ClNAC84 interacts with ClMIP to regulate the cell cycle and reduce the size of Chrysanthemum lavandulifolium organs
- 1State Key Laboratory of Crop Genetics and Germplasm Enhancement, Key Laboratory of Landscaping, Ministry of Agriculture and Rural Affairs. Key Laboratory of Biology of Ornamental Plants in East China, National Forestry and Grassland Administration, College of Horticulture, Nanjing Agricultural University, Nanjing, China
- 2College of Agricultural, Yanbian University, Yanji, Jilin, China
The NAC transcription factor is plant-specific proteins and one of the largest families of transcription factors in plants. NAC proteins are involved in various aspects of plant growth and development, but little is known about how NAC proteins regulate the cell cycle. Here, we characterized ClNAC84 from C. lavandulifolium (an NAC transcription factor). ClNAC84 overexpression in C. lavandulifolium resulted in a semi-dwarf phenotype with shorter plant height, smaller leaf size, and smaller flower size than wild-type plants. The number of cells in the S phase during the cell cycle was less in ClNAC84-overexpression transgenic C. lavandulifolium than in wild-type C. lavandulifolium. This indicates that ClNAC84 overexpression can induce cell cycle arrest at the S and G2 phases. To elucidate the ClNAC84 regulatory network, ClMIP protein was shown to interact with ClNAC84 in vitro and in vivo. ClMIP overexpression in C. lavandulifolium also resulted in dwarfism and decreased cell numbers, and the expression level of ClKRP5 was higher in transgenic C. lavandulifolium than in wild-type plants. We also found that ClMIP can bind to the promoter of ClKRP5. Our data indicate that the interaction between ClNAC84 and ClMIP may promote ClKRP5 expression and inhibit S and G2 phases of the cell cycling.
Introduction
Cell growth and division are essential processes in plant growth and development (Grandjean, 2004). The cell cycle includes the G1, S, and G2 phases which are affected by multiple genes including some cell division related genes (PBR-binding protein E2F, KIP-related proteins KRPs, cyclins CYCs and cyclin-dependent kinases CDKs) and some transcription factors such as NAC and v-myb avian myeloblastosis viral oncogene homolog (MYB), with the transcription factor NAC playing an important role. NAC (NAM, ATAF1/2, and CUC2) proteins are plant-specific transcription factors. The NAC genes are NO APICAL MERISTEM (NAM) and CUP-SHAPED-COTYLEDON 1 (CUC1), CUC2, and CUC3 in Petunia and Arabidopsis, respectively, all of which regulate the shoot apical meristem (Souer et al., 1996; Aida et al., 1997). Rice, Arabidopsis (Ooka et al., 2003), and Chrysanthemum (Huang et al., 2012) comprise 161, 138, and 44 NAC members, respectively, which are further classified into 6–18 subfamilies and are involved in regulating various biological processes involved in plant growth and development. A typical NAC transcription factor has a highly conserved NAC domain and a transcriptional regulatory (TR) region in their N-terminal and C-terminal regions, respectively. The NAC domain is further divided into five subdomains (A-E). Subdomain A is involved in the formation of a functional dimer, subdomains B and E make up the functional diversity of the NAC protein, and the highly conserved subdomains C and D can bind to DNA (Puranik et al., 2012). In addition, in rice, NAC proteins are divided into 15 types (A-O) according to the motifs: A-E contain NAC domain and transcriptional regulatory regions as the typical motifs, and the other types include NAC-like proteins as the distinct motifs (Fang et al., 2008).
The phenotype of a double mutant namcuc showed a fusion of cotyledons, flowers, floral organs, and ovules that formed a “cup” in Arabidopsis (Aida et al., 1997) through the restraining behaviors of boundary cells that block the fusion of organs (Aida and Tasaka, 2006). PIN-FORMED1 (PIN1) encoding the auxin efflux carrier was regulated by MONOPTEROS (MP) genes, and the corresponding mutants showed cotyledon fusion or defects in apical patterning and perturbations in embryo development, a process controlled by CUC genes (Aida et al., 2002), implying that CUC genes could respond to auxin signals. In addition, CUC can affect leaf morphology. In Arabidopsis, TCP (Teosinte branched1/cincinnata/proliferating cell factor) protein can interfere with the function of miR164 and CUC proteins, preventing the formation of serrated leaves at the seeding stage. In the aging pathway of plants, the accumulation of SPL mRNA is gradual. SPLs bind to TCP protein and reduce the formation of complex TCP and CUC. This instability causes activation of CUC protein complexes, increasing the complexity of leaf morphology (Rubio-Somoza et al., 2014). A similar phenomenon was also reported in tomatoes; atypical type II phosphatidylinositol 4-kinase (PI4KII) interacts with ANAC078 to suppress auxin synthesis and inhibit the formation of serrated leaves (Tang et al., 2016). NAC proteins not only regulate the development of cotyledons and leaf organs but also regulate plant height. The leaf size and plant height of ANAC036-overexpression transgenic plants were smaller than wild-type (WT) plants with a semi-dwarf phenotype. ANAC036 overexpression in rice resulted in smaller cell sizes in transgenic plant leaves than in WT plant leaves (Kato et al., 2010). XYLEM NAC DOMAIN1 (XND1) of NAC domain transcription factor members is mainly expressed in the plant xylem, and in plants overexpressing XND1, it reduced the number of tracheary elements compared to that in WT plants. Transmission electron microscopy showed that the secondary wall of parenchyma cells in transgenic plants did not develop well in the hypocotyls (Zhao et al., 2008). NTM, a membrane-bound NAC transcription factor, participates in the regulation of intramembrane proteolysis and mediates cytokine signaling during cell division. The Arabidopsis mutant ntm shows serrated leaves and dwarfism in plants with a significant reduction in the expression levels of histone H4 and CDK and an increase in the expression levels of CDK inhibitor genes (KIP-related proteins) (Kim et al., 2006). Willemsen et al. (2008) reported that the transcription factors FEZ and SMB (Sombrero) containing the NAC domain also regulate the orientation and timing of cell division in Arabidopsis stem cells. These data show that members of the NAC family are involved in cell cycle regulation in plants. The correlation between the changes in the G1, S, and G2 phases and NAC proteins, however, remains elusive.
Tetraploid C. lavandulifolium (Fisch. ex Trautv.) flowers are larger than diploid flowers because of enhanced cell numbers. A previous study in which the NAC fragment was screened by cDNA-AFLP and ClNAC84 was cloned showed that these may be involved in cell cycle regulation (Gao et al., 2016). In this study, we report that the ClNAC84 transcription factor gene from C. lavandulifolium plays a role in the cell cycle and the ClMIP protein potentially interacts with ClNAN84, which activates ClKRP5 by binding to its promoter resulting in inhibition of the cell cycle S and G2 phases.
Materials and methods
Plant materials and growth conditions
C. lavandulifolium for ClNAC84 cloning and expression analysis and transgenic lines were maintained by the Chrysanthemum Germplasm Resource Preserving Center, Nanjing Agricultural University, Nanjing, China. The plants were potted in a 1:1 (v/v) mixture of soil and vermiculite and grown in a greenhouse at day/night temperatures of 24°C/18°C and an 8/16 h light/dark photoperiod with a relative humidity of 70%.
Isolation of ClNAC84
Total RNA was extracted from the stem apex of C. lavandulifolium seedlings using RNAiso reagent (TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions. Based on C. nankingense expressed sequence tags (Wang et al., 2013), ClNAC84-fF/fR primers (Table S1) were designed to amplify full-length ClNAC84 by rapid amplification of cDNA ends (RACE) PCR. The resulting PCR product was purified and cloned into T-Vector pMD19 (Simple) (TaKaRa) for sequencing. The ClNAC84 amino acid sequence was aligned with those of other plant homologs using BLAST (http://www.ncbi.nlm.gov/blast) and DNAMAN 5.2.2. Phylogenetic trees were constructed using the neighbor-joining method and MEGA (version 5.0). Internal branching support was estimated using 1000 bootstrap replicates.
Subcellular localization
The ClNAC84 ORF containing BamHI and NotI sites and the pENTR™1A dual selection vector (Invitrogen) were digested (BamHI and NotI), ligated to generate a set of pENTR™1A-ClNAC84 fusions, and then subcloned into the pMDC43 vector with GFP for transient expression in onion epidermal cells (ClNAC84-pF/pR primer sequence in Table S1). After incubation for 20 h at 25°C in the dark, GFP expression was observed in onion epidermal cells via confocal laser microscopy (LeicaSP2, Germany).
Yeast transformation, growth, and observation
The coding region of ClNAC84 was inserted into the expression vector pESPM with T4 DNA ligase after double enzyme restriction (XhoІ and SalІ). The pESPM-ClNAC84 plasmid was transformed into the fission yeast strain SPQ-01(Leu-) according to the manufacturer’s transformation system 2 (TaKaRa). Cells were selected on Edinburgh Minimal Medium (EMM) with thiamine at 30°C. Yeast containing the pESPM empty vector was used as a control. The yeast was cultured in liquid EMM medium to the logarithmic phase stage, washed three times with EMM liquid medium to remove the thiamine for de-repressing the NMT1 promoter, which drives ClNAC84 expression; and cultured at 30°C for 20 h. Total RNA was isolated from pESPM-NAC84 overexpressing yeast strains selected on EMM medium with and without thiamine, using the yeast RNAiso Kit (TaKaRa). First-strand cDNA was synthesized from 1 μg of total RNA using M-MLV reverse transcriptase (TaKaRa). Gene-specific primers ClNAC84-oF/R were designed to amplify the transcripts, and tubulin was used as the reference gene (Table S1). PCR amplification was initially denatured (94°C for 4 min), followed by 29 cycles at 95°C for 30 s, 57°C for 30 s, and 72°C for 45 s. The transformed mid-exponential yeast cells (OD600 = 0.5) at 1-, 10-, 100-, and 1000-fold dilutions were inoculated on EMM media plates with or without thiamine, followed by incubation at 30°C for 60 h. To observe the cell density and shape, the yeast cells were fixed with 70% ethanol alcohol for 30 min, and to examine the DNA, the cells were stained with DAPI (1 mg/ml) and observed under a fluorescence microscope (Olympus Optical Co., Ltd).
