reference (RM1221) genome. Background colors represent different number of pairwise SNPs.
Supplementary Figure S2 | Distribution of virulence-related genes of 30 isolates.
References
Ahmed, T., Ameer, H. A., Javed, S. (2021). Pakistan’s backyard poultry farming initiative: Impact analysis from a public health perspective. Trop. Anim. Health Production 53 (2), 210. doi: 10.1007/s11250-021-02659-6
Aksomaitiene, J., Novoslavskij, A., Kudirkiene, E., Gabinaitiene, A., Malakauskas, M. (2021). Whole genome sequence-based prediction of resistance determinants in high-level multidrug-resistant campylobacter jejuni isolates in Lithuania. Microorganisms 9 (1), 66. doi: 10.3390/microorganisms9010066
Asuming-Bediako, N., Parry-Hanson Kunadu, A., Abraham, S., Habib, I. (2019). Campylobacter at the human–food interface: The african perspective. Pathogens 8 (2), 87. doi: 10.3390/pathogens8020087
Bai, J., Chen, Z., Luo, K., Zeng, F., Qu, X., Zhang, H., et al. (2021). Highly prevalent multidrug-resistant campylobacter spp. Isolated from a yellow-feathered broiler slaughterhouse in South China. Front. Microbiol. 12, 682741. doi: 10.3389/fmicb.2021.682741
Brena, M. C. (2013). Effect of different poultry production methods on Campylobacter incidence and transmission in the broiler meat food chain. (University of Liverpool). Available at: https://livrepository.liverpool.ac.uk/18837/1/BrenaMar_Nov2013_18837.pdf
Callahan Sean, M., Dolislager Carolina, G., Johnson Jeremiah, G. (2021). The host cellular immune response to infection by campylobacter Spp. and its role in disease. Infection Immun. 89 (8), e0011621. doi: 10.1128/iai.00116-21
Casagrande Proietti, P., Guelfi, G., Bellucci, S., De Luca, S., Di Gregorio, S., Pieramati, C., et al. (2020). Beta-lactam resistance in Campylobacter coli and Campylobacter jejuni chicken isolates and the association between blaOXA-61 gene expression and the action of β-lactamase inhibitors. Vet. 241, 108553. doi: 10.1016/j.vetmic.2019.108553
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), i884–i890. doi: 10.1093/bioinformatics/bty560
Cheng, Y., Zhang, W., Lu, Q., Wen, G., Zhao, Z., Luo, Q., et al. (2020). Point deletion or insertion in CmeR-Box, A2075G substitution in 23S rRNA, and presence of erm(B) are key factors of erythromycin resistance in campylobacter jejuni and campylobacter coli isolated from central China. Front. Microbiol. 11, 203. doi: 10.3389/fmicb.2020.00203
Dai, L., Lu, L.-M., Wu, C.-M., Li, B.-B., Huang, S.-Y., Wang, S.-C., et al. (2008). Characterization of antimicrobial resistance among Escherichia coli isolates from chickens in China between 2001 and 2006. FEMS Microbiol. Lett. 286 (2), 178–183. doi: 10.1111/j.1574-6968.2008.01272.x
Di Giannatale, E., Calistri, P., Di Donato, G., Decastelli, L., Goffredo, E., Adriano, D., et al. (2019). Thermotolerant Campylobacter spp. in chicken and bovine meat in Italy: Prevalence, level of contamination and molecular characterization of isolates. PLoS One 14 (12), e0225957. doi: 10.1371/journal.pone.0225957
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards), Koutsoumanis, K., Allende, A., Alvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., et al (2020). Update and review of control options for Campylobacter in broilers at primary production. EFSA J. 18 (4), e06090. doi: 10.2903/j.efsa.2020.6090
