- 1Institute of Biomedicine, Department of Medical Biochemistry and Cell Biology, University of Gothenburg, Gothenburg, Sweden
- 2Instituto de Investigación Sanitaria de Aragón (IIS Aragón), Zaragoza, Spain
- 3Departamento de Farmacología, Fisiología y Medicina Legal y Forense, Universidad de Zaragoza, Zaragoza, Spain
- 4Instituto Agroalimentario de Aragón—IA2 (Universidad de Zaragoza–CITA), Zaragoza, Spain
- 5Departamento de Bioquímica y Biología Molecular y Celular, Universidad de Zaragoza, Zaragoza, Spain
Disruption of the microbiota–gut–brain axis results in a wide range of pathologies that are affected, from the brain to the intestine. Gut hormones released by enteroendocrine cells to the gastrointestinal (GI) tract are important signaling molecules within this axis. In the search for the language that allows microbiota to communicate with the gut and the brain, serotonin seems to be the most important mediator. In recent years, serotonin has emerged as a key neurotransmitter in the gut–brain axis because it largely contributes to both GI and brain physiology. In addition, intestinal microbiota are crucial in serotonin signaling, which gives more relevance to the role of the serotonin as an important mediator in microbiota–host interactions. Despite the numerous investigations focused on the gut–brain axis and the pathologies associated, little is known regarding how serotonin can mediate in the microbiota–gut–brain axis. In this review, we will mainly discuss serotonergic system modulation by microbiota as a pathway of communication between intestinal microbes and the body on the microbiota–gut–brain axis, and we explore novel therapeutic approaches for GI diseases and mental disorders.
1 Introduction
The gastrointestinal (GI) tract is one of the major defensive organs in individuals because it is continuously exposed to the external environment. In this context, microbial colonization of the intestine during infancy is a major moment for the development of not only the GI tract (1) but also the brain (2) and the immune system (3). In the last years, numerous researchers have focused their efforts on understanding how intestinal microbiota have the ability to affect the brain and behavior, which has not yet been completely clarified. In this context, the neurotransmitter serotonin (5-hidroxytriptamine, 5-HT) could be the key to resolving this mystery.
The gut–brain axis is a bidirectional crosstalk between the central nervous system (CNS) and the gut. Recently, given the important role in the regulation of gut functions, microbiota are included in the axis. Then, the microbiota–gut–brain axis resides in a coordinated network composed of the CNS, enteric nervous system (ENS), hypothalamic–pituitary–adrenal axis, gut, and microbiota. Both clinical and experimental data suggest that intestinal microbiota play a crucial role in the axis, interacting not only locally with intestinal cells and the ENS but also directly with the CNS through neuroendocrine and metabolic pathways. In fact, germ-free mice studies have proven that the absence of microbial colonization leads to defects in neuron maturation at both CNS and ENS levels, altered expression of neurotransmitters, and gut sensory and motor dysfunctions (4). Intestinal microbiota dysbiosis has been extensively studied as one of the most important factors in the pathogenesis of inflammatory bowel diseases (IBDs) (5), including Crohn’s disease (CD) and ulcerative colitis (UC). In this context, several studies have described that intestinal serotonin may shape the microbiota composition that protects against the development of IBDs (6), suggesting the critical relation between the intestinal microbiota and serotonergic system in GI pathologies. However, the role of the microbiota–serotonin interaction would not be limited locally to the gut but also to the CNS. Germ-free mice studies have reported the importance of the microbiota control of the serotonergic system in the CNS (7) or how specific intestinal microorganisms, such as Akkermansia muciniphila, can increase serotonin production in the hippocampus (8). In this context, recent studies have described the involvement of microbiota in serotonin signaling in CNS disorders such as Alzheimer’s or schizophrenia (9).
Serotonin is a key neurotransmitter, which substantially coordinates the GI physiology and owns critical central functions. Interestingly, serotonin is involved in each component of the microbiota–gut–brain axis, acting as an ideal language for the crosstalk. Microbiota regulate the tryptophan metabolism involved in serotonin production, serotonin acts as a key neurotransmitter in the CNS and ENS, and serotonin receptors play a pivotal role in the hypothalamic–pituitary–adrenal axis.
Here, we highlight recent findings into how microbiota regulate the intestinal and central serotonergic systems, as well as novel clinical approaches to address GI pathologies and brain disorders through the microbiota–gut–brain axis.
2 Serotonergic System
In 1940, Vittorio Erspamer discovered serotonin (5-hydroxytryptamine, 5-HT) in the GI tract in rabbits (10) and it was later discovered in the CNS (11). There are two main serotonergic systems: the central serotonergic system located in the brain and the intestinal serotonergic system in the gut. Both share the same principles of synthesis (“ON mechanism”), internalization and degradation (“OFF mechanism”), and 5-HT signaling through its specific receptors (Figure 1).
● The “ON” mechanism is constituted in the gut by enterochromaffin cells and serotonergic neurons of the ENS, while in the CNS, 5-HT is produced only by serotonergic neurons. The primary source of 5-HT is the amino acid L-tryptophan that is catalyzed by the rate-limiting enzyme tryptophan hydroxylase (TPH) to synthesize 5-hydroxytryptophan (5-HTP), which then is converted into serotonin by aromatic amino acid decarboxylase (AAAD) (12). TPH reaction is a limitative step in the production of 5-HTP and, subsequently, serotonin. It has been described in two isoforms of TPH: TPH1 expressed in enterochromaffin cells and TPH2 in serotonergic neurons from both the ENS and CNS (13).
● The “OFF” mechanism in the gut is formed by enterocytes because these intestinal epithelial cells (IECs) internalize 5-HT from the extracellular compartment to the cytoplasm by means of the serotonin transporter (SERT) from the apical and the basolateral membranes. At the CNS level, the “OFF” mechanism is formed by the same serotonergic neurons that synthesize 5-HT because SERT is expressed at terminals and varicosities of serotonergic neurons (14). SERT is a transmembrane protein grouped in the solute carrier transporters of the SLC6 family that uptakes 5-HT from the extracellular space for subsequent catabolization, reuse, or storage, ending 5-HT effects. SERT is a classic secondary active transporter to which 5-HT binds together with a Na+ and a Cl-. Once extracellular serotonin is attached to SERT together with Na+ and Cl-, SERT undergoes a conformational change that allows SERT translocation with the release of 5-HT, Na+, and Cl- into the cytoplasm of the cell. Once 5-HT is transported inside the cell, intracellular K+ binds to SERT and is reoriented toward the extracellular direction, where K+ is released and the uptake of 5-HT continues. Then, SERT is not only a key component for the regulation of 5-HT levels, but also an important ion transporter (15).
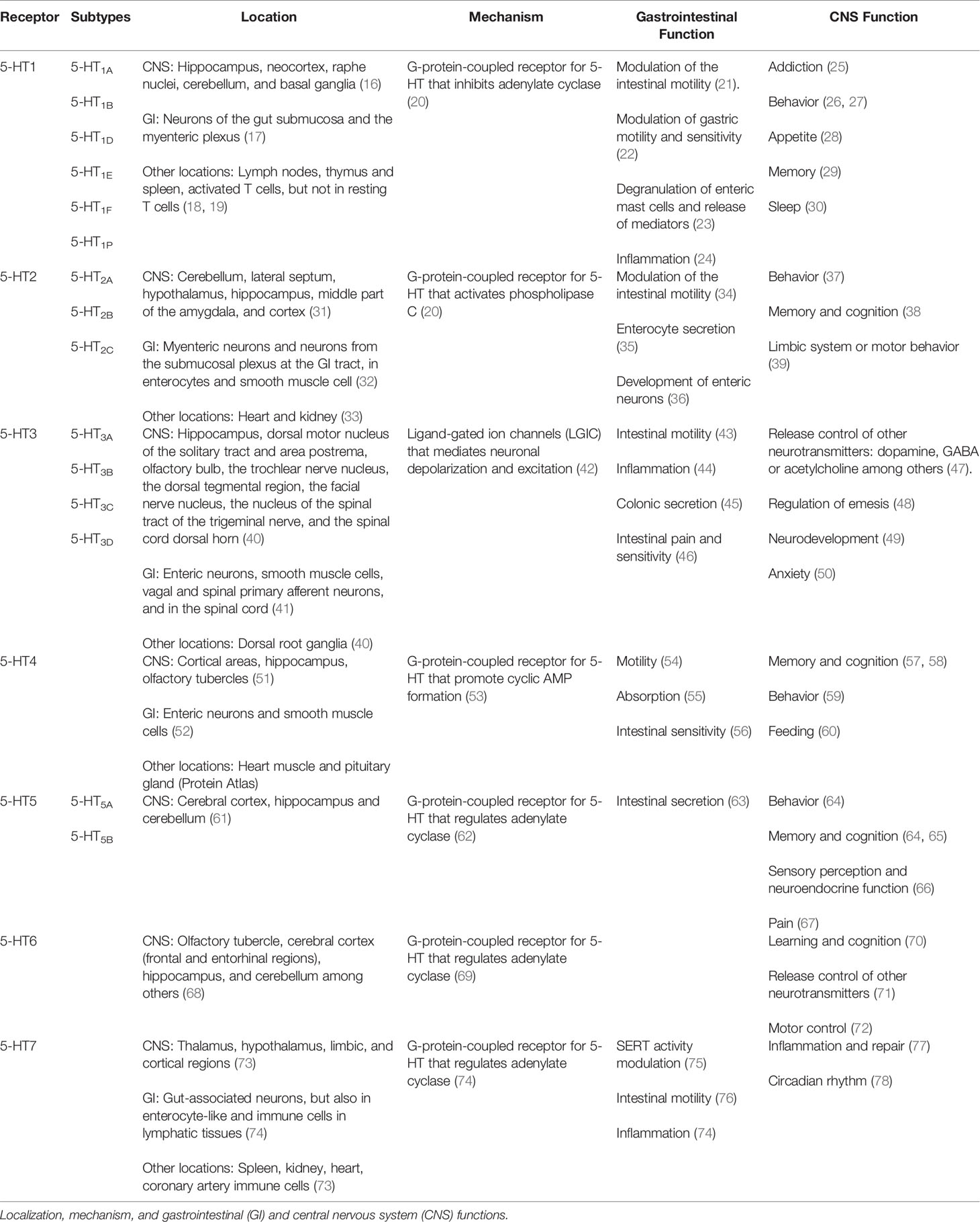
● 5-HT signaling is mediated by specific serotonin receptors that trigger intracellular 5-HT effects (Table 1). Scientific community studies on serotonin receptors have recently described a detailed work that classifies the 18 receptors grouped into seven families (5-HT1 to 5-HT7), which are widely expressed not only in the CNS and the GI tract but also in other systems such as the cardiovascular or immune system (79). As a short summary, the serotonin receptor family consists of G-protein-coupled receptors, with the exception of the 5-HT3 receptor family (80). 5-HT1 includes five subtypes: 5-HT1A, 5-HT1B, 5-HT1D, 5-HT1E, and 5-HT1F. They are fundamentally involved in CNS disorders such as anxiety. In the case of the GI tract, the 5-HT1 family is mainly expressed in neurons of the gut submucosa and the myenteric plexus, so their main function is the modulation of GI motility (18). The 5-HT2 family involves 5-HT2A, 5-HT2B, and 5-HT2C. 5-HT2A and 5-HT2B are expressed in myenteric neurons and neurons from the submucosal plexus in the GI tract, as well as in enterocytes and smooth muscle cells in the gut (36). Thus, the effect of these receptors is mainly in the GI tract through the regulation of GI motility (81). However, these receptors are expressed in the brain, where they may control central processes such as memory and cognition (82) or be implicated in CNS disorders such as depression (83). 5-HT2C is mainly expressed in the CNS and is involved in several central processes such as the limbic system and motor behavior (38). The 5-HT3 family includes five receptors (5-HT3A-D) and works as an ion channel similar to GABA receptors. 5-HT3 receptors are expressed in both the CNS and the GI tract and are involved in several GI processes such as intestinal motility (84), absorption and secretion (85), and even 5-HT release from enterochromaffin cells (86); in the brain, 5-HT3 receptors are related with cognition (87). In this context, 5-HT3 family dysfunction has been involved in a broad range of pathologies from brain disorders, including psychosis, anxiety, and eating disorders (43), to GI pathologies such as IBDs (88). 5-HT4 receptors are mainly expressed in the gut and participate in intestinal secretion (63) and motility (53). The 5-HT5 receptor is the least known from the serotonergic system as some researchers have referred to it for two decades as “the orphan serotonin receptor” (89). Despite the limited information about this 5-HT receptor, the scientific community has established two subtypes expressed exclusively in the nervous system: 5-HT5A and 5-HT5B (90). These receptors may be involved in several processes, including memory (65) or pain (67). 5-HT6 receptors, such as 5-HT5 receptors, have also been poorly studied. Previous studies in mice have highlighted that it may be important in the GI physiology; however, its importance is not clear (91). At the CNS level, 5-HT6 is involved in mental disorders, such as psychosis, and in cognition and learning (70). Finally, the 5-HT7 receptor is mainly expressed in the brain but is also located in peripheral organs such as the GI tract (73). The 5-HT7 receptor is involved in circadian rhythm (78), and its dysfunction is important in the onset of depression (92). In the GI tract, 5-HT7 modulates SERT activity (75) and intestinal motility (77).
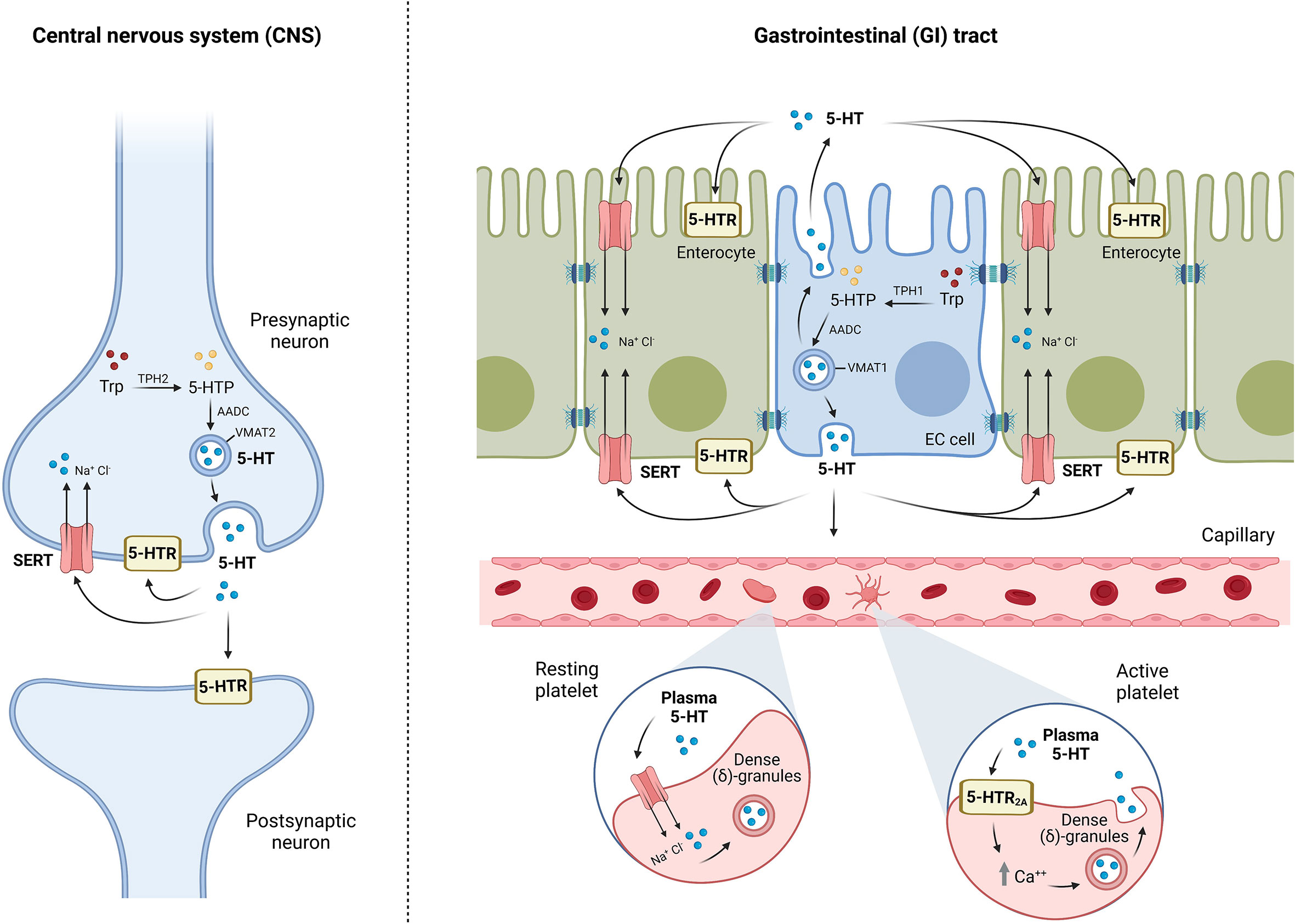
Figure 1 Schematic representation of brain and intestinal serotonergic systems: “ON/OFF” and signaling mechanisms. “ON” mechanism refers to the synthesis of 5-HT by enterochromaffin cells (EC) in the gut and serotonergic neurons both in the gut and in the central nervous system (CNS). Tryptophan (Trp) is catalyzed by the enzyme tryptophan hydroxylase (TPH), TPH1 in EC cells, and TPH2 in neurons, to synthesize 5-hydroxytryptophan (5-HTP), which is converted to 5-HT by aromatic amino acid decarboxylase (AADC). 5-HT is stored into vesicles through the vesicular monoamine transporter VMAT (VMAT1 in EC cells, and VMAT2 in neurons) and finally released into the extracellular space. 5-HT can bind to different serotonin receptors (5-HTR) or uptake into neurons, enterocytes, or platelets by the serotonin transporter (SERT), ending 5-HT effects (“OFF” mechanism). 5-HT is mostly stored in the dense (δ)-granules of platelets; however, the binding of plasma 5-HT to the platelet surface receptor 5-HT2A initiates the mobilization of intracellular calcium stores for platelet activation, which promotes platelet degranulation, resulting in 5-HT release. Serotonin exerts its effects by signaling mechanisms through the 5-HT receptors located in postsynaptic and presynaptic neurons at CNS and intestinal serotonergic neurons, and in different cell types of gastrointestinal (GI) tract, but also in other systems such as the cardiovascular or immune system.
3 Microbial Pattern Recognition Receptors: Effects on Serotonergic System
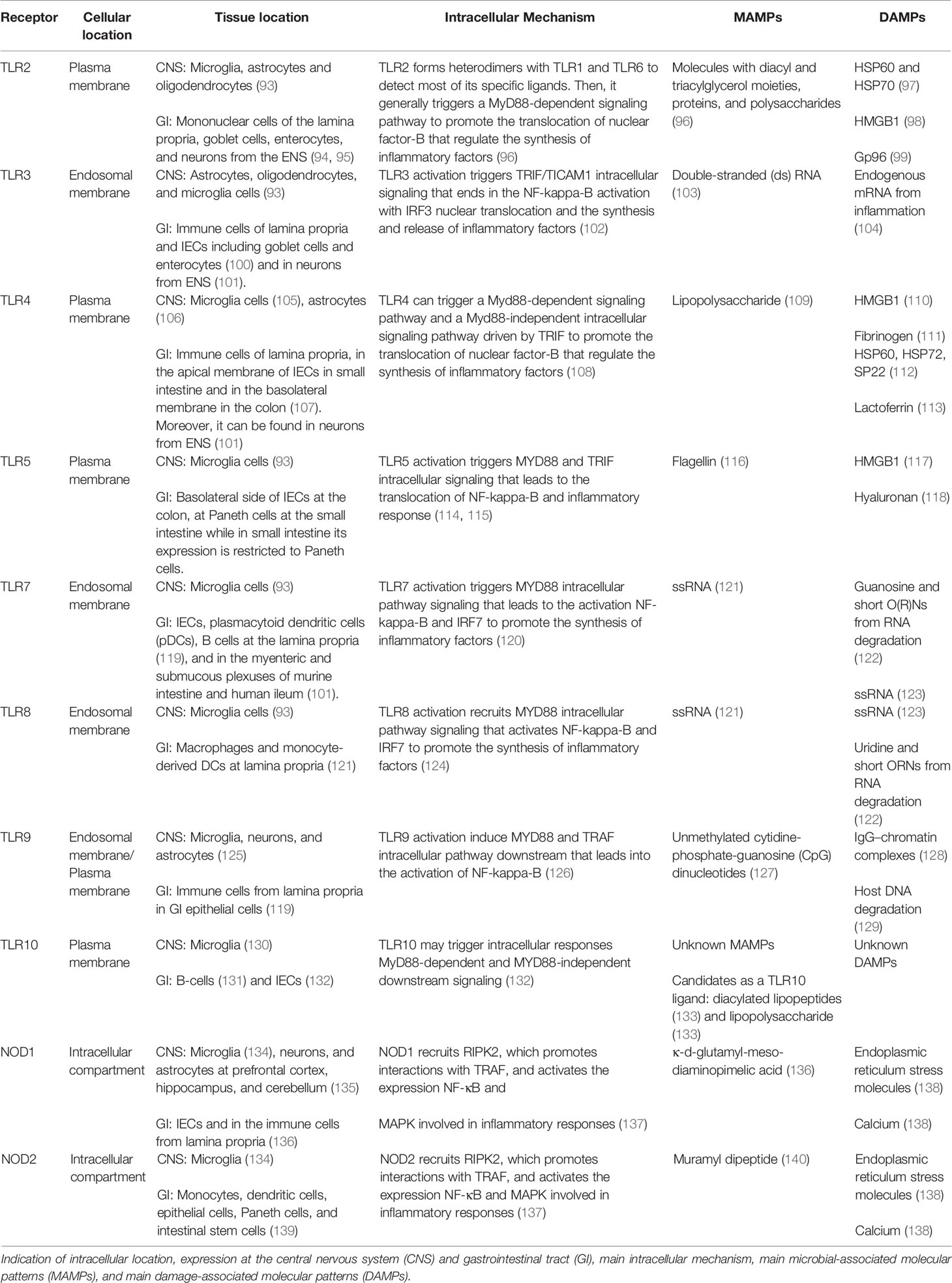
Defense mechanisms in the intestine are widely developed because external agents are in continuous contact with the intestinal epithelium. Innate immunity, throughout several detectors called pattern recognition receptors (PRRs), detects external factors, triggering either tolerant or defense responses to beneficial or pathogenic molecules, respectively. The most important and studied PRRs are microbial detectors: toll-like receptors (TLRs) and nucleotide oligomerization domain (NOD)-like receptors (NLRs) (Table 2). TLRs are transmembrane glycoproteins, whereas NLRs are cytosolic receptors. Until now, 11 different TLRs have been identified in humans (TLR1–TLR11) and expressed in both the endosomal membrane (TLR3, 7, 8, and 9) and cell membrane (TLR1, 2, 4, 5, 6, 9, and 10) (107). Regarding NLRs, 22 receptors have been discovered until now, which can be classified into five groups depending on their structure: NLRA, NLRB, NLRC, NLRP, and NLRX (141).
