DNA Analysis of Juvenile Scalloped Hammerhead Sharks Sphyrna lewini (Griffith, 1834) Reveals Multiple Breeding Populations and Signs of Adaptive Divergence in the South Pacific
- 1School of Marine Studies, Molecular Analytics Laboratory (MOANA LAB), Faculty of Science Technology and Environment, The University of the South Pacific, Laucala Campus, Suva, Fiji
- 2ESE, Ecology and Ecosystem Health, Agrocampus-Ouest, INRA, Rennes, France
- 3Instituto de Ciencias Marinas de Andalucía (ICMAN), Consejo Superior de Investigaciones Científicas, Campus Universitario Río San Pedro, Cádiz, Spain
Shark species have undergone drastic population declines in recent decades due to overfishing and habitat destruction; thus, establishing connectivity among the populations of various shark species is important to determine the appropriate units and spatial scale for conservation and management, particularly as this group is long-lived with late age of maturation. Consequently, we used DNA variation at 1,317 putatively neutral and 25 potentially adaptive single nucleotide polymorphisms (SNPs) to analyze population genetic structure among 174 unrelated individuals of scalloped hammerhead sharks (Sphyrna lewini) from the Rewa Delta and the Ba Estuary, where documented aggregations of neonates and young-of-the-year occur in the island of Viti Levu, Republic of Fiji. Results of the pairwise FST analysis for the neutral loci revealed a small but significant genetic differentiation (FST: 0.004; P-value = 0.0009). Furthermore, the 25 potentially adaptive loci (i.e., under putative selection) revealed a magnitude of differentiation four times bigger than the estimate obtained using neutral genetic diversity (FST: 0.017; P-value = 0.0009). Interestingly, population assignment tests, using the neutral SNP data set and two different software packages, Admixture and assignPOP, provided evidence for the existence of up to four genetically differentiated populations among our samples. Assignment probabilities ranged from 0.98 ± 0.01 to 0.81 ± 0.03. Admixture and assignPOP assigned the same individuals to the same putative populations for all sampled neonates. Thus, our results provide unequivocal evidence that adult females from multiple genetically differentiated breeding populations contribute to these juvenile aggregation sites.
Introduction
The conservation of elasmobranchs is essential as many shark species have undergone drastic population declines and are thus, some of the most threatened fishes worldwide (Dulvy et al., 2017). Ecologically, many shark species are dominant apex predators while others are at least tertiary consumers and their loss would likely result in catastrophic trophic cascades (Hussey et al., 2014). Some sharks possess traits such as large cosmopolitan ranges, long migrations and longevity, which enhance gene flow, resulting in low levels of genetic differentiation within ocean basins. Establishing connectivity among populations has important conservation implications for determining the appropriate units and spatial scale for management and conservation.
The scalloped hammerhead shark, Sphyrna lewini (Griffith, 1834), is a viviparous high trophic level predator, circumglobally distributed in tropical and warm temperate waters over both, neritic and adjacent pelagic waters, with fecundities ranging from 13 to 31 neonates of 42–55 cm total length per litter after a gestation period of 9–10 months (Compagno, 1984). The species is known to use coastal embayments and river mouths for parturition and juvenile development where they can grow and mature protected from large predators (Daly-Engel et al., 2012) and is known to form large female-dominated aggregations around oceanic islands with some degree of site fidelity and residency to these archipelagos (Nalesso et al., 2019). Juveniles are demersal and gregarious, while adult females form seasonal pelagic aggregations, and males appear to be more solitary and more common in open waters. In the Eastern Pacific Ocean, males, juvenile and neonate scalloped hammerhead sharks are uncommon at oceanic aggregations. Pupping grounds for the Cocos, Galapagos, Malpelo, Revillagigedo and Gulf of California island populations are thought to be located mainly along continental shores of southern-North, Central and South America (Nalesso et al., 2019). For example, important nursery grounds have been documented in the western Pacific in the Golf of California (Torres-Huerta et al., 2008) and in the Golf of Tehuantepec in Mexico (Bejarano-Álvarez et al., 2011), and in the Golf of Nicoya in Costa Rica (Zanella et al., 2009). Several scalloped hammerhead shark populations have been heavily exploited worldwide by both inshore and offshore fisheries and the species is considered endangered by the International Union for Conservation of Nature (IUCN) Red List (Nalesso et al., 2019 and references therein).
Studies on the population genetics of scalloped hammerhead sharks utilizing mitochondrial and nuclear microsatellite DNA markers detected, at the same geographical scale, significant structuring within ocean basins but male mediated gene flow and panmixia, respectively (Daly-Engel et al., 2012). Moreover, some intra-oceanic structuring has been detected using putatively neutral nuclear DNA markers. Nance et al. (2011) was able to detect a break between specimens from Mexico and Colombia, Spaet et al. (2015) found a break between the Red Sea and the Indian Ocean, and Castillo-Olguin et al. (2012) detected divergent populations in the Gulf of California, although no differentiation was observed among locations from the Mexican Pacific Ocean.
