- 1Ministry of Agriculture Key Laboratory of Biology and Genetic Resources of Rubber Tree/State Key Laboratory Breeding Base of Cultivation and Physiology for Tropical Crops, Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
- 2College of Agronomy and Biotechnology, China Agricultural University, Beijing, China
- 3Nextomics Biosciences Co., Ltd., Wuhan, China
Jasmonate signaling plays a vital role in the regulation of secondary laticifer differentiation and natural rubber biosynthesis in Hevea brasiliensis. Jasmonate ZIM-domain (JAZ) proteins are the master regulators of jasmonate signaling. Although several JAZs have been reported in the laticifer cells of H. brasiliensis, the genome-wide screening of HbJAZ members has not yet been explored. In the present study, 18 HbJAZs were identified based on the recent H. brasiliensis genome. Phylogenetic construction revealed that the HbJAZs were clustered into five subgroups and that members within the same subgroup shared highly conserved gene structures and protein motifs. Cis-element analysis of HbJAZ promoters suggested the presence of hormone, stress and development-related cis-elements. HbJAZ1.0, HbJAZ2.0, and HbJAZ5.0 interacted with CORONATINE INSENSITIVE1 (COI1) in the presence of coronatine (COR, a JA mimic). HbJAZ1.0, HbJAZ2.0, HbJAZ5.0, and HbJAZ12.0 could also interact with each other. Of the 18 HbJAZs, transcripts of 15 HbJAZs were present in the vascular cambium region except for that of HbJAZ7.0, HbJAZ8.0d, and HbJAZ13.0. Fourteen of the 15 HbJAZs were significantly up-regulated upon COR treatment. The transcripts of three genes that were absent from vascular cambium region were also absent from the latex. Among the 15 HbJAZs in the latex, the expression patterns of 13 HbJAZs were different between the tapping and ethrel treatments. Eight of the 14 COR-up-regulated HbJAZs in the vascular cambium region were also activated by tapping in latex. Of the eight tapping-activated HbJAZs, 5 HbJAZs were repressed by ethrel application. Based on the computational analyses and gene expression patterns described in this study, the HbJAZ5.0 and HbJAZ10.0b may be associated with laticifer differentiation while the HbJAZ8.0b is a negative regulator for natural rubber biosynthesis in H. brasiliensis.
Introduction
Jasmonates regulate various biological processes by the COI1-JAZ-MYC signaling pathway (Wasternack and Hause, 2013; Song et al., 2014; Yuan and Zhang, 2015). As repressors of jasmonate signaling, Jasmonate ZIM-domain (JAZ) proteins interact with MYC in the absence of active jasmonate (Singh et al., 2015). They are degraded by COI1-mediated ubiquitination in the presence of active jasmonate. This degradation releases MYC, which activates downstream gene transcription (Fonseca et al., 2009; Wager and Browse, 2012). The released MYC in turn activates the transcription of JAZ (Zhang et al., 2015). The JAZ proteins are a subgroup of the TIFY family, characterized by the N-terminal located ZIM domain and the C-terminal located Jas domain (Thines et al., 2007; Melotto et al., 2008; Li et al., 2017). The ZIM domain, which consists of 30 amino acids with a highly conserved TIFY motif (TIF[F/Y]XG), is required for the formation of homo- and hetero-dimers within the JAZ proteins (Vanholme et al., 2007). The Jas domain, containing 29 amino acids with a conserved motif (SLX2FX2KRX2RX5PY), is specific to JAZ members and interacts with a wide range of other proteins, such as COI1, and members of the MYC, MYB, and bHLH families (Yan et al., 2007; Sheard et al., 2010; Pauwels and Goossens, 2011). As a master regulator of jasmonate signaling, JAZ has been shown to directly regulate plant morphology, flower initiation, cotton fiber initiation, salvianolic acids, and tanshinone biosynthesis (Boter et al., 2015; Hu et al., 2016; Pei et al., 2018; Yu et al., 2018).
As an important industrial raw material, natural rubber is mainly produced by the commercialized tropical plant, the rubber tree (Hevea brasiliensis) (Chao et al., 2016). The secondary laticifers located in the bark of the trunk of H. brasiliensis are the site closely related to natural rubber production (Hao and Wu, 2000). Latex, a milky cytoplasm of laticifers, contains 20–40% of the rubber and is used as the raw material for refining natural rubber (Chao et al., 2017; Zhang et al., 2017). Although application of ethrel (an ethylene releaser) causes a significant increase in rubber yield per tapping via prolonging the duration of latex flow (Zhu and Zhang, 2009), it inhibits rubber biosynthesis (Deng et al., 2018). Activation of jasmonate signaling in laticifer cells is closely associated with the tapping-enhanced natural rubber biosynthesis (Zhao, 2011; Cao et al., 2017; Deng et al., 2018). Moreover, jasmonates are the key signal molecules for the secondary laticifer differentiation from vascular cambia in H. brasiliensis (Hao and Wu, 2000; Tian et al., 2015; Wu et al., 2016).
Until now, the core JA signaling components, such as HbCOI1 and HblMYC1-2, have been identified in the latex of H. brasiliensis, and a total of 10 HbJAZs have been reported in different studies (Peng et al., 2009; Tian et al., 2010; Zhao et al., 2011; Pirrello et al., 2014; Hong et al., 2015). The publication of several versions of the genome of H. brasiliensis (Rahman et al., 2013; Lau et al., 2016; Tang et al., 2016; Pootakham et al., 2017; Makita et al., 2018) facilitates the genome-wide identification of JAZ family members. In the present study, a total of 18 HbJAZs were identified on the basis of the latest version of the H. brasiliensis genome (Tang et al., 2016), analysis of the gene structures, phylogenetics, interactions, and expression patterns of HbJAZ family members were performed, and candidate members that may be related to laticifer differentiation and rubber biosynthesis were suggested.
Materials and Methods
Plant Materials and Treatments
Plantlets, 10-year-old virgin trees and regularly tapped trees of the H. brasiliensis clone CATAS7-33-97 were grown in the field of the Experimental Farm of the Chinese Academy of Tropical Agricultural Sciences (CATAS) on Hainan Island. The plantlets were developed from the latent buds on pruned trunks and thus called epicormic shoots. The COR treatment and collection of vascular cambia-containing samples of epicormic shoots were previously described in Wu et al. (2016). At each time interval, the bark tissues were collected from 10 epicormic shoots and combined as one sample. The regularly tapped trees were tapped by using a half spiral pattern every 3 days (S/2, d/3). The virgin trees had not been tapped until sampling. Application of 1.5% ethrel on the tapping panel of the regularly tapped rubber trees were performed. The latex samples were collected 24 h after ethrel treatment. The latex samples were also collected from regularly tapped rubber trees without ethrel treatment and virgin rubber trees. The latex samples from 10 tapped trees with or without ethrel treatment and 10 virgin trees were combined as one sample, respectively. Three biological replications were conducted.
RNA Isolation, DNA Isolation, and cDNA Synthesis
Total RNA was extracted from samples using RNAplant Plus reagent and evaluated by the NanoDrop 2000 (Thermo Scientific, Inc., United States). Approximately, 1 μg of RNA was used for reverse transcription and cDNA was synthesized using RevertAidTM First Strand cDNA Synthesis Kit (Thermo Scientific, Inc., United States). Genomic DNA was extracted from the leaves of epicormic shoots using DNAplant Plus reagent.
Computational Analysis
The whole genome sequence of H. brasiliensis was downloaded from NCBI1 (Tang et al., 2016). The Arabidopsis JAZ (AtJAZ) proteins were acquired from the TAIR database2. The HMM profiles of the TIFY domain (PF06200) and Jas domain (PF09425) were used as queries to search for predicted JAZ proteins in the H. brasiliensis genome using HMMER software (HMMER3). The BLAST algorithm was also used to identify the predicted H. brasiliensis JAZ proteins using AtJAZ as the queries. All putative HbJAZ proteins were further confirmed by the CDD database4. The AtJAZs and HbJAZs were aligned using the online software Multiple Sequence Alignment5, and then the phylogenetic tree was constructed using iTOL online software6. The molecular weight and isoelectric points of the HbJAZ proteins were predicted from the ExPASy database7. The genes structures and protein motif analysis were performed by GSDS2.0 software8 and MEME software9, respectively. For MEME analysis, the number of motifs was set as 12. The identified motifs were annotated by the InterProScan database10. Cis-acting regulatory elements within the HbJAZ promoters were analyzed by Plantcare11. The 1,500 bp genomic sequences upstream of the ATG of the HbJAZ genes were downloaded from the published H. brasiliensis genomes and used for the cis-regulatory element analysis.
