Sixteen Years of DNA Barcoding in China: What Has Been Done? What Can Be Done?
- College of Life Sciences, Capital Normal University, Beijing, China
Over the past 16 years, more than half (59.68%) of research papers in China on DNA barcoding have been published in Chinese rather than English. Using the records in the BOLD (Barcode of Life Data) system, we found Chinese scientists have contributed nearly 120,000 DNA barcodes for more than 16,000 species as of September 2019, with barcoded species distributed throughout China. Based on 2,624 articles and 494 dissertations published during the last 16 years, we reviewed the basic statistics of these studies as well as the type of articles contributed by Chinese scientists, the preference of taxonomic groups, the characteristic of barcoding studies in China, the current limitations, and potential future directions as well. We found that most barcode data pertain primarily to plants and animals. Most work in China has focused on verification of the authenticity of species used in traditional Chinese medicine, while other applications have paid more attention to food safety, inspection and quarantine, and the control of pests and invasive species. In methodology and technology, a number of new DNA barcoding methods have been developed by Chinese scientists. However, there are several significant limitations to research into DNA barcoding in China in general, such as the lack of leadership in pioneering international projects, the absence of an open bioinformatics infrastructure, and the fact that some Chinese journals do not clearly require data transparency and availability for DNA barcodes, impeding the further development of barcode libraries and research in China. In the future, Chinese scientists should build authoritative online libraries, while aiming for theoretical innovations for both concepts and methodology of DNA barcoding.
Introduction
Since the inception of DNA barcoding in 2003 (Hebert et al., 2003a, b), it has become widely used as a taxonomic tool (DeSalle and Goldstein, 2019). It is especially useful for species identification when accurate morphological information and taxonomic expertise are limiting factors (Ahrens et al., 2007; Valentini et al., 2009). With additional development and methodologies, barcoding is becoming increasingly useful outside of taxonomy (Hebert and Gregory, 2005), and it is becoming more popular in ecological (e.g., ecological interactions and food webs) studies, biodiversity surveys (Hajibabaei et al., 2007; Joly et al., 2013), conservation biology, biosecurity, and medicine and pharmacology (Pečnikar and Buzan, 2014).
In China, some taxonomists, such as those who work on plants, are deeply involved in the study of barcoding, providing many significant contributions to the international community of DNA barcoding. For example, 62 researchers from 19 research institutes and universities across the country have formed the “China Plant BOL (Barcode of Life) Group” to conduct in-depth research on the DNA barcoding of seed plants. Based on the barcode combinations recommended by the Consortium for Barcode of Life (CBOL), they proposed that ITS/ITS2 should be incorporated into the core barcode for seed plants after conducting a large number of tests on four DNA barcode candidate fragments of 6,286 specimens (China-Plant-BOL-Group et al., 2011). Their research not only solved the problem of low resolution using only rbcL + matK but also represented another step forward toward standardizing the routine use of DNA sequence data (Hollingsworth, 2011). Besides DNA barcoding of plants, other Chinese scientists have applied different DNA barcodes in their own taxonomic groups (Cheng et al., 2011). However, a systematic review on DNA barcoding research in China is lacking, especially in an international context, given the relative inaccessibility of this language to those who cannot read Chinese.
To this end, we systematically searched for articles published by Chinese scientists in both domestic and international journals from 2003 to August 2019 and summarized the contributions of Chinese scientists in DNA barcoding research in terms of their publications and data outputs. We have also pointed out severe limitations and potential future directions for barcoding research in China.
Literature Searching and Manual Data Mining
According to the ecological theory of species–area relations (Arrhenius, 1921; Gleason, 1922), countries with large land areas theoretically possess higher biodiversity. With a land area of more than 9.63 million square kilometers, China is the third largest country in the world. In the context of DNA barcoding, the Chinese scientific community is responsible for documenting an immense wealth of biodiversity and corresponding barcode sequences. To gauge the amount of barcode data generated and shared by China, the current number of records and related species were retrieved with the keyword “China” (incl. Taiwan) from the BOLD system (The Barcode of Life Data1; Ratnasingham and Hebert, 2007), which is one of the world’s most authoritative online barcode databases. The coordinates of those records were also downloaded to visualize their geographic distribution at the same time. Barcode data from the five other largest countries (excluding China), Russia, Canada, America, Brazil, and Australia were also downloaded for comparison.
To determine the proportion of the publications on DNA barcoding from Chinese scientists worldwide, a preliminary retrieval from the Web of Science (WOS1) database with the phrase “DNA barcode∗” (the asterisk was used to enable the return of results containing the words “barcode,” “barcodes” or “barcoding”) as the keyword was implemented. To make the results more general, we searched for publications where the keyword appeared throughout the full text of articles (with “topic” field tag in WOS) rather than just in title, which is slightly different from previous reviews (Taylor and Harris, 2012; DeSalle and Goldstein, 2019).
A final database was then assembled. To review the DNA barcoding studies contributed by Chinese scientists during the last 16 years, a comprehensive literature search was conducted from not only WOS1 but also China National Knowledge Infrastructure (CNKI2) for articles published during the period between January 2003 and August 2019. The latter database was generally ignored in previous studies by western researchers due to language issues. We searched for “DNA barcode∗” in the full text of the paper, with Chinese institutions/universities as the first research institute (Supplementary Data Sheet S1). Because of the partial overlap between these two online databases, we manually removed the duplicative records for subsequent analyses. Then, to summarize the problems and potential directions of DNA barcoding research in China in the future, information of each publication was listed, covering taxonomic groups, article types, journals, barcode selections, and research institutions.
DNA Barcoding and Its Current Situation in China
As the third largest country in the world, China possesses one of 25 global biodiversity hotspots (Myers et al., 2000). Based on a survey of the global DNA barcoding library BOLD system (The Barcode of Life Database3; Ratnasingham and Hebert, 2007), Chinese scientists have contributed 119,745 DNA barcodes belonging to 16,772 species as of September 2019 (Figure 1). Of the six largest countries examined, the only countries that have contributed more barcodes are Australia, Canada, and the United States. Geographically, studies have taken place throughout much of China, although fewer have been conducted in the northwest and northeast (Figure 2).
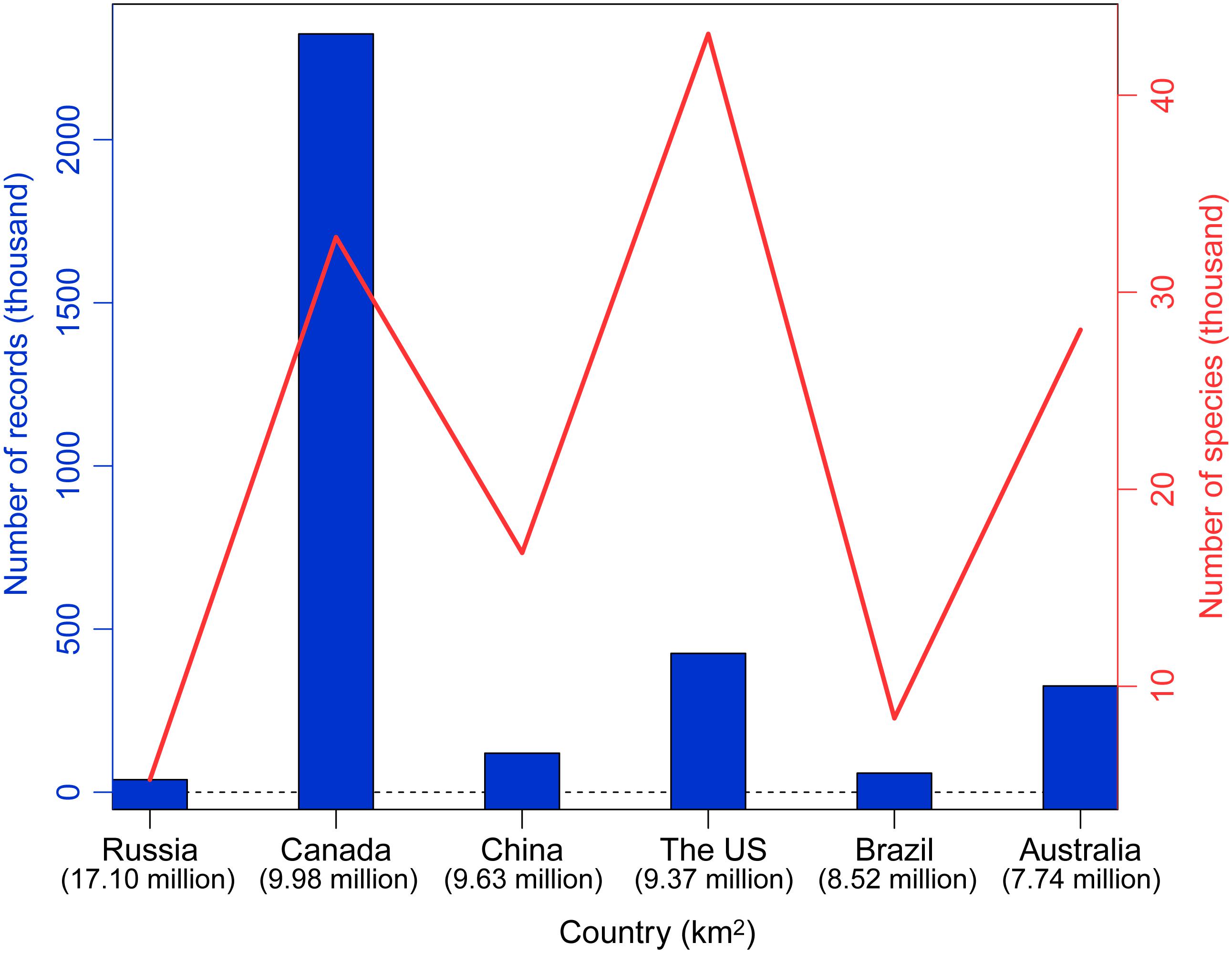
Figure 1. Numbers of barcoding records contributed by scientists from the six largest countries in the world. Countries are given on the X axis, and the Y axis indicates the number of barcode records (blue bar) and number of species (red solid line). Country area is given under the name.
