Odorant binding protein 18 increases the pathogen resistance of the imported willow leaf beetle, Plagiodera versicolora
- State Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Sciences, Hubei University, Wuhan, China
Background: Insect odorant-binding proteins (OBPs) are a class of small molecular weight soluble proteins. In the past few years, OBPs had been found to work as carriers of ligands and play a crucial role in olfaction and various other physiological processes, like immunity. A subset of insect OBPs had been found to be expressed differently and play a function in immunity of fungal infection. However, there are few studies on the role of OBPs in immunity of bacterial infection.
Methods: To identify the immune-related OBPs of Plagiodera versicolora after infected by Pseudomonas aeruginosa, we determined the mortality of P. versicolora to P. aeruginosa and selected the time point of 50% mortality of larvae to collect samples for RNA-seq. RNAi technology was used to investigate the function of immune-related OBPs after P. aeruginosa infection.
Results: RNA-seq data shows that PverOBP18 gene significantly up-regulated by 1.8-fold and further RT-qPCR affirmed its expression. Developmental expression profile showed that the expression of PverOBP18 was highest in the pupae, followed by the female adults, and lower in the 1st-3rd larvae and male adults with lowest in eggs. Tissue expression profiling showed that PverOBP18 was dominantly expressed in the epidermis. RNAi knockdown of PverOBP18 significantly reduced the expression of bacterial recognition receptor gene PGRP and antibacterial peptide gene Attacin and reduced the resistance of P. versicolora to P. aeruginosa infection.
Conclusion: Our results indicated that PverOBP18 gene increased the pathogen resistance of P. versicolora by cooperating with the immune genes and provided valuable insights into using OBPs as targets to design novel strategies for management of P. versicolora.
1 Introduction
In the process of long-term evolution, a complex chemosensory system is necessary for insects to find mating partners, locate oviposition sites, forage for foods, and to avoid predators or toxic compounds in their natural environments (Field et al., 2000). Several major steps of the insect chemosensory process are necessary, such as odorant transmission, odorant recognition and insect behavioral guidance. Odorant-binding proteins (OBPs) originally are considered to take the lead in transporting chemical odors from environment to odorant receptors (ORs) of sensory cells, which is the first step of odor recognition (Rihani et al., 2021).
Insect OBP is a small molecular weight odorant carrier protein (15 – 20 kDa), containing approximately 120 to 150 amino acids, belonging to a large family of ligand-binding proteins, and is mainly secreted in sensillum lymph (Pelosi et al., 2018a; Pelosi et al., 2018b). A pheromone binding protein (PBP) found in the antennae of the male Antheraea polyphemus is the first member of the OBP family (Vogt and Riddiford, 1981). In the past decades, due to the rapid advancement of genome projects and transcriptome sequencing technologies, a vast number of OBPs have been discovered in various insect species (Venthur and Zhou, 2018; Pelosi et al., 2018a). The quantity of genes encoding OBPs varies significantly across species of insect, ranging from as few as 13 in silkworm Bombyx mori to approximately 109 in German cockroach Blattella germanica (Sun et al., 2018). However, only a few OBPs have been proved to be exclusively related to olfactory sensing, the functions of most OBPs in insects are still unclear.
Recently, many more OBPs have been reported to be endowed with different functions besides chemo-detection, including insecticide resistance, anti-inflammation and reproduction, as well as immunity (Xiong et al., 2019). For example, several studies indicated that the OBPs in insects were significantly induced by exogenous toxicants and gradually enhanced the resistance of insects to poisons (Gao et al., 2021; Chen et al., 2022). Besides insecticide resistance, the OBPs secreted in mosquito saliva were shown high binding affinity to host biogenic amines and displayed an anti-inflammatory function (Calvo et al., 2009). In addition, some OBPs were identified to be expressed not only in olfactory tissues but also in insect immune tissues such as hemocytes and fat bodies, and participated in innate immunity of insects (Thomas et al., 2016). Certain types of OBPs were found to be induced in insects which may be involved in resistance for fungal infection (Zhang et al., 2017; Zhang et al., 2020; Zheng et al., 2021). For example, in Locusta migratoria, the expression of OBP11 was negatively correlated with larval resistance to Metarhizium anisopliae by counteracting the innate immunity of the Toll-pathway (Zhang et al., 2023). These findings suggested that the OBPs played a crucial role in facilitating the immune responses of insects to combat pathogenic fungus.
The Pseudomonas aeruginosa is a Gram-negative bacterium and ubiquitous in the environment. It is an opportunistic pathogen and causes a wide range of infectious diseases in diverse host organisms, including insects and humans (Admella and Torrents, 2023; Varponi et al., 2023). Plagiodera versicolora (Coleoptera: Chrysomelidae), is a well-known forest pest that poses a threat to plants in the Salicaceae family (Li et al., 2023). In previous study, the chemosensory genes of P. versicolora had been identified based on transcriptome of larvae, adult antennas and forelegs, respectively. Collectively, a total of 40 candidate OBPs of P. versicolora had been identified (Liu et al., 2021; Wu et al., 2022a; Wu et al., 2022b).
However, there has still been a lack of research on the involvement of OBPs in immunity, particularly concerning bacterial infections. The P. aeruginosa was used to infect P. versicolora, in order to examine whether OBPs were altering during pathogen infection and carrying out roles in immunity. We first determined the pathogenicity of P. aeruginosa to P. versicolora and identified the PverOBP18 gene was immune-induced based on RNA-seq data. RT-qPCR results indicated that PverOBP18 is highly and specifically expressed in the epidermis. The synergistic toxicity in larvae of P. aeruginosa infection and PverOBP18 knockdown was significantly increased.
2 Materials and methods
2.1 Insect rearing, pathogen strain and pathogenic infection bioassays
The P. versicolora larvae and adults were obtained from Sha Lake Park in Wuhan, China and reared with fresh leaves of willows in laboratory at 28 ± 1°C, under a photoperiod of 12:12 h (light: dark) and 70 ± 5% RH (relative humidity). Strain of P. aeruginosa was stored at – 80°C and cultured in Luria-Bertani sterile liquid medium at 37°C for 12 h. OD600 was measured to calculate the concentration of P. aeruginosa and adjusted it by sterile water to approximately 2.0 for feeding assay. Fresh willow leaves were cut into rectangle and areas were 6 cm2. The bacteria solution was added onto the surface of fresh willow leaves for 60 μl and painted repeatedly for several times by plastic coating bar. Fresh willow leaves were air dried with the bacteria concentration was 10 μl/cm2 and then fed to the 2nd instar P. versicolora larvae that had been deprived of food for 4 h. Synchronized groups of larvae were selected and divided into three groups (each group containing 20 individuals and serving as a biological replicate).
For all pathogenic infection bioassays, the concentrations of the bacteria solutions were consistent with those described above. The experiments were conducted in the artificial climate chamber under controlled conditions as previously described, with willow leaves replaced and survival rate recorded daily.
