Microbial Community Structure and Interannual Change in the Last Epishelf Lake Ecosystem in the North Polar Region
- 1Département de Biologie, Centre d'Études Nordiques and Takuvik Joint International Laboratory, Université Laval, Québec, QC, Canada
- 2Institut de Biologie Intégrative et des Systèmes, Université Laval, Québec, QC, Canada
- 3Québec-Océan, Université Laval, Québec, QC, Canada
- 4Faculty of Applied Science, Civil Engineering, University of British Columbia, Vancouver, BC, Canada
Climate warming is proceeding rapidly in the polar regions, posing a threat to ice-dependent ecosystems. Among the most vulnerable are microbial-dominated epishelf lakes, in which surface ice-damming of an embayment causes a freshwater layer to overlie the sea, creating an interface between distinct habitats. We characterized the physicochemical and biotic environment of Milne Fiord epishelf lake (82° N, Canada) in three successive summers (2010–2012), and on one date of profiling (5 July 2011) we collected samples for high through-put amplicon sequencing of variable regions of small subunit rRNA to characterize the microbial community (Eukarya, Bacteria and Archaea). Potentially active water column communities were investigated using reverse-transcribed rRNA, and phytoplankton were further characterized by accessory pigment analysis. Cluster analysis of pigment data showed a demarcation between freshwater and marine communities, which was also evident in the sequence data. The halocline community of Eukarya was more similar to the deeper marine sample than to the freshwater surface community, while the Archaea and Bacteria communities at this interface clustered more with surface communities. In 2012, conductivity-depth profiles indicated shallowing of the freshwater layer and mixing across the halocline, accompanied by lower picocyanobacteria and higher picoeukaryote concentrations. Picocyanobacteria cells were more evenly distributed throughout the water column in 2012, implying partial deep mixing. Several mixotrophic taxa of Eukarya were more abundant in the freshwater layer, where low nutrient concentrations may favor this lifestyle. Unusual features of Milne Fiord microbial communities included benthic taxa not previously reported in marine water columns (notably, the archaeon Halobacteriales), and dominance by taxa that are typically present in sparse concentrations elsewhere: for example, the Chlorophyte group Radicarteria and the betaproteobacterium Rhodoferax. Milne Fiord epishelf lake is the last known lake of this kind remaining in the Arctic, and the fate of this distinct microbial ecosystem may ultimately depend on the stability of the Milne Fiord ice shelf, which has experienced a negative mass balance over the past half century.
Introduction
Epishelf lakes form in high latitude environments when an ice-shelf blocks the mouth of a fiord or other embayment, allowing freshwater from precipitation and glacier run-off to accumulate atop the denser seawater (Veillette et al., 2008; Laybourn-Parry and Wadham, 2014). Numerous epishelf lakes occur in Antarctica (Gibson and Andersen, 2002; Laybourn-Parry et al., 2006), and many were once located in the Arctic, forming behind ice shelves along the northern coast of Ellesmere Island (Vincent et al., 2001). However, the break-up of these ice-shelves over the past century has caused the drainage of most of these lakes in the northern hemisphere. Most recently, the Disraeli Fiord epishelf lake disappeared in the early 2000s (Mueller et al., 2003), and the Petersen Bay epishelf lake was lost in 2005 (White et al., 2015).
The Milne Fiord epishelf lake, located behind the Milne Ice Shelf, is the last of its kind in the High Arctic. The present form of the lake was created between 1959 and 1984 by deterioration of the landward edge of the ice shelf, while the lake is bounded on its landward side by the floating tongue of Milne Glacier (Mortimer et al., 2012). There has been an ongoing warming and attrition of ice along northern Ellesmere Island over the last decade. For example, Ward Hunt Lake lost its thick perennial ice cover for possibly the first time in more than 50 years in 2011, and in 2012 was ice-free for almost 1 month (Paquette et al., 2015). These observations imply that the ice-bound epishelf lake ecosystem at Milne Fiord may also have experienced change over the same period.
The microbial diversity of Milne Fiord epishelf lake was studied in 2007 using clone libraries (Veillette et al., 2011); however, the sampling was restricted to dark incubated enrichment cultures and did not include the halocline. The study showed large differences in bacterial species, as well as in phytoplankton pigment composition between samples from the surface freshwater and deep saltwater. The zooplankton community in Milne Fiord was found to contain an unusual mix of freshwater and marine zooplankton species, as observed in the former Disraeli Fiord epishelf lake ecosystem (Van Hove et al., 2001).
Our objectives in the current study were twofold. First, we aimed to determine the interannual variability in the summer properties of the Milne Fiord lake ecosystem, including its primary producers, by way of a suite of limnological and biological measurements over 3 years. The sampling covered a period of warming along the northern Ellesmere coastline, 2010–2012, and we hypothesized that the physical, chemical and biological properties of this ecosystem would show evidence of change. Secondly, we aimed to undertake an in-depth characterization of all three domains of the microbial communities using high through-put SSU rRNA amplicon sequencing. We addressed this second objective by way of a more detailed water column sampling and molecular analysis in July 2011. We focused on RNA to provide an indication of the taxa more likely to be involved in protein synthesis (Blazewicz et al., 2013), and size-fractionated this “active community” by sequential filtration. We hypothesized that the microbial community structure of Milne Fiord epishelf lake would reflect its unique highly stratified water column structure.
Materials and Methods
Study Site and Sampling
Milne Fiord (lat. 82° 35.540′N; long. 80° 35.761′W) is located on the northern coastline of Ellesmere Island, Nunavut, Canada (Figure 1). It has a maximum recorded depth of 436 m, with the depth of the surface freshwater layer thought to correspond to the minimum thickness of the ice shelf. In 2009, the thickness of this epishelf freshwater layer was 14.3 m, slightly less the 16.6 m recorded 2 years previously (Veillette et al., 2011). The lake had an area of 64.4 km2 in May 2012 and is typically covered by perennial freshwater ice. However, satellite imagery showed that this ice partially broke up in August 2012, at the end of the warm 2012 season. The Milne Ice Shelf itself has been in a state of negative mass balance since at least 1953 (Mortimer et al., 2012).
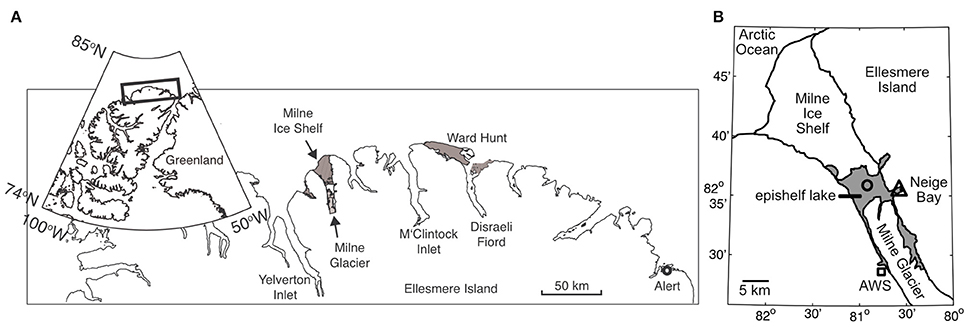
Figure 1. (A) General location of Milne Fiord on the northern coast of Ellesmere Island. The gray areas show the extent of the Milne and Ward Hunt Ice Shelves as of 2011. (B) Local geography of Milne Fiord. The gray area shows the extent of the Milne Fiord epishelf lake, the main sampling location in Neige Bay (triangle), and a secondary sampling location where oxygen samples were collected in 2012 (circle), as well as the location of the automated weather station (AWS; square). Ice shelf outlines are from Mueller et al. (2017).
The epishelf lake was sampled over 3 years on 9 July in 2010, 5 July 2011, and 5 July 2012, as part of the program “Northern Ellesmere Island in the Global Environment” (NEIGE). Access was by helicopter from our field laboratory at Ward Hunt Island, located 115 km east of the fiord. Samples were collected in Neige Bay (unofficial name), a shallow inlet on the eastern shore of the fiord, which has a depth of 84 m. At the time of sampling, the average ice thickness on the epishelf lake was 84 ± 16 cm over the 3 years.
Water Column Profiles
Physico-chemical profiles of the water column were made using a conductivity-temperature-depth profiler (XR-420 CTD-RBR profiler; RBR Ltd, Ottawa, Canada), and a Hydrolab DS5X profiler (Loveland, CO, USA). These data are archived in NEIGE (2016a). Owing to a problem with the profiler in 2012, oxygen data was instead measured using a YSI Pro Plus multiparameter handheld meter on discrete water samples collected in the main fiord, rather than in Neige Bay. These oxygen samples were collected the day before the other sampling activities occurred. Samples for chemical and biological analyses were taken using a Kemmerer bottle (Wildlife Supply Company, Yulee, FL, USA) from various depths (1.5–30 m). Twelve liters of water were transferred into acid- and fiord water rinsed 20-l propylene Cubitainers, and kept cold and dark until transport to the field laboratory on Ward Hunt Island. The water was subsampled for nutrients, flow cytometry and high-performance liquid chromatography (HPLC) pigment analyses within 4 h.
Chemical Analysis
Water samples for chemical analyses were collected from 1.5, 16, and 30 m depths in all years, and an additional sample at 10 m was obtained in 2010 and 2012. Aliquots of 40 ml were placed in brown bottles for total organic carbon analyses. For total phosphorus (TP) and total nitrogen (TN), 0.833 ml of H2SO4 (30%) was added to 125 ml of sample water and stored in the dark at 4°C until later analysis at the Institut National de Recherche Scientifique–Eau, Terre & Environnement, Québec, Canada (INRS-ETE). TN and dissolved organic carbon (DOC) were assessed using a Shimadzu VCPH Lachat with detection limits of 0.02 mg l−1 and 0.05 mg l−1 respectively. UV-VIS spectrophotometry was used for TP quantification with detection limits of 0.5 and 0.2 μg l−1, respectively. Dissolved inorganic carbon (DIC) was analyzed using a CombiPal 3800 Varian with a detection limit of 0.02 mM.
