Zero-Valent Iron Sand Filtration Can Reduce Human and Plant Pathogenic Bacteria While Increasing Plant Growth Promoting Bacteria in Reclaimed Water
- 1Maryland Institute for Applied Environmental Health, University of Maryland School of Public Health, College Park, MD, United States
- 2University of Maryland Institute for Advanced Computer Studies, College Park, MD, United States
- 3National Institute of Standards and Technology Biosystems and Biomaterials Division, Gaithersburg, MD, United States
- 4United Stated Department of Agriculture, Agricultural Research Service, Environmental Microbial and Food Safety Laboratory, Beltsville, MD, United States
- 5Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, MD, United States
- 6Genentech, Department of Biostatistics, Product Development, South San Francisco, CA, United States
- 7Department of Animal and Food Sciences, University of Delaware, Newark, DE, United States
- 8Department of Civil and Environmental Engineering, University of Delaware, Newark, DE, United States
The increasing use of reclaimed water for irrigation in areas lacking access to advanced wastewater treatment and reclaimed water distribution systems calls for an examination of irrigation-site-based treatment technologies that can improve the quality of this alternative water source. To address this need, we investigated the impact of zero-valent iron (ZVI)-sand filtration on the bacterial community structure and functional potential of conventionally treated reclaimed water utilized in downstream applications. Over a 2-month period, reclaimed water was collected from a tertiary wastewater treatment plant in the Mid-Atlantic, U.S. and trucked to our greenhouse facility. The water was stored in rain barrels and then filtered through one ZVI-sand filter every 5 days. Filtrate was then subjected to enumeration, phylotyping, shiga toxin screening, and antimicrobial susceptibility testing of Escherichia coli. Aliquots of filtrate were also DNA extracted, and purified DNA was subjected to 16S rRNA gene sequencing and metagenomic shotgun sequencing. The genera Dechloromonas, Desulfotomaculum, Leptonema, and Thermomonas, which contain denitrifying and sulfate reducing species, commonly used in bioremediation, and known to increase the inherent reactivity of ZVI, were significantly more relatively abundant in ZVI-sand filtered reclaimed water compared to reclaimed water. The concentration of E. coli in ZVI-sand filtered reclaimed water was significantly lower compared to that of reclaimed water, and cefoxitin- and tetracycline-resistant E. coli were undetectable after ZVI-sand filtration. ZVI-sand filtration reduced the occurrence of human as well as plant pathogenic genera (Aeromonas, Mycobacterium, Shewanella, Acidovorax, Agrobacterium, and Clavibacter) but increased the proportion of Azospira, a nitrogen fixing bacterial genera, in the microbial community. Our exploratory functional analysis showed a modest non-significant increase in the proportion of open reading frames for genes associated with iron uptake, oxidative stress, as well as defense and repair mechanisms after ZVI-sand filtration. These data indicate an iron rich environment in the filter causing an oxidative stress response by the bacterial community present in the reclaimed water. Our findings demonstrate that ZVI-sand filtration effectively filters conventionally treated reclaimed water. Longer-term, field-based studies are needed to evaluate the effectiveness of the filter in agricultural settings and inform the development of future agricultural water reuse regulations.
Introduction
Reclaimed water (treated municipal wastewater) has emerged as one of the most commonly used alternative sources of irrigation water in the United States (U.S.) (U.S. Environmental Protection Agency (EPA), 2012). Historically drought-prone states have been early adopters of reclaimed water, and have developed stringent treatment requirements (U.S. Environmental Protection Agency (EPA), 2012). For example, California requires oxidation, coagulation, filtration and disinfection of reclaimed water before use for irrigation [California Department of Public Health (CA DPH), 2009]. Climate change is beginning to compromise the quality and availability of groundwater and other freshwater resources in areas of the U.S. previously considered water-rich (e.g., the Mid-Atlantic region; U.S. Environmental Protection Agency (EPA), 2012; U.S. Global Change Research Program, 2015), and proactive water management, including the use of reclaimed water, is emerging in these areas (U.S. Environmental Protection Agency (EPA), 2012). However, the type of advanced level treatment performed in California is not typical of conventional wastewater treatment across the U.S., and in these emerging-use areas, the infrastructure is not in place to perform such advanced treatment (U.S. Environmental Protection Agency (EPA), 2012).
In areas with long-established use of reclaimed water for irrigation, advanced treatment is often performed at a central location and then reclaimed water is distributed, for use in irrigation, to areas with predominantly agricultural land-use (Monterey Regional Water Pollution Control Agency (MRWPCA), 2013; American Farmland Trust, 2017a). In contrast, emerging-use areas tend to be closer to municipal wastewater treatment plants (Thebo et al., 2017), which often perform more conventional wastewater treatment, and usually lack the infrastructure required for the type of centralized advanced treatment and distribution seen in established use regions (U.S. Environmental Protection Agency (EPA), 2012). Moreover, land use in emerging-use areas is more often a mixture of agricultural and residential applications, with agricultural irrigation performed on a much smaller scale compared to areas of established reclaimed water use, such as California (Monterey Regional Water Pollution Control Agency (MRWPCA), 2013; American Farmland Trust, 2017a,b).
Bacterial, viral, and protozoal pathogens have been shown to persist in reclaimed water after conventional wastewater treatment (Rose et al., 1996; Harwood et al., 2005; Brissaud et al., 2008; Jjemba et al., 2010; Rosenberg Goldstein et al., 2012, 2014). Specifically, indicator organisms and opportunistic pathogens have been detected in reclaimed water distribution systems after being non-detectable or present in low concentrations in conventionally treated wastewater (Jjemba et al., 2010). In addition, secondary treated, chlorinated, and dechlorinated reclaimed water can be a reservoir of antibiotic resistance genes (Fahrenfeld et al., 2013). Therefore, to facilitate the safe adoption of conventionally treated reclaimed water, it may be necessary to examine point-of-use treatment solutions that work within the existing infrastructure and scale of irrigation in emerging-use areas where centralized advanced treatment and dispersal may not be feasible.
A potential candidate for on-site treatment is zero-valent iron (ZVI) sand filtration. ZVI has been predominantly used for the remediation of groundwater contaminated with chlorinated compounds, but previous studies have shown it to be effective in the removal of viruses and bacteria as well (You et al., 2005; Ingram et al., 2012; Shi et al., 2012; Chiu, 2013; Shearer and Kniel, 2018; Marik et al., 2019). Furthermore, since ZVI does not generate potentially harmful by-products (United States Environmental Protection Agency (EPA), 2015), applications such as drinking water treatment and wastewater treatment are currently being explored (You et al., 2005; Ingram et al., 2012; Chiu, 2013).
Nevertheless, very few studies have been conducted on the effectiveness of ZVI treatment of reclaimed water and its potential as an on-site filtration system for conventionally treated reclaimed water use on small-scale farms. Therefore, the goal of this study was to examine the influence of ZVI-sand filtration on the bacterial community structure and functional potential of conventionally treated reclaimed water utilized in downstream applications.
Materials and Methods
Reclaimed Water Collection Site
Reclaimed water was collected from a tertiary wastewater treatment plant (WWTP) located in a rural town, in the Mid-Atlantic U.S., with land use including suburban developments and farmland (Maryland Department of Commerce, 2016). The WWTP has a maximum daily capacity of 1,900 m3 and treats between 1,100 and 1,400 m3 of domestic wastewater per day. This WWTP performs conventional wastewater treatment—large debris and grit removal followed by activated sludge treatment, secondary clarification and chlorination. The chlorinated effluent is used for groundwater recharge by spray irrigation. Chlorinated effluent used for this experiment was collected from an open-air lagoon prior to land application.
ZVI-Sand Filter
The container (approximate total volume of 55.5 L) from a commercially available biosand filter (HydrAid® BioSand Water Filter, NativeEnergy, Burlington, VT, USA) was adapted for this experiment. Fine filtration sand (crushed quartzite (silica) provided with the filter) (Manz, 2000; Triple Quest LLC, 2010) and ZVI (Peerless Metal Powders and Abrasives Company, Detroit, MI, USA) were sieved to achieve a particle size range of 400–625 μm. A schematic of the cross section of the ZVI-sand filter (Supplementary Figure 1) can be found in the supplementary material. The empty filter was first filled with 20 L of ultrapure water, and equal parts by volume (~25.2 L) of ZVI and sand were mixed thoroughly to the filter in batches by displacing the ultrapure water to prevent the formation of air gaps and ensure complete compaction of the ZVI-sand mixture. Preferential flow through the filter was avoided by using a gravel filled diffuser plate to manually pour reclaimed water into the filter to achieve gravity filtration. The approximate porosity of the filter was 0.52 [Center for Affordable Water and Sanitation Technology (CAWST), 2015], the approximate average flow rate through the filter was 5.6 L/min, and the filtration rate was 18 L/min/m2. The approximate contact time with the ZVI-sand filtration medium was 2.58 min, and was calculated using the following formula:
Collection of Chlorinated Effluent
The experiment was designed to simulate reuse site conditions in which reclaimed water would be delivered to the reuse site and stored until filtration and irrigation (Figure 1). Every fortnight, 240 L of chlorinated effluent was collected from the WWTP, driven to the reuse site (University of Maryland Research Greenhouse Complex, College Park, MD), divided equally (80 L each) into three 189 L rain barrels (Cat # 81313 Algreen Products Inc., Ontario, Canada) and stored until needed for filtration.
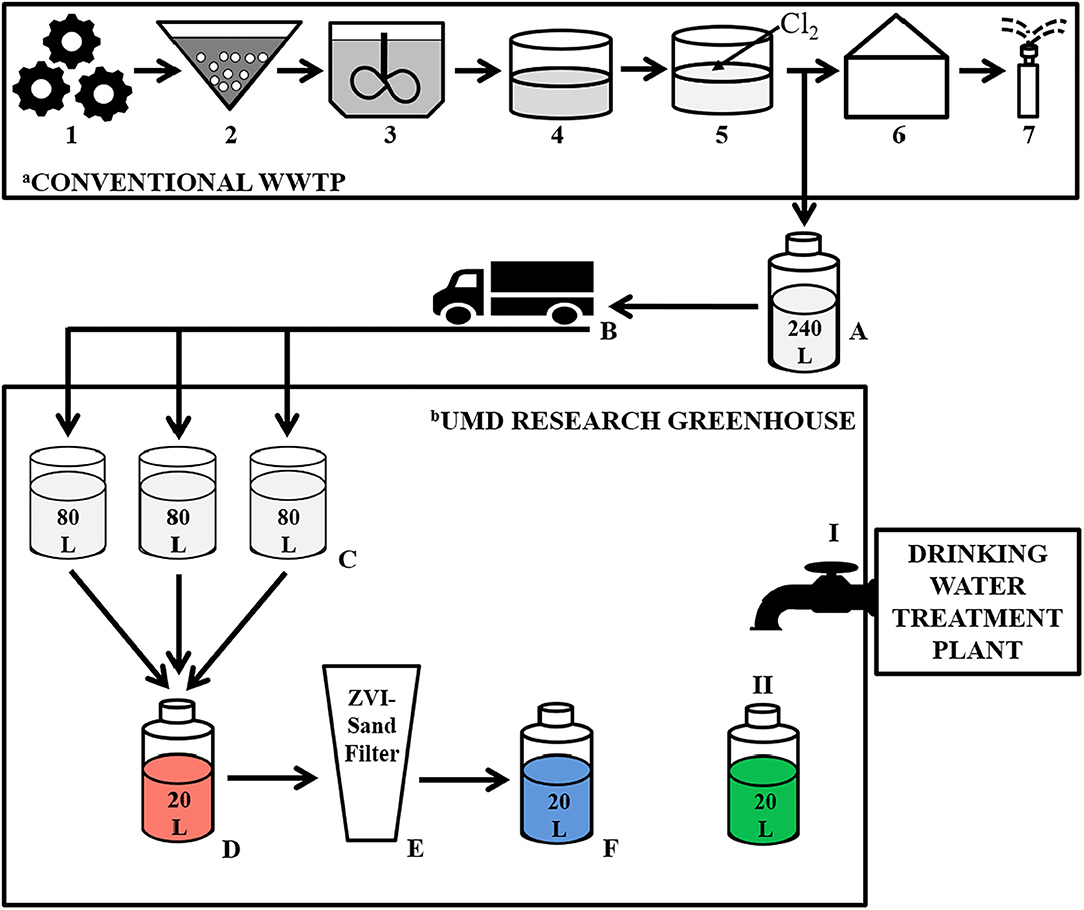
Figure 1. Schematic illustrating treatment steps at the wastewater treatment plant (WWTP), including the collection point of the chlorinated effluent used in the experiment, as well as the filtration process during our greenhouse-based experiment. aConventional WWTP: Primary Treatment: 1—Large object grinding and grit removal; Secondary Treatment: 2—Activated sludge reactor, 3—Secondary clarifier, 4—Storage lagoon; Tertiary Treatment: 5—Chlorination, 6–Pumphouse, 7–Spray irrigation, A—Collection of Chlorinated Effluent, B–Transportation of chlorinated effluent to the UMD Research Greenhouse. bUMD Research Greenhouse: C—Adding chlorinated effluent to the rain barrels; D—Generating a composite volume of reclaimed water; E—Filtering the reclaimed water through the ZVI-sand filter; F—ZVI-sand filtered reclaimed water; I—Tap supplied by a municipal drinking water treatment plant; II—Collecting tap water.
