Droplet digital PCR as alternative to microbiological culture for Mycobacterium tuberculosis complex detection in bovine lymph node tissue samples
- 1Department of Anatomy and Comparative Pathology and Toxicology, Pathology and Immunology Group (UCO-PIG), Unidad de Investigación Competitiva (UIC) Zoonosis y Enfermedades Emergentes ENZOEM, University of Córdoba, Córdoba, Spain
- 2Department of Animal Health, Unidad de Investigación Competitiva (UIC) Zoonosis y Enfermedades Emergentes (ENZOEM), University of Córdoba, University of Córdoba, Córdoba, Spain
- 3UK Health Security Agency (UKHSA), Salisbury, United Kingdom
Introduction: Bovine tuberculosis (bTB) caused by Mycobacterium tuberculosis complex (MTC) remains a significant concern for public health. Direct real-time PCR and droplet digital PCR (ddPCR) are proposed as alternative tools to enhance diagnostic precision and efficiency. This study aims to assess the diagnostic performance of a ddPCR assay targeting IS6110 for the detection of MTC DNA in both microbiological culture and fresh lymph node (LN) tissue samples obtained from cattle, in comparison with the established reference standard, the microbiological culture followed by real-time PCR.
Methods: The fresh LNs (N=100) were collected each from a different cattle carcass at the slaughterhouse. The limit of detection of ddPCR-IS6110 was set to 101 copies per 20 μl reaction.
Results: DdPCR-IS6110 detected 44 out of 49 reference-standard positive samples and yielded negative results in 47 out of 51 reference-standard negative samples, resulting in adjusted sensitivity (Se) and specificity (Sp) of 90.76% [95% confidence interval (CI): 82.58 - 98.96%)], and 100% (95% CI: 100%) respectively. The estimated adjusted false negative rate (FNR) was 9.23% (95% CI: 1.04 - 17.42%) and the false positive rate (FPR) was 0% (95% CI: 0%). When directly applied from fresh bovine LN tissues, ddPCR-IS6110 identified 47 out of 49 reference-standard positive samples as ddPCR-IS6110-positive and 42 out of 51 reference-standard negative samples as ddPCR-IS6110-negative, resulting in adjusted Se and Sp values of 94.80% [95% (CI): 88.52 - 100%] and 100% (95% CI: 100%), respectively. The adjusted FNR was 5.20% (95% CI: 0 - 11.50%) and the FPR was 0% (95% CI: 0%). Noteworthy, ddPCR-IS6110 disclosed as positive 9 samples negative to reference-standard.
Discussion: DdPCR-IS6110 proved to be a rapid, highly sensitive, and specific diagnostic tool as an alternative to reference-standard method.
1 Introduction
Bovine tuberculosis (bTB) is caused by bacteria belonging to Mycobacterium tuberculosis complex (MTC), mainly M. bovis and M. caprae (Aranaz et al., 2004; Domingo et al., 2014). bTB is still a zoonoses of major concern for public health, especially in developing countries (Dean et al., 2018). In the European Union (EU), even though the eradication is the main goal, this disease is still present in dairy and beef herds (MAPA, 2022). Current European approved surveillance systems are based on detection of a specific cellular immune reaction by single or comparative intradermal tuberculin testing (SIT or SCIT) and interferon gamma release assays (IGRA), followed by compulsory slaughter of reactor animals as well as post-mortem confirmation. Furthermore, this program includes abattoir surveillance for undetected bTB-infected animals, regular retesting and culling of infected animals and restrictions on the movement of livestock to prevent introduction of infected animals (EU, 2023).
These approaches have simplified the diagnosis of MTC-infected cattle at the early stage of the disease (de la Rua-Domenech et al., 2006), therefore, animals with clinical signs or gross tuberculosis-like lesions (TBL) are lack or rarely found at the slaughterhouse (de la Rua-Domenech et al., 2006; Ramos et al., 2015; Pozo et al., 2021). This success of surveillance systems has challenged the traditional examination of atypical or enlarged lymph nodes (LNs) or parenchymatous organs with gross TBLs, and/or culture of MTC in primary isolation medium followed by real-time PCR. In this scenario, several factors could play a role in hindering the eradication of bTB. One of the most relevant drawbacks is the poor diagnostic performance reported for the current diagnostic tools. As a consequence, truly infected animals are misclassified as bTB-free, which contribute to maintaining the chain of infection on the farm, but also in sharing pasture areas.
In order to make the diagnostic and confirmation procedure for bTB more reliable and swifter, several diagnostic tools have been proposed as alternatives to the reference standard (microbiological culture followed by confirmation by real-time PCR), which is considered an imperfect reference technique taking up to three months to obtain a confirmatory result (Courcoul et al., 2014; Lorente-Leal et al., 2021; Sánchez-Carvajal et al., 2021). Direct real-time PCR from tissue samples has been reported to be a potential first-line technique for the detection of MTC species in animal tissues worldwide (Courcoul et al., 2014; Lorente-Leal et al., 2019; Wang et al., 2019; Lorente-Leal et al., 2021; Sánchez-Carvajal et al., 2021). Nevertheless, although direct real-time PCR seems to be a simple, rapid and robust alternative to microbiological culture, PCR results could be affected mainly by the characteristics of the lesion (necrosis, calcification or fibrosis), a low mycobacterial load, the DNA isolation procedure and the presence of inhibitors (Lorente-Leal et al., 2019; Sánchez-Carvajal et al., 2021).
These limiting factors could be overcome by other molecular tools such as droplet digital PCR (ddPCR). The ddPCR is an emerging PCR assay, based on water–oil emulsion droplet technology, which have been described in several medical fields in recent years, including diagnosis of several infectious pathogens, DNA methylation determination, gene expression, and gene mutation analysis (Strain et al., 2013; Lillsunde Larsson and Helenius, 2017; Guil-Luna et al., 2023). Each sample is partitioned into approximately 20,000 droplets before being subjected to the PCR and, therefore, each droplet could contain one target molecule or none. This is a substantial advantage compared to real-time PCR, since ddPCR has been reported to be less sensitive to inhibitors due to sample partitioning (Dingle et al., 2013; Yang et al., 2014). Another key argument to bear in mind is that ddPCR is more sensitive and accurate than real-time PCR, especially in the case of low-copy acid nucleic (Devonshire et al., 2015; Yang et al., 2017; Nyaruaba et al., 2020). This technology has been already reported for the detection of Mycobacterium tuberculosis in human samples (Devonshire et al., 2015; Yang et al., 2017; Cao et al., 2020), or recently, for the detection of Mycobacterium avium subsp. paratuberculosis DNA in whole-blood and faecal samples from cattle (Badia-Bringué et al., 2022). However, ddPCR capability to detect MTC in fresh bovine tissue samples has not been yet evaluated. Thus, the primary aim of this research was to assess the diagnostic performance of a ddPCR assay targeting IS6110 for the rapid and sensitive detection of MTC DNA in both microbiological culture and fresh LN tissue samples obtained from cattle, in comparison with the established reference standard.
2 Materials and methods
2.1 Samples selection and processing
This study was part of a larger project focused on developing rapid and accurate diagnostic tools in the current framework of bTB surveillance and control programs. LN tissue samples (N=100) were collected each from a different cattle carcass at the slaughterhouse from 2018 to 2019 in the context of Spanish bTB eradication program. All samples were collected during routine post-mortem veterinary examination within an official context and according with national and European regulations. No purpose killing of animals was performed for this study, so no ethical or farmer’s consent approval was required.
LNs were independently sliced to confirm the presence of visible TBLs (N=19) or no visible lesions (NVLs) (N=81) and fixed in 10% neutral-buffered formalin for the histopathological analysis. Each LN was then individually homogenized using a tissue homogenizer (Fisherbrand, Fisher Scientific, Madrid, Spain) to obtain a uniform mixture. Tissue homogenate was divided into paired samples that were used for DNA isolation and selective microbiological culture.
