- Department of Radiation Oncology, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
Background: Locally advanced rectal cancers (LARC) show a highly variable response to neoadjuvant chemoradiotherapy (nCRT), and the impact of the tumor immune response in this process is poorly understood. This study aimed to characterize the immune-related gene expression profiles (GEP), pathways, and cell types associated with response or resistance to neoadjuvant chemoradiotherapy.
Methods: The transcriptomic and clinical data of Rectal carcinoma from the Gene Expression Omnibus database and Immune-related genes (IRGs) from ImmPort were downloaded to identify the differentially expressed immune-related genes (DEIRGs) between responder and non-responder to neoadjuvant chemoradiotherapy. Gene set enrichment analyses were performed to uncover significantly enriched GO terms and KEGG pathways. Immune cell infiltration was estimated from RNA-sequencing data using ImmuCellAI. Afterward, we constructed an immune-related gene-based predictive model (IRGPM) by Support Vector Machine and validated it in an external cohort.
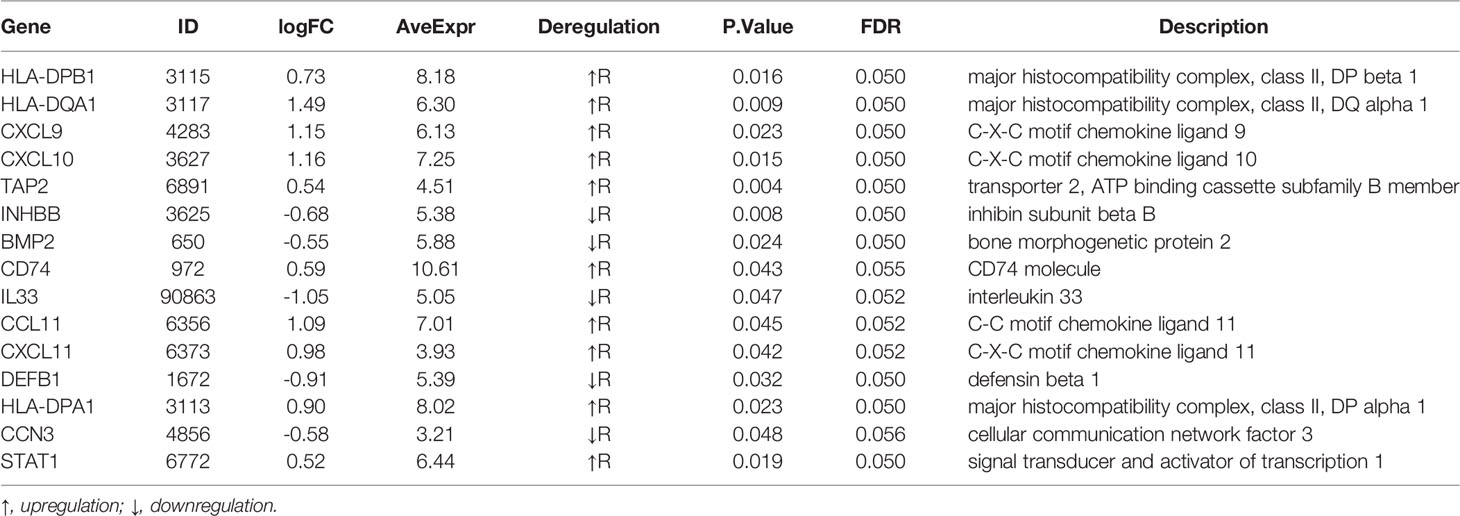
Result: A 15-gene signature (HLA-DPB1, HLA-DQA1, CXCL9, CXCL10, TAP2, INHBB, BMP2, CD74, IL33, CCL11, CXCL11, DEFB1, HLA-DPA1, CCN3, STAT1) was identified as DEIRGs and found to be significantly associated with nCRT outcomes. Gene set enrichment analyses indicated that the 15 genes play active roles in inflammation-related biological processes. In addition, ImmuCellAI revealed that CD4 naive T cells, Tex, Th1 were significantly up-regulated (p=0.035, p=0.02, p=0.0086, respectively), while Tfh were significantly down-regulated (p=0.015) in responder subgroup. Finally, a novel predictive model was developed by SVM based on DEIRGs with an AUC of 80% (internal validation) and 73.5% (external validation).
Conclusion: Our team conducted a genomic study of the relationship between gene expression profile and response to nCRT in LARC. Our data suggested that the DEIRGs signature could help predict the efficacy of nCRT. And a DEIRGs‐based SVM model was developed to monitor the outcomes of nCRT in LARC.
Introduction
Rectal cancer (RC) is one of the most common malignancies, amounts to one-third of colorectal cancer, which is viewed as the third leading cancer and the second cancer-related mortality in men and women worldwide (1). And the majority of the patients are diagnosed with invasive stage, should be referred for multidisciplinary treatment. For patients with locally advanced rectal cancer (LARC), neoadjuvant chemoradiotherapy (nCRT) followed by radical surgery with total mesorectal excision (TME) has become the standard of care according to the clinical practice guideline of NCCN (National Comprehensive Cancer Network) (2). As was demonstrated in previous research, nCRT has emerged as an indispensable component of curative therapy, which offers a higher probability to downsize and downstage tumors, to enhance tumor resectability and sphincter preservation, and to improve local control rate (3–6). However, LARC exhibits heterogeneity in response to nCRT, varying from nonresponse to complete pathologic response (7). Given the spectrum of response, tailored decisions should be made individually by physicians. However, there is a lack of effective methods to select rectal cancer patients who would or would not have a benefit from nCRT. Numerous efforts have been made, but none has currently reached the clinic (8).
The behavior of a tumor is not only governed by the epithelial component but also by the tumor environment (9). As an essential component of the tumor microenvironment, the tumor immune microenvironment (TIME) serves a vital role in tumor progression and treatment outcome, has gradually acquired accumulative attention. Mounting evidence indicates that characterizing TIME could enable the identification of new prognostic and predictive biomarkers and the possibility to guide first-line treatment algorithms (10). Some features of TIME have been quantified and incorporated into an index called ‘immunoscore,’ which can be used as a strong predictor of patient survival in colorectal carcinoma, providing information more robust than the standard AJCC/UICC-TNM staging system and microsatellite instability (11–14). Therefore, the contexture of TIME has the potential to predict response to nCRT, which has been confirmed in a recent study (15). But the preliminary findings need to be further corroborated in large patient cohorts. Herein, it is urgent to construct an immune-related gene signature closely related to TIME, aiming to predict nCRT efficacy.
This study aimed to characterize the immune-related gene expression profiles (GEP), pathways, and cell types associated with response or resistance to neoadjuvant chemoradiotherapy. Initially, we mainly focused on significant immune genes associated with poor tumor response through data mining in the Gene Expression Omnibus (GEO) databases. We found that differentially expressed immune-related genes (DEIRGs) were significantly associated with the efficacy of nCRT. Then, Gene set enrichment analyses were performed to uncover greatly enriched GO terms and KEGG pathways. Immune cell infiltration was estimated from RNA-sequencing data using ImmuCellAI. Finally, an immune-related gene-based prognostic model (IRGPM) was constructed by Support Vector Machine and validated in an external database.
Materials and Methods
Data Acquisition
The publicly available mRNA expression profiles and corresponding clinical information were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), with accession numbers GSE87211, GSE45404, and GSE35452. The gene expression profiles (ID, GSE87211) were previously produced using the Agilent-026652 Whole Human Genome Microarray 4x44K v2 (Probe Name version). Specimens in GSE87211, including 203 tumor samples and 160 mucosa control samples, were obtained from 243 patients. All patients were staged as locally advanced (cUICCII/III/IV) and treated according to the CAO/ARO/AIO-94 or the CAO/ARO/AIO-04 trial (16). The other two microarray datasets (ID, GSE45404, and GSE35452) were previously produced using Affymetrix HGU133 Plus 2.0 GeneChips (Affymetrix, Santa Clara, Ca).
