Transcriptomic Analysis of the Onchidium reevesii Central Nervous System in Response to Cadmium
- 1National Demonstration Center for Experimental Fisheries Science Education, Shanghai Ocean University, Shanghai, China
- 2Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai, China
- 3Shanghai Engineering Research Center of Aquaculture, Shanghai, China
- 4Shanghai Universities Key Laboratory of Marine Animal Taxonomy and Evolution, Shanghai, China
Shellfish are widely used in coastal environments for biomonitoring of heavy metal pollution. Research on the sea slug Onchidium reevesii, an economically important benthic gastropod, is limited due to a lack of genomic resources. This study, based on Illumina sequencing, compared the transcriptomes of O. reevesii before and after cadmium exposure, and used time-series expression analysis. The molecular level stress response mechanism operating in this organism in environmental stress conditions was investigated. De novo assembly of the O. reevesii transcriptome yielded 51,007 unigenes, of which 14,774 were annotated. Time-series expression analyses resulted in clustering of most responsive transcripts into 16 statistically significant expression profiles. Gene Ontology (GO) enrichment analysis based on time-series expression identified metal ion binding, synaptic transmission, and oxidative stress as important biological processes, and expression analysis identified genes related to invertebrate responses to environmental stress. Furthermore, we have generated the first transcriptome of O. reevesii, and the extensive sequence data produced generated will provide valuable molecular resources for the study of the response of O. reevesii response to cadmium exposure.
Introduction
The Onchidiidae species Onchidium reevesii is a typical benthic sea slug distributed across various coastal areas of China and other countries along the Indian-Pacific Ocean. The important economically shellfish has high nutritional and medicinal value (Huang and Wang, 2008). Coastal ecosystems are constantly threatened by pollution from human settlements and their associated agricultural and industrial activities. In the past few decades, a large number of studies have shown that shellfish can accumulate heavy metals from the environment, and the heavy metal content in their tissues is generally correlated with pollution levels in the habitat (Ren et al., 2007; Sun et al., 2014). Therefore, some benthic invertebrates can serve as useful biomonitors for assessing the quality of our marine ecosystems (Kiffney and Clements, 1993; Santoro et al., 2009).
Under laboratory conditions, only the toxicological effects of copper on adult O. reevesii have been reported, and the results indicated that cadmium (Cd) and copper (Cu) levels were beyond acceptable for aquatic products (Li, 2008). At the same time, it is interesting to note that the heavy metal content of adult O. reevesii collected in the reed beach of Chongming Island, Shanghai Municipality, China, found that the Cd content of heavy metals in the tissues (Muscle, Hepatopancreas, Digenetic gonad, Vitelline gland) exceeded the national standard (Wu et al., 2007). Tissue damage and enzyme parameters changes caused by heavy metals can be used as indicators to measure the degree of heavy metal pollution in a certain area (Li et al., 2009). In lower organisms, the central nervous system (CNS) acts as a regulatory hub in adults, it plays an essential role in the adaptive response caused by the change of external environment (Alex et al., 2008; Strickler and Soares, 2011). Invertebrates are particularly useful biological models because their nervous systems are relatively simple compared with those of mammals, and Functional genes that play an important role in signal transduction of CNS are highly conserved in the evolution of species. Invertebrates have the advantages of easy access, easy processing, strong reproductive ability and clear genetic background when they are used as experimental objects to study stress response.
Heavy metals such as Cd, Cu, lead (Pb), and mercury (Hg) are major environmental pollutants that cause serious metal contamination problems, especially in coastal tidal flats, where the aquaculture industry and biodiversity are vulnerable to accumulation of heavy metals. Cd is a non-essential element in animals (Friberg et al., 1985), exists in various valence states in water and soil, and has a strong tendency to accumulate in the tissues of animals inhabiting tidal flats. Hence it poses a health risk to humans and animals following the contaminated organisms entry into the food chain (Ren et al., 2007; Lawson and Lawson, 2011). Therefore, monitoring ecological security of tidal flats and establishing appropriate biological indicators are essential for analyzing environmental pollution. In the United States, the Environmental Protection Agency (EPA) published Rapid Bioassessment Protocols (1999) based on measuring macroinvertebrates, as well as periphyton and fish, for assessment of water quality (United States Environmental Protection Agency [US EPA], 1997; Office of Water and United States Environmental Protection Agency, 1999). In 2006, Europe used a remote online biomonitoring system for coastal water quality assessment based on bivalve mollusks, and the exchange of real-time data between a remote intelligent device in the field. However, there are few reports using invertebrates in intertidal zones to monitor environmental pollution in tidal flats.
In conventional biomonitoring, the body burden of common pollutants, but limited number of physiological and molecular biomarkers are used as end-points for assessing the effects of chemical pollution on Mollusca (Livingstone et al., 2000; Nicholson and Lam, 2005; Solé et al., 2009). To understand the toxic effects of chemical pollutants, and to acquire early warning signals of pollutant stress by biomonitoring, it is essential to have an in-depth knowledge of toxicity mechanisms at the molecular level. However, like many other non-model marine organisms, genomic data for O. reevesii is limited, which hinders the elucidation of molecular mechanisms related to specific toxic stress responses.
The underlying molecular mechanisms and phenotypic plasticity of an organism in response to environmental conditions can be interpreted from transcriptome dynamics analyses. RNA sequencing (RNA-Seq) has proved revolutionary (Ozsolak and Milos, 2010; McGettigan, 2013) since it can reveal the molecular basis of functional responses to environmental changes (Wang et al., 2009; Suárez-Ulloa et al., 2013) and the underlying molecular mechanisms of the responses to environmental stresses (Leung et al., 2014; Rebl et al., 2014). RNA-Seq analysis has been applied to several marine bivalve species, the Pacific oyster Crassostrea gigas (Zhang et al., 2012), the Manila clam Ruditapes philippinarum (Milan et al., 2011), and the Yesso scallop Mizuhopecten yessoensis (Hou et al., 2011). Some of these studies highlight the species specificity of the transcriptome, and also demonstrate differences in transcript profiles in different tissues. The increased availability of transcriptome data from non-model organisms has revealed many fundamental molecular mechanisms related to the responses of different environmental stresses (Zhao et al., 2012; Mun et al., 2017).
Herein, we performed Illumina sequencing and de novo assembly of the CNS transcriptome of O. reevesii under the experimental conditions of Cd stress. The results expand our understanding of Onchidiidae biology, with a focus on the nervous system, a tissue known to regulate metabolism of heavy metal ions. By comparing the relevant transcriptome data files obtained in this experiment, the regulatory expression of metal ion binding protein and synapse associated protein related genes can be revealed, and which genes are suitable as molecular markers under the stress of Cd can also be analyzed.
Materials and Methods
Experimental Treatments and Sample Collection
A total of 42 O. reevesii (body length = 6.3 ± 0.4 cm) were collected from Chongming, Shanghai, China, in June 2017. According to the previous experimental results, the semi-lethal concentration of Cd ion of O. reevesii was 1.64 mg/L, which was determined to be used in the follow-up experiments. Animals were acclimated in 1 m3 tanks (salinity = 11.5 ± 0.2 ppt) for 2 weeks at temperatures similar to those habitat. Animals were fed with corn starch once per day (12:00 am) and randomly divided into two tanks with 21 individuals per tank. In one tank, animals were maintained in NaCl solution (control group), and in the other tank 1.64 mg/L of Cd 2+ (CdCl2⋅2.5H2O; Sinopharm Chemical Reagent Co., Ltd., China) was prepared (experimental group). All animals were raised under the same conditions, apart from the difference in Cd concentration. O. reevesii was kept in Cd ionic saline solution. Sampling time points were set at 0, 2, 4, 12, 24, 48, and 96 h, respectively. Three individuals were randomly selected at each sampling point. CNS tissue was collected, flash-frozen in liquid nitrogen and stored at −80°C for total RNA extraction and transcriptome sequencing.
RNA Isolation
RNA extraction was extracted using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, United States) following the manufacturer’s protocol. The amount and quality of RNA from each sample were determined using a Nanodrop 2000C spectrophotometer (Nanodrop Technologies, United States), 1.2% agarose gel electrophoresis, and a Tanon 2500 gel imaging system (Tianneng, Shanghai, China). For high-purity RNA, the A260/A280 ratio was 1.8−2.1, and the brightness of 28S:18S RNA in gel images was close to 2:1. After the sample total RNA was digested with DNase, mRNA was enriched with magnetic beads containing Oligo (dT) to remove microRNAs.
cDNA Library Preparation and Sequencing
The RNA integrity of cadmium-exposed and control samples was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). Samples with an RNA Integrity Number (RIN) ≥ 7 were subjected to subsequent analysis. Libraries were constructed using a TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, United States) according to the manufacturer’s instructions. Libraries were sequenced on an Illumina HiSeq X Ten sequencing platform, and 150 bp paired-end reads were generated. All unigenes were also deposited in the NCBI database and can be accessed in the Sequence Read Archive (SRA) under the accession number PRJNA511644.
Transcriptome Assembly
Raw reads obtained from the Illumina HiSeq X Ten were converted to high-quality reads for subsequent analysis by quality filtering using NGS QC Tool kit software (Patel and Jain, 2012) for quality control and removing joints. On this basis, low-mass bases and N-bases were filtered out, and high-quality clean reads were obtained. The resulting high-quality clean reads comprised 250,000 pairs of reads (500,000 unpaired reads), and these were compared with the nt database1 for contamination detection. Reads with an E-value < 1e-10 and coverage >80% were retained.
Transcript sequences were obtained from paired-end reads using the splicing method of Trinity (version trinityrnaseq_r20131110) software (Grabherr et al., 2011). According to sequence similarity and length, the longest sequences were selected as unigenes, and TGICL (version: 2.1) software (Pertea et al., 2003) was used for clustering to remove redundant sequences, yielding a final set of unigenes as reference sequences for subsequent analysis.
