Chemosensory Receptors in the Larval Maxilla of Papilio hospiton
- 1University Institute of Biotechnology and Biomedicine, University of Valencia, Valencia, Spain
- 2Research and Innovation Centre, Fondazione Edmund Mach, San Michele All’adige, Italy
- 3Whitney Laboratory for Marine Bioscience, University of Florida, Gainesville, FL, United States
- 4Department of Biomedical Sciences, University of Cagliari, Cagliari, Italy
- 5Centre of Agriculture, Food and Environment, University of Trento, Trento, Italy
- 6Chemical Ecology, Department of Plant Protection Biology, Swedish University of Agricultural Sciences, Uppsala, Sweden
- 7Benton Lab, Center for Integrative Genomics, University of Lausanne, Lausanne, Switzerland
Among the butterflies of the genus Papilio (Lepidoptera: Papilionidae), Papilio hospiton (Géné) has a geographical distribution limited to the Mediterranean islands of Sardinia (Italy) and Corsica (France). This is mainly due to the host range that includes only a few plant species of Apiaceae and Rutaceae growing on these islands. In a previous electrophysiological investigation conducted on the maxillary gustatory system of larvae of P. hospiton and its closely phylogenetically related species Papilio machaon, a significantly higher spike activity was shown for the gustatory neurons of lateral and medial styloconic sensilla in P. hospiton when bitter compounds were tested. This effect was possibly correlated to the limited host choice range for P. hospiton. To shed light on the molecular aspects of this phenomenon, we investigated the expression pattern of sensory-related sequences by conducting a transcriptomic analysis from total RNA isolates of P. hospiton larval maxillae. We identified several transcripts that may be involved in taste (one gustatory receptor, one divergent ionotropic receptor, and several transient receptor potential channels, TRPs) as well as transcripts supporting an olfactory function for this appendage, including odorant receptors (ORs), antennal ionotropic receptors (A-IRs), sensory neuron membrane proteins (SNMPs), and odorant-binding proteins (OBPs). We used Human Embryonic Kidney (HEK293A) cells to heterologously express two of the identified receptors, PhospOR1 and PhospPain, together with their orthologs from P. machaon, for functional characterization. While our data suggest no activation of these two receptors by the ligands known so far to activate the electrophysiological response in larval maxillary neurons of Papilio species, nor temperature activation of both Papilio TRPA-channel Painless, they represent the first attempt in connecting neuronal activity with their molecular bases to unravel diet specialization between closely related Papilio species.
Introduction
External chemoreceptors in lepidopteran larvae are located on the antennae, on the maxillae, and in the epipharynx (Dethier and Schoonhoven, 1969; Laue, 2000). Together with this last, the maxillae are the main location of the sense of taste and mediate acceptance or rejection of host plants, therefore underlying diet specialization (Schoonhoven and van Loon, 2002). Each maxilla is formed by two parts: the maxillary galea and the maxillary palp, which generally harbor two or three styloconic and eight basiconic sensilla, respectively (Schoonhoven and van Loon, 2002). The role of the galean styloconic sensilla in taste has been widely studied in several species. In contrast, although it is known that basiconic sensilla on the palps contain both gustatory and olfactory receptor neurons, their role in chemoperception is poorly understood (Schoonhoven and van Loon, 2002). Only recently, in the model species Bombyx mori, a two-factor host acceptance system has been proposed. In this system, while the chemosensory neurons in the maxillary palps are involved in the detection of leaf-surface compounds and hence, in the induction of a test biting, the chemosensory neurons present in the galea are responsible for sensing the toxic compounds in leaf sap generated by test biting, and consequently mediate the following persistent biting (Tsuneto et al., 2020).
Papilio hospiton is an olygophagous butterfly species endemic to the Mediterranean islands of Sardinia and Corsica. It displays a more pronounced olygophagy compared to the Holarctic species Papilio machaon since the larvae of this latter feed on several Apiaceae and Rutaceae, whereas larvae of P. hospiton feed almost exclusively on a single Apiaceae species, the giant fennel Ferula communis (Sollai et al., 2014, 2018a). Due to this shift in diet specialization, P. hospiton and P. machaon have been extensively studied to identify the neuronal basis underlying their differential host plant ranges (Sollai et al., 2014, 2015, 2018b; Sollai and Crnjar, 2019). Studies targeting the lateral and the medial styloconic sensilla present on the maxillary galea have shown that both contain four gustatory receptor neurons (GRNs). In each sensillum, two GRNs respond to phagostimulants (primary plant metabolites such as sugars and amino acids), a third GRN is activated by deterrents (bitter secondary plant metabolites) and the fourth one responds to salt (Sollai et al., 2014, 2017). Comparison between P. machaon and P. hospiton highlighted in this latter an overall higher spike activity of the GRNs in response to mono- and disaccharides, bitter tastants, and salts (Sollai et al., 2014). This, together with observations that sap from P. hospiton non-host species, such as Seseli tortuosum, Foeniculum vulgare, and Daucus carota evoked a higher neural activity in phagostimulant and bitter sensing in P. hospiton GRNs suggests the existence of discriminant neurological bases in the maxillae of these two closely related papilionid species that may recapitulate diet specialization (Sollai et al., 2017, 2018a,b; Sollai and Crnjar, 2019).
In insects, the molecular basis of taste and olfaction mainly relies on several gene families that code for the chemoreceptors expressed on the membrane of sensory neurons and are responsible for the binding to specific volatiles or tastants (Joseph and Carlson, 2015). Odorant receptors (ORs) are seven-transmembrane domain proteins that are expressed in olfactory receptor neurons (ORNs) and have a wide spectrum of activity, ranging from narrowly tuned ORs, that recognize only one or few volatiles, to widely tuned receptors (Joseph and Carlson, 2015; de Fouchier et al., 2017). Ionotropic receptors (IRs) are three transmembrane domain proteins, expressed in ORNs and GRNs, and are involved in both olfaction and taste (Benton et al., 2009). Generally, antennal IRs (A-IRs) are expressed in the ORNs in the antennae, and they sense volatiles. In contrast, divergent IRs (D-IRs) are expressed in GRNs and have mainly a taste function (Koh et al., 2014). Gustatory receptors (GRs) are seven-transmembrane domain proteins evolutionarily related to ORs but expressed in GRNs and involved in taste (Agnihotri et al., 2016). They detect non-volatile compounds including sugars, bitter molecules as plant secondary metabolites, amino acids, via contact chemosensation. An exception is represented by a few GRs, which have been specialized in detecting airborne molecules such as CO2 (Joseph and Carlson, 2015; Agnihotri et al., 2016). Transient receptor potential (TRP) channels are a large family of proteins present among vertebrates and invertebrates involved in several functions (Matsuura et al., 2009; Himmel and Cox, 2020). In insects, some TRP-channels are involved in complex chemosensory mechanisms at the base of olfactory and taste transduction (Jörs et al., 2006; Xu et al., 2008; Zhang et al., 2013a,b) and in the avoidance of noxious chemicals (Al-Anzi et al., 2006; Kang et al., 2010; Kwon et al., 2010). Other insect TRP channels have a wider spectrum of activation, being reported as sensors for temperature, phototransduction, mechanoreception, light sensation, and gravity (Liu et al., 2007; Sun et al., 2009; Wei et al., 2015; Montell, 2021).
In the present study, we carried out a transcriptome profiling of larval P. hospiton maxillae (which include both galea and maxillary palps as well as other surrounding tissues) intending to identify the molecular basis responsible for chemosensation in this appendage. Results showed the presence of few chemoreceptors belonging to OR, IR, GR, and TRP families, as well as binding proteins and sensory neuron membrane proteins. Among the receptors, the coding sequences of one OR (PhospOR1) and one TRP channel (PhospPain) were the sole full-length ones that we were able to isolate in parallel with their orthologs from P. machaon, and that were heterologously expressed to be functionally characterized throughout Human Embryonic Kidney (HEK293A) cells. Among our experiments, we attempted to deorphanize Papilio OR1s to tastants to test the hypothesis of possible alternative roles of ORs from odorant-binding (Garczynski et al., 2017). We based this hypothesis also on the documented co-existence of complex neurosensory mechanisms for smell and taste in the maxilla of caterpillars (Solari et al., 2002), and more general pieces of evidence not harmonizing with a precise differentiation between the chemical senses of taste and smell (Mollo et al.–submitted to Quarterly Review of Biology).
Materials and Methods
Papilio hospiton Larval Collection, Nucleic Acid Extraction, and Illumina Sequencing
Gravid females of P. hospiton were left free to oviposit on potted host plants (F. communis) inside a butterfly egg-laying annex (3 m × 3 m × 3 m cage) at the Physiology laboratories of the University of Cagliari. After the eggs hatched, larvae were collected from plants and reared at the insectary annex of the same labs in 1,500-ml plastic cups (4–5 larvae per cup) and kept in an environmental growth chamber (24–25°C, 70% R.H., 16 h light/8 h dark photoperiod). Larvae were fed daily with fresh foliage of F. communis until they reached the last larval instar. Dissection was carried out in the morning; larvae were first anesthetized with CO2 and then both maxillae from a single larva were excised. The final sample consisted of maxillae from 12 larvae, and it was flash-frozen in liquid nitrogen at the end of the dissection. Total RNA was extracted using QIAamp® RNeasy Mini Kit (Qiagen, Hilden Germany) in accordance with the manufacturer’s instructions. Total RNA (∼3μg) was sent to Macrogen (Seoul, South Korea) for library preparation and sequencing. The sample was first quality checked using a BioAnalyzer (Agilent, Santa Clara, CA, United States), rendering a RNA integrity number (RIN) of 10, and then, a double-strand cDNA library was constructed with the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA, United States) starting from polyadenylated RNA. Finally, the library was paired-end sequenced using a HiSeq 2000 (Ilumina, San Diego, CA, United States) instrument with TruSeq SBS Kit v3 chemistry (Illumina, San Diego, CA, United States). Raw reads are available at Genbank sequence reads archive with accession number SRR14415798.
Assembly and Transcriptome Annotation
Raw sequenced reads were low-quality trimmed using Trimmomatic (Bolger et al., 2014). De novo transcriptome assembly was constructed using Trinity with default parameters (Grabherr et al., 2011). To estimate assembly quality, clean reads were then re-mapped using Bowtie (Langmead et al., 2009), and RSEM (Li and Dewey, 2011) extracted read count data. The de novo assembly was annotated with Trinotate v3.1.1 pipeline (Bryant et al., 2017) and with blastx searches using contigs as query and the predicted proteome of P. machaon (Li et al., 2015) or Drosophila melanogaster (Marygold et al., 2013) as database, using an E-value lower than 10–5. Blastx results against P. machaon were used to calculate the Ortholog hit ratio (OHR) to estimate transcript completeness (O’Neil et al., 2010). Transcriptome completeness was also evaluated with BUSCO (benchmarking universal single-copy orthologs) (Seppey et al., 2019). Blastx results against D. melanogaster proteome were used to retrieve Gene Ontology terms from Panther v.14 (Mi et al., 2019).
Putative transcripts related to chemical sensing and transcripts for TRP-channels were initially identified from blastx searches against P. machaon and D. melanogaster proteomes. Further iterative tblastn searches using the assembled contigs as database and curated chemosensory receptor repertoires from close species as queries were run to identify further candidates. Specifically, we used the whole chemosensory receptor repertoire annotated from the genome of the moth Spodoptera frugiperda (Gouin et al., 2017), the curated IR datasets from P. machaon, Papilio polytes, and Papilio xuthus (Liu et al., 2018), Papilio memnon and Papilio bianor (Yin et al., 2021), the curated GR dataset from Heliconius melpomene (Briscoe et al., 2013) and the curated OBP dataset from Lepidoptera (Vogt et al., 2015). Additional transcripts were searched in the database produced by Trinotate, particularly focusing on domain prediction results generated by hmmscan searches against the Pfam-A database.
Phylogenetic Analysis
Contigs identified as candidate transcripts related to chemical sensing and transcripts for TRP-channels were manually examined and their coding sequences (CDSs) (either partial or complete) were manually annotated using BioEdit (Hall, 1999). Translated amino acid sequences were aligned with annotated datasets from other Lepidoptera species using the MUSCLE algorithm implemented in MEGA X (Kumar et al., 2018). Alignments were manually inspected and used to build maximum-likelihood trees with RAxML (Stamatakis, 2014) using 500 bootstrap pseudoreplicates. The nomenclature of IRs was based on the closest ortholog in other Papilio species (Liu et al., 2018; Yin et al., 2021) and for TRPs, on the closest ortholog in H. melpomene (Macias-Muñoz et al., 2019). OBP nomenclature was based on the best ortholog on Danaus plexippus (Vogt et al., 2015), whereas the P. hospiton-specific OBPs had sequential names starting from OBP28.
Cloning the Coding Sequences of the Papilio hospiton and Papilio machaon Odorant Receptors and Transient Receptor Potential-Channels
The full-length CDSs of PhospOR1, PhospPain, and their orthologs PmachOR1 and PmachPain were cloned following protocols we described with more details in our previous investigations (Cattaneo et al., 2017a; Bobkov et al., 2021). We amplified retro-transcribed cDNA samples with primers shown in Supplementary Table 1, which were designed on the P. machaon sequences (GenBank XM_014512458.1 and XM_014500087.1). Primer sequences were integrated with 5’-attB-regions suitable for BP-cloning (Gateway Technology, Invitrogen Life Technologies, Grand Island, NY, United States). Amplifications for OR1 were performed using the GoTaq Green Master Mix (Promega, Fitchburg, WI, United States) with a temperature program of 95°C for 15 min, followed by 35 cycles of 95°C for 45 sec, 51°C for 1 min, 72°C for 90 s, and a final elongation of 72°C for 7 min. Amplifications for Pain were performed using Advantage RT-for-PCR kit (Clontech, Mountain View, CA, United States) with a temperature program of 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 59.2°C for 1 min, 68°C for 3 min, and a final elongation of 68°C for 7 min. Amplicons were separated in 1.5% agarose gel and visualized after staining with ethidium bromide in a Gel Doc XR (Bio-Rad, Hercules, CA, United States). From each PCR reaction, 4 μl were mixed with 1 μl of Gateway BP-clonase (Invitrogen, Thermo Fischer Scinetific, Waltham, MA, United States) and 150 ng of pDONR221 (Invitrogen) and incubated for 4 h at 25°C. Among these reaction volumes, 2 μl were used to transform TOP10 competent cells (Invitrogen, Thermo Fischer Scinetific, Waltham, MA, United States). After transformation, 50 μl of transformed cells were plated on Petri dishes containing 50 μg/ml kanamycin selective media and incubated overnight at 37°C. Colonies were sampled and diluted in 50 μl selective LB media with 50 μg/mL kanamycin, to be grown to pre-cultures for 2 h at 37°C and 225 rpm. Colony PCR was performed to confirm inserts testing colonies with the same primers and M13 universal primers. Pre-cultures producing relevant bands in colony PCR were scaled-up to 5 ml LB-media with 50 μg/ml kanamycin and grown at 37°C and 225 rpm overnight. The pDONR221 vectors containing CDSs of P. hospiton and P. machaon were purified using a QIAprep Spin Miniprep kit (Qiagen, Hilden Germany), quantified with Nanodrop 8000 (Thermo Fisher Scientific, Waltham, MA, United States), and Sanger sequenced. The complete CDS for the P. hospiton candidate OR1 and Pain were deposited at GenBank (MZ172417 and MZ172418, respectively). Topology prediction of the encoded proteins was tested with TopCons (Tsirigos et al., 2015).
