Perspective: Microbial hydrogen metabolism in rock-hosted ecosystems
- 1Department of Microbiology and Cell Biology, Montana State University, Bozeman, MT, United States
- 2Department of Geological Sciences, University of Colorado, Boulder, CO, United States
Hydrogen (H2) is among the most common and widely utilized electron donors in microbial metabolism. This is particularly true for microorganisms that inhabit subsurface environments where H2 concentrations can be high due to H2 generation via one or more abiotic and biotic processes, such as serpentinization, radiolysis, cataclasis, and microbial fermentation. A surge in interest in the exploration for and exploitation of geologic (i.e., white and orange) H2 as a clean low carbon fuel therefore necessitates an evaluation of the influence of microorganisms on its flux and potential recovery from subsurface systems. The widespread application of high throughput metagenomic sequencing approaches to rock-hosted ecosystems now makes it possible to readily identify microorganisms that harbor the potential to metabolize H2 and to predict their mode of coupling H2 oxidation with available oxidants using comparative genomic data from natural samples alone. When combined with several recent reports of measured rates of net microbial H2 consumption in rock-hosted ecosystems, such information provides new perspective on the potential for microorganisms to impact the economics of H2 recovery from geologic systems. In this perspective, the different classes of enzymes that microorganisms use to reversibly oxidize H2 to fuel their energy metabolism are introduced and their distribution in several rock-hosted ecosystems is discussed. A compilation of net microbial H2 oxidation activities in rock-hosted ecosystems is also presented to enable estimates of potential H2 loss from natural or stimulated geologic reservoirs during mining activities, with an example provided from the Samail Ophiolite that indicates >90% of geologic H2 produced could be lost to microbial consumption. Finally, avenues to guide future microbial research in environments where geologic H2 mining is planned are discussed.
Introduction
H2 is a common electron donor (reductant) for microorganisms in rock-hosted ecosystems (Lin et al., 2005; Nealson et al., 2005; Spear et al., 2005; Telling et al., 2015; Macdonald et al., 2018; Lindsay et al., 2019), due in part, to the variety of mechanisms by which it can be produced. H2 can be generated abiotically through the reduction of water by iron minerals in mafic (e.g., basalt) and ultramafic rock (e.g., peridotite) through reactions collectively referred to as serpentinization (Figure 1A (Stevens and McKinley, 2000; Kelley et al., 2005)). Likewise, in ecosystems hosted in crystalline basement rocks (e.g., granite) that are enriched with radionuclides such as uranium and thorium, radiolytic splitting of water can lead to formation of H2 (Lin et al., 2005). Similarly, in seismically active systems (fault zones), or in subsurface environments where active rock comminution occurs (e.g., underneath active glaciers), mechanical shearing (cataclasis) of silicate minerals can produce silica radicals that can interact with water to form H2 (Kita et al., 1982; Telling et al., 2015; Macdonald et al., 2018; Parkes et al., 2019). Finally, there is also the potential for deep seated or primordial H2 to be released into subsurface aquifers (Zgonnik, 2020; Milkov, 2022). Along with biological mechanisms for H2 production (discussed below), these processes are widely thought to lead to the high concentrations of H2 commonly detected in subsurface environments (Stevens and McKinley, 1995; Zgonnik, 2020; Dunham et al., 2021; Templeton et al., 2021; Milkov, 2022). Indeed, estimates of H2 production from the above processes taking place in the marine lithosphere and continental Precambrian lithosphere are estimated at −1011 mol yr−1 (as summarized in (Sherwood Lollar et al., 2014)). Far less is known of processes that influence the flux of this H2 to the near surface.
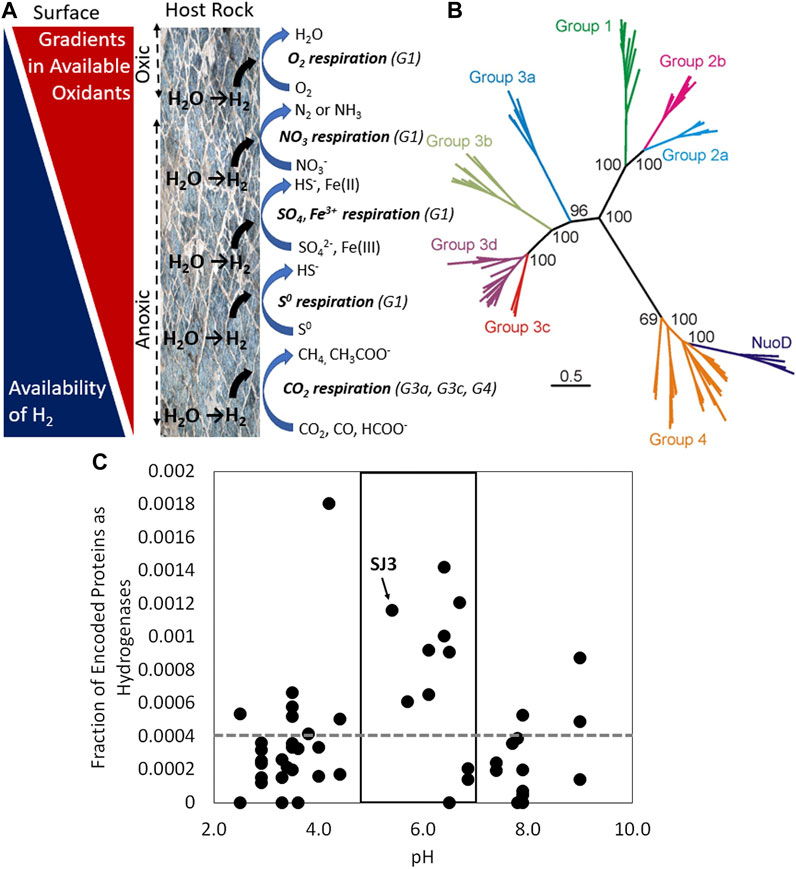
Figure 1. (A) Gradients in available oxidants and available hydrogen (H2) in a host rock unit (e.g., mafic or ultramafic rock) undergoing the geological process of serpentinization. H2-consuming microbial processes are depicted from the surface to the subsurface based on decreasing energetics associated with coupling with available oxidants. Subgroup designations (G) for [NiFe]-hydrogenases are depicted and correspond with those in panel (B). Image adapted from Boyd et al., 2014. (B) Phylogenetic reconstruction of representative large subunits of the various subgroups of [NiFe]-hydrogenases shows the general coherence of coupling of H2 oxidation to various oxidants, as indicated in panel (A). For context, representative homologs of the H subunit of the paralog of [NiFe]-hydrogenase, NADH dehydrogenase (Nuo or complex I), that are involved in various electron transport chains but that do not catalyze the reversible oxidation of H2 are included. Image adapted from Schut et al., 2013. (C) Fraction of total genes encoded in the metagenomes of 50 non-photosynthetic hot spring communities that encode hydrogenases as a function of the pH of those hot springs. [NiFe]- and [FeFe]-hydrogenase homologs were summed and are plotted together for each of the 50 metagenomes. The box highlights springs that predicted based on geochemical data to be sourced with volcanic vapor and that tend to be enriched in H2. The grey dashed line represents the average abundance of hydrogenase homologs across all 50 metagenomes. Smokejumper hot spring 3 (SJ3) is depicted. Data adapted from Lindsay et al., 2019. Abbreviations: CO2, carbon dioxide; CO, carbon monoxide; HCOO−, formate; CH4, methane, CH3COO−, acetate; S0, elemental sulfur; HS−, sulfide; SO42-, sulfate; Fe(III), ferric iron; Fe(II), ferrous iron; NO3−, nitrate; N2, dinitrogen; NH3, ammonia; O2, oxygen; H2O, water.
Microbial metabolism in subsurface environments is fuelled by chemical energy generated by dissipating gradients in electron donors and acceptors through redox reactions (Templeton and Caro, 2023). Among available electron donors, geologic H2 is often considered to play a central role in sustaining microbial metabolism in subsurface rock-hosted communities (Stevens and McKinley, 1995; Nealson et al., 2005; Brazelton et al., 2012; Gregory et al., 2019), including those recovered from deep (2.0 and 2.8 km) crystalline bedrock in Siberia and South Africa (Chivian et al., 2008; Karnachuk et al., 2019), a variety of subsurface serpentinizing environments (Brazelton et al., 2012; Schrenk et al., 2013; Templeton et al., 2021), deep (>1 km) basalt bedrock in the Columbia River Plain, U.S.A. (Stevens and McKinley, 1995), and beneath basalt-hosted glaciers and ice sheets (Macdonald et al., 2018; Dunham et al., 2021), among many others. A major reason for concluding that H2 is supporting these communities comes from high concentrations of the highly diffusive gas. For example, H2 has been measured at mM concentrations in fracture fluids from several environments undergoing serpentinization (Charlou et al., 2010; Rempfert et al., 2017). Importantly, that H2 can be measured in subsurface ecosystems indicates that the rate of its production exceeds the rate of its consumption. For microorganisms, this could be due to either the availability of reductant, such as H2, exceeding the availability of oxidants in those environments or that other nutrients (e.g., phosphorus, nitrogen, trace metals) are limiting the energy metabolism of cells. This is because microbial communities assemble to maximize the production of biomass given a finite supply of energy and/or nutrients (Konopka, 2006). In other words, microbial ecosystems are limited by a finite resource(s) (Konopka, 2006) and the availability of electron acceptors is chief among these in subsurface ecosystems (Lindsay et al., 2018). In soils, microbial activity controls the flux of trace gas to the atmosphere, including H2 (Conrad, 1996). This is accomplished due to strong gradients in the availability of electron acceptors to drive trace gas oxidation, such as atmospheric O2 (Ji et al., 2017; Giguere et al., 2021). To what extent do microorganisms oxidize H2 in more oxidant-limited subsurface environments (e.g., rock-hosted) and thereby influence the flux of H2 to the near surface? To begin to address this question, it is first necessary to understand how microorganisms metabolize H2.
Enzymes and processes involved in microbial hydrogen metabolism
Enzymes (proteins that catalyze chemical reactions) that enable the reversible oxidation of H2 to protons (H+) and electrons (e−; Reaction 1) can be traced back to the last common ancestor of Archaea and Bacteria (Boyd et al., 2014; Weiss et al., 2016), at least 3.8 billion years ago (Battistuzzi et al., 2004; Wolfe and Fournier, 2018).
Over the past −3.8 Ga, organisms with the ability to metabolize H2 have diversified to enable H2 oxidation to be coupled to the reduction of nearly all imaginable and available oxidants, including oxygen (O2), nitrate (NO3−), ferric iron (Fe(III)), sulfate (SO42−), and carbon dioxide (CO2), among others (Figures 1A,B). In this capacity, H2 can support the growth of aerobic (respire O2) or anaerobic (respire an oxidant other than O2) microorganisms and those that are autotrophic (fix inorganic carbon to biomass) or heterotrophic (use organic carbon as an electron donor and/or for biomass). In the case of heterotrophic microorganisms, H2 oxidation typically is used to supplement their energy metabolism (Peters et al., 2015; Greening et al., 2016). Alternatively, heterotrophic organisms that are oxidant limited often can ferment, leading to H2 production (discussed below).