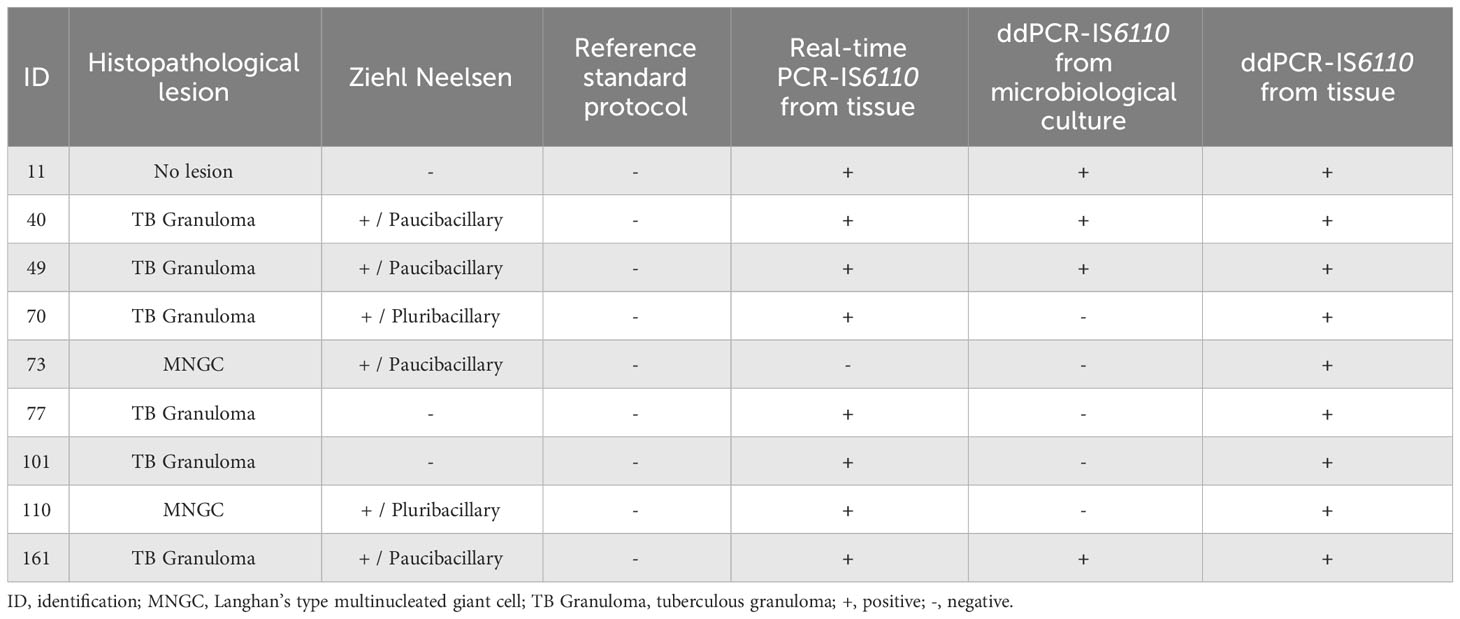
For histopathological evaluation, formalin fixed LNs were processed and embedded in paraffin following standard procedures. Four-micron sections were stained with haematoxylin-eosin (H&E) and Ziehl-Neelsen (ZN). Histopathological findings were classified as TBLs for those samples with a tuberculous granuloma, pyogranuloma, or scattered Langhans-type multinucleated giant cells (MNGCs), or as no histopathological lesion (NHLs), for the tissue with normal histological characteristics and no lesion compatible with TBL (Larenas-Muñoz et al., 2022). In addition, Ziehl-Neelsen (ZN) technique was performed to detect acid-fast bacilli (AFB). A sample was considered positive for ZN when one or more AFB were found in at least one high-power field magnification (HPF, 100x). The lesions were classified as paucibacillary if it was observed with 1 to 10 AFB bacilli, or pluribacillary if ≥ 11 AFB were observed per HPF (Larenas-Muñoz et al., 2022).
For the reference standard (microbiological culture followed by real-time PCR-IS6110), tissue homogenates were decontaminated with an equal volume of 0.75% (w/v: 1/1) hexadecyl pyridinium chloride solution in agitation for 30 min. Samples were centrifuged for 30 min at 1,500 × g. The pellets were collected with swabs and cultured in liquid media (MGIT™ 960, Becton Dickinson, Madrid, Spain) using an automatized BD Bacter™ MGIT™ System (Becton Dickinson) (Corner and Trajstman, 1988). DNA extraction was performed using the MagMAX Total Nucleic Acid Isolation Kit according to the manufacturer’s instructions (Thermo Fisher Scientific, Lissieu, France). DNA was eluted in 50 μl. Then, cultures were considered positive when isolates were confirmed as MTC using real-time PCR (Thierry et al., 1990). The cut-off value of real-time PCR-IS6110 assay was set at 10 to 100 genomic equivalents, and the cut-off set at Cq ≤ 38 (10 to 100 genomic equivalents/15 μL reaction mixture) (Sánchez-Carvajal et al., 2021).
2.2 DNA isolation from microbiological culture and LNs
Genomic DNA isolation was conducted from tissue homogenate according to Vera-Salmoral et al., 2023 with several modifications using NucleoSpin Tissue Kit® (Macherey-Nagel, Düren, Germany). In brief, 1 mL of homogenized tissue was centrifuged during 5 min at 9,000 g. The supernatant was discarded, and the resulting tissue pellet was added in a tube together with 250 µl of Sample Buffer T1, 150 mg of 0.5-mm glass beads and 50 mg of 0.1-mm glass beads. Then, samples were subjected to mechanical disruption (SI™ Disruptor Genie™, Scientific Industries, New York, USA) (2,850 rpm/50 Hz/20 min). After an overnight enzymatic digestion at 56°C with 30 µl proteinase K in a thermo-shaker (600 rpm/12 h), a new mechanical disruption step was conducted. Samples were centrifuged 2 min at 9,000 g, and pellets were again subjected to the steps described above. Then, samples were mixed with 200 µl of buffer T3, incubating the mixture for 10 min at 70°C. The lysate was transferred to a silica-based nucleic acid purification column and managed according to manufacturer’s instructions. Isolated DNA samples were stored at −20°C until used in downstream PCR assays. Positive and negative extraction controls were included.
2.3 Primers and probe targeting IS6110
Specific primers and probe were based on the homology region of the partial insertion sequence 6110 (IS6110), a repetitive mobile element specific for all the pathogens belonging to MTC widely used to diagnose and genotype this pathogen. The fluorogenic IS6110-probe was labelled with a fluorescent reporter dye [6-carboxyfluorescein (FAM)] at the 5′-end and a 3′-Black Hole Quencher 1 (BHQ1). Primers and probe used for real-time PCR and ddPCR are listed in Table 1 (Sánchez-Carvajal et al., 2021; Lorente-Leal et al., 2021).

Table 1 Sequences of MTC specific primers and TaqMan probe targeting IS6110 for real-time PCR and droplet digital PCR (ddPCR) assays.
2.4 Real-time PCR targeting IS6110
QuantiFast® Pathogen PCR + IC Kit (QIAGEN, Hilden, Germany) was used to conduct the real-time PCR-IS6110 evaluating each sample in duplicate in the MyiQ™2 Two-Color real-time PCR Detection System (Bio-Rad, Hercules, Ca, USA) under the following cycling conditions: 95°C for 5 min to activate the DNA polymerase followed by 42 amplification cycles that consisted of a denaturation step at 95°C for 15 s, an annealing-extension step at 60°C for 30 s. Following manufacturer’s guidelines, an exogenous inhibition heterologous control (internal amplification control, IAC) supplied with the kit was included. An inter-run calibrator with a known quantification cycle (Cq) value of 32 was introduced in each assay to self-control intra-assay. Complete inhibition of amplification was considered when IAC did not amplify and partial inhibition when it showed a Cq > 33. The analytical sensitivity or limit of detection (LOD) was estimated for the proposed primers and probes. LOD is defined as the lowest concentration in which 95% of replicates were positive, according to the Clinical and Laboratory Standard Institute guidelines. A serial 10-fold dilution series of M. bovis genomic DNA with known quantities ranging from 106 to 100 were used. The reactions were performed in triplicate for each dilution in three different assays. Thus, the LOD was determined to be ranging from 10 to 100 genomic equivalents, and the cut-off was established at Cq < 38 (Sánchez-Carvajal et al., 2021).
2.5 ddPCR targeting IS6110 for MTC detection in microbiological culture and LNs
For ddPCR assay targeting IS6110, QX200™ ddPCR™ Supermix for probe (No dUTP) (Bio-Rad, Hercules, CA, USA) was used according to Bio-Rad ddPCR system guidelines. Each sample was evaluated in duplicate in a reaction mix with a final volume of 21 μl as follows: 10.5 μl of 1x ddPCR Supermix for probe (No dUTP), 1.7 μl of IS6110-forward (900 nM), 0.85 μl of IS6110-reverse (600 nM), 0.65 μl of IS6110-probe (FAM-labelled, 200 mM), 3 μl of template, and 4.3 μl of nuclease-free water. It is important to mention that several protocols for ddPCR recommend performing a restriction digestion of DNA samples outside the amplicon in order to make the template more accessible reducing sample viscosity. Nevertheless, we decided to not use restriction enzymes due to the extraction protocol herein reported got an efficient reduction of host DNA ranging from 50 – 100 ng/μl. Afterwards, the droplets were generated on the QX200 Droplet Generator (Bio-Rad, USA) using 70 μl of droplet generation oil for Probes® (Bio-Rad, USA) dispatched into the bottom of the oil wells of the DG8™ Cartridge droplet generator (Bio-rad, USA) according to the manufacturer’s instructions. The droplets were carefully transferred to a specific 96-well ddPCR reaction plate (Bio-Rad, USA) using a RAININ Pipet-Lite Multi Pipette (Mettler Toledo, Columbus, Ohio, USA). After heat sealing by PX1™ PCR Plate Sealer (Bio-Rad, USA) at 180°C for 5 s, amplifications were run in the C1000 Touch thermal cycler (Bio-Rad, Hercules, CA, USA) under the following cycling conditions: 95°C for 10 min, followed by 40 cycles of 94°C for 30 s and 60°C (annealing/extension) for 1 min, and finally 98°C for 10 min. The temperature ramping rate was set at 2°C/s. Thereafter, the droplets were stored in darkness at 4°C for 12 h.
