Research progress in isolation and identification of rumen probiotics
- College of Veterinary Medicine, Gansu Agricultural University, Lanzhou, China
With the increasing research on the exploitation of rumen microbial resources, rumen probiotics have attracted much attention for their positive contributions in promoting nutrient digestion, inhibiting pathogenic bacteria, and improving production performance. In the past two decades, macrogenomics has provided a rich source of new-generation probiotic candidates, but most of these “dark substances” have not been successfully cultured due to the restrictive growth conditions. However, fueled by high-throughput culture and sorting technologies, it is expected that the potential probiotics in the rumen can be exploited on a large scale, and their potential applications in medicine and agriculture can be explored. In this paper, we review and summarize the classical techniques for isolation and identification of rumen probiotics, introduce the development of droplet-based high-throughput cell culture and single-cell sequencing for microbial culture and identification, and finally introduce promising cultureomics techniques. The aim is to provide technical references for the development of related technologies and microbiological research to promote the further development of the field of rumen microbiology research.
1 Introduction
The Food and Agriculture Organization of the United Nations and the World Health Organization defines probiotics as live microorganisms which provide a beneficial effect on the host when ingested in moderation (FAO/WHO, 2001). This definition is widely accepted by the International Scientific Association for Probiotics and Prebiotics (ISAPP) (Hill et al., 2014). Probiotics perform many biological functions in the ecosystem, such as aiding in digestion, inhibiting pathogenic bacteria, promoting growth and regulating immunity (Dasriya et al., 2024). The efficacy of probiotics has been demonstrated through in vivo experiments. Probiotics’ beneficial effects and safety are usually evaluated through in vivo experiments, while their beneficial potential and safety are characterized through in vitro studies or animal models (Reid, 2006). Therefore, the first step in evaluating probiotics for food use is in vitro studies, followed by double-blind, randomized, placebo-controlled human trials (Fijan, 2014; de Melo Pereira et al., 2018).General in vitro study properties of probiotics include resistance to gastric acid, bile acid resistance, adhesion to mucus or human epithelial cells, antimicrobial activity against potentially pathogenic bacteria or fungi, reduction of pathogen adhesion, bile salt hydrolase activity and enhancement of beneficial bacterial viability (Megur et al., 2023; Prakash et al., 2023; Sreepathi et al., 2023).
The rumen is one of the vital nutrient digestive organs in the digestive system of ruminants, and its internal microflora is very rich, including bacteria, fungi, archaea and a small number of phage viruses (Mizrahi et al., 2021). Rumen probiotics are probiotics that grow and multiply in the rumen of ruminants (Kmet et al., 1993) Rumen probiotics have specific functional roles in nutrition, digestion, immunity and health of ruminants. Numerous studies have shown that rumen probiotics can promote nitrite metabolism (Latham et al., 2018; Deng et al., 2021) promote intestinal development (Arshad et al., 2021) reduce methane production (Maake et al., 2021; Pittaluga et al., 2023), Fibre degradation (Chen et al., 2022), inhibition of pathogenic bacteria (Poothong et al., 2024), immunomodulation (Varada et al., 2022) and regulation of rumen acidosis (Lettat et al., 2012).
In recent years, high-throughput sequencing has provided a wealth of information on the composition, host specificity, and spatial and temporal dynamics of rumen-associated microbial communities (Li et al., 2023c). With the development of non-targeted and new high-throughput culture methods, culture genomics platforms using a range of media and high-throughput screening methods offer the potential to bring more “dark matter” into culture (Zhang et al., 2021). The genomics-based approach provides additional insights and suggests new hypotheses for most uncultured organisms (Gutleben et al., 2018). More importantly, with the elucidation of the beneficial mechanisms of some new strains, mining the next generation of candidate probiotics from the gut has become a new research hotspot (Wan et al., 2023). This paper provides an overview of the current applications of rumen probiotics in production, the classical techniques for culturing and identifying rumen probiotics, the culture strategies for cultivating “dark matter” from the rumen, and the technological tools for analyzing the diversity and dynamics of rumen bacteria (Figure 1).
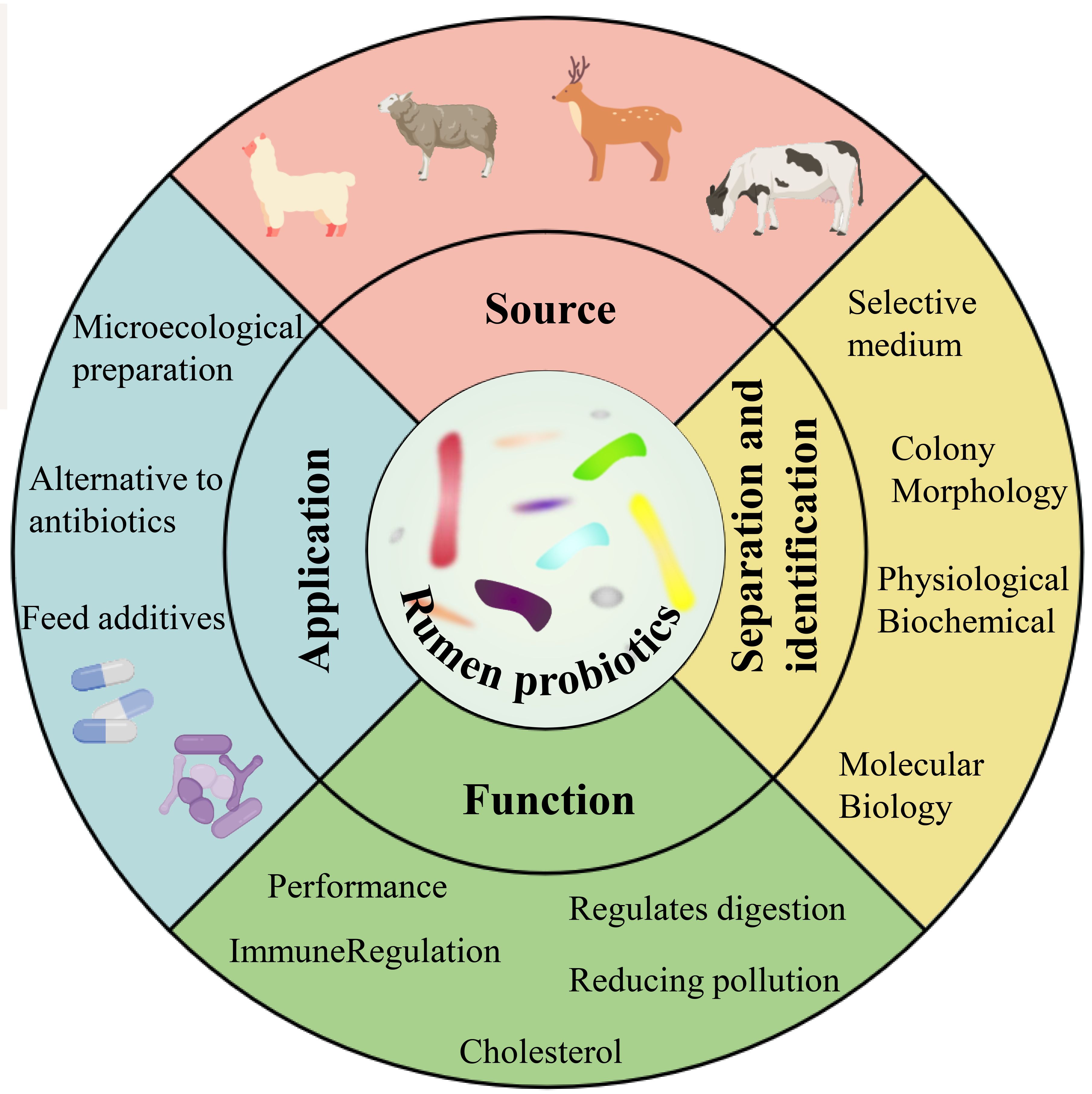
Figure 1 Sources, isolation and identification, functions and applications of rumen probiotics (Yuan et al., 2015; Noel et al., 2019; Xiong et al., 2020).
2 Probiotic bacteria in livestock and poultry breeding applications
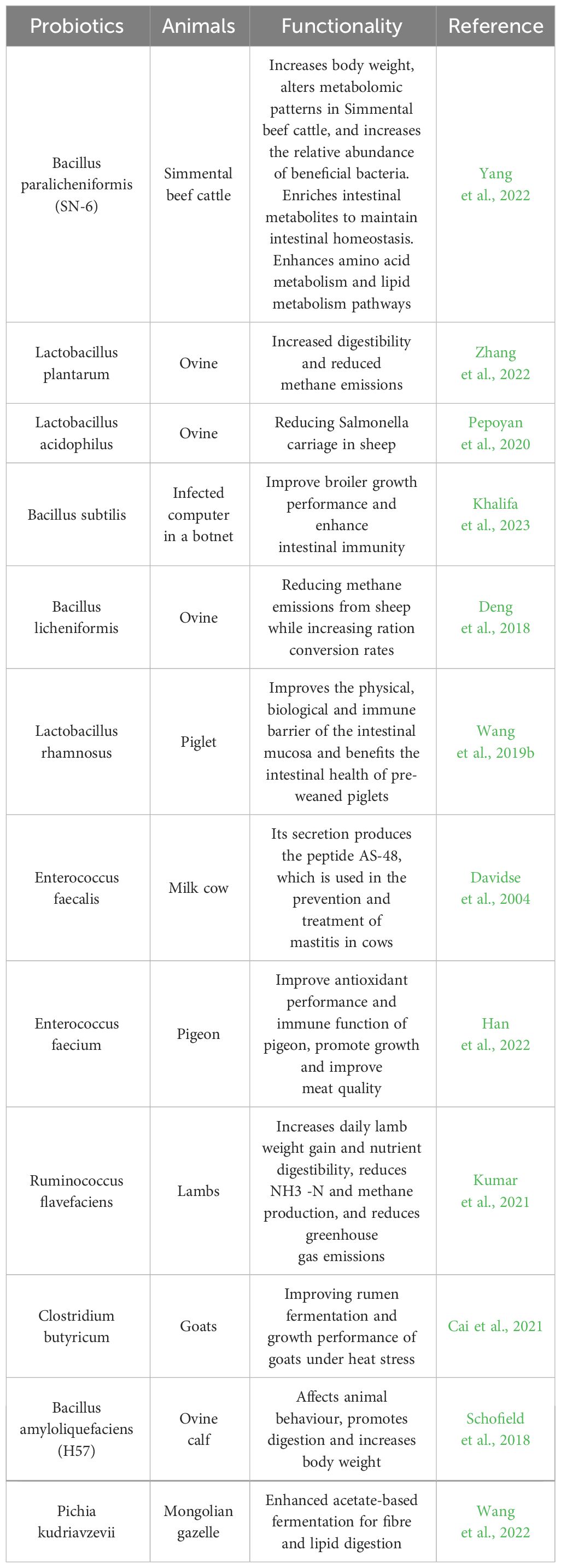
With the current research background of “forbidden resistance” on the feed side and “antibiotic reduction” and “substitute antibiotic” on the breeding side, probiotics are often used in the most common and widely used feed additives in the market because they can effectively improve the growth performance of livestock, enhance the immunity and regulate the gastrointestinal flora, and they are safe and environmentally friendly (Li et al., 2019; Wang et al., 2019a). Probiotic microecological preparations are prepared by isolating, identifying, and screening probiotics and their metabolites from different ecosystems and evaluating their safety in vitro, and through a series of technological treatments, they are prepared into biological preparations containing rich active bacteria (Xie et al., 2021). Current research has shown that enzyme preparations of cellulolytic bacteria isolated from the rumen of reindeer can increase average milk yield, enhance ciliate abundance, and improve the natural resistance of newborn calves (Litonina et al., 2021); The addition of Lactobacillus casei and Saccharomyces cerevisiae to diets increased the growth performance and apparent digestibility of nutrients, the relative abundance of short-chain fatty acid-producing microbiota, and the total short-chain fatty acid content of fattening pigs (Hassan et al., 2020). A strain of Bacillus denitrificans 79R4 (Paenibacillus 79R4) isolated from the bovine rumen enhanced rumen nitrite detoxification and reduced the risk of methemoglobinemia in cattle due to nitrate addition (Latham et al., 2019).
