- 1Affiliated Hospital of Shaoxing University, Shaoxing, China
- 2Shanghai Engineering Research Center of Pharmaceutical Translation, Shanghai, China
- 3Shanghai Institute of Nutrition and Health, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
- 4Department of Respiratory Medicine, Zhejiang Provincial People’s Hospital, People’s Hospital of Hangzhou Medical College, Hangzhou, China
Lung cancer, the most common of malignant tumors, is typically of the non-small cell (NSCLC) type. T-cell-based immunotherapies are a promising and powerful approach to treating NSCLCs. To characterize the CD8+ T cells of non-small cell lung cancer, we re-analyzed the published RNA-Seq gene expression profiles of 36 CD8+ T cell isolated from tumor (TIL) samples and 32 adjacent uninvolved lung (NTIL) samples. With an advanced Monte Carlo method of feature selection, we identified the CD8+ TIL specific expression patterns. These patterns revealed the key dysfunctional genes and pathways in CD8+ TIL and shed light on the molecular mechanisms of immunity and use of immunotherapy.
Introduction
Lung cancer, the most common of malignant tumors, is typically (∼80%) of the non-small cell (NSCLC) type (Zhan et al., 2017). Current therapies for NSCLC include surgery followed by adjuvant radiotherapy, chemoradiotherapy, and molecule-targeted therapy; these methods have produced excellent results (Antonicelli et al., 2013; Martinez et al., 2014; Nascimento et al., 2015). However, most patients with NSCLC are in the advanced or inoperable stage with limited treatment options, and the 5-year survival rate is still less than 20% (Siegel et al., 2012; van der Drift et al., 2012). Thus, innovative therapeutic approaches to achieve long-term disease control without obvious adverse reactions are needed.
Tumor-infiltrating lymphocytes are considered to play a critical role in the immune response to many human solid cancers. Most CD8+ T cells are cytotoxic T lymphocytes in the case of tumor-infiltrating lymphocytes (Farhood et al., 2019). In the immune response to cancer, these tumor-infiltrating CD8+ T cells have the potential to recognize specific antigens that are presented by the MHC class I receptor on cancer cells and target them for destruction. Studies have shown that immune infiltration by CD8+ cytotoxic T cells is significantly correlated with improved clinical outcome in non-small cell lung cancer (NSCLC) (Johnson et al., 2000; Welsh et al., 2005; Al-Shibli et al., 2008; Kawai et al., 2008). High density of tumor-infiltrating lymphocytes usually signified strong prognostic value (Hiraoka et al., 2006; Al-Shibli et al., 2008; Kawai et al., 2008; Schalper et al., 2015).
Currently, T-cell-based immunotherapies are a promising and innovative approach toward treating NSCLC. The development of anticancer drugs targeting T cells to enhance the immune response has shown great clinical benefit in NSCLC (Garon et al., 2015; Gettinger et al., 2015; Jia et al., 2015). Programmed death factor-1 (PD-1) expressed in tumor tissue plays a key role in downregulating T-cell activation and promoting tumor immune escape by binding to its ligand PD-L1, which is expressed on the surface of tumor cells (Pardoll, 2012; Dermani et al., 2019). Nivolumab, a PD-1 immune checkpoint inhibitor antibody, was recently approved by the United States Food and Drug Administration for treatment of patients with metastatic squamous NSCLC (Morgensztern and Herbst, 2016). It disrupts PD-1-mediated signaling and is linked to an anticancer immune response. Early clinical trials have indicated that PD-L1 expression on tumor-infiltrating lymphocytes and tumor cells may increase the response to PD-1-directed therapies in metastatic NSCLC (Herbst et al., 2014, 2016; Ramalingam et al., 2016). Therefore, tumor-infiltrating lymphocytes and the expression of PD-L1 are being considered as biomarkers capable of screening NSCLC patients most likely to respond to checkpoint antibody therapy (Johnson et al., 2014).
To identify the markers for CD8+ T cells in lung cancer, we compared the published RNA-Seq gene expression profiles of 36 CD8+ T cell isolated from tumor (TIL) samples and 32 adjacent uninvolved lung (NTIL) samples. With a Monte Carlo feature selection method, we identified the CD8+ TIL-specific expression patterns, which can accurately predict such cells. The original study of this published dataset identified 1,403 differentially expressed genes using DE-Seq with fold change greater than 1.5 and adjusted value of p < 0.05 (Ganesan et al., 2017). This number of genes is too numerous for use in a biomarker analysis along with the low expected utility of the set of statistically significant genes (Simon, 2008). Instead, we used a Monte Carlo feature selection method, which assembled a series of decision trees for classification of genes by importance (Draminski et al., 2008). The usefulness of this method has been evaluated by others (Li et al., 2019; Chen et al., 2020). The functional analysis of these genes and the CD8+ TIL signatures are presented in this study to help understand the molecular mechanisms of immunity and their possible relevance to immunotherapy.
Materials and Methods
The RNA-Seq Gene Expression Profiles of Non-Small Cell Lung Cancer
We downloaded the gene expression profiles of 36 CD8+ T cells isolated from tumor (TIL) samples and 32 adjacent uninvolved lung (NTIL) samples from the Gene Expression Omnibus (GEO) under accession number GSE90728 (Ganesan et al., 2017). All lung patients had non-small cell lung cancer (NSCLC). Other clinical details are available in Ganesan et al. (2017). The gene expression levels were quantified with HTSeq (Anders et al., 2015) after the RNA sequencing reads were mapped onto the human reference genome (hg19) using the TopHat software (Trapnell et al., 2009) by Ganesan et al. (2017). The processed matrix of 23,366 genes in 36 TIL samples and 32 NTIL samples was used to identify the key discriminative genes between TIL samples and 32 NTIL samples.
The Monte Carlo Feature Selection Method
There have been many methods for identifying differentially expressed genes, such as the t-test, significance analysis of microarrays (SAM) (Tusher et al., 2001), and DESeq2 (Love et al., 2014). However, they typically only consider the statistical significance even though the statistically significant genes do not have discriminative ability (Simon, 2008). Since they do not consider the relationship between genes, they may be redundant or without known biological functions. To overcome these problems, we used a Monte Carlo feature selection method (Draminski et al., 2008; Cai et al., 2018; Chen et al., 2018a; Pan et al., 2018) to extract the CD8+ T-cell-specific gene expression patterns. The Monte Carlo feature selection method is powerful in discriminating features in a data set and has been widely used (Chen et al., 2018a, 2020; Chen L. et al., 2019; Chen X. et al., 2019; Li et al., 2019; Pan et al., 2019).
The Monte Carlo Feature Selection Algorithm Works as Follows
Let us use d to denote the number of features, i.e., 23,366 genes in this study. To explain the feature selection algorithm, we used features instead of the expression level of genes since feature was a broader concept. The expression levels of genes can be features, but features can be any numerical vector.
First, m features (m≪d) are randomly selected for s times;
Then, t trees for each of the s subsets are constructed;
Last, s⋅t classification trees will be grouped to calculate a feature g’s relative importance (RI).
To be more specific, RI of feature g is based on how many times feature g is selected by the s⋅t trees and how much feature g contributes to the classification of the s⋅t trees. The equation of RI is
in which wAcc is the weighted classification accuracy of decision tree τ, IG(ng(τ)) is the information gain of node ng(τ), which is a decision rule of feature g, (no.inng(τ)) is the number of samples under node ng(τ), (no.inτ) is the number of samples in decision tree τ, and u and ν are additional tunable parameters, which adjust the influence of wAcc and , respectively (set to 1 by default).
The Monte Carlo feature selection method is a complex algorithm when the dataset is large. Therefore, a software called dmLab (Draminski et al., 2008), which can be downloaded from http://www.ipipan.eu/staff/m.draminski/mcfs.html was used to apply the Monte Carlo feature selection method.
