Gram-negative bacterial sRNAs encapsulated in OMVs: an emerging class of therapeutic targets in diseases
- 1Department of Molecular Genetics, Faculty of Biological Sciences, Tarbiat Modares University, Tehran, Iran
- 2Department of Molecular Biology of The Cell, Faculty of Bioscience, University of Milan, Milan, Italy
- 3Department of Biology, Faculty of Basic Sciences, Semnan University, Semnan, Iran
- 4Department of Genetics, Faculty of Advanced Science and Technology, Tehran Medical Sciences, Islamic Azad University, Tehran, Iran
Small regulatory RNAs (sRNAs) encapsulated in outer membrane vesicles (OMVs) are critical post-transcriptional regulators of gene expression in prokaryotic and eukaryotic organisms. OMVs are small spherical structures released by Gram-negative bacteria that serve as important vehicles for intercellular communication and can also play an important role in bacterial virulence and host-pathogen interactions. These molecules can interact with mRNAs or proteins and affect various cellular functions and physiological processes in the producing bacteria. This review aims to provide insight into the current understanding of sRNA localization to OMVs in Gram-negative bacteria and highlights the identification, characterization and functional implications of these encapsulated sRNAs. By examining the research gaps in this field, we aim to inspire further exploration and progress in investigating the potential therapeutic applications of OMV-encapsulated sRNAs in various diseases.
1 Introduction
Since the identification of bacteria as the primary cause of infectious diseases, researchers have been interested in the variety of interactions between these tiny organisms and their environment. In particular, bacterial interaction with eukaryotic host cells has gained significant attention during the past few decades (Kaparakis-Liaskos and Kufer, 2020). Bacterial cells release membrane vesicles known as extracellular vesicles (EV) to communicate with host cells and other bacteria (Hosseini-Giv et al., 2022). Depending on the cell structure, Gram-positive and Gram-negative bacteria use various ways to create extracellular vesicles, known as outer membrane vesicles (OMVs) and membrane vesicles (MVs), respectively (Bose et al., 2021). OMVs were first discovered in Escherichia coli (E. coli) in the 1960s, however, Gram-positive MVs were discovered much later because scientists believed that the thick cell wall that surrounds these bacteria would prevent the release of MVs (Brown et al., 2015; Srivastava and Kim, 2022; Huang et al., 2023). The first proof of vesiculation in Gram-positive bacteria was presented by Dorward and Garon in 1990 (Bose et al., 2021).
EVs are small biological structures that are released by cells that in both in physiological and pathological conditions, can play a central role in cell-cell communication and transfer a variety of cargos, including lipids, proteins, and nucleic acids, which are employed to interact with and have an impact on host cells, such as cytotoxicity and immunomodulation (Hosseini-Giv et al., 2022; Palazzolo et al., 2022). EVs have also been used as drug delivery systems since they have characteristics that made them ideal for this purpose, leading to interesting results in preclinical and clinical trials (Palazzolo et al., 2022). When EVs enter target cells, they release their contents, which include proteins, short RNAs (sRNAs), tRNA fragments, and microRNAs (miRNAs), which then control the gene expression and function of the recipient cell (Stanton, 2021). Recent studies have demonstrated that bacterial vesicles containing noncoding regulatory RNAs are released into the surrounding environment, and transferred to other microorganisms and host cells, as already reported by the protozoan pathogen Trypanosoma cruzi. However, intracellular bacterial pathogens can express sRNAs that have regulatory functions in a similar manner as miRNAs. The significance of microbial sRNAs as molecules that can mediate host-microbe interactions is highlighted by recent studies which have demonstrated that bacterial vesicles containing noncoding regulatory RNAs are released into the surrounding environment, and transferred to other microorganisms and host cells, as already reported for the protozoan pathogen Trypanosoma cruzi. However, intracellular bacterial pathogens can also express sRNAs that have regulatory functions in a similar manner as miRNAs (Ahmadi Badi et al., 2020; Stanton, 2021). Extracellular sRNAs can then be found in a variety of bodily fluids, such as serum, plasma, and urine, and they exhibit altered circulating levels in a wide range of disorders, making them potential biomarker possibilities for pathological states (Cucher et al., 2023).
EVs secreted by the microbiota have recently emerged as a new means of communication. The largest microbial ecosystem in the human body, the gut microbiota, carry the message of antibiotic resistance to the surrounding bacteria. Furthermore, they function as a powerful system for the detoxification of substances that are harmful to bacterial growth (Heydari et al., 2022). In the intercellular signaling system, MEVs have become important mediators that may play a crucial role in the communication between the microbiota and the host. Microbiota-derived EVs (MEVs) are tiny membrane-bound vesicles that contain a variety of biologically active substances, including proteins, mRNA, miRNA, DNA, carbohydrates, and lipids. These vesicles act as carriers for their payload when transported horizontally across cells (Sultan et al., 2021). In 2013, MEVs in mouse stools were identified by Kang et al. (2013). They demonstrated how the stool MEVs from an IBD mouse model displayed substantial dysbiosis in comparison to the difference in the microbiota composition between the inflammatory and control phenotypes. This study demonstrates that EVs play a regulating function in intestinal immunity and homeostasis, even though it was unclear whether the dysbiosis was an effect of the inflammation or its cause. For instance, mice were prevented from developing colitis by the EVs of the gut bacterium Akkermansia muciniphila, and the proinflammatory cytokine IL6 was reduced in response to E. coli treatment (Kang et al., 2013). Clinical research has shed light on how the microbiome affects immunity and a variety of disorders. For instance, Bacteroides thetaiotaomicron EVs contain hydrolytic enzymes that, when shared with bacteria lacking hydrolytic enzymes, boost the potential digestion of gut microbiota. Therefore, administering EVs produced from particular bacterial strains may alter host nutrition, immunological signaling pathways, and the generation of bacterial metabolites. The EV-based network most likely represents significant links that organize the gut microbiota’s ecological units (Badi et al., 2017).
2 Small RNA and its biological function
2.1 Eukaryotic sRNA
Non-coding RNAs (ncRNAs) are a large group of RNA molecules that cannot encode proteins. ncRNA comprise 98% of all transcriptional output, which can be divided into two main subgroups: housekeeping ncRNA and short (less than 200 nt) and long (more than 200 nt) ncRNA (Wang and Li, 2012). Small non-coding RNAs (sncRNAs) are key mediators of post-transcriptional regulators in bacteria and eukaryotes that control gene expression through a variety of mechanisms (Wang and Fu, 2019). These include micro-RNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs) (Orendain-Jaime et al., 2021), and tRNA-derived small RNAs (tsRNAs) (Li et al., 2018). Apart from these, which mostly act as silencers, in 2006 Long-Cheng Li and colleagues identified small RNAs that target gene promoter sequences to activate expression in a process called RNA activation (RNAa) (Li et al., 2006).
2.1.1 miRNA
miRNA is an endogenous sRNA with a length of about 22 nt (Bhaskaran and Mohan, 2014), which was first discovered by Lee and colleagues in the nematode Caenorhabditis elegans (Lee et al., 1993). They have the role of post-transcriptional regulators of gene expression and often lead to changes or prevent the production of protein products through binding to the complementary sequence of mRNA and interfering with the translation machines (Bhaskaran and Mohan, 2014. It should be noted that miRNAs, in addition to mRNA degradation, transcription silencing, and post-transcriptional silencing, play an important role in humans in modulating the process of apoptosis (Allmer and Yousef, 2014) and inhibiting the proliferation and migration of cancer cells (Ding et al., 2021). It also serves as diagnostic and prognostic biomarkers for diseases such as cancer (Saldanha et al., 2016; Liu et al., 2017; Yang et al., 2017), neurological disorders (Khoo et al., 2012), and type 2 diabetes (Mahjoob et al., 2022). In general, miRNAs in cancer can be classified into two categories: oncogenic (oncomiRs) and tumor suppressor (tsmiRs), which play a role in cancer progression and suppression, respectively (Wang and Li, 2012).
2.1.2 siRNA
siRNAs are a class of 22 nt double-stranded RNAs (dsRNAs) that are produced endogenously or synthetically and are known as activators of the RNA interference (RNAi) mechanism (Wang and Li, 2012). In 2006, Fire and Mello won the Nobel Prize for coining the term “RNA interference” and discovering the mechanics of its occurrence. They reported that dsRNAs induce gene silencing through Watson-Crick base pairing with a complementary sequence in mRNAs (Alshaer et al., 2021). This makes siRNAs superior to monoclonal antibody drugs and small molecule therapies, because theoretically, any gene of interest can be targeted with the help of siRNA, while these drugs must recognize the complex spatial composition of specific proteins (Hu et al., 2020). Considering that, many human diseases are caused by excessive production of specific gene products such as oncogenes, siRNAs can be used to target the active genes. To date, many siRNAs have been used to treat ocular (Nikam and Gore, 2018), liver (Zhao et al., 2019), kidney (Stokman et al., 2010), cancer (Hou et al., 2006), etc., and are modified with various chemical compounds, and are phosphonated for proper function.
2.1.3 piRNA
piRNA is an endogenous single-stranded sRNA with a length of ~27nt found in vertebrates and invertebrates (Wang and Li, 2012). The PIWI as a gene encoding piRNA was first identified in Drosophila in 1997, where mutants showed defects in germ cell maintenance (Cox et al., 1998). Due to the larger size of piRNAs compared to miRNAs, they bind to mRNA more tightly and it is thought that they can inhibit protein synthesis (Kamenova et al., 2023). Although the function of piRNA in humans is not fully understood, abnormal expression of Hiwi (ortholog humans Piwi) has been reported in a variety of cancers (Wang and Li, 2012). piRNAs have been identified as new prognostic and diagnostic tools for cancer (Guo et al., 2020) and MS (Kamenova et al., 2023).
