The Horizontal and Vertical Distribution of Deep-Sea Sediments Fungal Community in the South China Sea
- State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
Although diverse fungi have been found in the deep-sea habitats, the space distribution of fungi has not been well characterized. In this study, the fungal horizontal and vertical distribution of the deep-sea sediments, four locations, three depths each, in the South China Sea, were compared using ITS2 high-throughput sequencing. It was revealed that the South China Sea deep-sea sediments harbor diverse marine fungi, including 82.39% Ascomycota, 8.10% Basidiomycota, 0.55% Zygomycota and 8.96% unknown fungi. The results indicate that fungal community structure is not uniform among the different sediment habitats. Though surface sediments have similar fungal diversity across the 4 locations, the fungal abundance and diversity increase with the depth of the sediments from 0 to 2 m, and 1 and 2 m deep sediments show obvious location-dependent fungal community structure. This is the first time to compare the horizontal and vertical distribution of fungal community among different deep-sea sediments in the South China Sea by high-throughput sequencing, providing novel insights into the space distribution characteristics of deep-sea sediments fungi.
Introduction
The deep-sea fungal diversity was first explored by the isolation of deep-sea fungi from the Atlantic Ocean at a depth of 4,450 m in 1964 (Roth et al., 1964). Traditionally, studies on the deep-sea fungal diversity are based on cultivation strategy (Zhang et al., 2013). However, few fungi could be cultured from deep-sea sediments, and most of the culturable fungi were identified as facultative marine fungi, few fungi were identified as marine obligates, such as Dendriphiella (Khudyakova et al., 2000). With the development of high-throughput sequencing, molecular methods enable the detection of difficult-to-culture species and rare species, e.g., LKM11 clade (Bass et al., 2007), KD14 clade (Nagahama et al., 2011). To date, the diversity of fungi from various deep-sea habitats such as South China Sea sediments (Lai et al., 2007; Zhang et al., 2013), Pacific Ocean sediments (Burgaud et al., 2009; Xu et al., 2014), Indian Ocean sediments (Damare et al., 2006; Singh et al., 2010, 2011; Zhang et al., 2014a), Arctic sediments (Zhang et al., 2015) and Mediterranean Sea (Barone et al., 2018), has been explored using either culture-dependent or cultured-independent methods.
It is known that both 18S rRNA gene and ITS region can be used to reveal the fungal community structure. However, the 18S rRNA gene is problematic because the identification is commonly limited to genus or family. The ITS regions of fungi have been used as discriminative targets for molecular analysis of fungal community using high-throughput sequencing (Ihrmark et al., 2012; Lindahl et al., 2013; Gweon et al., 2015; Morgado et al., 2015), their high sequence variability relative to the flanking sequences makes them valuable for genus and species level identification.
Though increasing attention has been paid to deep-sea fungi, compared with deep-sea bacteria and archaea, little is known about the deep-sea fungal space distribution. In this study, the deep-sea (897–1,434 m) fungal communities in the sediments of the South China Sea, four locations and three depths each, were investigated using high-throughput sequencing, with the aim to describe the fungal horizontal and vertical distribution in the deep-sea sediments in the South China Sea.
Materials and Methods
Sample Information
Deep-sea sediment cores (10 cm in diameter, 2 m in length) samples were collected (three replicates for each site) at four different locations: A (1,138 m), B (897 m), C (1,000 m), and D (1,434 m) in the South China Sea (Supplementary Figure 1). 0.25 g (wet weight) of sediments were collected from the 0.05 m depth (surface), 1 m depth, and 2 m depth of each sediment core, respectively. These samples were frozen at −20°C before DNA extraction.
DNA Extraction, ITS2 Sequence Amplification and High-Throughput Sequencing and PCoA Analysis
DNA was extracted using a Power Soil DNA Isolation Kit (MO BIO) according to the manufacturer’s instructions (Ihrmark et al., 2012; Thaler et al., 2012). They ITS2 region was amplified using fungal-specific primers fITS7 (5′- GTGARTCATCGAATCTTTG-3′) and universal ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Wang et al., 2012). Approximately 100 ng of template DNA was used for PCR in a total volume of 25 ml reaction system. PCR was conducted using the following program: denaturation 5 min at 95°C, followed by 30 cycles of 1 min at 95°C, 30 s at 56°C and 1 min at 72°C. A final step of 10 min at 72°C was included to complete any partial polymerizations.
The PCR amplicons were sequenced using Illumina Platform at Genewiz Co., Ltd., Suzhou, China. First, software Trimmomatic was used to filter the raw data, and then the chimera was removed to get the final valid data (The raw data have been submitted to NCBI with BioProject number PRJNA637466). The operational taxonomic unit (OTU) was assigned using a BLAST-based method (Altschul et al., 1997) against the Greengenes database (v13_8) (McDonald et al., 2012). OTU cluster was defined by a 97% identity threshold using Quantitative Insights Into Microbial Ecology (QIIME v1.9.1) (Caporaso et al., 2010). Venn diagram showing the unique and shared OTUs among all the sediment samples was performed with the VennDiagram package (Chen and Boutros, 2011). To present the distribution feature of fungal communities among different locations, a Bray-Curtis dissimilarity-based principal coordinates analysis (PCoA) was performed with the vegan package (Oksanen et al., 2016). Both Venn and PCoA were finished in the R software environment.
