A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester
- 1Research and Innovation Centre, Fondazione Edmund Mach, San Michele all'Adige, Italy
- 2Chemical Ecology Unit, Department of Plant Protection Biology, Swedish University of Agricultural Sciences, Alnarp, Sweden
- 3Institute of Ecology and Environmental Sciences, UPMC Univ Paris 06, Sorbonne Universités, Paris, France
- 4INRA, Institute of Ecology and Environmental Sciences, Versailles, France
Plant volatiles mediate host discrimination and host finding in phytophagous insects. Understanding how insects recognize these signals is a current challenge in chemical ecology research. Pear ester, ethyl (E,Z)-2,4-decadienoate, is a powerful, bisexual attractant of codling moth Cydia pomonella (Lepidoptera, Tortricidae) and strongly synergizes the male response to female-produced sex pheromone. We show here that the codling moth odorant receptor (OR) CpomOR3 is dedicated to detecting this plant volatile. Heterologous expression of CpomOR3 in Drosophila T1 trichoid and ab3A basiconic sensilla, followed by a screening with codling moth pheromone compounds and known plant volatile attractants, confirms that CpomOR3 binds to pear ester. Although CpomOR3 does not respond to any of the pheromone components tested, a phylogenetic analysis of lepidopteran chemosensory receptor genes reveals a close relationship of CpomOR3 with pheromone receptors (PRs) in moths. This corroborates the interaction of ecological and social chemosensory cues during premating communication. The finding that a plant volatile compound, pear ester, is a specific ligand for a PR-like lepidopteran receptor adds to our understanding of insect-plant interactions and emphasizes the interaction of natural and sexual selection during the phylogenetic divergence of insect herbivores.
Introduction
Interactions between plants and insects shape many terrestrial ecosystems, and the primary mode of communication between plants and insects is chemical. Plant volatile chemicals mediate recognition of adult food sites, adequate oviposition sites and larval host plants (Bruce and Pickett, 2011) and accordingly play a prominent role in premating reproductive isolation and phylogenetic diversification of insect herbivores (Dres and Mallet, 2002; Smadja and Butlin, 2009; Matsubayashi et al., 2010). Decoding the plant volatile signatures that enable insects to discriminate between host and non-host plants is a long-standing research challenge in chemical ecology (Dethier, 1947, 1982; Ehrlich and Raven, 1964).
The identification of behaviorally active plant volatiles is a delicate and tedious task since plants release a large suite of volatiles, with no apparent correlation between the relative abundance of these compounds and their behavioral role in associated insects. Moreover, a behavioral response is frequently elicited by compound blends, where single compounds can often be exchanged with no apparent loss of activity (Bengtsson et al., 2006; Tasin et al., 2006, 2010; Pinero et al., 2008; Riffell et al., 2009; Cha et al., 2011; Schmidt-Busser et al., 2011; Thoming and Knudsen, 2014). This makes it particularly difficult to determine which plant volatiles encode host finding in phytophagous insects. In comparison, the identification of insect sex pheromones is facilitated by the production of few compounds in dedicated glands in one sex, together with a strong, distinctive behavioral response in the other.
The larvae of codling moth, Cydia pomonella (Lepidoptera, Tortricidae), feed on apple, pear, and walnut. The main sex pheromone compound codlemone, (E,E)-8,10-dodecadien-1-ol, was identified long ago (Roelofs et al., 1971; Beroza et al., 1974), but it is still open to question which compounds evoke attraction of egg-laying codling females to the plant host. Plant odorants obviously account for host attraction in codling moth, and several compounds from apple fruit and foliage elicit a strong antennal response. However, these compounds produce only a rather weak behavioral response (Bengtsson et al., 2001; Coracini et al., 2004; Hern and Dorn, 2004; Witzgall et al., 2005).
The strongest known kairomonal attractant is a pear ester, ethyl (E,Z)-2,4-decadienoate (Jennings et al., 1964; Berger and Drawert, 1984; Willner et al., 2013), which attracts codling moth adult males and females, as well as larvae (Knight and Light, 2001; Light et al., 2001; Light and Knight, 2005). This makes pear ester a versatile tool for sustainable insect control. It is used to monitor the seasonal abundance of codling moth (Knight and Light, 2012; Knight et al., 2013), as well as to enhance population control by mating disruption, in blends with codlemone (Knight et al., 2012). More recently, a microencapsulated formulation of pear ester has been developed for disruption of larval orientation and host finding (Light and Beck, 2012; Knight and Light, 2013).
Pear ester has been identified by screening codling moth antennal response to a wide range of apple and pear volatiles, followed by field trapping (Light et al., 2001; Light and Knight, 2005). Its biological significance is, however, not entirely clear, since it is found mainly in pear and only in some apple cultivars (Jennings et al., 1964; Berger and Drawert, 1984; Willner et al., 2013). The association of codling moth with cultivated apple is, on the other hand, recent and the response to pear ester may stem from an evolutionarily ancient host plant of codling moth.
Given the difficulties associated with completely assessing the pool of plant volatiles produced by the various host plants of codling moth, it is sensible to also investigate the response of single odorant receptors (ORs), many of which are likely dedicated to the perception of plant volatiles. ORs interface insects with their odor environment by binding odorants, and are expressed in olfactory sensory neurons (OSNs), which transmit olfactory information to the brain. The number of ORs expressed on the antenna and their compound-specificity determines the range of odorants an insect can detect. General ORs are tuned to environmental odors including plant volatiles, while pheromone receptors (PRs), a male-biased receptor clade, respond mainly to sex pheromones (Jacquin-Joly and Merlin, 2004; Ihara et al., 2013; Leal, 2013).
An emerging technique, which is quickly becoming an integral part of the toolbox for identification of behaviorally relevant plant odorants, is the functional characterization (“deorphanization”) of ORs, following expression in heterologous expression systems. The OR repertoire of Drosophila has been studied exhaustively (Hallem et al., 2004; Kreher et al., 2005; Hallem and Carlson, 2006) and current research aims at other insect groups. For moths, a number of ORs and PRs have been identified and functionally characterized, using various heterologous expression systems, including human embryonic kidney (HEK) cells (Grosse-Wilde et al., 2007), Xenopus oocytes (Sakurai et al., 2004; Nakagawa et al., 2005; Jiang et al., 2014), Sf9, a cell line derived from fall armyworm Spodoptera frugiperda ovaries (Jordan et al., 2009), and Drosophila OSNs (Syed et al., 2010; Montagné et al., 2012), which is an in vivo antennal expression approach.
Expressing ORs in single Drosophila neurons comprises two main advantages. The biochemical environment of Drosophila OSNs endogenously provides odorant binding proteins (OBPs) and Orco, a canonical receptor conserved across insects (Krieger et al., 2003; Jones et al., 2005; Leal, 2013), which may enhance response sensitivity and specificity of the expressed OR, compared with non-insect cell lines. In addition, electrophysiological techniques, namely single sensillum recordings (SSRs) are well established for Drosophila sensilla.
Two main systems are available for expression and deorphanization of ORs in Drosophila OSNs, the “empty neuron” (ab3A) in ab3 basiconic sensilla, which lacks its native OR (Dobritsa et al., 2003) and the Or67dGAL4 knock-in mutant line in trichoid T1 sensilla (Kurtovic et al., 2007). While the empty neuron system has been used mainly to functionally characterize general odorant receptors, pheromone receptors may respond more strongly when expressed in T1 rather than in ab3A (Syed et al., 2010; Montagné et al., 2012).
We have previously identified 43 candidate OR protein sequences in the antennal transcriptome of codling moth, five of which cluster within the conserved pheromone receptor clade of lepidopteran PRs (Bengtsson et al., 2012).
