- Brain Research Institute, Jeffrey Cheah School of Medicine and Health Sciences, Monash University Malaysia, Bandar Sunway, Malaysia
Gonadotropin-releasing hormone (GnRH) is a reproductive neuropeptide, which controls vertebrate reproduction. In most vertebrates, there are more than two GnRH orthologs in the brain. In cichlid fish, the Nile tilapia (Oreochromis niloticus), GnRH1 is the primary hypophysiotropic hormone, while GnRH2 and GnRH3 are non-hypophysiotropic but neuromodulatory in function. Hypophysiotropic GnRH neurons are thought to inter-communicate, while it remains unknown if hypophysiotropic and non-hypophysiotropic GnRH systems communicate with each other. In the present study, we examined interrelationship between three GnRH types using specific antibodies raised against their respective GnRH associated peptide (GAP) sequence. Double-immunofluorescence labeling coupled with confocal microscopy revealed that in sexually mature males, GnRH-GAP1-immunoreactive (-ir) processes are in proximities of GnRH-GAP3-ir cell somata in the terminal nerve, while GnRH-GAP1-ir cell somata were also accompanied by GnRH-GAP3-ir processes in the preoptic area. However, such interaction was not seen in immature males. Further, there was no interaction between GnRH-GAP2 and GnRH-GAP1 or GnRH-GAP3 neurons. Single cell gene expression analysis revealed co-expression of multiple GnRH receptor genes (gnrhr1 and gnrhr2) in three GnRH-GAP cell types. In mature males, high levels of gnrhr2 mRNA were expressed in GnRH-GAP1-ir cells. In immature males, gnrhr1 and gnrhr2 mRNAs are highly expressed in GnRH-GAP3-ir cells. These results suggest heterologous interactions between the three GnRH-GAP cell types and their potential functional interaction during different reproductive stages.
Introduction
Gonadotropin-releasing hormone (GnRH) is a neuropeptide that primarily plays an essential role in the control of reproduction in vertebrates (1–5). GnRH stimulates the release of gonadotropins: follicle-stimulating hormone (FSH for gamete growth) and luteinizing hormone (LH for gamete maturation and release) from the adenohypophysis or anterior pituitary (6). Most vertebrate species possess more than two different GnRH types and their receptor (GnRH receptor, GnRHR) orthologs (7, 8). In mammals, there are two GnRH types, GnRH1 (mammalian GnRH) and GnRH2 (chicken-GnRH-II). GnRH1 is primarily hypophysiotropic in function and is the species-specific form (7, 9). On the other hand, GnRH2 is structurally most conserved in vertebrates and is expressed in the midbrain region. In addition to these two GnRH types, the third type of GnRH (GnRH3 = salmon GnRH) is present in the forebrain of teleosts. Similarly, most vertebrates possess more than two GnRHR types. For example, mammalian species have two forms of GnRHR, while amphibian and reptilian species have three forms of GnRHR (10). In fish, GnRHR orthologs have been classified into three major lineages of GnRHR types that are further subdivide into five classes: non-mammalian type I (GnRHRn1 and GnRHRn1b), non-mammalian type II (GnRHRn2), and non-mammalian type III (GnRHRn3 and GnRHRn3b) (11).
Differential distribution of GnRH and GnRHR types in several brain regions indicate their involvement in a variety of functions that are directly or indirectly associated with reproduction. In mammals, GnRH1 neurons located within the hypothalamic regions send neuronal processes mainly to the median eminence to control gonadotropin release. However, GnRH1 neuronal processes as well as GnRHR are also found in extrahypothalamic regions such as the hippocampus, amygdala, striatum, septum, thalamic nuclei, midbrain tectum and periaqueductal gray region, and brainstem (12, 13). GnRH1-GnRHR pathway has been implicated in functions such as cognition and mood, which are associated with reproductive stages such as puberty and menopause in human, mice and sheep (14–16).
Although GnRH2 can pharmacologically stimulate LH release, GnRH2 has been speculated to have originated in the immune system and it has recently evolved as a neuromodulator (17). In several vertebrates, GnRH2 stimulates sexual behavior (18–20) and in the musk shrew, GnRH2 suppresses feeding behavior (21). Similarly, in the zebrafish, central administration of GnRH2 peptides reduce food intake (22), and GnRH2 gene knockout zebrafish exhibit increased feeding and growth (23). The fish specific form, GnRH3 is mainly localized in the terminal nerve region in the forebrain and is involved in a variety of roles depending on the fish species. In some fish species, such as the goldfish and zebrafish, GnRH3 is expressed in the terminal nerve and in the preoptic area, indicating its hypophysiotropic and neuromodulatory functions (24, 25). GnRH3 in the terminal nerve regions regulates social behaviors such as aggression, sexual behavior, and partner preference (26–28). Although the hypophysiotropic GnRH form is primarily essential for the LH secretion, but other functions such as social behaviors, food intake, and mood that are modulated by non-hypothalamic GnRH forms are also closely associated with reproductive conditions.
It is clear that socio-sexual behaviors and mental status that are known to be influenced by GnRH1, are closely associated with the reproductive status (14, 29). However, it remains unknown how GnRH neurons control non-hypophysiotropic functions in the extrahypothalamic regions. Do the same GnRH neurons send neuronal projections to the adenohypophysis and to other brain regions? How do GnRH neurons co-modulate reproduction (LH surge) and its associated functions? Is there a mechanism/pathway to connect these functions? Hypothalamic GnRH1 neurons coordinate amongst themselves through synaptic interactions and gap junctions (30), which suggest an autocrine/paracrine regulation (31). On the other hand, other functions that are affected by non-hypothalamic GnRH systems, such as metabolic status or sexual behaviors are also closely associated with hypophysiotropic GnRH function. However, it is unknown if hypophysiotropic GnRH form can communicate with non-hypothalamic GnRH forms.
To elucidate the possible functional interaction between different GnRH systems, we used a cichlid fish, Nile tilapia (Oreochromis niloticus), which possesses three GnRH and GnRHR types (32). In the brain of the Nile tilapia and other perciforms, GnRH1 is localized in the preoptic-hypothalamic regions, GnRH2 is in the midbrain tegmentum and GnRH3 is mainly in the terminal nerve, but there is some overlap in the distribution between GnRH1 and GnRH3 cell somata/processes in the forebrain and the terminal nerve-preoptic region (33–36). In tilapia, three GnRHR (GnRHR1, GnRHR2, and GnRHR3) types have been identified, among which, GnRHR1 and GnRHR3 are classified as GnRHRn1, while GnRHR2 is classified as GnRHRn3 based on genome synteny analysis and sequence similarity (11, 37). Interrelationship between the three GnRH cell types were examined using specific antisera for GnRH associated peptide (GAP) derived from GnRH precursor, which is coexpressed with GnRH (33, 35, 36) (and thus hereafter referred to as “GnRH-GAP”). Further, to investigate the expression of multiple GnRHR types in three GnRH-GAP cell types (GnRH-GAP1, GnRH-GAP2, and GnRH-GAP3) and their potential involvement under different reproductive stages, expression of three GnRHR types (gnrhr1, gnrhr2, and gnrhr3) mRNA levels were examined in microdissected three GnRH-GAP cell types in immature and mature males of tilapia.
Materials and Methods
Animals
Nile tilapia (Oreochromis niloticus) were maintained in freshwater aquaria at 27 ± 1°C with a controlled natural photo-regimen (14/10 h, light/dark). Juvenile fish (2–3 weeks after fertilization) were obtained from the fish facility at Tokyo University of Marine Science and Technology (Tokyo, Japan) and were reared until the experimental sizes [sexually immature: standard length (SL), 4.4 ± 0.07 cm; body weight (BW), 3.2 ± 0.2 g; gonado-somatic index (GSI), 0.032 ± 0.018%, and mature: SL, 14.6 ± 0.2 cm; BW, 92.6 ± 4.5 g, GSI, 1.6 ± 0.4%] in the fish facility at Nippon Medical School (Tokyo, Japan). Sexually mature males were placed in a 60 L tank with two polyvinyl chloride (PVC) pipes (10 cm in diameter and 15 cm in length) as shelters for subordinate males to reduce social stress, which also prevented territory holding and reduced territorial behavior. The fish were fed commercial fish diet once a day. Fish were anesthetized by immersion in a 0.01% solution of 3-aminobenzonic acid ethyl ester (MS222; Sigma, St. Louis, MO, USA) before they were killed by decapitation. The fish were maintained and used in accordance with the Guidelines of the Animal Ethics Committees of Nippon Medical School.
