- 1Nicholas School of the Environment, Duke University, Durham, NC, United States
- 2Department of Biology, Duke University, Durham, NC, United States
- 3Center for Genomic and Computational Biology, Duke University, Durham, NC, United States
Mitochondria are central players in host immunometabolism as they function not only as metabolic hubs but also as signaling platforms regulating innate immunity. Environmental exposures to mitochondrial toxicants occur widely and are increasingly frequent. Exposures to these mitotoxicants may pose a serious threat to organismal health and the onset of diseases by disrupting immunometabolic pathways. In this study, we investigated whether the Complex I inhibitor rotenone could alter C. elegans immunometabolism and disease susceptibility. C. elegans embryos were exposed to rotenone (0.5 µM) or DMSO (0.125%) until they reached the L4 larval stage. Inhibition of mitochondrial respiration by rotenone and disruption of mitochondrial metabolism were evidenced by rotenone-induced detrimental effects on mitochondrial efficiency and nematode growth and development. Next, through transcriptomic analysis, we investigated if this specific but mild mitochondrial stress that we detected would lead to the modulation of immunometabolic pathways. We found 179 differentially expressed genes (DEG), which were mostly involved in detoxification, energy metabolism, and pathogen defense. Interestingly, among the down-regulated DEG, most of the known genes were involved in immune defense, and most of these were identified as commonly upregulated during P. aeruginosa infection. Furthermore, rotenone increased susceptibility to the pathogen Pseudomonas aeruginosa (PA14). However, it increased resistance to Salmonella enterica (SL1344). To shed light on potential mechanisms related to these divergent effects on pathogen resistance, we assessed the activation of the mitochondrial unfolded protein response (UPRmt), a well-known immunometabolic pathway in C. elegans which links mitochondria and immunity and provides resistance to pathogen infection. The UPRmt pathway was activated in rotenone-treated nematodes further exposed for 24 h to the pathogenic bacteria P. aeruginosa and S. enterica or the common bacterial food source Escherichia coli (OP50). However, P. aeruginosa alone suppressed UPRmt activation and rotenone treatment rescued its activation only to the level of DMSO-exposed nematodes fed with E. coli. Module-weighted annotation bioinformatics analysis was also consistent with UPRmt activation in rotenone-exposed nematodes consistent with the UPR being involved in the increased resistance to S. enterica. Together, our results demonstrate that the mitotoxicant rotenone can disrupt C. elegans immunometabolism in ways likely protective against some pathogen species but sensitizing against others.
Introduction
Increasing evidence reveals that different classes of environmental toxicants can have substantial effects on the function and homeostasis of the immune system of multiple animal species [see reviews (1–7)]. The complexity of the immune system provides a multitude of possible mechanisms by which toxicants may impact immune-related responses. Immunotoxicity may occur through direct interactions with molecules implicated in canonic immunological pathways, or by disrupting the crosstalk between the immune system and other physiological systems (2).
Recent research has shown that energy metabolism and immune responses are intimately connected: Metabolic shifts may directly modulate immune function, and molecules and cells of the immune system regulate metabolic pathways (8). These connections are studied by the recently established research field of immunometabolism (9). Studies in immunometabolism have revealed mitochondria as a key modulator of immune responses (10), which can be also defined as mitoimmunity (11). Some of the main findings regarding the mitochondrial control of immune responses reveal that (i) mitochondrial components, such as mitochondrial DNA and cardiolipin, act as a danger-associated molecular pattern (DAMP) activating cell-autonomous and cell non-autonomous immune responses; (ii) molecules present at the mitochondrial outer membrane form a platform for anti-viral signaling and inflammasome activation; (iii) mitochondrial dynamics (biogenesis, fusion, and fission) have roles in immune-cell activation; and (iv) Krebs cycle intermediates are necessary for immune-related responses and inflammation, in both innate and adaptive immune cells (12). Therefore, it is reasonable to consider that toxicant-induced mitochondrial damage or disruption may have profound effects on the homeostasis of the immune system. Indeed, increasing evidence suggests that mitochondrial dysfunction caused by environmental toxicants is implicated in the onset of several diseases and immune-related disorders (13). Moreover, mitochondria can be particularly vulnerable targets to several classes of environmental toxicants, including metals, polycyclic aromatic hydrocarbons (PAHs), and pesticides (14, 15). Despite this awareness, little is known about how mitotoxicants affect immunometabolic processes.
Caenorhabditis elegans, a free-living nematode, has traditionally been utilized in the areas of neuroscience, aging, and developmental biology. Over the past few decades, this model has also been recognized as a valuable tool in revealing fundamental principles in innate immunity and host-pathogen interactions (16). Most recently, C. elegans was also established as an important model for unraveling key concepts in immunometabolism (17). Most of the success in the use of C. elegans in research relies on the fact that this species is a valuable representative of metazoans, including humans. For example, 83% of the worm proteome contains human homologs (18, 19). Importantly, various molecular pathways involved in immunometabolic responses in mammals are conserved in C. elegans (Figure 1) (17, 20, 21). Moreover, the key role of mitochondria as a signaling hub for innate immune responses also seems to be conserved between vertebrates and C. elegans (22, 23).
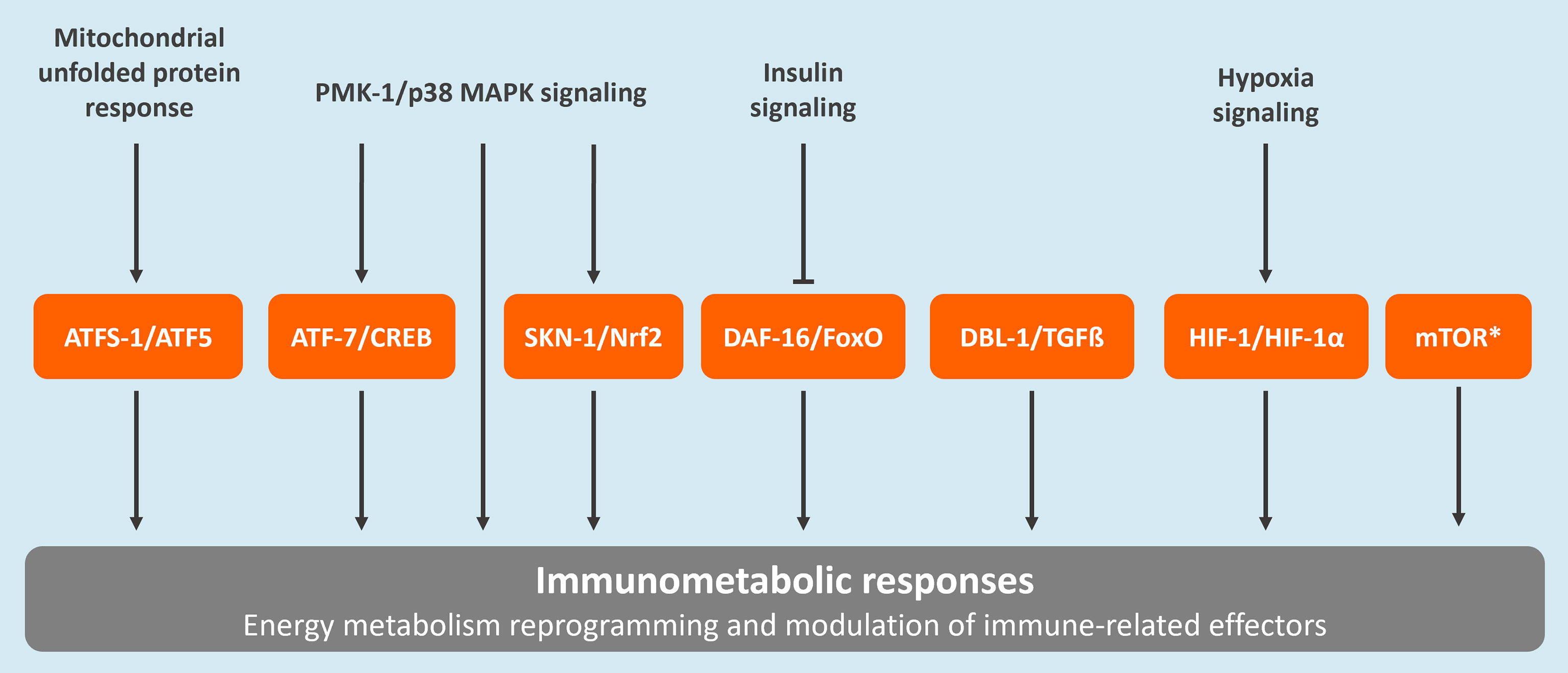
Figure 1 Conserved regulatory pathways associated to immunometabolism in C. elegans. These include the mitochondrial unfolded protein response (UPRmt), the p38 mitogen-activated protein kinase (MAPK) signaling pathway, the insulin/insulin-like growth factor signaling (IIS) pathway, the forkhead box transcription factor O (FoxO)-mediated signaling, the transforming growth factor-beta (TGFβ) signaling pathway, the nuclear factor erythroid 2-related factor 2 (Nrf2) pathway, the hypoxia-Inducible factor (HIF)-1 regulatory pathway, and the mammalian target of rapamycin (mTOR) pathway. The C. elegans protein names are provided followed by the corresponding human homolog. *The mTOR pathway is regulated by two complexes, TORC1 and TORC2. The main proteins composing these complexes, which are conserved between C. elegans and humans, are the following: LET-363/TOR, DAF-15/Raptor, and RICT-1/Rictor (20).
Over the past years, C. elegans has also emerged as a prevalent toxicological test organism, being successfully used in drug discovery screens (24) and shown to be predictive of chemical toxicity in mammals, with several reported cases of a conserved mode of toxic action (24). C. elegans has also been proposed as a prominent model in environmental toxicological studies (25, 26). Thus, research using this model may reveal valuable insights regarding immunometabolism toxicity mechanisms and environmental health.
In this study, we exposed nematodes to the pesticide rotenone, a classic complex I inhibitor, to investigate the role of specific mitochondrial disruption on modulating conserved immuno-metabolic molecular pathways and disease susceptibility. We found that rotenone disrupted nematode growth and mitochondrial bioenergetics, as expected, and modulated the expression of genes and pathways mostly involved in detoxification, energy metabolism, and pathogen defense, including genes related to conserved immunometabolic pathways such as UPRmt and HIF. Among the down-regulated DEG, most of the known genes were involved in immune defense, revealing a major role of rotenone in suppressing immune-related genes, which (according to the WormExp gene overlap analysis tool) included genes upregulated by Pseudomonas aeruginosa resistance. In line with these results, rotenone-exposed animals were more susceptible to P. aeruginosa (PA14) challenges. In contrast, rotenone rendered animals more resistant when challenged with another pathogen, Salmonella enterica (SL1344). Finally, by evaluating the expression of the UPRmt reporter gene hsp-6, we propose that the UPRmt pathway might mediate rotenone-induced resistance to S. enterica.