We here show that CpomOR3, belonging to the PR clade, is strictly tuned to pear ester. This result emphasizes the biological significance of pear ester (Light et al., 2001) and shows that the PR clade contains co-evolving receptors for sex pheromones and for host odorants. This corroborates the modulation of male sexual behavior by host plant odorants in codling moth (Trona et al., 2010, 2013), and adds to our understanding of the evolution of sexual communication and olfaction-driven speciation in insect herbivores.
Methods
Insects, Dissection, and RNA Extraction
Cydia pomonella pupae were obtained from a laboratory rearing center (Andermatt Biocontrol, Grossdietwil, Switzerland), and adults were allowed to emerge in cages kept at 23°C, 70 ± 5% relative humidity and a 16 h:8 h light:dark cycle, and fed with 10% sugar solution. For dissections, 2–3 day old female and male insects were used. Using sharp forceps, antennae were removed at the base of the pedicel, and legs at the coxa. For thorax samples, head, wings, legs, and abdomen were removed. Wings were removed at their base, and the abdomen removed at the connection to the thorax. All body parts were immediately flash-frozen using liquid nitrogen, and thereafter kept at −80°C. RNAs were extracted using the RNeasy kit (Qiagen, Hilden, Germany).
Rapid Amplification of cDNA Ends (RACE)-PCR
RACE-PCR was performed to obtain the complete open reading frame (ORF) for CpomOR3. A cDNA library for extension in the 5′ direction was created using the SMARTer kit (Clontech, Mountain View, CA, USA) on male antennal RNA. For the PCR reaction, the Advantage 2 kit (Clontech) was used, with a temperature program of 95°C for 2 min, then 30 cycles of 95°C for 1 min, 65°C for 90 s, 68°C for 2 min and a final elongation of 68°C for 7 min. A gene-specific primer (5′-CCCTAGAGCTTCGGTGTCCAATGTAGAGC-3′) was used together with the Universal primer mix (Clontech). The PCR product was analyzed by electrophoresis on an agarose gel, and the relevant band excised and purified by the Gel extraction kit (Qiagen). It was then cloned into the pGEM®-T Easy plasmid (Promega, Fitchburg, WI, USA), with which TOP10 cells were transformed (Invitrogen, Life Technologies, Carlsbad, CA, USA). Plasmids were subsequently purified using the Miniprep kit (Qiagen). Purified plasmids were quantified by nanodrop (Nanodrop 8000 UV-Vis Spectrophotometer, Thermo Scientific, Wilmington, DE, USA) and then Sanger sequenced (3730xl Applied Biosystems, Life Technologies) using the forward and reverse M13 universal primers. Transmembrane domains were predicted using TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), on sequence translated to protein using ExPASy (http://web.expasy.org/translate/).
Phylogenetic Analysis
Amino acid sequences of CpomORs clustering in the candidate PR clade (Bengtsson et al., 2012) were included in a dataset together with sequences of candidate PRs from the following Lepidoptera: Antheraea polyphemus (Forstner et al., 2009), Bombyx mori (Nakagawa et al., 2005), Danaus plexippus (Zhan et al., 2011), Diaphania indica (Mitsuno et al., 2008), Epiphyas postvittana (Jordan et al., 2009), Heliconius melpomene (Heliconius Genome Consortium, 2012), Helicoverpa armigera (Liu et al., 2012), Heliothis virescens (Grosse-Wilde et al., 2007; Wang et al., 2010), Manduca sexta (Grosse-Wilde et al., 2010), Mythimna separata (Mitsuno et al., 2008), Ostrinia furnacalis (Miura et al., 2010; Leary et al., 2012), O. nubilalis (Wanner et al., 2010; Leary et al., 2012), O. scapulalis (Miura et al., 2009, 2010), Plutella xylostella (Mitsuno et al., 2008; Sun et al., 2013), Spodoptera exigua (Liu et al., 2013) and S. littoralis (Legeai et al., 2011; Montagné et al., 2012). Sequences from B. mori (BmorOR6) and H. melpomene (HmelOR5, 6, and 7) were also included in the dataset as external groups, since they belong to the sister group to the PR clade (Poivet et al., 2013). The CpomOR1 sequence was not included in the dataset because of its short length (only 101 amino acid residues). The 74 amino acid sequences were aligned using the online version of MAFFT v.7 (Katoh and Standley, 2013), with the G-INS-i algorithm (Katoh et al., 2005) and default parameters.
Phylogenetic reconstruction was performed using the maximum likelihood method. The LG+I+G+F substitution model (Le and Gascuel, 2008) was determined as the best-fit model of protein evolution by ProtTest 2.4 (Abascal et al., 2005) following Akaike information criterion. Rate heterogeneity was set at four categories, and the gamma distribution parameter and the proportion of invariable sites were estimated from the dataset. Tree reconstruction was performed using PhyML 3.0 (Guindon et al., 2010), with both SPR (Subtree Pruning and Regrafting) and NNI (Nearest Neighbor Interchange) methods for tree topology improvement. Node support was estimated using a bootstrap procedure based on 100 replicates, and nodes supported by a bootstrap value below 70% were collapsed. The figure was created using the iTOL web server (Letunic and Bork, 2011) and Adobe Illustrator.
Reverse Transcription (RT)-PCR for CpomoR3 Expression Analysis
cDNAs were synthesized from RNAs extracted from different tissues using the RT-for-PCR kit (Clontech), following the recommended protocol. Integrity of cDNAs was tested by PCR, using degenerate primers for RPL8 (Forward primer 5′-GAGTCATCCGAGCTCARMGNAARGG-3′; Reverse primer 5′-CCAGCAGTTTCGCTTNACYTTRTA-3′) and GoTaq Green Master Mix (Promega) with an annealing temperature of 54°C. PCR reactions to screen for expression of CpomOR3 in different tissues used GoTaq Green Master Mix, and consisted of an initial 5-min step at 94°C, and then 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, and a final 7-min step at 72°C. Gene specific primers (GSP) for CpomOR3, 5′-AGATGAAGAGTATCGGAATTGCATGG-3′ (forward) and 5′-CCAACTGGGATCATGCCACAAGC-3′ (reverse), were used, giving a product of 436 bp. Product identity was confirmed by direct sequencing, following gel extraction (QIAquick Gel Extraction Kit, Qiagen). Each PCR reaction was repeated three times and control consisted of a no template PCR. PCR was performed in parallel on C. pomonella genomic DNA templates, extracted from larvae using PureLink Genomic DNA kit (Invitrogen). No amplification or amplification of larger size bands was observed, revealing specific cDNA amplification at the expected size. Products were analyzed on a 1.5% agarose gel and visualized after staining with ethidium bromide using a Gel Doc XR (Bio-Rad, Hercules, CA, USA).
Heterologous Expression of Putative ors in Drosophila melanogaster
The complete ORF encoding CpomOR3 was amplified by PCR (forward primer 5′-ATGTTTAGTTATGAAAATGAAGACAGC-3′, reverse primer 5′-TCAAGTCATTTCTTCAGTAGAGGT-3′), with antennal cDNA created by the RT-for-PCR kit (Invitrogen) as a template. The purified PCR product was then cloned into the PCR8/GW/TOPO plasmid (Invitrogen). The cassette with the insert was then transferred from the TOPO/GW/PCR8 plasmid to the destination vector (pUASg-HA.attB, constructed by E. Furger and J. Bischof, kindly provided by the Basler group, Zürich), using the Gateway LR Clonase II kit (Invitrogen). The integrity and orientation of the insert was confirmed by sequencing. A transformant UAS-CpomOR3 line was generated by BestGene (Chino Hills, CA, USA), using the PhiC31 integrase system. Briefly, recombinant pUASg-HA.attB-CpomOR3 plasmids were injected into embryos of a D. melanogaster line containing an attP insertion site within the second chromosome (genotype y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP'}ZH-51C), leading to non-random integration. To drive expression of CpomOR3 in OSNs housed in T1 sensilla, the transformant UAS-CpomOR3 line was crossed to the Or67dGAL4 strain (kindly provided by Barry Dickson) to generate a double homozygous line w+;UAS-CpomOR3;Or67dGAL4. To verify insertion of the UAS-CpomOR3 construct into the genome, gDNA was extracted and used as template in PCR with primers for the full ORF of CpomOR3.
