Microscopic and Genetic Characterization of Bacterial Symbionts With Bioluminescent Potential in Pyrosoma atlanticum
- 1Guy Harvey Oceanographic Center, Halmos College of Arts and Sciences, Nova Southeastern University, Dania Beach, FL, United States
- 2Center for Advanced Microscopy, College of Arts and Sciences, University of Miami, Coral Gables, FL, United States
The pelagic tunicate pyrosome, Pyrosoma atlanticum, is known for its brilliant bioluminescence, but the mechanism causing this bioluminescence has not been fully characterized. This study identifies the bacterial bioluminescent symbionts of P. atlanticum collected in the northern Gulf of Mexico using several methods such as light and electron microscopy, as well as molecular genetics. The bacteria are localized within the pyrosome light organs. Greater than 50% of the bacterial taxa present in the tunicate samples were the bioluminescent symbiotic bacteria Vibrionaceae as determined by utilizing current molecular genetics methodologies. A total of 396K MiSeq16S rRNA reads provided total pyrosome microbiome profiles to determine bacterial symbiont taxonomy. After comparing with the Silva rRNA database, a Photobacterium sp. r33-like bacterium (which we refer to as “Photobacterium Pa-1”) matched at 99% sequence identity as the most abundant bacteria within Pyrosoma atlanticum samples. Specifically designed 16S rRNA V4 probes for fluorescence in situ hybridization (FISH) verified the Photobacterium Pa-1 location as internally concentrated along the periphery of each dual pyrosome luminous organ. While searching for bacterial lux genes in two tunicate samples, we also serendipitously generated a draft tunicate mitochondrial genome that can be used for Pyrosoma atlanticum identification. Scanning (SEM) and transmission (TEM) electron microscopy confirmed the presence of intracellular rod-like bacteria in the light organs. This intracellular localization of bacteria may represent bacteriocyte formation reminiscent of other invertebrates.
Introduction
Bioluminescence is an important adaptive trait in ocean dwelling taxa that appears to be more prevalent than originally thought (Martini and Haddock, 2017). Over 700 animal genera are known to include luminous species, with more than 80% being marine organisms (Widder, 2010). Within this group, 90% of pelagic organisms that live between 200 and 1000 m are known to have bioluminescent capabilities. In addition, fishes, squid, and shrimp are able to modify aspects of their light production, such as the intensity, kinetics, wavelength, and angular distribution. The emission of light by organisms has evolved independently over 40 times in marine and terrestrial organisms (Haddock et al., 2010). This emphasizes the evolutionary importance of the bioluminescence mechanism. There are several critical ways bioluminescence can aid an organism’s survival, such as facilitating food location and capture, attracting mates, allowing for species recognition, and functioning as a defense mechanism (Widder, 2010).
Pyrosomes derive their name from the Greek words pyro (“fire”) and soma (“body”) from the “fiery” bioluminescence that is so visible at night (Sutherland et al., 2018). Pyrosomes were classified by Lamarck and Huxley under the subphylum Tunicata (previously known as Urochordata) due to the zooids, which compose these organisms, being encased by a tunic (Huxley, 1851; Lemaire and Piette, 2015). Pyrosomes are approximately 95% water and are extremely well adapted for rapid growth and efficient energy use. Transparency makes pyrosomes difficult to see at any depth, which is why they can be found throughout the pelagic realm. Aside from being transparent and of limited nutritional value, pyrosomes have few sensory or predator-avoidance adaptations. Most biological processes, such as feeding, respiration, and swimming occur simultaneously through contraction of the same muscles (Alldredge and Madin, 1982).
Pyrosomes, in the class Thaliacea, remain one of the least studied planktonic grazers, despite their widespread distribution and ecological significance. Pyrosomes are characterized as highly successful planktonic grazers, and swarms of colonies can consume substantial amounts of phytoplankton (Alldredge and Madin, 1982; Décima et al., 2019). They have been noted for their potential to restructure the food web when aggregating in large quantities (Sutherland et al., 2018). The species examined in our study, Pyrosoma atlanticum, have been observed since the 1840s worldwide, and can be found in tropical and temperate waters ranging from 45°N to 45°S (Sutherland et al., 2018).
Bioluminescence in planktonic colonial tunicates is not as common as in other pelagic organisms (Haddock et al., 2010). The most well-known example of colonial tunicate bioluminescence is the pyrosome. However, two other urochordate groups have recently been found to have luminous members: a deep-sea doliolid (Robison et al., 2005) and a shallow living benthic ascidian, Clavelina miniate (Aoki et al., 1989; Chiba et al., 1998; Hirose, 2009). The bioluminescence mechanisms are not well understood in these tunicates, but Appendicularians, another class of tunicates, secrete luminous inclusions or use a coelenterazine + luciferase system (Galt and Sykes, 1983; Galt and Flood, 1998). It has been proposed that pyrosome bioluminescence is unique compared to other pelagic organisms, such as fishes and ctenophores, as it is in the form of a steady glow and can be induced by light illumination (Mackie and Bone, 1978). The propagation of bioluminescence throughout the colony does not appear to be neural, as there is no evidence for innervation or epithelial conduction pathways (Mackie and Bone, 1978; Bowlby et al., 1990). Several authors have hypothesized that the bioluminescence is bacterial in nature, based on the presence of bacteria-like particles (Mackie and Bone, 1978; Haygood, 1993), but this has never been verified. It has also been hypothesized that symbionts in the dual light organs of each Pyrosoma atlanticum zooid are intracellular and thus will likely have undergone some metabolic integration with host cells (Mackie and Bone, 1978; Holland, 2016). Mackie and Bone based this statement on the fact that luminous bacteria glow continuously, and fishes that utilize luminous bacterial symbionts possess shutters to block the light. Pyrosoma appears able to directly control the light emissions from their putative bacterial symbionts by an as yet unidentified mechanism, but very likely involving the supply of an essential metabolite. However, since these symbionts have not been successfully isolated and cultivated, their role in pyrosome bioluminescence remains unconfirmed. The goal of the present study is to determine if the bioluminescence of Pyrosoma atlanticum, is from bacterial origin by identifying the location, anatomy, and classification of its symbiotic bacteria.
Materials and Methods
Sample Collection and Fixation
Tunicate samples were collected on cruises conducted by the Deep Pelagic Nekton Dynamics of the Gulf of Mexico (DEEPEND)1 consortium (Sutton et al., 2020). In 2017, midwater trawling was conducted on DEEPEND Cruise DP05, during which fishes, crustaceans, cephalopods, large gelatinous zooplankton, and other pelagic taxa were collected from the Gulf of Mexico, including Pyrosoma atlanticum. On shipboard, specimens were frozen or stored in dimethyl sulfoxide (DMSO), then transferred and stored in the Microbiology and Genetics Laboratory at Nova Southeastern University. Twenty-nine additional samples were collected from the Gulf of Mexico on the July 2018 DEEPEND Cruise DP06 and stored in DMSO or RNALater. Samples from both cruises were collected from depths of 0 to 1,500 m at multiple collection sites. Of the samples stored in RNALater, two samples were utilized for genetic sequencing (Table 1). In 2019, 12 additional P. atlanticum samples were collected in the Gulf of Mexico during a NOAA Ocean Exploration and Research-funded cruise aboard the R/V Point Sur (Supplementary Figure 1). Water samples used in comparative analysis were collected from the DEEPEND cruise DP05 using Niskin bottles deployed on a CTD (Baker et al., 2019; Easson and Lopez, 2019; Freed et al., 2019). These samples were taken at the same time and location as the DMSO samples were collected. Water samples were not collected for both2019 cruises for this study.
Light and Fluorescence Microscopy (Histology)
Samples were fixed for light microscopy in 2% glutaraldehyde with a sodium cacodylate-buffered seawater fixative, washed with a buffer rinse, placed in 70% EtOH overnight, processed through a graded series of ethanols, cleared, and infiltrated with molten Paraplast Plus®, and embedded in Paraplast Xtra®. Using a Leica RM 2125 microtome, 4 μm thick sections were cut, mounted on microscope slides, and stained with Harris’s hematoxylin and eosin. Slides were examined using an Olympus BX43 light microscope at 4–60 × magnification. Fluorescence microscopy was performed on an Olympus IX70 Fluorescence Microscope with green (500–570 nm) and red (610 ∼750 nm) filter cubes. Bacteriocytes were counted on light micrographs of the histology sections. We define bacteriocytes as membrane bound structures that surround more than one individual bacterium (Sen et al., 2018). A structure was considered a bacteriocyte if it was a dark cluster of multiple bacteria about 5 μm in size with a discernable membrane within the interior of the light organ. Additional estimation of the quantity of bacteria within the light organ was determined on TEM and SEM micrographs.
Scanning and Transmission Electron Microscopy
Samples utilized for SEM were stored in a 2% glutaraldehyde with a sodium cacodylate-buffered seawater fixative. Pyrosomes were dissected in the fixative and divided into three sections per sample. They were rinsed three times in sodium cacodylate-buffered seawater, postfixed in 1% osmium tetroxide, rinsed in the buffer, dehydrated through a graded series of ethanol (20, 50, 70, 95, and 100%), and dried in hexamethyldisilazane (HMDS). Dried samples were outgassed overnight, coated with palladium in a sputter coater, and examined in a Philips XL-30 Field Emission SEM at the University of Miami Center for Microscopy (UMCAM) located in the Chemistry Department at the University of Miami Coral Gables Campus.
