The Role of Platelet Microparticle Associated microRNAs in Cellular Crosstalk
- Institute of Pediatrics, Guangzhou Women and Children's Medical Centre, Guangzhou Medical University, Guangzhou, China
Platelet is an anucleate cell containing abundant messenger RNAs and microRNAs (miRNAs), and their functional roles in hemostasis and inflammation remain elusive. Accumulating evidence has suggested that platelets can actively transfer RNAs to hepatocytes, vascular cells, macrophages, and tumor cells. The incorporated mRNAs are translated into proteins, and miRNAs were found to regulate the gene expression, resulting in the functional change of the recipient cells. This novel intercellular communication opens up a new avenue for the pathophysiological role of platelet in platelet-associated vascular diseases. Therefore, understanding the underlying mechanism and identification of the platelet miRNAs involved in this biological process would provide novel diagnostic and therapeutic targets for cardiovascular diseases.
Introduction
Platelet plays a pivotal role in maintaining primary hemostasis, and contributes to the pathogenesis of thrombotic and occlusive vascular disorders such as acute coronary syndrome or stroke (1). Despite lack of nucleus and genomic DNA, there are diverse types of RNAs in platelet, such as protein coding messenger RNAs (mRNAs), microRNAs (miRNAs), YRNAs and circular RNAs, which are inherited from their parent megakaryocytes (2–5). Platelets contain the essential machinery for processing miRNAs, which regulate the expression of proteins through complementary sequence recognition, binding and post-translational repression of mRNA transcripts (6, 7). The function of platelet miRNAs has gained widespread interest in the field of platelet biology since the platelet miRNA expression profile is closely associated with platelet biogenesis and activation. Recent studies have shown that platelets transfer and incorporate specific miRNAs into the recipient cells (8–11), raising the exciting possibility of potential therapeutic targets and disease biomarkers. In this review, we summarize and discuss the mechanistic roles of platelet miRNAs in circulation.
Platelet mRNAs and miRNAs
MicroRNAs are generated either as transcripts that rely on transcription as the promoter for the host protein-encoding gene or by using their own promoter transcript in intergenic region of the genome (12). Anucleate platelets retain megakaryocyte-derived mRNAs (13) and their unique adaptive signals have evolved in maintaining the diversity of genes and proteins (14, 15).
With the use of high-throughput sequencing, Hélène Plé et al. showed that based on the size of RNA, most of small RNAs found in platelets were miRNAs (16). Platelets have miRNAs, Dicer and Argonaute protein complexes, so pre-RNAs can be processed into miRNAs and control reporter transcripts. Analysed by the deep sequencing techniques, there were about 9,500 transcripts in the platelet of healthy subject (5, 17). Subsequent analysis of microarray and RNA sequencing have focused on non-healthy individuals, which correlated RNA profiles to specific human diseases (18–23). The stability of platelet miRNAs is unique, especially for the most abundant transcripts, suggesting that the platelet miRNome may be more stable than the miRNomes of other circulating cells or blood pools, such as plasma (24). It is noteworthy that almost half of miRNAs in microparticles, the major proportion of miRNAs in plasma, are produced by platelet (25). Numerous miRNAs have been found to be expressed in platelets and associated with cardiovascular diseases (CVD). Duisters RF et al. (24) reported that miR-30c repressed CTGF expression which was associated with ventricular fibrosis and heart failure. Tang, Y. et al. (26) found that miRNA–150 protected the mouse heart from ischemic injury by regulating EGR2, MYB and P2R × 7. McManus DD et al. (27, 28) demonstrated that miR-328 regulated CACNA1C and CACNB1 expressions, leading to atrial fibrillation and electrical remodeling. Willeit P et al. (29) showed that miR-342 regulated AKT1, glucose metabolism, apoptosis and cell proliferation, resulting in inflammatory stimulation of macrophages. Over the past decade, emerging evidence suggested that platelet miRNAs were potential regulators of platelet protein translation and expression as well as biomarkers for hematologic disease and platelet reactivity (4). In addition, considerable attention has be paid to the horizontal transfer of platelet miRNAs to the recipient cells.