Additionally, to compare the similarity of results between expression sites (trichoid and basiconic sensilla) male flies with the genotype w;UAS-CpomOR3/CyO;+/+ were mate paired with female flies of the genotype w;delta-Halo/Cyo;Dmel-UAS-OR22a-Gal4. This cross drove ectopic expression of CpomOR3 in the A neuron of the ab3 sensilla, which also expressed the endogenous DmelOR22a receptor in the same neuron. SSR recordings in parental flies from the cross confirmed the absence of any response from DmelOR22a to pear ester (data not shown).
Single Sensillum Recordings
The D. melanogaster line expressing CpomOR3 in T1 OSNs, along with the flies expressing CpomOR3 in ab3A OSNs were tested by SSRs. In all cases, flies were restrained as described in Stensmyr et al. (2003). Briefly, flies were immobilized in 100 μl pipette tips with only the top half of the head protruding. The left antenna was pushed onto a piece of double-adhesive tape, and held firm by a capillary pressing down from above. Sensilla were contacted with tungsten electrodes (diameter 0.12 mm, Harvard Apparatus Ltd, Edenbridge, United Kingdom) electrolytically sharpened in a saturated KNO3 solution. A DC-3K micromanipulator equipped with a PM-10 piezo translator (Märzhäuser Wetzler GmbH, Wetzler, Germany) was used to gently maneuver the recording electrode into the base of a sensillum. The reference electrode was inserted through the eye using a DC-3K Rachts PM-10 piezo micromanipulator (Märzhäuser Wetzler GmbH, Wetzler, Germany). The signal from the OSNs was registered and amplified 10 times with a probe (INR-02, Syntech, Hilversum, the Netherlands), and transferred to a computer through an IDAC-4-USB (Syntech) interface, where it was visualized and analyzed with the software Autospike v. 3.4 (Syntech). A constant flow of 0.65 m/s of charcoal-filtered and humidified air was delivered through a glass tube with its outlet approximately 15 mm from the antenna. Stimuli were presented to the insect by inserting a stimulus pipette through a hole in the glass tube, and blowing an air puff of 2.5 ml during 0.5 s through the pipette into the air stream, using a stimulus controller (Syntech SFC-1/b).
Synthetic Compounds and Odor Stimuli
An array of pheromone compounds for C. pomonella and related species (Witzgall et al., 1996), as well as known pheromone synergists (El-Sayed, 2014), were tested on CpomOR3 (Table 1). Combinations of the C. pomonella main pheromone compound, codlemone, with the synergists were also tested, as they have previously been shown to create distinct activation patterns in the antennal lobe, the primary olfactory center, compared to either compound alone (Trona et al., 2013). Purity of compounds was estimated by GC-MS.
Stimuli were prepared by applying compounds to 1.5 × 1 cm pieces of filter paper that were placed in disposable glass Pasteur pipettes (VWR International, Stockholm, Sweden). Truncated 1 ml pipette tips were put on the wide end of the Pasteur pipettes, to reduce evaporation of the test compound(s). Compounds were diluted in hexane (redistilled from 95%, Lab-scan, Dublin, Ireland). A volume of 10 μl of a 1 μg/μl solution was applied to filter papers for a total amount of 10 μg per stimulus. The same dilution procedure was used in dose-response experiments, except that compounds were diluted to concentrations ranging from 0.1 ng/μl to 10 μg/μl in decadic steps, to achieve different concentrations when 10 μl of the diluted compound were applied to the filter paper in the stimulus pipette. Control stimuli with only solvent were also prepared. Fresh stimuli were prepared before each recording session, and kept at −18°C until the start of the recording session, to avoid evaporation. Only complete recording sessions of the entire set of test stimuli were evaluated, and only one screening or dose response session was performed from a single sensillum per individual. A total of 16 screenings were performed, while for dose response experiments, 10 replicates were performed.
Responses were quantified by counting the number of spikes for 500 ms starting from the onset of response (as determined by the earliest response for the recording session), subtracting the number of spikes during the 500 ms before response, and doubling this value to get the response in Hz (spikes/s). Responses of T1 sensilla to different pheromone and pheromone synergist compounds were compared using ANOVA with repeated measures, while responses to different doses of pear ester with the two types of sensilla evaluated were compared with Two-Way ANOVA. All statistical analyses were performed using SPSS Version 19.0 (IBM Corp., Armonk, NY, USA).
Results
Cloning of the Open Reading Frame of CpomoR3 and Sequence Analysis
The partial CpomOR3 sequence (Bengtsson et al., 2012), judged to be complete at the 3′ end based on the presence of a stop codon, but not at the 5′ end, was extended by 5′ RACE-PCR. Merging the sequence of the 1096 bp 5′RACE-PCR product we obtained together with the previous sequence led to a 1281 bp transcript, containing the complete ORF of CpomOR3, confirmed by alignment of the deduced protein with other lepidopteran ORs. The full ORF sequence for CpomOR3 was further amplified and sequenced to verify the absence of chimera. The full sequence has been submitted to Genbank (accession number KJ420588). The TMHMM2.0 model predicted 6 transmembrane domains for CpomOR3. CpomOR3 exhibits a mean sequence identity of 34.3% with other PRs, with a maximum identity of 41.4% with Diaphania indica OR1. Alignment with lepidopteran candidate PRs did not reveal any notable feature of CpomOR3, apart from a serine residue—also present in other tortricid sequences—located within the final transmembrane domain (position 296), instead of the glycine residue found in all the other lepidopteran PR sequences.
Phylogeny of Lepidopteran Candidate PRs
A maximum likelihood phylogeny was built from a large dataset containing CpomOR3 to 6—the putative C. pomonella PRs (Bengtsson et al., 2012)—and 70 other candidate PR full-length sequences. In this tree (Figure 1), the candidate PRs grouped within five large sub-clades within the PR clade. All the sequences from tortricid moths (C. pomonella and E. postvittana, green branches), including CpomOR3, clustered within one of these five clades (supported by a bootstrap value of 80), albeit the exact relationships between CpomOR3 and the other receptors of this clade were not resolved due to low bootstrap support values (to reflect lack of support, nodes with a bootstrap value lower than 70 were collapsed). Even if the CpomOR1 sequence was not part of this dataset because of its short length, it also clustered in the same clade during previous analyses, as a sister group to EposOR1 (data not shown). All the PR candidates from C. pomonella characterized to date thus have a relatively recent common origin, in spite of their low sequence identity levels.
