Development and application of one-step multiplex Real-Time PCR for detection of three main pathogens associated with bovine neonatal diarrhea
- 1State Key Laboratory for Animal Disease Control and Prevention, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, China
- 2College of Animal Science and Technology, Tarim University, Alar, Xinjiang, China
- 3Institute of Western Agriculture, The Chinese Academy of Agricultural Sciences, Changji, China
Introduction: Neonatal calf diarrhea (NCD) is one of the most common diseases in calves, causing huge economic and productivity losses to the bovine industry worldwide. The main pathogens include bovine rotavirus (BRV), bovine coronavirus (BCoV), and Enterotoxigenic Escherichia coli (ETEC) K99. Since multiple infectious agents can be involved in calf diarrhea, detecting each causative agent by traditional methods is laborious and expensive.
Methods: In this study, we developed a one-step multiplex Real-Time PCR assay to simultaneously detect BRV, BCoV, and E. coli K99+. The assay performance on field samples was evaluated on 1100 rectal swabs of diseased cattle with diarrhea symptoms and compared with the conventional gel-based RT-PCR assay detect BRV, BCoV, and E. coli K99+.
Results: The established assay could specifically detect the target pathogens without cross-reactivity with other pathogens. A single real-time PCR can detect ~1 copy/µL for each pathogen, and multiplex real-time PCR has a detection limit of 10 copies/µL. Reproducibility as measured by standard deviation and coefficient of variation were desirable. The triple real-time PCR method established in this study was compared with gel-based PT-PCR. Both methods are reasonably consistent, while the real-time PCR assay was more sensitive and could rapidly distinguish these three pathogens in one tube. Analysis of surveillance data showed that BRV and BCoV are major enteric viral pathogens accounting for calves’ diarrhea in China.
Discussion: The established assay has excellent specificity and sensitivity and was suitable for clinical application. The robustness and high-throughput performance of the developed assay make it a powerful tool in diagnostic applications and calf diarrhea research.
Introduction
Neonatal calf diarrhea (NCD) contributes significantly to calf mortality and morbidity worldwide. As reported, during the first 30 days after birth, the case fatality risk caused by NCD is approximately 5% (Svensson et al., 2006; Windeyer et al., 2014). NCD is mainly caused by infection with viruses, bacteria, and parasites. Of these, bovine rotavirus (BRV), bovine coronavirus (BCoV), and enterotoxigenic Escherichia coli K99 (ETEC) are the most frequently identified causative factors (Acres, 1985; Krogh and Henriksen, 1985; Tzipori, 1985; Snodgrass et al., 1986). These pathogens are often mixed and lead to several complications. Thus, early diagnosis and treatment facilitate timely management and control of NCD.
BRV is believed to be the leading cause of viral diarrhea disease of newborn calves. BRV is a non-enveloped and double-stranded RNA virus that belongs to the genus Rotavirus in the family Reoviridae. Its genome comprises 11 double-stranded RNA segments that encode six structural proteins (VP1−VP4, VP6−VP7) and six nonstructural proteins (NSP1−NSP6). Based on the antigenicity of the middle VP6 capsid antigen, rotavirus is classified into at least 10 groups (A-J) (Bányai et al., 2017). Among which, group A rotavirus cause disease in both humans and animals. Group A rotaviruses can be further divided into P and G types based on the genetic and antigenic characteristics of VP4 and VP7, respectively. In cattle, G6, G8, and G10 strains appear to be the dominant strain circulating in the herd (Snodgrass et al., 1990; Ciarlet et al., 1997; Rao et al., 2000). As reported, the pooled prevalence of BRV in cattle was approximately 46% in China (Chen et al., 2022).
In addition to BRV, BCoV is frequently identified from the calves with viral diarrhea disease. BCoV is a single-stranded positive-sense RNA virus with a lipid envelope that belongs to the genus Beta-coronavirus within the Coronaviridae family (Pfefferle et al., 2009; Yang and Leibowitz, 2015; Oma et al., 2016). There are 13 open reading frames (ORFs) in the approximately 31 kb-long RNA genome, bordered by 5′ caps and 3′ polyadenylated tails (Chouljenko et al., 2001). The nucleocapsid protein (N), membrane protein (M), minor capsid protein (E), spike protein (S), and hemagglutinin esterase protein (HE) are the five structural proteins inducing antibodies. In addition to severe diarrhea in newborn calves, BCoV infection causes respiratory illness in calves and feedlot cattle, and winter dysentery in adult cattle (Gomez and Weese, 2017). However, less systematic studies have been conducted to evaluate the prevalence of BCoV infection in China (Geng et al., 2023). Infections with either BRV or BCoV, usually result in intestinal injury and immune suppression. This could increase the likelihood of infection with E. coli K99+.
Since NCD is usually caused by mixed infections with multiple pathogens and has similar clinical symptoms, they are not easy to differentiate. In the complex chain of epidemiologic and therapeutic measures to prevent outbreaks, there is a need for rapid and sensitive diagnostic tools for early differential diagnosis of mixed infection. The traditional detection methods (e.g. virus isolation, ELISA, PCR, etc.) have some limitations (Decaro et al., 2008; Mijatovic-Rustempasic et al., 2013; Jesser and Levy, 2020). Therefore, single, and multiplex real-time PCR assays have been developed for the rapid detection of pathogens associated with NCD in the past years (West et al., 2007; Decaro et al., 2008; Cho et al., 2010; Mijatovic-Rustempasic et al., 2013). Single detection methods require separate amplification of the pathogens, which is a cumbersome and time-consuming process. Although, BCoV, BRV, and E. coli K99+ are reported to be the major cause of NCD (Brunauer et al., 2021). There are no multiplex methods that place these three pathogens in the same reaction for detection currently.
In this study, a multiplex real-time PCR assay based on TaqMan technology was developed for the rapid and sensitive diagnosis of BCoV, BRV, and E. coli K99+ infection and accurate quantification of nucleic acid in clinical samples. The method enabled the simultaneous resolution by using of three sets of specific primers and probes in a single reaction mixture. The assay performance was verified by the evaluation of specificity, sensitivity, reproducibility, and dynamic range. Consequently, this method can be effectively used in diagnostic, surveillance, and epidemiologic studies of NCD caused by BRV, BCoV, and E. coli K99+.
Materials and methods
Primer and probe design
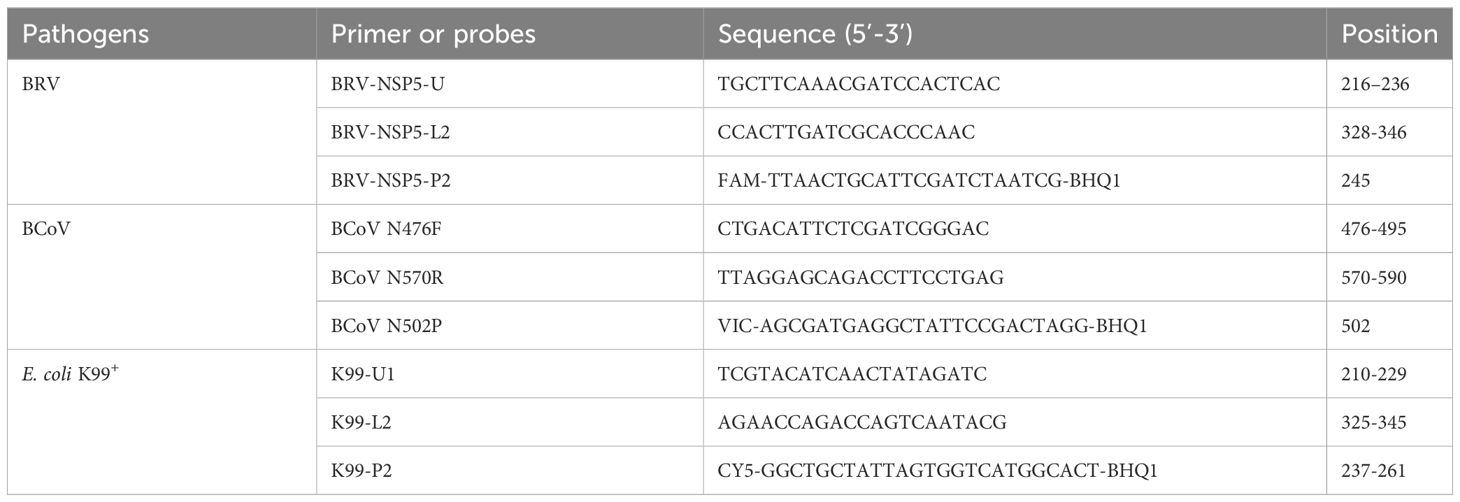
All available complete sequences of BRV (forty clinical isolates), BCoV(forty-one clinical isolates), and E. coli K99+ (eighteen clinical isolates) were retrieved from GenBank. The NSP5 gene sequences of BRV, the N gene sequences of BCoV and the K99(F5)pili sequences of E. coli K99+ were aligned with DNASTAR software for identifying the highly conserved regions of each gene. Primers and probes are then designed to target the conserved regions via the Primer Premier 5 program. Upon the filtration based on the design criteria such as hairpin configurations and primer dimer formation. Three pairs of primers and probes were picked for the synthesis by Comate Bioscience Company (Jilin, China), and the detailed information is shown in Table 1.