TEM samples were prepared similarly as for SEM, except that samples at the last dehydration step (100% EtOH) were embedded in Spurr resin and polymerized for 3 days at 60°C. Blocks were trimmed, sectioned, sections were floated onto grids, stained with either Uranyl Acetate and/or Lead Citrate and examined in a JEOL 1,400X TEM located at the University of Miami Miller School of Medicine TEM Core Lab. Semi-thin sections of TEM prepared samples were examined in an IX-70 fluorescent microscope to examine gross structures.
DNA Extraction and Polymerase Chain Reaction (PCR)
Total microbiome DNAs were extracted with the standard protocol for the Qiagen PowerLyzer PowerSoil kit. Whole body slices of the pyrosome with at least two or three zooids included in the section were used for extraction. This study focused on amplifying the 16S gene in order to identify luminescent bacteria the pyrosome. Once DNA extractions were completed, polymerase chain reactions (PCR) were run using Invitrogen Platinum Hot Start PCR Master Mix (2×) and the universal primers 515F and 806R. Each sample was given a unique 806R primer in order to identify and separate the samples during the post run analysis. The 515F and 806R primers were used to amplify the 200 bp sequence of the V3 and V4 region of the 16S gene (Caporaso et al., 2011; Easson and Lopez, 2019). The V3 and V4 region are two of nine hypervariable regions (V1-V9) found within the 16S gene (Gray et al., 1984). The V4 region provides the lowest error rates for identification (Wang et al., 2007). The PCR products were cleaned using AMPure XP beads. This process was used to purify the 16S V3 and V4 amplicon away from free primers and primer dimer species (Chakravorty et al., 2007). The final DNA concentration was checked using a Qubit 2.0 fluorometer (Life Technologies).
Illumina High-Throughput 16S Sequencing and Analysis
The 16S rRNA gene fragment was the target for bacterial identification via high throughput DNA sequencing (O’Connell et al., 2018; Easson and Lopez, 2019). Samples were prepared for sequencing following the 16S Illumina Amplicon Protocol per the Earth Microbiome Project (Kuczynski et al., 2011; Thompson et al., 2017). The final DNA concentrations from the PCR extractions were used to dilute each sample to a normalization of 4 pM. All DNA samples were library pooled and rechecked on the Qubit 2.0 fluorometer to make sure the concentration was between 4 and 6 ng/μL. A final quality check was done using an Agilent Bio analyzer Tapestation 2000, which reads the quality of the template DNA and identifies any possible contamination. The final product was loaded into an Illumina MiSeq system for 16S metagenomics DNA at 500 cycles using V2 chemistry. The sequencing followed a modified Illumina workflow protocol.
The Quantitative Insights into Microbial Ecology v.2 (QIIME2) pipeline was used to demultiplex, quality filter, assign taxonomy, reconstruct phylogeny, and produce diversity analysis and visualizations from the FASTQ DNA sequence files (Caporaso et al., 2010). The quality filtering and trimming of the data was conducted in DADA2, which was used to create a feature table that was analyzed in RStudio. The QIIME2-generated sequences were assigned taxonomy through a learned SILVA classifier (silva-132-99- 515-806-nb-classifier.qza). This feature table was used for SIMPER statistical analysis through the “vegan” package (Oksanen et al., 2013) in RStudio to determine which taxa are driving the differences in the water and pyrosome microbial assemblages (Rees et al., 2004). Additional comparisons were made in CosmosID®, a bioinformatic pipeline used for microbial analysis that employs a phylogenetic and k-mer based approach to metagenomics. FASTQ files were uploaded to CosmosID®, which provided relative abundance of the microbial community described in a heatmap comparison. Further data analysis used 16S rRNA multiple sequence alignments with MAFFT (Katoh and Standley, 2013), in order to generate a phylogeny to compare the extracted 16S sequence from the MiSeq run with known luminescent bacterial species. Pyrosome microbiome sequences have been deposited to the NCBI Sequence Read Archive (PRJNA636187).
Fluorescence in situ Hybridization (FISH)
Pyrosoma atlanticum samples were stored in paraformaldehyde and dehydrated through an ethanol series, cleared in xylenes and infiltrated with paraffin. Serial sections were cut at 4 and 8 μm, mounted on slides, and deparaffinized with xylene and ethanol series (100–70%). Specific probes were then added to localize the bacteria within the light organs. Optimal probes sequences were designed using MAFFT alignment (Katoh and Standley, 2013) of the new tunicate 16S rRNA sequences obtained from our Illumina MiSeq run, with previously determined 16S sequences from various bacterial species from NCBI database (DQ889917, DQ889916, DQ889915, DQ889914, DQ889913).
The 16S rRNA sequences from samples previously mentioned were aligned using MAFFT. The sequence we had generated, TTCAGGTGTAGCGGTGAAATGC, was chosen because it appeared to match closest to Photobacterium sequences and was also the most variable part of the V4 region alignment. This also indicated that it would be unique with less similarity to other sequences in the fixed tissues. The high specificity was required to specifically detect the Photobacterium in FISH prepared slides. The probes were then tested on the NCBI PROBE Database2 and Microbial Ribosomal Databases Probe Match3 (Negandhi et al., 2010).
Labeled probes for FISH were manufactured by IDT Inc. (Iowa, United States). The dye used for the Photobacterium-specific probe was Cy3, which is a standard orange-fluorescent label for nucleic acids and was attached at the 5′ end (Table 2). The control probe EUB338 is a universal bacteria probe and was dyed with 6-FAM (fluorescein) (Negandhi et al., 2010). FAM is the most commonly used fluorescent dye attachment for oligonucleotides and this particular dye was attached at the 3′ end and appears green. The probes attach to one end or the other to allow for overlap. This is possible because the two probes’ nucleotide sequences are at different location on the ribosome (either the 5′ or 3′ end). When imaging the samples, only orange and green fluorescence should appear, and red fluorescence should be excluded due to double binding. This means both probes should bind to the targeted Photobacterium sp. and present the orange fluorescence while the rest of the bacteria appear green.
FISH protocols utilized here followed previously described methods (Sharp et al., 2007; Negandhi et al., 2010). FISH hybridization buffer (35% formamide) contained 5 M NaCl, 40 μl 1 M Tris-HCl, 700 μl formamide, 900 μl H2O, and 2 μl 10% SDS, and applied as 45 μl mixed with 5 ng/μl of the desired probe, for a total of 50 μl per slide. Pyrosome tissues were then incubated inside a humidity chamber with a paper towel that was saturated with the hybridization buffer for 2 h at 46°C. After hybridization, slides were put in a buffer wash (700 μl 5 M NaCl, 1 ml 1 M Tris-HCl, 500 μl 0.5 EDTA, 50 ml H2O, and 50 μl 10% SDS) for 20 min at 48°C, then rinsed with dH2O and air dried.
FISH was performed on three samples with two sections each. The control runs utilized probe EUB338. In addition to the control, a slide with no probes as well as slides with both EUB338 and Photobacterium probes were hybridized. This allowed for an autofluorescence assessment and aided in eliminating background noise. Slides were examined using an Olympus IX70 Fluorescence Microscope with green (500–570 nm) and red (610 ∼750 nm) filter cubes.
Results
Structure and Morphology of the Light Organ: Light and Fluorescence Microscopy
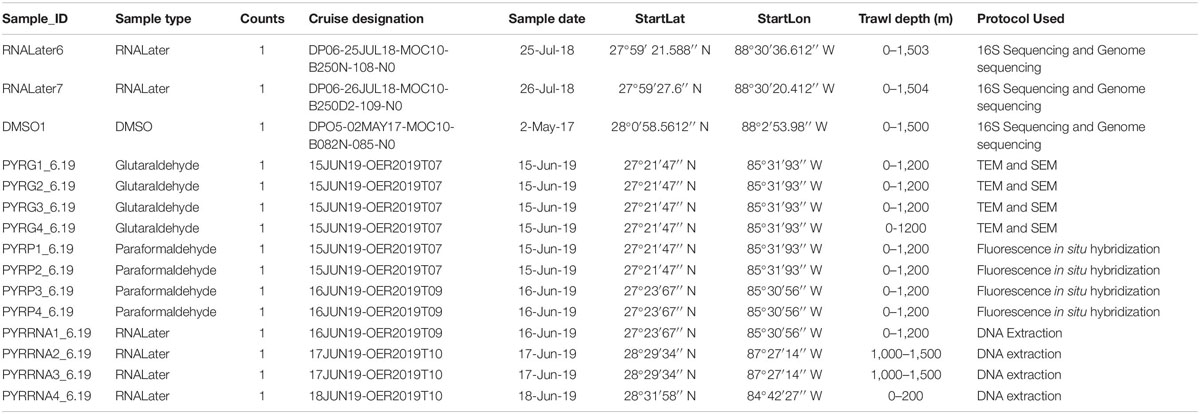
Two luminous light organs per Pyrosoma atlanticum zooid along with potential symbiotic microbes were identified with light and fluorescent microscopy. The bacteria within each light organ were well resolved and clearly identifiable on each side of the buccal siphon (Figure 1A). The left and right light organs (30–50 μm in diameter) were oval shaped structures, usually fully intact with each exhibiting a nodule capping the end (Figure 1A). Within the light organ, a clear, possibly open space in the center appeared surrounded by bacteria-filled cells delineating the boundaries of each light organ. At higher magnification, the bacteria clustered in the light organ were found to be bacteriocytes (Figure 1B). As many as 72 bacteriocytes were identified within a single light organ. This value was calculated by counting the number of dark structures within the light organ.