Cell Enumeration by Flow Cytometry (FCM)
In 2010, the water column was sampled at 10 depths: 1.5, 6, 8, 10, 12, 14, 16, 18, 22, and 30 m. In 2011 and 2012, an eleventh sample depth was added at 3 m. Aliquots of 4 ml were placed in Cryovials and fixed with 160 μl of glutaraldehyde 25%, Grade I, for a final concentration of 1%. While in the field, these samples were stored in a dry-shipper previously cooled with liquid nitrogen, and upon returning to the lab at −80°C. Cell analyses were performed using a BD FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) following the method of Casamayor et al. (2007). In the present study, two fluorescence channels were used: FL3 (670 nm) for picoeukaryotes, and a combination of FL3 and FL2 (585 nm) for picocyanobacteria. FL3 and FL2 detected the natural fluorescence of chlorophyll a (Chl a) and phycobilins respectively. Beads of 1-μm diameter (Fluoresbrite, Calibration Grade; Polysciences, Inc., Warrington, PA, USA) were added to samples to assess picoeukaryote and picocyanobacteria size. Trucount beads (BD Biosciences, Franklin Lakes, NJ, USA) were used to calibrate the 1-μm bead solutions and to assess population size. To ensure quality of cell detection, a minimum of 15,000 events were measured per analysis, with less than 1000 events s−1. Each sample was analyzed in triplicate.
Pigment Analysis
In 2010, samples for pigment analysis by HPLC were collected from the same four depths as for nutrient analyses, while in 2011 and 2012 they were collected from the same 11 depths as for FCM analyses. We filtered 0.3–2 l of water through 25-mm diameter Whatman GF/F glass fiber filters. These were then wrapped in aluminum foil and immediately placed into a dry shipper (previously cooled with liquid nitrogen) in the field, then transferred to −80°C for storage until pigment analysis at Laval University. Within 1 month of collection, the filter samples were extracted in the dark by sonication treatment for 30 s at 17 W in 3 ml of 95% methanol (by volume). The supernatant was recovered after vortexing and centrifugation (4150 rpm for 15 min at 4°C), filtered through a 0.2-μm pore size Polytetrafluoroethylene (PTFE) Acrodisc filter (PALL Corporation, Ann Arbor, MI, USA) and placed in a 2-ml amber vial with an argon gas atmosphere for immediate analysis by HPLC.
HPLC analyses in 2010 were performed using a ProStar HPLC system (Varian, Palo Alto, CA, USA) with a Symmetry C8 column (3.5-μm pore size, 4.6 × 150 mm; Waters Corporation, Milford, MA, USA) at 25°C, and a C8 guard column (5-μm pore size, 3.9 × 20 mm; Waters Corporation). In 2011 and 2012, HPLC analyses were performed using a Thermo Scientific system (Thermo Scientific, West Palm Beach, FL, USA) with a Hypersil Gold C8 HPLC column (3.0-μm pore size, 4.6 x 150 mm, Thermo Scientific) at 25°C, with a C8 guard column. Carotenoids were quantified by their absorbance at 450 nm in a Diode Array Detector (350–750 nm) and chlorophylls were detected by fluorescence (excitation at 400 nm and emission at 650 nm). The HPLC separation method followed Zapata et al. (2000). Pigments were identified based on retention time and spectral comparisons with standards from DHI (Water & Environments, Horsholm, Denmark). The HPLC data are archived in NEIGE (2016b). Clustering analysis was performed using Bray-Curtis distance, as implemented in the R package vegan (Oksanen et al., 2010). Pigments were interpreted as indicators of specific phytoplankton phyla following Wright and Jeffrey (2006).
RNA Collection and Extraction
Water samples for RNA extractions were collected from Milne Fiord epishelf lake (82° 35.479 N; 80° 35.824 W) on 5 July 2011 using a Kemmerer bottle at three depths: 1.5, 16, and 30 m. Samples were transported in 20-l polyethylene containers in darkness, and processed within 4 h. Four liters of water were filtered sequentially through a 47-mm diameter 3-μm pore size polycarbonate filter and a 0.22-μm Sterivex filter cartridge (Millipore, Etobicoke, ON, Canada). For the 1.5-m sample, the 3-μm filters were replaced as they became clogged with material, for a total of four filters, which were stored in a single 1.5-ml tube. The Sterivex cartridge for this sample was replaced after 2 l, and the two cartridges were preserved separately. Samples were preserved by adding 1 ml RLT buffer (Qiagen, Germantown, MD, USA), and frozen in liquid nitrogen for transport. Samples were stored at −80°C.
RNA was extracted from filters and Sterivex cartridges using a Qiagen AllPrep DNA/RNA Mini Kit. For the 1.5-m sample, all four 3-μm filters were used for the large size fraction, while only the first Sterivex cartridge was used for the small size fraction. The RLT buffer was removed before extraction and replaced with RLT-plus from the extraction kit. Cells were lysed by an initial incubation with 3 μg ml−1 lysozyme (Sigma-Aldrich, St. Louis, MO, USA) at 37°C for 45 min, and 0.78 mg ml−1 proteinase K (Bio Basic, Markham, ON, Canada) with 1.11% SDS at 65°C for 15 min. The lysate was then homogenized using the QiaShredder columns included with the extraction kit. An incubation with DNAse I (0.23 units μl−1) for 15 min at room temperature in RDD buffer (Qiagen, Hilden, Germany), was added between wash steps. To increase the final concentration of RNA, 35 μl of elution buffer was passed twice through the RNeasy column at the last step. The absence of DNA was immediately confirmed by a PCR of the 16S rRNA gene on the extracted RNA using the primers 8F and 1492R (Edwards et al., 1989; Stackebrandt and Liesack, 1993) in a 25-μl reaction volume containing High-Fidelity buffer 1X, 0.2 μM of each primer, 0.2 μM dNTPs, 0.2 mg ml−1 bovine serum albumin (BSA), and 1 U of Phusion High-Fidelity DNA polymerase (all reagents from New England Biolabs, Ipswich, MA, USA), using a 16S amplicon from a bacterial clone library as a positive control. The thermal cycling protocol was an initial denaturation at 98°C for 30 s, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, with a final extension of 72°C for 4.5 min. PCR products were checked on a 1% agarose gel stained with 4% v/v ethidium bromide. No band was observed on the gel. Extracted RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA).
High Throughput Multiplex Tag Sequencing
The V4 region of the eukaryotic 18S rRNA gene, and the V6-V8 and V3-V5 regions of the respective bacterial and archaeal 16S rRNA genes were amplified using the primers described in Comeau et al. (2011). PCR was carried out as above, but in a total volume of 20 μl and with BSA omitted. Three reactions were performed for each sample, using 0.5, 1, and 2 μl of template cDNA. For eukaryotes, the thermal cycling protocol was as described above. For Bacteria, the protocol was as described above but with 25 cycles instead of 30 and a final extension step of 7 min. For Archaea, the protocol was also as described above but with an annealing temperature of 52°C, 35 cycles, and a final extension step of 5 min. PCR products from the three different concentrations of template cDNA were pooled and purified by use of magnetic beads (Axygen, Corning, New York, NY, USA). PCR product bound to magnetic beads was washed twice with 80% ethanol and eluted in 10 mM Tris buffer (pH 8.13). A second PCR used primers provided by Illumina (San Diego, CA, USA) to add barcodes to the PCR products. Each sample received a unique combination of forward and reverse primers. The purified DNA template was diluted 10X, and concentrations of reagents and thermocycler protocol were the same as in the first PCR, except that the thermocycler was run for only 15 cycles. Two reaction volumes of 25 μl each were run for each sample and then pooled before PCR products were again purified using magnetic beads. The DNA concentration was measured spectrophotometrically using a Nanodrop 1000 (Nanodrop, Wilmington, DE, USA). Samples were mixed equimolarly, along with barcoded samples from other projects, and the amplicons were then paired-end sequenced on an Illumina MiSeq platform at the IBIS/Laval University Plate-forme d'Analyses Génomiques (Québec, QC, Canada). Raw reads have been deposited in the NCBI database under accession number SRP076684. An initial run was truncated because of an error in reagent volumes. The sequences from this run were analyzed together with the sequences from a completed run, resulting in 16% more total reads.
Sequence Processing
Sequences were analyzed using the UPARSE pipeline (Edgar, 2013) as implemented in QIIME (Caporaso et al., 2010). Reads with phred quality scores <20 were discarded. Chimeras were detected both de novo and by reference to the SILVA database (Pruesse et al., 2007) and the intersection of good sequences from these two methods were retained, before clustering into Operational Taxonomic Units (OTUs) at 97% similarity (98% for eukaryotes). Singleton OTUs were discarded. OTUs were assigned a taxonomic identity using the Mothur classifier (Schloss et al., 2009) with a 0.8 confidence threshold using an in-house curated taxonomic database (Lovejoy et al., 2016). Very abundant OTUs were manually checked for chimeras by performing separate BLAST searches on the front and back halves of the sequence, and a few abundant OTUs that could not be identified with Mothur were successfully identified using a BLAST search.
