Cytokinins Induce Transcriptional Reprograming and Improve Arabidopsis Plant Performance under Drought and Salt Stress Conditions
- 1Faculty of Biology, Technion – Israel Institute of Technology, Haifa, Israel
- 2Bioinformatics Knowledge Unit, The Lorry I. Lokey Interdisciplinary Center for Life Sciences and Engineering, Technion – Israel Institute of Technology, Haifa, Israel
In nature, annual plants respond to abiotic stresses by activating a specific genetic program leading to early flowering and accelerated senescence. Although, in nature, this phenomenon supports survival under unfavorable environmental conditions, it may have negative agro-economic impacts on crop productivity. Overcoming this genetic programing by cytokinins (CK) has recently been shown in transgenic plants that overproduce CK. These transgenic plants displayed a significant increase in plant productivity under drought stress conditions. We investigated the role of CK in reverting the transcriptional program that is activated under abiotic stress conditions and allowing sustainable plant growth. We employed 2 complementary approaches: Ectopic overexpression of CK, and applying exogenous CK to detached Arabidopsis leaves. Transgenic Arabidopsis plants transformed with the isopentyltransferase (IPT) gene under the regulation of the senescence associated receptor kinase (SARK) promoter displayed a significant drought resistance. A transcriptomic analysis using RNA sequencing was performed to explore the response mechanisms under elevated CK levels during salinity stress. This analysis showed that under such stress, CK triggered transcriptional reprograming that resulted in attenuated stress-dependent inhibition of vegetative growth and delayed premature plant senescence. Our data suggest that elevated CK levels led to stress tolerance by retaining the expression of genes associated with plant growth and metabolism whose expression typically decreases under stress conditions. In conclusion, we hypothesize that CK allows sustainable plant growth under unfavorable environmental conditions by activating gene expression related to growth processes and by preventing the expression of genes related to the activation of premature senescence.
Introduction
In nature, annual plants frequently encounter abiotic stresses that adversely affect growth, development, or productivity. Such stresses trigger a wide range of plant responses including a specific genetic program leading to growth retardation, early flowering, and accelerated senescence. Although, such responses may support survival in nature under severe stresses, they have negative agro-economic impact on crop productivity. Cytokinins (CKs; a class of phytohormones) have a role in maintaining plant health and their biosynthesis is decreased under water deficit stress (Chernyad'ev, 2005). Low CK levels are associated with inhibition of growth, and acceleration of senescence onset (Roitsch and Ehne, 2000; Gepstein and Glick, 2013), whereas applying exogenous CK delays leaf senescence (Chernyad'ev, 2005). Also, in transgenic plants that overproduce CK, the aforementioned stress-induced responses are attenuated (Rivero et al., 2007).
The first committed and rate-limiting step in the biosynthesis of CK is catalyzed by the enzyme isopentenyltransferase (IPT). In transgenic tobacco where IPT was expressed under the regulation of promoter of the senescence gene SARK from Phaseolus vulgaris (Hajouj et al., 2000), CK was maintained at high levels under water deficit stress, leading to improved drought tolerance (Rivero et al., 2007). These studies were later extended to other crops such as rice (Peleg et al., 2011). Notably, not only did these transgenic plants survive, they also displayed a significantly increased plant productivity under drought stress conditions (Rivero et al., 2007).
The genetic reprograming of plants under abiotic stresses involves reducing water demand by stomatal closure, decreased photosynthesis, reduced leaf evaporative area, as well as by premature termination of vegetative growth followed by a reproductive phase and plant senescence (Roitsch and Ehne, 2000; Gepstein and Glick, 2013). However, not reducing transpiration and maintaining photosynthetic capacity is advantageous under moderate stress conditions, since it allows sustainable vegetative growth.
Drought resistance involves multiple responses and the interactions thereof. Reprograming of genomic expression leads to developmental and morphological changes, osmotic adjustment, and biochemical responses, ultimately resulting in a new homoeostatic state. Signaling pathways activated in response to abiotic stress include components related to phosphorylation/dephosphrylation activities such as protein kinases, protein phosphatases, activation of reactive oxygen species (ROS), and signaling of various transcriptional regulators (Arnholdt-Schmitt, 2004). Phytohormones are also key factors in the regulation of adaptive responses to drought and other stresses.
Among the major hormones produced by plants, Abscisic acid (ABA) is known to playing a key role in mediating abiotic stresses. Typically, environmental conditions such as drought and salinity are known to trigger increase in ABA levels. Increased ABA levels lead to stomatal closure which, in turn, minimizes water loss by reducing transpiration, but also reduces photosynthetic activity and accelerates premature senescence (Verma et al., 2016). ABA elevation during abiotic stresses, such as drought, has been proven beneficial for plant survival (Fujita et al., 2011). Many genes associated with ABA synthesis and genes encoding receptors and downstream signaling components through ABA response elements (ABRE) and dehydration-response element-binding proteins (DRE) motifs, have been identified (Mishra et al., 2014). Gene regulatory networks derived from genomic studies revealed that ABA is a universal hormone involved in massive transcriptional reprograming events leading to stress adaptation responses (Sreenivasulu et al., 2012).
Until a decade ago, CKs were not considered important stress-response hormones. CKs and ABA have antagonistic effects on stomatal opening, transpiration, and photosynthesis. Most of the studies support the notion that during stress response, CK levels decrease and that stress response is controlled by ABA and ethylene (Wang et al., 2011). However, recent studies using genetic manipulation of CK demonstrate a more complicated picture. We previously reported on the development of transgenic plants carrying the autoregulatory system of IPT activation and showed that CK plays a role in delaying tobacco plant senescence (Hajouj et al., 2000). We later found that these transgenic plants also displayed a significant drought tolerance (Rivero et al., 2007). Not only did these plants survive longer under water deficit, they also grew throughout the growth period on just 30% of the optimal watering regimes with almost no loss of plant yield (Rivero et al., 2007). Thus, we hypothesized that induction of CK biosynthesis during stress could enhance stress tolerance. Indeed, studies using the aforementioned transgenic tobacco plants showed that during water deficit stress, the protection of biochemical processes associated with photosynthesis increased alongside the induction of photorespiration (Rivero et al., 2007). The transcriptome of these plants during water deficit revealed that the expression of genes encoding components of the carotenoid pathway leading to ABA biosynthesis was enhanced in wild-type (w.t.) plants and repressed in the transgenic plants, suggesting that the photosynthetic apparatus was degraded in w.t. plants and unaffected in the transgenic plants (Rivero et al., 2010). This specific CK induction approach was successfully demonstrated in dicots and monocots (Rivero et al., 2007; Peleg et al., 2011), and could potentially serve as a generic system. However, several studies pointed at a negative effect of CKs on the stress tolerance. Constitutive expression of CK dehydrogenase (CKX, which degrades CK in plants) or multiple mutation of IPT 1, 3, 5, and 7 in tobacco showed significant resistance to drought, which was attributed to changes in membrane durability and integrity as manifested by a decrease in electrolyte leakage. These CK deficient mutants displayed ABA hypersensitivity which suggests that decreased CK content increased ABA sensitivity and ABA response in the context of improved stress tolerance (Nishiyama et al., 2011). The effect of silencing CK receptors AHK3 and AHK2 on stress resistance in plants indicated an improved resistance to drought and salt stress (Tran et al., 2007). Many stress- and ABA-controlled genes were upregulated at the mutant plants. Furthermore, some ABA-controlled genes were upregulated even under non-stress conditions, suggesting that AHK3- and AHK2 negatively regulate plant response to osmotic stress (Tran et al., 2007). Combined, these studies demonstrate that during stress, high CK levels affect plant metabolism at different levels, depending on the stress severity.
CKs are involved in plant development and metabolism; yet, very little is known about the molecular nature of CK signaling that mediates abiotic stress tolerance. Identifying genes that are regulated by CK during stress is key for enhancing our molecular understanding the role of CK in stress response and drought tolerance. Analyses of drought-induced genes have demonstrated the existence of ABA-independent signal transduction cascades between the initial signal of water deficit and the expression of specific genes (Shinozaki and Yamaguchi-Shinozaki, 2000).