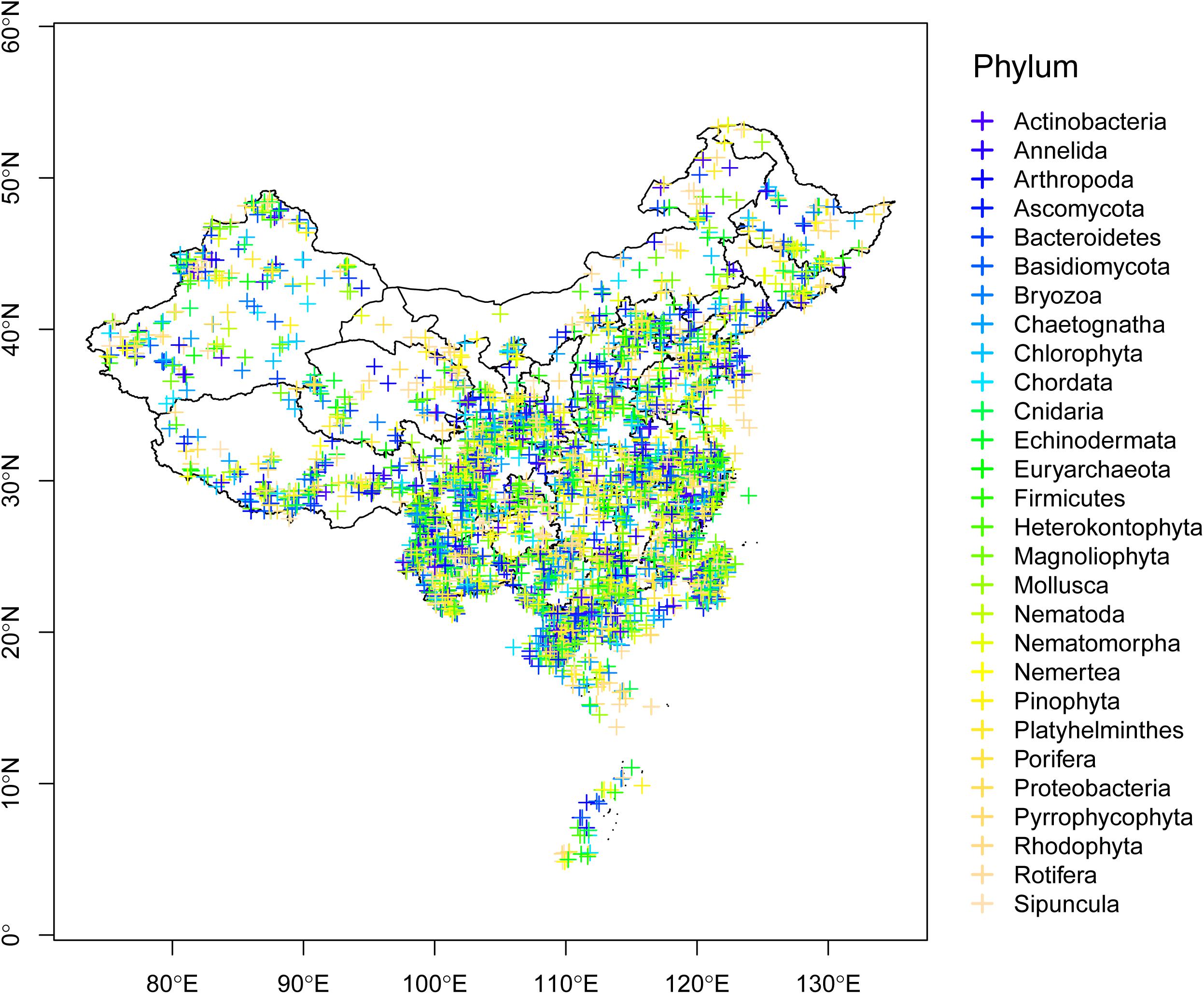
Figure 2. Geographical distribution of barcodes in the BOLD system contributed by Chinese scientists, with samples collected in China. Phyla are denoted by different colors.
According to the data from WOS1, 1,993 articles from China (incl. Taiwan) that were published between 2003 and 2018 include the phrase “DNA barcode∗” in their “topic” field tag. Following a review on barcoding published in a domestic journal in 2004, Chinese scientists started publishing their DNA barcoding research in international journals in 2006, and the number of articles began to increase in 2009. By the end of 2018, the total number of publications on DNA barcoding contributed from Chinese researchers reached 20.06% of the total number of DNA barcoding papers published throughout the world (Figure 3), indicating that China has become one of the major countries dedicated to research on DNA barcoding. However, this is only part of China’s contribution to DNA barcoding because more than half (59.68%) of their publications occur in internal Chinese journals (most are not databased in WOS).
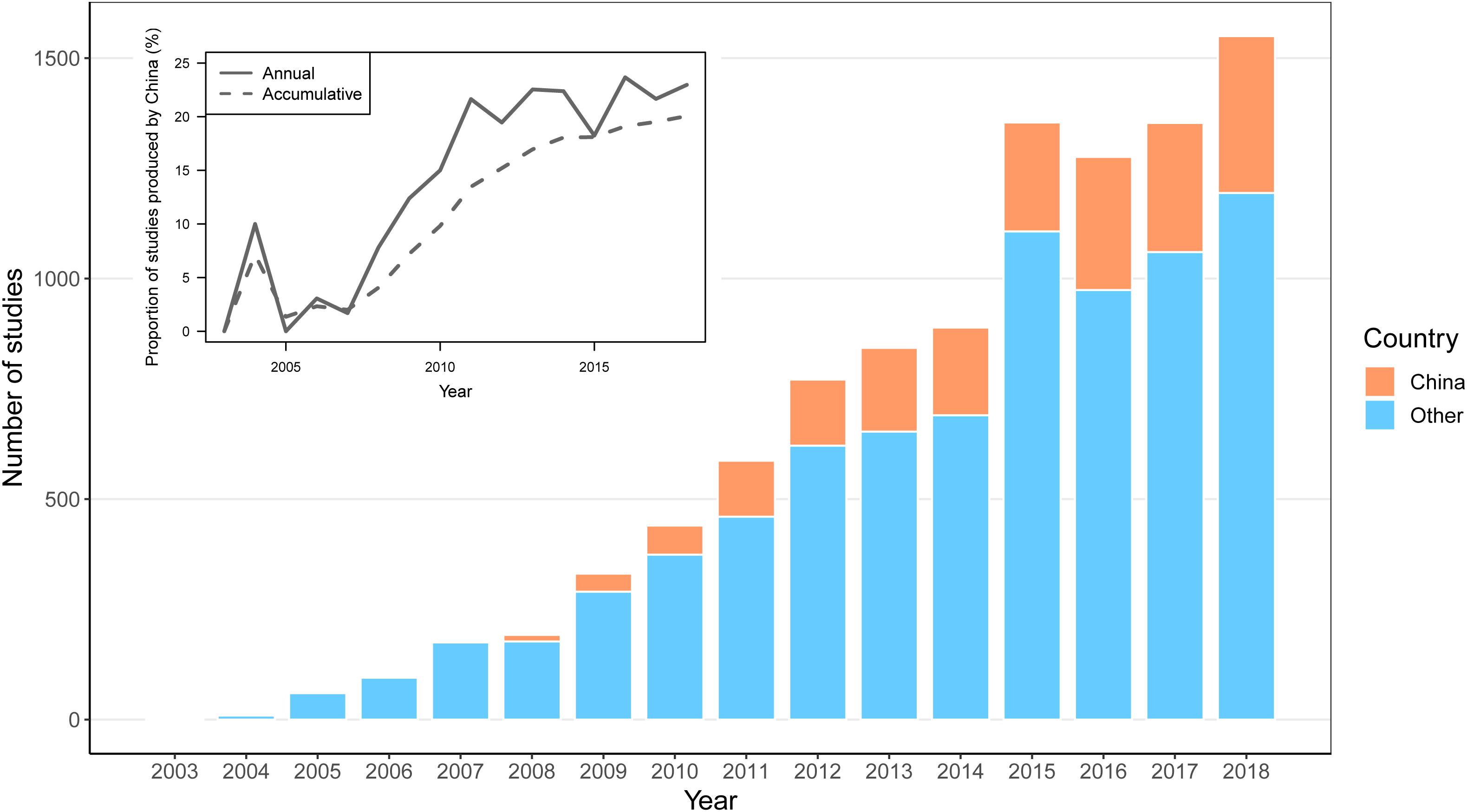
Figure 3. Numbers of articles published on DNA barcoding by Chinese scientists and researchers from all other countries between 2003 and 2018. The X axis gives the year of publication, and the Y axis indicates the number of studies. Orange bars represent the number of articles published by Chinese scientists, while blue bars indicate the number of articles published by other researchers from all other countries. The small figure in the upper left corner represents the proportion of the number of articles from Chinese studies to the total number of articles, where the solid line represents the proportion of articles contributed by Chinese scientists to all others each year, and the dotted line represents the proportion of accumulated articles to all others.
Publications Contributed by Chinese Scientists
More Empirical but Fewer Methodological Studies
In this study, all 2,624 articles were classified into four categories: Category 1 – basic studies, where one or more DNA barcodes are established for specific taxonomic groups; Category 2 – practical studies, where DNA barcodes are used to identify species or other ecologically related research; Category 3 – methodological studies, where new algorithms or methods of species identification are developed, computer programs are established, or comparisons are made between different DNA barcoding approaches; and Category 4 – reviews that summarize recent advances in DNA barcoding, including those focusing on certain groups of taxa.
Based on the statistics derived from these different types of articles, we found that the number of articles pertaining to Category 1 showed an annual increase, and, by August 2019, they represented nearly half (47.14%) of the total number of articles published (Figure 4); Category 2 showed a similar trend, with 28.24% of articles, and this result implies that China has a huge demand for DNA barcoding technology, including demand from traditional Chinese medicine and social needs related to food safety, inspection and quarantine, pest control, and other applications (see below).