The sequences amplified with Archaea-specific primers were found to contain a number of sequences from the other two domains even after processing. These were detected and removed using the Ribosomal Database Project Classifier (Wang et al., 2007). An Archaea reference tree was constructed using an alignment of 73 16S rRNA sequences obtained from GenBank and aligned using MUSCLE (Edgar, 2004). The alignment had 1496 characters. A maximum-likelihood tree was constructed using RAxML v7.2.7 (Stamatakis, 2006), with 100 bootstraps, and rooted using Korarchaeum cryptofilum. Short archaeal reads were mapped onto the reference tree using the Evolutionary Placement Algorithm (EPA) of RAxML (Berger et al., 2011). Only read placements with likelihood weight of >0.5 were retained.
The phylogenetic distance among communities from different depths and size fractions was analyzed for each of the three domains of life using the weighted Unifrac beta diversity metric (Lozupone and Knight, 2005) as implemented in Mothur.
Results
Air Temperature and Water Column Structure
Meteorological records from nearby Ward Hunt Island beginning in 1996 have shown that regional summer air temperatures were warmer during 2010–2012 in comparison to most of the preceding decade (Paquette et al., 2015; CEN, 2016). Air temperature records from an automated weather station in Milne Fiord are shorter, beginning in 2009, but reveal that 2011 and 2012 were particularly warm summers. For the period 2009–2015, the mean (standard deviation) number of melting degree days was 203 (76), compared to 185, 278, and 253 in 2010, 2011, and 2012, respectively. The maximum hourly air temperature for 2009–2015 was 19.9°C, occurring in July 2012.
Water column profiles showed a several meter thick relatively warm (up to 1.5°C) freshwater layer above colder (<−1.5°C) seawater in all years (Figure 2). Salinity increased from about 0.1 at the surface to >30 below 25 m depth, with a sharp salinity gradient at the base of the freshwater layer. The depth of the halocline, defined as the level of the maximum salinity gradient, varied from approximately 15 m in 2010, to 14 m in 2011, and 11 m in 2012. In each year, the depth of the thermocline and oxycline coincided with the halocline, with the warm freshwater layer relatively well-oxygenated compared to that of seawater below. The nutricline also appeared to coincide with the halocline, with the surface freshwater depleted in both nitrogen and phosphorus each year (Figure 3).
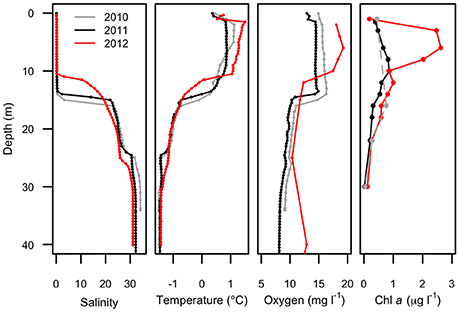
Figure 2. Physical profiles of Milne Fiord epishelf lake over 3 years (2010–2012) showing salinity, temperature, oxygen concentration, and chlorophyll a concentration. Note the fewer depths for chlorophyll a in 2010 (dashed line). Oxygen data in 2012 are for the main fiord; all other profiles are from Neige Bay.
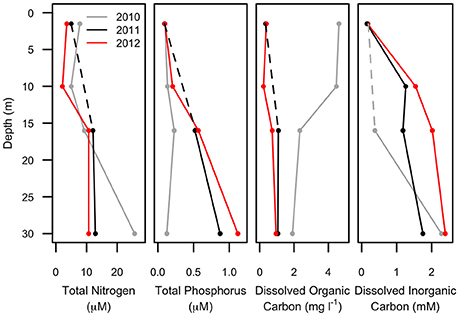
Figure 3. Chemical profiles of Milne Fiord epishelf lake over 3 years (2010–2012) showing total nitrogen, total phosphorus, dissolved organic and inorganic carbon. Note that the 10-m sample is missing for some variables in some years (dashed line).
Water properties above 23 m depth in Neige Bay were similar to measured water properties in the main fiord (not shown), while the properties of deep water in Neige Bay were distinct, and nearly homogenous with depth to the bottom. These observations suggested the existence of a 23 m-deep sill at the mouth of Neige Bay. The 2012 oxygen profile was collected in the main fiord and showed higher oxygen concentrations below 25 m compared to the other years when profiles were collected in Neige Bay. This suggests that the deep water in Neige Bay was slightly depleted in oxygen relative to the main fiord, but oxygen concentrations were >7 mg l−1 at all depths suggesting that the water was likely renewed annually or even more frequently.
Pigment Composition
Maximum Chl a (Figure 2) was observed in the surface freshwater layer and reached 0.72 μg l−1 at 10 m in 2011, and 2.53 μg l−1 at 6 m in 2012. In 2010, a peak was not observed in the upper layer, perhaps because only one sample was taken in this layer, at 1.5 m. In all years, zeaxanthin, likely associated with picocyanobacteria, was observed in the upper water column down to 22 m (Figure 4). Violaxanthin, chlorophyll b, alloxanthin and lutein were found as important pigments in the freshwater surface layer, indicating the presence of chlorophytes and cryptophytes. In 2011, the presence of chlorophyll c2, peridinin and diadinoxanthin just beneath the ice suggested the presence of dinoflagellates. In the deeper seawater layer, fucoxanthin and chlorophyll c3 were observed, indicating the presence of either diatoms or prymnesiophytes.
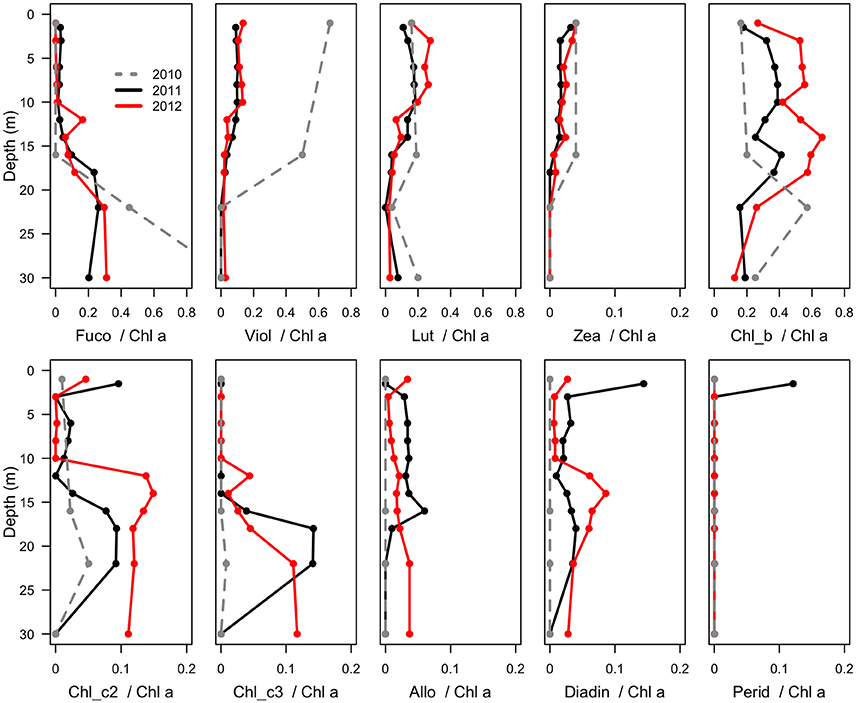
Figure 4. Mass ratios of photosynthetic pigments relative to chlorophyll a in the water column of Milne Fiord epishelf lake over 3 years (2010–2012), determined by HPLC analysis. Note the fewer sample depths in 2010 (dashed line). Fuco, fucoxanthin; Viol, violaxanthin; Lut, lutein; Zea, zeaxanthin; Chl, chlorophyll; Allo, alloxanthin; Diadin, diadinoxanthin; Perid, peridinin.
Flow Cytometry
Picocyanobacteria and Chl a-containing picoeukaryotes were mainly detected in the surface freshwater layer (Figure 5). Maximal abundance of picocyanobacteria was observed at different depths across years of sampling: at 8 m in 2010 with 2.1 × 104 cell ml−1, and at 14 m in 2011 with 2.8 × 103 cells ml−1. In 2012, no maximum was observed and the abundance was in general lower, ranging from 0.3 to 1 × 103 cells ml−1 down the water column. The picoeukaryotes were more evenly distributed through the water column, with minor peaks at 18 (2010), 16 (2011), and 14 m (2012). The average abundance in the freshwater layer was 3.4 (±0.8) × 103 cells ml−1 in 2010 and 2.7 (±0.5) × 103 cells ml−1 in 2011, rising to 6.6 (±1.9) × 103 cells ml−1 in 2012.
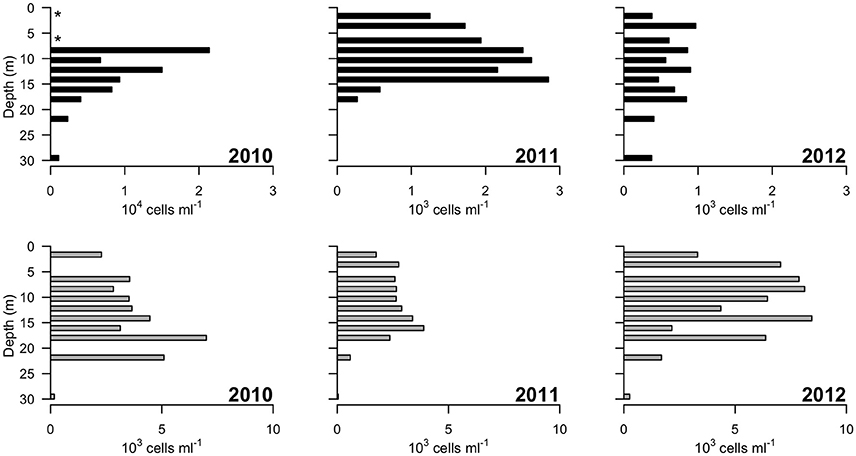
Figure 5. Concentrations of photosynthetic cells determined by flow cytometry over 3 years (2010–2012). Top panels: picocyanobacteria; bottom panels: picoeukaryotic cells containing Chlorophyll a. Note the order of magnitude difference in the x-axis of the leftmost top panel. “*” indicates depths at which values of zero cells were measured, while in 2011 picocyanobacteria data from 22 and 30 m were not available.