Despite the vast diversity of metabolic backgrounds that feature reversible H2 oxidation, they are all dependent on one of three classes of enzyme to catalyze its transformation: [FeFe]-hydrogenases, [NiFe]-hydrogenases, and [Fe]-hydrogenases (or Hmd) (Peters et al., 2015; Greening et al., 2016; Greening and Boyd, 2020). Hmd catalyzes the reduction of the substrate methenyltetrahydromethanopterin with H2 in methanogens (Shima et al., 2008), and won’t be discussed more here given its limited metabolic distribution. The least ambiguous way of examining whether microbial cultures or environmental samples can catalyze reversible H2 oxidation activity is to measure rates of H2 consumption in microcosm experiments. However, this is a time-consuming task and is infrequently done (discussed below). Fortunately, like other enzymes/proteins that a cell is dependent on, hydrogenase enzymes are encoded by genes in the genomes of cells. Therefore, organisms with the ability to metabolize H2 can be readily identified by sequencing the genome of the culture (or the entire community in the case of environmental samples) and subjecting those genomes to comparative bioinformatics analyses. Such studies show that [FeFe]-hydrogenases are widely distributed among Bacteria, anaerobic eukaryotes (ciliates, flagellates, and fungi), and algae (Peters et al., 2015) and were only recently discovered in a limited subset of Archaea (Peters et al., 2015; Greening et al., 2023). In contrast, [NiFe]-hydrogenases are widespread among Archaea and Bacteria but have yet to be detected in Eukarya (Peters et al., 2015). Paralogs (evolutionarily related proteins that have different functions) of [NiFe]-hydrogenases, however, are widespread in Archaea, Bacteria, and Eukarya where they function as entry points for reducing equivalents into electron transport chains (i.e., NADH dehydrogenase, Nuo) (Friedrich and Scheide, 2000; Schut et al., 2013). However, these proteins are derived from [NiFe]-hydrogenases and therefore H2 metabolism is a trait commonly associated with microbial life rather than higher forms of life.
Common among all characterized [NiFe]- and [FeFe]-hydrogenases, but distinct between these two classes, is an evolutionarily conserved large subunit that harbors the nickel-iron-sulfur or iron-sulfur active site, respectively, where reversible H2 oxidation takes place (Peters et al., 2015). Variation in how H2 is metabolized by cells via [NiFe]- and [FeFe]-hydrogenase has occurred through recruitment of genes that encode additional protein subunits that enable diverse redox coupling with intermediate electron carriers in the cell or in the cell membranes such as ferredoxin (Fd), NADH, flavins (e.g., F420), quinones (e.g., methanophenozine), and heterodisulfide (e.g., coenzyme M-coenzyme B), among others (Thauer et al., 2010). These intermediary electron carriers pass electrons to electron transport chains specific for reduction of O2, NO3−, Fe(III), SO42−, etc. allowing for energy to be conserved that can be used to power cells. The recruitment of these additional protein encoding genes to [NiFe]- and [FeFe]-hydrogenases was highly advantageous to cells since it allowed for electrons from H2 (via these intermediary electron carriers) to enter a variety of electron transport chains thereby enabling energy to be conserved from H2 to power cells. As such, these particular protein recruitments were under strong selective pressure to be maintained, allowing for the diversification and general phylogenetic coherence of large subunit sequences that reflect their mode of metabolic functioning (Figures 1A,B). This feature has been exploited bioinformatically such that is possible to predict the functionality and general directionality of hydrogenase enzymes using genomic data alone (Vignais and Billoud, 2007; Boyd et al., 2014; Peters et al., 2015; Greening et al., 2016; Poudel et al., 2016). This information can be combined with other genomic data to further examine modes by which H2 oxidation might be coupled with reduction of various oxidants such as O2, NO3−, Fe(III), SO42−, and CO2. Such information is powerful when trying to understand and predict the potential for microorganisms to influence the flux of H2 from the deep surface to near surface environments where it can be mined.
Abundance of hydrogenotrophs in subsurface ecosystems
The size of a population is generally scalable to the availability of resources to support that population, since communities assemble to maximize utilization of resources until one becomes limiting (i.e., carrying capacity). Using genome sequences from Archaea and Bacteria, it was shown that nearly 30% code for one or more [NiFe]- or [FeFe]-hydrogenase enzyme (Peters et al., 2015; Greening et al., 2016), consistent with a widespread ability to reversible oxidize H2 among microorganisms. While it is not atypical for an organismal genome to encode multiple copies of hydrogense enzymes, the genomes of several organisms from deep subsurface rock-hosted environments stand out, including that of the bacterium “Candidatus Desulforudis audaxviator” (Chivian et al., 2008). This organismal genome was reconstructed from metagenomic sequence obtained from fracture fluids from a 2.8 km depth in South African gold mines. The genome encodes four copies or homologs of [NiFe]-hydrogenase and six homologs of [FeFe]-hydrogenase, alluding to the central role that H2 has in the cell’s energy metabolism. More recent work has brought this organism into culture and has demonstrated that it can indeed grow via H2 oxidation (Karnachuk et al., 2019), as predicted from genomic data (Chivian et al., 2008). Multiple copies of hydrogenases in the genomes of organisms from taxonomic groups (phyla) that characteristically inhabit anoxic habitats is common (Peters et al., 2015), including in the Firmicutes, the group that “Ca. Desulforudis audaxviator” belongs to. Indeed, comparative metagenomic (shotgun sequencing) studies of natural microbial communities from surface environments relative to subsurface environments identified 5- and 2-fold enrichments in the genes encoding [NiFe]- and [FeFe]-hydrogenases in communities from subsurface environments relative to those obtained from surface environments (Colman et al., 2017).