Yeast Two-Hybrid Assay
The full-length CDSs of five HbJAZ proteins (HbJAZ1.0/2.0/5.0/10.0b/12.0) and HbCOI1 were amplified and inserted into the pGBKT7 vector to generate a bait plasmid. The CDS of HbJAZ genes were then fused into the pGADT7 vector to generate a prey plasmid. The bait vector and prey vector were transformed into Y187 and Y2H gold strains, respectively. The yeast Y187 and Y2H gold strains were mated, and protein interaction was assessed by the expression of different reporters under control of GAL4-responsive promoters. For the interactions between HbJAZs and HbCOI1, 20 or 60 μM COR and 10% ethanol were respectively added as treatment or MOCK, according to the reference (Melotto et al., 2008).
qRT-PCR Analysis
The primers for the HbJAZs were designed using the Primer Premier 5 software (Supplementary Table S1). The experiment was performed with the CFX384 real-time PCR system (Bio-Rad, United States) using the SYBR Prime Script RT-PCR Kit (TaKaRa, Dalian). HbUBC2b was used as the standard control for gene normalization (Chao et al., 2016). Three biological replicates were performed.
Statistical Analysis
The relative expression of HbJAZs were assessed by the 2−ΔΔCt method. Significant differences in the expression level were tested by Student’s t-test using GraphPad Prism 5 software.
Results
Identification and Isolation of JAZ Family Proteins in H. brasiliensis
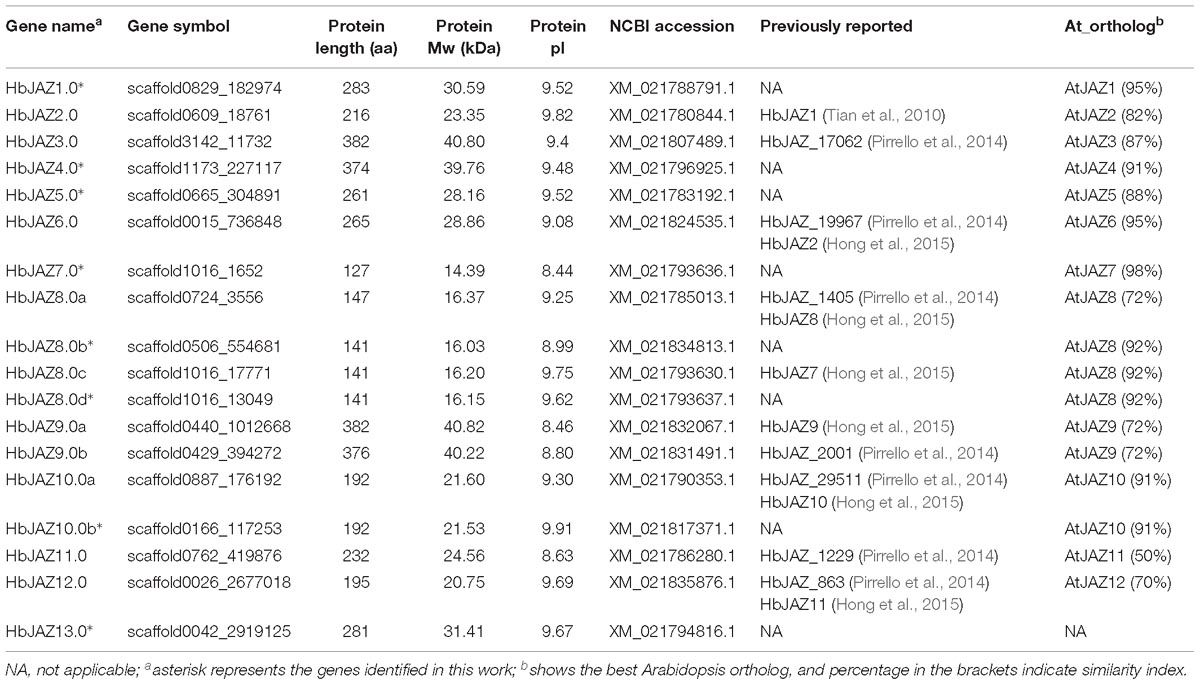
The HMM profiles of the TIFY domain (PF06200) and Jas domain (PF09425), as well as BLAST searches using Arabidopsis JAZ sequences as queries, were used to perform genome-wide identification of JAZ proteins in H. brasiliensis based on the most recent genome database. After validating the TIFY domain and Jas domain by NCBI Conserved Domain Search Service, a total of 18 HbJAZ proteins were identified. The identified HbJAZs were designated based on the names of their presumptive Arabidopsis orthologs (Figure 1). The 18 HbJAZs ranged from 127 (HbJAZ7.0) to 382 (HbJAZ9.0a) amino acid residues in length, with molecular weights of 14.39 kDa (HbJAZ7.0) to 40.82 kDa (HbJAZ9.0a) (Table 1 and Supplementary Table S2). The predicted isoelectric points of these proteins ranged from 8.44 (HbJAZ7.0) to 9.91 (HbJAZ10.0b) (Table 1).
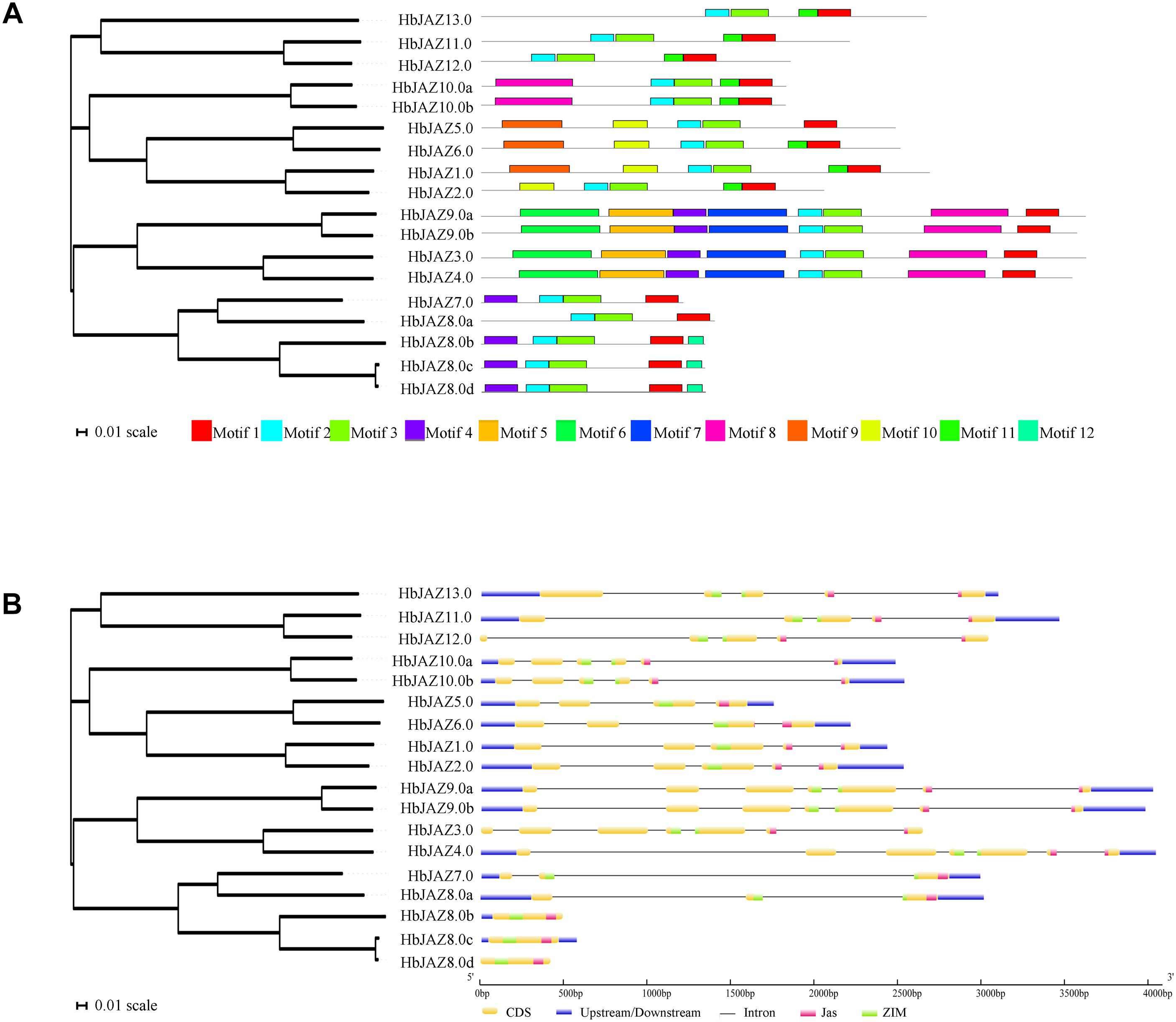
Figure 1. Structural analysis of HbJAZ family. (A) Motif analysis of of HbJAZ genes. 12 conserved motifs were identified using MEME program. (B) The exon/intron organization of HbJAZ genes. The yellow and blue boxes indicated coding sequence and untranslated region, respectively. Thin black lines indicated introns. The pink and green boxes respectively indicated Jas domain and ZIM domain.