Platelets and Platelet-Derived Microparticles
Platelets are fragments of cytoplasm, which are derived from the megakaryocytes (29) of the bone marrow and lung (30, 31), then entered into the circulation. Circulating blood platelets have a lifespan of 8–10 days, and play a central role in hemostasis (32, 33). At the site of injury, platelets contact with many adhesion molecules, including collagen, which interacts with platelet surface receptors, leading to the adhesion and activation of platelets. Once activated, platelets release their bioactive components, such as ADP, serotonin and thromboxane A2, leading to further platelet recruitment, aggregation and plug formation (7). Platelet hyperreactivity contributes to the pathogenesis of thrombotic and occlusive vascular disorders, such as atherosclerosis, thrombotic cardiovascular disorders, occlusive thrombus or inflammation and cancer (34–38).
Microparticles (MPs), also referred to as microvesicles or more rarely ectosomes, are submicron fragments shed from stimulated plasma membrane or apoptotic cells (39). Platelet-derived MPs (PMPs) generated from more than 45% of plasma-borne MPs (39, 40). Based on the size of exosomes (<0.1 µm) and apoptotic bodies (>1 µm), PMPs (0.1–1 µm) can be distinguished from them. PMPs are shed from cytoplasmic membrane of platelet express the surface antigens, such as CD41 and CD62p (P-selectin) which are different from exosomes released from intracellular compartments (41). Boilard et al. showed that platelets generated microparticles (0.1 µm) by stimulating the collagen receptor glycoprotein VI in arthritis pathophysiology (36). Moreover, recent studies have demonstrated that measurement of PMPs can be used as the marker for platelet activation (42, 43).
The Horizontal Transfer of Platelet-Derived mRNA/miRNA Into the Vascular Cells
Upon activation, platelets can regulate the function of vascular cells by releasing the bioactive molecules. Platelet mRNAs are only associated with low levels of protein translation; however, platelets have a unique membrane structure that allows small molecules to pass through, so their cytoplasmic RNA can be delivered to nucleated cells (44). Antonina Risitano et al. demonstrated that coculture of platelet-like particles (PLPs) derived from megakaryoblastic cell line Meg-01 and human umbilical vein endothelial cells (HUVECs) led to the horizontal transfer of platelet RNA into HUVECs, and those incorporated RNA were functional in the recipient cells (44). Interestingly, platelets were also found to transfer specific miRNA to endothelial cells and regulate their gene expression of the endothelial cells. Benoit Laffont et al. found that platelets stimulated with 0.1 U/ml thrombin preferentially released MPs containing miR-223, and those MPs were internalized by HUVECs (8). Also, PMPs contain Argonaute 2 (Ago2)·miR-223 complexes, suggesting that the incorporated miRNAs were functional in the recipient HUVECs. Indeed, they found that platelet-derived miR-223 regulated two endogenous endothelial genes FBXW7 and EFNA1 at mRNA and protein levels (8).
The horizontal transfer of platelet RNA has also been reported in THP-1 cells (44). Antonina Risitano et al. found that the RNA of PLPs derived from Meg-01 was transferred to THP-1 cells, and those incorporated RNAs were functional in the recipient cells (44). Using microarray analysis of THP-1 cells showed that the genes globins (HGB1/HGB2) and globins (HBA1/HBA2) involved in transfer were up-regulated, but not altered in the control cells (44). In addition, after infused with wildtype platelets, the presence of TLR2-positive monocytes was found in TLR2-deficient mice treated with LPS, indicating that specific platelet RNAs are transferred and incorporated into leukocytes in vivo (44).