Morphological investigation of transgenic C. lavandulifolium
To further investigate the functions of ClNAC84 and ClMIP, pMDC43-ClNAC84 and pMDC43-ClMIP were constructed and transformed into Agrobacterium tumefaciens strain EHA105 by the freeze-thaw method. Genetic transformation of C. lavandulifolium was performed according to the methods in a previous study (Gao et al., 2018). Each line was repeated 10 times, and the data are presented as the mean ± standard error. SPSS v17.0, and Microsoft Excel 2016 were used for the statistical analysis (p < 0.05).
Transcriptional activity analysis
ClNAC84 transcriptional activity assays were performed using the Match maker GAL4 One-Hybrid System (Clontech, Mountain View, CA, USA). The pGBKT7-ClNAC84 vector was constructed and introduced into yeast strain Y2HGold (Clontech) following the manufacturer’s protocol, and the empty pCL1 and pGBKT7 vectors were transformed into the yeast strain as positive and negative controls, respectively. pGBKT7-ClNAC84 and pGBKT7 were cultured on SD medium without tryptophan (SD/-Trp), while the pCL1 transformants were incubated on SD medium without leucine (SD/-Leu). After culturing at 30°C for 3 d, the transgenic cell lines were transferred onto SD medium lacking both histidine and adenine (hemisulfate salt) either in the presence or absence of β-galactosidase.
Y2H assay
Protein–protein interaction Y2H assays was performed using a GAL4-based Y2H system (Matchmaker Gold Systems; Clontech, Palo Alto, CA, USA). The full-length coding sequence of the candidate gene ClMIP was inserted into pGBKT7 (Clontech, Mountain View, CA, USA). The ClMIP-gF/gR primers were used to create the constructs and are listed in Table S1. The bait and prey plasmids were co-transformed into the yeast strain Y2HGold (Clontech, Palo Alto, CA, USA). Yeast transformation was performed according to the manufacturer’s instructions. The co-transformants were incubated in SD medium lacking Leu, Trp, His, and Ade (SD/-Leu/-Trp/-His/-Ade/+X-α-Gal) plates, after culturing at 30°C for 3 days to obtain a graph.
Bimolecular fluorescence complementation assay
The BiFC assay was used to study the interaction between ClNAC84 and ClMIP. To construct pSAT4A-ClNAC84-nEYFP-N1 and pSAT4A-ClMIP-cEYFP-N1 vectors, ClNAC84 and ClMIP ORF were amplified using ClNAC84-bF/R and ClMIP-bF/bR primers (Table S1); recombined with the N- and C-termini of YFP, respectively; and subsequently introduced into the pSAT4A vector. The two plasmids were mixed with gold particles and transformed as described above for transient expression analysis. Confocal laser microscopy was used to monitor the expression of YFP.
ClKRP5 promoter cloning and vector construction
Genomic DNA was isolated from C. lavandulifolium using the Plant Genomic DNA Rapid Extraction Kit (TaKaRa, Beijing, China). According to the ClKRP5 full-length cDNA, ClKRP5-fF/fR primers were designed to amplify intron-containing regions. The promoter region of ClKRP5 was amplified from the genomic DNA, purified, and cloned into the T-Vector pMD19 (Simple) (TaKaRa) for sequencing using the improved TAIL-PCR method (Li et al., 2015). Briefly, based on the genomic sequence of ClKRP5, three gene-specific primers (GSP1, GSP2, and GSP3) were designed, and the other four primers AP1, AP2, AP3, and AP4 were from the TaKaRa kit (Table S1). PCR conditions and procedures were performed according to the manufacturer’s instructions. The final PCR-amplified products (>1000 bp) were purified and cloned into the T-Vector pMD19 (Simple) (TaKaRa) for sequencing.
The ClKRP5 promoter (1479 bp) was amplified using LA Taq (TaKaRa) with the primer pair KRP5-pF/R containing BamH І and Hind Ш sites (Table S1). The PCR products were subcloned into the pMD19-T vector and confirmed by sequencing. pMD19-T harboring ClKRP5 and pCAMBIA1381Z were digested with BamH І and Hind Ш, respectively. The purified products were ligated with T4 DNA ligase to obtain the vector pCAMBIA1381Z-KRPpro.
DNA-binding assay of CmKRP5 using a yeast one-hybrid system
To examine whether ClMIP has DNA-binding ability to the ClKRP5 promoter, a yeast one-hybrid assay was performed using a Clontech system (Clontech, Mountain View, CA, USA). The ClKRP5 promoter sequence was constructed into a pHIS expression vector, and both pGADT7-ClMIP and pHIS-ClKRP5pro were transfected into the yeast strain Y1H (Clontech). The empty vectors pGADT7 and pHIS-ClKRP5pro were used as negative controls. The growth status of the transformed yeast cells was tested on SD/-His-Ura-Leu medium containing 50 mM 3-AT. The culture conditions were the same as those for the Y2H assay.
Split-LUC assay
The constructs were transformed into Agrobacterium strain 1301 with 35S::p19 and used to co-infiltrate 3-week-old tobacco leaves as follows: pCAMBIA1381Z-ClKRP5pro, pMDC43-ClNAC84, pCAMBIA1381Z-ClKRP5pro + pMDC43-ClNAC84, pCAMBIA1381Z-ClKRP5pro + pMDC43-ClMIP, and pCAMBIA1381Z-ClKRP5pro + pMDC43-ClMIP + pMDC43-ClNAC84. The infiltrated tobacco plants were grown for an additional 3 days in a growth chamber under a 16/8 h light/dark photoperiod at 21°C. LUC images were captured using a low-light cooled CCD imaging apparatus (DU934P Andor, UK). LUC activity was measured with 10 s integration periods (Promega, Madison, Wisconsin, USA). Counts of luminescence were quantified using a 20/20n luminometer (Turner BioSystems). Three independent experiments were performed for each assay.
Flow cytometric analysis
The ClNAC84 or ClMIP overexpression and WT C. lavandulifolium leaves were cultured on MS medium containing 6-benzylaminopurine 1.5 mg L-1 and naphthaleneacetic acid 0.8 mg L-1 for callus induction. Cells from the callus culture were resuspended in the above medium. This synchronous cell culture method was based on that of Menges (Menges and Murray, 2002). The synchronous cells were filtered with nylon mesh (pore size 300 mesh) and collected in microcentrifuge tubes (100 mmol L-1 citric acid monohydrate; 5 μg L-1 RNase; 0.5% (v) Tween20; pH 2.0–3.0) and stained with propidium iodide. The samples were then incubated at 4°C in an icebox for 30 min after flow cytometry analysis (Coulter Epics XL, Beckman Coulter, Miami, USA). The percentage of cells in different cell cycle phases was evaluated using ModfidLT4.1 program. The experiments were repeated at least ten times using independent apical bud samples. The data were evaluated by Student’s t-test (one-tailed distribution, two-sample equal variance) using SPSS v16.0 (SPSS, Inc., Chicago, IL, USA). The data were considered significant at p < 0.05.
qRT-PCR analysis
Total RNA was isolated from the different tissues of C. lavandulifolium various developmental periods by using RNAiso (TaKaRa) in accordance with the manufacturer’s instructions. cDNA was synthesized from 1000 ng of total RNA by using a reverse transcriptase FastQuant RT kit (Tiangeng). Gene-specific primers were designed for the 3′UTR sequences, and elongation factor 1a (ClEF1α) was used as the reference sequence (Table S1). Three replicates of qRT-PCR data were processed using the comparative CT method (Gao et al., 2018).
Results
Characterization of ClNAC84
The ClNAC84 sequence isolated from the cDNA of C. lavandulifolium contains an open reading frame (ORF) of 690 bp and encodes a protein of 229 amino acids. A BLASTP search showed that the deduced polypeptide is similar to NAC proteins from a range of plant species and contains a highly conserved NAC domain (a, b, c, d, and e) in the N-terminus and a transcriptional regulation domain (f) in the C terminus (Figure 1A). To investigate the potential role of ClNAC84 in regulating plant development, we evaluated its expression in different organs, including apical buds, flower buds, leaves, stems, and roots at different developmental stages. ClNAC84 transcripts were detected in all the investigated organs. Relative transcript abundance in different organs gradually decreased from young plants to the full flowering stage, but transcript abundance in flower buds increased at visible capitulum stages and then declined at the full flowering stage. Transcript abundance in apical buds and leaves of young plants was significantly higher than that in other organs at various stages of development (Figure 1B). These results suggest that during plant development, ClNAC84 was expressed at high levels in the apical buds and flower buds.