Tilapia GAP Antisera Production and Specificity Characterization
Possible association of neuronal processes (axon/dendrites) and cell somata between the three GnRH-GAP types in the brain were examined using specific antibodies against the tilapia GAP types (Antibody codes: #ISP105 for GAP1, #ISP205 for GAP2, and #ISP305 for GAP3) (38–40). The antisera were generated in rabbits against commercially synthesized tilapia GAP peptides (GAP1: amino acids 63-94; GAP2, amino acids 55-94; GAP3, amino acids 46-85; GenBank Accession nos., AB101665, AB101666, and AB101667, respectively) (T.K. Craft Co., Ltd, Japan). After final blood collection, the blood was centrifuged and the affinity-purified serum was stored at – 80°C.
Specificity of the GAP antisera were evaluated by the Western blot analysis (see below). In addition, the GAP antisera immunoreactivities were also verified by GnRH-immunoreactivities and gene expression patterns of three GnRH types (gnrh1, gnrh2, and gnrh3) in the brain (41, 42). For controls for immunohistochemistry, brain sections of adult tilapia were incubated with the GAP antisera that have been preabsorbed with respective antigen (see below).
Western Blot Analysis
Western blot analysis was performed according to Pandolfi et al. (36) with a slight modification. Sexually mature male tilapias (n = 3) were anesthetized by immersing into 0.01% tricaine methanesulfonate (MS222; Sigma, St. Louis, MO, USA) solution. The brains and pituitaries were separately dissected out and homogenized in 1 ml and 100 μl of 50 mM Tris-HCl buffer (pH 7.4) containing 10 μl per ml protease inhibitor mix (GE Healthcare Life Sciences, Piscataway, NJ), respectively. The soluble fractions were obtained by centrifugation at 6,000 G for 30 min at 4°C. The protein concentration was measured by the Lowry assay (Bio-Rad, Hercules, CA). An aliquot of tissue sample was diluted with an equal volume of Laemmli's sample buffer (Bio-Rad). Supernatants were separated by 15% polyacrylamide gel with 20 μg protein per lane along with the marker (ECL DualVue Western Blotting Markers; GE Healthcare Life Sciences). After electrophoresis, the proteins were transferred onto a polyvinylidene difluoride membrane (Hybond-P, GE Healthcare Life Sciences) in Tris/glycine buffer (25 mM Tris, 192 mM glycine), and 20% methanol for 30 min at 40 mA. The membranes were washed in Tris-buffered saline (pH 7.4) with 1% Tween-20 (TBST) and blocked with TBST containing 0.5% non-fat dry milk for 2 h at room temperature, and washed with TBST three times at 5 min intervals and then incubated with GAP antisera at dilution of 1:1000 (#ISP105 and #ISP205) and 1:3000 (#ISP306) for overnight at 4°C. The membrane was washed in TBST and then incubated with horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG (GE Healthcare Cat# NA934-100Ul) at room temperature for 1 h. ECL Plus Western Blotting Detection Reagents (GE Healthcare) was used to detect antibody binding and chemiluminofluorescence signals were visualized with the Light-Capture System attached to a cool CCD camera (ATTO, Tokyo, Japan).
Immunohistochemistry
The dissected brains (n = 3) were fixed in Bouin's solution, and dehydrated through graded series of ethanols, cleared in n-butanol and embedded in Paraplast Plus (Oxford Labware, St Louis, MO, USA). Serial brain sections were cut in the sagittal plane (15 μm) and processed for immunohistochemistry. The rabbit anti-tilapia GAP antiserum (dilution of 1:1000–3000) were prepared in PBS containing 2% normal goat serum, and 0.5% Triton X-100, and the control anti-GAP antisera were preabsorbed with 10 μg/ml of respective antigen at the working dilution for 24 h. Alternate brain sections were incubated with primary GAP antiserum and pre-absorbed primary antiserum (n = 1 for each antiserum) at 4°C for 24 h in a closed moist chamber. Diluted biotinylated anti-rabbit IgG and avidin-biotinylated horse radish peroxidase (HRP) complex (Vectastain ABC Elite kit, Vector Laboratories Cat# PK-6101) were then applied to the sections for 30 and 45 min, respectively. Development was achieved through application of 0.05% 3,3-diaminobenzidine tetrahydrochloride (Sigma) with 0.03% H2O2 in 0.05 M Tris-HCl (pH 7.5). After dehydration, stained sections were cleared in xylene, coverslipped, and sealed with DPX mounting medium (Fisher Scientific, Loughborough, UK). Sections were scanned and images were captured with a Carl Zeiss MIRAX slide scanning system (Zeiss GmbH, Göttingen, Germany) using the Mirax Viewer Image Software (3DTech, Budapest, Hungary) at a 230 nm resolution with a × 20 objective.
Double-Immunofluorescence of GnRH-GAP Types
Brain of sexually immature (n = 3) and mature tilapia (n = 3) was dissected and fixed in buffered 4% paraformaldehyde (pH 7.5) for 6–8 h, cryoprotected in 20% sucrose in 0.1 M phosphate buffer and embedded in Tissue Tek OCT compound (Sakura Finetechnical, Tokyo, Japan). Sagittal sections (30 μm) were cut on a cryostat and thaw-mounted onto APS-coated slide glass. Sections were incubated for 24 h at 4°C with GAP antisera and incubated for 2 h with Alexa Fluor 488- or 594-labeled anti-rabbit IgG (Invitrogen, dilution of 1:400). Since double-label immunohistochemistry using the antisera generated in the same animal species can potentially result in cross-reactivity, we used F(ab)2 fragments anti-rabbit IgG to block the free-binding sites of secondary antibody (43). After several washes in PBS, the sections were incubated for 2 h in 2% normal rabbit serum at room temperature, and incubated for 2 h with F(ab')2 fragment anti-rabbit IgG (Jackson Immunoresearch Laboratories, Inc., PA, USA; dilution of 1:100). Sections were then incubated for 24 h at 4°C with different types of GAP antisera to be paired with different GnRH-GAP types (GAP1-GAP2, GAP2-GAP3, and GAP3-GAP1), and incubated for 2 h with Alexa Fluore 488- or 594-labeled anti-rabbit IgG (Invitrogen). Coverslips were applied with Vectashield medium (Vector Laboratories, Burlingame, CA) and dual-label immunofluorescent images were obtained using a fluorescent microscope (Nikon, Eclips 90i, Nikon) attached with a cooled CCD camera.
Detailed structures were further analyzed under a laser confocal microscope (C1si, Nikon Instruments, Tokyo, Japan) using laser wavelength of 488 and 543 nm. The images were captured and superimposed automatically by an imaging software (NIS Elements, Nikon). The red channel was converted to magenta, and brightness and contrast adjustments were made in Adobe Photoshop CC (Adobe, San Jose, CA, USA).
Single Cell Dissection of GnRH-GAP Cells and RNA Extraction
Single cell harvesting of GnRH-GAP-immunoreactive cells and total RNA extraction was performed as described previously with slight modifications (38, 39, 42, 44). Briefly, the brain sections were immunoreacted with GAP antiserum for overnight followed by incubation with secondary antibodies (AlexaFluor 488- or 594-labeled anti-rabbit IgG) as described above in RNase-free buffer. GnRH-GAP-immunolabeled cells were microdissected by heat-pulled borosilicate glass micro capillary pipette (1.5 mm outer diameter, Harvard Apparatus Ltd., Edenbridge, Kent; micropipette puller; Type PE-2, Narishige, Tokyo, Japan) attached to a micromanipulator (Narishige). Then, single GnRH-GAP cells were harvested by the micropipette and collected into a sterile 1.5 ml reaction tube containing 50 μl of the lysis buffer. The harvested single GAP cell was digested with 1 μg of proteinase K (Gentra Systems, Minneapolis, MN) for 1 h at 53°C. The cell lysate was incubated for 1 h at 37°C with 1U RNase-free DNase I (Promega, Madison, WI) to eliminate genomic DNA. Total RNA was extracted from the cell lysate using ISOGEN (Nippon Gene, Tokyo, Japan) and reverse transcribed to cDNA in a reaction mixture (20 μl) with 100 pmol of random primers (Takara, Tokyo, Japan) and 40U SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA).