Materials and Methods
C. elegans and Bacterial Strains
Bristol N2 and SJ4100 (zcIs13[hsp-6p::gfp]) strains, were obtained from the Caenorhabditis Genetics Center (CGC). The following bacterial strains were used in this study: Escherichia coli OP50-1 [SmR] and HB101, which were obtained from the CGC, and P. aeruginosa (PA14) and S. enterica serovar Typhimurium 1344 (SL1344), which were provided by the laboratory of Alejandro Aballay. All C. elegans strains used were grown and maintained at 20°C on K-agar plates (51 mM sodium chloride, 32 mM potassium chloride, 1 mM calcium chloride, 1 mM magnesium sulfate, 10 µg/mL cholesterol, 0.25% peptone, 2% agar) seeded with OP50 E. coli bacteria.
Rotenone Exposures
K-agar 100 mm Petri plates containing a non-starved mixed population of nematodes and plenty of embryos were washed with K-medium (51 mM sodium chloride, 32 mM potassium chloride) a few times until no animals were observed in the plates, leaving only the embryos. These plates were then used to obtain roughly age-synchronized young gravid adults 3-4 days later, which were used to obtain intact, closely age-synchronized embryos by dissolving adults for 8-10 min in freshly prepared sodium hydroxide bleach solution (3.5 mL K-medium, 500 µL 5N NaOH, 1 mL sodium hypochlorite). Age-synchronized embryos were then added to Erlenmeyer exposure flasks containing complete K-medium (51 mM sodium chloride, 32 mM potassium chloride, 3 mM calcium chloride, 3 mM magnesium sulfate, 5 µg/mL cholesterol) and live HB101 E. coli at a final OD600 of 2.0. For all assays, embryos were added at a density of 1 embryo/100 µL, except for the RNA-seq assay in which embryos were added at a density of 1 embryo/µL (it is important to note that the increased nematode density did not significantly decrease HB101 concentration; data not shown). 1 mM rotenone frozen stocks (-80°C) were previously prepared in pure dimethyl sulfoxide (DMSO), then freshly diluted with pure DMSO to obtain 500 µM rotenone, and further diluted with complete K-medium to 100 µM rotenone in 25% DMSO. This rotenone solution was then used to add to the exposure flasks to a final concentration of 0.5 µM rotenone and 0.125% DMSO as a solvent. In rotenone groups (Rot), rotenone was added at the start of the exposure and, because of its short half-life, re-added after 24 and 48 h of exposure. In the solvent control groups (Ctrl), the solvent solution (25% DMSO diluted in complete K-medium to a final concentration of 0.125%) was added at the start of the exposure and re-added after 24 h. Exposure flasks were maintained inside a 20°C incubator under constant agitation. Nematodes were then harvested when most of the animals reached the L4 larval stage, which was after 52 h for the solvent control animals (and for the growth experiment with non-stage-synchronized rotenone-exposed animals) and was either 68 or 76 h for the L4 stage-synchronized rotenone-exposed animals (Figure 2A).
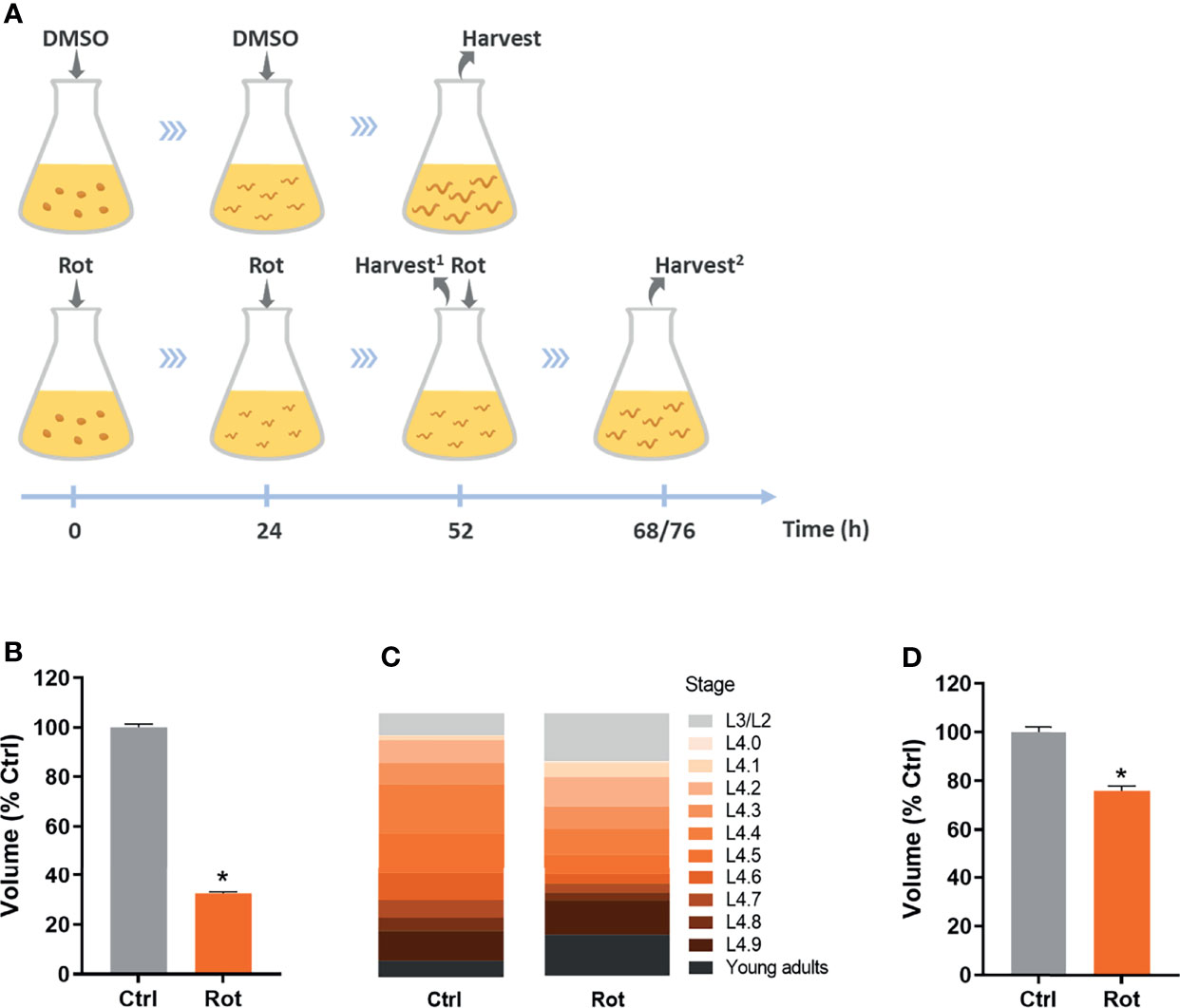
Figure 2 Rotenone impairs C. elegans growth and development. Solvent control (Ctrl) and rotenone-exposed (Rot) nematodes were assessed for size and developmental stage as described in the methods. (A) Representative image of the exposure design: dimethyl sulfoxide (DMSO) and 0.5 µM rotenone solutions containing 0.125% DMSO were added at the indicated time points and nematodes were harvested when most of the animals were at the L4 larval stage (Harvest2) for all the assays, except for results shown in Figure 1B, where animals were harvested at the same time point as the control animals (52 h; Harvest1) (see methods for further details). (B) Volume of nematodes exposed to rotenone for 52 h compared to solvent control (% Ctrl). (C) Sub-stage distribution and (D) volume of Ctrl and Rot-treated animals after L4 stage synchronization. * represents significant difference (p < 0.05) compared to control.
Nematode Growth and Developmental Stage
N2 nematodes exposed to rotenone or DMSO were harvested after 52 h and after 68/76 h (rotenone-exposed animals only) for size measurements. After exposure, animals were rinsed with K-medium and transferred to K-agar plates without any bacteria for imaging. Once the plates were air-dried, they were then placed on a Keyence BZ-X700 microscope for nematode imaging. Images were analyzed using the WormSizer (27) plugin for Fiji/ImageJ image software (28). A subset of the animals was transferred to 96 Well Microplates, µClear® (Greiner Bio-One) containing 100 µL of K-medium containing 50 mM sodium azide for immobilization, and vulval development was examined to precisely determine the larval stage and L4 sub-stages of the animals (29).
Mitochondrial Functional Parameters
N2 nematodes exposed to rotenone or DMSO were harvested once most of the animals reached the L4 stage. Nematodes oxygen consumption rate (OCR) was measured using the Seahorse Xfe24 Extracellular Flux Analyzer (Agilent Technologies, Inc.) as described by Luz and colleagues (30), except that we used the 24-well Seahorse XF24 V7 PS Cell Culture microplates, not islet plates. Pharmacological inhibitors of the electron transport chain (ETC) were used to assess different mitochondrial functional parameters. Dicyclohexylcarbodiimide (DCCD) was used to inhibit ATP synthase and provide a measure of the amount of oxygen consumption coupled to ATP production (ATP-linked OCR). Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) was used to uncouple ATP production from oxygen consumption, providing a measure of maximal OCR. By subtracting the basal OCR from the maximal OCR, we calculated the spare respiratory capacity as an indication of an organism’s ability to respond to increased energy demands. Finally, sodium azide was used to inhibit cytochrome c oxidase, providing a measure of non-mitochondrial OCR, and by subtracting the azide-inhibited OCR from the DCCD-inhibited OCR we calculated proton leak. Calculations were made according to Luz and colleagues (30), with the exception that all basal OCR readings, the lowest three OCR readings after DCCD injection, and the highest three OCR readings after FCCP injection were used. Moreover, the first OCR reading after any drug injection was not considered. After the OCR readings were finalized, the number of animals in each well was counted and OCR values were normalized to the average nematode volume (WormSizer) within each experiment. At least three replicate wells were used for each inhibitor within each treatment group. The experiment was repeated at least three times.
mRNA-seq Analysis
N2 nematodes exposed to rotenone or DMSO were harvested for mRNA-seq analysis once most of the animals reached the L4 stage. Samples containing approximately 1,000 nematodes from each group were rinsed several times with K medium (51 mM sodium chloride, 32 mM potassium chloride) until most of the bacteria were cleared out, then flash-frozen and maintained at -80°C. Only one exposure flask was used per group per experiment (no technical replicates) and the entire experiment was repeated four times using independent biological replicates (N=4 per group).