Viruses, bacterial and cell cultures
BRV G6 strain (C7-3), BRV G10 strain (HM26), BCoV strain (HM-XC), Bovine Viral Diarrhea Virus (BVDV NADL strain), Bovine Enterovirus (BEV strain BHM26 and BJ50) (Li et al., 2012), Bovine Parvovirus (BPV-1 strain HADEN) (Chang et al., 2019), Salmonella enterica (S. enterica) strain (FH10) (Lee et al., 2009) Clostridium perfringens (C. perfringens) type A strain CVCC C57–8 was purchased from the China Institute of Veterinary Drug Control (Beijing, China), and E. coli K99+ strain HLJ23 isolated from calf diarrhea. TCID50 of the BcoV, BEV, BPV and BRV were determined in HRT-18G (ATCC, CRL-3609), BHK-21 cell, primary bovine testicular cell and Marc-145 (a clone of the MA-104 cell line, ATCC CRL-2378), respectively. The colony-forming units (CFU) of E. coli K99+ strain was quantified by the traditional plate colony counting method. Both viruses and bacteria mentioned above were sequenced.
DNA/RNA was extracted from cell cultures, bacteria, and clinical samples according to the manufacturer’s instructions of the AxyPrep body fluid viral DNA/RNA miniprep kit (Axygen Biosciences, USA). The obtained nucleic acids are stored at -80°C until needed. To obtain the standard plasmid, the target fragments of BRV, BcoV and E. coli K99+ were amplified and cloned into the pMD18-T vector (Takara, China). The standard plasmids were then transformed into DH5α chemically competent cells, and the plasmids were extracted using the GeneJET Plasmid Miniprep Kit (ThermoFisher Scientific,USA). The concentration of the purified plasmid DNA was determined by measuring the optical density at 260 nm (OD 260) with a Nanodrop One (ThermoFisher Scientific, USA)
The copy number of the extracted plasmids was calculated according to the following formula.
Single real-time PCR was performed using serial 10-fold dilutions of the plasmid from 107 to 1 copies/µL, and the plasmid from 107 to 101 copies/µL were selected to construct the standard curve. Three standard curves were constructed, and corresponding equations were determined by taking amplification efficiency and correlation coefficients into account.
Optimization of the single and multiplex TaqMan real-time PCR assays
Single real-time PCR was performed using the HiScript II U+ One Step qRT-PCR Probe Kit from Vazyme. The total volume of the PCR system was set as 10 µL, including 2× One Step U+ MIX, One Step U+ Enzyme MIX, and 50× ROX Reference Dye 2, the amount of the primer addition was optimized from 0.1 to 0.8 µL, while the amount of the probe addition was optimized from 0.1 to 0.6 µL, which were both at a final concentration of 10 µM. Amplification was carried out using the following program: 55°C 15 min, 95°C, 30 s, 45 cycles of 95°C 10 s, annealing temperature was optimized between 54°C and 60°C. Amplification with Quant Studio™ Design & Analysis Software. Signals were automatically collected at the end of each cycle.
Multiplex real-time PCR was established based on three single methods by adjusting the reaction system and conditions to reduce interference among three different fluorescent molecules and to increase amplification efficiency. The final concentration for the primer and probe was both set as 10 µM, and the amount of primer additions was optimized between 0.1 µL and 0.5 µL, while the amount of probe additions was optimized from 0.1 µL to 0.4 µL. The reaction temperature was optimized between 55°C and 65°C.
Sensitivity and specificity of the multiplex real-time PCR assay
Multiplex real-time PCR was as performed using the HiScript II U+ One Step qRT-PCR Probe Kit from Vazyme. A 10-fold gradient dilution of 108−101 copies/µL of the standard plasmids served as the template. Multiplex real-time PCR was conducted in tube, with three replicates assigned to each reaction to examine the detection limits of multiplex reactions. The primers and probes used in this study were tested for specificity. To avoid false positives caused by other pathogens that may be present in the assay, the multiplex real-time PCR detection assay was used to detect BVDV, BEV, BPV, S. enterica, and C. perfringens and RNA-free H2O as negative control.
Repeatability test
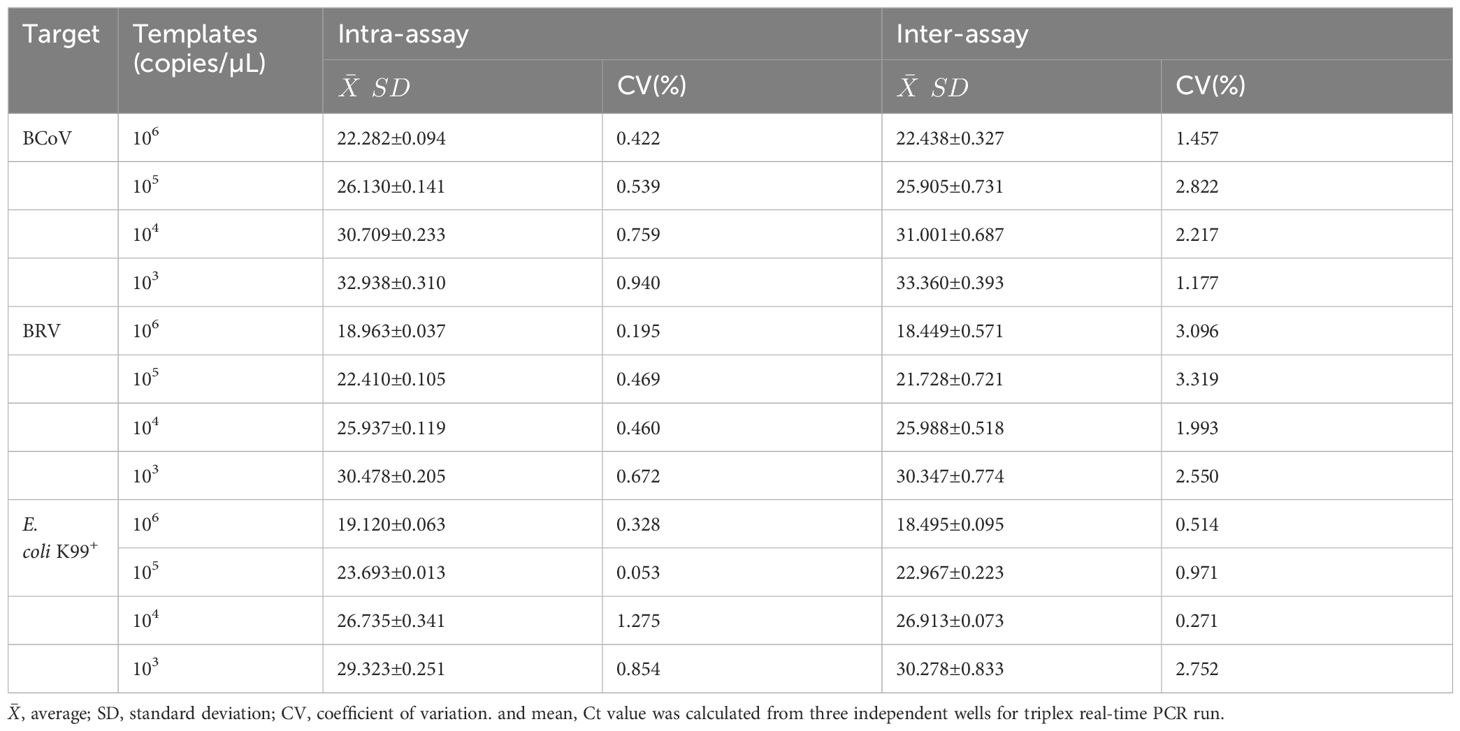
Standard plasmids with four different copy numbers (103−106) were used for the repeatability test of the real-time PCR assay. Three replicate experiments were set up in both intra- and inter-groups. The standard deviation and coefficient of variation were calculated for the three reactions.
