Variations in gut microbiota associated with changes in life-history traits of Daphnia galeata induced by fish kairomones
- Department of Biotechnology, Sangmyung University, Seoul, Republic of Korea
The gut microbiota plays a crucial role in host physiology and the disruption of host–microbiota relationships caused by environmental stressors can impact host growth and survival. In this study, we used Daphnia galeata as a model organism to investigate the interactive effects of fish kairomones on the life-history traits and gut microbiota alterations of D. galeata, as well as the relationship between life-history traits and gut microbiota composition. The presence of fish kairomones enhanced fecundity, decreased growth, and altered gut microbiota, with significant changes in alpha diversity but not in beta diversity in the genotype KB5 of D. galeata. Statistical analysis revealed that the relative abundance of the Pseudomonadaceae family significantly increased upon exposure to fish kairomone, while the relative abundance of the Comamonadaceae family significantly decreased. The decreased growth in genotype KB5 may be associated with a significant increase in Pseudomonas, a member of the family Pseudomonadaceae, which is generally deficient in essential fatty acids, potentially negatively impacting growth. Meanwhile, it is speculated that the significant decrease in Limnohabitans belonging to the Comamonadaceae family is associated with the reduction of body size and increased fecundity of KB5 when exposed to fish kairomones. Furthermore, the genus Candidatus Protochlamydia was observed only under the fish kairomones-treated condition. These data suggest that variations in host life-history traits related to reproduction and growth are potentially associated with the relative abundance or presence of these microbial genera. Our research findings provide valuable insights into understanding the impact of biotic stress on the interaction between hosts and microbiota.
1 Introduction
It is well known that complex microbial communities that reside in animal gastrointestinal tracts have crucial functions in various biological processes (Colston, 2017). The extent to which bacterial communities impact host life history, physiology, and behavior has been a long-standing subject of research, and there is currently a growing interest in understanding the variation in microbial communities associated with these changes (Akbar et al., 2020; Frankel-Bricker et al., 2020; Rajarajan et al., 2023). Understanding the feedback between hosts and their associated microbiomes and investigating whether environmental conditions affect microbe–host interactions is now widely considered to be an important area of study in ecology and evolution (Frankel-Bricker et al., 2020). Microbial communities, including symbiotic bacteria, fungi, protozoa, and others, are known as the microbiome. They can inhabit an animal’s digestive tract, skin, and other organs, playing a crucial role in host physiology and behavior (Frankel-Bricker et al., 2020). According to previous studies, variations in the gut microbiota have been shown to have various impacts, ranging from life-history traits to adaptation and ecological processes of the host, contributing to the general health of the host (Macke et al., 2017; Bang et al., 2018). Additionally, the gut bacteria of many arthropods have a preponderant impact on the reproduction, development, growth, and survival of the host (Akbar et al., 2020; Frankel-Bricker et al., 2020; Liu et al., 2022).
Predation risk may affect the diversity and composition of the host gut microbiome’s diversity and composition, which is critical for the fitness of individuals. A recent study has shown that the host–microbial composition varies under different stressors, including predation stress in Daphnia species (Sadeq et al., 2021; Liu et al., 2022; Rajarajan et al., 2023). Aquatic predators release kairomones into the water, which trigger various responses in the prey that can reduce the prey’s vulnerability to predators (Stibor and Lüning, 1994; Schoeppner and Relyea, 2009). These responses induced by predators represent typical instances of phenotypic plasticity and have been reported in various Daphnia species (Yin et al., 2011; Herzog et al., 2016). This genus links trophic levels from primary producers to consumers in freshwater ecosystems and is therefore vulnerable to a high risk of predation (Lampert, 2011). When Daphnia are exposed to fish kairomones, variations in life-history traits, morphological, physiological, and behavioral plasticity, as well as other effects, have been reported (Lampert, 1993; Carter et al., 2013). Among the Daphnia species, Daphnia galeata is a relatively large zooplankton found throughout the northern hemisphere and has been identified as a dominant plankton species in the Han River, Korea (Choi et al., 2023a). The freshwater crustacean D. galeata, which has a short life cycle, a rapid response to environmental stress, convenient clonal reproduction under laboratory conditions, and typical defense mechanisms induced by predators in life-history traits, is an appropriate organism for research in this field (Tams et al., 2018; Choi et al., 2023b).
The gut microbiota of Daphnia has been shown to influence its fitness and survival (Peerakietkhajorn et al., 2016; Callens et al., 2018). Recent advances in sequencing technology and other molecular techniques have revealed that the microbiota plays an essential role in host fitness, adaptation, and evolution (King et al., 2013; Foster et al., 2017). Metagenomic studies have utilized next-generation sequencing methods and bioinformatic workflows to survey these communities in various study systems (Frankel-Bricker et al., 2020). For example, when Daphnia pulex was exposed to two stressors, copper and predation, the composition of the D. pulex microbiome community exhibited distinct and unique patterns upon exposure to each stress factor or both (Sadeq et al., 2021). In a previous study that examined the impact of food ration and predation on the overall composition of the gut microbiota of Perca fluviatilis, it was found that Fusobacteria increased under conditions of low food ration and predation, indicating a preference for Fusobacteria in stressful situations (Zha et al., 2018). Here, we investigate the association between phenotypic variation and changes in the gut microbial community in response to fish kairomones in D. galeata. We used 16S rRNA gene amplicon sequencing to investigate the gut microbial communities of D. galeata. This is in line with the broader context of understanding the intricate relationships between host organisms and their associated microbiomes, shedding light on the impacts of environmental stressors on microbial communities and their implications for host fitness and survival.
2 Materials and methods
2.1 Sample collection and culture
D. galeata was collected from the Hoam Reservoir in Chungcheongbuk Province (36°42′48.70′′ N, 127°25′34.41′′ E), Korea, in March 2023, using a Kitahara Zooplankton net with a mesh size of 55 μm (Samjee-tech, Korea). Once the samples were transferred to the laboratory, each egg-bearing female was placed in a separate beaker for parthenogenetic reproduction. D. galeata individuals were cultured under laboratory conditions (20°C, a 16-h light/8-h dark cycle using ADaM (Aachener Daphnien medium)). The ADaM medium, composed of A (CaCl2 x 2H2O), B (NaHCO3), and C (SeO2) stocks, was prepared according to Klüttgen et al. (1994). D. galeata individuals were reared in 1 L beaker at densities of fewer than 30 neonates per beaker. We supplied 1 mL of Chlorella vulgaris per liter of D. galeata culture medium daily after diluting 1.0 mg of C L-1 Chlorella vulgaris cultured in the laboratory with 50 mL of distilled water. The genotype of D. galeata (KB5 clonal line) was identified through sequence analyses of mitochondrial cytochrome oxidase I (COX1) and NADH dehydrogenase subunit 2 (ND2) genes.
