Luciferins Under Construction: A Review of Known Biosynthetic Pathways
- 1Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry RAS, Moscow, Russia
- 2Pirogov Russian National Research Medical University, Moscow, Russia
Bioluminescence, or the ability of a living organism to generate visible light, occurs as a result of biochemical reaction where enzyme, known as a luciferase, catalyzes the oxidation of a small-molecule substrate, known as luciferin. This advantageous trait has independently evolved dozens of times, with current estimates ranging from the most conservative 40, based on the biochemical diversity found across bioluminescence systems (Haddock et al., 2010) to 100, taking into account the physiological mechanisms involved in the behavioral control of light production across a wide range of taxa (Davis et al., 2016; Verdes and Gruber, 2017; Bessho-Uehara et al., 2020a; Lau and Oakley, 2021). Chemical structures of ten biochemically unrelated luciferins and several luciferase gene families have been described; however, a full biochemical pathway leading to light emission has been elucidated only for two: bacterial and fungal bioluminescence systems. Although the recent years have been marked by extraordinary discoveries and promising breakthroughs in understanding the molecular basis of multiple bioluminescence systems, the mechanisms of luciferin biosynthesis for many organisms remain almost entirely unknown. This article seeks to provide a succinct overview of currently known luciferins’ biosynthetic pathways.
Introduction
A remarkable diversity of animals and microorganisms possess the ability to generate light, or bioluminescence, which has been adapted by thousands of species to serve the purpose of intra- and inter-species visual communication, to attract prey, escape predators, and in mating (Haddock et al., 2010; Widder, 2010; Shimomura and Yampolsky, 2019; Lau and Oakley, 2021). Various luciferins, the small molecules capable of light emission upon oxidation (Figure 1), have been derived by evolution from unrelated biochemical pathways. At the chemical level, in the majority of known bioluminescent systems light is produced as a result of the decomposition of a four-membered dioxetanone ring, although in some systems (such as bacterial and fungal) acyclic peroxide moieties or a six-membered trioxanone ring intermediates are formed (Vacher et al., 2018; Wang and Liu, 2021). Cleavage of intermediate peroxides requires relatively low energy, yielding compounds in electronically excited state, that decay radioactively to the ground state (Bastos et al., 2017). At the molecular level the luciferin biosynthetic routes remain largely understudied. In this work, we review bioluminescence systems for which the mechanisms of luciferin biosynthesis have been investigated.
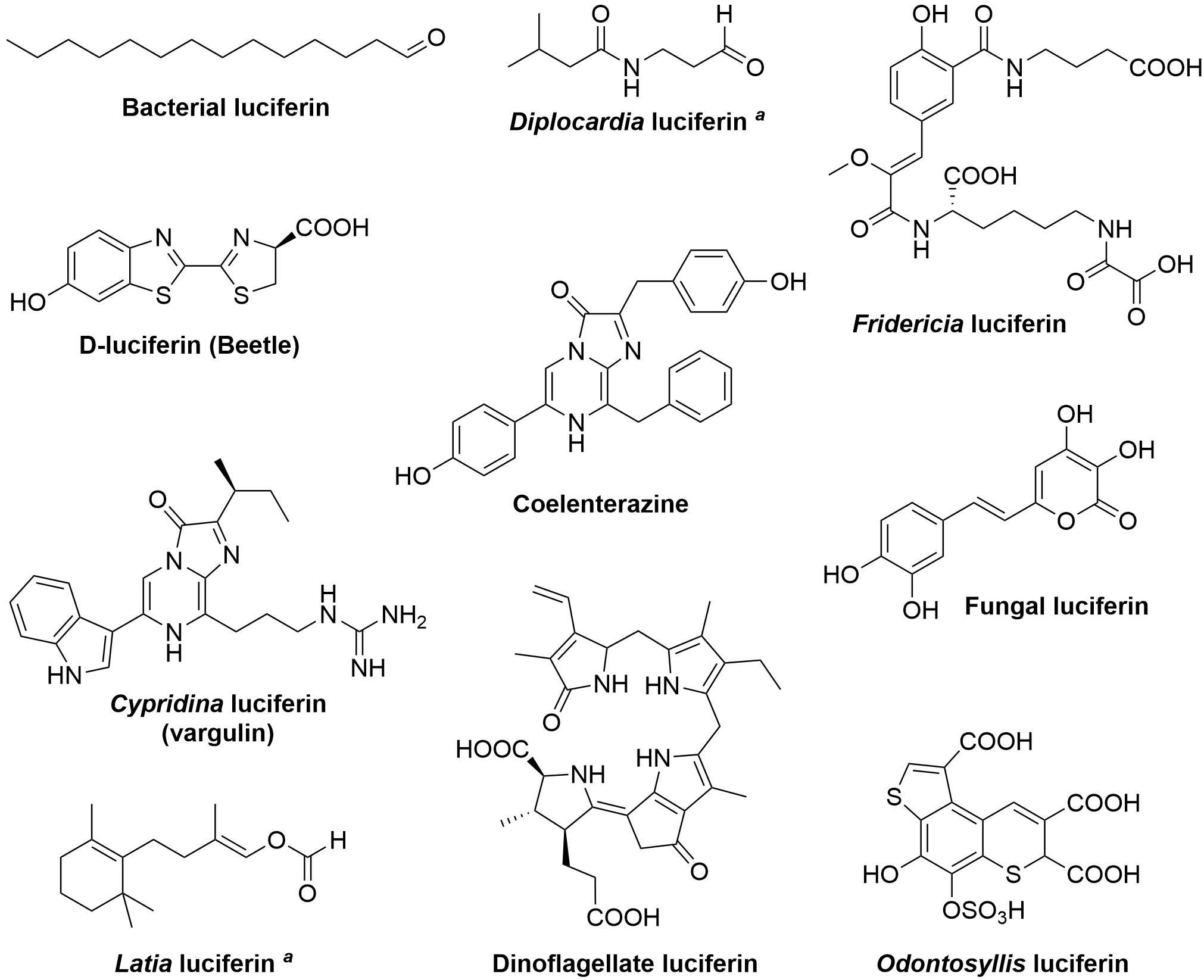
Figure 1. Structures of known luciferins. aThe structure of the light-emitting species is still unknown.
Bacterial Bioluminescent System
Biosynthetic pathway of bacterial bioluminescence is one of the most thoroughly investigated among all luminescent organisms. To date, 25 characterized luminous bacterial species belong in three families of the Gammaproteobacteria: Shewanellaceae (Shewanella), Enterobacteriaceae (Photorhabdus), and Vibrionaceae (Aliivibrio, Photobacterium, and Vibrio) (Vannier et al., 2020). All luminescent bacteria utilize a singular mechanism for light emission and employ similar luciferases (Marquette and Blum, 2010). Bacterial light emission results from the cleavage of the peroxyhemiacetal formed by the reaction among FMNH2, myristic aldehyde (Figure 1) and oxygen within the active site of luciferase (Cormier and Strehler, 1953) producing light at 490 nm, although in some cases the color of luminescence is altered by an energy transfer to fluorescent protein that interacts with the luciferase (Lee et al., 2019). Recycling of FMN is then catalyzed by NADH-dependent flavin reductase (Figure 2A).
