Heavy Metals in Soils and the Remediation Potential of Bacteria Associated With the Plant Microbiome
- Departamento de Ciencias Biológicas, Universidad Icesi, Cali, Colombia
High concentrations of non-essential heavy metals/metalloids (arsenic, cadmium, and lead) in soils and irrigation water represent a threat to the environment, food safety, and human and animal health. Microbial bioremediation has emerged as a promising strategy to reduce the concentration of heavy metals in the environment due to the demonstrated ability of microorganisms, especially bacteria, to sequester and transform these compounds. Although several bacterial strains have been reported to be capable of remediation of soils affected by heavy metals, published information has not been comprehensively analyzed to date to recommend the most efficient microbial resources for application in bioremediation or bacterial-assisted phytoremediation strategies that may help improve plant growth and yield in contaminated soils. In this study, we critically analyzed eighty-five research articles published over the past 15 years, focusing on bacteria-assisted remediation strategies for the non-essential heavy metals, arsenic, cadmium, and lead, and selected based on four criteria: i) The bacterial species studied are part of a plant microbiome, i.e., they interact closely with a plant species ii) these same bacterial species exhibit plant growth-promoting characteristics, iii) bacterial resistance to the metal(s) is expressed in terms of the Minimum Inhibitory Concentration (MIC), and iv) metal resistance is related to biochemical or molecular mechanisms. A total of sixty-two bacterial genera, comprising 424 bacterial species/strains associated with fifty plant species were included in our analysis. Our results showed a close relationship between the tolerance level exhibited by the bacteria and metal identity, with lower MIC values found for cadmium and lead, while resistance to arsenic was widespread and significantly higher. In-depth analysis of the most commonly evaluated genera, Agrobacterium, Bacillus, Klebsiella, Enterobacter, Microbacterium, Pseudomonas, Rhodococcus, and Mesorhizobium showed significantly different tolerance levels among them and highlighted the deployment of different biochemical and molecular mechanisms associated with plant growth promotion or with the presence of resistance genes located in the cad and ars operons. In particular, the genera Klebsiella and Enterobacter exhibited the highest levels of cadmium and lead tolerance, clearly supported by molecular and biochemical mechanisms; they were also able to mitigate plant growth inhibition under phytotoxic metal concentrations. These results position Klebsiella and Enterobacter as the best potential candidates for bioremediation and bacteria-assisted phytoremediation strategies in soils contaminated with arsenic, cadmium, and lead.
Introduction
Soil pollution by heavy metals represents a threat to the environment and food security due to the fast growth of industry and agriculture, and the disruption of natural ecosystems by anthropogenic pressure linked to the growth of human populations (Sarwar et al., 2017). Environmental pollution and human exposure associated with heavy metals are attributed to different anthropogenic activities that include mining, industrial production, and the use of metal-containing compounds in domestic and agricultural settings (Tchounwou et al., 2012). Throughout the world, there are 5 million sites of soil contaminated by heavy metals/metalloids with current concentrations above the regulatory levels (Li et al., 2019). Contamination by heavy metals poses many risks to the ecosystem and humans and affects food chain safety, food quality, and the ability to use the land for agricultural production, which in turn affects food security and exacerbates land tenure problems (Wuana and Okieimen, 2011).
Heavy metals are a group of elements with metallic properties that include transition metals, metalloids, lanthanides, and actinides (Singh et al., 2011). Some of them are essential cofactors for different enzymes, while others are non-essential (Theron et al., 2012). The first group includes the trace elements cobalt, copper, iron, manganese, molybdenum, selenium, and zinc, whose concentrations are closely regulated by interactions with binding proteins since they represent potential risks to cell function (Theron et al., 2012). The second group comprises arsenic, cadmium, lead, mercury, plutonium, tungsten, and vanadium; non-essential metals that constitute potent toxins and enter the cells/tissues thanks to their physicochemical properties, like ionic charge (Duce and Bush, 2010; Johri et al., 2010). The latter, deserve special attention since they do not have any essential function in living organisms, are highly toxic at low exposure levels, and are considered as the main threat to life forms (Atobatele and Olutona, 2015). According to the Environmental Protection Agency (EPA), arsenic, cadmium, lead, and mercury, are among the most toxic metals in the environment (Goyer, 2004). Moreover, the United States Agency for Toxic Substances and Disease Registry (ATSDR) lists more than 20 heavy metals with pronounced toxicity but four are of particular concern to human health; arsenic (As), lead (Pb), cadmium (Cd) and mercury (Hg); out of these four, arsenic is the most common cause of acute heavy metal poisoning and hence ranked number 1 on ATSDR’s “top 20 list”, lead is number 2 on the list and cadmium ranks in seventh place (Fay and Mumtaz, 1996; Flora et al., 2011); therefore, arsenic, cadmium and lead are objects of study in this paper.
Arsenic (As) is a natural and abundant element of the Earth’s crust that is chemically classified as a metalloid for its metallic and nonmetallic properties (Nriagu et al., 2007; Kesici, 2016). In the soil, As is present in both, organic and inorganic forms, being the latter, a highly toxic form (Shrivastava et al., 2015). In well-drained (oxic) surface soils, As occurs predominantly as oxyanions of As5+ (arsenate), whereas As3+ (arsenite) species are more abundant in reducing environments, such as waterlogged soils (Roberts et al., 2010). The relative abundance of As5+ and As3+ critically influences As mobility, toxicity and environmental behavior, with inorganic As3+ species generally considered to be more mobile and toxic than inorganic As5+ species (Shumlas et al., 2016; Johnston et al., 2019). As5+ is less mobile than As3+ because it forms complexes on mineral surfaces, especially on hydroxides and oxides of iron (III) as well as on calcite (Bowell, 1994; Guo et al., 2009). Under iron reducing conditions arsenic is released from those surfaces into groundwater and remains mobile after subsequent reduction to As3+ (Biswas et al., 2011; Maier et al., 2019). Moreover, by binding to thiol or sulfhydryl groups on proteins, arsenite can inactivate over 200 enzymes while arsenate can replace phosphate, which is involved in many biochemical pathways (Tchounwou et al., 2012).
Cadmium (Cd) constitutes a non-essential element, and a significant pollutant due to its high toxicity and solubility in water (Pinto et al., 2004; Benavides et al., 2005). It is a metal that tends to form stable dissolved complexes with inorganic and organic ligands, which inhibits its sorption and precipitation (Kubier et al., 2019). Furthermore, cadmium can interrupt enzyme activities and inhibit the DNA-mediated transformation in microorganisms; its primary anthropogenic sources in soils are the direct input of waste material from mining, industry, and agricultural application (Kabata-Pendias, 2010; Kubier et al., 2019).
Lead (Pb) is a toxic non-essential heavy metal, that is widely distributed and induces a wide range of negative effects on living organisms at morphological, physiological and biochemical level since it is highly persistent in water and soil, accumulates in the upper eight inches of the ground and is highly immobile (Zeng et al., 2007; Pourrut et al., 2011; Tangahu et al., 2011). Although it is a naturally occurring element, anthropogenic activities like mining, fossil fuel burning, and manufacturing, contribute to its increased concentrations in soils (Tchounwou et al., 2012).
Microorganisms play an important role in the biogeochemical cycle of metals and the remediation of environments contaminated by heavy metals (Spain and Alm, 2003). Despite the toxic effect they exert, microorganisms can survive in their presence thanks to several types of mechanisms to reduce or tolerate their toxicity (Spain and Alm, 2003). According to Bruins et al. (2000) and Choudhury and Srivastava (2001), there are five main mechanisms of resistance to heavy metals in bacteria: 1) Extracellular barriers: the cell wall, plasma membrane or capsule can prevent the entry of metal ions to the cell; 2) Active transport of metal ions (efflux): constitute a mechanism to export toxic metals from the cytoplasm that include proteins belonging to three families: P-type ATPases, CDF (Cation Diffusion Facilitator) and RND (Resistance, Nodulation, Cell Division); 3) Extracellular sequestration: consists on the accumulation of metal ions by cellular components in the periplasm, the outer membrane or complexation of metal ions as insoluble compounds; 4) Intracellular sequestration: is based on the accumulation of metals in not bioavailable forms within the cytoplasm to prevent exposure to essential cellular components, two examples exist for this form of metal resistance: metallothionein production in Synechococcus sp. and cysteine-rich proteins in Pseudomonas sp. (Rouch et al., 1995; Silver and Phung, 1996) and 5) Reoxidation of metal ions. Oxidation of metals such as Cu and As is also important detoxification mechanisms, for instance, oxidation of Cu(I) to Cu(II) by CueO, and oxidation of As(V) to As(III) then efflux A(III) out of cells (Bruins et al., 2000; Choudhury and Srivastava, 2001; Barkay et al., 2003; Viti et al., 2003; Ianeva, 2009).
The mechanisms used by bacteria can be classified into biochemical and molecular (Ma et al., 2016). Biochemical mechanisms include those by which microorganisms interact with extracellular soluble metal ions and have a role in microbe tolerance through metal detoxification, mobilization, immobilization, transformation, transport, and distribution (Ma et al., 2016; White et al., 2016). Molecular mechanisms constitute the genetic determinants of heavy metal resistance that can be localized either on the chromosome or extrachromosomal genetic elements (Ianeva, 2009). In addition to biochemical and molecular mechanisms, heavy metal tolerance by bacteria can be assessed in terms of the Minimum Inhibitory Concentration (MIC), defined as the lowest concentration of the metal that prevents the growth of bacteria (Parameswari et al., 2010). Many studies have assessed the tolerance of heavy metals in terms of the MIC of microorganisms (Mejias Carpio et al., 2018); therefore, the present study will not be the exception.
The remediation of heavy metals in soils is essential to preserve the environment and protect living organisms (Glick, 2010). Traditional remediation methods include physical and chemical techniques and engineering repair; which are fast but expensive and deficient since they cause secondary pollution and negative effects on soil properties (Singh and Prasad, 2015; Ullah et al., 2015). Conversely, biological methods are considered as an effective technique for heavy metal remediation, which include: bioremediation, a process that makes use of microorganisms to eliminate pollutants from soils, and phytoremediation, a process that uses plants that can remove heavy metals (Doble and Kumar, 2005; Adams et al., 2015; Nakbanpote et al., 2016). Microbial bioremediation is an efficient, economic, and environmentally friendly procedure that reduces the cost of the cleanup process associated with heavy metal contamination (Kumar Mishra, 2017). The mechanisms behind microbial remediation of heavy metals mainly encompass biosorption (which includes precipitation, chemical adsorption, and ion exchange, surface precipitation, the formation of complexes with organic ligands, and redox reactions), biomineralization (that covers both bioleaching, involved in the mobilization of heavy metal ions from insoluble ores by dissolution or complexation), and bio-oxidation (Jin et al., 2018). On the other hand, phytoremediation, often depends on the climate and water and soil conditions, proving to be a very slow and seasonally effective method (Salt et al., 1995; Chintakovid et al., 2008). Phytoremediation includes various processes in function of the plant-soil-atmosphere interactions; for heavy metal contaminated soil, four processes of phytoremediation are known: phytoextraction, phytostabilization, phytovolatilization, and rhizofiltration (Laghlimi et al., 2015). In this context, microbial methods represent an advantageous alternative for the remediation of heavy metals from soils.
Plant-microorganism interactions play a key role in the adaptation to heavy metal polluted environments and thus, can be investigated in depth to improve microbe-assisted phytoremediation methods (Ma et al., 2016). Plant associated microorganisms, particularly, Plant Growth-Promoting Rhizobacteria (PGPR), play an important role in the remediation of polluted soils and the enhancement of plant growth by different mechanisms that include biological nitrogen fixation, phosphate solubilization, phytohormone production [Indole-3-acetic acid (IAA), cytokinins and gibberellins], siderophore production, production of 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, among others (Goswami et al., 2016; El-Mehy et al., 2019). PGPR represent important metal resistant bacteria and phytoremediation enhancing agents in heavy metal contaminated soils, whose application can lead to a marked improvement in metal mobilization or immobilization in soils and plant biomass (Ma et al., 2011; Ma et al., 2016).
