The basic genetic toolkit to move in with your photosynthetic partner
- 1Biology Department, University of New Brunswick, Fredericton, NB, Canada
- 2Integrated Microbiology Program, Canadian Institute for Advanced Research, Toronto, ON, Canada
The origin of photosynthetic organelles via endosymbiosis more than 1 Gya ago was a major detonator of eukaryotic diversification. The evolution of a stable endosymbiotic relationship between eukaryotic cells and photosynthetic cyanobacteria involved series of cellular and molecular processes that are not entirely understood. Critical steps toward the evolution of plastids occurred when the host cell gained genetic and metabolic control over the captured cyanobacterium. Proteins recruited from the host repertoire had major roles initiating the metabolite exchange between both symbiotic partners. Concurrently, the relocation of certain cyanobacterial genes into the host nuclear genome was critical to coordinate the division of the endosymbiotic cells and the transit of nuclear-encoded proteins into the novel organelle. This review explores diverse studies that have identified key “endosymbiosis genes” and discusses the putative roles of the encoded proteins during the early evolution of plastids. The understanding of the regulation mechanisms and functions of the “endosymbiosis genes” will shed light on the design of genetic engineering approaches to facilitate endosymbiotic associations.
Introduction
Photosynthesis is the generic name for different photoautotrophic pathways (i.e., use of light energy and inorganic substrates to produce organic compounds) that vary depending on the types of light-harvesting systems, photosynthetic pigments, electron donors (e.g., H2O, H2, S2, H2S, S2) and released byproducts (e.g., O2, S2, H2S) (Bryant and Frigaard, 2006; Blankenship, 2010). The oxygen producing (i.e., oxygenic) photosynthesis likely evolved first in the ancestors of modern cyanobacteria between 3.8 and 2.8 Gya ago (Bryant and Frigaard, 2006; Buick, 2008; Blankenship, 2010). Geochemical evidence indicate that oxygenic photosynthesis became significant at planetary scale 2.5 Gya ago (Kopp et al., 2005; Buick, 2008), and since then it has been one of the major processes of primary production on Earth and pivotal for the evolution of Life (Farquhar et al., 2011).
Circa 2 Gya after the appearance of the oxygenic photosynthesis some eukaryotes acquired the photosynthetic metabolism through the establishment of a “permanent” endosymbiotic relationship with cyanobacteria (Hedges et al., 2004; Yoon et al., 2004). A widely accepted scenario suggests that those endosymbiotic cyanobacteria evolved into the photosynthetic organelles (i.e., primary plastids) of the first photoautotrophic eukaryotes, which presumably are the ancestors of the Archaeplastida supergroup (Palmer, 2003; Bhattacharya et al., 2004). Following the evolution of the first photosynthetic eukaryotes photosynthesis spread to diverse groups via numerous eukaryote-eukaryote (i.e., secondary and tertiary) endosymbioses (Palmer, 2003; Bhattacharya et al., 2004).
Escaping Digestion
The transformation of free-living cells into endosymbionts involves a series of foundational steps. Initially, the captured photosynthetic cells must survive the host digestive processes (Callieri, 2002; Xinyao et al., 2006). Engulfed cells are usually stored in food vacuoles and digested shortly after (Callieri, 2002; Gu et al., 2002; Xinyao et al., 2006; Kodama and Fujishima, 2012), but in certain cases the captured cells avoid digestion and survive metabolically active for some periods of time (Kodama and Fujishima, 2005, 2014; Escalera et al., 2011). Survival of digestion may occur by preventing lysosome fusion to the food vacuole, resistance to the hydrolytic enzymes of lysosomes, or escaping from the digestive vacuole into the host cytosol (Matz and Kjelleberg, 2005; Kodama et al., 2007, 2011).
Retention of viable captured cells is an obvious key step for the establishment of permanent endosymbiosis (Kodama and Fujishima, 2012, 2014), but another crucial event is the establishment of a steady flux of metabolites between the photosynthetic endosymbiont and the host cytoplasm (Weber et al., 2006; Reyes-Prieto et al., 2007). The stable exchange of compounds must have been critical as well for the evolution of metabolic interdependencies (e.g., complementary biochemical pathways) between the novel partners (Figure 1B). What molecular mechanisms may have facilitated the flux of metabolites during early stages of organelle evolution?