After the RI values for all 23,366 genes were calculated, all these gene features were ranked as
in which N is the total number of gene features, i.e., 23,366 in this study.
The gene features with smaller indices have greater RI value. In other words, the genes are sorted decreasingly. Since all the genes were ranked by importance, the top 500 genes are sufficient for identifying a potential biomarker for practical use. This set of genes was analyzed in the next step.
The Support Vector Machine Classifier for CD8+ T Cells
Although all gene features may be ranked by their RI values (Monte Carlo feature selection), it was difficult to discern how many top features to select as optimal CD8+ T cell biomarkers.
To determine the number of features required for accurate classifier, we adopted an incremental feature selection (IFS) method (Wang S. et al., 2017; Zhang Y. H. et al., 2017; Chen et al., 2018b, c; Li et al., 2018a). First, 500 different feature sets F1,F2⋯F500 were constructed. In these feature sets, feature set Fi = [f1,f2⋯fi] included the top i features of f in Eq. (2). As explained above, features with a smaller index were more important, and features with a larger index were less important. These less important genes were more likely to introduce noise in the classifier and, therefore, decrease the performance of the classifier. Therefore, we needed to find the balance between signal (important features with small index) and noise (unimportant features with large index). For each feature set Fi, a support vector machine (SVM) classifier was built based on these top i features, and their performance was evaluated with leave-one-out cross validation (LOOCV). An svm classifier can predict whether a cell was TIL based on its expression levels of the top i features/genes. Using the number of features as x-axis and their LOOCV accuracy as y-axis, an IFS curve can be plotted. The accuracy was the number of correctly predicted samples over the number of total samples. Based on the peak of IFS curve, the optimal number of gene features can be determined.
In this study, the SVM classifier was built using the R function svm from package e1017.1 The default parameters of R function svm were used to train the SVM models.
Results
The Relative Importance of Genes for CD8+ T Cells
As we described in the methods, the Monte Carlo feature selection method was adopted to analyze the gene expression profiles of 36 TIL samples and 32 NTIL samples. The goal was to identify the discriminative genes between TIL samples and NTIL samples. The 23,366 genes were ranked based on their relative importance calculated by the Monte Carlo feature selection algorithms.
The relative importance value reflected how well and how often this gene can be used to classify the TIL samples and NTIL samples in the resampling feature subsets on the decision trees. Since relative importance value integrates the information of many decision trees, it is a robust measurement that will not be easily influenced by noise. The genes can be ranked based on their relative importance values.
If a gene is important, it will rank at the top. All 23,366 genes were ranked, but only the top 500 genes were further analyzed for biomarker identification.
The Key Genes and Pathways of CD8+ T Cells
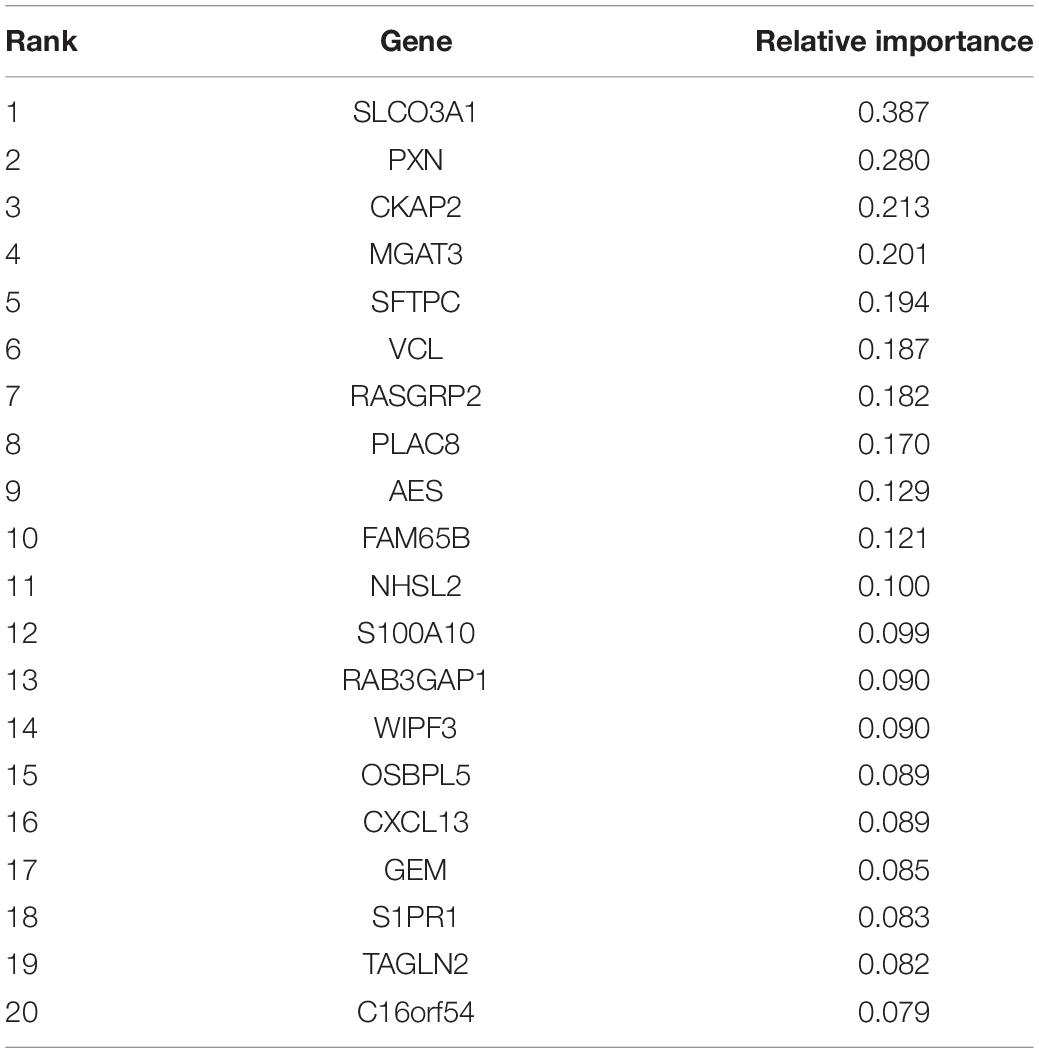
After the genes were ranked based on their relative importance by the Monte Carlo feature selection, we applied the IFS method to further optimize the final key gene set that pertains to CD8+ T cells. We constructed 500 gene sets in which each gene sets included top i genes in the ranked gene list. Based on the number of genes and their prediction accuracy, we plotted the IFS curve in Figure 1. It can be seen that with the top 20 genes, the LOOCV accuracy was the highest, 0.971. Therefore, these 20 genes were considered as the key gene set of CD8+ T cells, and they are listed in Table 1. Even with the top two genes, SLCO3A1 and PXN, the accuracy was 0.882. Since there was no similar CD8+ T cell dataset, we searched these two genes against the CellMarker database (Zhang et al., 2018a). This curated database has 13,605 cell markers of 467 cell types among 158 human tissue types, and 9,148 cell markers of 389 cell types among 81 mouse tissue types. Based on the CellMarker database, SLCO3A1 was a marker for the natural killer T (NKT) cell; PXN was a cell marker for natural killer, CD4+ cytotoxic T cell, and effector CD8+ memory T (Tem) cell. All 20 genes were included in the 1,403 differentially expressed genes as identified by Ganesan et al. (2017).