2.6 Limit of detection and limit of blank of ddPCR targeting IS6110 assay
The limit of detection (LOD) was determined to be the lowest concentration of IS6110 copies at which detection is possible (Armbruster and Pry, 2008). LOD for ddPCR-IS6110 was determined by measuring three concentrations around LOD. Reactions were run in triplicates for each concentration (M. bovis genomic DNA with known quantities ranging from 104 to 100), and LOD was defined as the lowest concentration in which 95% of replicates were positive according to the Clinical and Laboratory Standards Institute guidelines. The limit of blank (LOB) was defined as the highest number of IS6110 copies found in 12 blank samples (Armbruster and Pry, 2008).
2.7 ddPCR targeting IS6110 data analysis
The QX200 Droplet Reader (Bio-Rad, USA) was used to read and count the fluorescent positive and negative droplets. Then, the data were analyzed using the QuantaSoft™ Analysis Pro software (version 1.0.596) (Bio-Rad, USA). Data from samples with 12,000 – 16,000 droplets were used for concentration calculations. Samples with a low number of droplets (< 10,000) were excluded from the analysis. According with the results for LOB, those samples with fewer than two positive droplets were considered “MTC-negative”, in contrast, samples were considered as “MTC-positive” when more than two droplets were found (Whale et al., 2020). Thus, the cut-off value of ddPCR-IS6110 assay was set at 3 positive droplets in 20 μL reaction mixture.
2.8 Statistical analysis
The diagnostic performance of ddPCR targeting IS6110 was evaluated for the detection of MTC in microbiological culture and fresh LN tissue samples. The diagnostic accuracy was compared with microbiological culture as the refence standard, considered as an imperfect reference technique for bTB diagnosis (Corner and Trajstman, 1988; Courcoul et al., 2014; Pucken et al., 2017). The adjusted sensitivity and specificity, false positive rate, false negative rate, positive likelihood ratio (PLR), and negative likelihood ratio (NLR) were calculated using Epidat 3.1 software. The PLR and NLR were interpreted according to the criteria published by Sackett et al. (2001), where a PLR > 10 or NLR < 0.1 indicates a technique of high diagnostic value that can discriminate between healthy and diseased animals, 5 < PLR ≤ 10 or NLR = 0.1-0.2 indicates a technique involving moderate changes in probability, 2 < PLR ≤ 5 or 0.2 < NLR ≤ 0.5 indicates a technique involving small changes in probability, and PLR ≤ 2 and NLR > 0.5 indicates rarely discernible changes. Finally, agreement between microbiological culture and ddPCR from culture, microbiological culture and ddPCR from fresh tissue, and real-time PCR and ddPCR both from fresh tissue was assessed using Cohen’s kappa coefficient (κ): κ = 0 indicated no agreement; 0.01 ≤ κ ≤ 0.20, slight agreement; 0.21 ≤ κ ≤ 0.40, fair agreement; 0.41 ≤ κ ≤ 0.60, moderate agreement; 0.61 ≤ κ ≤ 0.80, substantial agreement; and, 0.81 ≤ κ ≤ 1.00, almost perfect agreement (McHugh, 2012) (WinEpi software 2.0, Faculty of Veterinary Medicine, University of Zaragoza, Spain).
3 Results
3.1 Optimization of the ddPCR assay targeting IS6110 for DNA isolated from microbiological culture and homogenized fresh tissue LNs
In order to optimize the ddPCR-IS6110 assay, the first step was to determine an optimal annealing temperature, considered as one of the most critical parameters. Thus, we tested a range of annealing temperatures ranging from 56°C to 64°C. A total of 50 ng of MTC DNA isolated from a selective microbiological bacterial culture, 900 nM of forward and reverse primers together with 500 nM of probe were used in the assay. No restriction digestion of the DNA samples was performed. A negative template control (NTC) containing sterile water instead of DNA was included. We were able to detect the IS6110 specific region in all the assessed temperatures (Figure 1A), however, the annealing temperature of 60°C (E08) showed a higher amplitude between positive and negative droplets compared with other temperatures and resulted in less non-specific amplification (rain). No positive droplets were observed in NTC for any of the temperatures (Figure 1B). Therefore, an annealing temperature of 60°C (E08) (Figure 1A) was selected as ideal temperature for further experiments.
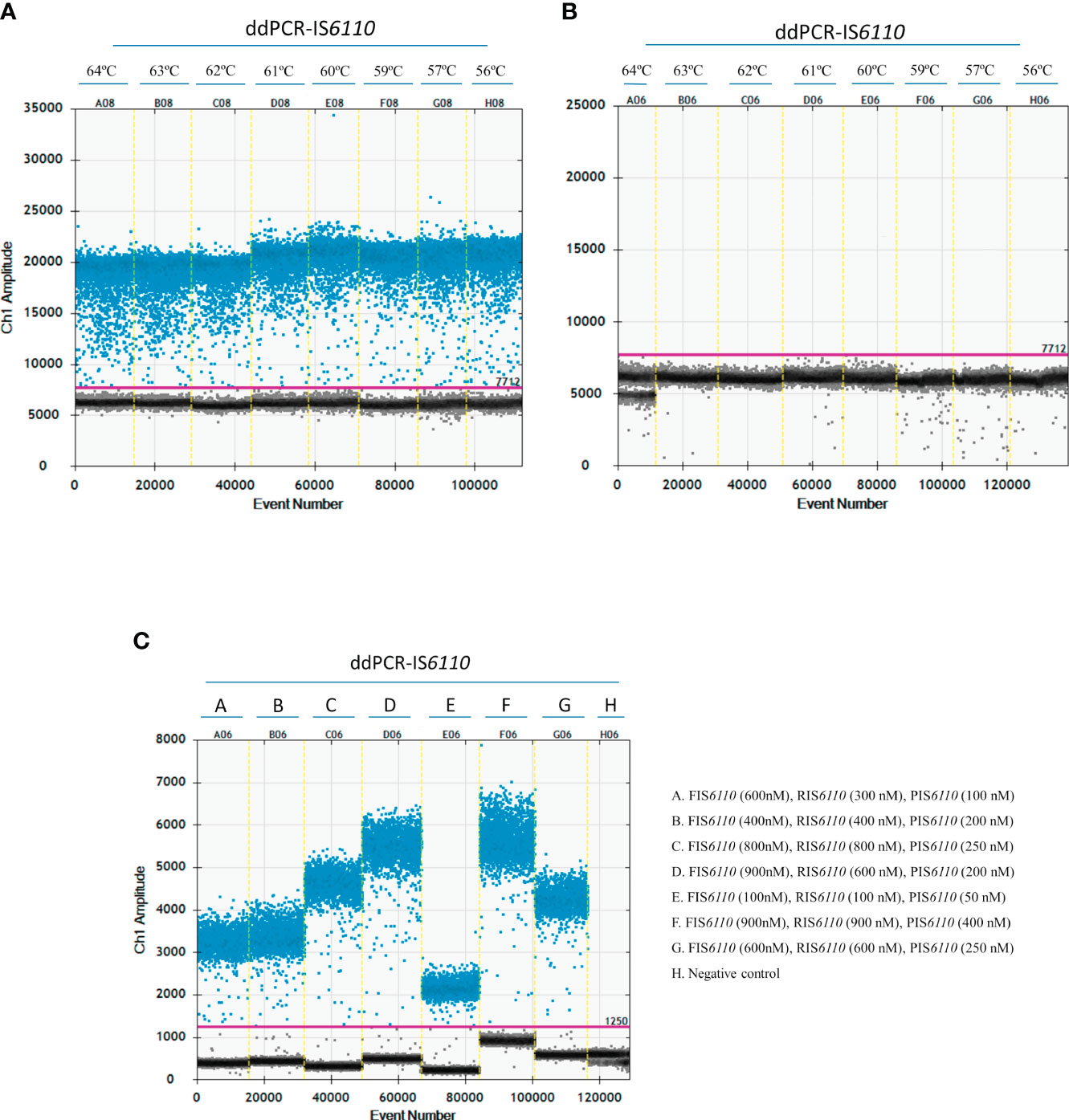
Figure 1 Optimization of the ddPCR-IS6110 assay using Mycobacterium tuberculosis complex (MTC) DNA isolated from LN tissue samples. The gradient of annealing temperatures (ranging from 64°C to 56°C) for a positive sample (A) and non-template control (NTC) (B) were plotted with positive (blue) and negative (grey) droplets. E08 plotted the optimal temperature of annealing and extension (60°C). (C) Descending concentrations of primers and probe. D06 plotted the optimal primers and probe concentration assay (F-IS6110 (900nM), R-IS6110 (600 nM), P-IS6110 (200 nM).