Transcriptome Annotation
Comparisons were made using the BLAST algorithm with a maximum E-value threshold of 1E-5 e. Unigene sequences were searched against the NCBI non-redundant (NR) database2 to obtain NR annotation information, and more comprehensive Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation information was obtained using KAAS3. Based on the annotation results of SwissProt4, Gene Ontology (GO) terms5 were mapped according to the UniProt ID to obtain protein function annotation information. Proteins were also classified using Clusters of Orthologous Groups for Eukaryotic Complete Genomes (KOG)6. Unigene information from different databases was combined and summarized.
Comparative Expression Analysis
To evaluate the effects of sequencing depth and gene length on fragment number, fragments per kb per million reads (FPKM) method of calculation was often used (Trapnell et al., 2010). However, we used a more precise method to assess expression trends in this study. Differentially expressed genes (DEGs) were identified using the negative binomial distribution test in DESeq software7 (Anders and Huber, 2013), and a p-value < 0.05 was considered statistically significant. Meanwhile, DEGs were further categorized based on expression fold change (FC) ≥ 1.5. FPKM values of DEGs were then considered as expression levels based on the number of reads aligned with each gene.
Short Time-series Expression Miner (STEM) software was used to cluster genes with coherent changes (Ziv and Jason, 2006). An FC value ≥ 1.5 was set as the threshold for expression pattern differences. Thereafter, expression patterns of time-series genes for experimental groups at six different time points were compared with the control group. Trends in expression were visually using a Summary Plot.
Gene Ontology enrichment analysis8 is the most common approach for functional annotation of large-scale genomic data (Harris et al., 2004). STEM is fully integrated with the GO database and supports GO category gene enrichment analysis of sets of genes sharing similar temporal expression patterns. Using this approach, functional enrichment analysis of time-series genes was carried out to identify significant gene expression patterns.
Corroboration of Illumina Sequencing Data by Reverse-Transcriptase Quantitative-PCR
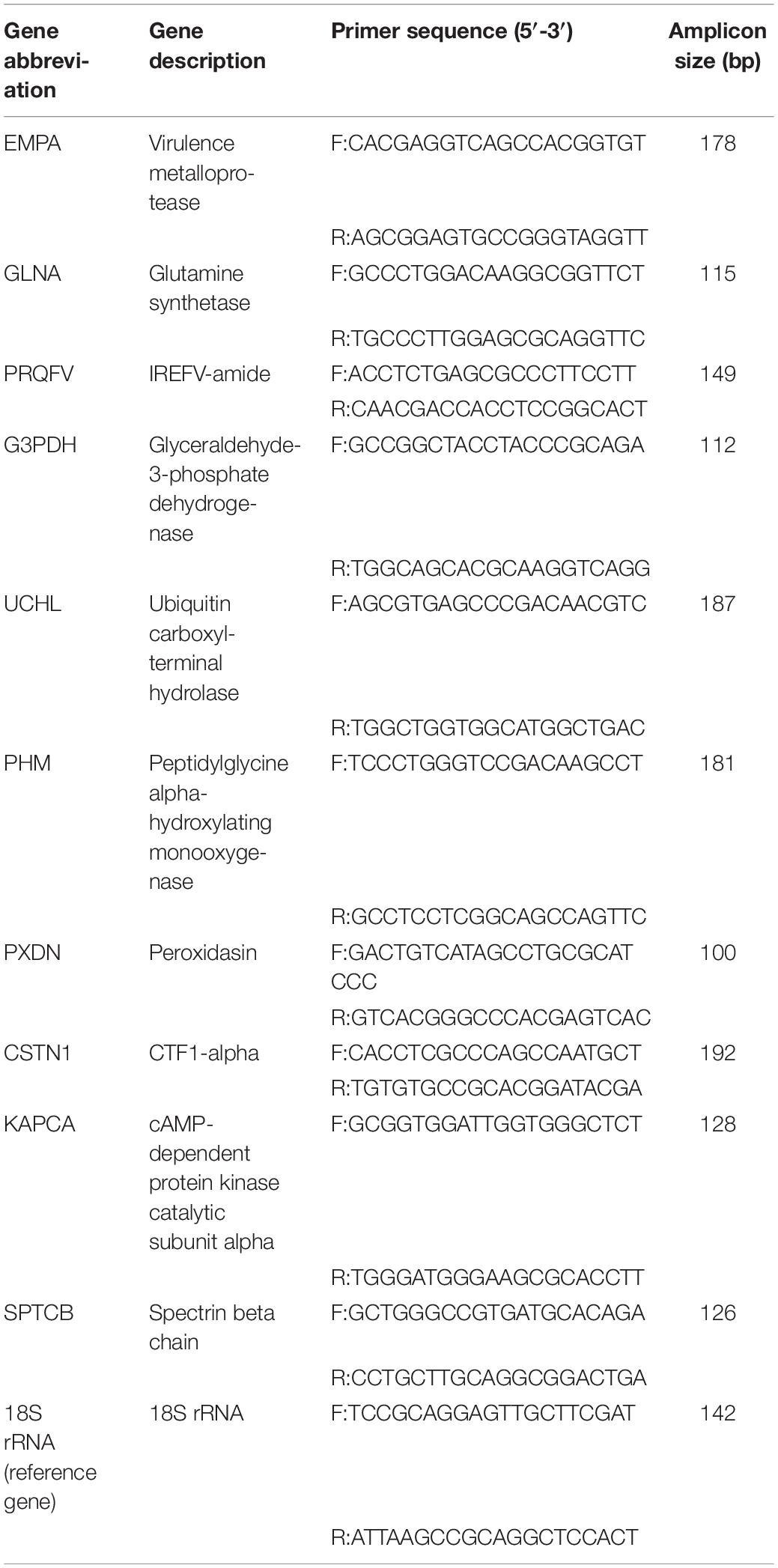
RNA extracted from control and experimental groups was used for real-time quantitative-PCR (RT-qPCR) verification of Illumina sequencing. First-strand cDNA synthesis was performed using 1 μg of total RNA and the GoScript Reverse Transcription System (TaKaRa, China) according to the manufacturer’s protocol. Subsequently, cDNA was diluted 3-fold and used as a template in qPCR experiments with qPCR primers designed by Primer 5 (Lalitha, 2000) to amplify the 10 selected genes (Table 1). All qPCR experiments (20 μl volume), including no-template controls containing nuclease-free water, were performed by using three technical repeats in 96-well microplates containing 10.0 μl of 2 × ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China), 0.4 μl of each primer (The final concentration of primers is 0.2 μM), and 2 μl of cDNA. All RT-qPCR experiments were performed using a QuantStudio 6-Flex Real-time PCR System (Applied Biosystems, Life Technologies Corporation, United States) with an initial denaturation at 95°C for 30 s, followed by 40 cycles at 95°C for 10 s, 56°C for 30 s, and 72°C for 30 s. After amplification, melting curve analysis (95°C for 15 s, 60°C for 1 min, 95°C for 15 s) was used to confirm whether the amplification product is specific amplification.
The standard curve of the primers was constructed by using cDNA gradient dilution (1:3) of CNS tissue to determine whether the primers can be used for subsequent qRT-PCR experiments (Larionov et al., 2005), wherein an amplification efficiency of 95–105% was selected for candidate primers (Supplementary File S2). All experiments were performed in triplicate technical repetition. Primer efficiency and cycle threshold (CT) values were calculated from the original amplification data, and the expression levels of different genes were analyzed using the comparative CT (2–ΔΔCT) method (Livak and Schmittgen, 2001).
Results
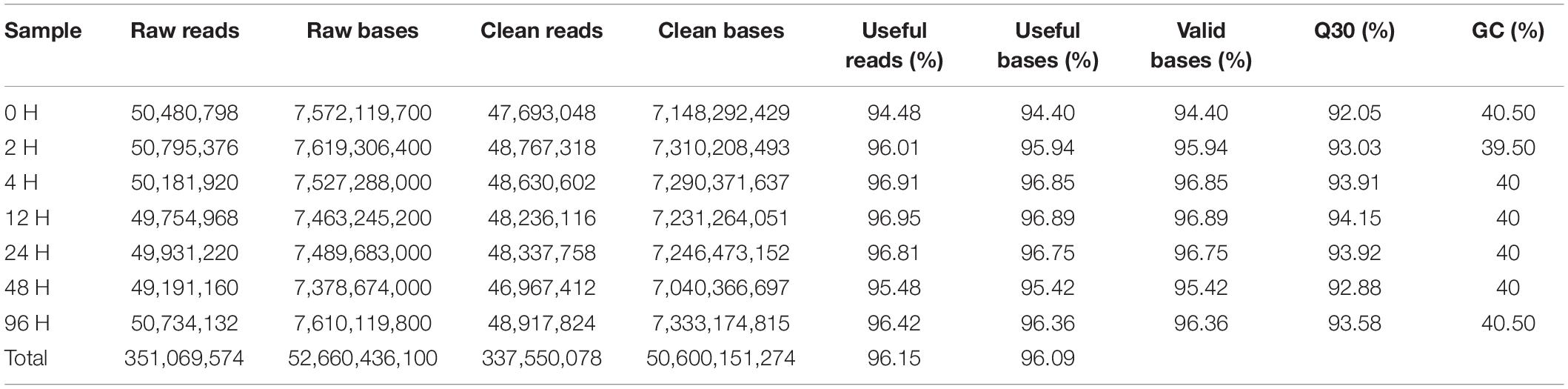
The RNA-Seq data from cadmium-exposed and control groups were generated from, yielding 351.06 million reads, and 337.55 million clean reads after trimming, of which 94.48−96.95% per sample were useful (Table 2). Clean reads were assembled into 51,007 unigenes (>300 bp), the longest unigene was 27,174 bp, the shortest was 301 bp, the average was 1,604.83 bp, and the N50 length was 2,426 bp (Table 3).
