- Department of Urology, The Second Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, China
Objective: To evaluate the associations between systemic inflammatory biomarkers—systemic immune-inflammation index (SII), systemic inflammation response index (SIRI), pan-immune inflammation value (PIV), neutrophil-to-lymphocyte ratio (NLR), lymphocyte-to-monocyte ratio (LMR), and platelet-to-lymphocyte ratio (PLR)—and prostate cancer (PCa) risk, and to assess their potential for risk in both general and clinical populations.
Methods: A dual-cohort study was conducted using data from the National Health and Nutrition Examination Survey (NHANES; 2001–2010; N=7,354 males, 514 were classified as PCa) and a clinical validation cohort from the second affiliated hospital of Nanchang University (N=353, 175 with biopsy-confirmed PCa). Multivariable logistic regression, restricted cubic spline (RCS) analysis, and receiver operating characteristic (ROC) curve analysis were employed to examine linear/nonlinear relationships and predictive performance of the biomarkers. Models were adjusted for demographic, clinical, and laboratory covariates.
Results: Elevated SII, NLR, PLR, SIRI, and PIV were significantly associated with increased PCa risk in both cohorts, while higher LMR was protective. In the clinical cohort, the highest quartile of SIRI (OR=6.265, 95% CI: 3.130–13.012) and PIV (OR=6.638, 95% CI: 3.343–13.665) showed the strongest risks. RCS analyses revealed nonlinear relationships between biomarkers and PCa risk, total PSA (tPSA), and free PSA (fPSA). Elevated SII, NLR, PLR, SIRI, and PIV were significantly associated with increased PCa risk in both cohorts, while a higher LMR was protective. In the clinical cohort, the highest quartile of SIRI (OR=6.265, 95% CI: 3.130–13.012) and PIV (OR=6.638, 95% CI: 3.343–13.665) exhibited the strongest risks. RCS analyses revealed nonlinear relationships between biomarkers and PCa risk, total PSA (tPSA), and free PSA (fPSA). ROC analysis indicated moderate discriminatory power for PIV (AUC=0.709, 95% CI: 0.655–0.763) and SIRI (AUC=0.704, 95% CI: 0.650–0.759) compared with tPSA in the clinical cohort. However, fPSA and SIRI did not demonstrate a clear advantage, and the DeLong test showed no significant statistical difference.
Conclusion: Systemic inflammatory biomarkers, particularly composite indices such as SIRI and PIV, are strongly associated with PCa risk and demonstrate nonlinear relationships with PSA parameters. These biomarkers may enhance risk stratification for PCa and serve as non-invasive tools to complement existing diagnostic approaches.
Introduction
Prostate cancer (PCa) is a leading cause of cancer-related mortality among men worldwide, and early detection is critical for improving outcomes, particularly in high-risk cases (1, 2). Current screening relies heavily on prostate-specific antigen (PSA) testing, but its limited specificity, especially in the PSA “gray zone“ (4–20 ng/mL), often results in unnecessary biopsies or missed diagnoses (3, 4). Chronic inflammation, a hallmark of cancer, orchestrates a pro-tumorigenic microenvironment through immune cell dysregulation and cytokine signaling, driving PCa initiation and progression (5–7). Systemic inflammatory biomarkers, such as the systemic immune-inflammation index (SII), lymphocyte-to-monocyte ratio (LMR), systemic inflammation response index (SIRI), neutrophil-to-lymphocyte ratio (NLR), pan-immune inflammation value (PIV), and platelet-to-lymphocyte ratio (PLR), serve as accessible proxies for immune dynamics and have emerged as promising tools for cancer risk assessment and prognosis. For instance, studies have identified a correlation between the expression levels of inflammatory markers, such as SII, SIRI, and NLR, in rectal cancer patients and the subsequent development and progression of the disease (8, 9). Further research highlights the predictive utility of these indices across various solid tumors, suggesting a generalized role of systemic inflammation in oncogenesis.
Evidence suggests that elevated systemic inflammation, as measured by these biomarkers, is associated with increased PCa risk. For instance, a study using the National Health and Nutrition Examination Survey (NHANES) reported a 168% increased PCa risk with elevated SII (odds ratio [OR] = 2.68, 95% confidence interval [CI]: 1.32-5.46) in U.S. men (10). While these biomarkers have shown prognostic value in malignancies such as colorectal cancer, their role in PCa, particularly high-risk cases, remains underexplored. Furthermore, the nature of their associations with PSA parameters (total PSA [tPSA] and free PSA [fPSA])—whether linear or nonlinear—and their clinical utility in PCa screening and risk stratification are not well-established.
This study employs a dual-cohort design, integrating population-based data from NHANES with retrospective clinical data from a tertiary hospital in China, to investigate the associations of six systemic inflammatory biomarkers (SII, SIRI, PIV, NLR, LMR, and PLR) with PCa risk. Using advanced statistical approaches, including receiver operating characteristic (ROC) curve analysis and restricted cubic spline (RCS) regression, we aim to elucidate linear and nonlinear relationships between these biomarkers and PCa risk, as well as their predictive potential. By bridging knowledge gaps in the role of systemic inflammation in PCa, this study seeks to inform early detection strategies and personalized immunotherapeutic approaches, aligning with the growing emphasis on immune-based interventions in cancer management.
Methods
Study populations
NHANES cohort
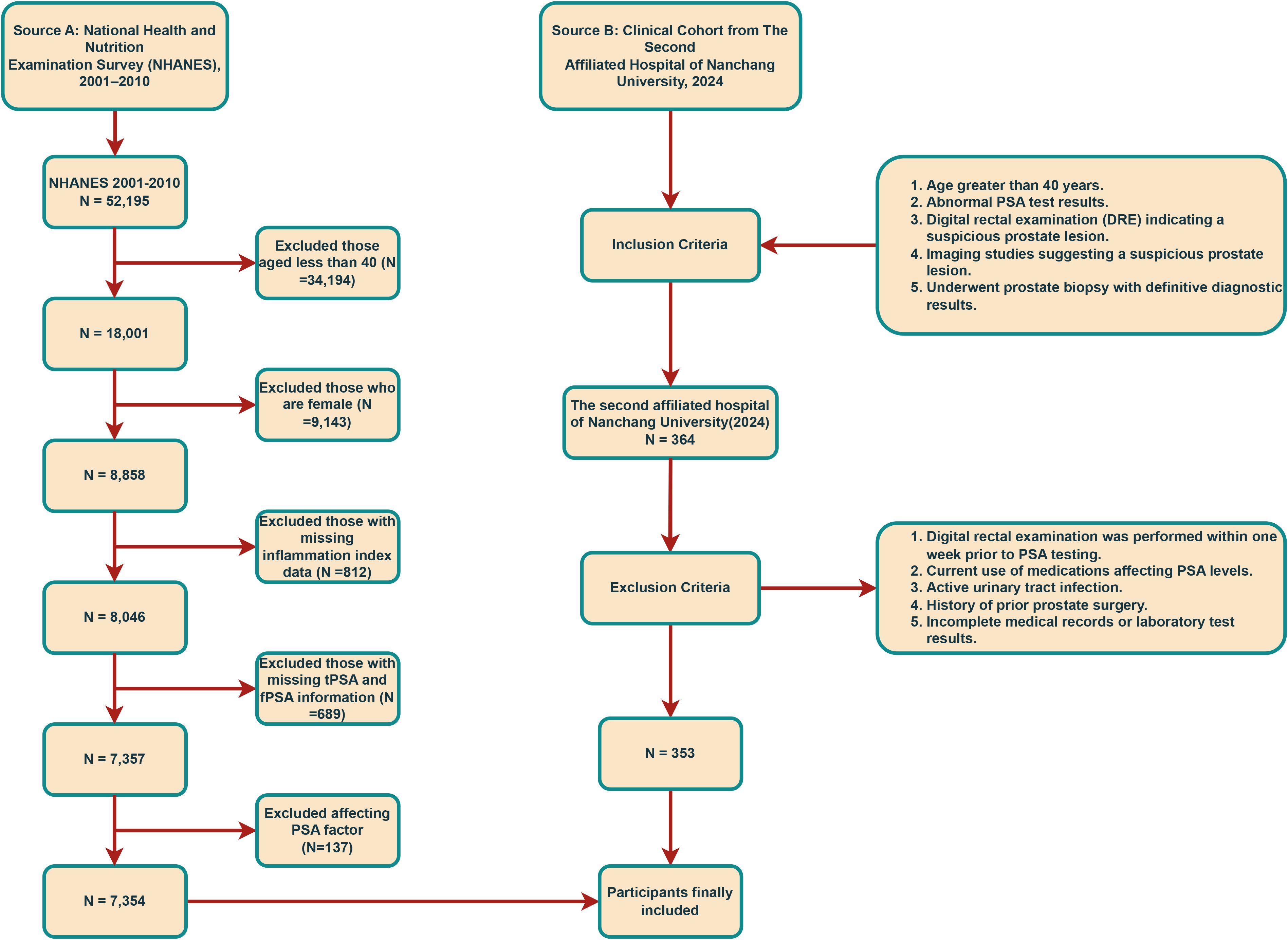
Data for this study were sourced from the NHANES, a comprehensive survey evaluating the health and nutritional status of individuals across the United States. NHANES ensures data quality through self-reported diagnoses, medical record validation, embedded validation questions, and regular quality control checks. We included 52,195 participants from the 2001–2010 cycles, excluding those with incomplete data (N=45,841), resulting in 7,354 male participants (514 were classified as PCa, 6,840 without). The participant selection process is detailed in Figure 1. The study protocol was approved by the NCHS Ethics Review Board, and written informed consent was secured from all participants. No additional external ethical approval was required. Data and study details are available at https://www.cdc.gov/nchs/nhanes.