Results
A total of 6,104,436 sequences were obtained by ITS sequencing. 5,188,297 sequences were obtained after quality trimming. The saturated rarefaction curves of these samples indicated that the sequencing could meet the requirement of fungal diversity analysis (Supplementary Figure 2). Fungal diversity and richness estimator were used to compare the difference among the different sediment samples (Supplementary Table 1). A total of 1,272 OTUs were obtained with 97% similarity. The most OTU number was found at location D, followed by location A, locations B and C, particularly 2 m deep sediments exhibited higher fungal diversity than the 1 m deep and surface layer sediments.
The 1,272 OTUs were divided into 3 phyla including 1,048 Ascomycota, 103 Basidiomycota, 7 Zygomycota. Meanwhile, there were a total of 387 unidentified OTUs (Figures 1, 2). Ascomycota sequences were grouped into 8 known classes, i.e., Xylonomycetes, Taphrinomycetes, Sordariomycetes, Saccharomycetes, Pezizomycetes, Leotiomycetes, Eurotiomycetes, and Dothideomycetes, among which Dothideomycetes was the most abundant, especially at locations C and D, followed by Sordariomycetes at location A, and Eurotiomycetes at location B. Sequences matching Basidiomycota were grouped into 9 known classes, i.e. Wallemiomycetes, Ustilaginomycetes, Tremelicomycetes, Microbotryomycetes, Maiasseziomycetes, Exobasidiomycetes, Entorrhizomycetes, Cystobasidiomycetes, and Agaricomycetes, among which Tremellomycetes was dominant (Figure 1). Locations A and C have much more diverse Basidomycota fungi than B and D. Zygomycota including Mucoromycotina and Mortierellomycotina was the rare fungal component in all the sediments.
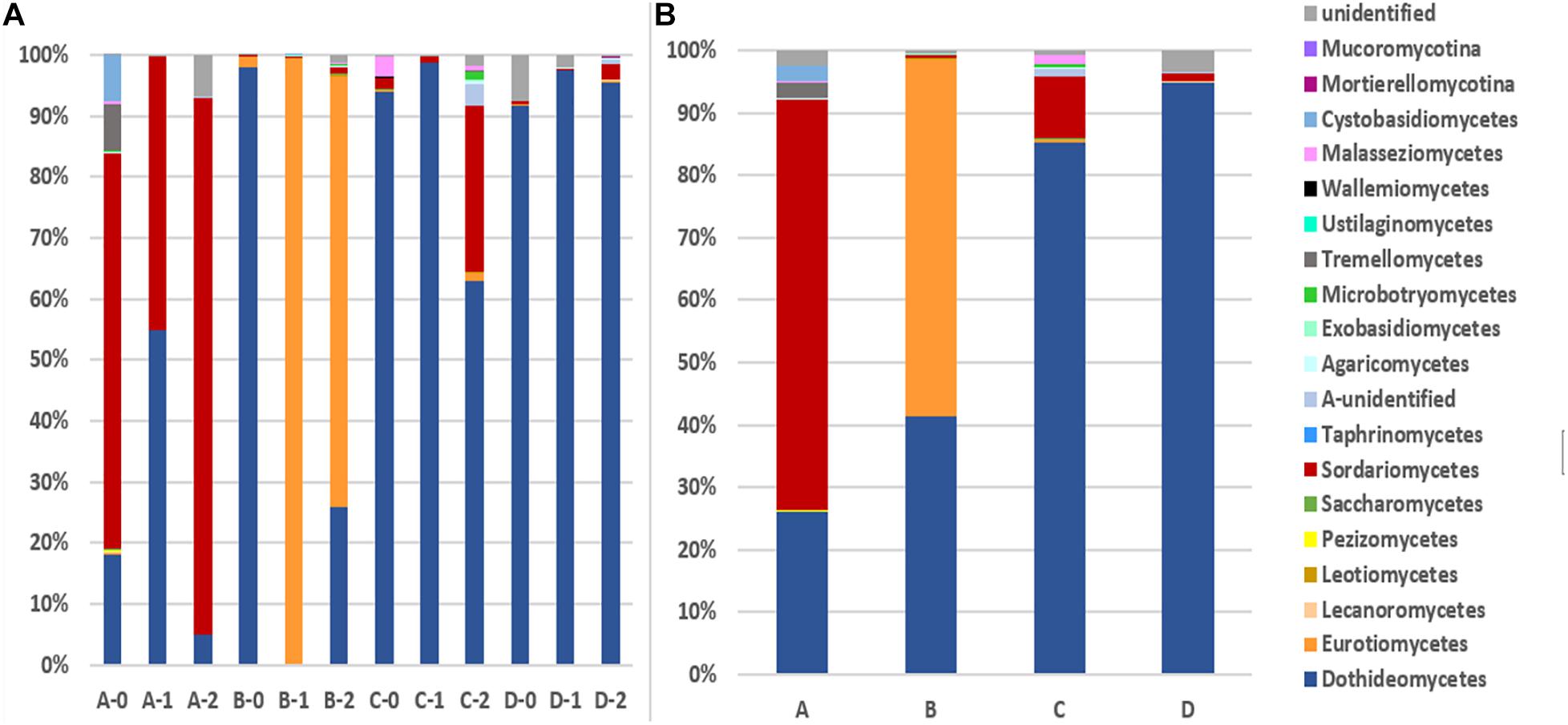
Figure 1. The taxonomic diversity of fungi at class level. (A): 12 depths at 4 locations (B): 4 locations. A, B, C, D: location; 0: 0.05 m depth sediment (surface); 1: 1 m depth sediment; 2: 2 m depth sediment.
