Effects of Phytoremediation on Microbial Biomass, Composition, and Function in a Sulphide-Rich Tailing From a Metal-Contaminated Region
- 1Biomolecular Sciences Program, Sudbury, ON, Canada
- 2Department of Biology, Sudbury, ON, Canada
- 3School of Environmental Sciences Laurentian University, Sudbury, ON, Canada
Mining activities lead to serious land deterioration and large scale mine waste generation. Reclamation has been carried out on several technogenic materials to encourage the development of soils. To date no detailed studies have been conducted to assess if soil developed in reclaimed tailings can be suitable for microbial community sustainability and associated plant population. This study investigated if 1) soil metal contamination affects microbial biomass and composition in sulphide tailings and 2) phytoremediation of tailing increases microbial abundance, diversity, and function. Microbial biomass was assessed using Phospholipid fatty acid analysis (PLFA). Soil bacterial and fungal microbiota was determined by high throughput sequencing of 16S rRNA gene for bacteria and internal transcribed spacer region for fungi using the Illumina platform. Total copper, nickel, iron, and titium were higher in unreclaimed sites compared to vegetated areas but the total microbial biomass was significantly higher in reclaimed sites compared to reference areas. More importantly, the levels of microbial biomass were not impacted by metals since the bioavailable Cu, Ni, and Ti were low in all the sites. Site-specific bacterial and fungal genera were identified. Proteobacteria was the most dominant bacterial phylum while Ascomicota was the predominant fungal phylum. Interestinlgy, Acidiferrobacter, an acidophilic, thermotolerant and facultatively anaerobic was the most predominant genus in unreclaimed site that is characterized by extreme acidity (pH = 2.8). Analysis of microbial diversity revealed higher Chao 1, # of OTUs, Shannon index, and species richness in bacterial and fungal populations from reclaimed sites compared to controls. The levels of β-glucosidase (BG), cellobiohydrolase (CBH), β-N-acetylglucosaminidase (NAGase), aryl sulfatase (AS), acid phosphatase (AP), alkaline phosphatase (AlP), glycine aminopeptidase (GAP), and leucine aminopeptidase (LAP) activities were significantly higher in vegetated sites compared to reference areas. Strong positive correlation coefficients were observed between soil organic matter and total microbial biomass (r = 0.99). These two factors were positively correlated with enzymatic activities and bacterial population diversity. Overall, newly developed soils can sustain diverse microbial communities and associated vegetations.
1 Introduction
The increase of metals in environment matrices is mainly caused by industrial and anthropogenic activities. This has been linked to harmful effects to biota including human (Sharma et al., 2022). Mining activities cause serious land deterioration and large scale mine waste generation, creating an essential importance for rehabilitation strategies in the mine planning and development Mine tailings possess many physical and chemical properties detrimental to plant and soil organisms (Lottermoser, 2010). As a result, they are nutrient deprived and highly contaminated with toxic metals, contributing to development of undesirable habitats that generates stress on organisms and restrict development of healthy biological processes (Khozhina and Sherriff, 2006; Kumar, 2013). It has been established that soil microbial communities are coupled to their associated vegetation. Hence, compromised soil communities may curtail particular plant assemblages from forming (Yiki and Zhou, 2010).
Phytoremediation is a bioremediation process that uses various types of grasses, shrubs, trees, aquatic plants, to remove, transfer, stabilize, and/or destroy contaminants in the soil, groundwater, and aquatic systems. The phytoremediation mechanisms include phytoextraction, phytostabilization, and phytodegradation. Details of phytoremediation applications in metal – contaminated lands are dicussed in Narendrula-Kotha et al. (2018, 2020) and Sharma et al. (2020). Phytostabilisation of tailings landscapes has been challenging (Mendez and Maier, 2008). But, it has been demonstrated that ecological restoration of mine tailings by revegetation provides a model system for investigating biotic interactions below and above ground. Microorganisms have been shown to rapidily respond to environmental change during the restoration process in an ecosystem of tailings (Mendez and Maier, 2008; Li et al., 2018). Strong correlations between soil microbial biomass, fertility and health have been demonstrated in non-tailing settings (Cheng et al., 2013). Londry and Sherriff (2005) reported a significant correlation between microbial biomass and biodiversity with physicochemical conditions in mine tailing in Manitoba (Canada). Their findings were consistent with data reported by Feketeová et al. (2021) investigating the recovery of soil ecosystem at tailings ponds affected by mining in Slovakia. Li et al. (2018) on the other hand reported microbial diversity increases and high activities of microorganisms induced by applications of zinc, lead, and sulfidic copper in tailing areas in Australia. Analysis of below ground communities, showed that fungi and bacteria play crucial roles in ecosystem function since there are the main source of enzymes in soils. Fungi are involved in ecosystem function, the breakdown of cellulose and tannins that are complex biomolecules in soils. Soil fungal respiration has been found to be two to four times greater than that of bacteria and fungi account for up to 90% of total living biomass in forest soils (Barron, 2003; Rousk et al., 2009; Nkongolo et al., 2016). The long-term stability of ecosystems is therefore dependent on the continued contribution of fungal activities.
A sulfide tailing located in Onaping Falls town at 30 km from the City of Greater Sudbury (Ontario, Canada) was developed by the Sudbury Nickel Integrated Mining Operations (Glencore). Corn (Zea mays), canola (Brassica napus), big bluestem (Andropogon gerardi), swich grass (Panicum virgatum), and several species of willows (Salix spp.) were planted within the last 15 years as part of a bioenergy trial (Coggans, 1992).
Studies on phytoremediation of mine tailings in the Boreal Regions with regards to soil quality, metal content, and their long-term ability to sustain both plant and associated microbiological populations are limited. This study investigated if 1) soil metal contamination affects microbial biomass in tailings and 2) phytoremediation of tailing increases microbial abaundance, diversity and function. We hypothesize that the abundance, composition, diversity, and activities of soil microbial communities are sensitive in detecting significant positive environmental changes in phytoremediated tailings.
2 Material and Methods
2.1 Site Characterization
The Sulfide tailings at Onaping Falls (46.61⁰N 81.43⁰W) was reclaimed by the Sudbury Integrated Mining Operations (Glencore–Sudbury) (Supplementary Figure S1). Five areas within the tailing were selected based on the plant species. These sites include an area covered with up to a metre of an organic technosol and planted with big bluestem (Andropogon gerardi), swich grass (Panicum virgatum), and willows (Salix spp.). Two unreclaimed tailing areas were used as references (pyrhotite and low south-unreclaimed areas).
2.2 Sample Collection
Soil samples were collected as composites at each area. Three transects of 10 m diameter each were used. Overall, 10 soil samples per replication/transects (total of three replicates) were collected from the organic layer (0–10 cm) in 2017. The samples were sieved in a 1 mm mesh to remove debris, rocks and plants. Plant materials, stones, and residues were removed.
2.3 Analysis of Soil pH, Organic Matter, and Metal Content
Soil pH was measured in water and organic matter was determined using the loss on ignition (LOI) analysis at Testmark Inc. (Sudbury), an ISO 17025 accredited facility. Total metal analysis in soil was performed as described by Nkongolo et al. (2013), Ghazouani et al. (2020), and Nkongolo et al. (2022). Soil pH was measured in water and a neutral salt solution pH (0.1 M CaCl2) (Carter and Gregorich, 2008). Likewise, the estimation of total and bioavailable, metal concentrations were performed as described by Nkongolo et al. (2013) and Nkongolo et al. (2022). The quality control program included Internal Reference Materials (IRM’s), Certified Reference Materials (CRM’s, analysis of duplicates), procedural and calibration blanks, with continuous calibration verification and use of internal standards (Sc, Y, Bi). All metal concentrations were estimated in mass/mass dry soil basis. The resulting data were within ±12% of the certified level (Nkongolo et al., 2022).
2.4 Phospholipid Fatty Acid Analysis
PLFA analysis of soil samples was performed as described by Buyer and Sasser (Buyer and Sasser, 2012; Narendrula et al., 2013). Total PLFA extracted from soil was used as an index of living microbial biomass. The targeted PLFAs for bacterial biomass include cy19:0, 18:1ω7, i17:0, cy17:0, 16:1ω7c, 16:1ω9, i16:0, a15., and i15:0. For fungi, PLFA 18:2ω6 and 18:1w9 were used. For identification of actinomycetes, protozoa, and eukaryotes, 10Me, 20:3ω6/20:4ω6 and 12:0/16:1ω7/18:2ω9/18:2ω12/18:3ω9/18:3ω12/18:3ω15/polyunsaturated fatty acids were used, respectively.