2.1.4 tsRNA
tRNAs comprise about 4-10% of all cellular RNAs, which are the most abundant type of small noncoding RNAs (sncRNAs). They are key components in the translation process and transport amino acids to the ribosome. Many studies have shown that many sncRNAs are derived from tRNAs that are ~15-50 nt in length (Watson et al., 2019) and play different roles in epigenetic regulation, gene expression, protein translation, and immune processes. tsRNAs are divided into two main types based on the length and site of tRNA cleavage, tRNA stress-induced RNA (tiRNA) and tRNA-derived fragment (tRF). Abnormal levels of tsRNAs have been reported in infectious, neurological, acquired metabolic diseases and cancer in humans. Researchers have examined the level of tsRNAs in serum as a biomarker in disease diagnosis (S. Li et al., 2018).
2.1.5 saRNA
saRNA is a class of 21nt long sncRNAs that have the same structure and chemical components as siRNA, although their biological functions are opposite (Kwok et al., 2019). In 2000, saRNAs were first identified by Li et al., who reported that saRNAs activate gene expression in mammalian cells by targeting gene promoters (Li et al., 2006). Considering that saRNA is a very powerful tool for gene activation, it has been used in regenerative medicine and it is predicted that it is possible to combine saRNA with other drugs to improve treatment (Kwok et al., 2019). In overall, in various reports, sRNAs have been used as non-invasive clinical diagnostic biomarkers for diseases such as acute myeloid leukemia (Xia et al., 2023), neurodegenerative disease (Watson et al., 2019), renal cell carcinoma (Ding et al., 2021), and multiple sclerosis (Piket et al., 2019).
2.2 Bacterial sRNA
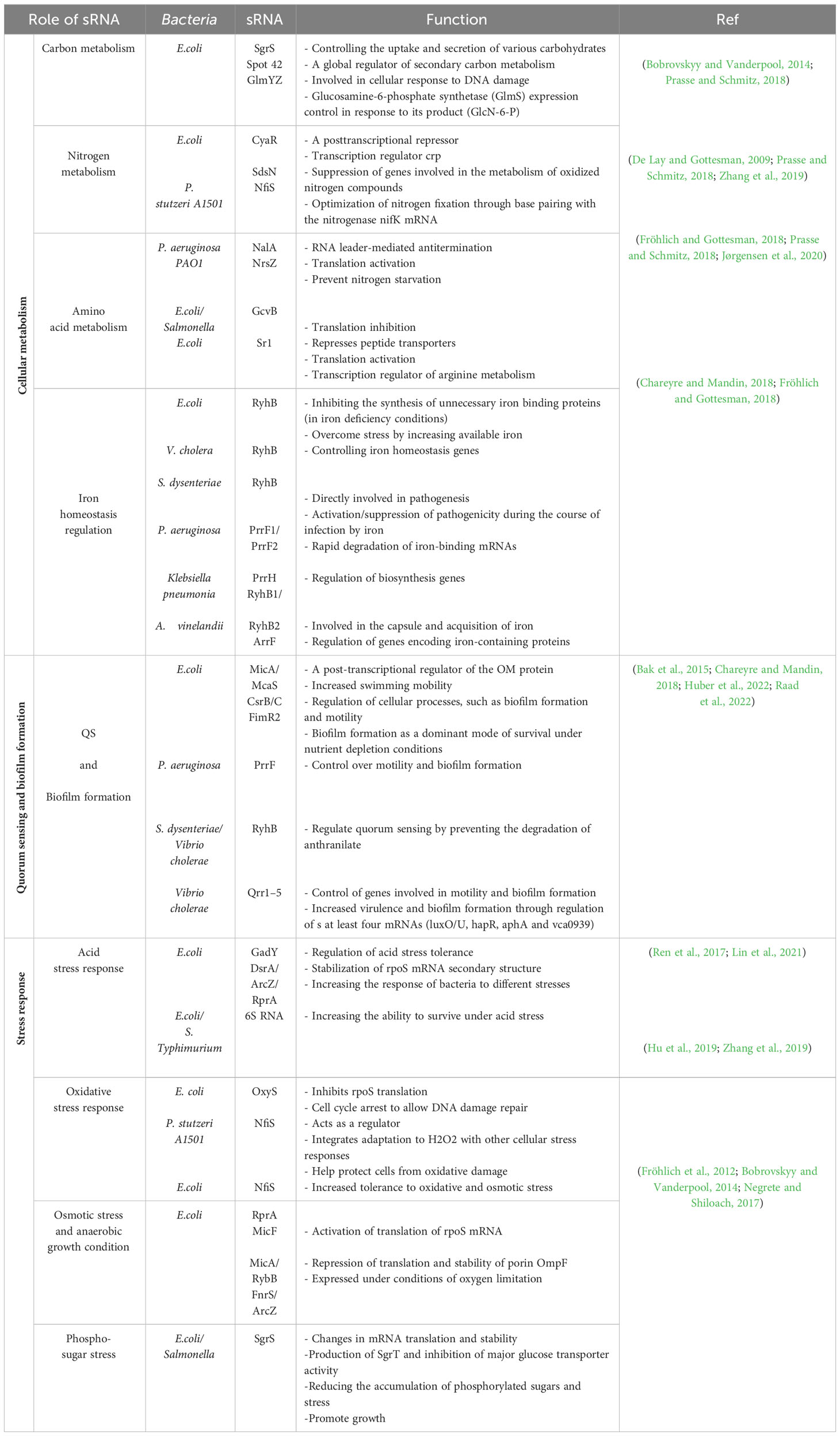
Bacterial sRNAs are transcribed from the intergenic regions of the bacterial genome, which are usually 50-400 nt in length (Wang and Fu, 2019) and are recognized as important elements of bacterial adaptation to environmental changes (Jørgensen et al., 2020). As bacteria are exposed to environmental fluctuations such as changes in pH, temperature, nutrient concentration, water and others, they use fast and flexible mechanisms for survival and reproduction (Raina et al., 2022). Bacterial sRNAs regulate various biological processes such as energy metabolism, quorum sensing (QS), biofilm formation, and stress response (Michaux et al., 2014). Gram-negative bacteria have unique structural features that require maintaining homeostasis in the inner membrane (IM), outer membrane (OM), and periplasmic space for microbial growth and cell proliferation. These cells use various stress response systems to monitor the status of membrane proteins, with sRNAs playing an essential role (Papenfort and Melamed, 2023). In addition, sRNAs are involved in modulating the stability or translation of mRNAs through short-base pair interactions and are among the main post-transcriptional regulators in bacteria (Raina et al., 2022). Here, pathways for bacterial metabolism and homeostasis are summarized, and specific roles for sRNA in the regulation of those pathways are presented in Table 1.
Cellular metabolism is essential in bacteria for optimizing nutrient utilization. Regulatory mechanisms ensure that bacteria preferentially use the most favorable carbon, nitrogen, and amino acid sources based on their availability, energy efficiency, and growth requirements. sRNAs regulate metabolic processes by influencing metabolic enzymes, transporters and regulators. (Michaux et al., 2014). Carbon metabolism is a versatile and essential process that helps bacteria use organic compounds for energy production and the biosynthesis of nucleic acids, amino acids, lipids, and carbohydrates (Noor et al., 2010). Nitrogen metabolism enables bacteria to utilize nitrogen in different forms such as ammonium, nitrate, and amino acids, and use them to synthesize nitrogen-containing compounds necessary for cell. Amino acids metabolism includes the synthesis, breakdown, and conversion of amino acids, which are essential for the adequate supply of these building blocks for protein synthesis and carrying out cellular processes (Prasse and Schmitz, 2018).
Iron plays a dual role in bacteria, serving as a crucial micronutrient for bacterial growth while also potentially harming bacterial cells by generating reactive oxygen species (ROS) during aerobic metabolism. Therefore, iron homeostasis is critical for bacterial survival, and bacteria use complex mechanisms to absorb, store, and utilize iron (Klebba et al., 2021). Bacterial sRNAs play a crucial role in these metabolic processes as post-transcriptional regulators, influencing gene expression and metabolic pathways (Papenfort and Melamed, 2023). Quorum sensing (QS) is a cellular communication mechanism in bacteria and fungi that indicates population density and plays an important role in pathogenicity and biofilm formation (Michaux et al., 2014). When the concentration of signaling molecules reaches a certain threshold and binds to receptor proteins, it triggers the activation of genes that promote biofilm formation (Williams, 2007). The primary function of QS is to enable bacteria to monitor their environment and adjust their behavior accordingly. As a result, it allows bacteria to assess the number of neighboring bacteria and coordinate their activities as a collective group (Wang et al., 2022). Bacterial sRNAs are responsible for transcription regulation, biofilm formation and information integration by QS systems (Michaux et al., 2014).
In the natural habitat, bacteria face various stressors such as temperature and pH fluctuations, nutrient limitation, exposure to toxins, oxidative stress, and physical damage. Stress response mechanisms enable bacteria to sense and respond to these stressors and increase their survival. sRNAs play an essential role in coordinating the stress response by regulating the expression of stress-related genes, thereby enabling the activation or suppression of stress response pathways to support bacterial survival (Holmqvist and Wagner, 2017). When bacteria are exposed to acidic conditions as a result of metabolic processes, acid stress is induced. This in turn triggers the activation of bacteria’s acid resistance systems to protect against cell damage and maintain homeostasis (Dawan and Ahn, 2022).
ROS are released upon exposure to oxygen or as byproducts of metabolic reactions. In these conditions, bacteria use antioxidant defense systems (such as catalase and superoxide dismutase enzymes and small molecule antioxidants) to neutralize ROS and protect cellular components against oxidative damage (Seixas et al., 2022). When the osmolarity of the surrounding environment changes, it leads to a change in water availability and ion concentration, which bacteria maintain osmotic balance through the synthesis and absorption of compatible solutes and prevent cell shrinkage or lysis (Krämer, 2010). Phosphosugar stress arises from an imbalance in the availability or use of phosphosugars, which are crucial for cellular processes like energy metabolism and cell wall biosynthesis. Bacteria use regulatory proteins and metabolic pathways to adapt and restore phospho-sugar homeostasis (Papenfort et al., 2013). Overall, sRNAs are a flexible and rapid tool for gene regulation in bacteria, allowing them to respond to changing environmental conditions and optimize their survival strategies (Papenfort and Melamed, 2023).