Quantitative Analysis of GnRH and GnRHR Types mRNAs in GnRH-GAP Cells
To quantify mRNA levels of gnrhr types in GnRH-GAP cells, single GnRH-GAP cell's cDNA samples were subjected to real-time PCR for gnrh1, gnrh2, gnrh3, gnrhr1, gnrhr2, and gnrhr3 with an ABI PRISM 7700 Sequence Detection System (PE Applied Biosystems). The PCR mixture (10 μl) contained 1 × TaqMan Universal PCR Master Mix (Applied Biosystems), 300 nM of forward and reverse primers (Supplementary Table 1), 200 nM of hybridization probe (Supplementary Table 1), and one 20th of a single cell's cDNA or absolute standard cDNA. All primers and probes were designed to span intron-exon junction, and thus, no amplification of genomic sequence was seen in the cDNA samples. The PCR conditions were 95°C for 10 min, 60 cycles at 95°C for 15 s and 60°C for 1 min. The absolute amounts of transcripts were determined by establishing a linear amplification curve from serial dilutions (101 to 106 fg) of a plasmid DNA containing a gnrh or a gnrhr partial cDNA sequence. For each animal, the average mRNA levels of gnrh types and gnrhr per cell were determined and these values were combined to give experimental group means. For GnRH-GAP cells, expression of glial fibrillary acidic protein (gfap) gene (Supplementary Table 1) was also examined, and only those cells that tested negative for gfap but positive for respective gnrh genes were included for data analyses. All values are presented as the means (±SEM). Correlation of gnrh and gnrhr types mRNA levels were examined by Pearson R, while the mean difference of gnrh and gnrhr types mRNA levels between mature and immature were analyzed by unpaired Student's t-test or non-parametric ANOVA followed by post-hoc Dunn's multiple comparison test. The Tukey's criterion applied to each group of data sets, allowing detection of outliers (45). P < 0.05 was considered statistically significant.
Results
Antibody Specificity
Specificity of antisera to GAP types, brain tissue extracts were analyzed by SDS-PAGE and Western blots. Distinct single bands were detected in the brain and pituitary homogenate reacted with the antisera to GAP types (Figure 1A). A band immunoreactive to GAP2 in the brain was absent in the pituitary. However, detected immunoreactive bands were seen in unexpected sizes ranging from 35 to 45 KDa for all antisera, which are far bigger than the expected sizes of tilapia GAP types (7, 5.7, and 6 kDa for GAP1, GAP2, and GAP3, respectively) or prepro-GnRH types (11.4, 9.6, and 10.1 KDa for tilapia prepro-GnRH1, GnRH2, and GnRH3, respectively).
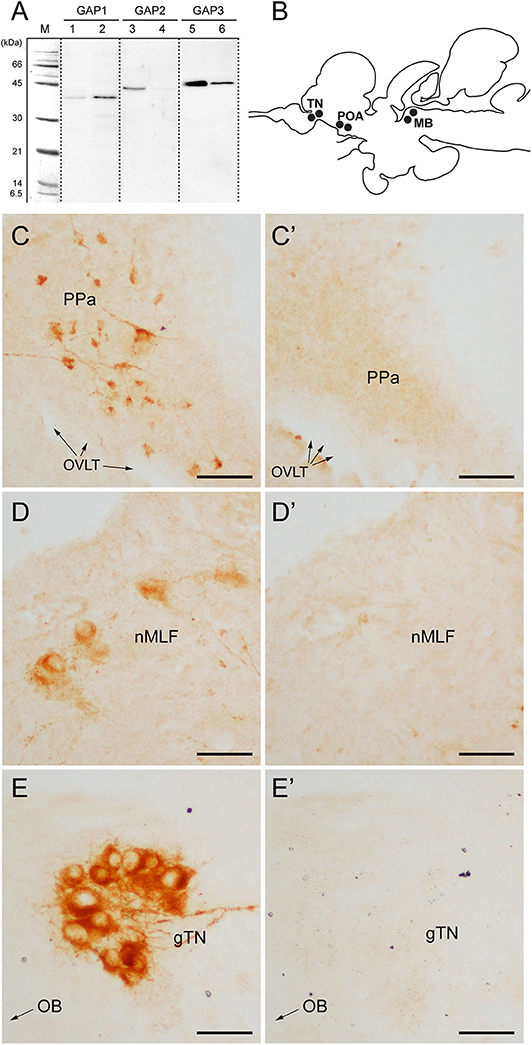
Figure 1. Antibody characterization and immunohistochemical distribution of three GAP-immunoreactivities in the brain of tilapia. (A) Western blot analysis of antisera to GAP types (#ISP105 for GAP1, #ISP205 for GAP2, and #ISP306 for GAP3) in the brain (lanes 1, 3 and 5) and pituitary (lanes 2, 4 and 6) homogenate. M, molecular mass marker. (B) Schematic diagram of distribution of three GnRH-GAP-immunoreactive cell types in the brain of tilapia: GnRH-GAP1 in the preoptic area (POA), GnRH-GAP2 in the midbrain (MB) and GnRH-GAP3 in the terminal nerve (TN). (C–E) Photomicrographs of three GAP-immunoreactivities (C–E) and their preaborption with respective GAP antigen (C'–E'). PPa, anterior part of the parvocellular preoptic nucleus; OVLT, organum vasculosum laminae terminalis; nMFL, nucleus of the medial longitudinal fascicle; gTN, terminal nerve ganglion; OB, olfactory bulb. Scale bars: 50 μm.
Using GAP antisera, three distinct GnRH-GAP neural populations were observed (Figure 1B); GnRH-GAP1 cell somata in the ventral telencephalon and anterior part of the parvocellular preoptic nucleus (Figure 1C), GnRH-GAP2 cell somata in the nucleus of the medial longitudinal fasciculus of the midbrain tegmentum (Figure 1D) and GnRH-GAP3 cell somata in the terminal nerve ganglion (also known as the nucleus olfacto-retinalis) (Figure 1E). Parhar et al. (34), Parhar et al. (41) all immunoreactivities in the brain were successfully blocked by the preabsorption with respective antigen peptide (Figures 1C'–E').
Association Between Neuronal Processes (Axons/Dendrites) and Cell Somata of Three GnRH-GAP Types
Using the GAP antisera, we examined possible association between neuronal processes and cell somata of three GnRH-GAP types in the brain. Since the discrimination between dendrites and axons is technically difficult, we define the labeled dendrites and axons collectively as neuronal processes in the entire manuscript.
In mature males, GnRH-GAP1-immunoreactive (ir) cell somata are accompanied by closely located GnRH-GAP3-ir neuronal processes in the anterior preoptic area (Figure 2A). Similarly, GnRH-GAP1-ir neuronal processes are in proximities of GnRH-GAP3-ir cell somata in the terminal nerve ganglion (Figure 2B). Confocal imaging further confirmed the proximities of GnRH-GAP1-ir and GnRH-GAP3-ir neuronal processes with cell somata (Figure 2a,b). However, such proximities of GnRH-GAP1 and GnRH-GAP3 neuronal processes with cell somata was not seen in immature fish (Figures 2C,D). There were no proximities of GnRH-GAP1 and GnRH-GAP2 processes/cell somata in mature and immature fish. On the other hand, GnRH-GAP2-ir neuronal processes are in proximities of GnRH-GAP3-ir cell somata in the terminal nerve in mature and immature males (Figures 3A,C), while no GnRH-GAP3-ir neuronal processes were seen in proximities of GnRH-GAP2-ir cell somata in the midbrain (Figures 3B,D).