Fabre, A., Oleastro, M., Nunes, A., Santos, A., Sifré, E., Ducournau, A., et al. (2018). Whole-genome sequence analysis of multidrug-resistant campylobacter isolates: A focus on aminoglycoside resistance determinants. J. Clin. Microbiol. 56 (9), e00390–e00318. doi: 10.1128/JCM.00390-18
Feng, Y., Zou, S., Chen, H., Yu, Y., Ruan, Z. (2021). BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 49 (D1), D644–D650. doi: 10.1093/nar/gkaa821
Fortuoso, B. F., dos Reis, JoãoH., Gebert, R. R., Barreta, M., Griss, L. G., Casagrande, R. A., et al. (2019). Glycerol monolaurate in the diet of broiler chickens replacing conventional antimicrobials: Impact on health, performance and meat quality. Microbial Pathogenesis 129, 161–167. doi: 10.1016/j.micpath.2019.02.005
Foster-Nyarko, E., Alikhan, N. F., Ravi, A., Thomson, N. M., Jarju, S., Kwambana-Adams, B. A., et al. (2021). Genomic diversity of Escherichia coli isolates from backyard chickens and Guinea fowl in the Gambia. Microb. Genom. 7 (1), mgen000484. doi: 10.1099/mgen.0.000484
Gomes, C. N., Frazão, M. R., Passaglia, J., Duque, S. S., Medeiros, M. I.C., Falcão, J. P. (2019). Molecular epidemiology and resistance profile of campylobacter jejuni and campylobacter coli strains isolated from different sources in Brazil. Microbial Drug Resistance 26 (12), 1516–1525. doi: 10.1089/mdr.2019.0266
Griggs, D. J., Peake, L., Johnson, M. M., Ghori, S., Mott, A., et al. (2009). β-lactamase-mediated β-lactam resistance in campylobacter species: Prevalence of cj0299 (blaOXA-61) and evidence for a novel β-lactamase in C. jejuni. Antimicrob. Agents Chemother. 53 (8), 3357–3364. doi: 10.1128/aac.01655-08
Guirado, P., Paytubi, S., Miró, E., Iglesias-Torrens, Y., Navarro, F., Cerdà-Cuéllar, M., et al. (2020). Differential Distribution of the wlaN and cgtB Genes, Associated with Guillain-Barré Syndrome, in Campylobacter jejuni Isolates from Humans, Broiler Chickens, and Wild Birds. Microorganisms 8 (3), 325. doi: 10.3390/microorganisms8030325
Habib, I., Ibrahim Mohamed, M.-Y., Ghazawi, A., Lakshmi, G. B., Khan, M., Li, D., et al. (2023). Genomic characterization of molecular markers associated with antimicrobial resistance and virulence of the prevalent Campylobacter coli isolated from retail chicken meat in the United Arab Emirates. Curr. Res. Food Sci. 6, 100434. doi: 10.1016/j.crfs.2023.100434
Hadiyan, M., Momtaz, H., Shakerian, A. (2022). Prevalence, antimicrobial resistance, virulence gene profile and molecular typing of Campylobacter species isolated from poultry meat samples. Vet. Med. Sci. 8 (6), 2482–2493. doi: 10.1002/vms3.944
Hawkey, P. M. (2008). The growing burden of antimicrobial resistance. J. Antimicrob. Chemother. 62 (suppl_1), i1–i9. doi: 10.1093/jac/dkn241
Hodges, L. M., Taboada, E. N., Koziol, A., Mutschall, S., Blais, B. W., Douglas Inglis, G., et al. (2021). Systematic evaluation of whole-genome sequencing based prediction of antimicrobial resistance in campylobacter jejuni and C. coli. Front. Microbiol. 12, 776967. doi: 10.3389/fmicb.2021.776967
Huang, J. L., Xu, H. Y., Bao, G. Y., Zhou, X. H., Ji, D. J., Zhang, G., et al. (2009). Epidemiological surveillance of Campylobacter jejuni in chicken, dairy cattle and diarrhoea patients. Epidemiol. Infection 137 (8), 1111–1120. doi: 10.1017/S0950268809002039
Humphrey, S., Chaloner, G., Kemmett, K., Davidson, N., Williams, N., Kipar, A., et al. (2014). Campylobacter jejuni is not merely a commensal in commercial broiler chickens and affects bird welfare. mBio 5 (4), e01364–e01314. doi: 10.1128/mBio.01364-14