Next step was to determine the optimal primer and probe concentrations for ddPCR-IS6110 assay. Thus, five different concentrations of forward primer (100, 400, 600, 800 and 900 nM), reverse primer (100, 300, 400, 600, 800 and 900 nM) and probe (50, 100, 200, 250 and 400 nM) were tested (Figure 1C). Similarly, as above mentioned, a total of 50 ng of MTC DNA isolated from a selective bacterial culture were used in the assay, with no restriction digestion of DNA samples and inclusion of a NTC with sterile water instead of DNA. Figure 1C showed that the overall fluorescence amplitude of positive droplets increased with primers and probe concentrations. On the other hand, a much better amplitude between positive and negative droplets was observed when we used a concentration of 900 nM for forward, 600 nM for reverse and 200 nM for probe (Figure 1C, D06). This set up of primers and probes were used for further experiments.
In the case of microbiological culture, we decided to use a sample concentration of 50 ng of DNA according to the concentration and volume available after DNA extraction. In the case of DNA isolated from fresh LN tissue, we were able to test the effect of sample quantity. A ddPCR assay was run using different concentrations of DNA from two different LNs that were IS6110-positive by real-time PCR with different Cq values (500 ng, 250ng, 50 ng and 10 ng) (Figure 2A, Cq = 26; and Figure 2B, Cq = 34). As illustrated in Figure 2A, a good separation between positive and negative droplets was observed at the four DNA concentrations (500 ng, 250ng, 50 ng and 10 ng) for DNA isolated from LN tissue samples. Because Poisson statistics test required enough negative droplets to be applied and calculate DNA concentration, we decided to use 50 ng (C12) of DNA isolated from tissue samples in further ddPCR assays. Also, this concentration could be a good fit to avoid cross-contamination in the case of samples with a high concentration of bacterial DNA.
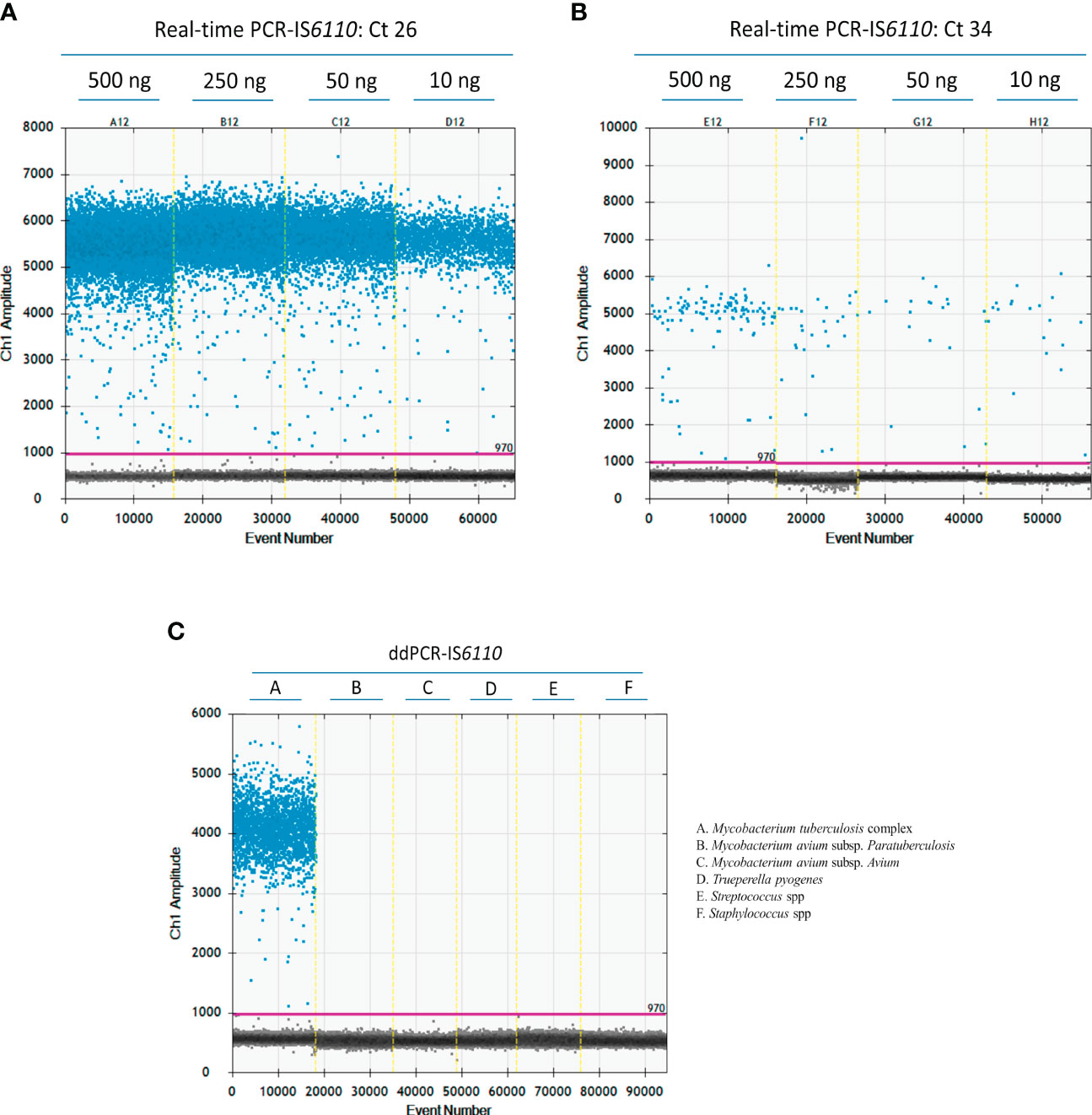
Figure 2 Optimization of the ddPCR-IS6110 assay using Mycobacterium tuberculosis complex (MTC) DNA isolated from LN tissue samples. Descending concentrations of DNA isolated from MTC-positive samples with a Cq value of 26 (A) or 34 (500 ng, 250 ng, 50 ng, and 10 ng) in duplicate (B). Analytical specificity of ddPCR-targeting IS6110 evaluating the most common bacterial agents found in tuberculosis like lesion (Mycobacterium avium subsp. paratuberculosis, Mycobacterium avium subsp. avium, Trueperella pyogenes, Streptococcus suis and Staphylococcus spp.) (C).
3.2 Analytical specificity
The analytical specificity of ddPCR-targeting IS6110 was tested against some of the most common microorganisms identified in TBL, such as Mycobacterium avium subsp. paratuberculosis, Mycobacterium avium subsp. avium, Trueperella pyogenes, Streptococcus suis, and Staphylococcus spp. (Cardoso-Toset et al., 2015). All these tests yielded negative results by ddPCR-IS6110, demonstrating the specificity of the primers and probe included in the study (Figure 2C).
3.3 Limit of blank and limit of detection
No positive droplets were found in 10 out of 12 blank samples (ddH2O instead of DNA sample), but one positive droplet per 20 μl reaction mix was detected in two of these blank samples. Accordingly, the limit of blank was set at 1 drop/20 μl reaction, and therefore, a sample was considered as “negative” when no more than two positive droplets were obtained.
To determine the LOD, 10-fold serial dilutions of M. bovis genomic DNA with known quantities ranging from 104 to 100 were used. The reactions were performed in triplicate for each dilution in three different assays. MTC IS6110 sequences were detected in 100% of 101 dilutions assayed; however, only 50% positivity was obtained at the level of 100. Thus, LOD of this ddPCR targeting IS6110 was set to 101 copies per 20 µl reaction.
3.4 Comparison of confirmatory IS6110 real time PCR and ddPCR from microbiological culture
As previously mentioned, microbiological culture positive samples need to be confirmed as MTC using real-time PCR (Thierry et al., 1990). Therefore, this part of the study compared both real-time PCR and ddPCR for the confirmation of culture positive samples. DNA was isolated from selective microbiological culture from 100 samples with gross TBL (N=19) or NVLs (N=81). Forty-nine out of 100 samples were tested as MTC-positive by real-time PCR (17 out of 19 with gross TBL) whereas 51 samples yielded a negative result and were classified as MTC-negative (Table 2). The Cq values for the real-time PCR targeting IS6110 ranged from 20.00 to 36.25 (average = 29.35). No partial or complete inhibition were found in DNA isolated from microbiological culture. In the case of ddPCR targeting IS6110, forty-eight samples were found to be MTC-positive (19 out of 19 with gross TBL) and 52 MTC-negative (Table 2). There was an association between real time-PCR and ddPCR, with the higher number of positive droplets, which ranged from 13,402 to 4 droplets, coinciding with those samples with a lower Cq value.
