It takes Two: Discovery of Spider Pax2 Duplicates Indicates Prominent Role in Chelicerate Central Nervous System, Eye, as Well as External Sense Organ Precursor Formation and Diversification After Neo- and Subfunctionalization
- 1Evolutionary Ecology, Faculty of Biology, Ludwig-Maximilians-University Munich, Planegg-Martinsried, Germany
- 2School of Biological and Chemical Sciences, Queen Mary University of London, London, United Kingdom
Paired box genes are conserved across animals and encode transcription factors playing key roles in development, especially neurogenesis. Pax6 is a chief example for functional conservation required for eye development in most bilaterian lineages except chelicerates. Pax6 is ancestrally linked and was shown to have interchangeable functions with Pax2. Drosophila melanogaster Pax2 plays an important role in the development of sensory hairs across the whole body. In addition, it is required for the differentiation of compound eyes, making it a prime candidate to study the genetic basis of arthropod sense organ development and diversification, as well as the role of Pax genes in eye development. Interestingly, in previous studies identification of chelicerate Pax2 was either neglected or failed. Here we report the expression of two Pax2 orthologs in the common house spider Parasteatoda tepidariorum, a model organism for chelicerate development. The two Pax2 orthologs most likely arose as a consequence of a whole genome duplication in the last common ancestor of spiders and scorpions. Pax2.1 is expressed in the peripheral nervous system, including developing lateral eyes and external sensilla, as well as the ventral neuroectoderm of P. tepidariorum embryos. This not only hints at a conserved dual role of Pax2/5/8 orthologs in arthropod sense organ development but suggests that in chelicerates, Pax2 could have acquired the role usually played by Pax6. For the other paralog, Pt-Pax2.2, expression was detected in the brain, but not in the lateral eyes and the expression pattern associated with sensory hairs differs in timing, pattern, and strength. To achieve a broader phylogenetic sampling, we also studied the expression of both Pax2 genes in the haplogyne cellar spider Pholcus phalangioides. We found that the expression difference between paralogs is even more extreme in this species, since Pp-Pax2.2 shows an interesting expression pattern in the ventral neuroectoderm while the expression in the prosomal appendages is strictly mesodermal. This expression divergence indicates both sub- and neofunctionalization after Pax2 duplication in spiders and thus presents an opportunity to study the evolution of functional divergence after gene duplication and its impact on sense organ diversification.
Introduction
Paired box (Pax) family transcription factors are among the oldest genetic toolkit components required for animal development, as they were likely already present in the last common ancestor of metazoans (Hoshiyama et al., 1998). Among their diverse functions, Pax genes are best known for their involvement in neurogenesis. Many studies have confirmed this role to be well conserved across metazoans (Kozmik et al., 2003; Kozmik, 2005; Scherholz et al., 2017). Seminal work in this area suggested that the Pax gene Pax6 is a key player in eye development in both vertebrates and insects (Quiring et al., 1994). Moreover, the high sequence conservation between these distant groups was pointedly proven when murine Pax6 was shown to be able to induce ectopic eye formation in the fruit fly Drosophila melanogaster (Halder et al., 1995). While these findings suggest that Pax6 was involved in sense organ development in the last common ancestor of bilaterians, the subsequent discovery of a Pax6-related gene expressed in the developing eyes of a cubozoan jellyfish points to an even deeper conservation (Kozmik et al., 2003). Indeed, phylogenetic studies suggest that all extant Pax genes evolved by duplication from a single Ur-Pax gene, whose singular descendent PaxB is still present in poriferans (Hill et al., 2010). Successive duplication events of PaxB then gave rise to the diversity of Pax genes present in cnidarians and bilaterians today. Current phylogenetic analyses group bilaterian Pax genes into seven main subfamilies: Pax1/9, Pax2/5/8, Pax3/7, Pax4/6/10, Pox neuro, Pax eyegone, and Paxa/β (Hill et al., 2010; Franke et al., 2015; Friedrich, 2015). They are all characterized by the presence of a unique N-terminal sequence, encoding the DNA-binding Paired domain (PD) (Bopp et al., 1986). Two other motifs, the homeodomain and octapeptide sequence, are exclusive to certain Pax gene subfamilies and thus serve as criteria for their classification. The C-terminal homeodomain acts as a second DNA-binding domain in Pax3/7 and Pax4/6 group genes (Frigerio et al., 1986). Severely truncated remnants of the homeodomain are also observed in some Pax2/5/8 group genes (Bopp et al., 1986; Plachov et al., 1990; Fu and Noll, 1997). Finally, the octapeptide sequence, a highly conserved stretch of eight amino acids of unknown function, is located in between the two binding domains of Pax1/9, Pax2/5/8, and Pax3/7 group genes (Burri et al., 1989; Dressler et al., 1990).
Previous studies showed that the two subfamilies Pax6 and Pax2/5/8 are closely related and have a shared ancestry to cnidarian PaxB (Hill et al., 2010). This close phylogenetic relationship between Pax6 and Pax2 is also reflected in their role during development, especially regarding a functionally conserved transcriptional regulation of crystallin genes required for eye development (recently discussed in Charlton-Perkins et al., 2021). A wealth of functional information on Pax2 and Pax6 stems from studies on vertebrates, where both genes were duplicated multiple times, giving rise to Pax5 and Pax8 (originating from Pax2) and Pax4 and Pax10 (duplicates of Pax6) (Wada et al., 1998; Feiner et al., 2014). Together, the vertebrate Pax2/5/8 genes are crucial for eye development, forming the optic stalk and enabling generation of glial cells along the optic nerve tract (Torres et al., 1996). Unlike Pax6, however, Pax2/5/8 group genes appear to be related to sense organs in general, as they are also implicated in development of the inner ear, where they control growth and morphology of the cochlea (Burton et al., 2004). Surprisingly, apart from their role in sense organ development, Pax2/5/8 genes also take part in forming the vertebrate excretory system. The ureters as well as the genital tract are dependent on Pax2 for their formation and as a result, Pax2-deficient mice fail to form kidneys altogether (Torres et al., 1995). Data from lophotrochozoan species support a conserved function of Pax2 in sense organ development. In the marine gastropod Haliotis asinina, Pax2 is expressed in the geosensory statocysts and the (adult) eyes (O’brien and Degnan, 2003). Similarly, the chiton Acanthochitona crinita shows expression in larval chemosensory organs, while in the cephalopod Idiosepius notoides, expression is found in the CNS and optic ganglia during development (Wollesen et al., 2015). In contrast, there are no peer-reviewed studies indicating a function or at least expression of Pax2 in the excretory system of any lophotrochozoan species.
Given the importance of Pax2 for sense organ formation in vertebrates, mollusks and even cnidarians and poriferans, data on its expression during arthropod development is surprisingly scarce and almost entirely exclusive to insects, especially D. melanogaster. In fruit fly embryos, Pax2 is expressed in the developing CNS (Hirth et al., 2003). Pax2 gene function during late embryogenesis was studied using the two mutant lines sparkling and shaven (sv), named in accordance with their phenotypes, as these lines carry mutations in the enhancer regions of the fruit fly Pax2 gene, which modularly control expression in two different tissues (Fu et al., 1998). During larval and pupal development, Pax2 is expressed in the eye imaginal discs, where it controls development of the cone and primary pigment cells of the ommatidia (Fu and Noll, 1997; Charlton-Perkins et al., 2011). Additionally, a spot-like epithelial expression of Pax2 during pupal development is related to sensory hair formation, in particular to the formation of the shaft bearing (trichogen) and glial sheath (thecogen) cell (Kavaler et al., 1999). Thus, functional studies of the Pax2 ortholog in D. melanogaster confirm a conserved dual role of Pax2 in the eye as well as external sense organ development. To date, there is only a single study reporting the developmental expression pattern of Pax2 in another arthropod, the parasitoid wasp Nasonia vitripennis (Keller et al., 2010). In late embryos of N. vitripennis, Pax2 is expressed anteriorly, in tissues that the authors speculate to be presumptive head structures. However, since neither larval nor pupal expression patterns were included in the publication, this statement remains hypothetical. The iBeetle-Base, an online database for RNAi-induced phenotypes in the red flour beetle Tribolium castaneum, reports larval bristles to be missing with a penetrance of over 80% following pupal injection of Pax2 RNAi-constructs (TC003570) into the mother (Dönitz et al., 2015). Unfortunately, developmental expression patterns for T. castaneum Pax2 are not available. A tissue-specific transcriptome search in developmental stages of the ostracod crustacean Euphilomedes carcharodonta revealed expression of Pax2 in developing eyes (Sajuthi et al., 2015). To our knowledge, this is the only publication reporting expression of Pax2 in a non-insect arthropod to date. Taken together, these results tentatively point to a possible functional conservation of Pax2 across arthropod CNS, sense hair and eye development, but not excretory system formation. While studies from non-insect arthropods are lacking to strengthen this hypothesis, there is information on embryonic Pax2 expression in tardigrades and velvet worms, which represent the closest living relatives to arthropods. In early embryos of the tardigrade Hypsibius exemplaris, Pax2 is expressed in the trunk ganglia and legs (Smith et al., 2018). During embryonic development of the velvet worm Euperipatoides rowelli, however, Pax2 is expressed along the ventrolateral ectoderm and later adjacent to the walking legs in a segmental fashion, which the authors interpret as nephridial openings (Franke et al., 2015). In summary, data from arthropods and sister groups suggest a role of Pax2 in the developing CNS and PNS, including eyes and sensory hairs. However, a potentially conserved role of Pax2 in the excretory system for most bilaterians remains to be investigated. Altogether, the sparse availability of data from arthropods, especially from non-insect species, impedes a definitive statement on how Pax2 function evolved in this phylum.
This is in stark contrast to other key developmental genes like Pax6, which has been intensively studied in a range of arthropods, including beetles, springtails, millipedes, spiders, and horseshoe crabs (Prpic, 2005; Blackburn et al., 2008; Yang et al., 2009; Luan et al., 2014; Samadi et al., 2015; Schomburg et al., 2015; Hou et al., 2016). As a member of the Chelicerata, the sister group to all other arthropods, spiders are particularly interesting in this regard. Moreover, they also evolved several unique morphological innovations, like silk glands, spinnerets, the patella, and several novel sense organs, like lyriform organs and trichobothria, which are in turn closely linked to key behavioral innovations like silk production, stridulation, web building, and vibrational communication. Interestingly, a previous genomic study found evidence for a whole genome duplication event in the lineage leading to arachnopulmonates, the group encompassing spiders and scorpions (Schwager et al., 2017). Gene duplications are predicted to fuel evolution by providing gene copies serving as new genetic material, which can then evolve via sub- or neofunctionalization. It is thus of particular interest to study the evolution of Pax family genes in a spider, to first observe whether paralogs are present and, if so, how they diverged from their original functions. With the establishment of the cobweb spider Parasteatoda tepidariorum as a model system for chelicerate development over the last years (Hilbrant et al., 2012), Pax2 should be an obvious candidate for expression analysis.
Unfortunately, several publications on the expression of key developmental genes neglected or failed the identification of Pax2 in P. tepidariorum. Although there are in-depth studies of spider eye development, both in P. tepidariorum and the large Mexican wandering spider Cupiennius salei, which included expression patterns of major eye transcription factors such as Pax6, Pax2 was left out entirely (Samadi et al., 2015; Schomburg et al., 2015). Even a large-scale transcriptomic search of homeobox-containing transcription factors across chelicerates, covering spiders, scorpions, pseudoscorpions, a harvestman and a tick, did not identify a Pax2 ortholog in any of the included species (Leite et al., 2018). The most likely explanation for this is that Pax2 orthologs escaped previous identification attempts because of their severely truncated or completely absent homeodomain, a typical feature of Pax2 genes (Bopp et al., 1986; Dressler et al., 1990; Plachov et al., 1990). Pax2 expression in chelicerates is thus currently entirely unknown, despite its significance for arthropod sense organ evolution and development. Its dual role in eye and external sense organ development, mainly studied in the fruit fly, makes it an even more interesting candidate to analyze in different spider species, especially since both eyes and sense organs show a great diversification in spiders in terms of their number, function, and distribution along the body (Foelix, 1996; Barth, 2004; Barth and Stagl, 2004; Ganske and Uhl, 2018; Schacht et al., 2020).
To fill this gap, we searched for Pax2 genes in two spider species from different groups, the entelegyne common house spider P. tepidariorum, and the haplogyne cellar spider Pholcus phalangioides. Pax2 is present as two paralogs in these species, which is, as suggested by phylogenetic analyses an ancestral feature of spiders and scorpions. We analyzed the expression of these two Pax2 paralogs during embryonic development in both spiders. Pax2.1 and Pax2.2 are both expressed in the ventral neuroectoderm (VNE), as well as in the embryonic appendages, where they are likely associated with developing sensory hairs. Expression in embryonic appendages differs between the two paralogs, with Pax2.1 showing a stronger and earlier expression than Pax2.2 in P. tepidariorum. Expression differences between the two paralogs are even more obvious in P. phalangioides. While Pp-Pax2.1 is ubiquitously expressed in the early developing appendages, before eventually becoming restricted to the characteristic spot-like pattern similar as in P. tepidariorum, Pp-Pax2.2 expression in appendages is restricted to the mesoderm. Furthermore, in both species Pax2.1, but not Pax2.2, is expressed in the developing lateral eyes, which are most likely homologous to the compound eyes of insects present in D. melanogaster (Paulus, 1979; Spreitzer and Melzer, 2003; Schomburg et al., 2015). Taken together, these expression differences between Pax2 paralogs argue for sub- or neofunctionalization following their proposed split in the last common ancestor of spiders and scorpions.
Materials and Methods
Animal Cultivation
P. tepidariorum embryos were raised in the lab from the original genome line stock (received from Göttingen). P. phalangioides embryos were obtained from females wild caught in lower saxony. Stock keeping was mainly performed as described previously (Turetzek et al., 2016; Turetzek and Prpic, 2016) except that stocks were kept at room temperature and with natural light in the laboratory.
Gene Cloning
Pax2 candidates were cloned with gene specific primers (Supplementary Table 1) from cDNA [either SMARTer RACE cDNA (P. phalangioides) or SMARTer cDNA (P. tepidariorum), both Clontech, Mountain View, CA, United States] synthesized from RNA of a combination of all embryonic stages isolated with either TRIzol (Invitrogen) or Quick-RNA Tissue/Insect Kit (Zymo Research). Gene fragments were cloned into the Pjet1.2 plasmid using the CloneJET PCR Cloning Kit (Thermo Fisher Scientific).
Phylogenetic Analysis
Pax2 ortholog candidates for the spiders P. tepidariorum and P. phalangioides were identified using the D. melanogaster sv protein sequence (CG11049). We performed this BLAST search in the previously published genome of P. tepidariorum (Schwager et al., 2017) available via the i5k BLAST database1 and the previously described P. phalangioides developmental transcriptome (Janssen et al., 2015; Leite et al., 2018). For both species we found two genes with high sequence similarity, aug3.g12456 and aug3.g762 for P. tepidariorum and c56107_g1 and c100802_g1 for P. phalangioides indicated by the two lowest e-value hits. In addition, we also used D. melanogaster protein sequences for Pax2, Pax6, Poxn, and Poxm [accessed from Uniprot (Q59DN9, O18381, P23758, and P23757) (The UniProt Consortium, 2015)] to BLAST against the P. tepidariorum genome. In total, we identified nine candidate sequences with an e-value lower than e-50, all of which were confirmed as Pax gene candidates for further analysis by a reciprocal BLAST (back-BLAST) against D. melanogaster and Danio rerio. These P. tepidariorum candidate sequences were then used to identify similar sequences in several other species, covering the arthropod phylogeny (Cao et al., 2013; Conzelmann et al., 2013; Schwager et al., 2017; Vizueta et al., 2017; Liao et al., 2019; Pechmann, 2020; Liu et al., 2021) as well as lophotrochozoan and vertebrate species. From these, only unique proteins with an e-value lower than e-50 were kept for the phylogenetic analysis. A full species list with source references is shown in Supplementary Table 2.