The goal of the current study was to investigate CK signaling under drought and salinity stress, to understand the overall effect of CK under such conditions, and to identify key genes involved in these processes.
Materials and Methods
Plant Material and Growing Conditions
Arabidopsis thaliana plants (Columbia ecotype) were grown on soaked peat pellets (Jiffy 7, Kappa Forenade Well) in temperature-controlled growth room at 23°C (±25°C) under fluorescent lamps (75 μE × m−2 × s−1) at long-day conditions (18 h light, 8 h dark).
Plasmid Construction and Plant Transformation
The pSARK:IPT vector contains the SARK:IPT:NOS cassette which carries the IPT gene under the regulation of the SARK promoter and the nopaline synthase (NOS) terminator as previously described (Rivero et al., 2007). The Arabidopsis transgenic lines were produced by infiltrating Col-0 using floral-dip method (Clough and Bent, 1998). Transgenic lines were screened and confirmed by PCR analysis.
Salinity and Drought Stress Experiments
Arabidopsis plants were grown for 40 days and irrigated with tap water. Then, plants were subjected to salinity stress by watering with 300 mM NaCl solution for 3 weeks. For the drought experiments, Arabidopsis plants were watered daily to keep the soil near 100% field capacity until 40 days after germination. Then, plants were divided into two groups: a control with daily irrigation and the stress group where water was withheld for 5 days. Applying exogenous CK under salinity stress was performed by excising fully expanded Arabidopsis leaves (non-yellowing) from the plant and incubating them in water for 1 h. After that, the leaves were divided into 4 treatments: Control (water), salinity (100–150 mM NaCl), synthetic CK (6-benzylaminopurine [BAP]; 10 μm), and salinity plus BAP. The exogenous application of CK's may suffer possible limitations such as penetration problems and using non-physiological concentration. However, this approach complements and supports the alternative approach of activation on endogenous CK's.
RNA Extraction and Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from ~150 mg plant material using SV Total RNA Isolation kit (Promega, Madison, WI), according to the manufacturer's instructions. Concentration, integrity, and extent of contamination by ribosomal RNA were monitored using the ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). cDNA was prepared from the total RNA with qScript cDNA Synthesis Kit (Quanta, Beverly, MA) as described by the manufacturer. qRT-PCR was performed using teal-time SYBR Green FastMix ROX (Quanta, Beverly, MA). Primers used are listed in Supplementary Table 1. In each case, three biological samples were analyzed in triplicates. The expression of candidate genes was normalized to cyclophilin (At2g36130). Comparative Ct values were determined using Applied Biosystems, User Bulletin #2, (http://home.appliedbiosystems.com/). Results were statistically processed by the One Way Analysis of Variance (ANOVA) method using SPSS Statistics 19.0. P ≤ 0.05 was considered significant. Error bars represent standard error (SE) of the mean of the 3 samples.
RNA Sequencing
Total RNA was extracted from 12 samples of rosette leaves of w.t plants, 3 samples for each of the 4 treatments: water, salt (NaCl 120 Mm), water plus BAP (10 μm), or salt (NaCl 120 Mm) plus BAP (10 μm). The 12 samples were treated with DNAse and sent to the Technion Genome Center (TGC) where cDNA libraries were constructed with TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA). High-throughput sequencing was performed using the Illumina HiSeq 2500 with 50 bp single-reads and barcoding. TGC personnel performed the initial analysis of results, and the quality of the reads was tested using the FastQC software with a phred score of +33. The short reads alignment/mapping was performed by the “TopHat2/Bowtie2” software (version 2.0.6) against the A. thaliana genome (version TAIR10) allowing 3 mismatches per read. Minimum and maximum intron sizes were set to 10 and 11,000, respectively, and the Bowtie2 mapping option –b2-sensitive was set. Furthermore, the aligned reads were processed by the Python package “HTSeq” version 0.5.3 p1 to get raw counts for each gene in TAIR10. The following A. thaliana reference genome was used: ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_chromosome_files/TAIR10_chr_all.fa. The annotation file ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_gff3/TAIR10_GFF3_genes.gff was modified to contain genes and not transcripts.
Differential Expression
The normalization of raw counts was performed using R-package DESeq2 with the default parameters and the Wald test (which serves in DESeq2 as a substitute for t-test and one-way ANOVA) was chosen for the detection of statistically significant differentially expressed genes. Results of DESeq2 were subject to advanced bioinformatics analysis by the Technion Bioinformatics Knowledge Unit.
Clustering Analysis and GO Functional Enrichment Analysis
The output of the initial analysis performed at the TGC was further analyzed with the EXPANDER 6.4 (EXpression Analyzer and DisplayER) suite of tools for bioinformatics analysis of microarray and RNA-seq data.
The SOM clustering algorithm was applied to produce a partition of the gene profiles into several clusters and to visualize the mean pattern of the cluster (clusters were then refined by manual inspection and re-clustering); the TANGO tool was used for GO (Group by pattern similarity) functional enrichment analysis that was employed to identify genes with coordinated behavior across experimental conditions. The latter analysis was also performed using the QuickGO tool at EBI (https://www.ebi.ac.uk/QuickGO).
Results
Drought Tolerance of Transgenic Sark::IPT Arabidopsis Plants
We cloned the IPT gene under senescence-induced promoter to produce SARK::IPT transgenic Arabidopsis plants. The presence of the IPT transgene in each of the lines was confirmed by PCR. After 5 days of water withholding, the w.t. plants showed symptoms of withering and senescence, whereas, the transgenic plants seemed to overcome the severe drought treatment as reflected by leaf turgidity and the greener color (Figure 1). Even after re-watering for a week, w.t. plants displayed senescence symptoms (yellowing and wilting), whereas, the transgenic plants seemed to overcome the severe drought treatment.
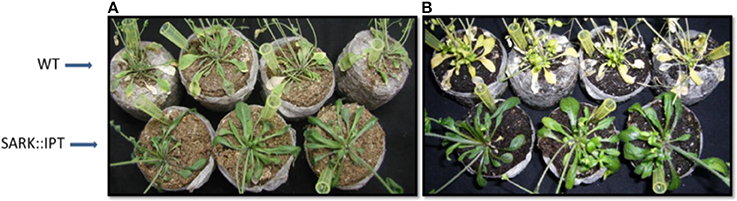
Figure 1. Responses of SARK::IPT transgenic Arabidopsis plants to drought conditions followed by re-watering. Arabidopsis plants were grown for 40 days under long day regimes (16 h light, 8 h darkness) and were then subjected to drought stress by withholding water for 5 days. (A) w.t. and transgenic plants SARK::IPT after 5 days of drought. (B) w.t. and transgenic plants SARK::IPT after 7 days of re-watering.
We then assessed, under normal and drought conditions (after 7 days of water withholding), the expression of candidate genes in w.t. vs. transgenic plants. First, we assessed 2 antioxidant genes: cat1 (catalase 1) and apx1 (ascorbate peroxidase 1; Figure 2). As expected, the expression of both genes increased under drought stress in the w.t. plants. In the transgenic plants, expression of these genes was lower relative to the w.t. plants under normal as well as drought conditions. To test whether the observed drought resistance is associated with known drought cell signals, we tested the expression of the early responsive dehydration (ERD1) gene, which is a known drought marker gene encoding a protein with homology to the ATP binding subunit of the Clp ATP-dependent protease of E. coli (Simpson et al., 2003), and is a known component of the ABA-independent pathway of drought response. Expression of the erd1 gene was upregulated under drought stress in w.t. plants suggesting a classical induction of the stress response associated with the ABA independent pathway (Figure 2). In contrast, the transgenic plants displayed reduced ERD1 expression compared to w.t. plants, with no upregulation under drought conditions.