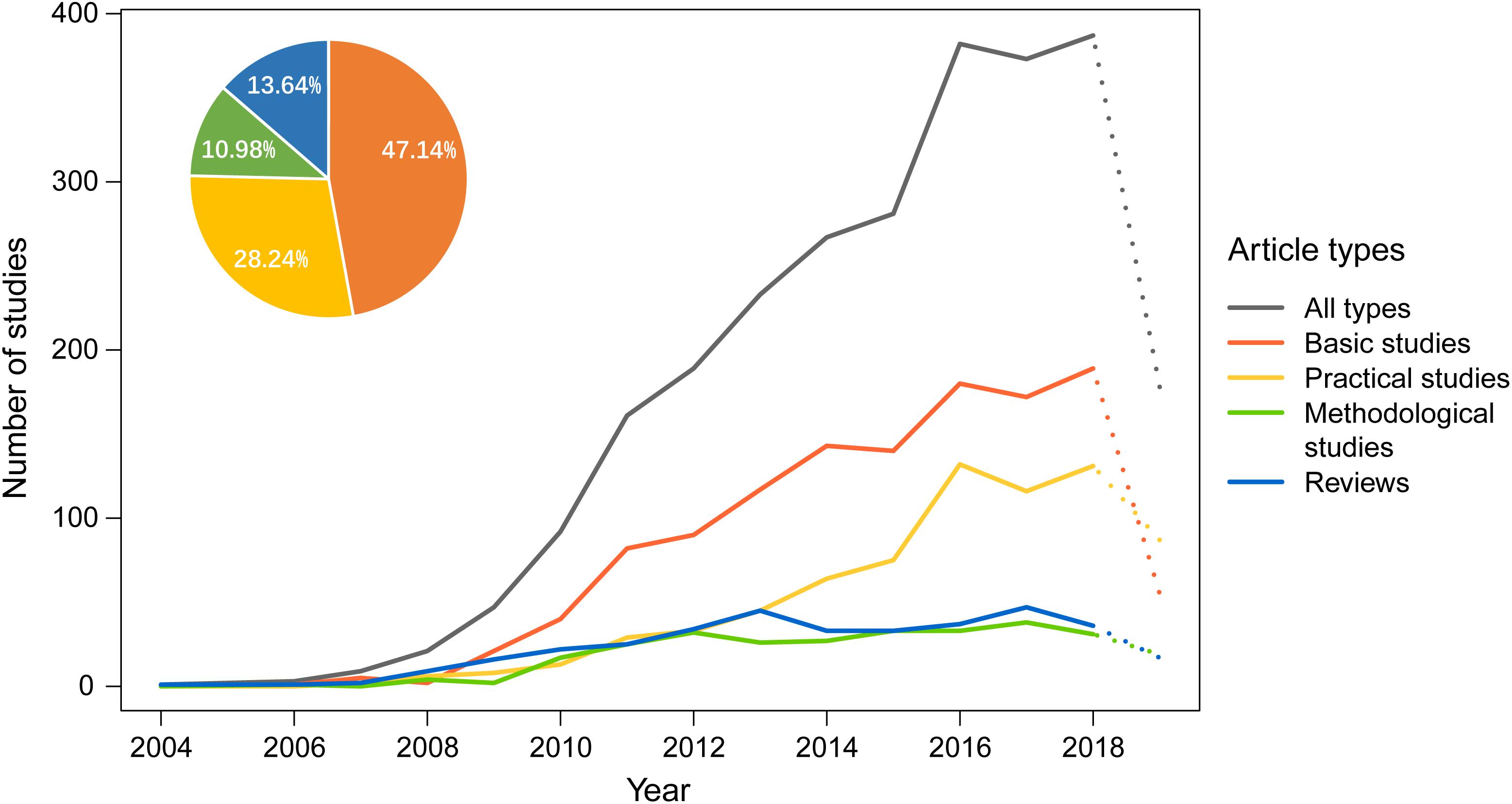
Figure 4. The number of different types of articles on DNA barcoding published by Chinese scientists each year. The X axis represents the year of publication, and the Y axis represents the number of studies. A complete dataset for 2019 is not yet available and is shown as a dotted line. The pie chart at the top left represents the proportion of the total number of research articles of each type derived over 16 years.
In contrast to Category 1 and 2, the number of articles pertaining to Category 3 has increased at a much slower rate (Figure 4). Methodological studies, accounting for 10.98% of the total studies, are considerably less common than practical studies (28.24%). Despite the small percentage of methodological studies, they may have comprehensive and profound effects on other DNA barcoding studies. Therefore, we have paid more attention to them here. In this category, internationally, one of the earliest algorithms for sequence assignment was the BLAST algorithm (Altschul et al., 1990), which relies on local similarity between sequences. However, the credibility of the assignment results can be questionable in DNA barcoding (Ross et al., 2008). Most tree-based methods, such as maximum parsimony (MP; Czelusniak et al., 1990), maximum-likelihood (ML; Felsenstein, 1981), and Bayesian approaches (Huelsenbeck and Ronquist, 2001; Munch et al., 2008), are probably more accurate, but they usually require long processing times and high-RAM (random access memory) when dealing with very large DNA datasets (Austerlitz et al., 2009) except neighbor joining (NJ; Saitou and Nei, 1987). Chinese scientists have used these approaches in their DNA barcoding studies. The last decade, however, has also witnessed significant progress in the methodology of DNA barcoding given many new approaches proposed by Chinese scientists (Zhang et al., 2008, 2012a,b, 2017; Yu et al., 2012; Liu et al., 2013, 2017; Jin et al., 2018; Shi et al., 2018). The main advances include both algorithm development and the optimization of sequencing strategies, as summarized below.
Artificial intelligence (AI) is used for industrialized applications in China, and Chinese scientists appear to be among the first to introduce AI into species identification algorithms (Zhang et al., 2008). The proposed method is used for identification of species with unknown barcodes based on referencing library trained back-propagation (BP) neural networks. The BP-based method appears to be superior to commonly used distance-based methods, particularly in cases involving incomplete lineage sorting (Zhang et al., 2008). Species identification algorithms for non-coding barcode sequences based on machine learning methods, such as DV-RBF and FJ-RBF, also performed well (Zhang et al., 2012a). The problem of species membership can also be solved by linking it to fuzzy-set-theory (Zadeh, 1965), which efficacy has been demonstrated by its successful application to empirical datasets (Zhang et al., 2012b). Compared with other methods, the fuzzy-set-theory-based approach has great efficacy in reducing false-positive species identification when conspecifics of the query are absent from the reference database (Zhang et al., 2012b). In addition, Shi et al. (2018) combined the Hidden Markov Model (HMM; Eddy, 1998) algorithm with the fuzzy membership function and further improved the processing speed of this approach for exploring large datasets. Naturally, the expanding number of available methods begets a need for an integrated toolkit for DNA barcoding. BarcodingR is one of the most useful software packages that provides a comprehensive implementation of species identification methods with additional new functions in R (Zhang et al., 2017). With the great facility of this package for DNA barcoding research, the high performance of machine learning approaches has been successfully applied in studies, such as wood barcoding (He et al., 2019).
Aside from analysis algorithms, in the optimization of sequencing strategies, scientists are also developing more efficient means of obtaining accurate metadata. Yu et al. (2012) proposed protocols for the extraction of ecological, taxonomic, and phylogenetic information from bulk samples by combining mass trapping, mass-PCR amplification, pyrosequencing, and bioinformatics analysis. They demonstrated that metabarcoding allows for a broad and efficient estimate of biodiversity for the first time, which can facilitate assessment of the state of current ecosystems worldwide. One problem with barcodes derived from next-generation-sequencing (NGS) analyses is the shorter maximum read lengths (typically < 150 bp) and consequent lost taxonomic information. To overcome this problem, Liu et al. (2013) presented a new Illumina-based pipeline (SOAPBarcode) that allows for the full-length recovery of COI barcodes from mixed samples. Their assemblage protocol involves the use of two libraries: the full-length library (insert size = 658 bp) and the shotgun library (insert size = 200 bp). This approach can deliver reliable and taxonomically informative metabarcoding outcomes for biodiversity-related research (Liu et al., 2013). Although the introduction and optimization of metabarcoding has applications for biodiversity studies, the most accurate approach for taxonomists is to obtain the complete barcode sequence by amplification from a single sample. Because Sanger sequencing is approaching its limits in terms of throughput and chemistry cost, Liu et al. (2017) developed an Illumina-based pipeline, HIFI-Barcode, to produce full-length COI barcodes from pooled PCR amplicons generated by individual specimens. The accuracy of barcode sequences generated by the new pipeline is comparable to sequences derived from the Sanger method and only requires about one-tenth of the current cost (Liu et al., 2017).
The ever-increasing number of DNA barcoding methods has led to many reviews on the subject. The number of reviews accounted for 13.64% of all articles. The first review of DNA barcoding was published in 2004 (Xiao et al., 2004), and it was the first to introduce Chinese scientists to the concept, basic principles, and potentials of DNA barcoding. The increase in the number of reviews came after 2010. Many papers summarized the application and methods of barcoding technology in different taxonomic groups (e.g., Cheng et al., 2011; Yao et al., 2013; Chen et al., 2014; Liang et al., 2015; Sun et al., 2016). Lately, some researchers have also reviewed DNA barcoding from the perspective of ecological communities, and they have proposed a “purpose-driven barcode” fit for multi-level applications (Pei et al., 2017).
The Vast Majority of Species Barcoded in China: Animals and Plants
As originally proposed in 2003, DNA barcoding largely focused on species of animals (Hebert et al., 2003a), thus indicating a taxonomic bias in that other groups were less studied (Taylor and Harris, 2012). This trend continues in China (Figure 5). As of August 2019, the total number of articles related to animal groups in China reached 1,104, nearly half (48.72%) of the total number of research papers (2,266, excluding review articles). Likewise, plant barcoding studies showed a trend of continuous and rapid increase similar to that of animal groups after 2009 (Figure 5). The rapid growth of DNA barcoding research on plant groups is probably related to Chinese traditional medicine culture (see below). At the same time, Chinese researchers have paid less attention to DNA barcoding of microorganisms. As of August 2019, only 147 research papers on other groups were published, and most are related to the classification and identification of fungi as well as viruses and pathogens (Figure 5).