Interestingly, adaptive DNA markers of the major histocompatibility complex (MHC) gene family have proven to be extremely informative for a wide ranging, highly migratory, and philopatric marine species, such as the endangered loggerhead sea turtle (Caretta caretta; Linnaeus, 1758). Genetically distinct nesting colonies (rockeries) were identified on islands separated by between 100 and 200 km (Stiebens et al., 2013). The aim of this study is to assess the connectivity between two recently documented aggregations of neonates and young-of-the-year (YOY) scalloped hammerhead sharks in Viti Levu, the largest island in the Republic of Fiji, which have been identified as critical habitats for the species (Marie et al., 2017; Vierus et al., 2018). Nursery and parturition grounds play a fundamental role in the survival of YOY, and their persistence is fundamental for stock recruitment. This is particularly significant for sharks, as their litters are generally small. Because variability in mortality and growth rates through YOY life stage is large, substantial research efforts have focused in understanding how populations compensate for the reduction in numbers experienced during this period and how the habitats where they occur can be adequately managed (Chambers and Trippel, 2012).
In this study, we tested the hypothesis that the distribution of potential adaptive genetic diversity may reflect significant population structure between samples of YOY captured in the Rewa Delta and the Ba Estuary, Fiji, where the above mentioned aggregations occur; just as found in loggerhead sea turtles rockeries (Stiebens et al., 2013). Fiji, as an oceanic and remote island, is isolated from the other suitable nursery grounds and provides adequate parturition areas given the number of large estuaries present in its topography. S. lewini is documented to migrate up to 1,671 km (Bessudo et al., 2011) and other recorded juvenile aggregations in Melanesia, Micronesia and Polynesia are found at greater distances (e.g., Hawaiian Islands). Furthermore, oceanic islands such as Cocos, Galapagos and Malpelo in the Eastern Pacific are known to harbor long-term resident aggregations for this species (Nalesso et al., 2019). Therefore, understanding the role of this critical habitat in the population dynamics of the species in Melanesia and beyond is fundamental for its protection.
Materials and Methods
Sampling Procedures and Sequencing
The study focused on the Rewa Delta and Ba Estuary (Rewa and Ba hereafter) (Figure 1). Samplings were conducted under a research permits provided by the Rewa and Ba Provincial Councils as well as the Department of Fisheries. All handling procedures of sharks were approved by the relevant ethical guidelines and regulations (Marie et al., 2017; Vierus et al., 2018). Each shark was measured, sexed and aged according to the umbilical scar status (Marie et al., 2017). In addition, two biopsy tissue samples were taken from the pelvic fins of each individual and preserved in 95% ethanol until DNA extraction. Due to financial constraints, we only genotyped a subsample of 166 individuals out of the 1054 captures from Rewa (Marie et al., 2019). All 80 samples captured in Ba were analyzed. For DNA sequencing and genotyping, we followed previously developed protocols available in Ren et al. (2015) and Marie et al. (2019).
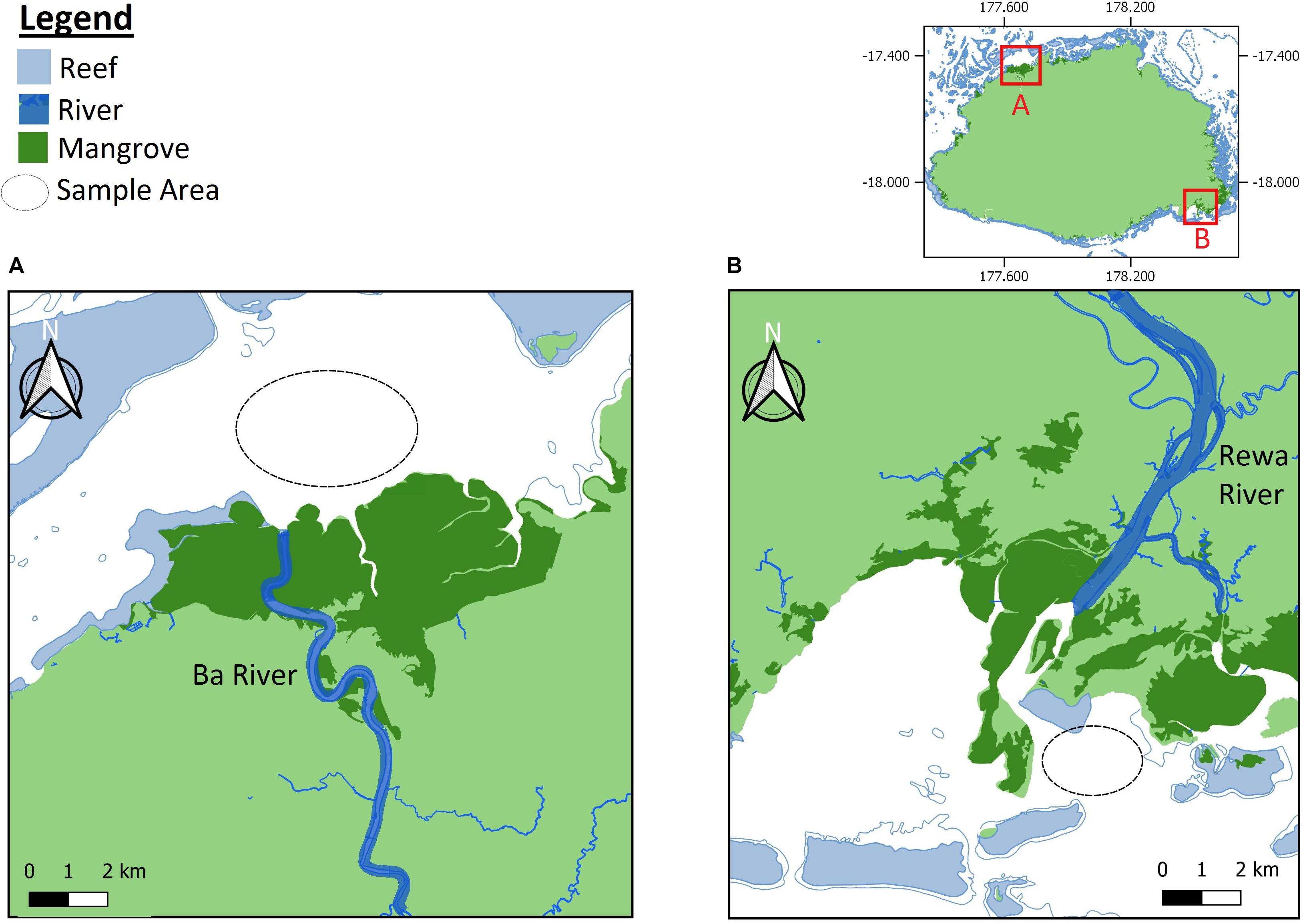
Figure 1. Geographic location of sampling sites in Viti Levu Island, Republic of Fiji. This figure was prepared using online resources from QGIS, version 3.6.0 https://www.qgis.org/en/site/.