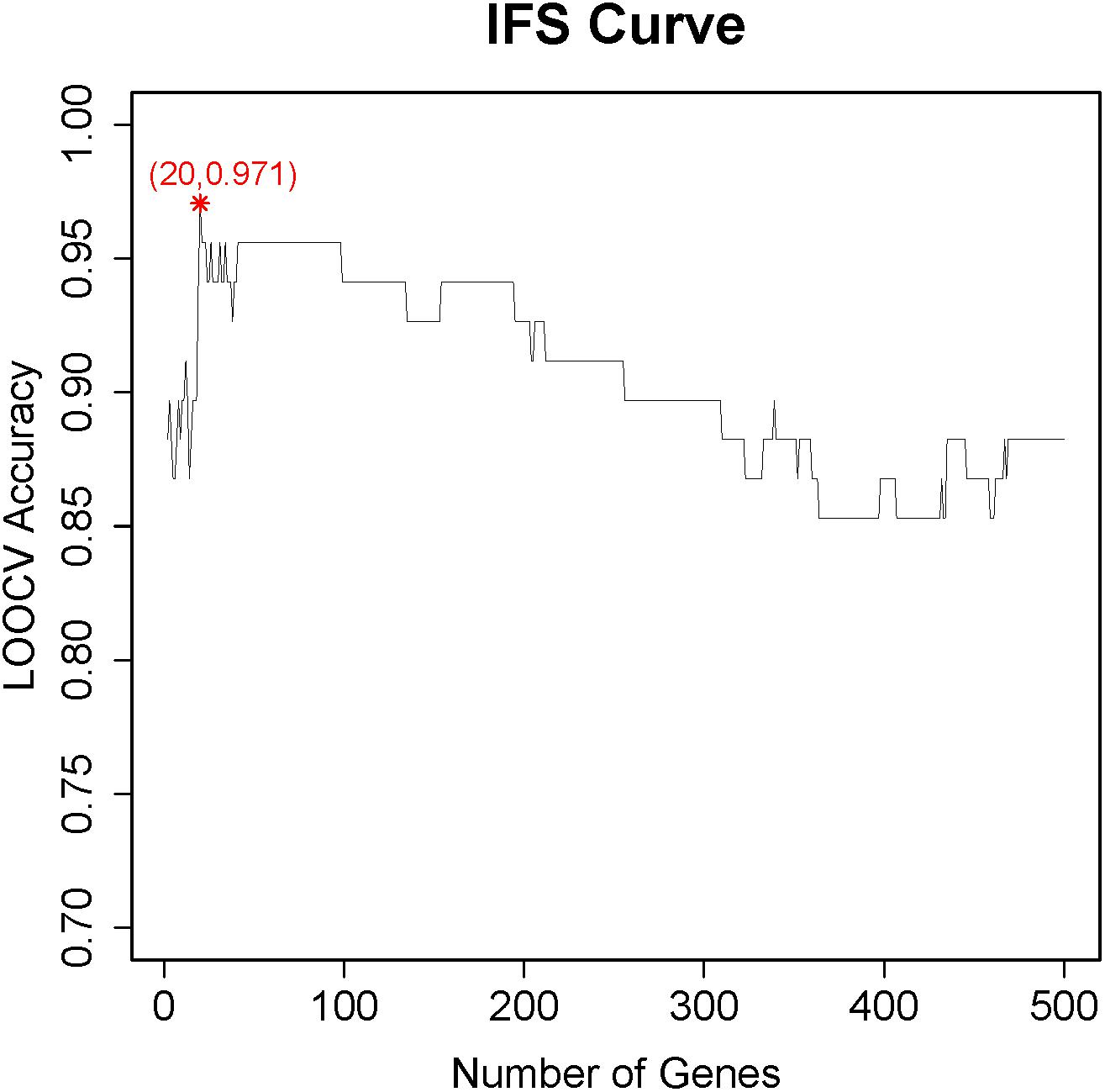
Figure 1. The incremental feature selection (IFS) curve to determine the optimal number of key genes for CD8+ T cells. The X-axis was the number of genes used to build the support vector machine (SVM) classifier, while the Y-axis was the prediction accuracy evaluated with leave-one-out cross validation (LOOCV). When the top 20 genes were used, the LOOCV accuracy was the highest, 0.971. These 20 genes were considered to be the key genes for CD8+ T cells.
To test whether the classification model can affect the feature selection, we used the decision tree (R package rpart) instead of SVM, and the peak also appeared at 18, 19, and 20 with the highest LOOCV accuracy of 0.824. The 20 genes still performed the best. These results suggested the genes selected by IFS were robust to classifiers.
To explore the expression pattern of these 20 genes, we plotted the heatmap of these 20 genes and two classes of samples in Figure 2. It can be seen that the NTIL samples and TIL samples were correctly clustered into two groups. Only two samples were not correctly clustered. Within the 20 genes, RAB3GAP1, WIPF3, GEM, CKAP2, and C-X-C motif chemokine ligand 13 (CXCL13) were highly expressed in TIL samples, while the other genes were lowly expressed in NTIL samples.
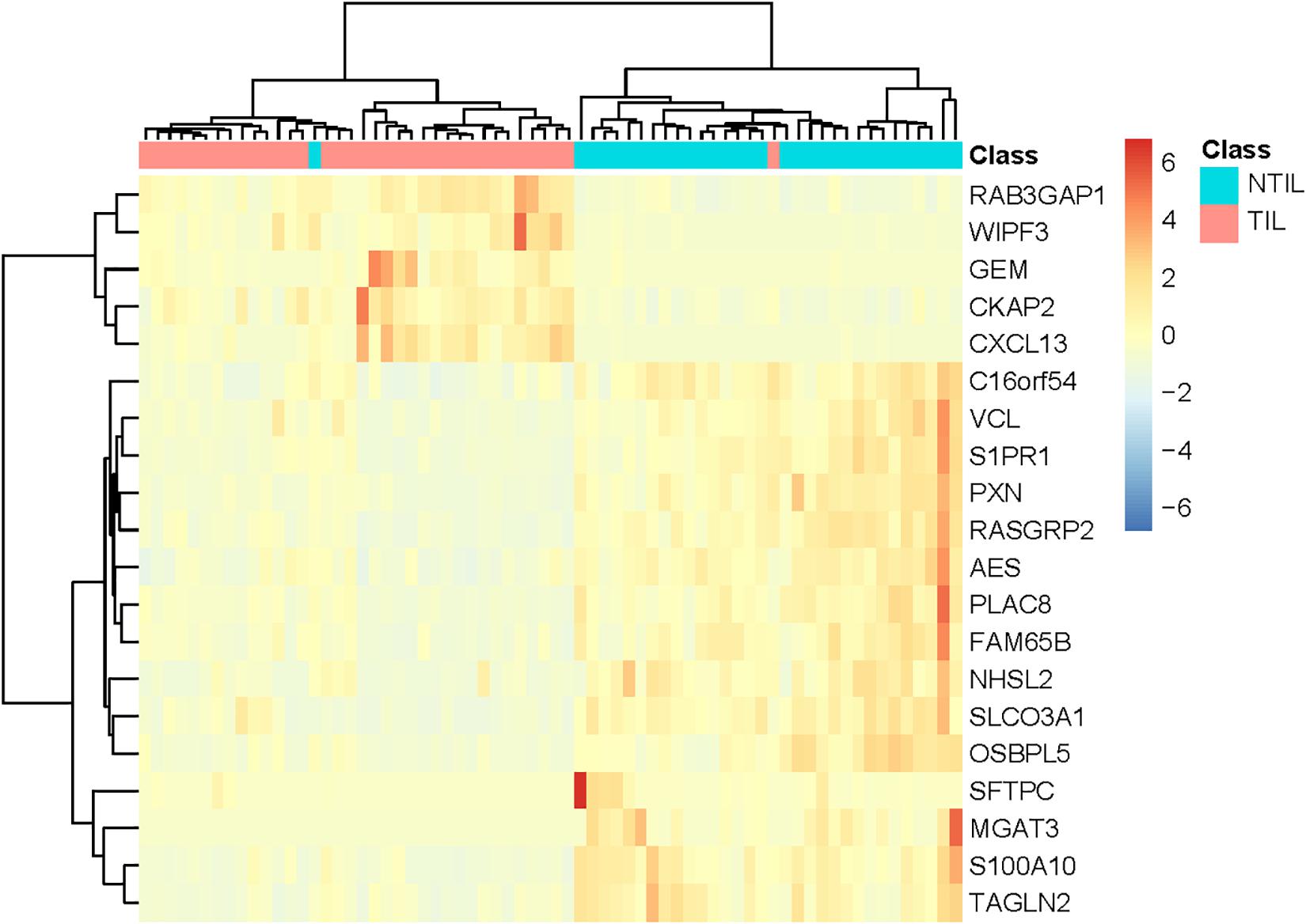
Figure 2. The heatmap of the 20 genes in TIL samples and NTIL samples. The sample classes were indicated on the first row: blue for NTIL samples and red for TIL samples. The two classes of samples were correctly clustered into two groups. Only two samples were not correctly clustered. It can be seen that RAB3GAP1, WIPF3, GEM, CKAP2, and C-X-C motif chemokine ligand 13 (CXCL13) were highly expressed in TIL samples, while other genes were lowly expressed in NTIL samples.