Nematode samples were then used for RNA extraction using 1 ml TRIzol Reagent (Invitrogen) according to the manufacturer’s protocol with the exception that approximately 100 µL of acid-washed sand (Sigma-Aldrich), measured with a graduated 1.7 mL microcentrifuge tube, was added to each sample at the beginning of the protocol to aid homogenization. RNA was eluted in nuclease-free water and stored at -80°C until further use. A nanodrop spectrophotometer (ThermoFisher) was used to assess the purity of the extracted RNA and the Qubit RNA HS Assay kit (ThermoFisher) was used to determine concentration. All RNA samples used for mRNA-Seq library preparation had a 260/280 absorbance ratio over 1.8. Libraries were prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs) starting with 500 ng total RNA per library and 9 cycles of PCR. Libraries were sequenced on Illumina NovaSeq 6000 to obtain 50 bp paired-end reads.
For mRNA-seq analysis, version WS273 of the C. elegans genome was used for mapping. Paired-end reads were mapped with bowtie (31) with the following settings: bowtie -I 0 -X 500 –chunkmbs 256 -k 1 -m 2 -S -p 2. The average mapping efficiency across all samples was 63.3% with a standard deviation of 0.74%. HTSeq version 0.11.2 (32) was used to count reads mapping to the WS273 canonical geneset. Count data was restricted to include only protein-coding genes (20,127), and then further restricted to include only genes with CPM > 1 in at least 4 libraries (14,426 genes). Differential expression analysis was then performed using edgeR version 3.24.3 (33). The web-based WormCat tool (34) was used to perform gene set enrichment analysis of the differentially expressed genes (DEG). For this analysis, we used the complete list of significantly DEG (FDR < 0.05) upon rotenone treatment which was mapped to the C. elegans “Whole genome v2”, following the standard WormCat Flow analysis parameters. The WormExp v 1.0 application for gene set enrichment analysis for C. elegans (https://wormexp.zoologie.uni-kiel.de/wormexp/) (35) and the “module-weighted annotations” analysis tool DEXICA (http://genemodules.org/) (36) were also used to further explore genes and pathways potentially involved in the rotenone-induced effects. For the WormExp analysis, only DEG annotated as immune or pathogen defense-related were used (19 genes) and query was run against the “microbes” category using the application’s default parameters. For the “module-weighted annotations” analysis, the whole transcriptomic data from solvent control vs. rotenone-exposed nematodes was used, following the creators’ instructions, using default parameters.
Preparation of Pathogen Plates
P. aeruginosa PA14 and S. enterica SL1344 bacteria were picked from a frozen 25% glycerol stock, spread on a Luria-Bertani (LB) agar plate, and allowed to grow at 37°C overnight. Colony plates were stored sealed with parafilm at 4°C and used for up to one week. Bacterial cultures were prepared by placing 3–4 colonies into a 4 mL LB broth and growing at 37°C under constant shaking for 8 h. To prepare pathogen slow killing (SK) plates, modified nematode growth medium (NGM) (37) (0.35% instead of 0.25% peptone) Petri plates (35 mm) were seeded with 50 μL of each bacterial culture, which was spread over the entire plate (full lawn) using a sterile glass rod, and then placed at 37°C overnight. Freshly prepared pathogen SK plates equilibrated to room temperature were then used for the hsp-6p::GFP expression and survival assays.
Survival
N2 nematodes exposed to rotenone or DMSO were harvested once most of the animals reached the L4 stage, placed into OP50 K-agar plates for 48 h, and then transferred to P. aeruginosa PA14 or S. enterica SL1344 SK plates and maintained at 25°C. Animals were transferred to fresh SK pathogen plates daily during the reproductive period. Survival was scored twice a day (early morning and late afternoon) for P. aeruginosa and once a day for S. enterica. Animals were considered dead when no pharyngeal pumping was observed, or nematodes did not respond to touch. Animals that crawled out of the agar were censored. Two or three replicate plates were used per exposure group per experiment, which was repeated three times.
hsp-6p::GFP Expression
SJ4100 nematodes exposed to rotenone or DMSO were harvested once most of the animals reached the L4 stage and placed into OP50 K-agar plates for at least 1 h and then transferred to fresh P. aeruginosa PA14, S. enterica SL1344, or E. coli OP50 SK plates for 24 h at 25°C. At the end of the exposure, nematodes were transferred to 96 Well Microplates, µClear® (Greiner Bio-One) containing 100 µL of K-medium containing 50 mM sodium azide for immobilization. Nematodes were imaged using a Keyence BZ-X700 microscope equipped with a GFP filter. Images were transformed to 32-bit and nematodes were manually outlined using the polygon selection tool using Fiji/ImageJ image software (28). Mean gray values within the selection were obtained from each nematode and subtracted from the average mean gray value from three selections of the image background. Results are shown as the fold-change of the E. coli OP50 solvent control group and represent 22-45 nematodes analyzed per experiment, which was repeated twice.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism 9 (GraphPad Software, LLC©). The size and mitochondrial functional parameters data were compared between groups using a Student’s T-test. Nematode survival was plotted as a nonlinear regression curve and the significance of differences (p<0.05) between survival curves were analyzed using the Mantel-Cox log-rank test. Survival scoring results of each experiment and statistical analysis of rotenone vs. solvent control (DMSO) animals are provided in the Supporting Information file. The time points at which mortality reaches 50% of the animals (LT50) were obtained from the best-fit IC50 value given for each curve. All curves presented R squared ≥ 0.86. The hsp-6p::GFP expression data were analyzed via two-way ANOVA followed by pairwise comparisons using Fisher’s LSD test to determine significant differences between groups (two independent variables: toxicant exposure and bacterial food source), adjusting the significance level to α = 0.05/8 = 0.00625 (Bonferroni correction) to control for multiple comparisons.
Results
Rotenone Impairs Growth and Developmental Rate
Age-synchronized C. elegans embryos were exposed to rotenone or DMSO throughout larval development (Figure 2A). As a first approach to assess if our rotenone developmental exposure protocol was promoting mitochondrial dysfunction, we performed nematode growth and development measurements, which are parameters commonly disturbed by mitochondrial inhibitors. As expected, nematodes exposed to rotenone presented a significantly smaller body size than animals exposed to DMSO only (Figure 2B). Nematode anatomic inspections using a stereo microscope allowed us to identify that rotenone-exposed animals were not only smaller but developmentally delayed (data not shown). By staging the vulval development of nematodes, we were able to determine that rotenone-exposed nematodes required on average 20 more hours to reach the mid-L4 larval stage and displayed a higher spread of animals at different stages than the solvent control group (Figure 2C). Nonetheless, even when rotenone-exposed nematodes were harvested at the time-point when the majority of the animals were at the L4 stage (stage-synchronized rotenone-exposed animals), they were still significantly smaller than solvent control animals (Figure 2D). These results suggest a rotenone-dependent impairment of mitochondrially derived energy to promote C. elegans growth and development.
Rotenone Disrupts Mitochondrial Bioenergetics
Real-time oxygen consumption of live nematodes was monitored in rotenone or DMSO-exposed animals at the L4 larval stage using different mitochondrial inhibitors (Figure 3A). Total basal, mitochondrial basal, and ATP-linked OCR were all significantly lower in rotenone-exposed animals (Figures 3B–D). Rotenone also caused an increase in proton leak (Figure 3E), and a decrease in maximal OCR (Figure 3F), while no significant changes were detected in the spare respiratory capacity (Figure 3G). The non-mitochondrial OCR was also significantly reduced in rotenone-exposed animals (Figure 3H). Together, these results reveal a rotenone-induced disruption of C. elegans mitochondrial bioenergetics towards a lower mitochondrial efficiency (mitochondrial respiration coupled to ATP production), which are in line with the abovementioned findings on growth impairment.

Figure 3 Rotenone disrupts C. elegans mitochondrial function. Solvent control (Ctrl) and rotenone-exposed (Rot) nematodes were assessed for oxygen consumption using a Seahorse XFe24 bioanalyzer as described in the methods. Oxygen consumption rates (OCR) were normalized to the average nematode volume within each experiment. (A) Representative image of the mitochondrial function parameters assessed using the inhibitors dicyclohexylcarbodiimide (DCCD), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), and sodium azide (NaAz). (B) Total basal OCR, (C) mitochondrial basal OCR, (D) ATP-linked OCR, (E) proton leak, (F) maximal OCR, (G) spare respiratory capacity, and (H) non-mitochondrial OCR are presented. Student’s T-test was performed for each parameter. Bars represent mean ± SEM. * represent significant difference (p < 0.05) between groups.
Rotenone Alters the Transcript Levels of Genes Related to Metabolism, Immune and Stress Response Pathways
To investigate the role of specific mitochondrial disruption on modulating conserved immunometabolic molecular pathways, we performed a whole transcriptome mRNA-seq analysis. Our results revealed that rotenone-treated animals displayed a total of 179 differentially expressed genes (DEG), among which 134 were up-regulated (Supplementary Table 1) and 45 were down-regulated (Supplementary Table 2). By applying the WormCat gene enrichment analysis tool, we detected an enrichment of genes involved in detoxification, energy metabolism, or pathogen defense (Figures 4A–C). Among the known genes which were up-regulated, we can highlight genes involved in detoxification, from the cytochrome P450 (CYP) and UDP-glucuronosyltransferase (UGT) families, as well as beta-oxidation and other lipid and mitochondrial metabolism pathways (Figure 4D). Despite a few genes associated with pathogen defense being represented within the up-regulated list of genes, interestingly, these comprised the majority of the down-regulated set of genes (Figure 4E). Some genes involved in lipid metabolism were also down-regulated, with fat-7 being found as the most largely down-regulated gene (Figure 4E). This gene-category enrichment analysis, thus, revealed a major role of rotenone in modulating the expression of several genes related to immune and metabolic function.
