Development of a real-time recombinase-aided amplification assay for rapid and sensitive detection of Edwardsiella piscicida
- 1School of Ocean, Yantai University, Yantai, China
- 2Yantai Marine Economic Research Institute, Yantai, China
Edwardsiella piscicida, a significant intracellular pathogen, is widely distributed in aquatic environments and causes systemic infection in various species. Therefore, it’s essential to develop a rapid, uncomplicated and sensitive method for detection of E. piscicida in order to control the transmission of this pathogen effectively. The recombinase-aided amplification (RAA) assay is a newly developed, rapid detection method that has been utilized for various pathogens. In the present study, a real-time RAA (RT-RAA) assay, targeting the conserved positions of the EvpP gene, was successfully established for the detection of E. piscicida. This assay can be performed in a one-step single tube reaction at a temperature of 39°C within 20 min. The RT-RAA assay exhibited a sensitivity of 42 copies per reaction at a 95% probability, which was comparable to the sensitivity of real-time quantitative PCR (qPCR) assay. The specificity assay confirmed that the RT-RAA assay specifically targeted E. piscicida without any cross-reactivity with other important marine bacterial pathogens. Moreover, when clinical specimens were utilized, a perfect agreement of 100% was achieved between the RT-RAA and qPCR assays, resulting a kappa value of 1. These findings indicated that the established RT-RAA assay provided a viable alternative for the rapid, sensitive, and specific detection of E. piscicida.
Introduction
Edwardsiella piscicida, previously known as E. tarda, is a Gram-negative, rod-shaped, facultatively anaerobic, intracellular pathogen, which is widely prevalent in aquatic environments and causes systemic infections in various species such as fish, amphibians, reptiles, and mammals (Leung et al., 2019). Moreover, E. piscicida also serves as an etiological agent for both extraintestinal and gastrointestinal infections in humans (Leung et al., 2019). The initial documentation of E. piscicida infection in aquaculture was observed in channel catfish (Meyer and Bullock, 1973), subsequently, severe infections caused by E. piscicida have been reported in numerous commercially significant marine and freshwater fish species, including eels, mullet, chinook salmon, Japanese flounder, turbot, tilapia, and striped bass, leading to large economic losses in aquaculture worldwide (Park et al., 2012). Similar to other invasive pathogens, E. piscicida exhibits a preference for infiltrating host cells as a means to evade immune system detection and facilitate replication within both phagocytes and nonphagocytes, thereby contributing to its pathogenicity (Xie et al., 2014; Zhang et al., 2016). In recent years, increasing number of E. piscicida isolates have been obtained from aquacultural environment (Ferrari et al., 2022; Kim et al., 2022; Sugiura et al., 2022). Consequently, the development of a prompt and highly sensitive diagnostic technique for E. piscicida is of utmost importance in the prevention of outbreaks.
Currently, the identification of E. piscicida primarily depends on conventional techniques, including culture, PCR, or immune-testing (Swain et al., 2001; Hong et al., 2009; Castro et al., 2010; Ferrari et al., 2022; Sugiura et al., 2022). While cultures offer undeniable proof of infection, their lengthy incubation period renders them impractical for clinical diagnosis, except for antimicrobial susceptibility analysis (Kim et al., 2022; Sugiura et al., 2022). Various methods that rely on antibodies, such as enzyme-linked immunosorbent assay (ELISA), and quartz crystal microbalance (QCM) biosensor, have been developed for E. piscicida detection (Swain et al., 2001; Hong et al., 2009). Over the past few years, several PCR-based molecular detection systems, and loop-mediated isothermal amplification (LAMP) have been developed for detection of E. piscicida (Castro et al., 2010; Xie et al., 2012; Reichley et al., 2015). However, the complex operation or expensive equipment requirements limit their application in clinical or environmental samples. Thus, it is necessary to establish a fast, simple, and sensitive method for E. piscicida detection.
The recombinase-aided amplification (RAA) assay is an innovative isothermal amplification technology, which offers distinct advantages such as quickness (within 20 min), simplicity, and low cost (Pang and Long, 2023). Consequently, RAA surpasses other methods and holds great potential for pathogen diagnosis (Fan et al., 2021). The RAA reaction system consists of three essential proteins: recombinase UvsX, which facilitates the annealing of primers to template DNA; single strand DNA binding protein (SSB); and DNA polymerase, which enhanced the speed and specificity nature of the reaction system. In recent time, RAA has proven effective in identifying different types of microbial pathogens (Wang et al., 2020; Qin et al., 2021; Tu et al., 2021).
The gene EvpP (E. tarda virulent protein P) is an important T6SS effector, which is essential for the virulence of E. piscicida (Hu et al., 2014). Interestingly, EvpP gene could not be found in other bacteria with T6SS (Ho et al., 2014). In this study, we effectively developed a real-time RAA (RT-RAA) based method for detecting E. piscicida, specifically targeting the EvpP gene. This assay exhibits accessibility, specificity, and sensitivity, with a completion time of just 20 minutes. The characteristics of the RT-RAA method make it a promising candidate for early diagnosis of E. piscicida infection.
Material and methods
Bacteria and clinical and environmental samples
Edwardsiella piscicida DC1 was isolated from diseased turbot (Scophthalmus maximus) reared in a commercial fish farm located in Qingdao, Shandong province, which also showed high pathogenicity to Lateolabrax maculatus. Another eight clinically-common pathogens were evaluated in this study, including Pseudomonas fluorescens, Leclercia adecarboxylata, Vibrio rotiferianus, Vibrio scophthalmi, Vibrio alginolyticus, Vibrio hyugaensis, Haemophilus piscium and Vibrio azureus, which were all isolated and stored in our lab. All strains were verified by 16S rDNA gene sequencing in Sangon Biotech Co., Ltd. (Shanghai, China). All strains were cultured in a Luria-Bertani broth (LB) medium at 28°C.
For clinical specimen preparation, 30 clinical samples were prepared. Briefly, clinically healthy L. maculatus (average weight 14.7 g) were purchased from a commercial fish farm located in Yantai, Shandong province. Before experiment, fish were randomly sampled for the examination of E. piscicida presence by plate counts. For experimental infection, E. piscicida DC1 were cultured in LB medium to an OD600 = 0.8, L. maculatus were challenged via intraperitoneal (i.p.) injection with 100 μl of E. piscicida DC1 (107 cfu/ml) and maintained at 22°C. At 0, 12, 24, 36, and 48 h post-challenge (hpc), the liver, spleen, and kidney were obtained aseptically from the fish (three fish/time point). For E. piscicida detection, the tissues obtained above were homogenated in PBS, spread onto the LB agar plates, and incubation at 28°C for 48 h, furthermore, the observed bacterial colonies were verified by 16S rDNA sequencing (Zhang et al., 2014). The samples were further confirmed by quantitative real-time PCR as reported previously (Reichley et al., 2015). A total of 24 positive and 6 negative fish samples were prepared for further RT-RAA analysis. For environmental samples preparation, nine sea water and nine sediment samples were collected from the intertidal zone of Yantai, China. Before experiment, these samples were randomly sampled for the examination of E. piscicida presence by plate counts. Different concentrations of E. piscicida cultured above were added, and verified as above. A total of 12 positive (6 sea water and 6 sediment) and 6 negative samples (3 sea water and 3 sediment) were prepared for further RT-RAA analysis.