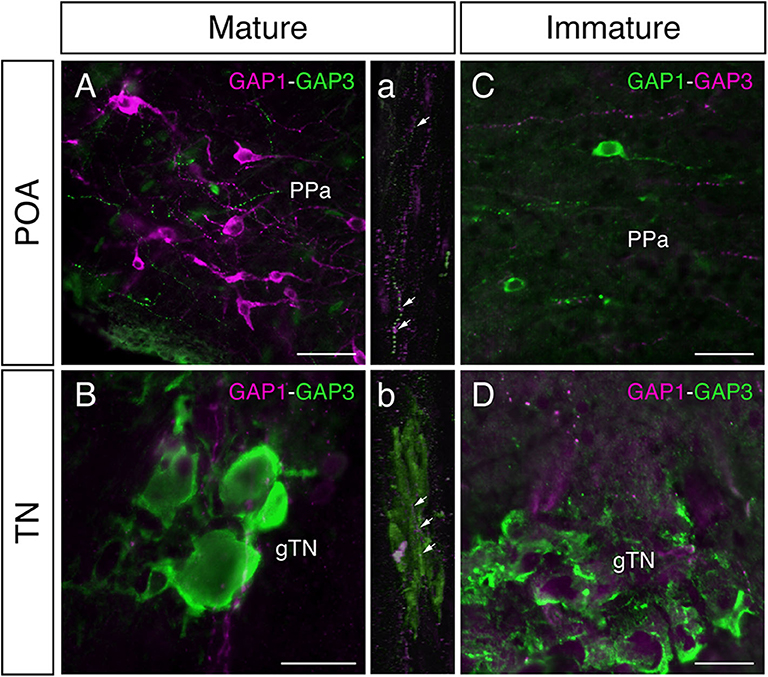
Figure 2. Photomicrographs of double immunofluorescence of GnRH-GAP1 and GnRH-GAP3 in the brain of sexually mature and immature male tilapia. In mature males, GnRH-GAP3-immunoreactive (ir) neuronal processes (green) are closely located to GnRH-GAP1-ir cell somata (magenta) in the anterior part of the parvocellular preoptic nucleus (PPa) of the preoptic area (POA, A), while in the terminal nerve ganglion (gTN), GnRH-GAP1-ir neuronal processes (magenta) are also closely located to GnRH-GAP3-ir cell somata (green) the (B). Confocal scanning of these regions further confirmed the proximities of GnRH-GAP1 neuronal processes with GnRH-GAP3 cell somata (a,b). In contrast, neuronal interaction between GnRH-GAP1 and GnRH-GAP3 were not seen in immature males (C,D). Scale bars = 20 μm.
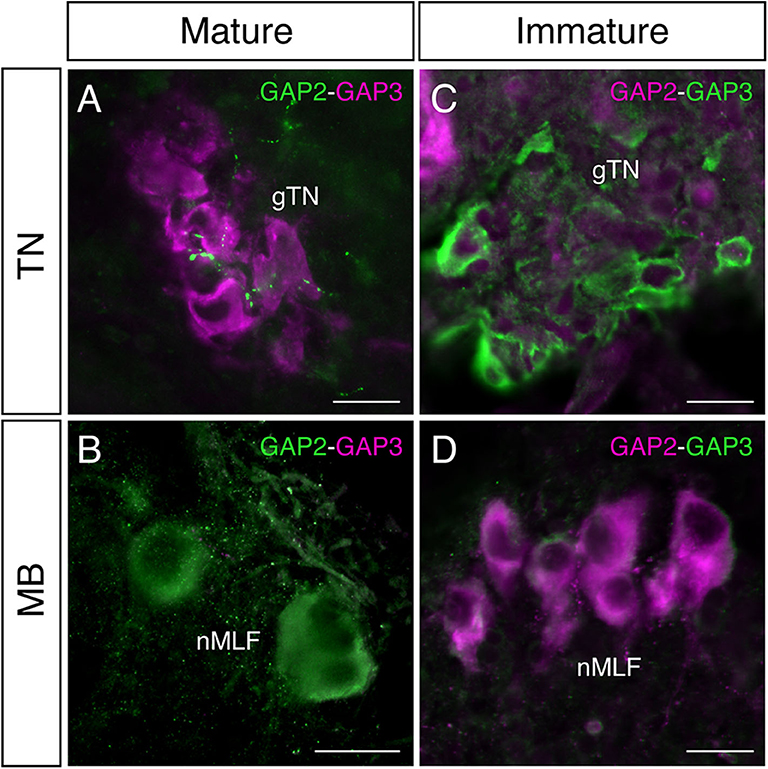
Figure 3. Photomicrographs of double immunofluorescence of GnRH-GAP2 and GnRH-GAP3 in the brain of sexually mature and immature male tilapia. In mature males, GnRH-GAP2-immunoreactive (ir) neuronal processes (green) are closely located to GnRH-GAP3-ir cell somata (magenta) in the terminal nerve ganglion (gTN, A), while such interaction between GnRH-GAP3-ir neuronal processes (magenta) and GnRH-GAP2-ir cell soma (green) was not seen in the nucleus of the medial longitudinal fascicle (nMLF) of the midbrain (MB, B). No proximities of GnRH-GAP2 (magenta) and GnRH-GAP3 (green) neuronal processes/cell somata were observed in immature males (C,D). Scale bars = 20 μm.
Expression of GnRH and GnRHR Types in Single GnRH-GAP Cells
To investigate functional interaction between different GnRH-GAP cell types, expression of GnRHR genes were examined in harvested single-cell GnRH-GAP neurons (Figure 4). All harvested GnRH cell types expressed respective gnrh types, gnrhr1 and gnrhr2 mRNA transcripts in mature and immature males, while gnrhr3 mRNA were only detected in few cells. Thus, we only proceeded for quantification of gnrhr1 and gnrhr2 mRNAs.
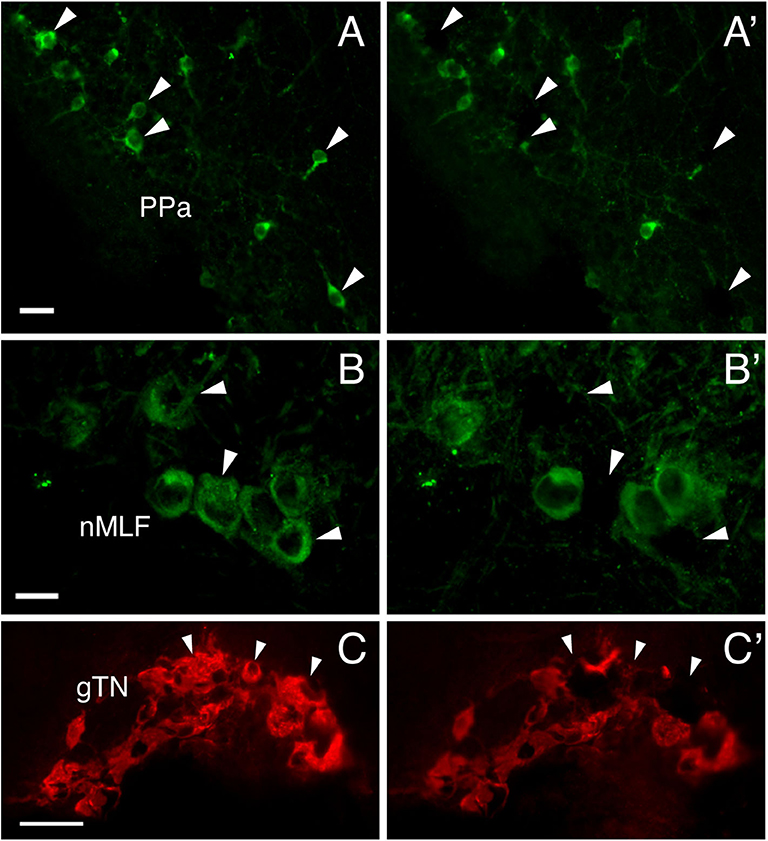
Figure 4. Photomicrographs of GnRH-GAP immunoreactive cell somata before (A–C) and after (A'–C') microdissection. Arrows indicate harvested cell somata of GnRH-GAP1 (A,A') in the anterior part of the parvocellular preoptic nucleus (PPa), GnRH-GAP2 (B,B') in the nucleus of the medial longitudinal fascicle (nMLF) and GnRH-GAP3 (C,C') in the terminal nerve ganglion (gTN). Scale bars, (A,B), 20 μm; (C) 50 μm.
GnRH-GAP1
At the single cell levels, gnrh1 mRNA levels were significantly (P < 0.01) higher in mature (30.4 ± 4.7 copies/cell) than those in immature males (15.5 ± 1.6 copies/cell) (Figure 5C). In mature and immature males, 90–100% of GnRH-GAP1 cells expressing single or multiple GnRHR types (mature, 33/33 cells; immature, 44/49 cells) (Table 1). There was no significant correlation between mRNA levels of gnrh1 and either gnrhr1 or gnrhr2 in immature and mature males (Figures 5A,B). There was no significant difference in gnrhr1 mRNA levels in GnRH-GAP1 cells between mature and immature males, while gnrhr2 mRNA levels were significantly (P < 0.001) higher in mature males (Figure 5C).
