Antimicrobials and Antibiotic Resistance Genes in Water Bodies: Pollution, Risk, and Control
- Centre of Experimental Medicine and Surgery, Institute of Medical Sciences, Banaras Hindu University, Varanasi, India
The manuscript endeavors to provide a perspective on the role of water bodies in the spread of antimicrobial (antibiotic) resistance (AMR), antimicrobial resistant bacteria (ARB), and antimicrobial resistance genes (ARGs) among pathogens, animals, and humans. We briefly indicate how the AMR problem is globally affecting public health, along with strategies and mechanisms to combat the dissemination of ARB and ARGs. A brief systematic survey of the literature (2015-onwards) for the presence of antimicrobial residues and the occurrence of ARGs and antimicrobial resistant microorganisms in different water bodies/sources indicates the gravity of the situation and suggests their important role in the occurrence and spread of AMR, ARB, and ARGs. The prevalent water treatment methods which tend to reduce ARB and ARGs from water resources are unable to remove them completely, allowing the problem of AMR to continue and spread to organisms of concern. In this opinion article, we attempt to underline the key role of controlling the release/discharge of antimicrobial contaminants in water bodies and their buildup in checking the development and spread of AMR. The reduction in the release of antibiotic residues in the environment, especially water bodies, combined with the development of improved surveillance means and efficacious treatment/removal/decomposition methods could help curb the menace of AMR effectively. We suggest the expansion of the ambit of ‘One Health Approach to AMR crises proposed by the World Bank, 2021 to include the ‘reduction of antimicrobial contamination of the environment’ as the ‘seventh domain’ of activity to effectively achieve its objective.
Introduction
Aquatic ecosystems are very important to maintain the high levels of biodiversity, livelihood, and productivity of the biosphere (Hossain et al., 2018; Vilca and Angeles, 2018; Irfan and Alatawi, 2019; Hassan et al., 2020). The presence of antimicrobials (antibiotics etc.), antimicrobialresistant bacteria (ARB), and antimicrobial resistance genes (ARGs) in the aquatic environment is becoming a cause of great concern as the possibility of development of antibiotic-resistant pathogens, even superbugs, is increasingly posing problems to the environment and human health (Ma et al., 2015; Wang et al., 2020; Zhuang et al., 2021). It is recognized that aquatic environments are one of the key reservoirs and transmission routes for the spread of antimicrobial/antibiotic resistance (AMR/AR) (Amarasiri et al., 2020). Antibiotics reach the environment via feces and urine of humans and animals, inappropriate disposal of unused drugs, and direct environmental contamination by waste material from antibiotic production units (Amaya et al., 2012; World Health Organization, 2017a; United Nations Environment Programme, 2022). Antimicrobials/antibiotics exert selection pressure, accelerating the development of ARB resistant to the used antimicrobial and related compounds (Kolář et al., 2001; Ayukekbong et al., 2017; Serwecinska, 2020). All antibiotics put to use consistently end up in the environment, further accelerating the pace of AMR development (Larsson and Flach, 2021). Globally, AR has been frequently reported from freshwater sources (Kumar et al., 2013; Abdel Rahim et al., 2015; Jabbar Ibrahim and Kareem Hameed, 2015; Guzman-Otazo et al., 2019; Singh et al., 2020; Subbiah et al., 2020), wastewater systems including but not limited to pharmaceutical industries, and wastewater treatment plants (WWTPs) (Ferreira Da Silva et al., 2007; Tesfaye et al., 2019; Adegoke et al., 2020; Obayiuwana and Ibekwe, 2020; Praveenkumarreddy et al., 2020). AMR has emerged as one of the key public health problems of the 21st century that overshadows the efficacy of available effective treatments against a large number of pathogens which are increasingly no longer susceptible to common antimicrobials (Prestinaci et al., 2015). The AMR problem is increasing rapidly and becoming more critical with each passing day. Pathogens causing different common infections have been consistently acquiring and displaying a varying degree of resistance to most of the new antibiotics within <5–10 years of their introduction into the market (Supplementary Table S1). AMR is observed in bacteria, fungi, viruses, and parasites as they get adapted to multiply in the presence of antimicrobials (Founou et al., 2017; Dadgostar, 2019). The infection with AMR pathogens is supposed to escalate healthcare costs and treatment failures and cause up to 10 million more deaths annually by 2050 (Dadgostar, 2019; Amarasiri et al., 2020). The World Health Organization (WHO) has declared that due to increasing AR, we are almost out of treatment options (World Health Organization, 2017b; World Health Organization, 2021). Seeing the growing threat of AMR, the WHO has proposed a six-point plan, that is, “One Health Approach” (OHA) (World Bank Group, 2018; Mazimba et al., 2021). The OHA is envisaged as “involvement of human health, animal health, and environmental health and focus on those infectious disease-related issues (including AMR) that undermine overall health and well-being” (World Bank Group, 2018; Mazimba et al., 2021).
Numerous studies across the globe have reported the prevalence of ARB in different water bodies (Kumar et al., 2013; Abdel Rahim et al., 2015; Jabbar Ibrahim and Kareem Hameed, 2015; Guzman-Otazo et al., 2019; Singh et al., 2020; Subbiah et al., 2020) and wastewater systems (Ferreira Da Silva et al., 2007; Tesfaye et al., 2019; Adegoke et al., 2020; Obayiuwana and Ibekwe, 2020; Praveenkumarreddy et al., 2020). The inability of different drinking water treatments and WWTPs to completely remove ARGs and ARBs from water allows for their buildup in large water bodies (Alexander et al., 2020; Amarasiri et al., 2020). Considering the central role of water bodies in the development and spread of AMR (Figure 1A), the active monitoring of antimicrobial residues in the environment and control of disposal into the environment are suggested to help reduce the rate of AMR emergence/development (Figure 1A, B).
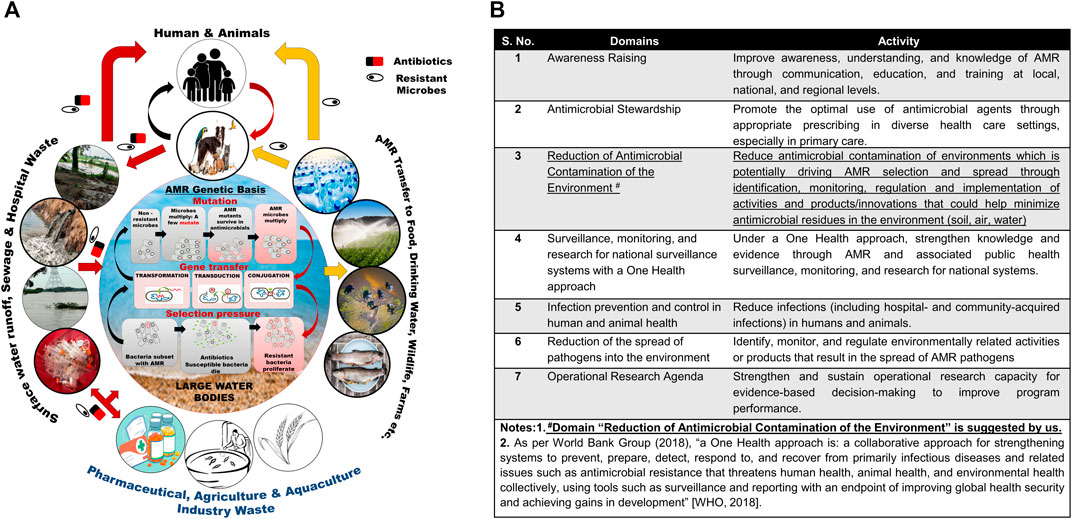
FIGURE 1. (A) Antimicrobial resistance development and transmission of ARGs and ARBs. (B) One health approach to AMR crises: Seven domains and activity for guiding the data collection, analyses, and action.
Antimicrobial Resistance Emergence and Spread
Microbial genome plasticity supported by numerous genetic mechanisms such as conjugation, transformation, and transduction enables them to evolve, adapt, and survive in environments contaminated with antibiotics. The development of AMR in microbes results from selection pressure, mutation, and gene transfer (Serwecinska, 2020; Larsson and Flach, 2021; Michael et al., 2014; Caniça et al., 2019; Van Hoek et al., 2011; Samreen et al., 2021; Kunhikannan et al., 2021; Amábile-Cuevas, 2021; Sriram et al., 2021; von Wintersdorff et al., 2016) (Supplementary Table S2), whereas conjugation supposedly remains the most frequently used mode of ARG transmission (Figure 1A) (von Wintersdorff et al., 2016).
Selection pressure determines the occurrence, amplification, and dissemination of ARGs in the environment and pathogens. Even low concentrations of antimicrobials/antibiotics can result in the selection of ARGs—making the establishment of a safe concentration of any antimicrobial compound in the environment a challenging task (Wang et al., 2020; Zhuang et al., 2021; Stanton et al., 2020; Yang et al., 2018). Several culture-independent studies on animals, food, humans, and environmental samples had shown the presence of huge reservoirs of ARGs (i.e., resistome) that could be potentially mobilized and transferred to other organisms (Abdel Rahim et al., 2015; Subbiah et al., 2020; Obayiuwana and Ibekwe, 2020; Adzitey, 2020; Forsberg et al., 2012; Hu et al., 2016; Abdel-Rahman et al., 2020; Meng et al., 2020; Morris and Cerceo, 2020; Balakrishna et al., 2017; D’Costa et al., 2011). Aquatic environments are identified as ideal settings for the acquisition and dissemination of AMR/AR. Human exposure to ARB and ARGs from aquatic environments poses an additional health risk (Karkman et al., 2018; Suzuki et al., 2017; Wellington et al., 2013; Leonard et al., 2018; Søraas et al., 2013; Leonard et al., 2015; O'Flaherty et al., 2018). Drinking water and wastewater treatment processes are mostly inadequate to remove ARGs (Li et al., 2015; Rodriguez-Mozaz et al., 2015; McGowan, 2007). The WWTP effluents, agriculture runoffs, etc. comprising ARB and ARGs can end up in aquatic environments such as lakes and rivers (Figure 1A). Usage of domestic wastewater in agricultural irrigation and recreational activities can introduce new ARB and ARGs to the specific environment (Leonard et al., 2018; Søraas et al., 2013; Leonard et al., 2015; O'Flaherty et al., 2018; Rodriguez-Mozaz et al., 2015; McGowan, 2007; Ben et al., 2017). In addition to drinking water, humans can be exposed to ARB and ARGs via different activities such as aquatic sports, bathing, occupational exposure during agricultural irrigation, and consumption of food produce from fields irrigated with reclaimed water (Leonard et al., 2018; Søraas et al., 2013; Leonard et al., 2015; O'Flaherty et al., 2018). However, the extent of human health risk resulting from exposure to ARB and ARGs present in aquatic environments remains poorly understood. It is primarily due to specific information such as the dose–response curves and exposure assessment data related to ARB and ARGs in different water usage scenarios being a prerequisite to conducting a quantitative microbial risk assessment (Ashbolt et al., 2013; Pepper et al., 2018).
Antimicrobial Resistance in Water Bodies: Cause and Impact
Antimicrobial Resistance in Aquaculture
The aquaculture field heavily relies on the application of antibiotics either directly in water or mixed with fish food to control infections, causing explosive growth of ARGs in farmed aquatic animals and environments (Heuer et al., 2009; Hinchliffe et al., 2018; Preena et al., 2020). Many antibiotics used in aquaculture are critically important for human treatment, for example, tetracycline, macrolides, and aminoglycosides (World Health Organization, 2021). Several studies indicated the indirect transfer of ARGs from fish origin antibiotic-resistant microbes to human pathogens such as E. coli, Salmonella spp., and Aeromonas spp through culture-independent studies (West et al., 2008; Heuer et al., 2009; Zou et al., 2012; Amarasiri et al., 2020). The presence of ARGs in S. enterica serotype Typhimurium DT104 isolates that caused salmonellosis outbreaks in Europe and the United States is also suspected to originate from the aquaculture system (Zou et al., 2012). The incidence of MDR in Vibrio alginolyticus and Vibrio parahaemolyticus isolates from farmed fishes in Korea further reaffirms the prevalent gene transfer phenomenon (Oh et al., 2011). Metagenomics or culture-independent studies have reported the occurrence of ARGs of different classes in marine sediments, suggesting a vital role for them in lateral gene transfer (Yang et al., 2013; Jiang et al., 2017; Karkman et al., 2018; Lerminiaux and Cameron, 2019; Sun et al., 2019).