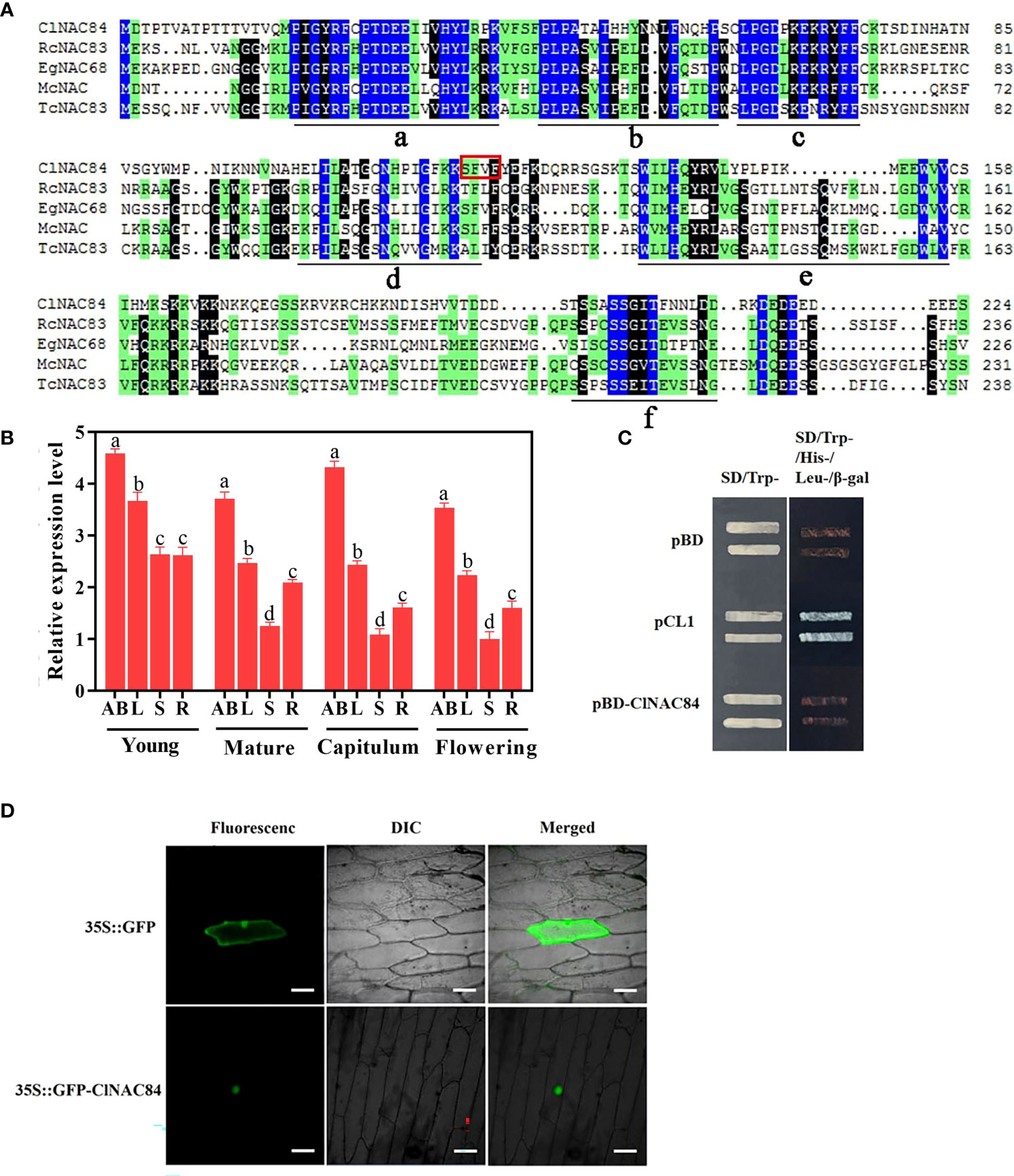
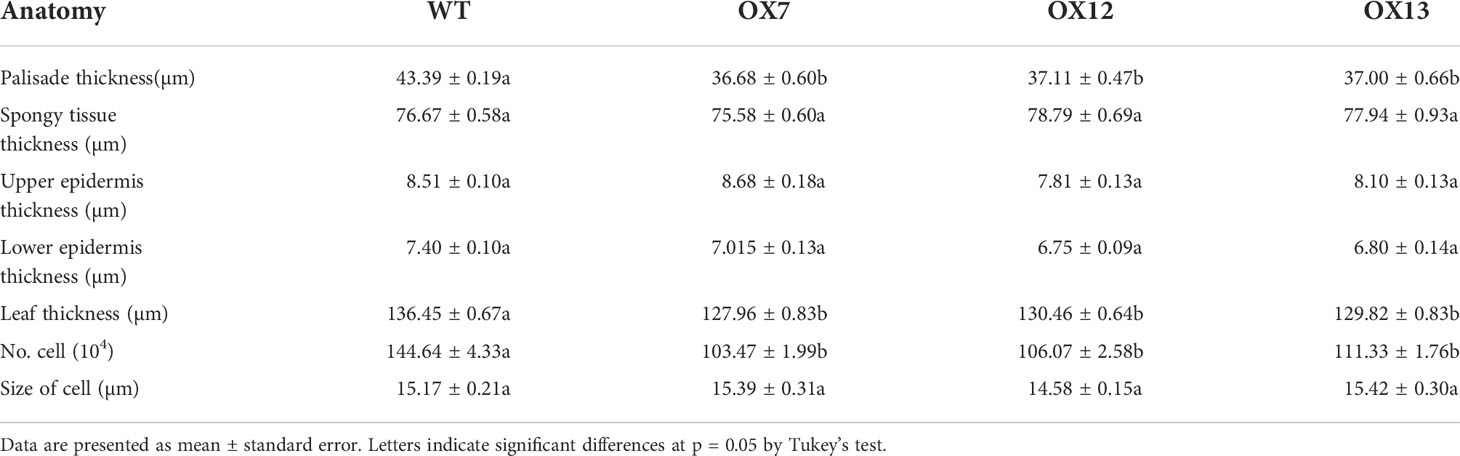
Figure 1 Characterization of ClNAC84. (A) The deduced amino acid sequence of ClNAC84 and other genes harboring a NAC domain. The NAC domain (a, b, c, d, e) and transcriptional regulation domain (f) are shown underlined. The GenBank accession numbers of the referred to genes are as follows: RcNAC83 (XP_002512886.1), EgNAC68 (XP_012844037.1), McNAC (AKO82482.1), TcNAC83 (XP_007031956.2). (B) ClNAC84 expression in different tissues at various developmental periods: young, young plant (height 20 cm); mature, mature plant before flower differentiation; capitulum, visible capitulum; flowering, full flowering; AB, apical buds; L, third leaves; S, stems of between third leaves and second leaves; R, roots; FB, flower buds. Uppercase letter represents significant differences in the analyses of apical (flower) buds at various developmental stages; lowercase letter indicates between the leaves. Error bars indicate SE (n = 3). Significant differences were analyzed by Tukey multiple range test (p < 0.05). (C) Transcriptional activation in yeast. pCL1 and pBD plasmids (pGBKT7) represent, respectively, the positive and negative controls. Left: SD medium lacking tryptophan; right: SD medium lacking tryptophan, histidine, and leucine and supplemented with β-Gal. (D) Subcellular localization of ClNAC84 transiently expressed by the 35S::GFP-ClNAC84 transgene in onion epidermal cells. Left: dark field image, center: bright field image, right: merged image. 35S::GFP transgene was used as a control. Bar: 50 μm.
The green fluorescent protein (GFP)-ClNAC84 fusion construct under the control of the 35S promoter was used to investigate the subcellular localization of ClNAC84. In onion epidermal cells, green fluorescence of 35S::GFP-ClNAC84 markedly accumulated in the nucleus, while the product of the control 35S::GFP transgene was deposited throughout the onion epidermal cells (Figure 1D). Next, we performed a transcription activation assay using yeast. The harboring the complete GAL4 domain (pCL1) and pGBK7 vector were used as positive and negative controls, respectively. Yeasts harboring ClNAC84 were not able to activate the reporter genes His, Leu, and X-α-Gal (Figure 1C). The results showed that ClNCA84 had no transcriptional activation activity in yeast cells.
ClNAC84 overexpression suppressed C. lavandulifolium growth by inhibition of cell cycle
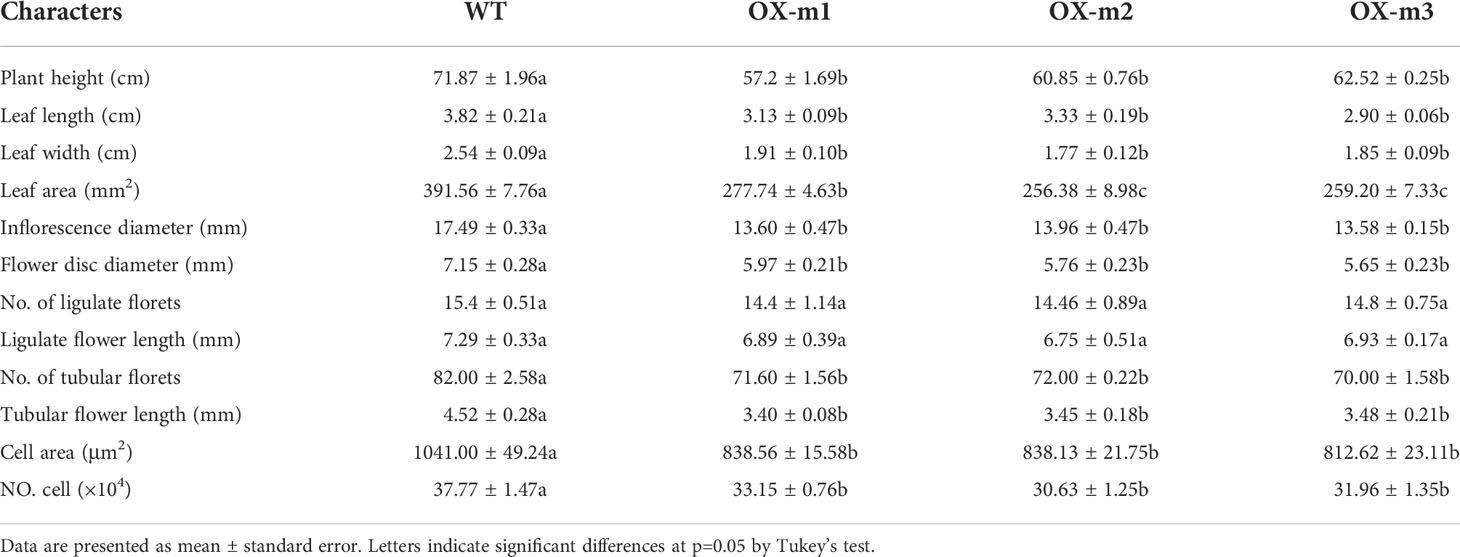
To investigate the function of ClNAC84, ClNAC84 was overexpressed in C. lavandulifolium (OX), and six transgenic lines were obtained; the presence of the gene was confirmed by PCR. The OX-7, OX-12, and OX-13 lines, which had the highest levels of ClNAC84 expression, were selected for further investigation (Figure S1). Morphological changes, such as changes in plant height, leaf length, leaf width, leaf area, inflorescence diameter, flower disk diameter, ligulate flower length, number of ligulate florets, and tubular flower length, were observed in each transgenic plant. Only the ligulate flower number showed no obvious differences compared with that in the WT plants (Figure 2A (a–j), Table 1).