2.5 Amplicon Sequencing - Illumina
Details of the protocol are described in (Nkongolo et al., 2022). “Soil microbial DNA was extracted from fresh samples. This was followed by purification of genomic DNA, fragmentation, ligation to sequencing adapters and purification. The Nextera DNA Sample Preparation Kit (Illumina) was used to prepare the libraries. Library insert size was determined by Experion Automated Electrophoresis Station (Bio-Rad). The insert size of the libraries ranged from 300 to 850 bp (average 500 bp). Pooled library (12p.m.) was loaded to a 600 Cycles v3 Reagent cartridge (Illumina) and the sequencing was performed on Miseq (Illumina). The ITS specific primers ITS1F-Bt1 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) were used to amplify 600 bp fragment of the fungal ITS region (Schoch et al., 2012). For bacteria, the 16S rRNA gene V4 variable region PCR primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) with barcode on the forward primer were used. This high thoughouput sequencing was performed at the MR DNA Molecular Research DNA laboratory (Shallowater, Texas, United States)”.
2.6 Data Processing
The processing of sequencing data was as described in Nkongolo et al. (2022): “The Q25 sequence data derived from the sequencing were processed using MR DNA ribosomal and functional gene analysis pipeline (www.mrdnalab.com, MR DNA, Shallowater, TX, United States). Sequences were depleted of barcodes and primers and short sequences <150 bp and sequences with ambiguous base calls and with homopolymer runs exceeding 6 bp were removed. Sequences were then denoised and operational taxonomic units (OTUs) were defined clustering at 3% divergence (97% similarity) followed by removal of singleton sequences and chimeras. Final OTUs were taxonomically classified using BLASTN (Basic Local Alignment Search Tool for Nucleotides) against a curated database derived from GreenGenes, the Ribosomal database Project (RDPII), and the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov, http://rdp.cme.msu.edu, (DeSantis et al., 2006)) and compiled into each taxonomic level into both “counts” and “percentage” files”.
2.7 Enzymatic Activities
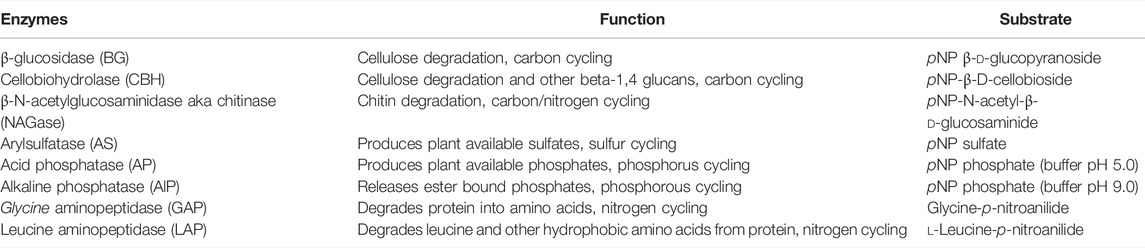
Leucine aminopeptidase (LAP), glycine aminopeptidase (GAP), alkaline phosphatase (ALP), arylsulfatase (AS), β-N-acetylglucosaminidase (NAGase), cellobiohydrolase (CBH), and β-glucosidase (BG), were selected for this study based on the ability for catalyzing reactions as well as their role in geochemical processes (Table 1). Enzymatic activities were performed as described in Narendrula-Kotha and Nkongolo (2017b).
2.8 Statistical Analyses
Statistic analyses were performed as described in Nkongolo et al. (2022). SPSS statistics version 20 for Window, was used to analyze the metal levels in soil samples, microbial biomass, fatty acid content, and enzymatic activities. Data were log10 transformed to assist in achieving a normal distribution when required. One-way analyses of variance, followed by Tukey’s HSD multiple comparison analysis, were performed to determine significant differences (p ≤ 0.05) among means. Variance-ratio test was done with an assumption of data normality in the underlying population distributions of the data. QIIME (version 1.8.0) was used to estimate species richness and evenness, Simpson index, Shannon index, and Chao1, (Kuczynski et al., 2011).
3 Results
3.1 Soil Organic Matters, pH, and Metal Contents
Soil pH, organic matters, total and bioavailable metal data are described in Tables 2 and 3. Reclamation of Onaping falls tailings increased significantly the levels of soil pH and organic matter compared to unreclaimed areas. The estimated total metal concentrations in the soil samples are illustrated in Table 2. There were 4 to 7x more Cu in unreclaimed areas compared to each of the areas reclaimed with willow grass, switch grass, and big bluestem (lili turf) grass. There were up to 47x more Ni in unclaimed low site area compared to the adjacent reclaimed areas (with big bluestem, switch, and willow grasses).
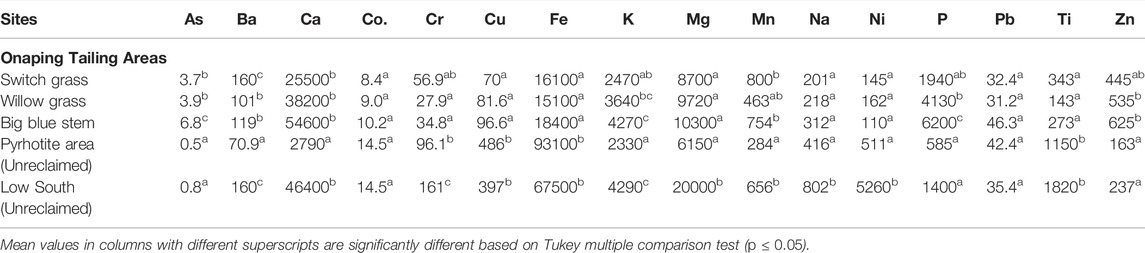
TABLE 2. Total metal concentration (mg/kg dry wt) in soil samples from Onaping Falls (Glencore) tailings in Northern Ontario.
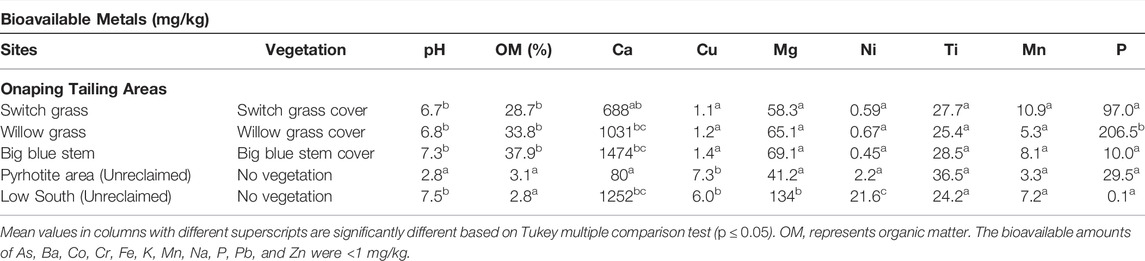
TABLE 3. Vegetation, pH, organic matters, and bioavailable metal (mg/kg dry wt) in soil samples from Onaping Falls tailings.
At each targeted area, the bioavailable amounts of As, Ba, Co, Cr, Fe, K, Na, Pb, and Zn were <1 mg/kg. The levels of other bioavailable elements are described in Table 4. The bioavailable Cu, Ni, and Mn varied between 0.45 and 11 mg/kg. The levels of bioavailable Ca and Mg were significantly higher than other elements. Bioavailable Ca ranged from 80 mg/kg to 1,474 mg/kg and Mg ranged varied from 30 mg/kg to 134 mg/kg (Table 3).
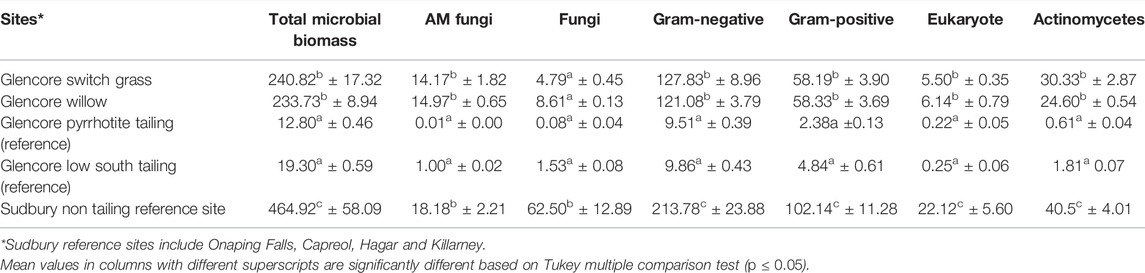
TABLE 4. Total microbial biomass, AM fungi, Fungi, bacteria, Eukaryote, anaerobe, and actinomycetes identified in soil samples from the Onaping Falls (Glencore) tailing (Sudbury) using phospholipid fatty acid (PLFA) analysis. Data in nMol/g.
