Genetic infrapopulation sizes in blood parasites: a pilot quantification of the bottleneck in louse fly vectors
- 1Department of Animal Behaviour, Bielefeld University, Bielefeld, Germany
- 2JICE, Joint Institute for Individualization in a Changing Environment, University of Münster and Bielefeld University, Bielefeld, Germany
Introduction: Bottleneck events are crucial for the strength of genetic drift, selection and speed of evolution. They are believed to play a particularly prominent role for parasitic infrapopulations, inhabiting single host individuals, which are often established by very few parasite individuals during transmission. In vector-borne pathogens, the bottlenecking effects can even be serialized through repeated filtering of parasitic stages at different tissues and organs of the vector. Using qPCR we aimed to quantify the number of potentially transmittable sporozoites of the hemosporidian blood parasite Haemoproteus columbae in the specialized vector louse flies Pseudolynchia canariensis which transmit these parasites between house pigeon hosts Columba livia.
Results: Based on qPCR measurements of organ-derived DNA of individual louse flies, we estimate that the midgut of these vectors contains on average 20 parasites, the hindgut and other intestines ca. 50 parasites and the salivary glands ca. 5 parasite cells. Nearly one third of all vector individuals appeared to lack parasite DNA, despite having only infected hosts as blood meal sources. The magnitude of parasite numbers in midgut and salivary glands tended to correlate positively.
Discussion: Our results indicate, potential severe bottlenecking of parasite populations during individual transmission events and a probable effect of individual vector immunity on this variable. However, this may be partly alleviated by the coloniality of house pigeons, the frequency of louse flies and their daily feeding events in most populations, leading to repeated transmission opportunities, decreased quasi-vertical transmission between parents and offspring and probable panmixia of Haemoproteus columbae lineages. Many of these mechanisms might not apply in other host-vector systems. We propose several additional molecular and microscopical tools to improve the accuracy of estimating parasite population sizes in vectors and call for more estimations in different vector species to better understand the co-evolution between malaria-like blood parasites and their avian and insect hosts.
Introduction
Population genetics examines the diversity of populations, as well as the demographic processes which contribute to it and lead to distinct evolutionary trajectories (Hamilton, 2021). A decisive mechanism shaping the genetic diversity of populations are bottleneck and founder events, where only a subset of individuals and the population-wide genetic diversity (re-) establishes a (new) population (Templeton, 1980). An inherent instance of a founder effect is the establishment of an infrapopulation, the population of parasite individuals which inhabit a single host individual. Although final infrapopulations of parasites can appear large, the founding events are often performed by few germinating propagules, spores or other infective stages (Huyse et al., 2005). The size of the founder population may therefore be a crucial variable determining the genetic diversity of incoming parasites in a new host, which may correspond to the final available and further transferrable population-wide diversity (Criscione et al., 2005). Meanwhile, the genetic diversity of parasites in single hosts can affect host health and the activation of the immune response (Ferguson et al., 2003). Consequently, the founding infrapopulation size can have fitness effects and affect the speed and direction of host-parasite co-evolution (Schmid-Hempel and Frank, 2007). In pathogenic bacteria, the concept of minimum infective dose (number of individual pathogens necessary to start an infection) has been intensively studied theoretically and experimentally, but in most infectious organisms and parasites, the mean inoculum dose occurring in natural systems is harder to establish and has rarely been quantified (Abel et al., 2015; Prentiss et al., 2022).
Vector-borne parasites can be particularly informative models for the evolutionary effects of variable population dynamics, bottlenecks and founder events (Weaver and Barrett, 2004). A vector transmits parasites to a larger host where the parasite population size often grows strongly (Schmid-Hempel, 2011). However, a vector can be viewed as a serial filter, which causes the parasite population and its genetic composition to change (Abel et al., 2015). This is particularly true for parasites with a complex life cycle whose population may have to pass several transitions between stages, which migrate through or develop in different tissues and organs within the vector, while facing its defences and immunity (Criscione and Blouin, 2005; Vaughan, 2007). While both clonal multiplication and sexual reproduction of parasites can happen in vectors, they often appear to receive more parasite individuals from a donor host than they pass on to a receptive one. Sometimes even more substantial genetic bottlenecking effects occur within the vector (Smith et al., 2014). Additionally, when initial infection intensity is too high this can reduce vector survival, thus adding one more limitation to the initial number of potentially transmissible parasites (Elliot et al., 2003; Witsenburg et al., 2015; Bukauskaitė et al., 2016). Although these processes are commonly understood, in most vector studies the trait reported about a vector is binary – the vector species is competent and with individuals having transmissible stages present, or the vector is not competent (Beerntsen et al., 2000; Weiss and Aksoy, 2011). Actual quantification of how many stages of different parasites pass through different organs of a specific vector, and then are available to finally be transferred to a new host, have been done rarely and for few model species. On the other hand, if done more commonly, such quantifications should help to understand the population-wide processes of some parasite clades and thus, potentially their evolution e.g. to become more speciose, virulent and/or specialized.