2.3 sRNA applications
Natural or synthesized sRNAs are used to silence or regulate gene expression related to disease pathways, after identifying the specific mRNA involved in the pathway. sRNAs complementary to target mRNA sequences are designed or identified to inhibit their activity. Using chemical modifications such as locked nucleic acids (LNAs) or phosphorothioate linkages increases the stability and protects sRNAs from degradation by cellular nucleases Finally, efficient delivery systems are used to deliver engineered sRNAs (Rupaimoole and Slack, 2017). For example, in a study, a new sRNA called EsrF was reported that through binding to flhB mRNA leads to increased bacterial motility and adhesion to host cells, which is ultimately associated with increased infection. Engineered sRNA is predicted to reduce infection by not binding to mRNA (Jia et al., 2021).
3 Bacterial outer membrane vesicles
Gram-negative bacteria produce ectosomes called outer membrane vesicles (OMVs), which are spherical lipid bilayer structures with sizes ranging from approximately 20 to 250 nm (Yang et al., 2018). OMVs are present in all stages of bacterial growth, and they represent the structure of the bacterial OM (Furuyama and Sircili, 2021). In 1967, Chatterjee and Das first identified OMVs during the natural growth of Vibrio cholera (Chatterjee, 1967. After that, the presence of OMVs was observed in different types of Gram-negative bacteria (Devoe and Gilchrist, 1973) and even in patients with meningococcal infection, which indicates the role of OMVs in bacterial pathogenesis (DeVoe and Gilchrist, 1975). The structure of OMVs allows them, in addition to carrying different cargoes (lipopolysaccharides (LPS), phospholipids, peptidoglycan (PG), proteins, nucleic acids, etc.), to be genetically engineered and chemically modified to increase efficiency (Xue et al., 2022).
Bacterial extracellular vesicles contain a significant amount of RNA, and among mRNA, tRNA, rRNA, and sRNA, sRNAs occupy a significant part (Luo et al., 2023). In the Gram-negative bacteria, which are characterized by an OM rich in lipopolysaccharide outside the thin layer of PG, the formation of OMVs is a highly spontaneous and conserved process (Xue et al., 2022). In general, the biogenesis of OMVs is based on three mechanisms (Figure 1): 1) Reducing the interaction between the OM and the underlying structures such as the PG layer and its associated lipoproteins (Pita et al., 2020). 2) Accumulating misfolded proteins, PG fragments, and LPS in a specific region of the bacterial periplasm, leading to deformation of the upper OM. 3) Altering membrane chemical-physical properties and asymmetric distribution of phospholipids, which modulates asymmetric membrane expansion, protrusion, and OMV biogenesis (Luo et al., 2023).
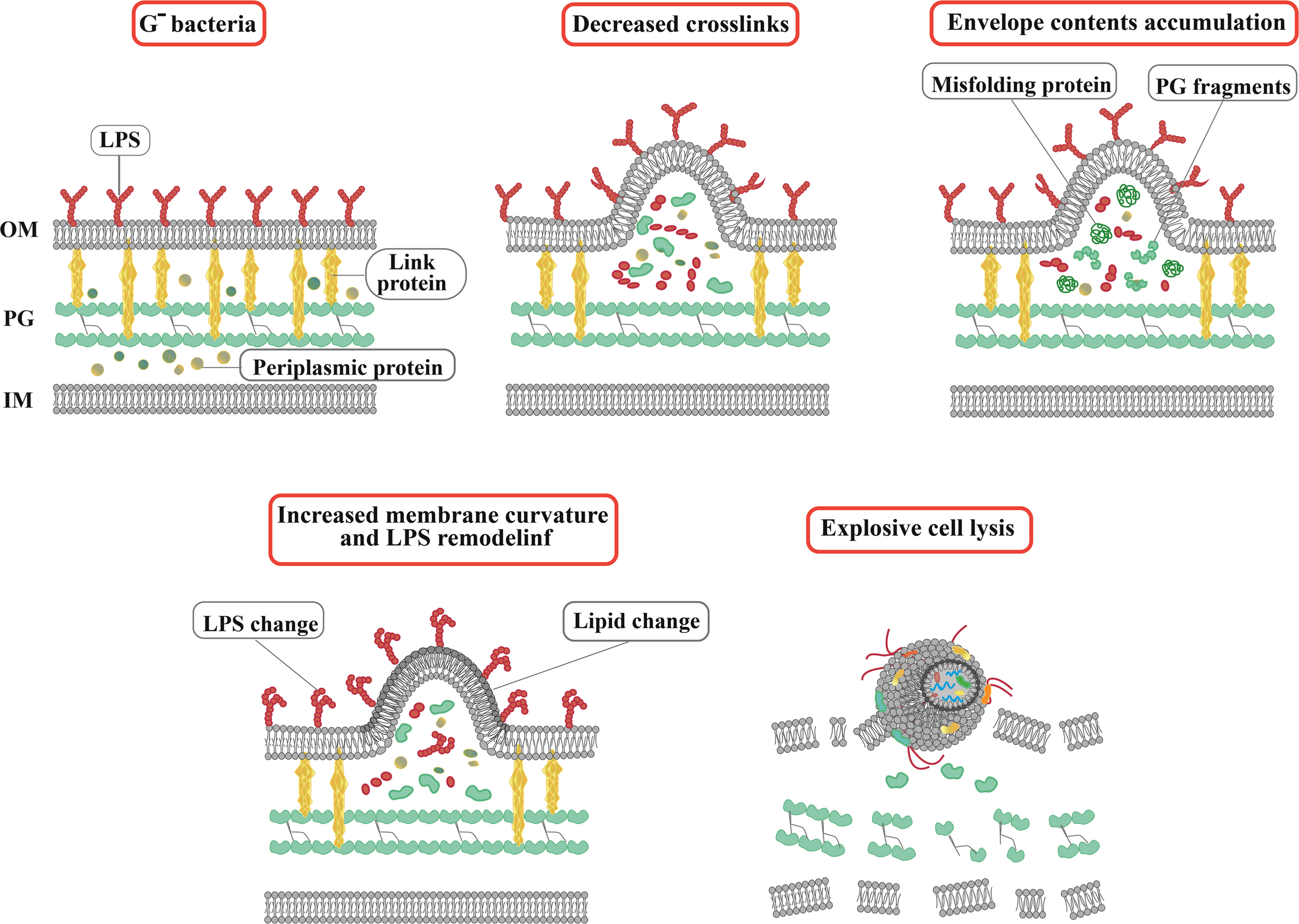
Figure 1 Actual and schematic structure of OMVs. Steps of OMVs biogenesis in Gram-negative bacteria. The arrows indicate OMVs. SEM, scanning electron microscope; TEM, transmission electron microscopy.
In addition to OMVs, membrane vesicles of Gram-negative bacteria can also be divided into outer-inner membrane vesicles (OIMVs) and explosive outer membrane vesicles (EOMVs) based on their formation pathways, structure, and composition (Toyofuku et al., 2019). In general, three functions have been considered for OMVs, including, bacterial survival (nutrient acquisition, stress response), regulation of interactions in bacterial communities (biofilm, quorum sensing), and induction of pathogenesis (immunomodulation) (Diallo et al., 2022). OMVs, as protectors and carriers of functional and gene-regulatory sRNAs, mediate direct contact-free transfer of sRNA fragments between bacterial and mammalian cells (Han et al., 2022). sRNAs in OMVs act as interspecies communication molecules to modulate gene expression in different cell types and species (Li et al., 2020).
3.1 Bacterial-derived nanoparticle
Nanoparticles (NP) have many potential properties that promote their use in various biomedical applications including diagnostics, cellular imaging, chemical assays, drug delivery and therapy. However, NPs have limitations such as cytotoxicity, low immunogenicity, low cellular uptake by target cells, non-selective targeting and increased clearance rate, which make their use challenging (Ajam-Hosseini et al., 2023). OMVs have gained attention by overcoming these limitations and are valuable to researchers in biomedical applications (Naskar et al., 2021). The use of bacterial OMVs as a therapeutic strategy to overcome the challenges of biocompatibility and large-scale production associated with synthetic nanocarriers has been made possible by advances in genetic engineering. Bacterial OMVs have a tough membrane that provides stability and reduces leakage into the systemic circulation (Gujrati et al., 2019), such that approximately 75% of the surface of E. coli is occupied by rigid lipopolysaccharides (Magennis et al., 2014).
OMVs have shown great potential in biomedical applications as they are used as nanocarriers for drug delivery, bioimaging, immune system modulation (Naskar et al., 2021) and various therapeutic strategies, including vaccine development 48, gene therapy, and cancer treatment (Pan et al., 2022). With the help of genetic engineering, membrane modification, and membrane coating bacteria can be manipulated as nanovesicles with non-toxic OM components (Liu L. et al., 2020). Genetic engineering is an ideal tool for designing nanoplatforms related to bacteria in the treatment of various diseases with the help of transferring drugs, genes, proteins, and enzymes.