SNPs Filtering
The genotype dataset was filtered to retain only the highly informative single nucleotide polymorphisms (SNP). We initially filtered the entire dataset to run kinship analysis within and between sites. From these results, we modified the dataset retaining only one individual per family to assess the genetic diversity and population genetic structure between the two areas avoiding consanguinity biases.
For each analysis, we first removed duplicate SNPs (possessing identical Clone IDs), and retained loci according to call rate of 98%, read depth > 7 and minor allele frequency (MAF) > 5%. A MAF > 2% was used for estimating genetic diversity and population genetic structure analysis. We then searched for candidate loci under selection using BayeScan v.2.1 (Foll and Gaggiotti, 2008; Foll, 2012) software at FDR = 0.05. Departure from Hardy-Weinberg Equilibrium was then assessed for each locus using the software Arlequin v.3.5.2.2 (Schneider et al., 2000) using an exact test with 10,000 steps in the Markov Chain method and 100,000 dememorizations. Tests for linkage disequilibrium was performed using the software PLINK v.1.07 (Chang et al., 2015).
Kinship Analyses
Kinship was analyzed using TrioML with COANCESTRY v. 1.0.1.7 (Wang, 2011). The data set was initially run to estimate the degree of relatedness within the samples. This new data set was then used in the analyses from this point onward. To determine cut-off r values for categorizing relatedness coefficients produced by the various algorithms into full-sib, half-sib, and less-related brackets, COANCESTRY was again used to simulate 200 dyads in each group based on the same allele frequencies as the original population samples. These values became the cut-offs for delineating full- and half-siblings in the empirical dataset.
Detecting Outlier Loci
FST outliers were identified using LOSITAN (Antao et al., 2008) at a confidence level of 95% and False Discovery Rate of 10%. Five analyses were run and a final loci list comprised of an 80% consensus was generated. The consensus outliers were extracted from the neutral dataset, resulting in three sets: neutral loci, loci under putative selection (LUPS), and those under balancing selection.
Genetic Diversity and Population Genetic Structure Analyses
The observed (HO), expected (HE) and sample size-corrected expected (Hnb) heterozygosity as well as the inbreeding coefficient (FIS – analyzed across 1,000 permutations) were determined for each sampling site using the program Genetix version 4.05 (Belkhir et al., 1996). The genetic differentiation was assessed using pairwise FST estimates, calculated across 10,000 permutations, and performed with the software Arlequin version 3.5.2.1 (Excoffier and Lischer, 2010). Admixture was used to detect population structuring, but only on the neutral dataset since this analysis requires a large dataset for accurate estimates (Alexander et al., 2009). First, the optimal number of groups (k) was assessed and then the probability of individual population assignment was calculated. AssignPOP (Chen et al., 2018) was also used to calculate population membership of individuals based on the k determined by Admixture.
Results
Kinship Analyses
Using 69,164 SNPs, we identified 1,871 unlinked loci following quality-filtering controls. After determining the cut-off values for full siblings (FS), 83 dyads including 88 individuals were detected for Rewa (Marie et al., 2019), while only 13 dyads and 18 individuals were detected for Ba, and no dyads involving individuals from each of the rivers was detected (Table 1). For half siblings (HS), larger numbers of both dyads (109) and individuals (99) were found within Rewa, while fewer dyads (4) and individuals (7) were found to be HS from Ba (Table 1). Four dyads including six individuals were found to be HS between the sites. In summary, roughly 75% of all neonates from Rewa were either part of a FS or HS dyad while only 25% of the pups from Ba were part of a related dyad (Table 1). For further analyses, only one individual from related dyads (both FS and HS) was retained resulting in 104 from Rewa and 70 individuals from Ba.