Other selected probiotic or potential probiotic application studies are shown in Table 1.
3 Isolation of rumen probiotics
The growth of microorganisms depends on various nutrient factors within the medium and various physicochemical factors in the culture environment, which are the primary conditions for the pure culture of microorganisms Ren et al. (2016). Therefore, understanding the nutritional requirements and metabolic properties is very important for the isolation and stable cultivation of microorganisms. The pure culture of rumen microorganisms is an important cornerstone for in-depth research on genes, proteins, and metabolic pathways, as well as a valuable resource for experimental research on microbial traits, enriching reference databases and biological classification frameworks (Stewart et al., 2019). The classical methods for isolation and culture of rumen probiotics include selective isolation and culture and the Heinz rolling tube method, which have inherent drawbacks such as low throughput, labor-intensive and high cost. The development of emerging technologies, such as microfluidic devices, offers great promise for high-throughput cultures (Figure 2).
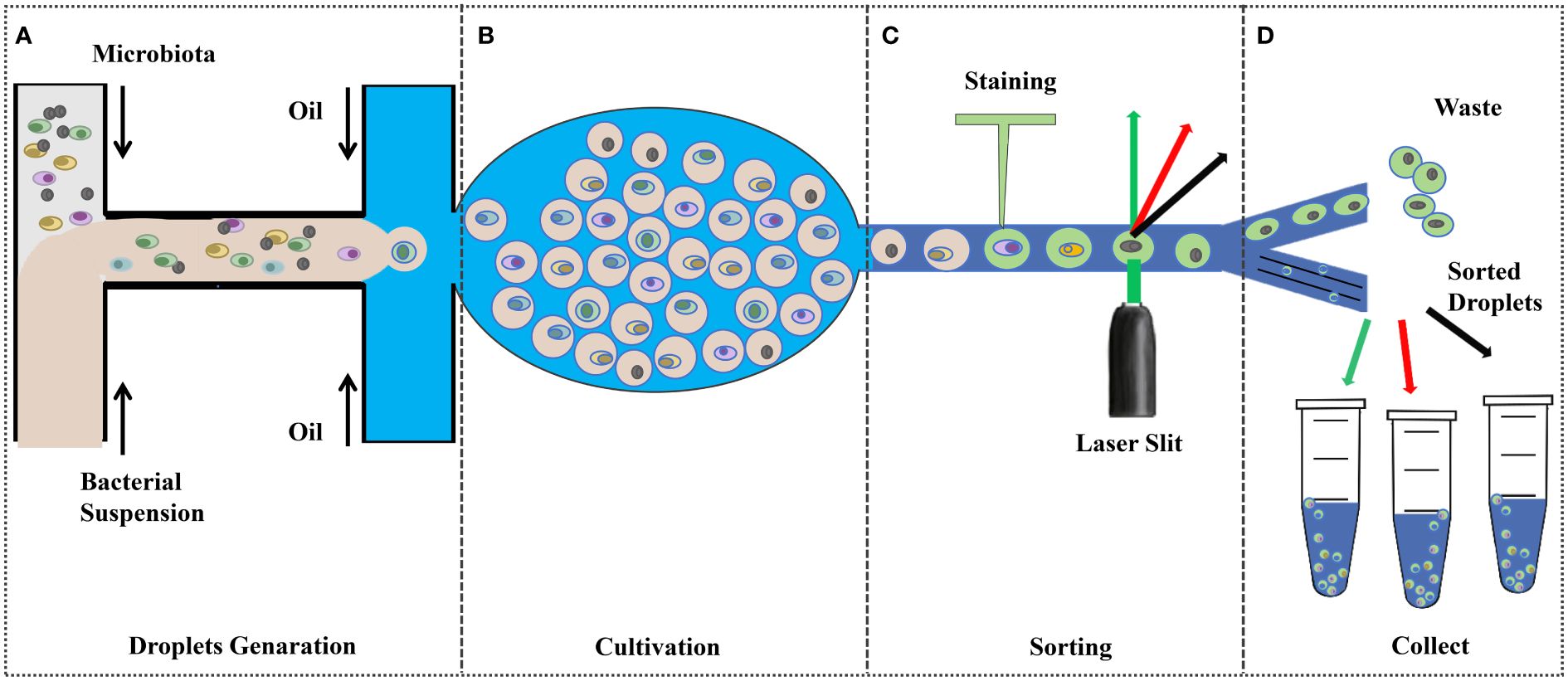
Figure 2 Droplet-based high-throughput culture (Tan et al., 2020; Anggraini et al., 2022; Yin et al., 2022; Zhao et al., 2016). (A) Pre-treated bacterial suspensions were injected into the microfluidic device to encapsulate the cells in the droplets; (B) Incubation under ideal conditions; (C);Fluorescence Signal Dependent Droplet Sorting; and (D) collect.
3.1 Selective isolation culture method
Selective isolation and culture method is to prepare the corresponding selection medium to enrich the target probiotics, which is essentially to eliminate the undesired microbial flora (Bonnet et al., 2019).The media used can be undefined and optimize the media using biochemical studies or one-factor methods according to experimental needs, which can reduce the complex effects between complex components and also allow easier detection of target metabolites (Hayek et al., 2019; Lewis et al., 2021). However, the number of rumen probiotics in the traditional culture strategy is limited, and the growth and metabolic characteristics of many rumen microorganisms are still unclear, which hinders the systematic study of rumen microorganisms. Currently, selective culture media among probiotics include Bifidobacterium selective medium, Lactobacillus selective medium, low carbon-resistant selective medium, and sodium carboxymethylcellulose, etc (Kumar et al., 2021; Jaglan et al., 2019; Kuppusamy et al., 2020). The addition of antioxidants (ascorbic acid, glutathione, and uric acid in the culture medium) enhances the culture of anaerobic bacteria (Tidjani Alou et al., 2020); It can also use the rumen fluid to simulate the natural environment of certain bacteria to promote their growth (Bonnet et al., 2019). Recently, strategies combining classical microbiological techniques with macro-barcoding methods have emerged to assess the selective enrichment efficacy of media for specific rumen microorganisms (Botero Rute et al., 2023). In addition, pharmaceuticals can be added to the medium as screening indicators according to experimental needs, e.g. Tyagi et al. (2020) to obtain lactic acid bacteria with conjugated linoleic acid production potential from the rumen fluid of lactating goats, linoleic acid was added to MRS broth medium; Zhong et al. (2023) to isolate novel urea-hydrolyzing bacteria from the rumen fluid of dairy cows, urea and phenol red were used as the screening indexes.
3.2 Hungate’s roll-tube technique
This method was proposed by American microbiologist Hungate in 1950 and applied to the isolation and culture of anaerobic bacteria (Hungate and Macy, 1973). After years of practice and improvement has been very mature. The basic steps of this method4~5 mL of dissolved agar culture solution were dispensed into Heinz tubes and inoculated when it cooled down to 43°C, immediately, the tubes were rolled rapidly at low temperatures to solidify the liquid agar. After agar curing, microbial colonies will be produced after several days of incubation under a constant temperature incubator at 39°C. Then, under an anaerobic environment, single colonies on the wall of the rolled tubes will be picked out by inoculation ring and put into fresh medium, and purified strains will be obtained through several times of rolling tubes and isolation of single colonies and the number of viable bacteria in the bacterial liquid will be measured, which will provide a basis for quantitative determination (Cheng et al., 2006; Jing et al., 2011). The Heinz tube rolling method can meet the growth requirements of different cultures, and isolation, purification, and morphological identification can be achieved in one tube. However, different microorganisms have different needs for the optimal medium, resulting in different growth rates, which has a greater impact on quantitative analysis (Guo et al., 2009). Hu et al. (2022) used the roll-tube technique to isolate Streptococcus equinus, Enterococcus avium, and Streptococcus lutetiensis, which have inhibitory effects on Escherichia coli and Staphylococcus aureus, from the rumen juice of Holstein cows as potential probiotics or silage inoculants. Kumar et al. (2021) screened three strains of cellulose-degrading rumen-producing Ruminococcus flavefaciens from buffalo rumen fluid by dispensing carboxymethylcellulose medium in Heinz tubes, and subsequent animal experiments showed that Ruminococcus flavefaciens significantly increased the number and activity of beneficial gut microorganisms and enhanced the digestive function of milk-producing buffaloes.
3.3 Droplet-based single-cell cultures
For microbial culture and screening, droplet-based microfluidic methods offer the advantages of single-cell, high-throughput, high-resolution, and low-cost, making them a promising approach for isolating uncultured microorganisms (Huys and Raes, 2018). Droplet-based microfluidics can provide a separate space for cells to divide and block the influence of competitors or predators, precisely control the microenvironment of single-cell cultures, manipulate individual cells through external interventions, and detect single-cell behaviors in real time for enzyme, antibody, or rare-cell screenings, and recover cells from the microfluidic system for a variety of downstream analyses, even after cultivation (Huys and Raes, 2018; Li et al., 2023a).
Microfluidic Stripe Plate (MSP) and SlipChip are two representative microfluidic methods for static droplets (Yu et al., 2022). MSP builds on traditional streak plate technology for high-throughput single-cell analysis and culture using nanolitre droplets that can be manually or robotically streaked onto Petri dishes for single-cell culture, as well as encoding chemical gradients in the droplet array for comprehensive dose-response analysis (Jiang et al., 2016). MSP can restore microbial diversity more than traditional agar plates. Currently, there are examples of applications of the MSP method in culturing and isolating Fastidous bacteria, such as the successful isolation of fluoranthene-degrading Blastococcus from the soil, studies of termite gut microbiota, and isolation of rare deep-sea biosphere members in long cultures (Jiang et al., 2016; Xu et al., 2018a; Zhou et al., 2019a). The SlipChip consists of two glass plates with several pre-filled reagent holes, the top plate is pre-filled with samples, and the bottom plate is pre-filled with reagents. Fluorocarbon compounds act as a lubricant layer between the two plates, and when the two plates slip, the complementary pattern of holes in the plates overlap to form tens of thousands of closed chambers or channels, the top plate sample-containing wells are exposed to the bottom plate reagent wells for reaction (Du et al., 2009). The enclosed microenvironment is particularly suitable for the cultivation of anaerobic microorganisms (Yu et al., 2022). Chen et al. (2019) A SlipChip device for chemotaxis sorting and a microfluidic streak plate device for bacterial culture were newly developed as new pipelines for screening and isolating microbial species that can degrade imidazolidinone as an imidazolidinone degrader.