In this regard, Gujrati et al. bioengineered E.coli to produce OMVs with less toxicity to deliver siRNA and antitumor drug melanin (Gujrati et al., 2019). Compared to Gram-positive bacteria, Gram-negative bacteria are easier to bioengineer, so most studies have focused on them (Liu L. et al., 2020). Some researchers have studied the genetic modification of bacterial protoplasts because they are easily manipulated to make nanovesicles with non-toxic OM components. Therefore, bacterial protoplast-derived nanovesicles (PDNVs) have been used for adjuvant-free vaccine delivery in bacterial infection, showing remarkable efficacy and safety (Kim et al., 2015). Coating NPs with bacterial OMVs (OMV-NPs) is a facile process that has good biocompatibility and alleviates some of the limitations of traditional surface modifications (Naskar et al., 2021). Due to the large number of immunogenic antigens and different pathogenic molecular patterns in the bacterial membrane that play an important role in creating innate and adaptive immunity, it makes them desirable in various research fields (Liu W. L. et al., 2020). By modifying properties of bacteria such as membrane proteins (Anwar et al., 2021), lipid compositions (Pichler and Emmerstorfer-Augustin, 2018), the inflammatory responses of the body against them and side effects can be reduced (Cao et al., 2019). Although, bacteria-derived nanoparticles have created a new frontier in medical treatment strategies, the use of sRNA encapsulated with OMV-NPs requires further investigation.
3.2 OMVs engineering
In therapeutic strategies, OMVs are engineered through immune modulation, size, surface modification and composition. Since OMVs have intrinsic immunomodulatory properties, it is possible to modulate the immune response for therapeutic purposes by modifying compounds such as immunostimulatory or immunosuppressive molecules (Kim et al., 2013). OMV size can be controlled during purification and can impact their stability, biodistribution, and cellular uptake. OMVs’ efficiency in binding to target cells can be improved by using specific ligands and peptides. Furthermore, genetic modification of the original bacterial strain can introduce the desired surface proteins. It should be noted that the engineering of OMVs is different based on the desired therapeutic application and desired cargo (Huang et al., 2022).
4 Clinical relevance of sRNA encapsulated in OMVs derived from Gram-negative bacteria
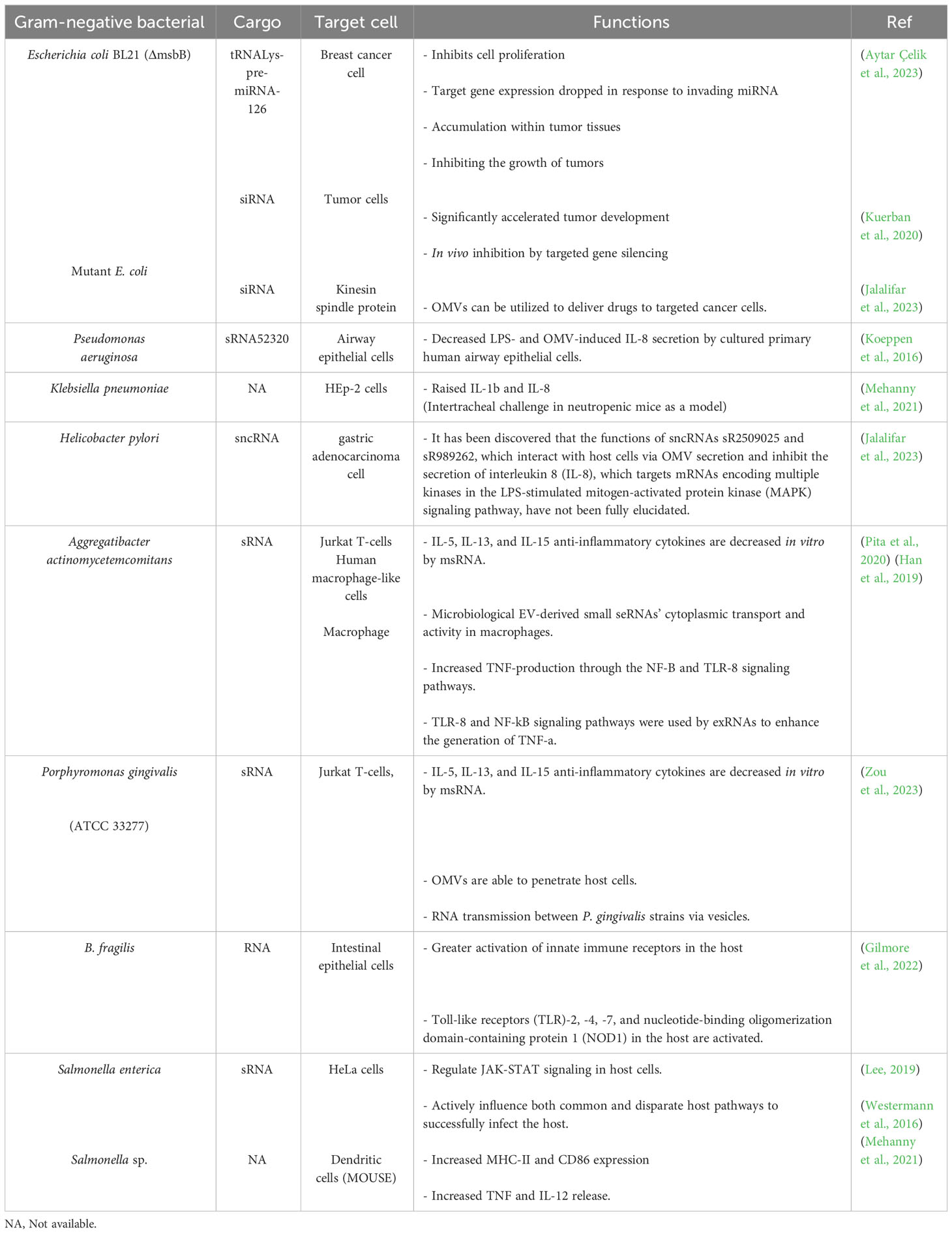
Due to the limited information available regarding the biological role of bacterial sRNAs within OMVs, few studies have been conducted so far. However, functional sRNAs derived from bacterial OMVs have been identified using high-throughput RNA sequencing. Recent studies have investigated the potential role of vesicular and bacterial sRNAs in host-pathogen interactions. For the first time, Koeppen et al. delivered sRNAs into host cells using OMVs from Pseudomonas aeruginosa bacteria (Koeppen et al., 2016). P. aeruginosa is a Gram-negative bacterium that is the main cause of bacterial colonization in chronic obstructive pulmonary disease (COPD) affected patients (Nakamoto et al., 2019). They also showed that the actual expression level of sRNAs in OMVs can lead to a decrease in host immune response by reducing cytokine secretion. The pathogen-associated molecular patterns (PAMP) on the outside of OMVs induce a proinflammatory host immune response (Koeppen et al., 2016). PAMPs, which includes LPS, peptidoglycan, flagellin, lipoproteins, and purines, bind to toll-like receptors (TLR) of host airway epithelial cells and lead to increased secretion of pro-inflammatory cytokines, especially IL-8, through the mitogen-activated protein kinase (MAPK) signaling pathway (Bauman and Kuehn, 2006). Cytokine secretion recruits and activates neutrophils to enhance the clearance of P. aeruginosa infection, and the increase of IL-8, a potent chemoattractant for neutrophils, leads to extensive neutrophil infiltration and the production of proteolytic enzymes such as elastase, leading to bacterial phagocytosing and tissue destruction (Huang et al., 2020) (Figure 2). In other words, sRNAs reduced OMV-stimulated IL-8 secretion by translocation from OMVs to host cells (Zhang et al., 2020).
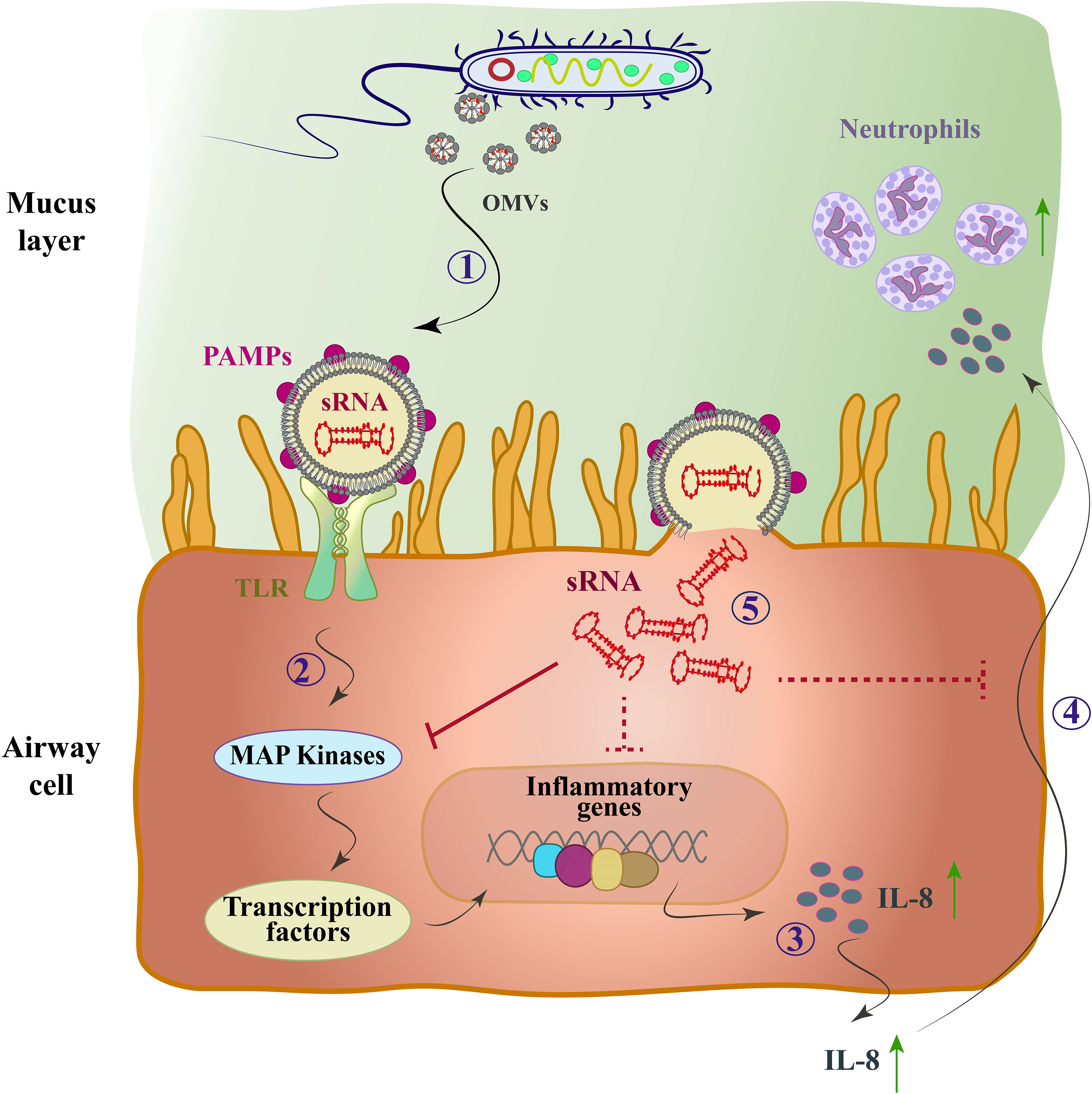
Figure 2 The mode of action of the OMV-encapsulated sRNA of P. aeruginosa in human airway epithelial cells. P. aeruginosa produces OMVs after entering the mucosal layer of the airways. (1) OMVs bind to TLRs through PAMPs. (2) The MAP-kinase (TLR/MAPK) signaling pathway is activated and induces the host’s innate immune response. (3) Transcription factors are activated and cause up-regulation of IL-8 mRNA and IL-8 secretion (4) IL-8 attracts neutrophils and they phagocytose P. aeruginosa by entering the lungs. (5) OMVs fuse with sRNA and enter cells, which by targeting mRNA of the MAPK signaling pathway upstream of IL-8 leads to reduced host IL-8 secretion and neutrophil recruitment. Solid arrows, direct inhibitor; Dashed arrows, indirect inhibitor.