Examination of clinical fecal samples using real-time PCR
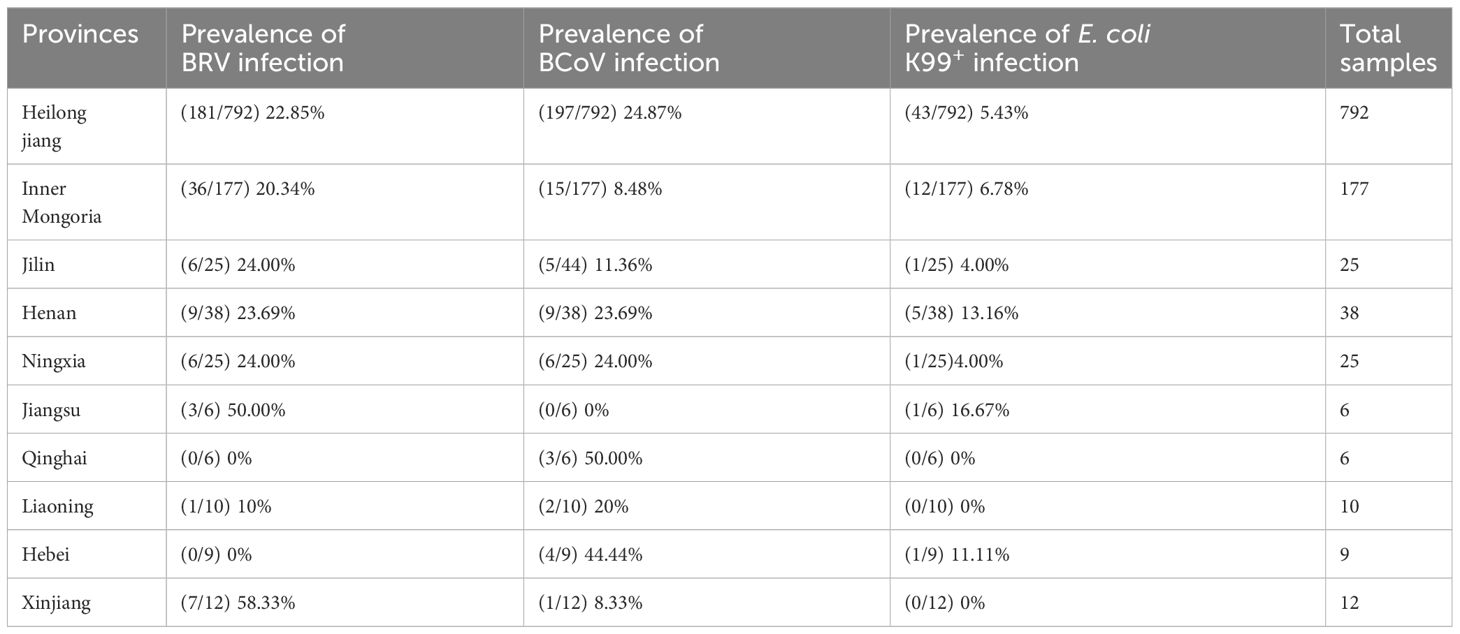
A total of 1100 rectal swabs were collected from cattle in different regions including Heilongjiang Province, Inner Mongolia Autonomous Region, Ningxia Hui Autonomous Region, Jilin Province, Henan Province, and Jiangsu Province, etc. The majority of calves included in the samples showed diarrheic symptoms. The swabs were preserved at -80°C until needed.
Results
Primers and probe for real-time PCR
By comparison and analysis of the retrieved sequences from GenBank, we found that the segment 11, which encodes NSP5 protein is most conserved among the 11 segments of BRV genome. Thus, the fragment ranging from 216 bp to 346 bp in segment 11 gene was selected as a target for primer and probe design. The N gene is the most conserved segment of BcoV, and we chose the 476-590 bases at the 5’ end of the N gene to design primers and probes, and the amplified fragment length was 115 bp. In addition, the E. coli K99+ (F5) pilis gene is often used as a target for detecting E. coli infections, and we chose the most conserved region 210−345 bases in E. coli K99+ (F5) pilis gene for the design. The primers and probes designed for real-time PCR were listed in Table 1. Theoretically, these primer/probe sets could recognize all BRV, BcoV, and E. coli K99+ strains prevalent in the herd. To establish a standard curve, each of the three amplicon fragments was cloned into the pMD18-T vector and used as a standard plasmid for further use.
Specificity analysis of primer/probe sets
To evaluate the specificity of our designed primers and probes, we selected 1×106 copies/µL of standard plasmids harboring the specific fragments of BRV, BcoV, and E. coli K99+ for the assay. As expected, all the primers and probes only bind to their target regions without any cross-reaction. To further verify the specificity of the designed primer/probe sets, the genomes were extracted from the strains of BRV, BcoV, BVDV, BEV, BPV, E. coli K99+, S. enterica, and C. perfringens for assessment. Consistently, the designed primers and probes for detecting BRV, BcoV, and E. coli K99+ could specifically recognize the genome isolated from BRV, BcoV, and E. coli K99+. There were no signals detected for other pathogens, confirming that the real-time PCR assay is specific for BRV, BcoV, and E. coli K99+. (Figure 1)
