Taxonomic Identification of Two Poorly Known Lantern Shark Species Based on Mitochondrial DNA From Wet-Collection Paratypes
- 1Evolutionary Adaptive Genomics, Department of Mathematics and Natural Sciences, Institute for Biochemistry and Biology, University of Potsdam, Potsdam, Germany
- 2Florida Museum of Natural History, University of Florida, Gainesville, FL, United States
- 3Centre for Taxonomy and Morphology, Zoological Museum, Leibniz Institute for the Analysis of Biodiversity Change (LIB), Hamburg, Germany
- 4Department of Biology, Biodiversity Research, University of Hamburg, Hamburg, Germany
- 5Elasmo-Lab, Elasmobranch Research Laboratory, Hamburg, Germany
- 6Department of Zoology, University of Cambridge, Cambridge, United Kingdom
- 7School of Natural Sciences, Bangor University, Bangor, United Kingdom
- 8Department of Natural History, University Museum of Bergen, Bergen, Norway
Etmopteridae (lantern sharks) is the most species-rich family of sharks, comprising more than 50 species. Many species are described from few individuals, and re-collection of specimens is often hindered by the remoteness of their sampling sites. For taxonomic studies, comparative morphological analysis of type specimens housed in natural history collections has been the main source of evidence. In contrast, DNA sequence information has rarely been used. Most lantern shark collection specimens, including the types, were formalin fixed before long-term storage in ethanol solutions. The DNA damage caused by both fixation and preservation of specimens has excluded these specimens from DNA sequence-based phylogenetic analyses so far. However, recent advances in the field of ancient DNA have allowed recovery of wet-collection specimen DNA sequence data. Here we analyse archival mitochondrial DNA sequences, obtained using ancient DNA approaches, of two wet-collection lantern shark paratype specimens, namely Etmopterus litvinovi and E. pycnolepis, for which the type series represent the only known individuals. Target capture of mitochondrial markers from single-stranded DNA libraries allows for phylogenetic placement of both species. Our results suggest synonymy of E. benchleyi with E. litvinovi but support the species status of E. pycnolepis. This revised taxonomy is helpful for future conservation and management efforts, as our results indicate a larger distribution range of E. litvinovi. This study further demonstrates the importance of wet-collection type specimens as genetic resource for taxonomic research.
Introduction
Shark diversity is poorly represented in the scientific literature. Shark biologists have tended to focus on a few easy-to-access taxa that are assumed to be representative of the groups to which they belong. For example, though there are more than 40 different species of deep-sea lantern sharks (genus Etmopterus), nearly a quarter of the 2082 publications devoted to Lantern shark biology (Pollerspöck and Straube, 2021) has focussed on a single species (Etmopterus spinax). Thus, most of the diversity of this group remains relatively unexplored (Figure 1). To make matters worse, a substantial fraction of lantern shark diversity is known only from formalin preserved type material that was collected prior to the advent of DNA sequencing. Hence, tissue sampling, common practice today for performing DNA sequence-based analysis such as DNA barcoding (Hebert et al., 2003), was not conducted and fixation in formaldehyde and preservation in ethanol causes DNA damage (Gilbert et al., 2007; Hoffman et al., 2015; Hykin et al., 2015; Stiller et al., 2016; McGuire et al., 2018; Hahn et al., 2021). This means that, while we know that the group has diversified extensively (e.g., Straube et al., 2011a; Ebert et al., 2016, 2021; White et al., 2017; Dolganov and Balanov, 2018), it has been hard to decipher how the different species are related to one another and how different ecological pressures have contributed to their diversification. Recently developed tools allow us to obtain DNA sequence data from formalin preserved animals (Gansauge et al., 2017; Hahn et al., 2021; Straube et al., 2021a). In the current contribution we have applied these tools to type material for two species of Etmopterus and show how the data collected have implications, not only for understanding their taxonomy and evolution, but also their ranges, which has consequences for their conservation and management. The genus is subdivided into four clades supported by both DNA sequence data and morphological characters (Straube et al., 2010). Morphological characters therefore allow for tentative assignments of species lacking DNA sequence information to one of the four clades. Our first target species, Etmopterus litvinovi (Smalleye lantern shark, Parin and Kotlyar, 1990) has been assigned to the E. spinax clade (Straube et al., 2010, 2011a) comprising 11 species today (Ebert et al., 2021). The presence and shape of flank markings, dark patterns above the pelvic fins, is a key character allowing for species-to-clade assignments in many Etmopterus species. While the character is not present in all species and ontogenetic stages, every species of the E. lucifer clade shows distinct flank markings characterised by anterior and posterior branches. Species of the E. lucifer clade can further be subdivided into three subclades based on length comparisons of the anterior and posterior flank mark branches (Ebert et al., 2021). The three subclades are the E. lucifer, the E. molleri and the E. burgessi subclades. The E. lucifer subclade includes the four species E. brosei, E. lailae, E. lucifer and E. sculptus. E. alphus, E. brachyurus, E. bullisi, E. decacuspidatus, E. dislineatus, E. molleri, and E. samadiae are the seven species assigned to the E. molleri subclade. The E. burgessi subclade comprises four species, namely E. burgessi, E. evansi, E. marshae, and E. pycnolepis (Ebert et al., 2021). Etmopterus pycnolepis (Dense-scale lantern shark, Kotlyar, 1990) is our second target species. Both E. litvinovi and E. pycnolepis are known from their type specimens only and little is known regarding their biology as they were hitherto sampled only once each in the Salas y Gómez and Nazca submarine ridges in the Southeast Pacific (Kotlyar, 1990; Ebert et al., 2013).