Is the enrichment of hydrogenases in the genomes of organisms from subsurface environments related to the availability of H2? Hot springs have been proposed as accessible windows into subsurface environments (Colman et al., 2017), and numerous community metagenomes have been generated from these environments (e.g. (Inskeep et al., 2013)). A comparative study of 50 metagenomes from 50 different high temperature hot springs revealed that hydrogenase homologs can be particularly enriched (2X higher than background) in microbial communities from springs that are sourced by reduced volcanic gases and condensed steam or vapor (Figure 1C (Lindsay et al., 2019)). Magmatic degassing of H2 and high temperature reactions between steam and reduced iron bearing minerals during the ascent of vapor to the surface leads to enrichment of H2 in this spring type (Lindsay et al., 2019). Indeed, a genomic analysis of one of those springs, termed SmokeJumper3 (SJ3), showed that >70% of the community members encoded at least one hydrogenase homolog (many genomes encoded multiple homologs), the vast majority of which were predicted using bioinformatics approaches to be involved in H2 oxidation (Lindsay et al., 2019). Further, the total number of cultivatable cells in SJ3 that used H2 as a reductant coupled to various oxidants (O2, NO3−, Fe(III), SO42−, thiosulfate, and elemental sulfur) was found to be one to two orders of magnitude greater than in two nearby springs (termed SJ1 and SJ2). Geochemical analyses indicate that SJ3 waters harbor the highest concentration of H2 (2.1 µM) measured in a hot spring in Yellowstone to date, including in SJ1 (1.2 µM) and SJ2 (1.0 µM (Lindsay et al., 2019)). This suggests that organisms in SJ3 are particularly tuned to metabolize H2 but are likely limited by availability of other nutrients, since H2 was still measured at high concentration. In other H2 supported subsurface biospheres such as hydrothermal vents, the number of hydrogenase homologs encoded by resident microbial communities can be even higher, in some cases 40-fold higher than from other surface habitats (Brazelton et al., 2012; Adam and Perner, 2018). Collectively, these observations point to a relationship between the availability of H2 in subsurface environments and the extent to which microbial inhabitants of those environments depend on H2 to support their energy metabolism. However, the nature of this relationship is likely to be nuanced by differences in other parameters in these systems, including variation in host rock that controls geochemical compositions, the availability of oxidants, and other nutrients (Adam and Perner, 2018).
Rates of net microbial oxidation in rock-supported ecosystems
The apparent relationship between the prevalence of hydrogenase encoding genes in community metagenomes from subsurface environments and the availability of H2 in those environments strongly suggests that many of the cells in those environments are dependent on H2. Yet, not all cells in natural environments are active and it is widely accepted that most (up to 80%) of them are inactive or in a stationary phase of growth (Kolter et al., 1993; Lennon and Jones, 2011), likely due to limitation of energy substrates, nutrients, water, or accumulation of metabolic by-products (waste) that feedback inhibits biochemical processes. Further, the presence of genes allowing for a particular metabolic function does not necessarily mean that function is being carried out. This is because genes must first be converted or transcribed to RNA before the RNA/transcripts can be read and proteins/enzymes be synthesized (i.e., the central dogma of biology). Yet, RNA is easily degraded and is difficult to recover from microbial cells in natural environments. This limitation of RNA, when combined with the added complexity of limited amount of biomass (and therefore RNA) in rock-hosted ecosystems, makes it advantageous to directly measure the activity of enzymes that are involved in H2 consumption activity. Measurements of H2 oxidation or consumption generally take one of three forms, each of which can provide different data. One way of measuring H2 net consumption (consumption-production) is by measuring H2 concentrations over time following a direct injection of gas into a formation (e.g. (Bagnoud et al., 2016)). This approach is labor-, time-, and cost-intensive but provides estimates that are less prone to “bottle” artifacts (described below). The other common approaches use microcosms, or small bioreactors, that can be sealed and their contents manipulated. A known volume of sediment or ground rock is added to the microcosm along with either filter-sterilized fracture water from the formation or synthetic medium designed to chemically mimic fracture waters. Then, a fixed amount of H2 is added to the headspace and its net consumption is quantified over time. Alternatively, tritiated hydrogen is added and its conversion to tritiated water is quantified. The difference in these later two approaches is that the former is reflective of net H2 consumption whereas the latter is measuring H2 oxidation only. Killed (autoclave or otherwise) controls allow for these activities to be attributed to microbial processes or abiotic processes. Both of the last two approaches, which tend to be carried out in sealed reactors, can lead to under- or over-estimates of activity, since nutrient limitation can develop, microbial communities can be stimulated due to added substrates, or disturbance can affect interactions among populations.
Due to these methodological differences as well as large-scale differences in environment (e.g., acidic pH versus circumneutral pH; impacted by photosynthesis or not), the results of H2 consumption measurements are often difficult to compare. For example, most measurements of net H2 oxidation rates in hydrothermal vent communities are reported on a per cell basis (e.g. (Adam and Perner, 2018)). This is done due to the comparative ease by which cells can be enumerated in aqueous samples. However, this assumes that all cells are similarly dependent on H2 and active, which is not likely to be true (see above). Further, this makes it difficult to compare to rock-matrix associated cells, since quantifying cells in such samples is difficult. Nonetheless, several measurements of net H2 oxidation have been reported from rock-hosted ecosystems that span a range of temperatures (4°C–70°C) and geologic settings and with circumneutral pH and freshwater salinities, and these were compiled here for comparison. This included a single measurement from a circumneutral hot spring (Lindsay et al., 2018), two measurements from basaltic and carbonate sediments in circumneutral glacial meltwaters (Dunham et al., 2021), and a single measurement from a subsurface clay aquifer/reservoir proposed for use in storing radioactive waste (Bagnoud et al., 2016). These data from rock-hosted, non-photosynthetic ecosystems were then compared to several rates reported from non-rock-hosted environments, including terrestrial soils (Ji et al., 2017; Giguere et al., 2021) and marine and soda lake sediments (Adhikari et al., 2016), both of which are potentially influenced directly or indirectly by photosynthesis. These were all normalized to per unit mass or volume of material that was then converted to mass (soil mass, sediment mass), permitting their comparison.