Figure 1. Orientation of both light organs on either side of the buccal siphon (A). Higher magnification of one individual light organ (LO) with bacteria (or more likely bacteriocytes) (B).
Sequencing of P. atlanticum Microbiomes and Partial Mitochondrial Genome
Microbiomes based on 16s rRNA amplicons were sequenced from three different Pyrosoma atlanticum individuals and compared to 10 seawater samples. The sampling design provided a background environmental microbial profile for comparison with the pyrosome microbiomes. Each pyrosome sample was matched to a previously sequenced seawater microbiome collected at two sites and depths of 1,500 m, corresponding to the pyrosome collections. The seawater samples were previously sequenced as a part of another DEEPEND consortium project (Easson and Lopez, 2019). A total of 396K MiSeq reads and 497 Amplicon Sequence Variant (ASV) were produced. In all three pyrosome samples, the most abundant 16S rRNA sequences matched Photobacterium sp. r33 in the pyrosome microbiomes (Table 3). In order to confirm the identity of the symbiont, the sequence derived from the MiSeq run was aligned with the sequence of Photobacterium sp. r33 from NCBI (Supplementary Figure 2). This was done through the NCBI BLAST program4. This alignment exhibited a 99% match, with only a single nucleotide difference. However, until bacterial identity is confirmed with deeper genomic sequencing, the most abundant bacterium taxon will be referred to as Photobacterium Pa-1.
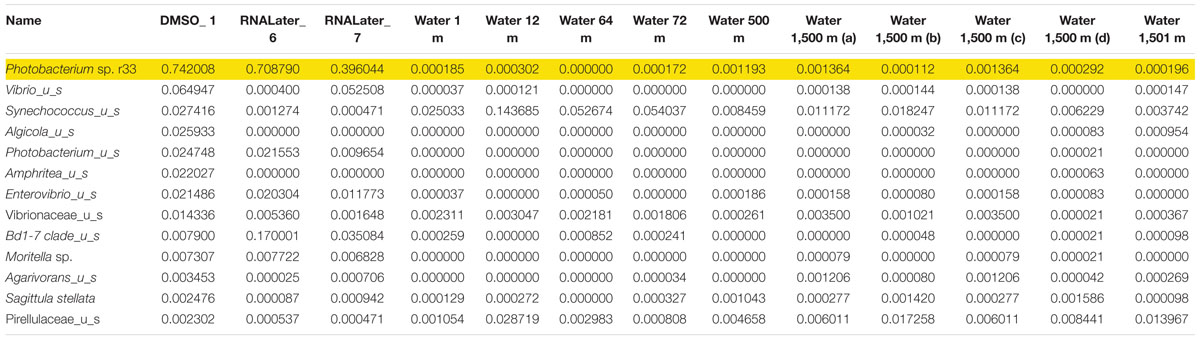
Table 3. Relative abundance of the top 13 bacterial 673 species found with Photobacterium sp. r33 highlighted.
In the samples analyzed, matches to a Photobacterium sp. r33- like had a range of abundances. In the 10 seawater samples, Photobacterium sp. r33 appeared less than 0.12% at the highest abundance, while in the tissue samples, the same Photobacterium appeared in DMSO1 at 74.20%, RNALater6 at 70.88%, and in RNALater7 at 39.60%. These abundances in the pyrosomes were calculated through the CosmosID® pipeline and showed significant differences in beta diversity of bacterial ASV’s between the pyrosome and water samples after an ANOSIM test (R = 0.8704, p = 0.0001) (Figure 2), with the water samples having a considerably more diverse bacterial community (Supplementary Figure 3). The top two bacterial taxa in the three pyrosome samples were Photobacterium sp. r33 and Vibrio_us (unidentified species). There were over 1,100 ASV’s found in the water samples.
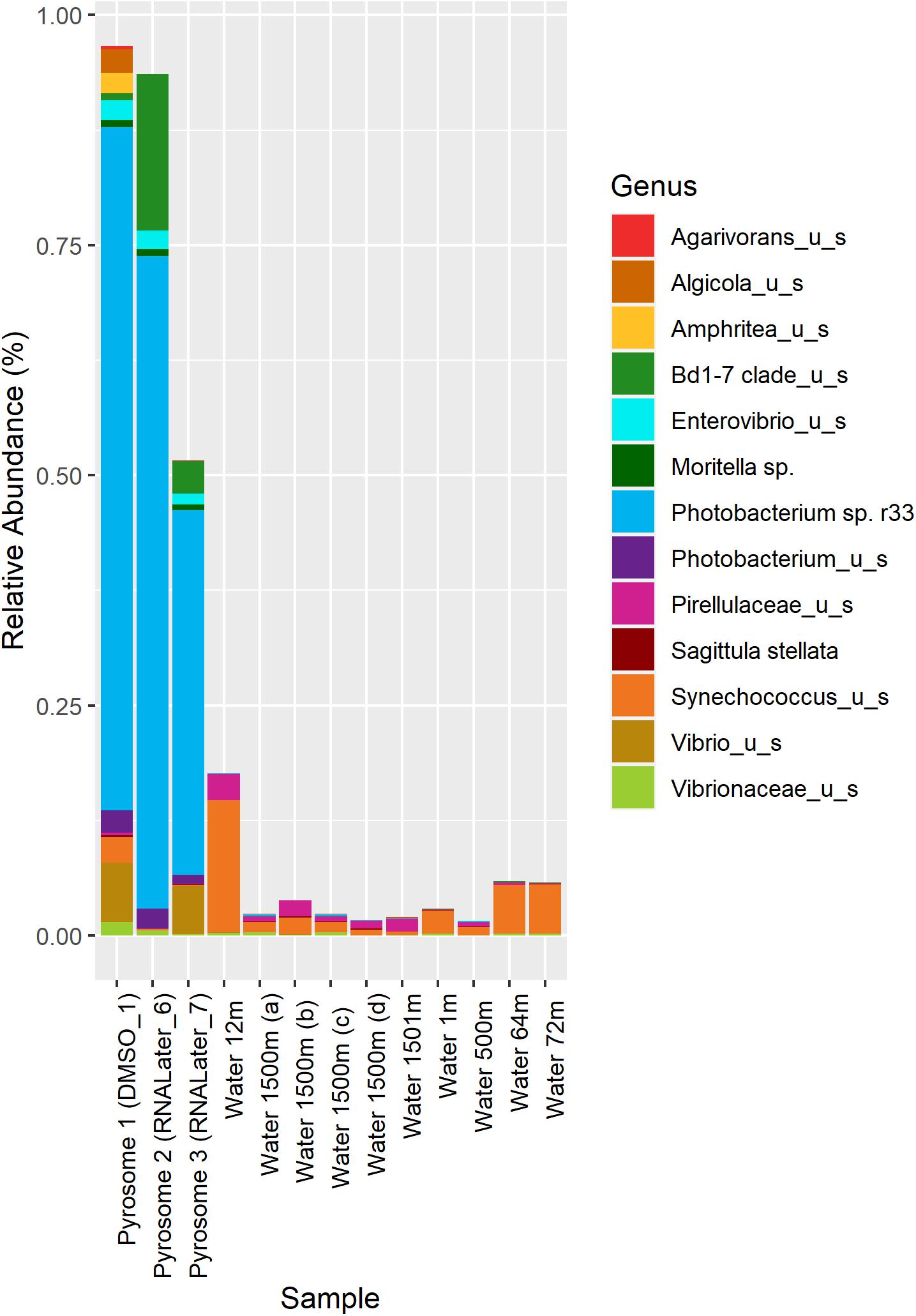
Figure 2. Relative abundance of the top 13 bacterial taxa across different samples. Columns 1–3 represent microbiomes derived from tunicate specimens preserved under different conditions. The next 10 samples show bacterial distributions in seawater samples from the same location (Easson and Lopez, 2019).
In an effort to detect and characterize lux genes of a bacterial photosymbiont producing bioluminescence, we conducted a whole-genome (metagenomic) Illumina sequencing run. This was based on the evidence that most of the Pyrosoma atlanticum 16S rRNA profiles appeared to be dominated by just a few bacterial taxa. Unfortunately, bacterial lux genes were not detected in the resulting assemblies, although a partial mitochondrial DNA contig of 14,302 base pairs (bp) was serendipitously generated with 26X coverage. The mtDNA sequences provided an opportunity to gain a genetic basis for the taxonomic identity of Pyrosoma atlanticum. The next closest sequence to P. atlanticum was another tunicate in class Thaliacea, pelagic order Doliolida, Doliolum nationalis. A preliminary phylogeny based on the mitochondrial cytochrome oxidase subunit 1 (COI) genes supports the affinity of P. atlanticum with Doliolum nationalis (Supplementary Figure 4).