Benoit Laffont et al. demonstrated that PMPs containing miR-126–3 p was internalized by primary human macrophages (10). The incorporated miR-126–3 p suppressed the expression of four predicted mRNA targets of miR-126–3 p, and two of them were determined at the protein level. The effect of miR-126–3 p was further confirmed by expressing a neutralising miR-126–3 p sponge (10). Furthermore, PMPs induced the upregulation of 34 miRNAs with concomitant downregulation of 367 RNAs, including mRNAs encoding cytokines/chemokines chemokine (C-C motif) ligand 4 (CCL4), colony stimulating factor 1 (CSF1) and tumor necrosis factor (TNF) (10). These changes induced by PMPs were accompanied by a marked increase in the phagocytic capacity of macrophages (10).
Vascular smooth muscle cells (VSMCs) are the major component of the vascular wall and endure both ongoing damage and repair during atherosclerosis (45). During repair, newly migrated VSMCs tend to differentiate and frequently undergo apoptosis (46). A recent study has found that platelets may transfer miRNAs to VSMCs via platelet-derived exosomes (47). They also showed that in the murine model of carotid tandem stenosis, the levels of miR-21, miR-223 and miR-339 were associated with platelet activation (47). These miRNAs were elevated in the pooled mouse plasma exosomes before thrombosis, and were also found to be enriched in thrombin-stimulated platelet-derived exosomes in vitro (47). After cocultured VSMCs with platelet-derived exosomes, the expression of platelet-derived growth factor receptor-beta (PDGFRβ) in VSMCs was decreased (47). Platelet-derived exosomes also inhibited PDGF-stimulated VSMC proliferation. Furthermore, a decrease in PDGFRβ expression was observed in VSMCs around thrombotic areas in vivo (46). Despite lack of direct evidence showing the horizontal transfer of platelet miRNAs into VSMCs in vivo, these findings provide some clues for the possible role of platelet-derived exosomes containing miR-21, miR-223 and miR-339 in regulating the phenotypic switching of VSMCs in diseased condition (46).
The Horizontal Transfer of Platelet-Derived mRNA Into the Hepatocytes
Marc Kirschbaum et al. showed that either platelets or PLPs derived from the MEG-01 stimulated the proliferation of HepG2 cells (9). They also observed platelet internalization into hepatocytes following a partial hepatectomy in mice (9). PLPs-derived RNA was detected in the cytoplasm of the HepG2 cells. The removal of PLPs-derived RNA by RNA-degrading enzymes partly blocked the stimulating effect of platelets on hepatocyte proliferation (9). Thus, platelets stimulate hepatocyte proliferation via the horizontal transfer of platelet RNA (9). Although no further evidence has been reported that miRNAs were involved in this process, it is plausible that activated platelet could transfer specific miRNAs to hepatocytes and participate into the regulation of liver regeneration.
The Horizontal Transfer of Platelet-Derived mRNA/miRNA Into the Tumor Cells
Tumor growth is a major pathophysiological condition that may be affected by platelet MPs and associated miRNAs (7, 48). PMPs can directly stimulate the tumor growth through the release of potent growth factors in the tumor micro-environment (49). In 2015, Liang et al. first reported that platelet-secreted miRNA exacerbated lung cancer cell invasion (49). They found that miR-223 significantly increased in MVs derived from platelets of Non-small cell lung cancer patients (49). After cocultured with A549 cells, MVs derived from platelets delivered miR-223 into the recipient cells (49). The incorporated miR-223 exacerbated A549 cells invasion via targeting EPB41L3 (49).
Recently, James V. Michael et al. reported that platelet MPs suppressed tumor growth and induced tumor cell apoptosis (11). Solid tumor vasculature is highly permeable, allowing the possibility of PMPs-tumor interaction. They showed that PMPs infiltrated solid tumors in human and mice, thereby transferring platelet-derived miRNAs into tumor cells in vitro and in vivo, resulting in tumor cell apoptosis (11). PMPs transfusion suppressed the growth of both lung and colon carcinoma ectopic tumors, whereas blockade of miR-24 in tumor cells accelerated tumor growth. Par4 KO mice had the phenotype of reduced PMPs generation, and the tumor growth in Par4 KO mice was accelerated compared with wild type mice, indicating that PMPs play a pivotal role in the inhibition of tumor growth (11). The direct targets of PMPs-derived miR-24 in tumor cells were found to be the mitochondrial mt-Nd2 and a small noncoding nucleolar RNA Snora75 (11). The inhibition of mt-Nd2 and Snora75 resulted in mitochondrial dysfunction and growth inhibition of tumor cells (11). Thus, further studies are needed to determine the mechanistic role of the incorporated PMPs-derived miRNAs in the tumor progression.