High Throughput Sequencing (HTS)
Approximately 925,000 reads were retained after processing for quality control (Table 1), consisting of a total of 3575 microbial OTUs. In all domains, OTU richness was lowest in freshwater sample immediately beneath the ice (1.5 m). This richness was similar in both size fractions, with the exception of 30 m, where Bacteria had a greater OTU richness in the >3 μm size fraction, pointing to a greater diversity of particle-attached bacteria at this depth. Archaeal sequences could not be identified below phylum level using sequence similarity, but with EPA approximately 70% of sequences could be placed on a reference tree at class level or below (Figure 6; Figure S1B). Most of the sequences whose placement fell below the Maximum Likelihood cut-off of 0.5 appeared related to Marine Benthic Group B (at 30 m) and Group I.1a (at the surface), both members of Thaumarchaeota.
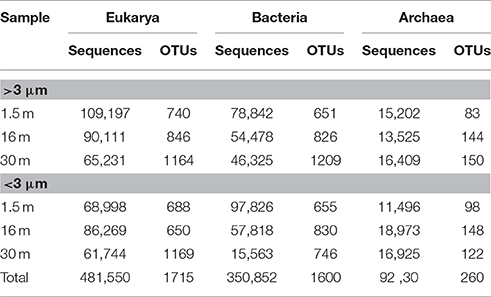
Table 1. Number of sequences and Operational Taxonomic Units (OTUs) for samples from Milne Fiord epishelf lake in July 2011, according to depth and size fraction, after quality screening.
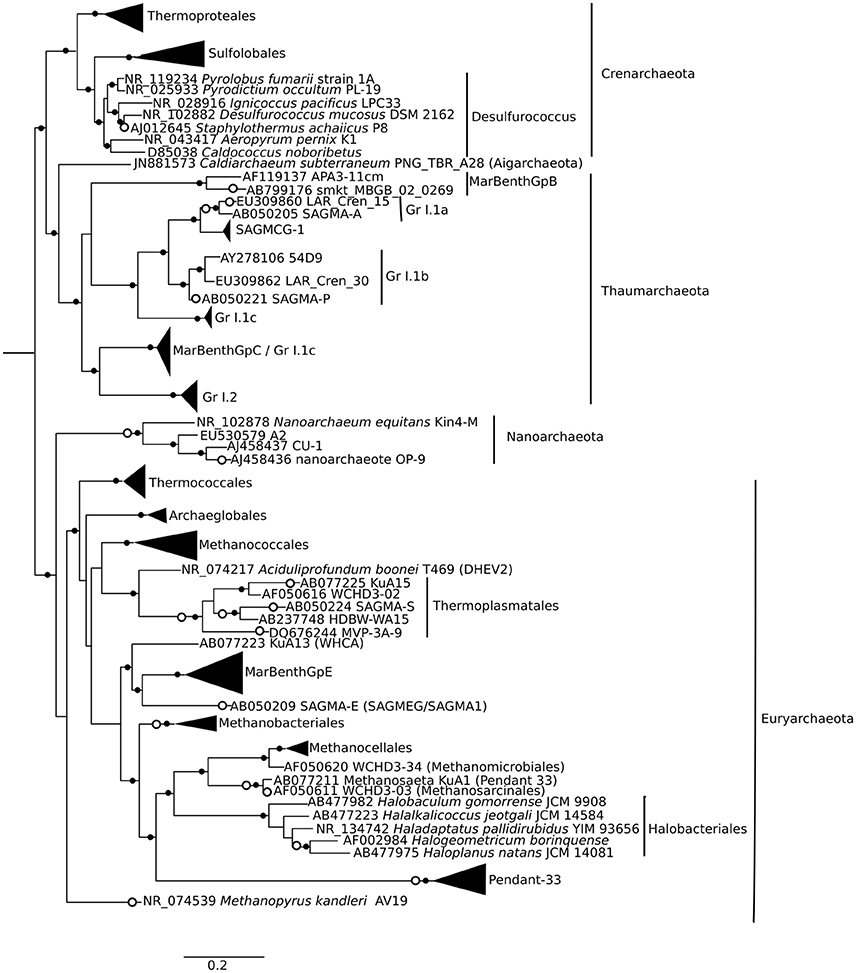
Figure 6. Phylogenetic mapping of archaeal short reads. The reference tree, constructed using maximum likelihood from an alignment of 73 sequences and 1496 characters, is rooted using Korarchaeum cryptofilum (not shown). Small closed circles indicate nodes with bootstrap support of >50 (of 100). Large open circles indicate where short reads from Milne Fiord samples in July 2011 mapped to nodes using Evolutionary Placement Algorithm (EPA). Scale bar indicates the number of substitutions per position.
Samples clustered based on their phylogenetic distance by depth rather than size fraction for all three domains of life, and surface samples were the most distinct from other depths (Figure 7). The surface community of Eukarya separated from the 16 and 30-m communities, while for the Bacteria and Archaea the 30-m sample separated from the cluster of surface and 16-m communities.
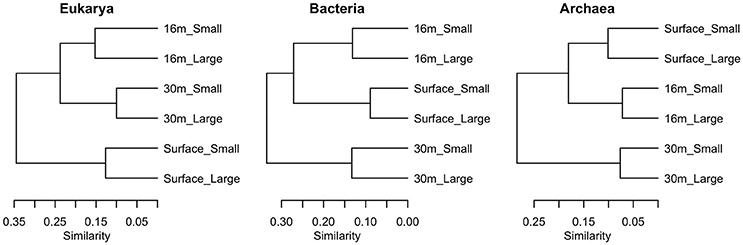
Figure 7. Clustering of samples using weighted Unifrac distance based on sequences from each of the three domains in July 2011, in two size fractions, Large (>3 μm) and Small (<3 μm).
With the exception of the 30-m sample, the bacterial community was dominated by the phylum Proteobacteria (Figure 8). A few taxa, such as Flavobacteria and the Roseobacter-group were more abundant in the >3 μm (particle-attached) fraction, while Sphingobacteria and Polaromonas were more abundant in the <3 μm (free-living) fraction, but the majority of taxa showed no distinction by size. Some bacterial taxa showed no clear pattern with depth when considered at a high taxonomic level (for example Gammaproteobacteria, which constituted 5–15% of reads in all samples), but evidence of vertical structure was observed when Unifrac distances were applied at the OTU level. The bacterial community at the surface was strongly dominated by a single betaproteobacterium OTU (45%) which was 99.5% identical to the corresponding V4 region of Rhodoferax saidenbachensis. At the halocline, there were noticeable peaks of the betaproteobacterium genus Polaromonas (17%) and the alphaproteobacterial Roseobacter-group (33% in the >3 μm size fraction), while several non-Proteobacteria taxa appeared restricted to 30-m samples, including Acidobacteria, Chloroflexi, Planctomycetes, and the environmental clade PAUC34f.
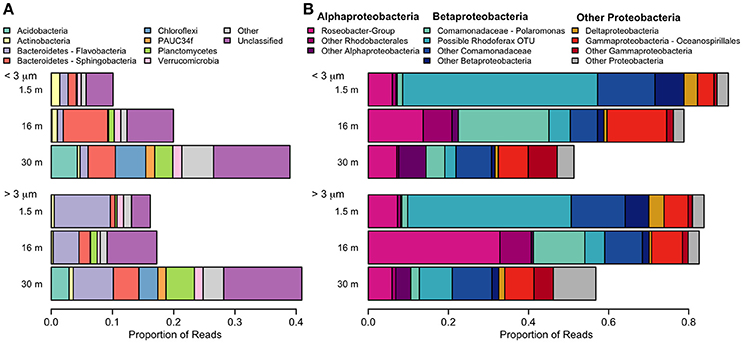
Figure 8. Relative abundance of taxa detected with small-subunit rRNA high through-put sequencing from July 2011. (A) Bacteria, excluding Proteobacteria. (B) Proteobacteria. Pink colors are alphabacteria, blue colors are betaproteobacteria.
The dominant microbial eukaryotes were the Chlorophyte group Radicarteria, dinoflagellates, and, in the freshwater layer, a species of cryptophyte (Figure 9). Fungi were not detectable in our sequences. Although the surface had the lowest OTU richness, it had the greatest evenness of high-level eukaryote groups, including the cryptophyte Teleaulax gracilis (23%), chrysophytes (3.5% of reads in surface samples), and the rhizarian Protaspis (4.7%) (Figure 9A). Ciliates at the surface included a higher proportion of Litostomatea (3.4%), and dinoflagellates included the photosynthetic genus Woloszynskia (4.0%). The community composition of photosynthetic taxa agreed well with pigment data from HPLC. All of these surface-associated taxa diminished markedly in relative abundance at greater depth, or disappeared entirely. In contrast, Radicarteria comprised a high proportion of reads at all depths, though it reached its greatest dominance at the halocline (57%; Figure 9A, Figure S1A), while a Unifrac analysis of this taxon did not separate the 16 and 30-m depths (analysis not shown). The distribution of several alveolate taxa showed vertical structure (Figure 9B), with the chloroplast-bearing dinoflagellate Prorocentrales becoming dominant at 16 and 30 m, while the genus Katodinium increased to 2.9% in the <3 μm fraction at 30 m. Marine Alveolates, mainly the parasitic MALV I, appeared restricted to the 30-m sample (4.1%).