Clinical cohort
To validate findings, we analyzed retrospective data from 353 patients undergoing prostate biopsy at the East Lake Campus of the Second Affiliated Hospital of Nanchang University in 2024 (175 with PCa, 178 without). Patients were classified as PCa or Non-PCa based on biopsy-confirmed pathology (prostate acinar adenocarcinoma or variants). Ethics approval for the study was granted by the Ethics Committee of the principal investigator’s institution, The Second Affiliated Hospital of Nanchang University (IIT-O-2025-275). Written informed consent was obtained from all participants at admission.
Inflammatory biomarker definitions
Inflammatory indices were calculated from peripheral blood data. For the NHANES cohort, complete blood counts provided neutrophil, monocyte, lymphocyte, and platelet counts. For the clinical cohort, indices were derived from routine blood tests at initial consultation. The six biomarkers were calculated as follows:
PIV: (Monocyte count × Platelet count × Neutrophil count) ÷ Lymphocyte count
SII: (Platelet count × Neutrophil count) ÷ Lymphocyte count
NLR: Neutrophil count ÷ Lymphocyte count
LMR: Lymphocyte count ÷ Monocyte count
SIRI: (Monocyte count × Neutrophil count) ÷ Lymphocyte count
PLR: Platelet count ÷ Lymphocyte count
Prostate cancer assessment
In NHANES, high-risk PCa was defined as tPSA >10 ng/mL or tPSA 4–10 ng/mL with free-to-total PSA ratio (f/t PSA) ≤25%, based on established risk stratification criteria (11). In the clinical cohort, PCa was confirmed by biopsy pathology (acinar adenocarcinoma or variants).
Covariates
NHANES covariates included biochemical markers (e.g., alanine aminotransferase, triglycerides), poverty income ratio (PIR), age, alcohol use, body mass index (BMI), race/ethnicity, education, blood pressure, smoking status, diabetes history and marital status. Clinical cohort covariates included age, BMI, Types of health insurance, white blood cell count, urea, creatinine, total calcium, estimated glomerular filtration rate (eGFR), potassium, uric acid, and glucose, obtained from routine blood and biochemical tests at initial consultation.
Statistical analysis
This study analyzed data from both a clinical cohort and NHANES database. The clinical cohort was derived from electronic medical records, with participants excluded if any data were missing. For the NHANES dataset, missing values were imputed using the “mice” R package, and sensitivity analyses confirmed no significant differences between imputed and complete-case datasets (P > 0.05; see Supplementary Materials). To assess baseline characteristics across different groups, categorical variables were analyzed with chi - square tests, and continuous variables were examined using T - tests. The Shapiro - Wilk test was employed to evaluate the normality of continuous variables. For normally distributed continuous variables, data were presented as the mean ± standard deviation (SD); for non - normally distributed ones, the median (interquartile range [IQR]) was reported. Categorical variables were described as counts (along with their corresponding percentages). Inflammatory indices were partitioned into quartiles: Q1 (values below the 25th percentile), Q2 (ranging from the 25th to 50th percentile), Q3 (from the 50th to 75th percentile), and Q4 (above the 75th percentile). Univariate logistic regression was used to explore the crude associations between inflammatory indices and PCa. Three models were developed for multivariable logistic regression: Model 1 (unadjusted), Model 2 (adjusted for age, race, and marital status), and Model 3 (adjusted for all covariates, including clinical and laboratory parameters). Due to data availability constraints in the retrospective clinical cohort, adjustments were limited to age, BMI, and select laboratory parameters, which may introduce residual confounding. RCS regression with four knots was utilized to investigate the dose - response relationships between inflammatory indices and PCa, tPSA, and fPSA. ROC curve analysis combined with DeLong’s test was used to assess the discriminatory ability of each index for PCa risk. All statistical analyses were carried out using R software (version 4.5.1), and a two - sided P - value less than 0.05 was regarded as statistically significant.
Results
Baseline characteristics
This dual-cohort study included 7,707 participants, comprising a population-based cohort from NHANES (N=7,354) and an independent clinical cohort (N=353) to validate findings and enhance generalizability.
In the NHANES cohort, 514 participants (7.0%) were classified as PCa. Table 1 summarizes baseline characteristics, revealing significant differences between Non-PCa and PCa groups. Participants with PCa were older (median age: 71.0 vs. 57.0 years, P<0.001). Five inflammatory indices were significantly increased in the PCa group compared to controls (all P<0.001): NLR (2.44 vs. 2.07), SII (549 vs. 484), PLR (136 vs. 122), SIRI (1.33 vs. 1.13), and PIV (299 vs. 265). Conversely, LMR was lower in the PCa group (3.12 vs. 3.50, P< 0.001), indicating a heightened inflammatory state. tPSA and fPSA levels were significantly higher in the PCa group (tPSA: 6.72 vs. 0.91 ng/mL; fPSA: 1.07 vs. 0.27 ng/mL, P<0.001).
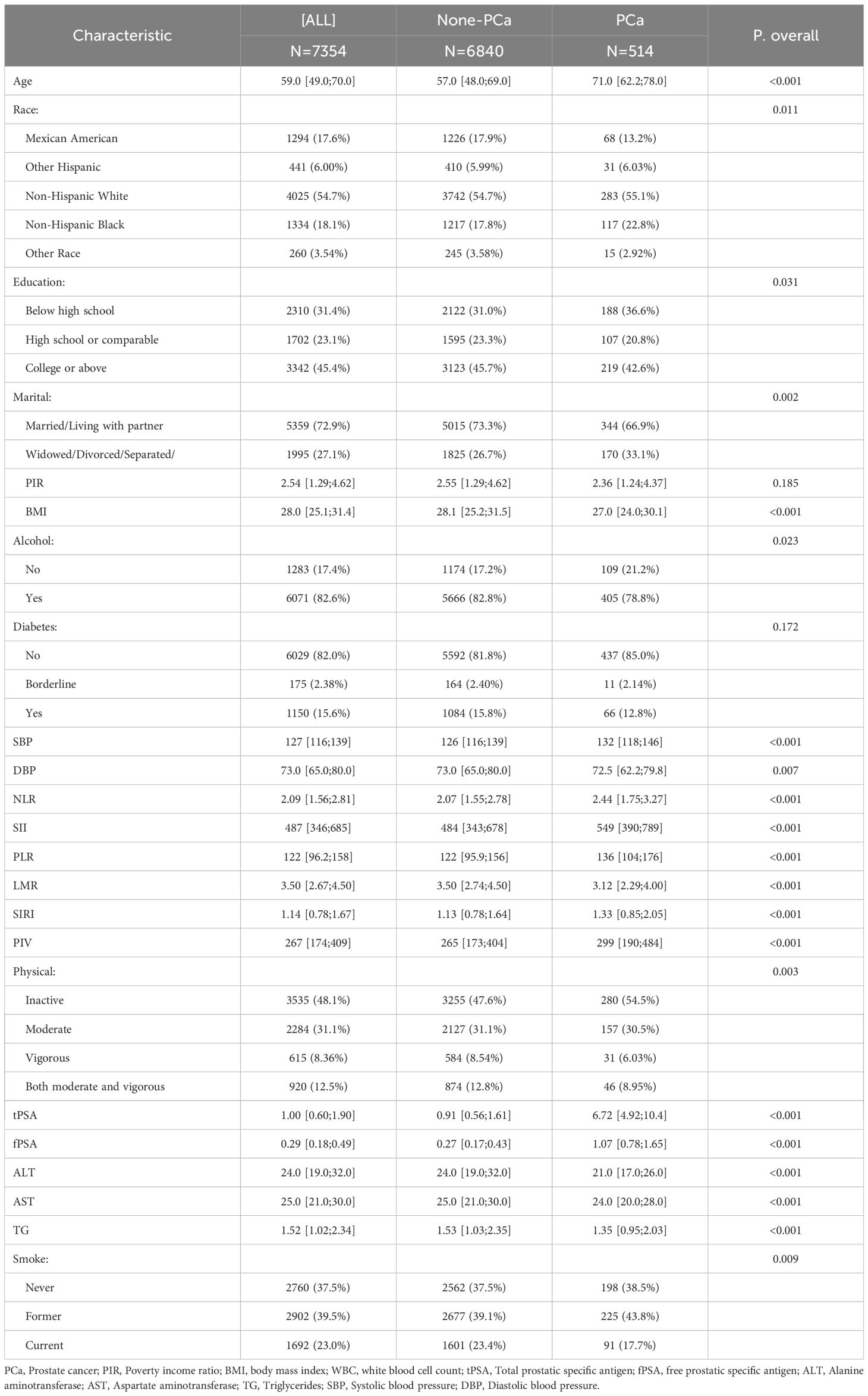
Table 1. Baseline characteristics of study participants based on the presence of prostate cancer from NHANES 2001-2010.