This preliminary phylogenetic analysis in total included 148 full-length protein sequences, nine P. tepidariorum Pax candidates identified in this study as well as previously published spider Pax family genes (Schoppmeier and Damen, 2005; Samadi et al., 2015; Schomburg et al., 2015) plus their potential homologs from a range of bilaterians (summarized in Supplementary Table 2). Alignment was performed using Clustal OMEGA (Sievers et al., 2011) with standard parameters (nexus file is available in Supplementary Data Sheet 1). Phylogenetic tree building was performed using MrBayes version 3.2.6 (Ronquist and Huelsenbeck, 2003). Mixed amino acid substitution models were tested, and the Jones model (Jones et al., 1992) was chosen and used to generate topological convergence using metropolis coupling. The analysis was stopped due to time limitations after 3,000,000 generations reaching an average standard deviation of split frequencies of 0.029. A total number of 12.002 trees were written in two files (6,001 trees per file of which 4,501 were sampled).
In a second phylogenetic analysis only a subset of closely related Pax subfamilies was used (Pax2/5/8, Pax6, and poxn). Using the candidates for the respective groups recovered from P. tepidariorum, D. rerio Pax2/5/8 as well as D. melanogaster Pax2, poxn, and Pax6 orthologs were used to search for similar sequences in more chelicerate genomes including the xiphosuran Limulus polyphemus, and the spiders Argiope bruennichi (Sheffer et al., 2021), Stegodyphus dumicola (Liu et al., 2019) and Araneus ventricosus (Kono et al., 2019). Again, only unique hits with an e-value threshold lower than e-50 were kept (except for the xiphosuran L. polyphemus, for which only hits lower than e-100 were kept because of the large number of candidate hits). The resulting candidate protein sequences were again confirmed by reciprocal BLAST. Full-length protein sequences of all chelicerate Pax2/5/8, Pax6, and poxn proteins were aligned (same parameters as above, nexus file available as Supplementary Data Sheet 2). Again, MrBayes version 3.2.6 (Ronquist and Huelsenbeck, 2003) was used to generate topological convergence using metropolis coupling after amino acid substitution models testing, while the Jones model (Jones et al., 1992) was chosen. After 514,000 generations, an average standard deviation of split frequencies below 0.01 was achieved. A total number of 2,058 trees were written in two files (1,029 trees per file of which 772 were sampled). The respective 50% majority rule consensus trees are shown in Figure 1 and Supplementary Figures 1, 2. Consensus trees were visualized from the MrBayes nex.con.tre output file using the software Geneious (version 11.1.5, Biomatters). The annotation of the respective subfamilies was done using Adobe Illustrator 2020 version 24.0 for Apple Macintosh.
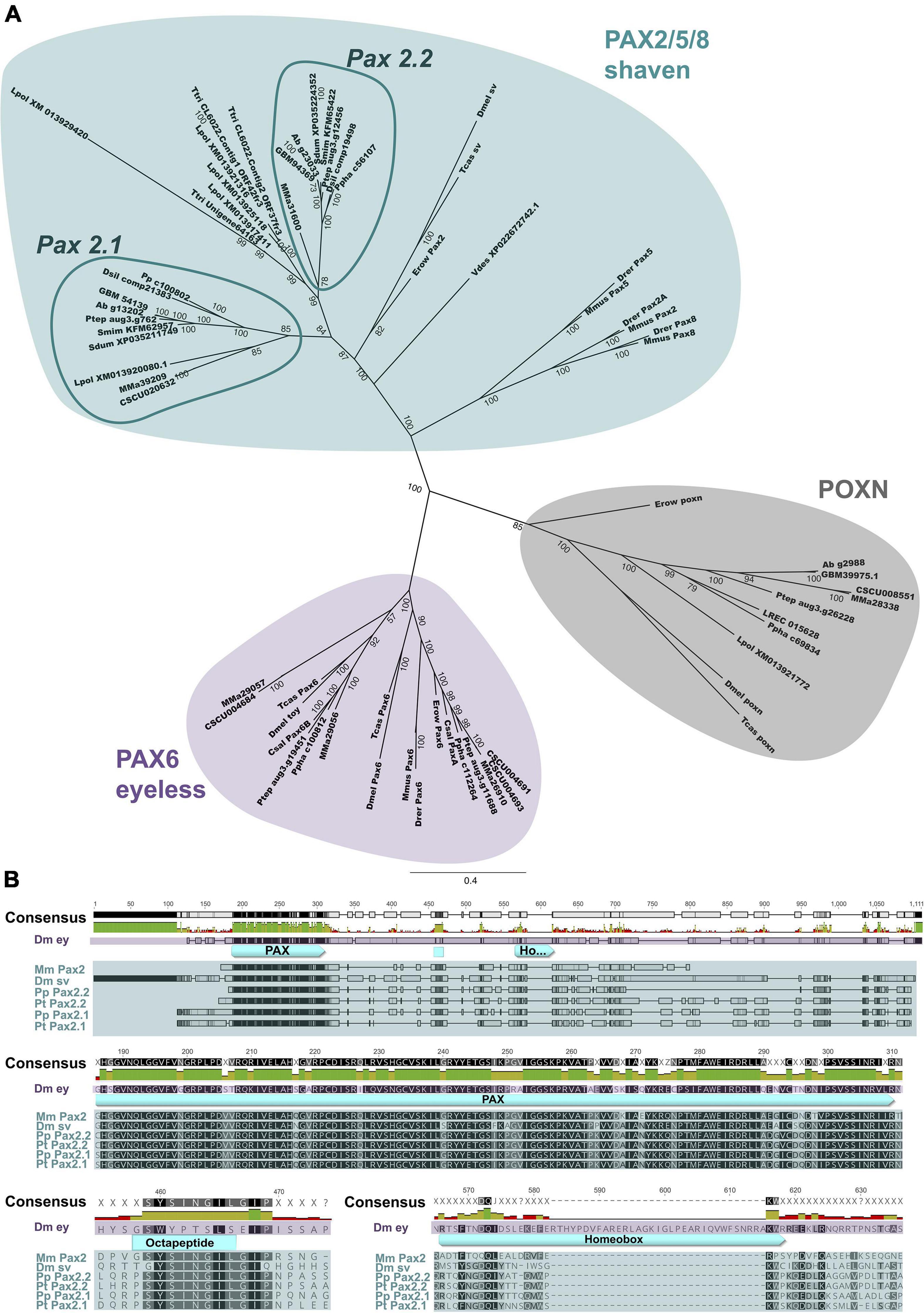
Figure 1. Phylogenetic analysis and protein alignment of selected Pax genes. (A) Bayesian Markov chain Monte Carlo analysis depicted as unrooted 50% majority rule consensus tree. Branch lengths indicate expected substitutions per site. Labels at the tree edges represent clade credibility values, which are a measure of the posterior probability of each clade in the tree. Turquoise indicates the PAX2/5/8 cluster (D. melanogaster ortholog is shaven), purple shading marks Pax6 ortholog group (D. melanogaster ortholog is eyeless), and the Poxn cluster is highlighted in gray. The arachnopulmonate specific Pax2.1 and Pax2.2 groups within the Pax2/5/8 cluster are highlighted with dark turquoise circles. (B) Protein alignment of selected sequences (Dm ey, D. melanogaster eyeless (purple); Mm Pax2, M. musculus Pax2; Dm sv, D. melanogaster shaven; Pp Pax2.1, P. phalangioides Pax2.1; Pt Pax2.1, P. tepidariorum Pax2.1; Pp Pax2.2, P. phalangioides Pax2.2; Pt Pax2.2, P. tepidariorum Pax2.2 (turquoise). PAX domain, as well as octapeptide and homeobox domains are highlighted and shown in detail. Species abbreviations and accession numbers or genomic and transcriptomic resources are summarized in Supplementary Table 2.
Whole Mount in situ Hybridization and Imaging
Whole mount in situ hybridization was performed as previously described (Prpic et al., 2008) with minor modifications explained below. For the staining we used fixed P. tepidariorum and P. phalangioides embryos of developmental stages coinciding with CNS and sense organ development (starting from germ band stage until late ventral closure, a few representatives of younger stages were also included per staining reaction). Staging of the two spider species was performed based on previously published descriptions of embryonic development (Mittmann and Wolff, 2012; Turetzek and Prpic, 2016). The published whole mount in situ hybridization protocol was slightly modified by omitting the treatment with proteinase K prior to hybridization. Another difference was that two different staining detection methods were used. One half of the embryos was stained with NBT/BCIP (ROTH) in the dark until a specific staining was detected, while the other half was stained overnight with FAST RED Tablets (Biotrend/Kem-En-Tec) according to manufacturer’s guidelines. After whole mount in situ hybridization using the colorimetric detection method with NBT/BCIP, embryos were additionally stained with SYTOX-Green (Invitrogen) to label cell nuclei. These embryos were then imaged using a combination of white light and green channel with a Leica M205 FA binocular equipped with a Qimaging Micropublisher 5.0 RTV camera. Images of dissected appendages stained with NBT/BCIP were captured using a Zeiss Axioplan with a Manta G609C camera (Alited Vision Technologies). Embryos stained with FAST RED after whole mount in situ hybridization were incubated with DAPI (Merck) to visualize cell nuclei. These embryos were either flat mounted as a whole by removing the yolk, or appendages were removed by using a fine pair of Dumont forceps (Fine Science Tools) and minutes/insect pins. Samples were then mounted on a slide with 90% glycerol (in PBS) containing DABCO ® (ROTH) as an antifading agent. FAST RED samples were imaged using the Leica Sp5 confocal laser scanning microscope performed at the “Center for Advanced Light Microscopy” (CALM). Maximum projections of confocal stacks were generated with FIJI (Schindelin et al., 2012). Brightness and contrast of the digital images were adjusted with Adobe Photoshop version 2020 for Apple Macintosh.
Results
Duplicated Pax2 Genes in Parasteatoda tepidariorum and Pholcus phalangioides
While the Pax family gene Pax6 is well studied in many arthropod species (Prpic, 2005; Blackburn et al., 2008; Yang et al., 2009; Luan et al., 2014; Samadi et al., 2015; Schomburg et al., 2015; Hou et al., 2016), the closely related Pax2 gene with equally interesting roles in neurogenesis is not well analyzed in arthropods. A previous study investigating the evolution of the homeobox genes in arachnids already showed that Pax3/7 and Pax4/6 group genes were duplicated multiple times in spiders and scorpions (Leite et al., 2018). Unfortunately, no chelicerate Pax2 gene was identified, which was also the case for two other studies focusing on the genetic basis of eye development in spiders (Samadi et al., 2015; Schomburg et al., 2015). We were able to identify two candidate homologs, aug3.g762 and aug3.g12456 in the current genome version of P. tepidariorum (Schwager et al., 2017). Due to their expression in the eye and appendage sense organ primordia comparable to insects, as well as their conservation between both spider species (explained below), Pt-aug3.g762 is referred to as Pt-Pax2.1, while the paralog Pt-aug3.g12456 is termed Pt-Pax2.2. Both proteins show high sequence similarity to the D. melanogaster Pax2 ortholog sv and the Pax2/5/8 proteins from the zebrafish D. rerio. These orthologs presumably escaped previous identification attempts due to their degraded homeodomain protein sequence (Figure 1B), a common feature of Pax2/5/8 group genes (Bopp et al., 1986; Dressler et al., 1990; Plachov et al., 1990). Furthermore, we identified two genes (c56107_g1 and c100802_g1) with high sequence similarity to the D. melanogaster Pax2 ortholog in the de novo assembled developmental embryonic transcriptome of the haplogyne cellar spider P. phalangioides (Janssen et al., 2015; Leite et al., 2018). These genes align well with the two Pax2 candidates from P. tepidariorum, with c56107_g1 (Pp-Pax2.2) being similar to Pt-Pax2.2 and c100802_g1 (Pp-Pax2.1) to Pt-Pax2.1. These four spider Pax2 candidates align well which each other and also with other Pax genes like Mus musculus Pax2 as well as the Pax6 ortholog eyeless and Pax2 ortholog sv from D. melanogaster (Figure 1B). The paired box domain shows the highest sequence similarity, with only some amino acid changes between mouse, fruit fly and spiders. Especially in the first half of the paired box domain, all Pax2 homologs share a higher sequence similarity compared to D. melanogaster Pax6. For the two Pax2 paralogs from both spiders, the entire paired box domain is identical with only one exception. We find a Pax2.1 specific change from Valine to Methionine following the highly conserved GRPLPD sequence (this exchange is also present in all other Pax2.1 paralogs from spiders and scorpions Supplementary Data Sheets 1–3). Also, the Pax2 specific octapeptide motif is 100% identical for all six Pax2 orthologs shown in the alignment (Figure 1B). The homeobox domain is, as expected, only fully present in D. melanogaster eyeless, but is truncated in all Pax2 orthologs. Overall, there is a higher sequence similarity between the four spider homologs, and to the corresponding paralog than there is to the Pax2 sequences from mouse and D. melanogaster. These results indicate that P. tepidariorum Pt-Pax2.1 (aug3.g762) and Pt-Pax2.2 (aug3.g12456) as well as P. phalangioides Pp-Pax2.2 (c56107_g1) and Pp-Pax2.1 (c100802_g1) represent duplicated Pax2 spider orthologs.
Phylogenetic Analysis Reveals Independent Pax2 Duplication Events in Chelicerates
To further confirm the duplication of Pax2 in spiders we performed a thorough phylogenetic analysis (nexus alignment file available as Supplementary Data Sheet 1, phylogenetic reconstruction in Supplementary Figures 1, 2 and summarized in Supplementary Table 2). In agreement with the BLAST results and the alignment, Pt-Pax2.1, Pt-Pax2.2 as well as Pp-Pax2.1 and Pp-Pax2.2 are grouped together with all other Pax2/5/8 orthologs from vertebrates, the annelid Platynereis dumerilii, onychophorans, the myriapod Glomeris marginata as well as insects (turquoise Supplementary Figures 1, 2). The tree also reveals that the chelicerate Pax2 duplication has most likely no common origin with the vertebrate duplication events, because the vertebrate Pax2, Pax5, and Pax8 genes are grouped together and are clearly separated from the spider Pax2 orthologs. Focusing on the chelicerate Pax2 family genes, it is apparent that their duplication most likely occurred before the split of entelegyne and haplogyne spiders, because Pt-Pax2.1 and Pp-Pax2.1 form one spider specific Pax2.1 cluster, grouping together with sequences from the entelegyne spider species Stegodyphus mimosarum and the haplogyne spider Dysdera silvatica. Interestingly, the two scorpion Pax2 homologs from Mesobuthus martensii (MMA39209) (Cao et al., 2013) and Centruroides sculpturatus (CSCU020632) form a group with all Pax2.1 spider orthologs. Like Pax2.1, all Pax2.2 sequences from haplogyne and entelegyne spiders are grouped together and also form a cluster with three sequences from the xiphosuran Tachypleus tridentatus (Liao et al., 2019). A second scorpion Pax2 sequence is only present for M. martensii (MMA31600) and does not cluster with any of the spider Pax2 groups. Thus, this large analysis strongly supports both P. tepidariorum as well as the P. phalangioides candidates as Pax2 orthologs, which arose from a shared duplication event in the lineage leading to spiders. Where and how many duplication events occurred in chelicerates, however, is difficult to resolve and more thorough sequencing, especially in scorpions, is needed.