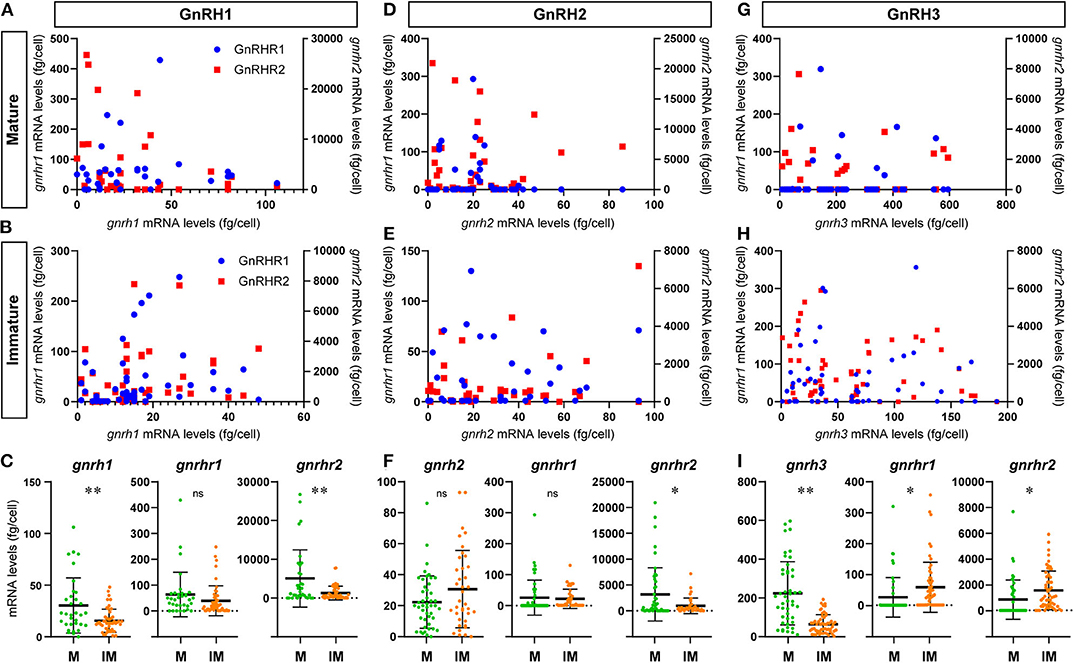
Figure 5. Graphs showing the distribution of GnRH receptor types mRNA transcripts in microdissected single GnRH-GAP neurons in mature and immature males. In the scatter blot diagrams (A,B,D,E,G,H), the x-axis represents the levels of each GnRH mRNAs in GnRH-GAP cell soma, while the 1st (left side) y-axis represents gnrhr1 mRNA levels and 2nd (right side) y-axis represents gnrhr2 mRNA levels in each GnRH-GAP cell soma. Red dots represent gnrhr1 and blue dots represent gnrhr2 mRNA levels. Column scatter blot diagrams in below (C,F,I) show the mRNA levels of GnRH (gnrh1, gnrh2, and gnrh3) and GnRH receptor types per GnRH-GAP cell soma in mature (M, green) and immature (IM, orange) males. *P < 0.05; **P < 0.001; ns, non-significant.
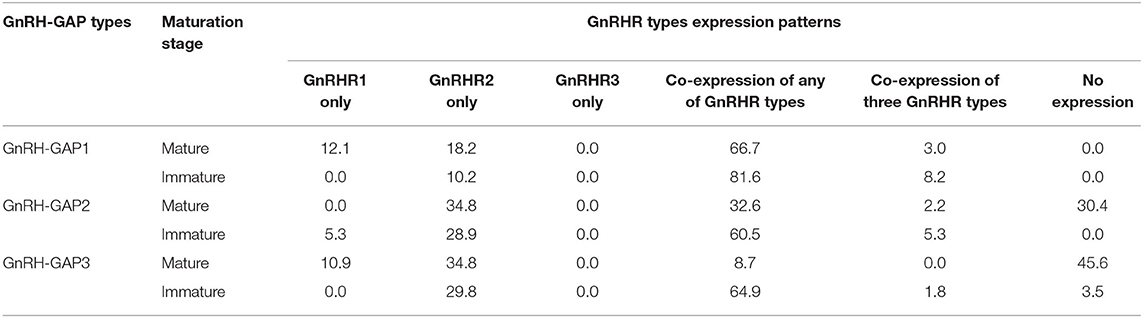
Table 1. Percentage of GnRH-GAP1-3 subtypes cells expressing GnRH receptor mRNA transcripts in mature and immature males.
GnRH-GAP2
There was no difference in gnrh2 mRNA levels between the two reproductive statuses (immature, 30.7 ± 4.1; mature, 22.3 ± 2.5 fg/cell) (Figure 5F). In GnRH-GAP2 cells, 73–95% of cells express single or multiple GnRHR types (mature, 32/46 cells; immature, 36/38 cells) (Table 1). There was no correlation between mRNA levels of gnrh2 and gnrhr types in immature and mature males (Figures 5D,E). There was no difference in gnrhr1 mRNA levels in GnRH-GAP2 cells between the two reproductive states, while gnrhr2 mRNA levels were significantly (P < 0.05) higher in mature male (3190.0 ± 760.2 fg/ cell) as compared with immature males (961.9 ± 242.2 fg/cell) (Figure 5F).
GnRH-GAP3
gnrh3 mRNA levels were significantly (P < 0.0001) higher in mature (221.9 ± 24.1 fg/cell) compared with immature males (66.9 ± 7.7 fg/cell) (Figure 5I). In immature males, high percentage of GnRH-GAP3 cells express gnrhr types (55/57 cells), while in mature males, gnrhr types expression was only detected in half of cells (24/46 cells) (Table 1). There was no correlation between mRNA levels of gnrh3 and either gnrhr1 or gnrhr2 in immature and mature males (Figures 5G,H). GnRH-GAP3 neurons express significantly (P < 0.05) higher mRNA levels of gnrhr1 and gnrhr2 in immature (gnrhr1, 57.9 ± 10.6 fg/cell; gnrhr2, 1525.6 ± 198.5 fg/cell) as compared with mature (gnrhr1, 26.7 ± 9.4 copies/cell; gnrhr2, 841.3 ± 222.9 copies/cell) males (Figure 5I).
Discussion
To examine potential functional interaction among GnRH types, we generated three tilapia GAP-specific antisera. Specificity of the three tilapia GAP antisera were characterized by Western blot analysis, which showed a single distinct band of different size for each GAP type, however, the molecular weight for each GAP types was much higher than predicted sizes for GAP types (6–7 kDa) or prepro-GnRH (9–11 KDa). The exact reason for the contradictory results of Western blot assays remains unknown, however, previous studies have also shown high molecular weight of GAP or prepro-GnRH-immunoreactive bands in some fish species. In a cichlid fish, Cichlasoma dimerus, Western blot assay for chicken II-GAP antiserum showed a band of 34 kDa in the brain extract (36). Similarly, in clownfish (Amphiprion melanopus), a band of 52 KDa was detected by LRH13, a monoclonal antibody to mammalian GnRH1 in Western blot assay (46). In the studies mentioned above, observation of bands of higher molecular weight than expected could be due to factors such as protein aggregation, incomplete denaturation or residual disulfide bonding. In fact, GAP is suggested to form a helix-loop-helix structure (47), which may affect Western blotting results. Hence, our results of Western blot assay are like those reported in other fish species. Further, our pre-absorption study using respective GAP antigens successfully abolished the GAP immunoreactivities in the brain. Our current and previous single cell RT-PCR showed expression of gnrh1 in GnRH-GAP1 cells, gnrh2 in GnRH-GAP2 cells and gnrh3 in GnRH-GAP3 cells (38, 39), confirming genetic characteristics of GAP-ir cells as GnRH cells. In addition, the localization patterns of GAP-ir cells completely overlapped with cells expressing respective GnRH mRNA in the in situ hybridization study and GnRH peptide synthesizing cells in the immunocytochemical study (34, 41). Although there remains a low possibility of cross-reaction of the three GAP antisera to some unknown proteins with high molecular weight, but our present and past studies strongly suggest that the immunoreactivity to the three GAP types is highly specific.
The three GAP antisera used in this study specifically immunoreacted with their respective GAP types without cross-reactivity in the brain. Double-immunofluorescence labeling showed the proximities of GAP1- and GAP3-ir neuronal processes and cell somata, which suggests possible functional interaction between GnRH-GAP1 and GnRH-GAP3 neurons. In fact, the density of GnRH-GAP1-ir and GnRH-GAP3-ir neuronal processes, closely located to GnRH-GAP1-ir and GnRH-GAP3-ir cell somata was higher in mature males as compared to immature males.