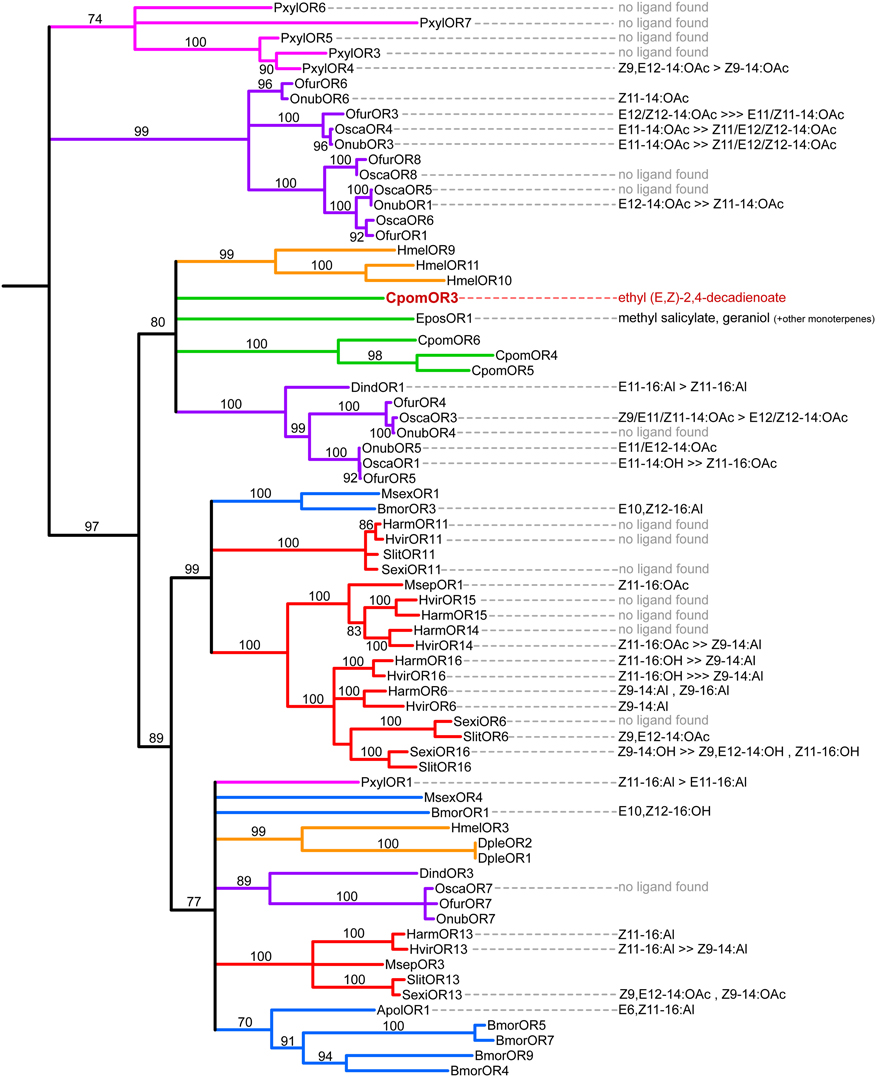
Figure 1. Maximum likelihood tree of lepidopteran candidate PRs, highlighting their corresponding ligand(s). Branch colors represent different lepidopteran lineages: blue for Bombycoidea, red for Noctuoidea, orange for Papilionoidea, purple for Pyraloidea, green for Tortricoidea, and pink for Yponomeutoidea. The outgroup (containing BmorOR6 and HmelOR5, 6, and 7) has been removed from the figure. Functional data has been compiled from the literature (see references in the Methods section). “No ligand found”: OR did not respond to any tested pheromone component. Cpom, Cydia pomonella, Apol, Antheraea polyphemus, Bmor, Bombyx mori, Dple, Danaus plexippus, Dind, Diaphania indica, Epos, Epiphyas postvittana, Hmel, Heliconius melpomene, Harm, Helicoverpa armigera, Hvir, Heliothis virescens, Msex, Manduca sexta, Msep, Mythimna separata, Ofur, Ostrinia furnacalis, Onub, O. nubilalis, Osca, O. scapulalis, Pxyl, Plutella xylostella, Sexi, Spodoptera exigua, Slit, S. littoralis.
Tissue-Related Expression of CpomoR3
Reverse transcription PCR showed a clear expression pattern for CpomOR3, with strong expression in antennae, but not in other body parts (Figure 2). Moreover, there was no sex-specific expression of CpomOR3, as it appeared to be expressed in antennae of both males and females.
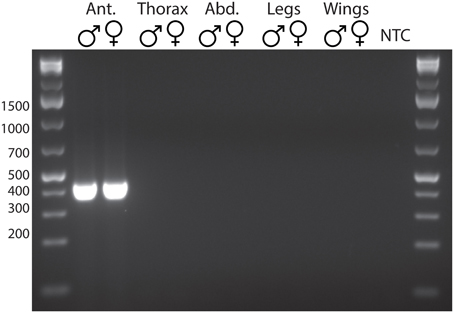
Figure 2. Reverse transcription PCR showing antennal specific expression of C. pomonella OR3 in both sexes. Ant., antennae, Abd., abdomen, NTC, no template control.
Response Spectrum of CpomoR3 to Putative Ligands
Single-sensillum recordings from transformed Drosophila line expressing CpomOR3 in T1 OSNs revealed that these neurons only responded to pear ester (41 spikes/s, N = 16) out of 15 compounds. Six different mixtures of different combinations of pheromone components and plant compounds were also tested, and only the one that contained pear ester and codlemone elicited a significant response (Figure 3). No synergy between these two compounds was observed (Bonferroni post-hoc test). Dose response experiments established the threshold of response to pear ester to be at 10 μg for both trichoid T1 and ab3A OSNs (Figure 4).
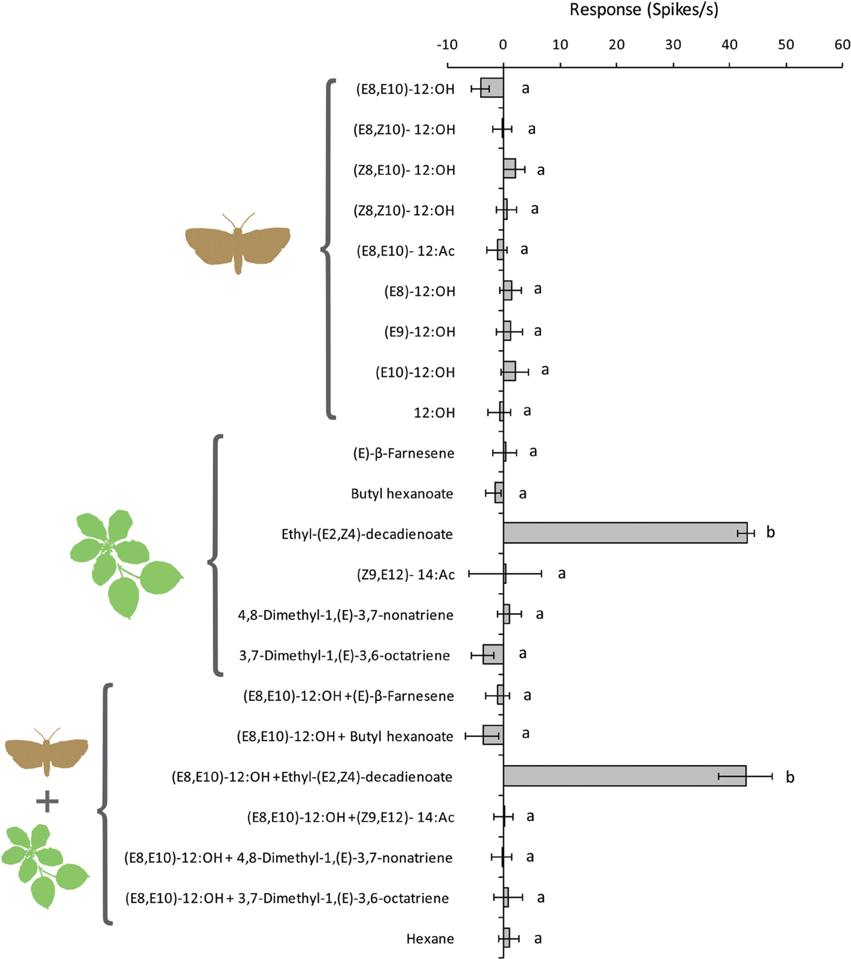
Figure 3. Response (in Hz) of D. melanogaster T1 OSNs expressing CpomOR3 to stimulation with known C. pomonella pheromone components, plant-related synergists, and combinations of the main pheromone component codlemone, with plant synergists. Letters denote subgroups with a statistically significant separation (Repeated measures ANOVA with Bonferroni post-hoc test, n = 16).