DNA extraction
For bacterial DNA isolation, 1 ml sample of pure cultured bacterial suspension was obtained. The DNA extraction was performed using the TIANamp Bacteria DNA Kit (Tiangen Biotech Co, Ltd, China) following to the manufacturer’s instructions. For clinical samples’ DNA extraction, 50 mg samples were homogenized in 200 μL PBS and the total DNA was extracted using CTAB method (Guerra et al., 2020). The DNA were eluted in 100 μL of nuclease-free water and stored at a temperature of -20°C until utilized.
Recombinant plasmid construction
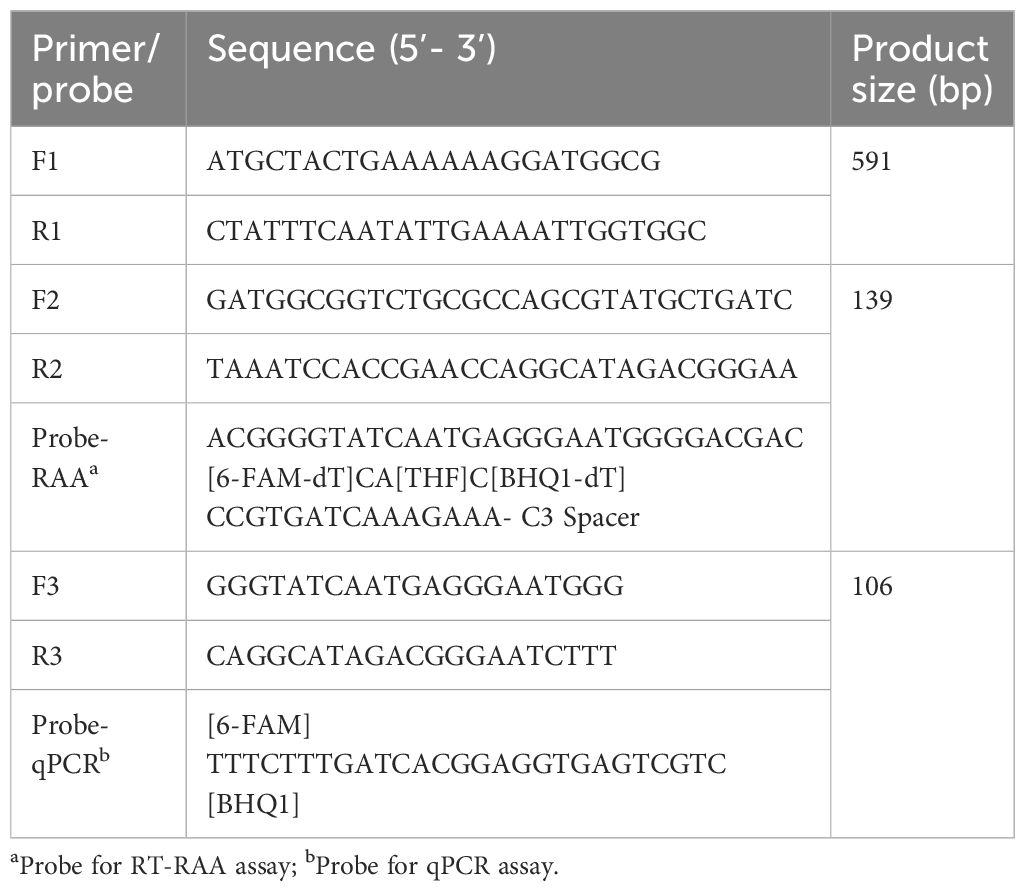
The EvpP gene was selected as a target gene for detecting E. piscicida. The EvpP gene sequence was obtained from E. piscicida DC1, and submitted into the GenBank (accession no. OR670479.1). Using primers EvpP-F1/R1 (Table 1), we amplified a 591 bp fragment of the EvpP gene and inserted it into the pMD-19T vector (TAKARA, Dalian, China). The recombinant plasmid was confirmed by sequencing in Sangon Biotech Co., Ltd. and named pEvpP. The recombinant plasmid’s concentration was quantified using Nano-500 micro-spectrophotometer (Allsheng, Hangzhou, China) and the DNA copy number was determined using the formula: DNA copy number (copies/μL) = [6.02 × 1023 × plasmid concentration (ng/μL) × 10−9]/[DNA length × 660]. Then, the standard recombinant plasmids diluted in a 10-fold manner ranging from 2.1×108 to 2.1×100 copies/μL and stored at −20°C until further use.
RAA primer design
Other nine EvpP genes of different E. piscicida strains were obtained from the National Center for Biotechnology Information (NCBI) database. The DNAMAN software was utilized to conduct multiple sequence alignment in order to identify the conserved regions of the EvpP gene. The primers and probe for RT-RAA were designed within the highly-conserved regions using an online tool “Primer & probe design for RPA/RAA” (https://ezassay.com/primer), adhering to the guidelines for designing RT-RAA primers and probe. To ensure specificity, a Primer-BLAST search was performed against the NCBI database to verify the sequences the primer and probe. Furthermore, the possibility of primer dimers and hairpins was evaluated by utilizing the online OligoEvaluator software (http://www.oligoevaluator.com). Finally, the primer and probe were synthesized and purified using high-performance liquid chromatography in Sangon Biotech Co., Ltd.
Screening the optimal primers for RT-RAA assay
First, an ideal probe (p308–358) was designed (Table 1, Figure 1). Then, six candidate primer pairs (F264–293/R405–434, F265–294/R404–433, F266–295/R403–432, F267–296/R402–431, F267–297/R401–431, F268–297/R401–430) were designed (Supplementary Table S1; Figure 1). The RT-RAA assay was performed with the above primers and the probe, and the primer pair with the highest end fluorescence intensity value (RFU value) and better “S” amplification curve was selected for further analysis. All reactions were performed three times.