Recently, Watterson et al. (2020) built a platform consisting of an image processing system and a droplet microfluidic device operating in an anaerobic chamber. This platform sorts slow-growers in microorganisms in droplets by density, speeding up their growth and enriching rare taxa in fecal microbiota samples,and realize high-throughput single-cell cultivation. However, as the vast majority of microorganisms colonizing the gastrointestinal tract are almost exclusively anaerobic, integrating a single-cell isolation platform into a standard anaerobic workstation is costly. Yin et al. (2022) improved a simple droplet-based method for isolation and enrichment of functional gut bacteria by encapsulation of single-cell suspensions, which was accomplished by using diluted bacterial suspensions as the dispersed phase mineral oil as the continuous phase and then transferring to agar plates in an anaerobic chamber for incubation to form discrete single-cell colonies. This method does not require sophisticated instrumentation to sort droplets and therefore can easily be operated in a conventional anaerobic chamber to successfully isolate anaerobic Lactobacilli and Bifidobacteria.
3.4 Droplet-based co-culture techniques
Droplet-based co-culture systems are likewise a promising approach for the discovery of natural microbial products and the isolation of uncultured microorganisms (Jian et al., 2020; Park et al., 2011). The system promotes continuous co-culture of colonies and cells by adjusting the concentration of microbial communities, developing microdroplets with different proportions of cells, simulating microenvironments to meet microbial growth requirements, and securing information exchange between strains (Qi et al., 2023). This process is expected to solve the problem that traditional pure culture methods can interrupt the ecological interactions between microorganisms, and provide a promising way to assess complex microbial communities in more detail (Anggraini et al., 2022; Kim et al., 2023; Baichman-Kass et al., 2023). Hua et al. (2022) created a rapid screening platform for actinomycetes——the whole-cell biosensor and producer co-cultivation-based microfluidic platform for screening actinomycetes (WELCOME), By combining an MphR-based Escherichia coli whole-cell biosensor sensitive to erythromycin with Saccharopolyspora erythraea co-cultivation, they isolated six high production erythromycin-producing strains from industrial strains within a short time. Tan et al. (2020) Complex human faecal samples were dissected into sub-communities for highly parallel co-culture using a droplet microfluidic device. Twenty-two individual droplets with strong bacterial symbiosis were then selected by microfluidics, in which a partial genome of a representative of a new genus of Neisseriaceae was found, highlighting the ability of microfluidic co-cultures to access and study uncharacterized microbial diversity.
4 Identification of rumen probiotics
In the early days, microbiological studies based on morphological features and physiological and biochemical traits provided insights into the microbial world, but today, they can only provide limited resolution (Escobar-Zepeda et al., 2015). Advances in molecular techniques have provided access to the “new uncultured world” of microbial communities. Among these techniques, polymerase chain reaction (PCR), denaturing gradient gel electrophoresis (DGGE), terminal restriction fragment length polymorphism (T-RFLP), fluorescence in-situ hybridization (FISH) and rRNA gene cloning and sequencing have had a significant impact (Escobar-Zepeda et al., 2015). However, while they describe the diversity of microorganisms, amplicon sequencing, macrogenomics, and single-cell genomics are the most widely used techniques for solving environmental microbiological problems (Xu and Zhao, 2018b).
4.1 Morphological identification
Morphological features are the important references for the classification and identification of microorganisms. Identification is carried out by observing both colony morphology and microscopic morphology. Colony morphology includes colony size, shape, surface, texture, transparency, degree of elevation, and medium color (Meruvu and Harsa, 2023). The bacterial morphology reflects the survival value of bacteria for acquiring nutrients, moving, and avoiding predators (Young, 2007). In microscopic morphology, commonly used types of microscopes are ordinary optical microscopes and phase contrast microscopes, for ultra-fine structures and complex structures can use scanning electron microscope and transmission electron microscope observation (Sun and Lin, 2009). With the rapid development of microimaging technologies and microfluidic chips, it becomes feasible to observe microbial germination and growth in real time (Zhou et al., 2019b). Zhang et al. (2019) used a gradient microfluidic chip to observe the morphological changes of various bacteria under the action of antibiotics. The rumen probiotics can also be classified with the help of Gram staining, which provides a reference basis for isolating rumen probiotics (Wang et al., 2021). Han et al. (2018) and Li et al. (2017) used colony morphology and Gram staining techniques for preliminary isolation and identification of probiotic bacteria in the samples, followed by 16SrRNA gene homology analysis to obtain probiotic strains of bovine origin.
4.2 Physiological and biochemical tests
The production and metabolic activities of microorganisms depend on extracellular enzymes to degrade macromolecular substances, the enzyme systems of different microorganisms will show significant differences in metabolic activities, which are confirmed by the changes in the substances in the vicinity of the microbial colonies, providing a basis for the identification and classification of microorganisms (Wu et al., 2018). At present, the physiological and biochemical tests that are more frequently used in identifying probiotics include the gelatin liquefaction test, methyl red test, glycolysis test, indole test, and starch hydrolysis test for the preliminary assessment of probiotics (Lin, 1999). The test is used for the preliminary evaluation of probiotics. However, due to the slow growth of some bacteria and the difficulty of cultivation, they cannot meet the requirements of biochemical reactions for microbial concentration and colony freshness, and the accuracy of biochemical tests for identifying anaerobic microorganisms in the rumen needs to be improved. Abid et al. (2022) in order to screen probiotic strains favorable for milk fermentation, four strains of Lactobacillus spp. were screened from 15 strains showing gas production in Durham tubes. These four probiotic strains were identified morphologically, identified by biochemical tests, and evaluated for their probiotic potential. It was found that Lactobacillus fermentum strains showed significant viability in the presence of pepsin, trypsin, and lysozyme.
4.3 16SrRNA-based amplicon sequencing
The advantage of amplicon sequencing lies in the contrasting biases generated by using only one phylogenetic marker (Escobar-Zepeda et al., 2015). Because 16SrRNAs are ubiquitous in all species and are functionally integral to the core genome, the composition and relative abundance of microbial communities in environmental samples are often investigated by amplifying and sequencing specific regions of the 16SrRNA gene (Daubin et al., 2003; Větrovský and Baldrian, 2013). The 16SrDNA gene sequence is divided into constant and variable regions. The constant region reflects the kinship between species (Head et al., 1998); The variable region reflects the specificity of the species and is used to classify them biologically (Chaudhary et al., 2015). Second-generation sequencing platforms, such as Illumina, can sequence amplicons of up to 600 bp with high precision (Bharti and Grimm, 2021). Third-generation amplicon sequencing platforms such as PacBio and Oxford Nanopore can sequence full-length 16S rRNA genes in a short period of time at the single-molecule level, which reduces the problems of amplification bias and short read lengths and makes it possible to annotate the microbiome at the species and strain level (Abellan-Schneyder et al., 2021). The 16SrRNA homology analysis has been successfully used to construct a gene library of Holstein cow rumen bacteria, which facilitates microbial species analysis (Yang et al., 2010; King et al., 2011). Although the 16SrRNA identification technique reflects the diversity of rumen microbes, it does not have a sufficient resolution at the species level, resulting in a loss of information on low-abundance members of the microbiota and an inability to understand the function of the community (Bowers et al., 2022). Abedini et al. (2023) reported for the first time the isolation of probiotics from camel rumen, preliminary screening of Gram-positive, catalase-negative colonies with white-colored colonies from the contents of camel rumen, identified as Enterococcus faecium96B4 by 16SrRNA, and subsequent evaluation of the probiotic activity and safety evaluation revealed good probiotic potential, reflecting the potential research value of camel rumen as a pristine environment.
4.4 Denaturing gradient gel electrophoresis
DGGE is an electrophoretic technique that separates DNA fragments based on differences in the order of DNA bases and is used to detect nucleic acid mutations and point mutations. Its basic principle is that in DNA molecules under the influence of specific temperature conditions and chemical denaturants, a region of double-stranded DNA starts to unlink, the unlinking region is related to the order of the base arrangement, and the unlinking can occur with the difference of only one base pair, and the difference of the base sequence of the DNA fragments will be denatured under the corresponding denaturing conditions in the process of the denaturing gradient gel swimming. When the ends of double-stranded DNA molecules are unstranded, their electrophoretic resistance increases greatly and their speed decreases significantly, which leads to the DNA fragments staying in different parts of the gel to achieve separation (Li et al., 2008; Ercolini, 2004). DGGE has the characteristics of good reproducibility, high detection rate, convenience and quickness, etc. It can be applied to the analysis of uncultured microorganisms (McGenity et al., 2010). DGGE was first applied to the structural analysis of rumen microorganisms by Kocherginskaya et al. (2001) analyzed the effect of two diets, hay, and maize, on the structure of the rumen bacterial community of castrated cows and showed that the bacterial populations in the rumen were relatively more diverse and numerous after feeding the maize diet. Through DGGE technology, Yu et al. (2020) found that supplementing Dihydropyridine (DHP) in the diet can promote the growth of Xanthomonadaceae and Xanthomonas and enhance the diversity of ruminal bacteria. Min et al. (2021) explored the effect of supplementing Condensed Tannins (CT) on calf rumen bacterial diversity and methane emissions. DGGE results showed that Firmicutes and Bacteroidetes populations seemed to increase as CT content increased., CT and can exert anti-methanogenic activity by directly inhibiting methanogens or indirectly through rumen fermentation, thereby reducing methane emissions.
4.5 Terminal restriction fragment length polymorphism
T-RFLP is an extension of RFLP. T-RFLP allows culture-independent assessment of subtle genetic differences between strains and provides a molecular approach to the evaluation of microbial community structure and function (Marsh, 1999). The technique uses PCR to amplify small subunit rRNA genes from total community DNA, where one or two primers are labeled with a fluorescent dye, followed by digestion of the PCR product with a restriction endonuclease with a four-base-pair recognition site and determination of the size and relative abundance of fluorescently-labeled T-RFs using a DNA sequencer. Because differences in T-RF size reflect sequence polymorphism, phylogenetically distinct populations of organisms can be resolved (Schütte et al., 2008). Although the use of T-RFLP is declining, it is still the method of choice for community dynamics studies (Prakash et al., 2014). Zhu et al. (2018) combined T-RFLP and clone library analysis to compare changes in rumen bacterial and archaeal communities in response to dietary disturbances before and after low-grain and high-grain production and found significant changes in the relative abundance of methane-producing communities in cows during the transition period, as well as a clear shift in rumen fermentation patterns.