The Structure of the HbJAZ Genes in H. brasiliensis
Using MEME software, 12 conserved motifs were identified among the 18 HbJAZs (Figure 1A and Supplementary Figure S1). Prediction by the InterProscan database annotated motif 1 as the Jas domain, motifs 2 and 3 as ZIM domains, and motifs 4 and 5 as EAR domains (Yang et al., 2018). Motifs 6, 7, 8, 9, 10, 11, 12 were not annotated in the database. All the HbJAZs comprised motifs 1, 2, and 3. Four of the HbJAZs (HbJAZ3.0/ 4.0/9.0a/9.0b) contained eight motifs (1, 2, 3, 4, 5, 6, 7, 8). Four HbJAZs (HbJAZ5.0/6.0/1.0/2.0) included motif 10; three of them (HbJAZ5.0/6.0/1.0) owned extra motif 9. Motif 11 existed in eight HbJAZs (HbJAZ1.0/2.0/6.0/10.0a/10.0b/11.0/12.0/13.0) while motif 12 was found in other three HbJAZs (HbJAZ8.0b/8.0c/8.0d).
Exon–intron organization analysis of the 18 HbJAZs uncovered that most of the HbJAZs had at least two introns, except for three genes (HbJAZ8.0b/8.0c/8.0d) which had no intron (Figure 1B). Both the ZIM and Jas domains were separated by several introns in nine genes (HbJAZ3.0/ 4.0/9.0a/9.0b/10.0a/10.0b/11.0/12.0/13.0). The ZIM domains of HbJAZ7.0 and HbJAZ8.0a and the Jas domains of HbJAZ1.0, HbJAZ2.0, and HbJAZ6.0 were however separated by one intron.
Several cis-elements were identified in the 1,500 bp upstream sequence of the 18 HbJAZs (Figure 2). They were classified into three categories: hormone responsive, stress responsive, and developmental regulation. For the hormone responsive category, eight HbJAZs (HbJAZ2.0/ 3.0/6.0/8.0a/8.0b/8.0c/10.0b/13) had JA responsive elements (TGACG-motif, CGTCA-motif), and five HbJAZs (HbJAZ1.0/ 3.0/5.0/7.0/8.0b) contained EREs. Additionally, the abscisic acid responsive element (ABRE) was present in the promoter regions of 12 HbJAZs (HbJAZ1.0/ 2.0/4.0/5.0/7.0/8.0a/8.0b/8.0c/9.0a/9.0b/10.0a/10.0b/11.0). Four HbJAZs (HbJAZ3.0/8.0c/8.0d/9.0b) contained auxin responsive elements (TGA-element, AuxRR-core), nine HbJAZs (HbJAZ1.0/2.0/7.0/8.0c/8.0d/9.0a/9.0b/11.0/12.0/13.0) had gibberellin responsive elements (GARE-motif, P-box), and eight HbJAZs contained salicylic acid responsive elements (HbJAZ1.0/2.0/3.0/5.0/6.0/7.0/8.0a/8.0b/8.0c/10.0b; TCA-element). For the stress responsive category, 11 HbJAZs (HbJAZ2.0/3.0/5.0/6.0/8.0a/8.0b/8.0c/9.0a/9.0b/11.0/13.0) had wound responsive elements (WUN-motif, W-box, Box-W1), 16 HbJAZs (HbJAZ1.0/2.0/3.0/4.0/7.0/8.0a/8.0b/8.0c/8.0d/9.0a/ 9.0b/10.0a/10.0b/11.0/12.0/13.0) contained HSEs, four HbJAZs (HbJAZ8.0b/8.0c/9.0b/10.0a) had LTRs, and 11 HbJAZs (HbJAZ1. 0/2.0/3.0/7.0/8.0a/8.0c/8.0d/9.0a/9.0b/10.0a/10.0b) contained drought response elements (MBS). For the developmental regulation category, six HbJAZs (HbJAZ4.0/ 5.0/7.0/8.0d/9.0a/10.0b) contained meristem specific elements (CAT-box, CCGTCC-box) and seven HbJAZs (HbJAZ1.0/2.0/ 3.0/4.0/8.0b/10.0b/12.0) had secondary metabolic biosynthesis regulation specific elements (MBSIs) (MBSI, MBSII, O2-site).

Figure 2. Cis-elements analysis of HbJAZ gene promoters. 1,500 bp genomic sequences upstream of the translational initiation codon of HbJAZ genes were analyzed by Plantcare. The yellow, blue, pink represented the elements related to hormones response, stresses response, tissues regulation, respectively. Number within the parentheses represented the number of cis-element.
Phylogenetic Analysis and Classification of the HbJAZ Proteins
The 18 identified HbJAZs and 12 Arabidopsis JAZs (AtJAZs) were used to construct a phylogenetic tree (Figure 3). All the JAZs distinctly grouped into five clusters (I, II, III, IV, and V). Cluster I included two AtJAZs (AtJAZ7 and AtJAZ8) and six HbJAZs (HbJAZ7.0/8.0a/8.0b/8.0c/8.0d), Cluster II contained three AtJAZs (AtJAZ3/4/9) and four HbJAZs (HbJAZ3.0/4.0,/9.0a,/9.0b), Cluster III included four AtJAZs (AtJAZ1/2/5/6) and four HbJAZs (HbJAZ1.0/2.0/5.0/6.0), Cluster IV contained AtJAZ10, HbJAZ10.0a and HbJAZ10.0b, and Cluster V included two AtJAZs (AtJAZ11 and AtJAZ12) and two HbJAZs (HbJAZ11.0 and HbJAZ12.0).