Haemosporidian blood parasites (Haemosporida, Apicomplexa) are vector-borne pathogens, including the very species-rich genera of Leucocytozoon, Haemoproteus and Plasmodium, which contains the causative agents of avian and human malaria (Valkiūnas, 2004). In the intermediate vertebrate hosts, such as mammals, reptiles and birds, the parasites reach greatest population sizes. There, they first reach and multiply asexually in diverse organs. Stages released from the organs infect the blood cells, where Plasmodium make additional asexually-reproducing stages but Haemoproteus and Leucocytozoon only have pre-gametic stages (Valkiūnas, 2004). After a vector of a dipteran family, specific to each hemosporidian genus, ingests a blood meal, the haploid blood-infecting gametocytes, develop into micro- and macrogametes (Iyer et al., 2007). These merge in the gut of the vector, producing a diploid zygote, which morphs into an meiotic ookinete, then penetrates the vector midgut wall to settle in its epithelium and form an oocyst (Baton and Ranford-Cartwright, 2005). Within the oocyst a massive endomitosis takes place and produces haploid sporozoites (sporogony) which burst the oocyst and migrate to the salivary glands (Vaughan, 2007). At each of these steps, apart from the sporogony, there is substantial mortality and reduction of the parasite population. This reduces the diversity to one or few oocysts (genetic individuals), dividing into several dozens to several thousand sporozoite clones, which would be available for transmission in the salivary glands of human-infective Anopheles mosquitoes (Rosenberg et al., 1990; Vaughan, 2007). However, comparable numbers from other taxa are very scarce. This can be attributed to several factors. Quantifying each of the filtering steps for a parasitic lineage in a vector demands the keeping of infected hosts and a representative number of vector individuals (Valkiūnas, 2004). The vectors have to be held at normal conditions before dissections at different time intervals to observe which and how many parasitic stages have developed (Bernotienė et al., 2019; Chagas et al., 2019; Ilgūnas et al., 2019). Some hemosporidian vectors are very difficult or logistically impossible to keep under species-appropriate conditions in captivity (Valkiūnas, 2004). Some Anopheline and Culicine mosquitoes transmitting Plasmodium have well-established keeping conditions and this has reinforced their status as model systems (Rose et al., 2023). Correct conditions have been also found for some Culicoides biting midges, which transmit Parahaemoproteus (e.g. Bukauskaitė et al., 2019). Keeping Leucocytozoon-transmitting ornithophilic simuliid blackflies in captivity is still a major obstacle to research (Edman and Simmons, 1985; Crosskey, 1990). Finally, analyzing Haemoproteus-transmitting hippoboscid flies, demands also the keeping of their avian hosts in captivity, which is feasible only for a small subset of bird species. However, even for these, quantifications of the vector filtering effects remain to be explored. In order to understand the serial bottleneck effects within vectors, the organs and tissues of these have to be separated through dissection and parasitic stages counted. The separation step remains challenging for very small insects such as biting midges, but even in larger insects the quantification through counting is rarely performed as it requires highly developed skills and potentially special staining for quantitative imaging (Valkiūnas, 2004). A more commonly accessible alternative is the quantification of DNA parasitic copies for the given organ through qPCR, assuming that this is representative of viable stages in this organ (Bell and Ranford-Cartwright, 2004; Medica and Sinnis, 2005).
In this study, we aim to quantify via qPCR the sporozoites available for transmission in the salivary glands of Pseudolynchia canariensis, the specialized hippoboscid fly species transmitting Haemoproteus columbae among house pigeons Columba livia (Cepeda et al., 2019). Hippoboscids are one of the smallest groups of hemosporidian vectors and only few species in the group are known to transmit blood parasites (Santiago-Alarcon et al., 2012). Hippoboscids transmit only members of the subgenus Haemoproteus (Haemoproteus) which is a small, rather species-poor branch of the hemosporidian phylogenety. The members of this subgenus are specialized and infect only few bird species, albeit sometimes with high prevalence and infection intensity (Valkiūnas, 2004). Especially because of these peculiarities and differences to the main vector groups and hemosporidian genera, hippoboscids and their transmitted blood parasites are an interesting case where the demographic effect of vector passage on the parasite populations should be established and compared with the more diversified hemosporidian branches. Additionally, house pigeons and their vectors are outstandingly manageable in captivity, allowing detailed research over each step of the parasitic development and making them one of the few model systems in avian hemosporidian research (Valkiūnas, 2004; Cepeda et al., 2019).
Materials and methods
A population of infected house pigeons Columba livia hosts and Pseudolynchia canariensis vectors was kept in isolation as a source population. Using blood smears, the infection rates of available hosts were determined. Three hosts with the highest infection intensity were isolated and served as blood meal sources for the test vector individuals. Vectors were collected from the main pigeon colony, marked with a non-toxic color pen and transferred to the three isolated hosts. Pseudolynchia are known to feed several times per day, so we assumed that the transferred individual hippoboscids fed on the newly available host individuals shortly after the transfer (Coatney, 1931). After a prepatent period of 16 days post transfer, allowing these vector individuals to digest the blood meals and Haemoproteus parasites to develop in them, the vector individuals were collected into vials. For negative control samples, pupae were collected and allowed to hatch in vials (Waite et al., 2012). Animal handling and sampling procedures were approved by the Animal Ethics Committee at Bielefeld University and conducted with permission from the federal state authority 81-02.04.2018.A258 accordance with German federal and state laws.
A blood smear was taken from the three blood meal supplying hosts, fixed in 100% ethanol for 10 minutes and stained with Giemsa. Infection intensity was determined by counting the number of infected cells among 8000 erythrocytes per smear. A serial dilution of DNA from the most intensely infected sample was created as a standard for the following quantitative PCR measurements.
Vectors were immobilized by transferring the vials into a freezer for 5 minutes. Dissection took place under a stereomicroscope in order to identify salivary glands, midgut and hindgut along with further intestines. Each of these organs was extracted and transferred into a separate vial with 0.5 ml DNA extraction buffer.
After digestion with proteinase K at 56°C for at least 24h, a standard Chloroform-Isoamylalkohol-Protocol was used to extract the DNA from the three different organ samples of each individual hippoboscid fly. The DNA concentration was measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Sample concentrations between 5 and 100 ng/µl were accepted as favorable and used directly for qPCR, as standardizing to absolute total insect or vertebrate DNA concentration might have diluted the parasite DNA below detection thresholds. Higher concentrations were diluted to 20 ng/µl using water. Quantitative real-time PCRs (qPCRs) were performed using triplicate measurements for each sample. For parasitic template amplification the primers 3524F (5′-AGG CAA AGA AAA TGA CCG G-3′) and 3655R (5′-ATG GCG AGA AGG GAA GTG TG-3′) were used which target a 131-bp fragment in the parasite mitochondrial genome (Huang et al., 2020). The standard sample row created from the DNA of the most intensely infected pigeon host, consisted of five DNA concentrations (25 ng/µl, 5 ng/µl, 1 ng/µl, 0.2 ng/µl, 0.04 ng/µl). This standard row was included in each measurement plate in triplicate to ensure the comparability between plates. All qPCR assays were performed with a CFX Connect™ real-time qPCR instrument using EvaGreen DNA-binding dye (my-Budget 5x EvaGreen QPCR-Mix II, Bio-Budget Technologies GmbH). Each 10 µl PCR reaction consisted of 2 µl template DNA (10-200 ng of DNA), 2 µl 5x EvaGreen, 1.25 µl of each primer (10 µM) and 3.5 µl water. After a 10 min initiation temperature of 95°C, 40 cycles were performed consisting of 10 sec at 95°C, 10 sec at 56°C and 30 sec at 72°C. For a plate of 96 wells, 26 samples were run in triplicate along with a 5-fold dilution series of the highly infected sample with known DNA concentrations used as standard (25 μM, 5 μM, 1 μM, 0.2 μM, 0.04 μM). DNA concentrations in the standard sample row consist of host and parasite DNA but estimates of concentrations for each can be discerned by multiplying with the ratio of the expected genome sizes of both species (1.5 Gb and 0.02 Gb respectively) with the microscopically counted infection rate. Amplification curves were inspected after the runs to verify the cycle threshold (Ct) of each sample. Default settings were used to calculate the plate-specific threshold cycle, indicating amplification-caused fluorescence to exceed background noise (Asghar et al., 2011). qPCR plates producing standard curves that were outside the range of 100 ± 15% qPCR efficiency were discarded, because this is indicative of errors in qPCR estimates (Asghar et al., 2011). The estimated number of cells was adjusted to the total volume of DNA extract.