We therefore performed a second phylogenetic analysis, containing Pax2 candidates from more chelicerate species (Figure 1A). In this analysis, chelicerate Pax2 candidates again cluster together with the insect, onychophoran and vertebrate Pax2 orthologs (turquoise Figure 1A), while all other candidates are grouped with either poxn or Pax6 (purple and gray Figure 1A, respectively). This supports the results from the reciprocal BLAST and first phylogenetic analysis. In Figure 1A, the chelicerate sequences are again distinctly separated from insect and onychophoran Pax2 orthologs. The only exception is XP022672742.1 from the mite Varroa destructor, which is positioned outside the chelicerate and insect clusters, but still shows an overall closer alliance to arthropods than to vertebrates. Similarly, as observed in the first phylogenetic analysis, the spider Pax2 homologs split into two clusters, Pax2.1 and Pax2.2. The xiphosuran sequences form a separate Pax2 group, containing three sequences from T. tridentatus and four from L. polyphemus, suggesting several independent duplication events in xiphosurans. The Pax2.1 cluster again contains both Pt-Pax2.1 and Pp-Pax2.1, which further divide into entelegyne and haplogyne Pax2.1 clusters, resolving all other spider sequences. Only a single xiphosuran sequence, originating from L. polyphemus (XM013920080.1) is grouped into this Pax2.1 cluster, but this is most likely caused by the partial and short protein sequence. The amino acid residue following the conserved GRPLPD sequence of L. polyphemus XM013920080.1 is an Isoleucine, which neither fits with the amino acid exchange of arachnopulmonates Pax2.1 nor Pax2.2. A similar exchange to Isoleucine is, however, observed in other L. polyphemus Pax2 candidate genes (XM013925118.1 and XM013929420.1) (see Supplementary Data Sheet 2) and thus more an indicator for complications by rapidly evolving xiphosuran sequences and the partial character of the protein than against for the position of this protein in the arachnopulmonate Pax2.1 cluster. Interestingly, in this new phylogenetic analysis, the two scorpion proteins from M. martensii (Mma39209 and Mma31600) do cluster together with the respective spider Pax2.1 and Pax2.2 groups. To further validate the grouping of spider and scorpion homologs into two separate Pax2 clusters we performed even another phylogenetic analysis using only well-alignable regions of the identified chelicerate Pax2 proteins (Supplementary Data Sheet 3 and Supplementary Figure 8). Fittingly, this additional phylogenetic analysis highly supports grouping of the scorpion MMa39209 and CSCU020632 proteins with the spider Pax2.1 cluster and MMa31600 with Pax2.2. This suggests that the Pax2 paralogs most likely evolved through the whole genome duplication (Schwager et al., 2017) ancestral to arachnopulmonates.
Conserved Spider Pax2.1 Expression Domains
As Pax2 genes are known for their important role in neurogenesis, we wanted to test if the spider Pax2 paralogs might also be involved in developing neural tissues and sense organs. Based on data from the two distinct mutant phenotypes shaven and sparkling in D. melanogaster (Fu and Noll, 1997; Kavaler et al., 1999; Hirth et al., 2003), RNAi phenotypes in the red flour beetle T. castaneum (Dönitz et al., 2015) and expression patterns in the jewel wasp N. vitripennis (Keller et al., 2010) Pax2 is known to be a conserved factor required for the development of arthropod sense organs, having a dual role in developing eyes and external sense organs. Therefore, we investigated the expression of the two Pax2 paralogs in the entelegyne spider P. tepidariorum and the haplogyne spider P. phalangioides. While both Pax2 paralogs showed expression during embryonic stages of both species, especially in the head region, the ventral nervous system and the developing appendages, timing of expression as well as expression pattern dynamics and localization differed between paralogs and between species.
Pt-Pax2.1 expression starts around the first signs of brain differentiation (stage 10). In the head, Pt-Pax2.1 is expressed bilaterally symmetric in the non-neurogenic ectoderm, lateral to the forming lateral furrow, presumably in the field which later gives rise to the lateral eyes (red arrowhead, Figures 2A,E and red ellipse Figure 3A). This lateral expression stays unaltered (Figures 2F,G, arrowhead) until the non-neurogenic ectoderm starts to overgrow the brain during dorsal closure (stage 13). At this stage, the initial single field of Pt-Pax2.1 expression broadens (red arrowhead, Figure 2H and red ellipse Figure 3B) and divides, giving rise to three spots of expression during prosomal ventral closure (stage 14) (red arrowheads Figure 2L). In the haplogyne spider, Pp-Pax2.1 is similarly expressed in the region of the developing lateral eyes (red arrowhead, Supplementary Figures 3D–F), similar to Pt-Pax2.1.
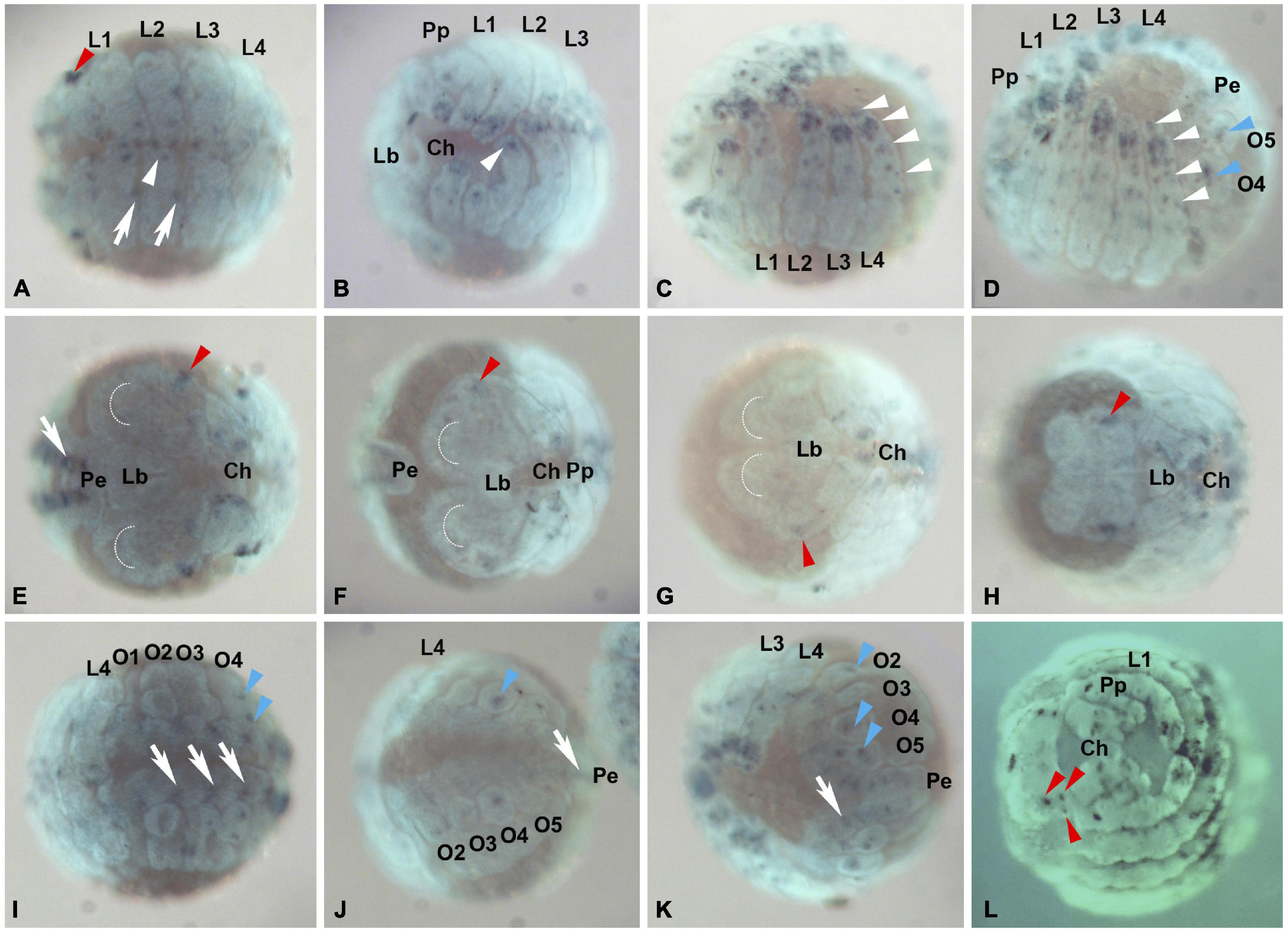
Figure 2. Pax2.1 (aug3.g762) expression in P. tepidariorum embryos. Expression of Pt-Pax2.1 is detected beginning with brain differentiation (Stage 10, A,E,I) and expression stays throughout inversion (Stage 11, B,F,J), dorsal (Stage 13, C,G,K) and early and late ventral closure (stage 14, D,H,L). (A–D) Ventral view of embryos in different developmental stages. Pt-Pax2.1 is expressed in prosomal and opisthosomal appendages (white and blue arrowheads, respectively) and the ventral neuroectoderm (white arrows). (E–H) Frontal view. Pt-Pax2.1 is expressed in the head lobes next to the lateral furrow (red arrowheads E–G) during brain differentiation, inversion, and dorsal closure (stages 10–13). Later during ventral closure (Stage 14) Pt-Pax2.1 is expressed in the lateral eye primordia [red arrowhead in (H) depicting the primordial cluster, three red arrowheads in (L) depicting the three distinct eye primordia]. (I–K) Ventral view of the developing opisthosoma. Pt-Pax2.1 is expressed in the opisthosomal ventral neuroectoderm (white arrows) and developing opisthosomal appendages (blue arrowheads). Anterior is to the left in all panels, all images show ventral aspect, except (E–H,L) which are in frontal aspect. Dotted half circles indicate the anterior furrow in the prechelceral head lobes. Lb, labrum; Pe, Posterior end; O1–O5, Opisthosomal segments 1–5; Prosomal appendages: Ch, chelicerae; Pp, pedipalps; L1–L4, 1st–4th walking leg. Dark blue, NBT/BCIP staining showing specific Pt-Pax2.1 expression; Cyan, nuclear Sytox staining.
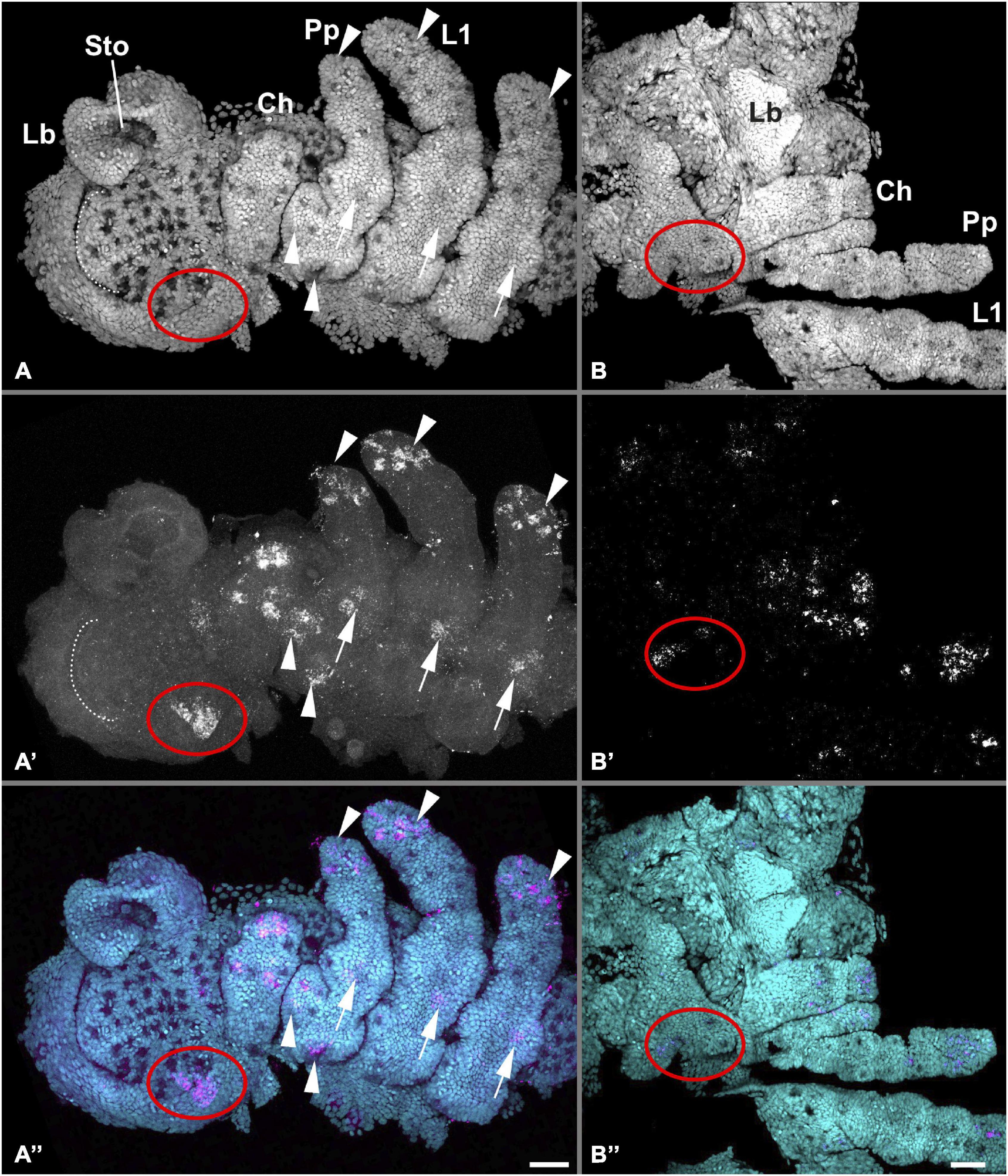
Figure 3. Detailed embryonic Pt-Pax2.1 expression in the head. Red ellipses mark Pt-Pax2.1 expression domains in the eye primordia throughout embryonic development, white dotted line marks the anterior furrow in the head lobes. Left panels (A–A”) show the right anterior half of an embryo including the head lobes and prosomal segments until the second leg segment during early embryogenesis coinciding with brain differentiation (stage 10). Pt-Pax2.1 is expressed in the developing lateral eye primordia next to the lateral furrow (red ellipse) ventral neuroectoderm (arrows, see also Supplementary Figure 7B for VNE specific Z-projection), and in proximal and distal spots in the appendages (arrowheads, see also Supplementary Figure 7A for appendage specific Z-projection). (B–B”) Head and right appendages (chelicera, pedipalp, L1) during mid-ventral closure (stage 13.1 late) showing diffused Pt-Pax2.1 gene expression in the appendages and as patches in the developing eyes. Flat mounted preparations; anterior is to the left in all panels. Cyan is nuclear DAPI staining; magenta is Pt-Pax2.1 specific expression detected with FAST RED. Lb, labrum; Sto, stomodeum. Prosomal appendages: Ch, chelicerae; Pp, pedipalps; L1, 1st walking leg. Scale bar represents 50 μm.
In addition, Pax2.1 paralogs in both spiders are expressed in the developing ventral neuroectoderm (white arrow in Figures 2, 3 and Supplementary Figures 3, 7), the only difference being that the prosomal ventral neuroectodermal expression is weaker and less prominent during later stages in P. tepidariorum compared to P. phalangioides. In the prosomal and opisthosomal appendages, Pt-Pax2.1 expression starts during brain differentiation (stage 10) (white and blue arrowheads in Figures 2–4 and Supplementary Figure 7). In the chelicerae, Pt-Pax2.1 is expressed in three distinct regions, most prominently in the distal tip and as two spots and later two clusters in the medial region (Figures 3, 4A,D). In the pedipalps, Pt-Pax2.1 expression is more dynamic. It also starts in three clusters, one at the distal tip, one in the gnathendite and one at the dorsal proximal rim (white arrowhead, Figures 3A, 4B and Supplementary Figure 7). While the initial distal expression cluster broadens until ventral closure (Stage 13), nearly covering a third of the distal pedipalp (Figure 4E), the proximal expression cluster is refined, giving rise to two single spots (white arrowheads, Figure 4E). In addition, two similar expression spots arise in the medial area of the pedipalp (magenta arrowhead, Figure 4E), while the expression in the gnathendite slightly broadens. In the legs, Pt-Pax2.1 is expressed in only one distal cluster early on (white arrowhead, Figures 2A,B, 3A, 4C and Supplementary Figure 7), while a spot-like expression is present later on in the four distal leg segments (tibia, metatarsus, tarsus, and the claw) as well as proximally, on the ventral side (white arrowheads, Figures 2C,D, 4F). A similar spot-like expression is present in the opisthosomal appendages (blue arrowheads, Figures 2D,I–K). For the cellar spider P. phalangioides, Pp-Pax2.1 expression in the appendages starts ubiquitously (Supplementary Figures 3A,I) and later resolves during ventral closure, showing a spot-like expression throughout the entire walking leg with clusters in the femur and distal thickening of the walking legs, as well as in the developing spinnerets (white arrowhead, Supplementary Figures 3C,J). Taken together, the Pax2.1 paralog is expressed in three distinct neural tissues, the lateral eyes, external sense organs on the appendages, as well as the developing ventral neuroectoderm in the prosoma and opisthosoma. Apart from slight differences in the expression dynamics and timing, the expression patterns (except for the P. phalangioides late femoral expression) are comparable for the two spiders analyzed here, suggesting a conserved role of Pax2 in the development of neural tissues, in a strikingly similar fashion to its dual role in sense organ development shown in D. melanogaster (Fu and Noll, 1997; Kavaler et al., 1999; Hirth et al., 2003).