3.2 Phospholipid Fatty Acid Analysis
Results of the PLFA analysis are described in Table 4, Supplementary Tables S1 and S2. There were more microbial biomass in reclaimed sites (with willow, switch, and big bluestem grasses) compared to the unreclaimed areas (Table 4).
Bacteria biomass represented 90% at the area reclaimed with switch grass, 87% for area restored with willow, 91% for area reclaimed with mixed grasses, 97% at unreclaimed area (pyrhhotite areas), and 77% at the reference site outside the tailing. There were twice more Gram negative than Gram positive bacteria in almost all the tailing and references samples.
Palmitic acid (18:1), palmitoleic acid (16:1), and palmitic acid (16:0) were among the most prevalent of all the fatty acids identified in all the soil samples as they account for 56% of all FAs in Glencore Onaping tailing samples (Supplementary Table S2). Specifically, the bioremediation of the Clencore Onaping tailing sites significantly improved the FA outcome. Oleic acid (18:1) was the most prevalent representing 21% of all FAs at the site followed by FA 16:0 (19%) and FA 17:0 (19.5%). As expected, the levels of these FAs and other types were significantly lower in samples from unreclaimed areas compared to revegetated areas (Supplementary Table S2). The area revegetated with either switch grass or willow showed similar levels of FA.
3.3 Bacterial and Fungal Community Composition and Diversity Analysis
The reads generated in this project have been deposited in the NCBI Short Read Archive database (Accession number: SRP071853). The total numbers of reads per assay varied between 15,000 and 20,000. Bacterial and fungal species, genera, families, classes, and phyla specific to each area were identified (Supplementary Tables S2–S8; Figure 1 and Supplementary Figures S2–S7). Venn diagrams show different bacterial and fungal families, classes, and phyla and their distribution (Figure 1 and Supplementary Figures S2–S7). Relative abundance for bacteria and fungi are described in Supplementary Tables S3, S5–S9, S11–S14) and microbial commuty diversities in Table 5, Supplementary Tables S15−S17.
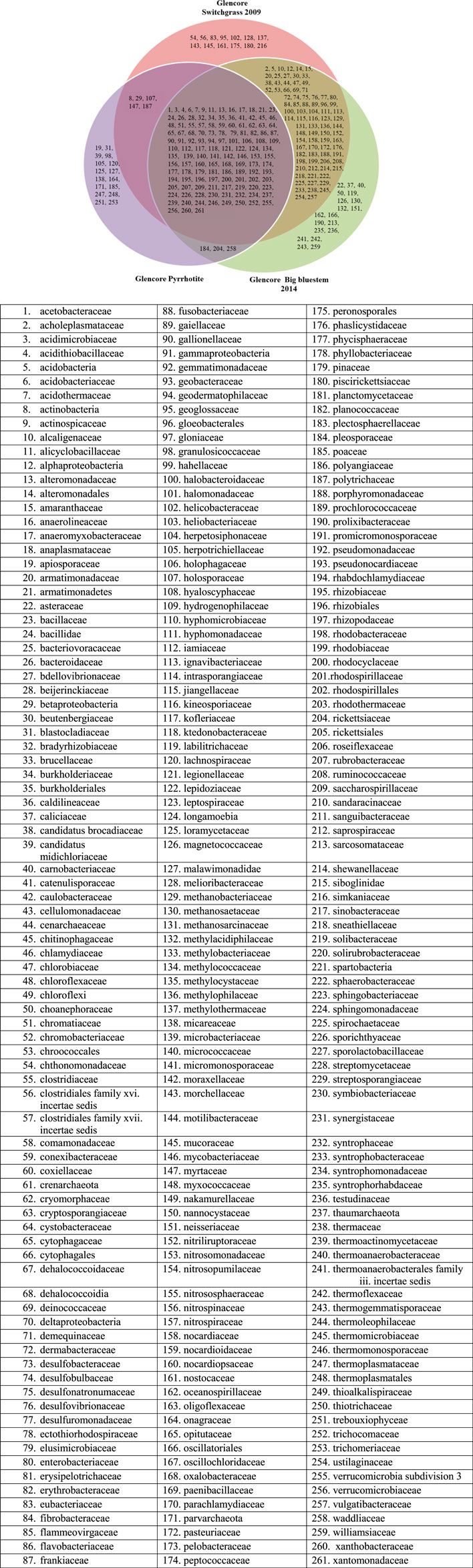
FIGURE 1. Venn diagram showing distribution of bacterial families identified in Onaping Falls (Glencore) tailing sites from the City of Greater Sudbury.
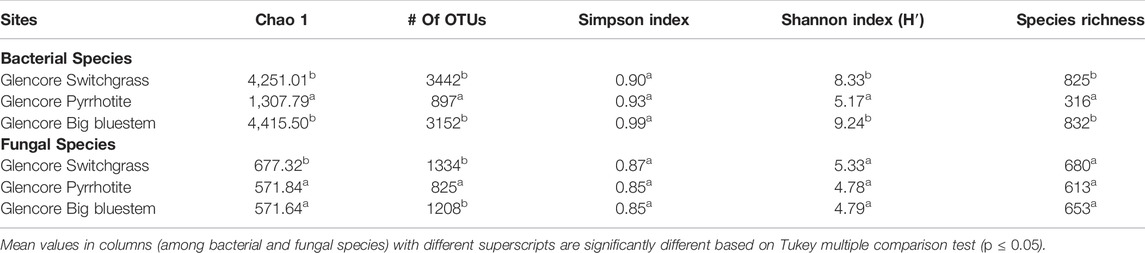
TABLE 5. Microbial diversity index values for bacterial and fungal communities from the Onaping falls (Glencore) tailing sites (City of Greater Sudbury).
3.3.1 Bacterial Populations
In total, 992 species, 639 genera, 261 families, 76 classes, and 36 phyla of bacteria were identified at the Onaping tailing. Overall, 9% of the bacterial species identified were specific to the switchgrass area, 10% to the big bluesteem area, and 5% to the unreclaimed pyrrhotite area. Among the 8 phyla, Proteobacteria (53% at switchgrass site, 52% at big bluesteen area, and 54% at the unreclaimed pyrrhotite area), and Actinobacteria (11% at the switchgrass area, 13% at big bluestem and 20% at the unreclaimed pyrrhotite site), were the most represented. Acidobacteria, bacteroidetes, and Firmicutes account for 6–10% each of the bacteria found in samples from the switchgrass, big bluestem, and unreclaimed pyrrhotite areas. Verrucomicrobia were the least identified with less than 3% at each targeted site (Supplementary Table S8). The most abundant bacterial genera at the switchgrass reclaimed site include Pelobacter (6%), Holophaga (5%), and Acidobacterium (4%). For the big bluesteem area, Pelobacter (6%) and Acidobacterium (3.4%) were the most represented. For the unreclaimed pyrrhotite site, Acidiferrobacter (22%), Acidiphilium (15%) and Acidithiobacillus (11%) were the most abundant genera (Supplementary Table S5; Figure 2).
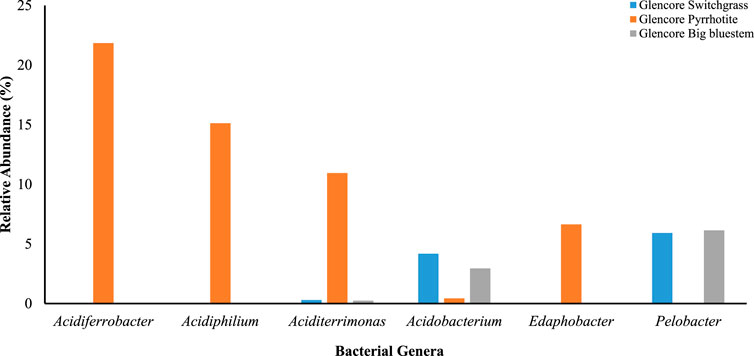
FIGURE 2. Top six bacterial genera and their relative abundance identified in soil samples from the Onaping Falls (Glencore) tailing (City of Greater Sudbury). Data in percentage.