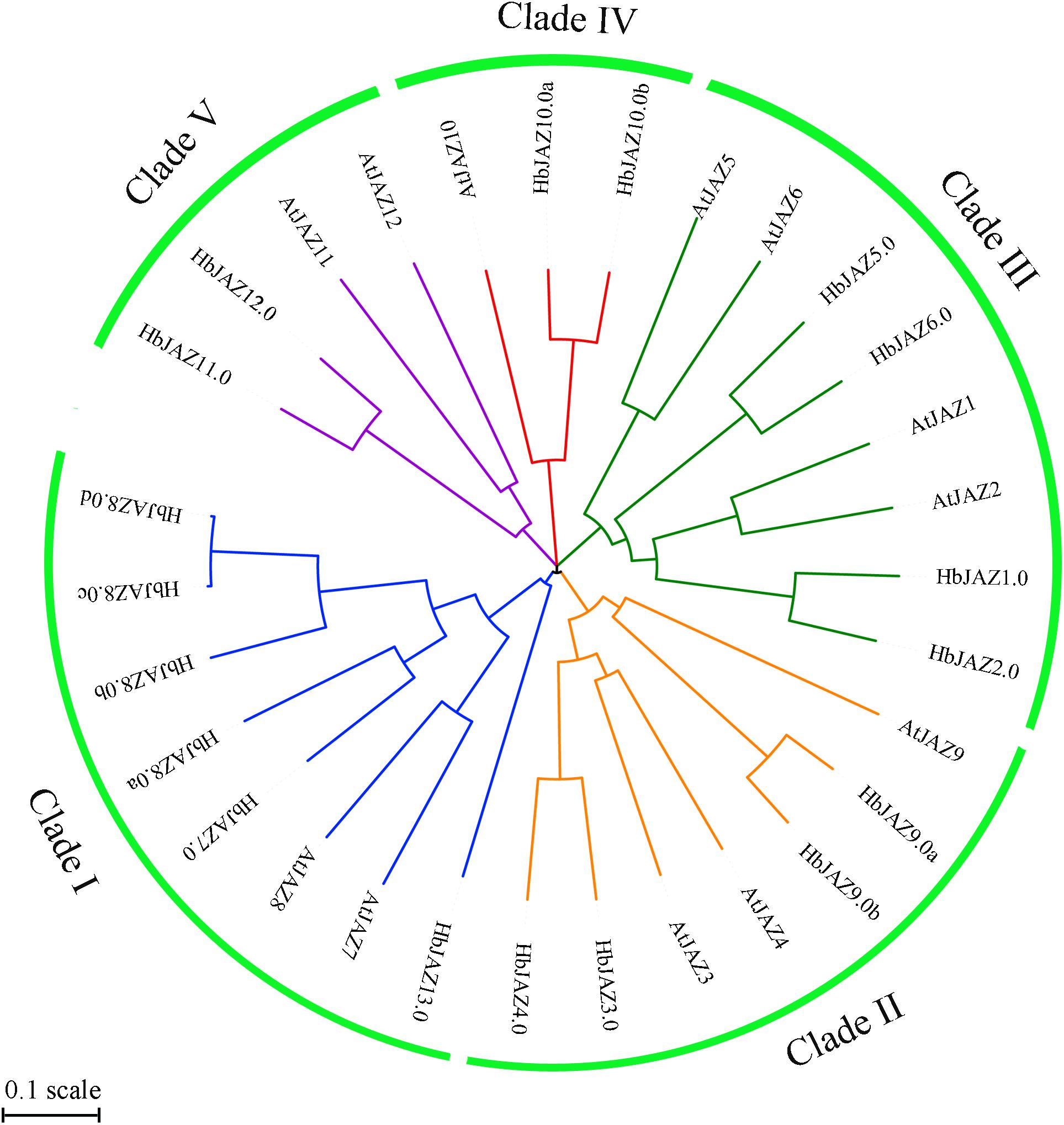
Figure 3. Phylogenetic tree of the HbJAZs and AtJAZs. The phylogenetic tree was calculated by Multiple Sequence Alignment software based on neighboring-joining phylogeny test, and displayed by iTOL software.
Interaction Among the HbJAZ Proteins
Five HbJAZs were selected for interaction analysis via yeast two-hybrid assay (Figure 4). The interactions among HbJAZs and HbCOI were COR-dependent (Figure 4A). In the presence of 60 μM COR, three HbJAZs (HbJAZ1.0/2.0/5.0) interacted with HbCOI1 (Figure 4A). These interactions also occurred in the presence of 20 μM COR (Supplementary Figure S2). The interactions among the HbJAZs were COR-independent. HbJAZ5.0 could respectively interact with HbJAZ1.0, HbJAZ2.0, HbJAZ5.0 and HbJAZ12.0, the HbJAZ1.0 interacted with itself and HbJAZ2.0, and HbJAZ2.0 interacted with itself (Figure 4B).
Figure 4. Interaction analysis of HbJAZ proteins. (A) Coronatine-dependent interaction between HbCOI1 and HbJAZs. (B) Homo- and hetero-meric interactions among HbJAZs. Controls for autoactivation were provided by transformation with the corresponding empty vector.
The Expression Patterns of HbJAZ Genes in Vascular Cambium Region in Response to COR
Among the 18 HbJAZs, the transcripts of HbJAZ7.0, HbJAZ8.0d, and HbJAZ13.0 were absent from the vascular cambium region (Figure 5). COR upregulated the 14 HbJAZs while had little influence on the expression of HbJAZ3.0. The duration of up-regulated expression was different among the 14 HbJAZs. The up-regulated expression lasted 2 h for HbJAZ8.0c, 4 h for six HbJAZs (HbJAZ1.0/6.0/8.0b/9.0a/10.0a/10.0b), 1 day for five HbJAZs (HbJAZ2.0/4.0/8.0a/9.0b/11.0) and 3 days for HbJAZ12.0 (Figure 5). Except for HbJAZ11.0 and HbJAZ12.0, the expression level reached peak at 2 h for 12 of the 14 HbJAZs. The expression level of HbJAZ11.0 and HbJAZ12.0 reached the peak at 4 h. The transcript levels of seven HbJAZs (HbJAZ1.0/5.0/8.0a/8.0b/8.0c/10.0a/10.0b) were more than 10 times of the control at 2 h (Figure 5).
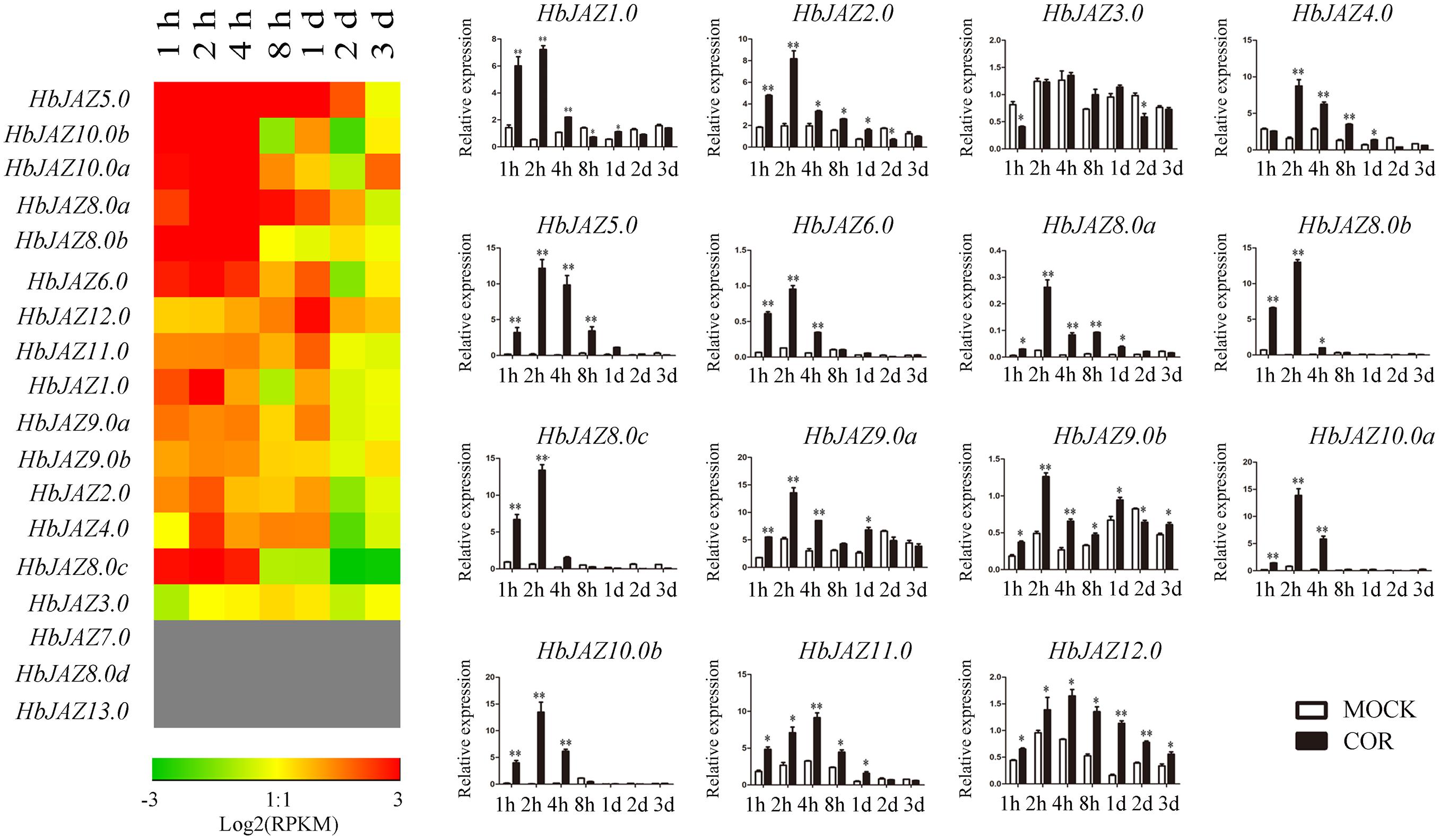
Figure 5. Expression profiles of HbJAZs in cambium upon COR treatment. The qRT-PCR results were displayed by heatmap (left) and bar chart (right). Red showed high expression and green showed low expression. Bar represented the Log 2 value. ∗, ∗∗ represented 0.05, 0.01 significant difference.
