The role of type VI secretion system genes in antibiotic resistance and virulence in Acinetobacter baumannii clinical isolates
- 1School of Public Health, Xi’an Jiaotong University Health Science Center, Xi’an, Shaanxi, China
- 2Department of Microbiology and Immunology, School of Basic Medical Sciences, Xi’an Jiaotong University Health Science Center, Xi’an, Shaanxi, China
- 3Department of Laboratory Medicine, Shaanxi Provincial People’s Hospital, Xi’an, China
- 4Department of Laboratory Medicine, the Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China
- 5Department of Laboratory Medicine, Xi’an Daxing Hospital, Xi’an, China
- 6Department of Respiratory and Critical Care Medicine, West China Hospital, Sichuan University, Chengdu, China
- 7Department of Laboratory Medicine, the First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China
- 8Department of Hepatobiliary Surgery, the First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China
Introduction: The type VI secretion system (T6SS) is a crucial virulence factor in the nosocomial pathogen Acinetobacter baumannii. However, its association with drug resistance is less well known. Notably, the roles that different T6SS components play in the process of antimicrobial resistance, as well as in virulence, have not been systematically revealed.
Methods: The importance of three representative T6SS core genes involved in the drug resistance and virulence of A. baumannii, namely, tssB, tssD (hcp), and tssM was elucidated.
Results: A higher ratio of the three core genes was detected in drug-resistant strains than in susceptible strains among our 114 A. baumannii clinical isolates. Upon deletion of tssB in AB795639, increased antimicrobial resistance to cefuroxime and ceftriaxone was observed, alongside reduced resistance to gentamicin. The ΔtssD mutant showed decreased resistance to ciprofloxacin, norfloxacin, ofloxacin, tetracycline, and doxycycline, but increased resistance to tobramycin and streptomycin. The tssM-lacking mutant showed an increased sensitivity to ofloxacin, polymyxin B, and furazolidone. In addition, a significant reduction in biofilm formation was observed only with the ΔtssM mutant. Moreover, the ΔtssM strain, followed by the ΔtssD mutant, showed decreased survival in human serum, with attenuated competition with Escherichia coli and impaired lethality in Galleria mellonella.
Discussion: The above results suggest that T6SS plays an important role, participating in the antibiotic resistance of A. baumannii, especially in terms of intrinsic resistance. Meanwhile, tssM and tssD contribute to bacterial virulence to a greater degree, with tssM being associated with greater importance.
1 Introduction
Acinetobacter baumannii, an opportunistic nosocomial pathogen, shows an extraordinary ability to acquire resistance to antimicrobials (AlAmri et al., 2020). An increasing number of multidrug-resistant (MDR), extremely-drug-resistant (XDR), and even pan-drug-resistant (PDR) strains impose a great burden on modern medicine (Ramirez et al., 2020). Aa a member of ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter) (Pogue et al., 2015), A. baumannii poses a great challenge in hospital-acquired infections, such as ventilator-associated pneumonia, bloodstream infections, surgical site or wound infections, urinary tract infections, and meningitis. It is particularly problematic for immunocompromised and debilitated patients confined to intensive care units (ICUs) (McConnell et al., 2013; Wong et al., 2017).
Different antibiotic-resistance mechanisms have been identified in A. baumannii. Moreover, adaptive resistance plays a key role in facilitating the rapid response of bacteria to antibiotic challenges and external stresses (Arzanlou et al., 2017; Christaki et al., 2020). This can result in decreased antimicrobial susceptibility by altering the expression of porins and efflux pumps (Fernández and Hancock, 2012). Similar to antibiotic resistance, A. baumannii can adapt to stresses such as desiccation and oxidation, which are dependent on its specific structure (Kwon et al., 2017). However, we have a limited understanding of the basic biological processes that drive A. baumannii to successfully infiltrate the healthcare environment, adapt to antibiotics and environmental stresses, and cause diseases. A deep understanding of this process may provide novel potential targets for the development of future antimicrobials and vaccines.
Protein secretion systems are critical virulence structures in A. baumannii and have received increased attention recently (Skariyachan et al., 2019). They can transport virulence factors into the surrounding environment or directly into neighboring cells (Gerlach and Hensel, 2007). Recent studies have shown that secretion systems are involved in the antibiotic resistance of A. baumannii (Elhosseiny et al., 2019). The type VI secretion system (T6SS) is the most frequently reported secretion system and is important for microbial communication within the environment and hosts; thus, it may contribute to antibiotic adaptation and drug resistance.
T6SS is a vital dynamic protein delivery apparatus that is analogous to the tail assembly of contractile bacteriophages that can inject toxic effector molecules into both prokaryotic and eukaryotic prey cells (Alcoforado Diniz et al., 2015). It is composed of a membrane complex, cytoplasmic baseplate, and contractile tail tube/sheath complex encoded by 13 well-conserved core genes (Nguyen et al., 2018). TssD (Hcp) is an important protein in the tail tube/sheath complex and is commonly used as a marker of T6SS activity (Weber et al., 2017). Hexameric TssD forms a tube-like structure arranged with helical symmetry similar to the T4 bacteriophage tail tube and is surrounded by TssB/TssC (Ruiz et al., 2015; Park et al., 2018). It acts as a virulence factor and chaperone that can transport effectors and interact with effector substrates via its ring-shaped structure (Silverman et al., 2013). TssB, in complex with TssC, assembles cytoplasmic structures resembling a bacteriophage sheath, which can change rapidly between contracted and extended states and generate a mechanical force to drive T6SS components outside the cell (Silverman et al., 2012). VgrG proteins, which participate in the composition of the baseplate complex, are structurally similar to the puncturing device of the T4 bacteriophage (Leiman et al., 2009). Another inner membrane protein, TssM, representing homologs of the T4SS IcmF, has been shown to be required for TssD secretion; therefore, TssM is required for T6SS activity (Weber et al., 2013).
Wang et al. (Wang et al., 2018) confirmed that VgrG is involved in both antimicrobial resistance and virulence in A. baumannii ATCC 19606. Therefore, markers of T6SS, e.g., TssB, TssD, and TssM, were investigated in this study to determine their importance in the drug resistance and bacterial virulence of A. baumannii.
2 Materials and methods
2.1 A. baumannii clinical isolates and cultural conditions
A total of 114 A. baumannii clinical strains isolated from inpatient specimens were collected from the Shaanxi Provincial People’s Hospital and First Hospital of Yulin in 2018. Isolates were confirmed as A. baumannii by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker Daltonik GmbH, Leipzig, Germany). All strains were grown on Luria-Bertani (LB, Beijing Land Bridge, Beijing, China) agar plates at 37°C for 18-24 h. When necessary, kanamycin (Aladdin, Shanghai, China) and carbenicillin (Aladdin) were added to the LB medium at final concentrations of 50 μg/mL and 100 μg/mL, respectively. The bacteria were cultured at 37°C and preserved in LB broth containing 20% glycerol at -80°C for further analysis.
2.2 Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using a VITEK 2 system (bioMérieux, Marcyl’ Étoile, France). The susceptibility results were interpreted according to the guidelines of the Clinical and Laboratory Standard Institute (CLSI) (CLSI, 2018). The resistance rate was used to reflect the resistance level of the strains to antibiotics, which is present as the percent of antibiotic resistant isolates in the total tested samples (ANRESIS, 2022).
Furthermore, the disk-diffusion method was used to detect the antibiotic sensitivity of the knockout and complementary mutants of the target genes (Wiegand et al., 2008). Fifteen categories containing thirty-five antimicrobial agents were tested, including penicillins (penicillin, PEN, 1 μg; oxacillin, OXA, 1 μg; ampicillin, AMP, 10 μg), extended-spectrum cephalosporins (ceftazidime, CAZ, 30 μg; cephalexin, LEX, 30 μg; cefazolin, CZO, 30 μg; cefradine, CED, 30 μg; cefuroxime, CXM, 30 μg; ceftriaxone, CRO, 30 μg; cefepime, FEP, 30 μg; cefotaxime, CTX, 30 μg; cefoperazone, CFP, 75 μg), carbapenems (meropenem, MEM, 10 μg; imipenem, IPM, 10 μg), aminoglycosides (gentamicin, GEN, 120 μg; tobramycin, TOB, 10 μg; amikacin, AMK, 30 μg; streptomycin, STR, 10 μg), fluoroquinolones (ciprofloxacin, CIP, 5 μg; norfloxacin, NOR, 10 μg; levofloxacin, LVX, 5 μg; ofloxacin, OFX, 5 μg), folate pathway inhibitors (trimethoprim/sulfamethoxazole, SXT, 25 μg), macrolides (erythromycin, ERY, 15 μg; midecamycin, MID, 30 μg), glycopeptide (vancomycin, VAN, 30 μg), nitrofurans (furazolidone, FRZ, 300 μg), chloramphenicols (chloramphenicol, CHL, 30 μg), lincosamides (clindamycin, CLI, 2 μg), nitroimidazoles (metronidazole, MTR), rifamycin (rifampin, RFP, 5 μg), tetracyclines (minocycline, MIN, 30 μg; tetracycline, TCY, 30 μg; doxycycline, DOX, 30 μg), and polymyxins (polymyxin B, PMB, 300 μg) (Hangzhou Microbial Reagent co., LTD, Hangzhou, China).
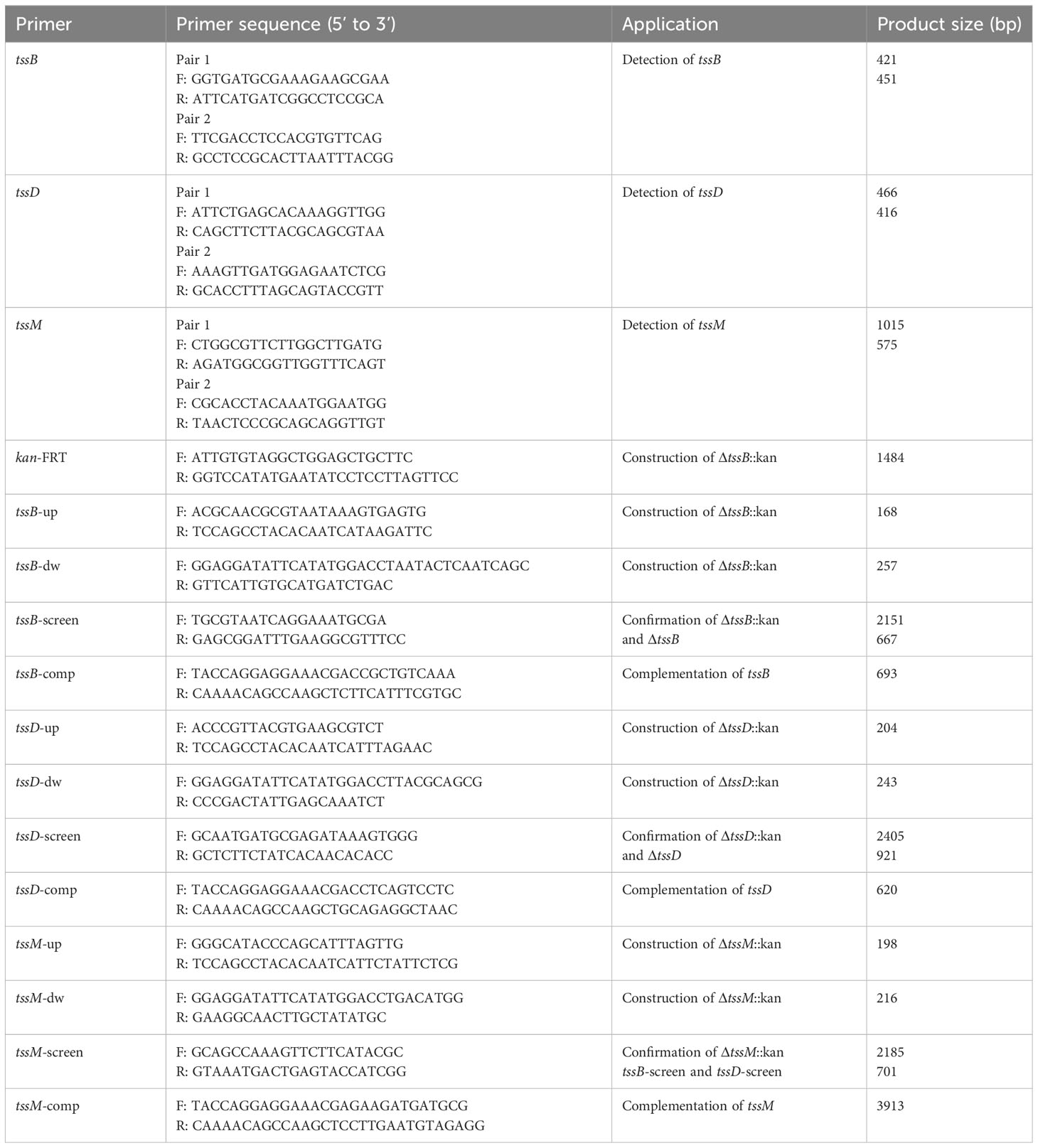
2.3 Identification of target T6SS core genes in A. baumannii clinical isolates
One hundred complete genomes of A. baumannii strains (Supplementary Table S2), listed in the NCBI Nucleotide database, were selected for comparative analysis of the T6SS gene cluster using Mauve. The sequences of the three T6SS core genes, tssB, tssD, and tssM, were aligned using SnapGene. After alignment, the phylogeny of the three core genes were constructed using the UPGMA model in MEGA, with 1000 Bootstrap replications. Two pairs of primers for each gene were designed based on the conservative clips of the selected genes. The gene targets, primer sequences, and expected amplicon sizes are shown in Table 1. Polymerase chain reaction (PCR) was performed to identify the three genes in 114 clinical A. baumannii isolates using 2 × Taq Master Mix (Novoprotein, Shanghai, China) with a total volume of 20 μL. This volume contained 10 μL of 2 × Taq Master Mix, 1 μL of each primer, and 8 μL of sterile water. The amplification program consisted of a pre-denaturation step at 94°C for 2 min, followed by 30 cycles of denaturation at 95°C for 20 s, annealing at 55°C for 20 s, and extension at 72°C for 1 min, with a final elongation at 72°C for 5 min.