The average estimated minimum number of operational taxonomic units (OTU), Chao1 index, Shannon’s index, Simpson index, and species richness at the three sites from the Onaping (Glencore) tailing are described in Table 5. There were significant differences for # of OTUs, Chao 1, Simpson index, Shannon index, and species richness between bacterial community from the switch grass compared to the unreclaimed sites as well between the samples from the big bluesteem and the reference pyrrhotite (p ≤ 0.05) (Table 5). In the present study, Jaccard similarity indices were low when the three sites were compared to each other (15S). Sorenson’s index was low as well when each of the reclaimed site was compared to the unreclaimed area, but intermediate when they were compared to each other (switch grass vs. big bluesteem). This was confirmed by the Bray-Curtis dissimilarity indices that were high when reclaimed sites were compared to unreclaimed areas. Whittaker dissimilarity indices were low to intermediate based on the pairwise analysis (15S). Further, we compared the sites with one another using clustering based on the weighed UniFrac (incorporates relative abundance) distances (17S). The distance values estimated were low between the two reclaimed sites (switch grass and big bluestem) and high between each of the reclaimed area compared to the unreclaimed area.
3.3.2 Fungal Populations
Analyis of Onaping (Glencore) tailing sites revealed 1,141 species, 588 genera, 284 families, 56 classes, and 10 phyla. Ascomycota phylum was the most abundant (58–69%). Overall, the two main phyla included Ascomycota (69% at switchgrass area, 63% at the area reclaimed with big bluestem, and 58% at pyrrhotite unreclaimed reference site) (Supplementary Table S14). Relative abundances of fungal species, genera, families, classes, and phyla are described in Supplementary Tables S9, S11−S14.
Overall, 12% of the 1,141 fungal species identified were specific to the areas reclaimed with switch grasses; 9.7% to the big bluesteem reclaimed areas, and 27% to unreclaimed pyrrhotite area. The most abundant fungal genera at the switchgrass area include Mortierella (8.2%), Cladosporium (7.8%), and Alternaria (7.6%). Microbothryozyma (11.8%), Alternaria (7.4%) and Epicoccum (7.0%) were the most represented at the unreclaimed pyrrhotite site. The most abundant genera at the big bluesteem reclaimed site include Myriococcum (17.8%), Mortierella (5.3%), and Alternaria (4%) (Supplementary Table S11; Figure 3).
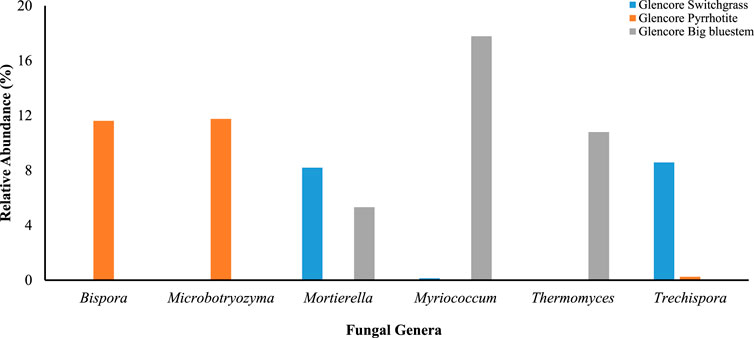
FIGURE 3. Top six fungal genera and their relative abundance identified in soil samples from the. Onaping Falls (Glencore) tailing (City of Greater Sudbury). Data in percentage.
There were significantly higher number of OTU at the two sites reclaimed with switch grass (1,334 OTUs) and big bluesteem (1,208 OTUs) compared to unreclaimed pyrrhotite site (825 OTUs) (Table 5). Chao index was significantly higher for the fungal population at the switch grass area compared to big bluesten reclaimed site and the unreclaimed pyrrhotite area. No significant difference was observed between the three sites based on Simpson index, Shannon index, and species richness (Table 5). Jaccard similarity indices were low when the three sites were compared among each other and the Sorenson’s similarlity index was low when the reclaimed sites were compared to the unreclaimed pyrrhotite site. This index was intermediate when the reclaimed sites were compared to each other. Bray-Curtis dissimilarity indices were low when switchgrass was compared to unreclaimed pyrrhotite and high when the two reclaimed sites were compared. Whittaker dissimilarity indices were low to intermediate (0.29–0.43) when the three sites were compared to each other (15S). Distance matrix values based on weighted variants showed high distances among the three sites (Supplementary Table S17).
3.4 Enzymatic Activities
Analysis of enzymatic activities was performed to determine if phytoremediation of Onaping tailings affects microbial functions. Soil samples from reclaimed areas (planted with grasses) exhibited significantly higher (p ≤ 0 0.05) enzymatic activity for β-glucosidase (BG), acid phosphatase (AP), alkaline phosphatase (ALP), and glycine aminopeptidase (GAP) compared to samples from the unreclaimed area (Figures 4–6 and Supplementary Figure S8). Overall, significant levels of enzymatic activities were observed in areas reclaimed with blue grass compared to the other sites followed by sites reclaimed with swittchgrass (Figures 4–6 and Supplementary Figure S8).
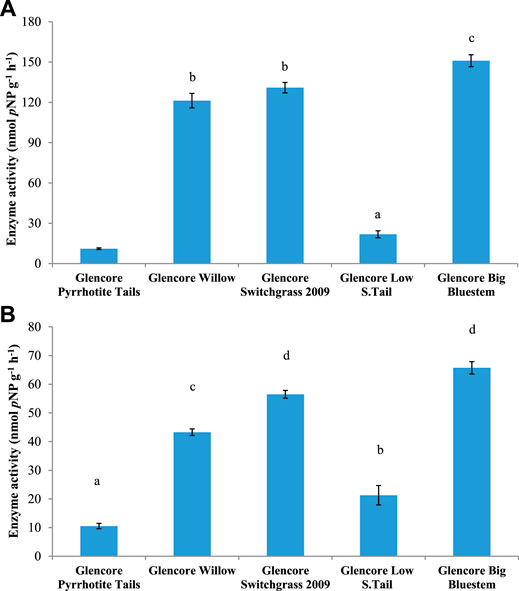
FIGURE 4. Activity of (A) β-glucosidase (BG) and (B) cellobiohydrolase (CBH) in soils from Onaping tailing sites in the City of Greater Sudbury. Mean values with different letters are significantly different based on Tukey multiple comparison test (p ≤ 0.05).
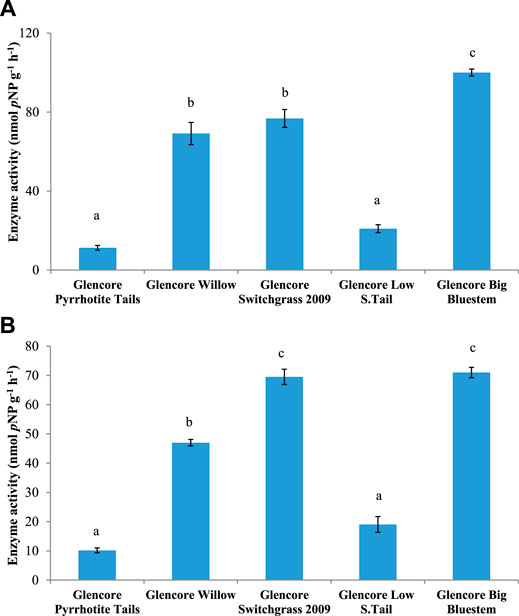
FIGURE 5. Activity of (A) β-N-acetylglucosaminidase (NAGase) and (B) arylsulfatase (AS) in soils from Onaping tailing sites (City of Greater Sudbury). Mean values with different letters are significantly different based on Tukey multiple comparison test (p ≤ 0.05).