The Clinical Significance, Challenges and Limitations
Accumulating evidence shed light on the use of platelet-derived miRNAs as diagnostic markers for various diseases. Platelet-derived miRNAs have been associated with atrial fibrillation, coronary disease, heart failure and vascular disease (50). For example, miR-30c was associated with heart failure (24), miR-126 was associated with myocardial angiogenesis (28), miR-197 was associated with metabolic syndrome (51), miR-328 was associated with atrial fibrillation (27). Since the blood samples from patients are easily accessible, the identification of platelet-derived miRNAs in diseases will open up a new avenue for the clinical diagnosis and treatment.
As discussed above, many studies have reported that platelets, MPs and associated miRNAs can be transferred and incorporated into the various cells, and regulate the function of the recipient cells. However, there are still a number of concerns to be addressed. First, it is important to understand how platelets secretes specific miRNAs in response to different stimuli and pathological conditions, in order to precisely deliver therapeutic RNAs to target sites. Second, it is important to know how platelets and PMPs were selectively internalised by the recipient cells, in order to accurately deliver the platelets or PMPs to the specific cells, tissues, or injured sites. If all these unanswered questions could be addressed, using platelets and MPs as drug deliver system would be an efficient and site-specific approach for the treatment of platelet-associated diseases.
Conclusion
Platelets participate into multiple biological processes, such as inflammation, wound healing, maintenance of blood-lymph barrier and blood clotting where they provide a first and crucial line of defense against injury, thus maintaining normal homeostasis. As discussed above, recent advance in platelet biology demonstrates that activated platelets release the MPs containing abundant mRNA and miRNA, which are internalised by the recipient cells. The incorporated mRNA or miRNA can modulate the gene expression and regulate the function of the recipient cells, e.g., macrophages, ECs, leukocytes, VSMCs, hepotacytes, and tumor cells (Figure 1). Since the horizontal transfer of platelet miRNA may represent a novel form of cell-cell communication, which may participate in many pathophysiological processes, growing concern has been received in this field. However, many unanswered questions remain to be addressed about the specificity and selectivity of the horizontal transfer of miRNAs. Therefore, understanding the mechanism by which platelet interacts with other cells, and identifying which miRNAs are involved in the horizontal transfer would provide novel therapeutic targets and diagnostic markers for platelet-associated diseases.