2.2 RNA extraction, cDNA library construction, and Illumina sequencing
Three time points were selected to collected samples, first day, time to reach 10% and 50% mortality. Time to reach 10% and 50% mortality were the time point that the mortality rates of larvae were nearly 10% and 50% (Ma et al., 2021). Considering the dynamics of pathogenicity and lethality in P. aeruginosa, the infected group was sampled at day 1, day 2 (time to reach 10% mortality), day 5 (time to reach 50% mortality) after infection and the H2O treatment group was sampled at the same time points as controls. Meanwhile, the samples of day 5 were prepared for RNA-seq and the samples of day 1, day 2 and day 5 were prepared for RT-qPCR. Five live larvae in each group were pooled as one biological replication. The whole body of larva was sampled in each treatment.
After collection, all samples were promptly frozen in liquid nitrogen and kept at a temperature of – 80°C for RNA extraction. The total RNA of all collected samples was extracted using Trizol Reagent (Invitrogen, USA) according to protocol. The quality of RNA was checked with a NanoDrop-2000 (Thermo Scientific, USA). The Illumina sequencing of the samples was performed by Majorbio (China). The cDNA library was synthetized with NEBNext ® Ultra mRNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s instructions and previous study (Liu et al., 2021).
2.3 Sequencing assembly, annotation and gene expression analysis
After removing the raw reads containing the adaptor sequences, low-quality reads (Quality score < 20), and repeated reads, the clean reads were obtained. The transcriptome was assembled according to these clean reads by using Trinity 2.8.5 to generate a set of transcripts. These transcripts were annotated according to the following databases: non-redundant (NR) protein database, Swiss-prot, KEGG, KOG/COG, and a search conducted in the National Center for Biotechnology Information (NCBI). A differentially expressed gene (DEG) analysis was performed based on the significance level using the R package, DEGseq2 (p-adjust < 0.05, fold change > 2).
2.4 Spatiotemporal gene expression analyses by RT-qPCR
Different stages of P. versicolora including eggs, 1st to 3rd instar larvae, pupae, male and female adults were collected respectively. Different tissues of P. versicolora including epidermis, foregut, midgut, hindgut and Malpighian tubule were dissected from 3rd instar larvae, respectively (Li et al., 2022). Each sample was studied using three biological replicates. All samples were immediately frozen in liquid nitrogen and stored at – 80°C until used. Following the extraction of RNA, a reverse transcription reaction was carried out using 1 µg of total RNA. The HiScript III RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China) was utilized for cDNA synthesis following the guidelines from the manufacturer. The housekeeping gene RPS18 was chosen as a reference gene for internal control. Reaction mixtures, reaction protocols and gene expression calculation methods for RT-qPCR have been reported before (Liao et al., 2023).
2.5 In vitro dsRNA synthesis, RNAi and synergistic toxic bioassays
First, dsRNA template was prepared by PCR using specific primers containing the T7 promoter sequence (Supplementary Table S1;10 µM) and purified with a gel extraction kit (Omega, China). Second, dsRNA was synthesized in vitro using the T7 RNAi Transcription Kit (Vazyme, China) according to the manufacturer’s instructions and previous study (He et al., 2020). Following the production of dsRNA, two continuous experiments were conducted for synergistic toxic bioassays. First, 1st P. versicolora larvae were deprived of food for 4 h before the start of RNAi. Identical amounts (96 ng, in 50 μl H2O) of dsRNAs were applied onto fresh willow leaves, covering a consistent area of 6 cm2, thus adjusting the concentration of dsRNA to 16 ng/cm2. The insects were provided with an adequate amount of fresh leaf coated with dsRNA for RNAi or H2O for control. The leaves were replaced daily for three days to ensure that the foliar-applied dsRNA remained stable and target genes were knocked down. Samples were collected on day 3 for RT-qPCR to evaluate the silencing efficiency of RNAi. Second, synchronized groups of 2nd larvae that dsRNA-treated or not were selected, divided into three groups (each group containing 10 individuals and serving as a biological replicate) for pathogenic infection bioassays, and the operations were the same as mentioned above. Samples were collected on day 4 for RT-qPCR to analysis the expression patterns of immune-related genes.
2.6 Statistical analysis
The Kaplan-Meier method was utilized to analyze survival curves. The log-rank test was employed to assess the significance of differences between the two groups. The t-test was utilized to analyze the data for bacteria induced genes expression and RNAi efficiency by RT-qPCR. The ANOVA was employed to analyze the data for the expression of immunity-related genes by RT-qPCR. A value of P < 0.05 meant significantly different. All statistical analyses were performed using SPSS version 22. Figures were drawn with GraphPad Prism 8.
3 Results
3.1 Survival analysis of P. versicolora larvae infected with P. aeruginosa
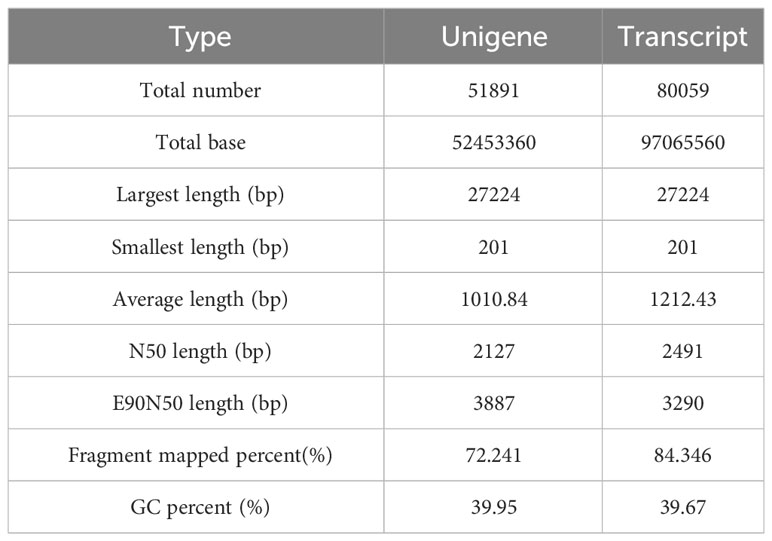
The survivorship of larvae infected by P. aeruginosa was significantly lower than that of control group (Figure 1A), demonstrating that P. versicolora is very sensitive to this pathogen. After P. aeruginosa infection, the mortality rates of P. versicolora on day 2 and day 5 were 8.3% and 56.7%, respectively, which were close to 10% and 50%. In comparison, the P. versicolora exhibited high survivorship (up to 81.4%) at day 5 in the control group (Figure 1A). Therefore, including the first day after P. aeruginosa infection, three time points (day 1, day 2, day 5) were selected for further study.