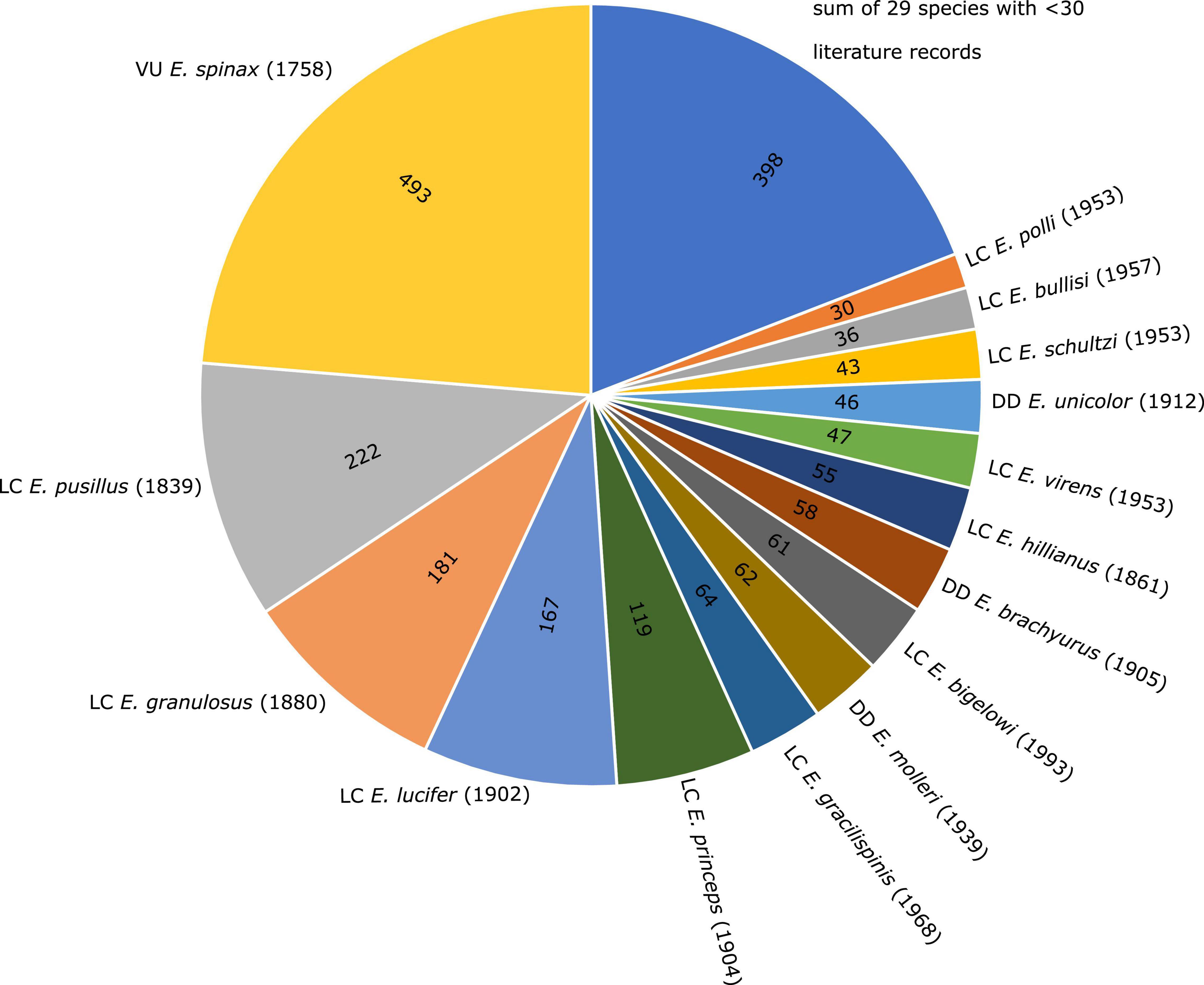
Figure 1. Pie chart showing the number of scientific publications listed per Etmopterus species in the bibliographic database Shark References. Species with a total number of publications below 30 are summarised. Global IUCN Red List of Threatened Species status (VU = vulnerable; LC = least concern; DD = data deficient) is given in front of species names, year of description in brackets.
Materials and Methods
Etmopterus litvinovi (Smalleye Lantern Shark)
This species is known from 32 type specimens housed in three different museum collections, the Laboratory of Ichthyology at the Zoological Institute of the Russian Academy of Sciences (ZIN), St. Petersburg, Russia (holotype: ZIN 49228; six paratypes: ZIN 49229–32), the Zoological Museum (ZMMU), Biological Faculty, M. V. Lomonosov Moscow State University, Moscow, Russia (21 paratypes ZMMU: P-17989–91; two paratypes P-18222) and the ichthyological collection of the Zoological Museum (ZMH) of the LIB in Hamburg, Germany [paratype ZMH 24994 (ex ISH 6-1989); paratype ZMH 24993 (ex ISH 5-1989)]. We sampled muscle tissue from the paratype specimen ZMH 24994 (Figure 2A) at the caudal peduncle using a biopsy needle for minimally invasive sampling. The tissue was preserved in the original preservation fluid of the storage container. The specimen was captured at 25°21′S and 85°8′W at a depth of 720 m on 24.04.1987. It is a juvenile male of 445 mm total length (Thiel et al., 2009; Straube et al., 2011a). Although not explicitly mentioned in the original description, or tested by us, the overall condition of the specimen indicates a fixation in formaldehyde: both body and eyes do not show bleaching of exclusively ethanol preserved samples. Furthermore, the common procedure during research cruises at the time of sampling was a fixation of specimens in 4% formaldehyde and long-term preservation in 70% ethanol.

Figure 2. Paratype images of (A) Etmopterus litvinovi (ZMH 24994) and (B) Etmopterus pycnolepis (ZMH 24995). Bars indicate 1 cm.
Laboratory steps and analysis of test-sequencing data of this specimen is described in detail in Straube et al. (2021a). The sample was incubated in a GuSCN-based buffer (Rohland et al., 2004) applying the protocol by Dabney et al. (2013) for DNA purification. A single-stranded DNA library was then constructed, and test-sequencing was performed to check for the ratio of target DNA and contamination. After detection of endogenous DNA in the test-sequencing dataset, target capture for mitochondrial DNA was performed using home-made baits. These were generated from long-range PCR products amplified from the DNA of Etmopterus cf. molleri tissue housed in the tissue sample collection of the Bavarian State Collection of Zoology (registration number: Ich-P-CH-0264). For the long-range PCR protocol and primers see Straube et al. (2021a). Hybridisation capture was then performed following the protocol of González Fortes and Paijmans (2019), where the single-stranded library is mixed with the denatured bait library after addition of blocking oligos. Hybridisation of target DNA to baits was carried out for 24 h at 65°C. The captured library was then amplified, and the capture procedure and amplification repeated. The resulting double captured library was then sequenced using custom sequencing and index 2 read primers (Gansauge and Meyer, 2013; Paijmans et al., 2017) on an Illumina® MiniSeq instrument. We used a mid-output kit in a pool of double indexed samples.
Paired-end raw reads were quality and adapter trimmed with Cutadapt v.1.16 (Martin, 2011) using default settings. The iterative mapping algorithm MitoBim v. 1.9.1 (Hahn et al., 2013) was then used to reconstruct the mitochondrial genome sequence, using default settings and Genbank entry KU892588 (Etmopterus pusillus; Chen et al., 2016) as reference for initial baiting. Annotation was performed by aligning the paratype consensus sequence to KU892588 in Geneious® Prime 2021.1 (Biomatters Ltd. Auckland, New Zealand), and checked for internal stop codons. Protein coding genes could not be fully reconstructed. The tRNA-Phe and tRNA-Val transfer RNAs, and the 12S and 16s ribosomal RNAs could be completely reconstructed and were therefore extracted for phylogenetic analysis (2676 bp in total). Reads used in the last iteration of Mitobim were mapped back to the mitochondrial genome consensus sequence as well as to the tRNA and rRNA sequences using BWA aln v.0.7.17 (Li and Durbin, 2009), with default settings, to check if the reads could be unambiguously mapped. Further, BWA was used to align the trimmed and quality filtered reads excluding duplicates to the full mitochondrial genome sequence as well as the tRNA-Phe, the 12S ribosomal RNA, the tRNA-Val and the 16S ribosomal RNA of KU892588 to assess coverage. Obtained sequences were aligned to the sequences of specimens listed in Supplementary Table 1, covering nine of the eleven species of the E. spinax group (Straube et al., 2010; Ebert et al., 2021). Sequences used to determine the phylogenetic placement of E. litvinovi were obtained from the Chondrichthyan Tree of Life (2016) project1 which are collected from vouchered and validated specimens, as described in White et al. (2018). A maximum likelihood tree was computed using RAxML v.8.2.4 (Stamatakis, 2014) under the general time reversible model. Heterogeneity of substitution rates among sites was modelled using a GAMMA distribution. To assess the statistical support for nodes, bootstrapping with 100 replicates was performed and plotted onto the maximum likelihood tree. A haplotype network was reconstructed with POPArt v. 1.7 (Leigh and Bryant, 2015) using the median joining network algorithm (Bandelt et al., 1999) under default settings. The RAxML tree served as a basis for calculating the p-distances between E. litvinovi and E. spinax clade species analysed herein using the Species Delimitation Plugin 1.4.5 (Masters et al., 2011) in Geneious®.