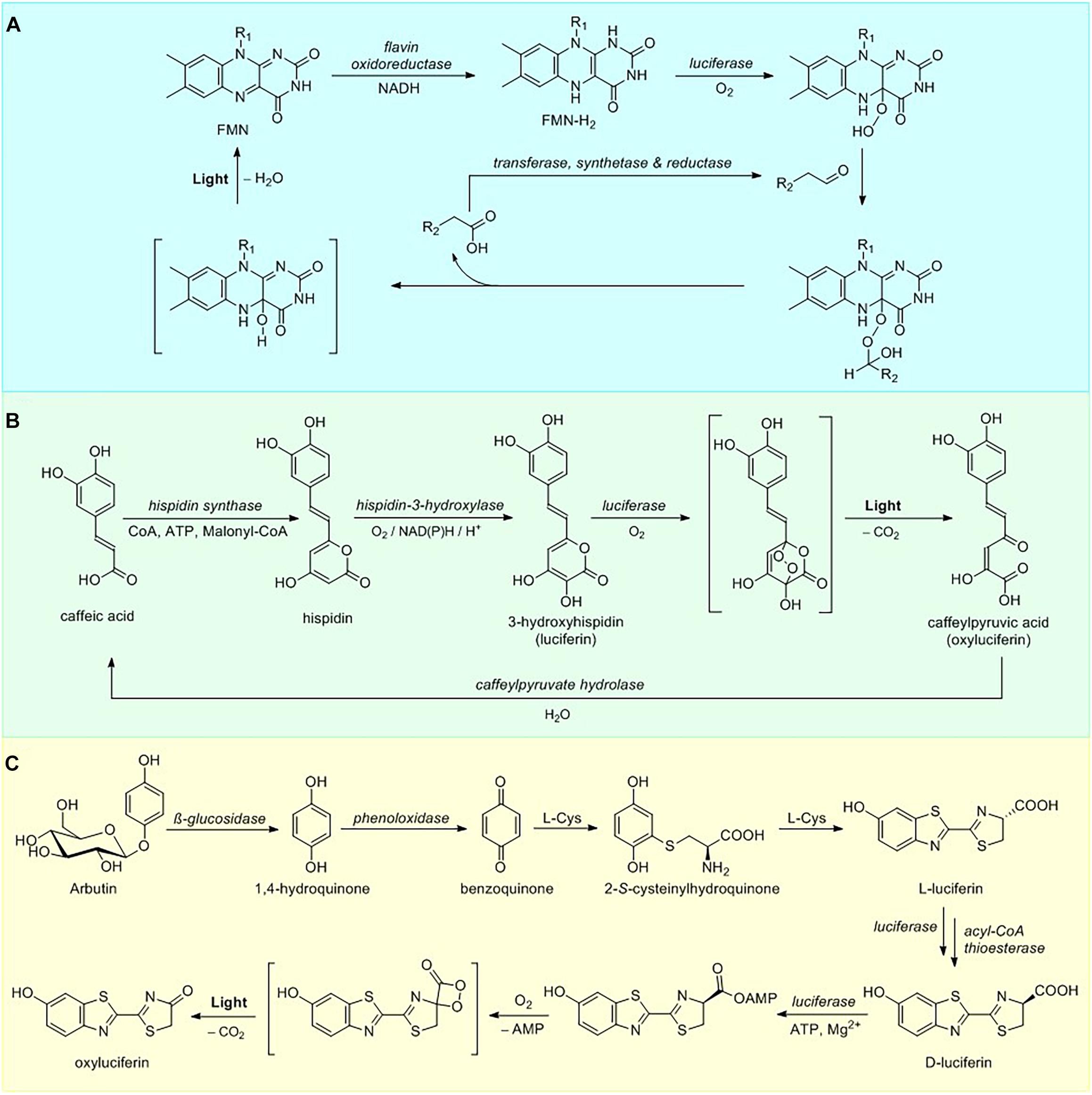
Figure 2. Luciferin metabolism. (A) Overview of bacterial luciferin bioluminescence and recycling pathways. (B) Pathway of fungal luciferin biosynthesis and recycling. (C) Partial D-luciferin biosynthetic pathway and bioluminescence reaction.
One of the most important features of the bacterial bioluminescence system lies in the fact that the full pathway of luciferin biosynthesis and bioluminescence is encoded by a single lux operon (Meighen, 1991), containing genes for bacterial luciferase, consisting of two polypeptide chains (heterodimers luxA and luxB), along with genes luxC, D, and E encoding fatty-acid luciferin reductase complex responsible for the synthesis of the long chain aldehyde substrate and luxG encoding a flavin oxidoreductase (Nijvipakul et al., 2008). In addition to luxCDABE(G) gene cluster, a number of bioluminescent Photobacteria carry an additional luxF gene, showing a limited identity to luxAB encoding bacterial luciferases (Brodl et al., 2020). The LuxF protein is able to scavenge an inhibitory byproduct of bacterial bioluminescence – 6-(3′-(R)-myristyl)-flavin mononucleotide (myrFMN), thus increasing the total intensity of bioluminescence over time. Due to oxygen-labile nature of FMNH2, its transfer mechanism from reductase to luciferase active site is a matter of considerable debate (Brodl et al., 2018), with some research supporting the formation of a transient luciferase-LuxG complex (Jeffers and Tu, 2001; Tu, 2008) and other favoring FMNH2 free diffusion mechanism (Tinikul et al., 2013).
Despite their wide distribution in marine environments, ranging from free-swimming to symbiotic species, bioluminescent bacteria all carry a highly conserved luxCDAB(F)E(G) core (Dunlap and Urbanczyk, 2013), with minor variations correlated with environmental parameters (Brodl et al., 2018) and species life-style, such as symbiotic associations with squid or fish (Davis et al., 2016; Schwartzman and Ruby, 2016), observed in the lux operon architecture. A recent study employing metagenomic data analysis revealed a much wider diversity of lux operon sequences organization with novel lux genes and operons being more abundant in the global ocean than the canonical CDAB(F)E(G) operon (Vannier et al., 2020). Using structural information from the newly discovered individual enzymes of the overarching lux family and addressing evolutionary conserved areas within these structures will eventually provide insight on the evolutionary function and origin of bioluminescence.
Fungal Bioluminescent System
To date the only genetically encodable eukaryotic bioluminescence system was described for luminous fungi. All of the light-emitting fungal species sharing a single bioluminescent system belong to the order Agaricales (Oliveira et al., 2012). In 2015 fungal bioluminescence substrate and its precursor were identified as simple styrylpyrones 3-hydroxyhispidin (Figure 1) and hispidin, respectively (Purtov et al., 2015). In contrast to the previous theory that luciferin is obtained via reduction of its precursor (Airth and McElroy, 1959), the 2015 paper has established that fungal luciferin is biosynthesized by oxidation of the hispidin, catalyzed by a soluble NADPH-dependent hydroxylase. Further research has shown that luciferin undergoes oxidation, catalyzed by an insoluble luciferase in a reaction requiring no other cofactors to produce green light (520 nm) (Kaskova et al., 2017). The fungal bioluminescence is initiated by the cycloaddition of oxygen to luciferin forming an α-pyrone endoperoxide high-energy intermediate, decarboxylation of which results in fungal oxyluciferin, caffeylpyruvate (Figure 2B). The enzymatic hydrolysis of oxyluciferin produces caffeic acid, which is then recycled in the biosynthesis of hispidin via the styrylpyrone pathway (Lee and Yun, 2011). Subsequent experiments showed this mechanism to be ubiquitous in bioluminescence reactions of different species of luminous fungi (Oba et al., 2017).