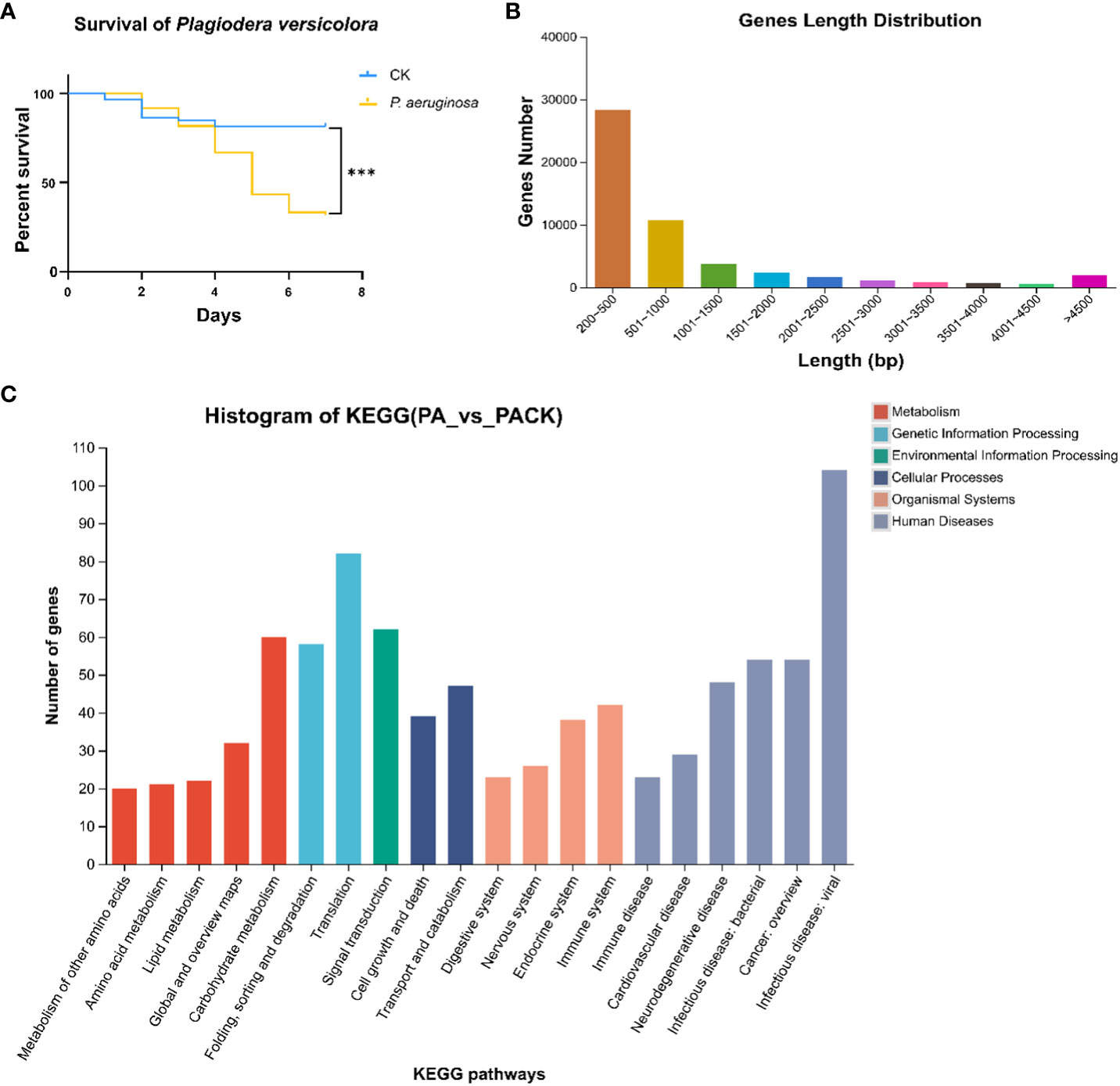
Figure 1 The survival curves and transcriptomic changes of P. versicolora larvae infected with P. aeruginosa (A) Kaplan–Meier survival curves of second-instar P. versicolora larvae treated with P. aeruginosa. The log-rank test was used to assess the significance of differences between two survival curves. *** P < 0.001. (B) Distribution of unigene size in the P. versicolora transcriptome assembly (C) KEGG classification of differentially expressed unigenes between control (PACK) group and P. aeruginosa infected (PA) group.
3.2 Overview of the sequence assembly
The cDNA library from P. versicolora larvae, either infected or not infected with P. aeruginosa on day 5, was constructed using the Illumina NovaSeq platform for next-generation sequencing. A total of 59,629,750 clean reads were acquired, with a Q20 percentage of 97.84%. About 51,891 unigenes were discovered, with a combined length of 52453360 bp and an N50 length of 2127 bp (Table 1). Statistics showed that 75.2% of the 51,891 unigenes were lower than 1000 bp in length (Figure 1B). In total, 20,439 unigenes were matched to entries in the NCBI NR protein database (http://www.ncbi.nlm.nih.gov/protein (accessed on 10 November 2023)) by a BLASTX search.
3.3 Functional classification of KEGG annotation
Overall, the differentially expressed unigenes were categorized into six functional groups based on their KEGG annotation, including environmental information processing (156), human diseases (667), genetic information processing (187), cellular processes (158), organismal systems (347), and metabolism (318). In the functional group of organismal systems, the most differentially expressed genes were found to be associated with the immune system (42) and the endocrine system (38). In the functional group of environmental information processing, the most differentially expressed genes were found to be associated with the signal transduction (62) (Figure 1C). The results indicating that some unigenes in these sub-categories might have a connection with chemosensory behavior and immunity in insects.
3.4 PverOBP15 and PverOBP18 expression were activated during P. aeruginosa infection
The transcriptomic studies revealed 11 candidate OBP genes, 2 of which were differentially expressed. PverOBP15 and PverOBP18 genes were up-regulated in larvae infected with P. aeruginosa, when compared to the control group (Figure 2A). In order to investigate whether the transcript level of PverOBP15 and PverOBP18 could be triggered by P. aeruginosa infection, the expression profiles of these genes were conducted using RT-qPCR at the three previously mentioned time points. The results revealed that the PverOBP15 gene expression was significantly increased by P. aeruginosa infection comparing to control only on day 2, but not day 1 and day 5 (Supplementary Figure S1). Unlike PverOBP15, the expression of PverOBP18 was significantly downregulated on day 1. As time went on, the expression of PverOBP18 was upregulated and significance showed on day 5 (Figure 2B).