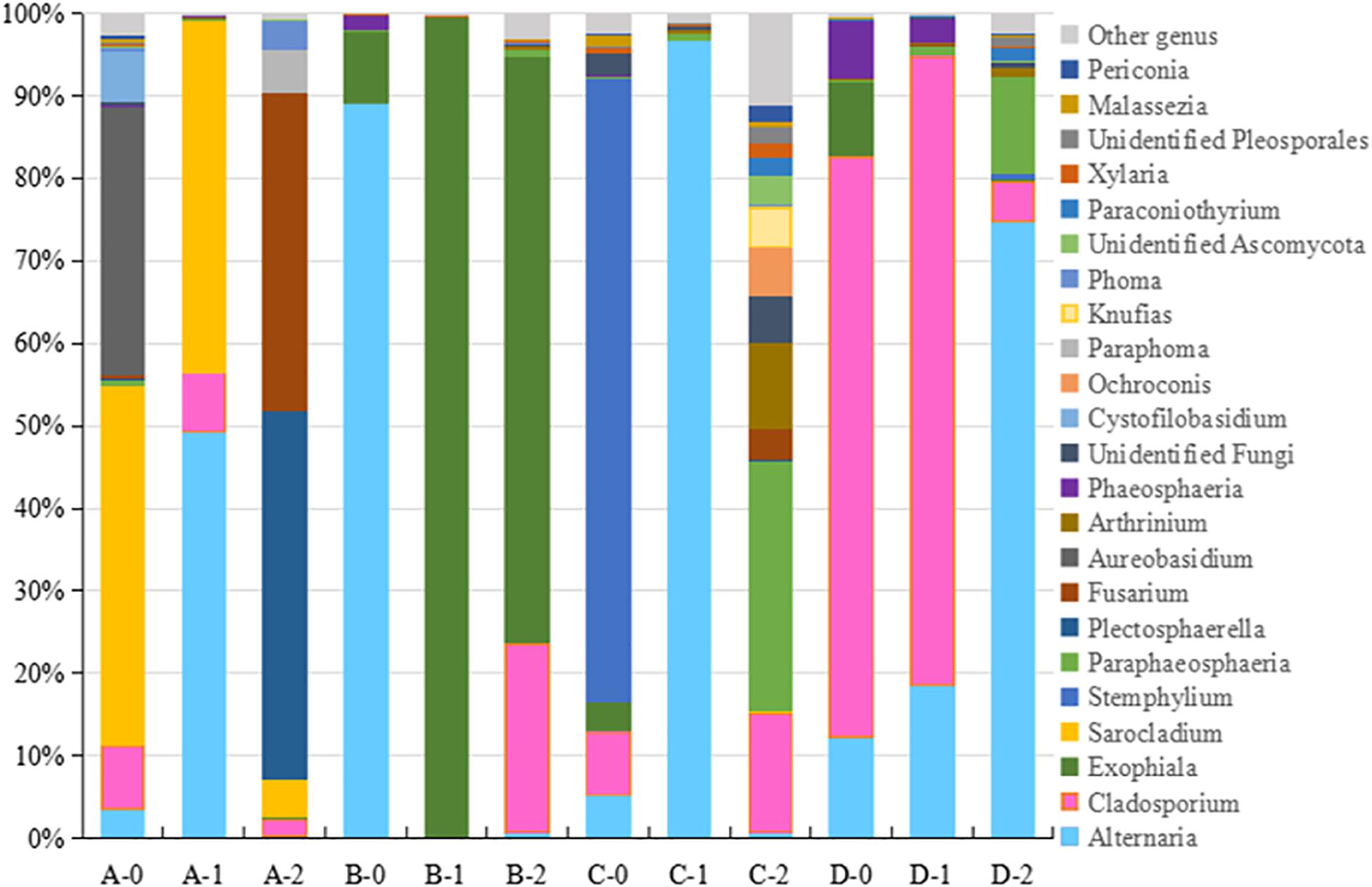
Figure 2. The taxonomic diversity of fungi at genus level 4 locations. A, B, C, D: location; 0: 0.05 m depth sediment (surface); 1: 1 m depth sediment; 2: 2 m depth sediment.