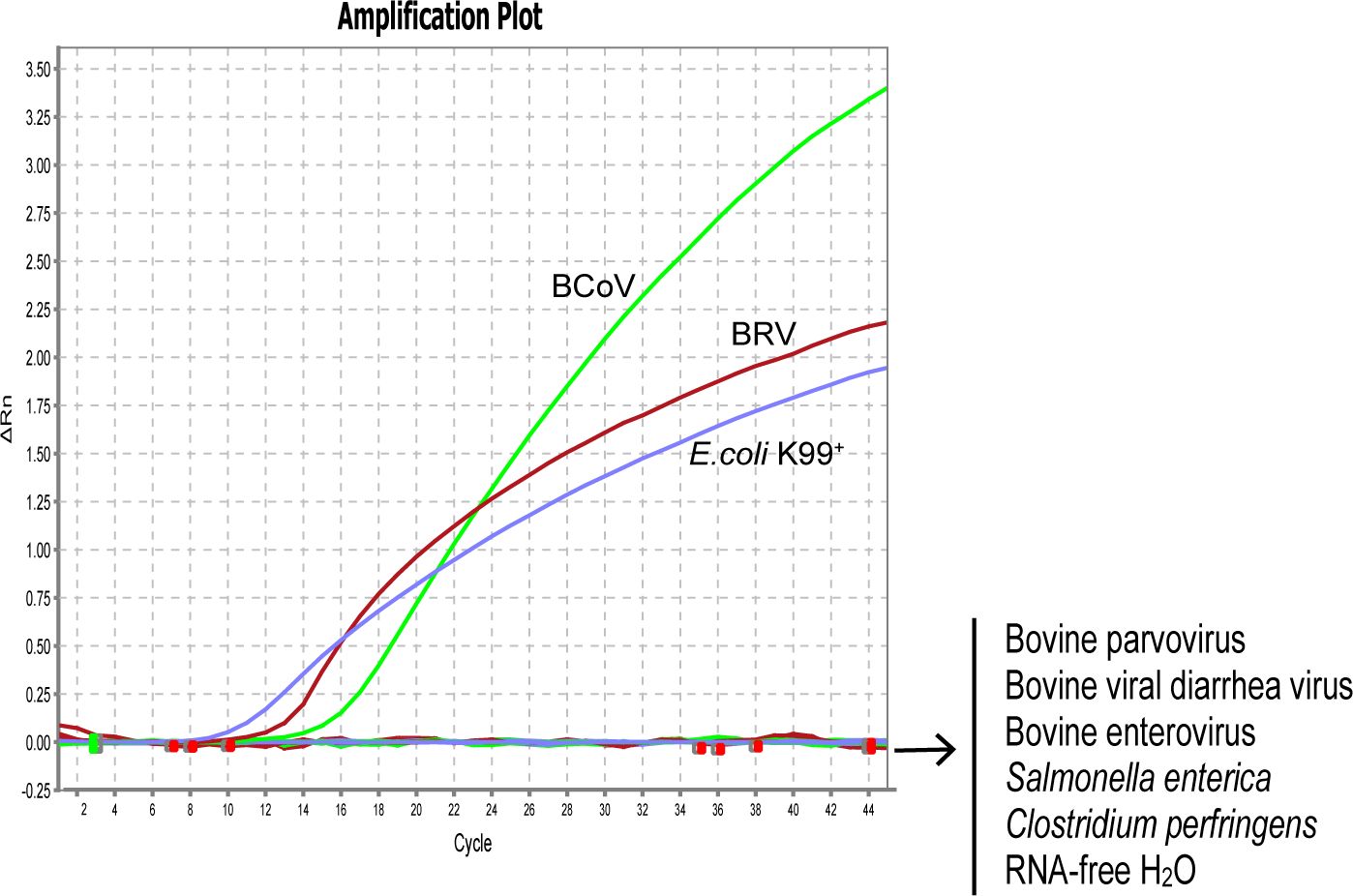
Figure 1 Analytical specificity of the primer/probe sets used in the one-step real-time RT-PCR assay: Amplification curves represent samples positive for BRV, BCoV, and E. coli K99+ detected by the real-time Negative samples include Bovine viral diarrhea virus, Bovine enterovirus, Bovine parvovirus, Salmonella enterica, Clostridium perfringens, and RNA-free H2O. Equal amount of isolated viral/bacterial genome were mixed for the assay. The non-detected samples are labeled on the right side of the figure.
Optimized single real-time PCR
Upon confirmation of the specificity of the designed primers and probes used in this study, a single real-time PCR assay was first generated for individual pathogen. The optimized reaction system for the single assay was 10 µL: 5 µL of 2× One Step U+ MIX, 0.5 µL of One Step U+ Enzyme MIX, 0.2 µL of 50× ROX Reference Dye 2, 0.2 µL of each primer (10 µM), 0.3 µL of each probe (10 µM), 2.6 µL of Rnase-free dd H2O and Template 1 µL. The single assay can detect a minimum of 1 copy of the standard plasmid, as shown in Figure 2 (A-C left panel). The real-time PCR amplifications of each standard plasmid DNA showed amplification plots corresponding to mean Ct values of 10.343−40.853 for BRV, 12.346−35.181 for BcoV, and 12.622−36.709 for E. coli K99+ (Figure 2). As the linear range includes 8 orders of magnitude (from 1 to 107 copies of the standard plasmid), we chose 101−107 copies to establish the standard equation (Figures 2A–C). The slope of the standard curve, correlation coefficient (R2), and amplification efficiency (Eff%) were calculated as follows −3.140, 0.999, and 96.463% for BRV; −3.415, 0.995, and 96.269% for BcoV; −3.477, 0.996, and 93.927% for E. coli K99+. (Figures 2D–F). Collectively, these results demonstrated that the optimized single real-time PCR were sensitive for the detection of BRV, BcoV, and E. coli K99+ individually.
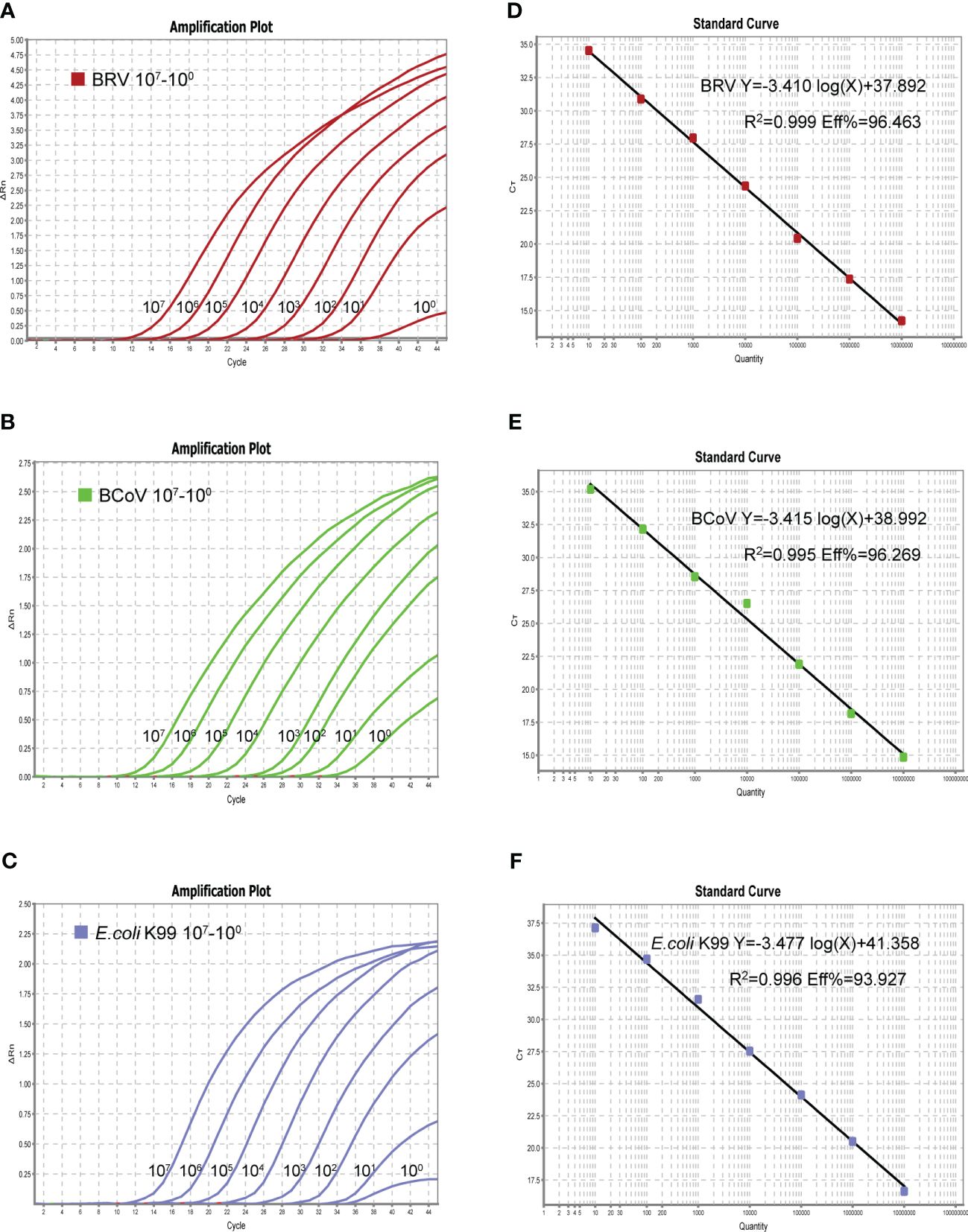
Figure 2 The amplification curves (X-axis: Cycle, Y-axis: △Rn) of the plasmid standards are shown in left panel (A–C). The plasmid was diluted in a concentration range of 107-1 copies/µL. Standard curves of 107-101 copies/µL plasmid standards for BRV, BCoV, and E. coli K99+ (D–F). BRV: Y=−3.410log(X)+37.892 R2 = 0.999 Eff%=96.463, BCoV: Y=−3.415log(X)+38.992 R2 = 0.995 Eff%=96.269, K99: Y=−3.477log(X)+ 41.358 R2 = 0.996 Eff%=93.927.