Fluorescence in situ Hybridization (FISH)
Under fluorescence microscopy, pyrosome tissue exhibited autofluorescence, but the bacteria were still clearly discernable (Figure 1). When histology sections were compared with those prepared for fluorescence in situ hybridization (FISH) analyses, similar orientation and morphology of the two light organs were evident (Figure 3). Both methods revealed putative “bacteriocytes” containing bacteria concentrated at the outer edges of the organ with a clear space in the center. FISH controls included pyrosome sections that were incubated with no fluorescent probes. The control sections reflected native background autofluorescence and did not display the same intensity (brightness) as seen in sections hybridized with probes (Figure 3A). This comparison demonstrated that the probes annealed specifically to their respective DNA targets and produced a signal after stringent washing. The pair of light organs fluoresced in the absence of a probe, but not as brilliantly as when the EUB338 and Photobacterium probes were used (Figure 3B). The signal produced with both probes was very intense and, as anticipated, bacteria surrounding the light organ as well as the light organ emitted a signal above background. This was likely a result of the universal EUB338 bacterial probe being designed to bind to almost all bacteria within the sample (Figure 3C). When the Photobacterium probe was employed, only the light organ emitted a signal (Figure 3D). Other areas of the tunic did not emit a signal, indicating that the photobacteria were concentrated in the light organ. The morphology of bacteria differed in the tissue throughout the section, ranging from coccoid in the light organ to bacterium with flagella-like structures in the tunic. Due to the high light intensity, the shutter on the microscope was partially closed for the sections hybridized with probes while the sections with no probes were imaged with the shutter remaining fully open.
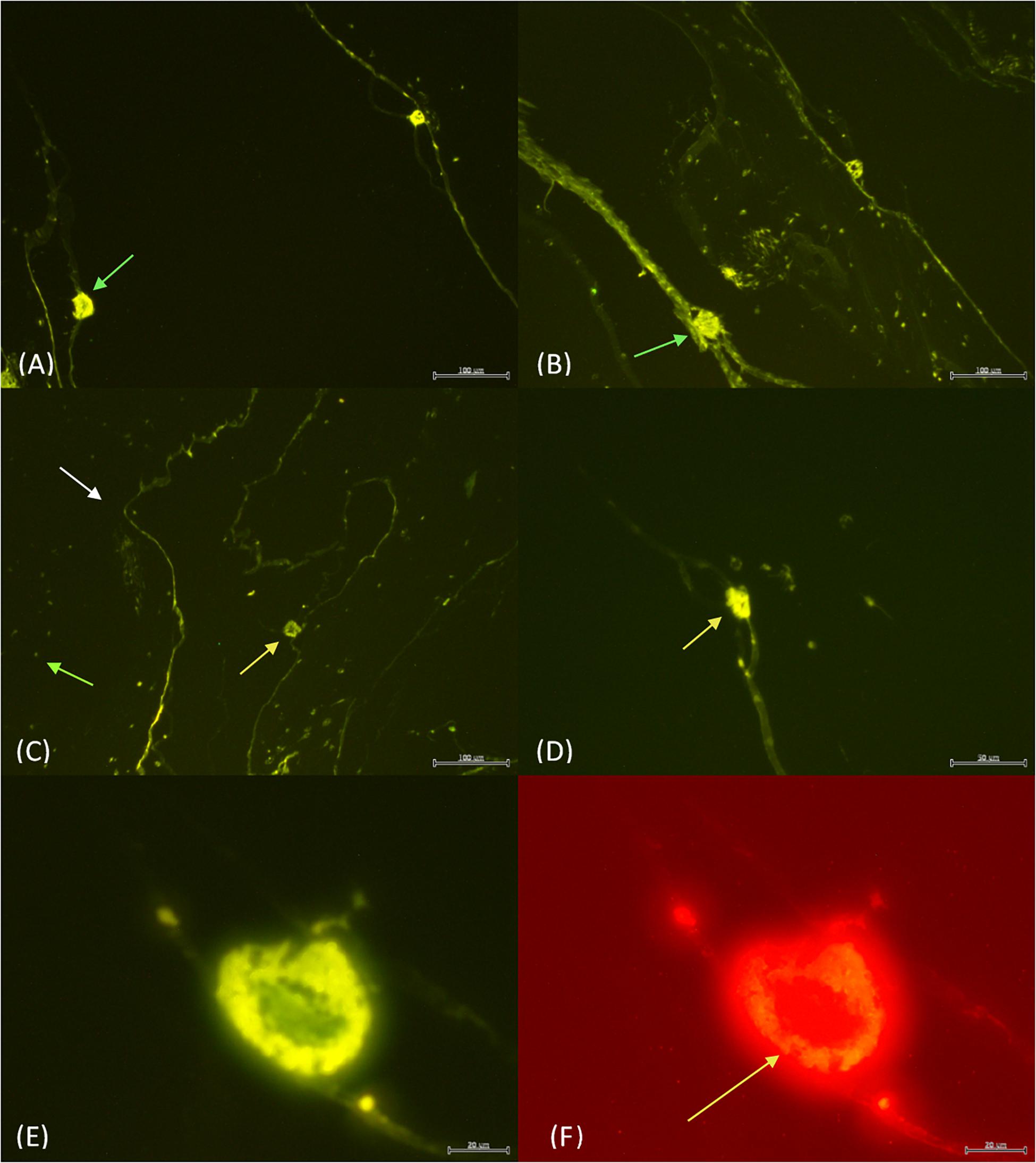
Figure 3. The light organs (green arrows) with no probe and the shutter wide open (A) vs. 4 μl of both EUB338 and Photobacterium probe with the shutter partially closed (B). The EUB338 probe (C) binds to many bacteria (green arrow) within the tunic (white arrow) and the light organ (yellow arrow). In contrast, the Photobacterium probe (D) only illuminated the light organ. EUB338 and Photobacterium probes in green (E) vs. red (F). The orange fluorescence in Photobacterium Pa-1 is found exclusively concentrated around the edges of the light organ (yellow arrow). Scale bar = 100 μm in (A–C), while the scale bar is 50 μm (D), and 20 μm in (E,F).
When the EUB338 and Photobacterium probes were combined, variability in the intensity of signal emission was evident. Photobacterium Pa-1 fluorescence was brightest when both probes were combined. For example, under the green filter the bacteria are seen concentrated around the outer edges of the light organ with a clear space in the center similarly to previous observations (Figure 3E). Under the red filter, the same outer edges are packed with Photobacterium Pa-1 fluorescing orange (Figure 3F). This filter and orange fluorescence strongly indicates the presence of Photobacterium Pa-1 in the light organ.
Fine Structure and Bacterial Cluster Location in P. atlanticum (SEM)
A three-dimensional view of the pyrosome light organ and associated microbes was obtained using SEM imaging (Figure 4). This imaging shows the relationship between the tissue surrounding the light organ with the intracellular bacteria present. The light organ was approximately 20 μm in diameter and confirms diameter estimates made using light microscopy. The light organ is divided into two distinct areas confirming our light microscope observations. In the organ shown, the bacteria were clustered in an area of the light organ and include approximately 25 spherical 1–2 μm cells. In this SEM image it is not possible to discern if the microbial cells were intracellular but was better addressed in the TEM images.