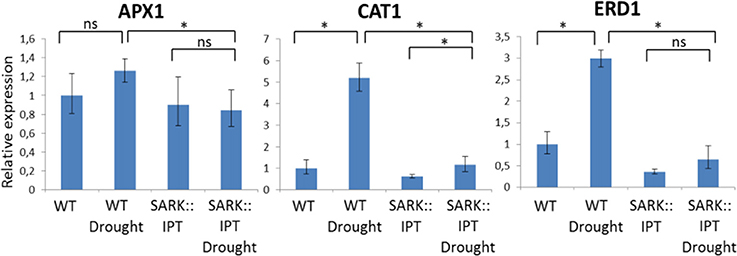
Figure 2. Relative expression ofAPX1, CAT1, and ERD1 genes in w.t. and transgenic SARK::IPT plants during drought stress. Arabidopsis plants were grown for 40 days in optimal conditions and were then transferred to drought stress for 7 days. Controls were well-watered plants (w.t. and transgenic). The qPCR assay reflects relative expression levels for each gene and was performed in triplicate for each of the three independent biological samples. Expression of each of the tested genes in the control treatment of w.t. plants was used as the basis for comparison (relative expression = 1). Horizontal brackets denote significance of P < 0.05 (*, statistical significance; ns, no significance).
Influence of Exogenous Application of CK on Expression of Candidate Genes in Response to Salinity Stress
Next, we tested expression of candidate genes (APX1, CAT1, ERD1) after adding exogenous CK to excised leaves of plants with and without salinity stress (100–150 nM NaCl). In leaves under salinity stress and without CK, increased expression of the 3 tested genes was observed (Figure 3). Adding CK (BAP) resulted in a significantly lower gene expression relative to the salinity treatment (Figure 3). We also tested the expression of the rd29A gene (which, like the cat1 gene is controlled by ABA; Yamaguchi-Shinozaki and Shinozaki, 1994; Xing et al., 2008). The expression pattern for rd29A was similar to that of APX1, CAT1, and ERD1 (Figure 3). These findings suggest that during stress, CK downregulated the expression of genes encoding components involved in the ABA dependent or independent plant stress responses.
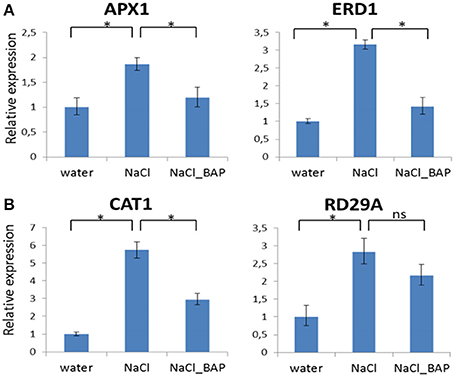
Figure 3. Effect of CK on relative expression of the APX1, ERD1 (A), and CAT1, RD29A (B) genes in Arabidopsis plants under salinity stress. Arabidopsis w.t. plants were grown for 30 days in optimal conditions and then leaves were detached and exposed to salinity stress of 100 mM NaCl or salinity plus 10 μm CK (BAP) for 24 h. Controls were detached leaves in water treatment. Samples (3 for each treatment) were collected from control and stressed leaves. Relative expression was measured by qRT- PCR. Expression of each gene in w.t. control plants was used for comparison (relative expression = 1). Horizontal brackets denote significance of P < 0.05 (*, statistical significance; ns, no significance).
We used the same experimental design to test the effect of exogenous CK on expression of components of the ethylene-signaling pathway (the MKK6-MKK9 kinase cascade) as ethylene also plays a role in plant stress response (Xu et al., 2008). Four genes were evaluated: mitogen kinase (MKK9), ACC synthase 6 (ACS6), and ethylene response factors (ERF1 and ERF5). The expression of all 4 genes increased under salinity stress, but this increase was attenuated with exogenous CK (Figure 4), suggesting that CK reduced ethylene signal transduction during stress. These findings may explain the delay of premature senescence known to be regulated by ethylene and ABA.
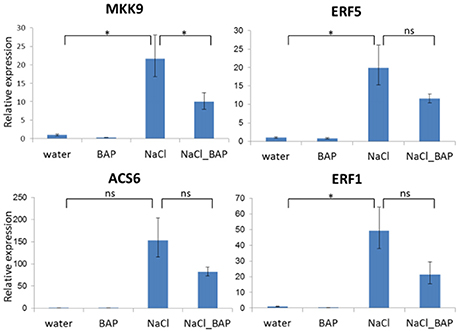
Figure 4. Influence of exogenous CK on expression of genes related to ethylene biosynthesis and signaling during salt stress. Arabidopsis w.t. plants were grown for 30 days in optimal conditions and then leaves were detached and exposed to salinity stress of 120 mM NaCl in the presence of CK (BAP) for 24 h. Controls were detached leaves in water or BAP treatments. Relative expression of MKK9, ACS6, ERF1, and ERF5 genes was assessed. Samples (3 for each treatment) were collected from controls and stressed leaves. Relative expression was measured by qRT-PCR. Expression of each gene in w.t. control plants was used for comparison (relative expression = 1). Horizontal brackets denote with significance of P < 0.05 (*, statistical significance; ns, no significance).
Transcriptomic Analysis of Arabidopsis Leaves Treated with CK under Salinity Stress
We performed a transcriptomic analysis using next-generation sequencing in order to gain an overall understanding of CK-induced transcription reprograming in Arabidopsis leaves under salinity stress. The initial RNA sequencing analysis revealed, 28,775 genes which were then filtered using four criteria: At least 3 out of 12 samples showing minimum 20 results; expression that was significantly different between control and salt treatment (P < 0.05); expression that differed by ≥1.5-fold between control and salt treatment; a change (increase or decrease) in gene expression of ≥30% due to the addition BAP. A total of 5084 genes met the selection criteria. The EXPANDER 6.4 tool was used for the division of clusters according to the expression patterns in the 4 treatment groups using the SOM clustering algorithm (Figure 5). Two clusters (2 and 8) were removed from the analysis as they included only few genes and did not yield additional results. The remaining clusters were divided into 2 super groups based on similarity in expression patters: Super-group 1 (clusters 1, 3, and 4) and super-group 2 (clusters 5, 6, and 7). We analyzed the enrichment of differentially expressed genes in the different treatments to search for over-represented Gene Ontology functional terms using the TANGO tool of the Expander 6.4 (Supplementary Tables 2–8). Supplementary Table 2 describes the functional groups in the 2 super-groups each with a typical and distinctive expression pattern. The first super-group displayed an expression pattern that included upregulation of gene expression under salt treatment as well as partial downregulation due to the addition of exogenous CK. In the context of functional categories as defined in GO, 20 functional groups (P < 0.05) were determined. This super-group included genes involved in “defense response,” “response to water stimulus,” “cellular response to abscisic acid stimulus,” and “cellular response to jasmonic acid stimulus” (Figure 6), (from the GO database or from published data; Supplementary Table 2). Although this super-group is characterized by down-regulation due to addition of CK during salinity stress, these results are consistent with our previous findings showing that CK decreased the levels of stress-protective factors. In the second super-group, which was characterized by downregulation of genes under salinity stress and upregulation by adding CK under salinity stress, 81 functional groups were identified (Supplementary Table 2). Most of these were not directly related to stress response, but rather to CK-dependent growth processes under normal conditions (i.e., genes involved in normal homeostasis in the context of cell energy, metabolism, growth, and photosynthesis; Figure 7A). This group also included genes related to the perception and responses to light in the context of photomorphogenesis (i.e., phytochrome and blue-light pigments; Figure 7B) and components associated with general gene expression and translation (Figure 7C). These findings suggest a promotive effect of CK on cell normal functioning during stress that may allow normal growth even under stress conditions. Thus, the contribution of CK to stress resistance may be related to sustaining growth capacity and maintaining metabolic processes that are otherwise disabled under stress conditions.