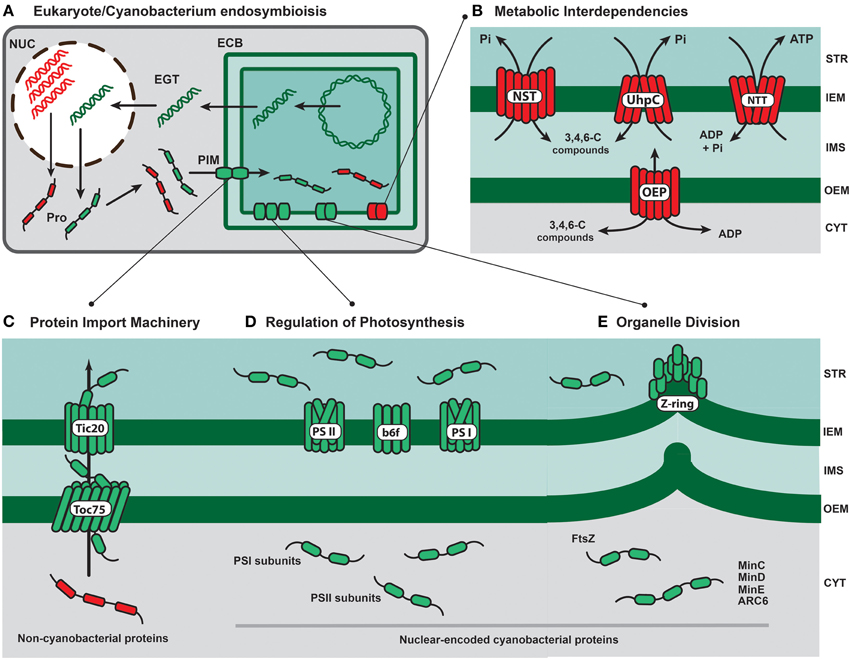
Figure 1. Key subcellular events and proteins involved in the evolution of primary photosynthetic organelles. (A) The genome reduction of the endosymbiotic cyanobacteria (ECB) was triggered by both gene losses via deleterious events and endosymbiotic gene transfer (EGT) into the host nucleus (NUC). Successfully transferred genes (green double strands) should have been expressed and translated in the host nucleocytoplasm system. Cytoplasm-translated proteins (Pro) of non-cyanobacterial origin (red ribbons) and cyanobacterial provenance (green ribbons) were imported into the cyanobacterial compartment via the ancestral protein import machinery (PIM). (B) The recruitment of cytoplasm-translated membrane transporters of host (e.g., members of the NST family and hexose transporters of the UhpC type) and diverse bacterial (e.g., the Chlamydiae-like ATP/ADP nucleotide translocator; NTT) sources was essential for the establishment of steady metabolite (e.g., 3-, 4-, or 6-carbon compounds; ATP) fluxes and further evolution of metabolic interdependencies. OEP represents diverse host-derived pore proteins (e.g., OEP24, OEP16 and OEP21) that facilitate the transport of a broad range of metabolites across the outer envelope membrane. (C) The evolution of the core plastid protein import system relied on the activity of the cyanobacterial proteins Toc75 and Tic20. These two pore-forming proteins were present originally in the plastid ancestor genome but the encoding genes were likely transferred early during endosymbiosis to the host genome (Tic20 are still plastid-encoded in some algal lineages). The ancestral organelle import system was able to translocate both cyanobacterial (green ribbons) and non-cyanobacterial (red ribbons) proteins translated in the host cytoplasm. (D) Other transferred cyanobacterial genes with putative key roles on plastid evolution were those encoding subunits of the Photosytems I (psaE, psaI, psaK) and II (psbO, psbP, and psbU) with important roles for the photosynthetic activity under stress conditions. The cytochrome b6f complex (b6f) is indicated. (E) Initial host control over the endosymbiont division likely relied on transferred cyanobacterial genes encoding cytokinesis-associated proteins, such as FtsZ, which is a tubulin-like protein fundamental to assemble the contractile Z-ring. The protein FtsZ is nuclear-encoded in most eukaryotes bearing plastids. Other cyanobacterial-derived genes frequently present in nuclear genomes encode the key plastid-division proteins ARC6 MinC, MinD, and MinE. Key code: STR (plastid stroma); IEM (inner envelope membrane); IMS (intermembrane space); OEM (outer envelope membrane); CYT (host cytoplasm). Red and green rounded rectangles represent non-cyanobacterial and cyanobacterial-derived proteins, respectively.
Sharing Resources with the Photosynthetic Partner
Photosynthetic organelles of plants and algae are bound by at least two membranes (e.g., the outer envelope membrane, OEM, and the inner envelope membrane, IEM, of primary plastids), with numerous examples of organelles delimited by three or four membranous layers (McFadden, 1999; Van Dooren et al., 2001). The membranes surrounding recently captured photosynthetic cells are significant physical barriers for the flux of metabolites between the potential endosymbiont and the host cytoplasm. The recruitment of membrane proteins able to translocate carbon compounds produced by the cyanobacterial photosynthesis was a critical step for the metabolic integration of the host and the endosymbionts, allowing the interconnection of the metabolite pools of both cells (Weber et al., 2006; Tyra et al., 2007; Weber and Linka, 2011).
It has been suggested that the diverse plastid-localized antiporters of the nucleotide-sugar/triose phosphate translocator (NST) family, which are essential to mobilize different carbon compounds (e.g., phosphorylated 3-, 5-, and 6-carbon sugars) across the plastid inner membrane, evolved from a “generalist” transporter selected from the host endomembrane system to extract carbohydrates from the endosymbiont (Weber et al., 2006; Facchinelli and Weber, 2011) (Figure 1B). Interestingly, alternative solutions may have evolved to take advantage of the endosymbiont photosynthates and different membrane transporters have been recruited to translocate metabolites from the photosynthetic compartment. For example, the glaucophyte Cyanophora paradoxa lacks plastid antiporters of the NST family, and UhpC-type transporters (Uhp: uptake of hexose phosphate) are possibly responsible for the carbohydrate extraction (Price et al., 2012) (Figure 1B). Other plastid-localized transporters that regulate the export and import of diverse metabolites, such as 2-oxoglutarate, glucose-6-P, ribose-5-P, pyruvate, and tetrahydrofolate, evolved from proteins encoded in the host genome. The evidence suggests that a basic set of metabolite membrane transporters has been fundamental to highjack photosynthetic endosymbionts and initiate the metabolic integration (Tyra et al., 2007; Facchinelli and Weber, 2011).