To investigate the dysfunctional pathways affected by these genes, we did KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis using WebGestalt (WEB-based GEneSeTAnaLysis Toolkit) (Wang J. et al., 2017) and found that hsa04062 chemokine signaling pathway was most significantly enriched with a p value of 6.12e-04 and odds ratio of 15.99.
Discussion
The Key Dysfunctional Genes in CD8+ T Cells
Biomarkers are of great significance for the diagnosis and treatment of cancer. Recent studies have found that in colorectal cancer, CHGA is more predictive of early diagnosis than other biomarkers, such as KRAS and TP53 (Zhang et al., 2019). High expression of DOCK4 is closely related to invasive breast cancer and subsequent bone metastasis, making it a potentially useful biomarker to predict the risk of tumor bone metastasis (Westbrook et al., 2019). In addition, the role of biomarkers in lung cancer has also been reported. UCK2 may be a biomarker for early diagnosis and prognosis of lung cancer (Wu et al., 2019). In NSCLC, the level of serum IDH2 can be regarded as an effective biomarker for the diagnosis and prognosis (Li et al., 2018b), and LRP12 DNA methylation can be used as predictive biomarker for carboplatin resistance (Grasse et al., 2018).
As shown in Table 1, many of the 20 key genes have shown significant potential as biomarkers for CD8+ T cells. We will discuss several high confidence genes and try to show potential mechanisms of these genes in CD8+ T cells.
Pro-inflammatory protein S100A10, a member of the S100 protein family, is a crucial plasminogen receptor and is reported to be involved in the regulation of various intracellular processes such as cell cycle progression, transcription, and differentiation (Kwon et al., 2005). S100A10 is overexpressed in various cancers and plays a role in facilitating cell invasiveness by regulating pericellular proteolysis (Choi et al., 2003; Ji et al., 2004; Zhang et al., 2004). S100A10 is mainly expressed in regions with strong proliferation capacity (Petersson et al., 2009). Yang et al. observed that the reduction of availability of S100A10 had negative impact on the growth of tumor cells in vitro (Yang et al., 2011), suggesting the role of S100A10 in regulating cell proliferation. The current study by Katono et al. (2016) showed that S100A10 expression was significantly associated with high TNM stage, poor overall prognosis, and frequent vascular invasion. Moreover, several studies have shown that up-regulation of S100A11 is significantly associated with lymph node metastasis in patients with NSCLC (Tian et al., 2007; Yang et al., 2011; Katono et al., 2016).
CXCL13 (C-X-C motif chemokine ligand 13) is an antimicrobial peptide and CXC chemokine strongly expressed in the follicles of the spleen and lymph node. Recent studies recognized that the CXCR5–CXCL13 axis is involved in tumor dissemination to lymph nodes (Meijer et al., 2006; Singh et al., 2014). Analysis of serum CXCL13 levels in both subtypes of NSCLCs, squamous cell carcinoma (SCC) and adenocarcinoma (AC), showed that serum CXCL13 levels in ACs were higher than that in SCCs; this may be associated with patient prognosis (Singh et al., 2014). These findings of Singh et al. (2014) indicated that CXCR5 and CXCL13 may be useful as prognostic biomarkers for NSCLC. Smoke and air pollution are well known to be associated with lung cancer (Li et al., 2013; Berrandou et al., 2018), both of which contain polycyclic aromatic hydrocarbons (PAHs), a carcinogenic substance (Baxter et al., 2014). The experiments conducted by Wang et al. (2015) showed that CXCL13 levels of lung epithelial cells, of cancer cells, and of mice exposed to PAHs led to increased rates of lung cancer in mice, demonstrating that CXCL13 has an important role in PAH-induced lung cancers. Thommen et al. (2018) reported that CXCL13 plays an important role in the recruitment of lymphocytes to tertiary lymphoid structures.
S1PR1 is a G-protein-coupled receptor of the bioactive lipid sphingosine-1-phosphate (S1P) that is abundantly expressed in endothelial cells and blood (Cantalupo et al., 2017; Meissner, 2017) and plays a vital role in angiogenesis (Liu et al., 2000). Angiogenesis is a key process in the early stage of tumor progression and spread (Metodieva et al., 2011). Sarkisyan et al. (2014) suggested that S1PR1 signaling could delay tumor progression by enhancing or destabilizing integrity of neovasculature. S1PR signaling pathways are also reported to be involved in the oncogenesis of various cancers including NSCLC (Zhang et al., 2018b; Zhu et al., 2018). Apolipoprotein M (ApoM) is a sphingosine 1-phosphate (S1P) carrier, which is involved in regulating S1P (Duan et al., 2001; Sevvana et al., 2009). Overexpression of ApoM could promote proliferation, invasion, and tumor growth of NSCLC cell via upregulation of S1PR1 (Zhu et al., 2018).
Transgelin 2 (TAGLN2), an actin-binding protein, is overexpressed in various tumors and thought to be a tumor suppressor (Zhang et al., 2010; Jin et al., 2016; Han et al., 2017). Studies suggest that high levels of TAGLN2 in NSCLC cells were significantly associated with tumor development, neural invasion, and metastasis (Jin et al., 2016; Kim et al., 2018). Therefore, it has been considered a crucial diagnostic biomarker for early diagnosis and treatment guidance of NSCLC (Rho et al., 2009). Recent studies have focused on investigating microRNAs targeting TAGLN2 for tumor suppression (Nohata et al., 2011; Yoshino et al., 2011; Du et al., 2016).
The Key Dysfunctional Pathway in CD8+ T Cells
As previously discussed, the most significantly enriched pathway of the 20-gene set is the hsa04062 chemokine signaling pathway. Three of these genes (CXCL13, RASGRP2, and PXN) are involved in this pathway.
Chemokines, a group of small molecular weight proteins, play an important role in cell migration, immune surveillance, and inflammation via binding to chemokine receptors on cell membranes (Raman et al., 2011). Numerous studies have shown the role of chemokine and chemokine receptors in the progression and metastasis of lung cancer (Cavallaro, 2013; Sarvaiya et al., 2013). The chemokine receptors, like CXCR4 and CCR7, are well studied (Mishan et al., 2016; Pozzobon et al., 2016). CXCR4 is the most commonly overexpressed and studied chemokine receptor in many different malignant tumors, including lung cancer (Balkwill, 2004). Differential expression of CXCR4 has been reported to be related to the metastatic potential of non-small cell lung cancer (NSCLC) (Phillips et al., 2003; Su et al., 2005). Wang et al. (2014) also revealed that the high-level CXCR4 expression was associated with brain-specific metastasis after complete resection of non-small cell lung cancer. In addition, CXCL12/CXCR4 axis is demonstrated to play a crucial role in migration and metastasis of NSCLC, and high expression of CXCL12/CXCR4 is related to poor prognosis in NSCLC (Suzuki et al., 2008). CCR7, a CC chemokine receptor, is mainly expressed on naive T cells, B cells, and mature dendritic cells (DCs) (Xu et al., 2012). Activation of CCR7 has been also proved to mediate the invasion and progression of NSCLC in most investigations (Cabioglu et al., 2007; Zhang L. et al., 2017). There was a correlation between tumor-infiltrating DC aggregation and apoptosis of NSCLC.
