- 1Yunnan Key Laboratory of Green Prevention and Control of Agricultural Transboundary Pests, Agricultural Environment and Resource Research Institute, Yunnan Academy of Agricultural Sciences, Kunming, China
- 2Flower Research Institute, Yunnan Academy of Agricultural Sciences, Kunming, China
- 3The Ministry of Education Key Laboratory for Agricultural Biodiversity and Pest Management, Yunnan Agricultural University, Kunming, China
Rice blast is caused by Magnaporthe oryzae (M. oryzae), which is considered one of the most serious pathogens of rice around the globe. It causes severe losses owing to its proven capability to disrupt the host resistance. Recently, its invasion of new hosts like the Musa species or banana plants has been noticed. To understand the possible level of genetic variation, we sequenced the genomes of eight different isolates of the Magnaporthe species infecting rice, Digitaria (a weed), finger millet, Elusine indica, and banana plants. Comparative genomic analysis of these eight isolates with the previously well-characterized laboratory strain M. oryzae 70-15 was made. The infectivity of the newly isolated strain from Musa species suggested that there is no resistance level in the host plants. The sequence analysis revealed that despite genome similarities, both the banana and Digitaria isolates have relatively larger genome sizes (∼38.2 and 51.1 Mb, respectively) compared to those of the laboratory reference strain M. oryzae 70-15 (∼37 Mb). The gene contraction, expansion, and InDel analysis revealed that during evolution, a higher number of gene insertions and deletions occurred in the blast fungus infecting Digitaria and banana. Furthermore, each genome shared thousands of genes, which suggest their common evolution. Overall, our analysis indicates that higher levels of genes insertion or deletions and gain in the total genome size are important factors in disrupting the host immunity and change in host selection.
Introduction
Rice blast is considered the most disastrous disease of rice owing to its global distribution (in more than 80 countries on all continents) and destructiveness under favorable conditions. It is caused by M. oryzae (also known as Pyricularia oryzae), which is a filamentous ascomycete fungus. Rice blast is usually responsible for about 10–30% of total rice yield losses annually (Wilson and Talbot, 2009; Ashkani et al., 2015; Sakulkoo et al., 2018). However, under favorable conditions, M. oryzae can destroy entire rice plants within 15–20 days post-infection leading up to 100% yield losses subsequently resulting in total crop failure (Musiime et al., 2005; Skamnioti and Gurr, 2009). It is reported that rice blast accounts for the annual rice production losses of above 50 million tonnes globally which are sufficient enough to feed 210–740 million people (Boddy, 2016). Since rice is a staple food, and provides almost 25% of the calories of the daily human diet, its decreased yield because of rice blast is a serious threat to global food security (Asibi et al., 2019).
Currently, synthetic fungicides are commonly used for the control of rice blast, but the use of these chemicals is gradually being reduced to ensure sustainable food production. Fungicides have attendant problems of cost, development of fungal resistance, and damage to ecosystems. Therefore, the use of resistant cultivars, antagonistic phylloplane bacteria, and fungi is also being tested as an alternative option. But the major issue with M. oryzae is its rapid ability to overcome resistance based on R-specific genes (Kiyosawa, 1982; Zeigler et al., 1994; Orbach et al., 2000). Understanding the pathogen’s biology is a prerequisite for developing disease control strategies and in the case of M. oryzae, it is more crucial to understand its biology as its host range is extended to wheat which is now an urgent problem in South America (Boddy, 2016). The spores of fungus have the inherent adhesive capability for their attachment to the hydrophobic surface of the leaves (Nilson and Talbot 2009). The molecular studies revealed that the Mpg1 gene coded by the fungal genome is responsible for the attachment of the fungus on the plant surface (Talbot et al., 1993). Once the fungus invades the host plant, it can change the hormonal activities of the host cells for its own aggressive growth.
The first genome sequence of M. oryzae for strains 70-15 was published in 2005 and now there are over 30 genomes available from different strains around the world. The genome is composed of seven chromosomes and has a size of ∼40 Mb. Its genome regulates the expression of about 1,500 proteins, many with unknown functions, but certainly, there are some effector proteins among them (Xu et al., 2007). Many novel genes affecting various metabolic networks have been identified in different isolates of the M. oryzae. Over 20% (2,154) of genes are differentially expressed, mostly up-regulated during the processes prior to invasion (Oh et al., 2008). Temporal analysis of the transcriptome during infection, together with gene replacement experiments, identifies virulence-related genes required for successful invasion. For example, avirulence in some strains of M. oryzae has been traced to the loss of function in several specific genes: two of these encode proteins in signaling pathways linked to G-protein-coupled receptors, both of which are required for appressorium formation (Boddy, 2016).