A tendency of fungal genes encoding specific secondary metabolites to cluster (Keller et al., 2005) has allowed the identification of a set of genes and enzymes involved in fungal luciferin bioluminescence cascade, thus making it the first fully characterized luminescence system from eukaryotes (Kotlobay et al., 2018). Luciferase was discovered to be a member of a conserved gene cluster, which included at least three other genes: h3h encoding hispidin-3-hydroxylase, hisps gene encoding a member of the polyketide synthase family – hispidin synthase, and cph gene encoding caffeylpyruvate hydrolase, belonging to the family of fumarylacetoacetate hydrolases involved in oxyluciferin recycling. A detailed analysis of the reconstructed Agaricales species phylogenetic tree for luz, h3h, and hisps genes revealed a single evolutionary event of the bioluminescence emergence in fungi, indicating that these genes emerged through gene cluster duplication. The gene cluster continued to evolve dynamically after the acquisition of bioluminescence with several independent partial or complete gene loss events leading to the secondary loss of bioluminescence (Ke et al., 2020). This mosaic pattern of bioluminescence in fungi may indicate that selective advantage conveyed by this trait depends on yet unknown specific environmental factors.
Though biological role of light emission in higher fungi remains unclear, further bioinformatic, genomic and evolutionary research of luz, h3h, hisps, and cph genes and their homologs from non-luminescent fungal taxa will provide clues to the origin and function of bioluminescence in fungi. Moreover, a set of 50 genes possessing regulatory function associated with fungal luminescence intensity at different developmental stages and tissues was recently identified (Ke et al., 2020). Future exploration of these might shed light on the ecological significance of fungal regulation of bioluminescence.
D-Luciferin-Dependent Systems
Beetle bioluminescence systems belong to one of the most practically important and well-understood groups of bioluminescence reactions. The ability to emit light has evolved in several Coleoptera families including fireflies (Lampyridae), click beetles (Elateridae) and railroad worms (Phengodidae), with the light emission wavelength ranging from green to red (540–640 nm) depending on the beetle species (Viviani et al., 2011; Kotlobay et al., 2020). All known beetle bioluminescence system depend on a common substrate (S)-2-(6-hydroxy-2-benzothiazolyl)-2-thiazoline-4-carboxylic acid, commonly known as D-luciferin (Figure 1), which consists of two structural units, benzothiazole and thiazoline rings (Shimomura and Yampolsky, 2019). D-luciferin bioluminescence reaction proceeds in two general steps (Figure 2C) shared by all the investigated coleopteran species: substrate adenylation and oxygenation, resulting in the formation of the high-energy dioxetanone moiety, further decomposition of which leads to oxyluciferin and the emission of a photon of light (Branchini et al., 2015). The luciferases catalyzing D-luciferin oxidation are a family of highly conserved homologous ATP-dependent enzymes evolved from the ubiquitous fatty acyl-CoA synthetases (Inouye, 2010; Fallon et al., 2018).
The synthetic D-luciferin is typically obtained via condensation of 2-cyano-6-hydroxybenzothiazole with D-cysteine (White et al., 1963). Although early research has suggested that in vivo biosynthesis might proceed via the same route, neither of these compounds has ever been isolated from beetle biomass. A number of recent publications has revealed the partial D-luciferin biosynthetic pathway using incorporation of stable isotope-labeled compounds into the adult lantern of the live firefly Aquatica lateralis (formerly Luciola lateralis) and mass-spectrometry experiments (Oba et al., 2013; Kanie et al., 2016, 2018). The proposed de novo biosynthetic pathway commences with hydrolysis of arbutin releasing 1,4-hydroquinone (Figure 2C), which is then oxidized to para-benzoquinone. A 1,4-addition of L-cysteine to the benzoquinone, followed by decarboxylation and carbon-sulfur rearrangement of cysteine leads to the formation of benzothiazole ring of the beetle luciferin (Kanie et al., 2016) in the process similar to that of late stage pheomelanogenesis (Wakamatsu et al., 2009; Napolitano et al., 2013). Subsequent addition of a second L-cysteine yields L-luciferin. Current evidence suggests that at the final step L-luciferin undergoes CoA esterification catalyzed by luciferase, followed by epimerization and thioester hydrolysis thus generating the D-form (Niwa et al., 2006; Maeda et al., 2017). These results were further supported by expression analysis of firefly luciferin biosynthetic pathway candidate genes (polyphenol oxidase, β-glucosidase, luciferase, and acyl-CoA thioesterase) in the luminous organs of Lamprigera yunnana and Abscondita terminalis and L-luciferin enzymatic deracemization experiments in vitro (Zhang et al., 2020), demonstrating that acyl-CoA thioesterases can efficiently convert L-luciferin to D-luciferin. An earlier hypothesis of possible luciferin storage mechanism in the form of sulfoluciferin (Fallon et al., 2016) was also corroborated by sulfotransferase expression analysis in the luminous organs of both species (Zhang et al., 2020).
Although great strides in the investigation of the metabolic biochemistry of the firefly bioluminescent system have been made, it should be noted, that the existing hypothesis of D-luciferin biosynthesis is based on the studies in Lampyridae family, while luciferin biosynthetic pathways in Elateridae and Phengodidae lineages are vastly underinvestigated. Moreover, the question of oxyluciferin recycling mechanism in Lampyridae also remains unanswered. Preliminary studies have proposed that in fireflies oxyluciferin could be enzymatically converted to luciferin by luciferin-regenerating enzyme (LRE) (Gomi and Kajiyama, 2001; Emamzadeh et al., 2010); however, more recent evidence suggests that LRE may perform other functions in cells and its role in D-luciferin recycling in vivo requires further clarification (Hosseinkhani et al., 2017).
Imidazopyrazinone-Based Systems
Despite the fact that the highest diversity of luminous organisms is found in marine ecosystems, a majority of known marine bioluminescence systems depend on two modified tripeptides, sharing the same imidazopyrazinone core: coelenterazine and Cypridina luciferin (vargulin) (Figure 1; Haddock et al., 2010). Only three animals have been shown to produce coelenterazine, the decapod shrimp Systellaspis debilis (Thomson et al., 1995), the copepods Metridia (Oba et al., 2009; Tessler et al., 2018) and ctenophores Bolinopsis infundibulum and Mnemiopsis leidyi (Bessho-Uehara et al., 2020a). Interestingly, hydromedusa Aequorea victoria, from which coelenterazine was first isolated, is unable to produce its own coelenterazine for bioluminescence but acquires it through the diet (Haddock et al., 2001). In fact, most species that use coelenterazine as a luciferin do not synthesize it themselves, and its widespread occurrence in both luminous and non-luminous organisms can be explained by its presence in marine food chains (Haddock et al., 2010; Widder, 2010; Mallefet et al., 2020). Moreover, a recent report showed that some species, namely semi-translucent luminous fish Parapriacanthus ransonneti, are able to obtain not only its luciferin (vargulin) but also its luciferase enzyme, aptly termed “kleptoprotein,” from bioluminescent ostracod prey (Bessho-Uehara et al., 2020b).
Several marine species use sulfated derivatives of imidazopyrazinone luciferins as substrates in bioluminescence reaction or as a form of luciferin storage (Lau and Oakley, 2021). To prevent non-specific oxidation of luciferins both Cypridina hilgendorfii and Renilla reniformis were proposed to utilize species-specific sulfotransferases to catalyze the reversible sulfation of substrates in the presence of PAPS to produce 3-enol sulfate derivatives of vargulin (Nakamura et al., 2014) and coelenterazine (Cormier et al., 1970; Inoue et al., 1977), respectively. Other species have been found to use modified forms of coelenterazine as luciferins: the firefly squid Watasenia scintillans utilizes disulfated coelenterazine (Inoue et al., 1976), while the squid Sthenoteuthis oualaniensis (Takahashi and Isobe, 1994) and bivalve mollusk Pholas dactylus (Tanaka et al., 2009) use dehydrocoelenterazine (Lau and Oakley, 2021).