At the single cell level, all three GnRH-GAP cell types expressed multiple GnRHR gene types, in particular, gnrhr1 and gnrhr2, which is in good agreement with our previous immunohistochemical localization of GnRHR types in GnRH3 cells in the tilapia (48) and mRNA expression of two GnRHR types in GnRH3 cells in the dwarf gourami (49). These results suggest the roles of GnRHR in autocrine/paracrine regulation of GnRH cell types as has demonstrated by electrophysiological studies (50). Although the ligand-receptor interaction of three GnRH and GnRHR types in the tilapia still remains unclear, in medaka, which possesses three GnRHR types that are structurally similar to three tilapia GnRHR types, GnRHR1 and GnRHR2 share similar potencies to GnRH ligands with GnRH2 > GnRH3 > GnRH1 (51). Based on the affinity of medaka GnRHR types against three GnRH types, it can be speculated that GnRH1 and GnRH3 cells could be sensitive to multiple GnRH types through GnRHR1 and GnRHR2 signaling. Although GnRH2 is the most potent ligand for all three GnRHR types in fish (52), there was no proximities of GnRH-GAP1 or GnRH-GAP3 neuronal processes with GnRH-GAP2 cell soma, except GnRH-GAP2 neuronal processes were closely located to GnRH-GAP3 cell somata. This suggests that functional interaction between GnRH1 or GnRH3 with GnRH2 could be limited. Alternatively, GnRH2 could reach its target GnRH cell types via non-synaptic and disparate delivery mechanism such as through the circulation as has been demonstrated in the pituitary of tilapia (53, 54).
In GnRH-GAP3 cells, gnrhr1 and gnrhr2 mRNA levels were higher in immature as compared to mature males. In contrast, in GnRH-GAP1 and GnRH-GAP2 cells, gnrhr2 mRNA levels were significantly higher in mature males, while GnRH-GAP1 neuronal processes with GnRH-GAP3 cell somata were more abundant in mature males as compared to immature males. These observations suggest that GnRHR1 and GnRHR2 could regulate different GnRH cell types under different reproductive conditions. In mature males, GnRHR types could be involved in para or endocrine control by different GnRH peptides, while in immature males, GnRHR types may be participating in autocrine control. In some fish species, GnRH neural activities and physiology are closely associated with social hierarchy. In male Astatotilapia burtoni, the size of GnRH1 cell soma changes reversibly depending on male social status (55). In medaka fish, the expressions of gnrh1 is higher in dominant than in subordinate males, while gnrh3 expression is higher in subordinate than in dominant males (56). In the tilapia, immuno-suppression of GnRH3 results in reduction of male social behaviors (27). However, social hierarchy in male cichlid fish is more for territory defense (aggressiveness), access to food resources (body mass) and securing future mating opportunity (57–60). Therefore, it is likely that GnRH-GAP1 is mainly responsible for the reproduction (61, 62), GnRH-GAP2 is for food intake/metabolism (22, 23), and GnRH-GAP3 is for socio-sexual behaviors (26–28), but they might coordinate with each other. Nevertheless, in the future, it will be interesting to study the three GnRH-GAP changes in one species at the same time.
Although the autocrine regulation of GnRH neurons has been considered a mechanism for the pulsatile secretion of GnRH (31), the physiological significance of expression of multiple GnRHR types in GnRH neurons as well as paracrine or autocrine regulation of GnRH neurons in other functions remains unclear. Some studies have implied a possible role of autocrine regulation of GnRH. For example, during developmental stages of zebrafish, GnRH3 peptides have been shown to regulate development of GnRH3 neurons in an autocrine fashion (63). In hypothalamic GT1-1 neuronal cells, GnRH has been shown to exert autocrine regulation at the level of GnRH gene transcription (64). Interestingly, in the zebrafish embryo, gene knockdown of gnrh3 affected regionalization of the brain and eye formation (65). Therefore, these non-reproductive functions of GnRH3 could also be regulated by multiple GnRH types in an autocrine and/or paracrine manner during sexual maturation.
The present study shows morphological and molecular evidences for the possibility of homogenous interaction within the GnRH-GAP neural group as well as heterogeneous interaction among different GnRH-GAP cell types. Co-expression of multiple GnRHR types in GnRH-GAP cell types may also suggest the possibility of heterodimerization of GnRHR types. In the protochordate, Ciona intestinalis, GnRHR heterodimer has been found to regulate various physiology of GnRH cells such as the elevation of intracellular calcium, time-extension of ERK phosphorylation, and up-regulation of cell proliferation (66). GPCR dimerization are also known to promote receptor folding, maturation and/or transport to the cell surface (67). Therefore, the presence of multiple GnRHR types and their functional interaction could also be important for not only the regulation of GnRH neuronal activity, but also the regulation of GnRHR cell-surface expression, which might influence on the probability of ligand-receptor binding under different physiological conditions. Autocrine/paracrine mechanisms have also been suggested to play an important role as a pulse generator (31). However, there is no direct evidence of coordinated release of GnRH by the different GnRH-GAP types in the non-mammalian species. We have previously demonstrated that inhibition of GnRH3 system influences the outcome of GnRH1 and GnRH2 in a cichlid (27). The present study provides a morphological evidence for the potential interrelationship between the hypophysiotropic and non-hypophysiotropic GnRH systems, which might be involved in co-modulation of reproductive and non-reproductive events.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by Animal Ethics Committee of Nippon Medical School.
Author Contributions
SO and IP: conceptualization, data analysis, and funding acquisition. SO: investigation and writing—original draft preparation. IP: writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Grants-in-Aid from Monash University Malaysia Research Grants [M-2-07 and M-4-08].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The initial part of this project was conducted at the Nippon Medical School, Tokyo, Japan. We thank Prof. Yasuo Sakuma for advice during the study. We are grateful to Dr. Masato Endo and Prof. Toshio Takeuchi (the Laboratory of Aquaculture, Tokyo University of Marine Science and Technology) for kindly providing us juvenile fish. We also thank Mr. Sho-Rei Choh and Mr. Akira Takahashi from Sankei Co., Tokyo, Japan for letting us to use the laser confocal microscopy together with their valuable technical supports. This study was conducted with a technical support provided by the Bioimaging Infrastructure Platform and the Drug Discovery Platform, Monash University Malaysia.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2020.00586/full#supplementary-material
References
1. Matsuo H, Baba Y, Nair RM, Arimura A, Schally AV. Structure of the porcine LH- and FSH-releasing hormone. I The proposed amino acid sequence. Biochem Biophys Res Commun. (1971) 43:1334–9. doi: 10.1016/S0006-291X(71)80019-0
2. Pfaff DW. Luteinizing hormone-releasing factor potentiates lordosis behavior in hypophysectomized ovariectomized female rats. Science. (1973) 182:1148–9. doi: 10.1126/science.182.4117.1148
3. Knobil E. The neuroendocrine control of ovulation. Hum Reprod. (1988) 3:469–72. doi: 10.1093/oxfordjournals.humrep.a136730
4. Sherwood NM, Lovejoy DA, Coe IR. Origin of mammalian gonadotropin-releasing hormones. Endocr Rev. (1993) 14:241–54. doi: 10.1210/edrv-14-2-241
5. Parhar IS. GnRH in tilapia: three genes, three origins and their roles. In: Parhar IS, Sakuma Y, editors. GnRH Neurons: Gene to Behavior. Tokyo: Brain Shuppan (1997). p. 99–122.
6. Mccann SM, Karanth S, Mastronardi CA, Dees WL, Childs G, Miller B, et al. Control of gonadotropin secretion by follicle-stimulating hormone-releasing factor, luteinizing hormone-releasing hormone, and leptin. Arch Med Res. (2001) 32:476–85. doi: 10.1016/S0188-4409(01)00343-5
7. Parhar IS. Cell migration and evolutionary significance of GnRH subtypes. Prog Brain Res. (2002) 141:3–17. doi: 10.1016/S0079-6123(02)41080-1
8. Millar RP. GnRHs and GnRH receptors. Anim Reprod Sci. (2005) 88:5–28. doi: 10.1016/j.anireprosci.2005.05.032
9. Okubo K, Nagahama Y. Structural and functional evolution of gonadotropin-releasing hormone in vertebrates. Acta Physiol. (2008) 193:3–15. doi: 10.1111/j.1748-1716.2008.01832.x
10. Williams BL, Akazome Y, Oka Y, Eisthen HL. Dynamic evolution of the GnRH receptor gene family in vertebrates. BMC Evol Biol. (2014) 14:215. doi: 10.1186/s12862-014-0215-y
11. Kim D-K, Cho EB, Moon MJ, Park S, Hwang J-I, Kah O, et al. Revisiting the evolution of gonadotropin-releasing hormones and their receptors in vertebrates: secrets hidden in genomes. Gen Comp Endocrinol. (2011) 170:68–78. doi: 10.1016/j.ygcen.2010.10.018
12. Muske LE. Evolution of gonadotropin-releasing hormone (GnRH) neuronal systems. Brain Behav Evol. (1993) 42:215–30. doi: 10.1159/000114156
13. Gore AC. Neuroanatomy of the GnRH-1 system. GnRH: The Master Molecule of Reproduction. Boston, MA: Springer (2002). p. 9–27. doi: 10.1007/978-1-4757-3565-9
14. Warnock JK, Bundren JC. Anxiety and mood disorders associated with gonadotropin-releasing hormone agonist therapy. Psychopharmacol Bull. (1997) 33:311.