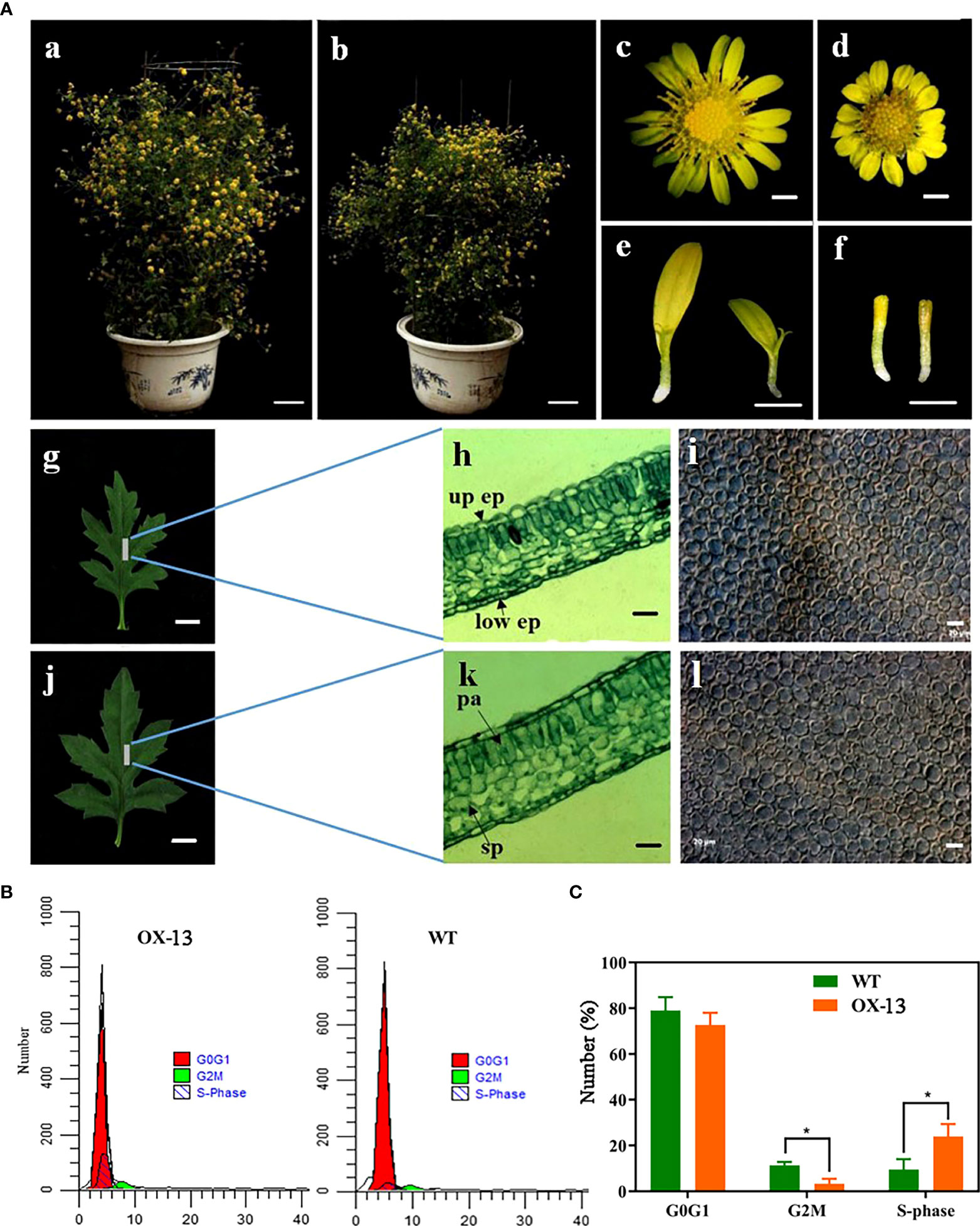
Figure 2 ClNAC84 overexpression suppressed C. lavandulifolium growth by inhibition of cell cycle. (A) Phenotypic characterization of ClNAC84 overexpression C. lavandulifolium. (a, c, g, h, and i) Wild-type C. lavandulifolium, flower, leaf, anatomical structure of leaf, and cells of palisade tissue first layer. (b, d, j, k, and l) ClNAC84-overexpressing plant (OX-13), flower, leaf, anatomical structure of leaf, and cells of palisade tissue first layer. (e) ligulate florets of wild-type plant (left) and ClNAC84-overexpressing plant (right). (f) tubular florets of wild-type plant (left) and ClNAC84-overexpressing plant (right). (h and k) up ep, upper epidermis; low ep, lower epidermis; sp, spongy tissue; pa, palisade tissue. (a and b) bars = 5 cm; (c, d, e, and f) bars = 2 mm; (g and j) bars = 5 mm; (h, k, i, and k) bars = 20 µm. (B) Comparison of the flow cytometric profiles of wild-type and ClNAC84 overexpression C. lavandulifolium. Red peaks indicate normal G0G1 cell number, green peaks indicate G2M cell number, and black peaks indicate S phase cell number. (C) The cell number at different phases of the cell cycle. The percentages were derived from (B).
Because the leaves of overexpression lines were thinner and smaller than those of WT plants, we observed the anatomical structure of the leaves and found that the palisade tissue thickness decreased by 14.89%, while the upper and lower epidermis thickness and spongy tissue thickness showed little change (Figure 2A (h–k), Table 2). To further understand the effect of ClNAC84 overexpression on C. lavandulifolium leaf development, epidermal cells of the leaves of overexpression lines and WT C. lavandulifolium were inspected using differential interference contrast microscopy. The number of palisade cells was significantly changed in the leaves of transgenic plants but decreased by 26.1% in overexpression lines; the cell size showed no change. These data indicated that the leaves of overexpression lines became smaller through the retardation of cell division (Figure 2A (i–l), Table 2).
To further reveal the potential role of ClNAC84 in cell division, we investigated the function of ClNAC84 in the yeast system. The NMT promoter in the pESPM vector drives gene expression in the absence of thiamine (-VB1); it also inhibits gene expression in the presence of thiamine (+VB1) (Mu et al., 2009). The transformants harboring ClNAC84 showed higher expression levels under -VB1 than under +VB1 (Figure S2A). Under inductive conditions (-VB1), the proliferation of transformants harboring ClNAC84 was inhibited in comparison with that of transformants harboring the control vector. Under surpressive conditions (+VB1), there was no difference in proliferation between transformants harboring ClNAC84 and those harboring the control vector (Figure S2B). The yeast transformants in the liquid medium under -VB1 were stained with 4,6-diamidino-2-phenylindole (DAPI) and observed under a fluorescence microscope. The cell morphology showed no difference between yeast cells containing ClNAC84 and those containing the pESPM vector. However, the percentage of yeast cells containing double nuclei was 5.2 times more in ClNAC84 cells than in pESPM cells (Figure S2C). In the yeast cell growth curve analysis, cell density (OD = 600) of the yeast cells containing ClNAC84 was consistently lower than that of yeast cells containing the pESPM vector in the -VB1 liquid medium after 4 h of culture; significant differences were noted in cell density 6 h later (Figure S2D). These results indicate that ClNAC84 decreases the proliferation of yeast cells and promotes the occurrence of double nuclei.
To further determine whether the phase of the cell cycle was blocked in C. lavandulifolium overexpressing ClNAC84, we collected synchronous cells of OX-13 lines and WT plants for cell cycle analysis. The number of S stage cells in ClNAC84 overexpression lines was more than twice that of the WT plants, but the number of G2M stage cells from WT plants was 3.28% lower than that of overexpression lines (Figures 2B, C). These results indicate inhibition of the cell cycle.
ClNAC84 interacts with ClMIP
To elucidate the mechanism of ClNAC84 in cell division, we identified cell division-related proteins including ClMIP, ClH2B1, and ClMafB transcription factors by screening the yeast library (Table S2). ClMIP is known as ClAE or FAM96 (family with sequence similarity 96, member B). To assess whether ClNAC84 and ClMIP proteins interact, we performed yeast two-hybrid (Y2H) assays. ClNAC84 grew well and appeared as a blue spot when co-transformed with ClMIP, and the positive control exhibited X-α-Gal activity, but the negative control did not (Figure 3A). Bimolecular fluorescence complementation (BiFC) experiments showed that a yellow fluorescent protein (YFP) signal was observed in onion epidermis cells co-expressing nEYFP-ClNAC84 and cEYFP-ClMIP. In contrast, there was no YFP signal in cells co-expressing nEYFP-ClNAC84 and cEYFP or nEYFP and in cEYFP-ClMIP as controls (Figure 3B). These results demonstrate that ClNCA84 interacts with ClMIP in plant cells.