Besides the translocation of photosynthates into the host cytoplasm, the metabolic integration must require also the flux of minerals, vitamins and nutrients into the endosymbiont. Inner plastid membranes harbor transporters that import substrates from the host pool, such as tetrahydrofolate (purine biosynthesis), ammonia (nitrogen fixation network), phosphoenolpyruvate (aromatic amino acid biosynthesis), and S-adenosylmethionine (methylation reactions) (Bräutigam et al., 2012). Among these diverse plastid membrane transporters, the ATP/ADP translocator (NTT) (Reiser et al., 2004), which imports ATP into the organelle in exchange for ADP, was likely a key player recruited from third-party sources (Chlamydiae bacteria; Linka et al., 2003) to provide the captured cyanobacteria with high-energy molecules to drive reactions during dark periods (Neuhaus et al., 1997) (Figure 1B).
A central event to facilitate the recruitment of host proteins for organelle functions is the relocation (i.e., import) of proteins translated in the host cytoplasm into the endosymbiotic compartment. What do we know about the ancestral mechanisms of protein import underlining the evolution of primary photosynthetic organelles?
Redecoration of the Endosymbiotic Compartment
Relocating Proteins: A Small Step for a Cell, One Big Leap for Organelle Origins
The import of cytoplasm-translated proteins into plastids depends on the activity of the TOC/TIC machinery, which is composed of two multimeric translocation channels localized in the organelle membranes: the TOC (translocon at the outer membrane) and the TIC (translocon at the inner membrane) complexes. Diverse comparative analyses suggest that the ancestral TOC/TIC machinery comprised just few key proteins that were able to translocate proteins into the endosymbiotic compartment (Steiner et al., 2005; Kalanon and McFadden, 2008; Bodył et al., 2009; Gross and Bhattacharya, 2009b).
The ancestral TOC/TIC system likely included the receptor/pore protein Toc75 (a member of the Omp85 superfamily) (Hsu et al., 2008; Day et al., 2014; Richardson et al., 2014) and the pore-forming protein Tic20 (Kikuchi et al., 2013; Nakai, 2015) (Figure 1C). Other TOC/TIC subunits, such as the receptors/GTPases Toc33/34 and Toc159, the presumed pore-forming Tic110, the plastid-encoded Tic214 (Ycf1) were incorporated later during plastid evolution in some eukaryote groups (Li and Chiu, 2010; Nakai, 2015; De Vries et al., 2015). The phylogenetic history of the TOC/TIC core subunits indicates that Toc75 and Tic20 were recruited from the cyanobacterial genetic repertoire, whereas other pieces of later addition, such as Tic 110, Toc33/34, and Toc159, are of non-cyanobacterial origin (Kalanon and McFadden, 2008; Gross and Bhattacharya, 2009b; Shi and Theg, 2013). Regardless of the increased molecular complexity (i.e., gaining of additional subunits) observed during evolution of the TOC/TIC machinery in some lineages, it seems that cyanobacterial proteins were the pivotal elements to form the primary membrane channels able to import proteins from the host cytoplasm (Figure 1C). But how did the primordial TOC/TIC machinery evolve?
A plausible scenario is that a gene encoding a protein of the Omp85 family (the ancestral Toc75) was transferred from the cyanobacterial genome into the host nucleus. Subsequently, the Omp85-like protein was translated in the host cytoplasm and relocated via the host endomembrane system (e.g., nuclear envelope, endoplasmic reticulum, Golgi apparatus, vesicle trafficking) into the cyanobacterial OEM (Gross and Bhattacharya, 2009a; Day et al., 2014). The evolution of the protein import function of the TOC pore involved an opposite topological orientation of Toc75 with respect to the orientation of the original Omp85-like protein in the cyanobacterial OEM (Steiner et al., 2005; Sommer et al., 2011). The ancestral TIC pore was likely assembled with the Tic20 protein, which has the ability to form a multimeric membrane channel (Kovács-Bogdán et al., 2011; Nakai, 2015).
Organelle import systems have evolved and been remodeled several times during eukaryote history (e.g., peroxisomes, mitochondria, and the diverse serial gains of plastids) (see Kunau et al., 2001; Wickner and Schekman, 2005; Sheiner and Striepen, 2013), but the case of the filose amoeba Paulinella chromatophora, which harbors cyanobacterial-derived organelles (i.e., chromatophores) that evolved independently from the plastids of plants and algae (Marin et al., 2005, 2007), has offered unique insights into the evolution of protein import systems in organelles of endosymbiotic origin. The chromatophores of Paulinella are able to import proteins translated in the cytoplasm, probably using both elements of the host endomembrane system and pore-forming proteins localized in the endosymbiont envelope (Mackiewicz et al., 2012; Nowack, 2014).
Overall, phylogenetic and functional data suggest that in principle the import of unfolded proteins into an endosymbiotic compartment can be achieved with a relatively simple set of pore-forming proteins and the participation of the host endomembrane system.
The Plastid Proteome is a Phylogenetic Mosaic
Plastid genomes contain, at the most, 210 protein-coding genes (Wang et al., 2013), but the collection of plastid-localized proteins, which varies among different biological groups, comprises between 2500 and 4000 proteins (Van Wijk and Baginsky, 2011). Therefore, hundreds of plastid proteins encoded in the nuclear genome have to be imported into the photosynthetic organelle via the TOC/TIC machinery (Keegstra and Cline, 1999; Leister, 2003) (Figure 1A). Remarkably, only between 600 and 1700 of the plastid-localized proteins are of cyanobacterial provenance (Price et al., 2012; Dagan et al., 2013), and a considerable proportion of the organelle protein collection has been recruited from the host repertoire and diverse bacterial sources during plastid evolution (Suzuki and Miyagishima, 2010; Qiu et al., 2013; Schönknecht et al., 2014).