2.2 Experimental design and a life-history experiment
The study used D. galeata, which had been cultured for 6 months prior to the experiment and had formed an established clonal line. The ADaM medium was used as the control group medium. In order to mimic fish predation, D. galeata was exposed to a kairomone-enriched medium. The medium was obtained by cultivating approximately 20 freshwater mandarin fish (Siniperca scherzeri), which are known for consuming Daphnia species (Wang et al., 2022), in 100 L of water within the aquarium where the fish were raised. Furthermore, the medium containing kairomones was filtered using a 0.45-μm-pore-sized cellulose acetate membrane (Hyundai-Micro, Korea) and then mixed at a ratio of 1:2 by combining 600 mL of ADaM medium with 300 mL of the medium containing kairomones. The control (fish-kairomone-nonexposed) and experimental (fish-kairomone-exposed) groups were cultured in 1 L beakers with less than 30 neonates in each. After several trials, the ratio of fish kairomone medium was determined to be the optimal ratio for inducing changes in life-history traits while also ensuring the survival rate of D. galeata. The experimental design and procedures were based on a previous study (Choi et al., 2023b). To minimize interindividual variances, D. galeata was bred for two generations in a kairomone-free medium (control group) and a fish kairomone medium (experimental group) before the experiment (Tams et al., 2018). The D. galeata individuals used in this experiment are the same age. We recorded the life-history traits of each experimental individual for 21 days (t21) to examine the long-term effects of fish kairomones. To record life history traits, we used a total of 60 D. galeata individuals, with 20 individuals used in each of the three replicates. Life-history traits related to reproduction, such as “Age at First Reproduction”, “Number of Offspring First Brood”, “Total Number of Broods”, and “Total Number of Offspring” were estimated. The “Age at First Reproduction” refers to the day when the first neonates were released from the brood pouch. The “Number of Offspring First Brood” indicates the number of neonates on the day of the first reproduction. The “Total Number of Broods” represents the total number of times each individual reproduced during the experiment. The “Total Number of Offspring” was calculated by summing up the number of offspring produced by each individual during the experiment. Furthermore, life-history traits related to growth were recorded, such as the “Somatic Growth Rate” and the “Body Length”. The “Somatic Growth Rate” of D. galeata was determined by dividing the difference in body length between the neonates at the start (t0) and the adults at the end (t21) of the experiment by the total experimental duration. Body length of D. galeata was measured from the top of the head, through the middle of the eye, to the base of the spine excluding the spine itself (Tams et al., 2018). The individuals were observed with the CX22LED microscope (Olympus, Japan) and photographed with the eXcope T500 microscope camera (DIXI Science, Korea). In order to assess variations in life-history traits between the presence and absence of fish kairomones, we performed one-way analysis of variance (ANOVA) statistical analyses using the aov() function in R version 4.2.2 (R Core Team, 2017; MacFarland, 2014).
2.3 DNA preparation and high-throughput sequencing
At the end of the experiment, D. galeata individuals were pooled (n = 20), and three biological replicates per experimental condition (control and exposed fish kairomones) were used, resulting in 120 guts (20 individuals × 3 biological replicates × two conditions). Gut extraction from D. galeata was performed following the method of Akbar et al. (2021). The individuals cultured under both conditions were placed in fresh sterile ADaM medium for 24 h to remove food particles and nonsymbiotic bacteria from the gut (Akbar et al., 2020). Subsequently, individuals were gently washed with sterile water to remove external bacterial communities and then dissected under a stereoscopic microscope to isolate their guts. Next, DNA was immediately extracted from the guts of all experimental replicates. DNA extraction was performed using the Qiagen DNeasy blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Then, a polymerase chain reaction (PCR) was performed using total DNA samples as the template. The V3–V4 hypervariable regions of the partial 16S rRNA gene, with amplicons of approximately 459 base pairs, were amplified using the primer pairs S-D-Bact-0341-b-S-17 and S-D-Bact-0785-a-A-21 (Klindworth et al., 2013). In order to amplify sequences from various taxa, gradient PCR was performed by dividing the annealing temperature into 12 temperatures ranged from 40°C to 60°C with intervals of 1°C to 2.4°C set automatically by the PCR Thermal Cycler Dice® Gradient (TaKaRa, Tokyo, Japan). The PCR procedure included an initial denaturation at 95°C for 2 min, followed by 35 cycles consisting of 30 s at 95°C, 45 s in the temperature range of 40°C–60°C, and 45 s at 72°C and completed with a final extension step of 5 min at 72°C. Negative controls for PCR were included in each PCR run to assess the possibility of contamination. The amplified PCR products were visualized by gel electrophoresis, and the gradient PCR products were pooled in the same quantity. Sequencing libraries were prepared using the TruSeq Nano DNA High Throughput Library Prep Kit (Illumina, CA, USA). The library was measured for quality and quantity using the Agilent Technologies 4200 TapeStation D1000 screentape (Agilent Technologies, California, USA) and sequenced using an Illumina MiSeq sequencing platform with 300 base pair (bp) paired-end reads.
2.4 Bioinformatic analyses of 16S rRNA gene sequencing
Bioinformatics analyses were conducted using the QIIME2 version 2022.2.1 pipeline (Bolyen et al., 2019). Read quality scores were validated using FastQC version 0.11.9 (Andrews, 2010). Based on the read quality score, raw reads were trimmed for bases below Q30, which was implemented in QIIME2 to produce amplicon sequence variants (ASVs). Subsequent examinations were conducted using ASVs that allow for the preservation of biological sequence variation in the output reads, unlike traditional operational taxonomic unit (OTU) calling, which relies on similarity and may fail to capture subtle biological variations by grouping sequences (An et al., 2023). Low-quality base call-containing or chimera sequences were removed using DADA2 (Callahan et al., 2016). For the taxonomic assignment of the processed ASVs, the consensus-BLAST function of QIIME2 was used (Camacho et al., 2009; Bolyen et al., 2019). Taxonomies were assigned to the resulting ASV with a q2-feature-classifier plug-in using consensus-BLAST based on a 16S rRNA silva 138 SILVA SSU gene database with 80% similarity (Quast et al., 2012). ASVs were normalized to the total number of reads for each sample. After normalization, the ASVs were analyzed at the phylum level up to the lowest assignable taxonomic levels. Nonmicrobiome sequences were excluded from downstream analyses and unassigned ASVs were removed. The significance of relative abundance between the control and experimental groups is calculated using one-way ANOVA statistical analyses by the aov() function in R version 4.2.2 (R Core Team, 2017). Alpha diversity was determined for the control and experimental samples using the QIIME2 diversity function. Alpha diversity measures included Chao1 richness, number of ASVs, Shannon index, and Faith’s phylogenetic diversity. Pairwise comparisons between alpha diversity under experimental conditions were assessed using the Kruskal–Wallis test (Kruskal and Wallis, 1952). Beta diversity was determined using Bray–Curtis distance measures, which were represented by exporting the QIIME2 output to the R package Phyloseq (McMurdie and Holmes, 2013). QIIME2 was used to determine the depth of sequencing required to accurately represent the species present in the sample by calculating rarefaction curves for ASV in microbiome metabarcoding with 16S rRNA markers. Gut bacterial taxa that exhibited significantly different abundances between the control and experimental groups were identified using DESeq2 (Love et al., 2014). The results were filtered post hoc to decrease the false discovery rate using an adjusted p-value <0.05 (Benjamini and Hochberg, 1995). Furthermore, the results were filtered using a fold change ≥of 2 (log2FC ≤ −1 or log2FC ≥ 1).
3 Results
3.1 Effect of fish kairomone on life-history traits
Six life-history traits related to reproduction and growth were measured to compare variations in life-history traits under predator stress in the genotype KB5 of D. galeata. The reaction norms of the life-history traits in D. galeata demonstrate adaptation strategies in response to predator kairomones (Figure 1). The results of one-way ANOVA indicate a significant effect of the “fish kairomones” (P < 0.001) on the six life-history traits, except for the “Total Number of Broods” (Figure 1). In the life-history traits related to reproduction, in the presence of fish kairomones, D. galeata reproduced at a younger age (Figure 1A; P < 0.001). In contrast, in the presence of fish kairomones, D. galeata produced more offspring in their first brood (Figure 1B; P < 0.001). Furthermore, a significant increase (P < 0.001) was observed in the “Total Number of Offspring (Figure 1C),” but the “Total Number of Broods (Figure 1D)” did not show a significant change in the presence of fish kairomones. In the life-history traits related to growth, the “Somatic Growth Rate” and “Body Length” significantly decreased (P < 0.001) when exposed to fish kairomones (Figures 1E, F).