Despite that some bacterial strains with the potential to remediate soils with heavy metals have been reported, three key aspects need to be addressed: i) new microbial resources are needed for application in bioremediation or bacteria-assisted phytoremediation strategies, ii), most studies include strains resistant to few metals, and polluted soils exhibit multi-contamination with heavy metals (Tirry et al., 2018) and iii) published information has not been comprehensively analyzed to date to recommend the most efficient microbial resources for remediation strategies in heavy metal contaminated soils. Therefore, this study aims to characterize and select genera associated with the plant microbiome with potential for bioremediation or bacteria-assisted phytoremediation strategies of soils contaminated with arsenic, cadmium, and lead, through a literature review and an analysis of the metal-tolerance level of bacteria in terms of the Minimum Inhibitory Concentration (MIC) and the biochemical and molecular mechanisms they use to deal with toxicity by these metals.
Materials and Methods
Bibliographic Research and Data Collection
Data were collected from research articles selected from the Science Direct, Taylor & Francis, Springer, and Google Scholar databases, the search results are highlighted as a PRISMA flowchart (Liberati et al., 2009) in Figure 1 together with exclusions, and the reasons for exclusion. The keywords used in the search were: “bacteria”, “plant growth-promoting bacteria” and “minimum inhibitory concentration”. “Arsenic”, “cadmium”, “lead”, and “heavy metals” were also added as independent variables in the keywords to refer to the stress factor. A total of eighty-five research articles from 2005 to May 2020, met our selection criteria:
i. The bacterial species studied are part of a plant microbiome, as this would allow its use in both bioremediation and phytoremediation. Additionally, these microorganisms could be considered as “naturally occurring” by associating with plants (in the rhizosphere or their tissues) as opposed to bacteria from other environments, for example, marine or extreme environments. Together, these aspects would enhance their potential use in heavy metal remediation strategies.
ii. These bacterial species exhibit plant growth-promoting characteristics because there is evidence that the capacity of bacteria to promote the growth of plant species to which they are associated is positively correlated with the efficiency of phytoremediation; and as described in the previous criterion, the interest of this work is to identify bacteria with utility in both bioremediation and phytoremediation.
iii. Bacterial resistance to the metal(s) is expressed in terms of the Minimum Inhibitory Concentration (MIC). Our goal is to analyze the remediation capabilities of bacteria quantitatively and for this we require a quantitative indicator of bacterial resistance to heavy metals. MIC has been extensively used in this area of study.
iv. Metal resistance is related to biochemical or molecular mechanisms. Only publications that have included this relationship were considered.
v. Studies had to be published as full research articles in indexed journals (grey literature was not considered).
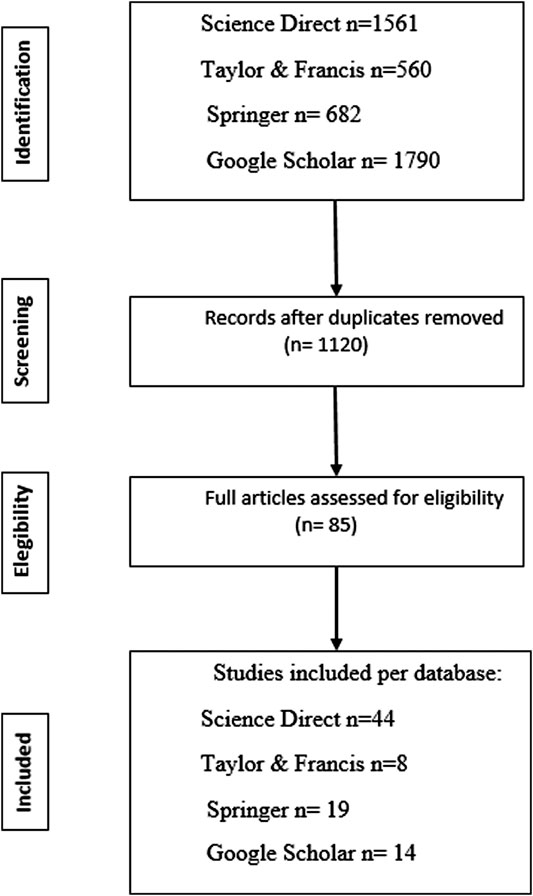
FIGURE 1. PRISMA Flowchart showing the selection of the research articles that met our selection criteria: The bacterial species studied are part of a plant microbiome, exhibit plant growth-promoting characteristics, bacterial resistance to the metals (arsenic, cadmium, and lead) is expressed in terms of the Minimum Inhibitory Concentration (MIC) and metal resistance is related to biochemical or molecular mechanisms.
From the exhaustive search of the aforementioned databases, a general report of the literature reviewed is presented in Supplementary Table S1.
In each study the MIC values differed in their units, therefore, it was necessary to convert them to a common unit: milliMolar (mM), using the ENDMEMO calculation tool.
Analysis of Bacteria Tolerance Level
The data collected from the literature were analyzed through a one-way analysis of variance (ANOVA) and Tukey’s post hoc test with a level of significance of 95% (α = 0.05) in RStudio 3.5.1 (Allaire, 2012). Differences at p ≤ 0.05 were considered statistically significant. Supplementary Table S2 presents the categorical and dependent parameters applied to the analyses.
Analysis of Metal Type and Minimum Inhibitory Concentration Values
The first analysis involved the total of data collected from the literature, 655 MIC values that belong to different strains/species of bacteria (Supplementary Table S3). Information of strains/species of bacteria used in the research articles is presented in Supplementary Table S4. As shown in Supplementary Table S2, these data were classified into four groups of heavy metals (arsenite, arsenate, cadmium, and lead); the type of metal represented the categorical parameter and the MIC data the dependent parameter. Before the ANOVA, the assumptions of normality and homogeneity of variances were corroborated through the Shapiro and Bartlett tests, respectively, and since they were not fulfilled, a natural logarithm conversion of the MIC data was carried out to get a normal distribution (Supplementary Figure S1). Finally, the ANOVA was performed to establish if there was a significant effect of the metal type variable on bacterial tolerance in terms of the MIC values, and the Tukey Post Hoc test was done to identify which metals had the highest and lowest MIC values.
Since a variety of culture media for MIC evaluation were reported in the studies finally included in the analyses (Supplementary Table S5), we evaluated the influence of media type on MIC. The influence of the growth medium on the MIC values is mainly determined by the chemical interactions that are established between heavy metals, in cationic form, and other components of the growth medium, which can reduce the concentration of free metals and consequently result in the overestimation of the MICs. These interactions are more common in rich media enriched with different supplements. Based on this, we collected information regarding the growth medium from the eighty-five research articles reviewed and classified the media as i) Minimum, those composed of salts and only one carbon source, without the addition of amino acids, and ii) Rich, those which provide salts, different sugars, and other biomolecules. We performed an ANOVA with the 655 MIC values, where the type of medium was the categorical parameter, and the MIC data the dependent parameter (Supplementary Table S6). As in the previous analyses, the assumptions of normality and homogeneity of variances were not fulfilled; therefore, a natural logarithm conversion of the MIC data was carried out to get a normal distribution (Supplementary Figure S2), and finally, the Tukey Post Hoc test was applied (Supplementary Table S7). Additionally, the influence of the type of medium on the reported MIC values for each metal was evaluated, discriminating the MIC values by type of medium and type of metal (arsenite, arsenate, cadmium, and lead). The discriminated data were analyzed with a two-way ANOVA (Supplementary Table S8), including again a natural logarithm conversion of the MIC data (Supplementary Figure S3), and the Tukey Post Hoc test (Supplementary Table S9). The results of these analyses did not show a significant influence of the type of media on the MICs for the heavy metals arsenite, arsenate, and cadmium, but for lead. The subsequent analyses (MIC and bacterial genotypes) were conducted with the full MIC data set and the influence of media type on bacterial resistance to lead discussed in detail.
Analysis of Metal Resistance by Bacterial Genus and Minimum Inhibitory Concentration Values
A second analysis that aimed to determine if different bacterial genera have different metal resistance, was done using a sample of the total data collected (655), in this case, 463 MIC values from different bacterial genera (Supplementary Table S3). Out of sixty-two genera collected from the literature, only those with the highest amount of MIC values were used for the analysis, which included: Acinetobacter, Agrobacterium, Arthrobacter, Bacillus, Enterobacter, Klebsiella, Mesorhizobium, Microbacterium, Pseudomonas, Rhizobium, Rhodococcus, and Variovorax. As in the first analysis, the assumptions of normality and homogeneity of variances were not fulfilled; therefore, a natural logarithm conversion of the MIC data was carried out to get a normal distribution (Supplementary Figure S4). Finally, the ANOVA was performed using the bacterial genera as the categorical parameter and the MIC values as the dependent variable, and the Tukey Post Hoc test was done to determine which genera had the highest MIC values.
Average Minimum Inhibitory Concentration of the Most Resistant Bacterial Genera
To establish against which specific metal (arsenite, arsenate, cadmium, and lead) each genus exhibited greater or less resistance, the average of the MIC values for each heavy metal was calculated for the genera that showed differential resistance to metals from the second statistical analysis.
Biochemical Mechanisms
Data Collection
We included all the reports regarding biochemical mechanisms that contributed to explain the bacterial metal tolerance presented in the selected articles. Most of the reported mechanisms were associated with plant growth-promoting (PGP) traits such as biological nitrogen fixation, phosphate solubilization, production of auxins, biosurfactants, siderophores, exopolysaccharides, hydrogen cyanide (HCN), indole acetic acid (IAA), and ACC deaminase, as well as enzymatic mechanisms that included: phosphatase, oxidase, catalase, amylase, pectinase, cellulase, chitinase, protease, and lipase.
Analysis of Biochemical Mechanisms
After data collection, only the bacterial genera that showed differential resistance to metals were included in the analysis, as well as the most reported mechanisms in the articles which included the PGP traits: nitrogen fixation, phosphate solubilization, ACC deaminase, production of siderophores, IAA, and enzymatic properties such as oxidase and catalase (Supplementary Table S10). For the analysis, the quantitative data were transformed into qualitative data (presence or absence) and the percentage of strains with activity for the selected mechanisms for each genus was calculated using Eq. 1.
Molecular Mechanisms
Gene Searching
Forty-five reference genome assemblies that included forty-nine chromosomal and plasmid sequences of different strains/species of bacteria from the most resistant genera were downloaded from GenBank NCBI (Supplementary Table S11). The analysis was carried out with the reference genomes given the lack of genomic information for several strains/species of the selected genera. For the gene analysis, the feature table for the chromosomal or plasmid sequence of each reference genome was downloaded from GenBank and imported into NCBI Genome Workbench (https://www.ncbi.nlm.nih.gov/tools/gbench/), where the Generic Tabular View tool was used to search and query the genes of interest belonging to the ars, cad and pbr operons for arsenic, cadmium and lead resistance, respectively. For the search, the keywords “ars”, “cad” and “pbr” were used and then filtered to show only the query results which included the genes of interest (Supplementary Table S12). Finally, the percentage of total genes corresponding to each operon by genus was calculated (Eq. 2), as well as the percentage of each gene in each genus (Eq. 3).
Sequence Identity Confirmation
We did an additional analysis to confirm the sequence identity of the ars and cad operon genes found in the 45 reference genomes. To do this, we manually downloaded different protein sequences corresponding to each gene for each genus (Agrobacterium, Bacillus, Enterobacter, Klebsiella, Microbacterium, Pseudomonas, and Rhodococcus) from the Uniprot database (including both manually curated and no-curated sequences) and uploaded them to the Galaxy Europe platform (Afgan et al., 2016); on this platform, we used the MEME tool (Motif Discovery Tool) (Bailey et al., 2006) to find a conserved region (a motif) (Supplementary Table S13). Then, to confirm the identity of the studied genes, we carry out a protein blast (BLASTP) with the consensus sequences on the protein sequences of the reference genomes (Supplementary Table S14). As the motifs are short sequences (between 24 and 50 nucleotides), we modified two parameters of the BLASTP algorithm: the expectation threshold (10) and the word size (3).