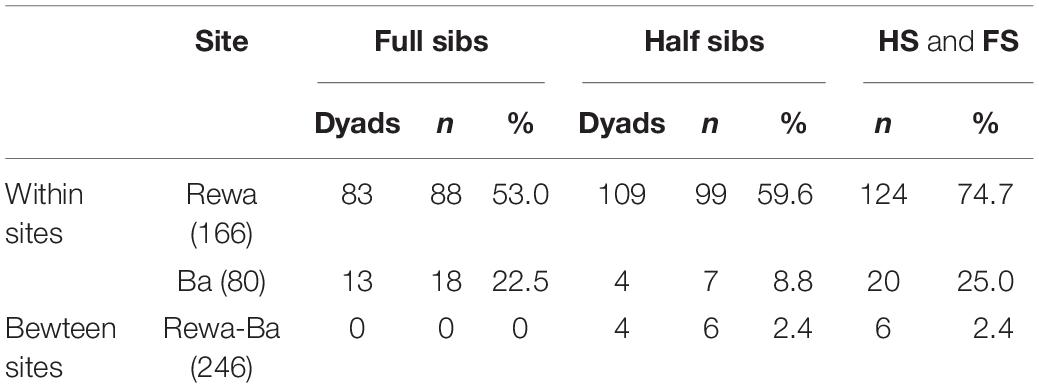
Table 1. Numbers of full sibling (FS) and half sibling (HS) dyads and numbers of individuals (n) represented in each of the related categories for each of the sites and related dyads identified between sites.
Genetic Diversity and Population Genetic Structure
One hundred seventy four unrelated individuals from Rewa and Ba were retained and re-filtered using the same parameters, but MAF, resulting in 1,402 SNPs. The five LOSITAN runs identified between 45 and 19 outlier loci under positive/directional selection and 57 to 89 SNP under balancing selection. An 80% consensus dataset produced 25 and 60 loci as potentially adaptive and under balancing selection, respectively. The remaining 1,317 loci were considered neutral.
Using the neutral dataset, individuals from Rewa showed a HO, HE, and Hnb of 0.289 ± 0.157, 0.227 ± 0.157 and 0.297 ± 0.158, respectively (Table 2). Similar results were observed for individuals from Ba: 0.284 ± 0.158, 0.230 ± 0.156 and 0.299 ± 0.157 for HO, HE, and Hnb, respectively. As well, the FIS assessed for each locality display similar results with a significant FIS of 0.028 and 0.050 (P-value < 0.05) for individuals from Rewa and Ba, respectively.

Table 2. Measures of diversity for each population sample, including the inbreeding coefficient FIS, adjusted expected heterozygosity Hnb, observed heterozygosity HO, and the difference between Hnb and Ho.
Results of the pairwise FST analysis for the neutral loci revealed a small but significant differentiation (FST: 0.004; P-value = 0.0009) between sampling sites. However, the pairwise FST values using the 25 LUPS resulted in a larger genetic differentiation (FST: 0.017; P-value = 0.0009) between the two sites.
With Admixture, an optimal k of four was selected for the neutral data set based on comparing cross validation values. We arbitrarily labeled these four putative populations as A, B, C, and D, because we do not know their geographic origins. The probability of individuals belonging to each of the four putative populations revealed that deme A was by far the most abundant for both sites (Figure 2). More specifically Ba was less diverse and only had two individuals identified as belonging to population D, while the Rewa samples had a handful of individuals identified as belonging to the other three demes (Figure 2). Results from assignPOP, given a k of four, were almost identical to that of Admixture (Figure 2). The best performing assignment model was Support Vector Machine with an assignment accuracy of 0.98 (s.d. = 0.01) for population A, 0.88 (s.d. = 0.04) for B, 0.89 (s.d. = 0.06) for C, and 0.81 (s.d. = 0.03) for D. Admixture and assignPOP allocated the same individuals to the same putative populations for all sampled individuals.