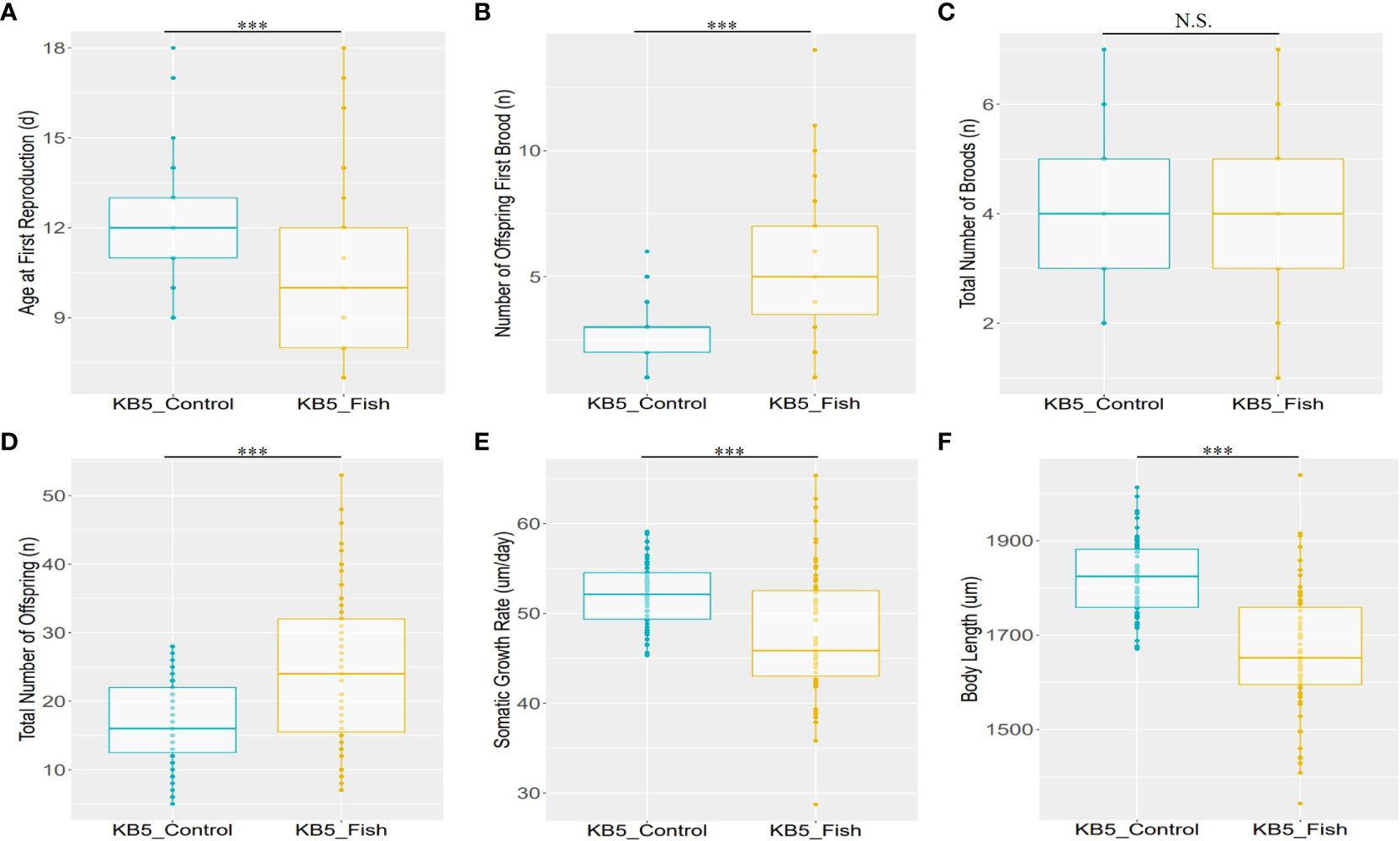
Figure 1 Box plots show the reaction norms for selected life-history traits of (D) galeata (median +/− SD). The experimental individuals were exposed for 21 days to record their life-history traits. (A) Age at First Reproduction, (B) Number of Offspring First Brood, (C) Total Number of Broods, (D) Total Number of Offspring, (E) Somatic Growth Rate, (F) Body Length. KB5_Control and KB5_Fish indicate the control (the absence of fish kairomones) and experimental (exposed to fish kairomones) groups of the genotype KB5, respectively. The dots indicate all individual datapoints. The stars denote significant differences (P.adjust*** < 0.001 and N.S., no significance) between the fish kairomones exposed and not exposed to the fish kairomones in the genotype KB5 of (D) galeata.
3.2 Summary of 16S rRNA sequence data and diversity of the gut microbiota
Metabarcoding sequences were generated for all experimental samples to analyze the gut microbiome of D. galeata. These sequencing efforts produced a total of 876,955 raw unpaired sequence reads from all samples. After applying stringent quality filtering, the sequences of 16S rRNA were obtained for 396,844 unpaired sequence reads from control samples and 377,905 unpaired sequence reads from experimental samples. Of these, 337,369 unpaired sequence reads from control samples and 322,156 unpaired sequence reads from experiment samples pair-merge sequences (reflecting 85.01% and 85.25% of filtered sequences) were subjected to denoising and merging in the 16S rRNA samples. Sequence chimeras were successfully removed and ASVs were extracted from the remaining sequences. The summary results of sequence processing are shown in Supplementary Table 1. The rarefaction curves showed that all samples reached near-saturation in species richness (Supplementary Figure 1). Therefore, it was considered that the sample had sufficient sequencing depth for this study. The ASVs extracted through this process were used for further analyses to compare the control and experimental samples. Diversity indices were calculated using rarefied ASV data for samples across fish kairomone exposure (Table 1). Exposure to fish kairomones increased the alpha diversity of the gut microbiome. This increase was particularly evident in the Chao1 richness, which accounts for rare species, the number of ASVs representing different variants, and the Shannon index, which measures richness and evenness. However, Faith’s phylogenetic diversity, which measures richness weighted by phylogeny, did not show a significant change (Table 1). These results indicate that while there is a significant difference in richness and abundance between KB5_Control and KB5_Fish, the phylogenetic diversity of the microbiomes that consist KB5_Control and KB5_Fish are not significantly different. Additionally, the Bray–Curtis dissimilarity metric used to measure beta diversity, considering dissimilarities in presence/absence and relative abundance of microbial species in fish-kairomone-exposed D. galeata versus fish-kairomone-nonexposed D. galeata, was insignificant (Table 1).
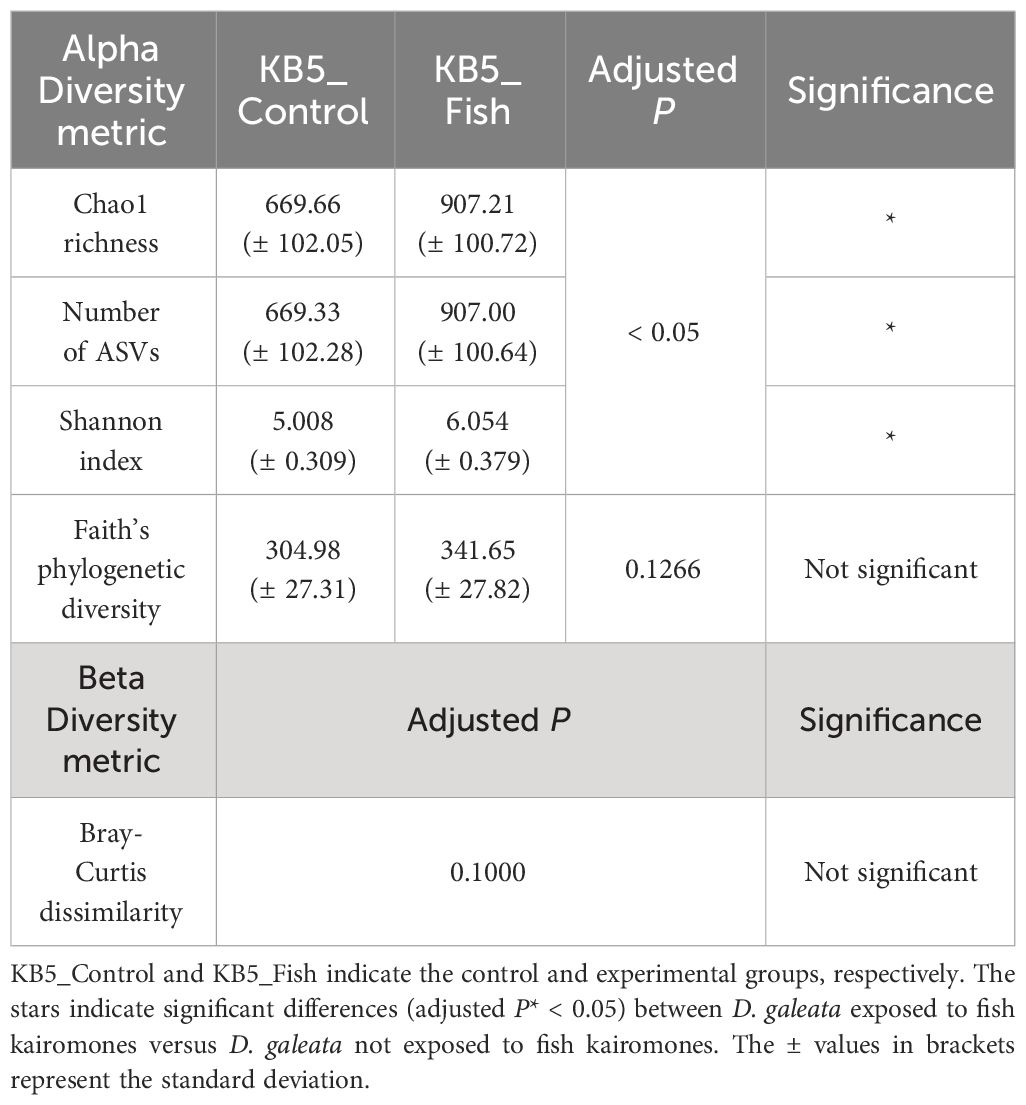
Table 1 Analysis of the alpha and beta diversity of the gut microbiome of the control (without fish kairomones) and experimental (exposed to fish kairomones) groups of D. galeata.