The detected fungal OTUs were grouped as 187 known genera (Supplementary Table 2), the dominant genera were Alternaria, Exophiala, Cladosporium, Plectosphaerella, Stemphylium, Sarocladium, and Fusarium (Figure 2). As shown in Figure 2, the sediments at different locations and depths show obvious different fungal community structures at genus level. For example Aureobasidium (A-0), Sarocladium (A-0, A-1 and A2) and Fusarium (A-2) at location A, Alternaria (B-0), Exophiala (B-1, B-2) at location B, Plectosphaerella (C-0), Alternaria (C-1), Paraphaeosphaeria and Fusarium (C-2) at location C, Cladosporium (D-0, D-1) at location D. Totally, B-1 B-2 sediments host different fungal diversity from others.
To gain further insights into the difference of fungal communities among the 4 sites and 3 depths, we applied Venn and PCoA analyses of the 1,272 OTUs to compare their space distribution difference (Figures 3, 4). Among locations B, C, and D, most OTUs were detected in the 2 m deep sediments (B-2, C-2, and D-2) (Figure 3), while most OTUs were detected in 1 m deep sediment A-1 at site A. Some OTUs were found to be specific with the location or the depth of the deep-sea sediments, while some OTUs were shared among different samples. But, totally, the number of the common OTUs is less than location or depth-specific OTUs number. According to Figure 4, 1 m and 2 m deep sediments host location-dependent fungal community, while the surface sediments have similar fungal diversity.
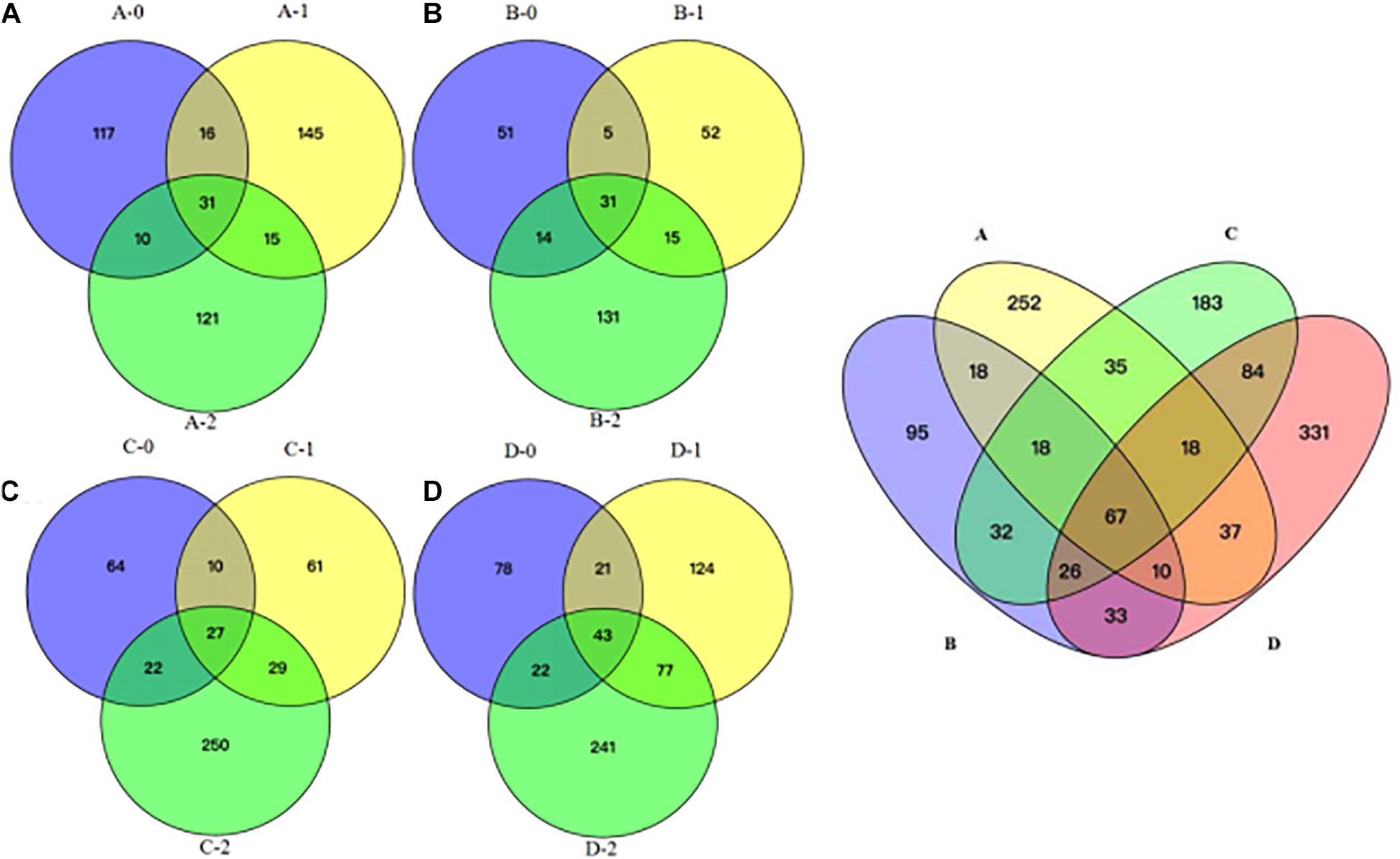
Figure 3. Venn diagram showing the unique and shared OTUs among all the sediment samples. (A) location A; (B) location B; (C) location C; (D) location D; A, B, C, D: location; 0: 0.05 m depth sediment (surface); 1: 1 m depth sediment; 2: 2 m depth sediment.