Etmopterus pycnolepis (Dense-Scale Lantern Shark)
This species is known from six specimens housed in three different museum collections, the ZIN (holotype: ZIN: 49226; two paratypes: ZIN: 49227); the ZMMU (paratype ZMMU: P-17992, paratype ZMMU P-17993) and the ZMH [paratype ZMH: 24995 (ex ISH 4-1989)]. We sampled tissue from the paratype specimen ZMH 24995 (Figure 2B) as described previously for the E. litvinovi paratype specimen. The specimen was captured at 25°56′ S and 88°33′ W at a depth of 580 m on 30.04.1987. It is an adult male of 426 mm total length (Thiel et al., 2009). As described for the E. litvinovi paratype, the overall condition and sampling date of the specimen suggests fixation with formaldehyde.
DNA extraction of the sample involved the same procedure as for E. litvinovi. Single stranded library preparation of E. pycnolepis DNA followed the protocol described in Gansauge et al. (2017). The E. pycnolepis sample underwent different laboratory procedures in comparison to the E. litvinovi sample, as the samples were processed with a considerable temporal gap, during which time the standard procedures in the historical laboratory at the University of Potsdam had been updated. Raw test-sequencing reads were analysed as in Straube et al. (2021a). FastQ Screen v0.14.0 (Wingett and Andrews, 2018) was used to check for unique hits to Etmopterus references and estimate contamination levels, before proceeding with target capture. After detection of target DNA in the test sequencing dataset, target capture was performed using an Arbor Bioscience myBaits® RNA bait kit. The baits were part of a multi-locus, multi species museum specimen barcoding approach described in Agne et al. (2022). NADH2 bait sequences were derived from representatives of all four Etmopterus clades (Straube et al., 2010) deposited in Genbank: E. lucifer (JQ518963), E. gracilispinis (JQ518960), E. granulosus (KF861686) and E. bigelowi (JQ518959). The four sequences were initially published in Naylor et al. (2012) and Straube et al. (2015). The single stranded DNA library was captured twice following the protocol described in Huang et al. (2021) using a hybridisation temperature of 65°C for 24 h. Sequencing of the double-captured, indexed library was performed on an Illumina NextSeq 500 System at the University of Potsdam as described in Paijmans et al. (2017). After quality filtering and adapter trimming using Cutadapt v. 2.10 (Martin, 2011) under default settings, reads were processed as described for E. litvinovi to reconstruct the NADH2 sequence of the paratype, using the NADH2 sequence of E. lucifer (JQ518963; Naylor et al., 2012) as reference.
The NADH2 consensus sequence (1044 bp) of the paratype was subsequently aligned with NADH2 sequences of other Etmopterus species with focus on the E. lucifer clade (Supplementary Table 2). Comparative sequences were obtained from the Chondrichthyan Tree of Life (2016) project (see text footnote 1). For details of NADH2 amplification and sequencing see Naylor et al. (2005, 2012). Forward and reverse sequences were aligned based on chromatograms and edited using Geneious® Pro v. 6.1.7 (Biomatters Ltd. Auckland, New Zealand). The consensus sequences were translated to amino acids and aligned with corresponding NADH2 sequences from representatives of closely related species using the MAFFT (Katoh et al., 2002, 2005) module in Geneious®. The aligned amino acid sequences were translated back in frame to their original nucleotide sequences, to yield a nucleotide alignment 1044 base pairs in length. Analysed samples are listed in Supplementary Table 2 including 13 of the 15 E. lucifer clade species. Phylogenetic inference and species delimitation was performed as described for E. litvinovi.
Results
Etmopterus litvinovi (Smalleye Lantern Shark)
A total of 3,734,481 trimmed and quality filtered reads were available after combining test-sequencing and target capture data, including duplicates. MitoBim ran for four iterations. 1,589,598 reads were used for baiting in the final iteration step. The consensus sequence shows 1.26% ambiguities scattered across the mitochondrial genome. Excluding duplicate sequences, 4985 reads map to the consensus sequence resulting from the Mitobim analysis providing an average coverage of 22 reads. The GC content is 40%. The mitochondrial tRNA and rRNA markers used for the phylogenetic analysis showed mapped read lengths mostly larger than 70 base pairs (Supplementary Figure 1A) and an average coverage of 58 reads, excluding duplicates (Supplementary Figure 2A). They did not show any ambiguous nucleotides.
The maximum likelihood phylogeny of the tRNA and rRNA sequences identifies lineages corresponding to species within the E. spinax clade. The relationships in the tree are mostly well-supported with many bootstrap values reaching 100% (Figure 3A). The E. litvinovi paratype sequence is sister to a sample identified as E. benchleyi. This clade also includes a specimen of E. benchleyi sampled in the Indian Ocean (GN4952). The clade as a whole is sister to the North Atlantic species E. princeps and E. spinax, which together form the sister clade to the Southern Hemisphere species E. viator (Figure 3A). The reconstructed haplotype network detected five haplotypes with 34 segregating sites and 16 parsimony-informative characters. Figure 3B shows that the haplotype sequence of the E. litvinovi paratype is identical to the E. benchleyi sample GN14570 and separated by a single mutational step from E. benchleyi sample GN4952. The species delimitation analysis shows that the interspecific K2P distance between two valid sister species within the E. spinax clade is on average 1.6% (Supplementary Table 3A), while the K2P distance between E. litvinovi and E. benchleyi is substantially smaller (K2P distance = 0.0518%; Supplementary Table 3A). Overall, our data does not support the validity of both species due to the phylogenetic placement of the E. litvinovi paratype sequence in the E. benchleyi clade and a very small K2P distance between both species.