The clinical validation cohort included 353 participants, with 175 (49.6%) classified as having PCa. The groups were balanced for BMI and insurance status (P > 0.05). Consistent with NHANES findings, all inflammatory indices were significantly dysregulated in the positive outcome group (Table 2). This group exhibited higher neutrophil counts (5.33 vs. 4.30 ×10³/µL, P<0.001), monocyte counts (0.57 vs. 0.45 ×10³/µL, P<0.001), and platelet counts (224 vs. 194 ×10³/µL, P=0.001), resulting in elevated SII (865 vs. 628), NLR (3.90 vs. 3.04), PLR (155 vs. 139), SIRI (2.22 vs. 1.25), and PIV (503 vs. 272) (all P ≤ 0.001). LMR was lower (2.33 vs. 3.30, P<0.001), and PSA levels were substantially higher (tPSA: 38.1 vs. 11.0 ng/mL; fPSA: 6.21 vs. 1.69 ng/mL, P<0.001).
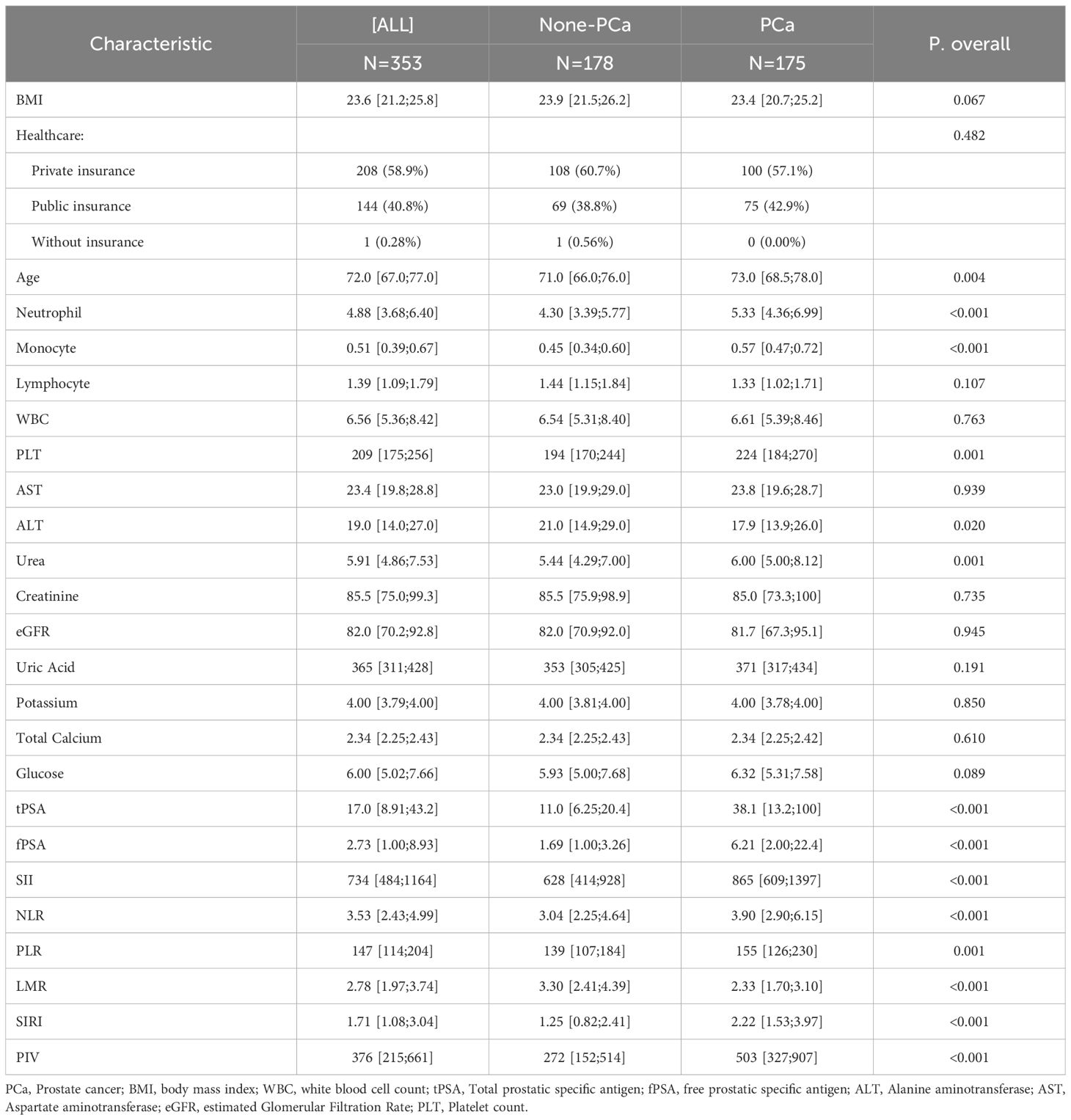
Table 2. Baseline characteristics of study participants based on the presence of prostate cancer from clinical cohort.
Associations of inflammatory indices with PCa risk
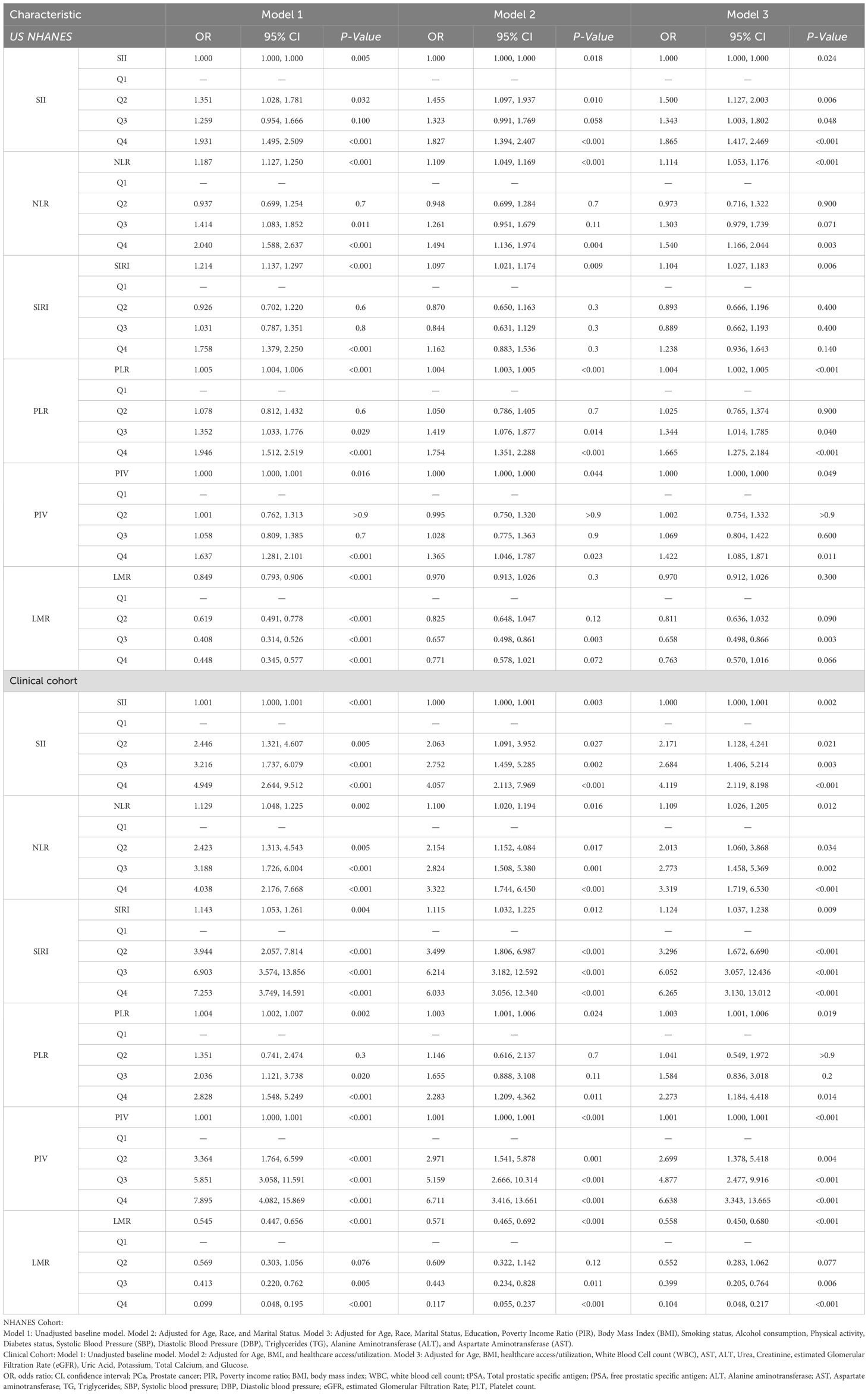
As demonstrated in Table 3, multivariable logistic regression (Model 3) revealed statistically significant associations between systemic inflammatory indices and PCa risk when evaluated as continuous variables within the NHANES cohort. Higher NLR (OR=1.114, 95% CI: 1.053–1.176, P<0.001), SIRI (OR=1.104, 95% CI: 1.027–1.183, P=0.006), and PLR (OR=1.004, 95% CI: 1.002–1.005, P<0.001) were associated with increased PCa risk. Quartile analysis (Q4 vs. Q1) revealed stronger associations: SII (OR=1.865, 95% CI: 1.417–2.469, P<0.001), NLR (OR=1.540, 95% CI: 1.166–2.044, P=0.003), PLR (OR=1.665, 95% CI: 1.275–2.184, P<0.001), and PIV (OR=1.422, 95% CI: 1.085–1.871, P=0.011) significantly increased PCa risk. The association for SIRI was attenuated in Q4 (P=0.140). Higher LMR was protective, with Q3 showing reduced PCa odds (OR=0.658, 95% CI: 0.498–0.866, P=0.003).