Average rates of net H2 oxidation in recently comminuted and fine grained carbonate sediments from Robertson Glacier and basaltic sediments from Kötlujökull Glacier (both −0.1°C), when incubated in microcosms at 4°C, revealed low rates of H2 oxidation (−1.1 and 7.6 nmol d−1 G dry mass (gdm)−1) (Dunham et al., 2021). In a clay-hosted aquifer (incubated in situ at −17.5°C), a net rate of microbial H2 oxidation of 1.53 μmol d−1 cm3 −1 was reported (Bagnoud et al., 2016). Assuming a density of clay of 1.7 g cm3 −1 (Bagnoud et al., 2016), this value equates to 0.9 μmol d−1 gdm-1. Finally, a rate of net H2 oxidation of 7.2 μmol d−1 gdm−1 was reported for fine-grained hot spring sediments sampled at the source of a circumneutral pH hot spring (pH, 6.8) when incubated in microcosms at 70°C (Lindsay et al., 2018). Despite the limited size of the dataset, they were plotted as a function of temperature (pseudo-Arrhenius plot), which revealed a linear relationship (R2 = 0.99) that has a slope of −10 and intercept of −5. This relationship yields a −1-2 fold increase in rate per 10°C increase in temperature, which is close to the expectation for a reaction with Arrhenius-like kinetics (2-fold increase in rate per 10°C increase). While this relationship is defined with only 4 datapoints, it is a bit unexpected due to large-scale expected differences in the organisms that inhabit these environments (e.g., archaeal-versus bacterial-dominated, aerobic versus anaerobic), the presumed differences in the type of hydrogenase enzymes they encode and their kinetics, differences in modes of redox coupling, and different amounts of biomass (e.g., 104–105 cells gdm−1 in subglacial sediments to 107–108 cells gdm−1 in hot spring sediments (Lindsay et al., 2019; Dunham et al., 2022)). This is in addition to differences in how the measurements were made (microcosm versus in situ measurements). To further explore the relationship between net H2 oxidation and temperature, measurements from non-rock-hosted ecosystems were compared to those above.
A net H2 oxidation rate of 0.84 μmol d−1 gdm−1 was reported for Antarctic soils when incubated at 10°C (Ji et al., 2017), which is close to what would be predicted (14°C) by the linear relationship. A net oxidation rate of 3.36 μmol d−1 gdm−1 was reported for a temperate soil; however, incubation temperature was not reported (Giguere et al., 2021). Applying the linear relationship from above yields a temperature of 40°C, which is ∼15°C higher than most temperate soils. We also note that rates of H2 oxidation (not net) in marine sediments from a variety of environments range from 0.08 to 17.85 μmol d−1 cm3. Assuming an average density for marine sediments (clay) of 1.7 g cm3 (Tenzer and Gladkikh, 2014), these rates translate to 0.05–10.5 μmol d−1 gdm−1 which are generally higher than for rock-hosted communities. This may be due to these measurements being direct H2 oxidation measurements (conversion of tritiated H2 to water), as opposed to net oxidation measurements, and these systems receiving organic carbon from the overlying water column that could further stimulate biomass production. While the relationship between net H2 oxidation potential and temperature needs to be further refined and evaluated using additional measurements from diverse subsurface systems, the limited data compiled here indicates that microorganisms consume H2 in rock-hosted ecosystems and that their net H2 consumption rates are at least partially dependent on temperature.
To begin to examine how microbial H2 oxidation could impact H2 recovery in a natural system, we consider a 1 km deep, 1 km diameter cylinder (0.785 km3) of harzburgite/dunite rock in the Samail Ophiolite, Oman. An average rock density for harzburgite/dunite of 2.55 g cm3 (measured in cores BA1B, BA3A, and BA4A (Kelemen et al., 2020)) was used to normalize this volume of rock to mass and a porosity of 3.8% cm3 (average in these same cores (Kelemen et al., 2020)) was used as a crude method to identify the percentage of this rock that could be inhabited by microorganisms. Applying the net H2 oxidation rate of 3.4 μmol d gdm at 40°C (extrapolated from Figure 2), which is a likely average for a 1 km depth (Nothaft et al., 2021), and assuming a constant H2 oxidation rate throughout the rock column for simplicity, a net microbial consumption rate is estimated at 52 × 103 tonnes per year. Assuming complete rock alteration of 1 kg of such rock, −0.3 mol H2 can be produced (Leong et al., 2023), equivalent to −1011 tonnes for the 0.785 km3 rock volume, which would support microbial hydrogenotrophs for −107 years at this level of activity. Yet, the annual outgassing rate (flux of H2) from the surface of the Samail Ophiolite is estimated to be 3.6 × 103 tonnes km3 (Leong et al., 2023), an order of magnitude lower than the estimated net microbial H2 oxidation potential. This suggests that up to >90% of H2 produced in the Samail Ophiolite could be lost through microbial consumption. Importantly, the estimates for net H2 oxidation for rock-hosted microbial communities were derived largely using samples from near surface rock-hosted ecosystems (e.g., subglacial sediments, hot spring sediments) which may differ markedly from those that inhabit subsurface rocks where the flux of oxidants and nutrients could be far lower.
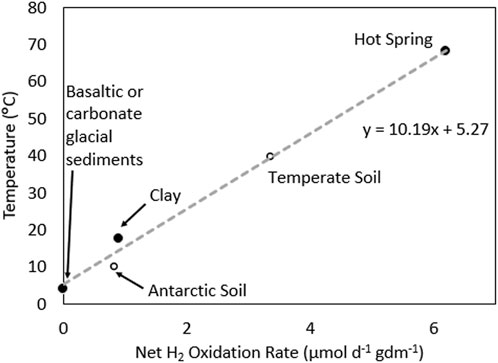
Figure 2. Net hydrogen (H2) oxidation rates for various samples plotted as a function of the temperature of the experiments where those measurements were made for those samples. The trendline was generated using the four datapoints (solid circles) from basaltic or carbonate glacial sediments, subsurface clay, and hot spring sediments. The net H2 oxidation rates from an Antarctic soil where temperature was reported and from a temporal soil where temperature was estimated from the depicted regression are plotted for reference in open circles. Data adapted from Bagnoud et al., 2016; Ji et al., 2017; Lindsay et al., 2018; Dunham et al., 2020; Giguere et al., 2021.