To transfer P. hospiton and P. machaon OR1 and Pain CDSs to the destination vector for HEK293A heterologous expression, 100 ng of pDONR221 containing CDS-inserts were mixed with 150 ng of pcDNA40-DEST (Invitrogen), 2 μl LR-clonase (Invitrogen), and TE-buffer to a final volume of 10 μl, and incubated overnight at 25°C. Then, 1 μl of proteinase K was added and samples were incubated at 37°C for 10 min. Aliquots of 1 μl final reaction volumes were used to transform TOP10 competent cells (Invitrogen, Thermo Fischer Scinetific, Waltham, MA, United States). After transformation, 50 μl of cells were plated on 100 μg/ml ampicillin selective media and incubated overnight at 37°C. Positive colonies were identified by colony PCR and grown in 5 ml LB-media with 100 μg/ml ampicillin for plasmid purification. Purified pcDNA40-DEST-vectors containing CDS of P. hospiton and P. machaon were Sanger sequenced.
Heterologous Expression in HEK293 Cells and Transient Transfection
Human Embryonic Kidney (HEK293A) cells were grown to semi-confluence in 35-mm Petri dishes containing HEK cell media [Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (MP Biomedicals, Solon, OH, United States), 2 mM L-glutamine, and 100 μg/ml penicillin/streptomycin (Invitrogen)] at 37°C and 5% CO2. Transient expression was conducted co-transfecting 1.2 μg of pcDNA40-DEST-PhospOR1 or pcDNA40-DEST-PmachOR1 with 0.6 μg of pcDNA5/TO (Invitrogen) carrying the CDS of the CpomOrco variant from the codling moth Cydia pomonella (Genbank accession number JN836672.1, Bengtsson et al., 2012). For control experiments, CpomOrco was co-transfected alone or combined with 1.2 μg of pcDNA5/TO carrying the CDS of CpomOR3 (Cattaneo et al., 2017a). To conduct experiments with painless TRP channels, 1.2 μg of pcDNA40-DEST-PhospPain or pcDNA40-DEST-PmachPain were used to transfect separate samples of HEK cells. For control experiments, HEK were transfected with pcDNA3-DmelTRPA1(B) [dTRPA1(B), a gift from Dr. Paul Garrity, Brandeis University, MA, United States: Kang et al., 2012]. To report expression, 0.6 μg of a separate plasmid DNA carrying the CDS for a blue fluorescent protein (EBFP) was co-transfected [pEBFP2-Nuc, a gift from Robert Campbell, University of Alberta, Alberta, Canada: Ai et al., 2007 (Addgene plasmid #14893)]. Expression of fluorescent reporter genes was under the regulation of the same promoter for Orco/OR/TRP-genes (CMV). In brief, transfection DNAs were dissolved in 100 μl sterile DMEM, mixed with 3 μl Calfectin (SignaGen, Rockville, MD, United States) following the recommended protocol to incubate cells overnight for up to 18 h. After incubation, HEK cell media was replaced with 2 ml fresh media to incubate cells at 37°C for up to 6–8 additional hours, at which point part of the cell culture was spread in the middle of a 35-mm plate as individual cells or small clusters and rinsed at the sides with 2 ml fresh HEK media. After splitting, cells were allowed to recover for at least 1 day prior to calcium imaging.
Imaging Experiments
Activation of HEK293A cells transfected with CpomOrco + PhospOR1, CpomOrco + PmachOR1, CpomOrco + CpomOR3, the sole CpomOrco, PhospPain, PmachPain, and dTRPA1(B) was tested using the same procedures we previously described (Cattaneo et al., 2017a,b, Bobkov et al., 2021). Petri dishes were incubated for 1 h at room temperature in 1.0 mL HEK Ca++ Ringer (mM: 140 NaCl, 5 KCl, 2 CaCl2, 10 HEPES, pH 7.4) containing the fluorescent calcium indicator Fluo-4AM (Invitrogen) at 5–15 μM prepared with 0.06–0.2% Pluronic F-127 (Invitrogen).
As reported by Cattaneo et al. (2017a), the buffer was removed after incubation, cells were rinsed with 4 ml fresh HEK Ca++ Ringer and placed on the stage of an inverted microscope (Olympus IX-71, Olympus Corp., Tokyo, Japan) equipped with a cooled CCD camera (ORCA R2, Hamamatsu, Hamamatsu City, Japan). Cells were continuously perfused with Ca++ Ringer using two gravity-fed perfusion contours. The stimulating contour washing of the cells (∼250 μl/min) was switched rapidly to the stimulus contour using a multi-channel rapid solution changer (RSC-160, Bio-Logic, Claix, France) under the software control of Clampex 9 (Molecular Devices, Sunnyvale, CA, United States).
Fluorescence imaging was performed using settings optimized for Imaging Workbench 6 software (INDEC BioSystems, Santa Clara, CA, United States) (Cattaneo et al., 2017a). The non-responsive cells were not included in these analyses. Each cell was assigned a region of interest (ROI) and changes in fluorescence intensity within each ROI were measured and expressed as the fractional change in fluorescence intensity (dF). Stored time-series image stacks were analyzed offline using Imaging Workbench 6, Clampfit 10.5 (Molecular Devices LLC, San Jose, CA, United States), and SigmaPlot 11 (Systat Software Inc., San Jose, CA, United States). Dose-response curves were approximated using the Hill equation. Constraints were applied in some cases to fit either limited or greatly scattered datasets. Continuous traces of multiple responses were compensated for slow drift of the baseline fluorescence when necessary. All recordings were performed at room temperature (22–25°C).
Dose-Response Characteristics to VUAA1
The non-specific Orco-agonist VUAA1: acetamide, N-(4-ethylphenyl)-2-[[4-ethyl-5-(3-pyridinyl)-4H-1,2,4-triazol-3- yl]thio]-, CAS 525582-84-7 (Glixx Laboratories, Southborough, MA, United States), selected among the ligands we previously reported active on CpomOrco/OR channels (Cattaneo et al., 2017a; Bobkov et al., 2021), was dissolved in dimethyl sulfoxide (DMSO, Sigma Aldrich, St. Louis, MO, United States) and stored as a stock solution (200 mM) at -20°C. The final working concentrations (10–1,000 μM) of VUAA1 were always prepared right before the experiments. Amplitudes of the calcium responses were used to generate dose-response characteristics and values were normalized to the response amplitude recorded at 1,000 μM VUAA1. EC50 values were compared using Welch’s t-test (Systat Software Inc., San Jose, CA, United States).
Screening of Ligand Candidates
Among the tested ligands, L-nicotine (CAS 54-11-5), caffeine (CAS 58-08-2), and D-(-)-salicin (CAS 138-52-3) were selected based on our previous pieces of evidence on their activation of P. machaon and P. hospiton gustatory neurons (Sollai et al., 2014, 2015). Quercetin (CAS 117-39-5) was included in the panel since methanolic extracts from different aerial parts of F. communis presented quantities of quercetin between 1 and 4% of dry content (Rahali et al., 2018). Additionally, we tested allyl-isothiocyanate (AITC, 3-Isothiocyanato-1-Propene, CAS 57-06-7), and only for HEK293A cells transfected with PhospPain and PmachPain, we tested hydrogen peroxide (CAS 7722-84-1) and an essential oil extracted from Ruta graveolens (kindly provided by Prof. Angela Bassoli, University of Milan, Italy). All ligands were purchased from Sigma Aldrich. Quercetin and AITC were diluted in DMSO; caffeine, D-(-)-salicin, and L-nicotine were diluted in water. Stock solutions (200–500 mM) were stored at 4°C. H2O2 was diluted at 3% in Ringer buffer and R. graveolens was water diluted at 150 μg/ml. Right before use, caffeine stocks were slightly warmed in hot water to be properly dissolved before the preparation of final concentrations. To test for any possible dependence of the agonist effect on the solvent, before the experiments non-transfected and transfected cells were tested to the maximum DMSO-solvent concentration used (1%): lack of effects demonstrated that their response was not solvent dependent (data not shown). VUAA1 was tested as a positive control for CpomOrco-based channels activation.
Thermal Experiments
In the temperature-controlled experiments conducted on PhospPain and PmachPain, solutions at different temperatures were applied using a gravity-fed perfusion system, following a similar protocol previously established by Sokabe et al. (2008). Two perfusion contours were used: one was constantly perfusing cells with room temperature solution (∼1.0 mL/min), while the stimulating contour (“temperature-controlled”) was set at 50 or 0°C (depending on if stimulating with heat or cold), which allowed reaching temperatures proximal to 42–44°C or 9–10°C on the preparation. Contours were switched rapidly using a rapid solution changer (RSC-160, Bio-Logic, Claix, France); the duration of the temperature pulse was set at 20 s. The temperature of the solutions was controlled by a bipolar temperature controller (model CL-100, Warner Instruments, Harvard Apparatus, Holliston, MA, United States) and an SC-20 dual in-line solution heater-cooler (Warner Instruments). Temperatures in the Petri dish were measured using a TA-29 thermistor (Warner instruments). Both perfusion output ports and the external thermistor were positioned in close proximity to the imaging area.
Results
Transcriptome Annotation and Identification of Sensory Receptor Transcripts Expressed in the Maxilla of Papilio hospiton
Ribonucleic acid sequencing (RNA-Seq) of mRNA extracted from maxillae of P. hospiton larvae resulted in approximately 23.5 million trimmed reads. The final de novo Trinity assembly consisted of 44,153 contigs, from which 20,870 putative protein sequences were predicted with TransDecoder. On average, approximately 83% of reads were successfully aligned to the assembly (Supplementary Table 2). Completeness of contig assembly was evaluated using the BUSCO Lepidoptera gene set (Seppey et al., 2019), which looked for 5,826 highly conserved genes, whose presence or absence indicated the degree of completeness of the transcriptome. Results showed that 81% of single-copy orthologs were present in P. hospiton transcriptome as full-length transcripts, whereas 15% was missing and 4% was present only as a fragment (Figure 1A). Of the full-length transcripts, 12% were present as duplicated isoforms, indicating some level of redundancy present in the final assembly (Figure 1A). The average OHR was 0.55, and 6,880 contigs (15.6% of the total) had an OHR comprised between 0.95 and 1.05, which indicates a correct full-length transcript assembly (Figure 1B). Searches against the D. melanogaster protein database returned 17,365 contigs showing sequence similarity to known fly proteins. We retrieved 7,347 and 3,105 gene ontology (GO) terms corresponding to biological process and molecular function, respectively. The most abundant GO term referred to basic cell functions; however, GO terms associated with olfaction (e.g., “response to stimulus,” “sensory perception,” and “signal transducer activity”) were also represented (Supplementary Figure 1). Notably, among the most expressed contigs, we found one candidate odorant-binding protein (TR13788_c0_g2_i4) (Figure 1C).
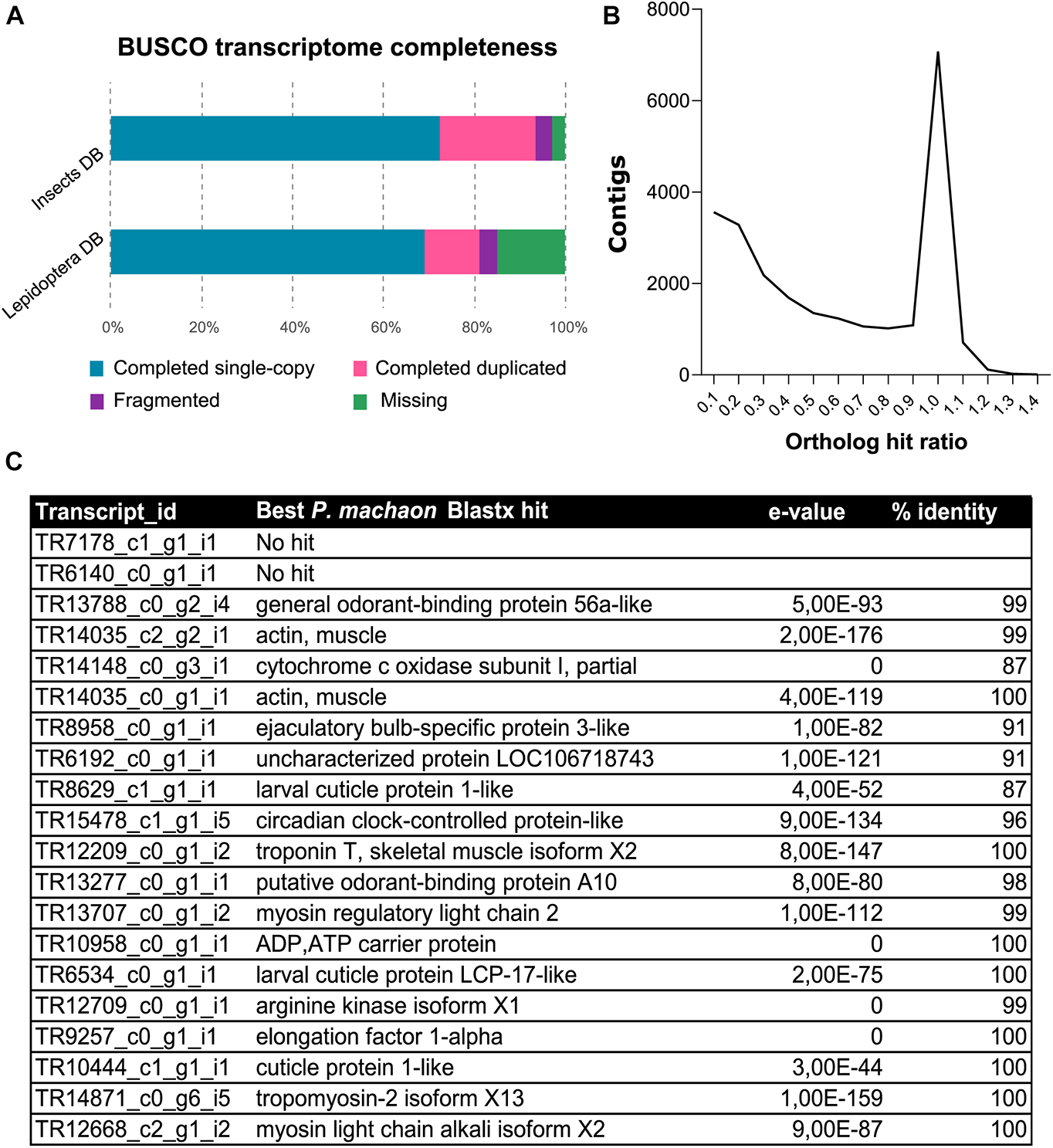
Figure 1. Annotation of de novo assembly from P. hospiton maxilla mRNA. (A) Results from BUSCO analysis against insect and Lepidoptera ortholog databases showing the frequencies of complete single-copy, complete duplicated, fragmented, and missing ortholog from P. hospiton transcriptome (B) ortholog hit ratio (OHR) that estimates contig completeness. OHR was calculated accordingly to O’Neil et al. (2010) using top blastx hit against P. machaon proteome. (C) Top twenty most expressed transcripts.