In the clinical cohort, associations were stronger and more consistent. All inflammatory indices, analyzed continuously in Model 3, were significantly associated with PCa: SII (OR=1.000, 95% CI: 1.000–1.001, P=0.002), NLR (OR=1.109, 95% CI: 1.026–1.205, P=0.012), SIRI (OR=1.124, 95% CI: 1.037–1.238, P=0.009), PLR (OR=1.003, 95% CI: 1.001–1.006, P=0.019), PIV (OR=1.001, 95% CI: 1.000–1.001, P<0.001), and LMR (OR=0.558, 95% CI: 0.450–0.680, P<0.001). Quartile analysis demonstrated a dose-response relationship, with Q4 showing notably elevated risks: SII (OR=4.119, 95% CI: 2.119–8.198, P<0.001), NLR (OR=3.319, 95% CI: 1.719–6.530, P<0.001), SIRI (OR=6.265, 95% CI: 3.130–13.012, P<0.001), PIV (OR=6.638, 95% CI: 3.343–13.665, P<0.001), and PLR (OR=2.273, 95% CI: 1.184–4.418, P=0.014). Higher LMR was strongly protective (Q4 vs. Q1: OR=0.104, 95% CI: 0.048–0.217, P<0.001).
Nonlinear associations with PCa risk
RCS analyses explored nonlinear associations between inflammatory indices and PCa risk (Figure 2). In the NHANES cohort (Figures 2A–F), PIV, NLR, SIRI, SII, and PLR showed positive, dose-dependent associations with PCa risk (all P-overall < 0.001). Nonlinearity was evident for PIV (P-nonlinear = 0.008) and SII (P-nonlinear = 0.002), while NLR (P-nonlinear = 0.149), SIRI (P-nonlinear = 0.254), and PLR (P-nonlinear = 0.981) were largely linear. LMR exhibited a U-shaped relationship (P-overall < 0.001, P-nonlinear < 0.001), with both low and high values associated with increased PCa risk and an optimal range linked to lower risk.
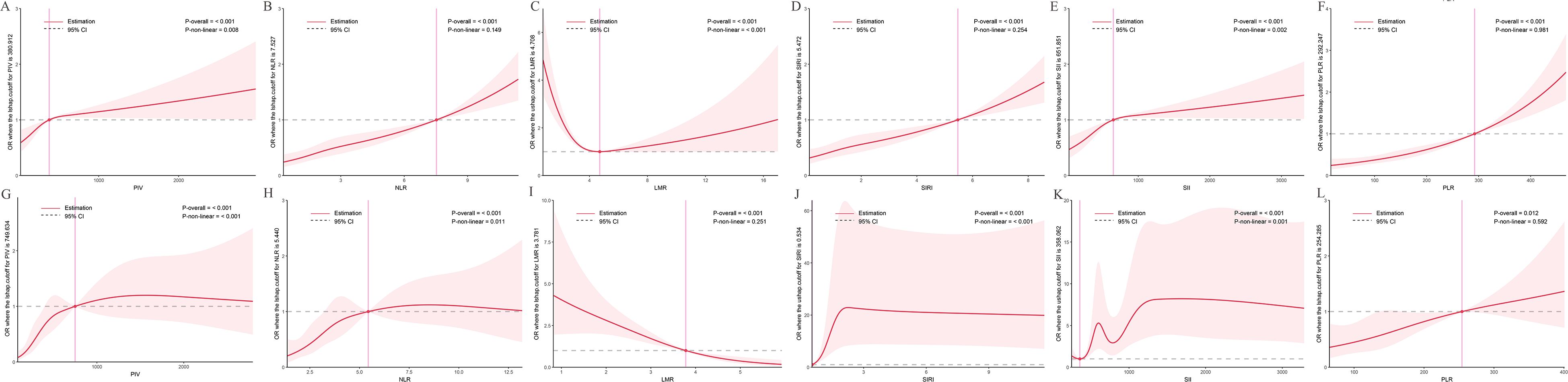
Figure 2. Dose-response relationships between inflammatory indices and PCa risk. The solid red line represents the estimated odds ratio (OR), and the shaded area indicates the 95% confidence interval (CI). The dashed horizontal gray line marks an OR of 1, indicating no association. The vertical pink line highlights a specific cutoff value or inflection point for each inflammatory index, as denoted in the Y-axis label. Statistical significance for the overall association and non-linearity is provided (P-overall and P-non-linear, respectively). (A-F) Data derived from the NHANES cohort. (G-L) Data derived from the independent clinical validation cohort. (A) PIV in NHANES; (B) NLR in NHANES; (C) LMR in NHANES; (D) SIRI in NHANES; (E) SII in NHANES; (F) PLR in NHANES; (G) PIV in clinical cohort; (H) NLR in clinical cohort; (I) LMR in clinical cohort; (J) SIRI in clinical cohort; (K) SII in clinical cohort; (L) PLR in clinical cohort.
In the clinical cohort (Figures 2G–L), similar patterns were observed, reinforcing NHANES findings. PIV (P-overall < 0.001, P-nonlinear < 0.001), NLR (P-overall < 0.001, P-nonlinear = 0.011), SIRI (P-overall < 0.001, P-nonlinear < 0.001), and SII (P-overall < 0.001, P-nonlinear = 0.001) showed increasing trends with PCa risk. PLR exhibited a weaker but significant association (P-overall = 0.012, P-nonlinear = 0.592). LMR displayed a negative, nonlinear relationship (P-overall < 0.001, P-nonlinear = 0.251), with lower values linked to higher risk, though the U-shape was less pronounced than in NHANES.
Nonlinear associations with tPSA and fPSA
RCS analyses assessed relationships between inflammatory indices and tPSA levels (Figure 3). In the NHANES cohort (Figures 3A–F), LMR (P-overall < 0.001, P-nonlinear < 0.001), NLR (P-overall < 0.001, P-nonlinear < 0.001), SIRI (P-overall < 0.001, P-nonlinear < 0.001), and SII (P-overall < 0.001, P-nonlinear < 0.001) exhibited U-shaped associations, with minimum tPSA levels at intermediate values (LMR ~4.070, NLR ~1.522, SIRI ~0.743, SII ~264.027). PIV (P-overall < 0.001, P-nonlinear < 0.001) showed a complex pattern, with tPSA initially decreasing, then rising steeply, and plateauing. PLR (P-overall < 0.001, P-nonlinear = 0.762) displayed a linear increase with tPSA.
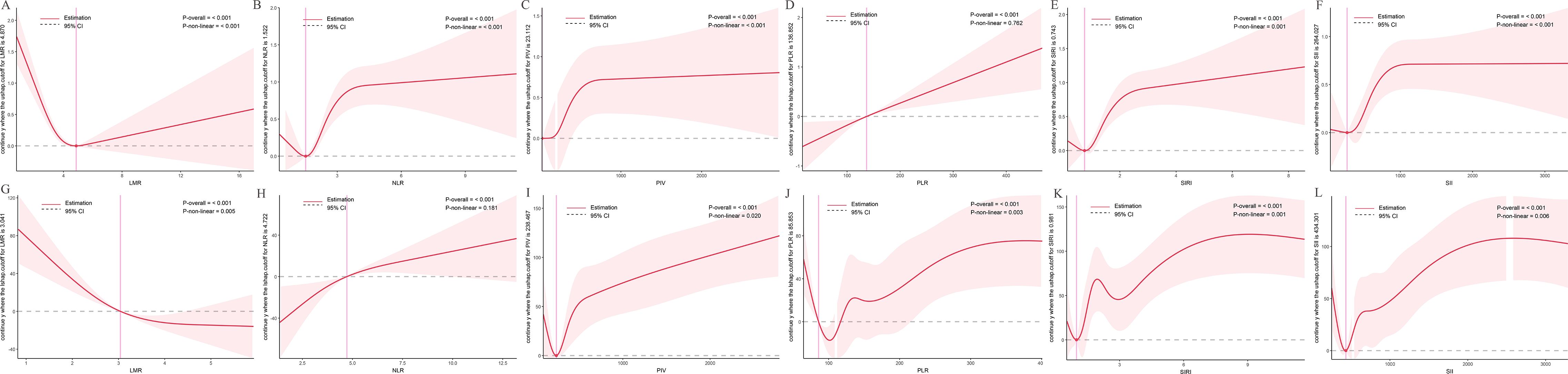
Figure 3. Dose-response relationships between inflammatory indices and total prostate-specific antigen (tPSA) levels. The solid red line represents the estimated spline function value, reflecting the continuous association with tPSA levels. The shaded area indicates the 95% confidence interval (CI). The dashed horizontal gray line at Y=0 indicates the reference point, meaning no estimated change in tPSA levels. The vertical pink line highlights a specific cutoff value or inflection point for each inflammatory index, with its corresponding estimated Y value denoted in the Y-axis label. Statistical significance for the overall association and non-linearity is provided (P-overall and P-non-linear, respectively). (A-F) Data derived from the NHANES cohort. (G-L) Data derived from the independent clinical validation cohort. (A) LMR in NHANES; (B) NLR in NHANES; (C) PIV in NHANES; (D) PLR in NHANES; (E) SIRI in NHANES; (F) SII in NHANES; (G) LMR in clinical cohort; (H) NLR in clinical cohort; (I) PIV in clinical cohort; (J) PLR in clinical cohort; (K) SIRI in clinical cohort; (L) SII in clinical cohort.