PRRs are widely expressed in immune cells (phagocytes, neutrophils, macrophages, or lymphocytes) and nonimmune ones, such as IECs in the GI tract, as well as microglia cells, neurons, or astrocytes in the CNS. PRRs trigger defense-related responses by the detection of specific microbial-associated molecular patterns from microorganisms (MAMPs) or damage-associated molecular patterns (DAMPs) from tissue injury, so we can consider the PRRs the caretakers of our body.
PRRs functioning in IECs are focused on the protection of the intestinal epithelium from potential harmful agents. Thus, and through PRR signaling, the intestine continuously develops the status of physiological inflammation to prevent possible damage and maintain intestinal homeostasis (142). In the brain, the main role of the PRRs is to detect dangerous molecules that can injure the tissue and trigger repair mechanisms. The brain is protected by the skull, the fluid cerebrospinal, the meninges, and the blood–brain barrier (BBB), which isolates the CNS from the general circulation. However, under pathological conditions, harmful microorganisms can breach the BBB and access the CNS, where the PRRs can trigger defense mechanisms to eliminate the pathogen and to repair the tissue (143).
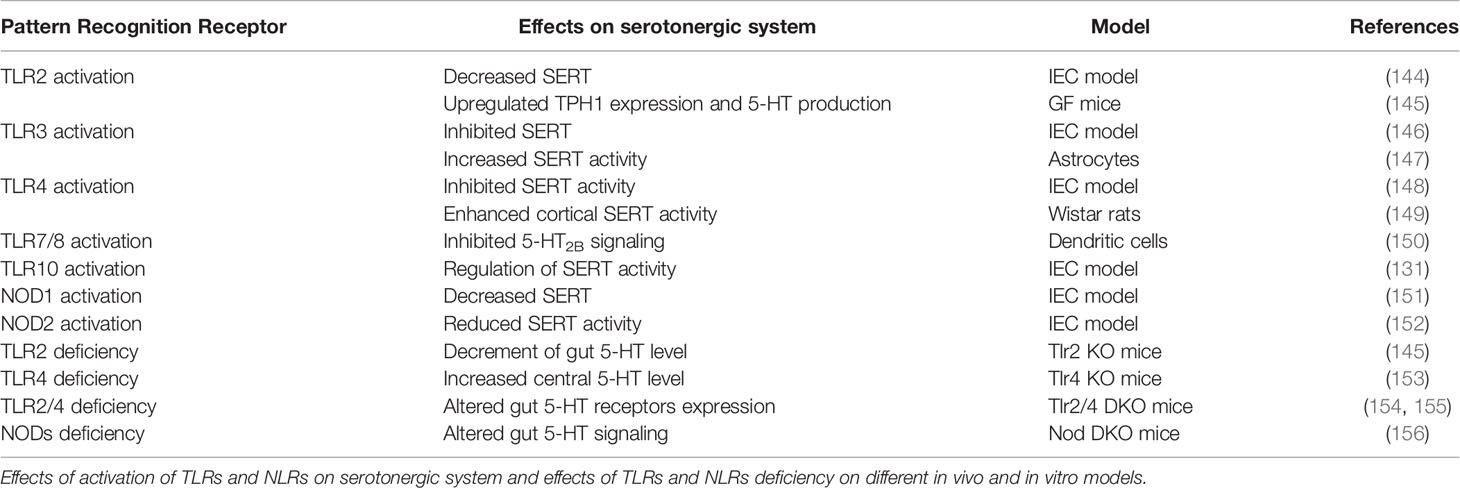
PRRs are widely expressed along the GI tract, which differs dramatically between the small intestine and colon (122). From all of them, TLR2, TLR3, TLR4, TLR5, and TLR9 seem to be critical in microbial detection and damage repair in the intestine. In the brain, the most studied PRRs, in relation with brain injury and pathogen infection, are TLR2, TLR3, TLR4, and TLR9. However, the scientific community does not discard the relevant importance of other TLRs in this location because they are expressed in several cells from the CNS (125). PRRs influence the serotonergic system activity and expression (Table 3).
3.1 Toll-Like Receptor 2
TLR2 is expressed in the GI tract in mononuclear cells of the lamina propria, goblet cells, and enterocytes (96), as well as neurons from the ENS (97). TLR2 is able to detect a broad range of MAMPs from several microorganisms, including Gram-positive bacteria through the formation of heterodimers with TLR1 (TLR2/1) and TLR6 (TLR2/6) (157), some fungi such as Candida albicans (158), viruses such as the hepatitis C virus (159), and some parasites such as Trypanosoma cruzi (160). At the CNS level, TLR2 is expressed in microglia, astrocytes, and oligodendrocytes (93). TLR2 in the brain mainly recognizes DAMPs as heat shock family proteins HSP60 and HSP70 (95) or high-mobility group box 1 proteins from dying tumor cells (HMGB1) (98), among others. However, the effect of TLR2 is not limited to immune responses. Previous results carried out in our laboratory have showed that TLR2 activation may modify the intestinal serotonergic system. TLR2 activation could decrease SERT activity due to a reduction in SERT protein expression, with cAMP/PKA and p38/MAPK intracellular pathways being implicated. Moreover, the expected increment of extracellular 5-HT will induce a negative feedback in TLR2 expression, supported by this cross-regulation between the TLR2 and serotonergic system (144). In fact, TLR2 and TLR4 activation may increase the production of IL-10 in the intestine (161), which in turn seems to modify SERT (162). In addition, TLR2 and TLR4 signaling seem to modulate GI motility mediated by 5-HT2, 5-HT3, 5-HT4, and 5-HT7 receptors (154). In line with these results, other researchers have found that TLR2 deficiency results in a decrement of gut 5-HT synthesis in vivo and that TLR2 activation upregulates the expression of TPH1 and 5-HT production in the gut (145). Serotonin-TLR2 relation is not limited to the GI tract, as previous data have highlighted that 5-HT2B receptor activation downregulates TLR2 expression and TLR3-induced proinflammatory factors in the brain (150). Selective 5-HT2A receptor antagonists activate glucocorticoid receptor nuclear translocation to upregulate TLR2 and TLR4 in response to microglial phagocytosis stimulation as a novel therapy in central pathologies such as Alzheimer’s disease (163).
3.2 Toll-Like Receptor 3
TLR3 is expressed in IECs, which mainly differentiates double-stranded RNA (dsRNA) from viruses. Surprisingly, TLR3 levels are age dependent because TLR3 expression increases after the suckling-to-weaning transition so as to give protection to the individuals against the virus as a rotavirus (164). In contrast, central TLR3 expression decreases during neurogenesis of the CNS in the embryo (165). TLR3 is also able to recognize endogenous mRNA as a DAMP from necrotic cells during intestinal inflammation (102). At the CNS level, TLR3 is expressed in a broad range of cells, including astrocytes, oligodendrocytes, and microglia cells (93), which is not surprising because viruses can easily reach the brain through other ways different from the BBB, such as neural pathways. Thus, TLR3 can detect dsRNA from the virus in the brain and trigger defense responses to protect the CNS against pathogens. Actually, TLR3 may protect the brain against some viruses such as the herpes simplex virus type 1 (HSV-1) (166). However, other microorganisms such as the Zika virus can activate TLR3 and induce an exacerbated inflammation and necrosis of the natural defenses of the brain, including the BBB (167). TLR3’s role in inflammatory responses may also be exacerbated by its potential pro-oxidant effect. In fact, TLR3 induces protein and lipid oxidation by reducing antioxidant enzymatic activity (168).
TLR3 activation is involved not only in inflammatory and oxidative damage–related responses but also in the modulation of the serotonergic system in the GI tract; TLR3 activation inhibits SERT activity and expression (146). In contrast, central TLR3 may have an opposite role because recent results have shown that TLR3 activation in a mice model with a brain infection increases SERT activity in astrocytes and therefore reduces extracellular 5-HT levels (147). In contrast to other TLRs, increased levels of 5-HT will not regulate TLR3 expression (146); meanwhile, other studies have reported that the activation of 5-HT2B receptors may reduce TLR3 expression (150).
3.3 Toll-Like Receptor 4
TLR4 is one of the most studied PRRs, and its expression can be found in the apical membrane of IECs in the small intestine and in the basolateral membrane in the colon (110). In the brain, TLR4 is an important PRR in the glia because several researchers have reported its expression (105); meanwhile, TLR4 is expressed less often in astrocytes (106) and may be absent in oligodendrocytes (93). TLR4 recognizes the lipopolysaccharide (LPS), which is the fundamental component of Gram-negative bacteria walls. In this process, the myeloid differentiation factor 2 (MD-2) protein is critical because several studies have found that MD-2 deletion yields to the lack of detection of LPS by TLR4 (169), suggesting that MD-2 retains TLR4 in the cellular surface to detect LPS due to changes in TLR4 glycosylation (170). Due to the broad microorganisms that TLR4 can identify through LPS detection, TLR4 has been defined as a gate keeper of microbial homeostasis in the intestine, where it is involved in several defense mechanisms, including the zoonotic Campylobacter (171), Helicobacter pylori (172), or Salmonella (173). TLR4 could also have a regulator role in the serotonergic system. TLR4 modulates contractile response in the intestine and is mediated by serotonin receptors (154). TLR4 activation inhibits SERT activity through post-transcriptional mechanisms, leading to an increase in extracellular 5-HT (148). In addition, melatonin, a molecule linked with 5-HT synthesis, may modify intestinal microbiota composition through TLR4 signaling (174). At the CNS level, TLR4 participates in the detection of pathogens that cause meningitis, such as Neisseria meningitidis (175), where some DAMPs linked to brain damage mediate TLR4 signaling (176). Interestingly, recent results have pointed out that microbiota and TLR4 signaling are key players in Parkinson’s disease, one of the most important degenerative brain pathologies (177). In this context, previous studies have shown that the lack of TLR4 in the CNS leads to an increase in the central 5-HT level, suggesting the critical regulatory role of TLR4, not only in the GI tract but also in the central serotonergic system (153).
3.4 Toll-Like Receptor 5
TLR5 seems to be one of the most important TLRs in the GI tract because its expression and activity has been reported in all intestinal segments (122). In this context, TLR5 is expressed in the basolateral side of IECs from the colon, while in the small intestine, its expression is restricted to Paneth cells. TLR5 recognizes flagellin, a component that enables the motility of several bacteria. Several studies have indicated that flagellin origin is determinant in the defense response against bacteria because flagellin from pathogenic Salmonella typhimurium triggers a more exacerbated immune response than does flagellin from the nonpathogenic bacteria E. coli (178). In this context, TLR5 is a critical gatekeeper because it may control the intestinal microbiota composition by maintaining a physiological low grade of inflammation in the GI tract (179). Previous studies have extensively described TLR5 expression in microglia cells, where its function may be involved in the inflammatory diseases in the brain comprising bacteria that cause meningitis (180). However, TLR5 is not only involved in bacterial infection but can also be related with depression. Previous works have described how TLR3, TLR4, TLR5, TLR7, TLR8, and TLR9 mRNA expressions in peripheral blood mononuclear cells seem to be increased in patients with depression. The improvement of these patients through the use of selective serotonin reuptake inhibitors (SSRIs) indicates the implication not only of TLR5 but also other PRRs in the modulation of the serotonergic system in brain disorders (181).
3.5 Toll-Like Receptor 7 and Toll-Like Receptor 8
TLR7 and TLR8 are closely related PRRs expressed in endosomal membranes that can detect single-stranded RNA (ssRNA) (120). Previous works have described the lack of TLR7 expression in IECs, being mainly expressed in plasmacytoid dendritic cells (pDCs), in B cells at the lamina propria (122), and in the myenteric and submucous plexuses of murine intestine and human ileum (104). Meanwhile, TLR8 can be found in macrophages and monocyte-derived DCs (120). In both cases, it seems that TLR7 and TLR8 could have more importance in other organs, such as the respiratory system, than in the GI tract by recognizing respiratory viruses and triggering inflammatory responses (182). At the CNS level, TLR7 and TLR8 are mainly expressed in microglia cells. TLR7 acts by regulating the inflammation (183) and modulation of TLR9 expression (184); meanwhile, TLR8 is related with the attenuation of the outgrowth of neurons and the induction of apoptosis (185). In the GI tract, 5-HT can act by regulating TLR7 in DC through the 5-HT2B receptor (150). Moreover, SSRIs seem to decrease the expression of both TLR7 and TLR8 in the CNS (181).
3.6 Toll-Like Receptor 9
TLR9 is included, together with TLR3, TLR7, and TLR8, in the group of TLRs that is classically expressed in membranes of intracellular organelles such as the endoplasmic reticulum, endosomes, and lysosomes. However, TLR9 can also be detected in endosomal locations (186). In the GI tract, TLR9 can be expressed in the apical and basolateral membrane of IECs to control homeostasis by means of various intracellular signaling (187). The intestinal map of TLRs describes TLR9 expression mainly in the lamina propria, and at low levels in GI epithelial cells (122). TLR9 recognizes unmethylated DNA found generally in microorganisms such as viruses and bacteria (127). However, TLR9 can also detect host DNA in aberrant locations, such as a DAMP of tissue damage (129), and it participates in the protection against GI damage and in GI repair (188). Moreover, TLR9 seems to act as an inhibitor of antimicrobial peptides in the intestine to avoid the colonization of pathogens (189). Because pathogen-free mice display a higher TLR9 expression in the intestine than germ-free mice do, it has been suggested that beneficial bacteria could modulate TLR9 expression in the GI tract (190). At the CNS level, TLR9 is expressed in microglia, neurons, and astrocytes (125), mediates immune responses related with brain infections, such as the herpes simplex virus (191), and attenuates brain injury (192). Little research has been carried out in the influence of TLR9 over the serotonergic system, and only a few works have indicated that SSRIs may modulate TLR9 mRNA expression in the peripheral blood mononuclear cells of depression patients (181) and will be implicated in the tryptophan catabolism (i.e., the main 5-HT resource) (193). In fact, preliminary data from our research group indicate that TLR9 could affect SERT activity and expression in an IECs model (194).
3.7 Toll-Like Receptor 10
TLR10 is the only PRR without known ligand specificity and biological function. Human TLR10 is encoded on chromosome 4 within the TLR2 gene cluster, together with TLR1, TLR2, and TLR6, suggesting a possible heterodimer TLR2/TLR10 (195). It has been described that TLR10 could act as an inhibitory receptor that essentially controls TLR2-driven signals (196). TLR10 is predominantly expressed in tissues rich in immune cells, such as the spleen, lymph node, thymus, tonsil, and lung (197). Genetic variations found in the TLR10 gene may cause a shift in the levels of pro- and anti-inflammatory responses and enhance the susceptibility to autoimmune diseases, cancers, and infections at the GI tract (198–200). Recently, TLR10 has been described in multiple mucosal sites, such as the small intestine, fallopian tubes, eyes, or stomach (198, 201, 202), suggesting a key role as a pathogen sensor in the mucosa. In the GI tract, TLR10 seems to be a chief component in the immune response to Listeria monocytogenes in IECs. In this context, previous studies have shown that L. monocytogenes affects SERT activity mediated by TLR10, which triggers the activation of a MyD88-dependent intracellular pathway (which may increase 5-HT uptake), and by a MyD88-independent downstream signaling (which may decrease 5-HT uptake), proving a deep involvement of TLRs in the serotonergic mechanism (131). At the CNS level, TLR10 could be critical for macrophage activity. In fact, microglial cells express TLR10, and this receptor inhibits M1 macrophage cytokines but promotes M2 cytokines, indicating that TLR10 may have a protective role in the brain (130).
3.8 NOD-Like Receptors
Like the TLRs, the NLRs are PRRs that detect both DAMPs and MAMPs triggering immune-related responses to protect the host. However, NLRs differentiate from TLRs with regard to the quality of being cytosolic receptors. NLRs can be classified into two big groups: the NLRC subfamily that encompasses the most popular, including NOD1, NOD2, and NLRC4, and the NLRP subfamily that includes up to 14 PRRs (203).
3.8.1 NOD1
NOD1 is an intracellular PRR widely expressed in the organism with special relevance in the IECs and in the immune cells from lamina propria in the GI tract, where this PRR detects κ-d-glutamyl-meso-diaminopimelic acid (iE-DAP) from bacterial peptidoglycan, which can be found in most of the bacterial wall (136). NOD1 has been involved in the protection of the GI tract against pathogens such as S. typhimurium (204), Citrobacter rodentium (205), or H. pylori (206), among others. Previous works have described the expression of NOD1 in the CNS but at a lower level compared with TLRs (207), where one of the main functions is the protection against bacterial infections (208). Interestingly, NOD1 and NOD2 defense effects are only related with immunity because an elegant study has demonstrated that the lack of both receptors in mice leads to signs of stress-induced anxiety, cognitive impairment, and depression, together with increased GI permeability and altered serotonin signaling in the gut, suggesting that NOD1 and NOD2 are novel therapeutic targets for gut–brain axis disorders (156). Supporting these results, NOD1 activation may decrease SERT activity in IECs due to the diminishment of SERT expression. In turn, 5-HT levels seem also to upregulate NOD1 expression. However, NOD1 could also regulate other PRR expression such as TLR2 and TLR4 (151).
3.8.2 NOD2
NOD2 is one of the most studied NLRs in the GI tract because polymorphisms in the gene that encodes NOD2 have been strongly associated with IBDs (209) and colorectal cancer (210). NOD2 is an intracellular PRR expressed in all IECs in the GI tract, which explains its implication in the protection of the intestine against the mentioned pathologies (211). NOD2 detects the bacterial peptidoglycan named muramyl dipeptide (MDP), which allows the identification of several pathogens, including Yersinia (212), Campylobacter (213), and Listeria (214). At the CNS level, NOD2 seems to play a similar role by detecting pathogens, triggering immune-related responses, and protecting the host (215). Like NOD1, NOD2 would be an important PRR in the gut–brain axis, especially because of its relation with the serotonergic system in both the CNS and the GI tract (156). In this sense, bacterial activation of NOD2 may decrease SERT activity and expression, thus leading to an increase in extracellular serotonin, and then serve as a negative feedback modulation of NOD2. In addition, NOD2 not only modulates the serotonergic system directly but also through its interdependence with TLR2 and TLR4 (152). In fact, the increase of extracellular 5-HT by NOD2 is not only for the downregulation of SERT but also for the increase of enterochromaffin cells that are responsible for 90% of the total 5-HT (216).
4 Intestinal Microbiota: Direct Effects on Serotonergic System
In recent years, intestinal microbiota involvement has gained high importance in numerous pathologies, including gut–brain disorders such as IBDs (217), depression (218), or Alzheimer’s disease (219). In this context, several studies have indicated that 5-HT and serotonergic system modulation by intestinal microbiota are critical in the maintenance of the gut–brain axis (220–222). Microbiota can produce tryptophan and tryptamine, directly affecting central 5-HT production (223). GF mice display a reduction in anxiety-like behavior compared with specific pathogen-free mice, showing a decreased expression of serotonin receptor 1A in the hippocampus (224). In the GI tract, microbiota increase the production of intestinal 5-HT by increasing TPH1 expression (225), and, more interestingly, microbiota can also synthesize 5-HT on their own (226). In agreement with this study, the alteration of microbiota composition and diversity seems to reduce host serotonin levels, increase tryptamine levels, and disrupt the GI immune system (227). However, it seems that microbiota not only influence 5-HT synthesis and SERT expression but also modulate the expression of some 5-HT receptors (228).