2.4 Construction of tssB, tssD, and tssM knockout mutant
For convenience of constructing the deletion mutants, the clinical strain AB795639 was selected as the wild-type strain. This strain was not only sensitive to all the tested antibiotics but also harbored T6SS genes. The tssB, tssD, or tssM genes were knocked out from the AB795639 genome by allelic replacement using the RecAb system, as previously described (Tucker et al., 2014), with minor modifications. Briefly, the genomic DNA of AB795639 isolates was extracted using a TIANamp Bacteria DNA Kit (TIANGEN, Beijing, China), according to the manufacturer’s instructions. The homologous upstream and downstream fragments flanking the coding sequences of tssB, tssD, and tssM were amplified from AB795639, and the kanamycin resistance (KmR) selection marker was amplified from pKD4 by PCR using 2 × Fast Pfu Master Mix (Novoprotein). The deletion cassette of the upstream–KmR–downstream fragment was linked by overlapping PCR. This was subsequently electroporated into pAT02-containing competent AB795639 cells, and bacteria were grown in LB medium supplemented with 2 mM IPTG. KanR-insertion mutants were selected on LB agar plates containing kanamycin and subsequently confirmed by PCR using 2 × Taq Master Mix (Novoprotein). The kanamycin cassette in successful recombinants was deleted by the pAT03 plasmid expressing the FLP recombinase. Furthermore, the target gene mutant strain was confirmed by PCR and Sanger sequencing. Deletion mutants are listed in Table 2.
2.5 Complementation of tssB, tssD, and tssM
For genetic complementation of the mutant strain, the target gene was amplified from AB795639 by PCR using 2 × Fast Pfu Master Mix (Novoprotein) and the linear pAT03 plasmid was amplified from pAT03 DNA using PrimeSTAR Max DNA polymerase (TaKaRa, Beijing, China). The fragment was cloned into pAT03 using the Seamless Cloning Kit (Beyotime) and further transformed into DH5α to obtain the recombinant plasmids pAT03-tssB, pAT03-tssD, or pAT03-tssM. The plasmids obtained were confirmed by sequencing, followed by transformation into competent target gene-deletion mutant cells. The positive cells were selected on LB agar containing carbenicillin and IPTG. Complementation of tssB, tssD, or tssM was confirmed via PCR. Complementary strains are presented in Table 2.
2.6 Bacterial morphology and growth curve analysis
The morphology of AB795639 and all of the mutants was observed by Gram staining. Furthermore, the effects of tssB, tssD, and tssM genes on bacterial growth were determined by growth curves (Han et al., 2022). Briefly, overnight cultures of the wild-type strain, the deletion mutants, and the complemented strains were inoculated into fresh LB broth at 37°C with a concentration of 5 × 105 CFU/mL. The optical density of cultured strains at 600 nm (OD600) was evaluated every hour from 0 to 24 h using an UNICO UV-2100 spectrophotometer (UNICO, Shanghai, China).
2.7 Biofilm formation assay
Biofilm production abilities of the deletion and complementary mutants, as well as their parental strains, were assayed using the crystal violet staining method as previously described (Ryu et al., 2017). Briefly, bacterial suspensions from overnight cultures were normalized to the OD600 of 0.2, which contained approximately 108 CFU/mL bacteria. Next, 200 μL of the bacterial suspensions were seeded into 96-well plates and cultured at 37°C for 24 h. After incubation, planktonic bacteria were removed, and the plates were gently washed with phosphate buffered saline (PBS) three times and air-dried. The biofilm remaining in the well was stained with 0.1% crystal violet for 20 min. After rinsing twice with PBS, the dye attached to the biofilm was solubilized in 95% ethanol. Experiments were performed with three independent cultures, and the results were recorded as the mean ± SD of OD570.
2.8 Serum resistance assay
The serum resistance assay was slightly modified according to a previous report (Shin and Ko, 2015). Normal human serum (NHS) was collected from 10 healthy human donors. The pooled NHS were aliquoted and stored at -70°C until further use. Briefly, the overnight A. baumannii cultures were adjusted to an OD600 of 0.1 and resuspended in 1 mL of PBS. Then, 10 μL of the bacterial suspension was mixed with 100 μL normal human serum (NHS). The mixtures were incubated at 37°C for 1 h, followed by serial dilutions, which were plated on LB agar plates. Bacterial colonies were counted, and the serum bactericidal effect was expressed as the ratio of the CFUs in the serum bacterial suspension to the CFUs in a bacterial suspension without NHS. All experiments were performed in triplicate.
2.9 In vitro interbacterial competitive growth assays
Assays to determine the ability of A. baumannii strains to out-compete rival bacteria were performed as described previously, with minor modifications (Weber et al., 2016). Briefly, overnight cultures of different A. baumannii and Escherichia coli strains with the pWH1266 plasmid carrying a tetracycline-resistance gene were adjusted to an OD600 of 1.0. Then, 100 μL of E. coli was mixed with 10 μL of A. baumannii, and 10 μL of the mixture was spotted onto 0.22 μm nitrocellulose membranes overlaid on an LB agar plate. After 12 h incubation at 37°C, the spots were excised from the agar and resuspended by mixing with 1 mL of LB. The cultures underwent 10-fold serial dilutions, then, 100 μL aliquots of the selected dilutions were plated onto 16 μg/mL tetracycline-containing LB agar plates to determine the number of E. coli remaining. The controls consisted of E. coli mixed with LB medium. The assays were repeated in triplicate, and a representative image is shown.
2.10 Galleria mellonella infection assay
The G. mellonella infection experiment was performed as described by Harding et al. (Harding et al., 2016) with minor modifications. The different A. baumannii strains tested were grown in LB medium overnight and then adjusted to a final OD600 of 0.2. A concentration of 105 CFU/mL was obtained by serial dilution of the suspensions. Groups of 20 randomly selected larvae were used in each assay. A Hamilton microliter syringe was used to inject 20 μL of the bacterial suspension into the hemolymph of each larva via the second last left proleg. As a control, one group of G. mellonella was injected with 20 μL sterile LB. After injection, the larvae were kept on glass plates at 37°C, and the number of dead individuals was scored approximately every 6 h until 96 h, depending on the response to physical stimuli using a sterilized tip.
2.11 Statistical analysis
Data analysis was performed using the GraphPad Prism software (version 8.0 (San Diego, CA, USA) and SPSS (version 22.0; SPSS Inc., Chicago, IL, USA). The relationship between the prevalence of T6SS core genes and antimicrobial resistance in A. baumannii clinical isolates was analyzed using the Pearson chi-square test. The survival rate of larvae was compared using the log-rank test. To determine the differences in the drug sensitivity, growth, ability to form biofilms, ability to kill rival bacterial strains, and resistance to human serum of the A. baumannii strains (wild type, deletion mutants, and complemented mutants), a one-way ANOVA was employed. Statistical significance was set at P < 0.05.
3 Results
3.1 The antimicrobial susceptibility profiles
The 114 A. baumannii clinical isolates were resistant to multiple antibiotics (Supplementary Table S1). The strains possessed the highest resistance rate against ciprofloxacin (88.6%). This was followed by meropenem, ceftazidime, piperacillin/tazobactam, imipenem, levofloxacin, cefepime, and tobramycin, with resistance rates of over 80%. The resistance rates of minocycline, trimethoprim/sulfamethoxazole, amikacin, and cefoperazone/sulbactam were slightly lower, that from 40.4% to 74.6%. Only tigecycline and polymyxin showed high activity against A. baumannii (Figure 1). Among the isolates, six were susceptible to all of the tested antimicrobial agents. Eleven strains were resistant to all antimicrobial agents except tigecycline and polymyxin. The distribution of minimum inhibitory concentrations (MICs) of the antibiotics tested in this study is shown in Figure 2.