To calculate the relative quantities (RQ) for each sample, the formula, , was used with a being the primer-specific slope of the standard dilution row, i.e. the log2 of the 5-fold standard concentrations against the Ct-values measured for these concentrations. The formula is based on Livak and Schmittgen (2001) formula to calculate qPCR data. Further extrapolations were based on the assumption that sporozoites contain the same abundance of mitochondrial DNA as gametocytes and that each pigeon blood cell contains 3 pg of DNA, corresponding approximately to twice the avian host haploid genome size. Thus, the parasitic cell number for the standard row was derived by microscopy and the Ct values corresponding to the number of parasitic cells was calculated and was further applied to the test samples where only Ct value was measured in order to estimate the number of parasite DNA templates and cells represented in 2 µl of DNA solution. All statistical analyses were done in R version 4.1.3.
Results
Organs were extracted from 24 individual Pseudolynchia flies, in two of these only salivary glands being successfully extracted. The infection intensity of the most highly infected standard sample (and potentially the donor host bird) was 0.89% of all erythrocytes infected. Sanger sequencing revealed that the infecting Haemoproteus columbae lineage was COLIV03.
The results of the qPCR showed amplification of parasitic DNA in all three organ sample types, however not for all flies. The estimated number of parasites in the midgut ranged between 0 and 1826.3 cells with arithmetic mean of 229 cells per individual and a median of 20 parasite cells. The highest numbers were estimated for the hindgut and intestines with 0 to 1484 parasitic cells and with arithmetic mean cell count estimate of 252 and median of 49. Lastly, the derived estimates for number of parasitic cells in the salivary glands showed a range of 0 to 476 parasites, arithmetic mean of 40 and a median of 5.6 parasites per individual salivary glands (Figure 1). For all three of the negative samples of non-exposed flies the estimates for each organ were under the detection threshold of 1 parasitic cell.
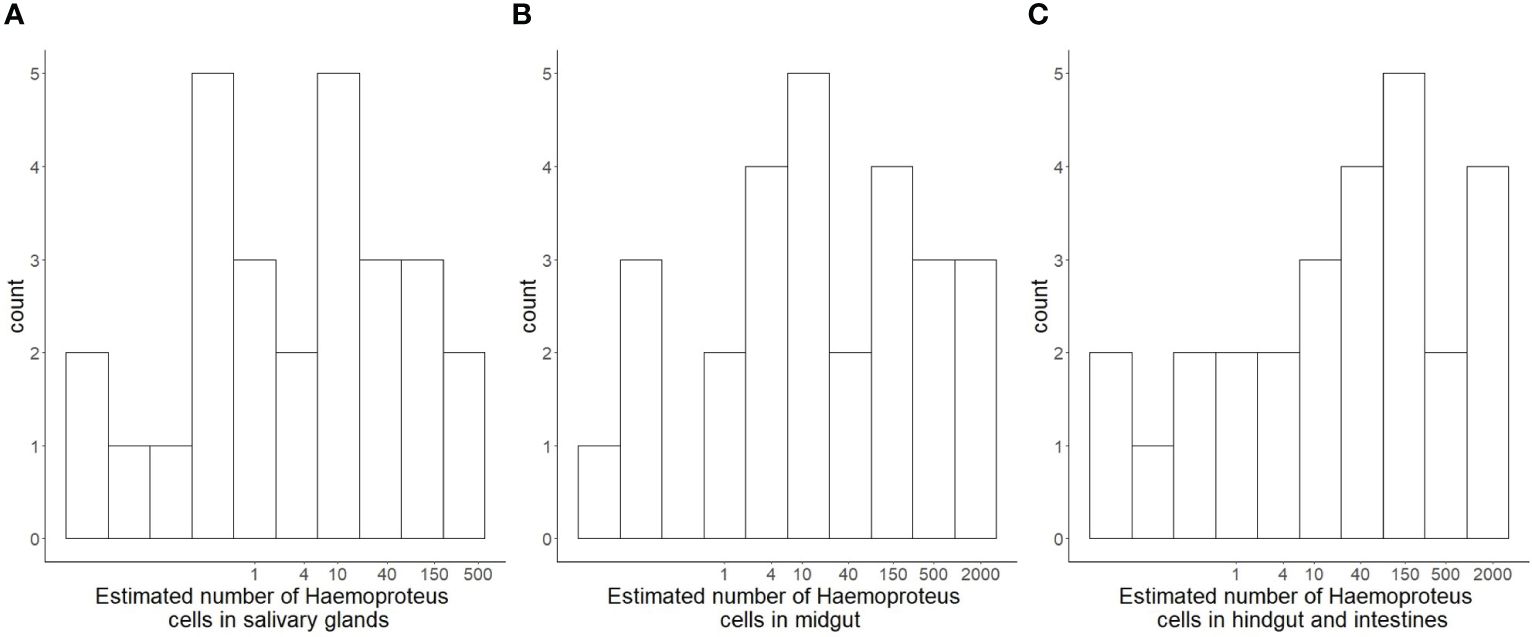
Figure 1 Histograms (log-transformed X-axis) of estimated numbers of parasitic Haemoproteus cells in different organs of hippoboscid vector flies Pseudolynchia canariensis. Estimates were derived through qPCR with complete organ-specific DNA extracts of individual flies, after a prepatent period of 16 days for the development and passage of parasites through the organs and reaching the salivary glands. (A) estimated number of cells in salivary glands. (B) estimated number of cells in midgut (C) estimated number of cells in hindgut and residual intestines.