Antimicrobial Resistance in Fresh and Wastewater
Freshwater bodies such as rivers, streams, springs, and lakes continuously receive antimicrobials/antibiotics, ARBs, and ARGs through different sources such as effluents from WWTPs, chemical manufacturing plants, animal husbandry, aquaculture, etc. Several studies had reported the presence of different antibiotics and ARGs in surface and groundwater sources (Liang et al., 2013; Shimizu et al., 2013; Ma et al., 2015; Matongo et al., 2015; Deng et al., 2016; Fernando et al., 2016; Madikizela et al., 2017; Danner et al., 2019) (Supplementary Table S3A). Even the bacterial communities of Pseudomonas, Acinetobacter, Bacillus, Arthrobacter, Xanthomonas, and Flavobacterium isolated from Eastern Siberian permafrost sediments had been shown to harbor several ARBs and ARGs by culture-dependent methods (Mindlin et al., 2008). Similarly, ARBs showing resistance to different classes of antibiotics had also been reported from freshwater samples of Antarctica and Siberian lakes (Lobova et al., 2011; Jara et al., 2020).
Wastewater remains a major reservoir of AMR in the environment as it allows ARBs with ARGs to persist and transfer the ARGs in the environment via different mechanisms (Rizzo et al., 2013; Fouz et al., 2020) (Figure 1A; Supplementary Table S2). The prevalent wastewater treatment methods that decrease the ARBs only have a limited impact on ARGs present in the environment (Ben et al., 2017; Hiller et al., 2019a; Singh, 2020). The ARGs present in the environment can get transmitted through horizontal gene transfer (HGT) to different organisms, including the medically important ones (Singh, 2020; Woolhouse et al., 2015). The environment assists the transfer of ARGs from one component to another, viz., animals, soil, water, sediments, and sewage (Figure 1A) (Wellington et al., 2013; Balcazar, 2014; Berglund, 2015; Fouz et al., 2020). ARGs are supposedly ubiquitous. However, their concentration may vary in different environments. The transfer of ARGs is not only limited to closely related species or genera but also occurs among phylogenetically distant species (Jiang et al., 2017). It leads to the ceaseless emergence of new variants of AMR organisms (Bouki et al., 2013; Hocquet et al., 2016; McKinney et al., 2018). The presence of ARGs in the environment represents a more complex and challenging problem concerning containment as they are not degradable and can be easily transmitted (Treangen and Rocha, 2011; Stecher et al., 2012; Beceiro et al., 2013; Hiller et al., 2019b; Ibrahim et al., 2019; Galhano et al., 2021; Koch et al., 2021; Zhuang et al., 2021; Grenni, 2022).
The major source of ARB and ARGs are human and veterinary clinical settings where intestinal bacteria encounter a high concentration of antibiotics, along with the associated WWTPs and land wastes (Karkman et al., 2018; Salyers et al., 2004; Ishikawa et al., 2018; Hendriksen et al., 2019). Bacteria passing via the intestinal tract can acquire AR through conjugation and transformation before ending up in human and animal feces (Figure 1A) (Salyers et al., 2004; Anderson et al., 2006). The effluent discharges from WWTPs to different environments where the environmental microorganisms can interact with ARB and ARGs may act as primary places of AMR development (Ben et al., 2017; Lee et al., 2017; Fouz et al., 2020; Manoharan et al., 2021).
Antimicrobial Resistance in Marine Environments
The mechanisms responsible for the occurrence of ARBs and ARGs in marine environments can be different from those of fresh water and wastewater. As per a report, marine environments contain about 28% of the ARGs (Hatosy and Martiny, 2015). The major source of increased AMR occurrence in the marine environment is the coastal runoff of the ARBs from the terrestrial environment (Hatosy and Martiny, 2015). In addition, anthropogenic activities are causing direct antibiotic residue outpourings into marine systems, for example, Chilean marine salmonid farms alone had used about 363.4 tons of antibiotics in 2016 that can act as a selection pressure for the development of AR in marine environments (Miranda et al., 2018). Metagenomics studies had reported the same ARGs in the intestines of Baltic Sea farm fishes and farm sediments; the possible reason suggested behind the observation is the usage of antibiotics during the hatching and rearing of juvenile fish or the acquisition of the ARGs by fishes from marine microorganisms in the farms (Rosenfeld and Zobell, 1947; Baam et al., 1966; Miranda et al., 2018; Tortorella et al., 2018). The potential bidirectional transfer of ARGs between these aquatic environments and humans cannot be ruled out.
Studies using both culture-dependent and culture-independent approaches suggest global contamination of the water environments including open oceans and widespread presence of ARBs (Shimizu et al., 2013; Hatosy and Martiny, 2015; Segura et al., 2015; Fekadu et al., 2019). In the natural aquatic environment, bacteria can develop AR due to induced mutagenesis at a low concentration of antibiotics (Kohanski et al., 2010). Although the fraction of resistant mutants is very low, the accelerated selection of ARBs could occur over generations (Gullberg et al., 2011) due to continued antimicrobial presence. Accordingly, attention should be paid to water environments as a key to the origin and spread of ARBs and ARGs.
Antimicrobial Resistance in Water and Public Health
It was observed that AMR, including multiresistance and pan-resistance, is rapidly spreading in bacteria, leading to severe infections untreatable with current antimicrobials (World Health Organization, 2014; World Health Organization, 2015; George, 2019). The spread of AMR in the environment had received comparatively less attention as compared to the spread of AMR pathogens in animals and humans (Baekkeskov et al., 2020). There are two types of AMR in bacteria, that is, acquired AMR and intrinsic AMR (World Health Organization, 2015; Baekkeskov et al., 2020) (Supplementary Table S2). The release of antimicrobial compounds into the environment allows it to come in direct contact with the naturally occurring microbes and act as a driving force for microbial evolution and the emergence of more resistant strains (European Centre for Disease Prevention and Control, 2019; Graham et al., 2019; Taneja and Sharma, 2019; Singh, 2020).
AMR is beginning to endanger public health worldwide (Prestinaci et al., 2015; Founou et al., 2017; Centers for Disease Control and Prevention, 2019; World Health Organization, 2021; The World Bank, 2021). Infection with AMR pathogens causes serious illnesses requiring longer hospital stays and increased healthcare costs due to the higher cost of second-line drugs and sometimes treatment failure (Llor and Bjerrum, 2014; Prestinaci et al., 2015; Centers for Disease Control and Prevention, 2018; Shrestha et al., 2018; European Centre for Disease Prevention and Control, 2019). AMR in common infections heavily impacts immunocompromised individuals and those undergoing treatments such as chemotherapy, dialysis, joint replacement, surgery, etc. (Centers for Disease Control and Prevention, 2018; Centres for Disease Control and Prevention, 2021).
Globally, the frequent incidences of infection with multidrug-resistant Gram-negative bacteria (MDR-GNB) and Gram-Positive bacteria are posing treatment challenges (Llor and Bjerrum, 2014; Bassetti et al., 2019; Ramírez-Castillo et al., 2018; Annavajhala et al., 2019; Centers for Disease Control and Prevention, 2019; World Health Organization, 2017a; Viney et al., 2021). The MDR cases are projected to become a serious issue by 2040 (Friedrich, 2017; Salvatore et al., 2019; Viney et al., 2021). Common pathogens of concern, namely, Enterococcus faecium, Helicobacter pylori, Neisseria gonorrhoeae, Campylobacter spp, etc. are currently included in the list of priority pathogens by the WHO for the development of new antibiotics due to rapid development of AMR in these pathogens (World Health Organization, 2017b).
The municipal wastewaters contain a high concentration of organic and inorganic matter that supports the growth of AMR microorganisms, further promoting the spread of ARGs and AMR (Da Silva et al., 2006; Baquero et al., 2008; Exner et al., 2017; Karkman et al., 2018). Unrestricted discharge of untreated urban waste had been contributing to an overall rise of ARBs and ARGs in the environment (Da Silva et al., 2006; Moura et al., 2009; Osińska et al., 2016). The WWTPs are the meeting point of most of the ARBs, especially in those processes in which activated sludge or percolator biological filter are used for biological treatment (Baquero et al., 2008; Karkman et al., 2018; Kumar and Pal, 2018). Some studies had shown a higher percentage of MDR bacteria in the effluent than the effluent of treated wastewater (Osińska et al., 2016; Exner et al., 2017). Generally, wastewater treatment regulates the level of bacterial count, but due to differences in treatment plant designs and operations, the fate of ARBs and ARGs may remain unaffected or amplified.
The bacterial communities proliferate in drinking water distribution system pipes even after chlorination (Korzeniewska et al., 2013; Razavi et al., 2017). As the process of chlorination initially lowers the total load of microbes, it may significantly increase the level of ARBs. Expectedly, the effect of chlorination on the secondary effluent of WWTPs had been found to cause reactivation of ARBs (Zhang et al., 2009). The plausible reason for ARB increase could be a decline of antibiotic-susceptible bacteria or the selective rise of the ARB population in wastewater. However, the potential threat to public health from ARBs, whether from reactivation or regrowth, calls for more intensive research on the phenomenon (Blasco et al., 2008; Martinez, 2009; Munir et al., 2011; Huang et al., 2012; Harnisz, 2013; Klanicova et al., 2013). A crucial point of intervention for environmental AMR management could be the removal of ARB and ARG contaminants from wastewater effluents that pose a direct threat to negatively impact other water resources. The enhancement of wastewater treatment technologies and rational use of antibiotics should be promoted to minimize the threat of pathogenic ARB emergence and infection.
The regular uptake of antibiotics through several environmental sources changes the composition of gut microbiota composition and induces the growth of ARB in human and animal gut (Cho and Blaser, 2012; Francino, 2016). This gut microbiota inequity leads to the growth of several AR pathogenic and opportunistic bacteria with the possibility of them evolving into superbugs whose infection could not respond to treatments and lead to untimely death (Cho and Blaser, 2012; Ben et al., 2019). There is a growing need to understand the relationship between antimicrobial/antibiotic exposure and the human microbiome, and its functional aspect related to health (Ben et al., 2019).
The extent of the growing global AMR problem can be gaged by a systematic search of the databases, viz., Web of Science, JSTOR, and PubMed using a combination of pertinent keywords such as antimicrobial, antibiotic resistance gene, water, environmental factors, antibiotics, heavy metals, water bodies, pollutants, etc. for the original research article and review articles. A brief systematic literature search performed for the antibiotic levels, ARBs, and ARGs in fresh and marine water on 17th February 2022 for articles published 2015 onwards (Supplementary Table S3B, S4) indicates the widespread presence of the residues of different antimicrobials (amoxicillin, penicillin tetracycline, ofloxacin, ciprofloxacin, etc.), a large number of ARGs (tetA, tetB, sulI, qnr, aadA, tetO, ampC, etc), and ARBs of concern (E. coli, Enterococcus, Salmonella, Shigella, Aeromonas, Vibrio, etc) in the water bodies (aquaculture, freshwater, wastewater, marine water) as presented in tabular form in Supplementary Table S3A, highlighting the severity of the AMR problem in water bodies.
The infections caused by ARBs increase the economic burden in terms of healthcare and associated costs. The infection caused by ARBs drastically inflates the cost of treatment and increases the chances of adverse outcomes, as compared to that caused by antibiotic-susceptible bacteria (World Health Organization, 2014; Centers for Disease Control and Prevention, 2018). The estimated deaths caused by AMR could rise from the current rate of about 0.7 million to 10 million annually by 2050, if comprehensive actions are not taken (World Health Organization, 2019; Samreen et al., 2021). It would further cause a 3–4% reduction in the annual gross domestic product (GDP) globally—translating into an economic cost of 1–6 trillion yearly from 2030–2050 onwards, based upon AMR (low to high) scenarios encountered and depending upon the measures undertaken now (World Bank, 2017).
Control of Antimicrobial Resistance Emergence and Dissemination in Water Bodies
Removal of Antimicrobial Resistant Bacteria and Antimicrobial Resistance Genes From Water Supply Systems and Wastewater Treatment Units
The WWTPs were designed for the removal of organic matter, nutrients, and solids, but now they need to be able to remove antimicrobials/antibiotics, ARBs, and ARGs as well. So far, very little is known about the effectiveness of the treatments in the removal of ARB and ARGs. Research is needed to fill a huge knowledge gap in this area to help improve the design of WWTPs and the used methodologies. Water supply systems and WWTPs use a single or a combination of different treatment processes to achieve many log reductions in the number of specific target microbes which show intra- and inter-process variations (Sano et al., 2016; Amarasiri et al., 2017). Specific comprehensive guidelines suggesting minimum reductions for antibiotics, ARB, and ARGs in water/wastewater are desired (Hong et al., 2018).