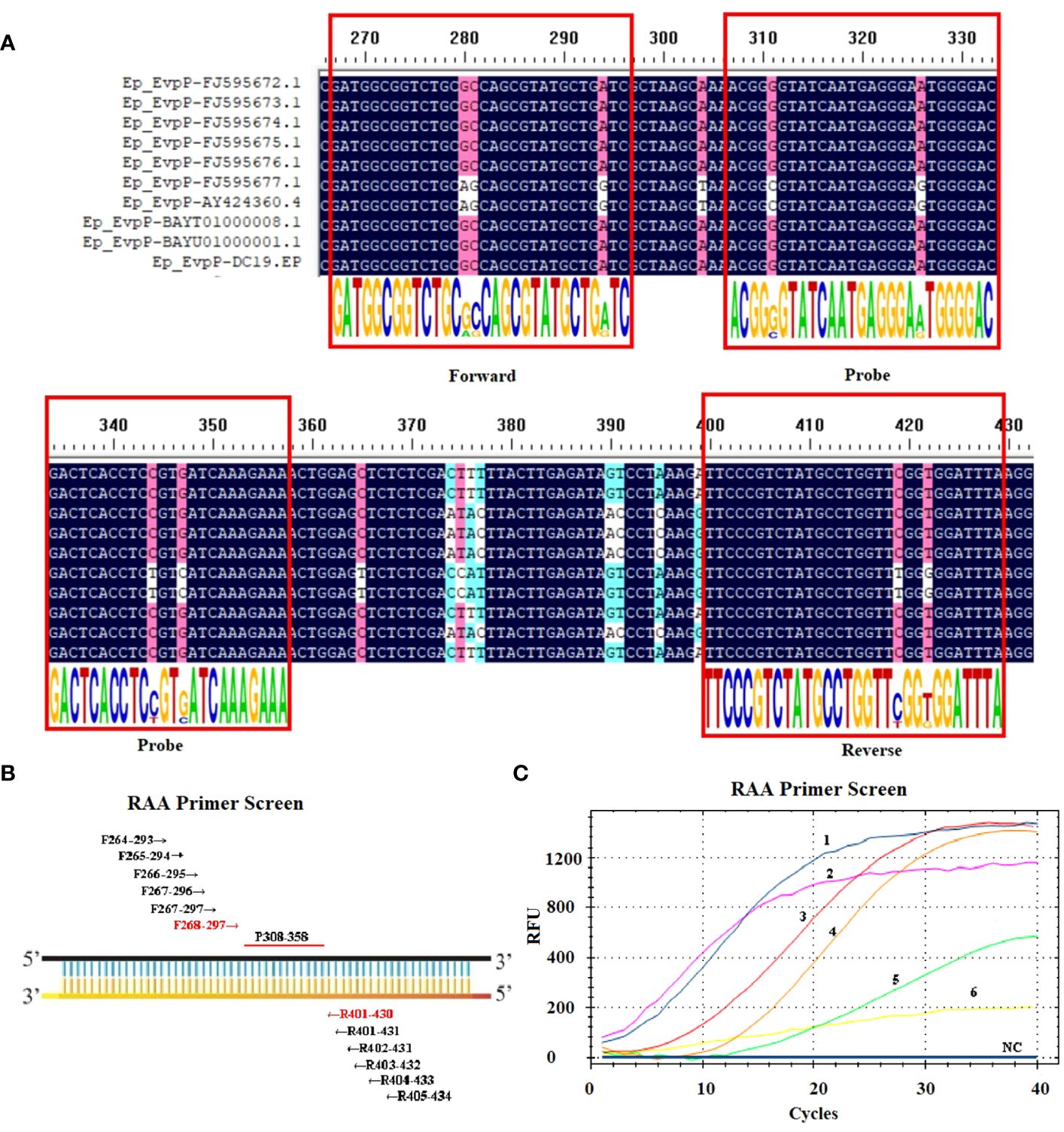
Figure 1 Screening the optical primers for RT-RAA assay. (A) Multiple sequence alignment of primers and probe with EvpP sequences of E. piscicida strains. The consensus residues are in black, and the residues that are ≥75% identical among the aligned sequences are in pink. (B) Sketch map of primary primers screening, the numbers indicate the position within the EvpP gene. (C) Screening results of the forward and reverse primers. 1: F268-297/R401-430; 2: F264-293/R405-434; 3: F266-295/R403-432; 4: F267-297/R401-431; 5: F267-296/R402-431; 6: F265-294/R404-433; NC, Negative Control.
RAA assay
A commercial RAA fluorescence method kit (Zhongce, Hangzhou, China) was used to perform the RT-RAA assay in a reaction volume of 20 μL according to the manufacturer’ s instructions. Each primer and probe described above was diluted to a final concentration of 10 μM. The reaction mixture contained 2.0 μL of DNA template that was extracted, 10 μL of reaction buffer, 5.56 μL of DNase-free water, 0.4 μL of each primer, 0.24 μL of probe, 1.0 μL of magnesium acetate and reactive dry powder. Following the addition of all remaining components to the reactive dry powder, magnesium acetate is subsequently introduced to the tube lids. Subsequently, the tube lids are securely closed and the reaction tubes are subjected to gentle agitation by inverting them 5-6 times, followed by centrifugation at a low speed for a duration of 10 seconds. Afterward, the resulting tubes are transferred to the Bio-rad CFX96 Touch Real-Time PCR Detection System (Bio-rad, Hercules, CA, USA) and maintained at a temperature of 39°C for 20 minutes. Each experimental run includes both a positive control (pEvpP plasmid) and a negative control.
Taqman real-time quantitative PCR assay
The qPCR assay was performed in 20 μL reaction volume using Pro Taq HS Premix Probe real-time PCR Kit III (Accurate Biotechnology Co., Ltd, Hunan, China) according to the manufacturer’ s instructions. The primers (F3/R3) and probe (Probe-qPCR) for aPCR assay were listed in Table 1. In the 20 μL reaction mixture, there was 2.0 μL of extracted DNA template, 10 μL of 2 × Pro Taq HS Probe Premix III, 4.4 μL of DNase-free water, 1.0 μL of each primer F3/R3 (10 μM), 1.6 μL of Probe-qPCR (10 μM). Next, the reactions were examined on the Bio-rad CFX96 Touch Real-Time PCR Detection System (Bio-rad, Hercules, CA, USA). The qPCR procedure involved an initial denaturation step at a temperature of 95°C for a duration of 30 s, followed by 40 cycles of amplification (with a denaturation step at 95°C for 5 s and an annealing step at 60°C for 30 s). Every trial includes both a positive control (pEvpP plasmid) and a negative control.
Specificity analysis
To assess the specificity of the RAA assay, eight additional bacterial pathogens were included: P. fluorescens, L. adecarboxylata, V. rotiferianus, V. scophthalmi, V. alginolyticus, V. hyugaensis, H. piscium and V. azureus. The genomic DNA of these bacteria were extracted as described above and then utilized as the template. The specificity test was performed under the aforementioned conditions and system, and repeated more than three times with each template and negative controls.