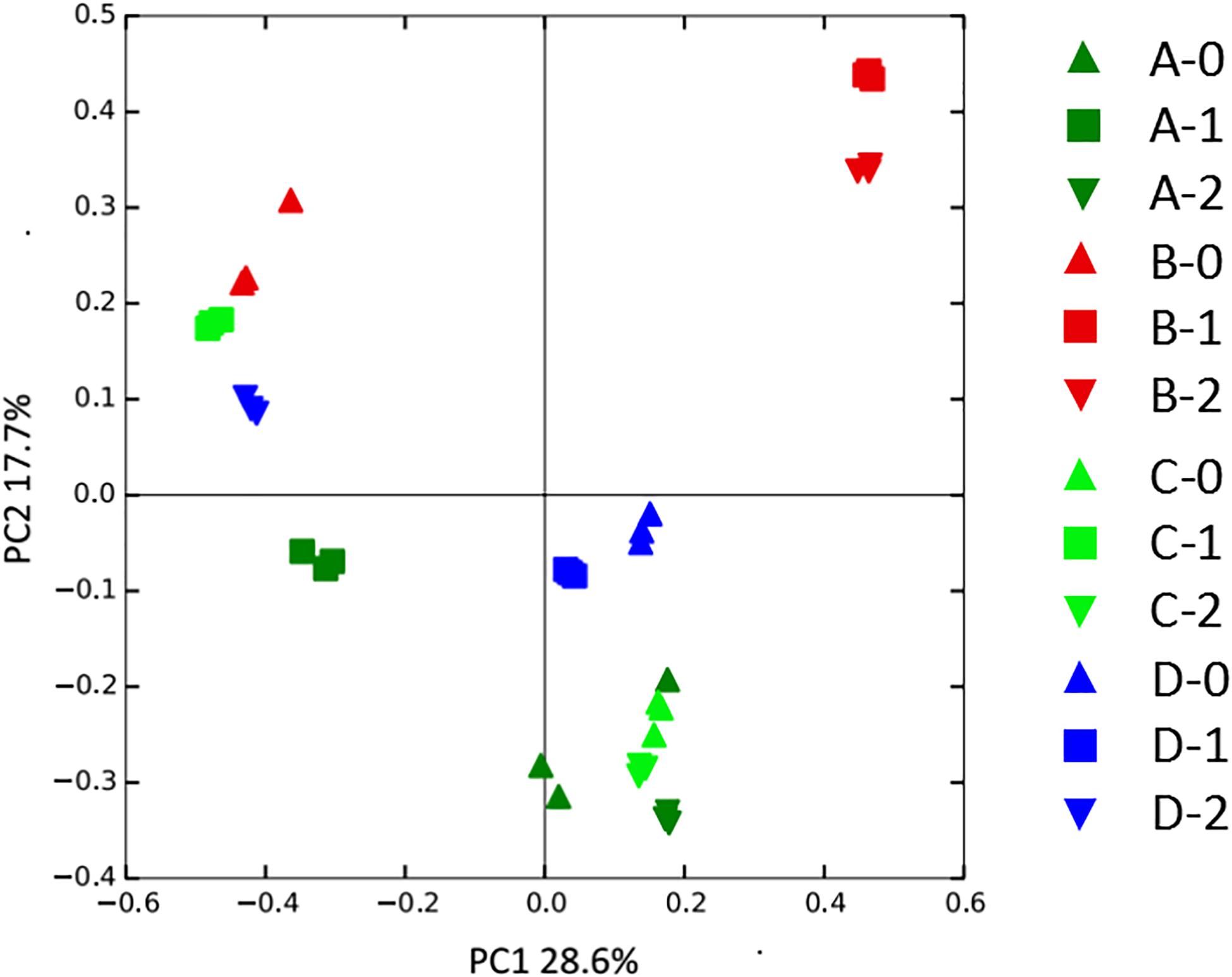
Figure 4. PCoA analysis of the fungal community variation among all sediment samples. A, B, C, D: location; 0: 0.05 m depth sediment (surface); 1: 1 m depth sediment; 2: 2 m depth sediment.
Discussion
Despite the extreme deep-sea environment, diverse fungi, e.g., 187 known genera, were detected in the deep-sea (897–1,434 m) sediments in this study. Some genera detected here have been reported in other deep-sea environments (Supplementary Table 3; Lai et al., 2007; Burgaud et al., 2009; Chen et al., 2011; Singh et al., 2011; Singh et al., 2012a,b; Li et al., 2013; Zhang et al., 2013; Rédou et al., 2014, 2015; Xu et al., 2014; Zhang et al., 2014a,b; Zhang et al., 2016), indicating their wide distribution in the deep-sea sediments.