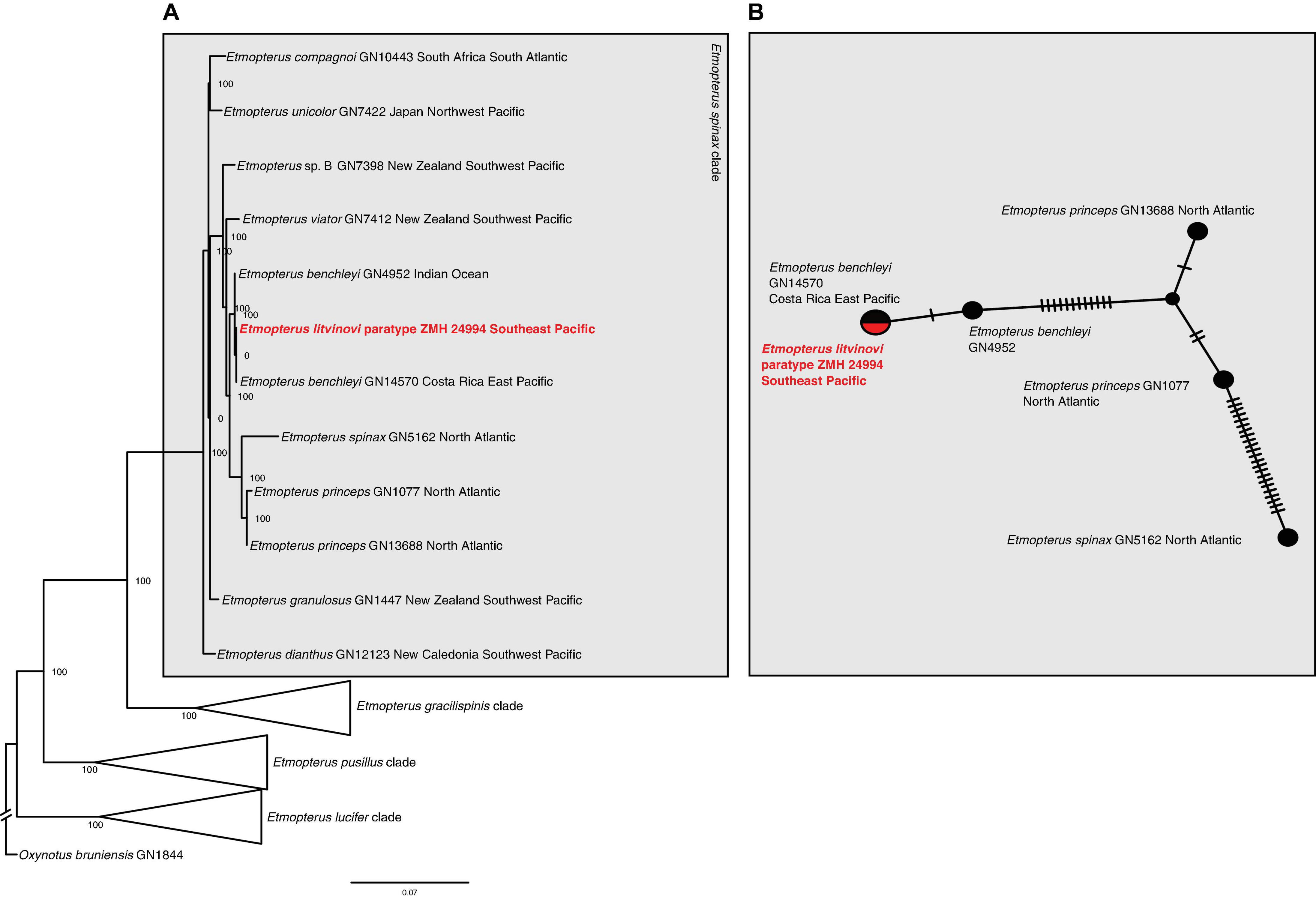
Figure 3. (A) Maximum likelihood analysis of Etmopterus 12s and 16s rRNA sequences. Numbers at nodes denote bootstrap support values. Oxynotus bruniensis was chosen as outgroup. (B) Haplotype network of 12s and 16s rRNA sequences including sequences from the E. litvinovi paratype, E. benchleyi and its sister clade comprising E. princeps and E. spinax. Crossbars indicate mutational steps. E. litvinovi and E. benchleyi GN14570 share a haplotype, GN4952 differs in a single mutational step.
Etmopterus pycnolepis (Dense-Scale Lantern Shark)
The test-sequencing dataset of 398,752 trimmed reads detected the presence of Etmopterus DNA, as indicated by 5.62% unique hits to the E. spinax transcriptome used as reference in the FastQscreen analysis. 82.83% of reads were un-assigned to any of the provided references, and contamination with other samples processed simultaneously was not detected. 4,029,200 raw reads were produced by sequencing of the target captured library. Quality filtering and trimming reduced this to 2,680,159 reads. Of these, 139,219 reads mapped to the NADH2 gene of E. lucifer (JQ518963). The complete NADH2 sequence was reconstructed after three iterations in Mitobim, using 115,548 reads in the final iteration. The mapped read length distribution is shown in Supplementary Figure 1B. The modal fragment length is around 50 base pairs. Mapping those reads back to the reconstructed NADH2 consensus sequence, showed that 277 reads mapped with an average coverage of 13 reads, excluding duplicate sequences (Supplementary Figure 2B). The maximum likelihood NADH2 phylogeny shows well-supported clades within the E. lucifer group. Several clades do not correspond to species: E. brosei clusters with E. sculptus, E. cf. molleri clusters with E. cf. decacuspidatus and forms a distinct clade not including E. molleri. Etmopterus molleri and E. dislineatus do not form two distinct clades. The E. pycnolepis paratype specimen is sister to a clade containing the southern hemisphere samples of E. lucifer and E. sculptus (Figure 4). The species delimitation analysis shows that the interspecific K2P distance of the NADH2 gene between sequences of two valid species in the E. lucifer clade, excluding clades not corresponding to species, is on average 4% (Supplementary Table 3B). Comparing the K2P distance of E. pycnolepis to its closest sister taxon, E. lucifer, the K2P distance is 3.4%. Our data therefore supports the species status of E. pycnolepis.