In the clinical cohort (Figures 3G–L), LMR (P-overall < 0.001, P-nonlinear = 0.005) showed an inverse relationship, with lower LMR linked to higher tPSA. NLR (P-overall < 0.001, P-nonlinear = 0.181) exhibited a U-shaped trend. PIV (P-overall < 0.001, P-nonlinear = 0.020), PLR (P-overall < 0.001, P-nonlinear = 0.003), SIRI (P-overall < 0.001, P-nonlinear = 0.001), and SII (P-overall < 0.001, P-nonlinear = 0.006) showed complex nonlinear patterns, with intermediate index values linked to lower tPSA and extremes to higher tPSA.
RCS analyses for fPSA (Figure 4) revealed similar patterns. In the NHANES cohort (Figures 4A–F), LMR (P-overall < 0.001, P-nonlinear < 0.001), NLR (P-overall < 0.001, P-nonlinear < 0.001), PIV (P-overall < 0.001, P-nonlinear < 0.001), SII (P-overall < 0.001, P-nonlinear < 0.001), and SIRI (P-overall < 0.001, P-nonlinear < 0.001) showed nonlinear associations, with minimum fPSA at intermediate values (LMR ~5.033, NLR ~1.354, PIV ~141.369, SII ~297.039, SIRI ~0.658). PLR (P-overall < 0.001, P-nonlinear = 0.980) showed a linear increase.
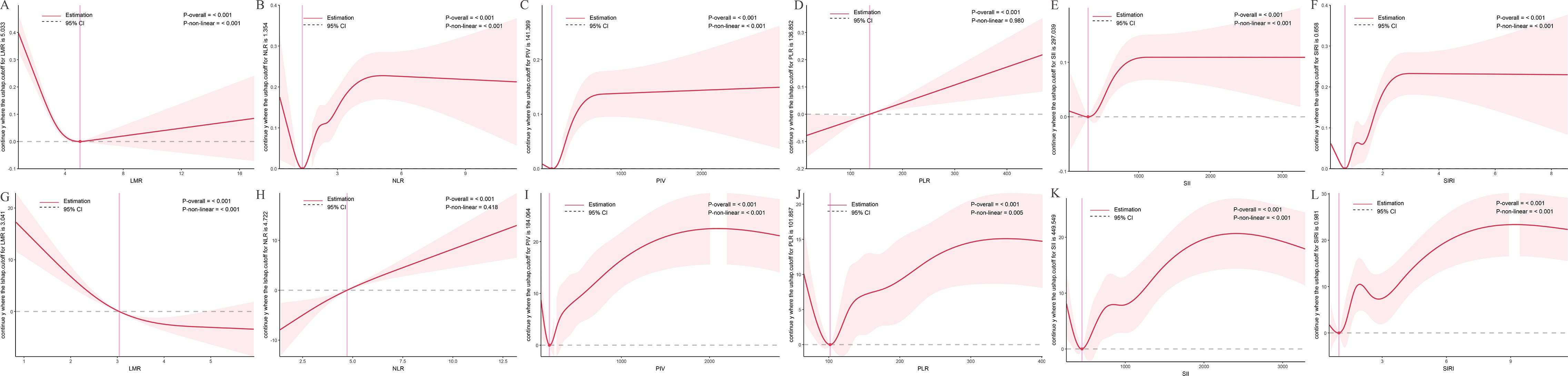
Figure 4. Dose-response relationships between inflammatory indices and free prostate-specific antigen (fPSA) levels. The solid red line represents the estimated spline function value, reflecting the continuous association with fPSA levels. The shaded area indicates the 95% confidence interval (CI). The dashed horizontal gray line at Y=0 indicates the reference point, meaning no estimated change in fPSA levels. The vertical pink line highlights a specific cutoff value or inflection point for each inflammatory index, with its corresponding estimated Y value denoted in the Y-axis label. Statistical significance for the overall association and non-linearity is provided (P-overall and P-non-linear, respectively). (A-F) Data derived from the NHANES cohort. (G-L) Data derived from the independent clinical validation cohort. (A) LMR in NHANES; (B) NLR in NHANES; (C) PIV in NHANES; (D) PLR in NHANES; (E) SII in NHANES; (F) SIRI in NHANES; (G) LMR in clinical cohort; (H) NLR in clinical cohort; (I) PIV in clinical cohort; (J) PLR in clinical cohort; (K) SII in clinical cohort; (L) SIRI in clinical cohort.
In the clinical cohort (Figures 4G–L), LMR (P-overall < 0.001, P-nonlinear < 0.001) exhibited an inverse relationship, with lower LMR linked to higher fPSA. NLR (P-overall < 0.001, P-nonlinear = 0.418) showed a largely linear increase. PIV (P-overall < 0.001, P-nonlinear < 0.001), PLR (P-overall < 0.001, P-nonlinear = 0.005), SII (P-overall < 0.001, P-nonlinear < 0.001), and SIRI (P-overall < 0.001, P-nonlinear < 0.001) displayed complex nonlinear patterns, with fPSA decreasing initially, then increasing and plateauing at higher values.
Predictive performance of inflammatory indices
ROC curve analysis evaluated the discriminatory ability of inflammatory indices for PCa risk (Figure 5). In the NHANES cohort (Figures 5A, B), where PCa diagnosis was derived from tPSA and fPSA levels, tPSA [area under the curve (AUC) = 0.991, 95% CI: 0.989–0.993] and fPSA (AUC=0.940, 95% CI: 0.933–0.948) demonstrated high discriminatory power. However, this might introduce statistical bias given the methods of PCa ascertainment. Among the inflammatory indices, AUC values ranged from 0.407 to 0.587, with NLR showing the highest discriminatory ability (AUC=0.587, 95% CI: 0.560–0.614), followed by PLR (0.581, 95% CI: 0.555–0.608), SII (0.570, 95% CI: 0.543–0.597), SIRI (0.569, 95% CI: 0.542–0.597), and PIV (0.558, 95% CI: 0.531–0.585). LMR had the lowest AUC (0.407, 95% CI: 0.380–0.434; P<0.001 vs. others, DeLong’s test). Significant differences included NLR vs. SII (P=0.0179) and NLR vs. PIV (P=0.0033).