The Expression Patterns of HbJAZ Genes in the Latex in Response to Tapping and Ethrel Treatment
The transcripts of three genes (HbJAZ7.0, HbJAZ8.0d, and HbJAZ13.0) which were absent from the vascular cambium region, were also absent from the latex. Of the 14 COR-upregulated HbJAZs in the vascular cambium region, eight genes (HbJAZ1.0/6.0/8.0a/9.0b/10.0a/10.0b/11.0/12.0) in the latex were up-regulated while two genes (HbJAZ8.0b and HbJAZ9.0a) were down-regulated by tapping. The other four genes (HbJAZ2.0/4.0/5.0/8.0c) had little response to tapping (Figure 6A). By contrast, nine genes (HbJAZ1.0/2.0/4.0/5.0/9.0a/9.0b/10.0a/10.0b/12.0) were down-regulated while three genes (HbJAZ6.0, HbJAZ8.0b, and HbJAZ8.0c) were up-regulated by ethrel treatment. The other two genes (HbJAZ8.0a and HbJAZ11.0) had little response to ethrel treatment (Figure 6B). Tapping also up-regulated the expression of HbJAZ3.0 which had little response to COR and ethrel treatment (Figure 6). Except for HbJAZ6.0 and HbJAZ9.0a, the expression patterns of 13 of the 15 HbJAZs which transcripts were present in the latex were different between tapping and ethrel treatment (Figure 6). The HbJAZ6.0 was up-regulated and HbJAZ9.0a was down-regulated by either tapping or ethrel treatment (Figure 6).
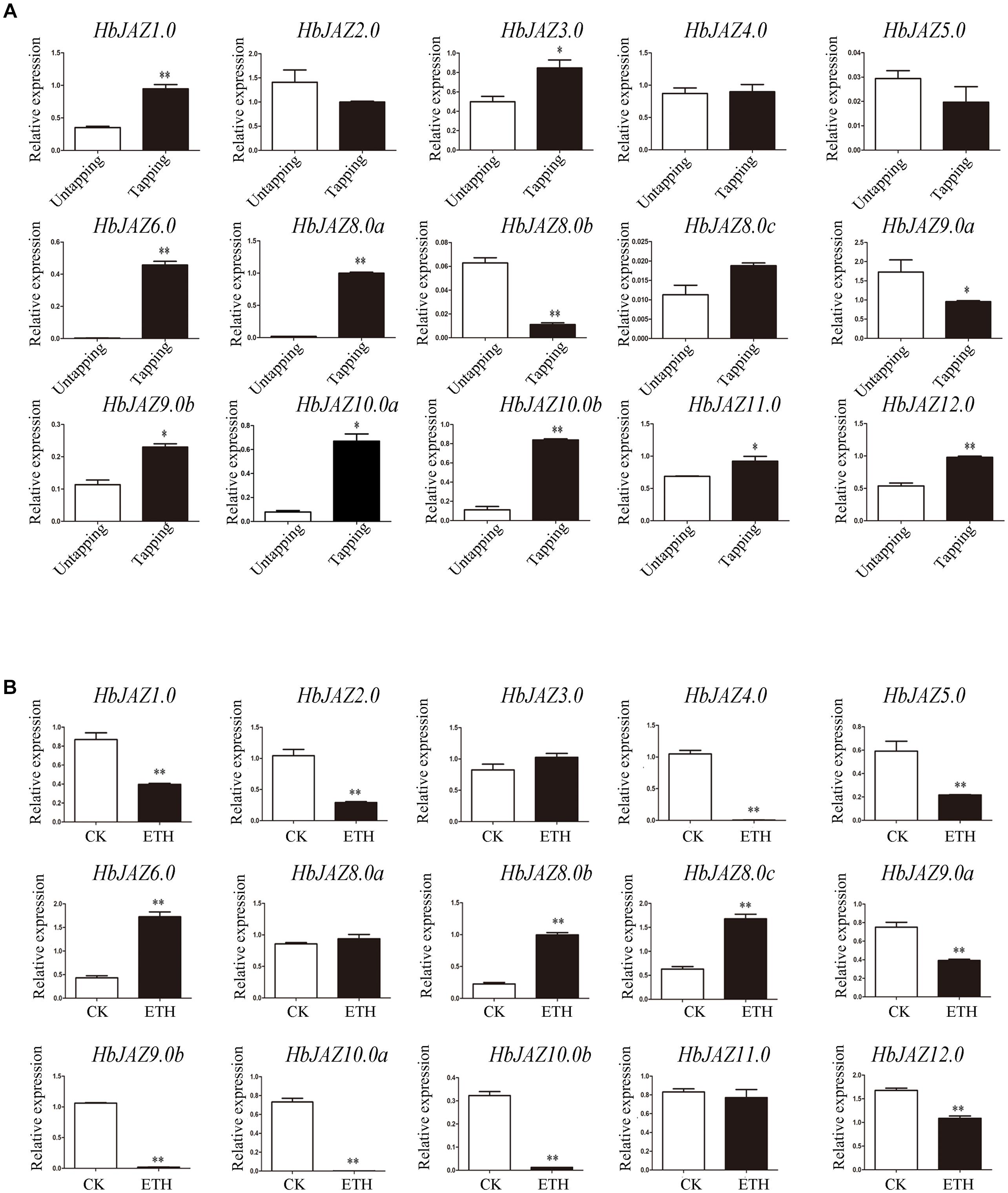
Figure 6. Expression profiles of HbJAZs in latex upon tapping (A) or ethrel treatment (B). Bar represented the Log 2 value of qRT-PCR results. ∗, ∗∗ represented 0.05, 0.01 significant difference.
Discussion
In the past decade, JAZ family has been widely identified in different plant species. There are 12 JAZs in Arabidopsis (Thines et al., 2007), 15 JAZs in rice (Ye et al., 2009), 18 JAZs in apple (Li et al., 2015), 30 JAZs in cotton (Li et al., 2017), 13 JAZs in tomato (Chini et al., 2017), and 11 JAZs in grape (Zhang et al., 2012). The following features could distinguish JAZs from other TIFY family proteins: (1) the C-terminal located Jas domain at, (2) the jasmonate-dependent interaction between JAZ and COI1, and (3) the formation of homo- and hetero-dimers within the JAZ subfamily. Several versions of the rubber tree genome have published in the past few years (Rahman et al., 2013; Lau et al., 2016; Tang et al., 2016; Pootakham et al., 2017; Makita et al., 2018). As Tang et al. (2016) version has less scaffolds number (7,453) and longer scaffolds N50 (1.28 Mb) (Supplementary Table S3), the genome-wide identification of JAZs was based on this version in the present study. As a result, 18 HbJAZs were identified in rubber tree. Ten of them (HbJAZ2.0/3.0/6.0/8.0a/8.0c/9.0a/9.0b/10.0a/11.0/12.0) are identical to those that have ever been reported (Tian et al., 2010; Pirrello et al., 2014; Hong et al., 2015). The other eight HbJAZs (HbJAZ1.0/4.0/5.0/7.0/8.0b/8.0d/10.0b/13.0) are novel (Table 1). In addition to the bioinformatic identification, evidence are provided for the interaction between some members and their interaction with HbCOI1 in the presence of COR (Figure 4).