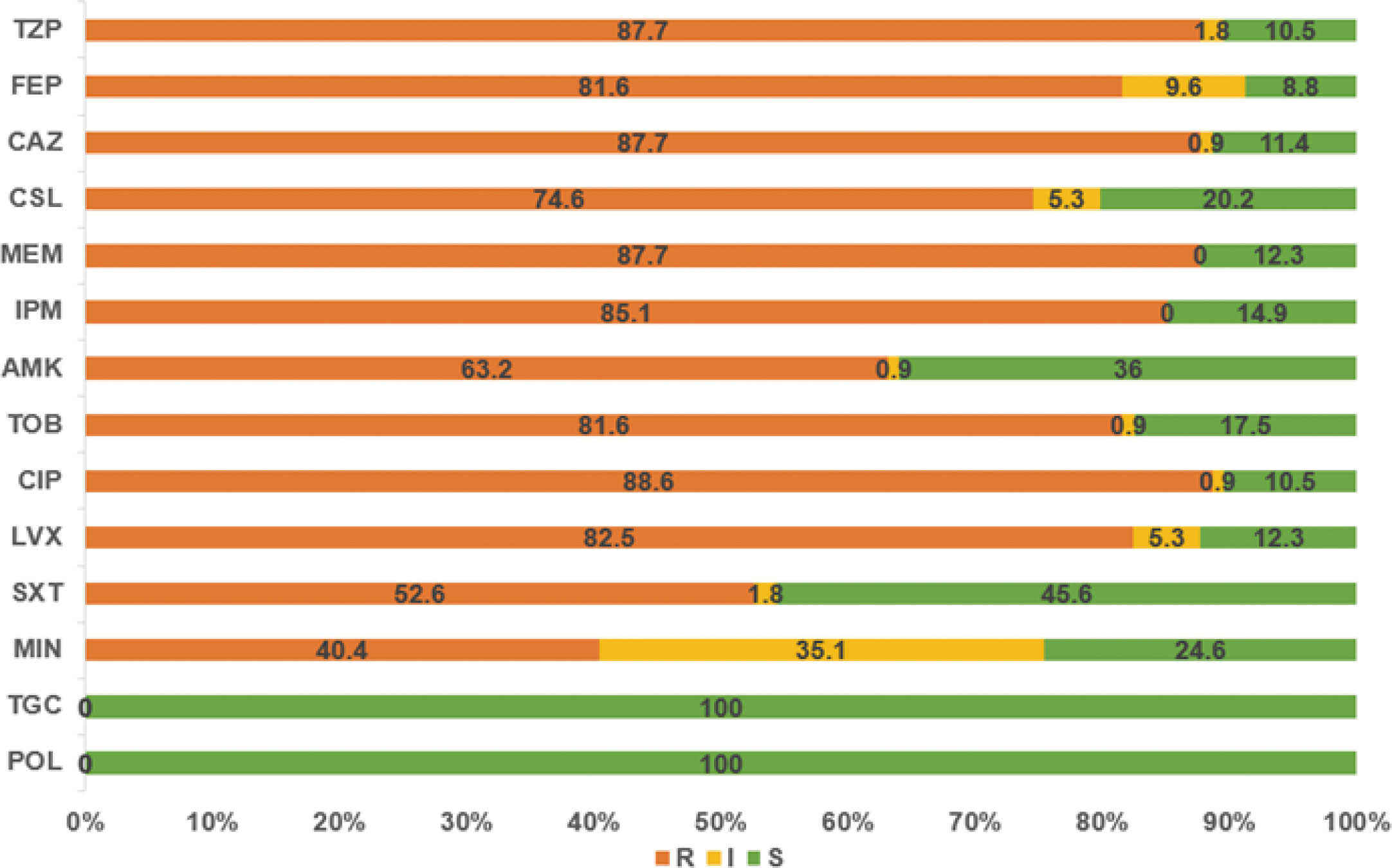
Figure 1 The resistance rates of A. baumannii strains to various antibiotics. TZP, piperacillin/tazobactam; FEP, cefepime; CAZ, ceftazidime; CSL, cefoperazone/sulbactam; MEM, meropenem; IPM, imipenem; AMK, amikacin; TOB, tobramycin; CIP, ciprofloxacin; LVX, levofloxacin; SXT, trimethoprim/sulfamethoxazole; MIN, minocycline; TGC, tigecycline; POL, polymyxin.
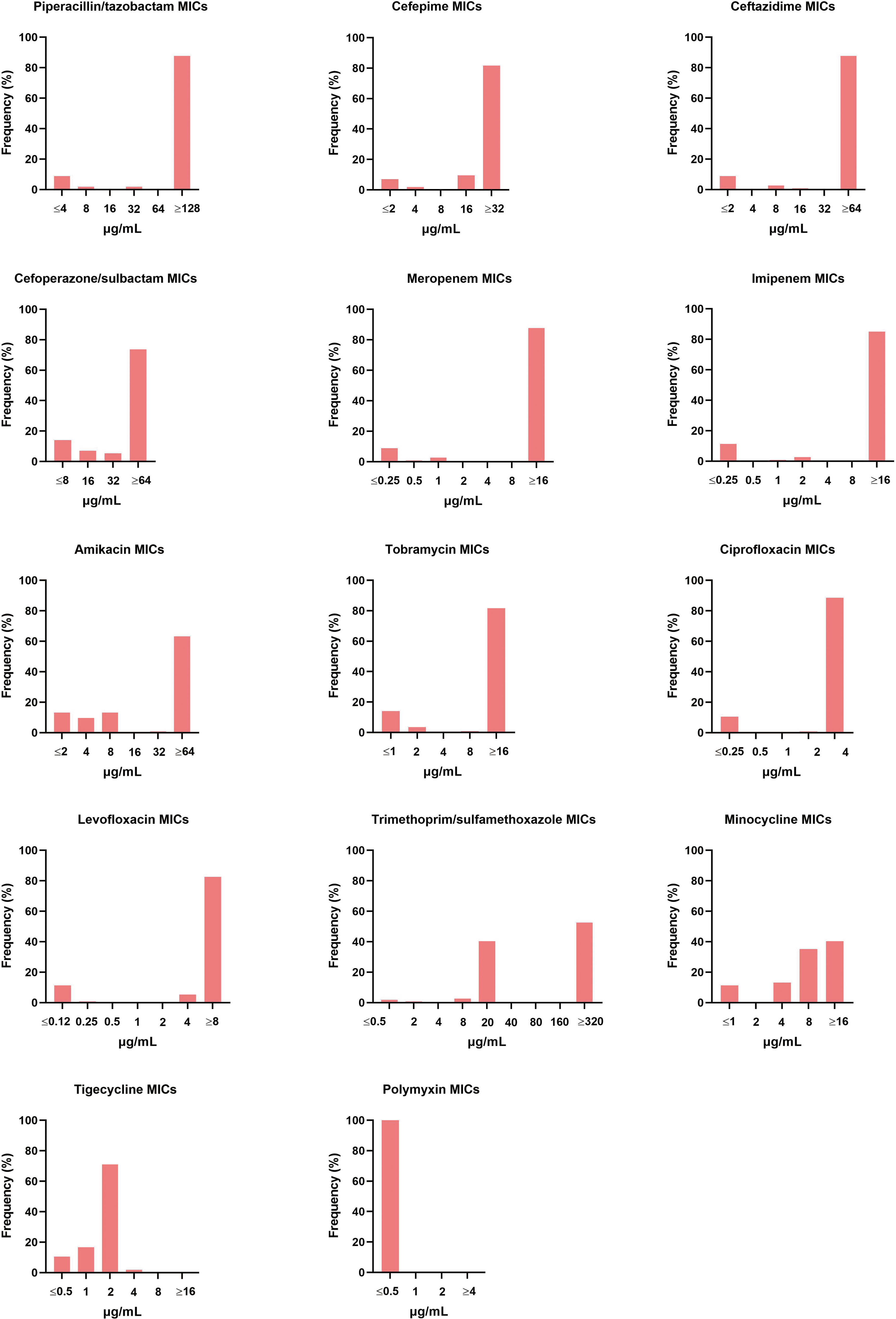
Figure 2 Distribution of different antibiotic MICs. The MIC breakpoints for each of the antibiotics tested was as per the guidelines of the Clinical and Laboratory Standard Institute.
3.2 Conservation of tssB, tssD (hcp), and tssM in A. baumannii genomes and their prevalence in A. baumannii clinical isolates.
We first selected 100 complete genome sequences of A. baumannii containing T6SS from GenBank and analyzed the T6SS locus. Comparative analysis using Mauve indicated that the T6SS gene cluster is effectively conserved among various A. baumannii isolates (Supplementary Figure S1). The positions of the three core genes, tssB, tssD, and tssM, in each genome are listed in Supplementary Table S2. Thereafter, the tssB, tssD, and tssM from the 100 genomes were aligned. According to the analysis using MEGA (Supplementary Figure S2) and SnapGene (Supplementary Figures S3–S5), the sequences of tssB, tssD, and tssM genes were highly conserved among these 100 A. baumannii strains.
The highly conservative nature of the core genes makes it possible to analyze their prevalence in clinical isolates. As is shown in Supplementary Table S1, 77.2% (88/114) of the clinical strains harbored all three genes. Moreover, 2.63% (3/114) and 8.77% (10/114) of the isolates were positive for two genes and one gene, respectively. For the remaining 11.4% (13/114) of isolates, the absence of T6SS was confirmed using both pairs of primers for each gene.
3.3 The relationship between T6SS core genes and antibiotic resistance in A. baumannii clinical isolates
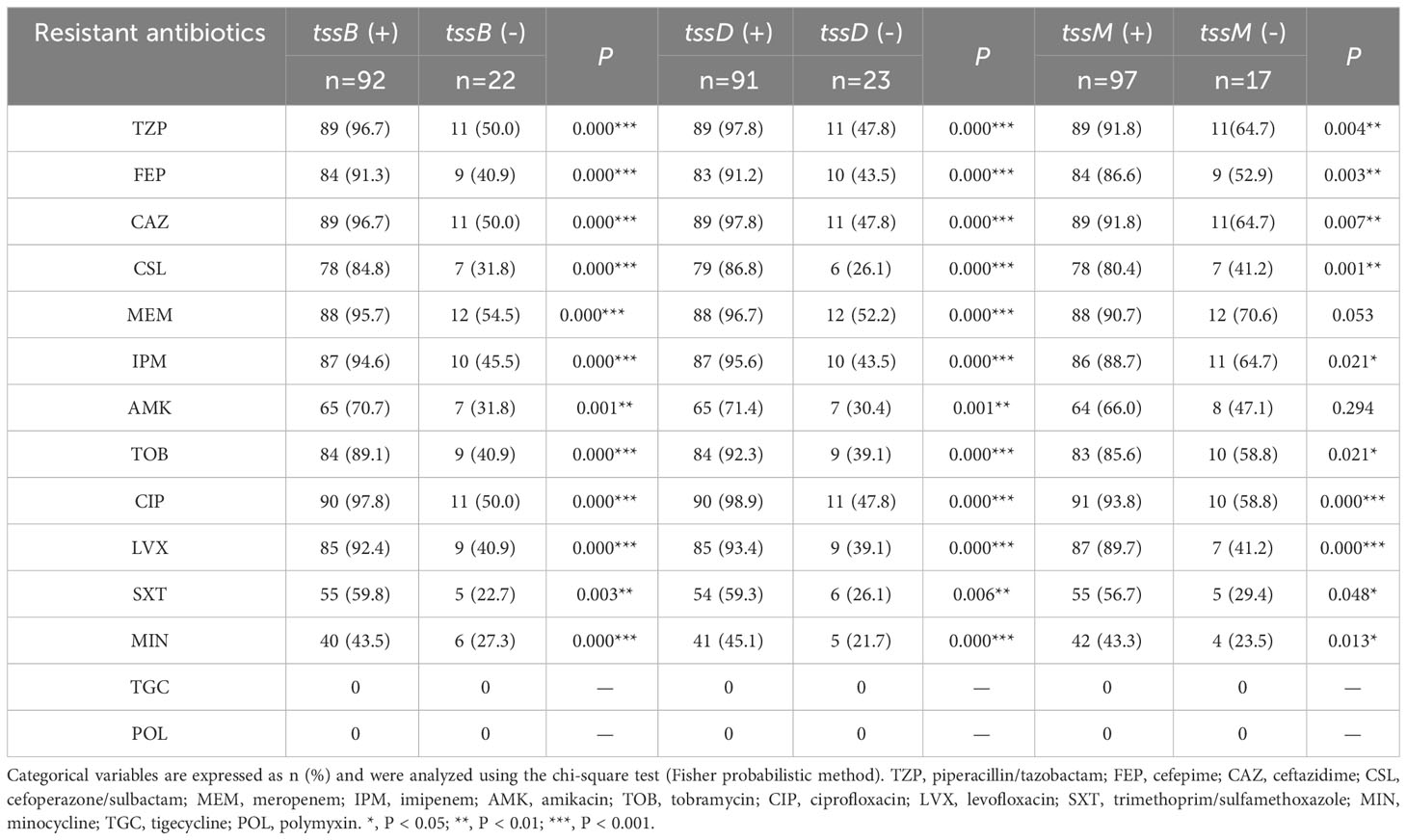
As T6SS has been found to be closely related to antibiotic resistance (Li et al., 2023), the correlation between each core gene and the antibiotic resistance data for the clinical strains was analyzed (Table 3). It was found that tssB correlated with resistance to all antibiotics (P < 0.05), except for tigecycline and polymyxin. Specifically, the resistance rate of strains with tssB was higher than that without tssB. Similar results were observed for tssD. In addition, tssM also showed correlation in terms of antibiotic resistance with all drugs except meropenem, amikacin, tigecycline, and polymyxin.