Analysis of the P. hospiton transcriptome identified 15 contigs likely encoding chemoreceptors (5 for ORs, 1 for a GR, and 9 for IRs) and others 22 contigs likely encoding TRPs (Table 1 and Supplementary Dataset 1). The five contigs encoding ORs were partial sequences. Phylogenetically based reconstruction identified two contigs (TRP2191_c0_g1_i1 and TR18932_c0_g1_i1) as part of the transcript named PhospOR1 whereas the others likely represented fragments of individual mRNAs. Thus, the total number of candidate PhospORs reported was four. The phylogenetic tree clustered them in four different monophyletic groups, each of them displaying a clear one-to-one ortholog with other Papilio species (Figure 2). We identified the ortholog of the insect OR co-receptor (named PhospORco) whose sequence was highly conserved among all the species included in the analysis. PhospOR2 retained a clear ortholog in B. mori (BmOR63) and in Spodoptera littoralis (SlitOR44). The other two PhospORs (named PhospOR1 and PhospOR3), had no clear orthologs in B. mori and S. littoralis.
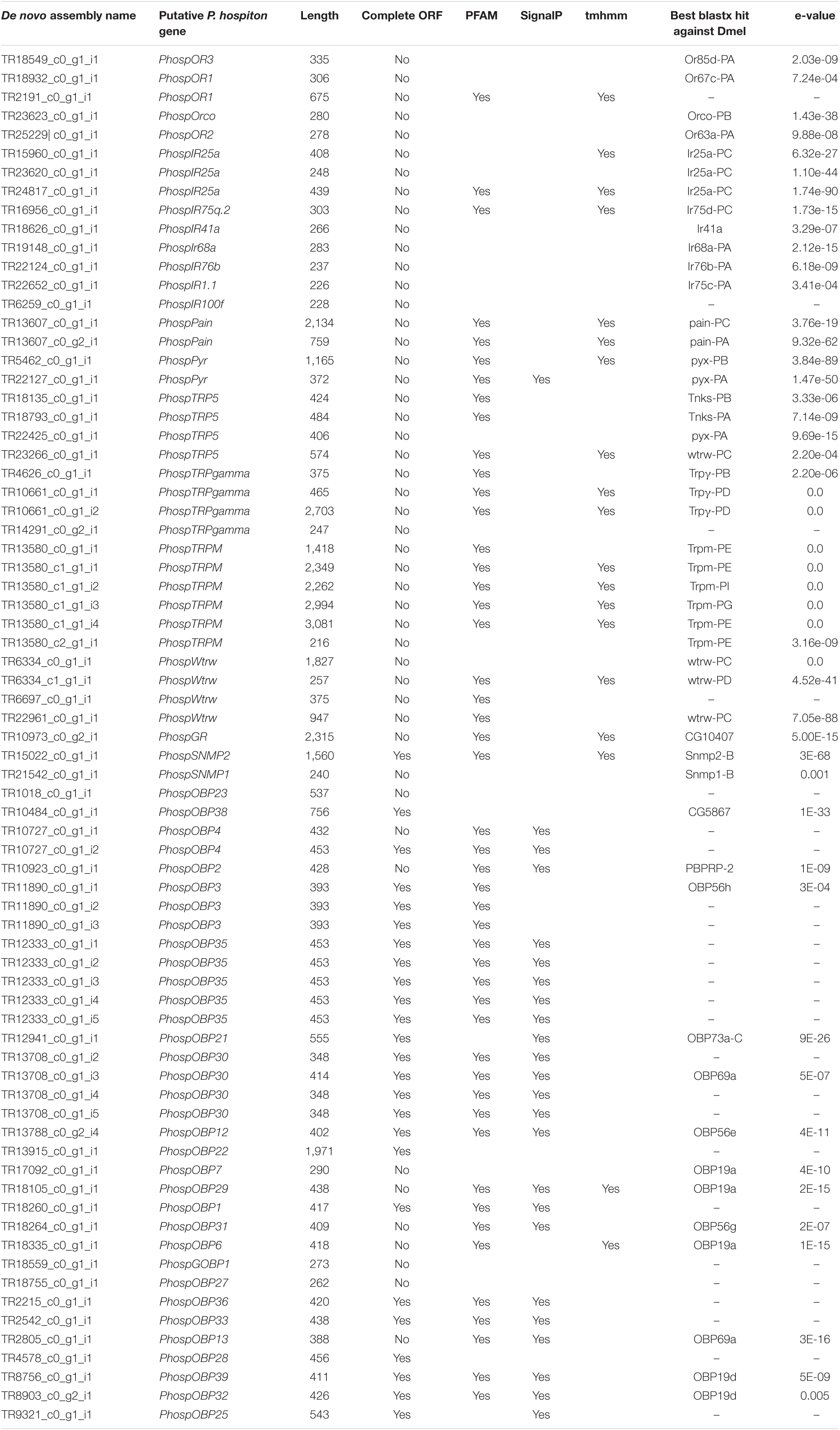
Table 1. Contigs identified as putative transcripts related to chemical sensing and transcripts for TRP-channels.
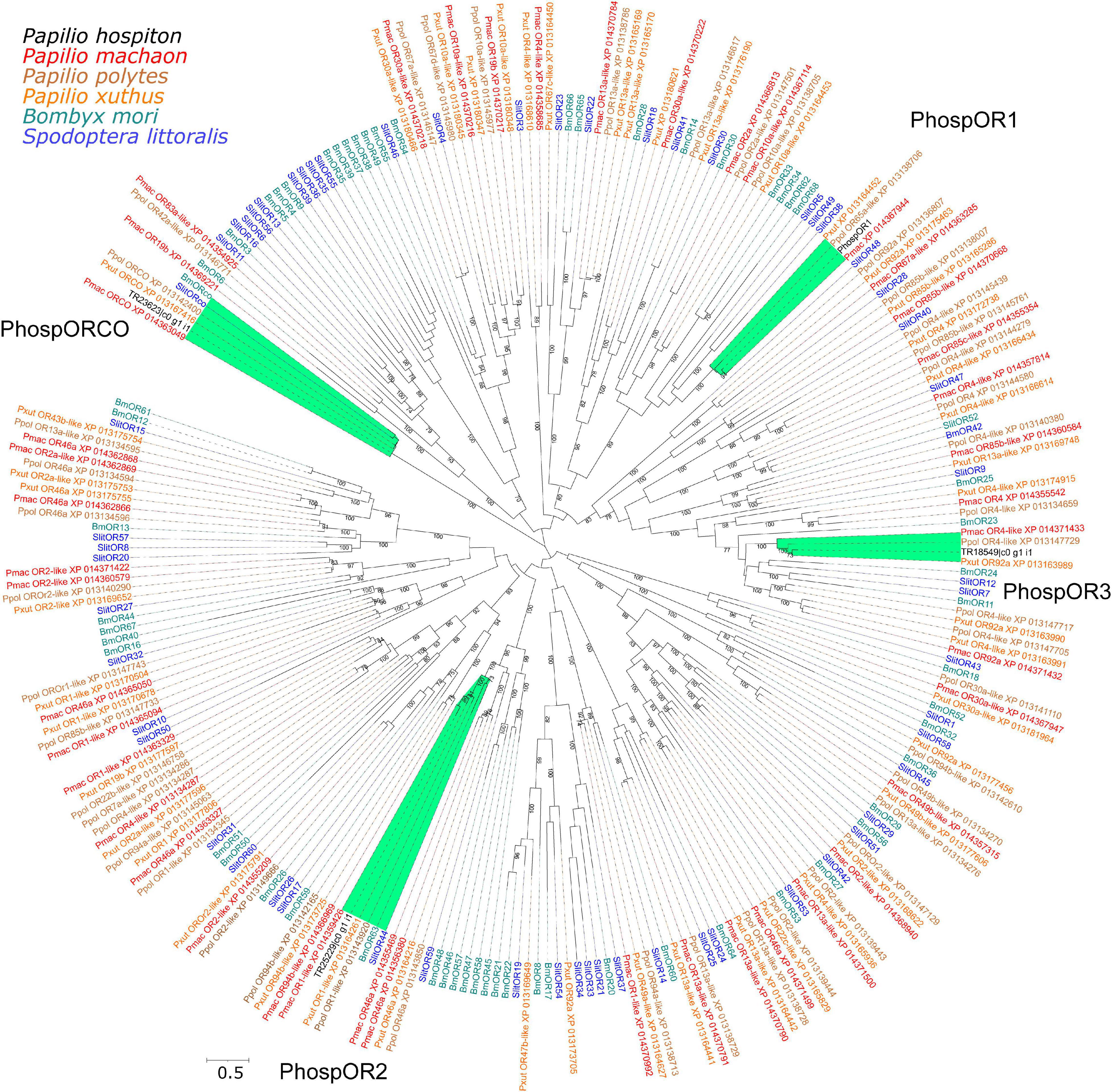
Figure 2. Phylogeny of P. hospiton maxilla-expressed odorant receptors (ORs). A maximum-likelihood tree was built with OR repertoires extracted from automatically annotated genomic datasets of P. machaon, P. xuthus, and P. polytes (Li et al., 2015) using RAxML. Manually curated B. mori (Nakagawa et al., 2005; Tanaka et al., 2009) and S. littoralis ORs repertoires (Walker et al., 2019) are included. For PhospOR1 we used the full-length coding sequence amplified from cDNA instead of the two fragments obtained from RNA-seq. The tree was re-rooted on Orco clade. Labels highlighted in green refer to sequences that form a monophyletic group together with P. hospiton sequences. Branch lengths are scaled by the number of nucleotide substitutions per site. Bootstrap values refer to 500 pseudo-replicates.
Only one-candidate GR transcript was identified in the P. hospiton transcriptome: TR10973_c0_g2_i1 (Table 1) that is a partial fragment of 2,315 bp, which topology prediction indicated to contain six transmembrane domains. The best blastx hit against H. melponene, which is one of the few butterfly species with a well-curated GR repertoire genomic annotation (Briscoe et al., 2013), showed that the closest ortholog in this species was HmelGr23 (E-value 3e-38, amino acid identity 40%). Among Papilio species, blastx searches found a candidate ortholog only in P. machaon (accession number XP_014361781.1, value 0.0, percentage of identity 93%, query cover 99%).
Nine contigs were identified as IRs (Table 1). All of them were partial fragments and phylogenetic analysis showed that three contigs (TR15960_c0_g1_i1, TR23620_c0_g1_i1, TR24817_c0_g1_i1) likely corresponded to different parts of PhospIR25a (Figure 3). Hence, the candidate maxilla-expressed PhospIRs detected in this work were in total seven. All of them had a clear one-to-one ortholog relationship with IRs from other Papilio species, whose IR repertoire had been manually curated from genomic datasets (Liu et al., 2018; Yin et al., 2021). Five maxilla-expressed PhospIRs belonged to the highly conserved antennal IRs (A-IRs) clade (PhospIR68a, PhospIR75q.2, PhospIR25a, PhospIR76b, and PhospIR41a), PhospIR1.1 belonged to the Lepidoptera specific IR (LS-IR) clade whereas PhospIR100f was a candidate divergent IR (D-IR) (Figure 3).