The natural resistance against rice blast fungus has been introduced into the rice cultivars, which is durable and long-lasting (Ashkani et al., 2014: Sharma et al., 2012). Due to the availability of complete genome sequences and studies on the interaction of the rice blast fungus with its hosts, the Magnaporthe species appeared as a model organism for genetic studies. The evolution of rice blast fungus is due to point mutations and insertions or deletions of complete genes (Fudal et al., 2009; Van de Wouw et al., 2010). Due to continuous breeding efforts, the fungal populations usually respond by regaining expelled avirulence genes. The gene-for-gene resistance concept is well established for rice blast fungus. It is a proven fact that deletion or point mutation of certain Avr genes like Avr-Pita is associated with resistance breakdown in the field conditions (Orbach et al., 2000; Zhou et al., 2007; Dai et al., 2010).
Due to its agricultural impact and tractability, M. oryzae has become a model fungus for studying host-pathogen interactions, at both the cell biology and molecular levels. To determine the genetic variation among the isolates of M. oryzae, we sequenced eight field isolates of this fungus collected from different hosts. We envisaged that a mechanistic understanding of the complex interactions among the pathogen, host, and environment will lead to accurate forecasts of pathogen distribution and greatly advance the management of rice blast for global food security under the present scenario of climate change.
Results
Characterization of Magnaporthe Fungi Isolated From Rice
Based on the host’s diversity, morphology, and molecular characterization, eight different strains of rice blast pathogens were identified. The Magnaporthe species infecting the above-mentioned hosts including Oryzae sativa were germinated for 15 days. The tissue samples obtained from different hosts plants were inoculated on a liquid PDA medium, which resulted in the development of white mycelia. The four strains isolated from Oryza sativa were named R01, R05, R06, and R08. While the strains isolated from, Eleusine coracana, Digitaria sanguinalis, Eleusine indica, and banana plants were designated as R02, R03, R04, and R07, respectively. The microscopic analysis confirmed the curve-shaped conidium (Figure 1).

FIGURE 1. Spore shape of rice blast fungus from a different host. R07, R03, R02, R04, R01, R05, R06, R08 were collected from Banana, Digitaria sanguinalis, Eleusine coracana, Eleusine indica, Oryza sativa, Oryza sativa, Oryza sativa, Oryza sativa, respectively.
In order to confirm the pathogenicity of Pyricularia angulate (afterward referred to as R07, originally isolated from banana samples), we inoculated it on the leaves of nine different varieties of Banana. Interestingly, all the banana varieties tested here were highly susceptible to the newly isolated Pyricularia strain. The infection resulted in angular necrotic lesions on the leaf surfaces (Figure 2). Subsequent confirmation was done through molecular methods. Total extracted DNA was subjected to PCR reaction with ITS1 and ITS4 primers. The ITS region was amplified and further sequencing was performed to confirm the presence of rice blast fungus.

FIGURE 2. Symptoms of R07 isolate on nine different banana varieties. The symptoms were observed at 12 days post-inoculation.
The phylogenetic tree constructed from the ITS region suggested that the five isolates (R01, R02, R04, R05, R06, and R08) tightly clustered together. While the two isolates (R03 and R07) isolated from Digitaria sanguinalis and banana branched out separately (Figure 3). These observations suggested that R03 and R07 are evolutionary different from other isolates of blast fungus infecting the Gramineae family.
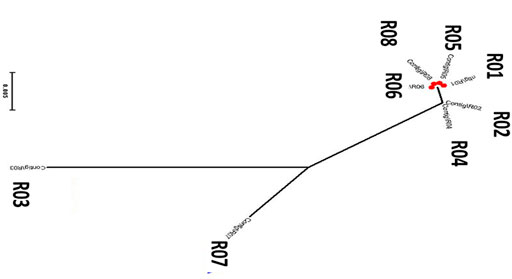
FIGURE 3. Phylogenetic tree of eight different Magnaporthe species isolated from different hosts in Yunnan Province of China. The sequences were derived from the PCR amplified products with ITS1 and ITS4 primers.