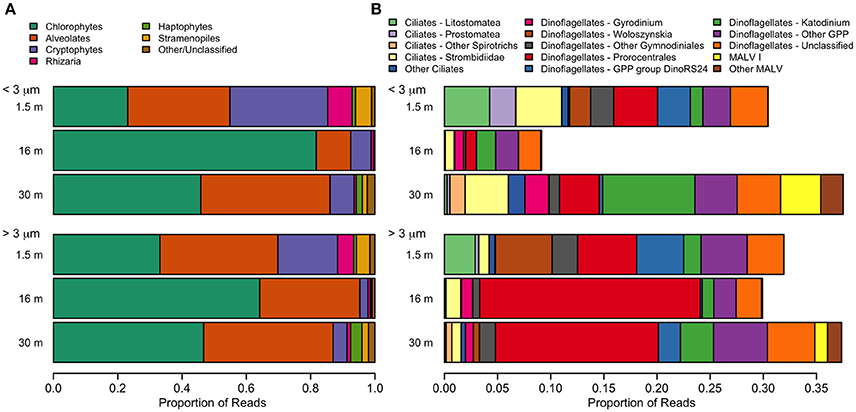
Figure 9. Relative abundance of taxa detected with small-subunit rRNA high through-put sequencing from July 2011. (A) Eukarya. The category “Other” includes small numbers of choanoflagellates, katablepharids, Picozoa, and Telonemia. (B) Alveolates. A small number of non-GPP dinoflagellates (<0.1%) were also detected.
Although there was a large number of unclassified reads in Archaea, some structuring by depth was apparent. The Archaea environmental clade Pendant-33 was only detected in the freshwater layer (3.8% in the small size fraction), while Halobacteriales was restricted to 30 m. At all depths, the most dominant taxon by far was Group I.1a Thaumarchaeota (Figure S1A).
Discussion
General Features of the Milne Fiord Epishelf Lake Ecosystem
Previous studies on Milne Fiord epishelf lake (Veillette et al., 2008, 2011) have drawn attention to the highly stratified nature of this ecosystem, with freshwater overlying saltwater and a vertical separation of freshwater and marine biota, as found in Antarctic epishelf lakes (Laybourn-Parry and Wadham, 2014). Our observations confirmed this vertical structure, with a strong halocline separating the upper layer, derived from snow and glacial meltwater, from the water below which was derived from the Arctic Ocean. Similarly, the HPLC results showed a separation between pigmented organisms above and below the halocline (Figure 10), although this transition was less sharply defined in the FCM data (Figure 5). The RNA sequences showed some taxa present at all depths, but in general there was strong separation between the upper and lower communities (Figures 7–9). Particularly for the eukaryotes, this separation was less evident at higher taxonomic levels (Figure 8A) than with OTU-level analysis (Figure 7), highlighting the importance of analyzing communities at the appropriate taxonomic scale.
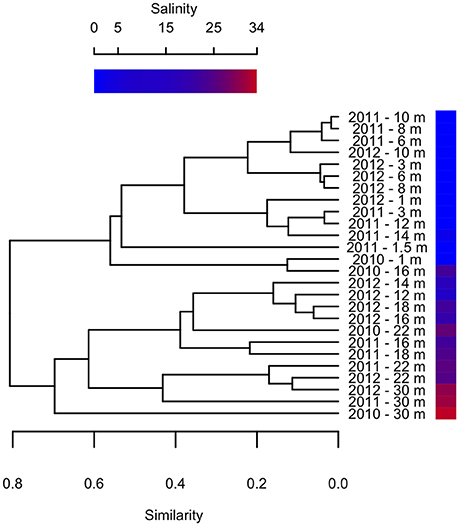
Figure 10. Bray-Curtis clustering of samples from different depths in 2010–2012 according to pigment composition. Sample salinity is shown by gradient.
Microbial Community Composition
Identifying the taxonomic affiliation of short reads generated by HTS requires comprehensive, curated taxonomic databases. While many excellent resources exist, such as SILVA and the Ribosomal Database Project (Pruesse et al., 2007; Cole et al., 2014), these large databases cannot keep pace with the current rapid rate of phylogenetic revision. To improve the taxonomic resolution of our reads, we developed and applied a curated database for Eukarya and Bacteria (Lovejoy et al., 2016), following the approach in Comeau et al. (2011) and incorporating the published taxonomic revisions for abundant groups in our northern lake and ocean datasets, including Milne Fiord epishelf lake. This tailored database allowed a high degree of taxonomic resolution for some groups, as far as species-level. On the other hand, the lack of reference sequences for Archaea made taxonomic identification by sequence similarity ineffective. We therefore used EPA as described in Thaler and Lovejoy (2015) to identify short reads by mapping them onto a reference phylogenetic tree (Figure 6). Since 16S rRNA gene analysis is known to be poor at resolving high-level taxonomy in Archaea, particularly in distinguishing Crenarchaeota and Thaumarchaeota (Brochier-Armanet et al., 2008), future environmental studies may choose to target other marker genes.
The microbial community in the Milne Fiord epishelf lake identified by HTS in 2011 was in broad agreement with FCM and HPLC results. In the same year, phytoplankton in the layer beneath the lake ice were dominated by a combination of the microplanktonic chlorophytes in the Radicarteria group that have a reported cell diameter of 30 μm (Balzano et al., 2012) and the nanoplanktonic cryptophyte Teleaulax gracilis, that has a reported cell diameter of 7–12 μm (Laza-Martínez et al., 2012; Figure 9). The dominance of these larger cells may account for the lower concentrations of pigmented picoeukaryotes detected by FCM in 2011 relative to other years (Figure 5). In 2009, a high proportion of prasinophytes was detected at the halocline from HPLC (Veillette et al., 2011), likely Micromonas sp. (Lovejoy et al., 2007); in contrast, this group was among the least abundant phytoplankton taxa in 2011. HTS did not indicate any particular taxon that could have accounted for the fucoxanthin and chlorophyll c3 detected at 30 m (Figure 4), although prymnesiophytes (2.7% of reads) seem more likely than diatoms (<1% of reads). One discrepancy between the approaches was the Cyanobacteria, which appeared as high as 3000 cells ml−1 in FCM, but were always <1% of sequences. This was likely because heterotrophic bacteria were present at a much higher concentration than cyanobacteria, competing for PCR primers. Overall, FCM, HPLC, and HTS provided complementary views of the communities.
Clustering analysis showed that pigment composition grouped primarily according to salinity of the sample, and to a lesser extent by sampling year (Figure 10), suggesting that different photosynthetic communities occurred above and below the halocline. However, at least one abundant taxon, the eukaryote Radicarteria, was abundant at all depths. Samples were clustered by depths based on the abundance and phylogenetic distances between different rRNA sequences, in contrast to other studies which found that community composition of particle-attached and free-living bacteria correlated with different environmental variables (Dupont et al., 2014). This may be because, in the Milne Fiord epishelf lake, most physico-chemical variables are so strongly correlated with depth (Figure 2) that their differing effects cannot be disentangled.
For Bacteria, the overall dominance of Betaproteobacteria is indicative of a freshwater influence, as found elsewhere in the Arctic (Galand et al., 2008b). We found Betaproteobacteria at a lower proportion in deeper, more saline waters (Figure 8). Finer taxonomic identification indicated the presence of both marine and freshwater groups in the halocline, with peaks in the relative abundance of the typically-marine alphaproteobacterial Roseobacter-group (Pujalte et al., 2014) and the sometimes freshwater or ice-associated betaproteobacterium Polaromonas (Darcy et al., 2011). This latter taxon has also been reported from the water column beneath Antarctic sea-ice (Irgens et al., 1996), and it dominated the 5-m enrichment sample from Milne Fiord epishelf lake in 2007 (Veillette et al., 2011). In addition to marine and freshwater influences, the lake also receives glacial inputs. To date, there have been few HTS studies of glacier microbial communities. One study of cryoconite holes in Svalbard (Edwards et al., 2014) found similar groups at a high taxonomic levels, but a detailed meta-analysis would be needed to determine whether glacial meltwater may have had an influence on the Milne Fiord epishelf lake community.
Two previous HTS studies of freshwater environments on northern Ellesmere Island have reported Fungi, although at relative abundances of <1% (Comeau et al., 2016). Their absence from our sequencing data indicates that their concentration in Milne Fiord Epishelf Lake must have been quite low. The MALV I found in the deep layer (Figure 9B) belonged to the Radiolarian-associated clade RAS1 identified by Bråte et al. (2012). Radiolarians, though found in many mesopelagic water columns, were very rare in our 30 m samples (<0.001%). However, these authors found that transitions between hosts may be common in the evolutionary history of this group, so these MALV could conceivably be infecting other protist groups in the seawater layer below the lake.
Mixotrophy may be a key survival strategy for many microbial flagellates that are at the base of food webs in nutrient-poor Arctic lakes (Charvet et al., 2014), and many of the taxa found in the freshwater layer of the lake were potential phagotrophic mixotrophs. For example, one species of the chloroplast-bearing woloszynskioid dinoflagellates is known to feed on nanoplanktonic algal prey (Kang et al., 2011), and has previously been reported to form a bloom in the freshwater that accumulates beneath sea-ice in the Baltic Sea (Spilling, 2007). Unrein et al. (2014) detected bacterivory in cryptophytes in a mixed natural community that included the genus Teleaulax. Finally, one of the ciliate groups associated with the surface, Litostomatea, includes species which can acquire phototrophy through a cryptophyte endosymbiont or through kleptoplasty of cryptophyte plastids (Stoecker et al., 2009). Given the relative abundance of available cryptophyte prey immediately below the ice, it may be speculated that the litostomate ciliates in the surface of the lake might use this strategy; however, microscopy combined with fluorescent in situ hybridization would be need to confirm this conjecture. Dinoflagellate mixotrophs, in the class Prorocentrales (Stoecker, 1999), also comprised a high proportion of eukaryote reads at 16 and 30 m (Figure 9B).