A number of coelenterazine-utilizing luciferases are well known, including those of soft corals (Renilla), copepods (Gaussia, Metridia) and decapods (Oplophorus) (Kotlobay et al., 2020), while vargulin-dependent luciferases have been derived only from an ostracod lineage Cypridinidae (Hensley et al., 2021). Coelenterazine also serves as the light emitter in the Ca2+-binding photoproteins, such as aequorin and obelin (Head et al., 2000). While imidazopyrazinone luciferins are conserved, the enzymes (luciferases and photoproteins) were thought to be unique and species-specific (Haddock et al., 2010). However, two recent publications have revealed a surprising similarity between luciferases of phylogenetically distant organisms: the luminous brittle star Amphiura filiformis (Echinodermata), tunicates Pyrosoma atlanticum (Chordata) and the sea pansy Renilla (Cnidaria), indicating that all three luciferases may have evolved convergently from homologous dehalogenases (Loening et al., 2006; Delroisse et al., 2017; Tessler et al., 2020). Bioluminescence maxima of all naturally occurring imidazopyrazinone-dependent enzymes lie in the blue region of the visible spectrum (450–490 nm), although in some cases the color of luminescence is biochemically altered by an energy transfer to fluorescent protein that interacts with the luciferase (Kotlobay et al., 2020).
The mechanism of imidazopyrazinones bioluminescence reaction follow classical pathway of luciferase-catalyzed luciferin oxidation, dioxetanone intermediate formation and decarboxylation yielding an electronically excited oxyluciferin, that relaxes to the ground state by photon emission. Apart from oxygen imidazopyrazinone-dependent luciferases typically do not require any additional cofactors for light emission (Kaskova et al., 2016), while photoproteins are triggered to produce light upon binding to di- or mono- valent metal ions, which cause a conformational change in the protein. Although, the exact biosynthetic routes of imidazopyrazinone luciferins are currently unknown, feeding studies with living animal specimens have shown that vargulin is biosynthesized from L-tryptophan, L-arginine, and L-isoleucine (Kato et al., 2004, 2007), while coelenterazine is derived from L-phenylalanine and two L-tyrosines (Oba et al., 2009; Tessler et al., 2018). It had been proposed that a sequence of cyclization–dehydration reactions of the corresponding tripeptides could potentially lead to the light-emitting imidazopyrazinones (McCapra and Roth, 1972). Recently transcriptome analysis has been employed to probe for coelenterazine biosynthesis pathway candidate genes, containing FYY motif (Francis et al., 2015). A set of FYY-containing genes encoding isopenicillin-N-synthase homologs were found in the transcriptomes of 24 luminous ctenophore species but were conspicuously absent in the transcriptomes of the non-luminous relatives, suggesting their importance in physiology of bioluminescent ctenophores. However, due to the lack of experimental evidence the role of isopenicillin-N-synthase, and the identity of other enzymes directly involved in coelenterazine biosynthetic pathway remains unknown. Discovery of the biosynthetic pathway of imidazopyrazinone luciferins would enable the development of a broad range of novel reporter systems and may ultimately provide insights into the evolution of bioluminescence in marine organisms.
Fridericia Bioluminescence System
Within the past decade another ATP-dependent bioluminescence system was discovered in the Siberian earthworm Fredericia heliota (Petushkov et al., 2014). The luciferin and luciferase of F. heliota bioluminescence system are structurally distinct from those of Coleoptera, but share a requirement for ATP, Mg2+ ions and oxygen to produce light at 478 nm. Fridericia luciferin was revealed to be an unusual peptide consisting of a modified tyrosine residue, γ-aminobutyric acid, lysine, and oxalic acid residues (Figure 1). The overall mechanism of Fridericia bioluminescence shares similarities with D-luciferin, proceeding through ATP-dependent oxidative decarboxylation of the lysine moiety (Dubinnyi et al., 2015a). However, unlike in the case of beetle oxyluciferin, the conjugated π-system of Fridericia light emitter (a modified tyrosine residue) remains unchanged during bioluminescence reaction.
An unusual number of modified peptides structurally analogous to Fridericia luciferin were found in F. heliota luminescent tissues allowing Dubinnyi et al. (2015b) to propose two putative luciferin biosynthesis pathways. Currently the luciferase, presumably containing an adenylation domain, necessary for oxidation of the F. heliota luciferin and the enzymes responsible for luciferin biosynthesis, remains unknown.
Odontosyllis Bioluminescence System
The small polychaetes Odontosyllis undecimdonta luciferin-luciferase system is the latest addition to the list of known bioluminescence reactions. Odontosyllis worms display green bioluminescence (510 nm), correlated with the lunar cycle. In 2018 and 2019, respectively, the structures of Odontosyllis luciferase (Mitani et al., 2018; Schultz et al., 2018), luciferin (Figure 1) and oxyluciferin, both containing tricyclic thieno[3,2-f]thiochromene core, were reported (Kotlobay et al., 2019). The newly discovered enzyme has proven to be non-homologous with other known luciferases, suggesting a distinct evolutionary origin of Odontosyllis bioluminescence.
Along with the identification of Odontosyllis luciferin Kotlobay et al. (2019) also reported the discovery of a tricyclic luciferin precursor, termed compound 476, in the Odontosyllis biomass, thus allowing the researchers to propose a potential biosynthetic pathway. The suggested pathway starts from a common tyrosine metabolite L-DOPA, which undergoes oxidative coupling with 2 cysteine molecules, sulfation, and transamination, followed by spontaneous dehydrative cyclization leading to the formation of luciferin (Kotlobay et al., 2019). Three enzymes: tyrosinase, phenol sulfotransferase, and transaminase, were proposed to play the key roles in each step of Odontosyllis luciferin biosynthesis (Kotlobay et al., 2019); however, no experimental evidence has yet been obtained to support these claims.
Under-Investigated Bioluminescence Systems
Apart from luminescent beetles (Coleoptera), the ability to emit visible light is also found in the insect family Keroplatidae (Diptera), including subfamilies Arachnocampininae and Keroplatinae (Viviani et al., 2020). These Keroplatidae subfamilies possess biochemically distinct bioluminescence systems both from each other and from that of Coleoptera, giving no cross-reactions of enzymes or substrates (Viviani et al., 2002). The plausible structure of Arachnocampa luminosa luciferin was been recently reported to be a derivative of xanthurenic acid and tyrosine, while the luciferase was found to belong to the CoA-ligase superfamily (Watkins et al., 2018). Within the Keroplatinae subfamily luciferin, termed keroplatin, was found in both luminous and non-luminous species thus suggesting additional biological functions for this compound, however, no chemical structure was determined as yet.
Dinoflagellates (protists) and Euphausiids (krill) utilize two structurally similar tetrapyrrole luciferins (Figure 1), suggesting dietary dependence on dinoflagellate luciferin in krill (Haddock et al., 2010). Moreover, chemical structure of tetrapyrrole luciferins is similar to that of chlorophyll, a molecule that has long been hypothesized to be the precursor for dinoflagellate luciferin (Topalov and Kishi, 2001; Wu et al., 2003). However, the discovery of the plastid tetrapyrrole biosynthetic pathway in both photosynthetic and non-photosynthetic bioluminescent dinoflagellate species implies that luciferin might not be derived from chlorophyll, but originates from an earlier intermediate in its biogenesis (Janouskovec et al., 2017). Dinoflagellate bioluminescence is triggered by electrical or mechanical stimulation and is thought to serve a defensive function (Haddock et al., 2010), while the exact ecological role of light emission in Euphausiids is yet unclear.