Optimization of the multiplex reaction system
Following the optimization of the single real-time PCR for each pathogen, the multiplex real-time PCR was developed through multiplex rounds of optimization. We found that a good amplification effect can be achieved when the total reaction system is 20 µL, and the addition of each component was multiplied twice compared to the single system, with optimized primer and probe additions: 0.1 µL NSP5-U/NSP5-L2 primer set (10 µM), 0.1 µL BRV-NSP5-P2 (10 µM), 0. 1 µL N476F/N570R primer set (10 µM), 0.1 µL BcoV N502P (10 µM), 0. 2 µL K99-U1/K99-L2 primer set (10 µM), 0. 2 µL K99-P2 (10 µM) The annealing temperature was optimized to be 55°C. Good correlation coefficients and amplification efficiencies for the standard curves were observed when the total volume was 20 µL (Figures 3A, B) BRV (R2 = 0.998 Eff%=89.028), BcoV (R2 = 0.995 Eff%=95.810), E. coli K99+. (R2 = 0.993 Eff%=101.021). The optimized multiple-reaction system was able to identify specific pathogens effectively, and the correlation coefficients of the three pathogens were able to reach more than 0.99. The amplification efficiencies of both BcoV and E. coli K99+ were excellent, although the amplification efficiency of BRV was slightly lower. Taken together, after the optimization, the multiplex real-time PCR was established for further assessment.
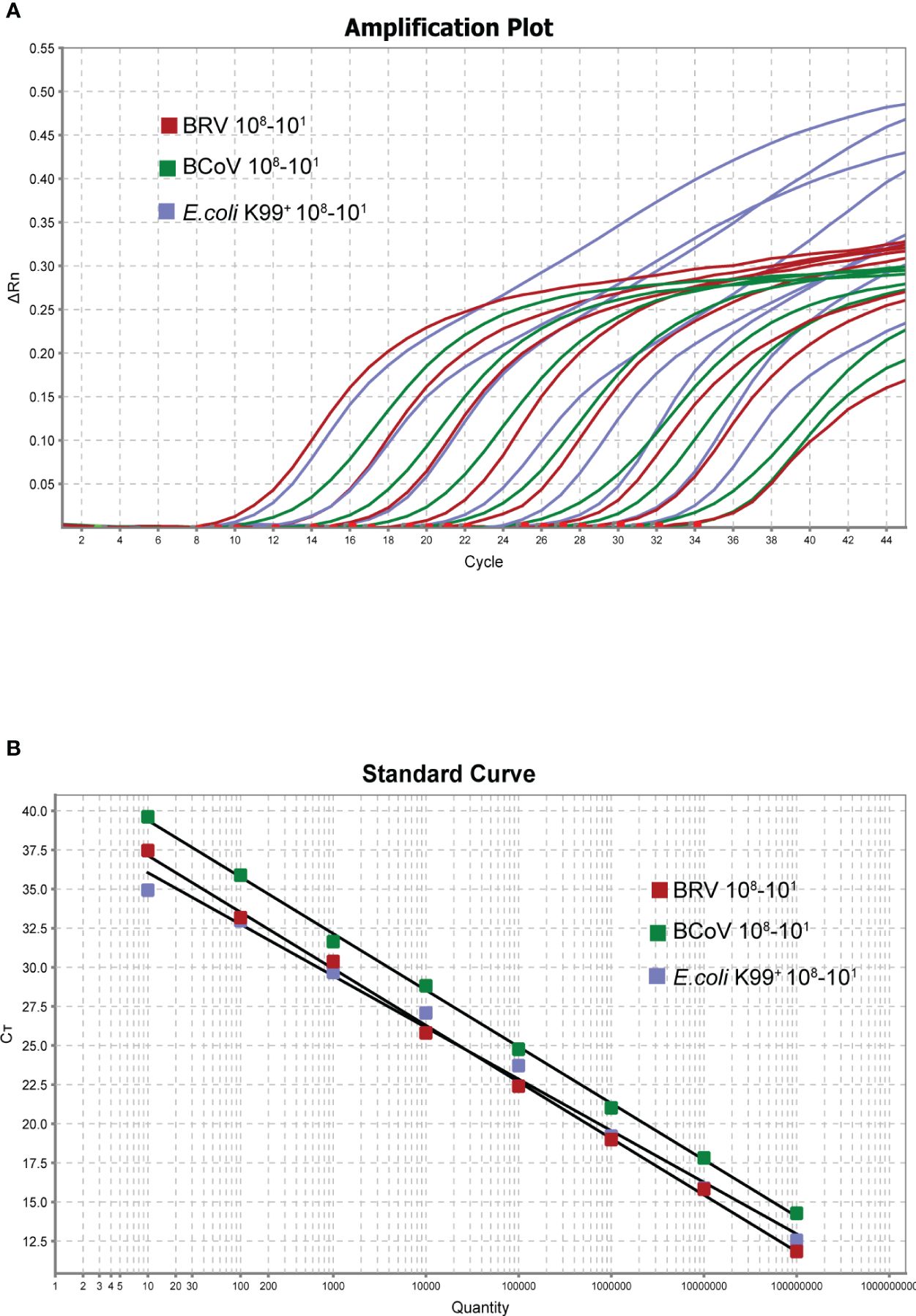
Figure 3 Amplification curve of triple reaction (X-axis: number of cycles, Y-axis: △Rn) (A). The copy number of each viral plasmid ranged from 108-101 copies/µL. Standard curve (X-axis: copy number, Y-axis: CT value) (B). BRV: Y=−3.616 log(X)+40.756 R2 = 0.998 Eff%=89.028, BCoV: Y=−3.427log(X)+41.866. R2 = 0.995 Eff%=95.810, E. coli K99+: Y=−3.298log(X)+39.343 R2 = 0.993 Eff%=101.021.
Sensitivity and limit of detection of real-time PCR
By using the 10-fold serial diluted plasmid standards, we further validated the amplification efficiency and detection limits. After extensive experiments, we found that the single real-time PCR can detect all three pathogens down to 1 copy/µL (Figures 2A–C), while the detection limit of multiplex real-time PCR only can reach 10 copies/µL (Figure 3A). To further verify the sensitivity and detection limits of the multiplex assay, the cultured viruses with specific titers were used for the quantification. The minimum value for detection of BRV in the multiplex real-time PCR assay was 0.5 TCID50 (50% tissue culture infected dose) and the minimum value for detection of BcoV was 0.14 TCID50. For E. coli K99+, 0.7433 CFU can be detected. Furthermore, we defined cutoff values for these three pathogens in the multiplex assay. The cutoff CT value for BRV positivity was defined at 36, which means the sample with a CT value less than or equal to 36 (≤ 36) was considered positive, but higher than 36(>36) is negative. The cutoff CT value for BcoV and E. coli K99+ was 36 and 35, respectively.
Repeatability of the multiplex real-time PCR
To evaluate the reproducibility of the multiplex real-time PCR, the standard plasmids at the concentrations of 1×106, 1×105, 1×104, and 1×103 copies/µL were used. As a result, the coefficients of variation of the intra and inter-assay ranged from 0.053% to 1.275% and 0.270% to 3.319%, respectively (Table 2), indicating good repeatability of the assay.