Microbial production of H2
While the focus of this perspective is on H2 oxidation via hydrogenases, such enzymes are reversible and, under certain conditions, microorganisms can produce H2. Microbial production of H2 is most commonly associated with the process of fermentation, whereby organic carbon molecules are broken down in the absence of O2 or other suitable oxidants. In the absence of other suitable oxidants, microbial cells turn to protons as a source of oxidant to regenerate oxidized intermediate electron carriers (Fd, NAD+, etc.) and enable organic carbon oxidation to continue. In the process of oxidizing intermediate electron carriers, protons are reduced to yield H2 (Reaction 1). Fermentation is a common process in environments with available organic carbon but where other more preferrable oxidants are limiting. Examples of subsurface environments meeting these characteristics are oil and natural gas reservoirs, which are also often replete with H2 (Zgonnik, 2020). Thus, the biological potential to produce H2 is highest in subsurface environments that have substantial organic carbon that drives the consumption of available oxidants.
Conclusions and future directions
Genomic and metagenomic studies reveal that the ability to reversibly oxidize H2 is a widespread trait that is integrated into the energy metabolism of nearly all types of microbial cells, ranging from aerobes to anaerobes, autotrophs to heterotrophs, and chemosynthetic to photosynthetic organisms. Metagenomic sequence from rock-hosted subsurface ecosystems reveal enrichment of hydrogenases in subsurface communities relative to those from the surface, alluding to the even greater importance for H2 cycling in subsurface rocky environments that exclude light as an available energy source. Rates of net microbial H2 oxidation in several rock-hosted habitats have been measured and a relationship between rate and temperature was established, with rates increasing by a factor of ∼1-2 per 10°C increase in temperature. This suggests that communities may have a significant impact on the flux of H2 from the subsurface and that this effect might be more pronounced at higher temperature.
Yet, the availability of data to evaluate the potential effect of microorganisms on the flux of H2 from subsurface environments remains highly limited, despite the recent surge in interest and investment in prospecting for and mining white and orange H2 as a component of the energy economy (Hand, 2023). This is partly attributable to logistics and costs associated with gaining access to subsurface samples and fluids and ensuring that they are collected in a manner that is conducive for downstream use in geomicrobiological work. We call for closer communication and coordination between prospectors interested in maximizing white and orange H2 recovery and geomicrobiologists interested in understanding the role of geologic H2 in sustaining subsurface biospheres. Specifically, coordination and communication between prospectors and geomicrobiologists is needed to ensure that samples can be collected in a manner that is suitable for downstream use in microbiological work. Through such coordination, prospectors and microbiologists will be in a unique position to not only develop a more robust understanding of H2 cycling activities of microbial communities in rock-hosted ecosystems but to also better guide prospecting efforts and to develop interventive strategies to mitigate H2 loss due to microbial activities. Further, obtaining such understanding through coordination of efforts could result in new technologies and approaches to engineer subsurface systems to poise microorganisms to enhance the production of H2 through fermentative or other biological activities.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
EB: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Supervision, Writing–original draft, Writing–review and editing. DC: Data curation, Writing–review and editing. AT: Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Aeronautics and Space Administration under Award 80NSSC21K0489 (EB and AT) and by the W. M. Keck Foundation (EB).
Acknowledgments
The authors are grateful for the numerous discussions with Peter Kelemen, James Leong, and Eric Ellison that significantly improved this paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adam, N., and Perner, M. (2018). Microbially mediated hydrogen cycling in deep-sea hydrothermal vents. Front. Microbiol. 9, 2873. doi:10.3389/fmicb.2018.02873
Adhikari, R. R., Glombitza, C., Nickel, J. C., Anderson, C. H., Dunlea, A. G., Spivack, A. J., et al. (2016). Hydrogen utilization potential in subsurface sediments. Front. Microbiol. 7, 8. doi:10.3389/fmicb.2016.00008
Bagnoud, A., Leupin, O., Schwyn, B., and Bernier-Latmani, R. (2016). Rates of microbial hydrogen oxidation and sulfate reduction in Opalinus Clay rock. Appl. Geochem. 72, 42–50. doi:10.1016/j.apgeochem.2016.06.011
Battistuzzi, F. U., Feijao, A., and Hedges, S. B. (2004). A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 4, 44. doi:10.1186/1471-2148-4-44
Boyd, E. S., Schut, G. J., Adams, M. W. W., and Peters, J. W. (2014). Hydrogen metabolism and the evolution of biological respiration. Microbe 9, 361–367. doi:10.1128/microbe.9.361.1
Brazelton, W. J., Nelson, B., and Schrenk, M. O. (2012). Metagenomic evidence for H2 oxidation and H2 production by serpentinite-hosted subsurface microbial communities. Front. Microbiol. 2, 268. doi:10.3389/fmicb.2011.00268
Charlou, J. L., Donval, J. P., Konn, C., Ondréas, H., Fouquet, Y., Jean-Baptiste, P., et al. (2010). “High production and fluxes of H2 and CH4 and evidence of abiotic hydrocarbon synthesis by serpentinization in ultramafic-hosted hydrothermal systems on the Mid-Atlantic Ridge,” in Diversity of hydrothermal systems on slow spreading ocean ridges (Washington, D.C., United States: American Geophysical Union), 265–296.