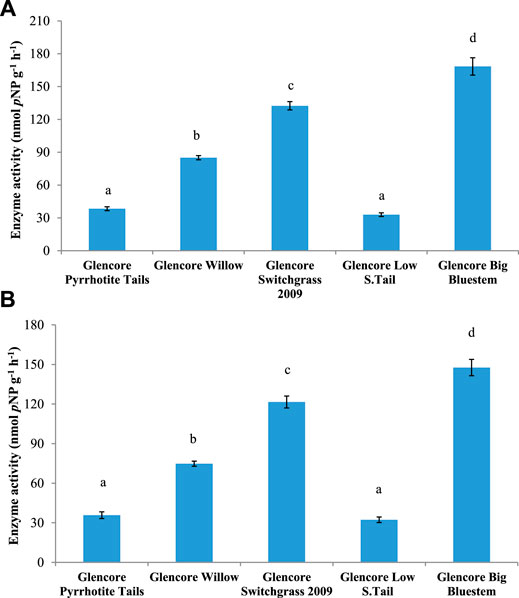
FIGURE 6. Activity of (A) acid phosphatase (AP) and (B) alkaline phosphatase (AlP) in soils from Onaping tailing sites (City of Greater Sudbury). Mean values with different letters are significantly different based on Tukey multiple comparison test (p ≤ 0.05).
3.5 Correlations Between pH, Organic Matter, Microbial Biomass, Microbial Diversity and Enzymatic Activities
A strong positive correlation (r = 0.99) was observed between organic matter and total microbial biomass. Likewise, the high correlation coefficients between total microbial biomass and the activities of each of the six enzyme tested (BG, CBH, NAGas, AS, Ap, and AIP) were high (r > 75). The correlation between the pH levels with the organic matter content and the the microbial biomass were positive and intermediate with coefficient values of 0.50 and 0.45, respectively. Chaos 1 diversity index and species richness were strongly correlated with microbial biomass and soil orgaic matter with correlation coefficient values varying between 0.86 and 0.88 for bacteria. No correction was found between these diversity parameter with microbial biomass and soil organic matter for fungi.
4 Discussion
4.1 Microbial Biomass and Fatty Acid Profile
PLFA analysis was used to determine soil microbial responses to tailing management. The results showed that metal contamination levels including cobalt, nickel, and copper have no effects on microbial biomass. In fact, at Onaping fall (Glencore) tailing, all the reclaimed sites covered with different grasses had significantly higher levels of total microbial biomass compared to unreclaimed sites without vegetation. This suggests that phytoremediation of tailing improve soil microbial community development. It should be pointed out that although the total metals were extremely high in some sites, over 90% of them were in the form that were not available to microorganisms and below the levels known to affect bacteria and fungi for example. This indicates that metals may have not been easily leachable from the tailing samples (Kwon et al., 2015; Ghazouani et al., 2020).
It is known that increased soil organic matters change PLFA composition. Several studies have reported a strong positive correlation between microbial biomass carbon (MBC) and soil organic carbon (SOC) (Dimitriu and Grayston, 2010; Nkongolo et al., 2016; Narendrula-Kotha R. and Nkongolo K. K., 2017; Narendrula-Kotha and Nkongolo, 2017b). Our data are consistent with previous findings since higher microbial biomass in sites was associated with high organic matter. Londry and Sherriff (Londry and Sherriff, 2005) found that the distribution of microbial biomass was correlated to physical/chemical condition. Changes in vegetation cover can also afftect soil microbial biomasses. For example, vegetation could retard soil degradation of mine tailing by stimulation of soil pH and alteration of bacterial communities in upper soil layer (Li et al., 2016). Sites with relatively higher vegetation cover and organic matter content had in some studies a positive association with enzymatic activities (Claassens et al., 2006; Narendrula-Kotha and Nkongolo, 2017c). In the present study, the microbial communities in all the sites were dominated by bacteria, suggesting that both phtoremediated and reference tailing areas are still under severe environmental stress. This is consistent with the low ratios between fungal and bacterial biomass. Gram-positive bacteria are thought to be more resistant to stress and harsh conditions than are Gram negative bacteria (De Vries and Shade, 2013).
4.2 Metal Contamination and Bacterial and Fungal Abundance and Composition
4.2.1 Bacteria
In the present study, the relative abundance expressing the proportion of each bacterial and fungal species, genera, and families out of the total number recorded were estimated. In previous studies, Li et al. (2016) reported a predominance of Proteobacteria and Bacteroidetes phyla in tailing reclaimed with soil mixtures compared to reference sites in Australia based on pyrosequencing analysis. In the present study, Proteobacteria was the most dominant phylum at Onaping tailings. Chung et al. (2019) analyzed microbial profile at two tailing basins from a tungsten mine in Portugal. They found that the distribution of the different microbial groups was associated with soil physicochemical characteristics of the mine tailings.
In this study, no predominant bacterial genus was identified in areas reclaimed with either switch grass or big bluesteem. Analysis of unreclaimed area (Glencore–pyrrhotite) revealed that Acidiferrobacter was the most dominant genus (with 22% of relative abundance). This area contains significantly higher levels of Cu, Fe, Ni, and Ti compared to the revegetated areas. Acidiferrobacter with only one identified species belong to the Ectothiorhodospiraceae family. It is an acidophilic, thermotolerant and facultatively anaerobic genus of bacteria (Hallberg et al., 2011). In other studies, Acidiferrobacter thiooxydans has been isolated from coal mining areas in the United States. This Acidiferrobacter genus has been also found abundant in leaching heap (LH) during copper ore bioleaching. The cycling and metabolism of S and Fe have been identified in mine tailings, and acidic river (Teruel et al., 2014; Y et al., 2015; Niu et al., 2016) which are comparable to subsystems in bioleaching system. The high abundance of Acidiferrobacter at the unreclaimed site appears to be associated with the elevated levels of Fe that was 4 to 5x more compared to the reclaimed sites (93,100 mg/kg vs. < 19,000 mg/kg). This area was also characterized by an extreme acidity with a pH of 2.8. Members of Acidiferrobacter have a potential role in the anaerobic oxidation of Iron and Sulfur radicals, hence contributing to the overall ecology of the environment. Other dominant genera found at this site include members of Acidiphilium (15%) and Aciditerrimonas (11%).
4.2.2 Fungi
Li et al. (2016) identified Ascomycota as the dominant phylum in reclaimed sulfidic copper (Cu)-lead (Pb)-zinc (Zn) tailings in Australia. In the present study, Myriococcum was predominant at the area covered with big bluestem (18%) and absent at the unreclaimed site. On the other hand, Microbotryozyma represented 12% of the fungal community at the unrelaimed area (Glencore pyrrhotite) and was absent in revegetated areas. Myriococcum are mycorrhizal thermophilic fungi that have a powerful ability to degrade polysaccharide. They are constituents of the microflora that develops in heaped masses of plant material, piles of agricultural and forestry products, and other accumulations of organic matter such as Glencore organic cover wherein the warm, humid, and aerobic environment provides the basic conditions for their development mass (Maheshwari et al., 2000). Microbotryozyma are basidiomycetous yeasts that can grow on a number of materials and areas. The predominance of these fungi in soils from the unreclaimed site without vegetation might be associated with appropriate yeast growing conditions.
4.3 Bacterial and Fungal Diversity in Mine Tailings
The comparison of relative abundance did help determine differences in species, genera, families, classes, and phyla. It did also help shed the light into the diversity. Microbial diversity is an important ecological indicator; it is believed that higher diversity resulted in higher sustainability of microbial communities. In this study, relative abundance and diversity were also estimated by the number of OTUs, Chao1, Shannon index and Simpson index. Detailed description of these parameters and their use in ecological studies are described in (Chao, 1984; Roesch et al., 2007; Clivot et al., 2012; Narendrula-Kotha and Nkongolo, 2017b). Previous studies revealed a significant increase of the number of OTUs and CHAO index in reclaimed compared to unreclaimed sites (Narendrula-Kotha and Nkongolo, 2017b). At Onaping tailing, significant differences were observed between reclaimed sites and unreclaimed areas for Chao 1, # of OTUs, Shannon index, and species richness. These differences are likely related to higher pH, cation exchange capacity (CEC), aboveground plant species diversity, and abundance and the overall ecosystem health in reclaimed and unreclaimed areas (Narendrula and Nkongolo, 2015; Nkongolo et al., 2016). In fact, strong correlations between soil organic matters with CHAO 1 index, # of OTU, Shannon index and Simpson index, and species richness for bacterial populations were observed in the present study. These findings are consistent with Wolińska et al. (2020) who reported changes in microbial structures and metabolic activity resulting from reclamation of technosols developed from mine wastes containing Fe sulphides. In their study, the analysis was performed over 45 years after soil restoration.