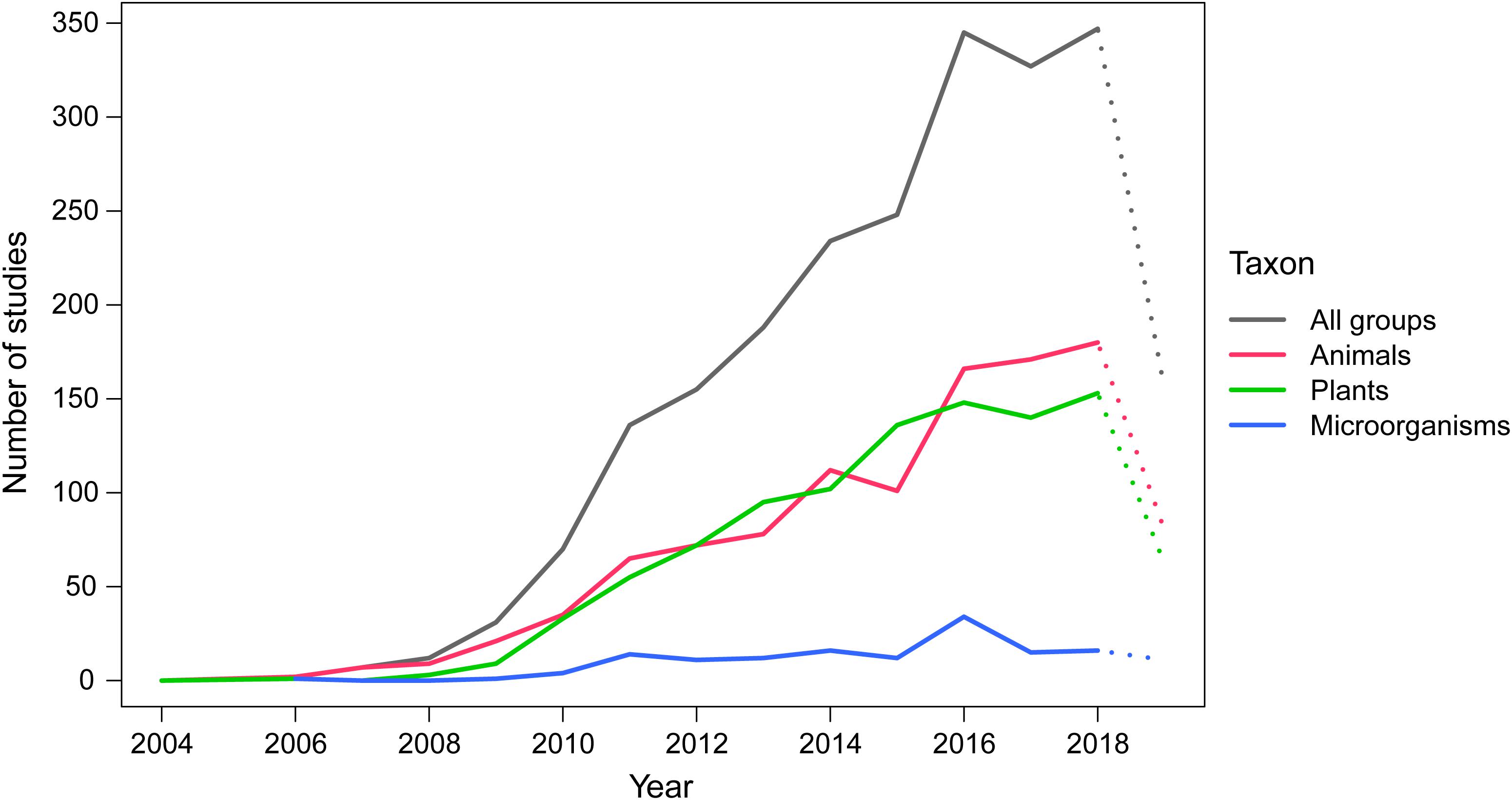
Figure 5. The number of DNA barcoding studies by Chinese scientists published over different years on different taxonomic groups. The X axis represents the year of publication, and the Y axis represents the number of studies. A complete dataset for 2019 is not yet available and is shown as a dotted line. The count of articles on different taxonomic groups is not completely mutually exclusive – some articles involve two or more taxonomic groups, and these articles were used to estimate the statistics for each taxonomic group respectively. Therefore, the total number of articles is not a simple sum of the number of articles of the three groups.
Internal Publication Chinese Barcoding Research
In order to present the contributions made by Chinese scientists to the worldwide efforts focused on DNA barcoding, we compared the 20 journals where Chinese scientists published their research most frequently over the last 16 years. As shown in Figure 6, two thirds of the publications were in Chinese journals. More than half of these domestic journals pertained to traditional Chinese medicine, indicating the great need of DNA barcoding technology for medically related studies. These types of studies are more likely to be of use to Chinese researchers than a global audience, so Chinese journals may be the most appropriate. The English journals PLoS One, Zootaxa, Scientific Reports, Molecular Ecology Resources, ZooKeys, Mitochondrial DNA Part A, Systematics and Evolution, and Ecology and Evolution comprised 46.2% of the publications contributed by Chinese scientists (Figure 6) and a majority of what could be considered systematics, evolution, ecology, and biodiversity studies. One potential benefit from publishing the research in Chinese journals is that access to the research is locally available, thus enhancing more general use of barcode data. One drawback from publishing primarily in Chinese journals is that contributions made by scientists from China are inaccessible to scientists from other countries. Therefore, the contributions made by Chinese scientists are underappreciated, but data transparency is less acute in the ecology and evolution literature rather than that in medical or pharmaceutical publications.
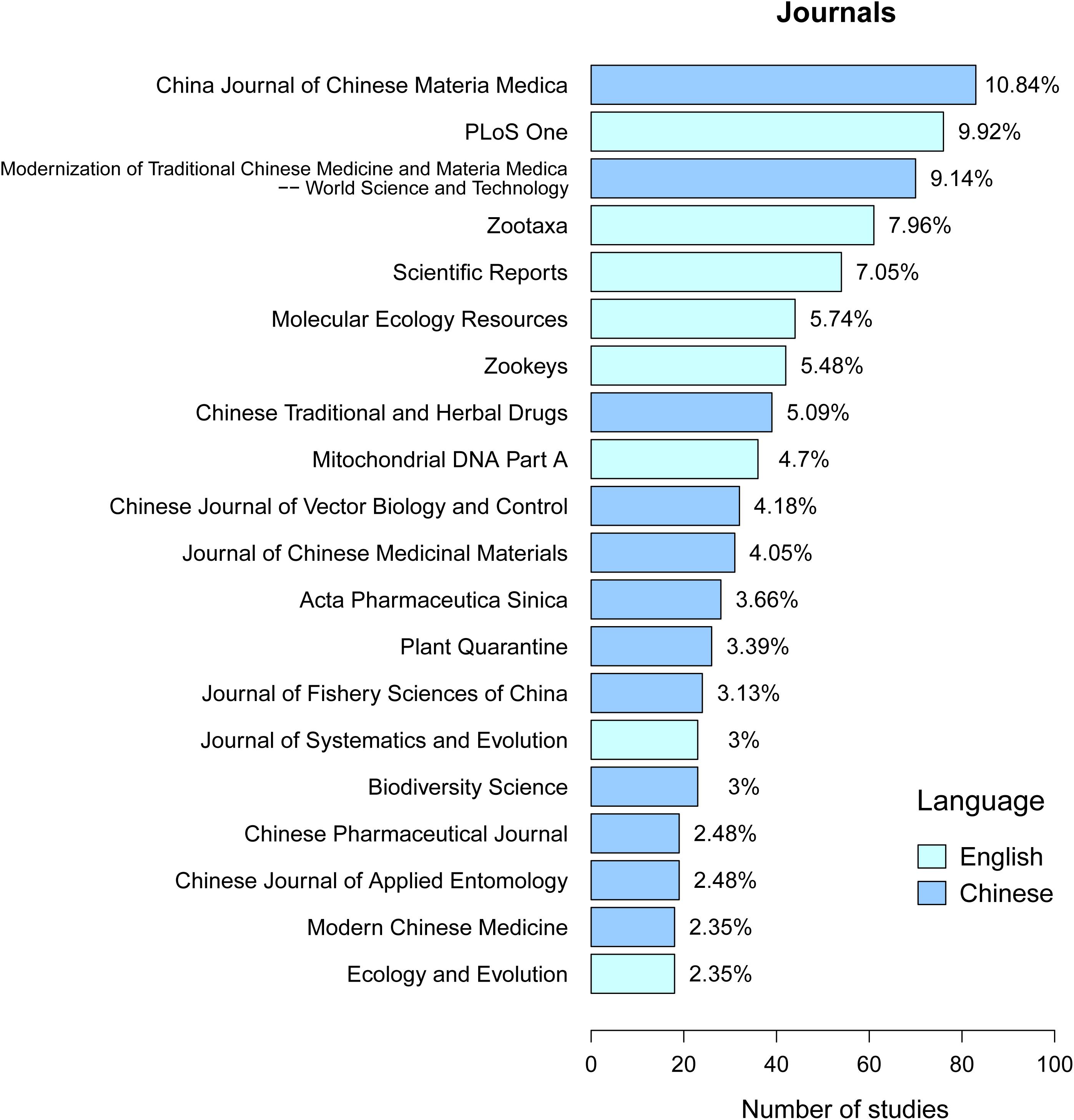
Figure 6. Top 20 journals that published DNA barcoding studies contributed by Chinese scientists. The X axis represents the number of studies, and the Y axis represents journal names. Percentages represent the number of articles published by each journal as a percentage of the total number of the top 20 journals.
DNA Barcoding-Related Research Areas in China
Species Identification and Diversity
DNA barcoding was firstly proposed to simplify the taxonomic identification of species by providing an efficient and accurate method that did not require taxonomic expertise (Hebert and Gregory, 2005). Based on the prevalence of specific “keywords” in articles published by Chinese scientists, the current application of DNA barcoding in China is primarily for species identification (Figure 7). More recently, the application of DNA barcodes for species identification has matured, and researchers have turned from the exploration and verification of barcode technology to the applications and solutions of practical problems in the taxonomic groups they specialize in.