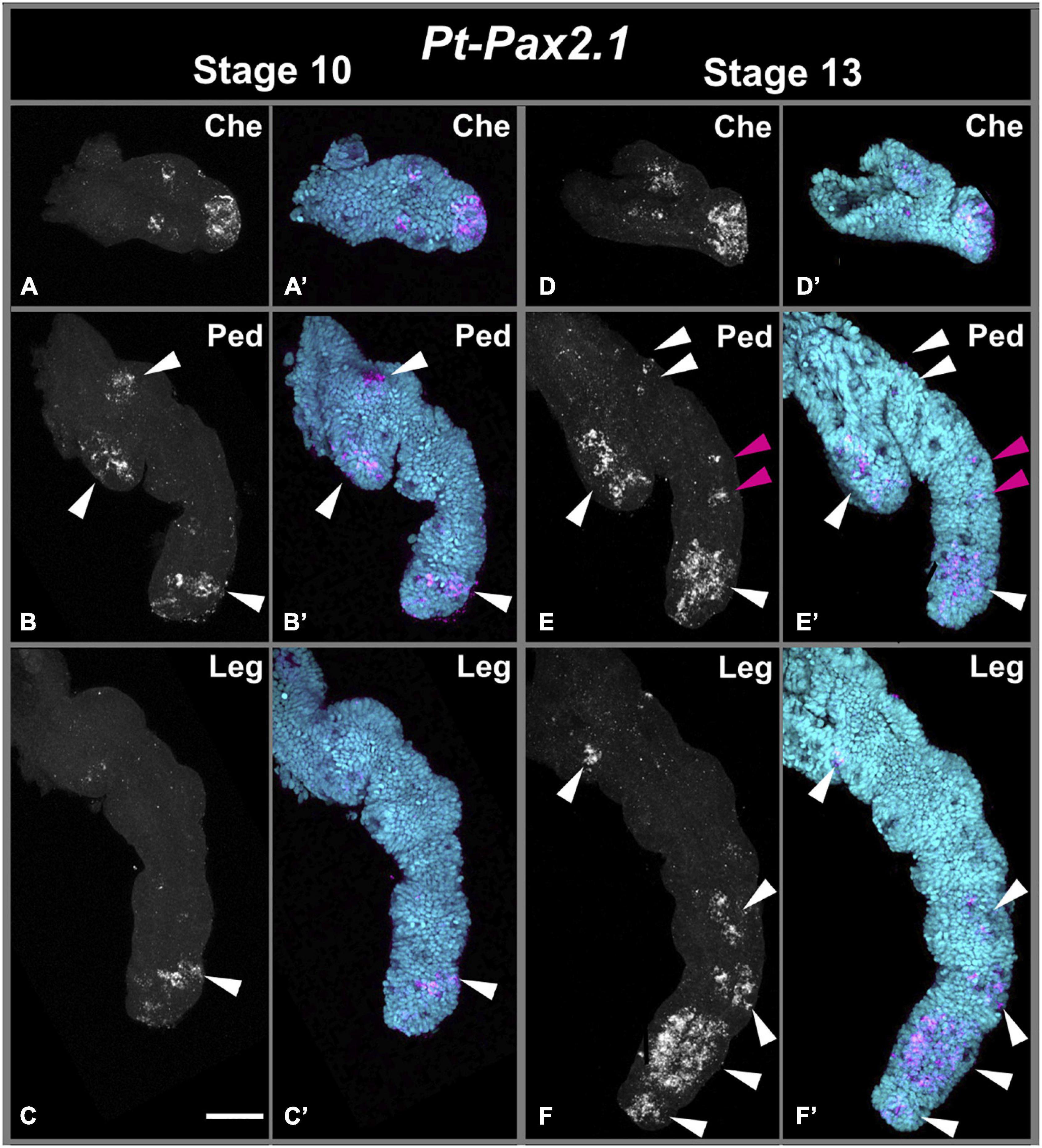
Figure 4. Embryonic Pt-Pax2.1 expression in prosomal appendages. Left panels show Pt-Pax2.1 expression during brain differentiation (stage 10). Panels on the right show expression during dorsal closure (stage 13). Top row (A,A’,D,D’) shows Pt-Pax2.1 domains in the dissected chelicerae (Che). Center row (B,B’,E,E’) shows Pt-Pax2.1 expression in dissected pedipalps (Ped). Bottom row (C,C’,F,F’) shows expression in dissected walking leg(s) (Leg). White arrowheads indicate expression clusters throughout development, and magenta arrowheads mark newly emerged pedipalpal medial expression domains. Early Pt-Pax2.1 expression in the chelicera is present as two medial spots and a distal domain. While the distal domain stays in later stages, the two medial spots slightly broaden and diffuse. In the pedipalp Pt-Pax2.1 expression starts in three patches, one in the gnathendite, one in the proximal ventral side and one domain in the distal tip, similar to the expression in the walking leg. Later the expression in the gnathendite and distal tip broadens, the proximal patch splits and two spot-like expression domains arise in the medial part of the pedipalp. In the walking leg expression starts in the distal leg region, which broadens during development. Later Pt-Pax2.1 expression covers all distal leg segments both as large domain but also spot-like patterns. In the proximal part a spot-like domain appears. Cyan is nuclear DAPI staining; magenta is Pt-Pax2.1 specific expression detected with FAST RED. Scale bar represents 50 μm.
Diverged Expression of Spider Pax2.2
Expression of Pt-Pax2.2 starts around the same time as Pt-Pax2.1 but is much weaker. The earliest expression of Pt-Pax2.2 is present in the brain (black arrowhead, Figure 5E) and the VNE (white arrow, Figures 5A,I) but no expression is detected just slightly before that stage (Supplementary Figure 5). Starting during inversion and retraction (stage 11 and 12), expression in the VNE and brain (Figures 5F,G,J,K) is getting stronger. Expression in the neurogenic region of the brain is dynamic. While in one lobe, two spots of Pt-Pax2.2 expression are present, the more posterior spot seems to have split in the other head lobe, giving rise to three expression spots (black arrowhead, Figure 6A). After the non-neurogenic tissue has overgrown the brain, Pt-Pax2.2 is again only expressed in two spots in the brain (black arrowhead Figure 6B’) and as a weak cluster on the left side of the head, not coinciding with the developing eyes (red ellipses Figures 6B–B”). In the haplogyne spider, Pp-Pax2.2 is expressed in faint half-circles in the ventral neuroectoderm (white arrow in Supplementary Figure 4), while no expression is present in the brain throughout development.
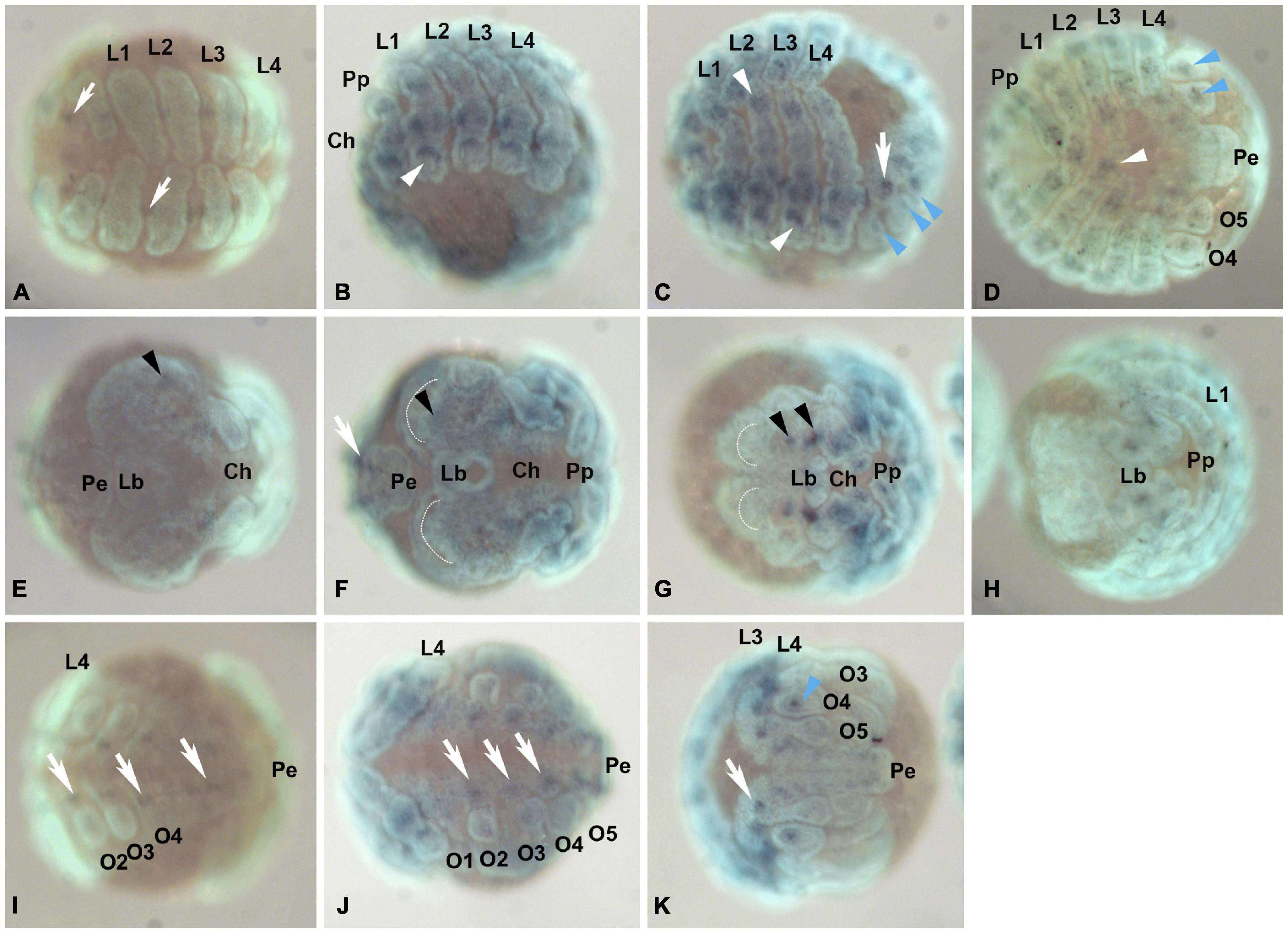
Figure 5. Pt-Pax2.2 (aug3.g12456) expression during embryogenesis. Leftmost panels (A,E,I) show embryos during early brain differentiation stages (Stage 10), the panels in the middle show embryos during inversion (Stage 11, B,F,J) and between retraction and dorsal closure (Stage 12/13, C,G,K), while the rightmost panels (D,H) show embryos during early ventral closure (Stage 14.1). (A–D) Embryos shown in ventral (or lateral ventral) aspect. Pt-Pax2.2 is expressed in the ventral neuroectoderm (white arrows), and later in the prosomal and opisthosomal appendages (white and blue arrowheads, respectively). (E–H) Pt-Pax2.2 expression domains in the head lobes (black arrowhead) shown in embryos in frontal view. (I–K) Opisthosomal view showing Pt-Pax2.2 expression in the opisthosomal ventral neuroectoderm (white arrows) and opisthosomal appendage, the developing spinneret on O4 (blue arrowhead). Anterior is to the left in all panels, all images show ventral aspects, except (E–H) which are in frontal aspect and (B) shown in ventral-lateral view. Dotted half circles indicate the anterior furrow in the prechericeral head lobes. Lb, labrum; Pe, Posterior end; O1–O5, Opisthosomal segments 1–5; Prosomal appendages: Ch, chelicerae; Pp, pedipalps; L1-L4, 1st to 4th walking leg. Dark blue, NBT/BCIP staining showing specific Pt-Pax2.2 expression; cyan, nuclear Sytox staining.
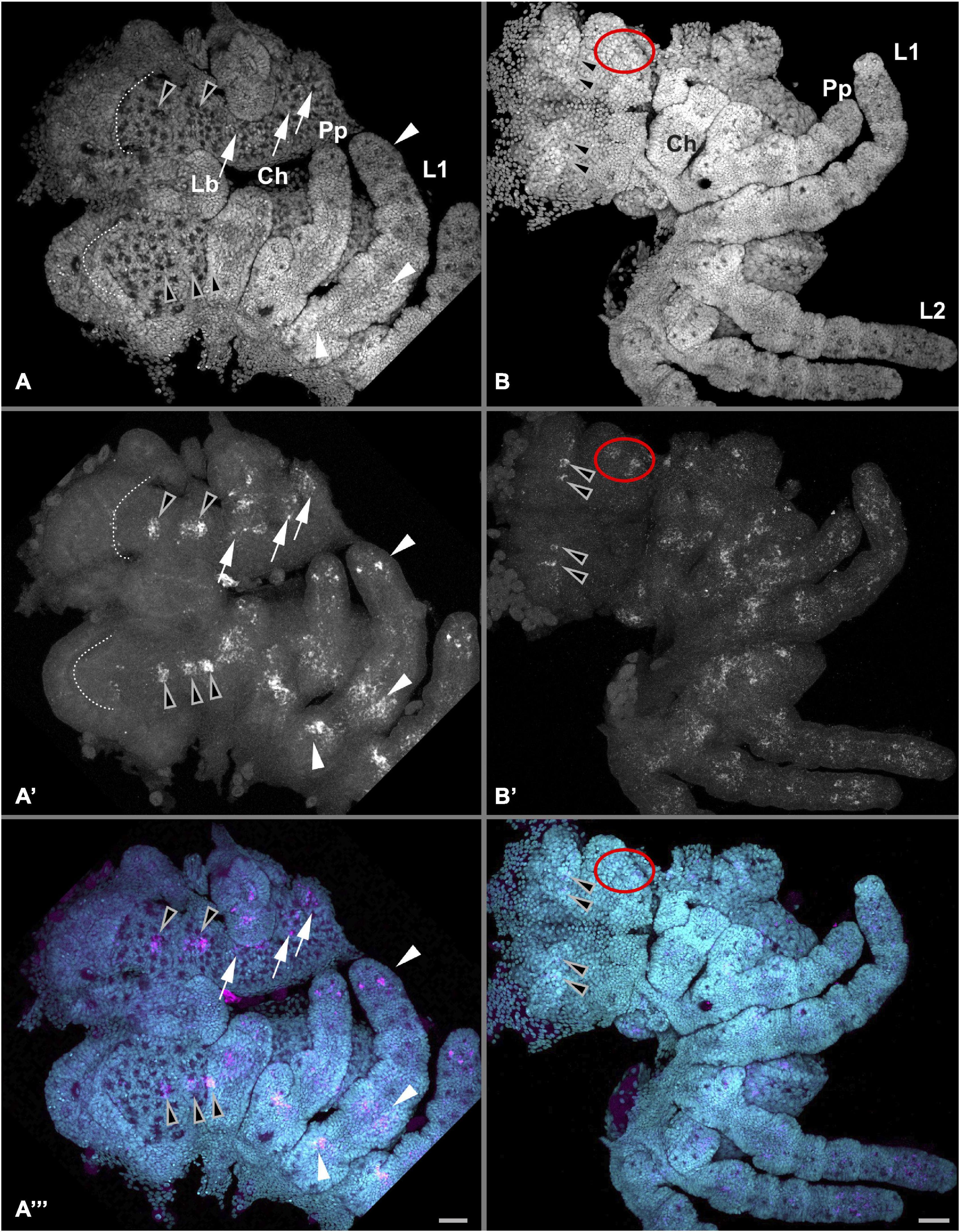
Figure 6. Detailed Pt-Pax2.2 expression in the anterior embryo. Anterior is to the left in all panels. Left panels (A–A”) show Pt-Pax2.2 expression during late retraction of the embryo (Stage 12) in the ventral neuroectoderm (white arrows), the appendages (arrowheads), and in the brain (black arrowheads). During this stage the dynamic expression in the neurogenic part of the head lobes later giving rise to the brain is detectable. While in one half of the head only two expression domains are visible in the other half already three distinct expression domains are present. Pt-Pax2.2 expression in the pedipalps and walking legs is present in one proximal and one medial domain as well as in a spot-like pattern in the distal tip. Dotted line indicates anterior furrow in the head lobes. Right panels (B–B”) show Pt-Pax2.2 expression later in embryos during ventral closure (Stage 14), present as two spots in the brain (black arrowheads), as well as in two clusters at the lateral rim of the head (red ellipses). Expression in the prosomal appendages is still present but more diffuse. Lb, labrum. Prosomal appendages: Ch, chelicerae; Pp, pedipalps; L1, 1st walking leg. Cyan is nuclear DAPI staining; magenta is Pt-Pax2.2 specific expression detected with FAST RED. Scale bar represents 50 μm.