The membrane bioreactor treatment plants had achieved significantly higher reductions of ARB and ARGs (log reduction range: 2.57–7.06) than conventional treatment methods such as sand filtration, sedimentation, activated sludge, and rotating biological contactor or oxidation ditch (log reduction range: 2.37–4.56; p < 0.05) (Munir et al., 2011; O'Flaherty and Cummins, 2017; Su et al., 2018). The retention in the sand filter medium with low nutrient conditions is supposed to cause ARB starvation, leading to plasmid degradation and permanent loss of antibiotic resistance (Griffiths et al., 1990; Tan et al., 2019). Drinking water treatment by conventional methods had achieved variable ARG log reductions (0.03–2.4) that differed with the types of ARGs evaluated (Hu et al., 2019; Zhang et al., 2019). Sulfamethoxazole had killed ARBs immediately, but a delay in ARGs reduction was reported, possibly resulting from a competitive consumption of free radicals by sulfamethoxazole and ARGs (Hu et al., 2019).
Several studies indicate the inability of the water treatment processes to eliminate ARB and ARGs. There had been increased incidences of specific ARGs in the WWTP effluents (Rizzo et al., 2013; Xu et al., 2015; Chu et al., 2018; Karkman et al., 2018). A study revealed no significant differences in the ARGs present in potable source water and treated water, indicating the nonremoval of ARGs during the process (Garner et al., 2018). The activated granular carbon filtration method even enhanced the abundance of ARGs in the filtered water due to the formation of biofilms on the biological activated carbon surfaces where ARB can adhere and grow (Xu et al., 2016; Su et al., 2018; Hu et al., 2019; Tan et al., 2019). The significant correlations (p < 0.05) between the effluent ARG concentration (2 μg/l) and residual antibiotic concentration (0.5–0.22 μg/l) suggest a role for selection pressure on ARG enrichment (Mao et al., 2015; Hu et al., 2019).
The search for local solutions to avoid environmental dissemination of these pollutants requires prior information on the specific residues and the AR determinants present in wastewater. There is an urgent need for public health research to increase its pace to keep up with water sustainability technologies and even go beyond. In addition, more research work is essential for the application of effective treatment and disinfection approaches for the complete removal of ARB in WWTPs as the associated immediate environmental and public health risks are high.
Tracking the Sources of Antimicrobial Resistance in Organisms
The modern molecular techniques for the characterization of bacterial organisms can readily increase our ability to track the source of AMR and ARGs. These could provide useful insights, including but not limited to a comprehensive understanding of the population biology of organisms and the genetic diversity of organisms entering water (Olivas and Faulkner, 2008). These techniques could provide fast and accurate AMR source tracking and other genetic mobile platforms involved in AMR dissemination, providing a much more accurate image of the real diversity and complexity of AR in water-borne bacteria, unlike cultivation-dependent approaches (Henriques et al., 2006).
Revision of Domains of “One Health Approach” for Tackling Antimicrobial Resistance
The “One Health Approach” suggested by the WHO that currently focuses on activities to reduce the contamination and usage of antimicrobials and ways to minimize the development and spread of AMR pathogens (World Bank Group, 2018; Mazimba et al., 2021) should consider including active pursuance of the reduction of antimicrobial contamination of the environment as the seventh domain of the OHA for AMR crises to curb the development and spread of AMR (See Figure 1B).
Discussion
Antimicrobials/antibiotics are used as both preventive and therapeutic agents in the treatment of animal diseases, human infections, aquaculture, agriculture, and the livestock industry (Baquero et al., 2008; Forsberg et al., 2012; Ben et al., 2019; Dadgostar, 2019; Schar et al., 2020; Schar et al., 2021). Antibiotic residues reach different environments through excretions (stool and urine of animals and human), improper disposal of unused drugs, waste stream from the antibiotic production unit, antibiotics used for plant production, etc. (Baquero et al., 2008; Ishikawa et al., 2018; Karkman et al., 2018; Pepper et al., 2018; Dadgostar, 2019; Larsson and Flach, 2021). Water bodies get contaminated by municipal sewage discharges, animal husbandry, landfill leachates of antibiotic disposal, manufacturing industries, and agricultural runoff (Hernando-Amado et al., 2019; Serwecinska, 2020). The increased frequency of ARGs in various ARBs of different environments is one of the concerning consequences of antimicrobial/antibiotic misuse and subsequent pollution (Kraemer et al., 2019). Studies indicate that aquatic environments act as a key reservoir and means of antibiotic resistance spread (Zhang et al., 2013; Matongo et al., 2015; Binh et al., 2018; Yang et al., 2018; Danner et al., 2019; Kraemer et al., 2019; Amarasiri et al., 2020; Schar et al., 2020; Larsson and Flach, 2021; Liyanage et al., 2021). In an aquatic environment, wastewater and WWTPs are considered one of the key potential hot spots for the spread of AR and transfer of ARGs (Matongo et al., 2015; Amarasiri et al., 2020; Ali et al., 2021; Buriánková et al., 2021; Guo et al., 2021; Markkanen et al., 2021; Obayiuwana et al., 2021; Yoo and Lee, 2021; Zhang et al., 2021). It was estimated that for the production of aquaculture animals, the global consumption of antimicrobials which was 10,259 tons in 2017 is projected to register an increase of 33% to 13,600 tons by 2030 (Schar et al., 2020). Different ARBs and ARGs had been frequently detected in groundwater (Singh et al., 2020; Kunhikannan et al., 2021), surface water (Deng et al., 2016; Binh et al., 2018), wastewater (Karkman et al., 2018; Nguyen et al., 2021), sediments (Liang et al., 2013; Xu et al., 2014; McInnes et al., 2021) and marine water (Buschmann et al., 2012; Shimizu et al., 2013; Vilca and Angeles, 2018). A brief systematic review of the literature aptly highlights the growing menace of ARBs and ARGs in fresh and marine water environments along with the contamination of different antibiotics (Supplementary Table S3A).
Both culture-dependent and culture-independent (metagenomics) studies have contributed to our understanding of the AMR problem. The combination of culture-dependent and culture-independent metagenomic techniques is reported to provide better retrieval of ARGs than either method alone (Korzeniewska and Harnisz, 2012; Fenske et al., 2020). Metagenomic studies provide an avenue to study the uncultivable total microorganisms (Forbes et al., 2017). A metagenomics study of municipal wastewater and hospital wastewater revealed the presence of tetracycline, beta-lactam, macrolide–lincosamide–streptogramin resistance gene and multidrug resistance genes ranging from 0.06–0.98 copy/cell, and biocide/metal resistance gene ranging from 0.30–1.99 copies/cell (Zhang et al., 2021). One of the studies reported the presence of different ARGs in the seawater sample at 1.7 × 102 copies/giga base (Zeng et al., 2019). The highest ARG levels of 1.57–700.58 × 102 copy/ml for penicillin were reported from surface water, whereas 0.37–312.7 × 102 copy/ml was reported from the groundwater of Sri Lanka. Among the penicillin resistance genes, the highest percentage of blaTEM (700.58 × 102 copy/ml) followed by ampicillin (0.37–371.7 × 102 copy/ml) and OPR D (1.57 × 102 copy/ml) resistance genes were reported from aquatic samples, whereas tetM and tetA resistance genes at the levels of 1.35–439.88 × 102 copy/ml were reported from the surface water samples. Only the tetM resistance gene was reported at 215.99 × 102 copy/ml from the groundwater sample of Sri Lanka (Liyanage et al., 2021). Zhang et al., 2021 reported that the number of ARGs is strongly correlated with the number of biocide/metal resistance genes in the WWTPs with more chemicals (Zhang et al., 2021). The municipal wastewaters had more abundant and diverse ARGs than hospital wastewater. From the urban canals and lakes of Vietnam, different levels of erythromycin, amoxicillin, sulfamethoxazole, ampicillin, clindamycin, tylosin, vancomycin, tetracycline, chloramphenicol, etc. were frequently detected (Tran et al., 2019). Metagenomics study of rural and urban water and sediments of Bangladesh reported a significant correlation between ARGs and human origin bacteria (R2 = 0.73; P= < 0.01), suggesting that the release of untreated sewage could act as a driver for the transmission of ARGs in the environment (McInnes et al., 2021). A recent metagenomics study of 79 WWTPs situated in 60 countries reported the differences in diversity and abundance of ARGs among Africa, Asia, North America, South America, Oceania, and Europe (Strange et al., 2021). The Oceanic cluster reported a limited number of ARGs encoding macrolides in high number, whereas Africa, Asia, and South America clusters harbored ARGs representing sulfonamides and chloramphenicol. A study has also reported Vietnam, India, and Brazil to have the most divergent ARG distribution and suggested them as possible hotspots for the emergence of new antibiotic resistance mechanisms (Hendriksen et al., 2019). Several studies reported the prevalence of antibiotics, ARBs, and ARGs in the aquatic environment that was correlated with environmental factors (Supplementary Table S3A) (Binh et al., 2018; Tran et al., 2019; Hanna et al., 2020; Anh et al., 2021; Azanu et al., 2021; Duong et al., 2021; Guo et al., 2021; Lai et al., 2021; McInnes et al., 2021; Zhang et al., 2021; Zhuang et al., 2021). A metagenomics study of wastewater in Benin and Burkina showed the prevalence of resistance genes van, blaOXA, blaGES, blaIMP, blaKPC, blaNDM, blaOXA, blaVIM, qnr, and mcr (Markkanen et al., 2021). Recently, a large number of ARGs subtypes, viz., blaNDM-1, blaCTX-M-15, mecA, blaTEM-1, sul1, vanA, blaKPC-2, sul2, blaCTX-M-14, and blaOXA-48 had also been reported in decreasing order from Asia, Europe, Africa, and North and South America (Zhuang et al., 2021).
The major problem to tackle the issue of AMR is existing knowledge gaps comprising incomplete knowledge or information and misperceptions about the use of antibiotics and the relative contribution of the release of ARB or ARGs in the environment from different sources (McCullough et al., 2016; Singh, 2020). Health professionals can play a major role in the prevention and spread of AMR by educating people about the possible risks of inappropriate usage and disposal of antibiotics and contaminated material containing ARBs with ARGs. The effective control of AMR development and spread of ARGs and ARBs can be facilitated by promoting the development of self-contained local wastewater treatment modules, use of antibiotics/antimicrobial degrading contraptions, and implementing strategies to minimize the concentration of antibiotics required for treatment, including the use of nanotechnology (Malakootian et al., 2019; Singh, 2020; Kaur et al., 2021; Singh et al., 2022).
AMR/AR has the potential to threaten human health and inflict huge blows to the economies of both developed and developing countries (Ventola, 2015; World Health Organization, 2021). Estimates for Europe, the United Kingdom, Thailand, and the United States, the project substantial increase in health costs from antibiotic-resistant bacterial infections (Da Silva et al., 2006; World Health Organization, 2015; Osińska et al., 2016; Abdel-Rahman et al., 2020; Obayiuwana and Ibekwe, 2020; Yoo and Lee, 2021). The waterborne AMR is causing an economic impact of $340-$680 billion annually on the health care system (World Economic Forum, 2021). The waterborne AMR is contributing about $1 to $5 billion per year in additional health care expenditure, and it is expected to increase as resistance develops further. The waterborne AMR could be responsible for about 3.5 million additional sicknesses annually at the cost of $300 million (World Economic Forum, 2021). The alarm of AMR crisis raised in recent times by various bodies such as WHO, FAO, CDC, World Bank, etc. (World Health Organization, 2018; Mulani et al., 2019; Centers for Disease Control and Prevention, 2019; Centres for Disease Control and Prevention, 2021) also calls for the strengthening of the synthesis and discovery pipeline of new antibiotics with better activities or activity against various antibiotic-resistant pathogens (Supplementary Table S5). Development of new more potent antibiotics with different or multiple modes of action, along with focused steps to curb AMR development and dissemination to pathogens, is required (Mulani et al., 2019; Singh, 2020; Léger et al., 2021). AMR, being a multidimensional problem, requires a proactive holistic, constructive, collaborative, and synergistic strategy and action by different stakeholders to comprehensively implement a One Health Approach to overcome the unfolding AMR crises.