3.3 Composition of gut microbiome communities
There were no significant differences in beta diversity, but notable differences in alpha diversity. Based on this, we compared the relative abundance of gut microbial community compositions according to the exposure of fish kairomones between the control and experimental groups (Supplementary Table 2). We compared the community compositions between the control and experimental groups using all reads and those representing ≥0.1% to explore differences in the community compositions (Supplementary Tables 2, 3). The reads were removed about 1.2% of reads at the genus level and less than 1% at the family and phylum level when we removed reads with a relative abundance of less than 0.1% among all reads. Therefore, following the results, we consider the difference to be insignificant. As a result, we selected ASVs that assigned taxa comprising at least 0.1% of the reads in the average relative abundance across all samples following a previous study (Callens et al., 2020). We compared the relative abundances of the gut microbiota from the phylum level to the lowest assignable taxonomic level, the genus level (Figure 2; Supplementary Table 3). Specifically, we selected taxonomic levels, phylum, family, and genus that exhibited clear differences to compare the compositions of gut microbiome communities between both groups. At the phylum level, bacteria belonging to Proteobacteria dominated the gut microbiota in all samples, accounting for approximately 68.21% and 62.46% relative abundances in the fish-kairomone-nonexposed D. galeata and the fish-kairomone-exposed D. galeata, respectively (Figure 2; Supplementary Table 3). Additionally, Bacteroidota represented approximately 31.44% and 33.21% relative abundance in the control and treatment groups, respectively. However, the significance of relative abundance between the control group and the experimental group of Proteobacteria and Bacteroidota was observed to be as not significant (Supplementary Table 3). At the family level, Comamonadaceae, followed by Flavobacteriaceae, was the dominant family in both groups (Figure 2; Supplementary Table 3). The relative abundances of Flavobacteriaceae were similar, with 31.57% in KB5_Control and 33.31% in the KB5_Fish, with no significant differences (Supplementary Table 3). However, notable differences in relative abundances were observed when comparing the KB5_Control and KB5_Fish. Comamonadaceae’s distribution was approximately 66.94% in KB5_Control and 42.12% in KB5_Fish, representing a significant difference (P-value < 0.01) (Supplementary Table 3). Additionally, bacteria belonging to the Pseudomonadaceae, Burkholderiaceae, Moraxellaceae, and Parachlamydiaceae families were detected at 0.51%, 0.07%, 0.04%, and 0.02% in the KB5_Control, respectivley. However, these taxa were identified in KB5_Fish as approximately 6.98%, 6.73%, 5.28%, and 1.79% of the mapped reads, respectively. Pseudomonadaceae did not show significant differences between the relative abundance of KB5_Control and KB5_Fish, whereas these differences were significant (P-value < 0.001) (Supplementary Table 3) in Burkholderiaceae, Moraxellaceae, and Parachlamydiaceae. Meanwhile, at the genus level, Limnohabitans and Flavobacterium were found in both groups (Figure 2C; Supplementary Table 3). Limnohabitans had a relative abundance of approximately 64.19% in the KB5_Control, while the KB5_Fish had a relative abundance of approximately 40.76%, with a significant difference (P-value < 0.01). Flavobacterium was represented at 32.51% and 34.89% in KB5_Control and KB5_Fish, respectively, with no significance (Supplementary Table 3). Pseudomonas, Polynucleobacter, and Acinetobacter were also higher in KB5_Fish at 7.31%, 6.94%, and 5.49% compared to KB5_Control with abundances of less than 1%. Despite this, Pseudomonas did not demonstrate notable differences in relative abundance between KB5_Control and KB5_Fish, whereas Polynucleobacter and Acinetobacter showed significance (P-value < 0.001) (Supplementary Table 3).
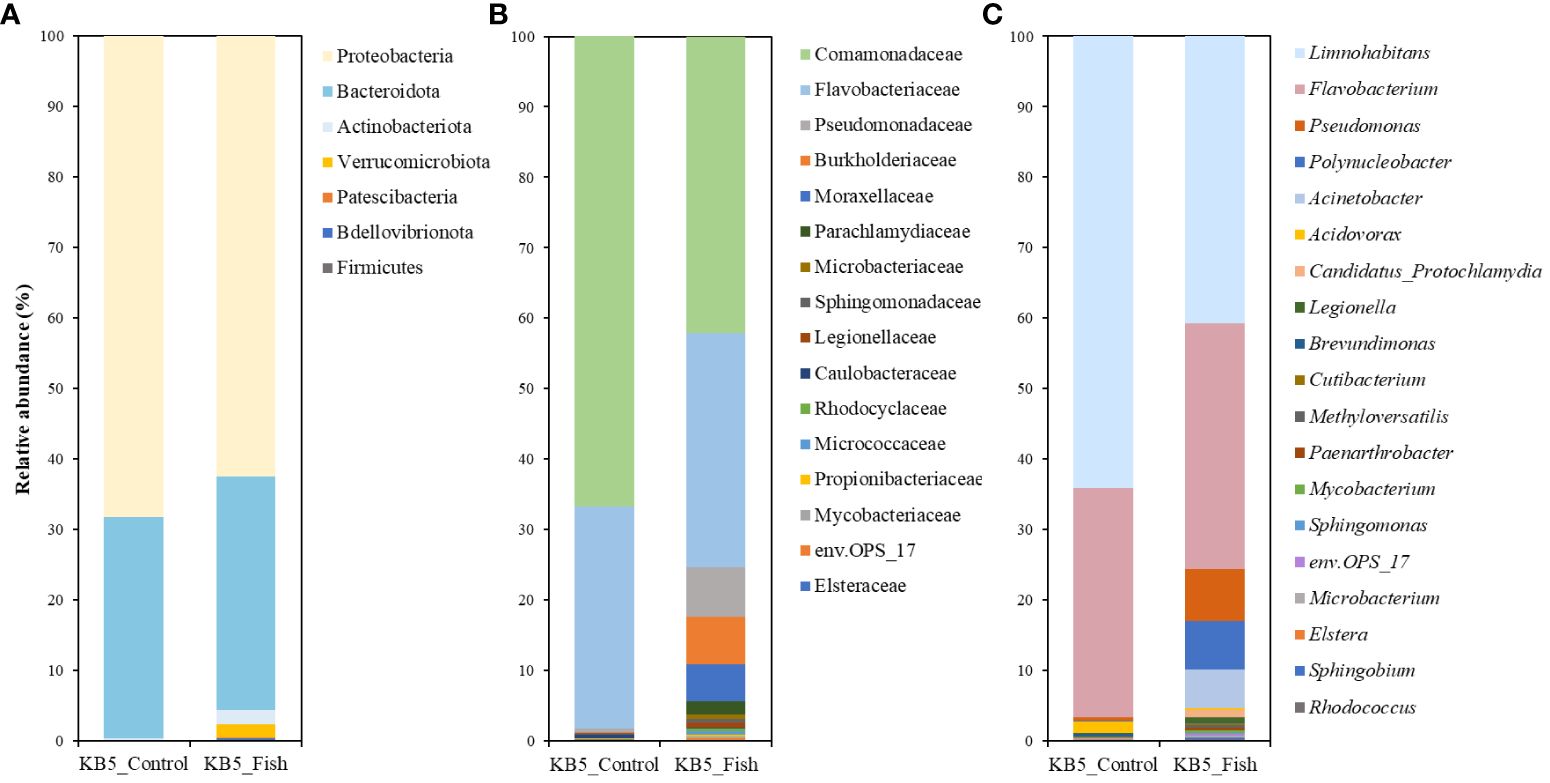
Figure 2 Relative abundance of the gut microbiota of (D) galeata under control and fish-kairomone-induced conditions at the levels of the phylum (A), family (B), and genus (C). KB5_Control indicates the control group (without fish kairomones), and KB5_Fish indicates the experimental group (exposed to fish kairomones).