Gene family expansion is a source of new gene generation (Jia et al., 2015). Tandem duplication and segmental duplication are the two strategies for expanding the number of genes in a family (Cannon et al., 2004; Jin et al., 2017). Compared to genes with tandem duplication, genes that come from segmental duplication at different chromosomes or scaffolds have been suggested to perform different functions (Dennis and Eichler, 2016). In rice, the clusters of OsJAZ9/10/11 and OsJAZ12/13/14 are from tandem duplication, while the clusters of OsJAZ3/4, OsJAZ2/15, and OsJAZ6/7 are from segmental duplication (Ye et al., 2009). In apple, Li et al. (2015) determine that the clusters of MdJAZ3/4/5/6/7/8, MdJAZ11/12, and MdJAZ13/14 are from tandem duplication, while the clusters of MdJAZ4/7, MdJAZ7/3, MdJAZ1/15, MdJAZ12/18, and MdJAZ18/11 are from segmental duplication. In grape, Zhang et al. (2012) report that VvJAZ5/6/7/8 are from tandem duplication, while the pairs of VvJAZ1/11 and VvJAZ4/9 are from segmental duplication. In rubber tree, the lack of high-density genetic map precludes the identification of paralogs derived from Whole Genome Duplication (WGD) or segmental duplication. However, Zou et al. (2016, 2017, 2019) identify several segmental duplication events in rubber tree genome, and firstly reveal that ρ WGD occurred in rubber tree is shared by cassava. In the present study, we identified at least three clusters (HbJAZ3.0/4.0, HbJAZ9.0a/9.0b, HbJAZ10.0a/10.0b) that were segmental duplications while one pair (HbJAZ8.0c/8.0d) was tandem duplicated, based on the scaffold location, gene structure, and protein conserved motifs (Figure 1 and Table 1). It is clear that the two segmentally duplicated clusters (HbJAZ3.0/4.0, HbJAZ9.0a/9.0b) have differential expression patterns in response to COR, ethrel or tapping treatments, suggesting the putative functional diversity of these genes. Phylogenetic tree analysis of JAZs from multiple species shows that the JAZ members that come from segmental duplication separate into different clusters while those from the tandem duplication are clustered together. The exception to this is JAZ members in rice (Supplementary Figure S3 and Supplementary Table S4), which may be attributed to the fact that rice is a monocot while the other species are dicots.
It has been suggested that JA signaling is involved in the regulation of plant development (Singh et al., 2015; Huang et al., 2017). In Arabidopsis, AtJAZ7 and AtJAZ10 are the key regulators in JA-mediated cambium differentiation (Sehr et al., 2010). Li et al. (2015) show knockdown of AsJAZ1 decreases the number of nodules in Astragalus sinicus. In cotton, over-expression of GhJAZ2 inhibits both lint and fuzz fiber initiation and reduces the fiber length (Hu et al., 2016). Over-expression of SlJAZ2 in tomato directly activates meristem-related genes and results in production of more leaves and flowers (Yu et al., 2018). To further explore the key members involved in secondary laticifer differentiation of rubber trees, we constructed a phylogenetic tree using the 18 HbJAZs together with AtJAZ7, AtJAZ10, AsJAZ1, GhJAZ2, SlJAZ2 (Supplementary Figure S4 and Supplementary Table S4). It is intriguing that 11 of the HbJAZs (HbJAZ1.0/2.0/5.0/6.0/7.0/8.0a/8.0b/8.0c/8.0d/10.0a/10.0b) are homologous to the plant development-related homologs, and five of them (HbJAZ5.0/8.0b/8.0c/10.0a/10.0b) are significantly up-regulated greater than 10 times upon COR treatment. Moreover, a meristem specific element (CAT-box) is present in the promoter regions of HbJAZ5.0 and HbJAZ10.0b. The CAT-box is a cis-regulatory element related to meristem expression. In Brassica juncea, mutation of the CAT-box in the promoter of BjuA07.CLV1 disturbs the CLAVATA (CLV)/WUSCHEL (WUS) signaling pathway, leading to the enlargement of the shoot and floral meristem, and ultimately multilocular siliques (Xiao et al., 2018). Thus, we speculate that HbJAZ5.0 and HbJAZ10.0b may play a key role in laticifer differentiation in H. brasiliensis.
Jasmonate is widely involved in the regulation of secondary metabolism in plants. Boter et al. (2015) show that degradation of JAZ3 activates a subset of JA-regulated genes in leaves, leading to anthocyanin accumulation in Arabidopsis. Mutation of the Jas motif of NtJAZ1 or NtJAZ3 reduces nicotine content in tobacco BY-2 cells (Shoji et al., 2008). In Salvia miltiorrhiza, over-expression of SmJAZ3 or SmJAZ8 notably reduces tanshinone production (Shi et al., 2016; Pei et al., 2018). Natural rubber is a typical secondary metabolite and its biosynthesis is significantly activated by tapping (Deng et al., 2018). The tapping-enhanced rubber biosynthesis is closely related to the activation of jasmonate signaling in laticifer cells (Deng et al., 2018). Exogenous methyl jasmonate enhances rubber biosynthesis while ethrel, inhibits rubber biosynthesis (Deng et al., 2018). In the present study, the expression patterns of HbJAZs in response to tapping were similar to those in response to COR, but generally opposite to the patterns in response to ethrel treatment (Figures 5, 6). The differences in the expression levels of 13 of the HbJAZs between the tapping and ethrel treatments may be associated with the different effects of methyl jasmonate and ethrel on rubber biosynthesis. Phylogenetic tree analysis reveals that 11 HbJAZs (HbJAZ1.0/2.0/3.0/4.0/7.0/8.0a/8.0b/8.0c/8.0d/9.0a/9.0b) are homologous to AtJAZ3, NtJAZ1, NtJAZ3, SmJAZ3, and SmJAZ8 (Supplementary Figure S4 and Supplementary Table S4), and tapping causes the expression level of HbJAZ8.0a and HbJAZ8.0b more than five times of control (Figure 6A). We also identified a secondary MBSI in the promoter region of HbJAZ8.0b. MBSI is present in the promoter of flavonoid biosynthetic genes, such as chsJ. In Petunia hybrida, the petal epidermis-specific MYB.Ph3 binds the MBSI element that regulates the biosynthesis of flavonoids (Solano et al., 1995). Based on the evidence described above, HbJAZ8.0b is likely a key regulator of natural rubber biosynthesis.
Conclusion
Jasmonate signaling plays a vital role in the regulation of secondary laticifer differentiation and natural rubber biosynthesis. In the present study, JAZs, the repressors of jasmonate signaling, are genome-wide identified. Based on the computational analyses and gene expression patterns, HbJAZ5.0 and HbJAZ10.0b might be key regulators of laticifer differentiation whereas HbJAZ8.0b was crucial for regulation of natural rubber biosynthesis in H. brasiliensis. The genome-wide identification of HbJAZs will facilitate the jasmonate signaling-mediated laticifer differentiation and natural rubber biosynthesis in rubber tree.
Author Contributions
JC designed and carried out the experiment of this study and wrote the manuscript. YZ, JJ, SW, XM, and YC participated and analyzed data in the experiment. W-MT planned the study and participated in the design of the experiment. All authors have read and approved the manuscript in its final form.
Funding
This work was supported by the earmarked Fund for Modern Agro-industry Technology Research System (Grant No. CARS-34-GW1), the National Natural Science Foundation of China (30872002), the National Key R&D Program of China (Grant No. 2018YFD1000502), the Fundamental Research Funds for Rubber Research Institute, CATAS (Grant Nos. 1630022013004 and 1630022015010).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00372/full#supplementary-material
FIGURE S1 | Conserved amino acid sequences of 12 motifs.