Conclusion
The continued antimicrobial overuse, misuse, and uncontrolled contamination of the environment throughout the world are turning the AMR issue into a global health crisis. The tackling of the AMR situation requires implementation of new policies that limit the release of antimicrobial residues into the environment and support appropriate monitoring to minimize their buildups and timely removal. More research efforts are needed toward understanding the extent and mechanistic underpinnings of AMR development and ARG transfer to other pathogenic bacteria to develop better control strategies. The involvement of the public to locally manage and dispose of the antimicrobials and AMB remains a potential area of collaboration and policy development to control the AMR crisis as it could promote both a sense of responsibility and awareness.
Author Contributions
SS conceptualized, supervised, prepared, and finalized the manuscript. RK, AKS, and SS prepared the first draft. SV provided critical inputs. RK, AKS, SV, and SS together revised the manuscript. All authors approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The laboratory of SS is supported by IoE grant from Banaras Hindu University. RK is a recipient of the UGC Non-NET fellowship by Banaras Hindu University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.830861/full#supplementary-material
References
Abdel Rahim, K. A. A., Hassanein, A. M., and Abd El Azeiz, H. A. E. H. (2015). Prevalence, Plasmids and Antibiotic Resistance Correlation of Enteric Bacteria in Different Drinking Water Resources in Sohag, Egypt. Jundishapur J. Microbiol. 8, 1–8. doi:10.5812/jjm.18648
Abdel-Rahman, M. A. A., Roshdy, H., Samir, A. H., and Hamed, E. A. (2020). Antibiotic Resistance and Extended-Spectrum β-lactamase in Escherichia coli Isolates from Imported 1-Day-Old Chicks, Ducklings, and turkey Poults. Vet. World 13, 1037–1044. doi:10.14202/vetworld.2020.1037-1044
Adegoke, A. A., Madu, C. E., Aiyegoro, O. A., Stenström, T. A., and Okoh, A. I. (2020). Antibiogram and Beta-Lactamase Genes Among Cefotaxime Resistant E. coli from Wastewater Treatment Plant. Antimicrob. Resist. Infect. Control. 9, 1–12. doi:10.1186/s13756-020-0702-4
Adzitey, F. (2020). Incidence and Antimicrobial Susceptibility of Escherichia coli Isolated from Beef (Meat Muscle, Liver and Kidney) Samples in Wa Abattoir, Ghana. Cogent Food Agric. 6, 1718269. doi:10.1080/23311932.2020.1718269
Alexander, J., Hembach, N., and Schwartz, T. (2020). Evaluation of Antibiotic Resistance Dissemination by Wastewater Treatment Plant Effluents with Different Catchment Areas in Germany. Sci. Rep. 10, 1–9. doi:10.1038/s41598-020-65635-4
Ali, O. S., Hozayen, W. G., Almutairi, A. S., Edris, S. A., Abulfaraj, A. A., Ouf, A. A., et al. (2021). Metagenomic Analysis Reveals the Fate of Antibiotic Resistance Genes in a Full-Scale Wastewater Treatment Plant in Egypt. Sustainability 13, 11131–11219. doi:10.3390/su132011131
Amábile-Cuevas, C. F. (2021). Antibiotic Resistance from, and to the Environment. AIMS Environ. Sci. 8, 18–35. doi:10.3934/environsci.2021002
Amarasiri, M., Kitajima, M., Nguyen, T. H., Okabe, S., and Sano, D. (2017). Bacteriophage Removal Efficiency as a Validation and Operational Monitoring Tool for Virus Reduction in Wastewater Reclamation: Review. Water Res. 121, 258–269. doi:10.1016/j.watres.2017.05.035
Amarasiri, M., Sano, D., and Suzuki, S. (2020). Understanding Human Health Risks Caused by Antibiotic Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARG) in Water Environments: Current Knowledge and Questions to Be Answered. Crit. Rev. Environ. Sci. Technol. 50, 2016–2059. doi:10.1080/10643389.2019.1692611
Amaya, E., Reyes, D., Paniagua, M., Calderón, S., Rashid, M.-U., Colque, P., et al. (2012). Antibiotic Resistance Patterns of Escherichia coli Isolates from Different Aquatic Environmental Sources in Leon, Nicaragua. Clin. Microbiol. Infect. 18, E347–E354. doi:10.1111/j.1469-0691.2012.03930.x
Anderson, M. A., Whitlock, J. E., and Harwood, V. J. (2006). Diversity and Distribution of Escherichia coli Genotypes and Antibiotic Resistance Phenotypes in Feces of Humans, Cattle, and Horses. Appl. Environ. Microbiol. 72, 6914–6922. doi:10.1128/AEM.01029-06
Anh, H. Q., Le, T. P. Q., Da Le, N., Lu, X. X., Duong, T. T., Garnier, J., et al. (2021). Antibiotics in Surface Water of East and Southeast Asian Countries: A Focused Review on Contamination Status, Pollution Sources, Potential Risks, and Future Perspectives. Sci. Total Environ. 764, 142865. doi:10.1016/j.scitotenv.2020.142865
Annavajhala, M. K., Gomez-Simmonds, A., and Uhlemann, A. C. (2019). Multidrug-resistant Enterobacter cloacae Complex Emerging as a Global, Diversifying Threat. Front. Microbiol. 10, 44. doi:10.3389/fmicb.2019.00044
Ashbolt, N. J., Amézquita, A., Backhaus, T., Borriello, P., Brandt, K. K., Collignon, P., et al. (2013). Human Health Risk Assessment (HHRA) for Environmental Development and Transfer of Antibiotic Resistance. Environ. Health Perspect. 121, 993–1001. doi:10.1289/ehp.1206316
Ayukekbong, J. A., Ntemgwa, M., and Atabe, A. N. (2017). The Threat of Antimicrobial Resistance in Developing Countries: Causes and Control Strategies. Antimicrob. Resist. Infect. Control. 6, 47–48. doi:10.1186/s13756-017-0208-x
Azanu, D., Adu-poku, D., Saah, S. A., and Appaw, W. O. (2021). Prevalence of Pharmaceuticals in Surface Water Samples in Ghana. J. Chem. 2021, 1–11. doi:10.1155/2021/7829477
Baam, R. B., Gandhi, N. M., and Freitas, Y. M. (1966). Antibiotic Activity of marine Microorganisms: the Antibacterial Spectrum. Helgolander Wiss. Meeresunters 13, 188–191. doi:10.1007/BF01612664
Baekkeskov, E., Rubin, O., Munkholm, L., and Zaman, W. (2020). “Antimicrobial Resistance as a Global Health Crisis,” in Oxford Research Encyclopedia of Politics. Editor E. Stern (Oxford University Press), 1–24. doi:10.1093/acrefore/9780190228637.013.1626
Balakrishna, K., Rath, A., Praveenkumarreddy, Y., Guruge, K. S., and Subedi, B. (2017). A Review of the Occurrence of Pharmaceuticals and Personal Care Products in Indian Water Bodies. Ecotoxicology Environ. Saf. 137, 113–120. doi:10.1016/j.ecoenv.2016.11.014
Balcazar, J. L. (2014). Bacteriophages as Vehicles for Antibiotic Resistance Genes in the Environment. Plos Pathog. 10, e1004219. doi:10.1371/journal.ppat.1004219
Baquero, F., Martínez, J. L., and Cantón, R. (2008). Antibiotics and Antibiotic Resistance in Water Environments. Curr. Opin. Biotechnol. 19, 260–265. doi:10.1016/j.copbio.2008.05.006
Bassetti, M., Peghin, M., Vena, A., and Giacobbe, D. R. (2019). Treatment of Infections Due to MDR Gram-Negative Bacteria. Front. Med. 6, 74. doi:10.3389/fmed.2019.00074
Beceiro, A., Tomás, M., and Bou, G. (2013). Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 26, 185–230. doi:10.1128/CMR.00059-12
Ben, W., Wang, J., Cao, R., Yang, M., Zhang, Y., and Qiang, Z. (2017). Distribution of Antibiotic Resistance in the Effluents of Ten Municipal Wastewater Treatment Plants in China and the Effect of Treatment Processes. Chemosphere 172, 392–398. doi:10.1016/j.chemosphere.2017.01.041
Ben, Y., Fu, C., Hu, M., Liu, L., Wong, M. H., and Zheng, C. (2019). Human Health Risk Assessment of Antibiotic Resistance Associated with Antibiotic Residues in the Environment: A Review. Environ. Res. 169, 483–493. doi:10.1016/j.envres.2018.11.040
Berglund, B. (2015). Environmental Dissemination of Antibiotic Resistance Genes and Correlation to Anthropogenic Contamination with Antibiotics. Infect. Ecol. Epidemiol. 5, 28564. doi:10.3402/iee.v5.28564
Binh, V. N., Dang, N., Anh, N. T. K., Ky, L. X., and Thai, P. K. (2018). Antibiotics in the Aquatic Environment of Vietnam: Sources, Concentrations, Risk and Control Strategy. Chemosphere 197, 438–450. doi:10.1016/j.chemosphere.2018.01.061
Blasco, M. D., Esteve, C., and Alcaide, E. (2008). Multiresistant Waterborne Pathogens Isolated from Water Reservoirs and Cooling Systems. J. Appl. Microbiol. 105, 469–475. doi:10.1111/j.1365-2672.2008.03765.x
Bouki, C., Venieri, D., and Diamadopoulos, E. (2013). Detection and Fate of Antibiotic Resistant Bacteria in Wastewater Treatment Plants: A Review. Ecotoxicology Environ. Saf. 91, 1–9. doi:10.1016/j.ecoenv.2013.01.016
Buriánková, I., Kuchta, P., Molíková, A., Sovová, K., Výravský, D., Rulík, M., et al. (2021). Antibiotic Resistance in Wastewater and its Impact on a Receiving River: A Case Study of WWTP Brno-Modřice, Czech Republic. Water 13, 2309. doi:10.3390/w13162309
Buschmann, A. H., Tomova, A., López, A., Maldonado, M. A., Henríquez, L. A., Ivanova, L., et al. (2012). Salmon Aquaculture and Antimicrobial Resistance in the marine Environment. PLoS One 7, e42724–28. doi:10.1371/journal.pone.0042724
Caniça, M., Manageiro, V., Abriouel, H., Moran-Gilad, J., and Franz, C. M. A. P. (2019). Antibiotic Resistance in Foodborne Bacteria. Trends Food Sci. Technol. 84, 41–44. doi:10.1016/j.tifs.2018.08.001
Centers for Disease Control and Prevention (2018). Antibiotic Resistance Threats in the United States, 2013. Atlanta, GA: CDC. Available at: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
Centers for Disease Control and Prevention (2019). Antibiotic Resistance Threats In The United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC. Available at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf and www.cdc.gov/DrugResistance/Biggest-Threats.html (Accessed November 22, 2021). doi:10.15620/cdc:82532
Centres for Disease Control and Prevention (2021). Antibiotic/Antimicrobial Resistance (AR/AMR). Atlanta, GA: U.S. Department of Health & Human Services. Available at: https://www.cdc.gov/drugresistance/food.html (Accessed November 22, 2021).
Cho, I., and Blaser, M. J. (2012). The Human Microbiome: At the Interface of Health and Disease. Nat. Rev. Genet. 13, 260–270. doi:10.1038/nrg3182
Chu, B. T. T., Petrovich, M. L., Chaudhary, A., Wright, D., Murphy, B., Wells, G., et al. (2018). Metagenomics Reveals the Impact of Wastewater Treatment Plants on the Dispersal of Microorganisms and Genes in Aquatic Sediments. Appl. Environ. Microbiol. 84, 2168. doi:10.1128/AEM.02168-17
Da Silva, M. F., Tiago, I., VerÃssimo, A., Boaventura, R. A. R., Nunes, O. C., and Manaia, C. l. M. (2006). Antibiotic Resistance of Enterococci and Related Bacteria in an Urban Wastewater Treatment Plant. FEMS Microbiol. Ecol. 55, 322–329. doi:10.1111/j.1574-6941.2005.00032.x
Dadgostar, P. (2019). Antimicrobial Resistance: Implications and Costs. Idr Vol. 12, 3903–3910. doi:10.2147/IDR.S234610
Danner, M. C., Robertson, A., Behrends, V., and Reiss, J. (2019). Antibiotic Pollution in Surface Fresh Waters: Occurrence and Effects. Sci. Total Environ. 664, 793–804. doi:10.1016/j.scitotenv.2019.01.406
D’Costa, V. M., King, C. E., Kalan, L., Morar, M., Sung, W. W. L., Schwarz, C., et al. (2011). Antibiotic Resistance Is Ancient. Nature 477, 457–461. doi:10.1038/nature10388
Deng, W., Li, N., Zheng, H., and Lin, H. (2016). Occurrence and Risk Assessment of Antibiotics in River Water in Hong Kong. Ecotoxicology Environ. Saf. 125, 121–127. doi:10.1016/j.ecoenv.2015.12.002
Duong, H. A., Phung, T. V., Nguyen, T. N., Phan Thi, L. A., and Pham, H. V. (2021). Occurrence, Distribution, and Ecological Risk Assessment of Antibiotics in Selected Urban Lakes of Hanoi, Vietnam. J. Anal. Methods Chem. 2021, 1–13. doi:10.1155/2021/6631797
European Centre for Disease Prevention and Control, (2019). Surveillance of Antimicrobial Resistance in Europe 2018. Stockholm, Sweden: ECDC.