3.4 Variations in the differential abundance of bacterial taxa in the gut of D. galeata
Based on the relative abundance of the compositions of the gut microbiome community, we identified specific ASVs that exhibited statistically significant differences concerning exposure to fish kairomones. Differentially abundant ASVs assigned to the SILVA database were found between the control and experimental groups (Figure 3). Differentially abundant ASVs were selected using DESeq2 with the criteria of p-value <0.05 and fold change ≥2. The complete taxonomic assignment results for all significantly differentially abundant ASVs are provided in Supplementary Table 4. Notably, a differential abundance of ASV was observed between the control and experimental groups at the family level (Figure 3; Table 2). Seventy-four differentially abundant ASVs were identified, with 59 ASVs showing a significant increase and 15 ASVs showing a significant decrease in the group exposed to fish kairomones compared to the group without fish kairomone treatment (Supplementary Table 4). Among the taxonomically assigned ASVs, Pseudomonadaceae (ASV14) and Comamonadaceae (ASV1) exhibited the highest log2 fold changes of 13.86 and −16.44, respectively, in the experimental group compared to the control group (Figure 3; Table 2). Furthermore, the relative abundances of Moraxellaceae (ASV24, log2 fold change: 12.63), Burkholderiaceae (ASV41, log2 fold change: 11.13), and Parachlamydiaceae (ASV44, log2 fold change: 11.06) increased in the experimental group compared to the control group (Figure 3; Table 2). Most of the ASVs that were significantly decreased between the control and experimental groups belong to the Comamonadaceae family (Figure 3; Table 2). In summary, exposure of fish kairomone to genotype KB5 had wide-ranging effects on the microbiota, leading to a decrease in the relative abundance of mainly Comamonadaceae, but an increase in relative abundances across multiple families.
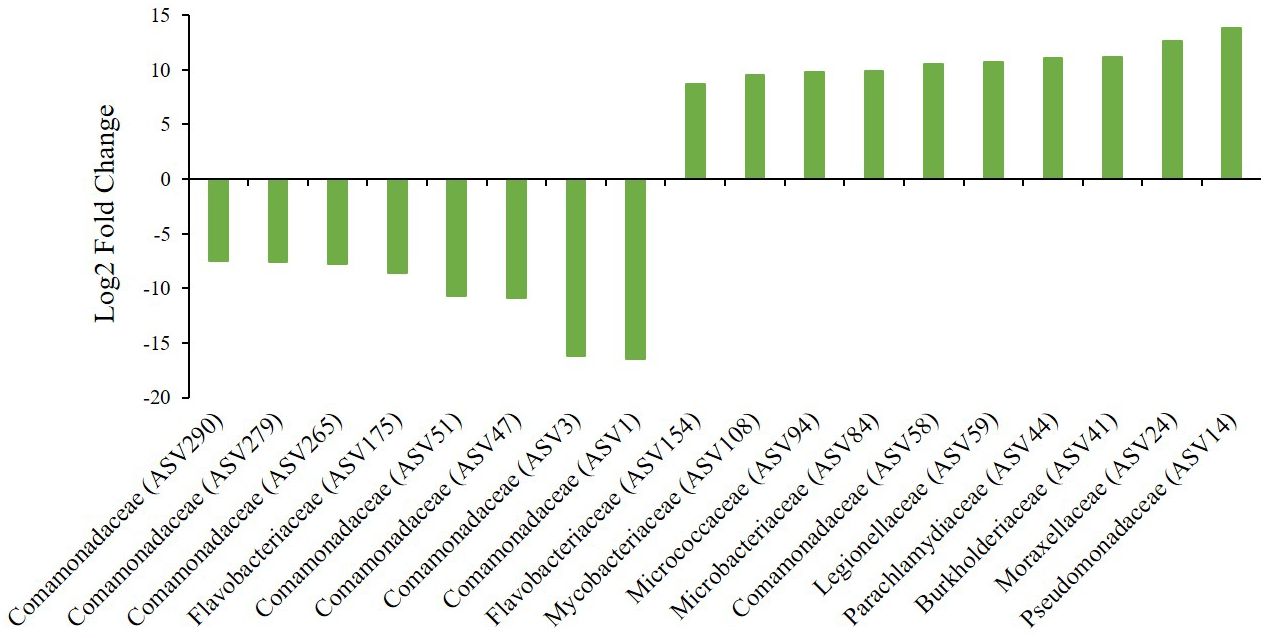
Figure 3 Gut bacterial taxa showing significant differences between the control group (without fish kairomones) and the experimental group (exposed to fish kairomones) in the genotype KB5 of D. galeata. Each bar represents an individual ASV identified at the family level, with the family name shown on the x axis. The presented ASVs are those with the highest or lowest log2FoldChange values among those belonging to the same family in Supplementary Table 3. Negative values of the log2-fold change exhibited a lower abundance of bacterial taxa in the experimental group compared to the control group, whereas positive values exhibited an increase in the abundance of a particular family in the experimental samples compared with the control group.
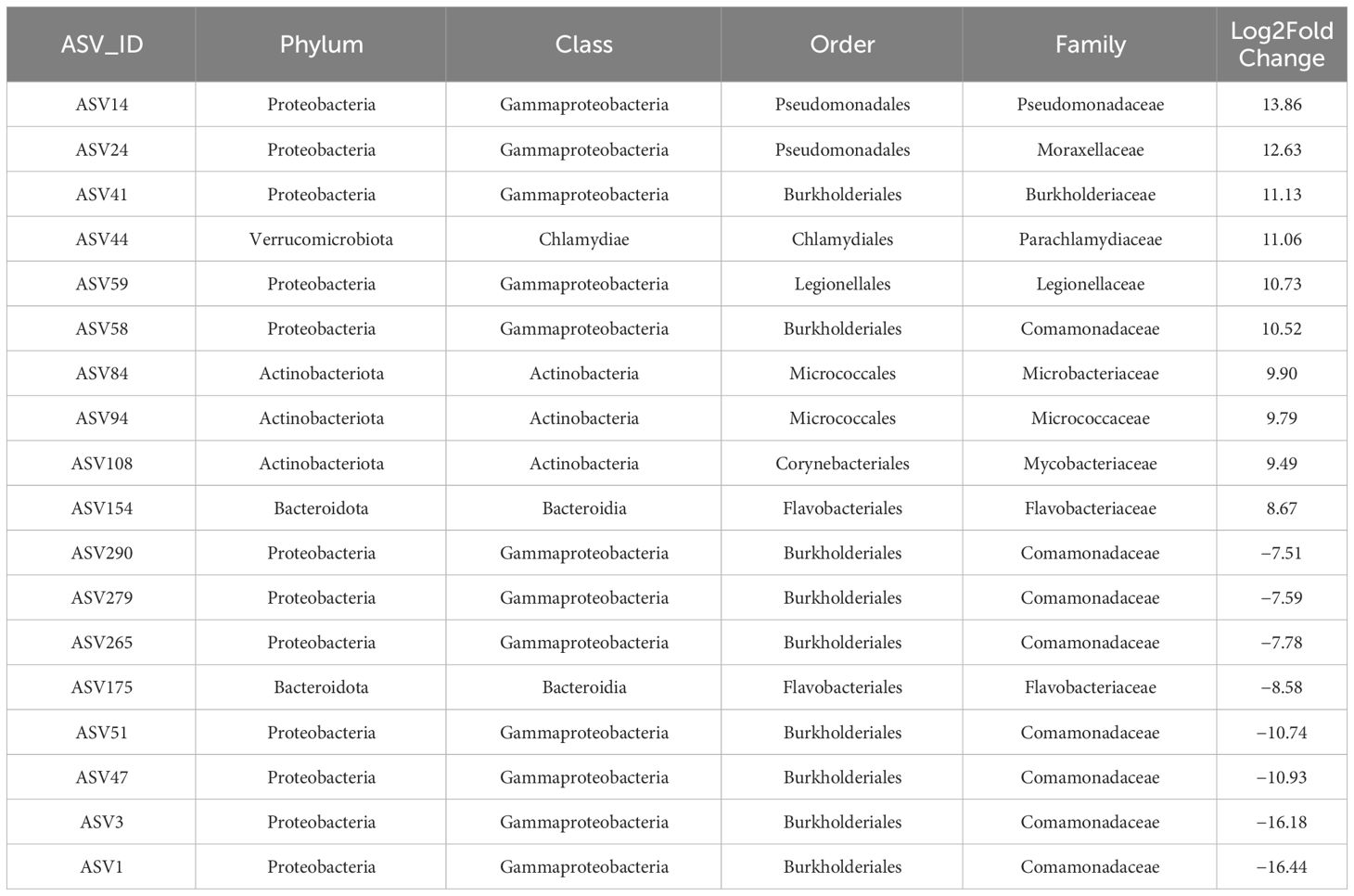
Table 2 The representative gut bacterial taxa identified as differentially abundant between the control group (without fish kairomones) and the experimental group (exposed to fish kairomones) in genotype KB5.