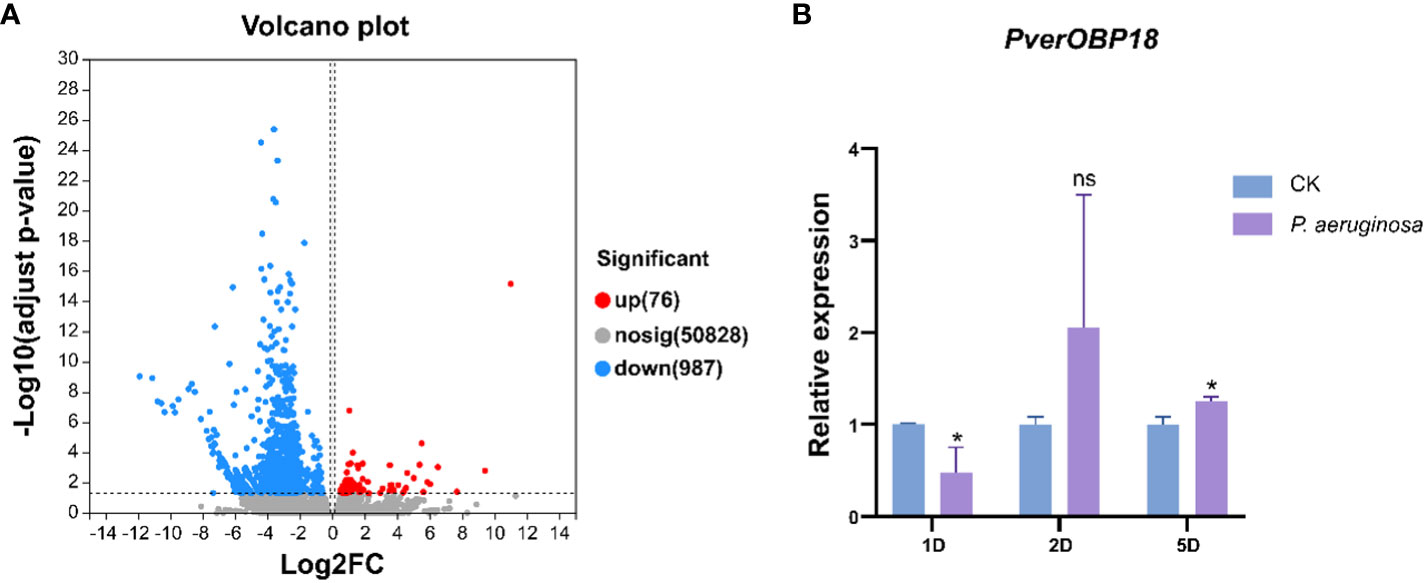
Figure 2 Differential expression analysis between control (PACK) group and P. aeruginosa infected (PA) group and the expression profiles of up-regulated gene PverOBP18. (A) Volcano plot. The log2 FC indicates the mean expression level for each gene. Each dot represents one gene. After P. aeruginosa infection, gray dots represent no significant unigenes between PACK and PA, the blue dots represent down-regulated genes and red dots represent up-regulated genes. (B) Expression levels of PverOBP18 gene at three time points, day 1 (1D), day 2 (2D) and day 5 (5D) assessed by RT-qPCR and normalized to the reference gene RPS18 expression level. Data are means ± SD (n = 3). The letters above the bar indicate the significance of differences as determined by t-test. ns, not significant; * P < 0.05.
3.5 Expression profiles of PverOBP15 and PverOBP18
The relative expression of PverOBP15 and PverOBP18 transcripts in different body segments and developmental stages were investigated using RT-qPCR. PverOBP15 transcript was expressed highest in 3rd larvae and female adults, lower in pupae, and lowest in eggs, 1st larvae, 2nd larvae and male adults (Supplemetary Figure S2A). Unlike PverOBP15, PverOBP18 transcript was expressed highest in pupae, followed by female adults, and lower in 1st-3rd larvae and male adults with lowest in eggs (P < 0.05, LSD test) (Figure 3A).
Figure 3 Temporal and spatial expression profiles of PverOBP18. (A) Expression levels of PverOBP18 gene at different development stages. L1 - L3, 1st instar larvae – 3rd instar larvae; MA, male adults; FA, female adults. (B) Expression levels of PverOBP18 gene at different tissues of 3rd instar larvae. EP, epidermis; FG, foregut; MG, midgut; HG, hindgut; FB, fat body; MT, Malpighian tubule; Relative expression levels were analyzed using RT-qPCR and normalized to the reference gene RPS18 expression level. Data are means ± SD (n = 3). The letters above the bar indicate the significance of differences as determined by one-way ANOVA (LSD, P < 0.05).
In different tissues, PverOBP15 transcript was expressed highest in the fat body, followed by the foregut, and lowest in the epidermis, midgut, hindgut, and Malpighian tubule (Supplemetary Figure S2B). Unlike PverOBP15, the expression of PverOBP18 transcripts was highest in the epidermis, lower in the midgut, hindgut, fat body, and Malpighian tubule, and lowest in foregut (P < 0.05, LSD test) (Figure 3B).
3.6 P. versicolora larvae were sensitive to P. aeruginosa infection after knockdown of PverOBP18
For the purpose of investigating the role of PverOBP18 and PverOBP15 in response to P. aeruginosa infection, the dsRNAs for gene knockdown of PverOBP18 and PverOBP15 were synthesized in vitro and fed to P. versicolora larvae. Analyzing the efficiency of RNAi in P. versicolora, the results revealed that both PverOBP15 and PverOBP18 expression were reduced by nearly 65% after fed dsRNA for 3 days (Supplementary Figure S3A; Figure 4A). Survival curves showed that the larval mortality of dsPverOBP18 + ; P. aeruginosa treated group was significantly higher than that of the P. aeruginosa treated group, while the mortality of dsPverOBP18 treated group and untreated group were relatively low. Unlike dsPverOBP18 treatment, no significant difference was found between the dsPverOBP15 + P. aeruginosa treated group and ; P. aeruginosa treated group. The results revealed a reduce in the resistance of P. versicolora to bacterial infection after the knockdown of PverOBP18, but not PverOBP15 (Supplementary Figure S3B; Figure 4B).
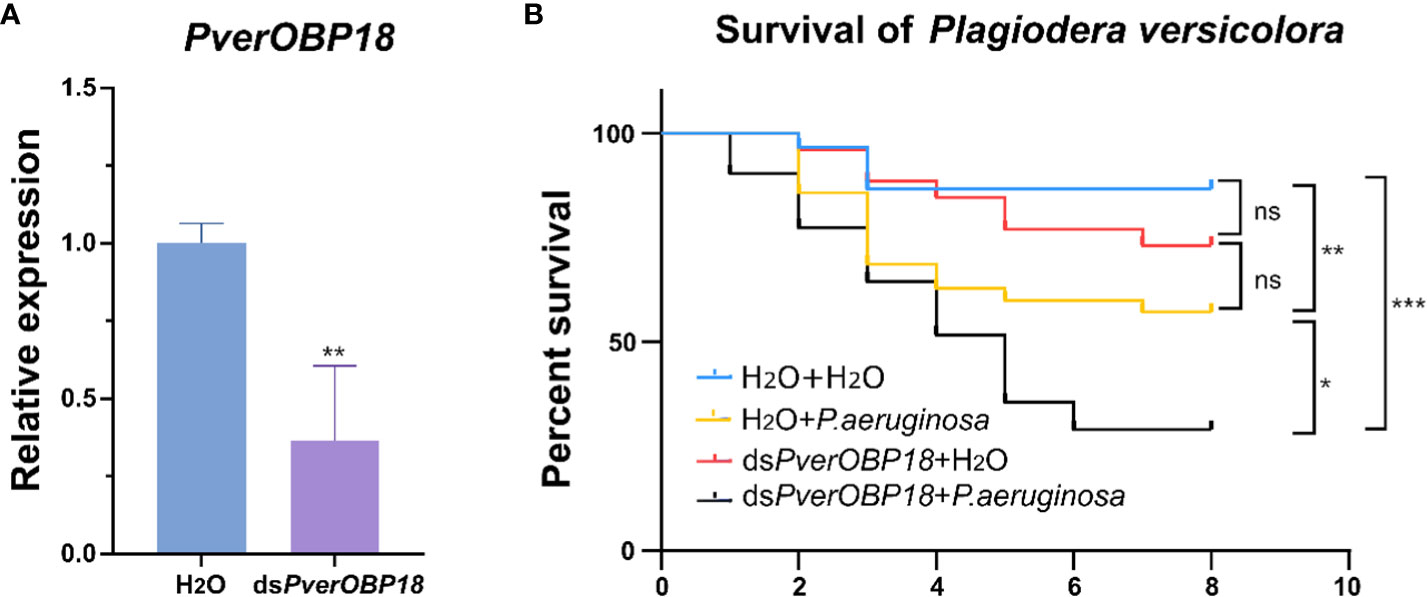
Figure 4 The synergistic effect of dsRNA and the entomopathogenic P. aeruginosa on P. versicolora larvae. (A) Silencing efficiency of PverOBP18 in P. versicolora treated with dsPverOBP18. RPS18 was used as a reference gene to normalize the relative expression of the indicated genes. Data are means ± SD (n = 3). The letters above the bar indicate the significance of differences as determined by t-test. ** P < 0.01. (B) Comparison of mortality of P. aeruginosa infected P. versicolora in response to the administration of dsPverOBP18. Kaplan–Meier survival curves of second-instar P. versicolora larvae. The log-rank test was used to assess the significance of differences between two survival curves. ns, not significant; *** P < 0.001; ** P < 0.01; * P < 0.05.