Besides CXCL13 as a chemokine, Paxillin (PXN) encodes a cytoskeletal protein, which contributes to actin-membrane attachment in the extracellular matrix. PXN is involved in signal transduction, which has been shown to be closely correlated with the oncogenesis and metastasis of NSCLC (Jagadeeswaran et al., 2008; Wu et al., 2010). Previous studies report that miR-137 suppressed cell migration and invasion by targeting PXN, therefore providing a potential therapy for NSCLC by targeting miRNA expression (Dacic et al., 2010; Bi et al., 2014). The expression of paxillin has also been observed as closely associated with the prognosis and the lymph node metastasis of NSCLC patients (Salgia et al., 1999; Zuo and Li, 2003; Wu et al., 2010). These studies strongly suggest the role of PXN in NSCLC, and thus, PXN is recommended as a potential target for NSCLC treatment.
Conclusion
For more effective immunotherapy in the case of non-small cell lung cancers (NSCLC), we require further knowledge of T-cell biology. Therefore, we collected the published RNA-Seq gene expression data of 36 T-cell samples isolated from tumor and 32 adjacent uninvolved lung samples from a publicly available database. By analyzing them with a Monte Carlo feature selection method, we identified the discriminative genes between tumor T cells and adjacent uninvolved lung cells. In addition, we investigated the expression pattern of these key genes for CD8+ T cells of non-small cell lung cancer and their biological functions and pathways. However, tumors are commonly heterogeneous at the cellular level, and therefore, there are different proportions of CD8+ T-cell types (Wagner et al., 2019). This is currently an unresolved question.
Data Availability Statement
We downloaded the gene expression profiles from Gene Expression Omnibus (GEO) under accession number GSE90728.
Author Contributions
DM and TH designed the experiment. XT and XW performed the experiment and analyzed the data. XT and TH wrote the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 81172221, 31701151), National Key R&D Program of China (2018YFC0910403), Shanghai Municipal Science and Technology Major Project (Grant No. 2017SHZDZX01), Shanghai Sailing Program (16YF1413800), and The Youth Innovation Promotion Association of Chinese Academy of Sciences (CAS) (2016245).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Al-Shibli, K. I., Donnem, T., Al-Saad, S., Persson, M., Bremnes, R. M., and Busund, L. T. (2008). Prognostic effect of epithelial and stromal lymphocyte infiltration in non-small cell lung cancer. Clin. Cancer Res. 14, 5220–5227. doi: 10.1158/1078-0432.ccr-08-0133
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Antonicelli, A., Cafarotti, S., Indini, A., Galli, A., Russo, A., Cesario, A., et al. (2013). EGFR-targeted therapy for non-small cell lung cancer: focus on EGFR oncogenic mutation. Int. J. Med. Sci. 10, 320–330. doi: 10.7150/ijms.4609
Balkwill, F. (2004). The significance of cancer cell expression of the chemokine receptor CXCR4. Semin. Cancer Biol. 14, 171–179. doi: 10.1016/j.semcancer.2003.10.003
Baxter, C. S., Hoffman, J. D., Knipp, M. J., Reponen, T., and Haynes, E. N. (2014). Exposure of firefighters to particulates and polycyclic aromatic hydrocarbons. J. Occup. Environ. Hyg. 11, D85–D91. doi: 10.1080/15459624.2014.890286
Berrandou, T., Mulot, C., Cordina-Duverger, E., Arveux, P., Laurent-Puig, P., Truong, T., et al. (2018). Association of breast cancer risk with polymorphisms in genes involved in the metabolism of xenobiotics and interaction with tobacco smoking: a gene-set analysis. Int. J. Cancer 144, 1896–1908. doi: 10.1002/ijc.31917
Bi, Y., Han, Y., Bi, H., Gao, F., and Wang, X. (2014). miR-137 impairs the proliferative and migratory capacity of human non-small cell lung cancer cells by targeting paxillin. Hum. Cell 27, 95–102. doi: 10.1007/s13577-013-0085-84
Cabioglu, N., Gong, Y., Islam, R., Broglio, K. R., Sneige, N., Sahin, A., et al. (2007). Expression of growth factor and chemokine receptors: new insights in the biology of inflammatory breast cancer. Ann. Oncol. 18, 1021–1029. doi: 10.1093/annonc/mdm060
Cai, Y.-D., Zhang, S., Zhang, Y.-H., Pan, X., Feng, K., Chen, L., et al. (2018). Identification of the Gene Expression Rules That Define the Subtypes in Glioma. J. Clin. Med. 7:350. doi: 10.3390/jcm7100350
Cantalupo, A., Gargiulo, A., Dautaj, E., Liu, C., Zhang, Y., Hla, T., et al. (2017). S1PR1 (Sphingosine-1-Phosphate Receptor 1) signaling regulates blood flow and pressure. Hypertension 70, 426–434. doi: 10.1161/hypertensionaha.117.09088
Cavallaro, S. (2013). CXCR4/CXCL12 in non-small-cell lung cancer metastasis to the brain. Int. J. Mol. Sci. 14, 1713–1727. doi: 10.3390/ijms14011713
Chen, L., Li, J., Zhang, Y. H., Feng, K., Wang, S., Zhang, Y., et al. (2018a). Identification of gene expression signatures across different types of neural stem cells with the Monte-Carlo feature selection method. J. Cell. Biochem. 119, 3394–3403. doi: 10.1002/jcb.26507
Chen, L., Pan, X., Hu, X., Zhang, Y. H., Wang, S., Huang, T., et al. (2018b). Gene expression differences among different MSI statuses in colorectal cancer. Int. J. Cancer 143, 1731–1740. doi: 10.1002/ijc.31554
Chen, L., Wang, S., Zhang, Y. H., Wei, L., Xu, X., Huang, T., et al. (2018c). Prediction of nitrated tyrosine residues in protein sequences by extreme learning machine and feature selection methods. Comb. Chem. High Throughput Screen. 21, 393–402. doi: 10.2174/1386207321666180531091619
Chen, L., Pan, X., Guo, W., Gan, Z., Zhang, Y.-H., Niu, Z., et al. (2020). Investigating the gene expression profiles of cells in seven embryonic stages with machine learning algorithms. Genomics 112, 2524–2534. doi: 10.1016/j.ygeno.2020.02.004
Chen, L., Pan, X., Zeng, T., Zhang, Y., Huang, T., and Cai, Y. (2019). Identifying essential signature genes and expression rules associated with distinctive development stages of early embryonic cells. IEEE Access. 7, 128570–128578. doi: 10.1109/ACCESS.2019.2939556
Chen, X., Jin, Y., and Feng, Y. (2019). Evaluation of plasma extracellular vesicle MicroRNA signatures for lung adenocarcinoma and granuloma with Monte-Carlo feature selection method. Front. Genet. 10:367. doi: 10.3389/fgene.2019.00367
Choi, K. S., Fogg, D. K., Yoon, C. S., and Waisman, D. M. (2003). p11 regulates extracellular plasmin production and invasiveness of HT1080 fibrosarcoma cells. FASEB J. 17, 235–246. doi: 10.1096/fj.02-0697com