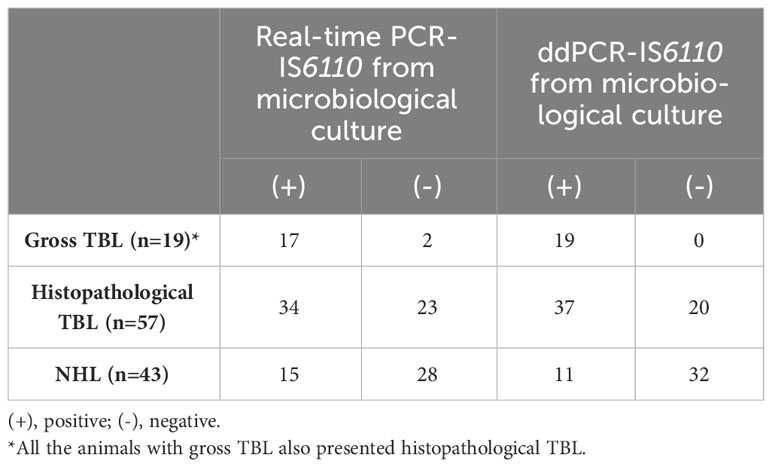
Table 2 Evaluation of confirmatory real-time PCR and ddPCR targeting IS6110 from microbiological culture DNA isolation according to the presence of gross tuberculosis-like lesions (TBL), histopathological TBL or no histopathological lesion (NHL).
Analyzing the histopathological evaluation, 34 out of 49 samples positive to the microbiological culture by real-time PCR-IS6110 were classified as TBL whereas 15 as NHL (Table 2). For the ddPCR-IS6110, histopathological TBL were found in 37 out of 48 ddPCR- IS6110-positive samples whereas 11 were classified as NHL (Table 2).
3.5 Comparison of IS6110 real time PCR and ddPCR from fresh LN tissue samples
DNA was isolated from homogenized fresh LN tissue samples (N=100) with gross TBL (N=19) or NVL (N=81) and subjected to both real-time PCR and ddPCR. All the samples with gross TBL were found to be MTC-positive by both real-time PCR or ddPCR targeting IS6110 (Table 3). For real-time PCR-IS6110, 53 out of 100 tested samples were detected as MTC-positive and 47 as negative. The Cq values ranged from 22.96 to 38.10 (average = 32.06). A partial inhibition of IAC was found in 6 out of 100 samples, with 2 out of 5 samples revealing a positive result after dilution 1:2 and re-evaluation. In the case of ddPCR, 55 samples were tested as MTC-positive and 45 as MTC-negative (Table 3). The number of positive droplets ranged from 12,618 to 3 droplets, with the highest number of positive droplets corresponding to those animals with pluribacillary lesions. As described before, an association was observed between real-time PCR and ddPCR, whereby a higher number of positive droplets corresponded to samples with higher Cq values.
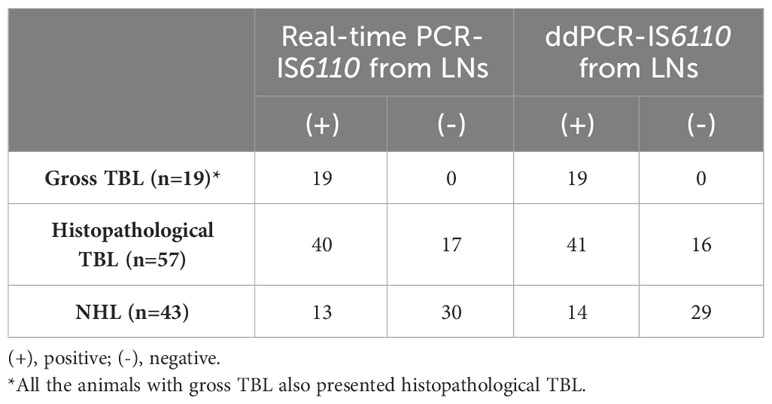
Table 3 Evaluation of real-time PCR and ddPCR targeting IS6110 from LNs DNA isolation according to the presence of gross tuberculosis-like lesions (TBL), histopathological TBL or no histopathological lesion (NHL).
According to histopathological evaluation, 40 out of 53 positive samples to real-time PCR presented histopathological TBL whereas 13 did not present microscopic lesion (Table 3). Regarding ddPCR, 41 out of 55 positive samples were disclosed as positive to histopathology with 14 samples presenting NHL (Table 3).
3.6 Diagnostic performance of ddPCR-IS6110 for the detection of MTC from microbiological culture and fresh LN tissue samples
In order to validate the ddPCR-IS6110 from microbiological culture and fresh LN tissue samples, these assays were compared with the reference standard assay (selective microbiological culture confirmed by real-time PCR-IS6110). Assuming that this reference assay is considered an imperfect assay for performing MTC diagnosis (Corner et al., 2012; Courcoul et al., 2014; Pucken et al., 2017) validation was carried out using EPIDAT 3.1 software. The ddPCR-IS6110 from microbiological culture detected 44 out of 49 reference standard positive samples [Se adjusted 90.76% (95% CI: 82.58-98.96%)], and 47 out of 51 reference standard negative samples resulted to be negative for ddPCR-IS6110 [Sp adjusted = 100% (95% CI: 100%)]. Thus, an adjusted FNR of 9.23% (95% CI: 1.04-17.42%) and FPR of 0% (95% CI: 0%) were estimated (Table 4). The PLR value (PLR = ∞) implies a high diagnostic value for the positive results discriminating between MTC-infected and non-infected animals. In addition, the NLR value was 0.07 meaning that it is a technique of a high diagnostic value to discriminate between healthy and diseased animals (Table 4). Of note, all reference standard-negative but ddPCR-positive samples were also classified as MTC positive by histopathological evaluation or real-time-IS6110 from fresh LNs tissue (Table 5 summaries the discordant results between different diagnostic techniques).
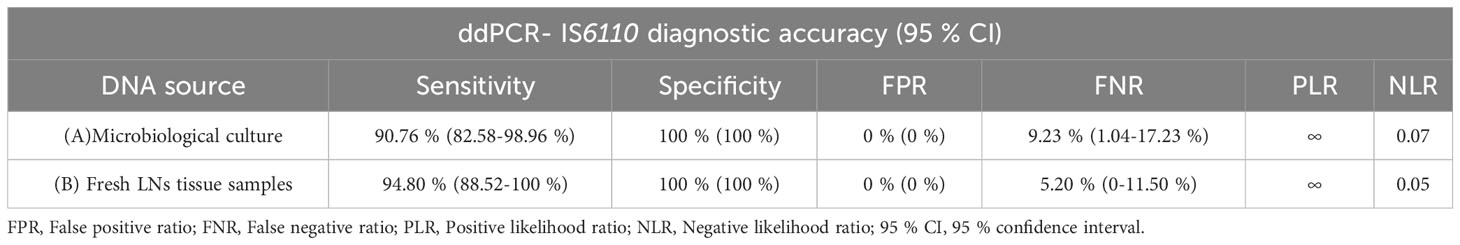
Table 4 Diagnostic performance of droplet digital PCR (ddPCR) targeting IS6110 from microbiological culture (A) and fresh lymph node (LN) tissue samples (B) in comparison with the established reference standard (selective microbiological culture confirmed by real-time PCR-IS6110) (N=100).
In the case of ddPCR-IS6110 carried out from fresh LN tissue samples, 47 out of 49 samples positive to the reference standard were found to be positive for ddPCR-IS6110 [adjusted Se 94.80% (95% CI: 88.52-100%), and 42 out of 51 reference standard-negative samples were tested as ddPCR-IS6110-negative [adjusted Sp = 100% (95% CI: 100%)] (Table 4). According to these results, ddPCR-IS6110 from fresh tissue presented an adjusted FNR of 5.20% (95% CI: 0-11.50%) and FPR of 0% (95 CI: 0%) (Table 4). Regardless of the true prevalence, ddPCR-IS6110 had a high diagnostic utility to confirm and discard MTC infection (PLR = ∞ and NLR = 0.05) (Table 4). Noteworthy, 9 reference standard-negative but ddPCR-IS6110-positive samples were found also as positive by histopathological evaluation (8 out of 9) or real-time PCR-IS6110 from fresh LN tissue (8 out of 9) (Table 5 summaries the discordant results between different diagnostic techniques).
Finally, Cohen’s kappa coefficient (κ) showed an almost perfect agreement between the ddPCR-IS6110 from culture and the reference standard (κ = 0.82) and a substantial agreement between the ddPCR-IS6110 from fresh LN tissue samples and the reference standard (κ = 0.76).