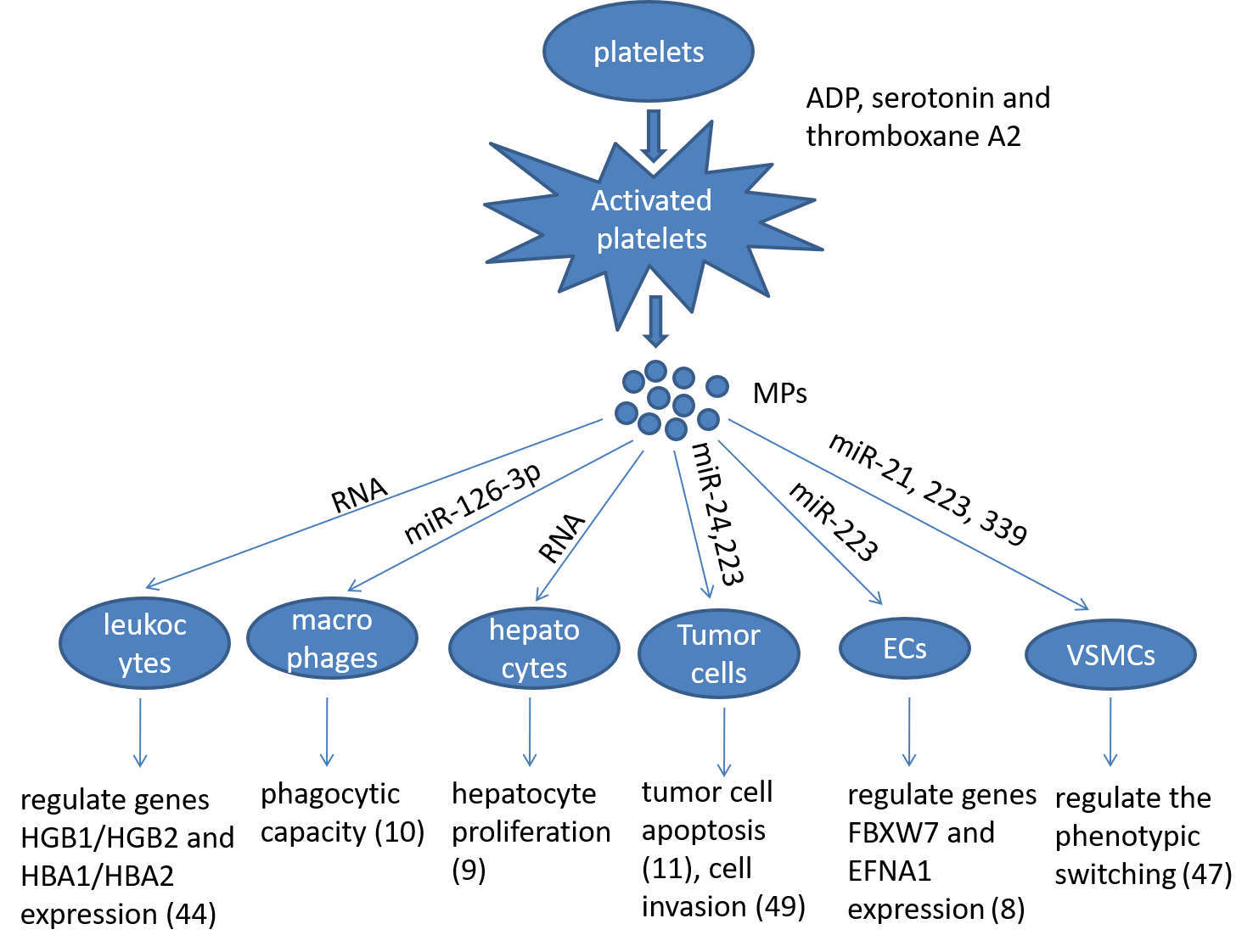
Figure 1. Summarizing the role of platelet microparticles (MPs) in cellular activation by the transfer of miRNAs. Ecs: endotheilal cells, VSMCs: vascular smooth muscle cells.
Author Contributions
LX wrote this manuscript. ZZ and WT were responsible for editing the manuscript. All authors accept responsibility for the entire content of this submitted manuscript and have approved its submission.