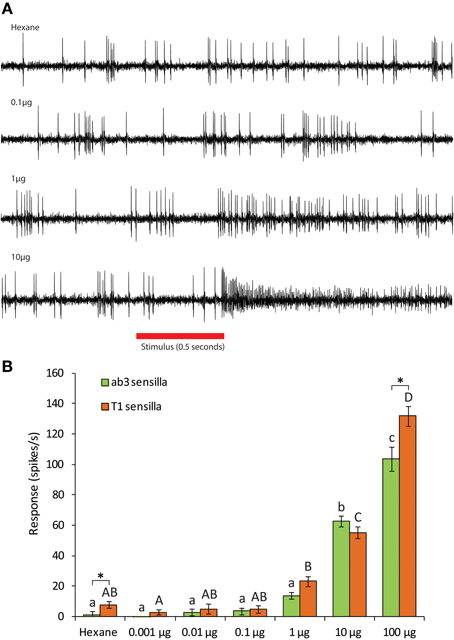
Figure 4. (A) Traces of single sensillum recordings from D. melanogaster T1 sensilla expressing CpomOR3 to pear ester at different doses. (B) Dose-dependent response of CpomOR3 to pear ester in different types of sensilla. Bars of the same color followed by different letters indicate subgroups with statistically significant differences. Asterisks denote significant differences among different types of sensilla for the dose indicated (Two-Way ANOVA with Tukey's post-hoc test, p < 0.05, n = 10).
Discussion
CpomoR3 is Tuned to the Plant Volatile Pear Ester
Electrophysiological recordings from Drosophila basiconic ab3 and trichoid T1 sensilla, housing OSNs heterologously expressing CpomOR3, demonstrate that CpomOR3 is tuned to pear ester, ethyl (E,Z)-2,4-decadienoate (Figures 3, 4). Reverse transcription PCR suggests that CpomOR3 is expressed without sex bias in the antennae of both males and females (Figure 2). This finding matches the behavioral evidence, since pear ester is a bisexual codling moth attractant (Light et al., 2001; Light and Knight, 2005). The existence of a dedicated receptor corroborates the significance of pear ester for host plant detection in codling moth males and females, and contributes to current research aiming at a complete identification of codling moth host plant attractants.
Results from these heterologous expression studies confirm previous recordings obtained from codling moth antennae, showing presence of OSNs responding to pear ester (De Cristofaro et al., 2004; Ansebo et al., 2005). However, a spatially tight arrangement of sensilla on codling moth antennae renders it difficult to obtain replicated recordings from the same sensillum type, and to differentiate between responses from co-localized OSNs in the same sensillum, or even from OSNs in adjacent sensilla (Lee and Baker, 2008). This further demonstrates the appreciable addition of heterologous OR expression in Drosophila to the toolbox for identification of behaviorally relevant plant odorants.
Intracellular recordings of axons of OSNs projecting to the antennal lobe (AL), the olfactory center of the insect brain, and functional imaging of AL glomeruli, receiving input from OSNs expressing the same ORs, support our finding that pear ester activates a dedicated olfactory channel and that interaction of pear ester with other compounds, including the sex pheromone codlemone, takes place in the AL, and not at the periphery (Figure 3; Trona et al., 2010, 2013).
CpomoR3 Belongs to the Pheromone Receptor Clade
CpomOR3 belongs to the conserved clade of lepidopteran pheromone receptors (Figure 1), although it binds to pear ester only and to none of the pheromonal compounds produced by C. pomonella females or closely related Cydia species (Witzgall et al., 1996, 2001). CpomOR3 was almost equally sensitive when expressed in trichoid T1 and basiconic ab3 sensilla, except at the highest dose of pear ester (Figure 4). Interestingly, the pheromone receptors BmorOR1 of silkmoth B. mori and SlitOR6 of cotton leafworm moth S. littoralis were more sensitive when expressed in T1 than in ab3 sensilla (Syed et al., 2010; Montagné et al., 2012). This indicates that T1 sensilla, containing an important PR partner, the sensory neuron membrane protein (Benton et al., 2007), are more adapted for correct PR functioning, whereas plant odorant ORs function equally well in T1 or ab3.
In addition, the demonstration that an OR clustering in the PR clade is a plant odorant receptor offers an explanation for the lack of a response of orphan lepidopteran PRs to pheromone compounds (Wang et al., 2010; Liu et al., 2013). Phylogenetic analysis confirms that the lepidopteran PR clade contains another co-evolved receptor for plant compounds, EposOR1, from another tortricid species, the light brown apple moth E. postvittana. The strongest ligand for EposOR1 is a common plant compound, methyl salicylate (Jordan et al., 2009), which has a behavioral effect in many insects (Figure 1; El-Sayed, 2014). With the currently available sequence and functional data, phylogenetic analysis cannot resolve if EposOR1 and CpomOR3 have a single ancestor, or if two unique evolutionary events gave rise to these plant volatile receptors within the PR clade (Figure 1). However, both CpomOR3 and EposOR1 belong to the same clade, which notably also contains the four other C. pomonella candidate PRs (Bengtsson et al., 2012; Garczynski et al., 2012). Further studies, using both pheromones and plant volatiles, will help to understand the functional divergence of the PR clade.
Interaction Between Pear Ester and Codling Moth Pheromone
The finding that a codling moth PR is tuned to pear ester is remarkable. It corroborates the interaction between pear ester and codlemone, which may play an important role in codling moth premating communication and reproductive isolation (Trona et al., 2013).
Axons of OSNs expressing the same OR or PR genes converge onto the same glomerulus in the antennal lobe (AL). Since each OR corresponds to a glomerulus in the AL, it follows that new glomeruli arise during OR repertoire expansion. Indeed, closely related ORs with high sequence similarity are often expressed in OSNs that project to neighboring glomeruli in the AL (Couto et al., 2005; Masse et al., 2009; Ramdya and Benton, 2010; Cande et al., 2013).
Accordingly, the architecture of the codling moth AL lends support to the hypothesis that the OR genes for pear ester and codlemone, the codling moth sex pheromone, are closely related—the glomeruli dedicated to pear ester and codlemone are adjacent glomeruli in the codling moth AL, where stimulation with a blend of codlemone and pear ester produces a very strong synergistic effect (Trona et al., 2010, 2013). Although the PR for codlemone has not yet been found, we can reasonably assume that it belongs to the PR clade, which contains the putative pheromone receptors CpomOR1, and CpomOR4 through 6 (Figure 1; Bengtsson et al., 2012).
Chemosensory receptor genes arise by gene duplication and progressively diverge following adaptive changes. In Drosophila, phylogenetically related chemosensory genes on a chromosome tend to be located closely together on a chromosome (Nei et al., 2008; Sanchez-Gracia et al., 2009). Physically neighboring chemosensory genes restrict genetic recombination and thus become a combined target for selection. Tight physical linkage between host performance and preference genes, leading to assortative mating through habitat choice, has been first discovered in pea aphids (Hawthorne and Via, 2001; Smadja et al., 2012). Key traits that are associated via linkage and which combine ecological and sexual selection are particularly powerful during phylogenetic divergence (Servedio et al., 2011; Merrill et al., 2012; Safran et al., 2013).