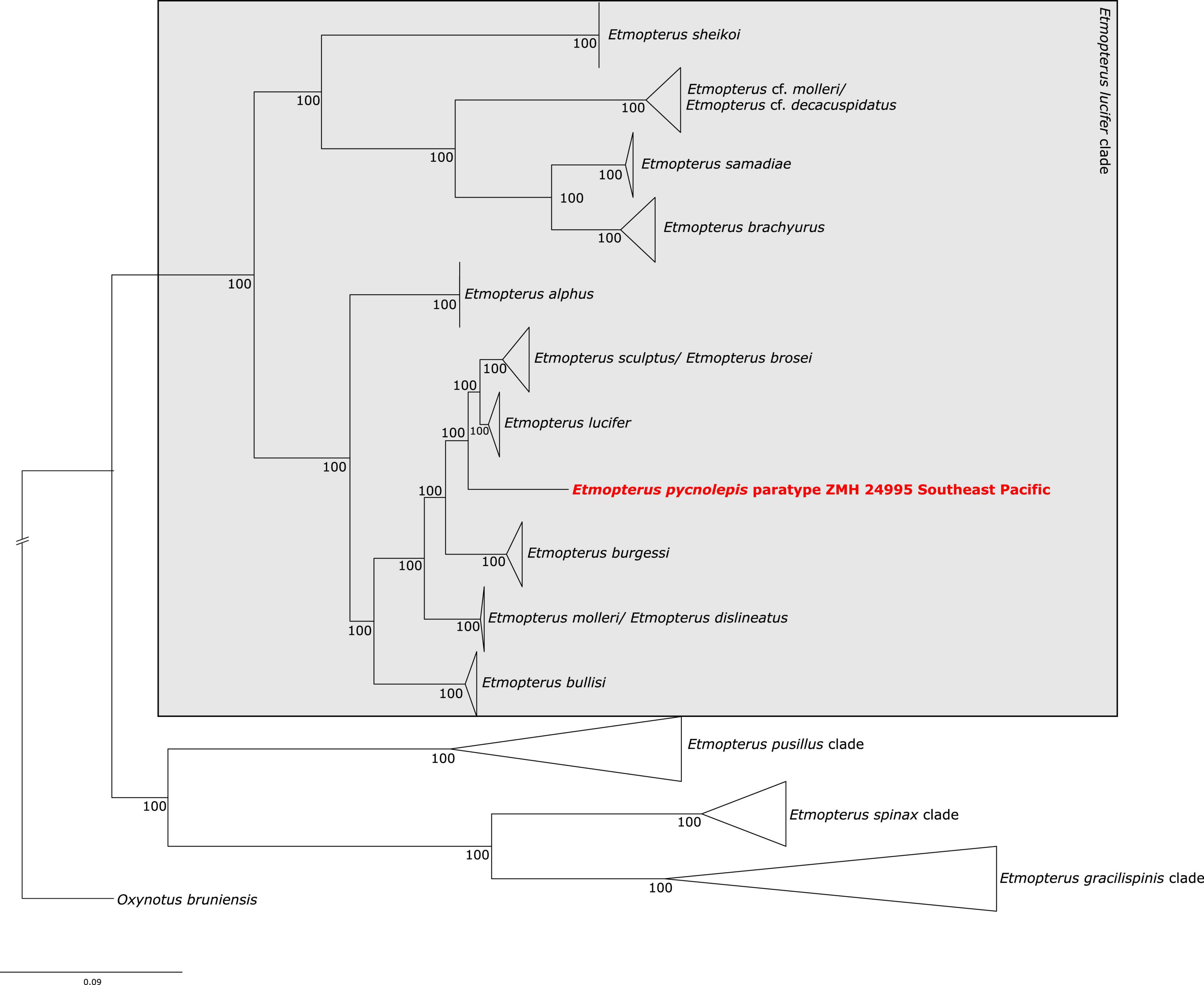
Figure 4. Maximum likelihood analysis of Etmopterus NADH2 sequences. Numbers at branches denote bootstrap support values. Oxynotus bruniensis was chosen as outgroup.
Discussion
Mitochondrial DNA Characteristics of the Two Paratype Specimens
The mitochondrial DNA we obtained from both paratype specimens, while fragmented (Supplementary Figure 1), was less degraded than other museum samples analysed in previous studies (Straube et al., 2021b). Fixation and preservation cause DNA damage (e.g., Stiller et al., 2016; Hahn et al., 2021; Straube et al., 2021a); however, the mitochondrial DNA of the two paratype samples analysed herein may be less affected due to the comparatively young age of 32 and 34 years, respectively, at the time of extraction. More comparative data is necessary to test if time is correlated with mitochondrial DNA fragmentation levels, and if fragmentation is ongoing under the current storage conditions.
Taxonomic Implications
Etmopterus litvinovi (Smalleye Lantern Shark)
The phylogenetic placement of the paratype sequence aligns with the morphology-based prediction that E. litvinovi is a member of the E. spinax species clade (Straube et al., 2010). A close relationship of E. litvinovi with morphological congeners, including cryptic species, was suggested in Straube et al. (2011a,b). This species complex was recently expanded with several species from which mitochondrial DNA sequence information is, however, available from only two species, E. benchleyi (Vásquez et al., 2015) and E. viator (Straube et al., 2011a). All analyses, i.e., the phylogenetic reconstruction, the haplotype network and the species delimitation analysis of the E. litvinovi paratype specimen suggest that E. litvinovi is conspecific with E. benchleyi, where E. benchleyi forms a junior synonym to E. litvinovi (Figure 3 and Supplementary Table 3A). Notably, the sequenced paratype specimen’s sampling locality is the Naszca Ridge in the Pacific Ocean, while another E. litvinovi haplotype (GN4952) is derived from a specimen sampled in the Indian Ocean (Figure 5). This suggests that E. litvinovi is widespread and occurs both in the Indian and Pacific oceans. Its overall distribution range may cover an even larger area of Southern Hemisphere oceans, and that its northern and southern distribution limits have yet to be identified. Similarly, wide distribution ranges are also documented for other closely related Etmopterus species in the E. spinax clade such as E. granulosus (Straube et al., 2011a,b, 2015) or E. viator (Straube et al., 2011a).
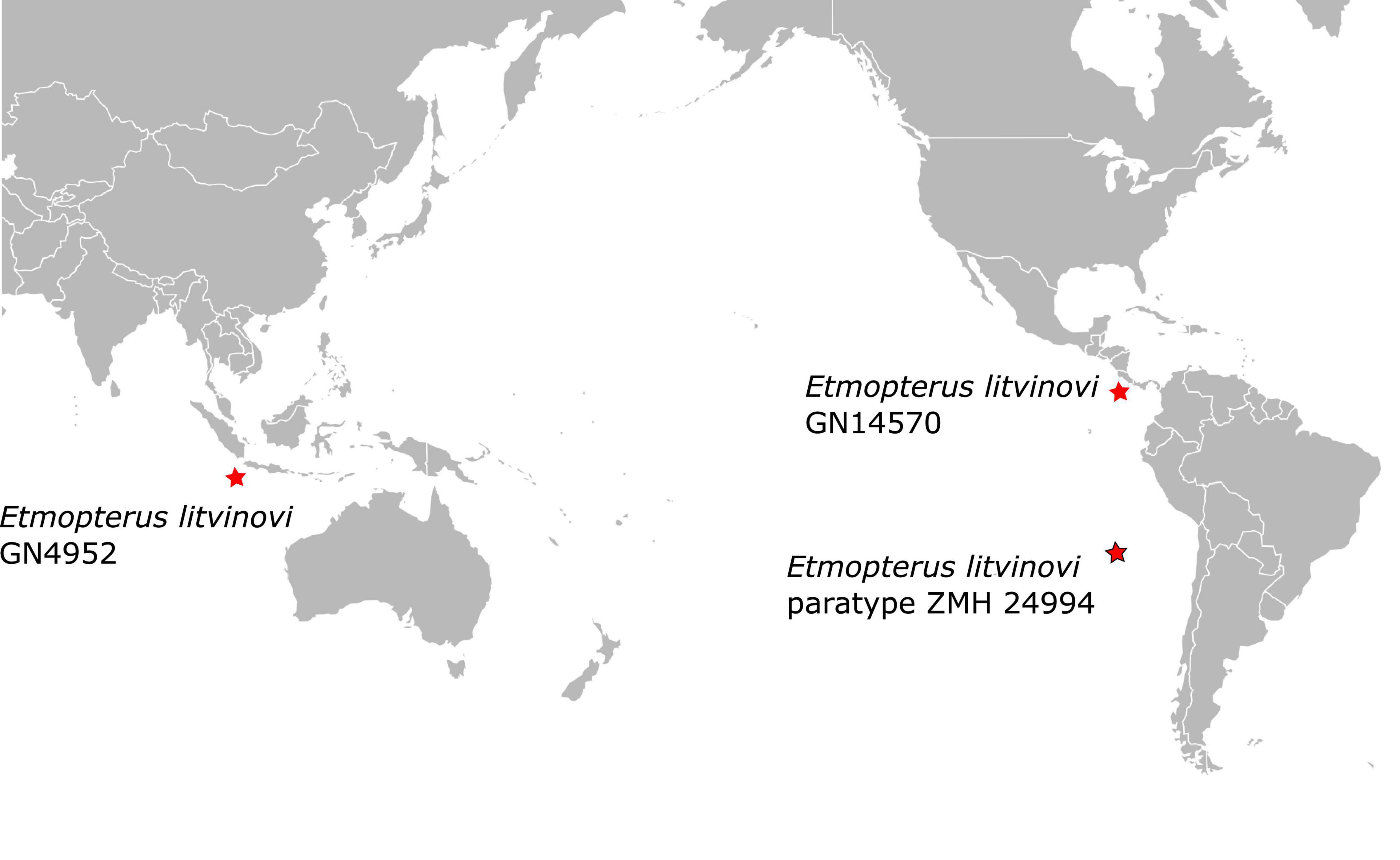
Figure 5. Paratype locality of Etmopterus litvinovi ZMH 24994 (red star with black frame) and sampling locations of the two other E. litvinovi specimens (red stars) initially labeled as E. benchleyi (Supplementary Table 1).