Funding
WT is supported by the Guangdong Outstanding Young Scientist Funding (Grant No. 2016A030306049). This work was supported by the National Natural Science Foundation of China (Grant No. 81670117), the Guangdong Province Science and Technology Project (Grant No. 2017A050506008) and by the Guangzhou Science and Technology Project (Grant No. 201707010270).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Clancy L, Freedman JE. New paradigms in thrombosis: novel mediators and biomarkers platelet RNA transfer. J Thromb Thrombolysis (2014) 37(1):12–6. doi: 10.1007/s11239-013-1001-1
2. Sunderland N, Skroblin P, Barwari T, Huntley RP, Lu R, Joshi A, et al. MicroRNA biomarkers and platelet reactivity: the clot thickens. Circ Res (2017) 120(2):418–35. doi: 10.1161/CIRCRESAHA.116.309303
3. Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L, et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood (2011) 117(19):5189–97. doi: 10.1182/blood-2010-09-299719
4. Edelstein LC, Bray PF. MicroRNAs in platelet production and activation. Blood (2011) 117(20):5289–96. doi: 10.1182/blood-2011-01-292011
5. Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, et al. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood (2011) 118(14):e101–11. doi: 10.1182/blood-2011-03-339705
6. Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol (2009) 16(9):961–6. doi: 10.1038/nsmb.1651
7. Provost P. The clinical significance of platelet microparticle-associated microRNAs. Clin Chem Lab Med (2017) 55(5):657–66. doi: 10.1515/cclm-2016-0895
8. Laffont B, Corduan A, Plé H, Duchez AC, Cloutier N, Boilard E, et al. Activated platelets can deliver mRNA regulatory Ago2·microRNA complexes to endothelial cells via microparticles. Blood (2013) 122(2):253–61. doi: 10.1182/blood-2013-03-492801
9. Kirschbaum M, Karimian G, Adelmeijer J, Giepmans BN, Porte RJ, Lisman T. Horizontal RNA transfer mediates platelet-induced hepatocyte proliferation. Blood (2015) 126(6):798–806. doi: 10.1182/blood-2014-09-600312
10. Laffont B, Corduan A, Rousseau M, Duchez AC, Lee CH, Boilard E, et al. Platelet microparticles reprogram macrophage gene expression and function. Thromb Haemost (2016) 115(2):311–23. doi: 10.1160/TH15-05-0389
11. Michael JV, Wurtzel JGT, Mao GF, Rao AK, Kolpakov MA, Sabri A, et al. Platelet microparticles infiltrating solid tumors transfer miRNAs that suppress tumor growth. Blood (2017) 130(5):567–80. doi: 10.1182/blood-2016-11-751099
12. Bauersachs J, Thum T. Biogenesis and regulation of cardiovascular microRNAs. Circ Res (2011) 109(3):334–47. doi: 10.1161/CIRCRESAHA.110.228676
13. Gnatenko DV, Dunn JJ, Mccorkle SR, Weissmann D, Perrotta PL, Bahou WF. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood (2003) 101(6):2285–93. doi: 10.1182/blood-2002-09-2797
14. Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell (2005) 122(3):379–91. doi: 10.1016/j.cell.2005.06.015
15. Weyrich AS, Dixon DA, Pabla R, Elstad MR, Mcintyre TM, Prescott SM, et al. Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. Proc Natl Acad Sci USA (1998) 95(10):5556–61. doi: 10.1073/pnas.95.10.5556
16. Plé H, Landry P, Benham A, Coarfa C, Gunaratne PH, Provost P. The repertoire and features of human platelet microRNAs. PLoS ONE (2012) 7(12):e50746. doi: 10.1371/journal.pone.0050746
17. Bray PF, Mckenzie SE, Edelstein LC, Nagalla S, Delgrosso K, Ertel A, et al. The complex transcriptional landscape of the anucleate human platelet. BMC Genomics (2013) 14:1. doi: 10.1186/1471-2164-14-1