Though some of the fungi are not marine obligate or generally grouped as terrestrial species, they probably have adapted to the deep-sea environments (Damare et al., 2006; Nagano et al., 2010; Rédou et al., 2014, 2015; Zhang et al., 2014a, 2015). For instance, antifreeze protein found in the yeast Rhodotorula mucilaginosa may play role in its adaptation to the deep-sea environments (Singh et al., 2012b; Rédou et al., 2015; Zhang et al., 2015). Penicillium-related and Hortaea-related phylotypes are haloduric (Khudyakova et al., 2000), and Cryptococcus-related phylotypes are cold tolerant (Orsi et al., 2013). Some fungi detected here maybe present in an inactive state e.g., spores (Lai et al., 2007; Rédou et al., 2015), because spores can survive at elevated hydrostatic pressure and low temperature i.e. deep-sea habitats.
The results of high-throughput sequencing show that most of the OTUs belong to Ascomycota (82.39%) followed by Basidomycota (8.10%). Ascomycota and Basidomycota may be the real dominant phyla in the deep-sea sediments, which is consistent with the results from the South China Sea sediments (Lai et al., 2007; Nagano and Nagahama, 2012; Zhang et al., 2013) and the Mediterranean Sea (Barone et al., 2018). Similar with the results from Barone et al. (2018), unknown fungi amount to ca. 8.96% in the deep-sea sediments of the South China Sea, suggesting that deep-sea sediments harbor novel fungal lineages. We have used the same sediment samples in this study to isolate fungi, only Ascomycota and Basidomycota fungi e.g., Cladosporium sp., Alternaria sp., Rhodotorula sp., Exophiala sp., Aspergillus sp., Stemphylium sp., Penicillium sp., and Sarocladium sp. were isolated successfully (Feng et al., 2017). So far, there is no report on the successful isolation of fungi belonging to other phylum e.g., Zygomycota from the deep-sea sediments. But Zygomycota which account to 0.55% of the OTUs was detected in this study, suggesting the necessity to seek new ways to isolate novel deep-sea fungi which is difficult to cultivate.
Marine fungi have been suggested to play roles in the nitrogen cycling, e.g., Aspergillus, Fusarium are probably involved in the denitrification of the deep sea (Cathrine and Raghukumar, 2009). Another emphasis of deep-sea fungal function is on the secondary metabolic products and their biological activities such as antimicrobial, antifouling, cytotoxic activities (Khudyakova et al., 2000; Wang et al., 2012; Li et al., 2013; Sun et al., 2013; Zhang et al., 2014b; Feng et al., 2017). On the whole, compared with the knowledge of fungal diversity, our understanding of the deep-sea fungal function is very limited. Thus, fungal function presents a frontier hot of deep-sea fungal research.
In summary, diverse fungi including some unknown species were detected in the deep-sea sediments in the South China Sea using high-throughput sequencing in this study. The results suggested that the sediment fungal community structures were not uniform across the different locations and depths. The fungal abundance and diversity were found to increase with the depth of the sediments from 0 to 2 m. Though the surface sediments exhibited similar fungal diversity, location and depth dependent fungal community structures were detected in 1 and 2 m deep sediments, which might be related to the various environments of the different deep-sea sediments. Barone et al. (2018) found that fungal biomass, abundance and diversity increased with increasing concentrations of carbohydrates. Thus, the detailed environment factors will provide more evidence for the location-dependent fungal communities in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
LF and ZL conceived and designed the study. LF, QS, and QJ performed data analyses. LF and ZL drafted the manuscript. All authors approved the final manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (2018YFC0309805, 2019YFC0312501, and 2018YFA0901901).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks for Fengping Wang, SJTU, to provide the deep-sea sediment samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.592784/full#supplementary-material
Supplementary Figure 1 | The sampling locations of deep-sea sediments in the South China Sea.
Supplementary Figure 2 | Rarefaction curves of fungal OTUs number. A, B, C, D: location; 0: 0.05 m depth sediment (surface); 1: 1 m depth sediment; 2: 2 m depth sediment.
Supplementary Table 1 | The comparison of fungal diversity and richness estimator among different deep-sea sediments.
Supplementary Table 2 | Fungal genera revealed by ITS high-throughput sequencing.
Supplementary Table 3 | Fungal genera detected in deep-sea environments.