Thus, the highest number of parasitic cells was estimated in the hindgut and intestine followed by the gut, and the lowest number was found in the salivary glands. There were no significant correlations between raw estimates of parasite numbers in different organs of individual flies (Figure 2). However, log-transformed estimates for parasite numbers in organs of individual flies were significantly positively correlated (for each salivary glands~midgut, midgut~intestines and salivary glands~intestines two-way correlations p<0.001). Restricting the dataset to Pseudolynchia individuals with probable Haemoproteus infection (RQ>5), only a trend of a correlation between the log-transformed parasite estimates of the salivary glands and the midgut remained (linear model, est. = 0.554, se=0.271, p=0.058, t-value = 2.041).
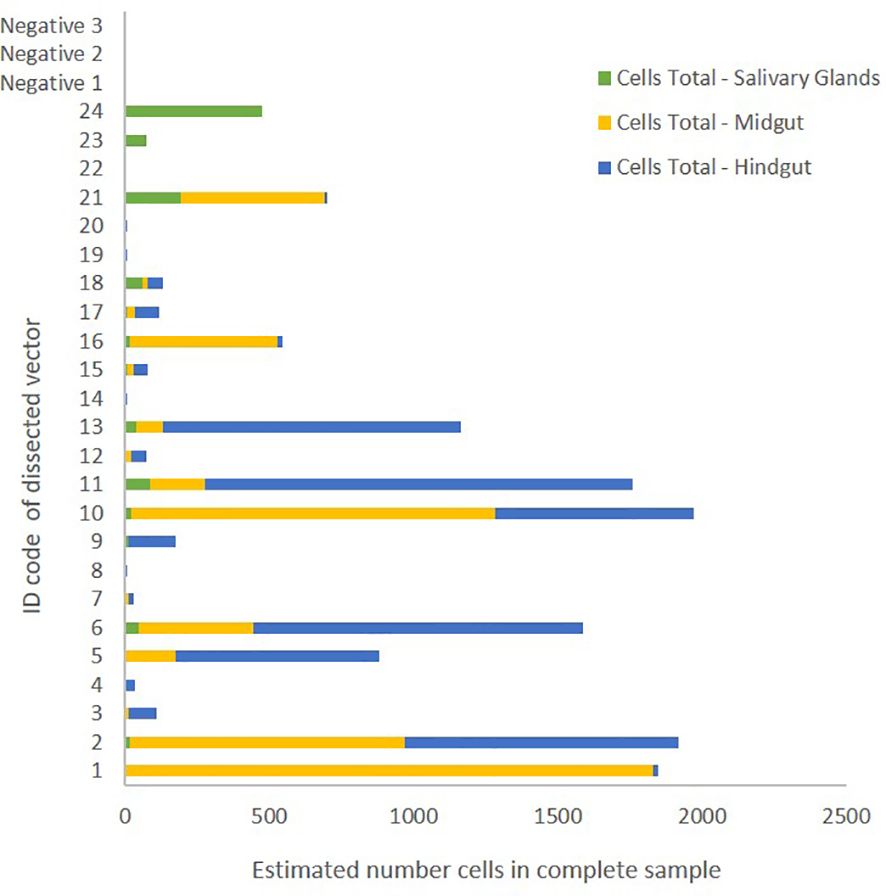
Figure 2 Estimated number of Haemoproteus cells in different organs of dissected Pseudolynchia canariensis individuals. Extracted and compared organs were salivary glands (green), midgut (yellow) and hindgut including residual intestines (blue). Negative samples 1-3 were derived from Ps. canarianesis individuals without prior contact to pigeon hosts.
Discussion
We estimated the number of Haemoproteus parasites in three parts of hippoboscid vectors, which play an important role in the life cycle of hemosporidian parasites on the way to being transmitted to a new host. We found that the number of parasite copies appeared to be highest in the hindgut and other intestines, while finding lower numbers in the midgut and substantially lower numbers in the salivary glands. Samples from one third of the hippobiscid flies showed no clear amplification of parasite DNA, despite the fly individuals having only infected hosts as a meal source. Vector individuals, which had many parasites in the midgut also had many in the salivary glands. The average number of sporozoites, transferrable via bites to further hosts was estimated to be between 5 and 20.
Classical fundamental studies have identified the midgut of vectors as the main arena for development of ookinetes and oocysts of hemosporidian parasites (Valkiūnas, 2004; Hajkazemian et al., 2021). Few additional studies suggest that development of oocysts is possible also in posterior parts of the dipteran digestive tract, such as the hindgut and the rectum, however without indications of completion of this development or effects on the number of produced sporozoites (Haraguchi et al., 2022). We find that the sections of the hippoboscid digestive tract after the midgut appear to contain more parasites than the midgut itself. In this respect, failure of macrogametocytes to reach the ookinete stage is substantial in Pl. falciparum in Anopheles mosquitoes (Vaughan et al., 1992, 1994; Vaughan, 2007). Similarly high failure rates, reaching more than 100-fold reductions in numbers at each stage, affect ookinetes failing to penetrate the gut wall and to form oocysts (Ghosh et al., 2000; Smith et al., 2014; Simões et al., 2018). This may indicate what proportions of the ingested gametocytes fail due to inhibition by the vector digestion and immunity and thus, get digested and excreted by the vector. A comparable estimate may be achievable through qPCR measurements in vector excretions. However, this should be compared with the number of gametocytes entering the digestive tract through a blood meal and corrected for the fraction of potentially developing free oocysts and released sporozoites which may inflate the estimates (Pichon et al., 2000; Haraguchi et al., 2023). Similarly, the number of parasites retained in the midgut appears high in comparison with the estimate of the number passed further for potential excretion. At this stage, only 0.5% of the ingested Pl.falciparum gametocytes persist as oocysts in the midgut and the other 99.5% are likely digested and excreted (Vaughan, 2007). Several factors may contribute to this apparent discrepancy. Firstly, in our setup individual flies were allowed to feed until shortly before dissection. Thus, both in the midgut and in the hindgut, the content was not restricted to oocysts or failed parasites but likely also included a mix of subsequently ingested gametocytes, ookinetes and failed stages resulting from them. Secondly, the endomitosis performed in oocysts in the lamina of the gut wall may inflate the impression of retained and successful ookinetes in comparison to the ones that have failed and are passed towards the rectum. Microscopic analyses may prove more suitable for quantifying the proportions of these stages. Alternatively, stage-specific expression factors could be used for qPCR of organ-derived RNA. However, developing such markers is still complicated, by the shortage of genomic data and gene characterization for non-Plasmodium hemosporidians (Bensch et al., 2016, Chakarov et al. in prep). Thirdly, similarly to gametocytes, oocysts and sporozoites contain mitochondria, but they might not be completely comparable since the amount of mitogenome copies in these organelles can vary between stages as it appears to vary between host environments (Brandler and Chakarov, 2024). A future useful step would be to compare mitogenome-based qPCR estimates, as performed in this study, with qPCRs based on nuclear markers, measures of within-lineage parasite genetic diversity in the DNA extracts and microscopic counts of different stages, which would enable the quantification of stage-specific differences in the copy numbers of the used DNA template. For identification and quantification of hemoparasites in avian hosts, a similar complementary combination of microscopy and molecular methods is already recommended (Valkiūnas et al., 2014).