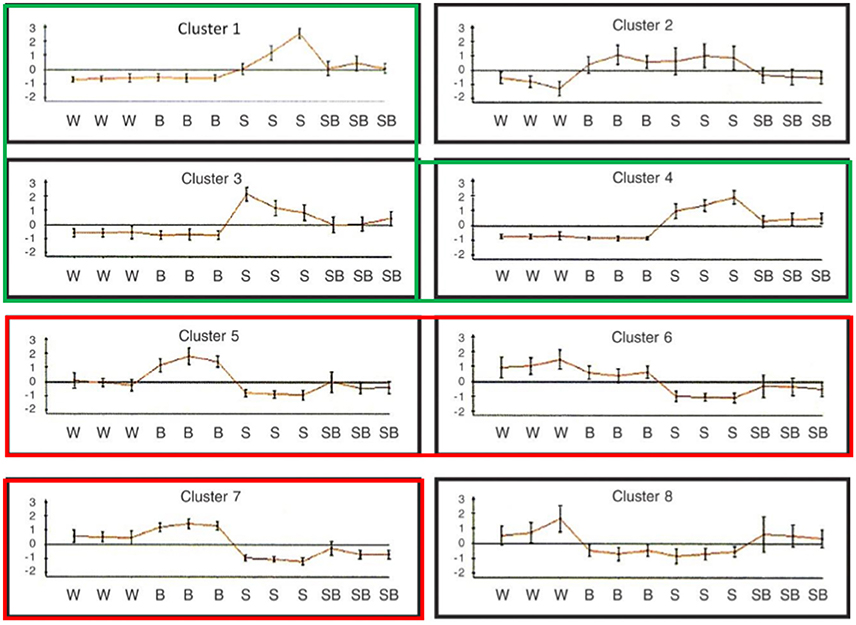
Figure 5. Graphic representation of the expression patterns of gene clusters under salinity stress with and without adding CK. Genes were divided into 8 clusters according to the expression pattern. Each graph presents one cluster with 12 samples: 4 treatments (W, water; B, 10 μm BAP; S, 120 mM NaCl; SB, NaCl plus BAP) in three biological replicates for each treatment. Gene clusters: cluster 1 contained 710 probes; cluster 2, 32 probes; cluster 3, 487 probes; cluster 4, 1405 probes; cluster 5, 342 probes; cluster 6, 553 probes; cluster 7, 1517 probes; and cluster 8, 38 probes. The red (clusters 1,3,4) and green (clusters 5,6,7) colors represent gene clusters displaying similar expression.
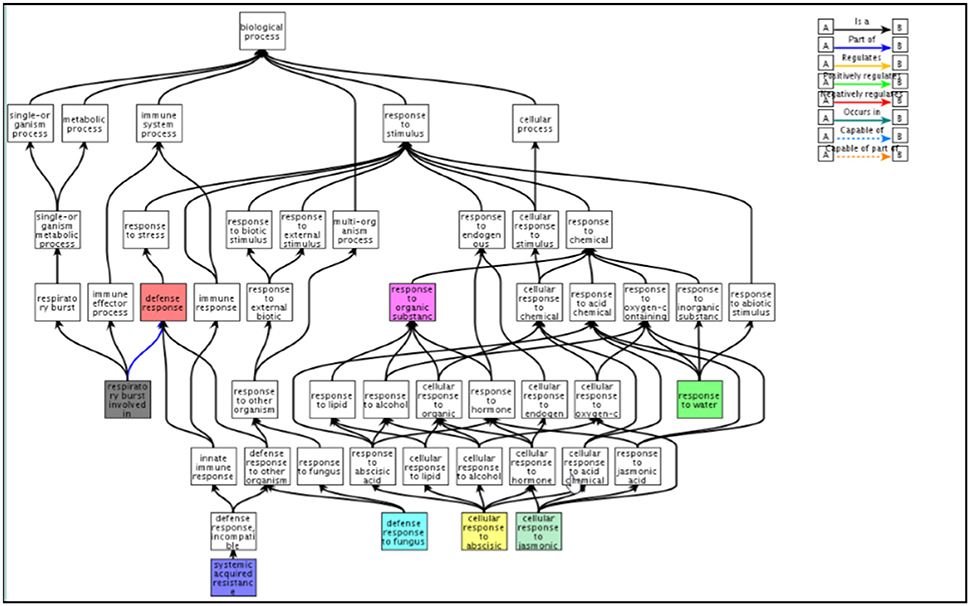
Figure 6. Functional classification of gene clusters displaying reduced expression with CK treatment under salinity stress. QuickGo representation of down-regulated Gene Ontology groups identified by the enrichment analysis of the RNA sequencing data. Clusters were characterized by upregulation of gene expression under salinity which was attenuated with CK (clusters 1,3,4). The functional classification was performed with the QuickGo platform https://www.ebi.ac.uk/QuickGO/GMultiTerm#a=64%2400ft01ie02J702MR02Sn0CQG0.HOl0HRZ&tab=chart&c=.
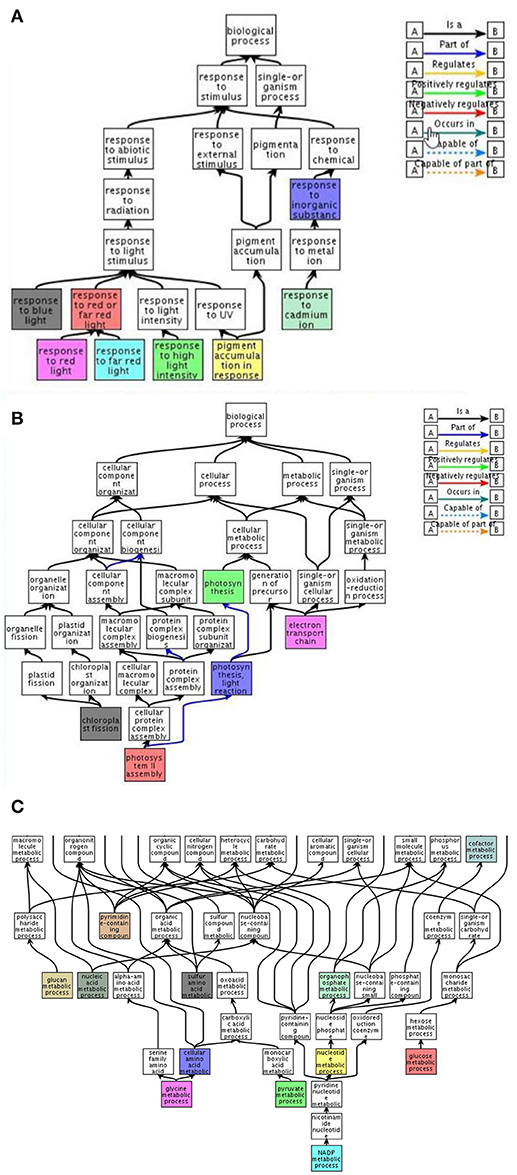
Figure 7. Functional classification of gene clusters displaying CK-dependent increased expression under salinity stress. QuickGo representation of up-regulated Gene Ontology groups identified by the enrichment analysis of the RNA sequencing data. These gene clusters are characterized by transcriptional suppression under salinity stress and moderate suppression with CK (clusters 5, 6, and 7). Functional identification was performed with the QuickGO platform (A) https://www.ebi.ac.uk/QuickGO/GMultiTerm#a=64%2402Mb02Md02Mi02Sp02U202Vg0AdM0BPU&tab=chart&c=; (C) https://www.ebi.ac.uk/QuickGO/GMultiTerm#a=64%24001W01Ts01VA01bu01cG01fJ01g602ET04or0AmA0CVo0HjF0M30&tab=chart&c=.
The Expression Patterns of the DEAD-Box ATP-Dependent RNA Helicases
We used a bioinformatics scan (DESeq 2) to identify functional groups that could contribute to the observed CK-induced stress resistance. Expression of 7 DEAD-box ATP-dependent RNA helicase genes was found to be significantly changed under salinity stress and upon CK addition (P < 0.05; Figure 8). This group of RNA helicases was previously demonstrated to have a positive or negative effect on stress resistance (Luo et al., 2009; Tuteja et al., 2014). We found that 3 out of the 7 genes were upregulated under salinity stress. All 7 genes had increased expression in salt plus BAP conditions compared to treatment with water; however, six had increased expression in salt plus BAP relative to salt treatment alone, suggesting that for one gene (AT3G58570), salt-induced activation was attenuated by CK.