4 Discussion
Selective microbiological culture followed by a confirmatory real-time PCR, despite being an imperfect assay with some limitations, is still considered the gold standard technique to confirm bTB infection (Taylor et al., 2007; Liebana et al., 2008; Courcoul et al., 2014). Thus, recovery rates for MTC culture oscillates between 30 and 95% (Hines et al., 2006; Corner et al., 2012; Courcoul et al., 2014; Yates et al., 2017;), depending on the preservation of samples until culture, the chemical decontamination process influencing the viability of MTC, the type of culture media chosen, and the extremely slow nature of MTC growth (Hines et al., 2006; Corner et al., 2012; Yates et al., 2017;). To address these challenges, the use of PCR for detecting MTC in animal tissue samples has been proposed as a rapid, accurate and sensitive alternative to conduct MTC confirmation (Lorente-Leal et al., 2019; Lorente-Leal et al., 2021; Sánchez-Carvajal et al., 2021; Vera-Salmoral et al., 2023). In this context, ddPCR, a third-generation PCR technology known for its ability to detect small amounts of nucleic acids with high precision and sensitivity, has been reported. Additionally, ddPCR has a high resistance to inhibitors due to sample partitioning (Baker, 2012; Pinheiro et al., 2012; Kuypers and Jerome, 2017). Therefore, ddPCR represents a promising alternative to other molecular diagnostic methods for instance real-time PCR (Baker, 2012; Pinheiro et al., 2012; Kuypers and Jerome, 2017). The present study aimed to develop and validate a ddPCR assay targeting IS6110 to detect MTC in microbiological culture and fresh tissue samples with distinct TBL.
There are several performance parameters considered as key players in ddPCR including the concentration of primers and probe, the annealing temperature, or the quantity of the template (Whale et al., 2020). Optimization of these parameters is important to ensure the separation of positive and negative droplets and maximize the accuracy and sensitivity of the assay. Our results showed that the overall fluorescence amplitude of positive droplets increased with primers and probe concentrations. Thus, although we were able to detect the IS6110 specific region in all the assessed setups, the annealing temperature of 60°C together with the primers and probe concentrations of 900 nM for forward, 600 nM for reverse and 200 nM for probe yielded a higher fluorescence amplitude between positive and negative droplets compared with other setups and resulted in less non-specific amplification. In the case of template concentration, no pre-digestion DNA steps were performed as the extraction protocol used in this study allows improving the detection of the MTC DNA with a minimal raining signal. In addition, a better diagnostic performance was demonstrated using this protocol for DNA isolation as we have previously reported for real-time PCR (Vera-Salmoral et al., 2023). Since the Poisson statistics test requires a sufficient number of negative droplets for accurate calculation of DNA concentration (Whale et al., 2020), we decided to proceed with a template DNA concentration of 50 ng from tissue sample. This concentration was chosen to ensure a suitable number of negative droplets for statistical analysis and to minimize the risk of cross-contamination in samples with high concentration of bacterial DNA.
Considering the multi-etiological nature of TBL (Cardoso-Toset et al., 2015a), we proceeded to assess the analytical specificity of ddPCR targeting IS6110. We tested several common microorganisms associated with TBL and observed that the primers and probe exhibited high specificity for MTC IS6110. The selection of an appropriate genetic target plays a key role for the accurate detection of MTC (Sevilla et al., 2015). Among the various targets available (Lorente-Leal et al., 2019; Wang et al., 2019), the insertion sequence IS6110 is reported as one of the primary choices for diagnosing MTC (Sevilla et al., 2015). This genetic target not only enables differentiation between MTC and other bacteria, including non-tuberculous mycobacteria (NTM), but also offers the advantage of being a multicopy gene, ensuring sensitive and reliable detection of MTC (Charles et al., 2022). However, recent studies have identified the presence of an IS6110-like element in the genomes of certain NTM species, reporting a potential cross-reactivity between NTM and specific IS6110 primer pairs or probes (Coros et al., 2008; Thacker et al., 2011; Michelet et al., 2018; Lorente-Leal et al., 2021). Nevertheless, the impact of these findings on the specificity of PCR-IS6110 is expected to be minimal, as demonstrated in the following analysis.
ddPCR-IS6110 demonstrated an adjusted Se of 90.77% (95% CI: 82.58 - 98.96%) and a Sp of 100% (95% CI: 100%) for the confirmation of MTC in selective bacterial culture when compared with the reference standard. The application of ddPCR in microbiological culture not only detected all samples with gross TBL but also increased the number of positive samples detected with NHL compared to real-time PCR-IS6110. However, it is noteworthy that five positive samples to the refence standard were classified as ddPCR-negative. It is possible that these cases represent false-negative results to ddPCR. One potential explanation could be the inhibition of the PCR reaction. Although the probability of this issue is low because ddPCR is known to be highly resistant to PCR inhibitors (Baker, 2012; Pinheiro et al., 2012; Kuypers and Jerome, 2017), ddPCR may remain to be susceptible to some inhibitors. To address the issue of uncertain results, including an IAC into the ddPCR assay can provide added reliability. By designing a duplex reaction, the IAC can be labelled in a separate channel during analysis using the Bio-Rad QX100/QX200™ Droplet Digital™ PCR system, which is capable of detecting duplex targets in two separate channels (FAM and VIC/HEX) when TaqMan hydrolysis probes are utilized. This approach allows for simultaneous detection of the target of interest, IS6110, and the IAC, providing an internal reference for assay performance and identifying any potential inhibition or technical issues during the analysis.
In the case of DNA isolated directly from fresh bovine LN tissue samples, ddPCR targeting IS6110 proved to be a rapid and effective diagnostic assay when compared to traditional selective microbiological culture confirmed by real-time PCR. This ddPCR assay allowed us to detect all samples with gross TBL but also identified additional positive samples with microscopic lesions that were missed at the postmortem visual inspection. The ddPCR-IS6110 assay showed an adjusted Se of 94.80% (95% CI: 88.52 - 100%) and a Sp of 100% (95% CI: 100%) demonstrating a significantly improved diagnostic performance and accuracy compared to the reference standard. Particularly, the ddPCR-IS6110 assay disclosed as positive 9 samples negative to reference standard. Among these samples, 8 exhibited positive results in Ziehl-Neelsen staining and/or presented characteristic microscopic lesion. The remaining sample disclosed to be positive for both real-time and ddPCR-IS6110 but negative to histopathology. These findings indicate the superior Se and Sp of ddPCR-IS6110 directly from fresh tissue sample in detecting MTC compared to the reference standard. Furthermore, our findings reveal that ddPCR-IS6110 exhibits significantly enhanced sensitivity and specificity when contrasted with real-time PCR-IS6110 (Sánchez-Carvajal et al., 2021).
Although there have been no previous studies evaluating the diagnostic performance of ddPCR for MTC in animal samples, our research group conducted a preliminary approach using formalin-fixed paraffin-embedded (FFPE) tissue samples (Larenas-Muñoz et al., 2022). Our findings are consistent when compared to other studies conducted on human clinical samples (Song et al., 2018; Cho et al., 2020), reporting the rapid detection of MTC DNA. Furthermore, ddPCR offers advantages for MTC diagnostics across several sample types, including whole blood from patients with pulmonary and extrapulmonary TB lesion (Yang et al., 2017), culture isolates (Nyaruaba et al., 2020) or FFPE samples (Larenas-Muñoz et al., 2022). Additionally, our results demonstrated higher adjusted Se and Sp considering previous real-time PCR studies (Costa et al., 2013; Courcoul et al., 2014; Cardoso-Toset et al., 2015b; Lorente-Leal et al., 2019; Wang et al., 2019).
ddPCR technology offers several advantages over real-time PCR, making it an ideal technique for the detection of MTC, particularly in cases with a low-copy-number of the target (Kuypers and Jerome, 2017). Pathogens belonging to MTC are characterized by a paucibacillar pattern, which together the early detection of infected animals with NVL and low mycobacterial load would be beyond the LOD of traditional assays (Lorente-Leal et al., 2019; Sánchez-Carvajal et al., 2021). Due to sample partitioning, one notable advantage is the ddPCR ability to overcome the limitations caused by sample inhibitors which are commonly challenged in MTC samples (Dingle et al., 2013; Yang et al., 2014; Kuypers and Jerome, 2017). Additionally, ddPCR is less affected by poor amplification efficiency, further contributing to its robust performance in MTC detection (Kuypers and Jerome, 2017). Overall, these findings highlight the potential of ddPCR-IS6110 as a valuable tool for accurate and sensitive MTC diagnosis which would be susceptible to be included in bTB routine confirmation procedure.
Nonetheless, this study has some limitations that also need to be addressed. Firstly, the number of samples might have been larger in order to increase the robustness of the results providing a more comprehensive evaluation. Also, the absence of a ring trial, which would involve multiple laboratories and diverse epidemiological scenarios, limits the external validation and applicability of the findings to broader contexts. Moreover, it is important to highlight some drawbacks of ddPCR system over other molecular techniques. In general, ddPCR is more time-consuming than real-time PCR. The chances of contamination are higher, the implementation of this assay also demands a higher level of technical expertise and specialized training for personnel involved in the procedure. In contrast, the cost per reaction in ddPCR is more cost-effective than other standard molecular methods, excluding the initial investment required to acquire the necessary equipment. Therefore, further work on the re-validation of the present protocol should be performed in the future.