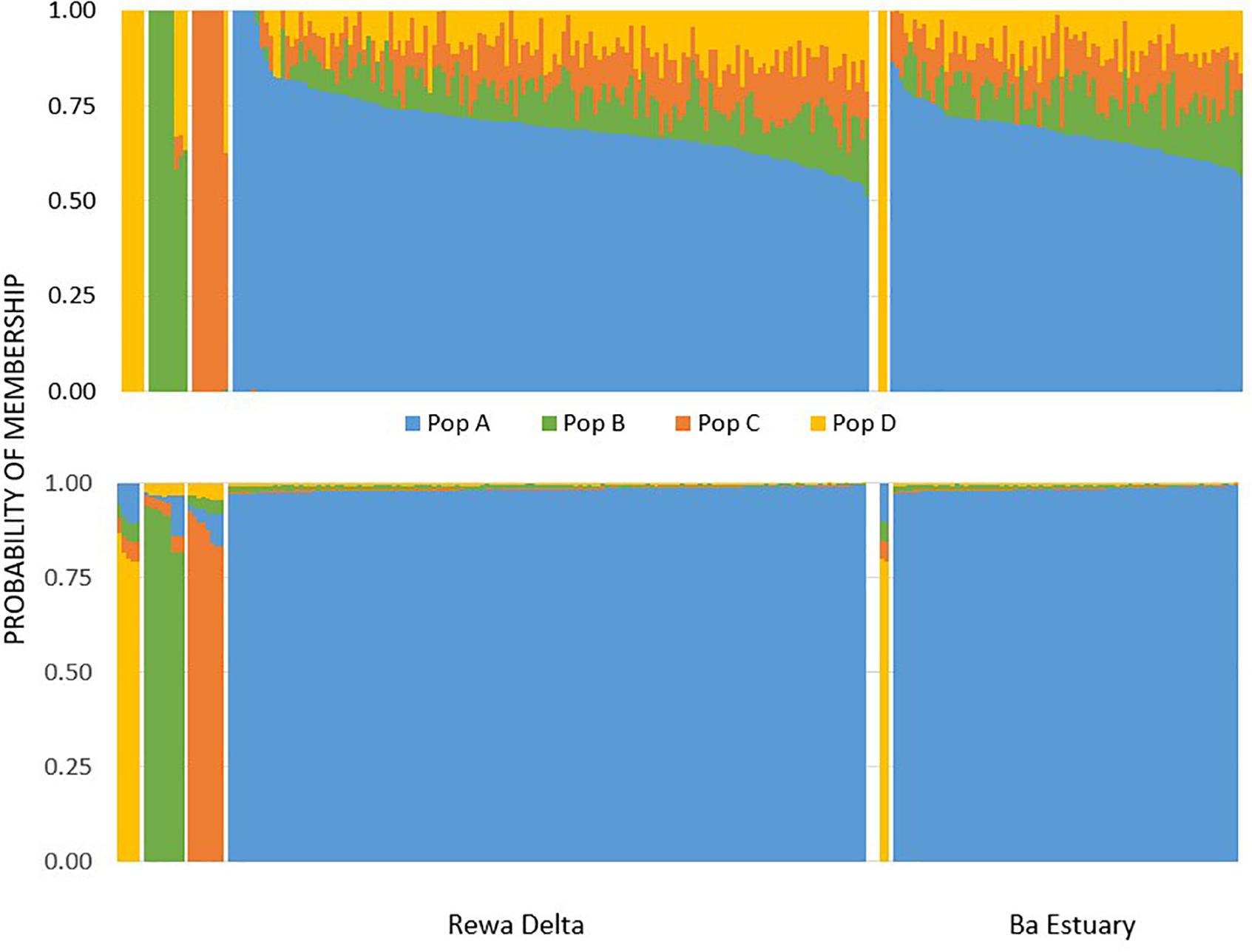
Figure 2. Probability of population assignment based on the optimal k of four for Admixture (top) and assignPOP (bottom) using the SVM model.
Discussion
Kinship
The four dyads of HS detected between sites included two neonates from Ba and four from Rewa. In more detail, #35 was determined to be a HS with two neonates from Rewa. #19 was a HS with two additional neonates from Rewa. These four neonates from Rewa were not found to be FS or HS with each other. This suggests a shared parent for one neonate from Ba and two from Rewa. It is hypothesized that scalloped hammerhead sharks are philopatric and that females of this species utilize only one parturition site per season (Daly-Engel et al., 2012). This hypothesis would suggest that the same male mated with three females and supports male mediated gene flow. The mating system, reproductive sex ratio and kinship relationships of the populations from Rewa are thoroughly analyzed in Marie et al. (2019).
Population Structure at Neutral Loci
Admixture identified four clusters as the optimal number of populations for the neutral data set. Moreover, both Admixture and assignPOP reliably allocated the same individuals to each of these four populations. This indicates that at least two and four genetically differentiated populations utilize the Ba and Rewa river mouths, respectively. The greater number of populations detected at Rewa may either be a result of the larger sample size or the greater use of the Delta by neonates and YOY sharks as suggested by the reported catches in two fisheries independent surveys (Marie et al., 2017; Vierus et al., 2018). This sampling protocol of targeting neonates at known spawning grounds was able to detect multiple populations. The large number of populations utilizing the rivers of Fiji may be a result of the lack of large suitable rivers in the South Pacific outside of this archipelago. The nearest islands with suitable rivers in Melanesia and Polynesia are over 1,200 km to the south-west (New Caledonia) and 4,500 km to the northeast (Hawaii), respectively. Thus, the rivers of Fiji are likely to be important nursery grounds for a large portion of Melanesia, Micronesia and Polynesia. Many South Pacific islands are in fact atolls and thus, large river mouths are simply non-existent.
Further evidence for population sub-structuring comes from the significant heterozygosity deficits. For both, Rewa and Ba adjusted expected multilocus heterozyosity (Hnb) was greater than the observed heterozygosity (HO) indicating an overall significant heterozygosity reduction. Genotyping artifacts are unlikely to be a major cause because the quality filtering steps used in this study are quite stringent and no single locus deviated from HWE. Therefore, admixture as an indicator for the presence of multiple populations at the local level through the Wahlund effect is the most likely explanation for the significant heterozygous deficits observed. Thus, these two independent lines of evidence suggest that sharks from multiple genetically differentiated populations engendered the neonates and YOY sampled at both river mouths. Because these results are based on neonates and YOY collected at known aggregation sites, it can only be assumed that the parents come from different regions of the Pacific Ocean.