Disease Reaction on Different Hosts
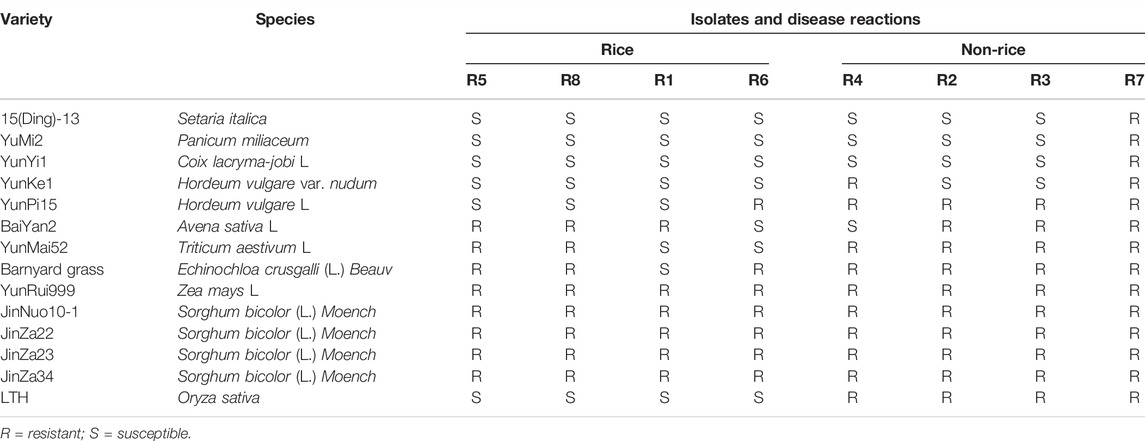
The four Magnaporthe species isolated from rice and four from non-rice plants were inoculated on 14 different hosts of the Gramineae family (Table 1). Interestingly, susceptible disease reaction was noticed for Setaria, Panicum, Coix, Hordeum, and rice fungal isolates against the strains isolated from Rice. Indeed, their infection resulted in typical leaf blast symptoms. While non-rice pathotypes were unable to infect Oryza varieties. These results confirmed the hypothesis that Oryzae sativa is immune to the blast fungus disease of bananas. Interestingly, none of the Graminae hosts showed susceptibility to the R07 strain isolated from bananas.
General Features of Genome Sequencing Assembly
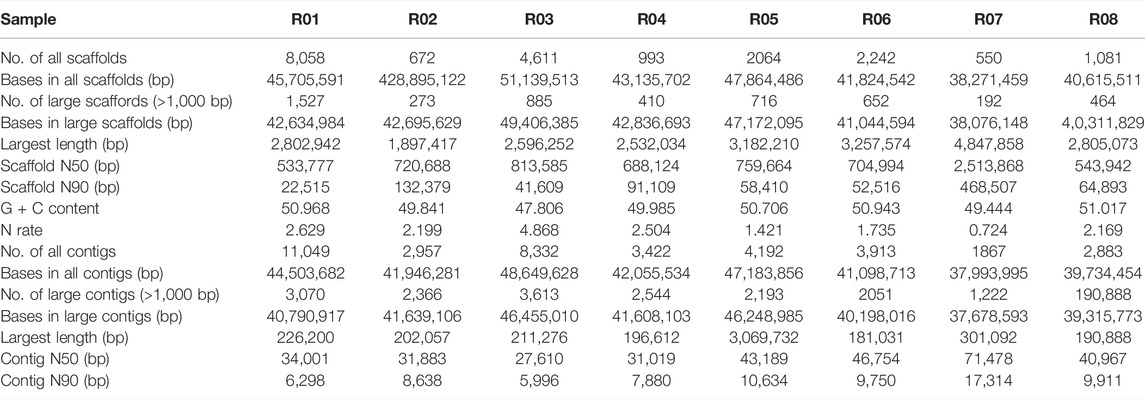
The genomes of blast fungus (R01 to R08) isolated from rice and other hosts were sequenced by the Illumina platform. The Illumina reads were assembled into contigs through SOAPdenovo (V2.04). The assembled genomes consisted of 8,058, 672, 4,611, 993, 2064, 2,242, 550 and 1,081 scaffolds for each strain, respectively (Table 2). The N50 and N90 scaffold lengths for the R07 strain were 251 Kb and 46Kb, respectively (Table 2). Approximately 30% of sequence reads were assembled from more than 1000bp reads for all the strains studied here.
To avoid the repetitive genome sequences which were ∼10% in the laboratory reference strain (70-15), the RepeatMasker program was used for genome comparative analysis. As a result, the number of bases in the genomes after masking the repetitive sequences were 45.7, 42.9, 51.1, 43.1, 47.9, 41.8, 38.2, and 40.6 Mb, respectively (Table 2). Whereas the genome size of 70-15 strain is 37.5 Mb (Dean et al., 2005). These results indicate that the size of core genomes of the eight isolates studied here vary significantly. Nevertheless, the genome size of R07 is very close to the reference strain (70-15). Despite genome size differences in different strains, the GC contents across the genomes were centered at 50% (Table 2).
InDel Length Distribution
InDel length distribution revealed that four isolates of M. oryzae from the rice plant (R01, R05, R06, and R08) have the same length distribution of short and long indels while two strains isolated from Eleusine showed the same number of indels of different lengths but higher than rice isolates (Supplementary Figure S1). The M. oryzae isolated from Digitaria sanguinalis showed a quite higher number of short and long inDels as compared to all other fungal isolates. Interestingly, M. oryzae isolate from Digitaria sanguinalis (R03) exhibited a quite similar InDel distribution with Pyricularia angulate isolate from Musa spp (R07). In both these species (R03 and R07), the length distribution of InDels was quite higher in non-coding regions as compared to their frequency observed in overall CDS (Supplementary Figure S1). A comparison of InDdel distribution between both isolates (R03 and R07) revealed a relatively higher number of short InDdels (1 to 4 bp) in R03 as compared to that of R07.