Transcriptome Annotation
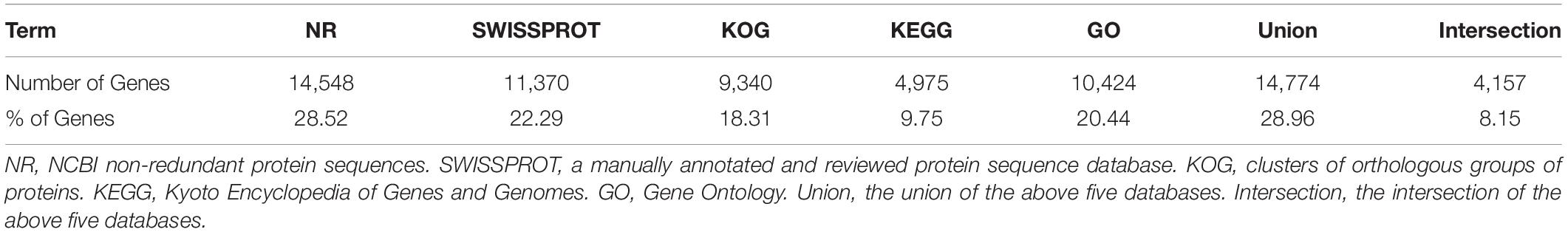
All unigene sequences were searched against the NR, SwissProt, KOG, KEGG, and GO databases, resulting in 14,548 (28.52%), 11,370 (22.29%), 9,340 (18.31%), 4,975 (9.75%), and 10,424 (20.44%) annotated unigenes, respectively (Table 4). Figure 1 shows the degree of overlap between unigenes annotated in different databases, and 4,157 (8.15%) unigenes overlap in all five databases, while 14,774 (28.96%) unigenes overlap in two or more databases.
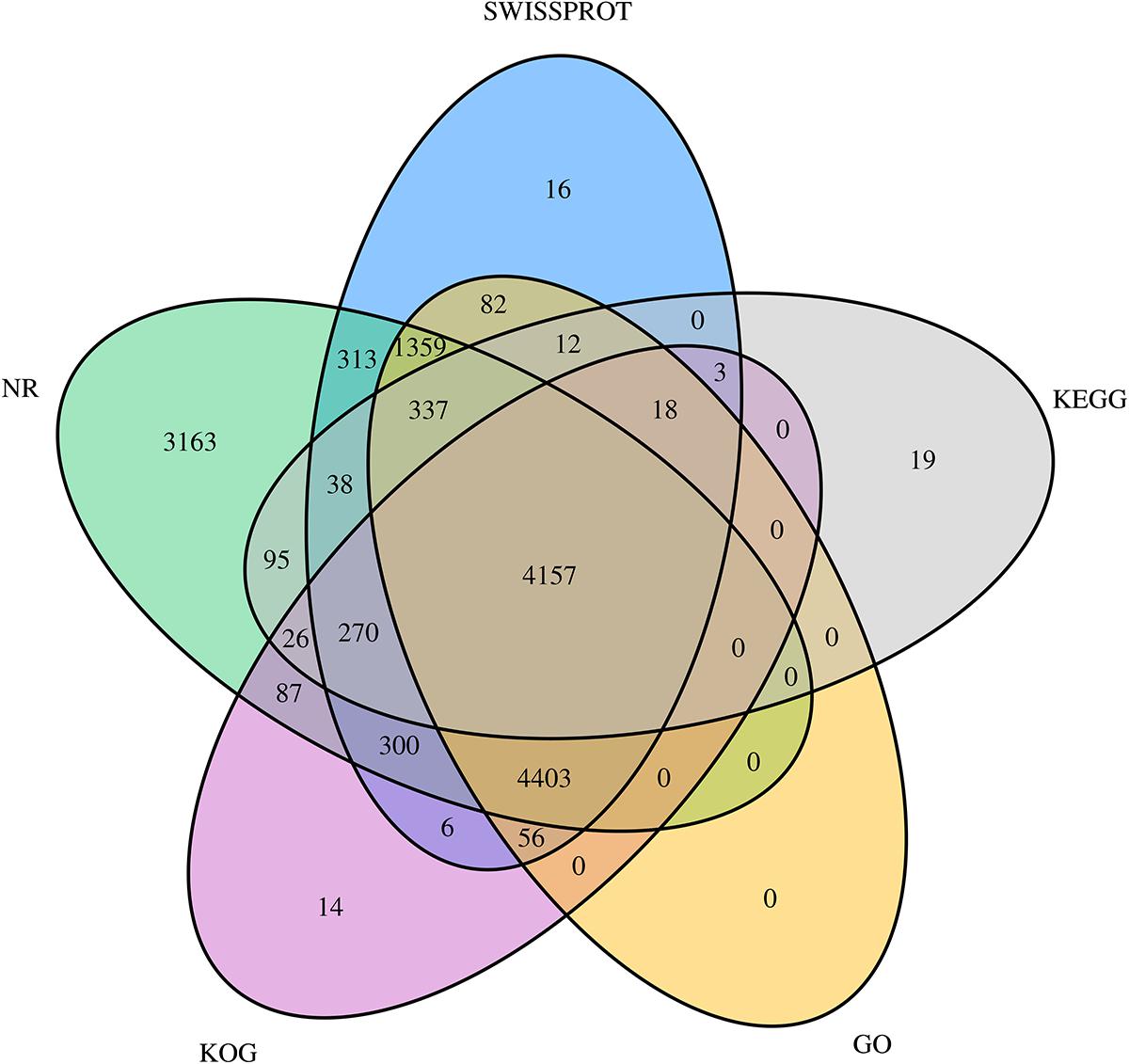
Figure 1. Venn diagram showing the degree of overlap of unigenes annotated using different databases. Different colors represent the number of overlaps of unigenes between different annotation libraries.
All unigenes were assigned to known protein sequences. As shown in Figure 2, 10,424 unigenes were classified based on GO biological process, molecular function, and cellular component categories. The biological process category (57,515 functional terms) was the most enriched, followed by cellular component (48,152) and molecular function (14,243) categories. A large number of putative unigenes were associated with binding (6,496), catalytic activity (4,246), response to stimulus (3,381), cell (8,915), and cell part (8,897) subcategories, indicating that stress responses in O. reevesii involve cellular metabolic activities. Of particular interest in the present study, various genes involved in oxidative stress or synaptic transmission were identified.
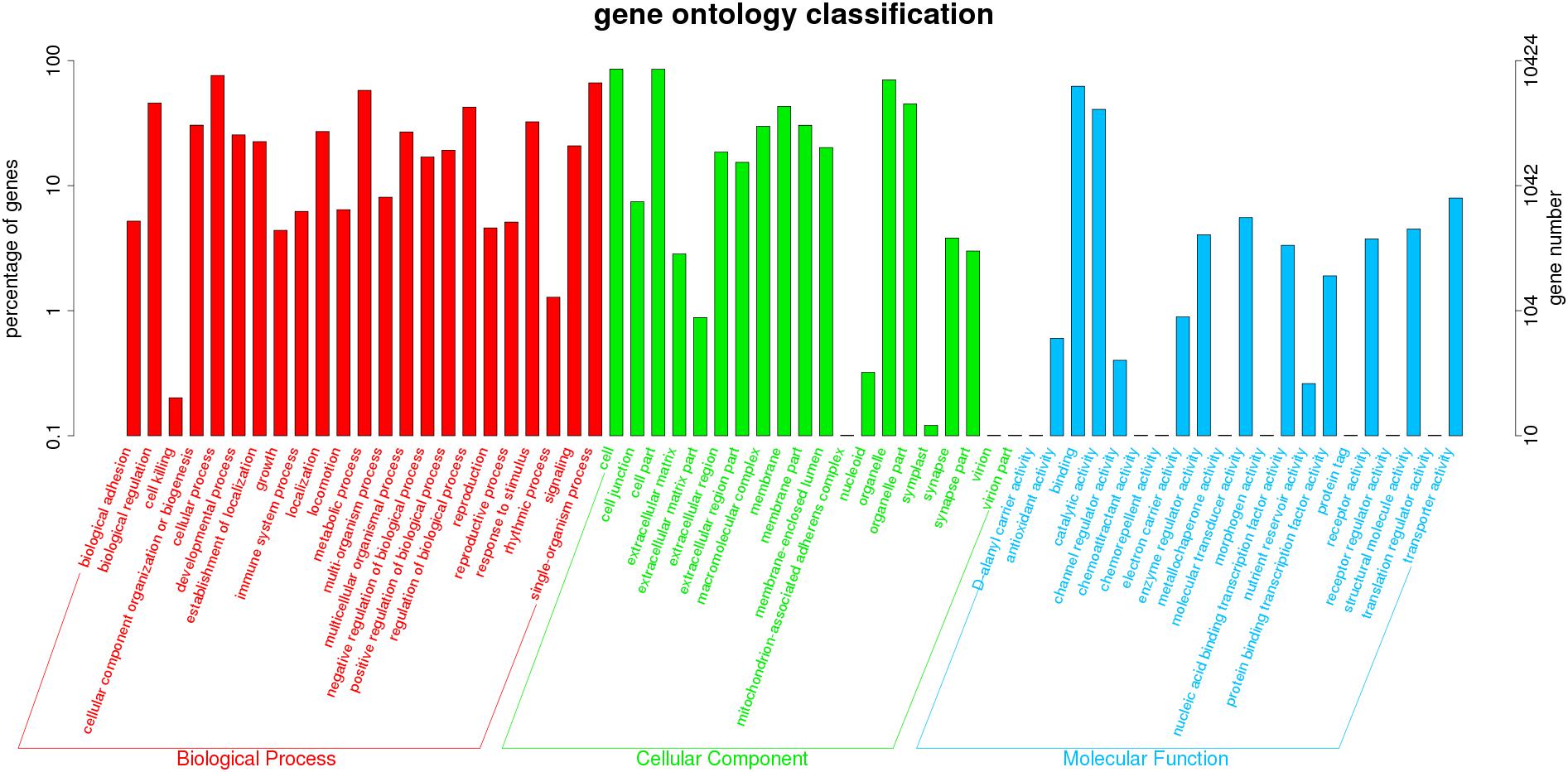
Figure 2. GO annotation of unigenes. Different colors represent annotation based on different categories (red = biological process, green = cellular component, blue = molecular function). The vertical axis represents the number of unigenes, and the horizontal axis represents the functional classes.