Interestingly, almost a third of all vector individuals did not show measurable parasite DNA, despite only having infected blood meal sources available and presumably having to feed often (Coatney, 1931). This suggests that some vector individuals have strong immunity, which in some cases may be sufficient to clear all parasites not long after the digestion of a blood meal. While many cellular and molecular resistance factors have been characterized in insects and specifically in malaria vectors, the functionality of many specific proteins and mechanisms should be verified in divergent vector groups such as hippoboscid flies (Simões et al., 2018). Congruent with immunity playing a strong role in the filtering effect, is the trend for vector individuals with many detectable parasites in the midgut to also have many parasites in the salivary glands. These insect individuals may have been particularly immunocompromised, weakened or tolerant to parasite passage (Rivero and Ferguson, 2003). We focused on the number of transmissible stages in a vector at any point after initial feeding and sufficient developmental time. However, exact quantification of the filtering effects at every tissue and organ of the vector will require a more sophisticated experiment with many more replicates, restricted access to blood meals, and the ability to distinguish parasites from different lineages or meal sources (e.g. Himmel et al., 2019). It would be particularly interesting to develop non-lethal methods for estimating parasite numbers at some filtering steps and use these to study host-switching and sucking behavior of vector individuals. Deducing the number and proportion of parasites in vector excrements through molecular methods may be one non-invasive method to achieve this.
The main aim of our study was to estimate the number of sporozoites available in the salivary glands of vector individuals to transmit to the next, possibly naïve hosts. This number appears to be less than 20 and thus most probably represent one or very few individual oocysts. This constitutes close to the maximal possible bottleneck for the genetic diversity of the parasite infrapopulation. Similar numbers apply for human malaria vectors and still need to be quantified for various other hemosporidia-transmitting dipterans as well as for other populations of our study system (Smith et al., 2014). The minimum conditions for such measurements are a) being able to sustain vectors alive for the developmental period after initial engorgement on an infected host and b) being able to extract the salivary glands without substantial contamination from the gut (Bukauskaitė et al., 2019, 2019; Chagas et al., 2019; Ilgūnas et al., 2019). Our estimates were based on mitogenome copies typical for gametocytes and thus, all cell number estimates are in effect expressed in gametocyte mitogenome density equivalents, without knowledge how well this corresponds to nuclear equivalents. The complementary methods suggested above should help to verify the number of sporozoites found in salivary glands. Such strong bottlenecks increase the effects of genetic drift in the overall population and allow the application of alternative evolutionary strategies by mutants without competition. Such genetic bottlenecks may be a main mechanisms leading to the impressive diversity of hemosporidian parasite lineages and their variable infection and coexistence strategies (Bergstrom et al., 1999; Alcala et al., 2017). Another mechanism with potentially comparable evolutionary consequences may result from high ratios of vertical-to-horizontal transmission, i.e. quasi-vertical transmission conducted by some vectors mainly between parents and offspring because of locally restricted foraging and/or other foraging patterns of vectors (Chakarov et al., 2015, 2020, 2021).
The low number of transmissible parasites per vector suggests either very high efficiency of invasion during every bite (which does not appear to be the case in human malaria (Beier et al., 1991)), or many repeatable chances for transmission. Both of these life history traits likely differ between lineages and between vector species and their feeding styles, as well as between core and edge distribution areas of hosts and vectors. The vector species used in our study, Pseudolynchia canariensis, is a louse fly specialized on house pigeons. As Pseudolynchia inhabit the skin under the feathers of birds, they are most likely transmitted to pigeon nestlings by their parents and potentially infect these with the parasites of the parents. Such a mechanism has been suggested for solitary hosts and may be effective but weaker in colonial pigeons (Chakarov et al., 2015). Despite having preferences for specific host traits, such as for dark plumage coloration, vector individuals specialized to colonial house pigeons likely have the opportunity to switch often between individual hosts within a colony and potentially transmit several times per day (Jacquin et al., 2013). While house pigeons have now colonized most of the global urbanized areas, their coloniality is an ancestral trait, which was also common in the original colonies in rocks around the Mediterranean. Additionally, among the hemosporidian vectors, only in hippoboscid flies both females and males feed on blood which can double the frequency of transmission (Waite et al., 2012). Under these colonial circumstances Haemoproteus columbae and other members of the subgenus Haemoproteus (Haemoproteus) may not require high sporozoite numbers in individual salivary glands, nor a high colonization efficiency after transmission and can still reach near-panmixia within the mobile host population. Nevertheless, with several described genetic lineages of H. columbae it would be very interesting to quantify the effects of competition or mutual enhancement between lineages in vertebrate and insect hosts (Palinauskas et al., 2018; Nebel et al., 2020; Palinauskas et al., 2022). Indeed, using a mixture of lineages may offer a further handle on tracing the development of effective population sizes of parasites at every step of their development in the invertebrate and vertebrate hosts, which can be a further development of the use of blood parasites as biological markers (Valkiūnas, 2004). Finer genetic markers of deeper population structure within a hemosporidian lineage will become available with advancing molecular methods and should improve the resolution of these mechanisms even more (Hellgren et al., 2015; Huang et al., 2018, Chakarov et al. in prep). These processes can be studied exceptionally well with the system used here, both in controlled experiments in captivity and by sampling of wild populations. The dynamics of these processes can be expected to change when the vectors become rare, e.g. at the edge of the distribution of the temperature-limited Pseudolynchia (Klei and Degiusti, 1975). Given the overall sturdiness of hippoboscid flies, they may be also relatively resistant (or tolerant) and not experience substantial damage after ingesting blood with high Haemoproteus infection intensity. This may constitute an additional difference to vectors from other groups but should be specifically tested and could co-determine the small number of sporozoites in the salivary glands (Bukauskaitė et al., 2016).