Results and Discussion
Effect of Media Composition on Minimum Inhibitory Concentration Values Exhibited by Bacteria Under Heavy Metal Stress
Different factors affect the form and toxicity of metals, these include pH, the concentration of chelating agents, the concentration of inorganic anions, and competition from other cations; moreover, many natural and synthetic chelating agents can reduce the toxicity of heavy metals and it has been theorized that MIC values vary with the type of media used, since components of bacterial growth media can form a complex with heavy metals, remove them from solution, and reduce their concentrations in the media (Kumar et al., 2013; Aljerf and AlMasri, 2018). Metals may also have their toxicity reduced by components of nutrient media, for example, Leitão and Sá-Correia (1997) found that growth of Pseudomonas aeruginosa in the presence of inhibitory concentrations of copper was improved by increasing the concentrations of nutrients in the medium. These interactions between the components of the medium and the heavy metals would cause the MIC values to be overestimated (as the bacteria would be subjected to lower concentrations of heavy metals than reported) and therefore the results obtained by different studies would not be comparable (Dean-Ross and Mills, 1989).
The ideal solution to this problem would be to establish international standards for the characterization of bacteria for heavy metal remediation. However, to date, such standards do not exist, and assessments are conducted under a wide variety of culture media. Although the culture media used varies in the identity of its components, the effects caused by these are common (complex formation, precipitation, etc.), and the interactions of the heavy metal-culture media depend on the pH and whether or not the media is rich in nutrients.
The research articles reviewed in this work used different types of media (Supplementary Table S5), therefore, to make valid comparisons, these were grouped into minimal and rich media; minimum media is composed of salts and one carbon source, generally without the presence of amino acids; whereas rich media contain salts, different sugars and other biomolecules from cell extracts (Mejias Carpio et al., 2018). We performed an analysis of the effect of the culture medium (minimal vs. rich) on the MIC values through statistical analysis and found that, as theorized, the culture medium affects bacterial heavy metal resistance (Figure 2). The results showed that there is a statistically significant difference between the growth media at a p-value of 1.03e-05. However, a detailed analysis of each heavy metal (arsenite, arsenate, cadmium, and lead) revealed that the effect is only significant for lead (Figure 3). Thus, for the rest of the analyses presented in this study, the conclusions for arsenate, arsenite, and cadmium are independent of the culture medium used in the studies, while for lead; there could be an influence of the growth medium on MIC.
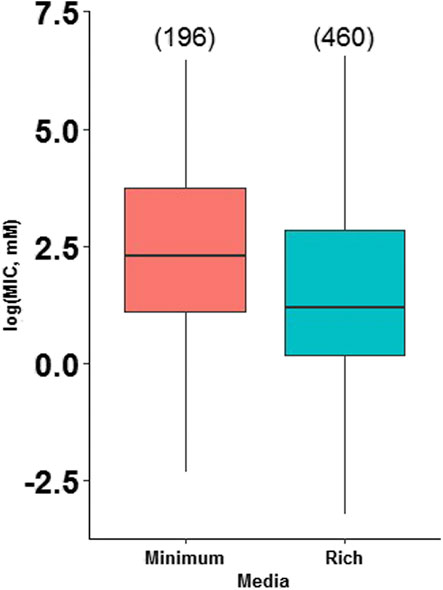
FIGURE 2. Bacterial metal tolerance in terms of the logarithm of the Minimum Inhibitory Concentration (MIC), as a function of the type of media (Minimum and Rich) analyzed with ANOVA in RStudio 3.5.1. We present an analysis of 655 MIC values obtained from different research articles of different strains/species of bacteria associated with the plant microbiome. The number of reports for each media is indicated in parentheses.
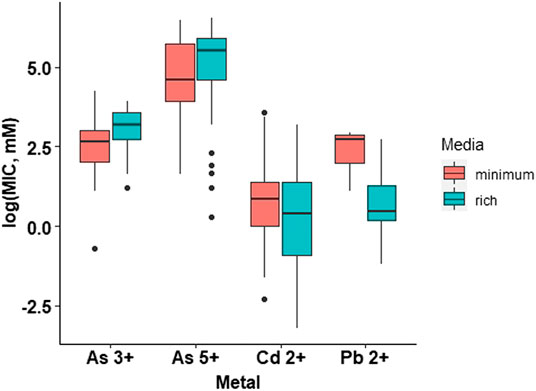
FIGURE 3. Bacterial metal tolerance in terms of the logarithm of the Minimum Inhibitory Concentration (MIC), as a function of the type of media (Minimum and Rich) and the type of metal (As3+. As5+. Cd2+, Pb2+) analyzed with a two way ANOVA in RStudio 3.5.1. We present an analysis of 655 MIC values obtained from different research articles of different strains/species of bacteria associated with the plant microbiome.
Relationship Between Bacterial Metal Tolerance and the Type of Metal
The ability of microorganisms, such as bacteria, fungi, or algae to remove heavy metals or to transform them into less toxic forms, has attracted the attention of environmental scientists and biotechnologists for years (Mustapha and Halimoon, 2015). It is important to identify if bacterial metal tolerance is associated with the type of metal evaluated since metals (cadmium and lead) and metalloids (arsenic) are being considered in this study.
The first analysis showed a statistically significant difference between the type of metal and bacterial tolerance with a p-value <2e-16 (Supplementary Table S15). As presented in Figure 4, cadmium and lead were the most toxic metals since they had the lowest MIC values, and according to Tukey’s test, there is a significant difference between them (p = 0.035). Conversely, the least toxic metal was arsenic, being arsenate less toxic than arsenite and showing a significant difference from each other (p = 0.00), as well as between cadmium and lead with both arsenic forms (Supplementary Table S16). These results coincide with the reported by Mejias Carpio et al. (2018), who showed a statistically significant difference between the metals studied (As, Cd, Cr, Cu, Ni, Pb, Zn) and the overall microbial tolerance, being cadmium one of the most toxic metals, while arsenic and lead grouped showing the least toxicity. Nevertheless, for this study, lead together with cadmium represented the most toxic metals.
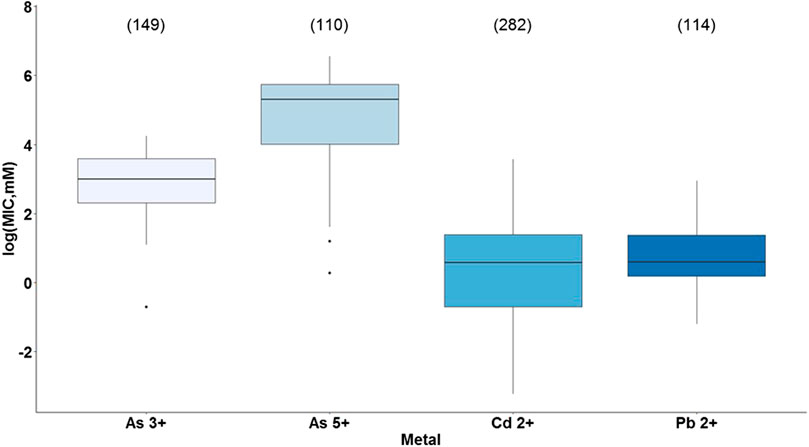
FIGURE 4. Bacterial metal tolerance in terms of the logarithm of the Minimum Inhibitory Concentration (MIC), as a function of the type of metal (As3+. As5+. Cd2+, Pb2+) analyzed with ANOVA in RStudio 3.5.1. We present an analysis of 655 MIC values obtained from different research articles of different strains/species of bacteria associated with the plant microbiome. The number of reports for each metal is indicated in parentheses.
Relationship Between Bacterial Resistance and Bacterial Genus
After the analysis of the type of metal, it was evaluated whether the bacterial genus affected the MIC values. According to the results, there is a significant statistical difference between bacterial metal tolerance and the bacterial genus (p = 0.000205) (Supplementary Table S17). Through Tukey’s post hoc test (Supplementary Table S18) we found that the genera Agrobacterium, Bacillus, Enterobacter, Klebsiella, Microbacterium, Pseudomonas, and Rhodococcus showed statistically significant differences with Mesorhizobium (Figure 5), a genus that has reported resistant species to only one or two heavy metals (Zn, Cd, or As), including M. metallidurans, Mesorhizobium sp. AH5, and M. amorphae (Fan et al., 2018). Among the genera evaluated, they were the most resistant to arsenic, cadmium, and lead, and were included in subsequent analyses of the average MIC values and biochemical and molecular mechanisms.
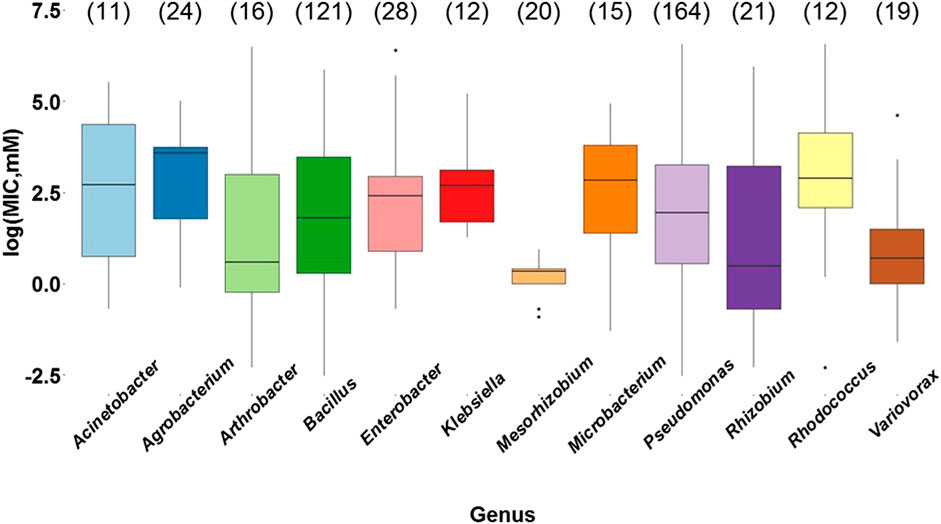
FIGURE 5. Bacterial metal tolerance expressed as the logarithm of the Minimum Inhibitory Concentration (MIC) as a function of the most commonly reported bacterial genera (Acinetobacter, Agrobacterium, Arthrobacter, Bacillus, Enterobacter, Klebsiella, Mesorhizobium, Microbacterium, Pseudomonas, Rhizobium, Rhodococcus, and Variovorax), analyzed with ANOVA in RStudio 3.5.1. The data presented correspond to a total of 463 MIC values obtained from the selected research articles. The number of reports for each genus is indicated in parentheses.
Analysis of the Average Minimum Inhibitory Concentration Values for the Most Resistant Genera
Once the statistical analyses were carried out, we proceeded to identify towards which metal the most resistant genera had greater or less tolerance by obtaining the average of the MIC values for arsenite, arsenate, cadmium, and lead for each genus. In the first place, for arsenite, Figure 6A shows that the genus with the highest resistance was Agrobacterium (38.5 mM) followed by Bacillus (25.3 mM) while the others exhibited MIC values ranging from 12.9 to 20.7 mM. The lowest MIC (12.9 mM) was exhibited by Pseudomonas. For arsenate (Figure 6B), Rhodococcus (423.3 mM) followed by Pseudomonas (328.6 mM) and Enterobacter (212.3 mM) presented the highest averages while the others exhibited values below 200 mM; the lowest MIC (89.8 mM) was exhibited by Bacillus.