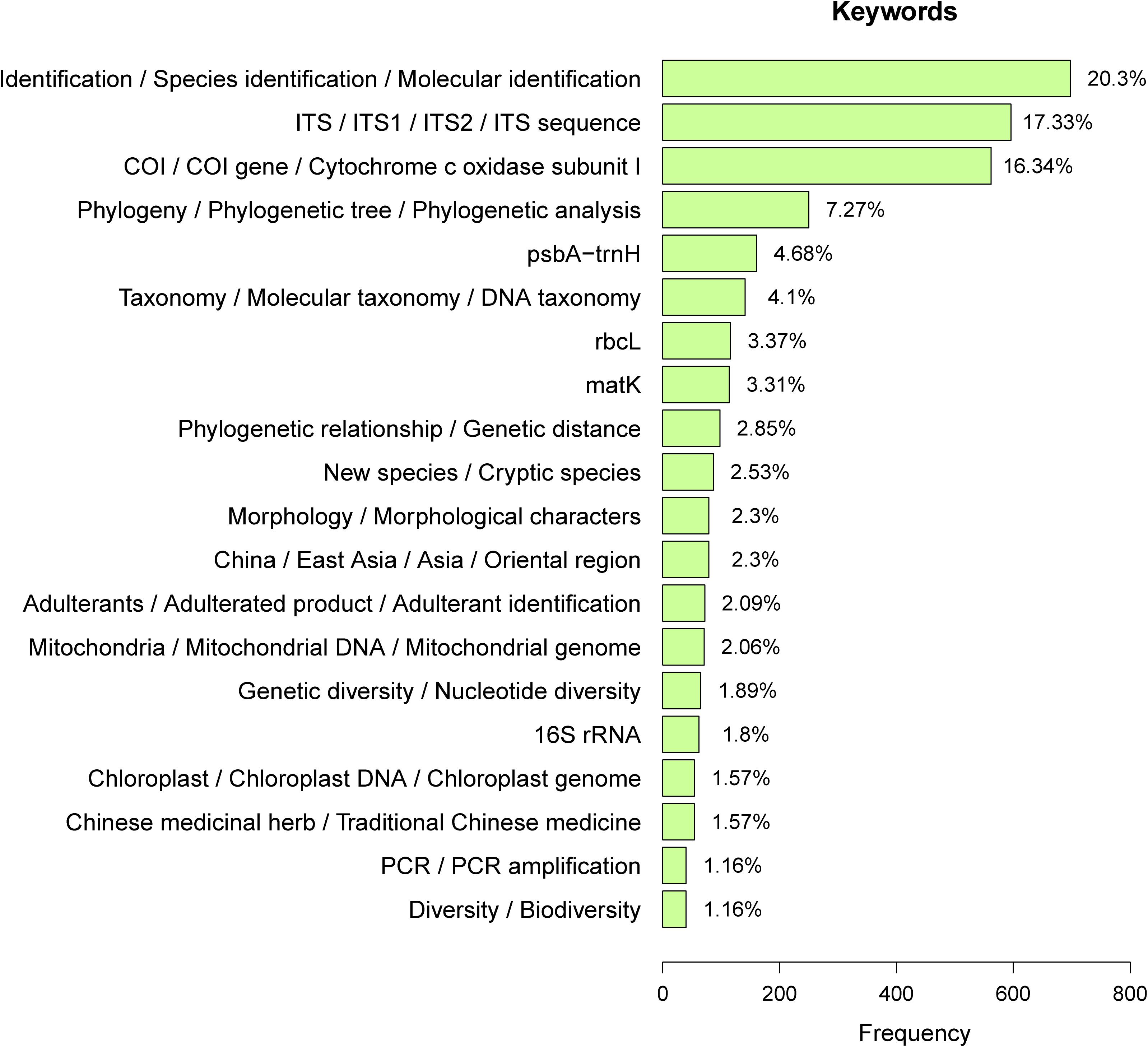
Figure 7. Frequency of key words in DNA barcoding studies published by Chinese researchers. The X axis represents the frequency of various keywords in the search results of all articles, and the Y axis represents similar categories of keywords. Percentages represent the ratio of the frequency of each keyword to the total number of terms in the top 20. There are no terms such as “DNA barcoding,” “DNA barcode,” “DNA barcodes,” and “barcoding” presented here because they are obviously the most frequent ones.
In addition to species identification, Chinese scientists are using DNA barcodes in phylogenetics (e.g., Feng et al., 2016; Liu et al., 2016; Chesters, 2017), the discovery of new or cryptic species (e.g., Liu et al., 2011a, b; Qin et al., 2018), and the evaluation of the levels of biodiversity (e.g., Chen et al., 2015; Chesters et al., 2015; Li et al., 2017; Sun et al., 2018; Figure 7).
Based on the statistics of keywords with the top 20 highest frequency in different literatures, 1.57% of the Chinese barcode research pertains to herbal medicine and 2.09% for identification of adulterants (identification of fraudulent products) in Chinese herbal medicine (Figure 7). Therefore, the emergence of DNA barcoding technology has indeed proven important for research on Chinese traditional medicine.
Standard DNA Barcodes for Plant Groups Need Further Exploration
Ideally, DNA barcodes should at least satisfy the following criteria: (1) specificity – the DNA fragment must be nearly identical in the same species but different between different species; (2) uniformity – the section must be standardized (the same section should be used in different taxonomic groups); and (3) robustness – the marker must have conservative primer binding sites that allow it to be amplified and sequenced from a large number of groups (Pečnikar and Buzan, 2014). Despite years of effort to find universal DNA barcodes for different taxonomic groups, people have to admit that searching for a universal barcode for all species is utopian. The top five most commonly used barcodes by Chinese scientists for their own taxonomic groups are listed in Figure 8. It was found that COI was used in nearly all studies involving the barcoding of animal groups in China (Figure 8A), indicating that the COI region has been consistently important for the general use of DNA barcodes of animal groups due to the fact that COI barcodes perform excellently in most animal groups (e.g., Hebert et al., 2003a, b; Rougerie et al., 2009; Steinke et al., 2009). Although other markers, such as 16S rRNA, Cytb, ITS2, etc., have also been used in some studies of animal groups, they were co-analyzed with COI in most cases (e.g., Li et al., 2010; Jin et al., 2018; Huang et al., 2019). Similarly, ITS genes are the most commonly used molecular markers (Figure 8C) in studies that focus on microorganisms, while other genes are used relatively infrequently and are generally used as auxiliary barcodes.
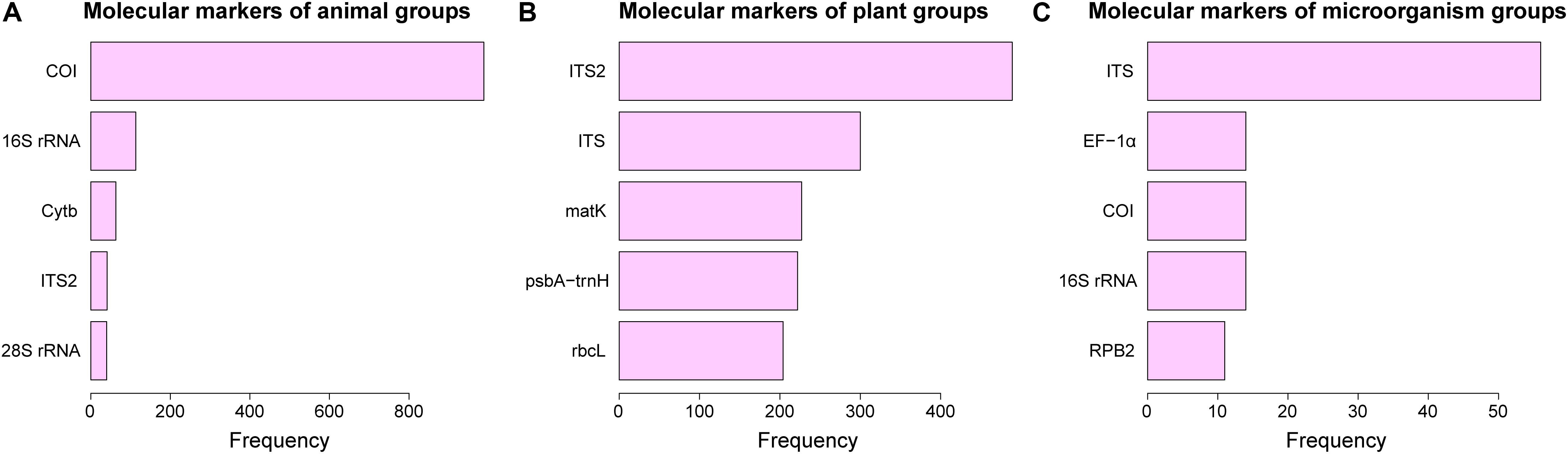
Figure 8. Frequency of DNA barcodes articles pertaining to animals, plants, and microorganisms. (A) animal, (B) plant, and (C) microorganism groups. The X axis represents the frequency of each molecular marker used in the article, and the Y axis represents the name of the gene used as a molecular marker.
However, in plant groups, the most frequently used molecular markers are not as obvious (Figure 8B). ITS2 and ITS are the most widely used markers in Figure 8B, which were proposed as novel barcodes for medicinal plants by Chen et al. (2010) and were suggested to be incorporated into the core barcode for seed plants by China-Plant-BOL-Group et al. (2011). MatK, psbA-trnH, and rbcL are high-frequency candidate barcodes for plants as well, which may be related to the joint use of multiple plant barcodes in most studies (e.g., Yang et al., 2012; Jin et al., 2014; Gong et al., 2016; Bao et al., 2018).
In fact, a large part of the studies on plant barcodes in China are carried out on Chinese medicinal herbs, and the barcodes selected for these studies are often different. For example, Li et al. (2014) identified the herbal medicinal materials from Aristolochia using the matK, rbcL, psbA-trnH, and trnL-trnF DNA regions. Guo et al. (2017) identified the herbal materials from Cynanchum using the ITS2 barcode; Gong et al. (2018) constructed a DNA barcode reference library for “Nan Yao” (crude drugs mainly produced in or imported through tropical and subtropical China, especially the Lingnan region, i.e., the territories south of the Nanling Mountains) using ITS2; and Jiao et al. (2018) identified the medicinal Polygonati Rhizoma (a traditional medicinal and edible product with Polygonatum polysaccharides, saponins, phenols, and flavonoids) efficiently and accurately using ITS2 and psbA-trnH sequences. This shows that the selection of molecular markers for plant groups in China still relies heavily on the combination of multiple markers.
Primary Research Institutions on DNA Barcoding
The Institute of Chinese Medicine Science and Marine Biology: Dominant Institutions Focusing on DNA Barcoding Research in China
As shown in Figure 9A, the top five Chinese institutions with the largest number of articles published on DNA barcodes include (in order of the most to fewest publications) the Institute of Medicinal Plant Development (Chinese Academy of Medical Sciences & Peking Union Medical College), the Ocean University of China, the Institute of Zoology (Chinese Academy of Sciences), the Kunming Institute of Botany (Chinese Academy of Sciences), and the South China Agricultural University. Research at these institutions mainly focuses on traditional Chinese medicine, marine organisms, and other animals and plants.