Igwaran, A., Okoh, A. I. (2020). Occurrence, virulence and antimicrobial resistance-associated markers in campylobacter species isolated from retail fresh milk and water samples in two district municipalities in the Eastern Cape Province, South Africa. Antibiotics 9 (7), 426. doi: 10.3390/antibiotics9070426
Iovine, N. M. (2013). Resistance mechanisms in Campylobacter jejuni. Virulence 4 (3), 230–240. doi: 10.4161/viru.23753
Jones, D. R., Guard, J., Gast, R. K., Buhr, R. J., Fedorka-Cray, P. J., Abdo, Z., et al. (2016). Influence of commercial laying hen housing systems on the incidence and identification of Salmonella and Campylobacter1. Poultry Sci. 95 (5), 1116–1124. doi: 10.3382/ps/pew036
Ju, C. Y., Zhang, M. J., Ma, Y. P., Lu, J. R., Yu, Mu H., Chen, H., et al. (2018). Genetic and antibiotic resistance characteristics of campylobacter jejuni isolated from diarrheal patients, poultry and cattle in Shenzhen. Biomed. Environ. Sci. 31 (8), 579–585. doi: 10.3967/bes2018.079
Lapierre, L., Gatica, MaríaA., Riquelme, Víctor, Vergara, C., Yañez, JoséM., San Martín, B., et al. (2016). Characterization of antimicrobial susceptibility and its association with virulence genes related to adherence, invasion, and cytotoxicity in campylobacter jejuni and campylobacter coli isolates from animals, meat, and humans. Microbial Drug Resistance 22 (5), 432–444. doi: 10.1089/mdr.2015.0055
Lawson, A. J., Shafi, M. S., Pathak, K., Stanley, J. (1998). Detection of campylobacter in gastroenteritis: comparison of direct PCR assay of faecal samples with selective culture. Epidemiol. Infection 121 (3), 547–553. doi: 10.1017/S0950268898001630
Li, B., Ma, L., Li, Y., Jia, H., Wei, J., Shao, D., et al. (2016). Antimicrobial resistance of campylobacter species isolated from broilers in live bird markets in Shanghai, China. Foodborne Pathog. Dis. 14 (2), 96–102. doi: 10.1089/fpd.2016.2186
Linton, D., Lawson, A. J., Owen, R. J., Stanley, J. (1997). PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35 (10), 2568–2572. doi: 10.1128/jcm.35.10.2568-2572.1997
Lopes, G. V., Ramires, T., Kleinubing, N. R., Scheik, LetíciaK., Fiorentini, ÂngelaM., Padilha da Silva, W. (2021). Virulence factors of foodborne pathogen Campylobacter jejuni. Microbial Pathogenesis 161, 105265. doi: 10.1016/j.micpath.2021.105265
Luangtongkum, T., Jeon, B., Han, J., Paul, P., Logue, C. M., Zhang, Q. (2009). Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4 (2), 189–200. doi: 10.2217/17460913.4.2.189
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1 (1), 2047–217X-1-18. doi: 10.1186/2047-217X-1-18
Ma, L., Wang, Y., Shen, J., Zhang, Q., Wu, C. (2014). Tracking Campylobacter contamination along a broiler chicken production chain from the farm level to retail in China. Int. J. Food Microbiol. 181, 77–84. doi: 10.1016/j.ijfoodmicro.2014.04.023
Mancuso, G., Midiri, A., Gerace, E., Biondo, C. (2021). Bacterial antibiotic resistance: The most critical pathogens. Pathogens 10 (10), 1310. doi: 10.3390/pathogens10101310
Metreveli, M., Bulia, S., Tevzadze, L., Tsanava, S., Zarske, M., Goenaga, J. C., et al. (2022). Comparison of antimicrobial susceptibility profiles of thermotolerant campylobacter spp. Isolated from human and poultry samples in Georgia (Caucasus). Antibiotics 11 (10), 1419. doi: 10.3390/antibiotics11101419
Modirrousta, S., Shapouri, R., Rezasoltani, S., Molaabaszadeh, H. (2016). Prevalence of campylobacter spp. and their common serotypes in 330 cases of red-meat, chicken-meat and egg-shell in Zanjan City, Iran. Infect. Epidemiol. Med. 2 (1), 8–10. doi: 10.18869/modares.iem.2.1.8