4 Discussion
Evidence suggests that the gut microbiome is essential for influencing life-history traits and adapting to changing environmental conditions. There are increasing attempts to predict the gut microbiome’s impact on the host by understanding the microbiome community and investigating associations between variations in the gut microbiome and host phenotypes (Liu et al., 2021). Similar to numerous eukaryotes, Daphnia hosts a diverse community of microbes externally and internally (Qi et al., 2009). Previous research has demonstrated that the Daphnia microbiome influences various aspects of host fitness, including lifespan, fecundity, and body size, suggesting that specific members within the community may have significant effects (Sison-Mangus et al., 2015; Peerakietkhajorn et al., 2016). In addition, previous studies have shown that environmental factors, such as biotic and abiotic stress, can influence the gut microbial community of Daphnia (Akbar et al., 2022). For example, a recent study showed that predation pressure can induce antipredation defense traits in Daphnia magna and reconfigure the composition of the gut microbiota, highlighting a complex correlation between induced defense traits and altered gut microbiota (Liu et al., 2021). In this study, we examined changes in life-history traits induced by fish kairomones, the diversity and composition of the gut microbiota and the differential abundance of bacteria. These findings have significant implications for understanding the interactions between D. galeata and its microbiota under predation stress.
Prey species demonstrate diverse adaptations to predation, including alterations in life history, behavior, and morphological and physiological traits (Lima, 1998). Consistent with previous studies (Tams et al., 2018; Choi et al., 2023b), the presence of fish kairomones significantly affected variations in life-history traits related to reproduction and growth of the examined genotype of D. galeata (Figure 1). The KB5 genotype exhibited defensive strategies by improving fertility to sustain generations, producing a substantial number of offspring, and minimizing their “Body Length” (Figure 1). Other studies have demonstrated that Daphnia will prioritize reproduction when exposed to fish kairomones, resulting in a higher rate of neonate growth to adulthood and increased offspring production. Initiating reproduction early, before the onset of predation, confers an ecological advantage in sustaining population numbers (Lampert, 1993). Individuals that produce a larger number of lower-quality offsprings are expected to be more advantageous in predation situations than those that produce fewer high-quality offsprings. Furthermore, a previous study suggested that Daphnia decreases in body size in response to vertebrate predators (Lampert, 1993). Daphnia can reduce their body size in response to predator-induced stress, which improves their chances of evading detection and predation (Tams et al., 2018).
The gut microbiota of KB5_Fish exposed to fish kairomones exhibited significantly higher alpha diversity indices except Faith’s phylogenetic diversity that indicates phylogenetic biodiversity than that of KB5_Control, which was not exposed to fish kairomones (Table 1). These results may be attributed to the interaction between D. galeata and the fish kairomone-rich environment. Predator-induced stress can influence Daphnia to adjust their gut microbiota diversity as a survival strategy to respond to environmental changes (Liu et al., 2022). However, the results of this study differed from previous findings in that the alpha diversity of the gut microbiome decreased in response to predator-induced stress in D. magna (Liu et al., 2022). These differences may be due to differences in the species, genotype, and developmental stage of Daphnia (Akbar et al., 2022). Changes in gut microbiota diversity in response to hypoxic stress in D. magna have been shown to be genotype-dependent (Coone et al., 2023). Therefore, future studies should investigate changes in gut microbiome diversity in response to predator-induced stress in different genotypes of D. galeata. Additionally, the gut microbiota of KB5_Fish and KB5_Control did not exhibit significant differences in beta diversity (Table 1). These results suggest that the KB5_Fish and KB5_Control gut microbiota are similar in the presence and abundance of key species. Previous studies have suggested that some bacterial groups and strains are found in the gut microbiota of Daphnia and that environmental changes are characterized by differences in the presence/absence or abundance of relatively less abundant minor bacterial species rather than major bacterial species (Akbar et al., 2020, 2022). Similarly, the beta diversity of the present study assumed that the differences between the gut microbiota of KB5_Fish and KB5_Control were characterized by the influence of a few consistently found bacterial groups and strains, resulting in insignificant variances. The dominant bacterial community composition identified in this study is similar to previous investigations of Daphnia guts exposed to several environmental stresses (Akbar et al., 2020; Frankel-Bricker et al., 2020; Liu et al., 2022), and the relative abundance of various bacterial taxa changed in response to fish kairomones (Figure 2).
An earlier study indicated that certain bacterial strains can provide essential elements for host reproduction and growth, contributing to the benefit of the host (Motiei et al., 2020). At the family level, statistically significant differences in relative abundance between the control and experimental groups were observed in Pseudomonadaceae (mainly the genus Pseudomonas) and Comamonadaceae (mainly the genus Limnohabitans) (Figure 3). The genus Pseudomonas belonging to the Pseudomonadaceae family exhibited a significant increase in relative abundance after exposure to fish kairomones (Supplementary Table 4). According to a previous study, when high-quality food (the phytoplankton Rhodomonas) was diluted with Pseudomonas and supplied to D. galeata, an increase in the addition of Pseudomonas resulted in a decrease in the growth rate of D. galeata (Wenzel et al., 2021). Pseudomonas, as heterotrophic bacteria in general, lacks essential fatty acids (FAs) and sterols, which likely negatively affects the growth of D. galeata. Therefore, it is speculated that the observed increase in “Body Length” and “Somatic Growth Rate” in the genotype KB5 under fish kairomone exposure conditions is associated with a significant increase in Pseudomonas. Conversely, the relative abundance of the Comamonadaceae family significantly decreased (P < 0.05) following exposure to fish kairomones (Figure 3; Supplementary Table 4). A previous study also reported a significant decrease in the relative abundance of Comamonadaceae in D. magna caused by fish kairomones (Liu et al., 2022). The increase in body size of D. magna when exposed to fish kairomones has been attributed to the high relative abundance of Comamonadaceae (Liu et al., 2022). This suggests a potential association between the decrease in body size in KB5 exposed to fish kairomones and the significant reduction in the relative abundance of Comamonadaceae. Furthermore, it has been suggested that the reproduction of Daphnia is associated with the relative abundance of the genus Limnohabitans, which belongs to the family Comamonadaceae (Peerakietkhajorn et al., 2015, 2016). Limnohabitans, which are key components of the gut microbiota of Daphnia, have been linked to obtaining essential amino acids, polyunsaturated fatty acids, and sterols that positively affect the reproduction and growth of Daphnia (Peerakietkhajorn et al., 2015). Sterols and polyunsaturated fatty acids restrict the growth and reproduction of D. galeata (Peerakietkhajorn et al., 2015). In this study, it was speculated that the increased relative abundance of the genus Limnohabitans induced by exposure to fish kairomone would enhance the fecundity of D. galeata. However, among the ASVs assigned to the genus Limnohabitans, there were ASVs with decreased relative abundance, which appeared to contradict previous findings that bacteria in the genus Limnohabitans are associated with higher fecundity in Daphnia (Peerakietkhajorn et al., 2015, 2016; Liu et al., 2022). According to previous research, only specific strains within the genus Limnohabitans are known for their unique ability to recover the fitness of germ-free Daphnia individuals after reinoculation, highlighting the specificity of host–microbiota interactions (Peerakietkhajorn et al., 2016). Furthermore, a previous study revealed substantial variations within the genus Limnohabitans on associations between microbiome, diet quality, and life history (Akbar et al., 2020). Therefore, in future research, it will be necessary to classify the various ASVs identified within the genus Limnohabitans at the species and/or strain levels and investigate the interactions with hosts based on these variations. Moreover, Candidatus Protochlamydia, a member of the Parachlamydiaceae family, was detected only in the gut of D. galeata when exposed to fish kairomones (Figure 2). This indicates that Candidatus Protochlamydia may be used as an indicator of predation risk. The function of Candidatus Protochlamydia as a possible defense strategy against predation in D. galeata is not yet known, and more research is needed to analyze its association with life-history traits in future studies.