The surveillance of NCD cases in China by using the established multiplex assay
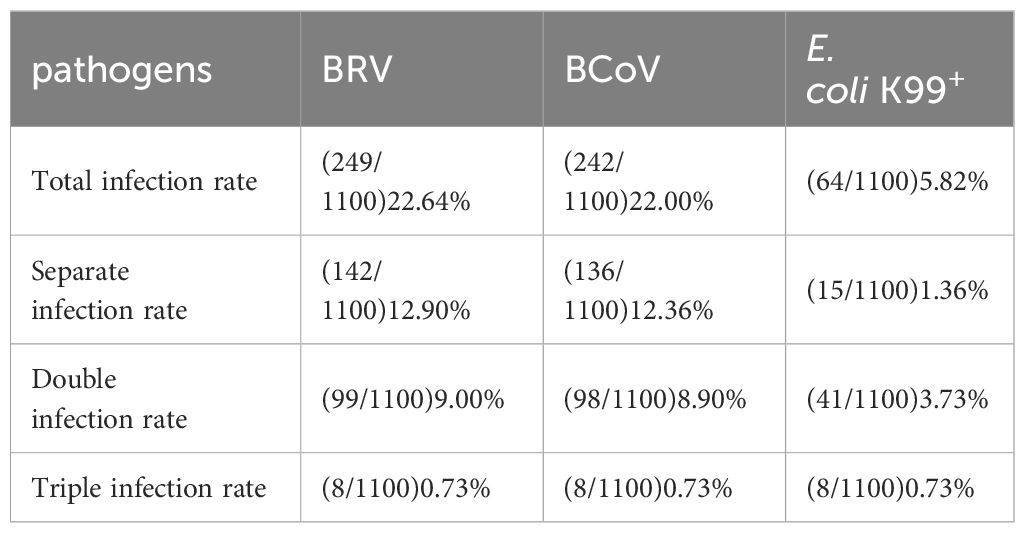
To evaluate the optimized assay in clinical application, 1100 rectal swabs of NCD cases were collected in 10 provinces as shown in Table 3, the average positive rate for BRV, BcoV, and E. coli K99+ was 22.64%, 22.00%, and 5.82%, respectively. Further analysis found that the highest positive rate of BRV(58.33%) was observed in Xinjiang region, while Qinghai had the highest positive rate of BcoV (50%) (Figure 4A). However, the overall infection rate of E. coli K99+ was low. In Qinghai, only BcoV infection was detected, while in the other 9 provinces, at least two pathogens were tested positive. Notably, 792/1100 samples were collected from Heilongjiang province, the analysis showed that both BRV and BcoV infections in calves was highly prevalence, which accounted for the occurrence of NCD cases in Heilongjiang province. In addition, we analyzed the co-infection rate in the detected samples, and found that the co-infection of BRV and BcoV was higher at a rate of 7.1% (78/1100), while the co-infection of BRV and E. coli K99+ as well as BcoV and E. coli K99+ was similar at 1.9% (21/1100), and 1.8% (20/1100), respectively. Only 8 out of 1100 samples were mix-infected with these three pathogens (Figure 4B and Table 4). We also picked 200 samples to analyze the CT values of BRV, BcoV, and E. coli K99+ detected via the multiplex PCR assay (Figure 4C), we found that the CT value for BRV and BcoV were lower, compared to that for E. coli K99+, further confirming that the co-infection of BRV and BcoV occurs frequently in the calves with diarrhea.
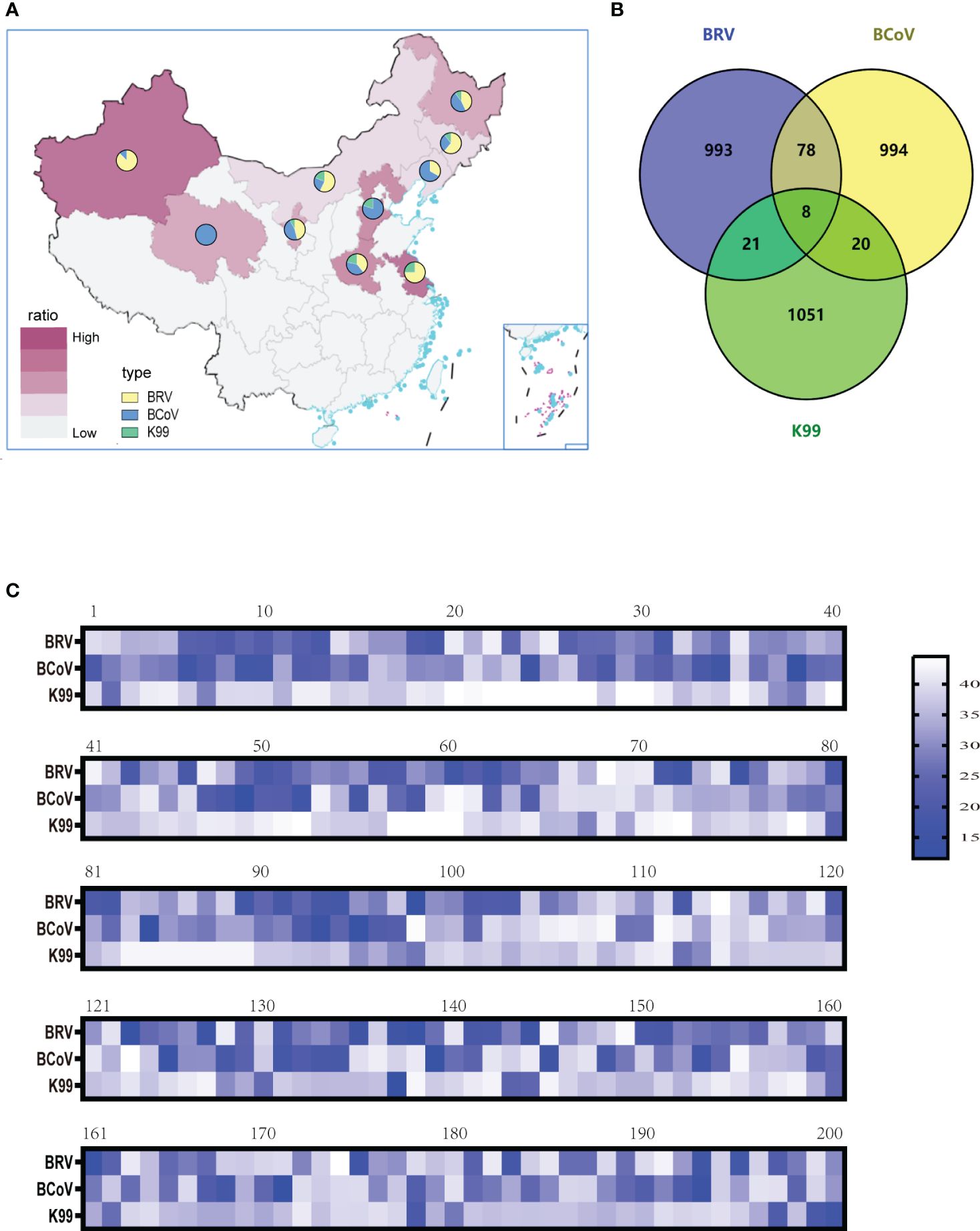
Figure 4 Clinical sample detection: (A) BRV, BCoV, and E. coli K99+ infection rates in ten provinces and analysis of the proportions of the three pathogens (B)The mix-infection incidence among BRV, BCoV, and E. coli K99+. (C) Selection of 200 clinical samples to analyze the level of pathogen contained (CT value).
To further confirm the results obtained from the multiplex assay, we performed the conventional gel-based RT-PCR assay in parallel for comparison. In 1100 samples, the multiplex PCR assay could detect 249 BRV positive samples, while gel-based RT-PCR only detected BRV 230 samples (Table 5). For BcoV detection, 242 samples were positive in the multiplex assay, while only 226 samples were positive in the gel-based RT-PCR assay. Similar results were also observed for E. coli K99+ (64 positive in the multiplex assay and 41 positive in the gel-based RT-PCR assay). These results demonstrated that our established multiplex PCR assay was more sensitive than the conventional gel-based RT-PCR assay for the clinical application.