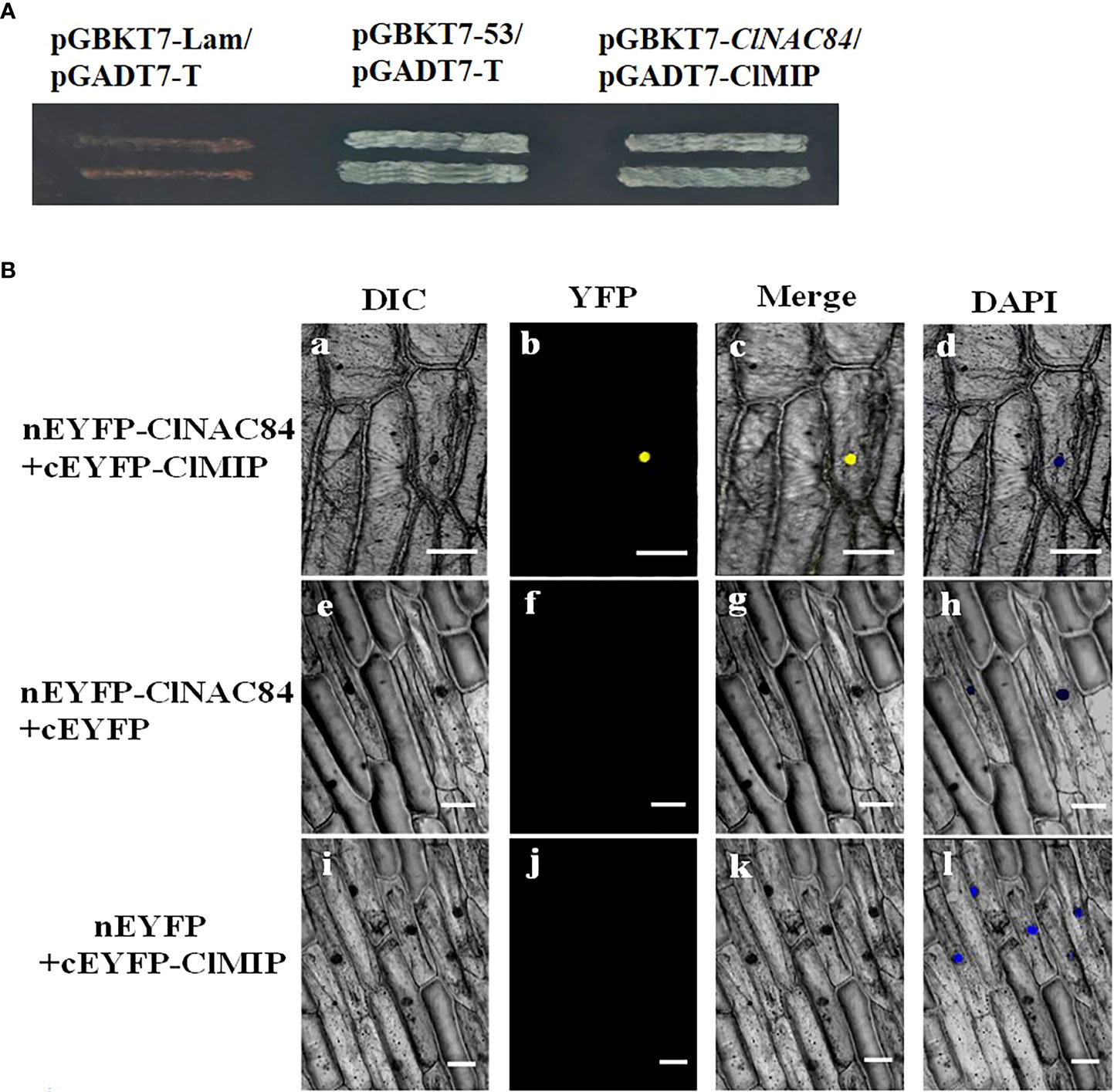
Figure 3 Interaction between ClNAC84 and ClMIP. (A) Interaction between ClNAC84 and ClMIP in vitro as demonstrated by yeast two-hybrid (Y2H) assay. pGBKT7-Lan/pGADT7-T and pGBKT7-53/pGADT7-T were used as negative and positive control, respectively. Selection of yeast colonies culture on SD medium lacking tryptophan, histidine, leucine, and adenine and containing X-α-Gal. (B) BiFC assay of ClNAC84 and ClMIP interaction in transiently transformed onion epidermal cells. Differential interference contrast (DIC), YFP fluorescence, merged images are shown for each transgenic combination and 4,6-diamidino-2-phenylindole staining (DAPI). The construct pairs of [ClNAC84-nEYFP and cEYPF] and [ClMIP-cEYFP and nEYFP] were used as negative controls. Bars = 100 mm.
Involvement of ClMIP in the regulation of cell cycle
To further clarify the function of ClMIP, the 35S::ClMIP vector was transformed into C. lavandulifolium, and three transgenic lines, OX-m1, OX-m2, and OX-m3, were obtained after qRT-PCR identification (Figure S3). Differences in phenotypes were observed between transgenic and WT plants. The plant height, leaf length and width, leaf area, inflorescence, flower disk diameter, tubular flower number, and tubular flower length of transgenic plants were less than those of WT plants (Figure 4A a–e, Table 3). Epidermal cell area and number decreased by 15.51% and 20.29%, respectively, in the transgenic lines (Figure 4A, f, g, Table 3). We investigated the number of G0G1, S, and G2M phase cells of WT and OX-m3 by flow cytometry. The number of S phase cells in OX-m3 was 33.29% higher than that in WT plants, and the number of G0G1 and G2M phase cells was higher in WT plants than in OX-m3 (Figures 4B, C).
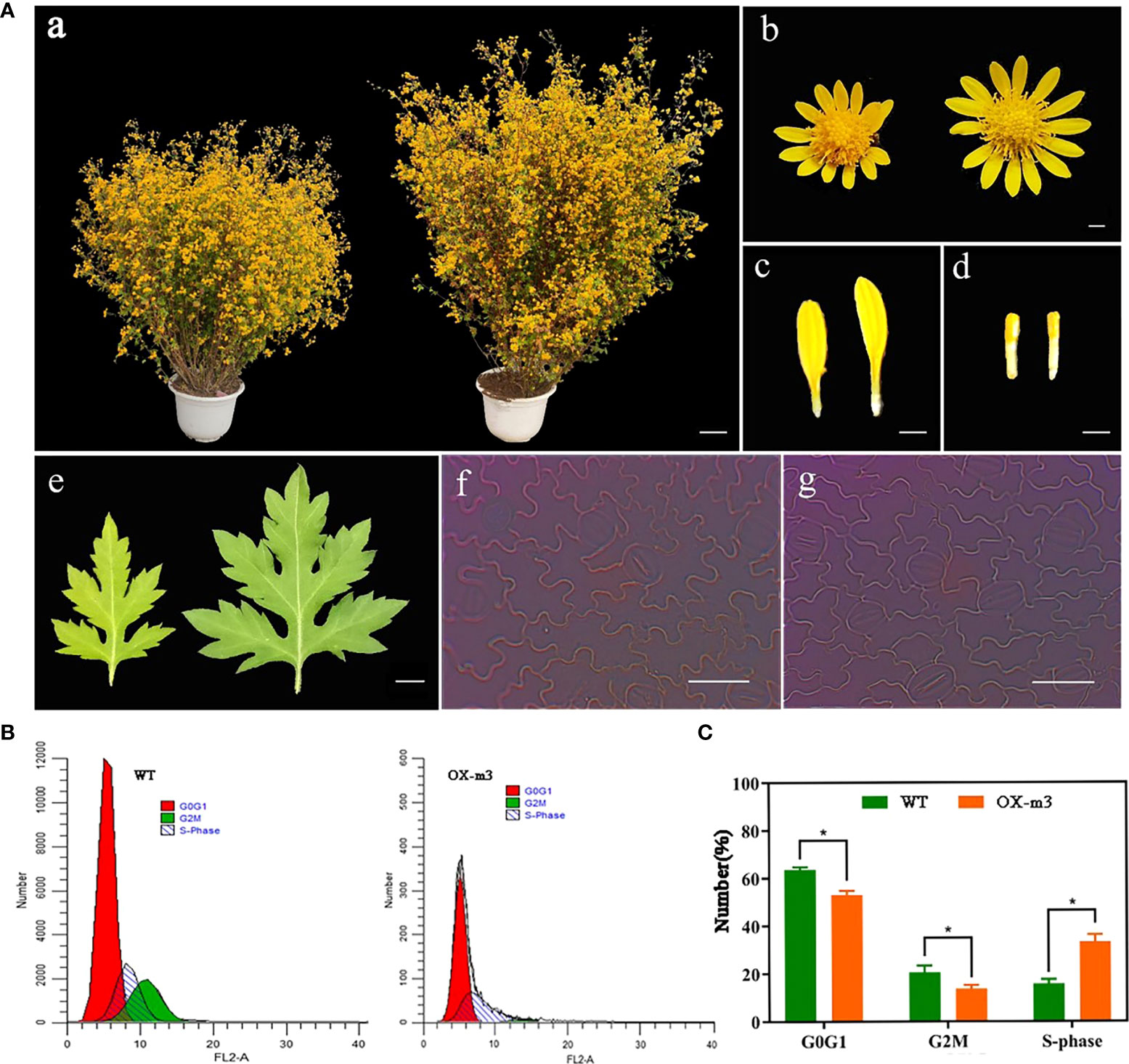
Figure 4 Involvement of ClMIP in the regulation of cell cycle. (A) Phenotypic characterization of ClMIP overexpression C. lavandulifolium. (a, b, c, d, and e) Plants, flowers, ligulate florets, tuber florets, and leaves of wild-type plants on the left and the overexpression line (OX-m3) on the right. a: bar = 5 cm; b, c, and d: bars = 0.2 cm; e: bar = 0.5 cm. (f and g) First layer epidermal cells of wild-type plants (f) and the overexpression line (g); bars = 50 μm. (B) Comparison of the flow cytometric profiles of wild-type and ClMIP overexpression C. lavandulifolium. Red peaks indicate normal G0-G1 cell number, green peaks indicate G2-M cell number, and black peaks indicate S phase cell number. (C) The cell number at different phases of the cell cycle. The percentages were derived from (B).