ZVI-Sand Filtration
ZVI-sand filtration took place at the University of Maryland Research Greenhouse Complex in the same room in which the chlorinated effluent was stored in the rain barrels (Figure 1). Reclaimed water (RW) was gravity filtered, every 5 days, by pouring it through the ZVI-sand filter. Specifically, equal volumes of chlorinated effluent stored in each of the three rain barrels were combined to generate a 20 L composite of reclaimed water. The ZVI-sand filter was kept submerged in reclaimed water between filtration events, and just prior to filtration, the 5-day old water held in the ZVI-sand filter was displaced, and thus completely flushed out, by pouring the aforementioned 20 L composite of reclaimed water through the filter. This displaced 5-day-old water was discarded. A new 20 L composite of reclaimed water was then generated, as described above, and poured through the ZVI-sand filter and the resulting 20 L filtrate (ZVI-sand filtered reclaimed water— “ZW”) was collected for analysis. One liter of tap water (TW) supplied to the greenhouse from a drinking water treatment plant was also collected at each filtration event. The tap water samples were used as a representation of safe, high quality irrigation water, to which the reclaimed water and ZVI-sand filtered reclaimed water could be compared. All the samples were filtered every 5 days, starting on 7/15/2016 and ending on 8/24/2016. However, a test sample that was filtered on 6/21/2016 was also included in the data analysis.
“In total there were 10 filtration events resulting in n = 10 20 L reclaimed water samples and n = 10 20 L ZVI-filtered water samples. Every time a filtration event occurred, a one-liter tap water sample was also collected as a control, as noted above. Prior to filtration, two 500 mL aliquots were taken from each 20 L reclaimed water sample. After filtration, two 500 mL aliquots were taken from each 20 L ZVI-filtered water sample. Two 500 mL aliquots were also taken from the one-liter tap water sample. These aliquots were collected for DNA extraction and enumeration of Escherichia coli. Immediately after collection, each 500 mL aliquot was taken to the laboratory on ice and held at 4°C and processed within 24 h of collection.”
DNA Extraction
Within 24 h of collection, each 500 mL aliquot was vacuum filtered through a 0.2 μm, 47 mm hydrophilic polyethersulfone (PES) filter (Pall Corporation, Port Washington, NY, USA). Total genomic DNA was extracted from the filters utilizing enzymatic and mechanical lyses using previously published procedures (Zupancic et al., 2012; Jackson et al., 2014). After each filter was aseptically placed in a sample lysis tube (Lysing Matrix B) (MP Biomedicals, Solon, OH, USA), ice-cold molecular biology grade 1X Phosphate Buffered Saline (PBS) (Gibco-Life Technologies, Grand Island, NY, USA), lysozyme from chicken egg white (10 mg/mL, Sigma-Aldrich, St. Louis, MO, USA), lysostaphin from Staphylococcus staphylolyticus (5 mg/mL Sigma-Aldrich, St. Louis, MO, USA), and mutanolysin from Streptomyces globisporus ATCC 21553 (1 mg/ml Sigma-Aldrich, St. Louis, MO, USA) were added, followed by incubation at 37°C for 30 min. A second enzymatic lysis step was conducted using Proteinase K (20 mg/mL, Invitrogen-Life Technologies, Grand Island, NY, USA) and 10% (w/v) sodium dodecyl sulfate (SDS) (BioRad, Hercules, CA, USA) followed by incubation at 55 °C for 45 min. Mechanical lysis (6.0 m/s for 40 s) was then performed using the FastPrep®-24 benchtop homogenizer (MP Biomedicals, Irvine, CA, USA). Further DNA purification was performed using the QIAmp DSP DNA mini kit 50, v2 (Qiagen, Valencia, CA, USA), according to the manufacturer's protocol. DNA quality was measured using a NanoDrop® spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and gel electrophoresis.
16S rRNA Gene Amplification and Sequencing
Previously published procedures were used to perform polymerase chain reaction (PCR) amplification of the V3-V4 hypervariable region of the 16S rRNA gene using universal primers 319F and 806R (Caporaso et al., 2012; Sellitto et al., 2012; Fadrosh et al., 2014). To allow for multiplexing samples in a single Illumina MiSeq (Illumina, San Diego, CA, USA) run, unique 12 base pair (bp) sequence tags were included with the 806R primer to barcode for each sample (Fadrosh et al., 2014). Phusion High-Fidelity DNA polymerase and mastermix (Thermo Fisher Scientific, Waltham, MA, USA) with 20 mg/mL additional bovine serum albumin (BSA) (to overcome PCR inhibition) (Sigma-Aldrich St. Louis, MO, USA) was used to perform PCR amplification in a DNA Engine Tetrad 2 thermal cycler (Bio-Rad, Hercules, CA, USA). The cycling parameters were: 30 s at 98°C, followed by 30 cycles of 10 s at 98°C, 15 s at 66°C, 15 s at 72°C, and 5 min at 72°C. For each primer pair, negative controls excluding templates were also processed. Gel electrophoresis was used to confirm amplicon presence, with quantification performed using a KAPA library quantification kit (KAPA Biosystems, Wilmington, MA, USA). From each sample, equimolar (25 ng) PCR amplicons, were mixed in a single tube and amplification primers and reaction buffers were removed using the AMPure kit (Agencourt Biosciences, Beverly, MA, USA). Amplicons were pooled and sequenced according to the manufacturer's protocol using the Illumina MiSeq (Illumina, San Diego, CA, USA).
16S rRNA Sequencing Analysis Pipeline and Data Normalization
Multiplexed 16S rRNA reads were screened for the removal of low-quality base calls and insufficient raw read lengths. PANDAseq (Masella et al., 2012) was used for assembly resulting in high-quality consensus sequences, which then underwent de-multiplexing followed by trimming of barcodes and 5′ and 3′ primer regions. UCHIME (Edgar et al., 2011) was used in de novo mode to assess for chimeras. Chloroplast and eukaryotic DNA were filtered out to reduce interference. The resulting dataset was analyzed for taxonomic and diversity analysis using the CloVR-16S pipeline (White et al., 2013) which uses two parallel protocols (a QIIME-based analysis and a Mothur/RDP-based analysis). The number of observed sequences compared to the estimated coverage can be seen in Supplementary Figure 2. The estimated coverage was determined using the Good's coverage metric (Hsieh et al., 2016). Sufficient sequencing depth was obtained and samples containing fewer than 100 sequences were excluded from downstream analysis. Data were normalized with cumulative sum scaling using metagenomeSeq (Paulson et al., 2013).
Shotgun Metagenomic Sequencing Analysis Pipeline
A subset of RW (n = 3) and ZW (n = 3) samples were selected for shotgun metagenomic sequencing which was performed by CosmosID (Rockville, MD, USA). An HS DNA Qubit fluorescent concentration assay was used to quantify each DNA sample. For each sample, all of the DNA was used in the tagmentation reaction. This was followed by 13 cycles of PCR amplification using Nextera i7 & i5 index primers & 2X KAPA master mix according to the modified Nextera XT protocol. The KAPA SYBR FAST qPCR kit was used to quantify the final libraries, with concentrations ranging from (0.1 to 212) ng/uL. Library concentrations were measured using KAPA qPCR prior to pooling. The pooled libraries were then loaded onto a high sensitivity (HS) chip run on the Caliper LabChipGX. The base pair size reported was in the range of 254–895 bp. Each pool of 84 samples was run across 8 lanes of an Illumina HiSeq 4000 flow cell targeting 100 bp paired end reads per sample. The CosmosID cloud bioinformatics platform (CosmosID Inc., Rockville, MD) was used to analyze unassembled metagenomic sequencing reads for identification at the species, subspecies, and/or strain level as well as for the quantification of relative abundance using previously described methods (Hasan et al., 2014; Lax et al., 2014; Ottesen et al., 2016; Ponnusamy et al., 2016).
Functional Annotation
Metagenomic reads were assembled into contigs. Initial quality filtering was performed using Trimmomatic (Bolger et al., 2014), paired end reads were merged using FLASH (Magoč and Salzberg, 2011), and merged reads were assembled using Spades (Bankevich et al., 2012). Individual assemblies were generated for each sample. Open reading frames (ORFs) were predicted from the assembled contigs using Metagene (Beauparlant et al., 2017). Functional annotation of ORFs was performed using the evolutionary genealogy of genes: non-supervised orthologous groups (eggNOG) database (Huerta-Cepas et al., 2016). ORFs were mapped to eggNOG using emapper-0.99.2-3-g41823b2 (Huerta-Cepas et al., 2017) and sequence searches using DIAMOND (Buchfink et al., 2014). Relative abundance values represent the number of ORFs assigned to a gene of interest and normalized by total sum scaling (TSS), dividing abundance values by total number of assigned ORFs.
Enumeration, Phylotyping, Shiga Toxin Screening, and Antimicrobial Susceptibility Testing of E. coli
Viable organisms were enumerated by using the most probable number (MPN) determination assay. One hundred milliliters aliquots were generated from each of the aforementioned 500 mL aliquots of reclaimed water and ZVI-sand-filtered reclaimed water. These 100 mL aliquots underwent serial dilution in Tryptic Soy Broth (TSB, Accumedia, Lansing, MI), followed by incubation at 42°C for 24 h, and isolation streaking on Tryptone Bile X-glucuronide (TBX, Accumedia, Lansing, MI) agar with incubation at 37°C for 24 h. MPN values of E. coli in the samples were calculated using an online MPN calculator (Curiale, 2016) and PCR confirmation of the presence of the uidA gene in presumptive E. coli was performed (Jefferson et al., 1986; Bej et al., 1991). PCR confirmed E. coli isolates were stored at −80°C until antibiotic susceptibility testing, phylotyping, and the detection of Shiga toxin-producing E. coli (STEC) were performed. Antibiotic susceptibility testing was performed based on methods determined by the United States Centers for Disease Control and Prevention (CDC) National Antimicrobial Resistance Monitoring System (NARMS) for E. coli (US Food Drug Administration, 2016). Isolates were tested on a panel of 14 antibiotics (CMV3AGNF) using a Sensititre automated microdilution system (Thermo Scientific, Waltham, MA, USA). Minimum inhibitory concentrations (MIC) were based on resistance breakpoints published by the Clinical and Laboratory Standards Institute (CLSI, 2018). E. coli phylo-typing was performed using the Clermont 2000 PCR method (Clermont et al., 2013). The detection of STEC was performed using an 11-gene multiplex polymerase chain reaction (mPCR) for detection of the presence of seven major serotypes (O26, O45, O103, O111, O121, O145, and O157) of enterohemorrhagic E. coli (EHEC), a subset of STEC, along with four major virulence factors (stx1, stx2, ehxA, and eae) (Bai et al., 2012).