Using kinship analyses Marie et al. (2019) suggest that at least one resident population may exist in the region. Residency is an important factor for conservation efforts as the effectiveness of a marine protected area is closely related with the time a species spends within it. Scalloped hammerhead sharks are known to aggregate in large numbers at Cocos Island where residency, defined as the number of days a shark tagged with a radio-acoustic device is detected by receivers from the tagging date, was estimated to be 0.52 ± 0.31, implying that overall the sharks are strongly associated with the island (Nalesso et al., 2019). This study demonstrates the importance of oceanic islands for this species, showing that these habitats are possibly part of a network of abundance hotspots in the Eastern Tropical Pacific. No adult aggregations have been formally reported in the scientific literature in Melanesia, but dive shops in the region often claim seasonal sightings of large numbers (Rico personal observations). Fiji is a Melanesian archipelago of volcanic origin comprised of more than 330 islands and 500 islets, atolls and seamounts and thus, constitutes an ideal habitat for harboring resident populations of scalloped hammerhead sharks such as those reported in the Eastern Pacific (Torres-Huerta et al., 2008; Zanella et al., 2009; Bejarano-Álvarez et al., 2011; Bessudo et al., 2011; Castillo-Olguin et al., 2012; Nalesso et al., 2019). The archipelago harbors a resident and geographically isolated population of bull sharks [Carcharhinus leucas (Müller and Henle, 1838–1841)], which also utilizes the Rewa River for parturition (Glaus et al., 2019). This resident population has a relatively large and healthy effective size (NE > 1000) (Frankham et al., 2014) and is genetically differentiated from other west Pacific and Indian Ocean demes (Glaus, 2019). The genetic structure, mating grounds and home ranges of the hypothetical populations of scalloped hammerhead sharks identified in this study remain to be determined with a Pacific-wide study, but the above studies substantiate the possibility that at least one of them may be local. Determining the residency of these populations also requires extensive radio-acoustic or satellite tagging studies to assist the design of marine protected areas sufficiently large to provide meaningful levels of protection. International collaboration would be essential in this important endeavor to inform management and conservation efforts.
Population Structuring: Outlier Loci
Despite the close proximity of the sampling sites (<300 km), a consensus of 25 LUPS was detected. The significant and larger pairwise FST value of 0.017 for these LUPS over the neutral data set indicate selection mediated isolation to gene flow between samples from four putative populations captured in these aggregations. Although these sites are located on the same island, the Rewa and Ba river mouths vary in both timing and strength of flood time (Catalog of Rivers for the Pacific Islands, 2012) due to the differing monsoon seasons and rainfall on the southeast and the northwest of Viti Levu (Marie et al., 2017; Vierus et al., 2018). However, most individuals from both aggregations were assigned to population A, and thus it is difficult to reconcile the signals of adaptive divergence based on environmental differences between these habitats. Unfortunately, given the short read lengths (30 bps), these sequences could not be annotated reliably, and thus we were unable to identify the genes involved. While highly informative molecular markers such as genes of the MHC have been able to differentiate nesting populations of loggerhead sea turtles separated at comparable distances to this study (Stiebens et al., 2013), we are unable to confirm the hypothesis of adaptive divergence between juvenile aggregation sites. This is because the source of selective constraints resulting in adaptive divergence is unknown. However, the presence of a handful of individuals from genetically differentiated populations present in Rewa appears to be sufficient to increase the FST between sites despite clear evidence of panmixia among individuals from putative population A present at both sites. As mentioned earlier, this species is hypothesized to be philopatric (Daly-Engel et al., 2012) in that females return to the same estuary to give birth. Therefore, the distribution of the adaptive genetic diversity might be sufficiently different to be detected even with male mediated gene flow as suggested by the presence of related individuals between estuaries.
The location of adult home ranges is likely to be hundreds of kilometers from these nursery grounds based on previous studies. Quintanilla et al. (2015) sampled juvenile and adult scalloped hammerhead sharks and found four juveniles on the coastline of Colombia whose parents were captured on an island 500 km offshore. Furthermore, a multidisciplinary study utilized catch size frequencies, tagging, and genetics to infer that estuaries in northern Australia serve as nursery grounds for the species in Indonesia and Papua New Guinea (Chin et al., 2017). Therefore, the non-lethal sampling of adults throughout the Pacific is necessary to determine which populations are utilizing Fijian estuaries as nursery grounds.
In conclusion, aside from genotyping adults from waters of the Pacific Ocean, further research determining the importance of the estuaries of Viti Levu is necessary. As stated above, currently the residency of these females is unknown and if they utilize these estuaries as a nursery grounds permanently or are just transients, remains to be determined. Future research in these estuaries is critical for the conservation of this endangered species in the Pacific since this study has discovered at least four distinct populations that rely on these essential habitats for juvenile survival.
Data Availability Statement
The metadata on shark samples and the raw SNP loci generated for this study are available from Open Science Framework repository at https://osf.io/ykp4h/.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of The University of the South Pacific.
Author Contributions
CR wrote the project proposal and secured the funding. AM, BS, and CR designed the study. AM and BS performed the data analysis. All authors wrote and revised the manuscript.