Altered expression of ClKRPs in ClMIP overexpression and ClNAC84 overexpression lines
The relative expression of ClKRPs was analyzed in ClMIP and ClNAC84 overexpression lines by qRT-PCR. The relative expression of ClKRP5 and ClKRP8 in the ClMIP overexpression lines was higher than that in WT C. lavandulifolium. Similarly, the relative expression of ClKRP3, ClKRP4, and ClKRP5 was also high in ClNAC84 overexpression lines. Therefore, the relative expression of ClKRP5 in both ClMIP overexpression and ClNAC84 overexpression lines was higher than that in WT (Figures 5A, B). This suggests that ClMIP may regulate the cell cycle through ClKRP5.
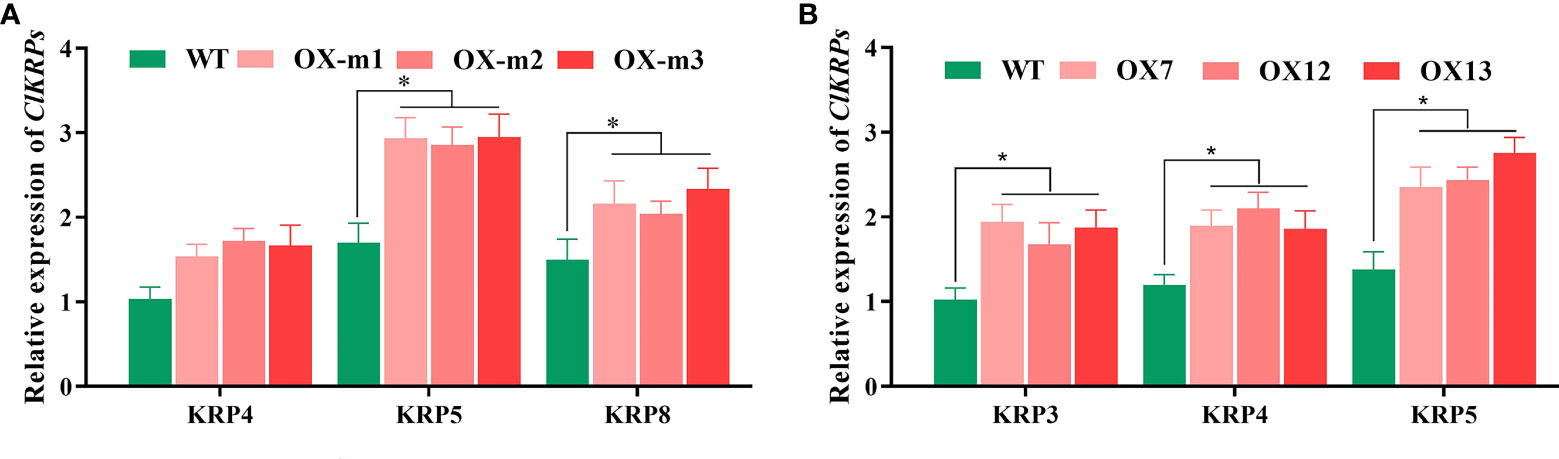
Figure 5 Relative expression of ClKRPs between (A) ClMIP and (B) ClNAC84 overexpression C. lavandulifolium. * p < 0.05 levels, based on t-test.
ClMIP proteins can activate ClKRP5 expression
To confirm whether ClMIP can bind to the ClKRP5 promoter, we performed a yeast one-hybrid and transient expression assay using the luciferase reporter system. The pHIS-ClKRP5pro vector containing the ClKRP5 promoter sequence (1479 bp) was co-transformed into yeast Y187 competent cells with an AD-ClMIP vector containing the ClMIP sequence and an empty pGADT7 vector (AD), respectively. The yeast cells of pHIS-ClKRP5pro and AD-ClMIP and pHIS-ClKRP5pro and AD groups grew well in the yeast medium without His, Trp, and Leu. The yeast cells containing pHIS-ClKRP5pro and AD-ClMIP grew normally, but those with pHIS-ClKRP5pro and empty pGADT7 did not grow on yeast medium lacking His, Trp, and Leu with the addition of 50 mM 3-aminotrizole (3-AT) (Figure 6A). For the luciferase reporter system, a construct containing the ClKRP5 promoter region fused to firefly luciferase (PClKRP5:LUC) was agroinfiltrated into tobacco (Nicotiana benthamiana) leaves together with 35S::ClMIP and (or) 35S::ClNAC84. LUC bioluminescence was induced in leaves co-infiltrated with ClKRP5:LUC + 35S::ClMIP + 35S::ClNAC84, followed by co-infiltration with ClKRP5:LUC + 35S::ClMIP. The intensity of bioluminescence was very weak in leaves infiltrated with P ClKRP5:LUC and P ClKRP5:LUC + 35S::ClNAC84 (Figures 6B, C). These results indicate that ClMIP activated the transcription of ClKRP5.
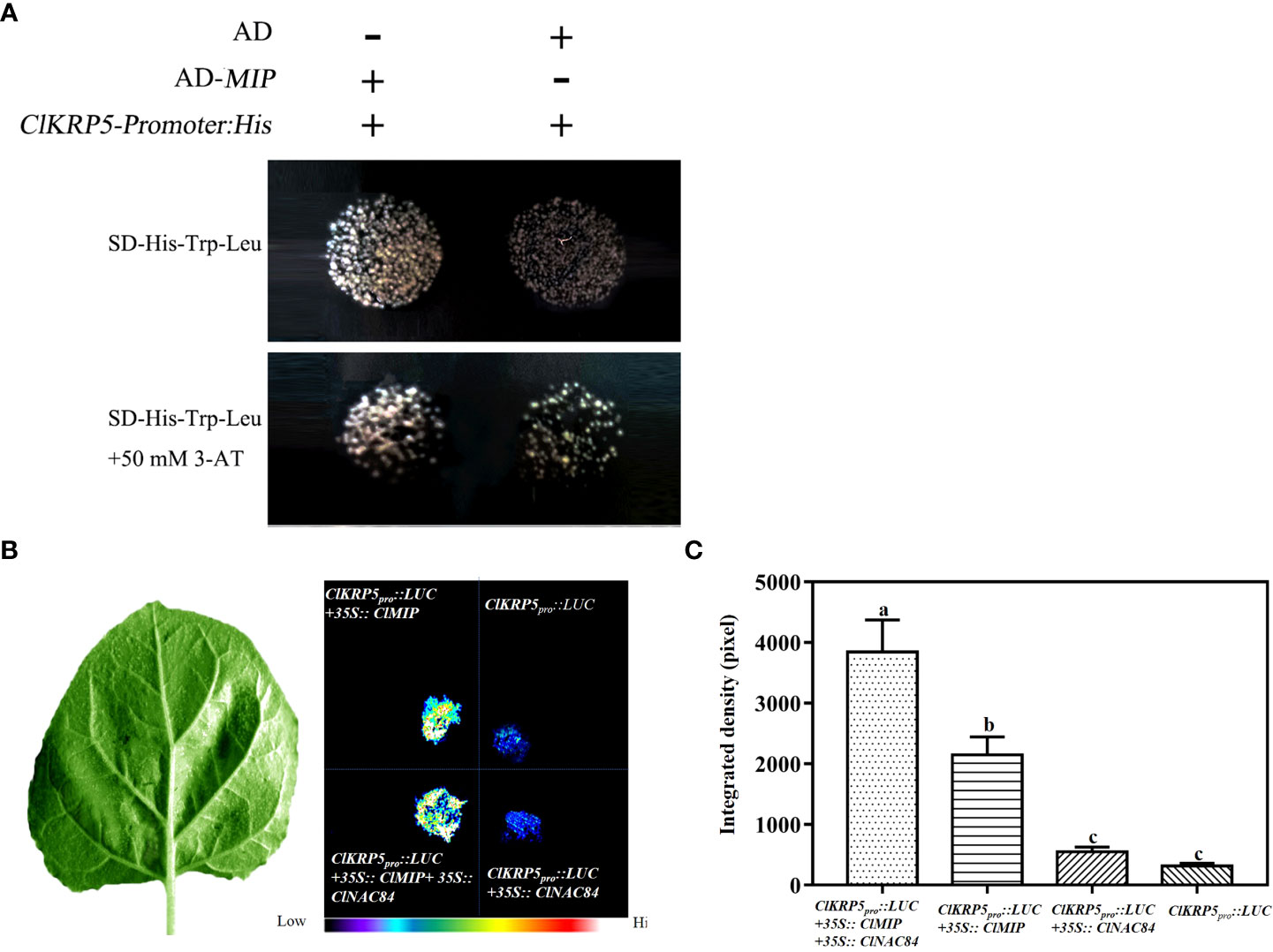
Figure 6 ClNAC84 interaction with ClMIP protein promotes ClKRP5 expression. (A) Yeast one-hybrid assay between ClMIP protein and the promoter of ClKRP5. The co-transfected plasmids were cultured in yeast medium lacking His, Trp, and Leu (above) and His, Trp, and Leu with 50 mM 3-AT (below). (B) Representative images of transient expression assays in Nicotiana benthamiana leaves expressing Pkrp5:LUC, Pkrp5:LUC +35S::ClMIP, Pkrp5:LUC +35S::ClMIP + 35S::ClNAC84, and Pkrp5:LUC + 35S::ClNAC84 as displayed by bright field (left) and dark field (right). (C) Intensities of the LUC bioluminescence presented in (B) measured using ImageJ analysis software. Data are shown as means + SE (n = 6). Different lowercase letters indicate significant differences (p < 0.05).