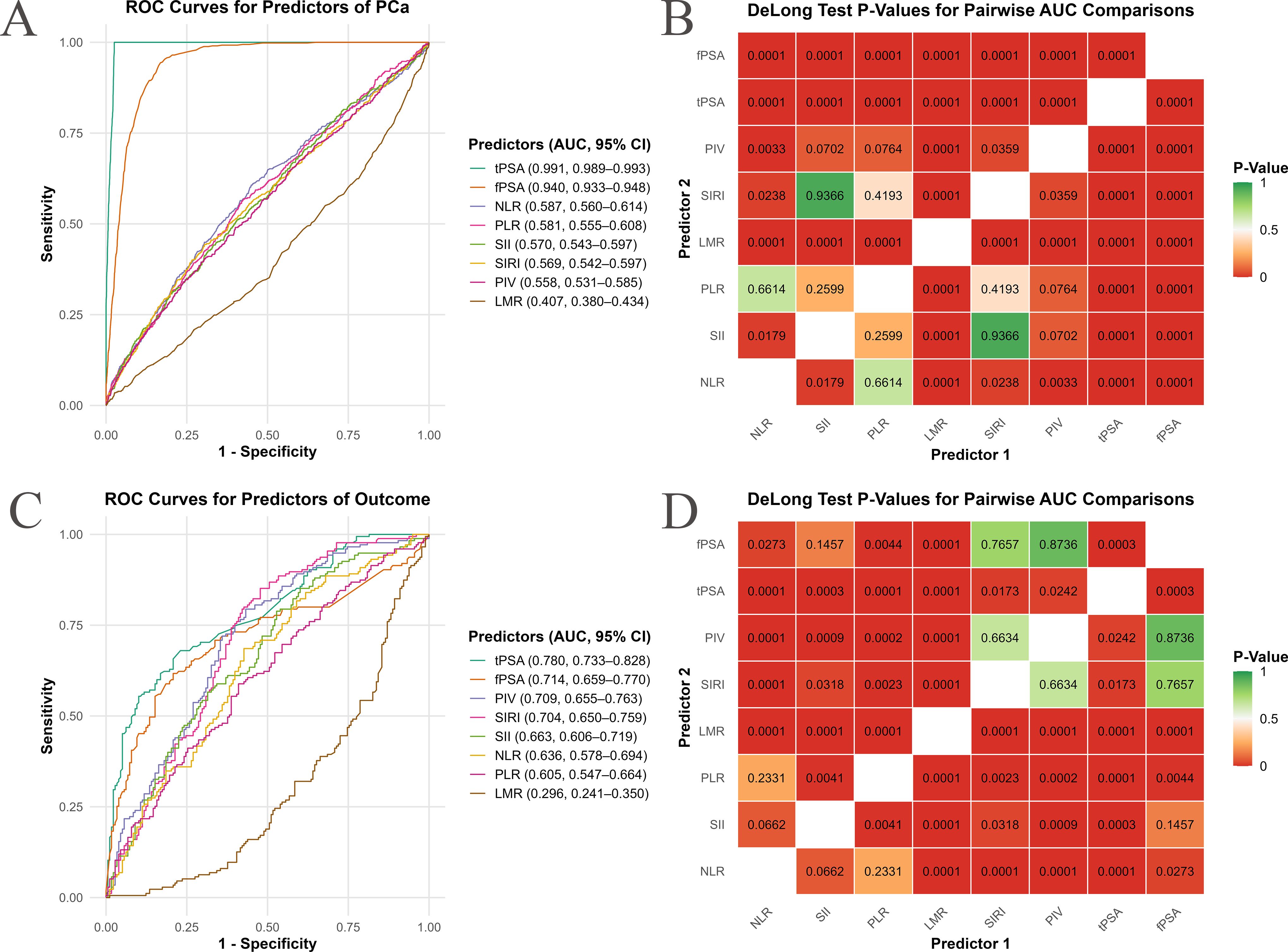
Figure 5. Receiver operating characteristic (ROC) curve analysis of inflammatory indices for PCa risk prediction. This figure assesses the predictive performance of inflammatory indices for PCa risk using ROC curve analysis and Area Under the Curve (AUC) comparisons. (A, C) show ROC curves for individual inflammatory indices, plotting sensitivity (true positive rate) against 1-specificity (false positive rate). Each colored line represents an inflammatory index, with its AUC value and 95% confidence interval (CI) listed in the subpanel legend. Curves shifted higher and to the left indicate superior discriminatory ability. (B, D) present heatmaps of P-values from DeLong’s test for pairwise AUC comparisons between inflammatory indices. Each cell displays the P-value for the comparison of their AUCs. The color scale ranges from green (P ≥ 0.05, non-significant difference) through orange (intermediate P-values) to red (P < 0.05, significant difference). (A, B) represent data from the NHANES cohort, while (C, D) are from an independent clinical validation cohort.
In the clinical cohort (Figures 5C, D), predictive performance was stronger, tPSA showed an AUC of 0.780 (95% CI: 0.733–0.828), and fPSA had an AUC of 0.714 (95% CI: 0.659–0.770). PIV had the highest AUC (0.709, 95% CI: 0.655–0.763), followed by SIRI (0.704, 95% CI: 0.650–0.759), SII (0.663, 95% CI: 0.606–0.719), NLR (0.636, 95% CI: 0.578–0.694), and PLR (0.605, 95% CI: 0.547–0.664). LMR again showed poor performance (AUC=0.296, 95% CI: 0.241–0.350; P<0.001 vs. others). PIV outperformed most indices except SIRI (P=0.6634), and SIRI outperformed SII (P=0.0318), NLR (P=0.0023), and PLR (P=0.0001). While tPSA demonstrated a higher AUC, it did not exhibit a distinctly superior advantage when compared to inflammatory indices such as PIV and SIRI. Furthermore, the performance of PIV and fPSA did not show a significant statistical difference (DeLong test P=0.7657).
Discussion
This dual-cohort study provides evidence linking systemic inflammatory biomarkers (SII, SIRI, PIV, NLR, LMR, and PLR) to PCa risk and PSA parameters (tPSA and fPSA). By integrating population-based data from the NHANES with a biopsy-confirmed clinical cohort from a tertiary hospital in China, we found that elevated pro-inflammatory indices (SII, PIV, SIRI, PLR, NLR) were associated with increased PCa risk, while higher LMR was protective. These associations were more pronounced in the clinical cohort, suggesting a stronger role for systemic inflammation in PCa. Crucially, RCS analyses unveiled intricate nonlinear relationships, offering a nuanced perspective on the dynamic interplay between systemic inflammation and PCa immunobiology, which extends beyond conventional linear assumptions and represents a key novel finding of our study.
In the clinical cohort, SIRI (OR=6.265, 95% CI: 3.130–13.012) and PIV (OR=6.638, 95% CI: 3.343–13.665) in the highest quartile showed the strongest associations with PCa risk, indicating a dose-response effect. Conversely, higher LMR was protective (Q4 vs. Q1: OR=0.104, 95% CI: 0.048–0.217). These findings align with a prior study that reported a 33% increased PCa risk with elevated SIRI (OR=2.57, 95% CI: 1.86–3.54) (12) and suggest that composite indices like SIRI and PIV, which incorporate monocyte counts, may reflect the critical role of monocyte-derived tumor-associated macrophages in PCa progression (13, 14). The protective effect of LMR underscores the importance of lymphocyte-mediated immune surveillance in counteracting pro-tumorigenic inflammation, consistent with the cancer-immunity cycle (6, 15). SIRI and PIV outperform simpler two-cell ratios (e.g., NLR, PLR) likely due to their integration of broader immune cell dynamics, particularly monocytes, which infiltrate the tumor microenvironment (TME) and differentiate into TAMs. These TAMs, often M2-like, promote tumor growth through pro-angiogenic factors, growth factors, and immunosuppressive cytokines, and are enriched in advanced PCa (16, 17). Specifically, M2-like TAMs orchestrate immune evasion by secreting arginase-1 and IL-10, thereby inhibiting T-cell function, and actively participate in extracellular matrix remodeling, facilitating PCa invasion and metastasis (18, 19). The CSF1/CSF1R signaling pathway, a key driver of monocyte recruitment and TAM differentiation, underscores the mechanistic basis for these associations (20, 21).
Systemic inflammation shapes the PCa tumor microenvironment through immune cell interactions and signaling pathways (22–26). Neutrophils, which contribute to NLR, SII, SIRI, and PIV, promote tumor progression via neutrophil extracellular traps and pro-angiogenic factors (27). In contrast, platelets, which contribute to PLR and SII, support tumor cell survival and metastasis (28, 29). Monocytes, reflected in SIRI, PIV, and LMR, differentiate into TAMs that secrete pro-inflammatory cytokines (e.g., IL-6, TNF-α) and immunosuppressive signals (e.g., PD-L1) (30, 31). The NF-κB pathway, activated by inflammatory cascades, drives expression of these cytokines and chemokines, sustaining a pro-tumorigenic TME and promoting epithelial-mesenchymal transition in PCa (32). Beyond its role in TME regulation, persistent activation of NF-κB in PCa cells themselves promotes their survival, proliferation, and resistance to apoptosis, forming a critical feedback loop with inflammatory mediators (33, 34). Additionally, emerging evidence suggests that microbial dysbiosis in the gut-prostate axis may exacerbate systemic inflammation via regulatory T cell modulation and androgen receptor signaling, offering a potential mechanistic link warranting further investigation (35, 36).
Moreover, inflammatory biomarkers such as SIRI and PIV may interact with genetic alterations, such as PTEN loss, which are prevalent in PCa. For instance, PTEN loss promotes an inflammatory tumor microenvironment via PI3K/AKT signaling, potentially amplifying the effects of TAM-derived cytokines (e.g., IL-6) (16, 37). Integrating these biomarkers with genomic profiling could enhance risk stratification and guide targeted therapies. However, our study lacked genomic data, precluding direct analysis of these interactions and necessitating future research.
The U-shaped association of LMR with PCa risk, revealed by RCS analyses, suggests dual roles for inflammation. Low LMR, often indicative of lymphopenia or elevated monocyte counts, may reflect indeed reflect a state of immune suppression, where reduced lymphocyte-mediated anti-tumor surveillance allows for PCa initiation and progression (38, 39). Conversely, an excessively high LMR, driven by significant lymphocyte proliferation or a severe reduction in monocytes, could also represent a dysregulated immune response (40). While lymphocytes are typically anti-tumorigenic, an extreme proliferation might indicate chronic immune activation or an ongoing inflammatory process (e.g., certain chronic infections or autoimmune conditions) that paradoxically fuels tumorigenesis (41, 42). Such persistent immune imbalances can lead to the generation of genotoxic substances, contribute to chronic tissue repair and proliferation, or even trigger compensatory immune responses that ultimately create a pro-tumorigenic environment (41). Similarly, nonlinear relationships with tPSA and fPSA (e.g., minimum tPSA at LMR ~4.07) suggest that inflammatory states modulate PSA dynamics, potentially complicating PSA-based screening. These findings underscore the need for context-specific biomarker thresholds in PCa risk assessment.