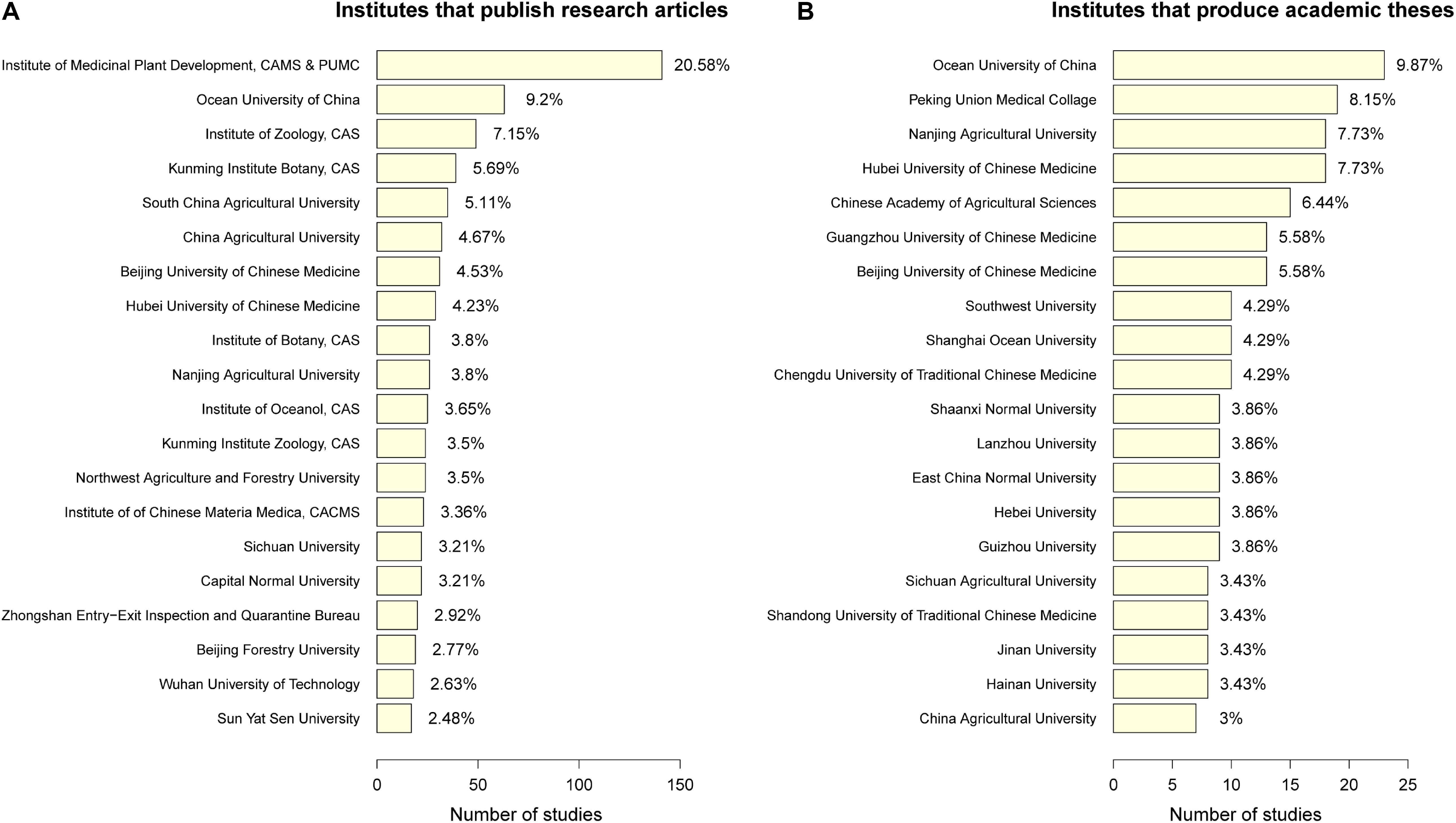
Figure 9. Top 20 Chinese institutions that published (A) research articles and (B) academic dissertations in China. The X axis represents the number of studies, and the Y axis represents the name of the institution. The abbreviations are as follows: CAMS & PUMC – Chinese Academy of Medical Sciences & Peking Union Medical College; CAS – Chinese Academy of Sciences; and CACMS – China Academy of Chinese Medical Sciences.
Comparatively, Figure 9B lists the top 20 universities or research institutions that have contributed the highest proportion of 494 dissertations related to DNA barcoding. The Ocean University of China has produced the most master’s and doctoral dissertations, followed by Peking Union Medical Collage, Nanjing Agricultural University, Hubei University Chinese Medicine, and the Chinese Academy of Agricultural Sciences. These dissertations focused primarily on marine organisms and traditional Chinese medicine. Together, these figures reveal which institutions have pioneered barcoding research in China.
Conclusion and Future Perspectives
Based on our analyses, the contribution over the last 16 years by Chinese scientists to research using DNA barcoding is underappreciated, primarily because of the bias in that over half of relevant articles were published in the Chinese rather than the international literature. In terms of the overall number of barcode entries, Chinese scientists have produced a considerable amount of information on plants and animals (Figure 9), and the amount of data is close to that produced by several other leading countries in the field (Figure 1). Yet some DNA barcode sequences are not totally publicly available due to Chinese journals not clearly requiring data transparency and accessibility for DNA barcodes. In addition to barcode information on a variety of species, Chinese scientists are involved in the development of new barcoding methods as well as the analysis of barcode data from a large amount of sequencing information.
During the inception of barcoding, research in China was less well developed than the rest of the world, but it has, since 2009, witnessed a rapid growth (Figure 3). This growth of DNA barcoding in China is continuously expanding from medicinal plants to including other plants and animals, but the primary focus is still on medically and economically important species in need of identification. Additionally, the application of barcode technology is expanding, with studies related to phylogenetics, population genetics, and biodiversity becoming more common.
There are several potential research directions for Chinese scientists:
(1). Developing integrated evolutionary and/or ecological projects implementing DNA barcoding. We must admit that most current barcoding studies in China represent follow-up research and lack conceptual originality. The main important concepts and initiatives of DNA barcoding were not proposed by Chinese scientists in general (Pei et al., 2017). Studies with barcode data that appear in western journals where data transparency is required are often concerned with solving important ecological and evolutionary problems. However, China has the funding for – and satisfies the conditions of – the development of comprehensive research projects and promotion of theoretical innovation. In China, there is still a lot of unsurveyed biodiversity, from rainforests to deserts, where both taxonomists and evolutionary biologists could conduct investigations via DNA barcoding. This technology may also be applied to studies on macroevolution, interactions and food webs, environmental monitoring (Valentini et al., 2009; Garlapati et al., 2019). To maximize the value of DNA barcoding data, the people who collect it must collaborate with ecologists and evolutionary biologists (Joly et al., 2013; Cristescu, 2014) to expand the usefulness of barcode data. In the process, Chinese scientists have the opportunity to come up with their own new ideas and approaches to barcoding by developing integrated evolutionary and/or ecological projects implementing DNA barcoding.
(2). Proposing new approaches and de novo assigning algorithms for NGS related DNA barcoding. The concept of metabarcoding (Taberlet et al., 2012) has greatly expanded the potential scope of applications of DNA barcoding in recent years. A few scientists from China have published important papers on metabarcoding (e.g., Yu et al., 2012; Zhou et al., 2013; Liu et al., 2017; Lang et al., 2019), showing great potential in this field. As DNA barcoding technology matures, we think Chinese scientists should make more contributions in metabarcoding. Currently, fewer methodological studies are optimizing sequencing procedures or proposing new assignment algorithms to better address the challenges of the big data era (Coissac et al., 2016). The need for biodiversity-related research also poses new challenges for barcode bioinformatics analysis (Taberlet et al., 2012; Wang et al., 2019). For example, neither PCR-based nor PCR-free metabarcoding protocol allows the accurate estimation of species abundance (Braukmann et al., 2019), several barriers are still exist in metabarcoding when solving quantitative ecological issues. As each method has its shortcomings in certain contexts (Paz and Crawford, 2012), no perfect DNA barcoding method has been proposed for all cases (Li et al., 2013). The direction of multi-gene, multi-method, and multi-discipline combinations will become a primary focus in the future (Yang et al., 2018), and that is why there is so much space for the development of methodological advances, given the high demand for biodiversity research in China.
(3). Constructing a national-level DNA barcoding reference library. This has also been suggested by some other scientists (Pei et al., 2017). Although there are a few local barcoding libraries constructed for specific taxa (e.g., Hou et al., 2017; Gong et al., 2018; Liu et al., 2018), few leading and international DNA barcoding libraries have been created or have been hosted by Chinese scientists. Chen et al. (2014) established and continually maintain an online DNA barcoding database for herbal materials4 with 78,847 barcode records belonging to 23,262 species, which shows the possibility of constructing national-level DNA barcode sequence libraries in China. Based on such efforts to build a foundation for barcoding, China can achieve far more toward documenting its immense biodiversity (Xu et al., 2015; Pei et al., 2017).
(4). Integrating into global research by making their DNA barcode data available to global barcoding research communities. Some Chinese journals do not clearly require authors to submit their DNA barcodes to a publicly available database (e.g., submission to GenBank), rendering these DNA barcodes invisible to the broader scientific community, impeding DNA barcoding research both globally and in China. Together with help from the global scientist community, Chinese scientists must further their efforts to close the gap with their international counterparts, especially in data standardization and disclosure. With the efforts made by the biodiversity committee of the Chinese Academy of Sciences since 2013, GBIF (Global Biodiversity Information Facility) has made a Chinese portal5. If a Chinese edition of GenBank can be established, as proposed in (3), and be accessible to the researchers all over the world, submitting the data (including but not only DNA barcodes) to the library should be equivalent to submitting to GenBank. Chinese and overseas researchers are to be encouraged to submit data to both of them simultaneously before publishing their works. Currently, the National Genomics Data Center6 may be the most appropriate candidate for a Chinese DNA barcode repository.
Author Contributions
AZ designed the study. CY performed the research. CY and QL analyzed the data. CY and AZ wrote the first draft of the manuscript. All authors contributed substantially to revisions.