Exner, M., Bhattacharya, S., Christiansen, B., Gebel, J., Goroncy-Bermes, P., Hartemann, P., et al. (2017). Antibiotic Resistance: What Is So Special about Multidrug-Resistant Gram-Negative Bacteria? GMS Hyg. Infect. Control. 12, Doc05. doi:10.3205/dgkh000290
Fekadu, S., Alemayehu, E., Dewil, R., and Van der Bruggen, B. (2019). Pharmaceuticals in Freshwater Aquatic Environments: A Comparison of the African and European challenge. Sci. Total Environ. 654, 324–337. doi:10.1016/j.scitotenv.2018.11.072
Fenske, G. J., Ghimire, S., Antony, L., Christopher-Hennings, J., and Scaria, J. (2020). Integration of Culture-dependent and Independent Methods Provides a More Coherent Picture of the Pig Gut Microbiome. FEMS Microbiol. Ecol. 96, 1–10. doi:10.1093/femsec/fiaa022
Fernando, D. M., Tun, H. M., Poole, J., Patidar, R., Li, R., Mi, R., et al. (2016). Detection of Antibiotic Resistance Genes in Source and Drinking Water Samples from a First Nations Community in Canada. Appl. Environ. Microbiol. 82, 4767–4775. doi:10.1128/AEM.00798-16
Ferreira Da Silva, M., Vaz-Moreira, I., Gonzalez-Pajuelo, M., Nunes, O. C., and Manaia, C. l. M. (2007). Antimicrobial Resistance Patterns in Enterobacteriaceae Isolated from an Urban Wastewater Treatment Plant. FEMS Microbiol. Ecol. 60, 166–176. doi:10.1111/j.1574-6941.2006.00268.x
Forbes, J. D., Knox, N. C., Ronholm, J., Pagotto, F., and Reimer, A. (2017). Metagenomics: The Next Culture-independent Game Changer. Front. Microbiol. 8, 1–21. doi:10.3389/fmicb.2017.01069
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O. A., and Dantas, G. (2012). The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 337, 1107–1111. doi:10.1126/science.1220761
Founou, R. C., Founou, L. L., and Essack, S. Y. (2017). Clinical and Economic Impact of Antibiotic Resistance in Developing Countries: a Systematic Review and Meta-Analysis. PLoS One 12, e0189621. doi:10.1371/journal.pone.0189621
Fouz, N., Pangesti, K. N. A., Yasir, M., Al-Malki, A. L., Azhar, E. I., Hill-Cawthorne, G. A., et al. (2020). The Contribution of Wastewater to the Transmission of Antimicrobial Resistance in the Environment: Implications of Mass Gathering Settings. TropicalMed 5, 33. doi:10.3390/tropicalmed5010033
Francino, M. P. (2016). Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Front. Microbiol. 6, 1–11. doi:10.3389/fmicb.2015.01543
Friedrich, M. J. (2017). Health Care Attacks Continue in 23 Countries Around the World. JAMA 318 (3), 231. doi:10.1001/jama.2017.8323
Galhano, B. S. P. B., Ferrari, R. G. R., Panzenhagen, P., de Jesus, A. C. S., and Conte-Junior, C. A. (2021). Antimicrobial Resistance Gene Detection Methods for Bacteria in Animal-Based Foods: A Brief Review of Highlights and Advantages. Microorganisms 9, 923. doi:10.3390/microorganisms9050923
Garner, E., Chen, C., Xia, K., Bowers, J., Engelthaler, D. M., McLain, J., et al. (2018). Metagenomic Characterization of Antibiotic Resistance Genes in Full-Scale Reclaimed Water Distribution Systems and Corresponding Potable Systems. Environ. Sci. Technol. 52, 6113–6125. doi:10.1021/acs.est.7b05419
George, A. (2019). Antimicrobial Resistance (AMR) in the Food Chain: Trade, One Health and Codex. TropicalMed 4, 54. doi:10.3390/tropicalmed4010054
Graham, D. W., Bergeron, G., Bourassa, M. W., Dickson, J., Gomes, F., Howe, A., et al. (2019). Complexities in Understanding Antimicrobial Resistance across Domesticated Animal, Human, and Environmental Systems. Ann. N.Y. Acad. Sci. 1441, 17–30. doi:10.1111/nyas.14036
Grenni, P. (2022). Antimicrobial Resistance in Rivers: A Review of the Genes Detected and New Challenges. Enviro Tox. Chem. 41, 687–714. doi:10.1002/etc.5289
Griffiths, R. P., Moyer, C. L., Caldwell, B. A., Ye, C., and Morita, R. Y. (1990). Long-Term Starvation-Induced Loss of Antibiotic Resistance in Bacteria. Microb. Ecol. 19, 251–257. doi:10.1007/bf02017169
Gullberg, E., Cao, S., Berg, O. G., Ilbäck, C., Sandegren, L., Hughes, D., et al. (2011). Selection of Resistant Bacteria at Very Low Antibiotic Concentrations. Plos Pathog. 7, e1002158–9. doi:10.1371/journal.ppat.1002158
Guo, X., Tang, N., Lei, H., Fang, Q., Liu, L., Zhou, Q., et al. (2021). Metagenomic Analysis of Antibiotic Resistance Genes in Untreated Wastewater from Three Different Hospitals. Front. Microbiol. 12. doi:10.3389/fmicb.2021.709051
Guzman-Otazo, J., Gonzales-Siles, L., Poma, V., Bengtsson-Palme, J., Thorell, K., Flach, C.-F., et al. (2019). Diarrheal Bacterial Pathogens and Multi-Resistant Enterobacteria in the Choqueyapu River in La Paz, Bolivia. PLoS One 14, e0210735. doi:10.1371/journal.pone.0210735
Hanna, N., Purohit, M., Diwan, V., Chandran, S. P., Riggi, E., Parashar, V., et al. (2020). Monitoring of Water Quality, Antibiotic Residues, and Antibiotic-Resistant escherichia Coli in the Kshipra River in india over a 3-year Period. Ijerph 17, 7706–7722. doi:10.3390/ijerph17217706
Harnisz, M. (2013). Total Resistance of Native Bacteria as an Indicator of Changes in the Water Environment. Environ. Pollut. 174, 85–92. doi:10.1016/j.envpol.2012.11.005
Hassan, B., Qadri, H., Ali, M. N., Khan, N. A., and Yatoo, A. M. (2020). Impact of Climate Change on Freshwater Ecosystem and its Sustainable Management. Fresh Water Pollut. Dyn. Remediat, 105–121. doi:10.1007/978-981-13-8277-210.1007/978-981-13-8277-2_7
Hatosy, S. M., and Martiny, A. C. (2015). The Ocean as a Global Reservoir of Antibiotic Resistance Genes. Appl. Environ. Microbiol. 81, 7593–7599. doi:10.1128/AEM.00736-15
Hendriksen, R. S., Munk, P., Munk, P., Njage, P., van Bunnik, B., McNally, L., et al. (2019). Global Monitoring of Antimicrobial Resistance Based on Metagenomics Analyses of Urban Sewage. Nat. Commun. 10. doi:10.1038/s41467-019-08853-3
Henriques, I. S., Fonseca, F., Alves, A., Saavedra, M. J., and Correia, A. (2006). Occurrence and Diversity of Integrons and β-lactamase Genes Among Ampicillin-Resistant Isolates from Estuarine Waters. Res. Microbiol. 157, 938–947. doi:10.1016/j.resmic.2006.09.003
Hernando-Amado, S., Coque, T. M., Baquero, F., and Martínez, J. L. (2019). Defining and Combating Antibiotic Resistance from One Health and Global Health Perspectives. Nat. Microbiol. 4, 1432–1442. doi:10.1038/s41564-019-0503-9
Heuer, O. E., Kruse, H., Grave, K., Collignon, P., Karunasagar, I., and Angulo, F. J. (2009). Human Health Consequences of Use of Antimicrobial Agents in Aquaculture. Clin. Infect. Dis. 49, 1248–1253. doi:10.1086/605667
Hiller, C. X., Hübner, U., Fajnorova, S., Schwartz, T., and Drewes, J. E. (2019). Antibiotic Microbial Resistance (AMR) Removal Efficiencies by Conventional and Advanced Wastewater Treatment Processes: A Review. Sci. Total Environ. 685, 596–608. doi:10.1016/j.scitotenv.2019.05.315
Hiller, C. X., Hübner, U., Fajnorova, S., Schwartz, T., and Drewes, J. E. (2019). Antibiotic Microbial Resistance (AMR) Removal Efficiencies by Conventional and Advanced Wastewater Treatment Processes: A Review. Sci. Total Environ. 685, 596–608. doi:10.1016/j.scitotenv.2019.05.315
Hinchliffe, S., Butcher, A., and Rahman, M. M. (2018). The AMR Problem: Demanding Economies, Biological Margins, and Co-producing Alternative Strategies. Palgrave Commun. 4, 1–12. doi:10.1057/s41599-018-0195-4
Hocquet, D., Muller, A., and Bertrand, X. (2016). What Happens in Hospitals Does Not Stay in Hospitals: Antibiotic-Resistant Bacteria in Hospital Wastewater Systems. J. Hosp. Infect. 93, 395–402. doi:10.1016/j.jhin.2016.01.010
Hong, P. Y., Julian, T., Pype, M.-L., Jiang, S., Nelson, K., Graham, D., et al. (2018). Reusing Treated Wastewater: Consideration of the Safety Aspects Associated with Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes. Water 10, 244. doi:10.3390/w10030244
Hossain, M. A. R., Ahmed, M., Ojea, E., and Fernandes, J. A. (2018). Impacts and Responses to Environmental Change in Coastal Livelihoods of South-West Bangladesh. Sci. Total Environ. 637-638, 954–970. doi:10.1016/j.scitotenv.2018.04.328
Hu, Y., Yang, X., Li, J., Lv, N., Liu, F., Wu, J., et al. (2016). The Bacterial Mobile Resistome Transfer Network Connecting the Animal and Human Microbiomes. Appl. Environ. Microbiol. 82, 6672–6681. doi:10.1128/AEM.01802-16
Hu, Y., Zhang, T., Jiang, L., Luo, Y., Yao, S., Zhang, D., et al. (2019). Occurrence and Reduction of Antibiotic Resistance Genes in Conventional and Advanced Drinking Water Treatment Processes. Sci. Total Environ. 669, 777–784. doi:10.1016/j.scitotenv.2019.03.143
Huang, J. J., Hu, H. Y., Lu, S. Q., Li, Y., Tang, F., Lu, Y., et al. (2012). Monitoring and Evaluation of Antibiotic-Resistant Bacteria at a Municipal Wastewater Treatment Plant in China. Environ. Int. 42, 31–36. doi:10.1016/j.envint.2011.03.001
Ibrahim, R. A., Cryer, T. L., Lafi, S. Q., Basha, E. A., Good, L., and Tarazi, Y. H. (2019). Identification of Escherichia coli from Broiler Chickens in Jordan, Their Antimicrobial Resistance, Gene Characterization and the Associated Risk Factors. BMC Vet. Res. 15, 159–216. doi:10.1186/s12917-019-1901-1
Irfan, S., and Alatawi, A. M. M. (2019). Aquatic Ecosystem and Biodiversity: A Review. Oje 09, 1–13. doi:10.4236/oje.2019.91001
Ishikawa, N. K., Touno, E., Higashiyama, Y., Sasamoto, M., Soma, M., Yoshida, N., et al. (2018). Determination of Tylosin Excretion from Sheep to Assess Tylosin Spread to Agricultural fields by Manure Application. Sci. Total Environ. 633, 399–404. doi:10.1016/j.scitotenv.2018.03.216
Jabbar Ibrahim, I. A., and Kareem Hameed, T. A. (2015). Isolation, Characterization and Antimicrobial Resistance Patterns of Lactose-Fermenter Enterobacteriaceae Isolates from Clinical and Environmental Samples. Ojmm 05, 169–176. doi:10.4236/ojmm.2015.54021
Jara, D., Bello-Toledo, H., Domínguez, M., Cigarroa, C., Fernández, P., Vergara, L., et al. (2020). Antibiotic Resistance in Bacterial Isolates from Freshwater Samples in Fildes Peninsula, King George Island, Antarctica. Sci. Rep. 10, 3145. doi:10.1038/s41598-020-60035-0
Jiang, X., Ellabaan, M. M. H., Charusanti, P., Munck, C., Blin, K., Tong, Y., et al. (2017). Dissemination of Antibiotic Resistance Genes from Antibiotic Producers to Pathogens. Nat. Commun. 8, 15784–15787. doi:10.1038/ncomms15784
Karkman, A., Do, T. T., Walsh, F., and Virta, M. P. J. (2018). Antibiotic-Resistance Genes in Waste Water. Trends Microbiol. 26, 220–228. doi:10.1016/j.tim.2017.09.005
Kaur, R., Kaur, A., Kaur, R., Singh, S., Bhatti, M. S., Umar, A., et al. (2021). Cu-BTC Metal Organic Framework (MOF) Derived Cu-Doped TiO2 Nanoparticles and Their Use as Visible Light Active Photocatalyst for the Decomposition of Ofloxacin (OFX) Antibiotic and Antibacterial Activity. Adv. Powder Technol. 32, 1350–1361. doi:10.1016/j.apt.2021.02.037
Klanicova, B., Seda, J., Slana, I., Slany, M., and Pavlik, I. (2013). The Tracing of Mycobacteria in Drinking Water Supply Systems by Culture, Conventional, and Real Time PCR. Curr. Microbiol. 67, 725–731. doi:10.1007/s00284-013-0427-1
Koch, N., Islam, N. F., Sonowal, S., Prasad, R., and Sarma, H. (2021). Environmental Antibiotics and Resistance Genes as Emerging Contaminants: Methods of Detection and Bioremediation. Curr. Res. Microb. Sci. 2, 100027. doi:10.1016/j.crmicr.2021.100027
Kohanski, M. A., DePristo, M. A., and Collins, J. J. (2010). Sublethal Antibiotic Treatment Leads to Multidrug Resistance via Radical-Induced Mutagenesis. Mol. Cel 37, 311–320. doi:10.1016/j.molcel.2010.01.003
Kolář, M., Urbánek, K., and Látal, T. (2001). Antibiotic Selective Pressure and Development of Bacterial Resistance. Int. J. Antimicrob. Agents 17, 357–363.