Some pathogenic bacteria such as E. coli can downregulate the activity and expression of SERT in the intestine (229), and an increase of extracellular 5-HT may induce an adherent-invasive E. coli colonization (230). Moreover, it has been described that E. coli can produce tryptophan, which will affect 5-HT production (231). Similarly, some beneficial bacteria such as Lactobacillus seem to degrade tryptophan, affecting central and intestinal 5-HT production (232). Several studies have shown that germ-free animals have a lower number of enterochromaffin cells compared to those with a standard microbiota (233). Specific pathogen-free mice display lower 5-HT levels (234), concluding that microbiota can regulate host 5-HT production not only at the intestinal level but also in the CNS (235). Apart from that, intestinal microbiota can produce tryptamine, the precursor of 5-HT, independently of the host (226), which introduces new strategies as to how microbiota will not only modify the intrinsic serotonergic system but also externally modify the levels of 5-HT in the host.
Moreover, intestinal microbiota can modify serotonergic systems by means of their metabolites and affect behavior through the modulation of 5-HT signaling (236). In this context, some metabolites, including the short-chain fatty acids (SCFAs), are a key component in this modulation and directly affect the gut–brain axis (237). SCFAs are metabolites from dietary fiber fermentation. They are characterized by having less than six carbon atoms, so they can easily cross membranes, including the BBB. Although studies on the physiological concentrations of SCFAs in the brain are scarce, the three main SCFAs—acetate, propionate, and butyrate—have been detected in cerebrospinal fluid (Human Metabolome Database. Available online at: http://www.hmdb.ca/). In fact, SCFAs could have a critical role in the maintenance and integrity of the BBB (238). SCFAs seem to regulate the expression levels of TPH1 in the intestine (239). In our lab, we have described that SCFAs can regulate intestinal SERT activity and expression (240). Similarly, other bacterial metabolites such as L-lactate seem to control the expression of 5-HT receptors 1B, 1D, and 4 in the CNS (241). In fact, there is a growing interest in the involvement of microbiota metabolites in the modulation of multiple neurochemical pathways through the highly interconnected gut–brain axis, which could be open novel approaches for gut–brain axis disorders (242).
5 Conclusions and Future Perspectives
The serotonergic system is the chief mechanism in the intestine that controls the GI tract (243) and the CNS physiology (244), with serotonin being one of the most important neurotransmitters in these organs. In addition, 5-HT modulates not only the GI tract and CNS functions, but also their interconnection (i.e., the gut–brain axis). In this context, numerous researchers have claimed that either 5-HT or tryptophan (main 5-HT resource) could be a key factor in gut–brain axis regulation (245) and that its imbalance could trigger pathologies in any of these organs (246). Interestingly, intestinal microbiota participate directly in 5-HT production, and by means of PRRs activation, microbiota can also affect SERT and regulate 5-HT levels. Moreover, changes in the extracellular 5-HT level may affect PRRs expression in a feedback regulation in order to maintain homeostasis (Figure 2).
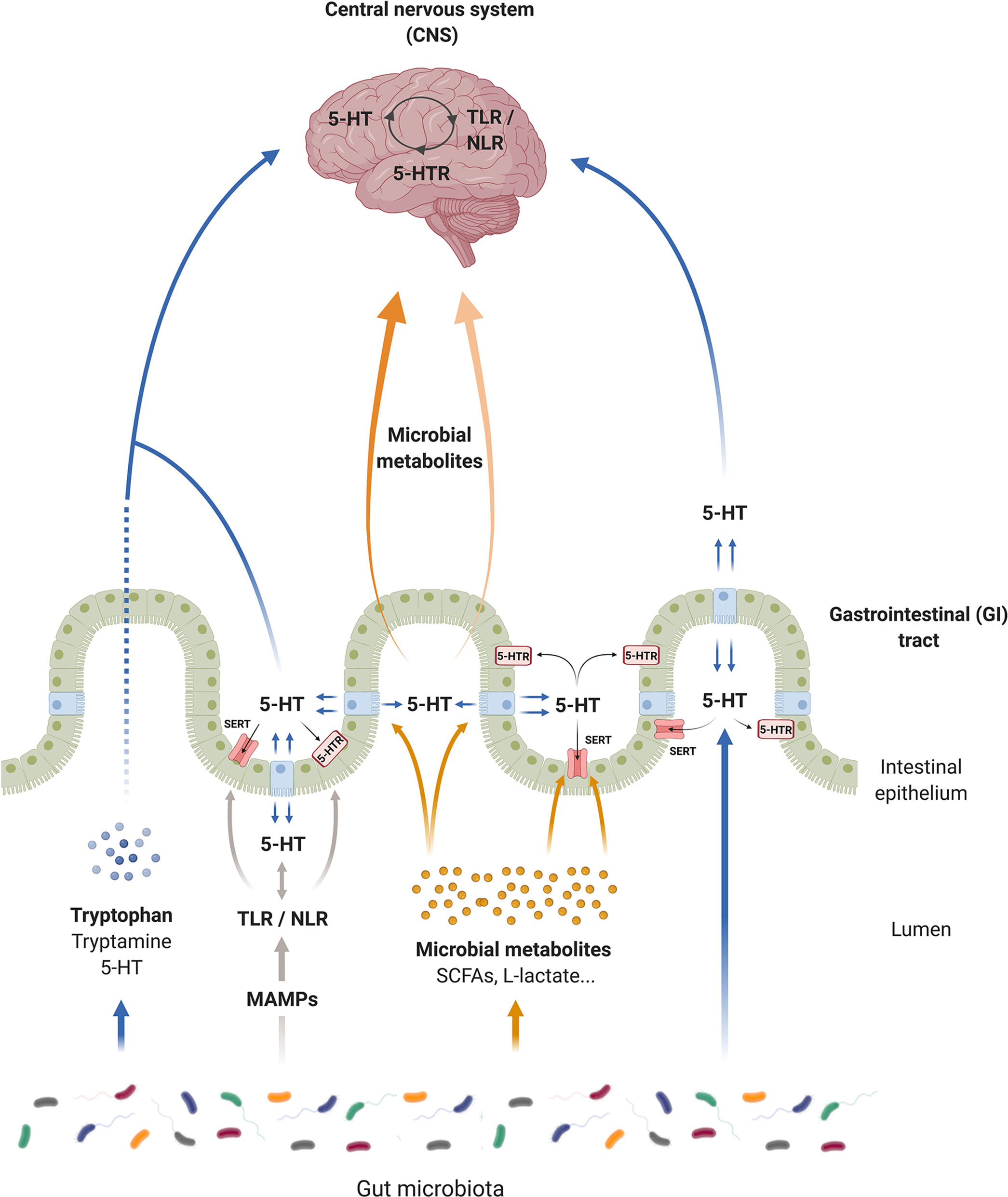
Figure 2 Serotonin (5-HT) communication pathways of the microbiota–gut–brain axis. Serotonin can modulate gastrointestinal (GI) and central nervous system (CNS) functions and is a key network for the gut–brain axis. Microorganisms produce tryptophan, and degrade tryptophan, affecting the central and intestinal 5-HT production. Intestinal microbiota modulate the synthesis of 5-HT and produce 5-HT independently of the host. Microbial associated molecular patterns from microorganisms (MAMPs) through toll-like receptors (TLRs) and nucleotide oligomerization domain (NOD)-like receptors (NLRs) affect directly the serotonergic system. TLR/NLR signaling seems to modulate the activity and the expression of serotonin transporter (SERT) and serotonin receptors (5-HTRs), as well as the 5-HT synthesis in the GI tract. However, this interconnection between TLRs/NLRs and serotonergic system exists in the CNS. In a feedback regulation, 5-HT affects pattern recognition receptor (PRR) expression. In addition, microbial metabolites, such as short chain fatty acids (SCFAs), can promote 5-HT synthesis by enterochromaffin (EC) cells and regulate SERT activity and expression. In the same way, these metabolites can migrate into the bloodstream to reach the brain, and some of them such as L-acetate can modulate the nervous serotonergic system, controlling the expression of 5-HT receptors.
Interestingly, various pathologies within the gut–brain axis that are apparently unrelated seem to have three common aspects: changes in intestinal microbiota, alterations of the intestinal serotonergic system, and dysfunction of the PRRs (Table 4). In the GI tract, IBDs, including CD and UC, are characterized by changes in the microbiota (345), alterations in the serotonergic system (346, 347), and dysfunction of the innate immune system, including TLRs (109) and NLRs (348). In recent years, novel IBD therapy has focused on treatment to reestablish these three components. Classical control of the intestinal microbiota has focused on antibiotics treatment. However, long-term use of antibiotics in IBDs seems not to resolve the inflammation and is associated with more harm than benefits (349). Fecal microbial transplantation is one of the most promising novel treatments in IBDs (350), together with the use of probiotics (351). In the last century, the use of anti-TNF has also been the most important drug intervention in IBD patients (352). However, this therapy may be insufficient, and novel studies have indicated that more treatments addressing innate immunity should be carried out. In this context, several studies have indicated that TLR (353) and NLR (354) modulation may help in the treatment of these chronic pathologies. Finally, serotonergic system modulation has been one of the main targets for IBD therapeutics in recent years. In this context, the inhibition of mucosal serotonin (355) or the use of inhibitors for SERT (356) may help in IBDs and thus be considered as a novel therapy for IBDs.
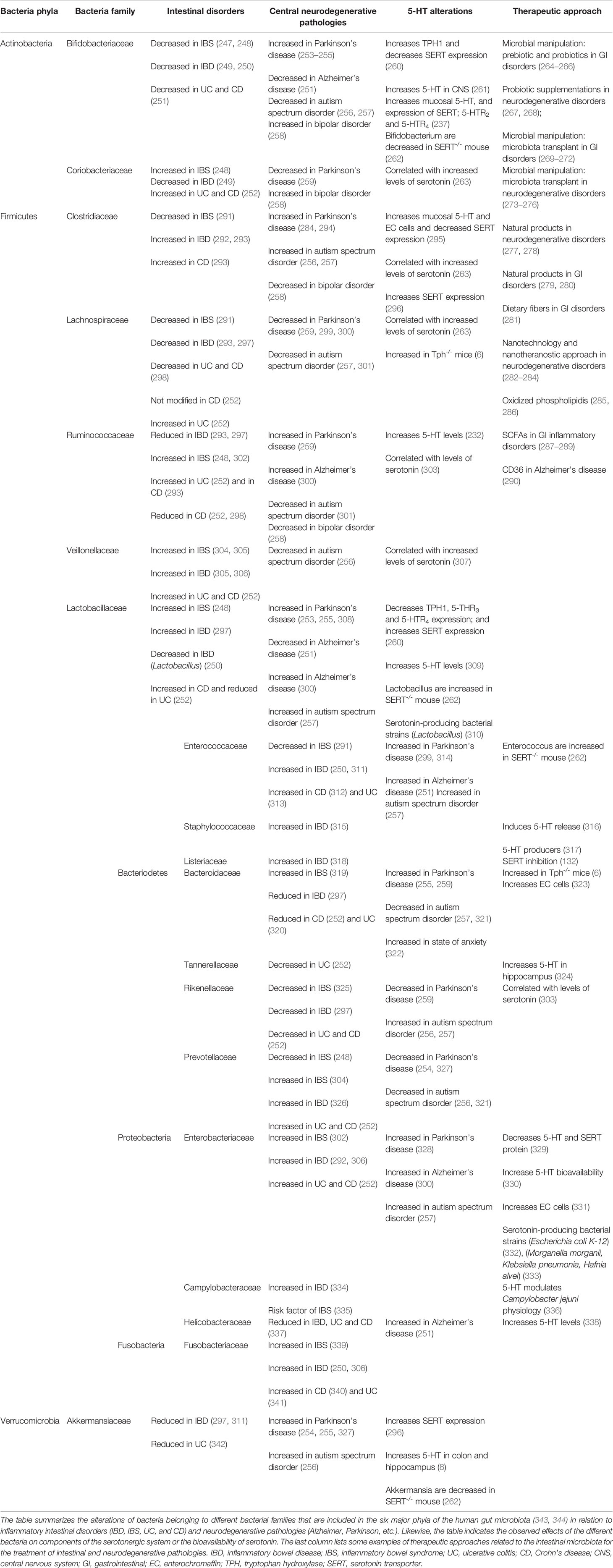
Table 4 Gut microbiota in intestinal disorders (IBD and IBS) and neurodegenerative pathologies and their interaction with serotonergic system.
Irritable bowel syndrome (IBS) has been described as a gut–brain disorder, where the serotonergic system may be altered in both the intestine and the CNS (357). Interestingly, intestinal microbiota (358), as well as SCFAs and 5-HT, are altered in IBS patients (359). In addition, TLRs and NLRs play a chief role in the pathogenesis of IBS. In fact, several studies have indicated that some PRRs serve as predictive markers for the disease (360) because their expression is increased in the mucosa from IBS patients (361). Thus, it is not surprising that gut–brain axis modulation in IBS seems to be the most effective therapy in this pathology. Previous studies have shown that SERT regulation (362, 363) and the synthesis of 5-HT (364) may be important in the treatment of IBS. Moreover, serotonin therapy efficiency in IBS is improved through the modulation of microbiota (365, 366), and previous studies have suggested the immunomodulation of PRRs in this GI disease (367).
Surprisingly, disorders in the CNS may share the same alterations as the GI pathologies. In this context, serotonergic system alteration may be involved not only in depression and anxiety (368) but also in Parkinson’s disease (369), multiple sclerosis (370), amyotrophic lateral sclerosis (370), and autism spectrum disorder (371), among others. In fact, conventional treatment for CNS disorders, especially depression, has focused on the modulation of the serotonergic system by means of SSRIs (372). Important findings have been published in the last years regarding the changes of intestinal microbiota in the CNS pathologies. Recent data have shown that intestinal microbiota may be a critical susceptibility factor in the development of neurological disorders such as Alzheimer’s disease, autism spectrum disorder, multiple sclerosis, Parkinson’s disease (373), and depression in particular, where the modulation of the intestinal serotonin by the microbiota seems to be an important trigger (138, 374). In fact, certain bacteria families modulate tryptophan levels in blood plasma that can cross the BBB and thus influence the central serotonergic system (375). Within this context, novel therapies of brain pathologies, such as Alzheimer’s disease, are focused on the modulation of intestinal microbiota to prevent and ameliorate the development of the pathology (376). These new studies have shown that the balance of the gut–brain axis is critical in preventing the development of GI and brain disorders mediated by 5-HT (377). Innate immune receptors, including TLRs and NLRs, could also be a key component in the correct function of the microbiota–gut–brain axis. Previous works have shown that TLR modulation by means of intestinal microbiota may be a critical factor in the development of brain disorders such as Parkinson’s disease (177, 378); in addition, NLRs may be involved in CNS inflammation and neurodegenerative diseases (379). New therapeutics have shown that the use of antidepressants may improve the negative regulation of PRRs in some CNS disorders such as depression (380), especially for TLR4 (381).
Based on the numerous studies focusing on the gut–brain axis, it is clear that the balance of this bidirectional communication may be important in the prevention of GI and CNS disorders, where the intermodulation of the microbiome, serotonergic system, and innate immunity is critical in maintaining homeostasis. However, more studies are needed to understand the implication of these elements, as well as their modulation as novel therapeutic targets, for the GI and CNS pathologies.
Author’s Note
In memoriam: This paper is dedicated to the memory of Professor Ana Isabel Alcalde, a brilliant and enthusiastic scientist, professor, and colleague, as well as our director and mentor, who dedicated her last 20 years to the study of the serotonergic system.
Author Contributions
Conceptualization: ElL and BB. Investigation: ElL and BB. Writing—Original Draft Preparation: ElL. Writing—Review and Editing: JM and EvL. Supervision: EvL. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by grants from the Foundation for the Study of Inflammatory Bowel Diseases in Aragón (ARAINF 2012/0567) and the Aragón Regional Government (A20_20 R).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Figures 1 and 2 were created with BioRender.com. We also acknowledge Papercheck Proofreading and Editing Services.
References
1. Obermajer T, Grabnar I, Benedik E, Tusar T, Robic Pikel T, Fidler Mis N, et al. Microbes in Infant Gut Development: Placing Abundance Within Environmental, Clinical and Growth Parameters. Sci Rep (2017) 7(1):11230. doi: 10.1038/s41598-017-10244-x
2. Al-Asmakh M, Anuar F, Zadjali F, Rafter J, Pettersson S. Gut Microbial Communities Modulating Brain Development and Function. Gut Microbes (2012) 3(4):366–73. doi: 10.4161/gmic.21287
3. Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Dejardin F, et al. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity (2019) 50(5):1276–88.e5. doi: 10.1016/j.immuni.2019.02.014
4. Carabotti M, Scirocco A, Maselli MA, Severi C. The Gut-Brain Axis: Interactions Between Enteric Microbiota, Central and Enteric Nervous Systems. Ann Gastroenterol (2015) 28(2):203–9.
5. Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut Microbiota in the Pathogenesis of Inflammatory Bowel Disease. Clin J Gastroenterol (2018) 11(1):1–10. doi: 10.1007/s12328-017-0813-5
6. Kwon YH, Wang H, Denou E, Ghia JE, Rossi L, Fontes ME, et al. Modulation of Gut Microbiota Composition by Serotonin Signaling Influences Intestinal Immune Response and Susceptibility to Colitis. Cell Mol Gastroenterol Hepatol (2019) 7(4):709–28. doi: 10.1016/j.jcmgh.2019.01.004
7. Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, et al. Normal Gut Microbiota Modulates Brain Development and Behavior. Proc Natl Acad Sci USA (2011) 108p(7):3047–52. doi: 10.1073/pnas.1010529108
8. Yaghoubfar R, Behrouzi A, Ashrafian F, Shahryari A, Moradi HR, Choopani S, et al. Modulation of Serotonin Signaling/Metabolism by Akkermansia Muciniphila and Its Extracellular Vesicles Through the Gut-Brain Axis in Mice. Sci Rep (2020) 10(1):22119. doi: 10.1038/s41598-020-79171-8
9. Zhuang Z, Yang R, Wang W, Qi L, Huang T. Associations Between Gut Microbiota and Alzheimer’s Disease, Major Depressive Disorder, and Schizophrenia. J Neuroinflamm (2020) 17(1):288. doi: 10.1186/s12974-020-01961-8
10. Erspamer V. Pharmakologische Studien Über Enteramin. Naunyn Schmiedebergs Arch Exp Pathol Pharmakol (1940) 196(2):343–65. doi: 10.1007/BF01861121
11. Twarog BM. Responses of a Molluscan Smooth Muscle to Acetylcholine and 5-Hydroxytryptamine. J Cell Comp Physiol (1954) 44(1):141–63. doi: 10.1002/jcp.1030440112
12. Lv J, Liu F. The Role of Serotonin Beyond the Central Nervous System During Embryogenesis. Front Cell Neurosci (2017) 11:74. doi: 10.3389/fncel.2017.00074
13. Walther DJ, Bader M. A Unique Central Tryptophan Hydroxylase Isoform. Biochem Pharmacol (2003) 66(9):1673–80. doi: 10.1016/s0006-2952(03)00556-2
14. Rudnick G. “SERT, Serotonin Transporter”, in xPharm: The Comprehensive Pharmacology Reference, eds. Enna SJ, Bylund DB. Ed. Elsevier; Amsterdam, The Netherlands: (2007) pp. 1–5. doi: 10.1016/B978-008055232-3.60442-8
15. Murphy DL, Lerner A, Rudnick G, Lesch KP. Serotonin Transporter: Gene, Genetic Disorders, and Pharmacogenetics. Mol Interv (2004) 4(2):109–23. doi: 10.1124/mi.4.2.8
16. Burnet PWJ, Eastwood SL, Lacey K, Harrison PJ. The Distribution of 5-HT1A and 5-HT2A Receptor mRNA in Human Brain. Brain Res (1995) 676(1):157–68. doi: 10.1016/0006-8993(95)00104-X
17. Pithadia AB, Jain SM. 5-Hydroxytryptamine Receptor Subtypes and Their Modulators With Therapeutic Potentials. J Clin Med Res (2009) 1(2):72–80. doi: 10.4021/jocmr2009.05.1237
18. Hoyer D. “5-HT-1 Receptors”, in xPharm: The Comprehensive Pharmacology Reference, eds. Enna SJ, Bylund DB, Ed. Elsevier; Amsterdam, The Netherlands: (2007) pp. 1–5. doi: 10.1016/B978-008055232-3.60123-0
19. Miszkiel J, Filip M, Przegalinski E. Role of Serotonin 5-HT1B Receptors in Psychostimulant Addiction. Pharmacol Rep (2011) 63(6):1310–5. doi: 10.1016/s1734-1140(11)70695-8
20. Albert PR, Vahid-Ansari F. The 5-HT1A Receptor: Signaling to Behavior. Biochimie (2019) 161:34–45. doi: 10.1016/j.biochi.2018.10.015
21. O’Neill MF, Conway MW. Role of 5-HT(1A) and 5-HT(1B) Receptors in the Mediation of Behavior in the Forced Swim Test in Mice. Neuropsychopharmacology (2001) 24(4):391–8. doi: 10.1016/S0893-133X(00)00196-2
22. Kirchgessner AL, Liu MT, Raymond JR, Gershon MD. Identification of Cells That Express 5-Hydroxytryptamine1a Receptors in the Nervous Systems of the Bowel and Pancreas. J Comp Neurol (1996) 364(3):439–55. doi: 10.1002/(SICI)1096-9861(19960115)364:3<439::AID-CNE5>3.0.CO;2-5
23. Tack J, Sarnelli G. Serotonergic Modulation of Visceral Sensation: Upper Gastrointestinal Tract. Gut (2002) 51(Suppl 1):i77–80. doi: 10.1136/gut.51.suppl_1.i77
24. Wauson SE, Sarkodie K, Schuette LM, Currie PJ. Midbrain Raphe 5-HT1A Receptor Activation Alters the Effects of Ghrelin on Appetite and Performance in the Elevated Plus Maze. J Psychopharmacol (2015) 29(7):836–44. doi: 10.1177/0269881115581981
25. Kobilka BK, Frielle T, Collins S, Yang-Feng T, Kobilka TS, Francke U, et al. An Intronless Gene Encoding a Potential Member of the Family of Receptors Coupled to Guanine Nucleotide Regulatory Proteins. Nature (1987) 329(6134):75–9. doi: 10.1038/329075a0
26. Aune TM, McGrath KM, Sarr T, Bombara MP, Kelley KA. Expression of 5HT1a Receptors on Activated Human T Cells. Regulation of Cyclic AMP Levels and T Cell Proliferation by 5-Hydroxytryptamine. J Immunol (1993) 151(3):1175–83.