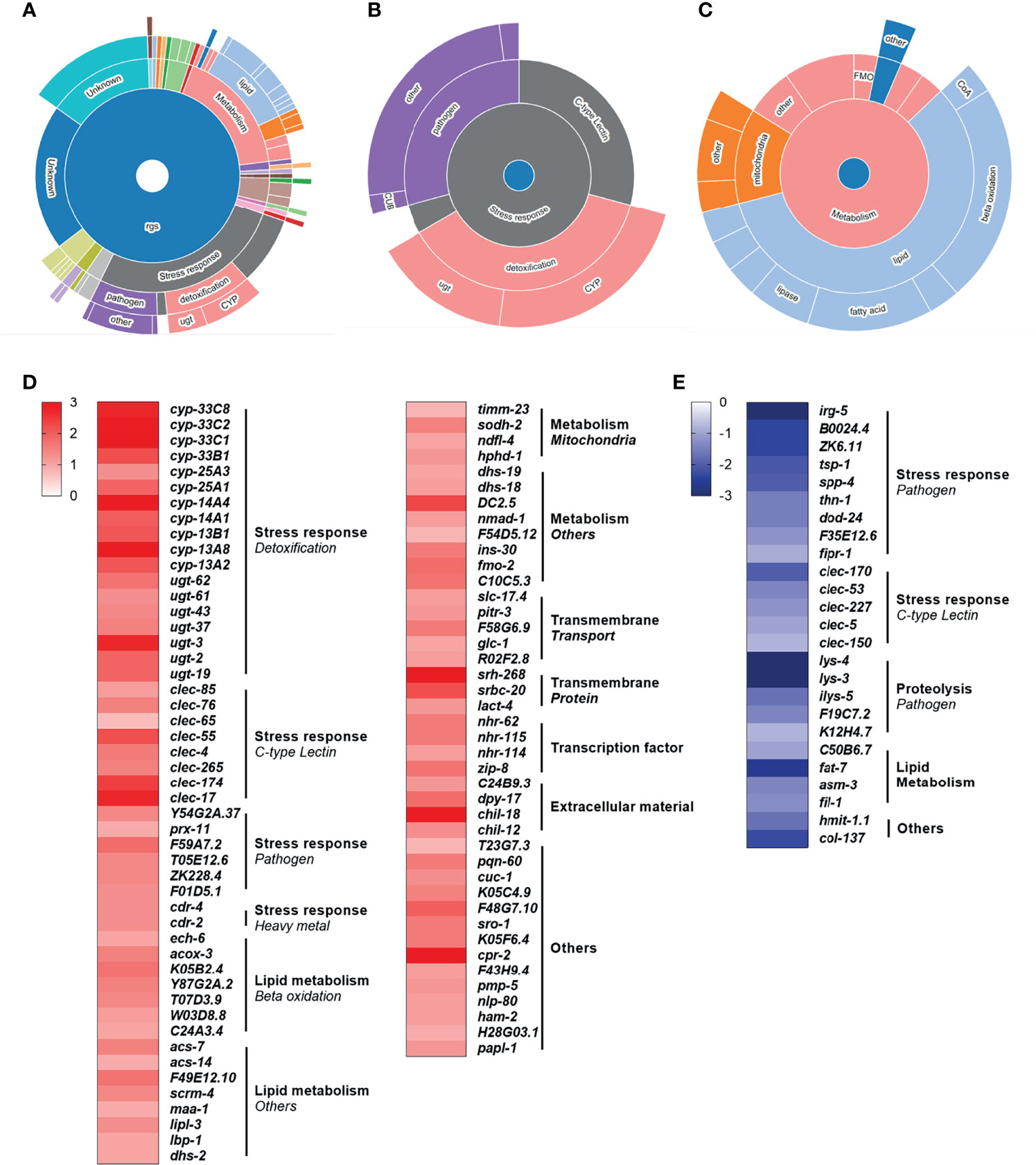
Figure 4 Rotenone modulates genes related to metabolism, immune and stress response pathways. All 179 differentially expressed genes (DEG) from the rotenone treatment were submitted to WormCat analysis for annotation and visualization of enrichment data. (A) Sunburst diagram showing significantly enriched categories, highlighting the two major broad enriched categories: (B) stress response and (C) metabolism. Heatmap representing the log fold-change of (D) up-regulated and (E) down-regulated genes with known function are shown grouped by gene categories (bold) and subcategories (italic).
We further applied our whole transcriptome data (log fold-change of all 14,426 quantified genes) to the “module-weighted annotations” analysis tool 1 (36). This analysis compares transcriptomic data to 209 modules that represent patterns of transcriptional regulation (co-expressed genes) in C. elegans, representing different conditions such as responses to changes in the environment (e.g., starvation, exposure to xenobiotics), genes regulated by transcriptions factors (e.g., ATFS-1, DAF-16), genes specific to tissues (e.g., neurons, muscle), genes that change during development, and other complex transcriptional responses to genetic, environmental and temporal perturbations. Results from our data set revealed four significantly active modules (Supplementary Figure 1A), representing: (i) activation of the UPRmt, evidenced by a significantly positive association with module #47; (ii) activation of neuronal genes and neuronal peptide signaling, evidenced by a significantly positive association with module #23; and (iii) suppression of hypoxia-inducible factor (HIF-1)-dependent genes, evidenced by a significantly negative association with modules #66 and #169. A list with the main gene ontology process, molecular function, and cellular component associated with each active module is provided in Supplementary Figure 1B. Thus, this analysis allowed us to identify two conserved immunometabolic pathways, the UPRmt and HIF-1 regulatory pathways, that are potentially being disturbed upon rotenone-induced mitochondrial dysfunction.
Rotenone Alters Nematode Susceptibility to Pathogens
To address if the rotenone-induced modulation of immuno-metabolic pathways observed at the gene expression level could translate to organismal-level effects on pathogen susceptibility, DMSO or rotenone-exposed animals were challenged with the gram-negative pathogens P. aeruginosa (PA14) or S. enterica (SL1344). Survival results revealed that while rotenone-exposed nematodes were more susceptible to P. aeruginosa (Figure 5A), they were overall more resistant to S. enterica (Figure 5B). Based on the curve fit data, we were able to obtain the time point at which mortality reaches 50% of the animals (LT50), which was 68.7h and 58.0h for solvent control and rotenone-exposed nematodes, respectively, challenged with P. aeruginosa. This corresponds to a 16% decrease in life expectancy for half of the population tested caused by the rotenone exposures. On the other hand, nematodes challenged with S. enterica presented a 12% increase in LT50 (95.3h and 107.8h for solvent control and rotenone-exposed nematodes, respectively). These results reveal that a specific mitochondrial dysfunction induced by rotenone can alter C. elegans pathogen susceptibility.
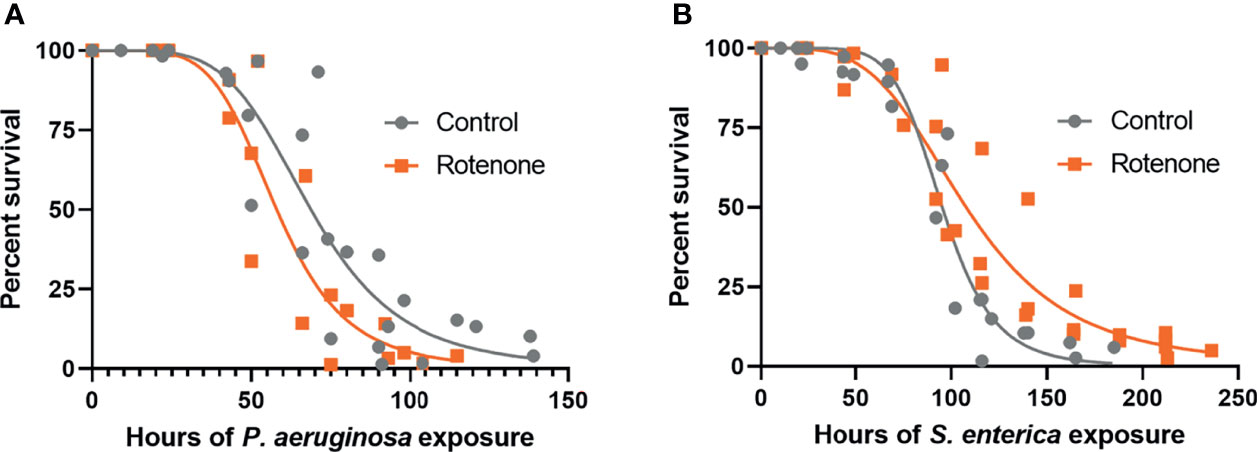
Figure 5 Rotenone alters C. elegans pathogen susceptibility. Solvent control (Control) and rotenone-exposed nematodes (Rotenone) were further continuously exposed to (A) Pseudomonas aeruginosa (PA14) or (B) Salmonella enterica (SL1344) and scored for survival. Curves were fit by nonlinear regression based on the results from three independent experiments for each pathogen exposure and represent a total of 148-197 animals scored (already excluding censored animals) per group. Details about the number of animals in each experiment and statistical results using the Mantel-Cox log-rank test are shown in Supplementary Table 1.
The Gene hsp-6 Is Upregulated by Rotenone Exposure During Pathogen Infections
The potential role of the mitochondrial unfolded protein response (UPRmt) pathway in the differential pathogen susceptibility of rotenone-exposed animals to P. aeruginosa or S. enterica was investigated by quantification of hsp-6p::GFP expression in DMSO and rotenone-treated nematodes further exposed for 24 h to E. coli (OP50), P. aeruginosa (PA14) and S. enterica (SL1344). hsp-6p::GFP expression was significantly upregulated in rotenone-treated animals at similar levels (1.4-1.5 fold-induction) in all bacterial exposures (Figure 6). Nonetheless, P. aeruginosa exposure alone (solvent control animals) suppressed hsp-6p::GFP expression in comparison to animals that were fed the control bacteria E. coli (OP50), and rotenone treatment was able to restore hsp-6p::GFP expression only to the level of solvent control OP50-fed nematodes (Figure 6). In line with the “module-weighted annotations” analysis, these results are also consistent with an activation of the UPRmt pathway by rotenone. Nonetheless, this apparent activation is limited in the presence of P. aeruginosa, but not S. enterica, which could partly explain our findings on differential pathogen susceptibility.