Two most mysterious known luciferins belong to bioluminescence systems of freshwater limpets Latia neritoides and Diplocardia longa earthworms, wherein the luciferins were identified as (E)-2-methyl-4-(2,6,6-trimethyl-1-cyclohex-1-yl)-1-buten-1-ol formate (Shimomura and Johnson, 1968) and N-isovaleryl-3-aminopropanal (Ohtsuka et al., 1976), respectively (Figure 1). Even though the structures of these compounds were identified the mechanisms of their bioluminescence and the nature of light-emitting species are yet to be determined.
Concluding Remarks
It is curious that chemically identical luciferins can be used as substrates by numerous independently evolved luciferases in phylogenetically distant organisms. The most prominent example of this phenomenon is coelenterazine, which is the most prevalent light emitter in marine ecosystems (Haddock et al., 2010; Kaskova et al., 2016; Lau and Oakley, 2021). One of the explanations for this convergence is that in some cases luciferin is not synthesized in situ but is acquired exogenously through the diet (Frank et al., 1984; Haddock et al., 2001; Mallefet et al., 2020). On the other hand, the significant mismatch between a small number of natural luciferins and a large number of unique luciferases highlights the inherent challenges in biosynthesizing luminogenic substrates. Evolution has contrived relatively few biochemical solutions to this problem, with a surprising number of luciferins belonging to a class of small modified peptides, often comprising aromatic amino acids (L-tyrosine, L-tryptophan, and L-phenylalanine) and L-cysteine (Figure 1).
The classical enzyme-centric hypothesis stipulates that bioluminescence might have arisen in relatively hypoxic marine environments as a way to remove oxygen on the cellular level, with ancestral luciferases originally acting as mixed-function oxygenases and light being a non-functional byproduct at the time (Widder, 2010). Current evidence revealed that in many instances the main function of luciferases is not that of efficient oxygenation, but rather to provide the environment for optimal luciferin chemiluminescence, with many organisms co-opting enzymes catalyzing non-related reactions as a starting point for the evolution of luciferases (Viviani, 2002; Loening et al., 2006; Dubinnyi et al., 2015a; Watkins et al., 2018; Delroisse et al., 2021). Although the enzymatic function of the ancestral proteins is often unknown, it is clear that not all luciferases originate from oxygenases but, rather, luciferins themselves drove the emergence of new oxygenase functions and shaped the evolution of bioluminescence.
Currently no definitive hypothesis exists on the origins of luciferins. What selective pressures might have driven the emergence of luciferin biosynthetic pathways, and were the luciferins and their biosynthetic precursors of use in ancestral organisms? One hypothesis suggests that luciferins evolved from detoxification systems as some substrates show characteristics of strong antioxidants (Rees et al., 1998; Timmins et al., 2001; Dubuisson et al., 2004). Derived from tyrosine and cysteine, biosynthetic pathways of D-luciferin and Odontosyllis luciferin might have arisen from a mutation-induced deviation of the melanogenic pathways to provide photoprotection against free radical species, particularly reactive oxygen species (Napolitano et al., 2013). No such evidence, however, exists for dinoflagellate, Fridericia, Diplocardia, and fungal luciferins and their precursors. Elucidation of complete luciferin biosynthetic pathways and progress in sequencing and characterization of luciferin metabolic pathways genes and luciferases is needed to explore the likelihood of the “oxidative stress protection” hypothesis in bioluminescent organisms.
Various luciferases have been incorporated into a diverse array of cell types and long been used as reporter tools for tracking gene expression patterns and cell movements in research animals (Love and Prescher, 2020). The knowledge of all enzymes participating in bioluminescence cascades opens the possibility of engineering organisms with self-sustained luminescence, thus allowing the development of substrate-independent bioluminescence-based reporter technologies (Syed and Anderson, 2021). Apart from practical importance the exploration of luciferin biosynthetic pathways promotes our understanding of the origins and function of bioluminescence and provides new insights into ecological significance of this trait.
Author Contributions
AT is the sole author of the mini-review.
Funding
This work was supported by the Russian Science Foundation grant 18-74-10102 (https://rscf.ru/en/project/18-74-10102/).
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Airth, R. L., and McElroy, W. D. (1959). Light emission from extracts of luminous fungi. J. Bacteriol. 77, 249–250. doi: 10.1128/jb.77.2.249-250.1959
Bastos, E. L., Farahani, P., Bechara, E. J. H., and Baader, W. J. (2017). Four-membered cyclic peroxides: carriers of chemical energy. J. Phys. Org. Chem. 30:e3725. doi: 10.1002/poc.3725
Bessho-Uehara, M., Huang, W., Patry, W. L., Browne, W. E., Weng, J. K., and Haddock, S. H. D. (2020a). Evidence for de novo biosynthesis of the luminous substrate coelenterazine in ctenophores. iScience 23:101859. doi: 10.1016/j.isci.2020.101859
Bessho-Uehara, M., Yamamoto, N., Shigenobu, S., Mori, H., Kuwata, K., and Oba, Y. (2020b). Kleptoprotein bioluminescence: parapriacanthus fish obtain luciferase from ostracod prey. Sci. Adv. 6:eaax4942. doi: 10.1126/sciadv.aax4942
Branchini, B. R., Behney, C. E., Southworth, T. L., Fontaine, D. M., Gulick, A. M., Vinyard, D. J., et al. (2015). Experimental support for a single electron-transfer oxidation mechanism in firefly bioluminescence. J. Am. Chem. Soc. 137, 7592–7595. doi: 10.1021/jacs.5b03820
Brodl, E., Csamay, A., Horn, C., Niederhauser, J., Weber, H., and Macheroux, P. (2020). The impact of LuxF on light intensity in bacterial bioluminescence. J. Photochem. Photobiol. B Biol. 207:111881. doi: 10.1016/j.jphotobiol.2020.111881
Brodl, E., Winkler, A., and Macheroux, P. (2018). Molecular mechanisms of bacterial bioluminescence. Comput. Struct. Biotechnol. J. 16, 551–564. doi: 10.1016/j.csbj.2018.11.003
Cormier, M. J., Hori, K., and Karkhanis, Y. D. (1970). Studies on the bioluminescence of Renilla reniformis. VII. Conversion of luciferin into luciferyl sulfate by luciferin sulfokinase. Biochemistry 9, 1184–1189. doi: 10.1021/bi00807a019
Cormier, M. J., and Strehler, B. L. (1953). The identification of KCF: requirement of long-chain aldehydes for bacterial extract luminescence. J. Am. Chem. Soc. 75, 4864–4865. doi: 10.1021/ja01115a534