The expression pattern in the appendages also differs between the two paralogs. Pt-Pax2.2 expression starts during inversion (stage 11) whereas Pt-Pax2.1 expression begins earlier during stage 9/10. Pt-Pax2.2 expression in the pedipalps and walking legs starts in three areas, two broad clusters in the proximal and medial region and as two or three distal spots (white arrowheads, Figures 5B, 6A, 7A,B). During later development, this expression remains in the same regions, but gets more diffuse (Figures 5D,H, 6B, 7C,D). Likewise, expression in the chelicerae is ubiquitous and diffuse (Figures 5, 6A,B). In the prosomal appendages of P. phalangioides, Pax2.2 is exclusively expressed in the mesoderm, starting once the appendages start to grow (white arrowhead, Supplementary Figures 4A,G). This mesodermal Pp-Pax2.2 expression stays throughout development (Supplementary Figures 4B,H) but slightly decreases during ventral closure (white arrowhead, Supplementary Figures 4C,I). In summary, in contrast to the conserved expression of Pax2.1, the Pax2.2 paralog has only kept the conserved expression in the ventral neuroectoderm but is not detected in the developing eyes in any of the spider species observed in this study. In addition, Pax2.2 not only diverged from its paralog, but also shows more differences between spider species, especially in the appendages.
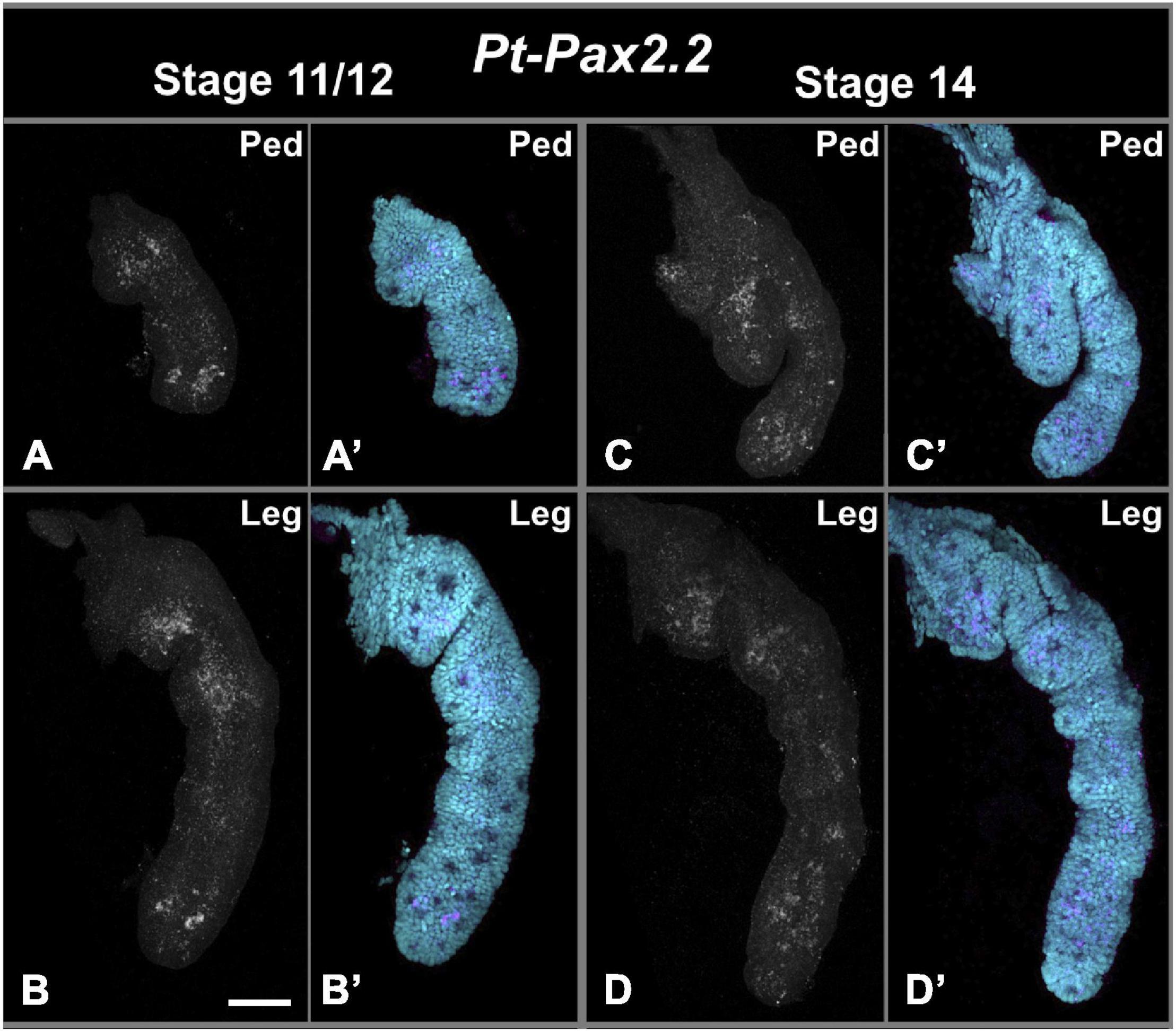
Figure 7. Pt-Pax2.2 expression in pedipalps and walking legs during late embryogenesis. Panels on the left (A,A’,B,B’) show dissected prosomal appendages during inversion and/or retraction stage (stage 11/12). Right panels (C,C’,D,D’) show appendage preparations from ventral closure stage (stage 14). Upper panels (A,A’,C,C’) show Pt-Pax2.2 expression in dissected pedipalps (Ped), note most proximal region of the pedipalp was lost during dissection. Lower panels (B,B’,D,D’) show Pt-Pax2.2 expression in dissected walking legs. During inversion and retraction of the embryo Pt- Pax2.2 is expressed three region of the pedipalps and walking legs. While the proximal (missing for the pedipalp, please see Figure 4A) and medial expression domain has a broad and more diffuse appearance, the expression in the distal tip of these prosomal appendages has a spot-like pattern. During later stages the expression fades and becomes even more diffuse also in the distal part. Cyan is nuclear DAPI staining; magenta is Pt-Pax2.2 specific expression detected with FAST RED. Scale bar represents 50 μm.
Discussion
Independent Pax2 Duplication Events in Chelicerates
As paired box transcription factors are some of the oldest genetic toolkit components and play conserved dominant roles in neurogenesis across metazoans, they represent prime candidates to study the evolution of gene regulatory networks involved in neurogenesis and sense organ development. Interestingly, a genomic study of the common house spider P. tepidariorum revealed that spiders most likely underwent a whole genome duplication (Schwager et al., 2017). Several studies on gene duplication in spiders showed its importance for evolving novel phenotypes or altering development by bringing new genetic material to key developmental processes like appendage development, eye formation, body patterning, and segmentation (Pechmann et al., 2011, 2015; Khadjeh et al., 2012; Schomburg et al., 2015; Turetzek et al., 2016, 2017). Since not a single Pax2 ortholog has been identified in any spider species so far, we analyzed several genomic and transcriptomic resources to investigate whether this Pax family member might have been entirely lost in spiders or if it diverged drastically after a duplication event. We identified two spider Pax2 orthologs that form a Pax2/5/8 specific cluster in majority consensus trees including several Pax family genes (Figure 1A and Supplementary Figures 1, 2). These analyses also provided information on the evolution of this family and the timeframe of duplication events. We found multiple Pax2 gene copies not only in the two spiders observed here but also in other haplogyne and entelegyne spiders, as well as in scorpions and horseshoe crabs. Because all vertebrate Pax2/5/8 proteins are forming a cluster separate from the insect and chelicerate orthologs, it is unlikely that the duplicated Pax2 genes in chelicerates and vertebrates share a common origin and were lost in insects (Figure 1A and Supplementary Figure 1). Except for one partial sequence from the horseshoe crab L. polyphemus, which might not contain sufficient sequence information to be grouped properly, all horseshoe crab sequences are forming a separate Pax2 cluster (Figure 1A and Supplementary Figure 8). All scorpion and spider Pax2 orthologs on the other hand group together in a Pax2.1 cluster and a Pax2.2 cluster, which is further supported by a shared amino acid exchange in the paired box domain (Figure 1B). Thus, the Pax2 gene duplications in horseshoe crabs seem to have evolved independently from other chelicerates. Our analysis predicts one duplication event in the last common ancestor of spiders and scorpions (and thus most likely at the base of Arachnopulmonata), as well as at least three duplication events in horseshoe crabs. Interestingly, within scorpions we only detected a Pax2.2 paralog in M. martensii, but not in C. sculpturatus. This is not necessarily contradicting our hypothesis, as the duplicated protein might have been lost in this scorpion species or escaped our analysis because of the quality of the C. sculpturatus genome assembly and annotation. In general, additional high-quality genomes of arachnopulmonates are needed to resolve duplication events in this group more comprehensively. Nevertheless, recent studies on whole genome duplication events in chelicerates report both a shared whole genome duplication in the lineage leading to scorpions and spiders (Schwager et al., 2017), and three whole genome duplications in xiphosurans (Nong et al., 2021), supporting our interpretation. Interestingly, the other Pax family member Pax6, for which several studies in other arthropods also identified two paralogs, seems not to be the product of an arachnopulmonate specific duplication but could indeed date back to the last common ancestor of arthropods. Our phylogenetic analysis supports this hypothesis by suggesting that the arthropod Pax6 orthologs eyeless (ey) and twin of eyeless (toy) predate the separation of the main arthropod groups alive today (Yang et al., 2009; Keller et al., 2010; Samadi et al., 2015). This is in line with the results inferred from the large consensus tree (Supplemental Figures 1, 2), showing separate clusters of the two Pax6 paralogs (note that the support values are not as high as for most of the other Pax clusters). Our second phylogenetic analysis (Figure 1A), which focuses on a subset of insect, vertebrate and arachnopulmonate Pax family members including Pax6 supports this hypothesis as well, as two separate Pax6 clusters are present both containing insect and arachnopulmonates Pax6 duplicates. It should, however, be noted that our analysis focused on the evolution of Pax2 paralogs, and that comprehensive studies including Pax6 orthologs from representatives of all major arthropod groups are needed to further elucidate the duplication events of other Pax genes. Nevertheless our results are also consistent with recent findings that ey and toy are genetically redundant and orchestrate an arthropod-wide conserved gene regulatory network module involved in arthropod ocular head segment formation (Friedrich, 2017).
Conserved Role of Pax2
Arthropods are extremely diverse and have adapted to a multitude of different ecological niches, which is prominently reflected in the large repertoire of sense organs present in this group. Despite this diversity, most sense organs develop in a very stereotypical fashion. This is especially true for external sense organs, which arise from a single precursor cell or cell cluster which subsequently undergoes a defined number of divisions controlled by a conserved genetic toolkit (Ghysen and Dambly-Chaudiere, 1989; Hartenstein, 2005; Biffar and Stollewerk, 2015; Klann et al., 2021). Thus, studying external sense organ development in various arthropods is important to answer the question how this great diversity could evolve under such restrictive developmental conditions. For instance, D. melanogaster Pax2 occupies a key developmental role in neurogenesis. In the fruit fly, Pax2 has a dual role in sense organ development, as it is both necessary for eye development (Fu and Noll, 1997) and involved in cell divisions required to form external sense organs (Fu et al., 1998; Kavaler et al., 1999). One of the most famous examples of a conserved role of Pax genes in sense organ development is certainly Pax6, which is required for brain and especially eye development. Pax6 orthologs from a wide range of species including sponges, cnidarians and vertebrates were shown to be able to rescue mutant Pax6 phenotypes in D. melanogaster or even induce development of ectopic eyes (Halder et al., 1995; Kozmik et al., 2003; Hill et al., 2010), while their expression is lacking in developing lateral eyes in chelicerates (Blackburn et al., 2008; Schomburg et al., 2015). Contrary to Pax6, functional studies of Pax2 orthologs from arthropods other than D. melanogaster are only available for the beetle T. castaneum (Dönitz et al., 2015), and data for non-insect arthropods are missing entirely. Spiders are especially fitting to amend this deficit, since many of their unique behaviors, like web building, catching prey, communication, courtship, and stridulation, are heavily dependent on mechanoreception and vision (Foelix, 1996). Consequently, spiders possess a wealth of mechanoreceptive sense organs all over their body, among them several unique structures like trichobothria, which detect minute airflows and the extremely sensitive groups of slit sense organs, the lyriform organs (Pringle, 1955; Foelix, 1996; Barth, 2004; Barth and Stagl, 2004; Patil et al., 2006; Morley et al., 2016; Ganske and Uhl, 2018). Spider eyes have diversified in a similar manner, as they were adapted for the varied environments and ecological niches occupied by these animals (Foelix, 1996; Miether and Dunlop, 2016; Morehouse et al., 2017; Morehouse, 2020). We were thus interested which role the Pax2 duplicates might play in the development of eyes and other sense organs in spiders, and if the two identified paralogs replace Pax6 in chelicerate eye development and might be linked to diversification of the spider sense organ repertoire. We were able to show that the two Pax2 paralogs are expressed in the developing ventral neuroectoderm, brain, eyes, and appendages in spiders. We identified prominent expression in the brain and ventral neuroectoderm, which suggests a conserved role in neurogenesis. In addition, the dual role in eye and external sense organ development initially described for the fruit fly Pax2 ortholog seems to be conserved in spiders as well. Both Pax2 paralogs from P. tepidariorum, as well as one paralog from P. phalangioides are expressed in the developing appendages in a manner most likely associated with sense organ development. While one copy of the gene shows a more diverged expression compared to the literature, expression of the other paralog is very conserved and seemingly linked to both eye and external sense organ development on the appendages. We therefore termed this paralog Pax2.1.