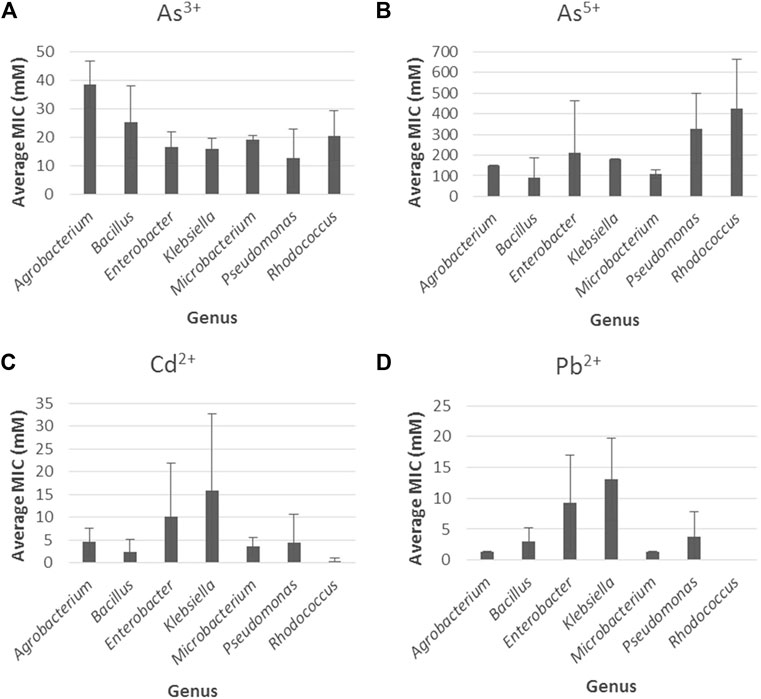
FIGURE 6. Bacterial metal tolerance expressed as the average of the MIC values (mM) with the standard error for (A) Arsenite (As3 +), (B) Arsenate (As5 +), (C) Cadmium (Cd2 +) and (D) lead (Pb2+) for the genera with the highest resistance against heavy metals (Agrobacterium, Bacillus, Klebsiella, Enterobacter, Microbacterium, Pseudomonas, Rhodococcus).
For cadmium (Figure 6C), the genera with the highest resistance were Klebsiella (15.8 mM) and Enterobacter (10.1 mM), while the others showed MIC values below 5 mM. Similarly, for lead (Figure 6D), Klebsiella (13.1 mM) followed by Enterobacter (9.2 mM) showed the highest values and the other genera exhibited average values below 5 mM. Among the genera, Rhodococcus had the lowest average for cadmium (0.5 mM), while for lead, no MIC values were reported for this genus according to the literature reviewed.
Analysis of Biochemical and Molecular Mechanisms
Regarding the biochemical mechanisms, the percentage of strains with activity for the different PGP traits (nitrogen fixation, phosphate solubilization, and production of ACC deaminase, siderophores, and IAA) and enzymatic properties (oxidase and catalase) is presented in Figure 7 for the most resistant genera.
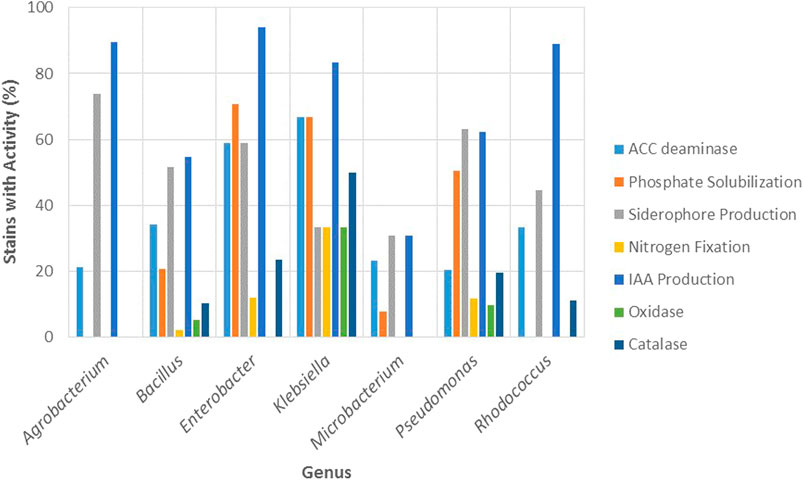
FIGURE 7. Percentage of strains with activity for the most commonly reported PGP traits (ACC deaminase, phosphate solubilization, siderophore production, nitrogen fixation and IAA production) and enzymatic properties (oxidase and catalase) in function of most resistant bacterial genera (Agrobacterium, Bacillus, Klebsiella, Enterobacter, Microbacterium, Pseudomonas and, Rhodococcus).
Plant growth-promoting traits are involved in plant growth promotion and mitigation of the toxic effects on plants exerted by heavy metals (Tirry et al., 2018). PGP mechanisms have been grouped traditionally into direct and indirect mechanisms: any mechanism that increases plant growth by producing growth regulators or providing nutrients are considered direct mechanisms (Goswami et al., 2016). On the contrary, mechanisms that help the plant to grow healthfully under different environmental stresses are portrayed as indirect mechanisms (Goswami et al., 2016). Among the five PGP traits studied, biological nitrogen fixation, phosphate solubilization, and IAA belong to the direct mechanisms; conversely, siderophore and ACC deaminase production are indirect mechanisms of PGPR (Goswami et al., 2016). The enzymatic mechanisms studied (oxidase and catalase), represent important properties in heavy metal resistance since these environmental pollutants produce oxidative stress in most soil bacteria causing an alteration of the cellular redox status; nonetheless, the antioxidant enzymes are capable of maintaining the cellular redox state and alleviate the damages by counteracting the detrimental effects of Reactive Oxygen Species (ROS) (Behera et al., 2014).
For the molecular mechanisms, Figure 8 shows that the genes associated with the ars operon were present in all the genera. The ubiquitous distribution of arsenic compounds in the environment has constituted a selective pressure for microorganisms, therefore, they have acquired or evolved multiple arsenic resistance genetic systems that include the ars operons; groups of genes widely present in species of bacteria and archaea (Ben Fekih et al., 2018). For this operon the best-characterized genes include arsR, arsA, arsB, arsC, arsD, arsH, and arsM: the arsR gene encodes an As(III)-responsive transcriptional repressor, arsA and arsB form an oxyanion-translocating complex, in which arsA functions as an ATPase, while arsB is a membrane efflux transporter, arsC encodes a arsenate reductase, that converts arsenate to arsenite, arsD functions as an arsenic metallochaperone transferring As(III) to arsA and increases the rate of arsenic extrusion, arsH encodes an NADPH-flavin mononucleotide oxidoreductase and arsM encodes an arsenite S-adenosylmethionine methyltransferase that contributes to arsenic detoxification in bacteria (Wang et al., 2009). Similarly, the cad operon was present in all genera studied (Figure 8). The cad operon is a two-component operon that essentially harbors two genes: cadA and cadC (Zheng et al., 2019). The CadA protein constitutes an ATPase that transports cadmium outside the cells, whereas the CadC protein is a transcriptional regulator that up-regulates cadA expression (Nucifora et al., 1989; Endo and Silver, 1995). In addition to cadA and cadC, the cadB and cadD genes contribute to cadmium resistance (Nies, 1992; Crupper et al., 1999; Zheng et al., 2019).
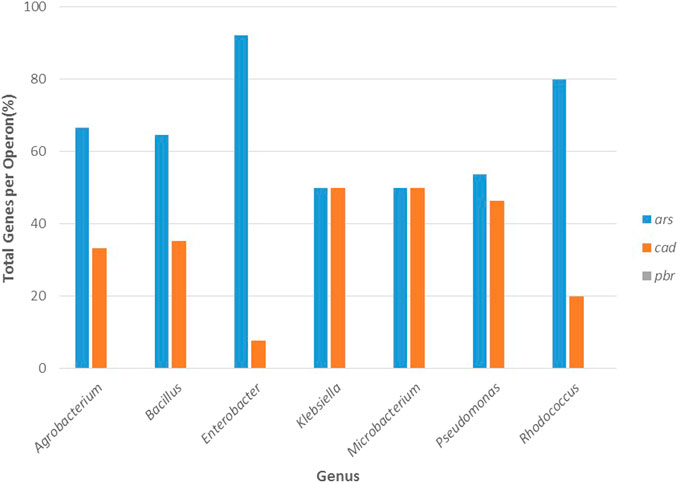
FIGURE 8. Total percentage of genes by operon: ars, cad, and pbr for arsenic, cadmium, and lead resistance, respectively, in function of the most resistant bacterial genera (Agrobacterium, Bacillus, Klebsiella, Enterobacter, Microbacterium, Pseudomonas, and Rhodococcus).
As presented in Figure 8, the lead operon was absent in all the genera. The absence of pbr genes in the bacterial genomes can be explained by the fact that the molecular mechanisms behind lead resistance have not been studied in depth (Borremans et al., 2001; Hynninen et al., 2009; Hynninen, 2010). One of the few lead resistance systems reported was found in Cupriavidus metallidurans CH34, and it is a system involved in uptake, efflux, and accumulation of Pb2+ that harbors four main resistance genes: 1) a pbrT gene, that encodes a Pb2+ uptake protein; 2) a pbrA gene, encoding for a Pb2+ efflux ATPase; 3) a pbrB, that encodes an integral membrane protein and 4) a pbrC, encoding for a prolipoprotein signal peptidase (Borremans et al., 2001). Hynninen et al. (2009) examined the metal specificity and functions of pbrABCD in a C. metallidurans strain DN440 and showed that the activity of pbrA (an efflux transporter for Zn2+, Cd2+, and Pb2+) and pbrB (a C55-PP phosphatase) is required to effectively detoxify Pb2+, which is accomplished by transporting Pb2+ ions out of the cell with pbrA and sequestering them with phosphate ions produced by pbrB. It has been suggested that lead detoxification by ATPases and phosphatases is a widespread mechanism for lead tolerance in these microorganisms (Hynninen et al., 2009), therefore, more studies are needed to elucidate these metal resistance genes and systems in different bacterial genera.
Figure 9, shows that most cadmium resistance genes corresponded to cadA, a gene encoding for a cadmium efflux pump (a Cd2+/ATPase protein transporter) that can be found in the plasmid or the chromosome (Oger et al., 2001); which according to the results presented by Oger et al. (2001), can increase its copy number in bacteria after exposure to a high concentration of dissolved cadmium. In addition to cadA, several genes belonging to the ars operon were found in the different genera studied (Figure 9); among them, the arsBC genes are of great importance since a main microbial arsenic detoxification mechanism involves the reduction of arsenate to arsenite through a cytoplasmic arsenate reductase (arsC) and a membrane-associated arsB efflux pump, which after reduction of arsenate, will extrude As3+ (Selvi et al., 2014).
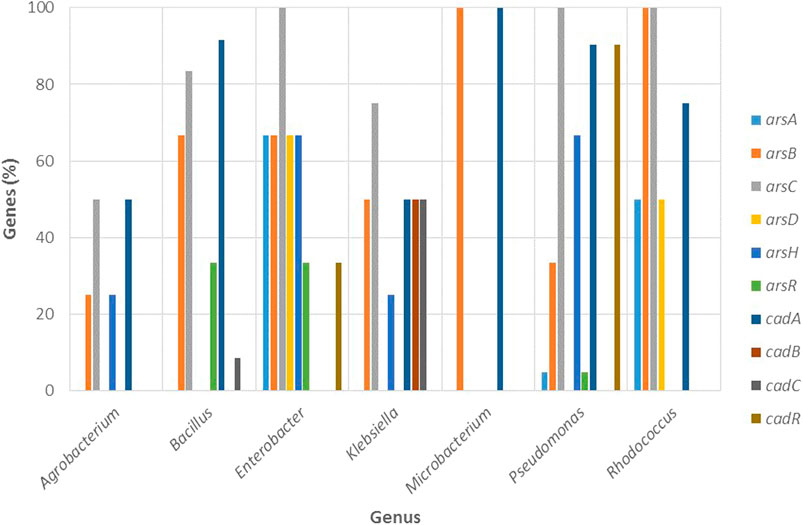
FIGURE 9. Percentage of genes involved in arsenic and cadmium resistance in function of the most resistant bacterial genera (Agrobacterium, Bacillus, Klebsiella, Enterobacter, Microbacterium, Pseudomonas, and Rhodococcus).