The specimens in the GSE45404 were obtained from 42 patients with LARC (91% and 88% of patients were clinically staged as T3–4 and lymph nodes positive, respectively). 38 (90%) patients received a total dose of radiotherapy higher than 50 Gy, and 15 out of these cases (36%), drugs other than 5-FU were administered (n=11, Oxaliplatin; n=4, Carboplatin). For 33 (79%) patients, 5-FU was administered by continuous venous infusion. The median (range) interval time between the completion of nCRT and surgery was 46 (30–66) days. The tumor response to nCRT was defined as the tumor regression grade (TRG) and was scored following the criteria proposed by Mandard et al. (17).
The specimens in the GSE35452 were obtained from 46 patients with LARC. All patients received a total dose of 50.4Gy of radiation, given in 28 fractions over six weeks. Tegafur-uracil (300-500mg/day) and leucovorin (75mg/day) was given concomitantly with radiotherapy. Standardized curative resection was performed six weeks after the completion of nCRT. Response to nCRT was determined by histopathological examination of surgically resected specimens based on a semiquantitative classification system defined by the Japanese Society for Cancer of the Colon and Rectum (18).
Screen Genes of Differential Expression
The rectal cancer-related gene expression profile was identified in the GSE87211 database. And GSE45404 database was used to select genes associated with nCRT efficacy by comparing responders and non-responders. We utilized the R package ‘limma’ to screen the differentially expressed genes (19). Those genes with P-value < 0.05 and the expression ratio lg |fold change| > 0.5 were picked out for further evaluation. A Benjamini-Hochberg FDR of 0.1 was used to identify highly significant associations to account for multiple hypothesis testing.
Generation of DEIRGs
1793 IRGs were retrieved from 17 categories after excluding the duplicates from the Immunology Database and Analysis Portal (ImmPort) website (https://www.immport.org) (20). The overlapping DEGs in GSE87211 and GSE45404 datasets were intersected with immune-related genes from ImmPort to obtain the target genes defined as the differentially expressed immune-related genes (DEIRGs).
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA), also called functional enrichment analysis, is a popular calculation method applied to analyze the slight changes in the expressions of genes that belong to a critical pathway. So GSEA can be used to understand the molecular mechanisms of complex disorders, lead to the discovery of new suspect genes and biological pathways, and help provide molecular evidence for the association of biological pathways with diseases (21). In our study, Gene set enrichment analysis was performed and illustrated by ‘Clusterprofiler’ (22) and ‘Pathview’ (23) packages in R. Gene Ontology (GO) classification, including GO-BP (biological process), GO-MF (molecular function), and GO-CC (cellular component), was used to uncover the functions of intersecting genes and further test the biological links in DEGs. GO terms with significant P values were selected according to the BP, CC, and MF results. The pathways with the following criteria were significantly enriched: nominal p-value < 0.05, false discovery rate (FDR) q-value < 0.25.
Estimation of Immune Cell Infiltration
To explore the associations between nCRT efficacy and immune cell infiltration, we employed Immune Cell Abundance Identifier (ImmuCellAI) (24), a valuable resource for comprehensive analysis of tumor-infiltrating immune cells. ImmuCellAI is a tool to estimate the abundance of 24 immune cells from gene expression datasets, including RNA-Seq and microarray data. It can be applied to estimate the difference in immune cell infiltration in diverse groups. The core algorithm of ImmuCellAI includes three main steps: 1) reference expression matrix and marker gene preparation, 2) enrichment score calculation, and 3) compensation matrix correction. The reference expression profiles of the immune cells were obtained from GEO, and marker genes per immune cell type were obtained from the literature and analytical methods. For each queried sample, the enrichment score of the total expression deviation of the signal gene sets was calculated and assigned to each immune cell type by the ssGSEA algorithm. The compensation matrix and least square regression were implemented to correct the bias caused by the shared marker genes among different immune cell types.
Construction of an SVM Classifier
Machine learning with maximization (support) of separating margin (vector), called support vector machine (SVM), is a powerful classification tool that has been used for cancer genomic classification or subtyping (25). SVM aims to create a decision boundary between two classes that enables the prediction of labels from one or more feature vectors (26). because our classification problem was nonlinear, the polynomial kernel was adopted, the formula as follows:
To evaluate the predictive accuracy of DEIRGs for “N” and “NR” samples, a supervised SVM classifier was constructed and trained via the SVM function in the ‘e1071’ package of the R software.
Validation Analysis
The efficacy of the SVM model was further verified in the GSE45404 dataset (internal validation set) and GSE35452 dataset (external validation set), respectively. Receiver operating characteristic (ROC) curves were used to evaluate the sensitivity and specificity of the predicting model by the ‘pROC’ package in R. The area under the curve (AUC) ≥ 0.7 indicated a good differentiation ability. To more detailedly confirm the prediction value of the SVM classifier model, an SVM with a confusion matrix was constructed.
Statistical Analysis
Most statistical analyses were performed using R software (Version 4.0.2; R Foundation for Statistical Computing, Vienna, Austria). Graphs related to R statistical analyses were drawn using the ggplot2 package in R.
Result
An Immune-Related Signature of Chemo-Radio Resistance in LARC
The flowchart of the procedures in this study is shown in Figure 1. Three independent datasets were enrolled in our study (Table 1). Initially, we focused on gene expression profiles (GEP) of rectal carcinoma to reveal the underlying mechanism of rectal cancer. DEGs analysis was performed between the tumor and normal tissue in the GSE87211 cohort. A total of 7941 deregulated genes were identified, which were defined as RC-related DEGs. A volcano plot and heatmap were generated to show the distribution of the DEGs (Figures 2B, C). Besides, the principal component analysis (PCA) was applied to analyze the whole gene expression, and the 2D-PCA plot showed significant differences between rectal cancer and normal tissue (Figure 2A).
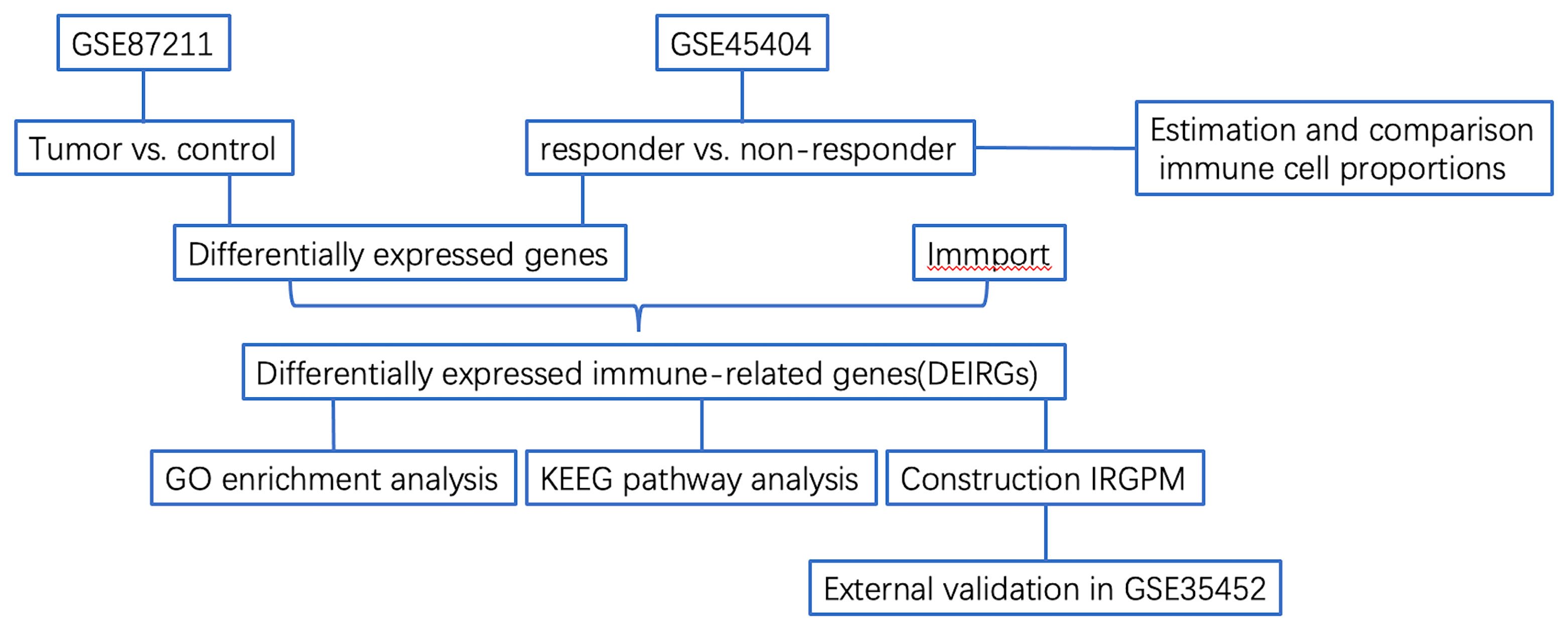
Figure 1 Flowchart describing the process used to identify DEIRGs and construct IRGPM of rectal carcinoma.