Chivian, D., Brodie, E. L., Alm, E. J., Culley, D. E., Dehal, P. S., Desantis, T. Z., et al. (2008). Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322, 275–278. doi:10.1126/science.1155495
Colman, D. R., Poudel, S., Stamps, B. W., Boyd, E. S., and Spear, J. R. (2017). The deep, hot biosphere: twenty-five years of retrospection. Proc. Natl. Acad. Sci. 114, 6895–6903. doi:10.1073/pnas.1701266114
Conrad, R. (1996). Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60, 609–640. doi:10.1128/mr.60.4.609-640.1996
Dunham, E. C., Dore, J. E., Skidmore, M. L., Roden, E. E., and Boyd, E. S. (2021). Lithogenic hydrogen supports microbial primary production in subglacial and proglacial environments. Proc. Natl. Acad. Sci. 118, e2007051117. doi:10.1073/pnas.2007051117
Dunham, E. C., Keller, L. M., Skidmore, M. L., Mitchell, K. R., and Boyd, E. S. (2022). Iron minerals influence the assembly of microbial communities in a basaltic glacial catchment. FEMS Microb. Ecol. 99, fiac155. doi:10.1093/femsec/fiac155
Friedrich, T., and Scheide, D. (2000). The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479, 1–5. doi:10.1016/s0014-5793(00)01867-6
Giguere, A. T., Eichorst, S. A., Meier, D. V., Herbold, C. W., Richter, A., Greening, C., et al. (2021). Acidobacteria are active and abundant members of diverse atmospheric H2-oxidizing communities detected in temperate soils. ISME J. 15, 363–376. doi:10.1038/s41396-020-00750-8
Greening, C., Biswas, A., Carere, C. R., Jackson, C. J., Taylor, M. C., Stott, M. B., et al. (2016). Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10, 761–777. doi:10.1038/ismej.2015.153
Greening, C., and Boyd, E. (2020). Editorial: microbial hydrogen metabolism. Front. Microbiol. 11, 56. doi:10.3389/fmicb.2020.00056
Greening, C., Cabotaje, P. R., Alvarado, L. E. V., Leung, P. M., Land, H., Senger, M., et al. (2023). Unique minimal and hybrid hydrogenases are active in anaerobic archaea. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4520792.
Gregory, S. P., Barnett, M. J., Field, L. P., and Milodowski, A. E. (2019). Subsurface microbial hydrogen cycling: natural occurrence and implications for industry. Microorganisms 7, 53. doi:10.3390/microorganisms7020053
Inskeep, W., Jay, Z., Tringe, S., Herrgard, M., and Rusch, D.YNP Metagenome Project Steering Committee and Working Group Members (2013). The YNP metagenome project: environmental parameters responsible for microbial distribution in the Yellowstone geothermal ecosystem. Front. Microbiol. 4, 67. doi:10.3389/fmicb.2013.00067
Ji, M., Greening, C., Vanwonterghem, I., Carere, C. R., Bay, S. K., Steen, J. A., et al. (2017). Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 552, 400–403. doi:10.1038/nature25014
Karnachuk, O. V., Frank, Y. A., Lukina, A. P., Kadnikov, V. V., Beletsky, A. V., Mardanov, A. V., et al. (2019). Domestication of previously uncultivated Candidatus Desulforudis audaxviator from a deep aquifer in Siberia sheds light on its physiology and evolution. ISME J. 13, 1947–1959. doi:10.1038/s41396-019-0402-3
Kelemen, P. B., Matter, J. M., Teagle, D. A. H., Coggon, J. A., and Team, T. O. D. P. S. (2020). Proceedings of the Oman drilling project. College Station, TX, USA: International Ocean Discovery Program. doi:10.14379/OmanDP.proc.117.2020
Kelley, D. S., Karson, J. A., Fruh-Green, G. L., Yoerger, D. R., Shank, T. M., Butterfield, D. A., et al. (2005). A serpentinite-hosted ecosystem: the lost city hydrothermal field. Science 307, 1428–1434. doi:10.1126/science.1102556
Kita, I., Matsuo, S., and Wakita, H. (1982). H2 generation by reaction between H2O and crushed rock: an experimental study on H2 degassing from the active fault zone. J. Geophys. Res. Sol. Earth 87, 10789–10795. doi:10.1029/JB087iB13p10789
Kolter, R., Siegele, D. A., and Tormo, A. (1993). The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47, 855–874. doi:10.1146/annurev.mi.47.100193.004231
Konopka, A. (2006). Microbial ecology: searching for principles. Microbe 1, 175–179. doi:10.1128/microbe.1.175.1
Lennon, J. T., and Jones, S. E. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130. doi:10.1038/nrmicro2504
Leong, J. A., Nielsen, M., Mcqueen, N., Karolytė, R., Hillegonds, D. J., Ballentine, C., et al. (2023). H2 and CH4 outgassing rates in the Samail ophiolite, Oman: implications for low-temperature, continental serpentinization rates. Geochim. Cosmochim. Acta 347, 1–15. doi:10.1016/j.gca.2023.02.008
Lin, L.-H., Hall, J., Lippmann-Pipke, J., Ward, J. A., Lollar, B. S., Deflaun, M., et al. (2005). Radiolytic H2 in continental crust: nuclear power for deep subsurface microbial communities. Geochem. Geophys. Geosyst. 6, 7. doi:10.1029/2004GC000907
Lindsay, M. R., Amenabar, M. J., Fecteau, K. M., Debes Ii, R. V., Fernandes Martins, M. C., Fristad, K. E., et al. (2018). Subsurface processes influence oxidant availability and chemoautotrophic hydrogen metabolism in Yellowstone hot springs. Geobiology 16, 674–692. doi:10.1111/gbi.12308
Lindsay, M. R., Colman, D. R., Amenabar, M. J., Fristad, K. E., Fecteau, K. M., Debes, R. V., et al. (2019). Probing the geological source and biological fate of hydrogen in Yellowstone hot springs. Environ. Microbiol. 21, 3816–3830. doi:10.1111/1462-2920.14730
Macdonald, M. L., Wadham, J. L., Telling, J., and Skidmore, M. L. (2018). Glacial erosion liberates lithologic energy sources for microbes and acidity for chemical weathering beneath glaciers and ice sheets. Front. Earth Sci. 6, 212. doi:10.3389/feart.2018.00212