Analysis of Biochemical and Molecular Mechanisms for the Most Resistant Genera
Agrobacterium and Bacillus
Biochemical Mechanisms
Agrobacterium and Bacillus displayed the highest resistance to arsenite and in general, low resistance to the other metals. Figure 7 shows that Agrobacterium exhibited the highest percentage of strains with activity for siderophore production (73.68%) among the genera studied, while Bacillus reported strains with activity for all the PGP and enzymatic properties, being particularly high the percentage of strains with activity for IAA (54.64%) and siderophores (51.55%).
According to the reported results by Gu et al. (2018), Agrobacterium and Bacillus were the most highly arsenite-resistant bacteria (MIC > 45 mM) among genera of bacterial endophytes isolated from the roots of Pteris vittata L, which also displayed high levels of IAA and siderophore production. IAA can directly promote root growth by increasing cell division or plant cell elongation, and therefore, can improve the absorption of arsenic by the roots (Wang et al., 2011). Siderophores, on the other hand, constitute organic molecules with a high affinity for Fe3+ ions that form complexes with other metals, and thus, are involved in the mobilization of nutrients and the availability of metals for plants (Schalk et al., 2011). Regarding these molecules, Román-Ponce et al. (2018) assayed the ligands production of Arsenic Resistant Endophytic Bacteria (AREB) and proposed the term “arsenophore” [arseno from (arsenic) + phore (carry)], referring to the ability of a ligand to chelate arsenite and arsenate.
Molecular Mechanisms
Regarding the molecular mechanisms (Figure 9), Agrobacterium reported the cadA gene and Bacillus the cadAC genes for cadmium resistance. For arsenic, both genera exhibited various genes: Agrobacterium showed the arsBCH genes, while Bacillus presented arsBCR.
In the literature, there are reports of Agrobacterium’s high resistance to arsenite as well as the presence of different ars genes (Cai et al., 2009). Cai et al. (2009) identified arsenite-oxidizing bacteria from Agrobacterium, Achromobacter, and Pseudomonas that showed high arsenite tolerance levels, and found out that the bacteria carrying an arsenite oxidase (aoxB) and an arsenite transporter gene (ACR3 or arsB) exhibited higher arsenite resistance than those with just an arsenite transporter gene. On the contrary, regarding the Bacillus genus, Bhat et al. (2011), reported the presence of two functional chromosomal encoded ars gene clusters in Bacillus sp. CDB3 strain. According to the authors, cluster 1 exerted a much higher resistance to arsenic than cluster 2, and it seems to be the largest cluster characterized so far in bacteria (Bhat et al., 2011). Moreover, in a related study that investigated the presence, diversity, and phylogenetic relationships of genes associated with arsenic resistance, the detection of the arsB and arsC genes in 21 of the 37 Bacillus strains analyzed was correlated with arsenic hyper resistance values that corresponded to 32 and 128 mM of arsenite and arsenate, respectively (Prieto-Barajas et al., 2018).
Enterobacter and Klebsiella
Biochemical Mechanisms
Enterobacter and Klebsiella were the genera with the highest resistance against cadmium and lead and reported a high percentage of strains with activity for all PGP traits and enzymatic properties (Figure 7). Particularly, among the genera studied, Enterobacter presented the highest percentage of strains for IAA production (94.12%), phosphate solubilization (70.59%), and catalase activity (23.53%). Conversely, Klebsiella displayed the highest percentage of strains with ACC deaminase (66.67%) and nitrogen fixation (33.33%), as well as the highest percentage for oxidase (33.33%) and catalase (50%) activity.
Regarding the Enterobacter genus, Pramanik et al. (2018) isolated a potent cadmium resistant PGPR bacteria identified as Enterobacter aerogenes K6; a strain that exhibited a high degree of resistance to cadmium, lead, and arsenite according to the MIC values reported: 36, 18 and 20 mM, respectively. Moreover, it showed several important PGP traits even under high cadmium stress, including IAA and phosphate solubilization activity (Pramanik et al., 2018). IAA improves nutrient acquisition by increasing root development and is capable to alleviate cadmium toxicity in plants by stimulating antioxidant enzymes (Chmielewska-Bak et al., 2014). On the other hand, the phosphate solubilizing activity constitutes an ability of bacteria that enhances plant growth in polluted soils by providing phosphorus; these bacteria dissolve inorganic phosphates by secreting organic acids that increase phosphate solubility and in particular, organic acid anions that can form complexes with metal cations like cadmium and release phosphate phosphorus (Ahemad, 2015; Yang et al., 2018; Belimov et al., 2019).
Pramanik et al. (2017) reported the most resistant PGPR strain to cadmium so far described: Klebsiella pneumoniae K5, a strain that displayed a MIC value of 36 mM for cadmium, exhibited several PGP traits and demonstrated multiple resistance to heavy metals such as lead and arsenite with MIC values of 19 and 20 mM, respectively. In the study, the strain showed high enzymatic hydrolysis of ACC by ACC deaminase, an essential property for promoting plant growth under cadmium stress and tolerance toward this metal (Pramanik et al., 2017). ACC is a precursor of ethylene, a plant hormone that under stress conditions, such as heavy metal pollution, increases its concentration and inhibits plant growth, however, microorganisms possessing the ACC deaminase enzyme, can hydrolyze ACC to ammonia and α-ketobutyrate and in this way, reduce the hormone’s concentration in plants and counteract its detrimental effect (Glick et al., 1994; Belimov et al., 2019). Like K. pneumoniae K5, Klebsiella michiganensis MCC3089 represents a strain with high resistance to cadmium and a considerable extent to lead and arsenic and exhibits various PGP traits such as nitrogen fixation capacity, a common mechanism in this genus since Klebsiella species are known to be free-living nitrogen fixers (Mitra et al., 2018). Furthermore, in the study, the authors reported that the strain enhanced the growth of rice seedlings under cadmium stress and decreased oxidative stress (Mitra et al., 2018). As noted before (Figure 7), this genus reported high enzymatic activity (oxidase and catalase), an advantageous mechanism that plays a fundamental role against the oxidative stress generated by heavy metals (Hussein and Joo, 2013).
Molecular Mechanisms
Regarding the molecular mechanisms (Figure 9), Enterobacter only showed the cadR gene (found in the plasmid), while Klebsiella reported the highest number of cad genes among the genera studied with cadABC (found on the bacterial chromosome).
In the literature search, we found very few articles that report molecular mechanisms regarding the cad operon to explain cadmium resistance in Enterobacter and Klebsiella. Conversely, mostly biochemical mechanisms such as those previously described, are reported as a source of resistance against cadmium toxicity in these genera. Among the few studies found, Haq et al. (1999) detected the presence of resistance genes for cadmium in plasmids of Klebsiella sp. and Enterobacter cloacae strains that displayed high removing capacity of cadmium; these results are by those reported in our study since the cadR gene was detected in the plasmid for the Enterobacter genus. In a related study, a heavy metal translocating ATPase gene (hmtp) was identified in an Enterobacter strain, a gene that may be involved in bacterium heavy metal resistance since its overexpression could increase zinc and cadmium tolerance (Chien et al., 2013). Moreover, for Klebsiella, the presence of the cad operon containing the cadABC genes has been reported in Klebsiella pneumonia participating in the organism adaptation to intestinal colonization and gastrointestinal stress (Hsieh et al., 2010).
As seen in Figure 9, Enterobacter and Klebsiella also reported multiple genes for the ars operon. For Enterobacter the arsABCDHR genes were identified at the chromosomal level while the arsBCH genes were reported for Klebsiella, and among them, arsCH were found in the plasmid. Unlike the cad operon, the ars operon is well established in these genera (Selvi et al., 2014). Selvi et al. (2014) proposed the use of Enterobacter strains as candidates for bioremediation processes and the efficient removal of arsenic from contaminated sites based on the presence of the ars operon encoding an arsenite oxidizing gene (aoxA) and arsenate reducing gene (arsC). For Klebsiella, the arsC gene has been detected on both, the chromosome as well as on the plasmid in Klebsiella pneumonia (Daware and Gade, 2015) which coincides with the results presented here.
It is worth noting that these genera were the only ones in which resistance genes for the heavy metals studied were found at plasmid level: arsCH for Klebsiella and cadR for Enterobacter, according to the available information. The localization of metal resistance genes on plasmids suggests that these genes could be transferred between bacteria by Horizontal Gene Transfer (HGT) (Ianeva, 2009). Plasmids play an important role in HGT since they increase bacterial adaptation by transferring important traits between strains and species (Harrison and Brockhurst, 2012). In the literature, it is suggested that when the oxygen appeared in the atmosphere, the arsC gene (arsenate reductase) was added to the original ars operon (arsRB) and that subsequent HGT and sequence divergence, gave rise to the different arsC classes that exist in the present day (Rosen, 2002; Jackson and Dugas, 2003).
Microbacterium
Biochemical Mechanisms
Microbacterium exhibited low resistance to cadmium, lead, and arsenate and moderate to arsenite. Moreover, this genus reported the least percentage of strains with activity for PGP traits, which only included ACC deaminase (23.08%), phosphate solubilization (7.69%), siderophore (30.77%), and IAA production (30.77%) and did not report enzymatic properties (Figure 7). Nevertheless, there is evidence that strains of this genus may exhibit different mechanisms of resistance to arsenate and arsenite, which would involve differences in the competition between PO43− and As5+ for the phosphate anion transporters, and also differences in the impact of PO43− on As5+ reduction. Román-Ponce et al. (2018) evaluated the reciprocal impact of PO43− and As5+ on their uptake rates in two endophytic strains of arsenate-resistant Microbacterium sp. called NE2E2 and NE2E3, which exhibited similar metal resistances (>100 mM As5+, >20 mM As3+); for NE2E2 they found that PO43− uptake decreased with the increase of As5+ concentration in the medium, while reduction of As5+ to As3+ was decreased as PO43− concentration increased, as reported for other arsenic resistant bacterial genera. For NE2E3, however, they observed completely opposite patterns. Although no mechanical explanation is given for these differences, the authors suggest that some strains of the genus Microbacterium mitigate the toxic effects of arsenite by increasing intracellular phosphate concentration.
Molecular Mechanisms
Due to the lack of genomic information for Microbacterium, it was only possible to study the presence of genes in one reference genome (M. paraoxydans), which had a single gene for arsenic: arsB, as well as for cadmium: cadA (Figure 9). Achour-Rokbani et al. (2010) reported a set of arsenic resistance genes in Microbacterium sp. strain A33, an ars system with notable characteristics that include: 1) the presence of genes required to provide electrons for the reduction of arsenate in a single operon, 2) an arsenate reductase and 3) an ArsRC2 fusion protein that functions as an arsenic-dependent transcriptional repressor.
Pseudomonas
Biochemical Mechanisms
Pseudomonas was the most commonly reported genus in the literature reviewed with 164 strains. It was the second genus most resistant to arsenate, the least resistance to arsenite, and showed low tolerance for cadmium and lead. As seen in Figure 7, this genus displayed a percentage of strains with activity for all the mechanisms represented (PGP and enzymatic). The results of this study coincide with the reported by Wevar Oller et al. (2013), where Pseudomonas strains were highly resistant to arsenate, displayed different PGP traits such as siderophores, phosphate solubilization, IAA production, and ACC utilization, and was the most represented genus, which is explained by the fact that Pseudomonas constitute ubiquitous bacteria with great adaptability to various environments (Wevar Oller et al., 2013). In a related study, the authors isolated Pseudomonas strains for their tolerance to different heavy metals (As3+, As5+, Cr2+, Hg2+, and Cd2+) and PGP properties, and showed that the isolates reported maximum tolerance to arsenate and least tolerance to Mercury (Adhikary et al., 2019).