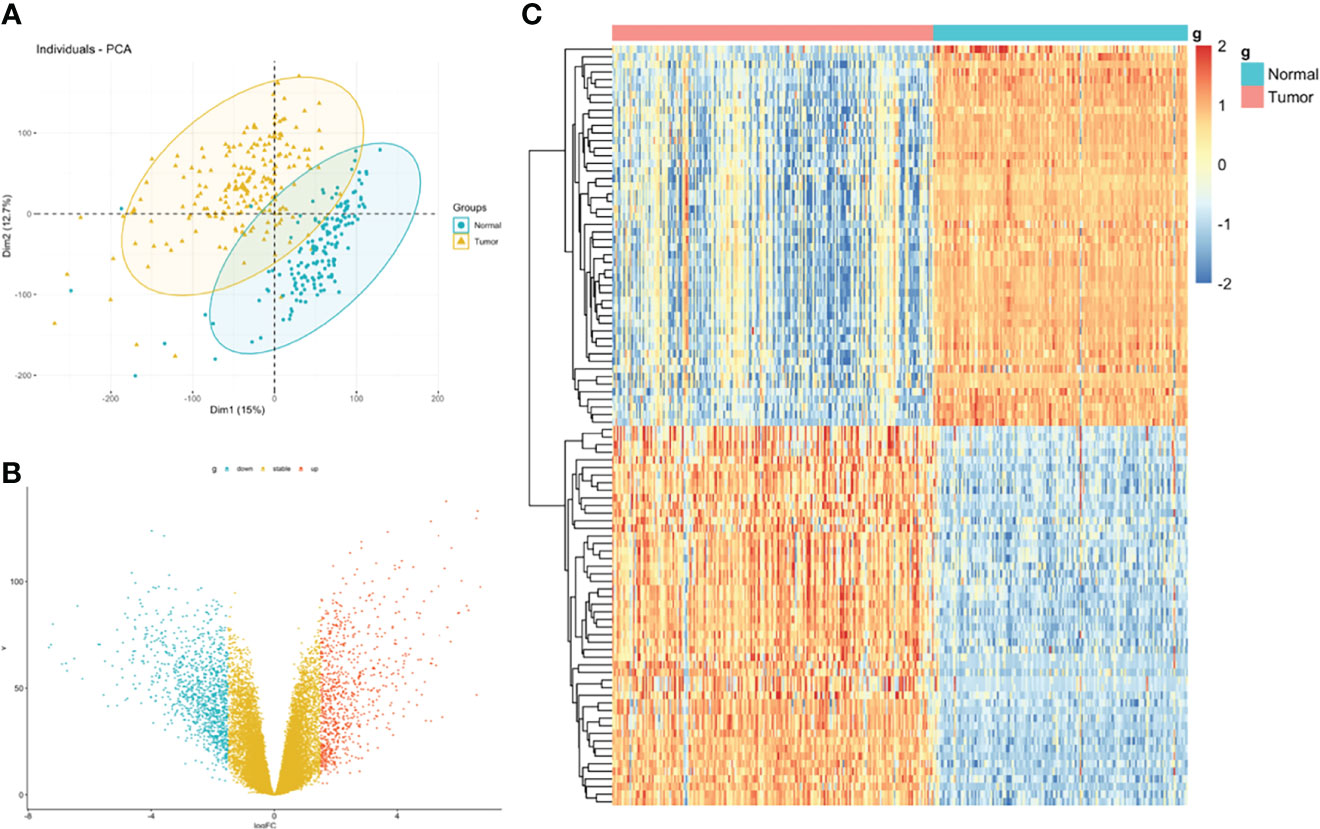
Figure 2 Identification of DEGs between rectal carcinoma tissue and mucosa control tissue. (A) Principal component analysis (PCA) of expression level of rectal cancer and normal tissue. (B) Volcano plot and (C) hot map illustrating differentially expressed genes (DEGs) between rectal carcinoma tissue and mucosa control tissue.
Then, we integrated gene expression signatures to search for differences in gene expression linked to nCRT response and obtain information on the molecular mechanisms involved in differential nCRT efficacy. DEGs analysis between responders and non-responders in the GSE45404 cohort was performed. A total of 73 upregulated and 40 downregulated genes were identified, defined as nCRT-related DEGs. A volcano plot and heatmap were generated to show the distribution of the DEGs (Figures 3B, C). Auxiliary, the principal component analysis (PCA) was applied to analyze the whole gene expression, and the difference between responders and non-responders was not significant in the 2D-PCA plot (Figure 3A).
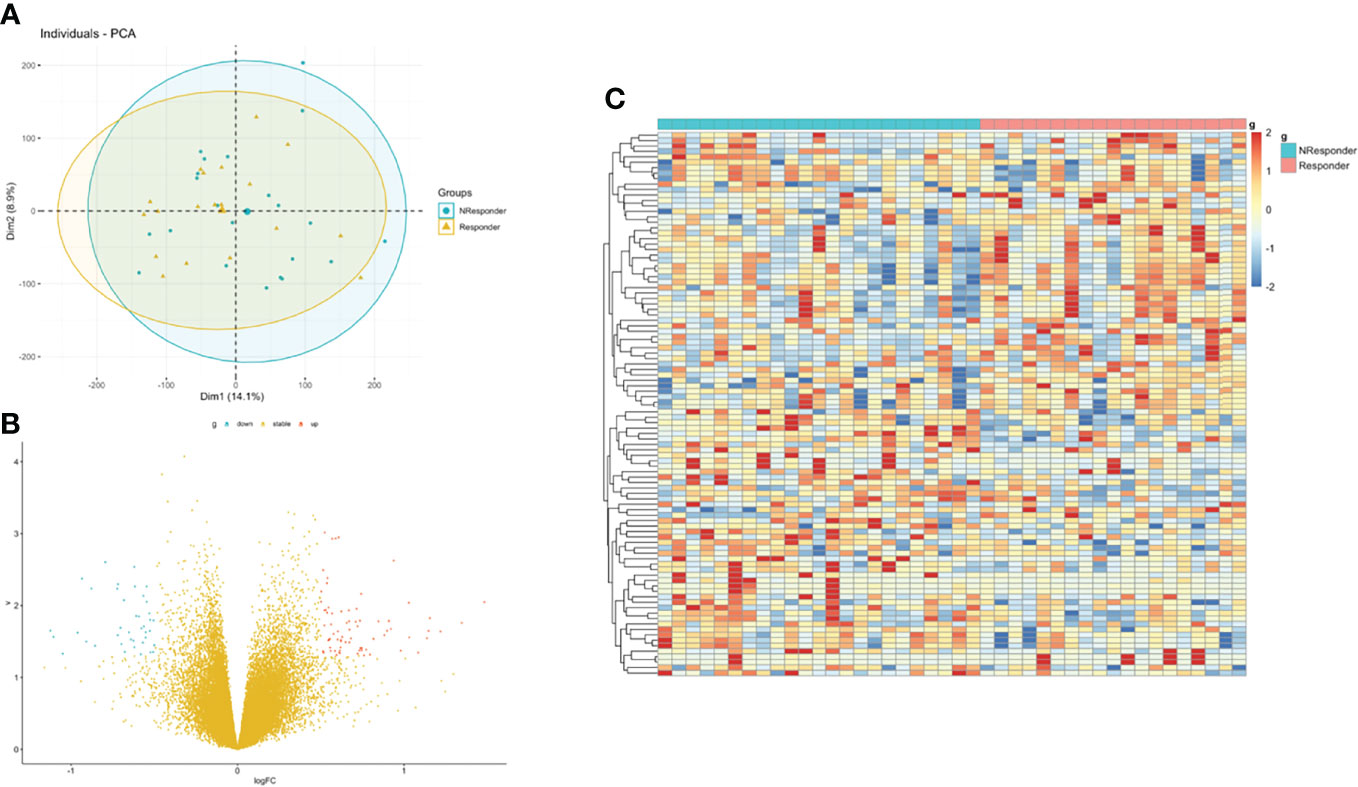
Figure 3 Identification of DEGs between rectal responder (R) subgroup and non-responder (NR) subgroup. (A) Principal component analysis (PCA) of expression level of rectal responder (R) subgroup and non-responder (NR). (B) Volcano plot and (C) hot map illustrating differentially expressed genes (DEGs) between rectal responder (R) subgroup and non-responder (NR).