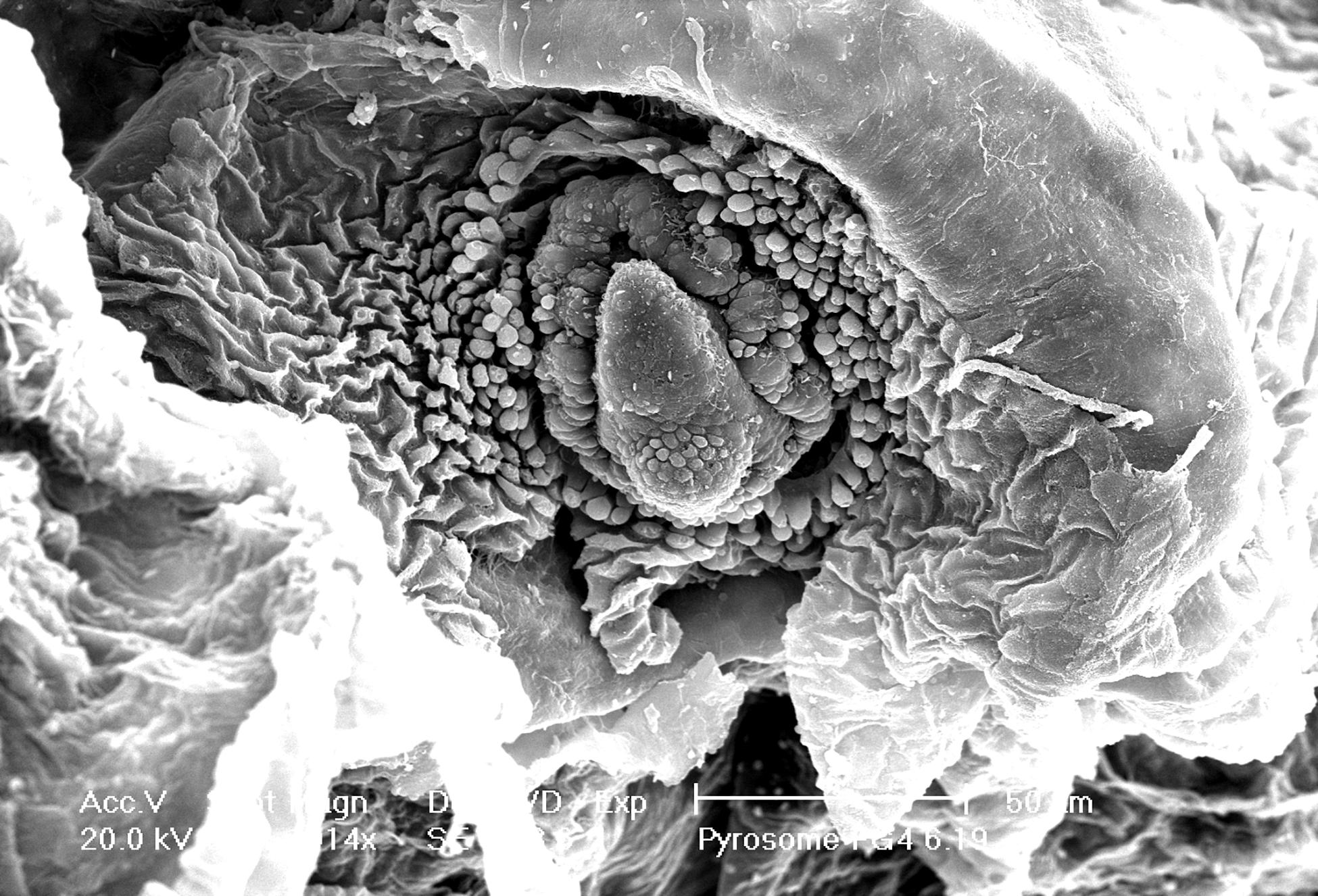
Figure 4. Intact light organ (∼30 μm diameter) semi encased in the tunic. Intracellular microbial cells were not discernable due to the intact light organ. Scale bar is set at 50 μm.
Ultrastructure of the Microbial Population in P. atlanticum (TEM)
The light organ bacterial symbionts were intracellular and associated with mitochondria (Figure 5A). Their intracellular location was confirmed by observation of a cell membrane that encased both bacterial symbionts and mitochondria (Figure 5A). Cells containing these bacteria and mitochondria were also associated with abundant endoplasmic reticulum (Figures 5A,B). The mitochondria were sometimes closely associated with the bacteria. However, when comparing clusters of microbes and mitochondria, the bacteria could be easily distinguished from the mitochondria by the presence of prominent cristae in the mitochondria and the bacterial cells resembling bacteriocytes (Figure 5A). In some cases, the bacteria were clustered around an “opening” that suggests excretion activity (Figure 5C). It appears that fluid filled vesicles may pinch off and move to the extracellular environment. The nature of these vesicles are unknown, or whether these excretory products are associated with bioluminescence. Within a single bacteriocyte, 5–7 bacteria were visualized (Figure 5) and therefore we estimated that a minimum of 480 ∼ 1,200 putative photosymbiotic bacteria could be found within a single Pyrosoma atlanticum light organ. These are approximate numbers since we cannot fully view all bacteriocytes and their contents in one micrograph.
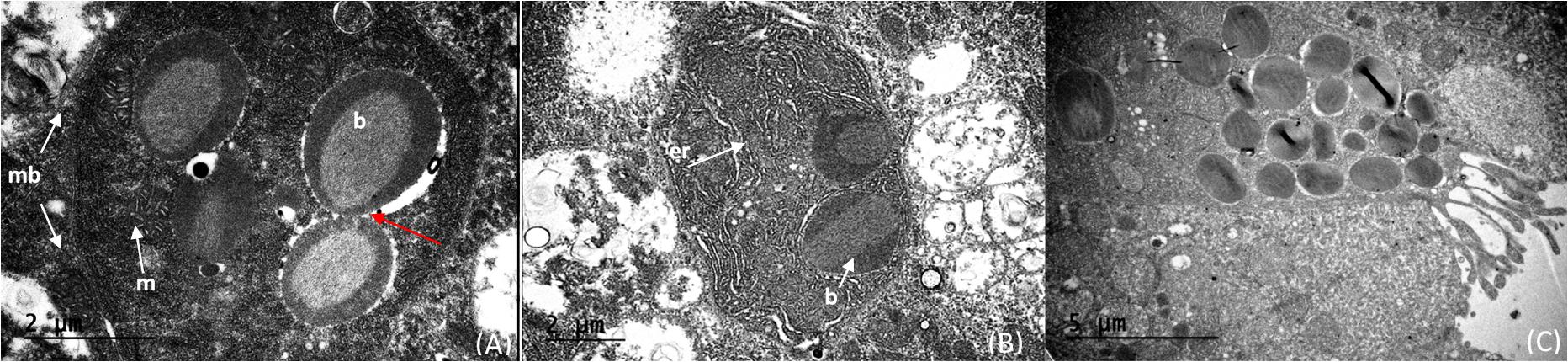
Figure 5. Cristae of the mitochondria (m) distinctly shown compared to the intracellular microbes (b) within the cell (membrane—mb). The cells on the right appear to have just divided (red arrow) (A). Intracellular microbes (b) with endoplasmic reticulum (er) distributed throughout the cell (B). Bacteria within the light organ suggest intracellular to extracellular excretion activity of the light organ (C).
Discussion
Structure of the Light Organ in P. atlanticum
Our imaging of the light organ of Pyrosoma atlanticum supported previous descriptions (Mackie and Bone, 1978; Nealson et al., 1981). Previous studies had micrographs of semi-thin sections showing “photic” organelles in the light organ (Mackie and Bone, 1978). This study is the first to combine detailed light and electron microscopic imaging of P. atlanticum with genetic analyses (light, TEM, SEM, FISH, and 16s rRNA sequencing) simultaneously. This combination demonstrated clearly that Photobacterium sp. r33-like bioluminescent symbionts are contained intracellularly within bacteriocyte resembling structures intracellularly in the light organ. The specialized adipocytes are heavily concentrated around the outer edges of the light organ and have intracellular vacuoles containing multiple bacteria, which have been found in several different marine holobionts, including tunicates (Kwan et al., 2012; Lopez, 2019). The intracellular location of the bacterial symbionts is highly unusual here, as most bioluminescent bacterial symbionts are extracellular (Mackie and Bone, 1978).
Source of Bioluminescence in P. atlanticum
We have demonstrated a likely bacterial source for bioluminescence in Pyrosoma atlanticum, a taxon that is genetically closest to Photobacterium sp. r33. However, we cannot unequivocally identify the species without additional genomic data, since 100% identity to a single gene such as 16s rRNA does not confirm species identity (Fox et al., 1992; Janda and Abbott, 2007). The genus Photobacterium is known to show substantial ecophysiological diversity, which includes free-living, symbiotic, and parasitic lifestyles (Labella et al., 2017). The bioluminescent Photobacterium species, in particular P. aquimaris, P. damselae, P. kishitanii, P. leiognathi, and P. phosphoreum, exhibit free-living and symbiotic lifestyles. They can be found in dense populations associated with tissues in the light organs of their selective hosts (Labella et al., 2017). These tissues could be reflectors, shutter lens, or other tissues that are used to control, focus, and/or diffuse the bacterial light produced from the organism’s body (Urbanczyk et al., 2011). Some of the hosts of P. kishitanii and P. leiognathi are marine fishes, cephalopods, and octopods. However, P. leiognathi has established a highly specific symbiosis with the fish families Leiognathidae, Acropomatidae, and Apogonidae, while P. damselae has been found to form a symbiosis only with damselfish (Labella et al., 2017). In deep sea anglerfish, Baker et al. (2019) found some bioluminescent Enterovibrio symbionts with variable and incongruent fidelities to specific fish host lineages. In our study, similar host specificity is exhibited by Photobacterium Pa-1, as indicated by the high relative abundance of Photobacterium sp. r33 from 16S sequencing as well as evidence from microscopy. SEM, TEM, and FISH confirm that Photobacterium Pa-1 inhabits the light organ of Pyrosoma atlanticum.
Photobacterium sp. hosts range from fishes to cephalopods and are found throughout the water column. The bacterially luminous fishes are widely distributed in coastal demersal, epibenthic, and pelagic waters (Urbanczyk et al., 2011). The fishes that house P. leiognathi and P. mandapamensis are more commonly found in shallower and warmer waters, whereas P. kishitanii can be found in fishes inhabiting deeper waters (Dunlap et al., 2007; Kaeding et al., 2007; Nelson et al., 2016). The pelagic tunicate, Pyrosoma atlanticum, can now be added now as a host of Photobacterium Pa-1 further diversifying the range of habitats encompassed by Photobacterium sp. hosts.