Gene Distribution in the Rice Blast Fungus
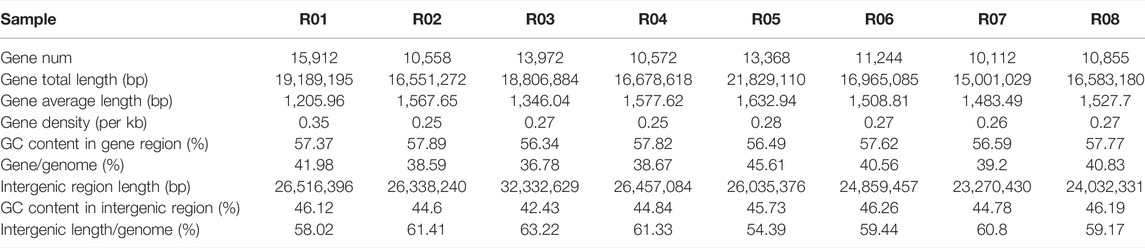
Results revealed significant differences in the genome sizes of different strains, and the number of genes also varied in each strain (Table 3). The number of genes was approximately 15.9, 10.6, 13.9, 10.4, 13.7, 11.2, 10.1, and 10.8 K, respectively (Table 3). The average gene size ranged from 1.2 Kb to 1.6 Kb for each strain, which is approximately 0.8 Kb less than the other rice fungus genomes (Fan et al., 2015). On average, genes covered 40% of the genome with the approximate gene density of ∼0.26 genes per Kb which is similar to rice hoppers fungi (Fan et al., 2015).
As compared with other rice fungal pathogens, these strains have gene families with variable gene expansion and contraction. Just like the reference strain (70-15) which is basal to the phylogenetic tree (Figure 3), R05 and R06 strains had 0 values for gene expansion and contraction, while R07 is only represented by gene contraction (−147). These values indicate that during the course of evolution, the R07 strain deleted gene families (Figure 4A). We also searched for orthologs in these strains. The protein data analysis suggested that these strains share 7,113 proteins with each other representing their common origin (Figure 4B).
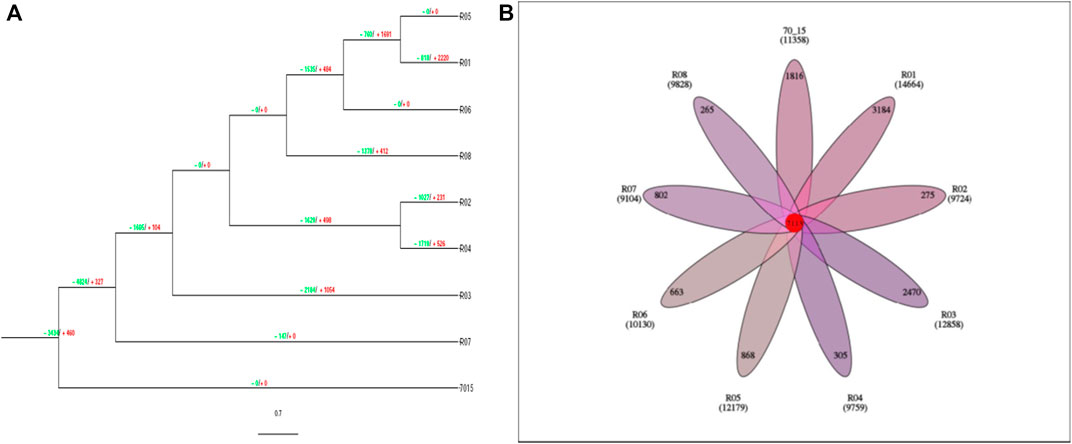
FIGURE 4. (A) Evolutionary branch diagram of gene family expansion and contraction. The tree is rooted on 70-15 strain. The green values represent the reduction while values in red represent the gene expression. (B). The Venn diagram reveals the common ortholog conserved among the rice blast fungus strains. The numbers highlighted in red color represent the common orthologs, while the branch numbers represent the endemic gene families.
The contribution of eight fungal strains to the core and pan-genome analysis of the strain 70-15 is presented in Figure 5. All the strains contributed to an increase in the pan-genome. The pan-genome curve demonstrated that all the strains contributed to a sharp increase in gene families. The core genome slightly increased for R01, R02, and R03. While it slightly changed for the R04, R05, R06, R07, and R08 strains. The gene families for the core genome of the R01 strain represented more than 11,000 gene families. However, the strains R03-R08 are presented between ∼6,000 and 8,000 gene families. Overall, there was an increasing trend in the pan-genome plot while a decreasing trend was observed for the core genome plot (Figure 5).