3.7 Validation of immunity-related genes expression patterns by RT-qPCR
To determine which immunity-related genes were involved in PverOBP18 mediated resistance to P. aeruginosa, genes in immune effectors (Attacin, Defensin, Lysosome), signal modulation (Serpin), recognition receptors (PGRP) and Toll pathway (Toll-1) were selected to analysis the expression patterns by RT-qPCR. The results revealed that PGRP gene expression in the P. aeruginosa treated group was significantly increased, compared with the dsPverOBP18 + P. aeruginosa treated group, dsPverOBP18 treated group and untreated group. Attacin gene showed a similar expression patten like PGRP gene, although there were no notable alterations detected between the P. aeruginosa treated group and the dsPverOBP18 + P. aeruginosa treated group. Furthermore, there were no notable alterations detected in the expression of other four immunity-related genes (Figure 5).
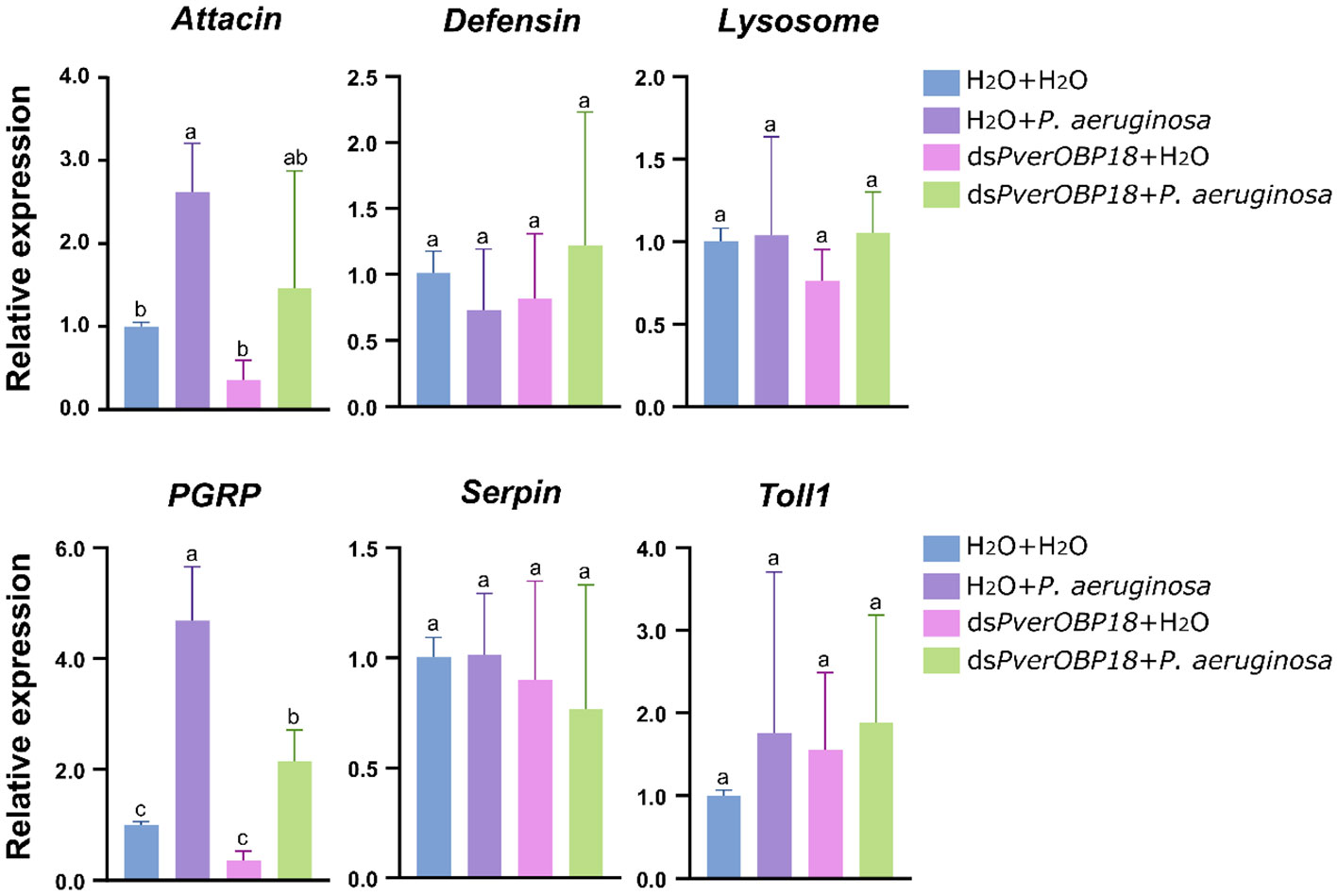
Figure 5 Immunity-related genes analysis of PverOBP18-silenced and H2O control P. versicolora larvae to P. aeruginosa infection by RT-qPCR. PGRP, peptidoglycan-recognition protein. RPS18 was used as a reference gene to normalize the relative expression of the indicated genes. Data are means ± SD (n = 3). The letters above the bar indicate the significance of differences as determined by one-way ANOVA (LSD, P < 0.05).