27. Wang GD, Wang XY, Zou F, Qu M, Liu S, Fei G, et al. Mast Cell Expression of the Serotonin1a Receptor in Guinea Pig and Human Intestine. Am J Physiol Gastrointest Liver Physiol (2013) 304(10):G855–63. doi: 10.1152/ajpgi.00421.2012
28. Ogren SO, Eriksson TM, Elvander-Tottie E, D’Addario C, Ekstrom JC, Svenningsson P, et al. The Role of 5-HT(1A) Receptors in Learning and Memory. Behav Brain Res (2008) 195(1):54–77. doi: 10.1016/j.bbr.2008.02.023
29. Liu N, Sun S, Wang P, Sun Y, Hu Q, Wang X. The Mechanism of Secretion and Metabolism of Gut-Derived 5-Hydroxytryptamine. Int J Mol Sci (2021) 22(15):7931. doi: 10.3390/ijms22157931
30. Boutrel B, Monaca C, Hen R, Hamon M, Adrien J. Involvement of 5-HT1A Receptors in Homeostatic and Stress-Induced Adaptive Regulations of Paradoxical Sleep: Studies in 5-HT1A Knock-Out Mice. J Neurosci (2002) 22(11):4686–92. doi: 10.1523/JNEUROSCI.22-11-04686.2002
31. Pompeiano M, Palacios JM, Mengod G. Distribution of the Serotonin 5-HT2 Receptor Family mRNAs: Comparison Between 5-HT2A and 5-HT2C Receptors. Brain Res Mol Brain Res (1994) 23(1-2):163–78. doi: 10.1016/0169-328x(94)90223-2
32. Fiorica-Howells E, Hen R, Gingrich J, Li Z, Gershon MD. 5-HT(2A) Receptors: Location and Functional Analysis in Intestines of Wild-Type and 5-HT(2A) Knockout Mice. Am J Physiol Gastrointest Liver Physiol (2002) 282(5):G877–93. doi: 10.1152/ajpgi.00435.2001
33. Doly S, Valjent E, Setola V, Callebert J, Herve D, Launay JM, et al. Serotonin 5-HT2B Receptors Are Required for 3,4-Methylenedioxymethamphetamine-Induced Hyperlocomotion and 5-HT Release In Vivo and In Vitro. J Neurosci (2008) 28(11):2933–40. doi: 10.1523/JNEUROSCI.5723-07.2008
34. Imada-Shirakata Y, Kotera T, Ueda S, Okuma M. Serotonin Activates Electrolyte Transport via 5-HT2A Receptor in Rat Colonic Crypt Cells. Biochem Biophys Res Commun (1997) 230(2):437–41. doi: 10.1006/bbrc.1996.5921
35. Meneses A. 5-HT System and Cognition. Neurosci Biobehav Rev (1999) 23(8):1111–25. doi: 10.1016/s0149-7634(99)00067-6
36. Borman RA, Tilford NS, Harmer DW, Day N, Ellis ES, Sheldrick RL, et al. 5-HT(2B) Receptors Play a Key Role in Mediating the Excitatory Effects of 5-HT in Human Colon In Vitro. Br J Pharmacol (2002) 135(5):1144–51. doi: 10.1038/sj.bjp.0704571
37. Fiorica-Howells E, Maroteaux L, Gershon MD. Serotonin and the 5-HT(2B) Receptor in the Development of Enteric Neurons. J Neurosci (2000) 20(1):294–305. doi: 10.1523/JNEUROSCI.20-01-00294.2000
38. Hensler JG. “Chapter 15 – Serotonin” in Basic Neurochemistry (Eighth Edition), eds. Brady ST, Siegel GJ, Albers RW, Price DL. Ed. Elsevier Academic Press: Amsterdam, The Netherlands: (2012) pp. 300–22. doi: 10.1016/B978-0-12-374947-5.00015-8
39. Foguet M, Hoyer D, Pardo LA, Parekh A, Kluxen FW, Kalkman HO, et al. Cloning and Functional Characterization of the Rat Stomach Fundus Serotonin Receptor. EMBO J (1992) 11(9):3481–7. doi: 10.1002/j.1460-2075.1992.tb05427.x
40. Tecott LH, Maricq AV and Julius D. Nervous System Distribution of the Serotonin 5-HT3 Receptor mRNA. Proc Natl Acad Sci USA (1993) 90(4):1430–4. doi: 10.1073/pnas.90.4.1430
41. Thompson AJ, Lummis SC. 5-HT3 Receptors. Curr Pharm Des (2006) 12(28):3615–30. doi: 10.2174/138161206778522029
42. Liu HN, Ohya S, Nishizawa Y, Sawamura K, Iino S, Syed MM, et al. Serotonin Augments Gut Pacemaker Activity via 5-HT3 Receptors. PloS One (2011) 6(9):e24928. doi: 10.1371/journal.pone.0024928
43. Thompson AJ, Lummis SC. The 5-HT3 Receptor as a Therapeutic Target. Expert Opin Ther Targets (2007) 11(4):527–40. doi: 10.1517/14728222.11.4.527
44. Browning KN. Role of Central Vagal 5-HT3 Receptors in Gastrointestinal Physiology and Pathophysiology. Front Neurosci (2015) 9:413. doi: 10.3389/fnins.2015.00413
45. Kato S. Role of Serotonin 5-HT(3) Receptors in Intestinal Inflammation. Biol Pharm Bull (2013) 36(9):1406–9. doi: 10.1248/bpb.b13-00363
46. Stasi C, Bellini M, Bassotti G, Blandizzi C, Milani S. Serotonin Receptors and Their Role in the Pathophysiology and Therapy of Irritable Bowel Syndrome. Tech Coloproctol (2014) 18(7):613–21. doi: 10.1007/s10151-013-1106-8
47. Bhattarai Y, Schmidt BA, Linden DR, Larson ED, Grover M, Beyder A, et al. Human-Derived Gut Microbiota Modulates Colonic Secretion in Mice by Regulating 5-HT3 Receptor Expression via Acetate Production. Am J Physiol Gastrointest Liver Physiol (2017) 313(1):G80–7. doi: 10.1152/ajpgi.00448.2016
48. Engel M, Smidt MP, van Hooft JA. The Serotonin 5-HT3 Receptor: A Novel Neurodevelopmental Target. Front Cell Neurosci (2013) 7:76. doi: 10.3389/fncel.2013.00076
49. Gershon MD. Serotonin and Its Implication for the Management of Irritable Bowel Syndrome. Rev Gastroenterol Disord (2003) 3(Suppl 2):S25–34.
50. O'Leary OF, Codagnone MG, Cryan JF. “Chapter 38 - Revisiting the Behavioral Genetics of Serotonin: Relevance to Anxiety and Depression” in Handbook of Behavioral Neuroscience, volume 31, eds. Müller CP, Cunningham KA. Ed. Elsevier: Amsterdam, The Netherlands: (2020) pp. 665–709. doi: 10.1016/B978-0-444-64125-0.00038-4
51. Manuel-Apolinar L, Rocha L, Pascoe D, Castillo E, Castillo C, Meneses A. Modifications of 5-HT4 Receptor Expression in Rat Brain During Memory Consolidation. Brain Res (2005) 1042(1):73–81. doi: 10.1016/j.brainres.2005.02.020
52. Hoyer D. “5-HT-4 Receptor”, in xPharm: The Comprehensive Pharmacology Reference, eds. Enna SJ, Bylund DB. Ed. Elsevier: Amsterdam, The Netherlands: (2008) pp. 1–16. doi: 10.1016/B978-008055232-3.63759-6
53. Hoffman JM, Tyler K, MacEachern SJ, Balemba OB, Johnson AC, Brooks EM, et al. Activation of Colonic Mucosal 5-HT(4) Receptors Accelerates Propulsive Motility and Inhibits Visceral Hypersensitivity. Gastroenterology (2012) 142(4):844–54.e4. doi: 10.1053/j.gastro.2011.12.041
54. Marchetti E, Dumuis A, Bockaert J, Soumireu-Mourat B, Roman FS. Differential Modulation of the 5-HT(4) Receptor Agonists and Antagonist on Rat Learning and Memory. Neuropharmacology (2000) 39(11):2017–27. doi: 10.1016/s0028-3908(00)00038-1
55. King MV, Marsden CA, Fone KC. A Role for the 5-HT(1A), 5-HT4 and 5-HT6 Receptors in Learning and Memory. Trends Pharmacol Sci (2008) 29(9):482–92. doi: 10.1016/j.tips.2008.07.001
56. Liu M, Geddis MS, Wen Y, Setlik W, Gershon MD. Expression and Function of 5-HT4 Receptors in the Mouse Enteric Nervous System. Am J Physiol Gastrointest Liver Physiol (2005) 289(6):G1148–63. doi: 10.1152/ajpgi.00245.2005
57. Park CJ, Armenia SJ, Zhang L, Cowles RA. The 5-HT4 Receptor Agonist Prucalopride Stimulates Mucosal Growth and Enhances Carbohydrate Absorption in the Ileum of the Mouse. J Gastrointest Surg (2019) 23(6):1198–205. doi: 10.1007/s11605-018-3907-6
58. Amigo J, Diaz A, Pilar-Cuellar F, Vidal R, Martin A, Compan V, et al. The Absence of 5-HT4 Receptors Modulates Depression- and Anxiety-Like Responses and Influences the Response of Fluoxetine in Olfactory Bulbectomised Mice: Adaptive Changes in Hippocampal Neuroplasticity Markers and 5-HT1A Autoreceptor. Neuropharmacology (2016) 111:47–58. doi: 10.1016/j.neuropharm.2016.08.037
59. Coffin B, Farmachidi JP, Rueegg P, Bastie A, Bouhassira D. Tegaserod, a 5-HT4 Receptor Partial Agonist, Decreases Sensitivity to Rectal Distension in Healthy Subjects. Aliment Pharmacol Ther (2003) 17(4):577–85. doi: 10.1046/j.1365-2036.2003.01449.x
60. Compan V, Charnay Y, Dusticier N, Daszuta A, Hen R, Bockaert J. Feeding Disorders in 5-HT4 Receptor Knockout Mice. J Soc Biol (2004) 198(1):37–49. doi: 10.1051/jbio/2004198010037
61. Pasqualetti M, Ori M, Nardi I, Castagna M, Cassano GB, Marazziti D. Distribution of the 5-HT5A Serotonin Receptor mRNA in the Human Brain. Mol Brain Res (1998) 56(1):1–8. doi: 10.1016/S0169-328X(98)00003-5
62. Francken BJ, Jurzak M, Vanhauwe JF, Luyten WH, Leysen JE. The Human 5-Ht5a Receptor Couples to Gi/Go Proteins and Inhibits Adenylate Cyclase in HEK 293 Cells. Eur J Pharmacol (1998) 361(2–3):299–309. doi: 10.1016/S0014-2999(98)00744-4
63. Tuo BG, Sellers Z, Paulus P, Barrett KE, Isenberg JI. 5-HT Induces Duodenal Mucosal Bicarbonate Secretion via cAMP- and Ca2+-Dependent Signaling Pathways and 5-HT4 Receptors in Mice. Am J Physiol Gastrointest Liver Physiol (2004) 286(3):G444–51. doi: 10.1152/ajpgi.00105.2003
64. Oliver KR, Kinsey AM, Wainwright A, Sirinathsinghji DJS. Localization of 5-Ht5a Receptor-Like Immunoreactivity in the Rat Brain. Brain Res (2000) 867(1):131–42. doi: 10.1016/S0006-8993(00)02273-3
65. Gonzalez R, Chavez-Pascacio K, Meneses A. Role of 5-HT5A Receptors in the Consolidation of Memory. Behav Brain Res (2013) 252:246–51. doi: 10.1016/j.bbr.2013.05.051
66. Kinsey AM, Wainwright A, Heavens R, Sirinathsinghji DJ, Oliver K. Distribution of 5-ht(5A), 5-ht(5B), 5-ht(6) and 5-HT(7) Receptor mRNAs in the Rat Brain. Brain Res Mol Brain Res (2001) 88(1):194–8. doi: 10.1016/S0169-328X(01)00034-1
67. Avila-Rojas SH, Velazquez-Lagunas I, Salinas-Abarca AB, Barragan-Iglesias P, Pineda-Farias JB, Granados-Soto V. Role of Spinal 5-HT5A, and 5-HT1A/1B/1D, Receptors in Neuropathic Pain Induced by Spinal Nerve Ligation in Rats. Brain Res (2015) 1622:377–85. doi: 10.1016/j.brainres.2015.06.043
68. Kohen R, Metcalf MA, Khan N, Druck T, Huebner K, Lachowicz JE, et al. Cloning, Characterization, and Chromosomal Localization of a Human 5-HT6 Serotonin Receptor. J Neurochem (1996) 66(1):47–56. doi: 10.1046/j.1471-4159.1996.66010047.x
69. Yun HM, Kim S, Kim HJ, Kostenis E, Kim JI, Seong JY, et al. The Novel Cellular Mechanism of Human 5-HT6 Receptor Through an Interaction With Fyn*. J Biol Chem (2007) 282(8):5496–505. doi: 10.1074/jbc.M606215200
70. Glennon RA, Siripurapu U, Roth BL, Kolanos R, Bondarev ML, Sikazwe D, et al. The Medicinal Chemistry of 5-HT6 Receptor Ligands With a Focus on Arylsulfonyltryptamine Analogs. Curr Top Med Chem (2010) 10(5):579–95. doi: 10.2174/156802610791111542
71. Lacroix LP, Dawson LA, Hagan JJ and Heidbreder CA. 5-HT6 Receptor Antagonist SB-271046 Enhances Extracellular Levels of Monoamines in the Rat Medial Prefrontal Cortex. Synapse (2004) 51(2):158–64. doi: 10.1002/syn.10288
72. Gerard C, Martres MP, Lefevre K, Miquel MC, Verge D, Lanfumey L, et al. Immuno-Localization of Serotonin 5-HT6 Receptor-Like Material in the Rat Central Nervous System. Brain Res (1997) 746(1-2):207–19. doi: 10.1016/s0006-8993(96)01224-3
73. Glennon RA. Higher-End Serotonin Receptors: 5-HT(5), 5-HT(6), and 5-HT(7). J Med Chem (2003) 46(14):2795–812. doi: 10.1021/jm030030n
74. Quintero-Villegas A, Valdés-Ferrer SI. Role of 5-HT7 Receptors in the Immune System in Health and Disease. Mol Med (2019) 26(1):2. doi: 10.1186/s10020-019-0126-x
75. Iceta R, Mesonero JE, Aramayona JJ, Alcalde AI. Expression of 5-HT1A and 5-HT7 Receptors in Caco-2 Cells and Their Role in the Regulation of Serotonin Transporter Activity. J Physiol Pharmacol (2009) 60(1):157–64.