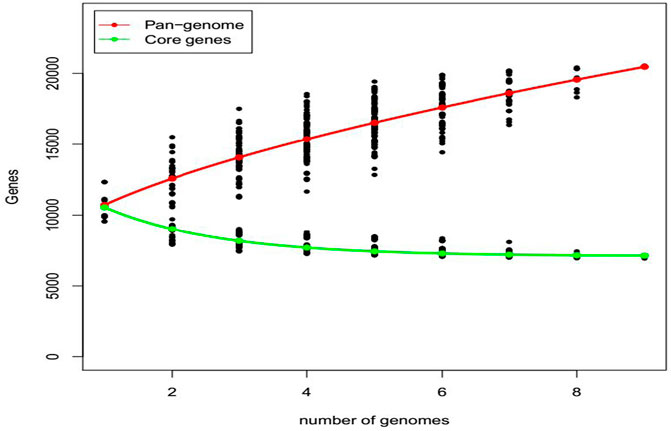
FIGURE 5. The pan and core genome analysis of eight fungal strains isolated from five different hosts. The red pan-genome curve represents the total number of gene families present in the fungal strains. While the core genome is presented by a green curve showing the conserved gene families.
Presence and Absence of AVR and MAT Genes in Banana Blast Fungus
So far, only 14 Avr genes have been reported from the blast fungus (Huang et al., 2014). In the present study, 14 Avr genes were compared for their presence and absence from the eight different strains sequenced (Supplementary Table S1). Only six Avr genes, namely AVR1-CO39, Avr-Pi54, MAT1-1, PWL1, ACE1, and AVR-Pi9 were reported from the banana blast fungus (Supplementary Table S1). Where, the Avr-Pi54, PWL1, ACE1, and AVR-Pi9 were found conserved among all the strains presented here (Supplementary Table S1). These results are in agreement with the previous reports that Avr genes are conserved and polymorphic in nature (Li et al., 2009). Interestingly, AVR1-CO39 gene was only present in non-rice host plants. However, Avr-Pi54, PWL1, ACE1, and AVR-Pi9 were present in all the strains. This data suggests that some gain of Avr genes (AVR1-CO39) occurred during the evolution of non-rice fungus. While some other genes were still found conserved among all the fungal strains under investigation. Furthermore, the presence of the MAT1 gene in banana blast fungus (Supplementary Table S2) suggest that sexual reproduction might potential also occur in the field conditions. Interestingly, in all these strains either the MAT1 or MAT2 gene was present, which supports the hypothesis that gain or loss of genes is possible due to sexual reproduction.
Discussion
The comparative fungal genome analysis is important to determine the genetic variation and host selection (Andersen et al., 2011; Xue et al., 2012). The natural genetic variation of rice blast fungus is well established. Rice blast fungus is a worldwide destructive fungal pathogen of many kinds of cereals (Talbot, 2003). Although its first genome sequence was published in 2005, subsequent studies have shown a great array of diversity. Due to the ever-increasing host range, it is important to make evolutionary comparisons based on complete genome sequencing. In this study, genomes of eight different strains of Magnaporthe species from five different hosts were sequenced and compared with 70-15 reference genomes. This is the first report on the genome resequencing of Magnaporthe species infecting bananas in the Peoples’ republic of China.
Our analysis confirmed the previous reports that Magnaporthe species infecting rice are closely related to each other. The sequencing of the ITS region indicated that blast fungus infecting banana is distinct from rice infecting species (Dean et al., 2005). The host pathogenicity test demonstrated that Magnaporthe isolates of rice are capable of infecting other cereals including millets and rye species. These results were in agreement with the previous findings that due to its capability to overcome the R genes resistance, the rice blast fungus poses a serious threat to global food security (Asibi et al., 2019).
Our pathogenicity test on nine banana varieties revealed that as a whole banana crop is susceptible to Magnaporthe isolates. There is a need to further test the banana germplasm for possible variation in resistance against blast fungus. The present study revealed that blast fungus infecting bananas has certain unique genome characteristics. The genome size of banana infecting blast fungus is relatively smaller than the other isolates of rice in this study and the 70-15 strain (∼38 Mb as compared to ∼41 Mb) (Dean et al., 2005; Xue et al., 2012). Similarly, the number of predicted genes in bananas infecting strain was ∼17% less than the reference strain 70-15 (∼10,112 for R07 compared to rice blast 12,440). In contrast, the R03 strain (isolated from Digitaria) had ∼11% more genes as compared to the 70-15 strain. Therefore, it can be concluded that several genes in the rice and non-rice fungus isolates may be different, which could have resulted in differential host selection. In the genomes of R03 and R07 strains, a higher number of InDels was observed, which suggests a possible reason for the host switching from rice to non-rice plants. Because sexual reproduction is not observed for Magnaporthe fungi, therefore it is plausible that the difference in genome sizes and a number of genes could be the result of strong selection pressure from infect different hosts. The frequent gain and loss of avirulence genes is an indication of adaptive evolution among fungal strains. The higher conservation of the ACE1 gene is plausible as it encodes a protein involved in the production of secondary metabolites (Fudal et al., 2005). Previous studies have revealed that Avr genes in M. oryzae might pass through several spontaneous deletions and insertions to increase their diversity and as a result, the avirulence may be lost or gained (Dai et al., 2010; Huang et al., 2014). The absence of AVR-Pita from banana blast fungus was surprising as, it is an important Avr gene and has previously been reported in non-rice fungal diseases (Chuma et al., 2011).