Statistical Analysis
Normalized data were used to estimate the Shannon Index (Shannon and Weaver, 1948) and Simpson's Diversity Index (Simpson, 1949) using R statistical software, version 3.3.0 (R Core Team, 2017) using packages phyloseq, version 1.16.2 (McMurdie and Holmes, 2013) and vegan, version 2.3.5 (Dixon, 2003). The Kruskal-Wallis test was used to evaluate differences in alpha-diversity estimates, while the breakaway betta model (Willis et al., 2017) was used to test for differences in richness (number of bacterial species) in samples collected before and after ZVI filtration. Beta diversity was estimated using Bray-Curtis dissimilarity (Bray and Curtis, 1957) and compared using analysis of similarities (ANOSIM) on the normalized data with 999 permutations; the pairwise differences were calculated using Tukey's test. Differential relative abundance of operational taxonomic units (OTUs) across samples was estimated using metagenomeSeq, version 1.14.2 (Paulson et al., 2013). For the comparison of differential relative abundance, OTUs present in fewer than half of the samples with counts at least equal to 1 were excluded from the analysis to reduce potential biases in the statistical test due to sparsity (high frequency of unobserved OTUs). In the case of comparison of differential relative abundance specifically across treatment processes, OTUs present in less than half the samples were excluded from the analysis. MetagenomeSeq was used to compare the log-fold changes in ORF relative abundances between reclaimed water and ZVI-filtered reclaimed water samples. MPN estimates of E. coli in reclaimed water and ZVI-filtered reclaimed water were compared using the Wilcox test. In all analyses, differences were considered statistically significant at p < 0.05. When comparing reclaimed water and ZVI-filtered reclaimed water samples, the paired nature of the samples was accounted for during significance testing. All visualizations were performed using ggplot2, version 2.1.0 (Wickham, 2009).
Results
Ten reclaimed water, 10 ZVI-sand filtered reclaimed water and nine tap water samples were sequenced. 573,152, 191,137, and 3,927 16S rRNA gene sequences were obtained for reclaimed water, ZVI-sand filtered reclaimed water and tap water samples, respectively. The estimated coverage using the Good's coverage metric (Hsieh et al., 2016) is illustrated in Supplementary Figure 2.
The total number of assembled contigs, mean contig length, percentage of reads recruited, ORFs and complete ORFs for the three reclaimed water, three ZVI-sand filtered reclaimed water and three tap water samples that were shotgun sequenced are listed in Table 1.
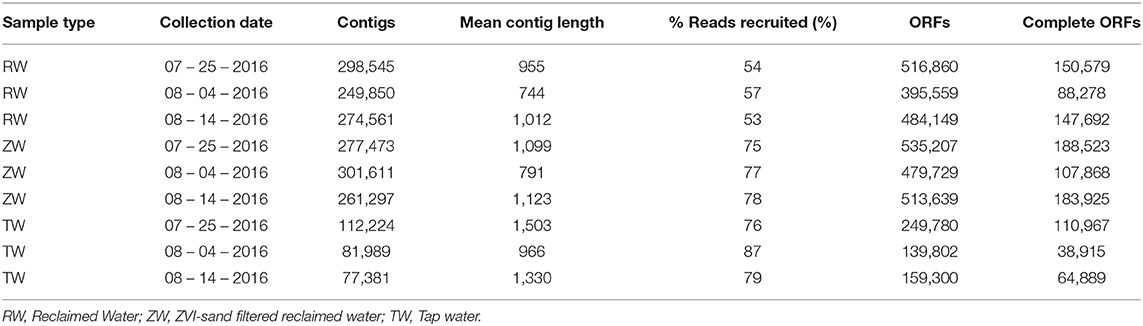
Table 1. Number of contigs, mean contig length, % reads recruited, open reading frames (ORFs), and complete ORFs per sample by collection date.
Bacterial Community Composition Differences Between Reclaimed Water and ZVI-Sand Filtered Reclaimed Water
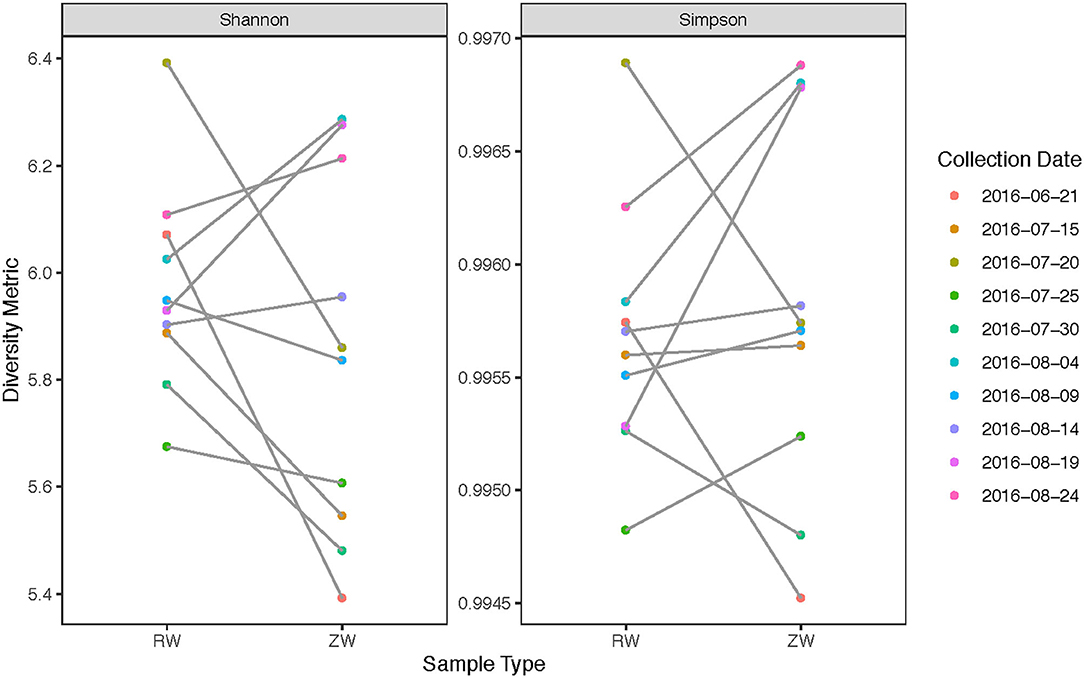
Figure 2 illustrates the relative abundance, at the phylum level, within each reclaimed water and corresponding ZVI-sand filtered reclaimed water sample collected during the experiment. Actinobacteria, Bacteroidetes, and Proteobacteria were the most relatively abundant phyla across all samples. A heatmap based on the relative abundance of cumulative sum scaling (CSS) normalized counts, at the genus level, within each reclaimed water and corresponding ZVI-sand filtered reclaimed water sample collected during the experiment is shown in Figure 3. Differences in richness and estimates of Simpson's and Shannon's alpha diversity between reclaimed water samples before and after ZVI-sand filtration were not significant (Figures 4, 5). When the reclaimed water, ZVI-sand-filtered reclaimed water and tap water samples were analyzed collectively, tap water clustered away from reclaimed water and ZVI-sand-filtered reclaimed water samples (Figure 6; ANOSIM statistic R: 0.6941, p < 0.01). Further analysis indicated that the differences between the tap water and the reclaimed water clusters and the differences between the tap water and the ZVI-sand-filtered reclaimed water clusters were significant (p < 0.05). The top five differentially relatively abundant genera between reclaimed water and ZVI-sand-filtered reclaimed water are displayed in Figure 7. The most dominant genus that was significantly (p < 0.01) higher in reclaimed water samples compared to ZVI-sand-filtered reclaimed water samples was Mycobacterium. Bacteria belonging to the genera Dechloromonas, Desulfotomaculum, Leptonema, and Thermomonas were significantly (p < 0.01) more relatively abundant in samples after ZVI-sand filtration compared to before.
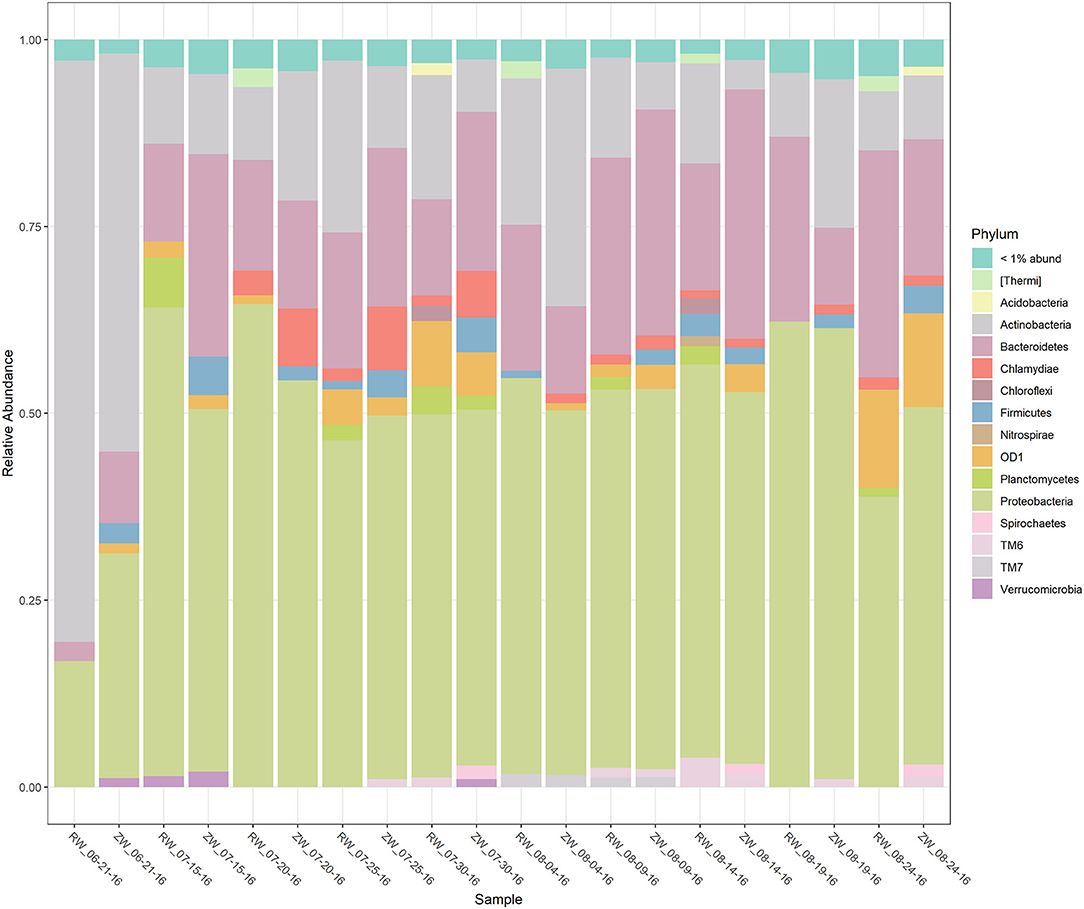
Figure 2. Stacked bar plot of the relative abundance of the bacterial phyla, within each reclaimed water (RW) and corresponding ZVI-sand filtered reclaimed water (ZW) sample.
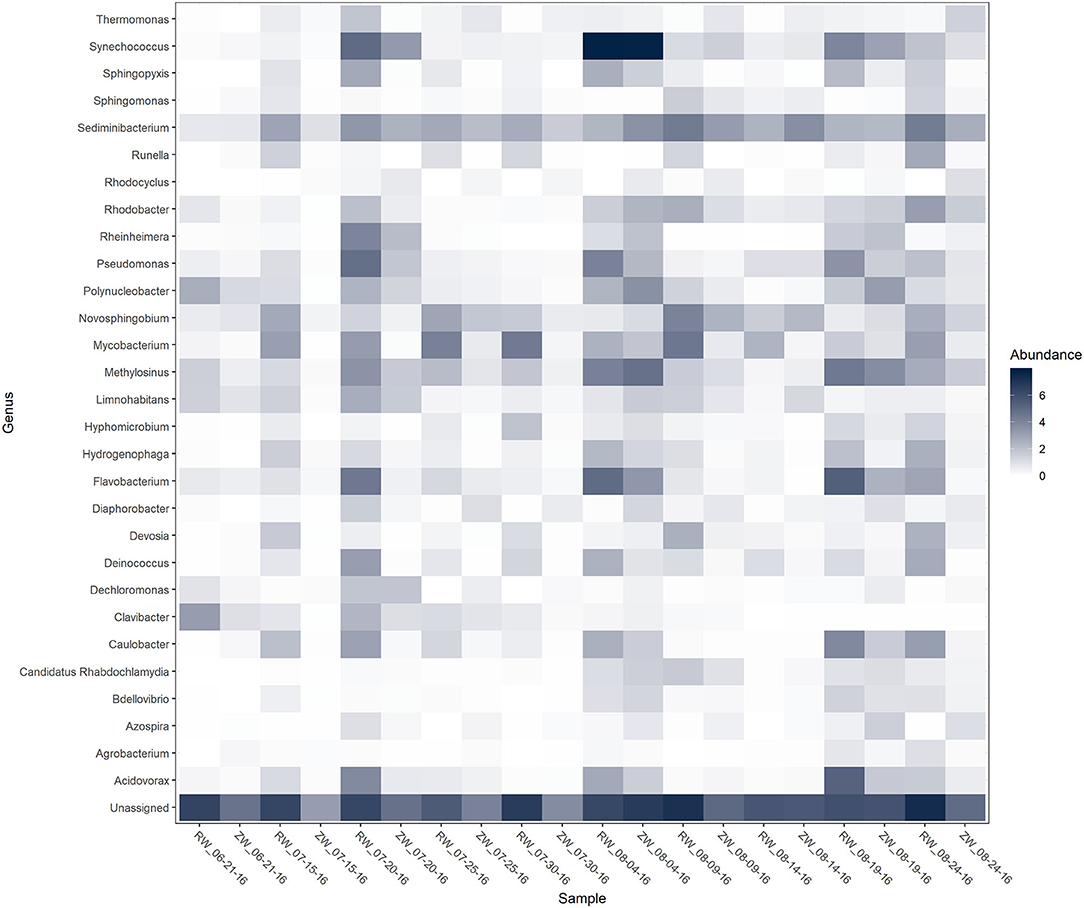
Figure 3. Heatmap of genera with >1% relative abundance [cumulative sum scaling (CSS) normalized counts] within each reclaimed water (RW) and corresponding ZVI-filtered reclaimed water (ZW) sample.