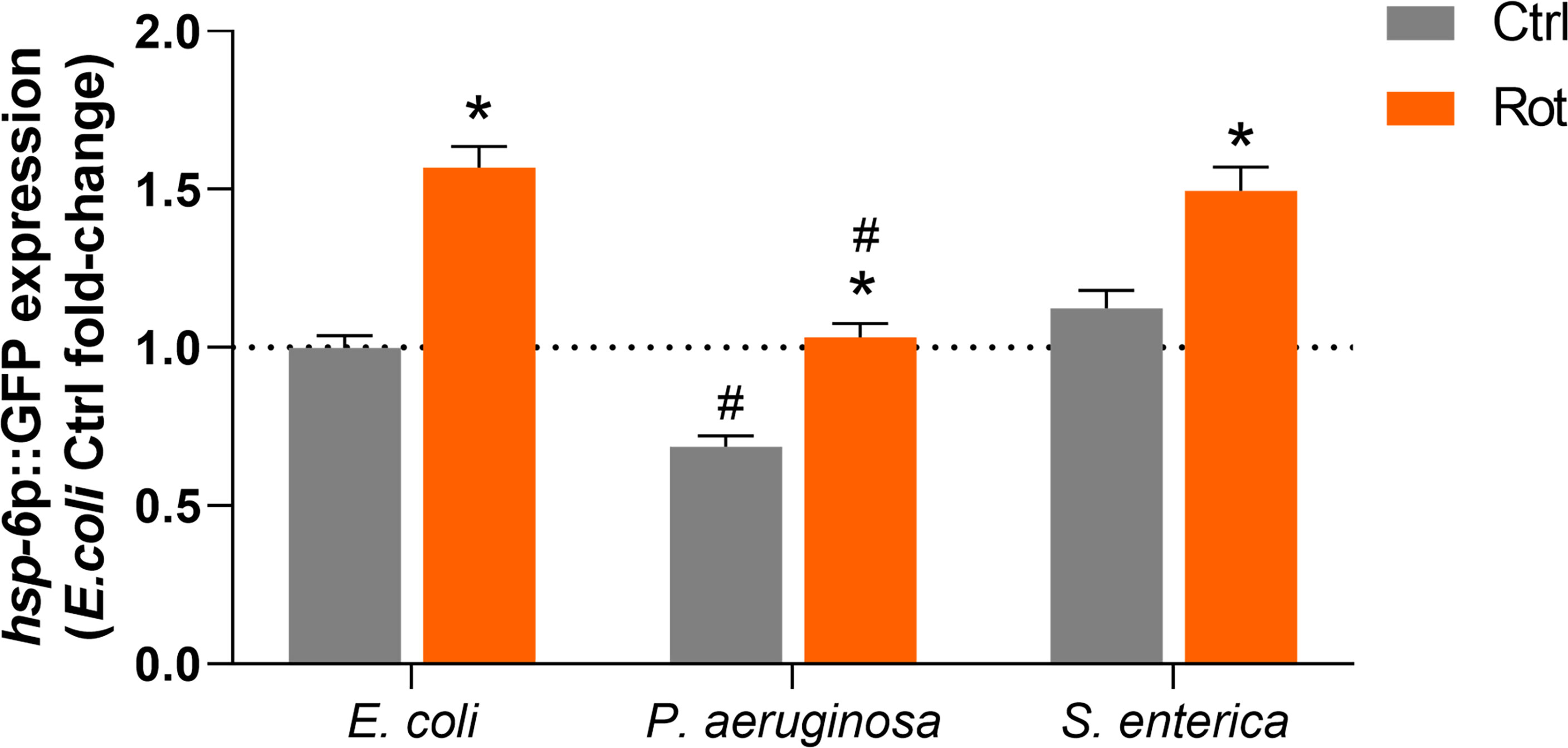
Figure 6 Activation of the mitochondrial unfolded protein response by rotenone. Solvent control (Ctrl) and rotenone-exposed (Rot) SJ4100 nematodes were further exposed for 24 h to Escherichia coli (OP50), Pseudomonas aeruginosa (PA14) or Salmonella enterica (SL1344) and the GFP intensity was measured. The presented hsp-6p::GFP fold-change results are relative to the E. coli OP50 solvent control. A two-way ANOVA with Bonferroni multiple comparison test was performed. Bars represent mean ± SEM. * represent significant difference (p < 0.05) between the rotenone and solvent control exposure groups within each bacterial feeding group. # represent significant difference (p < 0.05) comparing pathogenic bacterial exposures to E. coli within the solvent control or rotenone exposure groups. No significant interaction effect between the two independent variables – ”toxicant exposure” and “bacterial food source” – was detected (p = 0.09). However, each independent variable had a significant effect on hsp-6p::GFP expression (p < 0.0001 for both independent variables).
Discussion
Previous studies from our research group and data from the literature show that different classes of toxicants can promote physiological stress in organisms by changing energy metabolism (14, 38–41) and innate immune responses (1–7, 42–48). However, although the fields of immunometabolism and mitoimmunity have recently been the focus of extensive research, most studies use vertebrate models, and little attention is directed to mitoimmune dysfunction in the context of pollutant exposures.
To start tackling the hypothesis that mitochondrial toxicants can also act as immunometabolic disruptors and alter disease susceptibility, we exposed nematodes to the pesticide rotenone, which is a classic and specific mitochondrial complex I inhibitor. In this study, we opted to perform a developmental rotenone exposure because, at this stage, animals are usually more sensitive to mitochondrial toxicants. This greater sensitivity is because while early developmental stages and growth involve biological processes which require high levels of mitochondrially-derived energy, at these stages, several mitochondrial parameters are limited or greatly reduced (49–53). Moreover, mitochondrial parameters and function are highly variable with developmental stage; thus, it is likely that mitochondrial toxicants can have more profound effects on some developmental windows of susceptibility (14, 41). Furthermore, it is expected and frequently observed that defects should occur during exposures at these early life stages with mitochondrial toxicants (40, 54). Indeed, our results demonstrated that rotenone caused significant impairment in growth (Figures 2B–D) and bioenergetic efficiency (Figures 3A–E). Associations between growth and decreases in mitochondrial efficiency (respiration coupled to ATP production) have already been reported (55, 56). It is important to highlight, however, that despite these rotenone-induced effects in growth and mitochondrial function, no mortality was observed, and animals were able to fully develop to adulthood and reproductive maturation without affecting total brood size (pilot study data in Supplementary Figure 2). Sublethal rotenone exposures as well as mild inhibition of mitochondrial complex I genes have been reported to increase C. elegans lifespan (57–59). Therefore, it is reasonable to believe that, with our rotenone exposure protocol, we did not cause a severe and generalized organismal toxicity but rather a relatively mild mitochondrial stress.
Next, we investigated if this mild and specific mitochondrial stress in rotenone-exposed nematodes could lead to the modulation of immunometabolic pathways through transcriptomic analysis. Gene ontology analysis of the DEG revealed that the great majority of the known genes were implicated in energy metabolism and stress response (Figure 4A). Most of the DEG associated with energy metabolism were up-regulated and annotated as involved in fatty acid β-oxidation (Figure 4C). This result is consistent with a previous study from our group which also detected, through metabolomics analysis, increased fatty acid β-oxidation in adult nematodes exposed to rotenone (39). This increase might be related to a need to replenish acetyl-CoA levels or to boost ATP production upon Complex I inhibition, although the latter hypothesis was not confirmed in our previous work (39). Fatty acid β-oxidation plays a significant role in immunometabolism. For example, increased fatty acid β-oxidation is important for innate immune responses in higher vertebrates such as macrophage polarization towards an anti-inflammatory state (60). In C. elegans, although, to our knowledge, direct associations between fatty acid β-oxidation and innate immunity have not been yet explored, several studies describe the importance of certain fatty acids for pathogen resistance [see review (17)]. For example, the fatty acid oleate, which is synthesized by fat-6 and fat-7 desaturases, is required in C. elegans for the pathogen-mediated induction of immune defense genes, and disruption of oleate synthesis increase nematode susceptibility to P. aeruginosa (61). The fat-7 gene was highly down-regulated by our rotenone treatment, which could have contributed to our observations of increased susceptibility of the rotenone-exposed nematodes to this same pathogen (Figure 5A). Additionally, fat-7 down-regulation was reported to cause growth delay, reduced size, and increased expression of genes in β-oxidation (62, 63). Interestingly, all of these phenotypes were also observed in our rotenone-treated nematodes. These literature reports, together with our findings, indicate that fat-7 down-regulation –thus potentially oleate synthesis disruption– might be a central player governing the rotenone-induced effects on immunometabolic and pathogen susceptibility responses in C. elegans.
The stress-response DEGs were further separated into three subcategories: detoxification, C-type lectins, and pathogens (Figure 4B). Among the xenobiotic metabolism (“detoxification”)-related DEG are genes belonging to the Cytochrome P450 (CYP) and UDP-glucuronosyltransferase (UGT) families, which were identified as up-regulated by our rotenone treatment (Figure 4D). The enzymatic modification (biotransformation) of rotenone in rats and fish has already been shown to involve oxidation by CYPs (64). Evidence of rotenone detoxification by CYPs was also reported in invertebrates such as insects and helminths (65, 66). While we could not find studies describing the direct involvement of UGTs in rotenone detoxification, similarly to our findings, rotenone also induced several UGT genes in the fruit fly Drosophila melanogaster (67). These results suggest that the mechanisms of rotenone biotransformation might be conserved amongst vertebrates and invertebrates, although in general the C. elegans xenobiotic biotransformation processes, and in particular the substrate specificities of the relevant enzymes, are relatively poorly understood (68). Together with our growth and mitochondrial dysfunction observations, these findings strengthen the suitability of the C. elegans model to address conserved modes of action of mitotoxicants.
It is not surprising that exposure to a xenobiotic that inhibits mitochondrial function elicited a transcriptomic response fitting with metabolic restructuring and xenobiotic metabolism/detoxification. Perhaps more surprisingly, we also saw gene expression changes consistent with altered immune responses. For example, among the other prominent stress-response-related subcategories were “C-type lectins”. These pattern recognition receptors (PRRs) are well-recognized for their role in the innate immune response. They can detect pathogen-derived carbohydrate ligands and activate immune signaling pathways to protect against infection (69, 70). In C. elegans, to our knowledge, the specific function of proteins belonging to this family has not been characterized. Nonetheless, it has been suggested based on sequence analogy, that several C. elegans C-type lectins may have a role in recognizing non-self carbohydrates (71). Most of the C-type lectin DEG in our database were up-regulated (Figure 4D), but a few of them were also found to be down-regulated (Figure 4E). It would be interesting to investigate if some of these differentially-expressed C-type lectins could be specific to one or the other pathogen used in this study and potentially involved in the rotenone-induced differential pathogen resistance phenotype. Although, to our knowledge, the role of C-type lectins in immunometabolism has not been much explored, there is evidence of metabolic changes of vertebrate immune cells induced through C-type lectin-signaling pathways (72). Thus, future studies should further explore the role of this PRR family in the metabolic regulation of C. elegans.
The final of the three most represented stress-response-related subcategories was “Pathogens”. This subcategory comprised only a few genes that were up-regulated (Figure 4D) with a majority that were down-regulated (Figure 4E), suggesting a role of rotenone in shifting nematodes to a more immunosuppressive state. To compare how this set of genes is usually modulated during infections, we took advantage of another C. elegans bioinformatics tool, WormExp (35), and performed a gene enrichment analysis using a list comprising only our DEG related to innate immunity and/or pathogen defense (19 genes which are highlighted in Supplementary Tables 1, 2 and from now on described as “immune-related DEG”). We have identified significant overlap with different gene expression data sets related to P. aeruginosa infection and, interestingly, all of these data sets were described as up-regulated upon P. aeruginosa infection (Supplementary Table 2). By compiling all our immune-related DEG that overlapped with these data sets describing genes up-regulated upon P. aeruginosa infection, we obtained a list of fifteen genes in total and found that eleven of these were downregulated upon rotenone treatment. In fact, two of them were among the list of genes confirmed for P. aeruginosa resistance: dod-24 and irg-5 (73). This major role of rotenone in suppressing immune-related pathways and specific genes up-regulated by P. aeruginosa might contribute to the observed increased susceptibility to P. aeruginosa infection (Figure 5A). In another study, however, Chikka et al. (74) found that rotenone exposures induced a distinct immunomodulatory response in C. elegans. Rotenone activated the p38-MAPK pathway, which is a conserved immune-related pathway required for resistance to a diverse range of pathogens (75). Interestingly, none of the p38-MAPK-dependent genes that were up-regulated upon their rotenone exposures were differentially-expressed within our dataset. However, it is unknown whether this rotenone-induced p38-MAPK pathway activation altered the ability of nematodes to survive P. aeruginosa infections. Similarly to the findings from Chikka et al. (74), Campos et al. (59) reported that a point mutation in the complex I subunit gene nuo-6 which resulted in a mild impairment of mitochondrial function also activated the p38-MAPK pathway. Moreover, these mutants were more resistant to P. aeruginosa infections (59). Although the results from these two studies may seem to contradict our findings, it is important to note that the exposure conditions from Chikka et al. (74) were very distinct from our study including differences in rotenone concentration, developmental exposure window, and the nematode age at which these gene expressions were assessed. Similarly, in the study from Campos et al. (59), nuo-6 mutants were subjected to a continuous mitochondrial impairment throughout their lifetime and for several generations, thus probably eliciting a series of adaptive molecular mechanisms, which may not be present during a time-limited chemically-induced mitochondrial impairment. Most importantly, however, all these studies are in line with a key role of mitochondrial dysfunction in the modulation of immune pathways in the C. elegans model.