Materials and Methods
Source of Fungus, Morphology, and Hosts
The Magnaporthe fungus species namely, M. oryzae were isolated from four rice plants (designated as R01, R05, R06, and R08), finger millet (R02), Digitaria sanguinalis (R03), Eleusine indica (R04), and banana plants (R07) from the Yunnan province of China. The mycelium of fungal species was collected and the cultures were confirmed under the microscope (Nikon eclipse 80i, Nikon Corporation, Japan). The M. oryzae was inoculated on 11 different Graminae host species to check the differential response of each species. These 11 host species included, O. sativa, Setaria italic, Panicum miliaceum, Coix lacryma-jobi L., Avena chinensis, Hordeum vulgare L., Avena sativa L., Triticum aestivum L., Echinochloa crusgalli (L.) Beauv., Zea mays L,. and Sorghum bicolor (L.) Moench.
The pyricularia angulate (R07) symptoms severity was assessed on banana leaves. Briefly, the R07 strain was cultured on a PDA medium at 26°C (Peng and Shishiyama, 1988). The nine different varieties namely, Dwarf banana, Xing-3, 998, Baodao, NanTianHuan, Han11, Xinghong, 119, and Brazil were inoculated with R07 strain and the symptoms were evaluated and photographed after 12 days of inoculation. The disease reaction of different strains (R01 to R08) was recorded on 11 different hosts. The disease reaction was recorded as Susceptible (S) for angular necrotic spots appear on leaf surfaces, while resistance (R) was recorded for no discernable symptoms.
Sequencing of Genomes and Assembly
The fungal species M. Oryzae and Pyricularia angulate were grown on potato dextrose medium and total genomic DNA was isolated using the Omega Fungal DNA Kit according to the manufacturer’s protocol (Margulies et al., 2005). The fungal strains genome was sequenced according to the already established standard procedures of the paired-end library sequencing approach of Illumina GAII sequencing (Masella et al., 2012). The initial sequencing reads were trimmed and shorter reads (less than 20 nt) were removed. The filtered sequencing reads after Illumina sequencing were assembled using SOAPdenovo (V2.04) genome assembler (Luo et al., 2012). The multi-kmer integrated into SOAPdenovo was used to assemble the contigs. The GapCloser was used to reduce the size of gaps present in the scaffolds generated by SOAPdenovoe2.
Prediction of Genes and the Genome Annotation
The resulting genomes were annotated using the MAKER2 software (Holt and Yandell, 2011). The gene prediction was carried out using MAKER2 having integrated SNAP tool to predict the mRNA coding regions using prior information from NCBI data. The gene functions were predicted using the STRING database (STRING V11.0; https://string-db.org/). Briefly, the annotated genome of Magnaporthe and Pyricularia were added to the STRING database to know the proteins and their interaction with other proteins. Since, STRING contains protein information for thousands of genes, therefore adding a genome-wide dataset resulted in robust information on genes coding for different proteins in the newly sequenced genomes.
Genes Function Prediction Through Cluster of Ortholog Groups (COGs/KOGs)
For comprehensive gene annotation studies, we used three different methods, BLASTp, COGs/KOGs, and STRING database. Briefly, BLASTp was used to confirm the annotated genes’ functions already predicted through the STRING database. The COGs and KOGs were further screened to confirm the ortholog genes’ functions in the newly sequenced genomes.