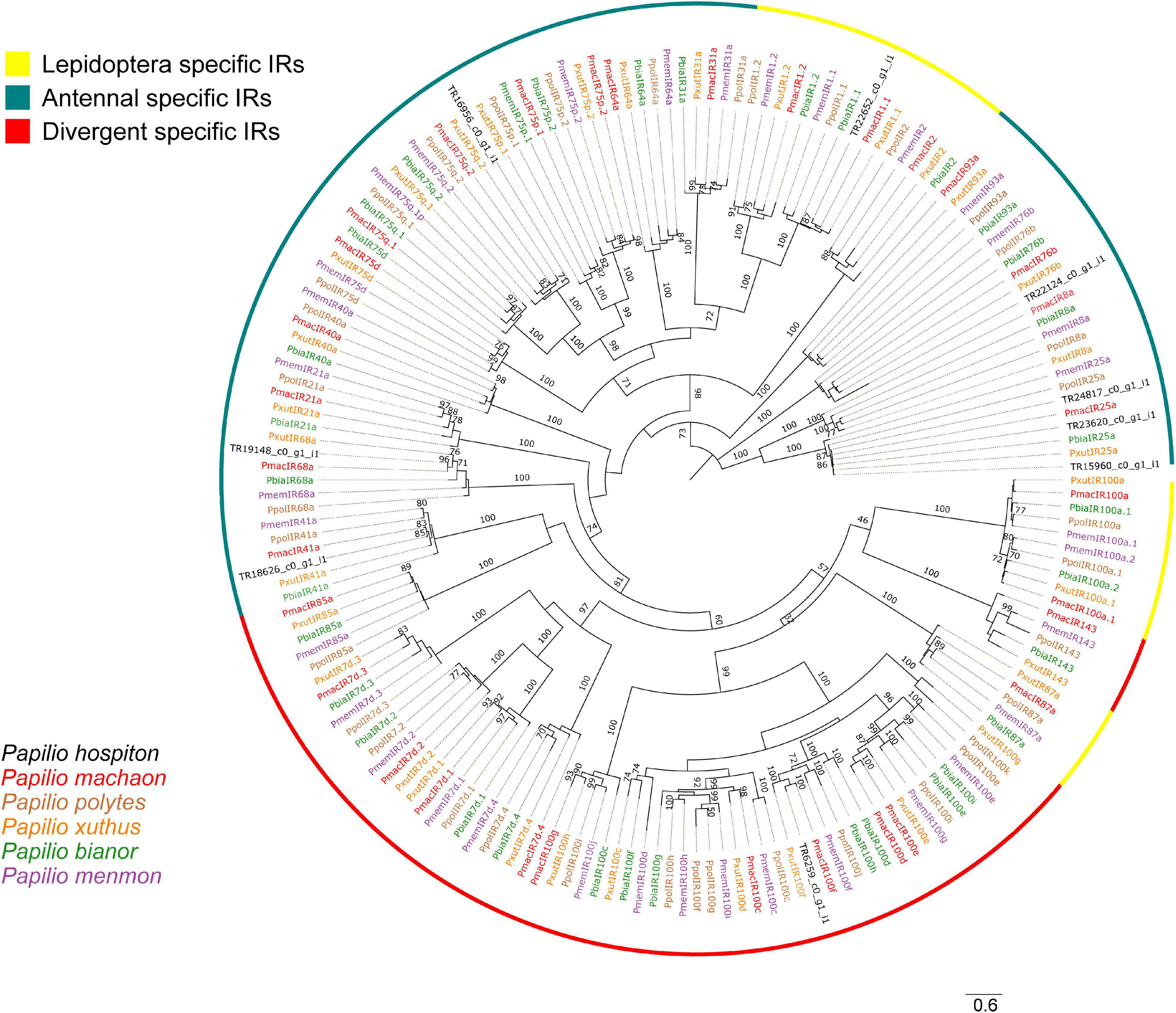
Figure 3. Phylogeny of P. hospiton maxilla-expressed ionotropic receptors (IRs). A maximum-likelihood tree was built with IR repertoires annotated from genomic datasets of P. machaon, P. xuthus, and P. polytes (Liu et al., 2018), and P. memnon and P. bianor (Yin et al., 2021) using RAxML. The tree was re-rooted on IR8/IR25 clade. Branch lengths are scaled by the number of nucleotide substitutions per site. Bootstrap values refer to 500 pseudo-replicates.
Twenty-two contigs were identified as candidate TRP channels (Table 1), and none of them contained a full-length CDS. Phylogenetic analysis showed that these 22 contigs clustered into six monophyletic groups; thus, they likely represented redundant isoforms of the same transcript (Figure 4). In total, TRP channels identified in the P. hospiton larval maxilla assembly were six: PhospTRPγ, which belongs to the TRPC subfamily, PhospTRPM, and four members of the TRPA subfamily, namely pyrexia (PhospPyx), water witch (PhospWtrw), PhospTRPA5, and painless (PhospPain).
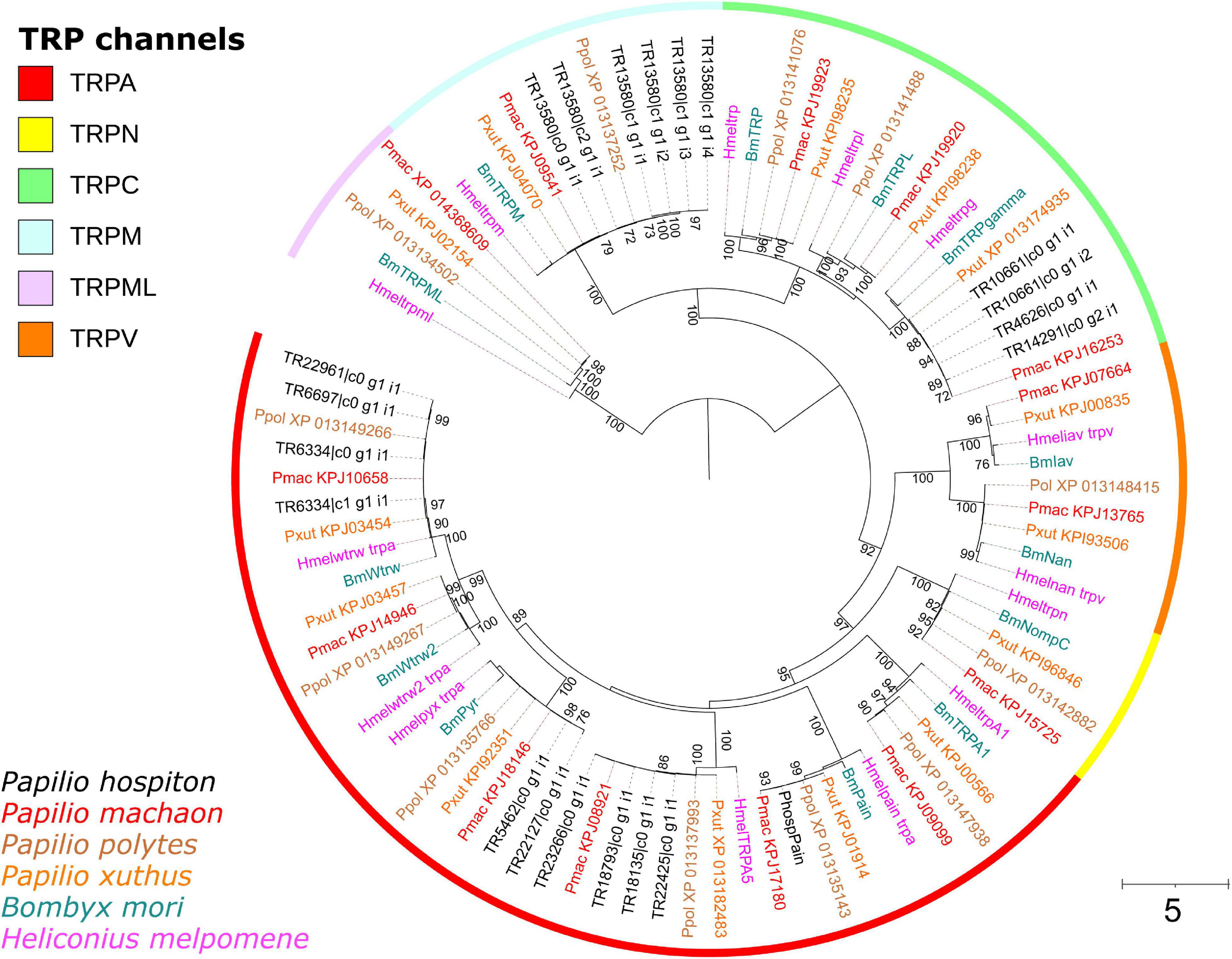
Figure 4. Phylogeny of P. hospiton maxilla-expressed transient receptor potential channels (TRPs). A maximum-likelihood tree was built with TRP repertoires from automatically annotated genomic datasets of P. machaon, P. xuthus, and P. polytes (Li et al., 2015) using RAxML. Manually curated genomic datasets of B. mori (Matsuura et al., 2009), and H. melpomene (Macias-Muñoz et al., 2019) are included. For PhospPain we used the full-length coding sequence amplified from cDNA instead of the two fragments obtained from RNA-seq. Branch lengths are scaled by the number of nucleotide substitutions per site. Bootstrap values refer to 500 pseudo-replicates.
Analysis of the P. hospiton larval maxilla transcriptome identified also two contigs encoding SNMPs (Table 1). One of them represented a full-length transcript that is ortholog to lepidopteran SNMP2 (Supplementary Figure 2) whereas the other contig was a partial fragment belonging to clade SNMP1.
Thirty-four candidate OBP contigs were identified in P. hospiton. Several contigs contained redundant sequences and the final number of total OBPs was 24 (Table 1 and Supplementary Figure 3). Fifteen of them contained full-length sequences. Of these, three PhospOBPs exhibited the classic arrangement of conserved six-cysteines, five displayed the Plus-C gene motif, six were Minus-C and one was a duplex. Phylogenetic analysis revealed that many candidate PhospOBPs clustered in accordance with Lepidopteran OBP sub-families (Supplementary Figure 3) and one of them belonged to the lepidopteran-specific GOBP1 clade. Six PhospOBPs seemed to represent P. hospiton specific OBP clade.
To functionally characterize the sensory proteins expressed in P. hospiton larval maxilla along with their orthologs in P. machaon, we tried to isolate full-length sequences. Our efforts resulted in the obtainment of a complete coding sequencing for the OR-candidates PhospOR1 and its ortholog PmachOR1, and the TRPA-channels PhospPain and its ortholog PmachPain. The respective polypeptides shared 96.46% identity and 98.48% similarity (OR1, 394 amino acids) and 98.33% identity and 99.37% similarity (Pain, 955 amino acids) between the two species (similarity matrix BLOSUM62, BioEdit). Topology of both PhospOR1 and PmachOR1 returned seven transmembrane domains (PhospOR1: TM1: 33-53, TM2: 67-87, TM3: 131-151, TM4: 186-206, TM5: 268-288, TM6: 298-318, TM7: 367-387; PmachOR1: TM1: 33-53, TM2: 67-87, TM3: 130-150, TM4: 184-204, TM5: 268-288, TM6: 299-319, TM7: 368-388) with intracellular N-terminals, suggesting complete polypeptide sequences for these ORs. Topology of both PhospPain and PmachPain returned six transmembrane domains with the same predicted amino acid positions (TM1: 536-556, TM2: 572-592, TM3: 605-625, TM4: 631-651, TM5: 671-691, TM6: 751-771), having both N- and C-terminal intracellular, suggesting complete polypeptide sequences also for these TRP channels.
Dose-Response Characteristics of Papilio OR1 for VUAA1
HEK293A cells transfected with CpomOrco alone or in combination with CpomOR3 and either PmachOR1 or PhospOR1 generated calcium signals upon application of VUAA1 in a concentration-dependent manner (10–1,000 μM, Figure 5). For both individual cells and cell populations, the response amplitude depended on the agonist concentration: in particular, response amplitudes for CpomOrco appeared to be substantially reduced when compared with CpomOrco + CpomOR3 and CpomOrco + PmachOR1. Although with similar amplitudes with CpomOrco + PhospOR1, the kinetics of the calcium responses of the homomeric CpomOrco to VUAA1 started from higher concentrations (250 μM VUAA1, Figure 5A). In addition, summary plot comparisons of the mean response amplitudes to VUAA1 proximal to saturation (500 μM VUAA1) resulted in shifted dose-dependences (Figure 5B).
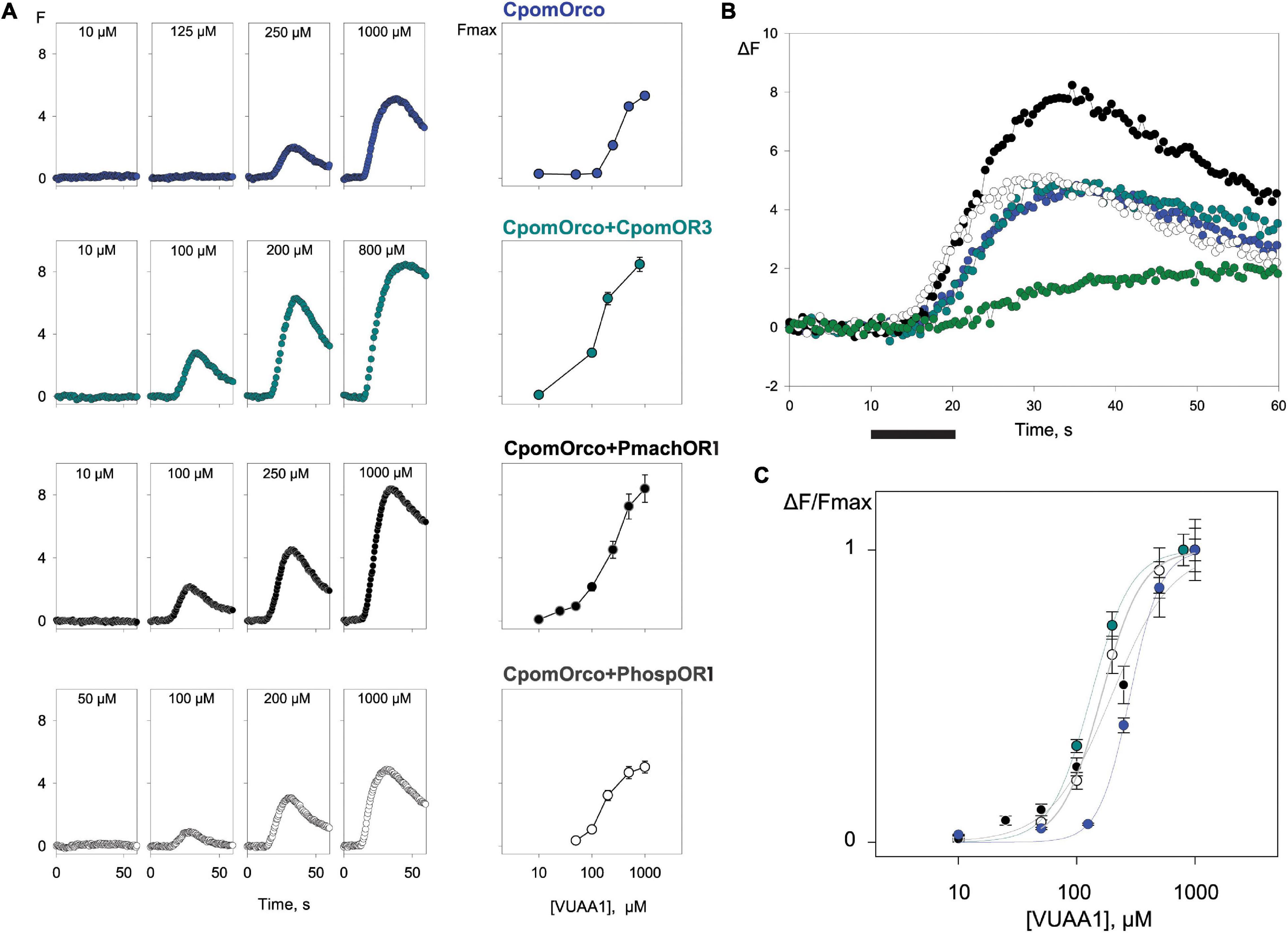
Figure 5. Comparison of HEK293A cell responses expressing CpomOrco, CpomOrco + OR3 and chimeric CpomOrco + PapilioOR1. (A) Universal Orco agonist, VUAA1, elicits dose-dependent Ca++i increase in HEK293A cells expressing homomeric or heteromeric CpomOrco. Data points represent the mean response amplitudes. Left: response to single doses of VUAA1 (as indicated within panels); right: dose-response plot. (B) Plot of the mean response to 500 μM VUAA1: blue, CpomOrco (N = 246); dark cyan, CpomOrco + OR3 (N = 240); black, CpomOrco + PmachOR1 (N = 55); white, CpomOrco + PhospOR1 (N = 63); green, CpomOrco + OR3 to 1,000 μM pear ester (N = 134). Black bar: stimulus (10 s). (C) Normalized VUAA1 concentration dependencies. Data were fit to a Hill equation for CpomOrco (blue, EC50 = 282 ± 0.13 μM, Hill coefficient ∼3.34 ± 0.47, N = 269), CpomOrco + OR3 (green, EC50 = 132.2 ± 3.15 μM; Hill coefficient ∼2.5 ± 0.15, N = 123), CpomOrco + PmachOR1 (dark gray, EC50 = 197.2 ± 34.5 μM, Hill coefficient ∼1.7 ± 0.33, N = 68) and CpomOrco + PhospOR1 (light gray, EC50 = 162 ± 6.64 μM, Hill coefficient ∼2.5 ± 0.23; N = 72).