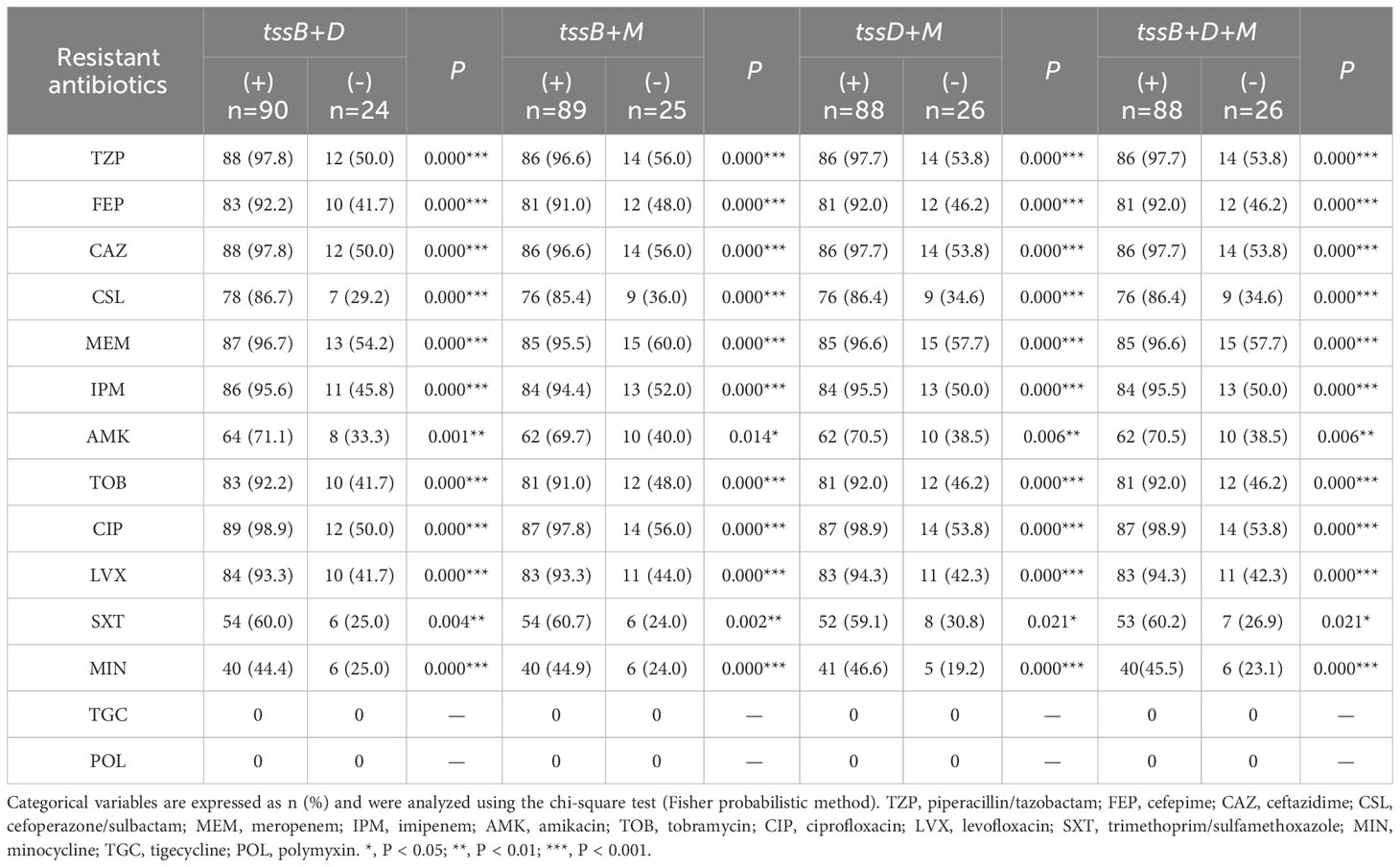
Further analysis was performed to explore the relationship between the core gene combinations and antimicrobial resistance (Table 4). Interestingly, every type of combination showed strong correlation with all antibiotics (P < 0.001), except for amikacin and trimethoprim/sulfamethoxazole (P < 0.05). Owing to the full susceptibility of all A. baumannii clinical isolates to tigecycline and polymyxin, there was no correlation between the three core genes and these two antibiotics.
3.4 Effects of tssB, tssD, and tssM on antibiotic resistance of A. baumannii
To better understand the antimicrobial roles that tssB, tssD, and tssM play, we constructed deletion mutants and complementary strains from the sensitive strain AB795639, which harbored all three core genes (Table 2). The sensitivity of the mutants to 35 antibiotics was determined to comprehensively determine the contribution of the three T6SS core genes to drug resistance in A. baumannii. The disk diffusion method was performed instead of MIC detection to better observe slight changes in drug resistance. Compared with the wild-type strain, the inhibition zone of ΔtssB by cefuroxime and ceftriaxone was reduced (P = 0.009 and P = 0.010, respectively); however, that of gentamicin was remarkably increased (P = 0.000) (Figure 3A).
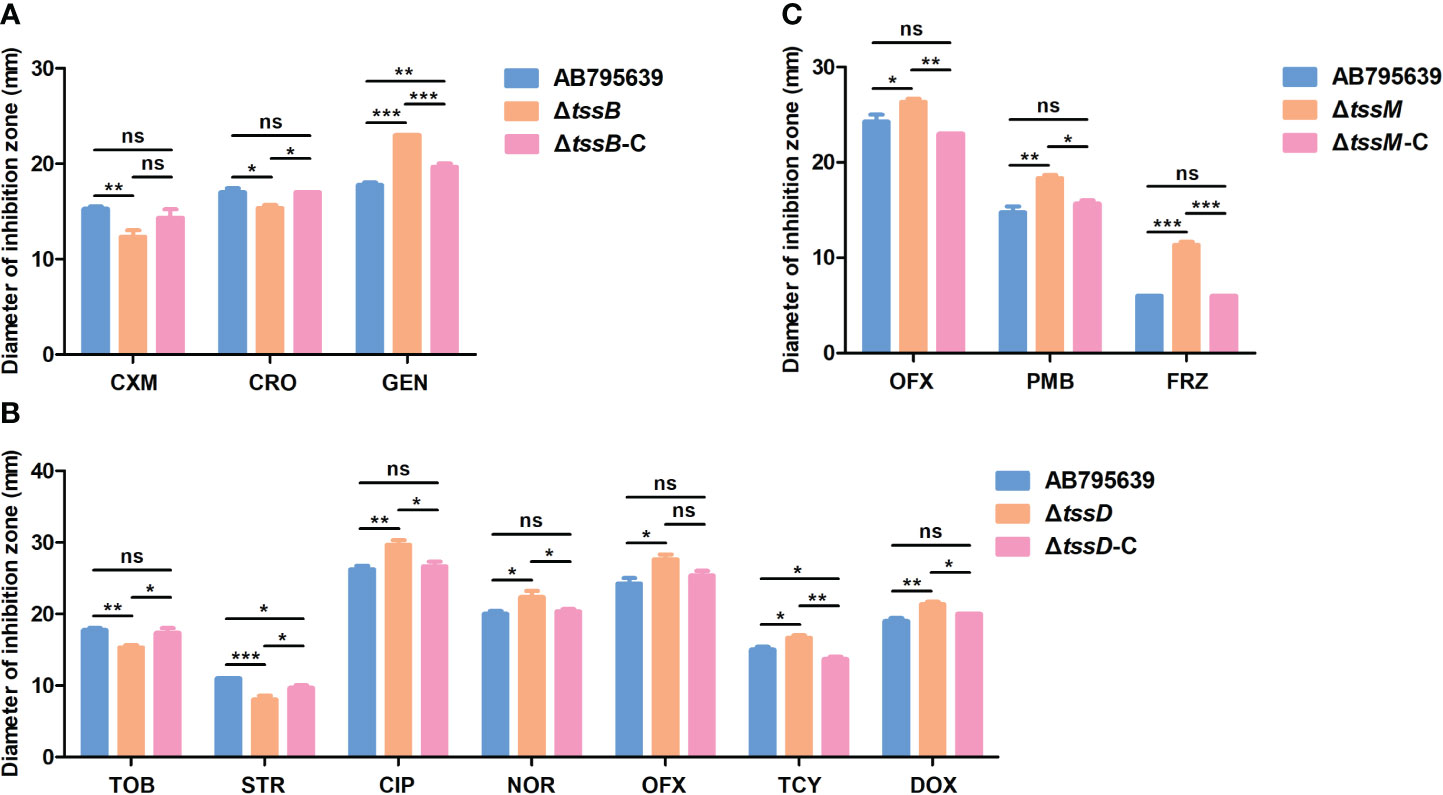
Figure 3 Antibiotic susceptibilities of AB795639 using the deletion and complementary mutants of three T6SS core genes as assessed by disk diffusion. The changed antibiotic susceptibility related to tssB (A), tssD (B), and tssM (C). CXM, cefuroxime; CRO, ceftriaxone; GEN, gentamicin; TOB, tobramycin; STR, streptomycin; CIP, ciprofloxacin; NOR, norfloxacin; OFX, ofloxacin; TCY, tetracycline; DOX, doxycycline; PMB, polymyxin B; FRZ, furazolidone. The experiment was repeated at least in triplicate, and the results are shown as means ± SD. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
As is shown in Figure 3B, tssD was the most relevant gene for antimicrobial susceptibility as the inhibition zone of the seven antibiotics changed. This demonstrated that the strain became more resistant to tobramycin (P = 0.004) and streptomycin (P = 0.000), but more sensitive to ciprofloxacin (P = 0.004), norfloxacin (P = 0.020), ofloxacin (P = 0.011), tetracycline (P = 0.016), and doxycycline (P = 0.002) when tssD was missing.
Meanwhile, the mutant strain ΔtssM displayed an increased susceptibility to ofloxacin (P = 0.033), polymyxin B (P = 0.002), and furazolidone (P = 0.000) (Figure 3C).
When the tssB, tssD, and tssM genes were complemented (Figure 3), the antimicrobial sensitivity of the complementary strains was recovered or partly restored to the level of the wild-type strain. The antimicrobial susceptibility results for all strains are shown in Supplementary Table S3.
3.5 The effects of three T6SS core genes on the bacterial morphology, growth, and biofilm formation in A. baumannii
Loss of tssB, tssD, or tssM did not affect either the morphology (Supplementary Figure S6) or the growth of AB795639 in LB broth (Figure 4A). The growth curves of three deletion mutants were similar to those of the wild type and the complementary strains.