18. Freedman JE, Larson MG, Tanriverdi K, O'Donnell CJ, Morin K, Hakanson AS, et al. Relation of platelet and leukocyte inflammatory transcripts to body mass index in the Framingham heart study. Circulation (2010) 122(2):119–29. doi: 10.1161/CIRCULATIONAHA.109.928192
19. Gnatenko DV, Cupit LD, Huang EC, Dhundale A, Perrotta PL, Bahou WF. Platelets express steroidogenic 17beta-hydroxysteroid dehydrogenases. Distinct profiles predict the essential thrombocythemic phenotype. Thromb Haemost (2005) 94(2):412–21. doi: 10.1160/TH05-01-0037
20. Gnatenko DV, Zhu W, Xu X, Samuel ET, Monaghan M, Zarrabi MH, et al. Class prediction models of thrombocytosis using genetic biomarkers. Blood (2010) 115(1):7–14. doi: 10.1182/blood-2009-05-224477
21. Lood C, Amisten S, Gullstrand B, Jönsen A, Allhorn M, Truedsson L, et al. Platelet transcriptional profile and protein expression in patients with systemic lupus erythematosus: up-regulation of the type I interferon system is strongly associated with vascular disease. Blood (2010) 116(11):1951–7. doi: 10.1182/blood-2010-03-274605
22. Nagalla S, Bray PF. Platelet RNA chips dip into thrombocytosis. Blood (2010) 115(1):2–3. doi: 10.1182/blood-2009-10-246405
23. Raghavachari N, Xu X, Harris A, Villagra J, Logun C, Barb J, et al. Amplified expression profiling of platelet transcriptome reveals changes in arginine metabolic pathways in patients with sickle cell disease. Circulation (2007) 115(12):1551–62. doi: 10.1161/CIRCULATIONAHA.106.658641
24. Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res (2009) 104(2):170–8. doi: 10.1161/CIRCRESAHA.108.182535
25. Diehl P, Fricke A, Sander L, Stamm J, Bassler N, Htun N, et al. Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res (2012) 93(4):633–44. doi: 10.1093/cvr/cvs007
26. Tang Y, Wang Y, Park KM, Hu Q, Teoh JP, Broskova Z, et al. MicroRNA-150 protects the mouse heart from ischaemic injury by regulating cell death. Cardiovasc Res (2015) 106(3):387–97. doi: 10.1093/cvr/cvv121
27. Mcmanus DD, Lin H, Tanriverdi K, Quercio M, Yin X, Larson MG, et al. Relations between circulating microRNAs and atrial fibrillation: data from the Framingham Offspring Study. Heart Rhythm (2014) 11(4):663–9. doi: 10.1016/j.hrthm.2014.01.018
28. Willeit P, Zampetaki A, Dudek K, Kaudewitz D, King A, Kirkby NS, et al. Circulating microRNAs as novel biomarkers for platelet activation. Circ Res (2013) 112(4):595–600. doi: 10.1161/CIRCRESAHA.111.300539
29. Machlus KR, Thon JN, Italiano JE. Interpreting the developmental dance of the megakaryocyte: a review of the cellular and molecular processes mediating platelet formation. Br J Haematol (2014) 165(2):227–36. doi: 10.1111/bjh.12758
30. Howell WH, Donahue DD. The production of blood platelets in the lungs. J Exp Med (1937) 65(2):177–203. doi: 10.1084/jem.65.2.177
31. Lefrançais E, Ortiz-Muñoz G, Caudrillier A, Mallavia B, Liu F, Sayah DM, et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature (2017) 544(7648):105–9. doi: 10.1038/nature21706
32. Harker LA, Finch CA. Thrombokinetics in man. J Clin Invest (1969) 48(6):963–74. doi: 10.1172/JCI106077
33. Hartwig JH. The platelet: form and function. Semin Hematol (2006) 43(Suppl 1):S94–100. doi: 10.1053/j.seminhematol.2005.11.004
34. Kottke-Marchant K. Importance of platelets and platelet response in acute coronary syndromes. Cleve Clin J Med (2009) 76(Suppl 1):S2–7. doi: 10.3949/ccjm.76.s1.01
35. Nurden AT, Platelets NAT. Platelets, inflammation and tissue regeneration. Thromb Haemost (2011) 105(Suppl 1):S13–33. doi: 10.1160/THS10-11-0720