Korzeniewska, E., and Harnisz, M. (2012). Culture-dependent and Culture-independent Methods in Evaluation of Emission of Enterobacteriaceae from Sewage to the Air and Surface Water. Water Air Soil Pollut. 223, 4039–4046. doi:10.1007/s11270-012-1171-z
Korzeniewska, E., Korzeniewska, A., and Harnisz, M. (2013). Antibiotic Resistant Escherichia coli in Hospital and Municipal Sewage and Their Emission to the Environment. Ecotoxicology Environ. Saf. 91, 96–102. doi:10.1016/j.ecoenv.2013.01.014
Kraemer, S. A., Ramachandran, A., and Perron, G. G. (2019). Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 7, 180. doi:10.3390/microorganisms7060180
Kumar, A., and Pal, D. (2018). Antibiotic Resistance and Wastewater: Correlation, Impact and Critical Human Health Challenges. J. Environ. Chem. Eng. 6, 52–58. doi:10.1016/j.jece.2017.11.059
Kumar, S., Tripathi, V. R., and Garg, S. K. (2013). Antibiotic Resistance and Genetic Diversity in Water-Borne Enterobacteriaceae Isolates from Recreational and Drinking Water Sources. Int. J. Environ. Sci. Technol. 10, 789–798. doi:10.1007/s13762-012-0126-7
Kunhikannan, S., Thomas, C. J., Franks, A. E., Mahadevaiah, S., Kumar, S., and Petrovski, S. (2021). Environmental Hotspots for Antibiotic Resistance Genes. Microbiologyopen 10, 1–11. doi:10.1002/mbo3.1197
Lai, F. Y., Muziasari, W., Virta, M., Wiberg, K., and Ahrens, L. (2021). Profiles of Environmental Antibiotic Resistomes in the Urban Aquatic Recipients of Sweden Using High-Throughput Quantitative PCR Analysis. Environ. Pollut. 287, 117651. doi:10.1016/j.envpol.2021.117651
Larsson, D. G. J., and Flach, C. F. (2021). Antibiotic Resistance in the Environment. Nat. Rev. Microbiol., 0123456789. doi:10.1038/s41579-021-00649-x
Lee, J., Jeon, J. H., Shin, J., Jang, H. M., Kim, S., Song, M. S., et al. (2017). Quantitative and Qualitative Changes in Antibiotic Resistance Genes after Passing through Treatment Processes in Municipal Wastewater Treatment Plants. Sci. Total Environ. 605-606, 906–914. doi:10.1016/j.scitotenv.2017.06.250
Léger, A., Lambraki, I., Graells, T., Cousins, M., Henriksson, P. J. G., Harbarth, S., et al. (2021). AMR-intervene: a Social-Ecological Framework to Capture the Diversity of Actions to Tackle Antimicrobial Resistance from a One Health Perspective. J. Antimicrob. Chemother. 76, 1–21. doi:10.1093/jac/dkaa394
Leonard, A. F. C., Zhang, L., Balfour, A. J., Garside, R., and Gaze, W. H. (2015). Human Recreational Exposure to Antibiotic Resistant Bacteria in Coastal Bathing Waters. Environ. Int. 82, 92–100. doi:10.1016/j.envint.2015.02.013
Leonard, A. F. C., Zhang, L., Balfour, A. J., Garside, R., Hawkey, P. M., Murray, A. K., et al. (2018). Exposure to and Colonisation by Antibiotic-Resistant E. coli in UK Coastal Water Users: Environmental Surveillance, Exposure Assessment, and Epidemiological Study (Beach Bum Survey). Environ. Int. 114, 326–333. doi:10.1016/j.envint.2017.11.003
Lerminiaux, N. A., and Cameron, A. D. S. (2019). Horizontal Transfer of Antibiotic Resistance Genes in Clinical Environments. Can. J. Microbiol. 65, 34–44. doi:10.1139/cjm-2018-0275
Li, J., Cheng, W., Xu, L., Strong, P. J., and Chen, H. (2015). Antibiotic-resistant Genes and Antibiotic-Resistant Bacteria in the Effluent of Urban Residential Areas, Hospitals, and a Municipal Wastewater Treatment Plant System. Environ. Sci. Pollut. Res. 22, 4587–4596. doi:10.1007/s11356-014-3665-2
Liang, X., Chen, B., Nie, X., Shi, Z., Huang, X., and Li, X. (2013). The Distribution and Partitioning of Common Antibiotics in Water and Sediment of the Pearl River Estuary, South China. Chemosphere 92, 1410–1416. doi:10.1016/j.chemosphere.2013.03.044
Liyanage, G. Y., Illango, A., and Manage, P. M. (2021). Prevalence and Quantitative Analysis of Antibiotic Resistance Genes (ARGs) in Surface and Groundwater in Meandering Part of the Kelani River Basin in Sri Lanka. Water Air Soil Pollut. 232, 1–11. doi:10.1007/s11270-021-05300-2
Llor, C., and Bjerrum, L. (2014). Antimicrobial Resistance: Risk Associated with Antibiotic Overuse and Initiatives to Reduce the Problem. Ther. Adv. Drug. Saf. 5 (6), 229–241. doi:10.1177/2042098614554919
Lobova, T. I., Feil, E. J., and Popova, L. Y. (2011). Multiple Antibiotic Resistance of Heterotrophic Bacteria Isolated from Siberian Lakes Subjected to Differing Degrees of Anthropogenic Impact. Microb. Drug Resist. 17, 583–591. doi:10.1089/mdr.2011.0044
Ma, Y., Li, M., Wu, M., Li, Z., and Liu, X. (2015). Occurrences and Regional Distributions of 20 Antibiotics in Water Bodies during Groundwater Recharge. Sci. Total Environ. 518-519, 498–506. doi:10.1016/j.scitotenv.2015.02.100
Madikizela, L. M., Tavengwa, N. T., and Chimuka, L. (2017). Status of Pharmaceuticals in African Water Bodies: Occurrence, Removal and Analytical Methods. J. Environ. Manage. 193, 211–220. doi:10.1016/j.jenvman.2017.02.022
Malakootian, M., Yaseri, M., and Faraji, M. (2019). Removal of Antibiotics from Aqueous Solutions by Nanoparticles: a Systematic Review and Meta-Analysis. Environ. Sci. Pollut. Res. 26, 8444–8458. doi:10.1007/s11356-019-04227-w
Manoharan, R. K., Srinivasan, S., Shanmugam, G., and Ahn, Y.-H. (2021). Shotgun Metagenomic Analysis Reveals the Prevalence of Antibiotic Resistance Genes and mobile Genetic Elements in Full Scale Hospital Wastewater Treatment Plants. J. Environ. Manage. 296, 113270. doi:10.1016/j.jenvman.2021.113270
Mao, D., Yu, S., Rysz, M., Luo, Y., Yang, F., Li, F., et al. (2015). Prevalence and Proliferation of Antibiotic Resistance Genes in Two Municipal Wastewater Treatment Plants. Water Res. 85, 458–466. doi:10.1016/j.watres.2015.09.010
Markkanen, M. A., Haukka, K., Parnanem, K. M. M., Dougnon, T. V., Bonkoungou, j. o. I., Garba, Z., et al. (2021). Metagenomic Analysis of Antimicrobial Resistance Genes in Wastewaters in Benin and Burkina Faso Indicates a Serious Health Risk from Untreated Hospital Wastewaters in Low-Income Countries. medRxiv 1–24.
Martinez, J. L. (2009). Environmental Pollution by Antibiotics and by Antibiotic Resistance Determinants. Environ. Pollut. 157, 2893–2902. doi:10.1016/j.envpol.2009.05.051
Matongo, S., Birungi, G., Moodley, B., and Ndungu, P. (2015). Pharmaceutical Residues in Water and Sediment of Msunduzi River, KwaZulu-Natal, South Africa. Chemosphere 134, 133–140. doi:10.1016/j.chemosphere.2015.03.093
Mazimba, A., Rupasinghe, N., Gomez, A. C. C., Zhao, F., and Pate, M. (2021). Landscape Analysis of Tools to Address Antimicrobial Resistance. Washington, D. C.: World Bank. Available: www.worldbank.org Some.