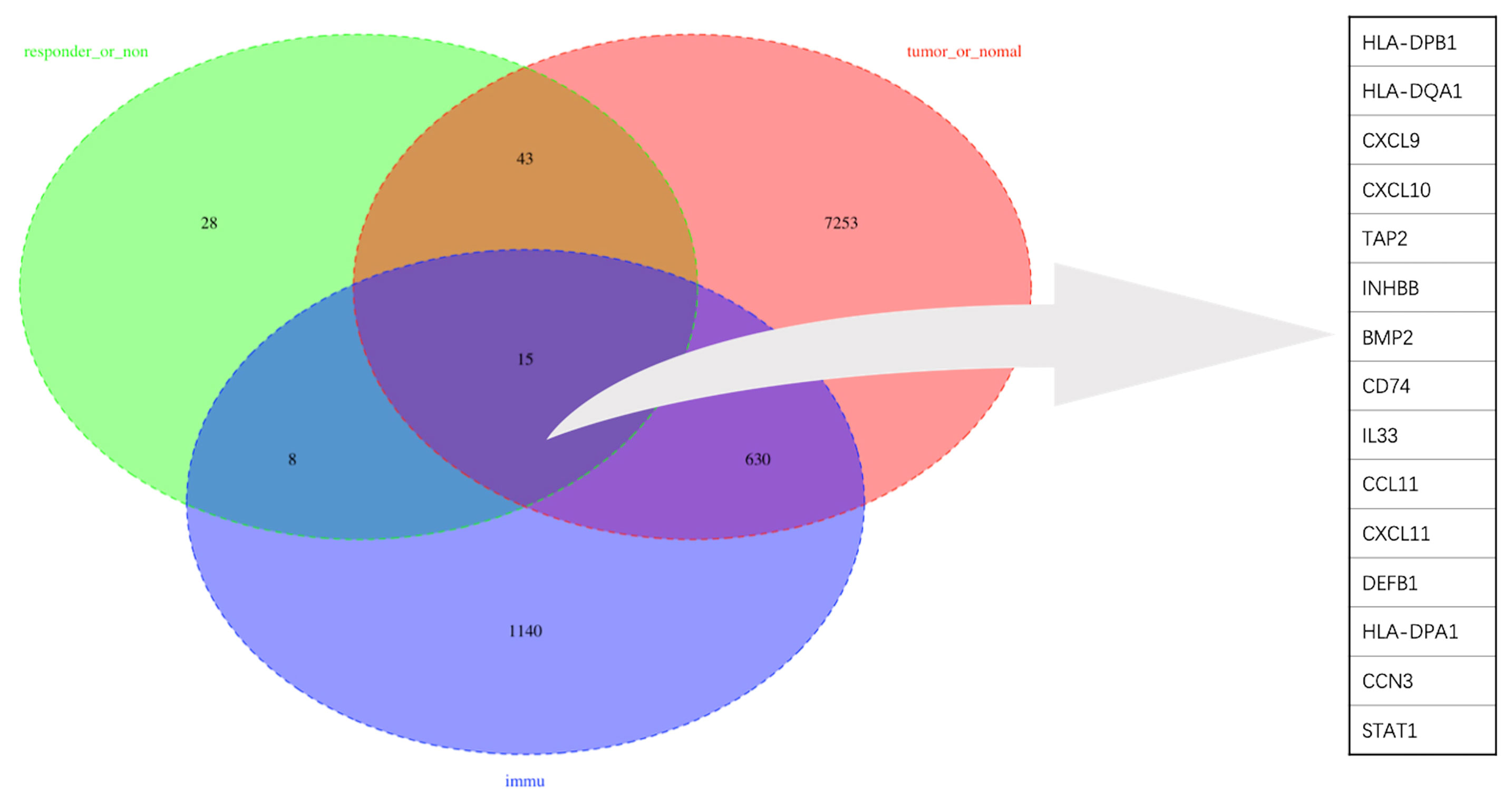
To obtain a robust gene signature to predict nCRT efficacy, GO enrichment analysis of biological processes was conducted in the overlapping DEGs in GSE87211 and GSE45404 datasets, which demonstrated “immune-related responses” were the most frequently used biological terms in biological processes (Supplementary Figure 1). In this regard, an immune-related gene signature may be able to predict chemo-radio resistance. To address the issue, the 58 overlapping DEGs were intersected with 1793 immune-related genes from ImmPort to obtain the target genes that were defined as the differentially expressed immune-related genes (DEIRGs) (Figure 4). In the end, 15 DEIRGs were generated, and the details were shown in Table 2.
GSEA Analysis of DEIRGs
GO, and KEGG functional enrichment analyses were performed on the fifteen selected genes. KEGG pathway analysis showed that the DEIRGs were mainly enriched in cytokine-cytokine receptor interactions (Figure 5A). Concerning biological processes, the DEGs were significantly enriched in leukocyte migration, myeloid leukocyte migration, leukocyte chemotaxis, and cell chemotaxis (Figure 5B). Meanwhile, the most enriched terms in the aspect of molecular function were cytokine receptor binding, receptor-ligand activity, and signaling receptor activator activity (Figure 5D). Enrichment analysis of cellular compartment and the corresponding distributions are shown in Figure 5C.