To our knowledge, this is the first attempt of quantification of the bottleneck of avian hemosporidian parasites in hippoboscid vectors, based on molecular methods. More such attempts would be valuable, as this variable may be key to understand the differences between hemosporidian species and lineages in terms of prevalence, specialization and virulence, as well as other aspects of host-pathogen co-evolution in general (Bergstrom et al., 1999; Valkiūnas, 2004; Alcala et al., 2017; Santiago-Alarcon and Marzal, 2020). We therefore call for its exploration in the great diversity of vector-transmitted parasites.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
KF: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. NC: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We acknowledge the financial support of the German Research Foundation (DFG) and the Open Access Publication Fund of Bielefeld University for the article processing charge. This work was supported by the DFG, as part of the SFB TRR 212 (NC 3) -Project numbers 316099922 and 396780709; DFG-project number 398434413; and DFG project number 433069365. The funding body played no role in the design of the study and collection, analysis, interpretation of data, and in writing the manuscript.
Acknowledgments
We would like to thank the animal caretakers of the department of Animal Behaviour and Oliver Krüger for help in managing the house pigeon population. We are grateful to Diego Santiago-Alarcon, Aneliya Bobeva, and an anonymous referee for their helpful comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1319829/full#supplementary-material
References
Abel S., Abel zur Wiesch P., Davis B. M., Waldor M. K. (2015). Analysis of bottlenecks in experimental models of infection. PloS Pathog. 11, e1004823. doi: 10.1371/journal.ppat.1004823
Alcala N., Jenkins T., Christe P., Vuilleumier S. (2017). Host shift and cospeciation rate estimation from co-phylogenies. Ecol. Lett. 20, 1014–1024. doi: 10.1111/ele.12799
Asghar M., Hasselquist D., Bensch S. (2011). Are chronic avian hemosporidian infections costly in wild birds? J. Avian Biol. 42, 530–537. doi: 10.1111/jav.2011.42.issue-6
Baton L. A., Ranford-Cartwright L. (2005). Spreading the seeds of million-murdering death*: metamorphoses of malaria in the mosquito. Trends Parasitol. 21, 573–580. doi: 10.1016/j.pt.2005.09.012
Beerntsen B. T., James A. A., Christensen B. M. (2000). Genetics of mosquito vector competence. Microbiol. Mol. Biol. Rev. 64, 115–137. doi: 10.1128/MMBR.64.1.115-137.2000
Beier J. C., Davis J. R., Vaughan J. A., Noden B. H., Beier M. S. (1991). Quantitation of Plasmodium falciparum sporozoites transmitted in vitro by experimentally infected Anopheles Gambiae and Anopheles stephensi. Am. J. Trop. Med. Hygiene. 44 (5), 564–570. doi: 10.4269/ajtmh.1991.44.564
Bell A., Ranford-Cartwright L. (2004). A real-time PCR assay for quantifying Plasmodium falciparum infections in the mosquito vector. Int. J. Parasitol. 34, 795–802. doi: 10.1016/j.ijpara.2004.03.008
Bensch S., Canbäck B., DeBarry J. D., Johansson T., Hellgren O., Kissinger J. C., et al. (2016). The genome of haemoproteus tartakovskyi and its relationship to human malaria parasites. Genome Biol. Evol. 8, 1361–1373. doi: 10.1093/gbe/evw081
Bergstrom C. T., McElhany P., Real L. A. (1999). Transmission bottlenecks as determinants of virulence in rapidly evolving pathogens. Proc. Natl. Acad. Sci. 96, 5095–5100. doi: 10.1073/pnas.96.9.5095
Bernotienė R., Žiegytė R., Vaitkutė G., Valkiūnas G. (2019). Identification of a new vector species of avian haemoproteids, with a description of methodology for the determination of natural vectors of hemosporidian parasites. Parasites Vectors 12, 1–10. doi: 10.1186/s13071-019-3559-8
Brandler T., Chakarov N. (2024). Mitochondrial and apicoplast genome copy abundances of hemosporidian parasites are explained by host species and parasitic lineage. Front. Ecol. Evol. 12, 1305418. doi: 10.3389/fevo.2024.1305418
Bukauskaitė D., Bernotienė R., Iezhova T. A., Valkiūnas G. (2016). Mechanisms of mortality in Culicoides biting midges due to Haemoproteus infection. Parasitology 143, 1748–1754. doi: 10.1017/S0031182016001426
Bukauskaitė D., Chagas C. R. F., Bernotienė R., Žiegytė R., Ilgūnas M., Iezhova T., et al. (2019). A new methodology for sporogony research of avian haemoproteids in laboratory-reared Culicoides spp., with a description of the complete sporogonic development of Haemoproteus pastoris. Parasites Vectors 12, 1–13. doi: 10.1186/s13071-019-3832-x