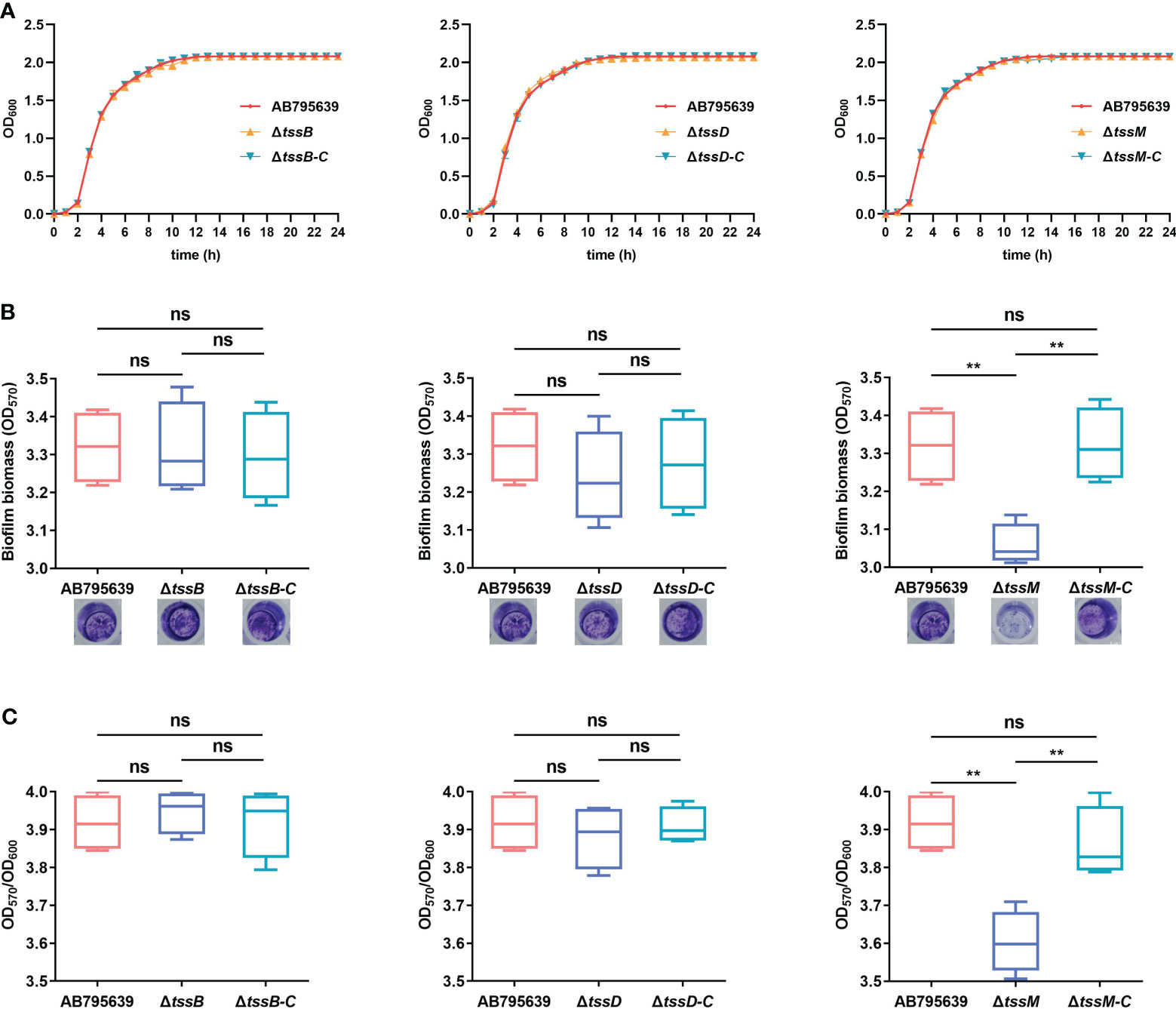
Figure 4 Effects of tssB, tssD, and tssM on bacterial growth and biofilm formation in A baumannii. (A) The growth curves of the wild-type strain, deletion mutants, and the respective complemented strains in LB broth. OD600 values of the bacterial cultures grown at 37°C with shaking at 200 rpm were recorded during 0-24 (h) The experiment was repeated in triplicate, and the results were presented as means ± SD. (B) Biofilm formation ability of AB795639, and the deletion and complementary mutants of three T6SS core genes. Biofilm biomass measured by crystal violet staining using the optical density at 570 nm (OD570) after 24 h incubation in strains expressing tssB, tssD, and tssM, in addition to a strain not expressing these genes. (C) The ratio of OD570/OD600 was evaluated for all the tested strains to avoid the bias caused by dissimilar bacterial numbers. The experiment was repeated in triplicate, and the results are shown as means ± SD. ns, P > 0.05; *, P < 0.05; **, P < 0.01.
In A. baumannii, biofilm formation is an important factor that involves in antibiotic resistance and bacterial virulence. Hence, the role of TssB, TssD, and TssM in biofilm formation by A. baumannii was investigated. Following 24 h of incubation, only the ΔtssM mutant exhibited a decreased ability to adhere to the well surfaces, suggesting a lower degree of biofilm production (P = 0.002). Complementation of tssM restored the biofilm-forming ability. In contrast, for tssB and tssD, no significant difference was observed for strains expressing these genes (P > 0.05) (Figures 4B, C). These results indicate that tssM is required for biofilm formation of this strain.
3.6 T6SS core genes tssD and tssM are required by A. baumannii for human serum resistance
By avoiding the bactericidal activity of serum, pathogenic bacteria can invade and survive in certain hosts, thus causing disease (Kim et al., 2009). Therefore, the effects of the three T6SS core genes on serum resistance were determined (Figure 5). The survival of A. baumannii in the presence of NHS differed significantly between the wild-type and mutant strains, except for ΔtssB (P > 0.05). Specifically, ΔtssD and ΔtssM mutant strains showed remarkably decreased survival after 1 h incubation with normal human serum compared with the wild-type strain (P = 0.033 and P = 0.020, respectively). The complementation of tssD and tssM restored this ability.
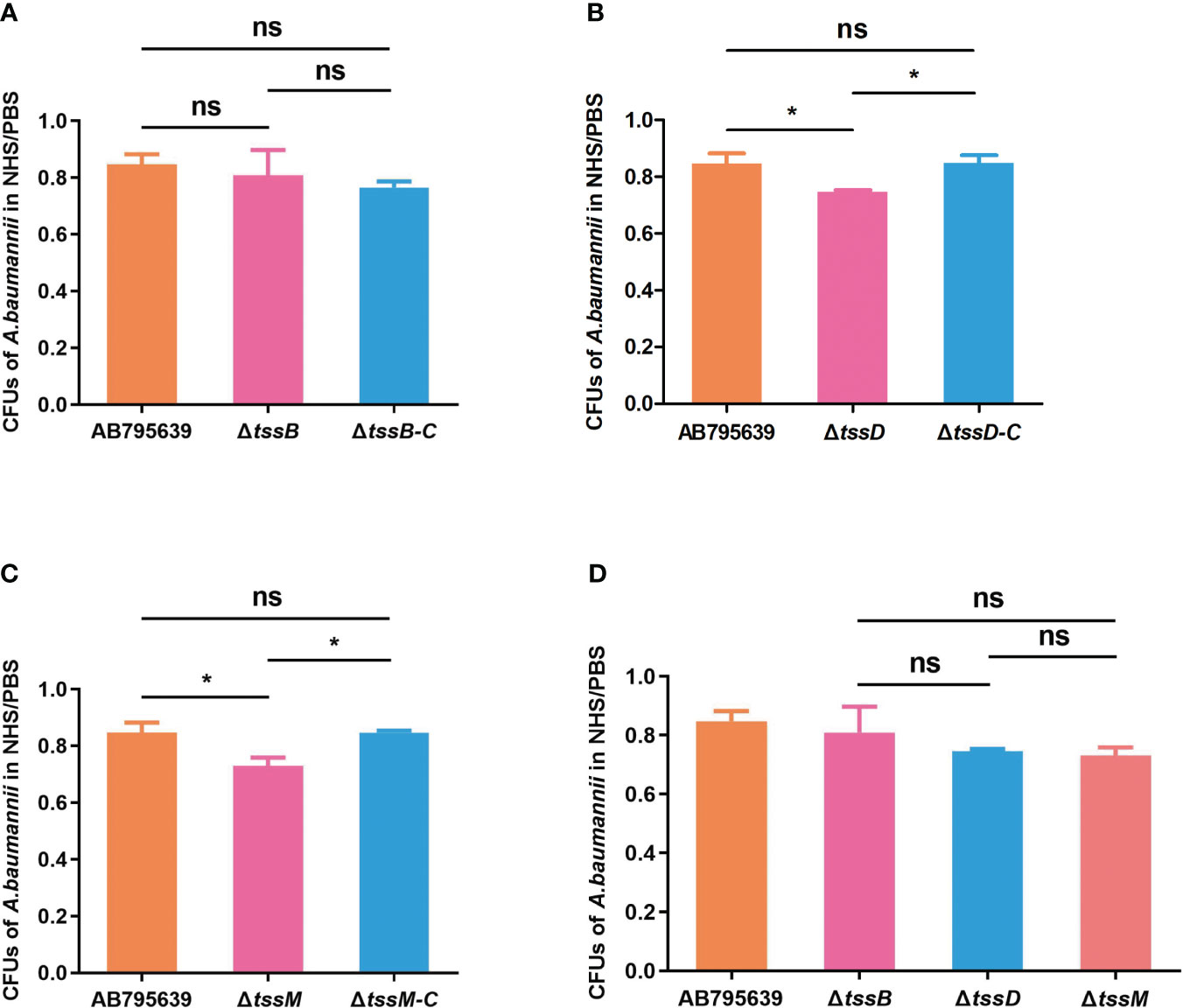
Figure 5 The human serum resistance ability of AB795639, the deletion and complementary mutants of three T6SS core genes. The viability ratios of strains for tssB (A), tssD (B) and tssM (C) were calculated (colony forming units (CFUs) of bacteria in serum/CFUs of bacteria in phosphate buffer saline (PBS)). (D) Comparison of viability ratios among deletion mutants of three core genes. The experiment was repeated in triplicate, and the results are shown as means ± SD. ns, P > 0.05; *, P < 0.05.
3.7 A. baumannii strain AB795639 outcompetes Escherichia coli that is more highly dependent on TssD and TssM
T6SS has been shown to mediate interbacterial antagonism and confer a competitive advantage in different bacteria (Basler et al., 2013; Russell et al., 2014; Zhao et al., 2018; Hsieh et al., 2019); however, less is known about the role of the three core genes in terms of these effects, especially in A. baumannii. The competition assays showed that when co-incubated with AB795639, the E. coli bacterial counts were remarkably reduced compared with those incubated in LB medium. This indicates a strong capability of this wild-type strain to outcompete other bacterial strains (Figure 6). The lack of tssB did not significantly reduce the competitive ability of AB795639 cells (Figure 6A). However, ΔtssD and ΔtssM weakened this ability, in contrast to AB795639 (P = 0.008 and P = 0.001, respectively). Complementation of tssD and tssM restored the ability of this bacterial to kill E. coli. This ability was at a similar level to the wild-type strain (Figures 6B, C). Among these core genes, strains expressing tssM showed the strongest competitive ability, followed by tssD (Figure 6D). In conclusion, killing of E. coli by AB795639 is dependent on a functional T6SS, and TssD and TssM play a predominant role in this process.
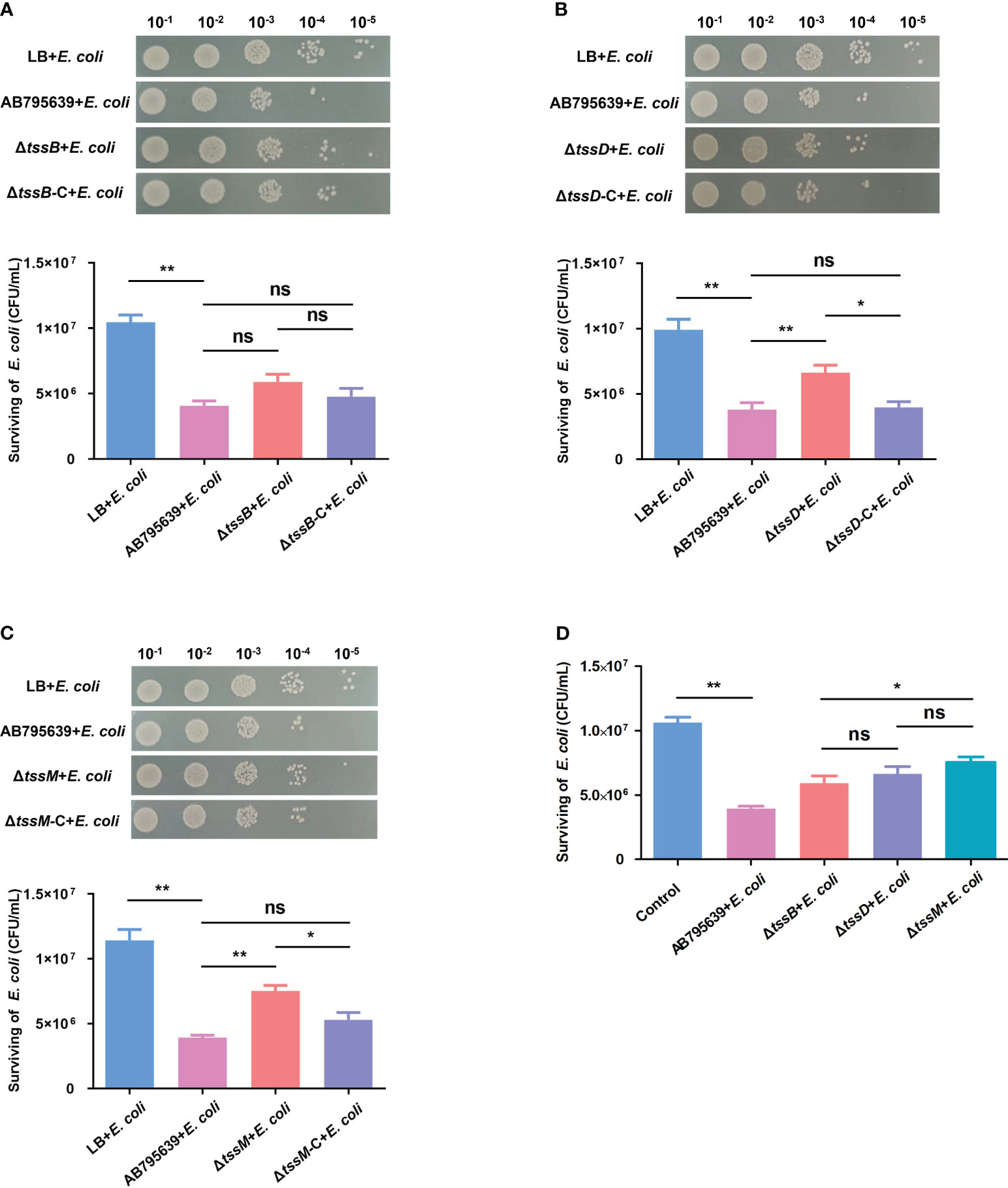
Figure 6 Effects of three T6SS core genes of A baumannii AB795639 in terms of outcompeting E coli. Representative images show the survival of E coli after incubation in LB medium (control) or with A baumannii strains: wild type, ΔtssB (A), ΔtssD (B), and ΔtssM (C), as well as respective complemented strains. (D) Comparison of E coli survival after competition with different mutants. The experiment was repeated in triplicate, and the results are shown as means ± SD. ns, P > 0.05; *, P < 0.05; **, P < 0.01.