76. Mahe C, Loetscher E, Dev KK, Bobirnac I, Otten U, Schoeffter P. Serotonin 5-HT7 Receptors Coupled to Induction of Interleukin-6 in Human Microglial MC-3 Cells. Neuropharmacology (2005) 49(1):40–7. doi: 10.1016/j.neuropharm.2005.01.025
77. Kim JJ, Khan WI. 5-HT7 Receptor Signaling: Improved Therapeutic Strategy in Gut Disorders. Front Behav Neurosci (2014) 8:396. doi: 10.3389/fnbeh.2014.00396
78. Monti JM, Jantos H. The Role of Serotonin 5-HT7 Receptor in Regulating Sleep and Wakefulness. Rev Neurosci (2014) 25(3):429–37. doi: 10.1515/revneuro-2014-0016
79. Barnes NM, Ahern GP, Becamel C, Bockaert J, Camilleri M, Chaumont-Dubel S, et al. International Union of Basic and Clinical Pharmacology. CX. Classification of Receptors for 5-Hydroxytryptamine; Pharmacology and Function. Pharmacol Rev (2021) 73(1):310–520. doi: 10.1124/pr.118.015552
80. Gresch PJ, Sanders-Bush E. “Serotonin Receptor Signaling” in Encyclopedia of Biological Chemistry, eds. Lennarz WJ, Lane MD. Ed. Elsevier: Amsterdam, The Netherlands: (2004). pp. 33–7. doi: 10.1016/B0-12-443710-9/00621-9
81. Gershon MD. 5-HT (Serotonin) Physiology and Related Drugs. Curr Opin Gastroenterol (2000) 16(2):113–20. doi: 10.1097/00001574-200003000-00004
82. Meneses A, Terrón JA, Hong E. Effects of the 5-HT Receptor Antagonists GR127935 (5-HT1B/1D) and MDL100907 (5-HT2A) in the Consolidation of Learning. Behav Brain Res (1997) 89(1-2):217–23. doi: 10.1016/s0166-4328(97)00055-7
83. Diaz SL, Narboux-Neme N, Boutourlinsky K, Doly S, Maroteaux L. Mice Lacking the Serotonin 5-HT2B Receptor as an Animal Model of Resistance to Selective Serotonin Reuptake Inhibitors Antidepressants. Eur Neuropsychopharmacol (2016) 26(2):265–79. doi: 10.1016/j.euroneuro.2015.12.012
84. Raybould HE, Glatzle J, Robin C, Meyer JH, Phan T, Wong H, et al. Expression of 5-HT3 Receptors by Extrinsic Duodenal Afferents Contribute to Intestinal Inhibition of Gastric Emptying. Am J Physiol Gastrointest Liver Physiol (2003) 284(3):G367–72. doi: 10.1152/ajpgi.00292.2001
85. Salvador MT, Rodriguez-Yoldi MC, Alcalde AI, odriguez-Yoldi MJ. 5-HT Receptor Subtypes Involved in the Serotonin-Induced Inhibition of L-Leucine Absorption in Rabbit Jejunum. Life Sci (1997) 61(3):309–18. doi: 10.1016/s0024-3205(97)00387-1
86. Schworer H, Ramadori G. Autoreceptors can Modulate 5-Hydroxytryptamine Release From Porcine and Human Small Intestine In Vitro. Naunyn Schmiedebergs Arch Pharmacol (1998) 357(5):548–52. doi: 10.1007/pl00005206
87. Arnsten AF, Lin CH, Van Dyck CH, Stanhope KJ. The Effects of 5-HT3 Receptor Antagonists on Cognitive Performance in Aged Monkeys. Neurobiol Aging (1997) 18(1):21–8. doi: 10.1016/s0197-4580(96)00162-5
88. Motavallian A, Minaiyan M, Rabbani M, Andalib S, Mahzouni P. Involvement of 5HT3 Receptors in Anti-Inflammatory Effects of Tropisetron on Experimental TNBS-Induced Colitis in Rat. Bioimpacts (2013) 3(4):169–76. doi: 10.5681/bi.2013.021
89. Hoyer D, Hannon JP, Martin GR. Molecular, Pharmacological and Functional Diversity of 5-HT Receptors. Pharmacol Biochem Behav (2002) 71(4):533–54. doi: 10.1016/s0091-3057(01)00746-8
90. Plassat JL, Boschert U, Amlaiky N, Hen R. The Mouse 5HT5 Receptor Reveals a Remarkable Heterogeneity Within the 5HT1D Receptor Family. EMBO J (1992) 11(13):4779–86. doi: 10.1002/j.1460-2075.1992.tb05583.x
91. Heal DJ, Smith SL, Fisas A, Codony X, Buschmann H. Selective 5-HT6 Receptor Ligands: Progress in the Development of a Novel Pharmacological Approach to the Treatment of Obesity and Related Metabolic Disorders. Pharmacol Ther (2008) 117(2):207–31. doi: 10.1016/j.pharmthera.2007.08.006
92. Guscott M, Bristow LJ, Hadingham K, Rosahl TW, Beer MS, Stanton JA, et al. Genetic Knockout and Pharmacological Blockade Studies of the 5-HT7 Receptor Suggest Therapeutic Potential in Depression. Neuropharmacology (2005) 48(4):492–502. doi: 10.1016/j.neuropharm.2004.11.015
93. Bsibsi M, Ravid R, Gveric D, van Noort JM. Broad Expression of Toll-Like Receptors in the Human Central Nervous System. J Neuropathol Exp Neurol (2002) 61(11):1013–21. doi: 10.1093/jnen/61.11.1013
94. Oliveira-Nascimento L, Massari P, Wetzler LM. The Role of TLR2 in Infection and Immunity. Front Immunol (2012) 3:79. doi: 10.3389/fimmu.2012.00079
95. Swaroop S, Sengupta N, Suryawanshi AR, Adlakha YK, Basu A. HSP60 Plays a Regulatory Role in IL-1beta-Induced Microglial Inflammation via TLR4-P38 MAPK Axis. J Neuroinflamm (2016) 13:27. doi: 10.1186/s12974-016-0486-x
96. Cario E. Microbiota and Innate Immunity in Intestinal Inflammation and Neoplasia. Curr Opin Gastroenterol (2013) 29(1):85–91. doi: 10.1097/MOG.0b013e32835a670e
97. Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, et al. Genetic Analysis of Host Resistance: Toll-Like Receptor Signaling and Immunity at Large. Annu Rev Immunol (2006) 24:353–89. doi: 10.1146/annurev.immunol.24.021605.090552
98. Ved R, Sharouf F, Harari B, Muzaffar M, Manivannan S, Ormonde C, et al. Disulfide HMGB1 Acts via TLR2/4 Receptors to Reduce the Numbers of Oligodendrocyte Progenitor Cells After Traumatic Injury In Vitro. Sci Rep (2021) 11(1):6181. doi: 10.1038/s41598-021-84932-0
99. Vabulas RM, Braedel S, Hilf N, Singh-Jasuja H, Herter S, Ahmad-Nejad P, et al. The Endoplasmic Reticulum-Resident Heat Shock Protein Gp96 Activates Dendritic Cells via the Toll-Like Receptor 2/4 Pathway. J Biol Chem (2002) 277(23):20847–53. doi: 10.1074/jbc.M200425200
100. Oshiumi H, Matsumoto M, Funami K, Akazawa T and Seya T. TICAM-1, an Adaptor Molecule That Participates in Toll-Like Receptor 3-Mediated Interferon-Beta Induction. Nat Immunol (2003) 4(2):161–7. doi: 10.1038/ni886
101. Bell JK, Askins J, Hall PR, Davies DR, Segal DM. The dsRNA Binding Site of Human Toll-Like Receptor 3. Proc Natl Acad Sci USA (2006) 103(23):8792–7. doi: 10.1073/pnas.0603245103
102. Cavassani KA, Ishii M, Wen H, Schaller MA, Lincoln PM, Lukacs NW, et al. TLR3 Is an Endogenous Sensor of Tissue Necrosis During Acute Inflammatory Events. J Exp Med (2008) 205(11):2609–21. doi: 10.1084/jem.20081370
103. Yu S, Gao N. Compartmentalizing Intestinal Epithelial Cell Toll-Like Receptors for Immune Surveillance. Cell Mol Life Sci (2015) 72(17):3343–53. doi: 10.1007/s00018-015-1931-1
104. Barajon I, Serrao G, Arnaboldi F, Opizzi E, Ripamonti G, Balsari A, et al. Toll-Like Receptors 3, 4, and 7 Are Expressed in the Enteric Nervous System and Dorsal Root Ganglia. J Histochem Cytochem (2009) 57(11):1013–23. doi: 10.1369/jhc.2009.953539
105. Lehnardt S, Lachance C, Patrizi S, Lefebvre S, Follett PL, Jensen FE, et al. The Toll-Like Receptor TLR4 Is Necessary for Lipopolysaccharide-Induced Oligodendrocyte Injury in the CNS. J Neurosci (2002) 22(7):2478–86. doi: 10.1523/JNEUROSCI.22-07-02478.2002
106. Gorina R, Font-Nieves M, Marquez-Kisinousky L, Santalucia T, Planas AM. Astrocyte TLR4 Activation Induces a Proinflammatory Environment Through the Interplay Between MyD88-Dependent NFkappaB Signaling, MAPK, and Jak1/Stat1 Pathways. Glia (2011) 59(2):242–55. doi: 10.1002/glia.21094
107. Kawasaki T, Kawai T. Toll-Like Receptor Signaling Pathways. Front Immunol (2014) 5:461. doi: 10.3389/fimmu.2014.00461
108. Park BS, Lee JO. Recognition of Lipopolysaccharide Pattern by TLR4 Complexes. Exp Mol Med (2013) 45(12):e66. doi: 10.1038/emm.2013.97
109. Aucott H, Sowinska A, Harris HE, Lundback P. Ligation of Free HMGB1 to TLR2 in the Absence of Ligand Is Negatively Regulated by the C-Terminal Tail Domain. Mol Med (2018) 24(1):19. doi: 10.1186/s10020-018-0021-x
110. Ortega-Cava CF, Ishihara S, Rumi MA, Kawashima K, Ishimura N, Kazumori H, et al. Strategic Compartmentalization of Toll-Like Receptor 4 in the Mouse Gut. J Immunol (2003) 170(8):3977–85. doi: 10.4049/jimmunol.170.8.3977
111. Al-ofi E, Coffelt SB, Anumba DO. Fibrinogen, an Endogenous Ligand of Toll-Like Receptor 4, Activates Monocytes in Pre-Eclamptic Patients. J Reprod Immunol (2014) 103:23–8. doi: 10.1016/j.jri.2014.02.004
112. Roelofs MF, Boelens WC, Joosten LA, Abdollahi-Roodsaz S, Geurts J, Wunderink LU, et al. Identification of Small Heat Shock Protein B8 (HSP22) as a Novel TLR4 Ligand and Potential Involvement in the Pathogenesis of Rheumatoid Arthritis. J Immunol (2006) 176(11):7021–7. doi: 10.4049/jimmunol.176.11.7021
113. Curran CS, Demick KP, Mansfield JM. Lactoferrin Activates Macrophages via TLR4-Dependent and -Independent Signaling Pathways. Cell Immunol (2006) 242(1):23–30. doi: 10.1016/j.cellimm.2006.08.006
114. Choi YJ, Im E, Chung HK, Pothoulakis C, Rhee SH. TRIF Mediates Toll-Like Receptor 5-Induced Signaling in Intestinal Epithelial Cells. J Biol Chem (2010) 285(48):37570–8. doi: 10.1074/jbc.M110.158394
115. Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting Edge: Bacterial Flagellin Activates Basolaterally Expressed TLR5 to Induce Epithelial Proinflammatory Gene Expression. J Immunol (2001) 167(4):1882–5. doi: 10.4049/jimmunol.167.4.1882
116. Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, et al. Structural Basis of TLR5-Flagellin Recognition and Signaling. Science (2012) 335(6070):859–64. doi: 10.1126/science.1215584
117. Das N, Dewan V, Grace PM, Gunn RJ, Tamura R, Tzarum N, et al. HMGB1 Activates Proinflammatory Signaling via TLR5 Leading to Allodynia. Cell Rep (2016) 17(4):1128–40. doi: 10.1016/j.celrep.2016.09.076
118. Hussain S, Johnson CG, Sciurba J, Meng X, Stober VP, Liu C, et al. TLR5 Participates in the TLR4 Receptor Complex and Promotes MyD88-Dependent Signaling in Environmental Lung Injury. Elife (2020) 9:e50458. doi: 10.7554/eLife.50458
119. Zhang Z, Ohto U, Shibata T, Krayukhina E, Taoka M, Yamauchi Y, et al. Structural Analysis Reveals That Toll-Like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity (2016) 45(4):737–48. doi: 10.1016/j.immuni.2016.09.011
120. Vierbuchen T, Stein K, Heine H. RNA Is Taking Its Toll: Impact of RNA-Specific Toll-Like Receptors on Health and Disease. Allergy (2019) 74(2):223–35. doi: 10.1111/all.13680
121. Roers A, Hiller B, Hornung V. Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity (2016) 44(4):739–54. doi: 10.1016/j.immuni.2016.04.002
122. Price AE, Shamardani K, Lugo KA, Deguine J, Roberts AW, Lee BL, et al. A Map of Toll-Like Receptor Expression in the Intestinal Epithelium Reveals Distinct Spatial, Cell Type-Specific, and Temporal Patterns. Immunity (2018) 49(3):560–75.e6. doi: 10.1016/j.immuni.2018.07.016
123. Piccinini AM, Midwood KS. DAMPening Inflammation by Modulating TLR Signalling. Mediators Inflammation (2010) 2010:672395. doi: 10.1155/2010/672395
124. Qin J, Yao J, Cui G, Xiao H, Kim TW, Fraczek J, et al. TLR8-Mediated NF-kappaB and JNK Activation Are TAK1-Independent and MEKK3-Dependent. J Biol Chem (2006) 281(30):21013–21. doi: 10.1074/jbc.M512908200
125. Hanke ML, Kielian T. Toll-Like Receptors in Health and Disease in the Brain: Mechanisms and Therapeutic Potential. Clin Sci (Lond) (2011) 121(9):367–87. doi: 10.1042/CS20110164
126. Takeshita F, Leifer CA, Gursel I, Ishii KJ, Takeshita S, Gursel M, et al. Cutting Edge: Role of Toll-Like Receptor 9 in CpG DNA-Induced Activation of Human Cells. J Immunol (2001) 167(7):3555–8. doi: 10.4049/jimmunol.167.7.3555
127. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-Like Receptor Recognizes Bacterial DNA. Nature (2000) 408(6813):740–5. doi: 10.1038/35047123
128. Christensen SR, Kashgarian M, Alexopoulou L, Flavell RA, Akira S, Shlomchik MJ. Toll-Like Receptor 9 Controls Anti-DNA Autoantibody Production in Murine Lupus. J Exp Med (2005) 202(2):321–31. doi: 10.1084/jem.20050338
129. Barton GM, Kagan JC, Medzhitov R. Intracellular Localization of Toll-Like Receptor 9 Prevents Recognition of Self DNA But Facilitates Access to Viral DNA. Nat Immunol (2006) 7(1):49–56. doi: 10.1038/ni1280
130. Verma R, Kim JY. 1,25-Dihydroxyvitamin D3 Facilitates M2 Polarization and Upregulates TLR10 Expression on Human Microglial Cells. Neuroimmunomodulation (2016) 23(2):75–80. doi: 10.1159/000444300
131. Latorre E, Pradilla A, Chueca B, Pagan R, Layunta E, Alcalde AI, et al. Listeria Monocytogenes Inhibits Serotonin Transporter in Human Intestinal Caco-2 Cells. Microb Ecol (2016) 72(3):730–9. doi: 10.1007/s00248-016-0809-6
132. Henrick BM, Yao XD, Zahoor MA, Abimiku A, Osawe S, Rosenthal KL. TLR10 Senses HIV-1 Proteins and Significantly Enhances HIV-1 Infection. Front Immunol (2019) 10:482. doi: 10.3389/fimmu.2019.00482
133. Fore F, Indriputri C, Mamutse J, Nugraha J. TLR10 and Its Unique Anti-Inflammatory Properties and Potential Use as a Target in Therapeutics. Immune Netw (2020) 20(3):e21. doi: 10.4110/in.2020.20.e21
134. Rosenzweig HL, Planck SR, Rosenbaum JT. NLRs in Immune Privileged Sites. Curr Opin Pharmacol (2011) 11(4):423–8. doi: 10.1016/j.coph.2011.07.002
135. Arentsen T, Qian Y, Gkotzis S, Femenia T, Wang T, Udekwu K, et al. The Bacterial Peptidoglycan-Sensing Molecule Pglyrp2 Modulates Brain Development and Behavior. Mol Psychiatry (2017) 22(2):257–66. doi: 10.1038/mp.2016.182
136. Muniz LR, Knosp C, Yeretssian G. Intestinal Antimicrobial Peptides During Homeostasis, Infection, and Disease. Front Immunol (2012) 3:310. doi: 10.3389/fimmu.2012.00310
137. Keestra-Gounder AM, Byndloss MX, Seyffert N, Young BM, Chavez-Arroyo A, Tsai AY, et al. NOD1 and NOD2 Signalling Links ER Stress With Inflammation. Nature (2016) 532(7599):394–7. doi: 10.1038/nature17631
138. Kuss-Duerkop SK, Keestra-Gounder AM. NOD1 and NOD2 Activation by Diverse Stimuli: A Possible Role for Sensing Pathogen-Induced Endoplasmic Reticulum Stress. Infect Immun (2020) 88(7):e00898–19. doi: 10.1128/IAI.00898-19
139. Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, et al. Nod2 Is a General Sensor of Peptidoglycan Through Muramyl Dipeptide (MDP) Detection. J Biol Chem (2003) 278(11):8869–72. doi: 10.1074/jbc.C200651200
140. Caruso R, Warner N, Inohara N, Nunez G. NOD1 and NOD2: Signaling, Host Defense, and Inflammatory Disease. Immunity (2014) 41(6):898–908. doi: 10.1016/j.immuni.2014.12.010
141. Saxena M, Yeretssian G. NOD-Like Receptors: Master Regulators of Inflammation and Cancer. Front Immunol (2014) 5:327. doi: 10.3389/fimmu.2014.00327
142. Fukata M, Arditi M. The Role of Pattern Recognition Receptors in Intestinal Inflammation. Mucosal Immunol (2013) 6(3):451–63. doi: 10.1038/mi.2013.13
143. Kigerl KA, de Rivero Vaccari JP, Dietrich WD, Popovich PG, Keane RW. Pattern Recognition Receptors and Central Nervous System Repair. Exp Neurol (2014) 258:5–16. doi: 10.1016/j.expneurol.2014.01.001
144. Latorre E, Layunta E, Grasa L, Castro M, Pardo J, Gomollon F, et al. Intestinal Serotonin Transporter Inhibition by Toll-Like Receptor 2 Activation. A Feedback Modulation. PloS One (2016) 11(12):e0169303. doi: 10.1371/journal.pone.0169303
145. Wang H, Kwon YH, Dewan V, Vahedi F, Syed S, Fontes ME, et al. TLR2 Plays a Pivotal Role in Mediating Mucosal Serotonin Production in the Gut. J Immunol (2019) 202(10):3041–52. doi: 10.4049/jimmunol.1801034
146. Mendoza C, Matheus N, Latorre E, Castro M, Mesonero JE, Alcalde AI. Toll-Like Receptor 3 Activation Affects Serotonin Transporter Activity and Expression in Human Enterocyte-Like Caco-2 Cells. Cell Physiol Biochem (2012) 30(1):187–98. doi: 10.1159/000339057
147. Noda M, Ifuku M, Hossain MS, Katafuchi T. Glial Activation and Expression of the Serotonin Transporter in Chronic Fatigue Syndrome. Front Psychiatry (2018) 9:589. doi: 10.3389/fpsyt.2018.00589
148. Mendoza C, Matheus N, Iceta R, Mesonero JE, Alcalde AI. Lipopolysaccharide Induces Alteration of Serotonin Transporter in Human Intestinal Epithelial Cells. Innate Immun (2009) 15(4):243–50. doi: 10.1177/1753425909104781
149. Schwamborn R, Brown E, Haase J. Elevation of Cortical Serotonin Transporter Activity Upon Peripheral Immune Challenge Is Regulated Independently of P38 Mitogen-Activated Protein Kinase Activation and Transporter Phosphorylation. J Neurochem (2016) 137(3):423–35. doi: 10.1111/jnc.13596
150. Szabo A, Gogolak P, Koncz G, Foldvari Z, Pazmandi K, Miltner N, et al. Immunomodulatory Capacity of the Serotonin Receptor 5-HT2B in a Subset of Human Dendritic Cells. Sci Rep (2018) 8(1):1765. doi: 10.1038/s41598-018-20173-y
151. Layunta E, Latorre E, Forcen R, Grasa L, Plaza MA, Arias M, et al. NOD1 Downregulates Intestinal Serotonin Transporter and Interacts With Other Pattern Recognition Receptors. J Cell Physiol (2018) 233(5):4183–93. doi: 10.1002/jcp.26229
152. Layunta E, Latorre E, Forcen R, Grasa L, Castro M, Arias MA, et al. NOD2 Modulates Serotonin Transporter and Interacts With TLR2 and TLR4 in Intestinal Epithelial Cells. Cell Physiol Biochem (2018) 47(3):1217–29. doi: 10.1159/000490218
153. Femenia T, Qian Y, Arentsen T, Forssberg H, Diaz Heijtz R. Toll-Like Receptor-4 Regulates Anxiety-Like Behavior and DARPP-32 Phosphorylation. Brain Behav Immun (2018) 69:273–82. doi: 10.1016/j.bbi.2017.11.022
154. Forcen R, Latorre E, Pardo J, Alcalde AI, Murillo MD, Grasa L. Toll-Like Receptors 2 and 4 Modulate the Contractile Response Induced by Serotonin in Mouse Ileum: Analysis of the Serotonin Receptors Involved. Neurogastroenterol Motil (2015) 27(9):1258–66. doi: 10.1111/nmo.12619
155. Forcen R, Latorre E, Pardo J, Alcalde AI, Murillo MD, Grasa L. Toll-Like Receptors 2 and 4 Exert Opposite Effects on the Contractile Response Induced by Serotonin in Mouse Colon: Role of Serotonin Receptors. Exp Physiol (2016) 101(8):1064–74. doi: 10.1113/EP085668
156. Pusceddu MM, Barboza M, Keogh CE, Schneider M, Stokes P, Sladek JA, et al. Nod-Like Receptors Are Critical for Gut-Brain Axis Signalling in Mice. J Physiol (2019) 597(24):5777–97. doi: 10.1113/JP278640
157. Farhat K, Riekenberg S, Heine H, Debarry J, Lang R, Mages J, et al. Heterodimerization of TLR2 With TLR1 or TLR6 Expands the Ligand Spectrum But Does Not Lead to Differential Signaling. J Leukoc Biol (2008) 83(3):692–701. doi: 10.1189/jlb.0807586
158. Netea MG, van der Meer JW, Kullberg BJ. Both TLR2 and TLR4 Are Involved in the Recognition of Candida Albicans. Reply to "TLR2, But Not TLR4, Triggers Cytokine Production by Murine Cells in Response to Candida Albicans Yeasts and Hyphae" by Gil and Gozalbo. Microbes Infection (2006) 8:2823–2824.; author reply 2823-4. doi: 10.1016/j.micinf.2006.07.021
159. Hoffmann M, Zeisel MB, Jilg N, Paranhos-Baccala G, Stoll-Keller F, Wakita T, et al. Toll-Like Receptor 2 Senses Hepatitis C Virus Core Protein But Not Infectious Viral Particles. J Innate Immun (2009) 1(5):446–54. doi: 10.1159/000226136
160. Gravina HD, Antonelli L, Gazzinelli RT, Ropert C. Differential Use of TLR2 and TLR9 in the Regulation of Immune Responses During the Infection With Trypanosoma Cruzi. PloS One (2013) 8(5):e63100. doi: 10.1371/journal.pone.0063100
161. Latorre E, Layunta E, Grasa L, Pardo J, Garcia S, Alcalde AI, et al. Toll-Like Receptors 2 and 4 Modulate Intestinal IL-10 Differently in Ileum and Colon. United Eur Gastroenterol J (2018) 6(3):446–53. doi: 10.1177/2050640617727180