Minimizing the Genetic Program to Run a Photosynthetic Endosymbiont
An inevitable consequence of the endosymbiotic life style is the reduction of the genome of the captured cells (Selosse et al., 2001; Reyes-Prieto et al., 2010). Besides the losses of genetic material, the transference of genes to the host nucleus (i.e., endosymbiotic gene transfer; EGT) is another mechanism leading the reduction of endosymbionts genomes (Martin and Herrmann, 1998). The transference of endosymbiont genetic material seems to be stochastic, relatively frequent and correlated with the amount of endosymbiotic genomic copies available (Stegemann and Bock, 2006). However, successful EGT implies not only the relocation of the genetic material to the nuclear genome, but the acquisition of elements necessary for expression (e.g., promoters regions and intron sequences) and translation (e.g., similar GC content, potential changes in codon usage) in the host genetic system (Martin and Herrmann, 1998). It is widely assumed that EGT has played a major role for the establishment of “permanent” endosymbionts. However, the magnitude and overall impact of EGT during early stages of primary plastid evolution is not entirely understood.
A pivotal issue in plastid evolution is the identification of those transferred cyanobacterial genes that were critical for the success of the early endosymbiosis. Most of the genomic evidence to answer that question in plants and algae with primary plastids has been diluted after more than 1 billion years of evolution (Yoon et al., 2009). However, comparative genomics studies including information from Archaeplastida lineages and P. chromatophora have provided valuable insights to better understand the scale, impact and tempo of EGT during early stages of endosymbiosis.
The idea that photosynthesis was the major driving force of plastid evolution correlates with the relatively high number of plastid genes related with the photosynthetic function and carbon fixation. Among the “typical” plastid genomes of plants and algae there are circa 40 genes encoding subunits of the Photosystems I and II, the Cytochrome b6f complex, light harvesting systems, photopigment biosynthesis and enzymes of the Calvin-Benson cycle. However, in all studied algal lineages some subunits of these key plastid pathways have been transferred, sometimes independently, to the nuclear genomes. In contrast to “typical” primary plastids, the chromatophore genome of P. chromatophora retains ~850 protein-coding genes (the number of genes varies between Paulinella strains), which represent approximately one third of the coding capacity of their cyanobacterial ancestors (Nowack et al., 2008; Reyes-Prieto et al., 2010), with most of the genes involved in photosynthesis and carbon fixation still present in the organelle gene collection. Unlike the case of plants and algae, where several hundreds of genes have been transferred via EGT from the plastid ancestor into the nuclear genome, only 32 chromatophore-derived genes have been identified among thousands of nuclear transcripts of P. chromatophora (Nowack et al., 2011).
This minimal estimate of 32 chromatophore-derived genes encodes mainly proteins of relative low molecular weight (≤250 amino acids), including 19 involved in photosynthesis and light-stress responses (e.g., several proteins of the high light-inducible protein family; Hli) (Nowack et al., 2011). These results revealed two trends: genes of relatively short length were prone to successful EGT and the encoded proteins, which participate in photosynthesis and light stress responses, were likely important to gain initial control over the endosymbiont autotrophic metabolism (Nowack et al., 2011). It has been suggested that the transference of sequences encoding proteins important for photosynthesis under stress conditions (e.g., oxidative or light stress; nutrient deprivation), such as the photosystems subunits encoded by psaE, psaK psaI, psbO, psbP and psbU, or the Hli proteins involved in photo-acclimation (Figure 1D), was important to incorporate key modulators of the photosynthetic activity into the host genetic system (Nowack et al., 2011; Dorrell and Howe, 2012).
Controlling Proliferation and Segregation of the Endosymbioitc Partner
Other key genetic mechanism that must have emerged early during organelle evolution is the host control over both the division of the endosymbiotic cells and the synchronized segregation of the divided endosymbionts into the next generation of host cells. How did the first genetic mechanisms to control the endosymbiont division evolve?
Certain clues have come from comparative studies about the origin of the plastid-division machinery and the localization of the corresponding protein-coding genes. For instance, some of the proteins that choreograph the plastid division are nuclear encoded and were likely selected from the host repertoire to control organellar functions. This is the case of the dynamin-like DRP5B, the glycogenin-like PDR1, the inner envelope protein MCD1 and the outer envelope proteins PDV1and PDV2 (Miyagishima et al., 2012; Vasetenkov and Koksharova, 2013). However, not all these host-derived components are universally present in the diverse plastid-bearing lineages and are likely independent additions. The other major component of the plastid division machinery comprises proteins of cyanobacterial provenance, such as FtsZ, FtsI, FtsW, SepF, ARC6, MINC, MINE and MIND, which are present in most algal and plant groups (Miyagishima et al., 2012). Similarly to the cases of the photosynthetic and carbon fixation pathways, a number of the cyanobacterial-derived proteins involved in plastid-division are still encoded in the organelle genome, but certain genes have been transferred to the nuclear genome (Miyagishima et al., 2012).