4 Discussion
In this study, PverOBP18 gene expression was significantly downregulated on day 1 after P. aeruginosa infection. As time went on, the expression of PverOBP18 was upregulated and the significance showed on day 5, suggesting that PverOBP18 may be involved in P. aeruginosa resistance (Figure 2B). In nature, because insects are exposed to finite resources and the energetic requirements to trigger immune defense are high, trade-offs often occur between the immune system and other life history traits, such as development, reproduction and physiology (Ardia et al., 2012; Dolezal et al., 2019). For example, in Drosophila melanogaster, a series of genes were characterized by a strong decrease in expression at the initial period followed by an increase after the pathogen infection (Schlamp et al., 2021). In Pieris napi, several types of immune responses were upregulated during the initial period of Micrococcus luteus infection, whereas all other non-immune processes were strongly downregulated (Keehnen et al., 2020). OBPs in insects play important role in binding and transporting chemical odors to odorant receptor neurons (ORNs) through the sensillum lymph during olfaction (Leal, 2013). Therefore, the expression patten of PverOBP18 induced by P. aeruginosa might be a tradeoff between olfactory and immune responses in P. versicolora.
In recent years, numerous OBPs in insects had been found to express in both larva and adult (Xue et al., 2016; Liu et al., 2020; Tian et al., 2023). In this study, our results showed that PverOBP18 gene expression was highest in the pupae, followed by the female adults, and lower in the 1st-3rd larvae and male adults with lowest in eggs. (Figure 3A). This indicating that PverOBP18 may play an important role in various biologic processes during the life stage of P. versicolora. Usually, the majority of OBP genes are found to be expressed in chemosensory organs of insectile adults, including the antennae, maxillary palp and proboscis, such as P. versicolora (Liu et al., 2021), Tribolium castaneum (Dippel et al., 2014) and Rhynchophorus ferrugineus (Huang et al., 2023). Previous study had found that PverOBP18 was mainly expressed in the antennae of adult P. versicolora when compared to other tissues, indicating the similar chemoreception function in P. versicolora (Liu et al., 2021). However, studies have shown that OBPs are present in various insect tissues and serve functions beyond chemoreception. For example, in T. castaneum, OBPC12 genes was dominantly expressed in the epidermis and displayed high binding affinity to exogenous toxic substances (Gao et al., 2021). Many OBPs in Lygus lineolaris demonstrated high levels of expression in the legs and proboscis, indicating their potential roles in chemical detection (Hull et al., 2014). In our study, PverOBP18 was dominantly expressed in epidermis of 3rd instar larvae, which is an integral part of immune systems and plays a vital role as the first line of defense in insects (Figure 3B). These might be the reason why PverOBP18 increases the pathogen resistance of P. versicolora larvae.
In this study, both PverOBP15 and PverOBP18 gene expression were activated during P. aeruginosa infection (Figure 2B; Supplementary Figure S1). And the expression of PverOBP15 and PverOBP18 in P. versicolora were both significantly suppressed after dsRNA treatment (Figure 4A; Supplementary Figure S3A). However, a significant reduction was showed in P. versicolora resistance to P. aeruginosa infection just followed by PverOBP18 RNAi knockdown, but not PverOBP15 (Figure 4B; Supplementary Figure S3B). In our comparative analysis, the expression levels of PverOBP18 were higher than PverOBP15 in almost all tested tissues and developmental stages (Figure 3; Supplementary Figure S2). In consideration of the expression level of PverOBP15 and PverOBP18, these might be the reason why the knockdown of PverOBP15 was useless.
In insects, immune resistance to foreign pathogenic microorganisms relies on the recognition of conserved microbial structures of the microorganisms. For example, peptidoglycan-recognition proteins (PGRP) in insects can recognize bacterial surfaces and activate relevant immune pathways to resist bacterial invasion. Recognition of the pathogen leads to downstream reactions, such as antimicrobial peptide synthesis (Eleftherianos et al., 2022). In this study, our results showed that the PGRP gene expression level of P. versicolora treated with dsPverOBP18 + P. aeruginosa could not reach as high as P. versicolora infected by P. aeruginosa alone (Figure 5). In addition, the Attacin gene expression showed a similar result like PGRP gene, which is an antimicrobial peptide gene. This means PverOBP18 gene might relate to PGRP gene to influence the recognition of P. versicolora to P. aeruginosa and further to influence the synthesis of antimicrobial peptide Attacin.
In conclusion, the RT-qPCR results showed that the PverOBP18 gene is highly and specifically expressed in the epidermis. P. aeruginosa infection and PverOBP18 knockdown significantly increased the synergistic mortality in larvae, indicating that the PverOBP18 gene was clearly triggered. We think these results are broadly relevant and significant, because they (i) show PverOBP18 is collaborating with immune-related genes to increase P. versicolora anti-bacterial resistance, (ii) offer insightful information about using OBPs as targets to develop creative P. versicolora management strategies, and (iii) deepen our understanding of OBPs’ functions beyond chemoreception (Figure 6).
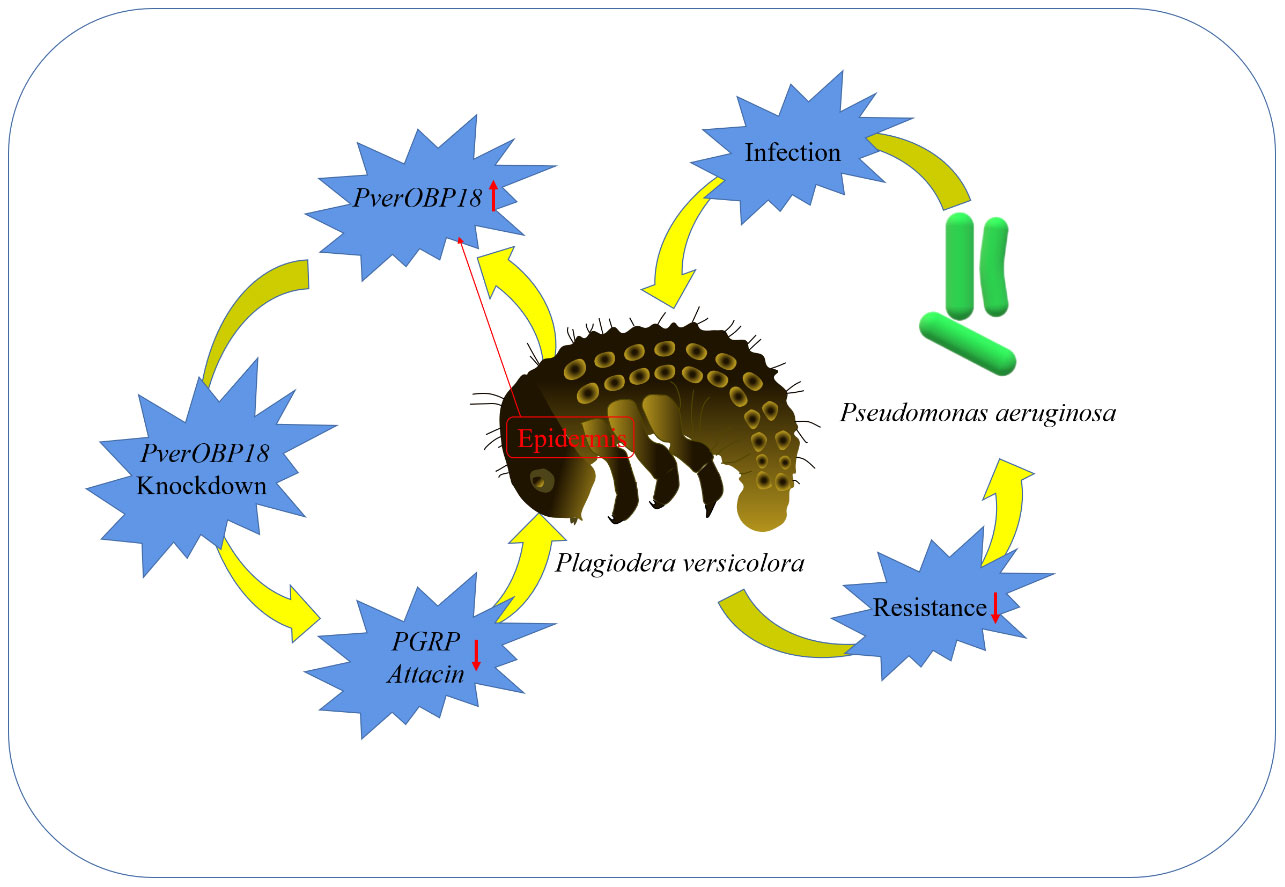
Figure 6 A simple diagram illustrating the mechanism and outcome of how the studied OBP increased the resistance of P. versicolora against P. aeruginosa..