In codling moth, chemosensory receptor genes encoding host preference and mate recognition, tuned to the plant volatile pear ester and sex pheromone, are expected to be associated to facilitate host adaptation and reproductive isolation in concert. This hypothesis can be tested after the receptor gene for codlemone has been found.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi: 10.1093/bioinformatics/bti263
Ansebo, L., Ignell, R., Löfqvist, J., and Hansson, B. S. (2005). Responses to sex pheromone and plant odours by olfactory receptor neurons housed in sensilla auricillica of the codling moth, Cydia pomonella (Lepidoptera: Tortricidae). J. Insect Physiol. 51, 1066–1074. doi: 10.1016/j.jinsphys.2005.05.003
Bengtsson, J. M., Trona, F., Montagné, N., Anfora, G., Ignell, R., Witzgall, P., et al. (2012). Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS ONE 7:e31620. doi: 10.1371/journal.pone.0031620
Bengtsson, M., Bäckman, A.-C., Liblikas, I., Ramirez, M. I., Borg-Karlson, A.-K., Ansebo, L., et al. (2001). Plant odor analysis of apple: antennal response of codling moth females to apple volatiles during phenological development. J. Agric. Food Chem. 49, 3736–3741. doi: 10.1021/jf0100548
Bengtsson, M., Jaastad, G., Knudsen, G., Kobro, S., Bäckman, A.-C., Pettersson, E., et al. (2006). Plant volatiles mediate attraction to host and non-host plant in apple fruit moth, Argyresthia conjugella. Entomol. Exp. Appl. 118, 77–85. doi: 10.1111/j.1570-7458.2006.00359.x
Benton, R., Vannice, K. S., and Vosshall, L. B. (2007). An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450, 289–293. doi: 10.1038/nature06328
Berger, R. G., and Drawert, F. (1984). Changes in the composition of volatiles by post-harvest application of alcohols to red delicious apples. J. Sci. Food Agric. 35, 1318–1325. doi: 10.1002/jsfa.2740351208
Beroza, M., Bierl, B. A., and Moffitt, H. R. (1974). Sex pheromones: (E,E)-8,10-dodecadien-1-ol in the codling moth. Science 183, 89–90. doi: 10.1126/science.183.4120.89
Bruce, T. J. A., and Pickett, J. A. (2011). Perception of plant volatile blends by herbivorous insects—finding the right mix. Phytochemistry 72, 1605–1611. doi: 10.1016/j.phytochem.2011.04.011
Cande, J., Prud'homme, B., and Gompel, N. (2013). Smells like evolution: the role of chemoreceptor evolution in behavioral change. Curr. Opin. Neurobiol. 23, 152–158. doi: 10.1016/j.conb.2012.07.008
Cha, D. H., Linn, C. E., Teal, P. E. A., Zhang, A. J., Roelofs, W. L., and Loeb, G. M. (2011). Eavesdropping on plant volatiles by a specialist moth: significance of ratio and concentration. PLoS ONE 6:e17033. doi: 10.1371/journal.pone.0017033
Coracini, M., Bengtsson, M., Liblikas, I., and Witzgall, P. (2004). Attraction of codling moth males to apple volatiles. Entomol. Exp. Appl. 110, 1–10. doi: 10.1111/j.0013-8703.2004.00124.x
Couto, A., Alenius, M., and Dickson, B. J. (2005). Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 15, 1535–1547. doi: 10.1016/j.cub.2005.07.034
De Cristofaro, A., Ioriatti, C., Pasqualini, E., Anfora, G., Germinara, G. S., Villa, M., et al. (2004). Electrophysiological responses of Cydia pomonella (L.) to codlemone and pear ester ethyl (E,Z)-2,4-decadienoate: peripheral interactions in their perception and evidences for cells responding to both compounds. Bull. Insectol. 57, 137–144.
Dethier, V. G. (1982). Mechanism of host plant recognition. Entomol. Exp. Appl. 31, 49–56. doi: 10.1111/j.1570-7458.1982.tb03118.x
Dobritsa, A. A., van der Goes van Naters, W., Warr, C. G., Steinbrecht, R. A., and Carlson, J. R. (2003). Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841. doi: 10.1016/S0896-6273(03)00094-1
Dres, M., and Mallet, J. (2002). Host races in plant-feeding insects and their importance in sympatric speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 471–492. doi: 10.1098/rstb.2002.1059
Ehrlich, P., and Raven, P. (1964). Butterflies and plants: a study in coevolution. Evolution 18, 586–608. doi: 10.2307/2406212
El-Sayed, A. M. (2014). The Pherobase: Database of Pheromones and Semiochemicals. Available online at: http://www.pherobase.com
Forstner, M., Breer, H., and Krieger, J. (2009). A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int. J. Biol. Sci. 5:745. doi: 10.7150/ijbs.5.745
Garczynski, S. F., Wanner, K. W., and Unruh, T. R. (2012). Identification and initial characterization of the 3′ end of gene transcripts encoding putative members of the pheromone receptor subfamily in Lepidoptera. Insect Sci. 19, 64–74. doi: 10.1111/j.1744-7917.2011.01423.x
Grosse-Wilde, E., Gohl, T., Bouche, E., Breer, H., and Krieger, J. (2007). Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci. 25, 2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x
Grosse-Wilde, E., Stieber, R., Forstner, M., Krieger, J., Wicher, D., and Hansson, B. S. (2010). Sex-specific odorant receptors of the tobacco hornworm Manduca sexta. Front. Cell. Neurosci. 4:22. doi: 10.3389/fncel.2010.00022
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hallem, E. A., and Carlson, J. R. (2006). Coding of odors by a receptor repertoire. Cell 125, 143–160. doi: 10.1016/j.cell.2006.01.050
Hallem, E. A., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the Drosophila Antenna. Cell 117, 965–979. doi: 10.1016/j.cell.2004.05.012
Hawthorne, D. J., and Via, S. (2001). Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412, 904–907. doi: 10.1038/35091062
Heliconius Genome Consortium. (2012). Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98. doi: 10.1038/nature11041
Hern, A., and Dorn, S. (2004). A female-specific attractant for the codling moth, Cydia pomonella, from apple fruit volatiles. Naturwissenschaften 91, 77–80. doi: 10.1007/s00114-003-0484-6
Ihara, S., Yoshikawa, K., and Touhara, K. (2013). Chemosensory signals and their receptors in the olfactory neural system. Neuroscience 254, 45–60. doi: 10.1016/j.neuroscience.2013.08.063
Jacquin-Joly, E., and Merlin, C. (2004). Insect olfactory receptors: contributions of molecular biology to chemical ecology. J. Chem. Ecol. 30, 2359–2397. doi: 10.1007/s10886-004-7941-3
Jennings, W. G., Heinz, D. E., and Creveling, R. K. (1964). Volatile esters of Bartlett pear. IV. Esters of trans-2-cis-4-decadienoic acid. J. Food Sci. 29, 730–734. doi: 10.1111/j.1365-2621.1964.tb00439.x
Jiang, X.-J., Guo, H., Di, C., Yu, S., Zhu, L., Huang, L.-Q., et al. (2014). Sequence similarity and functional comparisons of pheromone receptor orthologs in two closely related Helicoverpa species. Insect Biochem. Mol. Biol. 48, 63–74. doi: 10.1016/j.ibmb.2014.02.010