162. Latorre E, Mendoza C, Matheus N, Castro M, Grasa L, Mesonero JE, et al. IL-10 Modulates Serotonin Transporter Activity and Molecular Expression in Intestinal Epithelial Cells. Cytokine (2013) 61(3):778–84. doi: 10.1016/j.cyto.2013.01.012
163. Lu J, Zhang C, Lv J, Zhu X, Jiang X, Lu W, et al. Antiallergic Drug Desloratadine as a Selective Antagonist of 5HT2A Receptor Ameliorates Pathology of Alzheimer’s Disease Model Mice by Improving Microglial Dysfunction. Aging Cell (2021) 20(1):e13286. doi: 10.1111/acel.13286
164. Pott J, Stockinger S, Torow N, Smoczek A, Lindner C, McInerney G, et al. Age-Dependent TLR3 Expression of the Intestinal Epithelium Contributes to Rotavirus Susceptibility. PloS Pathog (2012) 8(5):e1002670. doi: 10.1371/journal.ppat.1002670
165. Lathia JD, Okun E, Tang SC, Griffioen K, Cheng A, Mughal MR, et al. Toll-Like Receptor 3 Is a Negative Regulator of Embryonic Neural Progenitor Cell Proliferation. J Neurosci (2008) 28(51):13978–84. doi: 10.1523/JNEUROSCI.2140-08.2008
166. Sato R, Kato A, Chimura T, Saitoh SI, Shibata T, Murakami Y, et al. Combating Herpesvirus Encephalitis by Potentiating a TLR3-Mtorc2 Axis. Nat Immunol (2018) 19(10):1071–82. doi: 10.1038/s41590-018-0203-2
167. Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-Like Receptor 3 Mediates West Nile Virus Entry Into the Brain Causing Lethal Encephalitis. Nat Med (2004) 10(12):1366–73. doi: 10.1038/nm1140
168. Latorre E, Mendoza C, Layunta E, Alcalde AI, Mesonero JE. TLR2, TLR3, and TLR4 Activation Specifically Alters the Oxidative Status of Intestinal Epithelial Cells. Cell Stress Chaperones (2014) 19(2):289–93. doi: 10.1007/s12192-013-0461-8
169. Schromm AB, Lien E, Henneke P, Chow JC, Yoshimura A, Heine H, et al. Molecular Genetic Analysis of an Endotoxin Nonresponder Mutant Cell Line: A Point Mutation in a Conserved Region of MD-2 Abolishes Endotoxin-Induced Signaling. J Exp Med (2001) 194(1):79–88. doi: 10.1084/jem.194.1.79
170. Ohnishi T, Muroi M, Tanamoto K. MD-2 Is Necessary for the Toll-Like Receptor 4 Protein to Undergo Glycosylation Essential for Its Translocation to the Cell Surface. Clin Diagn Lab Immunol (2003) 10(3):405–10. doi: 10.1128/cdli.10.3.405-410.2003
171. Klove S, Genger C, Mousavi S, Weschka D, Bereswill S, Heimesaat MM. Toll-Like Receptor-4 Dependent Intestinal and Systemic Sequelae Following Peroral Campylobacter Coli Infection of IL10 Deficient Mice Harboring a Human Gut Microbiota. Pathogens (2020) 9(5):386. doi: 10.3390/pathogens9050386
172. Ishihara S, Rumi MA, Kadowaki Y, Ortega-Cava CF, Yuki T, Yoshino N, et al. Essential Role of MD-2 in TLR4-Dependent Signaling During Helicobacter Pylori-Associated Gastritis. J Immunol (2004) 173(2):1406–16. doi: 10.4049/jimmunol.173.2.1406
173. Talbot S, Totemeyer S, Yamamoto M, Akira S, Hughes K, Gray D, et al. Toll-Like Receptor 4 Signalling Through MyD88 Is Essential to Control Salmonella Enterica Serovar Typhimurium Infection, But Not for the Initiation of Bacterial Clearance. Immunology (2009) 128(4):472–83. doi: 10.1111/j.1365-2567.2009.03146.x
174. Kim SW, Kim S, Son M, Cheon JH, Park YS. Melatonin Controls Microbiota in Colitis by Goblet Cell Differentiation and Antimicrobial Peptide Production Through Toll-Like Receptor 4 Signalling. Sci Rep (2020) 10(1):2232. doi: 10.1038/s41598-020-59314-7
175. Mogensen TH, Paludan SR, Kilian M, Ostergaard L. Live Streptococcus Pneumoniae, Haemophilus Influenzae, and Neisseria Meningitidis Activate the Inflammatory Response Through Toll-Like Receptors 2, 4, and 9 in Species-Specific Patterns. J Leukoc Biol (2006) 80(2):267–77. doi: 10.1189/jlb.1105626
176. Yao X, Liu S, Ding W, Yue P, Jiang Q, Zhao M, et al. TLR4 Signal Ablation Attenuated Neurological Deficits by Regulating Microglial M1/M2 Phenotype After Traumatic Brain Injury in Mice. J Neuroimmunol (2017) 310:38–45. doi: 10.1016/j.jneuroim.2017.06.006
177. Perez-Pardo P, Dodiya HB, Engen PA, Forsyth CB, Huschens AM, Shaikh M, et al. Role of TLR4 in the Gut-Brain Axis in Parkinson’s Disease: A Translational Study From Men to Mice. Gut (2019) 68(5):829–43. doi: 10.1136/gutjnl-2018-316844
178. Yang J, Zhang E, Liu F, Zhang Y, Zhong M, Li Y, et al. Flagellins of Salmonella Typhi and Nonpathogenic Escherichia Coli Are Differentially Recognized Through the NLRC4 Pathway in Macrophages. J Innate Immun (2014) 6(1):47–57. doi: 10.1159/000351476
179. Chassaing B, Ley RE, Gewirtz AT. Intestinal Epithelial Cell Toll-Like Receptor 5 Regulates the Intestinal Microbiota to Prevent Low-Grade Inflammation and Metabolic Syndrome in Mice. Gastroenterology (2014) 147(6):1363–77.e17. doi: 10.1053/j.gastro.2014.08.033
180. Ifuku M, Hinkelmann L, Kuhrt LD, Efe IE, Kumbol V, Buonfiglioli A, et al. Activation of Toll-Like Receptor 5 in Microglia Modulates Their Function and Triggers Neuronal Injury. Acta Neuropathol Commun (2020) 8(1):159. doi: 10.1186/s40478-020-01031-3
181. Hung YY, Huang KW, Kang HY, Huang GY, Huang TL. Antidepressants Normalize Elevated Toll-Like Receptor Profile in Major Depressive Disorder. Psychopharmacol (Berl) (2016) 233(9):1707–14. doi: 10.1007/s00213-015-4087-7
182. Khanmohammadi S, Rezaei N. Role of Toll-Like Receptors in the Pathogenesis of COVID-19. J Med Virol (2021) 93(5):2735–9. doi: 10.1002/jmv.26826
183. Olson JK, Miller SD. Microglia Initiate Central Nervous System Innate and Adaptive Immune Responses Through Multiple TLRs. J Immunol (2004) 173(6):3916–24. doi: 10.4049/jimmunol.173.6.3916
184. Butchi NB, Du M, Peterson KE. Interactions Between TLR7 and TLR9 Agonists and Receptors Regulate Innate Immune Responses by Astrocytes and Microglia. Glia (2010) 58(6):650–64. doi: 10.1002/glia.20952
185. Ma Y, Li J, Chiu I, Wang Y, Sloane JA, Lu J, et al. Toll-Like Receptor 8 Functions as a Negative Regulator of Neurite Outgrowth and Inducer of Neuronal Apoptosis. J Cell Biol (2006) 175(2):209–15. doi: 10.1083/jcb.200606016
186. Guerrier T, Pochard P, Lahiri A, Youinou P, Pers JO, Jamin C. TLR9 Expressed on Plasma Membrane Acts as a Negative Regulator of Human B Cell Response. J Autoimmun (2014) 51:23–9. doi: 10.1016/j.jaut.2014.02.005
187. Lee J, Mo JH, Katakura K, Alkalay I, Rucker AN, Liu YT, et al. Maintenance of Colonic Homeostasis by Distinctive Apical TLR9 Signalling in Intestinal Epithelial Cells. Nat Cell Biol (2006) 8(12):1327–36. doi: 10.1038/ncb1500
188. Rose WA 2nd, Sakamoto K, Leifer CA. TLR9 Is Important for Protection Against Intestinal Damage and for Intestinal Repair. Sci Rep (2012) 2:574. doi: 10.1038/srep00574
189. Yang H, Yu HB, Bhinder G, Ryz NR, Lee J, Yang H, et al. TLR9 Limits Enteric Antimicrobial Responses and Promotes Microbiota-Based Colonisation Resistance During Citrobacter Rodentium Infection. Cell Microbiol (2019) 21(7):e13026. doi: 10.1111/cmi.13026
190. Burgueno JF, Abreu MT. Epithelial Toll-Like Receptors and Their Role in Gut Homeostasis and Disease. Nat Rev Gastroenterol Hepatol (2020) 17(5):263–78. doi: 10.1038/s41575-019-0261-4
191. Sorensen LN, Reinert LS, Malmgaard L, Bartholdy C, Thomsen AR, Paludan SR. TLR2 and TLR9 Synergistically Control Herpes Simplex Virus Infection in the Brain. J Immunol (2008) 181(12):8604–12. doi: 10.4049/jimmunol.181.12.8604
192. Matsuda T, Murao N, Katano Y, Juliandi B, Kohyama J, Akira S, et al. TLR9 Signalling in Microglia Attenuates Seizure-Induced Aberrant Neurogenesis in the Adult Hippocampus. Nat Commun (2015) 6:6514. doi: 10.1038/ncomms7514
193. Fallarino F, Puccetti P. Toll-Like Receptor 9-Mediated Induction of the Immunosuppressive Pathway of Tryptophan Catabolism. Eur J Immunol (2006) 36(1):8–11. doi: 10.1002/eji.200535667
194. Layunta E, Latorre E, Gimeno A, Grasa L, Castro M, Plaza MA, et al. Toll-Like Receptor 9 Activation Affects Intestinal Serotonin Transporter Activity and Expression in Caco-2 Cells. Acta Physiol (Oxf) (2014) 214(Suppl. 701):15. doi: 10.1111/apha.12498.
195. Hasan U, Chaffois C, Gaillard C, Saulnier V, Merck E, Tancredi S, et al. Human TLR10 Is a Functional Receptor, Expressed by B Cells and Plasmacytoid Dendritic Cells, Which Activates Gene Transcription Through Myd88. J Immunol (2005) 174(5):2942–50. doi: 10.4049/jimmunol.174.5.2942
196. Oosting M, Cheng SC, Bolscher JM, Vestering-Stenger R, Plantinga TS, Verschueren IC, et al. Human TLR10 Is an Anti-Inflammatory Pattern-Recognition Receptor. Proc Natl Acad Sci USA (2014) 111(42):E4478–84. doi: 10.1073/pnas.1410293111
197. Chuang T, Ulevitch RJ. Identification of Htlr10: A Novel Human Toll-Like Receptor Preferentially Expressed in Immune Cells. Biochim Biophys Acta (2001) 1518(1-2):157–61. doi: 10.1016/s0167-4781(00)00289-x
198. Hart KM, Murphy AJ, Barrett KT, Wira CR, Guyre PM, Pioli PA. Functional Expression of Pattern Recognition Receptors in Tissues of the Human Female Reproductive Tract. J Reprod Immunol (2009) 80(1-2):33–40. doi: 10.1016/j.jri.2008.12.004
199. Ravishankar Ram M, Goh KL, Leow AH, Poh BH, Loke MF, Harrison R, et al. Polymorphisms at Locus 4p14 of Toll-Like Receptors TLR-1 and TLR-10 Confer Susceptibility to Gastric Carcinoma in Helicobacter Pylori Infection. PloS One (2015) 10(11):e0141865. doi: 10.1371/journal.pone.0141865
200. Tongtawee T, Bartpho T, Kaewpitoon S, Kaewpitoon N, Dechsukhum C, Leeanansaksiri W, et al. Genetic Polymorphisms in TLR1, TLR2, TLR4, and TLR10 of Helicobacter Pylori-Associated Gastritis: A Prospective Cross-Sectional Study in Thailand. Eur J Cancer Prev (2018) 27(2):118–23. doi: 10.1097/CEJ.0000000000000347
201. Nagashima H, Iwatani S, Cruz M, Jimenez Abreu JA, Uchida T, Mahachai V, et al. Toll-Like Receptor 10 in Helicobacter Pylori Infection. J Infect Dis (2015) 212(10):1666–76. doi: 10.1093/infdis/jiv270
202. Mohammed I, Abedin A, Tsintzas K, Abedin SA, Otri AM, Hopkinson A, et al. Increased Expression of Hepcidin and Toll-Like Receptors 8 and 10 in Viral Keratitis. Cornea (2011) 30(8):899–904. doi: 10.1097/ICO.0b013e31820126e5
203. Claes AK, Zhou JY, Philpott DJ. NOD-Like Receptors: Guardians of Intestinal Mucosal Barriers. Physiol (Bethesda) (2015) 30(3):241–50. doi: 10.1152/physiol.00025.2014
204. Le Bourhis L, Magalhaes JG, Selvanantham T, Travassos LH, Geddes K, Fritz JH, et al. Role of Nod1 in Mucosal Dendritic Cells During Salmonella Pathogenicity Island 1-Independent Salmonella Enterica Serovar Typhimurium Infection. Infect Immun (2009) 77(10):4480–6. doi: 10.1128/IAI.00519-09
205. Geddes K, Rubino SJ, Magalhaes JG, Streutker C, Le Bourhis L, Cho JH, et al. Identification of an Innate T Helper Type 17 Response to Intestinal Bacterial Pathogens. Nat Med (2011) 17(7):837–44. doi: 10.1038/nm.2391
206. Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, et al. Nod1 Responds to Peptidoglycan Delivered by the Helicobacter Pylori Cag Pathogenicity Island. Nat Immunol (2004) 5(11):1166–74. doi: 10.1038/ni1131
207. Ochoa-Cortes F, Ramos-Lomas T, Miranda-Morales M, Spreadbury I, Ibeakanma C, Barajas-Lopez C, et al. Bacterial Cell Products Signal to Mouse Colonic Nociceptive Dorsal Root Ganglia Neurons. Am J Physiol Gastrointest Liver Physiol (2010) 299(3):G723–32. doi: 10.1152/ajpgi.00494.2009
208. Woodhams KL, Chan JM, Lenz JD, Hackett KT, Dillard JP. Peptidoglycan Fragment Release From Neisseria Meningitidis. Infect Immun (2013) 81(9):3490–8. doi: 10.1128/IAI.00279-13
209. Horowitz JE, Warner N, Staples J, Crowley E, Gosalia N, Murchie R, et al. Mutation Spectrum of NOD2 Reveals Recessive Inheritance as a Main Driver of Early Onset Crohn’s Disease. Sci Rep (2021) 11(1):5595. doi: 10.1038/s41598-021-84938-8
210. Branquinho D, Freire P, Sofia C. NOD2 Mutations and Colorectal Cancer - Where do We Stand? World J Gastrointest Surg (2016) 8(4):284–93. doi: 10.4240/wjgs.v8.i4.284
211. Ferrand A, Al Nabhani Z, Tapias NS, Mas E, Hugot JP, Barreau F. NOD2 Expression in Intestinal Epithelial Cells Protects Toward the Development of Inflammation and Associated Carcinogenesis. Cell Mol Gastroenterol Hepatol (2019) 7(2):357–69. doi: 10.1016/j.jcmgh.2018.10.009
212. Netea MG, van der Leij F, Drenth JP, Joosten LA, te Morsche R, Verweij P, et al. Chronic Yersiniosis Due to Defects in the TLR5 and NOD2 Recognition Pathways. Neth J Med (2010) 68(10):310–5.
213. Sun X, Jobin C. Nucleotide-Binding Oligomerization Domain-Containing Protein 2 Controls Host Response to Campylobacter Jejuni in Il10-/- Mice. J Infect Dis (2014) 210(7):1145–54. doi: 10.1093/infdis/jiu148
214. Al Nabhani Z, Dietrich G, Hugot JP, Barreau F. Nod2: The Intestinal Gate Keeper. PloS Pathog (2017) 13(3):e1006177. doi: 10.1371/journal.ppat.1006177
215. Geyer S, Jacobs M, Hsu NJ. Immunity Against Bacterial Infection of the Central Nervous System: An Astrocyte Perspective. Front Mol Neurosci (2019) 12:57. doi: 10.3389/fnmol.2019.00057
216. Nabatov AA. The Vesicle-Associated Function of NOD2 as a Link Between Crohn’s Disease and Mycobacterial Infection. Gut Pathog (2015) 7(1):1. doi: 10.1186/s13099-015-0049-1
217. Sinagra E, Utzeri E, Morreale GC, Fabbri C, Pace F, Anderloni A. Microbiota-Gut-Brain Axis and Its Affect Inflammatory Bowel Disease: Pathophysiological Concepts and Insights for Clinicians. World J Clin cases (2020) 8(6):1013–25. doi: 10.12998/wjcc.v8.i6.1013
218. Clapp M, Aurora N, Herrera L, Bhatia M, Wilen E, Wakefield S. Gut Microbiota’s Effect on Mental Health: The Gut-Brain Axis. Clin Pract (2017) 7(4):987. doi: 10.4081/cp.2017.987
219. Kowalski K, Mulak A. Brain-Gut-Microbiota Axis in Alzheimer’s Disease. J Neurogastroenterol Motil (2019) 25(1):48–60. doi: 10.5056/jnm18087
220. Israelyan N, Margolis KG. Reprint of: Serotonin as a Link Between the Gut-Brain-Microbiome Axis in Autism Spectrum Disorders. Pharmacol Res (2019) 140:115–20. doi: 10.1016/j.phrs.2018.12.023
221. Martin CR, Osadchiy V, Kalani A, Mayer EA. The Brain-Gut-Microbiome Axis. Cell Mol Gastroenterol Hepatol (2018) 6(2):133–48. doi: 10.1016/j.jcmgh.2018.04.003
222. Chen Y, Xu J, Chen Y. Regulation of Neurotransmitters by the Gut Microbiota and Effects on Cognition in Neurological Disorders. Nutrients (2021) 13(6):2099. doi: 10.3390/nu13062099
223. Bhattarai Y, Williams BB, Battaglioli EJ, Whitaker WR, Till L, Grover M, et al. Gut Microbiota-Produced Tryptamine Activates an Epithelial G-Protein-Coupled Receptor to Increase Colonic Secretion. Cell Host Microbe (2018) 23(6):775–85.e5. doi: 10.1016/j.chom.2018.05.004
224. Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, et al. The Microbiome-Gut-Brain Axis During Early Life Regulates the Hippocampal Serotonergic System in a Sex-Dependent Manner. Mol Psychiatry (2013) 18(6):666–73. doi: 10.1038/mp.2012.77
225. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous Bacteria From the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell (2015) 161(2):264–76. doi: 10.1016/j.cell.2015.02.047
226. Williams BB, Van Benschoten AH, Cimermancic P, Donia MS, Zimmermann M, Taketani M, et al. Discovery and Characterization of Gut Microbiota Decarboxylases That Can Produce the Neurotransmitter Tryptamine. Cell Host Microbe (2014) 16(4):495–503. doi: 10.1016/j.chom.2014.09.001
227. Saraf MK, Piccolo BD, Bowlin AK, Mercer KE, LeRoith T, Chintapalli SV, et al. Formula Diet Driven Microbiota Shifts Tryptophan Metabolism From Serotonin to Tryptamine in Neonatal Porcine Colon. Microbiome (2017) 5(1):77. doi: 10.1186/s40168-017-0297-z
228. Chang EB, Rao MC. A New Role for Microbiota? Dulling the Thrust of Serotonin and 5-HT3 Signaling Cascade. Am J Physiol Gastrointest Liver Physiol (2017) 313(1):G14–5. doi: 10.1152/ajpgi.00166.2017
229. Esmaili A, Nazir SF, Borthakur A, Yu D, Turner JR, Saksena S, et al. Enteropathogenic Escherichia Coli Infection Inhibits Intestinal Serotonin Transporter Function and Expression. Gastroenterology (2009) 137(6):2074–83. doi: 10.1053/j.gastro.2009.09.002
230. Banskota S, Regmi SC, Gautam J, Gurung P, Lee YJ, Ku SK, et al. Serotonin Disturbs Colon Epithelial Tolerance of Commensal E. Coli by Increasing NOX2-Derived Superoxide. Free Radic Biol Med (2017) 106:196–207. doi: 10.1016/j.freeradbiomed.2017.02.034
231. Agus A, Planchais J, Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe (2018) 23(6):716–24. doi: 10.1016/j.chom.2018.05.003
232. Roager HM, Licht TR. Microbial Tryptophan Catabolites in Health and Disease. Nat Commun (2018) 9(1):3294. doi: 10.1038/s41467-018-05470-4
233. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics Analysis Reveals Large Effects of Gut Microflora on Mammalian Blood Metabolites. Proc Natl Acad Sci USA (2009) 106(10):3698–703. doi: 10.1073/pnas.0812874106
234. Hata T, Asano Y, Yoshihara K, Kimura-Todani T, Miyata N, Zhang XT, et al. Regulation of Gut Luminal Serotonin by Commensal Microbiota in Mice. PloS One (2017) 12(7):e0180745. doi: 10.1371/journal.pone.0180745
235. Gao K, Pi Y, Mu CL, Peng Y, Huang Z, Zhu WY. Antibiotics-Induced Modulation of Large Intestinal Microbiota Altered Aromatic Amino Acid Profile and Expression of Neurotransmitters in the Hypothalamus of Piglets. J Neurochem (2018) 146(3):219–34. doi: 10.1111/jnc.14333
236. Chen YC, Seyedsayamdost MR, Ringstad N. A Microbial Metabolite Synergizes With Endogenous Serotonin to Trigger C. Elegans Reproductive Behavior. Proc Natl Acad Sci USA (2020) 117(48):30589–98. doi: 10.1073/pnas.2017918117
237. Engevik MA, Luck B, Visuthranukul C, Ihekweazu FD, Engevik AC, Shi Z, et al. Human-Derived Bifidobacterium Dentium Modulates the Mammalian Serotonergic System and Gut-Brain Axis. Cell Mol Gastroenterol Hepatol (2021) 11(1):221–48. doi: 10.1016/j.jcmgh.2020.08.002
238. Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, et al. The Gut Microbiota Influences Blood-Brain Barrier Permeability in Mice. Sci Transl Med (2014) 6(263):263ra158. doi: 10.1126/scitranslmed.3009759
239. Reigstad CS, Salmonson CE, Rainey JF 3rd, Szurszewski JH, Linden DR, Sonnenburg JL, et al. Gut Microbes Promote Colonic Serotonin Production Through an Effect of Short-Chain Fatty Acids on Enterochromaffin Cells. FASEB J (2015) 29(4):1395–403. doi: 10.1096/fj.14-259598
240. Latorre E, Forcén A, Buey B, Grasa L, Mesonero JE. Microbiota-Derived Short-Chain Fatty Acids as Modulators of Intestinal Serotonin Transporter. FEBS Open Bio (2021) 11(S1):46. doi: 10.1002/2211-5463.13206
241. Carrard A, Elsayed M, Margineanu M, Boury-Jamot B, Fragniere L, Meylan EM, et al. Peripheral Administration of Lactate Produces Antidepressant-Like Effects. Mol Psychiatry (2018) 23(2):392–9. doi: 10.1038/mp.2016.179
242. Silva YP, Bernardi A, Frozza RL. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front Endocrinol (Lausanne) (2020) 11:25. doi: 10.3389/fendo.2020.00025
243. Cirillo C, Vanden Berghe P, Tack J. Role of Serotonin in Gastrointestinal Physiology and Pathology. Minerva Endocrinol (2011) 36(4):311–24.