In conclusion, our results showed that D. galeata not only developed adaptive traits related to reproduction and growth but also changed the gut microbiota in response to fish kairomones. Alpha diversity was significantly increased under the influence of fish kairomones in three out of the four examined alpha-diversity metrics (Table 1), and the change in the relative abundance of the microbiota was mainly manifested in the Pseudomonadaceae and Comamonadaceae families (Figures 2, 3). In this study, variations in relative abundance and the presence of several bacterial communities were assumed to be associated with observed life-history traits induced by fish kairomones. Although we starved D. galeata for 24 hours to remove as much of the C. vulgaris used during culture as possible from their gut, some of the remaining C. vulgaris may have interacted with the gut microbiota of D. galeata. Comprehending the interactions between Daphnia and its microbiome is challenging due to the dynamic nature of microbiome composition and structure, influenced by genotype, environmental factors, microbe interactions, and developmental stage (Akbar et al., 2022). This study focused on a KB5 D. galeata genotype associated with fish kairomone exposure, but there is a potential for variations in life-history trait characteristics and gut microbiota composition across additional D. galeata populations and genotypes. Therefore, to elucidate the general changes in life-history traits of D. galeata induced by exposure to fish kairomones and to investigate a precise association with the gut microbiota, it is crucial to collect additional genotypes of D. galeata and analyze the gut microbiota at various stages of development. To mimic fish predation, we used two different mediums: one consisting of ADaM as a control, and the other consisting of a mixture of ADaM and fish-exposed water as the experimental medium. This means that the media differed not only in terms of the presence or absence of fish kairomones, but also in terms of the amount of ADaM trace elements. This difference could also lead to changes in observed life-history traits and alterations in microbiota composition. Therefore, in future studies, should aim to use a control medium consisting of a mixture of ADaM and non-fish-exposed water, in order to conduct both conditions as similarly as possible. Additionally, we plan to extract DNA from the gut microbiota of D. galeata, as well as the control and experimental media where D. galeata was cultured, perform metabarcoding, and analyze the association between the media and the gut microbiota. Furthermore, additional research is required to understand not only the variations in the gut microbiota in response to aquatic vertebrate kairomones but also the associations between changes in life-history trait characteristics and gut microbiota alterations in invertebrate kairomones. Ultimately, more research is required on the holobiont, which encompasses various symbiotic microorganisms such as bacteria, fungi, and viruses, extending beyond the classification of gut bacteria (Bordenstein and Theis, 2015; Theis et al., 2016). This study is the first to examine the gut microbiota in response to fish kairomone exposure in D. galeata, which suggests that gut microbiota may play an important role in the induction of adaptive traits against fish predators.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1066084.
Author contributions
TC: Conceptualization, Data curation, Methodology, Resources, Writing – original draft. HA: Conceptualization, Data curation, Methodology, Writing – original draft. MM: Data curation, Methodology, Writing – original draft. SH: Data curation, Methodology, Writing – original draft. CK: Conceptualization, Funding acquisition, Methodology, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) [No. NRF-2022R1A2C3002750].
Acknowledgments
We acknowledge Jung-Il Kim (Sangmyung University, Korea) for help during Daphnia breeding and the experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1385972/full#supplementary-material
Supplementary Figure 1 | Rarefaction curves of D. galeata gut microbiome samples. Curves illustrate the predicted total diversity by sequencing depth. The X-axis represents sample sequencing depth, and the Y-axis represents species richness, as estimated by the observed features. KB5_Control and KB5_Fish indicate the control (the absence of fish kairomones) and experimental groups (exposed to fish kairomones) of genotype KB5, respectively. All replication samples of KB5_Control and KB5_Fish were calculated using the same method for sequencing depths.
References
Akbar S., Gu L., Sun Y., Zhang L., Lyu K., Huang Y., et al. (2022). Understanding host-microbiome-environment interactions: Insights from Daphnia as a model organism. Sci. Total Environ. 808, 152093. doi: 10.1016/j.scitotenv.2021.152093
Akbar S., Gu L., Sun Y., Zhou Q., Zhang L., Lyu K., et al. (2020). Changes in the life history traits of Daphnia magna are associated with the gut microbiota composition shaped by diet and antibiotics. Sci. Total Environ. 705, 135827. doi: 10.1016/j.scitotenv.2019.135827
Akbar S., Huang J., Zhou Q., Gu L., Sun Y., Zhang L., et al. (2021). Elevated temperature and toxic microcystis reduce Daphnia fitness and modulate gut microbiota. Environ. pollut. 271, 116409. doi: 10.1016/j.envpol.2020.116409
An H., Mun M., Kim C. (2023). Metabarcoding by combining environmental DNA with environmental RNA to monitor fish species in the Han river, korea. Fishes 8, 550. doi: 10.3390/fishes8110550
Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ Accessed 2 Nov 2018.