KOG analysis was performed to further assess the integrity of the transcriptome library and the validity of the annotation process. A total of 10,475 annotated putative proteins were classified into 25 molecular families based on function (Figure 3). Following the ‘general function prediction only’ category, ‘signal transduction mechanisms” was the second largest category (1425 unigenes, 13.60%). This suggests that after stimulation with Cd, many of the identified unigenes were primarily involved in the regulation of signal transduction during adaptation to environmental changes. Specifically, 1.03% of unigenes were assigned to the ‘defense mechanisms’ category, indicating that unigenes encoding stress-related proteins may be involved in the defense mechanism in O. reevesii. In addition, posttranslational modification, protein turnover, and chaperones were enriched (918, 8.76%), along with intracellular trafficking, secretion, and vesicular transport (557, 5.32%), translation, ribosomal structure and biogenesis (467, 4.46%) and cytoskeleton (439, 4.19%).
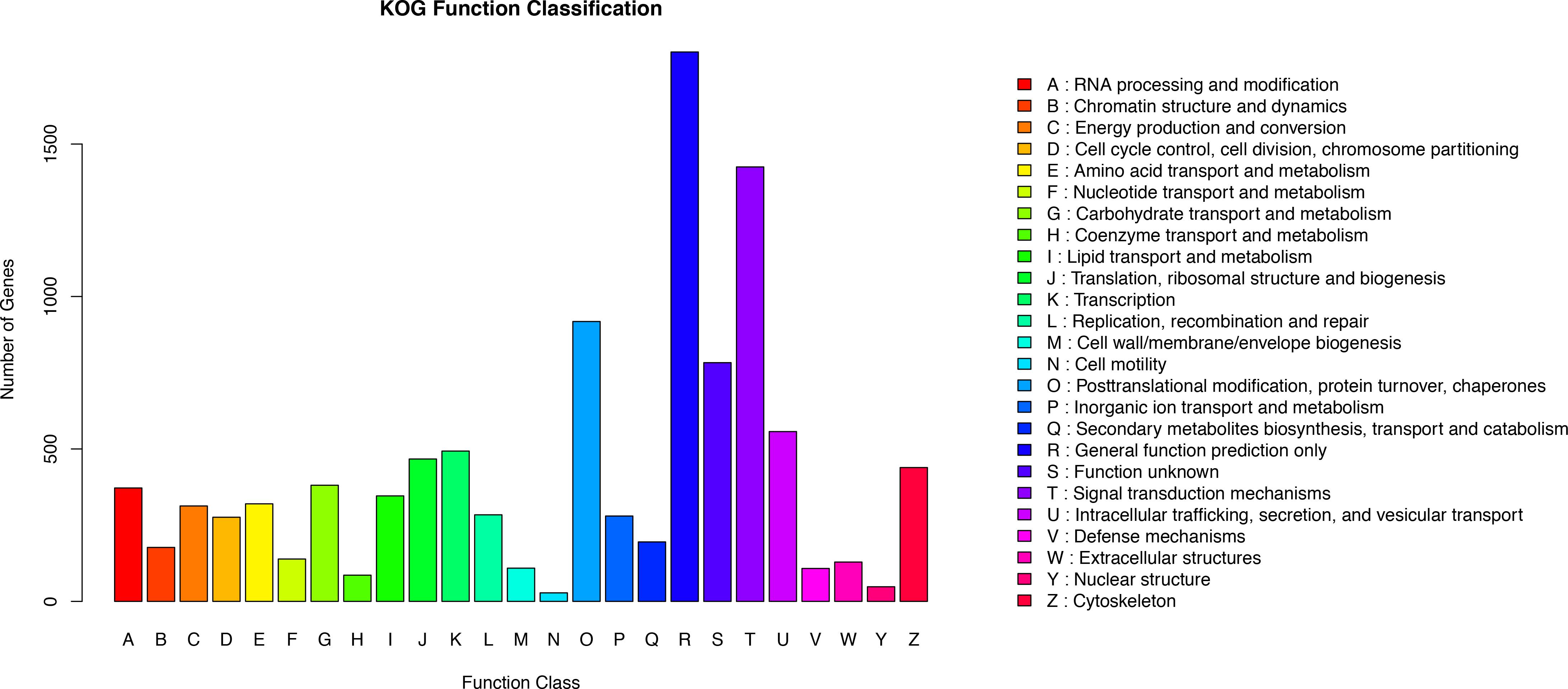
Figure 3. Functional annotation based on the KOG database. Different colors represent annotation based on different categories. The vertical axis represents the number of unigenes, and the horizontal axis represents the functional classes.
In addition to GO and KOG analyses, additional filtering and analysis of the resulting data were performed. In order to investigate different time points, data were subjected to STEM analysis to measure the FC in gene expression following Cd exposure. Supplementary File S1 shows the results of 16 STEM expression profiles in descending order of STEM clustering.
A flower plot of the degree of overlap between gene transcription at seven time points is shown in Figure 4 for unigenes with FC values ≥1.5. The flower plot shows the number of common and time point-specific genes that vary significantly at different time points. Time-series genes in the six experimental groups treated with Cd at different times were compared with control (S0) samples, and 5666 unigenes overlap in all six treatment groups, while 1212, 1597, 657, 939, 529, and 846 unigenes were specific to S2 h, S4 h, S12 h, S24 h, S48 h, and S96 h treatment groups respectively. Following STEM clustering, functional enrichment analysis based on p-value revealed that metal ion binding (31.48%, Figure 5A), intracellular signal transduction and synaptic transmission (29.79 and 8.51%, Figure 5B), and oxidative stress (6.94%, Figure 5C) were major functions of the identified DEGs (Figure 5).
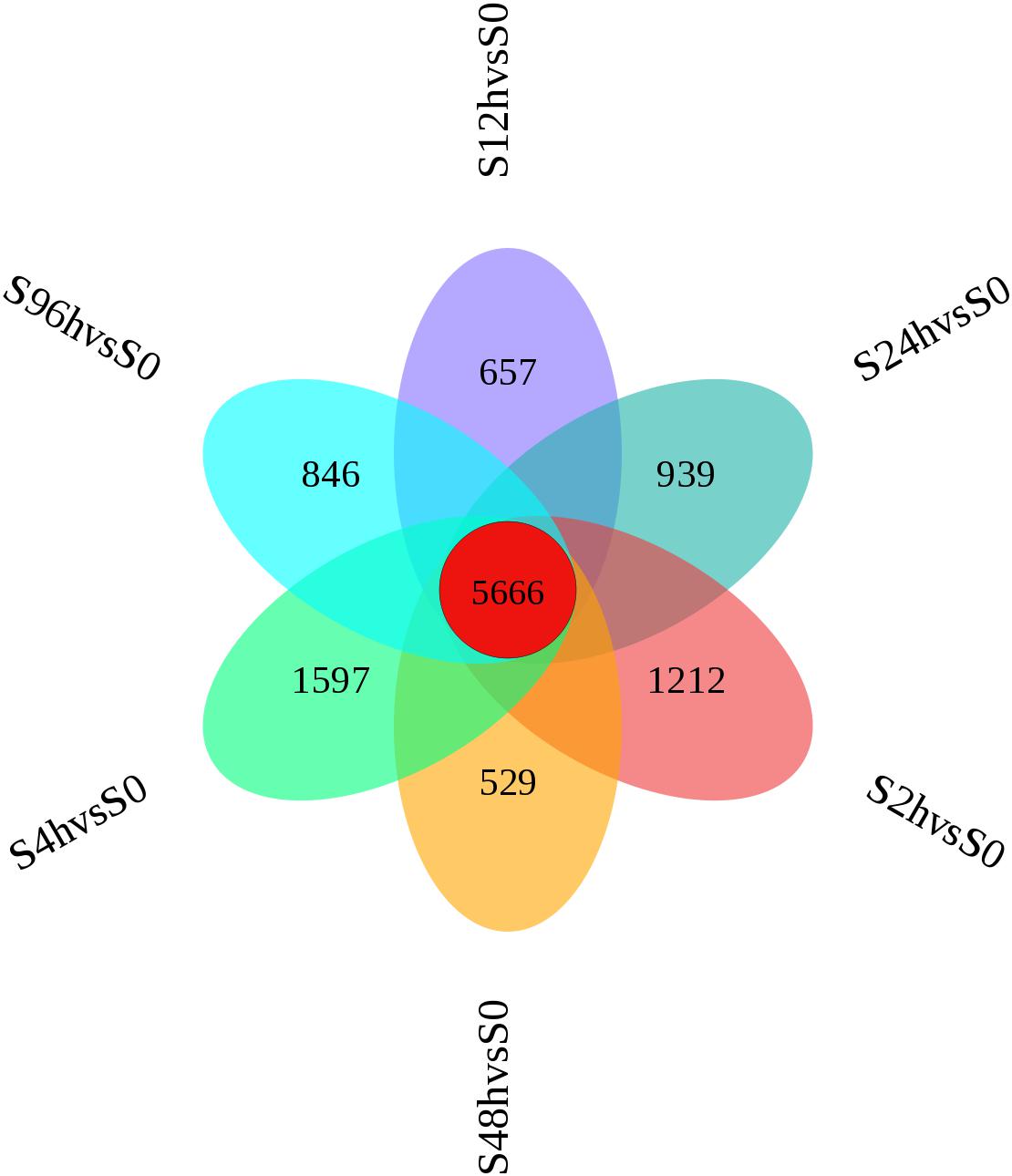
Figure 4. Flower plot of time-series genes in the O. reevesii CNS at different times following cadmium treatment. For unigenes with fold change ≥ 1.5, red represents the number of intersections of unigenes shared by the control group (0 h) and the six experimental groups (2, 4, 12, 24, 48, 96 h); the other six colors correspond to the number of intersections of the unigenes of the respective experimental group and the control group (0 h).