Jones, W. D., Nguyen, T. A. T., Kloss, B., Lee, K. J., and Vosshall, L. B. (2005). Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr. Biol. 15, R119–R121. doi: 10.1016/j.cub.2005.02.007
Jordan, M. D., Anderson, A., Begum, D., Carraher, C., Authier, A., Marshall, S. D. G., et al. (2009). Odorant receptors from the light brown apple moth (Epiphyas postvittana) recognize important volatile compounds produced by plants. Chem. Senses 34, 383–394. doi: 10.1093/chemse/bjp010
Katoh, K., Kuma, K.-I., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Knight, A., Light, D., and Chebny, V. (2013). Monitoring codling moth (Lepidoptera: Tortricidae) in orchards treated with pear ester and sex pheromone combo dispensers. J. Appl. Entomol. 137, 214–224. doi: 10.1111/j.1439-0418.2012.01715.x
Knight, A. L., and Light, D. M. (2001). Attractants from Bartlett pear for codling moth, Cydia pomonella (L.), larvae. Naturwissenschaften 88, 339–342. doi: 10.1007/s001140100244
Knight, A. L., and Light, D. M. (2012). Monitoring codling moth (Lepidoptera: Tortricidae) in sex pheromone-treated orchards with (E)-4,8-dimethyl-1,3,7-nonatriene or pear ester in combination with codlemone and acetic acid. Environ. Entomol. 41, 407–414. doi: 10.1603/EN11310
Knight, A. L., and Light, D. M. (2013). Adding microencapsulated pear ester to insecticides for control of Cydia pomonella (Lepidoptera: Tortricidae) in apple. Pest Manag. Sci. 69, 66–74. doi: 10.1002/ps.3363
Knight, A. L., Stelinski, L. L., Hebert, V., Gut, L., Light, D., and Brunner, J. (2012). Evaluation of novel semiochemical dispensers simultaneously releasing pear ester and sex pheromone for mating disruption of codling moth (Lepidoptera: Tortricidae). J. Appl. Entomol. 136, 79–86. doi: 10.1111/j.1439-0418.2011.01633.x
Kreher, S. A., Kwon, J. Y., and Carlson, J. R. (2005). The molecular basis of odor coding in the Drosophila larva. Neuron 46, 445–456. doi: 10.1016/j.neuron.2005.04.007
Krieger, J., Klink, O., Mohl, C., Raming, K., and Breer, H. (2003). A candidate olfactory receptor subtype highly conserved across different insect orders. J. Comp. Physiol. A 189, 519–526. doi: 10.1007/s00359-003-0427-x
Kurtovic, A., Widmer, A., and Dickson, B. J. (2007). A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446, 542–546. doi: 10.1038/nature05672
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Leary, G. P., Allen, J. E., Bunger, P. L., Luginbill, J. B., Linn, C. E., Macallister, I. E., et al. (2012). Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U.S.A. 109, 14081–14086. doi: 10.1073/pnas.1204661109
Lee, S. G., and Baker, T. C. (2008). Incomplete electrical isolation of sex-pheromone responsive olfactory receptor neurons from neighboring sensilla. J. Insect Physiol. 54, 663–671. doi: 10.1016/j.jinsphys.2008.01.006
Legeai, F., Malpel, S., Montagné, N., Monsempes, C., Cousserans, F., Merlin, C., et al. (2011). An expressed sequence tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC Genomics 12:86. doi: 10.1186/1471-2164-12-86
Letunic, I., and Bork, P. (2011). Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478. doi: 10.1093/nar/gkr201
Light, D. M., and Beck, J. J. (2012). Behavior of codling moth (Lepidoptera: Tortricidae) neonate larvae on surfaces treated with microencapsulated pear ester. Environ. Entomol. 41, 603–611. doi: 10.1603/EN11273
Light, D. M., and Knight, A. (2005). Specificity of codling moth (Lepidoptera: Tortricidae) for the host plant kairomone, ethyl (2E,4Z)-2,4-decadienoate: field bioassays with pome fruit volatiles, analogue, and isomeric compounds. J. Agric. Food Chem. 53, 4046–4053. doi: 10.1021/jf040431r
Light, D. M., Knight, A. L., Henrick, C. A., Rajapaska, D., Lingren, B., Dickens, J. C., et al. (2001). A pear-derived kairomone with pheromonal potency that attracts male and female codling moth, Cydia pomonella (L.). Naturwissenschaften 88, 333–338. doi: 10.1007/s001140100243
Liu, C., Liu, Y., Walker, W. B., Dong, S., and Wang, G. (2013). Identification and functional characterization of sex pheromone receptors in beet armyworm Spodoptera exigua (Hübner). Insect Biochem. Mol. Biol. 43, 747–754. doi: 10.1016/j.ibmb.2013.05.009
Liu, Y., Gu, S., Zhang, Y., Guo, Y., and Wang, G. (2012). Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PLoS ONE 7:e48260. doi: 10.1371/journal.pone.0048260
Masse, N. Y., Turner, G. C., and Jefferis, G. S. X. E. (2009). Olfactory information processing in Drosophila. Curr. Biol. 19, R700–R713. doi: 10.1016/j.cub.2009.06.026
Matsubayashi, K. W., Ohshima, I., and Nosil, P. (2010). Ecological speciation in phytophagous insects. Entomol. Exp. Appl. 134, 1–27. doi: 10.1111/j.1570-7458.2009.00916.x
Merrill, R. M., Wallbank, R. W. R., Bull, V., Salazar, P. C. A., Mallet, J., Stevens, M., et al. (2012). Disruptive ecological selection on a mating cue. Proc. Biol. Sci. 279, 4907–4913. doi: 10.1098/rspb.2012.1968
Mitsuno, H., Sakurai, T., Murai, M., Yasuda, T., Kugimiya, S., Ozawa, R., et al. (2008). Identification of receptors of main sex pheromone components of three Lepidopteran species. Eur. J. Neurosci. 28, 893–902. doi: 10.1111/j.1460-9568.2008.06429.x
Miura, N., Nakagawa, T., Tatsuki, S., Touhara, K., and Ishikawa, Y. (2009). A male-specific odorant receptor conserved through the evolution of sex pheromones in Ostrinia moth species. Int. J. Biol. Sci. 5:319. doi: 10.7150/ijbs.5.319
Miura, N., Nakagawa, T., Touhara, K., and Ishikawa, Y. (2010). Broadly and narrowly tuned odorant receptors are involved in female sex pheromone reception in Ostrinia moths. Insect Biochem. Mol. Biol. 40, 64–73. doi: 10.1016/j.ibmb.2009.12.011
Montagné, N., Chertemps, T., Brigaud, I., Francois, A., Francois, M. C., de Fouchier, A., et al. (2012). Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur. J. Neurosci. 36, 2588–2596. doi: 10.1111/j.1460-9568.2012.08183.x
Nakagawa, T., Sakurai, T., Nishioka, T., and Touhara, K. (2005). Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307, 1638–1642. doi: 10.1126/science.1106267
Nei, M., Niimura, Y., and Nozawa, M. (2008). The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 9, 951–963. doi: 10.1038/nrg2480
Pinero, J. C., Galizia, C. G., and Dorn, S. (2008). Synergistic behavioural responses of female oriental fruit moths (Lepidoptera: Tortricidae) to syntetic host plant-derived mixtures are mirrored by odor-evoked calcium activity in their antennal lobes. J. Insect Physiol. 54, 333–343. doi: 10.1016/j.jinsphys.2007.10.002
Poivet, E., Gallot, A., Montagné, N., Glaser, N., Legeai, F., and Jacquin-Joly, E. (2013). A comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis. PLoS ONE 8:e60263. doi: 10.1371/journal.pone.0060263