P. aeruginosa has received much attention as the first virus to use OMV-encapsulated sRNAs to suppress the host cell immune response, so we investigated it for this purpose. In Table 2, we reviewed more examples of the current application of OMV-encapsulated sRNAs of Gram-negative bacteria for biomedical purposes. Although, study of these processes remains complex and challenging and requires further investigations in the field of transmission mechanism, they provide a new model on host-pathogen dynamics.
5 A summary of research challenges
Although OMVs-encapsulated sRNAs have been widely used in research, there are still many challenges and gaps in this field that limit the practical applications of OMVs, including:
5.1 OMV toxicity
There are two main strategies to reduce LPS-induced OMV toxicity. 1) Modifying the genes responsible for LPS synthesis (msbA2, msbB, lpxL1, lpxM) in order to reduce the number of acyl chains or phosphate groups of LPS, which leads to the detoxification of molecules in bacteria (Simpson and Trent, 2019). However, there are G- mutant bacteria such as E. coli EMKV15 that do not contain LPS and may be a better choice for drug delivery (Liu et al., 2022). 2) Chemical modification of LPS species from Salmonella minnesota to produce a mixture of mono-phosphorylated lipid A species (MPL) with less toxicity as reported in 1982 by Riby et al. (Qureshi et al., 1982). In 2009, following FDA approval, mainly 3-O-deacyl-4’-monophosphoryl lipid A became an adjuvant in MPL vaccines (Casella and Mitchell, 2008).
5.2 Ambiguous mechanism
Although engineered BEVs can serve as a targeted drug delivery system, further research is still needed for more precise performance with fewer side effects (Gujrati et al., 2014). BEVs are internalized through various mechanisms such as clathrin/non-clathrin-mediated endocytosis, micropinocytosis, and membrane fusion (Liu et al., 2022). For example, the size of H. pylori OMVs can determine the host cell entry mechanism to be taken up by gastric epithelial cells through clathrin-mediated endocytosis and lipid raft-mediated endocytosis. Burkholderia pseudomallei is the causative agent of melioidosis, a severe infectious disease in which EVs are taken up by host cells through various mechanisms including clathrin-mediated endocytosis, macropinocytosis, and phagocytosis (Li et al., 2020).
OMVs from Legionella pneumophila can deliver contents to the host cell membrane by fusing with eukaryotic membrane systems. Membrane properties and the exact mode of interaction between L. pneumophila OMVs and host cell surfaces are not fully understood. Adhesion to plasma membrane proteins and subsequent uptake by phagocytosis are predicted to be effective (Jäger et al., 2015). These examples demonstrate that the effectiveness of these uptake mechanisms can vary depending on the characteristics of BEVs and the receptor cell type, which is crucial for their manipulation in research.
5.3 Heterogeneous contents
BEVs are rich in biological and functional molecules whose research field is still evolving. The specific cargo of BEVs can vary depending on factors such as the bacterial strain, environmental and host conditions, not all of which may be favorable for internalization into recipient cells (Taboada et al., 2019). This variability provides valuable insights into their functional roles and potential impact on host-microbe interactions.
5.4 Complicated isolation and purification
Presented BEV separation and purification methods, such as ultracentrifugation, are time-consuming and expensive and require advanced equipment. It also usually results in low yield due to loss of desired products (Huang et al., 2022). On the other hand, due to insufficient knowledge about specific (surface) markers in BEVs, the discovery of specific markers can be one of the most promising methods (Liu et al., 2022). Thus, new methods such as membrane bioreactors with separation functions and OMVs purification methods based on reversible phase separation, can be utilized to address these issues (Huang et al., 2022).
Additionally, there may be potential negative consequences associated with the use of OMV-encapsulated bacterial sRNA in treatment, such as unintended gene silencing or dysregulation in non-target cells or tissues (Choi et al., 2017).
Since OMVs originate from bacteria, they can also trigger immune responses that lead to inflammation or immunogenic reactions. Prolonged exposure to OMV-encapsulated sRNA may result in immune-related toxicities. Finally, the long-term effects of this treatment could potentially cause genomic instability or alterations in cellular processes (Kim et al., 2013).
6 Conclusion
New drug delivery systems use nanotechnology, biomaterials, and RNA to improve drug stability, increase targeting capabilities, and controlled release. Compared to conventional drug delivery methods, nanotechnology-based systems offer advantages such as controlled release, increased tissue penetration, and targeted delivery to specific cells or tissues (Ajam-Hosseini et al., 2023). Drug delivery systems based on biological materials such as hydrogels, microparticles, and scaffolds can improve the sustained release of drugs, local delivery, and therapeutic outcomes. This system has the potential to overcome limitations associated with conventional methods, such as repeated dosing and systemic side effects (Sun et al., 2023).
RNA-based therapies, including mRNA and small interfering RNA (siRNA), have attracted considerable attention in recent years as they enable modulation of gene expression compared to conventional methods and offer opportunities for personalized medicine (Kwok et al., 2019). Significant structural investigations have been conducted on RNA motifs and/or RNA-protein complexes, which aid drug development through structure-based virtual screening (Yang et al., 2023). The effectiveness of these new systems depends on the specific application, the nature of the drug, and the disease or condition in question.
For this purpose, this review provides sufficient insight into the clinical significance of Gram-negative bacterial sRNAs in biomedical applications. However, more studies are needed to solve the mentioned research challenges and use the structure of Gram-negative bacterial OMVs to diagnose human diseases with robust methods and develop new clinical applications.