Dacic, S., Kelly, L., Shuai, Y., and Nikiforova, M. N. (2010). miRNA expression profiling of lung adenocarcinomas: correlation with mutational status. Mod. Pathol. 23, 1577–1582. doi: 10.1038/modpathol.2010.152
Dermani, F. K., Samadi, P., Rahmani, G., Kohlan, A. K., and Najafi, R. (2019). PD-1/PD-L1 immune checkpoint: potential target for cancer therapy. J. Cell. Physiol. 234, 1313–1325. doi: 10.1002/jcp.27172
Draminski, M., Rada-Iglesias, A., Enroth, S., Wadelius, C., Koronacki, J., and Komorowski, J. (2008). Monte Carlo feature selection for supervised classification. Bioinformatics 24, 110–117. doi: 10.1093/bioinformatics/btm486
Du, Y. Y., Zhao, L. M., Chen, L., Sang, M. X., Li, J., Ma, M., et al. (2016). The tumor-suppressive function of miR-1 by targeting LASP1 and TAGLN2 in esophageal squamous cell carcinoma. J. Gastroenterol. Hepatol. 31, 384–393. doi: 10.1111/jgh.13180
Duan, J., Dahlback, B., and Villoutreix, B. O. (2001). Proposed lipocalin fold for apolipoprotein M based on bioinformatics and site-directed mutagenesis. FEBS Lett. 499, 127–132. doi: 10.1016/s0014-5793(01)02544-3
Farhood, B., Najafi, M., and Mortezaee, K. (2019). CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: a review. J. Cell. Physiol. 234, 8509–8521. doi: 10.1002/jcp.27782
Ganesan, A. P., Clarke, J., Wood, O., Garrido-Martin, E. M., Chee, S. J., Mellows, T., et al. (2017). Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat. Immunol. 18, 940–950. doi: 10.1038/ni.3775
Garon, E. B., Rizvi, N. A., Hui, R., Leighl, N., Balmanoukian, A. S., Eder, J. P., et al. (2015). Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 372, 2018–2028. doi: 10.1056/NEJMoa1501824
Gettinger, S. N., Horn, L., Gandhi, L., Spigel, D. R., Antonia, S. J., Rizvi, N. A., et al. (2015). Overall survival and long-term safety of nivolumab (Anti-Programmed Death 1 Antibody, BMS-936558, ONO-4538) in patients with previously treated advanced non-small-cell lung cancer. J. Clin. Oncol. 33, 2004–2012. doi: 10.1200/jco.2014.58.3708
Grasse, S., Lienhard, M., Frese, S., Kerick, M., Steinbach, A., Grimm, C., et al. (2018). Epigenomic profiling of non-small cell lung cancer xenografts uncover LRP12 DNA methylation as predictive biomarker for carboplatin resistance. Genome Med. 10:55. doi: 10.1186/s13073-018-0562-561
Han, M. Z., Xu, R., Xu, Y. Y., Zhang, X., Ni, S. L., Huang, B., et al. (2017). TAGLN2 is a candidate prognostic biomarker promoting tumorigenesis in human gliomas. J. Exp. Clin. Cancer Res. 36:155. doi: 10.1186/s13046-017-0619-619
Herbst, R. S., Baas, P., Kim, D. W., Felip, E., Perez-Gracia, J. L., Han, J. Y., et al. (2016). Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387, 1540–1550. doi: 10.1016/s0140-6736(15)01281-1287
Herbst, R. S., Soria, J. C., Kowanetz, M., Fine, G. D., Hamid, O., Gordon, M. S., et al. (2014). Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515, 563–567. doi: 10.1038/nature14011
Hiraoka, K., Miyamoto, M., Cho, Y., Suzuoki, M., Oshikiri, T., Nakakubo, Y., et al. (2006). Concurrent infiltration by CD8+ T cells and CD4+ T cells is a favourable prognostic factor in non-small-cell lung carcinoma. Br. J. Cancer 94, 275–280. doi: 10.1038/sj.bjc.6602934
Jagadeeswaran, R., Surawska, H., Krishnaswamy, S., Janamanchi, V., Mackinnon, A. C., Seiwert, T. Y., et al. (2008). Paxillin is a target for somatic mutations in lung cancer: implications for cell growth and invasion. Cancer Res. 68, 132–142. doi: 10.1158/0008-5472.can-07-1998
Ji, J., Zhao, L., Wang, X., Zhou, C., Ding, F., Su, L., et al. (2004). Differential expression of S100 gene family in human esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 130, 480–486. doi: 10.1007/s00432-004-0555-x
Jia, M., Feng, W., Kang, S., Zhang, Y., Shen, J., He, J., et al. (2015). Evaluation of the efficacy and safety of anti-PD-1 and anti-PD-L1 antibody in the treatment of non-small cell lung cancer (NSCLC): a meta-analysis. J. Thorac. Dis. 7, 455–461. doi: 10.3978/j.issn.2072-1439.2015.02.06
Jin, H., Cheng, X., Pei, Y., Fu, J., Lyu, Z., Peng, H., et al. (2016). Identification and verification of transgelin-2 as a potential biomarker of tumor-derived lung-cancer endothelial cells by comparative proteomics. J. Proteomics 136, 77–88. doi: 10.1016/j.jprot.2015.12.012
Johnson, D. B., Rioth, M. J., and Horn, L. (2014). Immune checkpoint inhibitors in NSCLC. Curr. Treat. Options. Oncol. 15, 658–669. doi: 10.1007/s11864-014-0305-305
Johnson, S. K., Kerr, K. M., Chapman, A. D., Kennedy, M. M., King, G., Cockburn, J. S., et al. (2000). Immune cell infiltrates and prognosis in primary carcinoma of the lung. Lung Cancer 27, 27–35. doi: 10.1016/s0169-5002(99)00095-1
Katono, K., Sato, Y., Jiang, S. X., Kobayashi, M., Saito, K., Nagashio, R., et al. (2016). Clinicopathological significance of S100A10 expression in lung adenocarcinomas. Asian Pac. J. Cancer Prev. 17, 289–294. doi: 10.7314/apjcp.2016.17.1.289
Kawai, O., Ishii, G., Kubota, K., Murata, Y., Naito, Y., Mizuno, T., et al. (2008). Predominant infiltration of macrophages and CD8(+) T Cells in cancer nests is a significant predictor of survival in stage IV nonsmall cell lung cancer. Cancer 113, 1387–1395. doi: 10.1002/cncr.23712
Kim, I. G., Lee, J. H., Kim, S. Y., Hwang, H. M., Kim, T. R., and Cho, E. W. (2018). Hypoxia-inducible transgelin 2 selects epithelial-to-mesenchymal transition and gamma-radiation-resistant subtypes by focal adhesion kinase-associated insulin-like growth factor 1 receptor activation in non-small-cell lung cancer cells. Cancer Sci. 109, 3519–3531. doi: 10.1111/cas.13791
Kwon, M., MacLeod, T. J., Zhang, Y., and Waisman, D. M. (2005). S100A10, annexin A2, and annexin a2 heterotetramer as candidate plasminogen receptors. Front. Biosci. 10:300–325. doi: 10.2741/1529
Li, H., Li, Q. D., Wang, M. S., Li, F. J., Li, Q. H., Ma, X. J., et al. (2013). Smoking and air pollution exposure and lung cancer mortality in Zhaoyuan County. Int. J. Hyg. Environ. Health 216, 63–70. doi: 10.1016/j.ijheh.2012.06.003
Li, J., Lan, C.-N., Kong, Y., Feng, S.-S., and Huang, T. (2018a). Identification and analysis of blood gene expression signature for osteoarthritis with advanced feature selection methods. Front. Genet. 9:246. doi: 10.3389/fgene.2018.00246
Li, J. J., Li, R., Wang, W., Zhang, B., Song, X., Zhang, C., et al. (2018b). IDH2 is a novel diagnostic and prognostic serum biomarker for non-small-cell lung cancer. Mol. Oncol. 12, 602–610. doi: 10.1002/1878-0261.12182