Discussion
Involvement of ClNAC84 in the regulation of cell cycle
The NAC gene family is a plant-specific transcription factor that is widely involved in plant growth and development (Jin et al., 2017; Yan et al., 2017). NAC protein has a consensus sequence at the NAC domain, which contains 160 amino acid residues and a highly divergent sequence in the transcriptional activation or repression region located in the C-terminal. The ClNAC84 protein contains a NAC domain (a-e subdomains) but has no transcriptional activation activity (Figure 1C). The hydrophobicity associated with FVFY residue interference with DNA binding may be responsible for repression (Puranik et al., 2012). Phylogenetic analysis of NAC proteins from Arabidopsis and rice revealed that the ClNAC84 protein belonged to the ONAC22 subgroup (Figure S4). In this novel subgroup, ANAC36-overexpressing transgenic plants showed a semi-dwarf phenotype in Arabidopsis (Hao et al., 2010). We found that ClNAC84 overexpression lines also showed a dwarf phenotype, including the flowers, leaves, and plant height. Microscopic observation of cell size and cell number revealed that cell number reduction was the main cause of dwarfing in overexpression lines (Figure 3A).
Yeast is a model system for studying gene function. Mu et al. (2019) found that AtMYB59 overexpressing yeast cells had two nuclei and appeared to be three times longer than the control cells. Wang et al. (2004) reported that TaRAN1I overexpression inhibits the cell cycle and changes the morphology of yeast cells. Here, the yeast cell cycle was blocked by ClNAC84 overexpression, which led to two-nuclei yeast cells, further confirming that ClNAC84 is involved in the cell cycle.
In plants, cell cycle arrest may lead to a reduction in cell numbers. The cell cycle includes four phases. The initial quiescent (G0-G1) phase is when cell cycle regulatory protein synthesis and activation begins. When the cells reach the checkpoint and activation occurs, DNA begins replication in the S-phase. After DNA replication, the cell enters the G2 and M phases and divides into two identical cells (Day et al., 1996; Ren et al., 2002; Sakaue-Sawano et al., 2008). Our data showed that cell number reduction in ClNAC84 overexpression C. lavandulifolium was due to stagnation of the S and G2M phases (Figures 2B, C). The G1-to-S-phase transition is a key regulatory point in the cell cycle, which requires the concerted action of specific proteins, such as E2F, CYCD3;1, CDK, and ICK/KRP (Zhang et al., 2016). The relative expression of ClE2F, ClCYCA3;1, ClCYCD6;1, and ClCKDA in ClNAC84 overexpression C. lavandulifolium was lower than that in WT, but the expression levels of ClKRP were higher than those in WT (Figure S5). CYCD3;1 expression in Arabidopsis results in a reduced proportion of cells in the G1 phase (Dewitte et al., 2003). These results suggest that ClNAC84 is involved in the KRP pathway.
Involvement of ClMIP in the regulation of cell cycle
ClMIP is also known as ClAE or FAM96 (family with sequence similarity 96, member B). ClMIP contained one DUF59 (domain of unknown function 59) and a signal peptide in the N-terminal (Figure S6A). Green fluorescence of 35S::GFP-ClMIP markedly accumulated in the onion epidermal cell nucleus (Figure S6B). The yeast assay showed that ClMIP promoted transcriptional activation in yeast cells (Figure S6C). ClMIP overexpression in C. lavandulifolium resulted in a small phenotype, which was similar to that of the ClNAC84 overexpression plants. The number of epidermal cells was reduced in the ClMIP overexpression lines relative to that in WT plants (Figure 4A). ClMIP overexpression in C. lavandulifolium resulted in smaller plant stem, flowers, and leaves than those of WT plants, which were similar to those of ClNAC84 overexpression C. lavandulifolium. Mip18 localized to the mitotic spindle and interacted with Ciao1, an ANT2 protein, regulating nucleotide excision repair and transcription; the siRNA-mediated knockdown of MIP18 results in improper chromosome segregation (Ito et al., 2010). The AE7 (AS1/2 ENHANCER7) gene encodes a protein of the DUC59 superfamily, which is required for leaf polarity. The ae7 mutant showed leaf adaxial–abaxial polarity defects (Yuan et al., 2010). Therefore, the DUF59 domain may be involved in cell division and changes in leaf morphological structure.
ClNAC84 interaction with ClMIP protein promotes ClKRP5 expression
The cell numbers of both ClNAC84 and ClMIP overexpression C. lavandulifolium were reduced and the cell cycle was inhibited (Figure 2, Figure 4), suggesting similar genetic functions. This further indicates that ClMIP interacts with ClNAC84. Luc bioluminescence experiments showed that ClNAC84 and ClMIP bind to ClKRP5 promoter elements (Figure 6). The relative expression of ClKPR5 was significantly increased in ClMIP overexpression and ClNAC84 overexpression lines when compared to that of WT plants (Figure 5). KRP plays an important role in the cell cycle. The G2/M transition most likely requires CYCA and CYCB proteins to form CYC/CDK complexes (Churchman et al., 2006; Francis, 2011; Li et al., 2016). These complexes were also inhibited by the expression of KRPs to regulate cell cycle progression (Inze and De Veylder, 2006). The KRP family has seven members in Arabidopsis, which has various levels in different tissues (Cheng et al., 2013), and KRP overexpression in Arabidopsis results in some common phenotypes, such as small plant size, serrated leaves, and reduced cell number (Wang et al., 2000; Schnittger et al., 2003; Li et al., 2016).
In summary, we propose a model to describe cell cycle regulation by ClNAC84 expression and its interaction with ClMIP protein to promote the ClKRP5 expression to restrict the cell cycle (Figure 7).
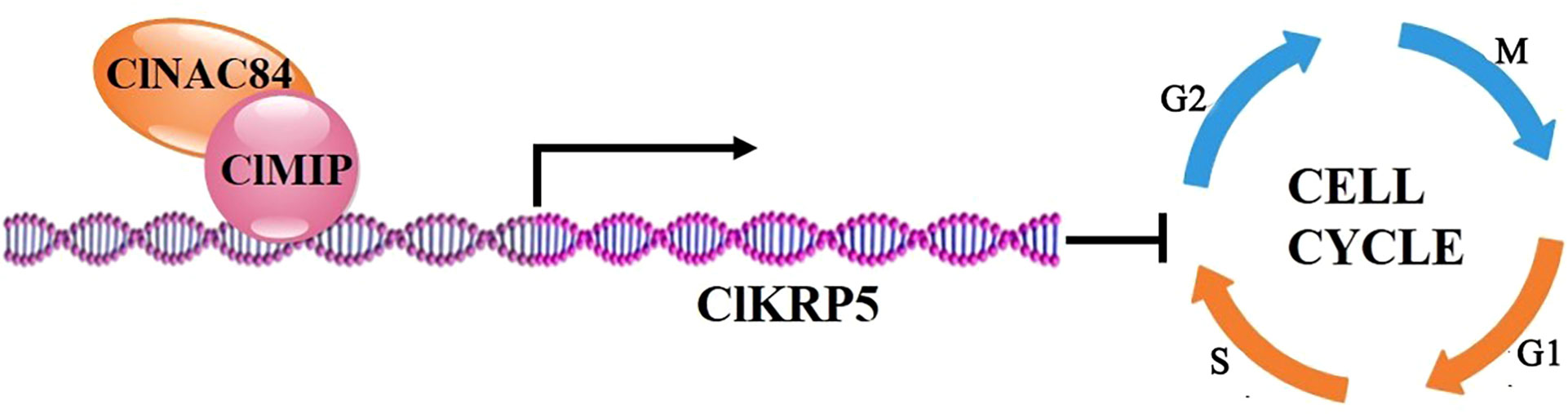
Figure 7 Model for ClNAC84, ClMIP, and ClKRP5 inhibiting cell cycle. Interaction between ClNAC84 and ClMIP promotes ClKRP5 expression and inhibits cells cycling through the G0 and S phases.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
Conceived and designed the experiments: FC. Performed the experiments: RG, HW, XQ, LZ, XY. Analyzed the data: RG, HW, XQ, LZ. Contributed reagents/materials/analysis tools: RG, HW, XQ. Wrote the paper: RG, SC, JJ, ZW, FC. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported grants from National Natural Science Foundation of China (31730081, 31872149, 32060696), Natural Science Foundation of Jiangsu Province (BK20190076), the earmarked fund for Jiangsu Agricultural Industry Technology System, and A project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. The foundation of Key Laboratory of Landscaping, and Ministry of Agriculture and Rural Affairs (KF201902).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fhort.2022.1042105/full#supplementary-material
Supplementary Figure 1 | Transcript abundance of ClNAC84 in wild type and transgenic C. lavandulifolium. WT: wild type. Different numbers representing for 2#, 7#, 8#, 12#, 13# and 17# transgenic lines.
Supplementary Figure 2 | ClNAC84 gene expression in pESPM-ClNAC transformants with VB1 (+VB1) or without. VB1 (–VB1) by RT-PCR analyses.
Supplementary Figure 3 | Transcript abundance of ClMIP in wild type and transgenic C. lavandulifolium. WT: wild type. Different numbers representing for OX-1m, OX-2m, OX-3m transgenic lines.