The present study describes a complete protocol including sample pre-processing, DNA purification and ddPCR analysis. According to our results, ddPCR-IS6110 demonstrated to be a rapid, highly sensitive and specific diagnostic tool as alternative to microbiological culture to confirm MTC infection shortening turnaround time for decision makers to be promptly informed. Comparing with real-time PCR, ddPCR has proved to be a potential first-choice molecular assay to detect MTC directly in fresh bovine tissue samples with increased Se and Sp. Therefore, ddPCR-IS6110 approach has the potential to be included in bTB surveillance and control programs.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because No purpose killing of animals was performed for this study, so no ethical or farmer’s consent approval was required.
Author contributions
JS-C: Conceptualization, Investigation, Methodology, Validation, Writing – original draft. EV-S: Data curation, Formal Analysis, Investigation, Methodology, Writing – review & editing. BH: Data curation, Formal analysis, Software, Validation, Writing – review & editing. ÁG-R: Data curation, Methodology, Writing – review & editing. IR-T: Investigation, Methodology, Writing – review & editing. FL-M: Formal analysis, Investigation, Methodology, Writing – review & editing. IL: Funding acquisition, Project administration, Resources, Writing – review & editing. LC: Funding acquisition, Resources, Supervision, Writing – review & editing. JG-L: Conceptualization, Funding acquisition, Methodology, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the investigation project New measures and techniques to control Bovine Tuberculosis in Andalusia (Financial support for Operational Groups of the European Innovation Partnership for Agricultural Productivity and Sustainability, EIP-AGRI; GOP2I-CO-16-0010). AG-R, IR-T and JS-C was supported by a Margarita Salas contract of the Spanish Ministry of Universities.
Acknowledgments
We appreciate the technical support offered by the Animal Health and Production Laboratory of Córdoba (Spain) and Alberto Alcántara and Marta Ordóñez-Martínez for their technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aranaz, A., De Juan, L., Montero, N., Sánchez, C., Galka, M., Delso, C., et al. (2004). Bovine tuberculosis (Mycobacterium bovis) in wildlife in Spain. J. Clin. Microbiol. 42, 2602–2608. doi: 10.1128/JCM.42.6.2602-2608.2004
Armbruster, D. A., Pry, T. (2008). Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 29, S49
Badia-Bringué, G., Canive, M., Casais, R., Blanco-Vázquez, C., Amado, J., Iglesias, N., et al. (2022). Evaluation of a droplet digital PCR assay for quantification of Mycobacterium avium subsp. paratuberculosis DNA in whole-blood and fecal samples from MAP-infected Holstein cattle. Front. Vet. Sci. 9, 944189. doi: 10.3389/fvets.2022.944189
Cao, Z., Wu, W., Wei, H., Gao, C., Zhang, L., Wu, C., et al. (2020). Using droplet digital PCR in the detection of Mycobacterium tuberculosis DNA in FFPE samples. Int. J. Infect. Dis. 99, 77–83. doi: 10.1016/j.ijid.2020.07.045
Cardoso-Toset, F., Gómez-Laguna, J., Amarilla, S. P., Vela, A. I., Carrasco, L., Fernández-Garayzábal, J. F., et al. (2015a). Multi-etiological nature of tuberculosis-like lesions in condemned pigs at the slaughterhouse. PloS One 10, e0139130. doi: 10.1371/journal.pone.0139130
Cardoso-Toset, F., Luque, I., Amarilla, S. P., Gómez-Gascón, L., Fernández, L., Huerta, B., et al. (2015b). Evaluation of rapid methods for diagnosis of tuberculosis in slaughtered free-range pigs. Vet. J. 204, 232–234. doi: 10.1016/j.tvjl.2015.01.022
Charles, C., Conde, C., Biet, F., Boschiroli, M. L., Michelet, L. (2022). IS6110 copy number in multi-host mycobacterium bovis strains circulating in bovine tuberculosis endemic french regions. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.891902
Cho, S. M., Shin, S., Kim, Y., Song, W., Hong, S. G., Jeong, S. H., et al. (2020). A novel approach for tuberculosis diagnosis using exosomal DNA and droplet digital PCR. Clin. Microbiol. Infect. 26, 942.e1–942.e5. doi: 10.1016/j.cmi.2019.11.012
Corner, L. A. L., Gormley, E., Pfeiffer, D. U. (2012). Primary isolation of Mycobacterium bovis from bovine tissues: Conditions for maximising the number of positive cultures. Vet. Microbiol. 156, 162–171. doi: 10.1016/j.vetmic.2011.10.016
Corner, L. A., Trajstman, A. C. (1988). An evaluation of 1-hexadecylpyridinium chloride as a decontaminant in the primary isolation of Mycobacterium bovis from bovine lesions. Vet. Microbiol. 18, 127–134. doi: 10.1016/0378-1135(88)90058-2
Coros, A., DeConno, E., Derbyshire, K. M. (2008). IS6110, a Mycobacterium tuberculosis Complex-Specific Insertion Sequence, Is Also Present in the Genome of Mycobacterium smegmatis, Suggestive of Lateral Gene Transfer among Mycobacterial Species. J. Bacteriol. 190, 3408. doi: 10.1128/JB.00009-08
Costa, P., Ferreira, A. S., Amaro, A., Albuquerque, T., Botelho, A., Couto, I., et al. (2013). Enhanced detection of tuberculous mycobacteria in animal tissues using a semi-nested probe-based real-time PCR. PloS One 8, e81337. doi: 10.1371/journal.pone.0081337
Courcoul, A., Moyen, J.-L., Brugère, L., Faye, S., Hénault, S., Gares, H., et al. (2014). Estimation of sensitivity and specificity of bacteriology, histopathology and PCR for the confirmatory diagnosis of bovine tuberculosis using latent class analysis. PloS One 9, e90334. doi: 10.1371/journal.pone.0090334.
Dean, A. S., Forcella, S., Olea-Popelka, F., El Idrissi, A., Glaziou, P., Benyahia, A., et al. (2018). A roadmap for zoonotic tuberculosis: a One Health approach to ending tuberculosis. Lancet Infect. Dis. 18, 137–138. doi: 10.1016/S1473-3099(18)30013-6
de la Rua-Domenech, R., Goodchild, A. T., Vordermeier, H. M., Hewinson, R. G., Christiansen, K. H., Clifton-Hadley, R. S. (2006). Ante mortem diagnosis of tuberculosis in cattle: A review of the tuberculin tests, γ-interferon assay and other ancillary diagnostic techniques. Res. Vet. Sci. 81 (2), 190–210. doi: 10.1016/j.rvsc.2005.11.005
Devonshire, A. S., Honeyborne, I., Gutteridge, A., Whale, A. S., Nixon, G., Wilson, P., et al. (2015). Highly reproducible absolute quantification of mycobacterium tuberculosis complex by digital PCR. Anal. Chem. 87 (7), 3706–3713. doi: 10.1021/ac5041617
Dingle, T. C., Sedlak, R. H., Cook, L., Jerome, K. R. (2013). Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin. Chem. 59, 1670–1672. doi: 10.1373/clinchem.2013.211045.
Domingo, M., Vidal, E., Marco, A. (2014). Pathology of bovine tuberculosis. Res. Vet. Sci. 97, S20–S29. doi: 10.1016/j.rvsc.2014.03.017
EU. (2023). EUR-lex - 32020R0689 - EN - EUR-lex. Available online at: https://eur-lex.europa.eu/eli/reg_del/2020/689/oj.
Guil-Luna, S., Sánchez-Céspedes, R., Rivas Crespo, A., Dolores Fernández, M., Fernández Sarmiento, J. A., Rodríguez-Ariza, A., et al. (2023). Analysis of cell-free DNA concentration, fragmentation patterns and TP53 gene expression in mammary tumor-bearing dogs: A pilot study. Front. Vet. Sci. 10, 1157878. doi: 10.3389/fvets.2023.1157878
Hines, N., Payeur, J. B., Hoffman, L. J. (2006). Comparison of the recovery of Mycobacterium bovis isolates using the BACTEC MGIT 960 system, BACTEC 460 system, and Middlebrook 7H10 and 7H11 solid media. J. Vet. Diagn. Investig. 18, 243–250. doi: 10.1177/104063870601800302
Kuypers, J., Jerome, K. R. (2017). Applications of digital PCR for clinical microbiology. J. Clin. Microbiol. 55, 1621–1628. doi: 10.1128/JCM.00211-17
Larenas-Muñoz, F., Sánchez-Carvajal, J. M., Galán-Relaño, Á., Ruedas-Torres, I., Vera-Salmoral, E., Gómez-Gascón, L., et al. (2022). The role of histopathology as a complementary diagnostic tool in the monitoring of bovine tuberculosis. Front. Vet. Sci. 9, 816190. doi: 10.3389/fvets.2022.816190
Liebana, E., Johnson, L., Gough, J., Durr, P., Jahans, K., Clifton-Hadley, R., et al. (2008). Pathology of naturally occurring bovine tuberculosis in England and Wales. Vet. J. 176, 354–360. doi: 10.1016/j.tvjl.2007.07.001
Lillsunde Larsson, G., Helenius, G. (2017). Digital droplet PCR (ddPCR) for the detection and quantification of HPV 16, 18, 33 and 45 - a short report. Cell Oncol. 40, 521–527. doi: 10.1007/s13402-017-0331-y
Lorente-Leal, V., Liandris, E., Castellanos, E., Bezos, J., Domínguez, L., de Juan, L., et al. (2019). Validation of a real-time PCR for the detection of mycobacterium tuberculosis complex members in Bovine tissue samples. Front. Vet. Sci. 6. doi: 10.3389/fvets.2019.00061
Lorente-Leal, V., Liandris, E., Pacciarini, M., Botelho, A., Kenny, K., Loyo, B., et al. (2021). Direct PCR on tissue samples to detect mycobacterium tuberculosis complex: An alternative to the bacteriological culture. J. Clin. Microbiol. 59, 10–1128. doi: 10.1128/JCM.01404-20
MAPA. (2022). Programa nacional de erradicación de tuberculosis bovina 2022 (Infección por el complejo Mycobacterium tuberculosis) Available at: https://www.mapa.gob.es/es/ganaderia/temas/sanidad-animal-higiene-ganadera/sanidad-animal/enfermedades/tuberculosis/Tuberculosis_bovina.aspx.