15. Umathe SN, Bhutada PS, Jain NS, Dixit PV, Wanjari MM. Effects of central administration of gonadotropin-releasing hormone agonists and antagonist on elevated plus-maze and social interaction behavior in rats. Behav Pharmacol. (2008) 19:308–16. doi: 10.1097/FBP.0b013e328308f1fb
16. Hough D, Bellingham M, Haraldsen IRH, Mclaughlin M, Rennie M, Robinson JE, et al. Spatial memory is impaired by peripubertal GnRH agonist treatment and testosterone replacement in sheep. Psychoneuroendocrinology. (2017) 75:173–82. doi: 10.1016/j.psyneuen.2016.10.016
17. Fernald RD, White RB. Gonadotropin-releasing hormone genes: phylogeny, structure, and functions. Front Neuroendocrinol. (1999) 20:224–40. doi: 10.1006/frne.1999.0181
18. Maney DL, Richardson RD, Wingfield JC. Central administration of chicken gonadotropin-releasing hormone-II enhances courtship behavior in a female sparrow. Horm Behav. (1997) 32:11–8. doi: 10.1006/hbeh.1997.1399
19. Kauffman AS, Rissman EF. A critical role for the evolutionarily conserved gonadotropin-releasing hormone II: mediation of energy status and female sexual behavior. Endocrinology. (2004) 145:3639–46. doi: 10.1210/en.2004-0148
20. Barnett DK, Bunnell TM, Millar RP, Abbott DH. Gonadotropin-releasing hormone ii stimulates female sexual behavior in marmoset monkeys. Endocrinology. (2005) 147:615–23. doi: 10.1210/en.2005-0662
21. Kauffman A, Wills A, Millar R, Rissman E. Evidence that the type-2 Gonadotrophin-Releasing Hormone (GnRH) receptor mediates the behavioural effects of GnRH-II on feeding and reproduction in musk shrews. J Neuroendocrinol. (2005) 17:489–97. doi: 10.1111/j.1365-2826.2005.01334.x
22. Nishiguchi R, Azuma M, Yokobori E, Uchiyama M, Matsuda K. Gonadotropin-releasing hormone 2 suppresses food intake in the zebrafish, Danio rerio. Front Endocrinol. (2012) 3:122. doi: 10.3389/fendo.2012.00122
23. Marvel MM, Spicer OS, Wong TT, Zmora N, Zohar Y. Knockout of Gnrh2 in zebrafish (Danio rerio) reveals its roles in regulating feeding behavior and oocyte quality. Gen Comp Endocrinol. (2019) 280:15–23. doi: 10.1016/j.ygcen.2019.04.002
24. Abe H, Oka Y. Neuromodulatory functions of terminal nerve-GnRH neurons. Fish Physiol. (2006) 25:455–503. doi: 10.1016/S1546-5098(06)25011-8
25. Abraham E, Palevitch O, Gothilf Y, Zohar Y. Targeted gonadotropin-releasing hormone-3 neuron ablation in zebrafish: effects on neurogenesis, neuronal migration, and reproduction. Endocrinology. (2010) 151:332–40. doi: 10.1210/en.2009-0548
26. Yamamoto N, Oka Y, Kawashima S. Lesions of gonadotropin-releasing hormone-immunoreactive terminal nerve cells: effects on the reproductive behavior of male dwarf gouramis. Neuroendocrinology. (1997) 65:403–12. doi: 10.1159/000127203
27. Ogawa S, Akiyama G, Kato S, Soga T, Sakuma Y, Parhar IS. Immunoneutralization of gonadotropin-releasing hormone type-III suppresses male reproductive behavior of cichlids. Neurosci Lett. (2006) 403:201–5. doi: 10.1016/j.neulet.2006.02.041
28. Okuyama T, Yokoi S, Abe H, Isoe Y, Suehiro Y, Imada H, et al. A neural mechanism underlying mating preferences for familiar individuals in medaka fish. Science. (2014) 343:91–4. doi: 10.1126/science.1244724
29. Georgopoulos NA, Armeni AK, Stamou M, Kentrou A, Tsermpini EE, Iconomou G, et al. Gonadotropin-releasing hormone (GnRH) deficiency under treatment: psychological and sexual functioning impacts. Hormones. (2018) 17:383–90. doi: 10.1007/s42000-018-0055-z
31. Krsmanovic LZ, Hu L, Leung PK, Feng H, Catt KJ. The hypothalamic GnRH pulse generator: multiple regulatory mechanisms. Trends Endocrinol Metab. (2009) 20:402–8. doi: 10.1016/j.tem.2009.05.002
32. Parhar IS, Ogawa S, Sakuma Y. Three GnRH receptor types in laser-captured single cells of the cichlid pituitary display cellular and functional heterogeneity. Proc Natl Acad Sci USA. (2005) 102:2204–9. doi: 10.1073/pnas.0409494102
33. Polkowska J, Przekop F. Effect of protein deficiency on luteinizing hormone releasing hormone (LHRH), gonadotropin releasing hormone associated peptide (GAP) and luteinizing hormone (LH) immunocytochemistry in the hypothalamus and pituitary gland of prepubertal ewes. Exp Clin Endocrinol Diabetes. (1993) 101:230–7. doi: 10.1055/s-0029-1211237
34. Parhar IS, Pfaff DW, Schwanzel-Fukuda M. Gonadotropin-releasing hormone gene expression in teleosts. Mol Brain Res. (1996) 41:216–27. doi: 10.1016/0169-328X(96)00099-X
35. Gonzalez-Martinez D, Zmora N, Mananos E, Saligaut D, Zanuy S, Zohar Y, et al. Immunohistochemical localization of three different prepro-GnRHs in the brain and pituitary of the European sea bass (Dicentrarchus labrax) using antibodies to the corresponding GnRH-associated peptides. J Comp Neurol. (2002) 446:95–113. doi: 10.1002/cne.10190
36. Pandolfi M, Munoz Cueto JA, Lo Nostro FL, Downs JL, Paz DA, Maggese MC, et al. GnRH systems of Cichlasoma dimerus (Perciformes, Cichlidae) revisited: a localization study with antibodies and riboprobes to GnRH-associated peptides. Cell Tissue Res. (2005) 321:219–32. doi: 10.1007/s00441-004-1055-7
37. Gopurappilly R, Ogawa S, Parhar IS. Functional significance of GnRH and kisspeptin, and their cognate receptors in teleost reproduction. Front Endocrinol. (2013) 4:24. doi: 10.3389/fendo.2013.00024
38. Phang Y, Soga T, Kitahashi T, Parhar I. Cloning and functional expression of novel cholesterol transporters ABCG1 and ABCG4 in gonadotropin-releasing hormone neurons of the tilapia. Neuroscience. (2012) 203:39–49. doi: 10.1016/j.neuroscience.2011.12.016
39. Ogawa S, Ng K, Xue X, Ramadasan P, Sivalingam M, Levavi-Sivan B, et al. Thyroid hormone upregulates hypothalamic kiss2 gene in the male Nile tilapia, Oreochromis niloticus. Front Endocrinol. (2013) 4:184. doi: 10.3389/fendo.2013.00184
40. Mizrahi N, Gilon C, Atre I, Ogawa S, Parhar IS, Levavi-Sivan B. Deciphering direct and indirect effects of neurokinin B and GnRH in the brain-pituitary axis of tilapia. Front Endocrinol. (2019) 10:469. doi: 10.3389/fendo.2019.00469
41. Parhar IS, Ogawa S, Hamada T, Sakuma Y. Single-cell real-time quantitative polymerase chain reaction of immunofluorescently identified neurons of gonadotropin-releasing hormone subtypes in cichlid fish. Endocrinology. (2003) 144:3297–300. doi: 10.1210/en.2003-0386
42. Parhar IS, Ogawa S, Sakuma Y. Laser-captured single digoxigenin-labeled neurons of gonadotropin-releasing hormone types reveal a novel g protein-coupled receptor (Gpr54) during maturation in cichlid fish. Endocrinology. (2004) 145:3613–8. doi: 10.1210/en.2004-0395