Conclusion
Overall, the present study indicated that differences in genome sizes and the number of genes of various Magnaporthe species can be attributed to insertions/deletions in the chromosomes and the gain or loss of new gene families. These chromosomal variations might result in the selection of different hosts like bananas or Digitaria other than rice. It is the first comprehensive genome study of a fungal pathogen infecting banana which is an economically important crop on the global scale.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Conceptualization, JL, LL, ZG; methodology, JL, ZS; software, JL, QW; validation, JL, LL, QW; formal analysis, ZS, CL, ZG; investigation, QW, ZS; resources, JL, LL; data curation, ZS, QW; writing—original draft preparation, JL, CL; writing—review and editing, JL, ZS; visualiza-tion, QW, JL; supervision, ZS, CL; project administration, ZG, QW; funding acquisition, JL. All authors have read and approved final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31860481), the Key Research and Development Program of Yunnan Province (2019IB007 and 202102AE090003), the program of the Innovative Research Team of Yunnan Province (202005AE160003), the Key Laboratory of Green Prevention and Control of Agricultural Transboundary Pests of Yunnan Province (202105AG070001), Key Laboratory of Prevention and Control of Biological Invasions, Ministry of Agriculture and Rural Affairs, P.R. China (Kunming), Innovative Science and Technology Institute of Green Prevention and Control of Agricultural Pests in South & Southeast Asia, Ministry of Agriculture and Rural Affairs, P.R. China. The funding organizations had no role in designing, data collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.861727/full#supplementary-material
References
Andersen, M. R., Salazar, M. P., Schaap, P. J., van de Vondervoort, P. J. I., Culley, D., Thykaer, J., et al. (2011). Comparative Genomics of Citric-Acid-Producing Aspergillus niger ATCC 1015 versus Enzyme-Producing CBS 513.88. Genome Res. 21 (6), 885–897. doi:10.1101/gr.112169.110
Ashkani, S., Yusop, M. R., Shabanimofrad, M., Harun, A. R., Sahebi, M., and Latif, M. A. (2015). Genetic Analysis of Resistance to rice Blast: a Study on the Inheritance of Resistance to the Blast Disease Pathogen in an F3 Population of rice. J. Phytopathol 163 (4), 300–309. doi:10.1111/jph.12323
Asibi, A. E., Chai, Q., and Coulter, J. A. (2019). Rice Blast: A Disease with Implications for Global Food Security. Agronomy 9 (8), 451. doi:10.3390/agronomy9080451
Boddy, L. (2016). “Pathogens of Autotrophs,” in The Fungi (Elsevier), 245–292. doi:10.1016/b978-0-12-382034-1.00008-6
Chuma, I., Isobe, C., Hotta, Y., Ibaragi, K., Futamata, N., Kusaba, M., et al. (2011). Multiple Translocation of the AVR-Pita Effector Gene Among Chromosomes of the rice Blast Fungus Magnaporthe Oryzae and Related Species. Plos Pathog. 7 (7), e1002147. doi:10.1371/journal.ppat.1002147
Dai, Y., Jia, Y., Correll, J., Wang, X., and Wang, Y. (2010). Diversification and Evolution of the Avirulence Gene AVR-Pita1 in Field Isolates of Magnaporthe Oryzae. Fungal Genet. Biol. 47 (12), 973–980. doi:10.1016/j.fgb.2010.08.003
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The Genome Sequence of the rice Blast Fungus Magnaporthe Grisea. Nature 434 (7036), 980–986. doi:10.1038/nature03449
Fan, H.-W., Noda, H., Xie, H.-Q., Suetsugu, Y., Zhu, Q.-H., and Zhang, C.-X. (2015). Genomic Analysis of an Ascomycete Fungus from the rice Planthopper Reveals How it Adapts to an Endosymbiotic Lifestyle. Genome Biol. Evol. 7 (9), 2623–2634. doi:10.1093/gbe/evv169
Fudal, I., Böhnert, H. U., Tharreau, D., and Lebrun, M.-H. (2005). Transposition of MINE, a Composite Retrotransposon, in the Avirulence Gene ACE1 of the rice Blast Fungus Magnaporthe Grisea. Fungal Genet. Biol. 42 (9), 761–772. doi:10.1016/j.fgb.2005.05.001
Fudal, I., Ross, S., Brun, H., Besnard, A.-L., Ermel, M., Kuhn, M.-L., et al. (2009). Repeat-induced point Mutation (RIP) as an Alternative Mechanism of Evolution toward Virulence in Leptosphaeria Maculans. Mpmi 22 (8), 932–941. doi:10.1094/mpmi-22-8-0932
Holt, C., and Yandell, M. (2011). MAKER2: an Annotation Pipeline and Genome-Database Management Tool for Second-Generation Genome Projects. BMC bioinformatics 12 (1), 491–514. doi:10.1186/1471-2105-12-491
Huang, J., Si, W., Deng, Q., Li, P., and Yang, S. (2014). Rapid Evolution of Avirulence Genes in rice Blast Fungus Magnaporthe Oryzae. BMC Genet. 15 (1), 45. doi:10.1186/1471-2156-15-45
Kiyosawa, S. (1982). Genetics and Epidemiological Modeling of Breakdown of Plant Disease Resistance. Annu. Rev. Phytopathol. 20 (1), 93–117. doi:10.1146/annurev.py.20.090182.000521
Li, W., Wang, B., Wu, J., Lu, G., Hu, Y., Zhang, X., et al. (2009). The Magnaporthe Oryzae Avirulence Gene AvrPiz-T Encodes a Predicted Secreted Protein that Triggers the Immunity in rice Mediated by the Blast Resistance Gene Piz-T. Mpmi 22 (4), 411–420. doi:10.1094/mpmi-22-4-0411
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an Empirically Improved Memory-Efficient Short-Read De Novo Assembler. Gigascience 1 (1), 18–217X118. doi:10.1186/2047-217X-1-18
Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S., Bemben, L. A., et al. (2005). Genome Sequencing in Microfabricated High-Density Picolitre Reactors. Nature 437 (7057), 376–380. doi:10.1038/nature03959
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D. (2012). PANDAseq: Paired-End Assembler for Illumina Sequences. BMC bioinformatics 13 (1), 31–37. doi:10.1186/1471-2105-13-31
Musiime, O., Tenywa, M. M., and Majaliwa, M. (2005). “Constraints to rice Production in Bugiri District,” in African Crop Science Conference Proceedings.