Author contributions
MA-H: Writing – original draft. FA: Writing – original draft. FP: Writing – review & editing. HF: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmadi Badi, S., Bruno, S. P., Moshiri, A., Tarashi, S., Siadat, S. D., Masotti, A. (2020). Small RNAs in outer membrane vesicles and their function in host-microbe interactions. Front. Microbiol. 11, 103980. doi: 10.3389/fmicb.2020.01209
Ajam-Hosseini, M., Akhoondi, F., Doroudian, M. (2023). Nano based-oncolytic viruses for cancer therapy. Crit. Rev. Oncology/Hematol. 185. doi: 10.1016/j.critrevonc.2023.103980
Allmer, J., Yousef, M. (2014). miRNomics microRNA biology and computational analysis second edition methods. Mol. Biol. 1107, 333. doi: 10.1007/978-1-62703-748-8
Alshaer, W., Zureigat, H., Al Karaki, A., Al-Kadash, A., Gharaibeh, L., Hatmal, M. M., et al. (2021). siRNA: Mechanism of action, challenges, and therapeutic approaches. Eur. J. Pharmacol. 905, 174178. doi: 10.1016/j.ejphar.2021.174178
Anwar, M., Muhammad, F., Akhtar, B., Anwar, M. I., Raza, A., Aleem, A. (2021). Outer membrane protein-coated nanoparticles as antibacterial vaccine candidates. Int. J. Pept. Res. Ther. 27 (3), 1689–1697. doi: 10.1007/s10989-021-10201-3
Aytar Çelik, P., Erdogan-Gover, K., Barut, D., Enuh, B. M., Amasya, G., Sengel-Türk, C. T., et al. (2023). Bacterial membrane vesicles as smart drug delivery and carrier systems: A new nanosystems tool for current anticancer and antimicrobial therapy. Pharmaceutics 15 (4), 1052. doi: 10.3390/pharmaceutics15041052
Badi, S. A., Moshiri, A., Fateh, A., Jamnani, F. R., Sarshar, M., Vaziri, F., et al. (2017). Microbiota-derived extracellular vesicles as new systemic regulators. Front. Microbiol. 8 (AUG). doi: 10.3389/fmicb.2017.01610
Bak, G., Lee, J., Suk, S., Kim, D., Lee, J. Y., Kim, K. S., et al. (2015). Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli. Sci. Rep. 5 (1), 15287. doi: 10.1038/srep15287
Bauman, S. J., Kuehn, M. J. (2006). Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 8 (9–10), 2400–2408. doi: 10.1016/j.micinf.2006.05.001
Bhaskaran, M., Mohan, M. (2014). MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 51 (4), 759–774. doi: 10.1177/0300985813502820
Bobrovskyy, M., Vanderpool, C. K. (2014). The small RNA SgrS: Roles in metabolism and pathogenesis of enteric bacteria. Front. Cell. Infect. Microbiol. 4 (MAY). doi: 10.3389/fcimb.2014.00061
Bose, S., Aggarwal, S., Singh, D. V., Acharya, N. (2021). Extracellular vesicles: An emerging platform in gram-positive bacteria. Microbial. Cell 7 (12), 312–322. doi: 10.15698/MIC2020.12.737
Brown, L., Wolf, J. M., Prados-Rosales, R., Casadevall, A. (2015). Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 13 (10), 620–630. doi: 10.1038/nrmicro3480
Cao, Z., Cheng, S., Wang, X., Pang, Y., Liu, J. (2019). Camouflaging bacteria by wrapping with cell membranes. Nat. Commun. 10 (1), 3452. doi: 10.1038/s41467-019-11390-8
Casella, C. R., Mitchell, T. C. (2008). Putting endotoxin to work for us: Monophosphoryl lipid a as a safe and effective vaccine adjuvant. Cell. Mol. Life Sci. 65 (20), 3231–3240. doi: 10.1007/s00018-008-8228-6
Chareyre, S., Mandin, P. (2018). Bacterial iron homeostasis regulation by sRNAs. Microbiol. Spectr. 6 (2), 349–366. doi: 10.1128/microbiolspec.rwr-0010-2017
Chatterjee, A N, S. N. (1967). Electron microscopic observations on the excretion of cell-wall material by vibriu chulerae. J. Gen. Microhiof. 49, 1–11. doi: 10.1099/00221287-49-1-1
Choi, J. W., Kim, S. C., Hong, S. H., Lee, H. J. (2017). Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J. Dental Res. 96 (4), 458–466. doi: 10.1177/0022034516685071
Cox, D. N., Chao, A., Baker, J., Chang, L., Qiao, D., Lin, H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12 (23), 3715–3727. doi: 10.1101/gad.12.23.3715
Cucher, M. A., Mariconti, M., Manciulli, T., Vola, A., Rosenzvit, M. C., Brehm, K., et al. (2023). Circulating small RNA profiling of patients with alveolar and cystic echinococcosis. Biology 12 (5), 715. doi: 10.3390/biology12050715
Dawan, J., Ahn, J. (2022). Bacterial stress responses as potential targets in overcoming antibiotic resistance. Microorganisms 10 (7), 1385. doi: 10.3390/microorganisms10071385
De Lay, N., Gottesman, S. (2009). The crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 191 (2), 461–476. doi: 10.1128/JB.01157-08
Devoe, I. W., Gilchrist, J. E. (1973). Release of endotoxin in the form of cell wall blebs during in vitro growth of Neisseria meningitidis. J. Exp. Med. 138 (5), 1156–1167. doi: 10.1084/jem.138.5.1156
DeVoe, I. W., Gilchrist, J. E. (1975). Pili on meningococci from primary cultures of nasopharyngeal carriers and cerebrospinal fluid of patients with acute disease. J. Exp. Med. 141 (2), 297–305. doi: 10.1084/jem.141.2.297
Diallo, I., Ho, J., Lalaouna, D., Massé, E., Provost, P. (2022). RNA sequencing unveils very small RNAs with potential regulatory functions in bacteria. Front. Mol. Biosci. 9. doi: 10.3389/fmolb.2022.914991
Ding, L., Jiang, M., Wang, R., Shen, D., Wang, H., Lu, Z., et al. (2021). The emerging role of small non-coding RNA in renal cell carcinoma. Trans. Oncol. 14 (1), 100974. doi: 10.1016/j.tranon.2020.100974
Fröhlich, K. S., Gottesman, S. (2018). Small regulatory RNAs in the enterobacterial response to envelope damage and oxidative stress. Regulating with RNA in Bacteria and Archaea. Microbiol Spectr Press , 211–228.
Fröhlich, K. S., Papenfort, K., Berger, A. A., Vogel, J. (2012). A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 40 (8), 3623–3640. doi: 10.1093/nar/gkr1156
Furuyama, N., Sircili, M. P. (2021). Outer membrane vesicles (OMVs) produced by gram-negative bacteria: Structure, functions, biogenesis, and vaccine application. BioMed. Res. Int. 2021. doi: 10.1155/2021/1490732
Gilmore, W. J., Johnston, E. L., Bitto, N. J., Zavan, L., O’Brien-Simpson, N., Hill, A. F., et al. (2022). Bacteroides fragilis outer membrane vesicles preferentially activate innate immune receptors compared to their parent bacteria. Front. Immunol. 13. doi: 10.3389/fimmu.2022.970725
Gujrati, V., Kim, S., Kim, S. H., Min, J. J., Choy, H. E., Kim, S. C., et al. (2014). Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 8 (2), 1525–1537. doi: 10.1021/nn405724x
Gujrati, V., Prakash, J., Malekzadeh-Najafabadi, J., Stiel, A., Klemm, U., Mettenleiter, G., et al. (2019). Bioengineered bacterial vesicles as biological nano-heaters for optoacoustic imaging. Nat. Commun. 10 (1), 1114. doi: 10.1038/s41467-019-09034-y
Guo, B., Li, D., Du, L., Zhu, X. (2020). piRNAs: biogenesis and their potential roles in cancer. Cancer Metastasis. Rev. 39 (2), 567–575. doi: 10.1007/s10555-020-09863-0
Han, E. C., Choi, S. Y., Lee, Y., Park, J. W., Hong, S. H., Lee, H. J. (2019). Extracellular RNAs in periodontopathogenic outer membrane vesicles promote TNF-α production in human macrophages and cross the blood-brain barrier in mice. FASEB J. 33 (12), 13412–13422. doi: 10.1096/fj.201901575R
Han, F., Wang, W., Shi, M., Zhou, H., Yao, Y., Li, C., et al. (2022). Outer membrane vesicles from bacteria: Role and potential value in the pathogenesis of chronic respiratory diseases. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.1093327
Heydari, A., Parvini, F., Fard, N. A. (2022). Functional foods and antioxidant effects: emphasizing the role of probiotics. In: N, Naofumi Shiomi, A, Savitskaya, Eds. Current Topics in Functional Food. IntechOpen: London, UK Press.
Holmqvist, E., Wagner, G. H. (2017). Impact of bacterial sRNAs in stress responses. Biochem. Soc. Trans. 45 (6), 1203–1212. doi: 10.1042/BST20160363
Hosseini-Giv, N., Basas, A., Hicks, C., El-Omar, E., El-Assaad, F., Hosseini-Beheshti, E. (2022). Bacterial extracellular vesicles and their novel therapeutic applications in health and cancer. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.962216
Hou, J., He, J., Wang, X., Wen, D., Chen, Z. (2006). Effect of small interfering RNA targeting survivin gene on biological behaviour of bladder cancer. Chin. Med. J. 119 (20), 1734–1739.