Sensitivity analysis
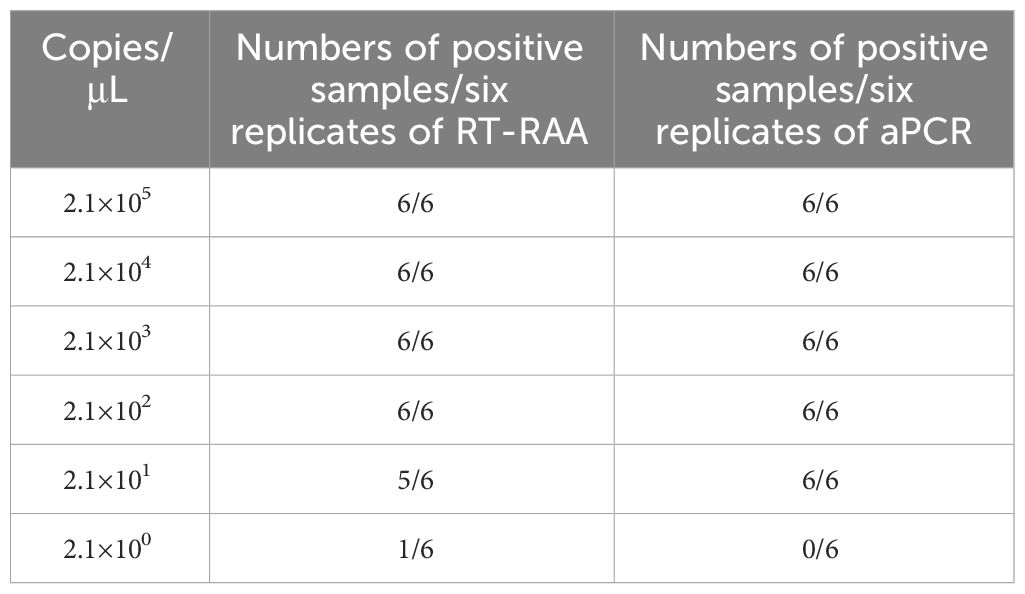
To determine the sensitivity of the RAA assay, plasmid pEvpP was diluted in 10-fold serial dilutions ranging from 2.1×105 to 2.1×100 copies/μL, with six replicates for each dilution. For comparison, the same templates were tested in parallel using the qPCR assay. The sensitivity assays were carried out utilizing the aforementioned conditions and systems, and were repeated more than three times with each template and negative controls.
Evaluation of the RAA assay using clinical samples
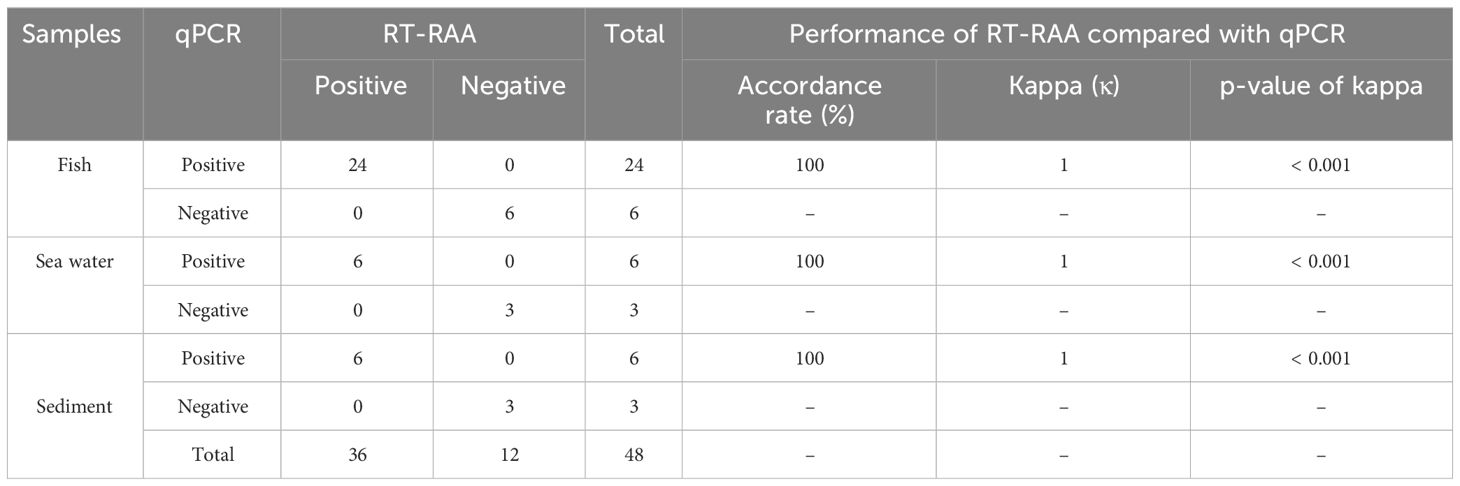
In order to assess applicability of the RT-RAA assay for E. piscicida detection in clinical and environmental samples, bacterial DNA were extracted from a total of 48 samples. Among these samples, 30 were clinical samples (24 positive and 6 negative), 9 were sea water samples (6 positive and 3 negative), and 9 were sediment samples (6 positive and 3 negative). Subsequently, the RAA assay was conducted on all 48 samples. For comparative purposes, the same samples were also subjected to the qPCR assay for E. piscicida in parallel.
Statistical analysis
Probit analysis for the detection limit of the RT-RAA assay and qPCR assay was performed at a 95% probability level. The kappa and p values of the RT-RAA and qPCR assays were calculated. All statistical analysis was carried out with SPSS 21.0 (IBM, Armonk, NY).
Results
Primers and probe designed for the RT-RAA assay
Ten EvpP gene sequences of E. piscicida were obtained from the GenBank database. Based on the multiple sequence alignment analysis, conserved nucleotide residues were identified and visually emphasized with a black background. In order to design primers and probe, 10 EvpP sequences were aligned with F2, R2, and Probe-RAA primers. The sequence alignment revealed high conserved regions for primers and probe design (Figure 1A). After primer screening, the optimal primer pair F268–297/R401–430 was screened out according to the amplification curve and renamed F2/R2 (Figure 1C, Table 1).
Specificity of RAA assay
The RAA assay showed a positive result for E. piscicida and negative results for P. fluorescens, L. adecarboxylata, V. rotiferianus, V. scophthalmi, V. alginolyticus, V. hyugaensis, H. piscium and V. azureus (Figure 2). The results showed that the established RT-RAA assay exhibited a considerable degree of specificity towards its intended targets, with no cases of cross-reactivity observed with other prevalent marine pathogenic bacteria.