Oh, Y., Donofrio, N., Pan, H., Coughlan, S., Brown, D. E., Meng, S., et al. (2008). Transcriptome Analysis Reveals New Insight into Appressorium Formation and Function in the rice Blast Fungus Magnaporthe Oryzae. Genome Biol. 9 (5), R85–R24. doi:10.1186/gb-2008-9-5-r85
Orbach, M. J., Farrall, L., Sweigard, J. A., Chumley, F. G., and Valent, B. (2000). A Telomeric Avirulence Gene Determines Efficacy for the rice Blast Resistance Gene Pi-Ta. Plant Cell 12 (11), 2019–2032. doi:10.1105/tpc.12.11.2019
Peng, Y.-L., and Shishiyama, J. (1988). Temporal Sequence of Cytological Events in rice Leaves Infected with Pyricularia Oryzae. Can. J. Bot. 66 (4), 730–735. doi:10.1139/b88-107
Sakulkoo, W., Osés-Ruiz, M., Oliveira Garcia, E., Soanes, D. M., Littlejohn, G. R., Hacker, C., et al. (2018). A Single Fungal MAP Kinase Controls Plant Cell-To-Cell Invasion by the rice Blast Fungus. Science 359 (6382), 1399–1403. doi:10.1126/science.aaq0892
Skamnioti, P., and Gurr, S. J. (2009). Against the Grain: Safeguarding rice from rice Blast Disease. Trends Biotechnology 27 (3), 141–150. doi:10.1016/j.tibtech.2008.12.002
Talbot, N. J. (2003). On the Trail of a Cereal Killer: Exploring the Biology of Magnaporthe Grisea. Annu. Rev. Microbiol. 57 (1), 177–202. doi:10.1146/annurev.micro.57.030502.090957
Van de Wouw, A. P., Cozijnsen, A. J., Hane, J. K., Brunner, P. C., McDonald, B. A., Oliver, R. P., et al. (2010). Evolution of Linked Avirulence Effectors in Leptosphaeria Maculans Is Affected by Genomic Environment and Exposure to Resistance Genes in Host Plants. Plos Pathog. 6 (11), e1001180. doi:10.1371/journal.ppat.1001180
Wilson, R. A., and Talbot, N. J. (2009). Under Pressure: Investigating the Biology of Plant Infection by Magnaporthe Oryzae. Nat. Rev. Microbiol. 7 (3), 185–195. doi:10.1038/nrmicro2032
Xu, J. R., Zhao, X., and Dean, R. A. (2007). From Genes to Genomes: a New Paradigm for Studying Fungal Pathogenesis in Magnaporthe Oryzae. Adv. Genet. 57, 175–218. doi:10.1016/s0065-2660(06)57005-1
Xue, M., Yang, J., Li, Z., Hu, S., Yao, N., Dean, R. A., et al. (2012). Comparative Analysis of the Genomes of Two Field Isolates of the rice Blast Fungus Magnaporthe Oryzae. Plos Genet. 8, e1002869. doi:10.1371/journal.pgen.1002869
Zeigler, R., Leong, S., and Teng, P. (1994). Rice Blast Disease. Manila, Philippines: International Rice Research Institute.
Keywords: rice blast, Magnaporthe oryzae, genome resequencing, indels, host-specific origin, avr genes
Citation: Li J, Lu L, Wang Q, Shi Z, Li C and Guo Z (2022) Genome Re-Sequencing Reveals the Host-Specific Origin of Genetic Variation in Magnaporthe Species. Front. Genet. 13:861727. doi: 10.3389/fgene.2022.861727
Received: 25 January 2022; Accepted: 11 April 2022;
Published: 16 May 2022.
Edited by:
Mohan Lal, North East Institute of Science and Technology (CSIR), IndiaReviewed by:
Kumar Paritosh, University of Delhi, IndiaNeelam Sheoran, Indian Agricultural Research Institute (ICAR), India
Copyright © 2022 Li, Lu, Wang, Shi, Li and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinbin Li, kmlijinbin@163.com
†These authors share first authorship
 Jinbin Li
Jinbin Li Lin Lu
Lin Lu Qun Wang
Qun Wang Zhufeng Shi1
Zhufeng Shi1 Chengyun Li
Chengyun Li