Cepeda A. S., Lotta-Arévalo I. A., Pinto-Osorio D. F., Macías-Zacipa J., Valkiūnas G., Barato P., et al. (2019). Experimental characterization of the complete life cycle of Haemoproteus columbae, with a description of a natural host-parasite system used to study this infection. Int. J. Parasitol. 49, 975–984. doi: 10.1016/j.ijpara.2019.07.003
Chagas C. R. F., Bukauskaitė D., Ilgūnas M., Bernotienė R., Iezhova T., Valkiūnas G. (2019). Sporogony of four Haemoproteus species (Haemosporida: Haemoproteidae), with report of in vitro ookinetes of Haemoproteus hirundinis: phylogenetic inference indicates patterns of hemosporidian parasite ookinete development. Parasites Vectors 12, 1–16. doi: 10.1186/s13071-019-3679-1
Chakarov N., Kampen H., Wiegmann A., Werner D., Bensch S. (2020). Blood parasites in vectors reveal a united blackfly community in the upper canopy. Parasites Vectors 13, 309. doi: 10.1186/s13071-020-04177-0
Chakarov N., Linke B., Boerner M., Goesmann A., Kruger O., Hoffman J. I. (2015). Apparent vector-mediated parent-to-offspring transmission in an avian malaria-like parasite. Mol. Ecol. 24, 1355–1363. doi: 10.1111/mec.13115
Chakarov N., Veiga J., Ruiz-Arrondo I., Valera F. (2021). Atypical behavior of a black fly species connects cavity-nesting birds with generalist blood parasites in an arid area of Spain. Parasites Vectors 14, 1–9. doi: 10.1186/s13071-021-04798-z
Coatney G. R. (1931). On the biology of the pigeon fly, Pseudolynchia maura Bigot (Diptera, Hippoboscidae). Parasitology 23, 525–532. doi: 10.1017/S0031182000013901
Criscione C. D., Blouin M. S. (2005). Effective sizes of macroparasite populations: a conceptual model. Trends Parasitol. 21, 212–217. doi: 10.1016/j.pt.2005.03.002
Criscione C. D., Poulin R., Blouin M. S. (2005). Molecular ecology of parasites: elucidating ecological and microevolutionary processes. Mol. Ecol. 14, 2247–2257. doi: 10.1111/j.1365-294X.2005.02587.x
Edman J. D., Simmons K. R. (1985). Rearing and colonization of black flies (Diptera: Simuliidae). J. Med. Entomol. 22, 1–17. doi: 10.1093/jmedent/22.1.1
Elliot S. L., Adler F. R., Sabelis M. W. (2003). How virulent should a parasite be to its vector? Ecology 84, 2568–2574. doi: 10.1890/02-8013
Ferguson H., Rivero A., Read A. (2003). The influence of malaria parasite genetic diversity and anaemia on mosquito feeding and fecundity. Parasitology 127, 9–19. doi: 10.1017/S0031182003003287
Ghosh A., Edwards M., Jacobs-Lorena M. (2000). The journey of the malaria parasite in the mosquito: hopes for the new century. Parasitol. Today 16, 196–201. doi: 10.1016/S0169-4758(99)01626-9
Hajkazemian M., Bossé C., Mozūraitis R., Emami S. N. (2021). Battleground midgut: The cost to the mosquito for hosting the malaria parasite. Biol. Cell 113, 79–94. doi: 10.1111/boc.202000039
Haraguchi A., Takano M., Hakozaki J., Nakayama K., Nakamura S., Yoshikawa Y., et al. (2022). Molecular mechanisms of Plasmodium development in male and female Anopheles mosquitoes. bioRxiv, 2022–2001. doi: 10.1101/2022.01.27.477980
Haraguchi A., Takano M., Hakozaki J., Nakayama K., Nakamura S., Yoshikawa Y., et al. (2023). Formation of free oocysts in Anopheles mosquitoes injected with Plasmodium ookinetes. J. Vet. Med. Sci. 85, 921–928. doi: 10.1292/jvms.23-0099
Hellgren O., Atkinson C. T., Bensch S., Albayrak T., Dimitrov D., Ewen J. G., et al. (2015). Global phylogeography of the avian malaria pathogen Plasmodium relictum based on MSP1 allelic diversity. Ecography 38, 842–850. doi: 10.1111/ecog.01158
Himmel T., Harl J., Kübber-Heiss A., Konicek C., Fernández N., Juan-Sallés C., et al. (2019). Molecular probes for the identification of avian Haemoproteus and Leucocytozoon parasites in tissue sections by chromogenic in situ hybridization. Parasites Vectors 12, 1–10. doi: 10.1186/s13071-019-3536-2
Huang X., Hansson R., Palinauskas V., Valkiūnas G., Hellgren O., Bensch S. (2018). The success of sequence capture in relation to phylogenetic distance from a reference genome: a case study of avian hemosporidian parasites. Int. J. Parasitol. 48, 947–954. doi: 10.1016/j.ijpara.2018.05.009
Huang X., Huang D., Liang Y., Zhang L., Yang G., Liu B., et al. (2020). A new protocol for absolute quantification of hemosporidian parasites in raptors and comparison with current assays. Parasites Vectors 13, 1–9. doi: 10.1186/s13071-020-04195-y
Huyse T., Poulin R., Theron A. (2005). Speciation in parasites: a population genetics approach. Trends Parasitol. 21, 469–475. doi: 10.1016/j.pt.2005.08.009
Ilgūnas M., Romeiro Fernandes Chagas C., Bukauskaitė D., Bernotienė R., Iezhova T., Valkiūnas G. (2019). The life-cycle of the avian hemosporidian parasite Haemoproteus majoris, with emphasis on the exoerythrocytic and sporogonic development. Parasites Vectors 12, 1–15.