Supplementary Figure 4 | Unrooted phylogenetic tree of NAC between Arabidopsis thaliana and rice.
Supplementary Figure 5 | Relative expression of genes between ClMIP and ClNAC84 overexpression C. lavandulifolium.
Supplementary Figure 6 | Comparison of deduced amino acid sequences of ClMIP with MIPs from other plant species (A), AaMIP18(PWA83908.1), CcAE7(XP_024994445.1), HaAE7(XP_021969890.1), LsAE7(XP_023750175.1) DcAE7(GEX21963.1). Subcellular localization (B) and transactivation of ClMIP (C).
References
Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Aida M., Tasaka M. (2006). Genetic control of shoot organ boundaries. Curr. Opin. Plant Biol. 9, 72–77. doi: 10.1016/j.pbi.2005.11.011
Aida M., Vernoux T., Furutani M., Traas J., Tasaka M. (2002). Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the arabidopsis embryo. Development 129, 3965–3974. doi: 10.1242/dev.129.17.3965
Cheng Y., Cao L., Wang S., Li Y., Shi X., Liu H., et al. (2013). Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in arabidopsis. Plant J. 75, 642–655. doi: 10.1111/tpj.12228
Churchman M. L., Brown M. L., Kato N., Kirik V., Hulskamp M., Inze D., et al. (2006). SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in arabidopsis thaliana. Plant Cell 18, 3145–3157. doi: 10.1105/tpc.106.044834
Day I. S., Reddy A., Golovkin M. (1996). Isolation of a new mitotic-like cyclin from arabidopsis: complementation of a yeast cyclin mutant with a plant cyclin. Plant Mol. Biol. 30, 565–575. doi: 10.1007/BF00049332
Dewitte W., Riou-Khamlichi C., Scofield S., Healy J. M., Jacqmard A., Kilby N. J., et al. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the d-type cyclin CYCD3. Plant Cell 15, 79–92. doi: 10.1105/tpc.004838
Fang Y., You J., Xie K., Xie W., Xiong L. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280, 547–563. doi: 10.1007/s00438-008-0386-6
Francis D. (2011). A commentary on the G(2)/M transition of the plant cell cycle. Ann. Botan 107, 1065–1070. doi: 10.1093/aob/mcr055
Gao R., Wang H., Dong B., Yang X., Chen S., Jiang J., et al. (2016). Morphological, genome and gene expression changes in newly induced autopolyploid Chrysanthemum lavandulifolium (Fisch. ex trautv.) makino. Int. J. Mol. Sci. 17, 1690–1702. doi: 10.3390/ijms17101690
Gao R., Yan Y., Yang X., Wang Y., Fang W., Chen S., et al. (2018). ClE2F1 overexpression enhances plant growth in Chrysanthemum lavandulifolium (Fisch. ex trautv.) makino. Plant Mol. Biol. Rep. 36, 341–349. doi: 10.1007/s11105-018-1084-0
Grandjean O. (2004). In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. Plant Cell 16 (1), 74–87. doi: 10.1105/tpc.017962
Hao Y. J., Song Q. X., Chen H. W., Zou H. F., Wei W., Kang X. S., et al. (2010). Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232, 1033–1043. doi: 10.1007/s00425-010-1238-2
Huang H., Wang Y., Wang S., Wu X., Yang K., Niu Y., et al. (2012). Transcriptome-wide survey and expression analysis of stress-responsive NAC genes in Chrysanthemum lavandulifolium. Plant Sci. 193-194, 18–27. doi: 10.1016/j.plantsci.2012.05.004
Inze D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77–105. doi: 10.1146/annurev.genet.40.110405.090431
Ito S., Tan L. J., Andoh D., Narita T., Seki M., Hirano Y., et al. (2010). MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol. Cell 39, 632–640. doi: 10.1016/j.molcel.2010.07.029
Jin C., Li K. Q., Xu X. Y., Zhang H. P., Chen H. X., Chen Y. H., et al. (2017). A novel NAC transcription factor, PbeNAC1, of pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant Sci. 8, 1049. doi: 10.3389/fpls.2017.01049
Kato H., Motomura T., Komeda Y., Saito T., Kato A. (2010). Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 167, 571–577. doi: 10.1016/j.jplph.2009.11.004
Kim Y. S., Kim S. G., Park J. E., Park H. Y., Lim M. H., Chua N. H., et al. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144. doi: 10.1105/tpc.106.043018
Li Q., Shi X., Ye S., Wang S., Chan R., Harkness T., et al. (2016). A short motif in Arabidopsis CDK inhibitor ICK1 decreases the protein level, probably through a ubiquitin-independent mechanism. Plant J. 87, 617–628. doi: 10.1111/tpj.13223
Li P., Song A., Gao C., Wang L., Wang Y., Sun J., et al. (2015). Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 34, 1365–1378. doi: 10.1007/s00299-015-1793-x
Menges M., Murray J. A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30, 203–212. doi: 10.1046/j.1365-313X.2002.01274.x
Mu R. L., Cao Y. R., Liu Y. F., Lei G., Zou H. F., Liao Y., et al. (2009). An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res. 19, 1291–1304. doi: 10.1038/cr.2009.83
Ooka H., Satoh K., Doi K., Nagata T., Otomo Y., Murakami K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. doi: 10.1093/dnares/10.6.239
Puranik S., Sahu P. P., Srivastava P. S., Prasad M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R. A., et al. (2002). E2F integrates cell cycle progression with DNA repair, replication, and G(2./M) checkpoints. Genes Dev. 16, 245–256. doi: 10.1101/gad.949802
Rubio-Somoza I., Zhou C. M., Confraria A., Martinho C., von Born P., Baena-Gonzalez E., et al. (2014). Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 24, 2714–2719. doi: 10.1016/j.cub.2014.09.058
Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., et al. (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498. doi: 10.1016/j.cell.2007.12.033
Schnittger A., Weinl C., Bouyer D., Schobinger U., Hulskamp M. (2003). Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15, 303–315. doi: 10.1105/tpc.008342
Souer E., Van A. H., Kloos D., Mol J., Koes R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundarie. Cell 85, 159–170. doi: 10.1016/S0092-8674(00)81093-4
Tang Y., Zhao C. Y., Tan S. T., Xue H. W. (2016). Arabidopsis type II phosphatidylinositol 4-kinase PI4Kgamma5 regulates auxin biosynthesis and leaf margin development through interacting with membrane-bound transcription factor ANAC078. PloS Genet. 12, e1006252. doi: 10.1371/journal.pgen.1006252
Wang H., Jiang J., Chen S., Qi X., Peng H., Li P., et al. (2013). Next-generation sequencing of the Chrysanthemum nankingense asteraceae. transcriptome permits large-scale unigene assembly and SSR marker discovery. PloS One 8, e62293. doi: 10.1016/j.plantsci.2004.03.0
Wang X., Xu W.-Z., Xu Y.-Y., Chong K., Xu Z.-H., Xia G.-X. (2004). Wheat RAN1, a nuclear small G protein, is involved in regulation of cell division in yeast. Plant Sci. 167, 1183–1190. doi: 10.1016/j.plantsci.2004.03.011
Wang H., Zhou Y., Gilmer S., Whitwill S., Fowke L. C. (2000). Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 24, 613–623. doi: 10.1046/j.1365-313x.2000.00899.x
Willemsen V., Bauch M., Bennett T., Campilho A., Wolkenfelt H., Xu J., et al. (2008). The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell 15, 913–922. doi: 10.1016/j.devcel.2008.09.019
Yan H., Zhang A., Ye Y., Xu B., Chen J., He X., et al. (2017). Genome-wide survey of switchgrass NACs family provides new insights into motif and structure arrangements and reveals stress-related and tissue-specific NACs. Sci. Rep. 7, 3056. doi: 10.1038/s41598-017-03435-z
Yuan Z. H., Luo D. X., Li G. A., Yao X. Z., Wang H., Zeng M. H., et al. (2010). Characterization of the AE7 gene in Arabidopsis suggests that normal cell proliferation is essential for leaf polarity establishment. Plant J. 64, 331–342. doi: 10.1111/j.1365-313X.2010.04326.x
Zhang W., Xie H. Y., Ding S. M., Xing C. Y., Chen A., Lai M. C., et al. (2016). CADM1 regulates the G1/S transition and represses tumorigenicity through the Rb-E2F pathway in hepatocellular carcinoma. Hepatob Pancreat Dis. 15, 289–296. doi: 10.1016/S1499-3872(16)60099-1
Keywords: Chrysanthemum lavandulifolium, ClNAC84, ClMIP, cell cycle, ClKRP5
Citation: Gao R, Wang H, Qi X, Zhu L, Yang X, Chen S, Jiang J, Wang Z and Chen F (2022) ClNAC84 interacts with ClMIP to regulate the cell cycle and reduce the size of Chrysanthemum lavandulifolium organs. Front. Hortic. 1:1042105. doi: 10.3389/fhort.2022.1042105
Received: 12 September 2022; Accepted: 31 October 2022;
Published: 16 November 2022.
Edited by:
Aamir Raina, Aligarh Muslim University, IndiaReviewed by:
Ayan Sadhukhan, Indian Institute of Technology Jodhpur, IndiaNasrin Moshtaghi, Ferdowsi University of Mashhad, Iran
Copyright © 2022 Gao, Wang, Qi, Zhu, Yang, Chen, Jiang, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fadi Chen, chenfd@njau.edu.cn
 Ri Gao
Ri Gao Haibin Wang
Haibin Wang Xiangyu Qi1
Xiangyu Qi1  Lu Zhu
Lu Zhu Sumei Chen
Sumei Chen Jiafu Jiang
Jiafu Jiang Zhenxing Wang
Zhenxing Wang Fadi Chen
Fadi Chen