36. Boilard E, Nigrovic PA, Larabee K, Watts GF, Coblyn JS, Weinblatt ME, et al. Platelets amplify inflammation in arthritis via collagen-dependent microparticle production. Science (2010) 327(5965):580–3. doi: 10.1126/science.1181928
37. Jain S, Harris J, Ware J. Platelets: linking hemostasis and cancer. Arterioscler Thromb Vasc Biol (2010) 30(12):2362–7. doi: 10.1161/ATVBAHA.110.207514
38. Dangwal S, Thum T. MicroRNAs in platelet physiology and pathology. Hamostaseologie (2013) 33(1):17–20. doi: 10.5482/HAMO-13-01-0002
39. Vanwijk MJ, Vanbavel E, Sturk A, Nieuwland R. Microparticles in cardiovascular diseases. Cardiovasc Res (2003) 59(2):277–87. doi: 10.1016/S0008-6363(03)00367-5
40. Horstman LL, Ahn YS. Platelet microparticles: a wide-angle perspective. Crit Rev Oncol Hematol (1999) 30(2):111–42. doi: 10.1016/S1040-8428(98)00044-4
41. Hristov M, Erl W, Linder S, Weber PC. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood (2004) 104(9):2761–6. doi: 10.1182/blood-2003-10-3614
42. Nomura S. Function and clinical significance of platelet-derived microparticles. Int J Hematol (2001) 74(4):397–404. doi: 10.1007/BF02982082
43. Italiano JE, Mairuhu AT, Flaumenhaft R. Clinical relevance of microparticles from platelets and megakaryocytes. Curr Opin Hematol (2010) 17(6):578–84. doi: 10.1097/MOH.0b013e32833e77ee
44. Risitano A, Beaulieu LM, Vitseva O, Freedman JE. Platelets and platelet-like particles mediate intercellular RNA transfer. Blood (2012) 119(26):6288–95. doi: 10.1182/blood-2011-12-396440
45. Uryga AK, Bennett MR. Ageing induced vascular smooth muscle cell senescence in atherosclerosis. J Physiol (2016) 594(8):2115–24. doi: 10.1113/JP270923
46. Silverman-Gavrila R, Silverman-Gavrila L, Bendeck MP. Cell division fidelity is altered during the vascular response to injury: its novel role in atherosclerosis progression. Am J Pathol (2013) 182(3):628–39. doi: 10.1016/j.ajpath.2012.11.007
47. Tan M, Yan HB, Li JN, Li WK, Fu YY, Chen W, et al. Thrombin stimulated platelet-derived exosomes inhibit platelet-derived growth factor receptor-beta expression in vascular smooth muscle cells. Cell Physiol Biochem (2016) 38(6):2348–65. doi: 10.1159/000445588
48. Goubran H, Sabry W, Kotb R, Seghatchian J, Burnouf T. Platelet microparticles and cancer: an intimate cross-talk. Transfus Apher Sci (2015) 53(2):168–72. doi: 10.1016/j.transci.2015.10.014
49. Liang H, Yan X, Pan Y, Wang Y, Wang N, Li L, et al. MicroRNA-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor EPB41L3. Mol Cancer (2015) 14:58. doi: 10.1186/s12943-015-0327-z
50. Mcmanus DD, Freedman JE. MicroRNAs in platelet function and cardiovascular disease. Nat Rev Cardiol (2015) 12(12):711–7. doi: 10.1038/nrcardio.2015.101
Keywords: platelet, microparticle, microRNA, RNA, cardiovascular disease
Citation: Xia L, Zeng Z and Tang WH (2018). The Role of Platelet Microparticle Associated microRNAs in Cellular Crosstalk. Front. Cardiovasc. Med. 5:29. doi: 10.3389/fcvm.2018.00029
Received: 24 January 2018; Accepted: 15 March 2018;
Published: 04 April 2018
Edited by:
Manfredi Tesauro, Università degli Studi di Roma Tor Vergata, ItalyReviewed by:
Paola Patrignani, Università degli Studi G. d'Annunzio Chieti e Pescara, ItalyTarun Tyagi, Yale University, United States
Copyright © 2018 Xia, Zeng and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wai Ho Tang, waiho.tang@gwcmc.org
 Luoxing Xia
Luoxing Xia Zhi Zeng
Zhi Zeng Wai Ho Tang
Wai Ho Tang