Iyer J., Gruner A. C., Renia L., Snounou G., Preiser P. R. (2007). Invasion of host cells by malaria parasites: a tale of two protein families. Mol. Microbiol. 65, 231–249. doi: 10.1111/j.1365-2958.2007.05791.x
Jacquin L., Haussy C., Bertin C., Laroucau K., Gasparini J. (2013). Darker female pigeons transmit more specific antibodies to their eggs than do paler ones. Biol. J. Linn. Soc 108, 647–657. doi: 10.1111/bij.12001
Klei T. R., Degiusti D. L. (1975). Observations on the bionomics of Pseudolynchia canariensis (Diptera: Hippoboscidae). Parasitology 70, 195–202. doi: 10.1017/S0031182000049660
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Medica D. L., Sinnis P. (2005). Quantitative dynamics of Plasmodium yoelii sporozoite transmission by infected anopheline mosquitoes. Infect. Immun. 73, 4363–4369. doi: 10.1128/IAI.73.7.4363-4369.2005
Nebel C., Harl J., Pajot A., Weissenböck H., Amar A., Sumasgutner P. (2020). High prevalence and genetic diversity of Haemoproteus columbae (Haemosporida: Haemoproteidae) in feral pigeons Columba livia in Cape Town, South Africa. Parasitol. Res. 119, 447–463. doi: 10.1007/s00436-019-06558-6
Palinauskas V., Žiegytė R., Šengaut J., Bernotienė R. (2018). Different paths–the same virulence: experimental study on avian single and co-infections with Plasmodium relictum and Plasmodium elongatum. Int. J. Parasitol. 48, 1089–1096. doi: 10.1016/j.ijpara.2018.08.003
Palinauskas V., Žiegytė R., Šengaut J., Bernotienė R. (2022). Experimental study on primary bird co-infection with two Plasmodium relictum lineages—pSGS1 and pGRW11. Animals 12, 1879. doi: 10.3390/ani12151879
Pichon G., Awono-Ambene H., Robert V. (2000). High heterogeneity in the number of Plasmodium falciparum gametocytes in the bloodmeal of mosquitoes fed on the same host. Parasitology 121, 115–120. doi: 10.1017/S0031182099006277
Prentiss M., Chu A., Berggren K. K. (2022). Finding the infectious dose for COVID-19 by applying an airborne-transmission model to superspreader events. PloS One 17, e0265816. doi: 10.1371/journal.pone.0265816
Rivero A., Ferguson H. (2003). The energetic budget of Anopheles stephensi infected with Plasmodium chabaudi: is energy depletion a mechanism for virulence? Proc. R. Soc. London. Ser. B.: Biol. Sci. 270, 1365–1371. doi: 10.1098/rspb.2003.2389
Rose N. H., Shepard J. J., Ayala D. (2023). Establishing colonies from field-collected mosquitoes: special accommodations for wild strains. Cold Spring Harbor Protoc. doi: 10.1101/pdb.top107654
Rosenberg R., Wirtz R. A., Schneider I., Burge R. (1990). An estimation of the number of malaria sporozoites ejected by a feeding mosquito. Trans. R. Soc. Trop. Med. Hygiene. 84, 209–212. doi: 10.1016/0035-9203(90)90258-G
Santiago-Alarcon D., Marzal A. (2020). Avian malaria and related parasites in the tropics. Ecol. Evol. Syst.. ( Springer), 575. doi: 10.1007/978-3-030-51633-8
Santiago-Alarcon D., Palinauskas V., Schaefer H. M. (2012). Diptera vectors of avian Haemosporidian parasites: untangling parasite life cycles and their taxonomy. Biol. Rev. 87, 928–964. doi: 10.1111/j.1469-185X.2012.00234.x
Schmid-Hempel P. (2011). Evolutoinary Parasitology. The integrated study of infections, immunology, ecology, and genetics (Oxford, UK: Oxford University Press).
Schmid-Hempel P., Frank S. A. (2007). Pathogenesis, virulence, and infective dose. PloS Pathog. 3, e147. doi: 10.1371/journal.ppat.0030147
Simões M. L., Caragata E. P., Dimopoulos G. (2018). Diverse host and restriction factors regulate mosquito–pathogen interactions. Trends Parasitol. 34, 603–616. doi: 10.1016/j.pt.2018.04.011
Smith R. C., Vega-Rodríguez J., Jacobs-Lorena M. (2014). The Plasmodium bottleneck: malaria parasite losses in the mosquito vector. Memórias. do Inst. Oswaldo. Cruz. 109, 644–661. doi: 10.1590/0074-0276130597
Templeton A. R. (1980). The theory of speciation via the founder principle. Genetics 94, 1011–1038. doi: 10.1093/genetics/94.4.1011
Valkiūnas G. (2004). Avian malaria parasites and other haemosporidia (Boca Raton, USA: CRC press). doi: 10.1201/9780203643792
Valkiūnas G., Palinauskas V., Ilgūnas M., Bukauskaitė D., Dimitrov D., Bernotienė R., et al. (2014). Molecular characterization of five widespread avian hemosporidian parasites (Haemosporida), with perspectives on the PCR-based detection of hemosporidians in wildlife. Parasitol. Res. 113, 2251–2263. doi: 10.1007/s00436-014-3880-2
Vaughan J. A. (2007). Population dynamics of Plasmodium sporogony. Trends Parasitol. 23, 63–70. doi: 10.1016/j.pt.2006.12.009
Vaughan J. A., Noden B. H., Beier J. C. (1992). Population dynamics of Plasmodium falciparum sporogony in laboratory-infected Anopheles Gambiae. J. Parasitol. 78(4), 716–724. doi: 10.2307/3283550
Vaughan J. A., Noden B. H., Beier J. C. (1994). Sporogonic development of cultured Plasmodium falciparum in six species of laboratory-reared Anopheles mosquitoes. Am. J. Trop. Med. Hygiene. 51 (2), 233–243. doi: 10.4269/ajtmh.1994.51.233
Waite J. L., Henry A. R., Adler F. R., Clayton D. H. (2012). Sex-specific effects of an avian malaria parasite on an insect vector: support for the resource limitation hypothesis. Ecology 93, 2448–2455. doi: 10.1890/11-2229.1
Weaver S. C., Barrett A. D. T. (2004). Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2, 789–801. doi: 10.1038/nrmicro1006
Weiss B., Aksoy S. (2011). Microbiome influences on insect host vector competence. Trends Parasitol. 27, 514–522. doi: 10.1016/j.pt.2011.05.001
Keywords: avian malaria, hemosporidian blood parasites, genetic bottleneck, host-parasite coevolution, vector-transmitted diseases, vertical transmission, louse flies, house pigeon
Citation: Fischer K and Chakarov N (2024) Genetic infrapopulation sizes in blood parasites: a pilot quantification of the bottleneck in louse fly vectors. Front. Ecol. Evol. 12:1319829. doi: 10.3389/fevo.2024.1319829
Received: 11 October 2023; Accepted: 27 March 2024;
Published: 09 April 2024.
Edited by:
Benjamin Jerry Ridenhour, University of Idaho, United StatesReviewed by:
Diego Santiago-Alarcon, University of South Florida, United StatesAneliya Borisova Bobeva, Bulgarian Academy of Sciences, Bulgaria
Copyright © 2024 Fischer and Chakarov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nayden Chakarov, nayden.chakarov@uni-bielefeld.de
 Kai Fischer1
Kai Fischer1  Nayden Chakarov
Nayden Chakarov