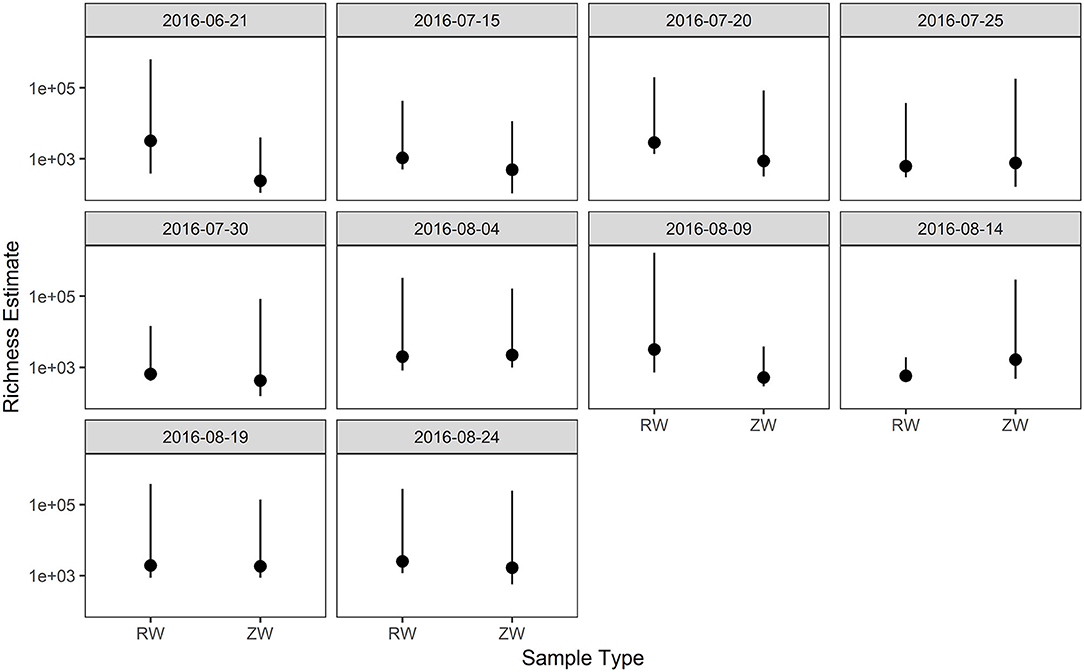
Figure 4. Comparison of richness estimates between samples collected before and after ZVI-sand filtration. There were no statistically significant differences in richness estimates between reclaimed water (RW) and ZVI-sand filtered reclaimed water (ZW).
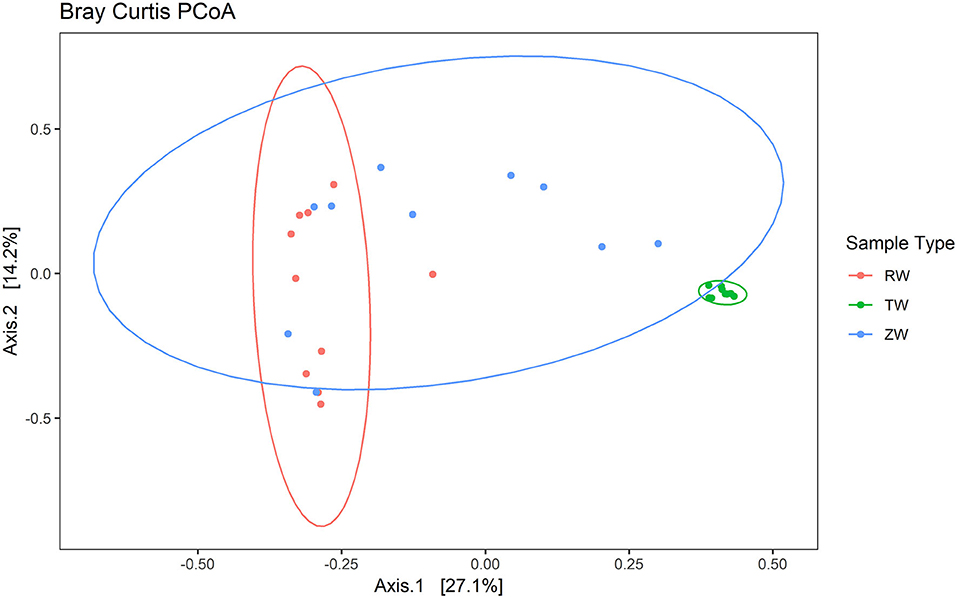
Figure 6. Principal Coordinates Analysis (PCoA) plot with Bray-Curtis dissimilarity illustrating the clustering of reclaimed water and ZVI-sand filtered reclaimed water samples with a distinct separation from the tap water group (ANOSIM statistic R: 0.6941, p < 0.01). The difference between tap water and reclaimed water sample clusters and the difference between the tap water and ZVI-sand-filtered reclaimed water sample clusters were significant (p < 0.05).
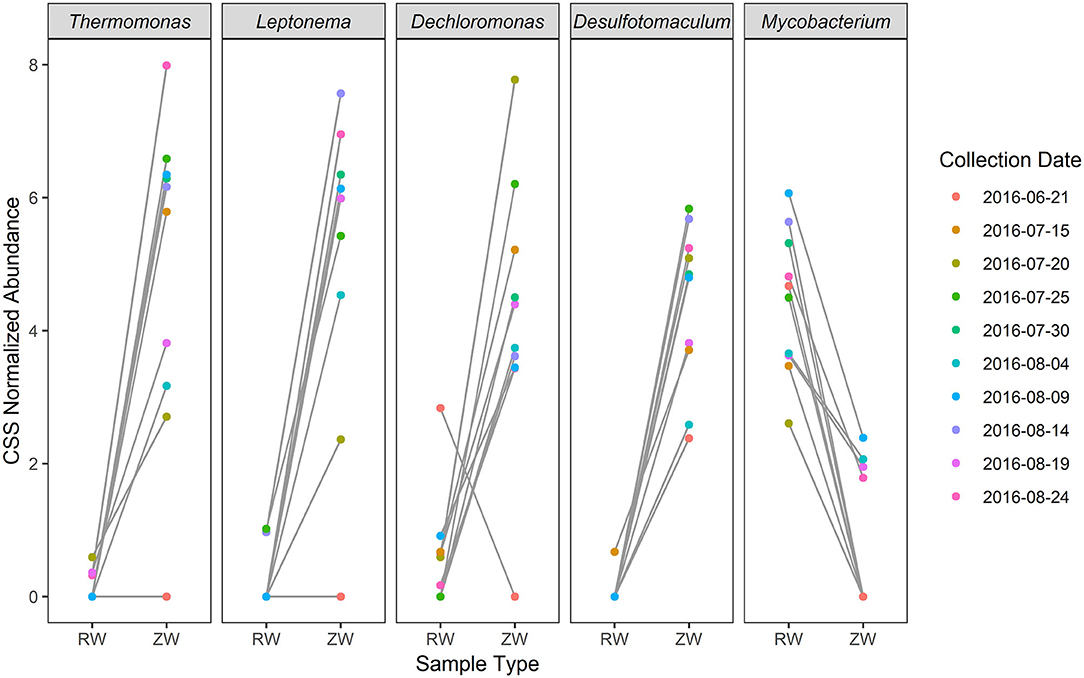
Figure 7. Top five differentially relatively abundant [cumulative sum scaling (CSS) normalized relative abundance] genera between reclaimed water and ZVI-sand filtered reclaimed water by collection date (p < 0.01).
Differences in the Concentration and Antimicrobial Susceptibility of E. coli Between Reclaimed Water and ZVI-Sand Filtered Reclaimed Water
MPN estimates of E. coli were significantly (p < 0.05) lower in all samples after ZVI-sand filtration (Figure 8). The logMPN estimates of E. coli of reclaimed water and ZVI-sand filtered reclaimed water are listed in Supplementary Table 7. Among all PCR confirmed E. coli isolates analyzed, no STEC were detected in either reclaimed water or ZVI-filtered reclaimed water samples. All PCR confirmed E. coli isolates analyzed were classified as belonging to phylogroup B1 and only two of all of the 20 E. coli isolates analyzed were found to be resistant to antibiotics. Both resistant isolates were recovered from reclaimed water samples—one collected on July 15th was found to be resistant to cefoxitin (MIC ≥ 32 μg/ml) and one collected on August 4th was found to be resistant to tetracycline (MIC ≥ 16 μg/ml) (CLSI, 2018). None of the E. coli isolates analyzed from corresponding ZVI-sand-filtered reclaimed water samples were found to be resistant to any of the antibiotics included in the panel, including isolates from ZVI-sand filtered samples corresponding to reclaimed water samples filtered on July 15th and August 4th.
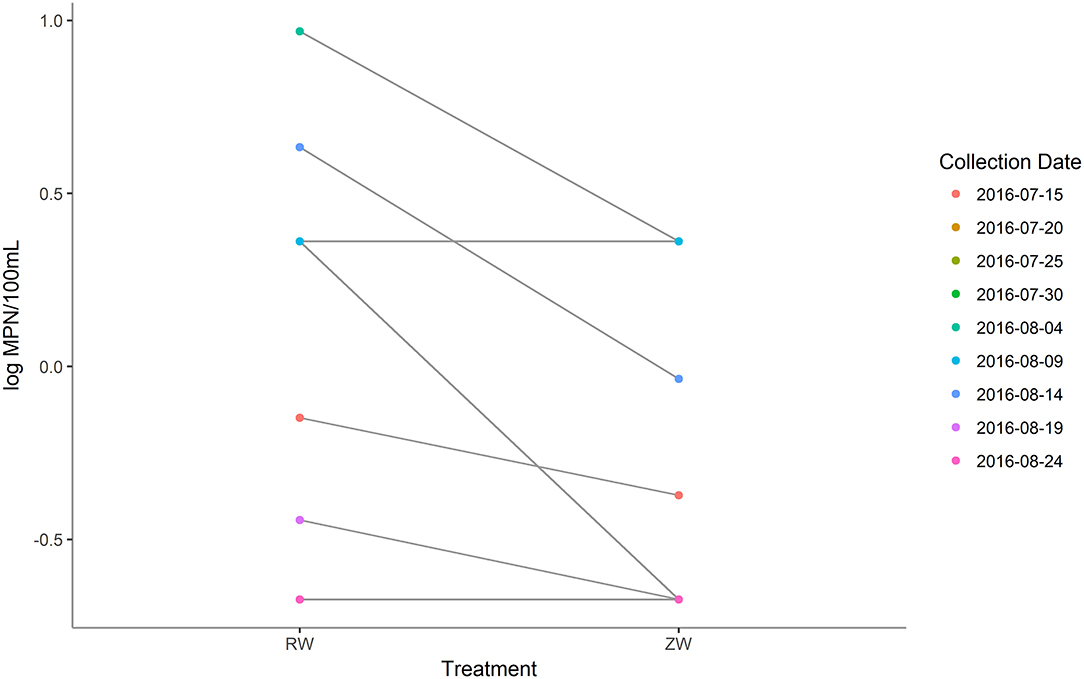
Figure 8. Most probable number (MPN) estimates (log MPN/100 mL) of Escherichia coli before and after ZVI-sand filtration of reclaimed water. MPN estimates of E. coli were significantly (p < 0.05) lower in all samples after ZVI-sand filtration.