4.6 Fluorescence in situ hybridization
FISH was introduced in 1980 (Bauman et al., 1980). FISH uses fluorescein oligonucleotide probes to bind complementarily to specific target nucleic acid sequences and detects the corresponding fluorescent signals for single-cell identification and quantification by fluorescence microscopy, whole-slide images, or flow cytometry (Amann and Fuchs, 2008). FISH has been widely used for the diagnosis of chromosomal aberrations in medicine, the identification of microorganisms in complex samples, and the identification of microorganisms (Ratan et al., 2017)., also provides a basis for in situ image-based spatial transcriptomics (Wen et al., 2022; Zhou et al., 2023). With improvements in fluorescence microscopy and fluorescent labeling of various nucleic acid probes, FISH has evolved to be used with other biotechnologies as a rapid and accurate biosensor system (Kuo et al., 2020). FISH combined with Raman spectroscopy can rapidly identify target microorganisms in complex samples by labeling the DNA of specific species (Cui et al., 2022). FISH can also be integrated into microfluidic microarray platforms to speed up the process of colony identification, reduce reagent consumption, and have the potential for automation (Rodrigues et al., 2021). Liu et al. (2009) developed a microfluidic device that integrates FISH identification and droplet-splitting modules for parallel high-throughput single-cell culture and identification. The single-cell droplet was split into two sub-droplets with the aid of a droplet-splitting chip, one of which was added to an agar plate, and the other was subjected to FISH identification, and the droplet that encapsulated the target species was finally selected from the agar plate based on the identification results. Liu et al. (2011) Seamlessly integrated two components, FISH and fluorescence-activated cell sorting (FACS), into a microfluidic device, which forms a hybridization chamber between two photopolymeric membranes in which cells and probes are electrophoretically loaded, incubated, and washed; a downstream cross structure is used to electrically focus the cells into a single flow for FACS analysis, providing a quantitative detection of microbial cells in complex samples automated platform. Batani et al. (2019) developed a new method to isolate live bacteria based only on their 16S rRNA gene sequences (Live-FISH), which combines FISH with FACS to enable the sorting and culturing of live bacteria. With the development of highly specific probes, Live-FISH has greater potential for targeted sorting of target microorganisms. However, FISH requires the rRNA content or the number of microorganisms in the probe target organisms, otherwise the fluorescence signal cannot be detected under the microscope (Hoshino et al., 2008). In addition, the FISH technique suffers from the problem of crosstalk of organic fluorescein excitation light (Waters, 2009). Meanwhile, FISH can only identify a small number of microorganisms with a high degree of certainty in a single experiment, and cannot elucidate the distribution of microorganisms in the overall microecosystem. The development of combinatorial labeling and spectroscopic imaging can increase the microspatial relationships of different species of microorganisms in a single field of view, but it is too costly (Valm et al., 2011).
4.7 Macrogenomics
Automated sequencing of DNA by Sanger sequencing in the late 1970s ushered in the era of genomics Sanger sequencing retrieves up to 96 sequences per run at an average length of 650 bp, which may be sufficient for phylogenetic marker analyses, but the emergence of what is known as next-generation sequencing (NGS) technology has enabled researchers to bypass the cultivation of parallel sequencing of millions of DNA molecules with varying yields and sequence lengths directly after extraction from highly diverse populations, i.e., macrogenome sequencing (Escobar-Zepeda et al., 2015; Hu et al., 2021). It is usually performed using birdshot sequencing methods that are non-discriminatory, allowing for the quantitative assignment of taxonomy and organisms to the species level, and with the help of databases of identified functional genes, such as KEGG, GO, COG, and eggNOG, the understanding of microbial communities in terms of their composition, function, evolution, and interactions in their natural environments, which directly contributes to the flourishing of microbial ecology (Tatusov et al., 1997; Kanehisa et al., 2008; Huerta-Cepas et al., 2016). However, macrogenome sequencing uses environmental DNA samples, which cannot link each functional gene to a specific microbial individual, and for high-diversity samples or low-abundance organisms, it is very difficult to assemble a single discrete genome to capture strain-level variation (Kaster and Sobol, 2020; Daliri et al., 2021). Li et al. (2023c) studied changes in bovine rumen microbes from pre-transport to 1-month post-transport by macrogenomics and found that the abundance of rumen bacteria and archaea was higher on day 16 post-transport than pre-transport, but eukaryotic abundance was highest on day 30 post-transport. Before transport, most bacteria were mainly involved in polysaccharide digestion. On day 4 post-transport, KEGG pathway enrichment was most notable for nucleotide metabolism. On day 16 post-transhipment, energy metabolism and rumen content of MCPs and VFAs increased significantly, but at the same time, energy loss due to methane production (Methanobrevibacter) and pathogenic bacteria (Saccharopolyspora rectivirgula) together induced inflammation and oxidative stress in cattle, which is important for the establishment of new management and nutritional specification strategies.
4.8 Single-cell genomics
Genomics for microorganisms in the environment, macro-genome sequencing describes the full range of genetic information, and single-cell genomics reveals individual genomes, and combining the two can compensate for their respective shortcomings (Nobu et al., 2015; Mende et al., 2016). Sequencing the microbiome at the resolution of individual microorganisms effectively improves the efficiency and accuracy of obtaining genome-wide information from complex microbial communities, and also allows for the study of individual cellular behaviors underlying the complexity of microbial ecosystems (Lloréns-Rico et al., 2022; Madhu et al., 2023). Single-cell isolation is essential to performing high-throughput single-cell genomics workflows (Xu et al., 2018a). Microfluidic devices offer advantages in terms of throughput, affordability, and automation for single-cell capture and retrieval applications (Han et al., 2023). Its subsequent steps include DNA extraction, 16S rRNA gene PCR phylogenetic identification, multiple displacement amplification (MDA), library construction, sequencing, and data analysis (Xu and Zhao, 2018b; Arikawa et al. (2021) developed a framework for integrating single-cell genomics and macrogenomics, integrating single-cell amplified genomes (SAG) and macrogenome assembled genomes (MAG) to reconstruct competent microbial genomes, and achieving high-quality recovery of strain-resolved genomes. Zheng et al. (2022) developed and validated a strain-resolved, high-throughput single-cell sequencing method (Microbe-seq) that uses a microfluidic platform to individually encapsulate microorganisms into droplets where whole-genome amplification and specific barcode coding are performed, followed by sequencing of merged labeled DNA to generate a single amplified genome (SAG), and then finally co-assembling the SAGs of the same bacterial species. SAGs from the same bacterial species are finally co-assembled to achieve single-strain level resolution. However, SAGs can introduce chimeric and biased sequences during genome amplification, leading to sequence incompleteness. To address this problem, Kogawa et al. (2023) developed a single-cell amplified genome long read-length assembly workflow to construct complete circular SAGs (cSAGs).
5 Culturography
Culturomics is a high-throughput method of isolating and culturing bacteria under as many combinations of culture conditions as possible, using a combination of matrix-assisted laser-resolved ionization time-of-flight mass spectrometry (MALDI-TOF MS) or other sequencing technologies for microbial identification (Diakite et al., 2021; Wan et al., 2023; Yu et al., 2023). It is a high-throughput method for bacterial isolation and culture. The technique was first used for the analysis of uncultured microorganisms identified in the human gut, and with the significant expansion of strain databases, many microorganisms previously overlooked or considered unculturable have been brought into the culture (Diakite et al., 2021). Classical cultureomics is an untargeted strategy that includes steps such as sample collection, sample processing, microbial isolation, culture, identification, and preservation (Wan et al., 2023). That is, the treated samples are dispersed and cultured into different media, and then the characteristic colonies are selected for identification using MALDI-TOF MS analysis if the reference profiles are lacking, 16SrRNA sequencing is required for further identification, and if the similarity is < 98.65%, it is considered to be a new species, and then describing new species using taxonomic genomics (Lagier et al., 2018; Peng et al., 2020). In addition, with the use of membrane diffusion culture (Nichols et al., 2010; Chaudhary et al., 2019) microfluidic devices (Anggraini et al., 2022; Luo et al., 2022) FACS, FISH (Waters, 2009) and reverse genomics (Cross et al., 2019), and other high-throughput culture technologies and targeted sorting techniques, the culture results of cultureomics will be greatly enriched (Figure 3). Combined with full-length 16S rRNA gene amplicons and birdshot macro-genome sequencing, culture-based metabonomics (CBM) will deeply explore untapped novel bacterial resources at high resolution (Li et al., 2023b). In addition, information specific to the target microorganisms obtained in advance from literature studies or macrogenomics data can also be used to reverse-guide the isolation and culture of the target microorganisms, providing additional opportunities to obtain pure cultures (Bellais et al., 2022; Liu et al., 2022). Of course, there are still some constraints in cultureomics: it is still at the beginning stage in the cultivation of rumen microorganisms, which is labor-intensive and will result in a waste of manpower and resources (Mordant and Kleiner, 2021); gut-microbiota interactions and symbiotic relationships between microorganisms are still unclear (Yadav et al., 2022).
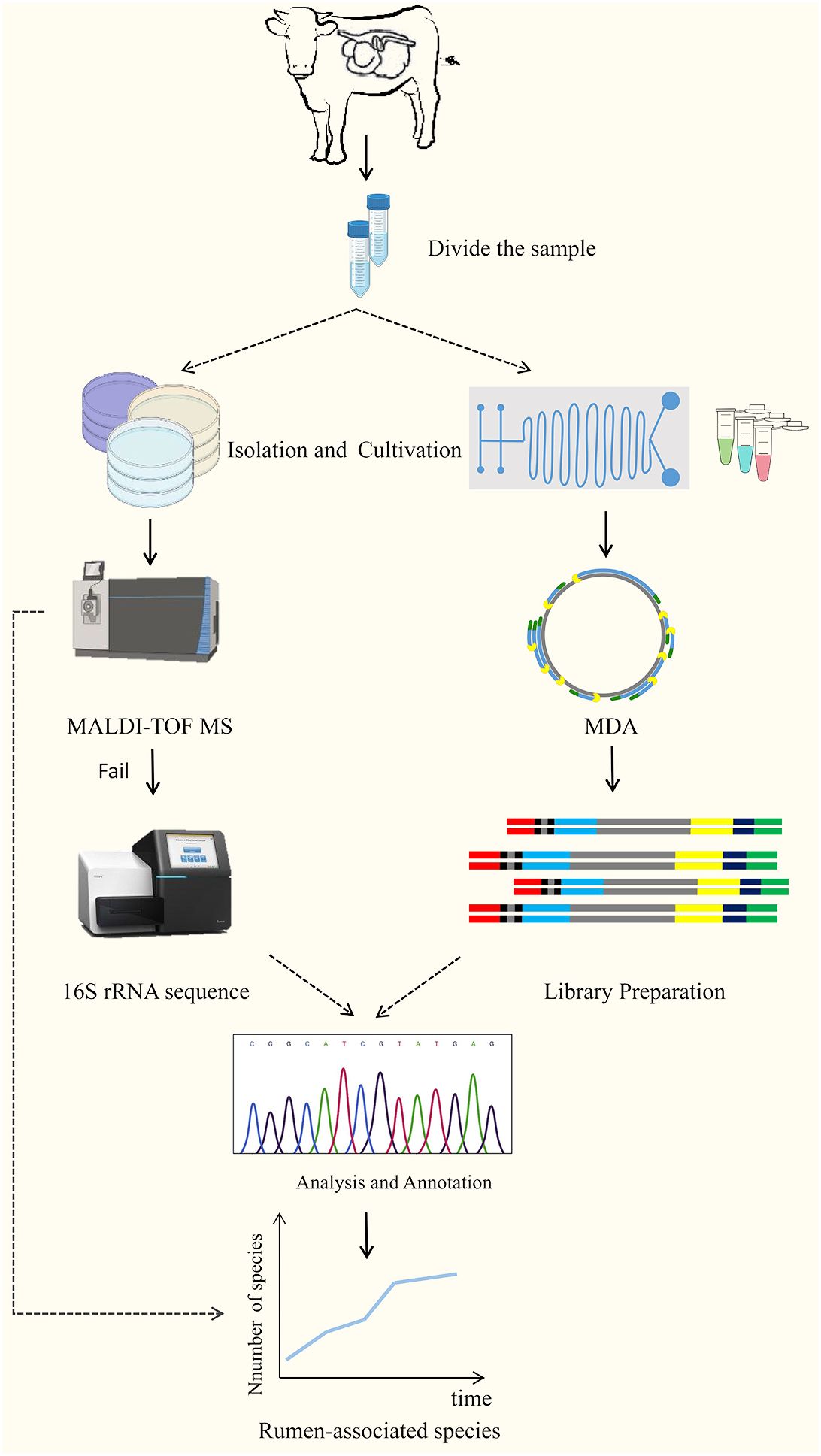
Figure 3 The technical route of culturomics (Wan et al., 2023).