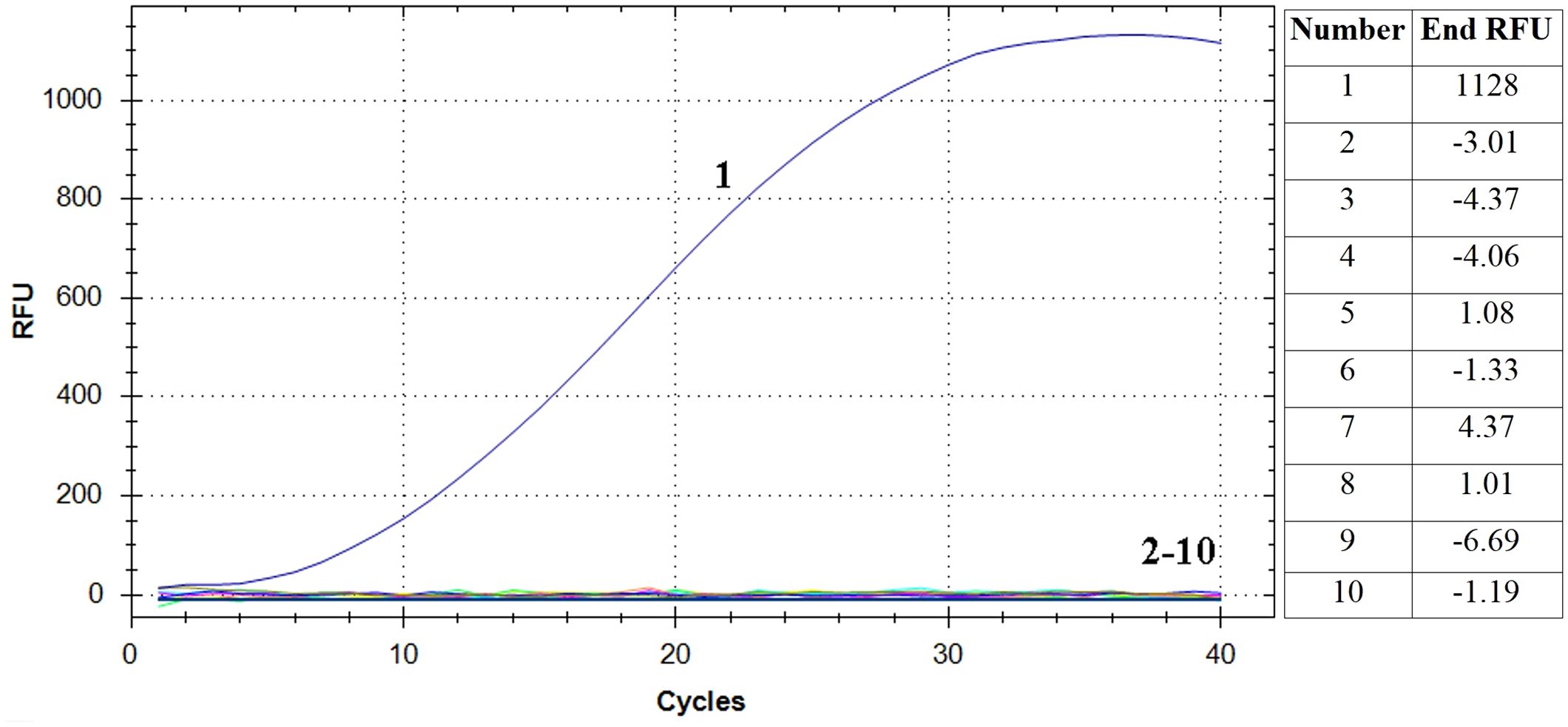
Figure 2 Specificity analysis of the RT-RAA assay. Only the reaction containing E. piscicida DC1 DNA as template produced a positive signal, while nucleic acid from all other eight bacteria and negative control did not. Curve 1: E. piscicida; Curve 2-10: Pseudomonas fluorescens, Leclercia adecarboxylata, Vibrio rotiferianus, Vibrio scophthalmi, Vibrio alginolyticus, Vibrio hyugaensis, Haemophilus piscium and Vibrio azureus and negative controls, respectively. The final relative fluorescence unit (RFU) values of each samples were shown in the right table.
Analytical sensitivity of RT-RAA assay
The pEvpP plasmid, which was serially diluted from 2.1×105 to 2.1×100 copies/μL, was subjected to testing using the RT-RAA assay. The results indicated that consistent signals were observed within the range of 2.1×100 and 2.1×105 copies/μL for each reaction. As a result, the detection limit of the RAA assay was determined to be 42 copies (0.14 fg) per reaction with a 95% probability (Table 2, Figure 3) (Probit analysis, p=0.015), which aligns with the detection limit of 42 copies per reaction for the qPCR method (Table 2, Figure 3) (Probit analysis, p=0.02).
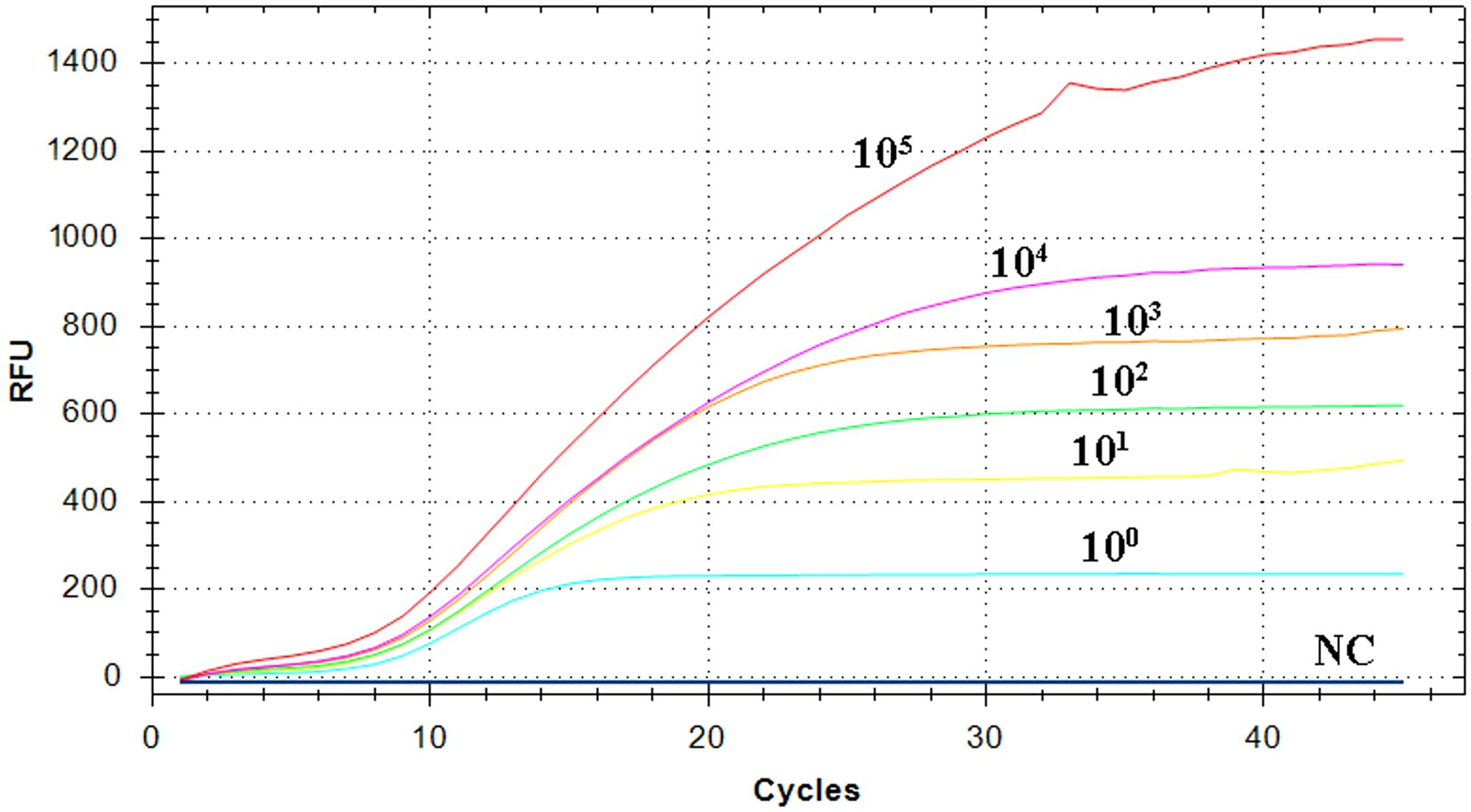
Figure 3 Sensitivity analysis of the RT-RAA assay. A panel of serially diluted recombinant plasmid pEvpP, from 105 to 100 copies per reaction and a negative control sample (NC), were used to determine the detection limit of the RT-RAA assay.