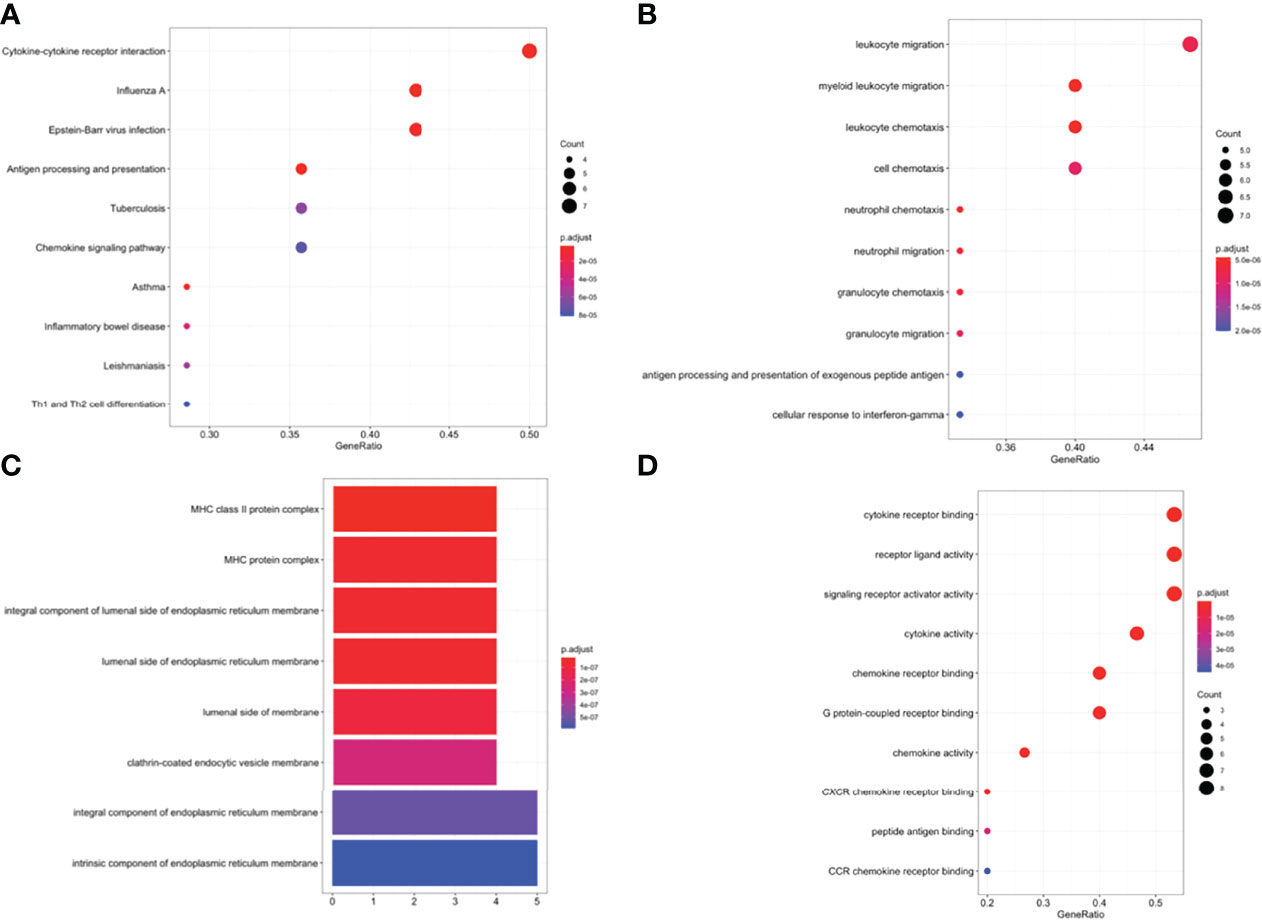
Figure 5 (A) Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of DEIRGs. Enriched Gene Ontology (GO) pathways of DEIRGs. (B) BP biological process, (C) CC cell component, (D) MF molecular function.
Immune Cell Infiltration Between Responders and Non-Responders
Given that the expression of immune genes was closely associated with nCRT efficacy, it is necessary to investigate the role of TIME on chemo-radio resistance. Twenty-four immune cell fractions were generated using a ssGSEA algorithm to confirm the correlation between TIME and nCRT efficacy. As shown in Figure 6, CD4 naive T cells, Tex (exhausted T cells), Th1 (type 1 CD4+ T helper cells) were significantly up-regulated (p=0.035, p=0.02, p=0.0086, respectively), while Tfh (T follicular helper cells) were significantly down-regulated (p=0.015) in responder subgroup of GSE45404 cohort. However, this difference in the distribution ratio of immune cells infiltration was not observed in GSE35452 (Supplementary Figure 2).
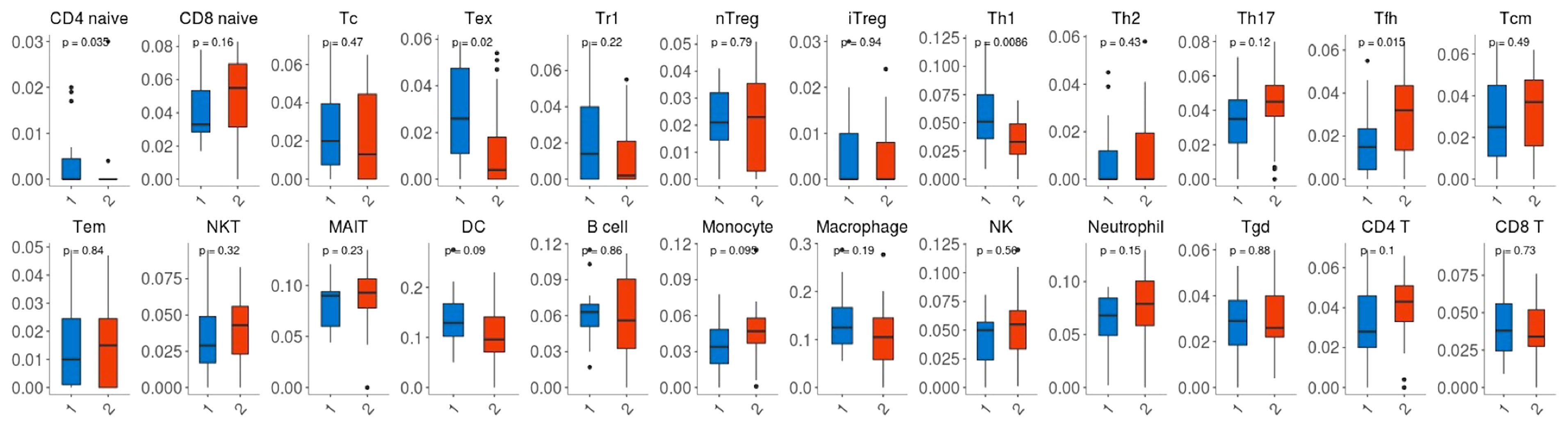
Figure 6 Comparison between the fractions of immune cells in the responder (R) and non-responder (NR) subgroup of the GSE45404 cohort via the ImmuCellAI method.
Construction of Predictive Model and External Validation
SVM, a method for building a classifier, aims to create a decision boundary, which is orientated in such a way that it is as far as possible from the closest data points from each of the classes. In our study, we addressed the outcome of nCRT as a two-class classification problem (“R” or “NR”) and the above 15 DEIRGs as features. The polynomial kernel was adopted, and the SVM classifier was developed using the GSE45404 dataset. We performed a set of experiments in the training cohort of the first 10 patients, tested the classifier with the remaining 32 samples. As a result, 80% prediction accuracy was found to be achieved using this developed SVM classifier (Kernel=polynomial, cost=1/32, degree=2, gamma =0.125, coef0 = 2) in GSE45404 dataset (Figure 7A). Furthermore, SVM with confusion matrix analysis also suggested an 80% prediction accuracy for response samples (sensitivity) and 80% prediction accuracy for nonresponse samples (specificity) (Figure 7C).