Studies in diverse algae have demonstrated that the expression of nuclear genes encoding proteins participating in plastid division, such as ftsZ, ARC6, minC is regulated by the host cell cycle (Miyagishima et al., 2012). This evidence would explain why in some eukaryote groups the photosynthetic organelles divide just once during the cell cycle. In contrast to the nuclear set, the expression of the plastid genes that encode elements of the organelle division machinery, for instance ftsI, ftsW, and sepF (minD and minE in certain algal groups), is not regulated by the cell cycle. Gene expression analyses suggest that the early transfer and integration of key cyanobacterial genes under the host cell cycle regulation was a crucial step to gain control over the endosymbiont division. The ftsZ gene encodes a GTPase (protein FtsZ) of the tubulin superfamily that participates in the formation of the cytokinetic contractile ring (i.e., FtsZ- or Z-ring; Figure 1E) underneath the inner membrane during organelle/bacterial division. The fact that ftsZ is nuclear-encoded in plants and algae suggests that that the cooption and regulation of this tubulin-like protein was important for the host to coordinate the organelle proliferation and segregation (Miyagishima et al., 2012).
The case of P. chromatophora seems to depict a different evolutionary solution to exert control over the endosymbiont division. It seems that the Paulinella host genetic machinery coordinates the chromatophore division (Nomura et al., 2014), but several genes typically involved in cyanobacterial division, such as minC, minD, minE, ftn2 (ARC6 homolog), cdv1, cdv2 and, interestingly, ftsZ are encoded in the genome of the Paulinella photosynthetic organelle (Nowack et al., 2008). The presence of the ftsZ gene in the chromatophore genome indicates that the Paulinella nucleus control over the organelle division probably relies on the recruitment of different protein players. For example, the gene sulA, which encodes a protein that inhibits the polymerization of FtsZ and prevents the formation of the Z-ring, is not present in the Paulinella chromatophore genome. It has to be explored if the sulA gene has been transferred into the Paulinella nuclear genome and if the encoded protein has any particular role regulating the chromatophore division (Nowack et al., 2008; Nowack, 2014). Overall, the evidence from independently evolved photosynthetic eukaryotes suggests that gaining control over the genes encoding proteins involved in cytokinesis is a key step to orchestrate the division of the photosynthetic endosymbionts.
Final Remarks
Vast amount of data has revealed the relevance of both EGT and the recruitment of host genes to integrate the cyanobacterial ancestor of plastids into the eukaryote cell context. Overall, the evolution of the initial metabolic interdependencies relied primarily on non-cyanobacterial elements (e.g., host-derived or from other bacterial sources). In contrast, the early evolution of the organelle protein import system, the regulation of the photosynthetic activity and the control over the endosymbiont division were facilitated mainly by the cooption of cyanobacterial-derived proteins.
Besides the relevance that the identification of the key “endosymbiosis genes” has to better understand the evolution and the astonishing diversification of photosynthetic eukaryotes, the information derived from the physiological roles of the “endosymbiotic proteins” provides fertile grounds for studies in the synthetic biology field. For example, genetic transformation of potential host or endosymbiotic cells with reengineered “endosymbiosis genes,” may facilitate survival of the partners, the exchange of metabolites, or the transit of key proteins between cells (see Weber and Osteryoung, 2010). This research avenue involves series of complex experimental and methodological difficulties, but some studies have provided interesting results. For instance, in vitro experimental investigations with murine macrophages (phagocytic cells) report that captured cyanobacterial cells (Synechococcus elongatus), previously transformed with bacterial genes encoding “cell invasion” proteins (e.g., invasins and hemolysins), are able to escape lysosome digestion, survive for several days and even divide at slow rate inside the mammalian cell host (Agapakis et al., 2011). These results suggest that relatively simple genetic engineering protocols can preset cells with basic gene kits useful to establish endosymbiotic associations.
A better understanding of the early evolution of photosynthetic organelles will provide reliable road maps to experimentally facilitate those critical stages for successful endosymbiosis. This may direct the design of artificial organelles to arm non-photosynthetic cells with light-powered compartments. In the other direction, investigations on artificial or assisted endosymbiosis have the potential to untangle crucial subcellular or genomic events not yet identified during the evolution of photosynthetic organelles.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The present work was supported by the Natural Sciences and Engineering Research Council of Canada (project 402421-2011), the Canada Foundation for Innovation (project 28276) and the New Brunswick Innovation Foundation (project RIF2012-006). I want to thank Oscar Juarez (Illinois Institute of Technology) for critical reading of the manuscript and valuable comments.