Funding
This work was supported by China National Funds for Distinguished Young Scientists (Grant Number 31425023), the Natural Science Foundation of China (Grant Number 31772501), Support Project of High-level Teachers in Beijing Municipal Universities (Grant Number IDHT20180518), and Academy for Multidisciplinary Studies, Capital Normal University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. Michael C. Orr and the editor, Dr. RH, whose comments have considerably improved the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00057/full#supplementary-material
DATA SHEET S1 | Summary of articles with Chinese institutions/universities as the first research institute during the period between 2003 and August 2019.
Footnotes
- ^ http://isiknowledge.com
- ^ https://www.cnki.net/
- ^ http://www.boldsystems.org
- ^ http://www.tcmbarcode.cn
- ^ http://www.gbifchina.org/
- ^ http://bigd.big.ac.cn/
References
Ahrens, D., Monaghan, M. T., and Vogler, A. P. (2007). DNA-based taxonomy for associating adults and larvae in multi-species assemblages of chafers (Coleoptera: Scarabaeidae). Mol. Phylogenet. Evol. 44, 436–449. doi: 10.1016/j.ympev.2007.02.024
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Austerlitz, F., David, O., Schaeffer, B., Bleakley, K., Olteanu, M., Leblois, R., et al. (2009). DNA barcode analysis: a comparison of phylogenetic and statistical classification methods. BMC Bioinformatics 10:S10. doi: 10.1186/1471-2105-10-S14-S10
Bao, W., Li, D., and Li, X. (2018). DNA barcoding of Actinidia (Actinidiaceae) using internal transcribed spacer, matK, rbcL and trnH-psbA, and its taxonomic implication. N. Z. J. Bot 56, 360–371. doi: 10.1080/0028825X.2018.1491009
Braukmann, T. W., Ivanova, N. V., Prosser, S. W., Elvrecht, V., Steinke, D., Ratnasingham, S., et al. (2019). Metabarcoding a diverse arthropod mock community. Mol. Ecol. Resour. 19, 711–727. doi: 10.1111/1755-0998.13008
Chen, S., Pang, X., Song, J., Shi, L., Yao, H., Han, J., et al. (2014). A renaissance in herbal medicine identification: from morphology to DNA. Biotechnol. Adv. 32, 1237–1244. doi: 10.1016/j.biotechadv.2014.07.004
Chen, S., Yao, H., Han, J., Liu, C., Song, J., Shi, L., et al. (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5:e8613. doi: 10.1371/journal.pone.0008613
Chen, W., Ma, X., Shen, Y., Mao, Y., and He, S. (2015). The fish diversity in the upper reaches of the Salween River, Nujiang River, revealed by DNA barcoding. Sci. Rep. 5:17437. doi: 10.1038/srep17437
Cheng, X., Wang, A., Gu, Z., Wang, Y., Zhan, X., and Shi, Y. (2011). Current progress of DNA barcoding. Genom. Appl. Biol. 30, 748–758. doi: 10.3969/gab.030.000748
Chesters, D. (2017). Construction of a species-level tree of life for the insects and utility in taxonomic profiling. Syst. Biol. 66, 426–439. doi: 10.1093/sysbio/syw099
Chesters, D., Zheng, W. M., and Zhu, C. D. (2015). A DNA barcoding system integrating multigene sequence data. Methods Ecol. Evol. 6, 930–937. doi: 10.1111/2041-210X.12366
China Plant Bol Group, Li, D. Z., Gao, L. M., Li, H. T., Wang, H., Ge, X. J., et al. (2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U.S.A. 108, 19641–19646. doi: 10.1073/pnas.1104551108
Coissac, E., Hollingsworth, P. M., Lavergne, S., and Taberlet, P. (2016). From barcodes to genomes: extending the concept of DNA barcoding. Mol. Ecol. 25, 1423–1428. doi: 10.1111/mec.13549
Cristescu, M. E. (2014). From barcoding single individuals to metabarcoding biological communities: towards an integrative approach to the study of global biodiversity. Trends Ecol. Evol. 29, 566–571. doi: 10.1016/j.tree.2014.08.001
Czelusniak, J., Goodman, M., Moncrief, N. D., and Kehoe, S. M. (1990). Maximum parsimony approach to construction of evolutionary trees from aligned homologous sequences. Methods Enzymol. 183, 601–615. doi: 10.1016/0076-6879(90)83039-C
DeSalle, R., and Goldstein, P. (2019). Review and interpretation of trends in DNA barcoding. Front. Ecol. Evol. 7:302. doi: 10.3389/fevo.2019.00302
Eddy, S. R. (1998). Profile hidden Markov models. Bioinformatics 14, 755–763. doi: 10.1093/bioinformatics/14.9.755
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Feng, S., Jiang, M., Shi, Y., Jiao, K., Shen, C., Lu, J., et al. (2016). Application of the ribosomal DNA ITS2 region of Physalis (Solanaceae): DNA barcoding and phylogenetic study. Front. Plant Sci. 7:1047. doi: 10.3389/fpls.2016.01047
Garlapati, D., Charankumar, B., Ramu, K., Madeswaran, P., and Murthy, M. V. R. (2019). A review on the applications and recent advances in environmental DNA (eDNA) metagenomics. Rev. Environ. Sci. Biotechnol. 28, 389–411. doi: 10.1007/s11157-019-09501-4
Gleason, H. A. (1922). On the relation between species and area. Ecology 3, 158–162. doi: 10.2307/1929150
Gong, L., Qiu, X. H., Huang, J., Xu, W., Bai, J. Q., Zhang, J., et al. (2018). Constructing a DNA barcode reference library for southern herbs in China: a resource for authentication of southern Chinese medicine. PLoS One 13:e0201240. doi: 10.1371/journal.pone.0201240
Gong, W., Liu, Y., Chen, J., Hong, Y., and Kong, H. H. (2016). DNA barcodes identify Chinese medicinal plants and detect geographical patterns of Sinosenecio (Asteraceae). J. Syst. Evol. 54, 83–91. doi: 10.1111/jse.12166
Guo, M., Ren, L., and Pang, X. (2017). Inspecting the true identity of herbal materials from Cynanchum using ITS2 barcode. Front. Plant Sci. 8:1945. doi: 10.3389/fpls.2017.01945
Hajibabaei, M., Singer, G. A., Hebert, P. D., and Hickey, D. A. (2007). DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 23, 167–172. doi: 10.1016/j.tig.2007.02.001
He, T., Jiao, L., Wiedenhoeft, A. C., and Yin, Y. (2019). Machine learning approaches outperform distance- and tree-based methods for DNA barcoding of Pterocarpus wood. Planta 249, 1617–1625. doi: 10.1007/s00425-019-03116-3
Hebert, P. D., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003a). Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hebert, P. D., and Gregory, T. R. (2005). The promise of DNA barcoding for taxonomy. Syst. Biol. 54, 852–859. doi: 10.1080/10635150500354886
Hebert, P. D., Ratnasingham, S., and deWaard, J. R. (2003b). Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. Ser. B Biol. Sci. 270, S96–S99.
Hollingsworth, P. M. (2011). Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 108, 19451–19452. doi: 10.1073/pnas.1116812108
Hou, G., Chen, W. T., Lu, H. S., Cheng, F., and Xie, S. G. (2017). Developing a DNA barcode library for perciform fishes in the South China Sea: species identification, accuracy and cryptic diversity. Mol. Ecol. Resour. 18, 137–146. doi: 10.1111/1755-0998.12718
Huang, X. C., Su, J. H., Ouyang, J. X., Ouyang, S., Zhou, C. H., and Wu, X. P. (2019). Towards a global phylogeny of freshwater mussels (Bivalvia: Unionida): species delimitation of Chinese taxa, mitochondrial phylogenomics, and diversification patterns. Mol. Phylogenet. Evol. 130, 45–59. doi: 10.1016/j.ympev.2018.09.019
Huelsenbeck, J. P., and Ronquist, F. (2001). MrBayes: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jiao, J., Huang, W., Bai, Z., Liu, F., Ma, C., and Liang, Z. (2018). DNA barcoding for the efficient and accurate identification of medicinal polygonati rhizoma in China. PLoS One 13:e0201015. doi: 10.1371/journal.pone.0201015
Jin, Q., Hu, X. M., Han, H. L., Chen, F., Cai, W. J., Ruan, Q. Q., et al. (2018). A two-step DNA barcoding approach for delimiting moth species: moths of Dongling Mountain (Beijing, China) as a case study. Sci. Rep. 8:14256. doi: 10.1038/s41598-018-32123-9
Jin, W. T., Jin, X. H., Schuiteman, A., Li, D. Z., Xiang, X. G., Huang, W. C., et al. (2014). Molecular systematics of subtribe Orchidinae and Asian taxa of Habenariinae (Orchideae, Orchidaceae) based on plastid matK, rbcL and nuclear ITS. Mol. Phylogenet. Evol. 77, 41–53. doi: 10.1016/j.ympev.2014.04.004
Joly, S., Davies, T. J., Archambault, A., Bruneau, A., Derry, A., Kembel, S. W., et al. (2013). Ecology in the age of DNA barcoding: the resource, the promise and the challenges ahead. Mol. Ecol. Resour. 14, 221–232. doi: 10.1111/1755-0998.12173
Lang, D. D., Tang, M., Hu, J. H., and Zhou, X. (2019). Genome-skimming provides accurate quantification for pollen mixtures. Mol. Ecol. Resour. 19, 1433–1446. doi: 10.1111/1755-0998.13061
Li, G. Q., Xue, X. F., Zhang, K. J., and Hong, X. Y. (2010). Identification and molecular phylogeny of agriculturally important spider mites (Acari: Tetranychidae) based on mitochondrial and nuclear ribosomal DNA sequences, with an emphasis on Tetranychus. Zootaxa 2647, 1–15. doi: 10.11646/zootaxa.2647.1.1
Li, J., Liu, X., Guo, H., Yue, B., and Li, J. (2013). Progress of analytic methods in animal DNA barcoding. Sichuan J. Zool. 32, 950–954. doi: 10.3969/j.issn.1000-7083.2013.06.030