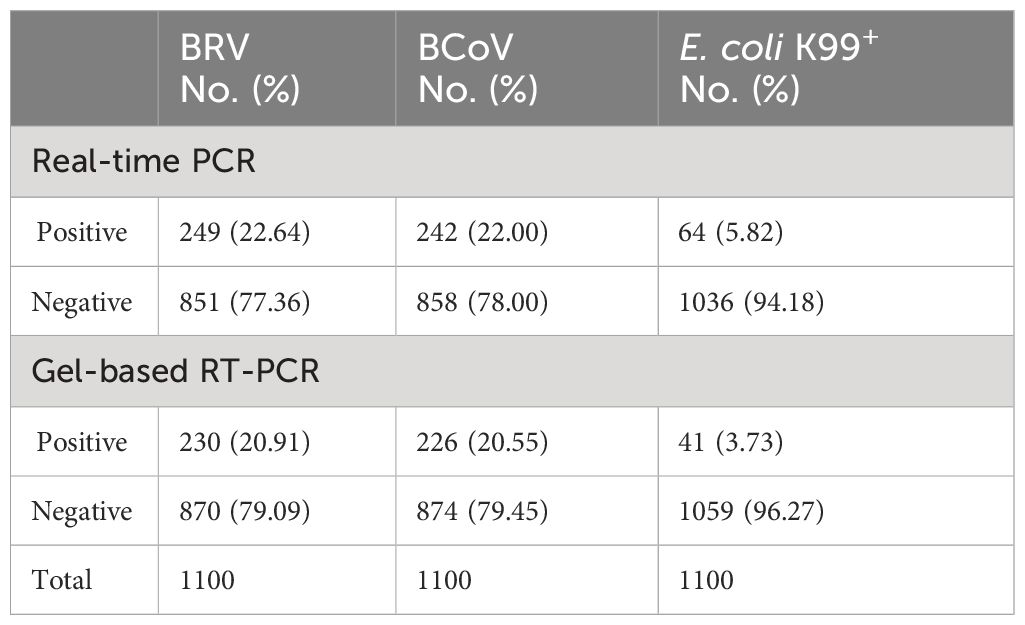
Table 5 Comparative analysis of bovine clinical samples using real-time PCR and gel-based RT-PCR assays.
Discussion
NCD is a significant global issue in the cattle industry, which deeply impact calves’ growth rate and lifetime productivity. As reported, it is a muti-factorial disease, involving both infectious and non-infectious factors (Oguejiofor et al., 2019). The high probability of mixed infections of these three pathogens makes the rapid detection of these pathogens of great importance. Among these etiological agents that cause calf diarrhea, BCoV and BRV account for about 27%–36% (Cho and Yoon, 2014), which is consistent with our detection results. Moreover, the main pathogenic bacteria causing NCD are reportedly enterotoxigenic E.coli K99+ (Cho and Yoon, 2014). However, in our detected clinical samples, the positive rate of E.coli K99+ is lower (0% to 16.67%), which might be due to the antibiotics usage in the clinical management prior to the detection.
At present, several diagnostic assays have been reported to individually detect BCoV, BRV, and E. coli K99+. A sensitive quantitative real-time PCR method for detecting BCoV in fecal samples could detect 102 copies/reaction (Decaro et al., 2008). In addition, one-step real-time reverse transcription-PCR assay was previously established to detect rotavirus A approximately 1 genome copy per reaction (Mijatovic-Rustempasic et al., 2013). In our single real-time PCR detection assay, through a series of optimizations, we could achieve a sensitivity of 1 copy/µL for the detection of BCoV and BCoV. More recently, a multiplex real-time PCR method was also reported by dividing six bovine diarrhea viruses into two systems for detection (Meng et al., 2023). However, the detection limits only reached to 16.4 copies/µL and 18.2 copies/µL for BRV and BCoV, respectively. In our study, the sensitivity was up to 10 copies/µL for all 3 detected pathogens, suggesting that our developed assay is more robust for clinical detection. In addition, different from previous reports using the segment 8 as the target for BRV detection, we chose the segment 11 gene in our single-weighted method. Similar to our assay, two panels of multiplex real-time PCR assays, one for viral agents (BCoV and BRV) and the other for bacterial/protozoan agents (E. coli K99+, Salmonella, and Cryptosporidium) were recently established (Cho et al., 2010). This assay can effectively perform multiplex detection of diarrhea pathogens, however, the separation of viruses and bacteria into two test tubes is still not convenient enough in the clinical application. In our study, BCoV, BRV, and E. coli K99+, the major causes of diarrhea were built into one multiplex assay, Pathogens whose genetic material is DNA and RNA can be tested in a single reaction tube without the need for an additional reverse transcription step. The performance of this assay showed high sensitivity and specificity. More importantly, the detection results can be acquired within less than 2 hours.
In our clinical study, we found that calf diarrhea is widespread throughout China, especially in northeastern China. In the 1100 feces collected from calves with diarrhea in 10 provinces (Table 3). BRV prevalence exceeds 20% in seven provinces, and six provinces had BCoV prevalence rates above 20%. It appears that BRV remained the main etiological agents causing NCD in China, which is consistent with previous reports (Cheng et al., 2021). Intriguingly, the E. coli K99+ detection rate in the herd is lower, which may be due to the current widespread use of antibiotics. In addition, the mixed infections occurred frequently, especially for BCoV and BRV(Table 4). However, currently, there are no commercial vaccines against BCoV and BRV available in China. Therefore, the further surveillance of BCoV and BRV circulating in China and the effective vaccine development are urgent.
The efficacy of the established multiplex real-time PCR assay was evaluated by analysis of 1100 rectal swab samples in comparison to the gel-based RT-PCR assay (Table 5). In general, the developed multiplex real-time PCR assay is more sensitive than the conventional gel-based RT-PCR assay. Some of the samples tested negative by gel-based RT-PCR were found to be positive by real-time PCR. While none of the samples tested negative by real-time PCR was found positive by the gel-based method. Based on the above conjecture, we selected 200 samples for statistical analysis of CT values (Figure 4C), Indeed, the positive samples confirmed by real-time PCR assay rather than conventional gel-based RT-PCR assay had higher CT values. Thus, the established multiplex PCR assay would provide the conventional tool to accurately clarify the etiology of NCD in China and help rapidly employ effective preventive and therapeutic approaches to reduce economic losses.
In summary, we have established a one-step multiplex detection assay for simultaneous detection and characterization of BRV, BCoV, and E. coli K99+. The method is highly sensitive, specific, and reproducible, which can rapidly recognize the diarrheal pathogens in the clinical samples with mixed infections.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
CW: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. FW: Conceptualization, Supervision, Writing – original draft, Writing – review & editing. JC: Methodology, Resources, Writing – review & editing. ZJ: Investigation, Methodology, Validation, Writing – review & editing. YH: Investigation, Methodology, Writing – review & editing. MW: Investigation, Methodology, Writing – review & editing. BJ: Supervision, Writing – review & editing. AZ: Supervision, Writing – review & editing. XY: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Inner Mongolia Autonomous Region Major Science and Technology Project (2020ZD0006 and 2021ZD0013).
Acknowledgments
The authors would like to thank the members from Yin Laboratory for their critical suggestions.