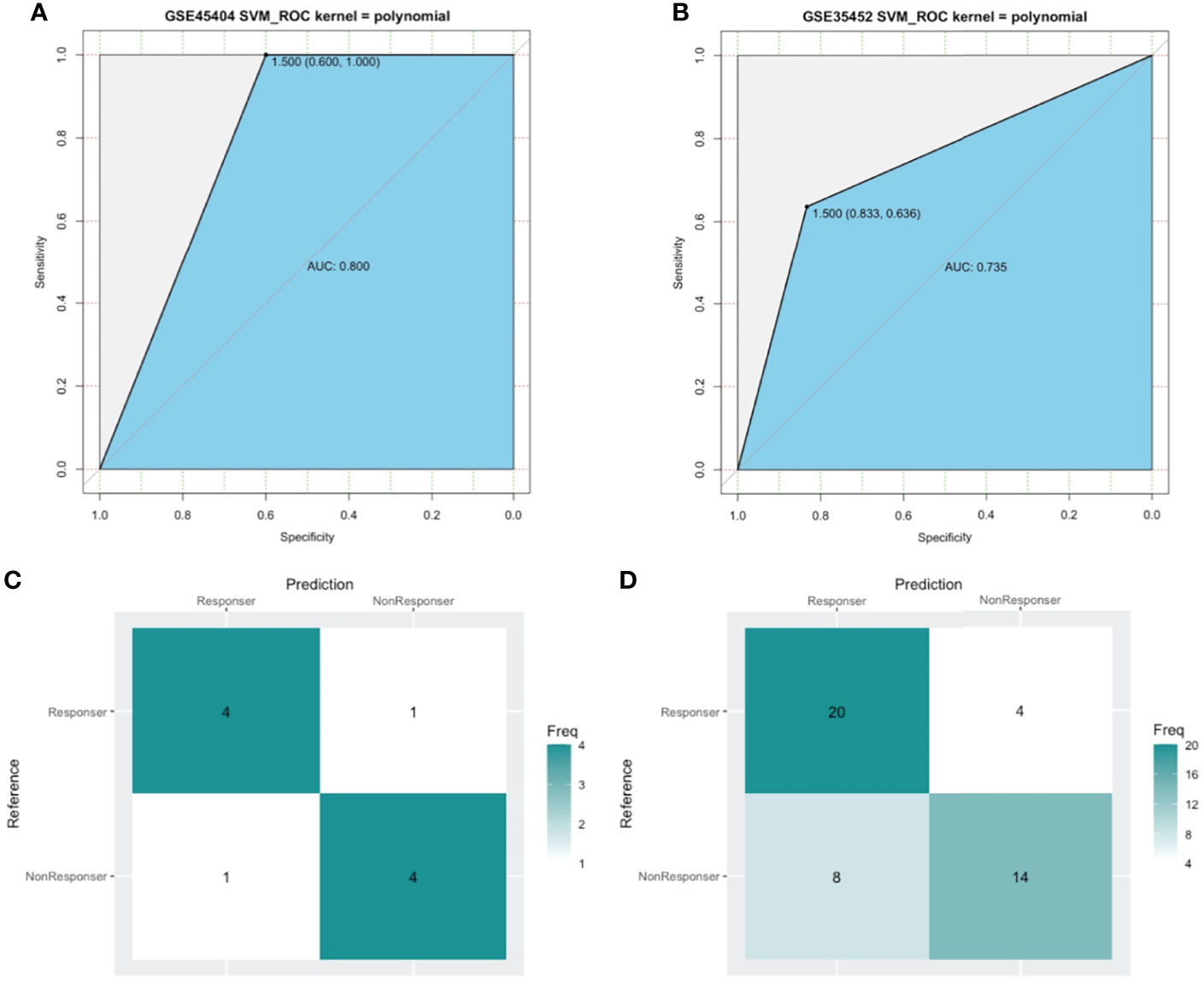
Figure 7 The model measured by receiver-operating characteristic (ROC) curves in GSE45404 dataset (internal validation) and GSE35452 dataset (external validation). (A, B) ROC curve. The horizontal axis indicated the specificity and the vertical axis indicated the sensitivity. (C, D) confusion matrix. The horizontal axis indicated the predictive case numbers and the vertical axis indicated the present case numbers.
To further confirm the classification reliability of the above DEIRGs, another microarray dataset GSE35452 was collected and underwent the SVM model analysis. As expected, the SVM exhibited a 73.5% prediction accuracy in the external cohort of 46 samples (Figure 7B). The confusion matrix suggested an 83.3% sensitivity, a 63.6% specificity, a 71.4% positive predictive value (PPV), and a 77.8 negative predictive value (NPV) (Figure 7D). These findings all proved an outstanding predictive value of this 15 DEIRGs signature.
Discussion
Response to nCRT varies among patients, and pathological complete response is associated with a better outcome. However, there is a lack of effective methods to select rectal cancer patients who would or would not have a benefit from nCRT. The utility of clinicopathological and radiological features is limited due to a lack of adequate sensitivity and specificity. Although the relationship of gene expression profile and response to nCRT has been well studied (27–29), Predictors of outcome have not been systematically explored in the context of spontaneous tumor immunity. To our knowledge, the predictive ability of the immune gene signature per se is confirmed for the first time.
This study sorted out the most influential immune-related genes affecting patients’ nCRT outcomes with a public database and bioinformatical method. Based on this immune gene signature, we generated an SVM classifier capable of stratifying patients with LARC. Notably, satisfactory AUCs of the SVM classifier imply that the LARC response to nCRT may be affected by the immune system. This conclusion echoes a series of ongoing phase II clinical trials combining nCRT and immunotherapy in LARC patients, such as the VOLTAGE-A trial in Japan, which have achieved better pCR rates (30). Our research found that the DEIRGs and DEIRGs-based SVM model have the potential to be a reference for whether or not to combine immunotherapy in LARC patients.
The immune gene signature consists of 10 upregulated genes and 5 down-regulated genes. The upregulated gene profile consisted of five MHC genes (HLA-DPB1, HLA-DQA1, HLA-DPB1, TAP2, CD74), four chemokine genes (CXCL9, CXCL10, CXCL11, CCL11), and STAT1.
HLA-DPA1, HLA-DQA1, and HLA-DPB1 belong to the HLA class II play a central role in the immune system by presenting peptides derived from extracellular proteins. TAP is essential for peptide delivery from the cytosol to the lumen of the endoplasmic reticulum in the major histocompatibility complex (MHC) class I antigen-presenting pathway (31). TAP expression level in tumor cells defines the nature and processing of MHC class I peptides for recognition by tumor-specific cytotoxic T lymphocytes (32). CD74 associates with class II major histocompatibility complex (MHC) and is a vital chaperone that regulates antigen presentation for immune response. The high expression of these genes indicates an active antigen presentation environment, which is conducive to the presentation of antigens generated during radiotherapy and chemotherapy to tumor killer cells, thereby activating anti-tumor immunity and suggesting better tumor efficacy. Some studies confirmed our conjecture. Restoration of the expression of TAP2 may increase tumor-specific immune responses and survival (33). Expression of CD74 associated with favorable survival for stage III melanoma (34).
CXCL9, CXCL10, CXCL11 are interferon-stimulated chemokines that attract T cells. The chemokine encoded by the CCL11 gene displays the chemotactic activity of eosinophils and shows anti-cancer characteristics in colorectal carcinoma (35). STAT1 signaling promotes T helper 1 (Th1) differentiation and interleukin-12 receptor (IL-12) expression, and thus, in some context, has a role in the anti-tumor immune response (36, 37). Overexpression of these genes may better recruit T cells, thereby activating more robust anti-tumor immune responses.
The five genes downregulated in the R group were involved in tumor progression to varying degrees. INHBB and BMP2 are TGF-β (transforming growth factor-beta) related genes. TGF-β is central to immune suppression within the tumor microenvironment, and recent studies have revealed roles in tumor immune evasion and poor responses to cancer immunotherapy (38). in vivo IL33 is involved in the maturation of Th2 cells and activating mast cells, basophils, eosinophils, and natural killer cells. IL33 recruited macrophages into the cancer microenvironment and stimulated them to produce prostaglandin E2, which supported colon cancer stemness and tumor growth (39). CCN3 is a member of the CNN family, which plays crucial roles in CRC progression, including cell migration, invasion, adhesion, and distal metastasis (40). DEFB1 is an antimicrobial peptide implicated in the resistance of epithelial surfaces to microbial colonization, which decreased in colon cancer specimens (41).