Davis, M. P., Sparks, J. S., and Smith, W. L. (2016). Repeated and widespread evolution of bioluminescence in marine fishes. PLoS One 11:e0155154. doi: 10.1371/journal.pone.0155154
Delroisse, J., Duchatelet, L., Flammang, P., and Mallefet, J. (2021). Leaving the dark side? Insights into the evolution of luciferases. Front. Mar. Sci. 8:673620. doi: 10.3389/FMARS.2021.673620
Delroisse, J., Ullrich-Lüter, E., Blaue, S., Ortega-Martinez, O., Eeckhaut, I., Flammang, P., et al. (2017). A puzzling homology: a brittle star using a putative cnidarian-type luciferase for bioluminescence. Open Biol. 7:160300. doi: 10.1098/rsob.160300
Dubinnyi, M. A., Kaskova, Z. M., Rodionova, N. S., Baranov, M. S., Gorokhovatsky, A. Y., Kotlobay, A., et al. (2015a). Novel mechanism of bioluminescence: oxidative decarboxylation of a moiety adjacent to the light emitter of fridericia luciferin. Angew. Chemie Int. Ed. 54, 7065–7067. doi: 10.1002/anie.201501668
Dubinnyi, M. A., Tsarkova, A. S., Petushkov, V. N., Kaskova, Z. M., Rodionova, N. S., Kovalchuk, S. I., et al. (2015b). Novel peptide chemistry in terrestrial animals: natural luciferin analogues from the bioluminescent earthworm Fridericia heliota. Chem. A Eur. J. 21, 3942–3947. doi: 10.1002/chem.201406498
Dubuisson, M., Marchand, C., and Rees, J. F. (2004). Firefly luciferin as antioxidant and light emitter: the evolution of insect bioluminescence. Luminescence 19, 339–344. doi: 10.1002/bio.789
Dunlap, P. V., and Urbanczyk, H. (2013). “Luminous bacteria,” in The Prokaryotes: Prokaryotic Physiology and Biochemistry, eds E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer-Verlag), 495–528. doi: 10.1007/978-3-642-30141-4_75
Emamzadeh, R., Hosseinkhani, S., Hemati, R., and Sadeghizadeh, M. (2010). RACE-based amplification of cDNA and expression of a luciferin-regenerating enzyme (LRE): an attempt towards persistent bioluminescent signal. Enzyme Microb. Technol. 47, 159–165. doi: 10.1016/J.ENZMICTEC.2010.05.008
Fallon, T. R., Li, F. S., Vicent, M. A., and Weng, J. K. (2016). Sulfoluciferin is biosynthesized by a specialized luciferin sulfotransferase in fireflies. Biochemistry 55, 3341–3344. doi: 10.1021/acs.biochem.6b00402
Fallon, T. R., Lower, S. E., Chang, C. H., Bessho-Uehara, M., Martin, G. J., Bewick, A. J., et al. (2018). Firefly genomes illuminate parallel origins of bioluminescence in beetles. eLife 7:e36495. doi: 10.1101/237586
Francis, W. R., Shaner, N. C., Christianson, L. M., Powers, M. L., and Haddock, S. H. D. (2015). Occurrence of isopenicillin-N-synthase homologs in bioluminescent ctenophores and implications for coelenterazine biosynthesis. PLoS One 10:e0128742. doi: 10.1371/journal.pone.0128742
Frank, T. M., Widder, E. A., Latz, M. I., and Case, J. F. (1984). Dietary maintenance of bioluminescence in a deep-sea mysid. J. Exp. Biol. 109, 385–389.
Gomi, K., and Kajiyama, N. (2001). Oxyluciferin, a luminescence product of firefly luciferase, is enzymatically regenerated into luciferin. J. Biol. Chem. 276, 36508–36513. doi: 10.1074/JBC.M105528200
Haddock, S. H., Rivers, T. J., and Robison, B. H. (2001). Can coelenterates make coelenterazine? Dietary requirement for luciferin in cnidarian bioluminescence. Proc. Natl. Acad. Sci. U.S.A. 98, 11148–11151. doi: 10.1073/pnas.201329798
Haddock, S. H. D., Moline, M. A., and Case, J. F. (2010). Bioluminescence in the Sea. Ann. Rev. Mar. Sci. 2, 443–493. doi: 10.1146/annurev-marine-120308-081028
Head, J. F., Satoshi, I., Shimomura, O., and Teranishi, K. (2000). The crystal structure of the photoprotein aequorin at resolution 2.3 A. Nature 405, 372–376. doi: 10.1038/35012659
Hensley, N. M., Ellis, E. A., Leung, N. Y., Coupart, J., Mikhailovsky, A., Taketa, D. A., et al. (2021). Selection, drift, and constraint in cypridinid luciferases and the diversification of bioluminescent signals in sea fireflies. Mol. Ecol. 30, 1864–1879. doi: 10.1111/MEC.15673
Hosseinkhani, S., Emamgholi Zadeh, E., Sahebazzamani, F., Ataei, F., and Hemmati, R. (2017). Luciferin-regenerating enzyme crystal structure is solved but its function is still unclear. Photochem. Photobiol. 93, 429–435. doi: 10.1111/PHP.12723
Inoue, S., Kakoi, H., and Goto, T. (1976). Squid bioluminescence III. Isolation and structure of Watasenia luciferin. Tetrahedron Lett. 17, 2971–2974. doi: 10.1016/S0040-4039(01)85503-9
Inoue, S., Kakoi, H., Murata, M., Goto, T., and Shimomura, O. (1977). Complete structure of renilla luciferin and luciferyl sulfate. Tetrahedron Lett. 18, 2685–2688. doi: 10.1016/S0040-4039(01)83046-X
Inouye, S. (2010). Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions. Cell. Mol. Life Sci. 67, 387–404. doi: 10.1007/s00018-009-0170-8
Janouskovec, J., Gavelis, G. S., Burki, F., Dinh, D., Bachvaroff, T. R., Gornik, S. G., et al. (2017). Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl. Acad. Sci. U.S.A. 114, E171–E180. doi: 10.1073/pnas.1614842114
Jeffers, C. E., and Tu, S. C. (2001). Differential transfers of reduced flavin cofactor and product by bacterial flavin reductase to luciferase. Biochemistry 40, 1749–1754. doi: 10.1021/bi0024310
Kanie, S., Nakai, R., Ojika, M., and Oba, Y. (2018). 2-S-cysteinylhydroquinone is an intermediate for the firefly luciferin biosynthesis that occurs in the pupal stage of the Japanese firefly, Luciola lateralis. Bioorg. Chem. 80, 223–229. doi: 10.1016/j.bioorg.2018.06.028
Kanie, S., Nishikawa, T., Ojika, M., and Oba, Y. (2016). One-pot non-enzymatic formation of firefly luciferin in a neutral buffer from p-benzoquinone and cysteine. Sci. Rep. 6, 1–8. doi: 10.1038/srep24794
Kaskova, Z. M., Dörr, F. A., Petushkov, V. N., Purtov, K. V., Tsarkova, A. S., Rodionova, N. S., et al. (2017). Mechanism and color modulation of fungal bioluminescence. Sci. Adv. 3:e1602847. doi: 10.1126/sciadv.1602847
Kaskova, Z. M., Tsarkova, A. S., and Yampolsky, I. V. (2016). 1001 lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev. 45, 6048–6077. doi: 10.1039/c6cs00296j
Kato, S. I., Oba, Y., Ojika, M., and Inouye, S. (2007). Biosynthesis of cypridina luciferin in Cypridina noctiluca. Heterocycles 72, 673–676. doi: 10.3987/com-06-s(k)27
Kato, S., Oba, Y., Ojika, M., and Inouye, S. (2004). Identification of the biosynthetic units of Cypridina luciferin in Cypridina (Vargula) hilgendorfii by LC/ESI-TOF-MS. Tetrahedron 60, 11427–11434. doi: 10.1016/j.tet.2004.09.080
Ke, H. M., Lee, H. H., Chan-Yi Ivy, Lin, Liu, Y. C., Lu, M. R., et al. (2020). Mycena genomes resolve the evolution of fungal bioluminescence. Proc. Natl. Acad. Sci. U.S.A. 117, 31267–31277. doi: 10.1073/pnas.2010761117