43. Negoescu A, Labat-Moleur F, Lorimier P, Lamarcq L, Guillermet C, Chambaz E, et al. F(ab) secondary antibodies: a general method for double immunolabeling with primary antisera from the same species. Efficiency control by chemiluminescence. J Histochem Cytochem. (1994) 42:433–7. doi: 10.1177/42.3.7508473
44. Ogawa S, Parhar IS. Single-cell gene profiling reveals social status-dependent modulation of nuclear hormone receptors in GnRH neurons in a male cichlid fish. Int J Mol Sci. (2020) 21:2724. doi: 10.3390/ijms21082724
45. Burns MJ, Nixon GJ, Foy CA, Harris N. Standardisation of data from real-time quantitative PCR methods - evaluation of outliers and comparison of calibration curves. BMC Biotechnol. (2005) 5:31. doi: 10.1186/1472-6750-5-31
46. Ogawa S, Soga T, Sakuma Y, Parhar IS. Modulation of GnRH subtypes by social stress and aggressive behavior. Fish Physiol Biochem. (2003) 28:49–50. doi: 10.1023/B:FISH.0000030474.32151.12
47. Kim NN, Shin HS, Habibi HR, Lee J, Choi CY. Expression profiles of three types of GnRH during sex-change in the protandrous cinnamon clownfish, Amphiprion melanopus: effects of exogenous GnRHs. Comp Biochem Physiol Part B Biochem Mol Biol. (2012) 161:124–33. doi: 10.1016/j.cbpb.2011.10.003
48. Pérez Sirkin DI, Lafont A-G, Kamech N, Somoza GM, Vissio PG, Dufour S. Conservation of three-dimensional helix-loop-helix structure through the vertebrate lineage reopens the cold case of gonadotropin-releasing hormone-associated peptide. Front Endocrinol. (2017) 8:207. doi: 10.3389/fendo.2017.00207
49. Soga T, Ogawa S, Millar RP, Sakuma Y, Parhar IS. Localization of the three GnRH types and GnRH receptors in the brain of a cichlid fish: insights into their neuroendocrine and neuromodulator functions. J Comp Neurol. (2005) 487:28–41. doi: 10.1002/cne.20519
50. Hajdu P, Ikemoto T, Akazome Y, Park MK, Oka Y. Terminal nerve Gonadotrophin-Releasing Hormone (GnRH) neurones express multiple GnRH receptors in a teleost, the dwarf gourami (Colisa lalia). J Neuroendocrinol. (2007) 19:475–9. doi: 10.1111/j.1365-2826.2007.01553.x
51. Abe H, Oka Y. Modulation of pacemaker activity by salmon gonadotropin-releasing hormone (sGnRH) in terminal nerve (TN)-GnRH neurons. J Neurophysiol. (2000) 83:3196–200. doi: 10.1152/jn.2000.83.5.3196
52. Okubo K, Nagata S, Ko R, Kataoka H, Yoshiura Y, Mitani H, et al. Identification and characterization of two distinct GnRH receptor subtypes in a teleost, the medaka Oryzias latipes. Endocrinology. (2001) 142:4729–39. doi: 10.1210/endo.142.11.8475
53. Kah O, Lethimonier C, Somoza G, Guilgur LG, Vaillant C, Lareyre JJ. GnRH and GnRH receptors in metazoa: a historical, comparative, and evolutive perspective. Gen Comp Endocrinol. (2007) 153:346–64. doi: 10.1016/j.ygcen.2007.01.030
54. Weber GM, Powell JF, Park M, Fischer WH, Craig AG, Rivier JE, et al. Evidence that gonadotropin-releasing hormone (GnRH) functions as a prolactin-releasing factor in a teleost fish (Oreochromis mossambicus) and primary structures for three native GnRH molecules. J Endocrinol. (1997) 155:121–32. doi: 10.1677/joe.0.1550121
55. Golan M, Zelinger E, Zohar Y, Levavi-Sivan B. Architecture of GnRH-gonadotrope-vasculature reveals a dual mode of gonadotropin regulation in fish. Endocrinology. (2015) 156:4163–73. doi: 10.1210/en.2015-1150
56. White S, Nquyen T, Fernald R. Social regulation of gonadotropin-releasing hormone. J Exp Biol. (2002) 205:2567–81.
57. Kagawa N, Hirose S, Fujimoto K, Nomura C, Fujita Y, Honda A, et al. Social rank-dependent expression of gonadotropin-releasing hormones and kisspeptin in the medaka brain. Gen Comp Endocrinol. (2017) 249:48–54. doi: 10.1016/j.ygcen.2017.03.001
58. Koebele BP. Growth and the size hierarchy effect: an experimental assessment of three proposed mechanisms; activity differences, disproportional food acquisition, physiological stress. Environ Biol Fishes. (1985) 12:181–8. doi: 10.1007/BF00005149
59. Oliveira RF, Hirschenhauser K, Carneiro LA, Canario AV. Social modulation of androgen levels in male teleost fish. Comp Biochem Physiol B Biochem Mol Biol. (2002) 132:203–15. doi: 10.1016/S1096-4959(01)00523-1
60. Alonso F, Honji RM, Moreira RG, Pandolfi M. Dominance hierarchies and social status ascent opportunity: anticipatory behavioral and physiological adjustments in a Neotropical cichlid fish. Physiol Behav. (2012) 106:612–8. doi: 10.1016/j.physbeh.2012.04.003
61. Karigo T, Kanda S, Takahashi A, Abe H, Okubo K, Oka Y. Time-of-day-dependent changes in GnRH1 neuronal activities and gonadotropin mRNA expression in a daily spawning fish, medaka. Endocrinology. (2012) 153:3394–404. doi: 10.1210/en.2011-2022
62. Ma Y, Juntti SA, Hu CK, Huguenard JR, Fernald RD. Electrical synapses connect a network of gonadotropin releasing hormone neurons in a cichlid fish. Proc Natl Acad Sci USA. (2015) 112:3805–10. doi: 10.1073/pnas.1421851112
63. Abraham E, Palevitch O, Ijiri S, Du SJ, Gothilf Y, Zohar Y. Early development of forebrain gonadotrophin-releasing hormone (GnRH) neurones and the role of GnRH as an autocrine migration factor. J Neuroendocrinol. (2008) 20:394–405. doi: 10.1111/j.1365-2826.2008.01654.x
64. Cho S, Han J, Sun W, Choi D, Kwon HB, Jarry H, et al. Evidence for autocrine inhibition of gonadotropin-releasing hormone (GnRH) gene transcription by GnRH in hypothalamic GT1-1 neuronal cells. Mol Brain Res. (1997) 50:51–8. doi: 10.1016/S0169-328X(97)00171-X
65. Wu S, Page L, Sherwood NM. A role for GnRH in early brain regionalization and eye development in zebrafish. Mol Cell Endocrinol. (2006) 257–258:47–64. doi: 10.1016/j.mce.2006.06.010
66. Sakai T, Aoyama M, Kusakabe T, Tsuda M, Satake H. Functions of a GnRH receptor heterodimer of the ascidian, Ciona intestinalis. Acta Biol Hung. (2008) 59:241–3. doi: 10.1556/ABiol.59.2008.Suppl.34
Keywords: GnRH, GAP, GnRHR, cichlid, reproduction
Citation: Ogawa S and Parhar I (2020) Morphological Evidence for Functional Crosstalk Between Multiple GnRH Systems in the Male Tilapia, Oreochromis niloticus. Front. Endocrinol. 11:586. doi: 10.3389/fendo.2020.00586
Received: 08 April 2020; Accepted: 17 July 2020;
Published: 02 September 2020.
Edited by:
Gustavo M. Somoza, CONICET Instituto Tecnológico de Chascomús (INTECH), ArgentinaReviewed by:
Matias Pandolfi, University of Buenos Aires, ArgentinaTomohiro Osugi, Suntory Foundation for Life Sciences, Japan
Copyright © 2020 Ogawa and Parhar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ishwar Parhar, ishwar@monash.edu
 Satoshi Ogawa
Satoshi Ogawa Ishwar Parhar
Ishwar Parhar