6 Perspectives and conclusions
The rumen microecological environment is extremely complex, and it is difficult to simulate its physicochemical parameters in vitro. Traditional in vitro techniques for isolation and culture of rumen probiotics have significant limitations and it is difficult to identify rare microorganisms by general identification techniques. Therefore, it is necessary to introduce new techniques for large-scale mining of probiotics in the rumen. This will help rationally define the beneficial flora, pinpoint microbial-derived metabolites with beneficial effects, and provide technical support for developing novel probiotic formulations. This is of great significance for animal nutrition and health, food safety, and ecological protection.
7 Future Direction
The first key feature of the state-of-the-art technologies presented in this review is high-throughput culture. Traditional laboratory microbiological cultures are usually inefficient and time-consuming, but the development of microfluidic devices has created conditions for high-throughput cultures. Therefore, instrument design, microfluidic chip design, and fabrication have become crucial. The integration of experimental functions such as dilution, separation, single-cell encapsulation, anaerobic incubation, and targeted sorting onto a small chip requires interdisciplinary collaboration, while reagent development, construction of equipment such as anaerobic devices, and sorting and collection devices requires an experienced R&D team. Second, genome sequencing at single-cell resolution is another key feature. Single-cell sequencing can reveal functional information about rare species, help understand inter-microbial or microbe-host interactions, and explore potential metabolic pathways. The development of various histological approaches will also provide information on the rumen microbiome and its metabolites, which will help guide the isolation and culture of target microorganisms. While cultureomics, with its attributes of high-throughput culture and single-cell resolution, will significantly increase the potential for isolating and culturing “dark matter” from the rumen, this endeavor has significant challenges. The adaptation of optimal media for microorganisms is time- and labor-intensive, and the design of co-culture or mono-culture systems may not be able to meet the growth needs of all microorganisms. In addition, targeted sorting devices such as FACS and FISH still suffer from problems such as crosstalk and insufficient number of stains. Overcoming these technological bottlenecks requires the unremitting efforts of the R&D team.
Rumen probiotics from cattle and sheep are relatively well documented, but there are fewer reports on probiotics from antelope, deer, and musk family sources. The rumen microbial composition is very rich and susceptible to factors such as feeding management, geographic location, and significant differences in microbial composition between individuals and species, so gastrointestinal microbiological studies in rare ruminants may yield unexpected findings. In addition, the research and application of rumen probiotics require the establishment of a standard system for probiotic isolation and culture, identification, evaluation of probiotic properties, safety evaluation, and application. Only by ensuring the maximum use of strain resources can the potential of rumen probiotics be better explored, their application in agriculture and industry be improved, and a greater contribution be made to the sustainable development of animal husbandry.
Author contributions
RW: Writing – original draft, Writing – review & editing. PJ: Conceptualization, Investigation, Project administration, Supervision, Writing – review & editing. YH: Data curation, Funding acquisition, Resources, Writing – review & editing. HL: Conceptualization, Investigation, Writing – review & editing. WZ: Data curation, Writing – review & editing. YW: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Work in our lab was funded by the National Natural Science Foundation of China (grant number U21A20262, 32260899, 32060812), Ningxia Key R&D Project (2023BCF01039); Gansu Province University Industry Support Project (2020C-14); Gansu Agricultural University Youth Tutor Fund Grant (GAU-QDFC-2021-04); Ministry of Finance and Ministry of Agriculture and Rural Development: National Modern Agricultural Industrial Technology System (CARS-37).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abedini, R., Zaghari, G., Jabbari, L., Salekdeh, G. H., Hashemi, M. (2023). A potential probiotic Enterococcus faecium isolated from camel rumen, fatty acids biotransformation, antilisteria activity and safety assessment. Int. Dairy J. 145, 105706. doi: 10.1016/j.idairyj.2023.105706
Abellan-Schneyder, I., MatChado, M. S., Reitmeier, S., Sommer, A., Sewald, Z., Baumbach, J., et al. (2021). Primer, pipelines, parameters: issues in 16S rRNA gene sequencing. mSphere 6, e01202–e01220. doi: 10.1128/mSphere.01202-20
Abid, S., Farid, A., Abid, R., Rehman, M. U., Alsanie, W. F., Alhomrani, M., et al. (2022). Identification, Biochemical Characterization, and Safety Attributes of Locally Isolated Lactobacillus fermentum from Bubalus bubalis (buffalo) Milk as a Probiotic. Microorganisms 10, 954. doi: 10.3390/microorganisms10050954
Amann, R., Fuchs, B. M. (2008). Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6, 339–348. doi: 10.1038/nrmicro1888
Anggraini, D., Ota, N., Shen, Y., Tang, T., Tanaka, Y., Hosokawa, Y., et al. (2022). Recent advances in microfluidic devices for single-cell cultivation: methods and applications. Lab. chip 22, 1438–1468. doi: 10.1039/D1LC01030A
Arikawa, K., Ide, K., Kogawa, M., Saeki, T., Yoda, T., Endoh, T., et al. (2021). Recovery of strain-resolved genomes from human microbiome through an integration framework of single-cell genomics and metagenomics. Microbiome 9, 202. doi: 10.1186/s40168-021-01152-4
Arshad, M. A., Hassan, F. U., Rehman, M. S., Huws, S. A., Cheng, Y., Din, A. U. (2021). Gut microbiome colonization and development in neonatal ruminants: Strategies, prospects, and opportunities. Anim. Nutr. (Zhongguo xu mu shou yi xue hui) 7 (3), 883–895. doi: 10.1016/j.aninu.2021.03.004
Baichman-Kass, A., Song, T., Friedman, J. (2023). Competitive interactions between culturable bacteria are highly non-additive. eLife 12, e83398. doi: 10.7554/eLife.83398.sa2
Batani, G., Bayer, K., Böge, J., Hentschel, U., Thomas, T. (2019). Fluorescence in situ hybridization (FISH) and cell sorting of living bacteria. Sci. Rep. 9, 18618. doi: 10.1038/s41598-019-55049-2
Bauman, J. G., Wiegant, J., Borst, P., van Duijn, P. (1980). A new method for fluorescence microscopical localization of specific DNA sequences by in situ hybridization of fluorochromelabelled RNA. Exp. Cell Res. 128, 485–490. doi: 10.1016/0014-4827(80)90087-7
Bellais, S., Nehlich, M., Ania, M., Duquenoy, A., Mazier, W., van den Engh, G., et al. (2022). Species-targeted sorting and cultivation of commensal bacteria from the gut microbiome using flow cytometry under anaerobic conditions. Microbiome 10, 24. doi: 10.1186/s40168-021-01206-7
Bharti, R., Grimm, D. G. (2021). Current challenges and best-practice protocols for microbiome analysis. Briefings Bioinf. 22, 178–193. doi: 10.1093/bib/bbz155
Bonnet, M., Lagier, J. C., Raoult, D., Khelaifia, S. (2019). Bacterial culture through selective and non-selective conditions: the evolution of culture media in clinical microbiology. New Microbes New Infect. 34, 100622. doi: 10.1016/j.nmni.2019.100622
Botero Rute, L. M., Caro-Quintero, A., Acosta-González, A. (2023). Enhancing the conventional culture: the evaluation of several culture media and growth conditions improves the isolation of ruminal bacteria. Microbial Ecol. 87, 13. doi: 10.1007/s00248-023-02319-2
Bowers, R. M., Nayfach, S., Schulz, F., Jungbluth, S. P., Ruhl, I. A., Sheremet, A., et al. (2022). Dissecting the dominant hot spring microbial populations based on community-wide sampling at single-cell genomic resolution. ISME J. 16, 1337–1347. doi: 10.1038/s41396-021-01178-4
Cai, L., Hartanto, R., Zhang, J., Qi, D. (2021). Clostridium butyricum improves rumen fermentation and growth performance of heat-stressed goats in vitro and in vivo. Animals: an Open Access J. MDPI 11, 3261. doi: 10.3390/ani11113261
Chaudhary, D. K., Khulan, A., Kim, J. (2019). Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 9, 6666. doi: 10.1038/s41598-019-43182-x
Chaudhary, N., Sharma, A. K., Agarwal, P., Gupta, A., Sharma, V. K. (2015). 16S classifier: a tool for fast and accurate taxonomic classification of 16S rRNA hypervariable regions in metagenomic datasets. PloS One 10, e0116106. doi: 10.1371/journal.pone.0116106
Chen, D., Liu, S. J., Du, W. (2019). Chemotactic screening of imidazolinone-degrading bacteria by microfluidic SlipChip. J. Hazard. Mater. 366, 512–519. doi: 10.1016/j.jhazmat.2018.12.029
Chen, X., Yan, F., Liu, T., Zhang, Y., Li, X., Wang, M., et al. (2022). Ruminal microbiota determines the high-fiber utilization of ruminants: evidence from the ruminal microbiota transplant. Microbiol. Spectr. 10, e0044622. doi: 10.1128/spectrum.00446-22
Cheng, Y. M., Deng, W. D., Mao, H. M. (2006). Research progress on traditional techniques and molecular biological techniques in modern times for rumen microbial diversity. Prog. Vet. Med. S1, 51–54. doi: 10.16437/j.cnki.1007-5038.2006.s1.013
Cross, K. L., Campbell, J. H., Balachandran, M., Campbell, A. G., Cooper, C. J., Griffen, A., et al. (2019). Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol. 37, 1314–1321. doi: 10.1038/s41587-019-0260-6
Cui, D., Kong, L., Wang, Y., Zhu, Y., Zhang, C. (2022). In situ identification of environmental microorganisms with Raman spectroscopy. Environ. Sci. ecotechnology 11, 100187. doi: 10.1016/j.ese.2022.100187
Daliri, E. B., Ofosu, F. K., Chelliah, R., Lee, B. H., Oh, D. H. (2021). Challenges and perspective in integrated multi-omics in gut microbiota studies. Biomolecules 11, 300. doi: 10.3390/biom11020300
Dasriya, V. L., Samtiya, M., Ranveer, S., Dhillon, H. S., Devi, N., Sharma, V., et al. (2024). Modulation of gut-microbiota through probiotics and dietary interventions to improve host health. J. Sci. Food Agric., 1–17. doi: 10.1002/jsfa.13370
Daubin, V., Moran, N. A., Ochman, H. (2003). Phylogenetics and the cohesion of bacterial genomes. Science 301, 829–832. doi: 10.1126/science.1086568
Davidse, E. K., Balla, E., Holzapfel, W. H., Muller, C. J. C., Cloete, S. W. P., Dicks, L. M. T. (2004). Peptide AS-48 (Enterococcus faecalis) for the prevention and treatment of mastitis in dairy cows. Online J. Vet. Res. 8, 22–32.
de Melo Pereira, G. V., de Oliveira Coelho, B., Magalhães Júnior, A. I., Thomaz-Soccol, V., Soccol, C. R. (2018). How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 36 (8), 2060–2076. doi: 10.1016/j.biotechadv.2018.09.003
Deng, B., Wang, L., Ma, Q., Yu, T., Liu, D., Dai, Y., et al. (2021). Genomics analysis of bacillus megaterium 1259 as a probiotic and its effects on performance in lactating dairy cows. Animals: an Open Access J. MDPI 11, 397. doi: 10.3390/ani11020397
Deng, K. D., Xiao., Y., Ma., T., Tu., Y., Diao., Q. Y., Chen., Y. H., et al. (2018). Ruminal fermentation, nutrient metabolism, and methane emissions of sheep in response to dietary supplementation with Bacillus licheniformis. Anim. Feed Sci. Technol. 241, 38–44. doi: 10.1016/j.anifeedsci.2018.04.014
Diakite, A., Dubourg, G., Raoult, D. (2021). Updating the repertoire of cultured bacteria from the human being. Microbial pathogenesis 150, 104698. doi: 10.1016/j.micpath.2020.104698
Du, W., Li, L., Nichols, K. P., Ismagilov, R. F. (2009). SlipChip. Lab. chip 9, 2286–2292. doi: 10.1039/b908978k
Ercolini, D. (2004). PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56 (3), 297–314. doi: 10.1016/j.mimet.2003.11.006
Escobar-Zepeda, A., Vera-Ponce de León, A., Sanchez-Flores, A. (2015). The road to metagenomics: from microbiology to DNA sequencing technologies and bioinformatics. Front. Genet. 6. doi: 10.3389/fgene.2015.00348
FAO/WHO (2001). Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Food and Agriculture Organization of the United Nations, 1–14.