Average responses were used to generate VUAA1-concentration dependencies (Figure 5C), where the average peak amplitudes of the responses of different cells were normalized to the responses elicited by application of 1,000 μM VUAA1 (saturating concentration). The comparison of the dose-response characteristics obtained for different CpomOrco + OR complexes suggested that homomeric CpomOrco (EC50∼282 μM) is less sensitive to the VUAA-agonist than its heteromeric combinations [CpomOrco + CpomOR3, EC50∼132 μM; CpomOrco + PmachOR1, EC50∼197 μM; CpomOrco + PhospOR1, EC50∼162 μM—Welch’s t-test p ≤ 0.017 (for all combinations)].
Screening of Possible Ligands on Papilio OR1
Among the ligands tested at 500 μM concentration (VUAA1, L-nicotine, caffeine, D-(-)-salicin, quercetin, AITC), only VUAA1 demonstrated an evident effect on HEK293A cells co-transfected with CpomOrco and Papilio OR1 (Figure 6A). Although a slight fluorescence variation was observed when AITC was perfused on HEK-cells transfected with CpomOrco + PmachOR1, AITC sensitivity for the chimeric CpomOrco + Papilio cation channel was excluded given the recorded effect to the same dose of ligand for the negative control of a non-transfected HEK293A cells sample. Interestingly, the effect on non-transfected cells seemed to not be repeatable when cells were stimulated for the second time, suggesting the existence of possible endogenous cation channels somehow responsive to AITC on HEK293A, which may be saturated upon the first application of the ligand (Figure 6B).
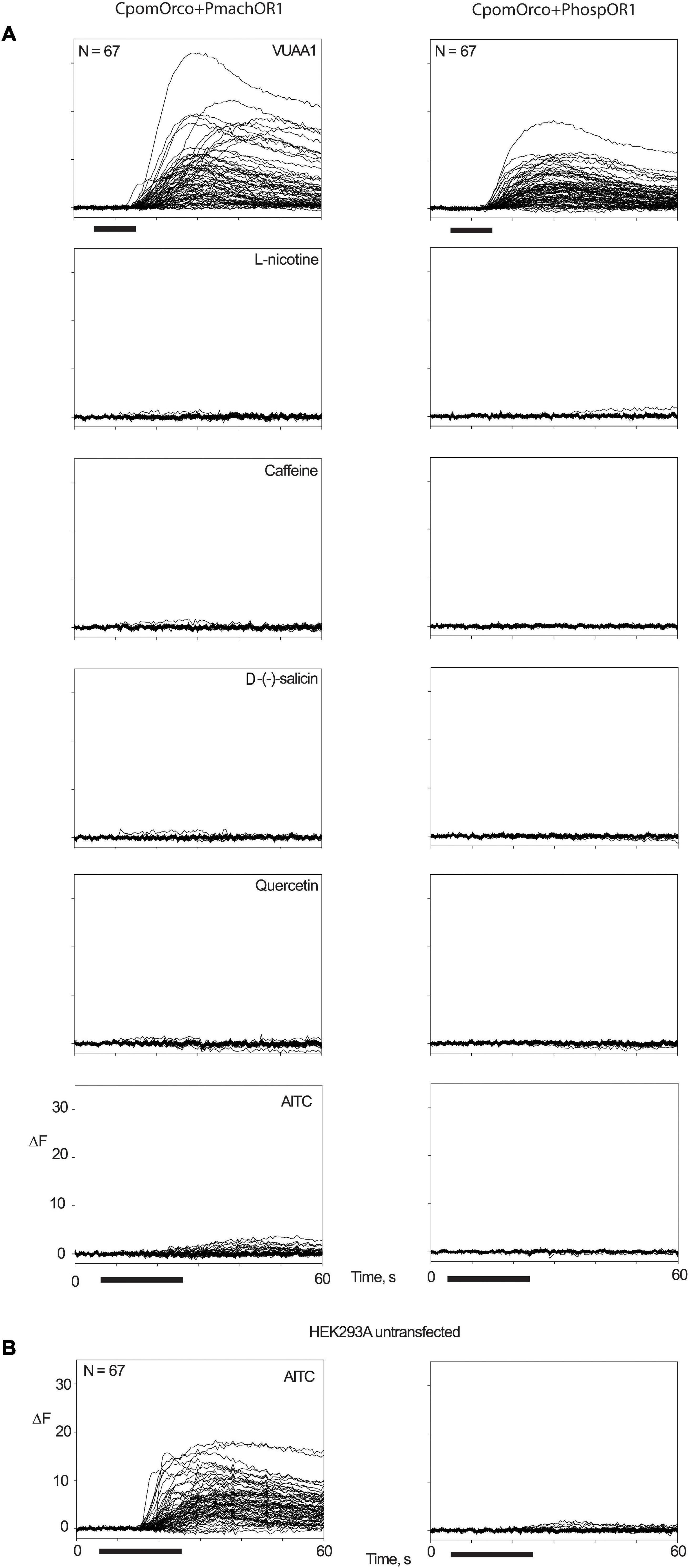
Figure 6. Screening of candidate ligands on OR1. Multiple traces of fluorescence variation associated with Ca++i increase in HEK293A cells transfected with CpomOrco + PmachOR1 (left) and CpomOrco + PhospOR1 (right). (A) Application of 500 μM doses of VUAA1, L-nicotine, caffeine, D-(-)-salicin, quercetin, and AITC. (B) Two consecutive applications of AITC (500 μM) to non-transfected HEK293A. A reversible effect on non-transfected cells was observed, excluding its relations with the human TRPA1 (Cattaneo et al., 2017b), which is in accordance with the documented absence of TRPA1-transcripts in HEK cells (BioGPS Cell Line Gene Expression Profiles—HEK293). N = 67 for all experiments; black bar: stimulus (10 s for VUAA1, 20 s for ligands).
Functional Studies on Papilio Painless
Testing AITC (Figure 7A), a repeatable fluorescent variation was recorded for a limited number of cells of the preparation for the sole HEK293A samples transfected with PhospPain (N = 33). When HEK293A cells were transfected with PmachPain, a reduced effect, which was not repeatable, suggested being rather associated with an artifact. Responsive HEK293A cells expressing PhospPain (N = 33) were tested to the same compounds screened on Papilio OR1 subunits (Figure 6A), with the exception of the Orco-specific VUAA1. No effects suggested the absence of activation for PhospPain to these ligands (Figure 7B). When the effects of PhospPain and PmachPain on the temperature stimulus (T ∼ 42–44°C) were compared with the positive control dTRPA1(B) (Figure 7C) an evident fluorescence variation was not observed, indicating the absence of temperature-related activation. Additionally, our results revealed a lack of activation on these subunits for hydrogen peroxide and the essential oil extracted from R. graveolens (Supplementary Figure 4).
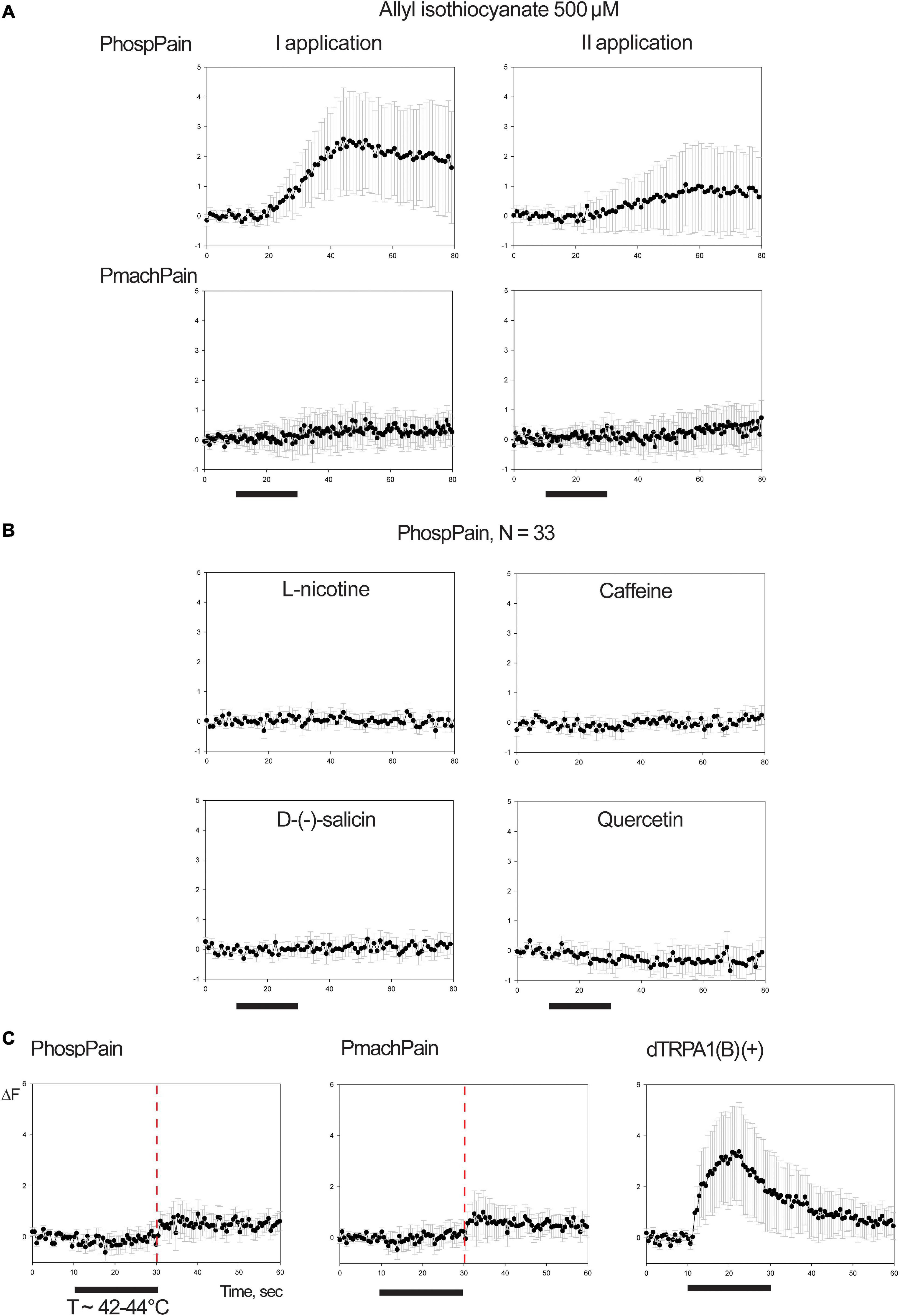
Figure 7. Functional studies on TRP Painless. (A) Mean ± SE of fluorescence variation associated with PhospPain (N = 33) and PmachPain (N = 28) upon stimulation with 500 μM allyl-isothiocyanate (AITC, two consecutive applications). (B) Application of 500 μM doses of L-nicotine, caffeine, D-(-)-salicin, quercetin on HEK-cells transfected with PhospPain that were found responsive to AITC. Black bars: stimulus (20 s). (C) Mean ± SE of fluorescence variation associated with PhospPain (N = 94) and PmachPain (N = 107) upon stimulation with heat (T ∼ 42–44°C). Right: positive control experiment stimulating HEK293A cells transfected with dTRPA1(B) (N = 101). Red line: time point for thermal experiments preceding an endogenous-effect observed in the phase of testing HEK-cells transfected for Papilio Painless. The demonstrated Ca++-dependency of this effect on HEK cells (Ong et al., 2015; Shalygin et al., 2015) may associate the activation of cation channels most probably belonging to the asset of thermal-gated channels selective for Calcium (Xiao et al., 2011).