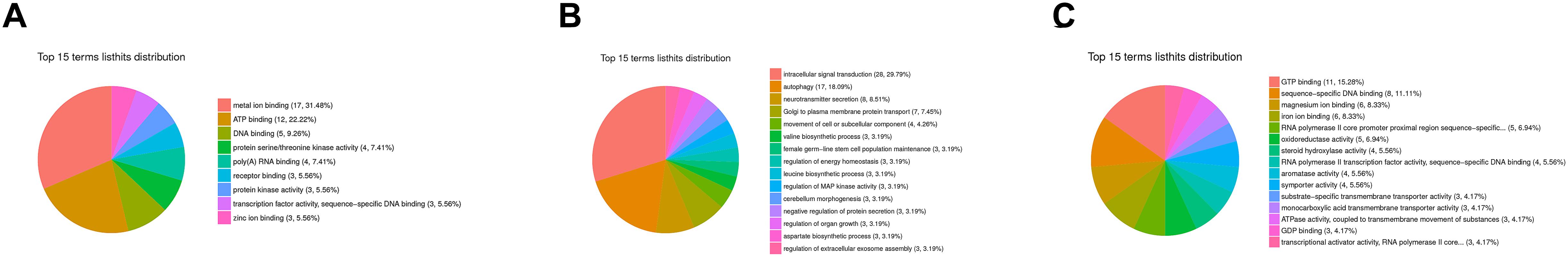
Figure 5. GO enrichment analysis based on STEM clustering. The summary plots show profiles for three representative trends; (A) the top 15 terms in profile 47; (B) the top 15 terms in profile 13; (C) the top 15 terms in profile 11. Among them, different colors represent the proportion of various types of molecular functions.
Corroboration of Illumina Sequencing by RT-qPCR
Expression levels determined by RT-qPCR analysis of 10 selected genes were found to be comparable to those obtained using the RNA-seq approach (Table 1). Furthermore, the expression profile trends obtained by RT-qPCR which were similar to our transcriptome database results, confirming that the validity and reliability of the RNA-Seq data (Figure 6).
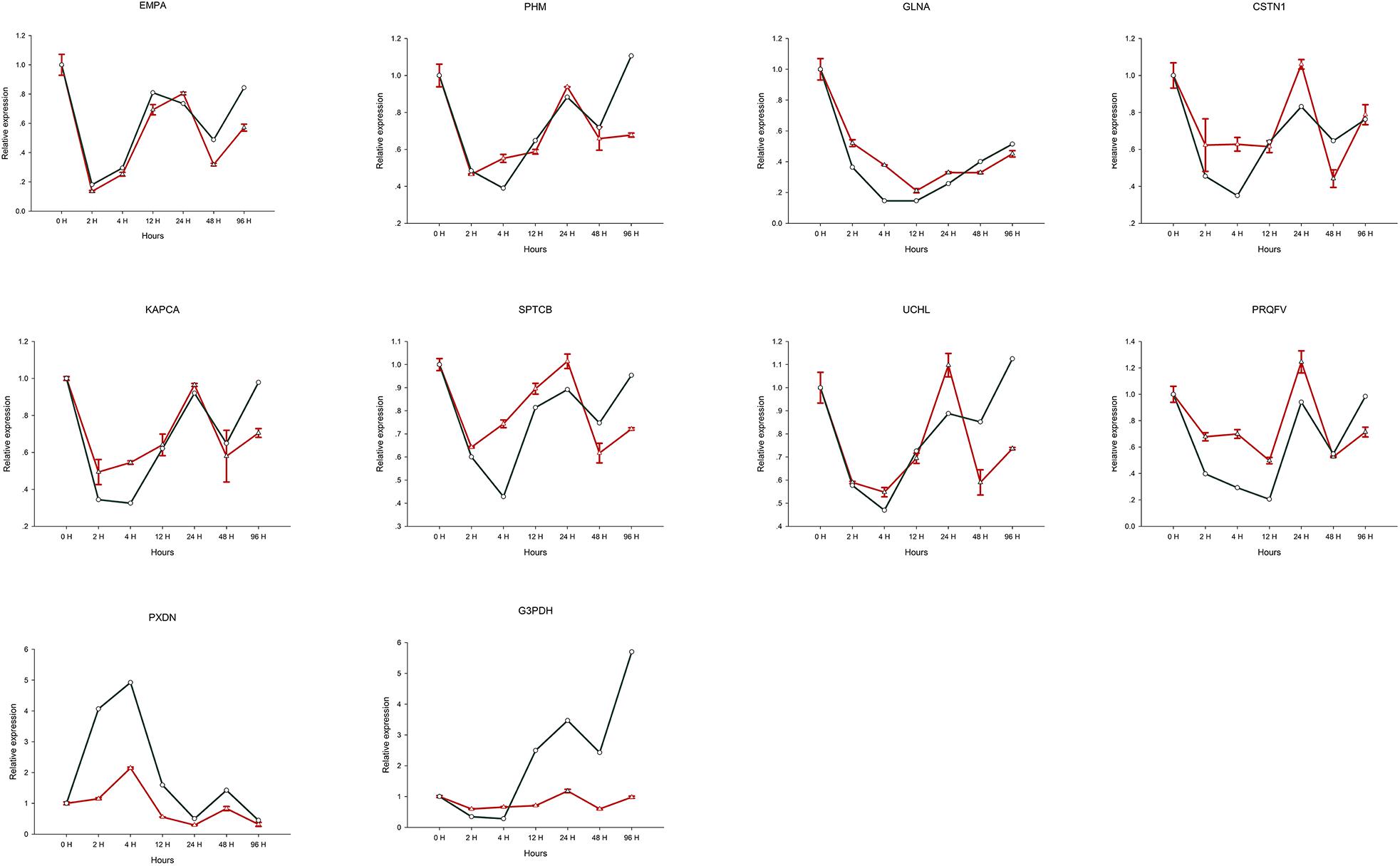
Figure 6. Relative expression of 10 selected genes differentially expressed after Cd treatment. The green line is the expression level of the assembled transcript, and the red line is the expression level of unigenes.
Discussion
Herein, we analyzed the transcriptome of O. reevesii in response to Cd, and present the first transcriptomic analysis of an Onchidiidae species. We functionally annotated 51,007 unigenes, and the results provide an important resource for future studies on the responses to environmental stress in O. reevesii. Numerous functional genes were differentially expressed in O. reevesii following Cd exposure, including metal ion binding proteins, synapse-associated proteins, and environmental stress markers.
Functional Genes Involved in Metal Ion Binding
Based on functional enrichment analysis, most enriched genes encode proteins involved in metal ion binding (Figure 5), and many were significantly differentially expressed in O. reevesii following Cd exposure, indicating that metal ion binding proteins are involved in regulating the responses to this environmental stress (Supplementary File S3). These results are consistent with those of other shellfish such as Elliptio complanata (Gagné et al., 2013), Mytilus galloprovincialis (Tania et al., 2011; Gomes et al., 2012), Crassostrea virginica (Ringwood et al., 2010), and Mizuhopecten yessoensis (Evtushenko et al., 1986) following exposure to Cd. In tissues, Cd is usually bound to high molecular weight proteins (HMWP) (Evtushenko et al., 1986), and a significant dose-dependent increase in metallothionein (MT) concentration was observed in Dreissena polymorpha following Cd exposure at different doses (Ivankovic et al., 2010). Ng et al. (2007) investigated the cytosolic distribution of Cd in mussels, and concluded that MTs (7000−20,000 Da) and HMWPs (>20,000 Da) are important for Cd binding and purification, and other studies have indicated that MT in aquatic invertebrates detoxifies non-essential Cd (Ng and Wang, 2004; Amiard et al., 2006).
In response to the toxicological threat of metal exposure, shellfish have evolved a range of defensive measures including barriers to uptake, entrapment and removal via mucus secretion, factors that neutralize or break down substances to aid elimination, and cells capable of metal ingestion, digestion and sequestration. Once dissolved in cells, soluble metals tend to bind wide range of host molecules. Metal uptake can be enhanced by the synthesis metal binding proteins and/or by increasing the formation of mineralized particles within membrane-bound vesicles, as is the case for soluble Cd. Lysosomes and related vesicles can reduce cytotoxicity via exocytosis in the blood and incorporation into blood cells. When blood cells participate in metal transport, they can be redistributed between tissues, hence the nervous system may be stimulated or damaged shortly after exposure. It can be seen from Figure 6, that among the 10 selected genes, only the expression level of PXDN gene was up-regulated after 4 h of heavy metal stress, and the remaining 9 genes showed a relatively obvious downregulation trend. Furthermore, continuous expression trend analysis at 7 sampling time points showed that the qRT-PCR results were consistent with the transcriptome data. We speculate that the obvious down-regulation of genes may be due to the influence on the biological activity and metabolism of individuals after heavy metal Cd ions entering the organism, thus inhibiting the expression of most genes.