ROC analyses demonstrated moderate discriminatory ability for PIV (AUC=0.709, 95% CI: 0.655–0.763) and SIRI (AUC=0.704, 95% CI: 0.650–0.759) in the clinical cohort, outperforming simpler indices like NLR and PLR. These composite biomarkers show promise for risk stratification, particularly for PCa, where PSA and multiparametric magnetic resonance imaging (mpMRI) have limitations, especially in the PSA “gray zone”. Integrating inflammatory biomarkers with PSA or mpMRI may enhance diagnostic accuracy and guide personalized immunotherapeutic strategies, such as targeting NF-κB or tumor-associated macrophages via CDK12/13 inhibition or CD47 blockade.
This study has limitations. The cross-sectional NHANES design limits causal inference, and reliance on PSA-based PCa definitions may introduce misclassification bias, as undiagnosed cases may be present in the non-PCa group, contributing to a milder disease spectrum. Furthermore, the exceptionally high AUCs for tPSA and fPSA in the NHANES cohort are closely tied to this cohort’s PCa definition, which may require validation with large-scale prospective data in the future. The stark PCa prevalence difference (7.0% in NHANES vs. 49.6% in the clinical cohort) reflects distinct populations, with the clinical cohort representing a pre-selected, high-risk group with more advanced disease, driving stronger associations and better predictive performance (spectrum effect). Inconsistent covariate adjustment—socioeconomic factors in NHANES versus clinical biochemical markers in the clinical cohort—may introduce residual confounding and limit direct comparisons. Ethnic differences between U.S. and Chinese populations may influence biomarker levels, and single-timepoint measurements may not capture dynamic inflammatory changes. Unmeasured confounders, such as infections or medications, could also affect results. Additionally, the clinical cohort did not fully collect all clinical variables, such as Gleason score, precluding in-depth exploration of associations between clinically significant PCa (Gleason Score ≥7) and inflammatory indices.
The strong associations of monocyte-inclusive indices (SIRI, PIV) with PCa risk and PSA levels suggest interplay with PCa’s endocrine drivers. Inflammatory cytokines (e.g., IL-6, TNF-α), often secreted by TAMs, modulate AR signaling, promoting AR overexpression in low-androgen environments and potentially driving castration-resistant PCa (39, 43). These findings suggest that targeting inflammatory pathways, particularly TAM-related mechanisms, could disrupt inflammation–endocrine crosstalk, offering novel therapeutic avenues.
Future research should pay more attention on prospective, multi-center studies to validate these biomarkers and establish clinical thresholds, accounting for ethnic variability and nonlinear associations. Integrating inflammatory biomarkers with genomic profiling or tumor microenvironment analyses could enhance risk stratification and personalization. To further elucidate the causal relationships and therapeutic potential of these pathways, interventional studies targeting inflammatory pathways are needed, which may include the administration of anti-inflammatory drugs, lifestyle modifications, or immunotherapies. Exploring the gut-prostate axis and its impact on systemic inflammation may further elucidate PCa pathogenesis and inform novel immunotherapeutic approaches.
Conclusion
This study demonstrates consistent associations between systemic inflammatory biomarkers (SII, SIRI, PIV, NLR, PLR, LMR) and PCa risk, with nonlinear relationships to tPSA and fPSA, highlighting their potential as non-invasive tools for risk stratification. The superior performance of SIRI and PIV in the clinical cohort underscores the value of composite indices in capturing complex immune dynamics. These findings advance our understanding of inflammation-driven PCa and support the integration of inflammatory biomarkers into diagnostic and therapeutic strategies. Future research should focus on prospective validation, mechanistic elucidation, and targeted interventions to modulate inflammation for PCa prevention and management.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
All procedures of the National Health and Nutrition Examination Survey (NHANES) involving human participants, materials, or data were conducted in accordance with the Declaration of Helsinki and approved by the National Center for Health Statistics (NCHS) Ethics Review Board (Protocol #2011-17). Written informed consent was obtained from all participants. The study did not require clinical trial registration, and the survey data are publicly accessible for researchers and data users worldwide at https://www.cdc.gov/nchs/nhanes. Ethics approval for the study was granted by the Ethics Committee of the principal investigator's institution, The Second Affiliated Hospital of Nanchang University (IIT-O-2025-275). Written informed consent was obtained from all participants at admission.
Author contributions
GH: Writing – original draft, Data curation, Visualization, Formal Analysis, Methodology, Conceptualization, Writing – review & editing. KX: Writing – original draft, Writing – review & editing, Methodology. SL: Writing – review & editing, Conceptualization, Writing – original draft, Project administration, Formal Analysis, Methodology. XL: Validation, Conceptualization, Writing – review & editing, Writing – original draft, Supervision, Project administration.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was funded by the National Natural Science Foundation of China (No. 82060465) and the Natural Science Foundation of Jiangxi Province (No. 20212ACB206023).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2025.1697617/full#supplementary-material
References
1. Siegel RL, Miller KD, Wagle NS, and Jemal A. Cancer statistics, 2023. CA Cancer J Clin. (2023) 73:17–48. doi: 10.3322/caac.21763
2. He R, Ye Y, Zhu Q, and Xie C. Systemic immune-inflammation index is associated with high risk for prostate cancer among the U.S. elderly: Evidence from NHANES 2001-2010. Front Oncol. (2024) 14:1441271. doi: 10.3389/fonc.2024.1441271
3. Grönberg H, Adolfsson J, Aly M, Nordström T, Wiklund P, Brandberg Y, et al. Prostate cancer screening in men aged 50–69 years (STHLM3): A prospective population-based diagnostic study. Lancet Oncol. (2015) 16:1667–76. doi: 10.1016/S1470-2045(15)00361-7
4. Sequeira JP, Salta S, Freitas R, López-López R, Díaz-Lagares Á, Henrique R, et al. Biomarkers for pre-treatment risk stratification of prostate cancer patients: A systematic review. Cancers (Basel). (2024) 16:1363. doi: 10.3390/cancers16071363
5. Keibel A, Singh V, and Sharma MC. Inflammation, microenvironment, and the immune system in cancer progression. Curr Pharm Des. (2009) 15:1949–55. doi: 10.2174/138161209788453167
6. Wen Y, Zhu Y, Zhang C, Yang X, Gao Y, Li M, et al. Chronic inflammation, cancer development and immunotherapy. Front Pharmacol. (2022) 13:1040163. doi: 10.3389/fphar.2022.1040163
7. Schwartsburd PM. Chronic inflammation as inductor of pro-cancer microenvironment: Pathogenesis of dysregulated feedback control. Cancer Metastasis Rev. (2003) 22:95–102. doi: 10.1023/a:1022220219975
8. Patrzałek P, Froń A, Mielczarek M, Karwacki J, Lesiuk G, Janczak D, et al. Inflammatory-based prognostic indicators in prostate cancer: Evaluating NLR, PLR, and SII in relation to cambridge and ISUP classifications. Front Oncol. (2025) 15:1595000. doi: 10.3389/fonc.2025.1595000
9. Zhou M, Gu Q, Zhou M, Yang S, Liu Y, Guan B, et al. Extensive study on the associations of 12 composite inflammatory indices with colorectal cancer risk and mortality a cross-sectional analysis of NHANES 2001-2020. Int J Surg. (2025). doi: 10.1097/JS9.0000000000002996
10. Yao W, Wu J, Kong Y, Xu F, Zhou Y, Sun Q, et al. Associations of systemic immune-inflammation index with high risk for prostate cancer in middle-aged and older US males: A population-based study. Immun Inflammation Dis. (2024) 12:e1327. doi: 10.1002/iid3.1327
11. Flynn-Evans EE, Mucci L, Stevens RG, and Lockley SW. Shiftwork and prostate-specific antigen in the national health and nutrition examination survey. J Natl Cancer Inst. (2013) 105:1292–7. doi: 10.1093/jnci/djt169