Many of the dominant taxa in Milne Fiord epishelf lake have previously been reported from Arctic water columns, and some are known to be ice-associated, such as the eukaryotes Teleaulax (Piwosz et al., 2013) and Protaspis (Werner et al., 2007), the bacterium Polaromonas (Irgens et al., 1996), and the archaeon Halobacteriales (Collins et al., 2010). Teleaulax gracilis has only been described recently, with accompanying revision of the genus (Laza-Martínez et al., 2012), and therefore there are no historic records for this species. However, the related species T. acuta, has been reported to form a bloom in an Atlantic-influenced fiord in Svalbard (Kubiszyn et al., 2014). Similarly, among the Archaea, Thaumarchaeota are generally thought of as marine in Arctic estuarine systems (Galand et al., 2008a), and Group I.1a has been reported as the dominant archaeon in first-year sea-ice (Collins et al., 2010). However, Group I.1a is also reported from diverse freshwater environments worldwide (Bomberg et al., 2008), and Galand et al. (2008b) identified a freshwater clade within the group. The increase in relative abundance of this group at 16-m depth (Figure S1B) suggests they were primarily marine, though able to tolerate euryhaline conditions. Since Veillette et al. (2011) found that Group I.1a also dominated archaea in the enrichments from Milne Fiord epishelf lake in 2007, they may be a consistent feature of this community.
Other taxa identified in Milne Fiord epishelf lake have either not been previously reported from the Arctic, or have never been observed as dominant in a fiord environment. For example, the Radicarteria clade, which includes Carteria radiosa and Carteria obtusa from the polyphyletic chlorophyte genus Carteria (Nakada et al., 2008), dominated the eukaryote community at all depths and size fractions (Figure 9A). Historically, these two species have mainly been reported by researchers in Russia and the Ukraine, perhaps because of a greater familiarity with their taxonomic descriptions in those countries. Nearly all microscopic records are from terrestrial, mainly freshwater systems (Compère, 1966; Khondker et al., 2007; Tsarenko, 2008; Demchenko, 2011; Hindáková and Hindák, 2012; Gorokhov, 2013; Tarasova et al., 2013), but also one hypersaline lake (Gheorghievici et al., 2015). However, two marine strains have been brought into culture, both from the Arctic: one from the ice algae community in Baffin Bay (Bigelow National Center for Marine Algae and Microbiota, CCMP1189), and one from the deep chlorophyll maximum of the Beaufort Sea (Balzano et al., 2012). The identities of both these cultures were confirmed by phylogenetic analysis relating them to cultured freshwater strains of C. radiosa and C. obtusa. This suggests that members of this clade are euryhaline, or have diversified to fill different ecological niches, similar to the dinoflagellate Polarella (Rengefors et al., 2015) or the Peridinium-Scrippsiella complex, which is found in both seawater and cold water lakes (Logares et al., 2009). The dominance of this eukaryote phytoplankton in Milne Fiord epishelf lake is in distinct contrast to other aquatic Arctic communities studied by HTS. For example, in meromictic lakes located near Milne Fiord, the dominant phytoplankton taxa were chrysophytes (Charvet et al., 2014), while coastal marine waters in the Arctic are dominated by haptophytes, diatoms, and picochlorophytes, depending on the season (Pedrós-Alió et al., 2015).
Protaspis, the most abundant genus among OTUs of the Phylum Rhizaria, has been detected in Arctic and Baltic sea-ice (Werner et al., 2007; Majaneva et al., 2012), but its presence here in the water column may be unusual. The only planktonic record to date is a parasitoid of diatoms, Protaspis longipes (= Cryothecomonas longipes; Hoppenrath and Leander, 2006); however, the low frequency of diatom sequences in this layer (<1%) suggests that these Protaspis were either bacterivorous or parasitic on non-diatom taxa.
The betaproteobacterium OTU that dominated the surface water of the lake (Figure 8B) had closest sequence similarity to R. saidenbachensis, a non-photosynthetic, strictly aerobic, psychrotolerant species which has been described from the sediments of a temperate lake (Kaden et al., 2014). Based on related environmental sequences, these researchers speculated that it has an affinity for low-nutrient environments, which would be the case for Milne Fiord epishelf lake. The genus has also been recovered in a clone library from sediment in the perennial ice of an Antarctic lake (Gordon et al., 2000). However, our phylogenetic analysis of the whole 16S rRNA gene using reference sequences obtained from NCBI GenBank suggested there is a high level of undescribed diversity in the genus Rhodoferax (Figure S2), which may conceal a large amount of phenotypic variability. Zwart et al. (2002) describes an environmental clade they call “Rhodoferax sp. Bal47,” which they report as widespread in freshwater, estuarine and coastal habitats worldwide, and which was also detected in the Mackenzie River outflow in the Arctic (Galand et al., 2008b). However, our analysis shows that their clade was misidentified and actually corresponds to the well-studied freshwater genus Limnohabitans (Figure S2), which occurred at very low relative abundance in our samples (<0.001% in the >3 μm fraction only). The present study may therefore be the first to demonstrate that Rhodoferax can be a dominant component of a bacterial community in a freshwater environment.
The Archaea order Halobacteriales was originally believed to be obligately halophilic, but they have since been detected in tidal sediments of low to moderate salinity (Purdy et al., 2004). Members of this taxonomic order have also been detected by T-RFLP fingerprinting in first-year Arctic sea ice, under conditions of extreme salinity many times that of seawater (Collins et al., 2010). To our knowledge, ours is the first study that shows them to be a major fraction of the archaeal community in a marine water column.
Ecosystem Effects of Deteriorating Milne Ice Shelf
The multi-annual time series presented in this study is for only 3 years of study, reflecting the great difficulty of accessing this remote site in the extreme high Arctic. However, in the context of other Arctic epishelf lakes that have experienced abrupt, non-linear changes right before total break-up (Mueller et al., 2003; White et al., 2015), the shorter time-scale may nevertheless be sufficient to capture regime-changing events. The freshwater layer of the Milne Fiord epishelf lake had a recorded thickness of 17.5 m in 1983, and similar values were measured in 2004 (Veillette et al., 2008). However, in 2007 the halocline was at 16.6 m, rising further to 14.3 m by 2009 (Veillette et al., 2011). Our observations in the present study indicated a similar halocline depth in 2010 and 2011, shoaling to 14 m (Figure 2), but the chemical data imply some degree of mixing between these 2 years, with a pronounced increase in TP and DIC at 16 m, accompanied by a drop in DOC (Figure 3). The presence of the most abundant taxa in the RNA sequence data at all sampling depths in 2011 is also consistent with halotolerant species in a partially mixed layer.
Much greater physico-chemical changes were observed in 2012, by which time the halocline had risen to 11 m. The salinity profile was particularly striking in that the halocline had lost the extreme sharpness of transition from freshwater to saltwater observed in previous years, and there was a pronounced increase in TP and decrease in DOC at 10 m (Figure 3), possibly as a result of this mixing. In part these changes may relate to the long term thinning of the ice shelf that is evident over timescales of decades. However, short term variations have also been observed, including strong seasonal changes in the thickness of the lake (by several meters), and episodic mixing events in the fiord that can change the depth of the lake, including in the winter of 2011–2012 (Hamilton, A.K. et al., unpublished data).
The biological observations from 2012 showed marked differences relative to the previous years. The picocyanobacteria declined by an order of magnitude, and were distributed throughout the water column, consistent with mixing, and the picoeukaryotes showed a more than twofold increase (Figure 5). Chl a concentrations were higher in 2012 than 2011, while oxygen concentrations also indicate potentially higher net photosynthetic rates in 2012 than in either of the two previous years (Figure 2). There was a rise in the ratio of some accessory pigments per unit Chl a, notably Chl b (Figure 4), implying an increased proportional abundance of green algae. In part these changes may be the result of year-to-year variability, but the trend of decreasing freshwater layer thickness over the period 2004–2012 and the weakening of the density gradient at the halocline implies that this ice-dammed ecosystem is in a period of transition associated with the meltdown of its ice shelf. If the Milne Ice Shelf thins further and fractures, the freshwater layer would completely drain from the lake, as observed in Disraeli Fiord (Mueller et al., 2003), and the microbial community would shift to an exclusively marine composition. Although the ultimate ecosystem of Milne Fiord remains uncertain, it may come to resemble the stratified, flagellate-dominated fiords observed in Svalbard by Carrier (2016).
Conclusions
The epishelf lake in Milne Fiord has many features that appear unique among Arctic aquatic ecosystems, including a combination of marine and freshwater taxa, the predominance of the eukaryotic phytoplankton Radicarteria, and a number of other taxa rarely reported from marine water columns. Our multi-annual observations cover a limited period, but they imply that 2010–2012 corresponded to a period of transition in terms of community structure as inferred from pigments, as well as in the physical and chemical environment. If the ice cover of the lake continues to break up seasonally, increased light and the mixing of nutrients and salt into the nutrient-depleted surface freshwaters will likely have further effects on microbial community structure.
Author Contributions
WV and ML designed the study; ML, WV, and AH undertook the field sampling and profiling; ML and MT performed laboratory analyses; MT and CL analyzed and interpreted sequence data; MT and ML led the drafting of the manuscript, and all authors contributed to and have approved the final version.