Molecular Mechanisms
Since it was the most representative genera, it was possible to evaluate the presence of resistance genes in various reference genomes, such as cadA and cadR for the cad operon (Figure 9). Sequence analysis carried out by Lee et al. (2001), revealed two divergently transcribed genes, cadA, and cadR, from Pseudomonas putida 06909. Furthermore, the mutational analysis revealed that cadA and cadR were completely responsible for cadmium resistance, and cadmium transporter ATPase homologs were detected in many Pseudomonas species (Lee et al., 2001).
Pseudomonas also showed a high percentage of arsenic resistant genes: arsABCHR (Figure 9). In Pseudomonas fluorescens MSP3, the authors observed an ATP-dependent efflux mechanism of detoxification encoded from an operon consisting of an arsenite inducible repressor that regulates arsC expression and an inner membrane arsenite export system encoded by arsB (Prithivirajsingh et al., 2001). Cánovas et al. (2003) reported two highly homologous ars operons in different locations in the genome of Pseudomonas putida KT2440, specifically, two copies of an arsRBCH operon, whose presence suggests that each copy could be active over different arsenic exposure levels. Similarly, in a related study, the authors presented evidence that the operons of P. putida KT2440 (ars1 and ars2) encode arsenic regulatory genes and extrusion systems that confer resistance to arsenite and arsenate, and identified that the ars1 operon, seems to only occur in the KT2440 strain, whereas the ars2 operon, is part of the core genome of P. putida (Fernández et al., 2014).
Rhodococcus
Biochemical Mechanisms
Rhodococcus showed high resistance to arsenic since it was the most resistant genus to arsenate and the third most resistant to arsenite. As seen in Figure 7, this genus reported a high percentage of strains with activity for IAA production (88.89%) and in lower percentage for ACC deaminase (33.33%) and siderophores (44.44%), while for the enzymatic mechanisms only reported strains with catalase activity (11.11%). Wevar Oller et al. (2013), showed that Enterobacter, Pseudomonas, and Rhodococcus strains grew in the presence of high concentrations of arsenite and arsenate; arsenite was used at concentrations from 12 to 38 mM, whereas arsenate resistance was tested in the range of 400–700 mM and the bacterial strains showed resistance to concentrations of up to 24 mM for arsenite and up to 400 mM of arsenate. Rhodococcus erythropolis AW3 was the most arsenic resistant strain since it was able to remove and accumulate the greatest amounts of the metalloid; moreover, the strain reported catalase activity but did not display PGP traits (Wevar Oller et al., 2013). In a related study that investigated the potential of arsenic resistant bacteria to improve arsenic phytoremediation, a Rhodococcus strain exhibited IAA, ACC deaminase, and siderophore production, and was resistant to a high concentration of arsenite (Mesa et al., 2017).
Molecular Mechanisms
Figure 9 shows that Rhodococcus displayed various arsenic resistance genes: arsABCD, while for cadmium, only reported the cadA gene. Bacterial strains of Rhodococcus can cope with increased concentrations of toxic metalloids like arsenic, however, the molecular systems behind arsenic resistance have not been studied in depth (Firrincieli et al., 2019). Among the few studies, Firrincieli et al. (2019) reported that Rhodococcus aetherivorans BCP1 tolerated high concentrations of arsenate (up to 240 mM); a remarkable feature attributed to three gene clusters implicated in arsenic resistance that include three arsenate reductase genes (arsC1/2/3).
A Caution About Minimum Inhibitory Concentration Values, Culture Media, and In Vitro Studies of Heavy Metal Tolerance in Bacteria
Several biotic and abiotic factors can affect the toxicity of metals to microorganisms; biotic factors include tolerance, size and life stages, species, and nutrition-related to the test organisms, whereas abiotic factors include organic substances, pH, temperature, alkalinity and hardness, inorganic ligands, interactions, sediments, and others (Wang, 1987). Also, the chemical species and the availability of metal can be influenced markedly by physical and chemical conditions, which include conditions in the field and of the experimental assay under controlled conditions. Thus, researchers using microorganisms to assess the toxicity of heavy metals should specify the experimental conditions in detail, and they should be very similar to in situ conditions (Cooney and Pettibone, 1986).
As we have warned in the initial sections of this article, the studies analyzed used different culture media and this variability could influence the reported MIC values, specifically for the heavy metal lead, for which our analyses showed an influence of the type of media (minimum vs. rich). Beyond the context of this study, such variability in experimental conditions invites researchers in this area to reflect on the need to work together in establishing standards for the characterization of microorganisms with bioremediation and phytoremediation potential. This type of effort would contribute to the generation of valuable information for the design of bioprospecting studies, but above all for the effective use of microorganisms. Examples of such joint efforts exist for different areas of scientific research, including microbiology.
Conclusion
The Minimum Inhibitory Concentration (MIC) is an important parameter in the evaluation of bacterial heavy metal tolerance, however, when studying this parameter, it is relevant to take into account the culture medium since it can influence bacterial heavy metal resistance. In this study, after an analysis of the MIC values retrieved from the literature, it was shown that the most resistant bacterial genera against arsenic, cadmium, and lead were Agrobacterium, Bacillus, Klebsiella, Enterobacter, Microbacterium, Pseudomonas, and Rhodococcus. Among these, Klebsiella and Enterobacter exhibited the highest resistance to cadmium and lead, Rhodococcus showed the highest resistance to arsenate, and Agrobacterium to arsenite. Moreover, the genera displayed different biochemical and molecular mechanisms that included the presence of PGP properties and enzymatic mechanisms, as well as groups of genes associated with the cad operon for cadmium resistance and a high diversity of ars genes in the different genera, showing that arsenic is among the non-essential heavy metals most tolerated by the bacteria studied.
Microorganisms have evolved different strategies to cope with heavy metal toxicity, and their multiple mechanisms of microbial detoxification can be applied to design different remediation strategies. In the present study, based on the high resistance to the most toxic non-essential heavy metals studied (cadmium and lead) and the presence of various biochemical and molecular mechanisms, we propose Klebsiella and Enterobacter as the best candidates for bioremediation and bacteria-assisted phytoremediation strategies in soils contaminated with arsenic, cadmium, and lead. Additionally, more studies are needed to elucidate the molecular mechanisms behind cadmium and lead resistance in these genera.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
TG-H conceived the idea, designed, and contributed to the writing of the manuscript. SG performed the data collection, statistical analysis, and contributed to the writing of the manuscript. Both authors read and approved the final version of the manuscript to be published.
Funding
This study was financially supported by a grant from Universidad Icesi and the CIBioFi Project, funded by the Colombian Science, Technology and Innovation Fund-General Royalties System (Fondo CTeI-SGR), Gobernación del Valle del Cauca, and COLCIENCIAS, under Contract No. BPIN 2013000100007.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the contributions of specific colleagues, institutions, or agencies that aided the efforts of the authors.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.604216/full#supplementary-material.
References
Achour-Rokbani, A., Cordi, A., Poupin, P., Bauda, P., and Billard, P. (2010). Characterization of the ars gene cluster from extremely arsenic-resistant Microbacterium sp. strain A33. Aem 76 (3), 948–955. doi:10.1128/aem.01738-09
Adams, G., Tawari-Fufeyin, P., Okoro, S., and Ehinomen, I. (2015). Bioremediation, biostimulation and bioaugmention: a review. Int. J. Environ. Bioremediation Biodegradation 3, 28–39. doi:10.12691/ijebb-3-1-5
Adhikary, A., Kumar, R., Pandir, R., Bhardwaj, P., Wusirika, R., and Kumar, S. (2019). Pseudomonas citronellolis; a multi-metal resistant and potential plant growth promoter against arsenic (V) stress in chickpea. Plant Physiol. Biochem. 142, 179–192. doi:10.1016/j.plaphy.2019.07.006
Afgan, E., Baker, D., Van den Beek, M., Blankenberg, D., Bouvier, D., Čech, M., et al. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10. doi:10.1093/nar/gkw343
Ahemad, M. (2015). Phosphate-solubilizing bacteria-assisted phytoremediation of metalliferous soils: a review. 3 Biotech. 5 (2), 111–121. doi:10.1007/s13205-014-0206-0
Aljerf, L., and AlMasri, N. (2018). A gateway to metal resistance: bacterial response to heavy metal toxicity in the biological environment. Ann. Adv. Chem. 2 (1), 32–44. doi:10.18689/mjcrs-1000123
Atobatele, O. E., and Olutona, G. O. (2015). Distribution of three non-essential trace metals (cadmium, mercury and lead) in the organs of fish from aiba reservoir, iwo, Nigeria. Toxicol. Rep. 2, 896–903. doi:10.1016/j.toxrep.2015.06.003
Bailey, T. L., Williams, N., Misleh, C., and Li, W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 44, W369–W373. doi:10.1093/nar/gkl198
Barkay, T., Miller, S. M., and Summers, A. O. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27 (2-3), 355–384. doi:10.1016/s0168-6445(03)00046-9
Behera, M., Dandapat, J., and Rath, C. C. (2014). Effect of heavy metals on growth response and antioxidant defense protection inBacillus cereus. J. Basic Microbiol. 54 (11), 1201–1209. doi:10.1002/jobm.201300805
Belimov, A. A., Zinovkina, N. Y., Safronova, V. I., Litvinsky, V. A., Nosikov, V. V., Zavalin, A. A., et al. (2019). Rhizobial ACC deaminase contributes to efficient symbiosis with pea (Pisum sativum L.) under single and combined cadmium and water deficit stress. Environ. Exp. Bot. 167, 103859. doi:10.1016/j.envexpbot.2019.103859
Ben Fekih, I., Zhang, C., Li, Y. P., Zhao, Y., Alwathnani, H. A., Saquib, Q., et al. (2018). Distribution of arsenic resistance genes in prokaryotes. Front. Microbiol. 9, 2473. doi:10.3389/fmicb.2018.02473
Benavides, M. P., Gallego, S. M., and Tomaro, M. L. (2005). Cadmium toxicity in plants. Braz. J. Plant Physiol. 17 (1), 21–34. doi:10.1590/s1677-04202005000100003
Bhat, S., Luo, X., Xu, Z., Liu, L., and Zhang, R. (2011). Bacillus sp. CDB3 isolated from cattle dip-sites possesses two ars gene clusters. J. Environ. Sci. 23 (1), 95–101. doi:10.1016/s1001-0742(10)60378-6
Biswas, A., Majumder, S., Neidhardt, H., Halder, D., Bhowmick, S., Mukherjee-Goswami, A., et al. (2011). Groundwater chemistry and redox processes: depth dependent arsenic release mechanism. Appl. Geochem. 26 (4), 516–525. doi:10.1016/j.apgeochem.2011.01.010
Borremans, B., Hobman, J. L., Provoost, A., Brown, N. L., and van Der Lelie, D. (2001). Cloning and functional analysis of thepbr lead resistance determinant of Ralstonia metallidurans CH34. J. Bacteriol. 183 (19), 5651–5658. doi:10.1128/jb.183.19.5651-5658.2001
Bowell, R. J. (1994). Sorption of arsenic by iron oxides and oxyhydroxides in soils. Appl. Geochem. 9 (3), 279–286. doi:10.1016/0883-2927(94)90038-8
Bruins, M. R., Kapil, S., and Oehme, F. W. (2000). Microbial resistance to metals in the environment. Ecotoxicology Environ. Saf. 45 (3), 198–207. doi:10.1006/eesa.1999.1860
Cai, L., Liu, G., Rensing, C., and Wang, G. (2009). Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 9 (1), 4. doi:10.1186/1471-2180-9-4
Cánovas, D., Cases, I., and De Lorenzo, V. (2003). Heavy metal tolerance and metal homeostasis inPseudomonas putidaas revealed by complete genome analysis. Environ. Microbiol. 5 (12), 1242–1256. doi:10.1111/j.1462-2920.2003.00463.x
Chien, C.-C., Huang, C.-H., and Lin, Y.-W. (2013). Characterization of a heavy metal translocating P-type ATPase gene from an environmental heavy metal resistance Enterobacter sp. isolate. Appl. Biochem. Biotechnol. 169 (6), 1837–1846. doi:10.1007/s12010-012-0047-4
Chintakovid, W., Visoottiviseth, P., Khokiattiwong, S., and Lauengsuchonkul, S. (2008). Potential of the hybrid marigolds for arsenic phytoremediation and income generation of remediators in Ron Phibun District, Thailand. Chemosphere 70 (8), 1532–1537. doi:10.1016/j.chemosphere.2007.08.031
Chmielewska-Bak, J., Lefèvre, I., Lutts, S., Kulik, A., and Deckert, J. (2014). Effect of cobalt chloride on soybean seedlings subjected to cadmium stress. Acta Societatis Botanicorum Poloniae 83 (3), 201-207. doi:10.5586/asbp.2014.027
Choudhury, R., and Srivastava, S. (2001). Zinc resistance mechanisms in bacteria. Curr. Sci. 81 (7), 768–775.