Evaluation of the RT-RAA assay using clinical and environmental samples
In this RT-RAA assay, a total of 48 samples, consisting of 24 positive clinical samples, 6 positive sea water samples, 6 positive sediment samples, and 12 negative samples (6 clinical samples, 3 sea water samples, 3 sediment samples), were utilized, and the results were comparable with those obtained by qPCR. Using qPCR, 36 out of the 48 samples were identified as positive. The RT-RAA assay successfully detected and distinguished all 36 positive samples with complete accordance, sensitivity, and specificity (Table 3, Figure 4). Notably, no significant disparities were observed between the detection outcomes of the RT-RAA assay and qPCR. The RT-RAA assay yielded a kappa value of 1.0 (p < 0.001).
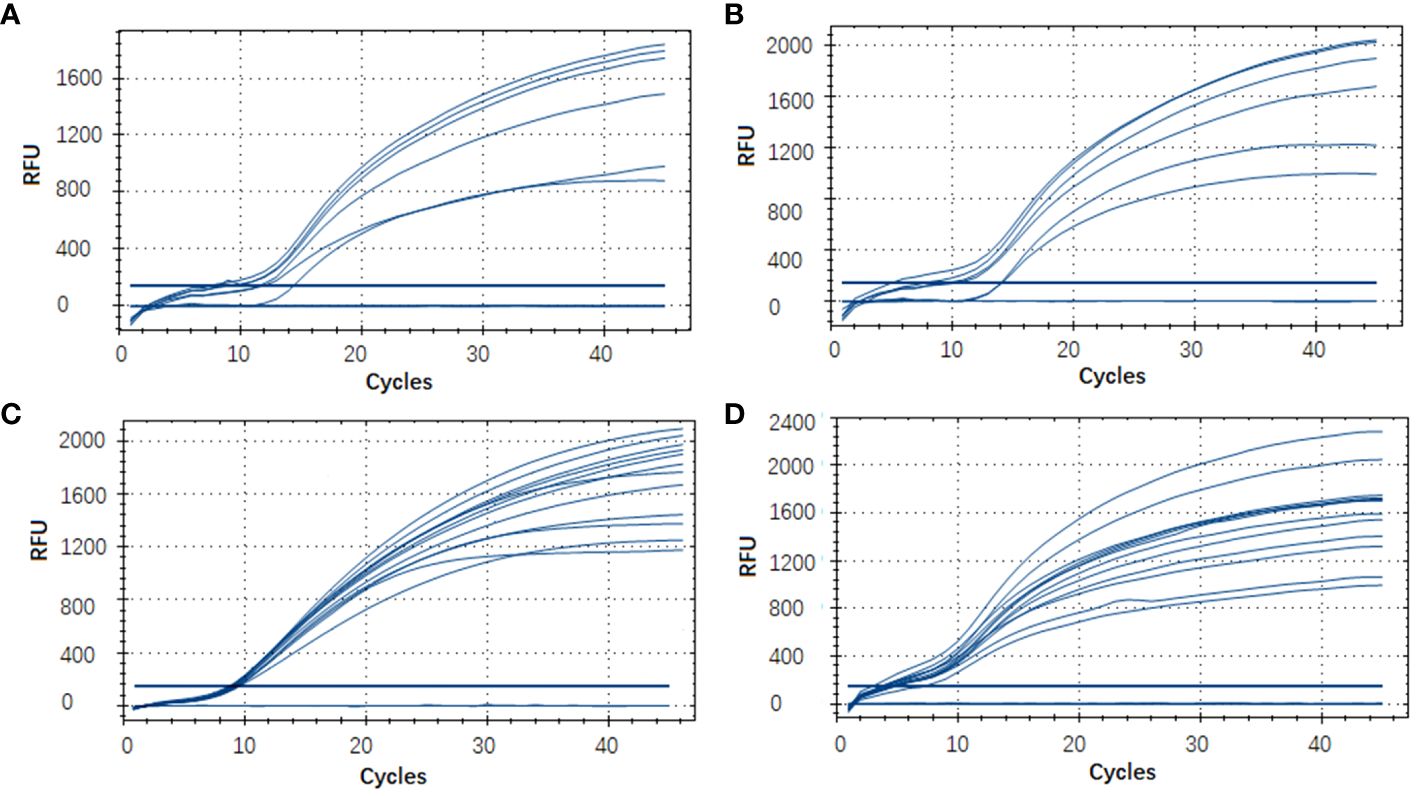
Figure 4 Evaluation of the RT-RAA assay using clinical and environmental samples. A total of 48 samples, consisting of 9 sea water samples (A), 9 sediment samples (B), 30 clinical fish samples (C, D) were analyzed with the RT-RAA assay.