References
Agapakis, C. M., Niederholtmeyer, H., Noche, R. R., Lieberman, T. D., Megason, S. G., Way, J. C., et al. (2011). Towards a synthetic chloroplast. PLoS ONE 6:e18877. doi: 10.1371/journal.pone.0018877
Bhattacharya, D., Yoon, H. S., and Hackett, J. D. (2004). Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays 26, 50–60. doi: 10.1002/bies.10376
Blankenship, R. E. (2010). Early evolution of photosynthesis. Plant Physiol. 154, 434–438. doi: 10.1104/pp.110.161687
Bodył, A., Mackiewicz, P., and Stiller, J. W. (2009). Early steps in plastid evolution: current ideas and controversies. Bioessays 31, 1219–1232. doi: 10.1002/bies.200900073
Bräutigam, A., Facchinelli, F., and Weber, A. P. M. (2012). Metabolic integration of the chloroplast during endosymbiosis—molecular characterization of envelope transport proteins. J. Endocytobiosis Cell Res. 23, 96–102. Available online at: http://zs.thulb.uni-jena.de/receive/jportal_jparticle_00270546
Bryant, D. A., and Frigaard, N.-U. (2006). Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 14, 488–496. doi: 10.1016/j.tim.2006.09.001
Buick, R. (2008). When did oxygenic photosynthesis evolve? Philos. Trans. R. Soc. Lond. B. Biol. Sci. 363, 2731–2743. doi: 10.1098/rstb.2008.0041
Callieri, C. (2002). Grazing by ciliates and heterotrophic nanoflagellates on picocyanobacteria in Lago Maggiore, Italy. J. Plankton Res. 24, 785–796. doi: 10.1093/plankt/24.8.785
Dagan, T., Roettger, M., Stucken, K., Landan, G., Koch, R., Major, P., et al. (2013). Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 5, 31–44. doi: 10.1093/gbe/evs117
Day, P. M., Potter, D., and Inoue, K. (2014). Evolution and targeting of Omp85 homologs in the chloroplast outer envelope membrane. Front. Plant Sci. 5:535. doi: 10.3389/fpls.2014.00535
De Vries, J., Sousa, F. L., Bölter, B., Soll, J., and Gould, S. B. (2015). YCF1: a green TIC? Plant Cell 27, 1827–1833. doi: 10.1105/tpc.114.135541
Dorrell, R. G., and Howe, C. J. (2012). What makes a chloroplast? Reconstructing the establishment of photosynthetic symbioses. J. Cell Sci. 125, 1865–1875. doi: 10.1242/jcs.102285
Escalera, L., Reguera, B., Takishita, K., Yoshimatsu, S., Koike, K., and Koike, K. (2011). Cyanobacterial endosymbionts in the benthic dinoflagellate Sinophysis canaliculata (Dinophysiales, Dinophyceae). Protist 162, 304–314. doi: 10.1016/j.protis.2010.07.003
Facchinelli, F., and Weber, A. P. M. (2011). The metabolite transporters of the plastid envelope: an update. Front. Plant Sci. 2:50. doi: 10.3389/fpls.2011.00050
Farquhar, J., Zerkle, A. L., and Bekker, A. (2011). Geological constraints on the origin of oxygenic photosynthesis. Photosynth. Res. 107, 11–36. doi: 10.1007/s11120-010-9594-0
Gross, J., and Bhattacharya, D. (2009a). Mitochondrial and plastid evolution in eukaryotes: an outsiders' perspective. Nat. Rev. Genet. 10, 495–505. doi: 10.1038/nrg2610
Gross, J., and Bhattacharya, D. (2009b). Revaluating the evolution of the Toc and Tic protein translocons. Trends Plant Sci. 14, 13–20. doi: 10.1016/j.tplants.2008.10.003
Gu, F., Chen, L., Ni, B., and Zhang, X. (2002). A comparative study on the electron microscopic enzymo-cytochemistry of Paramecium bursaria from light and dark cultures. Eur. J. Protistol. 38, 267–278. doi: 10.1078/0932-4739-00875
Hedges, S. B., Blair, J. E., Venturi, M. L., and Shoe, J. L. (2004). A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 4:2. doi: 10.1186/1471-2148-4-2
Hsu, S.-C., Patel, R., Bédard, J., Jarvis, P., and Inoue, K. (2008). Two distinct Omp85 paralogs in the chloroplast outer envelope membrane are essential for embryogenesis in Arabidopsis thaliana. Plant Signal. Behav. 3, 1134–1135. doi: 10.4161/psb.3.12.7095
Kalanon, M., and McFadden, G. I. (2008). The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics 179, 95–112. doi: 10.1534/genetics.107.085704
Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11, 557–570. doi: 10.1105/tpc.11.4.557
Kikuchi, S., Bédard, J., Hirano, M., Hirabayashi, Y., Oishi, M., Imai, M., et al. (2013). Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339, 571–574. doi: 10.1126/science.1229262
Kodama, Y., and Fujishima, M. (2005). Symbiotic Chlorella sp. of the ciliate Paramecium bursaria do not prevent acidification and lysosomal fusion of host digestive vacuoles during infection. Protoplasma 225, 191–203. doi: 10.1007/s00709-005-0087-5
Kodama, Y., and Fujishima, M. (2012). Characteristics of the digestive vacuole membrane of the alga-bearing ciliate Paramecium bursaria. Protist 163, 658–670. doi: 10.1016/j.protis.2011.10.004
Kodama, Y., and Fujishima, M. (2014). Symbiotic Chlorella variabilis incubated under constant dark conditions for 24 hours loses the ability to avoid digestion by host lysosomal enzymes in digestive vacuoles of host ciliate Paramecium bursaria. FEMS Microbiol. Ecol. 90, 946–955. doi: 10.1111/1574-6941.12448