Cooney, J. J., and Pettibone, G. W. (1986). Metals and microbes in toxicity testing. Environ. Toxicol. Water Qual. 1 (4), 487–499. doi:10.1002/tox.2540010408
Crupper, S. S., Worrell, V., Stewart, G. C., and Iandolo, J. J. (1999). Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J. Bacteriol. 181 (13), 4071–4075. doi:10.1128/jb.181.13.4071-4075.1999
Daware, V., and Gade, W. (2015). Mechanism of arsenic tolerance in Klebsiella pneumoniae (HQ857583). Int. J. Curr. Microbiol. App Sci. 4 (11), 457–469.
Dean-Ross, D., and Mills, A. L. (1989). Bacterial community structure and function along a heavy metal gradient. Appl. Environ. Microbiol. 55 (8), 2002–2009. doi:10.1128/aem.55.8.2002-2009.1989
Duce, J. A., and Bush, A. I. (2010). Biological metals and Alzheimer’s disease: implications for therapeutics and diagnostics. Prog. Neurobiol. 92 (1), 1–18. doi:10.1016/j.pneurobio.2010.04.003
El-Mehy, R., Abou-Aly, H., Youssef, A., Tewfike, T., and Elakshar, E. (2019). Efficiency of heavy metals-tolerant plant growth promoting bacteria for alleviating heavy metals toxicity on sorghum. Environ. Exp. Bot. 162. doi:10.1016/j.envexpbot.2019.03.005
Endo, G., and Silver, S. (1995). CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol. 177 (15), 4437–4441. doi:10.1128/jb.177.15.4437-4441.1995
Fan, M., Liu, Z., Nan, L., Wang, E., Chen, W., Lin, Y., et al. (2018). Isolation, characterization, and selection of heavy metal-resistant and plant growth-promoting endophytic bacteria from root nodules of Robinia pseudoacacia in a Pb/Zn mining area. Microbiol. Res. 217, 51–59. doi:10.1016/j.micres.2018.09.002
Fay, R. M., and Mumtaz, M. M. (1996). Development of a priority list of chemical mixtures occurring at 1188 hazardous waste sites, using the HazDat database. Food Chem. Toxicol. 34 (11-12), 1163–1165. doi:10.1016/s0278-6915(97)00090-2
Fernández, M., Udaondo, Z., Niqui, J. L., Duque, E., and Ramos, J. L. (2014). Synergic role of the two ars operons in arsenic tolerance in P seudomonas putida KT 2440. Environ. Microbiol. Rep. 6 (5), 483–489. doi:10.1111/1758-2229.12167
Firrincieli, A., Presentato, A., Favoino, G., Marabottini, R., Allevato, E., Stazi, S. R., et al. (2019). Identification of resistance genes and response to arsenic in Rhodococcus aetherivorans BCP1. Front. Microbiol. 10, 888. doi:10.3389/fmicb.2019.00888
Flora, S. J. S., Pachauri, V., and Saxena, G. (2011). Arsenic, cadmium and lead. Reproductive and developmental toxicology. London: Academic Press.
Glick, B. R., Jacobson, C. B., Schwarze, M. M. K., and Pasternak, J. J. (1994). 1-Aminocyclopropane-1-carboxylic acid deaminase mutants of the plant growth promoting rhizobacterium Pseudomonas putida GR12-2 do not stimulate canola root elongation. Can. J. Microbiol. 40 (11), 911–915. doi:10.1139/m94-146
Glick, B. R. (2010). Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 28 (3), 367–374. doi:10.1016/j.biotechadv.2010.02.001
Goswami, D., Thakker, J. N., and Dhandhukia, P. C. (2016). Portraying mechanics of plant growth promoting rhizobacteria (PGPR): a review. Cogent Food Agric. 2 (1), 1127500. doi:10.1080/23311932.2015.1127500
Goyer, R. (2004). Issue paper on the human health effects of metals. Washington, DC: US Environmental Protection Agency.
Gu, Y., Wang, Y., Sun, Y., Zhao, K., Xiang, Q., Yu, X., et al. (2018). Genetic diversity and characterization of arsenic-resistant endophytic bacteria isolated from Pteris vittata, an arsenic hyperaccumulator. BMC Microbiol. 18 (1), 42. doi:10.1186/s12866-018-1184-x
Guo, H., Stüben, D., Berner, Z., and Yu, Q. (2009). Characteristics of arsenic adsorption from aqueous solution: effect of arsenic species and natural adsorbents. Appl. Geochem. 24 (4), 657–663. doi:10.1016/j.apgeochem.2008.12.017
Haq, R., Zaidi, S. K., and Shakoori, A. R. (1999). Cadmium resistant Enterobacter cloacae and Klebsiella sp. isolated from industrial effluents and their possible role in cadmium detoxification. World J. Microbiol. Biotechnol. 15 (2), 283–290. doi:10.1023/a:1008986727896
Harrison, E., and Brockhurst, M. A. (2012). Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiology 20 (6), 262–267. doi:10.1016/j.tim.2012.04.003
Hsieh, P. F., Lin, H. H., Lin, T. L., and Wang, J. T. (2010). CadC RegulatescadandtdcOperons in response to gastrointestinal stresses and enhances intestinal colonization ofKlebsiella pneumoniae. J. Infect. Dis. 202 (1), 52–64. doi:10.1086/653079
Hussein, K. A., and Joo, J. H. (2013). Heavy metal resistance of bacteria and its impact on the production of antioxidant enzymes. Afr. J. Microbiol. Res. 7 (20), 2288–2296. doi:10.5897/AJMR12.1764
Hynninen, A., Touzé, T., Pitkänen, L., Mengin-Lecreulx, D., and Virta, M. (2009). An efflux transporter PbrA and a phosphatase PbrB cooperate in a lead-resistance mechanism in bacteria. Mol. Microbiol. 74 (2), 384–394. doi:10.1111/j.1365-2958.2009.06868.x
Hynninen, A. (2010). Zinc, cadmium and lead resistance mechanisms in bacteria and their contribution to biosensing.
Jackson, C. R., and Dugas, S. L. (2003). Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase. BMC Evol. Biol. 3 (1), 18. doi:10.1186/1471-2148-3-18
Jin, Y., Luan, Y., Ning, Y., and Wang, L. (2018). Effects and mechanisms of microbial remediation of heavy metals in soil: a critical review. Appl. Sci. 8 (8), 1336. doi:10.3390/app8081336
Johnston, S. G., Karimian, N., and Burton, E. D. (2019). Fire promotes arsenic mobilization and rapid arsenic (III) formation in soil via thermal alteration of arsenic-bearing iron oxides. Front. Earth Sci. 7, 139. doi:10.3389/feart.2019.00139
Johri, N., Jacquillet, G., and Unwin, R. (2010). Heavy metal poisoning: the effects of cadmium on the kidney. Biometals 23 (5), 783–792. doi:10.1007/s10534-010-9328-y
Kubier, A., Wilkin, R. T., and Pichler, T. (2019). Cadmium in soils and groundwater: a review. Appl. Geochem. 108, 104388. doi:10.1016/j.apgeochem.2019.104388
Kumar Mishra, G. (2017). Microbes in heavy metal remediation: a review on current trends and patents. Recent patents Biotechnol. 11 (3), 188–196. doi:10.2174/1872208311666170120121025
Kumar, R., Nongkhlaw, M., Acharya, C., and Joshi, S. R. (2013). Growth media composition and heavy metal tolerance behaviour of bacteria characterized from the sub-surface soil of uranium rich ore bearing site of Domiasiat in Meghalaya.
Laghlimi, M., Baghdad, B., Hadi, H. E., and Bouabdli, A. (2015). Phytoremediation mechanisms of heavy metal contaminated soils: a review. Oje 05 (08), 375. doi:10.4236/oje.2015.58031
Lee, S.-W., Glickmann, E., and Cooksey, D. A. (2001). Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl. Environ. Microbiol. 67 (4), 1437–1444. doi:10.1128/aem.67.4.1437-1444.2001
Leitão, J. H., and Sá-Correia, I. (1997). Effects of growth-inhibitory concentrations of copper on alginate biosynthesis in highly mucoid Pseudomonas aeruginosa. Microbiology 143 (2), 481–488. doi:10.1099/00221287-143-2-481
Li, C., Zhou, K., Qin, W., Tian, C., Qi, M., Yan, X., et al. (2019). A review on heavy metals contamination in soil: effects, sources, and remediation techniques. Soil Sediment. Contamination: Int. J. 28 (4), 380–394. doi:10.1080/15320383.2019.1592108
Liberati, A., Altman, D. C., Tetzlaff, J., Mulrow, C., Gotzsche, P., and Ioannidis, J. J. B. (2009). The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. J. Clin. Epidemiol. 339, b2700–b2700. doi:10.1136/bmj.b2700
Ma, Y., Oliveira, R. S., Freitas, H., and Zhang, C. (2016). Biochemical and molecular mechanisms of plant-microbe-metal interactions: relevance for phytoremediation. Front. Plant Sci. 7, 918. doi:10.3389/fpls.2016.00918
Ma, Y., Prasad, M. N. V., Rajkumar, M., and Freitas, H. (2011). Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol. Adv. 29 (2), 248–258. doi:10.1016/j.biotechadv.2010.12.001
Maier, M. V., Wolter, Y., Zentler, D., Scholz, C., Stirn, C. N., and Isenbeck-Schröter, M. (2019). Phosphate induced arsenic mobilization as a potentially effective in-situ remediation technique-preliminary column tests. Water 11 (11), 2364. doi:10.3390/w11112364
Mejias Carpio, I. E., Ansari, A., and Rodrigues, D. F. (2018). Relationship of biodiversity with heavy metal tolerance and sorption capacity: a meta-analysis approach. Environ. Sci. Technol. 52 (1), 184–194. doi:10.1021/acs.est.7b04131
Mesa, V., Navazas, A., González-Gil, R., González, A., Weyens, N., Lauga, B., et al. (2017). Use of endophytic and rhizosphere bacteria to improve phytoremediation of arsenic-contaminated industrial soils by autochthonous Betula celtiberica. Appl. Environ. Microbiol. 83 (8). doi:10.1128/aem.03411-16
Mitra, S., Pramanik, K., Ghosh, P. K., Soren, T., Sarkar, A., Dey, R. S., et al. (2018). Characterization of Cd-resistant Klebsiella michiganensis MCC3089 and its potential for rice seedling growth promotion under Cd stress. Microbiol. Res. 210, 12–25. doi:10.1016/j.micres.2018.03.003
Mustapha, M. U., and Halimoon, N. (2015). Microorganisms and biosorption of heavy metals in the environment: a review paper. J. Microb. Biochem. Technol. 7, 253–256. doi:10.4172/1948-5948.1000219
Nakbanpote, W., Meesungnoen, O., and Prasad, M. N. V. (2016). “Chapter 9 - potential of ornamental plants for phytoremediation of heavy metals and income generation,” in Bioremediation and bioeconomy. Editor M. N. V. Prasad (Elsevier), 179–217.