Funding
The work was supported by the University of the South Pacific, SRT project F1006-R1001-71502-621.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Semi Nagatalevu, Inoke Nagatalevu, Diego Cardeñosa, Gauthier Mescam, Ron Ronaivakulua, Sydney Shier, Kristian Miles, Brittany Waters, Niklas Komoss, Timoci Varinava, and Ramesh Prasad for helping during fieldwork. We are particularly indebt to Celso Cawich for sample collection and fieldwork coordination in the Rewa Delta and to Tom Vierus and Andrew Paris for the same tasks in the Ba Estuary. We are also grateful to Sione Kaitu’u for preparing Figure 1. We also acknowledged Aisake Batibasaga (Director of the Fisheries Department of the Ministry of Fisheries and Forest from 2014 to 2016) and George Madden (Director of the Fisheries Department of the Ministry of Fisheries and Forest since 2016), Joji Kalounivalu (Roko Tui, Rewa Provincial Council), Lary Kemueli (Roko Tui, Ba Provincial Council), Ian Campbell (Shark Conservation Program, WWF-Fiji), and Mike Neuman (Beqa Adventure Divers) for their continued advice and/or support during the project.
References
Alexander, D. H., Novembre, J., and Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Antao, T., Lopes, A., Lopes, R. J., Beja-Pereira, A., and Luikart, G. (2008). LOSITAN: a workbench to detect molecular adaptation based on a F(st)-outlier method. BMC Bioinformatics 9:323. doi: 10.1186/1471-2105-9-323
Bejarano-Álvarez, M., Galván-Magaña, F., and Ochoa-Báez, R. I. (2011). Reproductive biology of the scalloped hammerhead shark Sphyrna lewini (Chondrichthyes: Sphyrnidae) off southwest Mexico. Aqua Int. J. Ichthyol. 17, 11–22.
Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N., and Bonhomme, F. (1996). GENETIX 4.05, Logiciel Sous Windows TM Pour la Génétique Des Populations. Laboratoire Genome, Populations, Interactions, CNRS UMR 5000. Montpellier: Universite’ de Montpellier II
Bessudo, S., Soler, G. A., Klimley, A. P., Ketchum, J. T., Hearn, A., and Arauz, R. (2011). Residency of the scalloped hammerhead shark (Sphyrna lewini) at Malpelo Island and evidence of migration to other islands in the eastern tropical Pacific. Environ. Biol. Fishes 91, 165–176. doi: 10.1007/s10641-011-9769-3
Castillo-Olguin, E., Uribe-Alcocer, M., and Diaz-Jaimes, P. (2012). Assessment of the population genetic structure of Sphyrna lewini to identify conservation units in the Mexican Pacific. Cienc. Mar. 38, 635–652. doi: 10.7773/cm.v38i4.2110
Chambers, R. C., and Trippel, E. A. (2012). Early Life History and Recruitment in Fish Populations. Springer: Science & Business Media.
Chang, C. C., Chow, C. C., Tellier, L., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. doi: 10.1186/s13742-015-0047-8
Chen, K.-Y., Marschall, E. A., Sovic, M. G., Fries, A. C., Gibbs, H. L., and Ludsin, S. A. (2018). AssignPOP: an r package for population assignment using genetic, non-genetic, or integrated data in a machine-learning framework. Methods Ecol. Evol. 9, 439–446. doi: 10.1111/2041-210x.12897
Chin, A., Simpfendorfer, C. A., White, W. T., Johnson, G. J., McAuley, R. B., and Heupel, M. R. (2017). Crossing lines: a multidisciplinary framework for assessing connectivity of hammerhead sharks across jurisdictional boundaries. Sci. Rep. 7:46061. doi: 10.1038/srep46061
Compagno, L. J. V. (1984). FAO Species Catalogue. Sharks of the World. An Annotated and Illustrated Catalogue of Shark Species Known to Date. Rome: FAO Fish Synop.
Daly-Engel, T. S., Seraphin, K. D., Holland, K. N., Coffey, J. P., Nance, H. A., Toonen, R. J., et al. (2012). Global phylogeography with mixed-marker analysis reveals male-mediated dispersal in the endangered scalloped hammerhead shark (Sphyrna lewini). PLoS One 7:e29986. doi: 10.1371/journal.pone.0029986
Dulvy, N. K., Simpfendorfer, C. A., Davidson, L. N., Fordham, S. V., Bräutigam, A., Sant, G., et al. (2017). Challenges and priorities in shark and ray conservation. Curr. Biol. 27, R565–R572. doi: 10.1016/j.cub.2017.04.038
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Foll, M., and Gaggiotti, O. (2008). A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a bayesian perspective. Genetics 180, 977–993. doi: 10.1534/genetics.108.092221
Frankham, R., Bradshaw, C. J. A., and Brook, B. W. (2014). Genetics in conservation management: revised recommendations for the 50/500 rules, red list criteria and population viability analyses. Biol. Conserv. 170, 56–63. doi: 10.1016/j.biocon.2013.12.036
Glaus, K. (2019). Biology and Conservation of the Bull Shark (Carcharhinus leucas) in Fiji. PhD. Thesis, Suva: The University of the South Pacific.
Glaus, K. B., Brunnschweiler, J. M., Piovano, S., Mescam, G., Genter, F., Fluekiger, P., et al. (2019). Essential waters: young bull sharks in Fiji’s largest riverine system. Ecol. Evol. 9, 7574–7585. doi: 10.1002/ece3.5304
Griffith, E. (1834). The Class Pisces, Arranged by the Baron Cuvier, with Supplementary Additions by Edward Griffit and Lieut. Col. Charles Hamilton Smith (and PB Lord), 680. London: Animal Kingdom.