Li, J., Lu, L., Zhang, Y.-H., Xu, Y., Liu, M., Feng, K., et al. (2019). Identification of leukemia stem cell expression signatures through Monte Carlo feature selection strategy and support vector machine. Cancer Gene Ther. 27, 56–69. doi: 10.1038/s41417-019-0105-y
Liu, Y., Wada, R., Yamashita, T., Mi, Y., Deng, C. X., Hobson, J. P., et al. (2000). Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J. Clin. Invest. 106, 951–961. doi: 10.1172/jci10905
Love, M. I., Huber, W., and Anders, S. J. G. B. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-558
Martinez, P., Martinez-Marti, A., Navarro, A., Cedres, S., and Felip, E. (2014). Molecular targeted therapy for early-stage non-small-cell lung cancer: will it increase the cure rate? Lung Cancer 84, 97–100. doi: 10.1016/j.lungcan.2014.01.018
Meijer, J., Zeelenberg, I. S., Sipos, B., and Roos, E. (2006). The CXCR5 chemokine receptor is expressed by carcinoma cells and promotes growth of colon carcinoma in the liver. Cancer Res. 66, 9576–9582. doi: 10.1158/0008-5472.can-06-1507
Meissner, A. (2017). S1PR (Sphingosine-1-Phosphate Receptor) signaling in the regulation of vascular tone and blood pressure: is S1PR1 doing the Trick? Hypertension 70, 232–234. doi: 10.1161/hypertensionaha.117.09200
Metodieva, S. N., Nikolova, D. N., Cherneva, R. V., Dimova, I. I., Petrov, D. B., and Toncheva, D. I. (2011). Expression analysis of angiogenesis-related genes in Bulgarian patients with early-stage non-small cell lung cancer. Tumori 97, 86–94. doi: 10.1177/030089161109700116
Mishan, M. A., Ahmadiankia, N., and Bahrami, A. R. (2016). CXCR4 and CCR7: two eligible targets in targeted cancer therapy. Cell Biol. Int. 40, 955–967. doi: 10.1002/cbin.10631
Morgensztern, D., and Herbst, R. S. (2016). Nivolumab and Pembrolizumab for non-small cell lung cancer. Clin. Cancer Res. 22, 3713–3717. doi: 10.1158/1078-0432.ccr-15-2998
Nascimento, A. V., Bousbaa, H., Ferreira, D., and Sarmento, B. (2015). Non-Small cell lung carcinoma: an overview on targeted therapy. Curr. Drug Targets 16, 1448–1463. doi: 10.2174/1389450115666140528151649
Nohata, N., Sone, Y., Hanazawa, T., Fuse, M., Kikkawa, N., Yoshino, H., et al. (2011). miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget 2, 29–42. doi: 10.18632/oncotarget.213
Pan, X., Hu, X., Zhang, Y.-H., Chen, L., Zhu, L., Wan, S., et al. (2018). Identification of the copy number variant biomarkers for breast cancer subtypes. Mol. Genet. Genomics 294, 95–110. doi: 10.1007/s00438-018-1488-1484
Pan, X., Hu, X., Zhang, Y.-H., Chen, L., Zhu, L., Wan, S., et al. (2019). Identification of the copy number variant biomarkers for breast cancer subtypes. Mol. Genet. Genomics 294, 95–110. doi: 10.1007/s00438-018-1488-1484
Pardoll, D. M. (2012). The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264. doi: 10.1038/nrc3239
Petersson, S., Shubbar, E., Enerback, L., and Enerback, C. (2009). Expression patterns of S100 proteins in melanocytes and melanocytic lesions. Melanoma Res. 19, 215–225. doi: 10.1097/CMR.0b013e32832c6358
Phillips, R. J., Burdick, M. D., Lutz, M., Belperio, J. A., Keane, M. P., and Strieter, R. M. (2003). The stromal derived factor-1/CXCL12-CXC chemokine receptor 4 biological axis in non-small cell lung cancer metastases. Am. J. Respir. Crit. Care Med. 167, 1676–1686. doi: 10.1164/rccm.200301-071OC
Pozzobon, T., Goldoni, G., Viola, A., and Molon, B. (2016). CXCR4 signaling in health and disease. Immunol. Lett. 177, 6–15. doi: 10.1016/j.imlet.2016.06.006
Ramalingam, S., Hui, R., Gandhi, L., Carcereny, E., Felip, E., Ahn, M. J., et al. (2016). P2.39: long-term OS for patients with advanced NSCLC enrolled in the KEYNOTE-001 study of pembrolizumab: track: immunotherapy. J. Thorac. Oncol. 11, S241–S242. doi: 10.1016/j.jtho.2016.08.110
Raman, D., Sobolik-Delmaire, T., and Richmond, A. (2011). Chemokines in health and disease. Exp. Cell Res. 317, 575–589. doi: 10.1016/j.yexcr.2011.01.005
Rho, J. H., Roehrl, M. H., and Wang, J. Y. (2009). Tissue proteomics reveals differential and compartment-specific expression of the homologs transgelin and transgelin-2 in lung adenocarcinoma and its stroma. J. Proteome Res. 8, 5610–5618. doi: 10.1021/pr900705r
Salgia, R., Li, J. L., Ewaniuk, D. S., Wang, Y. B., Sattler, M., Chen, W. C., et al. (1999). Expression of the focal adhesion protein paxillin in lung cancer and its relation to cell motility. Oncogene 18, 67–77. doi: 10.1038/sj.onc.1202273
Sarkisyan, G., Gay, L. J., Nguyen, N., Felding, B. H., and Rosen, H. (2014). Host endothelial S1PR1 regulation of vascular permeability modulates tumor growth. Am. J. Physiol. Cell Physiol. 307, C14–C24. doi: 10.1152/ajpcell.00043.2014
Sarvaiya, P. J., Guo, D., Ulasov, I., Gabikian, P., and Lesniak, M. S. (2013). Chemokines in tumor progression and metastasis. Oncotarget 4, 2171–2185. doi: 10.18632/oncotarget.1426
Schalper, K. A., Brown, J., Carvajal-Hausdorf, D., McLaughlin, J., Velcheti, V., Syrigos, K. N., et al. (2015). Objective measurement and clinical significance of TILs in non-small cell lung cancer. J. Natl. Cancer Inst. 107:dju435. doi: 10.1093/jnci/dju435
Sevvana, M., Ahnstrom, J., Egerer-Sieber, C., Lange, H. A., Dahlback, B., and Muller, Y. A. (2009). Serendipitous fatty acid binding reveals the structural determinants for ligand recognition in apolipoprotein M. J. Mol. Biol. 393, 920–936. doi: 10.1016/j.jmb.2009.08.071
Siegel, R., DeSantis, C., Virgo, K., Stein, K., Mariotto, A., Smith, T., et al. (2012). Cancer treatment and survivorship statistics, 2012. CA Cancer J. Clin. 62, 220–241. doi: 10.3322/caac.21149
Simon, R. (2008). Development and validation of biomarker classifiers for treatment selection. J Stat. Plan Inference 138, 308–320. doi: 10.1016/j.jspi.2007.06.010
Singh, R., Gupta, P., Kloecker, G. H., Singh, S., and Lillard, J. W. Jr. (2014). Expression and clinical significance of CXCR5/CXCL13 in human nonsmall cell lung carcinoma. Int. J. Oncol. 45, 2232–2240. doi: 10.3892/ijo.2014.2688
Su, L., Zhang, J., Xu, H., Wang, Y., Chu, Y., Liu, R., et al. (2005). Differential expression of CXCR4 is associated with the metastatic potential of human non-small cell lung cancer cells. Clin. Cancer Res. 11, 8273–8280. doi: 10.1158/1078-0432.ccr-05-0537
Suzuki, M., Mohamed, S., Nakajima, T., Kubo, R., Tian, L., Fujiwara, T., et al. (2008). Aberrant methylation of CXCL12 in non-small cell lung cancer is associated with an unfavorable prognosis. Int. J. Oncol. 33, 113–119.