Differences in Functional Potential of the Bacterial Community of Reclaimed Water Before and After ZVI-Sand Filtration
The log-fold changes in expression of ORFs for genes after ZVI-sand filtration (classified by function) are listed in Supplementary Table 1 through Supplementary Table 6 provided in the Supplementary Material.
A modest non-significant decrease in log-fold expression levels of ORFs for genes encoding siderophores including aerobactin, enterobactin, and mycobactin was observed in ZVI-sand-filtered reclaimed water (Supplementary Tables 1–6). Siderophores are ferric ion specific chelating agents synthesized and secreted by microorganisms under conditions of stress caused by low iron (Neilands, 1995). However, log-fold expression levels of ORFs for genes encoding membrane proteins responsible for binding and uptake of various iron compounds including ferrous iron ions and iron dicitrate, heme binding, and ferric uptake regulation were slightly, but non-significantly, higher after ZVI-sand-filtration (Supplementary Tables 1–6). The log-fold expression levels of ORFs for genes associated with detoxification (hydrogen peroxide and superoxide removal and redox homeostasis) were also higher but non-significantly in ZVI-sand-filtered reclaimed water compared to reclaimed water (Supplementary Tables 1–6). A non-significant, increase in the log-fold expression levels of ORFs for genes associated with general stress responses (heat shock, cold shock, pH imbalance, starvation, radiation, and osmoregulation response) was observed after ZVI-sand filtration (Supplementary Tables 1–6). ORFs for genes associated with other defense mechanisms (biofilm formation, cell adhesion, dormancy, cell wall biogenesis, and peptidoglycan biosynthesis) were non-significantly higher in ZVI-sand-filtered reclaimed water compared to reclaimed water (Supplementary Tables 1–6).
Data Deposition—Datasets Are in a Publicly Accessible Repository
Sequence data generated and analyzed in this study were deposited with GenBank and linked to BioProject number PRJNA522745 in the NCBI BioProject database https://www.ncbi.nlm.nih.gov/bioproject/.
Discussion
Our analysis revealed that the dominant bacterial communities detected within the ZVI-sand filtered reclaimed water were denitrifying and sulfate-reducing bacteria, commonly used in bioremediation applications, and known to increase the inherent reactivity of ZVI (van Nooten et al., 2008, 2010). Our functional analysis revealed that a potential abundance of iron ions in the extracellular environment within the ZVI-sand filter may have resulted in a state of oxidative stress within the bacterial cells in the reclaimed water, resulting in several stress response genes being expressed by the reclaimed water bacterial community. ZVI-sand filtration was able to reduce the E. coli concentration in reclaimed water and eliminate antibiotic-resistant E. coli detected in reclaimed water.
Total Bacterial Community Composition of ZVI-Sand Filtered Reclaimed Water
Aerobic as well as anaerobic corrosion reactions within the ZVI filter give rise to highly-reducing, oxygen-diminished and hydrogen gas-rich conditions (Rowland, 2003), which are ideal for both sulfate-reducing (Rowland, 2003) and denitrifying (van Nooten et al., 2010) bacteria. These conditions may have resulted in Dechloromonas, Desulfotomaculum, Leptonema, and Thermomonas being the dominant genera in the ZVI-sand filtered reclaimed water. Conversely, the most relatively abundant bacteria in reclaimed water (pre-treatment) belonged to Mycobacterium, a genus of aerobic organisms (Grange, 1996), prolific in aquatic environments, and previously detected in chlorinated reclaimed water (Grange, 1996; Jjemba et al., 2010).
Thermomonas spp., Dechloromonas spp., and Leptonema spp. have all been isolated from an H2-dependent (hydrogenotrophic) denitrification reactor used for groundwater remediation (Long-Bohanon, 2010). Dechloromonas and Thermomonas are gram-negative, non-fermenting, non-spore forming, heterotrophic, facultative anaerobic denitrifying bacteria (Achenbach et al., 2001; Busse et al., 2002; Mcilroy et al., 2016). Thermomonas spp. perform phenol and nitrate reduction under low-oxygen conditions (Baek et al., 2003) and Dechloromonas spp. perform chlorate or perchlorate reduction (Bruce et al., 1999; Chakraborty et al., 2005), can use sulfide as an electron donor (Bruce et al., 1999) and are utilized for phosphate removal in enhanced biological phosphorus removal processes (Mcilroy et al., 2016). The genus Desulfotomaculum, is comprised of gram-positive spore-forming sulfate-reducing obligate anaerobic autotrophic bacteria which oxidize molecular hydrogen in anaerobic conditions (Liamleam and Annachhatre, 2007; Aüllo et al., 2013).
Sulfate-reducing and denitrifying bacteria are both known to increase ZVI reactivity by the generation of ferrous sulfides, which can be more reactive than iron, and by converting NO3-, which competes with ZVI for reactive sites, into N2O and N2 (van Nooten et al., 2008). Moreover, van Nooten et al. (2008) showed that ZVI reactivity, and subsequently, contaminant reduction, may be achieved, not only through physical-chemical processes, but also through the contributions of the specialized microbial community generated within ZVI columns (van Nooten et al., 2008). Both denitrifying and sulfate-reducing bacteria have been used for the anaerobic bioremediation of potentially harmful inorganic as well as organic compounds (Casella and Payne, 1996; Anderson and Lovley, 2000; Hussain et al., 2016), including those that have been detected in conventionally treated reclaimed water (National Research Council, 1996, 1998; Miège et al., 2008; U.S. Environmental Protection Agency (EPA), 2012). The ZVI-sand filter examined in this study was able to reduce both potentially harmful inorganic contaminants (Kulkarni et al., 2019) as well as potentially pathogenic bacterial species detected in reclaimed water.
In a companion study, the concentrations of azithromycin, ciprofloxacin, oxolinic acid, penicillin G, sulfamethoxazole, linezolid, pipemidic acid, vancomycin, nitrate ( -N) and nitrite () found in conventionally treated reclaimed water were significantly reduced after ZVI-sand filtration (Kulkarni et al., 2019). Azithromycin, the antimicrobial with the highest median concentration (320 ng/L), was reduced to below the limit of detection after ZVI-sand filtration (Kulkarni et al., 2019). Although not statistically significant, our ZVI-sand filtration system also achieved reductions in concentrations of antimony, cadmium, lead, and selenium found in reclaimed water (Kulkarni et al., 2019).
ZVI-sand filtration was able to reduce the relative abundance of bacterial genera, containing several potential human pathogenic species, detected in our reclaimed water samples. Specifically, genera containing species which were implicated in foodborne and enteric infections (Aeromonas, Arcobacter, Comamonas, and Vibrio) (Collado and Figueras, 2011; Igbinosa et al., 2012; Farooq et al., 2017; Department of Health Human Services, 2019) respiratory infections (Achromobacter, Cupriavidus, Delftia, Klebsiella, Legionella, Mycobacterium, and Sphingobacterium) (Bagley, 1985; World Heath Organization, 2007; Kalka-Moll et al., 2009; Lambiase et al., 2009; Neonakis et al., 2010; Bilgin et al., 2015; Swenson and Sadikot, 2015; Al Hamal et al., 2016), sepsis and bacteremia (Gordonia, Lysinibacillus, Myroides, Shewanella, and Sphingomonas) (Ryan and Adley, 2010; Sharma and Kalawat, 2010; Ramanan et al., 2013; Wenzler et al., 2015; Beharrysingh, 2017), opportunistic infections (Citrobacter, Chryseobacterium, Morganella, and Stenotrophomonas) (Ranjan and Ranjan, 2013; Liu et al., 2016; Imataki and Uemura, 2017; National Institutes of Health, 2018), and genera containing several antibiotic- resistant species (Pedobacter) (Viana et al., 2018) were reduced after ZVI-sand filtration. At the species level, Aeromonas hydrophila, Arcobacter cryaerophilus, Bacillus cereus, and Plesiomonas shigelloides, which cause diarrhea, vomiting, and gastroenteritis (Janda and Abbott, 1998; Janda et al., 2016; Barboza et al., 2017; United States Department of Health Human Services, 2019), Mycobacterium arupense responsible for pulmonary infections (Neonakis et al., 2010; Al Hamal et al., 2016), Eggerthella lenta and Elizabethkingia meningoseptica which cause bacteremia (Gardiner et al., 2015; Shinha and Ahuja, 2015), and pathogens causing other severe infections—Brevundimonas diminuta (antibiotic-resistant opportunistic infections) (Han and Andrade, 2005), Clostridium bifermentans (necrotizing endometritis and empyema) (Edagiz et al., 2015; Hale et al., 2016), Propionibacterium acnes (opportunistic infections of the bones and joints, mouth, eye, and brain) (Perry and Lambert, 2011), and Pseudomonas alcaligenes (endocarditis and bloodstream infections) (Valenstein et al., 1983)—were detected at lower levels in ZVI-sand filtered reclaimed water samples compared to reclaimed water samples.
ZVI-sand filtration was also able to reduce the relative abundance of bacterial genera containing phytopathogenic species detected in our reclaimed water samples. Specifically, Acidovorax (bacterial fruit blotch on cucurbits) (Shetty et al., 2005), Agrobacterium (crown gall disease) (Moore et al., 1997), Clavibacter (bacterial wilt and canker in tomatoes and potatoes) (Gartemann et al., 2003), and Erwinia (fire blight on apples and pears, bacterial wilt in cucurbits, and wound infections in pea plants) (Huang et al., 2004) were reduced after ZVI-sand filtration.
A higher relative abundance of plant growth promoting bacteria were detected in ZVI-sand filtered reclaimed water compared to reclaimed water. Specifically, the nitrogen-fixing species Azospirillum massiliensis and Pseudomonas stutzeri (Lalucat et al., 2006; Cassán et al., 2015), and phosphate solubilizing species Acinetobacter rhizosphaerae (Gulati et al., 2009) were detected after ZVI-sand filtration. Bacterial belonging to the genus Azospira, consisting of nitrogen fixing species of bacteria (Bae et al., 2007), were much more relatively abundant in ZVI-sand filtered water samples. Furthermore, Rhodospirillaceae, purple non-sulfur bacteria that contain species (belonging to the genus Azospirillum) which have the potential to promote plant growth (Baldani et al., 2014), were detected in both reclaimed water and ZVI-sand-filtered reclaimed water samples. Therefore, the retention of, and in some cases, the increase in relative abundance of, plant growth-promoting bacteria after ZVI-sand filtration showed that ZVI may not necessarily reduce any potentially positive impacts that reclaimed water might have on plant growth. However, further analysis of the impact of ZVI-sand filtration on plant beneficial bacteria is required.
Concentration of E. coli in ZVI-Sand Filtered Reclaimed Water
The reduction of E. coli levels after ZVI-sand filtration were consistent with the findings of other studies of bacterial reduction by ZVI (Lee et al., 2008; Diao and Yao, 2009; Mudd et al., 2011; Ingram et al., 2012; Marik et al., 2019). Micro-scale ZVI filtration achieved up to 4–5 log CFU E. coli O157:H7 inactivation in inoculated water (Mudd et al., 2011), and a significantly higher (6 log CFU/100 mL) reduction in the concentration of E. coli O157:H7 compared to sand filtration (0.49 CFU/100 mL) (Ingram et al., 2012).
Functional Potential of Bacterial Community in ZVI-Sand Filtered Reclaimed Water
ZVI compromises bacterial cell membrane permeability allowing Fe2+ to enter the bacterial cell and react with intracellular H2O2 resulting in the formation of highly reactive oxygen species (ROS) generating oxidative stress and denaturing macromolecules resulting in cell death (Lee et al., 2008). Bacterial cells can defend against ZVI toxicity by global (DNA repair, maintenance of metabolic robustness etc.) and oxidative stress response (cellular detoxification and iron homeostasis etc.) mechanisms and by reducing contact with ZVI through sporulation and biofilm formation (Lefevre et al., 2015; Seo et al., 2015). All of these mechanisms may have taken place within our ZVI-sand filter as described below.