Ramdya, P., and Benton, R. (2010). Evolving olfactory systems on the fly. Trends Genet. 26, 307–316. doi: 10.1016/j.tig.2010.04.004
Riffell, J. A., Lei, H., and Hildebrand, J. G. (2009). Neural correlates of behavior in the moth Manduca sexta in response to complex odors. Proc. Natl. Acad. Sci. U.S.A. 106, 19219–19226. doi: 10.1073/pnas.0910592106
Roelofs, W. L., Comeau, A., Hill, A., and Milicevic, G. (1971). Sex attractant of the codling moth: characterization with electroantennogram technique. Science 174, 297–299. doi: 10.1126/science.174.4006.297
Safran, R. J., Scordato, E. S. C., Symes, L. B., Rodriguez, R. L., and Mendelson, T. C. (2013). Contributions of natural and sexual selection to the evolution of premating reproductive isolation: a research agenda. Trends Ecol. Evol. 28, 643–650. doi: 10.1016/j.tree.2013.08.004
Sakurai, T., Nakagawa, T., Mitsuno, H., Mori, H., Endo, Y., Tanoue, S., et al. (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 101, 16653–16658. doi: 10.1073/pnas.0407596101
Sanchez-Gracia, A., Vieira, F. G., and Rozas, J. (2009). Molecular evolution of the major chemosensory gene families in insects. Heredity 103, 208–216. doi: 10.1038/hdy.2009.55
Schmidt-Busser, D., von Arx, M., Connetable, S., and Guerin, P. M. (2011). Identification of host-plant chemical stimuli for the European grape berry moth Eupoecilia ambiguella. Physiol. Entomol. 36, 101–110. doi: 10.1111/j.1365-3032.2010.00766.x
Servedio, M. R., Van Doorn, G. S., Kopp, M., Frame, A. M., and Nosil, P. (2011). Magic traits in speciation: “magic” but not rare? Trends Ecol. Evol. 26, 389–397. doi: 10.1016/j.tree.2011.04.005
Smadja, C., and Butlin, R. K. (2009). On the scent of speciation: the chemosensory system and its role in premating isolation. Heredity 102, 77–97. doi: 10.1038/hdy.2008.55
Smadja, C. M., Canback, B., Vitalis, R., Gautier, M., Ferrari, J., Zhou, J. J., et al. (2012). Large-scale candidate gene scan reveals the role of chemoreceptor genes in host plant specialization an speciation in the pea aphid. Evolution 66, 2723–2738. doi: 10.1111/j.1558-5646.2012.01612.x
Stensmyr, M. C., Dekker, T., and Hansson, B. S. (2003). Evolution of the olfactory code in the Drosophila melanogaster subgroup. Proc. Biol. Sci. 270, 2333–2340. doi: 10.1098/rspb.2003.2512
Sun, M., Liu, Y., Walker, W. B., Liu, C., Lin, K., Gu, S., et al. (2013). Identification and characterization of pheromone receptors and interplay between receptors and pheromone binding proteins in the diamondback moth, Plutella xyllostella. PLoS ONE 8:e62098. doi: 10.1371/journal.pone.0062098
Syed, Z., Kopp, A., Kimbrell, D. A., and Leal, W. S. (2010). Bombykol receptors in the silkworm moth and the fruit fly. Proc. Natl. Acad. Sci. U.S.A. 107, 9436–9439. doi: 10.1073/pnas.1003881107
Tasin, M., Bäckman, A.-C., Anfora, G., Carlin, S., Ioriatti, C., and Witzgall, P. (2010). Attraction of female grapevine moth to common and specific olfactory cues from 2 host plants. Chem. Senses 35, 57–64. doi: 10.1093/chemse/bjp082
Tasin, M., Bäckman, A.-C., Bengtsson, M., Varela, N., Ioriatti, C., and Witzgall, P. (2006). Wind tunnel attraction of grapevine moth females, Lobesia botrana, to natural and artificial grape odour. Chemoecology 16, 87–92. doi: 10.1007/s00049-005-0332-6
Thoming, G., and Knudsen, G. K. (2014). Attraction of pea moth Cydia nigricana to pea flower volatiles. Phytochemistry 100, 66–75. doi: 10.1016/j.phytochem.2014.01.005
Trona, F., Anfora, G., Balkenius, A., Bengtsson, M., Tasin, M., Knight, A., et al. (2013). Neural coding merges sex and habitat chemosensory signals in an insect herbivore. Proc. Biol. Sci. 280:20130267. doi: 10.1098/rspb.2013.0267
Trona, F., Anfora, G., Bengtsson, M., Witzgall, P., and Ignell, R. (2010). Coding and interaction of sex pheromone and plant volatile signals in the antennal lobe of the codling moth Cydia pomonella. J. Exp. Biol. 213, 4291–4303. doi: 10.1242/jeb.047365
Wang, G., Vásquez, G., Schal, C., Zwiebel, L., and Gould, F. (2010). Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect Mol. Biol. 20, 125–133. doi: 10.1111/j.1365-2583.2010.01045.x
Wanner, K. W., Nichols, A. S., Allen, J. E., Bunger, P. L., Garczynski, S. F., Linn, C. E., et al. (2010). Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS ONE 5:e8685. doi: 10.1371/journal.pone.0008685
Willner, B., Granvogl, M., and Schieberle, P. (2013). Characterization of the key aroma compounds in Bartlett pear brandies by means of the sensomics concept. J. Agric. Food Chem. 61, 9583–9593. doi: 10.1021/jf403024t
Witzgall, P., Ansebo, L., Yang, Z., Angeli, G., Sauphanor, B., and Bengtsson, M. (2005). Plant volatiles affect oviposition by codling moths. Chemoecology 15, 77–83. doi: 10.1007/s00049-005-0295-7
Witzgall, P., Bengtsson, M., Rauscher, S., Liblikas, I., Bäckman, A.-C., Coracini, M., et al. (2001). Identification of further sex pheromone synergists in the codling moth, Cydia pomonella. Entomol. Exp. Appl. 101, 131–141. doi: 10.1046/j.1570-7458.2001.00898.x
Witzgall, P., Chambon, J.-P., Bengtsson, M., Unelius, C. R., Appelgren, M., Makranczy, G., et al. (1996). Sex pheromones and attractants in the Eucosmini and Grapholitini (Lepidoptera, Tortricidae). Chemoecology 7, 13–23. doi: 10.1007/BF01240633
Keywords: olfaction, odorant receptor, heterologous expression, semiochemical, sex pheromone, plant volatile, insect control
Citation: Bengtsson JM, Gonzalez F, Cattaneo AM, Montagné N, Walker WB, Bengtsson M, Anfora G, Ignell R, Jacquin-Joly E and Witzgall P (2014) A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front. Ecol. Evol. 2:33. doi: 10.3389/fevo.2014.00033
Received: 20 May 2014; Paper pending published: 09 June 2014;
Accepted: 19 June 2014; Published online: 08 July 2014.
Edited by:
Sharon Rose Hill, Swedish University of Agricultural Sciences, SwedenReviewed by:
Maria Carolina Blassioli Moraes, Embrapa Genetic Resources and Biotechnology, BrazilRonald Jason Pitts, Vanderbilt University, USA
Copyright © 2014 Bengtsson, Gonzalez, Cattaneo, Montagné, Walker, Bengtsson, Anfora, Ignell, Jacquin-Joly and Witzgall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonas M. Bengtsson, Department of Zoology, Stockholm University, Svante Arrhenius v. 18b, 106 91 Stockholm, Sweden e-mail: jonas.bengtsson@zoologi.su.se
 Jonas M. Bengtsson
Jonas M. Bengtsson Francisco Gonzalez
Francisco Gonzalez Alberto M. Cattaneo
Alberto M. Cattaneo Nicolas Montagné
Nicolas Montagné William B. Walker
William B. Walker Marie Bengtsson
Marie Bengtsson Gianfranco Anfora
Gianfranco Anfora Rickard Ignell
Rickard Ignell Emmanuelle Jacquin-Joly
Emmanuelle Jacquin-Joly Peter Witzgall
Peter Witzgall