Pax2.1 Is a Promising Candidate for Spider Eye Development
Previous studies on the retinal determination network in P. tepidariorum found that the four pairs of eyes develop from two initial eye primordial fields near the anterior and lateral furrow in the non-neurogenic head region during early embryogenesis (Schomburg et al., 2015). While the medial eyes arise from an anterior field, the three lateral eyes develop from one unique field of cells, which splits into three primordia during later embryonic development. Similar to Pax2, many of the other retinal determination network genes were found to be duplicated and as every eye type expresses a unique combination of these transcription factors, it was hypothesized that these duplications might indeed have been beneficial for the diversification of spider eyes. We find that the spatial and temporal expression pattern of Pax2.1 in P. tepidariorum is comparable to that of previously identified retinal determination genes. This suggests that the lateral eyes are not only defined by dachshund 2 and six3.2, but also by Pax2.1, while sine oculis 2 and dachshund 1 further define only the anterior lateral eyes and sine oculis 1 and eyes absent are required for the development of all eyes. Comparisons between the two entelegyne spiders P. tepidariorum and C. salei revealed subtle differences in the use of transcription factor combinations required for the development of the different eye types (Samadi et al., 2015; Morehouse et al., 2017). Our finding that the Pax2.1 paralog shows a comparable expression in all three lateral eyes of the more distantly related haplogyne spider P. phalangioides and P. tepidariorum thus hints at a conserved role in the specification of lateral eyes in spiders. All other genes with specific expression in lateral eyes, namely six3, otx or dachshund are either absent in the lateral eyes of one of the entelegyne spiders previously studied or were shown to express different paralogs (Samadi et al., 2015; Schomburg et al., 2015; Morehouse et al., 2017). Thus Pax2.1 is so far the first retinal determination candidate gene with a conserved expression in all lateral eyes in more than one spider species. This conserved expression is especially interesting since the lateral eyes of spiders are suggested to be homologous to the compound eyes of other arthropods (Paulus, 1979; Spreitzer and Melzer, 2003; Schomburg et al., 2015). Curiously, Pax6 seems to play a less prominent role in spider eye development, which might further point to the importance of Pax2. In C. salei the secondary eyes (lateral eyes) develop in absence of Pax6 expression (Samadi et al., 2015) and only the median eyes (principal) were hypothesized to depend on Pax6. Even more striking, in P. tepidariorum Pax6 expression is exclusively observed in the neurogenic ectoderm of the developing brain but never in any eye primordial field (Schomburg et al., 2015). Thus, it was assumed that the only function of Pax6 in this species might be the specification of the non-neurogenic ectoderm, indicated by a short expression observed very early in the anterior head rim before germband formation. A study in the horseshoe crab L. polyphemus also found that the late embryonic lateral eyes develop from areas free of Pax6 expression (Blackburn et al., 2008). Therefore, it would be interesting to study the role of Pax2 and Pax6 genes, their interaction with the other conserved retinal determination genes like six3 and eyes absent regarding eye diversification in chelicerates in more detail using functional studies. The absence of Pax6 expression from developing eyes and its expression in the developing brain of chelicerates nicely aligns with the hypothesis that the role of Pax6 in eye development might be the result of a conserved role in brain differentiation which was then co-opted for eye development. This is in contrast to the previous hypothesis about an overall conserved role of Pax6 in bilaterian eyes, which are morphologically and developmentally clearly not homologous structures (Harris, 1997; Luan et al., 2014). Our finding that Pax2 might be the predominant transcription factor for chelicerate lateral eye development indeed fits very well to previous conclusions that Pax6 and Pax2 transcription factors have interchangeable functions, especially for crystallin gene expression (Kozmik et al., 2003; Suga et al., 2010; Charlton-Perkins et al., 2021). Our findings on the Pax2 duplication and its expression bridge the gap and show that Pax2 and Pax6, as descendants of the ancestral PaxB gene, can indeed change their role in eye and brain development and furthermore provide evidence for a high level of divergence after duplication.
Pax2 Expression in External Sense Organs
Comparatively little is known about the development of non-visual sense organs in arachnids, as published data is restricted to a few studies on proneural gene function in the entelegyne spider C. salei. As explained above, hair-like external sense organs and lyriform organs cover the whole spider body, especially the appendages and are of major importance for spider specific behaviors. Previous studies revealed a great diversity in number, distribution, morphology, and function of these appendage sense organs (Foelix, 1996; Barth, 2004; Barth and Stagl, 2004; Ganske and Uhl, 2018; Schacht et al., 2020). We thus started to investigate candidate genes with known roles in sense organ development. While most of the genes required for the specification of neural clusters and lateral inhibition seem to be well conserved in C. salei (Stollewerk, 2002; Gold et al., 2009; Doeffinger et al., 2010) and P. tepidariorum (data not shown), many downstream factors did not reveal a sense organ specific expression in the appendages (data not shown). Pax2 is unique in this regard, as it acts downstream of early sense organ development and yet shows a very distinct expression pattern in the developing appendages. In both species, at least one of the Pax2 paralogs is first expressed in clusters of cells along the appendages, which later refine to a more spot-like pattern which likely represents single sense organs. This expression suggests a role in both the pre-determination of the neural sense organ clusters usually performed by proneural genes (achaete scute and atonal complex genes), as well as an involvement in the specification of the different sense organ cells (sensory neuron and accessory cells). The latter role would fit to its function in D. melanogaster, where it is required downstream of the Notch signaling pathway involved in the asymmetric cell division of the sense organ precursor cell and controls cell fate determination and differentiation toward the shaft and sheath cells (Kavaler et al., 1999). In P. tepidariorum, the strongest expression of Pax2.1 is detected in the distalmost segments of the pedipalps, chelicerae and walking legs. A previous study on the distribution of sense organs in this spider revealed that chemosensory setae aggregate on the distal segments of the appendages (especially on tibia, metatarsus and tarsus of 1st walking leg), while mechanosensory setae and slit sense organs are present on every segment (Schacht et al., 2020). This suggests that Pax2 might be involved in chemosensory hair specification in P. tepidariorum.
Taken together, the observed expression in the two spiders presented here hints to a conserved role of Pax2 in neurogenesis, especially in the development of eyes, other sense organs and the ventral nervous system in arthropods. The high level of conservation is in line with results from tardigrades (Smith et al., 2018) and insects (Fu et al., 1998; Kavaler et al., 1999; Keller et al., 2010), where Pax2 is also shown to have a role in the development of eyes, brain, and nervous system.
Pax2 Is Not Linked to the Excretory System in Spiders
In contrast to the conserved expression in the CNS and peripheral sensory nervous system, we could not find evidence for a role of Pax2 in the excretory organ system in spiders. This is in stark contrast to findings in the velvet worm E. rowelli (Franke et al., 2015) that neither identified Pax2 expression in the eyes nor the developing ventral nervous system. Although the authors report a segmental expression pattern at the base of each leg pair and later also in the terminal anal segment, they interpret this as ectodermal expression surrounding the segmental nephridial anlagen. This is especially noteworthy as velvet worms, together with tardigrades, are the sister group to arthropods and thus occupy an especially interesting phylogenetic position. In vertebrates, Pax2/5/8 genes are involved in the development of the excretory system, as has been shown in the zebrafish D. rerio, the African claw frog Xenopus laevis and the mouse M. musculus (Nornes et al., 1990; Krauss et al., 1991; Heller and Brändli, 1999). Since its role in nervous system development seems to be well conserved throughout metazoans, this raises the question, whether Pax2 plays a role in excretory system development in invertebrate animals as well. Expression studies in the lancelet Branchiostoma floridae, a non-vertebrate chordate, have confirmed Pax2 expression in the developing metanephridia (Kozmik et al., 1999). Tunicates are now known to be more closely related to vertebrates than lancelets, but do not possess an elaborate excretory system, and hence it is unsurprising that studies on Pax2 expression in the tunicates Ciona intestinalis and Oikopleura dioica mostly focus on nervous system expression (Mazet et al., 2003; Bassham et al., 2008). Thus, it seems that the involvement of Pax2 in excretory system development at least dates back to the last common ancestor of chordates. Outside of the phylum Chordata, however, the study of E. rowelli is the only peer reviewed publication reporting Pax2 expression in developing nephridial tissue (Franke et al., 2015). A somewhat comparable segmental embryonic expression of Pax2 was reported in an unidentified species of the leech genus Helobdella (Lang, 2010), but there are presently no studies following up this finding and discussing it more in detail. In addition, especially in onychophorans it is hard to distinguish if the segmental expression close to the limbs is indeed linked to the development of the nephridial anlagen. Functional studies would be required to confirm that this expression is not involved in neurogenesis of the ventral neuroectoderm. Furthermore, the Pax2/5/8 expression shown in Franke et al. (2015) seems to be very weak and images showing the area of the developing eyes, as shown for the other Pax genes, are missing. Since Pax2 is involved in neurogenesis and eye development in nearly all species studied so far, but its role in excretory system development is apparently restricted to chordates, the most parsimonious hypothesis regarding its expression in velvet worms is that Pax2 was lost from neurogenesis and eye development in this group and gained an independent role in nephridia development. Another explanation could be that the expression of Pax2 in E. rowelli is not correlated with the developing nephridia but the nervous system. Another Pax gene, pox neuro, was analyzed in E. rowelli as well and its expression was also interpreted to be in the precursors of nephridial anlagen (Franke et al., 2015). Therefore, we investigated the expression of pox neuro in P. tepidariorum as well. Similar to Pax2, the spider pox neuro ortholog (aug3.g26228) is expressed as segmental spots especially in the developing opisthosoma (Supplementary Figure 6). Since spiders do not possess any excretory systems coinciding with such a segmental pattern in the posterior body half, we would interpret this expression, as suggested for Pax2, to be associated with the developing ventral neuroectoderm. This interpretation is favorable as it has been shown that pox neuro plays a role in CNS and VNS development in D. melanogaster (Dambly-Chaudière et al., 1992). Thus, our study supports the hypothesis that Pax2 orthologs have a conserved role in neurogenesis and eye development across metazoans, while its role in excretory system formation needs to be studied further.
Diverged Expression After Duplication
Similar to other studies in spiders covering genes for eye formation, appendage development, body patterning and segmentation, we found Pax2 to be duplicated in spiders and the resulting paralogs to be divergently expressed (Pechmann et al., 2010, 2015; Schomburg et al., 2015; Turetzek et al., 2016, 2017; Schwager et al., 2017; Leite et al., 2018; Paese et al., 2018; Oda and Akiyama-Oda, 2020). Following a duplication, one copy of the gene usually keeps the conserved function, while the other copy either evolves an entirely novel function or takes over part of the initial function. This is in line with the theory that gene duplications are a probable explanation for the generation of novel genes and that division of different functions between genes ultimately facilitates evolution of novel phenotypes (Ohno, 1970). The two outcomes of this scenario are termed sub- and neofunctionalization. In the case of Pax2, the role of the paralogs is especially interesting as this gene was shown to have pleiotropic functions in several important developmental processes. As for many other examples we found that only one copy, Pt-Pax2.1, kept the conserved expression in the developing lateral eyes and appendage sense organs, which is conserved in both spiders studied. Previous analyses found that most of the retinal determination genes are duplicated, which nicely fits to the duplication of Pax2 observed in this study and is also in line with the existence of a whole genome duplication, which probably occurred in the last common ancestor of arachnopulmonates (Schwager et al., 2017). Interestingly, for most of the retinal determination genes both paralogs are still expressed in the developing eyes but are expressed in different eye types, which paralog is expressed in which eye type, however, varies between species (Samadi et al., 2015; Schomburg et al., 2015). This hints at sub- or neofunctionalization after duplication, which is most probably also linked to the evolution of eye diversification observed in arachnids. In contrast to the other retinal determination genes, we did not find any eye specific expression of the second Pax2 paralog. While Pax2.2 in the haplogyne spider P. phalangioides is only weakly expressed in the head, Pt-Pax2.2 has distinct brain expression domains. Thus, the Pax2 paralogs from the common house spider have diverged expression in the head, while Pt-Pax2.1 most likely kept the conserved role being required for eye development. Nevertheless, both paralogs still showed expression patterns in the developing ventral neuroectoderm and the developing appendages. The exact expression in these tissues, however, again diverged between paralogs in both species. This suggests a subfunctionalization of the several original Pax2 pleiotropic functions in spiders, which are split between paralogs after duplication, especially regarding its conserved function in the eyes, which is kept by only one copy, while the expression in the ventral neuroectoderm is kept by both paralogs. Although a general expression in the ventral neuroectoderm is found for the two paralogs in both spider species, the concrete pattern differs between paralogs and also between species. The expression divergence in the appendages is even more drastic. In P. tepidariorum Pax2.1 is expressed in several domains, starting in the proximal and distal most segments (coxa and tarsus) and later becoming stronger, also being present in the metatarsus and tibia of the walking legs. Its paralog, Pt-Pax2.2, is expressed weaker and starting already with 3–4 domains and thus in more segments (the developing coxa, femur, and tarsus and later also metatarsus and tibia), however this expression quickly is getting diffuse. The early expression of Pt-Pax2.2 is thus more similar to the early Pp-Pax2.1 expression which is also present in four broad domains. Later Pp-Pax2.1 expression, however, is comparable to Pt-Pax2.1, and shows spot like pattern suggesting a role in developing sense organs. Interestingly, Pp-Pax2.1 has a strong expression in the femur, which has not been observed for the other species so far. A study on the lyriform organs on the pedipalps of different spiders showed that haplogyne pholcid spiders like P. phalangioides only have two of these organs on the femur of the pedipalps compared to entelegyne spiders, which have several, especially in the distal part (Patil et al., 2006). The hypothesis emerges that Pax2 might be required for the specification of different sense organs in the different species, chemosensory setae in P. tepidariorum and, among others, lyriform organs in P. phalangioides.
Although the P. tepidariorum Pax2 paralogs are not identical in their expression, the early expression in the appendages suggests a role in sense organ development for both paralogs. This is different for the Pax2 paralogs in the haplogyne spider P. phalangioides. Pp-Pax2.2 is never expressed in a spot-like pattern, but instead exclusively in the mesoderm of the developing appendages. Such a specific mesodermal expression is in other species usually observed for the Pax family member pox meso. Previous studies showed that pox meso has a conserved expression in the developing somatic mesoderm of fly embryos and in the appendages of the onychophoran E. rowelli (Bopp et al., 1989; Franke et al., 2015). Interestingly, P. phalangioides seems to have lost the pox meso ortholog (Supplementary Figures 1, 2). Even an additional search in the P. phalangioides transcriptome using insect pox meso protein sequences did not reveal any pox meso candidate (data not shown). This loss of pox meso seems to have only occurred in P. phalangioides and not in other haplogyne or entelegyne spiders. It would thus be interesting to study Pax2 paralog expression in more spider species to reveal if this novel mesodermal expression is a species- specific neofunctionalization or if the roles of the different Pax genes are less fixed than originally thought. Functional studies will help to elucidate if Pax2 represents an example of a gene which is still evolving after duplication and to reveal a potential role of these paralogs for species specific differences in eye morphology, appendage development, and sense organ repertoire.
In general, similarly to previously studied duplications of important developmental transcription factors, our study suggests that paralogs are free to evolve somewhat distinct or even novel functions. A known example of neofunctionalization after gene duplication in spiders is the gene dachshund. The dachshund duplication happened ancestrally in arachnopulmonates and its new role in specifying the patella segment is at least conserved among spiders (Turetzek et al., 2016). As another example, the gene homothorax was duplicated in a more recent spider specific event and the new paralog shows rapid diversification of expression, different for all spider species investigated (Turetzek et al., 2017). With the duplication of Pax2 we report yet another scenario. The Pax2 paralogs most likely duplicated before the split of arachnopulmonates and thus represent an evolutionary old duplication, similar to dachshund. By comparing paralog expression patterns within and between species we revealed signs of both sub- and neofunctionalization, indicated by losing expression in the eye, but on the other hand gaining different novel expression domains (in the brain in P. tepidariorum and mesodermally in the appendages of P. phalangioides). Interestingly, in contrast to dachshund, these novel Pax2 expression domains are not conserved between species.
Conclusion
In conclusion, Pax2 seems to have a conserved role for CNS, eye, and sense organs formation in arthropods and might have replaced Pax6 as the key gene to induce lateral eye development in chelicerates. In addition, the level of paralog divergence and its conservation between different spider species does not necessarily depend on the age of the duplication event.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MJ, MIS, and FP performed the experiments. MJ and NT performed the phylogenetic analyses and wrote the manuscript. MIS contributed to writing the manuscript. NT designed and supervised the experiments and illustrated the schemes and figures. All authors have read and approved of the final version of the manuscript.