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Barone, G., Rastellib, E., Corinaldesic, C., Tangherlinib, M., Danovaro, R., and Antonio Dell’Anno, A. (2018). Benthic deep-sea fungi in submarine canyons of the Mediterranean Sea. Progr. Oceanogr. 168, 57–64. doi: 10.1016/j.pocean.2018.09.011
Bass, D., Howe, A., Brown, N., Barton, H., Demidova, M., Michelle, H., et al. (2007). Yeast forms dominate fungal diversity in the deep oceans. Proc. R. Soc. B. 274, 3069–3077. doi: 10.1098/rspb.2007.1067
Burgaud, G., Le Calvez, T., Arzur, D., Vandenkoornhuyse, P., and Barbier, G. (2009). Diversity of culturable marine filamentous fungi from deep−sea hydrothermal vents. Environ. Microbiol. 11, 1588–1600. doi: 10.1111/j.1462-2920.2009.01886.x
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, D. F., Costello, K. E., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Cathrine, S. J., and Raghukumar, C. (2009). Anaerobic denitrification in fungi from the coastal marine sediments off Goa. India Mycol. Res. 113, 100–109. doi: 10.1016/j.mycres.2008.08.009
Chen, H., and Boutros, C. P. (2011). Venn Diagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformat. 12, 1–7. doi: 10.1186/1471-2105-12-35
Chen, Z., Huang, H., Chen, Y., Wang, Z., Ma, J., Wang, B., et al. (2011). New cytochalasins from the marine−derived fungus Xylaria sp. SCSIO 156. Helvetica Chim. Acta 94, 1671–1676. doi: 10.1002/hlca.201100051
Damare, S., Raghukumar, C., and Raghukumar, S. (2006). Fungi in deep-sea sediments of the Central Indian Basin. Deep Sea Res. Part I Oceanogr. Res. Papers 53, 14–27. doi: 10.1016/j.dsr.2005.09.005
Feng, L., Jiang, Q., Sun, W., Li, Y., Zhang, F., and Li, Z. (2017). Diversity and antimicrobial activity of fungi isolated from deep-sea sediments in South China Sea. Acta Microbiol. Sin. 57, 1332–1341.
Gweon, H. S., Oliver, A., Taylor, J., Booth, T., Gibbs, M., Read, D. S., et al. (2015). PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol. Evol. 6, 973–980. doi: 10.1111/2041-210x.12399
Ihrmark, K., Bodeker, I. T., Cruzmartinez, K., Friberg, H., Kubartova, A., Schenck, J., et al. (2012). New primers to amplify the fungal ITS2 region–evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 82, 666–677. doi: 10.1111/j.1574-6941.2012.01437.x
Khudyakova, Y. V., Pivkin, M. V., Kuznetsova, T. A., and Svetashev, V. I. (2000). Fungi in sediments of the sea of Japan and their biologically active metabolites. Microbiology 69, 608–611. doi: 10.1007/bf02756817
Lai, X., Cao, L., Tan, H., Fang, S., Huang, Y., and Zhou, S. (2007). Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 1, 756–762. doi: 10.1038/ismej.2007.51
Li, C., Li, X., Gao, S., Lu, Y., and Wang, B. (2013). Cytotoxic anthranilic acid derivatives from deep sea sediment-derived fungus Penicillium paneum SD-44. Mar. Drugs 11, 3068–3076. doi: 10.3390/md11083068
Lindahl, B. D., Nilsson, R. H., Tedersoo, L., Abarenkov, K., Carlsen, T., Kjoller, R., et al. (2013). Fungal community analysis by high−throughput sequencing of amplified markers–a user’s guide. N. Phytol. 199, 288–299. doi: 10.1111/nph.12243
McDonald, D., Price, N. M., Goodrich, J., Nawrocki, P. E., DeSantis, Z. T., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Morgado, L. N., Semenova, T. A., Welker, J. M., Walker, M. D., Smets, E., and Geml, J. (2015). Summer temperature increase has distinct effects on the ectomycorrhizal fungal communities of moist tussock and dry tundra in Arctic Alaska. Global. Change Biol. 21, 959–972. doi: 10.1111/gcb.12716
Nagahama, T., Takahashi, E., Nagano, Y., Abdelwahab, M. A., and Miyazaki, M. (2011). Molecular evidence that deep-branching fungi are major fungal components in deep-sea methane cold-seep sediments. Environ. Microbiol. 13, 2359–2370. doi: 10.1111/j.1462-2920.2011.02507.x
Nagano, Y., and Nagahama, T. (2012). Fungal diversity in deep-sea extreme environments. Fungal Ecol. 5, 463–471. doi: 10.1016/j.funeco.2012.01.004
Nagano, Y., Nagahama, T., Hatada, Y., Nunoura, T., Takami, H., Miyazaki, J., et al. (2010). Fungal diversity in deep-sea sediments–the presence of novel fungal groups. Fungal Ecol. 3, 316–325. doi: 10.1016/j.funeco.2010.01.002
Oksanen, J., Blanchet, F. G., Friendly, M., and Kindt, R. (2016). Vegan: Community Ecology Package. R Package Version 2.3– 5. Available at. http://CRAN.R-project.org/package=vegan (accessed April 9, 2016).