The new data presented herein will be helpful for future assessments of the species in the IUCN Red List of Threatened Species. As of today, the species is evaluated and listed under the “least concern” category justified due to limited fisheries in the area from which the species (i.e., the type material of E. litvinovi) was hitherto recorded (Ebert et al., 2020a). Based on our results, a notably larger distribution range should be considered in future evaluations taking different fishing pressure in other regions of occurrence into account. Our results do not confirm endemic occurrence (Kotlyar, 1990) but support its occurrence in the eastern Pacific as well as the Indian Ocean. The paratype sequence data analysed herein adds important alpha-level taxonomic information on the species, which will ease the collection of data on population size, as well as accurate geographic and depth distribution ranges, in the future. This forms the basis for conservation and management efforts for this poorly known deep-sea shark species.
Etmopterus pycnolepis (Dense-Scale Lantern Shark)
As already indicated by the distinct shape of its flank marking, our analysis further supports the assignment of E. pycnolepis as a distinct species (Supplementary Table 3B) within the E. lucifer clade (Figure 4; Straube et al., 2010). Its assignment to the E. burgessi subclade (Ebert et al., 2021) is not supported, however. The morphologically defined E. lucifer clade subclades described in Ebert et al. (2021) are generally not recovered in our molecular analysis (Figure 4). The phylogenetic inference displays to some extent geographic patterns of sampling locations instead. E. lucifer, E. pycnolepis, E. brosei and E. sculptus are represented by samples exclusively collected in Southern Hemisphere oceans (Supplementary Table 2), while E. burgessi samples stem from the Northwest Pacific. E. brachyurus, E. cf. molleri, and E. samadiae samples were also collected in the Northwest Pacific. E. alphus samples are from the Indian Ocean off Mauritius and E. bullisi was sampled in the Northwest Atlantic (Supplementary Table 2). Some species seem therefore confined to certain oceanic areas. The three different flank mark shapes characterising the E. lucifer clade subclades occur in three different ocean regions in parallel. In our study, E. brachyurus E. samadiae E. cf. molleri, E. cf. decacuspidatus, and E. burgessi represent the flank mark diversity of all three subclades in the Northwest Pacific; E. lucifer, E. molleri, E. dislineatus, E. brosei, E. alphus, E. pycnolepis and E. sculptus in the Indian and South Pacific oceans. In the Atlantic Ocean, only the E. molleri subclade type flank marking (Ebert et al., 2021) is represented by a single species (E. bullisi); however, E. bullisi is the only Atlantic species from the E. lucifer clade in general. A denser sampling is necessary to identify detailed species distribution boundaries and clarify indicated synonymies.
The IUCN Red List of Threatened species lists E. pycnolepis as least concern under the assumption that the area of origin of the six type specimens representing the species is not exposed to extensive fishing pressure as also in E. litvinovi. As mentioned in the evaluation justification, the species may be distributed in Chilean waters as well (Ebert et al., 2020b), which would amount to a large expansion of its distribution area. By providing the first DNA sequences for this species, newly collected samples available for NADH2 sequencing can be correctly assigned to the species and will therefore be useful for documenting its distribution range in the future.
Conclusion
Our results demonstrate the importance of archival DNA sequence information from type material for molecular based taxonomy. This is especially true for species which are known from few specimens only, and where re-sampling is hindered by remote sampling localities, as is the case for the two species analysed herein. Our results support the synonymy of E. benchleyi with E. litvinovi, and consequently suggest a notably larger distribution range than previously known, since the species was assumed to be endemic to the Salas y Gómez and Nazca Submarine Ridges (Kotlyar, 1990). The species status of E. pycnolepis is supported by our data, which is now available as reference for future molecular species-level identification of newly collected samples. This will help clarify the distribution of this species. Our results further show that genetic information from collection material can assist in the evaluation of species in a conservation and management context. While it is standard to evaluate morphological characters of wet-collection type material for descriptions of species new to science, the usage of wet-collection specimen DNA sequence information has only recently been established as such (Beermann et al., 2018; Lyra et al., 2020; Rancilhac et al., 2020; Scherz et al., 2020; Straube et al., 2021b) and our work is a further contribution to this.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Figshare (doi: 10.6084/m9.figshare.19446992) and GenBank (accession numbers ON185623-ON185724).
Ethics Statement
Ethical review and approval was not required for the animal study because no living animals were collected or examined. Tissue samples for DNA sequencing were taken solely from museum specimens and combined with existing data for analysis.