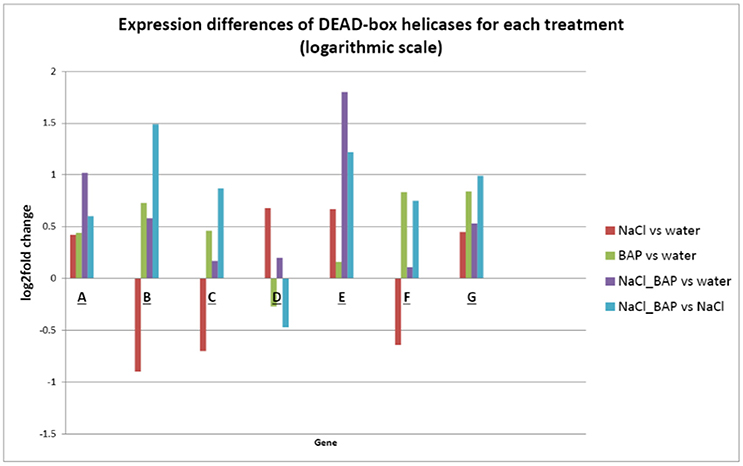
Figure 8. Effect of CK on differential expression of DEAD-box helicases (logarithmic scale). A bioinformatics scan (DESeq 2 software) was employed to identify functional groups related to the CK-induced stress resistance mechanism. The expression of 7 DEAD-box ATP-dependent RNA helicases genes were significantly changed by CK under salt stress (P < 0.05). (A) DEAD-box RNA helicase family protein (AT4G16630); (B) DEAD-box RNA helicase family protein (AT3G18600); (C) STRS1 (AT1G31970); (D) DEAD-box RNA helicase family protein (AT3G58570); (E) AtRH9 (AT3G22310); (F) PRH75 (AT5G62190); and (G) AtRH2/ STRS2 (AT5G08620).
The Expression Patterns of Type A and B Response Regulators (ARR)
Signal transduction of CK includes a 2-component system (TCS), by which a histidine protein kinase is auto-phosphorylated and the phosphate is transferred to a response regulator protein (Ishida et al., 2008). Bioinformatics enrichment analysis of our RNA sequencing data showed 8 TCS genes (type A and type B response regulator proteins) whose expression was significantly changed (P < 0.05) with salt treatment with or without exogenous CK treatment. Type A genes were found to be upregulated with salt treatment and by CK addition (Figure 9A), whereas type B genes were downregulated by CK (as their expression with salt treatment plus CK was decreased compared to their expression with salt treatment alone; Figure 9B).
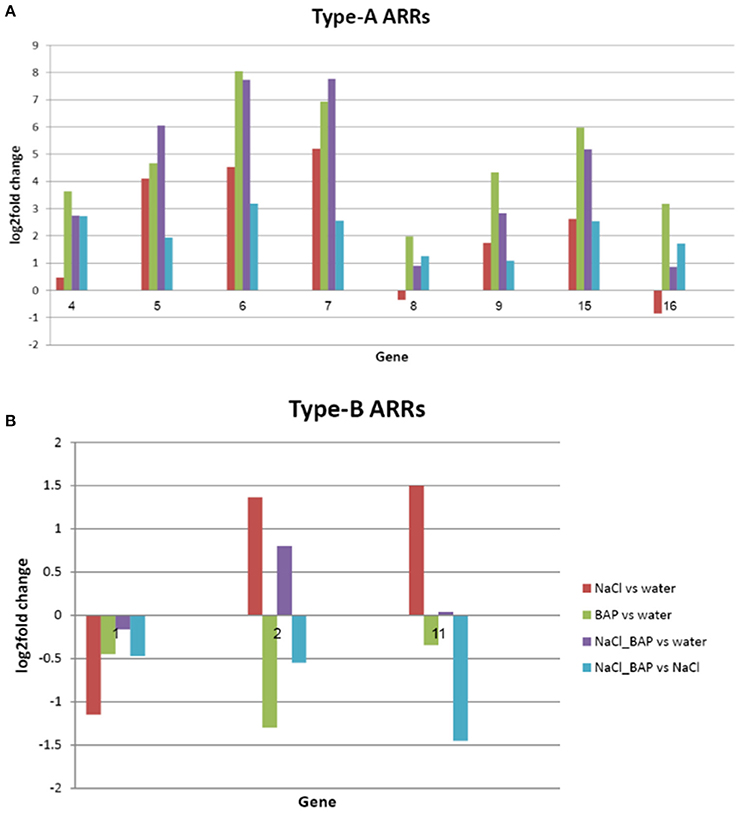
Figure 9. Effect of CK on differential expression of (A) type-A and (B) type-B ARRs in plants under salinity stress (logarithmic scale). The bioinformatics enrichment analysis of our RNA sequencing data results, showed that 8 TCS genes (type A and type B response regulator proteins) had significantly different gene expression in salinity and salinity plus CK t (P < 0.05). 4. ARR4 (AT1G10470); 5. ARR5 (AT3G48100); 6. ARR6 (AT5G62920); 7. ARR7 (AT1G19050); 8. ARR8 (AT2G41310); 9. ARR9 (AT3G57040); 15. ARR15 (AT1G74890);16. ARR16 (AT2G40670); 1. ARR1 (AT3G16857); 2. ARR2 (AT4G16110); and 11. ARR11 (AT1G67710).
Expression of Cell Wall Modification Genes
We scanned the transcriptome for other genes whose expression pattern suggest regulation by CK, by looking for genes with an expression pattern similar to that of ARR5, a known important negative regulator of CK signal transduction (To et al., 2007). In CK addition experiments on detached leaves, ARR5 was indeed shown to have increased levels under salt conditions and with CK (Figure 10). Overall, 1000 genes were selected based on similarity in expression to ARR5 and its homologs. We then searched for functional groups (GO database) in these 1000 genes and identified 5 such groups (P < 0.01; Supplementary Table 3). One of these groups was the “cell wall organization or biogenesis” group (Supplementary Table 4), that can be divided into three families: plant invertase/pectin methylesterase inhibitor superfamily, expansins, and xyloglucan endotransglusylase/hydrolases (XTH). In Arabidopsis, there are 33 genes in the XTH group (Rose et al., 2002), which are involved in softening and increasing the flexibility of the cell wall. We identified 11 of these genes in our analysis and divided them into 3 subgroups based on their gene expression pattern: Genes that were induced more under salt conditions than by CK plus salt conditions (XTH2, XTH11, XTH14, XTH24, and XTH26, Figure 11A); genes that were induced more by CK in water than by salt alone or by CK plus salt conditions (XTH8, XTH31, and XTH32; Figure 11B); and genes that were induced more by CK plus salt conditions than by either CK or salt conditions alone (XTH19, XTH29, and XTH33; Figure 11C). Expansins also participate in softening the cell wall. There are 38 expansins in Arabidopsis (Li et al., 2012), of which 13 were identified in our analysis. These 13 genes were also divided into 3 subgroups based on their expression pattern: Genes that were induced in salt treatment and had a reduced induction in salt conditions plus CK (EXPA2, EXPA20, EXPB1; Figure 12A); genes that were strongly induced with CK and had a reduced induction in salt conditions plus CK (EXPA1, EXPA3, EXPA4, EXPA11, EXPA15; Figure 12B); and genes that were induced in salt conditions and further induced by salt conditions plus CK (EXPA5, EXPA10, EXPA12, EXPA14, and EXPA16; Figure 12C). In Arabidopsis, there are >100 genes related to the plant invertase/pectin methylesterase inhibitor superfamily, although data on their function are lacking. We identified 7 genes from this family and divided them into 3 subgroups based on their expression pattern: Genes that were induced in salt conditions and had a reduced induction with salt conditions plus CK (AT3G05620, AT3G47380, and AT1G62760; Figure 13A); genes that were strongly induced in CK in water and were upregulated in salt conditions plus CK (AT3G10720 and AT5G20740; Figure 13B); and genes that were slightly upregulated in salt conditions and strongly induced upon adding CK (AT3G43270 and AT5G64640; Figure 13C).
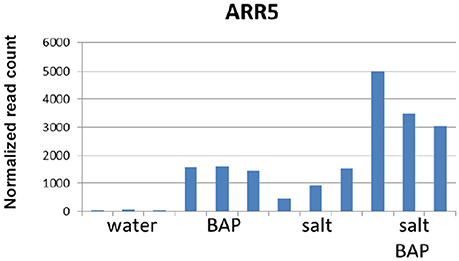
Figure 10. Effect of CK on the differential expression of ARR5 under salinity stress. Arabidopsis w.t. plants were grown for 30 days in optimal conditions and then leaves were detached and exposed to10 μm BAP, 120 mM NaCl or salt plus BAP treatments for 24 h. Controls were detached leaves in water. Samples (3 for each treatment) were collected from controls and stressed leaves.