Hussey, N. E., MacNeil, M. A., McMeans, B. C., Olin, J. A., Dudley, S. F., Cliff, G., et al. (2014). Rescaling the trophic structure of marine food webs. Ecol. lett. 17, 239–250. doi: 10.1111/ele.12226
Marie, A. D., Herbinger, C., Fullsack, P. M., and Rico, C. (2019). First reconstruction of kinship in a scalloped hammerhead shark aggregation reveals the mating patterns and breeding sex ratio. Front. Mar. Sci. 6:676. doi: 10.3389/fmars.2019.00676
Marie, A. D., Miller, C., Cawich, C., Piovano, S., and Rico, C. (2017). Fisheries-independent surveys identify critical habitats for young scalloped hammerhead sharks (Sphyrna lewini) in the Rewa Delta. Fiji. Sci. Rep. 7:17273. doi: 10.1038/s41598-017-17152-0
Müller, J., and Henle, F. G. J. (1838–1841). Systematische Beschreibung der Plagiostomen. Berlin: Veit.
Nalesso, E., Hearn, A., Sosa-Nishizaki, O., Steiner, T., Antoniou, A., Reid, A., et al. (2019). Movements of scalloped hammerhead sharks (Sphyrna lewini) at Cocos Island, Costa Rica and between oceanic islands in the eastern tropical Pacific. PLoS One 14:e0213741. doi: 10.1371/journal.pone.0213741
Nance, H. A., Klimley, P., Galván-Magaña, F., Martínez-Ortíz, J., and Marko, P. B. (2011). Demographic processes underlying subtle patterns of population structure in the scalloped hammerhead shark. Sphyrna lewini. PLoS One 6:e21459. doi: 10.1371/journal.pone.0021459
Quintanilla, S., Gómez, A., Mariño-Ramírez, C., Sorzano, C., Bessudo, S., Soler, G., et al. (2015). Conservation genetics of the scalloped hammerhead shark in the pacific coast of colombia. J. Hered. 106, 448–458. doi: 10.1093/jhered/esv050
Ren, R., Ray, R., Li, P., Xu, J., Zhang, M., Liu, G., et al. (2015). Construction of a high-density DArTseq SNP-based genetic map and identification of genomic regions with segregation distortion in a genetic population derived from a cross between feral and cultivated-type watermelon. Mol. Genet. Genomics 290, 1457–1470. doi: 10.1007/s00438-015-0997-7
Schneider, S., Roessli, D., and Excoffier, L. (2000). Arlequin Ver. 2.0: A Software for Population Genetics Data Analysis. Switzerland: Genetics and Biometry Laboratory.
Spaet, J. L. Y., Jabado, R. W., Henderson, A. C., Moore, A. B. M., and Berumen, M. L. (2015). Population genetics of four heavily exploited shark species around the Arabian Peninsula. Ecol. Evol. 5, 2317–2332. doi: 10.1002/ece3.1515
Stiebens, V. A., Merino, S. E., Roder, C., Chain, F. J. J., Lee, P. L. M., and Eizaguirre, C. (2013). Living on the edge: how philopatry maintains adaptive potential. Proc. Biol. sci. 280:20130305. doi: 10.1098/rspb.2013.0305
Torres-Huerta, A. M., Villavicencio-Garayzar, C., and Corro-Espinosa, D. (2008). Reproductive biology of the scalloped hammerhead shark Sphyrna lewini griffith & smith (Sphyrnidae) in the Gulf of California. Hidrobiologica 18, 227–237.
Vierus, T., Gehrig, S., Brunnschweiler, J. M., Glaus, K., Zimmer, M., Marie, A. D., et al. (2018). Discovery of a multispecies shark aggregation and parturition area in the ba Estuary, Fiji Islands. Ecol. Evol. 8, 7079–7093. doi: 10.1002/ece3.4230
Wang, J. (2011). COANCESTRY: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol. Ecol. Resour. 11, 141–145. doi: 10.1111/j.1755-0998.2010.02885.x
Keywords: scalloped hammerhead shark, population structure, genetic diversity, kinship analyses, SNPs
Citation: Marie AD, Stockwell BL and Rico C (2019) DNA Analysis of Juvenile Scalloped Hammerhead Sharks Sphyrna lewini (Griffith, 1834) Reveals Multiple Breeding Populations and Signs of Adaptive Divergence in the South Pacific. Front. Mar. Sci. 6:718. doi: 10.3389/fmars.2019.00718
Received: 25 July 2019; Accepted: 06 November 2019;
Published: 21 November 2019.
Edited by:
Jeffrey C. Mangel, Pro Delphinus, PeruReviewed by:
Gail Schofield, Queen Mary University of London, United KingdomFrancisco Concha, University of Valparaíso, Chile
Sebastian Alberto Klarian, Andres Bello University, Chile
Copyright © 2019 Marie, Stockwell and Rico. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amandine D. Marie, amandine.marie@inra.fr; Ciro Rico, ciro.rico@usp.ac.fj
†These authors have contributed equally to this work
 Amandine D. Marie
Amandine D. Marie Brian L. Stockwell
Brian L. Stockwell Ciro Rico
Ciro Rico