3.8 The ΔtssD and ΔtssM mutants have attenuated virulence in a G. mellonella infection model
To assess the role of tssB, tssD, and tssM in the virulence of A. baumannii, G. mellonella was infected with variants of these core genes as well as the wild-type strain. Larval viability was monitored every 6 h, and death was recorded. By the end of the 4-day study, the survival rate of larvae infected by ΔtssD and ΔtssM strains was significantly higher than that of the wild type strain AB795639, demonstrating remarkably attenuated virulence (P = 0.039 and P = 0.012, respectively). When the worms were inoculated with the corresponding complementary strains, larval survival was reduced or partly reduced to the level of the wild-type strain, indicating the restoration of virulence. However, no obvious difference in G. mellonella viability was observed for ΔtssB with respect to gene expression in the isolates. The control group of worms injected with sterile LB medium displayed 100% survival throughout the course of the study. Based on these data, we found that T6SS-dependent virulence of A. baumannii AB795639 exhibited a greater reliance on TssD and TssM (Figure 7).
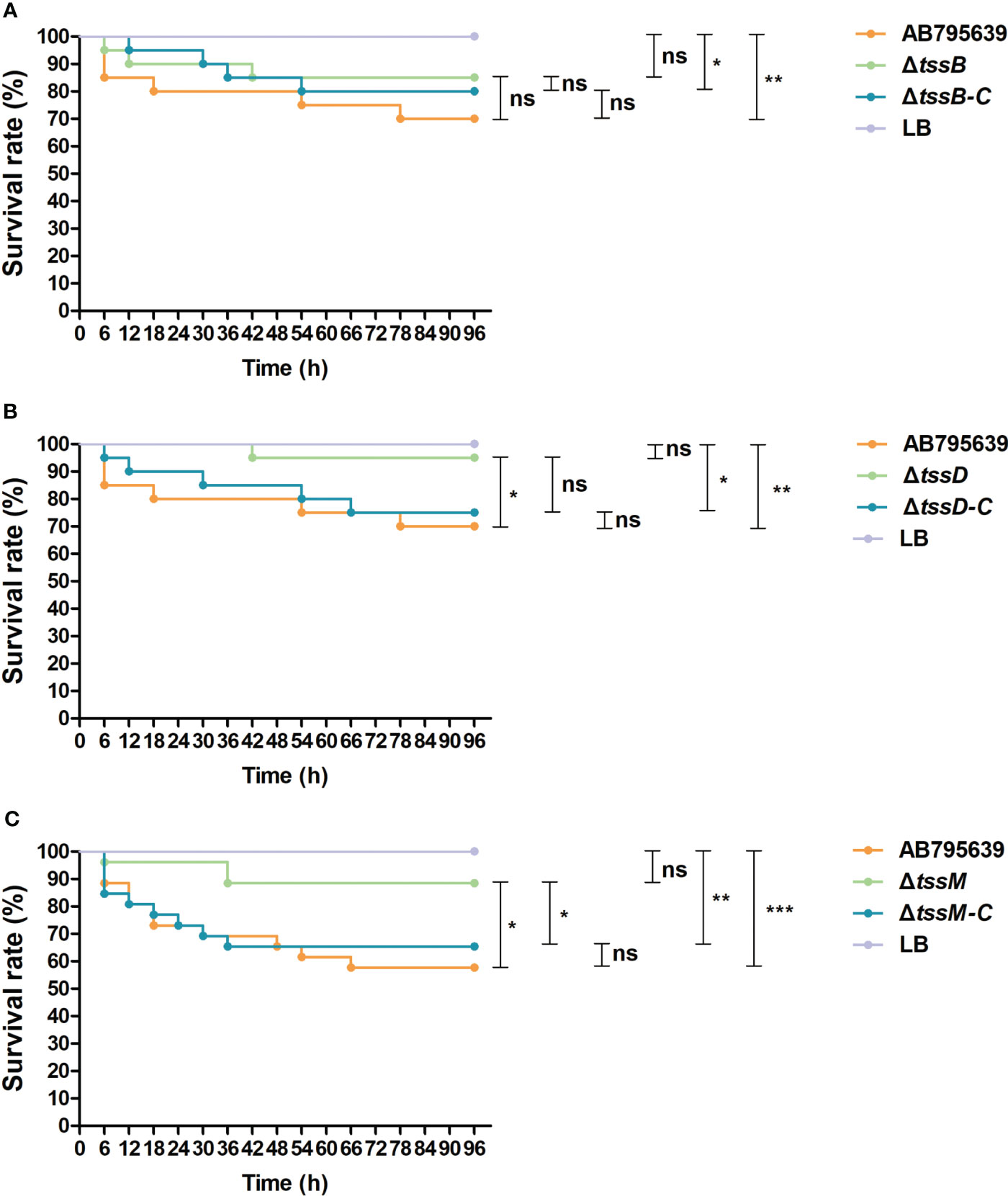
Figure 7 Survival of Galleria mellonella larvae infected with AB795639, the deletion and complementary mutants of three T6SS core genes. Kaplan–Meier survival curves of G mellonella infected with 1.0 × 105 CFU of different A baumannii strains expressing tssB (A), tssD (B) and tssM (C), and without these genes, are shown. ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
4 Discussion
The challenge in treating A. baumannii infections is primarily attributed to the ability of A. baumannii to develop resistance to the last line of antibacterial agents, including carbapenems. As expected, 114 clinical isolates of A. baumannii showed high rates of resistance to various antimicrobials. The resistance rate of A. baumannii to carbapenems is still high. Most studies on the mechanisms employed by A. baumannii to combat antibiotics have focused on permeability defects, efflux pumps, resistance genes harbored on genetic islands, and target mutations (Elhosseiny and Attia, 2018). Secondary to these well-known mechanisms, the important virulence factor T6SS had once been reported to be related to antibiotic resistance (Liao et al., 2022). In this study, a close relationship between the presence of T6SS and antimicrobial resistance in our clinical strains was found, with a higher presence of T6SS in drug-resistant A. baumannii strains than in sensitive isolates. This was consistent with the latest report by Dong et al. (Dong et al., 2022).
Few studies have investigated the role of T6SS core genes in drug resistance. Wang et al. reported that a lack of vgrG in A. baumannii increased antimicrobial resistance to ampicillin/sulbactam, but reduced resistance to chloramphenicol (Wang et al., 2018). We further determined the roles of other core genes in drug resistance by constructing deletion mutants and complementary strains from a representative strain. Initially, there was a concern that the resistance mechanisms employed by drug-resistant A. baumannii mainly work in a high activity that may offset the effect of missing a single T6SS core gene. Indeed, according to our previous detection (Zhang et al., 2023), the majority of our tested strains had hydrolysis enzymes including carbapenemases, which remarkably increased the MIC values. In addition, since the drug adaption will provide more opportunity for bacteria to survive and further become resistant strains, T6SS may contribute to this process according to our observation that there was a correlation between drug resistance and T6SS existence. In these cases, the sensitive strain AB795639 was chosen rather than a resistant strain to conduct the following experiments for observing positive results. To comprehensively understand the role of different T6SS core genes contributing to drug resistance in A. baumannii, many more antibiotics (35 antibiotics) than those recommended by the CLSI (CLSI, 2018) were tested. Generally speaking, the sensitivity of AB795639 to 12 antibiotics was, to some extent, affected by the loss of individual T6SS core genes. Among these antimicrobials, cefuroxime and ceftriaxone belong to the cephalosporins class of antibiotics; gentamicin, tobramycin, and streptomycin are aminoglycosides antibiotics; ciprofloxacin, norfloxacin, and ofloxacin are fluoroquinolones; tetracycline and doxycycline are tetracyclines; polymyxin B is a member of the polymyxin family; and furazolidone belongs to the nitrofuran family. Strains lacking the T6SS core genes became more sensitive to the majority of these antibiotics, with the exception of resistance to cefuroxime, ceftriaxone, tobramycin, and streptomycin, Specifically, although extreme changes in sensitivity and resistance were not observed in the three mutants, obvious differences among inhibition zones were recorded. The ΔtssD strain showed the most significant changes; it was more resistant to aminoglycosides and more sensitive to fluoroquinolones and tetracyclines. This may be due to its unique inner-tube-like structure, which assembles onto the base of VgrG and extends into the cytoplasm (Li et al., 2023), and this may facilitate interactions with more antibiotics. The strain without tssB became more resistant to cephalosporins. However, in contrast to ΔtssD, ΔtssB became more sensitive to aminoglycosides. This difference may be related with the tail tube/sheath complex structure of Hcp/TssB. For ΔtssM, the mutant strain demonstrated an increased susceptibility to fluoroquinolones, polymyxins and nitrofurans. This might be because TssM is involved in Hcp recruitment and secretion. Based on these results, the syringe-shaped apparatus of T6SS may also be primarily employed by A. baumannii to adapt to environmental antibiotic stress.
Biofilm formation is another crucial ability that contributes to both antibiotic resistance and bacterial persistence in healthcare settings (Di Domenico et al., 2017). Apart from the commonly reported factors such as porins, capsular polysaccharides, quorum sensing, etc. (Gaddy et al., 2009; Harding et al., 2018), the association of secretion systems such as T1SS (Bap) and T5SS (Ata) with biofilm formation has also been identified (Harding et al., 2017; Elhosseiny and Attia, 2018). A few T6SS components have been reported to be associated with the biofilm formation of other pathogens (Aschtgen et al., 2008; Zhang et al., 2014). However, the role of T6SS in the biofilm formation of A. baumannii remains unclear. As reported by Kim et al., A. baumannii T6SS+ isolates formed a significantly greater biofilm mass than the T6SS− isolates (Kim et al., 2017); however, the opposite result was observed by Dong et al. (Dong et al., 2022). The biofilm forming ability of A. baumannii may be strain specific. Although a remarkable increased biofilm was obtained when hcp was absent from T6SS+ A. baumannii A152 (Dong et al., 2022), our results showed that hcp was not involved in biofilm formation in the AB795639 strain. Similarly, no difference in biofilm quantity was observed between tssM-lacking and harboring strains in either 17978 or DSM30011 (Weber et al., 2013; Repizo et al., 2015). However, in our study, a significant reduction in biofilm biomass was observed in the ΔtssM mutant compared to that in the tssM-expressing strains. TssM, which is involved in Hcp recruitment and secretion, is important for the activity of T6SS (Li et al., 2023). Thus, it is rational to assume that this wound exhibits a stronger biofilm-forming activity than the other two components. The contribution of the tssB gene to biofilm formation in A. baumannii has not yet been reported. In this study, we demonstrated that tssB did not affect biofilm formation in AB795639.