Funding
This work was supported by the postgraduate research fund paid by Queen Mary University of London to MIS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Ralf Janssen for sharing the transcriptome of Euperipatoides kanangrensis and Glomeris marginata. We also thank Felix Quade and Anna Schönauer for helpful suggestions improving staining and imaging. We thank Nikola-Michael Prpic-Schäper and Ralf Janssen for helpful discussions. We thank Nicolas Gompel for support by providing logistics, equipment and reagents. We thank Sabine Radetzki and Alina Schreiner for maintaining the spider culture. We also thank the “Center for Advanced Light Microscopy (CALM)” at the LMU Munich. Furthermore, we would like to thank Gregor Bucher for providing the microscope set-up for imaging the whole mounts.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.810077/full#supplementary-material
Supplementary Figure 1 | Unrooted 50% majority rule consensus tree calculated by MrBayes to identify spider Pax2 orthologs. Six main clusters can be identified, namely PAX2/5/8 (D. melanogaster shaven ortholog, turquoise), PAX6 (D. melanogaster eyeless ortholog, purple), PAX3/7 (D. melanogaster gooseberry ortholog, blue), PAXɑ (orange), POXN (gray), and PAX1/9 (D. melanogaster pox meso ortholog, yellow). Branch lengths indicate expected substitutions per site. Labels at the tree edges represent clade credibility values, which are a measure of the posterior probability of each clade in the tree. A full list with species names and accession number is available in Supplementary Table 2. Note: Pt-Pax6.2 is clustering with the Paxɑ group (orange) because the used accession number only represented a partial sequence of the protein and hence does not cluster with the other Pax6 orthologs. In fact, Pt-aug3.g19451 is the protein sequence of the previously identified gene (see Schomburg et al., 2015) and does indeed cluster with the other Pax6 orthologs. Species abbreviations and accession numbers or genomic and transcriptomic resources are summarized in Supplementary Table 2.
Supplementary Figure 2 | Rooted 50% majority rule consensus tree calculated by MrBayes to identify spider Pax2 orthologs similar as in Supplementary Figure 1 provided for better visibility of posterior probabilities and names. Species abbreviations and accession numbers or genomic and transcriptomic resources are summarized in Supplementary Table 2.
Supplementary Figure 3 | Embryonic expression of Pp-Pax2.1 in P. phalangioides. (A–C) Ventro-lateral view; Pp-Pax2.1 is expressed in the ventral neuroectoderm (white arrows), and in the tip of walking legs, as well as opisthosomal segments (white arrowheads). (D–F) Frontal view; the red arrowheads depict the expression of Pp-Pax2.1 in the eye primordia. (G–J) Light micrographs of flat mounted appendages. (G) Ubiquitous expression in chelicera, and (H) dot-like expression in pedipalp. (I,J) Walking legs at different developmental stages. (J) At later stages, when the legs are fully elongated, expression is still dot-like throughout the leg with two prominent clusters in the femur and the distal tip. Asterisks depict non-specific staining of the developing cuticle. Ch, chelicerae; Pp, pedipalps; L1-L4, 1st to 4th walking leg. Dark blue is NBT/BCIP Pp-Pax2.1 specific staining, Cyan is nuclear staining (Sytox signal and white light).
Supplementary Figure 4 | Pp-Pax2.2 is expressed in the appendages and the ventral neuroectoderm in P. phalangioides. (A–C) White arrowheads indicate Pp-Pax2.2 expression in the walking legs. (A,D) In younger stages (during prosomal appendage growth) and later during inversion (B,E) Pp-Pax2.2 is expressed throughout the mesoderm of the developing appendages (C,F) Pp-Pax2.2 expression weakens in the distal appendages during dorsal closure. (D–F) Pp-Pax2.2 is expressed in half circles in the ventral neuroectoderm (white arrows in E), but not in the brain at any point during embryogenesis. (G–I) Prosomal appendages flat mounted highlighting the mesodermal expression of Pp-Pax2.2. Asterisks depict non-specific staining. (A–F) Anterior is to the left; (A–C) orientation is ventro-lateral; (D–F) depicts the same embryos (for each developmental stage, respectively) in frontal aspects. Ch, chelicerae; Pp, pedipalps; L1-L4, 1st to 4th walking leg. Dark blue is NBT/BCIP Pp-Pax2.1 specific staining, Cyan is nuclear staining (Sytox signal and white light).
Supplementary Figure 5 | No Pt-Pax2.2 expression in head lobes and appendages during very early brain differentiation (onset of stage 10). Neural invagination sites are clearly visible in head lobes (A) and VNE (B). No expression can be detected in head lobes (A’). (A,B) Is nuclear DAPI staining, (A’,B’) is FAST RED signal (no signal) staining Pt-Pax2.2. (A”,B”) Are overlays of (A,A’,B,B’), respectively.
Supplementary Figure 6 | Whole mount embryos showing Pt-poxn (aug3.g26228) expression. Pt-poxn is expressed in the VNE of opisthosomal segments, starting in the first two opisthosomal segments during brain differentiation (stage 10) (white arrows in A,D). During inversion and retraction (stage 11 and 12) Pt-poxn is expressed as small dots in the VNE of each opisthosomal segment which continues during dorsal and ventral closure (C,F). Ch, chelicerae; Pp, pedipalps; L1-L4, 1st to 4th walking leg. Dark blue is NBT/BCIP Pt-poxn specific staining, Cyan is nuclear staining (Sytox signal and white light).
Supplementary Figure 7 | Separated Z-projections of Pt-Pax2.1 expression during brain differentiation (Stage 10). Red ellipses mark Pt-Pax2.1 expression domains in the eye primordia white dotted line marks the anterior furrow in the head lobes for orientation. Left panels (A–A”) show the upper anterior half of the same embryo as shown in Figure 3A Pt-Pax2.1 is expression is visible in the developing lateral eye primordia next to the lateral furrow (red ellipse) and in proximal and distal spots in the appendages (arrowheads). (B–B”) Same embryo as in Figures 3A–A” this time the lower half Z-stack was projected to visualize Pt-Pax2.1 the expression in the VNE (arrows). Flat mounted preparations; anterior is to the left in all panels. Cyan is nuclear DAPI staining; magenta is Pt-Pax2.1 specific expression detected with FAST RED. Scale bar represents 50 μm.
Supplementary Figure 8 | Unrooted 50% majority rule consensus tree calculated by MrBayes. For this phylogenetic analysis only alignable regions of the protein sequences of the previously identified chelicerate Pax2 candidates were used (accessible as nexus file in Supplementary Data Sheet 3). The Jones amino acid substitution model was used to calculate 18 trees in 2 files of which 14 were sampled after reaching an average standard deviation of split frequencies of 0.00918 after 4,000 generations. Similar to the phylogenetic analysis in Figure 1, using the full-length protein sequences, this phylogenetic analysis also groups the spider and scorpion Pax2 candidates into two separate arachnopulmonate Pax2 clusters, Pax2.1, and Pax2.2.
Supplementary Table 1 | List of gene specific primer used to clone gene fragments of Pax2.1 and Pax2.2 of P. tepidariorum and P. phalangioides.
Supplementary Table 2 | Full lists of sequence information including species, accession number/Transcript ID and source used for phylogenetic analysis in Supplementary Figures 1, 2 (Sheet1:PAX148) and Figure 1 (Sheet2:PAX65). Ordered for Pax subfamilies indicated by same colour code as in used in Supplementary Figures 1, 2.
Footnotes
References
Barth, F., and Stagl, J. (2004). The slit sense organs of arachnids. Zoomorphologie 86, 1–23. doi: 10.1007/bf01006710
Bassham, S., Cañestro, C., and Postlethwait, J. H. (2008). Evolution of developmental roles of Pax2/5/8paralogs after independent duplication in urochordate and vertebrate lineages. BMC Biol. 6:35. doi: 10.1186/1741-7007-6-35
Biffar, L., and Stollewerk, A. (2015). Evolutionary variations in the expression of dorso-ventral patterning genes and the conservation of pioneer neurons in Tribolium castaneum. Dev. Biol. 400, 159–167. doi: 10.1016/j.ydbio.2015.01.025
Blackburn, D. C., Conley, K. W., Plachetzki, D. C., Kempler, K., Battelle, B.-A., and Brown, N. L. (2008). Isolation and expression of Pax6 and atonal homologues in the American horseshoe crab, Limulus polyphemus. Dev. Dyn. 237, 2209–2219. doi: 10.1002/dvdy.21634
Bopp, D., Burri, M., Baumgartner, S., Frigerio, G., and Noll, M. (1986). Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell 47, 1033–1040. doi: 10.1016/0092-8674(86)90818-4
Bopp, D., Jamet, E., Baumgartner, S., Burri, M., and Noll, M. (1989). Isolation of two tissue-specific Drosophila paired box genes, Pox meso and Pox neuro. EMBO J. 8, 3447–3457.
Burri, M., Tromvoukis, Y., Bopp, D., Frigerio, G., and Noll, M. (1989). Conservation of the paired domain in metazoans and its structure in three isolated human genes. EMBO J. 8, 1183–1190.
Burton, Q., Cole, L. K., Mulheisen, M., Chang, W., and Wu, D. K. (2004). The role of Pax2 in mouse inner ear development. Dev. Biol. 272, 161–175. doi: 10.1016/j.ydbio.2004.04.024
Cao, Z., Yu, Y., Wu, Y., Hao, P., Di, Z., He, Y., et al. (2013). The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat. Commun. 4:2602. doi: 10.1038/ncomms3602
Charlton-Perkins, M., Whitaker, S. L., Fei, Y., Xie, B., Li-Kroeger, D., Gebelein, B., et al. (2011). Prospero and Pax2 combinatorially control neural cell fate decisions by modulating Ras- and Notch-dependent signaling. Neural Dev. 6:20. doi: 10.1186/1749-8104-6-20
Charlton-Perkins, M. A., Friedrich, M., and Cook, T. A. (2021). Semper’s cells in the insect compound eye: insights into ocular form and function. Dev. Biol. 479, 126–138. doi: 10.1016/j.ydbio.2021.07.015
Conzelmann, M., Williams, E. A., Krug, K., Franz-Wachtel, M., Macek, B., and Jékely, G. (2013). The neuropeptide complement of the marine annelid Platynereis dumerilii. BMC Genomics 14:906. doi: 10.1186/1471-2164-14-906
Dambly-Chaudière, C., Jamet, E., Burri, M., Bopp, D., Basler, K., Hafen, E., et al. (1992). The paired box gene pox neuro: a determiant of poly-innervated sense organs in Drosophila. Cell 69, 159–172. doi: 10.1016/0092-8674(92)90127-x
Doeffinger, C., Hartenstein, V., and Stollewerk, A. (2010). Compartmentalisation of the precheliceral neuroectoderm in the spider Cupiennius salei: development of the arcuate body, the optic ganglia and the mushroom body. J. Comp. Neurol. 518, 2612–2632.
Dönitz, J., Schmitt-Engel, C., Grossmann, D., Gerischer, L., Tech, M., Schoppmeier, M., et al. (2015). iBeetle-Base: a database for RNAi phenotypes in the red flour beetle Tribolium castaneum. Nucleic Acids Res. 43, D720–D725. doi: 10.1093/nar/gku1054
Dressler, G. R., Deutsch, U., Chowdhury, K., Nornes, H. O., and Gruss, P. (1990). Pax2, a new murine paired-box-containing gene and its expression in the developing excretory system. Development 109, 787–795. doi: 10.1242/dev.109.4.787
Feiner, N., Meyer, A., and Kuraku, S. (2014). Evolution of the vertebrate Pax4/6 class of genes with focus on its novel member, the Pax10 gene. Genome Biol. Evol. 6, 1635–1651. doi: 10.1093/gbe/evu135
Franke, F. A., Schumann, I., Hering, L., and Mayer, G. (2015). Phylogenetic analysis and expression patterns of Pax genes in the onychophoran Euperipatoides rowelli reveal a novel bilaterian Pax subfamily: Pax genes in Onychophora (velvet worms). Evol. Dev. 17, 3–20. doi: 10.1111/ede.12110
Friedrich, M. (2015). Evo-Devo gene toolkit update: at least seven Pax transcription factor subfamilies in the last common ancestor of bilaterian animals. Evol. Dev. 17, 255–257. doi: 10.1111/ede.12137
Friedrich, M. (2017). Ancient genetic redundancy of eyeless and twin of eyeless in the arthropod ocular segment. Dev. Biol. 432, 192–200. doi: 10.1016/j.ydbio.2017.10.001
Frigerio, G., Burri, M., Bopp, D., Baumgartner, S., and Noll, M. (1986). Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell 47, 735–746. doi: 10.1016/0092-8674(86)90516-7
Fu, W., Duan, H., Frei, E., and Noll, M. (1998). shaven and sparkling are mutations in separate enhancers of the Drosophila Pax2 homolog. Development 125, 2943–2950. doi: 10.1242/dev.125.15.2943
Fu, W., and Noll, M. (1997). The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 11, 2066–2078. doi: 10.1101/gad.11.16.2066
Ganske, A.-S., and Uhl, G. (2018). The sensory equipment of a spider – A morphological survey of different types of sensillum in both sexes of Argiope bruennichi (Araneae, Araneidae). Arthropod Struct. Dev. 47, 144–161. doi: 10.1016/j.asd.2018.01.001
Ghysen, A., and Dambly-Chaudiere, C. (1989). Genesis of the Drosophila peripheral nervous system. Trends Genet. 5, 251–255.
Gold, K., Cotton, J. A., and Stollewerk, A. (2009). The role of Notch signalling and numb function in mechanosensory organ formation in the spider Cupiennius salei. Dev. Biol. 327, 121–131. doi: 10.1016/j.ydbio.2008.12.004
Halder, G., Callaerts, P., and Gehring, W. (1995). Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267, 1788–1792. doi: 10.1126/science.7892602
Harris, W. A. (1997). Pax-6: where to be conserved is not conservative. PNAS 94, 2098–2100. doi: 10.1073/pnas.94.6.2098
Hartenstein, V. (2005). “1.11 - development of insect sensilla*,” in Comprehensive Molecular Insect Science, ed. L. I. Gilbert (Amsterdam: Elsevier), 379–419. doi: 10.1016/b0-44-451924-6/00012-0
Heller, N., and Brändli, A. W. (1999). Xenopus Pax-2/5/8 orthologues: novel insights into Pax Gene evolution and identification of Pax-8 as the earliest marker for otic and pronephric cell lineages. Dev. Genet. 24, 208–219. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<208::AID-DVG4>3.0.CO;2-J
Hilbrant, M., Damen, W. G. M., and McGregor, A. P. (2012). Evolutionary crossroads in developmental biology: the spider Parasteatoda tepidariorum. Development 139, 2655–2662. doi: 10.1242/dev.078204
Hill, A., Boll, W., Ries, C., Warner, L., Osswalt, M., Hill, M., et al. (2010). Origin of Pax and Six gene families in sponges: single PaxB and Six1/2 orthologs in Chalinula loosanoffi. Dev. Biol. 343, 106–123. doi: 10.1016/j.ydbio.2010.03.010
Hirth, F., Kammermeier, L., Frei, E., Walldorf, U., Noll, M., and Reichert, H. (2003). An urbilaterian origin of the tripartite brain: developmental genetic insights from Drosophila. Development 130, 2365–2373. doi: 10.1242/dev.00438
Hoshiyama, D., Suga, H., Iwabe, N., Koyanagi, M., Nikoh, N., Kuma, K., et al. (1998). Sponge Pax cDNA related to Pax-2/5/8 and ancient gene duplications in the Pax family. J. Mol. Evol. 47, 640–648. doi: 10.1007/pl00006421
Hou, Y.-N., Li, S., and Luan, Y.-X. (2016). Pax6 in collembola: adaptive evolution of eye regression. Sci. Rep. 6:20800. doi: 10.1038/srep20800
Janssen, R., Schönauer, A., Weber, M., Turetzek, N., Hogvall, M., Goss, G. E., et al. (2015). The evolution and expression of panarthropod frizzled genes. Front. Ecol. Evol. 3:96. doi: 10.3389/fevo.2015.00096
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Kavaler, J., Fu, W., Duan, H., Noll, M., and Posakony, J. W. (1999). An essential role for the Drosophila Pax2 homolog in the differentiation of adult sensory organs. Development 126, 2261–2272.