In spite of the decrease observed in the expression of ORFs for genes associated with several siderophores and iron uptake proteins (Caspi et al., 2016; Bateman et al., 2017), the overexpression of ORFs for the fur gene (involved in iron-sensing transcription regulation), and genes associated with ferrous iron binding and transport (feoB, galT, mntH) (Caspi et al., 2016), was also observed after ZVI-sand filtration (Supplementary Tables 1, 2). Therefore, the potential abundance of iron ions may have resulted in their incorporation into the intracellular environment leading to the production of ROS within bacterial cells.
The potential uptake of iron into the intracellular environment may have resulted in the generation of H2O2 and superoxides (Supplementary Table 3). The log-fold expression of ORFs for oxyR (H2O2 sensor) and H2O2-inducible genes (ahpC, ahpF, dps, fur, grxA, katG) (Seo et al., 2015; Caspi et al., 2016) was higher after ZVI-sand filtration. ORFs for genes encoding thioredoxins and glutathiones (grxA, trxA, trxB, trxC), endogenous H2O2 scavengers (ahpC, ahpF), peroxidases (ccp, katG, yfcg), oxidoreductases (fpr, glpA, glpB, glpC, wrbA), hydrogenases (hyaA, hyaB, hyaC, hyfC, hyfG, lpd), ubiquinol oxidases (cydA, cydB, cydC), and ORFs for superoxide dismutase (sodB), superoxide response (fumC, ribA, yggE) and superoxide and H2O2 removal genes (ccmA, ccmB, ccmC, ccmD, ccmE, ccmF, ccmG) (Caspi et al., 2016; Bateman et al., 2017) were overexpressed in ZVI-sand filtered reclaimed water.
Contact with ZVI may have resulted in altered environmental conditions, resulting in the activation of the global stress response system (Supplementary Table 4). The log-fold expression levels of ORFs for genes encoding proteins involved in general stress response regulation (rpoS), temperature shock (cspA, cspD, rbfA, clpS, dnaK, grpE, oppA, pncB, pnp, rpoH, rseC), osmoregulation (envZ, fixX, gshA, hchA, mglA, osmY, proV, proX, yciF, yiaO), pH imbalance (adiA, apaH, gadC, hdeA, nhaA), and starvation (cstA, yhhY) (Caspi et al., 2016; Bateman et al., 2017) increased after ZVI-sand filtration.
An overexpression of mechanisms involving repair and metabolic response was also observed after ZVI-sand contact (Supplementary Table 5). Specifically, ORFs for genes involved with DNA repair (cas1, dam, dsdA, mutM, mutS, nfo, nth, phr, polA, recA, uvrA, uvrB, uvrC, xthA), DNA synthesis and replication (priB, purA, nrdB, nrdF, nrdH), and metabolic response (glgA, glpK, malE, malF, malG, pfkB, puuP) (Caspi et al., 2016; Gamma-Castro et al., 2016; Bateman et al., 2017) were overexpressed after ZVI-sand filtration. The expression levels of genes which allow for H2O2 stress adaptability, namely, mazF (blocks protein synthesis), dps and yaaA (sequester iron to protect against DNA damage), dmsB and dmsC, (reverse oxidative stress damage), and erpA, napB, napC, napF, napG, pgi, uxuA (increases growth and metabolic processes) were higher after ZVI-sand filtration (Caspi et al., 2016; Gamma-Castro et al., 2016; Bateman et al., 2017).
Finally, ORFs for genes involved in processes that reduce ZVI contact (Supplementary Table 6) were also overexpressed in ZVI-sand filtered reclaimed water, namely, cell adhesion and biofilm formation (ackA, bolA, csgD, csgE, csgF, csgG, fimA, glnA, htrE, nlpE, znuA), dormancy process (fau), and cell wall synthesis (ampD, blc, envC, fadD, lpxC, manA, mltB, mltF, mpl, ygeA) (Gamma-Castro et al., 2016).
Study Summary, Limitations, and Future Research
Our findings demonstrate the ZVI-sand filter to be a potentially effective on-site treatment for conventionally treated reclaimed water for use in small-scale agricultural irrigation. The organic and inorganic contaminant remediation achieved by the ZVI-sand filter may have been due to a combined impact of chemical reduction and adsorption due to ZVI, the reactive oxygen species generated upon contact of bacterial cells with ZVI as well as the specialized community of denitrifying and sulfate reducing bacteria that may have developed within the ZVI-sand filter. Our ZVI-sand filter reduced or eliminated potentially pathogenic bacterial species in reclaimed water. E. coli populations were significantly reduced after ZVI-sand filtration and ZVI-sand filtration was successful in filtering cefoxitin- and tetracycline-resistant E. coli isolates from reclaimed water. The scope of our bacterial community analysis was fairly narrow due to the small sample size and the absence of a sand-only filter comparison. Our findings can serve as a basis toward more in-depth investigations that could address the longevity of ZVI-based filters as well as the effects of filtration on human and plant pathogenic bacteria in treated water, to name a few. Long-term, field-based studies that evaluate the effectiveness of the filter given fluctuating levels of contaminants over time should also be conducted before ZVI-sand filters can be adopted to provide point-of-use filtration of conventionally treated reclaimed water for agricultural use.
Data Availability Statement
Sequence data generated and analyzed in this study were deposited with GenBank and linked to BioProject number PRJNA522745 in the NCBI BioProject database https://www.ncbi.nlm.nih.gov/bioproject/.
Author Contributions
PK, AS, and MS conceived and planned the experiments. PK, AB, RB, EH, CE, and LD performed the experiments. PK, NO, and JP analyzed the data. EM, LH, JG, and DN assisted with data analysis. PK wrote the manuscript. AS and MS supervised the project. NO, AB, RB, LD, LH, EH, JP, JG, DN, CE, JVK, KK, PC, EM, MS, and AS provided critical feedback. All authors have approved the final article.
Funding
This work was supported by the United States Department of Agriculture-National Institute of Food and Agriculture, Grant No. 2016-68007-25064, awarded to the University of Maryland School of Public Health, that established CONSERVE: a Center of Excellence at the Nexus of Sustainable Water Reuse, Food and Health.
Disclaimer
Certain commercial equipment, instruments, or materials are identified in this paper only to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the NIST, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose. Official contribution of NIST; not subject to copyrights in USA. Mention of trade names or commercial products does not imply recommendation or endorsement to the exclusion of other products by the U.S. Department of Agriculture.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank the employees of the wastewater treatment plant, and the Town Administrator, for access to chlorinated effluent; Dr. Shirley Micallef for access to greenhouse space; Robert Barra, Cary Coppock, Meghan Fisher Holbert, and Russell Reynells for assistance with experiments and analysis; Dr. Poorani Subramanian and Dr. Nur Hasan for conducting whole genome sequencing analysis and Dr. Todd Treangen for assistance with data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2020.541921/full#supplementary-material
References
Achenbach, L. A., Michaelidou, U., Bruce, R. A., Fryman, J., and Coates, J. D. (2001). Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. Evol. Microbiol. 51, 527–533. doi: 10.1099/00207713-51-2-527
Al Hamal, Z., Jordan, M., Hachem, R. Y., Alawami, H. M., Alburki, A. M., Yousif, A., et al. (2016). Mycobacterium arupense in cancer patients: an emerging infection or a commensal organism. Medicine. 95:e2691. doi: 10.1097/MD.0000000000002691
American Farmland Trust (2017a). American Farmland Trust - California. Available online at: https://www.farmland.org/our-work/where-we-work/california (accessed August 25, 2017).
American Farmland Trust (2017b). American Farmland Trust - Mid-Atlantic. Available online at: https://www.farmland.org/our-work/where-we-work/mid-atlantic (accessed August 25, 2017).
Anderson, R. T., and Lovley, D. R. (2000). Anaerobic bioremediation of benzene under sulfate-reducing conditions in a petroleum-contaminated aquifer. Environ. Sci. Technol. 34, 2261–2266. doi: 10.1021/es991211a
Aüllo, T., Ranchou-Peyruse, A., Ollivier, B., and Magot, M. (2013). Desulfotomaculum spp. and related gram-positive sulfate-reducing bacteria in deep subsurface environments. Front. Microbiol. 4:362. doi: 10.3389/fmicb.2013.00362
Bae, H.-S., Rash, B. A., Rainey, F. A., Nobre, M. F., Tiago, I., da Costa, M. S., et al. (2007). Description of Azospira restricta sp. nov., a nitrogen-fixing bacterium isolated from groundwater. Int. J. Syst. Evol. Microbiol. 57, 1521–1526. doi: 10.1099/ijs.0.64965-0
Baek, S. H., Kim, K. H., Yin, C. R., Jeon, C. O., Im, W. T., Kim, K. K., et al. (2003). Isolation and characterization of bacteria capable of degrading phenol and reducing nitrate under low-oxygen conditions. Curr. Microbiol. 47, 462–466. doi: 10.1007/s00284-003-4058-9
Bagley, S. T. (1985). Habitat association of Klebsiella species. Infect. Control 6, 52–58. doi: 10.1017/S0195941700062603
Bai, J., Paddock, Z. D., Shi, X., Li, S., An, B., and Nagaraja, T. G. (2012). Applicability of a multiplex PCR to detect the seven major shiga toxin–producing Escherichia coli based on genes that code for serogroup-specific O-antigens and major virulence factors in cattle feces. Foodborne Pathog. Dis. 9, 541–548. doi: 10.1089/fpd.2011.1082
Baldani, J. I., Videira, S. S., Dos Santos Teixeira, K. R., Reis, V. M., De Oliveira, A. L. M., Schwab, S., et al. (2014). “The family Rhodospirillaceae,” in The Prokaryotes: Alphaproteobacteria and Betaproteobacteria, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer), 533–618. doi: 10.1007/978-3-642-30197-1_300
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barboza, K., Cubillo, Z., Castro, E., Redondo-Solano, M., Fernández-Jaramillo, H., and Echandi, M. L. A. (2017). First isolation report of Arcobacter cryaerophilus from a human diarrhea sample in Costa Rica. Rev. Inst. Med. Trop. São Paulo 59:e72. doi: 10.1590/s1678-9946201759072
Bateman, A., Martin, M. J., O'Donovan, C., Magrane, M., Alpi, E., Antunes, R., et al. (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. doi: 10.1093/nar/gkw1099
Beauparlant, C., Lamaze, F., Samb, R., Lippens, C., Deschenes, A., and Droit, A. (2017). metagene: A Package to Produce Metagene Plots.
Beharrysingh, R. (2017). Myroides bacteremia: a case report and concise review. IDCases 8, 34–36. doi: 10.1016/j.idcr.2017.02.012
Bej, A. K., DiCesare, J. L., Haff, L., and Atlas, R. M. (1991). Detection of Escherichia coli and Shigella spp. in water by using the polymerase chain reaction and gene probes for uid. Appl. Environ. Microbiol. 57, 1013–1017. doi: 10.1128/AEM.57.4.1013-1017.1991
Bilgin, H., Sarmis, A., Tigen, E., Soyletir, G., and Mulazimoglu, L. (2015). Delftia acidovorans: a rare pathogen in immunocompetent and immunocompromised patients. Can. J. Infect. Dis. Med. Microbiol. 26, 277–279. doi: 10.1155/2015/973284
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bray, J. R., and Curtis, J. T. (1957). An ordination of upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Brissaud, F., Blin, E., Hemous, S., and Garrelly, L. (2008). Water reuse for urban landscape irrigation: aspersion and health related regulations. Water Sci. Technol. 57, 781–788. doi: 10.2166/wst.2008.162
Bruce, R. A., Achenbach, L. A., and Coates, J. D. (1999). Reduction of (per)chlorate by a novel organism isolated from paper mill waste. Environ. Microbiol. 1, 319–329. doi: 10.1046/j.1462-2920.1999.00042.x
Buchfink, B., Xie, C., and Huson, D. H. (2014). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Busse, H. J., Kämpfer, P., Moore, E. R. B., Nuutinen, J., Tsitko, I. V., Denner, E. B. M., et al. (2002). Thermomonas haemolytica gen. nov., sp. nov., a gamma-proteobacterium from kaolin slurry. Int. J. Syst. Evol. Microbiol. 52, 473–483. doi: 10.1099/00207713-52-2-473
California Department of Public Health (CA DPH) (2009). California Department of Public Health. Water Recycling Criteria. California Code of Regulations. Available online at: http://www.cdph.ca.gov/certlic/drinkingwater/Documents/Lawbook/RWregulations-01-2009.pdf (accessed January 27, 2015).