McHugh, M. L. (2012). Interrater reliability: the kappa statistic. Biochem. Med. 22, 276. doi: 10.11613/issn.1846-7482
Michelet, L., de Cruz, K., Karoui, C., Tambosco, J., Moyen, J. L., Hénault, S., et al. (2018). Second line molecular diagnosis for bovine tuberculosis to improve diagnostic schemes. PloS One 13, e0207614. doi: 10.1371/journal.pone.0207614
Nyaruaba, R., Xiong, J., Mwaliko, C., Wang, N., Kibii, B. J., Yu, J., et al. (2020). Development and evaluation of a single dye duplex droplet digital PCR assay for the rapid detection and quantification of mycobacterium tuberculosis. Microorganisms. 8(5), 701. doi: 10.3390/microorganisms8050701
Pinheiro, L. B., Coleman, V. A., Hindson, C. M., Herrmann, J., Hindson, B. J., Bhat, S., et al. (2012). Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 84, 1003–1011. doi: 10.1021/ac202578x
Pozo, P., Cardenas, N. C., Bezos, J., Romero, B., Grau, A., Nacar, J., et al. (2021). Evaluation of the performance of slaughterhouse surveillance for bovine tuberculosis detection in Castilla y Leon, Spain. Prev. Vet. Med. 189, 105307. doi: 10.1016/j.prevetmed.2021.105307
Pucken, V.-B., Knubben-Schweizer, G., Döpfer, D., Groll, A., Hafner-Marx, A., Hörmansdorfer, S., et al. (2017). Evaluating diagnostic tests for bovine tuberculosis in the southern part of Germany: A latent class analysis PloS One 12, e0179847. doi: 10.1371/journal.pone.0179847
Ramos, D. F., Silva, P. E. A., Dellagostin, O. A. (2015). Diagnosis of bovine tuberculosis: review of main techniques. Braz. J. Biol. 75, 830–837. doi: 10.1590/1519-6984.23613
Sackett, D. L., Richardson, W. S., Rosenberg, W., Haynes, R. B.. (2001). Diagnóstico y Cribado. edicina basada en la evidencia. Elsevier: España. p 62–68.
Sánchez-Carvajal, J. M., Galán-Relaño, Á., Ruedas-Torres, I., Jurado-Martos, F., Larenas-Muñoz, F., Vera, E., et al. (2021). Real-time PCR validation for mycobacterium tuberculosis complex detection targeting IS6110 directly from bovine lymph nodes. Front. Vet. Sci. 8, 643111. doi: 10.3389/fvets.2021.643111
Sevilla, I. A., Molina, E., Elguezabal, N., Pérez, V., Garrido, J. M., Juste, R. A. (2015). Detection of mycobacteria, Mycobacterium avium subspecies, and Mycobacterium tuberculosis complex by a novel tetraplex real-time PCR assay. J. Clin. Microbiol. 53, 930–940. doi: 10.1128/JCM.03168-14
Song, N., Tan, Y., Zhang, L., Luo, W., Guan, Q., Yan, M. Z., et al. (2018). Detection of circulating Mycobacterium tuberculosis-specific DNA by droplet digital PCR for vaccine evaluation in challenged monkeys and TB diagnosis. Emerg. Microbes Infect. 7, 1–9. doi: 10.1038/s41426-018-0076-3
Strain, M. C., Lada, S. M., Luong, T., Rought, S. E., Gianella, S., Terry, V. H., et al. (2013). Highly precise measurement of HIV DNA by droplet digital PCR. PloS One 8, e55943. doi: 10.1371/journal.pone.0055943
Taylor, G. M., Worth, D. R., Palmer, S., Jahans, K., Hewinson, R. G. (2007). Rapid detection of Mycobacterium bovis DNA in cattle lymph nodes with visible lesions using PCR. BMC Vet. Res. 3, 12. doi: 10.1186/1746-6148-3-12
Thacker, T. C., Harris, B., Palmer, M. V., Waters, W. R. (2011). Improved specificity for detection of Mycobacterium bovis in fresh tissues using IS6110 real-time PCR. BMC Vet. Res. 7, 50. doi: 10.1186/1746-6148-7-50
Thierry, D., Brisson-Noel, A., Vincent-Levy-Frebault, V., Nguyen, S., Guesdon, J. L., Gicquel, B. (1990). Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J. Clin. Microbiol. 28, 2668–2673. doi: 10.1128/jcm.28.12.2668-2673.1990
Vera-Salmoral, E., Gómez-Laguna, J., Galán-Relaño, Á., Ruedas-Torres, I., Carrasco, L., Luque, I., et al. (2023). Optimization of real-time PCR protocols from lymph node bovine tissue for direct detection of Mycobacterium tuberculosis complex. Microbiol. Spectr. 11 (5), e00348-23. doi: 10.1128/spectrum.00348-23
Wang, H. Y., Lu, J. J., Chang, C. Y., Chou, W. P., Hsieh, J. C. H., Lin, C. R., et al. (2019). Development of a high sensitivity TaqMan-based PCR assay for the specific detection of Mycobacterium tuberculosis complex in both pulmonary and extrapulmonary specimens. Sci. Rep. 9, 1–12. doi: 10.1038/s41598-018-33804-1
Whale, A. S., De Spiegelaere, W., Trypsteen, W., Nour, A. A., Bae, Y. K., Benes, V., et al. (2020). The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem. 66, 1012–1029. doi: 10.1093/clinchem/hvaa125
Yang, Han, X., Liu, A., Bai, X., Xu, C., Bao, F., et al. (2017). Use of digital droplet PCR to detect mycobacterium tuberculosis DNA in whole blood-derived DNA samples from patients with pulmonary and extrapulmonary tuberculosis. Front. Cell Infect. Microbiol. 7, 369. doi: 10.3389/fcimb.2017.00369
Yang, Paparini, A., Monis, P., Ryan, U., Yang, R., Paparini, A., et al. (2014). Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int. J. Parasitol. 44, 1105–1113. doi: 10.1016/j.ijpara.2014.08.004
Keywords: droplet digital PCR, bovine tuberculosis, Mycobacterium tuberculosis complex, molecular diagnosis, IS6110, lymph node, ddPCR
Citation: Sánchez-Carvajal JM, Vera-Salmoral E, Huerta B, Galán-Relaño Á, Ruedas-Torres I, Larenas-Muñoz F, Luque I, Carrasco L and Gómez-Laguna J (2024) Droplet digital PCR as alternative to microbiological culture for Mycobacterium tuberculosis complex detection in bovine lymph node tissue samples. Front. Cell. Infect. Microbiol. 14:1349999. doi: 10.3389/fcimb.2024.1349999
Received: 05 December 2023; Accepted: 07 February 2024;
Published: 26 February 2024.
Edited by:
Adela Oliva Chavez, Texas A and M University, United StatesReviewed by:
Marta Alonso-Hearn, Basque Research and Technology Alliance (BRTA), SpainSante Roperto, University of Naples Federico II, Italy
Copyright © 2024 Sánchez-Carvajal, Vera-Salmoral, Huerta, Galán-Relaño, Ruedas-Torres, Larenas-Muñoz, Luque, Carrasco and Gómez-Laguna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: José María Sánchez-Carvajal, v42sancj@uco.es; Belén Huerta, sa2hulob@uco.es
 José María Sánchez-Carvajal
José María Sánchez-Carvajal Eduardo Vera-Salmoral1,2
Eduardo Vera-Salmoral1,2  Ángela Galán-Relaño
Ángela Galán-Relaño Inés Ruedas-Torres
Inés Ruedas-Torres Fernanda Larenas-Muñoz
Fernanda Larenas-Muñoz Inmaculada Luque
Inmaculada Luque Librado Carrasco
Librado Carrasco Jaime Gómez-Laguna
Jaime Gómez-Laguna