Funding
This research was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC); the Networks of Centres of Excellence program ArcticNet; the Canada Research Chair program; the Northern Scientific Training Program; the Canadian Foundation for Innovation; the Canadian Northern Studies Trust; Centre d'études nordiques (CEN); Fonds de Recherche du Québec-Nature et Technologie (FRQNT); and the Association of Canadian Universities for Northern Studies (ACUNS).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sébastian Bourget, Sophie Charvet, and Denis Sarrazin for assistance in the field and Pierre Galand for taxonomic advice. Logistical support was provided by the Polar Continental Shelf Program (PCSP) and we are also grateful to Parks Canada for the use of their facilities at Ward Hunt Island.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fmars.2016.00275/full#supplementary-material
References
Balzano, S., Gourvil, P., Siano, R., Chanoine, M., Marie, D., Lessard, S., et al. (2012). Diversity of cultured photosynthetic flagellates in the northeast Pacific and Arctic Oceans in summer. Biogeosciences 9, 4553–4571. doi: 10.5194/bg-9-4553-2012
Berger, S. A., Krompass, D., and Stamatakis, A. (2011). Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst. Biol. 60, 291–302. doi: 10.1093/sysbio/syr010
Blazewicz, S. J., Barnard, R. L., Daly, R. A., and Firestone, M. K. (2013). Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 7, 2061–2068. doi: 10.1038/ismej.2013.102
Bomberg, M., Montonen, L., Münster, U., and Jurgens, G. (2008). Diversity and function of archaea in freshwater habitats. Curr. Trends Microbiol. 4, 1–89.
Bråte, J., Krabberød, A. K., Dolven, J. K., Ose, R. F., Kristensen, T., Bjørklund, K. R., et al. (2012). Radiolaria associated with large diversity of marine alveolates. Protist 163, 767–777. doi: 10.1016/j.protis.2012.04.004
Brochier-Armanet, C., Boussau, B., Gribaldo, S., and Forterre, P. (2008). Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6, 245–252. doi: 10.1038/nrmicro1852
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carrier, V. (2016). Structure et Biogéographie des Communautés de Pico- et de Nanoeucaryotes Pélagiques autour de l'Archipel l'Archipel du Svalbard. [MSc Thesis], Université Laval.
Casamayor, E. O., Ferrera, I., Cristina, X., Borrego, C. M., and Gasol, J. M. (2007). Flow cytometric identification and enumeration of photosynthetic sulfur bacteria and potential for ecophysiological studies at the single-cell level. Environ. Microbiol. 9, 1969–1985. doi: 10.1111/j.1462-2920.2007.01313.x
CEN (2016). Climate station data from Northern Ellesmere Island in Nunavut, Canada, v. 1.3 (2002–2015). Nordicana D1. doi: 10.5885/44985SL-8F203FD3ACCD4138
Charvet, S., Vincent, W. F., and Lovejoy, C. (2014). Effects of light and prey availability on Arctic freshwater protist communities examined by high-throughput DNA and RNA sequencing. FEMS Microbiol. Ecol. 88, 550–564. doi: 10.1111/1574-6941.12324
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Collins, R. E., Rocap, G., and Deming, J. W. (2010). Persistence of bacterial and archaeal communities in sea ice through an Arctic winter. Environ. Microbiol. 12, 1828–1841. doi: 10.1111/j.1462-2920.2010.02179.x
Comeau, A. M., Li, W. K., Tremblay, J.-É., Carmack, E. C., and Lovejoy, C. (2011). Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS ONE 6:e27492. doi: 10.1371/journal.pone.0027492
Comeau, A. M., Vincent, W. F., Bernier, L., and Lovejoy, C. (2016). Novel chytrid lineages dominate fungal sequences in diverse marine and freshwater habitats. Nat. Sci. Reports 6:30120. doi: 10.1038/srep30120
Compère, P. (1966). Observations sur les algues des groupement a Sphaignes des Hautes-Fagnes de Belgique. Bull. Jardin Bot. l'État a Bruxelles 36, 5–51.
Darcy, J. L., Lynch, R. C., King, A. J., Robeson, M. S., and Schmidt, S. K. (2011). Global distribution of Polaromonas phylotypes—evidence for a highly successful dispersal capacity. PLoS ONE 6:e23742. doi: 10.1371/journal.pone.0023742
Demchenko, E. N. (2011). Water bloom of ephemeral water bodies from city Kiev (Ukraine) [In Russian]. Flora Geogr. 21, 87–105.
Dupont, C. L., Larsson, J., Yooseph, S., Ininbergs, K., Goll, J., Asplund-Samuelsson, J., et al. (2014). Functional tradeoffs underpin salinity-driven divergence in microbial community composition. PLoS ONE 9:e89549. doi: 10.1371/journal.pone.0089549
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edwards, A., Mur, L. A., Girdwood, S. E., Anesio, A. M., Stibal, M., Rassner, S. M., et al. (2014). Coupled cryoconite ecosystem structure-function relationships are revealed by comparing bacterial communities in alpine and Arctic glaciers. FEMS Microbiol. Ecol. 89, 222–237. doi: 10.1111/1574-6941.12283
Edwards, U., Rogall, T., Blöcker, H., Emde, M., and Böttger, E. C. (1989). Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17, 7843–7853. doi: 10.1093/nar/17.19.7843
Galand, P. E., Lovejoy, C., Pouliot, J., Garneau, M.-È., and Vincent, W. F. (2008b). Microbial community diversity and heterotrophic production in a coastal Arctic ecosystem: a stamukhi lake and its source waters. Limnol. Oceanogr. 53, 813–823. doi: 10.4319/lo.2008.53.2.0813
Galand, P. E., Lovejoy, C., Pouliot, J., and Vincent, W. F. (2008a). Heterogenous archaeal communities in the particle-rich environment of an arctic shelf ecosystem. J. Mar. Syst. 74, 774–782. doi: 10.1016/j.jmarsys.2007.12.001
Gheorghievici, L., Gheorghievici, G., and Tănase, I. (2015). The phytoplankton composition features of five Romanian pelogenous ecosystems. Environ. Eng. Manag. J. 14, 975–984.
Gibson, J. A. E., and Andersen, D. T. (2002). Physical structure of epishelf lakes of the southern Bunger Hills, East Antarctica. Antarct. Sci. 14, 253–261. doi: 10.1017/S0954102002000100
Gordon, D. A., Priscu, J., and Giovannoni, S. (2000). Origin and phylogeny of microbes living in permanent Antarctic lake ice. Microb. Ecol. 39, 197–202. doi: 10.1007/s002480000016
Gorokhov, O. G. (2013). Taxonomical structure of the phytoplankton marsh reservoirs on Nature Sanctuary Territories “Racheysky Pine Forest” (Samara Region). Samara Bend: regional and global environmental problems. [In Russian]. 22, 63–79.
Hindáková, A., and Hindák, F. (2012). Diversity of the phytoplankton of a fishpond at Devínska Nová Ves (Bratislava, W. Slovakia). [in Slovak]. Bull. Slov. Bot. Spoločn. 34, 11–18.
Hoppenrath, M., and Leander, B. S. (2006). Dinoflagellate, euglenid or cercomonad? The ultrastructure and molecular phylogenetic position of Protaspis grandis n. sp. J. Eukaryot. Microbiol. 53, 327–342. doi: 10.1111/j.1550-7408.2006.00110.x
Irgens, R. L., Gosink, J. J., and Staley, J. T. (1996). Polaromonas vacuolata gen. nov., sp. nov., a psychrophilic, marine, gas vacuolate bacterium from Antarctica. Int. J. Syst. Bacteriol. 46, 822–826.
Kaden, R., Spröer, C., Beyer, D., and Krolla-Sidensteirn, P. (2014). Rhodoferax saidenbachensis sp. nov., a psychrotolerant, very slowly growing bacterium within the family Comamonadaceae, proposal of appropriate taxonomic position of Albidiferax ferrireducens strain T118T in the genus Rhodoferax and emended description of the genus Rhodoferax. Int. J. Syst. Evol. Micr. 64, 1186–1193. doi: 10.1099/ijs.0.054031-0
Kang, N. S., Jeong, H. J., Yoo, Y. D., Yoon, E. Y., Lee, K. H., Lee, K., et al. (2011). Mixotrophy in the newly described phototrophic dinoflagellate Woloszynskia cincta from Western Korean waters: feeding mechanism, prey species and effect of prey concentration. J. Eukaryot. Microbiol. 58, 152–170. doi: 10.1111/j.1550-7408.2011.00531.x
Khondker, M., Bhuiyan, R. A., Yeasmin, J., Alam, M., Sack, R. B., Huq, A., et al. (2007). New records of phytoplankton for Bangladesh. 3. Volvocales. Bangl. J. Plant Taxon. 14, 1–12. doi: 10.3329/bjpt.v14i1.518
Kubiszyn, A. M., Piwosz, K., Wiktor, J. M. Jr., and Wiktor, J. M. (2014). The effect of inter-annual Atlantic water inflow variability on the planktonic protist community structure in the West Spitsbergen waters during the summer. J. Plankton Res. 36, 1190–1203. doi: 10.1093/plankt/fbu044
Laybourn-Parry, J., Madan, N. J., Marshall, W. A., Marchant, H. J., and Wright, S. W. (2006). Carbon dynamics in an ultra-oligotrophic epishelf lake (Beaver Lake, Antarctica) in summer. Freshwater Biol. 51, 1116–1130. doi: 10.1111/j.1365-2427.2006.01560.x
Laza-Martínez, A., Arluzea, J., Miguel, I., and Orive, E. (2012). Morphological and molecular characterization of Teleaulax gracilis sp. nov. and T. minuta sp. nov. (Cryptophyceae). Phycologia 51, 649–661. doi: 10.2216/11-044.1
Logares, R., Boltovskoy, A., Bensch, S., Laybourn-Parry, J., and Rengefors, K. (2009). Genetic diversity patterns in five protist species occurring in lakes. Protist. 160, 301–317. doi: 10.1016/j.protis.2008.10.004
Lovejoy, C., Comeau, A., and Thaler, M. (2016). Curated reference database of SSU rRNA for Northern marine and freshwater communities of Archaea, Bacteria and microbial eukaryotes, v. 1.1 (2002–2008). Nordicana D23. doi: 10.5885/45409XD-79A199B76BCC4110
Lovejoy, C., Vincent, W. F., Bonilla, S., Roy, S., Martineau, M.-J., Terrado, R., et al. (2007). Distribution, phylogeny, and growth of cold-adapted picoprasinophytes in arctic seas. J. Phycol. 43, 78–89. doi: 10.1111/j.1529-8817.2006.00310.x
Lozupone, C., and Knight, R. (2005). Unifrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microb. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Majaneva, M., Rintala, J.-M., Piisilä, M., Fewer, D. P., and Blomster, J. (2012). Comparison of wintertime eukaryotic community from sea ice and open water in the Baltic Sea, based on sequencing of the 18S rRNA gene. Polar Biol. 35, 875–889. doi: 10.1007/s00300-011-1132-9
Mortimer, C. A., Copland, L., and Mueller, D. R. (2012). Volume and area changes of the Milne Ice Shelf, Ellesmere Island, Nunavut, Canada, since 1950. J. Geophys. Res. Earth. 117, F04011. doi: 10.1029/2011JF002074
Mueller, D. R., Copland, L., and Jeffries, M. (2017). “Changes in Canadian Arctic ice shelf extent since 1906,” in Arctic Ice Shelves and Ice Islands, eds L. Copland and D. R. Mueller (Dordrecht: Springer SBM).