Moffatt, C. R.M., Fearnley, E., Bell, R., Wright, R., Gregory, J., Sloan-Gardner, T., et al. (2019). Characteristics of campylobacter gastroenteritis outbreaks in Australia 2001 to 2016. Foodborne Pathog. Dis. 17 (5), 308–315. doi: 10.1089/fpd.2019.2731
Neal-McKinney, J. M., Liu, K. C., Lock, C. M., Wu, W.-H., Hu, J. (2021). Comparison of MiSeq, MinION, and hybrid genome sequencing for analysis of Campylobacter jejuni. Sci. Rep. 11 (1), 5676. doi: 10.1038/s41598-021-84956-6
Newell, D. G., Fearnley, C. (2003). Sources of campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69 (8), 4343–4351. doi: 10.1128/AEM.69.8.4343-4351.2003
Painset, Anaïs, Day, M., Doumith, M., Rigby, J., Jenkins, C., Grant, K., et al. (2020). Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Campylobacter jejuni and Campylobacter coli isolated from cases of diarrhoeal disease in England and Wales 2015–16. J. Antimicrob. Chemother. 75 (4), 883–889. doi: 10.1093/jac/dkz539
Panzenhagen, P., Portes, A. B., dos Santos, A. M.P., Duque, S. D., Conte Junior, C. A. (2021). The distribution of campylobacter jejuni virulence genes in genomes worldwide derived from the NCBI pathogen detection database. Genes 12 (10), 1538. doi: 10.3390/genes12101538
Parisi, A., Lanzilotta, S. G., Addante, N., Normanno, G., Di Modugno, G., Dambrosio, A., et al. (2007). Prevalence, molecular characterization and antimicrobial resistance of thermophilic campylobacter isolates from cattle, hens, broilers and broiler meat in south-eastern Italy. Vet. Res. Commun. 31 (1), 113–123. doi: 10.1007/s11259-006-3404-3
Parker, C. T., Quiñones, B., Miller, W. G., Horn, S. T., Mandrell, R. E. (2006). Comparative genomic analysis of campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in c. jejuni strain RM1221. J. Clin. Microbiol. 44 (11), 4125–4135. doi: 10.1128/jcm.01231-06
Pham, N. T. K., Thongprachum, A., Tran, D. N., Nishimura, S., Shimizu-Onda, Y., Duy Trinh, Q., et al. (2016). Antibiotic resistance of campylobacter jejuni and c. coli isolated from children with diarrhea in Thailand and Japan. J. Japanese J. Infect. Dis. 69 (1), 77–79. doi: 10.7883/yoken.JJID.2014.582
Poirel, L., Castanheira, M., Carrër, Amélie, Rodriguez, C. P., Jones, R. N., Smayevsky, J., et al. (2011). OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 55 (6), 2546–2551. doi: 10.1128/aac.00022-11
Quetz, J. da S., Lima, I. F.N., Havt, A., Prata, M. M.G., Cavalcante, P. A., Medeiros, P. H.Q.S., et al. (2012). Campylobacter jejuni infection and virulence-associated genes in children with moderate to severe diarrhoea admitted to emergency rooms in northeastern Brazil. J. Med. Microbiol. 61 (4), 507–513. doi: 10.1099/jmm.0.040600-0
Rangaraju, V., Malla, B. A., Milton, A. A. P., Madesh, A., Balasaheb Madhukar, K., Kadwalia, A., et al. (2022). Occurrence, antimicrobial resistance and virulence properties of thermophilic Campylobacter coli originating from two different poultry settings. Gene Rep. 27, 101618. doi: 10.1016/j.genrep.2022.101618
Rokney, A., Valinsky, L., Vranckx, K., Feldman, N., Agmon, V., Moran-Gilad, J., et al. (2020). WGS-based prediction and analysis of antimicrobial resistance in campylobacter jejuni isolates from Israel. Front. Cell. Infect. Microbiol. 10, 365. doi: 10.3389/fcimb.2020.00365