McCullough, A. R., Parekh, S., Rathbone, J., Del Mar, C. B., and Hoffmann, T. C. (2016). A Systematic Review of the Public's Knowledge and Beliefs about Antibiotic Resistance. J. Antimicrob. Chemother. 71, 27–33. doi:10.1093/jac/dkv310
McGowan, E. (2007). Comment on "Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado". Environ. Sci. Technol. 41, 2651–2652. doi:10.1021/es0680156
McInnes, R. S., uz-Zaman, M. H., Alam, I. T., Ho, S. F. S., Moran, R. A., Clemens, J. D., et al. (2021). Metagenome-Wide Analysis of Rural and Urban Surface Waters and Sediments in Bangladesh Identifies Human Waste as a Driver of Antibiotic Resistance. mSystems 6. doi:10.1128/msystems.00137-21
McKinney, C. W., Dungan, R. S., Moore, A., and Leytem, A. B. (2018). Occurrence and Abundance of Antibiotic Resistance Genes in Agricultural Soil Receiving Dairy Manure. FEMS Microbiol. Ecol. 94, fiy010. doi:10.1093/femsec/fiy010
Meng, L., Liu, H., Lan, T., Dong, L., Hu, H., Zhao, S., et al. (2020). Antibiotic Resistance Patterns of Pseudomonas spp. Isolated from Raw Milk Revealed by Whole Genome Sequencing. Front. Microbiol. 11. doi:10.3389/fmicb.2020.01005
Michael, C. A., Dominey-Howes, D., and Labbate, M. (2014). The Antimicrobial Resistance Crisis: Causes, Consequences, and Management. Front. Public Health 2, 145. doi:10.3389/fpubh.2014.00145
Mindlin, S. Z., Soina, V. S., Petrova, M. A., and Gorlenko, Z. M. (2008). Isolation of Antibiotic Resistance Bacterial Strains from Eastern Siberia Permafrost Sediments. Russ. J. Genet. 44, 27–34. doi:10.1134/S1022795408010043
Miranda, C. D., Godoy, F. A., and Lee, M. R. (2018). Current Status of the Use of Antibiotics and the Antimicrobial Resistance in the Chilean salmon Farms. Front. Microbiol. 9, 1–14. doi:10.3389/fmicb.2018.01284
Morris, S., and Cerceo, E. (2020). Trends, Epidemiology, and Management of Multi-Drug Resistant Gram-Negative Bacterial Infections in the Hospitalized Setting. Antibiotics 9, 196–220. doi:10.3390/antibiotics9040196
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: A Database and Search Engine for Integrons, Integrases and Gene Cassettes. Bioinformatics 25, 1096–1098. doi:10.1093/bioinformatics/btp105
Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., and Pardesi, K. R. (2019). Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 10, 539. doi:10.3389/fmicb.2019.00539
Munir, M., Wong, K., and Xagoraraki, I. (2011). Release of Antibiotic Resistant Bacteria and Genes in the Effluent and Biosolids of Five Wastewater Utilities in Michigan. Water Res. 45, 681–693. doi:10.1016/j.watres.2010.08.033
Nguyen, A. Q., Vu, H. P., Nguyen, L. N., Wang, Q., Djordjevic, S. P., Donner, E., et al. (2021). Monitoring Antibiotic Resistance Genes in Wastewater Treatment: Current Strategies and Future Challenges. Sci. Total Environ. 783, 146964. doi:10.1016/j.scitotenv.2021.146964
O'Flaherty, E., Borrego, C. M., Balcázar, J. L., and Cummins, E. (2018). Human Exposure Assessment to Antibiotic-Resistant Escherichia coli through Drinking Water. Sci. Total Environ. 616-617, 1356–1364. doi:10.1016/j.scitotenv.2017.10.180
O'Flaherty, E., and Cummins, E. (2017). Antibiotic Resistance in Surface Water Ecosystems: Presence in the Aquatic Environment, Prevention Strategies, and Risk Assessment. Hum. Ecol. Risk Assess. Int. J. 23, 299–322. doi:10.1080/10807039.2016.1247254
Obayiuwana, A., and Ibekwe, A. M. (2020). Antibiotic Resistance Genes Occurrence in Wastewaters from Selected Pharmaceutical Facilities in Nigeria. Water 12, 1897. doi:10.3390/w12071897
Obayiuwana, A., Ogunjobi, A., and Ibekwe, A. (2021). Prevalence of Antibiotic Resistance Genes in Pharmaceutical Wastewaters. Water 13, 1731–1811. doi:10.3390/w13131731
Oh, E.-G., Son, K. T., Yu, H., Lee, T. S., Lee, H. J., Shin, S., et al. (2011). Antimicrobial Resistance of Vibrio parahaemolyticus and Vibrio alginolyticus Strains Isolated from Farmed Fish in Korea from 2005 through 2007. J. Food Prot. 74, 380–386. doi:10.4315/0362-028x.jfp-10-307
Olivas, Y., and Faulkner, B. R. (2008). Fecal Source Tracking by Antibiotic Resistance Analysis on a Watershed Exhibiting Low Resistance. Environ. Monit. Assess. 139, 15–25. doi:10.1007/s10661-007-9805-0
Osińska, A., Harnisz, M., and Korzeniewska, E. (2016). Prevalence of Plasmid-Mediated Multidrug Resistance Determinants in Fluoroquinolone-Resistant Bacteria Isolated from Sewage and Surface Water. Environ. Sci. Pollut. Res. 23, 10818–10831. doi:10.1007/s11356-016-6221-4
Pepper, I. L., Brooks, J. P., and Gerba, C. P. (2018). Antibiotic Resistant Bacteria in Municipal Wastes: Is There Reason for Concern? Environ. Sci. Technol. 52, 3949–3959. doi:10.1021/acs.est.7b04360
Praveenkumarreddy, Y., Akiba, M., Guruge, K. S., Balakrishna, K., Vandana, K. E., and Kumar, V. (2020). Occurrence of Antimicrobial-Resistant Escherichia coli in Sewage Treatment Plants of South India. J. Water Sanit Hyg. Dev. 10, 48–55. doi:10.2166/washdev.2020.051
Preena, P. G., Arathi, D., Raj, N. S., Arun Kumar, T. V., Arun Raja, S., Reshma, R. N., et al. (2020). Diversity of Antimicrobial‐resistant Pathogens from a Freshwater Ornamental Fish Farm. Lett. Appl. Microbiol. 71, 108–116. doi:10.1111/lam.13231
Prestinaci, F., Pezzotti, P., and Pantosti, A. (2015). Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 109, 309–318. doi:10.1179/2047773215Y.0000000030
Ramírez-Castillo, F. Y., Moreno-Flores, A. C., Avelar-González, F. J., Márquez-Díaz, F., Harel, J., and Guerrero-Barrera, A. L. (2018). An Evaluation of Multidrug-Resistant Escherichia coli Isolates in Urinary Tract Infections from Aguascalientes, Mexico: Cross-Sectional Study. Ann. Clin. Microbiol. Antimicrob. 17, 1–13. doi:10.1186/s12941-018-0286-5
Razavi, M., Marathe, N. P., Gillings, M. R., Flach, C.-F., Kristiansson, E., and Joakim Larsson, D. G. (2017). Discovery of the Fourth mobile Sulfonamide Resistance Gene. Microbiome 5, 160. doi:10.1186/s40168-017-0379-y
Rizzo, L., Fiorentino, A., and Anselmo, A. (2013). Advanced Treatment of Urban Wastewater by UV Radiation: Effect on Antibiotics and Antibiotic-Resistant E. coli Strains. Chemosphere 92, 171–176. doi:10.1016/j.chemosphere.2013.03.021
Rodriguez-Mozaz, S., Chamorro, S., Marti, E., Huerta, B., Gros, M., Sànchez-Melsió, A., et al. (2015). Occurrence of Antibiotics and Antibiotic Resistance Genes in Hospital and Urban Wastewaters and Their Impact on the Receiving River. Water Res. 69, 234–242. doi:10.1016/j.watres.2014.11.021
Rosenfeld, W. D., and Zobell, C. E. (1947). Antibiotic Production by Marine Microorganisms. J. Bacteriol. 54, 393–398. doi:10.1128/jb.54.3.393-398.1947
Salvatore, P. P., Kendall, E. A., Seabrook, D., Brown, J., Durham, G. H., and Dowdy, D. W. (2019). Projecting the Impact of Variable MDR-TB Transmission Efficiency on Long-Term Epidemic Trends in South Africa and Vietnam. Sci. Rep. 9, 18099. doi:10.1038/s41598-019-54561-9
Salyers, A., Gupta, A., and Wang, Y. (2004). Human Intestinal Bacteria as Reservoirs for Antibiotic Resistance Genes. Trends Microbiol. 12, 412–416. doi:10.1016/j.tim.2004.07.004
Samreen, Ad. I., Ahmad, I., Malak, H. A., and Abulreesh, H. H. (2021). Environmental Antimicrobial Resistance and its Drivers: a Potential Threat to Public Health. J. Glob. Antimicrob. Resist. 27, 101–111. doi:10.1016/j.jgar.2021.08.001
Sano, D., Amarasiri, M., Hata, A., Watanabe, T., and Katayama, H. (2016). Risk Management of Viral Infectious Diseases in Wastewater Reclamation and Reuse: Review. Environ. Int. 91, 220–229. doi:10.1016/j.envint.2016.03.001
Schar, D., Klein, E. Y., Laxminarayan, R., Gilbert, M., and Van Boeckel, T. P. (2020). Global Trends in Antimicrobial Use in Aquaculture. Sci. Rep. 10, 1. doi:10.1038/s41598-020-78849-3
Schar, D., Zhao, C., Wang, Y., Larsson, D. G. J., Gilbert, M., and Van Boeckel, T. P. (2021). Twenty-year Trends in Antimicrobial Resistance from Aquaculture and Fisheries in Asia. Nat. Commun. 12, 6. doi:10.1038/s41467-021-25655-8
Segura, P. A., Takada, H., Correa, J. A., El Saadi, K., Koike, T., Onwona-Agyeman, S., et al. (2015). Global Occurrence of Anti-infectives in Contaminated Surface Waters: Impact of Income Inequality between Countries. Environ. Int. 80, 89–97. doi:10.1016/j.envint.2015.04.001
Serwecinska, L. (2020). Antimicrobials and Antibiotic-Resistant Bacteria. Water 12, 1–17. doi:10.3390/w12123313
Shimizu, A., Takada, H., Koike, T., Takeshita, A., Saha, M., Rinawati, , et al. (2013). Ubiquitous Occurrence of Sulfonamides in Tropical Asian Waters. Sci. Total Environ. 452-453, 108–115. doi:10.1016/j.scitotenv.2013.02.027
Shrestha, P., Cooper, B. S., Coast, J., Oppong, R., Do Thi Thuy, N., Phodha, T., et al. (2018). Enumerating the Economic Cost of Antimicrobial Resistance Per Antibiotic Consumed to Inform the Evaluation of Interventions Affecting Their Use. Antimicrob. Resist. Infect. Control. 7, 98–99. doi:10.1186/s13756-018-0384-3
Singh, A. K., Das, S., Kumar, S., Gajamer, V. R., Najar, I. N., Lepcha, Y. D., et al. (2020). Distribution of Antibiotic-Resistant Enterobacteriaceae Pathogens in Potable Spring Water of Eastern Indian Himalayas: Emphasis on Virulence Gene and Antibiotic Resistance Genes in Escherichia coli. Front. Microbiol. 11, 1–17. doi:10.3389/fmicb.2020.581072
Singh, S. (2020). Conventional Infection Prevention and Control Practices in Post-Antibiotic Era: A Perspective. J. Sci. Res. 64, 167–174. doi:10.37398/jsr.2020.640124
Singh, S., Kishore, D., and Singh, R. K. (2022). Potential for Further Mismanagement of Fever During COVID-19 Pandemic: Possible Causes and Impacts. Front. Med. (Lausanne) 9, 751929. doi:10.3389/fmed.2022.751929
Søraas, A., Sundsfjord, A., Sandven, I., Brunborg, C., and Jenum, P. A. (2013). Risk Factors for Community-Acquired Urinary Tract Infections Caused by ESBL-Producing Enterobacteriaceae -A Case-Control Study in a Low Prevalence Country. PLoS One 8, e69581–7. doi:10.1371/journal.pone.0069581
Sriram, A., Kalanxhi, E., Kapoor, G., Craig, J., Balasubramanian, R., Brar, S., et al. (2021). The State of the World’s Antibiotics in 2021: A Global Analysis of Antimicrobial Resistance and its Drivers. Washington, DC: Center for Disease Dynamics, Economics & Policy. Available at: https://cddep.org/blog/posts/the-state-of-the-worlds-antibiotics-report-in-2021/ and https://cddep.org/wp-content/uploads/2021/02/The-State-of-the-Worlds-Antibiotics-in-2021.pdf (Accessed November 20, 2021).