Bang C., Dagan T., Deines P., Dubilier N., Duschl W. J., Fraune S., et al. (2018). Metaorganisms in extreme environments: Do microbes play a role in organismal adaptation? Zoology 127, 1–19. doi: 10.1016/j.zool.2018.02.004
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc Ser. B Methodol 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bordenstein S. R., Theis K. R. (2015). Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PloS Biol. 13, e1002226. doi: 10.1371/journal.pbio.1002226
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Callens M., De Meester L., Muylaert K., Mukherjee S., Decaestecker E. (2020). The bacterioplankton community composition and a host genotype dependent occurrence of taxa shape the Daphnia magna gut bacterial community. FEMS Microbiol. Ecol. 96, fiaa128. doi: 10.1093/femsec/fiaa128
Callens M., Watanabe H., Kato Y., Miura J., Decaestecker E. (2018). Microbiota inoculum composition affects holobiont assembly and host growth in Daphnia. Microbiome 6, 1–12. doi: 10.1186/s40168-018-0444-1
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST: architecture and applications. BMC Bioinform. 10, 1–9. doi: 10.1186/1471-2105-10-421
Carter M. J., Silva-Flores P., Oyanedel J. P., Ramos-Jiliberto R. (2013). Morphological and life-history shifts of the exotic cladoceran Daphnia exilis in response to predation risk and food availability. Limnologica 43, 203–209. doi: 10.1016/j.limno.2012.09.004
Choi T., Do T. D., Kim J., Kim C. (2023a). Analysis of the complete mitogenome of Daphnia galeata from the Han River, South Korea: Structure comparison and control region evolution. Funct. Integr. Genomics 23, 65. doi: 10.1007/s10142-023-00986-5
Choi T., Han S., Malik A., Kim C. (2023b). Comparative transcriptome analysis of two Daphnia galeata genotypes displaying contrasting phenotypic variation induced by fish kairomones in the same environment of the Han River, Korea. BMC Genom 24, 580. doi: 10.1186/s12864-023-09701-x
Colston T. J. (2017). Gut microbiome transmission in lizards. Mol. Ecol. 26, 972–974. doi: 10.1111/mec.13987
Coone M., Vanoverberghe I., Houwenhuyse S., Verslype C., Decaestecker E. (2023). The effect of hypoxia on Daphnia magna performance and its associated microbial and bacterioplankton community: A scope for phenotypic plasticity and microbiome community interactions upon environmental stress? Front. Ecol. Evol. 11. doi: 10.3389/fevo.2023.1131203
Foster K. R., Schluter J., Coyte K. Z., Rakoff-Nahoum S. (2017). The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51. doi: 10.1038/nature23292
Frankel-Bricker J., Song M. J., Benner M. J., Schaack S. (2020). Variation in the microbiota associated with Daphnia magna across genotypes, populations, and temperature. Microb. Ecol. 79, 731–742. doi: 10.1007/s00248-019-01412-9
Herzog Q., Rabus M., Wolfschoon Ribeiro B., Laforsch C. (2016). Inducible defenses with a” twist”: Daphnia barbata abandons bilateral symmetry in response to an ancient predator. PloS One 11, e0148556. doi: 10.1371/journal.pone.0148556
King K. C., Auld S. K., Wilson P. J., James J., Little T. J. (2013). The bacterial parasite Pasteuria ramosa is not killed if it fails to infect: Implications for coevolution. Ecol. Evol. 3, 197–203. doi: 10.1002/ece3.438
Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1–e1. doi: 10.1093/nar/gks808
Klüttgen B., Dülmer U., Engels M., Ratte H. T. (1994). ADaM, an artificial freshwater for the culture of zooplankton. Water Res. 28, 743–746. doi: 10.1016/0043-1354(94)90157-0
Kruskal W. H., Wallis W. A. (1952). Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47, 583–621. doi: 10.1080/01621459.1952.10483441
Lampert W. (1993). Phenotypic plasticity of the size at first reproduction in Daphnia: The importance of maternal size. Ecology 74, 1455–1466. doi: 10.2307/1940074
Lampert W. (2011). Daphnia: development of a model organism in ecology and evolution. Excellence in Ecology. 21. 1–275
Lima S. L. (1998). Nonlethal effects in the ecology of predator-prey interactions. Bioscience 48, 25–34. doi: 10.2307/1313225
Liu Q., Akbar S., Ding Z., Zhou Q., Gu L., Sun Y., et al. (2021). Anti-predation defense traits of daphnia are associated with the gut microbiota composition shaped by fish kairomone. Preprint. doi: 10.21203/rs.3.rs-757585/v1
Liu Q., Akbar S., Ding Z., Zhou Q., Gu L., Sun Y., et al. (2022). Induced anti-predation defense traits of Daphnia are associated with the gut microbiota composition. Preprint. doi: 10.21203/rs.3.rs-1732132/v1
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
MacFarland T. W. (2014). Oneway analysis of variance (ANOVA). In: Introduction to Data Analysis and Graphical Presentation in Biostatistics with R. Cham: Springer
Macke E., Tasiemski A., Massol F., Callens M., Decaestecker E. (2017). Life history and eco-evolutionary dynamics in light of the gut microbiota. Oikos 126, 508–531. doi: 10.1111/oik.03900
McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Motiei A., Brindefalk B., Ogonowski M., El-Shehawy R., Pastuszek P., Ek K., et al. (2020). Disparate effects of antibiotic-induced microbiome change and enhanced fitness in Daphnia magna. PloS One 15, e0214833. doi: 10.1371/journal.pone.0214833
Peerakietkhajorn S., Kato Y., Kasalický V., Matsuura T., Watanabe H. (2016). Betaproteobacteria Limnohabitans strains increase fecundity in the crustacean Daphnia magna: symbiotic relationship between major bacterioplankton and zooplankton in freshwater ecosystem. Environ. Microbiol. 18, 2366–2374. doi: 10.1111/1462-2920.12919
Peerakietkhajorn S., Tsukada K., Kato Y., Matsuura T., Watanabe H. (2015). Symbiotic bacteria contribute to increasing the population size of a freshwater crustacean, Daphnia magna. Environ. Microbiol. Rep. 7, 364–372. doi: 10.1111/1758-2229.12260
Qi W., Nong G., Preston J. F., Ben-Ami F., Ebert D. (2009). Comparative metagenomics of Daphnia symbionts. BMC Genom 10, 1–21. doi: 10.1186/1471-2164-10-172
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rajarajan A., Wolinska J., Walser J., Dennis S. R., Spaak P. (2023). Host-associated bacterial communities vary between Daphnia galeata genotypes but not by host genetic distance. Microb. Ecol. 85, 1578–1589. doi: 10.1007/s00248-022-02011-x
R Core Team (2017) R: A Language and Environment for Statistical Computing. Available online at: https://www.R-project.org/.
Sadeq S. A., Mills R. L., Beckerman A. P. (2021). The microbiome mediates the interaction between predation and heavy metals. Sci. Total Environ. 775, 145144. doi: 10.1016/j.scitotenv.2021.145144
Schoeppner N. M., Relyea R. A. (2009). Interpreting the smells of predation: How alarm cues and kairomones induce different prey defences. Funct. Ecol. 23, 1114–1121. doi: 10.1111/j.1365-2435.2009.01578.x
Sison-Mangus M. P., Mushegian A. A., Ebert D. (2015). Water fleas require microbiota for survival, growth and reproduction. ISME J. 9, 59–67. doi: 10.1038/ismej.2014.116
Stibor H., Lüning J. (1994). Predator-induced phenotypic variation in the pattern of growth and reproduction in Daphnia hyalina (crustacea: Cladocera). Funct. Ecol., 8 (1), 97–101. doi: 10.2307/2390117
Tams V., Lüneburg J., Seddar L., Detampel J., Cordellier M. (2018). Intraspecific phenotypic variation in life history traits of Daphnia galeata populations in response to fish kairomones. PeerJ 6, e5746. doi: 10.7717/peerj.5746
Theis K. R., Dheilly N. M., Klassen J. L., Brucker R. M., Baines J. F., Bosch T. C., et al. (2016). Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. Msystems 1, e00028–e00016. doi: 10.1128/mSystems.00028-16
Wang Y. Y., Liang X. F., He S., Tang S. L., Peng D. (2022). The potential use of Artemia for larval rearing of mandarin fish (Siniperca chuatsi). Aquac. Rep. 25, 101216. doi: 10.1016/j.aqrep.2022.101216
Wenzel A., Vrede T., Jansson M., Bergström A. K. (2021). Daphnia performance on diets containing different combinations of high-quality algae, heterotrophic bacteria, and allochthonous particulate organic matter. Freshw. Biol. 66, 157–168. doi: 10.1111/fwb.13626
Yin M., Laforsch C., Lohr J. N., Wolinska J. (2011). Predator-induced defense makes Daphnia more vulnerable to parasites. Evolution 65, 1482–1488. doi: 10.1111/j.1558-5646.2011.01240.x
Keywords: gut microbiota, life-history traits, predator-induced response, 16S rRNA metabarcoding, Daphnia galeata, fish kairomones
Citation: Choi T-J, An H-E, Mun M-H, Han S-M and Kim C-B (2024) Variations in gut microbiota associated with changes in life-history traits of Daphnia galeata induced by fish kairomones. Front. Ecol. Evol. 12:1385972. doi: 10.3389/fevo.2024.1385972
Received: 14 February 2024; Accepted: 08 May 2024;
Published: 22 May 2024.
Edited by:
Peter Schausberger, University of Vienna, AustriaReviewed by:
Stefania Elisavet Kapsetaki, Tufts University, United StatesJos Kramer, ETH Zürich, Switzerland
Copyright © 2024 Choi, An, Mun, Han and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang-Bae Kim, evodevo@smu.ac.kr
 Tae-June Choi
Tae-June Choi Hyung-Eun An
Hyung-Eun An  Chang-Bae Kim
Chang-Bae Kim