Hu, G., Hu, T., Zhan, Y., Lu, W., Lin, M., Huang, Y., et al. (2019). NfiS, a species-specific regulatory noncoding RNA of Pseudomonas stutzeri, enhances oxidative stress tolerance in Escherichia coli. AMB. Express. 9 (1), 1–10. doi: 10.1186/s13568-019-0881-7
Hu, B., Zhong, L., Weng, Y., Peng, L., Huang, Y., Zhao, Y., et al. (2020). Therapeutic siRNA: state of the art. Signal Transduction. Targeted. Ther. 5 (1), 101. doi: 10.1038/s41392-020-0207-x
Huang, F. C., Lu, Y. T., Liao, Y. H. (2020). Beneficial effect of probiotics on Pseudomonas aeruginosa–infected intestinal epithelial cells through inflammatory IL-8 and antimicrobial peptide human beta-defensin-2 modulation. Innate. Immun. 26 (7), 592–600. doi: 10.1177/1753425920959410
Huang, Y., Nieh, M. P., Chen, W., Lei, Y. (2022). Outer membrane vesicles (OMVs) enabled bio-applications: A critical review. Biotechnol. Bioeng. 119 (1), 34–47. doi: 10.1002/bit.27965
Huang, J., Wang, X., Wang, Z., Deng, L., Wang, Y., Tang, Y., et al. (2023). Extracellular vesicles as a novel mediator of interkingdom communication. Cytokine Growth Factor. Rev. doi: 10.1016/j.cytogfr.2023.08.005
Huber, M., Lippegaus, A., Melamed, S., Siemers, M., Wucher, B. R., Hoyos, M., et al. (2022). An RNA sponge controls quorum sensing dynamics and biofilm formation in Vibrio cholerae. Nat. Commun. 13 (1), 7585. doi: 10.1038/s41467-022-35261-x
Jäger, J., Keese, S., Roessle, M., Steinert, M., Schromm, A. B. (2015). Fusion of Legionella pneumophila outer membrane vesicles with eukaryotic membrane systems is a mechanism to deliver pathogen factors to host cell membranes. Cell. Microbiol. 17 (5), 607–620. doi: 10.1111/cmi.12392
Jalalifar, S., Morovati Khamsi, H., Hosseini-Fard, S. R., Karampoor, S., Bajelan, B., Irajian, G., et al. (2023). Emerging role of microbiota derived outer membrane vesicles to preventive, therapeutic and diagnostic proposes. Infect. Agents Cancer 18 (1), 3. doi: 10.1186/s13027-023-00480-4
Jia, T., Liu, B., Mu, H., Qian, C., Wang, L., Li, L., et al. (2021). A novel small rna promotes motility and virulence of enterohemorrhagic escherichia coli o157:H7 in response to ammonium. MBio 12 (2), 1–19. doi: 10.1128/mBio.03605-20
Jørgensen, M. G., Pettersen, J. S., Kallipolitis, B. H. (2020). sRNA-mediated control in bacteria: An increasing diversity of regulatory mechanisms. Biochim. Biophys. Acta - Gene Regul. Mech. 1863 (5), 194504. doi: 10.1016/j.bbagrm.2020.194504
Kamenova, S., Sharapkhanova, A., Akimniyazova, A., Kuzhybayeva, K., Kondybayeva, A., Rakhmetullina, A., et al. (2023). piRNA and miRNA can suppress the expression of multiple sclerosis candidate genes. Nanomaterials 13 (1), 22. doi: 10.3390/nano13010022
Kang, C.s., Ban, M., Choi, E. J., Moon, H. G., Jeon, J. S., Kim, D. K., et al. (2013). Extracellular vesicles derived from gut microbiota, especially akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PloS One 8 (10), e76520. doi: 10.1371/journal.pone.0076520
Kaparakis-Liaskos, M., Kufer, T. A. (2020). “Bacterial membrane vesicles: biogenesis, functions and applications,” in Bacterial membrane vesicles: biogenesis, functions and applications (Cham Press: Springer International Publishing), 119–129. doi: 10.1007/978-3-030-36331-4
Khoo, S. K., Petillo, D., Kang, U. J., Resau, J. H., Berryhill, B., Linder, J., et al. (2012). Plasma-based circulating microRNA biomarkers for Parkinson’s disease. J. Parkinson’s. Dis. 2 (4), 321–331. doi: 10.3233/JPD-012144
Kim, O. Y., Choi, S. J., Jang, S. C., Park, K. S., Kim, S. R., Choi, J. P., et al. (2015). Bacterial protoplast-derived nanovesicles as vaccine delivery system against bacterial infection. Nano. Lett. 15 (1), 266–274. doi: 10.1021/nl503508h
Kim, O. Y., Hong, B. S., Park, K.-S., Yoon, Y. J., Choi, S. J., Lee, W. H., et al. (2013). Immunization with Escherichia coli Outer Membrane Vesicles Protects Bacteria - Induced Lethality via Th1 and Th17 Cell Responses. J. Immunol. 190 (8), 4092–4102. doi: 10.4049/jimmunol.1200742
Klebba, P. E., Newton, S. M. C., Six, D. A., Kumar, A., Yang, T., Nairn, B. L., et al. (2021). Iron acquisition systems of gram-negative bacterial pathogens define tonB-dependent pathways to novel antibiotics. Chem. Rev. 1219, 5193–5239. doi: 10.1021/acs.chemrev.0c01005
Koeppen, K., Hampton, T. H., Jarek, M., Scharfe, M., Gerber, S. A., Mielcarz, D. W., et al. (2016). A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PloS Pathog. 12 (6), e1005672. doi: 10.1371/journal.ppat.1005672
Krämer, R. (2010). Bacterial stimulus perception and signal transduction: Response to osmotic stress. Chem. Rec. 10 (4), 217–229. doi: 10.1002/tcr.201000005
Kuerban, K., Gao, X., Zhang, H., Liu, J., Dong, M., Wu, L., et al. (2020). Doxorubicin-loaded bacterial outer-membrane vesicles exert enhanced anti-tumor efficacy in non-small-cell lung cancer. Acta Pharm. Sin. B 10 (8), 1534–1548. doi: 10.1016/j.apsb.2020.02.002
Kwok, A., Raulf, N., Habib, N. (2019). Developing small activating RNA as a therapeutic: Current challenges and promises. Ther. Deliv. 10 (3), 151–164. doi: 10.4155/tde-2018-0061
Lee, H. J. (2019). Microbe-host communication by small RNAs in extracellular vesicles: Vehicles for transkingdom RNA transportation. Int. J. Mol. Sci. 20 (6), 1487. doi: 10.3390/ijms20061487
Lee, R. C., Feinbaum, R. L., Ambrost, V. (1993). The C. elegans Heterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to &II-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-y
Li, L.-C., Okino, S. T., Zhao, H., Pookot, D., Place, R. F., Urakami, S., et al. (2006). Small dsRNAs induce transcriptional activation in human cells. PNAS 103 (46), 17337–17342. doi: 10.1073pnas.0607015103
Li, S., Xu, Z., Sheng, J. (2018). tRNA-derived small RNA: A novel regulatory small non-coding RNA. Genes 9 (5), 246. doi: 10.3390/genes9050246
Li, M., Zhou, H., Yang, C., Wu, Y., Zhou, X., Liu, H., et al. (2020). Bacterial outer membrane vesicles as a platform for biomedical applications: An update. J. Controlled Release. 323, 253–268. doi: 10.1016/j.jconrel.2020.04.031
Lin, Z., Li, J., Yan, X., Yang, J., Li, X., Chen, P., et al. (2021). Engineering of the Small Noncoding RNA (sRNA) DsrA Together with the sRNA Chaperone Hfq Enhances the Acid Tolerance of Escherichia coli. Appl. Environ. Microbiol. 87 (10), e02923–20. doi: 10.1128/AEM.02923-20
Liu, L., Wu, J., Gao, J., Lu, X. (2020). Bacteria-derived nanoparticles: multifunctional containers for diagnostic and therapeutic applications. Adv. Healthcare. Mater. 9 (22), 2000893. doi: 10.1002/adhm.202000893
Liu, Q., Yu, Z., Yuan, S., Xie, W., Li, C., Hu, Z., et al. (2017). Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 8 (8), 13048. doi: 10.18632/oncotarget.14369
Liu, H., Zhang, Q., Wang, S., Weng, W., Jing, Y., Su, J. (2022). Bacterial extracellular vesicles as bioactive nanocarriers for drug delivery: Advances and perspectives. Bioactive. Mater. 14, 169–181. doi: 10.1016/j.bioactmat.2021.12.006
Liu, W. L., Zou, M. Z., Qin, S. Y., Cheng, Y. J., Ma, Y. H., Sun, Y. X., et al. (2020). Recent advances of cell membrane-coated nanomaterials for biomedical applications. Adv. Funct. Mater. 30 (39), 2003559. doi: 10.1002/adfm.202003559
Luo, R., Chang, Y., Liang, H., Zhang, W., Song, Y., Li, G., et al. (2023). Interactions between extracellular vesicles and microbiome in human diseases: New therapeutic opportunities. iMeta 2 (2), e86. doi: 10.1002/imt2.86
Magennis, E. P., Fernandez-Trillo, F., Sui, C., Spain, S. G., Bradshaw, D. J., Churchley, D., et al. (2014). Bacteria-instructed synthesis of polymers for self-selective microbial binding and labelling. Nat. Mater. 13 (7), 748–755. doi: 10.1038/nmat3949
Mahjoob, G., Ahmadi, Y., Fatima rajani, H., khanbabaei, N., Abolhasani, S. (2022). Circulating microRNAs as predictive biomarkers of coronary artery diseases in type 2 diabetes patients. J. Clin. Lab. Anal. 36 (5), e24380. doi: 10.1002/jcla.24380
Mehanny, M., Lehr, C. M., Fuhrmann, G. (2021). Extracellular vesicles as antigen carriers for novel vaccination avenues. In Adv. Drug Deliv. Rev. 173, 164–180. doi: 10.1016/j.addr.2021.03.016
Michaux, C., Verneuil, N., Hartke, A., Giard, J. C. (2014). Physiological roles of small RNA molecules. Microbiol. (United. Kingdom). 160 (6), 1007–1019. doi: 10.1099/mic.0.076208-0
Nakamoto, K., Watanabe, M., Sada, M., Inui, T., Nakamura, M., Honda, K., et al. (2019). Pseudomonas aeruginosa-derived flagellin stimulates IL-6 and IL-8 production in human bronchial epithelial cells: A potential mechanism for progression and exacerbation of COPD. Exp. Lung Res. 45 (8), 255–266. doi: 10.1080/01902148.2019.1665147
Naskar, A., Cho, H., Lee, S., Kim, K. S. (2021). Biomimetic nanoparticles coated with bacterial outer membrane vesicles as a new-generation platform for biomedical applications. Pharmaceutics 13 (11), 1887. doi: 10.3390/pharmaceutics13111887
Negrete, A., Shiloach, J. (2017). Improving E. coli growth performance by manipulating small RNA expression. Microbial. Cell Factories. 16 (1), 1–12. doi: 10.1186/s12934-017-0810-x
Nikam, R. R., Gore, K. R. (2018). Journey of siRNA: clinical developments and targeted delivery. Nucleic Acid Ther. 28 (4), 209–224. doi: 10.1089/nat.2017.0715