Romanescu, M., Oprean, C., Lombrea, A., Badescu, B., Teodor, A., Constantin, G. D., et al(2023). Current state of knowledge regarding WHO high priority pathogens—resistance mechanisms and proposed solutions through candidates such as essential oils: a systematic review. Int. J. Mol. Sci. 24 (11), 9727. doi: 10.3390/ijms24119727
Schirone, M., Visciano, P. (2021). Trends of major foodborne outbreaks in the european union during the years 2015–2019. Hygiene 1 (3), 106–1195. doi: 10.3390/hygiene1030010
Sheppard, S. K., McCarthy, N. D., Falush, D., Maiden, M. C.J. (2008). Convergence of campylobacter species: Implications for bacterial evolution. Science 320 (5873), 237–239. doi: 10.1126/science.1155532
Snipen, L., Wassenaar, T. M., Altermann, E., Olson, J., Kathariou, S., Lagesen, K., et al. (2012). Analysis of evolutionary patterns of genes in Campylobacter jejuni and C. coli. Microbial Inf. Experimentation 2 (1), 8. doi: 10.1186/2042-5783-2-8
Šoprek, S., Duvnjak, S., Kompes, G., Jurinović, L., Tambić Andrašević, A. (2022). Resistome analysis of campylobacter jejuni strains isolated from human stool and primary sterile samples in Croatia. Microorganisms 10 (7), 1410. doi: 10.3390/microorganisms10071410
Soro, A. B., Whyte, P., Bolton, D. J., Tiwari, B. K. (2020). Strategies and novel technologies to control Campylobacter in the poultry chain: A review. Compr. Rev. Food Sci. Food Saf. 19 (4), 1353–1377. doi: 10.1111/1541-4337.12544
St. Charles, J. L., Brooks, P. T., Bell, J. A., Ahmed, H., Allen, M. V., Manning, S. D., et al. (2022). Zoonotic transmission of campylobacter jejuni to caretakers from sick pen calves carrying a mixed population of strains with and without guillain barré Syndrome-associated lipooligosaccharide loci. Front. Microbiol. 13, 800269. doi: 10.3389/fmicb.2022.800269
Tang, Y., Jiang, Q., Tang, H., Wang, Z., Yin, Yi, Ren, F., et al. (2020b). Characterization and prevalence of campylobacter spp. From broiler chicken rearing period to the slaughtering process in Eastern China. Front. Vet. Sci. 7, 227. doi: 10.3389/fvets.2020.00227
Tang, M., Zhou, Q., Zhang, X., Zhou, S., Zhang, J., Tang, X., et al. (2020a). Antibiotic resistance profiles and molecular mechanisms of campylobacter from chicken and pig in China. Front. Microbiol. 11, 592496. doi: 10.3389/fmicb.2020.592496
Wang, Y., Dong, Y., Deng, F., Liu, D., Yao, H., Zhang, Q., et al. (2016). Species shift and multidrug resistance of Campylobacter from chicken and swine, China 2008–14. J. Antimicrob. Chemother. 71 (3), 666–669. doi: 10.1093/jac/dkv382
Wang, Y., Taylor, D. E. (1990). Natural transformation in Campylobacter species. J. Bacteriology 172 (2), 949–955. doi: 10.1128/jb.172.2.949-955.1990
Wang, J., Wang, Z., Zhang, J., Ding, Yu, Ma, Z., Jiang, F., et al. (2021). Prevalence, antibiotic susceptibility and genetic diversity of Campylobacter jejuni isolated from retail food in China. LWT 143, 111098. doi: 10.1016/j.lwt.2021.111098
Wang, T., Zhao, W., Li, S., Yao, H., Zhang, Q., Yang, L. (2022). Characterization of erm(B)-carrying Campylobacter Spp. of retail chicken meat origin. J. Global Antimicrobial Resistance 30, 173–177. doi: 10.1016/j.jgar.2022.05.029
Wardak, S., Szych, J., Aleksandra, A. Z., Gierczyński, Rafał (2007). Antibiotic Resistance of Campylobacter jejuni and Campylobacter coli Clinical Isolates from Poland. Antimicrob. Agents Chemother. 51 (3), 1123–1125. doi: 10.1128/aac.01187-06
WHO. (2022). World Food Safety Day 2022 - Safer food, better health. Available at: https://www.who.int/news/item/07-06-2022-world-food-safety-day-2022-safer-food-better-health.