Despite the down-regulation of several immune-related genes induced by rotenone, exposed nematodes did not display increased susceptibility to S. enterica. In fact, in two out of our three experiments, nematodes displayed a clear and significant increased resistance (Figure 5B and Supplementary Table 4). Unfortunately, we were unable to gain any mechanistic insights using the WormExp tool, as we did not identify any significant overlap with gene data sets related to S. enterica infection. This is probably because, to our knowledge, to this date, the WormExp database lacks gene sets with this pathogen species alone (there are some data sets available, but they represent combined treatments, e.g., “UP by S. enterica SL2048 under fer-1 mutant”).
In C. elegans, S. enterica and P. aeruginosa trigger distinct pathogen-defense mechanisms. For example, the ZIP-2 transcription factor is required for resistance against PA14 and regulates the expression of irg-1. irg-1 is up-regulated by PA14, but not S. enterica and other less pathogenic strains of P. aeruginosa (76). Moreover, in C. elegans, the pathogenicity of S. enterica relies on disseminated oxidative stress during infection (77), whereas such prominent oxidative stress levels, to our knowledge, have not been described during P. aeruginosa infections. With such distinct mechanisms of pathogenicity, it is reasonable to expect that host survival will also be dependent on different defense-related pathways.
One mechanism evolved in C. elegans to survive pathogens is an evasion strategy, also known as avoidance behavior, which is chiefly mediated by chemosensory neurons (78). Pathogen avoidance plays a significant role in the C. elegans defense response to P. aeruginosa but not to S. enterica (79). Therefore, one could argue that the differential pathogen susceptibility response caused by rotenone in this study could be related to effects on avoidance behavior. Indeed, chemicals may impact chemosensory behavior controlling the C. elegans ability to be attracted or avoid certain natural products. For example, we have recently shown that a sublethal dose of silver nanoparticles can completely block the C. elegans ability to be attracted to a food-like odor (80). Contributing to this idea is the fact that rotenone is also well known for causing neurotoxic effects (81). Having this in mind, all our pathogen challenges were conducted in Petri plates seeded with each bacterial culture spread over the entire plate (full lawn). With this approach, potential effects on pathogen avoidance are not likely to have affected our survival results.
Colonization and proliferation of bacterial pathogens in the C. elegans intestine causes luminal distension and correlates with C. elegans death (82). For example, S. enterica typically causes a remarkable luminal distention in nematodes, and S. enterica mutants that are less lethal to nematodes exhibit reduced colonization capabilities (82). Thus, it would be interesting to address in future studies if the increased resistance of rotenone-exposed animals to S. enterica could be related to an enhanced ability of the rotenone-exposed worms to destroy and eliminate bacterial cells and prevent gut colonization. Similarly, it would be interesting to test whether the increased susceptibility to P. aeruginosa could be related to a decreased capability of C. elegans exposed to rotenone in controlling intestinal bacterial colonization and proliferation.
Our results suggest that the UPRmt pathway could be involved in our rotenone-induced differential pathogen susceptibility. This pathway is probably the most studied in C. elegans linking mitochondria and innate immunity. Upon mitochondrial stress, the ATFS-1 transcription factor is translocated to the nucleus and triggers the expression of mitochondrial protective and immune-related genes (22, 59, 83). In our study, this pathway was shown to be up-regulated by rotenone which was evidenced in one of our transcriptomic dataset analyses (“module-weighted annotations” analysis; Supplementary Figure 1). This induction was sustained even after 24 h of S. enterica exposure. However, the UPRmt was suppressed by P. aeruginosa, and rotenone was only able to restore it to the same levels of solvent control animals (not exposed to rotenone) (Figure 6). Indeed, the role of P. aeruginosa in suppressing the C. elegans UPRmt was already reported (84). Therefore, it is possible that the rotenone-induced activation of UPRmt was involved in the higher resistance of nematodes to S. enterica but was not sufficiently up-regulated during the P. aeruginosa infection to promote protection. Moreover, P. aeruginosa is known to produce mitotoxic compounds (85, 86) and cause mitochondrial dysfunction (84, 87, 88), which suggests that rotenone-exposed nematodes further infected with P. aeruginosa might be suffering from a double-hit in mitochondria, possibly contributing to high energy deficits and death. Corroborating to this hypothesis, Revtovich et al. (87) showed that C. elegans fed with an E. coli strain that disrupts mitochondrial homeostasis are more susceptible to P. aeruginosa. Nonetheless, future studies are needed to address the double-hit hypothesis between mitotoxicant chemical exposures and pathogens that target mitochondrial function and its role in promoting premature death.
Pesticides are one of the most common and important classes of pollutants in the environment, mostly due to their intensive use in agriculture worldwide (89). Although these substances have societal benefits, their use and limitations in describing their negative effects pose serious risks to human, animal, and environmental health. Importantly, besides pesticides, several other classes of environmental pollutants are also known to cause mitochondrial toxicity (14, 15). Here, we provide evidence that mitotoxicant compounds have the potential to act as immunometabolic disruptors and affect host-pathogen interactions altering disease susceptibility, highlighting the importance of careful regulation of the production and use of mitotoxic chemicals. This work also opens new roads for future studies to further characterize C. elegans immunometabolic pathways, which are currently understudied. It also provides potential pathways implicated in pathogen resistance and susceptibility, which can be further investigated, shedding light on potential therapeutic targets.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: GEO database; accessions GSE195584; GSM5841177-GSM5841184.
Author Contributions
DM designed the experiment, performed the experiments and data analysis, prepared the graphical art, and wrote the main manuscript. CB contributed with experimental design and with performing experiments and data analysis. KF, RC, SB, YW, and AC contributed with experiments and data analysis. LB and JM contributed with experimental design, reagents, materials, and analysis tools. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Institute of Health (R01ES028218 and P42ES010356). Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Ian Ryde and Luiza Perez for technical assistance, and the laboratory of Alejandro Aballay for providing the bacterial strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.840272/full#supplementary-material
References
1. Germolec D, Luebke R, Rooney A, Shipkowski K, Vandebriel R, van Loveren H. Immunotoxicology: A Brief History, Current Status and Strategies for Future Immunotoxicity Assessment. Curr Opin Toxicol (2017) 5:55. doi: 10.1016/j.cotox.2017.08.002
2. Suzuki T, Hidaka T, Kumagai Y, Yamamoto M. Environmental Pollutants and the Immune Response. Nat Immunol (2020) 21(12):1486–95. doi: 10.1038/s41590-020-0802-6
3. Luster MI. A Historical Perspective of Immunotoxicology. J Immunotoxicol (2014) 11(3):197–202. doi: 10.3109/1547691X.2013.837121
4. Renault T. Immunotoxicological Effects of Environmental Contaminants on Marine Bivalves. Fish Shellfish Immunol (2015) 46(1):88–93. doi: 10.1016/j.fsi.2015.04.011
5. Mydlarz LD, Jones LE, Harvell CD. Innate Immunity, Environmental Drivers, and Disease Ecology of Marine and Freshwater Invertebrates. Annu Rev Ecology Evolution Systematics (2006) 37(1):251–88. doi: 10.1146/annurev.ecolsys.37.091305.110103
6. Descotes J. Immunotoxicity and Wildlife. In: Immunotoxicol. Drugs Chem. An Exp. Clin. Approach. Amsterdam, Netherlands: Elsevier (2004). p. 355–62.
7. Galloway TS, Depledge MH. Immunotoxicity in Invertebrates: Measurement and Ecotoxicological Relevance. Ecotoxicol (London England) (2001) 10(1):5–23. doi: 10.1023/A:1008939520263
8. Hotamisligil GS. Foundations of Immunometabolism and Implications for Metabolic Health and Disease. Immunity (2017) 47(3):406–20. doi: 10.1016/j.immuni.2017.08.009
9. Mathis D, Shoelson SE. Immunometabolism: An Emerging Frontier. Nat Rev Immunol (2011) 11:2. doi: 10.1038/nri2922
10. Mehta MM, Weinberg SE, Chandel NS. Mitochondrial Control of Immunity: Beyond ATP. Nat Rev Immunol (2017) 17:608–20.
11. Corrêa-da-Silva F, Pereira JAS, de Aguiar CF, de Moraes-Vieira PMM. Mitoimmunity—when Mitochondria Dictates Macrophage Function. Cell Biol Int (2018) 42(6):651–5. doi: 10.1002/cbin.10921
12. Mills EL, Kelly B, O’Neill LAJ. Mitochondria are the Powerhouses of Immunity. Nat Immunol (2017) 18(5):488–98. doi: 10.1038/ni.3704
13. West AP. Mitochondrial Dysfunction as a Trigger of Innate Immune Responses and Inflammation. Toxicology (2017) 391:54–63. doi: 10.1016/j.tox.2017.07.016
14. Meyer JN, Hartman JH, Mello DF. Mitochondrial Toxicity. Toxicol Sci (2018) 162(1):15–23. doi: 10.1093/toxsci/kfy008
15. Meyer JN, Leung MCK, Rooney JP, Sendoel A, Hengartner MO, Kisby GE, et al. Mitochondria as a Target of Environmental Toxicants. Toxicol Sci (2013) 134(1):1–17. doi: 10.1093/toxsci/kft102
16. Apfeld J, Alper S. What can We Learn About Human Disease From the Nematode C. Elegans? In: DiStefano J, editor. Dis. Gene Identification. Methods Mol. Biol. New York, NY: Humana Press (2018). p. 53–75.