For beta measures, Jaccard and Sorenson indices are based on the presence/absence data (Koleff et al., 2003; Kuczynski et al., 2011). Jaccard and Sorenson indices, for both bacteria and fungi were low indicating high dissimilarity (high beta diversity) among the three areas (Vale 1, Vale 2, and Vale 3). Unlike the Sorenson indices, the Brady-Curtis analysis is based on abundance data. For the Onaping Falls tailing, Jaccard’s index, Sorenson’s index, and Wittaker indices were low suggesting high dissimilary for bacterial and fungal commnuties.
Overall, for this investigation, both alpha and beta diversity indices were used. The results of the studies provide further evidence that ameliorating soil chemistry even at tailings through soil amendment and revegetation alters the composition and abundance of soil microbial communities.
4.4 Enzymatic Activities
Phytoremediation improved both microbial biomass and function. Specific enzymes were targeted in this study to measure microbial function. Overall, higher enzymatic acitivies were associated with an elevated level of microbial biomass observed in the targeted areas at Onaping tailing. Narendrula-Kotha and Nkongolo (2017c) reported a significantly higher activities levels of BG, CBH, NAGase, AS, AP, AlP, GAP, and LAP in non-tailing sites limed >35 years ago compared to adjacent unlimed area in the CGS. McKergow et al. (2021) on the other end showed an increase of BG, CBH and AS but a decrease of GAP activities in recently limed areas compared to untreated areas. They observed no significant change in AlP, AP, LAP, and NAGase activities when limed and unlimed soils were compared.
4.4.1 β-glucosidase
β-glucosidase (BG) plays an important role in nutrient cycling of carbon ions and in the degradation of starch (Ketudat Cairns et al., 2015; Xue et al., 2018). BG activities are high in non acidic soils (neutral pH) after limestone soil treatment (Lupwayi et al., 2018). Waldrop et al. (2000) reported a strong correlation between BG activity and microbial community composition. A high activity of BG is associated with a large soil starch content, as well as cellobiose, glucose, and other cellulose metabolites (Veres et al., 2015).
4.4.2 Activity of Cellobiohydrolase
Cellobiohydrolase (CBH) is involved in the degradation of cellulose (Acosta-Martinez and Tabatabai, 2000; Verchot and Borelli, 2005; Sinsabaugh et al., 2008). A decrease in the microbial biomass and diversity, or changes to the soil carbon content can significantly affect CBH activities (Brockett et al., 2012). An increase of CBH activity in soils with high organic N content has been reported in Saiya-Cork et al. (2002). The high level of CBH activities in revegetated sites compared to untreated areas in the present study is consistent with previous reports that showed an elevated levels of CBH in limed or fertilized lands compared to untreated areas with lower organic matters (McKergow et al., 2021; Narendrula-Kotha R. and Nkongolo K. K., 2017, 2017b, 2017c).
4.4.3 Activity of β-N-acetylglucosaminidase
β-N-acetylglucosaminidase is involved in the cycling of carbon and nitrogen and the degradation of chitin by hydrolyzing β-1,4-glycosidic bonds of chitooligosaccharides into N-acetylglucosamine (Verchot and Borelli, 2005; Sinsabaugh et al., 2008). Thus, NAGase is very important to ecosystem health. Chitin is found primarily in fungal walls and also in exoskeletons of some arthropods. It has been also found higher in reclaimed areas with higher level of organic matters. Previous studies revealed significantly higher level of NAGase activity in limed and reference soils contaminated with metals compared to metal contaminated unlimed sites (McKergow et al., 2021; Narendrula-Kotha R. and Nkongolo K. K., 2017, 2017c, 2017b).
4.4.4 Activity of Arylsulfatase
Arylsulfatse (AS) hydrolases organic sulfate esters (organic S form) by splitting the O–S bond and releasing sulfates into the environment. Previous reports have shown that AS activity is positively correlated with soil pH, organic C, total S, and microbial biomass (Ekenler and Tabatabai, 2003).
4.4.5 Activity of Acid and Alkaline Phosphatase
Acid and alkaline phosphatase (AP/AlP) are useful in phosphorous cycling. There is a strong evidence that phosphatase enzymes are significantly correlated with all functional genes involved in the phosphorous nutrient cycling (Xue et al., 2018). Phosphatases can help increase the bioavailable amount of mineralized phosphorous (Acosta-Martinez and Tabatabai, 2000; McKergow et al., 2021). They are also used to catalyze the hydrolysis of both esters and anhydrides of phosphoric acid.
4.4.6 Activity of Aminopeptidase
Glycine aminopeptidase (GAP) hydrolyzes Gly-X (amino acids, peptide or arylamide) bonds (Ito et al., 2003). Leucine aminopeptidase (LAP) hydrolyzes leucine and other hydrophobic amino acids from N terminus of polypeptides (Sinsabaugh et al., 2008). LAP and GAP activities are strongly associated with soil pH (Sinsabaugh et al., 2008; Narendrula-Kotha and Nkongolo, 2017c; McKergow et al., 2021).
4.4.7 Correlation Between Soil Enzymatic Activities and Organic Matter Content
The present study showed a significant positive relationship between the levels of soil organic matter and total biomass with soil enzyme activities. This is consistent with data reported by Xu et al. (2015) studying microbial communities in Changbai Mountain (China) and McKergow et al. (2021) comparing limed and unlimed sites in Northern Ontario, Canada.
5 Conclusion
In the present study, soil chemistry and microbial communities in phytoremediated and untreated tailing areas were characterized. Only <15% of total metals were phytoavailable. The level of total microbial biomass was significantly higher in recently reclaimed sites compared to unreclaimed. Palmitic acid (18.1), palmitoleic acid (16.1), and palmitic acid (16:0) were the most prevalent of all the fatty acids identified in all the soil samples. The phytoremediated sites also exhibited significantly higher levels (p ≤ 0.05) of organic matter and enzymatic activity (microbial function) compared to unreclaimed areas. More importantly, the high level of metals such as copper, nickel, and iron did not affect microbial biomass. Acidiferrobacter, an acidophilic, thermotolerant and facultatively anaerobic was the most predominant genus in unreclaimed site characterized by extremely elevated level of iron (93,100 mg/kg) and higher content of Ni, Cu, and Ti compared to vegetated areas. Soil samples from reclaimed areas (planted with grasses) exhibited significantly higher β-glucosidase (BG), cellobiohydrolase (CBH), β-N-acetylglucosaminidase (NAGase), arylsulfatase (AS), acid phosphatase (AP), alkaline phosphatase (ALP), glycine aminopeptidase (GAP), and leucine aminopeptidase (LAP) activities compared to samples from unreclaimed area. Overall, newly developed soils at recent (Onaping Falls) can sustain diverse microbial communities and associated vegetations.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRP071853.
Author Contributions
KN: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Resources; Supervision; Validation; Visualization; Roles/Writing – original draft; Writing – review; editing. GS: Investigation; Methodology; Validation; Writing – review; editing. PB: Investigation; Methodology; Validation; Writing – review; editing. R N-K: Data curation; Formal analysis; Methodology; Validation; Writing – review; editing.
Funding
This study received funding from the Natural Sciences and Engineering Research Council of Canada (NSERC), Vale, and Sudbury Nickel Operations (Glencore Limited), Canada (Grant numbers NSERC-CRD-470535-14). The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Natural Sciences and Engineering Research Council of Canada (NSERC), Vale and Sudbury Integrated Nickel Operations (Glencore Limited) for their financial support. Thanks to MR DNA Molecular Research DNA laboratory (Shallowater, Texas, United States) for amplicon sequencing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.908633/full#supplementary-material
References
Acosta-Martínez, V., and Tabatabai, M. A. (2000). Enzyme Activities in a Limed Agricultural Soil. Biol. Fertil. Soils 31, 85–91. doi:10.1007/s003740050628
Barron, G. L. (2003). Predatory Fungi, Wood Decay, and the Carbon Cycle. Biodiversity 4 (1), 3–9. doi:10.1080/14888386.2003.9712621
Brockett, B. F. T., Prescott, C. E., and Grayston, S. J. (2012). Soil Moisture Is the Major Factor Influencing Microbial Community Structure and Enzyme Activities across Seven Biogeoclimatic Zones in Western Canada. Soil Biol. Biochem. 44, 9–20. doi:10.1016/j.soilbio.2011.09.003
Buyer, J. S., and Sasser, M. (2012). High Throughput Phospholipid Fatty Acid Analysis of Soils. Appl. Soil Ecol. 61, 127–130. doi:10.1016/j.apsoil.2012.06.005
Carter, M. R., and Gregorich, E. G. (Editors) (2008). Soil Sampling and Methods of Analysis. Boca Raton, FL: CRC Press, Taylor & Francis Group.