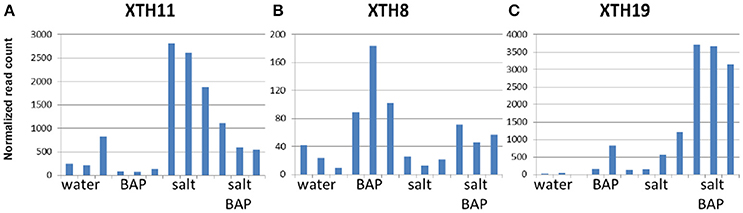
Figure 11. Effect of CK on differential expression of XTH11, XTH8, and XTH19 genes under salinity stress. (A) XTH11 gene expression (B) XTH8 gene expression (C) XTH19 gene expression. Arabidopsis w.t. plants were grown for 30 days in optimal conditions and then leaves were detached and exposed to BAP 10 μm, 120 mM NaCl, or salt plus BAP for 24 h. Controls were detached leaves in water. Three samples were collected from the controls and stressed leaves.
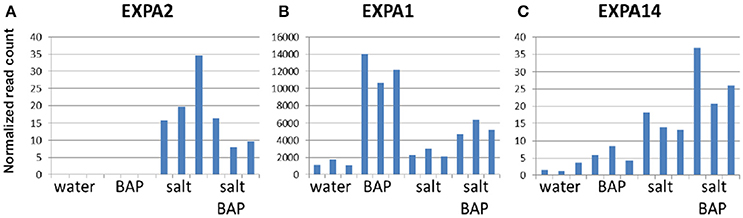
Figure 12. Effect of CK on differential expression of expansins genes under salinity stress. (A) EXPA2 gene expression (B) EXPA1 gene expression (C) EXPA14 gene expression. Arabidopsis w.t. plants were grown for 30 days in optimal conditions and then leaves were detached and exposed to BAP 10μm, 120mM NaCl, or salt plus BAP for 24h. Controls were detached leaves in water. Three samples were collected from the controls and stressed leaves.
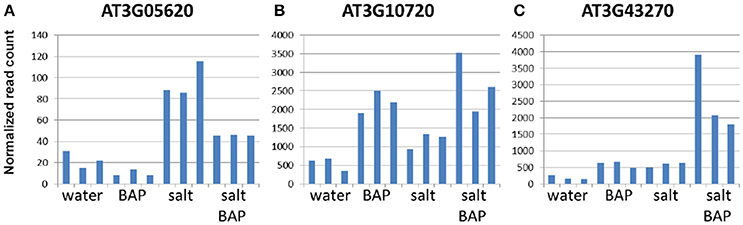
Figure 13. Effect of CK on differential expression of plant invertase/pectin methylesterase inhibitor superfamily genes under salinity stress. (A) AT3G05620 gene expression. (B) AT3G10720 gene expression. (C) AT3G43270 gene expression. Arabidopsis w.t. plants were grown for 30 days in optimal conditions and then leaves were detached and exposed to BAP 10 μm, 120 mM NaCl, or salt plus BAP for 24 h. Controls were detached leaves in water. Three samples were collected from the controls and stressed leaves.
qRT-PCR Validation of Differentially Expressed Transcripts from RNA Sequencing
We confirmed the RNA sequencing results by performing qRT-PCR on select genes. The relative gene expression of RT-PCR was calculated using the 2−ΔΔCt method. The expression trends of these genes agreed with the RNA sequencing data (Supplementary Figure 1). A significant correlation of the gene fold change was observed between BAP treatment (Pearsol = 0.74; Supplementary Figure 1A), salinity treatment (Pearsol = 0.78, Supplementary Figure 1B) and salt plus BAP experiment (Pearsol = 0.83, Supplementary Figure 1C; all the results were calculated in comparison to water). Furthermore, in qRT-PCR analyses of 2 representative genes (EXPA5 and XTH19) using the same experimental design as in the RNA sequencing experiments, we found expression patterns that were similar to those observed in the RNA sequencing analysis (Figure 14).
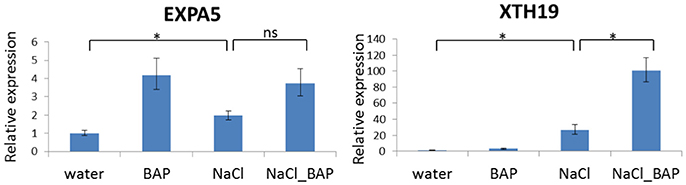
Figure 14. Effect of CK on differential expression of the EXPA5 and XTH19 genes under salinity stress. Arabidopsis w.t. plants were grown for 30 days in optimal conditions and then leaves were detached and exposed to BAP 10 μm, 120 mM NaCl, or salt plus BAP for 24 h. Controls were detached leaves in water. Three samples were collected from the controls and stressed leaves. Expression of each gene in w.t. control plants was used for comparison (relative expression = 1). Horizontal brackets denote t-test with significance of P < 0.05 (*, statistical significance; ns, no significance).
Discussion
CKs, Homeostasis, and Drought Tolerance
Studies in recent years have shown that CKs regulate responses to environmental stresses. This regulation involves interaction and crosstalk with other phytohormones. CKs have both negative and positive influence on drought tolerance. CK levels may decrease or increase depending on stress severity and duration (Zwack and Rashotte, 2015). The positive effect of CKs on drought tolerance has been demonstrated by increasing endogenous CK levels in transgenic (SARK::IPT) tobacco plants and showing that it led to improved drought tolerance mainly through delaying plant senescence and enhancing photosynthesis activity (Rivero et al., 2007). The negative influence of CKs is demonstrated by studies showing that reduced CK levels improve drought tolerance (Werner et al., 2010; Nishiyama et al., 2011, 2013). The contradicting evidence suggests that a complex CK-controlled network is involved in drought tolerance. The negative effect of CKs can derive from its effects on shoot and root growth. A decrease in the content of cytokinin, a hormone favoring shoot growth while inhibiting lateral root proliferation, can lead to increased water absorption in roots and decreased water loss under drought conditions (Ha et al., 2012).
This study focused on the molecular mechanisms underlying the positive influence of CKs on abiotic stress tolerance. We showed that CKs reverse the transcriptional programing that usually occurs in response to unfavorable abiotic stress conditions. We also showed that transgenic Arabidopsis with elevated CK levels display a significant drought resistance and maintain growth under unfavorable conditions without yield penalties. To understand how CKs influence yield under such conditions, we performed a transcriptomic analysis and showed that elevated CK levels led to stress tolerance by activating large set of genes that together maintain growth under unfavorable conditions while preventing premature senescence. All phytohormones are important for regulating every aspect of plant growth/development and in its adaptation to environmental stresses. However, phytohormones do not act in isolation and are interrelated by synergistic or antagonistic cross talk. Our data suggest the existence of cross talk between CKs and other phytohormones in the context of responses to abiotic stresses.
CK and ABA Cross-Talk
CK is antagonist of ABA: Exposing plants to water deficiency results in decreased CK levels. Exogenous ABA treatment leads to downregulation of the CK biosynthesis gene IPT (Wang et al., 2011). We demonstrated inhibition in the expression of the ABA responsive genes CAT1 and RD29A in transgenic plants that are overproducing CK. This observation suggests that CK may have an inhibitory effect on ABA biosynthesis or ABA signaling. Our transcriptomic analysis also supports this notion, as expression of genes in the Gene Ontology group associated with “cellular response to abscisic acid stimulus,” which is normally increased during stress, was inhibited by CK. Among the genes whose expression increased under salinity stress and was attenuated by CK we found the major regulatory ABA signaling component SnRK2.3 (Fujita et al., 2009) and ABF3 (Yoshida et al., 2010).