244. Jenkins TA, Nguyen JC, Polglaze KE, Bertrand PP. Influence of Tryptophan and Serotonin on Mood and Cognition With a Possible Role of the Gut-Brain Axis. Nutrients (2016) 8(1):56. doi: 10.3390/nu8010056
245. O’Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, Tryptophan Metabolism and the Brain-Gut-Microbiome Axis. Behav Brain Res (2015) 277:32–48. doi: 10.1016/j.bbr.2014.07.027
246. Del Colle A, Israelyan N, Gross Margolis K. Novel Aspects of Enteric Serotonergic Signaling in Health and Brain-Gut Disease. Am J Physiol Gastrointest Liver Physiol (2020) 318(1):G130–43. doi: 10.1152/ajpgi.00173.2019
247. Kerckhoffs AP, Samsom M, van der Rest ME, de Vogel J, Knol J, Ben-Amor K, et al. Lower Bifidobacteria Counts in Both Duodenal Mucosa-Associated and Fecal Microbiota in Irritable Bowel Syndrome Patients. World J Gastroenterol (2009) 15(23):2887–92. doi: 10.3748/wjg.15.2887
248. Barandouzi ZA, Lee J, Maas K, Starkweather AR, Cong XS. Altered Gut Microbiota in Irritable Bowel Syndrome and Its Association With Food Components. J Pers Med (2021) 11(1):35. doi: 10.3390/jpm11010035
249. Petrov VA, Saltykova IV, Zhukova IA, Alifirova VM, Zhukova NG, Dorofeeva YB, et al. Analysis of Gut Microbiota in Patients With Parkinson’s Disease. Bull Exp Biol Med (2017) 162(6):734–7. doi: 10.1007/s10517-017-3700-7
250. Vidal-Martinez G, Chin B, Camarillo C, Herrera GV, Yang B, Sarosiek I, et al. A Pilot Microbiota Study in Parkinson’s Disease Patients Versus Control Subjects, and Effects of FTY720 and FTY720-Mitoxy Therapies in Parkinsonian and Multiple System Atrophy Mouse Models. J Parkinsons Dis (2020) 10(1):185–92. doi: 10.3233/JPD-191693
251. Tan AH, Chong CW, Lim SY, Yap IKS, Teh CSJ, Loke MF, et al. Gut Microbial Ecosystem in Parkinson Disease: New Clinicobiological Insights From Multi-Omics. Ann Neurol (2021) 89(3):546–59. doi: 10.1002/ana.25982
252. Taverniti V, Cesari V, Gargari G, Rossi U, Biddau C, Lecchi C, et al. Probiotics Modulate Mouse Gut Microbiota and Influence Intestinal Immune and Serotonergic Gene Expression in a Site-Specific Fashion. Front Microbiol (2021) 12:706135. doi: 10.3389/fmicb.2021.706135
253. Ford AC, Harris LA, Lacy BE, Quigley EMM, Moayyedi P. Systematic Review With Meta-Analysis: The Efficacy of Prebiotics, Probiotics, Synbiotics and Antibiotics in Irritable Bowel Syndrome. Aliment Pharmacol Ther (2018) 48(10):1044–60. doi: 10.1111/apt.15001
254. Liu CY, Cham CM, Chang EB. Epithelial Wound Healing in Inflammatory Bowel Diseases: The Next Therapeutic Frontier. Transl Res (2021) 236:35–51. doi: 10.1016/j.trsl.2021.06.001
255. Di Pierro F, Pane M. Bifidobacterium Longum W11: Uniqueness and Individual or Combined Clinical Use in Association With Rifaximin. Clin Nutr ESPEN (2021) 42:15–21. doi: 10.1016/j.clnesp.2020.12.025
256. Maukonen J, Kolho KL, Paasela M, Honkanen J, Klemetti P, Vaarala O, et al. Altered Fecal Microbiota in Paediatric Inflammatory Bowel Disease. J Crohns Colitis (2015) 9(12):1088–95. doi: 10.1093/ecco-jcc/jjv147
257. Kowalska-Duplaga K, Kapusta P, Gosiewski T, Sroka-Oleksiak A, Ludwig-Slomczynska AH, Wolkow PP, et al. Changes in the Intestinal Microbiota Are Seen Following Treatment With Infliximab in Children With Crohn’s Disease. J Clin Med (2020) 9(3):687. doi: 10.3390/jcm9030687
258. Zhang W, Zhang X, Zhang Y, Wu H, Liu Q, Zhou X, et al. Analysis of Changes of Intestinal Flora in Elderly Patients With Alzheimer’s Disease and Liver Cancer and Its Correlation With Abnormal Gastrointestinal Motility. J Oncol (2021) p:7517379. doi: 10.1155/2021/7517379
259. Agusti A, Moya-Perez A, Campillo I, Montserrat-de la Paz S, Cerrudo V, Perez-Villalba A, et al. Bifidobacterium Pseudocatenulatum CECT 7765 Ameliorates Neuroendocrine Alterations Associated With an Exaggerated Stress Response and Anhedonia in Obese Mice. Mol Neurobiol (2018) 55(6):5337–52. doi: 10.1007/s12035-017-0768-z
260. Fattorusso A, Di Genova L, Dell’Isola GB, Mencaroni E, Esposito S. Autism Spectrum Disorders and the Gut Microbiota. Nutrients (2019) 11(3):521. doi: 10.3390/nu11030521
261. Lasheras I, Seral P, Latorre E, Barroso E, Gracia-Garcia P, Santabarbara J. Microbiota and Gut-Brain Axis Dysfunction in Autism Spectrum Disorder: Evidence for Functional Gastrointestinal Disorders. Asian J Psychiatr (2020) 47:101874. doi: 10.1016/j.ajp.2019.101874
262. Painold A, Morkl S, Kashofer K, Halwachs B, Dalkner N, Bengesser S, et al. A Step Ahead: Exploring the Gut Microbiota in Inpatients With Bipolar Disorder During a Depressive Episode. Bipolar Disord (2019) 21(1):40–9. doi: 10.1111/bdi.12682
263. Singhal M, Turturice BA, Manzella CR, Ranjan R, Metwally AA, Theorell J, et al. Serotonin Transporter Deficiency Is Associated With Dysbiosis and Changes in Metabolic Function of the Mouse Intestinal Microbiome. Sci Rep (2019) 9(1):2138. doi: 10.1038/s41598-019-38489-8
264. Zhu Y, Huan F, Wang J, Xie X, Yu G, Wang X, et al. 1-Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine Induced Parkinson’s Disease in Mouse: Potential Association Between Neurotransmitter Disturbance and Gut Microbiota Dysbiosis. ACS Chem Neurosci (2020) 11(20):3366–76. doi: 10.1021/acschemneuro.0c00475
265. Donato L, Alibrandi S, Scimone C, Castagnetti A, Rao G, Sidoti A, et al. Gut-Brain Axis Cross-Talk and Limbic Disorders as Biological Basis of Secondary TMAU. J Pers Med (2021) 11(2):87. doi: 10.3390/jpm11020087
266. Kesika P, Suganthy N, Sivamaruthi BS, Chaiyasut C. Role of Gut-Brain Axis, Gut Microbial Composition, and Probiotic Intervention in Alzheimer’s Disease. Life Sci (2021) 264:118627. doi: 10.1016/j.lfs.2020.118627
267. Talani G, Biggio F, Mostallino MC, Locci V, Porcedda C, Boi L, et al. Treatment With Gut Bifidobacteria Improves Hippocampal Plasticity and Cognitive Behavior in Adult Healthy Rats. Neuropharmacology (2020) 165:107909. doi: 10.1016/j.neuropharm.2019.107909
268. Alam MT, Amos GCA, Murphy ARJ, Murch S, Wellington EMH, Arasaradnam RP. Microbial Imbalance in Inflammatory Bowel Disease Patients at Different Taxonomic Levels. Gut Pathog (2020) 12:1. doi: 10.1186/s13099-019-0341-6
269. Lee SM, Kim N, Yoon H, Kim YS, Choi SI, Park JH, et al. Compositional and Functional Changes in the Gut Microbiota in Irritable Bowel Syndrome Patients. Gut Liver (2021) 15(2):253–61. doi: 10.5009/gnl19379
270. Satapathy MK, Yen TL, Jan JS, Tang RD, Wang JY, Taliyan R, et al. Solid Lipid Nanoparticles (SLNs): An Advanced Drug Delivery System Targeting Brain Through BBB. Pharmaceutics (2021) 13(8):1183. doi: 10.3390/pharmaceutics13081183
271. Qian Y, Yang X, Xu S, Wu C, Song Y, Qin N, et al. Alteration of the Fecal Microbiota in Chinese Patients With Parkinson’s Disease. Brain Behav Immun (2018) 70:194–202. doi: 10.1016/j.bbi.2018.02.016
272. Mandic AD, Woting A, Jaenicke T, Sander A, Sabrowski W, Rolle-Kampcyk U, et al. Clostridium Ramosum Regulates Enterochromaffin Cell Development and Serotonin Release. Sci Rep (2019) 9(1):1177. doi: 10.1038/s41598-018-38018-z
273. El-Salhy M, Hatlebakk JG, Gilja OH, Brathen Kristoffersen A, Hausken T. Efficacy of Faecal Microbiota Transplantation for Patients With Irritable Bowel Syndrome in a Randomised, Double-Blind, Placebo-Controlled Study. Gut (2020) 69(5):859–67. doi: 10.1136/gutjnl-2019-319630
274. Blaser MJ. Fecal Microbiota Transplantation for Dysbiosis - Predictable Risks. N Engl J Med (2019) 381(21):2064–6. doi: 10.1056/NEJMe1913807
275. Schierova D, Brezina J, Mrazek J, Fliegerova KO, Kvasnova S, Bajer L, et al. Gut Microbiome Changes in Patients With Active Left-Sided Ulcerative Colitis After Fecal Microbiome Transplantation and Topical 5-Aminosalicylic Acid Therapy. Cells (2020) 9(10):2283. doi: 10.3390/cells9102283
276. Kang YB, Cai Y. Faecal Microbiota Transplantation Enhances Efficacy of Immune Checkpoint Inhibitors Therapy Against Cancer. World J Gastroenterol (2021) 27(32):5362–75. doi: 10.3748/wjg.v27.i32.5362
277. Muniz Pedrogo DA, Chen J, Hillmann B, Jeraldo P, Al-Ghalith G, Taneja V, et al. An Increased Abundance of Clostridiaceae Characterizes Arthritis in Inflammatory Bowel Disease and Rheumatoid Arthritis: A Cross-Sectional Study. Inflammation Bowel Dis (2019) 25(5):902–13. doi: 10.1093/ibd/izy318
278. Sankarasubramanian J, Ahmad R, Avuthu N, Singh AB, Guda C. Gut Microbiota and Metabolic Specificity in Ulcerative Colitis and Crohn’s Disease. Front Med (Lausanne) (2020) 7:606298. doi: 10.3389/fmed.2020.606298
279. Shabbir U, Arshad MS, Sameen A and Oh DH. Crosstalk Between Gut and Brain in Alzheimer’s Disease: The Role of Gut Microbiota Modulation Strategies. Nutrients (2021) 13(2):690. doi: 10.3390/nu13020690
280. Zebrowska P, Laczmanska I, Laczmanski L. Future Directions in Reducing Gastrointestinal Disorders in Children With ASD Using Fecal Microbiota Transplantation. Front Cell Infect Microbiol (2021) 11:630052. doi: 10.3389/fcimb.2021.630052
281. Lorente-Picon M, Laguna A. New Avenues for Parkinson’s Disease Therapeutics: Disease-Modifying Strategies Based on the Gut Microbiota. Biomolecules (2021) 11(3):433. doi: 10.3390/biom11030433
282. Vendrik KEW, Ooijevaar RE, de Jong PRC, Laman JD, van Oosten BW, van Hilten JJ, et al. Fecal Microbiota Transplantation in Neurological Disorders. Front Cell Infect Microbiol (2020) 10:98. doi: 10.3389/fcimb.2020.00098
283. Yaghoubfar R, Behrouzi A, Fateh A, Nojoumi SA, Vaziri F, Khatami S, et al. Effects of Akkermansia Muciniphila and Faecalibacterium Prausnitzii on Serotonin Transporter Expression in Intestinal Epithelial Cells. J Diabetes Metab Disord (2021) 20(1):1–5. doi: 10.1007/s40200-020-00539-8
284. Lin A, Zheng W, He Y, Tang W, Wei X, He R, et al. Gut Microbiota in Patients With Parkinson’s Disease in Southern China. Parkinsonism Relat Disord (2018) 53:82–8. doi: 10.1016/j.parkreldis.2018.05.007
285. Garcez ML, Jacobs KR, Guillemin GJ. Microbiota Alterations in Alzheimer’s Disease: Involvement of the Kynurenine Pathway and Inflammation. Neurotox Res (2019) 36(2):424–36. doi: 10.1007/s12640-019-00057-3
286. Rahman MH, Bajgai J, Fadriquela A, Sharma S, Trinh TT, Akter R, et al. Therapeutic Potential of Natural Products in Treating Neurodegenerative Disorders and Their Future Prospects and Challenges. Molecules (2021) 26(17):5327. doi: 10.3390/molecules26175327
287. Les F, Valero MS, Moliner C, Weinkove D, Lopez V, Gomez-Rincon C. Jasonia Glutinosa (L.) DC., a Traditional Herbal Tea, Exerts Antioxidant and Neuroprotective Properties in Different In Vitro and In Vivo Systems. Biol (Basel) (2021) 10(5):443. doi: 10.3390/biology10050443
288. Lo Presti A, Zorzi F, Del Chierico F, Altomare A, Cocca S, Avola A, et al. Fecal and Mucosal Microbiota Profiling in Irritable Bowel Syndrome and Inflammatory Bowel Disease. Front Microbiol (2019) 10:1655. doi: 10.3389/fmicb.2019.01655
289. Liu S, Li E, Sun Z, Fu D, Duan G, Jiang M, et al. Altered Gut Microbiota and Short Chain Fatty Acids in Chinese Children With Autism Spectrum Disorder. Sci Rep (2019) 9(1):287. doi: 10.1038/s41598-018-36430-z
290. Vacca M, Celano G, Calabrese FM, Portincasa P, Gobbetti M, De Angelis M. The Controversial Role of Human Gut Lachnospiraceae. Microorganisms (2020) 8(4):573. doi: 10.3390/microorganisms8040573
291. Cao F, Liu J, Sha BX, Pan HF. Natural Products: Experimental Efficient Agents for Inflammatory Bowel Disease Therapy. Curr Pharm Des (2019) 25(46):4893–913. doi: 10.2174/1381612825666191216154224
292. Valero MS, Gonzalez M, Ramon-Gimenez M, Andrade PB, Moreo E, Les F, et al. Jasonia Glutinosa (L.) DC., a Traditional Herbal Medicine, Reduces Inflammation, Oxidative Stress and Protects the Intestinal Barrier in a Murine Model of Colitis. Inflammopharmacology (2020) 28(6):1717–34. doi: 10.1007/s10787-019-00626-0
293. Rodino-Janeiro BK, Vicario M, Alonso-Cotoner C, Pascua-Garcia R, Santos J. A Review of Microbiota and Irritable Bowel Syndrome: Future in Therapies. Adv Ther (2018) 35(3):289–310. doi: 10.1007/s12325-018-0673-5
294. Kim YM, Snijders AM, Brislawn CJ, Stratton KG, Zink EM, Fansler SJ, et al. Light-Stress Influences the Composition of the Murine Gut Microbiome, Memory Function, and Plasma Metabolome. Front Mol Biosci (2019) 6:108. doi: 10.3389/fmolb.2019.00108
295. So D, Gibson PR, Muir JG, Yao CK. Dietary Fibres and IBS: Translating Functional Characteristics to Clinical Value in the Era of Personalised Medicine. Gut (2021) gutjnl-2021-324891. doi: 10.1136/gutjnl-2021-324891
296. Chung CS, Chang PF, Liao CH, Lee TH, Chen Y, Lee YC, et al. Differences of Microbiota in Small Bowel and Faeces Between Irritable Bowel Syndrome Patients and Healthy Subjects. Scand J Gastroenterol (2016) 51(4):410–9. doi: 10.3109/00365521.2015.1116107
297. Bonder MJ, Tigchelaar EF, Cai X, Trynka G, Cenit MC, Hrdlickova B, et al. The Influence of a Short-Term Gluten-Free Diet on the Human Gut Microbiome. Genome Med (2016) 8(1):45. doi: 10.1186/s13073-016-0295-y
298. Li Y, Li ZX, Xie CY, Fan J, Lv J, Xu XJ, et al. Gegen Qinlian Decoction Enhances Immunity and Protects Intestinal Barrier Function in Colorectal Cancer Patients via Gut Microbiota. World J Gastroenterol (2020) 26(48):7633–51. doi: 10.3748/wjg.v26.i48.7633
299. Zoey FL, Palanivel M, Padmanabhan P, Gulyas B. Parkinson’s Disease: A Nanotheranostic Approach Targeting Alpha-Synuclein Aggregation. Front Cell Dev Biol (2021) 9:707441. doi: 10.3389/fcell.2021.707441
300. Nguyen TT, Dung Nguyen TT, Vo TK, Tran NM, Nguyen MK, Van Vo T, et al. Nanotechnology-Based Drug Delivery for Central Nervous System Disorders. BioMed Pharmacother (2021) 143:112117. doi: 10.1016/j.biopha.2021.112117
301. Stange EF, Schroeder BO. Microbiota and Mucosal Defense in IBD: An Update. Expert Rev Gastroenterol Hepatol (2019) 13(10):963–76. doi: 10.1080/17474124.2019.1671822
302. Baldini F, Hertel J, Sandt E, Thinnes CC, Neuberger-Castillo L, Pavelka L, et al. Parkinson’s Disease-Associated Alterations of the Gut Microbiome Predict Disease-Relevant Changes in Metabolic Functions. BMC Biol (2020) 18(1):62. doi: 10.1186/s12915-020-00775-7
303. Zhivaki D, Kagan JC. Innate Immune Detection of Lipid Oxidation as a Threat Assessment Strategy. Nat Rev Immunol (2021) 2021:1–9. doi: 10.1038/s41577-021-00618-8
304. Park J, Choi J, Kim DD, Lee S, Lee B, Lee Y, et al. Bioactive Lipids and Their Derivatives in Biomedical Applications. Biomol Ther (Seoul) (2021) 29(5):465–82. doi: 10.4062/biomolther.2021.107
305. Karen C, Shyu DJH, Rajan KE. Lactobacillus Paracasei Supplementation Prevents Early Life Stress-Induced Anxiety and Depressive-Like Behavior in Maternal Separation Model-Possible Involvement of Microbiota-Gut-Brain Axis in Differential Regulation of MicroRNA124a/132 and Glutamate Receptors. Front Neurosci (2021) 15:719933. doi: 10.3389/fnins.2021.719933
306. Gill PA, van Zelm MC, Muir JG, Gibson PR. Review Article: Short Chain Fatty Acids as Potential Therapeutic Agents in Human Gastrointestinal and Inflammatory Disorders. Aliment Pharmacol Ther (2018) 48(1):15–34. doi: 10.1111/apt.14689
307. Russo E, Giudici F, Fiorindi C, Ficari F, Scaringi S, Amedei A. Immunomodulating Activity and Therapeutic Effects of Short Chain Fatty Acids and Tryptophan Post-Biotics in Inflammatory Bowel Disease. Front Immunol (2019) 10:2754. doi: 10.3389/fimmu.2019.02754
308. So D, Yao CK, Gill PA, Pillai N, Gibson PR, Muir JG. Screening Dietary Fibres for Fermentation Characteristics and Metabolic Profiles Using a Rapid In Vitro Approach: Implications for Irritable Bowel Syndrome. Br J Nutr (2021) 126(2):208–18. doi: 10.1017/S0007114520003943
309. Özoğul F, Kuley E. The Function of Lactic Acid Bacteria on Biogenic Amines Production by Food-Borne Pathogens in Arginine Decarboxylase Broth. Food Sci Technol Res (2012) 18(6):795–804. doi: 10.3136/fstr.18.795
310. Hopfner F, Kunstner A, Muller SH, Kunzel S, Zeuner KE, Margraf NG, et al. Gut Microbiota in Parkinson Disease in a Northern German Cohort. Brain Res (2017) 1667:41–5. doi: 10.1016/j.brainres.2017.04.019