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Casella, S., and Payne, W. J. (1996). Potential of denitrifiers for soil environment protection. FEMS Microbiol. Lett. 140, 1–8. doi: 10.1111/j.1574-6968.1996.tb08306.x
Caspi, R., Billington, R., Ferrer, L., Foerster, H., Fulcher, C. A., Keseler, I. M., et al. (2016). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 44, D471–D480. doi: 10.1093/nar/gkv1164
Cassán, F. D., Okon, Y., and Creus, C. M. (2015). Handbook for Azospirillum: Technical Issues and Protocols. Switzerland: Springer. doi: 10.1007/978-3-319-06542-7
Center for Affordable Water Sanitation Technology (CAWST) (2015). How was the Biosand Filter Pore Volume Determined. Center for Affordable Water and Sanitation Technology. Biosand Filters Knowledge Base. Available online at: http://biosandfilters.info/faq/how-was-biosand-filter-pore-volume-determined (accessed November 1, 2017).
Chakraborty, R., Oandapos;Connor, S. M., Chan, E., and Coates, J. D. (2005). Anaerobic degradation of benzene, toluene, ethylbenzene, and xylene compounds by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71, 8649–8655. doi: 10.1128/AEM.71.12.8649-8655.2005
Chiu, P. C. (2013). “Applications of zero-valent iron (ZVI) and nanoscale ZVI to municipal and decentralized drinking water systems—a review,” in ACS Symposium Series (Washington, DC), 237–249. doi: 10.1021/bk-2013-1123.ch014
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
CLSI (2018). Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI Supplement M100, 28th Edn. Waye, PA: Clinical and Laboratory Standards Institute.
Collado, L., and Figueras, M. J. (2011). Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 24, 174–192. doi: 10.1128/CMR.00034-10
Curiale, M. (2016). MPN Calculator. Available online at: http://i2workout.com/mcuriale/mpn/index.html (accessed January 23, 2017).
Department of Health Human Services (2019). Vibrio Infections. Available online at: https://www.foodsafety.gov/poisoning/causes/bacteriaviruses/vibrio_infections/index.html (accessed February 14, 2019).
Diao, M., and Yao, M. (2009). Use of zero-valent iron nanoparticles in inactivating microbes. Water Res. 43, 5243–5251. doi: 10.1016/j.watres.2009.08.051
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Edagiz, S., Lagace-Wiens, P., Embil, J., Karlowsky, J., and Walkty, A. (2015). Empyema caused by Clostridium bifermentans: a case report. Can. J. Infect. Dis. Med. Microbiol. 26, 105–107. doi: 10.1155/2015/481076
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fadrosh, D. W., Ma, B., Gajer, P., Sengamalay, N., Ott, S., Brotman, R. M., et al. (2014). An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2:6. doi: 10.1186/2049-2618-2-6
Fahrenfeld, N., Ma, Y., O'Brien, M., and Pruden, A. (2013). Reclaimed water as a reservoir of antibiotic resistance genes: distribution system and irrigation implications. Front. Microbiol. 4:130. doi: 10.3389/fmicb.2013.00130
Farooq, S., Farooq, R., and Nahvi, N. (2017). Comamonas testosteroni: is it still a rare human pathogen? Case Rep. Gastroenterol. 11, 42–47. doi: 10.1159/000452197
Gamma-Castro, S., Salgado, H., Santos-Zavaleta, A., Ledezma-Tejeida, D., Muniz-Rascado, L., Garcia-Sotelo, J., et al. (2016). RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 44, D133–D143. doi: 10.1093/nar/gkv1156
Gardiner, B. J., Tai, A. Y., Kotsanas, D., Francis, M. J., Roberts, S. A., Ballard, S. A., et al. (2015). Clinical and microbiological characteristics of eggerthella lenta bacteremia. J. Clin. Microbiol. 53, 626–635. doi: 10.1128/JCM.02926-14
Gartemann, K.-H., Kirchner, O., Engemann, J., Gräfen, I., Eichenlaub, R., and Burger, A. (2003). Clavibacter michiganensis subsp. michiganensis: first steps in the understanding of virulence of a Gram-positive phytopathogenic bacterium. J. Biotechnol. 106, 179–191. doi: 10.1016/j.jbiotec.2003.07.011
Grange, J. M. (1996). The biology of the genus Mycobacterium. Soc. Appl. Bacteriol. Symp. Ser. 25, 1S−9S. doi: 10.1111/j.1365-2672.1996.tb04827.x
Gulati, A., Vyas, P., Rahi, P., and Kasana, R. C. (2009). Plant growth-promoting and rhizosphere-competent acinetobacter rhizosphaerae strain BIHB 723 from the cold deserts of the himalayas. Curr. Microbiol. 58, 371–377. doi: 10.1007/s00284-008-9339-x
Hale, A., Kirby, J. E., and Albrecht, M. (2016). Fatal spontaneous clostridium bifermentans necrotizing endometritis: a case report and literature review of the pathogen. Open forum Infect. Dis. 3:ofw095. doi: 10.1093/ofid/ofw095
Han, X. Y., and Andrade, R. A. (2005). Brevundimonas diminuta infections and its resistance to fluoroquinolones. J. Antimicrob. Chemother. 55, 853–859. doi: 10.1093/jac/dki139
Harwood, V. J., Levine, A. D., Scott, T. M., Chivukula, V., Lukasik, J., Farrah, S. R., et al. (2005). Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 71, 3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005
Hasan, N. A., Young, B. A., Minard-Smith, A. T., Saeed, K., Li, H., Heizer, E. M., et al. (2014). Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS ONE 9:e97699. doi: 10.1371/journal.pone.0097699
Hsieh, T. C., Ma, K. H., and Chao, A. (2016). iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 7, 1451–1456. doi: 10.1111/2041-210X.12613
Huang, H. S. C., Hsieh, T. F., and Erickson, R. S. (2004). Biology and Epidemiology of Erwinia rhapontici, Causal Agent of Pink Seed and Crown Rot of Plants. Available online at: https://www.semanticscholar.org/paper/Biology-and-Epidemiology-of-Erwinia-rhapontici-%2C-of-Huang-Hsieh/e06186199b3720175b8ebbbbec8ac91dea98d881 (accessed February 14, 2019).
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Hussain, A., Hasan, A., Javid, A., and Qazi, J. I. (2016). Exploited application of sulfate-reducing bacteria for concomitant treatment of metallic and non-metallic wastes: a mini review. 3 Biotech 6:119. doi: 10.1007/s13205-016-0437-3
Igbinosa, I. H., Igumbor, E. U., Aghdasi, F., Tom, M., and Okoh, A. I. (2012). Emerging Aeromonas species infections and their significance in public health. Sci. World J. 2012:625023. doi: 10.1100/2012/625023
Imataki, O., and Uemura, M. (2017). Chryseobacterium indologenes, a possible emergent organism resistant to carbapenem antimicrobials after stem cell transplantation. Clin. Case Rep. 5, 22–25. doi: 10.1002/ccr3.753
Ingram, D. T., Callahan, M. T., Ferguson, S., Hoover, D. G., Chiu, P. C., Shelton, D. R., et al. (2012). Use of zero-valent iron biosand filters to reduce Escherichia coli O157:H12 in irrigation water applied to spinach plants in a field setting. J. Appl. Microbiol. 112, 551–560. doi: 10.1111/j.1365-2672.2011.05217.x
Jackson, H. T., Mongodin, E. F., Davenport, K. P., Fraser, C. M., Sandler, A. D., and Zeichner, S. L. (2014). Culture-independent evaluation of the appendix and rectum microbiomes in children with and without appendicitis. PLoS ONE 9:e95414. doi: 10.1371/journal.pone.0095414
Janda, J. M., and Abbott, S. L. (1998). Evolving concepts regarding the genus Aeromonas: an expanding Panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27, 332–244. doi: 10.1086/514652
Janda, J. M., Abbott, S. L., and McIver, C. J. (2016). Plesiomonas shigelloides revisited. Clin. Microbiol. Rev. 29, 349–374. doi: 10.1128/CMR.00103-15
Jefferson, R. A., Burgess, S. M., and Hirsh, D. (1986). Beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. U.S.A. 83, 8447–8451. doi: 10.1073/pnas.83.22.8447
Jjemba, P. K., Weinrich, L. A., Cheng, W., Giraldo, E., and Lechevallier, M. W. (2010). Regrowth of potential opportunistic pathogens and algae in reclaimed-water distribution systems. Appl. Environ. Microbiol. 76, 4169–4178. doi: 10.1128/AEM.03147-09
Kalka-Moll, W. M., LiPuma, J. J., Accurso, F. J., Plum, G., van Koningsbruggen, S., and Vandamme, P. (2009). Airway infection with a novel Cupriavidus species in persons with cystic fibrosis. J. Clin. Microbiol. 47, 3026–3028. doi: 10.1128/JCM.00846-09
Kulkarni, P., Raspanti, G. A., Bui, A. Q., Bradshaw, R. N., Kniel, K. E., Chiu, P. C., et al. (2019). Zerovalent iron-sand filtration can reduce the concentration of multiple antimicrobials in conventionally treated reclaimed water. Environ. Res. 172, 301–309. doi: 10.1016/j.envres.2019.02.012
Lalucat, J., Bennasar, A., Bosch, R., García-Valdés, E., and Palleroni, N. J. (2006). Biology of Pseudomonas stutzeri. Microbiol. Mol. Biol. Rev. 70, 510–547. doi: 10.1128/MMBR.00047-05
Lambiase, A., Rossano, F., Del Pezzo, M., Raia, V., Sepe, A., de Gregorio, F., et al. (2009). Sphingobacterium respiratory tract infection in patients with cystic fibrosis. BMC Res. Notes 2:262. doi: 10.1186/1756-0500-2-262
Lax, S., Smith, D. P., Hampton-Marcell, J., Owens, S. M., Handley, K. M., Scott, N. M., et al. (2014). Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052. doi: 10.1126/science.1254529
Lee, C., Kim, J. Y., Lee, W. Il., Nelson, K. L., Yoon, J., and Sedlak, D. L. (2008). Bactericidal effect of zero-valent iron nanoparticles on Escherichia coli. Environ. Sci. Technol. 42, 4927–4933. doi: 10.1021/es800408u
Lefevre, E., Bossa, N., Wiesner, M. R., and Gunsch, C. K. (2015). A review of the environmental implications of in situ remediation by nanoscale zero valent iron (nZVI): Behavior, transport and impacts on microbial communities. Sci. Total Environ. 565, 889–901. doi: 10.1016/j.scitotenv.2016.02.003
Liamleam, W., and Annachhatre, A. P. (2007). Electron donors for biological sulfate reduction. Biotechnol. Adv. 25, 452–463. doi: 10.1016/j.biotechadv.2007.05.002
Liu, H., Zhu, J., Hu, Q., and Rao, X. (2016). Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 50, 10–17. doi: 10.1016/j.ijid.2016.07.006
Long-Bohanon, C. N. (2010). Biological Removal of Nitrate from Groundwater: A Comparative Assessment of Two Intermediate-Scale Systems. Beersheba: Ben Gurion University of the Negev.
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Manz, D. H. (2000). Slow Sand Filter for Use with Intermittently Flowing Water Supply and Method of Use Thereof US6123858A. Calgary, AB.
Marik, C. M., Anderson-Coughlin, B., Gartley, S., Craighead, S., Bradshaw, R., Kulkarni, P., et al. (2019). The efficacy of zero valent iron-sand filtration on the reduction of Escherichia coli and Listeria monocytogenes in surface water for use in irrigation. Environ. Res. 173, 33–39. doi: 10.1016/j.envres.2019.02.028
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31
Mcilroy, S. J., Starnawska, A., Starnawski, P., Saunders, A. M., Nierychlo, M., Nielsen, P. H., et al. (2016). Identification of active denitrifiers in full-scale nutrient removal wastewater treatment systems. Environ. Microbiol. 18, 50–64. doi: 10.1111/1462-2920.12614
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. doi: 10.1371/journal.pone.0061217
Miège, C., Choubert, J. M., Ribeiro, L., Eusèbe, M., and Coquery, M. (2008). Removal efficiency of pharmaceuticals and personal care products with varying wastewater treatment processes and operating conditions - Conception of a database and first results. Water Sci. Technol. 57, 49–56. doi: 10.2166/wst.2008.823
Monterey Regional Water Pollution Control Agency (MRWPCA) (2013). Recycled Water. Monterey Regional Water Pollution Control Agency. Turning Wastewater into Safe Water. Monterey Reg. Water Pollut. Control Agency. Turn. Wastewater into Safe Water. Available online at: http://www.mrwpca.org/about_facilities_water_recycling.php (accessed March 23, 2016).