12. Xiao Y, Tang B, Wang J, Cai Z, An H, and Tao N. Study of the interaction between cardiometabolic index and inflammatory index on the risk of prostate cancer development. Front Immunol. (2025) 16:1591879. doi: 10.3389/fimmu.2025.1591879
13. Shirotake S, Miyajima A, Kosaka T, Tanaka N, Kikuchi E, Mikami S, et al. Regulation of monocyte chemoattractant protein-1 through angiotensin II type 1 receptor in prostate cancer. Am J Pathol. (2012) 180:1008–16. doi: 10.1016/j.ajpath.2011.11.027
14. Zhou J-W, Mao Y-H, Liu Y, Liang H-T, Samtani CC, Fu Y-W, et al. A novel robust nomogram based on peripheral monocyte counts for predicting lymph node metastasis of prostate cancer. Asian J Androl. (2021) 23:409–14. doi: 10.4103/aja.aja_89_20
15. Akkız H, Şimşek H, Balcı D, Ülger Y, Onan E, Akçaer N, et al. Inflammation and cancer: Molecular mechanisms and clinical consequences. Front Oncol. (2025) 15:1564572. doi: 10.3389/fonc.2025.1564572
16. Fang B, Lu Y, Li X, Wei Y, Ye D, Wei G, et al. Targeting the tumor microenvironment, a new therapeutic approach for prostate cancer. Prostate Cancer Prostatic Dis. (2025) 28:260–9. doi: 10.1038/s41391-024-00825-z
17. Yang S, Wang M, Hua Y, Li J, Zheng H, Cui M, et al. Advanced insights on tumor-associated macrophages revealed by single-cell RNA sequencing: The intratumor heterogeneity, functional phenotypes, and cellular interactions. Cancer Lett. (2024) 584:216610. doi: 10.1016/j.canlet.2024.216610
18. Yang Y, Li S, To KKW, Zhu S, Wang F, and Fu L. Tumor-associated macrophages remodel the suppressive tumor immune microenvironment and targeted therapy for immunotherapy. J Exp Clin Cancer Res. (2025) 44:145. doi: 10.1186/s13046-025-03377-9
19. Liu R, Lu J, Liu J, Liao Y, Guo Y, Shi P, et al. Macrophages in prostate cancer: Dual roles in tumor progression and immune evasion. J Transl Med. (2025) 23:615. doi: 10.1186/s12967-025-06519-x
20. Cencini E, Sicuranza A, Ciofini S, Fabbri A, Bocchia M, and Gozzetti A. Tumor-associated macrophages in multiple myeloma: Key role in disease biology and potential therapeutic implications. Curr Oncol. (2023) 30:6111–33. doi: 10.3390/curroncol30070455
21. Strachan DC, Ruffell B, Oei Y, Bissell MJ, Coussens LM, Pryer N, et al. CSF1R inhibition delays cervical and mammary tumor growth in murine models by attenuating the turnover of tumor-associated macrophages and enhancing infiltration by CD8+ T cells. Oncoimmunology. (2013) 2:e26968. doi: 10.4161/onci.26968
22. Hao Q, Vadgama JV, and Wang P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun Signal. (2020) 18:82. doi: 10.1186/s12964-020-00589-8
23. Chen H, Pang B, Zhou C, Han M, Gong J, Li Y, et al. Prostate cancer-derived small extracellular vesicle proteins: The hope in diagnosis, prognosis, and therapeutics. J Nanobiotechnology. (2023) 21:480. doi: 10.1186/s12951-023-02219-0
24. Hashemi Karoii D, Bavandi S, Djamali M, and Abroudi AS. Exploring the interaction between immune cells in the prostate cancer microenvironment combining weighted correlation gene network analysis and single-cell sequencing: An integrated bioinformatics analysis. Discov Oncol. (2024) 15:513. doi: 10.1007/s12672-024-01399-x
25. Murase K, Kawase M, Ebara S, Tatenuma T, Sasaki T, Ikehata Y, et al. The negative impact of inflammation-related parameters in prostate cancer after robot-assisted radical prostatectomy: A retrospective multicenter cohort study in Japan (the MSUG94 group). J Clin Med. (2023) 12:7732. doi: 10.3390/jcm12247732
26. Cao H, Zhang D, Wang P, Wang Y, Shi C, Wu H, et al. Gut microbiome: A novel preventive and therapeutic target for prostatic disease. Front Cell Infect Microbiol. (2024) 14:1431088. doi: 10.3389/fcimb.2024.1431088
27. Zhou Y, Shen G, Zhou X, and Li J. Therapeutic potential of tumor-associated neutrophils: Dual role and phenotypic plasticity. Signal Transduct Target Ther. (2025) 10:178. doi: 10.1038/s41392-025-02242-7
28. Braun A, Anders H-J, Gudermann T, and Mammadova-Bach E. Platelet-cancer interplay: Molecular mechanisms and new therapeutic avenues. Front Oncol. (2021) 11:665534. doi: 10.3389/fonc.2021.665534
29. Liao K, Zhang X, Liu J, Teng F, He Y, Cheng J, et al. The role of platelets in the regulation of tumor growth and metastasis: The mechanisms and targeted therapy. MedComm (2020). (2023) 4:e350. doi: 10.1002/mco2.350
30. Li Y, Liu H, Zhao Y, Yue D, Chen C, Li C, et al. Tumor-associated macrophages (TAMs)-derived osteopontin (OPN) upregulates PD-L1 expression and predicts poor prognosis in non-small cell lung cancer (NSCLC). Thorac Cancer. (2021) 12:2698–709. doi: 10.1111/1759-7714.14108
31. Zhang W, Liu Y, Yan Z, Yang H, Sun W, Yao Y, et al. IL-6 promotes PD-L1 expression in monocytes and macrophages by decreasing protein tyrosine phosphatase receptor type O expression in human hepatocellular carcinoma. J Immunother Cancer. (2020) 8:e000285. doi: 10.1136/jitc-2019-000285
32. Chen L, De Menna M, Groenewoud A, Thalmann GN, Kruithof-de Julio M, and Snaar-Jagalska BE. A NF-ĸB-activin a signaling axis enhances prostate cancer metastasis. Oncogene. (2020) 39:1634–51. doi: 10.1038/s41388-019-1103-0
33. Cao Y, Yi Y, Han C, and Shi B. NF-κB signaling pathway in tumor microenvironment. Front Immunol. (2024) 15:1476030. doi: 10.3389/fimmu.2024.1476030
34. Khan A, Zhang Y, Ma N, Shi J, and Hou Y. NF-κB role on tumor proliferation, migration, invasion and immune escape. Cancer Gene Ther. (2024) 31:1599–610. doi: 10.1038/s41417-024-00811-6
35. Matsushita M, Fujita K, Hatano K, Hayashi T, Kayama H, Motooka D, et al. High-fat diet promotes prostate cancer growth through histamine signaling. Int J Cancer. (2022) 151:623–36. doi: 10.1002/ijc.34028
36. Zhong W, Wu K, Long Z, Zhou X, Zhong C, Wang S, et al. Gut dysbiosis promotes prostate cancer progression and docetaxel resistance via activating NF-κB-IL6-STAT3 axis. Microbiome. (2022) 10:94. doi: 10.1186/s40168-022-01289-w
37. Niu X, Ma J, Li J, Gu Y, Yin L, Wang Y, et al. Sodium/glucose cotransporter 1-dependent metabolic alterations induce tamoxifen resistance in breast cancer by promoting macrophage M2 polarization. Cell Death Dis. (2021) 12:509. doi: 10.1038/s41419-021-03781-x
38. Wu W, Zhao L, Wang Y, Chen P, Yuan X, Miao L, et al. Prognostic value of the peripheral blood lymphocyte/monocyte ratio combined with 18F-FDG PET/CT in patients with diffuse large B-cell lymphoma. Curr Probl Cancer. (2024) 48:101066. doi: 10.1016/j.currproblcancer.2024.101066
39. Peng H and Luo X. Prognostic significance of elevated pretreatment systemic inflammatory markers for patients with prostate cancer: A meta-analysis. Cancer Cell Int. (2019) 19:70. doi: 10.1186/s12935-019-0785-2
40. Carracedo J, Alique M, Vida C, Bodega G, Ceprián N, Morales E, et al. Mechanisms of cardiovascular disorders in patients with chronic kidney disease: A process related to accelerated senescence. Front Cell Dev Biol. (2020) 8:185. doi: 10.3389/fcell.2020.00185
41. Caro J, Braunstein M, Williams L, Bruno B, Kaminetzky D, Siegel A, et al. Inflammation and infection in plasma cell disorders: how pathogens shape the fate of patients. Leukemia. (2022) 36:613–24. doi: 10.1038/s41375-021-01506-9
42. Ponomarev AV and Shubina IZ. Insights into mechanisms of tumor and immune system interaction: association with wound healing. Front Oncol. (2019) 9:1115. doi: 10.3389/fonc.2019.01115
Keywords: prostate cancer, systemic inflammation, NHANES, PSA, NLR, SIRI
Citation: Huang G, Xiao K, Lin S and Lu X (2025) Association of systemic inflammatory biomarkers with prostate cancer risk: a population-based (NHANES) and clinical validation study. Front. Endocrinol. 16:1697617. doi: 10.3389/fendo.2025.1697617
Received: 03 September 2025; Accepted: 23 October 2025;
Published: 10 November 2025.
Edited by:
Yousheng Yao, Sun Yat-Sen Memorial Hospital, ChinaReviewed by:
Xiaoshuai Gao, Sichuan University, ChinaWenshuang Li, Third Affiliated Hospital of Sun Yat-Sen University, China
Copyright © 2025 Huang, Xiao, Lin and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuangquan Lin, ZG91YmxlbGluNDMwQDEyNi5jb20=; Xiongbing Lu, bmRlZnkwNTAyNkBuY3UuZWR1LmNu
 Guoqiang Huang
Guoqiang Huang Kaiwen Xiao
Kaiwen Xiao Shuangquan Lin
Shuangquan Lin Xiongbing Lu
Xiongbing Lu