Chao, A. (1984). Nonparametric Estimation of the Number of Classes in a Population. Scand. J. Stat. 11, 265–270.
Cheng, F., Peng, X., Zhao, P., Yuan, J., Zhong, C., Cheng, Y., et al. (2013). Soil Microbial Biomass, Basal Respiration and Enzyme Activity of Main Forest Types in the Qinling Mountains. PLOS 8 (6), e67353. doi:10.1371/journal.pone.0067353
Chung, A. P., Coimbra, C., Farias, P., Francisco, R., Branco, R., Simão, F. V., et al. (2019). Tailings Microbial Community Profile and Prediction of its Functionality in Basins of Tungsten Mine. Sci. Rep. 9 (9), 19596. doi:10.1038/s41598-019-55706-6
Claassens, S., Van Rensburg, P. J. J., and Van Rensburg, L. (2006). Soil Microbial Community Structure of Coal Mine Discard Under Rehabilitation. Water Air Soil Pollut. 174, 355–366. doi:10.1007/s11270-006-9125-y
Clivot, H., Pagnout, C., Aran, D., Devin, S., Bauda, P., Poupin, P., et al. (2012). Changes in Soil Bacterial Communities Following Liming of Acidified Forests. Appl. Soil Ecol. 59, 116–123. doi:10.1016/j.apsoil.2011.09.010
Coggans, C. (1992). Hydrogeology and Geochemistry of the INCO Ltd., Copper Cliff, Ontario, Mine Tailings Impoundment. Waterloo: City of Waterloo.
De Vries, F. T., and Shade, A. (2013). Controls on Soil Microbial Community Stability Under Climate Change. Front. Microbiol. 4, 265. doi:10.3389/fmicb.2013.00265
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi:10.1128/aem.03006-05
Dimitriu, P. A., and Grayston, S. J. (2010). Relationship Between Soil Properties and Patterns of Bacterial β-Diversity Across Reclaimed and Natural Boreal Forest Soils. Microb. Ecol. 59, 563–573. doi:10.1007/s00248-009-9590-0
Ekenler, M., and Tabatabai, M. A. (2003). Responses of Phosphatases and Arylsulfatase in Soils to Liming and Tillage Systems. Z. Pflanzenernähr. Bodenk. 166, 281–290. doi:10.1002/jpln.200390045
Feketeová, Z., Hrabovský, A., and Šimkovic, I. (2021). Microbial Features Indicating the Recovery of Soil Ecosystem Strongly Affected by Mining and Ore Processing. Int. J. Environ. Res. Public Health 18, 1–16. doi:10.3390/ijerph18063240
Ghazouani, S., Béjaoui, Z., Spiers, G., Beckett, P., Gtari, M., and Nkongolo, K. K. (2020). Effects of Rhizobioaugmentation with N-Fixing Actinobacteria Frankia on Metal Mobility in Casuarina Glauca-Soil System Irrigated with Industrial Wastewater: High Level of Metal Exclusion of C. glauca. Water. Air. Soil Pollut. 231, 1–17. doi:10.1007/s11270-020-04783-9
Hallberg, K. B., Hedrich, S., and Johnson, D. B. (2011). Acidiferrobacter Thiooxydans, Gen. Nov. Sp. nov.; An Acidophilic, Thermo-Tolerant, Facultatively Anaerobic Iron- and Sulfur-Oxidizer of the Family Ectothiorhodospiraceae. Extremophiles 15, 271–279. doi:10.1007/s00792-011-0359-2
Ito, K., Ma, X., Azmi, N., Huang, H.-S., Fujii, M., and Yoshimoto, T. (2003). Novel Aminopeptidase Specific for Glycine from Actinomucor Elegans. Biosci. Biotechnol. Biochem. 67, 83–88. doi:10.1271/bbb.67.83
Ketudat Cairns, J. R., Mahong, B., Baiya, S., and Jeon, J.-S. (2015). β-Glucosidases: Multitasking, Moonlighting or Simply Misunderstood? Plant Sci. 241, 246–259. doi:10.1016/j.plantsci.2015.10.014
Khozhina, E. I., and Sherriff, B. (2006). Background Research of the Tailings Area of a Ni-Cu Mine for the Determination of an Optimal Method of Revegetation. Geology 80, 367–386.
Koleff, P., Gaston, K. J., and Lennon, J. J. (2003). Measuring Beta Diversity for Presence-Absence Data. J. Anim. Ecol. 72, 367–382. doi:10.1046/j.1365-2656.2003.00710.x
Kuczynski, J., Stombaugh, J., Walters, W. A., González, A., Caporaso, J. G., and Knight, R. (2011). Using QIIME to Analyze 16S rRNA Gene Sequences from Microbial Communities. Curr. Protoc. Bioinforma. Chapter 10, Unit–28. doi:10.1002/0471250953.bi1007s36
Kumar, B. M. (2013). Mining Waste Contaminated Lands: An Uphill Battle for Improving Crop Productivity. J. Degrad. Min. Lands Manag. 1, 43–50. doi:10.15243/jdmlm.2013.011.043
Kwon, M. J., Yang, J.-S., Lee, S., Lee, G., Ham, B., Boyanov, M. I., et al. (2015). Geochemical Characteristics and Microbial Community Composition in Toxic Metal-Rich Sediments Contaminated with Au-Ag Mine Tailings. J. Hazard. Mater. 296, 147–157. doi:10.1016/j.jhazmat.2015.04.035
Li, Y., Carraro, N., Yang, N., Liu, B., Xia, X., Feng, R., et al. (2018). Genomic Islands Confer Heavy Metal Resistance in Mucilaginibacter Kameinonensis and Mucilaginibacter Rubeus Isolated from a Gold/Copper Mine. Genes 9, 573. doi:10.3390/genes9120573
Li, Y., Jia, Z., Sun, Q., Zhan, J., Yang, Y., and Wang, D. (2016). Ecological Restoration Alters Microbial Communities in Mine Tailings Profiles. Sci. Rep. 6, 1–11. doi:10.1038/srep25193
Londry, K. L., and Sherriff, B. L. (2005). Comparison of Microbial Biomass, Biodiversity, and Biogeochemistry in Three Contrasting Gold Mine Tailings Deposits. Geomicrobiol. J. 22, 237–247. doi:10.1080/01490450590947797
Lottermoser, B. G. (2010). Mine Wastes: Characterization, Treatment and Environmental Impacts | Request PDF. 3rd ed. Berlin Heidelberg: Springer-Verlag.
Lupwayi, N. Z., Kanashiro, D. A., Eastman, A. H., and Hao, X. (2018). Soil Phospholipid Fatty Acid Biomarkers and β-Glucosidase Activities After Long-Term Manure and Fertilizer N Applications. Soil Sci. Soc. Am. J. 82, 343–353. doi:10.2136/sssaj2017.09.0340
Maheshwari, R., Bharadwaj, G., and Bhat, M. K. (2000). Thermophilic Fungi: Their Physiology and Enzymes. Microbiol. Mol. Biol. Rev. 64, 461–488. doi:10.1128/mmbr.64.3.461-488.2000
McKergow, M., Narendrula-Kotha, R., Beckett, P., and Nkongolo, K. K. (2021). Microbial Biomass and Activity Dynamics in Restored Lands in a Metal Contaminated Region. Ecotoxicol. 2021, 1–12. doi:10.1007/s10646-021-02464-9
Mendez, M. O., and Maier, R. M. (2008). Phytostabilization of Mine Tailings in Arid and Semiarid Environments—An Emerging Remediation Technology. Environ. Health Perspect. 116, 283. doi:10.1289/ehp.10608
Narendrula, R., Nkongolo, K. K., Beckett, P., and Spiers, G. (2013). Total and Bioavailable Metals in Two Contrasting Mining Regions (Sudbury in Canada and Lubumbashi in DR-Congo): Relation to Genetic Variation in Plant Populations. Chem. Ecol. 29, 111–127. doi:10.1080/02757540.2012.696617
Narendrula, R., and Nkongolo, K. K. (2015). Fatty Acids Profile of Microbial Populations in a Mining Reclaimed Region Contaminated with Metals: Relation with Ecological Characteristics and Soil Respiration. J. Bioremediation Biodegr. 6, 1–9. doi:10.4172/2155-6199.1000274
Narendrula-Kotha, R., Mehes-Smith, M., and Nkongolo, K. K. (2018). Critical Analysis of Remediation Methods of Metal Contaminated Lands. J. Bioremed. Biodegrad. 6 (1), 1–7. doi:10.12691/ijebb-6-1-1
Narendrula-Kotha, R., and Nkongolo, K. K. (2017a). Bacterial and Fungal Resilience to Long-Term Metal Exposure in a Mining Region Revealed by Metagenomics Sequencing. Ecol. Genet. Genomics 2, 12–24. doi:10.1016/j.egg.2016.11.001
Narendrula-Kotha, R., and Nkongolo, K. K. (2017b). Microbial Response to Soil Liming of Damaged Ecosystems Revealed by Pyrosequencing and Phospholipid Fatty Acid Analyses. PLoS One 12, e0168497. doi:10.1371/journal.pone.0168497
Narendrula-Kotha, R., and Nkongolo, K. K. (2017c). Changes in Enzymatic Activities in Metal Contaminated and Reclaimed Lands in Northern Ontario (Canada). Ecotoxicol. Environ. Saf. 140, 241–248. doi:10.1016/j.ecoenv.2017.02.040
Narendrula-Kotha, R., Theriault, G., Mehes-Smith, M., Kalubi, K., and Nkongolo, K. K. (2020). Metal Toxicity and Resistance in Plants and Microorganisms in Terrestrial Ecosystems. Rev. Environ. Contam. Toxicol. 249, 1–27.