Functional Genes Involved in Responses to Environmental Stress
In many shellfish species, responses to environmental stress are used as a biomarker to assess the biological effects of contaminants in aquatic organisms, examples of which include the specific oxidative stress biomarkers acetylcholinesterase (AChE) and MT (Bodin et al., 2004; Leiniö and Lehtonen, 2005; Damiens et al., 2007), non-specific biomarkers such as glutathione S-transferase (GST) (Benoit and Thierry, 2002), catalase (CAT) (Leiniö and Lehtonen, 2005), glutathione-dependent oxidoreductases (Regoli and Principato, 1995), and lipid peroxidation (LPO) (Wang et al., 2011). In addition, due to the lack of immunoglobulins, invertebrates possess unique host defenses to cope with environmental changes. Thus, the expression levels and catalytic activities of the above genes may be sensitive biomarkers of oxidative stress induced by biotic and abiotic factors in aquatic organisms. Among the 10 selected genes, six (GLNA, PRQFV, UCHL, CSTN1 KAPCA, SPTCB) are involved in synaptic transmission, two (EMPA, PHM) are involved in metal dissociation, and the remaining two (G3PDH, PXDN) may be at work in Oxidative stress. A reliable and efficient environmental assessment may be possible by monitoring such indicators.
Synaptic Transmission in Response to Environmental Stress
Six unigenes (GLNA, PRQFV, UCHL, CSTN1 KAPCA, SPTCB) were identified which related to synaptic transmission (Figure 5). Thus, following exposure to Cd, various neurotransmitters appear to work together to cope with this stress. Neurotransmitters are classified as neuromodulators and hormones that regulate many physiological processes and behaviors. Moreover, the binding of neurotransmitters can affect postsynaptic neurons in an inhibitory or excitatory manner. Binding of neurotransmitters to postsynaptic neuron receptors can trigger short-term changes in membrane and postsynaptic potential, or long-term changes caused by activation of signaling cascades. At present, there are few studies on synapse-associated proteins (SAPs) in aquatic invertebrates. Aplysia synapse-associated protein (ApSAP), which is expressed in sensory and motor neurons in the CNS of Aplysia sea slugs, selectively binds to a Shaker-type AKv1.1 channel, but not to glutamate receptor ApGluR1/4 (Reissner et al., 2010). Meanwhile, Klose et al. (2010) hypothesized that neuropeptides induce behavioral responses to environmental stress in Drosophila via two G protein-coupled receptors.
Antioxidant Defenses
Through the enrichment of gene function and annotation, we found in the selected 10 genes, there are two (EMPA, PHM) possible involved in antioxidant defenses. Reactive oxygen species (ROS) are stimulated by contaminants, and the resulting oxidative damage may be a mechanism of toxicity in aquatic invertebrates exposed to pollutants (Livingstone, 1991; Livingston et al., 1994). Detoxification of ROS is a common defense mechanisms in aquatic organisms (Winston et al., 1991; Di Giulio et al., 1995). The primary antioxidant systems in aquatic organisms comprise reduced glutathione (GSH), and specific antioxidant enzymes including CAT and glutathione reductase (GR). Antioxidant defenses have been studied in Mytilus edulis (Benoit and Thierry, 2002), Macoma balthica (Leiniö and Lehtonen, 2005), Chlamys islandica (Baussant et al., 2009), and Pecten jacobaeus (Viarengo et al., 1995). Antioxidant defenses in aquatic organisms are often investigated by determining the total oxidant scavenging capacity of a particular ROS, which can illuminate the antioxidant function and regulation in aquatic organisms.
In the present study, unigenes related to ‘response to stimulus’ (32.43% in the biological process) and ‘catalytic activity’ (40.73% in the molecular function) categories were clearly enriched in the GO analysis. CAT is known to be upregulated in marine organisms following exposure to contaminants, possibly because CAT may catalyze the oxidation of various metabolites and toxins via hydrogen peroxide. Heavy metal ions can act as non-competitive inhibitors of CAT (Berra et al., 2004), which is a potential biomarker in benthic invertebrates exposed to contaminants. CAT activity in aquatic invertebrates is generally higher following heavy metal exposure, suggesting that activation of antioxidant defenses is associated with heavy metal exposure (de Oliveira and Martinez, 2007). Recent studies have shown that environmental changes can affect the accumulation of Cd in coastal benthic organisms, and regulate the toxicity of Cd-induced oxidative stress in mollusks (Ivanina et al., 2013).
Similarly, studies have found that in the aquatic invertebrate Tegillarca granosa (Wei et al., 2018), an evident downregulation of Ca2+-regulation-related genes (CaM, CaMK2, and CaMKK2) was detected after exposure to waterborne Cd2+. This is consistent with the KAPCA results determined in this study, and, interestingly, suggests that Cd2+ has a competitive inhibitory effect on intracellular Ca2+ regulation. Cd2+ toxicity may manifest, at least in part, by affecting Ca2+ signaling. Moreover, once it enters the cell, since CaM fails to distinguish between Ca2+ and Cd2+, Cd2+ can activate the synthesis of numerous enzymes through CaM-dependent systems (Sutoo et al., 1990), causing cell disorders. We speculate that additional Ca2+ supply may increase the activity of antioxidant enzymes and subsequently decrease the production of ROS, thus helping to maintain the normal structure and function of the cell.
Oxidative Stress, Disease, and Animal Fitness
Currently, information on the relationships between oxidative stress, disease, and fitness in aquatic organisms in clean and polluted environments is limited. Short-term studies using cells exposed to pro-oxidants indicate both structural and functional changes at the subcellular and cellular levels following exposure. An early study on the Eastern oyster (Crassostrea virginica), a marine bivalve, showed that elevated pro-oxidants led to a strong upregulation of antioxidant proteins (Lars et al., 2011). A similar mechanism may contribute to the observed reduction in ROS levels in cells exposed to pro-oxidants; under certain conditions, pro-oxidants can affect the uptake and potential toxicity of metal ions, and oxidative stress has been implicated in the toxicity of metals in cells (Ivanina et al., 2013).
Gastropoda such as O. reevesii have a high capacity for regeneration and repair (Gorbushin and Levakin, 2001). Herein, we identified unigenes related to environmental stress, and explored their potential roles in oxidative stress. In glyceraldehyde-3-phosphate dehydrogenase (G3PDH), which was verified by RT-qPCR, cysteine residue C152 in the enzyme active site is required for induction of apoptosis by oxidative stress (Nicholls et al., 2012), and G3PDH acts as a reversible metabolic switch under oxidative stress conditions (Agarwal et al., 2012). Peroxidasin (PXDN), a novel protein that binds to peroxidase and is involved in phagocytosis and defense in extracellular matrix formation, was upregulated in the present study, and is also known to function in hydrogen catabolism via hydrogen peroxide (Nelson et al., 1994; Tindall et al., 2005). PXDN exists as a homotrimer stabilized by conserved intermolecular disulfide bridges (Lázár et al., 2015). PXDN is found in the basement membrane and appears to catalyze the formation of sulfilimine bonds via the production of hypohalous acid as a reaction intermediate (Bhave et al., 2012). Although information on PXDN in shellfish remains limited, it appears to be stimulated by Cd, and is upregulated in apoptotic cells (Horikoshi et al., 1999). As can be seen from Figure 6, the expression level of G3PDH gene in the CNS tissue of the organism increases with the longer time exposure to Cd solution. We speculate that the biological function of G3PDH gene is most affected when organisms are subjected to heavy metal stress.
Conclusion
Onchidium reevesii is an economically important marine shellfish, and a convenient organism for biomonitoring of aquatic pollution. Herein, we report the first transcriptome analysis of O. reevesii, a first for an Onchidiidae species. We also report the first RNA-Seq analysis of O. reevesii in response to the heavy metal Cd. Many of the identified DEGs were metal ion binding proteins, synapse-associated proteins, and markers associated with environmental stress. Time-series expression analysis was used to investigate the CNS in the transcriptome of O. reevesii in response to Cd stimulation, and GO enrichment analysis was performed on genes displaying a significant FC in expression following Cd exposure. Metal ion binding, synaptic transmission, and oxidative stress were identified as important biological processes in the early stages of Cd exposure. These findings provide a comprehensive overview of the responses of O. reevesii to Cd stimulation, and identify potential molecular targets related to marine pollutants in shellfish. We believe that G3PDH gene could serve as biomarkers for marine environmental pollution monitoring.
Data Availability
The datasets generated for this study can be found in the Sequence Read Archive (SRA), the accession number PRJNA511644.
Ethics Statement
The study was approved by the ethical committee of Shanghai Ocean University (Ethical Application Ref: SHOU-DW-2019-024).
Author Contributions
BG was responsible for the experimental design, carrying out the experiments, data processing, and article writing. XL performed the sample collection. TY performed the sample processing and extraction. HS provided experimental guidance and supervision.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 41276157).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Part of this work was performed using the resources of the Computational Biology Service of Shanghai OE Biotech. Co., Ltd. (Shanghai, China). Thanks to Dr. Maoxiao Peng for his help in this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00547/full#supplementary-material
FILE S1 | The results of 16 STEM expression profiles in descending order of STEM clustering.
FILE S2 | Information related to the construction of the standard curve of the corresponding primer for the selected 10 genes. R2>0.99, and the amplification efficiency of primers was within the acceptable range.
FILE S3 | Through gene function annotation and enrichment, 17 related gene information of metal ion binding were identified.