Stanton, I. C., Murray, A. K., Zhang, L., Snape, J., and Gaze, W. H. (2020). Evolution of Antibiotic Resistance at Low Antibiotic Concentrations Including Selection below the Minimal Selective Concentration. Commun. Biol. 3, 467. doi:10.1038/s42003-020-01176-w
Stecher, B., Denzler, R., Maier, L., Bernet, F., Sanders, M. J., Pickard, D. J., et al. (2012). Gut Inflammation Can Boost Horizontal Gene Transfer between Pathogenic and Commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. U.S.A. 109, 1269–1274. doi:10.1073/pnas.1113246109
Strange, J. E. S., Leekitcharoenphon, P., Møller, F. D., and Aarestrup, F. M. (2021). Metagenomics Analysis of Bacteriophages and Antimicrobial Resistance from Global Urban Sewage. Sci. Rep. 11, 1–11. doi:10.1038/s41598-021-80990-6
Su, H.-C., Liu, Y.-S., Pan, C.-G., Chen, J., He, L.-Y., and Ying, G.-G. (2018). Persistence of Antibiotic Resistance Genes and Bacterial Community Changes in Drinking Water Treatment System: From Drinking Water Source to Tap Water. Sci. Total Environ. 616-617, 453–461. doi:10.1016/j.scitotenv.2017.10.318
Subbiah, M., Caudell, M. A., Mair, C., Davis, M. A., Matthews, L., Quinlan, R. J., et al. (2020). Antimicrobial Resistant Enteric Bacteria Are Widely Distributed Amongst People, Animals and the Environment in Tanzania. Nat. Commun. 11, 13995. doi:10.1038/s41467-019-13995-5
Sun, D., Jeannot, K., Xiao, Y., and Knapp, C. W. (2019). Editorial: Horizontal Gene Transfer Mediated Bacterial Antibiotic Resistance. Front. Microbiol. 10, 1933. doi:10.3389/fmicb.2019.01933
Suzuki, S., Pruden, A., Virta, M., and Zhang, T. (2017). Editorial: Antibiotic Resistance in Aquatic Systems. Front. Microbiol. 8, 14–23. doi:10.1264/jsme2.ME1512210.3389/fmicb.2017.00014
Tan, Q., Li, W., Zhang, J., Zhou, W., Chen, J., Li, Y., et al. (2019). Presence, Dissemination and Removal of Antibiotic Resistant Bacteria and Antibiotic Resistance Genes in Urban Drinking Water System: A Review. Front. Environ. Sci. Eng. 13. doi:10.1007/s11783-019-1120-9
Taneja, N., and Sharma, M. (2019). Antimicrobial Resistance in the Environment: The Indian Scenario. Indian J. Med. Res. 149, 119–128. doi:10.4103/ijmr.IJMR_331_18
Tesfaye, H., Alemayehu, H., Desta, A. F., and Eguale, T. (2019). Antimicrobial Susceptibility Profile of Selected Enterobacteriaceae in Wastewater Samples from Health Facilities, Abattoir, Downstream Rivers and a WWTP in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control. 8, 134. doi:10.1186/s13756-019-0588-1
The World Bank, (2021). Antimicrobial Resistance (AMR). The World Bank [Internet]. 2021. Available: https://www.worldbank.org/en/topic/health/brief/antimicrobial-resistance-amr (Accessed November 12, 2021).
Tortorella, E., Tedesco, P., Palma Esposito, F., January, G., Fani, R., Jaspars, M., et al. (2018). Antibiotics from Deep-Sea Microorganisms: Current Discoveries and Perspectives. Mar. Drugs 16, 355–416. doi:10.3390/md16100355
Tran, N. H., Hoang, L., Nghiem, L. D., Nguyen, N. M. H., Ngo, H. H., Guo, W., et al. (2019). Occurrence and Risk Assessment of Multiple Classes of Antibiotics in Urban Canals and Lakes in Hanoi, Vietnam. Sci. Total Environ. 692, 157–174. doi:10.1016/j.scitotenv.2019.07.092
Treangen, T. J., and Rocha, E. P. C. (2011). Horizontal Transfer, Not Duplication, Drives the Expansion of Protein Families in Prokaryotes. Plos Genet. 7, e1001284. doi:10.1371/journal.pgen.1001284
United Nations Environment Programme (2022). Antibiotic Resistance: A Global Threat. Stockholm: United Nations. Available at: https://www.unep.org/explore-topics/chemicals-waste/what-we-do/emerging-issues/antimicrobial-resistance-global-threat#:∼:text=The%20key%20reasons%20contributing%20to,human%20and%20veterinary%20healthcare%20settings (Accessed April 1, 2022).
Van Hoek, A. H. A. M., Mevius, D., Guerra, B., Mullany, P., Roberts, A. P., and Aarts, H. J. M. (2011). Acquired Antibiotic Resistance Genes: An Overview. Front. Microbio. 2, 1–27. doi:10.3389/fmicb.2011.00203
Ventola, C. L. (2015). The Antibiotic Resistance Crisis: Part 1: Causes and Threats. P T 40, 277–283.
Vilca, F. Z., and Angeles, W. G. (2018). Occurrence of Antibiotics Residues in the Marine Environment. Examines Mar. Biol. Ocean 2, 12–14. doi:10.31031/EIMBO.2018.02.000536
Viney, K., Linh, N. N., Gegia, M., Zignol, M., Glaziou, P., Ismail, N., et al. (2021). New Definitions of Pre-extensively and Extensively Drug-Resistant Tuberculosis: Update from the World Health Organization. Eur. Respir. J. 57, 2100361. doi:10.1183/13993003.00361-2021
von Wintersdorff, C. J. H., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 7, 173. doi:10.3389/fmicb.2016.00173
Wang, J., Chu, L., Wojnárovits, L., and Takács, E. (2020). Occurrence and Fate of Antibiotics, Antibiotic Resistant Genes (ARGs) and Antibiotic Resistant Bacteria (ARB) in Municipal Wastewater Treatment Plant: An Overview. Sci. Total Environ. 744, 140997. doi:10.1016/j.scitotenv.2020.140997
Wellington, E. M., Boxall, A. B., Cross, P., Feil, E. J., Gaze, W. H., Hawkey, P. M., et al. (2013). The Role of the Natural Environment in the Emergence of Antibiotic Resistance in Gram-Negative Bacteria. Lancet Infect. Dis. 13, 155–165. doi:10.1016/S1473-3099(12)70317-1
West, N. J., Obernosterer, I., Zemb, O., and Lebaron, P. (2008). Major Differences of Bacterial Diversity and Activity inside and outside of a Natural Iron-Fertilized Phytoplankton Bloom in the Southern Ocean. Environ. Microbiol. 10, 738–756. doi:10.1111/j.1462-2920.2007.01497.x
Woolhouse, M., Ward, M., Van Bunnik, B., and Farrar, J. (2015). Antimicrobial Resistance in Humans, Livestock and the Wider Environment. Phil. Trans. R. Soc. B 370, 20140083. doi:10.1098/rstb.2014.0083
World Bank (2017). Drug-Resistant Infections: A Threat to Our Economic Future. Washington, DC: World Bank Available at: https://documents1.worldbank.org/curated/en/323311493396993758/pdf/final-report.pdf (Accessed November 18, 2021).
World Bank Group (2018). One Health - Operational Framework for Strengthening Human, Animal, and Environment Public Health System at Their Interface. Washington, D. C.: World Bank Publications. Available: http://documents.worldbank.org/curated/en/703711517234402168/pdf/123023-REVISED-PUBLIC-World-Bank-One-Health-Framework-2018.pdf.
World Economic Forum (2021). Antimicrobial Resistance and Water: The Risks and Costs for Economies and Societies. Available at: http://www3.weforum.org/docs/WEF_Antimicrobial_Resistance_and_Water_2021.pdf (Accessed February 10, 2022).
World Health Organization (2014). Antimicrobial Resistance Global Report on Surveillance. Geneva: World Health Organization. Available at: https://apps.who.int/iris/handle/10665/112642 (Accessed November 12, 2021).
World Health Organization (2015). Global Action Plan on Antimicrobial Resistance. Geneva: World Health Organization. Available at: https://www.who.int/publications/i/item/9789241509763.
World Health Organization (2017a). National Action Plan on Antimicrobial Resistance (NAP-AMR) 2017-2021. Available at: https://www.who.int/publications/m/item/india-national-action-plan-on-antimicrobial-resistance-(nap-amr)-2017-2021 (Accessed November 15, 2021).
World Health Organization (2017b). WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. World Health Organization [Internet]Available: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (Accessed November 15, 2021).
World Health Organization (2018). Critically Important Antimicrobials for Human Medicine. 6th revision. Geneva: World Health Organization. Available at: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf (Accessed November 15, 2021).
World Health Organization (2019). New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. Geneva: World Health Organization. Available at: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (Accessed November 15, 2021).
World Health Organization (2021). Antimicrobial Resistance. World Health Organization [Internet]. 2021. Available at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accessed November 15, 2021).
Xu, J., Xu, Y., Wang, H., Guo, C., Qiu, H., He, Y., et al. (2015). Occurrence of Antibiotics and Antibiotic Resistance Genes in a Sewage Treatment Plant and its Effluent-Receiving River. Chemosphere 119, 1379–1385. doi:10.1016/j.chemosphere.2014.02.040
Xu, J., Zhang, Y., Zhou, C., Guo, C., Wang, D., Du, P., et al. (2014). Distribution, Sources and Composition of Antibiotics in Sediment, Overlying Water and Pore Water from Taihu Lake, China. Sci. Total Environ. 497-498, 267–273. doi:10.1016/j.scitotenv.2014.07.114
Xu, L., Ouyang, W., Qian, Y., Su, C., Su, J., and Chen, H. (2016). High-throughput Profiling of Antibiotic Resistance Genes in Drinking Water Treatment Plants and Distribution Systems. Environ. Pollut. 213, 119–126. doi:10.1016/j.envpol.2016.02.013
Yang, J., Wang, C., Shu, C., Liu, L., Geng, J., Hu, S., et al. (2013). Marine Sediment Bacteria Harbor Antibiotic Resistance Genes Highly Similar to Those Found in Human Pathogens. Microb. Ecol. 65, 975–981. doi:10.1007/s00248-013-0187-2
Yang, Y., Song, W., Lin, H., Wang, W., Du, L., and Xing, W. (2018). Antibiotics and Antibiotic Resistance Genes in Global Lakes: A Review and Meta-Analysis. Environ. Int. 116, 60–73. doi:10.1016/j.envint.2018.04.011
Yoo, K., and Lee, G. (2021). Investigation of the Prevalence of Antibiotic Resistance Genes According to the Wastewater Treatment Scale Using Metagenomic Analysis. Antibiotics 10, 188–213. doi:10.3390/antibiotics10020188
Zeng, J., Pan, Y., Yang, J., Hou, M., Zeng, Z., and Xiong, W. (2019). Metagenomic Insights into the Distribution of Antibiotic Resistome between the Gut-Associated Environments and the Pristine Environments. Environ. Int. 126, 346–354. doi:10.1016/j.envint.2019.02.052
Zhang, D., Peng, Y., Chan, C. L., On, H., Wai, H. K. F., Shekhawat, S. S., et al. (2021). Metagenomic Survey Reveals More Diverse and Abundant Antibiotic Resistance Genes in Municipal Wastewater Than Hospital Wastewater. Front. Microbiol. 12. doi:10.3389/fmicb.2021.712843
Zhang, M., Chen, S., Yu, X., Vikesland, P., and Pruden, A. (2019). Degradation of Extracellular Genomic, Plasmid DNA and Specific Antibiotic Resistance Genes by Chlorination. Front. Environ. Sci. Eng. 13, 1124. doi:10.1007/s11783-019-1124-5
Zhang, R., Tang, J., Li, J., Cheng, Z., Chaemfa, C., Liu, D., et al. (2013). Occurrence and Risks of Antibiotics in the Coastal Aquatic Environment of the Yellow Sea, North China. Sci. Total Environ. 450-451, 197–204. doi:10.1016/j.scitotenv.2013.02.024
Zhang, X. X., Zhang, T., and Fang, H. H. P. (2009). Antibiotic Resistance Genes in Water Environment. Appl. Microbiol. Biotechnol. 82, 397–414. doi:10.1007/s00253-008-1829-z
Zhuang, M., Achmon, Y., Cao, Y., Liang, X., Chen, L., Wang, H., et al. (2021). Distribution of Antibiotic Resistance Genes in the Environment. Environ. Pollut. 285, 117402. doi:10.1016/j.envpol.2021.117402
Keywords: antibiotic misuse, antimicrobial resistance genes, antimicrobial resistance, pollution, environment, wastewater, water bodies
Citation: Singh AK, Kaur R, Verma S and Singh S (2022) Antimicrobials and Antibiotic Resistance Genes in Water Bodies: Pollution, Risk, and Control. Front. Environ. Sci. 10:830861. doi: 10.3389/fenvs.2022.830861
Received: 07 December 2021; Accepted: 25 March 2022;
Published: 28 April 2022.
Edited by:
Surindra Suthar, Doon University, IndiaReviewed by:
Jagat Rathod, National Cheng Kung University, TaiwanCopyright © 2022 Singh, Kaur, Verma and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samer Singh, samer.singh10@bhu.ac.in
†These authors have contributed equally to this work
 Ashish Kumar Singh
Ashish Kumar Singh Rajinder Kaur
Rajinder Kaur Shashikala Verma
Shashikala Verma Samer Singh
Samer Singh