Keller, R. G., Desplan, C., and Rosenberg, M. I. (2010). Identification and characterization of Nasonia Pax genes. Insect Mol. Biol. 19, 109–120. doi: 10.1111/j.1365-2583.2009.00921.x
Khadjeh, S., Turetzek, N., Pechmann, M., Schwager, E. E., Wimmer, E. A., Damen, W. G., et al. (2012). Divergent role of the Hox gene Antennapedia in spiders is responsible for the convergent evolution of abdominal limb repression. Proc. Natl. Acad. Sci. U.S.A. 109, 4921–4926. doi: 10.1073/pnas.1116421109
Klann, M., Schacht, M. I., Benton, M. A., and Stollewerk, A. (2021). Functional analysis of sense organ specification in the Tribolium castaneum larva reveals divergent mechanisms in insects. BMC Biol. 19:22. doi: 10.1186/s12915-021-00948-y
Kono, N., Nakamura, H., Ohtoshi, R., Moran, D. A. P., Shinohara, A., Yoshida, Y., et al. (2019). Orb-weaving spider Araneus ventricosus genome elucidates the spidroin gene catalogue. Sci. Rep. 9:8380. doi: 10.1038/s41598-019-44775-2
Kozmik, Z., Daube, M., Frei, E., Norman, B., Kos, L., Dishaw, L. J., et al. (2003). Role of Pax genes in eye evolution: a cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev. Cell 5, 773–785. doi: 10.1016/s1534-5807(03)00325-3
Kozmik, Z., Holland, N. D., Kalousova, A., Paces, J., Schubert, M., and Holland, L. Z. (1999). Characterization of an amphioxus paired box gene, AmphiPax2/5/8: developmental expression patterns in optic support cells, nephridium, thyroid-like structures and pharyngeal gill slits, but not in the midbrain-hindbrain boundary region. Development 126, 1295–1304. doi: 10.1242/dev.126.6.1295
Krauss, S., Johansen, T., Korzh, V., and Fjose, A. (1991). Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development 113, 1193–1206. doi: 10.1242/dev.113.4.1193
Lang, B. (2010). Expression of Pax-2/5/8 in Helobdella sp. (Austin). Thesis. Austin, TX: The University of Texas.
Leite, D. J., Baudouin-Gonzalez, L., Iwasaki-Yokozawa, S., Lozano-Fernandez, J., Turetzek, N., Akiyama-Oda, Y., et al. (2018). Homeobox gene duplication and divergence in arachnids. Mol. Biol. Evol. 35, 2240–2253. doi: 10.1093/molbev/msy125
Liao, Y. Y., Xu, P. W., Kwan, K. Y., Ma, Z. Y., Fang, H. Y., Xu, J. Y., et al. (2019). Draft genomic and transcriptome resources for marine chelicerate Tachypleus tridentatus. Sci. Data 6:190029. doi: 10.1038/sdata.2019.29
Liu, F., Li, Y., Yu, H., Zhang, L., Hu, J., Bao, Z., et al. (2021). MolluscDB: an integrated functional and evolutionary genomics database for the hyper-diverse animal phylum Mollusca. Nucleic Acids Res. 49, D988–D997. doi: 10.1093/nar/gkaa918
Liu, S., Aagaard, A., Bechsgaard, J., and Bilde, T. (2019). DNA methylation patterns in the social spider, Stegodyphus dumicola. Genes 10:137. doi: 10.3390/genes10020137
Luan, Q., Chen, Q., and Friedrich, M. (2014). The Pax6 genes eyeless and twin of eyeless are required for global patterning of the ocular segment in the Tribolium embryo. Dev. Biol. 394, 367–381. doi: 10.1016/j.ydbio.2014.08.005
Mazet, F., Hutt, J. A., Millard, J., and Shimeld, S. M. (2003). Pax gene expression in the developing central nervous system of Ciona intestinalis. Gene Expr. Patterns 3, 743–745. doi: 10.1016/s1567-133x(03)00137-6
Miether, S. T., and Dunlop, J. A. (2016). Lateral eye evolution in the arachnids. Arachnology 17, 103–119.
Mittmann, B., and Wolff, C. (2012). Embryonic development and staging of the cobweb spider Parasteatoda tepidariorum C. L. Koch, 1841 (syn.: Achaearanea tepidariorum; Araneomorphae; Theridiidae). Dev. Genes Evol. 222, 189–216. doi: 10.1007/s00427-012-0401-0
Morehouse, N. I., Buschbeck, E. K., Zurek, D. B., Steck, M., and Porter, M. L. (2017). Molecular evolution of spider vision: new opportunities, familiar players. Biol. Bull. 233, 21–38. doi: 10.1086/693977
Morley, E. L., Sivalinghem, S., and Mason, A. C. (2016). Developmental morphology of a lyriform organ in the Western black widow (Latrodectus hesperus). Zoomorphology 135, 433–440. doi: 10.1007/s00435-016-0324-9
Nong, W., Qu, Z., Li, Y., Barton-Owen, T., Wong, A. Y. P., Yip, H. Y., et al. (2021). Horseshoe crab genomes reveal the evolution of genes and microRNAs after three rounds of whole genome duplication. Commun. Biol. 4, 1–11. doi: 10.1038/s42003-020-01637-2
Nornes, H. O., Dressler, G. R., Knapik, E. W., Deutsch, U., and Gruss, P. (1990). Spatially and temporally restricted expression of Pax2 during murine neurogenesis. Development 109, 797–809.
O’brien, E. K., and Degnan, B. M. (2003). Expression of Pax258 in the gastropod statocyst: insights into the antiquity of metazoan geosensory organs. Evol. Dev. 5, 572–578. doi: 10.1046/j.1525-142x.2003.03062.x
Oda, H., and Akiyama-Oda, Y. (2020). The common house spider Parasteatoda tepidariorum. EvoDevo 11:6. doi: 10.1186/s13227-020-00152-z
Paese, C. L. B., Schoenauer, A., Leite, D. J., Russell, S., and McGregor, A. P. (2018). A SoxB gene acts as an anterior gap gene and regulates posterior segment addition in a spider. eLife 7:e37567. doi: 10.7554/eLife.37567
Patil, B., Prabhu, S., and Rajashekhar, K. P. (2006). Lyriform slit sense organs on the pedipalps and spinnerets of spiders. J. Biosci. 31, 75–84. doi: 10.1007/BF02705238
Paulus, H. F. (1979). Eye structure and the monophyly of the arthropod eye. Arthropod Phylogeny 5, 299–383.
Pechmann, M. (2020). Embryonic development and secondary axis induction in the Brazilian white knee tarantula Acanthoscurria geniculata, C. L. Koch, 1841 (Araneae; Mygalomorphae; Theraphosidae). Dev. Genes Evol. 230, 75–94. doi: 10.1007/s00427-020-00653-w
Pechmann, M., Khadjeh, S., Sprenger, F., and Prpic, N.-M. (2010). Patterning mechanisms and morphological diversity of spider appendages and their importance for spider evolution. Arthropod Struct. Dev. 39, 453–467. doi: 10.1016/j.asd.2010.07.007
Pechmann, M., Khadjeh, S., Turetzek, N., McGregor, A. P., Damen, W. G. M., and Prpic, N.-M. (2011). Novel function of distal-less as a gap gene during spider segmentation. PLoS Genet. 7:e1002342. doi: 10.1371/journal.pgen.1002342
Pechmann, M., Schwager, E. E., Turetzek, N., and Prpic, N.-M. (2015). Regressive evolution of the arthropod tritocerebral segment linked to functional divergence of the Hox gene labial. Proc. R. Soc. B 282:20151162. doi: 10.1098/rspb.2015.1162
Plachov, D., Chowdhury, K., Walther, C., Simon, D., Guenet, J. L., and Gruss, P. (1990). Pax8, a murine paired box gene expressed in the developing excretory system and thyroid gland. Development 110, 643–651. doi: 10.1242/dev.110.2.643
Pringle, J. W. S. (1955). The function of the lyriform organs of arachnids. J. Exp. Biol. 32, 270–278. doi: 10.1242/jeb.32.2.270
Prpic, N.-M. (2005). Duplicated Pax6 genes in Glomeris marginata (Myriapoda: Diplopoda), an arthropod with simple lateral eyes. Zoology 108, 47–53. doi: 10.1016/j.zool.2004.11.003
Prpic, N.-M., Schoppmeier, M., and Damen, W. G. M. (2008). Whole-mount in situ hybridization of spider embryos. CSH Protoc. 2008:5068. doi: 10.1101/pdb.prot5068
Quiring, R., Walldorf, U., Kloter, U., and Gehring, W. J. (1994). Homology of the eyeless gene of Drosophila to the small eye gene in mice and aniridia in humans. Science 265, 785–789. doi: 10.1126/science.7914031
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sajuthi, A., Carrillo-Zazueta, B., Hu, B., Wang, A., Brodnansky, L., Mayberry, J., et al. (2015). Sexually dimorphic gene expression in the lateral eyes of Euphilomedes carcharodonta (Ostracoda, Pancrustacea). EvoDevo 6:34. doi: 10.1186/s13227-015-0026-2
Samadi, L., Schmid, A., and Eriksson, B. J. (2015). Differential expression of retinal determination genes in the principal and secondary eyes of Cupiennius salei Keyserling (1877). EvoDevo 6:16. doi: 10.1186/s13227-015-0010-x
Schacht, M. I., Francesconi, M., and Stollewerk, A. (2020). Distribution and development of the external sense organ pattern on the appendages of postembryonic and adult stages of the spider Parasteatoda tepidariorum. Dev. Genes Evol. 230, 121–136. doi: 10.1007/s00427-020-00655-8
Scherholz, M., Redl, E., Wollesen, T., de Oliveira, A. L., Todt, C., and Wanninger, A. (2017). Ancestral and novel roles of Pax family genes in mollusks. BMC Evol. Biol. 17:81. doi: 10.1186/s12862-017-0919-x
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schomburg, C., Turetzek, N., Schacht, M. I., Schneider, J., Kirfel, P., Prpic, N.-M., et al. (2015). Molecular characterization and embryonic origin of the eyes in the common house spider Parasteatoda tepidariorum. EvoDevo 6:15.
Schoppmeier, M., and Damen, W. G. M. (2005). Expression of Pax group III genes suggests a single-segmental periodicity for opisthosomal segment patterning in the spider Cupiennius salei. Evol. Dev. 7, 160–169. doi: 10.1111/j.1525-142X.2005.05018.x
Schwager, E. E., Sharma, P. P., Clarke, T., Leite, D. J., Wierschin, T., Pechmann, M., et al. (2017). The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 15:62. doi: 10.1186/s12915-017-0399-x
Sheffer, M. M., Hoppe, A., Krehenwinkel, H., Uhl, G., Kuss, A. W., Jensen, L., et al. (2021). Chromosome-level reference genome of the European wasp spider Argiope bruennichi: a resource for studies on range expansion and evolutionary adaptation. GigaScience 10:giaa148. doi: 10.1093/gigascience/giaa148
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Smith, F. W., Cumming, M., and Goldstein, B. (2018). Analyses of nervous system patterning genes in the tardigrade Hypsibius exemplaris illuminate the evolution of panarthropod brains. EvoDevo 9:19. doi: 10.1186/s13227-018-0106-1
Spreitzer, A., and Melzer, R. R. (2003). The nymphal eyes of Parabuthus transvaalicus Purcell, 1899 (Buthidae): an accessory lateral eye in a scorpion. Zool. Anzeiger J. Comp. Zool. 242, 137–143.
Stollewerk, A. (2002). Recruitment of cell groups through Delta/Notch signalling during spider neurogenesis. Development 129, 5339–5348. doi: 10.1242/dev.00109
Suga, H., Tschopp, P., Graziussi, D. F., Stierwald, M., Schmid, V., and Gehring, W. J. (2010). Flexibly deployed Pax genes in eye development at the early evolution of animals demonstrated by studies on a hydrozoan jellyfish. PNAS 107, 14263–14268. doi: 10.1073/pnas.1008389107
The UniProt Consortium (2015). UniProt: a hub for protein information. Nucl. Acids Res. 43, D204–D212. doi: 10.1093/nar/gku989
Torres, M., Gomez-Pardo, E., Dressler, G. R., and Gruss, P. (1995). Pax-2 controls multiple steps of urogenital development. Development 121, 4057–4065. doi: 10.1242/dev.121.12.4057
Torres, M., Gomez-Pardo, E., and Gruss, P. (1996). Pax2 contributes to inner ear patterning and optic nerve trajectory. Development 122, 3381–3391. doi: 10.1242/dev.122.11.3381
Turetzek, N., Khadjeh, S., Schomburg, C., and Prpic, N.-M. (2017). Rapid diversification of homothorax expression patterns after gene duplication in spiders. BMC Evol. Biol. 17:168. doi: 10.1186/s12862-017-1013-0
Turetzek, N., Pechmann, M., Schomburg, C., Schneider, J., and Prpic, N.-M. (2016). Neofunctionalization of a duplicate dachshund gene underlies the evolution of a novel leg segment in arachnids. Mol. Biol. Evol. 33, 109–121. doi: 10.1093/molbev/msv200
Turetzek, N., and Prpic, N.-M. (2016). Observations on germ band development in the cellar spider Pholcus phalangioides. Dev. Genes Evol. 226, 413–422. doi: 10.1007/s00427-016-0562-3
Vizueta, J., Frías-López, C., Macías-Hernández, N., Arnedo, M. A., Sánchez-Gracia, A., and Rozas, J. (2017). Evolution of chemosensory gene families in arthropods: insight from the first inclusive comparative transcriptome analysis across spider appendages. Genome Biol. Evol. 9, 178–196. doi: 10.1093/gbe/evw296
Wada, H., Saiga, H., Satoh, N., and Holland, P. W. (1998). Tripartite organization of the ancestral chordate brain and the antiquity of placodes: insights from ascidian Pax-2/5/8. Hox and Otx genes. Development 125, 1113–1122.
Wollesen, T., Rodríguez Monje, S. V., Todt, C., Degnan, B. M., and Wanninger, A. (2015). Ancestral role of Pax2/5/8 in molluscan brain and multimodal sensory system development. BMC Evol. Biol. 15:231. doi: 10.1186/s12862-015-0505-z
Keywords: gene duplication, Pax genes, EvoDevo, eyes, neo-/subfunctionalization, spiders
Citation: Janeschik M, Schacht MI, Platten F and Turetzek N (2022) It takes Two: Discovery of Spider Pax2 Duplicates Indicates Prominent Role in Chelicerate Central Nervous System, Eye, as Well as External Sense Organ Precursor Formation and Diversification After Neo- and Subfunctionalization. Front. Ecol. Evol. 10:810077. doi: 10.3389/fevo.2022.810077
Received: 06 November 2021; Accepted: 05 January 2022;
Published: 17 February 2022.
Edited by:
Linda C. Weiss, Ruhr University Bochum, GermanyReviewed by:
Yasuko Akiyama-Oda, JT Biohistory Research Hall, JapanMarkus Friedrich, Wayne State University, United States
Copyright © 2022 Janeschik, Schacht, Platten and Turetzek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natascha Turetzek, zhang@bio.lmu.de
†These authors have contributed equally to this work
‡ORCID: Matthias Janeschik, orcid.org/0000-0001-9689-7059; Magdalena Ines Schacht, orcid.org/0000-0002-9689-3864; Fabian Platten, orcid.org/0000-0003-4000-5079; Natascha Turetzek, orcid.org/0000-0002-5681-2177
 Matthias Janeschik
Matthias Janeschik Magdalena Ines Schacht
Magdalena Ines Schacht Fabian Platten
Fabian Platten Natascha Turetzek
Natascha Turetzek