P. atlanticum Bacterial Symbiont Location
Several probe controls were used to demonstrate that Photobacterium Pa-1 is located in the light organ of Pyrosoma atlanticum. Although the light organ exhibits some variability in morphology as well as orientation in the sections, in the FISH analyses the signals produced remain essentially the same. The EUB338 probe bound to more bacteria than the Photobacterium probe due to having a more universal and conserved rRNA sequence than the variable region V4 of the Photobacterium. The conserved regions of 16S rRNA has similar or identical sequences across many bacterial groups, while the variable region consists of clade or taxon-specific sequences (Negandhi et al., 2010). The EUB338 probe fluoresced a greenish tint under the green filter cube (500–570 nm) and produced more signals than the Photobacterium probe, binding to a wide variety of bacteria throughout the zooids. The red filter cube (610∼750 nm) served as the defining filter for the Photobacterium probe. With the red filter cube, the EUB338 probe showed that all bacteria fluoresced red while the slides with both probes or just the Photobacterium probe showed the probe-specific bacteria fluorescing orange. The orange fluorophores confirmed that Photobacterium Pa-1 was located in the light organ.
Light microscopy revealed microbial localization within the luminous organ, and the bacterial symbionts were identified by FISH. TEM clearly indicated intracellular bacteria concentrated in the organ. There were approximately 60 bacteriocytes found in a single light organ in light microscopy. Precise estimates of bacteriocyte numbers are likely affected by the plane in which tissues were sectioned, so there may be more Photobacterium sp. per cell than that observed using EM. In each micrograph, regardless of the type of microscopy used, the bacteria were concentrated on the interior border of the cells, while the bacteriocytes made up the periphery of light organ itself, surrounding a non-cellular space in the center. This begs the question as to why bacteria concentrate at the edges. To determine if the orientation of bacteria in the luminous organ plays a role, or stages, in the production of luminescence would pose an interesting question for future research.
P. atlanticum Bacterial Symbiont Morphology
Bacterial symbionts have been described in many marine invertebrates (McFall-Ngai et al., 2013; Lopez, 2019); however, only two papers have produced a description of the ultrastructure of photogenic organelles assumed to be bacteria in pyrosomes (Mackie and Bone, 1978; Nealson et al., 1981), and one paper describes bacterial luciferase activity similar to that of Photobacterium in Pyrosoma sp. (Leisman et al., 1980). There is precedence for bacterial symbionts to be contained intracellularly or within bacteriocytes, especially in tunicates such as Lissoclinum patella (Kwan et al., 2012). Bioluminescent bacterial symbionts, however, are often found to be extracellular.
Extracellular and free living bacterial symbionts are typically rod-shaped and more elongated (Nealson et al., 1981) than the bacteria present in pyrosomes. With the morphological similarities to gram-negative bacteria, thick cell walls and double membranes around each cell, our results provide strong support that the cells identified in the light organ are of microbial origin, aiding in the validation of the hypothesis that Pyrosoma atlanticum uses bacterial symbiosis as their bioluminescence mechanism (Dunlap and Schaechter, 2009). Gram staining could not be applied directly on the bacteria because neither they, nor the light organ, could be isolated. Nonetheless, the proposed intracellular luminescent bacteria in Pyrosoma differ morphologically and biochemically from almost all other bioluminescent bacteria, which are typically longer than oval or as subspherical rods and without granules (Mackie and Bone, 1978). These photobacteria in Pyrosoma atlanticum were exclusively coccoid in morphology and 1–2 μm in diameter, in agreement with previous bacterial ultrastructural descriptions of Pyrosoma atlanticum (Nealson et al., 1981).
The microscopy images in the current study produced a more detailed description of the bacteria found in Pyrosoma atlanticum than any previous work done on pyrosomes. Moreover, the current results can be added and contrasted to the continuing debate over whether P. atlanticum bioluminescence stems from bacterial or eukaryotic sources. For example, Tessler et al. (2020) recently described a potential luciferase, an Rluc-like protein in Pyrosoma. They provide compelling phylogenetic support that this enzyme is endogenous, decreasing the likelihood of contamination from orthologs. It is known that several non-luminous organisms such as Strongylocentrotus, Ciona, Trichoplax, or Saccoglossus, and likely other unidentified pelagic organisms have orthologs to Rluc luciferase which could also bind to the antibody (Tessler et al., 2020). However, the specific luciferase function of the proteins in these other organisms and pyrosomes was not confirmed, and there are many examples of “gene or protein sharing” throughout molecular evolution (Shimizu et al., 1998; Horwitz, 2000). This scenario indicates that a conserved luciferase protein sequence may be present, but still may have different functions that depend on tissue or taxonomic context. For example, although Tessler et al. (2020) did express the recombinant pyrosome Rluc homolog and show that it generated light with coelenterazine, the binding of their antibody did not unequivocally confirm that the target is the native luciferase. Lastly Tessler et al. (2020) did not unequivocally demonstrate that there is any coelenterazine in the native system in P. atlanticum that would be used by the endogenous luciferase. These represent major missing pieces that could support a primary host role in bioluminescence.
Moreover, Tessler et al.’s immunostained confocal micrographs indicated the presence of a strong signal to Renilla luciferase antibody in each zooid, in a circular region below the incurrent siphon containing nuclei. This strongly stained region was in the same regions of the luminous organ described by Mackie and Bone (1978). These nucleated cells may be from the eukaryotic pyrosome bacteriocytes in which the bacteria are localized. However, when the micrographs from Tessler et al. (2020) are compared with those from this study (Figures 3E,F), it is evident that different areas of the tissue and cells are examined. The incurrent siphons of the zooids are near the light organs, but the light organs are not shown in these figures. The immunostained confocal localization with some observations of nuclei (Tessler et al., 2020), does not preclude the presence of intracellular bacteria.
Continuing controversy exists in the literature concerning the relationship between bacteria and pyrosome luminescence. This may persist with the recent results and until unequivocal evidence appears [e.g., direct observation of bioluminescence, vertical transmission of photosymbionts (Sharp et al., 2007)]. For example, some suggest that pyrosome luminescence is due to an “intrinsic system” not related to bacterial symbionts, with the source of light being a type of tunic cell (Aoki et al., 1989). Our ultrastructural characterization of the putative bacterial cells for the most part coincided with published TEM work on Pyrosoma atlanticum (Mackie and Bone, 1978; Nealson et al., 1981; Aoki et al., 1989). The overall size and morphology of the microbial cells found in our study are similar to those in the literature in terms of ultrastructural characteristics and are found in the tunic cells packed with organelles and not innervated (Mackie and Bone, 1978).
Distribution and Acquisition of Bacteria in Organisms Related to Bioluminescence
Previous work on Pyrosoma atlanticum had not determined whether the bacteria are intra- or extracellular, and only two studies have hypothesized an intracellular organization for pyrosome bacterial symbionts (Mackie and Bone, 1978; Nealson et al., 1981). Our genetic and imaging data is consistent with the hypothesis that intracellular bacteria are present in the pyrosome light organs and provide strong evidence that intracellular bacterial symbionts are potentially responsible for light production in Pyrosoma atlanticum. The visualizations of the light organs in light, fluorescence, and electron microscopy showed that bacterial symbionts belonging to the bioluminescent genera Photobacterium were found in same region where the bacteriocytes were observed, encased by a cell membrane with the mitochondria. Intracellular organization, in conjunction with host mediated bacteriocyte structure, indicates a highly interdependent and specialized biochemical relationship between the bacteria and host cells (Nealson et al., 1981). Intracellular symbionts represent the most highly adapted of bacterial symbionts (Nealson et al., 1981; Shigenobu et al., 2000) and our current microscopy data provides the first evidence of Pyrosoma atlantcium harboring these highly adapted intracellular symbionts.
There is much to be learned on how and when the hosts of Photobacterium initiate symbioses. The intracellular occurrence of these symbionts brings up the question of whether Photobacterium Pa-1 symbiont may be acquired via horizontal or vertical transmission. Horizontal transmission is the acquisition of symbionts from the environment, while vertical transmission is the inheritance of symbionts from previous generations (Bright and Bulgheresi, 2010). In deep-sea ceratioid fishes it is reported that the bioluminescent symbionts are acquired from the environment during the larval migration of the fish from surface waters to the bathypelagic water, albeit in low levels of abundance (Freed et al., 2019). These symbionts were found in low levels of abundance in both mesopelagic and bathypelagic zones, which suggest that the microbes are not obligately dependent on the hosts for growth. Anglerfishes appear to not acquire their photosymbionts from the environment until they mature and move to lower depths (Freed et al., 2019). In one of the best examples of horizontal transmission, bioluminescent Vibrio fischeri symbionts appear to move freely from the environment to a residence within the Hawaiian bobtail squid via specialized ducts (Nyholm and McFall-Ngai, 2004). The light organ crypts have a small window for V. fischeri to inoculate –between 30 and 60 min after hatching when these crypts open (Nyholm and McFall-Ngai, 2004).