Author Contributions
NS, MH, JP, and AB designed the study. SA, MP, and NS conducted laboratory work. GN and LY provided comparative sequences for phylogenetics. SA, NS, LY, and GN analysed the data. RT and SW researched type specimen history, provided and helped sampling the type specimens. NS, SA, and MH wrote the manuscript with contributions from all authors. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the German Research Foundation (DFG; project number 351649567 to NS and MH within the DFG SPP 1991 “Taxon-Omics”). This project was further supported by the National Science Foundation (NSF), grant DEB-01132229 to GN.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to express our sincere thanks to Irina Eidus [Zoological Museum (ZMH) of the LIB] for her assistance during sampling of the paratype specimens. Jürgen Pollerspöck at shark references (www.shark-references.com) is thanked for literature search. We are also grateful to Mikhail Nazarkin (ZIN) and Matthias Stehmann (ICHTHYS) for information regarding handling of the specimens. Mikhail Nazarkin also kindly provided images of the holotypes of both species. Nataliya Budaeva (UM) is thanked for her help extracting collection information from the original description. We would also like to thank the HPC team at the University of Potsdam for creating and maintaining the university’s server on which we ran some of our analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.910009/full#supplementary-material
Footnotes
References
Agne, S., Straube, N., Preick, M., and Hofreiter, M. (2022). Simultaneous barcode sequencing of diverse museum collection specimens using a mixed RNA bait set. Front. Ecol. Evol. doi: 10.3389/fevo.2022.909846
Bandelt, H., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Beermann, J., Westbury, M. V., Hofreiter, M., Hilgers, L., Deister, F., Neumann, H., et al. (2018). Cryptic species in a well-known habitat: applying taxonomics to the amphipod genus Epimeria (Crustacea, Peracarida). Sci. Rep. 8:6893. doi: 10.1038/s41598-018-25225-x
Chen, H., Chen, X., Gu, X., Wan, H., Chen, X., and Ai, W. (2016). The phylogenomic position of the Smooth lanternshark Etmopterus pusillus (Squaliformes: Etmopteridae) inferred from the mitochondrial genome. Mitochondrial DNA B Resour. 1, 341–342. doi: 10.1080/23802359.2016.1172274
Chondrichthyan Tree of Life (2016). Chondrichthyan Tree of Life. Available online at: https://sharksrays.org/(accessed March 07, 2022).
Dabney, J., Knapp, M., Glocke, I., Gansauge, M. T., Weihmann, A., Nickel, B., et al. (2013). Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 110, 15758–15763. doi: 10.1073/pnas.1314445110
Dolganov, V. N., and Balanov, A. A. (2018). Etmopterus parini sp. n. (Squaliformes: Etmopteridae), a new shark species from the northwestern Pacific Ocean. Biol. Morya 44, 427–430.
Ebert, D. A., Concha, F., Herman, K., and Kyne, P. M. (2020a). Etmopterus litvinovi. The IUCN Red List of Threatened Species 2020. Gland: IUCN. doi: 10.2305/IUCN.UK.2020-3.RLTS.T63159A124463835.en
Ebert, D. A., Kyne, P. M., Concha, F., and Herman, K. (2020b). Etmopterus pycnolepis. The IUCN Red List of Threatened Species 2020. Gland: IUCN. doi: 10.2305/IUCN.UK.2020-3.RLTS.T63160A124463919.en
Ebert, D. A., Fowler, S. L., and Compagno, L. J. (2013). Sharks of the World: a Fully Illustrated Guide. Princeton, NJ: Wild Nature Press.
Ebert, D. A., Leslie, R. W., and Weigmann, S. (2021). Etmopterus brosei sp. nov.: a new lanternshark (Squaliformes: Etmopteridae) from the southeastern Atlantic and southwestern Indian oceans, with a revised key to the Etmopterus lucifer clade. Mar. Biodivers. 51, 1–17. doi: 10.1007/s12526-021-01173-0
Ebert, D. A., Straube, N., Leslie, R. W., and Weigmann, S. (2016). Etmopterus alphus n. sp.: a new lanternshark (Squaliformes: Etmopteridae) from the south-western Indian Ocean. Afr. J. Mar. Sci. 38, 329–340. doi: 10.2989/1814232X.2016.1198275
Gansauge, M. T., Gerber, T., Glocke, I., Korleviæ, P., Lippik, L., Nagel, S., et al. (2017). Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase. Nucleic Acids Res. 45:e79. doi: 10.1093/nar/gkx033
Gansauge, M. T., and Meyer, M. (2013). Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protocols 8, 737–748. doi: 10.1038/nprot.2013.038
Gilbert, M. T. P., Haselkorn, T., Bunce, M., Sanchez, J. J., Lucas, S. B., Jewell, L. D., et al. (2007). The Isolation of Nucleic Acids from Fixed, Paraffin-Embedded Tissues-Which Methods Are Useful When? PLoS One 2:e537. doi: 10.1371/journal.pone.0000537
Hahn, C., Bachmann, L., and Chevreux, B. (2013). Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - A baiting and iterative mapping approach. Nucleic Acids Res. 41:e129. doi: 10.1093/nar/gkt371
Hahn, E. E., Alexander, M. R., Grealy, A., Stiller, J., Gardiner, D. M., and Holleley, C. E. (2021). Unlocking inaccessible historical genomes preserved in formalin. Mol. Ecol. Resour. 1–18. doi: 10.1111/1755-0998.13505
Hebert, P. D., Cywinska, A., Ball, S. L., and DeWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hoffman, E. A., Frey, B. L., Smith, L. M., and Auble, D. T. (2015). Formaldehyde crosslinking: a tool for the study of chromatin complexes. J. Biol. Chem. 290, 26404–26411. doi: 10.1074/jbc.R115.651679
Huang, J. M., Yuan, H., and Li, C. H. (2021). Protocol for Cross-species Target-gene Enrichment. Bio-protocol 101:e1010606. doi: 10.21769/BioProtoc.1010606
Hykin, S. M., Bi, K., and McGuire, J. A. (2015). Fixing formalin: A method to recover genomic-scale DNA sequence data from formalin-fixed museum specimens using high-throughput sequencing. PLoS One 10:e0141579. doi: 10.1371/journal.pone.0141579
Katoh, K., Kuma, K. I., Toh, H., and Miyata, T. (2005). MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Katoh, K., Misawa, K., Kuma, K. I., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier Transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kotlyar, A. N. (1990). Dogfish sharks of the genus Etmopterus Rafinesque from the Nazca and Sala y Gómez submarine ridges. Tr. Inst. Okeanol. AN USSR 125, 127–147.
Leigh, J. W., and Bryant, D. (2015). PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Lyra, M. L., Lourenço, A. C. C., Pinheiro, P. D., Pezzuti, T. L., Baêta, D., Barlow, A., et al. (2020). High-throughput DNA sequencing of museum specimens sheds light on the long-missing species of the Bokermannohyla claresignata group (Anura: Hylidae: Cophomantini). Zool. J. Linn. Soc. 190, 1235–1255. doi: 10.1093/zoolinnean/zlaa033
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. Tech. Notes 7, 2803–2809. doi: 10.14806/ej.17.1.200
Masters, B. C., Fan, V., and Ross, H. A. (2011). Species delimitation-a geneious plugin for the exploration of species boundaries. Mol. Ecol. Resour. 11, 154–157. doi: 10.1111/j.1755-0998.2010.02896.x
McGuire, J. A., Cotoras, D. D., O’Connell, B., Lawalata, S. Z. S., Wang-Claypool, C. Y., Stubbs, A., et al. (2018). Squeezing water from a stone: High- throughput sequencing from a 145-year old holotype resolves (barely) a cryptic species problem in flying lizards. PeerJ 6:e4470. doi: 10.7717/peerj.4470
Naylor, G. J., Ryburn, J. A., Fedrigo, O., and Lopez, J. A. (2005). Phylogenetic relationships among the major lineages of modern elasmobranchs. Reprod. Biol. Phylogeny 3:25.