Although T6SS is a well-known multifunctional virulence apparatus in A. baumannii, so far, the most comprehensively studied core proteins are Hcp and VgrG. Little is known about the roles of other core proteins in terms of bacterial virulence.
Resistance to human serum is related to the mortality of patients infected with A. baumannii. This mechanism makes this bacterium a successful pathogen (Liao et al., 2007). Improved survival of A. baumannii clinical isolates harboring T6SS in the presence of NHS has been demonstrated (Repizo et al., 2017; Kim et al., 2017). However, only Hcp has been shown to play a role in serum resistance (Dong et al., 2022). In this study, we not only confirmed the importance of Hcp contributing to NHS resistance but also confirmed the role of TssM in this phenomenon. In contrast, TssB does not display this function in A. baumannii.
In order to colonize medical device surfaces, thrive, and eventually cause diseases in patients, a mixed population of bacteria would compete against co-existing bacteria to maintain the species. As a “molecular syringe”, T6SS is an efficient weapon with regard to the competition among bacterial species through the contact-dependent injection of effectors, interfering with vital cellular processes (Hachani et al., 2016). For A. baumannii, T6SS is involved in the competition with not only E. coli, but also with some nosocomial pathogens, e.g., P. aeruginosa and K. pneumoniae (Repizo; Repizo et al., 2015; Kim et al., 2017). This function relies on the core genes tssD and tssM; AB795639 lacking these two genes showed reduced ability to outcompete E. coli. This finding is consistent with that of previous studies. Carruthers et al. (Carruthers et al., 2013) demonstrated that the ability of A. nosocomialis M2 to kill E. coli is dependent on hcp. Furthermore, the Hcp-secreting variant of a clinical A. baumannii strain Ab04 can kill E. coli and K. pneumoniae; whereas, the non-Hcp-secreting variant from the same strain loses this ability (Weber et al., 2015). The importance of Hcp in competitive virulence might be dependent on its structure, namely, the inner tube of T6SS (Li et al., 2023). In addition, as a key component involving in the recruitment and secretion of Hcp (Ma et al., 2012), the positive effect of TssM on bacterial competition is expected. Similar to our results, tssM-dependent competition against E. coli was observed in various A. baumannii strains (Repizo et al., 2015; Fitzsimons et al., 2018). Interestingly, due to strain specificity, tssM was not implicated in terms of competition in the ATCC17978 strain (Weber et al., 2013). For TssB, losing its encoding gene resulted in an attenuated trend toward competition, while the difference was not significant. This may be due to the special structure of TssB, which encloses the Hcp tube that has less of an effect on direct virulence.
To verify the virulence of the three T6SS core genes beyond bacterial killing, we infected G. mellonella with various mutants. G. mellonella is the caterpillar of the greater wax moth and has been widely used to study the virulence mechanisms of several human pathogens, including A. baumannii (Jander et al., 2000; Peleg et al., 2009; Harding et al., 2012; Iwashkiw et al., 2012; Heindorf et al., 2014; Jacobs et al., 2014; Tsai et al., 2016). The innate immune system of G. mellonella shares a high degree of homology with that of other mammals. Consistent with our in vitro results, TssD and TssM contributed to the virulence of A. baumannii, and the loss of these two components dramatically increased the survival of the infected G. mellonella. Nevertheless, no significant difference was observed between the tssB-deletion mutant and the expressing strains. This demonstrated that TssB, which forms a helical sheath with TssC around the Hcp tube, plays a less important role in host colonization.
Consequently, we determined the effects of tssB, tssD, and tssM in terms of drug resistance and virulence in the sensitive clinical strain AB795639. However, additional studies are needed to confirm the roles of the three T6SS core genes in A. baumannii due to strain specificity. In addition, further research will be required to explore the roles of other T6SS core genes, as well as to determine the mechanisms of action and regulation of tssB, tssD, and tssM.
5 Conclusion
The prevalence of T6SS is high among clinical A. baumannii isolates. This study explored the finding indicating there was a high correlation between T6SS carriage and multidrug resistance in A. baumannii. T6SS may help bacteria to demonstrate increased drug resistance by enhancing horizontal gene transfer among bacteria (Dong et al., 2022). More interestingly, the existence of T6SS could contribute to intrinsic resistance to specific drugs such as gentamicin, ciprofloxacin, norfloxacin, ofloxacin, tetracycline, doxycycline, polymyxin B, and furazolidone.
Furthermore, we evaluated the roles of three T6SS core components, TssB, TssD (Hcp), and TssM, in terms of biofilm formation, bacterial competition, normal human serum resistance, and host colonization. TssM was the most important factor that participated in all of these virulence phenomena. This was followed by TssD, which was negative for biofilm formation. TssB has less of an effect on either biofilm formation or bacterial virulence.
This information may provide potential therapeutic and vaccine targets to control A. baumannii infections. Further studies are warranted to explore the roles of other T6SS core genes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Author contributions
PL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. SZ: Methodology, Writing – review & editing. JW: Methodology, Writing – review & editing. MMA: Methodology, Writing – review & editing. KL: Methodology, Writing – review & editing. SL: Methodology, Writing – review & editing. PM: Resources, Writing – review & editing. XW: Resources, Writing – review & editing. HL: Methodology, Writing – review & editing. HT: Methodology, Writing – review & editing. BH: Project administration, Supervision, Writing – review & editing. JL: Funding acquisition, Writing – review & editing. SH: Funding acquisition, Writing – review & editing. LH: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the National Natural Science Foundation of China (81702043), the Natural Science Foundation of Shaanxi Province (2024JC-YBMS-732) the Fundamental Research Funds for the Central Universities (xzy012023014), the “Basic-Clinical” Fusion Innovation Project of Xi’an Jiaotong University (YXJLRH2022020), the Clinical Research Award of the First Affiliated Hospital of Xi’an Jiaotong University (XJT1AF-CRF-2019-015), and the Key Research and Development Program of Shaanxi Province (2017SF-198).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1297818/full#supplementary-material
Supplementary Figure 1 | Comparative analysis of the T6SS gene cluster from the A. baumannii genomes collected from Genbank.
Supplementary Figure 2 | Phylogeny of three T6SS core genes tssB (A), tssD (B) and tssM (C) from the A. baumannii genomes collected from Genbank were constructed by UPGMA model in MEGA, with the Bootstrap replications as 1000.
Supplementary Figure 3 | The sequence alignment of tssB gene from the A. baumannii genomes collected from Genbank.
Supplementary Figure 4 | The sequence alignment of tssD gene from the A. baumannii genomes collected from Genbank.
Supplementary Figure 5 | The sequence alignment of tssM gene from the A. baumannii genomes collected from Genbank.
Supplementary Figure 6 | Representative Gram-stain results of AB795639, the deletion and complementary mutants of three T6SS core genes. Scale bars: 100 pixels.
References
AlAmri, A. M., AlQurayan, A. M., Sebastian, T., AlNimr, A. M. (2020). Molecular surveillance of multidrug-resistant acinetobacter baumannii. Curr. Microbiol. 77 (3), 335–342. doi: 10.1007/s00284-019-01836-z
Alcoforado Diniz, J., Liu, Y. C., Coulthurst, S. J. (2015). Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cell Microbiol. 17 (12), 1742–1751. doi: 10.1111/cmi.12532
ANRESIS (2022). Swiss antibiotic resistance report 2022. Usage of antibiotics and occurrence of antibiotic resistance in Switzerland (Switzerland: Federal Office of Public Health and Federal Food Safety and Veterinary Office).
Arzanlou, M., Chai, W. C., Venter, H. (2017). Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem. 61 (1), 49–59. doi: 10.1042/EBC20160063
Aschtgen, M. S., Bernard, C. S., De Bentzmann, S., Lloubès, R., Cascales, E. (2008). SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol 190 (22), 7523–7531. doi: 10.1128/JB.00945-08
Basler, M., Ho, B. T., Mekalanos, J. J. (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152 (4), 884–894. doi: 10.1016/j.cell.2013.01.042
Carruthers, M. D., Nicholson, P. A., Tracy, E. N., Munson, R. S., Jr (2013). Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PloS One 8 (3), 19. doi: 10.1371/annotation/7aa1688c-56c8-46ca-82ea-f86697f3c4fe
Christaki, E., Marcou, M., Tofarides, A. (2020). Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J. Mol. Evol. 88 (1), 26–40. doi: 10.1007/s00239-019-09914-3
CLSI (2018). Performance standards for antimicrobial susceptibility testing (Wayne, PA, USA: Clinical and Laboratory Standards Institute).