Discussion
E. piscicida, a significant intracellular pathogen, is found extensively in aquatic surroundings and causes systemic infections in different species. Until now, several methods, including ELISA, ordinary PCR, qPCR, and QCM biosensor, have been employed for the detection of E. piscicida (Swain et al., 2001; Hong et al., 2009; Castro et al., 2010; Reichley et al., 2015). However, these methods pose challenges in their implementation in underfunded laboratories with limited resources and expertise. Currently, the detection of E. piscicida can be accomplished within 45 minutes using a relatively fast detection technique called loop-mediated isothermal amplification (LAMP) (Xie et al., 2013). Nevertheless, the current LAMP methods suffer from unwanted nonselective amplification, leading to significant background signals due to the increasing number of target sites. This nonspecific amplification significantly undermines the dependability of LAMP and imposes constraints on its utilization in clinical diagnostics (Hardinge and Murray, 2019). Therefore, it is crucial to establish a rapid and sensitive diagnostic method for clinical diagnosis of E. piscicida.
RAA is a novel isothermal amplification technique that has been developed in recent years. Compared with the general molecular detection methods, RAA possesses several advantages, such as low-cost, simplicity of operation (as it does not necessitate complex equipment or highly skilled personnel, can be conducted using portable device), low and constant reaction temperature (39°C), and rapid reaction speed (within 20 min) (Xie et al., 2012). As a result, RAA has been widely used in identifying viruses, bacteria, and parasites (Cui et al., 2022; Fu et al., 2022; Lin et al., 2022; Cui H et al., 2023; Cui X et al., 2023). Consequently, it represents a rapid and user-friendly method suitable for clinical use, particularly in primary laboratories.
Although the genomes of E. piscicida from various isolates exhibit a relatively conserved nature, genetic diversity can be observed among different strains. It is important to mention that sequence variations within the target region can significantly affect the detection results of molecular diagnostic techniques. Previous studies have identified 16S rRNA, gyrB, hlyb, and EvpP as potential targets for E. piscicida detection (Swain et al., 2001; Hong et al., 2009; Castro et al., 2010; Reichley et al., 2015). Among these, the EvpP gene is exclusive to E. piscicida, offering high specificity and diagnostic effectiveness. In the present study, the RT-RAA primers and probe were, therefore, designed based on the conserved region of EvpP gene.
Previous studies on viral and bacterial pathogens have indicated the detection limit of the RAA assay varied from 10 to 142 copies per reaction when tested against standard plasmids (Zhang et al., 2017; Cui et al., 2022; Fu et al., 2022; Lin et al., 2022; Cui H et al., 2023; Cui X et al., 2023). In this study, a novel and simple method for E. piscicida detection was proposed based on RAA technique. The well-established RT-RAA method exhibited excellent sensitivity, capable of detecting as few as 42 (0.14 fg) copies of standard plasmids per reaction, indicating a high level of sensitivity and consistent with the detection limit of the qPCR method. Additionally, specificity analysis confirmed that the established RT-RAA assay exhibited a notable level of specificity for its intended targets, as it did not exhibit any instances of cross-reactivity with other commonly found marine pathogenic bacteria. To evaluate the clinical feasibility of the RAA-based method, a total of 48 clinical and environmental samples were collected for E. piscicida detection. Both RT-RAA and qPCR techniques successfully identified 36 out of the 48 samples as positive, demonstrating a sensitivity and specificity of 100%. A strong agreement between the RT-RAA and qPCR assays was indicated by a κ value of 1.0 (p < 0.001). These findings indicate that the RT-RAA technique established in this research is highly appropriate for detecting E. piscicida infections in fish.
Taken together, in this study, we have successfully developed an RT-RAA assay that shows outstanding specificity and sensitivity. This assay offers a straightforward, expeditious, and dependable approach for detecting E. piscicida. The distinctive attributes of this RT-RAA assay render it particularly suitable for implementation in under-resourced diagnostic laboratories, thereby potentially facilitating prompt clinical detection and treatment in both clinical and onsite settings.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OR670479.1.
Ethics statement
The animal study was approved by Ethics Committee of Yantai University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YD: Investigation, Methodology, Writing – original draft. DZ: Writing – original draft, Formal analysis, Software. BZ: Writing – original draft, Data curation, Resources. XX: Resources, Funding acquisition, Supervision, Validation, Writing – review & editing. JZ: Funding acquisition, Writing – review & editing, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work is supported by the Municipal Planning Agriculture-related Projects of Yantai (2023), Key Laboratory of Mariculture of Ministry of Education, Ocean University of China, and the Development Plan of Youth Innovation Team in Colleges and Universities of Shandong Province (2022KJ269).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1355056/full#supplementary-material
References
Castro, N., Toranzo, A. E., Núñez, S., Osorio, C. R., Magariños, B. (2010). Evaluation of four polymerase chain reaction primer pairs for the detection of Edwardsiella tarda in turbot. Dis. Aquat. Organ. 90, 55–61. doi: 10.3354/dao02203
Cui, X., Du, B., Feng, J., Feng, Y., Cui, J., Yan, C., et al. (2023). Rapid detection of mpox virus using recombinase aided amplification assay. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1008783
Cui, H., Tu, F., Zhang, C., Zhang, C., Zhao, K., Liu, J., et al. (2022). Real-time reverse transcription recombinase-aided amplification assay for rapid amplification of the N gene of SARS-CoV-2. Int. J. Mol. Sci. 23, 15269. doi: 10.3390/ijms232315269
Cui, H., Zhang, C., Tu, F., Zhao, K., Kong, Y., Pu, J., et al. (2023). Rapid detection of influenza A viruses using a real-time reverse transcription recombinase-aided amplification assay. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.1071288