Yu, H., Elbediwi, M., Zhou, X., Shuai, H., Lou, X., Wang, H., et al. (2020). Epidemiological and Genomic Characterization of Campylobacter jejuni Isolates from a Foodborne Outbreak at Hangzhou, China. Int. J. Mol. Sci. 21 (8), 3001. doi: 10.3390/ijms21083001
Zang, X., Lv, H., Tang, H., Jiao, X., Huang, J. (2021). Capsular genotype and lipooligosaccharide class associated genomic characterizations of campylobacter jejuni isolates from food animals in China. Front. Microbiol. 12, 775090. doi: 10.3389/fmicb.2021.775090
Zeng, X., Brown, S., Gillespie, B., Lin, J. (2014). A single nucleotide in the promoter region modulates the expression of the β-lactamase OXA-61 in Campylobacter jejuni. J. Antimicrob. Chemother. 69 (5), 1215–1223. doi: 10.1093/jac/dkt515
Zhang, L., Li, Yi, Shao, Y., Hu, Y., Lou, H., Chen, X., et al. (2020a). Molecular characterization and antibiotic resistant profiles of campylobacter species isolated from poultry and diarrheal patients in Southeastern China 2017–2019. Front. Microbiol. 11, 1244. doi: 10.3389/fmicb.2020.01244
Zhang, T., Luo, Q., Chen, Y., Li, T., Wen, G., Zhang, R., et al. (2016). Molecular epidemiology, virulence determinants and antimicrobial resistance of campylobacter spreading in retail chicken meat in central China. Gut Pathog. 8 (1), 48. doi: 10.1186/s13099-016-0132-2
Zhang, P., Zhang, X., Liu, Y., Cui, Q., Qin, X., Niu, Y., et al. (2022). Genomic insights into the increased occurrence of campylobacteriosis caused by antimicrobial-resistant campylobacter coli. mBio 13 (6), e02835–e02822. doi: 10.1128/mbio.02835-22
Zhang, P., Zhang, X., Liu, Y., Jiang, J., Shen, Z., Chen, Q., et al. (2020b). Multilocus sequence types and antimicrobial resistance of campylobacter jejuni and c. coli isolates of human patients from Beijing, China 2017–2018. Front. Microbiol. 11, 554784. doi: 10.3389/fmicb.2020.554784
Zhao, S., Tyson, G. H., Chen, Y., Li, C., Mukherjee, S., Young, S., et al. (2016). Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in campylobacter spp. Appl. Environ. Microbiol. 82 (2), 459–466. doi: 10.1128/AEM.02873-15
Zhong, C., Qu, B., Hu, G., Ning, K. (2022). Pan-genome analysis of campylobacter: Insights on the genomic diversity and virulence profile. Microbiol. Spectr. 10 (5), e01029-22. doi: 10.1128/spectrum.01029-22
Keywords: Campylobacter, antibiotic resistance, whole-genome sequencing, antibiotic-resistance gene, virulence factor
Citation: Xiao J, Cheng Y, Zhang W, Lu Q, Guo Y, Hu Q, Wen G, Shao H, Luo Q and Zhang T (2023) Genetic characteristics, antimicrobial susceptibility, and virulence genes distribution of Campylobacter isolated from local dual-purpose chickens in central China. Front. Cell. Infect. Microbiol. 13:1236777. doi: 10.3389/fcimb.2023.1236777
Received: 08 June 2023; Accepted: 18 August 2023;
Published: 07 September 2023.
Edited by:
Elvira Garza González, Autonomous University of Nuevo León, MexicoReviewed by:
Marja-Liisa Hänninen, University of Helsinki, FinlandLeonardo Gabriel Panunzi, CEA Saclay, France
Copyright © 2023 Xiao, Cheng, Zhang, Lu, Guo, Hu, Wen, Shao, Luo and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tengfei Zhang, tfzhang23@163.com; Qingping Luo, qingping0523@163.com
†These authors have contributed equally to this work
 Jia Xiao1†
Jia Xiao1†  Yunqing Guo
Yunqing Guo Guoyuan Wen
Guoyuan Wen Huabin Shao
Huabin Shao Tengfei Zhang
Tengfei Zhang