Mueller, D. R., Vincent, W. F., and Jeffries, M. O. (2003). Break-up of the largest Arctic ice shelf and associated loss of an epishelf lake. Geophys. Res. Lett. 30, 20. doi: 10.1029/2003GL017931
Nakada, T., Misawa, K., and Nozaki, H. (2008). Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol. Phylogenet. Evol. 48, 281–291. doi: 10.1016/j.ympev.2008.03.016
NEIGE (2016a). Water column physico-chemical profiles of lakes and fiords along the Northern coastline of Ellesmere Island, v. 1.0. Nordicana D27. doi: 10.5885/45445CE-7B8194DB81754841
NEIGE (2016b). Phytoplankton pigments of lakes and fiords of Northern Ellesmere Island, Nunavut, Canada, v. 1.0. Nordicana D26. doi: 10.5885/45436CE-0E0A89CB98C148F4
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., O'Hara, R. F., Simpson, G. L., et al. (2010). Vegan: Community Ecology Package. R package version 1.17-0. Available online at: http://CRAN.R-project.org/package=vegan
Paquette, M., Fortier, D., Mueller, D. R., Sarrazin, D., and Vincent, W. F. (2015). Rapid disappearance of perennial ice on Canada's most northern lake. Geophys. Res. Lett. 42, 1433–1440. doi: 10.1002/2014GL062960
Pedrós-Alió, C., Potvin, M., and Lovejoy, C. (2015). Diversity of planktonic microorganisms in the Arctic Ocean. Prog. Oceanogr. 139, 233–243. doi: 10.1016/j.pocean.2015.07.009
Piwosz, K., Wiktor, J. M., Niemi, A., Tatarek, A., and Michel, C. (2013). Mesoscale distribution and functional diversity of picoeukaryotes in the first-year sea ice of the Canadian Arctic. ISME J. 7, 1461–1471. doi: 10.1038/ismej.2013.39
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Pujalte, M. J., Lucena, T., Ruvira, M. A., Arahal, D. R., and Macián, M. C. (2014). “The Family Rhodobacteraceae,” in The Prokaryotes – Alphaproteobacteria and Betaproteobacteria, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer-Verlag), 439–512.
Purdy, K. J., Cresswell-Maynard, T. D., Nedwell, D. B., McGenity, T. J., Grant, W. D., Timmis, K. N., et al. (2004). Isolation of haloarchaea that grow at low salinities. Environ. Microbiol. 6, 591–595. doi: 10.1111/j.1462-2920.2004.00592.x
Rengefors, K., Logares, R., Laybourn-Parry, J., and Gast, R. J. (2015). Evidence of concurrent local adaptation and high phenotypic plasticity in a polar microeukaryote. Environ. Microbiol. 17, 1510–1519. doi: 10.1111/1462-2920.12571
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Spilling, K. (2007). Dense sub-ice bloom of dinoflagellates in the Baltic Sea, potentially limited by high pH. J. Plankton Res. 29, 895–901. doi: 10.1093/plankt/fbm067
Stackebrandt, E., and Liesack, W. (1993). “Nucleic acids and classification,” in Handbook of New Bacterial Systematics, eds M. Goodfellow and A. G. O'Donnell (London: Academic Press), 152–189.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stoecker, D. K. (1999). Mixotrophy among dinoflagellates. J. Eukaryot. Microbiol. 46, 397–401. doi: 10.1111/j.1550-7408.1999.tb04619.x
Stoecker, D. K., Johnson, M. D., de Vargas, C., and Not, F. (2009). Acquired phototrophy in aquatic protists. Aquat. Microb. Ecol. 57, 279–310. doi: 10.3354/ame01340
Tarasova, N. G., Burkova, T. N., Bykova, S. V., Mukhortova, O. V., Unkovskaya, E. N., and Sabitova, R. Z. (2013). Plankton community (phytoplankton, protozooplankton and zooplankton) in phytal zone of Lake Beloe (Volzhsko-Kamskiy State Natural Biospheric Reserve, Republic of Tatarstan, Russian Federation). Nat. Montenegrina Podgorica. 12, 1045–1064.
Thaler, M., and Lovejoy, C. (2015). Biogeography of heterotrophic flagellate populations indicates the presence of generalist and specialist taxa in the Arctic Ocean. Appl. Environ. Microbiol. 81, 2137–2148. doi: 10.1128/AEM.02737-14
Tsarenko, E. P. (2008). Peculiarities of the plankton diversity in the Southern Bug River in the region of the city of Vinnitsa (Ukraine). Int. J. Algae 10, 226–239. doi: 10.1615/InterJAlgae.v10.i3.20
Unrein, F., Gasol, J. M., Not, F., Forn, I., and Massana, R. (2014). Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J. 8, 164–176. doi: 10.1038/ismej.2013.132
Van Hove, P., Swadling, K. M., Gibson, J. A. E., Belzile, C., and Vincent, W. F. (2001). Farthest north lake and fjord populations of calanoid copepods Limnocalanus macrurus and Drepanopus bungei in the Canadian high Arctic. Polar Biol. 24, 303–307. doi: 10.1007/s003000000207
Veillette, J., Lovejoy, C., Potvin, M., Harding, T., Jungblut, A. D., Antoniades, D., et al. (2011). Milne Fiord epishelf lake: a coastal Arctic ecosystem vulnerable to climate change. Écoscience 18, 304–316. doi: 10.2980/18-3-3443
Veillette, J., Mueller, D. R., Antoniades, D., and Vincent, W. F. (2008). Arctic epishelf lakes as sentinel ecosystems: past, present and future. J. Geophys. Res. Biogeo. 113:G04014. doi: 10.1029/2008JG000730
Vincent, W. F., Gibson, J. A. E., and Jeffries, M. O. (2001). Ice-shelf collapse, climate change, and habitat loss in the Canadian high Arctic. Polar Rec. 37, 133–142. doi: 10.1017/S0032247400026954
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new Bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Werner, I., Ikävalko, J., and Schünemann, H. (2007). Sea-ice algae in Arctic pack ice during late winter. Polar Biol. 30, 1493–1504. doi: 10.1007/s00300-007-0310-2
White, A., Mueller, D., and Copland, L. (2015). Reconstructing hydrographic change in Petersen Bay, Ellesmere Island, Canada, inferred from SAR imagery. Remote Sens. Environ. 165, 1–13. doi: 10.1016/j.rse.2015.04.017
Wright, S. W., and Jeffrey, S. W. (2006). “Pigment markers for phytoplankton production,” in Marine Organic Matter: Biomarkers, Isotopes and DNA, ed J. K. Volkman (Berlin; Heidelberg: Spring), 71–104.
Zapata, M., Rodriguez, F., and Garrido, J. L. (2000). Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar. Ecol. Prog. Ser. 195, 29–45. doi: 10.3354/meps195029
Keywords: Arctic, Radicarteria, climate change, cyanobacteria, Halobacteriales, microbial biodiversity, picoeukaryotes, Rhodoferax
Citation: Thaler M, Vincent WF, Lionard M, Hamilton AK and Lovejoy C (2017) Microbial Community Structure and Interannual Change in the Last Epishelf Lake Ecosystem in the North Polar Region. Front. Mar. Sci. 3:275. doi: 10.3389/fmars.2016.00275
Received: 05 July 2016; Accepted: 12 December 2016;
Published: 10 January 2017.
Edited by:
Marcelino T. Suzuki, Université Pierre-et-Marie-Curie and CNRS, FranceReviewed by:
Sara Beier, Leibniz Institute for Baltic Sea Research (LG), GermanyAntonio Quesada, Autonomous University of Madrid, Spain
Copyright © 2017 Thaler, Vincent, Lionard, Hamilton and Lovejoy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mary Thaler, mary.thaler.1@ulaval.ca
 Mary Thaler
Mary Thaler Warwick F. Vincent
Warwick F. Vincent Marie Lionard
Marie Lionard Andrew K. Hamilton
Andrew K. Hamilton Connie Lovejoy
Connie Lovejoy