17. Anderson SM, Pukkila-Worley R. Immunometabolism in Caenorhabditis Elegans. PloS Pathogens (2020) 16(10):e1008897. doi: 10.1371/journal.ppat.1008897
18. Fitch DHA, Thomas WK. Phylogenetic Context for C. Elegans. 2nd. New York, United States: Cold Spring Harbor Laboratory Press (1997).
19. Lai CH, Chou CY, Ch’ang LY, Liu CS, Lin WC. Identification of Novel Human Genes Evolutionarily Conserved in Caenorhabditis Elegans by Comparative Proteomics. Genome Res (2000) 10(5):703–13. doi: 10.1101/gr.10.5.703
20. Keith Blackwell T, Sewell AK, Wu Z, Han M. TOR Signaling in Caenorhabditis Elegans Development, Metabolism, and Aging. Genetics (2019) 213(2):329. doi: 10.1534/genetics.119.302504
21. Penkov S, Mitroulis I, Hajishengallis G, Chavakis T. Immunometabolic Crosstalk: An Ancestral Principle of Trained Immunity? Trends Immunol (2019) 40(1):1. doi: 10.1016/j.it.2018.11.002
22. Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. Mitochondrial UPR-Regulated Innate Immunity Provides Resistance to Pathogen Infection. Nature (2014) 516(7531):414–7. doi: 10.1038/nature13818
23. Kwon S, Kim EJE, Lee S.-J.J. V., Ji E, Kim EJE, Lee S.-J.J. V. Mitochondria-Mediated Defense Mechanisms Against Pathogens in Caenorhabditis Elegans. BMB Rep (2018) 51(6):274–9. doi: 10.5483/BMBRep.2018.51.6.111
24. Hunt PR. The C. Elegans Model in Toxicity Testing. J Appl Toxicol: JAT (2017) 37(1):50–9. doi: 10.1002/jat.3357
25. Queirós L, Pereira JL, Gonçalves FJM, Pacheco M, Aschner M, Pereira P. Caenorhabditis Elegans as a Tool for Environmental Risk Assessment: Emerging and Promising Applications for a “Nobelized Worm.” Crit Rev Toxicol (2019) 49(5):411–29. doi: 10.1080/10408444.2019.1626801
26. Leung MCK, Williams PL, Benedetto A, Au C, Helmcke KJ, Aschner M, et al. Caenorhabditis Elegans: An Emerging Model in Biomedical and Environmental Toxicology. Toxicol Sci (2008) 106(1):5–28. doi: 10.1093/toxsci/kfn121
27. Moore BT, Jordan JM, Baugh LR. WormSizer: High-Throughput Analysis of Nematode Size and Shape. PloS One (2013) 8(2):57142. doi: 10.1371/journal.pone.0057142
28. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat Methods (2012) 9:7. doi: 10.1038/nmeth.2019
29. Mok DZL, Sternberg PW, Inoue T. Morphologically Defined Sub-Stages of C. Elegans Vulval Development in the Fourth Larval Stage. BMC Dev Biol (2015) 15(1):26. doi: 10.1186/s12861-015-0076-7
30. Luz AL, Smith LL, Rooney JP, Meyer JN. Seahorse Xfe 24 Extracellular Flux Analyzer-Based Analysis of Cellular Respiration in Caenorhabditis Elegans. Curr Protoc Toxicol (2015) 66:25.7.1–15. doi: 10.1002/0471140856.tx2507s66
31. Langmead B. Aligning Short Sequencing Reads With Bowtie. Curr Protoc Bioinf (2010) 32:11.7.1–14. doi: 10.1002/0471250953.bi1107s32
32. Anders S, Pyl PT, Huber W. HTSeq-A Python Framework to Work With High-Throughput Sequencing Data. Bioinformatics (2015) 31(2):166–9. doi: 10.1093/bioinformatics/btu638
33. Robinson MD, McCarthy DJ, Smyth GK. Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics (2009) 26(1):139–40. doi: 10.1093/bioinformatics/btp616
34. Holdorf AD, Higgins DP, Hart AC, Boag PR, Pazour GJ, Walhout AJM, et al. WormCat: An Online Tool for Annotation and Visualization of Caenorhabditis Elegans Genome-Scale Data. Genetics (2020) 214(2):279–94. doi: 10.1534/genetics.119.302919
35. Yang W, Dierking K, Schulenburg H. WormExp: A Web-Based Application for a Caenorhabditis Elegans-Specific Gene Expression Enrichment Analysis. Bioinformatics (2016) 32(6):943–5. doi: 10.1093/bioinformatics/btv667
36. Cary M, Podshivalova K, Kenyon C. Application of Transcriptional Gene Modules to Analysis of Caenorhabditis Elegans’ Gene Expression Data. G3: Genes Genomes Genet (2020) 10(10):3623–38. doi: 10.1534/g3.120.401270
37. Tan M, Mahajan-miklos S, Ausubel FM. Killing of Caenorhabditis Elegans by Pseudomonas Aeruginosa Used to Model Mammalian Bacterial Pathogenesis. Proc Natl Acad Sci USA (1999) 96:715–20. doi: 10.1073/pnas.96.2.715
38. Luz AL, Godebo TR, Smith LL, Leuthner TC, Maurer LL, Meyer JN. Deficiencies in Mitochondrial Dynamics Sensitize Caenorhabditis Elegans to Arsenite and Other Mitochondrial Toxicants by Reducing Mitochondrial Adaptability. Toxicology (2017) 387:81–94. doi: 10.1016/j.tox.2017.05.018
39. Gonzalez-Hunt CP, Luz AL, Ryde IT, Turner EA, Ilkayeva OR, Bhatt DP, et al. Multiple Metabolic Changes Mediate the Response of Caenorhabditis Elegans to the Complex I Inhibitor Rotenone. Toxicology (2021) 447:152630. doi: 10.1016/j.tox.2020.152630
40. Dreier DA, Mello DF, Meyer JN, Martyniuk CJ. Linking Mitochondrial Dysfunction to Organismal and Population Health in the Context of Environmental Pollutants: Progress and Considerations for Mitochondrial Adverse Outcome Pathways. Environ Toxicol Chem (2019) 38(8):1625–34. doi: 10.1002/etc.4453
41. Meyer JN, Leuthner TC, Luz AL. Mitochondrial Fusion, Fission, and Mitochondrial Toxicity. Toxicology (2017) 391:42–53. doi: 10.1016/j.tox.2017.07.019
42. Mello DF, Proença LAO, Barracco MA. Comparative Study of Various Immune Parameters in Three Bivalve Species During a Natural Bloom of Dinophysis Acuminata in Santa Catarina Island, Brazil. Toxins (2010) 2(5):1166–78. doi: 10.3390/toxins2051166
43. Mello DF, Trevisan R, Danielli NM, Dafre AL. Vulnerability of Glutathione-Depleted Crassostrea Gigas Oysters to Vibrio Species. Marine Environ Res (2020) 154:104870. doi: 10.1016/j.marenvres.2019.104870
44. Mello DF, Bergemann C, Fisher K, Chitrakar R, Bijwadia S, Wang Y, et al. Rotenone Modulates the C. Elegans Immunometabolism and Pathogen Susceptibility. To Be Submitted Front Immunol (2021).
45. Pipe RK, Coles JA. Environmental Contaminants Influencing Immunefunction in Marine Bivalve Molluscs. Fish Shellfish Immunol (1995) 5(8):581–95. doi: 10.1016/S1050-4648(95)80043-3
46. Mello DF, Silva PM, Barracco MA, Soudant P, Hégaret H. Effects of the Dinoflagellate Alexandrium Minutum and its Toxin (Saxitoxin) on the Functional Activity and Gene Expression of Crassostrea Gigas Hemocytes. Harmful Algae (2013) 26:45–51. doi: 10.1016/j.hal.2013.03.003
47. Luz Cunha AC, Pontinha V, de A, de Castro MAM, Sühnel S, Medeiros SC, et al. Two Epizootic Perkinsus Spp. Events in Commercial Oyster Farms at Santa Catarina, Brazil. J Fish Dis (2019) 42(3):455–63. doi: 10.1111/jfd.12958
48. Mello DF, Arl M, Trevisan R, Dafre AL. How Important are Glutathione and Thiol Reductases to Oyster Hemocyte Function? Fish Shellfish Immunol (2015) 46(2):566–72. doi: 10.1016/j.fsi.2015.07.017
49. Tsang WY, Lemire BD. The Role of Mitochondria in the Life of the Nematode, Caenorhabditis Elegans. Biochim Biophys Acta Mol Basis Dis (2003) 1638(2):91–105. doi: 10.1016/S0925-4439(03)00079-6
50. Hershberger KA, Rooney JP, Turner EA, Donoghue LJ, Bodhicharla R, Maurer LL, et al. Early-Life Mitochondrial DNA Damage Results in Lifelong Deficits in Energy Production Mediated by Redox Signaling in Caenorhabditis Elegans. Redox Biol (2021) 43:102000. doi: 10.1016/j.redox.2021.102000
51. Bess AS, Crocker TL, Ryde IT, Meyer JN. Mitochondrial Dynamics and Autophagy Aid in Removal of Persistent Mitochondrial DNA Damage in Caenorhabditis Elegans. Nucleic Acids Res (2012) 40(16):7916–31. doi: 10.1093/nar/gks532
52. Tsang WY, Sayles LC, Grad LI, Pilgrim DB, Lemire BD. Mitochondrial Respiratory Chain Deficiency in Caenorhabditis Elegans Results in Developmental Arrest and Increased Life Span. J Biol Chem (2001) 276(34):32240–6. doi: 10.1074/jbc.M103999200
53. Tsang WY, Lemire BD. Mitochondrial Genome Content is Regulated During Nematode Development. Biochem Biophys Res Commun (2002) 291(1):8–16. doi: 10.1006/bbrc.2002.6394
54. Bolser DG, Dreier DA, Li E, Kroll KJ, Martyniuk CJ, Denslow ND. Toward an Adverse Outcome Pathway for Impaired Growth: Mitochondrial Dysfunction Impairs Growth in Early Life Stages of the Fathead Minnow (Pimephales Promelas). Comp Biochem Physiol Part C: Toxicol Pharmacol (2018) 209:46–53. doi: 10.1016/j.cbpc.2018.03.009
55. Brookes PS. Mitochondrial H+ Leak and ROS Generation: An Odd Couple. Free Radical Biol Med (2005) 38(1):12–23. doi: 10.1016/j.freeradbiomed.2004.10.016
56. Salin K, Villasevil EM, Anderson GJ, Lamarre SG, Melanson CA, McCarthy I, et al. Differences in Mitochondrial Efficiency Explain Individual Variation in Growth Performance. Proc R Soc B (2019) 286(1909):20191466. doi: 10.1098/rspb.2019.1466