Footnotes
- ^ ftp://ftp.ncbi.nih.gov/blast/db
- ^ ftp://ftp.ncbi.nlm.nih.gov/blast/db/
- ^ http://www.genome.jp/kaas-bin/kaas_main
- ^ http://www.uniprot.org/downloads
- ^ http://www.geneontology.org/
- ^ ftp://ftp.ncbi.nih.gov/pub/COG/KOG/kyva
- ^ http://bioconductor.org/packages/release/bioc/html/DESeq.html
- ^ http://www.geneontology.org/
References
Agarwal, A. R., Liqin, Z., Harsh, S., Sundar, I. K., Irfan, R., and Enrique, C. (2012). Short-term cigarette smoke exposure induces reversible changes in energy metabolism and cellular redox status independent of inflammatory responses in mouse lungs. Am. J. Physiol. Lung Cell. Mol. Physiol. 303, 889–898. doi: 10.1152/ajplung.00219.2012
Alex, P., Jane, W., Yuriy, Z., Nataliya, K., Weiss, K. R., and Vladimir, B. (2008). Predicting adaptive behavior in the environment from central nervous system dynamics. PLoS One 3:e3678. doi: 10.1371/journal.pone.0003678
Amiard, J. C., Amiard-Triquet, C., Barka, S., Pellerin, J., and Rainbow, P. S. (2006). Metallothioneins in aquatic invertebrates: their role in metal detoxification and their use as biomarkers. Aquat. Toxicol. 76, 160–202. doi: 10.1016/j.aquatox.2005.08.015
Anders, S., and Huber, W. (2013). Differential Expression of RNA-Seq Data at the Gene Level – the DESeq Package. Heidelberg: European Molecular Biology Laboratory.
Baussant, T., Bechmann, R. K., Taban, I. C., Larsen, B. K., Tandberg, A. H., Bjørnstad, A., et al. (2009). Enzymatic and cellular responses in relation to body burden of PAHs in bivalve molluscs: a case study with chronic levels of North Sea and Barents Sea dispersed oil. Mar. Pollut. Bull. 58, 1796–1807. doi: 10.1016/j.marpolbul.2009.08.007
Benoit, B., and Thierry, B. (2002). Integrated biomarker response: a useful tool for ecological risk assessment. Environ. Toxicol. Chem. 21, 1316–1322. doi: 10.1002/etc.5620210629
Berra, E., Forcella, M., Giacchini, R., Marziali, L., Rossaro, B., and Parenti, P. (2004). Evaluation of enzyme biomarkers in freshwater invertebrates from Taro and Ticino River. Italy. Ann. Limnol. Int. J. Limnol. 40, 169–180. doi: 10.1051/limn/2004015
Bhave, G., Cummings, C. F., Vanacore, R. M., Kumagai-Cresse, C., Ero-Tolliver, I. A., Rafi, M., et al. (2012). Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat. Chem. Biol. 8, 784–790. doi: 10.1038/nchembio.1038
Bodin, N., Burgeot, T., Stanisière, J. Y., Bocquené, G., Menard, D., Minier, C., et al. (2004). Seasonal variations of a battery of biomarkers and physiological indices for the mussel Mytilus galloprovincialis transplanted into the northwest Mediterranean Sea. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 138, 411–427. doi: 10.1016/j.cca.2004.04.009
Damiens, G., Gnassia-Barelli, M., Loquès, F., Roméo, M., and Salbert, V. (2007). Integrated biomarker response index as a useful tool for environmental assessment evaluated using transplanted mussels. Chemosphere 66, 574–583. doi: 10.1016/j.chemosphere.2006.05.032
de Oliveira, L. F., and Martinez, S. (2007). 16.P.3. Aquatic invertebrates under waterborne lead exposure: catalase activity. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 148, S66–S79.
Di Giulio, R. T., Benson, W. H., Sanders, B. M., and Van Veld, P. A. (1995). “Biochemical mechanisms: metabolism, adaptation and toxicity,” in Fundamentals of Aquatic Toxicology:Effects, Environmental Fate and Risk Assessment, 2nd Edn, ed. G. M. Rand (Washington DC: Taylor & Francis), 523–561.
Evtushenko, Z. S., Belcheva, N. N., and Lukyanova, O. N. (1986). Cadmium accumulation in organs of the scallop Mizuhopecten yessoensis–II. Subcellular distribution of metals and metal-binding proteins. Comp. Biochem. Physiol. Part C Comp. Pharmacol. 83, 377–383. doi: 10.1016/0742-8413(86)90139-8
Friberg, L., Elinder, C., Kjellstrom, T., and Nordberg, G. F. (1985). Cadmium and Health: A Toxicological and Epidemiological Appraisal Volume 1: Exposure, dose, and Metabolism. Boca Raton, FL: CRC Press.
Gagné, F., Turcotte, P., and Auclair, J. (2013). The effects of zinc oxide nanoparticles on the metallome in freshwater mussels. Comp. Biochem. Physiol. Toxicol. Pharmacol. 158, 22–28. doi: 10.1016/j.cbpc.2013.04.001
Gomes, T., Pereira, C. G., Cardoso, C., Pinheiro, J. P., Cancio, I., and Bebianno, M. J. (2012). Accumulation and toxicity of copper oxide nanoparticles in the digestive gland of Mytilus galloprovincialis. Aquat. Toxicol. 118–119, 72–79. doi: 10.1016/j.aquatox.2012.03.017
Gorbushin, A. M., and Levakin, I. A. (2001). Hydrobia ulvae (Gastropoda: Prosobranchia): a new model for regeneration studies. J. Exp. Biol. 204, 283–289.
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1186/1471-2105-12-S14-S2
Harris, M. A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., et al. (2004). The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 32, 258–261.
Horikoshi, N., Cong, J., Kley, N., and Shenk, T. (1999). Isolation of differentially expressed cDNAs from p53-dependent apoptotic cells: activation of the human homologue of the Drosophila peroxidasin gene. Biochem. Biophys. Res. Commun. 261, 864–869. doi: 10.1006/bbrc.1999.1123
Hou, R., Bao, Z., Wang, S., Su, H., Li, Y., Du, H., et al. (2011). Transcriptome sequencing and de novo analysis for yesso scallop (Patinopecten yessoensis) using 454 GS FLX. PLoS One 6:e21560. doi: 10.1371/journal.pone.0021560
Huang, J. T., and Wang, A. M. (2008). Determination of the nutrients of Onchidium struma and evaluation of its quality. Mar. Sci. 32, 29–35.
Ivanina, A. V., Beniash, E., Etzkorn, M., Meyers, T. B., Ringwood, A. H., and Sokolova, I. M. (2013). Short-term acute hypercapnia affects cellular responses to trace metals in the hard clams Mercenaria mercenaria. Aquat. Toxicol. 140–141, 123–133. doi: 10.1016/j.aquatox.2013.05.019
Ivankovic, D., Pavicic, J., Beatovic, V., Klobucar, R., and Klobucar, G. (2010). Inducibility of metallothionein biosynthesis in the whole soft tissue of zebra mussels Dreissena polymorpha exposed to cadmium, copper, and pentachlorophenol. Environ. Toxicol. 25, 198–211. doi: 10.1002/tox.20489
Kiffney, P. M., and Clements, W. H. (1993). Bioaccumulation of heavy metals by benthic invertebrates at the arkansas river, colorado. Environ. Toxicol. Chem. 12, 1507–1517. doi: 10.1002/etc.5620120818
Klose, M. K., Dason, J. S., Atwood, H. L., Boulianne, G. L., and Mercier, A. J. (2010). Peptide-induced modulation of synaptic transmission and escape response in Drosophila requires two G-protein-coupled receptors. J. Neurosci. 30, 14724–14734. doi: 10.1523/JNEUROSCI.3612-10.2010
Lalitha, S. (2000). Primer Premier 5. Biotech Softw. Intern. Rep. 1, 270–272. doi: 10.1089/152791600459894
Larionov, A., Krause, A., and Miller, W. (2005). A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6:62. doi: 10.1186/1471-2105-6-62
Lars, T., Zuzow, M. J., Ivanina, A. V., Elia, B., and Sokolova, I. M. (2011). Proteomic response to elevated PCO2 level in eastern oysters, Crassostrea virginica: evidence for oxidative stress. J. Exp. Biol. 214, 1836–1844. doi: 10.1242/jeb.055475
Lawson, O. E., and Lawson, E. O. (2011). Physico-chemical parameters and heavy metal contents of water from the mangrove swamps of lagos lagoon, lagos, Nigeria. Adv. Biol. Res. 5, 8–21.
Lázár, E., Péterfi, Z., Sirokmány, G., Kovács, H. A., Klement, E., Medzihradszky, K. F., et al. (2015). Structure–function analysis of peroxidasin provides insight into the mechanism of collagen IV crosslinking. Free Radic. Biol. Med. 83, 273–282. doi: 10.1016/j.freeradbiomed.2015.02.015
Leiniö, S., and Lehtonen, K. K. (2005). Seasonal variability in biomarkers in the bivalves Mytilus edulis and Macoma balthica from the northern Baltic Sea. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 140, 408–421. doi: 10.1016/j.cca.2005.04.005
Leung, P. T., Ip, J. C., Mak, S. S., Qiu, J. W., Lam, P. K., Wong, C. K., et al. (2014). De novo transcriptome analysis of Perna viridis highlights tissue-specific patterns for environmental studies. BMC Genomics 15:804. doi: 10.1186/1471-2164-15-804
Li, X. B. (2008). Studies on the toxicity of copper on adult Onchidium struma. master’s thesis, East China Normal University, Shanghai.
Li, X. B., Hou, X. L., Qian, M., Zhao, Y. L., Cheng, Y. X., and Wang, Q. (2009). Toxic effects of copper on antioxidative and metabolic enzymes of the marine gastropod, Onchidium struma. Arch. Environ. Contam. Toxicol. 56, 776–784. doi: 10.1007/s00244-009-9290-2
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Livingston, D. R., Förlin, L., and George, S. G. (1994). Molecular Biomarkers and Toxic Consequences of Impact by Organic Pollution in Aquatic Organisms. Ambleside: Freshwater Biological Association, 154–171.