FIGURE S2 | Interactions between HbCOI1 and three HbJAZs in the presence of 20 μM COR.
FIGURE S3 | Phylogenetic relationships of HbJAZs with other species homolog. The trees were constructed by neighboring-joining phylogeny test, and 1,000 bootstrap replicates. The accession numbers for the genes are provided in Supplementary Table S4. Red color represented tandem duplication, while blue color represented segmental duplication.
FIGURE S4 | Phylogenetic relationships of HbJAZs with other species homolog related to plant development or secondary metabolites biosynthesis. The trees were constructed by neighboring-joining phylogeny test, and 1,000 bootstrap replicates. The accession numbers for the genes are provided in Supplementary Table S4. Blue circle represented pigments biosynthesis related TFs in other species. Red circle represented genes identified in this study.
TABLE S1 | Primers used in this paper.
TABLE S2 | The full amino acid sequences of the 18 HbJAZs.
TABLE S3 | Evaluation of four rubber tree genome versions.
TABLE S4 | Gene accession numbers used in this paper.
Abbreviations
CDSs, coding DNA sequences; COI1, CORONATINE INSENSITIVE1; COR, coronatine; EAR, ERF-associated amphiphilic repression; EREs, ethylene responsive elements; HMM, hidden Markov model; HSEs, heat responsive elements; JAZ, jasmonate ZIM-domain; LTR, low-temperature response elements; qRT-PCR, quantitative real-time polymerase chain reaction.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/genome/?term=rubber%20tree%20genome
- ^ http://www.arabidopsis.org/
- ^ http://hmmer.org/
- ^ http://www.ncbi.nlm.nih.gov/cdd/
- ^ https://www.ebi.ac.uk/Tools/msa/
- ^ http://itol.embl.de/
- ^ http://expasy.org/
- ^ http://gsds.cbi.pku.edu.cn/
- ^ http://meme-suite.org/tools/meme
- ^ http://www.ebi.ac.uk/Tools/pfa/iprscan/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Boter, M., Golz, J. F., Giménez-Ibañez, S., Fernandez-Barbero, G., Franco-Zorrilla, J. M., and Solano, R. (2015). FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell 27, 3160–3174. doi: 10.1105/tpc.15.00220
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Cao, X., Yan, J., Lei, J., Li, J., Zhu, J., and Zhang, H. (2017). De novo transcriptome sequencing of meja-induced taraxacum koksaghyz rodin to identify genes related to rubber formation. Sci. Rep. 7:15697. doi: 10.1038/s41598-017-14890-z
Chao, J., Yang, S., Chen, Y., and Tian, W. M. (2016). Evaluation of reference genes for quantitative real-time pcr analysis of the gene expression in laticifers on the basis of latex flow in rubber tree (Hevea brasiliensis muell. arg.). Front. Plant Sci. 7:1149. doi: 10.3389/fpls.2016.01149
Chao, J., Yang, S., Chen, Y., and Tian, W. M. (2017). Transcript Profiling of Hevea brasiliensis during latex flow. Front. Plant Sci. 8:1904. doi: 10.3389/fpls.2017.01904
Chini, A., Ben-Romdhane, W., Hassairi, A., and Aboul-Soud, M. A. M. (2017). Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS One 12:e0177381. doi: 10.1371/journal.pone.0177381
Deng, X., Guo, D., Yang, S., Shi, M., Chao, J., Li, H., et al. (2018). Jasmonate signalling in regulation of rubber biosynthesis in laticifer cells of rubber tree (Hevea brasiliensis muell. arg.). J. Exp. Bot. 69, 3559–3571. doi: 10.1093/jxb/ery169
Dennis, M. Y., and Eichler, E. E. (2016). Human adaptation and evolution by segmental duplication. Curr. Opin. Genet. Dev. 41, 44–52. doi: 10.1016/j.gde.2016.08.001
Fonseca, S., Chini, A., Hamberg, M., Adie, B., Porzel, A., Kramell, R., et al. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. doi: 10.1038/nchembio.161
Hao, B. Z., and Wu, J. L. (2000). Laticifer differentiation in Hevea brasiliensis: induction by exogenous jasmonic acid and linolenic acid. Ann. Bot. 85, 37–43. doi: 10.1006/anbo.1999.0995
Hong, H., Xiao, H., Yuan, H., Zhai, J., and Huang, X. (2015). Cloning and characterisation of JAZ gene family in Hevea brasiliensis. Plant Biol. 17, 618–624. doi: 10.1111/plb.12288
Hu, H., He, X., Tu, L., Zhu, L., Zhu, S., Ge, Z., et al. (2016). GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J. 88, 921–935. doi: 10.1111/tpj.13273
Huang, H., Liu, B., Liu, L., and Song, S. (2017). Jasmonate action in plant growth and development. J. Exp. Bot. 68, 1349–1359. doi: 10.1093/jxb/erw495
Jia, Y., Yuan, Y., Zhang, Y., Yang, S., and Zhang, X. (2015). Extreme expansion of NBS-encoding genes in Rosaceae. BMC Genet. 16:48. doi: 10.1186/s12863-015-0208-x
Jin, X., Zhu, L., Yao, Q., Meng, X., Ding, G., Wang, D., et al. (2017). Expression profiling of mitogen-activated protein kinase genes reveals their evolutionary and functional diversity in different rubber tree (Hevea brasiliensis) cultivars. Genes 8:261. doi: 10.3390/genes8100261
Lau, N. S., Makita, Y., Kawashima, M., Taylor, T. D., Kondo, S., Othman, A. S., et al. (2016). The rubber tree genome shows expansion of gene family associated with rubber biosynthesis. Sci. Rep. 6:28594. doi: 10.1038/srep28594
Li, W., Xia, X. C., Han, L. H., Ni, P., Yan, J. Q., Tao, M., et al. (2017). Genome-wide identification and characterization of JAZ gene family in upland cotton (Gossypium hirsutum). Sci. Rep. 7:2788. doi: 10.1038/s41598-017-03155-4
Li, X., Yin, X., Wang, H., Li, J., Guo, C., Gao, H., et al. (2015). Genome-wide identification and analysis of the apple (Malus × domestica Borkh.) TIFY gene family. Tree Genet. Genomes 11:808. doi: 10.1007/s11295-014-0808-z
Makita, Y., Kawashima, M., Lau, N. S., Othman, A. S., and Matsui, M. (2018). Construction of Pará rubber tree genome and multi-transcriptome database accelerates rubber researches. BMC Genomics 19(Suppl. 1):922. doi: 10.1186/s12864-017-4333-y
Melotto, M., Mecey, C., Niu, Y., Chung, H. S., Katsir, L., Yao, J., et al. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55, 979–988. doi: 10.1111/j.1365-313X.2008.03566.x
Pauwels, L., and Goossens, A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100. doi: 10.1105/tpc.111.089300
Pei, T., Ma, P., Ding, K., Liu, S., Jia, Y., Ru, M., et al. (2018). SmJAZ8 acts as a core repressor regulating JA-induced biosynthesis of salvianolic acids and tanshinones in Salvia miltiorrhiza hairy roots. J. Exp. Bot. 69, 1663–1678. doi: 10.1093/jxb/erx484
Peng, S. Q., Xu, J., Li, H. L., and Tian, W. M. (2009). Cloning and molecular characterization of HbCOI1 from Hevea brasiliensis. Biosci. Biotechnol. Biochem. 73, 665–670. doi: 10.1271/bbb.80721
Pirrello, J., Leclercq, J., Dessailly, F., Rio, M., Piyatrakul, P., Kuswanhadi, K., et al. (2014). Transcriptional and post-transcriptional regulation of the jasmonate signalling pathway in response to abiotic and harvesting stress in Hevea brasiliensis. BMC Plant Biol. 14:341. doi: 10.1186/s12870-014-0341-0
Pootakham, W., Sonthirod, C., Naktang, C., Ruang-Areerate, P., Yoocha, T., Sangsrakru, D., et al. (2017). De novo hybrid assembly of the rubber tree genome reveals evidence of paleotetraploidy in Hevea species. Sci. Rep. 7:41457. doi: 10.1038/srep41457