Conflict of interest
The authors declare that there is a patent related to this work (application no. 2024102313502).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acres, S. D. (1985). Enterotoxigenic Escherichia coli infections in newborn calves: a review. J. Dairy Sci. 68, 229–256. doi: 10.3168/jds.S0022-0302(85)80814-6
Bányai, K., Kemenesi, G., Budinski, I., Földes, F., Zana, B., Marton, S., et al. (2017). Candidate new rotavirus species in Schreiber's bats, Serbia. Infect. Genet. Evol. 48, 19–26. doi: 10.1016/j.meegid.2016.12.002
Brunauer, M., Roch, F. F., Conrady, B. (2021). Prevalence of Worldwide Neonatal Calf Diarrhoea Caused by Bovine Rotavirus in Combination with Bovine Coronavirus, Escherichia coli K99 and Cryptosporidium spp.: A Meta-Analysis. Anim. (Basel) 11 (4), 1014. doi: 10.3390/ani11041014
Chang, J., Zhang, Y., Yang, D., Jiang, Z., Wang, F., Yu, L. (2019). Potent neutralization activity against type O foot-and-mouth disease virus elicited by a conserved type O neutralizing epitope displayed on bovine parvovirus virus-like particles. J. Gen. Virol. 100, 187–198. doi: 10.1099/jgv.0.001194
Chen, S., Zhang, W., Zhai, J., Chen, X., Qi, Y. (2022). Prevalence of bovine rotavirus among cattle in mainland China: A meta-analysis. Microb. Pathog. 170, 105727. doi: 10.1016/j.micpath.2022.105727
Cheng, X., Wu, W., Teng, F., Yan, Y., Li, G., Wang, L., et al. (2021). Isolation and characterization of bovine RVA from Northeast China, 2017-2020. Life (Basel) 11. doi: 10.3390/life11121389
Cho, Y. I., Kim, W. I., Liu, S., Kinyon, J. M., Yoon, K. J. (2010). Development of a panel of multiplex real-time polymerase chain reaction assays for simultaneous detection of major agents causing calf diarrhea in feces. J. Vet. Diagn. Invest. 22, 509–517. doi: 10.1177/104063871002200403
Cho, Y. I., Yoon, K. J. (2014). An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J. Vet. Sci. 15, 1–17. doi: 10.4142/jvs.2014.15.1.1
Chouljenko, V. N., Lin, X. Q., Storz, J., Kousoulas, K. G., Gorbalenya, A. E. (2001). Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 82, 2927–2933. doi: 10.1099/0022-1317-82-12-2927
Ciarlet, M., Piña, C. I., García, O., Liprandi, F. (1997). Identification of bovine rotaviruses in Venezuela: antigenic and molecular characterization of a bovine rotavirus strain. Res. Virol. 148, 289–297. doi: 10.1016/S0923-2516(97)88366-5
Decaro, N., Elia, G., Campolo, M., Desario, C., Mari, V., Radogna, A., et al. (2008). Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay. J. Virol. Methods 151, 167–171. doi: 10.1016/j.jviromet.2008.05.016
Geng, H. L., Meng, X. Z., Yan, W. L., Li, X. M., Jiang, J., Ni, H. B., et al. (2023). Prevalence of bovine coronavirus in cattle in China: A systematic review and meta-analysis. Microb. Pathog. 176, 106009. doi: 10.1016/j.micpath.2023.106009
Jesser, K. J., Levy, K. (2020). Updates on defining and detecting diarrheagenic Escherichia coli pathotypes. Curr. Opin. Infect. Dis. 33, 372–380. doi: 10.1097/QCO.0000000000000665
Krogh, H. V., Henriksen, S. A. (1985). Bovine cryptosporidiosis in Denmark. 2. Cryptosporidia associated with neonatal calf diarrhea. Nord Vet. Med. 37, 42–47.
Lee, K., Iwata, T., Shimizu, M., Taniguchi, T., Nakadai, A., Hirota, Y., et al. (2009). A novel multiplex PCR assay for Salmonella subspecies identification. J. Appl. Microbiol. 107, 805–811. doi: 10.1111/jam.2009.107.issue-3
Li, Y., Chang, J., Wang, Q., Yu, L. (2012). Isolation of two Chinese bovine enteroviruses and sequence analysis of their complete genomes. Arch. Virol. 157, 2369–2375. doi: 10.1007/s00705-012-1424-6
Meng, W., Chen, Z., Jiang, Q., Chen, J., Guo, X., Ma, Z., et al. (2023). A multiplex real-time fluorescence-based quantitative PCR assay for calf diarrhea viruses. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1327291
Mijatovic-Rustempasic, S., Tam, K. I., Kerin, T. K., Lewis, J. M., Gautam, R., Quaye, O., et al. (2013). Sensitive and specific quantitative detection of rotavirus A by one-step real-time reverse transcription-PCR assay without antecedent double-stranded-RNA denaturation. J. Clin. Microbiol. 51, 3047–3054. doi: 10.1128/JCM.01192-13
Oguejiofor, C. F., Thomas, C., Cheng, Z., Wathes, D. C. (2019). Mechanisms linking bovine viral diarrhea virus (BVDV) infection with infertility in cattle. Anim. Health Res. Rev. 20, 72–85. doi: 10.1017/S1466252319000057
Oma, V. S., Tråvén, M., Alenius, S., Myrmel, M., Stokstad, M. (2016). Bovine coronavirus in naturally and experimentally exposed calves; viral shedding and the potential for transmission. Virol. J. 13, 100. doi: 10.1186/s12985-016-0555-x
Pfefferle, S., Oppong, S., Drexler, J. F., Gloza-Rausch, F., Ipsen, A., Seebens, A., et al. (2009). Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 15, 1377–1384. doi: 10.3201/eid1509.090224
Rao, C. D., Gowda, K., Reddy, B. S. (2000). Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology 276, 104–113. doi: 10.1006/viro.2000.0472
Snodgrass, D. R., Fitzgerald, T., Campbell, I., Scott, F. M., Browning, G. F., Miller, D. L., et al. (1990). Rotavirus serotypes 6 and 10 predominate in cattle. J. Clin. Microbiol. 28, 504–507. doi: 10.1128/jcm.28.3.504-507.1990
Snodgrass, D. R., Terzolo, H. R., Sherwood, D., Campbell, I., Menzies, J. D., Synge, B. A. (1986). Aetiology of diarrhoea in young calves. Vet. Rec 119, 31–34. doi: 10.1136/vr.119.2.31
Svensson, C., Linder, A., Olsson, S. O. (2006). Mortality in Swedish dairy calves and replacement heifers. J. Dairy Sci. 89, 4769–4777. doi: 10.3168/jds.S0022-0302(06)72526-7
Tzipori, S. (1985). The relative importance of enteric pathogens affecting neonates of domestic animals. Adv. Vet. Sci. Comp. Med. 29, 103–206.
West, D. M., Sprigings, K. A., Cassar, C., Wakeley, P. R., Sawyer, J., Davies, R. H. (2007). Rapid detection of Escherichia coli virulence factor genes using multiplex real-time TaqMan PCR assays. Vet. Microbiol. 122, 323–331. doi: 10.1016/j.vetmic.2007.01.026
Windeyer, M. C., Leslie, K. E., Godden, S. M., Hodgins, D. C., Lissemore, K. D., LeBlanc, S. J. (2014). Factors associated with morbidity, mortality, and growth of dairy heifer calves up to 3 months of age. Prev. Vet. Med. 113, 231–240. doi: 10.1016/j.prevetmed.2013.10.019
Keywords: neonatal calf diarrhea, bovine rotavirus, bovine coronavirus, Escherichia coli k99, real-time PCR
Citation: Wang C, Wang F, Chang J, Jiang Z, Han Y, Wang M, Jing B, Zhao A and Yin X (2024) Development and application of one-step multiplex Real-Time PCR for detection of three main pathogens associated with bovine neonatal diarrhea. Front. Cell. Infect. Microbiol. 14:1367385. doi: 10.3389/fcimb.2024.1367385
Received: 08 January 2024; Accepted: 14 March 2024;
Published: 02 April 2024.
Edited by:
Jiangchao Zhao, University of Arkansas, United StatesReviewed by:
Kuan Zhao, Hebei Agricultural University, ChinaLiqian Zhu, Yangzhou University, China
Guojun Wang, Inner Mongolia University, China
Copyright © 2024 Wang, Wang, Chang, Jiang, Han, Wang, Jing, Zhao and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Jing, jingbodky@126.com; Aiyun Zhao, zaydky@126.com; Xin Yin, yinxin@caas.cn
†These authors have contributed equally to this work and share first authorship
 Chaonan Wang1,2†
Chaonan Wang1,2†  Fang Wang
Fang Wang Jitao Chang
Jitao Chang Xin Yin
Xin Yin