Thommen, D. S., Koelzer, V. H., Herzig, P., Roller, A., Trefny, M., Dimeloe, S., et al. (2018). A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 24, 994–1004. doi: 10.1038/s41591-018-0057-z
Tian, T., Hao, J., Xu, A., Hao, J., Luo, C., Liu, C., et al. (2007). Determination of metastasis-associated proteins in non-small cell lung cancer by comparative proteomic analysis. Cancer Sci. 98, 1265–1274. doi: 10.1111/j.1349-7006.2007.00514.x
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Tusher, V. G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121. doi: 10.1073/pnas.091062498
van der Drift, M. A., Karim-Kos, H. E., Siesling, S., Groen, H. J., Wouters, M. W., Coebergh, J. W., et al. (2012). Progress in standard of care therapy and modest survival benefits in the treatment of non-small cell lung cancer patients in the Netherlands in the last 20 years. J. Thorac. Oncol. 7, 291–298. doi: 10.1097/JTO.0b013e31823a01fb
Wagner, J., Rapsomaniki, M. A., Chevrier, S., Anzeneder, T., Langwieder, C., Dykgers, A., et al. (2019). A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell 177, 1330–1345.e18. doi: 10.1016/j.cell.2019.03.005
Wang, G. Z., Cheng, X., Zhou, B., Wen, Z. S., Huang, Y. C., Chen, H. B., et al. (2015). The chemokine CXCL13 in lung cancers associated with environmental polycyclic aromatic hydrocarbons pollution. eLife 4:e09419. doi: 10.7554/eLife.09419
Wang, J., Vasaikar, S., Shi, Z., Greer, M., and Zhang, B. (2017). WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137. doi: 10.1093/nar/gkx356
Wang, S., Wang, D., Li, J., Huang, T., and Cai, Y.-D. (2017). Identification and analysis of the cleavage site in signal peptide by using SMOTE, Dagging, and feature selection methods. Mol. Omics 14, 64–73. doi: 10.1039/c7mo00030h
Wang, L., Wang, Z., Liu, X., and Liu, F. (2014). High-level C-X-C chemokine receptor type 4 expression correlates with brain-specific metastasis following complete resection of non-small cell lung cancer. Oncol. Lett. 7, 1871–1876. doi: 10.3892/ol.2014.1979
Welsh, T. J., Green, R. H., Richardson, D., Waller, D. A., O’Byrne, K. J., and Bradding, P. (2005). Macrophage and mast-cell invasion of tumor cell islets confers a marked survival advantage in non-small-cell lung cancer. J. Clin. Oncol. 23, 8959–8967. doi: 10.1200/jco.2005.01.4910
Westbrook, J. A., Wood, S. L., Cairns, D. A., McMahon, K., Gahlaut, R., Thygesen, H., et al. (2019). Identification and validation of DOCK4 as a potential biomarker for risk of bone metastasis development in patients with early breast cancer. J. Pathol. 247, 381–391. doi: 10.1002/path.5197
Wu, D. W., Cheng, Y. W., Wang, J., Chen, C. Y., and Lee, H. (2010). Paxillin predicts survival and relapse in non-small cell lung cancer by microRNA-218 targeting. Cancer Res. 70, 10392–10401. doi: 10.1158/0008-5472.can-10-2341
Wu, Y., Jamal, M., Xie, T., Sun, J., Song, T., Yin, Q., et al. (2019). Uridine-cytidine kinase 2 (UCK2): a potential diagnostic and prognostic biomarker for lung cancer. Cancer Sci. 110, 2734–2747. doi: 10.1111/cas.14125
Xu, Y., Liu, L., Qiu, X., Liu, Z., Li, H., Li, Z., et al. (2012). CCL21/CCR7 prevents apoptosis via the ERK pathway in human non-small cell lung cancer cells. PLoS one 7:e33262. doi: 10.1371/journal.pone.0033262
Yang, X., Popescu, N. C., and Zimonjic, D. B. (2011). DLC1 interaction with S100A10 mediates inhibition of in vitro cell invasion and tumorigenicity of lung cancer cells through a RhoGAP-independent mechanism. Cancer Res. 71, 2916–2925. doi: 10.1158/0008-5472.can-10-02158
Yoshino, H., Chiyomaru, T., Enokida, H., Kawakami, K., Tatarano, S., Nishiyama, K., et al. (2011). The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br. J. Cancer 104, 808–818. doi: 10.1038/bjc.2011.23
Zhan, Y., Zang, H., Feng, J., Lu, J., Chen, L., and Fan, S. (2017). Long non-coding RNAs associated with non-small cell lung cancer. Oncotarget 8, 69174–69184. doi: 10.18632/oncotarget.20088
Zhang, L., Fogg, D. K., and Waisman, D. M. (2004). RNA interference-mediated silencing of the S100A10 gene attenuates plasmin generation and invasiveness of Colo 222 colorectal cancer cells. J. Biol. Chem. 279, 2053–2062. doi: 10.1074/jbc.M310357200
Zhang, L., Xiao, X., An, H., Wang, J., Ma, Y., and Qian, Y. H. (2017). Inhibition of CCR7 promotes NF-kappaB-dependent apoptosis and suppresses epithelial-mesenchymal transition in non-small cell lung cancer. Oncol. Rep. 37, 2913–2919. doi: 10.3892/or.2017.5524
Zhang, Y. H., Huang, T., Chen, L., Xu, Y., Hu, Y., Hu, L. D., et al. (2017). Identifying and analyzing different cancer subtypes using RNA-seq data of blood platelets. Oncotarget 8, 87494–87511. doi: 10.18632/oncotarget.20903
Zhang, X., Lan, Y., Xu, J., Quan, F., Zhao, E., Deng, C., et al. (2018a). CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 47, D721–D728. doi: 10.1093/nar/gky900
Zhang, X., Liu, Y., Huang, W. C., and Zheng, L. C. (2018b). MiR-125b-1-3p exerts antitumor functions in lung carcinoma cells by targeting S1PR1. Chin. Med. J. 131, 1909–1916. doi: 10.4103/0366-6999.238135
Zhang, X., Zhang, H., Shen, B., and Sun, X. F. (2019). Chromogranin-A expression as a novel biomarker for early diagnosis of colon cancer patients. Int. J. Mol. Sci. 20:2919. doi: 10.3390/ijms20122919
Zhang, Y., Ye, Y., Shen, D., Jiang, K., Zhang, H., Sun, W., et al. (2010). Identification of transgelin-2 as a biomarker of colorectal cancer by laser capture microdissection and quantitative proteome analysis. Cancer Sci. 101, 523–529. doi: 10.1111/j.1349-7006.2009.01424.x
Zhu, Y., Luo, G., Jiang, B., Yu, M., Feng, Y., Wang, M., et al. (2018). Apolipoprotein M promotes proliferation and invasion in non-small cell lung cancers via upregulating S1PR1 and activating the ERK1/2 and PI3K/AKT signaling pathways. Biochem. Biophys. Res. Commun. 501, 520–526. doi: 10.1016/j.bbrc.2018.05.029
Keywords: gene, CD8+ T cell, non-small cell lung cancer, RNA sequencing, feature selection, dysfunctional pathways
Citation: Tao X, Wu X, Huang T and Mu D (2020) Identification and Analysis of Dysfunctional Genes and Pathways in CD8+ T Cells of Non-Small Cell Lung Cancer Based on RNA Sequencing. Front. Genet. 11:352. doi: 10.3389/fgene.2020.00352
Received: 02 January 2020; Accepted: 23 March 2020;
Published: 08 May 2020.
Edited by:
Robert Friedman, University of South Carolina, United StatesReviewed by:
Yuan Tian, Fred Hutchinson Cancer Research Center, United StatesJose Sullivan Lopez-Gonzalez, National Institute of Respiratory Diseases, Mexico
Copyright © 2020 Tao, Wu, Huang and Mu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deguang Mu, mudeguang0314@163.com; Tao Huang, tohuangtao@126.com
 Xuefang Tao1
Xuefang Tao1 Tao Huang
Tao Huang