Rahman, A. Y., Usharraj, A. O., Misra, B. B., Thottathil, G. P., Jayasekaran, K., Feng, Y., et al. (2013). Draft genome sequence of the rubber tree Hevea brasiliensis. BMC Genomics 14:75. doi: 10.1186/1471-2164-14-7
Sehr, E. M., Agusti, J., Lehner, R., Farmer, E. E., Schwarz, M., and Greb, T. (2010). Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 63, 811–822. doi: 10.1111/j.1365-313X.2010.04283.x
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Shi, M., Zhou, W., Zhang, J., Huang, S., Wang, H., and Kai, G. (2016). Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by JASMONATE ZIM-DOMAIN repressor proteins. Sci. Rep. 6:20919. doi: 10.1038/srep20919
Shoji, T., Ogawa, T., and Hashimoto, T. (2008). Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant Cell Physiol. 49, 1003–1012. doi: 10.1093/pcp/pcn077
Singh, A. P., Pandey, B. K., Deveshwar, P., Narnoliya, L., Parida, S. K., and Giri, J. (2015). JAZ repressors: potential involvement in nutrients deficiency response in rice and chickpea. Front. Plant Sci. 6:975. doi: 10.3389/fpls.2015.00975
Solano, R., Nieto, C., Avila, J., Cañas, L., Diaz, I., and Paz-Ares, J. (1995). Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 14, 1773–1784. doi: 10.1002/j.1460-2075.1995.tb07166.x
Song, S., Qi, T., Wasternack, C., and Xie, D. (2014). Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 21, 112–119. doi: 10.1016/j.pbi.2014.07.005
Tang, C., Yang, M., Fang, Y., Luo, Y., Gao, S., Xiao, X., et al. (2016). The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2:16073. doi: 10.1038/nplants.2016.73
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Tian, W. M., Huang, W. F., and Zhao, Y. (2010). Cloning and characterization of HbJAZ1 from the laticifer cells in rubber tree (Hevea brasiliensis muell. arg.). Trees 24, 771–779. doi: 10.1016/j.plaphy.2015.10.023
Tian, W. M., Yang, S. G., Shi, M. J., Zhang, S. X., and Wu, J. L. (2015). Mechanical wounding-induced laticifer differentiation in rubber tree: an indicative role of dehydration, hydrogen peroxide, and jasmonates. J. Plant Physiol. 182, 95–103. doi: 10.1016/j.jplph.2015.04.010
Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., and Gheysen, G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12, 239–244. doi: 10.1016/j.tplants.2007.04.004
Wager, A., and Browse, J. (2012). Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front. Plant Sci. 3:41. doi: 10.3389/fpls.2012.00041
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. an update to the 2007 review in annals of botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Wu, S., Zhang, S., Chao, J., Deng, X., Chen, Y., Shi, M., et al. (2016). Transcriptome analysis of the signalling networks in coronatine-induced secondary laticifer differentiation from vascular cambia in rubber trees. Sci. Rep. 6:36384. doi: 10.1038/srep36384
Xiao, L., Li, X., Liu, F., Zhao, Z., Xu, L., Chen, C., et al. (2018). Mutations in the CDS and promoter of BjuA07.CLV1 cause a multilocular trait in Brassica juncea. Sci. Rep. 8:5339. doi: 10.1038/s41598-018-23636-4
Yan, Y., Stolz, S., Chételat, A., Reymond, P., Pagni, M., Dubugnon, L., et al. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483. doi: 10.1105/tpc.107.050708
Yang, J., Liu, Y., Yan, H., Tian, T., You, Q., Zhang, L., et al. (2018). PlantEAR: functional analysis platform for plant ear motif-containing proteins. Front. Genet. 9:590. doi: 10.3389/fgene.2018.00590
Ye, H., Du, H., Tang, N., Li, X., and Xiong, L. (2009). Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 71, 291–305. doi: 10.1007/s11103-009-9524-8
Yu, X., Chen, G., Tang, B., Zhang, J., Zhou, S., and Hu, Z. (2018). The Jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 267, 65–73. doi: 10.1016/j.plantsci.2017.11.008
Yuan, Z., and Zhang, D. (2015). Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 27, 44–51. doi: 10.1016/j.pbi.2015.05.024
Zhang, F., Yao, J., Ke, J., Zhang, L., Lam, V. Q., Xin, X. F., et al. (2015). Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273. doi: 10.1038/nature14661
Zhang, Y., Gao, M., Singer, S. D., Fei, Z., Wang, H., and Wang, X. (2012). Genome-wide identification and analysis of the TIFY gene family in grape. PLoS One 7:e44465. doi: 10.1371/journal.pone.0044465
Zhang, Y., Leclercq, J., and Montoro, P. (2017). Reactive oxygen species in Hevea brasiliensis latex and relevance to tapping panel dryness. Tree Physiol. 37, 261–269. doi: 10.1093/treephys/tpw106
Zhao, Y. (2011). Involvement of Jasmonate Signaling Pathway Regulating Rubber Biosynthesis in Laticifer Cells of Hevea brasiliensis. Ph.D. thesis. Hainan University, Haikou.
Zhao, Y., Zhou, L. M., Chen, Y. Y., Yang, S. G., and Tian, W. M. (2011). MYC genes with differential responses to tapping, mechanical wounding, ethrel and methyl jasmonate in laticifers of rubber tree (Hevea brasiliensis muell. arg.). J. Plant Physiol. 168, 1649–1658. doi: 10.1016/j.jplph.2011.02.010
Zhu, J., and Zhang, Z. (2009). Ethylene stimulation of latex production in Hevea brasiliensis. Plant Signal. Behav. 4, 1072–1074. doi: 10.4161/psb.4.11.9738
Zou, Z., Xie, G. S., and Yang, L. (2017). Papain-like cysteine protease encoding genes in rubber (Hevea brasiliensis): comparative genomics, phylogenetic, and transcriptional profiling analysis. Planta 246, 999–1018. doi: 10.1007/s00425-017-2739-z
Zou, Z., Yang, J. H., and Zhang, X. (2019). Insights into genes encoding respiratory burst oxidase homologs (RBOHs) in rubber tree (Hevea brasiliensis muell. arg.). Ind. Crop Prod. 128, 126–139. doi: 10.1016/j.indcrop.2018.11.005
Zou, Z., Yang, L. F., Gong, J., Mo, Y., Wang, J., Cao, J., et al. (2016). Genome-Wide identification of Jatropha curcas aquaporin genes and the comparative analysis provides insights into the gene family expansion and evolution in Hevea brasiliensis. Front. Plant Sci. 7:395. doi: 10.3389/fpls.2016.00395
Keywords: Hevea brasiliensis Muell. Arg., natural rubber biosynthesis, JAZ identification, laticifer differentiation, vascular cambium region
Citation: Chao J, Zhao Y, Jin J, Wu S, Deng X, Chen Y and Tian W-M (2019) Genome-Wide Identification and Characterization of the JAZ Gene Family in Rubber Tree (Hevea brasiliensis). Front. Genet. 10:372. doi: 10.3389/fgene.2019.00372
Received: 28 December 2018; Accepted: 09 April 2019;
Published: 01 May 2019.
Edited by:
Mehar Hasan Asif, National Botanical Research Institute (CSIR), IndiaReviewed by:
Zhi Zou, Chinese Academy of Tropical Agricultural Sciences, ChinaJitender Giri, National Institute of Plant Genome Research (NIPGR), India
Susheng Song, Capital Normal University, China
Copyright © 2019 Chao, Zhao, Jin, Wu, Deng, Chen and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Min Tian, wmtian@163.com
 Jinquan Chao
Jinquan Chao Yue Zhao
Yue Zhao Jie Jin
Jie Jin Shaohua Wu
Shaohua Wu Xiaomin Deng
Xiaomin Deng Yueyi Chen
Yueyi Chen Wei-Min Tian
Wei-Min Tian