Discussion
In this work we conducted a transcriptomic analysis of the larval maxillae of P. hospiton, a species endemic of the Mediterranean islands of Corsica and Sardinia that has been used as a model for the comparative analysis of maxilla taste responses due to its strict olygophagy juxtaposed with the relaxed olygophagy of the close-related species P. machaon (Sollai et al., 2014, 2015, 2018b; Sollai and Crnjar, 2019). To date, although other Papilio genomes have been sequenced, no genomic data for P. hospiton are available. Our study rendered a handful of sequences encoding for putative ORs, IRs, a single GR, TRPs as well as SNMP and OBPs, which may represent the key genes involved in neuronal functions in the maxillae and constitute the first sensory-related available sequences for this species. We further used data obtained from RNA-seq experiments to full-length clone two sensory proteins, PhospOR1 and PhospPain along with their orthologs from P. machaon, for functional characterization in HEK293A cells. Although functional assays failed to identify ligands within the panel used in this study, our results pave the avenue for further assays aimed at a finer characterization of the site of action and the molecular substrates recognized by the receptors.
Ribonucleic acid sequencing (RNA-seq) is a powerful tool that has been widely employed to identify transcripts expressed in chemosensory organs of adult Lepidoptera, and to less extent, in larvae (Montagné et al., 2015). Data of larval maxilla-expressed chemosensory receptors are available for a handful of species, and show the presence of OR, GR, and IR mRNAs in this appendage (Tanaka et al., 2009; Poivet et al., 2013; Di et al., 2017; Guo et al., 2017). In two moth model species, B. mori and S. littoralis, the number of larval maxilla-expressed ORs are somewhat similar (20 and 16, respectively) whereas there is a dramatic difference in GRs (46 and 3, respectively) (Tanaka et al., 2009; Poivet et al., 2013). Our results showed the expression of a single candidate GR and four ORs, along with seven IRs and six TRPs. This reduced number of sensory-related transcripts may be due to several factors, including a lack of sequencing depth that hindered the detection of low expressed transcripts, such as GRs (Vosshall and Stocker, 2007). BUSCO analysis with the insect database of single-copy orthologs, identified as complete 93% of the queried sequences (81% when using Lepidoptera database, Figure 1A), indicating that the completeness of the assembly was satisfactory. However, the use of a composite tissue as the whole maxilla might have diluted mRNAs expressed in a few cells, such as ORNs and GRNs, resulting in fragmented transcripts and several sequences missing from the final assembly.
The larval maxilla has been extensively studied for its contribution to taste, whereas its role in olfaction has so far only been suggested based on structural data (Schoonhoven and van Loon, 2002). Among the identified sequences, two may have a canonical role in taste: the PhospGR, and the putative PhospIR100f that belong to the divergent IR subfamily. This subfamily has been widely detected in GRNs but not in the antennae, and experimental evidence in model insects points to a major role in taste reception (Koh et al., 2014; Stewart et al., 2015; Tauber et al., 2017). Other candidate taste receptors expressed in P. hospiton larvae maxilla may be PhospIR76b-based cation channels and TRP channels. In D. melanogaster, the IR76b subunit is co-expressed in both the olfactory and taste systems with other IRs mediating response to salt (Zhang et al., 2013a), polyamine (Silbering et al., 2011; Hussain et al., 2016), amino acids (Croset et al., 2016; Ganguly et al., 2017), fatty acids (Ahn et al., 2017), carbonation (Sánchez-Alcañiz et al., 2018), or regulating the response to sucrose and acids (Chen and Amrein, 2017). In particular, the sensory mechanism tuned to polyamines is based on the combination IR25a + IR76b + IR41a (Silbering et al., 2011; Hussain et al., 2016). The fact that these three IRs are among the ones detected in the maxillae of P. hospiton larvae may suggest the existence of a similar sensory mechanism in this appendage.
Transcriptomic analysis demonstrated the expression of several TRP channels (Table 1 and Supplementary Dataset 1). TRPs are responsible for detecting multiple types of stimuli in insect sensory systems (Fowler and Montell, 2013). In Drosophila, some TRP channels function in cold-avoidance (Rosenzweig et al., 2008) and thermotaxis (Lee et al., 2005; Sokabe et al., 2008; Sokabe and Tominaga, 2009), as well as in negative geotaxis (Sun et al., 2009). A specific TRP, water witch (Wtrw), is involved in hygrosensation (Liu et al., 2007), whereas another isoform, TRPM, is involved in magnesium homeostasis (Hofmann et al., 2010). Interestingly, some insect TRP channels are also involved in chemosensation (Kang et al., 2010; Kwon et al., 2010) or play a role in taste (Montell, 2021). For example, in D. melanogaster TRPL is directly activated by camphor (Zhang et al., 2013b), TRP-γ by polyunsaturated fatty acids (Jörs et al., 2006), and the TRPA Painless, initially identified as a nociceptive heat sensor (Tracey et al., 2003), was later found to be involved in the detection of allyl-isothiocyanates (Al-Anzi et al., 2006) and fructose (Xu et al., 2008). This raises the possibility that some TRP-channels function as ionotropic receptors in GRNs in insect species other than Drosophila, and for larvae of P. hospiton in particular, orthologs of TRP-γ and Painless may play a role in the maxillar sense of taste.
Apart from the various subunits with possible involvement in taste perception, we found transcripts encoding for olfactory receptors (such as ORs and A-IRs). We also demonstrated the presence of transcripts encoding for two SNMPs and several OBPs, which are two families indicated to play a critical role in the detection of certain odorants in insects (Cassau and Krieger, 2021). These observations support an olfactory function for P. hospiton larval maxilla. Accordingly, although no functional assays have been so far carried out for characterizing maxilla olfaction in P. hospiton (and neither in other Lepidoptera species), it is conceivable that some of the less characterized basiconic sensilla present in maxillary palps may sense odors (Schoonhoven and van Loon, 2002). The presence of the transcripts encoding the OR co-receptor Orco and two out of the three main IR co-receptors IR25a and IR76b (Abuin et al., 2019) (while IR8a was not identified) suggests the presence of both heteromeric OR and IR channels that likely mediate olfaction in the maxilla of P. hospiton larvae.
We cloned the full-length sequence of PhospOR1 and PhospPain, together with their orthologs from P. machaon, to test for their role in the differential spike activity previously recorded from maxillary GRNs in response to bitter tastants between these two species (Sollai et al., 2014, 2015, 2018b; Sollai and Crnjar, 2019). While some previous data from D. melanogaster support a likely role of Painless in taste (Xu et al., 2008), no data are available for supporting the involvement of an OR in taste. However, previous data from the morphological, electrophysiological, and behavioral investigation on moths, among Noctuidae, Cambridae, Plutellidae, Pyralidae, and Yponomeutidae families, underlined the flexibility of the chemosensory systems associated with larval maxilla of Lepidoptera (Roessingh et al., 2007; Rana and Mohankumar, 2017). For example, in Lymantria dispar, a remarkable influence of maxillary palps in facilitating feeding by the organization of a complex neurosensory mechanism composed by gustatory sensilla together with the olfactory ones has been proposed (Solari et al., 2002). This, together with some pieces of evidence not harmonizing with a precise differentiation between the chemical senses of taste and smell such as the fact that insect ORs can be functional in true taste neurons of Drosophila, producing responses to odorants that are similar to those obtained with tastants (Hiroi et al., 2008; Malik et al., 2019) or the role of some GRs in an olfactory function such as CO2 detection (Joseph and Carlson, 2015) inspired us to test the function of these two receptors with the tastants so far known to stimulate a physiological response in P. hospiton larval maxillae.
Both PhospOR1 and PmachOR1 subunits were expressed with CpomOrco, a Lepidoptera OR co-receptor that we have already functionally characterized (Cattaneo et al., 2017a). We used this latter because, despite the identification of the PhospOrco subunit within the P. hospiton transcriptome (Figure 2), we failed to amplify its full-length CDS. We are aware of the possibility to perform functional trials in HEK cells using synthetic codon-optimized sequences (Roberts et al., 2021). However, the choice of CpomOrco was based on the conservation of this subunit with orthologs within and outside the order of Lepidoptera that allows for the generation of functional chimeric Orco/OR channels (Bobkov et al., 2021). For example, many Lepidoptera ORs have been deorphanized through the heterologous expression of insect ORs in empty neurons of D. melanogaster, where dipteran Orco/lepidoteran OR channels are generated (Gonzalez et al., 2016). For this reason, we did not consider possible issues to re-propose the same approach in vitro.
To test the expression of Papilio OR1 subunits before testing candidate ligands, we used the main Orco agonist VUAA1, which has been reported for activating both homomeric and heteromeric Orco-based receptor channels (Jones et al., 2011). Dose-response characteristics to VUAA1 (Figure 5), which are consistent with the pharmacologic parameters reported in our previous investigations using CpomOrco + ORs (Cattaneo et al., 2017a; Bobkov et al., 2021) suggested the heterologous co-expression of Papilio OR1-subunits with CpomOrco. Despite expression pieces of evidence of both PhospOR1 and PmachOR1 in HEK-cells, we did not identify any ligand able to activate these subunits within the panel we used (Figure 6). This might mainly be due to our choice of only including compounds known to activate physiological responses in P. hospiton larval maxilla, which are tastants. However, we should also be aware that for both chimeric CpomOrco + PapilioOR1 combinations we observed a general reduction in the percentage of VUAA1-responsive cells (Supplementary Table 3). This phenomenon was previously associated with the malfunctional expression/assemblage of a mutagenized CpomOrco (Bobkov et al., 2021). In the mentioned study, the possibility of a malfunctional expression due to malfunctional assembly of an Orco + OR cation channel rather than to the reduction in its overall expression was observed to be more likely if the general spatial and quantitative EBFP expression overlapped with the response to VUAA1. In every experiment we conducted with CpomOrco + PapilioOR1 the predominant majority of the cells responding to VUAA1 were also EBFP positive (Supplementary Table 3), thus suggesting that lack of activation of CpomOrco + PapilioOR1 combinations might be also due to a compromised functional assembly of the cation channel.
In a consecutive set of experiments (Figure 7) we attempted activation of Papilio Painless to AITC and bitter tastants used with Papilio OR1 (Figure 5). AITC is known among the degradation products of plant glucosinolates in the foraging activity of larvae (Wittstock et al., 2003) and it is one of the most common chemosensory active noxious chemicals for various organisms, including insects of the order of Lepidoptera (Wei et al., 2015). In D. melanogaster, Painless is required for AITC avoidance response (Al-Anzi et al., 2006). In the case of Papilio Painless, testing AITC 500 μM (Figure 7) unveiled a repeatable activation for a limited number of cells for the sole subunit of P. hospiton (N = 33), although with evident desensitization in the response to the ligand upon the second application (Figure 7A), which suggests for these cells possible PhospPain functional expression. Oppositely, when cells were transfected with PmachPain and tested to AITC, only a slight fluorescent variation was recorded (N = 28). The fact that for the P. machaon subunit this effect was not repeatable seems to be far from suggesting its functional expression. In addition, analyzing EBFP fluorescence (CMV-promoter functionality) and TRP-activation (functional expression), cells transfected with PhospPain reported similar fluorescence overlap compared with the positive control (Supplementary Table 3). When transfected with PmachPain, only a few cells among the AITC-responsive were both EBFP-positive and fluorescent, suggesting their response was rather associated with an artifact. Indeed, we observed the existence of an occasional endogenous effect in the phase of testing AITC, which was present also when testing CpomOrco + PapilioOR1 (Figure 6). Even if such effect was reduced when cells were transfected with Papilio subunits, the existence of this effect and the evident desensitization in the response of Painless to the ligand (Figure 7A) caused difficulties to plan dose-response studies. Additionally, our results revealed a lack of activation on Painless subunits for the bitter tastants tested on OR1 (Figure 7B), and to hydrogen peroxide and the essential oil extracted from R. graveolens (Supplementary Figure 4), known among the plant hosts of P. machaon (Fagot, 1996).
To further investigate the function of PhospPain and its ortholog PmachPain, we evaluated their activation by high temperature. In D. melanogaster, Painless is known as the insect TRP-sensor activated by temperatures above 40°C (Sokabe and Tominaga, 2009). We used dTRPA1(B) as positive control and the same protocol adopted by Sokabe et al. (2008), in which we perfused Ringer buffer upon heating to reach the preparation temperatures proximal to 42–44°C. Activation of the positive control dTRPA1(B) to the thermal stimulus demonstrated the suitability of our cell system to express thermal TRP-channels (Figure 7C). However, when transfected with Papilio painless, HEK293A did not report any effect of activation to heat (Figure 7C) and cold (Supplementary Figure 4) suggesting that in Papilio, Painless subunits are not involved in thermosensation.
In conclusion, in this work we explored the asset of chemosensory receptors expressed in the larval maxillae of P. hospiton, identifying key genes expressed in this appendage that may underlie the sense of taste and olfaction. Although functional characterization of two of the transcripts detected, PhospOR1 and PhospPain, together with the respective orthologs of P. machaon, did not find any ligand within the panel used, which was formed by the compounds known so far to activate Papilio maxillary neurons, nor heat and cold activation of Painless, our study sets the basis for further studies aimed to decipher the role of the transcripts described here. Lack of activation of both OR1 and Painless to bitter compounds may indicate such chemicals being most probably detected by GRs rather than ORs or TRPs. Unfortunately, no GR could be tested because of the lack of full-length sequences that we could amplify from our transcriptome. Expanding the panel of chemicals used to test for receptor activation as well as the elucidation of the site of action of the encoded proteins (i.e., gustatory styloconic sensilla on the galea or basiconic sensilla on the palps) through fluorescent immunocytochemistry or single-cell resolution transcriptomics will give us important hints on their role in olfaction or taste, hence allowing us to leverage their contribution to diet specialization.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
AMC conceived and designed the experiments, cloned CpomOrco CDS and the full-length CDS of Papilionid subunits. GS reared and sampled larvae of Papilio hospiton and extracted total RNA from the larval maxilla. CC performed transcriptomic analysis, assembling, and phylogenetic analysis. AMC and YB optimized HEK293 cells transfection and protocols and performed calcium imaging on HEK293 cells. AMC, RC, and GA provided material support. AMC and RC supervised the research. AMC and CC wrote the manuscript. All authors edited and approved the final version of the manuscript.