Kodama, Y., Inouye, I., and Fujishima, M. (2011). Symbiotic Chlorella vulgaris of the ciliate Paramecium bursaria plays an important role in maintaining perialgal vacuole membrane functions. Protist 162, 288–303. doi: 10.1016/j.protis.2010.06.005
Kodama, Y., Nakahara, M., and Fujishima, M. (2007). Symbiotic alga Chlorella vulgaris of the ciliate Paramecium bursaria shows temporary resistance to host lysosomal enzymes during the early infection process. Protoplasma 230, 61–67. doi: 10.1007/s00709-006-0193-z
Kopp, R. E., Kirschvink, J. L., Hilburn, I. A., and Nash, C. Z. (2005). The Paleoproterozoic snowball Earth: a climate disaster triggered by the evolution of oxygenic photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 102, 11131–11136. doi: 10.1073/pnas.0504878102
Kovács-Bogdán, E., Benz, J. P., Soll, J., and Bölter, B. (2011). Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 11:133. doi: 10.1186/1471-2229-11-133
Kunau, W.-H., Agne, B., and Girzalsky, W. (2001). The diversity of organelle protein transport mechanisms. Trends Cell Biol. 11, 358–361. doi: 10.1016/S0962-8924(01)02083-9
Leister, D. (2003). Chloroplast research in the genomic age. Trends Genet 19, 47–56. doi: 10.1016/S0168-9525(02)00003-3
Li, H. M., and Chiu, C.-C. (2010). Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61, 157–180. doi: 10.1146/annurev-arplant-042809-112222
Linka, N., Hurka, H., Lang, B. F., Burger, G., Winkler, H. H., Stamme, C., et al. (2003). Phylogenetic relationships of non-mitochondrial nucleotide transport proteins in bacteria and eukaryotes. Gene 306, 27–35. doi: 10.1016/S0378-1119(03)00429-3
Mackiewicz, P., Bodył, A., and Gagat, P. (2012). Possible import routes of proteins into the cyanobacterial endosymbionts/plastids of Paulinella chromatophora. Theory Biosci. 131, 1–18. doi: 10.1007/s12064-011-0147-7
Marin, B., Nowack, E. C. M., Glöckner, G., and Melkonian, M. (2007). The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium. BMC Evol. Biol. 7:85. doi: 10.1186/1471-2148-7-85
Marin, B., Nowack, E. C. M., and Melkonian, M. (2005). A plastid in the making: evidence for a second primary endosymbiosis. Protist 156, 425–432. doi: 10.1016/j.protis.2005.09.001
Martin, W., and Herrmann, R. (1998). Gene transfer from organelles to the nucleus: how much, what happens, and Why? Plant Physiol. 118, 9–17. doi: 10.1104/pp.118.1.9
Matz, C., and Kjelleberg, S. (2005). Off the hook–how bacteria survive protozoan grazing. Trends Microbiol. 13, 302–307. doi: 10.1016/j.tim.2005.05.009
McFadden, G. I. (1999). Plastids and protein targeting. J. Eukaryot. Microbiol. 46, 339–346. doi: 10.1111/j.1550-7408.1999.tb04613.x
Miyagishima, S.-Y., Suzuki, K., Okazaki, K., and Kabeya, Y. (2012). Expression of the nucleus-encoded chloroplast division genes and proteins regulated by the algal cell cycle. Mol. Biol. Evol. 29, 2957–2970. doi: 10.1093/molbev/mss102
Nakai, M. (2015). The TIC complex uncovered: the alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts. Biochim. Biophys. Acta. 1847, 957–967. doi: 10.1016/j.bbabio.2015.02.011
Neuhaus, H. E., Thom, E., Möhlmann, T., Steup, M., and Kampfenkel, K. (1997). Characterization of a novel eukaryotic ATP/ADP translocator located in the plastid envelope of Arabidopsis thaliana L. Plant J. 11, 73–82. doi: 10.1046/j.1365-313X.1997.11010073.x
Nomura, M., Nakayama, T., and Ishida, K. (2014). Detailed process of shell construction in the photosynthetic testate amoeba Paulinella chromatophora (euglyphid, Rhizaria). J. Eukaryot. Microbiol. 61, 317–321. doi: 10.1111/jeu.12102
Nowack, E. C. M. (2014). Paulinella chromatophora—rethinking the transition from endosymbiont to organelle. Acta Soc. Bot. Pol. 83, 387–397. doi: 10.5586/asbp.2014.049
Nowack, E. C. M., Melkonian, M., and Glöckner, G. (2008). Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 18, 410–418. doi: 10.1016/j.cub.2008.02.051
Nowack, E. C. M., Vogel, H., Groth, M., Grossman, A. R., Melkonian, M., and Glöckner, G. (2011). Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora. Mol. Biol. Evol. 28, 407–422. doi: 10.1093/molbev/msq209
Palmer, J. D. (2003). The symbiotic birth and spread of plastids: how many times and whodunit? J. Phycol. 39, 4–12. doi: 10.1046/j.1529-8817.2003.02185.x
Price, D. C., Chan, C. X., Yoon, H. S., Yang, E. C., Qiu, H., Weber, A. P. M., et al. (2012). Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 335, 843–847. doi: 10.1126/science.1213561
Qiu, H., Price, D. C., Weber, A. P. M., Facchinelli, F., Yoon, H. S., and Bhattacharya, D. (2013). Assessing the bacterial contribution to the plastid proteome. Trends Plant Sci. 18, 680–687. doi: 10.1016/j.tplants.2013.09.007
Reiser, J., Linka, N., Lemke, L., Jeblick, W., and Neuhaus, H. E. (2004). Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiol. 136, 3524–3536. doi: 10.1104/pp.104.049502
Reyes-Prieto, A., Weber, A. P., and Bhattacharya, D. (2007). The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41, 147–168. doi: 10.1146/annurev.genet.41.110306.130134