Prior studies also support the existence of cross-talk between CK and ABA signaling. For example, Rivero et al. demonstrated an increased expression of genes involved in ABA biosynthesis in w.t. plants under stress conditions but only moderate expression of the same genes in transgenic plants overproducing CK (Rivero et al., 2010). Another study reported that plants carrying 2 mutations in the CK signaling pathway (in ahk2 and ahk3) had increased expression of ABA-inducible genes (Tran et al., 2007), and a more recent study showed that CK-deficient Arabidopsis plants had increased sensitivity to exogenous ABA, causing downregulation of key ABA biosynthetic genes, and a significant reduction in endogenous ABA levels (Nishiyama et al., 2011). Furthermore, recent data have shown that the ABA-to-CK ratio in xylem sap is important for stress signaling (Nishiyama et al., 2011), suggesting that modifying CK levels can change this ratio and consequently affect the expression of ABA-induced genes. Indeed, consistent with this notion, in our experiments, when endogenous CK levels were elevated or exogenous CK (BAP) was added, many ABA inducible genes known to inhibit growth were not activated.
The known antagonistic effects of ABA and CK (Gepstein and Thimann, 1980) on plant senescence are reflected by the delayed premature senescence of our SARK::IPT transgenic plants under stress. Also, in experiments where exogenous CK was added under salinity conditions, a reduced expression of the erd1 gene, a known marker of the drought-induced senescence process (Simpson et al., 2003) was observed. Furthermore, our transcriptomic analysis revealed the Gene Ontology group designated as “defense response” whose expression was upregulated in salt conditions but downregulated when exogenous CK was added. Among the identified genes in this group were WRKY53, an important senescence regulator, and 2 senescence-associated genes (SAG13 and SAG101).
CK and Ethylene Cross-Talk
Ethylene and CK are antagonists: ethylene inhibits growth under stress and accelerates premature senescence and CKs reverse these 2 processes. Our observation that genes involved in ethylene biosynthesis and signal transduction are upregulated in salt conditions and that this upregulation is attenuated with CK, may raise a question regarding a previous study demonstrating a CK-promotive effect on ethylene biosynthesis and signal transduction under stress conditions (Jones et al., 1995). The apparent discrepancy may stem from using plants at different developmental stages (matured plants in our study vs. young plants in prior studies). The inhibitory effect of CK on ethylene biosynthesis and signal transduction is supported by our transcriptomic analysis. The ethylene response factors ERF1 and ERF13 genes (involved in transcriptional regulation of several immune responsive genes; Cheng et al., 2013), and the WRK33 gene (which encodes a transcription factor associated with plant responses to microbial infection (Mao et al., 2011), were significantly upregulated in salt conditions but were only moderately induced by exogenous CK. Our findings suggest a possible direct inhibitory effect of CK on ethylene–dependent responses, or an indirect effect on other hormones that are also related to the group associated with “cellular response to abscisic and jasmonic acids.” Other findings supporting the cross talk between CK and ethylene include reduced expression of MKK9 (a key regulator of ethylene biosynthesis (Kudryakova et al., 2001; Xu et al., 2008; An et al., 2010) and MPK3 with CK treatment under salinity stress. Furthermore, ARR2 (a type B response regulator that regulates both ethylene and CK signal transduction pathways; Hass et al., 2004), was upregulated under salinity stress, but this upregulation was attenuated with CK, suggesting that high CK levels during stress may antagonize the negative effect of ethylene through the inhibition of ARR2 expression.
Transcriptional Re-Programing by CK under Salinity Stress
We performed a transcriptomic analysis to elucidate the molecular mechanisms underlying CK-induced abiotic stress tolerance, looking at both approaches of candidate genes and the entire Arabidopsis genome. Although candidate genes represent a select sample of genes, the 2 approaches yielded similar expression patterns reflecting the effects of CKs on plant growth under abiotic stress conditions. We used the EXPANDER tool to analyze gene expression, as it integrates all analysis steps and found it to be a superior platform. In the genes identified as having low expression under salinity stress and moderate expression with CK, we found genes involved in cell normal metabolism such as gene expression, translation, metabolism of nucleic and amino acids, and metabolism of mitochondria and chloroplasts. In addition, this group included genes related to the perception and response of plants to light. These results represent the normal influence of CK on growth processes under non-stress conditions. The second super-group of genes clusters included genes whose upregulation under salinity stress was attenuated by CKs. Overall, these genes are associated with stress responses, such as “defense response” mediated by plant hormones, “cellular response to abscisic acid stimulus,” “response to water stimulus,” and “cellular response to jasmonic acid stimulus.” Interestingly, although reducing stress response, as observed for CK, could potentially diminish stress tolerance, the overall effect of CK is better tolerance as reflected by growth improvement. This observation may be partially explained by the anti-senescence action of CK. CK prevents premature senescence in stressed plants resulting in wilted but not dead leaves despite losing osmotic turgor, which facilitates recovery after the stress period ends (Zwack and Rashotte, 2013). However, the delay of premature senescence does not explain the observed improvement in plant growth under harsh conditions. Our results suggest that the known role of CK in promoting sustainable growth and photosynthetic activity during normal plant development also exists under stress conditions.
Interestingly, the genes in the clusters characterized by upregulation by CK included genes associated with “photosynthesis, light reaction,” “photosystem II assembly,” “chlorophyll biosynthetic process,” and “protein targeting to chloroplast,” suggesting that CK plays a role in reconstiting chloroplast components under stress conditions. This observation is supported by previous studies such as those demonstrating that CK has a role in increasing electron donation capacity of photosynthesis system II in drought conditions (Shao et al., 2010). The clusters also included cell wall proteins such as expansions and XTH, that may provide increased growth under stress conditions (Cho et al., 2006; Lü et al., 2013), and genes in the family of DEAD-box ATP-dependent RNA helicases, which are known to be involved in plant stress resistance (Luo et al., 2009; Tuteja et al., 2014).
Conclusions
We have shown that CK confers improved performance in Arabidopsis plants grown under abiotic stress by overcoming the genetic program that facilitates survival by growth retardation, early flowering, and accelerated senescence. Transcriptomic analysis suggested that CK triggers transcriptional reprograming leading to attenuation of the stress-dependent inhibition of vegetative growth and delaying the premature plant senescence under salt stress. We therefore hypothesize that CK allows sustainable plant growth under unfavorable environmental conditions by activating the expression of genes related to photosynthetic activity and growth. The agricultural applications of this study are under advanced developmental stages for biotechnology initiatives focused on producting transgenic crops that may grow under unfavorable water and salinit regimes.