Naylor, G. J. P., Caira, J. N., Jensen, K., Rosana, K. A. M., White, W. T., and Last, P. R. (2012). A DNA sequence-based approach to the identification of shark and ray species and its implications for global elasmobranch diversity and parasitology. Bull. Am. Mus. Nat. Hist. 367, 1–262.
Paijmans, J. L. A., Baleka, S., Henneberger, K., Taron, U., Trinks, A., Westbury, M., et al. (2017). Sequencing single-stranded libraries on the Illumina NextSeq 500 platform. arXiv [Preprint]. doi: 10.48550/arXiv.1711.11004
Pollerspöck, J., and Straube, N. (2021). Bibliography Database of Living/Fossil Sharks, Rays and Chimaeras (Chondrichtyes: Elasmobranchii, Holocephali) – List of Valid Extant Species; List of Described Extant Species; Statistic, World Wide Web Electronic Publication, Version 03/2021. Available online at: www.shark-references.com
Rancilhac, L., Bruy, T., Scherz, M. D., Pereira, E. A., Preick, M., Straube, N., et al. (2020). Target-enriched DNA sequencing from historical type material enables a partial revision of the Madagascar giant stream frogs (genus Mantidactylus). J. Nat. Hist. 54, 87–118. doi: 10.1080/00222933.2020.1748243
Rohland, N., Siedel, H., and Hofreiter, M. (2004). Nondestructive DNAextraction method for mitochondrial DNA analyses of museum specimens. BioTechniques 36, 814–821. doi: 10.2144/04365ST05
Scherz, M. D., Rasolonjatovo, S. M., Köhler, J., Rancilhac, L., Rakotoarison, A., Raselimanana, A. P., et al. (2020). ‘Barcode fishing’for archival DNA from historical type material overcomes taxonomic hurdles, enabling the description of a new frog species. Sci. Rep. 10:19109. doi: 10.1038/s41598-020-75431-9
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stiller, M., Sucker, A., Griewank, K., Aust, D., Baretton, G. B., Schadendorf, D., et al. (2016). Single-strand DNA library preparation improves sequencing of formalin-fixed and paraffin-embedded (FFPE) cancer DNA. Oncotarget 7, 59115–59128. doi: 10.18632/oncotarget.10827
Straube, N., Duhamel, G., Gasco, N., Kriwet, J., and Schliewen, U. K. (2011a). “Description of a new deep-sea lantern shark Etmopterus viator sp. nov.(Squaliformes: Etmopteridae) from the Southern Hemisphere,” in The Kerguelen Plateau: Marine Ecosystem and Fisheries, eds G. Duhamel & D.C. Welsford (Paris: Société Française d’Ichtyologie). doi: 10.13140/2.1.1107.4248
Straube, N., Kriwet, J., and Schliewen, U. K. (2011b). Cryptic diversity and species assignment of large lantern sharks of the Etmopterus spinax clade from the Southern Hemisphere (Squaliformes, Etmopteridae). Zool. Scr. 40, 61–75. doi: 10.1111/j.1463-6409.2010.00455.x
Straube, N., Iglésias, S. P., Sellos, D. Y., Kriwet, J., and Schliewen, U. K. (2010). Molecular phylogeny and node time estimation of bioluminescent Lantern Sharks (Elasmobranchii: Etmopteridae). Mol. Phylogenet. Evol. 56, 905–917. doi: 10.1016/j.ympev.2010.04.042
Straube, N., Leslie, R. W., Clerkin, P. J., Ebert, D. A., Rochel, E., Corrigan, S., et al. (2015). On the occurrence of the Southern Lanternshark, Etmopterus granulosus, off South Africa, with comments on the validity of E. compagnoi. Deep Sea Res. II Top. Stud. Oceanogr. 115, 11–17. doi: 10.1016/j.dsr2.2014.04.004
Straube, N., Lyra, M. L., Paijmans, J. L. A., Preick, M., Basler, N., Penner, J., et al. (2021a). Successful application of ancient DNA extraction and library construction protocols to museum wet collection specimens. Mol. Ecol. Resour. 21, 2299–2315. doi: 10.1111/1755-0998.13433
Straube, N., Preick, M., Naylor, G. J. P., and Hofreiter, M. (2021b). Mitochondrial DNA sequencing of a wet-collection syntype demonstrates the importance of type material as genetic resource for lantern shark taxonomy (Chondrichthyes: Etmopteridae). R. Soc. Open Sci. 8:210474. doi: 10.1098/rsos.210474
Thiel, R., Eidus, I., and Neumann, R. (2009). The Zoological Museum Hamburg (ZMH) fish collection as a global biodiversity archive for elasmobranchs and actinopterygians as well as other fish taxa. J. Appl. Ichthyol. 25(Suppl. 1), 9–32. doi: 10.1111/j.1439-0426.2009.01296.x
Vásquez, V. E., Ebert, D. A., and Long, D. J. (2015). Etmopterus benchleyi n. sp., a new lanternshark (Squaliformes: Etmopteridae) from the central eastern Pacific Ocean. J. Ocean Sci. Found. 17, 43–55.
White, W. T., Corrigan, S., Yang, L. E. I., Henderson, A. C., Bazinet, A. L., Swofford, D. L., et al. (2018). Phylogeny of the manta and devilrays (Chondrichthyes: Mobulidae), with an updated taxonomic arrangement for the family. Zool. J. Linn. Soc. 182, 50–75. doi: 10.1093/zoolinnean/zlx018
White, W. T., Ebert, D. A., Mana, R. R., and Corrigan, S. (2017). Etmopterus samadiae n. sp., a new lanternshark (Squaliformes: Etmopteridae) from Papua New Guinea. Zootaxa 4244, 339–354. doi: 10.11646/zootaxa.4244.3.3
Keywords: type specimens, Etmopterus litvinovi, Etmopterus pycnolepis, deep-sea sharks, archival DNA
Citation: Agne S, Naylor GJP, Preick M, Yang L, Thiel R, Weigmann S, Paijmans JLA, Barlow A, Hofreiter M and Straube N (2022) Taxonomic Identification of Two Poorly Known Lantern Shark Species Based on Mitochondrial DNA From Wet-Collection Paratypes. Front. Ecol. Evol. 10:910009. doi: 10.3389/fevo.2022.910009
Received: 31 March 2022; Accepted: 25 May 2022;
Published: 30 June 2022.
Edited by:
Anchalee Aowphol, Kasetsart University, ThailandReviewed by:
Kieren James Mitchell, University of Otago, New ZealandVladimir A. Trifonov, Institute of Molecular and Cellular Biology (RAS), Russia
Copyright © 2022 Agne, Naylor, Preick, Yang, Thiel, Weigmann, Paijmans, Barlow, Hofreiter and Straube. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicolas Straube, Nicolas.Straube@uib.no
 Stefanie Agne
Stefanie Agne Gavin J. P. Naylor2
Gavin J. P. Naylor2  Ralf Thiel
Ralf Thiel Axel Barlow
Axel Barlow Nicolas Straube
Nicolas Straube