Di Domenico, E. G., Farulla, I., Prignano, G., Gallo, M. T., Vespaziani, M., Cavallo, I., et al. (2017). Biofilm is a major virulence determinant in bacterial colonization of chronic skin ulcers independently from the multidrug resistant phenotype. Int. J. Mol. Sci. 18 (5), 1077. doi: 10.3390/ijms18051077
Dong, J. F., Liu, C. W., Wang, P., Li, L., Zou, Q. H. (2022). The type VI secretion system in Acinetobacter baumannii clinical isolates and its roles in antimicrobial resistance acquisition. Microb. Pathog. 169 (105668), 8. doi: 10.1016/j.micpath.2022.105668
Elhosseiny, N. M., Attia, A. S. (2018). Acinetobacter: an emerging pathogen with a versatile secretome. Emerg. Microbes Infect. 7 (1), 018–0030. doi: 10.1038/s41426-018-0030-4
Elhosseiny, N. M., Elhezawy, N. B., Attia, A. S. (2019). Comparative proteomics analyses of Acinetobacter baumannii strains ATCC 17978 and AB5075 reveal the differential role of type II secretion system secretomes in lung colonization and ciprofloxacin resistance. Microb. Pathog. 128, 20–27. doi: 10.1016/j.micpath.2018.12.039
Fernández, L., Hancock, R. E. (2012). Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25 (4), 661–681. doi: 10.1128/CMR.00043-12
Fitzsimons, T. C., Lewis, J. M., Wright, A., Kleifeld, O., Schittenhelm, R. B., Powell, D., et al. (2018). Identification of novel acinetobacter baumannii type VI secretion system antibacterial effector and immunity pairs. Infect. Immun. 86 (8), 00297–00218. doi: 10.1128/IAI.00297-18
Gaddy, J. A., Tomaras, A. P., Actis, L. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77 (8), 3150–3160. doi: 10.1128/IAI.00096-09
Gerlach, R. G., Hensel, M. (2007). Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297 (6), 401–415. doi: 10.1016/j.ijmm.2007.03.017
Hachani, A., Wood, T. E., Filloux, A. (2016). Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 29, 81–93. doi: 10.1016/j.mib.2015.11.006
Han, L., Gao, Y., Liu, Y., Yao, S., Zhong, S., Zhang, S., et al. (2022). An outer membrane protein yiaD contributes to adaptive resistance of meropenem in acinetobacter baumannii. Microbiol. Spectr. 10 (2), 00173–00122. doi: 10.1128/spectrum.00173-22
Harding, C. M., Hennon, S. W., Feldman, M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16 (2), 91–102. doi: 10.1038/nrmicro.2017.148
Harding, C. M., Kinsella, R. L., Palmer, L. D., Skaar, E. P., Feldman, M. F. (2016). Medically relevant acinetobacter species require a type II secretion system and specific membrane-associated chaperones for the export of multiple substrates and full virulence. PloS Pathog. 12 (1), e1005391. doi: 10.1371/journal.ppat.1005391
Harding, C. M., Pulido, M. R., Di Venanzio, G., Kinsella, R. L., Webb, A. I., Scott, N. E., et al. (2017). Pathogenic Acinetobacter species have a functional type I secretion system and contact-dependent inhibition systems. J. Biol. Chem. 292 (22), 9075–9087. doi: 10.1074/jbc.M117.781575
Harding, C. R., Schroeder, G. N., Reynolds, S., Kosta, A., Collins, J. W., Mousnier, A., et al. (2012). Legionella pneumophila pathogenesis in the Galleria mellonella infection model. Infect. Immun. 80 (8), 2780–2790. doi: 10.1128/IAI.00510-12
Heindorf, M., Kadari, M., Heider, C., Skiebe, E., Wilharm, G. (2014). Impact of Acinetobacter baumannii superoxide dismutase on motility, virulence, oxidative stress resistance and susceptibility to antibiotics. PloS One 9 (7), e101033. doi: 10.1371/journal.pone.0101033
Hsieh, P. F., Lu, Y. R., Lin, T. L., Lai, L. Y., Wang, J. T. (2019). Klebsiella pneumoniae type VI secretion system contributes to bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization. J. Infect. Dis. 219 (4), 637–647. doi: 10.1093/infdis/jiy534
Iwashkiw, J. A., Seper, A., Weber, B. S., Scott, N. E., Vinogradov, E., Stratilo, C., et al. (2012). Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PloS Pathog. 8 (6), 7. doi: 10.1371/journal.ppat.1002758
Jacobs, A. C., Thompson, M. G., Black, C. C., Kessler, J. L., Clark, L. P., McQueary, C. N., et al. (2014). AB5075, a highly virulent isolate of acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5 (3), 01076–01014. doi: 10.1128/mBio.01076-14
Jander, G., Rahme, L. G., Ausubel, F. M. (2000). Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol 182 (13), 3843–3845. doi: 10.1128/JB.182.13.3843-3845.2000
Kim, S. W., Choi, C. H., Moon, D. C., Jin, J. S., Lee, J. H., Shin, J. H., et al. (2009). Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol. Lett. 301 (2), 224–231. doi: 10.1111/j.1574-6968.2009.01820.x
Kim, J., Lee, J. Y., Lee, H., Choi, J. Y., Kim, D. H., Wi, Y. M., et al. (2017). Microbiological features and clinical impact of the type VI secretion system (T6SS) in Acinetobacter baumannii isolates causing bacteremia. Virulence 8 (7), 1378–1389. doi: 10.1080/21505594.2017.1323164
Kwon, H. I., Kim, S., Oh, M. H., Na, S. H., Kim, Y. J., Jeon, Y. H., et al. (2017). Outer membrane protein A contributes to antimicrobial resistance of Acinetobacter baumannii through the OmpA-like domain. J. Antimicrob. Chemother. 72 (11), 3012–3015. doi: 10.1093/jac/dkx257
Leiman, P. G., Basler, M., Ramagopal, U. A., Bonanno, J. B., Sauder, J. M., Pukatzki, S., et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106 (11), 4154–4159. doi: 10.1073/pnas.0813360106
Li, P., Zhang, S., Wang, J., Al-Shamiri, M. M., Han, B., Chen, Y., et al. (2023). Uncovering the secretion systems of acinetobacter baumannii: structures and functions in pathogenicity and antibiotic resistance. Antibiotics 12 (2), 195. doi: 10.3390/antibiotics12020195
Liao, W., Huang, H. H., Huang, Q. S., Fang-Ling, D., Dan Wei, D., La-Gen, W., et al. (2022). Distribution of type VI secretion system (T6SS) in clinical Klebsiella pneumoniae strains from a Chinese hospital and its potential relationship with virulence and drug resistance. Microb. Pathog. 162 (105085), 9. doi: 10.1016/j.micpath.2021.105085
Liao, C. H., Sheng, W. H., Chen, Y. C., Hung, C. C., Wang, J. T., Chang, S. C. (2007). Predictive value of the serum bactericidal test for mortality in patients infected with multidrug-resistant Acinetobacter baumannii. J. Infect. 55 (2), 149–157. doi: 10.1016/j.jinf.2007.01.015
Ma, L. S., Narberhaus, F., Lai, E. M. (2012). IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J. Biol. Chem. 287 (19), 15610–15621. doi: 10.1074/jbc.M111.301630
McConnell, M. J., Actis, L., Pachón, J. (2013). Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 37 (2), 130–155. doi: 10.1111/j.1574-6976.2012.00344.x
Nguyen, V. S., Douzi, B., Durand, E., Roussel, A., Cascales, E., Cambillau, C. (2018). Towards a complete structural deciphering of Type VI secretion system. Curr. Opin. Struct. Biol. 49, 77–84. doi: 10.1016/j.sbi.2018.01.007
Park, Y. J., Lacourse, K. D., Cambillau, C., DiMaio, F., Mougous, J. D., Veesler, D. (2018). Structure of the type VI secretion system TssK-TssF-TssG baseplate subcomplex revealed by cryo-electron microscopy. Nat. Commun. 9 (1), 018–07796. doi: 10.1038/s41467-018-07796-5
Peleg, A. Y., Jara, S., Monga, D., Eliopoulos, G. M., Moellering, R. C., Jr., Mylonakis, E. (2009). Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob. Agents Chemother. 53 (6), 2605–2609. doi: 10.1128/AAC.01533-08
Pogue, J. M., Kaye, K. S., Cohen, D. A., Marchaim, D. (2015). Appropriate antimicrobial therapy in the era of multidrug-resistant human pathogens. Clin. Microbiol. Infect. 21 (4), 302–312. doi: 10.1016/j.cmi.2014.12.025
Ramirez, M. S., Bonomo, R. A., Tolmasky, M. E. (2020). Carbapenemases: Transforming Acinetobacter baumannii into a Yet More Dangerous Menace. Biomolecules 10 (5), 720. doi: 10.3390/biom10050720
Repizo, G. D. (2017). Prevalence of Acinetobacter baumannii strains expressing the Type 6 secretion system in patients with bacteremia. Virulence 8 (7), e0138265, 1099–1101. doi: 10.1080/21505594.2017.1346768
Repizo, G. D., Gagné, S., Foucault-Grunenwald, M. L., Borges, V., Charpentier, X., Limansky, A. S., et al. (2015). Differential role of the T6SS in acinetobacter baumannii virulence. PloS One 10 (9), e0138265. doi: 10.1371/journal.pone.0138265
Ruiz, F. M., Santillana, E., Spínola-Amilibia, M., Torreira, E., Culebras, E., Romero, A. (2015). Crystal structure of hcp from acinetobacter baumannii: A component of the type VI secretion system. PloS One 10 (6), e0129691. doi: 10.1371/journal.pone.0129691
Russell, A. B., Peterson, S. B., Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12 (2), 137–148. doi: 10.1038/nrmicro3185
Ryu, S. Y., Baek, W. K., Kim, H. A. (2017). Association of biofilm production with colonization among clinical isolates of Acinetobacter baumannii. Korean J. Intern. Med. 32 (2), 345–351. doi: 10.3904/kjim.2015.287
Shin, J., Ko, K. S. (2015). Effect of plasmids harbouring blaCTX-M on the virulence and fitness of Escherichia coli ST131 isolates. Int. J. Antimicrob. Agents 46 (2), 214–218. doi: 10.1016/j.ijantimicag.2015.04.012
Silverman, J. M., Agnello, D. M., Zheng, H., Andrews, B. T., Li, M., Catalano, C. E., et al. (2013). Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol. Cell 51 (5), 584–593. doi: 10.1016/j.molcel.2013.07.025
Silverman, J. M., Brunet, Y. R., Cascales, E., Mougous, J. D. (2012). Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66, 453–472. doi: 10.1146/annurev-micro-121809-151619
Skariyachan, S., Taskeen, N., Ganta, M., Venkata Krishna, B. (2019). Recent perspectives on the virulent factors and treatment options for multidrug-resistant Acinetobacter baumannii. Crit. Rev. Microbiol. 45 (3), 315–333. doi: 10.1080/1040841X.2019.1600472
Tsai, C. J., Loh, J. M., Proft, T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7 (3), 214–229. doi: 10.1080/21505594.2015.1135289
Tucker, A. T., Nowicki, E. M., Boll, J. M., Knauf, G. A., Burdis, N. C., Trent, M. S., et al. (2014). Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5 (4), 01313–01314. doi: 10.1128/mBio.01313-14
Wang, J., Zhou, Z., He, F., Ruan, Z., Jiang, Y., Hua, X., et al. (2018). The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PloS One 13 (2), e0192288. doi: 10.1371/journal.pone.0192288
Weber, B. S., Hennon, S. W., Wright, M. S., Scott, N. E., de Berardinis, V., Foster, L. J., et al. (2016). Genetic dissection of the type VI secretion system in acinetobacter and identification of a novel peptidoglycan hydrolase, tagX, required for its biogenesis. mBio 7 (5), 01253–01216. doi: 10.1128/mBio.01253-16
Weber, B. S., Kinsella, R. L., Harding, C. M., Feldman, M. F. (2017). The secrets of acinetobacter secretion. Trends Microbiol. 25 (7), 532–545. doi: 10.1016/j.tim.2017.01.005
Weber, B. S., Ly, P. M., Irwin, J. N., Pukatzki, S., Feldman, M. F. (2015). A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc. Natl. Acad. Sci. U.S.A. 112 (30), 9442–9447. doi: 10.1073/pnas.1502966112
Weber, B. S., Miyata, S. T., Iwashkiw, J. A., Mortensen, B. L., Skaar, E. P., Pukatzki, S., et al. (2013). Genomic and functional analysis of the type VI secretion system in Acinetobacter. PloS One 8 (1), 24. doi: 10.1371/journal.pone.0055142
Wiegand, I., Hilpert, K., Hancock, R. E. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3 (2), 163–175. doi: 10.1038/nprot.2007.521
Wong, D., Nielsen, T. B., Bonomo, R. A., Pantapalangkoor, P., Luna, B., Spellberg, B. (2017). Clinical and pathophysiological overview of acinetobacter infections: a century of challenges. Clin. Microbiol. Rev. 30 (1), 409–447. doi: 10.1128/CMR.00058-16
Zhang, S., Mi, P., Wang, J., Li, P., Luo, K., Liu, S., et al. (2023). The optimized carbapenem inactivation method for objective and accurate detection of carbapenemase-producing Acinetobacter baumannii. Front. Microbiol. 14 (1185450). doi: 10.3389/fmicb.2023.1185450
Zhang, L., Xu, J., Xu, J., Zhang, H., He, L., Feng, J. (2014). TssB is essential for virulence and required for type VI secretion system in Ralstonia solanacearum. Microb. Pathog. 74, 1–7. doi: 10.1016/j.micpath.2014.06.006
Keywords: Acinetobacter baumannii, T6SS, TssB, TssD, TSSM, antimicrobial resistance, virulence
Citation: Li P, Zhang S, Wang J, Al-Shamiri MM, Luo K, Liu S, Mi P, Wu X, Liu H, Tian H, Han B, Lei J, Han S and Han L (2024) The role of type VI secretion system genes in antibiotic resistance and virulence in Acinetobacter baumannii clinical isolates. Front. Cell. Infect. Microbiol. 14:1297818. doi: 10.3389/fcimb.2024.1297818
Received: 20 September 2023; Accepted: 23 January 2024;
Published: 07 February 2024.
Edited by:
Brankica Filipić, University of Belgrade, SerbiaReviewed by:
Roberto Rosales-Reyes, National Autonomous University of Mexico, MexicoMichelle Qiu Carter, United States Department of Agriculture (USDA), United States
Copyright © 2024 Li, Zhang, Wang, Al-Shamiri, Luo, Liu, Mi, Wu, Liu, Tian, Han, Lei, Han and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Han, lei.han@xjtu.edu.cn
 Pu Li1
Pu Li1  Mona Mohamed Al-Shamiri
Mona Mohamed Al-Shamiri Huohuan Tian
Huohuan Tian Shaoshan Han
Shaoshan Han Lei Han
Lei Han