Fijan, S. (2014). Microorganisms with claimed probiotic properties: an overview of recent literature. Int. J. Environ. Res. Public Health 11, 4745–4767. doi: 10.3390/ijerph110504745
Guo, J. T., Wang, J. Q., Wang, J. P., Huo, X. K., Bo, D. P., Wei, H. Y., et al. (2009). Research progress on quantitative method for rumen microorganism. China Anim. Husbandry Vet. Med. 36, 19–24.
Gutleben, J., Chaib De Mares, M., van Elsas, J. D., Smidt, H., Overmann, J., Sipkema, D. (2018). The multi-omics promise in context: from sequence to microbial isolate. Crit. Rev. Microbiol. 44, 212–229. doi: 10.1080/1040841X.2017.1332003
Han, Y., Li, X., Guo, P. F., Guan, G. Q., Liu, J. L., Liu, A. H., et al. (2018). lsolation and ldentification of probiotics bacterium in calve faeces. China Dairy Cattle 07, 21–24.
Han, X., Xu, X., Yang, C., Liu, G. (2023). Microfluidic design in single-cell sequencing and application to cancer precision medicine. Cell Rep. Methods 3, 100591. doi: 10.1016/j.crmeth.2023.100591
Han, P. M., Zhang, R., Bian, S. X., Li, L., Ni, A. X., Wang, Y. M., et al. (2022). Effects of Enterococcus faeciumand Bacillus subtilis on Growth Performance,Carcass Traits and lmmune Function of Squabs. Acta Veterinaria Zootechnica Sin. 53, 3880–3891. doi: 10.5555/20230041782
Hassan, A., Gado, H., Anele, U. Y., Berasain, M. A. M., Salem, A. Z. M. (2020). Influence of dietary probiotic inclusion on growth performance, nutrient utilization, ruminal fermentation activities and methane production in growing lambs. Anim. Biotechnol. 31, 365–372. doi: 10.1080/10495398.2019.1604380
Hayek, S. A., Gyawali, R., Aljaloud, S. O., Krastanov, A., Ibrahim, S. A. (2019). Cultivation media for lactic acid bacteria used in dairy products. J. Dairy Res. 86, 490–502. doi: 10.1017/S002202991900075X
Head, I. M., Saunders, J. R., Pickup, R. W. (1998). Microbial evolution, diversity, and ecology: A decade of ribosomal RNA analysis of uncultivated microorganisms. Microbial Ecol. 35, 1–21. doi: 10.1007/s002489900056
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Hoshino, T., Yilmaz, L. S., Noguera, D. R., Daims, H., Wagner, M. (2008). Quantification of target molecules needed to detect microorganisms by fluorescence in situ hybridization (FISH) and catalyzed reporter deposition-FISH. Appl. Environ. Microbiol. 74, 5068–5077. doi: 10.1128/AEM.00208-08
Hu, G., Jiang, H., Zong, Y., Datsomor, O., Kou, L., An, Y., et al. (2022). Characterization of lactic acid-producing bacteria isolated from rumen: growth, acid and bile salt tolerance, and antimicrobial function. Fermentation 8, 385. doi: 10.3390/fermentation8080385
Hu, B., Xu, P., Ma, L., Chen, D., Wang, J., Dai, X., et al. (2021). One cell at a time: droplet-based microbial cultivation, screening and sequencing. Mar. Life Sci. Technol. 3, 169–188. doi: 10.1007/s42995-020-00082-8
Hua, E., Zhang, Y., Yun, K., Pan, W., Liu, Y., Li, S., et al. (2022). Whole-cell biosensor and producer co-cultivation-based microfludic platform for screening saccharopolyspora erythraea with hyper erythromycin production. ACS synthetic Biol. 11, 2697–2708. doi: 10.1021/acssynbio.2c00102
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Hungate, R. E., Macy, J. (1973). The roll-tube method for cultivation of strict anaerobes. Bulletins Ecol. Res. committee 17), 123–126. Available at: http://www.jstor.org/stable/20111550.
Huys, G. R., Raes, J. (2018). Go with the flow or solitary confinement: a look inside the single-cell toolbox for isolation of rare and uncultured microbes. Curr. Opin. Microbiol. 44, 1–8. doi: 10.1016/j.mib.2018.05.002
Jaglan, N., Kumar, S., Choudhury, P. K., Tyagi, B., Tyagi, A. K. (2019). Isolation, characterization and conjugated linoleic acid production potential of bifidobacterial isolates from ruminal fluid samples of Murrah buffaloes. Anaerobe 56, 40–45. doi: 10.1016/j.anaerobe.2019.02.001
Jian, X., Guo, X., Wang, J., Tan, Z. L., Xing, X. H., Wang, L., et al. (2020). Microbial microdroplet culture system (MMC): An integrated platform for automated, high-throughput microbial cultivation and adaptive evolution. Biotechnol. Bioeng. 117, 1724–1737. doi: 10.1002/bit.27327
Jiang, C. Y., Dong, L., Zhao, J. K., Hu, X., Shen, C., Qiao, Y., et al. (2016). High-throughput single-cell cultivation on microfluidic streak plates. Appl. Environ. Microbiol. 82, 2210–2218. doi: 10.1128/AEM.03588-15
Jing, Y. Q., Song, E. L., Yang, W. R., Liu, G. F., Tan, X. W., Wan, F. C. (2011). Progress on quantification methods of bovine rumen microbes. Prog. Vet. Med. 32, 93–96. doi: 10.16437/j.cnki.1007-5038.2011.07.002
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kaster, A. K., Sobol, M. S. (2020). Microbial single-cell omics: the crux of the matter. Appl. Microbiol. Biotechnol. 104, 8209–8220. doi: 10.1007/s00253-020-10844-0
Khalifa, A., Ibrahim, H. M., Sheikh, A. (2023). Bacillus subtilis PM5 from Camel Milk Boosts Chicken Immunity and Abrogates Salmonella entertitidis Infections. Microorganisms 11, 1719. doi: 10.3390/microorganisms11071719
Kim, S., Moon, J. H., Jung, G. Y. (2023). Recent progress in the development of droplet-based microfluidic technologies for phenotypic screening using cell-cell interactions. Biotechnol. Bioprocess Eng. 28 (6), 929–935. doi: 10.1007/s12257-022-0081-1
King, E. E., Smith, R. P., St-Pierre, B., Wright, A. D. (2011). Differences in the rumen methanogen populations of lactating Jersey and Holstein dairy cows under the same diet regimen. Appl. Environ. Microbiol. 77, 5682–5687. doi: 10.1128/AEM.05130-11
Kmet, V., Flint, H. J., Wallace, R. J. (1993). Probiotics and manipulation of rumen development and function. Archiv fur Tierernahrung 44, 1–10. doi: 10.1080/17450399309386053
Kocherginskaya, S. A., Aminov, R. I., White, B. A. (2001). Analysis of the Rumen Bacterial Diversity under two Different Diet Conditions using Denaturing Gradient Gel Electrophoresis, Random Sequencing, and Statistical Ecology Approaches. Anaerobe 7, 1–16. doi: 10.1006/anae.2001.0378
Kogawa, M., Nishikawa, Y., Saeki, T., Yoda, T., Arikawa, K., Takeyama, H., et al. (2023). Revealing within-species diversity in uncultured human gut bacteria with single-cell long-read sequencing. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1133917
Kumar, B., Kumar, D., Mahesh, M. S., Sheel, R. (2021). Effect of oral dosing of live and autoclaved culture of Ruminococcus flavefaciens FD-1 on rumen bacterial and fungal populations in Murrah buffaloes. Indian J. Dairy Sci. 74 (5), 458–461. doi: 10.33785/IJDS.2021.v74i05.014
Kuo, J. T., Chang, L. L., Yen, C. Y., Tsai, T. H., Chang, Y. C., Huang, Y. T., et al. (2020). Development of fluorescence in situ hybridization as a rapid, accurate method for detecting coliforms in water samples. Biosensors 11 (1), 8. doi: 10.3390/bios11010008
Kuppusamy, P., Kim, D., Soundharrajan, I., Park, H. S., Jung, J. S., Yang, S. H., et al. (2020). Low-carbohydrate tolerant LAB strains identified from rumen fluid: investigation of probiotic activity and legume silage fermentation. Microorganisms 8, 1044. doi: 10.3390/microorganisms8071044
Lagier, J. C., Dubourg, G., Million, M., Cadoret, F., Bilen, M., Fenollar, F., et al. (2018). Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 16, 540–550. doi: 10.1038/s41579-018-0041-0
Latham, E. A., Pinchak, W. E., Trachsel, J., Allen, H. K., Callaway, T. R., Nisbet, D. J., et al. (2018). Isolation, characterization and strain selection of a Paenibacillus species for use as a probiotic to aid in ruminal methane mitigation, nitrate/nitrite detoxification and food safety. Bioresource Technol. 263, 358–364. doi: 10.1016/j.biortech.2018.04.116
Latham, E. A., Pinchak, W. E., Trachsel, J., Allen, H. K., Callaway, T. R., Nisbet, D. J., et al. (2019). Paenibacillus 79R4, a potential rumen probiotic to enhance nitrite detoxification and methane mitigation in nitrate-treated ruminants. Sci. total Environ. 671, 324–328. doi: 10.1016/j.scitotenv.2019.03.390
Lettat, A., Nozière, P., Silberberg, M., Morgavi, D. P., Berger, C., Martin, C. (2012). Rumen microbial and fermentation characteristics are affected differently by bacterial probiotic supplementation during induced lactic and subacute acidosis in sheep. BMC Microbiol. 12, 142. doi: 10.1186/1471-2180-12-142
Lewis, W. H., Tahon, G., Geesink, P., Sousa, D. Z., Ettema, T. J. G. (2021). Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 19, 225–240. doi: 10.1038/s41579-020-00458-8
Li, Q. L., Li, Y. P., Wang, S. Z. (2008). The Application of DGGE technology in polymorphism analysis of rumen microorganisms. China Cattle Sci. 01, 12–14. doi: 10.3969/j.issn.1673-1182.2008.03.024
Li, S., Lian, W. H., Han, J. R., Ali, M., Lin, Z. L., Liu, Y. H., et al. (2023b). Capturing the microbial dark matter in desert soils using culturomics-based metagenomics and high-resolution analysis. NPJ biofilms microbiomes 9 (1), 67. doi: 10.1038/s41522-023-00439-8
Li, B., Ma, X., Cheng, J., Tian, T., Guo, J., Wang, Y., et al. (2023a). Droplets microfluidics platform-A tool for single cell research. Front. Bioeng. Biotechnol. 11. doi: 10.3389/fbioe.2023.1121870
Li, Y., Mao, K., Zang, Y., Lu, G., Qiu, Q., Ouyang, K., et al. (2023c). Revealing the developmental characterization of rumen microbiome and its host in newly received cattle during receiving period contributes to formulating precise nutritional strategies. Microbiome 11, 238. doi: 10.1186/s40168-023-01682-z
Li, Q. F., Zhou, Z. H., Zie, Y. X. (2019). Application of Micro-ecological agents in ruminant Feed. Anim. Husbandry Feed Sci. 40, 29–30. doi: 10.5555/20193267181
Li, L. P., Zhu, H. J., Jing, X. Q., Huang, S., Qu, L. (2017). lsolation and identification of four bovine-derived probiotics. Heilongjiang Anim. Sci. Vet. Med. 23, 69–71, 294. doi: 10.13881/j.cnki.hljxmsy.2017.2156
Lin, D. W. (1999). Taxonomic identification and experimental methods of lactic acid bacteria. (Beijing: China Light Industry Press).