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/biosample/39742880, SAMN39742880 https://www.ncbi.nlm.nih.gov/biosample/39742881, SAMN39742881 https://www.ncbi.nlm.nih.gov/biosample/39742882, SAMN39742882 https://www.ncbi.nlm.nih.gov/biosample/39742883, SAMN39742883 https://www.ncbi.nlm.nih.gov/biosample/39742884, SAMN39742884 https://www.ncbi.nlm.nih.gov/biosample/39742885, SAMN39742885 https://www.ncbi.nlm.nih.gov/biosample/39742886, SAMN39742886 https://www.ncbi.nlm.nih.gov/biosample/39742887, SAMN39742887 https://www.ncbi.nlm.nih.gov/biosample/39742888, SAMN39742888 https://www.ncbi.nlm.nih.gov/biosample/39742889, SAMN39742889 https://www.ncbi.nlm.nih.gov/biosample/39742890, SAMN39742890 https://www.ncbi.nlm.nih.gov/biosample/39742891, SAMN39742891.
Author contributions
HR: Writing – review & editing, Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft. XH: Investigation, Writing – original draft. YL: Resources, Writing – original draft. MeL: Resources, Writing – original draft. XL: Funding acquisition, Methodology, Writing – review & editing. MiL: Funding acquisition, Writing – review & editing, Project administration.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (32301593) and the Hubei University national talent project (1070017364).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1360680/full#supplementary-material
References
Admella, J., Torrents, E. (2023). Investigating bacterial infections in Galleria mellonella larvae: Insights into pathogen dissemination and behavior. J. Invertebr Pathol. 200, 107975. doi: 10.1016/j.jip.2023.107975
Ardia, D. R., Gantz, J., Brent, C. S., Schneider, Strebel, S. (2012). Costs of immunity in insects: an induced immune response increases metabolic rate and decreases antimicrobial activity. Funct. Ecol. 26, 732–739. doi: 10.1111/j.1365-2435.2012.01989.x
Calvo, E., Mans, B. J., Ribeiro, J. M., Andersen, J. F. (2009). Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc. Natl. Acad. Sci. U.S.A. 106, 3728–3733. doi: 10.1073/pnas.0813190106
Chen, X., Yang, H., Wu, S., Zhao, W., Hao, G., Wang, J., et al. (2022). BdorOBP69a is involved in the perception of the phenylpropanoid compound methyl eugenol in oriental fruit fly (Bactrocera dorsalis) males. Insect Biochem. Mol. Biol. 147, 103801. doi: 10.1016/j.ibmb.2022.103801
Dippel, S., Oberhofer, G., Kahnt, J., Gerischer, L., Opitz, L., Schachtner, J., et al. (2014). Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genomics 15, 1141. doi: 10.1186/1471-2164-15-1141
Dolezal, T., Krejcova, G., Bajgar, A., Nedbalova, P., Strasser, P. (2019). Molecular regulations of metabolism during immune response in insects. Insect Biochem. Mol. Biol. 109, 31–42. doi: 10.1016/j.ibmb.2019.04.005
Eleftherianos, I., Tafesh-Edwards, G., Mohamed, A. (2022). Pathogen infection routes and host innate immunity: Lessons from insects. Immunol. Lett. 247, 46–51. doi: 10.1016/j.imlet.2022.05.006
Field, L. M., Pickett, J. A., Wadhams, L. J. (2000). Molecular studies in insect olfaction. Insect Mol. Biol. 9, 545–551. doi: 10.1046/j.1365-2583.2000.00221.x
Gao, S., Lu, R., Zhang, Y., Sun, H., Li, S., Zhang, K., et al. (2021). Odorant binding protein C12 is involved in the defense against eugenol in Tribolium castaneum. Pestic Biochem. Physiol. 179, 104968. doi: 10.1016/j.pestbp.2021.104968
He, W., Xu, W., Xu, L., Fu, K., Guo, W., Bock, R., et al. (2020). Length-dependent accumulation of double-stranded RNAs in plastids affects RNA interference efficiency in the Colorado potato beetle. J. Exp. Bot. 71, 2670–2677. doi: 10.1093/jxb/eraa001
Huang, Y., Hu, W., Hou, Y. M. (2023). Host plant recognition by two odorant-binding proteins in Rhynchophorus ferrugineus (Coleoptera: Curculionidae). Pest Manag. Sci. 79 (11), 4521–4534. doi: 10.1002/ps.7654
Hull, J. J., Perera, O. P., Snodgrass, G. L. (2014). Cloning and expression profiling of odorant-binding proteins in the tarnished plant bug, Lygus lineolaris. Insect Mol. Biol. 23, 78–97. doi: 10.1111/imb.12064
Keehnen, N. L. P., Kučerová, L., Nylin, S., Theopold, U., Wheat, C. W. (2020). Physiological tradeoffs of immune response differs by infection type in Pieris napi. Front. Physiol. 11. doi: 10.3389/fphys.2020.576797
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Li, B., Liu, F., He, X., Liu, X., Lu, M. (2023). Temporal transcriptomic changes in willow leaves oviposited by Plagiodera versicolora. Integr. Zool. 00, 1–4. doi: 10.1111/1749-4877.12797
Li, Y., Ze, L. J., Liu, F. J., Liao, W., Lu, M., Liu, X. L. (2022). RNA interference of vATPase subunits A and E affects survival of larvae and adults in Plagiodera versicolora (Coleoptera: Chrysomelidae). Pestic Biochem. Physiol. 188, 105275. doi: 10.1016/j.pestbp.2022.105275
Liao, J., Rong, H., You, L., Xia, K., Wang, M., Han, P., et al. (2023). Identification of leaf chloroplast-specific promoter to efficiently control of Colorado potato beetle with reduced dsRNA accumulation in potato tubers. Pest Manag Sci. 79, 3326–3333. doi: 10.1002/ps.7516
Liu, X., Tong, N., Wu, Z., Li, Y., Ma, M., Liu, P., et al. (2021). Identification of chemosensory genes based on the antennal transcriptomic analysis of Plagiodera versicolora. Insects 13 (1), 36. doi: 10.3390/insects13010036