Milkov, A. V. (2022). Molecular hydrogen in surface and subsurface natural gases: abundance, origins and ideas for deliberate exploration. Earth-Sci. Rev. 230, 104063. doi:10.1016/j.earscirev.2022.104063
Nealson, K. H., Inagaki, F., and Takai, K. (2005). Hydrogen-driven subsurface lithoautotrophic microbial ecosystems (SLiMEs): do they exist and why should we care? Trends Microbiol. 13, 405–410. doi:10.1016/j.tim.2005.07.010
Nothaft, D. B., Templeton, A. S., Boyd, E. S., Matter, J. M., Stute, M., Paukert Vankeuren, A. N., et al. (2021). Aqueous geochemical and microbial variation across discrete depth intervals in a peridotite aquifer assessed using a packer system in the Samail Ophiolite, Oman. J. Geophys. Res. Biogeosci. 126, e2021JG006319. doi:10.1029/2021JG006319
Parkes, R. J., Berlendis, S., Roussel, E. G., Bahruji, H., Webster, G., Oldroyd, A., et al. (2019). Rock-crushing derived hydrogen directly supports a methanogenic community: significance for the deep biosphere. Environ. Microbiol. Rep. 11, 165–172. doi:10.1111/1758-2229.12723
Peters, J. W., Schut, G. J., Boyd, E. S., Mulder, D. W., Shepard, E. M., Broderick, J. B., et al. (2015). [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim. Biophys. Acta 1853, 1350–1369. doi:10.1016/j.bbamcr.2014.11.021
Poudel, S., Tokmina-Lukaszewska, M., Colman, D. R., Refai, M., Schut, G. J., King, P. W., et al. (2016). Unification of [FeFe]-hydrogenases into three structural and functional groups. Biochim. Biophys. Acta 1860, 1910–1921. doi:10.1016/j.bbagen.2016.05.034
Rempfert, K. R., Miller, H. M., Bompard, N., Nothaft, D., Matter, J. M., Kelemen, P., et al. (2017). Geological and geochemical controls on subsurface microbial life in the Samail Ophiolite, Oman. Front. Microbiol. 8, 56. doi:10.3389/fmicb.2017.00056
Schrenk, M. O., Brazelton, W. J., and Lang, S. Q. (2013). Serpentinization, carbon, and deep life. Rev. Mineral. Geochem. 75, 575–606. doi:10.2138/rmg.2013.75.18
Schut, G. J., Boyd, E. S., Peters, J. W., and Adams, M. W. (2013). The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol. Rev. 37, 182–203. doi:10.1111/j.1574-6976.2012.00346.x
Sherwood Lollar, B., Onstott, T. C., Lacrampe-Couloume, G., and Ballentine, C. J. (2014). The contribution of the Precambrian continental lithosphere to global H2 production. Nature 516, 379–382. doi:10.1038/nature14017
Shima, S., Pilak, O., Vogt, S., Schick, M., Stagni, M. S., Meyer-Klaucke, W., et al. (2008). The crystal structure of [Fe]-hydrogenase reveals the geometry of the active site. Science 321, 572–575. doi:10.1126/science.1158978
Spear, J. R., Walker, J. J., Mccollom, T. M., and Pace, N. R. (2005). Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. 102, 2555–2560. doi:10.1073/pnas.0409574102
Stevens, T. O., and Mckinley, J. P. (1995). Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science 270, 450–455. doi:10.1126/science.270.5235.450
Stevens, T. O., and Mckinley, J. P. (2000). Abiotic controls on H2 production from basalt-water reactions and implications for aquifer biogeochemistry. Environ. Sci. Tech. 34, 826–831. doi:10.1021/es990583g
Telling, J., Boyd, E. S., Bone, N., Jones, E. L., Tranter, M., Macfarlane, J. W., et al. (2015). Erratum: rock comminution as a source of hydrogen for subglacial ecosystems. Nat. Geosci. 8, 981. doi:10.1038/Ngeo2604
Templeton, A. S., and Caro, T. A. (2023). The rock-hosted biosphere. Ann. Rev. Earth Planet. Sci. 51, 493–519. doi:10.1146/annurev-earth-031920-081957
Templeton, A. S., Ellison, E. T., Glombitza, C., Morono, Y., Rempfert, K. R., Hoehler, T. M., et al. (2021). Accessing the subsurface biosphere within rocks undergoing active low-temperature serpentinization in the Samail Ophiolite (Oman Drilling Project). J. Geophy. Res. Biogeosci. 126, e2021JG006315. doi:10.1029/2021JG006315
Tenzer, R., and Gladkikh, V. (2014). Assessment of density variations of marine sediments with ocean and sediment depths. Sci. World J. 2014, 1–9. doi:10.1155/2014/823296
Thauer, R. K., Kaster, A. K., Goenrich, M., Schick, M., Hiromoto, T., and Shima, S. (2010). Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 79, 507–536. doi:10.1146/annurev.biochem.030508.152103
Vignais, P. M., and Billoud, B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272. doi:10.1021/cr050196r
Weiss, M. C., Sousa, F. L., Mrnjavac, N., Neukirchen, S., Roettger, M., Nelson-Sathi, S., et al. (2016). The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 1, 16116. doi:10.1038/nmicrobiol.2016.116
Wolfe, J. M., and Fournier, G. P. (2018). Horizontal gene transfer constrains the timing of methanogen evolution. Nat. Ecol. Evol. 2, 897–903. doi:10.1038/s41559-018-0513-7
Keywords: geologic hydrogen, white hydrogen, orange hydrogen, hydrogenotroph, [NiFe]-hydrogenase, [FeFe]-hydrogenase, subsurface
Citation: Boyd ES, Colman DR and Templeton AS (2024) Perspective: Microbial hydrogen metabolism in rock-hosted ecosystems. Front. Energy Res. 12:1340410. doi: 10.3389/fenrg.2024.1340410
Received: 17 November 2023; Accepted: 22 January 2024;
Published: 21 March 2024.
Edited by:
Dariusz Strapoc, Schlumberger, FranceReviewed by:
Abhijeet Singh, Swedish University of Agricultural Sciences, SwedenCopyright © 2024 Boyd, Colman and Templeton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eric S. Boyd, eric.boyd@montana.edu
 Eric S. Boyd
Eric S. Boyd Daniel R. Colman
Daniel R. Colman Alexis S. Templeton
Alexis S. Templeton