Given that Immune-related genes expressed differentially in the subgroups of neoadjuvant radiotherapy and chemotherapy, we suggested that there may be different tumor immune microenvironments between the subgroups. Our data revealed a significant difference in the distribution of some immune cells between the responder group and the non-responder group in GSE45404. In the responder group, CD4+ naive T cells, Th1(type 1 CD4+ T helper cells) increased significantly. It is consistent with the current understanding, as CD4+ naive T cells differentiate into effector T cells (CD4+ helper cells) after stimulation, and Th1 cells produce IL-2 and IFNγ and favor cellular immunity (acting on CD8+ cytotoxic T cell, NK cells, and macrophages). This immune contexture predicts better outcomes in patients with colorectal cancer (42, 43). Previous studies have shown that Pre-nCRT CD8+TILs, CD4+TILs, and MDSC-TILs are sensitive predictors of response to CRT, and high CD8+TILs are associated with a better prognosis (44). We did not observe this phenomenon. In addition, the results of GSE45404 could not be confirmed in GSE35452. Perhaps limited by the limited sample size, the differences observed in GSE45404 are not generalizable. Or different nCRT schemes and different assessment protocols between GSE45404 and GSE35452 databases caused this result. In this regard, we revised the paper.
There were several limitations in our study that should be acknowledged. First, our study was based on publicly available data sets, and it was not possible to obtain complete clinical information (like pCR, recurrence, survival) and demographic data for each patient. Because of insufficient data, our model cannot be used to predict pCR, survival time, and prediction of disease recurrence. Second, as it was a retrospective study, the potential bias relating to unbalanced clinical-pathological features and treatment heterogeneity like not wholly same nCRT scheme and assessment protocol cannot be ignored. Prospective studies are further required to validate the results. Third, among the 15 DEIRG, there were 3 HLA class II genes. These are highly variable genes, and their expression couldn’t be appropriately addressed using microarrays. This factor affected the predictive performance of our model. Forth, Specimens used in this study were obtained from biopsy, and samples from the biopsy could not reflect the tumor because of tumor heterogeneity. Finally, we have not yet added experiments to explore the mechanism behind the immune-related gene signature, and experimental studies on these immune genes are greatly needed. Even so, our findings might provide some reference value for further research in the functional roles of these immune-related genes.
Conclusion
Overall, our data revealed that the DEIRGs signature could help predict the outcomes of nCRT, and the DEIRGs‐based SVM model could monitor the efficacy of nCRT in LARC. These findings may suggest that immune response plays a vital role in the therapeutic effects of chemoradiation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
LQ is mainly responsible for the layout of research ideas, data collection and processing, and the writing of papers. BG and XL participated in data collection and revision of the paperwork. XS made critical revisions to this paper and approved the final version of the article. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.784479/full#supplementary-material
Supplementary Figure 1 | Enriched Gene Ontology (GO) pathways of the overlapping DEGs in GSE87211 and GSE45404 dataset in biological processes.
Supplementary Figure 2 | Comparison between the fractions of immune cells in the responder (R) and non-responder (NR) subgroup of the GSE35452 cohort via the ImmuCellAI method.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71:7–33. doi: 10.3322/caac.21654
2. Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen YJ, Ciombor KK, et al. Rectal Cancer, Version 2.2018, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw (2018) 16:874–901. doi: 10.6004/jnccn.2018.0061
3. Roh MS, Colangelo LH, O'Connell MJ, Yothers G, Deutsch M, Allegra CJ, et al. Preoperative Multimodality Therapy Improves Disease-Free Survival in Patients With Carcinoma of the Rectum: NSABP R-03. J Clin Oncol (2009) 27:5124–30. doi: 10.1200/JCO.2009.22.0467
4. van Gijn W, Marijnen CA, Nagtegaal ID, Kranenbarg EM, Putter H, Wiggers T, et al. Preoperative Radiotherapy Combined With Total Mesorectal Excision for Resectable Rectal Cancer: 12-Year Follow-Up of the Multicentre, Randomised Controlled TME Trial. Lancet Oncol (2011) 12:575–82. doi: 10.1016/S1470-2045(11)70097-3
5. Rödel C, Martus P, Papadoupolos T, Füzesi L, Klimpfinger M, Fietkau R, et al. Prognostic Significance of Tumor Regression After Preoperative Chemoradiotherapy for Rectal Cancer. J Clin Oncol (2005) 23:8688–96. doi: 10.1200/JCO.2005.02.1329
6. Cercek A, Roxburgh CSD, Strombom P, Smith JJ, Temple LKF, Nash GM, et al. Adoption of Total Neoadjuvant Therapy for Locally Advanced Rectal Cancer. JAMA Oncol (2018) 4:e180071. doi: 10.1001/jamaoncol.2018.0071
7. Fokas E, Allgäuer M, Polat B, Klautke G, Grabenbauer GG, Fietkau R, et al. Randomized Phase II Trial of Chemoradiotherapy Plus Induction or Consolidation Chemotherapy as Total Neoadjuvant Therapy for Locally Advanced Rectal Cancer: CAO/ARO/AIO-12. J Clin Oncol (2019) 37:3212–22. doi: 10.1200/JCO.19.00308
8. Dayde D, Tanaka I, Jain R, Tai MC, Taguchi A. Predictive and Prognostic Molecular Biomarkers for Response to Neoadjuvant Chemoradiation in Rectal Cancer. Int J Mol Sci (2017) 18(3):573. doi: 10.3390/ijms18030573
9. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
10. Taube JM, Galon J, Sholl LM, Rodig SJ, Cottrell TR, Giraldo NA, et al. Implications of the Tumor Immune Microenvironment for Staging and Therapeutics. Mod Pathol (2018) 31:214–34. doi: 10.1038/modpathol.2017.156
11. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, et al. Type, Density, and Location of Immune Cells Within Human Colorectal Tumors Predict Clinical Outcome. Science (2006) 313:1960–4. doi: 10.1126/science.1129139
12. Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, et al. Towards the Introduction of the 'Immunoscore' in the Classification of Malignant Tumours. J Pathol (2014) 232:199–209. doi: 10.1002/path.4287
13. Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity (2016) 44:698–711. doi: 10.1016/j.immuni.2016.02.025
14. Mlecnik B, Tosolini M, Kirilovsky A, Berger A, Bindea G, Meatchi T, et al. Histopathologic-Based Prognostic Factors of Colorectal Cancers are Associated With the State of the Local Immune Reaction. J Clin Oncol (2011) 29:610–8. doi: 10.1200/JCO.2010.30.5425
15. El Sissy C, Kirilovsky A, Van den Eynde M, Muşină AM, Anitei MG, Romero A, et al. A Diagnostic Biopsy-Adapted Immunoscore Predicts Response to Neoadjuvant Treatment and Selects Patients With Rectal Cancer Eligible for a Watch-And-Wait Strategy. Clin Cancer Res (2020) 26:5198–207. doi: 10.1158/1078-0432.CCR-20-0337