Funding
Mobility between Sweden and the United States, costs for chemicals, and part of other running costs were financed by Martha och Dagny Larssons fond-Swedish University of Agricultural Sciences (Protokoll 172-174, Sammanträdesdatum 2018-04-24). Functional studies and other parts of running costs were financed by the division of Chemical Sensing, Whitney Laboratory for Marine Bioscience, University of Florida and by the Crafoord Foundation (Crafoordska Stiftelsen, referens nummer: 20180954–Sweden). Costs for NGS sequencing and cloning work were financed by the Department of Biomedical Sciences, University of Cagliari, project title: “Identificazione dei recettori gustativi nei Lepidotteri: la specie sardo-corsa Papilio hospiton e la specie olartica congenerica Papilio machaon,” by the use of grants from the autonomous region of Sardegna (Italy) (L.R. 7/2007–year 2012, CUP F71J12000950002, title “L’endemismo sardo-corso Papilio hospiton e sue piante-ospite: studio integrato per la conservazione”). Costs for cloning of Papilionid genes were financed by the Agricultural Entomology group of Foundation Edmund Mach: Centre of Agriculture, Food and Environment, University of Trento. Data analysis and manuscript preparation were undertaken in the course of the FORMAS Swedish research council project number 2018-00891, title “Control of fruit pests by targeting larval chemical sensing,” and in the course of the CDEIGENT founding, Conselleria de Innovación of Generalitat Valenciana, reference number: CDEIGENT/2019/009. Submission costs have been covered by Martha och Dagny Larssons fond-Swedish University of Agricultural Sciences (Protokoll 212-214, Sammanträdesdatum 2020-04-21).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge Barry Ache, Whitney Laboratory for Marine Bioscience – University of Florida, for lab and equipment availability. We thank William B. Walker III for the general discussion, personal communications, and general availability. This manuscript is part of the research topic: “An integrated model of the olfactory system through phylogenetic, evolutionary, and clinical aspects: from invertebrates to humans, through sensory, perceptual, and cognitive olfactometry studies.” We also thank the topic editors for their invitation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2021.795994/full#supplementary-material
Supplementary Figure 1 | Gene ontology of P. hospiton assembly. Gene Ontology (GO) terms retrieved for biological process (upper panel) and molecular function (lower panel) using top blastx hit against D. melanogaster and Panther v14.0. Lower level GO terms were selected to illustrate sensory perception-related functions present in the assembly.
Supplementary Figure 2 | Phylogeny of P. hospiton maxilla-expressed sensory neuron membrane proteins (SNMPs). A maximum-likelihood tree was built with selected SNMP sequences from Zhang et al. (2020) using RAxML. Branch lengths are scaled by the number of nucleotide substitutions per site. Dple Danaus plexippus; Bmor Bombyx mori, Harm Helicoverpa armigera; Sexi Spodoptera exigua.
Supplementary Figure 3 | Phylogeny of P. hospiton maxilla-expressed odorant-binding proteins (OBPs). A maximum-likelihood tree was built with selected OBP sequences from Vogt et al. (2015) using RAxML. Branch lengths are scaled by the number of nucleotide substitutions per site. Bootstrap values refer to 500 pseudo-replicates.
Supplementary Figure 4 | Supplementary functional studies on TRP-Painless. Mean ± SE of fluorescence variation associated with PhospPain and PmachPain upon testing different Petri dishes to a set of stimulus, including cold (T ∼ 9–10°C; PhospPain, N = 110; PmachPain, N = 108), hydrogen peroxide (H2O2; PhospPain, N = 88; PmachPain, N = 111) and an essential oil extracted from Ruta graveolens diluted in Ringer buffer at 150 μg/mL (PhospPain, N = 92; PmachPain, N = 94). The existence of a similar effect on un-transfected HEK293A cells recorded upon the cold stimulation suggested a possible involvement of other endogenous mechanisms, most probably based on the effects of cold-associated Calcium release from the endoplasmic reticulum (Reddish et al., 2021). 3.0% H2O2 was the maximum concentration used. Below: negative control testing untransfected HEK293A cells with cold (N = 80). Black bars: stimulus (20 s).
Supplementary Table 1 | List of primers to amplify Papilio OR1 and Pain.
Supplementary Table 2 | De novo assembly statistics.
Supplementary Table 3 | Cell percentages based on fluorescent features. Percentages of fluorescent cells were based on EBFP expression (EBFP+) and fluorescence induced by 500 μM VUAA1 application (Fluo4+) from different HEK293A-samples transfected with CpomOrco and CpomOrco+OR (PmachOR1, PhospOR1, and CpomOR3). For CpomOrco+OR3, plot of the mean response to 1000 μM pear ester (N = 134; 26,02%) is shown in Figure 5B. Data from previous analysis conducted in the frame of parallel experiments are reported for the wild-type CpomOrco, the mutagenized CpomOrco (CpomOrcoQ417H), and their heteromers with CpomOR6a (Bobkov et al., 2021). Data from the TRP-study conducted in this work are reported, comparing experiments between preparations of PhospPain and PmachPains tested to AITC, and of dTRPA1(B) tested to heat. Stimulus (AITC or heat) is indicated in brackets.
Supplementary Dataset 1 | Nucleotide sequences of contigs identified as candidate chemosensory receptor transcripts.
References
Abuin, L., Prieto-Godino, L. L., Pan, H., Gutierrez, C., Huang, L., Jin, R., et al. (2019). In vivo assembly and trafficking of olfactory Ionotropic Receptors. BMC Biol. 17:34. doi: 10.1186/s12915-019-0651-7
Agnihotri, A. R., Roy, A. A., and Joshi, R. S. (2016). Gustatory receptors in Lepidoptera: chemosensation and beyond. Insect Mol. Biol. 25, 519–529. doi: 10.1111/imb.12246
Ahn, J.-E., Chen, Y., and Amrein, H. (2017). Molecular basis of fatty acid taste in Drosophila. eLife 6:e30115. doi: 10.7554/eLife.30115
Ai, H. W., Shaner, N. C., Cheng, Z., Tsien, R. Y., and Campbell, R. E. (2007). Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry 46, 5904–5910. doi: 10.1021/bi700199g
Al-Anzi, B., Tracey, W. D., and Benzer, S. (2006). Response of Drosophila to wasabi is mediated by painless, the fly homolog of mammalian TRPA1/ANKTM1. Curr. Biol. 16, 1034–1040. doi: 10.1016/j.cub.2006.04.002
Bengtsson, J. M., Trona, F., Montagne, N., Anfora, G., Ignell, R., Witzgall, P., et al. (2012). Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS One 7:e31620. doi: 10.1371/journal.pone.0031620
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009) Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Bobkov, Y. V., Walker, W. B. III, and Cattaneo, A. M. (2021). Altered functional properties of the codling moth Orco mutagenized in the intracellular loop-3. Sci. Rep. 11:3893. doi: 10.1038/s4
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Briscoe, A. D., Macias-Muñoz, A., Kozak, K. M., Walters, J. R., Yuan, F., Jamie, G. A., et al. (2013). Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genetics 9:e1003620. doi: 10.1371/journal.pgen.1003620
Bryant, D. M., Johnson, K., DiTommaso, T., Tickle, T., Couger, M. B., Payzin-Dogru, D., et al. (2017). A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 18, 762–776. doi: 10.1016/j.celrep.2016.12.063
Cassau, S., and Krieger, J. (2021). The role of SNMPs in insect olfaction. Cell Tissue Res. 383, 21–33. doi: 10.1007/s00441-020-03336-0
Cattaneo, A. M., Gonzalez, F., Bengtsson, J. M., Corey, E. A., Jacquin-Joly, E., Montagné, N., et al. (2017a). Candidate pheromone receptors of codling moth Cydia pomonella respond to pheromones and kairomones. Sci. Rep. 7:41105.
Cattaneo, A. M., Bobkov, Y. V., Corey, E. A., Borgonovo, G., and Bassoli, A. (2017b). Perilla derived compounds mediate human TRPA1 channel activity. Med. Aromat Plants (Los Angel) 6:1. doi: 10.4172/2167-0412.1000283
Chen, Y., and Amrein, H. (2017). Ionotropic receptors mediate Drosophila oviposition preference through sour gustatory receptor neurons. Curr. Biol. 27, 2741–2750.e4. doi: 10.1016/j.cub.2017.08.003.
Croset, V., Schleyer, M., Arguello, J. R., Gerber, B., and Benton, R. (2016). A molecular and neuronal basis for amino acid sensing in the Drosophila larva. Sci. Rep. 6:34871. doi: 10.1038/srep34871
de Fouchier, A., Walker, W. B. III, Montagné, N., Steiner, C., Binyameen, M., Schlyter, F., et al. (2017). Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat. Comm. 8:15709. doi: 10.1038/ncomms15709
Dethier, V. G., and Schoonhoven, L. M. (1969). Olfactory coding by lepidopterous larvae. Entomol. Exp. Appl. 12, 535–543. doi: 10.1111/j.1570-7458.1969.tb02551.x
Di, C., Ning, C., Huang, L.-Q., and Wang, C.-Z. (2017). Design of larval chemical attractants based on odorant response spectra of odorant receptors in the cotton bollworm. Insect Biochem. Mol. Biol. 84, 48–62. doi: 10.1016/j.ibmb.2017.03.007
Fagot, J. (1996). Papilio machaon L. (Papilionidae) observed on Ruta graveolens L. (Rutaceae) at Gembloux. development of his diet. Nat. Mosana 49, 32–39.
Fowler, M. A., and Montell, C. (2013). Drosophila TRP channels and animal behavior. Life Sci. 92, 394–403. doi: 10.1016/j.lfs.2012.07.029
Ganguly, A., Pang, L., Duong, V.-K., Lee, A., Schoniger, H., Varady, E., et al. (2017). A molecular and cellular context-dependent role for Ir76b in detection of amino acid taste. Cell Rep. 18, 737–750. doi: 10.1016/j.celrep.2016.12.071
Garczynski, S. F., Martin, J. A., Griset, M., Willett, L. S., Cooper, W. R., Swisher, K. D., et al. (2017). CRISPR/Cas9 editing of the codling moth (Lepidoptera: Tortricidae) CpomOR1 gene affects egg production and viability. J. Econ. Entomol. 110, 1847–1855. doi: 10.1093/jee/tox166
Gonzalez, F., Witzgall, P., Walker, W. B., and Iii. (2016). Protocol for heterologous expression of insect odourant receptors in Drosophila. Front. Ecol. 4:24. doi: 10.3389/fevo.2016.00024
Gouin, A., Bretaudeau, A., Nam, K., Gimenez, S., Aury, J.-M., Duvic, B., et al. (2017). Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci. Rep. 7:11816. doi: 10.1038/s41598-017-10461-4
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., and Amit, I. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 29, 644–652. doi: 10.1038/nbt.1883
Guo, H., Cheng, T., Chen, Z., Jiang, L., Guo, Y., and Liu, J. (2017). Expression map of a complete set of gustatory receptor genes in chemosensory organs of Bombyx mori. Insect Biochem. Mol. Biol. 82, 74–82. doi: 10.1016/j.ibmb.2017.02.001
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Series 41, 95–98.
Himmel, N. J., and Cox, D. N. (2020). Transient receptor potential channels: current perspectives on evolution, structure, function and nomenclature. Proc. R. Soc. B. 287:20201309. doi: 10.1098/rspb.2020.1309
Hiroi, M., Tanimura, T., and Marion-Poll, F. (2008). Hedonic taste in Drosophila revealed by olfactory receptors expressed in taste neurons. PLoS One 3:e2610. doi: 10.1371/journal.pone.0002610
Hofmann, T., Chubanov, V., Chen, X., Dietz, A. S., Gudermann, T., and Montell, C. (2010). Drosophila TRPM channel Is essential for the control of extracellular magnesium levels. PLoS One 5:e10519. doi: 10.1371/journal.pone.0010519
Hussain, A., Zhang, M., Üçpunar, H. K., Svensson, T., Quillery, E., Gompel, N., et al. (2016). Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol. 14:e1002454. doi: 10.1371/journal.pbio.1002454
Jones, P. L., Pask, G. M., Rinkerb, D. C., and Zwiebel, L. J. (2011). Functional agonism of insect odorant receptor ion channels. PNAS 108, 8821–8825. doi: 10.1073/pnas.1102425108
Jörs, S., Kazanski, V., Foik, A., Krautwurst, D., and Harteneck, C. (2006). Receptor-induced activation of Drosophila TRP gamma by polyunsaturated fatty acids. J. Biol. Chem. 281, 29693–29702. doi: 10.1074/jbc.M602215200
Joseph, R. M., and Carlson, J. M. (2015). Drosophila chemoreceptors: a molecular interface between the chemical world and the brain. Trends Genet. 31, 683–695. doi: 10.1016/j.tig.2015.09.005
Kang, K., Panzano, V. C., Chang, E. C., Ni, L., Dainis, A. M., Jenkins, A. M., et al. (2012). Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature 481, 76–80. doi: 10.1038/nature10715
Kang, K., Pulver, S. R., Panzano, V. C., Chang, E. C., Griffith, L. C., Theobald, D. L., et al. (2010). Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature 464, 597–600. doi: 10.1038/nature08848
Koh, T. W., He, Z., Gorur-Shandilya, S., Menuz, K., Larter, N. K., Stewart, S., et al. (2014). The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865. doi: 10.1016/j.neuron.2014.07.012
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. E 35, 1547–1549. doi: 10.1093/molbev/msy096
Kwon, Y., Kim, S. H., Ronderos, D. S., Lee, Y., Akitake, B., Woodward, O. M., et al. (2010). Drosophila TRPA1 channel is required to avoid the naturally occurring insect repellent citronellal. Curr. Biol. 20, 1672–1678. doi: 10.1016/j.cub.2010.08.016
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Laue, M. (2000). Immunolocalization of general odorant-binding protein in antennal sensilla of moth caterpillars. Arthropod Struct. Dev. 29, 57–73. doi: 10.1016/s1467-8039(00)00013-x
Lee, Y., Lee, Y., Lee, J., Bang, S., Hyun, S., Kang, J., et al. (2005). Pyrexia is a new thermal transient receptor potential channel endowing tolerance to high temperatures in Drosophila melanogaster. Nat. Genet. 37, 305–310. doi: 10.1038/ng1513
Li, B., and Dewey, C. N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323.