Orsi, W., Biddle, J. F., and Edgcomb, V. (2013). Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS One 8:e56335. doi: 10.1371/journal.pone.0056335
Rédou, V., Ciobanu, M. C., Pachiadaki, M. G., Edgcomb, V. P., Alain, K., Barbier, G., et al. (2014). In-depth analyses of deep subsurface sediments using 454-pyrosequencing reveals a reservoir of buried fungal communities at record-breaking depths. FEMS Microbiol. Ecol. 90, 908–921. doi: 10.1111/1574-6941.12447
Rédou, V., Navarri, M., Mesletcladiere, L., Barbier, G., and Burgaud, G. (2015). Marine fungi from deep subseafloor sediments: species richness and adaptation. Appl. Environ. Microbiol. 81, 3571–3583. doi: 10.1128/aem.04064-14
Roth, F. J. Jr., Orpurt, P. A., and Ahearn, D. G. (1964). Occurrence and distribution of fungi in a subtropical marine environment. Can. J. Bot. 42, 375–383. doi: 10.1139/b64-037
Singh, P., Raghukumar, C., Meena, R. M., Verma, P., and Shouche, Y. S. (2012a). Fungal diversity in deep-sea sediments revealed by culture-dependent and culture-independent approaches. Fungal Ecol. 5, 543–553. doi: 10.1016/j.funeco.2012.01.001
Singh, P., Raghukumar, C., Verma, P., and Shouche, Y. S. (2012b). Assessment of fungal diversity in deep-sea sediments by multiple primer approach. World J. Microbiol. Biotechnol. 28, 659–667. doi: 10.1007/s11274-011-0859-3
Singh, P., Raghukumar, C., Verma, P., and Shouche, Y. S. (2010). Phylogenetic diversity of culturable fungi from the deep-sea sediments of the Central Indian Basin and their growth characteristics. Fungal Div. 40, 89–102. doi: 10.1007/s13225-009-0009-5
Singh, P., Raghukumar, C., Verma, P., and Shouche, Y. S. (2011). Fungal community analysis in the deep-sea sediments of the Central Indian Basin by culture-independent approach. Microbial. Ecol. 61, 507–517. doi: 10.1007/s00248-010-9765-8
Sun, L., Li, D., Tao, M., Chen, Y., Zhang, Q., Dan, F., et al. (2013). Two new polyketides from a marine sediment-derived fungus Eutypella scoparia FS26. Nat. Product. Res. 27, 1298–1304.
Thaler, A. D., Van Dover, C. L., and Vilgalys, R. (2012). Ascomycete phylotypes recovered from a Gulf of Mexico methane seep are identical to an uncultured deep-sea fungal clade from the Pacific. Fungal Ecol. 5, 270–273. doi: 10.1016/j.funeco.2011.07.002
Wang, F., Huang, Z., Shi, X., Chen, Y., Zhang, W., Tian, X., et al. (2012). Cytotoxic indole diketopiperazines from the deep sea-derived fungus Acrostalagmus luteoalbus SCSIO F457. Bioorg. Med. Chem. Lett. 22, 7265–7267. doi: 10.1016/j.bmcl.2012.08.115
Xu, W., Pang, K. L., and Luo, Z. H. (2014). High fungal diversity and abundance recovered in the deep-sea sediments of the Pacific Ocean. Microbial. Ecol. 68, 688–698. doi: 10.1007/s00248-014-0448-8
Zhang, T., Wang, N. F., Zhang, Y. Q., Liu, H. Y., and Yu, L. (2015). Diversity and distribution of fungal communities in the marine sediments of Kongsfjorden, Svalbard (High Arctic). Scient. Rep. 5:14524.
Zhang, X., Tang, G., Xu, X., Nong, X., and Qi, S. (2014a). Insights into deep-sea sediment fungal communities from the East Indian Ocean using targeted environmental sequencing combined with traditional cultivation. PLoS One 9:e109118. doi: 10.1371/journal.pone.0109118
Zhang, X., Xu, X., Peng, J., Ma, C., Nong, X., Bao, J., et al. (2014b). Antifouling potentials of eight deep-sea-derived fungi from the South China Sea. J. Industr. Microbiol. Biotechnol. 41, 741–748. doi: 10.1007/s10295-014-1412-9
Zhang, X., Wang, G., Xu, X., Nong, X., Wang, J., Amin, M., et al. (2016). Exploring fungal diversity in deep-sea sediments from Okinawa Trough using high-throughput Illumina sequencing. Deep Sea Res. Part I Oceanogr. Res. Papers 116, 99–105. doi: 10.1016/j.dsr.2016.08.004
Keywords: fungal community, high-throughput sequencing, space distribution, deep sea, sediment
Citation: Feng L, Song Q, Jiang Q and Li Z (2021) The Horizontal and Vertical Distribution of Deep-Sea Sediments Fungal Community in the South China Sea. Front. Mar. Sci. 8:592784. doi: 10.3389/fmars.2021.592784
Received: 08 August 2020; Accepted: 07 January 2021;
Published: 25 January 2021.
Edited by:
Cinzia Corinaldesi, Marche Polytechnic University, ItalyReviewed by:
Samir R. Damare, National Institute of Oceanography (CSIR), IndiaMichael Tangherlini, University of Naples Federico II, Italy
Copyright © 2021 Feng, Song, Jiang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyong Li, zyli@sjtu.edu.cn
 Li Feng
Li Feng  Zhiyong Li
Zhiyong Li