Nies, D. H. (1992). CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J. Bacteriol. 174 (24), 8102–8110. doi:10.1128/jb.174.24.8102-8110.1992
Nriagu, J. O., Bhattacharya, P., Mukherjee, A. B., Bundschuh, J., Zevenhoven, R., and Loeppert, R. H. (2007). Arsenic in soil and groundwater: an overview. Trace Met. other Contaminants Environ. 9, 3–60. doi:10.1016/s1875-1121(06)09001-8
Nucifora, G., Chu, L., Misra, T. K., and Silver, S. (1989). Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc. Natl. Acad. Sci. 86 (10), 3544–3548. doi:10.1073/pnas.86.10.3544
Oger, C., Berthe, T., Quillet, L., Barray, S., Chiffoleau, J.-F., and Petit, F. (2001). Estimation of the abundance of the cadmium resistance gene cadA in microbial communities in polluted estuary water. Res. Microbiol. 152 (7), 671–678. doi:10.1016/s0923-2508(01)01246-3
Parameswari, E., Lakshmanan, A., and Thilagavathi, T. (2010). Biosorption and metal tolerance potential of filamentous fungi isolated from metal polluted ecosystem. Electron. J. Environ. Agric. Food Chem. 9 (4)
Pinto, A. P., Mota, A. M. d., De Varennes, A., and Pinto, F. C. (2004). Influence of organic matter on the uptake of cadmium, zinc, copper and iron by sorghum plants. Sci. total Environ. 326 (1-3), 239–247. doi:10.1016/j.scitotenv.2004.01.004
Pourrut, B., Shahid, M., Dumat, C., Winterton, P., and Pinelli, E. (2011). Lead uptake, toxicity, and detoxification in plants. Rev. Environ. Contam. Toxicol. 213, 113–136. doi:10.1007/978-1-4419-9860-6_4
Pramanik, K., Mitra, S., Sarkar, A., and Maiti, T. K. (2018). Alleviation of phytotoxic effects of cadmium on rice seedlings by cadmium resistant PGPR strain Enterobacter aerogenes MCC 3092. J. Hazard. Mater. 351, 317–329. doi:10.1016/j.jhazmat.2018.03.009
Pramanik, K., Mitra, S., Sarkar, A., Soren, T., and Maiti, T. K. (2017). Characterization of cadmium-resistant Klebsiella pneumoniae MCC 3091 promoted rice seedling growth by alleviating phytotoxicity of cadmium. Environ. Sci. Pollut. Res. 24 (31), 24419–24437. doi:10.1007/s11356-017-0033-z
Prieto-Barajas, C. M., Elorza-Gómez, J. C., Loeza-Lara, P. D., Sánchez-Yáñez, J. M., Valencia-Cantero, E., and Santoyo, G. (2018). Identificación y análisis de genes ars en cepas de Bacillus hipertolerantes al arsénico, aisladas de pozas termales en Araró, México. TIP Revista Especializada en Ciencias Químico-Biológicas 21, 22–29. doi:10.22201/fesz.23958723e.2018.0.145
Prithivirajsingh, S., Mishra, S. K., and Mahadevan, A. (2001). Detection and analysis of chromosomal arsenic resistance in Pseudomonas fluorescens strain MSP3. Biochem. biophysical Res. Commun. 280 (5), 1393–1401. doi:10.1006/bbrc.2001.4287
Roberts, L. C., Hug, S. J., Dittmar, J., Voegelin, A., Kretzschmar, R., Wehrli, B., et al. (2010). Arsenic release from paddy soils during monsoon flooding. Nat. Geosci 3 (1), 53–59. doi:10.1038/ngeo723
Román-Ponce, B., Ramos-Garza, J., Arroyo-Herrera, I., Maldonado-Hernández, J., Bahena-Osorio, Y., Vásquez-Murrieta, M. S., et al. (2018). Mechanism of arsenic resistance in endophytic bacteria isolated from endemic plant of mine tailings and their arsenophore production. Arch. Microbiol. 200 (6), 883–895. doi:10.1007/s00203-018-1495-1
Rosen, B. P. (2002). Biochemistry of arsenic detoxification. FEBS Lett. 529 (1), 86–92. doi:10.1016/s0014-5793(02)03186-1
Rouch, D. A., Lee, B. T. O., and Morby, A. P. (1995). Understanding cellular responses to toxic agents: a model for mechanism-choice in bacterial metal resistance. J. Ind. Microbiol. 14 (2), 132–141. doi:10.1007/bf01569895
Salt, D. E., Blaylock, M., Kumar, N. P. B. A., Dushenkov, V., Ensley, B. D., Chet, I., et al. (1995). Phytoremediation: a novel strategy for the removal of toxic metals from the environment using plants. Nat. Biotechnol. 13 (5), 468–474. doi:10.1038/nbt0595-468
Sarwar, N., Imran, M., Shaheen, M. R., Ishaque, W., Kamran, M. A., Matloob, A., et al. (2017). Phytoremediation strategies for soils contaminated with heavy metals: modifications and future perspectives. Chemosphere 171, 710–721. doi:10.1016/j.chemosphere.2016.12.116
Schalk, I. J., Hannauer, M., and Braud, A. (2011). New roles for bacterial siderophores in metal transport and tolerance. Environ. Microbiol. 13 (11), 2844–2854. doi:10.1111/j.1462-2920.2011.02556.x
Selvi, M. S., Sasikumar, S., Gomathi, S., Rajkumar, P., Sasikumar, P., and Govindan, S. (2014). Isolation and characterization of arsenic resistant bacteria from agricultural soil, and their potential for arsenic bioremediation. Int. J. Agric. Pol. Res. 2 (11), 393–405. doi:10.1016/j.btre.2016.02.002
Shrivastava, A., Ghosh, D., Dash, A., and Bose, S. (2015). Arsenic contamination in soil and sediment in India: sources, effects, and remediation. Curr. Pollut. Rep 1 (1), 35–46. doi:10.1007/s40726-015-0004-2
Shumlas, S. L., Singireddy, S., Thenuwara, A. C., Attanayake, N. H., Reeder, R. J., and Strongin, D. R. (2016). Oxidation of arsenite to arsenate on birnessite in the presence of light. Geochemical Trans. 17 (1), 1–10. doi:10.1186/s12932-016-0037-5
Silver, S., and Phung, L. T. (1996). Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50 (1), 753–789. doi:10.1146/annurev.micro.50.1.753
Singh, A., and Prasad, S. M. (2015). Remediation of heavy metal contaminated ecosystem: an overview on technology advancement. Int. J. Environ. Sci. Technol. 12 (1), 353–366. doi:10.1007/s13762-014-0542-y
Singh, R., Gautam, N., Mishra, A., and Gupta, R. (2011). Heavy metals and living systems: an overview. Indian J. Pharmacol. 43 (3), 246. doi:10.4103/0253-7613.81505
Tangahu, B. V., Abdullah, S., Rozaimah, S., Basri, H., Idris, M., Anuar, N., et al. (2011). A review on heavy metals (As, Pb, and Hg) uptake by plants through phytoremediation. Int. J. Chem. Eng. 2011, 1687–806X. doi:10.1155/2011/939161
Tchounwou, P. B., Yedjou, C. G., Patlolla, A. K., and Sutton, D. J. (2012). Heavy metal toxicity and the environment. Exp. Suppl. 101, 133–164. doi:10.1007/978-3-7643-8340-4_6
Theron, A. J., Tintinger, G. R., and Anderson, R. (2012). Harmful interactions of non-essential heavy metals with cells of the innate immune system
Tirry, N., Tahri Joutey, N., Sayel, H., Kouchou, A., Bahafid, W., Asri, M., et al. (2018). Screening of plant growth promoting traits in heavy metals resistant bacteria: prospects in phytoremediation. J. Genet. Eng. Biotechnol. 16 (2), 613–619. doi:10.1016/j.jgeb.2018.06.004
Ullah, A., Heng, S., Munis, M. F. H., Fahad, S., and Yang, X. (2015). Phytoremediation of heavy metals assisted by plant growth promoting (PGP) bacteria: a review. Environ. Exp. Bot. 117, 28–40. doi:10.1016/j.envexpbot.2015.05.001
Viti, C., Pace, A., and Giovannetti, L. (2003). Characterization of Cr(VI)-Resistant bacteria isolated from chromium-contaminated soil by tannery activity. Curr. Microbiol. 46 (1), 1–5. doi:10.1007/s00284-002-3800-z
Wang, L., Jeon, B., Sahin, O., and Zhang, Q. (2009). Identification of an arsenic resistance and arsenic-sensing system in Campylobacter jejuni. Aem 75 (15), 5064–5073. doi:10.1128/aem.00149-09
Wang, Q., Xiong, D., Zhao, P., Yu, X., Tu, B., and Wang, G. (2011). Effect of applying an arsenic-resistant and plant growth-promoting rhizobacterium to enhance soil arsenic phytoremediation by Populus deltoides LH05-17. J. Appl. Microbiol. 111 (5), 1065–1074. doi:10.1111/j.1365-2672.2011.05142.x
Wang, W. (1987). Factors affecting metal toxicity to (and accumulation by) aquatic organisms - Overview. Environ. Int. 13 (6), 437–457. doi:10.1016/0160-4120(87)90006-7
Wevar Oller, A. L., Talano, M. A., and Agostini, E. (2013). Screening of plant growth-promoting traits in arsenic-resistant bacteria isolated from the rhizosphere of soybean plants from Argentinean agricultural soil. Plant Soil 369 (1), 93–102. doi:10.1007/s11104-012-1543-6
White, G. F., Edwards, M. J., Gomez-Perez, L., Richardson, D. J., Butt, J. N., and Clarke, T. A. (2016). Mechanisms of bacterial extracellular electron exchange. Adv. Microb. Physiol. 68, 87–138. doi:10.1016/bs.ampbs.2016.02.002
Wuana, R. A., and Okieimen, F. E. (2011). Heavy metals in contaminated soils: a review of sources, chemistry, risks and best available strategies for remediation. Isrn Ecol. 2011, 2090–4614. doi:10.5402/2011/402647
Yang, P., Zhou, X.-F., Wang, L.-L., Li, Q.-S., Zhou, T., Chen, Y.-K., et al. (2018). Effect of phosphate-solubilizing bacteria on the mobility of insoluble cadmium and metabolic analysis. Ijerph 15 (7), 1330. doi:10.3390/ijerph15071330
Zeng, L. S., Liao, M., Chen, C. L., and Huang, C. Y. (2007). Effects of lead contamination on soil enzymatic activities, microbial biomass, and rice physiological indices in soil-lead-rice (Oryza sativa L.) system. Ecotoxicology Environ. Saf. 67 (1), 67–74. doi:10.1016/j.ecoenv.2006.05.001
Keywords: bioremediation, bacteria-assisted phytoremediation, heavy metals, minimum inhibitory concentration (MIC), plant microbiome
Citation: González Henao S and Ghneim-Herrera T (2021) Heavy Metals in Soils and the Remediation Potential of Bacteria Associated With the Plant Microbiome. Front. Environ. Sci. 9:604216. doi: 10.3389/fenvs.2021.604216
Received: 25 September 2020; Accepted: 13 January 2021;
Published: 12 April 2021.
Edited by:
Markus Puschenreiter, University of Natural Resources and Life Sciences Vienna, AustriaReviewed by:
Xiuli Hao, Huazhong Agricultural University, ChinaLucia Cavalca, University of Milan, Italy
Copyright © 2021 González Henao and Ghneim-Herrera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thaura Ghneim-Herrera, tghneim@icesi.edu.co
 Sarah González Henao
Sarah González Henao Thaura Ghneim-Herrera
Thaura Ghneim-Herrera