Li, X., Fan, D., Zhang, W., Liu, G., Zhang, L., Zhao, L., et al. (2015). Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat. Comm. 6:8212. doi: 10.1038/ncomms9212
Liu, L., Li, Y., Wang, R., Yin, C., Dong, Q., Hing, H., et al. (2007). Drosophila hygrosensation requires the TRP channels water witch and nanchung. Nature 450, 294–298. doi: 10.1038/nature06223
Liu, N.-Y., Xu, W., Dong, S.-L., Zhu, J.-Y., Xu, Y.-X., and Anderson, A. (2018). Genome-wide analysis of ionotropic receptor gene repertoire in Lepidoptera with an emphasis on its functions of Helicoverpa armigera. Insect Biochem. Mol. Biol. 99, 37–53. doi: 10.1016/j.ibmb.2018.05.005
Macias-Muñoz, A., Rangel Olguin, A. G., and Briscoe, A. D. (2019). Evolution of phototransduction genes in Lepidoptera. Gen Biol. Evol. 11, 2107–2124. doi: 10.1093/gbe/evz150
Malik, B., Elkaddi, N., Turkistani, J., Spielman, A. I., and Ozdener, M. H. (2019). Mammalian taste cells express functional olfactory receptors. Chem. Senses 44, 289–301. doi: 10.1093/chemse/bjz019
Marygold, S. J., Leyland, P. C., Seal, R. L., Goodman, J. L., Thurmond, J., Strelets, V. B., et al. (2013). FlyBase: improvements to the bibliography. Nucleic Acids Res. 41, D751–D757. doi: 10.1093/nar/gks1024
Matsuura, H., Sokabe, T., Kohno, K., Tominaga, M., and Kadowaki, T. (2009). Evolutionary conservation and changes in insect TRP channels. BMC EBiol. 9:228. doi: 10.1186/1471-2148-9-228
Mi, H., Muruganujan, A., Huang, X., Ebert, D., Mills, C., Guo, X., et al. (2019). Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Prot. 14, 703–721. doi: 10.1038/s41596-019-0128-8
Montagné, N., De Fouchier, A., Newcomb, R. D., and Jacquin-Joly, E. (2015). Advances in the identification and characterization of olfactory receptors in insects. Prog. Mol. Biol. Transl. Sci. 130, 55–80. doi: 10.1016/bs.pmbts.2014.11.003
Montell, C. (2021). Drosophila sensory receptors - a set of molecular Swiss Army Knives. Genetics 217, 1–34. doi: 10.1093/genetics/iyaa011
Nakagawa, T., Sakurai, T., Nishioka, T., and Touhara, K. (2005). Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307, 1638–1642. doi: 10.1126/science.1106267
O’Neil, S. T., Dzurisin, J. D. K., Carmichael, R. D., Lobo, N. F., Emrich, S. J., and Hellmann, J. J. (2010). Population-level transcriptome sequencing of nonmodel organisms Erynnis propertius and Papilio zelicaon. BMC Genomics 11:310. doi: 10.1186/1471-2164-11-310
Ong, H. L., de Souza, L. B., Zheng, C., Cheng, K. T., Liu, X., Goldsmith, C., et al. (2015). STIM2 enhances receptor-stimulated Ca2+ signaling by promoting recruitment of STIM1 to the endoplasmic reticulum-plasma membrane junctions. Sci. Signal 8:ra3. doi: 10.1126/scisignal.2005748
Poivet, E., Gallot, A., Montagné, N., Glaser, N., Legeai, F., and Jacquin-Joly, E. (2013). A comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis. PLoS One 8:e60263. doi: 10.1371/journal.pone.0060263
Rahali, F. Z., Kefi, S., Rebey, I. B., Hamdaoui, G., Tabart, J., Kevers, C., et al. (2018). Phytochemical composition and antioxidant activities of different aerial parts extracts of Ferula communis L. Plant Biosyst. 153, 213–221. doi: 10.1080/11263504.2018.1461696
Rana, S., and Mohankumar, S. (2017). Comparison of sensory structures present on larval antennae and mouthparts of Lepidopteran crop pests. Florida Entomol. 100, 230–250. doi: 10.1653/024.100.0217
Reddish, F. N., Miller, C. L., Deng, X., Dong, B., Patel, A. A., Ghane, M. A., et al. (2021). Rapid subcellular calcium responses and dynamics by calcium sensor G-CatchER+. iScience 24:102129. doi: 10.1016/j.isci.2021.102129
Roberts, R. E., Yuvaraj, J. K., and Andersson, M. N. (2021). Codon optimization of insect odorant receptor genes may increase their stable expression for functional characterization in HEK293 cells. Front. Cell. Neurosci. 15:744401. doi: 10.3389/fncel.2021.744401
Roessingh, P., Xu, S., and Menken, S. B. J. (2007). Olfactory receptors on the maxillary palps of small ermine moth larvae: evolutionary history of benzaldehyde sensitivity. J. Comp.Physiol. A. 193, 635–647. doi: 10.1007/s00359-007-0218-x
Rosenzweig, M., Kang, K., and Garrity, P. A. (2008). Distinct TRP channels are required for warm and cool avoidance in Drosophila melanogaster. PNAS 105, 14668–14673. doi: 10.1073/pnas.0805041105
Sánchez-Alcañiz, J. A., Silbering, A. F., Croset, V., Zappia, G., Sivasubramaniam, A. K., Abuin, L., et al. (2018). An expression atlas of variant ionotropic glutamate receptors identifies a molecular basis of carbonation sensing. Nat. Comm. 9:4252. doi: 10.1038/s41467-018-06453-1
Schoonhoven, L. M., and van Loon, J. J. A. (2002). An inventory of taste in caterpillars: each species its own key. Act. Zool. Acad. Sci. Hung. 48, 215–263.
Seppey, M., Manni, M., and Zdobnov, E. M. (2019). BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245. doi: 10.1007/978-1-4939-9173-0_14
Shalygin, A., Skopin, A., Kalinina, V., Zimina, O., Glushankova, L., Mozhayeva, G. N., et al. (2015). STIM1 and STIM2 proteins differently regulate endogenous store-operated channels in HEK293 cells. J. Biol. Chem. 290, 4717–4727. doi: 10.1074/jbc.M114.601856
Silbering, A. F., Rytz, R., Grosjean, Y., Abuin, L., Ramdya, P., Jefferis, G. S. X. E., et al. (2011). Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 31, 13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011
Sokabe, T., and Tominaga, M. (2009). A temperature-sensitive TRP ion channel, painless, functions as a noxious heat sensor in fruit flies. Comm. Integr. Biol. 2, 170–173. doi: 10.4161/cib.7708
Sokabe, T., Tsujiuchi, S., Kadowaki, T., and Tominaga, M. (2008). Drosophila Painless is a Ca2+-requiring channel activated by noxious heat. J. Neurosci. 28, 9929–9938. doi: 10.1523/JNEUROSCI.2757-08.2008
Solari, P., Cerboneschi, A., Masala, C., Crnjar, R., and Liscia, A. (2002). Chemoreception in larvae of the moth Lymantria dispar. Italian J. Zool. 69, 305–312. doi: 10.1080/11250000209356474
Sollai, G., and Crnjar, R. (2019). The contribution of gustatory input to larval acceptance and female oviposition choice of potential host plants in Papilio hospiton (Géné). Arch. Insect Biochem. Physiol. 100:e21521. doi: 10.1002/arch.21521
Sollai, G., Barbarossa, I. T., Masala, C., Solari, P., and Crnjar, R. (2014). Gustatory sensitivity and food acceptance in two phylogenetically closely related Papilionid species: Papilio hospiton and Papilio machaon. PLoS One 9:e100675. doi: 10.1371/journal.pone.0100675
Sollai, G., Biolchini, M., and Crnjar, R. (2018a). Taste receptor plasticity in relation to feeding history in two congeneric species of Papilionidae (Lepidoptera). J. Ins. Physiol. 107, 41–56. doi: 10.1016/j.jinsphys.2018.02.007
Sollai, G., Biolchini, M., and Crnjar, R. (2018b). Taste sensitivity and divergence in host plant acceptance between adult females and larvae of Papilio hospiton. Insect Sci. 25, 809–822. doi: 10.1111/1744-7917.12581
Sollai, G., Biolchini, M., Solari, P., and Crnjar, R. (2017). Chemosensory basis of larval performance of Papilio hospiton on different host plants. J. Insect Physiol. 99, 47–57. doi: 10.1016/j.jinsphys.2017.02.007
Sollai, G., Tomassini Barbarossa, I., Solari, P., and Crnjar, R. (2015). Taste discrimination capability to different bitter compounds by the larval styloconic sensilla in the insect herbivore Papilio hospiton (Gené). J. Ins. Physiol. 74, 45–55. doi: 10.1016/j.jinsphys.2015.02.004
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stewart, S., Koh, T. W., Ghosh, A. C., and Carlson, J. R. (2015). Candidate ionotropic taste receptors in the Drosophila larva. Proc. Natl. Acad. Sci. U S A. 112, 4195–4201. doi: 10.1073/pnas.1503292112
Sun, Y., Liu, L., Ben-Shahar, Y., Jacobs, J. S., Eberl, D. F., and Welsh, M. J. (2009). TRPA channels distinguish gravity sensing from hearing in Johnston’s organ. PNAS 106, 13606–13611. doi: 10.1073/pnas.0906377106
Tanaka, K., Uda, Y., Ono, Y., Nakagawa, T., Suwa, M., Yamaoka, R., et al. (2009). Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr. Biol. 19, 881–890. doi: 10.1016/j.cub.2009.04.035
Tauber, J. M., Brown, E. B., Li, Y., Yurgel, M. E., Masek, P., and Keene, A. C. (2017). A subset of sweet-sensing neurons identified by IR56d are necessary and sufficient for fatty acid taste. PLoS Genet 13:e1007059. doi: 10.1371/journal.pgen.1007059
Tracey, W. D. Jr., Wilson, R. I., Laurent, G., and Benzer, S. (2003). Painless, a Drosophila gene essential for nociception. Cell 113, 261–273. doi: 10.1016/s0092-8674(03)00272-1
Tsirigos, K. D., Peters, C., Shu, N., Käll, L., and Elofsson, A. (2015). The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407. doi: 10.1093/nar/gkv485
Tsuneto, K., Endo, H., Shii, F., Sasaki, K., Nagata, S., and Sato, R. (2020). Diet choice: the two-factor host acceptance system of silkworm larvae. PLoS Biol. 18:e3000828. doi: 10.1371/journal.pbio.3000828
Vogt, R. G., Grosse-Wilde, E., and Zhou, J.-J. (2015). The Lepidoptera odorant binding protein gene family: gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 62, 142–153. doi: 10.1016/j.ibmb.2015.03.003
Vosshall, L. B., and Stocker, R. F. (2007). Molecular architecture of smell and taste in Drosophila. Ann. Reviof Neurosci. 30, 505–533. doi: 10.1146/annurev.neuro.30.051606.094306
Walker, W. B. III, Roy, A., Anderson, P., Schlyter, F., Hansson, B. S., and Larsson, M. C. (2019). Transcriptome analysis of gene families involved in chemosensory function in Spodoptera littoralis (Lepidoptera: Noctuidae). BMC Genomics 20:428. doi: 10.1186/s12864-019-5815-x
Wei, J. J., Fu, T., Yang, T., Liu, Y., and Wang, G. R. (2015). A TRPA1 channel that senses thermal stimulus and irritating chemicals in Helicoverpa armigera. Insect Mol. Biol. 24, 412–421. doi: 10.1111/imb.12168
Wittstock, U., Kliebenstein, D. J., Lambrix, V., Reichelt, M., and Gershenzon, J. (2003). “Glucosinolate hydrolysis and its impact on generalist and specialist insect herbivores,” in Integrative Phytochemistry: From Ethnobotany To Molecular Ecology. 1. 8, ed. J. T. Romeo (Amsterdam: Elsevier), doi: 10.1016/S0079-9920(03)80020-5
Xiao, B., Coste, B., Mathur, J., and Patapoutian, A. (2011). Temperature-dependent STIM1 activation induces Ca2+ influx and modulates gene expression. Nat. Chem. Biol. 6, 351–358. doi: 10.1038/nchembio.558
Xu, J., Sornborger, A. T., Lee, J. K., and Shen, P. (2008). Drosophila TRPA channel modulates sugar-stimulated neural excitation, avoidance and social response. Nat. Neurosci. 11, 676–682. doi: 10.1038/nn.2119
Yin, N. N., Nuo, S. M., Xiao, H.-Y., Zhao, Y.-J., Zhu, J.-Y., and Liu, N.-Y. (2021). The ionotropic receptor gene family in Lepidoptera and Trichoptera: annotation, evolutionary and functional perspectives. Genomics 113, 601–612. doi: 10.1016/j.ygeno.2020.09.056
Zhang, H. J., Xu, W., Chen, Q. M., Sun, L. N., Anderson, A., Xia, Q. Y., et al. (2020). A phylogenomics approach to characterizing sensory neuron membrane proteins (SNMPs) in Lepidoptera. Insect Biochem. Mol. Biol. 118:103313. doi: 10.1016/j.ibmb.2020.103313
Zhang, Y. V., Ni, J., and Montell, C. (2013a). The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338. doi: 10.1126/science.1234133
Keywords: papilionid butterflies, larval maxilla, RNA-seq analysis, odorant receptors, transient receptor potential (TRP) channels, heterologous expression, human embryonic kidney cells
Citation: Crava CM, Bobkov YV, Sollai G, Anfora G, Crnjar R and Cattaneo AM (2022) Chemosensory Receptors in the Larval Maxilla of Papilio hospiton. Front. Ecol. Evol. 9:795994. doi: 10.3389/fevo.2021.795994
Received: 15 October 2021; Accepted: 30 December 2021;
Published: 03 February 2022.
Edited by:
Wayne Iwan Lee Davies, Umeå University, SwedenReviewed by:
Nicolas Montagné, Sorbonne Universités, FranceGabriele Rondoni, University of Perugia, Italy
Copyright © 2022 Crava, Bobkov, Sollai, Anfora, Crnjar and Cattaneo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alberto Maria Cattaneo, albertomaria.cattaneo@gmail.com
†These authors jointly supervised this work
 Cristina M. Crava
Cristina M. Crava Yuriy V. Bobkov
Yuriy V. Bobkov Giorgia Sollai
Giorgia Sollai Gianfranco Anfora
Gianfranco Anfora Roberto Crnjar
Roberto Crnjar Alberto Maria Cattaneo
Alberto Maria Cattaneo