Moore, L. W., Chilton, W. S., and Canfield, M. L. (1997). Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors. Appl. Environ. Microbiol. 63, 201–207. doi: 10.1128/AEM.63.1.201-207.1997
Mudd, C., Callahan, M. T., Ferguson, S., Ingram, D. T., Shelton, D., Patel, J., et al. (2011). “The Use of Zero-valent Iron and Biosand Filtration to Inactivate Escherichia coli O157:H7 in Irrigation Water,” in International Association for Food Protection. Annual Meeting (Milwaukee, WI). Available online at: http://jfoodprotection.org/doi/pdf/10.4315/0362-028X-74.sp1.1?code=FOPR-site.
National Institutes of Health (2018). Stenotrophomonas Maltophilia Infection | Genetic and Rare Diseases Information Center (GARD) – An NCATS Program. Available online at: https://rarediseases.info.nih.gov/diseases/9772/stenotrophomonas-maltophilia-infection (accessed February 14, 2019).
National Research Council (1996). Use of Reclaimed Water and Sludge in Food Crop Production. Washington DC: National Academy Press
National Research Council (1998). Issues in Potable Reuse: The Viability of Augmenting Drinking Water Supplies with Reclaimed Water. Washington, DC: National Academies Press.
Neilands, J. B. (1995). Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–6. doi: 10.1074/jbc.270.45.26723
Neonakis, I., Gitti, Z., Kontos, F., Baritaki, S., Petinaki, E., Baritaki, M., et al. (2010). Mycobacterium arupense pulmonary infection: Antibiotic resistance and restriction fragment length polymorphism analysis. Indian J. Med. Microbiol. 28:173. doi: 10.4103/0255-0857.62502
Ottesen, A., Ramachandran, P., Reed, E., White, J. R., Hasan, N., Subramanian, P., et al. (2016). Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol. 16:275. doi: 10.1186/s12866-016-0894-1
Paulson, J. N., Stine, O. C., Bravo, H. C., and Pop, M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–2. doi: 10.1038/nmeth.2658
Perry, A., and Lambert, P. (2011). Propionibacterium acnes : infection beyond the skin. Expert Rev. Anti. Infect. Ther. 9, 1149–1156. doi: 10.1586/eri.11.137
Ponnusamy, D., Kozlova, E. V., Sha, J., Erova, T. E., Azar, S. R., Fitts, E. C., et al. (2016). Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc. Natl. Acad. Sci,. 113, 722–727. doi: 10.1073/pnas.1523817113
Ramanan, P., Deziel, P. J., and Wengenack, N. L. (2013). Gordonia bacteremia. J. Clin. Microbiol. 51, 3443–3447. doi: 10.1128/JCM.01449-13
Ranjan, K. P., and Ranjan, N. (2013). Citrobacter: an emerging health care associated urinary pathogen. Urol. Ann. 5, 313–314.
Rose, J., Dickson, L., Farrah, S., and Carnahan, R. (1996). Removal of pathogenic and indicator microorganisms by a full-scale water reclamation facility. Water Res. 30, 2785–2797. doi: 10.1016/S0043-1354(96)00188-1
Rosenberg Goldstein, R. E., Micallef, S. A., Gibbs, S. G., Davis, J. A., He, X., George, A., et al. (2012). Methicillin-resistant Staphylococcus aureus (MRSA) detected at four U.S. wastewater treatment plants. Environ. Health Perspect. 120, 1551–8. doi: 10.1289/ehp.1205436
Rosenberg Goldstein, R. E., Micallef, S. A., Gibbs, S. G., George, A., Claye, E., Sapkota, A. R. A., et al. (2014). Detection of vancomycin-resistant enterococci (VRE) at four U.S. wastewater treatment plants that provide effluent for reuse. Sci. Total Environ. 466–467, 404–411. doi: 10.1016/j.scitotenv.2013.07.039
Rowland, R. C. (2003). “Sulfate-reducing bacteria in the zero-valent iron permeable reactive barrier at Fry Canyon, Utah, USA,” in Handbook of Groundwater Remediation using Permeable Reactive Barriers, eds N. David, S. J. Morrison, C. C. Fuller, and J. A. Davis (San Diego, CA: Academic Press), 281–304. doi: 10.1016/B978-012513563-4/50014-1
Ryan, M. P., and Adley, C. C. (2010). Sphingomonas paucimobilis: a persistent Gram-negative nosocomial infectious organism. J. Hosp. Infect. 75, 153–157. doi: 10.1016/j.jhin.2010.03.007
Sellitto, M., Bai, G., Serena, G., Fricke, W. F., Sturgeon, C., Gajer, P., et al. (2012). Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS ONE 7:e33387. doi: 10.1371/journal.pone.0033387
Seo, S. W., Kim, D., Szubin, R., and Palsson, B. O. O. (2015). Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in Escherichia coli K-12 MG1655. Cell Rep. 12, 1289–1299. doi: 10.1016/j.celrep.2015.07.043
Shannon, C., and Weaver, W. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423 and 623–656. doi: 10.1002/j.1538-7305.1948.tb01338.x
Sharma, K. K., and Kalawat, U. (2010). Emerging infections: shewanella - a series of five cases. J. Lab. Phys. 2, 61–65. doi: 10.4103/0974-2727.72150
Shearer, A. E. H., and Kniel, K. E. (2018). Enhanced removal of norovirus surrogates, murine norovirus and tulane virus, from aqueous systems by zero-valent iron. J. Food Prot. 81, 1432–1438. doi: 10.4315/0362-028X.JFP-18-054
Shetty, A., Barnes, R. A., Healy, B., and Groves, P. (2005). A case of sepsis caused by Acidovorax. J. Infect. 51, e171–e172. doi: 10.1016/j.jinf.2004.12.014
Shi, C., Wei, J., Jin, Y., Kniel, K. E., and Chiu, P. C. (2012). Removal of viruses and bacteriophages from drinking water using zero-valent iron. Separat. Purif. Technol. 84, 72–78. doi: 10.1016/j.seppur.2011.06.036
Shinha, T., and Ahuja, R. (2015). Bacteremia due to Elizabethkingia meningoseptica. IDCases 2, 13–15. doi: 10.1016/j.idcr.2015.01.002
Swenson, C. E., and Sadikot, R. T. (2015). Achromobacter respiratory infections. Ann. Am. Thorac. Soc. 12, 252–258. doi: 10.1513/AnnalsATS.201406-288FR
Thebo, A., Nelson, K., and Sheikh, B. (2017). Agricultural Water Reuse: Impediments and Incentives WRRF 15-08. Available online at: https://watereuse.org/wp-content/uploads/2016/09/Thebo_CentralValleyReuse_19Oct2017.pdf (accessed August 14, 2018).
Triple Quest LLC (2010). Hydraid Biosand Water Filter Handbook. Available online at: www.HydrAid.org/resources (accessed November 18, 2016).
U.S. Environmental Protection Agency (EPA) (2012). Guidelines for Water Reuse. 600/R-12/618. Washington, DC: Environmental Protection Agency.
U.S. Global Change Research Program (2015). National Climate Change Assessment. Available online at: http://nca2014.globalchange.gov/ (accessed November 18, 2016).
United States Department of Health and Human Services (2019). Bacillus Cereus. Available online at: https://www.foodsafety.gov/poisoning/causes/bacteriaviruses/bcereus/index.html (accessed February 14, 2019).
United States Environmental Protection Agency (EPA) (2015). In Situ Chemical Reduction. Contam. Site Clean-Up Inf. Available online at: https://clu-in.org/techfocus/default.focus/sec/In_Situ_Chemical_Reduction/cat/Overview/ (accessed March 18, 2016).
US Food and Drug Administration (2016). The National Antimmicrobial Resistance Monitoring System Manual of Laboratory Methods.
Valenstein, P., Bardy, G. H., Cox, C. C., and Zwadyk, P. (1983). Pseudomonas alcaligenes endocarditis. Am. J. Clin. Pathol. 79, 245–247. doi: 10.1093/ajcp/79.2.245
van Nooten, T., Springael, D., and Bastiaens, L. (2008). Positive impact of microorganisms on the performance of laboratory-scale permeable reactive iron barriers. Environ. Sci. Technol. 42, 1680–1686. doi: 10.1021/es071760d
van Nooten, T., Springael, D., and Bastiaens, L. (2010). Microbial community characterization in a pilot-scale permeable reactive iron barrier. Environ. Eng. Sci. 27, 287–292. doi: 10.1089/ees.2009.0271
Viana, A. T., Caetano, T., Covas, C., Santos, T., and Mendo, S. (2018). Environmental superbugs: the case study of Pedobacter spp. Environ. Pollut. 241, 1048–1055. doi: 10.1016/j.envpol.2018.06.047
Wenzler, E., Kamboj, K., and Balada-Llasat, J.-M. (2015). Severe sepsis secondary to persistent lysinibacillus sphaericus, lysinibacillus fusiformis and paenibacillus amylolyticus bacteremia. Int. J. Infect. Dis. 35, 93–95. doi: 10.1016/j.ijid.2015.04.016
White, J. R., Maddox, C., White, O., Angiuoli, S. V., and Fricke, W. F. (2013). CloVR-ITS: Automated internal transcribed spacer amplicon sequence analysis pipeline for the characterization of fungal microbiota. Microbiome 1:6. doi: 10.1186/2049-2618-1-6
Wickham, H. (2009). Elegant graphics for data analysis. Media 35:211. doi: 10.1007/978-0-387-98141-3
Willis, A., Bunge, J., and Whitman, T. (2017). Improved detection of changes in species richness in high diversity microbial communities. J. R. Stat. Soc. Ser. C Appl. Stat. 66, 963–977. doi: 10.1111/rssc.12206
World Heath Organization (2007). Legionella and the Prevention of Legionellosis, eds. J. Bartram, Y. Chartier, J. V. Lee, K. Pond, and S.-L. Susanne (Geneva).
You, Y., Han, J., Chiu, P. C., and Jin, Y. (2005). Removal and inactivation of waterborne viruses using zerovalent iron. Environ. Sci. Technol. 39, 9263–9269. doi: 10.1021/es050829j
Keywords: zero-valent iron, reclaimed water, point-of-use treatment, small-scale agriculture, metagenomics, 16S rRNA sequence analysis
Citation: Kulkarni P, Olson ND, Bui AQ, Bradshaw RN, Del Collo LP, Hittle LE, Handy ET, Paulson JN, Ghurye J, Nasko DJ, East C, Kessel JAV, Kniel KE, Chiu PC, Mongodin EF, Pop M, Sharma M and Sapkota AR (2020) Zero-Valent Iron Sand Filtration Can Reduce Human and Plant Pathogenic Bacteria While Increasing Plant Growth Promoting Bacteria in Reclaimed Water. Front. Environ. Sci. 8:541921. doi: 10.3389/fenvs.2020.541921
Received: 10 March 2020; Accepted: 30 September 2020;
Published: 02 November 2020.
Edited by:
Prosun Bhattacharya, Royal Institute of Technology, SwedenReviewed by:
Konstantinos Plakas, Centre for Research and Technology Hellas (CERTH), GreeceBrandon Kyle Winfrey, Monash University, Australia
Copyright © 2020 Kulkarni, Olson, Bui, Bradshaw, Del Collo, Hittle, Handy, Paulson, Ghurye, Nasko, East, Kessel, Kniel, Chiu, Mongodin, Pop, Sharma and Sapkota. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amy R. Sapkota, ars@umd.edu
 Prachi Kulkarni
Prachi Kulkarni Nathan D. Olson
Nathan D. Olson Anthony Q. Bui
Anthony Q. Bui Rhodel N. Bradshaw4
Rhodel N. Bradshaw4  Laura P. Del Collo
Laura P. Del Collo Joseph N. Paulson
Joseph N. Paulson Daniel J. Nasko
Daniel J. Nasko Cheryl East
Cheryl East Kalmia E. Kniel
Kalmia E. Kniel Pei C. Chiu
Pei C. Chiu Emmanuel F. Mongodin
Emmanuel F. Mongodin Mihai Pop
Mihai Pop Manan Sharma
Manan Sharma Amy R. Sapkota
Amy R. Sapkota