Keller, N. P., Turner, G., and Bennett, J. W. (2005). Fungal secondary metabolism - From biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. doi: 10.1038/nrmicro1286
Kotlobay, A. A., Dubinnyi, M. A., Purtov, K. V., Guglya, E. B., Rodionova, N. S., Petushkov, V. N., et al. (2019). Bioluminescence chemistry of fireworm Odontosyllis. Proc. Natl. Acad. Sci. U.S.A. 116, 18911–18916. doi: 10.1073/pnas.1902095116
Kotlobay, A. A., Kaskova, Z. M., and Yampolsky, I. V. (2020). Palette of luciferases: natural biotools for new applications in biomedicine. Acta Nat. 12, 15–27. doi: 10.32607/actanaturae.10967
Kotlobay, A. A., Sarkisyan, K. S., Mokrushina, Y. A., and Marcet-, M. (2018). A genetically encodable bioluminescent system from fungi. PNAS 115, 12728–12732. doi: 10.1073/pnas.1803615115
Lau, E. S., and Oakley, T. H. (2021). Multi-level convergence of complex traits and the evolution of bioluminescence. Biol. Rev. 96, 673–691. doi: 10.1111/brv.12672
Lee, I.-K., and Yun, B.-S. (2011). Styrylpyrone-class compounds from medicinal fungi Phellinus and Inonotus spp., and their medicinal importance. J. Antibiot. 64, 349–359. doi: 10.1038/ja.2011.2
Lee, J., M€ Uller, F., and Visser, A. J. W. G. (2019). Invited review the sensitized bioluminescence mechanism of bacterial luciferase. Photochem. Photobiol. 95, 679–704. doi: 10.1111/php.13063
Loening, A. M., Fenn, T. D., Wu, A. M., and Gambhir, S. S. (2006). Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 19, 391–400. doi: 10.1093/protein/gzl023
Love, A. C., and Prescher, J. A. (2020). Seeing (and Using) the light: recent developments in bioluminescence technology. Cell Chem. Biol. 27, 904–920. doi: 10.1016/j.chembiol.2020.07.022
Maeda, J., Kato, D. I., Okuda, M., Takeo, M., Negoro, S., Arima, K., et al. (2017). Biosynthesis-inspired deracemizative production of D-luciferin by combining luciferase and thioesterase. Biochim. Biophys. Acta Gen. Subj. 1861, 2112–2118. doi: 10.1016/j.bbagen.2017.04.010
Mallefet, J., Duchatelet, L., and Coubris, C. (2020). Bioluminescence induction in the ophiuroid Amphiura filiformis (Echinodermata). J. Exp. Biol. 223:jeb218719. doi: 10.1242/jeb.218719
Marquette, C. A., and Blum, L. J. (2010). “Chemiluminescent and bioluminescent biosensors,” in Chemiluminescence and Bioluminescence: Past, Present and Future, ed. A. Roda (Cambridge: Royal Society of Chemistry), 488–510.
McCapra, F., and Roth, M. (1972). Cyclisation of a dehydropeptide derivative: a model for cypridina luciferin biosynthesis. J. Chem. Soc. Chem. Commun. 15, 894–895. doi: 10.1039/C39720000894
Meighen, E. A. (1991). Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55, 123–141. doi: 10.1128/mr.55.1.123-142.1991
Mitani, Y., Yasuno, R., Isaka, M., Mitsuda, N., Futahashi, R., Kamagata, Y., et al. (2018). Novel gene encoding a unique luciferase from the fireworm Odontsyllis undecimdonta. Sci. Rep. 8:12789. doi: 10.1038/s41598-018-31086-1
Nakamura, M., Suzuki, T., Ishizaka, N., Sato, J. I., and Inouye, S. (2014). Identification of 3-enol sulfate of Cypridina luciferin, Cypridina luciferyl sulfate, in the sea-firefly Cypridina (Vargula) hilgendorfii. Tetrahedron 70, 2161–2168. doi: 10.1016/j.tet.2014.01.075
Napolitano, A., Panzella, L., Leone, L., and D’Ischia, M. (2013). Red hair benzothiazines and benzothiazoles: mutation-inspired chemistry in the quest for functionality. Acc. Chem. Res. 46, 519–528. doi: 10.1021/ar300219u
Nijvipakul, S., Wongratana, J., Suadee, C., Entsch, B., Ballou, D. P., and Chaiyen, P. (2008). LuxG is a functioning flavin reductase for bacterial luminescence. J. Bacteriol. 190, 1531–1538. doi: 10.1128/JB.01660-07
Niwa, K., Nakamura, M., and Ohmiya, Y. (2006). Stereoisomeric bio-inversion key to biosynthesis of firefly d-luciferin. FEBS Lett. 580, 5283–5287. doi: 10.1016/j.febslet.2006.08.073
Oba, Y., Kato, S. I., Ojika, M., and Inouye, S. (2009). Biosynthesis of coelenterazine in the deep-sea copepod, Metridia pacifica. Biochem. Biophys. Res. Commun. 390, 684–688. doi: 10.1016/j.bbrc.2009.10.028
Oba, Y., Suzuki, Y., Martins, G. N. R., Carvalho, R. P., Pereira, T. A., Waldenmaier, H. E., et al. (2017). Identification of hispidin as a bioluminescent active compound and its recycling biosynthesis in the luminous fungal fruiting body. Photochem. Photobiol. Sci. 32, 27–29. doi: 10.1039/C7PP00216E
Oba, Y., Yoshida, N., Kanie, S., Ojika, M., and Inouye, S. (2013). Biosynthesis of firefly luciferin in adult lantern: decarboxylation of L-cysteine is a key step for benzothiazole ring formation in firefly luciferin synthesis. PLoS One 8:e84023. doi: 10.1371/journal.pone.0084023
Ohtsuka, H., Rudie, N. G., and Wampler, J. E. (1976). Structural identification and synthesis of luciferin from the bioluminescent earthworm, Diplocardia longa. Biochemistry 15, 1001–1004. doi: 10.1021/bi00650a009
Oliveira, A. G., Desjardin, D. E., Perry, B. A., and Stevani, C. V. (2012). Evidence that a single bioluminescent system is shared by all known bioluminescent fungal lineages. Photochem. Photobiol. Sci. 11, 848–852. doi: 10.1039/c2pp25032b
Petushkov, V. N., Dubinnyi, M. A., Tsarkova, A. S., Rodionova, N. S., Baranov, M. S., Kublitski, V. S., et al. (2014). A novel type of luciferin from the siberian luminous earthworm Fridericia heliota : structure elucidation by spectral studies and total synthesis. Angew. Chemie Int. Ed. 53, 5566–5568. doi: 10.1002/anie.201400529
Purtov, K. V., Petushkov, V. N., Baranov, M. S., Mineev, K. S., Rodionova, N. S., Kaskova, Z. M., et al. (2015). The chemical basis of fungal bioluminescence. Angew. Chemie Int. Ed. 54, 8124–8128. doi: 10.1002/anie.201501779
Rees, J. F., De Wergifosse, B., Noiset, O., Dubuisson, M., Janssens, B., and Thompson, E. M. (1998). The origins of marine bioluminescence: turning oxygen defence mechanisms into deep-sea communication tools. J. Exp. Biol. 201, 1211–1221. doi: 10.1242/jeb.201.8.1211
Schultz, D. T., Kotlobay, A. A., Ziganshin, R., Bannikov, A., Markina, N. M., Chepurnyh, T. V., et al. (2018). Luciferase of the Japanese syllid polychaete Odontosyllis umdecimdonta. Biochem. Biophys. Res. Commun. 502, 318–323. doi: 10.1016/j.bbrc.2018.05.135
Schwartzman, J. A., and Ruby, E. G. (2016). A conserved chemical dialog of mutualism: lessons from squid and vibrio. Microbes Infect. 18, 1–10. doi: 10.1016/j.micinf.2015.08.016
Shimomura, O., and Johnson, F. H. (1968). The structure of Latia luciferin. Biochemistry 7, 1734–1738. doi: 10.1021/bi00845a017
Shimomura, O., and Yampolsky, I. (2019). Bioluminescence: Chemical Principles and Methods, 3rd Edn. Singapore: World Scientific.