57. Schmeisser S, Priebe S, Groth M, Monajembashi S, Hemmerich P, Guthke R, et al. Neuronal ROS Signaling Rather Than AMPK/sirtuin-Mediated Energy Sensing Links Dietary Restriction to Lifespan Extension. Mol Metab (2013) 2(2):92. doi: 10.1016/j.molmet.2013.02.002
58. Maglioni S, Mello DF, Schiavi A, Meyer JN, Ventura N. Mitochondrial Bioenergetic Changes During Development as an Indicator of C. Elegans Health-span Aging (2019) 11(16):6535–54. doi: 10.18632/aging.102208
59. Campos JC, Wu Z, Rudich PD, Soo SK, Mistry M, Ferreira JC, et al. Mild Mitochondrial Impairment Enhances Innate Immunity and Longevity Through ATFS-1 and P38 Signaling. EMBO Rep (2021) 22(12):e52964. doi: 10.15252/embr.202152964
60. Angajala A, Lim S, Phillips JB, Kim JH, Yates C, You Z, et al. Diverse Roles of Mitochondria in Immune Responses: Novel Insights Into Immuno-Metabolism. Front Immunol (2018) 9(JUL):1605. doi: 10.3389/fimmu.2018.01605
61. Anderson SM, Cheesman HK, Peterson ND, Salisbury JE, Soukas AA, Pukkila-Worley R. The Fatty Acid Oleate is Required for Innate Immune Activation and Pathogen Defense in Caenorhabditis Elegans. PloS Pathogens (2019) 15(6):e1007893. doi: 10.1371/journal.ppat.1007893
62. Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear Hormone Receptor NHR-49 Controls Fat Consumption and Fatty Acid Composition in C. Elegans. PloS Biol (2005) 3(2):0301–12. doi: 10.1371/journal.pbio.0030053
63. Horikawa M, Nomura T, Hashimoto T, Sakamoto K. Elongation and Desaturation of Fatty Acids are Critical in Growth, Lipid Metabolism and Ontogeny of Caenorhabditis Elegans. J Biochem (2008) 144(2):149–58. doi: 10.1093/jb/mvn055
64. Caboni P, Sherer TB, Zhang N, Taylor G, Na HM, Greenamyre JT, et al. Rotenone, Deguelin, Their Metabolites, and the Rat Model of Parkinson’s Disease. Chem Res Toxicol (2004) 17(11):1540–8. doi: 10.1021/tx049867r
65. Fukami JI, Yamamoto I, Casida JE. Metabolism of Rotenone In Vitro by Tissue Homogenates From Mammals and Insects. Science (1967) 155 (3763):713–6. doi: 10.1126/science.155.3763.713
66. Kotze AC, Dobson RJ, Chandler D. Synergism of Rotenone by Piperonyl Butoxide in Haemonchus Contortus and Trichostrongylus Colubriformis In Vitro: Potential for Drug-Synergism Through Inhibition of Nematode Oxidative Detoxification Pathways. Veterinary Parasitol (2006) 136(3–4):275–82. doi: 10.1016/j.vetpar.2005.11.001
67. Ahn SJ, Marygold SJ. The UDP-Glycosyltransferase Family in Drosophila Melanogaster: Nomenclature Update, Gene Expression and Phylogenetic Analysis. Front Physiol (2021) 12:300. doi: 10.3389/fphys.2021.648481
68. Hartman JH, Widmayer SJ, Bergemann CM, King DE, Morton KS, Romersi RF, et al. Xenobiotic Metabolism and Transport in Caenorhabditis Elegans. J Toxicol Environ Health Part B: Crit Rev (2021) 24(2):51–94. doi: 10.1080/10937404.2021.1884921
69. Dambuza IM, Brown GD. C-Type Lectins in Immunity: Recent Developments. Curr Opin Immunol (2015) 32:21–7. doi: 10.1016/j.coi.2014.12.002
70. Chiffoleau E. C-Type Lectin-Like Receptors as Emerging Orchestrators of Sterile Inflammation Represent Potential Therapeutic Targets. Front Immunol (2018) 9227(FEB). doi: 10.3389/fimmu.2018.00227
71. Drickamer K, Dodd RB. C-Type Lectin-Like Domains in Caenorhabditis Elegans: Predictions From the Complete Genome Sequence. Glycobiology (1999) 9(12):1357–69. doi: 10.1093/glycob/9.12.1357
72. Domínguez-Andrés J, Arts RJW, ter Horst R, Gresnigt MS, Smeekens SP, Ratter JM, et al. Rewiring Monocyte Glucose Metabolism via C-Type Lectin Signaling Protects Against Disseminated Candidiasis. PloS Pathogens (2017) 13(9):e1006632. doi: 10.1371/journal.ppat.1006632
73. Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. P38 MAPK Regulates Expression of Immune Response Genes and Contributes to Longevity in C. Elegans. PloS Genet (2006) 2(11):1725–39. doi: 10.1371/journal.pgen.0020183
74. Chikka MR, Anbalagan C, Dvorak K, Dombeck K, Prahlad V. The Mitochondria-Regulated Immune Pathway Activated in the C. Elegans Intestine Is Neuroprotective. Cell Rep (2016) 16(9):2399–414. doi: 10.1016/j.celrep.2016.07.077
75. Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, et al. A Conserved P38 MAP Kinase Pathway in Caenorhabditis Elegans Innate Immunity. Science (2002) 297(5581):623–6. doi: 10.1126/science.1073759
76. Estes KA, Dunbar TL, Powell JR, Ausubel FM, Troemel ER. bZIP Transcription Factor Zip-2 Mediates an Early Response to Pseudomonas Aeruginosa Infection in Caenorhabditis Elegans. Proc Natl Acad Sci USA (2010) 107(5):2153–8. doi: 10.1073/pnas.0914643107
77. Sem XH, Rhen M. Pathogenicity of Salmonella Enterica in Caenorhabditis Elegans Relies on Disseminated Oxidative Stress in the Infected Host. PloS One (2012) 7(9):e45417. doi: 10.1371/journal.pone.0045417
78. Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and Avoidance of a Natural Product From the Pathogenic Bacterium Serratia Marcescens by Caenorhabditis Elegans. Proc Natl Acad Sci (2007) 104(7):2295–300. doi: 10.1073/pnas.0610281104
79. Sun J, Singh V, Kajino-Sakamoto R, Aballay A. Neuronal GPCR Controls Innate Immunity by Regulating Noncanonical Unfolded Protein Response Genes. Sci (New York NY) (2011) 332(6030):729–32. doi: 10.1126/science.1203411
80. Mello DF, Maurer LL, Ryde IT, Songr DH, Marinakos SM, Jiang C, et al. In Vivo Effects of Silver Nanoparticles on Development, Behavior, and Mitochondrial Function are Altered by Genetic Defects in Mitochondrial Dynamics. Environ Sci Technol (2022) 56(2):1113–24. doi: 10.1021/acs.est.1c05915
81. Sherer TB, Betarbet R, Testa CM, Seo BB, Richardson JR, Kim JH, et al. Mechanism of Toxicity in Rotenone Models of Parkinson’s Disease. J Neurosci (2003) 23(34):10756–64. doi: 10.1523/JNEUROSCI.23-34-10756.2003
82. Fuhrman LE, Shianna KV, Aballay A. High-Throughput Isolation and Mapping of C. Elegans Mutants Susceptible to Pathogen Infection. PloS One (2008) 3(8):e2882. doi: 10.1371/journal.pone.0002882
83. Wang S, Gao K, Liu Y. UPRmt Coordinates Immunity to Maintain Mitochondrial Homeostasis and Animal Fitness. Mitochondrion (2018) 41:9–13. doi: 10.1016/j.mito.2017.11.004
84. Deng P, Naresh NU, Du Y, Lamech LT, Yu J, Julie Zhu L, et al. Mitochondrial UPR Repression During Pseudomonas Aeruginosa Infection Requires the bZIP Protein ZIP-3. Proc Natl Acad Sci USA (2019) 116(13):6146–51. doi: 10.1073/pnas.1817259116
85. Kang D, Kirienko DR, Webster P, Fisher AL, Kirienko NV. Pyoverdine, a Siderophore From Pseudomonas Aeruginosa, Translocates Into C. Elegans, Removes Iron, and Activates a Distinct Host Response. Virulence (2018) 9(1):804–17. doi: 10.1080/21505594.2018.1449508
86. Kirienko NV, Ausubel FM, Ruvkun G. Mitophagy Confers Resistance to Siderophore-Mediated Killing by Pseudomonas Aeruginosa. Proc Natl Acad Sci USA (2015) 112(6):1821. doi: 10.1073/pnas.1424954112
87. Revtovich AV, Lee R, Kirienko NV. Interplay Between Mitochondria and Diet Mediates Pathogen and Stress Resistance in Caenorhabditis Elegans. PloS Genet (2019) 15(3):e1008011. doi: 10.1371/journal.pgen.1008011
88. Mir DA, Balamurugan K. A Proteomic Analysis of Caenorhabditis Elegans Mitochondria During Bacterial Infection. Mitochondrion (2019) 48:37–50. doi: 10.1016/j.mito.2019.03.002
Keywords: immunotoxicity, metabolism, innate immunity, mitoimmunity, invertebrate, mitochondrial toxicants, pesticides
Citation: Mello DF, Bergemann CM, Fisher K, Chitrakar R, Bijwadia SR, Wang Y, Caldwell A, Baugh LR and Meyer JN (2022) Rotenone Modulates Caenorhabditis elegans Immunometabolism and Pathogen Susceptibility. Front. Immunol. 13:840272. doi: 10.3389/fimmu.2022.840272
Received: 20 December 2021; Accepted: 31 January 2022;
Published: 22 February 2022.
Edited by:
Celine Ann Beamer, University of Montana, United StatesReviewed by:
Parisa Norouzitallab, Uppsala University, SwedenAmit Sinha, New England Biolabs, United States
Mark Pellegrino, University of Texas at Arlington, United States
Copyright © 2022 Mello, Bergemann, Fisher, Chitrakar, Bijwadia, Wang, Caldwell, Baugh and Meyer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danielle F. Mello, danielle.mello@ifremer; danielleferrazmello@gmail.com; Joel N. Meyer, joel.meyer@duke.edu
†These authors share first authorship
‡Present address: Danielle F. Mello, Équipe soutenue par la fondation ARC, Ifremer, Univ Brest, CNRS, IRD, LEMAR, Plouzané, France
 Danielle F. Mello
Danielle F. Mello Christina M. Bergemann1†
Christina M. Bergemann1† Kinsey Fisher
Kinsey Fisher Shefali R. Bijwadia
Shefali R. Bijwadia Alexis Caldwell
Alexis Caldwell Larry Ryan Baugh
Larry Ryan Baugh Joel N. Meyer
Joel N. Meyer