In the case of Pyrosoma atlanticum, the data suggest (Table 3) vertical transmission for Photobacterium Pa-1. A relative paucity of Photobacterium Pa-1 sequences in our large pelagic GoM dataset (Easson and Lopez, 2019) supports this hypothesis. The seawater samples showed virtually no Photobacterium sp. r33-like sequences (0.0–0.12%) compared to pyrosome samples, which contained a large concentration of 40–74% of these sequences. Since Pyrosoma atlanticum is specifically known to reproduce both sexually and asexually through internal fertilization and budding (Holland, 2016), vertical transmission of the Photobacterium Pa-1 symbiont is plausible. The 16S rRNA analyses and micrographs support the concept that the acquisition of symbionts is through vertical transmission due to the complexity of intracellular symbiont location as well as low abundances in surrounding seawater compared to high abundances in the pyrosome. Due to the possibility of either horizontal and vertical transmission (or both), further studies are required analyzing the entire lifecycle of the Pyrosoma atlanticum, tracking putative symbionts, in conjunction with seawater samples collected at the same time and location of the pyrosome.
Photobacterium Pa-1 Mitochondrial Association
Microbial symbionts occur in almost every metazoan, although many partnerships have not been sufficiently studied (McFall-Ngai et al., 2013; Apprill, 2017). With most microbial mutualisms, the host relies nutritionally on the microbial symbiont, such as chemosynthetic bacterium or photosynthetic algae, and without these symbionts, the host growth suffers significantly. Bioluminescent symbiosis differs from other types of symbiotic associations, as lumen production from the microbe is the currency of exchange rather than metabolic nutrients for the host (Dunlap and Schaechter, 2009). Some bioluminescent symbioses also appear facultative as the host or bacterial symbionts can survive without each other ex situ and in laboratory settings (Dunlap and Schaechter, 2009). Many bacterial bioluminescent symbionts appear to be extracellular, suggesting a facultative association, whereas in obligate symbiosis the bacteria are found intracellularly. Luminous bacteria are characterized as Gram-negative, non-spore-forming, motile, have cell walls difficult to penetrate, and are generally chemoorganotrophic.
Photobacteria have previously been found to be associated with mitochondria inside pyrosome cells (Nealson et al., 1981). It has been noted that there are several similarities between the respiratory chain of mitochondria and bioluminescent bacteria (Rees et al., 1998; Bourgois et al., 2001). Bacterial luciferase has previously been viewed as “an alternative” electron transport pathway, however, it is more recently considered an “alternative” oxidase (Bourgois et al., 2001). This is why the entire photogenic system of bioluminescent bacteria scavenges not only reducing equivalents (luciferase), but also ATP and NADPH. The close association also ties into the fact that the organism needs to consume a certain amount of energy to produce the visible spectrum of the bioluminescent light (Rees et al., 1998). In most cases it would be the blue photon (∼470 nm), which requires about 255 J mol –1. The fact that bioluminescence requires a lot of energy and mitochondria produce ATP, might explain why the mitochondria and microbes are so closely associated and densely packed into the cells (Bourgois et al., 2001).
Conclusion
This study provides new insights into the bioluminescent mechanism of Pyrosoma atlanticum, a dominant colonial tunicate found in tropical to temperate pelagic waters worldwide. Our findings support the hypothesis that bioluminescence in these tunicates is bacterial-based, specifically associated with Photobacterium Pa-1. The Family Vibrionaceae is known to contain three genera of bioluminescent bacteria, including Photobacterium. Photobacterium Pa-1 are found intracellularly and within the light organs of Pyrosoma atlanticum. They were found dominating the microbiome in greater relative abundances of about 40–74% in these pyrosomes. Future studies should focus on bioluminescent mechanisms of the light organ itself: regulation of bioluminescence, comparing the microbiome of the whole tunicate to that of the light organ to show the selectivity of its environment, and finally the acquisition and retention of bacterial symbionts if they proven to be major source of bioluminescence (Guindon and Gascuel, 2003).
Data Availability Statement
The datasets presented in this study can be found at NCBI repositories https://www.ncbi.nlm.nih.gov/. This includes Sequence Read Archives under PRJNA636187 and PRJNA635880.
Author Contributions
AB and JL were responsible for project and experimental design. AB conducted the above research. PB lead the electron microscopy work as well as imaging for all micrographs. TF guided and directed the bioluminescence work, and analysis. JL carried out 16S primer selection for FISH and phylogenetic analysis. TS, NP, and NS collected samples and metadata from the various research cruises for this project. JL, PB, TF, and TS edited manuscript drafts and helped prepare for press. All authors contributed to the article and approved the submitted version.
Funding
This research was partially made possible by a grant from the BP/The Gulf of Mexico Research Initiative to support the consortium research entitled “Deep Pelagic Nekton Dynamics of the Gulf of Mexico” administered by Nova Southeastern University (NSU). Other components were funded by the JL’s and internal NSU research funds.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the PIs and scientists of the DEEPEND consortium, especially the LUMCON crew of the R/V Point Sur for their invaluable help and support throughout this project. Microbial community sequence data are publicly available through the Gulf of Mexico Research Initiative Information and Data Cooperative—GRIIDC (R4.x257.228:0009). We would also like to thank Dr. Lauren Krausfeldt for her invaluable knowledge and advice and Colleen McMaken for lending her support, time, R code, and eyes throughout this project. In addition, many thanks to Dr. D. Abigail Renegar and Dawn Bickham for their patience and guidance through the histological part of this project. One last thanks to Dr. Rosanna Milligan for lending us her mapping expertise in RStudio.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.606818/full#supplementary-material
Footnotes
- ^ www.deependconsortium.org
- ^ www.ncbi.nlm.nih.gov/probe
- ^ http://rdp.cme.msu.edu/probematch/search.jsp
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Alldredge, A., and Madin, L. (1982). Pelagic tunicates: unique herbivores in the marine plankton. Bioscience 32, 655–663. doi: 10.2307/1308815
Aoki, M., Hashimoto, K., and Watanabe, H. (1989). The intrinsic origin of bioluminescence in the ascidian. Clavelina miniata. Biolog. Bull. 176, 57–62. doi: 10.2307/1541889
Apprill, A. (2017). Marine animal microbiomes: toward understanding host–microbiome interactions in a changing ocean. Front. Mar. Sci. 4:222. doi: 10.3389/fmars.2017.00222
Baker, L. J., Freed, L. L., Easson, C. G., Lopez, J. V., Fenolio, D., Sutton, T. T., et al. (2019). Diverse deep-sea anglerfishes share a genetically reduced luminous symbiont that is acquired from the environment. eLife 8:e47606.
Bourgois, J.-J., Sluse, F., Baguet, F., and Mallefet, J. (2001). Kinetics of light emission and oxygen consumption by bioluminescent bacteria. J. Bioenerget. Biomembr. 33, 353–363.
Bowlby, M. R., Widder, E. A., and Case, J. F. (1990). Patterns of Stimulated Bioluminescence in 2 Pyrosomes (Tunicata, Pyrosomatidae). Biol. Bull. 179, 340–350. doi: 10.2307/1542326
Bright, M., and Bulgheresi, S. (2010). A complex journey: transmission of microbial symbionts. Nat. Rev. Microbiol. 8, 218–230. doi: 10.1038/nrmicro2262
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335.
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108(Suppl. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Chakravorty, S., Helb, D., Burday, M., Connell, N., and Alland, D. (2007). A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69, 330–339. doi: 10.1016/j.mimet.2007.02.005
Chiba, K., Hoshi, M., Isobe, M., and Hirose, E. (1998). Bioluminescence in the tunic of the colonial ascidian, Clavelina miniata: identification of luminous cells in vitro. J. Exp. Zool. 281, 546–553. doi: 10.1002/(sici)1097-010x(19980815)281:6<546::aid-jez2>3.0.co;2-n
Décima, M., Stukel, M. R., López−López, L., and Landry, M. R. (2019). The unique ecological role of pyrosomes in the Eastern Tropical Pacific. Limnol. Oceanogr. 64, 728–743. doi: 10.1002/lno.11071
Dunlap, P., and Schaechter, M. (2009). Microbial bioluminescence. Encyclop. Microbiol. 2009, 45–61. doi: 10.1016/b978-012373944-5.00066-3
Dunlap, P. V., Ast, J. C., Kimura, S., Fukui, A., Yoshino, T., and Endo, H. (2007). Phylogenetic analysis of host–symbiont specificity and codivergence in bioluminescent symbioses. Cladistics 23, 507–532. doi: 10.1111/j.1096-0031.2007.00157.x
Easson, C. G., and Lopez, J. V. (2019). Depth-Dependent environmental drivers of microbial plankton community structure in the Northern Gulf of Mexico. Front. Microbiol. 9:3175. doi: 10.3389/fmicb.2018.03175
Fox, G. E., Wisotzkey, J. D., and Jurtshuk Jr. (1992). How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Evol. Microbiol. 42, 166–170. doi: 10.1099/00207713-42-1-166
Freed, L. L., Easson, C., Baker, L. J., Fenolio, D., Sutton, T. T., Khan, Y., et al. (2019). Characterization of the microbiome and bioluminescent symbionts across life stages of Ceratioid Anglerfishes of the Gulf of Mexico. FEMS Microb. Ecol. 95:fiz146.
Galt, C., and Flood, P. (1998). Bioluminescence in the Appendicularia. Biol. Pelagic Tunic. 1998, 215–229.