Syed, A. J., and Anderson, J. C. (2021). Applications of bioluminescence in biotechnology and beyond. Chem. Soc. Rev. 50, 5668–5705. doi: 10.1039/d0cs01492c
Takahashi, H., and Isobe, M. (1994). Photoprotein of luminous squid, symplectoteuthis oualaniensis and reconstruction of the luminous system. Chem. Lett. 23, 843–846. doi: 10.1246/cl.1994.843
Tanaka, E., Kuse, M., and Nishikawa, T. (2009). Dehydrocoelenterazine is the organic substance constituting the prosthetic group of Pholasin. ChemBioChem 10, 2725–2729. doi: 10.1002/cbic.200900503
Tessler, M., Gaffney, J. P., Crawford, J. M., Trautman, E., Gujarati, N. A., Alatalo, P., et al. (2018). Luciferin production and luciferase transcription in the bioluminescent copepod Metridia lucens. PeerJ 2018:e5506. doi: 10.7717/peerj.5506
Tessler, M., Gaffney, J. P., Oliveira, A. G., Guarnaccia, A., Dobi, K. C., Gujarati, N. A., et al. (2020). A putative chordate luciferase from a cosmopolitan tunicate indicates convergent bioluminescence evolution across phyla. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-73446-w
Thomson, C. M., Herring, P. J., and Campbell, A. K. (1995). Evidence for de Novo biosynthesis of coelenterazine in the bioluminescent midwater shrimp, systellaspis debilis. J. Mar. Biol. Assoc. United Kingdom 75, 165–171. doi: 10.1017/S0025315400015277
Timmins, G. S., Jackson, S. K., and Swartz, H. M. (2001). The evolution of bioluminescent oxygen consumption as an ancient oxygen detoxification mechanism. J. Mol. Evol. 52, 321–332. doi: 10.1007/s002390010162
Tinikul, R., Pitsawong, W., Sucharitakul, J., Nijvipakul, S., Ballou, D. P., and Chaiyen, P. (2013). The transfer of reduced flavin mononucleotide from luxg oxidoreductase to luciferase occurs via free diffusion. Biochemistry 52, 6834–6843. doi: 10.1021/bi4006545
Topalov, G., and Kishi, Y. (2001). Chlorophyll catabolism leading to the skeleton of dinoflagellate and krill luciferins: hypothesis and model studies. Angew. Chem. Int. Ed. Engl. 40, 3892–3894. doi: 10.1002/1521-3773(20011015)40:20<3892::AID-ANIE3892<3.0.CO;2-H
Tu, S. C. (2008). Activity coupling and complex formation between bacterial luciferase and flavin reductases. Photochem. Photobiol. Sci. 7, 183–188. doi: 10.1039/b713462b
Vacher, M., Fdez Galván, I., Ding, B. W., Schramm, S., Berraud-Pache, R., Naumov, P., et al. (2018). Chemi- and bioluminescence of cyclic peroxides. Chem. Rev. 118, 6927–6974. doi: 10.1021/acs.chemrev.7b00649
Vannier, T., Hingamp, P., Turrel, F., Tanet, L., Lescot, M., and Timsit, Y. (2020). Diversity and evolution of bacterial bioluminescence genes in the global ocean. NAR Genomics Bioinform. 2:lqaa018. doi: 10.1093/nargab/lqaa018
Verdes, A., and Gruber, D. F. (2017). Glowing worms: biological, chemical, and functional diversity of bioluminescent annelids. Integr. Comp. Biol. 57, 18–32. doi: 10.1093/icb/icx017
Viviani, V. R. (2002). The origin, diversity, and structure function relationships of insect luciferases. Cell. Mol. Life Sci. 59, 1833–1850. doi: 10.1007/PL00012509
Viviani, V. R., Amaral, D., Prado, R., and Arnoldi, F. G. C. (2011). A new blue-shifted luciferase from the Brazilian Amydetes fanestratus (Coleoptera: Lampyridae) firefly: molecular evolution and structural/functional properties. Photochem. Photobiol. Sci. 10, 1879–1886. doi: 10.1039/c1pp05210a
Viviani, V. R., Hastings, J. W., and Wilson, T. (2002). Two bioluminescent diptera: the North American Orfelia fultoni and the Australian Arachnocampa flava. Similar Niche, different bioluminescence systems. Photochem. Photobiol. 75:22. doi: 10.1562/0031-8655(2002)0750022TBDTNA2.0.CO2
Viviani, V. R., Silva, J. R., Amaral, D. T., Bevilaqua, V. R., Abdalla, F. C., Branchini, B. R., et al. (2020). A new brilliantly blue-emitting luciferin-luciferase system from Orfelia fultoni and Keroplatinae (Diptera). Sci. Rep. 10:9608. doi: 10.1038/s41598-020-66286-1
Wakamatsu, K., Ohtara, K., and Ito, S. (2009). Chemical analysis of late stages of pheomelanogenesis: conversion of dihydrobenzothiazine to a benzothiazole structure. Pigment. Cell Melanoma Res. 22, 474–486. doi: 10.1111/j.1755-148X.2009.00580.x
Wang, M.-Y., and Liu, Y.-J. (2021). Chemistry in fungal bioluminescence: a theoretical study from luciferin to light emission. J. Org. Chem. 86, 1874–1881. doi: 10.1021/acs.joc.0c02788
Watkins, O. C., Sharpe, M. L., Perry, N. B., and Krause, K. L. (2018). New Zealand glowworm (Arachnocampa luminosa) bioluminescence is produced by a firefly-like luciferase but an entirely new luciferin. Sci. Rep. 8:3278. doi: 10.1038/s41598-018-21298-w
White, E. H., McCapra, F., and Field, G. F. (1963). The structure and synthesis of firefly luciferin. J. Am. Chem. Soc. 85, 337–343. doi: 10.1021/ja00886a019
Widder, E. A. (2010). Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science 328, 704–708. doi: 10.1126/science.1174269
Wu, C., Akimoto, H., and Ohmiya, Y. (2003). Tracer studies on dinoflagellate luciferin with [15N]-glycine and [15N]-l-glutamic acid in the dinoflagellate Pyrocystis lunula. Tetrahedron Lett. 44, 1263–1266. doi: 10.1016/S0040-4039(02)02815-0
Keywords: bioluminescence, biochemistry, luciferin, biosynthesis, metabolism, evolution
Citation: Tsarkova AS (2021) Luciferins Under Construction: A Review of Known Biosynthetic Pathways. Front. Ecol. Evol. 9:667829. doi: 10.3389/fevo.2021.667829
Received: 14 February 2021; Accepted: 27 August 2021;
Published: 20 September 2021.
Edited by:
Jerome Mallefet, Catholic University of Louvain, BelgiumReviewed by:
Laurent Duchatelet, Catholic University of Louvain, BelgiumTodd H. Oakley, University of California, Santa Barbara, United States
Manabu Bessho-Uehara, Nagoya University, Japan
Copyright © 2021 Tsarkova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aleksandra S. Tsarkova, altsarkova@gmail.com
 Aleksandra S. Tsarkova
Aleksandra S. Tsarkova