Noor, E., Eden, E., Milo, R., Alon, U. (2010). Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol. Cell 39 (5), 809–820. doi: 10.1016/j.molcel.2010.08.031
Orendain-Jaime, E. N., Serafín-Higuera, N., Leija-Montoya, A. G., Martínez-Coronilla, G., Moreno-Trujillo, M., Sánchez-Muñoz, F., et al. (2021). Micrornas encoded by virus and small rnas encoded by bacteria associated with oncogenic processes. Processes 9 (12), 2234. doi: 10.3390/pr9122234
Palazzolo, S., Canzonieri, V., Rizzolio, F. (2022). The history of small extracellular vesicles and their implication in cancer drug resistance. Front. Oncol. 12. doi: 10.3389/fonc.2022.948843
Pan, J., Li, X., Shao, B., Xu, F., Huang, X., Guo, X., et al. (2022). Self-blockade of PD-L1 with bacteria-derived outer-membrane vesicle for enhanced cancer immunotherapy. Adv. Mater. 34 (7), 2106307. doi: 10.1002/adma.202106307
Papenfort, K., Melamed, S. (2023). Small RNAs, large networks: posttranscriptional regulons in gram-negative bacteria. Annu. Rev. Microbiol. 77. doi: 10.1146/annurev-micro-041320
Papenfort, K., Sun, Y., Miyakoshi, M., Vanderpool, C. K., Vogel, J. (2013). Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153 (2), 426–437. doi: 10.1016/j.cell.2013.03.003
Pichler, H., Emmerstorfer-Augustin, A. (2018). Modification of membrane lipid compositions in single-celled organisms – From basics to applications. Methods 147, 50–65. doi: 10.1016/j.ymeth.2018.06.009
Piket, E., Zheleznyakova, G. Y., Kular, L., Jagodic, M. (2019). Small non-coding RNAs as important players, biomarkers and therapeutic targets in multiple sclerosis: A comprehensive overview. J. Autoimmun. 101, 17–25. doi: 10.1016/j.jaut.2019.04.002
Pita, T., Feliciano, J. R., Leitão, J. H. (2020). Extracellular RNAs in bacterial infections: From emerging key players on host-pathogen interactions to exploitable biomarkers and therapeutic targets. Int. J. Mol. Sci. 21 (24), 1–22. doi: 10.3390/ijms21249634
Prasse, D., Schmitz, R. A. (2018). Small RNAs involved in regulation of nitrogen metabolism. Microbiol. Spectr. 6 (4), 10–1128. doi: 10.1128/microbiolspec.rwr-0018-2018
Qureshi, N., Takayama, K., Ribi, E. (1982). Purification and structural determination of nontoxic lipid A obtained from the lipopolysaccharide of salmonella ep?zimurium*. THE. J. OF. Biol. Chem. Printed. U. S. A. 257 (19), 11808–11815. doi: 10.1016/S0021-9258(18)33836-5
Raad, N., Tandon, D., Hapfelmeier, S., Polacek, N. (2022). The stationary phase-specific sRNA FimR2 is a multifunctional regulator of bacterial motility, biofilm formation and virulence. Nucleic Acids Res. 50 (20), 11858–11875. doi: 10.1093/nar/gkac1025
Raina, M., Aoyama, J. J., Bhatt, S., Paul, B. J., Zhang, A., Updegrove, T. B., et al. (2022). Dual-function AzuCR RNA modulates carbon metabolism. PNAS 119 (10), e2117930119. doi: 10.1073/pnas.2117930119/-/DCSupplemental
Ren, J., Sang, Y., Qin, R., Cui, Z., Yao, Y. F. (2017). 6S RNA is involved in acid resistance and invasion of epithelial cells in Salmonella enterica serovar Typhimurium. Future Microbiol. 12 (12), 1045–1067. doi: 10.2217/fmb-2017-0055
Rupaimoole, R., Slack, F. J. (2017). MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discovery 16 (3), 203–221. doi: 10.1038/nrd.2016.246
Saldanha, G., Elshaw, S., Sachs, P., Alharbi, H., Shah, P., Jothi, A., et al. (2016). microRNA-10b is a prognostic biomarker for melanoma. Modern. Pathol. 29 (2), 112–121. doi: 10.1038/modpathol.2015.149
Seixas, A. F., Quendera, A. P., Sousa, J. P., Silva, A. F. Q., Arraiano, C. M., Andrade, J. M. (2022). Bacterial response to oxidative stress and RNA oxidation. Front. Genet. 12. doi: 10.3389/fgene.2021.821535
Simpson, B. W., Trent, M. S. (2019). Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 17 (7), 403–416. doi: 10.1038/s41579-019-0201-x
Srivastava, P., Kim, K. S. (2022). Membrane vesicles derived from gut microbiota and probiotics: cutting-edge therapeutic approaches for multidrug-resistant superbugs linked to neurological anomalies. Pharmaceutics 14 (11), 2370. doi: 10.3390/pharmaceutics14112370
Stanton, B. A. (2021). Extracellular vesicles and host–pathogen interactions: A review of inter-kingdomsignaling by small noncoding rna. Genes 12 (7), 1010. doi: 10.3390/genes12071010
Stokman, G., Qin, Y., Rácz, Z., Hamar, P., Price, L. S. (2010). Application of siRNA in targeting protein expression in kidney disease. Adv. Drug Deliv. Rev. 62 (14), 1378–1389. doi: 10.1016/j.addr.2010.07.005
Sultan, S., Mottawea, W., Yeo, J., Hammami, R. (2021). Gut microbiota extracellular vesicles as signaling molecules mediating host-microbiota communications. Int. J. Mol. Sci. 22 (23), 13166. doi: 10.3390/ijms222313166
Sun, S., Cui, Y., Yuan, B., Dou, M., Wang, G., Xu, H., et al. (2023). Drug delivery systems based on polyethylene glycol hydrogels for enhanced bone regeneration. Front. Bioeng. Biotechnol. 11. doi: 10.3389/fbioe.2023.1117647
Taboada, H., Meneses, N., Dunn, M. F., Vargas-Lagunas, C., Buchs, N., Castro-Mondragón, J. A., et al. (2019). Proteins in the periplasmic space and outer membrane vesicles of Rhizobium etli CE3 grown in minimal medium are largely distinct and change with growth phase. Microbiol. (United. Kingdom). 165 (6), 638–650. doi: 10.1099/mic.0.000720
Toyofuku, M., Nomura, N., Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17 (1), 13–24. doi: 10.1038/s41579-018-0112-2
Wang, Y., Bian, Z., Wang, Y. (2022). Biofilm formation and inhibition mediated by bacterial quorum sensing. Appl. Microbiol. Biotechnol. 106 (19–20), 6365–6381. doi: 10.1007/s00253-022-12150-3
Wang, Y. F., Fu, J. (2019). Secretory and circulating bacterial small RNAs: A mini-review of the literature. ExRNA 1 (April-May-June), 1–5. doi: 10.1186/s41544-019-0015-z
Wang, J., Li, L. C. (2012). Small RNA and its application in andrology and urology. Trans. Androl. Urol. 1 (1), 33–43. doi: 10.3978/j.issn.2223-4683.2011.12.04
Watson, C. N., Belli, A., Di Pietro, V. (2019). Small non-coding RNAs: New class of biomarkers and potential therapeutic targets in neurodegenerative disease. Front. Genet. 10 (APR). doi: 10.3389/fgene.2019.00364
Westermann, A. J., Förstner, K. U., Amman, F., Barquist, L., Chao, Y., Schulte, L. N., et al. (2016). Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529 (7587), 496–501. doi: 10.1038/nature16547
Williams, P. (2007). Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153 (12), 3923–3938. doi: 10.1099/mic.0.2007/012856-0
Xia, L., Guo, H., Wu, X., Xu, Y., Zhao, P., Yan, B., et al. (2023). Human circulating small non-coding RNA signature as a non-invasive biomarker in clinical diagnosis of acute myeloid leukaemia. Theranostics 13 (4), 1289–1301. doi: 10.7150/thno.80054
Xue, K., Wang, L., Liu, J. (2022). Bacterial outer membrane vesicles and their functionalization as vehicles for bioimaging, diagnosis and therapy. Mater. Adv. 3 (19), 7185–7197. doi: 10.1039/d2ma00420h
Yang, J., Kim, E. K., McDowell, A., Kim, Y. K. (2018). Microbe-derived extracellular vesicles as a smart drug delivery system. Trans. Clin. Pharmacol. 26 (3), 103–110. doi: 10.12793/tcp.2018.26.3.103
Yang, J., Ma, D., Fesler, A., Zhai, H., Leamniramit, A., Li, W., et al. (2017). Expression analysis of microRNA as prognostic biomarkers in colorectal cancer. Oncotarget 8 (32), 52403. doi: 10.18632/oncotarget.14175
Yang, Y., Wang, L., Liu, L., Zou, J., Huang, D., Li, Y. (2023). Small noncoding RNA in Streptococci: From regulatory functions to drug development. Molecular Oral Microbiology. John Wiley and Sons Inc, 1–8. doi: 10.1111/omi.12411
Zhang, H., Zhan, Y., Yan, Y., Liu, Y., Hu, G., Wang, S., et al. (2019). The pseudomonas stutzeri-specific regulatory noncoding RNA nfis targets katb mRNA encoding a catalase essential for optimal oxidative resistance and nitrogenase activity. J. Bacteriol. 201 (19), 10–1128. doi: 10.1128/JB.00334-19
Zhang, H., Zhang, Y., Song, Z., Li, R., Ruan, H., Liu, Q., et al. (2020). sncRNAs packaged by Helicobacter pylori outer membrane vesicles attenuate IL-8 secretion in human cells. Int. J. Med. Microbiol. 310 (1), 151356. doi: 10.1016/j.ijmm.2019.151356
Zhao, Z., Lin, C. Y., Cheng, K. (2019). siRNA- and miRNA-based therapeutics for liver fibrosis. Trans. Res. 214, 17–29. doi: 10.1016/j.trsl.2019.07.007
Keywords: gram-negative bacteria, sRNA, extracellular vesicles, therapeutics, diseases
Citation: Ajam-Hosseini M, Akhoondi F, Parvini F and Fahimi H (2024) Gram-negative bacterial sRNAs encapsulated in OMVs: an emerging class of therapeutic targets in diseases. Front. Cell. Infect. Microbiol. 13:1305510. doi: 10.3389/fcimb.2023.1305510
Received: 01 October 2023; Accepted: 26 December 2023;
Published: 30 January 2024.
Edited by:
Bindu Subhadra, Long Island University, United StatesReviewed by:
Cuncong Zhong, University of Kansas, United StatesMary Bosch Farone, Middle Tennessee State University, United States
Copyright © 2024 Ajam-Hosseini, Akhoondi, Parvini and Fahimi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Farshid Parvini, f.parvini@semnan.ac.ir; Hossein Fahimi, h.fahimi@iaups.ac.ir
 Mobarakeh Ajam-Hosseini1
Mobarakeh Ajam-Hosseini1  Farshid Parvini
Farshid Parvini Hossein Fahimi
Hossein Fahimi