311. Dobri AM, Dudau M, Enciu AM, Hinescu ME. CD36 in Alzheimer’s Disease: An Overview of Molecular Mechanisms and Therapeutic Targeting. Neuroscience (2021) 453:301–11. doi: 10.1016/j.neuroscience.2020.11.003
312. Lo Sasso G, Khachatryan L, Kondylis A, Battey JND, Sierro N, Danilova NA, et al. Inflammatory Bowel Disease-Associated Changes in the Gut: Focus on Kazan Patients. Inflammation Bowel Dis (2021) 27(3):418–33. doi: 10.1093/ibd/izaa188
313. Kowalska-Duplaga K, Gosiewski T, Kapusta P, Sroka-Oleksiak A, Wedrychowicz A, Pieczarkowski S, et al. Differences in the Intestinal Microbiome of Healthy Children and Patients With Newly Diagnosed Crohn’s Disease. Sci Rep (2019) 9(1):18880. doi: 10.1038/s41598-019-55290-9
314. Seishima J, Iida N, Kitamura K, Yutani M, Wang Z, Seki A, et al. Gut-Derived Enterococcus Faecium From Ulcerative Colitis Patients Promotes Colitis in a Genetically Susceptible Mouse Host. Genome Biol (2019) 20(1):252. doi: 10.1186/s13059-019-1879-9
315. Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, et al. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens (2019) 8(3):126. doi: 10.3390/pathogens8030126
316. Hu DL, Zhu G, Mori F, Omoe K, Okada M, Wakabayashi K, et al. Staphylococcal Enterotoxin Induces Emesis Through Increasing Serotonin Release in Intestine and it Is Downregulated by Cannabinoid Receptor 1. Cell Microbiol (2007) 9(9):2267–77. doi: 10.1111/j.1462-5822.2007.00957.x
317. Luqman A, Nega M, Nguyen MT, Ebner P, Gotz F. SadA-Expressing Staphylococci in the Human Gut Show Increased Cell Adherence and Internalization. Cell Rep (2018) 22(2):535–45. doi: 10.1016/j.celrep.2017.12.058
318. Azimi T, Nasiri MJ, Chirani AS, Pouriran R, Dabiri H. The Role of Bacteria in the Inflammatory Bowel Disease Development: A Narrative Review. APMIS (2018) 126(4):275–83. doi: 10.1111/apm.12814
319. Duan R, Zhu S, Wang B, Duan L. Alterations of Gut Microbiota in Patients With Irritable Bowel Syndrome Based on 16S rRNA-Targeted Sequencing: A Systematic Review. Clin Transl Gastroenterol (2019) 10(2):e00012. doi: 10.14309/ctg.0000000000000012
320. Dan Z, Mao X, Liu Q, Guo M, Zhuang Y, Liu Z, et al. Altered Gut Microbial Profile Is Associated With Abnormal Metabolism Activity of Autism Spectrum Disorder. Gut Microbes (2020) 11(5):1246–67. doi: 10.1080/19490976.2020.1747329
321. Modasia A, Parker A, Jones E, Stentz R, Brion A, Goldson A, et al. Regulation of Enteroendocrine Cell Networks by the Major Human Gut Symbiont Bacteroides Thetaiotaomicron. Front Microbiol (2020) 11:575595. doi: 10.3389/fmicb.2020.575595
322. Zakerska-Banaszak O, Tomczak H, Gabryel M, Baturo A, Wolko L, Michalak M, et al. Dysbiosis of Gut Microbiota in Polish Patients With Ulcerative Colitis: A Pilot Study. Sci Rep (2021) 11(1):2166. doi: 10.1038/s41598-021-81628-3
323. Peter J, Fournier C, Durdevic M, Knoblich L, Keip B, Dejaco C, et al. A Microbial Signature of Psychological Distress in Irritable Bowel Syndrome. Psychosom Med (2018) 80(8):698–709. doi: 10.1097/PSY.0000000000000630
324. Deng Y, Zhou M, Wang J, Yao J, Yu J, Liu W, et al. Involvement of the Microbiota-Gut-Brain Axis in Chronic Restraint Stress: Disturbances of the Kynurenine Metabolic Pathway in Both the Gut and Brain. Gut Microbes (2021) 13(1):1–16. doi: 10.1080/19490976.2020.1869501
325. Taverniti V, Guglielmetti S. Methodological Issues in the Study of Intestinal Microbiota in Irritable Bowel Syndrome. World J Gastroenterol (2014) 20(27):8821–36. doi: 10.3748/wjg.v20.i27.8821
326. Lin CH, Chen CC, Chiang HL, Liou JM, Chang CM, Lu TP, et al. Altered Gut Microbiota and Inflammatory Cytokine Responses in Patients With Parkinson’s Disease. J Neuroinflamm (2019) 16(1):129. doi: 10.1186/s12974-019-1528-y
327. Zhang Y, Tan L, Li C, Wu H, Ran D, Zhang Z. Sulforaphane Alter the Microbiota and Mitigate Colitis Severity on Mice Ulcerative Colitis Induced by DSS. AMB Express (2020) 10(1):119. doi: 10.1186/s13568-020-01053-z
328. Unger MM, Spiegel J, Dillmann KU, Grundmann D, Philippeit H, Burmann J, et al. Short Chain Fatty Acids and Gut Microbiota Differ Between Patients With Parkinson’s Disease and Age-Matched Controls. Parkinsonism Relat Disord (2016) 32:66–72. doi: 10.1016/j.parkreldis.2016.08.019
329. Sivamaruthi BS, Madhumita R, Balamurugan K, Rajan KE. Cronobacter Sakazakii Infection Alters Serotonin Transporter and Improved Fear Memory Retention in the Rat. Front Pharmacol (2015) 6:188. doi: 10.3389/fphar.2015.00188
330. Nzakizwanayo J, Dedi C, Standen G, Macfarlane WM, Patel BA, Jones BV. Escherichia Coli Nissle 1917 Enhances Bioavailability of Serotonin in Gut Tissues Through Modulation of Synthesis and Clearance. Sci Rep (2015) 5:17324. doi: 10.1038/srep17324
331. Kim HS, Lim JH, Park H, Lee SI. Increased Immunoendocrine Cells in Intestinal Mucosa of Postinfectious Irritable Bowel Syndrome Patients 3 Years After Acute Shigella Infection–an Observation in a Small Case Control Study. Yonsei Med J (2010) 51(1):45–51. doi: 10.3349/ymj.2010.51.1.45
332. Shishov VA, Kirovskaia TA, Kudrin VS, Oleskin AV. [Amine Neuromediators, Their Precursors, and Oxidation Products in the Culture of Escherichia Coli K-12]. Prikl Biokhim Mikrobiol (2009) 45(5):550–4. doi: 10.1134/S0003683809050068
333. Özoğul F. Production of Biogenic Amines by Morganella Morganii, Klebsiella Pneumoniae and Hafnia Alvei Using a Rapid HPLC Method. Eur Food Res Technol (2004) 219(5):465–9. doi: 10.1007/s00217-004-0988-0
334. Castano-Rodriguez N, Kaakoush NO, Lee WS, Mitchell HM. Dual Role of Helicobacter and Campylobacter Species in IBD: A Systematic Review and Meta-Analysis. Gut (2017) 66(2):235–49. doi: 10.1136/gutjnl-2015-310545
335. Lyte JM, Shrestha S, Wagle BR, Liyanage R, Martinez DA, Donoghue AM, et al. Serotonin Modulates Campylobacter Jejuni Physiology and Invitro Interaction With the Gut Epithelium. Poult Sci (2021) 100(3):100944. doi: 10.1016/j.psj.2020.12.041
336. Berumen A, Lennon R, Breen-Lyles M, Griffith J, Patel R, Boxrud D, et al. Characteristics and Risk Factors of Post-Infection Irritable Bowel Syndrome After Campylobacter Enteritis. Clin Gastroenterol Hepatol (2021) 19(9):1855–63.e1. doi: 10.1016/j.cgh.2020.07.033
337. Yu Y, Zhu S, Li P, Min L, Zhang S. Helicobacter Pylori Infection and Inflammatory Bowel Disease: A Crosstalk Between Upper and Lower Digestive Tract. Cell Death Dis (2018) 9(10):961. doi: 10.1038/s41419-018-0982-2
338. Meng WP, Wang ZQ, Deng JQ, Liu Y, Deng MM, Lu MH. The Role of H. Pylori CagA in Regulating Hormones of Functional Dyspepsia Patients. Gastroenterol Res Pract (2016) 2016:7150959. doi: 10.1155/2016/7150959
339. Gu X, Song LJ, Li LX, Liu T, Zhang MM, Li Z, et al. Fusobacterium Nucleatum Causes Microbial Dysbiosis and Exacerbates Visceral Hypersensitivity in a Colonization-Independent Manner. Front Microbiol (2020) 11:1281. doi: 10.3389/fmicb.2020.01281
340. Guo S, Lu Y, Xu B, Wang W, Xu J, d Zhang G. A Simple Fecal Bacterial Marker Panel for the Diagnosis of Crohn’s Disease. Front Microbiol (2019) 10:1306. doi: 10.3389/fmicb.2019.01306
341. Porter RJ, Kalla R, Ho GT. Ulcerative Colitis: Recent Advances in the Understanding of Disease Pathogenesis. F1000Res (2020) 9:F1000 Faculty Rev-294. doi: 10.12688/f1000research.20805.1
342. Bian X, Wu W, Yang L, Lv L, Wang Q, Li Y, et al. Administration of Akkermansia Muciniphila Ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis in Mice. Front Microbiol (2019) 10:2259. doi: 10.3389/fmicb.2019.02259
343. Bliss ES, Whiteside E. The Gut-Brain Axis, the Human Gut Microbiota and Their Integration in the Development of Obesity. Front Physiol (2018) 9:900. doi: 10.3389/fphys.2018.00900
344. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem Across Age, Environment, Diet, and Diseases. Microorganisms (2019) 7(1):14. doi: 10.3390/microorganisms7010014
345. Glassner KL, Abraham BP, Quigley EMM. The Microbiome and Inflammatory Bowel Disease. J Allergy Clin Immunol (2020) 145(1):16–27. doi: 10.1016/j.jaci.2019.11.003
346. Khan WI. The Role of 5-HT Dysregulation in Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y) (2013) 9(4):259–61.
347. Jorandli JW, Thorsvik S, Skovdahl HK, Kornfeld B, Saeterstad S, Gustafsson BI, et al. The Serotonin Reuptake Transporter Is Reduced in the Epithelium of Active Crohn’s Disease and Ulcerative Colitis. Am J Physiol Gastrointest Liver Physiol (2020) 319(6):G761–8. doi: 10.1152/ajpgi.00244.2020
348. Davis BK, Philipson C, Hontecillas R, Eden K, Bassaganya-Riera J, Allen IC. Emerging Significance of NLRs in Inflammatory Bowel Disease. Inflammation Bowel Dis (2014) 20(12):2412–32. doi: 10.1097/MIB.0000000000000151
349. Ledder O, Turner D. Antibiotics in IBD: Still a Role in the Biological Era? Inflammation Bowel Dis (2018) 24(8):1676–88. doi: 10.1093/ibd/izy067
350. Caldeira LF, Borba HH, Tonin FS, Wiens A, Fernandez-Llimos F, Pontarolo R. Fecal Microbiota Transplantation in Inflammatory Bowel Disease Patients: A Systematic Review and Meta-Analysis. PloS One (2020) 15(9):e0238910. doi: 10.1371/journal.pone.0238910
351. Derwa Y, Gracie DJ, Hamlin PJ, Ford AC. Systematic Review With Meta-Analysis: The Efficacy of Probiotics in Inflammatory Bowel Disease. Aliment Pharmacol Ther (2017) 46(4):389–400. doi: 10.1111/apt.14203
352. D’Haens GR, van Deventer S. 25 Years of Anti-TNF Treatment for Inflammatory Bowel Disease: Lessons From the Past and a Look to the Future. Gut (2021) 70(7):1396–405. doi: 10.1136/gutjnl-2019-320022
353. Tam JSY, Coller JK, Hughes PA, Prestidge CA, Bowen JM. Toll-Like Receptor 4 (TLR4) Antagonists as Potential Therapeutics for Intestinal Inflammation. Indian J Gastroenterol (2021) 40(1):5–21. doi: 10.1007/s12664-020-01114-y
354. Geddes K, Magalhaes JG, Girardin SE. Unleashing the Therapeutic Potential of NOD-Like Receptors. Nat Rev Drug Discovery (2009) 8(6):465–79. doi: 10.1038/nrd2783
355. Levin AD, van den Brink GR. Selective Inhibition of Mucosal Serotonin as Treatment for IBD? Gut (2014) 63(6):866–7. doi: 10.1136/gutjnl-2013-305283
356. Hall BJ, Hamlin PJ, Gracie DJ, Ford AC. The Effect of Antidepressants on the Course of Inflammatory Bowel Disease. Can J Gastroenterol Hepatol (2018) p:2047242. doi: 10.1155/2018/2047242
357. Crowell MD. Role of Serotonin in the Pathophysiology of the Irritable Bowel Syndrome. Br J Pharmacol (2004) 141(8):1285–93. doi: 10.1038/sj.bjp.0705762
358. Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, et al. Gut Microbiota in Patients With Irritable Bowel Syndrome-A Systematic Review. Gastroenterology (2019) 157(1):97–108. doi: 10.1053/j.gastro.2019.03.049
359. Luo M, Zhuang X, Tian Z, Xiong L. Alterations in Short-Chain Fatty Acids and Serotonin in Irritable Bowel Syndrome: A Systematic Review and Meta-Analysis. BMC Gastroenterol (2021) 21(1):14. doi: 10.1186/s12876-020-01577-5
360. Kim HJ. Do Toll-Like Receptors Play a New Role as a Biomarker of Irritable Bowel Syndrome? J Neurogastroenterol Motil (2018) 24(4):510–1. doi: 10.5056/jnm18153
361. Dlugosz A, Zakikhany K, Acevedo N, D’Amato M, Lindberg G. Increased Expression of Toll-Like Receptors 4, 5, and 9 in Small Bowel Mucosa From Patients With Irritable Bowel Syndrome. BioMed Res Int (2017) p:9624702. doi: 10.1155/2017/9624702
362. Jin DC, Cao HL, Xu MQ, Wang SN, Wang YM, Yan F, et al. Regulation of the Serotonin Transporter in the Pathogenesis of Irritable Bowel Syndrome. World J Gastroenterol (2016) 22(36):8137–48. doi: 10.3748/wjg.v22.i36.8137
363. Bundeff AW, Woodis CB. Selective Serotonin Reuptake Inhibitors for the Treatment of Irritable Bowel Syndrome. Ann Pharmacother (2014) 48(6):777–84. doi: 10.1177/1060028014528151
364. Tack J, Janssen P, Wouters M, Boeckxstaens G. Targeting Serotonin Synthesis to Treat Irritable Bowel Syndrome. Gastroenterology (2011) 141(2):420–2. doi: 10.1053/j.gastro.2011.06.024
365. Herndon CC, Wang YP, Lu CL. Targeting the Gut Microbiota for the Treatment of Irritable Bowel Syndrome. Kaohsiung J Med Sci (2020) 36(3):160–70. doi: 10.1002/kjm2.12154
366. Harris LA, Baffy N. Modulation of the Gut Microbiota: A Focus on Treatments for Irritable Bowel Syndrome. Postgrad Med (2017) 129(8):872–88. doi: 10.1080/00325481.2017.1383819
367. Belmonte L, Beutheu Youmba S, Bertiaux-Vandaele N, Antonietti M, Lecleire S, Zalar A, et al. Role of Toll Like Receptors in Irritable Bowel Syndrome: Differential Mucosal Immune Activation According to the Disease Subtype. PloS One (2012) 7(8):e42777. doi: 10.1371/journal.pone.0042777
368. Zmudzka E, Salaciak K, Sapa J, Pytka K. Serotonin Receptors in Depression and Anxiety: Insights From Animal Studies. Life Sci (2018) 210:106–24. doi: 10.1016/j.lfs.2018.08.050
369. Kohl Z, Winkler J. “Chapter 50 - Serotonin in Parkinson's Disease” in Handbook of Behavioral Neuroscience, volume 31, eds. Müller CP, Cunningham KA. Ed. Elsevier: Amsterdam, The Netherlands: (2020). pp. 969–79. doi: 10.1016/B978-0-444-64125-0.00050-5
370. Malinova TS, Dijkstra CD, de Vries HE. Serotonin: A Mediator of the Gut-Brain Axis in Multiple Sclerosis. Mult Scler (2018) 24(9):1144–50. doi: 10.1177/1352458517739975
371. Abdulamir HA, Abdul-Rasheed OF, Abdulghani EA. Serotonin and Serotonin Transporter Levels in Autistic Children. Saudi Med J (2018) 39(5):487–94. doi: 10.15537/smj.2018.5.21751
372. Morrissette DA, Stahl SM. Modulating the Serotonin System in the Treatment of Major Depressive Disorder. CNS Spectr (2014) 19 Suppl 1:57–67; quiz 54-7, 68. doi: 10.1017/S1092852914000613
373. Cryan JF, O'Riordan KJ, Sandhu K, Peterson V, Dinan TG. The Gut Microbiome in Neurological Disorders. Lancet Neurol (2020) 19(2):179–94. doi: 10.1016/S1474-4422(19)30356-4
374. Huang F, Wu X. Brain Neurotransmitter Modulation by Gut Microbiota in Anxiety and Depression. Front Cell Dev Biol (2021) 9:649103. doi: 10.3389/fcell.2021.649103
375. Maiuolo J, Gliozzi M, Musolino V, Carresi C, Scarano F, Nucera S, et al. The Contribution of Gut Microbiota-Brain Axis in the Development of Brain Disorders. Front Neurosci (2021) 15:616883. doi: 10.3389/fnins.2021.616883
376. Bonfili L, Cecarini V, Gogoi O, Gong C, Cuccioloni M, Angeletti M, et al. Microbiota Modulation as Preventative and Therapeutic Approach in Alzheimer’s Disease. FEBS J (2021) 288(9):2836–55. doi: 10.1111/febs.15571
377. Margolis KG, Cryan JF, Mayer EA. The Microbiota-Gut-Brain Axis: From Motility to Mood. Gastroenterology (2021) 160(5):1486–501. doi: 10.1053/j.gastro.2020.10.066
378. Caputi V, Giron MC. Microbiome-Gut-Brain Axis and Toll-Like Receptors in Parkinson’s Disease. Int J Mol Sci (2018) 19(6):1689. doi: 10.3390/ijms19061689
379. Singh S, Jha S. NLRs as Helpline in the Brain: Mechanisms and Therapeutic Implications. Mol Neurobiol (2018) 55(10):8154–78. doi: 10.1007/s12035-018-0957-4
380. Hung YY. Antidepressants Improve Negative Regulation of Toll-Like Receptor Signaling in Monocytes From Patients With Major Depression. Neuroimmunomodulation (2018) 25(1):42–8. doi: 10.1159/000489562
Keywords: serotonin, 5-HT, tryptophan, microorganisms, PRRs, TLRs, NLRs
Citation: Layunta E, Buey B, Mesonero JE and Latorre E (2021) Crosstalk Between Intestinal Serotonergic System and Pattern Recognition Receptors on the Microbiota–Gut–Brain Axis. Front. Endocrinol. 12:748254. doi: 10.3389/fendo.2021.748254
Received: 27 July 2021; Accepted: 05 October 2021;
Published: 08 November 2021.
Edited by:
Cristina Giaroni, University of Insubria, ItalyReviewed by:
Elisabetta Barocelli, University of Parma, ItalyMaria Cecilia Giron, University of Padua, Italy
Copyright © 2021 Layunta, Buey, Mesonero and Latorre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elena Layunta, elena.layunta@medkem.gu.se
 Elena Layunta
Elena Layunta Berta Buey
Berta Buey Jose Emilio Mesonero
Jose Emilio Mesonero Eva Latorre
Eva Latorre