Livingstone, D. R. (1991). Organic Xenobiotic Metabolism in Marine Invertebrates. Berlin: Springer Berlin Heidelberg.
Livingstone, D. R., Chipman, J. K., Lowe, D. M., Minier, C., and Pipe, R. K. (2000). Development of biomarkers to detect the effects of organic pollution on aquatic invertebrates: recent molecular, genotoxic, cellular and immunological studies on the common mussel (Mytilus edulis L.) and other mytilids. Int. J. Environ. Pollut. 13, 56–91.
McGettigan, P. A. (2013). Transcriptomics in the RNA-seq era. Curr. Opin. Chem. Biol. 17, 4–11. doi: 10.1016/j.cbpa.2012.12.008
Milan, M., Coppe, A., Reinhardt, R., Cancela, L. M., Leite, R. B., Saavedra, C., et al. (2011). Transcriptome sequencing and microarray development for the Manila clam, Ruditapes philippinarum: genomic tools for environmental monitoring. BMC Genomics 12:234. doi: 10.1186/1471-2164-12-234
Mun, S. K., Yun, J. M., Kesavan, S., Wonseok, O., Sumin, W., Jiyoung, Y., et al. (2017). The whole-genome and transcriptome of the Manila Clam (Ruditapes philippinarum). Genome Biol. Evol. 9, 1487–1498. doi: 10.1093/gbe/evx096
Nelson, R. E., Fessler, L. I., Takagi, Y., Blumberg, B., Keene, D., Olson, P. F., et al. (1994). Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J. 13, 3438–3447. doi: 10.1002/j.1460-2075 1994.tb06649.x
Ng, T., and Wang, W. X. (2004). Detoxification and effects of Ag, Cd, and Zn pre-exposure on metal uptake kinetics in the clam Ruditapes philippinarum. Mar. Ecol. Prog. 268, 161–172. doi: 10.3354/meps268161
Ng, Y. T., Rainbow, P. S., Amiard-Triquet, C., Amiard, J. C., and Wang, W. X. (2007). Metallothionein turnover, cytosolic distribution and the uptake of Cd by the green mussel Perna viridis. Aquat. Toxicol. 84, 153–161. doi: 10.1016/j.aquatox.2007.01.010
Nicholls, C., Li, H., and Liu, J. P. (2012). GAPDH: a common enzyme with uncommon functions. Clin. Exp. Pharmacol. Physiol. 39, 674–679. doi: 10.1111/j.1440-1681.2011.05599.x
Nicholson, S., and Lam, P. K. S. (2005). Pollution monitoring in Southeast Asia using biomarkers in the mytilid mussel Perna viridis (Mytilidae: Bivalvia). Environ. Int. 31, 121–132. doi: 10.1016/j.envint.2004.05.007
Office of Water, and United States Environmental Protection Agency, (1999). Rapid Bioassessment Protocols for Use in Wadeable Streams and Rivers: Periphyton, Benthic Macroinvertebrates, and Fish. Washington, D.C.: United States Environmental Protection.
Ozsolak, F., and Milos, P. M. (2010). RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87–98. doi: 10.1038/nrg2934
Patel, R. K., and Jain, M. (2012). NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003). TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. doi: 10.1093/bioinformatics/btg034
Rebl, A., Verleih, M., Krasnov, A., Jaros, J., and Goldammer, T. (2014). Transcriptome profiling reveals insight into distinct immune responses to Aeromonas salmonicida in gill of two rainbow trout strains. Mar. Biotechnol. 16, 333–348. doi: 10.1007/s10126-013-9552-x
Regoli, F., and Principato, G. (1995). Glutathione, glutathione-dependent and antioxidant enzymes in mussel, Mytilus galloprovincialis, exposed to metals under field and laboratory conditions: implications for the use of biochemical biomarkers. Aquat. Toxicol. 31, 143–164. doi: 10.1016/0166-445x(94)00064-w
Reissner, K. J., Boyle, H. D., Xiaojing, Y., and Carew, T. J. (2010). Aplysia synapse associated protein (APSAP): identification, characterization, and selective interactions with Shaker-type potassium channels. J. Neurochem. 105, 1006–1018. doi: 10.1111/j.1471-4159.2007.05202.x
Ren, X. U., Yang, Y., and Zhien, L. I. (2007). Diffusion of heavy metals from marine environment to shellfish. Mar. Sci. Bull. 26, 117–120.
Ringwood, A. H., Mccarthy, M., Bates, T. C., and Carroll, D. L. (2010). The effects of silver nanoparticles on oyster embryos. Mar. Environ. Res. 69, S49–S51. doi: 10.1016/j.marenvres.2009.10.011
Santoro, A., Blo, G., Mastrolitti, S., and Fagioli, F. (2009). Bioaccumulation of heavy metals by aquatic macroinvertebrates along the Basento river in the South of Italy. Water Air Soil Pollut. 201, 19–31. doi: 10.1007/s11270-008-9923-5
Solé, M., Kopecka-Pilarczyk, J., and Blasco, J. (2009). Pollution biomarkers in two estuarine invertebrates, Nereis diversicolor and Scrobicularia plana, from a Marsh ecosystem in SW Spain. Environ. Int. 35, 523–531. doi: 10.1016/j.envint.2008.09.013
Strickler, A. G., and Soares, D. (2011). Comparative genetics of the central nervous system in epigean and hypogean Astyanax mexicanus. Genetica 139, 383–391. doi: 10.1007/s10709-011-9557-1
Suárez-Ulloa, V., Fernández-Tajes, J., Manfrin, C., Gerdol, M., Venier, P., and Eirín-López, J. M. (2013). Bivalve omics: state of the art and potential applications for the biomonitoring of harmful marine compounds. Mar. Drugs 11, 4370–4389. doi: 10.3390/md11114370
Sun, P. X., Hao, L. H., Bei-Bei, D. U., Wang, X. Q., Lian, Y., Dai, X. X., et al. (2014). Assesement of the heavy metals impact on marine environment and on the health risk of diatary shellfish in songgou bay. Adv. Mar. Sci. 32, 249–258.
Sutoo, D., Akiyama, K., and Imamiya, S. (1990). A mechanism of cadmium poisoning: the cross effect of calcium and cadmium in the calmodulin-dependent system. Arch. Toxicol. 64, 161–164. doi: 10.1007/bf01974404
Tania, G., Pinheiro, J. P., Ibon, C., Pereira, C. G., Cátia, C., and Maria João, B. (2011). Effects of copper nanoparticles exposure in the mussel Mytilus galloprovincialis. Environ. Sci. Technol. 45, 9356–9362. doi: 10.1021/es200955s
Tindall, A. J., Pownall, M. E., Morris, I. D., and Isaacs, H. V. (2005). Xenopus tropicalis peroxidasin gene is expressed within the developing neural tube and pronephric kidney. Dev. Dyn. 232, 377–384. doi: 10.1002/dvdy.20226
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
United States Environmental Protection Agency [US EPA], (1997). Volunteer Stream Monitoring: A Methods Manual. Washington, D.C: Office of Water.
Viarengo, A., Canesi, L., Garcia Martinez, P., Peters, L. D., and Livingstone, D. R. (1995). Pro-oxidant processes and antioxidant defence systems in the tissues of the Antarctic scallop (Adamussium colbecki) compared to the Mediterranean scallop (Pecten jacobaeus). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 111, 119–126. doi: 10.1016/0305-0491(94)00228-m
Wang, X. Y., Yang, H. S., Liu, G. P., and Wang, Q. (2011). Enzyme responses and lipid peroxidation in gills and hepatopancreas of clam Mactra vereformis, following cadmium exposure. Chinese J. Oceanol. Limnol. 29, 981–989. doi: 10.1007/s00343-011-0088-5
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wei, S., Xiaofan, G., Yu, H., Cheng, G., Jiahuan, R., Wenhao, S., et al. (2018). Waterborne Cd2+ weakens the immune responses of blood clam through impacting Ca2+ signaling and Ca2+ related apoptosis pathways. Fish Shellf. Immunol. 77, 208–213. doi: 10.1016/j.fsi.2018.03.055
Winston, G. W., Giulio, R. T., and Giulio, D. (1991). Prooxidant and antioxidant mechanisms in aquatic organisms. Aquat. Toxicol. 19, 137–161. doi: 10.1016/0166-445x(91)90033-6
Wu, X. G., Liu, F. P., Tang, B. P., Cheng, Y. X., Wang, J. H., Xu, R., et al. (2007). Study on the heavy metal content and its evaluation in the different tissues of adult Onchidium struma. Mar. Fish. 29, 319–324.
Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., Xu, F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zhao, X., Yu, H., Kong, L., and Li, Q. (2012). Transcriptomic responses to salinity stress in the pacific oyster Crassostrea gigas. PLoS One 7:e46244. doi: 10.1371/journal.pone.0046244
Keywords: Onchidium reevesii, transcriptome, time-series analysis, heavy metal exposure, environmental stress, metal ion binding, synaptic transmission, oxidative stress
Citation: Gu B, Yang T, Liu X and Shen H (2019) Transcriptomic Analysis of the Onchidium reevesii Central Nervous System in Response to Cadmium. Front. Mar. Sci. 6:547. doi: 10.3389/fmars.2019.00547
Received: 18 February 2019; Accepted: 20 August 2019;
Published: 03 September 2019.
Edited by:
Wan-Xi Yang, Zhejiang University, ChinaReviewed by:
Qiu-Ning Liu, Yancheng Teachers University, ChinaBoping Tang, Yancheng Teachers University, China
Copyright © 2019 Gu, Yang, Liu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heding Shen, hdshen@shou.edu.cn
†These authors have contributed equally to this work
 Bingning Gu
Bingning Gu Tiezhu Yang
Tiezhu Yang Xin Liu1,2,3,4
Xin Liu1,2,3,4  Heding Shen
Heding Shen