Galt, C., and Sykes, P. (1983). Sites of bioluminescence in the appendicularians Oikopleura dioica and O. labradoriensis (Urochordata: Larvacea). Mar. Biol. 77, 155–159. doi: 10.1007/bf00396313
Gray, M. W., Sankoff, D., and Cedergren, R. J. (1984). On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 12, 5837–5852. doi: 10.1093/nar/12.14.5837
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Haddock, S. H., Moline, M. A., and Case, J. F. (2010). Bioluminescence in the sea. Ann. Rev. Mar. Sci. 2, 443–493.
Haygood, M. G. (1993). Light organ symbioses in fishes. Crit. Rev. Microbiol. 19, 191–216. doi: 10.3109/10408419309113529
Hirose, E. (2009). Ascidian tunic cells: morphology and functional diversity of free cells outside the epidermis. Invert. Biol. 128, 83–96. doi: 10.1111/j.1744-7410.2008.00153.x
Horwitz, J. (2000). The function of alpha-crystallin in vision. Semin. Cell Dev. Biol. 2000, 53–60. doi: 10.1006/scdb.1999.0351
Huxley, T. H. (1851). XXIV. Observations upon the anatomy and physiology of salpa and pyrosoma. Philosop. Transac. R Soc. Lon. 141, 567–593. doi: 10.1098/rstl.1851.0027
Janda, J. M., and Abbott, S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microb. 45, 2761–2764. doi: 10.1128/jcm.01228-07
Kaeding, A. J., Ast, J. C., Pearce, M. M., Urbanczyk, H., Kimura, S., Endo, H., et al. (2007). Phylogenetic diversity and cosymbiosis in the bioluminescent symbioses of “Photobacterium mandapamensis”. Appl. Environ. Microbiol. 73, 3173–3182. doi: 10.1128/aem.02212-06
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30, 772–780. doi: 10.1093/molbev/mst010
Kuczynski, J., Stombaugh, J., Walters, W. A., González, A., Caporaso, J. G., and Knight, R. (2011). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protocols Bioinform. 36, 11–10.
Kwan, J. C., Donia, M. S., Han, A. W., Hirose, E., Haygood, M. G., and Schmidt, E. W. (2012). Genome streamlining and chemical defense in a coral reef symbiosis. Proc. Natl. Acad. Sci. 109, 20655–20660. doi: 10.1073/pnas.1213820109
Labella, A. M., Arahal, D. R., Castro, D., Lemos, M. L., and Borrego, J. J. (2017). Revisiting the genus Photobacterium: taxonomy, ecology and pathogenesis. Int. Microbiol. 20, 1–10. doi: 10.1007/978-94-017-2901-7_1
Leisman, G., Cohn, D. H., and Nealson, K. H. (1980). Bacterial Origin of Luminescence in Marine Animals. Science 208, 1271–1273. doi: 10.1126/science.208.4449.1271
Lemaire, P., and Piette, J. (2015). Tunicates: exploring the sea shores and roaming the open ocean. A tribute to Thomas Huxley. Open Biol. 5:150053. doi: 10.1098/rsob.150053
Lopez, J. V. (2019). After the taxonomic identification phase: addressing the functions of symbiotic communities within marine invertebrates in Symbiotic Microbiomes of Coral Reefs Sponges and Corals. Berlin: Springer. 105–144.
Mackie, G. O., and Bone, Q. (1978). “Luminescence and Associated Effector Activity in Pyrosoma (Tunicata Pyrosomida),” in Proceedings of the Royal Society Series B-Biological Sciences, Vol. 202:0081. doi: 10.1098/rspb.1978.0081
Martini, S., and Haddock, S. H. (2017). Quantification of bioluminescence from the surface to the deep sea demonstrates its predominance as an ecological trait. Sci. Rep. 7:45750.
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. 110, 3229–3236.
Nealson, K., Cohn, D., Leisman, G., and Tebo, B. (1981). Co−evolution of luminous bacteria and their eukaryotic hosts. Ann. N Y Acad. Sci. 361, 76–91. doi: 10.1111/j.1749-6632.1981.tb46512.x
Negandhi, K., Blackwelder, P. L., Ereskovsky, A. V., and Lopez, J. V. (2010). Florida reef sponges harbor coral disease-associated microbes. Symbiosis 51, 117–129. doi: 10.1007/s13199-010-0059-1
Nyholm, S. V., and McFall-Ngai, M. (2004). The winnowing: establishing the squid–Vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642. doi: 10.1038/nrmicro957
O’Connell, L., Gao, S., McCorquodale, D., Fleisher, J., and Lopez, J. V. (2018). Fine grained compositional analysis of Port Everglades Inlet microbiome using high throughput DNA sequencing. PeerJ 6:e4671.
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P., O’hara, R., et al. (2013). Community ecology package. R package version 2.0-2.
Rees, G. N., Baldwin, D. S., Watson, G. O., Perryman, S., and Nielsen, D. L. (2004). Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie van Leeuwenhoek 86, 339–347. doi: 10.1007/s10482-005-0498-5
Rees, J.-F., De Wergifosse, B., Noiset, O., Dubuisson, M., Janssens, B., and Thompson, E. M. (1998). The origins of marine bioluminescence: turning oxygen defence mechanisms into deep-sea communication tools. J. Exp. Biol. 201, 1211–1221.
Robison, B. H., Raskoff, K. A., and Sherlock, R. E. (2005). Ecological substrate in midwater: Doliolula equus, a new mesopelagic tunicate. J. Mar. Biol. Assoc. U K 85, 655–663. doi: 10.1017/s0025315405011586
Sen, A., Duperron, S., Hourdez, S., Piquet, B., Léger, N., Gebruk, A., et al. (2018). Cryptic frenulates are the dominant chemosymbiotrophic fauna at Arctic and high latitude Atlantic cold seeps. PLoS One 13:e0209273. doi: 10.1371/journal.pone.0209273
Sharp, K. H., Eam, B., Faulkner, D. J., and Haygood, M. G. (2007). Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl. Environ. Microbiol. 73, 622–629. doi: 10.1128/aem.01493-06
Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y., and Ishikawa, H. (2000). Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nat. 407, 81–86. doi: 10.1038/35024074
Shimizu, K., Cha, J., Stucky, G. D., and Morse, D. E. (1998). Silicatein α: cathepsin L-like protein in sponge biosilica. Proc. Natl. Acad. Sci. 95, 6234–6238. doi: 10.1073/pnas.95.11.6234
Sutherland, K. R., Sorensen, H. L., Blondheim, O. N., Brodeur, R. D., and Galloway, A. W. (2018). Range expansion of tropical pyrosomes in the northeast Pacific Ocean. Ecology 99, 2397–2399. doi: 10.1002/ecy.2429
Sutton, T. T., Frank, T., Judkins, H., and Romero, I. C. (2020). “As gulf oil extraction goes deeper, who is at risk? community structure, distribution, and connectivity of the deep-pelagic fauna,” in Scenarios and Responses to Future Deep Oil Spills, (Berlin: Springer), 403–418. doi: 10.1007/978-3-030-12963-7_24
Tessler, M., Gaffney, J. P., Oliveira, A. G., Guarnaccia, A., Dobi, K. C., Gujarati, N. A., et al. (2020). A putative chordate luciferase from a cosmopolitan tunicate indicates convergent bioluminescence evolution across phyla. Sci. Rep. 10, 1–11.
Thompson, L. R., Sanders, J. G., McDonald, D., Amir, A., Ladau, J., Locey, K. J., et al. (2017). A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457.
Urbanczyk, H., Ast, J. C., and Dunlap, P. V. (2011). Phylogeny, genomics, and symbiosis of Photobacterium. FEMS Microbiol. Rev. 35, 324–342. doi: 10.1111/j.1574-6976.2010.00250.x
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 73, 5261–5267. doi: 10.1128/aem.00062-07
Keywords: symbiosis, bioluminescence, microscopy, high throughput sequencing, 16S rRNA, tunicate
Citation: Berger A, Blackwelder P, Frank T, Sutton TT, Pruzinsky NM, Slayden N and Lopez JV (2021) Microscopic and Genetic Characterization of Bacterial Symbionts With Bioluminescent Potential in Pyrosoma atlanticum. Front. Mar. Sci. 8:606818. doi: 10.3389/fmars.2021.606818
Received: 15 September 2020; Accepted: 11 January 2021;
Published: 03 February 2021.
Edited by:
Severine Martini, UMR7294 Institut Méditerranéen d’Océanographie (MIO), FranceReviewed by:
Sophie Sanchez-Brosseau, Oceanographic Observatory in Banyuls-sur-Mer, FranceWarren Russell Francis, University of Southern Denmark, Denmark
Copyright © 2021 Berger, Blackwelder, Frank, Sutton, Pruzinsky, Slayden and Lopez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexis Berger, a.j.berger@outlook.com
 Alexis Berger1*
Alexis Berger1*  Patricia Blackwelder
Patricia Blackwelder Tamara Frank
Tamara Frank Tracey T. Sutton
Tracey T. Sutton Nina M. Pruzinsky
Nina M. Pruzinsky Jose V. Lopez
Jose V. Lopez