Reyes-Prieto, A., Yoon, H. S., Moustafa, A., Yang, E. C., Andersen, R. A., Boo, S. M., et al. (2010). Differential gene retention in plastids of common recent origin. Mol. Biol. Evol. 27, 1530–1537. doi: 10.1093/molbev/msq032
Richardson, L. G. L., Paila, Y. D., Siman, S. R., Chen, Y., Smith, M. D., and Schnell, D. J. (2014). Targeting and assembly of components of the TOC protein import complex at the chloroplast outer envelope membrane. Front. Plant Sci. 5:269. doi: 10.3389/fpls.2014.00269
Schönknecht, G., Weber, A. P. M., and Lercher, M. J. (2014). Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. Bioessays 36, 9–20. doi: 10.1002/bies.201300095
Selosse, M.-A., Albert, B., and Godelle, B. (2001). Reducing the genome size of organelles favours gene transfer to the nucleus. Trends Ecol. Evol. 16, 135–141. doi: 10.1016/S0169-5347(00)02084-X
Sheiner, L., and Striepen, B. (2013). Protein sorting in complex plastids. Biochim. Biophys. Acta 1833, 352–359. doi: 10.1016/j.bbamcr.2012.05.030
Shi, L.-X., and Theg, S. M. (2013). The chloroplast protein import system: from algae to trees. Biochim. Biophys. Acta 1833, 314–331. doi: 10.1016/j.bbamcr.2012.10.002
Sommer, M. S., Daum, B., Gross, L. E., Weis, B. L. M., Mirus, O., Abram, L., et al. (2011). Chloroplast Omp85 proteins change orientation during evolution. Proc. Natl. Acad. Sci. U.S.A. 108, 13841–13846. doi: 10.1073/pnas.1108626108
Stegemann, S., and Bock, R. (2006). Experimental reconstruction of functional gene transfer from the tobacco plastid genome to the nucleus. Plant Cell 18, 2869–2878. doi: 10.1105/tpc.106.046466
Steiner, J. M., Yusa, F., Pompe, J. A., and Löffelhardt, W. (2005). Homologous protein import machineries in chloroplasts and cyanelles. Plant J. 44, 646–652. doi: 10.1111/j.1365-313X.2005.02559.x
Suzuki, K., and Miyagishima, S. Y. (2010). Eukaryotic and eubacterial contributions to the establishment of plastid proteome estimated by large-scale phylogenetic analyses. Mol. Biol. Evol. 27, 581–590. doi: 10.1093/molbev/msp273
Tyra, H. M., Linka, M., Weber, A. P. M., and Bhattacharya, D. (2007). Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 8:R212. doi: 10.1186/gb-2007-8-10-r212
Van Dooren, G. G., Schwartzbach, S. D., Osafune, T., and McFadden, G. I. (2001). Translocation of proteins across the multiple membranes of complex plastids. Biochim. Biophys. Acta 1541, 34–53. doi: 10.1016/S0167-4889(01)00154-9
Van Wijk, K. J., and Baginsky, S. (2011). Plastid proteomics in higher plants: current state and future goals. Plant Physiol. 155, 1578–1588. doi: 10.1104/pp.111.172932
Vasetenkov, A. E., and Koksharova, O. A. (2013). Comparative and evolutionary aspects of cyanobacteria and plant plastid division study. Russ. J. Plant Physiol. 60, 453–464. doi: 10.1134/S1021443713040171
Wang, L., Mao, Y., Kong, F., Li, G., Ma, F., Zhang, B., et al. (2013). Complete sequence and analysis of plastid genomes of two economically important red algae: Pyropia haitanensis and Pyropia yezoensis. PLoS ONE 8:e65902. doi: 10.1371/journal.pone.0065902
Weber, A. P. M., Linka, M., and Bhattacharya, D. (2006). Single, ancient origin of a plastid metabolite translocator family in Plantae from an endomembrane-derived ancestor. Eukaryot. Cell 5, 609–612. doi: 10.1128/EC.5.3.609-612.2006
Weber, A. P. M., and Linka, N. (2011). Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu. Rev. Plant Biol. 62, 53–77. doi: 10.1146/annurev-arplant-042110-103903
Weber, A. P. M., and Osteryoung, K. W. (2010). From endosymbiosis to synthetic photosynthetic life. Plant Physiol. 154, 593–597. doi: 10.1104/pp.110.161216
Wickner, W., and Schekman, R. (2005). Protein translocation across biological membranes. Science 310, 1452–1456. doi: 10.1126/science.1113752
Xinyao, L., Miao, S., Yonghong, L., Yin, G., Zhongkai, Z., Donghui, W., et al. (2006). Feeding characteristics of an amoeba (Lobosea: Naegleria) grazing upon cyanobacteria: food selection, ingestion and digestion progress. Microb. Ecol. 51, 315–325. doi: 10.1007/s00248-006-9031-2
Yoon, H. S., Hackett, J. D., Ciniglia, C., Pinto, G., and Bhattacharya, D. (2004). A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818. doi: 10.1093/molbev/msh075
Keywords: gene transfer, endosymbiosis, membrane transporters, organelle genome, plastid protein import, photosynthesis, plastid division
Citation: Reyes-Prieto A (2015) The basic genetic toolkit to move in with your photosynthetic partner. Front. Ecol. Evol. 3:100. doi: 10.3389/fevo.2015.00100
Received: 20 June 2015; Accepted: 11 August 2015;
Published: 01 September 2015.
Edited by:
Arturo Becerra, Universidad Nacional Autónoma de México, MexicoReviewed by:
Andres Moya, University of Valencia, SpainNatalia Ivanova, Lawrence Berkeley National Laboratory, USA
Copyright © 2015 Reyes-Prieto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adrian Reyes-Prieto, Biology Department, University of New Brunswick, 10 Bailey Drive, Fredericton, NB E3B 5A3, Canada, areyes@unb.ca
 Adrian Reyes-Prieto
Adrian Reyes-Prieto