Niu, J., Rang, Z., Zhang, C., Chen, W., Tian, F., Yin, H., et al. (2016). The Succession Pattern of Soil Microbial Communities and its Relationship with Tobacco Bacterial Wilt. BMC Microbiol. 16, 233. doi:10.1186/s12866-016-0845-x
Nkongolo, K. K., Michael, P., Theriault, G., Narendrula, R., Castilloux, P., Kalubi, K. N., et al. (2016). Assessing Biological Impacts of Land Reclamation in a Mining Region in Canada: Effects of Dolomitic Lime Applications on Forest Ecosystems and Microbial Phospholipid Fatty Acid Signatures. Water, Air, Soil Pollut. 227, 1–13. doi:10.1007/s11270-016-2803-5
Nkongolo, K. K., Spiers, G., Beckett, P., Narendrula, R., Theriault, G., Tran, A., et al. (2013). Long-Term Effects of Liming on Soil Chemistry in Stable and Eroded Upland Areas in a Mining Region. Water, Air, Soil Pollut. 224 (7), 1618. doi:10.1007/s11270-013-1618-x
Nkongolo, K. K., Spiers, G., Beckett, P., and Narendrula-Kotha, R. (2022). Inside Old Reclaimed Mine Tailings in Northern Ontario, Canada: A Microbial Perspective. Ecol. Genet. Genomics 23, 100118. doi:10.1016/j.egg.2022.100118
Roesch, L. F. W., Fulthorpe, R. R., Riva, A., Casella, G., Hadwin, A. K. M., Kent, A. D., et al. (2007). Pyrosequencing Enumerates and Contrasts Soil Microbial Diversity. ISME J. 1, 283–290. doi:10.1038/ismej.2007.53
Rousk, J., Brookes, P. C., and Bååth, E. (2009). Contrasting Soil pH Effects on Fungal and Bacterial Growth Suggest Functional Redundancy in Carbon Mineralization. Appl. Environ. Microbiol. 75 (6), 1589–1596. doi:10.1128/aem.02775-08
Saiya-Cork, K. R., Sinsabaugh, R. L., and Zak, D. R. (2002). The Effects of Long Term Nitrogen Deposition on Extracellular Enzyme Activity in an Acer Saccharum Forest Soil. Soil Biol. Biochem. 34, 1309–1315. doi:10.1016/s0038-0717(02)00074-3
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear Ribosomal Internal Transcribed Spacer (ITS) Region as a Universal DNA Barcode Marker for Fungi. Proc. Natl. Acad. Sci. U. S. A. 109, 6241–6246. doi:10.1073/pnas.1117018109
Sharma, P., Dutta, D., Udayan, A., Nadda, A. K., Lam, S. S., and Kumar, S. (2022). Role of Microbes in Bioaccumulation of Heavy Metals in Municipal Solid Waste: Impacts on Plant and Human Being. Environ. Pollut. 305. doi:10.1016/j.envpol.2022.119248
Sharma, P., Tripathi, S., and Chandra, R. (2020). Phytoremediation Potential of Heavy Metal Accumulator Plants for Waste Management in the Pulp and Paper Industry. Heliyon 6 (7). doi:10.1016/j.heliyon.2020.e04559
Sinsabaugh, R. L., Lauber, C. L., Weintraub, M. N., Ahmed, B., Allison, S. D., Crenshaw, C., et al. (2008). Stoichiometry of Soil Enzyme Activity at Global Scale. Ecol. Lett. 11, 1252–1264. doi:10.1111/j.1461-0248.2008.01245.x
Teruel, M., Ruíz-Ruano, F. J., Marchal, J. A., Sánchez, A., Cabrero, J., Camacho, J. P., et al. (2014). Disparate Molecular Evolution of Two Types of Repetitive DNAs in the Genome of the Grasshopper Eyprepocnemis Plorans. Heredity 112, 531–542. doi:10.1038/hdy.2013.135
Verchot, L., and Borelli, T. (2005). Application of -nitrophenol (NP) Enzyme Assays in Degraded Tropical Soils. Soil Biol. Biochem. 37, 625–633. doi:10.1016/j.soilbio.2004.09.005
Veres, Z., Kotroczó, Z., Fekete, I., Tóth, J. A., Lajtha, K., Townsend, K., et al. (2015). Soil Extracellular Enzyme Activities Are Sensitive Indicators of Detrital Inputs and Carbon Availability. Appl. Soil Ecol. 92, 18–23. doi:10.1016/j.apsoil.2015.03.006
Waldrop, M. P., Balser, T. C., and Firestone, M. K. (2000). Linking Microbial Community Composition to Function in a Tropical Soil. Soil Biol. Biochem. 32, 1837–1846. doi:10.1016/s0038-0717(00)00157-7
Wolińska, A., Włodarczyk, K., Kuźniar, A., Marzec-Grządziel, A., Grządziel, J., Gałązka, A., et al. (2020). Soil Microbial Community Profiling and Bacterial Metabolic Activity of Technosols as an Effect of Soil Properties Following Land Reclamation: A Case Study from the Abandoned Iron Sulphide and Uranium Mine in Rudki (South-Central Poland). Agronomy 10, 1795.
Xu, Z., Yu, G., Zhang, X., Ge, J., He, N., Wang, Q., et al. (2015). The Variations in Soil Microbial Communities, Enzyme Activities and Their Relationships With Soil Organic Matter Decomposition Along the Northern Slope of Changbai Mountain. Appl. Soil Ecol. 86, 19–29.
Xue, K., Zhou, J., Van Nostrand, J., Mench, M., Bes, C., Giagnoni, L., et al. (2018). Functional Activity and Functional Gene Diversity of a Cu-Contaminated Soil Remediated by Aided Phytostabilization Using Compost, Dolomitic Limestone and a Mixed Tree Stand. Environ. Pollut. 242, 229–238. doi:10.1016/j.envpol.2018.06.057
Keywords: mine tailings, northern ontario, metal contamination, microbial biomass and function, bacterial and fungal abundance and diversity
Citation: Nkongolo KK, Spiers G, Beckett P and Narendrula-Kotha R (2022) Effects of Phytoremediation on Microbial Biomass, Composition, and Function in a Sulphide-Rich Tailing From a Metal-Contaminated Region. Front. Environ. Sci. 10:908633. doi: 10.3389/fenvs.2022.908633
Received: 30 March 2022; Accepted: 15 June 2022;
Published: 22 July 2022.
Edited by:
Luiz Fernando Wurdig Roesch, University of Florida, United StatesReviewed by:
Pooja Sharma, National University of Singapore, SingaporeYunus Effendi, Leibniz University Hannover, Germany
Copyright © 2022 Nkongolo, Spiers, Beckett and Narendrula-Kotha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: K. K. Nkongolo, knkongolo@laurentian.ca
 K. K. Nkongolo
K. K. Nkongolo G. Spiers
G. Spiers P. Beckett
P. Beckett R. Narendrula-Kotha1
R. Narendrula-Kotha1