Litonina, A. S., Smirnova, Y. M., Platonov, A. V., Laptev, G. Y., Dunyashev, T. P., Butakova, M. V. (2021). Application of enzyme probiotic drug developed based on microorganisms of the rumen of reindeer (Rangifer tarandus) in feeding cows. Regul. Mech. Biosyst. 12, 109–115. doi: 10.15421/022117
Liu, W., Kim, H. J., Lucchetta, E. M., Du, W., Ismagilov, R. F. (2009). Isolation, incubation, and parallel functional testing and identification by FISH of rare microbial single-copy cells from multi-species mixtures using the combination of chemistrode and stochastic confinement. Lab. chip 9, 2153–2162. doi: 10.1039/b904958d
Liu, P., Meagher, R. J., Light, Y. K., Yilmaz, S., Chakraborty, R., Arkin, A. P., et al. (2011). Microfluidic fluorescence in situ hybridization and flow cytometry (μFlowFISH). Lab. chip 11, 2673–2679. doi: 10.1039/c1lc20151d
Liu, S., Moon, C. D., Zheng, N., Huws, S., Zhao, S., Wang, J. (2022). Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome 10, 76. doi: 10.1186/s40168-022-01272-5
Lloréns-Rico, V., Simcock, J. A., Huys, G. R. B., Raes, J. (2022). Single-cell approaches in human microbiome research. Cell 185, 2725–2738. doi: 10.1016/j.cell.2022.06.040
Luo, X., Chen, J. Y., Ataei, M., Lee, A. (2022). Microfluidic compartmentalization platforms for single cell analysis. Biosensors 12, 58. doi: 10.3390/bios12020058
Maake, T. W., Aiyegoro, O. A., Adeleke, M. A. (2021). Effects of Lactobacillus rhamnosus and Enterococcus faecalis Supplementation as Direct-Fed Microbials on Rumen Microbiota of Boer and Speckled Goat Breeds. Vet. Sci. 8, 103. doi: 10.3390/vetsci8060103
Madhu, B., Miller, B. M., Levy, M. (2023). Single-cell analysis and spatial resolution of the gut microbiome. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1271092
Marsh, T. L. (1999). Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2, 323–327. doi: 10.1016/S1369-5274(99)80056-3
McGenity, T., van der Meer, J. R., de Lorenzo, V. (2010). Handbook of hydrocarbon and lipid microbiology. Ed. Timmis, K. N. (Berlin: Springer), 4716.
Megur, A., Daliri, E. B., Balnionytė, T., Stankevičiūtė, J., Lastauskienė, E., Burokas, A. (2023). In vitro screening and characterization of lactic acid bacteria from Lithuanian fermented food with potential probiotic properties. Front. Microbiol. 14, 1213370. doi: 10.3389/fmicb.2023.1213370
Mende, D. R., Aylward, F. O., Eppley, J. M., Nielsen, T. N., DeLong, E. F. (2016). Improved environmental genomes via integration of metagenomic and single-cell assemblies. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00143
Meruvu, H., Harsa, S. T. (2023). Lactic acid bacteria: isolation-characterization approaches and industrial applications. Crit. Rev. Food Sci. Nutr. 63, 8337–8356. doi: 10.1080/10408398.2022.2054936
Min, B. R., Pinchak, W. E., Hume, M. E., Anderson, R. C. (2021). Effects of condensed tannins supplementation on animal performance, phylogenetic microbial changes, and in vitro methane emissions in steers grazing winter wheat. Animals: an Open Access J. MDPI 11, 2391. doi: 10.3390/ani11082391
Mizrahi, I., Wallace, R. J., Moraïs, S. (2021). The rumen microbiome: balancing food security and environmental impacts. Nat. Rev. Microbiol. 19, 553–566. doi: 10.1038/s41579-021-00543-6
Mordant, A., Kleiner, M. (2021). Evaluation of sample preservation and storage methods for metaproteomics analysis of intestinal microbiomes. Microbiol. Spectr. 9, e0187721. doi: 10.1128/Spectrum.01877-21
Nichols, D., Cahoon, N., Trakhtenberg, E. M., Pham, L., Mehta, A., Belanger, A., et al. (2010). Use of ichip for high-throughput in situ cultivation of "uncultivable" microbial species. Appl. Environ. Microbiol. 76, 2445–2450. doi: 10.1128/AEM.01754-09
Nobu, M. K., Narihiro, T., Rinke, C., Kamagata, Y., Tringe, S. G., Woyke, T., et al. (2015). Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J. 9, 1710–1722. doi: 10.1038/ismej.2014.256
Noel, S. J., Olijhoek, D. W., Mclean, F., Løvendahl, P., Lund, P., Højberg, O. (2019). Rumen and fecal microbial community structure of holstein and Jersey dairy cows as affected by breed, diet, and residual feed intake. Animals: an Open Access J. MDPI 9, 498. doi: 10.3390/ani9080498
Park, J., Kerner, A., Burns, M. A., Lin, X. N. (2011). Microdroplet-enabled highly parallel co-cultivation of microbial communities. PloS one 6 (2), e17019. doi: 10.1371/journal.pone.0017019
Peng, N., Peng, X. Q., Le, M. (2020). The application of microbiome culturomics in veterinary medicine. Acta Veterinaria Zootechnica Sin. 51, 2942–2953. doi: 10.11843/j.issn.0366-6964.2020.12.004
Pepoyan, A. Z., Tsaturyan, V. V., Badalyan, M., Weeks, R., Kamiya, S., Chikindas, M. L. (2020). Blood protein polymorphisms and the gut bacteria: impact of probiotic Lactobacillus acidophilus Narine on Salmonella carriage in sheep. Beneficial Microbes 11, 183–189. doi: 10.3920/BM2019.0138
Pittaluga, A. M., Yang, F., Gaffney, J. R., Embree, M., Relling, A. E. (2023). Effect of supplementation with ruminal probiotics on growth performance, carcass characteristics, plasma metabolites, methane emissions, and the associated rumen microbiome changes in beef cattle. J. Anim. Sci. 101, skac308. doi: 10.1093/jas/skac308
Poothong, S., Tanasupawat, S., Chanpongsang, S., Kingkaew, E., Nuengjamnong, C. (2024). Anaerobic flora, Selenomonas ruminis sp. nov., and the bacteriocinogenic Ligilactobacillus salivarius strain MP3 from crossbred-lactating goats. Sci. Rep. 14, 4838. doi: 10.1038/s41598-024-54686-6
Prakash, V., Madhavan, A., Veedu, A. P., Babu, P., Jothish, A., Nair, S. S., et al. (2023). Harnessing the probiotic properties and immunomodulatory effects of fermented food-derived Limosilactobacillus fermentum strains: implications for environmental enteropathy. Front. Nutr. 10, 1200926. doi: 10.3389/fnut.2023.1200926
Prakash, O., Pandey, P. K., Kulkarni, G. J., Mahale, K. N., Shouche, Y. S. (2014). Technicalities and glitches of terminal restriction fragment length polymorphism (T-RFLP). Indian J. Microbiol. 54, 255–261. doi: 10.1007/s12088-014-0461-0
Qi, P., Lv, J., Yan, X., Bai, L., Zhang, L. (2023). Microfluidics: insights into intestinal microorganisms. Microorganisms 11, 1134. doi: 10.3390/microorganisms11051134
Ratan, Z. A., Zaman, S. B., Mehta, V., Haidere, M. F., Runa, N. J., Akter, N. (2017). Application of fluorescence in situ hybridization (FISH) technique for the detection of genetic aberration in medical science. Cureus 9, e1325. doi: 10.7759/cureus.1325
Reid, G. (2006). Safe and efficacious probiotics: what are they? Trends Microbiol. 14 (8), 348–352. doi: 10.1016/j.tim.2006.06.006
Ren, A., Zhang, B., Zhou, C. S. (2016). Research progress in separation and ldentification of ruminal microorganisms in ruminants. J. Domest. Anim. Ecol. 37, 85–89. doi: 10.3969/j.issn.1673-1182.2016.06.016
Rodrigues, C. F., Azevedo, N. F., Miranda, J. M. (2021). Integration of FISH and microfluidics. Methods Mol. Biol. 2246, 249–261. doi: 10.1007/978-1-0716-1115-9_16
Schofield, B. J., Lachner, N., Le, O. T., McNeill, D. M., Dart, P., Ouwerkerk, D., et al. (2018). Beneficial changes in rumen bacterial community profile in sheep and dairy calves as a result of feeding the probiotic Bacillus amyloliquefaciens H57. J. Appl. Microbiol. 124, 855–866. doi: 10.1111/jam.13688
Schütte, U. M., Abdo, Z., Bent, S. J., Shyu, C., Williams, C. J., Pierson, J. D., et al. (2008). Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 80, 365–380. doi: 10.1007/s00253-008-1565-4
Sreepathi, N., Kumari, V. B. C., Huligere, S. S., Al-Odayni, A. B., Lasehinde, V., Jayanthi, M. K., et al. (2023). Screening for potential novel probiotic Levilactobacillus brevis RAMULAB52 with antihyperglycemic property from fermented Carica papaya L. Front. Microbiol. 14, 1168102. doi: 10.3389/fmicb.2023.1168102
Stewart, R. D., Auffret, M. D., Warr, A., Walker, A. W., Roehe, R., Watson, M. (2019). Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 37 (8), 953–961. doi: 10.1038/s41587-019-0202-3
Sun, J. F., Lin, Y. T. (2009). Isolation, identification and quantitative analysis of ruminal microorganisms. China Dairy Cattle 04), 18–23. doi: 10.3969/j.issn.1004-4264.2009.04.008
Tan, J. Y., Wang, S., Dick, G. J., Young, V. B., Sherman, D. H., Burns, M. A., et al. (2020). Co-cultivation of microbial sub-communities in microfluidic droplets facilitates high-resolution genomic dissection of microbial 'dark matter'. Integr. biology: quantitative Biosci. nano to macro 12, 263–274. doi: 10.1093/intbio/zyaa021
Tatusov, R. L., Koonin, E. V., Lipman, D. J. (1997). A genomic perspective on protein families. Science 278 (11), 631–637. doi: 10.1126/science.278.5338.631
Tidjani Alou, M., Naud, S., Khelaifia, S., Bonnet, M., Lagier, J. C., Raoult, D. (2020). State of the art in the culture of the human microbiota: new interests and strategies. Clin. Microbiol. Rev. 34, e00129–e00119. doi: 10.1128/CMR.00129-19