Fan, G., Zhang, R., He, X., Tian, F., Nie, M., Shen, X., et al. (2021). RAP: A novel approach to the rapid and highly sensitive detection of respiratory viruses. Front. Bioeng. Biotechnol. 9. doi: 10.3389/fbioe.2021.766411
Ferrari, N. A., Takashe, J. V. G., Aburjaile, F. F., Azevedo, V., da Costa, M. M., Brenig, B., et al. (2022). Genome report of emergent fish pathogen edwardsiella piscicida recovered from Pseudoplatystoma corruscans in Brazil. Microbiol. Resour. Announc. 11, e0079622. doi: 10.1128/mra.00796-22
Fu, H., Gan, L., Tian, Z., Han, J., Du, B., Xue, G., et al. (2022). Rapid detection of Burkholderia cepacia complex carrying the 16S rRNA gene in clinical specimens by recombinase-aided amplification. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.984140
Guerra, V., Beule, L., Lehtsaar, E., Liao, H. L., Karlovsky, P. (2020). Improved protocol for DNA extraction from subsoils using phosphate lysis buffer. Microorganisms 8, 532. doi: 10.3390/microorganisms8040532
Hardinge, P., Murray, J. A. H. (2019). Lack of specificity associated with using molecular beacons in loop mediated amplification assays. BMC. Biotechnol. 19, 55. doi: 10.1186/s12896-019-0549-z
Ho, B. T., Dong, T. ,. G., Mekalanos, J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21. doi: 10.1016/j.chom.2013.11.008
Hong, S. R., Choi, S. J., Jeong, H. D., Hong, S. (2009). Development of QCM biosensor to detect a marine derived pathogenic bacteria Edwardsiella tarda using a novel immobilisation method. Biosens. Bioelectron. 24, 1635–1640. doi: 10.1016/j.bios.2008.08.027
Hu, W. T., Anand, G., Sivaraman, J., Leung, K. Y., Mok, Y. K. (2014). A disordered region in the EvpP protein from the Type VI scretion system of Edwardsiella tarda is essential for EvpC binding. PloS One 9, e110810. doi: 10.1371/journal.pone.0110810
Kim, N., Lee, Y., Tho, H. D., Park, J. Y., Lee, J. Y., Kang, Y. R., et al. (2022). Whole-Genome Sequences of three Edwardsiella piscicida isolates from diseased fish in South Korea. Microbiol. Resour. Announc. 11, e0009722. doi: 10.1128/mra.00097-22
Leung, K. Y., Wang, Q., Yang, Z., Siame, B. A. (2019). Edwardsiella piscicida: A versatile emerging pathogen of fish. Virulence 10, 555–567. doi: 10.1080/21505594.2019.1621648
Lin, H., Zhao, S., Ye, Y., Shao, L., Jiang, N., Yang, K. (2022). A fluorescent recombinase aided amplification assay for detection of Babesia microti. Korean. J. Parasitol. 60, 201–205. doi: 10.3347/kjp.2022.60.3.201
Meyer, F. P., Bullock, G. L. (1973). Edwardsiella tarda, a new pathogen of channel catfish (Ictalurus punctatus). Appl. Microbiol. 25, 155–156. doi: 10.1128/am.25.1.155-156.1973
Pang, F., Long, Q. (2023). Recent advances in diagnostic approaches for orf virus. Appl. Microbiol. Biotechnol. 107, 1515–1523. doi: 10.1007/s00253-023-12412-8
Park, S. B., Aoki, T., Jung, T. S. (2012). Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet. Res. 43, 67. doi: 10.1186/1297-9716-43-67
Qin, Z., Xue, L., Cai, W., Gao, J., Jiang, Y., Yang, J., et al. (2021). Development of a recombinase-aided amplification assay for rapid detection of Human Norovirus GII.4. BMC. Infect. Dis. 21, 248. doi: 10.1186/s12879-021-05942-x
Reichley, S. R., Ware, C., Greenway, T. E., Wise, D. J., Griffin, M. J. (2015). Real-time polymerase chain reaction assays for the detection and quantification of Edwardsiella tarda, Edwardsiella piscicida, and Edwardsiella piscicida-like species in catfish tissues and pond water. J. Vet. Diagn. Invest. 27, 130–139. doi: 10.1177/1040638714566672
Sugiura, H., Fukunishi, K., Kawakami, H., Imajoh, M. (2022). Phenotypic differences between Edwardsiella piscicida and Edwardsiella Anguillarum isolates in Japan. J. Aquat. Anim. Health 34, 197–207. doi: 10.1002/aah.10169
Swain, P., Mukherjee, S. C., Sahoo, P. K., Das, B. K., Pattnaik, P., Murjani, G., et al. (2001). Dot-enzyme-linked immunosorbent assay (Dot-ELISA) for the diagnosis of Edwardsiella tarda infection in fish. Asian. Fish. Sci. 14, 89–93. doi: 10.33997/j.afs.2001.14.1.010
Tu, F., Yang, X., Xu, S., Chen, D., Zhou, L., Ge, X., et al. (2021). Development of a fluorescent probe-based real-time reverse transcription recombinase-aided amplification assay for the rapid detection of classical swine fever virus. Transbound Emerg. Dis. 68, 2017–2027. doi: 10.1111/tbed.13849
Wang, S., Huang, B., Ma, X., Liu, P., Wang, Y., Zhang, X., et al. (2020). Reverse-transcription recombinase-aided amplification assay for H7 subtype avian influenza virus. Transbound Emerg. Dis. 67, 877–883. doi: 10.1111/tbed.13411
Xie, G., Huang, J., Zhang, Q. L., Shi, C. Y., Wang, X. H., Liu, Q. H. (2013). Specific and rapid diagnosis of Edwardsiella tarda by a novel loop-mediated isothermal amplification targeting the upstream region of hlyb gene. J. Aquat. Anim. Health 25, 110–118. doi: 10.1080/08997659.2013.781555
Xie, H. X., Lu, J. F., Rolhion, N., Holden, D. W., Nie, P., Zhou, Y., et al. (2014). Edwardsiella tarda-induced cytotoxicity depends on its type III secretion system and flagellin. Infect. Immun. 82, 3436–3445. doi: 10.1128/IAI.01065-13
Xie, G., Zhang, Q., Han, N., Shi, C., Wang, X., Liu, Q., et al. (2012). An improved method for detection of Edwardsiella tarda by loop-mediated isothermal amplification by targeting the EsrB gene. Chin. J. Oceanol. Limnol. 30, 595–603. doi: 10.1007/s00343-012-1293-6
Zhang, X., Guo, L., Ma, R., Cong, L., Wu, Z., Wei, Y., et al. (2017). Rapid detection of Salmonella with recombinase aided amplification. J. Microbiol. Methods 139, 202–204. doi: 10.1016/j.mimet.2017.06.011
Zhang, L., Ni, C., Xu, W., Dai, T., Yang, D., Wang, Q., et al. (2016). Intramacrophage infection reinforces the virulence of edwardsiella tarda. J. Bacteriol. 198, 1534–1542. doi: 10.1128/JB.00978-15
Keywords: E. piscicida, aquaculture, recombinase-aided amplification, rapid diagnosis, isothermal amplification
Citation: Dong Y, Zhou D, Zhang B, Xu X and Zhang J (2024) Development of a real-time recombinase-aided amplification assay for rapid and sensitive detection of Edwardsiella piscicida. Front. Cell. Infect. Microbiol. 14:1355056. doi: 10.3389/fcimb.2024.1355056
Received: 13 December 2023; Accepted: 14 March 2024;
Published: 28 March 2024.
Edited by:
Banikalyan Swain, University of Florida, United StatesReviewed by:
Sukalyani Banik, Rutgers University, United StatesZhaocai Li, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Dong, Zhou, Zhang, Xu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Zhang, zhangjian@ytu.edu.cn; Xiaoying Xu, xiaoyingxu810052@163.com
 Yuchen Dong1
Yuchen Dong1  Dandan Zhou
Dandan Zhou Jian Zhang
Jian Zhang