Li, L., Josef, B. A., Liu, B., Zheng, S., Huang, L., and Chen, S. (2017). Three-dimensional evaluation on ecotypic diversity of traditional Chinese medicine: a case study of Artemisia annua L. Front. Plant Sci. 8:1225. doi: 10.3389/fpls.2017.01225
Li, M., Au, K. Y., Lam, H., Chen, L., But, P. P., and Shaw, P. C. (2014). Molecular identification and cytotoxicity study of herbal medicinal materials that are confused by Aristolochia herbs. Food Chem. 147, 332–339. doi: 10.1016/j.foodchem.2013.09.146
Liang, F., Dai, Y., Yue, L., Li, F., and Liu, X. (2015). DNA barcoding and taxonomic review of the barklouse genus Stenopsocus (Psocoptera: Stenopsocidae) from Taiwan. Zootaxa 4057, 191–209. doi: 10.11646/zootaxa.4057.2.2
Liu, J., Li, Q., Kong, L., and Zheng, X. (2011a). Cryptic diversity in the pen shell Atrina pectinata (Bivalvia: Pinnidae): high divergence and hybridization revealed by molecular and morphological data. Mol. Ecol. 20, 4332–4345. doi: 10.1111/j.1365-294X.2011.05275.x
Liu, J., Möller, M., Gao, L. M., Zhang, D. Q., and Li, D. Z. (2011b). DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. Mol. Ecol. Resour. 11, 89–100. doi: 10.1111/j.1755-0998.2010.02907.x
Liu, J. X., Wei, M. J., Li, G., Cheng, S. H., Zhao, C. Y., Borjigidai, A., et al. (2018). Construction of ITS2 barcode database of Scutellariae radix and establishment of DNA barcode identification method for its seeds. Chin. J. Exper. Tradi. Med. Form. 24, 37–45. doi: 10.13422/j.cnki.syfjx.20180906
Liu, S., Li, Y., Lu, J., Su, X., Tang, M., Zhang, R., et al. (2013). SOAPBarcode: revealing arthropod biodiversity through assembly of Illumina shotgun sequences of PCR amplicons. Methods Ecol. Evol. 4, 1142–1150. doi: 10.1111/2041-210X.12120
Liu, S., Yang, C., Zhou, C., and Zhou, X. (2017). Filling reference gaps via assembling DNA barcodes using high-throughput sequencing—moving toward barcoding the world. Gigascience 6, 1–8. doi: 10.1093/gigascience/gix104
Liu, X., Liang, M., Etienne, R. S., Gilbert, G. S., and Yu, S. (2016). Phylogenetic congruence between subtropical trees and their associated fungi. Ecol. Evol. 6, 8412–8422. doi: 10.1002/ece3.2503
Munch, K., Boomsma, W., Huelsenbeck, J. P., Willerslev, E., and Nielsen, R. (2008). Statistical assignment of DNA sequences using Bayesian phylogenetics. Syst. Biol. 57, 750–757. doi: 10.1080/10635150802422316
Myers, N., Mittermeier, R. A., Mittermeier, C. G., da Fonseca, G. A. B., and Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature 403, 853–858. doi: 10.1093/acrefore/9780199389414.013.95
Paz, A., and Crawford, A. J. (2012). Molecular-based rapid inventories of sympatric diversity: a comparison of DNA barcode clustering methods applied to geography-based vs clade-based sampling of amphibians. J. Biosci. 37, 887–896. doi: 10.1007/s12038-012-9255-x
Pečnikar, F. Z., and Buzan, E. V. (2014). 20 years since the introduction of DNA barcoding: from theory to application. J. Appl. Genet. 55, 43–52. doi: 10.1007/s13353-013-0180-y
Pei, N., Chen, B., and Kress, W. J. (2017). Advances of community-level plant DNA barcoding in China. Front. Plant Sci. 8:225. doi: 10.3389/fpls.2017.00225
Qin, Y. G., Zhou, Q. S., Yu, F., Wang, X. B., Wei, J. F., Zhu, C. D., et al. (2018). Host specificity of parasitoids (Encyrtidae) toward armored scale insects (Diaspididae): untangling the effect of cryptic species on quantitative food webs. Ecol. Evol. 8, 7879–7893. doi: 10.1002/ece3.4344
Ratnasingham, S., and Hebert, P. D. N. (2007). BOLD: the barcode of life data system (https://www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364. doi: 10.1111/j.1471-8286.2007.01678.x
Ross, H. A., Murugan, S., and Li, W. L. S. (2008). Testing the reliability of genetic methods of species identification via simulation. Syst. Biol. 57, 216–230. doi: 10.1080/10635150802032990
Rougerie, R., Decaëns, T., Deharveng, L., Porco, D., James, S. W., Chang, C. H., et al. (2009). DNA barcodes for soil animal taxonomy. Pesqui. Agropecu. Bras. 44, 789–801. doi: 10.1590/S0100-204X2009000800002
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Shi, Z. Y., Yang, C. Q., Hao, M. D., Wang, X. Y., Ward, R. D., and Zhang, A. B. (2018). FuzzyID2: a software package for large data set species identification via barcoding and metabarcoding using hidden Markov models and fuzzy set methods. Mol. Ecol. Resour. 18, 666–675. doi: 10.1111/1755-0998.12738
Steinke, D., Zemlak, T. S., and Hebert, P. D. (2009). Barcoding nemo: DNA-based identifications for the ornamental fish trade. PLoS One 4:e6300. doi: 10.1371/journal.pone.0006300
Sun, W., Li, J. J., Xiong, C., Zhao, B., and Chen, S. L. (2016). The potential power of Bar-HRM technology in herbal medicine identification. Front. Plant Sci. 7:367. doi: 10.3389/fpls.2016.00367
Sun, X., Bedos, A., and Deharveng, L. (2018). Unusually low genetic divergence at COI barcode locus between two species of intertidal Thalassaphorura (Collembola: Onychiuridae). PeerJ 6:e5021. doi: 10.7717/peerj.5021
Taberlet, P., Coissac, E., Pompanon, F., Brochmann, C., and Willerslev, E. (2012). Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. doi: 10.1111/j.1365-294x.2012.05470.x
Taylor, H. R., and Harris, W. E. (2012). An emergent science on the brink of irrelevance: a review of the past 8 years of DNA barcoding. Mol. Ecol. Resour. 12, 377–388. doi: 10.1111/j.1755-0998.2012.03119.x
Valentini, A., Pompanon, F., and Taberlet, P. (2009). DNA barcoding for ecologists. Trends Ecol. Evol. 24, 110–117. doi: 10.1016/j.tree.2008.09.011
Wang, X., Hua, F., Wang, L., Wilcove, D. S., and Yu, D. W. (2019). The biodiversity benefit of native forests and mixed-species plantations over monoculture plantations. Divers. Distrib. 25, 1721–1735. doi: 10.1111/ddi.12972
Xiao, J. H., Xiao, H., and Huang, D. W. (2004). DNA barcoding: new approach of biological taxonomy. Acta Zool. Sin. 50, 852–855. doi: 10.3969/j.issn.1674-5507.2004.05.023
Xu, C., Dong, W., Shi, S., Cheng, T., Li, C., Liu, Y., et al. (2015). Accelerating plant DNA barcode reference library construction using herbarium specimens: improved experimental techniques. Mol. Ecol. Resour. 15, 1366–1374. doi: 10.1111/1755-0998.12413
Yang, H. Q., Dong, Y. R., Gu, Z. J., Liang, N., and Yang, J. B. (2012). A preliminary assessment of matK, rbcL and trnH-psbA as DNA barcodes for Calamus (Arecaceae) species in China with a note on ITS. Ann. Bot. Fenn. 49, 319–330. doi: 10.5735/085.049.0603
Yang, Q. Q., Liu, S. W., and Yu, X. P. (2018). Research progress on DNA barcoding analysis methods. Chin. J. Appl. Ecol. 29, 1006–1014. doi: 10.13287/j.1001-9332.201803.032
Yao, X. N., Liu, Y., Xue, K., Feng, X. X., Zhang, S. X., Ma, X., et al. (2013). Review of domestic research progress on animal taxonomy DNA barcoding. J. Agric. Sci. Technol. 15, 99–106.
Yu, D. W., Ji, Y., Emerson, B. C., Wang, X., Ye, C., Yang, C., et al. (2012). Biodiversity soup: metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring. Methods Ecol. Evol. 3, 613–623. doi: 10.1111/j.2041-210X.2012.00198.x
Zhang, A. B., Feng, J., Ward, R. D., Wan, P., Gao, Q., Wu, J., et al. (2012a). A new method for species identification via protein-coding and non-coding DNA barcodes by combining machine learning with bioinformatic methods. PLoS One 7:e30986. doi: 10.1371/journal.pone.0030986
Zhang, A. B., Muster, C., Liang, H. B., Zhu, C. D., Crozier, R., Wan, P., et al. (2012b). A fuzzy-set-theory-based approach to analyse species membership in DNA barcoding. Mol. Ecol. 21, 1848–1863. doi: 10.1111/j.1365-294X.2011.05235.x
Zhang, A. B., Hao, M. D., Yang, C. Q., and Shi, Z. Y. (2017). BarcodingR: an integrated R package for species identification using DNA barcodes. Methods Ecol. Evol. 8, 627–634. doi: 10.1111/2041-210X.12682
Zhang, A. B., Sikes, D. S., Muster, C., and Li, S. Q. (2008). Inferring species membership using DNA sequences with back-propagation neural networks. Syst. Biol. 57, 202–215. doi: 10.1080/10635150802032982
Keywords: DNA barcode, sequence assignment, COI, ITS, matK, BOLD
Citation: Yang C, Lv Q and Zhang A (2020) Sixteen Years of DNA Barcoding in China: What Has Been Done? What Can Be Done? Front. Ecol. Evol. 8:57. doi: 10.3389/fevo.2020.00057
Received: 12 November 2019; Accepted: 26 February 2020;
Published: 06 April 2020.
Edited by:
Rodney L. Honeycutt, Pepperdine University, United StatesReviewed by:
Zhi Chao, Southern Medical University, ChinaKaren Leanne Bell, The University of Western Australia, Australia
Xiaohui Pang, Chinese Academy of Medical Sciences, China
Paul Z. Goldstein, United States Department of Agriculture, United States
Copyright © 2020 Yang, Lv and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ai-bing Zhang, zhangab2008@cnu.edu.cn; zhangab2008@mail.cnu.edu.cn
†These authors have contributed equally to this work
‡ORCID: Ai-bing Zhang orcid.org/0000-0003-3450-5421
 Cai-qing Yang
Cai-qing Yang Qing Lv
Qing Lv Ai-bing Zhang
Ai-bing Zhang