Tyagi, A. K., Kumar, S., Choudhury, P. K., Tyagi, B., Tyagi, N. (2020). Conjugated linoleic acid producing potential of lactobacilli isolated from goat (AXB) rumen fluid samples. Asian-Australasian J. Anim. Sci. 33 (8), 1233–1241. doi: 10.5713/ajas.19.0080
Valm, A. M., Mark Welch, J. L., Rieken, C. W., Hasegawa, Y., Sogin, M. L., Oldenbourg, R., et al. (2011). Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. U.S.A. 108, 4152–4157. doi: 10.1073/pnas.1101134108
Varada, V. V., Tyagi, A. K., Banakar, P. S., Das, A., Tyagi, N., Mallapa, R. H., et al. (2022). Autochthonous Limosilactobacillus reuteri BFE7 and Ligilactobacillus salivarius BF17 probiotics consortium supplementation improves performance, immunity, and selected gut health indices in Murrah buffalo calves. Vet. Res. Commun. 46, 757–767. doi: 10.1007/s11259-022-09896-6
Větrovský, T., Baldrian, P. (2013). The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PloS One 8, e57923. doi: 10.1371/journal.pone.0057923
Wan, X., Yang, Q., Wang, X., Bai, Y., Liu, Z. (2023). Isolation and cultivation of human gut microorganisms: A review. Microorganisms 11 (4), 1080. doi: 10.3390/microorganisms11041080
Wang, R., Cai, W. T., Wang, X. L., Gao, J., Huang, M. (2019a). Progress in Chinese medicine-probiotics compound microecological preparations for livestock and poultry. Chin. J. Biotechnol. 35, 972–987. doi: 10.13345/j.cjb.180443
Wang, Y., Gong, L., Wu, Y. P., Cui, Z. W., Wang, Y. Q., Huang, Y., et al. (2019b). Oral administration of Lactobacillus rhamnosus GG to newborn piglets augments gut barrier function in pre-weaning piglets. J. Zhejiang Univ. Sci. B 20, 180–192. doi: 10.1631/jzus.B1800022
Wang, Y., Li, Z., Jin, W., Mao, S. (2022). Isolation and characterization of ruminal yeast strain with probiotic potential and its effects on growth performance, nutrients digestibility, rumen fermentation and microbiota of Hu sheep. J. fungi 8, 1260. doi: 10.3390/jof8121260
Wang, S. J., Li, X., Liu, L. Y., Dan, Y. L., Xiao, Z. T., Wang, Q. Y., et al. (2021). Screening and identification of probiotics from yak dung and milk in Tibet. China Brewing 40, 43–48. doi: 10.11882/j.issn.0254-5071.2021.07.009
Waters, J. C. (2009). Accuracy and precision in quantitative fluorescence microscopy. J. Cell Biol. 185 (7), 1135–1148. doi: 10.1083/jcb.200903097
Watterson, W. J., Tanyeri, M., Watson, A. R., Cham, C. M., Shan, Y., Chang, E. B., et al. (2020). Droplet-based high-throughput cultivation for accurate screening of antibiotic resistant gut microbes. eLife 9, e56998. doi: 10.7554/eLife.56998.sa2
Wen, L., Li, G., Huang, T., Geng, W., Pei, H., Yang, J., et al. (2022). Single-cell technologies: From research to application. Innovation 3, 100342. doi: 10.1016/j.xinn.2022.100342
Wu, H. H., Wang, M. G., Xu, H. W., Wang, D. M., Yang, D. (2018). Improvement of experimental methods for the biological and biochemical identification of microorganisms. Experiment Teach. Apparatus 35 (05), 34–40.
Xie, H. W., Weng, B., Li, X. Y., Chen, M., Guo, M. N., Zheng, L. L., et al. (2021). Research on the application of microecological preparations. Guangdong Chem. Industry 48, 17–19. doi: 10.3969/j.issn.1007-1865.2021.24.008
Xiong, R. Y., Yang, Q. N., Shang, Y., Wang, L. M. (2020). Screening and function of probiotics and their application in animal feed. Feed Res. 43, 147–150. doi: 10.13557/j.cnki.issn1002-2813.2020.03.038
Xu, B., Hu, B., Wang, J., Lan, Y., Zhu, Y., Dai, X., et al. (2018a). Virgibacillus indicus sp. nov. and Virgibacillus profundi sp. nov, two moderately halophilic bacteria isolated from marine sediment by using microfluidic streak plates. Int. J. Syst. Evol. Microbiol. 68, 2015–2023. doi: 10.1099/ijsem.0.002782
Xu, Y., Zhao, F. (2018b). Single-cell metagenomics: challenges and applications. Protein Cell 9, 501–510. doi: 10.1007/s13238-018-0544-5
Yadav, K. K., Nimonkar, Y., Poddar, B. J., Kovale, L., Sagar, I., Shouche, Y., et al. (2022). Two-dimensional cell separation: a high-throughput approach to enhance the culturability of bacterial cells from environmental samples. Microbiol. Spectr. 10, e0000722. doi: 10.1128/spectrum.00007-22
Yang, S., Luo, J., Chen, Y., Wu, R., Liu, H., Zhou, Z., et al. (2022). A buffalo rumen-derived probiotic (SN-6) could effectively increase simmental growth performance by regulating fecal microbiota and metabolism. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.935884
Yang, S., Ma, S., Chen, J., Mao, H., He, Y., Xi, D., et al. (2010). Bacterial diversity in the rumen of Gayals (Bos frontalis), Swamp buffaloes (Bubalus bubalis) and Holstein cow as revealed by cloned 16S rRNA gene sequences. Mol. Biol. Rep. 37, 2063–2073. doi: 10.1007/s11033-009-9664-6
Yin, J., Chen, X., Li, X., Kang, G., Wang, P., Song, Y., et al. (2022). A droplet-based microfluidic approach to isolating functional bacteria from gut microbiota. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.920986
Young, K. D. (2007). Bacterial morphology: why have different shapes? Curr. Opin. Microbiol. 10, 596–600. doi: 10.1016/j.mib.2007.09.009
Yu, Y., Wen, H., Li, S., Cao, H., Li, X., Ma, Z., et al. (2022). Emerging microfluidic technologies for microbiome research. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.906979
Yu, M. F., Zhao, X. M., Cai, H., Yi, J. M., Hua, G. H. (2020). Dihydropyridine enhances the antioxidant capacities of lactating dairy cows under heat stress condition. Animals: an Open Access J. MDPI 10, 1812. doi: 10.3390/ani10101812
Yu, X., Zhao, F. Y., Zhao, Z. H. (2023). Research progress in culturomics of beneficial bacteria in the intestines. Food Sci. 44 (23), 365–371. doi: 10.7506/spkx1002-6630-20221210-103
Yuan, C. L., Yu, Z. Y., Lin, Y. T., Wang, L. H. (2015). Effects of probiotics on the calves and ts affecting factors. China Dairy Cattle 08, 9–12. doi: 10.3969/j.issn.1004-4264.2015.08.002
Zhang, X., Ke, W., Ding, Z., Xu, D., Wang, M., Chen, M., et al. (2022). Microbial mechanisms of using feruloyl esterase-producing Lactobacillus plantarum A1 and grape pomace to improve fermentation quality and mitigate ruminal methane emission of ensiled alfalfa for cleaner animal production. J. Environ. Manage. 308, 114637. doi: 10.1016/j.jenvman.2022.114637
Zhang, J., Liu, Y. X., Guo, X., Qin, Y., Garrido-Oter, R., Schulze-Lefert, P., et al. (2021). High-throughput cultivation and identification of bacteria from the plant root microbiota. Nat. Protoc. 16, 988–1012. doi: 10.1038/s41596-020-00444-7
Zhang, D., Yang, Y., Qin, Q., Xu, J., Wang, B., Chen, J., et al. (2019). MALDI-TOF characterization of protein expression mutation during morphological changes of bacteria under the impact of antibiotics. Anal. Chem. 91, 2352–2359. doi: 10.1021/acs.analchem.8b05080
Zhao, Z., Fan, B., Chen, D., Men, D., Wang, J., Chen, J. (2016). Development of droplet microfluidics enabling high-throughput single-cell analysis. Molecules 21, 881. doi: 10.3390/molecules21070881
Zheng, W., Zhao, S., Yin, Y., Zhang, H., Needham, D. M., Evans, E. D., et al. (2022). High-throughput, single-microbe genomics with strain resolution, applied to a human gut microbiome. Science 376, eabm1483. doi: 10.1126/science.abm1483
Zhong, H., Zheng, N., Wang, J., Zhao, S. (2023). Isolation and pan-genome analysis of Enterobacter hormaechei Z129, a ureolytic bacterium, from the rumen of dairy cow. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1169973
Zhou, W., Le, J., Chen, Y., Cai, Y., Hong, Z., Chai, Y. (2019b). Recent advances in microfluidic devices for bacteria and fungus research. TrAC Trends Anal. Chem. 112, 175–195. doi: 10.1016/j.trac.2018.12.024
Zhou, N., Sun, Y. T., Chen, D. W., Du, W., Yang, H., Liu, S. J. (2019a). Harnessing microfluidic streak plate technique to investigate the gut microbiome of Reticulitermes chinensis. Microbiol. Open 8, e00654. doi: 10.1002/mbo3.654
Zhou, R., Yang, G., Zhang, Y., Wang, Y. (2023). Spatial transcriptomics in development and disease. Mol. Biomed. 4, 32. doi: 10.1186/s43556-023-00144-0
Keywords: rumen probiotics, microorganisms, isolation, culture, identification
Citation: Wu R, Ji P, Hua Y, Li H, Zhang W and Wei Y (2024) Research progress in isolation and identification of rumen probiotics. Front. Cell. Infect. Microbiol. 14:1411482. doi: 10.3389/fcimb.2024.1411482
Received: 03 April 2024; Accepted: 30 April 2024;
Published: 21 May 2024.
Edited by:
Aoyun Li, Henan Agricultural University, ChinaCopyright © 2024 Wu, Ji, Hua, Li, Zhang and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanming Wei, weiym@gsau.edu.cn; Peng Ji, jip@gsau.edu.cn
 Runmin Wu
Runmin Wu Peng Ji
Peng Ji Yongli Hua
Yongli Hua Hongya Li
Hongya Li Wenfei Zhang
Wenfei Zhang Yanming Wei
Yanming Wei