Author Contributions
All authors contributed extensively to the work presented in this paper. YG performed all the experiments and collected the data and did the analysis of the results; NS carried out with the data interpretation and wrote the manuscript; AA assisted with the Real time PCR experiments. MS assisted with the RNAseq data analysis. SG provided direction and guidance and did the critical revision of the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Nancy and Stephen Grand Technion Energy Program (GTEP), and comprises part of The Leona M. and Harry B. Helmsley Charitable Trust reports on Alternative Energy series of the Technion, Israel Institute of Technology, and the Weizmann Institute of Science. This research was also supported by the I-CORE (Israeli Centers for Research Excellence) Program of the Planning and Budgeting Committee and The Israel Science Foundation. The contribution of the KAMIN funding of the Office of the Chief Scientist (OCS) in the Ministry of Economy of Israel is also acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fenvs.2016.00063
References
An, F., Zhao, Q., Ji, Y., Li, W., Jiang, Z., Yu, X., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22, 2384–2401. doi: 10.1105/tpc.110.076588
Arnholdt-Schmitt, B. (2004). Stress-induced cell reprogramming. A role for global genome regulation? Plant Physiol. 136, 2579–2586. doi: 10.1104/pp.104.042531
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Chernyad'ev, I. I. (2005). Effect of water stress on the photosynthetic apparatus of plants and the protective role of cytokinins: a review. Appl. Biochem. Microbiol. 41, 115–128. doi: 10.1007/s10438-005-0021-9
Cho, S. K., Kim, J. E., Park, J. A., Eom, T. J., and Kim, W. T. (2006). Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 580, 3136–3144. doi: 10.1016/j.febslet.2006.04.062
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 doi: 10.1046/j.1365-313x.1998.00343.x
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132. doi: 10.1093/pcp/pcp147
Gepstein, S., and Glick, B. R. (2013). Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol. Biol. 82, 623–633. doi: 10.1007/s11103-013-0038-z
Gepstein, S., and Thimann, K. V. (1980). Changes in the abscisic acid content of oat leaves during senescence. Proc. Natl. Acad. Sci. U.S.A. 77, 2050–2053. doi: 10.1073/pnas.77.4.2050
Ha, S., Vankova, R., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. P. (2012). Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 17, 172–179. doi: 10.1016/j.tplants.2011.12.005
Hajouj, T., Michelis, R., and Gepstein, S. (2000). Cloning and characterization of a receptor-like protein kinase gene associated with senescence. Plant Physiol. 124, 1305–1314. doi: 10.1104/pp.124.3.1305
Hass, C., Lohrmann, J., Albrecht, V., Sweere, U., Hummel, F., Yoo, S. D., et al. (2004). The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J. 23, 3290–3302. doi: 10.1038/sj.emboj.7600337
Ishida, K., Yamashino, T., Yokoyama, A., and Mizuno, T. (2008). Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 49, 47–57. doi: 10.1093/pcp/pcm165
Jones, M. L., Larsen, P. B., and Woodson, W. R. (1995). Ethylene-regulated expression of a carnation cysteine proteinase during flower petal senescence. Plant Mol. Biol. 28, 505–512. doi: 10.1007/BF00020397
Kudryakova, N. V., Burkhanova, E. A., Rakitin, V. Y., Yakovleva, L. A., Smith, A., Hall, M. A., et al. (2001). Ethylene and cytokinin in the control of senescence in detached leaves of Arabidopsis thaliana eti-5mutant and wild-type plants. Russ. J. Plant Physiol. 48, 624–627. doi: 10.1023/A:1016708002899
Li, G., Meng, X., Wang, R., Mao, G., Han, L., Liu, Y., et al. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8:e1002767. doi: 10.1371/journal.pgen.1002767
Lü, P., Kang, M., Jiang, X., Dai, F., Gao, J., and Zhang, C. (2013). RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 237, 1547–1559. doi: 10.1007/s00425-013-1867-3
Luo, Y., Liu, Y. B., Dong, Y. X., Gao, X. Q., and Zhang, X. S. (2009). Expression of a putative alfalfa helicase increases tolerance to abiotic stress in Arabidopsis by enhancing the capacities for ROS scavenging and osmotic adjustment. J. Plant Physiol. 166, 385–394. doi: 10.1016/j.jplph.2008.06.018
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., and Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653. doi: 10.1105/tpc.111.084996
Mishra, S., Shukla, A., Upadhyay, S., Sanchita, Sharma, P., Singh, S., et al. (2014). Identification, occurrence, and validation of DRE and ABRE Cis-regulatory motifs in the promoter regions of genes of Arabidopsis thaliana. J. Integr. Plant Biol. 56, 388–399. doi: 10.1111/jipb.12149
Nishiyama, R., Watanabe, Y., Fujita, Y., Le, D. T., Kojima, M., Werner, T., et al. (2011). Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell Online 23, 2169–2183. doi: 10.1105/tpc.111.087395
Nishiyama, R., Watanabe, Y., Leyva-Gonzalez, M. A., Ha, C. V., Fujita, Y., Tanaka, M., et al. (2013). Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. U.S.A. 110, 4840–4845. doi: 10.1073/pnas.1302265110
Peleg, Z., Reguera, M., Tumimbang, E., Walia, H., and Blumwald, E. (2011). Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol. J. 9, 747–758. doi: 10.1111/j.1467-7652.2010.00584.x
Rivero, R. M., Gimeno, J., Van Deynze, A., Walia, H., and Blumwald, E. (2010). Enhanced cytokinin synthesis in tobacco plants expressing PSARK::IPT prevents the degradation of photosynthetic protein complexes during drought. Plant Cell Physiol. 51, 1929. doi: 10.1093/pcp/pcq143
Rivero, R. M., Kojima, M., Gepstein, A., Sakakibara, H., Mittler, R., Gepstein, S., et al. (2007). Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. U.S.A. 104, 19631. doi: 10.1073/pnas.0709453104
Roitsch, T., and Ehne, R. (2000). Regulation of source/sink relations by cytokinins. Plant Growth Regul. 32, 359–367. doi: 10.1023/A:1010781500705
Rose, J. K., Braam, J., Fry, S. C., and Nishitani, K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43, 1421–1435. doi: 10.1093/pcp/pcf171
Shao, R., Wang, K., and Shangguan, Z. (2010). Cytokinin-induced photosynthetic adaptability of Zea mays L. to drought stress associated with nitric oxide signal: Probed by ESR spectroscopy and fast OJIP fluorescence rise. J. Plant Physiol. 167, 472–479. doi: 10.1016/j.jplph.2009.10.020
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. doi: 10.1016/S1369-5266(00)00067-4
Simpson, S. D., Nakashima, K., Narusaka, Y., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 33, 259–270. doi: 10.1046/j.1365-313X.2003.01624.x
Sreenivasulu, N., Harshavardhan, V. T., Govind, G., Seiler, C., and Kohli, A. (2012). Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506, 265–273. doi: 10.1016/j.gene.2012.06.076
To, J. P. C., Deruère, J., Maxwell, B. B., Morris, V. F., Hutchison, C. E., Ferreira, F. J., et al. (2007). Cytokinin regulates type-A arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19, 3901–3914. doi: 10.1105/tpc.107.052662
Tran, L. S. P., Urao, T., Qin, F., Maruyama, K., Kakimoto, T., Shinozaki, K., et al. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 20623. doi: 10.1073/pnas.0706547105
Tuteja, N., Banu, M. S., Huda, K. M., Gill, S. S., Jain, P., Pham, X. H., et al. (2014). Pea p68, a DEAD-box helicase, provides salinity stress tolerance in transgenic tobacco by reducing oxidative stress and improving photosynthesis machinery. PLoS ONE 9:e98287. doi: 10.1371/journal.pone.0098287
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 1–10. doi: 10.1186/s12870-016-0771-y
Wang, Y., Li, L., Ye, T., Zhao, S., Liu, Z., Feng, Y. Q., et al. (2011). Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J. 68, 249–261. doi: 10.1111/j.1365-313X.2011.04683.x
Werner, T., Nehnevajova, E., Kollmer, I., Novák, O., Strnad, M., Krämer, U., et al. (2010). Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22, 3905–3920. doi: 10.1105/tpc.109.072694
Xing, Y., Jia, W., and Zhang, J. (2008). AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54, 440–451. doi: 10.1111/j.1365-313X.2008.03433.x
Xu, J., Li, Y., Wang, Y., Liu, H., Lei, L., Yang, H., et al. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283, 26996–27006. doi: 10.1074/jbc.M801392200
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell Online 6, 251–264. doi: 10.1105/tpc.6.2.251
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Zwack, P. J., and Rashotte, A. M. (2013). Cytokinin inhibition of leaf senescence. Plant Signal. Behav. 8:e24737. doi: 10.4161/psb.24737
Keywords: abiotic stress, cytokinins, RNA-sequencing, stress tolerance, transgenic plants
Citation: Golan Y, Shirron N, Avni A, Shmoish M and Gepstein S (2016) Cytokinins Induce Transcriptional Reprograming and Improve Arabidopsis Plant Performance under Drought and Salt Stress Conditions. Front. Environ. Sci. 4:63. doi: 10.3389/fenvs.2016.00063
Received: 06 June 2016; Accepted: 21 September 2016;
Published: 07 October 2016.
Edited by:
Urs Feller, University of Bern, SwitzerlandReviewed by:
Autar Krishen Mattoo, United States Department of Agriculture, USAJose M. Garcia-Mina, University of Navarra, Spain
Copyright © 2016 Golan, Shirron, Avni, Shmoish and Gepstein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shimon Gepstein, gepstein@tx.technion.ac.il
†These authors have contributed equally to this work.
 Yelena Golan
Yelena Golan Natali Shirron
Natali Shirron Avishi Avni1
Avishi Avni1  Michael Shmoish
Michael Shmoish