16. Hu Y, Gaedcke J, Emons G, Beissbarth T, Grade M, Jo P, et al. Colorectal Cancer Susceptibility Loci as Predictive Markers of Rectal Cancer Prognosis After Surgery. Genes Chromosomes Cancer (2018) 57:140–9. doi: 10.1002/gcc.22512
17. Agostini M, Zangrando A, Pastrello C, D'Angelo E, Romano G, Giovannoni R, et al. A Functional Biological Network Centered on XRCC3: A New Possible Marker of Chemoradiotherapy Resistance in Rectal Cancer Patients. Cancer Biol Ther (2015) 16:1160–71. doi: 10.1080/15384047.2015.1046652
18. Watanabe T, Kobunai T, Akiyoshi T, Matsuda K, Ishihara S, Nozawa K. Prediction of Response to Preoperative Chemoradiotherapy in Rectal Cancer by Using Reverse Transcriptase Polymerase Chain Reaction Analysis of Four Genes. Dis Colon Rectum (2014) 57:23–31. doi: 10.1097/01.dcr.0000437688.33795.9d
19. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res (2015) 43:e47. doi: 10.1093/nar/gkv007
20. Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, et al. ImmPort: Disseminating Data to the Public for the Future of Immunology. Immunol Res (2014) 58:234–9. doi: 10.1007/s12026-014-8516-1
21. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc Natl Acad Sci USA (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
22. Yu G, Wang LG, Han Y, He QY. Clusterprofiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics (2012) 16:284–7. doi: 10.1089/omi.2011.0118
23. Luo W, Brouwer C. Pathview: An R/Bioconductor Package for Pathway-Based Data Integration and Visualization. Bioinformatics (2013) 29:1830–1. doi: 10.1093/bioinformatics/btt285
24. Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, et al. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv Sci (Weinh) (2020) 7:1902880. doi: 10.1002/advs.201902880
25. Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genomics Proteomics (2018) 15:41–51. doi: 10.21873/cgp.20063
26. Noble WS. What Is a Support Vector Machine? Nat Biotechnol (2006) 24:1565–7. doi: 10.1038/nbt1206-1565
27. Guo Y, Cheng J, Ao L, Li X, Guan Q, Zhang J, et al. Discriminating Cancer-Related and Cancer-Unrelated Chemoradiation-Response Genes for Locally Advanced Rectal Cancers. Sci Rep (2016) 6:36935. doi: 10.1038/srep36935
28. Gim J, Cho YB, Hong HK, Kim HC, Yun SH, Wu HG, et al. Predicting Multi-Class Responses to Preoperative Chemoradiotherapy in Rectal Cancer Patients. Radiat Oncol (2016) 11:50. doi: 10.1186/s13014-016-0623-9
29. Kamran SC, Lennerz JK, Margolis CA, Liu D, Reardon B, Wankowicz SA, et al. Integrative Molecular Characterization of Resistance to Neoadjuvant Chemoradiation in Rectal Cancer. Clin Cancer Res (2019) 25:5561–71. doi: 10.1158/1078-0432.CCR-19-0908
30. Satoshi Y, Hideaki B, Yuichiro T, Koji I, Yoshito K, Shigenori H, et al. Short-Term Results of VOLTAGE-A: Nivolumab Monotherapy and Subsequent Radical Surgery Following Preoperative Chemoradiotherapy in Patients With Microsatellite Stable and Microsatellite Instability-High Locally Advanced Rectal Cancer. J Clin Oncol (2020) 38(15_suppl):4100–0. doi: 10.1200/JCO.2020.38.15_suppl.4100
31. Abele R, Tampé R. The ABCs of Immunology: Structure and Function of TAP, the Transporter Associated With Antigen Processing. Physiol (Bethesda) (2004) 19:216–24. doi: 10.1152/physiol.00002.2004
32. El Hage F, Durgeau A, Mami-Chouaib F. TAP Expression Level in Tumor Cells Defines the Nature and Processing of MHC Class I Peptides for Recognition by Tumor-Specific Cytotoxic T Lymphocytes. Ann NY Acad Sci (2013) 1283:75–80. doi: 10.1111/j.1749-6632.2012.06777.x
33. Durgeau A, El Hage F, Vergnon I, Validire P, de Montpréville V, Besse B, et al. Different Expression Levels of the TAP Peptide Transporter Lead to Recognition of Different Antigenic Peptides by Tumor-Specific CTL. J Immunol (2011) 187:5532–9. doi: 10.4049/jimmunol.1102060
34. Ekmekcioglu S, Davies MA, Tanese K, Roszik J, Shin-Sim M, Bassett RL, et al. Inflammatory Marker Testing Identifies CD74 Expression in Melanoma Tumor Cells, and Its Expression Associates With Favorable Survival for Stage III Melanoma. Clin Cancer Res (2016) 22:3016–24. doi: 10.1158/1078-0432.CCR-15-2226
35. Korbecki J, Kojder K, Simińska D, Bohatyrewicz R, Gutowska I, Chlubek D, et al. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of the Ligands of Receptors CCR1, CCR2, CCR3, and CCR4. Int J Mol Sci (2020) 21(21):8412. doi: 10.3390/ijms21218412
36. Verhoeven Y, Tilborghs S, Jacobs J, De Waele J, Quatannens D, Deben C, et al. The Potential and Controversy of Targeting STAT Family Members in Cancer. Semin Cancer Biol (2020) 60:41–56. doi: 10.1016/j.semcancer.2019.10.002
37. Ivashkiv LB. Ifnγ: Signalling, Epigenetics and Roles in Immunity, Metabolism, Disease and Cancer Immunotherapy. Nat Rev Immunol (2018) 18:545–58. doi: 10.1038/s41577-018-0029-z
38. Batlle E, Massagué J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity (2019) 50:924–40. doi: 10.1016/j.immuni.2019.03.024
39. Fang M, Li Y, Huang K, Qi S, Zhang J, Zgodzinski W, et al. IL33 Promotes Colon Cancer Cell Stemness via JNK Activation and Macrophage Recruitment. Cancer Res (2017) 77:2735–45. doi: 10.1158/0008-5472.CAN-16-1602
40. Chang CC, Lin BR, Wu TS, Jeng YM, Kuo ML. Input of Microenvironmental Regulation on Colorectal Cancer: Role of the CCN Family. World J Gastroenterol (2014) 20:6826–31. doi: 10.3748/wjg.v20.i22.6826
41. Bonamy C, Sechet E, Amiot A, Alam A, Mourez M, Fraisse L, et al. Expression of the Human Antimicrobial Peptide β-Defensin-1 is Repressed by the EGFR-ERK-MYC Axis in Colonic Epithelial Cells. Sci Rep (2018) 8:18043. doi: 10.1038/s41598-018-36387-z
42. Zeitoun G, Sissy CE, Kirilovsky A, Anitei G, Todosi AM, Marliot F, et al. The Immunoscore in the Clinical Practice of Patients With Colon and Rectal Cancers. Chirurgia (Bucur) (2019) 114:152–61. doi: 10.21614/chirurgia.114.2.152
43. Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The Immune Contexture in Human Tumours: Impact on Clinical Outcome. Nat Rev Cancer (2012) 12:298–306. doi: 10.1038/nrc3245
44. Teng F, Meng X, Kong L, Mu D, Zhu H, Liu S, et al. Tumor-Infiltrating Lymphocytes, Forkhead Box P3, Programmed Death Ligand-1, and Cytotoxic T Lymphocyte-Associated Antigen-4 Expressions Before and After Neoadjuvant Chemoradiation in Rectal Cancer. Transl Res (2015) 166:721–732.e1. doi: 10.1016/j.trsl.2015.06.019
Keywords: rectal carcinoma, neoadjuvant chemoradiotherapy, immune-related genes, Bioinformatics analysis, prognosis
Citation: Qian L, Lai X, Gu B and Sun X (2022) An Immune-Related Gene Signature for Predicting Neoadjuvant Chemoradiotherapy Efficacy in Rectal Carcinoma. Front. Immunol. 13:784479. doi: 10.3389/fimmu.2022.784479
Received: 27 September 2021; Accepted: 07 April 2022;
Published: 06 May 2022.
Edited by:
Nicolas Dulphy, Université de Paris, FranceReviewed by:
Zhen Zhang, Fudan University, ChinaShenghui Huang, Fujian Medical University Union Hospital, China
Anamaria Aranha Camargo, Hospital Sirio Libanes, Brazil
Copyright © 2022 Qian, Lai, Gu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaonan Sun, sunxiaonan@zju.edu.cn
 Liwen Qian
Liwen Qian Xiaojing Lai
Xiaojing Lai Benxing Gu
Benxing Gu Xiaonan Sun
Xiaonan Sun