Liu, X. Q., Jiang, H. B., Liu, Y., Fan, J. Y., Ma, Y. J., Yuan, C. Y., et al. (2020). Odorant binding protein 2 reduces imidacloprid susceptibility of Diaphorina citri. Pestic Biochem. Physiol. 168, 104642. doi: 10.1016/j.pestbp.2020.104642
Ma, M., Guo, L., Tu, C., Wang, A., Xu, L., Luo, J. (2021). Comparative analysis of Adelphocoris suturalis jakovlev (Hemiptera: Miridae) immune responses to fungal and bacterial pathogens. Front. Physiol. 12. doi: 10.3389/fphys.2021.646721
Pelosi, P., Iovinella, I., Zhu, J., Wang, G., Dani, F. R. (2018a). Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb Philos. Soc. 93, 184–200. doi: 10.1111/brv.12339
Pelosi, P., Zhu, J., Knoll, W. (2018b). Odorant-binding proteins as sensing elements for odour monitoring. Sensors (Basel) 18 (10), 3248. doi: 10.3390/s18103248
Rihani, K., Ferveur, J. F., Briand, L. (2021). The 40-year mystery of insect odorant-binding proteins. Biomolecules 11 (4), 509. doi: 10.3390/biom11040509
Schlamp, F., Delbare, S. Y. N., Early, A. M., Wells, M. T., Basu, S., Clark, A. G. (2021). Dense time-course gene expression profiling of the Drosophila melanogaster innate immune response. BMC Genomics 22, 304. doi: 10.1186/s12864-021-07593-3
Sun, J. S., Xiao, S., Carlson, J. R. (2018). The diverse small proteins called odorant-binding proteins. Open Biol. 8, 180208. doi: 10.1098/rsob.180208
Thomas, T., De, T. D., Sharma, P., Lata, S., Saraswat, P., Pandey, K. C., et al. (2016). Hemocytome: deep sequencing analysis of mosquito blood cells in Indian malarial vector Anopheles stephensi. Gene 585, 177–190. doi: 10.1016/j.gene.2016.02.031
Tian, Z., Li, R., Cheng, S., Zhou, T., Liu, J. (2023). The Mythimna separata general odorant binding protein 2 (MsepGOBP2) is involved in the larval detection of the sex pheromone (Z)-11-hexadecenal. Pest Manag Sci. 79, 2005–2016. doi: 10.1002/ps.7373
Varponi, I., Ferro, S., Menilli, L., Grapputo, A., Moret, F., Mastrotto, F., et al. (2023). Fighting Pseudomonas aeruginosa infections: antibacterial and antibiofilm activity of D-Q53 cecB, a synthetic analog of a silkworm natural cecropin B variant. Int. J. Mol. Sci. 24 (15), 12496. doi: 10.3390/ijms241512496
Venthur, H., Zhou, J. J. (2018). Odorant receptors and odorant-binding proteins as insect pest control targets: A comparative analysis. Front. Physiol. 9. doi: 10.3389/fphys.2018.01163
Vogt, R. G., Riddiford, L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293, 161–163. doi: 10.1038/293161a0
Wu, Z. R., Fan, J. T., Tong, N., Guo, J. M., Li, Y., Lu, M., et al. (2022b). Transcriptome analysis and identification of chemosensory genes in the larvae of Plagiodera versicolora. BMC Genomics 23, 845. doi: 10.1186/s12864-022-09079-2
Wu, Z., Tong, N., Li, Y., Guo, J., Lu, M., Liu, X. (2022a). Foreleg transcriptomic analysis of the chemosensory gene families in plagiodera versicolora (Coleoptera: chrysomelidae). Insects 13 (9), 763. doi: 10.3390/insects13090763
Xiong, W., Gao, S., Lu, Y., Wei, L., Mao, J., Xie, J., et al. (2019). Latrophilin participates in insecticide susceptibility through positively regulating CSP10 and partially compensated by OBPC01 in. Tribolium castaneum. Pestic Biochem. Physiol. 159, 107–117. doi: 10.1016/j.pestbp.2019.06.005
Xue, W., Fan, J., Zhang, Y., Xu, Q., Han, Z., Sun, J., et al. (2016). Identification and expression analysis of candidate odorant-binding protein and chemosensory protein genes by antennal transcriptome of sitobion avenae. PloS One 11, e0161839. doi: 10.1371/journal.pone.0161839
Zhang, W., Chen, J., Keyhani, N. O., Jin, K., Wei, Q., Xia, Y. (2017). Central Nervous System Responses of the Oriental migratory, Locusta migratoria manilensis, to Fungal Infection. Sci. Rep. 7, 10340. doi: 10.1038/s41598-017-10622-5
Zhang, W., Xie, M., Eleftherianos, I., Mohamed, A., Cao, Y., Song, B., et al. (2023). An odorant binding protein is involved in counteracting detection-avoidance and Toll-pathway innate immunity. J. Adv. Res. 48, 1–16. doi: 10.1016/j.jare.2022.08.013
Zhang, W., Zheng, X., Chen, J., Keyhani, N. O., Cai, K., Xia, Y. (2020). Spatial and temporal transcriptomic analyses reveal locust initiation of immune responses to Metarhizium acridum at the pre-penetration stage. Dev. Comp. Immunol. 104, 103524. doi: 10.1016/j.dci.2019.103524
Keywords: odorant binding protein, insect immunity, pathogen infection, pathogen resistance, Plagiodera versicolora, Pseudomonas aeruginosa
Citation: Rong H, He X, Liu Y, Liu M, Liu X and Lu M (2024) Odorant binding protein 18 increases the pathogen resistance of the imported willow leaf beetle, Plagiodera versicolora. Front. Cell. Infect. Microbiol. 14:1360680. doi: 10.3389/fcimb.2024.1360680
Received: 23 December 2023; Accepted: 30 January 2024;
Published: 27 February 2024.
Edited by:
Xin-Ru Wang, Upstate Medical University, United StatesReviewed by:
Yingjun Cui, Yale University, United StatesKaiying Chen, North Carolina State University, United States
Amr A. Mohamed, Cairo University, Egypt
Copyright © 2024 Rong, He, Liu, Liu, Liu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolong Liu, bruceliu2021@outlook.com; Min Lu, lumin@hubu.edu.cn
 Haoling Rong
Haoling Rong  Yipeng Liu
Yipeng Liu Mei Liu
Mei Liu Xiaolong Liu
Xiaolong Liu Min Lu
Min Lu